Abstract
Fusarium spp. are among the most important plant pathogens in the world. A survey on maize leaf blight was carried out in Heilongjiang province from 2019 to 2021. Based on morphological characteristics and a phylogenetic analysis on translation elongation factor (tef1) and second-largest subunit of RNA polymerase II (rpb2) genes, 146 Fusarium isolates were obtained and grouped into 14 Fusarium species, including F. ipomoeae (20.5%), F. compactum (17.1%), F. sporotrichioides (9.59%), F. graminearum (9.59%), F. citri (8.9%), F. asiaticum (6.85%), F. verticillioides (6.85%), F. acuminatum (5.48%), F. glycines (5.48%), F. temperatum (2.74%), F. armeniacum (2.74%), Fusarium sp. (2.05%), F. flagelliforme (1.4%), and F. annulatum (0.68%). The Fusarium incarnatum-equiseti species complex (FIESC, including F. ipomoeae, F. compactum, F. citri, and F. flagelliforme) was the most prevalent, indicating an evolving occurrence of the Fusarium species causing maize leaf blight. The typical symptoms observed on the maize leaves were oval to long strip lesions, with a gray to dark gray or brownish red coloration in the center and a chlorotic area at the edges. Based on the tef1 gene, seven haplotypes of FIESC were identified in Heilongjiang province, suggesting a population expansion. This is the first report of F. ipomoeae, F. compactum, F. flagelliforme, F. citri, F. sporotrichioides, F. graminearum, F. asiaticum, F. acuminatum, F. glycines, F. temperatum, F. armeniacum, Fusarium sp., and F. annulatum causing maize leaf blight in Heilongjiang province, China. The current research is informative for managing disease, exploring the phylogenetic relationship among Fusarium species, and clarifying the diversity of Fusarium species associated with maize leaf blight.
Keywords: Fusarium spp., maize, haplotype analysis, genetic diversity
1. Introduction
Fusarium spp. can cause several diseases in maize, such as Fusarium ear rot [1,2,3], Fusarium stalk rot and root rot [2,4], seedling blight [5], and maize leaf blight [6]. Regarding maize leaf blight, Fusarium verticillioides was the first pathogen, reported in 1968 [6], to cause the disease, and the only reported one up to now. However, the pathogenicity and diversity of Fusarium spp. causing maize leaf blight are still unclarified. Maize leaf blight is characterized by symptoms of irregular or spindle lesions, with gray to reddish brown coloration in the lesions’ center surrounded by a chlorotic halo. Sometimes, this disease is misjudged as northern corn leaf spot due to the similar symptoms in the field. Thus, the identification of the pathogens based only on disease symptoms in the field is difficult.
To our knowledge, the genus Fusarium includes more than 300 phylogenetic species [7] and is one of the most important plant pathogens in the world [8]. Most species within the genus can produce a diverse range of mycotoxins, causing varying degrees of acute or chronic toxic effects [1]. Therefore, the accurate identification of these mycotoxin producers is a considerable endeavor [9]. For the identification of fungi and the investigation of molecular ecology, the internal transcribed spacer (ITS) is the most sequenced DNA region [10]. However, the ITS region cannot distinguish the species complex of Fusarium due to its conservation [11]. By contrast, the tef1 gene can be used to discriminate Fusarium species at the species or subspecies level [11,12], and the rpb2 gene is also more informative and frequently employed, so it has been recommended that they are sequenced for Fusarium species identification. However, although the partial beta-tubulin gene has been used to identify several Fusarium species, it was not universally informative within Fusarium [13].
The members of Fusarium incarnatum-equiseti species complex (FIESC) are considered important plant pathogens. FIESC is rarely considered the major pathogen of disease epidemics, but it has been identified as a co-occurring fungal pathogen during an infection [14]. Thirty phylogenetic species within the FIESC (FIESC 1 through FIESC 30) were recognized through Multi-locus Sequence Typing (MLST) [15,16], and the species containing multiple haplotypes are designated by the addition of a lowercase letter to the phylogenetic species designation [9].
Phylogenetic and genetic diversity analyses based on multiple sequences can reveal evolutionary relationships associated with geographical regions [9]. High genetic diversity indicates greater adaptability to changing environmental conditions. In some complex evolutionary scenarios, appropriate and sufficient information may not be obtained from phylogenetic trees [17,18]. By comparison, haplotype networks can be employed to analyze the intraspecific diversity of populations, genetic processes, and the biogeography and history of populations [18,19].
To date, there has been little research on pathogenicity, genetic diversity, and the haplotype groups of pathogenic Fusarium species isolated from symptomatic maize leaves in China. Hence, the purposes of the present study were to: (i) describe the morphological characterization and phylogenetic relationships based on tef1 and rpb2 genes of Fusarium species responsible for maize leaf blight in Heilongjiang province, (ii) evaluate the pathogenicity of different Fusarium species, and (iii) determine the haplotype diversity of FIESC based on tef1 associated with maize leaf blight.
2. Materials and Methods
2.1. Fusarium Isolates Collection
From 2019 to 2021, a total of 132 symptomatic maize leaves were collected from 10 different maize-growing counties or cities in Heilongjiang province. The symptomatic maize leaves were cut with a sterilized scalpel, superficially disinfected with a 2% solution of sodium hypochlorite for 1 min and 75% ethanol for 30 s, rinsed thrice with sterile distilled water, and air-dried on sterile filter papers under aseptic conditions. Pure cultures were obtained by single-spore isolation and maintained on PDA (potato dextrose agar) at 25 °C for 7 days. Fusarium isolates were obtained and preserved on PDA slants at 4 °C and 20% glycerol at −80 °C for temporary storage and long-term storage, respectively.
2.2. Morphological Characterization
All Fusarium isolates were incubated on PDA plate in the dark at 25 °C for 7 days. Colony color and colony texture were observed for each isolate. To determine the size of well-developed macroconidia (n = 30) and the number of septa, these Fusarium isolates were incubated on PDA plates at 25°C for 7 days with light/dark cycle of 8/16 h. The macroconidia were observed under light microscopy (Zeiss Axiolab5 equipped with an Axiocam 208 color industrial digital camera).
2.3. DNA Extraction and Sequence Analysis
Fresh mycelia were harvested from cultures grown on PDA supplemented with streptomycin (50 mg/L) and tetracycline (50 mg/L) for 7 days at 28 °C. The extraction of fungal genomic DNA was performed as Ramdial et al. described [9]. The sequences of the translation elongation factor 1-alpha (tef1) gene, second-largest subunit of RNA polymerase II gene (rpb2), and partial beta-tubulin gene were amplified by the primers EF-1/EF-2, RPB2-5f2/RPB2-7cr, and Bt2a/Bt2b [13,20], respectively. The PCR products were sent to Jilin Comate Bioscience Co. Ltd. for purification and sequencing. Sequences of 146 Fusarium isolates were searched against GenBank and FUSARIOID-ID database (www.fusarium.org, accessed date: 1 September 2022) [21] by Basic Local Alignment Search Tool (BLAST) analysis and then deposited into the NCBI GenBank (Table 1).
Table 1.
List of GenBank accession numbers of Fusarium isolates obtained from symptomatic maize leaves collected from Heilongjiang province and reference strains used in this study.
| Isolates. | Latitude and Longitude | Species | GenBank Accession Nos. | ||
|---|---|---|---|---|---|
| tef1 | rpb2 | Beta-Tubulin | |||
| HA-z142 | 126.738196, 45.753014 | F. ipomoeae | OM985077 | OP436018 | OP642121 |
| HA-z11 | 126.738196, 45.753014 | F. ipomoeae | OM985078 | OP436019 | OP642120 |
| HA-z12 | 126.738196, 45.753014 | F. ipomoeae | OM985079 | OP436020 | OP642119 |
| HA-z13 | 126.738196, 45.753014 | F. ipomoeae | OM985080 | OP436021 | OP642118 |
| HA-z14 | 126.738196, 45.753014 | F. ipomoeae | OM985081 | OP436022 | OP642117 |
| HA-z15 | 126.738196, 45.753014 | F. ipomoeae | OM985082 | OP436023 | OP642116 |
| HA-z16 | 126.738196, 45.753014 | F. ipomoeae | OM985083 | OP436024 | OP642115 |
| HA-z17 | 126.738196, 45.753014 | F. ipomoeae | OM985084 | OP436025 | OP642114 |
| HA-z18 | 126.738196, 45.753014 | F. ipomoeae | OM985085 | OP436026 | OP642113 |
| HA-z19 | 126.738196, 45.753014 | F. ipomoeae | OM985086 | OP436027 | OP642112 |
| HA-z20 | 126.738196, 45.753014 | F. ipomoeae | OM985087 | OP436028 | OP642111 |
| HA-z21 | 126.738196, 45.753014 | F. ipomoeae | OM985088 | OP436029 | OP642110 |
| HA-z22 | 126.738196, 45.753014 | F. ipomoeae | OM985089 | OP436030 | OP642109 |
| HA-x22 | 126.868024, 45.850128 | F. ipomoeae | OM985106 | OP436031 | OP642108 |
| HA-xy82 | 126.933932, 45.769353 | F. ipomoeae | OM985109 | OP436032 | OP642122 |
| HA-xy83 | 126.933932, 45.769353 | F. ipomoeae | OM985110 | OP436033 | OP642123 |
| HA-31 | 126.868024, 45.850128 | F. ipomoeae | OM985118 | OP436034 | OP642107 |
| SH-11 | 127.270457, 46.64457 | F. ipomoeae | OM985119 | OP436035 | OP642106 |
| SH-63 | 127.270457, 46.64457 | F. ipomoeae | OM985120 | OP436036 | OP642105 |
| WC-31 | 127.22506, 44.93996 | F. ipomoeae | OM985124 | OP436037 | OP642104 |
| QQ-41 | 124.340195, 47.29158 | F. ipomoeae | OM985125 | OP436038 | OP642103 |
| SH-62 | 127.270457, 46.64457 | F. ipomoeae | OM985126 | OP436039 | OP642124 |
| HA-z201 | 126.738196, 45.753014 | F. ipomoeae | OM985127 | OP436040 | OP642125 |
| HA-21 | 126.868024, 45.850128 | F. ipomoeae | OM985128 | OP436041 | OP642126 |
| HA-22 | 126.868024, 45.850128 | F. ipomoeae | OM985129 | OP436042 | OP642127 |
| HA-x21 | 126.868024, 45.850128 | F. ipomoeae | OM985130 | OP436043 | OP642102 |
| HA-212 | 126.868024, 45.850128 | F. ipomoeae | OM985140 | OP436044 | OP642101 |
| DQ-n22 | 125.835845, 46.329205 | F. ipomoeae | OM985182 | OP436045 | OP642100 |
| JX-21 | 132.477436, 46.339951 | F. ipomoeae | OM985183 | OP436046 | OP642098 |
| DQ-n31 | 125.835845, 46.329205 | F. ipomoeae | OM985184 | OP436047 | OP642099 |
| HA-61 | 126.868024, 45.850128 | F. compactum | OM985144 | OP435951 | OP642130 |
| HA-111 | 126.868024, 45.850128 | F. compactum | OM985102 | OP435952 | OP642131 |
| JX-y11 | 132.477436, 46.339951 | F. compactum | OM985123 | OP435953 | OP642132 |
| HA-621 | 126.868024, 45.850128 | F. compactum | OM985145 | OP435975 | OP642128 |
| SYS-31 | 132.768479, 46.215238 | F. compactum | OM985146 | OP435954 | OP642129 |
| HA-z152 | 126.738196, 45.753014 | F. compactum | OM985147 | OP435955 | OP642133 |
| HA-z31 | 126.738196, 45.753014 | F. compactum | OM985148 | OP435956 | OP642134 |
| HA-z32 | 126.738196, 45.753014 | F. compactum | OM985149 | OP435957 | OP642135 |
| HA-z33 | 126.738196, 45.753014 | F. compactum | OM985150 | OP435958 | OP642136 |
| HA-z34 | 126.738196, 45.753014 | F. compactum | OM985151 | OP435959 | OP642137 |
| HA-z35 | 126.738196, 45.753014 | F. compactum | OM985152 | OP435960 | OP642138 |
| HA-z36 | 126.738196, 45.753014 | F. compactum | OM985153 | OP435961 | OP642139 |
| HA-z37 | 126.738196, 45.753014 | F. compactum | OM985154 | OP435962 | OP642140 |
| HA-z38 | 126.738196, 45.753014 | F. compactum | OM985155 | OP435963 | OP642141 |
| HA-z39 | 126.738196, 45.753014 | F. compactum | OM985156 | OP435964 | OP642142 |
| HA-z310 | 126.738196, 45.753014 | F. compactum | OM985157 | OP435965 | OP642143 |
| HA-z311 | 126.738196, 45.753014 | F. compactum | OM985158 | OP435966 | OP642144 |
| HA-z312 | 126.738196, 45.753014 | F. compactum | OM985159 | OP435967 | OP642145 |
| HA-xy151 | 126.933932, 45.769353 | F. compactum | OM985160 | OP435968 | OP642146 |
| HA-xy31 | 126.933932, 45.769353 | F. compactum | OM985161 | OP435969 | OP642147 |
| HA-a11 | 126.868024, 45.850128 | F. compactum | OM985162 | OP435970 | OP642152 |
| HA-42 | 126.868024, 45.850128 | F. compactum | OM985163 | OP435971 | OP642148 |
| JX-52 | 132.477436, 46.339951 | F. compactum | OM985164 | OP435972 | OP642149 |
| JX-121 | 132.477436, 46.339951 | F. compactum | OM985165 | OP435973 | OP642150 |
| JX-31 | 132.477436, 46.339951 | F. compactum | OM985166 | OP435974 | OP642151 |
| HA-x12 | 126.868024, 45.850128 | F. citri | OM985167 | OP435950 | OP642166 |
| QTH-21 | 131.139405, 45.733699 | F. citri | OM985168 | OP435949 | OP642167 |
| HA-z1125 | 126.738196, 45.753014 | F. citri | OM985169 | OP435948 | OP642158 |
| HA-z171 | 126.738196, 45.753014 | F. citri | OM985170 | OP435947 | OP642165 |
| HA-z172 | 126.738196, 45.753014 | F. citri | OM985171 | OP435946 | OP642164 |
| HA-z173 | 126.738196, 45.753014 | F. citri | OM985172 | OP435945 | OP642163 |
| HA-z174 | 126.738196, 45.753014 | F. citri | OM985173 | OP435944 | OP642162 |
| HA-z175 | 126.738196, 45.753014 | F. citri | OM985174 | OP435943 | OP642161 |
| HA-z176 | 126.738196, 45.753014 | F. citri | OM985175 | OP435942 | OP642160 |
| HA-z177 | 126.738196, 45.753014 | F. citri | OM985176 | OP435941 | OP642159 |
| HA-z1126 | 126.738196, 45.753014 | F. citri | OM985177 | OP435940 | OP642157 |
| HA-xy141 | 126.933932, 45.769353 | F. citri | OM985178 | OP435939 | OP642156 |
| HA-z203 | 126.738196, 45.753014 | F. citri | OM985179 | OP435938 | OP642155 |
| HA-x11 | 126.868024, 45.850128 | F. flagelliforme | OM985104 | OP435921 | OP642153 |
| HA-x51 | 126.868024, 45.850128 | F. flagelliforme | OM985105 | OP435920 | OP642154 |
| HA-a31 | 126.868024, 45.850128 | F. graminearum | OM985090 | OP435980 | OP642200 |
| HG-11 | 130.440826, 47.312952 | F. graminearum | OM985091 | OP435981 | OP642201 |
| QTH-23 | 131.139405, 45.733699 | F. graminearum | OM985103 | OP435982 | OP642202 |
| SH-x72 | 127.270457, 46.64457 | F. graminearum | OM985108 | OP435983 | OP642203 |
| SYS-y21 | 132.768479, 46.215238 | F. graminearum | OM985111 | OP435984 | OP642204 |
| SYS-21 | 132.768479, 46.215238 | F. graminearum | OM985199 | OP435985 | OP642205 |
| SYS-141 | 132.768479, 46.215238 | F. graminearum | OM985200 | OP435986 | OP642206 |
| SYS-142 | 132.768479, 46.215238 | F. graminearum | OM985201 | OP435987 | OP642207 |
| SYS-143 | 132.768479, 46.215238 | F. graminearum | OM985202 | OP435988 | OP642208 |
| SYS-144 | 132.768479, 46.215238 | F. graminearum | OM985203 | OP435989 | OP642209 |
| SYS-145 | 132.768479, 46.215238 | F. graminearum | OM985204 | OP435990 | OP642210 |
| SYS-146 | 132.768479, 46.215238 | F. graminearum | OM985205 | OP435991 | OP642211 |
| SYS-147 | 132.768479, 46.215238 | F. graminearum | OM985206 | OP435992 | OP642212 |
| HA-a142 | 126.868024, 45.850128 | F. graminearum | OM985138 | OP435993 | OP642213 |
| SYS-x71 | 131.583118, 46.462499 | F. asiaticum | OM985092 | OP436053 | OP642088 |
| SYS-x91 | 131.583118, 46.462499 | F. asiaticum | OM985093 | OP436054 | OP642089 |
| HA-x72 | 126.868024, 45.850128 | F. asiaticum | OM985094 | OP436055 | OP642090 |
| HG-x62 | 130.440826, 47.312952 | F. asiaticum | OM985095 | OP436056 | OP642091 |
| SYS-x62 | 131.583118, 46.462499 | F. asiaticum | OM985096 | OP436057 | OP642092 |
| SYS-x131 | 131.583118, 46.462499 | F. asiaticum | OM985097 | OP436058 | OP642093 |
| SYS-x132 | 131.583118, 46.462499 | F. asiaticum | OM985098 | OP436059 | OP642094 |
| SYS-x133 | 131.583118, 46.462499 | F. asiaticum | OM985099 | OP436060 | OP642095 |
| SYS-x134 | 131.583118, 46.462499 | F. asiaticum | OM985100 | OP436061 | OP642096 |
| SYS-x135 | 131.583118, 46.462499 | F. asiaticum | OM985101 | OP436062 | OP642097 |
| HA-zh142 | 126.738196, 45.753014 | F. temperatum | OM985107 | OP436049 | OP642174 |
| QTH-X332 | 131.139405, 45.733699 | F. temperatum | OM985131 | OP436050 | OP642171 |
| QTH-X331 | 131.139405, 45.733699 | F. temperatum | OM985132 | OP436051 | OP642173 |
| QTH-X33 | 131.139405, 45.733699 | F. temperatum | OM985133 | OP436052 | OP642172 |
| HA-z113 | 126.738196, 45.753014 | Fusarium sp. | OM985112 | OP436063 | OP642168 |
| HA-b113 | 126.738196, 45.753014 | Fusarium sp. | OM985113 | OP436064 | OP642170 |
| HA-Z1131 | 126.738196, 45.753014 | Fusarium sp. | OM985143 | OP436065 | OP642169 |
| SYS-x11 | 131.583118, 46.462499 | F. sporotrichioides | OM985209 | OP436017 | OP642176 |
| SYS-x61 | 131.583118, 46.462499 | F. sporotrichioides | OM985210 | OP436016 | OP642177 |
| SYS-x1 | 131.583118, 46.462499 | F. sporotrichioides | OM985211 | OP436015 | OP642178 |
| SYS-x2 | 131.583118, 46.462499 | F. sporotrichioides | OM985212 | OP436014 | OP642179 |
| HG-12 | 130.440826, 47.312952 | F. sporotrichioides | OM985213 | OP436013 | OP642180 |
| SYS-33 | 132.768479, 46.215238 | F. sporotrichioides | OM985214 | OP436012 | OP642181 |
| SYS-101 | 132.768479, 46.215238 | F. sporotrichioides | OM985215 | OP436011 | OP642182 |
| SYS-102 | 132.768479, 46.215238 | F. sporotrichioides | OM985216 | OP436010 | OP642183 |
| SYS-103 | 132.768479, 46.215238 | F. sporotrichioides | OM985217 | OP436009 | OP642184 |
| SYS-104 | 132.768479, 46.215238 | F. sporotrichioides | OM985218 | OP436008 | OP642185 |
| SYS-105 | 132.768479, 46.215238 | F. sporotrichioides | OM985219 | OP436007 | OP642186 |
| SYS-51 | 132.768479, 46.215238 | F. sporotrichioides | OM985220 | OP436006 | OP642187 |
| HG-y102 | 130.440826, 47.312952 | F. sporotrichioides | OM985121 | OP436005 | OP642188 |
| HG-DBy101 | 130.440826, 47.312952 | F. sporotrichioides | OM985122 | OP436004 | OP642189 |
| SH-z61 | 127.270457, 46.64457 | F. acuminatum | OM985115 | OP435923 | OP642072 |
| SH-61 | 127.270457, 46.64457 | F. acuminatum | OM985116 | OP435922 | OP642073 |
| SH-41 | 127.270457, 46.64457 | F. acuminatum | OM985117 | OP435924 | OP642074 |
| HA-a72 | 126.868024, 45.850128 | F. acuminatum | OM985221 | OP435925 | OP642075 |
| HA-a161 | 126.868024, 45.850128 | F. acuminatum | OM985222 | OP435926 | OP642076 |
| HA-a162 | 126.868024, 45.850128 | F. acuminatum | OM985223 | OP435927 | OP642077 |
| HA-a163 | 126.868024, 45.850128 | F. acuminatum | OM985224 | OP435928 | OP642078 |
| HA-a164 | 126.868024, 45.850128 | F. acuminatum | OM985225 | OP435929 | OP642079 |
| HA-a1211 | 126.868024, 45.850128 | F. armeniacum | OM985134 | OP435979 | OP642214 |
| HA-13 | 126.868024, 45.850128 | F. armeniacum | OM985135 | OP435978 | OP642215 |
| HA-a121 | 126.868024, 45.850128 | F. armeniacum | OM985136 | OP435976 | OP642216 |
| HA-a122 | 126.868024, 45.850128 | F. armeniacum | OM985137 | OP435977 | OP642217 |
| HL-42 | 132.943466, 45.768947 | F. verticillioides | OM985139 | OP435994 | OP642190 |
| DQ-n32 | 125.835845, 46.329205 | F. verticillioides | OM985141 | OP435995 | OP642191 |
| SH-n12 | 127.270457, 46.64457 | F. verticillioides | OM985142 | OP435996 | OP642192 |
| JX-123 | 132.477436, 46.339951 | F. verticillioides | OM985181 | OP435997 | OP642193 |
| SH-n11 | 127.270457, 46.64457 | F. verticillioides | OM985187 | OP435998 | OP642194 |
| SH-n201 | 127.270457, 46.64457 | F. verticillioides | OM985188 | OP435999 | OP642195 |
| SH-n202 | 127.270457, 46.64457 | F. verticillioides | OM985189 | OP436000 | OP642197 |
| SH-n203 | 127.270457, 46.64457 | F. verticillioides | OM985190 | OP436001 | OP642197 |
| SH-n204 | 127.270457, 46.64457 | F. verticillioides | OM985191 | OP436002 | OP642199 |
| SH-n205 | 127.270457, 46.64457 | F. verticillioides | OM985192 | OP436003 | OP642199 |
| JX-3352 | 132.477436, 46.339951 | F. glycines | OM985193 | OP435937 | OP642080 |
| JX-335 | 132.477436, 46.339951 | F. glycines | OM985194 | OP435930 | OP642081 |
| HA-171 | 126.868024, 45.850128 | F. glycines | OM985195 | OP435936 | OP642082 |
| HA-172 | 126.868024, 45.850128 | F. glycines | OM985196 | OP435935 | OP642083 |
| HA-173 | 126.868024, 45.850128 | F. glycines | OM985197 | OP435934 | OP642084 |
| HA-174 | 126.868024, 45.850128 | F. glycines | OM985198 | OP435933 | OP642085 |
| WC-b53 | 127.22506, 44.93996 | F. glycines | OM985208 | OP435932 | OP642086 |
| HA-z1412 | 126.738196, 45.753014 | F. glycines | OM985180 | OP435931 | OP642087 |
| WC-22 | 127.22506, 44.93996 | F. annulatum | OM985207 | OP436048 | OP642175 |
| NRRL 34034 | - | F. ipomoeae | GQ505636 | GQ505814 | - |
| LC0455 | - | F. ipomoeae | MK289580 | MK289734 | - |
| NRRL 45996 | - | F. ipomoeae | GQ505671 | GQ505849 | - |
| CBS 140909 | - | F. ipomoeae | MN170479 | MN170412 | - |
| NRRL 28029 | - | F. compactum | GQ505602 | GQ505780 | - |
| NRRL 36318 | - | F. compactum | GQ505646 | GQ505824 | - |
| NRRL 6548 | - | F. flagelliforme | GQ505589 | GQ505767 | - |
| CBS 731.87 | - | F. flagelliforme | GQ505600 | GQ505778 | - |
| LC12147 | - | F. arcuatisporum | MK289584 | MK289739 | - |
| NRRL 32997 | - | F. arcuatisporum | GQ505624 | GQ505802 | - |
| NRRL 45997 | - | F. clavus | GQ505672 | GQ505850 | - |
| NRRL 34037 | - | F. clavus | GQ505638 | GQ505638 | - |
| LC7937 | - | F. citri | MK289640 | GQ505816 | - |
| LC7922 | - | F. citri | MK289634 | MK289788 | - |
| NRRL 66939 | - | Fusarium sp. | MW233217 | MW233561 | - |
| FRC R-9121 | - | Fusarium sp. | MW233213 | MW233557 | - |
| CBS 462.94 | - | F. sporotrichioides | MN120771 | MN120750 | - |
| NRRL 53430 | - | F. sibiricum | HM744684 | MW233474 | - |
| NRRL 6227 | - | F. armeniacum | HM744692 | JX171560 | - |
| FRC R-09335 | - | F. armeniacum | GQ915501 | GQ915485 | - |
| NRRL 13818 | - | F. asiaticum | AF212451 | MW233412 | - |
| NRRL 46738 | - | F. asiaticum | FJ240299 | - | - |
| NL19-100008 | - | F. graminearum | MZ921906 | MZ921775 | - |
| CBS 136009 | - | F. graminearum | MW928838 | MW928826 | - |
| NRRL 54216 | - | F. acuminatum | HM068314 | HM068334 | - |
| JW 289003 | - | F. acuminatum | MZ921908 | MZ921777 | - |
| CBS 130180 | - | F. verticillioides | MW402024 | MW402740 | - |
| CBS 131389 | - | F. verticillioides | MN534047 | MN534288 | - |
| CBS 135541 | - | F. temperatum | MW402051 | KU604284 | - |
| CBS 130323 | - | Fusarium sp. | MH485018 | MH484927 | - |
| CBS 214.49 | - | F. glycines | MH484960 | MH484869 | - |
| CBS 127316 | - | F. annulatum | MW402021 | MW402738 | - |
| CBS 100001 | - | Macroconia leptosphaeriae | KM231959 | HQ728164 | - |
Bold accession numbers were generated from other studies.
2.4. Phylogenetic Relationships among Fusarium Isolates
The rpb2 (794–896 bp), tef1(546–686 bp), and β-tubulin (332–356 bp) gene sequences of Fusarium isolates were also compared to the sequences available in the FUSARIOID-ID database (www.fusarium.org, accessed date: 1 September 2022) to collect related sequences for inclusion in phylogenetic analysis. Multiple sequence alignments were correspondingly inferred in Molecular Evolutionary Genetics Analysis (MEGA) 7 software [22] using the MUSCLE (multiple sequence comparison by log-expectation) program [23] and refined manually if necessary. To generate concatenated datasets, single gene sequences (tef1 and rpb2) were manually combined utilizing BioEdit [24]. Phylogenetic tree based on the concatenated sequences of tef1 and rpb2 genes was built using the maximum likelihood (ML) method in MEGA 7, respectively. ML tree was generated from bootstrapping 1000 replicates. Bootstrap values ≥ 70% were shown in phylogenetic trees. The sequences from the Fusarium spp. type strains, initially identified as closely related to the sequences herein, were finally included by the preliminary BLAST searches.
2.5. Pathogenicity Tests
All Fusarium isolates were used to evaluate their pathogenicity based on the method described by Xu et al. [25]. To fulfill Koch’s postulates, 10 healthy, surface-sterilized, and four to five leaf-stage maize seedlings (var. Demeiya 3) for each Fusarium isolate were inoculated with Fusarium spore suspension (1 × 106 spores/mL). Twenty maize seedlings sprayed with sterile distilled water served as controls. All seedlings sealed with plastic bags were maintained in a greenhouse at 25 °C with 90% relative humidity and a light/dark cycle of 12/12 h.
Disease severity (DS) and disease incidence (DI) were assessed 14 days post-inoculation. DS was measured based on a 0–9 scale described by Rafael et al. [26] and Xu et al. [25]: 0 (no visible symptoms), 1 (0 up to 0.5%), 2 (0.5–1.6%), 3 (1.6–5.0%), 4 (5.0–15%), 5 (15–37%), 6 (37–66%), 7 (66–87%), 8 (87% to 96%), and 9 (96–100%). DI was computed by following formula: DI = [100 × ∑ (n × corresponding DS)]/(N × 9), where n is the number of infected inoculation leaves corresponding to each disease rating, and N is the total number of inoculation leaves. Disease incidence was computed by following formula: disease incidence = number of diseased leaves/total number of inoculated leaves of living maize plants. A least significant difference (LSD) test was used for statistical analysis at a significance level of p < 0.05 with the Statistical Package for Social Sciences (SPSS) software (v. 20.0; SPSS Inc., Wacker Drive, Chicago, IL, USA, Illinois.IBM Corp., 2012. IBM). All re-isolated pathogens from inoculated maize leaves were identified using morphological and molecular methods mentioned above. Each experiment was repeated two times.
2.6. DNA Polymorphism
DNA Sequence Polymorphism software version 6 was used to individually determine the DNA polymorphism relative degree of the tef1 gene sequences [27]. Furthermore, Tajima’s D, Fu and Li’s D, and Fu and Li’s F were used to determine neutrality test statistics. Significant values of these tests indicate the presence of population changes [28,29]. DNA polymorphism analyses were only performed on FIESC and not on other Fusarium species on account of the limited number of isolates from those species obtained in the current study.
2.7. Haplotype Analysis
Haplotype networks were individually generated based on the tef1 gene sequences of 70 FIESC isolates (including 30 F. ipomoeae isolates, 25 F. compactum isolates, 13 F. citri isolates, and 2 F. flagelliforme isolates in the present study) using PopART v. 1.7 (Allan Wilson Centre Imaging Evolution Initiative) to evaluate genealogy pattens of the haplotypes [19]. The aligned haplotype sequences were used to construct a TCS network [30,31].
3. Results
3.1. Fungal Isolation and Morphological Characterization
In this study, 146 Fusarium isolates were obtained from symptomatic maize leaves in China (Table 1), which were initially classified into 11 groups based on their morphological features, including the Fusarium incarnatum-equiseti species complex (FIESC, including F. ipomoeae, F. compactum, F. citri, and F. flagelliforme in this study), F. sporotrichioides, F. armeniacum, F. asiaticum, F. graminearum, Fusarium sp., F. acuminatum, F. glycines, F. annulatum, F. temperatum, and F. verticillioides (Table 2).
Table 2.
Geographic origins and number of Fusarium isolates recovered from symptomatic maize leaves with macroscopic symptoms of leaf blight collected from 10 locations in Heilongjiang province, China.
| Geographic Origins | Number of Fusarium Isolates | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FIESC | F. sporotrichioides | F. armeniacum | F. asiaticum | F. graminearum | Fusarium sp. | F. acuminatum | F. glycines | F. annulatum | F. temperatum | F. verticillioides | |
| Daqing city | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Harbin city | 56 | 0 | 4 | 1 | 2 | 3 | 5 | 5 | 0 | 1 | 0 |
| Hegang city | 0 | 3 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Jixi city | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 |
| Qiqihar city | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Qitaihe city | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 0 |
| Shuangyashan city | 1 | 11 | 0 | 8 | 9 | 0 | 0 | 0 | 0 | 0 | 0 |
| Suihua city | 3 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 7 |
| Hulin country | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Wuchang city | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| Total | 70 | 14 | 4 | 10 | 14 | 3 | 8 | 8 | 1 | 4 | 10 |
| Percentage a | 47.95 | 9.59 | 2.74 | 6.85 | 9.59 | 2.05 | 5.48 | 5.48 | 0.68 | 2.74 | 6.85 |
a Percentage = n/N × 100%, where n is the number of isolates for one species of Fusarium, and N is the total number of isolates for all Fusarium species.
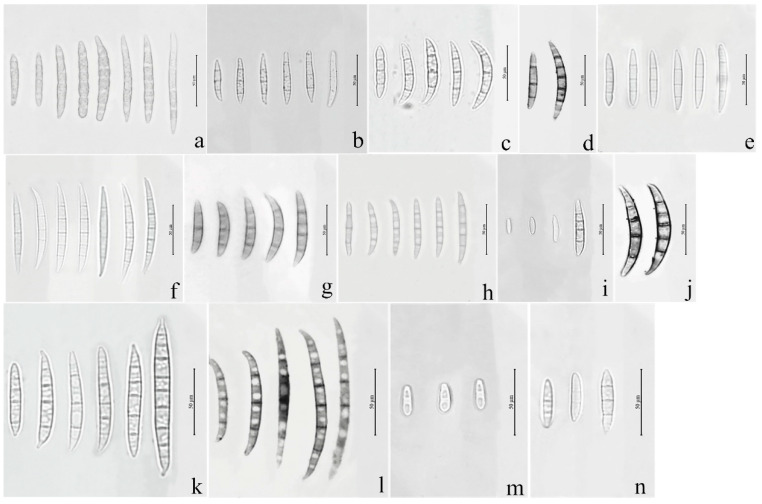
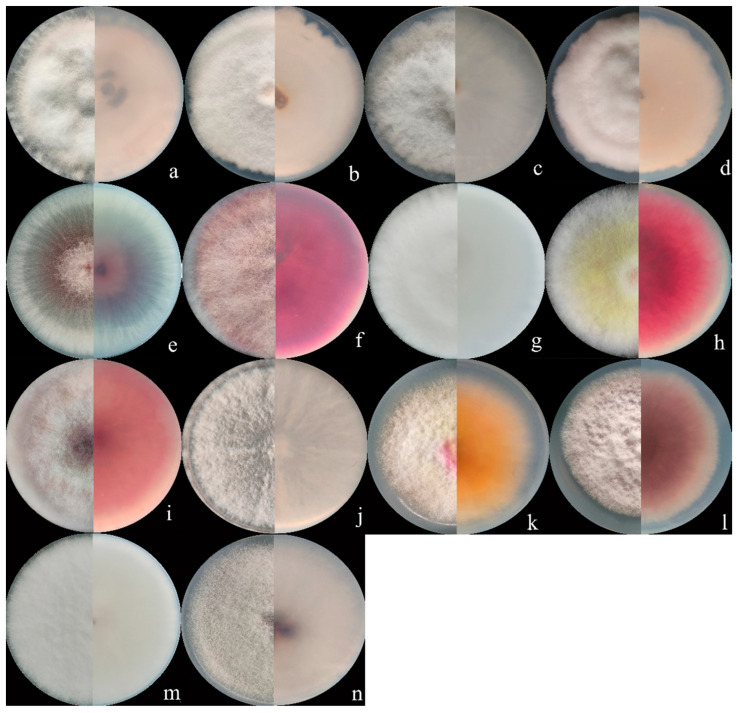
Seventy isolates were identified as the members of FIESC and produced white to light yellow aerial mycelia. The bottom of the plate turned white to pale brown with time. The macroconidia were slightly curved at the apex with three to five septa and ranged from 39.6 to 83.5 × 3.9 to 5.2 μm (n = 30, Figure 1a–d and Figure 2a–d) in size.
Figure 1.
Macroconidia or microconidia of representative isolates of 14 Fusarium species. (a) F. compactum; (b) F. ipomoeae; (c) F. citri; (d) F. flagelliforme; (e) F. temperatum; (f) F. acuminatum; (g) F. armeniacum; (h) F. asiaticum; (i) F. annulatum; (j) Fusarium sp.; (k); F. graminearum; (l) F. glycines; (m) F. verticillioides; (n) F. sporotrichioides.
Figure 2.
Colony appearance of representative isolates of 14 Fusarium species. (a) F. compactum; (b) F. ipomoeae; (c) F. citri; (d) F. flagelliforme; (e) F. verticillioides (f) F. sporotrichioides; (g) F. armeniacum; (h) F. asiaticum; (i) F. graminearum; (j) Fusarium sp.; (k) F. acuminatum; (l) F. glycines; (m) F. annulatum; (n) F. temperatum.
Fourteen F. sporotrichioides isolates produced dense, pinkish white to carmine red aerial mycelia, whose macroconidia were moderately curved to straight with three to five septa, but mostly three-septate, and measured 20.5 to 47.3µm × 2.8 to 4.2 µm (n = 30, Figure 1n and Figure 2f).
The colonies of four F. armeniacum isolates were white to light pink. The macroconidia were prominently curved with three to five septa and had sizes ranging from 35.6 to 59.3 μm × 4 to 4.6 μm (n = 30, Figure 1g and Figure 2g).
Ten isolates producing pink to fluffy dark red aerial mycelia, and red to aubergine pigmentation with age, were classified under F. asiaticum. Their macroconidia were falcate with three to five septa and measured 25.2 to 61.5 × 3.9 to 4.7 μm (n = 30, Figure 1h and Figure 2h).
Fourteen F. graminearum isolates produced white-pink aerial mycelia and had dark red pigmentation. Their macroconidia were straight or slightly curved with five to seven septa and measured 25.4 to 97.7 × 3.4 to 5.8 µm (n = 30, Figure 1k and Figure 2i).
Three Fusarium sp. isolates produced white to yellow colonies and red pigmentation. Their macroconidia were curved with three to five septa and measured 34.0 to 71.6 × 3.2 to 4.7 μm (n = 30, Figure 1j and Figure 2j).
The colonies of eight F. acuminatum isolates were whitish-pink or carmine to rose red. Their macroconidia were slender with a distinct curve of the apical cell, mostly three- to five-septate, and measured 31.3 to 65.3 × 4.0 to 6.5 µm (n = 30, Figure 1f and Figure 2k).
The colonies of eight F. glycines isolates produced fluffy, white aerial hyphae and a dark red pigment. Their macroconidia were three- to seven-septate, slightly curved, and ranged from 53.3 to 117.9 μm × 3.3 to 4.5 μm (n = 30, Figure 1l and Figure 2l) in size.
The aerial mycelia of the F. annulatum isolates were white to cream-colored and turned violet with age, and their macroconidia were straight or slightly curved and contained three to five septa, with sizes of 21.5 to 58.3 × 2.1 to 3.6 µm (n = 30, Figure 1i and Figure 2m).
The colonies of four F. temperatum isolates were pinkish-white and produced mostly three-septate macroconidia. Their macroconidia measured 34.5 to 60.8 × 3.2 to 4.1 µm (n = 30, Figure 1e and Figure 2n).
Ten F. verticillioides isolates formed cottony white to greyish-purple colonies with a dark yellow to purple-gray underside. Their microconidia were abundant and mainly showed clavate shapes measuring 4.2 to 7.5 × 2.1 to 3.8 μm (n = 30, Figure 1m and Figure 2e). However, there were no macroconidia of the F. verticillioides isolates observed in this study.
3.2. Phylogenetic Analysis
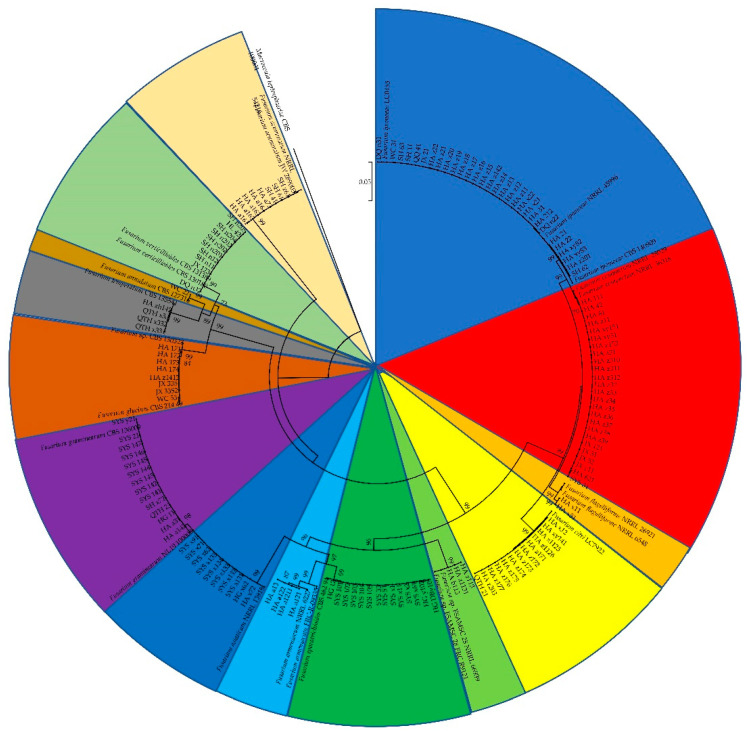
The sequences of the tef1, rpb2, and beta-tubulin genes of all the Fusarium isolates obtained in this study were searched against the FUSARIOID-ID database (www.fusarium.org, accessed date: 1 September 2022) using a BLAST analysis (Table S1). For further molecular verification, a multilocus phylogenetic analysis (MLSA) was further performed based on the concatenated sequences (tef1 and rpb2 genes) of all the Fusarium isolates (Figure 3). These results indicated that all the Fusarium isolates could be grouped into 14 clades, including F. ipomoeae, F. compactum, F. sporotrichioides, F. citri, F. graminearum, F. asiaticum, F. verticillioides, F. acuminatum, F.glycines, F. temperatum, F. armeniacum, Fusarium sp., F. flagelliforme, and F. annulatum.
Figure 3.
Phylogenetic tree obtained from maximum likelihood analysis based on the concatenated sequences of tef1 and rpb2 genes. Support values at nodes representing RA × ML bootstrap percentages with values ≥70 are shown above the branches.
3.3. Pathogenicity Tests
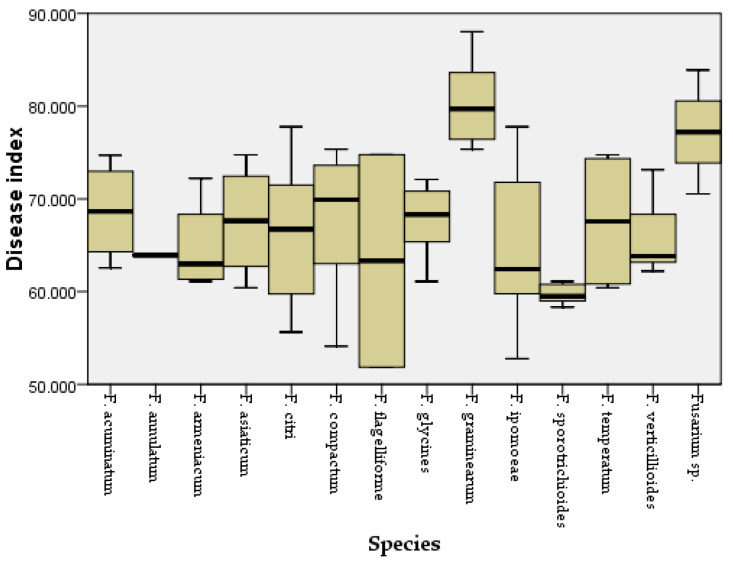
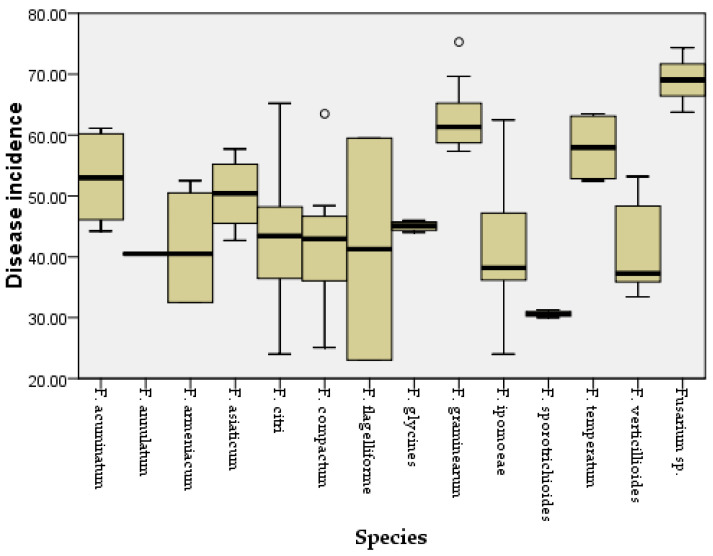
Two weeks after inoculation, the pathogenicity test revealed that all the Fusarium species could cause similar maize leaf blight symptoms (Figure 4). Small oval to fusiform or long striped spots initially appeared on the maize leaves three days post-inoculation, in which the lesions’ centers were gray to reddish brown and surrounded by a chlorotic area. The lesions gradually enlarged with time and merged into each other. In a severe case, the infected leaves were withered. The symptoms observed under greenhouse conditions were similar to the symptoms of maize leaf blight in the field (Figure 4a). No symptoms were observed in the control group. In addition, all the Fusarium species were consistently re-isolated and confirmed based on morphological and molecular methods, while no Fusarium isolates were obtained from the control group, thus fulfilling Koch’s postulates. The average disease incidence and average disease index caused by the Fusarium species ranged from 23 to 74% and from 52 to 85, respectively (Figure 5 and Figure 6; Table S2). Moreover, all the Fusarium isolates were pathogenic towards maize leaves (var. Demeiya 3) and caused maize leaf blight in the inoculation study. In addition, F. graminearum showed the highest virulence, followed by Fusarium sp., F. glycines, F. acuminatum, F. compactum, F. temperatum, F. asiaticum, F. citri, F. verticillioides, F. armeniacum, F. ipomoeae, F. annulatum, F. sporotrichioides, and F. flagelliforme.
Figure 4.
(a) Leaf blight symptoms on maize leaves caused by Fusarium species in the field; (b–o) Typical symptoms observed in greenhouse on maize leaves after inoculation with: (b) F. ipomoeae; (c) F. compactum; (d) F. flagelliforme; (e) F. asiaticum; (f) F. armeniacum; (g) F. citri; (h) F. sporotrichioides; (i) Fusarium sp.; (j) F. glycines; (k) F. graminearum; (l) F. annulatum; (m) F. temperatum; (n) F. verticillioides; (o) F. acuminatum.
Figure 5.
Disease index for maize leaves inoculated with different Fusarium species.
Figure 6.
Disease incidence for maize leaves inoculated with different Fusarium species. Outliers are represented by a hollow circle.
3.4. Haplotype Analyses and DNA Polymorphism
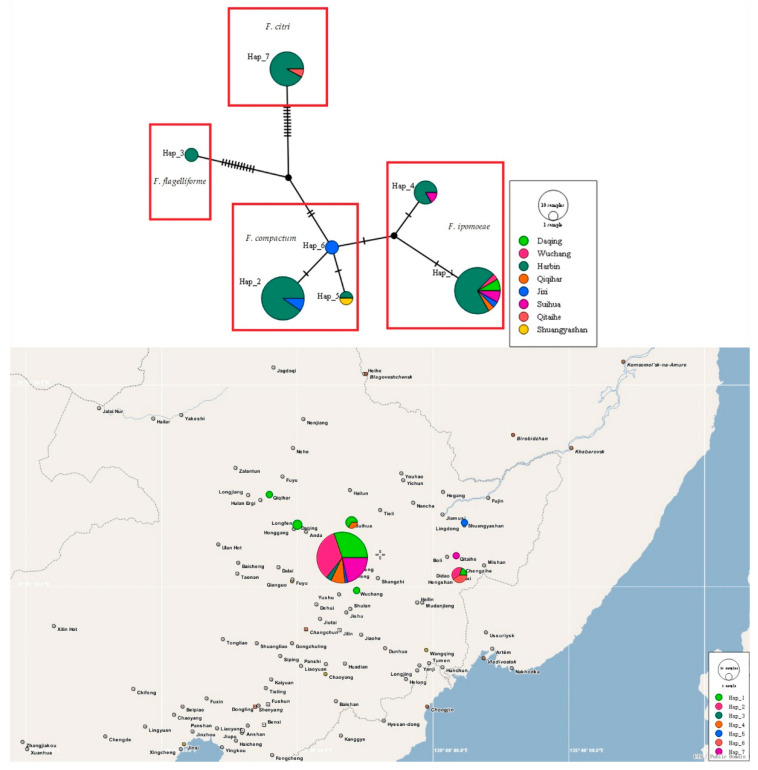
The haplotype networks based on the tef1 gene sequences of 70 FIESC isolates (including 30 F. ipomoeae isolates, 25 F. compactum isolates, 2 F. flagelliforme isolates, and 13 F. citri isolates) obtained in this study were used to determine evolutionary relationships among the haplotypes. Most haplotypes within one species were closely related and separated by one to three mutations.
A total of seven haplotypes were identified: the F. ipomoeae isolates were assigned to Hap 1 and 4; F. compactum isolates were assigned to Hap 2, 5, and 6; F. flagelliforme isolates were assigned to Hap 3; and F. citri isolates were assigned to Hap 7 (Figure 7).
Figure 7.
TCS analyses and the haplotype distribution based on the tef1 gene sequences of 70 FIESC isolates obtained in this study. Each haplotype is represented by a circle, the size of which is proportional to the haplotype frequency.
Meanwhile, Hap 1, 2, 4, 5, and 7 were shared haplotypes (Figure 7). Hap 1 was the most predominant haplotype, and presented in six locations (Harbin city, Wuchang city, Daqing city, Suihua city, Jixi city, and Qiqihar city). Hap 2 was found in Harbin city and Jixi city. Hap 4 was found in Harbin city and Wuchang city. Hap 5 was distributed in Harbin city and Shuangyashan city. Hap 7 was detected in Harbin city and Qitaihe city. Furthermore, two private haplotypes (Hap 3 and 6) were present in Harbin city and Jixi city, respectively. However, there was no obvious center between these predominant haplotypes. In addition, A low degree of nucleotide diversity (0.02706) and a high degree of haplotype diversity (Hd) (0.778) were found. Tajima’s D, Fu and Li’s D, and Fu and Li’s F tests were negative with no significance (p > 0.10, Table S3).
4. Discussion
As far as we know, this is the first systematic study of the Fusarium species associated with maize leaf blight. In this study, 146 Fusarium isolates delimited to 14 Fusarium species were obtained from symptomatic maize leaves in Heilongjiang province. To analyze the genetic relationship between these Fusarium isolates obtained in the current study, phylogenetic trees were constructed only based on the concatenated sequences of tef1 and rpb2 genes because these two genes were more informative and frequently employed, while the beta-tubulin gene was not universally informative in Fusarium [13]. A total of 14 Fusarium species were identified, including F. ipomoeae, F. compactum, F. sporotrichioides, F. citri, F. graminearum, F. asiaticum, F. verticillioides, F. acuminatum, F. glycines, F. temperatum, F. armeniacum, Fusarium sp., F. flagelliforme, and F. annulatum. Except for F. verticillioides, which was the only reported pathogen inciting maize leaf blight [6], the remaining Fusarium species were all first reported in Heilongjiang province, China, suggesting that the composition of Fusarium species causing maize leaf blight may have changed.
Furthermore, considerable pathogenicity differences were found among the different Fusarium species. F. graminearum showed significantly greater average disease incidence and average disease indices than those of other Fusarium species, followed by Fusarium sp., F. glycines, F. acuminatum, F. compactum, F. temperatum, F. asiaticum, F. citri, F. verticillioides, F. armeniacum, F. ipomoeae, F. annulatum, F. sporotrichioides, and F. flagelliforme. Members of FIESC are generally considered co-occurring pathogens [32,33], and the moderate aggressiveness of FIESC in this study seems to confirm the previous conclusion. FIESC was the most predominant in this study. Members of FIESC have been frequently isolated from maize, soybean, rice, barley, wheat, and so on [34,35,36,37,38,39] and have also been reported to cause leaf blight in peanut plants [40] and Cyperus iria [41].
The haplotype groups of FIESC associated with maize leaf blight were first identified in this work. The predominant haplotype (Hap 1) represented multiple locations (Harbin city, Wuchang city, Daqing city, Suihua city, Jixi city, and Qiqihar city). It is well-known that older haplotypes may have a wider geographic distribution, which suggests that Hap 1 has lasted in the population for a long time [42]. The rest of the haplotypes may represent recently evolved lineages [4]. Furthermore, haplotypes 2, 5, and 6 belonged to the F. compactum clade; haplotypes 1 and 4 belonged to the F. ipomoeae clade; haplotype 3 belonged to the F. flagelliforme clade; and haplotype 7 belonged to the F. citri clade. These FIESC isolates were distributed in different clades in the haplotype network, which suggests that the haplotype network could effectively differentiate the Fusarium species complex and further confirmed our identification results. Moreover, the F. flagelliforme haplotype (Hap 3) and F. citri haplotype (Hap 7) were observed in external parts of the haplotype network and showed more mutation events from their nearest haplotypes, which indicated that these two species have an older evolutionary relationship. In addition, the high haplotype diversity and low nucleotide diversity indicated a population expansion [43].
In conclusion, the current study focused on the pathogenicity and genetic diversity of Fusarium species causing maize leaf blight in Heilongjiang province, China, and is the first to report F. ipomoeae, F. compactum, F. flagelliforme, F. citri, F. sporotrichioides, F. graminearum, F. asiaticum, F. verticillioides, F. acuminatum, F. glycines, F. temperatum, F. armeniacum, Fusarium sp., and F. annulatum as the causal agents. Fusarium can cause various maize diseases; therefore, clarifying the population composition of Fusarium spp. on maize leaves will provide information for the overall control of maize diseases.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8111170/s1, Table S1. Tef1 gene sequences similarity to reference strain; Table S2. Disease index and disease incidence on maize leaves inoculated with different Fusarium isolates; Table S3. DNA polymorphism data for FIESC isolates based on tef1 gene sequences.
Author Contributions
X.X., L.Z., X.Y., G.S. and S.W. performed the experiments. H.T. and C.Y. prepared the figures and tables. X.X. and X.L. analyzed the data. X.W., W.X. and J.Z. designed the experiments and reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Sequences have been deposited in GenBank. The data presented in this study are openly available in NCBI. Publicly available datasets were analyzed in this study. These data can be found here: https://www.ncbi.nlm.nih.gov/, accessed on 3 September 2022.
Conflicts of Interest
The authors declare that there are no conflict of interest.
Funding Statement
This work was supported in part by grants from the Key Program of the National Natural Science Foundation of China (No. 32030090), the Outstanding Youth Project of Natural Science Foundation of Heilongjiang Province (YQ2021C012), the Postdoctoral research fund of Heilongjiang Province (LBH-Q21072), the Academic Backbone Project of Northeast Agricultural University (20XG33), and the National Natural Youth Science Foundation of China (No. 31701858).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Munkvold G.P. Epidemiology of Fusarium diseases and their mycotoxins in maize ears. Eur. J. Plant Pathol. 2003;109:705–713. doi: 10.1023/A:1026078324268. [DOI] [Google Scholar]
- 2.Leslie J., Summerell B.A. The Fusarium Laboratory Manual. Blackwell Publishing; Oxford, UK: 2006. [Google Scholar]
- 3.Machado F.J., de Barros A.V., McMaster N., Schmale D.G., 3rd, Vaillancourt L.J., Del Ponte E.M. Aggressiveness and mycotoxin production by Fusarium meridionale compared with F. graminearum on maize ears and stalks in the field. Phytopathology. 2022;112:271–277. doi: 10.1094/PHYTO-04-21-0149-R. [DOI] [PubMed] [Google Scholar]
- 4.Gai X.T., Yang R.X., Pan X.J., Yuan Y., Wang S.N., Liang B.B., Gao Z.G. First report of Fusarium incarnatum causing stalk rot on maize in China. Plant Dis. 2016;100:1010. doi: 10.1094/PDIS-07-15-0766-PDN. [DOI] [Google Scholar]
- 5.Dong H., Qin P., Gao Z., Xu J., Xu X. First report of seedling blight of maize caused by Fusarium asiaticum in Northeast China. Plant Dis. 2021;105:1206. doi: 10.1094/PDIS-08-20-1727-PDN. [DOI] [PubMed] [Google Scholar]
- 6.Schieber R. A leaf blight of corn (Zea mays) incited by Fusarium moniliforme. Phytopathology. 1968;58:554. [Google Scholar]
- 7.Jacobs-Venter A., Laraba I., Geiser D.M., Busman M., Vaughan M.M., Proctor R.H., McCormick S.P., O’Donnell K. Molecular systematics of two sister clades, the Fusarium concolor and F. babinda species complexes, and the discovery of a novel microcycle macroconidium-producing species from South Africa. Mycologia. 2018;110:1189–1204. doi: 10.1080/00275514.2018.1526619. [DOI] [PubMed] [Google Scholar]
- 8.Dean R., van Kan J.A., Pretorius Z.A., Hammond-Kosack K.E., Di Pietro A., Spanu P.D., Rudd J.J., Dickman M., Kahmann R., Ellis J., et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramdial H., Latchoo R.K., Hosein F.N., Rampersad S.N. Phylogeny and haplotype analysis of fungi within the Fusarium incarnatum-equiseti species complex. Phytopathology. 2017;107:109–120. doi: 10.1094/PHYTO-05-16-0209-R. [DOI] [PubMed] [Google Scholar]
- 10.White T.J., Bruns T., Lee S., Taylor J. PCR Protocols: A Guide to Methods and Applications. Academic Press; London, UK: 1990. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics; pp. 315–322. [Google Scholar]
- 11.O’Donnell K., Ward T.J., Robert V., Crous P.W., Geiser D.M., Kang S. DNA sequence-based identification of Fusarium: Current status and future directions. Phytoparasitica. 2015;43:583–595. doi: 10.1007/s12600-015-0484-z. [DOI] [Google Scholar]
- 12.Geiser D.M., Jiménez-Gasco M.D.M., Kang S., Makalowska I., Veeraraghavan N., Ward T.J., Zhang N., Kuldau G.A., O’Donnell K. Fusarium-ID v. 1.0: A DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 2004;110:473–479. doi: 10.1023/B:EJPP.0000032386.75915.a0. [DOI] [Google Scholar]
- 13.O’Donnell K., Whitaker B.K., Laraba I., Proctor R.H., Brown D.W., Broders K., Kim H.S., McCormick S.P., Busman M., Aoki T., et al. DNA Sequence-Based Identification of Fusarium: A Work in Progress. Plant Dis. 2022;106:1597–1609. doi: 10.1094/PDIS-09-21-2035-SR. [DOI] [PubMed] [Google Scholar]
- 14.Villani A., Moretti A., Saeger S.D., Han Z., Mavungu J., Soares C., Proctor R., Venâncio A., Limac N., Gaetano S., et al. A polyphasic approach for characterization of a collection of cereal isolates of the Fusarium incarnatum-equiseti species complex. Int. J. Food Microbiol. 2016;234:24–35. doi: 10.1016/j.ijfoodmicro.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 15.O’Donnell K., Sutton D.A., Rinaldi M.G., Gueidan C., Crous P.W., Geiser D.M. Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-equiseti and F. chalmydosporum species complexes within the United States. J. Clin. Microbiol. 2009;47:3851–3861. doi: 10.1128/JCM.01616-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Donnell K., Humber R.A., Geiser D.M., Kang S., Park B., Robert V.A.R.G., Crous P.W., Johnston P.R., Aoki T., Rooney A.P., et al. Phylogenetic diversity of insecticolous Fusariam inferred from multilocus DNA sequence data and their molecular identification via FUSARIUM-ID and Fusarium MLST. Mycologia. 2012;104:427–445. doi: 10.3852/11-179. [DOI] [PubMed] [Google Scholar]
- 17.Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 18.Blagojević J., Vukojevi J., Ivanovi B., Ivanovi A. Characterization of Alternaria species associated with leaf spot disease of Armoracia rusticana in Serbia. Plant Dis. 2010;104:1378–1389. doi: 10.1094/PDIS-02-19-0289-RE. [DOI] [PubMed] [Google Scholar]
- 19.Leigh J.W., Bryant D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015;6:1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
- 20.Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crous P., Lombard L., Sandoval-Denis M., Seifert K., Schroers H.-J., Chaverri P., Gené J., Guarro J., Hirooka Y., Bensch K., et al. Fusarium: More than a node or a foot-shaped basal cell. Stud. Mycol. 2021;98:100116. doi: 10.1016/j.simyco.2021.100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raza M., Zhang Z.-F., Hyde K.D., Diao Y.-Z., Cai L. Culturable plant pathogenic fungi associated with sugarcane in southern China. Fungal Divers. 2019;99:1–104. doi: 10.1007/s13225-019-00434-5. [DOI] [Google Scholar]
- 25.Xu X., Zhang L., Yang X., Cao H., Li J., Cao P., Guo L., Wang X., Zhao J., Xiang W. Alternaria spp. associated with leaf blight of maize in Heilongjiang Province, China. Plant Dis. 2022;106:572–584. doi: 10.1094/PDIS-06-21-1151-RE. [DOI] [PubMed] [Google Scholar]
- 26.Vieira R.A., Mesquini R.M., Silva C.N., Hata F.T., Tessmann D.J., Scapim C.A. A new diagrammatic scale for the assessment of northern corn leaf blight. Crop Prot. 2014;56:55–57. doi: 10.1016/j.cropro.2011.04.018. [DOI] [Google Scholar]
- 27.Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J.C., Guirao-Rico S., Librado P., Ramos-Onsins S.E., Sánchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 28.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu Y.X., Li W.H. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Templeton A.R., Crandall K.A., Sing C.F. A cladistics analysis of phenotypic associations with haplotypes inferred from restriction endo nuclease mapping and DNA sequence data III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clement M., Snell Q., Walke P., Posada D., Crandall K.A. TCS: Estimating gene genealogies; Proceedings of the 16th International Parallel and Distributed Processing Symposium (IPDPS 2002); Fort Lauderdale, FL, USA. 15–19 April 2002. [Google Scholar]
- 32.Summerell B.A., Salleh B., Leslie J.F. A utilitarian approach to Fusarium identification. Plant Dis. 2003;87:117–128. doi: 10.1094/PDIS.2003.87.2.117. [DOI] [PubMed] [Google Scholar]
- 33.Jurado M., Vázquez C., Patiño B., González-Jaén M.T. PCR detection assays for the trichothecene-producing species Fusarium graminearum, Fusarium culmorum, Fusarium poae, Fusarium equiseti and Fusarium sporotrichioides. Syst. Appl. Microbiol. 2005;28:562–568. doi: 10.1016/j.syapm.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Wang M.M., Chen Q., Diao Y.Z., Duan W.J., Cai L. Fusarium incarnatum-equiseti complex from China. Pers. Mol. Phylogeny Evol. Fungi. 2019;43:70–89. doi: 10.3767/persoonia.2019.43.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marín P., Moretti A., Ritieni A., Jurado M., Vázquez C., González-Jaén M.T. Phylogenetic analyses and toxigenic profiles of Fusarium equiseti and Fusarium acuminatum isolated from cereals from southern Europe. Food Microbiol. 2012;31:229–237. doi: 10.1016/j.fm.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 36.Barros G., Zanon M.S., Palazzini J.M., Haidukowski M., Pascale M., Chulze S. Trichothecenes and zearalenone production by Fusarium equiseti and Fusarium semitectum species isolated from Argentinean soybean. Food Addit. Contam.-Chem. Anal. Control Expo. Risk Assess. 2012;29:1436–1442. doi: 10.1080/19440049.2012.698397. [DOI] [PubMed] [Google Scholar]
- 37.Avila C.F., Moreira G.M., Nicolli C.P., Gomes L.B., Abreu L.M., Pfenning L.H., Haidukowski M., Moretti A., Logrieco A., Del Ponte E.M. Fusarium incarnatum-equiseti species complex associated with Brazilian rice: Phylogeny, morphology and toxigenic potential. Int. J. Food Microbiol. 2019;306:108267. doi: 10.1016/j.ijfoodmicro.2019.108267. [DOI] [PubMed] [Google Scholar]
- 38.Piacentini K.C., Rocha L.O., Savi G.D., Carnielli-Queiroz L., De Carvalho Fontes L., Correa B. Assessment of toxigenic Fusarium species and their mycotoxins in brewing barley grains. Toxins. 2019;11:31. doi: 10.3390/toxins11010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castellá G., Cabañes F.J. Phylogenetic diversity of Fusarium incarnatum-equiseti species complex isolated from Spanish wheat. Antonie Van Leeuwenhoek. 2014;106:309–317. doi: 10.1007/s10482-014-0200-x. [DOI] [PubMed] [Google Scholar]
- 40.Thirumalaisamy P.P., Dutta R., Jadon K.S., Nataraja M.V., Padvi R.D., Rajyaguru R., Yusufzai S. Association and characterization of the Fusarium incarnatum-F. equiseti species complex with leaf blight and wilt of peanut in India. J. Gen. Plant Pathol. 2018;85:83–89. doi: 10.1007/s10327-018-0827-y. [DOI] [Google Scholar]
- 41.Gupta V.S., Razdan V.K., John D.T., Sharma B.C. First report of leaf blight of Cyperus iria caused by Fusarium equiseti in India. Plant Dis. 2013;97:838. doi: 10.1094/PDIS-07-12-0690-PDN. [DOI] [PubMed] [Google Scholar]
- 42.Posada D., Crandall K.A. Evaluation of methods for detecting recombination from DNA sequences: Computer simulations. Proc. Natl. Acad. Sci. USA. 2001;98:13757–13762. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matić S., Tabone G., Garibaldi A., Gullino M.L. Alternaria leaf spot caused by Alternaria species: An emerging problem on ornamental plants in Italy. Plant Dis. 2020;104:2275–2287. doi: 10.1094/PDIS-02-20-0399-RE. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences have been deposited in GenBank. The data presented in this study are openly available in NCBI. Publicly available datasets were analyzed in this study. These data can be found here: https://www.ncbi.nlm.nih.gov/, accessed on 3 September 2022.