Abstract
Starch and storage proteins are the main components of rice (Oryza sativa L.) grains. Despite their importance, the molecular regulatory mechanisms of storage protein and starch biosynthesis remain largely elusive. Here, we identified a rice opaque endosperm mutant, opaque3 (o3), that overaccumulates 57-kDa proglutelins and has significantly lower protein and starch contents than the wild type. The o3 mutant also has abnormal protein body structures and compound starch grains in its endosperm cells. OPAQUE3 (O3) encodes a transmembrane basic leucine zipper (bZIP) transcription factor (OsbZIP60) and is localized in the endoplasmic reticulum (ER) and the nucleus, but it is localized mostly in the nucleus under ER stress. We demonstrated that O3 could activate the expression of several starch synthesis-related genes (GBSSI, AGPL2, SBEI, and ISA2) and storage protein synthesis-related genes (OsGluA2, Prol14, and Glb1). O3 also plays an important role in protein processing and export in the ER by directly binding to the promoters and activating the expression of OsBIP1 and PDIL1-1, two major chaperones that assist with folding of immature secretory proteins in the ER of rice endosperm cells. High-temperature conditions aggravate ER stress and result in more abnormal grain development in o3 mutants. We also revealed that OsbZIP50 can assist O3 in response to ER stress, especially under high-temperature conditions. We thus demonstrate that O3 plays a central role in rice grain development by participating simultaneously in the regulation of storage protein and starch biosynthesis and the maintenance of ER homeostasis in endosperm cells.
Key words: grain development, ER stress, OPAQUE3, high temperature, rice
OPAQUE3 (O3) encodes a transmembrane bZIP transcription factor that relocates to the nucleus under endoplasmic reticulum (ER) stress. O3 plays a central role in rice grain development by participating in the regulation of storage protein and starch biosynthesis and the maintenance of ER homeostasis in endosperm cells.
Introduction
Rice is a staple food for more than half of the world’s population. Starch and storage proteins determine the yield and quality of rice grains; they account for approximately 80% and 10% of rice grain weight, respectively. Starch is composed of linear amylose and multi-branched, semi-crystalline amylopectin. Its biosynthesis in plant endosperm involves a series of key enzymes that function coordinately. ADP-glucose pyrophosphorylase is responsible for the synthesis of ADP-glucose, which is the major substrate for starch synthesis (Beckles et al., 2001; Nakamura, 2002). Amylose is synthesized by granule-bound starch synthase I (GBSSI) through progressive addition of ADP-glucose to linear chains of α(1-4)-linked glucose residues. Amylopectin synthesis requires synergistic cooperation of soluble starch synthases, branching enzymes, and debranching enzymes (Ball and Morell, 2003). Many natural variations or artificial mutations of these genes cause abnormal endosperm phenotypes or grain-filling defects (Nishi et al., 2001; Umemoto et al., 2004; Tian et al., 2009; Wei et al., 2017; Zhang et al., 2019a). Rice grain storage proteins also determine rice quality and are classified into glutelin, globulin, prolamin, and albumin according to their solubility. Prolamins are synthesized and processed in the endoplasmic reticulum (ER) lumen, and they then aggregate and bud off to form the spherical protein body I (PBI) (Li et al., 1993). The 57-kDa proglutelins are synthesized in the ER and later trafficked to protein storage vacuoles (PSVs) through dense vesicles (DVs) to be processed into mature acidic and basic subunits, eventually forming the irregularly shaped protein body II (PBII) together with α-globulin (Yamagata et al., 1982; Washida et al., 2012). The genes that participate in processing and trafficking of storage proteins have been well investigated, including GPA1–GPA8. Loss of function of these genes often results in floury grains with abnormal protein bodies (Wang et al., 2010, 2016; Liu et al., 2013; Ren et al., 2014, 2020; Zhu et al., 2019, 2021; Pan et al., 2021).
Some transcription factors (TFs) have been reported to play important roles in regulation of rice endosperm starch and storage protein biosynthesis. Rice starch regulator 1 (RSR1) negatively regulates expression of type I starch synthesis genes. The knockout mutant rsr1 has a larger grain size with increased amylose content and altered fine structure of amylopectin and starch morphology compared with the wild type (Fu and Xue, 2010). NF-YB1 can directly bind to the “CCAAT boxes” of OsSUT1, OsSUT3 and OsSUT4 to activate their expression, regulating sucrose loading to the developing endosperm for starch biosynthesis (Bai et al., 2016). NF-YB1 can also interact with NF-YC12, bHLH144, or OsERF115 to form a transcription complex that directly binds to the “G box” or “GCC box” in promoters of genes involved in starch synthesis and sugar and amino acid transport (Xu et al., 2016; Bello et al., 2019; Xiong et al., 2019). OsbZIP58 regulates starch synthesis and aleurone layer number by directly binding to the promoters of starch synthesis-related genes (OsAGPL3, Wx, OsSSIIa, SBE1, OsBEIIb, and ISA2) and activating their expression (Wang et al., 2013). OsbZIP58 can also interact with rice prolamin box binding factor (RPBF) and activate expression of the rice grain storage protein synthesis-related genes GluA1, GluA2, GluA3, GluB1, GluD1, 10-kDa prolamin, 13-kDa prolamin, and 16-kDa prolamin (Kawakatsu et al., 2009). In maize, ZmNAC128 and ZmNAC130 can bind to the “ACGCAA” motif to activate transcription of the Bt2 and 16-kDa γ-zein genes, thus having a pleiotropic effect on biosynthesis of grain storage protein and starch (Zhang et al., 2019b). A similar mechanism has been reported for rice OsNAC20 and OsNAC26; simultaneous knockout of these two TFs significantly decreases starch and protein accumulation and grain weight and alters starch granule formation (Wang et al., 2020).
The ER is the major organelle for storage protein synthesis, folding, and export (Sun et al., 2021). Overaccumulation of unfolded proteins in the ER causes ER stress and activates the unfolded protein response (UPR), which subsequently increases expression of ER chaperone genes to maintain ER homeostasis (Howell, 2013). The UPR signaling pathway has been well elucidated in Arabidopsis; the bZIP TF AtbZIP60 is normally localized in the ER lumen through the transmembrane domain (TMD) in its C-terminal region. When stress is sensed in the ER, the TMD is cleaved by the ribonuclease AtIRE1, and the cytoplasmic N-terminal activation domain is transferred to the nucleus to upregulate ER stress-related chaperone genes (Iwata and Koizumi, 2005; Deng et al., 2011). Otherwise, AtbZIP17 and AtbZIP28 are cleaved by Golgi apparatus-localized S2P proteases under ER stress, and their cytoplasmic portion, including a bZIP domain, is then released to the nucleus to activate UPR genes for maintenance of ER homeostasis (Liu et al., 2007; Iwata et al., 2017). In rice, OsbZIP39 and OsbZIP50, the orthologs of Arabidopsis AtbZIP17 and AtbZIP60, are also cleaved and transferred to the nucleus from the ER to upregulate chaperone genes under ER stress (Hayashi et al., 2012; Takahashi et al., 2012). Binding proteins (BiPs) are ER chaperones that can interact with nascent immature secretory proteins and assist with protein folding in concert with other chaperones such as protein disulfide isomerase-like (PDIL) proteins in the ER lumen (Bertolotti et al., 2000; Pobre et al., 2019). Severe suppression or extreme overexpression of OsBiP1 can result in floury endosperm with abnormal protein bodies and reductions in grain weight, endosperm storage protein content, and starch accumulation (Yasuda et al., 2009; Wakasa et al., 2011). PDIL1-1 participates in the maturation of proglutelin in the ER in rice endosperm, and absence of PDIL1-1 also leads to the UPR and the formation of floury endosperm (Satoh-Cruz et al., 2010; Han et al., 2012). High temperature during the rice grain-filling stage can induce more serious ER stress, resulting in a higher grain chalkiness rate with a lower yield (Howell, 2013; Ren et al., 2021). Although bZIP TFs are known to play important roles in response to ER stress, the molecular mechanisms by which bZIP TFs respond to high temperature and regulate grain starch and storage protein biosynthesis in rice remain unclear.
Here, we identified a rice opaque endosperm mutant, opaque 3 (o3). OPAQUE3 (O3) encodes a transmembrane bZIP TF (OsbZIP60) that can directly bind to and activate the ER chaperone genes OsBiP1 and PDIL1-1 and endosperm storage protein and starch biosynthesis-related genes. It thus regulates ER homeostasis and affects grain starch and storage protein biosynthesis, especially under high-temperature conditions. These findings enhance our understanding of the genetic basis of ER stress responses and grain filling in rice, providing useful information for genetic improvement of grain yield and quality.
Results
The o3 mutant accumulates 57-kDa proglutelins and displays defects in grain filling
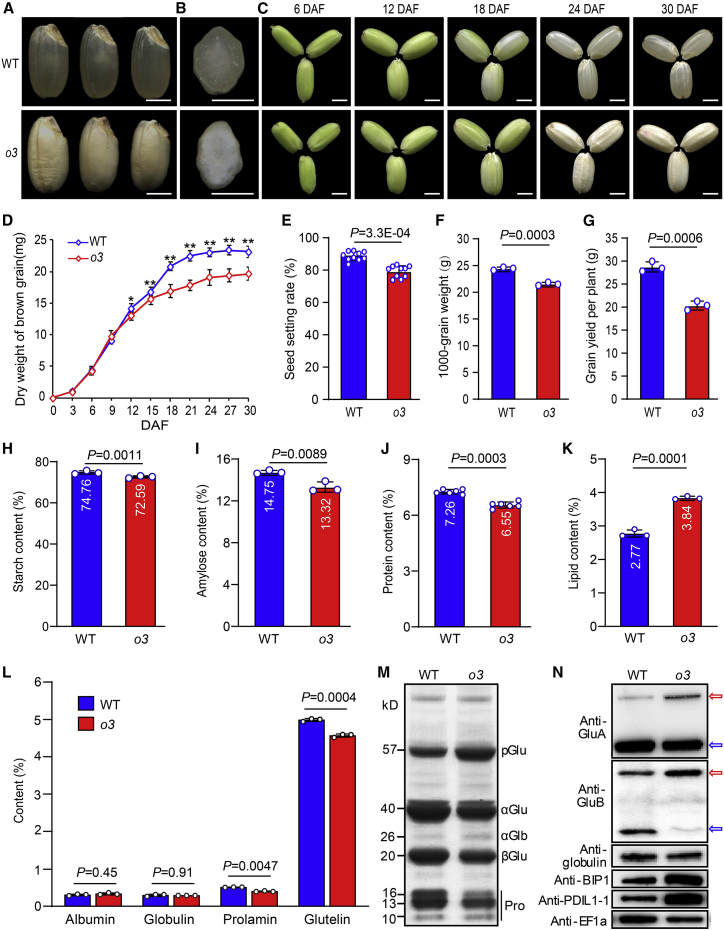
To understand the molecular mechanisms of grain filling and quality formation, we identified a rice mutant, o3, with opaque and floury endosperm from a rice mutant library induced by ethyl methanesulfonate (EMS) treatment of japonica ‘Zhonghua11’ (Figure 1A and 1B). The o3 mutant had a slower grain-filling rate than the wild type; the weight of developing grain from 12 days after fertilization (DAF) and the grain setting rate, 1000-grain weight, and grain yield per plant were significantly lower in the o3 mutant compared with the wild type (Figure 1C–1G). Except for significantly reduced grain thickness, there were no visible differences in plant morphology and grain shape between the o3 mutant and the wild type (Supplemental Figure 1). Next, we measured the contents of storage substances in mature endosperm of the wild type and the o3 mutant. The total starch, amylose, and total protein contents were significantly lower in o3 endosperm compared with wild-type endosperm, whereas the total lipid content was higher (Figure 1H–1K). Storage protein contents, including those of glutelin and prolamin, were also significantly lower in o3 endosperm (Figure 1L). An SDS-PAGE assay of total protein showed that 57-kDa proglutelins were overaccumulated, whereas the amounts of 20-kDa basic and 40-kDa acidic subunits of mature glutelins were reduced. Prolamins were also notably decreased in endosperm of o3 compared with the wild type (Figure 1M). The higher amount of 57-kDa proglutelins in o3 endosperm was confirmed by immunoblotting with isoform-specific antibodies (GluA and GluB). The protein levels of two key molecular chaperones, BIP1 and PDIL1-1, were greatly elevated in the o3 mutant (Figure 1N). These chaperones are known to be induced in 57H rice mutants defective in maturation and export of proglutelins from the ER (Takemoto et al., 2002; Wang et al., 2016). These results indicate that o3 is a 57-kDa proglutelin overaccumulation mutant and may be defective in the maturation and exit of proglutelins from the ER.
Figure 1.
Characterization of the opaque3 (o3) mutant.
(A) Appearance of wild-type (WT) and o3 mutant brown rice.
(B) Transverse sections of WT and o3 endosperm.
(C) Fresh WT and o3 grains at various stages of development. DAF, days after fertilization. Scale bars, 2 mm in (A–C).
(D) Dry weight of WT and o3 grains at various stages of grain filling.
(E–G) WT and o3 seed setting rate (E), 1000-grain weight (F), and grain yield per plant (G).
(H–K) The percent content of total starch (H), amylose (I), total protein (J), and lipid (K) in WT and o3 endosperm.
(L) Storage protein content in WT and o3 milled rice.
(M) SDS-PAGE profiles of total storage proteins in WT and o3 dry grain. pGlu, 57-kDa proglutelin; αGlu, 40-kDa glutelin acidic subunit; αGlb, 26-kDa α-globulin; βGlu, 20-kDa glutelin basic subunit; Pro, prolamin.
(N) Immunoblot analysis of GluA, GluB, globulin, and the molecular chaperones BiP1 and PDIL1-1. Red arrows denote 57-kDa proglutelins. Blue arrows denote the glutelin acidic subunits. EF1α was used as a loading control.
Data in (D–L) are means ± SD from at least three biological replicates. Statistically significant differences were determined using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). P values are shown in (E–L) when statistically significant.
O3 is involved in storage protein processing and export from the ER in developing endosperm cells
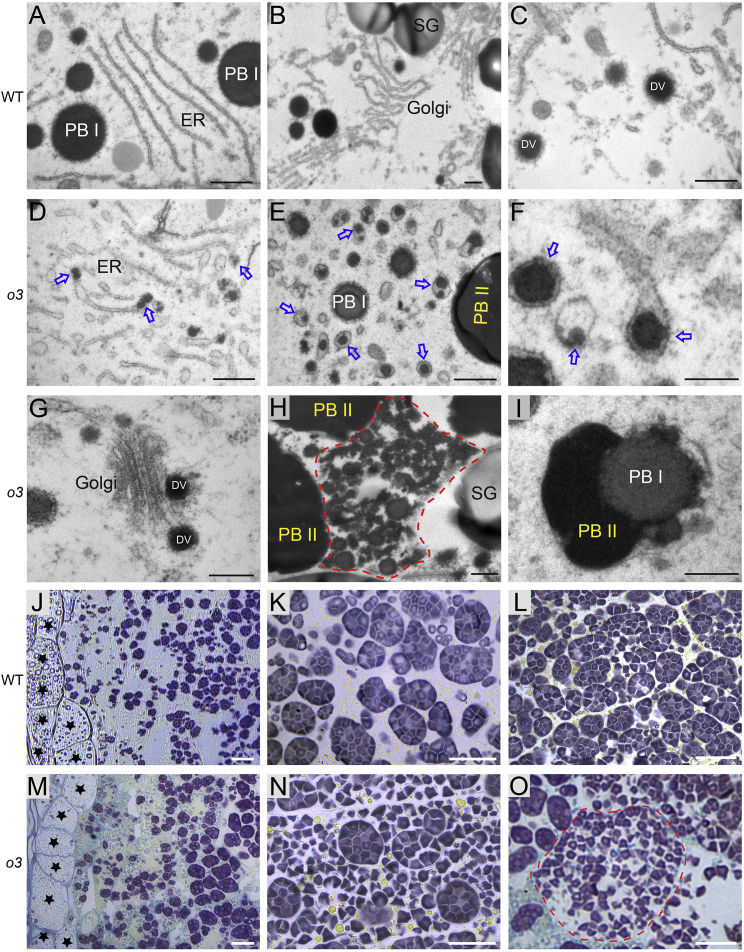
We prepared semi-thin sections (0.5 μm) of 12-DAF endosperm from o3 mutant and wild-type plants and subjected them to Coomassie blue staining to explore the cellular basis of abnormal accumulation of the glutelin precursor in the o3 mutant. Two types of protein bodies, irregularly shaped PBIIs and round PBIs, were observed in o3 and wild-type endosperm, and there were no significant differences in the numbers of PBI and PBII between o3 and the wild type (Supplemental Figure 2A and 2B). Transmission electron microscopy assays were performed to observe the structure of protein bodies in the 12-DAF endosperm. In o3 endosperm cells, the ER morphology was more bent and irregular, some small protein bodies remained in the ER terminal, and many ER-derived vesicles with or without protein bodies were observed (Figure 2A–2F). Unusual fusion of PBII and PBI as well as fragmented protein bodies were observed in the developing endosperm of o3 (Figure 2H and 2I and Supplemental Figure 2C and 2D). Otherwise, there were no obvious differences in the morphology or structure of the Golgi apparatus or DVs between o3 and wild-type endosperm (Figure 2B, 2C, and 2G). These results suggest that O3 plays an important role in processing and export of storage proteins from the ER in rice endosperm cells.
Figure 2.
Protein body and starch grain development in WT and o3 mutant endosperm cells.
(A) Endoplasmic reticulum (ER) and protein body I (PBI) were observed in WT endosperm cells at 12 DAF.
(B and C) The Golgi apparatus and dense vesicles (DVs) in WT endosperm cells.
(D–F) Small protein bodies (blue arrows) remaining in the terminal of ER or ER-derived vesicles and PBI or PBII were observed in o3 endosperm cells.
(G) The Golgi apparatus and DVs in o3 endosperm cells.
(H) The structure of fragmentized protein bodies (red dotted box) was observed in o3 endosperm cells.
(I) The structure of the fusion of PBI and PBII was observed in o3 endosperm cells.
(J–O) Semi-thin sections of WT (J–L) and o3(M–O) endosperm at 9 DAF.
(J and M) The peripheral region of the endosperm. (K, L, N, and O) The central region of the endosperm.
Stars indicate aleurone cells in (J) and (M). Red dotted lines indicate an endosperm cell of o3 filled with abundant single and dispersed starch granules (SGs). Scale bars, 1 μm (A–E), (H), and (I), 0.2 μm (F) and (G), and 20 μm (J–O).
O3 regulates formation of compound starch grains in endosperm cells
We next observed the morphology of starch grains in endosperm cells of o3 and the wild type. Scanning electron microscopy (SEM) of transverse sections from mature endosperm showed that wild-type endosperm cells were filled with densely packed and irregular polyhedral starch granules (SGs), whereas o3 endosperm cells contained spherical and loosely packed SGs (Supplemental Figure 2E and 2F). Semi-thin sections of developing endosperm at 9 DAF were prepared to observe the formation of compound starch grains in o3 and the wild type. Fewer well-developed compound grains and some immature compound grains were observed in peripheral endosperm cells of the wild type and o3 mutant (Figure 2J and 2M). In the center of wild-type endosperm, most amyloplasts produced compound grains consisting of several dozen SGs (Figure 2K and 2L). However, fewer compound grains and more single and dispersed SGs were observed in central cells of o3 endosperm (Figure 2N and 2O and Supplemental Figure 2S and 2T). We then used transmission electron microscopy to observe the development of compound starch grains. Wild-type amyloplasts were filled with polyhedral SGs, which quickly occupied the interior space during grain development, eventually forming a typical complex structure (Supplemental Figure 2G–2I). Similar to the wild type, there were few well-developed amyloplasts in endosperm cells of the o3 mutant (Supplemental Figure 2J–2L). However, the o3 mutant had many disintegrated amyloplasts during the early developmental stage, and the SGs were dispersed throughout the central endosperm cells (Supplemental Figure 2M–2O). There were abundant round and small single SGs instead of compound grains in developing endosperm cells of the o3 mutant (Supplemental Figure 2P–2R), consistent with the results observed in the semi-thin sections (Figures 2N–2O). These results suggest that O3 regulates the formation of starch compound grains in endosperm cells during rice grain development.
We also examined more starch physicochemical properties and the fine structure of wild-type and o3 endosperm. First, the chain length distribution of amylopectin was tested, and the results showed that the proportion of short chains with degree of polymerization values between 6 and 12 was higher in the o3 mutant compared with the wild type, whereas the proportion of intermediate chains with degree of polymerization values in the range of 14–26 was lower (Supplemental Figure 3A). The viscosity of o3 starch was lower and the gelatinization enthalpy (ΔH) of o3 starch was significantly higher than that of wild-type starch (Supplemental Figure 3B and 3C). The o3 endosperm starch was slightly harder to gelatinize in 4–6 mol/L urea compared with wild-type starch (Supplemental Figure 3D). These results show that the physicochemical properties of endosperm starch were altered in the o3 mutant.
O3 encodes a transmembrane bZIP TF
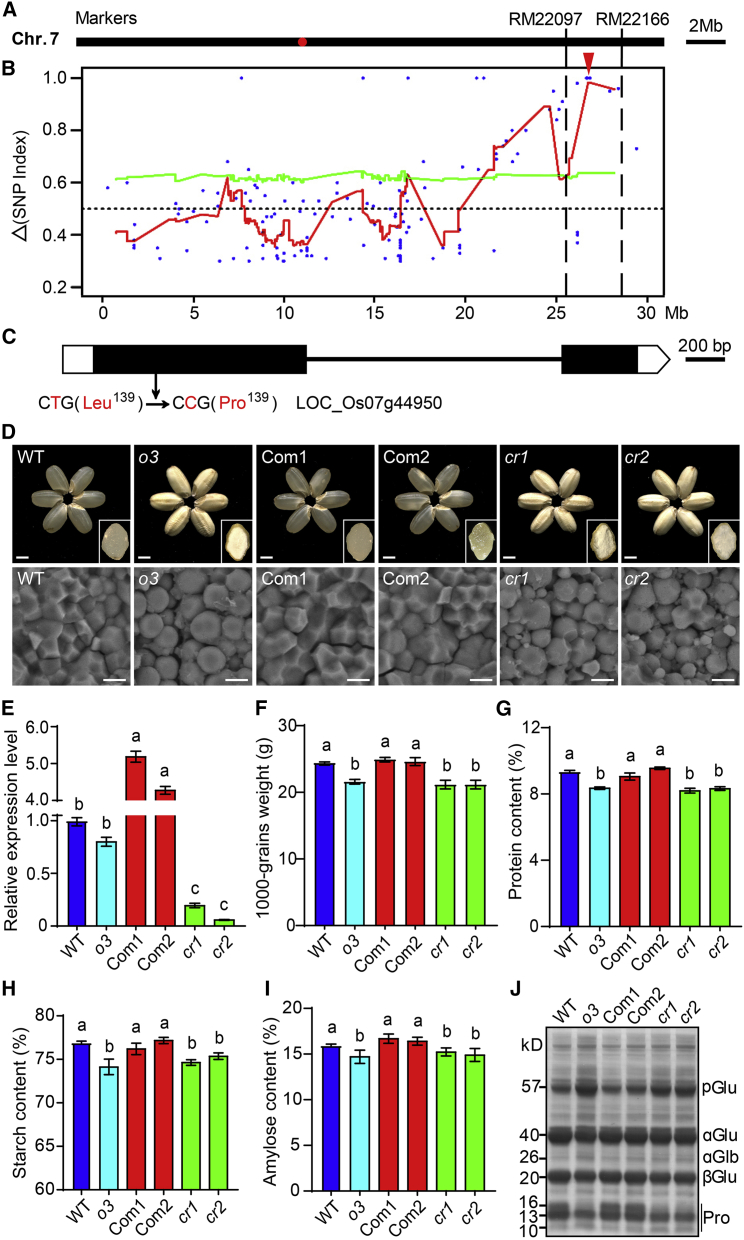
We used a marker mapping and MutMap combination method to clone the gene responsible for the o3 mutation phenotype. First, we selected 207 individuals showing the o3 phenotype from the F2 population derived from a cross between o3 and Nanjing11 (indica). The mutation locus was first mapped to the long arm of chromosome 7 between the markers RM22097 and RM22166 (Figure 3A). Next, we selected 40 individuals with opaque grains from the F2 population of a cross between o3 and the wild type and then sequenced pooled DNA from the selected individuals and wild-type plants using the Illumina HiSeq PE150 platform. Using the MutMap approach described by Abe et al. (2012), we detected a single peak in the primary mapping region on chromosome 7 that contained 13 SNPs with a SNP index greater than 0.8 (Figure 3B and Supplemental Figure 4). Among these SNPs, a nonsynonymous SNP (C to T) in the first exon of LOC_Os07g44950 in the o3 mutant caused leucine139 to be replaced by proline139 in the encoded protein (Figure 3C).
Figure 3.
Identification of Opaque3 (O3) using Map-based cloning and the MutMap method.
(A) The O3 locus was first mapped to the long arm of chromosome 7 between the markers RM22097 and RM22166.
(B) Identification of genomic regions possibly harboring causal mutations for o3 mutants using MutMap. The red curves represent SNP index plots on chromosome 7; the red arrowhead indicates a single peak detected in the primary mapping region.
(C) Gene structure and mutation site in O3 (LOC_Os07g44950). A SNP (T to C) caused a change from Leu-139 to Pro-139 in the encoded protein.
(D) Grain appearance and SG morphology of the WT, o3 mutant, complementation lines (o3 mutant expressing O3; Com1 and Com2), and O3 knockout lines (cr1 and cr2). Insets show transverse sections of representative grains. Scale bars, 2 mm (top) and 5 μm (bottom).
(E) Relative expression level of O3 in developing endosperm of WT, o3, Com1, Com2, cr1, and cr2.
(F–I) 1000-grain weight (F), total protein content (G), total starch content (H), and amylose content (I) of WT, o3, Com1, Com2, cr1, and cr2 grains.
Data in (E–I) are means ± SD from at least three biological replicates. Significant differences are indicated by different letters according to Student’s t-test.
(J) SDS-PAGE profiles of total storage proteins of WT, o3, Com1, Com2, cr1, and cr2 dry grains.
To verify whether LOC_Os07g44950 was the gene responsible for the mutant phenotype, we transformed the o3 mutant with a complementation vector carrying the O3 genomic sequence, including its native promoter. Grains harvested from independent T1 complementation lines showed phenotypes similar to the wild type (Figure 3D). The 1000-grain weight and the contents of total starch, amylose, and total protein in endosperm of complementation lines were similar to those in the wild type (Figure 3E–3I). We also knocked out LOC_Os07g44950 in the wild type using CRISPR-Cas9 editing and successfully obtained four independent homozygous mutants (Supplemental Figure 5A). These mutants exhibited opaque grains with reduced grain weight, and the contents of total starch, amylose, and total protein were similar to those of the o3 mutant (Figure 3D–3I and Supplemental Figure 5B–5D). SGs in endosperm cells of complementation and knockout mutant lines also exhibited granule morphologies similar to those of the wild type and o3 mutant, respectively (Figure 3D). SDS-PAGE showed that the level of storage protein subunits in complementation lines was restored to that in the wild type, and the level in knockout mutant lines was similar to that in the o3 mutant (Figure 3J). Therefore, LOC_Os07g44950 (O3) is the gene responsible for the o3 phenotype. We also performed O3 overexpression (OE) in the wild type; however, the positive OE lines showed no significant differences in plant architecture, grain appearance and weight, and storage substance contents in the endosperm compared with the wild type (Supplemental Figure 6).
O3 (LOC_Os07g44950) encodes a protein with 568 amino acids that was predicted to be a bZIP TF, OsbZIP60 (Nijhawan et al., 2008). O3 contains a bZIP DNA-binding domain followed by a TMD in the middle (Supplemental Figure 7A). A traditional transactivation test using yeast two-hybrid assays revealed that O3 has strong transcriptional activation ability and that the N-terminal region containing 82 amino acids is the critical transactivation domain (Supplemental Figure 7A). Phylogenetic analysis revealed that O3 shared high homology with other bZIP TFs, such as OsbZIP39, OsbZIP50, AtbZIP17, and AtbZIP28, in rice or Arabidopsis (Supplemental Figure 7B and 7C).
O3 is highly expressed in grains, and the encoded bZIP TF is transferred to the nucleus under ER stress
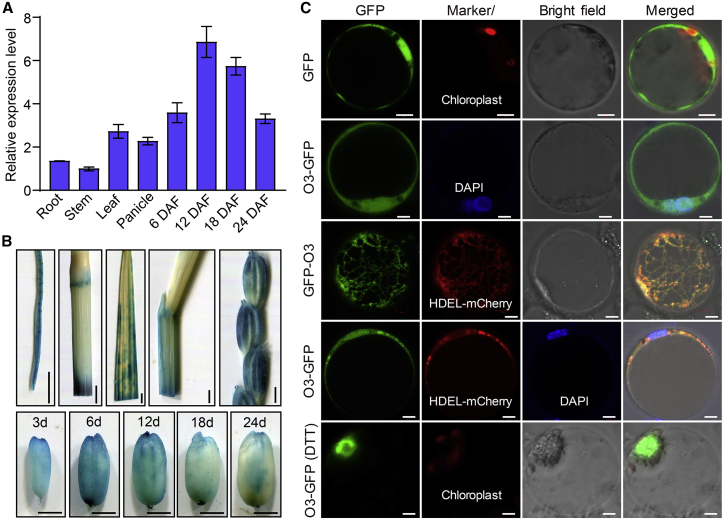
We investigated the temporal and spatial expression patterns of O3 using quantitative reverse transcriptase PCR (qRT–PCR). O3 was constitutively expressed in all examined tissues, with higher levels in developing grains, young panicles, and leaves (Figure 4A). We then ectopically expressed the reporter gene GUS driven by the O3 promoter in rice and examined GUS activity using histochemical analysis. GUS activity was more prominent in developing grains and young panicles, consistent with the qRT–PCR results (Figure 4B).
Figure 4.
Expression pattern and subcellular localization of O3.
(A)O3 expression levels in various tissues and in developing endosperm at 6, 12, 18, and 24 DAF. Values are means ± SD from three biological replicates.
(B) GUS staining in roots, stems, leaves, leaf sheaths, spikelets, and developing grains (3, 6, 12, 18, and 24 DAF) driven by the O3 promoter. Scale bars, 2 mm.
(C) Subcellular localization of O3. Free green fluorescent protein (GFP) and the full-length O3 fusion protein (O3-GFP or GFP-O3) were transiently expressed in rice protoplasts. O3-GFP or GFP-O3 co-localized with DAPI and HDEL-mCherry signals of the nucleus and ER, respectively. O3-GFP was mostly expressed in the nucleus after 2 h of dithiothreitol (DTT) treatment. GFP signals, various organelle marker signals, bright-field images, and merged images of GFP and marker signals are shown in each panel. Scale bars, 5 μm.
We next examined the subcellular localization of O3 by transiently expressing an O3-GFP fusion construct (35S::O3:GFP) in rice protoplasts and tobacco leaves. The GFP control protein was evenly distributed in the cytoplasm and nucleus. However, the O3-GFP fusion protein was present only in the nucleus and ER, based on co-localization with the DAPI signal of the nucleus and the HDEL-mCherry signal of the ER (Figure 4C and Supplemental Figure 7D and 7E). Most of the O3-GFP proteins were relocated to the nucleus under ER stress after 2 h of treatment with dithiothreitol (Figure 4C). The GFP signals of a fusion protein containing truncated O3 without a TMD and the subsequent C-terminal region (O31–240aa-GFP) showed the typical nuclear localization pattern. By contrast, the GFP signals of the fusion protein without the O3 N-terminal region containing the bZIP DNA-binding domain (O3241–568aa-GFP) were observed in the ER (Supplemental Figure 7F). These results suggest that O3 is a transmembrane bZIP TF with an ER and nucleus dual-localization signal that can be relocated to the nucleus under ER stress. Similar results have been reported for OsbZIP39 and OsbZIP50, homologous bZIP TFs in rice that can be cleaved and transferred from the ER to the nucleus under ER stress (Hayashi et al., 2012; Takahashi et al., 2012).
O3 regulates ER protein-processing genes, and OsBiP1 OE partly rescues the phenotype of o3
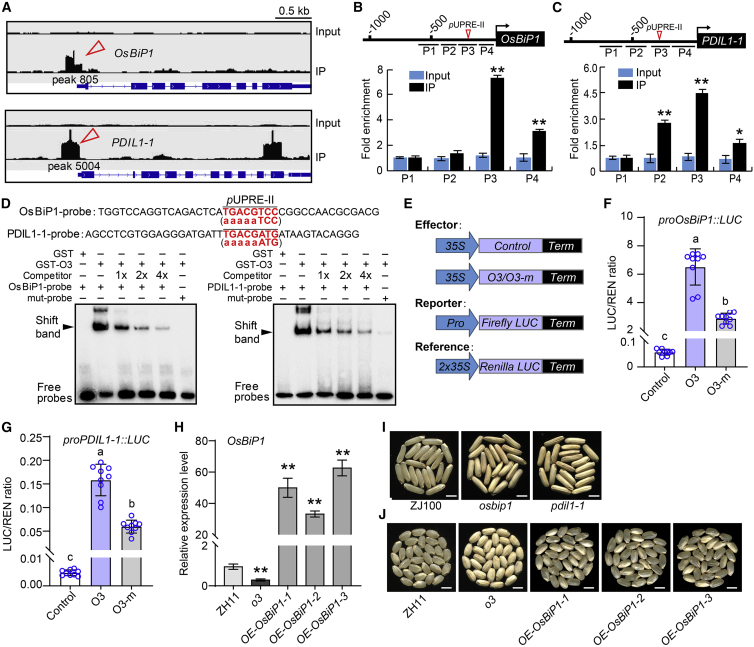
We used an RNA sequencing (RNA-seq)-based transcriptome analysis of 12-DAF grains to clarify the function of O3 in the regulation of endosperm development. A total of 810 and 665 genes were significantly up- and downregulated in o3, respectively (Supplemental Data 1). Gene Ontology analysis showed that the differentially expressed genes (DEGs) were mainly involved in protein processing in the ER, protein export, and starch and sucrose metabolism (Supplemental Figure 8A and 8B). Many ER lumen-localized chaperones and associated genes, such as BIPs, PDILs, OsFes1C, OsEro1, and OsSarld, were upregulated in o3 endosperm (Supplemental Figures 8C and 10A), suggesting that ER stress may occur in developing endosperm cells of the o3 mutant. To determine which DEGs were directly regulated by O3, chromatin immunoprecipitation sequencing (ChIP-seq) was performed (Supplemental Figure 9). A total of 5524 binding sites distributed on 1374 genes were identified (Supplemental Data 2), showing that O3 can bind to the promoters of the chaperone genes OsBiP1 and PDIL1-1 (Figure 5A). ChIP-qPCR and an electrophoresis mobility shift assay (EMSA) verified that O3 can directly bind to the unfolded protein response element (pUPRE)-II motif (TGACG) of the OsBiP1 and PDIL1-1 promoters (Figure 5B–5D). We used a dual luciferase (LUC) reporter system in rice protoplasts to investigate the difference in transcriptional activation activity between O3 and mutated O3. O3 had strong LUC activity, but the mutated O3 had significantly lower LUC activation (Figure 5E–5G), suggesting that O3 could bind to the promoters of OsBiP1 and PDIL1-1 to activate their expression, whereas mutated O3 had significantly lower transactivation activity.
Figure 5.
O3 directly binds to the promoters of OsBIP1 and PDIL1-1 to activate their transcription.
(A) ChIP-seq results showing the distribution of O3 binding sites for OsBIP1 and PDIL1-1 loci, as shown in Integrative Genomics Viewer. Red arrowheads indicate significant peaks calculated by MACS2; positions of peaks are shown in Attachment data II. The input sample was used as a negative control.
(B and C) ChIP-qPCR assay showing the enrichment of O3 at promoter regions of OsBIP1(B) and PDIL1-1(C). DNA samples acquired before immunoprecipitation were used as the input.
(D) EMSA showing that O3 can bind to probes of OsBIP1 and PDIL1-1. The 5-bp consensus pUPRE-II sequence (TGACG) in the promoters of OsBIP1 and PDIL1-1 is indicated by red text. The mutated pUPRE-II motif in mut probes is showed in brackets.
(E–G) LUC transient transactivation assay in rice protoplasts. Constructs used in the transient expression assays are shown in (E). O3 significantly activated transcription of OsBIP1(F) and PDIL1-1(G). O3-m represents the mutant form with a Leu-139 to Pro-139 substitution in the coding region.
(H) qRT–PCR analysis of OsBIP1 transcript level in OE (OE-OsBIP1) lines in the o3 mutant background.
(I) The grain appearance of osbip1 and pdil1-1 mutants in the ZhongJian100 (ZJ100) background. Scale bars, 5 mm.
(J) The grain appearance of OE-OsBIP1 lines in the o3 background. Scale bars, 5 mm.
Data in (B), (C), and (F–H) are means ± SD from at least three biological replicates. Statistically significant differences were determined using Student’s t-test (indicated by different lowercase letters (P < 0.05); ∗P < 0.05, ∗∗P < 0.01).
We had access to the osbip1 and pdil1-1 mutants in the indica ‘ZJ100’ background. Similar to o3, these two mutants had opaque and floury grains and showed 57-kDa proglutelin overaccumulation in the endosperm (Figure 5I and Supplemental Figure 10B–10D). Next, we overexpressed OsBiP1 and PDIL1-1 in the o3 mutant background. The PDIL1-1 OE lines and excessively overexpressed OsBiP1 lines (relative expression levels raised ∼300–900 times) failed to rescue the opaque endosperm phenotype of the o3 mutant (Supplemental Figure 10E–10H). However, the grain appearance qualities of moderately overexpressed OsBiP1 lines (relative expression levels raised ∼30–80 times) were considerably improved: many grains showed a transparent endosperm phenotype similar to the wild type (Figure 5H and 5J and Supplemental Figure 10G and 10H), and the expression levels of genes related to ER stress and starch and protein biosynthesis had returned to approximately wild-type levels in moderately overexpressed OsBiP1 lines (Supplemental Figure 10I and 10J). These results suggest that O3 plays an important role in response to ER stress and regulation of ER protein processing and export by activating the expression of OsBIP1 and PDIL1-1.
O3 directly regulates genes involved in starch and storage protein biosynthesis
Transcriptome analysis and qRT–PCR showed that the expression levels of many starch and storage protein biosynthesis-related genes were also significantly downregulated in developing endosperm of o3 (Supplemental Figures 8C and 11A). ChIP-seq, yeast one-hybrid, and dual-LUC reporter assays showed that O3 can directly target and activate the endosperm starch synthesis-associated genes GBSS1, AGPL2, SBEI, and ISA2 (Supplemental Figure 11B–11D). EMSA confirmed that O3 can directly bind to the GCN4 and CCGTCC motifs in the promoter of GBSSI (Supplemental Figure 11E). qRT–PCR analysis showed that the storage protein synthesis-related genes OsGluA2, Prol14, and Glb were dramatically downregulated in the developing endosperm of the o3 mutant (Supplemental Figures 8C and 12A). A dual-LUC reporter assay showed that O3 can directly target the promoters and activate expression of OsGluA2, Prol14, and Glb in rice protoplasts (Supplemental Figure 12B). EMSA confirmed that O3 can directly bind to the O2 motif in the promoter of OsGluA2 (Supplemental Figure 12C). These results show that O3 directly participates in regulation of endosperm starch and storage protein biosynthesis, thus regulating the grain yield and quality of rice.
The o3 mutant is more susceptible to ER stress under high-temperature conditions
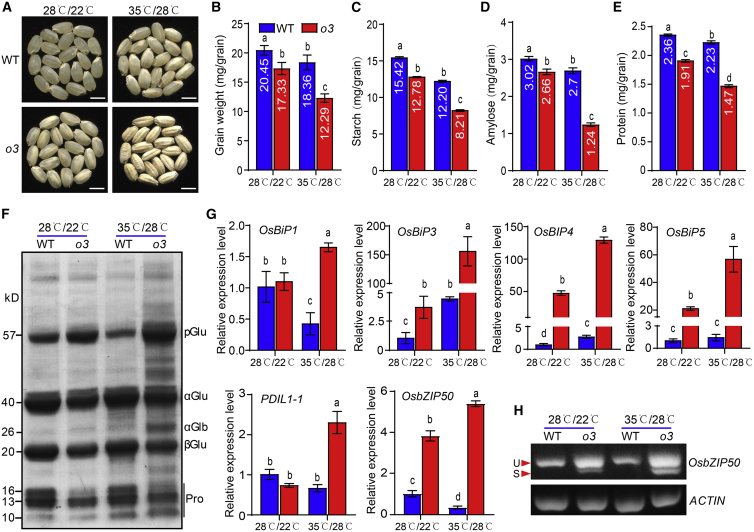
High temperatures during the rice grain-filling stage are known to induce ER stress (Howell, 2013). We found that expression of O3 and its homologous gene OsbZIP50 increased when seedlings were moved to high temperature (42°C) from normal temperature (28°C) (Supplemental Figure 13A). We next grew wild-type and o3 plants under field conditions until flowering and then moved the plants to artificial high temperature (35°C, 12 h light/28°C, 12 h dark) and normal temperature (28°C, 12 h light/22°C, 12 h dark) conditions. Under high-temperature conditions, mature grains of the wild type were slightly chalky with slightly reduced weight, but mature o3 grains were very floury and shrunken with significantly reduced size and weight (Figure 6A and 6B and Supplemental Figure 13B). Compared with normal-temperature conditions, the wild type and o3 mutant had significantly lower amylose and total protein content under high-temperature conditions; however, the differences between conditions were greater for the o3 mutant than for the wild type (Figure 6C–6E). An SDS-PAGE assay showed that the o3 mutant had a higher endosperm accumulation of 57-kDa proglutelin and a lower accumulation of 20-kDa basic and 40-kDa acidic subunits of mature glutelins and 13-kDa and 16-kDa prolamin under both normal- and high-temperature conditions; however, differences between the o3 mutant and the wild type were greater under high-temperature conditions (Figure 6F). These results indicate that endosperm development is more susceptible to heat stress in the o3 mutant than in the wild type.
Figure 6.
The o3 mutant is more prone to ER stress under high-temperature conditions.
(A) Appearance of WT and o3 grains under high-temperature conditions (35°C, 12 h light/28°C, 12 h dark) and normal-temperature conditions (28°C, 12 h light/22°C, 12 h dark). Scale bars, 5 mm.
(B–E) The grain weight (B), total starch content (C), amylose content (D), and total protein content (E) of WT and o3 grains under high- and normal-temperature conditions.
(F) SDS-PAGE profiles of total storage proteins of WT and o3 dry grains.
(G) Transcript levels of genes related to ER stress in WT and o3 grains at 9 DAF under high- and normal-temperature conditions.
(H) Semi-quantitative PCR analysis of OsbZIP50 transcripts in WT and o3 endosperm cells under high- and normal-temperature conditions. U and S, the unspliced and spliced forms of the OsbZIP50 mRNA, respectively.
Data in (B–E) and (G) are means ± SD from at least three biological replicates. Significant differences are indicated by different letters according to Student’s t-test.
We next examined the expression profiles of ER stress-associated genes in the wild type and o3 mutant under high-temperature and normal-temperature conditions. In the o3 mutant, genes that responded to ER stress, such as OsBIPs, PDIL1-1, OsbZIP50, PDIL2-3, Calnexin, OsFes1, and OsEro, had higher expression levels under high-temperature than under normal-temperature conditions (Figure 6G and Supplemental Figure 13C). Because OsbZIP50 can be cleaved and transferred from the ER to the nucleus to upregulate expression of chaperone genes under ER stress (Supplemental Figure 14A; Hayashi et al., 2012), we performed a semi-quantitative RT-PCR analysis of OsbZIP50 transcripts under high-temperature and normal-temperature conditions. OsbZIP50 mRNA was present in an unspliced (U) form in developing endosperm of the wild type under high-temperature and normal-temperature conditions; however, the spliced (S) form of OsbZIP50 mRNA was detected in the o3 mutant, and the S form was more abundant under high-temperature conditions (Figure 6H). This suggests that developing endosperm of the o3 mutant experienced more severe ER stress than that of the wild type under heat stress. In the wild type, the expression levels of OsBiP1, PDIL1-1, and OsbZIP50 were lower under high-temperature conditions than under normal-temperature conditions (Figure 6G and Supplemental Figure 13C), which suggests that O3 participates in maintaining ER homeostasis in developing endosperm of the wild type under heat stress. The expression levels of genes related to storage protein and starch synthesis were lower in developing endosperm of the o3 mutant under high-temperature conditions compared with normal-temperature conditions, but expression levels of genes related to starch degradation were higher (Supplemental Figure 13D and 13E). These results suggest that O3 plays an important role in maintaining ER homeostasis and regulating storage substance accumulation and endosperm development under heat stress.
Discussion
O3 affects rice grain filling by simultaneously regulating starch and storage protein biosynthesis
Starch and storage proteins determine the yield and quality of cereal grains. Their biosynthesis in grains requires a series of enzymes that are accurately regulated by a group of spatiotemporally expressed TFs (Kawakatsu et al., 2009; Wang et al., 2013; Bai et al., 2016; Xu et al., 2016; Bello et al., 2019; Xiong et al., 2019). In maize, many TFs have been reported to synergistically regulate starch and protein biosynthesis in endosperm. O2 is an endosperm-specific bZIP TF that mainly regulates expression of the α- and β-zein genes by recognizing the O2 box in their promoters. O2 can directly transactivate PPDK1, PPDK2, SSIII, SUS1, and SUS2 to regulate starch biosynthesis and sucrose synthase-mediated endosperm filling (Zhang et al., 2016; Deng et al., 2020). Prolamin-box binding factor (PBF), an endosperm-specific DNA binding with one finger (DOF) TF, can also regulate expression of zein genes by recognizing the prolamin-box; it can also interact with O2 and enhance its transcriptional activation activity (Vicente-Carbajosa et al., 1997). ZmNAC128 and ZmNAC130 are homologous and functionally redundant endosperm-specific NAC TFs; they bind to the ACGCAA motif on the promoter of 16-kDa γ-zein and the starch synthesis gene Bt2 to activate their expression. Simultaneous knockdown of ZmNAC128 and ZmNAC130 expression with RNAi causes a shrunken kernel phenotype with significantly reduced starch and protein contents (Zhang et al., 2019b). The upstream regulator ZmABI19 can directly regulate multiple key grain-filling TFs, including O2, PBF1, ZmbZIP22, NAC130, Opaque11, and SWEET4c, by binding to the RY motif in the early endosperm development stage of maize. The kernels of the zmabi19 mutant are opaque and reduced in size with lower accumulation of zeins, starch, and lipids (Zhang et al., 2021).
Few regulators that can simultaneously regulate starch and protein biosynthesis in rice endosperm have been reported. A bZIP TF homolog of maize O2, OsbZIP58, has been reported to regulate both starch and storage protein biosynthesis (Kawakatsu et al., 2009; Wang et al., 2013). The rice NAC TFs OsNAC20 and OsNAC26, orthologs of maize ZmNAC128 and ZmNAC130, respectively, also play an essential role in regulation of starch and storage protein synthesis by directly transactivating expression of SSI, Pul, GluA1, GluB4/5, α-globulin, and 16-kDa prolamin, and the osnac20/26 double mutant has floury seeds with significantly lower starch and storage protein contents (Wang et al., 2020). In this study, we identified the rice opaque endosperm mutant o3. O3 encodes a transmembrane bZIP TF, OsbZIP60 (Figure 3 and Supplemental Figure 7A–7C; Yang et al., 2022). ChIP-seq and LUC assays demonstrated that O3 could directly bind to the promoters and activate the expression of the starch synthesis-related genes GBSSI, AGPL2, SBEI, and ISA2 as well as the storage protein synthesis-related genes OsGluA2, Prol14, and Glb1 (Supplemental Figures 8, 11, and 12). The expression levels of these starch and storage protein synthesis-related genes were significantly downregulated in the o3 mutant, which may lead to the lower starch content, protein content, and grain weight of the o3 mutant. This also resulted in alterations of starch fine structure, such as amylopectin chain length distributions and developmental defects of compound starch grains in the endosperm of o3 compared with the wild type (Figures 1H–1J and 2J–2O and Supplemental Figures 2 and 3). EMSA further clarified that O3 directly binds to the GCN4 (TGA(G/C)TCA) and CCGTCC motifs in the GBSSI promoter and the O2 motif (GATGACATAG) in the OsGluA2 promoter (Supplemental Figures 11 and 12). In maize, the O2 motif (TGACGTGGC) is the key element for regulation of zein and starch synthesis-related genes (Zhang et al., 2016; Deng et al., 2020). Therefore, we believe that the O2 motif is the core element for regulation of starch and storage protein-related gene expression by bZIP TFs in maize and rice. O3 (OsbZIP60) can form homodimers (Hayashi et al., 2013). These results suggest that O3 is the core regulatory factor in rice endosperm development; it can simultaneously regulate biosynthesis of endosperm starch and storage protein, thus affecting rice quality and grain yield, similar to maize O2 (Figure 7; Zhang et al., 2016; Deng et al., 2020).
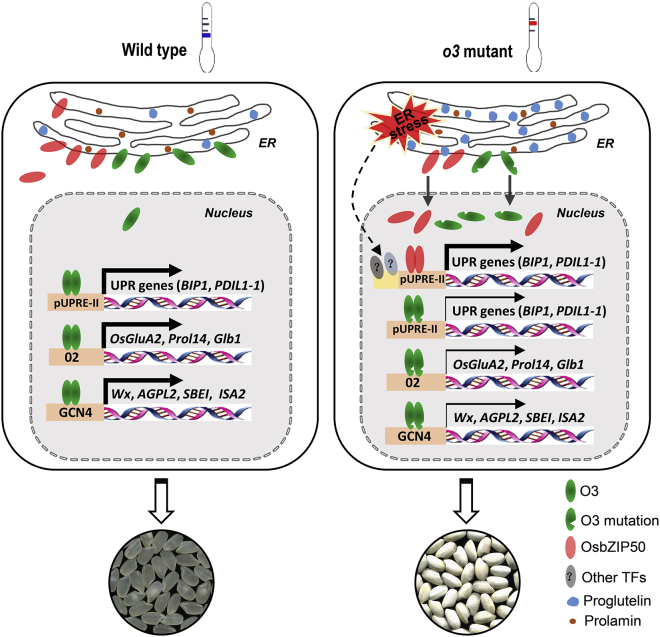
Figure 7.
Proposed model of the role of O3 in maintaining ER homeostasis and regulating endosperm storage protein and starch biosynthesis in rice.
Under normal conditions in the WT, O3 is located in the ER and the nucleus. It simultaneously regulates ER protein processes and secretion as well as storage protein and starch biosynthesis in endosperm cells by binding to specific motifs, such as pUPRE-II, O2, and the GCN4 box, to activate transcription of UPR genes and storage protein and starch biosynthesis genes, ultimately ensuring normal development of rice grains. However, mutation of O3 leads to downregulated expression of ER stress-related genes, such as OsBIP1 and PDIL1-1, as well as genes related to storage protein and starch biosynthesis, resulting in ER stress and an impaired protein folding process in the ER. This leads to excessive accumulation of the 57-kDa glutelin precursor and reduces the contents of starch and storage proteins. High temperature can aggravate ER stress and lead to more abnormal grain development in the o3 mutant. As physiological feedback, more O3 (mutated) is transferred to the nucleus from the ER, together with OsbZIP50 and other unknown TFs, to activate expression of key UPR genes to maintain ER homeostasis, especially under high-temperature conditions.
O3 plays an important role in protein processing and export in the ER of rice endosperm cells
Rice grain storage proteins are initially synthesized on the ER membrane and translocated into the ER lumen in endosperm cells. They are then stored as morphologically distinct protein bodies in different subcellular compartments (Yamagata et al., 1982). Prolamins are stored as intracisternal inclusion granules within the ER lumen and bud off from the ER to form PBIs. Glutelins are synthesized as 57-kDa precursors on the rough ER and are transported to PSVs to be cleaved into mature acidic and alkaline subunits to form PBII by the DV-mediated post-Golgi apparatus trafficking pathway or ER-derived precursor accumulating compartments that bypass the Golgi apparatus (Krishnan et al., 1986; Takahashi et al., 2005). Mutations in the key genes that regulate storage protein folding, processing, and transport lead to 57-kDa proglutelin overaccumulation (57H) in endosperm cells. The esp2 mutants, resulting from knockout of ER-localized PDIL1-1, develop many ER-derived, small, PBI-like vesicles containing cross-linked glutelins and prolamins in developing endosperm (Takemoto et al., 2002; Han et al., 2012). Other rice 57H mutants, such as rab5a, vps9a, gpa3, gpa5, osnhx5, and calcium caffeine zinc1, are defective in post-Golgi apparatus trafficking and produce many abnormal protein granules and paramural body structures at the apoplast (Wang et al., 2010; Liu et al., 2013; Zhu et al., 2019; Ren et al., 2020; Pan et al., 2021). OsVPE1 is responsible for processing and maturation of glutelins in PSV, and the osvpe1 mutants develop many round PBIIs (Wang et al., 2009).
In this study, we found that mutation of O3 caused overaccumulation of 57-kDa proglutelins and significantly lower accumulation of mature 20-kDa basic and 40-kDa acidic subunits. 13-kDa and 16-kDa prolamins were also notably decreased in the o3 mutant compared with the wild type (Figure 1M). Cytological studies showed that many protein bodies were retained in the ER lumen of o3, similar to the small PBI-like vesicles observed in the pdil1-1 mutant (Takemoto et al., 2002). Many ER-derived vesicles with or without protein bodies were observed in endosperm cells of the o3 mutant (Figure 2D–2F). The ER-derived vesicles containing protein bodies have certain similarities to the aberrant vesicle structures in the gpa4 mutant (Wang et al., 2016). These results demonstrate that grain storage protein processing and export in the ER were disturbed in o3 endosperm cells. In addition to directly binding and activating genes related to starch and storage protein biosynthesis, O3 also activated expression of OsBIP1 and PDIL1-1 (Figure 5A–5G), two major chaperones that assist with folding of immature secretory proteins in the ER. Therefore, the protein folding and processing system in the ER may be impaired in the o3 mutant, causing unfolded protein accumulation and production of abnormal protein body structures in endosperm cells. Otherwise, the vesicle structures that contain protein bodies in o3 endosperm cells may be precursor accumulating compartment vesicles formed by expansion of the terminal of rough ER containing glutelin precursors. These results show that O3 plays an important role in protein processing and export in the ER of rice endosperm cells.
O3 is essential for maintaining ER homeostasis in rice endosperm cells under heat stress
RNA-seq analysis showed that many UPR genes related to protein folding and processing were upregulated in the o3 mutant (Supplemental Figure 8), indicating that the o3 mutant experiences ER stress. Subcellular localization assays revealed that O3 is localized in the ER and nucleus under normal conditions. Under ER stress conditions, O3 was mostly transferred to the nucleus (Figure 4C and Supplemental Figure 7). O3 can directly bind to the promoters and activate the expression of OsBIP1 and PDIL1-1. However, the expression level and protein abundance of OsBIP1 and PDIL1-1, as well as the expression levels of many UPR genes, were significantly upregulated in the o3 mutant (Figure 1N and Supplemental Figures 8 and 10). This result suggests that there may be physiological feedback; to maintain ER homeostasis in o3 mutants, the expression levels of genes related to protein folding and processing were upregulated (Figure 7). The pdil1-1 and osbip1 mutants show an opaque endosperm phenotype similar to that of o3 (Han et al., 2012; Supplemental Figure 10C). Moderate OE of OsBIP1 in the o3 mutant could cause expression levels of genes related to ER stress and starch and protein biosynthesis to return to approximately wild-type levels, thus partly restoring the transparent endosperm phenotype; by contrast, excessive overexpression of OsBiP1 and OE of PDIL1-1 failed to rescue the o3 phenotype (Figure 5H–5J and Supplemental Figure 10E–10J). These results indicate that O3 plays an important role in maintaining ER homeostasis by activating the key stress response gene OsBIP1 rather than PDIL1-1. OsbZIP50, a homolog of O3, is a well-known regulator specifically responsive to ER stress. Under ER stress conditions, unconventional splicing of OsbZIP50 mRNA occurs, and the shorter OsbZIP50 protein is relocated to the nucleus to activate expression of UPR genes (Supplemental Figure 14A; Hayashi et al., 2012). We found that OsbZIP50 was present mainly in the U form in the wild type. However, OsbZIP50 was significantly upregulated and occurred in the S form in the o3 mutant (Figure 6G and 6H and Supplemental Figure 14B). We found that the TMD of O3 was responsible for its ER localization (Figure 4C and Supplemental Figure 7), whereas semi-quantitative RT-PCR analysis showed that the O3 mRNA did not have alternative splicing in developing endosperm, which suggested that the TMD of O3 may be cleaved by protease under ER stress, consistent with Arabidopsis AtbZIP17 and AtbZIP28 (Supplemental Figure 14B; Iwata et al., 2017). A LUC reporter assay showed that OsbZIP50 can enhance transient transactivation of OsBIP1 and PDIL1-1 together with O3 in rice protoplasts (Supplemental Figure 14C). This suggests that OsbZIP50 activates UPR genes related to protein folding to help maintain ER homeostasis in the o3 mutant (Figure 7).
High temperatures during the rice grain-filling stage can induce ER stress and lead to grain chalkiness (Howell, 2013; Ren et al., 2021). We found that the o3 mutant was more sensitive to high-temperature stress than the wild type (Figure 6A–6E). Genes that respond to ER stress, such as OsBIPs, PDILs, Erdj3B, Calnexin, OsFes1, and OsEro1, were significantly upregulated in the o3 mutant under high-temperature compared with normal-temperature conditions (Figure 6G and Supplemental Figure 13C). These results indicate that high-temperature conditions induce higher levels of ER stress in the endosperm cells of the o3 mutant. OsbZIP50 was also significantly upregulated with higher accumulation of the S form in the o3 mutants compared with normal-temperature conditions (Figure 6G and 6H). Although the transcriptional activation function of mutated O3 was weaker, its target genes, OsBIPs and PDIL1-1, and other UPR genes were upregulated rather than downregulated in the o3 mutant (Supplemental Figures 8 and 10 and Figure 6G). It is possible that these genes were activated by OsbZIP50 and other TFs induced by ER stress to maintain ER homeostasis in endosperm cells of the o3 mutant, especially under high-temperature conditions (Figure 7 and Supplemental Figure 14C). The expression levels of genes related to storage protein and starch biosynthesis were lower, but those of genes related to starch degradation were higher in developing endosperm of the o3 mutant under high-temperature conditions compared with normal-temperature conditions (Supplemental Figure 13D and 13E), which presumably led to the more pronounced abnormal grain phenotype of the o3 mutant under high-temperature conditions (Figure 7). These results suggest that O3 is essential for maintaining ER homeostasis in endosperm cells under heat stress. Our research indicates that rice O3 (OsbZIP60) participates simultaneously in regulating storage protein and starch biosynthesis and maintaining ER homeostasis in grain endosperm and therefore plays a central role in rice grain development. Our study provides useful information for potential genetic improvement of yield and grain quality in rice.
Methods
Plant materials and growth conditions
The o3 mutant was identified from an ethyl methanesulfonate-induced mutant pool of japonica rice ‘Zhonghua11’ (ZH11). Two F2 populations from the crosses of o3 × Nanjing11 (indica) and o3 × ZH11 were used to map the mutant gene. All plants used in this study were grown in paddy fields in Hangzhou, Zhejiang Province, China, during the normal growing seasons. At the flowering stage, some well-grown wild-type and o3 plants were moved to plant growth chambers under normal-temperature (28°C, 12 h light/22°C, 12 h dark) and high-temperature (35°C, 12 h light/28°C, 12 h dark) conditions.
Microscopy analysis
To observe starch grain and protein body development, transverse sections of wild-type and o3 endosperm at 9 DAF were used to prepare semi-thin sections (0.5 μm). Samples were stained with I2–KI or Coomassie brilliant blue for 5 s and subsequently examined under a light microscope. The brown rice was cut transversely to prepare samples for scanning electron microscopy analysis. Images were obtained with an S3400N scanning electron microscope (Hitachi, Tokyo, Japan). To observe the ultrastructure of amyloplasts, PBI, and PBII, 6–12-DAF grains were observed with a transmission electron microscope.
SDS-PAGE and immunoblot analysis
SDS-PAGE analysis was performed as described by Ren et al. (2014). In brief, total dry grain proteins were extracted using an extraction buffer. The proteins were resolved by SDS-PAGE on a 4%–20% (w/v) graded gel, followed by Coomassie brilliant blue staining and photography. For immunoblotting, fresh rice grains were used for protein extraction. Total proteins were then separated on 10% SDS-PAGE gels and immunoblotted with specific antibodies. The specific bands were detected using an enhanced chemiluminescence detection kit (Thermo Fisher Scientific).
Analyses of physicochemical properties of endosperm starch
The total starch and amylose content was measured using a starch assay kit (Megazyme, Wicklow, Ireland). Lipid and total protein contents in the grains were measured according to the method described by Kang et al. (2005). The glutelin, prolamin, globulin, and albumin in the mature endosperm (milled rice) were extracted and measured according to the methods described by Yang et al. (2015) and Kang et al. (2005). To determine the starch pasting properties, 3 g of milled rice powder was transferred to a container with 25 mL of distilled water. The sample was mixed and measured with a Rapid Visco Analyzer (Newport Scientific, Narrabeen, NSW, Australia). To determine the chain length distribution of amylopectin, 5 mg of rice flour was digested with isoamylase and then analyzed by capillary electrophoresis.
Mapping of the O3 gene
To map the O3 locus, 207 individuals with opaque endosperm were selected from the F2 population derived from o3 × NJ11. Polymorphic simple sequence repeats (SSR) markers were selected to link to O3. Next, the MutMap method was used to map O3 according to the description in Abe et al. (2012). A recessive pool containing 40 individuals with opaque endosperm was selected from the F2 population derived from o3 × ZH11; ZH11 was the dominant pool. Both bulked DNA samples were subjected to whole-genome sequencing using the Illumina HiSeq PE150 platform (Novogene, Beijing, China). The short reads were aligned to the Nipponbare reference sequence to enable identification of reliable SNPs. The regression lines of SNP index plots were generated by averaging scores in a sliding 2-Mb window with 10-kb increments. Candidate SNPs with a SNP index greater than 0.8 from the mutagenesis were screened further. The primers used for mapping are listed in Supplemental Table 1.
Vector construction and plant transformation
To create the complementation vector, the O3 genomic fragment, including its native promoter, was amplified from ZH11 and cloned into the binary expression vector pCAMBIA1300. The O3 cDNA sequence driven by the UBIQUTIN1 promoter was cloned into the binary vectors pCAMBIA1390 and pRHVnGFP to generate OE vectors. To knock out O3 using the CRISPR-Cas9 system, single guide RNAs targeting sites were constructed into the BGK03 vector (Biogle, Hangzhou, China). Complementation and OE vectors were transformed into o3, and the CRISPR-Cas9 vectors were transformed into ZH11. Sequences of the primers used for vector construction and detection are listed in Supplemental Table 1.
RNA-seq and qRT–PCR analysis
For transcriptome analysis, total RNA samples from wild-type and o3 grains at 9 DAF were sequenced on the HiSeq 4000 platform (Illumina) by Novogene Technology (Beijing, China) to obtain clean reads. DEGs were identified by a false discovery rate ≤0.05 and an absolute value of the log2 ratio ≥1. Gene Ontology enrichment analysis was performed based on the hypergeometric distribution using R. Pathway analysis was performed in MapMan (https://mapman.gabipd.org/) by searching against the Oryza sativa TIGR7 database.
Total RNA was extracted from different plant tissues using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA was reverse transcribed into cDNA using a ReverTra Ace qPCR RT Kit (Toyobo, Osaka, Japan). qPCR was performed using SYBR Green Real-Time PCR Master Mix (Toyobo). The rice Ubiquitin gene (Os03g0234200) was used as an internal control. The primer sequences used in this analysis are listed in Supplemental Table 1.
Transcriptional activity, yeast one-hybrid, and LUC activity assays
To test the transactivation activity of O3, the coding sequence (CDS) of O3 was amplified from ZH11, cloned into the pGBKT7 vector, and transformed into yeast strain AH109. The transformed yeast strains were examined on SD/-Trp and SD/-Trp/-His/-Ade/X-α-GAL plates for 4 days at 30°C. For the yeast one-hybrid assay, the CDS of O3 was cloned into the pB42AD vector, and the promoter regions of GBSS1, AGPL2, SBE1, and ISA2 were cloned into the pLacZi2μ vector. Bait and prey plasmids were co-transformed into yeast EGY48. The assay was performed using the Clontech Yeast One-Hybrid System (Takara, Beijing, China) following the manufacturer’s instructions.
For the LUC activity assay, the promoter regions of GBSS1, AGPL2, SBEI, ISA2, OsGluA2, Prol14, and Glb1 were inserted into the 190LUC vector as reporters. The coding sequences of O3 and the O3 mutant form (O3-m) were inserted into the expression vector as effectors. The LUC activity assay was then performed as described by Wang et al. (2020). The primers used in this assay are listed in Supplemental Table 1.
Subcellular localization of O3 and OsbZIP50
To verify the subcellular localization of O3 and OsbZIP50, coding regions without a termination codon were cloned into the pAN580 and pCAMBIA1305-GFP vectors. The fusion of GFP and different truncated forms of O3 together with marker vectors (HDEL-mCherry, D53-mCherry, GHD7-CFP, and PHT4-RFP) were co-transformed into rice protoplasts. DAPI was used as a nuclear marker. The pCAMBIA1305-O3-GFP vector was infiltrated into 3-week-old leaf epidermal cells of tobacco via Agrobacterium.
ChIP-seq and ChIP-qPCR
ChIP assays were performed following a method described previously with slight modifications (Xiong et al., 2019). In brief, 9-DAF endosperm was harvested from the GFP-O3 OE lines in the o3 background and crosslinked in 1% formaldehyde. The cross-linked endosperm was dried and ground into powder in liquid N2, followed by nuclear isolation and sonication. Protein A and G agarose beads (Beyotime, Shanghai, China) and a GFP antibody were used to precipitate the protein and DNA complex, which was digested by Proteinase K and recovered using a PCR purification kit. DNA samples acquired without immunoprecipitation by the GFP antibody were used as the input. The immunoprecipitated DNA and input DNA were sequenced on the HiSeq 2500 platform (Novogene, Beijing, China). ChIP-seq raw sequencing data were mapped to the rice reference genome using the Burrows–Wheeler Alignment tool. Model-based analysis of ChIP-seq data 2 (MACS2) was used for peak calling, and significantly enriched peaks (binding sites) were identified using a corrected P value of less than 0.05 in the IP libraries compared with input DNA (Supplemental Data 2). Visual analysis was performed using Integrative Genomics Viewer (v.2.3.26). To validate the specific regions of target genes bound by O3, the immunoprecipitated DNA and input DNA were used in ChIP-qPCR analysis. The enrichment value of each fragment was normalized to that of the input sample. The primers used in this assay are listed in Supplemental Table 1.
EMSA
The GST-O3104–176aa protein containing the bZIP domain structure was purified. The EMSA probes of BIP1, PDIL1-1, GBSS1, and OsGluA2 were commercially synthesized and labeled using the EMSA Probe Biotin Labeling Kit (Beyotime, Shanghai, China), and non-labeled probes were used as competitors. Probe sequences are listed in Supplemental Table 1. The DNA binding reaction was performed for 20 min at 25°C, and the products were then electrophoresed using 6% acrylamide gels. The DNA probes were transferred to a nylon membrane. Finally, the oligo bands were detected using the LightShift Chemiluminescent EMSA Kit and streptavidin-horseradish peroxidase (Beyotime).
Funding
This work was supported by the National Natural Science Foundation of China (31971925 and 32172080), the Natural Science Foundation of Zhejiang Province (LR20C13002), the Special Support Plan for High-Level Talents in Zhejiang Province (2019R52032), and the International Science & Technology Innovation Program of the Chinese Academy of Agricultural Sciences, China (CAAS-ZDRW202109).
Author contributions
X.W. and R.C. designed experiments and analyzed data. R.C., S.Z., G.J., Y.D., L.M., N.D., F.L., M.Z., G.S., S.H., Z.S., and S.T. performed the experiments. R.C., X.W., and J.Z. wrote the manuscript and prepared the illustrations. X.W. and P.H. conceived the idea and supervised the project. All authors read and approved the final manuscript.
Acknowledgments
We thank Prof. Jianmin Wan for providing the antibodies for PDIL1-1, OsBiP1, OsGluA, OsGluB, and Globulin. No conflict of interest is declared.
Published: November 14, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Xiangjin Wei, Email: weixiangjin@caas.cn.
Peisong Hu, Email: hupeisong@caas.cn, hupeisong@caas.cn.
Supplemental information
Supplemental Data 1. DEGs between ZH11 and the o3 mutant
References
- Abe A., Kosugi S., Yoshida K., Natsume S., Takagi H., Kanzaki H., Matsumura H., Yoshida K., Mitsuoka C., Tamiru M., et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012;30:174–178. doi: 10.1038/nbt.2095. [DOI] [PubMed] [Google Scholar]
- Bai A.N., Lu X.D., Li D.Q., Liu J.X., Liu C.M. NF-YB1-regulated expression of sucrose transporters in aleurone facilitates sugar loading to rice endosperm. Cell Res. 2016;26:384–388. doi: 10.1038/cr.2015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball S.G., Morell M.K. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 2003;54:207–233. doi: 10.1146/annurev.arplant.54.031902.134927. [DOI] [PubMed] [Google Scholar]
- Beckles D.M., Smith A.M., ap Rees T. A cytosolic ADP-glucose pyrophosphorylase is a feature of graminaceous endosperms, but not of other starch-storing organs. Plant Physiol. 2001;125:818–827. doi: 10.1104/pp.125.2.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello B.K., Hou Y., Zhao J., Jiao G., Wu Y., Li Z., Wang Y., Tong X., Wang W., Yuan W., et al. NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.) Plant Biotechnol. J. 2019;17:1222–1235. doi: 10.1111/pbi.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A., Zhang Y., Hendershot L.M., Harding H.P., Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- Deng Y., Humbert S., Liu J.X., Srivastava R., Rothstein S.J., Howell S.H. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2011;108:7247–7252. doi: 10.1073/pnas.1102117108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Wang J., Zhang Z., Wu Y. Transactivation of Sus1 and Sus2 by Opaque2 is an essential supplement to sucrose synthase-mediated endosperm filling in maize. Plant Biotechnol. J. 2020;18:1897–1907. doi: 10.1111/pbi.13349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu F.F., Xue H.W. Co-expression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 2010;154:927–938. doi: 10.1104/pp.110.159517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Wang Y., Liu X., Jiang L., Ren Y., Liu F., Peng C., Li J., Jin X., Wu F., et al. The failure to express a protein disulphide isomerase-like protein results in a floury endosperm and an endoplasmic reticulum stress response in rice. J. Exp. Bot. 2012;63:121–130. doi: 10.1093/jxb/err262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Takahashi H., Wakasa Y., Kawakatsu T., Takaiwa F. Identification of a cis-element that mediates multiple pathways of the endoplasmic reticulum stress response in rice. Plant J. 2013;74:248–257. doi: 10.1111/tpj.12117. [DOI] [PubMed] [Google Scholar]
- Hayashi S., Wakasa Y., Takahashi H., Kawakatsu T., Takaiwa F. Signal transduction by IRE1-mediated splicing of bZIP50 and other stress sensors in the endoplasmic reticulum stress response of rice. Plant J. 2012;69:946–956. doi: 10.1111/j.1365-313X.2011.04844.x. [DOI] [PubMed] [Google Scholar]
- Howell S.H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 2013;64:477–499. doi: 10.1146/annurev-arplant-050312-120053. [DOI] [PubMed] [Google Scholar]
- Iwata Y., Ashida M., Hasegawa C., Tabara K., Mishiba K.I., Koizumi N. Activation of the Arabidopsis membrane-bound transcription factor bZIP28 is mediated by site-2 protease, but not site-1 protease. Plant J. 2017;91:408–415. doi: 10.1111/tpj.13572. [DOI] [PubMed] [Google Scholar]
- Iwata Y., Koizumi N. An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. USA. 2005;102:5280–5285. doi: 10.1073/pnas.0408941102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.G., Park S., Matsuoka M., An G. White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB) Plant J. 2005;42:901–911. doi: 10.1111/j.1365-313X.2005.02423.x. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T., Yamamoto M.P., Touno S.M., Yasuda H., Takaiwa F. Compensation and interaction between RISBZ1 and RPBF during grain filling in rice. Plant J. 2009;59:908–920. doi: 10.1111/j.1365-313X.2009.03925.x. [DOI] [PubMed] [Google Scholar]
- Krishnan H.B., Okita T.W. Structural Relationship among the Rice Glutelin Polypeptides. Plant Physiol. 1986;81:748–753. doi: 10.1104/pp.81.3.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wu Y., Zhang D.Z., Gillikin J.W., Boston R.S., Franceschi V.R., Okita T.W. Rice prolamine protein body biogenesis: a BiP-mediated process. Science. 1993;262:1054–1056. doi: 10.1126/science.8235623. [DOI] [PubMed] [Google Scholar]
- Liu F., Ren Y., Wang Y., Peng C., Zhou K., Lv J., Guo X., Zhang X., Zhong M., Zhao S., et al. OsVPS9A functions cooperatively with OsRAB5Ato regulate post-Golgi dense vesicle-mediated storage protein trafficking to the protein storage vacuole in rice endosperm cells. Mol. Plant. 2013;6:1918–1932. doi: 10.1093/mp/sst081. [DOI] [PubMed] [Google Scholar]
- Liu J.X., Srivastava R., Che P., Howell S.H. An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell. 2007;19:4111–4119. doi: 10.1105/tpc.106.050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y. Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue. Plant Cell Physiol. 2002;43:718–725. doi: 10.1093/pcp/pcf091. [DOI] [PubMed] [Google Scholar]
- Nijhawan A., Jain M., Tyagi A.K., Khurana J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008;146:333–350. doi: 10.1104/pp.107.112821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi A., Nakamura Y., Tanaka N., Satoh H. Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol. 2001;127:459–472. [PMC free article] [PubMed] [Google Scholar]
- Pan T., Wang Y., Jing R., Wang Y., Wei Z., Zhang B., Lei C., Qi Y., Wang F., Bao X., et al. Post-Golgi trafficking of rice storage proteins requires the small GTPase Rab7 activation complex MON1-CCZ1. Plant Physiol. 2021;187:2174–2191. doi: 10.1093/plphys/kiab175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobre K.F.R., Poet G.J., Hendershot L.M. The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: getting by with a little help from ERdj friends. J. Biol. Chem. 2019;294:2098–2108. doi: 10.1074/jbc.REV118.002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Huang Z., Jiang H., Wang Z., Wu F., Xiong Y., Yao J. A heat stress responsive NAC transcription factor heterodimer plays key roles in rice grain filling. J. Exp. Bot. 2021;72:2947–2964. doi: 10.1093/jxb/erab027. [DOI] [PubMed] [Google Scholar]
- Ren Y., Wang Y., Liu F., Zhou K., Ding Y., Zhou F., Wang Y., Liu K., Gan L., Ma W., et al. GLUTELIN PRECURSOR ACCUMULATION3 encodes a regulator of post-Golgi vesicular traffic essential for vacuolar protein sorting in rice endosperm. Plant Cell. 2014;26:410–425. doi: 10.1105/tpc.113.121376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Wang Y., Pan T., Wang Y., Wang Y., Gan L., Wei Z., Wang F., Wu M., Jing R., et al. GPA5 encodes a Rab5a effector required for post-golgi trafficking of rice storage proteins. Plant Cell. 2020;32:758–777. doi: 10.1105/tpc.19.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Cruz M., Crofts A.J., Takemoto-Kuno Y., Sugino A., Washida H., Crofts N., Okita T.W., Ogawa M., Satoh H., Kumamaru T. Protein disulfide isomerase like 1-1 participates in the maturation of proglutelin within the endoplasmic reticulum in rice endosperm. Plant Cell Physiol. 2010;51:1581–1593. doi: 10.1093/pcp/pcq098. [DOI] [PubMed] [Google Scholar]
- Sun S., Tang X., Guo Y., Hu J. Endoplasmic reticulum composition and form: proteins in and out. Curr. Opin. Cell Biol. 2021;71:1–6. doi: 10.1016/j.ceb.2021.01.008. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Kawakatsu T., Wakasa Y., Hayashi S., Takaiwa F. A rice transmembrane bZIP transcription factor, OsbZIP39, regulates the endoplasmic reticulum stress response. Plant Cell Physiol. 2012;53:144–153. doi: 10.1093/pcp/pcr157. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Saito Y., Kitagawa T., Morita S., Masumura T., Tanaka K. A novel vesicle derived directly from endoplasmic reticulum is involved in the transport of vacuolar storage proteins in rice endosperm. Plant Cell Physiol. 2005;46:245–249. doi: 10.1093/pcp/pci019. [DOI] [PubMed] [Google Scholar]
- Takemoto Y., Coughlan S.J., Okita T.W., Satoh H., Ogawa M., Kumamaru T. The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol. 2002;128:1212–1222. doi: 10.1104/pp.010624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z., Qian Q., Liu Q., Yan M., Liu X., Yan C., Liu G., Gao Z., Tang S., Zeng D., et al. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Acad. Sci. USA. 2009;106:21760–21765. doi: 10.1073/pnas.0912396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto T., Aoki N., Lin H., Nakamura Y., Inouchi N., Sato Y., Yano M., Hirabayashi H., Maruyama S. Natural variation in rice starch synthase IIa affects enzyme and starch properties. Funct. Plant Biol. 2004;31:671–684. doi: 10.1071/FP04009. [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa J., Moose S.P., Parsons R.L., Schmidt R.J. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc. Natl. Acad. Sci. USA. 1997;94:7685–7690. doi: 10.1073/pnas.94.14.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasa Y., Yasuda H., Oono Y., Kawakatsu T., Hirose S., Takahashi H., Hayashi S., Yang L., Takaiwa F. Expression of ER quality control-related genes in response to changes in BiP1 levels in developing rice endosperm. Plant J. 2011;65:675–689. doi: 10.1111/j.1365-313X.2010.04453.x. [DOI] [PubMed] [Google Scholar]
- Wang J., Chen Z., Zhang Q., Meng S., Wei C. The NAC transcription factors OsNAC20 and OsNAC26 regulate starch and storage protein synthesis. Plant Physiol. 2020;184:1775–1791. doi: 10.1104/pp.20.00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.C., Xu H., Zhu Y., Liu Q.Q., Cai X.L. OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot. 2013;64:3453–3466. doi: 10.1093/jxb/ert187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu F., Ren Y., Wang Y., Liu X., Long W., Wang D., Zhu J., Zhu X., Jing R., et al. GOLGI TRANSPORT 1B regulates protein export from the endoplasmic reticulum in rice endosperm cells. Plant Cell. 2016;28:2850–2865. doi: 10.1105/tpc.16.00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ren Y., Liu X., Jiang L., Chen L., Han X., Jin M., Liu S., Liu F., Lv J., et al. OsRab5a regulates endomembrane organization and storage protein trafficking in rice endosperm cells. Plant J. 2010;64:812–824. doi: 10.1111/j.1365-313X.2010.04370.x. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhu S., Liu S., Jiang L., Chen L., Ren Y., Han X., Liu F., Ji S., Liu X., et al. The vacuolar processing enzyme OsVPE1 is required for efficient glutelin processing in rice. Plant J. 2009;58:606–617. doi: 10.1111/j.1365-313X.2009.03801.x. [DOI] [PubMed] [Google Scholar]
- Washida H., Sugino A., Doroshenk K.A., Satoh-Cruz M., Nagamine A., Katsube-Tanaka T., Ogawa M., Kumamaru T., Satoh H., Okita T.W. RNA targeting to a specific ER sub-domain is required for efficient transport and packaging of α-globulins to the protein storage vacuole in developing rice endosperm. Plant J. 2012;70:471–479. doi: 10.1111/j.1365-313X.2011.04880.x. [DOI] [PubMed] [Google Scholar]
- Wei X., Jiao G., Lin H., Sheng Z., Shao G., Xie L., Tang S., Xu Q., Hu P. GRAIN INCOMPLETE FILLING 2 regulates grain filling and starch synthesis during rice caryopsis development. J. Integr. Plant Biol. 2017;59:134–153. doi: 10.1111/jipb.12510. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Ren Y., Li W., Wu F., Yang W., Huang X., Yao J. NF-YC12 is a key multi-functional regulator of accumulation of grain storage substances in rice. J. Exp. Bot. 2019;70:3765–3780. doi: 10.1093/jxb/erz168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.J., Zhang X.F., Xue H.W. Rice aleurone layer specific OsNF-YB1 regulates grain filling and endosperm development by interacting with an ERF transcription factor. J. Exp. Bot. 2016;67:6399–6411. doi: 10.1093/jxb/erw409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H., Sugimoto T., Tanaka K., Kasai Z. Biosynthesis of storage proteins in developing rice grains. Plant Physiol. 1982;70:1094–1100. doi: 10.1104/pp.70.4.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Xu P., Zhang J., Zhang S., Li Z., Yang K., Chang X., Li Y. OsbZIP60-mediated unfolded protein response regulates grain chalkiness in rice. Journal of Genetics and Genomics. 2022;49:414–426. doi: 10.1016/j.jgg.2022.02.002. [DOI] [PubMed] [Google Scholar]
- Yang Y., Guo M., Li R., Shen L., Wang W., Liu M., Zhu Q., Hu Z., He Q., Xue Y., et al. Identification of quantitative trait loci responsible for rice grain protein content using chromosome segment substitution lines and fine mapping of qPC-1 in rice (Oryza sativa L.) Mol. Breeding. 2015;35:130. [Google Scholar]
- Yasuda H., Hirose S., Kawakatsu T., Wakasa Y., Takaiwa F. Overexpression of BiP has inhibitory effects on the accumulation of grain storage proteins in endosperm cells of rice. Plant Cell Physiol. 2009;50:1532–1543. doi: 10.1093/pcp/pcp098. [DOI] [PubMed] [Google Scholar]
- Zhang C., Zhu J., Chen S., Fan X., Li Q., Lu Y., Wang M., Yu H., Yi C., Tang S., et al. Wx-lv, the ancestral allele of rice waxy gene. Mol. Plant. 2019;12:1157–1166. doi: 10.1016/j.molp.2019.05.011. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Dong J., Ji C., Wu Y., Messing J. NAC-type transcription factors regulate accumulation of starch and protein in maize grains. Proc. Natl. Acad. Sci. USA. 2019;116:11223–11228. doi: 10.1073/pnas.1904995116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Guo L., Ji C., Wang H., Wang J., Zheng X., Xiao Q., Wu Y. The B3 domain-containing transcription factor ZmABI19 coordinates expression of key factors required for maize grain development and grain filling. Plant Cell. 2021;33:104–128. doi: 10.1093/plcell/koaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zheng X., Yang J., Messing J., Wu Y. Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis. Proc. Natl. Acad. Sci. USA. 2016;113:10842–10847. doi: 10.1073/pnas.1613721113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Ren Y., Wang Y., Liu F., Teng X., Zhang Y., Duan E., Wu M., Zhong M., Hao Y., et al. OsNHX5-mediated pH homeostasis is required for post-Golgi trafficking of grain storage proteins in rice endosperm cells. BMC Plant Biol. 2019;19:295. doi: 10.1186/s12870-019-1911-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Ren Y., Zhang Y., et al. Subunit E isoform 1 of vacuolar H+-ATPase OsVHA enables post-Golgi trafficking of rice seed storage proteins. Plant Physiol. 2021;187:2192–2208. doi: 10.1093/plphys/kiab099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data 1. DEGs between ZH11 and the o3 mutant