Abstract
In December 2019, a new betacoronavirus, known as severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), caused an outbreak at the Wuhan seafood market in China. The disease was further named coronavirus disease 2019 (COVID-19). In March 2020, the World Health Organization (WHO) announced the disease to be a pandemic, as more cases were reported globally. SARS-CoV-2, like many other viruses, employs diverse strategies to elude the host immune response and/or counter immune responses. The infection outcome mainly depends on interactions between the virus and the host immune system. Inhibiting IFN production, blocking IFN signaling, enhancing IFN resistance, and hijacking the host's translation machinery to expedite the production of viral proteins are among the main immune evasion mechanisms of SARS-CoV-2. SARS-CoV-2 also downregulates the expression of MHC-I on infected cells, which is an additional immune-evasion mechanism of this virus. Moreover, antigenic modifications to the spike (S) protein, such as deletions, insertions, and also substitutions are essential for resistance to SARS-CoV-2 neutralizing antibodies. This review assesses the interaction between SARS-CoV-2 and host immune response and cellular and molecular approaches used by SARS-CoV-2 for immune evasion. Understanding the mechanisms of SARS-CoV-2 immune evasion is essential since it can improve the development of novel antiviral treatment options as well as vaccination methods.
Keywords: Immune evasion, Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), Coronavirus disease 2019 (COVID-19), Molecular, Interferon
1. Introduction
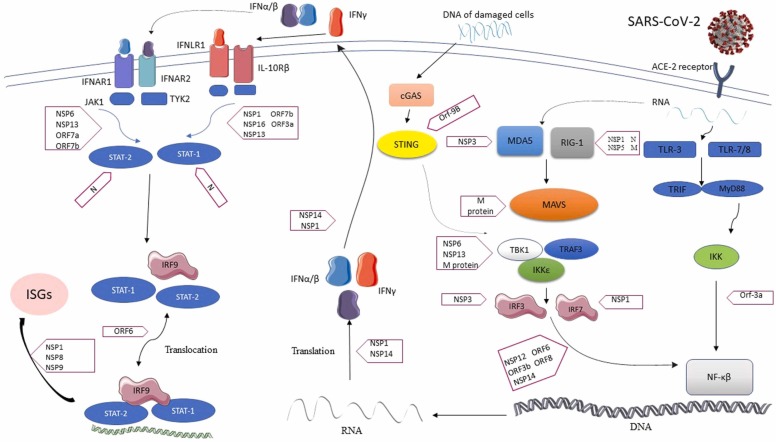
In December 2019, an outbreak began at the seafood market in Wuhan, China, induced by a novel coronavirus, named severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) (Bazargan et al., 2022). Since more cases were reported worldwide, in March 2020, World Health Organization (WHO) announced the disease a pandemic (Elahi et al., 2022). The main presentations of coronavirus disease 2019 (COVID-19) consist of fever or chills, cough, headache, and loss of taste or smell. In about 20% of cases, the infection can develop into acute respiratory distress syndrome (ARDS), and even death (Wang et al., 2020). SARS-CoV-2 belongs to the betacoronoviridea family of coronaviruses. Severe acute respiratory syndrome coronavirus (SARS-CoV) and middle east respiratory syndrome coronavirus (MERS-COV) are two other members of the betacoronavirus genus, which caused the SARS epidemic in 2003, followed by MERS in 2012, respectively (Hajjar et al., 2013, Cherry and Krogstad, 2004). The Coronoviridea family consists of single-stranded positive-sense RNA-enveloped viruses. The SARS-CoV-2 genome is about 30 kb in length, and it has 79% similarity to the SARS-CoV genome. It has at least 14 Open Reading Frames (ORF) encoding 16 non-structural proteins (Nsp1-Nsp16), four structural proteins (nucleocapsid (N), envelope (E), membrane (M), and spike (S)) (Mohamadian et al., 2021), and seven accessory proteins that regulate the host response to pave the way for infection and pathogenesis (V'Kovski et al., 2021). ( Fig. 1 and Table 1).
Fig. 1.
Mechanisms of inhibition of IFN induction and action by SARS-COV-2. The virus enters to host cells via angiotensin-converting enzyme 2 receptor (ACE-2R). IFN induction is initiated by host recognition of viral PAMPs (mainly specific viral nucleic acids or some other particular products of viral infection) via cellular pattern recognition receptors (PRRs), such as melanoma differentiation-associated protein 5 (MDA5), retinoic acid-inducible gene I (RIG-I), and toll-like receptors (TLRs). These pathways activate IFN-regulatory factor (IRF3) and/or nuclear factor kappa B (NF-kB), resulting in the production of type I/III IFN recognized by IFN receptors and later induction of the IFN-stimulated genes (ISGs) and proteins, many of which have potent anti-viral activities. Several structural and non-structural proteins of SARS-COV-2 block different parts of these pathways.
Table 1.
SARS-CoV-2 Proteins involved in immune evasion.
| Gene | Viral Protein | Function | Impact on immune response action |
|---|---|---|---|
| ORF1a | Nsp1 | Host translational shut off (Amor et al., 2020) | Reduces STAT-1 phosphorylation Inhibits ATF2/c-Jun, IRF3, IRF7, and NF-κB, (Amor et al., 2020). Interference in RIG-1 pathway (Amor et al., 2020). Binds to the 40 S ribosomal subunit by inserting its C-terminal domain containing two helices into the entrance region of the ribosomal mRNA channel, blocking host mRNA translation. Interacts with the host mRNA export receptor NXF1-NXT1, leading to nuclear retention of cellular mRNAs (Min et al., 2021). |
| Nsp3 | Papain-like protease (PLpro) Processes pp1a and pp1ab (Amor et al., 2020) Complexes with nsp4 and nsp6 (Amor et al., 2020) |
De-ubiquitinates and deISGylates host proteins. Blocks IFN-α, IFN-β, CXCL10, and CCL5. Inhibits TLR-7 signaling by removing Lys63-linked polyubiquitination of TRAF3 and 6 (Amor et al., 2020). Antagonizes IRF3 and stabilizes IκBα, thereby blocking NF-κB signaling (Amor et al., 2020). |
|
| Nsp5 | Chymotrypsin-like protease (3CLpro) (Amor et al., 2020) | Inhibition of K63 polyubiquitination of RIG-I (Beyer and Forero, 2022). | |
| Nsp6 | Complexes with nsp3 and nsp4 to form DMV (Amor et al., 2020) Membrane rearrangements (Beyer and Forero, 2022) |
Inhibition of IRF3, STAT1/2 phosphorylation (Beyer and Forero, 2022). | |
| Nsp7 | RdRp Subunit(non-enzymatic) (Beyer and Forero, 2022) | Inhibition of type I IFN signaling (Beyer and Forero, 2022). | |
| Nsp8 | RdRp Subunit(primase) (Beyer and Forero, 2022) | Bind to the 7SL RNA in the SRP and interfere with protein integration into the cell membrane and trafficking (Min et al., 2021). |
|
| Nsp9 | non-enzymatic RBP (Beyer and Forero, 2022) | Bind to the 7SL RNA in the SRP and interfere with protein integration into the cell membrane and trafficking (Min et al., 2021). |
|
| Nsp10 | RNA-capping (Beyer and Forero, 2022) | Aids RNA capping thus evades RIG-1 and MDA-5 recognition (Amor et al., 2020). Enhancement of NSP14 inhibition (Beyer and Forero, 2022). |
|
| ORF1b | Nsp12 | RNA-dependent RNA polymerase (RdRp) (Beyer and Forero, 2022) |
Targeting mitochondria limits host cellular responses and inhibit IRF3 nuclear import (Bazargan et al., 2022, Elahi et al., 2022). |
| Nsp13 | Helicase key for efficient replication of viral genome (Amor et al., 2020) | RLR evasion (Beyer and Forero, 2022). Inhibition of TBK1, IRF3, and STAT1/STAT2 phosphorylation (Beyer and Forero, 2022). |
|
| Nsp14 | Exons 3–5′ exonucleases play a crucial role in viral RNA synthesis and capping (Amor et al., 2020) Complexes with nsp10 (Amor et al., 2020) |
Involved in the capping through its function as a guanine-N7 Methyltransferase Blocks host mRNA translation (Bazargan et al., 2022, Wang et al., 2020). Helping nsp16 evade RIG-1 and MDA-5recognition (Amor et al., 2020) Inhibit IRF3 nuclear localization (Bazargan et al., 2022, Elahi et al., 2022). IFNAR1 Antagonism (Beyer and Forero, 2022). |
|
| Nsp15 | Uridylate-specific endoribonuclease (EndU) (Amor et al., 2020) | Limits exposure of viral dsRNA to the sensors MDA-5, PKR and OAS/RNaseL (Amor et al., 2020). Inhibits poly U, thereby evading MDA-5 thus antagonizing IFN-a/β production (Amor et al., 2020). Inhibit IRF3 nuclear localization (Min et al., 2021). |
|
| Nsp16 | 2′-O-ribose methyl transferase involved in RNA capping (Amor et al., 2020) Complexes with nsp10 (Amor et al., 2020) | Caps RNA, thus evades RIG-I and MDA-5 signaling (Amor et al., 2020). Binds pre-mRNA recognition domains of U1/U2 snRNAs and disrupts mRNA splicing and mature (Min et al., 2021). |
|
| Spike | Heavily glycosylated with 22 glycans ACE/ACE-2 interaction Requires priming to expose membrane fusion (Amor et al., 2020) |
Masks immunogenic protein epitopes (Amor et al., 2020). Induced misbalanced in RAS that triggers inflammation (Amor et al., 2020). |
|
| ORF3a | ORF3a | Interact with SARS-CoV M, S, E and 7a Proteins (Amor et al., 2020) Forms viroporins (Amor et al., 2020) |
Interacts with STING and blocks the nuclear accumulation of NF-kB, thus likely impeding IFN promoter activation (Min et al., 2021). Inhibition of STAT1 phosphorylation (Beyer and Forero, 2022). |
| ORF3b | ORF3b | Inhibits IRF3 nuclear localization (Elahi et al., 2022, Wang et al., 2020). | |
| Envelope | Essential for viral assembly and budding (Amor et al., 2020) Forms viroporins (Amor et al., 2020) |
Induces ROS and activates inflammasome (Amor et al., 2020). | |
| Membrane | Important for viral assembly (Amor et al., 2020) | Inhibits type I interferon production by impeding the formation of TRAF3. TANK. TBK1/IKKε complex (Amor et al., 2020). Blocks MAVS aggregation (Beyer and Forero, 2022). |
|
| ORF6 | ORF6 | Plays a role in viral pathogenesis, interacts with ORF8 (Amor et al., 2020) |
Inhibits STAT-1 nuclear import (Amor et al., 2020). Interacts with TBK1 to inhibit IRF3 activation (Min et al., 2021). Suppresses STAT1 and STAT2 phosphorylation (Min et al., 2021). |
| ORF7a | ORF7a | Interaction with S protein and p3a (Amor et al., 2020) Not essential for replication |
Inhibits BST-2 glycosylation leading to a loss of function of BST-2 (Amor et al., 2020). SARS-CoV ORF7a induces caspase-dependent apoptosis (Amor et al., 2020). Suppresses STAT2 phosphorylation (Elahi et al., 2022, Wang et al., 2020) |
| ORF7b | ORF7b | Not essential for viral replication but structural component of the virion (Amor et al., 2020) | Suppresses STAT1 and STAT2 phosphorylation (Elahi et al., 2022, Wang et al., 2020) |
| ORF8 | ORF8 | Differs from other HCoVs (Amor et al., 2020) | Interact and down-regulates MHC-I (Amor et al., 2020). Inhibits IRF3 nuclear localization (Min et al., 2021). |
| Nucleocapsid | Stabilizes viral RNA Interacts with stress granules G3BP1 (Amor et al., 2020) |
Targets MAVS–TRAF3–TRAF6 and antagonizes IFN-β (Amor et al., 2020). Binds to the DExD/H domain of RIG-I, thus impeding RIG-I signaling (Amor et al., 2020). Might bind to STAT1 and STAT2, suppressing STAT1 and STAT2 phosphorylation (Amor et al., 2020). |
|
| ORF9b | – | – | Interacts with TOM70, thus inhibiting type I IFN induction. Targets IKKγ and specifically interrupts IKKγ K63-linked polyubiquitination, thereby inhibiting NF-kB signaling and IFN promoter activation (Min et al., 2021). Blocks TOM70-HSP90 interaction (Beyer and Forero, 2022). Inhibition of TBK1 phosphorylation (Beyer and Forero, 2022). |
| ORF10 | ORF10 | Ubiquitin ligase (Amor et al., 2020) | Induces an autophagy pathway which leads to MAVS degradation. Induces the mitophagy process by expanding the accumulation of LC3 in mitochondria (Zandi, 2022). |
ACE 2: angiotensin-converting enzyme 2, DMV: double membrane vesicles, HCoV: human coronavirus, IFN: interferon, IFNR: IFN receptor, IKK: Inhibitor of κB kinase, IRF: IFN-regulatory factor, MAV: mitochondrial antiviral signaling protein, MDA: melanoma differentiation-associated, MHC: major histocompatibility complex antigen, NF-Kb: Nuclear Factor Kappa B, ORF: open reading frame, RAS: renin–angiotensin system, RIG-1: retinoic acid-inducible gene, RLR: Retinoic acid-inducible gene-I-Like Receptors, ROS: reactive oxygen species, STAT: signal transducer and activator of transcription, STING: Stimulator of Interferon Genes, TBK1: TANK-binding kinase-1, TRAF: Tumor Necrosis Factor Receptor -associated Factor
Attachment of the receptor-binding domain (RBD) of S protein to the cellular receptor angiotensin-converting enzyme 2 (ACE2) initiates the infection with SARS-CoV-2. The subsequent step is the merging of the virus with the cell membrane, followed by the deployment of the genome into the host cell. Then, viral components contribute to viral transcription, replication, translation, and protein synthesis. The final step is the assembly and release of the virion (Min et al., 2021, Esmaeilzadeh et al., 2021a).
During the infection phase of SARS-CoV-2, the virus employs its S protein to bind with the cell surface. After this interaction, the virus undergoes rapid endocytosis (Bayati et al., 2021). Removing clathrin-heavy chains from host cells has been shown to hinder clathrin-mediated endocytosis and reduce viral infectivity (Gu et al., 2022a). As demonstrated by these studies, the spike protein of SARS-CoV-2 is absorbed by clathrin-mediated endocytosis. In addition to ACE2, numerous other molecules engage in SARS-CoV-2's endocytosis pathway. NRP1 was also discovered as a SARS-CoV-2 receptor and it has an essential role in C-end rule-related endocytosis (Cantuti-Castelvetri et al., 2020). Prior studies indicated that SARS-CoV-2 targets the lungs and enters the body primarily through ACE2 receptors. Non-muscle myosin heavy chain IIA)NMMHC-IIA (was recognized as an ACE2 coreceptor which is colocalized with SARS-CoV-2 S-protein at the membrane and facilitates SARS-CoV-2 endocytosis entry (Chen et al., 2021). Another mechanism of endocytosis occurs when antibodies on the virus's surface connect with Fc receptors on target cells. In contrast to viral particle endocytosis, the destiny of SARS-CoV-2 within a cell is largely regulated by the type of receptor and intracellular transport system (Knyazev et al., 2021, Gu et al., 2022b).
Following viral infection, the innate immune system of mammals quickly identifies and prevents viral infection throughout the whole viral life cycle. The inherent immunity of humans defends against coronavirus, playing an essential role in maintaining health. Researchers in one study hypothesized that SARS-CoV-2 infection elicits two phases of immune responses. The initial phase of immunological-based protection against SARS-CoV-2 infection is noteworthy and is strongly tied to innate immune responses. Before administering effective treatments and immunizations, its concentration is evaluated as an alternative solution (Chowdhury et al., 2020, Shi et al., 2020). The innate immune cells detect the virus and launch a local viral clearance response. It consists of dendritic cells, macrophages, neutrophils, and natural killer (NK) cells and is the first line of defense for initiating an immune response. More monocytes, macrophages, and neutrophils were seen in the Bronchoalveolar Lavage Fluid of individuals infected with SARS-CoV-2. Patients with severe COVID-19 also showed lower amounts of mDC, pDC, T, and NK cells and higher levels of macrophages and neutrophils (Gu et al., 2022b, Liao et al., 2020).
Pathogen-associated molecular patterns (PAMPs), including viral ssRNA and intermediate replication dsRNA domains, are distinguished by pattern recognition receptors (PRRs) to initiate the activation of innate immune responses. The arms of innate immunity, such as interferon (IFN) systems, restrict viral replication and spread. Immunity to SARS-CoV-2 is comparable to that against SARS-CoV and MERS-CoV, including the generation of IgG and IgM (Shah et al., 2020).
It has been shown that coronaviruses employ various strategies to evade innate immunity, including inhibiting IFN production, blocking IFN signaling, and enhancing IFN resistance (Xia et al., 2020, Lei et al., 2020a, Lei et al., 2020b). Moreover, according to research, most coronaviruses, such as SARS-CoV-2, hijack the host's translation machinery to expedite the production of viral proteins. In addition, viruses, such as Kaposi sarcoma-associated herpesvirus and Human Immunodeficiency Virus, are capable of interfering with the antigen-presentation mechanism by downregulating MHC-I on cell surfaces, thereby causing immune evasion (Zhang et al., 2021a). According to studies, SARS-CoV-2 also inhibits MHC-I expression in infected cells (Xia et al., 2020, Zhang et al., 2021a). A precise understanding of the interactions between the immune system and SARS-CoV-2 is critical for developing novel and efficient therapeutic strategies against COVID-19. This review describes the interaction between SARS-CoV-2 and hosts' antiviral responses focusing on molecular mechanisms used by SARS-CoV-2 for immune evasion.
2. Immune evasion through inhibition of host protein synthesis
The cellular ribosomes are essential for the translation of viral mRNAs into polypeptides. As a frequent viral approach, this is accomplished by disrupting cellular mRNA translation in a process called "host shutoff." Both translational resources and ribosomes, especially, are assumed to be redirected toward viral mRNAs by host shutdown, and the capacity of infected cells to generate an effective antiviral response is blocked (Fisher et al., 2022). Suppression of translational mechanisms is an effective method of virus defense against the host immunity by which it can inhibit the synthesis of proteins enrolled in the innate immune response of the host. Furthermore, during an acute viral infection setting, the translation system is shut down by host cells to deal with infection stress, which is considered a total stress response. On the other hand, viruses have developed strategies, such as eukaryotic initiation factor (eIF) phosphorylation, cellular mRNA degradation, etc, to maintain viral replication (Shen et al., 2021). Although numerous SARS-CoV-2 proteins have been previously linked to the suppression of host protein expression, a comprehensive understanding of the mechanisms by which SARS-CoV-2 suppresses host cell gene expression is still lacking.
Similar in structure to host mRNAs, the mRNAs of coronaviruses are 5′-capped and 3′-polyadenylated. Due to translational competition between cells and viruses, coronaviruses must co-opt the host's translational machinery to produce their proteins (Shen et al., 2021). Gene 1 of the 5′-capped and 3′-polyadenylated RNA genome of the coronavirus encodes two enormous overlapping open reading frames (ORF1b and ORF1a). ORF1a and ORF1b are involved in viral replication, following host cell infection. SARS-CoV-2 proteins interact directly with host RNAs, inhibiting protein synthesis and interferon production (V'Kovski et al., 2021, Finkel et al., 2020).
Nsp1 K164 is the central residue linked to host gene expression inhibition (Shen et al., 2021) and is connected to the 40 S subunit of the ribosome's two distinct components (Molaei et al., 2021). SARS-CoV-2 Nsp1 reveals a considerably higher capability to interfere with the host proteins production, which may lead to the enhanced pathogenicity of SARS-CoV-2 (Shen et al., 2021, Thoms et al., 2020). Through the interaction of messenger RNAs (mRNAs) and the translation of their nucleotide sequences into amino acid sequences, ribosomes can synthesize proteins (Shyu et al., 2008). The amino acid sequence of a protein determines both its structure and biological activity. First, the mRNA binds to the small subunit, which then interacts with the 60 S subunit to form the cavity through which the mRNA is threaded. One end of the Nsp1 protein binds competitively with the 40 S subunit, preventing mRNA binding (Thoms et al., 2020). Consistently, subsequent research has demonstrated that Nsp1 can also interact with specific configurational states of the fully formed ribosome (Zhang et al., 2021b).
Coronavirus Nsp14 proteins are both exoribonucleases (3' to 5') and guanine-N7 methyltransferases. A portion of the C-terminal domain of Nsp14 that functions as a methyltransferase are essential for viral RNA 5' capping, which promotes the stability of viral mRNA and translation. There is evidence that overexpression of Nsp14 completely halts protein synthesis in the host cell. Inhibition of the host translation system by Nsp14 prevents the production of antiviral proteins such as interferons (IFNs), which aid immune escape by evading host innate immune responses. This is one of the significant features of SARS-CoV-2 compared to other coronaviruses, such as MERS-CoV (Finkel et al., 2020, Hsu et al., 2021).
NSP8 and NSP9 are two additional viral proteins implicated in the immune evasion of SARS-CoV-2. By attaching to Signal Recognition Particle (SRP) and inhibiting protein trafficking, NSP8 and NSP9 play a significant role in viral replication. Upon infection, NSP8 and NSP9 bind to the 7SL RNA in the SRP and prevent protein transport to the cell membrane. In addition, Nsp8/9-mediated viral suppression of SRP leads to IFN response suppression. To conclude, the extensive suppression of host gene expression, referred to as host shutdown, mainly through Nsp1, 8, 9, and 14, is a crucial step that provides the reallocation of cellular resources required for viral duplication and evading host immune response (Gu et al., 2022b).
3. Immune evasion through MHC-I down-regulation
Immune responses to viral infections involve intricate interactions among target cells, viruses, and immune cells (van Montfoort et al., 2014). Virus-specific CD8+ cytotoxic T lymphocytes (CTL) are central to the immune response against viral infections and virus clearance is primarily dependent on these cells. These cells can eliminate the virus-infected cells through direct and indirect mechanisms (van Montfoort et al., 2014). The presentation of major histocompatibility complex (MHC) class I–peptide complexes, concurrent with co-stimulation and the presence of cytokines, such as IFNs and IL-12, are essential for the activation of naïve CD8+ T cells and their transformation into effective CTL (van Montfoort et al., 2014, Schiavoni et al., 2013).
MHC-I and II play a critical role in adaptive immune responses. They display peptides on the cell surface for T-cell detection. The binding portion of MHC I and MHCII consists of two domains derived from a single heavy chain (HC) in MHC-I and two chains in MHC-II. The two domains constructed a base consisting of a curved-sheet and two-helices at the correct distance to accommodate a peptide chain (Wieczorek et al., 2017). MHC-I is the structural protein on antigen-presenting cells (APCs) that presents peptides to the adaptive immune cells. MHCI is necessary for viral antigen presentation and subsequent cytotoxic T lymphocyte-mediated lysis of infected cells. Viruses have developed proteins that block antigen presentation by MHC class I molecules, perhaps in response to selection pressure from the immune system (Hewitt, 2003).
When peptides are loaded, MHC-I molecules detach from TAP and assemble at the ER membrane's export sites. At these sites, they are packaged into cargo vesicles for transfer to the Golgi apparatus (Hewitt, 2003). The Golgi apparatus transports the protein to the plasma membrane. As previously stated, the antigen presentation pathway mediated by MHC class I is essential for antiviral immunity (Yoo et al., 2021). Cytotoxic CD8+ T Cells also have a significant role in the clearance of viruses from the respiratory tract in several viral infections. Antigen-presenting cells (APCs) are necessary for the activation of cytotoxic CD8+ T cells by presenting the viral antigen through MHC-I (Schmidt and Varga, 2018, Ghaebi et al., 2021).
In virus-infected cells, viral proteins are presented by MHC-Ι proteins. Once the T cell receptor on CD8+ T cell detects the particular signal exhibited by the MHC-Ι peptide complex, CTLs release several toxic substances such as perforins, granzyme, and membrane-bound fasL and finally lead to the death of virus-infected cells. SARS-CoV-2, similar to human immunodeficiency virus–1 (HIV-1) and Kaposi's sarcoma-associated herpesvirus (KSHV), can evade immune surveillance by inhibiting MHC-I expression on the cell surface, thereby interfering with antigen presentation. The main protein in down-regulating MHC-I is ORF-8 which can precisely interplay with MHC-I molecules and conduct their down-regulation (Zhang et al., 2021a, Flower et al., 2020). It has been indicated that the cells expressing ORF-8 and those infected with SARS-CoV-2 are more resistant to CTL lysis (Flower et al., 2020).
Studies analyzing the gene expression profile of COVID-19 patients have shown that SARS-CoV-2 infection represses the initiation of the MHC class I pathway (Yoo et al., 2021). ORF-6 represses NOD, LRR, and CARD-containing 5 (NLRC5), an MHC class I transactivator, by inhibiting the karyopherin complex-dependent transfer of NLRC5 to the nucleus. It also inhibits type II interferon-induced STAT-1 signaling, resulting in decreased NLRC5 and interferon regulatory factor 1 (IRF1) gene expression. In conclusion, one of SARS-CoV-2's immune evasion mechanisms targets essential MHC class I transcriptional regulators, STAT1-IRF-NLRC5, which leads to the downregulation of MHC-I and therefore reduces the identification of infected cells by the immune cells (Yoo et al., 2021, Vijayan et al., 2019).
4. Immune evasion through Spike (S) protein alterations
The spike (S) protein has two subunits, an S1 N-terminal and an S2 C-terminal membrane-proximal. The S1 subunit of spike protein includes the RBD and N-terminal domain (NTD). Therefore, escape mutations in these regions is significant for immune evasion and resistance against neutralizing antibodies (nAbs). Many SARS-CoV-2 variants consist of small deletions in the NTD regions (Harvey et al., 2021). The S1 component comprises four domains, including S1A, S1B, S1C, and S1D. The S1A domain identifies carbohydrates, such as sialic acid, which is essential for virus binding to host cells. The RBD is the S1B domain of the S protein. RBD interacts with the SARS-CoV-2 receptor ACE2 (Bayarri-Olmos et al., 2021). The S protein mutation is extremely significant because it affects virus replication and immune responses to the virus (Ding et al., 2021).
Between the S1 and S2 subunits, the PRRA sequence motif contains a furin-cleavage region. The other proteolytic site, S2', is located upstream of the fusion peptide in the S2 subunit. These cleavage sites contribute to the entrance of SARS-CoV-2 into the cell. The spike protein (S) moderates the attachment of viruses to cell surface receptors. Virus-cell membrane fusion is also mediated by the S protein. It is also a significant target for developing treatment strategies against SARS-CoV-2 (Letko et al., 2020). The S protein is the central target of neutralizing antibodies produced in response to SARS-CoV-2 infection. It is also the SARS-CoV-2 component of both licensed adenovirus-based and mRNA vaccines. Therefore, mutations that influence the antigenicity of the S protein are significantly important since they can reduce the SARS-CoV-2-specific immune response (Esmaeilzadeh et al., 2021b). Based on structural analysis, neutralizing antibodies are divided into four classes. The first class consists of Abs that bind to the S protein in its open conformation which inhibits ACE-2. The second class consists of Abs binding to RBD in both closed and open conformations which inhibit ACE2 in the same manner as class I. In the third category, Abs do not inhibit ACE2 and bind to RBD in both the closed and open conformations. Finally, the fourth-grade neutralizing Abs bind outside of the ACE2 binding region and only in the open conformation (Harvey et al., 2021). Therefore, antigenic alterations and mutations in the S protein are a common immune evasion mechanism of SARS-CoV-2, which are described in this section.
4.1. Mutation of S protein
Spike protein mutations have been observed in various variants of SARS-CoV-2. In November 2020, the B1.1.7 (Alpha) variant appeared in the United Kingdom which contained numerous RBD and NTD mutations (Harvey et al., 2021). Since then, several variants with S protein mutations have been detected, including the P.1 (Gamma variant), which was reported for the first time in Japan, and the B.1.351 (Beta) variant, which emerged in South Africa (Harvey et al., 2021). The B.1.351 and P.1 variants contain two crucial mutations in the RBD site (N501Y and E484K) and the deletion of three amino acids in the ORF1-ab gene (Rees-Spear et al., 2021). Based on early reports, the mutation in RBD N501Y of the B.1.1.7 variant does not interrupt serum neutralization after vaccination; however, the extra variations in B.1.351 variant interfere with neutralization (Rees-Spear et al., 2021).
Mutations in the S gene have altered the efficiency of protein attachment and immunogenicity, leading to the emergence of more invasive and adaptive strains (Ou et al., 2022). The highly infectious Omicron variant (B.1.1.529) variant, was first reported in South Africa in November 2021. Multiple mutations (more than 30) were identified in the spike gene of the B.1.1.529 variant, which, compared to the B.1.617.1 and B.1.1.7 variants (generally fewer than 15), raised concerns about increased infectivity and immune evasion potential (Ou et al., 2022). More than thirty mutations in genes encoding amino acids have been identified in the S protein of the Omicron variant, some of which are also present in other variants (Saxena et al., 2022). According to analyses of Omicron S gene sequences, the majority of Omicron spike mutations are stable. Approximately eighteen key mutations in BA.1 variant are present in the NTD, SD (underpinning domain), and S2 regions close to the S1/S2 cleavage site (Ou et al., 2022). BA.1 Subvariant shares nine amino acid mutations in the S protein with other Variants of Concern (VOCs), indicating that omicron's origin may be traced back to these variants. BA.2 subvariant has fewer mutations in common with other VOCs than BA.1 subvariant. According to studies on mutations in S protein that aid SARS-CoV-2 in evading the immune system, E484 has been identified as the most crucial residue of the RBD site of S protein whose substitution influences recognition by Abs (Ou et al., 2022).
4.2. Different mechanisms result in antigenic alterations
Some mechanisms have roles in antigenic change, which finally lead to the variation of antigenic features of glycoprotein. Amino acid substitution is one of these mechanisms which leads to the alteration of biophysical characteristics of an epitope and therefore Ab attachment decreases. For instance, a neutralizing Ab 48 A constructs salt bridges with K147 and K150 residues of S protein. Therefore, substitution at these sites leads to Ab attachment inhibition. A substitution in E484K amino acid also influences the neutralizing activity of the antibody. It has been reported that a change in charge due to the exchange of glutamate residue with lysin residue decreases Ab binding (Harvey et al., 2021). The other mechanism is an increase in receptor binding affinity which alters the binding balance between neutralizing Abs and glycoprotein. For example, the N501Y amino acid substitution in the S protein increases the binding affinity of ACE-2 (Harvey et al., 2021).
Over the past two years, the NTD region of the S protein has been recognized as a hotspot for deletions, consistent with S protein phylogenetic trees. These deletions are located within NTD at residues 69–70, 141–143, 156–159, and 242–245, and they occur independently in numerous unrelated lineages (Harvey et al., 2021). N2, N3, and N5 loops have a significant capacity for deletions, in addition to the ability to remove N1 via the deletion in the 15–136 disulfide bonds (DS15–136) mechanism. These alterations finally result in the rearrangement of all adjacent loops. This restructuring permits the total redesign of the NTD supersite. The reshaping of the loops through DS15–136 appears to have progressed in distinct variants of the SARS-CoV-2 phylogenetic tree, indicating that it will play a crucial role in future VOCs, as well (Harvey et al., 2021).
Other mechanisms that lead to epitope conformational changes that decrease Ab binding are residue deletions and insertions. Deletions in the NTD of S protein affect the neutralizing antibodies' ability to detect the protein. According to reports of in-vitro experiments, certain NTD insertions aid in the evasion of polyclonal Abs (Harvey et al., 2021). In addition to deletion and insertion, amino acid exchange outside of an epitope space can result in altered epitope expression and protein conformation. It has been demonstrated that modification of disulfide bonds in the NTD of S protein reduces the attachment of several monoclonal Abs (Harvey et al., 2021). E484K is a known escape mutation that develops in response to monoclonal Abs C121 and C144 and convalescent plasma. In E484, amino acid substitutions for K, Q, or P reduce neutralization titers. In a study of escape mutations, E484 substitutions occurred more frequently than any other residue (Greaney et al., 2021a). Among mAb escape mutations, mutations at position 477 of the S protein (S477R, S477N, and S477G) are notable. The N439K mutation raises affinity for ACE2 and is anticipated to lead to an extra salt bridge at the ACE2-RBM border and is believed to decrease the neutralization capacity of plasma which has a lower neutralizing capacity (Ou et al., 2022, Greaney et al., 2021a, Thomson et al., 2021).
5. Immune evasion through inhibiting IFN induction and function
SARS-CoV-2 infection begins as the viral S protein binds to its cell surface receptor, ACE2. In DCs, epithelial cells, monocytes, and macrophages, membrane-associated Toll-like receptor 2 (TLR-2) detects S protein and activates the Nuclear Factor K-B (NF-kB) cascade following the attachment of the virus to ACE-2. Once SARS-CoV-2 has entered a cell, ssRNA activates intracellular TLR-7/8 (Mabrey et al., 2021). After viral replication, other cytosolic PRRs, primarily MDA-5 and PKR, detect dsRNA domains produced during replication, thereby facilitating antiviral gene expression. Consequently, TLRs and RLRs prompt transcription factors necessary for producing proinflammatory cytokines and IFN-I via the NF-κB and IRF3/7 pathways. IFNs bind to the IFN receptor and activate the JAK/STAT cascades, thereby limiting viral replication (Lei et al., 2020a). In addition, NK cells produce IFN-gamma (IFN-γ) and certain chemokines in response to infection with respiratory viruses, such as SARS-CoV-2 (Xia et al., 2020).
SARS-CoV-2, MERS, and SARS-CoV use distinct strategies to evade host IFN responses, and these strategies vary among coronaviruses (Cao et al., 2017). The evasion of SARS-CoV-2 by IFNs can be classified as inhibition of IFN induction by masking or decreasing PAMPs to elude host PRR sensing, interfering with the IFN induction signaling pathway, suppression of IFN activity, and inhibition of host protein production by IFNs (Min et al., 2021). Here, these pathways are discussed in detail.
5.1. Inhibition of IFN induction
Diverse types of signaling PRRs activate immune responses, involving IFN-I and inflammatory molecules, through intracellular signaling pathways (Schneider et al., 2014). TLR in the endosome, RIG-I-like receptors (RLR) such as retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated protein 5 (MDA5) in the cytoplasm, and cytosolic protein kinase R (PKR) are the essential PRRS that play a crucial role in the detection of viral RNA (Lei et al., 2020a, Taefehshokr et al., 2020, Esmaeilzadeh and Elahi, 2021). The transcriptional activation, translation, and secretion of type I and type III IFNs result in late antiviral responses. SARS-CoV-2 inhibits the production of antiviral IFNs by two main mechanisms. The first is the prevention of PAMPs from being recognized by PRRs and the second is through interfering with IFN induction signaling pathways.
5.1.1. Prevention of PAMPs from being recognized by PRRs
The initial mechanism of inhibition of IFN induction is the concealment or reduction of PAMPs by SARS-CoV-2. SARS-CoV-2, like other positive-sense RNA viruses, conceals its PAMPS by replicating in double-membrane vesicles and remodeling the endomembrane of the host cell to exclude PRRs (Min et al., 2021, Taefehshokr et al., 2020, Sa Ribero et al., 2020). Nsp16, Nsp14, and Nsp13 function as the viral capping machinery to regulate viral mRNA and reduce PRR recognition. Nsp13 is a viral helicase that functions as an RNA triphosphatase, plays a role in cap formation, and decreases 5' triphosphorylated viral RNA and PAMPS recognized by PRRs (Min et al., 2021, Viswanathan et al., 2020, Yan et al., 2021). NSP16 is a component of the replication and transcription complex and plays an essential role in SARS-CoV-2 replication, mRNA translation, and immune evasion. Nsp16 is the viral S-adenosyl-L-methionine (SAM)-dependent 2'-O-methyltransferase (2'-O-MTase). Its function is to initiate the formation of the Cap-1 structure (Vithani et al., 2021). Nsp16 generates a cap-1 structure by methylating ribose at the 2'-O position, thereby preventing viral RNA detection. Nsp10 has a role in stabilizing the SAM-binding pockets of Nsp16 and Nsp14 to end the viral RNA casting process. Additionally, the Nsp10-Nsp16 complex converts Cap-0 mRNAs to the Cap-1 (me7GopppA1m) structure (Viswanathan et al., 2020). Nsp15, a highly conserved uridylate-specific endoribonuclease (NendoU), plays a role in virus evasion from MDA5 sensing via VRNA processing. Nsp15 trims and inhibits the buildup of 5-PolyU-containing, negative-sense (PUN) RNA, recognized by MDA5 (Frazier et al., 2021, Klein et al., 2020). Nsp12 is another viral protein that contributes to SARS-CoV-2 immune evasion by preventing viral RNA from being detected. Nsp 12 is an RdRp-associated nucleotidyltransferase (NiRAN) that possesses guanylyltransferase activity and catalyzes the formation of cap core structure (GpppA) (Yan et al., 2021).
5.1.2. Interfering with the IFN induction signaling pathway
In addition to the inhibition of IFN induction by hiding PAMPs, SARS-CoV-2 has multiple strategies to interfere with IFN induction pathways. Various viral proteins play a role in interfering with IFNs signaling pathways. At the onset of viral infection, triple-stranded viral RNA is detected by three major mechanisms. PKR pathway, 2',5'oligoadenylate synthetase (OAS)/RNAseL, and RLRs are examples of these pathways. RLRs bind viral RNA through the C-terminal helicase domain. RNA binding results in conformational changes and the exposure of the N-terminal caspase activation and recruitment domains (CARD). CARD binds to the CARD of the mitochondrially-localized adaptor molecule mitochondrial antiviral signaling (MAVS), also known as Cardif (CARD adaptor inducing IFN), VISA (virus-induced signaling adaptor), and IPS-1 (IFN promoter stimulator 1) (Thoms et al., 2020, Ou et al., 2022, Saxena et al., 2022, Amor et al., 2020). This attachment enhances the oligomerization of MAVS on mitochondrial membranes. Recruiting inhibitor of k-B kinase e (IKKe) or TANK binding kinase 1 (TBK1) to the RLR/MAVS signalosome improves kinase autophosphorylation and subsequent triggering of the NF-κB and IRF3 (Harvey et al., 2021, Walsh and Mohr, 2011). Phosphorylation of IRF-3 results in homodimerization and transport to the nucleus, where it can collaborate with NF-κB to express IFNs and IFN genes.
Various components of this pathway are inhibited by viral proteins. SARS-CoV-2 N protein binds to the ATPase-active DExD/H domain of RIG-I, required for the attachment of PAMP RNAs, thereby inhibiting RIG-I signaling (Oh and Shin, 2021). The M protein is an additional structural SARS-CoV-2 protein that suppresses IFN induction. M protein may interact with RIGI, MDA5, MAVS, and TBK1 to inhibit TRAF3–TBK1, MAVS-TBK1, and RIG-I–MAVS interactions, thereby preventing nuclear translocation and phosphorylation of IRF3 (Zheng et al., 2020). Nsp13 of SARS-CoV-2 binds to TBK1 and inhibits the linkage of TBK1 with TRAFs and MAVS, which facilitate the recruitment of TBK1 to MAVS, thereby inhibiting IRF3 and TBK1 activation and IFN production (Vazquez et al., 2021). Moreover, ORF3b of SARS-CoV-2 is involved in immune evasion by inhibiting IRF3 nuclear translocation.
Other non-structural SARS-CoV-2 proteins, Nsp12, Nsp14, and Nsp15, interfere with IFN induction by inhibiting IRF3 nuclear translocation (Yuen et al., 2020). OAS and PKR were identified as antiviral response regulators by analyzing the synthesis of proteins in cell-free lysates from dsRNA and IFN-treated cells (Drappier and Michiels, 2015, Cesaro and Michiels, 2021). The RLR pathway is independent of these two pathways. PKR and the OAS proteins are genes stimulated by IFNs (ISGs). Activation of the RLR pathway and induction of IFN led to the expansion of alternative host innate immune responses, resulting in the death of infected cells (Wang et al., 2016). RLR antagonism by the M protein is another immune evasion mechanism of SARS-CoV-2. Orf-9b, NSP13, 14, 15, and 16 have been observed to contribute to this process (Frazier et al., 2021, Yuen et al., 2020, Kouwaki et al., 2021). Through targeting multiple pathways, includingTLR3-TRIF and RIG-I/MDA-5-MAVS, Orf-9b antagonizes the RLR downstream IFN-I and III induction (Han et al., 2021).
5.2. Inhibiting IFN action
As IFNs are secreted, they serve as both paracrine and autocrine factors. Attachment of type I and type III IFNs to their receptors results in the phosphorylation of tyrosine residues on the transcription factors, STAT2 and 1, by Janus kinases JAK1 and TYK2. The transcription complex composed of IRF9 (ISGF3), STAT-2, and STAT-1 attaches IFN-stimulated response elements (ISREs) in the uperhand promoter regulatory regions of ISGs (Lei et al., 2020a).
ISGylation promotes host resistance to viral infections by combining a protein with a ubiquitin-like protein, ISG15 (Beyer and Forero, 2022). ISGylation is required for subsequent IFN secretion and ISG expression, activation of IRF3 phosphorylation (S396), and MDA5 oligomerization. Nsp3 primarily targets ISGylated substrates and isolates ISG15 from host protein substrates, especially MDA5 and IRF3, thereby reducing IFN induction (Beyer and Forero, 2022). In addition, SARS-CoV-2 Nsp3 inhibits IFN production by direct cleaving of IRF3 (Moustaqil et al., 2021). Nsp6 decreases STAT1 and STAT2 phosphorylation and inhibits IFN type I induction (Min et al., 2021, Hsu et al., 2021). In addition to other immune evasion activities of Nsp13, it has been demonstrated that Nsp13 decreases the phosphorylation of STAT1 and STAT2 as Nsp6 (Xia et al., 2020). ORF3a, ORF7a, and ORF7b decline STAT1 and STAT2 phosphorylation, and therefore they demonstrate an innate immune antagonistic role (Xia et al., 2020, Hayn et al., 2021).
It has been shown that the NSP3 macrodomain reverses interferon-induced PARP9/ DTX3L-dependent ADP-ribosylation to suppress the host's innate immune responses (Russo et al., 2021). It has also been suggested that the ORF6 and ORF9b proteins have IFN-antagonistic effects (Wu et al., 2021, Miorin et al., 2020). ORF6 protein inhibits immune responses at multiple phases. Several studies demonstrated that ORF6 is predominantly cytoplasmic and partially colocalized with Golgi and ER markers (Zhou et al., 2010, Schaecher et al., 2008). ORF6 protein has an inhibitory influence on IFN-β, MAVs, and IRF-3 activation (Gori Savellini et al., 2022). ORF6 interrupts the nuclear transfer of STAT1, and 2 and leads to the inhibition of ISG expression. Moreover, ORF6 serves a crucial function in inhibiting the host's antiviral response and in viral replication. By decreasing the phosphorylation and nuclear translocation of IRF3, ORF6 reduces the Sendai virus (SeV)-mediated IFN induction. Moreover, IRF3 nuclear translocation is prevented by ORF6 (Fung et al., 2021). ORF6 also suppresses the nuclear export of mRNA encoding RIG-I, inhibiting the detection of coronavirus-produced dsRNA. This ORF reduces the activity of IRF1/3/7 and the subsequent transcription of IFNα/β (Hall et al., 2022).
As previously described, once IFNs bind to their receptor, they activate the JAK-STAT pathway. Moreover, JAK1 and TYK2 kinases phosphorylate STAT1 and STAT2, promoting their translocation and dimerization into the nucleus. According to cell studies STAT1 phosphorylation, which is induced by IFN, remains intact in the presence of SARS-CoV and SARS-CoV-2 ORF6. These results indicate that ORF6 suppresses ISRE/ISG56 promoter activation, but does not affect STAT1 phosphorylation (Lei et al., 2020a, Park and Cho, 2022). ORF9b inhibits mitochondrial recruitment of IRF3 and TBK1 through translocase of outer membrane 70 (TOM70) to suppress IFN induction (Lei et al., 2020a, Park and Cho, 2022).
It has also been demonstrated that ORF10 interferes with the interferon signaling pathway by attaching to the mitochondrial antiviral signaling protein (MAVS). In addition, a rise in ORF10 expression is reported to cause the mitophagy process by expanding the accumulation of LC3 in mitochondria (Li et al., 2022). The ORF9c protein of SARS-CoV-2 interferes with both antigen presentation and interferon signaling (Li et al., 2022, Greaney et al., 2021b).
6. Immune evasion of the Omicron variant
Currently, Omicron is the predominant Trusted Source variant of SARS-CoV-2 worldwide. In comparison to earlier SARS-CoV-2 strains, Omicron has been quickly distributed across the globe. Changes to Omicron’s spike protein allow it to evade immunization by preventing antibodies from binding (Bazargan et al., 2022). The majority of studies concur that serum from convalescent and fully immunized patients contain very low levels of anti-Omicron nAbs. Vaccine boosters using the third dose of mRNA vaccine appear to restore neutralizing efficacy, possibly by enhancing cross-reactivity against variants and humoral immunity (Cameroni et al., 2022, Liu et al., 2022, Cele et al., 2022). In addition, some research suggests that double vaccination accompanied by infection with Delta variant, or previous infection followed by mRNA vaccine double immunization, results in higher neutralizing antibody levels that may be protective. Numerous studies indicate that Omicron is resistant to binding and neutralization by the vast majority of monoclonal antibodies (mAbs) against SARS-CoV-2, except for a few broadly neutralizing mAbs such as Sotrovimab (Hoffmann et al., 2022). Even though viral escape from nAbs may cause relapse infections in convalescent and vaccinated patients, it is important to note that residual neutralizing antibodies, non-neutralizing antibodies, and preexisting innate and cellular immunity are likely to protect against severe infection (Dejnirattisai et al., 2022, Carreño et al., 2022).
As stated previously, genomic sequence analysis uncovered numerous nonsynonymous mutations in Omicron variants. The majority of these mutations are associated with disease severity, spread, and immune evasion as most of them have occurred in spike proteins. In addition, over 60 insertions, substitutions, and deletions have been identified in Omicron, making it the SARS-CoV-2 variant with the largest mutation area described to date (Cameroni et al., 2022). It can evade previous immune responses to both infection and vaccination, necessitating widespread vaccination campaigns in multiple nations. Spike glycoproteins of Omicron, which aid in viral entry into cells, contain 37 residue mutations compared to the wild Wuhan-Hu-1 S variant, whereas Alpha and Delta contain 10 mutations (Alexandar et al., 2021). These mutations confer a tighter binding to human ACE2, whereas other mutations promote immunological escape (Cao et al., 2022).
Numerous dangerous mutations exist in Omicron's RBD. One of the pathogenic characteristics of the Omicron virus appears to be its ability to link to ACE2. It has been shown that two Omicron mutations, such as the N439K and N501Y mutations, may significantly enhance the affinity of the S protein for ACE2 (Bayarri-Olmos et al., 2021, Greaney et al., 2021a). In addition, three modifications near the Omicron cleavage site may increase transmissibility, whereas other modifications may result in immune evasion. Changing a hydrophilic amino acid to a hydrophobic amino acid in the most recent SARS-CoV-2 variant could disrupt the link between hACE2 and RBD. The mutation rate of the receptor-binding motif (RBM) in Omicron is approximately 6–10 times that of other VOC variants. Consequently, the virulence characteristics of the Omicron variation may be superior to those of the other mutual variations (Cameroni et al., 2022, Liu et al., 2022, Cele et al., 2022).
It is essential to emphasize that the majority of "concerns" associated with the above-listed mutations are hypothetical. Interestingly, they also permit binding to ACE2 in a broader range of species. However, the exact mechanisms of Omicron evasion remain a mystery. Based on recent findings, new subvariants of Omicron, known as BA.4, BA.5, and BA2.12.1. are spreading in the United States. It has been reported that these variants have an exceptional capacity to evade vaccination or previous infection-induced immunity (Vogel, 2022). However, further studies are still required to elucidate the exact immune escape mechanisms of these subvariants. In one study on specimens obtained from patients who recovered from COVID-19, it has been indicated that the neutralization titer value is about 50% lower in the Omicron variant in comparison to the Wuhan variant. This reveals that the Omicron variant poses a substantial risk of neutralizing antibody evasion from convalescent patients (Silva et al., 2022).
Based on recent findings, several lineages of the Omicron (B.1.1.529) strain of SARS-CoV-2 have evolved in recent months, with subvariants BA.1 and BA.2 exhibiting significant resistance to neutralizing antibodies (Bazargan et al., 2022). BA.2.12.1 is now the main variant in the United States, whereas BA.4 and BA.5 are the major strains in South Africa, but they also have an increasing distribution in the US. The spike protein sequences of subvariants BA.4 and BA.5 are comparably similar (Vogel, 2022). Furthermore, neutralizing antibody titers against the BA.4 or BA.5 subvariants, as well as (to a lesser extent) the BA.2.12.1 subvariant, were lower than titers against the BA.1 and BA.2 subvariants, indicating that the omicron variant has evolved with greater neutralization escape (Hachmann et al., 2022). These findings give an immunologic framework for the present outbreaks produced by the BA.2.12.1, BA.4, and BA.5 subvariants in populations with high vaccination rates and BA.1 or BA.2 infection. However, further studies are still required to elucidate the exact immune escape mechanisms of these subvariants.
7. Conclusion
Based on research on SARS-CoV-2 immune evasion mechanisms, it has been demonstrated that this virus employs multiple strategies to evade the host immune system. Several of these mechanisms are shared between different viruses, but a few of them are highlighted in SARS-CoV-2, which results in the emergence of novel variants and the persistence of infection. Evaluation of these strategies could be beneficial not only for COVID-19 management but also for future possible pandemics and other emerging infections. In addition to the development of new drugs and vaccines, the COVID-19 pandemic continues to evolve. We still require more effective and practical methods of prevention and treatment based on new findings on SARS-COV2 and host immune system interactions.
Funding
Not applicable.
CRediT authorship contribution statement
Sh.A. and M.B. contributed to data gathering and writing the primary manuscript. Sh.A. contributed to designing the figure and table. R.E. contributed to the scientific and grammatical revising of the manuscript. A.E. contributed to the hypothesis, correspondence, and editing manuscript before submission.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
As healthcare providers, we acknowledge all scholars for their precious efforts in overcoming the COVID-19 pandemic.
Data Availability
No data was used for the research described in the article.
References
- Alexandar S., Ravisankar M., Kumar R.S., Jakkan K. A comprehensive review on Covid-19 Delta variant. Int. J. Pharmacol. Clin. Res. (IJPCR) 2021;5(83–85):7. [Google Scholar]
- Amor S., Fernández Blanco L., Baker D. Innate immunity during SARS-CoV-2: evasion strategies and activation trigger hypoxia and vascular damage. Clin. Exp. Immunol. 2020;202(2):193–209. doi: 10.1111/cei.13523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayarri-Olmos R., Jarlhelt I., Johnsen L.B., Hansen C.B., Helgstrand C., Rose Bjelke J., et al. Functional effects of receptor-binding domain mutations of SARS-CoV-2 B.1.351 and P.1 variants. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.757197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayati A., Kumar R., Francis V., McPherson P.S. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazargan M., Elahi R., Esmaeilzadeh A. OMICRON: virology, Immunopathogenesis, and laboratory diagnosis. J. Gene Med. 2022 doi: 10.1002/jgm.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer D.K., Forero A. Mechanisms of antiviral immune evasion of SARS-CoV-2. J. Mol. Biol. 2022;434(6) doi: 10.1016/j.jmb.2021.167265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602(7898):664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Sci. (N. Y., NY) 2020;370(6518):856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Dhungel P., Yang Z. Going against the tide: selective cellular protein synthesis during virally induced host shutoff. J. Virol. 2017;91(17) doi: 10.1128/JVI.00071-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602(7898):657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreño J.M., Alshammary H., Tcheou J., Singh G., Raskin A.J., Kawabata H., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602(7898):682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602(7898):654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesaro T., Michiels T. Inhibition of PKR by viruses. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.757238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Fan J., Chen Z., Zhang M., Peng H., Liu J., et al. Nonmuscle myosin heavy chain IIA facilitates SARS-CoV-2 infection in human pulmonary cells. Proc. Natl. Acad. Sci. USA. 2021;118(50) doi: 10.1073/pnas.2111011118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J.D., Krogstad P. SARS: the first pandemic of the 21st century. Pedia Res. 2004;56(1):1–5. doi: 10.1203/01.PDR.0000129184.87042.FC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury M.A., Hossain N., Kashem M.A., Shahid M.A., Alam A. Immune response in COVID-19: a review. J. Infect. Public Health. 2020;13(11):1619–1629. doi: 10.1016/j.jiph.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185(3):467–484.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C., He J., Zhang X., Jiang C., Sun Y., Zhang Y., et al. Crucial mutations of spike protein on SARS-CoV-2 evolved to variant strains escaping neutralization of convalescent plasmas and rbd-specific monoclonal antibodies. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.693775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drappier M., Michiels T. Inhibition of the OAS/RNase L pathway by viruses. Curr. Opin. Virol. 2015;15:19–26. doi: 10.1016/j.coviro.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi R., Karami P., Heidary A.H., Esmaeilzadeh A. An updated overview of recent advances, challenges, and clinical considerations of IL-6 signaling blockade in severe coronavirus disease 2019 (COVID-19) Int. Immunopharmacol. 2022 doi: 10.1016/j.intimp.2022.108536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeilzadeh A., Elahi R. Immunobiology and immunotherapy of COVID-19: a clinically updated overview. J. Cell. Physiol. 2021;236(4):2519–2543. doi: 10.1002/jcp.30076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeilzadeh A., Jafari D., Tahmasebi S., Elahi R., Khosh E. Coronavirus Disease-COVID-19. Springer; 2021. Immune-based therapy for COVID-19; pp. 449–468. [DOI] [PubMed] [Google Scholar]
- Esmaeilzadeh A., Rostami S., Yeganeh P.M., Tahmasebi S., Ahmadi M. Recent advances in antibody-based immunotherapy strategies for COVID-19. J. Cell. Biochem. 2021;122(10):1389–1412. doi: 10.1002/jcb.30017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel Y., Gluck A., Winkler R., Nachshon A., Mizrahi O., Lubelsky Y., et al. SARS-CoV-2 utilizes a multipronged strategy to suppress host protein synthesis. bioRxiv. 2020:2020.11.25.398578.
- Fisher T., Gluck A., Narayanan K., Kuroda M., Nachshon A., Hsu J.C., et al. Parsing the role of NSP1 in SARS-CoV-2 infection. bioRxiv. 2022. [DOI] [PMC free article] [PubMed]
- Flower T.G., Buffalo C.Z., Hooy R.M., Allaire M., Ren X., Hurley J.H. Structure of SARS-CoV-2 ORF8, a rapidly evolving coronavirus protein implicated in immune evasion. bioRxiv. 2020. [DOI] [PMC free article] [PubMed]
- Frazier M.N., Dillard L.B., Krahn J.M., Perera L., Williams J.G., Wilson I.M., et al. Characterization of SARS2 Nsp15 nuclease activity reveals it's mad about U. Nucleic Acids Res. 2021;49(17):10136–10149. doi: 10.1093/nar/gkab719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung S.Y., Siu K.L., Lin H., Yeung M.L., Jin D.Y. SARS-CoV-2 main protease suppresses type I interferon production by preventing nuclear translocation of phosphorylated IRF3. Int J. Biol. Sci. 2021;17(6):1547–1554. doi: 10.7150/ijbs.59943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaebi M., Tahmasebi S., Jozghorbani M., Sadeghi A., Thangavelu L., Zekiy A.O., et al. Risk factors for adverse outcomes of COVID-19 patients: possible basis for diverse responses to the novel coronavirus SARS-CoV-2. Life Sci. 2021;277 doi: 10.1016/j.lfs.2021.119503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori Savellini G., Anichini G., Gandolfo C., Cusi M.G. Nucleopore traffic is hindered by SARS-CoV-2 ORF6 protein to efficiently suppress IFN-β and IL-6 secretion. Viruses. 2022;14(6) doi: 10.3390/v14061273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29(3):463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H., Starr T.N., Malone K.D., Chu H.Y., et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29(3):463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Gan H., Ma Y., Xu L., Cheng Z.J., Li B., et al. The molecular mechanism of SARS-CoV-2 evading host antiviral innate immunity. Virol. J. 2022;19(1):1–11. doi: 10.1186/s12985-022-01783-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Gan H., Ma Y., Xu L., Cheng Z.J., Li B., et al. The molecular mechanism of SARS-CoV-2 evading host antiviral innate immunity. Virol. J. 2022;19(1):49. doi: 10.1186/s12985-022-01783-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachmann N.P., Miller J., Collier A.Y., Ventura J.D., Yu J., Rowe M., et al. Neutralization Escape by SARS-CoV-2 Omicron Subvariants BA.2.12.1, BA.4, and BA.5. The New England journal of medicine. 2022. [DOI] [PMC free article] [PubMed]
- Hajjar S.A., Memish Z.A., McIntosh K. Middle East Respiratory Syndrome Coronavirus (MERS-CoV): a perpetual challenge. Ann. Saudi Med. 2013;33(5):427–436. doi: 10.5144/0256-4947.2013.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R., Guedán A., Yap M.W., Young G.R., Harvey R., Stoye J.P., et al. SARS-CoV-2 ORF6 disrupts innate immune signalling by inhibiting cellular mRNA export. PLoS Pathog. 2022;18(8) doi: 10.1371/journal.ppat.1010349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Zhuang M.W., Deng J., Zheng Y., Zhang J., Nan M.L., et al. SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5-MAVS, TLR3-TRIF, and cGAS-STING signaling pathways. J. Med Virol. 2021;93(9):5376–5389. doi: 10.1002/jmv.27050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayn M., Hirschenberger M., Koepke L., Nchioua R., Straub J.H., Klute S., et al. Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities. Cell Rep. 2021;35(7) doi: 10.1016/j.celrep.2021.109126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt E.W. The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology. 2003;110(2):163–169. doi: 10.1046/j.1365-2567.2003.01738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Krüger N., Schulz S., Cossmann A., Rocha C., Kempf A., et al. The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell. 2022;185(3):447–456.e11. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.C., Laurent-Rolle M., Pawlak J.B., Wilen C.B., Cresswell P. Translational shutdown and evasion of the innate immune response by SARS-CoV-2 NSP14 protein. Proc. Natl. Acad. Sci. USA. 2021;118(24) doi: 10.1073/pnas.2101161118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11(1):5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knyazev E., Nersisyan S., Tonevitsky A. Endocytosis and Transcytosis of SARS-CoV-2 Across the Intestinal Epithelium and Other Tissue Barriers. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.636966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouwaki T., Nishimura T., Wang G., Oshiumi H. RIG-I-Like Receptor-Mediated Recognition of Viral Genomic RNA of Severe Acute Respiratory Syndrome Coronavirus-2 and Viral Escape From the Host Innate Immune Responses. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.700926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11(1):3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11(1):3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Hou P., Ma W., Wang X., Wang H., Yu Z., et al. SARS-CoV-2 ORF10 suppresses the antiviral innate immune response by degrading MAVS through mitophagy. Cell Mol. Immunol. 2022;19(1):67–78. doi: 10.1038/s41423-021-00807-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F., Wang M., Liu L., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602(7898):676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- Mabrey F.L., Morrell E.D., Wurfel M.M. TLRs in COVID-19: how they drive immunopathology and the rationale for modulation. Innate Immun. 2021;27(7–8):503–513. doi: 10.1177/17534259211051364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min Y.Q., Huang M., Sun X., Deng F., Wang H., Ning Y.J. Immune evasion of SARS-CoV-2 from interferon antiviral system. Comput. Struct. Biotechnol. J. 2021;19:4217–4225. doi: 10.1016/j.csbj.2021.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. USA. 2020;117(45):28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamadian M., Chiti H., Shoghli A., Biglari S., Parsamanesh N., Esmaeilzadeh A. COVID‐19: virology, biology and novel laboratory diagnosis. J. Gene Med. 2021;23(2) doi: 10.1002/jgm.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaei S., Dadkhah M., Asghariazar V., Karami C., Safarzadeh E. The immune response and immune evasion characteristics in SARS-CoV, MERS-CoV, and SARS-CoV-2: vaccine design strategies. Int. Immunopharmacol. 2021;92 doi: 10.1016/j.intimp.2020.107051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustaqil M., Ollivier E., Chiu H.P., Van Tol S., Rudolffi-Soto P., Stevens C., et al. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerg. Microbes Infect. 2021;10(1):178–195. doi: 10.1080/22221751.2020.1870414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.J., Shin O.S. SARS-CoV-2 nucleocapsid protein targets RIG-I-like receptor pathways to inhibit the induction of interferon response. Cells. 2021;10(3) doi: 10.3390/cells10030530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J., Lan W., Wu X., Zhao T., Duan B., Yang P., et al. Tracking SARS-CoV-2 Omicron diverse spike gene mutations identifies multiple inter-variant recombination events. Signal Transduct. Target. Ther. 2022;7(1):138. doi: 10.1038/s41392-022-00992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park U., Cho N.H. Protective and pathogenic role of humoral responses in COVID-19. J. Microbiol. (Seoul., Korea) 2022;60(3):268–275. doi: 10.1007/s12275-022-2037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees-Spear C., Muir L., Griffith S.A., Heaney J., Aldon Y., Snitselaar J.L., et al. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. 2021;34(12) doi: 10.1016/j.celrep.2021.108890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo L.C., Tomasin R., Matos I.A., Manucci A.C., Sowa S.T., Dale K., et al. The SARS-CoV-2 Nsp3 macrodomain reverses PARP9/DTX3L-dependent ADP-ribosylation induced by interferon signaling. J. Biol. Chem. 2021;297(3) doi: 10.1016/j.jbc.2021.101041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa Ribero M., Jouvenet N., Dreux M., Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16(7) doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S.K., Kumar S., Ansari S., Paweska J.T., Maurya V.K., Tripathi A.K., et al. Characterization of the novel SARS-CoV-2 Omicron (B.1.1.529) variant of concern and its global perspective. J. Med. Virol. 2022;94(4):1738–1744. doi: 10.1002/jmv.27524. [DOI] [PubMed] [Google Scholar]
- Schaecher S.R., Diamond M.S., Pekosz A. The transmembrane domain of the severe acute respiratory syndrome coronavirus ORF7b protein is necessary and sufficient for its retention in the Golgi complex. J. Virol. 2008;82(19):9477–9491. doi: 10.1128/JVI.00784-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavoni G., Mattei F., Gabriele L. Type I interferons as stimulators of DC-mediated cross-priming: impact on anti-tumor response. Front. Immunol. 2013;4:483. doi: 10.3389/fimmu.2013.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M.E., Varga S.M. The CD8 T cell response to respiratory virus infections. Front. Immunol. 2018;9:678. doi: 10.3389/fimmu.2018.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah V.K., Firmal P., Alam A., Ganguly D., Chattopadhyay S. Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Zhang G., Yang Y., Li M., Yang S., Peng G. Lysine 164 is critical for SARS-CoV-2 Nsp1 inhibition of host gene expression. J. Gen. Virol. 2021;102:1. doi: 10.1099/jgv.0.001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., et al. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27(5):1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu A.B., Wilkinson M.F., van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27(3):471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S., Kohl A., Pena L., Pardee K. Recent insights into SARS-CoV-2 omicron variant. Rev. Med. Virol. 2022 doi: 10.1002/rmv.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taefehshokr N., Taefehshokr S., Hemmat N., Heit B. Covid-19: perspectives on innate immune evasion. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.580641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Sci. (N. Y., NY) 2020;369(6508):1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184(5):1171–1187.e20. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Montfoort N., van der Aa E., Woltman A.M. Understanding MHC Class I presentation of viral antigens by human dendritic cells as a basis for rational design of therapeutic vaccines. Front. Immunol. 2014;5 doi: 10.3389/fimmu.2014.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez C., Swanson S.E., Negatu S.G., Dittmar M., Miller J., Ramage H.R., et al. SARS-CoV-2 viral proteins NSP1 and NSP13 inhibit interferon activation through distinct mechanisms. PloS One. 2021;16(6) doi: 10.1371/journal.pone.0253089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan S., Sidiq T., Yousuf S., van den Elsen P.J., Kobayashi K.S. Class I transactivator, NLRC5: a central player in the MHC class I pathway and cancer immune surveillance. Immunogenetics. 2019;71(3):273–282. doi: 10.1007/s00251-019-01106-z. [DOI] [PubMed] [Google Scholar]
- Viswanathan T., Arya S., Chan S.-H., Qi S., Dai N., Misra A., et al. Structural basis of RNA cap modification by SARS-CoV-2. Nat. Commun. 2020;11(1):3718. doi: 10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vithani N., Ward M.D., Zimmerman M.I., Novak B., Borowsky J.H., Singh S., et al. SARS-CoV-2 Nsp16 activation mechanism and a cryptic pocket with pan-coronavirus antiviral potential. Biophys. J. 2021;120(14):2880–2889. doi: 10.1016/j.bpj.2021.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel G. New subvariants are masters of immune evasion. Sci. (N. Y., NY) 2022:679–680. doi: 10.1126/science.adc9448. 13 May 2022. [DOI] [PubMed] [Google Scholar]
- Walsh D., Mohr I. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 2011;9(12):860–875. doi: 10.1038/nrmicro2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Wang W.H., Azadzoi K.M., Su N., Dai P., Sun J., et al. Activation of innate antiviral immune response via double-stranded RNA-dependent RLR receptor-mediated necroptosis. Sci. Rep. 2016;6:22550. doi: 10.1038/srep22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek M., Abualrous E.T., Sticht J., Álvaro-Benito M., Stolzenberg S., Noé F., et al. Major histocompatibility complex (MHC) Class I and MHC Class II proteins: conformational plasticity in antigen presentation. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Shi Y., Pan X., Wu S., Hou R., Zhang Y., et al. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 2021;34(7) doi: 10.1016/j.celrep.2021.108761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Cao Z., Xie X., Zhang X., Chen J.Y., Wang H., et al. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020;33(1) doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Ge J., Zheng L., Zhang Y., Gao Y., Wang T., et al. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell. 2021;184(1):184–193.e10. doi: 10.1016/j.cell.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J.-S., Sasaki M., Cho S.X., Kasuga Y., Zhu B., Ouda R., et al. SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nat. Commun. 2021;12(1):6602. doi: 10.1038/s41467-021-26910-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen C.K., Lam J.Y., Wong W.M., Mak L.F., Wang X., Chu H., et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020;9(1):1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi M. ORF9c and ORF10 as accessory proteins of SARS-CoV-2 in immune evasion. Nat. Rev. Immunol. 2022;22(5):331. doi: 10.1038/s41577-022-00715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Miorin L., Makio T., Dehghan I., Gao S., Xie Y., et al. Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression. Sci. Adv. 2021;7(6) doi: 10.1126/sciadv.abe7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen Y., Li Y., Huang F., Luo B., Yuan Y., et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc. Natl. Acad. Sci. USA. 2021;118(23) doi: 10.1073/pnas.2024202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Zhuang M.W., Han L., Zhang J., Nan M.L., Zhan P., et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct. Target Ther. 2020;5(1):299. doi: 10.1038/s41392-020-00438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Ferraro D., Zhao J., Hussain S., Shao J., Trujillo J., et al. The N-terminal region of severe acute respiratory syndrome coronavirus protein 6 induces membrane rearrangement and enhances virus replication. J. Virol. 2010;84(7):3542–3551. doi: 10.1128/JVI.02570-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.