Background.
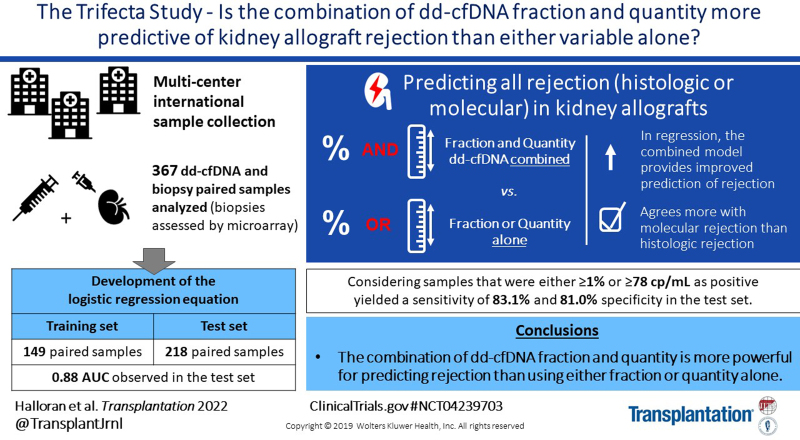
Donor-derived cell-free DNA (dd-cfDNA) fraction and quantity have both been shown to be associated with allograft rejection. The present study compared the relative predictive power of each of these variables to the combination of the two, and developed an algorithm incorporating both variables to detect active rejection in renal allograft biopsies.
Methods.
The first 426 sequential indication biopsy samples collected from the Trifecta study (ClinicalTrials.gov # NCT04239703) with microarray-derived gene expression and dd-cfDNA results were included. After exclusions to simulate intended clinical use, 367 samples were analyzed. Biopsies were assessed using the molecular microscope diagnostic system and histology (Banff 2019). Logistic regression analysis examined whether combining dd-cfDNA fraction and quantity adds predictive value to either alone. The first 149 sequential samples were used to develop a two-threshold algorithm and the next 218 to validate the algorithm.
Results.
In regression, the combination of dd-cfDNA fraction and quantity was found to be significantly more predictive than either variable alone (P = 0.009 and P < 0.0001). In the test set, the area under the receiver operating characteristic curve of the two-variable system was 0.88, and performance of the two-threshold algorithm showed a sensitivity of 83.1% and specificity of 81.0% for molecular diagnoses and a sensitivity of 73.5% and specificity of 80.8% for histology diagnoses.
Conclusions.
This prospective, biopsy-matched, multisite dd-cfDNA study in kidney transplant patients found that the combination of dd-cfDNA fraction and quantity was more powerful than either dd-cfDNA fraction or quantity alone and validated a novel two-threshold algorithm incorporating both variables.
INTRODUCTION
Donor-derived cell-free DNA (dd-cfDNA) has been established as a noninvasive biomarker that can detect organ allograft rejection.1–5 These studies used histology of kidney transplant (KTx) biopsies with standardized grading according to Banff criteria,6 which has some limitations.7,8 In the recent analysis of the Trifecta study,9 we showed that dd-cfDNA has strong correlations with molecular changes of rejection using the molecular microscope diagnostic system (MMDx), which interrogates KTx biopsies status using genome-wide transcript measurements and machine-learning derived algorithms.10–12
Although most clinical tests use dd-cfDNA fraction (measured as the proportion of dd-cfDNA to total cfDNA in the plasma) to detect graft injury and rejection, recent reports indicate that the absolute concentration of dd-cfDNA in copies/mL may provide a better estimate of the allograft rejection and injury.13–15 We hypothesized that both variables could add value based on results from unpublished data showing a lesser or greater variability of the recipient-derived cfDNA compared with dd-cfDNA levels under various physiological and/or pathological conditions. A preliminary report on an independent cohort16 explored an algorithm combining these two variables and suggested that the combination of the quantity of dd-cfDNA threshold and the previously validated dd-cfDNA fraction threshold (1%) holds promise for improved sensitivity in the detection of acute rejection (AR) in patients receiving a renal allograft while maintaining high specificity. That report, used thresholds derived in the training set analysis described in this manuscript; however, the small cohort size and limited confirmatory biopsy data required definitive analysis.
The present study comprehensively explored the potential utility of the two-variable algorithm using a large dataset derived from the Trifecta study, selected to simulate the intended clinical use of dd-cfDNA. We studied 367 biopsy-matched plasma samples from KTx patients using both histological (Banff 2019) and molecular pathology (MMDx) biopsy assessments. The large sample size provides a definitive analysis of the preliminary conclusions presented previously,16 exploring the ability of the two-threshold algorithm to detect active rejection (AR) with enhanced sensitivity while maintaining high specificity.
MATERIALS AND METHODS
Study Population and Samples
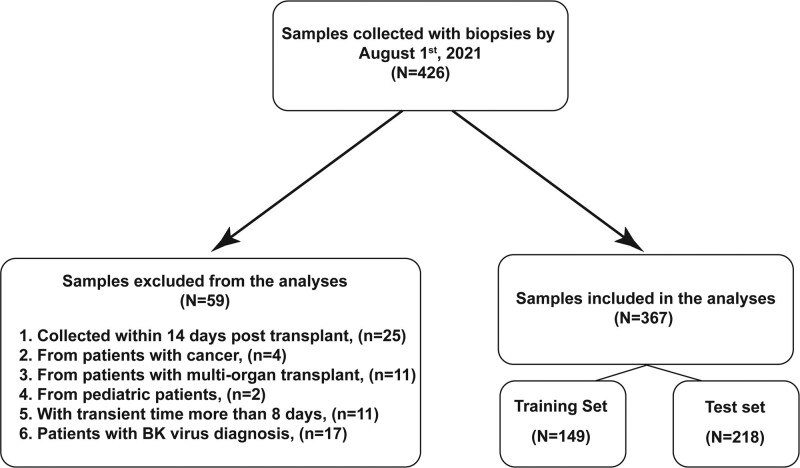
Trifecta is an ongoing, prospective study enrolling consenting KTx recipient patients at >25 participating clinics in Europe and the US (ClinicalTrials.gov Identifier: NCT04239703). The samples used in this manuscript were consecutive samples from the Trifecta study collected between December 1, 2019‚ and July 27, 2021. At the time of indication biopsy, KTx patients had blood drawn for donor-specific antibodies (DSAs) and dd-cfDNA testing. Criteria for sample exclusions from this study were (1) samples from patients with active cancer, (2) samples from patients with multiorgan transplant, (3) samples from pediatric patients, (4) samples drawn <14 d post-KTx, (5) patients with BK virus (polyoma virus nephropathy) (BKV) infection, (6) samples with transit time outside of test specifications (>8 d from blood draw to lab). The remaining samples were then sequentially divided into training and test sets.
KTx Biopsy Processing
The clinical indications and data for renal transplant biopsies for each patient were collected. Biopsies were scored for rejection using both histology per Banff 2019 criteria17 and MMDx. For the histological assessment, local pathologists graded the biopsies for rejection per Banff criteria and the local standard of care. Study case reports were generated by the site investigator using the official biopsy diagnostic reports. Data entered into the case reports were centrally reviewed and updated by transplant nephrologists to confirm the adherence of the findings to the most recent Banff working group classification criteria. MMDx is a central biopsy diagnostic system that measures genome-wide mRNA expression to assign molecular diagnoses and was performed as previously described.11 Briefly, MMDx was performed on a portion of the biopsy sample (mean length 3 mm), stabilized in RNAlater (Life Technologies, Carlsbad, CA), and shipped to the Alberta Transplant Applied Genomics Centre (http://atagc.med.ualberta.ca) at ambient temperature for RNA extraction and processing. All biopsy reads were made while blinded to cfDNA results.
dd-cfDNA Analysis
All blood samples collected for dd-cfDNA testing using the Prospera test (Natera, Inc., San Carlos, CA) were drawn in two 10-mL quantities in DNA Streck tubes and shipped to the processing laboratory. The amplification of cfDNA was performed using massively multiplexed-polymerase chain reaction, targeting 13 926 single-nucleotide polymorphisms designed to maximize the number of informative single-nucleotide polymorphisms across ethnicities followed by next-generation sequencing of the resultant amplicons on the Illumina NextSeq 500 on rapid run with an average of 8 million reads per sample.18 The samples were processed according to standard standard operating procedures used in the Clinical Laboratory Improvement Amendments laboratory responsible for running the Prospera test. For all samples, the dd-cfDNA fraction (analyzed as the percentage of total cfDNA) and quantity (genomic copies per milliliter [cp/mL]) were measured. Blood samples were collected on the same day as or before the allograft biopsy procedure to avoid any confounding effects on dd-cfDNA measurements.
Training of the New Algorithm
The two-threshold algorithm was configured by including a new cutoff based on dd-cfDNA quantity (cp/mL) such that samples that had either ≥1% dd-cfDNA fraction or ≥ the new dd-cfDNA quantity threshold were considered “at increased risk for rejection” for clinical purposes and positive for performance calculation purposes. The numerical cutoff value of the dd-cfDNA quantity was chosen at the inflection point at which the improvement in the sensitivity–specificity trade-off showed diminishing returns, using molecular pathology as the standard comparator.
Statistical Analysis
The area under the receiver operating characteristic curve (AUC) of the combination of dd-cfDNA fraction and quantity was calculated by logistic regression19 using the log-transformed values of these two continuous variables. Likelihood ratio tests were used to compare the performance of the logistic regression model using both variables (dd-cfDNA fraction and quantity) to either dd-cfDNA fraction or quantity alone. Two-tailed P < 0.05 were considered significant. All the statistical analyses were performed in Python 3.8 (https://www.python.org/psf/) and R 4.1 (https://www.r-project.org/) programming languages using SciPy (https://scipy.org/), statsmodels (https://www.statsmodels.org/stable/index.html), scikit-learn version (https://scikit-learn.org/stable/), rms (https://cran.r-project.org/web/packages/rms/index.html), ROCR (https://cran.r-project.org/web/packages/ROCR/readme/README.html),20 and Hmisc packages (https://cran.r-project.org/web/packages/Hmisc/index.html).
Performance Analyses
Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated for dd-cfDNA using the prior algorithm in which samples with ≥1% dd-cfDNA were considered at high risk for rejection and also using the new two-threshold algorithm. AUCs were calculated for individual continuous variables, dd-cfDNA fraction and quantity, from the receiver operating curves using the trapezoidal rule. For the combination of dd-cfDNA fraction and quantity, separate AUCs were calculated for MMDx and histology using scores generated from logistic regression. Each logistic regression model was trained using the respective rejection classification standard.
Performance metrics were calculated separately based on MMDx and histology. Samples signed out as antibody-mediated rejection (AMR), T-cell–mediated rejection (TCMR), or mixed rejection by MMDx were classified as “AR”. All other samples, including those signed out as inactive AMR or probable AMR/TCMR‚ were classified as non-AR. AR in histology was defined as samples classified as TCMR, active AMR, chronic active AMR, or mixed rejection using the Banff 2019 criteria; all other samples were considered to be non-AR. A separate analysis was performed on a subset of samples to test the performance of the two-threshold algorithm in discriminating between biopsies with AR versus quiescence (normal biopsy), with quiescent defined as samples with no major histological abnormalities, no detected DSAs, and nonrejection per MMDx.
RESULTS
Patient Demographics and Study Samples
The Trifecta study enrolled 426 consenting patients between December 1, 2019, and July 27, 2021, at 22 European and US participating clinics. Of the 426 biopsy-matched dd-cfDNA samples drawn before July 27, 2021, 59 were excluded (primarily to simulate the intended clinical use). Exclusions included (1) active cancer patients (N = 4), (2) multiorgan transplant recipients (N = 11), (3) patients with BKV infection (N = 17), (4) pediatric patients (N = 2), (5) samples drawn <14 d post-KTx (N = 25), and (6) samples with transit time >8 d from blood withdraw to laboratory (N = 11) (Figure 1). The remaining biopsy-matched dd-cfDNA samples (N = 367) were included in this analysis and divided sequentially into training set (N = 149 samples) and test set (N = 218 samples).
FIGURE 1.
Study flowchart. BK, polyoma virus.
Table 1 summarizes the population and biopsies, including 229 males and 136 females, with an average weight and height of 78.7 kg (66.7–91.6 kg) and 172.7 cm (165–179 cm), respectively. Seventy-two patients had a prior organ transplant; 45 patients received kidneys from living donors and 304 from deceased donors; 35.4% (79/223) of patients with local DSA results were DSA-positive at the time of biopsy.
TABLE 1.
Patients’ demographic, clinical, and biopsy characteristics
| Criteria | Options | All samples (N = 367) | Training set (N = 149) | Test set (N = 218) |
|---|---|---|---|---|
| Kidney recipients | ||||
| Age, median (IQR) | 52 (39–62) | 52 (40–63.25) | 51 (38–61) | |
| Mean time posttransplant in days (median) | 1440 (616) | 1440 (519) | 1439 (642) | |
| Sex | Male | 229 | 94 | 135 |
| Female | 136 | 54 | 82 | |
| Unknown | 2 | 1 | 1 | |
| Height (cm) | 172.7 | 173.5 | 172 | |
| Median (IQR) | (165–179) | (167–181) | (164–178) | |
| Weight (kg), median (IQR) | 78.7 (66.7–91.6) | 81.0 (65.9–93.0) | 78 (66.9–91.1) | |
| African American ethnicity | Yes | 39 | 3 | 36 |
| No | 325 | 146 | 179 | |
| Unknown | 3 | – | 3 | |
| Prior organ transplants | 72 | 28 | 44 | |
| Primary disease | Chronic pyelonephritis | 15 | 6 | 9 |
| Glomerulonephritis | 102 | 36 | 66 | |
| Diabetes | 41 | 20 | 21 | |
| Unknown cause | 43 | 23 | 20 | |
| Focal glomerulosclerosis | 33 | 11 | 22 | |
| Hypertension | 23 | 10 | 13 | |
| IgA nephropathy | 37 | 17 | 20 | |
| systemic lupus erythematosus nephritis | 12 | 1 | 11 | |
| Polycystic kidney disease | 36 | 18 | 18 | |
| Others | 83 | 13 | 70 | |
| Donor-specific antibodies at blood draw/biopsy | Negative | 144 | 58 | 86 |
| Positive | 116 | 37 | 79 | |
| Unknown | 107 | 54 | 53 | |
| Panel reactive antibodies at blood draw/biopsy | Negative | 52 | 11 | 41 |
| Positive | 84 | 24 | 60 | |
| Unknown | 231 | 114 | 117 | |
| Graft status at last follow-up | Functioning | 299 | 120 | 179 |
| Failed before patient death | 29 | 16 | 13 | |
| Failed after patient death | 3 | 3 | 26 | |
| Unknown | 36 | 7 | – | |
| Kidney donors | ||||
| Age, median (IQR) | Age, median (IQR) | 48 (36–58) | 49 (39.2–60.0) | 46.5 (35.2–55) |
| Sex | Males | 185 | 74 | 111 |
| Females | 159 | 67 | 92 | |
| Unknown | 23 | 8 | 15 | |
| Type | Unrelated | 336 | 137 | 199 |
| Related | 31 | 12 | 19 | |
| Status | Deceased donors | 314 | 130 | 184 |
| Living donors | 53 | 19 | 34 | |
| Molecular and histological diagnoses | ||||
| MMDx diagnosis | No rejection | 242 | 95 | 147 |
| AMR | 89 | 36 | 53 | |
| Mixed | 18 | 8 | 10 | |
| TCMR | 18 | 10 | 8 | |
| Histological diagnosis | AMR | 98 | 40 | 58 |
| TCMR | 32 | 17 | 15 | |
| Mixed | 12 | 2 | 10 | |
| Stable | 56 | 21 | 35 | |
| Other injury | 161 | 66 | 95 | |
- indicates that no samples fit this criteria for this particular cell. AMR, antibody-mediated rejection; IQR, interquartile range; MMDx, molecular microscope diagnostic system; TCMR, T-cell-mediated rejection.
The test data set biopsies were classified as rejection using MMDx and histology assessment. Of 218 biopsies in the test set, 71 AR episodes were identified by MMDx: 53 AMR, 8 TCMR, and 10 mixed rejections; the remaining 147 biopsies were non-AR biopsies. Histology assessment of 213 biopsies with valid diagnoses in the test set diagnosed 83 with AR: 58 AMR, 15 TCMR, and 10 mixed rejections. Of the remaining 130 non-AR biopsies, 35 were relatively normal, 21 were borderline, and 74 were “other injury”, including interstitial fibrosis and atrophy, recurrent disease, and acute tubular injury.
Assessment of Predictive Power of dd-cfDNA Fraction and Quantity
In the full cohort, we evaluated logistic regression models based on the continuous dd-cfDNA fraction, quantity, and the combination of the two. By comparing likelihood ratios, the two variables combined resulted in a significantly better model than either dd-cfDNA fraction (P < 0.0001) or dd-cfDNA quantity alone (P = 0.009). Therefore, the model combining both variables was superior to using either variable alone.
Using logistic regression to predict histological rejection in the entire dataset (N = 359, 8 of the 367 lack histological diagnoses), adding quantity to fraction improved the model, P = 3.7 × 10−5 (likelihood ratio test), whereas adding fraction to quantity was not significant (P = 0.10).
Selecting a dd-cfDNA Quantity Cutoff for the New Two-threshold Algorithm
In the training set, using MMDx to determine rejection status, we selected an optimal dd-cfDNA quantification cutoff of ≥78 cp/mL for the new two-threshold algorithm (Figure S1, SDC, http://links.lww.com/TP/C457). This new algorithm considered samples either the previously validated ≥1% dd-cfDNA cutoff or the ≥78 cp/mL quantity dd-cfDNA cutoff as rejecting, or for clinical purposes, as “at-risk for rejection.” In the training set, the two-threshold algorithm showed a sensitivity of 81.5% and specificity of 77.9%. For comparison, a sensitivity of 74.1% and specificity of 78.9% were calculated using the 1% dd-cfDNA fraction cutoff alone. Four rejection samples classified as not rejecting using the 1% threshold were newly interpreted as rejecting by the new algorithm.
Performance of dd-cfDNA in an Independent Test Set
The test set was used to validate the performance of the newly developed two-threshold algorithm using either MMDx or histology as comparator (Table 2). Using MMDx as comparator, the new two-threshold algorithm had a sensitivity of 83.1%, specificity of 81.0%, PPV of 67.8%, and NPV of 90.8%, with a 32.6% prevalence of AR in the test set. For comparison, when using histology as a comparator, the two-threshold algorithm showed a sensitivity of 73.5%, specificity of 80.8%, PPV of 70.9%, and NPV of 82.7%, with 39.0% prevalence of AR.
TABLE 2.
Performance of the two-threshold algorithm in the test set
| Two-threshold algorithm performance in the test cohort | ||||
|---|---|---|---|---|
| Comparator | Sensitivity (%; X/X) | Specificity (%; X/X) | PPV (%) | NPV (%) |
| Molecular pathology (N = 218) | 83.1 | 81.0 | 67.8 | 90.8 |
| 59/71 | 119/147 | 59/87 | 119/131 | |
| Histology (N = 213) | 73.5 | 80.8 | 70.9 | 82.7 |
| 61/83 | 105/130 | 61/86 | 105/127 | |
X/X indicates the table entries in that column provide a percentage, as well as a whole number fraction. NPV, negative predictive value; PPV, positive predictive value.
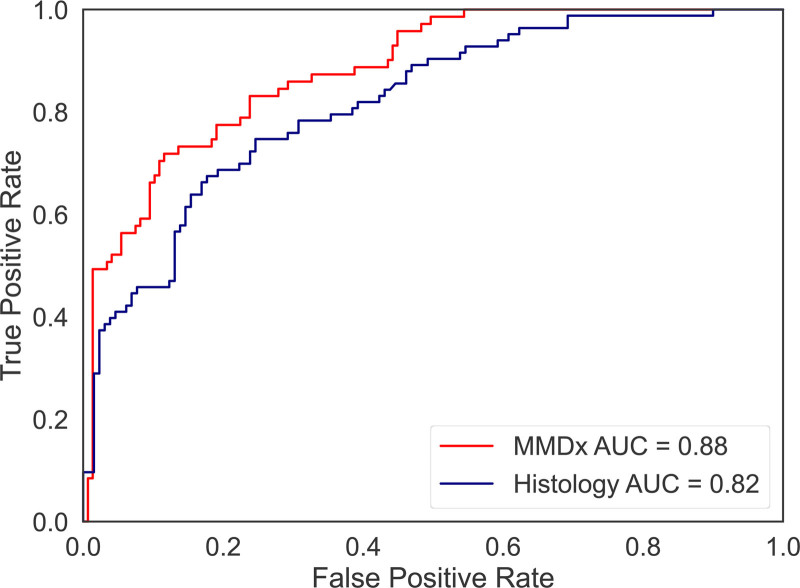
The logistic regression model built in the training set by combining the dd-cfDNA fraction and quantity to predict rejection produced AUCs of 0.88 and 0.82 for molecular and histological rejection, respectively, in the test set (Figure 2). The stronger correlation of dd-cfDNA with molecular rejection versus histological rejection is consistent with previous findings.9,21
FIGURE 2.
receiver operating characteristic curves predicting molecular and histological rejection in the test set, based on test set predictions from the logistic regression model built using dd-cfDNA fraction and quantity in the training set. MMDx shows better performance than traditional histology in rejection diagnosis of KTx. AUC, area under the receiver operating characteristic curve; dd-cfDNA, donor-derived cell-free DNA; KTx, kidney transplant; MMDx, molecular microscope diagnostic system.
Table S1, SDC, http://links.lww.com/TP/C457, shows the performance characteristics of both the two-threshold algorithm across the training and test and combined cohorts, compared with both histology and molecular pathology. Table S2, SDC, http://links.lww.com/TP/C457, shows the PPV and NPV for the test set projected to different cohort AR prevalences, using molecular pathology as a comparator. Note that Trifecta is an indication biopsy cohort with 32% incidence of rejection. If applied to populations with lower incidence of rejection, the PPV is reduced as expected.
In the test set, two additional molecular rejection samples were correctly identified using the two-threshold algorithm that were missed when solely using a 1% cutoff. Furthermore, using the new two-threshold algorithm and comparing it with molecular pathology in the test set, we found that the sensitivity for AMR, TCMR, and mixed rejection detection was 81.1% (43/53), 100% (8/8), and 80% (8/10), respectively (Figure 3, Figure S2B, SDC, http://links.lww.com/TP/C457, and Table 3); when comparing with histology, these sensitivities were 77.6% (45/58), 53.3% (8/15), and 80% (8/10), respectively (Figure S3, SDC, http://links.lww.com/TP/C457). In the entire dataset (N=367), the proportion of molecular TCMRs missed by the two-threshold method was slightly higher than the proportion of AMRs missed (22% versus 17%), but this difference was not significant (P = 0.59).
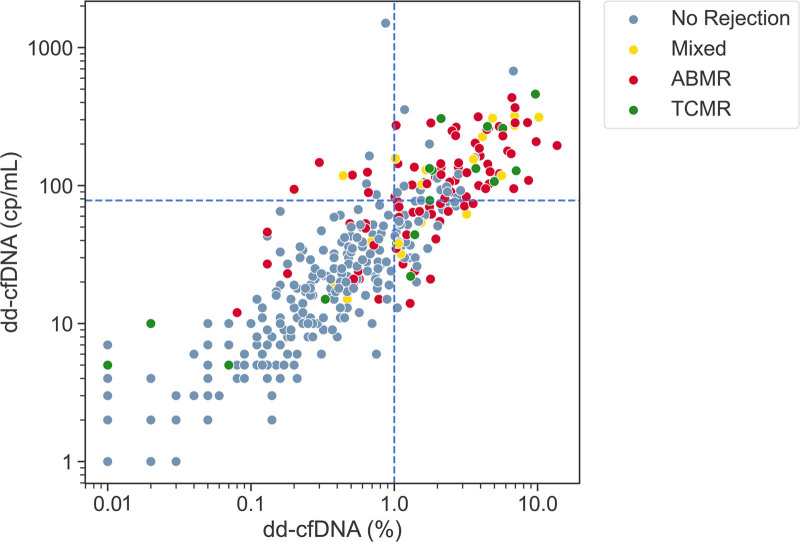
FIGURE 3.
Plot of dd-cfDNA fraction (%) and quantity (cp/mL) for the full cohort (n = 367). The blue dashed horizontal and vertical lines indicate the dd-cfDNA quantity (78 cp/mL) and fraction (1%) thresholds, respectively. Patients with biopsy-proven rejection: AMR, TCMR, and Mixed, as adjudicated by MMDx, are depicted as red, green and yellow dots, respectively. Patients with biopsies that show nonrejection are represented by gray dots. The two-threshold algorithm considers samples in the lower-left quadrant as low risk for rejection, and samples in the remaining 3 quadrants, those with either dd-cfDNA quantity or fraction above the relevant thresholds, as high risk for rejection. AMR, antibody-mediated rejection; dd-cfDNA, donor-derived cell-free DNA; MMDx, molecular microscope diagnostic system; TCMR, T-cell-mediated rejection.
TABLE 3.
The sensitivity of the two-threshold algorithm for detecting rejection types as determined by both MMDx and histology in the test set
| Diagnostic modality | Sample set | Two-threshold algorithm |
|---|---|---|
| MMDx | Test set (n = 218) | Sensitivity (%; X/X) |
| TCMR | 100 | |
| 8/8 | ||
| AMR | 81.1 | |
| 43/53 | ||
| Mixed | 80 | |
| 8/10 | ||
| Banff histology | Test set (n = 213) | Sensitivity (%; X/X) |
| TCMR | 53.3 | |
| 8/15 | ||
| AMR | 77.6 | |
| 45/58 | ||
| Mixed | 80 | |
| 8/10 |
X/X indicates the table entries in that column provide a percentage, as well as a whole number fraction. AMR, antibody-mediated rejection; MMDx, molecular microscope diagnostic system; TCMR, T-cell-mediated rejection.
In addition, a separate logistic regression with 10-fold cross-validation showed that using the combination of two variables discriminated between AR (n = 142) and quiescent samples (n = 41) with an AUC of 0.91. We provide this subcohort analysis because a similar approach has been used in a number of recent publications9,19,22–23: excluding samples with injury but not biopsy-proven rejection demonstrates the clear differentiation power of dd-cfDNA with respect to these two states. We note that because such cohorts exclude abnormal pathologies only detectable by biopsy, these analyses are not applicable to a clinical cohort.
Relating dd-cfDNA to Concordance Between Molecular and Histological Diagnoses
We assessed %dd-cfDNA or quantity of dd-cfDNA in instances in which either molecular or histology agreed (molecular and histological rejections or molecular and histological no rejections) and contrasted these scenarios against those cases in which molecular and histological rejections disagreed (Table 4). In cases of discrepancies, dd-cfDNA agrees more with MMDx than with histology, both by %dd-cfDNA and by the quantity of dd-cfDNA.
Table 4.
Geometric means of percentage dd-cfDNA and quantity of cfDNA when MMDx and histology diagnostic calls agree vs disagree (N = 359)
| MMDx call | Central histology call | Mean %dd-cfDNA | Mean quantity of dd-cfDNA |
|---|---|---|---|
| No rejection | No rejection | 0.27 | 15.6 |
| No rejection | Rejection | 0.52 | 30.8 |
| Rejection | No rejection | 0.87 | 46.5 |
| Rejection | Rejection | 1.78 | 91.7 |
dd-cfDNA, donor-derived cell-free DNA; MMDx, molecular microscope diagnostic system.
dd-cfDNA and Time Posttransplant
The dd-cfDNA is somewhat higher in the first weeks posttransplant, then declines, then increases over time posttransplant, as reported in our first Trifecta analysis.9 The first 13 d are excluded in the present population, but allowing for that, we are finding similar relationships to time in this expanded population. Most of the increase with time is due to rejection, which tends to occur late.
Within the test set, we examined 3 time periods and found that the predictive accuracy did not change appreciably over time: <6 mo (0.821), 6–12 mo (0.813), and >12 mo (0.815).
DISCUSSION
This prospective, biopsy-matched study aimed to determine the performance of dd-cfDNA in distinguishing AR from non-AR in KTx patients. This study represents a comprehensive analysis of the suggestion in a preliminary report from another population that the combination of dd-cfDNA fraction and quantity might improve the detection of rejection.16 The present analysis developed and validated a novel two-threshold algorithm incorporating both variables, using two diagnostic modalities as a comparator: traditional histology, scored using Banff 2019 criteria, and molecular analysis as recently used in Trifecta.9 We assessed the relative predictive power of the combination of dd-cfDNA fraction and quantity to each of these variables alone. We found that the combination of dd-cfDNA fraction and dd-cfDNA quantity was a significantly better predictor of rejection in KTx patients than either of the two variables by comparing the likelihood ratios of the models. The improved likelihood ratio from the model using both variables infers a greater posttest probability of rejection status and utility of the test to support an accurate diagnosis. We hypothesize the different information embedded in each variable potentially leads to the odds improvement.22 In particular, we expect dd-cfDNA quantity to be more sensitive to kidney rejection in cases in which significant systemic inflammation causes high total cfDNA levels, whereas we expect dd-cfDNA fraction to be more sensitive in cases in which organ rejection is the primary driver of inflammation in the body. This approach also follows the general principle that “ensembles” of estimates (even if correlated) are usually more accurate than single estimates.
The two-threshold algorithm was based on our observation in prior datasets that when using 1% dd-cfDNA as a cutoff, false negatives were overrepresented with samples with high dd-cfDNA quantity (Figure 3, upper left quadrant). To correctly classify these samples as high risk, we added a second threshold based on dd-cfDNA quantity, determined in the training set of this study to be 78 cp/mL. In the training set, implementation of the two-threshold algorithm detected 4 additional rejections, resulting in a 7.4% improvement in sensitivity along with a 1.0% decrease in specificity. In the full cohort, this quantification-based two-threshold algorithm detected 6 additional cases of rejection compared with when using the 1% cutoff alone. The dd-cfDNA result serves as a screening test but does not distinguish between AMR and TCMR and is not adequate on its own for determining the treatment of these major conditions without a biopsy in most circumstances.
The strengths of this study are the prospective study design, the large number of biopsy-matched samples, and the large number of sites involved, both in the US and internationally. The study was adequately powered to demonstrate that the combination of dd-cfDNA fraction and quantity is significantly better than either variable alone. We believe that AUC/receiver operating characteristic and, particularly, PPV/NPV should not be compared across different protocols and populations, and diagnostic criteria. For the record and with this caveat, we do include some published data in Table S3, SDC, http://links.lww.com/TP/C457. Moreover, this study used two different orthogonal methods for diagnosing rejection, molecular pathology, and histology. This was especially important as recent studies have indicated that molecular pathology provides a more consistent representation of rejection than histology.11,23
In addition, this study involved an analysis of sequential samples from a multicenter study, with no exclusions other than those relevant to the intended use: (1) patients who were not part of the intended use population (<14 d post-Tx, pediatric patients, multiorgan transplant patients, and cancer patients), (2) samples that were insufficient for analysis (not biopsy-matched or in transit for >8 d), and (3) patients with complicating conditions that are easily detectable with other noninvasive means (eg, BKV infection). Samples were not excluded based on the presence of rejection only detectable by biopsy. The result is a cohort from which clinically useful performance measurements could be calculated.
The study had several limitations. Our algorithm training only allowed variation of one of the two metrics (dd-cfDNA quantity), whereas the other was held constant (dd-cfDNA fraction, at 1%). Also, all biopsies in this study were indication biopsies, and thus, the performance of the two-threshold algorithm in protocol surveillance biopsies was not assessed. A previous study demonstrated the performance of this two-threshold algorithm in a clinical cohort that included the use of the assay in a surveillance setting16 and another study showed that dd-cfDNA fraction performs similarly in surveillance and indication biopsies. Given the high time-dependency of the rejection states, the application of these results to protocol surveillance biopsies cannot be assessed without knowing the details of the protocol and patient selection, which differs from center to center. We suggest that the present results anticipate the findings that will be obtained in protocol biopsies in terms of the relationship between quantity and fraction of dd-cfDNA in the rejection states, once the prior probabilities of rejection in the population being tested are taken into account.
In conclusion, the combination of dd-cfDNA fraction and quantity as continuous variables is a better predictor of molecular rejection in KTx patients than either variable individually. Moreover, we validated a two-threshold algorithm for discriminating AR from non-AR using dd-cfDNA in KTx patients in a large biopsy-matched cohort. The two-threshold algorithm performed well within a population of indication biopsies. It is clear that considering fraction plus quantity offers only incremental improvement, but this increment is free because the test already provides both variables and, in our opinion, is worth considering by the clinician.
Acknowledgments
*The Trifecta Investigators are Justyna Fryc, Beata Naumnik, Jonathan Bromberg, Matt Weir, Nadiesda Costa, Daniel Brennan, Sam Kant, Vignesh Viswanathan, Milagros Samaniego-Picota, Iman Francis, Anita Patel, Alicja Dębska-Ślizień, Joanna Konopa, Andrzej Chamienia, Andrzej Więcek, Grzegorz Piecha, Željka Veceric-Haler, Miha Arnol, Nika Kojc, Maciej Glyda, Katarzyna Smykal-Jankowiak, Ondrej Viklicky, Petra Hruba, Silvie Rajnochová Bloudíčkova, Janka Slatinská, Marius Miglinas, Marek Myślak, Joanna Mazurkiewicz, Marta Gryczman, Leszek Domański, Mahmoud Kamel, Agnieszka Perkowska-Ptasińska, Dominika Dęborska-Materkowska, Michal Ciszek, Magdalena Durlik, Leszek Pączek, Ryszard Grenda, Miroslaw Banasik, Mladen Knotek, Ksenija Vucur, Zeljka Jurekovic, Thomas Müller, Thomas Schachtner, Andrew Malone, Tarek Alhamad, Arksarapuk Jittirat, Emilio Poggio, Richard Fatica, Ziad Zaky, Kevin Chow, Peter Hughes, Sanjiv Anand, Gaurav Gupta, Layla Kamal, Dhiren Kumar, Irfan Moinuddin, and Sindhura Bobba.
The authors would like to thank Ashish Solanki and the rest of the valued clinicians in the Trifecta study group who partnered with us for this study by contributing biopsies and feedback. The authors would also like thank Dr Martina Mackova and Mrs Anna Hutton for biopsy processing for the microarray biopsy assessment component of this study and Natera Inc. for blood sample processing and Prospera results.
Supplementary Material
Footnotes
Clinical trial investigation number: NCT04239703
P.F.H. reports having shares in Transcriptome Sciences Inc, a University of Alberta research company with an interest in molecular diagnostics, and is a consultant to Natera. Inc. K.S.M.-T. is an employee of Transcriptome Sciences Inc. A.P. reports current employment with, and an ownership interest in, Natera, Inc, N.K., E.A., C.C., N.A.H.B., Z.D., N.L., R.K.S., B.G.Z., P.V.H., M.R., H.T., and P.G. are all employees of Natera, Inc, with stocks or options to buy stocks in the company. The other authors declare no conflicts of interest.
The Trifecta study is an investigator-initiated study supported by a grant from Natera to Transcriptome Sciences Inc/Alberta Transplant Applied Genomics Centre. The Microarray biopsy assessment project is supported in part by a licensing agreement with One Lambda/Thermo Fisher Scientific.
P.F.H. was the principal investigator, edited and reviewed the manuscript, and was responsible for data interpretation and study design. J.R. and K.S.M.-T. edited and reviewed the manuscript, and were responsible for data analysis and interpretation. N.K. analyzed data and helped write the first draft and reviewed and edited the final manuscript. E.A. trained the algorithm, analyzed data, and helped editing the manuscript. C.C. trained the algorithm, and analyzed data. B.G.Z., P.V.H., N.L., and R.K.S. were responsible for measurement of dd-cfDNA % (the Prospera test), M.R. contributed to technology development, N.A.H.B. drafted the original manuscript, reviewed and edited the final draft. Z.D. and A.P. were responsible for manuscript preparation, discussion of results, and interpretation and draft preparation and editing. H.T. and P.G. helped in data interpretation and discussion. The Trifecta Investigators were responsible for biopsy and blood sample collection.
CEL files will be available on Gene Expression Omnibus upon publication.
Supplemental Visual Abstract; http://links.lww.com/TP/C458.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
Contributor Information
Collaborators: Justyna Fryc, Beata Naumnik, Jonathan Bromberg, Matt Weir, Nadiesda Costa, Daniel Brennan, Sam Kant, Vignesh Viswanathan, Milagros Samaniego-Picota, Iman Francis, Anita Patel, Alicja Dębska-Ślizień, Joanna Konopa, Andrzej Chamienia, Andrzej Więcek, Grzegorz Piecha, Željka Veceric-Haler, Miha Arnol, Nika Kojc, Maciej Glyda, Katarzyna Smykal-Jankowiak, Ondrej Viklicky, Petra Hruba, Silvie Rajnochová Bloudíčkova, Janka Slatinská, Marius Miglinas, Marek Myślak, Joanna Mazurkiewicz, Marta Gryczman, Leszek Domański, Mahmoud Kamel, Agnieszka Perkowska-Ptasińska, Dominika Dęborska-Materkowska, Michal Ciszek, Magdalena Durlik, Leszek Pączek, Ryszard Grenda, Miroslaw Banasik, Mladen Knotek, Ksenija Vucur, Zeljka Jurekovic, Thomas Müller, Thomas Schachtner, Andrew Malone, Tarek Alhamad, Arksarapuk Jittirat, Emilio Poggio, Richard Fatica, Ziad Zaky, Kevin Chow, Peter Hughes, Sanjiv Anand, Gaurav Gupta, Layla Kamal, Dhiren Kumar, Irfan Moinuddin, and Sindhura Bobba
References
- 1.Beck J, Bierau S, Balzer S, et al. Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin Chem. 2013;59:1732–1741. [DOI] [PubMed] [Google Scholar]
- 2.De Vlaminck I, Valantine HA, Snyder TM, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med. 2014;6:241ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigdel TK, Archila FA, Constantin T, et al. Optimizing detection of kidney transplant injury by assessment of donor-derived cell-free DNA via massively multiplex PCR. J Clin Med. 2018;8:E19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filippone EJ, Farber JL. The monitoring of donor-derived cell-free DNA in kidney transplantation. Transplantation. 2021;105:509–516. [DOI] [PubMed] [Google Scholar]
- 5.Qazi Y, Patel A, Fajardo M, et al. Incorporation of donor-derived cell-free DNA into clinical practice for renal allograft management. Transplant Proc. 2021;53:2866–2872. [DOI] [PubMed] [Google Scholar]
- 6.Roufosse C, Simmonds N, Clahsen-van Groningen M, et al. A 2018 reference guide to the Banff classification of renal allograft pathology. Transplantation. 2018;102:1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furness PN, Taub N, Assmann KJ, et al. International variation in histologic grading is large, and persistent feedback does not improve reproducibility. Am J Surg Pathol. 2003;27:805–810. [DOI] [PubMed] [Google Scholar]
- 8.Loupy A, Haas M, Solez K, et al. The Banff 2015 kidney meeting report: current challenges in rejection classification and prospects for adopting molecular pathology. Am J Transplant. 2017;17:28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halloran PF, Reeve J, Madill-Thomsen KS, et al. ; Trifecta Investigators. The Trifecta study: comparing plasma levels of donor-derived cell-free DNA with the molecular phenotype of kidney transplant biopsies. J Am Soc Nephrol. 2022;33:387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halloran PF, Famulski K, Reeve J. The molecular phenotypes of rejection in kidney transplant biopsies. Curr Opin Organ Transplant. 2015;20:359–367. [DOI] [PubMed] [Google Scholar]
- 11.Halloran PF, Reeve J, Akalin E, et al. Real time central assessment of kidney transplant indication biopsies by microarrays: the INTERCOMEX study. Am J Transplant. 2017;17:2851–2862. [DOI] [PubMed] [Google Scholar]
- 12.Reeve J, Böhmig GA, Eskandary F, et al. ; INTERCOMEX MMDx-Kidney Study Group. Generating automated kidney transplant biopsy reports combining molecular measurements with ensembles of machine learning classifiers. Am J Transplant. 2019;19:2719–2731. [DOI] [PubMed] [Google Scholar]
- 13.Oellerich M, Shipkova M, Asendorf T, et al. Absolute quantification of donor-derived cell-free DNA as a marker of rejection and graft injury in kidney transplantation: results from a prospective observational study. Am J Transplant. 2019;19:3087–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitlam JB, Ling L, Skene A, et al. Diagnostic application of kidney allograft-derived absolute cell-free DNA levels during transplant dysfunction. Am J Transplant. 2019;19:1037–1049. [DOI] [PubMed] [Google Scholar]
- 15.Osmanodja B, Akifova A, Budde K, et al. Absolute or relative quantification of donor-derived cell-free DNA in kidney transplant recipients: case series. Transplant Direct. 2021;7:e778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bunnapradist S, Homkrailas P, Ahmed E, et al. Using both the fraction and quantity of donor-derived cell-free DNA to detect kidney allograft rejection. J Am Soc Nephrol. 2021;32:2439–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loupy A, Haas M, Roufosse C, et al. The Banff 2019 kidney meeting report (I): updates on and clarification of criteria for T cell- and antibody-mediated rejection. Am J Transplant. 2020;20:2318–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altuğ Y, Liang N, Ram R, et al. Analytical validation of a single-nucleotide polymorphism-based donor-derived cell-free DNA assay for detecting rejection in kidney transplant patients. Transplantation. 2019;103:2657–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park S, Guo K, Heilman RL, et al. Combining blood gene expression and cellfree DNA to diagnose subclinical rejection in kidney transplant recipients. Clin J Am Soc Nephrol. 2021;16:1539–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sing T, Sander O, Beerenwinkel N, et al. ROCR: visualizing classifier performance in R. Bioinform. 2005;21:3940–3941. [DOI] [PubMed] [Google Scholar]
- 21.Gupta G, Moinuddin I, Kamal L, et al. Correlation of donor-derived cell-free DNA with histology and molecular diagnoses of kidney transplant biopsies. Transplantation. 2022;106:1061–1070. [DOI] [PubMed] [Google Scholar]
- 22.John GE. Clinical utility of liklihood ratios. Ann Emerg Med. 1998;31:391–397. [DOI] [PubMed] [Google Scholar]
- 23.Halloran PF. Integrating molecular and histologic interpretation of transplant biopsies. Clin Transplant. 2021;35:e14244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.