Abstract
LAG3 (CD223) is an inhibitory receptor that is highly upregulated on exhausted T cells in tumors and chronic viral infection. Consequently, LAG3 is now a major immunotherapeutic target for the treatment of cancer and many monoclonal antibodies (mAbs) against human LAG3 (hLAG3) have been generated to block its inhibitory activity. However, little or no information is available on the epitopes they recognize. We selected a panel of seven therapeutic mAbs from the patent literature for detailed characterization. These mAbs were expressed as Fab or single-chain Fv fragments and shown to bind hLAG3 with nanomolar affinities, as measured by biolayer interferometry. Using competitive binding assays, we found that the seven mAbs recognize four distinct epitopes on hLAG3. To localize the epitopes, we carried out epitope mapping using chimeras between human and mouse LAG3. All seven mAbs are directed against the first immunoglobulin-like domain (D1) of hLAG3, despite their different origins. Three mAbs almost exclusively target a unique 30-residue loop of D1 that forms at least part of the putative binding site for MHC class II, whereas four mainly recognize D1 determinants outside this loop. However, since all the mAbs block binding of hLAG3 to MHC class II, each of the epitopes they recognize must at least partially overlap the MHC class II binding site.
Introduction
High expression of immune inhibitory receptors (IRs) on exhausted T cells in the tumor microenvironment limits their anti-tumor activity (1, 2). T cell IRs are now major therapeutic targets in cancer, with monoclonal antibodies (mAbs) that block PD1/PDL1 and CTLA4 in the clinic since 2010 (3-5). Although these mAbs have dramatically improved the outcomes of patients with some cancers, notably metastatic melanoma, many patients do not exhibit long-term durable responses to these therapies. Consequently, there is an urgent need to identify additional therapeutic targets to combine with anti-PD1/PDL1 and anti-CTLA4 mAbs to boost the efficacy of immunotherapy (6, 7). This in turn has generated considerable interest in lymphocyte activation gene-3 protein (LAG3; CD223), which is now the third IR to be targeted in the clinic (8).
Lymphocyte activation gene 3 protein (LAG3) is a type I transmembrane glycoprotein comprising four extracellular immunoglobulin (Ig)-like domains (D1–D4) (9, 10), a connecting peptide that is cleaved by the metalloproteases ADAM10 and ADAM17 (11), and a cytoplasmic tail that contains a repeating glutamic acid proline amino acid motif which is critical for the disruption of co-receptor–p56Lck association (12). It is expressed on activated CD4+ and CD8+ T cells, regulatory T cells (Tregs), and plasmacytoid dendritic cells (13-15). LAG3 downregulates T cell activation, proliferation, and cytokine production, rendering T cells dysfunctional (16). LAG3 resembles CD4 in that it has the same exon/intron organization, is ~25% identical at the amino acid level, and binds to MHC class II (9, 17). However, the affinity of LAG3 for MHC class II is ~1000-fold higher than that of CD4 (18). In addition to MHC class II, LAG3 also associates with the TCR–CD3 complex as well as liver and lymph node sinusoidal endothelial cell C-type lectin (LSECtin), galectin-3, and fibrinogen-like protein 1 (FGL1) (12, 19-21).
The importance of LAG3 as an IR on both effector T cells and Tregs has been demonstrated in multiple disease models, including type I diabetes (22), allogeneic bone marrow transplant (23), Parkinson’s disease (24), and cancer (25). Preclinical studies targeting LAG3 combined with PD1 showed significant increases in tumor clearance and survival in several mouse tumor models (26, 27). As a result, nearly 20 anti-human LAG3 (hLAG3) mAbs are already in clinical trials for cancer immunotherapy or will enter soon (28). In a phase 1–2 trial, the anti-LAG3 mAb relatlimab, in combination with the anti-PD1 mAb nivolumab, showed durable objective responses in patients with melanoma that relapsed after, or was refractory to, PD1 inhibition (29). In a subsequent phase 2–3 trial, inhibition of LAG3 and PD1 provided significantly greater benefit in terms of progression-free survival than inhibition of PD1 alone in patients with previously untreated metastatic melanoma (8).
Due to its potential in cancer immunotherapy, a large number of mAbs targeting hLAG3 have been generated (30-34). Although these mAbs generally block the binding of hLAG3 to MHC class II, little or no information is available on the epitopes they recognize. Accordingly, we selected seven therapeutic mAbs from the patent literature (30-34) for detailed characterization. We found that these mAbs target four distinct epitopes on hLAG3 and further localized these epitopes to the D1 domain. This domain includes a proline-rich 30-residue loop (9, 10), not found in CD4, that connects the C and C’ β-strands of D1 and is required for the binding of some, but not all, the tested mAbs.
Materials and Methods
Protein expression and purification
The VL and VH sequences of anti-hLAG3 mAbs were obtained from the patent literature: 4A10 (31), 496G6 (32), 22D2 (31), BAP050 (33), 11C9 (31), 13E2 (34), and relatlimab (30) (Table 1). To produce recombinant Fab fragments, VL and VH regions were linked to human Cκ and IgG1 CH1 regions, respectively. For relatlimab, a single-chain Fv (scFv) construct was designed by connecting VL and VH sequences through an 18-residue linker (GSTGGGGSGGGGSGGGGS). Codon-optimized genes encoding VLCκ, VHCH1, and scFv chains were synthesized chemically (GeneArt) and cloned into the mammalian expression vector pcDNA3.4-TOPO. All chains included an N-terminal Ig κ signal sequence for secretion. A streptavidin tag (WSHPQFEK) was attached to the C-terminus of the CH1 domain for affinity purification. Fab fragments were produced by co-transfecting equimolar amounts of VLCκ and VHCH1 chain plasmids into Expi293 cells with Expifectamine (ThermoFisher). After 96 h incubation, cultures were harvested by centrifugation and supernatants dialyzed against 100 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 1 mM EDTA. Recombinant Fabs were purified using sequential streptavidin affinity (IBA Lifesciences) and Superdex 200 (GE Healthcare) columns.
Table 1.
CDR3 sequences and germline genes of mouse anti-hLAG3 antibodies
| Antibody | Company | HCDR3 | Length | IGHV | LCDR3 | Length | IGKV | Reference |
|---|---|---|---|---|---|---|---|---|
| 4A10 | Merck Sharp & Dohme | FYDGLYFAF | 9 | 14-4*02 | MQHLEYPFT | 9 | 2-137*01 | 31 |
| 496G6 | Boehringer Ingelheim | IEDYGVSYYFDY | 12 | 8-8*01 | QQHYSIPLT | 9 | 6-17*01 | 32 |
| 22D2 | Merck Sharp & Dohme | NYRWFGAMDH | 10 | 1-18*01 | QQSTEDPRT | 9 | 3-4*01 | 31 |
| BAP050 | Novartis/Immutep | NPPYYYGTNNAEAMDY | 16 | 9-3*02 | QQYYNLPWT | 9 | 10-94*04 | 33 |
| 11C9 | Merck Sharp & Dohme | SPFYNSRGGNYFDY | 14 | 1-69*01 | QQGDTLPPWT | 10 | 10-96*02 | 31 |
| 13E2 | Immutep | IVEGSYSSSYFDV | 13 | 8-8*01 | QQHYSTPYT | 9 | 6-17*01 | 34 |
| relatlimab | Bristol-Myers Squibb | GYSDYEYNWFDP | 12 | 3-2*02 | QQRSNWPLT | 9 | 5-45*01 | 30 |
Germline genes were identified by IgBLAST (https://www.ncbi.nlm.nih.gov/igblast/) of nucleotide sequences against a mouse germline gene database.
A codon-optimized gene encoding the four Ig-like extracellular domains (D1–D4) of hLAG3 (residues 1–449) was cloned into pcDNA3.4-TOPO with a C-terminal His6 tag for affinity purification. The construct was transfected into Expi293 cells with Expifectamine (ThermoFisher). Culture was harvested after 72 h and hLAG3 protein was purified from supernatants using consecutive Ni2+-NTA and Superdex 200 columns. Soluble mLAG3 (D1–D4; residues 1–442) was produced similarly.
Biolayer interferometry
Binding of the Fab fragments of antibodies 4A10 (31), 496G6 (32), 22D2 (31), BAP050 (33), 11C9 (31), 13E2 (34), and scFv relatlimab (30) to hLAG3 was measured using an Octet RED96 instrument and Ni2+-NTA biosensors (Pall ForteBio). All assays were performed at 200 μl/well in PBS with 0.05% Tween 20 and 0.1% BSA. The biosensors were loaded with 5 μg/ml of purified His6-tagged hLAG3 for 300 sec. Association for 300 sec followed by dissociation for 300 sec against a 2-fold concentration dilution series of each Fab was performed. Data analysis was carried out using Octet Data Analysis 12.0 software. Curves were globally fitted based on association and dissociation rates to obtain KD values.
Epitope binning
Epitope binning was performed with an Octet RED96 instrument. All six anti-hLAG3 Fabs and scFv relatlimab were tested in a pairwise combinatorial manner, and those that competed for the same binding site were grouped together into bins. His6-tagged hLAG3 was immobilized on Ni2+-NTA biosensors, as described above, followed by coating with the first Fab at saturating concentration (200 nM). Binding of the second Fab was tested by dipping the coated biosensor into a concentration 5 × KD of the second Fab for a 900 sec association phase, followed by a 600 sec dissociation phase. Raw data were processed using Octet Data Analysis 12.0 software. Additional binding by the second Fab indicated an unoccupied epitope (non-competitor), while no additional binding indicated epitope blocking (competitor). To further validate epitope binning results, we prepared triple complexes by co-transfecting Expi293 cells with vectors encoding hLAG3 and two mAbs from different bins.
Epitope mapping
293T cells were incubated in 6 well plates at 2 × 105 cells/well overnight at 37 °C. On day 1, 250 μl of Opti-Mem media (Gibco) was mixed with 6 μl TransIT-LT1 (Mirus), while 3 μg of plasmid DNA (pCI-Neo based plasmid containing an IRES.Thy1.1 reporter) containing the hLAG3/mLAG3 chimeras was mixed with 250 μl of Opti-Mem media. The DNA containing mixture was added dropwise to the TransIT-LT1 containing Opti-Mem media mixture while vortexing, incubated at room temperature for 15 min, and added to 293T cells. On day 3, cells were analyzed for surface expression of the hLAG3 mutants. For cell surface staining, transfected cells were incubated with 10 μg/ml of Fabs or scFv for 30 min on ice. For detection of Fabs, scFv, and transfection efficiency, cells were stained with mouse PE-Cy5 anti-human IgG (Biolegend), APC-streptavidin (Biolegend), and rat BV421 anti-Thy1.1 (Biolegend), respectively, for 30 min on ice. The cells were washed and analyzed by flow cytometry using a BD LSR Fortessa instrument with analysis done using FlowJo software.
Results
Production of Fabs, hLAG3, and Fab–hLAG3 complexes
The germline genes and CDR3 sequences of the antibodies used in this study are shown in Table 1. The Fab fragments of anti-hLAG3 antibodies 4A10 (31), 496G6 (32), 22D2 (31), BAP050 (33), 11C9 (31), and 13E2 (34) were expressed by secretion from transfected Expi293 mammalian cells. The anti-hLAG3 antibody relatlimab (30) was produced as a single-chain Fv (scFv) because the corresponding Fab was not secreted by Expi293 cells for unknown reasons. Recombinant Fab and scFv fragments were purified from culture supernatants by streptavidin affinity chromatography. Purified Fabs migrated in reducing SDS-PAGE as two bands of molecular weight ~25 kDa corresponding to the VLCκ and VHCH1 chains (Fig. 1A). Purified relatlimab scFv appeared as a single band of ~25 kDa corresponding to the covalently linked VL and VH chains. Size exclusion chromatography profiles of Fabs showed a single peak at ~50 kDa, as expected, while relatlimab scFv eluted at ~25 kDa (Fig. 1B).
Figure 1.
Purification of Fabs and Fab–hLAG3 and scFv–hLAG3 complexes. (A) Reducing 16% SDS-PAGE gel of purified recombinant Fab and scFv fragments used for affinity measurements and epitope binning (Coomassie blue G-250 staining). Lane 1, markers; lane 2, Fab 496G6; lane 3, Fab 4A10; lane 4, Fab 22D2; lane 5, Fab BAP050; lane 6, Fab 11C9; lane 7, Fab 13E2; lane 8, scFv relatlimab. (B) Superdex 200 size exclusion chromatography profiles of Fabs and scFv following streptavidin affinity chromatography. The column was previously calibrated with thyroglobulin, ferritin, aldolase, conalbumin, and ovalbumin as molecular mass standards. (C) Reducing 16% SDS-PAGE gel of purified Fab–hLAG3 and scFv–hLAG3 complexes. The complexes were prepared by co-transfecting vectors encoding hLAG3 and Fab or scFv fragments into Expi293 cells. Lane 1, markers; lane 2, Fab 496G6–hLAG3; lane 3, Fab 4A10–hLAG3; lane 4, Fab 22D2–hLAG3; lane 5, Fab BAP050–hLAG3; lane 6, Fab 11C9–hLAG3; lane 7, Fab 13E2–hLAG3; lane 8, scFv relatlimab–hLAG3. (D) Superdex 200 size exclusion chromatography profiles of Fab–hLAG3 and scFv–hLAG3 complexes following streptavidin affinity chromatography.
To functionally validate our recombinant Fab and scFv proteins, we prepared Fab–hLAG3 complexes by co-expression. Vectors encoding hLAG3 and the VLCL and VHCH1 chains of antibodies 4A10, 496G6, 22D2, BAP050, 11C9, and 13E2 were co-transfected into Expi293 cells. The relatlimab scFv–hLAG3 complex was prepared similarly. Purification of Fab–hLAG3 and scFv–hLAG3 complexes from culture supernatants was performed using streptavidin affinity chromatography followed by size exclusion. Each purified Fab–hLAG3 or scFv–hLAG3 complex showed an additional band of ~55 kDa in reducing SDS-PAGE corresponding to hLAG3, which implies tight binding (Fig. 1C). In size exclusion profiles, the Fab–hLAG3 and scFv–hLAG3 complexes eluted as single peaks of ~100 kDa and ~75 kDa, respectively (Fig. 1D). Of note, yields of co-secreted Fab–hLAG3 and scFv–hLAG3 complexes were typically ~15 mg per liter of culture, compared to ~3 mg per liter for hLAG3 alone, suggesting stabilization of hLAG3 by the bound antibody.
Affinity of anti-hLAG3 antibodies
We used biolayer interferometry (BLI) to measure the affinities of our Fab and scFv fragments for hLAG3 (Fig. 2). Purified hLAG3 was directionally coupled to a Ni2+-nitrilotriacetic acid (Ni2+-NTA) biosensor surface through its C-terminal His6 tag. All seven antibodies bound hLAG3 with dissociation constants (KDs) in the nanomolar range (in order of decreasing affinity): relatlimab (1.8 nM), 13E2 (2.3 nM), 22D2 (5.0 nM), BAP050 (9.0 nM), 11C9 (11 nM), 496G6 (13 nM), and 4A10 (41 nM) (Table 2). These KD values are similar to those reported in the patent literature (30-34), even though we used monovalent Fab or scFv fragments instead of bivalent IgG constructs as in the patents. Thus, bivalent IgG binding does not appear to confer a large avidity effect on these anti-hLAG3 mAbs, at least not according to the assays used (30-34). None of the seven mAbs recognized mLAG3 in our BLI assays (not shown), demonstrating their exquisite specificity for hLAG3, in agreement with the corresponding patents (30-34).
Figure 2.
BLI analysis of Fab and scFv binding to hLAG3. (A) Sensograms (left) for Fab 4A10 binding to immobilized hLAG3. Fab 4A10 concentrations were 100, 50, 25, 12.5, 6.25, and 3.15 nM. Steady-state analysis graph (right) gave a KD of 41 ± 4.8 nM. (B) Sensograms (left) for Fab 496G6 binding to immobilized hLAG3. Steady-state analysis graph (right) gave a KD of 13 ± 1.8 nM. (C) Sensograms (left) for Fab 22D2 binding to immobilized hLAG3. Steady-state analysis graph (right) gave a KD of 5.0 ± 1.7 nM. (D) Sensograms (left) for Fab BAP050 binding to immobilized hLAG3. Steady-state analysis graph (right) gave a KD of 9.0 ± 0.07 nM. (E) Sensograms (left) for Fab 11C9 binding to immobilized hLAG3. Steady-state analysis graph (right) gave a KD of 11 ± 0.06 nM. (F) Sensograms (left) for Fab 13E2 binding to immobilized hLAG3. Steady-state analysis graph (right) gave a KD of 2.3 ± 0.1 nM. (G) Sensograms (left) for scFv relatlimab binding to immobilized hLAG3. Steady-state analysis graph (right) gave a KD of 1.8 ± 0.18 nM.
Table 2.
Equilibrium and kinetic measurements of mAb binding to hLAG3
| Equilibrium | Kinetic | |||
|---|---|---|---|---|
| mAb | KD (nM) | KD (nM) | kon (1/Ms) | koff (1/s) |
| 4A10 | 41 ± 0.50 | 10 ± 0.14 | (7.79 ± 0.08) × 104 | (8.08 ± 0.06) × 10−4 |
| 496G6 | 13 ± 0.17 | 12 ± 0.12 | (2.67 ± 0.23) × 105 | (3.32 ± 0.13) × 10−3 |
| 22D2 | 5.0 ± 0.17 | 0.90 ± 0.14 | (1.82 ± 0.06) × 105 | (1.81 ± 0.03) × 10−4 |
| BAP050 | 9.0 ± 0.07 | 0.80 ± 0.16 | (1.45 ± 0.03) × 105 | (1.27 ± 0.02) × 10−4 |
| 11C9 | 11 ± 0.06 | 12.0 ± 0.52 | (1.84 ± 0.07) × 105 | (2.22 ± 0.04) × 10−3 |
| 13E2 | 2.3 ± 0.10 | 2.50 ± 0.15 | (3.64 ± 0.01) × 105 | (9.22 ± 0.03) × 10−4 |
| relatlimab | 1.8 ± 0.18 | 5.16 ± 0.10 | (7.49 ± 0.04) × 104 | (3.87 ± 0.07) × 10−4 |
Kinetic parameters (on- and off-rates) for the binding of Fab and scFv fragments to hLAG3 are presented in Table 2. KDs calculated from kon and koff values agree well with KDs from equilibrium analysis for 496G6, 11C9, 13E2, and relatlimab, but are ~5-fold different for 4A10 and 22D2 and ~10-fold different for BAP050. We do not know the reason for these differences, but they could reflect complexities in binding. It is nevertheless clear from both equilibrium and kinetic analysis that all seven mAbs in this study are nanomolar binders.
Epitope binning of anti-hLAG3 antibodies
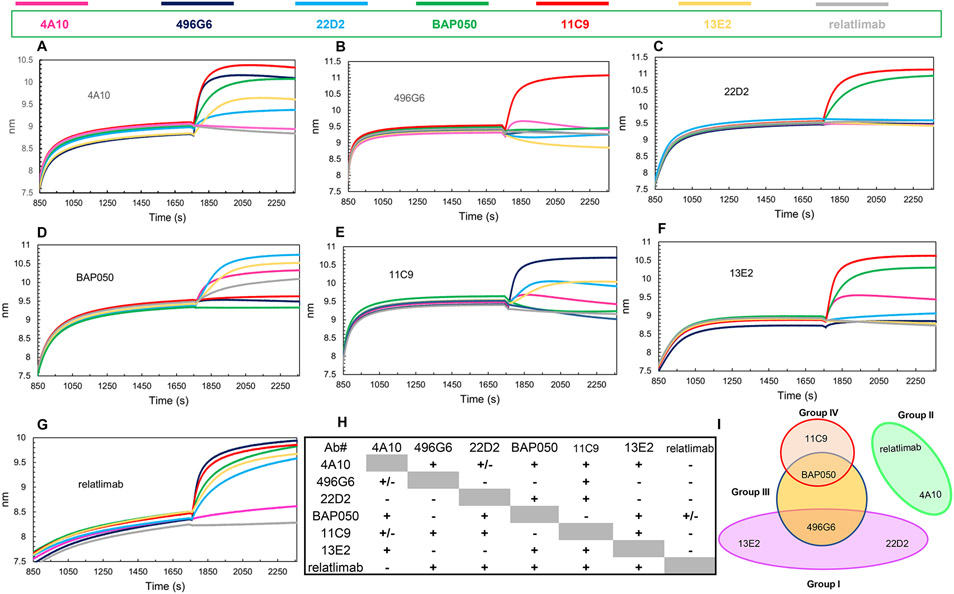
We carried out competitive binding assays using BLI to classify the seven anti-hLAG3 mAbs (as Fab or scFv fragments) according to the epitopes they recognize (epitope binning) (35). Competitions may reflect specific epitope overlap or steric hindrance from nearby epitopes but this is the common first step in assigning epitope targeting groups. In these assays, hLAG3 was immobilized on a Ni2+-NTA biosensor surface and presented with two antibodies, one after another: a saturating mAb followed by a competing one. The competing mAb should bind mAb-saturated hLAG3 only if its epitope does not overlap that of the saturating mAb. The experiment was repeated with each mAb as saturating antibody and the other six mAbs as competing mAbs to determine the ability of each pair to simultaneously bind hLAG3 (Fig. 3).
Figure 3.
Epitope binning of anti-LAG3 antibodies by BLI. Binning experiments were performed with His6-tagged hLAG3 immobilized on Ni2+-NTA biosensors. The binding step with saturating antibody followed by binding/blocking antibodies is presented for each indicated saturating antibody. In each set of experiments, the background signal was obtained from a parallel sensor incubated with the saturating antibody as control. (A) Sensograms with 4A10 as saturating antibody. (B) Sensograms with 496G6 as saturating antibody. (C) Sensograms with 22D2 as saturating antibody. (D) Sensograms with BAP050 as saturating antibody. (E) Sensograms with 11C9 as saturating antibody. (F) Sensograms with 13E2 as saturating antibody. (G) Sensograms with relatlimab as saturating antibody. (H) Matrix with relative binding values calculated by Octet Analysis Software 12.0. Saturating antibodies are listed in the left column and blocking antibodies in the top row. (I) Four noncompeting hLAG3 binding epitopes were identified and classified into four groups: I (496G6, 22D2, and 13E2); II (4A10 and relatlimab); III (BAP050 and 496G6); and IV (11C9 and BAP050).
In binning with 4A10 as the saturating mAb, 496G6, BAP050, 11C9, and 13E2 gave strong binding responses, indicating that they target epitopes different from 4A10 (Fig. 3A). 22D2 also bound to the 4A10–hLAG3 complex, albeit weakly. Relatlimab, however, showed no interaction. In the reverse setup, with relatlimab as the saturating mAb, 4A10 gave negligible binding to the relatlimab–hLAG3 complex (Fig. 3G), suggesting that 4A10 and relatlimab belong to one bin, different from the bins containing 496G6, BAP050, 11C9, 13E2, 496G6, 13E2, and 22D2.
With 496G6 as the saturating mAb, only 4A10 and 11C9 displayed binding responses (Fig. 3B). Inversely, 496G6 bound to the 4A10–hLAG3 (Fig. 3A) and 11C9–hLAG3 (Fig. 3E) complexes, thereby placing these three mAbs in different bins.
With 22D2 as the saturating mAb, only 11C9 and BAP050 showed strong binding (Fig. 3C). Inversely, 22D2 interacted strongly with hLAG3 saturated with BAP050 (Fig. 3D) and hLAG3 saturated with 11C9 (Fig. 3E), indicating that the 22D2 epitope is distinct from the BAP050 and 11C9 epitopes. With 13E2 as the saturating mAb, binding was observed with 4A10, BAP050, and 11C9 (Fig. 3F). Inversely, 13E2 bound to 4A10–hLAG3 (Fig. 3A), BAP050–hLAG3 (Fig. 3D), and 11C9–hLAG3 (Fig. 3E) complexes, assigning 13E2 to a different bin from 4A10, BAP050, or 11C9. The lack of interaction of 496G6 and 22D2 with the 13E2–hLAG3 complex (Fig. 3F) indicates that these three mAbs recognize the same or overlapping epitopes.
With BAP050 as the saturating mAb, binding was observed with 4A10, 22D2, 13E2, and relatlimab (Fig. 3D). Inversely, BAP050 bound to hLAG3 saturated with 4A10 (Fig. 3A), 22D2 (Fig. 3C), 13E2 (Fig. 3F), and relatlimab (Fig. 3G), which places BAP050 in a separate bin from these four mAbs. Moreover, although 496G6 and 11C9 recognize non-overlapping epitopes (Fig. 3B), these epitopes overlap with the BAP050 epitope, as neither 496G6 nor 11C9 bound to the BAP050–hLAG3 complex (Fig. 3D).
In binning experiments with 11C9 as the saturating mAb, 496G6, 22D2, and 13E2 gave clear binding responses (Fig. 3E). Inversely, 11C9 bound to the 496G6–hLAG3 (Fig. 3B), 22D2–hLAG3 (Fig. 3C), and 13E2–hLAG3 (Fig. 3F) complexes. Thus, the 11C9 epitope is different from the 496G6, 22D2, and 13E2 epitopes.
Finally, hLAG3 saturated with relatlimab interacted with 496G6, 22D2, BAP050, 11C9, and 13E2 (Fig. 3G). Inversely, relatlimab bound to BAP050–hLAG3 (Fig. 3D), as expected. However, a surprising lack of reciprocity was observed for 496G6 (Fig. 3B), 22D2 (Fig. 3C), 11C9 (Fig. 3E), and 13E2 (Fig. 3F). As saturating mAbs, each blocked binding of relatlimab to hLAG3. This discrepancy, which we encountered only for antibody pairs containing relatlimab, might be because relatlimab was expressed as a scFv (see above), whereas all other antibodies were expressed as Fabs. The small size of relatlimab scFv (~25 kDa) may be insufficient to sterically block binding of Fabs (~50 kDa) to the relatlimab–hLAG3 complex, unless the epitopes overlap completely, as appears to be the case for 4A10 (Fig. 3G). We therefore placed relatlimab and 4A10 in the same bin, separate from 496G6, 22D2, BAP050, 11C9, and 13E2. Lack of reciprocity has also been reported in other epitope binning studies, and is usually ascribed to affinity differences (35-38). However, relatlimab (KD = 1.8 nM) and 4A10 (41 nM) are at the opposite ends of the affinity range of our mAbs, yet still compete effectively for binding to hLAG3 in a reciprocal manner, whereas the other mAbs, whose KDs fall between 2.3 nM and 13 nM, do not reciprocate with relatlimab, which favors a steric explanation for its anomalous behavior in binning experiments. It should also be noted that even antibodies which recognize non-overlapping epitopes may nevertheless compete for binding if their angles of attack cause clashes between their constant domains (39, 40).
An antibody binding matrix based on relative binding values calculated by Octet Analysis Software 12.0 is shown in Fig. 3H. Antibody pairs with a “+” sign have binding values >0.5, indicating little or no competition for binding to hLAG3 (i.e. mAbs recognize distinct epitopes). Antibody pairs with a “−” sign have binding values <0.25, indicating complete, or nearly complete, competition (i.e the mAbs recognize the same epitope). Antibody pairs with a “+/−” sign have binding values between 0.25 and 0.5, indicating partial competition (i.e. the mAbs recognize partially overlapping epitopes). Based on this classification, the seven anti-hLAG3 mAbs were assigned to four groups: I (496G6, 22D2, and 13E2); II (4A10 and relatlimab); III (BAP050 and 496G6); and IV (11C9 and BAP050) (Fig. 3I).
To validate our epitope binning results, we co-transfected Expi293 cells with vectors encoding hLAG and two mAbs from different bins. If these mAbs indeed recognize non-overlapping epitopes on hLAG3, we expect to see formation of triple complexes (Fab2–hLAG3). In one combination, hLAG3 was co-expressed with Fab 22D2 (Group I) and Fab 11C9 (Group IV). In another combination, hLAG3 was co-expressed with Fab 22D2 (Group I) and Fab BAP050 (Group III). Complexes were purified from culture supernatants using consecutive Ni2+-NTA (hLAG3 has a C-terminal His6 tag) and Superdex 200 columns. For both combinations, a single major peak of ~150 kDa was observed in size exclusion profiles (Fig. 4A), as expected for an Fab2–hLAG3 triple complex. Reducing SDS-PAGE of these peaks showed a band of ~55 kDa corresponding to hLAG3 and four additional bands of ~25 kDa each corresponding the VLCL and VHCH1 chains of Fab 22D2 and Fab 11C9, or of Fab 22D2 and Fab BAP050 (Fig. 4B). Therefore, 22D2 and 11C9 bind non-overlapping epitopes of hLAG3, as do 22D2 and 11C9, in agreement with epitope binning (Fig. 3I).
Figure 4.
Preparation of Fab2–hLAG3 triple complexes. (A) Superdex 200 size exclusion chromatography profiles of Fab 22D2–Fab 11C9–hLAG3 and Fab 22D2–Fab BAP050–hLAG3 complexes following Ni2+-NTA affinity chromatography. (B) Reducing 16% SDS-PAGE gel of purified Fab2–hLAG3 complexes. Lane 1, Fab 22D2–Fab 11C9–hLAG3; lane 2, markers; lane 3, Fab 22D2–Fab BAP050–hLAG3. (C) Superdex 200 profile of mixture of purified Fab 22D2–hLAG3 complex and purified Fab 13E2. The vertical arrow indicates expected position of an Fab2–hLAG3 triple complex. (D) Reducing 16% SDS-PAGE of mixture of Fab 22D2–hLAG3 and Fab 13E2 that was loaded on Superdex 200 column (lane 1), major peak in size exclusion profile (lane 2), Fab 13E2 (lane 3), and markers (lane 4).
We also tested two mAbs (22D2 and 13E2) from the same bin (Group I), which should not be able to form triple complexes with hLAG3 because they recognize the same or overlapping epitopes. We first co-expressed hLAG3 with Fab 22D2 in Expi293 cells and purified Fab 22D2–hLAG3 complexes from culture supernatants using a Ni2+-NTA column (hLAG3 has a C-terminal His6 tag). We then added purified Fab 13E2 to the Fab 22D2–hLAG3 complex and applied the mixture to a Superdex 200 column. A major peak of ~100 kDa was observed in the size exclusion profile (Fig. 4C), as expected for a double Fab–hLAG3 complex, with no peak of ~150 kDa that would indicate formation of an Fab2–hLAG3 triple complex. Reducing SDS-PAGE of this peak showed a band of 55 kDa corresponding to hLAG3 and two additional bands of ~25 kDa each corresponding to the VLCL and VHCH1 chains of 22D2 (Fig. 4D). Therefore, 22D2 and 13E2 bind the same or overlapping epitopes of hLAG3, in agreement with BLI results.
Epitope mapping of anti-hLAG3 antibodies
Epitope binning provides information on whether antibody pairs bind the same or distinct epitopes on a protein antigen. However, it does not reveal the location of these epitopes. Thus, we carried out epitope mapping using a panel of hLAG3/mLAG3 chimeras expressed on the surface of transiently transfected 293T cells (Fig. 5A). We chose to use hLAG3/mLAG3 rather than, for example, hLAG3/hCD4 chimeras for this purpose because exchanging domains or other structural elements between hLAG3 and mLAG3 should be minimally disruptive beyond the site of exchange, given the considerably higher sequence homology between hLAG3 and mLAG3 (68% identity) than between hLAG3 and hCD4 (25% identity). In agreement with BLI affinity measurements (Fig. 2), all seven mAbs bound wild-type hLAG3, whereas none bound wild-type mLAG3 (Fig. 5A). Such high specificity for hLAG3 is essential for unambiguous epitope mapping using hLAG3/mLAG3 chimeras.
Figure 5.
Epitope mapping of anti-hLAG3 antibodies. 293T cells were transfected with plasmids containing deletions and domain swap mutants of hLAG3 and mLAG3, stained with the panel of anti-hLAG3 mAbs, and analyzed by flow cytometry. (A) Schematic diagrams on left depict deletion and domain-swapped mutants of hLAG3 and mLAG3. Heat map on right shows binding of mAbs to the mutants. Numbers indicate fold-increase of mean fluorescence intensity over vector only and are averages of 6 experiments ± SE. (B) Comparison of the amino acid sequences of D1 loop region of hLAG3 and mLAG3 with conserved residues highlighted in red.
All seven mAbs targeted the D1 domain of hLAG3, despite their different origins (30-34). As measured by flow cytometry (Supplemental Figure 1), the mAbs bound all hLAG3/mLAG3 chimeras containing D1 of hLAG3, including a chimera in which D2–D4 derived from mLAG3 (hLAG3D1.mLAG3D2–4) (Fig. 5A). Deletion of the 30-residue loop (residues 48–77) (Fig. 5B) in the hLAG3 D1 domain (hLAG3ΔD1loop) eliminated binding of 4A10, 22D2, and relatlimab, and greatly reduced binding of 496G6, BAP050, 11C9, and 13E2 relative to wild-type hLAG3. This suggests that the D1 loop forms at least part of the epitopes recognized by all seven mAbs, although we could not exclude that deletion of this long loop might perturb the structure of D1 beyond the deletion site, thereby indirectly impacting the binding of mAbs whose epitopes may not actually include the D1 loop. To address this possibility, we replaced the D1 loop of mLAG3 with that of hLAG3 (mLAG3D1–4hLAG3D1loop). These loops differ in both length (30 residues in hLAG3 versus 25 residues in mLAG3) and sequence (40% identity) (Fig. 5B). Remarkably, 4A10 and relatlimab bound mLAG3D1–4hLAG3D1loop better than hLAG3 (Fig. 5A), demonstrating that the determinants recognized by these mAbs reside primarily in the D1 loop. 22D2 bound mLAG3D1–4hLAG3D1loop somewhat less well than hLAG3, implicating D1 determinants outside the loop as also contributing to recognition. Epitope binning placed 4A10 and relatlimab in Group II and 22D2 in Group I (Fig. 3I), which suggests that 22D2 binds to a different region of the D1 loop than do 4A10 and relatlimab. In contrast to 4A10, 22D2, and relatlimab, 496G6 (Group I), BAP050 (Group III), 11C9 (Group IV), and 13E2 (Group I) did not bind mLAG3D1–4hLAG3D1loop. This indicates that their epitopes are composed mainly of D1 determinants that do not include the D1 loop, as manifested by strong binding to a chimera in which the D1 loop of hLAG3 was replaced by that of mLAG3 (hLAG3D1–4mLAG3D1loop) (Fig. 5A). Indeed, BAP050, 11C9, and 13E2 bound hLAG3D1–4mLAG3D1loop better than hLAG3. Stronger binding was also observed for 22D2, which implies that this mAb recognizes a distinct epitope (see Discussion).
Discussion
All seven anti-hLAG3 mAbs studied here were generated in a similar way using standard hybridoma technology, albeit in different companies (30-34). Mice were immunized with recombinant hLAG3–Fc fusion proteins and hybridoma supernatants screened for binding to hLAG3 expressed on the surface of CHO cells. The anti-hLAG3 mAbs bound to primary human T cells and blocked the binding of hLAG3–Fc to Daudi or Raji cells expressing MHC class II. Even though these mAbs had different origins (30-34), we found that they all targeted the D1 domain of hLAG3, as demonstrated using hLAG3/mLAG3 chimeras. This focus on D1 could be due to greater immunogenicity of D1 than D2–D4 in mice immunized with hLAG3–Fc. Alternatively, and more likely, mAbs against D2–D4 might have been discarded during the selection process because they did not inhibit binding of hLAG3–Fc to cells expressing MHC class II. Indeed, cell–cell adhesion assays using hLAG3 mutants previously localized the MHC class II binding site to the D1 domain and, more specifically, to the proline-rich 30-residue loop, not found in CD4, that connects the C and C’ β-strands of D1 (17).
Inhibition of MHC class II binding to hLAG3 by anti-hLAG3 antibodies may occur in three possible ways: 1) the antibody epitope completely overlaps the MHC class II binding site, 2) the epitope partially overlaps the MHC class II binding site, or 3) the antibody epitope does not overlap the MHC class II binding site at all, but the antibody binds to LAG3 in such a way that its constant region clashes with MHC class II. However, we cannot distinguish among these possibilities in the absence of structural information on LAG3–MHC class II and LAG3–antibody complexes. The recent crystal structures of human and mouse LAG3 (10) do not include a LAG3–MHC class II complex or a LAG3–antibody complex involving an antibody that blocks MHC class II binding.
Although the seven anti-hLAG3 mAbs we examined all target D1, they recognize four distinct epitopes, as demonstrated by competitive binding assays using BLI: I (496G6, 22D2, and 13E2), II (4A10 and relatlimab), III (BAP050 and 496G6), and IV (11C9 and BAP050). We confirmed our epitope binning results by showing that two mAbs from different bins can form stable triple complexes with hLAG3 (Fab2–hLAG3) that can be purified biochemically.
Using hLAG3/mLAG3 chimeras, we showed that 4A10 and relatlimab almost exclusively target the 30-residue loop of D1, whereas 496G6, BAP050, 11C9, and 13E2 mainly recognize D1 determinants outside this loop. 22D2 belongs to a different category. On the one hand, the fact that 22D2 binds mLAG3D1–4hLAG3D1loop nearly as well as hLAG3 implicates the D1 loop. On the other hand, the finding that 22D2 binds hLAG3D1–4mLAG3D1loop even better than hLAG3 suggests that D1 determinants outside the loop are even more important for recognition. However, since all seven mAbs block binding of hLAG3 to MHC class II (30-34), each of the epitopes they recognize must at least partially overlap the MHC class II binding site (17). Future studies assessing the combined ability of two or more mAbs as opposed to one alone to block the inhibitory activity of LAG3 will be important from a therapeutic perspective. More specifically, the likely increased efficacy of preventing LAG3 binding to MHC class II using multiple mAbs that target different epitopes may enhance their immunotherapeutic potential. As targeting LAG3 becomes standard of care in oncology, knowledge of the distinct binding characteristics including epitope targeting of these mAbs may contribute to treatment algorithms.
Supplementary Material
Key Points.
Seven therapeutic anti-LAG3 antibodies bind four distinct epitopes.
Three antibodies target a unique 30-residue loop in the D1 Ig-like domain of LAG3.
Four antibodies primarily recognize D1 determinants outside this loop.
Acknowledgments
We wish to thank everyone in the Vignali Lab (Vignali-lab.com; @Vignali_Lab) and the Mariuzza Lab for all their constructive comments and advice during this project.
This work was supported by National Institutes of Health Grants R01 AI144422 (to D.A.A.V., R.A.M. and C.J.W.) and P01 AI108545 (to D.A.A.V.) and by National Cancer Institute Comprehensive Cancer Center Support CORE Grant CA047904 (to D.A.A.V.).
Abbreviations used in this article:
- IR
immune inhibitory receptor
- mAb
monoclonal antibody
- LAG3
lymphocyte activation gene 3 protein
- Ig-like
immunoglobulin-like
- scFv
single-chain Fv
- BLI
biolayer interferometry
Footnotes
Disclosures
The authors declare the following competing financial interests: D.A.A.V. and C.J.W. have patents covering LAG3, with others pending, and are entitled to a share in net income generated from licensing of these patent rights for commercial development. D.A.A.V. – cofounder and stock holder for Novasenta, Potenza, Tizona, and Trishula; stock holder for Oncorus, Werewolf, and Apeximmune; patents licensed and royalties for Astellas, BMS, and Novasenta; scientific advisory board member for Tizona, Werewolf, F-Star, Bicara, Apeximmune, and T7/Imreg Bio; consultant for Astellas, BMS, Almirall, Incyte, Inzen Therapeutics and G1 Therapeutics; and received research funding from BMS, Astellas and Novasenta.
References
- 1.Topalian SL, Drake CG, and Pardoll DM. 2015. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 13: 450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Driessens G, Kline J, and Gajewski TF. 2009. Costimulatory and coinhibitory receptors in anti-tumor immunity. Immunol. Rev 229: 126–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. 2010. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med 363: 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topalian SL, Sznol M, McDermott DF, Kluger HM, Carvajal RD, Sharfman WH, Brahmer JR, Lawrence DP, Atkins MB, Powderly JD, et al. 2014. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J. Clin. Oncol 32: 1020–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Lao CD, Cowey CL, Schadendorf D, Wagstaff J, Dummer R, et al. 2019. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med 381: 1535–1546. [DOI] [PubMed] [Google Scholar]
- 6.Anderson AC, Joller N, and Kuchroo VK. 2016. Lag-3, Tim-3, and TIGIT: co-inhibitory receptors with specialized functions in immune regulation. Immunity 44: 989–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puhr HC, and Ilhan-Mutlu A. 2019. New emerging targets in cancer immunotherapy: the role of LAG3. ESMO Open 4: e000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tawbi HA, Schadendorf D, Lipson EJ, Ascierto PA, Matamala L, Castillo Gutiérrez E, Rutkowski P, Gogas HJ, Lao CD, et al. 2022. Relatlimab and nivolumab versus nivolumab in untreated advanced melanoma. N. Engl. J. Med 386: 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Triebel F, Jitsukawa S, Baixeras E, Roman-Roman S, Genevee C, Viegas-Pequignot E, and Hercend T. 1990. LAG-3, a novel lymphocyte activation gene closely related to CD4. J. Exp. Med 171: 1393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ming Q, Celias DP, Wu C, Cole AR, Singh S, Mason C, Dong S, Tran TH, Amarasinghe GK, Ruffell B, et al. 2022. LAG3 ectodomain structure reveals functional interfaces for ligand and antibody recognition. Nat. Immunol 23: 1031–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li N, Wang Y, Forbes K, Vignali KM, Heale BS, Saftig P, Hartmann D, Black RA, Rossi JJ, Blobel CP, et al. 2007. Metalloproteases regulate T-cell proliferation and effector function via LAG-3. EMBO J. 26: 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guy C, Mitrea DM, Chou PC, Temirov J, Vignali KM, Liu X, Zhang H, Kriwacki R, Bruchez MP, Watkins SC, et al. 2022. LAG3 associates with TCR-CD3 complexes and suppresses signaling by driving co-receptor-Lck dissociation. Nat. Immunol 23: 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huard B, Gaulard P, Faure F, Hercend T, and Triebel F. 1994. Cellular expression and tissue distribution of the human LAG-3-encoded protein, an MHC class II ligand. Immunogenetics 39: 213–217. [DOI] [PubMed] [Google Scholar]
- 14.Workman CJ, and Vignali DAA. 2005. Negative regulation of T cell homeostasis by lymphocyte activation gene-3 (CD223). J. Immunol 174: 688–695. [DOI] [PubMed] [Google Scholar]
- 15.Huang CT, Workman CJ, Flies D, Pan X, Marson AL, Zhou G, Hipkiss EL, Ravi S, Kowalski J, Levitsky HI, et al. 2004. Role of LAG-3 in regulatory T cells. Immunity 21: 503–513. [DOI] [PubMed] [Google Scholar]
- 16.Andrews LP, Yano H, and Vignali DAA. 2019. Inhibitory receptors and ligands beyond PD-1, PD-L1 and CTLA-4: breakthroughs or backups. Nat. Immunol 20: 1425–1434. [DOI] [PubMed] [Google Scholar]
- 17.Huard B, Mastrangeli R, Prigent P, Bruniquel D, Donini S, El-Tayar N, Maigret B, Dréano M, and Triebel F. 1997. Characterization of the major histocompatibility complex class II binding site on LAG-3 protein. Proc. Natl. Acad. Sci. USA 94: 5744–5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacLachlan BJ, Mason GH, Greenshields-Watson A, Triebel F, Gallimore A, Cole DK, and Godkin A. 2021. Molecular characterization of HLA class II binding to the LAG-3 T cell co-inhibitory receptor. Eur. J. Immunol 51: 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu F, Liu J, Liu D, Liu B, Wang M, Hu Z, Du X, Tang L, and He F. 2014. LSECtin expressed on melanoma cells promotes tumor progression by inhibiting antitumor T-cell responses. Cancer Res. 74: 3418–3428. [DOI] [PubMed] [Google Scholar]
- 20.Kouo T, Huang L, Pucsek AB, Cao M, Solt S, Armstrong T, and Jaffee E. 2015. Galectin-3 shapes antitumor immune responses by suppressing CD8+ T cells via LAG-3 and inhibiting expansion of plasmacytoid dendritic cells. Cancer Immunol. Res 3: 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Sanmamed MF, Datar I, Su TT, Ji L, Sun J, Chen L, Chen Y, Zhu G, Yin W, et al. 2019. Fibrinogen-like protein 1 is a major immune inhibitory ligand of LAG-3. Cell 176: 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Q, Chikina M, Szymczak-Workman AL, Horne W, Kolls JK, Vignali KM, Normolle D, Bettini M, Workman CJ, and Vignali DAA. 2017. LAG3 limits regulatory T cell proliferation and function in autoimmune diabetes. Sci. Immunol 31: eaah4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucas CL, Workman CJ, Beyaz S, LoCascio S, Zhao G, Vignali DAA, and Sykes M. 2011. LAG-3, TGF-β, and cell-intrinsic PD-1 inhibitory pathways contribute to CD8 but not CD4 T-cell tolerance induced by allogeneic BMT with anti-CD40L. Blood 117: 5532–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao X, Ou MT, Karuppagounder SS, Kam TI, Yin X, Xiong Y, Ge P, Umanah GE, Brahmachari S, Shin JH, et al. 2016. Pathological α-synuclein transmission initiated by binding lymphocyte-activation gene 3. Science 353: aah3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldberg MV, and Drake CG. 2011. LAG-3 in cancer immunotherapy. Curr. Top. Microbiol. Immunol 344: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woo SR, Turnis ME, Goldberg MV, Bankoti J, Selby M, Nirschl CJ, Bettini ML, Gravano DM, Vogel P, Liu CL, et al. 2012. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res. 72: 917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson AC, Joller N, and Kuchroo VK. 2016. Lag-3, Tim-3, and TIGIT: co-inhibitory receptors with specialized functions in immune regulation. Immunity 44: 989–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrews LP, Cillo AR, Karapetyan L, Kirkwood J, Workman CJ, and Vignali DAA. 2022. Molecular pathways and mechanisms of LAG3 in cancer therapy. Clin. Cancer Res clincanres.2390.2022. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ascierto PA, Melero I, Bhatia S, Bono P, Sanborn RE, Lipson EJ, Callahan MK, Gajewski T, Gomez-Roca CA, Hodi SF, et al. 2017. Initial efficacy of anti-lymphocyte activation gene-3 (anti-LAG-3; BMS-986016) in combination with nivolumab (nivo) in pts with melanoma (MEL) previously treated with anti-PD-1/PD-L1 therapy. J. Clin. Oncol 35: Suppl 15: 9520. Abstract. [Google Scholar]
- 30.Patent WO 2015/116539A1. Anti-LAG3 antibodies to treat hematological malignancies. Bristol-Myers Squibb Co. 6 August 2015.
- 31.Patent WO 2016/028672A1. Anti-LAG3 antibodies and antigen-binding fragments. Merck Sharp & Dohme Corp. 25 February 2016.
- 32.Patent WO 2017/198741A1. Anti-PD1 and anti-LAG3 antibodies for cancer treatment. Boehringer Ingelheim International GmbH. 23 November 2017.
- 33.Patent WO 2015/1389201A1. Antibody molecules to LAG3 and uses thereof. Novartis Ag, Immutep S.A. 17 September 2015.
- 34.Patent WO 2017/037203A1. Anti-LAG3 antibodies. Immutep S.A. 9 March 2017.
- 35.Abdiche YN, Malashock DS, Pinkerton A, and Pons J. 2009. Exploring blocking assays using Octet, ProteOn, and Biacore biosensors. Anal. Biochem 386: 172–180. [DOI] [PubMed] [Google Scholar]
- 36.Abdiche YN, Miles A, Eckman J, Foletti D, Van Blarcom TJ, Yeung YA, Pons J, and Rajpal A. 2014. High-throughput epitope binning assays on label-free array-based biosensors can yield exquisite epitope discrimination that facilitates the selection of monoclonal antibodies with functional activity. PLoS One 9: e92451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noy-Porat T, Alcalay R, Epstein E, Sabo T, Kronman C, and Mazor O. 2017. Extended therapeutic window for post-exposure treatment of ricin intoxication conferred by the use of high-affinity antibodies. Toxicon 127: 100–105. [DOI] [PubMed] [Google Scholar]
- 38.Noy-Porat T, Makdasi E, Alcalay R, Mechaly A, Levy Y, Bercovich-Kinori A, Zauberman A, Tamir H, Yahalom-Ronen Y, Israeli M, et al. 2020. A panel of human neutralizing mAbs targeting SARS-CoV-2 spike at multiple epitopes. Nat. Commun 11: 4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran K, Poulsen C, Guenaga J, de Val N, Wilson R, Sundling C, Li Y, Stanfield RL, Wilson IA, Ward AB, et al. 2014. Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign. Proc. Natl. Acad. Sci. USA 111: E738–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barnes CO, Jette CA, Abernathy ME, Dam KA, Esswein SR, Gristick HB, Malyutin AG, Sharaf NG, Huey-Tubman KE, Lee YE, et al. 2020. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588: 682–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.