Abstract
Transgenerational epigenetic inheritance (TEI) is mostly discussed in the context of physiological or environmental factors. Here, we show intergenerational and transgenerational inheritance of transcriptional adaptation (TA), a process whereby mutant messenger RNA (mRNA) degradation affects gene expression, in nematodes and zebrafish. Wild-type offspring of animals heterozygous for mRNA-destabilizing alleles display increased expression of adapting genes. Notably, offspring of animals heterozygous for nontranscribing alleles do not display this response. Germline-specific mutations are sufficient to induce TA in wild-type offspring, indicating that, at least for some genes, mutations in somatic tissues are not necessary for this process. Microinjecting total RNA from germ cells of TA-displaying heterozygous zebrafish can trigger TA in wild-type embryos and in their progeny, suggesting a model whereby mutant mRNAs in the germline trigger a TA response that can be epigenetically inherited. In sum, this previously unidentified mode of TEI reveals a means by which parental mutations can modulate the offspring’s transcriptome.
Ancestral mutations can influence the transcriptome of wild-type offspring in C. elegans and zebrafish.
INTRODUCTION
Inheritance is the process by which traits or information is transmitted from one generation to the next. The classical view of genetic inheritance involves the transmission of DNA from parents to their offspring (1). However, a number of studies have reported examples of heritable phenotypic variations that cannot be explained solely by the genetic makeup of the parents (2–4). Multigenerational “nongenetic” inheritance in mammals can refer to bioactive substances, such as hormones and cytokines, being passed across generations without the involvement of the gametes (5); however, it more commonly refers to intergenerational epigenetic inheritance (IEI) or transgenerational epigenetic inheritance (TEI), which describe the transmission of epigenetic information from the gametes to the zygote (3, 6). By definition, epigenetic inheritance occurs independently of the progeny’s genomic sequence (5). When the effects can be detected in the second and subsequent generations after the initial trigger (i.e., P0 to F1 to F2 and so on), this phenomenon is referred to as transgenerational inheritance; when the inheritance mode only lasts for one generation, it is referred to as intergenerational inheritance (i.e., P0 to F1) (7). Nevertheless, several studies have suggested that intergenerational and transgenerational effects share underlying mechanisms [reviewed in (7–9)]. A range of studies have provided evidence that physiological and environmental factors can modulate the inheritance of transgenerational traits through epigenetic mechanisms including DNA methylation, histone modifications, and noncoding RNAs (8, 10–15). Indeed, RNA-induced epigenetic modifications are now a well-documented phenomenon (16–18). Intriguingly, several studies in mouse have reported heterozygous traits in wild-type offspring of heterozygous parents (19, 20), a phenomenon attributed to paramutations, i.e., interactions between two alleles at a single locus whereby one allele induces a heritable change in the other allele. While studies reporting paramutations in plants (i.e., originally in maize) (21), Caenorhabditis elegans (22), and Drosophila melanogaster (23) suggest that parental genotype can influence the offspring’s traits independent of the offspring’s genotype, the underlying mechanisms remain largely unknown. Despite the increased discussion of the role of physiologically and environmentally induced IEI/TEI and their potential influence on genetic robustness and plasticity (3, 11, 15, 24–27), it remains unclear how often inter-/transgenerational effects are adaptive. Furthermore, the potential role of IEI/TEI to achieve robustness in the face of parental genetic alterations has not yet been reported.
Transcriptional adaptation (TA), a newly identified cellular response to some genetic perturbations, refers to the phenomenon whereby a mutation in one gene triggers the transcriptional modulation of other genes, termed adapting genes (28–30). In zebrafish, in mouse cells in culture, and in C. elegans, mutations leading to mutant mRNA degradation, including those leading to a premature termination codon (PTC), can induce the up-regulation of adapting genes (28–30), whereas mutations leading to the absence of mRNA (i.e., promoter deletions or full-locus deletions that lead to nontranscribing, or RNA-less, alleles) do not (28, 30), indicating that transcripts from the mutated locus are required for TA (28–30). Further data indicate that mutant mRNA degradation is required for TA (28, 30). Changes in chromatin modifications have also been observed during the TA response, as increased levels of the H3K4me3 histone mark were detected at the promoter region of the adapting genes in a mouse cell line TA model (28) and in a zebrafish TA model (29), suggesting that chromatin alteration is involved in the up-regulation of the adapting genes. Thus, the mutant mRNA (29), its degradation products (28), or their derivatives, could lead to a modified chromatin state at the adapting gene loci.
In this study, we investigated how a mutation in an ancestral genome can influence the offspring’s transcriptional landscape and thus possibly promote genetic robustness. We found that wild-type offspring of TA-displaying heterozygous animals exhibit increased expression of the adapting genes and term this phenomenon intergenerational and transgenerational inheritance of TA (IGTA and TGTA). TGTA in zebrafish can be observed for at least two generations and in C. elegans for at least nine generations. Microinjections into one-cell stage zebrafish embryos of total RNA from germ cells of TA-displaying heterozygotes can induce TA and IGTA. These data suggest that germline transmission of RNA, potentially the mutant mRNA and/or its degradation products (or their derivatives), can underlie the inheritance of TA. We also observed TA in the wild-type progeny of animals with a germline-specific aldh1a2 PTC mutation, further highlighting the key role of the germline in IGTA.
RESULTS
Transgenerational inheritance of the transcriptional adaptation response in C. elegans and zebrafish
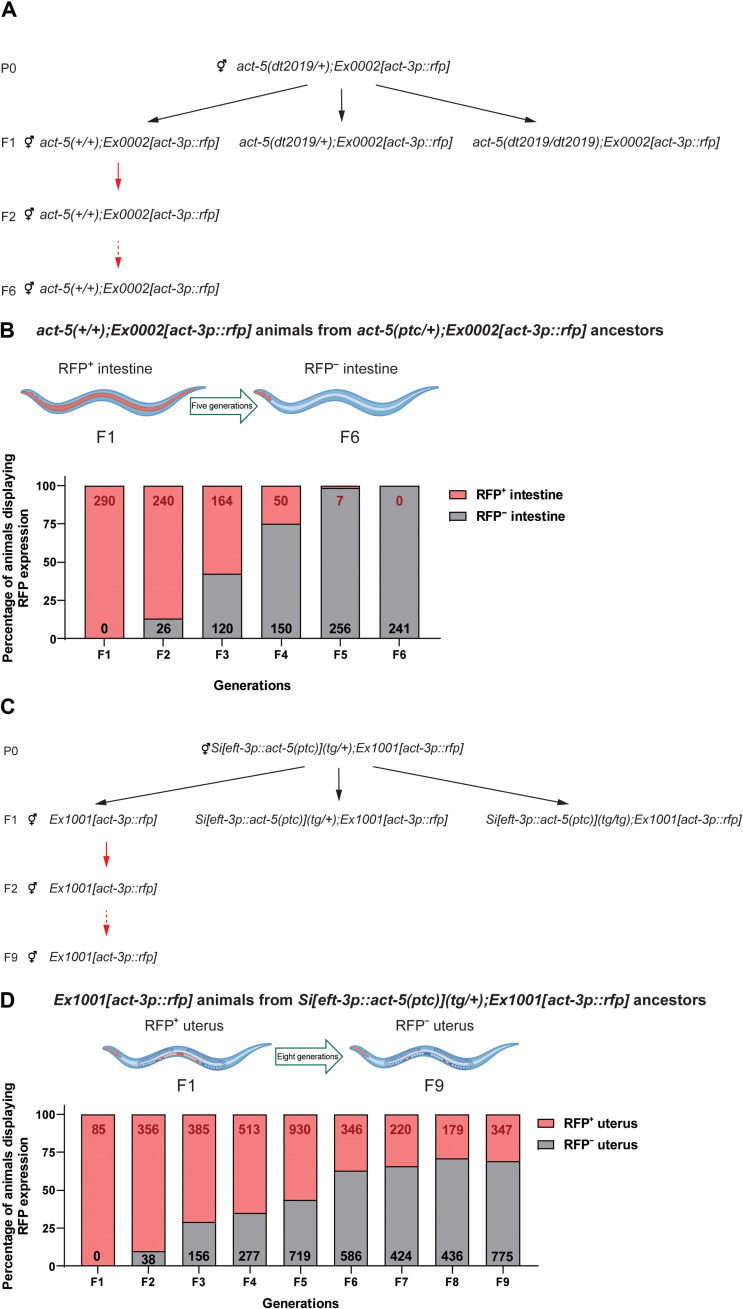
We previously reported that, in C. elegans, act-3 mRNA levels were up-regulated in act-5(dt2019) mutants, hereafter referred to as act-5(ptc) (30). In addition, the expression pattern of an extrachromosomal act-3p::rfp reporter (i.e., a 4.5-kb act-3 enhancer/promoter fragment driving rfp expression) in act-5(ptc) mutants was altered compared with that in wild-type nematodes or in an act-5(deletion) mutant allele that does not display TA (30). We observed red fluorescent protein (RFP) expression in the pharynx in act-5(+/+);Ex[act-3p::rfp] nematodes and in the pharynx and intestine in act-5(ptc/ptc);Ex[act-3p::rfp] nematodes (30). As we attempted to use this phenotype [RFP expression in the intestine of act-5(ptc) mutants] in a forward genetic screen for regulators of TA, we observed that 100% of the act-5(+/+);Ex[act-3p::rfp] offspring from act-5(ptc) heterozygous nematodes displayed intestinal RFP expression (Fig. 1, A and B). To further analyze the inheritance mode of this phenotype, we sequentially self-fertilized wild-type hermaphrodite offspring from act-5(ptc) heterozygous nematodes and observed the intestinal expression of the act-3p::rfp reporter up to six generations (Fig. 1B).
Fig. 1. TGTA in C. elegans.
(A) Genetic crosses used to obtain Ex0002[act-3p::rfp] wild-type offspring from act-5(dt2019);Ex0002[act-3p::rfp] heterozygous nematodes. Black arrows indicate the self-fertilization of hermaphrodites; red arrows indicate subsequent wild-type self-crosses. (B) Percentage of adult Ex0002[act-3p::rfp] wild-type nematodes displaying ectopic red fluorescent protein (RFP) expression in their intestine through six generations from act-5(dt2019/+);Ex0002[act-3p::rfp] ancestors. Red numbers indicate the total number of Ex0002[act-3p::rfp] wild-type nematodes displaying ectopic RFP expression in their intestine in each generation; black numbers indicate the total number of Ex0002[act-3p::rfp] wild-type nematodes displaying pharynx-only RFP expression in each generation. (C) Genetic crosses used to obtain Ex1001[act-3p::rfp] offspring from Si[eft-3p::act-5(ptc)](tg/+);Ex1001[act-3p::rfp] nematodes. Black arrows indicate the self-fertilization of hermaphrodites; red arrows indicate subsequent wild-type self-crosses. (D) Percentage of adult Ex1001[act-3p::rfp] wild-type nematodes displaying ectopic RFP expression in their uterus through nine generations from Si[eft-3p::act-5(ptc)](tg/+);Ex1001[act-3p::rfp] ancestors. Red numbers indicate the total number of Ex1001[act-3p::rfp] wild-type nematodes displaying ectopic RFP expression in their uterus in each generation; black numbers indicate the total number of Ex1001[act-3p::rfp] wild-type nematodes displaying only pharynx and spermatheca RFP expression (fig. S1D) in each generation. Illustrations were created with BioRender.com.
In addition, we generated an allele (Si[eft-3p::act-5(ptc)]) that expresses an act-5(ptc) transgene under the control of an eft-3 promoter via Mos1-mediated single-copy insertion (31). We observed increased act-3 mRNA levels in these transgenic animals (fig. S1A), and when we generated an Ex[act-3p::rfp] reporter line in this background (i.e., Si[eft-3p::act-5(ptc)](tg/tg);Ex[act-3p::rfp]), we observed ectopic RFP expression in their uterus, a tissue that expresses eft-3 (32). These data indicate that overexpressing an act-5(ptc) transgene can trigger TA. Similar to our observations with act-5(+/+);Ex[act-3p::rfp] nematodes, we observed that 100% of the wild-type Ex[act-3p::rfp] offspring from Si[eft-3p::act-5(ptc)](tg/+);Ex[act-3p::rfp] nematodes displayed ectopic RFP expression in their uterus (fig. S1, B and C), and this phenotype was observed across nine generations (Fig. 1, C and D). Together, these data show that the TA-induced ectopic RFP expression in both act-5(ptc);Ex[act-3p::rfp] and Si[eft-3p::act-5(ptc)](tg/+);Ex[act-3p::rfp] nematodes is inherited transgenerationally, revealing TGTA in C. elegans.
In zebrafish, the previously reported alcama, vcla, and egfl7 PTC-bearing mutants exhibit a decrease in mutant mRNA levels and an increase in mRNA levels of the adapting genes, namely, alcamb, vclb, and emilin2a/emilin3a, respectively (28, 33). In addition to these published models, we also introduce here an aldh1a2 PTC-containing allele (34) as another TA model that exhibits decreased mutant mRNA levels and increased aldh1a3 mRNA levels (fig. S2A). Injection of wild-type aldh1a2 mRNA into aldh1a2 mutants did not affect the up-regulation of aldh1a3 (fig. S2B), indicating that it is triggered independently of the loss of Aldh1a2 protein and thus a TA model. In addition, a similar increase in aldh1a3 mRNA levels was observed in aldh1a2floxed/+ embryos upon Cre mRNA-mediated recombination (fig. S2, C and D), which leads to the same PTC as in the aldh1a2ptc allele (34).
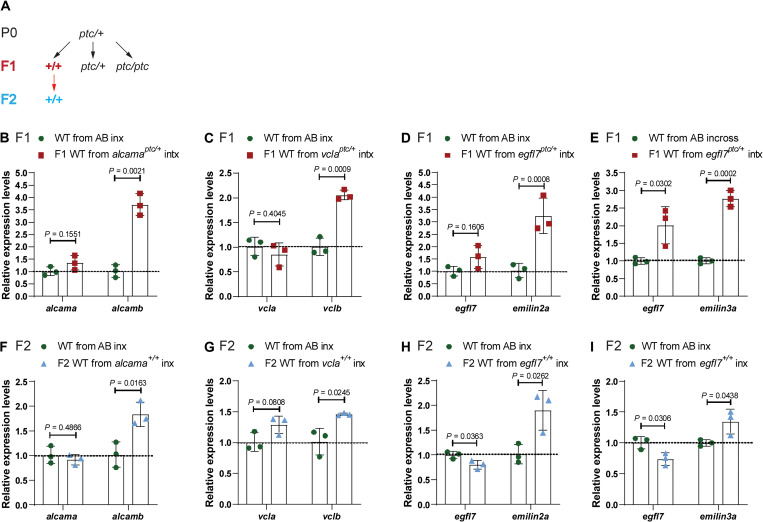
To investigate whether zebrafish exhibit IGTA, we first determined the mRNA levels of the adapting genes in wild-type offspring from PTC-allele heterozygous intercrosses (Fig. 2A) (see Materials and Methods) compared with their levels in embryos obtained from incrosses of the corresponding wild-type strain (AB or TL). Notably, we observed increased mRNA levels of the adapting genes, alcamb, vclb, emilin2a/emilin3a, and adh1a3, in the wild-type offspring from intercrosses of alcama, vcla, egfl7, and aldh1a2 heterozygous zebrafish, respectively (Fig. 2, B to E, and fig. S2E). To test whether this increase in mRNA levels of the adapting genes was triggered by the inheritance of the TA response, we analyzed wild-type offspring from alcama and egfl7 RNA-less allele heterozygous intercrosses and observed no significant up-regulation of the adapting genes (fig. S3, A and B). In addition, we generated an RNA-less allele of vcla (a 72.44-kb deletion that removes the vcla locus; fig. S3, C and D) and found that it does not display TA (fig. S3E). We also analyzed wild-type offspring from vcla RNA-less allele heterozygous intercrosses and observed no increased expression of the adapting gene when compared with embryos obtained from AB incrosses (fig. S3F). Together, these data indicate that the parents need to experience TA for the inheritance of the TA response to occur. Furthermore, we observed a significant increase in the mRNA levels of the adapting genes in second-generation wild-type embryos (F2) from F1 alcama+/+, vcla+/+, egfl7+/+, and aldh1a2+/+ incrosses (Fig. 2, F to I, and fig. S2F), indicating that, similar to the situation in C. elegans, TGTA is also observed in zebrafish. To begin to examine the specificity of TGTA, we measured alcamb mRNA levels in second-generation wild-type embryos (F2) from F1 egfl7+/+ incrosses and observed no significant difference compared with wild-type embryos from AB incrosses (fig. S3G).
Fig. 2. TGTA in zebrafish.
(A) Genetic crosses used to obtain wild-type (WT) embryos from TA-displaying heterozygous zebrafish and from subsequent incrosses. Black arrows indicate heterozygous intercrosses (intx); red arrow indicates subsequent wild-type incrosses (inx). (B) Relative mRNA levels of alcama and alcamb in 28 hpf wild-type embryos from AB incrosses and from alcamaptc/+ intercrosses. (C) Relative mRNA levels of vcla and vclb in 24 hpf wild-type embryos from AB incrosses and vclaptc/+ intercrosses. (D) Relative mRNA levels of egfl7 and emilin2a in 24 hpf wild-type embryos from AB incrosses and egfl7ptc/+ intercrosses. (E) Relative mRNA levels of egfl7 and emilin3a in 24 hpf wild-type embryos from AB incrosses and egfl7ptc/+ intercrosses. (F) Relative mRNA levels of alcama and alcamb in 28 hpf wild-type embryos obtained from AB incrosses and F1 alcama+/+ (i.e., wild types from alcamaptc/+ intercrosses) incrosses. (G) Relative mRNA levels of vcla and vclb in 24 hpf wild-type embryos obtained from AB incrosses and F1 vcla+/+ (i.e., wild types from vclaptc/+ intercrosses) incrosses. (H) Relative mRNA levels of egfl7 and emilin2a in 24 hpf wild-type embryos obtained from AB incrosses and F1 egfl7+/+ (i.e., wild types from egfl7ptc/+ intercrosses) incrosses. (I) Relative mRNA levels of egfl7 and emilin3a in 24 hpf wild-type embryos obtained from AB incrosses and F1 egfl7+/+ (i.e., wild types from egfl7ptc/+ intercrosses) incrosses. n = 3 biologically independent samples. Control expression levels were set at 1. Data are means ± SD, and a two-tailed Student’s t test was used to calculate P values. Ct values are listed in table S1.
Next, we investigated whether parental gender was an important factor in IGTA. We crossed male and female heterozygous zebrafish to wild types and examined the mRNA levels of the adapting genes. We observed an increase in alcamb mRNA levels in the wild-type offspring from alcamaptc/+ male outcrosses but not from alcamaptc/+ female outcrosses at 28 hours postfertilization (hpf) (fig. S4, A and B); however, when we analyzed pre-mRNA levels in such embryos at 6 hpf, we observed significant up-regulation of alcamb in the wild-type offspring from both male and female outcrosses (fig. S4, C and D), suggesting an increase in transcription of the adapting gene and an involvement of gender-specific posttranscriptional regulation. However, for the egfl7ptc model, we observed, at 24 hpf, an increase in emilin2a mRNA levels in the wild-type offspring from both male and female outcrosses (fig. S4, E and F).
To determine whether the increase in mRNA levels observed during TGTA was due to increased transcription or increased mRNA stability, we performed metabolic labeling of newly synthesized transcripts. We observed significantly increased transcription of the adapting genes alcamb, emilin2a, vclb, and aldh1a3 in second-generation wild-type embryos (F2) obtained from F1 alcama+/+, egfl7+/+, vcla+/+, and aldh1a2+/+ incrosses (fig. S5), respectively. Thus, TGTA, like TA, occurs due to increased transcription, not increased mRNA stability.
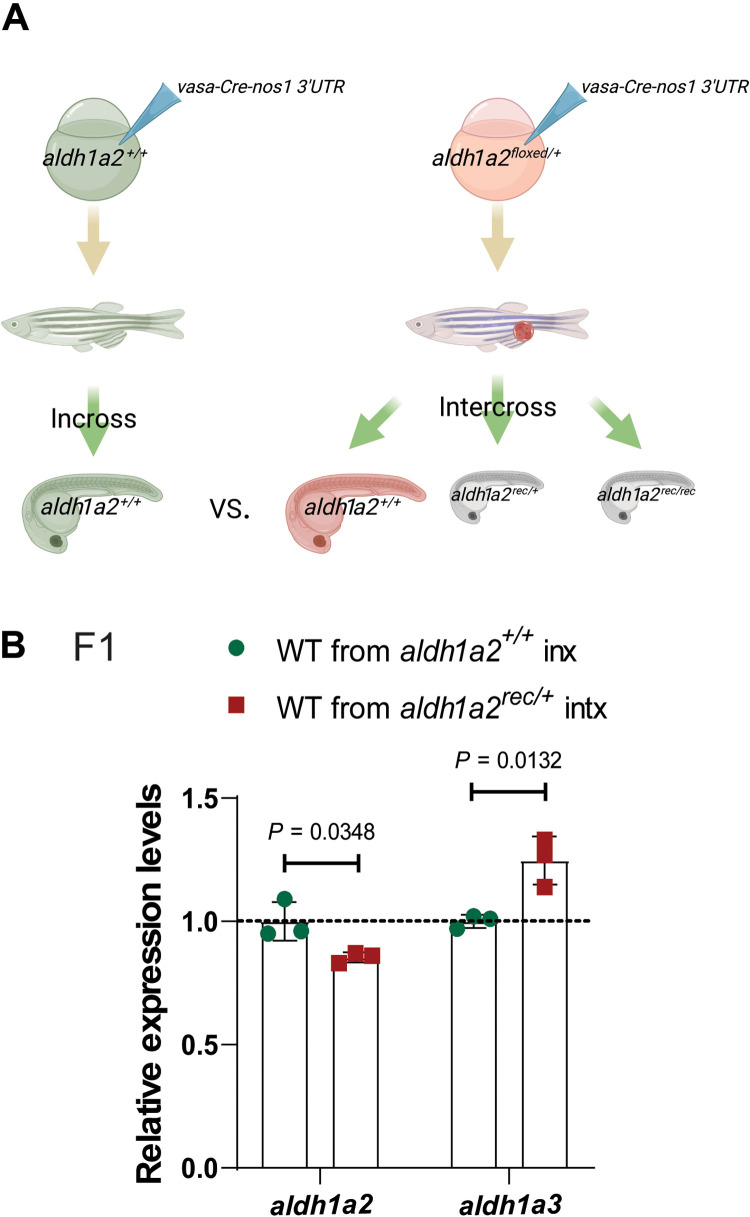
Given the importance of the germline in the transmission of information from one generation to the next, we first examined and found germline expression of the mutated gene for all four zebrafish TGTA models (fig. S6), in agreement with a recent publication (35), leading us to test the role of the germline in TA and its inheritance. To generate a germline-specific mutation, we used the aldh1a2-floxed allele in combination with vasa-GFP-nos1 3′UTR and vasa-Cre-nos1 3′UTR. These fusions of vasa and GFP/Cre followed by the 3′ untranslated region (3′UTR) of nos1 stabilize the mRNA in perinuclear granules (36), which are only present in primordial germ cells (37) (fig. S7, A to C’). Analysis of vasa-GFP-nos1 3′UTR and vasa-Cre-nos1 3′UTR mRNA-coinjected embryos from aldh1a2floxed/+ intercrosses at 24 hpf confirmed that recombination occurred in germ cells and not in somatic cells (fig. S7, D to F), and the embryos were then raised to adulthood (Fig. 3A). Wild-type offspring from intercrosses of vasa-Cre-nos1 3′UTR mRNA-injected aldh1a2floxed/+ zebrafish displayed significant up-regulation of aldh1a3 mRNA levels (Fig. 3B), indicating that a germline mutation alone can be sufficient to trigger TA in wild-type offspring. To determine whether IGTA occurs in somatic cells or whether it is confined to germ cells, a dnd1 morpholino, used to block primordial germ cell development (38), and vasa-GFP-nos1 3′UTR mRNA were coinjected into wild-type embryos and embryos from alcamaptc/+ intercrosses (fig. S8, A to D). Significant up-regulation of the adapting gene was observed in the dnd1 morpholino–injected wild-type embryos from alcamaptc/+ intercrosses at 28 hpf (fig. S8E), indicating that IGTA can be detected in somatic cells.
Fig. 3. The germline plays a key role in IGTA in zebrafish.
(A) Schematic representation of experimental setup and genetic crosses used to obtain wild-type offspring; red cell indicates a germline-specific mutation. (B) Relative mRNA levels of aldh1a2 and aldh1a3 in 24 hpf wild-type embryos from aldh1a2+/+ incrosses (inx) and aldh1a2rec/+ intercrosses (intx). Control expression levels were set at 1. n = 3 biologically independent samples. Data are means ± SD, and a two-tailed Student’s t test was used to calculate P values. Ct values are listed in table S1. (A) was created with BioRender.com.
Total RNA from germ cells of heterozygous zebrafish can trigger the transcriptional adaptation response
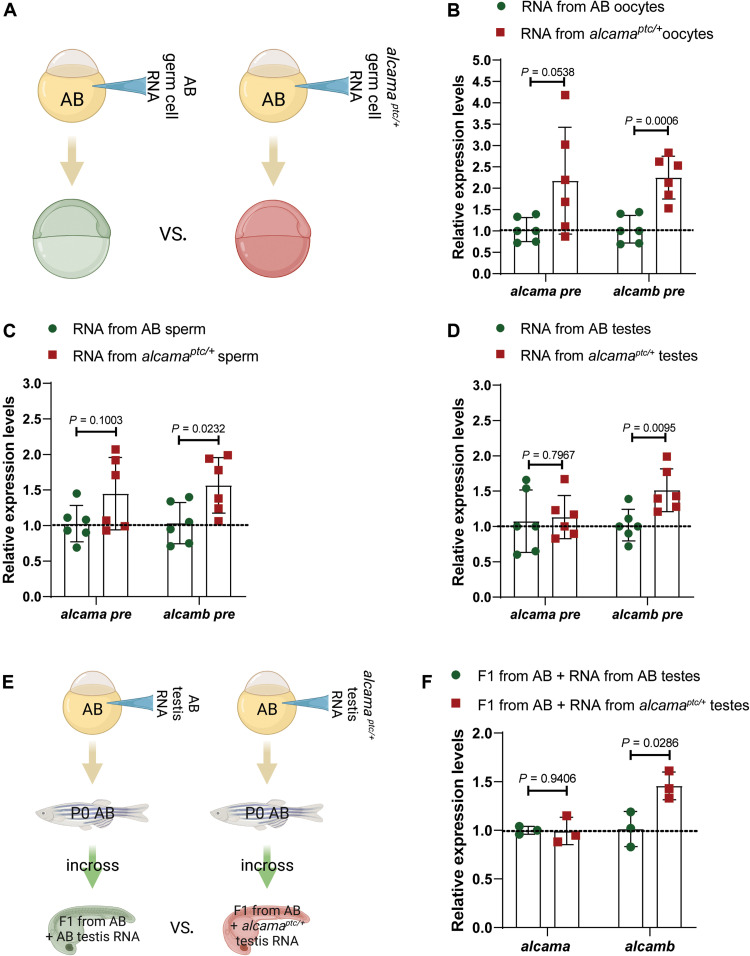
Because the mutated genes are expressed in the germline, we considered the possibility that transfer of RNA to the next generations could be involved in IGTA/TGTA. Previous studies have indeed suggested that small RNAs (including transfer RNA fragments) (39, 40) as well as long RNAs (41) can influence the progeny’s phenotype. To investigate whether IGTA/TGTA occurs in a similar manner, for example, through mRNA degradation products and/or their derivatives, we isolated total RNA from oocytes, sperm, and testes of wild-type strain and alcamaptc/+ zebrafish and injected them into one-cell stage zebrafish embryos (Fig. 4A). [We chose the alcama model because zebrafish oocytes appear to lack alcamb pre-mRNA expression (fig. S6, E and F), thereby facilitating the analysis of the effects of total RNA injections on alcamb expression.] Notably, we observed significantly elevated alcamb pre-mRNA levels 6 hours after injections of total RNA obtained from oocytes, sperm, or testes of alcamaptc/+ zebrafish compared with injections of total RNA obtained from oocytes, sperm, and testes of wild-type zebrafish, respectively (Fig. 4, B to D). [Because we were analyzing expression levels at 6 hpf, we focused on pre-mRNA rather than mRNA in order to avoid looking at any remaining maternal mRNA]. Together, these data suggest that germline transmission of mutant mRNAs, their degradation products, and/or their derivatives could underlie IGTA.
Fig. 4. Injection of total RNA from alcamaptc/+ germ cells triggers TA, which is also observed in the next generation.
(A) Schematic representation of the experimental setup. (B) Relative pre-mRNA levels of alcama and alcamb in 6 hpf AB embryos injected with total RNA isolated from oocytes of wild types and alcamaptc/+ zebrafish. (C) Relative pre-mRNA levels of alcama and alcamb in 6 hpf AB embryos injected with total RNA isolated from sperm of wild types and alcamaptc/+ zebrafish. (D) Relative pre-mRNA levels of alcama and alcamb in 6 hpf AB embryos injected with total RNA isolated from testes of wild types and alcamaptc/+ zebrafish. (E) Schematic representation of the experimental setup. (F) Relative mRNA levels of alcama and alcamb in 28 hpf F1 offspring from AB zebrafish injected at the one-cell stage with total RNA isolated from testes of wild types and alcamaptc/+ zebrafish. n ≥ 3 biologically independent samples. Control expression levels were set at 1. Data are means ± SD, and a two-tailed Student’s t test was used to calculate P values. Ct values are listed in table S1. (A) and (E) were created with BioRender.com.
Furthermore, we raised the embryos injected with total testis RNA to adulthood and analyzed gene expression levels in their offspring (Fig. 4E). We observed significantly increased alcamb mRNA levels in 28 hpf embryos from animals injected with total RNA obtained from alcamaptc/+ testes compared with 28 hpf embryos from animals injected with total RNA obtained from AB testes (Fig. 4F). These data indicate that the acquired TA traits from total testis RNA injections can be epigenetically inherited.
Inheritance of epigenetic marks is associated with the transgenerational inheritance of transcriptional adaptation in zebrafish
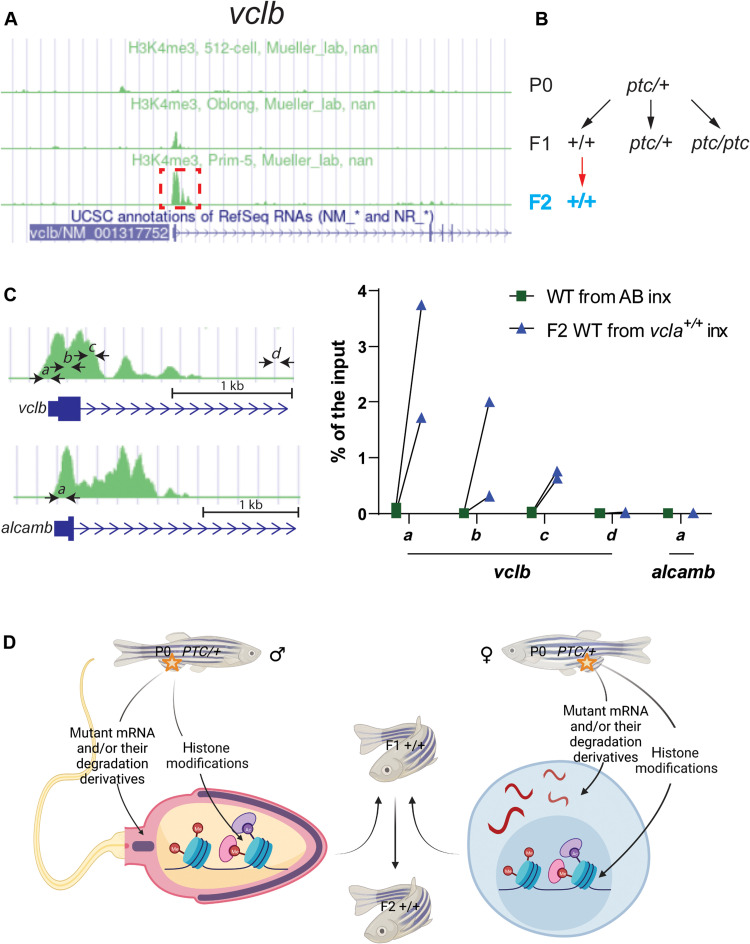
To begin to test the role of epigenetic modifications in IGTA/TGTA, we followed up on previous observations that WDR5-mediated deposition of H3K4me3 appears to be involved in TA (28, 29). To examine whether TA-related H3K4me3 modifications are inherited in the next generations, we first checked the available H3K4me3 chromatin immunoprecipitation sequencing (ChIP-seq) dataset in DANIO-CODE (https://danio-code.zfin.org) and found that, at the 512-cell stage (2.75 hpf) (i.e., prior to midblastula transition), vclb does not exhibit H3K4me3 enrichment at its promoter (Fig. 5A). We thus performed ChIP coupled with quantitative polymerase chain reaction (qPCR) (ChIP-qPCR) at this stage and observed the enrichment of H3K4me3 at the promoter of the adapting gene vclb in second-generation (F2) wild-type embryos from F1 vcla+/+ incrosses compared with wild types (Fig. 5, B and C). We also performed ChIP-qPCR with the alcama model after the onset of zygotic transcription and observed the enrichment of H3K4me3 at the promoter region of the adapting gene alcamb in second-generation (F2) wild-type embryos from F1 alcama+/+ incrosses compared with wild types (fig. S9). Thus, in addition to the possible germline transmission of mutant mRNAs, their degradation products, and/or their derivatives, IGTA/TGTA could also, in principle, be achieved via the inheritance of epigenetic marks (Fig. 5D), and additional studies will be required to determine the role of these epigenetic marks in IGTA/TGTA.
Fig. 5. TGTA in zebrafish is associated with H3K4me3 histone marks.
(A) Peaks of H3K4me3 ChIP-seq at the vclb locus at different stages (obtained from DANIO-CODE). Red boxed area was enlarged in (C) to show the relative location of the ChIP–qPCR primers marked by black arrows (also at the alcamb locus, which is used as an external/specificity control). (B) Genetic crosses were used to obtain F2 wild-type offspring from heterozygous zebrafish. Black arrows indicate heterozygous intercrosses (intx); red arrow indicates subsequent wild-type incrosses (inx). (C) ChIP-qPCR analysis of H3K4me3 occupancy near exon 1 of vclb in 512 cell stage wild-type embryos from incrosses of F1 vcla+/+ (i.e., wild types from vclaptc/+ intercrosses) compared with wild-type embryos from AB incrosses. Green peaks in vclb and alcamb loci represent prim-5 stage (24 hpf) H3K4me3 ChIP-seq data obtained from DANIO-CODE. Scale bars, 1 kb. n = 2 biologically independent samples. Ct values are listed in table S1. (D) Proposed model of intergenerational and transgenerational inheritance of TA (IGTA and TGTA). Mutant mRNAs, their degradation products (or their derivatives), and/or histone modifications are transmitted through the germline, leading to TA in the offspring. Additional mechanisms are likely to be involved in IGTA and TGTA. PTC, premature termination codon. (D) was created with BioRender.com.
DISCUSSION
This study shows that the TA response can be inherited over multiple generations. We report that a mutation in a parental genome can lead to transcriptional modulation in wild-type offspring. Observing TGTA in both C. elegans and zebrafish models indicates that this phenomenon, as well as its underlying mechanisms, may be common among metazoans. In addition to the well-documented impact of physiological and environmental factors on TEI (3, 11, 15, 24–27), these data indicate that genetic alterations in a parental genome can also influence the transcriptional landscape of the offspring. Thus, our study calls for even greater attention when designing experiments to examine phenotypic changes in mutants, as the transcriptome of their wild-type siblings may also be affected by mutations in their ancestors’ genome.
TEI is most often transmitted through the germline as a direct consequence of physiological or environmental factors, although an alternative model, one that has received increasing attention, postulates that somatic cells are capable of transferring heritable information to the gametes and thereby to progeny (7). In the past decade, studies in several model organisms have provided evidence of RNA-mediated soma-to-germline communication during spermatogenesis (42–47). In mammals, for instance, subpopulations of epididymis small RNAs are selectively packaged into epididymosomes, microvesicles present in the epididymal fluid with functions in sperm maturation and storage (48); however, the underlying mechanisms are currently unknown (49–52). In zebrafish and amphibians, spermatogenesis happens through Sertoli-germ cell interactions (53), which could allow for soma-to-germline communication. In this study, we showed that a germline-specific mutation in aldh1a2 in zebrafish is sufficient to induce TA in wild-type offspring (Fig. 3), indicating that, at least for this gene, which is expressed in germ cells, mutations in somatic tissues are not necessary for this process. It is of course likely that for genes not expressed in germ cells, mutations in somatic cells are necessary to induce IGTA. Additional experiments will be required to investigate this question and also to determine how the potential transfer of information occurs between the soma and the germline.
Paramutation is an IEI/TEI-like phenomenon observed in plants (21, 53), C. elegans (22), and Drosophila (23); yet, examples in vertebrates are limited. One example of paramutation in mammals is provided by the Kittm1Alf mouse. Wild-type progeny of Kittm1Alf/+ mice display the same white tail tip phenotype as their heterozygous parents (19). Notably, this phenotype has been attributed to the inheritance of RNA decay fragments from the Kit locus. In addition, another KitW-v missense mutation (which is unlikely to cause mutant mRNA degradation) does not lead to the white tail tip phenotype in the wild-type progeny (19, 55). The apparent requirement of mutant mRNA degradation in this paramutation example suggests that IGTA/TGTA might be at play.
We have previously observed that TA is triggered by mutant mRNA degradation in zebrafish, mouse cells in culture, and nematodes (28, 30). Furthermore, factors involved in small RNA biogenesis and transport are required for the TA response in C. elegans (30). In the current study, we observed TA in wild-type animals injected with total RNA from germ cells of TA-displaying heterozygous zebrafish as well as in their offspring (Fig. 4). These data are consistent with a model whereby the intergenerational inheritance of the TA response from P0 to F1 animals requires the mutant mRNA, its degradation products, and/or their derivatives, that are inherited through the germline (Fig. 5D). Since we observed an enrichment of H3K4me3 at the promoter region of the adapting genes vclb and alcamb in F2 wild-type offspring (Fig. 5C and fig. S9), an additional model involves the inheritance of histone modifications (Fig. 5D), themselves possibly induced by mutant mRNA degradation products, and/or their derivatives.
MATERIALS AND METHODS
C. elegans resource table
All C. elegans strains used in this study are found in Table 1.
Table 1. C. elegans resource table.
| Reagent type | Designation | Source or reference | Additional information |
| Strain, strain background | N2 | Caenorhabditis Genetics Center, Bristol strain |
Wild type |
| Strain, strain background | DYS0005 | (30) | act-5(dt2019) |
| Strain, strain background | DYS0014 | (30) | Ex0002[act-3p::rfp] |
| Strain, strain background | DYS0015 | (30) | act-5(dt2019);Ex0002[act-3p::rfp] |
| Strain, strain background | EG6699 | Caenorhabditis Genetics Center | Wild type for Mos1 integration |
| Strain, strain background | DYSSi1000 | This study | Si1000[eft-3p::act-5(ptc)::tbb-2 3′UTR] |
| Strain, strain background | DYSSi1001 | This study |
Si1000[eft-3p::act-5(ptc)::tbb-2 3′UTR];Ex1001[act-3p::rfp] |
| Strain, strain background | DYSEx1002 | This study | Ex1000[act-3p::rfp] |
C. elegans culture conditions and strains
All C. elegans strains were kept on 6-cm plates with nematode growth medium agar (56) and fed with a lawn of OP50 Escherichia coli grown in 500 μl of LB, except for mating plates where the worms were kept on nematode growth medium plates with a lawn of OP50 E. coli grown in 150 μl of LB. All C. elegans strains used in this study are listed in Table 1. Cultures were maintained at 18° to 20°C. In addition, to minimize the potential for laboratory evolution of the trait, a new culture of the strains was revived at least annually from frozen stocks. All plates with fungal or bacterial contamination were excluded from the experiments.
C. elegans transgenic generation
The DYSSi1000 Mos1-mediated single-copy insertion line was generated as described (31) using the EG6699 strain for integration of the eft-3p::act-5(ptc)::tbb-2 3′UTR transgene into chromosome 2. Injections to generate the RFP reporter lines DYSSi1001 and DYSEx1002 were performed as described (57, 58), with the following modifications: Plasmids were purified twice using the FastGene Plasmid Mini Kits (Nippon Genetics, FG-90402) and injected at a final total concentration of 100 ng/μl. DYSSi1001 and DYSEx1002 lines were generated by injecting a mixture of act-3p::rfp (90 ng/μl) and sur-5::GFP (10 ng/μl) plasmids into DYSSi1000 and N2 worms, respectively.
C. elegans construct generation
The overexpression vector [eft-3p::act-5(ptc)::tbb-2 3′UTR] was designed to express a PTC-bearing transcript matching the endogenous dt2019 mutant transcript and was generated by standard restriction enzyme and Gibson cloning methods using N2 genomic DNA as the template. The following primers 5′-ATCGATGCACCTTTGGTCTTTTATTGTCAAC-3′ and 5′-GTTAACTGTTTCCCAACTGAAAAAAAAACAATTTAAT-3′ were used to amplify the eft-3 promoter region, primers 5′-TTTTTCAGTTGGGAAACAGTTAACGATGGAAGAAGAAATCGCCGC-3′ and 5′-GAAAGGATCTTGCATTTATCAACTAGTCTAAGCCTAAAAAACAAAAACTCCACACG-3′ were used to amplify the complete act-5 sequence from the ATG to the end of the 3′UTR including introns, and primers 5′-ACTAGTTGATAAATGCAAGATCCTTTCAAGCATTC-3′ and 5′-TTACCGGTGGGAAAAGTTAATTAAGACTTTTTTCTTGGCGGCAC-3′ were used to amplify the tbb2 3′UTR to ensure proper termination of the resulting transcript. These amplicons were subsequently assembled into an intermediate vector. The act-5(PTC) single-nucleotide polymorphism matching the original dt2019 mutation (30) was generated by point mutagenesis using primers 5′-GAGAAAATCTGGCATCACACATTCTAC-3′ and 5′-CATATCATCtCAGTTGGTGACG-3′. The complete p-eft-3:act-5(ptc):tbb2 3′UTR sequence was subsequently cloned into the Mos1 integration vector pBN449 (Addgene, plasmid no. 129555) between the Nhe I and Xma I restriction sites.
C. elegans crossing scheme
N2 males were crossed with DYS0015 (act-5(dt2019);Ex0002[act-3p::rfp]) virgin hermaphrodites. The following day, males were removed from the mating plates, and the progeny was subsequently screened for RFP-positive individuals. A subset of the RFP-positive hermaphrodites was isolated for a day to lay eggs prior to genotyping. After identifying the wild-type nematodes, the plates with the eggs from these individuals were kept for further screening. After each generation, single nematodes were isolated to new plates, and their progeny were screened in the same manner. Crosses with DYSSiEx1001 animals were performed in the same manner: Si[eft-3p::act-5(ptc)](tg/+);Ex1001[act-3p::rfp] nematodes were obtained from crossing Si[eft-3p::act-5(ptc)](tg/tg);Ex1001[act-3p::rfp] virgin hermaphrodites with N2 males. The primer sequences used for genotyping are listed in table S2.
Zebrafish husbandry
All zebrafish (Danio rerio, wild-type strains: TL and AB) were maintained under standard conditions according to institutional (Max Planck Society) and national ethical and animal welfare guidelines approved by the Ethics Committee for Animal Experiments at the Regierungspräsidium Darmstadt, Germany (permit numbers: B2/1218 and B2/2023) as well as the Federation of European Laboratory Animal Science Associations guidelines (59). Most experiments were performed on zebrafish embryos between 6 and 28 hpf. We used the PTC-bearing alleles [alcamabns201 (28), egfl7s981 (33), aldh1a2tpl137 (34), and vclabns241 (28)] and the RNA-less alleles [alcamabns244 (28) and egfl7bns302 (28)]. All alleles were maintained in the AB background with the exception of aldh1a2tpl137, which was generated and maintained in the TL background, and alcamabns201, which was in a mixed background. As previously reported, mild up-regulation of emilin2a was observed in the egfl7RNA-less allele when compared with egfl7+/+ siblings (28), likely a result of the loss of Egfl7 protein function; a similar up-regulation of emilin2a was observed in wild-type offspring from egfl7 RNA-less allele heterozygous intercrosses. Most zebrafish embryos were obtained from single pair matings. To help keep track of the various matings performed, we refer to matings between animals with the same heterozygous genotype as intercrosses and matings between animals with the same homozygous genotype as incrosses (e.g., https://zfin.org/zf_info/glossary.html#i).
Engineering and validation of the zebrafish aldh1a2tpl146 (floxed) allele
We established a homozygous line for the intron 7 loxP site (aldh1a2tpl139; fig. S2C) (34) and injected embryos with aldh1a2 single-guide RNA (sgRNA), Cas9 mRNA, and an Homology-directed repair (HDR) template oligonucleotide using a published methodology (34). We identified an F0 that transmitted the integration of the second loxP site in the germline and verified the allele by screening individual F1 embryos (two embryos out of six screened were positive). Sibling F1s were raised to adulthood and screened for intron 8 loxP site by fin biopsy PCR and sequencing. One of the 13 zebrafish screened was found to contain a precise integration of the additional loxP site. To validate this newly engineered floxed aldh1a2tpl146 allele, this F1 zebrafish was crossed to the previously reported exon 8 deletion mutant aldh1a2tpl137 (34), and embryos were injected with Cre mRNA to induce excision of the eighth exon. All uninjected larvae were phenotypically normal at 72 hpf (n = 174), indicating that the floxed allele is functionally wild type. Approximately one-quarter of the Cre-injected larvae (38 of 136, 27.9%) displayed phenotypes consistent with the complete loss of aldh1a2 function, including the loss of mesodermal head tissue, pericardial edema, tail curvature, and the absence of pectoral fins. Sixteen Cre-injected larvae (eight phenotypically mutant and eight phenotypically wild type) were genotyped by PCR. All eight phenotypically mutant larvae were found to be transheterozygous for the aldh1a2tpl137 deletion allele and Cre-deleted floxed aldh1a2tpl146 allele, while all eight phenotypically wild-type larvae were found to contain at least one wild-type allele. Cre-mediated excision of exon 8 was confirmed by sequencing the PCR fragment. The aldh1a2tpl146 allele was generated and maintained in the TL background. The sequences of the oligos used for mutagenesis and genotyping are listed in table S2.
Genome editing by CRISPR-Cas9
CRISPR gRNA design was performed using the online tools (https://eu.idtdna.com/site/order/designtool/index/CRISPR_SEQUENCE and http://chopchop.cbu.uib.no/). gRNAs were generated with gRNA-specific primers (Thermo Fisher Scientific) as described (60). The gRNAs were transcribed with the MEGAshortscript T7 Transcription Kit (Thermo Fisher Scientific), followed by purification with the RNA Clean & Concentrator Kit (Zymo Research). The vcla full locus deletion allele (vclabns605) was generated with a pair of gRNAs as illustrated in fig. S3C. All gRNAs used for CRISPR-Cas9 genome editing are listed in table S2.
Genotyping
DNA and RNA were extracted from at least 24 individual embryos using TRIzol (Invitrogen) followed by phenol-chloroform extraction. In brief, single embryos were lysed and homogenized in 100 μl of TRIzol using a Next Advance Bullet Blender Homogenizer (Scientific Instrument Services). Twenty microliters of chloroform was then added, and phase separation was obtained following vortexing and centrifugation. The aqueous phase (containing RNA) was isolated and stored at −80°C, and the organic phase (containing DNA) was subjected to ethanol purification to precipitate the DNA. Purified DNA was then dissolved in Tris-EDTA buffer containing 1% proteinase K (Thermo Fisher Scientific) for genotyping. RNAs from genotyped zebrafish were then pooled for further purification and cDNA synthesis with the exception of the experiments shown in figs. S2D, S3E, and S8E, which were performed with a single-embryo RNA. For each experiment, these steps were performed on embryos from at least three different crosses. High-resolution melt analysis (61) was used to genotype all zebrafish alleles, with the exception of the aldh1a2tpl137 and aldh1a2tpl146 alleles, which were genotyped by PCR. The primer sequences used for genotyping are listed in table S2.
RT-qPCR analysis
Reverse transcription (RT)–qPCR was performed using a CFX Connect Real-Time System (Bio-Rad). C. elegans RNA was isolated as described (62). RNAs from five synchronized early larval stage worms were pooled. For RT, SuperScript III reverse transcriptase (Invitrogen) was used following the manufacturer’s instructions. Zebrafish RNA was isolated using TRIzol, and at least 500 ng of RNA was used for RT. cDNA synthesis was performed using the Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). All reactions were performed in at least technical duplicates, and the results usually represent biological triplicates; when the biological replicates were too variable to assess TGTA, additional ones were analyzed. RT-qPCR was performed at the embryonic stage when the wild-type version of the mutated gene exhibits its highest level of expression. rpl13a was used as a reference gene for all zebrafish experiments; cdc42 was used as a reference gene for all C. elegans experiments. Primer sequences used for the RT-qPCR experiments are listed in table S2. Fold changes were calculated using the 2−ΔΔCt method. All Ct values are listed in table S1. Ct values for the reference genes ranged between 12 and 26.
In vitro transcription and microinjections
sgRNAs were transcribed with the MEGAshortscript T7 Transcription Kit, followed by purification using the RNA Clean & Concentrator Kits. The pT3TSnCas9n (Addgene, plasmid no. 46757) plasmid (63) was linearized using Xba I [New England Biolabs (NEB)]; capped Cas9 mRNA was synthesized using the mMESSAGE mMACHINE T3 Transcription Kit (Thermo Fisher Scientific) and purified using the RNA Clean & Concentrator Kits. A cDNA corresponding to aldh1a2 full-length mRNA was amplified using whole-embryo cDNA as a template. PCR fragments were ligated into a pCS2+ vector between the Bam HI and Xba I (NEB) sites. Zebrafish codon-modified Cre recombinase was amplified from pCS2-Cre (gift from H. Burgess; Addgene, plasmid no. 61391) (64). Plasmids were linearized using Not I–HF (NEB) and in vitro transcribed using the mMESSAGE mMACHINE SP6 Transcription Kit. RNA was then purified using the RNA Clean & Concentrator Kits. A total of 75 pg of aldh1a2 full-length mRNA was injected into embryos from heterozygous intercrosses at the one-cell stage. A total of 100 pg of Cre mRNA was injected into embryos from heterozygous intercrosses at the one-cell stage. To generate the vasa-Cre-nos1 3′UTR plasmid, zebrafish codon-modified Cre recombinase was amplified from pCS2-Cre (63) and cloned into a vasa-mGFP-nos1 3′UTR plasmid (gift from E. Raz) (65). Plasmids were linearized using Not I–HF (NEB) and in vitro transcribed using the mMESSAGE mMACHINE SP6 Transcription Kit. RNA was then purified using the RNA Clean & Concentrator Kits. A total of 100 pg of vasa-GFP-nos1 3′UTR mRNA plus 0.6 ng of standard control morpholino or dnd1 morpholino were injected into embryos from AB incrosses and from alcamabns201/+ intercrosses at the one-cell stage. A total of 100 pg of vasa-GFP-nos1 3′UTR mRNA plus 10 pg of vasa-cre-nos1 3′UTR mRNA were injected into aldh1a2tpl146/+ intercrosses at the one-cell stage to create a germline-specific mutation.
Germline RNA preparation
Egg and sperm isolation and testis dissection were performed as described (66). Total RNA was extracted using TRIzol, followed by phenol-chloroform extraction. A total of 100 pg of total RNA was injected into embryos from AB incrosses at the one-cell stage. We raised embryos injected with total RNA from oocytes, sperm, and testes but only had enough surviving adults from the testis RNA-injected ones to carry out the experiments shown in Fig. 4E.
Sample preparation and cell sorting
Zebrafish embryos were obtained from aldh1a2tpl146/+ intercrosses, followed by 100 pg of vasa-GFP-nos1 3′UTR mRNA plus 10 pg of vasa-Cre-nos1 3′UTR mRNA injections at the one-cell stage. At 24 hpf, dechorionated embryos were dissociated with Liberase Blendzyme (Roche) and prepared as described (67) with the following modifications: Incubation was performed at 30°C with gentle shaking for 40 min, followed by careful resuspension in 1× Hanks’ balanced salt solution, 0.25% bovine serum albumin, and 10 mM Hepes buffer. The resuspended cells were immediately sorted using a BD FACSAria III (BD) sorter for green fluorescent protein (GFP)+/4′,6-diamidino-2-phenylindole (DAPI)− cells. Debris and clumps were excluded using the FSC/SSC parameters. Dead cells were excluded by selecting against DAPI-positive cells, which were detected using laser excitation at 405 nm and emission at 450/40 nm. GFP-labeled cells were detected using laser excitation at 488 nm and emission at 530/30 nm. The sort gate for live GFP-positive cells was defined on the basis of the GFP-negative control.
RNA metabolic labeling
Metabolic labeling was performed as described (28, 68, 69). In brief, at 24 and 28 hpf, dechorionated zebrafish embryos were treated with 25 ml of 200 μM 4-thiouridine (4sU; Sigma-Aldrich) for 1 hour, followed by phenol-chloroform total RNA extraction. A total of 80 μg of RNA was incubated with biotin-HPDP (Thermo Fisher Scientific) to specifically biotinylate the newly transcribed 4sU-labeled RNAs. Biotinylated RNAs were then pulled down using the μMACS Streptavidin Kit (Miltenyi), and the same amount of pulled-down RNA was used for RT and downstream qPCR analysis. This experiment was performed three times.
Chromatin immunoprecipitation
ChIP was performed using the truChIP Chromatin Shearing Reagent Kit (Covaris) using 500 embryos per immunoprecipitation according to the manufacturer’s instructions. Chromatin was sheared using a Bioruptor (Diagenode) to generate fragments of 200 to 400 base pairs in size. Immunoprecipitation was then performed as described (70). The following antibodies were used: rabbit immunoglobulin G (3 μl per immunoprecipitation; Thermo Fisher Scientific, 026102) and H3K4me3 (4 μl per immunoprecipitation; Cell Signaling Technology, no. 9751, lot 14). Following immunoprecipitation and reverse crosslinking, samples were purified using ethanol purification. Because of the challenges associated with collecting enough embryos for the ChIP-qPCR experiments, they were only performed twice.
Confocal microscopy
Fluorescence images of C. elegans were acquired using a Zeiss LSM 700 confocal microscope (Plan-Apochromat 10×/0.45 objective lens). Worms were mounted immobilized in polystyrene microbeads as described (71). Fluorescence images of zebrafish embryos were acquired using a Zeiss LSM 700 confocal microscope (Plan-Apochromat 10×/0.45 objective lens). Embryos were mounted in 1% UltraPure Low Melting Point Agarose (Thermo Fisher Scientific) in egg water with tricaine in a glass-bottomed petri dish (MatTek). Obtained images were subsequently processed with the ZEN software (black edition). All figures were prepared using Adobe Photoshop 2022 and Adobe Illustrator 2022.
Statistics and reproducibility
No statistical methods were used to predetermine the sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. All experiments were performed at least twice with three biological replicates each time/in total of six biological replicates, unless otherwise noted. Statistical analysis was performed using GraphPad Prism 9. Data are means ± SD, and a two-tailed Student’s t test was used to calculate P values with the exception of the data shown in fig. S5, which were analyzed with a two-tailed paired t test of the ΔΔCt. P < 0.05 was considered as statistically significant.
Acknowledgments
We thank S. Capon, C. Cirzi, A. Rossi, T. Bertozzi, Z. Kontarakis, S. Perathoner, L. Xie, and P. Krishnaraj for discussion and comments on the manuscript. We also thank A. Atzberger and K. Kikhi at the MPI Fluorescence-Activated Cell Sorting service group for the support with cell sorting. We thank S. Allanki, K. Mattonet, and S. Howard for technical advice and support and H. Burgess, E. Raz, and P. Askjaer for providing plasmids. The DYSSi1000 Mos1-mediated single-copy insertion line was generated in the Genome Engineering Facility, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany. We thank the DANIO-CODE consortium (https://danio-code.zfin.org) for providing zebrafish epigenomics data.
Funding: This research was supported by an award from the Cardio-Pulmonary Institute (EXC 2026, project ID: 390649896) to Z.J.; an Otto Bayer Fellowship from the Bayer Foundation to M.A.E.-B.; and funds from the Max Planck Society, the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (AdG 694455-ZMOD and AdG 101021349-TAaGC), and the Leducq Foundation to D.Y.R.S.
Author contributions: Conceptualization: Z.J., M.A.E.-B., V.S., and D.Y.R.S. Methodology: Z.J., M.A.E.-B., and V.S. Investigation: All authors. Resources: D.Y.R.S. Writing: Z.J., M.A.E.-B., V.S., and D.Y.R.S, with inputs from all authors. Supervision: D.Y.R.S. Project administration and funding acquisition: D.Y.R.S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
REFERENCES AND NOTES
- 1.Skvortsova K., Iovino N., Bogdanovic O., Functions and mechanisms of epigenetic inheritance in animals. Nat. Rev. Mol. Cell Biol. 19, 774–790 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Richards E. J., Inherited epigenetic variation–Revisiting soft inheritance. Nat. Rev. Genet. 7, 395–401 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Daxinger L., Whitelaw E., Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat. Rev. Genet. 13, 153–162 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Miska E. A., Ferguson-Smith A. C., Transgenerational inheritance: Models and mechanisms of non-DNA sequence-based inheritance. Science 354, 59–63 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Toth M., Mechanisms of non-genetic inheritance and psychiatric disorders. Neuropsychopharmacology 40, 129–140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heard E., Martienssen R. A., Transgenerational epigenetic inheritance: Myths and mechanisms. Cell 157, 95–109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez M. F., Lehner B., Intergenerational and transgenerational epigenetic inheritance in animals. Nat. Cell Biol. 21, 143–151 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Tuscher J. J., Day J. J., Multigenerational epigenetic inheritance: One step forward, two generations back. Neurobiol. Dis. 132, 104591 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Burton N. O., Greer E. L., Multigenerational epigenetic inheritance: Transmitting information across generations. Semin. Cell Dev. Biol. 127, 121–132 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwanam N., Lawir D. F., Sikora K., O’Meara C., Takeshita K., Schorpp M., Boehm T., Transgenerational inheritance of impaired larval T cell development in zebrafish. Nat. Commun. 11, 4505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serobyan V., Sommer R. J., Developmental systems of plasticity and trans-generational epigenetic inheritance in nematodes. Curr. Opin. Genet. Dev. 45, 51–57 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Lev I., Seroussi U., Gingold H., Bril R., Anava S., Rechavi O., MET-2-dependent H3K9 methylation suppresses transgenerational small RNA inheritance. Curr. Biol. 27, 1138–1147 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Duempelmann L., Skribbe M., Buhler M., Small RNAs in the transgenerational inheritance of epigenetic information. Trends Genet. 36, 203–214 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Cavalieri V., Spinelli G., Environmental epigenetics in zebrafish. Epigenetics Chromatin 10, 46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan Q. L., Meng X., Dai W., Luo Z., Wang C., Fu X., Yang J., Ye Q., Zhou Q., N6-methyldeoxyadenine and histone methylation mediate transgenerational survival advantages induced by hormetic heat stress. Sci. Adv. 7, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paroo Z., Liu Q., Wang X., Biochemical mechanisms of the RNA-induced silencing complex. Cell Res. 17, 187–194 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Holoch D., Moazed D., RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16, 71–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang D. L., Zhang G., Tang K., Li J., Yang L., Huang H., Zhang H., Zhu J. K., Dicer-independent RNA-directed DNA methylation in Arabidopsis. Cell Res. 26, 66–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rassoulzadegan M., Grandjean V., Gounon P., Vincent S., Gillot I., Cuzin F., RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 441, 469–474 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Yuan S., Oliver D., Schuster A., Zheng H., Yan W., Breeding scheme and maternal small RNAs affect the efficiency of transgenerational inheritance of a paramutation in mice. Sci. Rep. 5, 9266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandler V. L., Paramutation: From maize to mice. Cell 128, 641–645 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Sapetschnig A., Sarkies P., Lehrbach N. J., Miska E. A., Tertiary siRNAs mediate paramutation in C. elegans. PLOS Genet. 11, e1005078 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Vanssay A., Bougé A. L., Boivin A., Hermant C., Teysset L., Delmarre V., Antoniewski C., Ronsseray S., Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature 490, 112–115 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Kishimoto S., Uno M., Okabe E., Nono M., Nishida E., Environmental stresses induce transgenerationally inheritable survival advantages via germline-to-soma communication in Caenorhabditis elegans. Nat. Commun. 8, 14031 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seong K. H., Li D., Shimizu H., Nakamura R., Ishii S., Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell 145, 1049–1061 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Brink R. A., Styles E. D., Axtell J. D., Paramutation: Directed genetic change. Paramutation occurs in somatic cells and heritably alters the functional state of a locus. Science 159, 161–170 (1968). [DOI] [PubMed] [Google Scholar]
- 27.Constantinof A., Boureau L., Moisiadis V. G., Kostaki A., Szyf M., Matthews S. G., Prenatal glucocorticoid exposure results in changes in gene transcription and DNA methylation in the female juvenile guinea pig hippocampus across three generations. Sci. Rep. 9, 18211 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El-Brolosy M. A., Kontarakis Z., Rossi A., Kuenne C., Günther S., Fukuda N., Kikhi K., Boezio G. L., Takacs C. M., Lai S. L., Fukuda R., Gerri C., Giraldez A. J., Stainier D. Y. R., Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Z., Zhu P., Shi H., Guo L., Zhang Q., Chen Y., Chen S., Zhang Z., Peng J., Chen J., PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 568, 259–263 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Serobyan V., Kontarakis Z., El-Brolosy M. A., Welker J. M., Tolstenkov O., Saadeldein A. M., Retzer N., Gottschalk A., Wehman A. M., Stainier D. Y., Transcriptional adaptation in Caenorhabditis elegans. eLife 9, e50014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frokjaer-Jensen C., Wayne Davis M., Hopkins C. E., Newman B. J., Thummel J. M., Olesen S. P., Grunnet M., Jorgensen E. M., Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40, 1375–1383 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathies L. D., Ray S., Lopez-Alvillar K., Arbeitman M. N., Davies A. G., Bettinger J. C., mRNA profiling reveals significant transcriptional differences between a multipotent progenitor and its differentiated sister. BMC Genomics 20, 427 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossi A., Kontarakis Z., Gerri C., Nolte H., Hölper S., Krüger M., Stainier D. Y., Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230–233 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Burg L., Palmer N., Kikhi K., Miroshnik E. S., Rueckert H., Gaddy E., MacPherson Cunningham C., Mattonet K., Lai S. L., Marin-Juez R., Waring R. B., Stainier D. Y., Balciunas D., Conditional mutagenesis by oligonucleotide-mediated integration of loxP sites in zebrafish. PLOS Genet. 14, e1007754 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y., Kossack M. E., McFaul M. E., Christensen L. N., Siebert S., Wyatt S. R., Kamei C. N., Horst S., Arroyo N., Drummond I. A., Juliano C. E., Draper B. W., Single-cell transcriptome reveals insights into the development and function of the zebrafish ovary. eLife 11, e76014 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doitsidou M., Reichman-Fried M., Stebler J., Köprunner M., Dörries J., Meyer D., Esguerra C. V., Leung T., Raz E., Guidance of primordial germ cell migration by the chemokine SDF-1. Cell 111, 647–659 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Wolke U., Weidinger G., Koprunner M., Raz E., Multiple levels of posttranscriptional control lead to germ line-specific gene expression in the zebrafish. Curr. Biol. 12, 289–294 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Ciruna B., Weidinger G., Knaut H., Thisse B., Thisse C., Raz E., Schier A. F., Production of maternal-zygotic mutant zebrafish by germ-line replacement. Proc. Natl. Acad. Sci. U.S.A. 99, 14919–14924 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conine C. C., Sun F., Song L., Rivera-Perez J. A., Rando O. J., Small RNAs gained during epididymal transit of sperm are essential for embryonic development in mice. Dev. Cell 46, 470–480.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gapp K., Miska E. A., tRNA fragments: Novel players in intergenerational inheritance. Cell Res. 26, 395–396 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gapp K., van Steenwyk G., Germain P. L., Matsushima W., Rudolph K. L., Manuella F., Roszkowski M., Vernaz G., Ghosh T., Pelczar P., Mansuy I. M., Miska E. A., Alterations in sperm long RNA contribute to the epigenetic inheritance of the effects of postnatal trauma. Mol. Psychiatry 25, 2162–2174 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cunningham A. M., Walker D. M., Ramakrishnan A., Doyle M. A., Bagot R. C., Cates H. M., Peña C. J., Issler O., Lardner C. K., Browne C., Russo S. J., Shen L., Nestler E. J., Sperm transcriptional state associated with paternal transmission of stress phenotypes. J. Neurosci. 41, 6202–6216 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bourc’his D., Voinnet O., A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science 330, 617–622 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Martinez G., Panda K., Kohler C., Slotkin R. K., Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat. Plants 2, 16030 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Sharma U., Conine C. C., Shea J. M., Boskovic A., Derr A. G., Bing X. Y., Belleannee C., Kucukural A., Serra R. W., Sun F., Song L., Carone B. R., Ricci E. P., Li X. Z., Fauquier L., Moore M. J., Sullivan R., Mello C. C., Garber M., Rando O. J., Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma U., Sun F., Conine C. C., Reichholf B., Kukreja S., Herzog V. A., Ameres S. L., Rando O. J., Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev. Cell 46, 481–494.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morgan C. P., Chan J. C., Bale T. L., Driving the next generation: Paternal lifetime experiences transmitted via extracellular vesicles and their small RNA cargo. Biol. Psychiatry 85, 164–171 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trigg N. A., Eamens A. L., Nixon B., The contribution of epididymosomes to the sperm small RNA profile. Reproduction 157, R209–R223 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Sullivan R., Epididymosomes: A heterogeneous population of microvesicles with multiple functions in sperm maturation and storage. Asian J. Androl. 17, 726–729 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Belleannee C., Calvo E., Caballero J., Sullivan R., Epididymosomes convey different repertoires of microRNAs throughout the bovine epididymis. Biol. Reprod. 89, 30 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Reilly J. N., McLaughlin E. A., Stanger S. J., Anderson A. L., Hutcheon K., Church K., Mihalas B. P., Tyagi S., Holt J. E., Eamens A. L., Nixon B., Characterisation of mouse epididymosomes reveals a complex profile of microRNAs and a potential mechanism for modification of the sperm epigenome. Sci. Rep. 6, 31794 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuster A., Tang C., Xie Y., Ortogero N., Yuan S., Yan W., SpermBase: A database for sperm-borne RNA contents. Biol. Reprod. 95, 99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leal M. C., de Waal P. P., García-López Á., Chen S. X., Bogerd J., Schulz R. W., Zebrafish primary testis tissue culture: An approach to study testis function ex vivo. Gen. Comp. Endocrinol. 162, 134–138 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Hollick J. B., Paramutation and related phenomena in diverse species. Nat. Rev. Genet. 18, 5–23 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Nocka K., Tan J. C., Chiu E., Chu T. Y., Ray P., Traktman P., Besmer P., Molecular bases of dominant negative and loss of function mutations at the murine c-kit/white spotting locus: W37, Wv, W41 and W. EMBO J. 9, 1805–1813 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brenner S., The genetics of Caenorhabditis elegans. Genetics 1, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mello C., Fire A., Chapter 19 DNA transformation. Methods Cell Biol. 48, 451–482 (1995). [PubMed] [Google Scholar]
- 59.Alestrom P., D’Angelo L., Midtlyng P. J., Schorderet D. F., Schulte-Merker S., Sohm F., Warner S., Zebrafish: Housing and husbandry recommendations. Lab Anim. 54, 213–224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., Sander J. D., Peterson R. T., Yeh J. R., Joung J. K., Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samarut E., Lissouba A., Drapeau P., A simplified method for identifying early CRISPR-induced indels in zebrafish embryos using high resolution melting analysis. BMC Genomics 17, 547 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ly K., Reid S. J., Snell R. G., Rapid RNA analysis of individual Caenorhabditis elegans. MethodsX 2, 59–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jao L. E., Wente S. R., Chen W., Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. U.S.A. 110, 13904–13909 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Horstick E. J., Jordan D. C., Bergeron S. A., Tabor K. M., Serpe M., Feldman B., Burgess H. A., Increased functional protein expression using nucleotide sequence features enriched in highly expressed genes in zebrafish. Nucleic Acids Res. 43, e48 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dumstrei K., Mennecke R., Raz E., Signaling pathways controlling primordial germ cell migration in zebrafish. J. Cell Sci. 117, 4787–4795 (2004). [DOI] [PubMed] [Google Scholar]
- 66.M. Westerfield, M. Westernfield, in The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) (University of Oregon, 2007). [Google Scholar]
- 67.Traver D., Paw B. H., Poss K. D., Penberthy W. T., Lin S., Zon L. I., Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol. 4, 1238–1246 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Radle B., Rutkowski A. J., Ruzsics Z., Friedel C. C., Koszinowski U. H., Dölken L., Metabolic labeling of newly transcribed RNA for high resolution gene expression profiling of RNA synthesis, processing and decay in cell culture. J. Vis. Exp. , 50195 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun W., Chen W., Metabolic labeling of newly synthesized RNA with 4sU to in parallel assess RNA transcription and decay. Methods Mol. Biol. 1720, 25–34 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Blecher-Gonen R., Barnett-Itzhaki Z., Jaitin D., Amann-Zalcenstein D., Lara-Astiaso D., Amit I., High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat. Protoc. 8, 539–554 (2013). [DOI] [PubMed] [Google Scholar]
- 71.Kim E., Sun L., Gabel C. V., Fang-Yen C., Long-term imaging of Caenorhabditis elegans using nanoparticle-mediated immobilization. PLOS ONE 8, e53419 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 and S2