Abstract
Biomolecular condensates compartmentalize and regulate assemblies of biomolecules engaged in vital physiological processes in cells. Specific proteins and nucleic acids engaged in shared functions occur in any one kind of condensate, suggesting that these compartments have distinct chemical specificities. Indeed, some small molecule drugs concentrate in specific condensates due to chemical properties engendered by particular amino acids in the proteins in those condensates. Here we argue that the chemical properties that govern molecular interactions between a small molecule and biomolecules within a condensate can be ascertained for both the small molecule and the biomolecules. We propose that learning this chemical grammar, the rules describing the chemical features of small molecules that engender attraction or repulsion by the physicochemical environment of a specific condensate, should enable design of drugs that with improved efficacy and reduced toxicity.
Editor summary:
This Perspective discussed selective partitioning behaviors of biomolecules and small molecules and proposed that understanding the chemical properties that control their interactions within the condensates would promote drug development.
Introduction
Biomolecular condensates are dynamic assemblies that typically are not enclosed by membranes, and they compartmentalize and concentrate biomolecules involved in shared regulatory processes in cells. Early cytologists observed the largest and most stable of these compartments over a century ago, the nucleolus and Cajal bodies.1–5 There is now evidence that myriad cellular processes, including DNA repair, transcription, signaling, ribosome biogenesis, synaptic transmission, innate immune recognition and others involve condensates.6–15 Condensates thus help segregate subsets of the billions of molecules in a cell into compartments with specific functions.
Transient and multivalent noncovalent interactions among biopolymers drive the formation and behavior of condensates.16–19 The nature of these interactions is diverse and driven by contributions from dispersion, electrostatic, ionic, and electron donating interactions alongside the hydrophobic effect. Weak multivalent interactions are thought to drive formation of condensates through phase separation 16–18, 20–24, although additional mechanisms have been proposed.25–26 Biomolecular condensates form and dissolve in the face of external and internal stimuli, and the nature of regulatory stimuli dictate their life-time and size.6, 13, 27–35 These attributes are conferred by each condensate’s mesoscopic nature, wherein both bulk and microscopic physical and chemical behavior are important.
Here we discuss the chemical mechanisms that contribute to biomolecular condensate formation and regulation, summarize evidence that small molecules can concentrate in specific condensates, and argue that deeper insights into condensate chemistry will lead to improvements in therapeutic molecules (Figure 1). Condensates formed with different biomolecules have distinct chemical specificities that cause other biomolecules or small molecules to selectively associate with the internal condensate chemical environment. Anticancer drugs have been observed to selectively partition into particular condensates and there is evidence that the chemical interactions that govern small molecule-biomolecule interactions can be separated from the biomolecular interactions that govern condensate formation.36 Thus, the chemical properties that govern molecular interactions between a small molecule and biomolecules within a condensate can be ascertained for both the small molecule and biomolecules, and distinguished as contributing to either the small molecule’s interaction with the condensate environment or with its target biomolecule.
Figure 1.
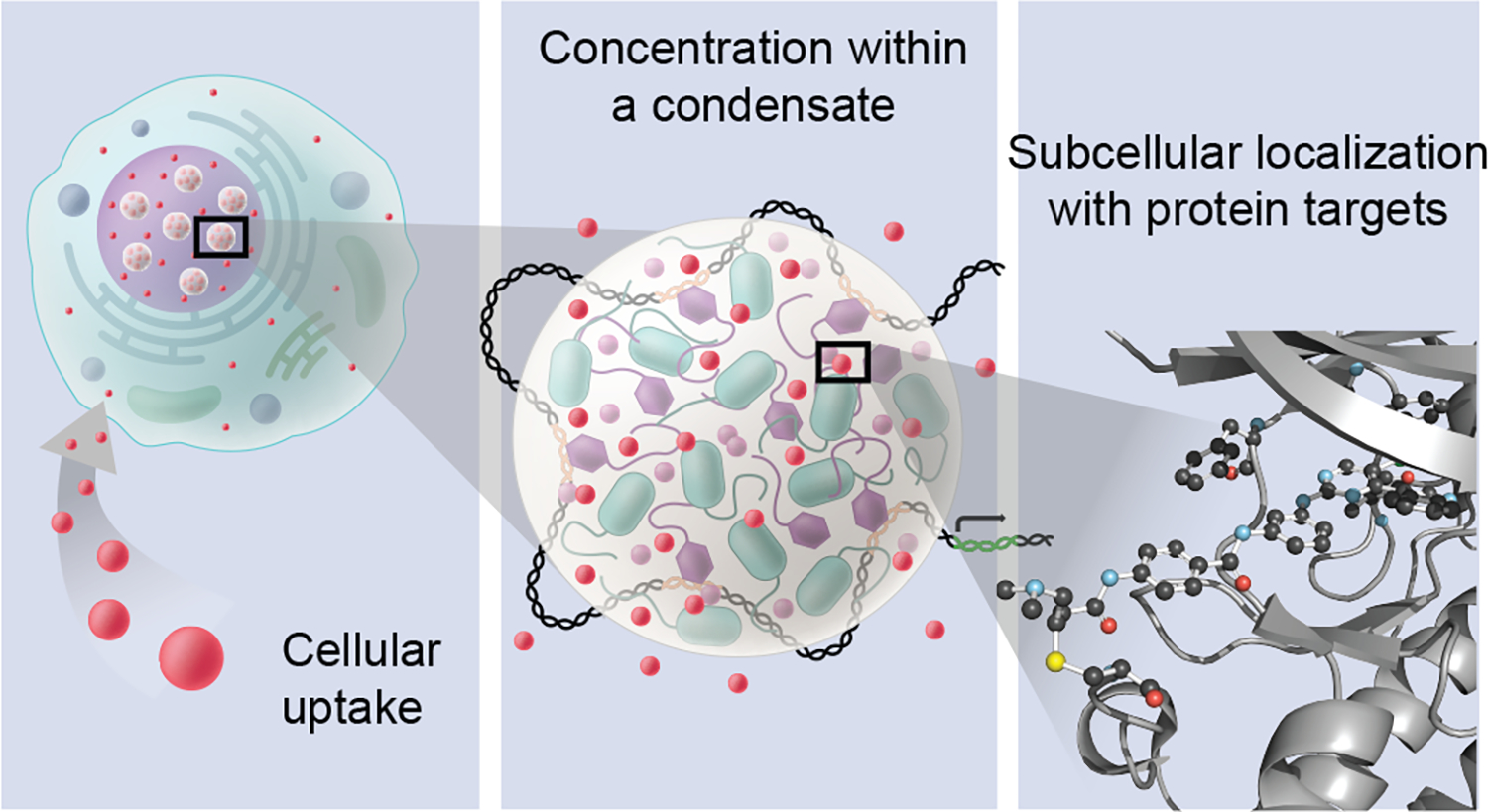
Small molecules can concentrate selectively within specific biomolecular condensates, and may do so both through interactions with the chemical environment of the condensate and through interactions with target proteins or nucleic acids within the condensate. Condensates are mesoscopic bodies and interaction of a molecule with the bulk solvation and microscopic chemical environments is determined by a condensate’s chemical specificity. This includes both specific interactions with target binding sites and interactions with other parts of biomolecules not constituting the target site that may contribute to the concentration of small molecules in a condensate. Left panel: small molecules (red spheres) that enter cells can be distributed unequally among diverse membrane-bound and non-membrane compartments. Middle panel: small molecules (red spheres) concentrating in a transcriptional condensate consisting of transcription apparatus assembled at a DNA locus; this can occur when the interaction of the small molecule with the chemical environment is favored over its interaction with the environment outside the condensate. Right panel: interaction of the small molecule is depicted with a specific biomolecular target, shown here: THZ1 bound to CDK7, PDB ID: 6xd3.
Chemical mechanisms in condensate assembly and behavior
Biomolecular condensates generally consist of assemblies of protein and RNA molecules but may also contain DNA and other biomolecules. To gain insights into the chemical mechanisms that contribute to these complex assemblies, investigators have typically studied how purified protein or RNA molecules contribute to condensate assemblies in vitro. For example, purified proteins that can form condensates in vitro have been studied for the contributions of specific amino acids to condensate formation and behavior. 16–17, 20, 37 RNA molecules have also been observed to form condensates, doing so by base pairing with one another or through interactions with RNA-binding proteins in vitro. 29–30, 38–42 Thus, diverse features of the complex environment of cellular condensates have often been modeled with simple systems in vitro and, where feasible, these findings have been extended to the more complex condensate environment in cells.
A “stickers and spacers” model, where polymers contain blocks with strong interaction potentials separated by blocks with weak interaction potentials, has provided a valuable coarse grain approach to describe the features of diverse biomolecules that promote condensate formation and that influence their material properties (Figure 2A). 17–18, 20, 43–49 The strength of sticker-sticker interaction potentials, their frequency, and the patterning of stickers and spacers has been shown with FET-family and other proteins to govern the saturation concentration of a condensate.16–18 Above this threshold concentration, a system of associative polymers will undergo a phase transition forming dense and dilute phases. Dense phases are characterized by satisfying the majority of sticker-sticker interactions and dilute phases are defined by satisfaction of fewer sticker-sticker interactions; these potentials vary with space and in response to different chemical and biological phenomena. This remarkably simple model has proven useful in interpreting experiments with model protein and nucleic acids and predicting the ability of sequences to engender condensates.
Figure 2.
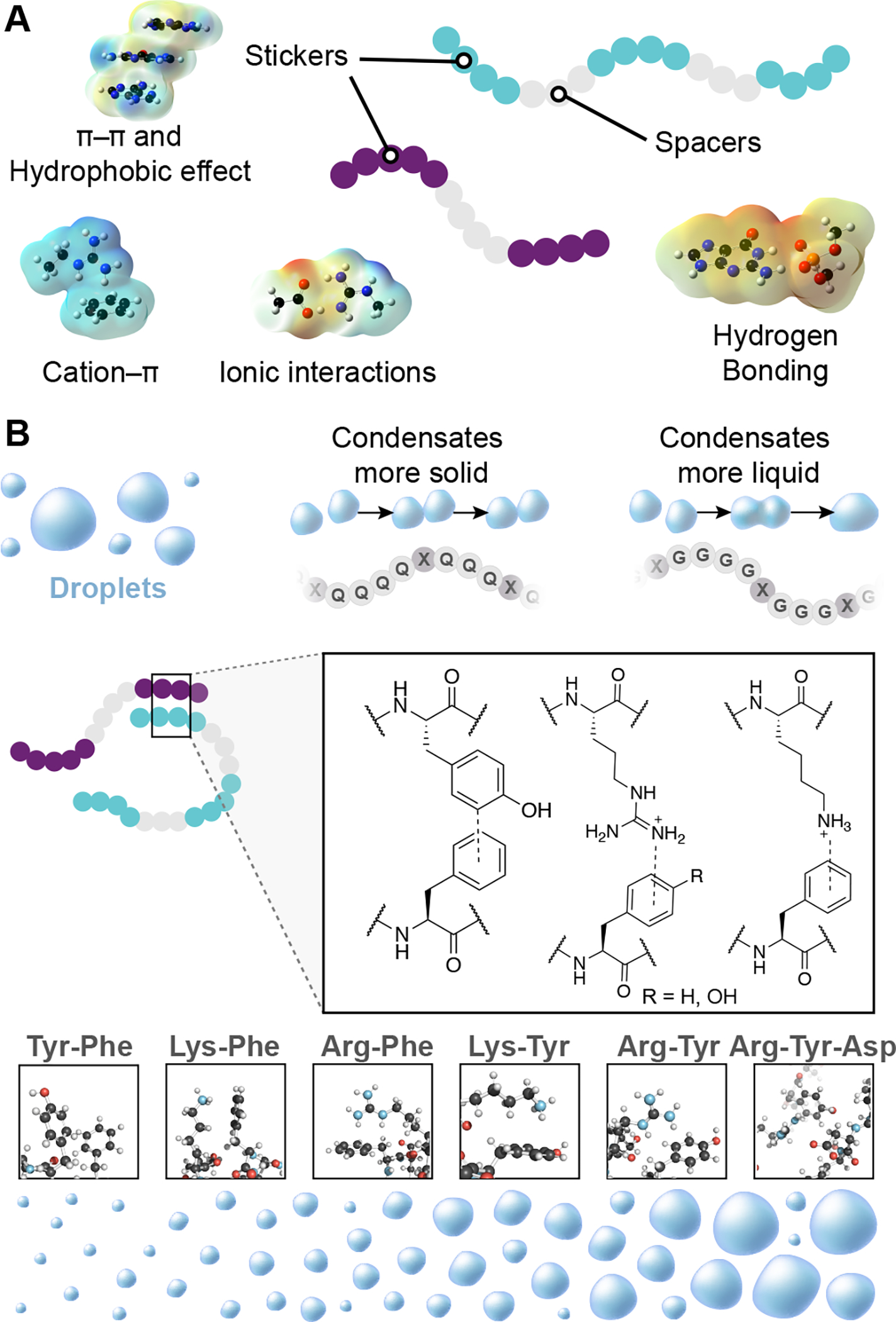
Chemical mechanisms in biomolecular condensate assembly. A) The stickers and spacers model suggests how polymers may associate to form condensates; amino acid side chains, nucleobases, and folded domains may produce interactions that qualify as stickers or spacers. 17–18, 20–21, 23, 38, 44–45, 51–53Examples of noncovalent interactions mediating sticker interactions is displayed with electrostatic surface potentials, computed at the M06-2x/6-311+g(d,p) level of theory.10 B) Condensates form droplets due to the association of biopolymers through noncovalent interactions between sticker regions which are separated by spacer regions in the same biopolymer. In the model system Fused in Sarcoma (FUS), glutamine rich spacers produced more solid-like behavior compared to the liquid-like character of glycine rich spacer regions.18,45 Noncovalent interactions between amino acids with π-systems, and other π-systems or cationic amino acids influence the formation and dissolution of FUS and other protein condensates by creating ‘sticker’ domains. 17–18, 20–21, 23, 38, 44–45, 51–53 These interactions are a consequence of a molecular grammar, rules by which specific amino acids influence condensate formation and behavior.17
The roles of diverse amino acids in the formation and behavior of simple protein condensates, which has been called the “molecular grammar” of these condensates, has come predominantly from studies of the Fused in Sarcoma (FUS) protein (Figure 2B).17, 50–51 FUS forms multimolecular condensates at low micromolar concentrations, driven by transient and multivalent noncovalent interactions between its arginine and tyrosine residues. In diverse proteins, multivalent noncovalent interactions between acidic, basic, and aromatic amino acids have been described that contribute to condensate formation and behavior. 17–18, 20–21, 23, 38, 44–45, 51–53 In FUS, spacer regions composed predominantly of glycine, serine, and glutamine residues occur between the blocks of strongly interacting groups. Spacer regions with a higher content of glycine residues endowed a more dynamic and liquid-like condensate, while a more solid-like condensate was observed with a higher glutamine content (Figure 2B). Analysis of how and where these substitutions endowed different physical properties was facilitated by the stickers and spacers framework.17, 38, 43, 45 Thus, the FET family proteins have provided useful systems for interrogating how condensate behavior is produced by their amino acid sequences and compositions.17, 38
Condensates can also be generated by interactions between structured domains that are bridged by a ligand, enabling a cell to achieve thermodynamic control over phase separation. A theoretical framework put forth by Wyman and Gill54 described how condensate scaffolding protein ligands may exert control over phase separation, in a process termed polyphasic linkage, and recent studies have provided additional experimental confirmation of their predictions.33, 55–56 Bivalent small molecules that have the effect of enhancing or reducing phase separation may prove to be a creative application of this concept.57–58
Evidence for chemical specificities in condensates
The chemical milieu of condensates has been proposed to be chemically distinct microenvironment where certain protein and RNA molecules are densely concentrated, and where these biomolecules will together solvate and enrich for specific sets of additional molecules while excluding others.59–64 Thus, a condensate that contains proteins and nucleic acids engaged in a specific function would be expected to have a microenvironment with chemical features that are distinct from those of condensates with different biomolecules engaged in other functions. Condensates involved in different functions can be visualized in cells by imaging proteins that are specific to these bodies (Figure 3A). These condensates exhibit a variety of properties that are consistent with the notion that they harbor different physicochemical environments; for example, some form and dissolve in short time frames (e.g., transcriptional condensates) whereas others remain assembled for much longer times (e.g., nucleoli).34, 65
Figure 3.
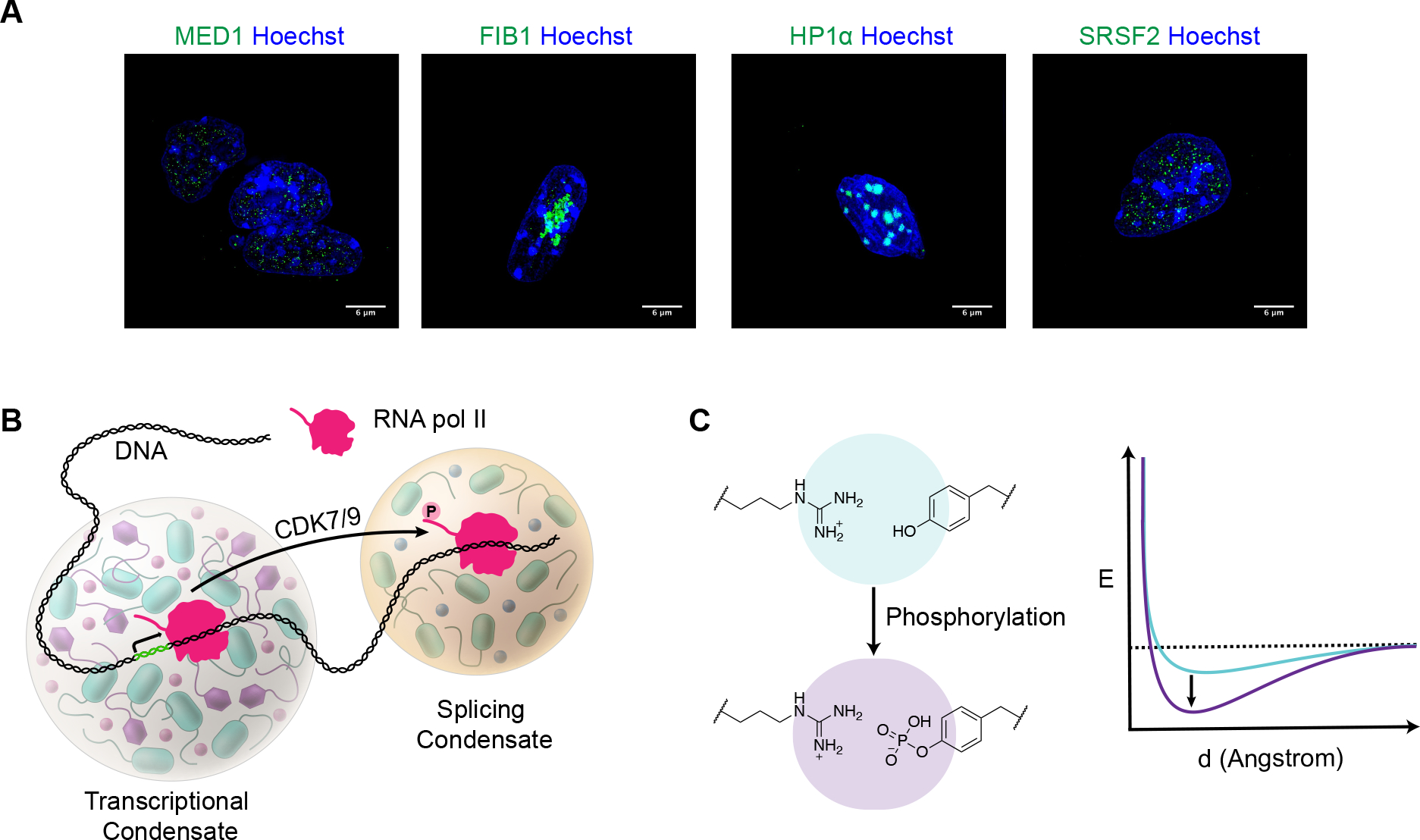
Biomolecular condensates composed of different protein components have different chemical environments that engender selective partitioning of biomolecules.
A) Condensates involved in different functions can be visualized in cells by imaging proteins that are specific to these bodies (e.g., MED1 in transcriptional activation, FIB1 in nucleolar ribosome biosynthesis, HP1a in heterochromatic gene silencing, and SRSF2 in RNA splicing). Images of murine embryonic stem cells with GFP-tagged proteins (green) and Hoechst staining (blue) acquired with a DeltaVision-OM Super resolution microscope.
B) Model illustrating how chemical modification of a protein molecule can cause that molecule to change its condensate partitioning behavior. The RNA polymerase II C-Terminal Domain (CTD) becomes hyperphosphorylated during the transition to elongation, reducing the enzyme’s affinity for transcriptional condensates and increasing its affinity for splicing condensates. The RNA polymerase II CTD kinases CDK7 and CDK9 play well-established regulatory roles in transcription.28
C) Energy diagram showing how posttranslational modifications such as phosphorylation can alter the strength of noncovalent interactions among amino acid side chains. For example, phosphorylation of tyrosine residues will increase the interaction potential with an arginine residue.
Selective partitioning of biomolecules.
Does the collection of biomolecules that distinguish one type of condensate from another create substantially different chemical microenvironments? The selective condensate partitioning behavior of chemically modified biomolecules provides one line of evidence consistent with this possibility. The chemical modification of a protein molecule can cause that molecule to change its condensate partitioning behavior, that is, to exit one condensate and enter another with different components.28, 53 The behavior of RNA Polymerase II during two different stages of transcription, initiation and elongation, provides an example of this type of modification-dependent condensate partitioning (Figure 3B).28, 66–67 RNA Polymerase II can be recruited into transcriptional condensates—compartments marked by the presence of the MED1 protein—during transcription initiation in a form that is minimally phosphorylated. The heptapeptide repeat domain of the largest subunit of RNA polymerase becomes hyperphosphorylated during the transition to elongation, reducing the enzyme’s affinity for transcriptional condensates and increasing its affinity for condensates containing the RNA splicing apparatus. This provides a form of spatiotemporal regulation of transcription, where the apparatus involved in initiation of RNA synthesis and that involved in RNA processing occur in a slightly different space and time.
What chemical features drive the change in condensate partitioning of the polymerase molecule described above? The ‘interaction potentials’ of stickers and spacers can be altered, and the behavior of the condensate assembly changed, with amino acid modification, a common event in biological regulation.17, 23–24, 27–28, 42–43, 50–51, 56, 68 For example, the effective interaction potential of a phosphotyrosine and an arginine residue will be significantly greater than that between a tyrosine and an arginine (Figure 3C). This variation arises because the ionic bond between the phosphate anion and arginine cation is an order of magnitude greater than the cation-π and hydrogen bonding interactions present in the unphosphorylated case (Figure 3C). Phosphorylation also endows a greater capacity to engage in hydrogen bonding interactions on a phosphotyrosine residue, drastically influencing its hydrophilicity and likely the local chemical structure of water and inorganic ions. Biological regulatory modifications that alter the pKa of a side chain, modulate the electrostatic surface potential, ablate hydrogen bonding capacity, alter the hydrophobicity of chemical groups, or alter the flexibility of the amino acids and nucleobases will directly influence the effective interaction potential of a spacer or sticker block.
There are other models that can account for selective partitioning of biomolecules such as that observed with RNA polymerase II modification. It is possible, for example, that proteins resident in one condensate have high-affinity binding sites for unmodified polymerase molecules, whereas proteins resident in the other have high-affinity sites for the modified enzyme. In the case of RNA polymerase II, however, experiments have shown that the heptapeptide repeat domain subjected to phosphorylation exhibits modification-dependent partitioning into simple condensates consisting of proteins representative of transcriptional and splicing condensates that do not have strong binding interactions with the heptapeptide repeat domain (Figure 3B). Thus, differences in condensate chemical environments can be exploited by cells to evolve regulatory mechanisms that involve selective partitioning of biomolecules due to chemical modification.
Selective partitioning of small molecules.
Diverse small molecule drugs have been observed to concentrate in biomolecular condensates. Mitoxantrone, a chemotherapy used in the treatment of some forms of cancer, was long ago observed to concentrate within the nucleolus.69–70 Additional anticancer drugs, such as cisplatin and tamoxifen, have now been demonstrated to concentrate in transcriptional condensates and act on DNA or protein targets that occur within those condensates.36 This selective concentrating behavior in transcriptional condensates is not dependent on an interaction with the ultimate target of these drugs—DNA for cisplatin and for the Estrogen Receptor (ER) for tamoxifen—but rather appears to be due to interactions with specific amino acid residues in the MED1 protein, a key coactivator protein that is a defining component of transcriptional condensates.34, 71 Thus, drug molecules can exploit both condensate partitioning properties and those involved in target engagement to concentrate in the same compartment as their target.
The chemical properties responsible for selective partitioning of small molecules into specific condensates are likely to be the same as those that enable selective partitioning of biomolecules in condensates (Figure 2A). As with biomolecules, in vitro droplet models have proven powerful for interrogating the chemical partitioning behavior of small molecules within condensates. These assays can be conducted with wild-type and mutant forms of protein molecules to identify amino acids that are essential for the partitioning properties of drugs (Figure 4A). Such an approach led to the observation that cisplatin partitioning into MED1 condensates depends on aromatic amino acid residues, and thus cation–π and π–π interactions associated with these residues, and revealed that the amino acids necessary for MED1 phase separation (conserved serine patches) are different from those necessary for the interactions with cisplatin (Figure 4B).36 Furthermore, experiments with MED1 in vitro droplets have suggested that structure-activity relationships between small molecules and biomolecules in condensates can be deduced with a limited range of fluorescent probes.36 These results suggest the chemical properties that govern molecular interactions between a small molecule and biomolecules within a condensate can be ascertained for both the small molecules and the biomolecules of interest (Figure 4C), and distinguished as contributing to either the small molecule’s interaction with the condensate environment or with its target biomolecule.
Figure 4.
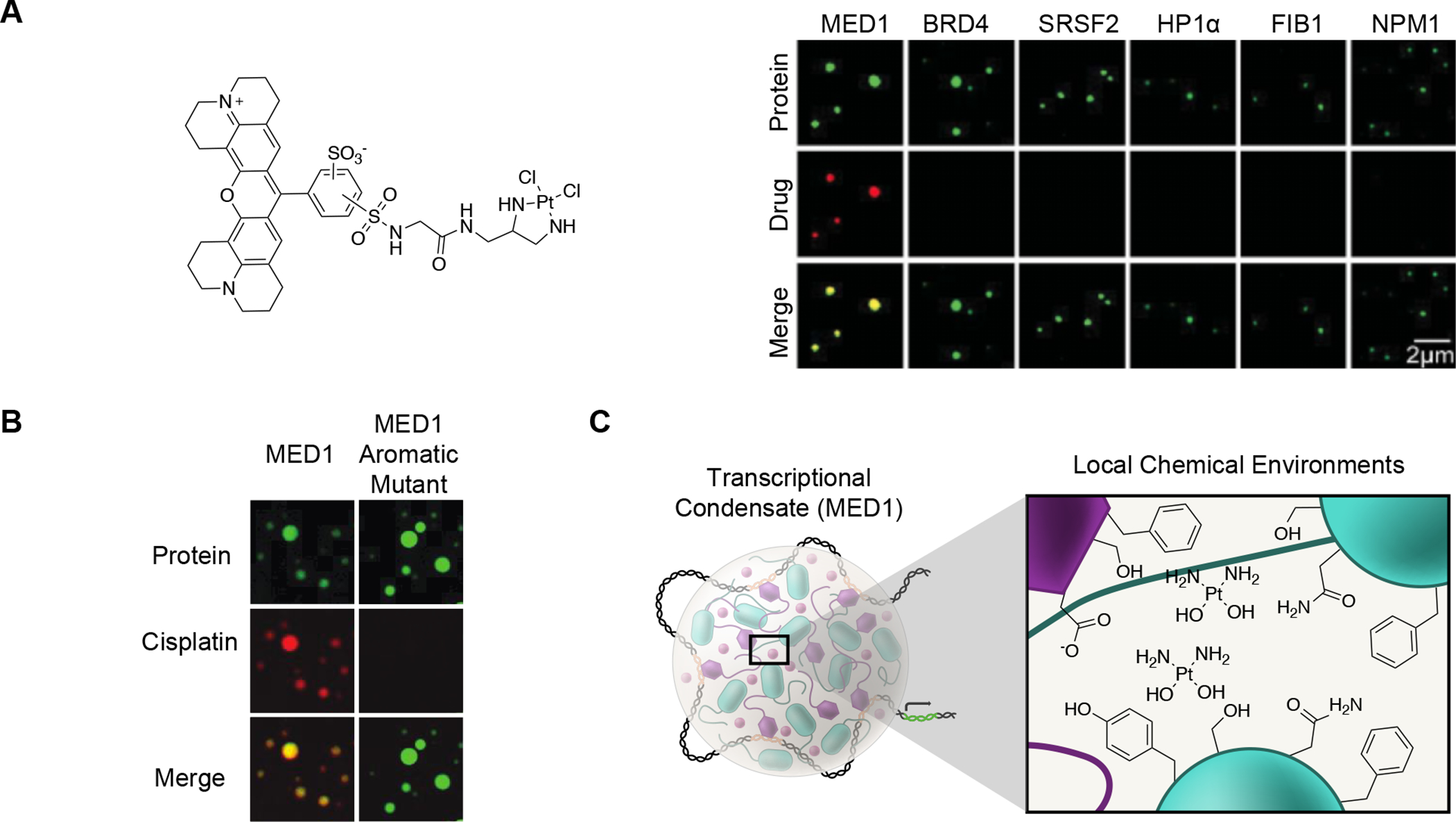
Chemical specificity for small molecules in condensates.
A) Cisplatin-TR selectively concentrates in specific condensates in droplet partitioning assays.36
B) Changes in amino acid composition of a condensate forming protein can abrogate small molecule partitioning behavior without affecting condensate formation. The ability of droplets formed by the MED1 protein of the Mediator complex to concentrate cisplatin is reduced with the replacement of aromatic residues with alanine residues.36.
C) Model depicting how specific local chemical environments within a condensate may influence cisplatin partitioning and concentration within a transcriptional condensate.
Novel models for drug action and resistance
The targets of many commonly used drugs are now known to occur in condensates, so it might be expected that efficacious drugs can readily access these compartments to engage their targets. Nonetheless, an understanding of the interaction of these drugs with the physicochemical environment of diverse condensates—currently lacking for most drugs—may present opportunities for improved therapeutics. As examples of the insights that can emerge from such understanding, we describe below concepts that have emerged from recent studies of drug-condensate interaction that suggest new models to account for therapeutic efficacy and resistance for widely used antineoplastic drugs.
Enhanced pharmacological specificity and activity.
Prior to the observation of selective partitioning in transcriptional condensates, it was widely assumed that the efficacy of cisplatin was due to random platination of the genome, thus selectively affecting tumor cells because they must repair their damaged DNA before continuing to replicate. However, the evidence that cisplatin concentrates in large transcriptional condensates at driver oncogenes, where it selectively platinates oncogene regulatory DNA, suggests a very different model for the drug’s anticancer activities (Figure 5). In this model, the reason that cisplatin has efficacy against a broad spectrum of cancers is due to the fact that each cancer evolves large and stable transcriptional condensates at its driver oncogenes, and that cisplatin concentrates and acts on the DNA in these condensates, thus ultimately destroying the ability of the transcription apparatus to operate specifically at those oncogenes. By contrast, the transcriptional condensates in normal cells are much smaller and have shorter lifetimes, and thus accumulate less of the drug (Figure 5). 72–75 71–74 71–74
Figure 5.
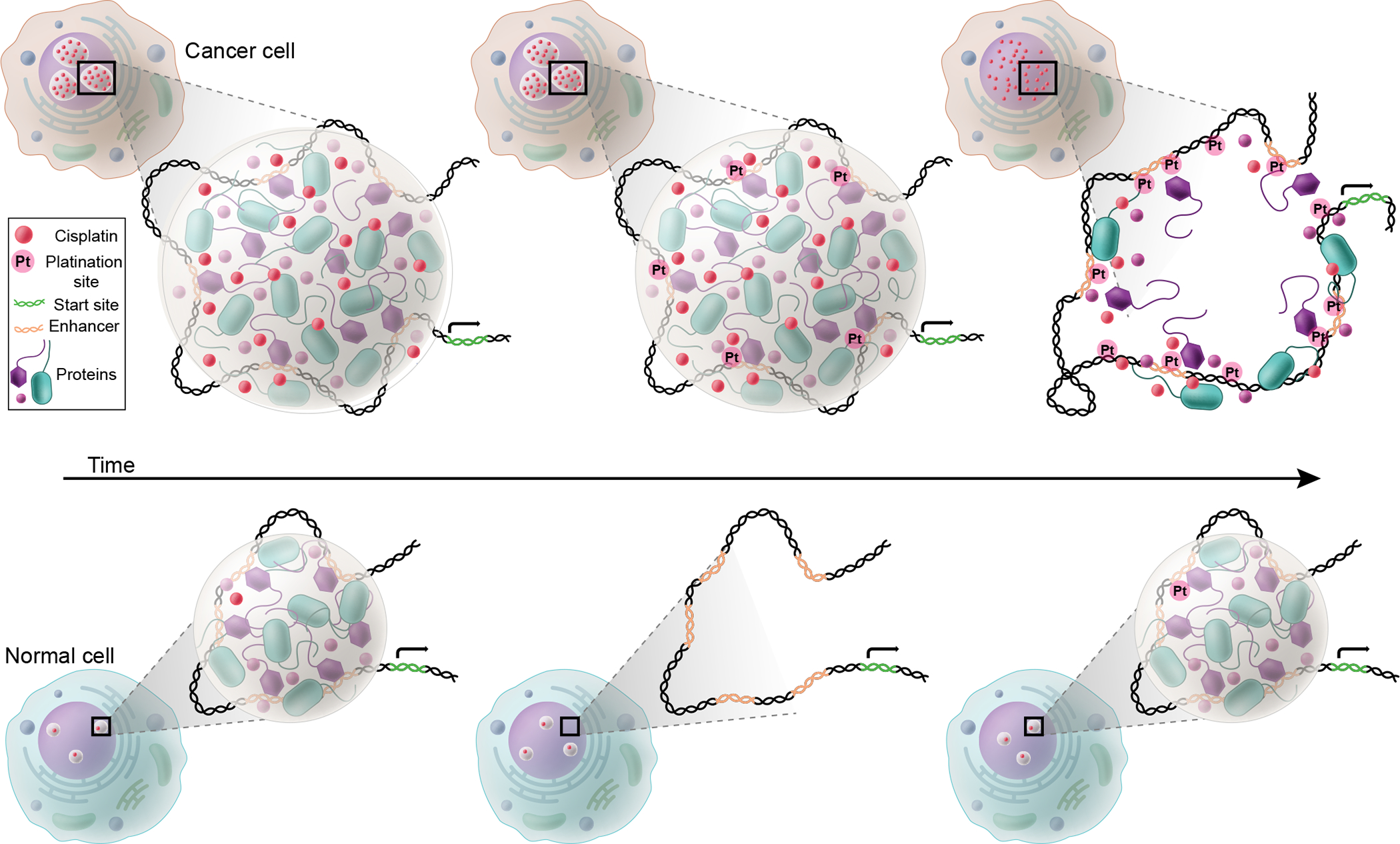
Cisplatin concentrates in large transcriptional condensates at driver oncogenes, where it selectively platinates oncogene regulatory DNA.36 Transcriptional condensates have been shown to occur at loci that contain clusters of enhancer regulatory elements called super-enhancers.34,71 In normal cells, super-enhancers typically span 5–20kb of DNA, but in metastatic tumor cells, driver oncogenes acquire super-enhancers that can span as much as 500 kb.72–75 The larger super-enhancers are associated with larger amounts of assembled transcription apparatus, and thus larger condensates. Larger transcriptional condensates have longer half-lives and produce more transcription from their associated genes.33–34 Thus, the continuous high concentration of cisplatin within the more long-lived oncogenic transcriptional condensates leads to robust DNA-platination at tumor-specific oncogenes, and ultimately this permanently disrupts the condensate at the oncogene, leading to tumor cell death. In contrast, the smaller short-lived condensates at normal genes accumulate far less cisplatin and suffer far less DNA damage.
There are now many examples of small molecule drugs and tool compounds that selectively partition into specific condensates in the absence of their defined target proteins. This includes the drugs cisplatin, mitoxantrone, and tamoxifen, as well as chemical probes that bind the transcriptional cofactors BRD4 and CDK7.36 Prior to these observations, there was a conundrum: BRD4 and CDK7 are present at all active genes and necessary for their transcription, yet BRD4 and CDK7 inhibitors selectively disrupted the tumor-specific oncogenes that engendered oncogenic properties of these diverse cancer cells.73–75 Again, the fact that most cancers evolve large and stable transcriptional condensates at their driver oncogenes, and that these BRD4 and CDK7 inhibitors are selectively concentrated in such condensates, together explain why these inhibitors have oncogene-selective inhibitory activities and that they have far less deleterious effects in normal cells, which have smaller and more transient transcriptional condensates (Figure 5).
Novel mechanisms of drug resistance.
Tamoxifen is an anti-estrogen that is highly effective drug in the treatment of estrogen receptor (ER) -positive breast cancer. Tamoxifen resistance can be conferred by ER mutations that reduce drug affinity, as might be expected, but can also be conferred by MED1 overexpression, which until recently did not have a mechanistic explanation.76–77 We found that ER partitions selectively into MED1-containing transcriptional condensates in a manner that is dependent on its binding to estradiol, but when Tamoxifen is present, the drug partitions selectively into the same transcriptional condensates and competes for ER binding with estradiol, tamoxifen binding leads to eviction of ER from the transcriptional condensate.36 MED1 overexpression was found to cause an expansion of the volume of transcriptional condensates, thereby diluting Tamoxifen in the condensate, and rendering Tamoxifen less efficient in evicting ER from the condensate. These results suggest that misregulation of genes, a hallmark of cancer, can lead to condensate alterations that contribute to drug resistance in cancer cells.
Chemical grammar and condensate compartments
We suggest that learning the chemical grammar of molecules with respect to condensates, which we define as the rules describing the chemical features of molecules that engender attraction to or repulsion by the physicochemical environment of a specific condensate, should enable design of small molecule drugs with three types of condensate-associated properties. It should be possible to endow small molecule drugs with chemical properties that 1) concentrate these molecules to higher levels in condensates where their targets occur and lower levels in condensates where toxic effects might be obtained (Figure 6A), 2) modulate the phase behavior of specific condensates (Figure 6B) and 3) modify the material properties of condensates (Figure 6C, D). Some small molecules may impact more than one of these properties, so these are not necessarily mutually exclusive.
Figure 6.
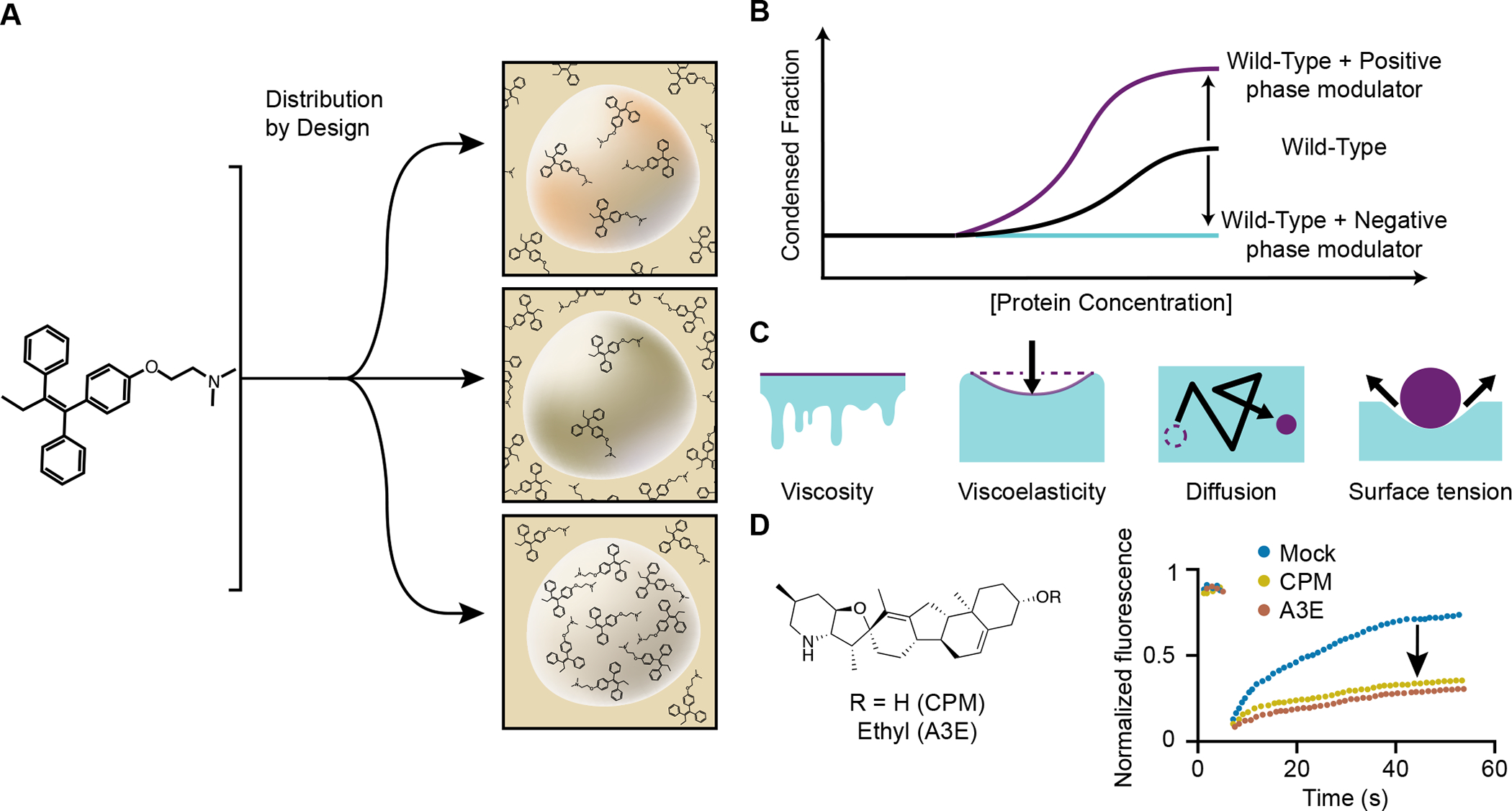
Small molecules and peptides may be designed to have chemical properties that interact with and influence condensates to improve therapeutic efficacy.
A) It should be possible to endow different classes of small molecules and peptides with features that concentrate these molecules in condensates where their targets occur and cause them to avoid partitioning into condensates where toxic effects might be obtained.
B) Alteration of a condensate’s condensed fraction and phase behavior with a positive (increasing) or negative (decreasing) phase modulator.
C) Material properties of condensate may be altered with small molecules; viscosity, viscoelasticity, surface tension, and diffusivity of molecules may be augmented, corrected, or depleted by drugs.
D) Cyclopamine is an example of a small molecule that can induce changes in respiratory syncytial virus condensates by hardening (decreased viscoelasticity).90
Condensate partitioning.
It should be possible to engineer small molecules that not only bind selectively to their target proteins, but also selectively concentrate in the particular condensates where their targets occur, thereby improving their pharmacological efficacy. Where the targets of small molecules involve families of proteins with similar sequences, such as kinases, and where such proteins reside in different condensates, some degree of target specificity might be obtained through selective condensate partitioning. Such an approach would benefit from an optimal balance between ligand binding and condensate interaction.
It is also possible that small molecule partitioning into inappropriate condensates contributes to toxicity. For example, a DNA-modifying drug that concentrates in nucleoli will disrupt the process of ribosome biosynthesis. Mitoxantrone concentrates in nucleoli as well as other condensates, and it is possible that its impact on nucleoli contributes to its toxicity, whereas its impact in other condensates contributes to its efficacy.78
Although we do not yet know of an instance where chemical features that influence partitioning have been purposely incorporated into the design of a small molecule drug, there is evidence supporting the notion that this can be accomplished. Investigators gained insights into the chemical features of small molecules that contribute to selective concentration in MED1 protein condensates by screening a fluorescent probe library of boron-dipyrromethene (BODIPY) dyes diversified with different chemical functional groups.36 Derivatization of the common fluorescent scaffold showed aromatic rings were found to preferentially engender concentration into these condensates, suggesting that π–π or cation–π interactions are among the noncovalent interactions leading to small-molecule partitioning in this compartment. Thus, a small molecule that targets a biomolecule resident in a MED1-dominated condensate might be modified to include an aromatic ring to enhance its ability to be selectively concentrated in this condensate.
Modulating phase behavior.
Condensate dysregulation due to pathogenic mutations in condensate-associated proteins is now thought to contribute to diverse diseases.9–10, 12, 14 In diseases caused by condensate dysregulation, it should be possible to use small molecules to selectively modulate the saturation concentration of specific condensate-forming proteins. In this manner, both noncovalent and covalent small molecule interactions with proteins have the potential to rescue the phase behavior of a condensate that has become dysregulated due to a pathological mutation.
Solutions of condensate-forming biopolymers will “demix”, separating into biopolymer dense and dilute phases, at their saturation concentration. Noncovalent interactions between side chains and molecules in solution can impact condensate size and dynamics by providing interaction partners. 24, 42, 56, 79–82 This concept is directly portable to small molecules that interact directly with a protein to change its saturation concentration (Figure 6B).80–81, 83 Specific chemical features have been shown to engage in the transforming of a condensate’s Csat, and may consist of a mixture of strong/weak ions, hydrophobic regions, and electron rich π-systems.81 These attributes are complicit with strong nonspecific interactions with proteins, driving the formation of transient noncovalent interactions between components of proteins important to condensate formation, mechanisms that are reminiscent of the ‘molecular grammar’ of condensate forming proteins, which are the rules describing how amino acid chemistries influence protein phase separation and condensate material properties.17
Modulating material properties.
Condensates have material properties - viscosity, viscoelasticity, surface tension, and diffusivity of molecules within condensates - that result from the chemical properties of polymers from which they’re composed (Figure 6C), and altered material properties can produce dysregulated condensates in diverse diseases.7, 12, 17, 84 Targeting these different physical properties in therapeutic design is an emergent approach for addressing diseases thought to involve condensate dysregulation.
Condensates can be described as liquids, gels or solids, which are terms that reflect the relative diffusivity of molecules within these assemblies. A hallmark of neurodegenerative diseases, which include amyotrophic lateral sclerosis, frontotemporal dementia, Alzheimer’s disease, and Parkinson’s disease, is the formation of protein or RNA aggregates that appear to be dysregulated condensates. Four well-studied proteins found aggregated in different neurodegenerative diseases —α-synuclein, FUS, tau, and TDP-43—have each been shown to participate in phase separation and the formation of aggregates is postulated to represent a solid or glass-like pathogenic condensate. In this context, small molecules may prove to have advantages over biomolecules as therapeutics for such neurodegenerative diseases, as they retain a greater capacity to penetrate into disease-state condensates with solid or glass-like material properties.
Condensate assemblies are often employed by viruses to compartmentalize essential life cycle functions and recent studies have shown that small molecules can provide efficacy against viral replication by modulating the properties of viral condensates. Viruses can hijack host cell compartments to provide a protected and metabolite rich microenvironment for replication while minimizing stimulation of innate immune responses.40, 85–89 Compartmentalization strategies include hijacking host condensates, the formation of phase separated inclusion bodies of viral components, and reengineering of the endoplasmic reticulum. Improved understanding of the properties of viral condensates could lead to new classes of antiviral drugs. Indeed, modification of the viscoelasticity of respiratory syncytial virus (RSV) condensates by a small molecule has proven possible and can arrest viral replication (Figure 6D). 90 New condensate driven strategies for the development of antiviral drugs may yield much needed new directions in this classically challenging field of medicinal chemistry.
Simple and cellular condensates.
Scientists have a long tradition of studying the complex environment of a cell by using reductionist approaches with purified components in vitro. The simple systems produced in this fashion are more amenable to theoretical and experimental analysis than the more complex systems in living cells, and most of our understanding of the fundamental behaviors of biological molecules has emerged from this approach. Thus, the study of simple homotypic protein condensates will continue to reveal fundamental insights such as the chemical features of small molecules that engender attraction or repulsion to condensate microenvironments and the ability of small molecules to modulate condensate phase behavior and material properties.
Once the fundamentals are established in simple systems, the question of relevance to the more complex living system arises. In this context, the simple condensates produced with a single type of protein are unlikely to have the same internal chemical environment that occurs in a cellular condensate where that protein functions with diverse other molecules in a nonequilibrium environment. This leads us to ask to what extent are insights gained from studies of simple condensates in vitro predictive of biochemical behaviors of more complex condensates in living cells? The answer is that we don’t know, but based on a limited amount of data with biomolecules and small molecules in studies of transcriptional condensates, observations with simple systems can be predictive of behaviors in vivo.27, 34, 36, 71 For example, the selective cisplatin and tamoxifen concentrating behaviors of simple MED1 protein condensates extend to condensates formed by the 30 subunit Mediator complex in vitro and transcriptional condensates in vivo.25
How is it possible that observations with simple condensate systems can be predictive of behaviors in the much more complex microenvironments of condensates in vivo? Certain proteins in cellular condensates have been proposed to play dominant roles by acting as “scaffolds” for other “client” proteins.15, 55 Proteins that have been proposed to act as scaffolds include MED1 in transcriptional activation, FIB1 in nucleolar ribosome biosynthesis, HP1a in heterochromatic gene silencing, and SRSF2 in RNA splicing. Despite being assemblies of many different biomolecules, it is possible that internal chemical microenvironment of some condensates is dominated by the chemical features of their scaffolds, and if so, this could account for the ability of some small molecules to concentrate selectively in both simple condensates containing a scaffold and in the more complex condensate with that scaffold in cells. We suspect, however, that the diverse population of molecules in any one type of cellular condensates creates a chemical microenvironment that is not well replicated in homotypic in vitro condensates.
We imagine that small molecules are not distributed such that they concentrate primarily in a single type of favored condensates, but rather are distributed such that they concentrate to different levels in diverse cellular condensates. Furthermore, the physicochemistry of a cellular condensate can be modulated dynamically by diverse clients such as proteins, nucleic acids, metabolites and ions, and such modulation is likely to alter small molecule partitioning. Thus, the chemical grammar of the cellular condensates will be more challenging to discern that that of simple in vitro condensates.
Perspective
We envision a time when molecules can be engineered to selectively enrich in any one type of compartment where a target is contributing to a disease phenotype, thus producing therapeutic molecules with improved efficacy and reduced toxicity. To reach that point and have optimal impact, several important advances are needed. The diverse types of condensate compartments that exist in cells will need to be further catalogued, their components described, and their physicochemical properties deduced. The rules describing the chemical features of molecules that engender attraction to or repulsion by the physicochemical environment of a specific condensate, which we call chemical grammar, will need to be learned for these diverse condensates. Where disease mutations cause pathological dysfunction by altering the material properties of condensates, it will be useful to obtain a deeper understanding of the means by which small molecules can modify the viscosity, viscoelasticity, surface tension, and diffusivity of condensates.
Conceptual and experimental innovation has led to a revolution in our understanding of the compartmental features of cells in the past decade. The conceptual innovations have come from introducing concepts from polymer chemistry and soft matter physics into regulatory biology. We suggest that further conceptual advances will come from treating cells as highly dynamic non-equilibrium environments. Similarly, there have been experimental innovations employing engineered cells and molecules. We suggest that learning condensate chemical grammar will be enhanced and accelerated by combining experimental strategies of chemical biology with modern computational approaches, such as deep learning. This should lead to new insights into the mechanics of how molecules are compartmentalized in and affect the materials properties of condensates, and ultimately enable the development of more potent therapeutics.
Acknowledgements:
We thank Alessandra Dall’agnese for providing the images presented in Figure 3A and Ann Boija and Kalon Overholt for helpful discussions.
Funding:
H.R.K is support by a fellowship from the Damon Runyon Cancer Research Foundation (Grant number: 2458-22). RAY is supported by NIH grant R01 GM123511, NCI grant CA155258, NSF grant PHY2044895.
Footnotes
Competing Interests:
R.A.Y is a founder and shareholder of Syros Pharmaceuticals, Camp4 Therapeutics, Omega Therapeutics, and Dewpoint Therapeutics. H.R.K is a consultant of Dewpoint Therapeutics.
Peer review information:
Nature Chemical Biology thanks Tingting Li and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
References
- 1. Wagner R, Einige Bemerkungen und Fragen über das Keimbläschen (vesicular germinativa). Müller’s Archiv Anat Physiol Wissenschaft Med 1835, 373–377. Wagner was apparently the first to describe the nucleolus, the largest and best characterized condensate.
- 2.Valentin G, Repertorium für Anatomie und Physiologie. Verlag von Veit und Comp.: Berlin, 1836; Vol. 1. [Google Scholar]
- 3.Valentin G, Repertorium für Anatomie und Physiologie. Verlag von Veit und Comp.: Berlin, 1839; Vol. 4. [Google Scholar]
- 4.Cajal S. R. y., Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab. Lab. Invest. Biol. (Madrid) 1903, 2, 129–221. [Google Scholar]
- 5.Vincent WS, The isolation and chemical properties of the nucleoli of starfish oocytes. Proc. Natl. Acad. Sci. USA 1952, 38, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brangwynne CP; et al. , Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324 (5935), 1729–1732. This pioneering study noted that the behavior of a cellular body was not that of a standard membrane-bound organelle, but rather more like liquid droplets that arise by phase separation.
- 7.Alberti S; Gladfelter A; Mittag T, Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 2019, 176 (3), 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Case LB; Ditlev JA; Rosen MK, Regulation of transmembrane signaling by phase separation. Annu. Rev. Biophys 2019, 48 (1), 465–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alberti S; Dormann D, Liquid–liquid phase separation in disease. Annu. Rev. Genetics 2019, 53 (1), 171–194. [DOI] [PubMed] [Google Scholar]
- 10.Boija A; Klein IA; Young RA, Biomolecular condensates and cancer. Cancer Cell 2021, 39 (2), 174–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabari BR; Dall’Agnese A; Young RA, Biomolecular condensates in the nucleus. Trends in Biochem. Sci 2020, 45 (11), 961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin Y; Brangwynne CP, Liquid phase condensation in cell physiology and disease. Science 2017, 357 (6357), eaaf4382. [DOI] [PubMed] [Google Scholar]
- 13.Chen X; Wu X; Wu H; Zhang M, Phase separation at the synapse. Nat. Neurosci 2020, 23 (3), 301–310. [DOI] [PubMed] [Google Scholar]
- 14.Nedelsky NB; Taylor JP, Bridging biophysics and neurology: aberrant phase transitions in neurodegenerative disease. Nat. Rev. Neurol 2019, 15 (5), 272–286. [DOI] [PubMed] [Google Scholar]
- 15.Banani SF; Lee HO; Hyman AA; Rosen MK, Biomolecular condensates: organizers of cellular biochemistry. Nat Rev. Mol. Cell Biol 2017, 18 (5), 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pak CW; et al. , Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell 2016, 63 (1), 72–85. This study provides a powerful example of combining theory and experimentation to enhance understanding of the roles of amino acid patterns in protein phase separation.
- 17. Wang J; et al. , A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins Cell 2018, 174 (3), 688–699.e16. The first study to explore the roles of diverse amino acids in promoting phase separation and influencing the material properties of condensates formed by a class of proteins.
- 18.Martin EW; et al. , Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 2020, 367 (6478), 694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banani SF; et al. , Composition control of phase-separated bodies. Cell 2016, 166 (3), 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abyzov A; Blackledge M; Zweckstetter M, Conformational dynamics of intrinsically disordered proteins regulate biomolecular condensate chemistry. Chem. Rev 2022, 122 (6), 6719–6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greig JA; et al. , Arginine-enriched mixed-charge domains provide cohesion for nuclear speckle condensation. Mol. Cell 2020, 77 (6), 1237–1250.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Case LB; Zhang X; Ditlev JA; Rosen MK, Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 2019, 363 (6431), 1093–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shrinivas K; et al. , Enhancer features that drive foramtion of transcriptional condensates. Mol. Cell 2019, 75 (3), 549–561.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi J-M; Holehouse AS; Pappu RV, Physical principles underlying the complex biology of intracellular phase transitions Annu. Rev. Biophys 2020, 49 (1), 107–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McSwiggen DT; Mir M; Darzacq X; Tjian R, Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev. 2019, 33 (23–24), 1619–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng A; Weber SC, Evidence for and against liquid-liquid phase separation in the nucleus. Non-Coding RNA 2019, 5 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boija A; et al. , Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 2018, 175 (7), 1842–1855.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo YE; et al. , Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 2019, 572 (7770), 543–548. This work establishes that posttranslational modifications of the transcription apparatus can regulate its partitioning into different condensates involved in RNA synthesis and splicing.
- 29.Henninger JE; et al. , RNA-Mediated feedback control of transcriptional condensates. Cell 2021, 184 (1), 207–225.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu S; et al. , The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat. Commun 2021, 12 (1), 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu X; et al. , The STING phase-separator suppresses innate immune signalling. Nat. Cell Biol 2021, 23 (4), 330–340. [DOI] [PubMed] [Google Scholar]
- 32.Su X; et al. , Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 2016, 352 (6285), 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dao TP; Yang Y; Cosgrov MS; Hopkins JB; Ma W; Castañeda CA, Mechanistic insights into the enhancement or inhibition of phase separation by polyubiquitin chains of different lengths or linkages. BioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho W-K; et al. , Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018, 361 (6400), 412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho W-K; et al. , RNA Polymerase II cluster dynamics predict mRNA output in living cells. eLife 2016, 5, e13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klein IA; et al. , Partitioning of cancer therapeutics in nuclear condensates. Science 2020, 368 (6497), 1386. This study showed that selective partitioning and concentration of small molecules within condensates contributes to drug pharmacodynamics, suggesting that further understanding of condensate chemical grammar may facilitate advances in disease therapy.
- 37.Schuster BS; et al. , Identifying sequence perturbations to an intrinsically disordered protein that determine its phase-separation behavior. Proc. Natl. Acad. Sci. USA 2020, 117 (21), 11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alshareedah I; Moosa MM; Pham M; Potoyan DA; Banerjee PR, Programmable viscoelasticity in protein-RNA condensates with disordered sticker-spacer polypeptides. Nat. Commun 2021, 12 (1), 6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roden C; Gladfelter AS, RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol 2021, 22 (3), 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savastano A; Ibáñez de Opakua A; Rankovic M; Zweckstetter M, Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun 2020, 11 (1), 6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guillén-Boixet J; et al. , RNA-Iinduced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 2020, 181 (2), 346–361.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanders DW; et al. , Competing protein-RNA interaction networks control multiphase intracellular organization. Cell 2020, 181 (2), 306–324.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi J-M; Holehouse AS; Pappu RV, Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys 2020, 49 (1), 107–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boeynaems S; et al. , Spontaneous driving forces give rise to protein–RNA condensates with coexisting phases and complex material properties. Proc. Natl. Acad. Sci. USA 2019, 116 (16), 7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holehouse AS; Ginell GM; Griffith D; Böke E, Clustering of aromatic residues in prion-like domains can tune the formation, state, and organization of biomolecular condensates. Biochemistry 2021, 60 (47), 3566–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kar M; et al. , Glycine-rich peptides from FUS have an intrinsic ability to self-assemble into fibers and networked fibrils. Biochemistry 2021, 60 (43), 3213–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rubinstein M; Semenov AN, Thermoreversible gelation in solutions of associating polymers. 2. linear dynamics. Macromolecules 1998, 31 (4), 1386–1397. [Google Scholar]
- 48.Tanaka F; Ishida M, Microphase formation in mixtures of associating polymers. Macromolecules 1997, 30 (6), 1836–1844. [Google Scholar]
- 49.Leibler L, Theory of microphase separation in block copolymers. Macromolecules 1980, 13 (6), 1602–1617. [Google Scholar]
- 50.Murthy AC; et al. , Molecular interactions underlying liquid–liquid phase separation of the FUS low-complexity domain. Nat. Struct. Mol. Biol 2019, 26 (7), 637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qamar S; et al. , FUS phase separation Is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell 2018, 173 (3), 720–734.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim TH; Payliss BJ; Nosella ML; Lee ITW; Toyama Y; Forman-Kay JD; Kay LE, Interaction hot spots for phase separation revealed by NMR studies of a CAPRIN1 condensed phase. Proceedings of the National Academy of Sciences 2021, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim TH; Tsang B; Vernon RM; Sonenberg N; Kay EL; Forman-Kay JD, Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 2019, 365 (6455), 825–829. [DOI] [PubMed] [Google Scholar]
- 54.Wyman J; Gill SJ, Ligand-linked phase changes in a biological system: applications to sickle cell hemoglobin. Proc. Natl. Acad. Sci. USA 1980, 77 (9), 5239–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruff KM; Dar F; Pappu RV, Polyphasic linkage and the impact of ligand binding on the regulation of biomolecular condensates. Biophys. Rev 2021, 2 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruff KM; Dar F; Pappu RV, Ligand effects on phase separation of multivalent macromolecules. Proc. Natl. Acad. Sci. USA 2021, 118 (10), e2017184118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burslem GM; Crews CM, Proteolysis-targeting chimeras as therapeutics and tools for biological discovery. Cell 2020, 181 (1), 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gerry CJ; Schreiber SL, Unifying principles of bifunctional, proximity-inducing small molecules. Nat. Chem. Biol 2020, 16 (4), 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forman-Kay JD; Ditlev JA; Nosella ML; Lee HO, What are the distinguishing features and size requirements of biomolecular condensates and their implications for RNA-containing condensates? RNA 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andersen JS; et al. , Nucleolar proteome dynamics. Nature 2005, 433 (7021), 77–83. [DOI] [PubMed] [Google Scholar]
- 61.Su X; et al. , Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 2016, 352 (6285), 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jain S; Wheeler JR; Walters RW; Agrawal A; Barsic A; Parker R, ATPase-Modulated stress granules contain a diverse proteome and substructure. Cell 2016, 164 (3), 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langdon EM; Gladfelter AS, A new Lens for RNA localization: liquid-liquid phaseseparation. Annu. Rev. Microbiol 2018, 72 (1), 255–271. [DOI] [PubMed] [Google Scholar]
- 64.Woodruff JB; Hyman AA; Boke E, Organization and function of non-dynamic biomolecular condensates. Trends in Biochem. Sci 2018, 43 (2), 81–94. [DOI] [PubMed] [Google Scholar]
- 65.Lafontaine DLJ; Riback JA; Bascetin R; Brangwynne CP, The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol 2020. [DOI] [PubMed] [Google Scholar]
- 66.Boehning M; et al. , RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol 2018, 25 (9), 833–840. [DOI] [PubMed] [Google Scholar]
- 67.Nojima T; et al. , RNA polymerase II phosphorylated on CTD serine 5 interacts with the spliceosome during co-transcriptional Splicing. Mol. Cell 2018, 72 (2), 369–379.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hnisz D; Shrinivas K; Young RA; Chakraborty AK; Sharp PA, A phase separation model for transcriptional control. Cell 2017, 169 (1), 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vibet S; et al. , Differential subcellular distribution of mitoxantrone in relation to chemosensitization in two human breast cancer cell lines. Drug. Metab. Disp 2007, 35 (5), 822. [DOI] [PubMed] [Google Scholar]
- 70.Smith PJ; Sykes HR; Fox ME; Furlong IJ, Subcellular distribution of the anticancer drug mitoxantrone in human and drug-resistant murine cells analyzed by flow cytometry and confocal microscopy and Its relationship to the induction of DNA damage. Cancer Res. 1992, 52 (14), 4000. [PubMed] [Google Scholar]
- 71.Sabari BR; et al. , Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361 (6400), eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bradner JE; Hnisz D; Young RA, Transcriptional addiction in cancer. Cell 2017, 168 (4), 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Filippakopoulos P; et al. , Selective inhibition of BET bromodomains. Nature 2010, 468 (7327), 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Christensen CL; et al. , Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell 2014, 26 (6), 909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kwiatkowski N; et al. , Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014, 511 (7511), 616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fanning SW; et al. , Estrogen receptor alpha somatic mutations Y537S and D538G confer breast cancer endocrine resistance by stabilizing the activating function-2 binding conformation. eLife 2016, 5, e12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nagalingam A; Tighiouart M; Ryden L; et al. , Med1 plays a critical role in the development of tamoxifen resistance. Carcinogenesis 2012, 33 (4), 918–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pommier Y; Leo E; Zhang H; Marchand C, DNA Topoisomerases and Their Poisoning by Anticancer and Antibacterial Drugs. Chem. Biol 2010, 17 (5), 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bielskutė S; Garacia-Cabau C; Frigolé-Vivas M; et al. , Low amounts of heavy water increase the phase separation propoensity of a fragment of the androgen receptor activation domain. Protein Sci. 2021, 30 (7 ), 1427–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Petronilho EC; Pedrote MM; Marques MA; et al. , Phase separation of p53 precedes aggregation and is affected by oncogenic mutations and ligands. Chem. Sci 2021, 12 (21), 7334–7349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Babinchak MW; et al. , Small molecules as potent biphasic modulators of protein liquid-liquid phase separation. Nat. Commun 2020, 11 (1), 5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krainer G; et al. , Reentrant liquid condensate phase of proteins is stabilized by hydrophobic and non-ionic interactions. Nat. Commun 2021, 12 (1), 1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fang MY; et al. , Small-molecule modulation of TDP-43 recruitment to stress granules prevents persistent TDP-43 accumulation in ALS/FTD. Neuron 2019, 103 (5), 802–819.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alshareedah I; Kaur T; Banerjee PR, Chapter Six - Methods for characterizing the material properties of biomolecular condensates. In Methods in Enzymology, Keating CD, Ed. Academic Press: 2021; Vol. 646, pp 143–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang S; et al. , Targeting liquid–liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nat. Cell. Biol 2021, 23 (7), 718–732. [DOI] [PubMed] [Google Scholar]
- 86.Hadjadj J; et al. , Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369 (6504), 718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Y; et al. , SARS-CoV-2 induces double-stranded RNA-mediated innate immune responses in respiratory epithelial-derived cells and cardiomyocytes. Proc. Natl. Acad. Sci. USA 2021, 118 (16), e2022643118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu J; et al. , SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 2021, 34 (7), 108761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zotta A; Hooftman A; O’Neill LAJ, SARS-CoV-2 targets MAVS for immune evasion. Nat. Cell Biol 2021, 23 (7), 682–683. [DOI] [PubMed] [Google Scholar]
- 90.Risso-Ballester J; et al. , A condensate-hardening drug blocks RSV replication in vivo. Nature 2021, 595 (7868), 596–599. [DOI] [PubMed] [Google Scholar]


