Abstract
Gonadal somatic cells are the main players in gonad development and are important for sex determination and germ cell development. Here, using a time-series single-cell RNA sequencing (scRNA-seq) strategy, we analyzed fetal germ cells (FGCs) and gonadal somatic cells in human embryos and fetuses. Clustering analysis of testes and ovaries revealed several novel cell subsets, including POU5F1+SPARC+ FGCs and KRT19+ somatic cells. Furthermore, our data indicated that the bone morphogenetic protein (BMP) signaling pathway plays cell type-specific and developmental stage-specific roles in testis development and promotes the gonocyte-to-spermatogonium transition (GST) in late-stage testicular mitotic arrest FGCs. Intriguingly, testosterone synthesis function transitioned from fetal Sertoli cells to adult Leydig cells in a stepwise manner. In our study, potential interactions between gonadal somatic cells were systematically explored and we identified cell type-specific developmental defects in both FGCs and gonadal somatic cells in a Turner syndrome embryo (45, XO). Our work provides a blueprint of the complex yet highly ordered development of and the interactions among human FGCs and gonadal somatic cells.
Keywords: Human gonad, scRNA-seq, Turner syndrome, Leydig-Sertoli cell–cell interaction, Gonocyte-to-spermatogonium transition
Introduction
Development of reproductive systems begins at 4–5 weeks (W) of gestation in humans and completes until puberty with establishment of secondary sexual characteristics and fertility [1]. The normal sex development depends on the appropriate and timely regulations and interactions between genes, signaling pathways, and different types of cells. As the first germ cell population, primordial germ cells (PGCs) are the origin of mature gametes, ensuring that genetic and epigenetic information perpetuates across the generations [2]. Specified from the epiblast, PGCs then migrate to and reside in the developing gonadal ridges at 5–6 W of gestation. During this developmental period, PGCs are bi-potential and can differentiate either to oogonia or gonocytes independent of their chromosome sexes [3], [4]. Instead, signals in the surrounding somatic environment of PGCs determine their sex determination, which reflects that the behaviors of PGCs are tightly regulated by the surrounding gonadal microenvironments [4], [5], [6]. Once PGCs arrive in the gonad, they are enclosed by the supporting cell progenitors and undergo multiple cycles of mitotic proliferation, and sex specification starts to occur. In the testis, after several cycles of mitotic division, PGCs enter mitotic arrest and meiosis will not occur until puberty. On the contrary, in the developing ovary, oogonia undergo a successive process of mitotic expansion (5–24 W of gestation), meiotic division (8–36 W), and meiotic arrest [1].
Despite PGCs playing important roles in transmitting genetic information across the generations, they are not essential for the establishment of somatic cell lineage in the gonads. During human gonad development, gonadal somatic cells first initiate sex-specific differentiation with the specification of two major cell lineages: the supporting cell lineage (Sertoli cells in the testes and granulosa cells in the ovaries) and the steroidogenic cell lineage (Leydig cells in the testes and theca cells in the ovaries) [7]. In XY embryos, initiated by the expression of the SRY gene, the supporting cells differentiate into Sertoli cells, which consequently leads to organization of distinct testis structures, appearance of a steroidogenic cell population (Leydig cells), and production of male hormones [testosterone and Anti-Müllerian hormone (AMH)]. As the main source of androgens, fetal Leydig cells appear shortly after the Sertoli cells at around 7–8 W of gestation [7]. In mammals, two distinct populations of Leydig cells, fetal Leydig cells (fLCs) and adult Leydig cells (aLCs), develop sequentially in the fetal and adult testes, respectively, and exhibit distinct characteristics [8]. The developmental relationship and the origins of these two distinct types of Leydig cells have not been fully understood and remain a matter of debate in humans. Because of the SRY absence in XX embryos, supporting cells differentiate into granulosa cells, which facilitates germ cell meiosis and folliculogenesis. Compared with the well-defined separation mechanisms of supporting Sertoli cells and Leydig cells in the testis, the lineage divergence mechanisms between granulosa cells and the theca cells remain a mystery in the ovary [7].
In humans, abnormal gonadal cell lineage specification and gonadal somatic cell dysfunction can result in disorders of sexual development (DSDs) [7]. Turner syndrome, a DSD caused by loss or structural alteration of the X chromosome in females, occurs in approximately 1 in 2500 newborn girls worldwide, and approximately half of the affected individuals have monosomy X (45, XO) [7]. Losing one copy of X chromosome can cause ovarian dysgenesis in girls with Turner syndrome. The mechanism of the development of germ cells and gonadal somatic cells in Turner syndrome is poorly understood. Here, we explore whether X monosomy has different effects on the gene expression of germ cells and gonadal somatic cells [1], [9].
Insights gained from mouse models have largely advanced our understanding of gonad development, sex-specific differentiation, and DSDs [7], [8], [10], [11], [12], [13], [14], [15]. However, these topics remain poorly explored in humans, especially regarding the development of gonadal somatic cells. Because of species differences, such as the time-spans of specification and differentiation, as well as differences in gene expression and growth factors, it is not feasible to make one-to-one translations between mouse models and humans. Recently, developments in single-cell transcriptomes have facilitated the developmental trajectory exploration, cellular characterization, and novel cell type identification of human gonads [16]. We and other groups described the developmental transcriptomic landscapes of human fetal germ cells (FGCs) and spermatogenesis in studies focused mainly on the development of germ cells [17], [18], [19], [20], [21], [22], [23], [24], [25], [26]. A variety of germ cell types and states have been well demonstrated throughout the development in human fetal, neonatal, infant, and adult stages [18], [20], [21], [27]. Although the major focus of RNA-seq in gonadal tissues is still on the germ cell population, some investigations have begun to focus on the somatic cell types in order to better understanding the somatic cell trajectories and interactions between germ cell populations during gonadal development and maturation. Based on single-cell RNA sequencing (scRNA-seq) data of the human adult testis, it revealed that Leydig cells and myoid cells have a common progenitor before puberty [19], [28]. As for females, the molecular and cellular characteristics of different granulosa and theca cell populations have been revealed in the human adult ovary with single-cell transcriptome data [22]. In addition, the interactions between somatic cells and germ cells through key signaling pathway components during human fetal stages were also predicted by “CellPhoneDB” software, and part of the interactions were further verified through immunofluorescent staining [29]. However, the lineage differentiation, the cell origin of human fetal gonadal somatic cells, and the cell–cell interactions between gonadal somatic cells during human gonad development remain to be studied.
To further expand our understanding of gonadal somatic cell specification and sex differentiation in humans, we performed large-scale scRNA-seq of gonadal cells in both sexes throughout gonad development from 6 W to as late as 23 W post conception. Our results provide an in-depth characterization of the transcriptomic landscapes as well as cellular compositions of human normal gonads and key insights into the transcriptomic features of germ cells and gonadal somatic cells in a human embryo with monosomy X.
Results
Classification of gonadal cell types and identification of embryo sexes
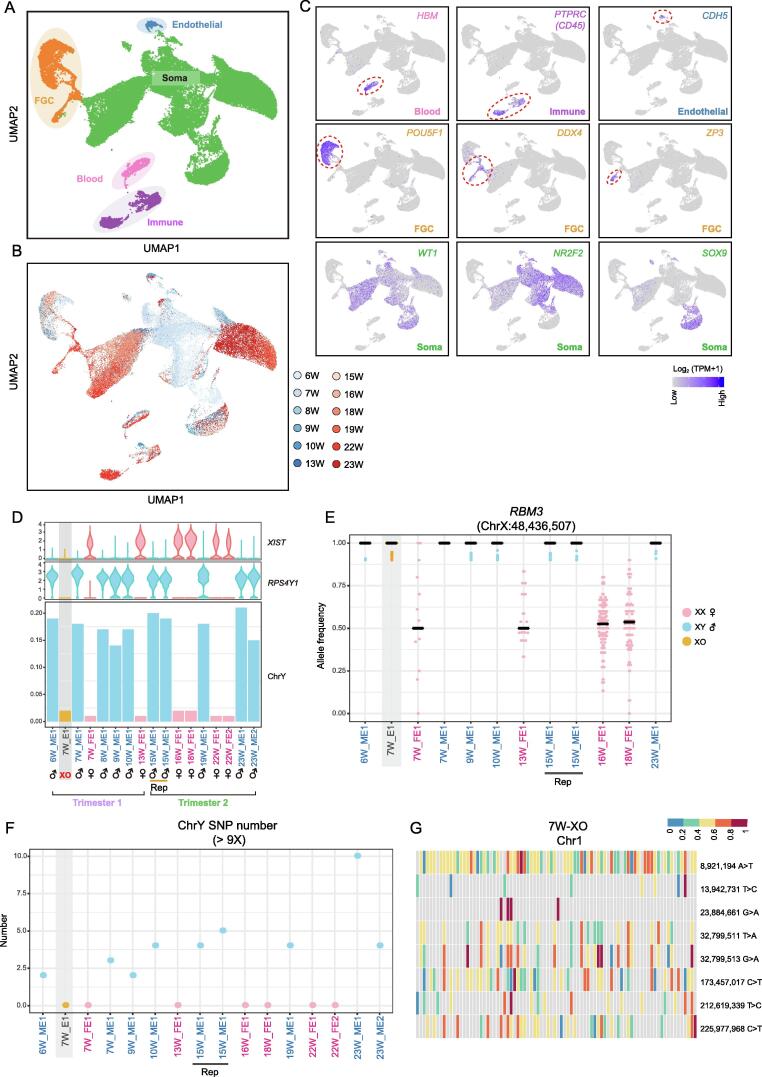
In this study, we used scRNA-seq to analyze all cells in the testes and ovaries from human embryos (before 8 W post conception) and fetuses (after 8 W post conception) at different developmental stages. This approach allowed us to identify all major FGC and somatic cell subsets and to trace the developmental trajectories of these subsets. After filtering out low-quality cells, we retained a total of 56,399 gonadal cells from 16 embryos and fetuses ranging from 6 W to 23 W of age for subsequent analyses (Figure 1A and B; Table S1). Based on uniform manifold approximation and projection (UMAP) clustering, we classified the major cell populations present in the developing gonads, including FGCs (5866), gonadal somatic cells (45,660), blood cells (1267), immune cells (3043), and endothelial cells (563) (Figure 1A–C, Figure S1A and B; Table S1). The clustering results for two technical replicates of a 15 W embryo revealed minimal batch influences (Figure S1A). In addition, the clustering result of two fetuses at the same developmental stage revealed minimal individual differences (Figure S1A). We also estimated the influence of doublet on the data and found that it has not impacted on subsequent analyses (Figure S1C). On average, 1694 genes were detected in each individual cell, and the mean percentage of reads mapped to mitochondria was just 3.2%, which reflected the high quality of our dataset (Figure S1D).
Figure 1.
Identification of cell types and sex determination for human fetal gonads
A. UMAP cluster map revealing the major human fetal gonadal cell types. In this study, all germ cells collected from embryos and fetuses were called FGCs, while germ cells collected from embryos before 8 W post conception were also called PGCs. Different colors represent different cell types. Detailed information is shown in Table S1. B. UMAP cluster map revealing the developmental stages of fetal gonadal cells. C. UMAP cluster maps showing the expression of cell type-specific genes. Colors from gray to purple represent low to high expression levels. Dashed lines highlight the cells that highly express specific genes. D. Violin plots showing the expression of sex-linked genes (XIST on the X chromosome and RPS4Y1 on the Y chromosome) for each embryo. Bar plot showing the ratio of reads mapped to the Y chromosome. Pink represents female embryos, sky blue represents male embryos, and orange represents the XO embryo. E. Dot plot showing the allele frequency of the X chromosome inactivation escape gene RBM3 in each embryo. One dot represents one cell. Only cells for which at least 9 reads were detected at this location are shown. The Y-axis represents the allele frequency of position ChrX:48,436,507, and the X-axis indicates different embryos/fetuses. The color of each dot represents the karyotype of the cell. If the embryo is female with two X chromosomes, then it should have a heterozygous SNP in the X chromosome inactivation escape region. In contrast, a male embryo with only one X chromosome should have a homozygous SNP. Inactivation of the X chromosome in female embryos will also lead to detection of a homozygous SNP. Therefore, it is necessary to determine sex by evaluating SNPs in the X chromosome inactivation escape region. F. Dot plot showing the number of SNPs located on the Y chromosome for each embryo. If an embryo does not contain a Y chromosome, then theoretically no such SNPs will be detected, i.e., SNPs on the Y chromosome will not detected in female or XO embryos. Only SNPs for which at least 9 reads were detected at this location are shown. G. Heatmap showing the detected SNP frequency on chromosome 1 of a 7 W (45, XO) embryo. Colors from blue to red represent low to high allele frequencies. Only positions with SNPs in at least 5 individual cells with more than 9 sequence reads are shown. Most of cells exhibited heterozygous SNPs on chromosome 1. Each column represents a single cell. FGC, fetal germ cell; PGC, primordial germ cell; Soma, the supporting cell lineage and steroidogenic cell lineage in gonads; W, week; UMAP, uniform manifold approximation and projection; SNP, single nucleotide polymorphism; Rep, technical replicate; XO, a Turner syndrome embryo with monosomy X (45, XO); E, embryo; ME, male embryo; FE, female embryo; Chr, chromosome.
At early developmental stages, such as 6 W to 10 W, it is relatively hard to distinguish female and male gonads based on sexual gland morphology. Therefore, we calculated the ratio of reads mapped to the Y chromosome and the expression of genes located on sex chromosomes (XIST on the X chromosome and RPS4Y1 on the Y chromosome) to resolve the sex of each embryo and fetus (Figure 1D). The results determined by this method were highly consistent (with 100% accuracy) with those determined by morphology for late-stage fetuses, verifying the accuracy of our strategy (Figure 1D, Figure S1E). Female embryos and fetuses strongly expressed XIST but barely expressed RPS4Y1, and few reads mapped to the Y chromosome (Figure 1D, Figure S1E). However, these genes were abnormally expressed in a 7 W embryo; this embryo expressed neither XIST nor RPS4Y1, and few reads mapped to the Y chromosome (Figure 1D). These results implied that this embryo exhibited monosomy X (45, XO). The gonads of this XO embryo did not show a significant morphological difference with those from normal XX embryos. To verify this preliminary determination of (45, XO), we performed single nucleotide polymorphism (SNP) calling in the scRNA-seq dataset (Figure 1E–G); the allele frequency of the X chromosome inactivation escape gene (RBM3) was approximately 50% in the female embryos but approximately 100% in the male embryos and the 7 W XO embryo (Figure 1E). We also counted the number of SNPs on the Y chromosome and found none in all female embryos and the 7 W XO embryo, as expected (Figure 1F). To confirm the technical accuracy on our findings, we investigated the allele frequency of SNPs on chromosome 1 of the 7 W XO embryo and found that most SNP loci were heterozygous, as expected (Figure 1G). Therefore, the 7 W embryo was confirmed as an XO embryo.
Comparison of the transcriptome profiles of PGCs and gonadal somatic cells among XO, XX, and XY embryos
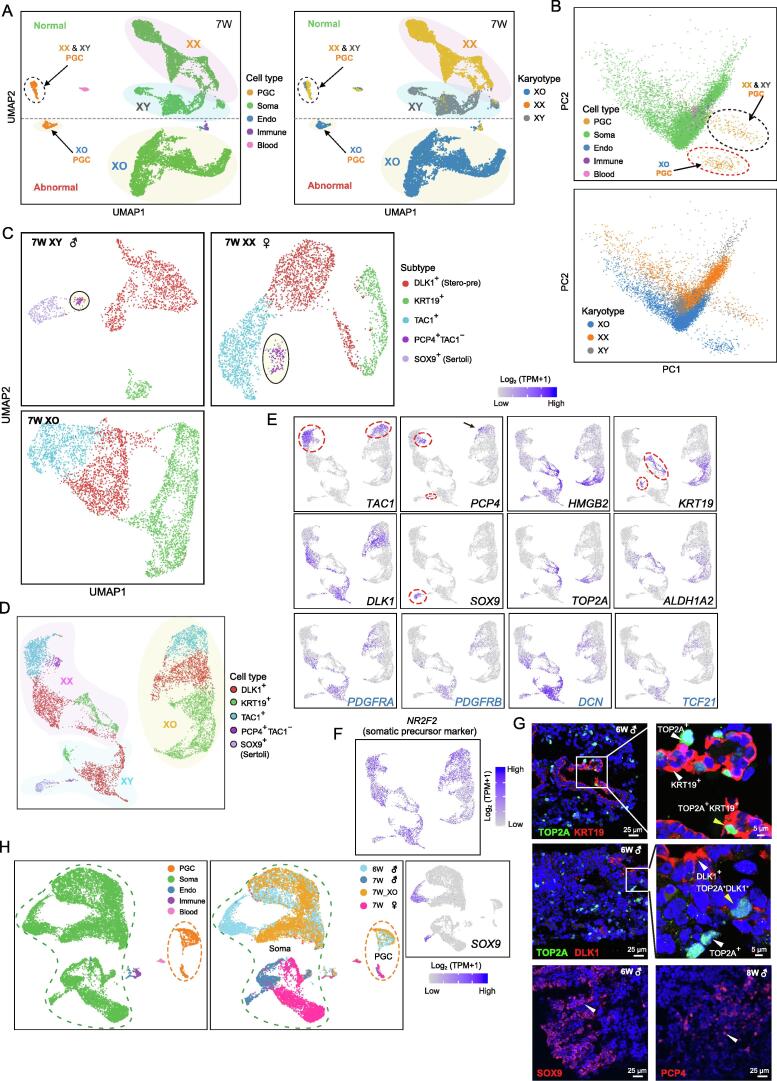
To characterize the phenotype of the monosomy X female embryo, we further clustered cells from XO, normal XX, and normal XY embryos at the same developmental stage (7 W). PGCs from normal female and male embryos clustered together, whereas those from the XO embryo clustered separately (Figure 2A and B, Figure S1F and G; Table S2). In addition, gonadal somatic cells clustered according to karyotype (XX, XY, and XO), but the transcriptomes of normal embryos (XX and XY) were more similar to each other than to that of the XO embryo (Figure 2A and B). Next, we identified differentially expressed genes (DEGs) in gonadal somatic cells from these embryos (Figure 2A and B, Figure S1F–J; Table S2). The highly expressed genes in the normal XX ovarian somatic cells were enriched for genes involved in the regulation of the estrogen signaling pathway [FOS and JUN, P = 1.5 × 10−5, false discovery rate (FDR) = 5.1 × 10−4]. In contrast, the XO gonadal somatic cells highly expressed genes such as APOE, RAC1, and IGFBP2. We also compared the expression profiles of PGCs between XO and normal XX and XY embryos (Figure S1G and I; Table S2). Similar to the results for gonadal somatic cells, PGCs in the XO embryo also highly expressed genes such as RAC1 and IGFBP2 (Figure S1H and I). On the other hand, unlike gonadal somatic cells, PGCs in the XO embryo showed a lack of expression of genes associated with cell cycle (P = 3.6 × 10−7, FDR = 3.5 × 10−5) and DNA replication (P = 7.7 × 10−6, FDR = 3.4 × 10−4). Specifically, PGCs in the XO embryo displayed specific down-regulation of HAT1 and PSMD12, genes that participate in replication-dependent chromatin assembly and cell cycle progression, and up-regulation of UTF1, PDCD2, and FRAT2, genes that play critical roles in the WNT signaling pathway, regulation of cell proliferation, and embryo differentiation (Figure S1H and I). Notably, these five up-/down-regulated genes did not show expression differences in gonadal somatic cells between the XO embryo and normal XX embryos, indicating that monosomy X embryo has cell type-specific defects in PGCs and somatic cells in the gonad microenvironment. PGCs and gonadal somatic cells did share changes in the expression of same genes, such as PDIA3, RAC1, ATP5D, and IGFBP2 (Figure S1H and I).
Figure 2.
Comparison of the transcriptome profiles of 7 W XO and normal XX and XY embryos
A. UMAP cluster maps revealing the major cell types and karyotypes of three 7 W-old embryos (45, XO; 46, XX; and 46, XY). Different colors represent different cell types (left) and karyotypes (right). The dashed black line separates the single cells from normal embryos (46, XX and 46, XY) and abnormal embryo (45, XO). Arrows indicate the location of PGCs, and normal embryo PGCs are highlighted with a dashed black circle. B. PCA cluster maps revealing the major cell types and karyotypes of cells. Different colors represent different cell types (top) and karyotypes (bottom). C. UMAP cluster maps revealing the major somatic cell types in each 7 W embryo. The black circles highlight the cell population that exists in only normal embryos (46, XX and 46, XY). D. UMAP clustering of all 7 W gonadal somatic cells, including one abnormal embryo (45, XO), one male embryo (46, XY), and one female embryo (46, XX). Colors represent the different cell types identified in (C). Detailed information is shown in Table S2. E. UMAP cluster maps showing the expression of genes used to identify cell types. In the XO embryo, PCP4 was expressed by some TAC1+ cells, but the PCP4+TAC1− cells were not observed. Genes with names in blue are steroidogenic cell lineage markers. Colors from gray to purple represent low to high expression levels. F. UMAP cluster map showing the expression of somatic precursor marker NR2F2 in all three 7 W embryonic somatic cells. Colors from gray to purple represent low to high expression levels. G. Immunostaining for TOP2A, KRT19, DLK1, ALDH1A2, SOX9, and PCP4 in early-stage fetal testes. H. UMAP clustering of all 7 W embryos and one 6 W embryo. Different colors represent different cell types (left) and embryos (right). The expression of the Sertoli cell marker SOX9 was projected onto the UMAP. Colors from gray to purple represent low to high expression levels. SOX9+ Sertoli cells existed in only male embryos (6 W and 7 W male embryos). PCA, principal component analysis; PC, principal component; Endo, endothelial.
Next, we classified the somatic cell populations present in all 7 W embryos using UMAP clustering (Figure 2C–E, Figure S2A; Table S2). Three cell clusters were identified in the XO embryo; two of these clusters were also detected in both normal XX and XY embryos (DLK1+ and KRT19+), whereas the TAC1+ cluster was only shared with the XX embryo. Notably, the cell clusters identified in all three 7 W embryos highly expressed the somatic progenitor cell marker LHX9 and NR2F2 (Figure 2F, Figure S1J) [7], [28]. The cell cluster that highly expressed TAC1 was shared by only XX and XO embryos, which may imply that this cell type is unique to female embryos at an early developmental stage (Figure 2E; Table S2). Interestingly, TAC1 was also reported to play important roles in control of sexual maturation and/or fertility in male mice [30]. Specifically, at this embryonic stage, some of the XY male gonadal somatic cells had differentiated into Sertoli cells, a supporting cell lineage, indicating that specification of the supporting cell lineage occurs earlier in male embryos than in female embryos (Figure 2C and D). In addition, both XX and XY embryos contained a subset of cells that highly expressed PCP4, which encodes a modulator of calmodulin activity (Figure 2C–E). Different from the fetal stages during which PCP4 is expressed by both sexes, PCP4 was mainly expressed by cells in male reproductive organs, indicating that PCP4 might be involved in the sex differentiation of gonads [31], [32] (Figure 2D and E). Immunostaining was performed to verify all the cell populations newly identified in the early-stage testes (Figure 2G). Interestingly, DLK1+ cells in all 7 W embryos (XX, XY, and XO) expressed marker genes of the steroidogenic cell lineage, such as PDGFRA, PDGFRB, DCN, and TCF21. Specifically, TCF21 has been reported as a marker gene for Leydig progenitor cells in mice [33], [34], [35], perhaps indicating that DLK1+ cells are steroidogenic progenitor cells (Figure 2E). Next, to explore whether the lack of a sex chromosome in the XO embryo delays embryonic development, we clustered the cells from 7 W embryos with those from a 6 W male embryo. The results showed that except for male-specific Sertoli cells (SOX9+), gonadal somatic and germ cells from the 7 W XO embryo clustered closely with those from the 6 W male (XY) embryo, indicating that the lack of an X chromosome delays embryonic development in XO embryos (Figure 2H). To summarize, XO embryo exhibited abnormal gene expression profiles in both germ cells and gonadal somatic cells, and lacked the PCP4+TAC1− cell populations present in normal XX embryos.
Identification of novel FGC subtypes
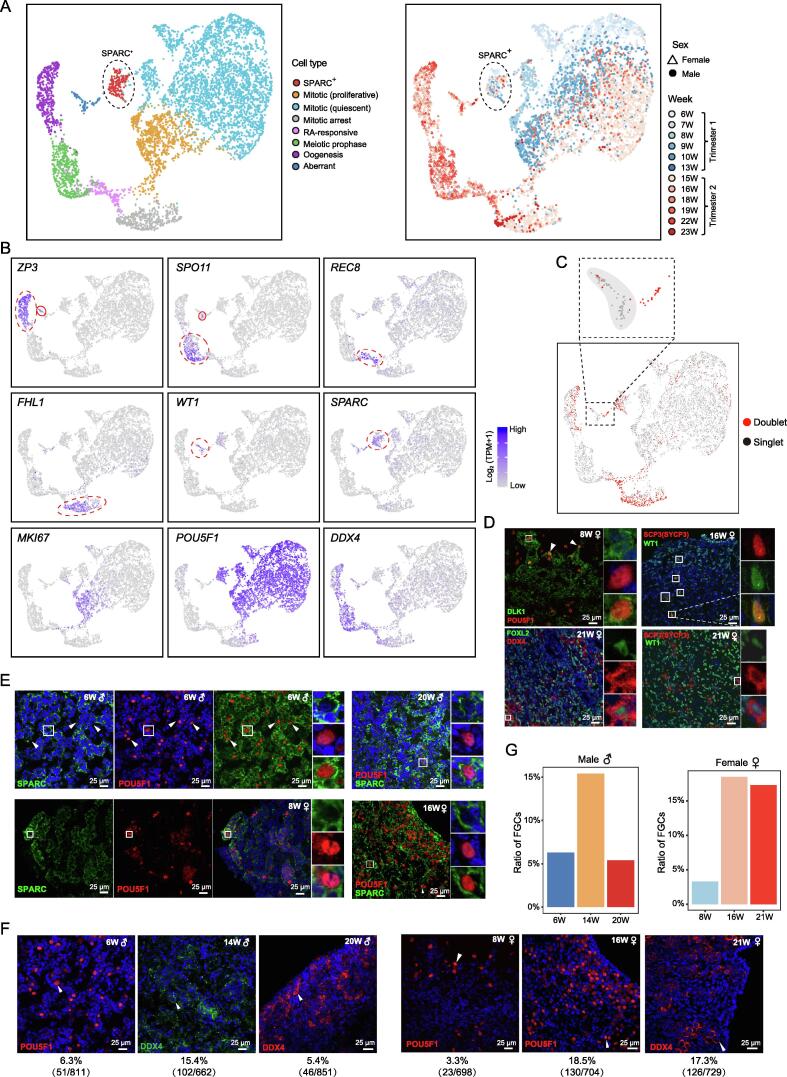
Next, we analyzed the transcriptional characteristics of FGCs. By performing UMAP unsupervised clustering, we clearly identified 8 clusters of FGCs (Figure 3A and B; Table S3). Based on the expression of well-known FGC markers, these clusters were classified as mitotic (proliferative and quiescent), mitotic arrest, and SPARC+ FGCs in male gonads as well as mitotic, meiotic prophase, retinoic acid (RA)-responsive, oogenesis, and aberrant FGCs in female gonads (Figure 3A and B). The cluster of aberrant FGCs simultaneously expressed somatic cell markers and FGC markers (Figure 3A and B). Since technical doublet might also exhibit similar patterns, we first used “DoubletFinder” to predict the doublet and found that most of these aberrant FGCs were identified as “Singlet” (Figure 3C) [36]. To further explore whether this group of cells really exists or is just an artifact due to cell debris contamination, we performed immunofluorescence staining of three pairs of well-known somatic and germ cell marker genes in three fetal ovaries at the early, middle, and late stages (Figure 3D). The results showed that a small fraction of gonadal cells in all three embryos expressed both germ cell and somatic cell marker genes at the protein levels, suggesting that the aberrant FGCs we identified do exist in human gonads in vivo (Figure 3C and D). Notably, we observed that the germ cell-specific marker SCP3/SYCP3 was expressed in different subcellular locations in 16 W and 21 W ovarian tissues. In addition, we found that there were a small number of gonadal somatic cells that also simultaneously expressed some germ cell markers, such as DDX4, POU5F1, and ZP3 (Figure 1A and C). The transcriptomic patterns of early- and late-stage FGCs differed dramatically in females but showed milder differences in males (Figure 3A). Most of these clusters were identified in our previous study [17], verifying the accuracy of our analyses. Due to the increased number of cells analyzed, we identified a new FGC subtype, SPARC+ cells, in both male and female gonads. SPARC+ cells were mainly found in first trimester embryos and fetuses (Figure 3A and B). Through immunofluorescence staining of early- and late-stage human fetal testes and ovaries, we verified the existence of these newly identified SPARC+POU5F1+ FGCs in both sexes (Figure 3E). Since SPARC plays an important role in cell migration, we speculate that this group of cells may retain the ability to migrate locally within the gonads at the later stages, such as migration between the cortex and medulla in the ovary [37]. To further investigate the expression profiles of these FGC subtypes, we performed the DEG analysis (Figure S2A; Table S3). As expected, previously reported FGC subtypes expressed cell type-specific marker genes; for example, mitotic FGCs highly expressed POU5F1, NANOG, and MKI67 (mitotic proliferative FGCs), whereas mitotic arrest FGCs highly expressed BMP signaling pathway target genes such as ID1, ID3, and ID4 (Figure S2A; Table S3) [17].
Figure 3.
FGC subtype identification and verification
A. Two-dimensional embedding of all fetal gonadal germ cells using UMAP. The cells are colored according to cell type (left) and gestational age (right). The shape indicates cells from male or female embryos. Dashed black circles highlight SPARC+ FGCs. B. UMAP cluster maps showing the expression patterns of known FGC markers. Red circles highlight cells that express specific marker genes. Colors from gray to purple represent low to high expression levels. C. UMAP cluster map showing the distribution of doublet. D. Immunostaining of markers of somatic cells and germ cells in fetal ovaries to validate the existence of cells expressing both somatic cell and germ cell markers. DLK1, FOXL2, and WT1 are somatic cell markers. POU5F1, SCP3 (also known as SYCP3), and DDX4 are germ cell markers. E. Immunostaining of male testes (6 W and 20 W) and female ovaries (8 W and 16 W) to verify the existence of the SPARC+POU5F1+ FGC subtype and its spatial distribution. Cells inside the white square are magnified and shown on the right of the corresponding image. F. Immunostaining of 8 W, 14 W, and 20 W testes and 8 W, 16 W, and 21 W ovaries. The percentage of FGCs for each stage is shown. G. Bar plots showing the ratios of FGCs at several developmental ages calculated by immunofluorescence showed in (F). RA, retinoic acid.
We then assessed the relative ratios of the defined FGC subtypes across different developmental stages in testes and ovaries (Figure S2B). In 6–7 W gonads, both male and female FGCs consisted mainly of mitotic FGCs (Figure S2B). From 13 W onward, RA-responsive, meiotic prophase, and oogenesis FGCs began to appear in ovaries. By 22 W, more than half of the FGCs in females were in meiosis, and the rest were mainly RA-responsive and oogenetic FGCs. In 8–10 W testes, more than 25% of FGCs were in the active proliferation phase, consistent with the significant increase in the total number of FGCs in testes from 8 W onward (Figure S2B). Mitotic arrest FGCs appeared at 10 W and became the major FGC type at 23 W, accounting for 68.1% of FGCs in testes.
Relative proliferation of human FGCs in males and females
Since we sequenced all the gonadal cells without fluorescence-activated cell sorting (FACS) enrichment, we could explore the dynamic changes in the percentages of FGCs during gonad development. The results showed that male FGCs underwent rapid proliferation and expansion at as early as 8 W and their percentages increased rapidly until 15 W and then sharply decreased after 19 W. However, the proportion of female FGCs was only 1.3% at 13 W, and the proportion increased 10 times to 13.4% at 16 W, indicating that female FGCs might experience a rapid increase phase during this period (Figure S2C). We then performed immunofluorescence staining of FGC markers in early-, middle-, and late-stage human fetal testes and ovaries to verify the changes in the FGC percentages (Figure 3F and G). Consistent with the sequencing results, the percentage of FGCs in male gonads first increased and then decreased sharply, while this percentage increased and then remained relatively constant at the sampling times in females. These lines of evidence suggest sex-specific proliferation patterns of FGCs in the human gonads. Since we measured the proliferation pattern of FGCs in different sexes by the ratio of FGCs to the total number of detected cells, the ratio remained unchanged when FGCs and somatic cells had comparable proliferation speed, so it only reflected the relative proliferation rate of FGCs (Figure S2C).
Transcriptomic features of prespermatogonia
Spermatogenesis is the continuous and highly coordinated process of sperm production [15], [20], [21], [24]. In mice, it has been reported that gonocyte-to-spermatogonium transition (GST) is initiated in a subset of gonocytes at as early as embryonic day 18.5 (E18.5) [38]. In humans, the initiation of spermatogenesis is believed to occur during puberty. Due to the scarcity of biological materials and technological limitations, there is no clear definition of GST, and the precise timing of GST remains unclear in human testes [38].
In males, after arrival at the gonadal anlage, FGCs usually give rise to prespermatogonia (gonocytes), which form a homogenous population of single, round cells in embryonic testes. During the first trimester, gonocytes continue to express markers of early FGCs such as OCT4 (POU5F1), NANOG, and DDX4, despite showing some morphological differences. During the second trimester, most but not all gonocytes gradually lose their mitotic activity along with early FGC marker expression. At this time, a new type of mitotic arrest germ cell begins to appear, called the transitional (T)-spermatogonia, which is similar to the f0 or gonia cells that were mentioned in other studies based on the scRNA-seq data of human fetal testes [28], [29], [39], [40], [41], [42].
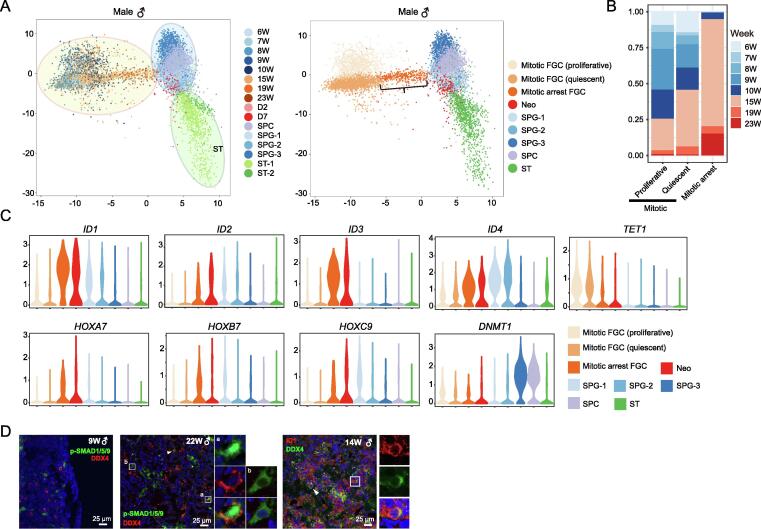
To characterize GST progression, we combined expression data for human fetal, neonatal, and adult germ cells and performed clustering by principal component analysis (PCA) (Figure 4A, Figure S2D and E; Table S4) [21]. According to the PCA and gene expression data, FGCs were grouped into mitotic FGCs and mitotic arrest FGCs, with the former group further divided into the proliferative and quiescent subtypes as we previously reported [17] (Figure 4A, Figure S2F). Mitotic arrest FGCs mainly originated from testes in the second trimester, accounting for 85% of FGCs in this group, while proliferative mitotic FGCs were predominantly derived from testes in the first trimester (Figure 4B). According to the PCA, mitotic arrest FGCs represent a transition state from gonocytes to spermatogonia (Figure 4A and C). The BMP signaling pathway and HOX family genes may play roles in GST, since ID1–4, as well as HOXA7, HOXB7, and HOXC9, was highly expressed from the mitotic arrest FGC stage to the postnatal stages (Figure 4C, Figures S2F, S3A and B). Through immunostaining of the BMP signaling pathway component p-SMAD1/5/9 and its target ID1, as well as the late-stage FGC marker DDX4 in human testes (9 W, 14 W, and 22 W), we verified that the BMP signaling pathway was activated in mitotic arrest FGCs (Figure 4D). Through immunofluorescent staining of SMAD proteins in the BMP signaling pathway, another study also verified the activated BMP singling pathway in mitotic arrest male FGCs, which further confirmed the accuracy of our study [29]. A previous study has reported that ALDH1A2 is most likely responsible for RA biosynthesis in developing germ cells [43]. We found that ALDH1A2 was expressed in a small fraction of mitotic arrest FGCs, indicating the initiation of GST in some of these FGCs in fetal late-stage testes (Figure S2G and H).
Figure 4.
GST characterization of male gonadal cells
A. Combined analysis of fetal, neonatal, and adult male germ cells by PCA. The cells are colored by cell type and developmental stage. B. Bar plot showing the developmental stage distribution of three male FGCs. The color represents developmental stage. C. Violin plot showing the trajectory expression patterns in male germ cells from fetal (6–23 W), neonatal, and adult testes. D. Immunostaining of the germ cell marker DDX4, the BMP signaling pathway component p-SMAD1/5/9, and a BMP signaling pathway target ID1 in fetal testes (9 W, 14 W, and 22 W). Cells inside the white squares are magnified and shown on the right of the corresponding images. The white square with symbol “a” highlights the germ cells that expressed p-SMAD1/5/9 in the nucleus, while the white square with symbol “b” highlights the germ cells that expressed p-SMAD1/5/9 in the cytoplasm. Neo, neonatal; D2, day 2 after birth; D7, day 7 after birth; SPG, spermatogonium; SPC, spermatocyte; ST, spermatid; GST, gonocyte-to-spermatogonium transition.
The BMP signaling pathway plays cell type- and developmental stage-specific roles in human gonad development
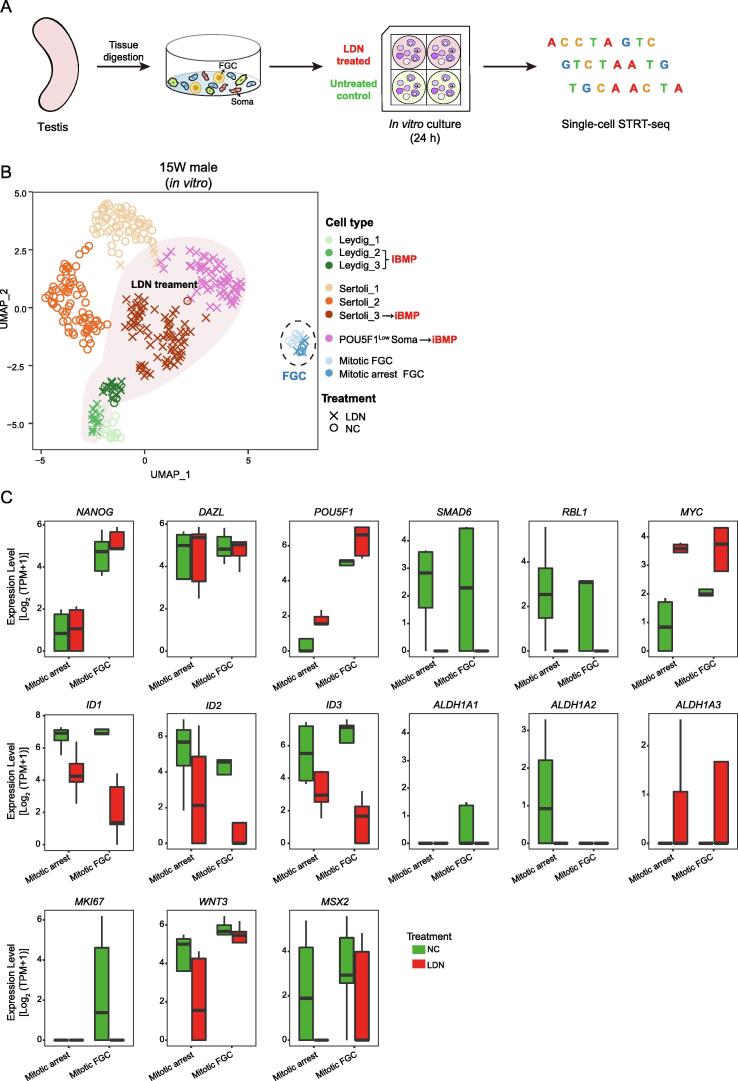
To further explore the roles of the BMP signaling pathway in GST, we blocked this pathway in cultured 15 W testicular cells with LDN-19318 (LDN) and then performed scRNA-seq analysis using a modified single-cell tagged reverse transcription sequencing (STRT-seq) method (Figure 5A). We captured all major cell types of late-stage fetal testes, including Leydig cells, Sertoli cells, mitotic FGCs, and mitotic arrest FGCs (Figure 5B, Figure S3C; Table S5). We first evaluated the inhibitory effect of LDN on the BMP signaling pathway (Figure 5C, Figure S3D) and found a dramatic decrease in the expression of the canonical BMP target genes ID1–3 in LDN-treated testicular cells. LDN treatment completely suppressed SMAD6 and RBL1 expression, resulting in increased expression of their downstream target gene MYC, a key cell cycle regulator. LDN treatment also blocked the proliferation of mitotic FGCs and decreased WNT3 and MSX2 expression, as we previously reported [44] (Figure S3D). Notably, inhibiting the BMP signaling pathway resulted in dramatically decreased ALDH1A2 expression in mitotic arrest FGCs (Figure 5C, Figure S3D). In addition, we further explored the expression of BMP signaling pathway ligand-coding genes in testicular somatic cells, and it showed that only the BMP signaling pathway ligand-coding gene AMH was highly expressed by Sertoli cells, which indicated that through secreting AMH, Sertoli cells might be involved in BMP signaling pathway activation in germ cells (Figure S5B). These data further verified that the BMP signaling pathway is activated in mitotic arrest FGCs and promotes the expression of the RA-synthesizing gene ALDH1A2 in mitotic arrest FGCs.
Figure 5.
Transcriptional changes after blockage of the BMP signaling pathway
A. The schematic illustrating the in vitro culture and blockade of the BMP signaling pathway in male testicular cells. B. Two-dimensional embedding of cultured 15 W testicular cells using UMAP. The cells are colored according to cell type. The shapes represent cells subjected to different treatments. FGCs are highlighted with a black dashed circle. The shadow roughly frames the LDN-treated cells. C. Boxplots showing the expression level [log2 (TPM + 1)] of germ cell marker genes (NANOG, DAZL, and POU5F1), BMP signaling pathway-related genes (SMAD6, RBL1, MYC, ID1, ID2, and ID3), RA-synthesizing genes (ALDH1A1, ALDH1A2, and ALDH1A3), and a WNT signaling pathway-related gene (WNT3) in mitotic arrest FGCs and mitotic FGCs. Different colors represent different experimental treatments. LDN, BMP signaling pathway inhibitor; STRT-seq, single-cell tagged reverse transcription sequencing.
Because ALDH1A2 was only expressed by a fraction of mitotic arrest FGCs at the late fetal stage, we speculated that the BMP signaling pathway may play developmental stage-specific roles in germ cells. To verify this hypothesis, we conducted a similar in vitro culture experiment involving testicular cells from a 7 W embryo (Figure 5A, Figure S4A; Table S5). Similar to the case for early testes in vivo, mitotic FGCs (both proliferative and quiescent) were captured in vitro (Figure S4A and B). The expression of BMP signaling pathway genes, such as ID1–3 and SMAD6/7/9, drastically deceased in LDN-treated 7 W FGCs (Figure S4B). However, the RA-synthesizing genes ALDH1A1–3 were not expressed in FGCs, and BMP signaling pathway inhibition had no effect on the expression of these genes as expected (Figure S4B). Taken together, these results showed that the BMP signaling pathway plays developmental stage-specific roles in male germ cells and specifically promotes the expression of the RA-synthesizing gene ALDH1A2 in late-stage testicular mitotic arrest FGCs.
In addition to germ cells, we also explored the effects of the BMP signaling pathway on early- and late-stage gonadal somatic cells (Figures S3D, S4A, C, and D; Table S5). The BMP signaling pathway inhibited the expression of RA-synthesizing genes in gonadal somatic cells, in contrast to the effects on germ cells. When the BMP signaling pathway was inhibited, ALDH1A3 expression dramatically increased in both early- and late-stage gonadal somatic cells. In addition, the expression of RBL1, MYC, MKI67, and MSX2 did not significantly change in the LDN-treated group, indicating that the BMP signaling pathway has a limited effect on the proliferation and apoptosis of gonadal somatic cells (Figure S4D). Interestingly, inhibition of the BMP signaling pathway resulted in the marginal expression of Sertoli cell markers such as AMH and SOX9 in Leydig cells (Figures S3D and S4B), indicating that the BMP signaling pathway may also play roles in the differentiation and maintenance of testicular somatic cells (Figure S4C and D).
Male gonadal somatic cells can be classified into three transcriptionally distinct cell populations
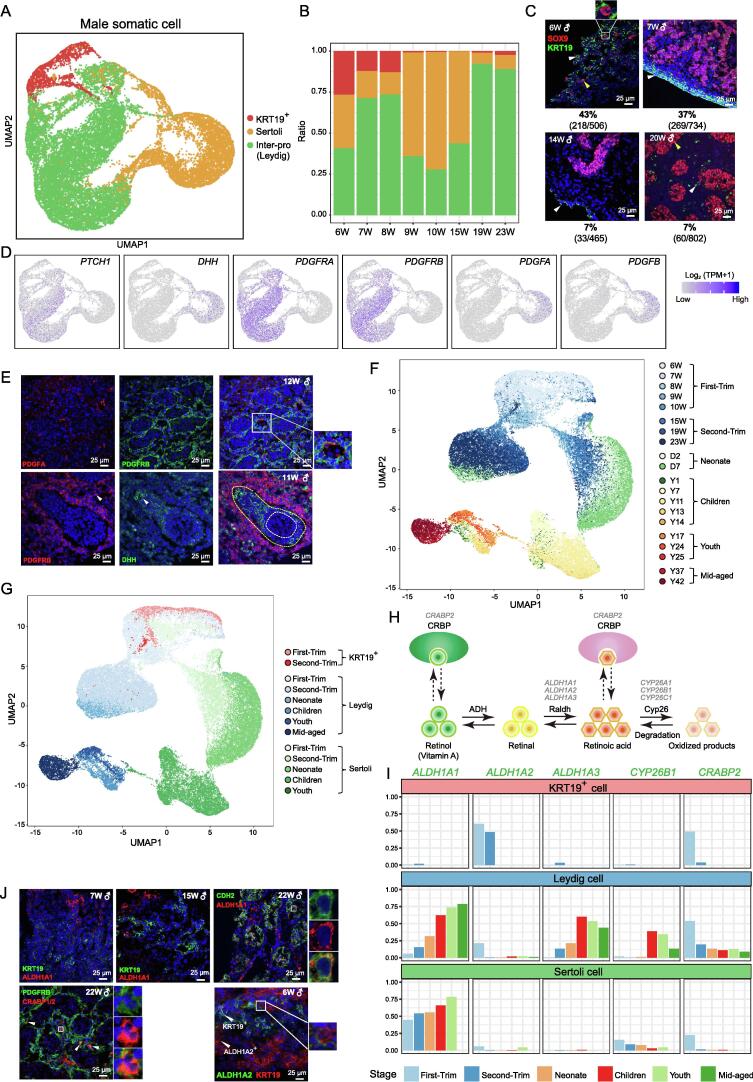
To investigate the transcriptomic features of male gonadal somatic cells, we first classified these cells by UMAP clustering and identified three distinct cell populations (Figure 6A, Figure S5A–C; Table S6). In addition to the established groups of male gonadal somatic cells, Leydig cells and Sertoli cells, we identified a new group of cells that highly expressed KRT19, and a small fraction of these KRT19+ cells also expressed the Sertoli cell marker SOX9 at an early stage (Figure 6B and C, Figure S5B). Notably, KRT19+ cells were mainly detected in gonads of embryos from 6 W to 8 W, with minimal detection after 9 W (Figure 6B). Through immunostaining of early-stage testes, we verified that approximately 43% of gonadal cells at 6 W were KRT19+, and a small fraction of these cells were also SOX9+ (2/218 at 6 W and 47/269 at 7 W); these results were consistent with our scRNA-seq data (Figure 6B and C, Figure S5B). Starting from 7 W, only a small fraction of cells located around the Sertoli cells expressed KRT19 (Figure 6C). To enrich specific somatic cells for further study, we first identified DEGs in these three gonadal somatic cell types, selected cell surface markers for them, and verified the specificity of some of the markers by immunostaining (Figure S5C–F). To analyze the structural changes in germ cells and somatic cells in the testes, we performed immunostaining of early- and late-stage testes with FGC and Leydig cell markers (Figure S5F). In the 6–7 W testes, germ cells, Leydig cells, and KRT19+ cells were scattered in the testis. In some regions, Sertoli cells had already formed small tubular-like structures surrounding the germ cells. At later developmental stages, Sertoli cells formed clear tubular structures surrounding the germ cells, with Leydig cells scattered outside the Sertoli cells (Figure S5F).
Figure 6.
Characteristics of male somatic cells and analysis of cell–cell interactions
A. Identification of fetal male testicular somatic cell subsets by clustering analysis using UMAP. Different colors represent different cell populations. B. Bar plot showing the cell type distributions at different developmental stages. C. Immunostaining of SOX9 and KRT19 at different stages. SOX9+ and KRT19+ cells are indicated by yellow and white triangles, respectively. SOX9+KRT19+ cells are framed with a white square and the corresponding image is magnified. The ratio of KRT19+ cells at each week is shown below the corresponding image. D. UMAP cluster maps showing the expression of the receptor-coding genes PTCH1, PDGFRA, and PDGFRB in Leydig cells and the ligand-coding genes DHH, PDGFA, and PDGFB in Sertoli cells. E. Immunostaining of PDGFRB, PDGFA, and DHH in male testes. Cells in the white square are magnified and shown on the right. FGCs are located inside the white dotted line. Sertoli cells are located in the area between the white and yellow dotted lines. Leydig cells are located outside the yellow dotted line. F. Combined analysis of male somatic cells from fetal, neonatal, pubertal, and adult individuals by UMAP cluster mapping. Color represents age. G. Combined analysis of male somatic cells from fetal, neonatal, pubertal, and adult individuals by UMAP cluster mapping. Color represents cell type and age. H. Diagrammatic representation of genes involved in RA synthesis and metabolism in gonads. I. Bar plot showing the ratio of cells expressing RA-synthesizing and RA-metabolizing genes specific to different cell populations at different stages. J. Immunostaining of RA signaling pathway-related proteins (ALDH1A1, ALDH1A2, and CRABP1/2), and KRT19 (KRT19+ cell marker), and PDGFRB (Leydig cell marker) in fetal testes at different stages. The areas in the white squares are magnified and arrows indicate PDGFRB+CRABP1/2+ cells, KRT19+ cells, and ALDH1A2+ cells. Inter-pro, interstitial progenitor cell.
Interactions between Leydig cells and Sertoli cells
In mammals, sex-specific differentiation is initiated by gonadal somatic cells with specification of the supporting cell lineages (Sertoli cells in males and granulosa cells in females) [45]. Then supporting cells facilitate the differentiation of the steroidogenic cell lineages (Leydig cells in males and theca cells in females). To elucidate how this facilitation step occurs, we explored the interactions between Sertoli cells and Leydig cells in the testes (Figure 6D and E, Figure S5G and H) [7]. We used NicheNet software to predict cell–cell interactions between Sertoli and Leydig cells; this analysis predicted interactions through ligand–receptor gene pairs such as DHH–PTCH1/2, PDGFA/B–PDGFRA/B, and GAS6–TYRO3 (Figure 6D, Figure S5G and H; Table S6) [46].
According to previous studies, DHH/PTCH1 signaling might function in Leydig cell differentiation and as signaling between Sertoli and peritubular myoid cells in mice [47]. Here, we found that the DHH–PTCH1 gene pair was expressed by Sertoli cells and Leydig cells, indicating the potential interactions between these two cell types in human fetal gonads, rather than the interactions between Sertoli cells and peritubular myoid cells seen in mice (Figure 6D, Figure S5G; Table S6).
The platelet-derived growth factor (PDGF) family participates primarily in migration, proliferation, and differentiation in various organ systems; genes in this family have been reported to be important downstream targets of Sry during testicular organogenesis and Leydig cell differentiation in mice. In our study, we found that Sertoli cells and Leydig cells may interact with each other through PDGF family members (Figure 6D, Figure S5G) [48]. Sertoli cells expressed the ligand PDGFA/PDFGB and Leydig cells expressed the receptor PDGFRA/PDGFRB (Figure 6D, Figure S5G). We verified the spatial expression patterns of the ligands (DHH, PDGFA) and receptor (PDGFRB) through immunofluorescence, and these spatial patterns were consistent with the sequencing data (Figure 6E).
In addition, we found that Leydig cells may interact with Sertoli cells through DHH–PTCH1 and PDGFA/B–PDGFRA/B at the neonatal, pubertal, and adult stages in the testes. The cell type-specific markers we identified in fetal testicular data were also universally applicable to neonatal, pubertal, and adult testicular cells (Figure 6F and G, Figure S6A and B).
Differences between antenatal and postnatal gonadal somatic cells
Androgens are critical components of the development and homeostasis of male reproductive function. It is well known that Leydig cells in testes are the primary source of physiologically active androgens. In mammals, testosterone, the most effective androgen, is synthesized from cholesterol through several reactions mediated by steroidogenic enzymes. To explore the relationship between fLCs and aLCs, we clustered male gonadal somatic cells from fetal (6–23 W, 8 stages), neonatal (2–7 days, 2 stages), pubertal (1–14 years old, 5 stages), and adult (17–42 years old, 5 stages) tissues by UMAP clustering (Figure 6F and G, Figure S6A; Table S4) [18], [19], [21]. The results showed that both Leydig and Sertoli cells followed continuous developmental trajectories from the first trimester to the second trimester and then through the neonatal, pubertal, and adult periods. Globally, neonatal gonadal somatic cells were most similar to cells at the fetal stages, while gonadal cells originating from individuals older than 1 year clustered together, indicating that the transcriptomes of gonadal somatic cells perhaps change greatly during the first year after birth (Figure 6F and G).
aLCs can produce testosterone to maintain male reproductive function [8]. However, it remains unknown whether fLCs can produce testosterone. Generally, Sertoli cells have been accepted as nonsteroidogenic cells [8], [49]. However, we found that at the fetal stage, Sertoli cells rather than Leydig cells expressed HSD17B3, which encodes the enzyme that mediates the final step of testosterone synthesis (Figure S6C–F) [49]. Shima et al. [49] observed similar patterns in mice during the fetal period and concluded that androstenedione produced by fLCs is transferred to fetal Sertoli cells and then converted to testosterone in mice. In addition, a small fraction of neonatal Leydig cells expressed HSD17B3, consistent with the findings of a study on postnatal fetal rats (P8: postnatal day 8) [50]. Moreover, the ratio of Leydig cells expressing HSD17B3 gradually increased from the neonatal stages; unexpectedly, the ratio of Sertoli cells that expressed HSD17B3 remained high after birth (Figure S6C–E). The expression pattern of CYP11A1, which encodes cholesterol sidechain cleavage P450 that catalyzes the formation of an intermediate product during testosterone synthesis, was similar to that of HSD17B3 [8]. To further validate this hypothesis, we performed immunostaining of CYP11A1, HSD17B3, and known Sertoli cell markers (SOX9 and PDGFA) in fetal and adult testes (Figure S6F–H). The Sertoli cell markers showed nice overlapping expression with CYP11A1 and HSD17B3 in fetal testes, especially CYP11A1, which indicated that CYP11A1 and HSD17B3 were indeed expressed by Sertoli cells but not by Leydig cells in fetal testes (Figure S6F). At the adult stage, the expression of HSD17B3 and the Sertoli cell marker PDGFA was generally mutually exclusive, as expected (Figure S6G). Consistent with the scRNA-seq data, some Sertoli cells (SOX9+) at the adult stage still expressed HSD17B3 (Figure S6H). Taken together, the data suggest that even after birth, Sertoli cells still marginally contribute to testosterone synthesis, which is gradually taken over by Leydig cells.
Both testosterone and RA signaling are important for spermatogenesis, and dysregulation of either RA or androgens can cause testicular dysfunction and result in male infertility. RA is tightly regulated by a group of RA-synthesizing enzymes (ALDH1A1, ALDH1A2, and ALDH1A3) and RA-metabolizing enzymes (CYP26A1, CYP26B1, and CYP26C1) (Figure 6H) [51], [52], [53], [54]. Here, we found that the three RA-synthesizing enzymes were expressed by different types of gonadal somatic cells in male fetuses (Figure 6I and J, Figure S6D). ALDH1A1 was expressed by fetal and neonatal Sertoli cells and aLCs but not by KRT19+ cells (Figure 6I). ALDH1A2 was expressed mainly by part of KRT19+ gonadal somatic cells and a small fraction of Leydig cells in the first trimester (Figure 6I and J). ALDH1A3 was expressed primarily in Leydig cells during the second trimester, neonatal period, and adulthood (Figure 6I). In addition, cellular retinoic acid binding protein II (CRABP2) was expressed by some Leydig cells in the second trimester and part of all types of male gonadal somatic cells in the first trimester (Figure 6I and J). Once the fetal supporting cell lineage (Sertoli cells in the testes and granulosa cells in the ovaries) was determined, FGCs in the ovaries began to undergo meiosis because of exposure to RA, while FGCs in the testes entered mitotic arrest because of expression of the RA-metabolizing gene CYP26B1 in Sertoli cells. Consistent with the results of previous studies, we found that a small fraction of fetal and neonatal Sertoli cells expressed CYP26B1 (Figure 6I, Figure S6D).
Female gonadal somatic cells can be classified into seven transcriptionally distinct cell populations
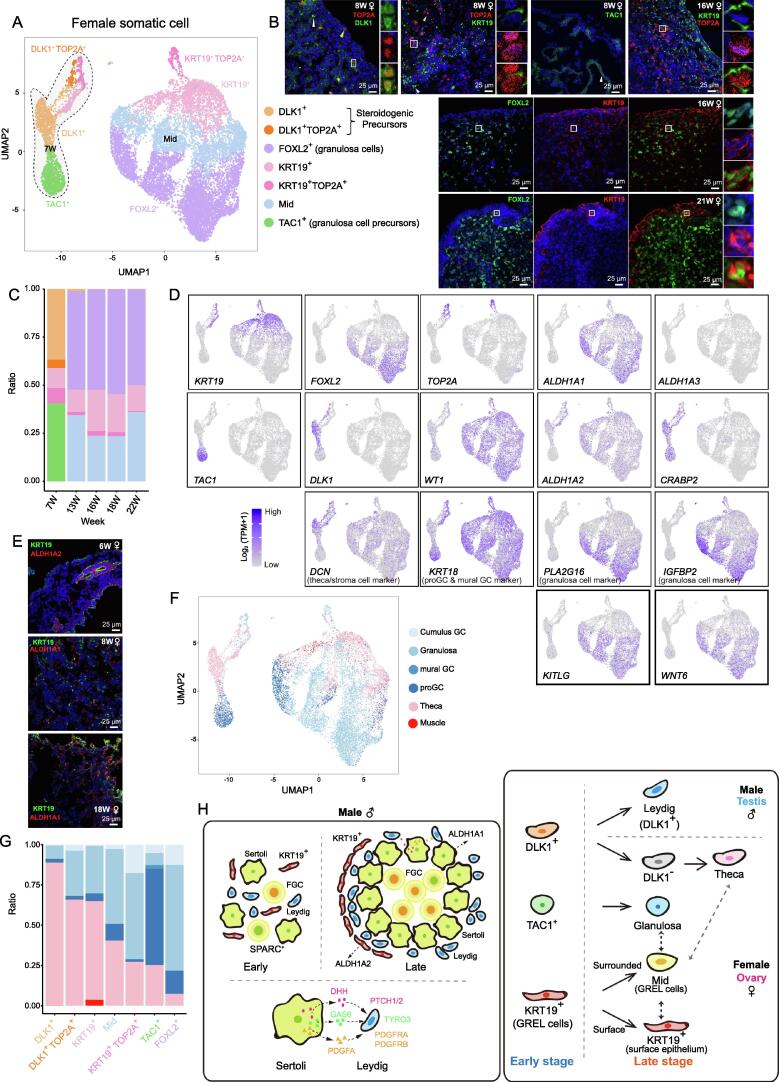
Having characterized somatic cells in male gonads, we next focused on gonadal somatic cells in female embryos and fetuses. We identified seven somatic cell types in fetal ovaries and verified the existence of these cell types through immunofluorescence staining (Figure 7A–D, Figure S7A; Table S7). Globally, early-stage female gonadal somatic cells showed expression patterns distinct from those of the cells derived from fetuses after 13 W (Figure 7A and C, Figure S7A). In detail, DLK1+, DLK1+TOP2A+, and TAC1+ cells were detected in only the gonads of 7 W embryos, and granulosa cells (FOXL2+) appeared after 13 W (Figure 7B). In addition to granulosa cells, KRT19+ cells were found in the ovaries of embryos and fetuses at all the analyzed developmental stages. To further evaluate these somatic cells, we first explored the expression patterns of somatic progenitor cell markers and genital ridge (GR)-associated genes [7], [28], [55] in these cells. Unexpectedly, WT1, GATA4, CITED2, and MAP3K1 were mainly expressed by ovarian somatic cells from 13 W onward; less than 20% of these cells expressed WT1 at 7 W, which may indicate the existence of other somatic progenitor cells that appear much earlier than WT1+ somatic progenitor cells. Furthermore, 61.2% of gonadal somatic cells in 7 W expressed another somatic precursor marker gene NR2F2 which revealed that NR2F2+ cells appeared earlier than WT1+ cells in the human ovary (Figure S7A and B) [28], [56]. EMX2 was expressed by both early- and late-stage somatic cells at similar ratios (Figure 7D, Figure S7A and B). In addition, LHX9 was expressed by all somatic cells except 7 W TAC1+ cells (Figure S7A and B). Similar to the results for KRT19+ cells in the testes, KRT19+ cells in early ovaries expressed the RA-synthesizing gene ALDH1A2 but barely expressed ALDH1A1 (Figure 6I and J, Figure 7A, D, and E, Figure S7C). The proportion of KRT19+ cells in both early- and late-stage fetal ovaries remained stable, whereas KRT19+ cells predominated in early-stage fetal testes (Figure 6B, Figure 7C). In addition, KRT19+ cells in late-stage fetal ovaries expressed both ALDH1A1 and ALDH1A2 (Figure 7D). ALDH1A2 was also expressed by other somatic cells in late-stage ovaries, such as granulosa cells. In contrast, other testicular somatic cells, such as Sertoli cells and Leydig cells, did not express ALDH1A2 in late-stage fetal testes (Figure 6I, Figure 7D, Figure S7C). Moreover, ALDH1A1 was expressed by gonadal somatic cells beginning at 13 W. In addition, similar to its expression pattern in somatic cells in fetal testes, CRABP2 was highly expressed by somatic cells in 7 W ovaries (Figure 7D, Figure S7C). Starting at 13 W, a group of KRT19+FOXL2+ cells (Mid cells) was identified by UMAP. Interestingly, immunofluorescence showed that Mid cells localized between KRT19+ cells and FOXL2+ cells (Figure 7B and D). Originally, the granulosa cells were thought to be derived from the mesonephros, but recently they were considered to be derived from the ovarian surface epithelia [57], [58]. It is reported that both ovarian surface epithelial cells and granulosa cells arise from a common type of precursor cells named the gonadal ridge epithelial-like (GREL) cells that express KRT19 [59]. Combined with the model of ovary and follicle development raised by Hummitzach et al. [60], we speculated that the KRT19+ cells at early stages (∼ 7 W) are GREL cells. At the later development, GREL cells located at the surface begin to differentiate into typical ovarian surface epithelia (KRT19+ cells at the later stages), while GREL cells surrounded by a basal lamina (Mid cells) have the ability to give rise to surface epithelia and granulosa cells (FOXL2+ cells), exhibiting non-proliferating and mildly FOXL2 expression features. In mice, some theca cells also expressed FOXL2, so we speculated that Mid cells might have the potential to differentiate into thecal cells (Figure 7F and G) [60], [61]. In addition, KRT19+ cells also show the potential to give rise to theca cells. However, these are speculated based on scRNA-seq data, and the actual differentiation direction needs to be further verified by experiments such as genetic lineage tracing. Then, we identified DEGs within these ovarian somatic cell subtypes (Figure S7D; Table S7). As expected, FOXL2+ cells highly expressed the granulosa cell marker PLA2G16, and TAC1+ cells highly expressed the known granulosa cell markers SERPINE2 and GSTA1 (Figure 7D, Figure S7D; Table S7), whereas DLK1+ cells highly expressed the theca cell markers, such as COL1A1, COL1A2, and CXCL12 (Figure S7D; Table S7) [22], [62]. In addition, we also showed the expression patterns of mice pre-granulosa cell markers WNT6 and KITLG on the UMAP (Figure 7D) [63]. It showed that instead of highly expressed by predicted pre-granulosa cells, these two genes were highly expressed by FOLX2+ granulosa cells, which may be due to differences between mouse and human.
Figure 7.
Identification of divergent populations of female somatic cells and their precursor cells
A. Identification of fetal female ovarian somatic cell subsets by UMAP clustering analysis. The different colors represent different cell populations. Cells from 7 W ovary are indicated with the black dashed line. Mid, cells co-expressing low levels of FOXL2 and KRT19. B. Immunostaining of cell type-specific markers to validate the existence of the ovarian somatic subtypes shown in (A). Cells inside the white squares are magnified and shown on the right of the corresponding images. C. Bar plot showing the cell type distributions at different stages. Different colors represent different cell types. The color corresponding to each cell type is the same as that shown in (A). D. UMAP cluster map showing the expression patterns of cell type-specific genes and RA signaling-associated genes. Colors from gray to purple represent low to high expression levels. E. Immunostaining of ALDH1A1/ALDH1A2 and KRT19 in ovaries of different stages. F. UMAP cluster map showing the cell types that map to adult ovarian cells. G. Bar plot revealing the relationship between fetal and adult ovarian cell types. H. Summary diagram of male and female gonadal development. The solid arrows represent our hypothesis, and the dotted arrows indicate uncertainty based on current knowledge. proGC, progenitor granulosa cell; GREL, gonadal ridge epithelial-like.
DLK1+ cells might be progenitors of the steroidogenic cell lineage
To explore the similarities between fetal and adult gonadal somatic cell types in females, we mapped our data to single-cell transcriptome data of adult ovaries (Figure 7F, Figure S7E) [22]. As expected, the transcriptome of FOXL2+ cells in fetal ovaries most closely resembled that of adult granulosa cells (Figure 7F and G). Specifically, TAC1+ cells in 7 W ovaries were most similar to adult progenitor granulosa cells (proGCs), indicating that TAC1+ cells may be progenitors of granulosa cells (Figure 7F and G). However, since TAC1+ cells disappeared in the later-stage ovaries, further experiments are needed to explore the relationship between TAC1+ cells and mature ovarian somatic cells. DLK1+ cells, another group detected in 7 W ovaries, were most similar to adult theca cells (Figure 7F and G). DLK1+ cells were detected in both 7 W male and female gonads and expressed steroidogenic cell markers, such as TCF21 and PDGFRA, indicating that they might be steroidogenic progenitor cells in both sexes (Figure 2E, Figure S7F). However, some differences were observed in later gonad development. In the male steroidogenic cell lineage, DLK1 and steroidogenic cell markers (TCF21 and PDFGRA) continued to be expressed in Leydig cells. By contrast, DLK1 was expressed at only 7 W in females and no ovarian somatic cells at later stages expressed DLK1 (Figure 7D, Figure S7D). Notably, KRT19+ cells seemed to have the potential to differentiate into both theca and granulosa cells (Figure 7F and G).
Discussion
We previously performed an scRNA-seq analysis on 2486 (319 with the Tang protocol and 2167 with a modified STRT-seq protocol) human FGCs and somatic cells in their microenvironment from 44 embryos between 4 W and 26 W [17]. We comprehensively and extensively analyzed the transcriptomic features of FGCs and clearly identified four and two types of FGCs in females and males, respectively, which were the mitotic phase, RA signaling response phase, meiotic prophase, and oogenesis phase FGCs in females and the mitotic phase and mitotic arrest phase FGCs in males. However, insufficient numbers of gonadal somatic cells were analyzed; thus, the developmental characteristics of gonadal somatic cells were not fully explored in our previous study [44].
In this study, we created a comprehensive map of cell populations in human ovaries and testes, including both FGCs and somatic cells in their niche. To characterize the differentiation and maturation of FGCs and gonadal somatic cells, we generated developmental expression profiles of 56,399 gonadal cells from 16 human embryos and fetuses between 6 W and 23 W. Specifically, we identified seven groups of FGCs, including a newly identified group that simultaneously expressed POU5F1 and SPARC. Notably, a novel group of abnormal cells that simultaneously expressed markers of somatic cells and germ cells was identified, and this expression pattern was verified at the protein levels in fetal gonads through immunofluorescence. In addition to immune cells, endothelial cells, and blood cells, we identified three and seven types of somatic cells in male gonads (testes) and female gonads (ovaries), respectively. In both testes and ovaries, we identified a new KRT19+ cell population that highly expressed the RA-synthesizing gene ALDH1A2 at early developmental stages (Figure 7H; Table S8). In both sexes, KRT19+ cells were scattered in early gonads and then neatly arranged outward of the gonads at later stages (Figure 7H). We identified and verified surface protein markers that can be used to isolate specific somatic cell types in future studies, such as CDH2 for Sertoli cells and PDGFRA for Leydig cells. In addition, we combined data for 55,404 postnatal gonadal single cells (20,676 ovarian single cells and 34,728 testicular single cells) from four published datasets, and comprehensively and systematically explored the gene expression patterns of gonadal cells [18], [19], [21], [22]. For the testis, we analyzed single-cell transcriptomic data for male individuals from 4 W to 25 W before birth and from 2 days to 42 years of age after birth, and conducted in-depth analyses on the expression patterns of testicular somatic and germ cells [17], [19], [21].
Second, we assessed the developmental origins of granulosa cells in females and those of steroidogenic lineages in both sexes. Our data indicated that DLK1+ cells in the gonads of 7 W male and female embryos may further differentiate into steroidogenic cell lineages, Leydig cells in the testes and theca cells in the ovaries. However, DLK1 expression remained in Leydig cells at subsequent developmental stages but disappeared in theca-like cells of the ovaries [22], [62]. A human fetus does not undergo sex differentiation until approximately 7 W [55]. Our data indicated that in 7 W embryos, the expression features and cell types of both germ cells and gonadal somatic cells were very similar, if not identical in male and female embryos (Figure S8A and B). Next, by mapping our ovarian somatic cell data to published adult ovarian scRNA-seq data, we found that the expression profiles of TAC1+ cells and proGCs are very similar [22]; thus, we speculate that TAC1+ cells in 7 W female gonads may differentiate into granulosa cells in later developmental stages. Interestingly, a few gonadal somatic cells (<20%) in 7 W ovaries expressed WT1 but more than 60% cells expressed NR2F2 at this stage, indicating that NR2F2+ somatic progenitor cell type appears earlier than WT1+ cells (Figure S7B). In contrast, starting at 13 W, nearly half of ovarian cells expressed somatic marker genes such as LHX9, WT1, and GATA4, which may explain why KRT19+ cells showed the potential to differentiate into both theca and granulosa cells (Figure 7G, Figure S7A). However, these preliminary conclusions based on the scRNA-seq analysis must be confirmed in further experiments, such as lineage tracing and in vitro differentiation. Although our data provide valuable insights into the differentiation of ovarian somatic cells, many questions remain that cannot be answered by the current study, and further research is needed. For example, studies are needed to determine the function of the Mid cell subtype in female ovaries and the relationships between Mid cells and KRT19+ cells and granulosa cells.
Notably, our data indicated that GST may have already been initiated in a subset of mitotic arrest FGCs in the second trimester and that the BMP signaling pathway regulates GST by promoting the expression of the RA-synthesizing gene ALDH1A2. It is reported that these transitional prespermatogonia appear at around 14 W and multiple signaling pathways, such as BMP, Notch, and WNT, participate in their development and interactions with surrounding microenvironmental somatic cells [29]. Our data revealed that the BMP signaling pathway plays developmental stage-dependent and cell type-specific roles in male germ cells (Figure S8C). This pathway regulated ALDH1A2 expression in only late-stage mitotic arrest FGCs; it did not affect the expression of RA-synthesizing enzymes in either early- or late-stage mitotic FGCs. On the other hand, the BMP signaling pathway inhibited the expression of the RA-synthesizing gene ALDH1A3 in both early- and late-stage testicular somatic cells. Interestingly, during the in vitro culture of testicular cells, we observed that inhibiting the BMP signaling pathway evoked low expression of Sertoli cell marker genes, such as SOX9, in Leydig cells. In addition, after testicular cells were treated with LDN, a group of special somatic cells appeared that started to express the germ cell marker POU5F1. This finding may indicate that the BMP signaling pathway is involved in cell identity determination and differentiation, and that inhibiting this pathway may cause the dedifferentiation of the gonadal cells. Furthermore, these data verified the existence of abnormal cell types that simultaneously express markers of gonadal somatic cells and germ cells. However, unlike the aberrant FGCs in vivo (Figure 3A), this group of cells (POU5F1Low Soma in Figure 5B) had a gene expression profile that was more similar to that of gonadal somatic cells than germ cells, perhaps indicating potential aberrant mutual transitions between a minor proportion of gonadal somatic cells and germ cells. In fact, there were also a small number of gonadal somatic cells that expressed germ cell markers in vivo, which can rule out the possibility that such germ cell-like somatic cells were by-product of in vitro culture (Figure 1A and C).
Although the molecular characteristics and cellular origins of fLCs and aLCs, as well as their developmental and functional links, have been extensively studied in rodents, we are just beginning to unravel these details in humans due to the scarcity of samples and technical limitations [12], [49], [50]. In mammals, there are two main types of Leydig cells, fLCs and aLCs. Leydig cells are the main source of androgens, while Sertoli cells are generally accepted as nonsteroidogenic [8]. However, in this study, we found that Sertoli cells, rather than Leydig cells, expressed HSD17B3, a gene encoding the enzyme that mediates the final step of testosterone synthesis, in the fetal period (18.5% in the first trimester and 34.5% in the second trimester) and the neonatal period (30%). Leydig cells did not express HSD17B3 until the neonatal stages (6.8%), and the ratio of cells that expressed HSD17B3 gradually increased in later stages. This finding indicates that androstenedione produced by fLCs is transferred to fetal Sertoli cells and then converted to testosterone. Then, during the neonatal stages, testosterone production is gradually taken over by Leydig cells. Immunofluorescence analysis of CYP11A1 and HSD17B3, which are associated with androstenedione production, revealed their expression by Sertoli cells instead of by Leydig cells before birth. At the adult stage, Leydig cells started to produce HSD17B3. Through performing CYP17A1 staining in human 10 W fetal testes in another study, it showed that CYP17A1 seemed to be expressed by interstitial cells, which seems to conflict with our conclusions [28]. However, a similar study in mice showed that fLCs can produce androstenedione but not testosterone due to lack of HSD18B3 expression. And androstenedione was converted to testosterone in the fetal Sertoli cells, in which HSD17B3 was expressed [49]. It suggested that compared to the CYP17A1, the downstream product HSD17B3 can better characterizing Leydig cell testosterone biosynthesis function. Additionally, we elucidated the potential cell–cell interactions between Leydig cells and Sertoli cells through the computational method NicheNet [46]. First, we found that the Hh ligand gene DHH and its downstream receptor gene PTCH1 were expressed in Sertoli cells and Leydig cells, respectively. However, the DHH–PTCH1 ligand–receptor pair mainly regulates interactions between Sertoli and peritubular myoid cells in mice [47]. Second, we found that Sertoli cells and Leydig cells likely interacted with each other through PDGFA/B–PDGFRA/B ligand–receptor pairs. Through immunofluorescence, we verified the spatial expression patterns of these ligand–receptor pairs. Notably, these potential interactions also likely occur in neonatal and adult testes. Further experiments, such as co-cultures and ligand/receptor knockouts, are needed to validate the regulatory potential of these ligand–receptor pairs.
For most of the time points, we sequenced only one sample, but all our findings were verified in independent embryos and fetuses. In total, we performed immunofluorescence with 16 additional embryos or fetuses at 11 different time points, and all these results were consistent with our sequencing data (Tables S8 and S9). In addition, we performed scRNA-seq of in vitro cultured gonadal cells from another 7 W embryo and one 15 W fetus using a modified STRT-seq method. Consistent with our 10X Genomics sequencing data, only early-stage testes contained KRT19+ somatic cells, and only mitotic FGCs existed in early-stage testes, while both mitotic FGCs and mitotic arrest FGCs existed in middle- and late-stages of testes.
Conclusion
To the best of our knowledge, this study is the first to systematically and comprehensively compare the gene expression patterns and cell type compositions among monosomy X (45, XO), normal female (46, XX), and normal male (46, XY) embryos at the same developmental stage (7 W). Globally, monosomy X caused abnormalities in the transcriptomes of not only FGCs, but also gonadal somatic cells. More importantly, monosomy X was associated with cell type-specific developmental defects in FGCs and gonadal somatic cells and caused the loss of PCP4+ cells in XO gonads compared with normal XX gonads. According to the human protein atlas, the expression of PCP4 has strong tissue-specific features and it is mainly expressed by brain and seminal vesicle, a male secondary sex organ. Secondary sex characteristics are usually determined by hormones secreted from gonads. In response to the testosterone, the seminal vesicle sprouts during the 10th week of human testis development under the influence of the Y chromosome. These may infer that PCP4 may be involved in the secondary sex determination of early stage of human gonads, but the underlying mechanism of PCP4 in gonad development still needs to be further studied [64].
Altogether, we identified and characterized new types of FGCs and gonadal somatic cells. We speculate that the DLK1+ cell population is a progenitor population of the steroidogenic cell lineage and that the TAC1+ cell population is a progenitor population of granulosa cells. Notably, the role of the BMP signaling pathway in the development of both testicular somatic cells and germ cells was elaborated in our study. In addition, we illustrated the transcriptomic changes in monosomy X embryos. Our data also have potential applications for the isolation of specific gonadal somatic cell populations in vivo. These results deepen our understanding of both germ cell and gonadal somatic cell development in the testes and ovaries. Finally, our work provides insights and valuable data resources for further elucidation of the molecular mechanisms of infertility.
Although most of our conclusions have been verified on independent embryos and fetuses using immunofluorescent staining, the limited number of embryos and fetuses we sequenced is still one limitation of our study, since there may be individual differences between embryos and the developmental stages were not fully covered. For example, although we have identified seven types of ovarian somatic cells, the lineage trajectories among these cells have not been clearly explored due to the failure to collecting female embryos between 7 W and 13 W. In addition, cell–cell interactions and cell origins were speculated only by expression data and computational predictions, so additional experiments are needed to further verify their existence. Finally, the BMP signaling pathway not only plays an important role in testicular cells, but also plays an indispensable role during the development of ovarian cells [29]. However, we only performed in vitro culture of testicular cells with BMP inhibitor. Further studies in ovarian cells or using in vivo animal models are still needed to better characterize the human gonad development.
Materials and methods
Key resources
All the key resources are listed in Table S8. Further information and requests for reagents and resources may be directed to, and will be fulfilled by, the Lead Contact, Fuchou Tang (tangfuchou@pku.edu.cn).
Sample collection
In this study, 9 male embryos/fetuses and 6 female embryos/fetuses from 6 W to 23 W were obtained, and the stages of the human embryos/fetuses were determined by clinicians based on the days from the last menstruation date. To remove the potential contamination of mesonephros, we removed the mesonephros based on the structural characteristic during sampling process. Notably, one 15 W male fetus had two technical replicates, and the 22 W (female) and 23 W (male) samples had two biological replicates. To compare prenatal and postnatal transcriptomic features in humans, neonatal and adult data for both testes and ovaries were downloaded from the Gene Expression Omnibus (GEO) database (GEO: GSE124263 [21], GSE134144 [19], GSE120508 [18], and GSE118127 [22]).
We focused on the following research aspects: 1) the role of somatic cells in key events during gonad development, such as sex determination and initiation of germ cell meiosis; 2) lineage separation and gene expression dynamics of the gonadal somatic cells during fetal gonad development; and 3) the interactions between different types of somatic cells. By the beginning of the 5th embryonic week, a distinct cell population progressively separates to form the adrenal primordium and gonadal ridge. In the 6th week after conception, the gonadal ridge has bipotential and can develop into an ovary or a testis, irrespective of its chromosome constitution [65]. Then by the beginning of the 7th embryonic week, testis differentiation was initiated in the XY fetus. In the testis, Sertoli cells and Leydig cells are present by approximately 7–9 W. In the ovary, the first primary follicles appear during 15–16 W, so granulosa cells start to appear at approximately 15 W, and theca cells undergo differentiation in the third trimester. According to the recommendations of the Ethics Committee of Third Hospital, Peking University, we only use embryos/fetuses before 26 W. Therefore, we mainly explored the gene expression profiles of embryos between 4 W and 26 W in this study.
Human fetal gonad sample preparation and single-cell isolation
Gonadal biopsies were isolated and washed twice with L15 medium (Catalog No. L1518, Sigma-Aldrich, Saint Louis, MO) supplemented with 10% fetal bovine serum (FBS; Catalog No. SH30084.03, Hyclone, Logan, UT). Then, the samples were cut into small pieces under controlled cooling conditions, digested with 600 µl of Accutase Cell Detachment Solution (Catalog No. SCR005, Millipore, Billerica, MA) at 37 °C, and centrifuged at 850 r/min for 15 min. To ensure as thorough digestion as possible, we pipetted the digestion mixture at least 20 times. Then, 500 µl of L15 medium with 10% phosphate-buffered saline (PBS) was added, and single gonadal cells were obtained by filtering through 30 mm Pre-Separation Filters (Catalog No. 130-041-407, Miltenyi Biotec, Bergisch Gladbach, Germany). Then, the cells were pelleted by centrifugation at 300 g for 8 min and resuspended in 500 µl of L15 medium with 10% PBS, at which point they were prepared for scRNA-seq. The cells were counted with a hemocytometer.
Human male gonad cell culture
Our culture system is optimized according to the existing culture conditions [44], [66]. The testes from a 7 W embryo and a 15 W fetus were separated from surrounding tissues and then washed twice with Dulbecco’s Modified Eagle Medium (DMEM; Catalog No. 12491015, Gibco, Carlsbad, CA) supplemented with 10% FBS (Catalog No. SH30084.03, Hyclone). After digesting the testes with Accutase Cell Detachment Solution (Catalog No. A6964, Sigma-Aldrich) at 37 °C for 15 min, approximately half of the cells were resuspended in Glasgow’s minimum essential medium (GMEM; Catalog No. 11710035, Gibco) supplemented with 15% (v/v) KnockOut Serum Replacement (Catalog No. 10828028, Gibco), 0.1 mM nonessential amino acids (Catalog No. 11140050, Invitrogen, New York, NY), 2 mM L-glutamine (Catalog No. 25030149, Invitrogen), 0.1 mM β-mercaptoethanol (Catalog No. M3148, Sigma-Aldrich), 1 mM sodium pyruvate (Catalog No. S8636, Sigma-Aldrich), 100 ng/ml leukemia inhibitory factor (LIF; Catalog No. 300–05-250, PeproTech, Rocky Hill, NJ), 500 ng/ml Bone Morphogenetic Protein 4 (BMP4; Catalog No.314-BP-500, R&D, Shanghai, China), 100 ng/ml stem cell factor (SCF; Catalog No. 255-sc-010, R&D), 50 ng/ml Epidermal Growth Factor (EGF; Catalog No. AF-100-15-100, PeproTech), and 10 μM Y27632 (Catalog No. 1254, Tocris, Shanghai, China). Then we seeded 50,000 cell per well into 24-well plates and cultured the cells at 37 °C with 5% CO2. After 4 h of culture, LDN-193189 2HCl (Catalog No. S7507, Selleckchem, Shanghai, China) was added to the LDN-treated group at a final concentration of 150 nM. Then the LDN-treated group and control group [treated with the same volume of dimethyl sulfoxide (DMSO)] were cultured for 24 h. Then, we performed scRNA-seq on the cultured cells with a modified STRT-seq method as previously described [44].
scRNA-seq library preparation and sequencing
In this study, scRNA-seq was performed with a 10X Genomics system. Cell capture and library preparation were performed according to the kit instructions (Single Cell 3′ Reagent Kits v2). In brief, cells were first diluted following the manufacturer’s recommendations and then mixed with a single cell master mix buffer before being loaded into a 10X Chromium controller. Then, RNA was reversely transcribed at 53 °C for 45 min and 85 °C for 5 min. After purifying the reverse transcription products, a sequencing library was prepared with 14 cycles of PCR for cDNA amplification. Then, the purified cDNA was used for library amplification, and the resulting library was sequenced on an Illumina HiSeq 4000 instrument.
scRNA-seq data processing
For the data acquired by the 10X Genomics system, the raw sequencing data were first demultiplexed by Cell Ranger (v1.3.1) software with the ‘mkfastq’ function. Then, the cellranger count, alignment, filtering, cell barcode, and unique molecular identifier (UMI) counting functions were run on the FASTQ files. Finally, we used the ‘cellranger aggr’ function to aggregate the outputs from multiple runs of cellranger count by normalizing those runs to the same sequencing depth. The combined cell count matrix was used for subsequent analysis.
For the data acquired by the modified STRT-seq method, paired-end sequencing reads were first split based on the cell barcodes in read 2. Then, the 8 bp UMI sequences located on read 2 were extracted and attached to the names of read 1. Next, low-quality reads and template switch oligo (TSO)-, polyA-, and adapter-contaminated sequences were removed. Cleaned reads were mapped to the human genome (hg19) using TopHat (v.2.0.14) [67], and unique mapped reads were kept for further analysis. HTSeq (v0.6.1) [68] was used to calculate the transcript abundance of each gene, and duplicated reads of each reads were removed based on the UMIs. The detailed code can be found at GitHub (https://github.com/WRui/Post_Implantation/tree/master/scRNA_UMI).
Quality control, cell identification, and clustering analysis
The Seurat [69] package (v.3.0.2) was applied for subsequent analyses, such as cell filtering, clustering, and DEG identification. For scRNA-seq data acquired by the 10X Genomics system, the Seurat object structure was initialized by running CreateSeuratObject with the settings min.cells = 3 and min.features = 200. Cells that expressed fewer than 200 genes and genes expressed in fewer than 3 cells were filtered out. The mean percentage of mitochondrial genes was 3.2%, whereas the mean number of expressed genes was 1694. For data acquired by the modified STRT-seq method, we used only cells that expressed at least 2000 genes and genes expressed in at least 3 cells for further analyses (Table S1). The mean numbers of expressed genes were 7672 and 5762 for 7 W and 15 W testes, respectively.
We next normalized the data with the “NormalizedData” function with the settings normalization.method = “LogNormalized” and scale.factor = 10,000. Then, a set of features was selected by running FindVariableFeatures with the parameters selection.method = “vst” and nfeatures = 2000. Next, we applied linear transformation with the function ScaleData. PCA was then performed on the scaled data with the RunPCA function. We then clustered and visualized the data with the RunUMAP function with dims = 1:10. According to well-established cell markers, we identified cells as endothelial cells (CDH5), immune cells (PTPRC, also known as CD45), blood cells (HBM), soma (WT1, SOX9, TCF21, and FOXL2), and FGCs (POU5F1, DDX4, and ZP3).
Doublet identification
The DoubletFinder software was used to identify the doublets [36]. As for scRNA-seq experiments with the 10X Genomics kit, we routinely targeted the retention of 5000 cells. According to the 10X Genomics User Guide, the multiplet rate is 3.9% when 5000 cells were recovered. Thus, the doublet rate was set as 3.9% when performing the DoubletFinder, and it would classify 2200 cells (equal to total number × 3.9% = 56,399 × 0.039 = 2200) as doublets. Since the identification of doublets prefers specific cell types and has no effect on our conclusions, we just put the results of DoubletFinder in Table S1, but did not remove these cells in the subsequent analyses.
Sex determination of embryos
For fetuses with a gestational age less than 10 W, it is hard to identify sex based on secondary sexual gland development. For embryos/fetuses less than 10 W post conception, we judged the sex through other methods. First, we calculated the ratio of reads mapped to the Y chromosome. In female embryos/fetuses, the ratio of reads mapped to the Y chromosome was less than 0.02%; while in male embryos/fetuses, the ratio was greater than 0.14% (at least six times higher than that in female embryos/fetuses). The number of reads mapped to each chromosome was calculated by running the SAMtools “idxstats” function on binary alignment and map (BAM) files. Second, we compared the expression of XIST and RPS4Y1, which are located on the X and Y chromosomes, respectively. Female embryos/fetuses highly expressed the XIST gene but minimally expressed the RPS4Y1 gene, while the opposite trend was seen in male embryos/fetuses. Third, sex was assessed by X chromosome SNP analysis. Since male embryos have only one X chromosome per cell, the SNPs on the X chromosome of a male embryo are all homozygous. We identified SNPs with the software “snpclust” following the manual; the specific code can be found at GitHub (https://github.com/10XGenomics/single-cell-3prime-snp-clustering).
Identification of DEGs
We used the FindAllMarkers function in the Seurat package to identify DEGs among more than three clusters and the FindMarkers function to identify DEGs between two clusters [69].
Combined analysis of prenatal and postnatal scRNA-seq data
We used the subset function in the Seurat package to extract specific cell type data from the Seurat object. We used the merge function to merge the raw count matrices of two Seurat objects and create a new Seurat object. We compared the transcriptomes of somatic cells and germ cells in the ovaries and testes before and after birth. For the different sexes, we combined somatic cells and germ cells in the fetal period with the corresponding cell types in the adult period. To identify the precursor cells of ovarian somatic cells, we used adult ovarian cells as a reference and mapped the fetal data to it by identifying anchors between these datasets with the Seurat package. The results showed that single-cell transcriptome data from different laboratories can overlap very well and be mutually verified.
Prediction of cell–cell interactions
We used the NicheNet (v.1.0.0) package to predict ligand–receptor interactions between Leydig cells and Sertoli cells based on single-cell expression data and prior knowledge of gene regulatory networks. NicheNet analysis was performed following the online vignette (https://github.com/saeyslab/nichenetr/blob/master/vignettes/seurat_steps.md). First, DEGs between Leydig cells and Sertoli cells were identified by the Seurat package with the FindAllMarkers function, and these DEGs were used as a gene set of interest for the following NicheNet analysis. We defined the Sertoli cells and Leydig cells as “sender/niche” and “receiver/target” cell populations, respectively. To ensure the accuracy of the results, only bona fide ligand–receptor interactions documented in the literature and publicly available databases were used to plot the heatmap showing potential interactions between Leydig cells and Sertoli cells.
Cell type annotations of ovarian somatic cells
To annotate the cell types of fetal ovarian somatic cells and to explore the relationship between fetal and adult ovarian cell types, Seurat package was used for mapping and annotating the fetal ovarian somatic cell dataset. We used a published adult ovarian dataset as the reference dataset and predicted the cell type annotations for our fetal dataset following the online Seurat manual (https://satijalab.org/seurat/articles/integration_mapping.html). To estimate the accuracy of the annotations, we calculated the ratio of FOXL+ cells that were successfully predicted as granulosa cells, and it showed that 92.6% of the FOXL+ cells were classified as granulosa cells, which reflected the high accuracy of this annotation method.
Gene Ontology analysis of DEGs
The DAVID website (v.6.8; https://david.ncifcrf.gov/tools.jsp) was used to perform Gene Ontology analysis. DEGs were uploaded as a gene list with “Official gene symbol” as the identifier and “Homo sapiens” as the species.
Immunostaining of human fetal gonad tissues
The gonads from human fetuses were washed three times with Dulbecco’s Phosphate-Buffered Saline (DPBS; Catalog No. CC011.1, Macgene, Beijing, China) supplemented with 10% FBS and fixed with 4% paraformaldehyde until they sank to the bottom of the container. Then, Tissue-Tek optimal cutting temperature (OCT) Compound (Catalog No. 4583, Sakura, Torrance, CA) was used to embed the fixed tissues, and 10 µm cryosections were prepared. Immunofluorescence assays were performed as described previously. A Leica confocal microscope (Catalog No. TCS SP8, Leica Microsystems, Wetzlar, Germany) was used to obtain confocal images.
In total, we performed immunofluorescence with 27 antibodies on 16 additional embryos/fetuses from 11 time points (Table S9).
Cell counting in immunofluorescence images
The number of cells in immunofluorescence images was counted by Image J. The immunofluorescence image was imported into Image J “Cell Counter” with the “Analyze” feature selected, and the cell number was counted manually.
Ethical statement
All the experiments with human materials were approved by the Ethics Committee of Third Hospital, Peking University, China (No. 2017SZ-043), and written informed consent was obtained from all the involved donors before surgery.
Code availability
Codes that support the findings of this study are available at GitHub (https://github.com/WRui/Human-Gonad-10X/).
Data availability
Our scRNA-seq data have been deposited in the Genome Sequence Archive [70] at the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation (GSA: HRA000344), and are publicly accessible at https://ngdc.cncb.ac.cn/.
CRediT author statement
Rui Wang: Conceptualization, Software, Validation, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization, Project administration. Xixi Liu: Methodology, Validation, Investigation, Writing - review & editing. Li Li: Methodology, Investigation. Ming Yang: Validation, Resources. Jun Yong: Resources. Fan Zhai: Resources. Lu Wen: Conceptualization, Funding acquisition. Liying Yan: Resources. Jie Qiao: Conceptualization, Validation, Resources, Supervision, Funding acquisition. Fuchou Tang: Conceptualization, Validation, Writing - review & editing, Supervision, Project administration, Funding acquisition. All authors have read and approved the final manuscript.
Competing interests
The authors have declared no competing financial interests.
Acknowledgments
This work was supported by the National Key R&D Program of China (Grant No. 2018YFA0107601). Part of analysis was performed on the High-Performance Computing Platform of the Center for Life Science, China. We thank all the patients for tissue donation in this study. We thank Chunyan Shan from the National Center for Protein Sciences at Peking University in Beijing, China for help with immunofluorescent staining training. We thank Yuqiong Hu, Xiaoying Fan, and Xinyi Ma for providing helps in experiments.
Handled by Feng Liu
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2022.04.002.
Contributor Information
Jie Qiao, Email: jie.qiao@263.net.
Fuchou Tang, Email: tangfuchou@pku.edu.cn.
Supplementary material
The following are the Supplementary material to this article:
Basic information and comparison of expression patterns between an abnormal 45, XO embryo and normal 46, XX/XY embryos A. UMAP cluster map showing the cells from the two technical replicates of a 15 W male embryo and from fetuses at same developmental stages. B. UMAP showing the clusters identified by Seurat package. Different colors represent different clusters. C. UMAP cluster map revealing the distribution of doublet identified by DoubletFinder R package, which was related to Table S1. D. Box plot showing the number of genes detected and the ratio of reads mapped to mitochondria in different cell types. E. UMAP clustering map showing the distribution of male and female cells and XIST expression. Female cells, pink; male cells, sky blue. Colors from gray to purple represent low to high expression levels. F. Heatmap showing the expression profile of somatic cell marker genes in normal embryos (46, XX/XY) and an XO embryo (45, XO). G. Heatmap showing the expression profiles of PGC marker genes in XO, normal female (XX), and normal male (XY) embryos, which was related to Table S2. H. Violin plot showing several genes that were highly expressed by somatic cells or PGCs in normal embryos. I. Violin plot showing highly expressed genes in gonadal somatic cells and/or PGCs from an abnormal XO embryo. J. UMAP cluster map showing the genes differentially expressed between a normal female embryo (XX) and a Turner syndrome embryo (XO). Endo, endothelial.
Expression characteristics for different FGC subtypes and characterization of GST A. Heatmap showing the expression profiles of FGC markers. The gene list can be found in Table S3. B. Bar plot showing the ratio of different subtypes of FGCs at different developmental stages for male and female embryos. C. Dot plot showing the ratio of germ cells at different gestational ages for the different sexes. Pink represents female and sky blue represents male. The X-axis shows different developmental stages, and the Y-axis represents the percentage of FGCs, which was calculated using scRNA-seq data. D. UMAP plot of an adult human testicular cell cluster that was defined according to scRNA-seq analysis. Others, cell types such as endothelial cells, blood cells, and immune cells. E. UMAP plot of neonatal human testicular cells and cell types defined by scRNA-seq analysis. F. Violin plot showing the expression profiles of pluripotent markers in the different cell populations defined in Figure 4A. G. Dot plot showing the expression of RA-synthesizing enzyme ALDH1A2 along PCA1 of Figure 4A. H. PCA plot showing the expression of RA-synthesizing enzymes (ALDH1A1, ALDH1A2, and ALDH1A3) in fetal testicular germ cells. Colors from gray to purple represent low to high expression levels.
Characterization of testicular FGCs in vivo and cultured testicular cells in vitro A. UMAP clustering of male FGCs that previously published [17]. B. Violin plot showing the expression profiles of pluripotent markers and genes involved in the BMP signaling pathway. C. Boxplot showing the expression of cell type-specific markers in the in vitro cultured 15 W testicular cells. PDGFRA, Leydig cell marker; SOX9, Sertoli cell marker; and DDX4 and POU5F1, germ cell marker. The arrow indicates a group of cultured cells that expressed low levels of POU5F1. D. Boxplot showing the expression of BMP signaling pathway-related genes (ID1, ID2, ID3, ID4, SMAD9, SMAD6, SMAD7, RBL1, and MYC), RA-synthesizing enzyme (ALDH1A1, ALDH1A2, and ALDH1A3) and cell type-specific markers (DAZL, PDGFRA, SOX9, and AMH) in different cell types with different experimental treatments.
Characterization of in vitro cultured 7 W and 15 W testicular cells A. Two-dimensional embedding of cultured 7 W testicular cells using UMAP. The cells are colored according to cell type. The shapes represent cells subjected to different treatments. The dashed line roughly frames the LDN-treated somatic cells. B. Boxplots showing the expression levels (log2(TPM+1)) of cell type-specific marker genes (DDX4, POU5F1, SOX9, PDGFRA, and KRT19), BMP signaling pathway-related genes (ID1, ID2, ID3, SMAD6, SMAD7, and SMAD9), and RA-synthesizing enzymes (ALDH1A1, ALDH1A2, and ALDH1A3) in cultured 7 W testicular cells. Different colors represent different experimental treatments. C. Dot plot showing the expression of cell type-specific genes in different cell types from 15 W fetal testes under different experimental conditions. NC is negative control, which means no LDN treatment. Dot size represents the percentage of cells expressing the specific genes. Dot color from blue to red represents low to high expression levels. D. Dot plot showing the expression of cell type-specific genes in different cell types of 7 W embryo under different experimental conditions. NC is negative control, which means no LDN treatment. Dot size represents the percentage of cells expressing specific genes. Dot color from blue to red represents low to high expression levels. Qui, quiescent; Pro, proliferative.
Testicular somatic subtype identification and verification A. UMAP plot of male gonadal somatic cells showing the developmental stages. B. UMAP plot of male gonadal somatic cells showing the expression profiles of genes used to identify male somatic cell subtypes. C. UMAP plot of male gonadal somatic cells showing the expression of cell type-specific genes. Genes in blue represent cell surface markers. D. Immunostaining of the Sertoli cell marker SOX9 and the Leydig cell marker PDGFRB. E. Immunostaining of a known Sertoli cell marker (SOX9) and a newly identified surface marker (CDH2) at several developmental stages. SOX9 and CDH2 have good colocalization. Scale bar, 25 μm. F. Immunostaining of KRT19, the Leydig cell marker PDGFRB, DCN, the Sertoli cell marker SOX9, and the FGC markers POU5F1/DDX4 in human fetal testes. Scale bar, 25 μm. G. Heatmap showing the predicted ligand–receptor gene pairs that facilitate interactions between Leydig cells and Sertoli cells. H. UMAP showing the expression levels of GAS6 and TYRO3 in testicular somatic cells.
Comparison of fetal, neonatal, and adult testicular somatic cells A. UMAP clustering of human pubertal testicular cells by scRNA-seq analysis. B. UMAP plot of fetal, neonatal, and adult male somatic cells showing the expression of cell type-specific genes. Genes in blue represent cell surface markers. C. Flow chart showing the genes involved in HSD17B3 synthesis. D UMAP plot showing the expression profiles of genes involved in RA and androgen metabolism E. Bar plot showing the ratio of testicular somatic cells that expressed RA and androgen metabolism-associated genes at different stages. F. Immunostaining of the Sertoli cell markers SOX9, PDGFA, and the androgen metabolism-related factors CYP11A1 and HSD17B3 in fetal testes. G. Immunostaining of the Sertoli cell marker PDGFA and the androgen metabolism-related factors CYP11A1 and HSD17B3 in adult testes. H. Immunostaining of the Sertoli cell markers SOX9 and HSD17B3 in adult testes. Regions inside the white square are magnified and shown below the corresponding figure. Scale bar, 25 μm.
Characteristics of fetal ovarian somatic cells A. UMAP plot of female ovarian somatic cells showing the expression of somatic progenitor cell markers. Colors from gray to purple represent low to high expression levels. B. Bar plot showing the ratio of ovarian somatic cells that expressed somatic progenitor cell markers at different stages. C. Bar plot showing the ratio of ovarian somatic cells that expressed RA signaling pathway and estrogen metabolism-related genes in different cell types at different stages. D. Heatmap showing ovarian somatic cell-type-specific genes. E. UMAP clustering of adult ovarian cells. The different colors represent different cell types. F. UMAP plot of female ovarian somatic cells showing the expression of the steroidogenic cell lineage markers TCF21 and PDGFRA. granulosa cell; atrGC, atretic GC; proGC, progenitor GC.
Summary diagram of male and female gonadal development A. Summary chart comparing 7 W female (XX) and 7 W male (XY) embryos. B. Summary chart comparing 7 W normal female (XX) and 7 W Turner syndrome (XO) embryos. C. Summary chart showing the cell-type-specific roles of BMP signaling pathway in testicular germ cells and somatic cells. Our results also reveal the potential transitions between germ cells and somatic cells. Cell–cell interactions between Sertoli cells and Leydig cells through several receptor–ligand gene pairs.
References
- 1.Achermann JC, Hughes IA. Disorders of sex development. In: Melmed S, Polonsky KS, Larsen PR, Kronenberg HM, editors. Williams Textbook of Endocrinology. 12th ed. Amsterdam: Elsevier Inc; 2011, p.868–934.
- 2.Saitou M., Yamaji M. Primordial germ cells in mice. Cold Spring Harb Perspect Biol. 2012;4:a008375. doi: 10.1101/cshperspect.a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taketo T. The role of sex chromosomes in mammalian germ cell differentiation: can the germ cells carrying X and Y chromosomes differentiate into fertile oocytes? Asian J Androl. 2015;17:360–366. doi: 10.4103/1008-682X.143306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurek M., Albalushi H., Hovatta O., Stukenborg J.B. Human pluripotent stem cells in reproductive science — a comparison of protocols used to generate and define male germ cells from pluripotent stem cells. Int J Mol Sci. 2020;21:1028. doi: 10.3390/ijms21031028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.She Z.Y., Yang W.X. Sry and SoxE genes: how they participate in mammalian sex determination and gonadal development? Semin Cell Dev Biol. 2017;63:13–22. doi: 10.1016/j.semcdb.2016.07.032. [DOI] [PubMed] [Google Scholar]
- 6.Childs A.J., Kinnell H.L., Collins C.S., Hogg K., Bayne R.A., Green S.J., et al. BMP signaling in the human fetal ovary is developmentally regulated and promotes primordial germ cell apoptosis. Stem Cells. 2010;28:1368–1378. doi: 10.1002/stem.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rotgers E., Jorgensen A., Yao H.H.C. At the crossroads of fate-somatic cell lineage specification in the fetal gonad. Endocr Rev. 2018;39:739–759. doi: 10.1210/er.2018-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shima Y. Development of fetal and adult Leydig cells. Reprod Med Biol. 2019;18:323–330. doi: 10.1002/rmb2.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh R.P., Carr D.H. The anatomy and histology of XO human embryos and fetuses. Anat Rec. 1966;155:369–383. doi: 10.1002/ar.1091550309. [DOI] [PubMed] [Google Scholar]
- 10.Sarraj M.A., Drummond A.E. Mammalian foetal ovarian development: consequences for health and disease. Reproduction. 2012;143:151–163. doi: 10.1530/REP-11-0247. [DOI] [PubMed] [Google Scholar]
- 11.Bowles J., Koopman P. Retinoic acid, meiosis and germ cell fate in mammals. Development. 2007;134:3401–3411. doi: 10.1242/dev.001107. [DOI] [PubMed] [Google Scholar]
- 12.Su D.M., Feng Y., Wang L., Wu Y.L., Ge R.S., Ma X. Influence of fetal Leydig cells on the development of adult Leydig cell population in rats. J Reprod Dev. 2018;64:223–231. doi: 10.1262/jrd.2017-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wen Q., Zheng Q.S., Li X.X., Hu Z.Y., Gao F., Cheng C.Y., et al. Wt1 dictates the fate of fetal and adult Leydig cells during development in the mouse testis. Am J Physiol Endocrinol Metab. 2014;307:E1131–E1143. doi: 10.1152/ajpendo.00425.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stevant I., Neirijnck Y., Borel C., Escoffier J., Smith L.B., Antonarakis S.E., et al. Deciphering cell lineage specification during male sex determination with single-cell RNA sequencing. Cell Rep. 2018;22:1589–1599. doi: 10.1016/j.celrep.2018.01.043. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y., Zheng Y., Gao Y., Lin Z., Yang S., Wang T., et al. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 2018;28:879–896. doi: 10.1038/s41422-018-0074-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen L., Tang F. Recent advances in single-cell sequencing technologies. Precis Clin Med. 2022;5:pbac002. doi: 10.1093/pcmedi/pbac002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L., Dong J., Yan L., Yong J., Liu X., Hu Y., et al. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell. 2017;20:891–892. doi: 10.1016/j.stem.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Guo J., Grow E.J., Mlcochova H., Maher G.J., Lindskog C., Nie X., et al. The adult human testis transcriptional cell atlas. Cell Res. 2018;28:1141–1157. doi: 10.1038/s41422-018-0099-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo J., Nie X., Giebler M., Mlcochova H., Wang Y., Grow E.J., et al. The dynamic transcriptional cell atlas of testis development during human puberty. Cell Stem Cell. 2020;26:262–276. doi: 10.1016/j.stem.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hermann B.P., Cheng K., Singh A., Roa-De La Cruz L., Mutoji K.N., Chen I.C., et al. The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 2018;25:1650–1667. doi: 10.1016/j.celrep.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sohni A., Tan K., Song H.W., Burow D., de Rooij D.G., Laurent L., et al. The neonatal and adult human testis defined at the single-cell level. Cell Rep. 2019;26:1501–1517. doi: 10.1016/j.celrep.2019.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan X., Bialecka M., Moustakas I., Lam E., Torrens-Juaneda V., Borggreven N.V., et al. Single-cell reconstruction of follicular remodeling in the human adult ovary. Nat Commun. 2019;10:3164. doi: 10.1038/s41467-019-11036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner M., Yoshihara M., Douagi I., Damdimopoulos A., Panula S., Petropoulos S., et al. Single-cell analysis of human ovarian cortex identifies distinct cell populations but no oogonial stem cells. Nat Commun. 2020;11:1147. doi: 10.1038/s41467-020-14936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M., Liu X., Chang G., Chen Y., An G., Yan L., et al. Single-cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell. 2018;23:599–614. doi: 10.1016/j.stem.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y., Yan Z., Qin Q., Nisenblat V., Chang H.M., Yu Y., et al. Transcriptome landscape of human folliculogenesis reveals oocyte and granulosa cell interactions. Mol Cell. 2018;72:1021–1034. doi: 10.1016/j.molcel.2018.10.029. [DOI] [PubMed] [Google Scholar]
- 26.Jan S.Z., Vormer T.L., Jongejan A., Roling M.D., Silber S.J., de Rooij D.G., et al. Unraveling transcriptome dynamics in human spermatogenesis. Development. 2017;144:3659–3673. doi: 10.1242/dev.152413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo F., Yan L., Guo H., Li L., Hu B., Zhao Y., et al. The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell. 2015;161:1437–1452. doi: 10.1016/j.cell.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 28.Guo J., Sosa E., Chitiashvili T., Nie X., Rojas E.J., Oliver E., et al. Single-cell analysis of the developing human testis reveals somatic niche cell specification and fetal germline stem cell establishment. Cell Stem Cell. 2021;28:764–778. doi: 10.1016/j.stem.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Overeem A.W., Chang Y.W., Spruit J., Roelse C.M., De Sousa Lopes S.M.C. Ligand-receptor interactions elucidate sex-specific pathways in the trajectory from primordial germ cells to gonia during human development. Front Cell Dev Biol. 2021;9:661243. doi: 10.3389/fcell.2021.661243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maguire C.A., Song Y.B., Wu M., Leon S., Carroll R.S., Alreja M., et al. Tac1 signaling is required for sexual maturation and responsiveness of GnRH neurons to kisspeptin in the male mouse. Endocrinology. 2017;158:2319–2329. doi: 10.1210/en.2016-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 32.Thul P.J., Akesson L., Wiking M., Mahdessian D., Geladaki A., Blal H.A., et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 33.Wen Q., Cheng C.Y., Liu Y.X. Development, function and fate of fetal Leydig cells. Semin Cell Dev Biol. 2016;59:89–98. doi: 10.1016/j.semcdb.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen Y.C., Shami A.N., Moritz L., Larose H., Manske G.L., Ma Q., et al. TCF21+ mesenchymal cells contribute to testis somatic cell development, homeostasis, and regeneration in mice. Nat Commun. 2021;12:3876. doi: 10.1038/s41467-021-24130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui S., Ross A., Stallings N., Parker K.L., Capel B., Quaggin S.E. Disrupted gonadogenesis and male-to-female sex reversal in Pod1 knockout mice. Development. 2004;131:4095–4105. doi: 10.1242/dev.01266. [DOI] [PubMed] [Google Scholar]
- 36.McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329–337. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hung J.Y., Yen M.C., Jian S.F., Wu C.Y., Chang W.A., Liu K.T., et al. Secreted protein acidic and rich in cysteine (SPARC) induces cell migration and epithelial mesenchymal transition through WNK1/snail in non-small cell lung cancer. Oncotarget. 2017;8:63691–63702. doi: 10.18632/oncotarget.19475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pui H.P., Saga Y. Gonocytes-to-spermatogonia transition initiates prior to birth in murine testes and it requires FGF signaling. Mech Dev. 2017;144:125–139. doi: 10.1016/j.mod.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 39.Abe S. Differentiation of spermatogenic cells from vertebrates in vitro. Int Rev Cytol. 1987;109:159–209. doi: 10.1016/s0074-7696(08)61722-2. [DOI] [PubMed] [Google Scholar]
- 40.Dym M., Fawcett D.W. Further observations on the numbers of spermatogonia, spermatocytes, and spermatids connected by intercellular bridges in the mammalian testis. Biol Reprod. 1971;4:195–215. doi: 10.1093/biolreprod/4.2.195. [DOI] [PubMed] [Google Scholar]
- 41.Culty M. Gonocytes, the forgotten cells of the germ cell lineage. Birth Defects Res C Embryo Today. 2009;87:1–26. doi: 10.1002/bdrc.20142. [DOI] [PubMed] [Google Scholar]
- 42.McCarrey JR. Transition of prenatal prospermatogonia to postnatal spermatogonia. In: Oatley JM, Griswold MD, editors. In the biology of mammalian spermatogonia. New York: Springer; 2017, p.23–38.
- 43.Amory J.K., Arnold S., Lardone M.C., Piottante A., Ebensperger M., Isoherranen N., et al. Levels of the retinoic acid synthesizing enzyme ALDH1A2 are lower in testicular tissue from men with infertility. Fertil Steril. 2014;101:960–966. doi: 10.1016/j.fertnstert.2013.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li L., Li L., Li Q., Liu X., Ma X., Yong J., et al. Dissecting the epigenomic dynamics of human fetal germ cell development at single-cell resolution. Cell Res. 2021;31:463–477. doi: 10.1038/s41422-020-00401-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia-Moreno S.A., Futtner C.R., Salamone I.M., Gonen N., Lovell-Badge R., Maatouk D.M. Gonadal supporting cells acquire sex-specific chromatin landscapes during mammalian sex determination. Dev Biol. 2019;446:168–179. doi: 10.1016/j.ydbio.2018.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Browaeys R., Saelens W., Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods. 2020;17:159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 47.Yao H.H.C., Whoriskey W., Capel B. Desert Hedgehog/Patched 1 signaling specifies fetal Leydig cell fate in testis organogenesis. Genes Dev. 2002;16:1433–1440. doi: 10.1101/gad.981202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brennan J., Tilmann C., Capel B. Pdgfr-α mediates testis cord organization and fetal Leydig cell development in the XY gonad. Genes Dev. 2003;17:800–810. doi: 10.1101/gad.1052503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shima Y., Miyabayashi K., Haraguchi S., Arakawa T., Otake H., Baba T., et al. Contribution of Leydig and Sertoli cells to testosterone production in mouse fetal testes. Mol Endocrinol. 2013;27:63–73. doi: 10.1210/me.2012-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weisser J., Landreh L., Soder O., Svechnikov K. Steroidogenesis and steroidogenic gene expression in postnatal fetal rat Leydig cells. Mol Cell Endocrinol. 2011;341:18–24. doi: 10.1016/j.mce.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 51.Childs A.J., Cowan G., Kinnell H.L., Anderson R.A., Saunders P.T. Retinoic acid signalling and the control of meiotic entry in the human fetal gonad. PLoS One. 2011;6:e20249. doi: 10.1371/journal.pone.0020249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jauregui E.J., Mitchell D., Topping T., Hogarth C.A., Griswold M.D. Retinoic acid receptor signaling is necessary in steroidogenic cells for normal spermatogenesis and epididymal function. Development. 2018;145:dev160465. doi: 10.1242/dev.160465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minkina A., Lindeman R.E., Gearhart M.D., Chassot A.A., Chaboissier M.C., Ghyselinck N.B., et al. Retinoic acid signaling is dispensable for somatic development and function in the mammalian ovary. Dev Biol. 2017;424:208–220. doi: 10.1016/j.ydbio.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vernet N., Dennefeld C., Rochette-Egly C., Oulad-Abdelghani M., Chambon P., Ghyselinck N.B., et al. Retinoic acid metabolism and signaling pathways in the adult and developing mouse testis. Endocrinology. 2006;147:96–110. doi: 10.1210/en.2005-0953. [DOI] [PubMed] [Google Scholar]
- 55.Yang Y., Workman S., Wilson M. The molecular pathways underlying early gonadal development. J Mol Endocrinol. 2019;62:R47–R64. doi: 10.1530/JME-17-0314. [DOI] [PubMed] [Google Scholar]
- 56.Zhao F., Franco H.L., Rodriguez K.F., Brown P.R., Tsai M.J., Tsai S.Y., et al. Elimination of the male reproductive tract in the female embryo is promoted by COUP-TFII in mice. Science. 2017;357:717–720. doi: 10.1126/science.aai9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilhelm D., Palmer S., Koopman P. Sex determination and gonadal development in mammals. Physiol Rev. 2007;87:1–28. doi: 10.1152/physrev.00009.2006. [DOI] [PubMed] [Google Scholar]
- 58.Maheshwari A., Fowler P.A. Primordial follicular assembly in humans — revisited. Zygote. 2008;16:285–296. doi: 10.1017/S0967199408004802. [DOI] [PubMed] [Google Scholar]
- 59.Hummitzsch K., Hatzirodos N., Irving-Rodgers H.F., Hartanti M.D., Perry V.E.A., Anderson R.A., et al. Morphometric analyses and gene expression related to germ cells, gonadal ridge epithelial-like cells and granulosa cells during development of the bovine fetal ovary. PLoS One. 2019;14:e0214130. doi: 10.1371/journal.pone.0214130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hummitzsch K., Irving-Rodgers H.F., Hatzirodos N., Bonner W., Sabatier L., Reinhardt D.P., et al. A new model of development of the mammalian ovary and follicles. PLoS One. 2013;8:e55578. doi: 10.1371/journal.pone.0055578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uhlenhaut N.H., Jakob S., Anlag K., Eisenberger T., Sekido R., Kress J., et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell. 2009;139:1130–1142. doi: 10.1016/j.cell.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 62.Aoyama M., Shiraishi A., Matsubara S., Horie K., Osugi T., Kawada T., et al. Identification of a new theca/interstitial cell-specific gene and its biological role in growth of mouse ovarian follicles at the gonadotropin-independent stage. Front Endocrinol (Lausanne) 2019;10:553. doi: 10.3389/fendo.2019.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niu W., Spradling A.C. Two distinct pathways of pregranulosa cell differentiation support follicle formation in the mouse ovary. Proc Natl Acad Sci U S A. 2020;117:20015–20026. doi: 10.1073/pnas.2005570117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McKay AC, Odeluga N, Jiang J, Sharma S. Anatomy, abdomen and pelvis, seminal vesicle. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2022. [PubMed]
- 65.Greeley S.A.W., Littlejohn E., Husain A.N., Waggoner D., Gundeti M., Rosenfield R.L. The effect of the testis on the ovary: structure-function relationships in a neonate with a unilateral ovotestis (ovotesticular disorder of sex development) Horm Res Paediatr. 2017;87:205–212. doi: 10.1159/000455142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamashiro C., Sasaki K., Yabuta Y., Kojima Y., Nakamura T., Okamoto I., et al. Generation of human oogonia from induced pluripotent stem cells in vitro. Science. 2018;362:356–360. doi: 10.1126/science.aat1674. [DOI] [PubMed] [Google Scholar]
- 67.Ghosh S., Chan C.K.K. Analysis of RNA-Seq data using TopHat and Cufflinks. Methods Mol Biol. 2016;1374:339–361. doi: 10.1007/978-1-4939-3167-5_18. [DOI] [PubMed] [Google Scholar]
- 68.Anders S., Pyl P.T., Huber W. HTSeq — a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen T., Chen X., Zhang S., Zhu J., Tang B., Wang A., et al. The Genome Sequence Archive Family: toward explosive data growth and diverse data types. Genomics Proteomics Bioinformatics. 2021;19:578–583. doi: 10.1016/j.gpb.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Basic information and comparison of expression patterns between an abnormal 45, XO embryo and normal 46, XX/XY embryos A. UMAP cluster map showing the cells from the two technical replicates of a 15 W male embryo and from fetuses at same developmental stages. B. UMAP showing the clusters identified by Seurat package. Different colors represent different clusters. C. UMAP cluster map revealing the distribution of doublet identified by DoubletFinder R package, which was related to Table S1. D. Box plot showing the number of genes detected and the ratio of reads mapped to mitochondria in different cell types. E. UMAP clustering map showing the distribution of male and female cells and XIST expression. Female cells, pink; male cells, sky blue. Colors from gray to purple represent low to high expression levels. F. Heatmap showing the expression profile of somatic cell marker genes in normal embryos (46, XX/XY) and an XO embryo (45, XO). G. Heatmap showing the expression profiles of PGC marker genes in XO, normal female (XX), and normal male (XY) embryos, which was related to Table S2. H. Violin plot showing several genes that were highly expressed by somatic cells or PGCs in normal embryos. I. Violin plot showing highly expressed genes in gonadal somatic cells and/or PGCs from an abnormal XO embryo. J. UMAP cluster map showing the genes differentially expressed between a normal female embryo (XX) and a Turner syndrome embryo (XO). Endo, endothelial.
Expression characteristics for different FGC subtypes and characterization of GST A. Heatmap showing the expression profiles of FGC markers. The gene list can be found in Table S3. B. Bar plot showing the ratio of different subtypes of FGCs at different developmental stages for male and female embryos. C. Dot plot showing the ratio of germ cells at different gestational ages for the different sexes. Pink represents female and sky blue represents male. The X-axis shows different developmental stages, and the Y-axis represents the percentage of FGCs, which was calculated using scRNA-seq data. D. UMAP plot of an adult human testicular cell cluster that was defined according to scRNA-seq analysis. Others, cell types such as endothelial cells, blood cells, and immune cells. E. UMAP plot of neonatal human testicular cells and cell types defined by scRNA-seq analysis. F. Violin plot showing the expression profiles of pluripotent markers in the different cell populations defined in Figure 4A. G. Dot plot showing the expression of RA-synthesizing enzyme ALDH1A2 along PCA1 of Figure 4A. H. PCA plot showing the expression of RA-synthesizing enzymes (ALDH1A1, ALDH1A2, and ALDH1A3) in fetal testicular germ cells. Colors from gray to purple represent low to high expression levels.
Characterization of testicular FGCs in vivo and cultured testicular cells in vitro A. UMAP clustering of male FGCs that previously published [17]. B. Violin plot showing the expression profiles of pluripotent markers and genes involved in the BMP signaling pathway. C. Boxplot showing the expression of cell type-specific markers in the in vitro cultured 15 W testicular cells. PDGFRA, Leydig cell marker; SOX9, Sertoli cell marker; and DDX4 and POU5F1, germ cell marker. The arrow indicates a group of cultured cells that expressed low levels of POU5F1. D. Boxplot showing the expression of BMP signaling pathway-related genes (ID1, ID2, ID3, ID4, SMAD9, SMAD6, SMAD7, RBL1, and MYC), RA-synthesizing enzyme (ALDH1A1, ALDH1A2, and ALDH1A3) and cell type-specific markers (DAZL, PDGFRA, SOX9, and AMH) in different cell types with different experimental treatments.
Characterization of in vitro cultured 7 W and 15 W testicular cells A. Two-dimensional embedding of cultured 7 W testicular cells using UMAP. The cells are colored according to cell type. The shapes represent cells subjected to different treatments. The dashed line roughly frames the LDN-treated somatic cells. B. Boxplots showing the expression levels (log2(TPM+1)) of cell type-specific marker genes (DDX4, POU5F1, SOX9, PDGFRA, and KRT19), BMP signaling pathway-related genes (ID1, ID2, ID3, SMAD6, SMAD7, and SMAD9), and RA-synthesizing enzymes (ALDH1A1, ALDH1A2, and ALDH1A3) in cultured 7 W testicular cells. Different colors represent different experimental treatments. C. Dot plot showing the expression of cell type-specific genes in different cell types from 15 W fetal testes under different experimental conditions. NC is negative control, which means no LDN treatment. Dot size represents the percentage of cells expressing the specific genes. Dot color from blue to red represents low to high expression levels. D. Dot plot showing the expression of cell type-specific genes in different cell types of 7 W embryo under different experimental conditions. NC is negative control, which means no LDN treatment. Dot size represents the percentage of cells expressing specific genes. Dot color from blue to red represents low to high expression levels. Qui, quiescent; Pro, proliferative.
Testicular somatic subtype identification and verification A. UMAP plot of male gonadal somatic cells showing the developmental stages. B. UMAP plot of male gonadal somatic cells showing the expression profiles of genes used to identify male somatic cell subtypes. C. UMAP plot of male gonadal somatic cells showing the expression of cell type-specific genes. Genes in blue represent cell surface markers. D. Immunostaining of the Sertoli cell marker SOX9 and the Leydig cell marker PDGFRB. E. Immunostaining of a known Sertoli cell marker (SOX9) and a newly identified surface marker (CDH2) at several developmental stages. SOX9 and CDH2 have good colocalization. Scale bar, 25 μm. F. Immunostaining of KRT19, the Leydig cell marker PDGFRB, DCN, the Sertoli cell marker SOX9, and the FGC markers POU5F1/DDX4 in human fetal testes. Scale bar, 25 μm. G. Heatmap showing the predicted ligand–receptor gene pairs that facilitate interactions between Leydig cells and Sertoli cells. H. UMAP showing the expression levels of GAS6 and TYRO3 in testicular somatic cells.
Comparison of fetal, neonatal, and adult testicular somatic cells A. UMAP clustering of human pubertal testicular cells by scRNA-seq analysis. B. UMAP plot of fetal, neonatal, and adult male somatic cells showing the expression of cell type-specific genes. Genes in blue represent cell surface markers. C. Flow chart showing the genes involved in HSD17B3 synthesis. D UMAP plot showing the expression profiles of genes involved in RA and androgen metabolism E. Bar plot showing the ratio of testicular somatic cells that expressed RA and androgen metabolism-associated genes at different stages. F. Immunostaining of the Sertoli cell markers SOX9, PDGFA, and the androgen metabolism-related factors CYP11A1 and HSD17B3 in fetal testes. G. Immunostaining of the Sertoli cell marker PDGFA and the androgen metabolism-related factors CYP11A1 and HSD17B3 in adult testes. H. Immunostaining of the Sertoli cell markers SOX9 and HSD17B3 in adult testes. Regions inside the white square are magnified and shown below the corresponding figure. Scale bar, 25 μm.
Characteristics of fetal ovarian somatic cells A. UMAP plot of female ovarian somatic cells showing the expression of somatic progenitor cell markers. Colors from gray to purple represent low to high expression levels. B. Bar plot showing the ratio of ovarian somatic cells that expressed somatic progenitor cell markers at different stages. C. Bar plot showing the ratio of ovarian somatic cells that expressed RA signaling pathway and estrogen metabolism-related genes in different cell types at different stages. D. Heatmap showing ovarian somatic cell-type-specific genes. E. UMAP clustering of adult ovarian cells. The different colors represent different cell types. F. UMAP plot of female ovarian somatic cells showing the expression of the steroidogenic cell lineage markers TCF21 and PDGFRA. granulosa cell; atrGC, atretic GC; proGC, progenitor GC.
Summary diagram of male and female gonadal development A. Summary chart comparing 7 W female (XX) and 7 W male (XY) embryos. B. Summary chart comparing 7 W normal female (XX) and 7 W Turner syndrome (XO) embryos. C. Summary chart showing the cell-type-specific roles of BMP signaling pathway in testicular germ cells and somatic cells. Our results also reveal the potential transitions between germ cells and somatic cells. Cell–cell interactions between Sertoli cells and Leydig cells through several receptor–ligand gene pairs.
Data Availability Statement
Our scRNA-seq data have been deposited in the Genome Sequence Archive [70] at the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation (GSA: HRA000344), and are publicly accessible at https://ngdc.cncb.ac.cn/.