Abstract
Salmonella enterica serotype Enteritidis (S. enteritidis) is a major food-borne pathogen, and its incidence among all Salmonella serotypes has increased dramatically in the last two decades. To study the virulence characteristics of clinical isolates of S. enteritidis, we determined the 50% lethal doses (LD50) in mice of isolates of two major phage types (4 and 8). Isolates of both phage types showed a wide range of LD50 after oral inoculation, varying from under 102 organisms to over 108 organisms. No significant difference in LD50 was observed between the phage types. These observations indicated that clinical isolates of S. enteritidis are highly heterogeneous in their ability to cause death in mice. We compared the LD50s of these isolates to the results observed from in vitro pathogenicity assays. We also analyzed these isolates for recognized Salmonella virulence loci (spv, sodCI, sopE, and sef). The in vitro phenotypes of the isolates showed no obvious correlation with their LD50 in any given assay, and the virulence genes tested were present in all isolates. However, the isolate with the lowest LD50 (isolate 97A 2472) was resistant to acidified sodium nitrite (ASN). Moreover, the most acid-susceptible, macrophage-susceptible, and ASN-susceptible isolates were attenuated for virulence in mice. These results, based on extensive analysis of clinical isolates of S. enteritidis, demonstrate the complex nature of Salmonella pathogenesis in mice. Our results also indicate the limitation of in vitro assays in predicting in vivo virulence.
Salmonella is one of the leading causes of food-borne illnesses worldwide (5). It is estimated that 800,000 to 4,000,000 human cases of salmonellosis occur each year in the United States and that about 1,000 people die of the disease each year (2). In recent years, Salmonella enterica serotype Enteritidis (S. enteritidis) surpassed S. enterica serotype Typhimurium (S. typhimurium) as the most common serotype reported in the United States (2). While most of the Salmonella pathogenesis studies to date have focused on S. typhimurium, the pathogenesis of S. enteritidis is poorly understood.
S. enteritidis can be divided into at least 27 subtypes by a phage-typing method described by Ward et al. (42). Among them, phage type 4 and phage type 8 are the most common subtypes of S. enteritidis reported in the United Kingdom and the United States, respectively (1, 3, 29, 42). The reasons for the disproportionate representation of phage types 4 and 8 in reported cases might include differences in their reservoirs, in the distribution of food products contaminated with them, or in their virulence characteristics. If infection with isolates of these phage types leads to more cases of clinically overt diseases, they are more likely to be reported.
In this work, we studied the virulence characteristics of clinical isolates of phage type 4 and 8 S. enteritidis. We first determined the 50% lethal doses (LD50) of these isolates in mice and then compared the doses to the results of in vitro assays to further characterize their virulence phenotypes. These analyses of the virulence characteristics of clinical isolates of S. enteritidis should provide insight into the organism's biological properties associated with the mechanisms of its pathogenesis that may be missed by studying laboratory strains.
MATERIALS AND METHODS
Bacterial isolates and culture.
The phage type 4 S. enteritidis isolates, 97A 2472, 97A 6782, 96A 8464, 96A 8743, and 96A 10871, were generously provided by the Department of Health Services, State of California. They were isolated from human gastroenteritis outbreaks that occurred in California during 1996 and 1997. The phage type 8 isolates, H4052, H4081, H4191, H4241, and H4386, were obtained from the Centers for Disease Control and Prevention. They were isolated from human gastroenteritis outbreaks that occurred across the United States. The S. enteritidis isolates used in this study will be referred to by the last four digits (or the last five digits in the case of 96A 10871) of their identification numbers. All of the S. enteritidis isolates were propagated in Luria-Bertani (LB) medium.
Purification of plasmid DNA.
The plasmid DNAs of the S. enteritidis isolates were purified by the alkaline lysis method (35). Purified plasmid DNAs were visualized by electrophoresis on agarose gels, and their molecular weights were determined by comparison to those plasmids in Escherichia coli V517 and 39R861 (27, 40).
Determination of LD50 in mice.
S. enteritidis was cultured overnight in LB medium at 37°C with shaking. Tenfold dilutions were prepared in phosphate-buffered saline (PBS) (pH 7.4) and used to infect 6- to 8-week-old female BALB/c mice. The dilutions were also plated on Hektoen enteric agar plates (Difco, Detroit, Mich.) for quantification and to confirm that the culture represented Salmonella. Mice were infected with 0.25 ml of diluted bacteria intragastrically via a feeding needle (43). Initially, three concentrations of approximately 5 × 105, 1 × 106, and 1 × 107 bacteria/ml were used to infect groups of two mice to obtain an estimate of the LD50. The infected mice were observed daily, and their mortality over the following 2 weeks was recorded. If the mortality of all of the groups was over or under 50% in the pilot experiment, additional pilot experiments were carried out with increased or reduced inoculum sizes. Once the inoculum that caused approximately 50% mortality (LD50est) was determined, three inocula at 0.1×, 1×, and 10× the LD50est were used to infect groups of six mice. When the LD50est of an isolate was less than 103 organisms, five concentrations of bacteria from approximately 5 × 101 to 5 × 105 organisms/ml were used. The mortality in each group was recorded over a 2-week period, and the LD50 was calculated by the method of Reed and Muench (32). As controls, mice given only PBS were used.
Invasion assay with HeLa cells.
HeLa cells were plated in 24-well plates at 6 × 104 cells/well and incubated at 37°C in 5% CO2 overnight. S. enteritidis was inoculated into 2 ml of LB medium and incubated at 37°C overnight without shaking. Five microliters of overnight culture was diluted in 5 ml of Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS) (Gibco BRL, Gaithersburg, Md.). A sample was taken from the diluted bacteria and plated on LB plates to determine the number of input bacteria. The medium (0.5 ml) containing Salmonella was added to each well at a multiplicity of infection of approximately 5 to 10. The plates were then centrifuged at 1,000 rpm (Sorvall RT7) for 5 min and incubated at 37°C in 5% CO2 for 1 or 2 h. At each time point, the cells were washed five times with PBS, and 1 ml of fresh medium containing 50 μg of gentamicin per ml was added. This concentration of gentamicin was used because it was determined in a separate experiment to be sufficient to kill the S. enteritidis isolates used in this study. After incubation at 37°C in 5% CO2 for an additional 1.5 h, the cells were washed three times with PBS and lysed in 0.5 ml of PBS with 0.1% Triton X-100. The lysates were pipetted vigorously to ensure the release of intracellular bacteria. The lysates were then diluted, plated on LB plates, and incubated overnight at 37°C. The CFU on the plates were counted and compared to those of the input bacteria (25).
Survival of S. enteritidis under acidic conditions.
S. enteritidis isolates were inoculated into 2 ml of LB medium and cultured overnight at 37°C with shaking. The cultures were spun down and resuspended in 2 ml of fresh LB medium at pH 4.0. Samples were taken at 0, 1, and 2 h and plated onto LB agar plates after appropriate dilutions. The ratio of survival was determined by comparison of the bacterial CFU recovered at 1 or 2 h to that obtained at 0 h (16).
Resistance of S. enteritidis isolates to ROI and RNI.
To determine the resistance of S. enteritidis isolates to reactive oxygen intermediates (ROI), the isolates were inoculated into 2 ml of LB medium and cultured overnight at 37°C with shaking. The overnight culture was plated at 106 organisms in 100 μl onto M9 minimal plates (35). The resistance of the bacteria to hydrogen peroxide and paraquat was assayed as described by De Groote et al. (9). Briefly, paper discs of 1/4-in. diameter were loaded with 30 μl of 3% hydrogen peroxide or 1.9% paraquat and placed in the center of plates onto the bacterial lawn. The plates were incubated overnight at 37°C, and the diameter of the inhibitory zone was measured.
To determine the resistance of S. enteritidis isolates to reactive nitrogen intermediates (RNI), we added 20 μl of an overnight culture to 2 ml of fresh LB medium (pH 5) containing 20 mM sodium nitrite. The bacteria were cultured at 37°C with shaking, and samples were removed at 0, 3, and 6 h. The samples were diluted, plated on LB agar plates, and incubated overnight at 37°C. Colonies were counted the next day, and the survival rate of Salmonella at 3 or 6 h was calculated by comparing the bacterial CFU at 3 or 6 h to that recovered at 0 h (12).
Survival of S. enteritidis isolates in activated peritoneal macrophages.
Survival of S. enteritidis isolates in activated mouse peritoneal macrophages was assayed as described previously (12). Female BALB/c mice 6 to 8 weeks old were injected intraperitoneally with 1 ml of freshly prepared 5 mM sodium periodate 4 days before harvest of peritoneal macrophages. We harvested the macrophages by flushing the peritoneal cavity with Hanks' balanced salt solution (Gibco BRL) supplemented with 2% FBS. The cells were then centrifuged at 1,000 rpm (Sorvall RT7) for 5 min and resuspended in RPMI 1640 (Gibco BRL) supplemented with 10% FBS, 2 mM glutamine, and 5 μM 2-mercaptoethanol. Approximately 3 × 105 cells were added to each well in a 96-well plate, and the plate was incubated for 30 min at 37°C in 5% CO2 to allow macrophages to adhere. The plates were then washed once with Hanks' balanced salt solution plus 2% FBS, and 0.1 ml of fresh medium containing 10 μg of gentamicin per ml was added. After overnight incubation, the cells were washed three times with PBS, and 0.1 ml of fresh medium containing 50 U of gamma interferon per ml was added. The cells were then incubated for 24 h before being used for the assay.
S. enteritidis isolates were inoculated into 2 ml of LB medium and incubated overnight at 37°C with shaking. Twenty microliters of overnight culture was diluted in 2 ml of fresh LB medium and cultured for 2 h at 37°C with shaking to obtain a log-phase culture. Subsequently, bacteria were opsonized with mouse serum by mixing 10 μl of log-phase culture, 80 μl of RPMI 1640 medium, and 10 μl of mouse serum and incubated at 37°C for 30 min. The peritoneal macrophages were washed twice with PBS, and 100 μl of fresh medium was added to each well. Five microliters of opsonized bacteria was added to each well, and each sample was tested in triplicate. The cells were centrifuged at 1,000 rpm (Sorvall RT7) for 5 min and incubated at 37°C in 5% CO2. After 30 min of incubation, the cells were washed three times with PBS, and 150 μl of fresh medium with 50 μg of gentamicin per ml was added to each well. At 0.5, 1.5, or 3 h after the addition of gentamicin-containing medium, the infected cells were washed three times with PBS and lysed in PBS with 0.1% Triton X-100 for 10 min. The cells were pipetted vigorously to release the intracellular bacteria, and the lysates were diluted and plated onto LB agar plates. After overnight incubation, the number of CFU was determined. The time point after 0.5 h of incubation in gentamicin was considered to have 0 h of intracellular exposure, while those after 1.5 and 3 h were considered to have 1 and 2.5 h of exposure, respectively.
Survival at the stationary growth phase.
S. enteritidis isolates were inoculated into 2 ml of minimal medium prepared with M9 minimal salts and supplemented with 0.1 mM CaCl2, 2 mM MgSO4, 0.05% NaCl, 4 mg of glucose per ml, 5 μg of thiamine per ml, and 0.1% Casamino Acids (35). The cultures were incubated at 37°C with shaking, and samples were removed after 1- and 6-day incubation periods. The cultures were plated onto LB agar plates, and the recovered CFU were enumerated. The survival rate after 6 days of incubation was determined by comparing the concentration of culture at day 6 to that at day 1 (16).
Southern hybridization.
The presence of specific Salmonella virulence loci was examined by Southern hybridization essentially as previously described (14, 35). Briefly, genomic DNAs from each S. enteritidis isolate and from S. typhimurium ATCC 14028s (17) and Escherichia coli W3110 (20) controls were isolated, digested with EcoRV (New England Biolabs, Beverly, Mass.), and electrophoresed through 0.8% agarose prior to transfer to a Nytran membrane (Schleicher and Schuell, Keene, N.H.) and immobilization by UV cross-linking. DNA probes, as described below, were labeled with digoxigenin by using a DIG DNA labeling and detection kit (Boehringer Mannheim, Indianapolis, Ind.) and hybridized at 42°C overnight. The membrane was washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.1% sodium dodecyl sulfate at room temperature and twice with 0.2× SSC with 0.1% sodium dodecyl sulfate at 55°C prior to immunological detection of the digoxigenin and detection of the chemiluminescent signal by using X-ray film.
A 2.4-kb SalI-EcoRI fragment of the plasmid pFF18 (15) was used as a probe for the detection of the spv Salmonella plasmid virulence genes. Oligonucleotide primers 5′CTTGCAAACATATACCTGC3′ and 5′GACTATCTGAATGCTTAC3′ were used to PCR amplify a 912-bp probe for the sodCI gene (8), 5′TCAGGGAGTGTTTTGTATATATTTA3′ and 5′GTGACAAAAATAACTTTATCTCCCC3′ were used to PCR amplify a 722-bp probe for the sopE gene (19), and 5′ATGCGTAAATCAGCATCTGCAGTAG3′ and 5′TTAGTTTTGATACTGCTGAACGTAG3′ were used to PCR amplify a 498-bp probe for the sef fimbrial locus (41).
RESULTS
LD50 of the clinical isolates of S. enteritidis.
To compare the pathogenicities of the clinical S. enteritidis isolates in mice, we determined the LD50 of the 10 isolates after intragastric challenge in BALB/c mice. Five isolates were of phage type 4, and the other five were of phage type 8. Isolates of both phage types exhibited a wide range of LD50 (Table 1). The LD50 ranged from 1.6 × 101 to 3 × 105 organisms for the phage type 4 isolates and from 2.3 × 101 to over 1 × 108 organisms for the phage type 8 isolates. Therefore, there was no apparent correlation between virulence in mice and phage type.
TABLE 1.
LD50 of S. enteritidis isolates in mice after oral infection
| Strain | Phage type | LD50 (no. of organisms) | Virulence rank |
|---|---|---|---|
| 2472 | 4 | 16 | 1 |
| 6782 | 4 | 128 | 4 |
| 8464 | 4 | 7.0 × 104 | 6 |
| 8743 | 4 | 1.9 × 105 | 7 |
| 10871 | 4 | 2.1 × 105 | 8 |
| 4052 | 8 | >1.0 × 108 | 10 |
| 4081 | 8 | 2.1 × 104 | 5 |
| 4191 | 8 | 23 | 2 |
| 4241 | 8 | 4.2 × 105 | 9 |
| 4386 | 8 | 80 | 3 |
Mice infected with isolates of either phage type 4 or 8 showed a similar progression of infection. Among mice infected with isolates with low LD50 (<103 organisms), such as 2472, 6782, and 4191, mortality occurred rapidly. Over 50% of the mortality occurred within 48 h postinfection. For those mice that survived the first 48 h, a second peak of mortality was observed at around day 7 after infection. For the mice infected with isolates with higher LD50, such as 8743 and 4241, no significant mortality was observed until day 5 or later. Although the direct cause of death is not completely understood, bacteremia seemed to play a role. Mice that exhibited clinically overt symptoms (ruffled fur, lethargy, and slight shivering) had much higher bacterial counts in their livers and spleens than those that did not appear to be as sick (data not shown). The infected mice displayed similar symptoms before death occurred, regardless of the length of survival after infection. No diarrhea was observed in any infected mouse. No fatality was observed in mice that were given PBS only.
Morphology and plasmid profile.
All S. enteritidis isolates formed smooth colonies on LB agar plates. No difference in colony morphology was observed among the isolates exhibiting different LD50 in mice. A single large plasmid of approximately 50 kb was found in all isolates (data not shown).
Presence of Salmonella virulence loci.
Each of the 10 S. enteritidis isolates was found to harbor the spv, sodCI, sopE, and sef loci by Southern hybridization. S. typhimurium ATCC 14028s contained only the spv, sodCI, and sopE loci, and E. coli W3110 did not contain any of these loci.
In vitro assays of virulence.
To further characterize phenotypes associated with bacterial virulence, we carried out a battery of in vitro assays to compare the phenotypes of the S. enteritidis isolates that may contribute to their virulence in vivo. After oral inoculation, Salmonella causes disease in mice in a multistep process that includes surviving the acidic environment in the stomach, entering the small intestine, and infecting the M cells, which are specialized phagocytic cells located under the intestinal lining in the Peyer's patch. After surviving the hostile environment of the intestinal phagocytes, Salmonella spreads to other tissues and lymphatics and enters the bloodstream (22, 23, 31, 38, 44). To evaluate in vitro the activity of the S. enteritidis isolates at each of these steps, we performed assays to measure their ability to (i) survive in acidic (pH 4) medium, (ii) invade epithelial cells (with HeLa cells as a model), (iii) survive inside activated mouse peritoneal macrophages, (iv) survive in the presence of ROI and RNI, and (v) survive in prolonged stationary-phase culture. For ease of description, the LD50 rank of an isolate is indicated in parentheses after the isolate identification number; the lower the rank, the more virulent the isolate was in mice (Table 1).
(i) Survival of the S. enteritidis isolates under acidic conditions.
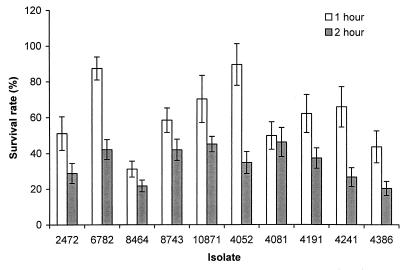
The first major stressful environment that Salmonella encounters after an oral infection is exposure to acidic gastric contents. Acid tolerance may contribute to virulence of S. enteritidis (21). Therefore, we tested the ability of the S. enteritidis isolates to survive in acidic medium at pH 4. Their survival rates varied widely; 1-h survival rates varied from 30 to 90% (Fig. 1). Isolate 8464 (rank, 6) was clearly more susceptible to acidic medium than the others after 1 h of incubation, and after 2 h of incubation, isolate 4386 (rank, 3) was found to be equally susceptible. After 1 h of incubation, the more resistant isolates included 4052 (rank, 10) and 6782 (rank, 4). For all isolates, the survival rates after 2 h of incubation were significantly lower than those after 1 h of incubation. No group difference between the phage type 4 and 8 isolates was observed in this assay.
FIG. 1.
Comparison of the survival rates of S. enteritidis isolates in pH 4 medium. Bars represent the survival rate, which corresponds to (bacterial CFU after the indicated incubation period in pH 4 medium/CFU before incubation) × 100. The results were from a single representative experiment performed in triplicate. Standard deviations (error bars) were calculated as described by Rice (33).
(ii) Invasion of epithelial cells.
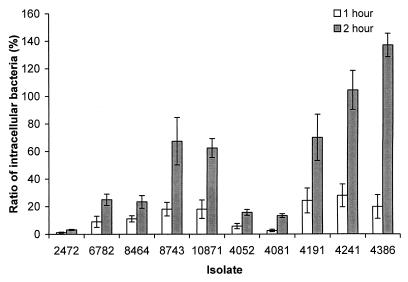
Invasion of epithelial cells is a characteristic of Salmonella associated with virulence (18, 23, 26, 31, 38). The ability of the S. enteritidis isolates to invade and multiply in the epithelial cells was tested with HeLa cells as a model system. We used the S. enteritidis cultures grown in limited oxygen, since the organisms grown under these conditions may be more invasive than those in aerobically grown cultures (13, 24, 36). Isolates 4241 (rank, 9), 4386 (rank, 3), and 10871 (rank, 8) were more invasive after both 1- and 2-h incubation periods, while isolate 2472 (rank, 1) was significantly less invasive (Fig. 2). Isolates 4052 (rank, 10) and 4081 (rank, 5) showed low invasion ability as well (Fig. 2). The more invasive isolates did not necessarily have a lower LD50 in mice, indicating that other factors may play more important roles in the disease outcome after oral infections in mice. There was no significant difference between the phage type 4 and phage type 8 isolates.
FIG. 2.
Comparison of the abilities of S. enteritidis isolates to invade and replicate in HeLa cells. Bars represent the ratio of intracellular bacteria to the input bacteria, calculated as (CFU of intracellular bacteria/CFU of input bacteria) × 100. Incubation was for 1 or 2 h as indicated. Values of over 100% indicate intracellular replication. Results from a single representative experiment performed in triplicate are presented. Error bars indicate standard deviations.
(iii) Survival in activated peritoneal macrophages.
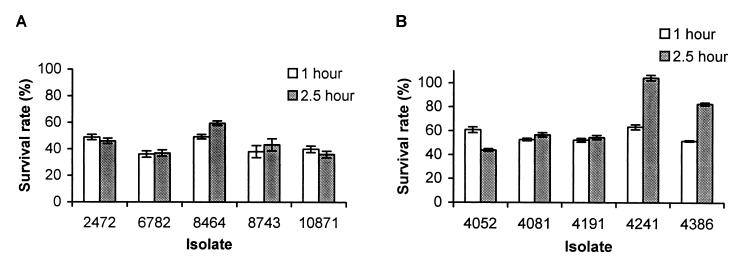
Since survival and replication in activated peritoneal macrophages are believed to be critical for the pathogenesis of S. typhimurium (4, 17), we tested the ability of the S. enteritidis isolates to survive in activated murine macrophages. Because of the large number of isolates, it was not feasible to assay all 10 isolates at the same time. Therefore, the phage type 4 and 8 isolates were assayed separately. Among the phage type 4 isolates, 8464 (rank, 7) and 2472 (rank, 2) were slightly more successful in surviving and multiplying inside macrophages (Fig. 3A). Among the phage type 8 isolates, 4241 (rank, 9) was more successful in surviving inside the macrophages than the other isolates (Fig. 3B). The rest of the isolates did not show any significant difference in their ability to survive and proliferate in macrophages. Therefore, the resistance of the isolates against murine peritoneal macrophages did not correlate with their LD50. In both assays, E. coli K-12 was much more susceptible to the killing inside the macrophages than any of the S. enteritidis isolates (data not shown).
FIG. 3.
Comparison of the resistances of S. enteritidis phage type 4 (A) and 8 (B) isolates to the killing of activated murine macrophages. Resistance to killing was measured by the survival rate after 1 or 2.5 h of incubation, which corresponds to (CFU of intracellular bacteria/CFU of input bacteria) × 100. Results from a single representative experiment performed in triplicate are presented. Error bars indicate standard deviations.
(iv) Resistance to killing by RNI and ROI.
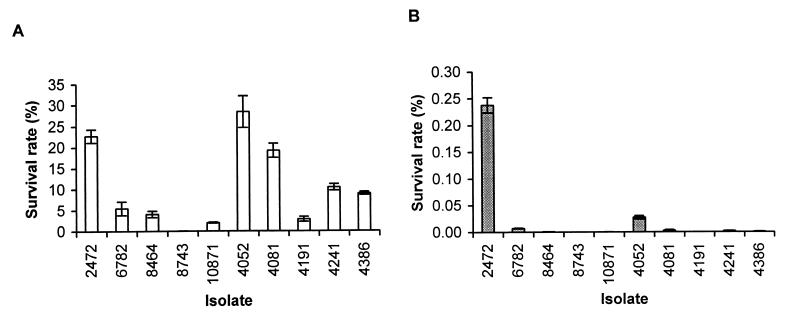
Resistance to killing by ROI and RNI is associated with increased virulence of S. typhimurium (6, 7, 9, 30). We tested the resistance of the S. enteritidis isolates to acidified sodium nitrite (ASN). Sodium nitrite generates RNI, including nitrous acid and nitric oxide, in acidic medium (37, 39). Stationary-phase cultures of S. enteritidis were diluted in acidic medium containing sodium nitrite. We used 20 mM sodium nitrite in the assay because lower concentrations were not effective in killing the S. enteritidis isolates. After 3 h of culture, isolates 2472 (rank, 1), 4081 (rank, 5), and 4052 (rank, 10) were found to be more resistant to ASN than the other isolates (Fig. 4A). Isolate 8743 (rank, 7) was significantly more susceptible to killing by RNI. After 6 h of incubation, 2472 (rank, 1) was markedly more resistant to ASN than the rest of isolates (Fig. 4B). Not only did 2472 have a much higher survival rate, but the colonies formed by this isolate were much larger than those formed by the other isolates, suggesting that they were less damaged by the RNI or able to repair this damage more rapidly.
FIG. 4.
Resistance of S. enteritidis isolates to ASN. Resistance was measured by the survival rate, which equals (CFU of bacteria after incubation in ASN/CFU of bacteria before incubation) × 100. (A) Survival of bacteria after 3 h of incubation; (B) survival of bacteria after 6 h of incubation. Results from a single representative experiment performed in triplicate are presented. Error bars indicate standard deviations.
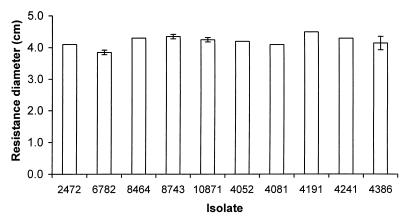
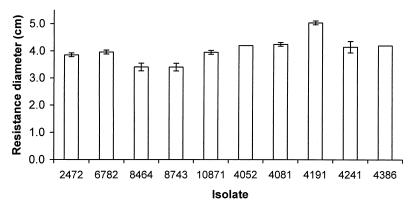
No significant difference was detected in the resistance of S. enteritidis isolates to hydrogen peroxide and paraquat (Fig. 5 and 6). Isolate 6782 (rank, 4) was slightly more resistant to hydrogen peroxide, which may contribute to its low LD50 of 128 organisms. Interestingly, isolate 4191 (rank, 2) has a low LD50 of 23 organisms despite its relatively high susceptibility to both hydrogen peroxide and paraquat.
FIG. 5.
Resistance of S. enteritidis isolates to hydrogen peroxide. Resistance was assayed by the ability to grow in the presence of hydrogen peroxide, and more resistant isolates show a smaller diameter of inhibition. Bars indicate the diameters of inhibitory zones of the S. enteritidis isolates on plates when a source of hydrogen peroxide was placed in the center. Error bars indicate standard deviations; no error bar is shown for data with a standard deviation of 0.
FIG. 6.
Resistance of S. enteritidis isolates to paraquat. Resistance was assayed by the ability to grow in the presence of paraquat, and more resistant isolates show a smaller diameter of inhibition. Bars indicate the diameters of inhibitory zones of the S. enteritidis isolates on plates when a source of paraquat was placed in the center. Error bars indicate standard deviations; no error bar is shown for data with a standard deviation of 0.
(v) Survival during prolonged stationary phase.
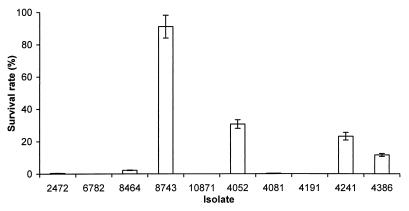
The ability to survive under conditions of nutrient deprivation is considered to contribute to bacterial persistence, which may be important for virulence. We tested the ability of the S. enteritidis isolates to resist starvation. We cultured the S. enteritidis isolates at prolonged stationary phase in M9 minimal medium supplemented with Casamino Acids and determined their survival rates (Fig. 7). Isolate 8743 (rank, 7) has a significantly higher survival rate than the other isolates. Isolates 4052 (rank, 10), 4241 (rank, 9), and 4386 (rank, 3) showed modest survival rates, while the rest of the isolates showed low or no survival. Therefore, no correlation was observed between the LD50 and the ability to survive at prolonged stationary phase.
FIG. 7.
Survival of S. enteritidis isolates during prolonged stationary phase. The ability of S. enteritidis isolates to withstand starvation was measured by the percentage of surviving bacteria after they were cultured at 37°C for 6 days. Bars represent the survival rate, which equals (CFU of culture after 6 days of incubation/CFU of culture after 1 day of incubation) × 100. Results from a single representative experiment performed in triplicate are presented. Error bars indicate standard deviations.
The results of our analysis of the virulence of the S. enteritidis isolates are summarized in Table 2. The 10 isolates were ranked for their resistance and susceptibility to each stress condition. With respect to invasiveness, they were designated invasive or noninvasive. The virulence in mice was ranked as follows: virulent for isolates with an LD50 of <103 organisms, intermediate for isolates with an LD50 of >103 organisms but <107 organisms, and avirulent for isolate with an LD50 of >107 organisms.
TABLE 2.
Summary of in vitro virulence assays of S. enteritidis isolates and comparison to virulence in mice
| Phage type | Isolate | Phenotypea
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Acid resistance | Invasion | Survival in macrophages | NO resistance | H2O2 resistance | Paraquat resistance | Stationary-phase survival | Virulence in mice | ||
| 4 | 2472 | M | N | M | R | M | M | S | V |
| 6782 | R | N | M | M | R | M | S | V | |
| 8464 | S | N | M | M | M | R | S | M | |
| 8743 | M | I | M | S | M | R | R | M | |
| 10871 | R | I | M | M | M | M | S | M | |
| 8 | 4052 | R | N | M | R | M | M | M | A |
| 4081 | M | N | M | R | M | M | S | M | |
| 4191 | M | I | M | M | S | S | S | V | |
| 4241 | M | I | R | M | M | M | M | M | |
| 4386 | M | I | R | M | M | M | M | V | |
The phenotypes of the isolates are assigned arbitrarily according to their relative activities. A, avirulent; V, virulent; R, resistant; M, intermediate; S, susceptible; I, invasive; N, noninvasive.
DISCUSSION
To compare the abilities of clinical isolates of S. enteritidis to produce disease in mice, and to compare the in vivo virulence to results of the in vitro assays often used as correlates of bacterial virulence phenotypes, we carried out a variety of assays in mice, cells, and culture. We chose phage type 4 and 8 isolates because they are responsible for the majority of S. enteritidis outbreaks in the United States and in Europe (1–3, 29). The central conclusions that can be made from this analysis are as follows: (i) clinical isolates of S. enteritidis are highly heterogeneous in their ability to cause death in mice and in their in vitro phenotypes associated with pathogenicity; (ii) phenotypes do not correlate with phage type; (iii) differences in virulence cannot be accounted for by the presence or absence of S. enterica virulence loci that are known to be found only in specific strains (spv, sodCI, sopE, and sef), because all S. enteritidis isolates examined in this study contained these loci; (iv) in vitro tests, especially susceptibility to H2O2 and paraquat and HeLa cell invasion, do not always correlate with pathogenicity in mice, although the most acid-susceptible, macrophage killing-susceptible, and ASN-susceptible isolates were also attenuated for virulence; and (v) the most ASN-resistant isolate (2472) was the most virulent in mice.
The LD50 in BALB/c mice ranged from under 102 organisms to over 108 organisms (Table 1). Our results indicate that isolates of both phage types caused the same spectrum of symptoms in mice and followed approximately the same course of infection. Both phage types included highly virulent isolates (LD50 of <102 organisms) as well as ones with low virulence (LD50 of >105 organisms). The predominance of phage types 4 and 8 in the United States and the United Kingdom, respectively, may be related to clonal differences in their pathogenicities in animals and humans rather than to the phage type per se.
A small number of virulence loci have been found in some isolates of S. enterica and not in others (14, 19, 34). The S. enteritidis isolates used in this study were probed for the presence of these loci to exclude the possibility that the absence of some or all of these loci might account for the reduced virulence of some of the isolates. No difference among the 10 isolates was identified by hybridization analysis; each was found to harbor the spv, sodCI, sopE, and sef loci. However, it is still possible that allelic differences in these genes account for the differences in virulence.
It has been shown that ASN is an important gastric barrier to bacteria (10, 11, 28). Isolate 2472 (rank, 1) was found to be more resistant to ASN than the other isolates after 6 h of incubation. This isolate had a much higher survival rate after ASN exposure, and the colonies formed by the surviving bacteria appeared to be more robust than those of other isolates tested in the same assay. The high resistance to ASN may be a possible reason for the high virulence of 2472 in mice. On the other hand, the isolates with relative low mouse virulence, 8743 (rank, 7) and 10871 (rank, 8), were more susceptible to ASN. However, isolate 4052, which had the highest LD50, was also resistant to ASN. The low in vivo pathogenicity of this isolate could be related to its low level of invasiveness.
Our results demonstrate the complex nature of pathogenesis of Salmonella in mice and the limitations of in vitro assays in predicting relevant phenotypes associated with pathogenesis. In the assays of invasion of epithelial cells and survival at pH 4 and inside activated peritoneal macrophages, the more virulent isolates did not necessarily show greater invasiveness or survival than the less virulent isolates. For example, the highly virulent isolate 2472 (rank, 1) was much less efficient in invading HeLa cells than the other isolates. Moreover, the less virulent isolates, such as 4241 (rank, 9), did not necessarily show any obvious defect in the in vitro assays. These results indicate that a single measure of virulence phenotype is not predictive of ability of S. enteritidis to cause disease and death in mice. Of course, it is possible that other factors not assessed in this study, such as the differences in the expression of endotoxin, serum resistance, or resistance to antimicrobial peptides of neutrophils, could account for the in vivo differences observed. Interestingly, the low-LD50 strains exhibited an unexpected mortality pattern in which half of the mortality in mice occurred in the first 48 h of oral infection. We are currently studying aspects of this phenomenon, such as the role of serum endotoxins. It is also possible that as-yet-unrecognized pathogenesis mechanisms are involved in these highly virulent S. enteritidis strains and that such an observation was made possible by the use of clinical isolates.
Many studies of Salmonella pathogenesis have been conducted by comparing a specific phenotype measured in an in vitro assay of a mutant and its parental strain. This approach has generated much useful information about the possible functions of individual genes in pathogenesis. Our present study suggests that there are as-yet-uncharacterized features of clinical isolates of S. enteritidis that determine their lethality in mice.
ACKNOWLEDGMENTS
We thank Duc Vugia and Sharon Abbott of the Department of Health Services, State of California, for providing the clinical isolates of phage type 4 S. enteritidis used in this study and Tim Barrett of the Centers for Disease Control and Prevention for providing the clinical isolates of phage type 8 S. enteritidis used in this study. We also thank Sabine Ehrt for suggestions and discussions.
This study was supported by grants AI43032 (to L.W.R.) and AI39557 (to F.C.F.) and by the James Biundo Foundation (F.C.F.).
REFERENCES
- 1.Altekruse S, Koehler J, Hickman-Brenner F, Tauxe R V, Ferris K. A comparison of Salmonella enteritidis phage types from egg-associated outbreaks and implicated laying flocks. Epidemiol Infect. 1993;110:17–22. doi: 10.1017/s0950268800050639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altekruse S F, Swerdlow D L. The changing epidemiology of foodborne diseases. Am J Med Sci. 1996;311:23–29. doi: 10.1097/00000441-199601000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Boyce T G, Koo D, Swerdlow D L, Gomez T M, Serrano B, Nickey L N, Hickman-Brenner F W, Malcolm G B, Griffin P M. Recurrent outbreaks of Salmonella enteritidis infections in a Texas restaurant: phage type 4 arrives in the United States. Epidemiol Infect. 1996;117:29–34. doi: 10.1017/s0950268800001096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchmeier N A, Heffron F. Intracellular survival of wild-type Salmonella typhimurium and macrophage-sensitive mutants in diverse populations of macrophages. Infect Immun. 1989;57:1–7. doi: 10.1128/iai.57.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Summary of notifiable diseases, United States—1997. Morbid Mortal Weekly Rep. 1998;46:1–87. [PubMed] [Google Scholar]
- 6.De Groote M A, Fang F C. NO inhibitions: antimicrobial properties of nitric oxide. Clin Infect Dis. 1995;21(Suppl. 2):S162–165. doi: 10.1093/clinids/21.supplement_2.s162. [DOI] [PubMed] [Google Scholar]
- 7.De Groote M A, Granger D, Xu Y, Campbell G, Prince R, Fang F C. Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proc Natl Acad Sci USA. 1995;92:6399–6403. doi: 10.1073/pnas.92.14.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Groote M A, Ochsner U A, Shiloh M U, Nathan C, McCord J M, Dinauer M C, Libby S J, Vazquez-Torres A, Xu Y, Fang F C. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc Natl Acad Sci USA. 1997;94:13997–14001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Groote M A, Testerman T, Xu Y, Stauffer G, Fang F C. Homocysteine antagonism of nitric oxide-related cytostasis in Salmonella typhimurium. Science. 1996;272:414–417. doi: 10.1126/science.272.5260.414. [DOI] [PubMed] [Google Scholar]
- 10.Duncan C, Dougall H, Johnston P, Green S, Brogan R, Leifert C, Smith L, Golden M, Benjamin N. Chemical generation of nitric oxide in the mouth from the enterosalivary circulation of dietary nitrate. Nat Med. 1995;1:546–551. doi: 10.1038/nm0695-546. [DOI] [PubMed] [Google Scholar]
- 11.Dykhuizen R S, Frazer R, Duncan C, Smith C C, Golden M, Benjamin N, Leifert C. Antimicrobial effect of acidified nitrite on gut pathogens: importance of dietary nitrate in host defense. Antimicrob Agents Chemother. 1996;40:1422–1425. doi: 10.1128/aac.40.6.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrt S, Shiloh M U, Ruan J, Choi M, Gunzburg S, Nathan C, Xie Q, Riley L W. A novel antioxidant gene from Mycobacterium tuberculosis. J Exp Med. 1997;186:1885–1896. doi: 10.1084/jem.186.11.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst R K, Dombroski D M, Merrick J M. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect Immun. 1990;58:2014–2016. doi: 10.1128/iai.58.6.2014-2016.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang F C, DeGroote M A, Foster J W, Baumler A J, Ochsner U, Testerman T, Bearson S, Giard J C, Xu Y, Campbell G, Laessig T A. Virulent Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc Natl Acad Sci USA. 1999;96:7502–7507. doi: 10.1073/pnas.96.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang F C, Krause M, Roudier C, Fierer J, Guiney D G. Growth regulation of a Salmonella plasmid gene essential for virulence. J Bacteriol. 1991;173:6783–6789. doi: 10.1128/jb.173.21.6783-6789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang F C, Libby S J, Buchmeier N A, Loewen P C, Switala J, Harwood J, Guiney D G. The alternative sigma factor KatF (RpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fields P I, Swanson R V, Haidaris C G, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-del Portillo F, Finlay B B. Invasion and intracellular proliferation of Salmonella within non-phagocytic cells. Microbiologia. 1994;10:229–238. [PubMed] [Google Scholar]
- 19.Hardt W D, Urlaub H, Galan J E. A substrate of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc Natl Acad Sci USA. 1998;95:2574–2579. doi: 10.1073/pnas.95.5.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill C W, Harnish B W. Inversions between ribosomal RNA genes of Escherichia coli. Proc Natl Acad Sci USA. 1981;78:7069–7072. doi: 10.1073/pnas.78.11.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humphrey T J, Williams A, McAlpine K, Lever M S, Guard-Petter J, Cox J M. Isolates of Salmonella enterica Enteritidis PT4 with enhanced heat and acid tolerance are more virulent in mice and more invasive in chickens. Epidemiol Infect. 1996;117:79–88. doi: 10.1017/s0950268800001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones B D, Falkow S. Salmonellosis: host immune responses and bacterial virulence determinants. Annu Rev Immunol. 1996;14:533–561. doi: 10.1146/annurev.immunol.14.1.533. [DOI] [PubMed] [Google Scholar]
- 23.Kohbata S, Yokoyama H, Yabuuchi E. Cytopathogenic effect of Salmonella typhi GIFU 10007 on M cells of murine ileal Peyer's patches in ligated ileal loops: an ultrastructural study. Microbiol Immunol. 1986;30:1225–1237. doi: 10.1111/j.1348-0421.1986.tb03055.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee C A, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci USA. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C A, Jones B D, Falkow S. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci USA. 1992;89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leung K Y, Finlay B B. Intracellular replication is essential for the virulence of Salmonella typhimurium. Proc Natl Acad Sci USA. 1991;88:11470–11474. doi: 10.1073/pnas.88.24.11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macrina F L, Kopecko D J, Jones K R, Ayers D J, McCowen S M. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid. 1978;1:417–420. doi: 10.1016/0147-619x(78)90056-2. [DOI] [PubMed] [Google Scholar]
- 28.McKnight G M, Smith L M, Drummond R S, Duncan C W, Golden M, Benjamin N. Chemical synthesis of nitric oxide in the stomach from dietary nitrate in humans. Gut. 1997;40:211–214. doi: 10.1136/gut.40.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishu B, Koehler J, Lee L A, Rodrigue D, Brenner F H, Blake P, Tauxe R V. Outbreaks of Salmonella enteritidis infections in the United States, 1985–1991. J Infect Dis. 1994;169:547–552. doi: 10.1093/infdis/169.3.547. [DOI] [PubMed] [Google Scholar]
- 30.Nathan C F, Hibbs J B., Jr Role of nitric oxide synthesis in macrophage antimicrobial activity. Curr Opin Immunol. 1991;3:65–70. doi: 10.1016/0952-7915(91)90079-g. [DOI] [PubMed] [Google Scholar]
- 31.Pospischil A, Wood R L, Anderson T D. Peroxidase-antiperoxidase and immunogold labeling of Salmonella typhimurium and Salmonella choleraesuis var kunzendorf in tissues of experimentally infected swine. Am J Vet Res. 1990;51:619–624. [PubMed] [Google Scholar]
- 32.Reed L J, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 33.Rice J. Mathematical statistics and data analysis. 2nd ed. Belmont, Calif: Wadsworth Publishing Co.; 1994. [Google Scholar]
- 34.Roudier C, Krause M, Fierer J, Guiney D G. Correlation between the presence of sequences homologous to the vir region of Salmonella dublin plasmid pSDL2 and the virulence of 22 Salmonella serotypes in mice. Infect Immun. 1990;58:1180–1185. doi: 10.1128/iai.58.5.1180-1185.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Schiemann D A, Shope S R. Anaerobic growth of Salmonella typhimurium results in increased uptake by Henle 407 epithelial and mouse peritoneal cells in vitro and repression of a major outer membrane protein. Infect Immun. 1991;59:437–440. doi: 10.1128/iai.59.1.437-440.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stuehr D J, Nathan C F. Nitric oxide. A macrophage product responsible for cytostasis and respiratory inhibition in tumor target cells. J Exp Med. 1989;169:1543–1555. doi: 10.1084/jem.169.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeuchi A. Electron microscope studies of experimental Salmonella infection. I. Penetration into the intestinal epithelium by Salmonella typhimurium. Am J Pathol. 1967;50:109–136. [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor T W J, Wignall E W, Cowley J F. The decomposition of nitrous acid in acqueous solutions. J Chem Soc. 1927;11:1923. [Google Scholar]
- 40.Threlfall E J, Rowe B, Ferguson J L, Ward L R. Characterization of plasmids conferring resistance to gentamicin and apramycin in strains of Salmonella typhimurium phage type 204c isolated in Britain. J Hyg. 1986;97:419–426. doi: 10.1017/s0022172400063609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turcotte C, Woodward M J. Cloning, DNA nucleotide sequence and distribution of the gene encoding the SEF14 fimbrial antigen of Salmonella enteritidis. J Gen Microbiol. 1993;139:1477–1485. doi: 10.1099/00221287-139-7-1477. [DOI] [PubMed] [Google Scholar]
- 42.Ward L R, de Sa J D, Rowe B. A phage-typing scheme for Salmonella enteritidis. Epidemiol Infect. 1987;99:291–294. doi: 10.1017/s0950268800067765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welkos S, O'Brien A. Determination of median lethal and infectious doses in animal model systems. Methods Enzymol. 1994;235:29–39. doi: 10.1016/0076-6879(94)35128-7. [DOI] [PubMed] [Google Scholar]
- 44.Worton K J, Candy D C, Wallis T S, Clarke G J, Osborne M P, Haddon S J, Stephen J. Studies on early association of Salmonella typhimurium with intestinal mucosa in vivo and in vitro: relationship to virulence. J Med Microbiol. 1989;29:283–294. doi: 10.1099/00222615-29-4-283. [DOI] [PubMed] [Google Scholar]