Summary
Background
Neoadjuvant chemoradiotherapy (neoCRT) followed by surgery is the most common approach for locally advanced resectable esophageal squamous cell carcinoma (ESCC) patients. How neoCRT impacts ESCC tumor immune microenvironment (TIME) has not been fully understood.
Methods
Single-cell RNA sequencing (scRNA-seq) was conducted to examine the neoCRT-driven cellular and molecular dynamics in 8 pre- and 7 post-neoCRT ESCC samples from 8 male patients.
Findings
scRNA-seq data of about 112,000 cells were obtained. Expression programs of cell cycle, epithelium development, immune response, and extracellular structure in pre-treatment tumor cells were related to neoCRT response. Spearman correlation between CD8+ T cells' cytotoxicity and expression of checkpoint molecules was prominent in pre-neoCRT intermediate activated/exhausted CD8+ T cells. NeoCRT increased CD8+ T cells’ infiltration but promoted their exhaustion in both major and minor responders. NeoCRT promoted differentiation of Th but demoted that of Treg cells in major responders. Maturation of cDC1s and expression of M2 macrophage markers increased while the number of cDC2s decreased after neoCRT. Higher activities of immune-related pathways in pre-neoCRT CD8+ T cells and macrophages, as well as a pronounced decrease of them after neoCRT, correlated with better neoCRT response. Interactions between intermediate activated/exhausted CD8+ T and macrophages, cDC1s, and LAMP3+ cDCs decreased after neoCRT.
Interpretation
Our comprehensive picture of the neoCRT-related immune changes provides deeper insights into immunological mechanisms associated with ESCC response to neoCRT, which may aid in future development of immune-strategies for improving ESCC treatment.
Funding
This work was supported by the National Natural Science Foundation of China (82072607).
Keywords: Esophageal squamous cell carcinoma, Neoadjuvant chemoradiotherapy, Tumor microenvironment, Single-cell RNA sequencing
Research in context.
Evidence before this study
A repertoire of non-malignant cells contributes to the tumor microenvironment (TME), and implicates in tumor development and treatment response. Although the heterogeneity of TME has been studied in esophageal squamous cell carcinomas (ESCCs), the impacts of chemoradiotherapy on ESCC TME at single-cell level have not been fully explored.
Added-value of this study
Expression programs of cell cycle, epithelium development, immune response, and extracellular structure in pre-treatment tumor cells were related to neoadjuvant chemoradiotherapy (neoCRT) response. The activation-dependent exhaustion expression program was prominent in pre-neoCRT intermediate activated/exhausted CD8+ T cells. Chemoradiotherapy altered the immune environment of ESCCs. NeoCRT increased CD8+ T cell infiltration but promoted its exhaustion. NeoCRT promoted differentiation of Th cells but demoted that of Treg cells in major responders, leading to a lower ratio of Treg to Th cells in major responders as compared to minor ones after neoCRT.
Implications of all the available evidence
Understanding the response of ESCC to chemoradiotherapy contributed by immune cells in TME may provide evidence for developing and applying immune-strategies for improving ESCC treatment. Utilizing PD-1-based immunotherapy as well as targeting Treg cells in neoadjuvant setting might improve ESCC neoCRT responses.
Introduction
Tumors have increasingly been recognized as organs whose complexity approaches containing a repertoire of recruited, ostensibly normal cells that contribute to the tumor microenvironment (TME). As a major component of TME, the immune cells are highly specialized, transcriptionally dynamic and extremely heterogeneous in regards to their phenotypes and functions, and have been implicated in each step of tumor development and response to treatment.1
Esophageal cancer is one of the most aggressive malignancies of the gastrointestinal tract.2 In China, esophageal squamous cell carcinoma (ESCC), a major pathological type of esophageal cancer, ranks as the third most common malignancy, accounting for more than half of the global burden.3 Preoperative neoadjuvant chemoradiotherapy (neoCRT) followed by surgery significantly improved the recurrence and survival for resectable ESCCs at locally advanced stage.4 However, the outcomes of neoCRT are heterogeneous, and survival benefit was observed in neoCRT major responders, including both pathological complete and nearly complete responders.5 The underlying mechanisms of heterogeneous neoCRT responses have been explored but not fully explained. Previous studies carried out at the whole-population level averaged out underlying differences of various components in the bulk tissue.6,7 Unbiased assessment of many heterogeneous tumor and immune cells at single-cell level would help to decipher the complex ecosystem of ESCC TME and their response to neoCRT and provide potential predictive biomarkers.
Here, we performed single-cell RNA sequencing (scRNA-seq) on pre- and post-neoCRT tumor samples from ESCC patients who received neoCRT and surgery, so as to characterize the cellular and molecular dynamics driven by neoCRT and their relationship to differential responses.
Methods
Ethics
The study was proved by the Research Ethics Committee of Sun Yat-sen University Cancer Center, Guangzhou, China (GZR2020-275). Patients gave informed consent at hospitalization.
Patients and sample collection
Pre- and post-neoCRT ESCC samples were obtained from pathologically diagnosed ESCC patients, who were clinically staged as T1-4N1-3M0 potentially resectable thoracic ESCCs, and received neoCRT and complete esophagectomy after neoCRT. The neoCRT was administrated as cisplatin (75 mg/m2) intravenous on Days 1 and 22, and vinorelbine (25 mg/m2) intravenous on Days 1, 8, 22 and 29 or cisplatin (25 mg/m2) intravenous and docetaxel (25 mg/m2) intravenous on Days 1, 8, 15 and 22, concurrently with 4 weeks of radiation therapy (44Gy/20 fractions). McKeown, Ivor Lewis or minimally invasive esophagectomy and two-field lymphadenectomy with total mediastinal lymph node dissection was performed 4–8 weeks after neoCRT.
Tumor response to neoCRT was evaluated with tumor regression grade (TRG) based on the estimated percentage of vital residual tumor cells (VRTCs): grade 0, pathological complete response; grade 1, nearly complete response with <10% VRTCs; grade 2, partial response with 10%–50% VTRCs; and grade 3, >50% VTRCs. Patients with TRG 0–1 was recorded as major responders, while others as minor responders.5
Up to 10 pre-neoCRT cancer biopsies were collected during endoscopy and pooled together. Post-neoCRT primary tumor specimens with vital tumor macroscopically or pre-existent tumor areas with erosive, ulcerated or fibrotic alteration were obtained during surgery. A tissue biopsy or specimen adjacent to each sample was formalin-fixed paraffin-embedded (FFPE) and hematoxylin and eosin stained and assessed for the presence of cancer cells. Biopsies or specimens left after single-cell preparation for RNA-sequencing were snap-frozen in liquid nitrogen and stored at −80 °C.
scRNA-seq and data processing
Immediately following biopsy or resection, tumor samples were minced and dissociated into single-cell suspensions in Leibovitz L-15 (Thermo Fisher Scientific, Waltham, MA) medium containing 0.375 U/ml Liberase TM (Roche, Indianapolis, IN). Erythrocyte in single-cell suspensions were lysed with red blood cell lysis buffer (Thermo Fisher Scientific). Then the single cell suspensions were stained with LIVE/DEAD Viability/Cytotoxicity Assay Kit (Thermo Fisher Scientific), filtered over Flowmi CellStrainer (Sigma–Aldrich, St. Louis, MO), and selected by flow cytometer MoFlo Astrios (Beckman Coulter, Brea, CA) for live cells.
Single-cell suspensions with more than 80% live cells counted by an automated cell counter (Bio-Rad, Hercules, CA) were loaded into Chromium Controller (10× Genomics, Pleasanton, CA) according to the standard protocol of the Chromium Single Cell 3′GEM, Library & Gel Bead Kit (v3, 10× Genomics) in order to capture 8000 cells of each tumor sample. Briefly, the cells were partitioned into Gel Beads-in-emulsion, then co-partitioned cell is lysed and barcoded reverse transcription of RNA occurred, followed by amplification, and adaptor and sample index attachment. Libraries were sequenced on a NovaSeq 6000 (Illumina, San Diego, CA) with paired-end 150 bp to achieve a sequencing saturation close to 80% and mapped to the human genome (GRCh 38) using CellRanger (v3.1.0, 10× Genomics).
Single-cell gene expression quantification and determination of the major cell types
The raw gene expression count matrix of each sample obtained using CellRanger were analyzed using the Seurat R package (v3.2.1).8 For each sample, low quality cells with unique feature less than <200 and mitochondrial mapping percentage >0.25 were removed. Also, cells exhibit an aberrantly high gene count were removed from count matrix of each sample. For the remaining cells of each sample, the count data was normalized and scaled using the function “SCTransform” with mitochondrial mapping percentage being regressed out as a confounding source of variant. Then, linear dimensional reduction, cell clustering, and non-linear dimensional reduction uniform manifold approximation and projection (UMAP) was performed with the number of principal components determined by elbow plot to cluster and visualize the data. The differentially expressed markers of each cluster were identified using the function “FindAllMarkers” with default setting. Clusters with high KRT5 expression were annotated as epithelial cells, while others non-epithelial cells. The non-epithelial cells of each sample were further integrated with 3000 features and 50 dimensions using Seurat “Reciprocal PCA” integration workflow to correct batch effect, while the epithelial cells were merged using the function “merge” so as to keep patient-specific tumor heterogeneity.9,10 All the integrated non-epithelial cells and merged epithelial cells were further clustered and visualized with proper settings, respectively. The cell clusters of all non-epithelial cells were classified into several major cell types based on the expression of known marker genes, including CD3D (T cells), CD8A (CD8+ T cells), CD4 [CD4+ helper T (Th) cells], FOXP3 [regulatory T (Treg) cells], TRDC (natural killer cells), MS4A1, CD19 (B cells), MZB1 (plasma cells), CSF1R [macrophages and conventional dendritic cells (cDCs)], LILRA4 [plasmacytoid DCs (pDCs)], CSF3R (neutrophils), TPSB2 (mast cells), ENG and CDH5 (endothelial cells), and COL1A1 (fibroblasts). To identify subclusters within each major cell types, we reanalyzed cells belongs to each of the major cell types separately. Linear dimensional reduction, cell clustering and UMAP were performed as described above with proper parameters for each major cell type. Cell clusters with an extremely low number of unique molecular identifiers and genes, an extremely high expression of mitochondrial genes, or multiple well-defined marker genes of different cell types were removed for further analysis. The differentially expressed genes of specific cell clusters or types between major and minor responders or between pre- and post-samples were identified using the function “FindAllMarkers” with Mann–Whitney U test and default setting.
Inferring copy number alterations from scRNA-seq data
The count matrix exported form Seurat object was used for copy number variation (CNV) analysis. For each sample, the initial CNV values of each single cell were estimated by inferCNV R package (v1.4.0) (https://github.com/broadinstitute/inferCNV) based on averaged expression profiles across chromosomal intervals. Cells were further clustered to identify clusters of malignant epithelial cells with higher CNV changes. A CNV score was calculated as the mean squares of the CNV values across the genome to evaluate the CNV level of each single cell quantitatively as described previously.11
Expression signature of intra-tumoral heterogeneity
For each sample, consensus non-negative matrix factorization (cNMF) analysis was applied to decipher potential signatures consisting of coexpressed genes with cNMF python pipeline (v1.2) based on the count matrix of malignant cells.12 Pearson correlation coefficient of these expression programs were calculated and clustered to identify meta-programs which were common across multiple samples. For each cluster, cNMF gene score were log2-transformed and then averaged across the programs in the clusters, and genes were ranked by their average scores. Enrichment of the Gene Ontology (GO) biological process was analyzed using g:Profiler13 with the top 60 genes of each meta-program. The top 30 genes for each meta-program were defined as the meta-signature that was used to define cell scores.
Gene set variation analysis (GSVA)
Pathway analyses were performed on the 50 hallmark pathways described in the Molecular Signature Database (v7.1),14 exporting using the GSEABase R package (v1.50.1). Pathway activity of individual cells were estimated using the GSVA R package (v1.36.2)15 with the normalized expression matrix exported from Seurat. To assess differential activities of pathways between sets of cells, we contrasted the activity scores of each cells by moderated t-test using a linear model and an empirical Bayes shrinkage method with limma R package (v3.44.3), and t value was calculated for differences between cells from one cluster and cells from all other clusters.16
Gene regulatory network analysis
In order to infer the gene regulatory network, we utilized pySCENIC (v0.10.3), a python-based algorithm for reconstructing regulons [transcription factors (TFs) and their target genes] of each individual cells from the scRNA-seq data.17 The input matrix was the raw count matrix, output from Seurat with low quality cells and low expression genes (genes expressed in fewer than 1% cells) filtered. Differences of the activities of the regulons between sets of cells were compared by moderated t-test using a linear model and an empirical Bayes shrinkage method with limma R package (v3.44.3), and t value was calculated for differences between cells from one cluster and cells from all other clusters.16
RNA velocity-based cell fate tracing
The spliced reads and unspliced reads were recounted by the velocyto python package (v0.17.15) based on previous aligned bam files of scRNA-seq data of each sample.18 The RNA velocity values for each gene in each cell of each major cell type were calculated using a likelihood-based dynamical model following the scVelo python pipeline (v0.2.2),19 which was further embedded into low-dimension space based on the Seurat UMAP coordinates and cluster information. The underlying gene-shared latent time was inferred in an efficient expectation-maximization framework to reconstruct the temporal sequence of transcriptomic events and cellular fates. The velocity-based cell transition matrix was calculated from scVelo, of which the element was the cosine correlation coefficient between the velocity vector and cell state difference vectors of the column cell. The destination of a cell was estimated by identifying the highest correlation value. Then Chi-square test was performed to test the fate destinations of interested cell clusters as described previously.20
Cell–cell interaction analysis
For systematic analysis of cell–cell interactions, we used CellPhoneDB (v2.1.4)21 with default augments to reveal ligand–receptor pairs between the major cell subtypes of T cells (intermediate activated/exhausted CD8+ T, exhausted CD8+ T, Th, and Treg cells) and macrophages (CD163+ and CD163− macrophages) and DCs (cDC1, cDC2, LAMP3+ cDCs and pDCs) from scRNA-seq data of each pre- and post-neoCRT sample. The significant ligand–receptor pairs were filtered with a P value <0.05. Only significant interactions existed in at least one sample were counted when calculating the number of all possible interactions between any two types of cells.
Bulk whole exome sequencing and copy number alternation analysis
Four pre-neoCRT biopsies left after scRNA-seq were used for bulk whole exome sequencing. Tissue and blood genomic DNA was extracted using the AllPrep DNA Mini Kit (QIAGEN, Germantown, MD) and QIAamp DNA blood mini kit (QIAGEN), respectively, and then randomly broken into fragments with a length of 180–280 bp. The library construction and capture were performed with the SureSelect Human All Exon V6 kit (Agilent, Santa Clara, CA) according to the manufactures’ protocol. Qualified captured libraries were sequenced at 150 paired-end cycles on a HiSeq4000 system (Illumina). Raw fastq data were quality-trimmed, and adapter sequences were removed, and aligned to the human reference genome (human_g1k_v37) using Burrows-Wheeler Aligner (v0.7.8-r455). Samblaster (v0.1.21)22 was used to select split reads and discord reads from the initial comparison results. Duplicate reads were marked with sambamba (v0.6.3).23 FastQC (v0.11.5) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Qualimap (v2.2)24 were used to perform quality checks on the fastq and bam files, respectively. Tissue copy number variation analysis calls were made by Controlfreec (v11.6)25 with matched blood genomic as normal control. Significant regions of copy number alteration were selected using default parameters at a q value threshold of 0.25 with GISTIC2 (v2.0.23).26
Multiplex fluorescent immunohistochemistry (mfIHC)
FFPE slides were subjected to mfIHC using the PANO Multiplex IHC kit (Panovue Biotechnology, BJ, China) to examine specific cell markers with antibodies of CD8 (ZSGB-Bio, Cat# ZA-0508, RRID:AB_2890107), GZMB (Abcam, Cat# ab4059, RRID:AB_304251), PDCD1 (Abcam, Cat# ab52587, RRID:AB_881954), and panCK (Abcam, Cat# ab215838, RRID:AB_2922672) in panel A, CD4 (Abcam, Cat# ab133616, RRID:AB_2750883), CCR7 (Abcam, Cat# ab253187, RRID:AB_2922673), CD45RO (Cell Signaling Technology, Cat# 55618, RRID:AB_2799491), FOXP3 (Abcam, Cat# ab215206, RRID:AB_2860568), and panCK (Abcam, Cat# ab215838, RRID:AB_2922672) in panel B, CD68 (Abcam, Cat# ab213363, RRID:AB_2801637), CD163 (Abcam, Cat# ab182422, RRID:AB_2753196), CD1C (Abcam, Cat# ab156708, RRID:AB_2889187), and panCK (Abcam, Cat# ab215838, RRID:AB_2922672) in panel C. For each panel, after each primary antibody was sequentially applied, the slides were incubated with horseradish peroxidase-conjugated secondary antibody (Panovue Biotechnology, Cat# 10013001050, RRID: AB_2924778) and tyramide signaling amplification (TSA). The slides were microwave heat-treated after each TSA operation. Nuclei were stained with 4′-6′-diamidino-2-phenylindole (DAPI, Sigma–Aldrich, St. Louis, MO) after all the antigens of each panel had been labelled. To obtain multispectral images, the stained slides were scanned using a Vectra Polaris Automated Quantitative Pathology Imaging System (PerkinElmer, Waltham, MA), which captured the fluorescence excitation spectrum at 10-nm wave length intervals (440–780 nm) within the same exposure time. HALO imaging analysis software (Indica Labs, Corrales, NM) were used to batch analyze all images. Areas of the whole pre-neoCRT biopsies' slides and areas of residual tumors and pre-existent tumors on the post-neoCRT specimens' slides were used for mfIHC analysis.
Statistical analysis
Statistical analysis was carried out using SPSS 20.0 software. The difference of TRG between the two different neoCRT regimens was analyzed by the Chi-square test. Wilcoxon signed-rank test was used for comparison of the differences of cell percentages between paired pre- and post-neoCRT samples, and Mann–Whitney U test for comparison between major and minor responders in either pre- or post-neoCRT samples. P value was adjusted with Benjamini-Hochberg correction for multiple tests. A significant difference was declared if the P value from a two-tailed test was <0.05.
Role of funders
The funding sources played no role in study design, collection, analysis, and interpretation of data, writing the report, or the decision of paper submission.
Results
ScRNA-seq and cell typing of cells in pre- and post-neoCRT ESCC samples
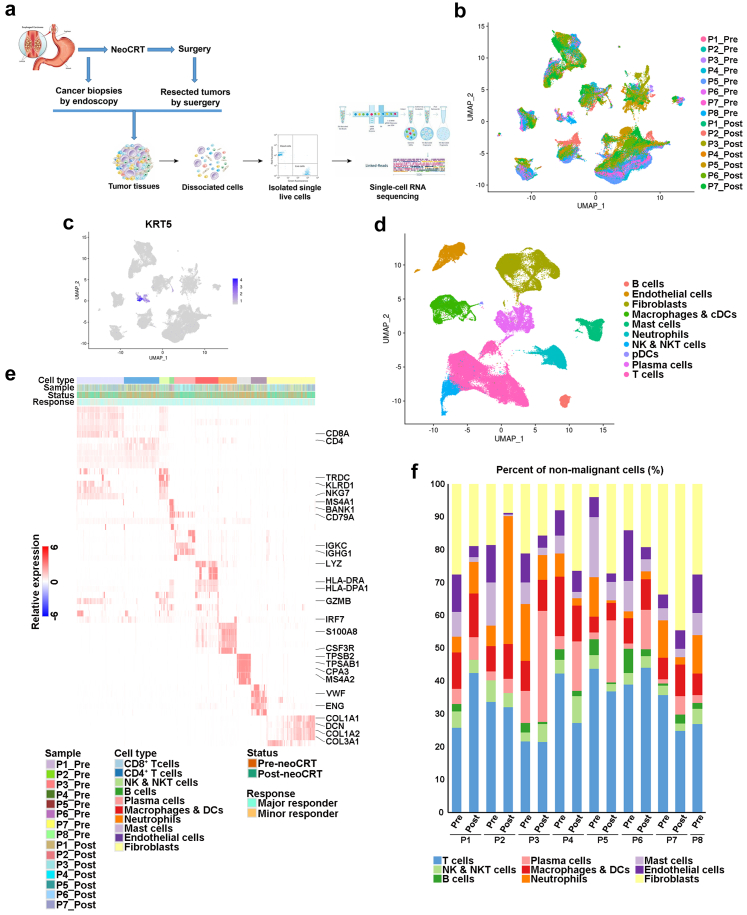
ScRNA-seq data of 111784 cells of 8 pre- and 7 post-neoCRT samples were obtained from 8 ESCC patients received neoCRT and surgery, 4 of whom were treated with docetaxel plus cisplatin and the other 4 with vinorelbine plus cisplatin in neoCRT (Fig. 1a and b, Supplementary Table S1). There was no significant difference of TRG between the two chemotherapy regimens (P = 0.629 by Chi-square test).
Fig. 1.
Single-cell transcriptional profiling of pre-neoCRT and post-neoCRT cells in ESCC. (a) Workflow showing collection and processing of fresh pre-neoCRT endoscopic biopsies and post-neoCRT surgical resected specimens for scRNA-seq. (b) UMAP plot of 111,784 cells in 8 pre- and 7 post-neoCRT ESCC samples. (c) UMAP plot showing expression of epithelial cell marker KRT5. (d) UMAP plot of non-epithelial cells to visualize major cell types. (e) Heatmap showing the signature gene expression of the major cell types. (f) The proportion of cells that contributed to each major cell type by each sample, colored by cell type.
The clusters of epithelial cells could be distinguished from those of non-epithelial cells according to the expression of epithelial marker gene KRT5 (Fig. 1c, Supplementary Fig. S1). Based on above analysis, 2970 epithelial cells from 8 pre-neoCRT and 1 post-neoCRT (P4_Post) samples were obtained in total.
The 108814 non-epithelial cells from 15 samples were further integrated, visualized and clustered as several major cell types based on canonical marker genes (Fig. 1d and e, Supplementary Fig. S2a). The major cell types exhibited different proportions in the samples, partially reflecting differences in disease etiology or progression related to neoCRT (Fig. 1f). There was no significant difference of the major cell type proportion in post-neoCRT samples between the two chemotherapy regimens (P > 0.05 by Mann–Whitney U test with Benjamini-Hochberg correction, Supplementary Fig. S2b).
Gene expression programs of tumor cells correlated with neoCRT responses
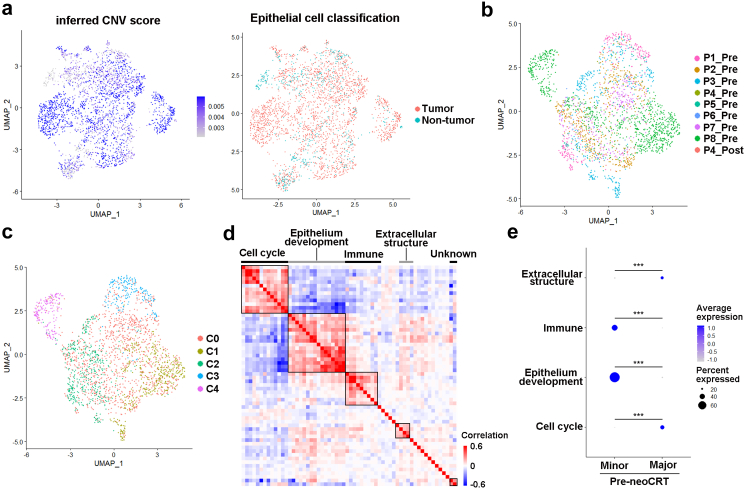
The 2970 epithelial cells were extracted and analyzed. Inferred large-scale CNVs confirmed the abnormal karyotypes of most epithelial cells, which were consistent with CNVs from whole exosome sequencing of bulk tissue samples from the same patients (Supplementary Fig. S3a and b). However, low CNV changes were observed in a few epithelial cells (Supplementary Fig. S3a), suggesting the existence of non-malignant esophageal squamous epithelial cells. Clustering of cells based on CNV changes identified a total of 2403 epithelial cells with higher CNV scores than others, therefore, these cells were considered as tumor cells (Fig. 2a, Supplementary Fig. S3c). There was no obvious expression of epithelial markers (Supplementary Fig. S1) and CNV changes (Supplementary Fig. S3a) in 6 out of 7 post-neoCRT samples, indicating no normal or malignant epithelial cells were detected in these 6 samples. The lower percentages and viabilities of tumor cells in post-neoCRT samples rendered them more vulnerable to sample processing and difficult to be captured by scRNA-seq, making it impossible to observe the change of tumor cells’ transcriptome induced by neoCRT. Clustering of the 2403 tumor cells from 9 samples revealed 5 clusters (Fig. 2b and c, Supplementary Fig. S4a, Supplementary Table S2).
Fig. 2.
Expression programs of tumor cells correlated with neoCRT responses. (a) UMAP plot showing the inferred CNV score and classification of 2970 epithelial cells. (b) UMAP plot of 2403 tumor cells in 8 pre- and 1 post-neoCRT ESCC samples. (c) UMAP plot of 5 clusters of tumor cells. (d) Heatmap depicting pairwise correlation of 60 intra-tumoral expression programs, and clustering identifying 5 coherent meta-programs across tumor cells from 8 pre-neoCRT samples. (e) Differences in the top 30-gene cell scores of the 4 major meta-programs between pre-neoCRT malignant cells from major or minor responders. Major, major responders; minor, minor responders. ∗∗∗, P < 0.001 by Mann–Whitney U test with Benjamini-Hochberg correction.
cNMF was applied to the tumor cells from 8 pre-neoCRT samples so as to decipher potential coherent sets of genes that were preferentially co-expressed by subsets of the cells. A total of 60 gene expression programs were identified. Hierarchical clustering of the programs’ correlation identified 5 meta-programs preferentially expressed by subset of malignant cells (Fig. 2d, Supplementary Table S3). GO analysis showed that the 4 major meta-programs were primarily involved in cell cycle, epithelium development, immune response, and extracellular structure (Supplementary Fig. S4b). The fifth meta-program was expressed by subsets of malignant cells from only two samples, and no enrichment of known GO biological process terms was identified. The top 30-gene cell score of each meta-program varied within tumor cells from each patient, demonstrating a pronounced intra-tumor heterogeneity (Supplementary Fig. S4c). The scores of the identified 4 major meta-program were compared between major and minor responders, and found that scores of cell cycle and extracellular structure were higher in pre-neoCRT tumor cells from major responders, while scores of epithelium development and immune response were higher in pre-neoCRT tumor cells from minor responders (P < 0.05 by Mann–Whitney U test with Benjamini-Hochberg correction, Fig. 2e).
NeoCRT increased CD8+ T cell infiltration but promoted its exhaustion
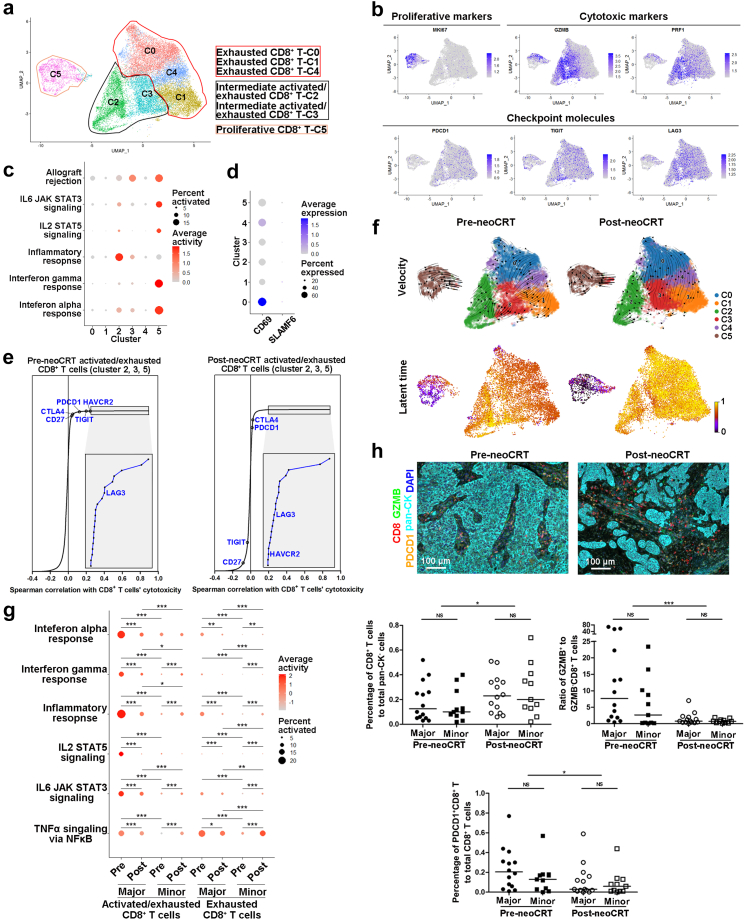
With 22244 cells detected, CD8+ T cells could be clustered into 6 clusters with differential gene expression patterns in both pre- and post-neoCRT cells (Fig. 3a, Supplementary Fig. S5a and b, Supplementary Table S2). Cluster 5 was proliferative cells with high expression of genes, as well as high activities of pathways and TFs, related to cell proliferation (Fig. 3b, Supplementary Fig. S5c and d). Cytotoxic markers GZMB and PRF1 were highly expressed in cluster 2, 3, and 5 (Fig. 3b), accordingly, immune-related pathways, such as inflammatory response, interferon alpha and gamma responses, allograft rejection, as well as IL-2 and IL-6 signalings were enriched in these clusters with varied degrees (Fig. 3c), indicating CD8+ T cells' activation and cytotoxicity towards cancer cells. However, a series of checkpoint molecules were co-expressed in these activated CD8+ T clusters (Fig. 3b, Supplementary Fig. S5e), curtailing CD8+ T cells' cytotoxic activities and suggesting a broadly exhausted state of CD8+ T cells in ESCC TME.9 Therefore, these activation- and exhaustion-associated genes co-expressed cells were defined as intermediate activated/exhausted CD8+ T cells as in other types of cancers, such as basal cell carcinomas.27 Cells in cluster 0, 1, and 4 were exhausted CD8+ T cells, with immune checkpoints' expression but few cytotoxic markers' expression and no T-cell activation pathways’ enrichment (Fig. 3b and c, Supplementary Fig. S5c and e). Among the exhausted CD8+ T cells, cluster 0 cells were with the highest expression of CD69 but negative expression of SLAMF6 (Ly108) (Fig. 3d), as well as high activity of TF EOMES (Supplementary Fig. S5d), characteristics of terminally exhausted CD8+ T cells as described by Beltra et al. previously.28 These terminally exhausted CD8+ T cells might regulate trafficking of immune cells in ESCC TME via secreting chemokines like CCL3 and CCL4,29,30 although their cytotoxicity was lost. Spearman correlation between cytotoxicity of CD8+ T cells, as measured by the average of GZMB and PRF1 expression, and the expression of checkpoint molecules was stronger in pre-neoCRT intermediate activated/exhausted CD8+ T cells (Fig. 3e, Supplementary Fig. S5f), suggesting the activation-dependent exhaustion expression program9 was prominent in these cells. RNA-velocity analysis showed a transition out from cluster 5 proliferative CD8+ T cells in both pre- and post-neoCRT CD8+ T cells. For pre-neoCRT cells, two transition directions, one to cluster 2 and 3 intermediate activated/exhausted CD8+ T cells and the other to exhausted CD8+ T cells, were observed. For post-neoCRT cells, transition to exhausted CD8+ T cells, especially to cluster 0 terminally exhausted ones, was more obvious (Fig. 3f). Furthermore, the latent time of post-neoCRT exhausted CD8+ T cells (cluster 0, 1, and 4) was later than that of pre-neoCRT ones, suggesting that the exhausted CD8+ T cells were produced during or after neoCRT (Fig. 3f), that is, neoCRT promoted CD8+ T cell exhaustion.
Fig. 3.
Characterization of CD8+T cells in pre- and post-neoCRT ESCCs. (a) UMAP plot of 6 clusters of CD8+ T cells. (b) UMAP plots showing expression of proliferative and cytotoxic markers and checkpoint molecules. (c) Dotplot showing activities of immune-related pathways in different CD8+ T clusters. (d) Dotplot showing expression of CD69 and SLAMF6 in different CD8+ T clusters. (e) Spearman correlation between the cytotoxicity and expression of immune checkpoint molecules in pre- and post-neoCRT intermediate activated/exhausted CD8+ T cells. (f) RNA velocities and latent time visualized on the UMAP projection of different CD8+ T clusters. (g) Dotplot showing differences in immune-related pathway activities in intermediate activated/exhausted and exhausted CD8+ T cells. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001 by moderated t-test with Benjamini-Hochberg correction. (h) Representative images of mfIHC staining of CD8, GZMB, PDCD1, and pan-CK in pre- and post-neoCRT ESCC slides. Differences in the percentages of total CD8+ and PDCD1+CD8+, and ratio of GZMB+ to GZMB− CD8+ T cells were compared. Major, major responders; minor, minor responders. ∗, P < 0.05; ∗∗∗, P < 0.001 by Wilcoxon signed-rank test. NS, not significant by Mann–Whitney U test.
The percentage of total CD8+ T to total non-malignant cells increased significantly in post-neoCRT samples compared with that in paired pre-neoCRT samples by scRNA-seq analysis (P = 0.018 by Wilcoxon signed-rank test, Supplementary Fig. S5g). Expression of cytotoxic marker GZMB in major responders but not in minor ones decreased, while expression of checkpoint molecules decreased after neoCRT in both major and minor responders (P < 0.05 by Mann–Whitney U test with Benjamini-Hochberg correction, Supplementary Fig. S5h). Consequently, activities of CD8+ T cells’ cytotoxicity-related pathways decreased in major responders but increased in minor responders (P < 0.05 by moderated t-test with Benjamini-Hochberg correction, Fig. 3g). Higher GZMB and PRF1 expression (P < 0.05 by Mann–Whitney U test with Benjamini-Hochberg correction, Supplementary Fig. S5h) and enrichment of cytotoxicity-related pathways (P < 0.05 by moderated t-test with Benjamini-Hochberg correction, Fig. 3g) were observed in major than that in minor responders of both pre- and post-neoCRT samples. In order to validate above findings by scRNA-seq analysis, additional 25 pairs of pre- and post-neoCRT ESCC samples from patients who received the same neoCRT regimens as those for scRNA-seq (Supplementary Table S4) were stained with CD8, GZMB, and PDCD1 using mfIHC. Results showed that CD8+ T cells increased after neoCRT (P = 0.014 by Wilcoxon signed-rank test, Fig. 3h). Among the CD8+ T cells, the ratio of GZMB+ (as the intermediate activated/exhausted CD8+ T cells in scRNA-seq) to GZMB− CD8+ T cells (as the exhausted CD8+ T cells in scRNA-seq) decreased significantly after neoCRT in both major and minor responders (P = 0.001 by Wilcoxon signed-rank test, Fig. 3h), suggesting that the increased CD8+ T cells after neoCRT were mainly exhausted ones. The proportion of PDCD1+CD8+ T to total CD8+ T cells decreased after neoCRT in both major and minor responders (P = 0.048 by Wilcoxon signed-rank test, Fig. 3h). There was no significant difference in total CD8+ T infiltration, the ratio of GZMB+ to GZMB− CD8+ T cells, and the proportion of PDCD1+CD8+ T cells between major and minor responders in either pre- and post-neoCRT ESCCs (P > 0.05 by Mann–Whitney U test, Fig. 3h).
NeoCRT promoted differentiation of Th but demoted that of Treg cells
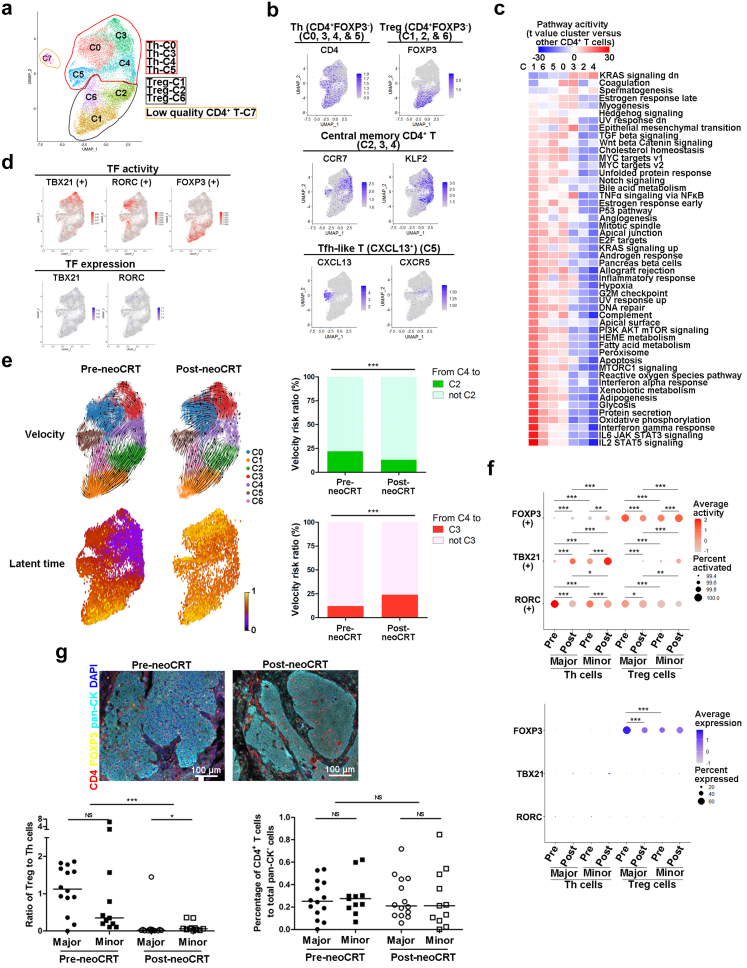
Clustering of the CD4+ T cells revealed 8 clusters with distinct marker genes’ expression in both per- and post-neoCRT samples (Fig. 4a, Supplementary Fig. S6a and b, Supplementary Table S2). Cells in cluster 7 were considered as low quality cells due to lower number of unique molecular identifiers and genes than other clusters and were not included for further analysis. CD4+ T cells were composed of two major cell types, Th (CD4+ FOXP3−, cluster 0, 3, 4, and 5) and Treg (CD4+FOXP3+, cluster 1, 2, and 6, Fig. 4b) cells. IFNG (a marker of Th1 cells) were primarily expressed in parts of cluster 0, 3, and 5, IL17A/F and IL22 (markers of Th17 cells) were in part of cluster 0, while no expression of IL4/5 (markers of Th2 cells) were observed (Supplementary Fig. S6c). Cluster 5 Th cells exhibited a relatively clear separation from other clusters on UMAP (Fig. 4a). These cells highly expressed CXCL13 like follicular helper T (Tfh) cells, but little CXCR5 expression (Fig. 4b). Cells in cluster 4 and parts of cluster 2, 3 were featured with the high expression of KLF2, a marker of quiescent cells, and CCR7, a marker of naïve or central memory T cells (Fig. 4b).31,32 mfIHC staining of ESCC tissues showed that CCR7+CD4+ cells were all with positive expression of CD45RO (Supplementary Fig. S6d), suggesting that they were central memory CD4+ T cells. These central memory CD4+ T cells were in a quiescent state with enrichment of Kras signaling downregulation and less enrichment of pathways related to immune effector activities (Fig. 4c). Th and Treg cells exhibited different patterns of TF activation (Fig. 4d, Supplementary Fig. S6e). Activity of TBX21, the classical TF regulating Th1 cells, was highest in cluster 3 and moderate in cluster 0 Th cells, while that of RORC, the TF regulating Th17 cells, was primarily observed in cluster 0 Th cells. Varied activities of FOXP3 were observed in cluster 1, 2, and 6 Treg cells. Differentiation of cluster 5 CXCL13+CXCR5− Tfh-like cells was driven mainly by SREBP2 and IRF8, different from master TFs for classical Tfh cells.33 ETV7, RELA, and FOS were main TFs for cluster 4 central memory CD4+ T cells.
Fig. 4.
Characterization of CD4+T cells in pre- and post-neoCRT ESCCs. (a) UMAP plot of 8 clusters of CD4+ T cells. (b) UMAP plots showing expression of markers for Th, Treg, Tfh-like and quiescent memory cells. (c) Heatmap showing the differences in pathway activities among different CD4+ T clusters, with t value calculated for differences between cells from one cluster and cells from all other clusters. (d) UMAP showing the TF activities and gene expression of TBX21, RORC, and FOXP3. (e) RNA velocities and latent time visualized on the UMAP projection of different CD4+ T clusters. Bar plots showing the ratios of RNA velocity links from cluster 4 to cluster 2 and 3 CD4+ T cells, respectively. ∗∗∗, P < 0.001 by Chi-square test. (f) Differences in transcription activities and gene expression of TBX21, RORC, and FOXP3 in Th and Treg cells. ∗P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001; or NS, not significant by moderated t-test for transcription activity comparison with Benjamini-Hochberg correction and Mann–Whitney U test for gene expression comparison with Benjamini-Hochberg correction. (g) Representative images of mfIHC staining of CD4, FOXP3, and pan-CK in pre- and post-neoCRT ESCC slides. Differences in the percentages of CD4+ T and ratio of Treg to Th cells were compared. Major, major responders; minor, minor responders. ∗P < 0.05; ∗∗∗, P < 0.001; or NS, not significant by Mann–Whitney U test for unpaired samples or Wilcoxon signed-rank test for paired samples.
Using RNA velocity, we found that the quiescent central memory T cells (cluster 4 and part of cluster 2, 3) could differentiate into effector Th (cluster 0, 3 and 5) and Treg cells (cluster 1, 2, and 6) in both pre- and post-neoCRT samples. However, transition from cluster 4 to cluster 2 Treg cells decreased while that from cluster 4 to cluster 3 Th cells increased after neoCRT (P < 0.001 by Chi-square test, Fig. 4e). Furthermore, the latent time of post-neoCRT Th (cluster 0, 3, 4, and 5) cells were obviously later than that of the pre-neoCRT ones (Fig. 4e), suggesting that Th cells, at least parts of, were differentiated during or after neoCRT. Meanwhile, transcription activities of the classical TFs for CD4+ T differentiation changed after neoCRT (Fig. 4f). Activity of TBX21 increased but that of RORC decreased in both major and minor responders after neoCRT (P < 0.05 by moderated t-test with Benjamini-Hochberg correction). For FOXP3, its activity decreased obviously in major responders but increased slightly in minor responders after neoCRT (P < 0.05 by moderated t-test with Benjamini-Hochberg correction). Furthermore, higher activity of RORC but lower that of TBX21 was observed in major responders of both pre- and post-neoCRT CD4+ T cells as compared to minor responders (P < 0.05 by moderated t-test with Benjamini-Hochberg correction). And CD4+ T cells from major responders exhibited high activity of FOXP3 than those from minor responders in pre-neoCRT samples, which was reversed in post-neoCRT samples (P < 0.001 by moderated t-test with Benjamini-Hochberg correction). Different from transcription activity, expression of either TBX21 or RORC was poor in CD4+ T cells, and no expression change was observed after neoCRT or between samples with different neoCRT responses. FOXP3 expression exhibited the same change tendency with its TF activity (P < 0.001 by Mann–Whitney U test with Benjamini-Hochberg correction, Fig. 4f).
The percentage of total CD4+ T to all non-malignant cells decreased significantly after neoCRT in paired scRNA-seq samples (P = 0.018 by Wilcoxon signed-rank test Supplementary Fig. S6f). Particularly, the ratio of Treg to Th cells decreased significantly after neoCRT (P = 0.018 by Wilcoxon signed-rank test, Supplementary Fig. S6f). mfIHC staining of 25 pairs of pre- and post-neoCRT ESCC samples (Supplementary Table S4) with CD4 and FOXP3 verified the significant decrease of Treg to Th ratio after neoCRT in both major and minor responders (P < 0.001 by Wilcoxon signed-rank test), although the percentage of total CD4+ T cells did not change after neoCRT (P = 0.840 by Wilcoxon signed-rank test, Fig. 4g). Furthermore, higher Treg to Th ratio was observed in minor responders than that in major responders in post-neoCRT (P = 0.021 by Mann–Whitney U test) but not pre-neoCRT samples (P = 0.202 by Mann–Whitney U test, Fig. 4g).
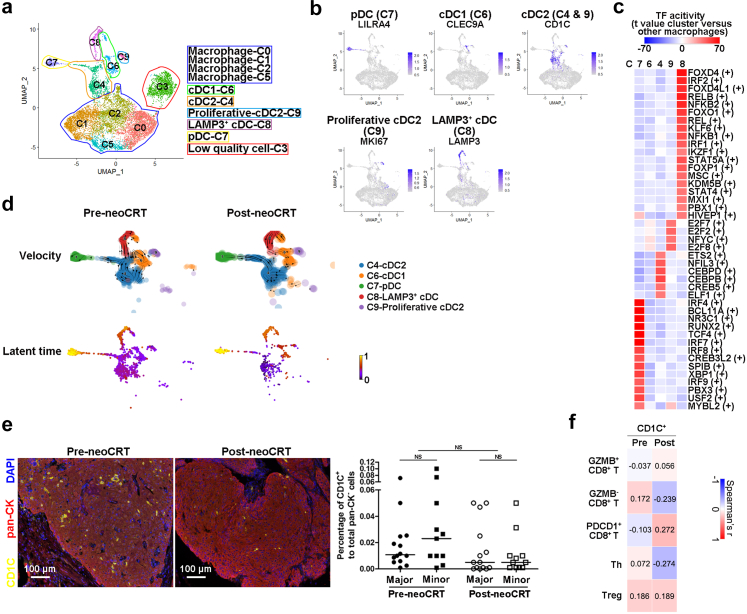
NeoCRT promoted the maturation of cDC1s but decreased the number of cDC2s
Clustering of the 10399 macrophages and DCs revealed 10 clusters with different marker genes’ expression in both pre- and post-neoCRT samples (Fig. 5a, Supplementary Fig. S7a and b, Supplementary Fig. S8a, Supplementary Table S2). Cells in cluster 3 were considered as low quality cells due to lower number of unique molecular identifiers and genes than other clusters and were not included for further analysis.
Fig. 5.
Characterization of DCs in pre- and post-neoCRT ESCCs. (a) UMAP plot of 10 clusters of macrophages and DCs. (b) UMAP plots showing expression of markers for pDC, cDC1, cDC2, proliferative cDC2, and LAMP3+ cDC. (c) Heatmap showing the differences in TF activities among different DC clusters, with t value calculated for differences between cells from one cluster and cells from all other clusters. (d) RNA velocities and latent time visualized on the UMAP projection of different DC clusters. (e) Representative images of mfIHC staining of CD1C and pan-CK in pre- and post-neoCRT ESCC slides. Differences in the percentages of CD1C+ cells were compared. Major, major responders; minor, minor responders. NS, not significant by Mann–Whitney U test for unpaired samples or Wilcoxon signed-rank test for paired samples. (f) Spearman correlation analyses between the percentages of CD1C+ cells and different subtypes of T cells in 25 pairs of pre- and post-neoCRT ESCCs stained by mfIHC. Correlation coefficient was labelled.
DCs in the ESCC TME includes pDCs (LILRA4+IRF7+TCF4+, cluster 7), cDC1s (CLEC9A+, cluster 6), and cDC2s (CD1C+CLEC10A+FCER1A+, cluster 4 and 9) (Fig. 5b, Supplementary Fig. S7b).33 Cluster 9 cDC2s highly expressed a series of proliferative markers (Fig. 5b, Supplementary Fig. S7b). Cluster 8 cells expressed the comparable levels of cDC marker HLA-DRs, but were negative for CLEC9A and CD1C expression (Fig. 5b, Supplementary Fig. S7b and c). These DCs highly expressed maturation marker LAMP3, the co-stimulatory molecules CD40 and CD80, the cell migration and motility markers CCR7 and FSCN1, and the lymphocyte recirculation chemokines CCL19 and CCL22, as the same as the previously reported activated cDCs in lung cancers10 and LAMP3+ cDCs in hepatocellular carcinoma.34 Meanwhile, expression of checkpoints IDO1, CD274 (PD-L1) and PDCD1LG2 (PD-L2) was also observed in these cells, suggesting the tolerogenicity of them at a certain extent (Supplementary Fig. S7c).35 Distinct groups of active TFs were observed for different clusters of DCs (Fig. 5c). Cluster 4 cDC2s were with highly activity of the cDC epigenetic fate-determining TF CEBPB.36 The IRF family members IRF7, IRF4, and IRF8 were with transcription activity at high levels in cluster 7 pDC cells. Cluster 8 LAMP3+ cDCs were mainly driven by TFs FOXD4, RELB, IRF2, and so on. Few activated TFs were observed in cluster 6 cDC1s.
To further examine the lineage relationship of different subtype DCs, we performed RNA velocity analysis of DC clusters and observed that both cluster 4 cDC2s and cluster 6 cDC1s could transit to cluster 8 LAMP3+ cDCs in pre-neoCRT samples, consistent with a previous report that LAMP3+ cDCs originated from both cDC1s and cDC2s in hepatocellular carcinoma.20 However, in post-neoCRT samples, LAMP3+ cDCs originated mainly from cDC1s but few from cDC2s. The analysis of latent time showed that the cDC1s were produced later than cDC2s in post-neoCRT samples, contradictory to the results in pre-neoCRT samples (Fig. 5d). As a consequence, cluster 4 and 9 cDC2s decreased significantly after neoCRT (P < 0.05 by Wilcoxon signed-rank test with Benjamini-Hochberg correction, Supplementary Fig. S7d), which was verified in another 25 pairs of pre- and post-neoCRT ESCC samples (Supplementary Table S4) by staining cDC2 marker CD1C, although statistical significance was not achieved (P = 0.059 by Wilcoxon signed-rank test, Fig. 5e). There was no significant difference of cDC2s infiltration between major and minor responders in either pre- (P = 0.322 by Mann–Whitney U test) or post-neoCRT samples (P = 0.782 by Mann–Whitney U test, Fig. 5e). Correlation analysis of the percentages of cDC2s (CD1C+ cells) and different subtypes of T cells in the 25 pairs of pre- and post-neoCRT ESCC samples stained by mfIHC identified no significant correlation between cDC2s and any subtypes of T cells (P > 0.05 by Spearman correlation analysis, Fig. 5f).
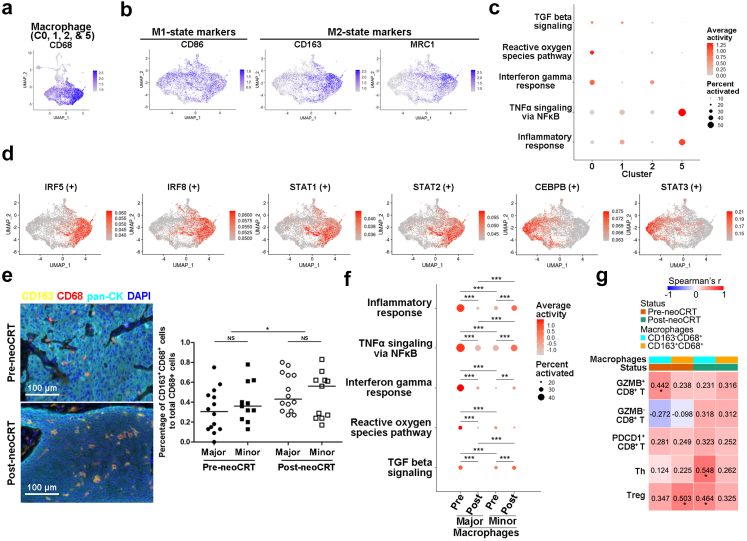
NeoCRT increased expression of M2 macrophage markers
Cluster 0, 1, 2, and 5 of macrophages and DCs were positive for CD68 (Fig. 5, Fig. 6a), corresponding to macrophages. Cluster 0 macrophages expressed a set of genes previously found in tumor associated macrophages, such as APOE, APOC1, and GPNMB.20 Cluster 1 and 2 cells were with the similar gene expression patterns as myeloid-derived suppressor cell-like macrophages in hepatocellular carcinoma, such as S100A family genes, FCN1, VCAN and THBS1.20 Cluster 5 was characterized by high expression of chemokines, such as CXCL5, CCL20, and CXCL8, suggesting possible roles of them on cancer progression by remodeling the TME through immune cell recruitment (Supplementary Fig. S8a).37,38
Fig. 6.
Characterization of macrophages in pre- and post-neoCRT ESCCs. (a, b) UMAP plots showing expression markers for macrophages (a), and the classical M1- and M2-macrophages (b). (c) Dotplot showing the activities of M1- and M2-related pathways among different macrophage clusters. (d) UMAP showing the classical activities of M1- and M2-related TFs. (e) Representative images of mfIHC staining of CD163 and CD68 in pre- and post-neoCRT ESCC slides. Differences in the percentages of CD163+CD68+ in CD68+ cells were compared. ∗, P < 0.05 by Wilcoxon signed-rank test. NS, not significant by Mann–Whitney U test. (f) Different activities of M1- and M2-related pathway in macrophages. Major, major responders; minor, minor responders. ∗∗, P < 0.01; ∗∗∗, P < 0.001 by moderated t-test with Benjamini-Hochberg correction. (g) Spearman correlation analyses between the percentages of different subtypes of macrophages and T cells in 25 pairs of pre- and post-neoCRT ESCCs stained by mfIHC. Correlation coefficient was labelled. ∗, P < 0.05 by spearman correlation analyses.
In ESCC TME, classically activated M1 macrophages' marker CD86 was widely expressed in all macrophage clusters like CD68. Alternatively activated M2 macrophages’ markers CD163 and MRC1 were expressed highly in cluster 0 and moderately in cluster 2 and 5 (Fig. 6b). Pathways related to pro-inflammatory response such as TNFalpha signaling and inflammatory response were activated in CD163lowMRC1low cluster 1 but inactivated in CD163highMRC1high cluster 0, supporting the pro- and anti-inflammatory function of M1-and M2-like macrophages in cluster 1 and 0, respectively. However, high enrichment of M1-related pathways interferon gamma response and reactive oxygen species producing39 in the CD163+MRC1+ clusters 0 and 2, while M2-related pathway TGFbeta signaling39 in CD163lowMRC1low cluster 1 were also observed (Fig. 6c, Supplementary Fig. S8b). TF analysis showed that IRF5, IRF8, STAT1, and STAT2, which favored M1-polarization,40 were highly activated in cluster 0 CD163highMRC1high macrophages, whereas STAT3 and CEBPB, which favored M2-polarizaion,40 were in cluster 1 CD163lowMRC1low macrophages (Fig. 6d, Supplementary Fig. S8c). These results suggested the co-existence of M1 and M2-states in ESCC TME, which could not be simplified as the classical M1/M2 model.
There was no significant difference in the proportion of any macrophage cluster between pre- and post-neoCRT samples (P > 0.05 by Wilcoxon signed-rank test with Benjamini-Hochberg correction, Supplementary Fig. S8d). However, the overall expression of M2 markers CD163 and MRC1 increased after neoCRT in both major and minor responders, which was more obvious in cluster 0 and 2 (P < 0.05 by Mann–Whitney U test with Benjamini-Hochberg correction, Supplementary Fig. S8e). Staining of another 25 pairs of pre- and post-neoCRT ESCC patients (Supplementary Table S4) with CD163 and CD68 using mfIHC verified the increase of CD163 expression in CD68+ macrophages after neoCRT (P = 0.021 by Wilcoxon signed-rank test, Fig. 6e). Despite increased expression of M2-markers, activities of both M1-and M2-related pathways decreased overall in macrophages from major responders but increased in those from minor responders after neoCRT (P < 0.05 by moderated t-test with Benjamini-Hochberg correction, Fig. 6f).
Correlation analysis of the percentages of CD163− and CD163+ macrophages and different subtypes of T cells in the 25 pairs of pre- and post-neoCRT ESCC samples stained by mfIHC identified significant correlations between CD163−CD68+ macrophages and GZMB+ CD8+ T cells (r = 0.442, P = 0.027 by Spearman correlation analysis) as well as between CD163+CD68+ macrophages and FOXP3+CD4+ Treg cells (r = 0.503, P = 0.010 by Spearman correlation analysis) in pre-neoCRT ESCC. And significant correlations were observed between CD163−CD68+ macrophages and FOXP3−CD4+Th and FOXP3+CD4+ Treg cells in post-neoCRT ESCCs (r = 0.548 and 0.464, P = 0.005 and 0.020 by Spearman correlation analysis, respectively, Fig. 6g). Presence of these positive significant correlations suggests possible interplays between the cells, which might affect cell recruitment and function further.39
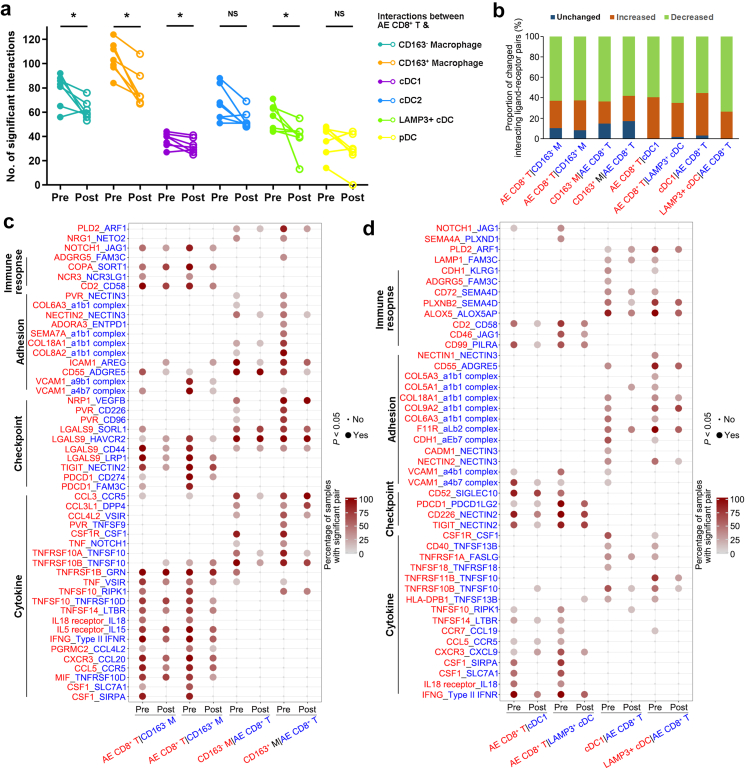
NeoCRT decreased interactions between the intermediate activated/exhausted CD8+ T and subtypes of macrophages and cDCs
Complex cell–cell interactions were observed between T cells (intermediate activated/exhausted CD8+ T, exhausted CD8+ T, Th, and Treg cells) and subtypes of macrophages (CD163− and CD163+ macrophages) and DCs (cDC1, cDC2, LAMP3+ cDCs and pDCs) in both pre- and post-neoCRT samples (Supplementary Fig. S9a). The number of significant interactions between intermediate activated/exhausted CD8+ T and CD163− and CD163+ macrophages, cDC1, and LAMP+ cDCs decreased after neoCRT (P < 0.05 by Wilcoxon signed-rank test, Fig. 7a). However, there was no difference in the number of significant interactions of other T and macrophage and DC subtypes between pre- and post-neo-CRT samples (P > 0.05 by Wilcoxon signed-rank test, Supplementary Fig. S9b, c, and d)
Fig. 7.
Characterization of predicted T-macrophage/DC interactions in pre- and post-neoCRT ESCCs. (a) Differences in the number of significant interactions between intermediate activated/exhausted CD8+ T and subtypes of macrophages and DCs in pre- and post-neoCRT ESCCs. ∗, P < 0.05; NS, not significant by Wilcoxon signed-rank test. (b) Bar plot showing the proportions of significant interacting pairs between intermediate activated/exhausted CD8+ T cells and subtypes of macrophages and DCs with increased, decreased, or unchanged percentages after neoCRT. Ligand (red) and receptor (blue) cells are shown on the x-axis. (c, d) Dot plot showing the top 50 (c) and 45 (d) interacting ligand–receptor pairs between intermediate activated/exhausted CD8+ T and subtypes of macrophages (c) and cDCs (d) with decreased percentages after neoCRT. Cell subtypes are shown on the x-axis; ligand (red) and receptor (blue) pairs are shown on the y-axis. The color of the circle denotes the percentages of samples with a significant interaction (P < 0.05 by CellphoneDB which uses a one-side permutation test to compute significant interactions) in the total samples with these interacting cell subsets. AE, intermediate activated/exhausted; M, macrophage.
Next, we focused on the changes of interacting ligand–receptor pairs between intermediate activated/exhausted CD8+ T and CD163− and CD163+ macrophages, cDC1, and LAMP3+ cDCs. The proportions of significant interacting pairs with decreased percentages after neoCRT were higher than those of interacting pairs with increased or unchanged percentages (Fig. 7b). The percentages of a series of interactions involving cytokines, immune checkpoints, immune response and cell adhesion between intermediate activated/exhausted CD8+ T and subtypes of macrophages and cDCs decreased after neoCRT (Fig. 7c and d). Alterations of interactions between cytokines and their receptors would affect immune cells' recruitment, function and response against tumor cells. Take CSF1 for example, it was expressed by the intermediate activated/exhausted CD8+ T cells to interact with its receptors CSF1R on CD163− and CD163+ macrophages, cDC1s, and LAMP3+ cDCs to affect the receptor cells’ proliferation, differentiation, and survival.41 Decreased percentages of samples with significant interactions between CSF1 and its receptors might alter the activities of these myeloid cells, especially macrophages in ESCCs after neoCRT.
A series of inhibitory checkpoint pairs, such as PDCD1_CD274, PDCD1_FAM3C, TIGIT_NECTIN2 between the intermediate activated/exhausted CD8+ T cells and macrophages (Fig. 7c), as well as PDCD1_PDCD1LG2 and TIGIT_NECTIN2 between the intermediate activated/exhausted CD8+ T cells and cDC1s as well as LAMP3+ cDCs (Fig. 7d) showed decreased percentages after neoCRT, consistently with decreased expression of checkpoints PDCD1 and TIGIT in the intermediate activated/exhausted CD8+ T cells (Supplementary Fig. S5h). Interactions of checkpoints LGALS9 and its receptors HAVCR2, SORL1, and CD44 were observed between the intermediate activated/exhausted CD8+ T cells and macrophages, and vice versa (Fig. 7c). It has been reported that binding of LGALS9 to HAVCR2 will lead to the HAVCR2-mediated immune anergic or apoptosis.42 The decreased percentage of samples with significant LGALS9_HAVCR2 interaction between the intermediate activated/exhausted CD8+ T cells and CD163+ but not CD163- macrophages might partly account for the reason why proportion of CD163+ macrophages increased after neoCRT (Fig. 6e). Decreased percentage of samples with significant interacting pair CD52_SIGLEC10 between the intermediate activated/exhausted CD8+ T cell and LAMP3+ cDCs after neoCRT (Fig. 7d) might alleviate the SIGLEC10-mediated inhibition of antigen presentation by cDCs.43
Discussion
A few researches have been conducted on tumor-infiltrating immune cells in ESCC.44, 45, 46, 47 The immunosuppressive status in ESCC TME was primarily due to the large amount of exhausted CD8+ T infiltration. Although activated CD8+ T cell existed, their cytotoxicity was ubiquitously curtailed by checkpoint expression. Infiltration of Treg cells further impairs the CD8+ T cells' antitumor immunity. However, there were some immune cells functioning more than immunosuppression. Firstly, a cluster of CXCL13+CXCR5− Tfh-like cells were identified. Lacking of CXCR5 hampered their localization to B cell follicles. However, expression of CXCL13, the B lymphocyte chemoattractant, suggested a novel role of these Tfh-like cells in orchestrating ESCC TME by promoting the migration of B cells.48 Secondly, the LAMP3+ cDCs were previously considered tolerogenic due to their expression of checkpoints and abilities in inhibiting CD8+ T cell proliferation46 or inducing FOXP3 expression on naïve CD4+ T cells47 in vitro. However, evidence from in vivo experiment is still lacking.49 CCR7 expression on LAMP3+ cDCs enables them to migrate to draining lymph nodes, and chemokine secretion enables them to recruit lymphocyte.10,20 Actually, the LAMP3+ cDCs in TME has been shown to orchestrate a Th1 cell-polarized and cytotoxic CD8+ T cell intra-tumoral immune response, usually correlating with improved cancer survivals.50 Lastly, most macrophages co-expressed both M1 and M2 markers in ESCC TME. And M1- or M2-markers’ expression was discordant with M1- or M2-related pathways or TFs’ activation, suggesting macrophages in the ESCC TME does not comport with the canonical polarization model, either as discrete states or along a spectrum of activation states like in several other types of cancers.34,51 Activation of pathways related to pro-inflammatory response in parts of macrophages indicated that macrophages in the ESCC TME were not completely immunosuppressive.
Chemo- or radio-therapy have been developed into clinically useful agents based on their ability to preferentially kill malignant cells. Characteristics of tumor cells directly affect chemo- and radio-sensitivity. As we shown in this study, pre-neoCRT malignant cells’ gene expression programs involving cell cycle, epithelium development, immune response, and extracellular structure were related to ESCC neoCRT responses, consistent with previous studies on chemo- and radio-sensitivity in the perspective of tumor cells.52,53 However, chemo- and radio-therapy could also modulate the tumor immune environment directly or indirectly by modulating the antigenicity or adjuvanticity of cancer cells.54,55 Park et al.56 previously showed that the immune scores and enrichment of immune signaling pathways increased after neoCRT in ESCCs by whole-transcriptome sequencing. However, detailed changes of specific immune cells were not achieved due to the limitation of bulk sequencing. In a recent study, Chen et al.57 analyzed the single-cell transcriptome in ESCC patients receiving neoadjuvant chemotherapy (neoCT) in comparison with those receiving surgery only. Elevated levels of exhausted CD8+ T cells and enrichment of interferon gamma pathway in macrophages were observed in post-neoCT ESCC samples, similar to the changes in our post-neoCRT ESCC samples. However, in lack of the information of tumor response to neoCT, it is difficult to correlate the cellular and molecular changes with neoCT sensitivity. Future conjoint analysis of ESCC samples receiving different treatment regimens including neoCT and neoCRT with tumor response evaluation would contribute to the understanding of differential treatment effects on ESCC TME and their relationship to treatment sensitivity.
In this study, we found increased infiltration of CD8+ T cells in ESCC TME after neoCRT in both major and minor responders, making the tumors “hotter”, a predictor of improved response and treatment outcome of immune checkpoint blockade-based immunotherapy, which supports the study that nivolumab adjuvant therapy significantly improved disease-free survival of ESCC patients who had received neoCRT.58 However, the increased CD8+ T cells after neoCRT were mainly exhausted ones, which might be more resistant to checkpoint inhibition due to the permanent and less reversible exhausted stage where they were in.59 It has been proved that the intermediate activated/exhausted tumor-infiltrated CD8+ T cells with co-expression of both T cell activation and exhaustion gene signatures could be increased by PD-1-based therapies.27,34,60 Therefore, utilizing PD-1-based immunotherapy in neoadjuvant setting, which have been shown to improve neoadjuvant treatment response by clinical trials,61,62 might counteract the increase of exhausted CD8+ T cells induced by neoCRT. Furthermore, higher expression of PDCD1 and higher correlation between PDCD1 expression and cytotoxicity were observed in pre-neoCRT intermediate activated/exhausted CD8+ T cells as compared to post-neoCRT ones. It rationalizes PD-1-based therapies before or in combination with neoCRT, as high expression of PDCD1 in CD8+ T cells was strongly predictive for both response and survival for cancer patients treated with PD-1 blockade.9,63 In addition to CD8+ T cells, neoCRT altered CD4+ T cell differentiation reflected by the activities of classical CD4+ T subset-regulating TFs, leading to the overall increase of Th but decrease of Treg cells after neoCRT. Furthermore, higher FOXP3 activity as well as higher Treg to Th cell ratio in post-neoCRT minor responders but not in pre-neoCRT ones suggested that increased Treg differentiation in minor responders during neoCRT might play an important role in neoCRT resistance. Therefore, targeting Treg during or after neoCRT in minor responders would be of importance in improving response or survival of these ESCC patients. The changes of CD8+ T infiltration and CD4+ T subset differentiation after neoCRT may be partially contributed by the changes of DCs, which have a critical role in that they cross-present tumor-derived antigens that become available in the context of CRT-driven tumor cell death.55,64 cDC1 could prime CD8+ T cells and induce Th1 cell differentiation, cDC2 are a crucial source of pro-Th17 differentiation cytokines, and cDC1 and cDC2 both can promote Treg cells.33,35 In addition to the changes of immune infiltration, neoCRT weakened the cell–cell interactions between intermediate activated/exhausted CD8+ T and CD163− and CD163+ macrophages, cDC1, and LAMP3+ cDCs. Decreased percentages of samples with significant interacting pairs of inhibitory checkpoints between these cells after neoCRT would alleviate the immune cells’ inhibitions of each other.
Although the immune cell infiltration was not different between major and minor responders in either pre- or post-neoCRT samples except Treg cells, the activities of infiltrated immune cells significantly different between major and minor responders and were affected by neoCRT. For pre-neoCRT samples, the overall activity of immune-related pathways in CD8+ T cells and macrophages were higher in major responders than that in minor responders. After neoCRT, immune activity decreased in major responders, probably because none or few tumor cells left after neoCRT in major responders could not elicit high immune response. However, for minor responders, immune activities significantly increased in post-neoCRT CD8+ T and macrophages.
There were some limitations in this study. First, the sample size used for scRNA-seq is relatively small, due to the difficulty in obtaining enough fresh samples for scRNA-seq, especially endoscopic biopsies. This might introduce sampling errors. For example, 8 ESCC patients for scRNA-seq analyses were all males probably because the high incidence of ESCC in males, however, sex did not affect neoCRT response.6,7 And some results of scRNA-seq might not be verified in other cohorts with larger sample size. Besides, gene expression of given transcripts determined by scRNA-seq may be different from surface protein level measured by mfIHC. Therefore, the decrease of CD4+ T cells identified in 7 pairs of pre- and post-neoCRT ESCCs by scRNA-seq could not be verified in another 25 paired samples by mfIHC staining. Future studies with larger sample size and different experimental techniques would help in the identification of more solid results.
Taken together, we deciphered the cellular and molecular alterations of ESCC immune ecosystem in the setting of neoCRT at single-cell resolution. Our work will help in understanding the response of ESCC to CRT contributed by immune cells in TME, and provide evidence for developing and applying immune-strategies for improving ESCC treatment.
Contributors
Jing Wen, Hong Yang, Jianhua Fu, and Qianwen Liu designed the study; Yi Hu, Mian Xi, Kongjia Luo, Qianwen Liu, Renchun Lai, Xiuying Xie, Xiaodan Lin, Ting Lin, Jiyang Chen, Jianhua Fu, and Hong Yang collected the patients’ information and samples; Jing Wen and Zelin Weng performed the bioinformatics analysis; Jing Wen, Shuogui Fang, Chuqing Pan, and Yihong Ling performed the experiment and data analysis; Jing Wen, Shuogui Fang, Yi Hu, and Mian Xi drafted the manuscript; Jing Wen, Shuogui Fang, Yi Hu, Mian Xi, Qianwen Liu, Jianhua Fu, and Hong Yang revised the manuscript; Jing Wen, Yi Hu, and Jianhua Fu provided funding and supervision. All authors read and approved the final manuscript. Jing Wen, Shuogui Fang, and Hong Yang have verified the underlying data reported in the manuscript.
Data sharing statement
The sequencing data has been deposited in GSA-Human (https://bigd.big.ac.cn/gsa-human) under the accession number HRA001861. The data used in this study can be obtained by request to J.W. (wenjing@sysucc.org.cn).
Declaration of interests
The authors declare no conflict of interest relevant to the present manuscript.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82072607 and 81871975); the Natural Science Foundation of Guangdong Province (2019A1515010715); the Fundamental Research Funds for the Central Universities (19ykzd44); the National Key R&D Program of China (2018YFC0910600); Guangdong Esophageal Cancer Institute Science and Technology Program (M202004).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2022.104371.
Contributor Information
Jing Wen, Email: wenjing@sysucc.org.cn.
Qianwen Liu, Email: liuqw@sysucc.org.cn.
Jianhua Fu, Email: fujh@sysucc.org.cn.
Hong Yang, Email: yanghong@sysucc.org.cn.
Appendix A. Supplementary data
References
- 1.Lawson D.A., Kessenbrock K., Davis R.T., Pervolarakis N., Werb Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat Cell Biol. 2018;20:1349–1360. doi: 10.1038/s41556-018-0236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Chen W., Zheng R., Baade P.D., et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Yang H., Liu H., Chen Y., et al. Long-term efficacy of neoadjuvant chemoradiotherapy plus surgery for the treatment of locally advanced esophageal squamous cell carcinoma: the NEOCRTEC5010 randomized clinical trial. JAMA Surg. 2021;156:721–729. doi: 10.1001/jamasurg.2021.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider P.M., Baldus S.E., Metzger R., et al. Histomorphologic tumor regression and lymph node metastases determine prognosis following neoadjuvant radiochemotherapy for esophageal cancer: implications for response classification. Ann Surg. 2005;242:684–692. doi: 10.1097/01.sla.0000186170.38348.7b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen J., Luo K., Liu H., et al. MiRNA expression analysis of pretreatment biopsies predicts the pathological response of esophageal squamous cell carcinomas to neoadjuvant chemoradiotherapy. Ann Surg. 2016;263:942–948. doi: 10.1097/SLA.0000000000001489. [DOI] [PubMed] [Google Scholar]
- 7.Wen J., Yang H., Liu M.Z., et al. Gene expression analysis of pretreatment biopsies predicts the pathological response of esophageal squamous cell carcinomas to neo-chemoradiotherapy. Ann Oncol. 2014;25:1769–1774. doi: 10.1093/annonc/mdu201. [DOI] [PubMed] [Google Scholar]
- 8.Stuart T., Butler A., Hoffman P., et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambrechts D., Wauters E., Boeckx B., et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018;24:1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 10.Zilionis R., Engblom C., Pfirschke C., et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. 2019;50:1317–1334. doi: 10.1016/j.immuni.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng J., Sun B.F., Chen C.Y., et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29:725–738. doi: 10.1038/s41422-019-0195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotliar D., Veres A., Nagy M.A., et al. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. Elife. 2019;8 doi: 10.7554/eLife.43803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raudvere U., Kolberg L., Kuzmin I., et al. Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritchie M.E., Phipson B., Wu D., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van de Sande B., Flerin C., Davie K., et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc. 2020;15:2247–2276. doi: 10.1038/s41596-020-0336-2. [DOI] [PubMed] [Google Scholar]
- 18.La Manno G., Soldatov R., Zeisel A., et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergen V., Lange M., Peidli S., Wolf F.A., Theis F.J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020;38:1408–1414. doi: 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q., He Y., Luo N., et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179:829–845. doi: 10.1016/j.cell.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 22.Faust G.G., Hall I.M. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics. 2014;30:2503–2505. doi: 10.1093/bioinformatics/btu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarasov A., Vilella A.J., Cuppen E., Nijman I.J., Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okonechnikov K., Conesa A., Garcia-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32:292–294. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boeva V., Popova T., Bleakley K., et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28:423–425. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yost K.E., Satpathy A.T., Wells D.K., et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med. 2019;25:1251–1259. doi: 10.1038/s41591-019-0522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beltra J.C., Manne S., Abdel-Hakeem M.S., et al. Developmental relationships of four exhausted CD8(+) T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity. 2020;52:825–841. doi: 10.1016/j.immuni.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukaida N., Sasaki S.I., Baba T. CCL4 signaling in the tumor microenvironment. Adv Exp Med Biol. 2020;1231:23–32. doi: 10.1007/978-3-030-36667-4_3. [DOI] [PubMed] [Google Scholar]
- 30.Ntanasis-Stathopoulos I., Fotiou D., Terpos E. CCL3 signaling in the tumor microenvironment. Adv Exp Med Biol. 2020;1231:13–21. doi: 10.1007/978-3-030-36667-4_2. [DOI] [PubMed] [Google Scholar]
- 31.Haaland R.E., Yu W., Rice A.P. Identification of LKLF-regulated genes in quiescent CD4+ T lymphocytes. Mol Immunol. 2005;42:627–641. doi: 10.1016/j.molimm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 32.Golubovskaya V., Wu L. Different subsets of T cells, memory, effector functions, and CAR-T immunotherapy. Cancers. 2016;8:36–47. doi: 10.3390/cancers8030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin X., Chen S., Eisenbarth S.C. Dendritic cell regulation of T helper cells. Annu Rev Immunol. 2021;39:759–790. doi: 10.1146/annurev-immunol-101819-025146. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y., Chen H., Mo H., et al. Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell. 2021;39:1578–1593. doi: 10.1016/j.ccell.2021.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Patente T.A., Pinho M.P., Oliveira A.A., Evangelista G.C.M., Bergami-Santos P.C., Barbuto J.A.M. Human dendritic cells: their heterogeneity and clinical application potential in cancer immunotherapy. Front Immunol. 2018;9:3176. doi: 10.3389/fimmu.2018.03176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bornstein C., Winter D., Barnett-Itzhaki Z., et al. A negative feedback loop of transcription factors specifies alternative dendritic cell chromatin states. Mol Cell. 2014;56:749–762. doi: 10.1016/j.molcel.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadomoto S., Izumi K., Mizokami A. The CCL20-CCR6 axis in cancer progression. Int J Mol Sci. 2020;21:5186. doi: 10.3390/ijms21155186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W., Zhang X., Wu F., et al. Gastric cancer-derived mesenchymal stromal cells trigger M2 macrophage polarization that promotes metastasis and EMT in gastric cancer. Cell Death Dis. 2019;10:918. doi: 10.1038/s41419-019-2131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeNardo D.G., Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol. 2019;19:369–382. doi: 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mantovani A., Locati M. Tumor-associated macrophages as a paradigm of macrophage plasticity, diversity, and polarization: lessons and open questions. Arterioscler Thromb Vasc Biol. 2013;33:1478–1483. doi: 10.1161/ATVBAHA.113.300168. [DOI] [PubMed] [Google Scholar]
- 41.Trus E., Basta S., Gee K. Who's in charge here? Macrophage colony stimulating factor and granulocyte macrophage colony stimulating factor: competing factors in macrophage polarization. Cytokine. 2020;127 doi: 10.1016/j.cyto.2019.154939. [DOI] [PubMed] [Google Scholar]
- 42.Wolf Y., Anderson A.C., Kuchroo V.K. TIM3 comes of age as an inhibitory receptor. Nat Rev Immunol. 2020;20:173–185. doi: 10.1038/s41577-019-0224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stanczak M.A., Laubli H. Siglec receptors as new immune checkpoints in cancer. Mol Aspects Med. 2022 doi: 10.1016/j.mam.2022.101112. [DOI] [PubMed] [Google Scholar]
- 44.Chen Z., Zhao M., Liang J., et al. Dissecting the single-cell transcriptome network underlying esophagus non-malignant tissues and esophageal squamous cell carcinoma. EBioMedicine. 2021;69 doi: 10.1016/j.ebiom.2021.103459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dinh H.Q., Pan F., Wang G., et al. Integrated single-cell transcriptome analysis reveals heterogeneity of esophageal squamous cell carcinoma microenvironment. Nat Commun. 2021;12:7335. doi: 10.1038/s41467-021-27599-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X., Peng L., Luo Y., et al. Dissecting esophageal squamous-cell carcinoma ecosystem by single-cell transcriptomic analysis. Nat Commun. 2021;12:5291. doi: 10.1038/s41467-021-25539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng Y., Chen Z., Han Y., et al. Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment. Nat Commun. 2020;11:6268. doi: 10.1038/s41467-020-20019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J.P., Wu C.Y., Chen M.Y., et al. PD-1(+)CXCR5(-)CD4(+) Th-CXCL13 cell subset drives B cells into tertiary lymphoid structures of nasopharyngeal carcinoma. J Immunother Cancer. 2021;9 doi: 10.1136/jitc-2020-002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Terness P., Chuang J.J., Opelz G. The immunoregulatory role of IDO-producing human dendritic cells revisited. Trends Immunol. 2006;27:68–73. doi: 10.1016/j.it.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 50.Sautes-Fridman C., Petitprez F., Calderaro J., Fridman W.H. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat Rev Cancer. 2019;19:307–325. doi: 10.1038/s41568-019-0144-6. [DOI] [PubMed] [Google Scholar]
- 51.Azizi E., Carr A.J., Plitas G., et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174:1293–1308. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim B.M., Hong Y., Lee S., et al. Therapeutic implications for overcoming radiation resistance in cancer therapy. Int J Mol Sci. 2015;16:26880–26913. doi: 10.3390/ijms161125991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mosca L., Ilari A., Fazi F., Assaraf Y.G., Colotti G. Taxanes in cancer treatment: activity, chemoresistance and its overcoming. Drug Resist Updat. 2021;54 doi: 10.1016/j.drup.2020.100742. [DOI] [PubMed] [Google Scholar]
- 54.Li J.Y., Chen Y.P., Li Y.Q., Liu N., Ma J. Chemotherapeutic and targeted agents can modulate the tumor microenvironment and increase the efficacy of immune checkpoint blockades. Mol Cancer. 2021;20:27. doi: 10.1186/s12943-021-01317-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McLaughlin M., Patin E.C., Pedersen M., et al. Inflammatory microenvironment remodelling by tumour cells after radiotherapy. Nat Rev Cancer. 2020;20:203–217. doi: 10.1038/s41568-020-0246-1. [DOI] [PubMed] [Google Scholar]
- 56.Park S., Joung J.G., Min Y.W., et al. Paired whole exome and transcriptome analyses for the Immunogenomic changes during concurrent chemoradiotherapy in esophageal squamous cell carcinoma. J Immunother Cancer. 2019;7:128. doi: 10.1186/s40425-019-0609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Z., Huang Y., Hu Z., et al. Dissecting the single-cell transcriptome network in patients with esophageal squamous cell carcinoma receiving operative paclitaxel plus platinum chemotherapy. Oncogenesis. 2021;10:71. doi: 10.1038/s41389-021-00359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelly R.J., Ajani J.A., Kuzdzal J., et al. Adjuvant nivolumab in resected esophageal or gastroesophageal junction cancer. N Engl J Med. 2021;384:1191–1203. doi: 10.1056/NEJMoa2032125. [DOI] [PubMed] [Google Scholar]
- 59.Pauken K.E., Sammons M.A., Odorizzi P.M., et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016;354:1160–1165. doi: 10.1126/science.aaf2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bassez A., Vos H., Van Dyck L., et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat Med. 2021;27:820–832. doi: 10.1038/s41591-021-01323-8. [DOI] [PubMed] [Google Scholar]
- 61.Li C., Zhao S., Zheng Y., et al. Preoperative pembrolizumab combined with chemoradiotherapy for oesophageal squamous cell carcinoma (PALACE-1) Eur J Cancer. 2021;144:232–241. doi: 10.1016/j.ejca.2020.11.039. [DOI] [PubMed] [Google Scholar]
- 62.Yang W., Xing X., Yeung S.J., et al. Neoadjuvant programmed cell death 1 blockade combined with chemotherapy for resectable esophageal squamous cell carcinoma. J Immunother Cancer. 2022;10 doi: 10.1136/jitc-2021-003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thommen D.S., Koelzer V.H., Herzig P., et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med. 2018;24:994–1004. doi: 10.1038/s41591-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Galluzzi L., Humeau J., Buque A., Zitvogel L., Kroemer G. Immunostimulation with chemotherapy in the era of immune checkpoint inhibitors. Nat Rev Clin Oncol. 2020;17:725–741. doi: 10.1038/s41571-020-0413-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.