Abstract
Wood is an abundant and renewable feedstock for the production of pulp, fuels, and biobased materials. However, wood is recalcitrant toward deconstruction into cellulose and simple sugars, mainly because of the presence of lignin, an aromatic polymer that shields cell-wall polysaccharides. Hence, numerous research efforts have focused on engineering lignin amount and composition to improve wood processability. Here, we focus on results that have been obtained by engineering the lignin biosynthesis and branching pathways in forest trees to reduce cell-wall recalcitrance, including the introduction of exotic lignin monomers. In addition, we draw general conclusions from over 20 years of field trial research with trees engineered to produce less or altered lignin. We discuss possible causes and solutions for the yield penalty that is often associated with lignin engineering in trees. Finally, we discuss how conventional and new breeding strategies can be combined to develop elite clones with desired lignin properties. We conclude this review with priorities for the development of commercially relevant lignin-engineered trees.
Key words: lignin, genetic engineering, breeding, CRISPR, field trial, forest trees
Lignin causes lignocellulosic biomass to be recalcitrant to enzymatic hydrolysis. This review overviews the results obtained by engineering lignin amount and composition in plantation forest trees and draws conclusions from comparisons between greenhouse-grown and field-grown trees. It further discusses possible causes and solutions for the yield penalty that is often associated with lignin modification and provide perspectives for the development of commercially relevant lignin-engineered trees.
Introduction
Fossil resources are currently the main feedstock for energy and organic compound synthesis. However, considering the high carbon footprint of the petrochemical industry, the development of renewable and carbon-neutral feedstocks is a critical societal challenge (Vanholme et al., 2013b; Galán-Martín et al., 2021; Huang et al., 2021; Yang et al., 2021). Lignocellulosic biomass is the most abundant renewable carbon source on earth and therefore a promising candidate. Wood is an important source of lignocellulosic biomass. It is mainly composed of secondary cell walls that are primarily made from cellulose, hemicellulose, and lignin. Cellulose can be used in the pulp and paper industry or can, together with hemicellulose, be hydrolyzed into monosaccharides that can be further converted into high-value compounds such as liquid biofuels, building blocks for bioplastics, cosmetics, etc. (Bozell et al., 2007; Corma et al., 2007; Vanholme et al., 2013b; Isikgor and Becer, 2015; de Vries et al., 2021b). The additional valorization of lignin has recently been recognized as essential to enable the economic viability of lignocellulosic biorefining (Liao et al., 2020; Abu-Omar et al., 2021).
Lignin in wood of forest trees is predominantly built from monolignols that are biosynthesized by a series of enzymatic conversions starting from the amino acid phenylalanine (Figure 1) (Freudenberg, 1959; Boerjan et al., 2003; Vanholme et al., 2019). After their biosynthesis in the cytosol, monolignols are translocated to the cell wall (Pesquet et al., 2010, 2013; Derbyshire et al., 2015; Serk et al., 2015; Vermaas et al., 2019; Blaschek et al., 2020; Perkins et al., 2022). Notably, monolignols are translocated not only to the cell walls of the cells that produced them but also to those of neighboring cells (Hosokawa et al., 2001; Tokunaga et al., 2005; Pesquet et al., 2013; Smith et al., 2013, 2017; De Meester et al., 2018, 2021). In the cell wall, monolignols are oxidized by peroxidases (PERs) and/or laccases (LACs) and subsequently polymerized into the lignin polymer through radical coupling, thereby generating a range of chemical bonds such as aryl ether (β-O-4), resinol (β-β), phenylcoumaran (β-5), biphenyl ether (5-O-4), spirodienone (β-1), and dibenzodioxocin (5-5/4-O-β) (Berthet et al., 2011, 2012; Lu et al., 2013; Ralph et al., 2019; Tobimatsu and Schuetz, 2019). Lignin amount and composition vary greatly within and among plant species, tissues, cell types, and cell-wall layers, and are influenced by developmental and environmental factors (Vanholme et al., 2019). Hardwood (angiosperm) lignin is mainly built from the monolignols coniferyl, sinapyl, and (traces of) p-coumaryl alcohol, producing guaiacyl (G), syringyl (S), and p-hydroxyphenyl (H) units, respectively. By contrast, softwood (gymnosperm) lignin is composed mostly of G units with a minor fraction of H units. Depending on the plant species, other (less traditional) phenolic metabolites also act as monomers for lignification (Figure 1) (Ralph et al., 2019; Vanholme et al., 2019; del Río et al., 2022). For instance, poplar lignin typically incorporates coniferyl acetate, sinapyl acetate, and sinapyl p-hydroxybenzoate, as well as traces of coniferyl p-hydroxybenzoate, sinapyl benzoate and coniferyl ferulate (Figure 1) (Morreel et al., 2004a, 2004b; Lu et al., 2004; Karlen et al., 2016; Zhao et al., 2021b; de Vries et al., 2022).
Figure 1.
Metabolic pathways leading to lignin monomers.
Only the route toward lignin monomers is shown; side branches of the pathway that result in soluble metabolites are not shown, with the exception of ferulic acid conjugates (see footnote2). As enzymes may have different substrates and pathway perturbation may induce branching pathways, the figure includes only metabolic conversions for which strong evidence exists of involvement in biosynthesis of the shown end products. A question mark “?” means that the enzymes catalyzing the metabolic conversion(s) are not known. C3H, p-COUMARATE 3-HYDROXYLASE; CHI, CHALCONE ISOMERASE; other abbreviations can be found in the manuscript text. 1The exact structures of the 4-dihydroxybenzoate conjugates are not known. 2Only a small portion of the ferulic acid pool is integrated into the lignin polymer; the majority of the pool is metabolized to soluble ferulic acid conjugates. 3The conversion of coniferin to coniferyl alcohol (which subsequently serves as a monomer for lignification) and glucose occurs in the apoplast of gymnosperm trees. Studies in Ginkgo biloba, Pinus contorta, Picea abies (Norway spruce), and Chamaecyparis obtusa (Japanese cypress) indicate that coniferin is a transport form of coniferyl alcohol in these species (Samuels et al., 2002; Tsuyama et al., 2013; Aoki et al., 2016; Väisänen et al., 2020).
Currently, narrow profit margins limit the economic feasibility of employing fast-growing woody feedstocks, such as poplar, willow, and eucalyptus, as dedicated energy and specialty chemical crops at an industrial scale (Mahon and Mansfield, 2019). Because lignin is the main contributor to the recalcitrance of lignocellulosic biomass toward deconstruction, numerous research efforts have focused on altering lignin amount and/or composition, aiming to increase wood processing efficiency (Chanoca et al., 2019; Bryant et al., 2020). However, the in planta effects of pathway perturbations are often unpredictable because the flux through the lignin biosynthesis pathway is regulated at multiple levels (Figure 2): (i) at the transcriptional level (Zhong et al., 2010; Bonawitz et al., 2014; Ohtani and Demura, 2019); (ii) by post-translational protein modifications (Wang et al., 2015); (iii) by the abundance of enzyme-inhibiting pathway intermediates (Wang et al., 2014; Eudes et al., 2016; Yokoyama et al., 2021); (iv) by the compartmentalization of substrates (Widhalm et al., 2015; Eudes et al., 2016; Guo et al., 2018; Vanholme et al., 2019); (v) by the different activities and spatial expression patterns of gene family members (Li et al., 2015; Vanholme et al., 2019); (vi) by the formation of protein complexes (Chen et al., 2011; Widhalm et al., 2015; Gou et al., 2018; Yan et al., 2019); (vii) by the availability of co-factors and -substrates (Tang et al., 2014; Vanholme et al., 2019; Hu et al., 2022); (viii) by pathway intermediate-triggered proteolysis of pathway enzymes (Guan et al., 2022); (ix) by branching pathways diverting the flux away from monolignols toward other sinks and products, such as flavonoids, benzenoids, phenylpropanoids, and monomer-glucosides (Vanholme et al. (2012); and (x) by the different parallel routes through which the pathway flux can flow via enzymes that often accept multiple substrates (Vanholme et al., 2019; Tsai et al., 2020). In addition, the outcome of the same pathway perturbation(s) in different species can differ significantly (Vanholme et al., 2019); e.g., CAFFEOYL SHIKIMATE ESTERASE (CSE)-knockout Arabidopsis, Medicago, and poplar display huge differences in the frequency of H units incorporated into their lignin and the severity of the associated yield penalty (Vanholme et al., 2013c; Ha et al., 2016; de Vries et al., 2021a).
Figure 2.
Schematic overview of multiple levels of lignin pathway regulation in trees.
Arrows with filled arrowheads represent metabolic conversions, and arrows with hollow arrowheads represent translocation, protein modification, or signaling. Pac-man-like spheres represent enzymes, and polygons represent metabolites.
For poplar, a mathematical model was generated that estimates how changing the expression of pathway genes affects protein abundance, metabolite concentrations, metabolic flux, and various wood traits such as lignin amount and composition, biomass yield, and saccharification efficiency (Wang et al., 2018). Even so, this model also has its limitations; to generate the model, the analysis was restricted to one specific developmental stage, environmental condition, and poplar accession. Moreover, some factors that affect flux through the pathway, such as the formation of C3′H-C4H protein complexes, spatial distribution of substrates and enzymes (all xylary cell types were pooled to generate the model), and further metabolization of compounds (e.g., glycosylation), were not (yet) taken into account. It will therefore be crucial to assess the value of a lignin-engineering strategy and evaluate it under relevant forestry practices in economically relevant biomass germplasm such as (elite) poplar or eucalyptus clones.
In this review, we give an overview of the results obtained by engineering the levels of endogenous and exotic lignin building blocks in forest trees. In addition, based on data derived from greenhouse- and field-grown lignin-engineered trees, we discuss the link between lignin reduction and growth phenotypes on the one hand and between lignin reduction and the age of the tree and the environment in which it was grown on the other hand. We explore the possible causes of the frequently observed yield penalty in lignin-engineered trees and discuss strategies to avoid this yield penalty. We also address breeding strategies for the modification of lignin in forest trees, including traditional breeding with genomic selection and modern breeding techniques such as transgene expression and gene editing. We conclude this article with perspectives on the development and use of commercially relevant lignin-engineered trees.
Altering lignin amount and composition via pathway engineering
For industrial purposes aimed at the use of polysaccharides from wood (e.g., for pulping or for fermentable sugar-based biorefinery), a lower amount of lignin or lignin that is more easily extracted is a desired trait. A reduction in lignin amount has typically been observed in trees with reduced activity of PHENYLALANINE AMMONIA LYASE (PAL), CINNAMATE 4-HYDROXYLASE (C4H), 4-COUMARATE:CoA LIGASE (4CL), p-COUMAROYLSHIKIMATE 3′-HYDROXYLASE (C3′H), p-HYDROXYCINNAMOYL-CoA:SHIKIMATE p-HYDROXYCINNAMOYLTRANSFERASE (HCT), CAFFEOYL-CoA O-METHYLTRANSFERASE (CCoAOMT), and CINNAMOYL-CoA REDUCTASE (CCR) (Wang et al., 2018; reviewed in Chanoca et al., 2019). In these trees, lignin composition is often also affected. For instance, HCT- or C3′H-deficient trees deposit lignin with relatively large increases in the frequency of H units (Coleman et al., 2008; Vanholme et al., 2013a). Downregulation of CAFFEIC ACID O-METHYLTRANSFERASE (COMT) or CINNAMYL ALCOHOL DEHYDROGENASE (CAD) and overexpression of FERULATE 5-HYDROXYLASE (F5H) generally have less severe effects on lignin amount and mostly affect lignin composition. Reducing the efficiency of COMT and CAD boosts the incorporation of monomers that are typically present in trace amounts or below the detection limit in lignin of wild-type (WT) trees: 5-hydroxyconiferyl alcohol and 5-hydroxyconiferaldehyde are new units in COMT-deficient trees, whereas hydroxycinnamaldehydes are incorporated in higher amounts in CAD-deficient trees (Van Doorsselaere et al., 1995; Baucher et al., 1996; Lapierre et al., 1999; Jouanin et al., 2000; Morreel et al., 2004a, 2004b; Van Acker et al., 2014; Yamamoto et al., 2020). The increased incorporation of these monomers into lignin is potentially desirable from an applied perspective because 5-hydroxyconiferyl alcohol gives rise to benzodioxane units that are anticipated to prevent covalent linkages between lignin and polysaccharide hydroxyl groups (Ralph et al., 2004), whereas incorporation of hydroxycinnamaldehydes gives rise to lignin that is more cleavable and soluble in alkaline conditions (Lapierre et al., 2004; Van Acker et al., 2017). Overexpression of F5H results in an increased fraction of S units (Franke et al., 2000; Stewart et al., 2009). Lignin with a high S/G ratio is interesting for both the pulp and paper industry and the lignin-first biorefinery because high-S lignin has a high fraction of beta-aryl ether bonds that are cleaved during alkaline pulping and via catalytic reductive fractionation, respectively (Baucher et al., 2003; Mansfield et al., 2012; Van den Bosch et al., 2015). For a thorough overview of lignin-engineering results achieved up to mid-2019 and from which the above-mentioned conclusions were drawn, we refer to Wang et al. (2018) and Chanoca et al. (2019) (Figure 1). Below, we focus on research published since then (Table 1; Figure 1).
Table 1.
Overview of greenhouse-grown trees with modified expression of lignin biosynthesis genes published since mid-2019.
| Species | Gene | Method | Effect on total lignin | Effect on lignin composition | Saccharification efficiency | Pulping efficiency | Biomass yield | Reference |
|---|---|---|---|---|---|---|---|---|
| P. trichocarpa | C4H1 | suppression via RNAi | ↓13% | ↓S/G | ↑ | n.d. | ↓ | Kim et al. (2020) |
| P. trichocarpa | C3H3 | suppression via RNAi | ↓42% | ↑S/G | ↑ | n.d. | ↓ | Kim et al. (2020) |
| P. trichocarpa | C4H1/C4H2/C3H3 | suppression via RNAi | ↓50% | ↑S/G | ↑ | n.d. | ↓ | Kim et al. (2020) |
| Populus deltoides × Populus euramericana | 4CL1 | fiber-specific suppression | ↓21% | n.d. | n.d. | n.d. | WT | Cao et al. (2020) |
| P. deltoides × P. euramericana | 4CL1 | vessel-specific suppression | ↓26% | n.d. | n.d. | n.d. | WT | Cao et al. (2020) |
| P. tremula × P. alba | 4CL1 | CRISPR-Cas9 | ↓19% | ↓S/G | WT | n.d. | WT | Tsai et al. (2020) |
| P. tremula × P. alba | 4CL5 | CRISPR-Cas9 | WT | WT | n.d. | n.d. | WT | Tsai et al. (2020) |
| P. tremula × P. alba | CSE1 | CRISPR-Cas9 | WT | n.d. | n.d. | n.d. | WT | de Vries et al. (2021a) |
| P. tremula × P. alba | CSE2 | CRISPR-Cas9 | WT | n.d. | n.d. | n.d. | WT | de Vries et al. (2021a) |
| P. tremula × P. alba | CSE1 and CSE2 | CRISPR-Cas9 | ↓35% | ↓S/G | ↑ | n.d. | ↓ | de Vries et al. (2021a) |
| P. alba × P. glandulosa | CSE1 | CRISPR-Cas9 | ↓16% | n.d. | n.d. | n.d. | WT | Jang et al. (2021) |
| P. alba × P. glandulosa | CSE2 | CRISPR-Cas9 | ↓16% | n.d. | n.d. | n.d. | WT | Jang et al. (2021) |
| P. alba × P. glandulosa | HCT | suppression via RNAi | ↓11% | ↓S/G | n.d. | n.d. | n.a. | Su et al. (2021) |
| P. tremula × P. alba | CCR2 | CRISPR-Cas9 | ↓10% | WT S/G, incorporation of ferulic acid | ↑ | n.d. | WT | De Meester et al. (2020) |
| P. tremula × P. alba | CCR2 | CRISPR-Cas9 knockout + vessel- and ray-specific overexpression of AtCCR1 | ↓18% | WT S/G, incorporation of ferulic acid | ↑ | n.d. | ↓ | De Meester et al. (2021) |
| P. trichocarpa | CCR2 | suppression via RNAi | ↓32% | ↓S/G, incorporation of ferulic acid | n.d. | n.d. | ↓ | Yan et al., 2019 |
| P. tomentosa | mF5H2a | overexpression | n.d. | ↑S/G | ↑ | n.d. | n.d. | Fan et al. (2020) |
| P. tremula × P. alba | AtF5H1 | overexpression | WT | ↑S/G | ↑ | n.d. | WT | Lapierre et al. (2021) |
| P. trichocarpa | CAD1 | suppression via RNAi | ↓9% | ↑S/G, incorporation of aldehyde components | n.d. | n.d. | WT | Yan et al., 2019 |
| P. deltoides × P. euramericana | LTF1 | fiber-specific suppression | ↓43% | ↓S/G | n.d. | n.d. | WT | Gui et al. (2020) |
| P. deltoides × P. euramericana | LTF1 | vessel-specific suppression | ↓16% | ↑S/G | n.d. | n.d. | ↓ | Gui et al. (2020) |
| P. tremula × P. alba | ClDCS and ClCURS2 | overexpression | ↑23% | ↓S/G | WT | n.d. | ↓ | De Meester et al. (2022a) |
| P. alba × Populus grandidentata | MdCHS3 | overexpression | ↓10% | WT S/G | ↑ | n.d. | WT | Mahon et al. (2022) |
| P. tremula × P. alba | PHBMT1 | CRISPR-Cas9 | WT | WT S/G, no detectable p-hydroxybenzoates | n.d. | n.d. | WTb | Zhao et al. (2021a); Zhao et al. (2021b) |
| P. tremula × P. alba | PHBMT1 | overexpression | WT | WT S/G, increased p-hydroxybenzoates | n.d. | n.d. | WT | Zhao et al. (2021a); Zhao et al. (2021b) |
| P. alba × P. grandidentata | PHBMT1 | overexpression | WT | WT S/G, increased p-hydroxybenzoates | n.d. | n.d. | WT | de Vries et al. (2022) |
| P. alba × P. grandidentata | CPL | overexpression | ↓5% | ↓S/G, increased p-hydroxybenzoates | ↑ | n.d. | ↓ | Mottiar et al. (2022) |
| P. alba × P. glandulosa | miR393 | suppression via STTM | slightly ↑ | n.d. | n.d. | n.d. | ↑ | Chu et al. (2021) |
| P. tremula × Populus tremuloides | HpSK | overexpression | WT | ↑S/G, increased H units | ↑ | n.d. | ↓ | Hu et al. (2022) |
| P. alba × P. grandidentata | QsuB | overexpression | ↓33% | ↑S/G | ↑ | n.d. | WT | Unda et al. (2022) |
| P. tremula × P. alba | BdPMT1 | overexpression | WT | WT S/G, incorporation of p-coumarates | ↑ | n.d. | WT | Lapierre et al. (2021) |
| P. tremula × P. alba | AtF5H1 and BdPMT1 | overexpression | WT | ↑S/G, incorporation of p-coumarates | ↑ | n.d. | WT | Lapierre et al. (2021) |
| P. trichocarpa | HSFB3-1 | CRISPR-Cas9 | ↓17% | n.d. | n.d. | n.d. | ↑ | Liu et al. (2021a) |
| P. trichocarpa | MYB092 | CRISPR-Cas9 | ↓27% | n.d. | n.d. | n.d. | WT | Liu et al. (2021a) |
| P. trichocarpa | HSFB3-1 | overexpression | ↑10% | n.d. | n.d. | n.d. | ↓ | Liu et al. (2021a) |
| P. trichocarpa | MYB092 | overexpression | ↑18% | n.d. | n.d. | n.d. | ↓ | Liu et al. (2021a) |
| P. tomentosa | miR828 | overexpression | ↓13% | n.d. | n.d. | n.d. | ↓ | Wang et al. (2022) |
| P. tomentosa | miR828 | suppression via STTM | ↑15% | n.d. | n.d. | n.d. | ↑ | Wang et al. (2022) |
| P. tomentosa | MYB171 | overexpression | ↑12% | n.d. | n.d. | n.d. | ↑ | Wang et al. (2022) |
| P. tomentosa | miR6443 | overexpression | WT | ↓S/G | ↓ | n.d. | WT | Fan et al. (2020) |
| P. tomentosa | miR6443 | suppression via STTM | WT | ↑S/G | ↑ | n.d. | WT, lodging phenotype | Fan et al. (2020) |
| Populus alba × P. glandulosa | MYB120 | dominant suppression via SDRX | ↓59% | ↓S/G | n.d. | n.d. | ↓ | Kim et al. (2021) |
| P. deltoides | EPSPSc | overexpression | ectopic lignificationd | n.d. | n.d. | n.d. | n.d. | Xie et al. (2018) |
| P. deltoides | EPSPSc | suppression via RNAi | n.d. | n.d. | n.d. | n.d. | n.d. | Xie et al. (2018) |
| P. tomentosa | LAC14 | overexpression | ↑15% | ↓S/G | n.d. | n.d. | ↓ | Qin et al. (2020) |
| P. tomentosa | LAC14 | CRISPR-Cas9 | ↓7% | ↑S/G | ↑ | n.d. | ↑ | Qin et al. (2020) |
For an overview of lignin-engineering results in greenhouse-grown trees achieved up to mid-2019, see Wang et al. (2018) and Chanoca et al. (2019). Since mid-2019, only lignin-engineering results from greenhouse-grown trees of Populus species have been published. n.d., not determined; S/G, syringyl/guaiacyl ratio; effect on total lignin is given as a percentage of the total lignin analyzed via different methods; see the corresponding reference for specific information. If multiple lines were tested, only the line with the most extreme change in lignin is shown. For abbreviations of gene names, see the manuscript text. RNAi, RNA interference; STTM, short tandem target mimic; SRDX refers to the protein domain LDLELRL; CRISPR, clustered regularly interspaced short palindromic repeats; At, Arabidopsis thaliana; Md, Malus domestica; Bd, Brachypodium distachyon; Cl, Curcuma longa.
mF5H2 is an engineered miR6443-resistant form of F5H2.
phbmt1 mutants showed a decrease in biomass until 2 months, when the plantlets were transferred from tissue culture to soil. When they grew for 3 or more months in soil, their growth appeared normal, although some mutant lines had a twisted trunk.
EPSPS is named EPSP in Xie et al. (2018) and EPSP-TF in Xie et al. (2020).
Overexpression under control of the CaMV35S promoter resulted in the ectopic deposition of lignin in cell walls of epidermis, phloem fiber, and pith cells as judged from microscopy sections stained with phloroglucinol-HCl. Lignin levels in the xylem were not quantified.
CSE is a relatively newly discovered enzyme of the lignin biosynthetic pathway (Vanholme et al., 2013c; Ha et al., 2016; Saleme et al., 2017; Wang et al., 2021). Two independent studies investigated CRISPR-Cas9-induced mutations of CSE1 and CSE2 in poplar. One study found lignin and biomass yield reductions in cse1 cse2 double mutants in Populus tremula × Populus alba cv 717-1B4 but no effects on lignin and growth in the respective single mutants (de Vries et al., 2021a). By contrast, the other study found that both cse1 and cse2 single mutants in P. alba × P. tremula var. glandulosa cv 84K showed up to 16% reduced lignin amount without biomass reduction (Jang et al., 2021). These different findings hint that the growth conditions or genetic background may interact with the cse1 and cse2 mutations, even though the two poplar genotypes used are interspecific hybrids of the same species.
Another recently discovered enzyme, p-HYDROXYBENZOYL-CoA MONOLIGNOL TRANSFERASE (PHBMT1), catalyzes the last step in the biosynthesis of coniferyl and sinapyl p-hydroxybenzoates in poplar (Zhao et al., 2021b; de Vries et al., 2022). Coniferyl and sinapyl p-hydroxybenzoates make up about 0.01% and 2.4%–6.5% of the lignin monomers in WT poplar, respectively, and might function in the regulation of the poplar gravitropic response (Regner et al., 2018; Kim et al., 2020; Zhao et al., 2021a). CRISPR-Cas9-induced knockout mutations in PHBMT1 in poplar result in lignin with no detectable p-hydroxybenzoate decorations, whereas overexpression results in an increase in p-hydroxybenzoates, all without affecting biomass yield (Zhao et al., 2021b; de Vries et al., 2022). Lignin of phbmt1 knockout poplar lines has a faster dissolution rate in acetyl bromide, whereas lignin dissolution of PHBMT1 overexpression lines is slower than that of the WT (Zhao et al., 2021b). Also, the introduction of a bacterial CHORISMATE PYRUVATE LYASE (CPL) gene into poplar results in increased amounts of cell-wall-bound p-hydroxybenzoates. These plants have a reduction of up to 6% in lignin amount and a small yield penalty (Mottiar et al., 2022). The increase in p-hydroxybenzoate decorations on lignin is a potentially interesting trait, as saponification results in increased release of p-hydroxybenzoic acid that can be used in cosmetics, in polyester plastics, and as a precursor to a wide range of pharmaceutical compounds (de Vries et al., 2022).
In addition to genes involved in monolignol biosynthesis, genes involved in the polymerization of monolignols in the cell wall are also interesting targets for lignin pathway engineering. For example, overexpression of LAC14—which encodes a laccase enzyme with a high oxidizing efficiency toward coniferyl alcohol—in poplar results in 15% more lignin, a relative increase in G units, and decreased biomass yield compared with the WT (Qin et al., 2020). On the other hand, CRISPR-Cas9-induced mutation of LAC14 in poplar results in lines with about 7% less lignin, a higher S/G ratio, and increased biomass yield (Qin et al., 2020).
Besides core genes of the lignin biosynthetic pathway, metabolic flux through the pathway can also be engineered via the depletion of essential co-factors. The expression of a bacterial SHIKIMATE KINASE (SK) gene in poplar results in a reduced shikimate pool in the cytoplasm, thereby hindering the HCT reaction for which shikimate is an essential substrate (Hu et al., 2022). The resulting transgenic poplars have a lignin content comparable to that of WT plants, but an increased amount of H units, a reduction in ether-linked G and S units, and a biomass yield penalty (Hu et al., 2022). The expression of another bacterial gene, QsuB, which encodes a 3-dehydroshikimate dehydratase, in poplar enables the plants to convert 3-dehydroshikimic acid into 3,4-dihydroxybenzoic acid (Unda et al., 2022). Transgenic poplars expressing QsuB partly divert the carbon flux away from the production of shikimate. These lines have up to 33% less lignin, incorporate ester-linked 3,4-dihydroxybenzoates in the lignin backbone, and have no biomass penalty.
Another strategy to divert carbon flux away from lignin biosynthesis has been achieved by expressing MONOLIGNOL 4-O-METHYLTRANSFERASE 4 (MOMT4) in poplar (Cai et al., 2016). MOMT4 is a methyltransferase engineered to methylate the para-hydroxylic function of lignin monomeric precursors, preventing the derived methylated products from participating in the lignin coupling process. Compared with the WT, MOMT4-expressing lines display a 13% reduction in lignin content, a 75% reduction in S/G ratio, and a 50% reduction in p-hydroxybenzoates (predominantly acylating S units) in the cell wall, without affecting growth and biomass yield.
Another strategy for steering metabolic flux in the lignin biosynthesis pathway is via transcriptional regulation. 5-ENOLPYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE (EPSPS) is a key enzyme in the shikimate biosynthetic pathway and has recently been identified as a transcriptional regulator of the phenylpropanoid pathway in poplar (Xie et al., 2018, 2020; Yao et al., 2021). The transcriptional regulatory functionality of EPSPS is defined by a single-nucleotide polymorphism (SNP), and natural poplar variants carrying this SNP have reduced lignin content (the amount of lignin reduction and the biomass yield were not reported) (Xie et al., 2018, 2020). Another example in which an allelic form of a transcription factor results in a reduced lignin content is the expression of a phosphorylation-null mutant of LIGNIN BIOSYNTHESIS-ASSOCIATED TRANSCRIPTION FACTOR 1 (LTF1) in poplar (Gui et al., 2019, 2020). LTF1 is a MYB transcription factor that functions as a transcriptional repressor of lignin biosynthesis in its unphosphorylated state. When LTF1 becomes phosphorylated in response to external stimuli, it undergoes degradation via the proteasome, resulting in the activation of lignification. Transgenic lines with constitutive expression of the LTF1 phosphorylation-null mutant (LTF1AA) display a decrease of up to 56% in lignin content but also a decrease in plant height (Gui et al., 2019, 2020), whereas expression of LTF1AA under the control of a fiber-specific promoter results in a decrease of up to 43% in lignin content and a plant height similar to that of controls (Gui et al., 2020). MYB120 is a transcription factor involved in the activation of lignin and anthocyanin biosynthesis. Transgenic poplars with dominant suppression of PtrMYB120 showed up to 59% less lignin, reduced anthocyanin levels, and a lower biomass yield (Kim et al., 2021).
Transcript levels can be post-transcriptionally regulated via microRNAs. MicroRNA393 (miR393) is a conserved miRNA family in plants that can regulate root growth, plant architecture, leaf development, and stress resistance (Chu et al., 2021). Poplar lines in which the miR393 family was blocked via short tandem target mimic (STTM) show an increased growth compared with WT trees, as well as a higher expression of lignin biosynthetic genes and a higher stem lignin content. How miR393 regulates lignin biosynthetic genes remains unknown (Chu et al., 2021). Overexpression of another microRNA, miR828, in poplar results in a reduced lignin content (−13%) in wood and a decreased biomass yield (Wang et al., 2022). Conversely, suppression of miR828 in poplar via STTM increases lignin content (+15%) and biomass yield (Wang et al., 2022). In addition, overexpression of MYB171, one of the direct targets of miR828, results in higher lignin amounts (+12%) and taller trees (Wang et al., 2022). Overexpression of miR6443, a microRNA that post-transcriptionally regulates F5H2, in poplar results in decreased transcript levels of F5H2 and consequently a 36% reduction in S units together with normal growth (Fan et al., 2020). On the other hand, blocking miR6443 via STTM results in poplar with increased F5H2 transcript levels, lignin with a 122% increase in S units, and a lodging phenotype (Fan et al., 2020).
Of special interest from a biorefinery perspective is an increase in the amount of tension wood in angiosperm trees. Tension wood is a kind of reaction wood that is typically formed on the upper side of tilted stems or branches and has a high cellulose and low lignin content compared with normal wood (Sawada et al., 2018). PtrHSFB3-1 and PtrMYB092 have been identified as repressors of tension wood formation in poplar (Liu et al., 2021a). CRISPR-Cas9-induced mutations in PtrHSFB3-1 or PtrMYB092 result in a stem wood composition resembling that of tension wood (i.e., high cellulose and low lignin), whereas overexpression of PtrHSFB3-1 or PtrMYB092 in poplar results in low cellulose and high lignin contents (Liu et al., 2021a). PtrHSFB3-1-knockout poplars are about 9% taller than the WT, whereas the height of PtrMYB092-knockout poplars is similar to that of the WT. On the other hand, overexpression of PtrHSFB3-1 or PtrMYB092 results in a 10% or 67% reduction in plant height compared with WT poplar, respectively.
Most of the engineering strategies mentioned above that resulted in wood with reduced lignin amounts also resulted in a higher saccharification yield (in cases where saccharification was performed; Table 1). This is in line with the general finding that lignin amount is negatively correlated with saccharification and pulping yield (Baucher et al., 2003; Chen and Dixon, 2007; Van Acker et al., 2013). The notable exceptions are the CRISPR-Cas9-generated 4cl1 poplar mutants that, despite having a 19% reduction in lignin amount, do not show a significantly increased saccharification yield (Tsai et al., 2020). The authors suggested that the expected gain in saccharification yield from the reduced lignin amount was offset by changes in lignin structure in 4cl1 poplar mutants (Tsai et al., 2020). Interestingly, an altered xylem coloration (brown, red, pink, or orange) has frequently been observed in lignin-engineered trees, such as those with reduced expression of 4CL (Wagner et al., 2009; Voelker et al., 2010; Zhou et al., 2015), CCoAOMT (Meyermans et al., 2000; Zhong et al., 2000; Jing et al., 2004), CCR (Leplé et al., 2007; Van Acker et al., 2014; De Meester et al., 2020), COMT (Van Doorsselaere et al., 1995; Tsai et al., 1998; Jouanin et al., 2000; Pilate et al., 2002), and CAD (Baucher et al., 1996; MacKay et al., 1997; Pilate et al., 2002; Van Acker et al., 2017; Yamamoto et al., 2020). In CCR-deficient wood, the reddish coloration has been hypothesized to be caused by the incorporation of ferulic acid, the de-esterification product derived from the CCR substrate feruloyl-coenzyme A (CoA), into the lignin polymer; synthetic lignins made from coniferyl alcohol, sinapyl alcohol, and ferulic acid are reddish (Leplé et al., 2007; Ralph et al., 2008). In CAD-deficient wood, the red xylem coloration is potentially caused by the formation of S′(8-8)S′ (or β-β′ di-sinapyl aldehyde), a reddish dimeric coupling product made from sinapaldehyde, one of the substrates of CAD (Fournand et al., 2003).
Exotic lignin
Basically, any molecule with a phenolic function that can be oxidized via the activity of laccases and/or peroxidases can serve as a lignin monomer, on the condition that the molecule can be translocated to the cell wall (Ralph, 2006; Mottiar et al., 2016; del Río et al., 2020). Although it has been suggested that ATP-binding cassette (ABC) transporter family proteins could be involved in the transport of monomers across the plasma membrane (reviewed in Perkins et al., 2019), more recent molecular dynamics simulations and experimental observations support a passive permeation mechanism for monomers and dimers across the plasma membrane lipid bilayer (Vermaas et al., 2019; Perkins et al., 2022). However, carboxylated or glycosylated phenolic monomers have been predicted to not diffuse easily through the plasma membrane and to require transporters for translocation across the membrane (Vermaas et al., 2019). These predictions are in agreement with experimental observations. For instance, only minor levels of ferulic acid are integrated into the lignin of Arabidopsis and poplar with compromised CCR activity, whereas soluble ferulic acid conjugates accumulate to high levels in these plants (Figure 1) (Leplé et al., 2007; Ralph et al., 2008; Van Acker et al., 2014; De Meester et al., 2018, 2020). The apparent absence of a need for specific transporters for non-glycosylated and non-carboxylated monomers (Vermaas et al., 2019; Perkins et al., 2022) suggests possibilities for the incorporation of non-canonical monomers that are normally not part of—or are present at very low levels in—the lignin polymer, through the introduction of novel metabolic routes (Figure 1).
An increasing number of studies demonstrate that lignin can be engineered through the coupling of non-canonical monomers (Figure 1). A first example of such a non-canonical monomer is coniferyl ferulate, a specialized metabolite that accumulates in Angelica sinensis but is naturally only found at trace levels in lignin (Karlen et al., 2016). When FERULOYL-COA:MONOLIGNOL TRANSFERASE (FMT) from A. sinensis is expressed in poplar, coniferyl ferulate copolymerizes with natural lignin monomers in the cell wall (Wilkerson et al., 2014). With the integration of coniferyl ferulate, ester linkages are introduced into the lignin backbone, rendering the lignin more easily extractable in alkaline conditions (Wilkerson et al., 2014). Second, curcumin is a soluble metabolite in Curcuma longa (turmeric) that is not known to be part of the natural lignin of tree species. By overexpression of two genes from turmeric, DIKETIDE-CoA SYNTHASE (DCS) and CURCUMIN SYNTHASE (CURS), curcumin has been successfully biosynthesized and integrated into the cell walls of Arabidopsis and poplar (Oyarce et al., 2019; De Meester et al., 2022a). However, in contrast to transgenic Arabidopsis plants, transgenic poplars have 23% more lignin, an altered lignin composition that resembles stress lignin, no increase in saccharification efficiency after alkaline pretreatment, and a yield penalty. These data highlight the importance of translating research from model organisms into crops. Third, the flavonoid tricin is naturally present in lignin of grasses (Lan et al., 2016), but flavonoids have not yet been naturally found in the lignin of tree species. However, the expression of CHALCONE SYNTHASE (CHS) from Malus domestica (apple) in poplar results in incorporation of the flavonoid naringenin into the lignin polymer, reduced total lignin content, and increased cell-wall carbohydrate content, with no effect on biomass yield (Mahon et al., 2022). Wood from these CHS-overexpressing poplars is substantially easier to saccharify after no pretreatment or dilute acid pretreatment (Mahon et al., 2022). Fourth, sinapyl p-coumarate and, to a lesser extent, coniferyl p-coumarate are common natural monomers of grass lignin (Grabber et al., 1996) but are absent or close to the detection limit in tree species. Overexpression of BdPMT1, a Brachypodium distachyon gene that encodes a p-COUMAROYL-CoA MONOLIGNOL TRANSFERASE, in poplar results in lignin with an even higher frequency of p-coumarates compared with Brachypodium, without affecting growth (Lapierre et al., 2021). The lignin of the transgenic trees has a lower polymerization degree and an improved solubility in cold NaOH solution, resulting in an improved saccharification yield of alkali-pretreated wood (Lapierre et al., 2021). The p-coumaroylation of lignin could not be further boosted by stacking the overexpression of BdPMT1 with the overexpression of F5H1 from Arabidopsis, even though the stacked lines had a higher frequency of syringyl units and the p-coumaroylation of sinapyl alcohols is favored (Lapierre et al., 2021).
Examples of alternative lignin monomers that have been tested in non-tree species are disinapoyl glucose and the coumarin scopoletin. Because disinapoyl glucose has two ester bonds, its incorporation into the lignin polymer could potentially result in a weaker backbone and easier degradability in alkaline conditions. Engineering of the phenylpropanoid pathway in Arabidopsis resulted in the successful accumulation of disinapoyl glucose (Lee et al., 2017). However, in line with the low efficiency of passive diffusion of glycosylated monomers into the cell wall (Vermaas et al., 2019), disinapoyl glucose was not integrated into the lignin of the transgenic plants (Lee et al., 2017). In two different studies, Arabidopsis has been engineered to boost scopoletin production by overexpressing FERULOYL-CoA 6′-HYDROXYLASE (F6′H) alone or together with COUMARIN SYNTHASE (COSY) (Sakamoto et al., 2020; Hoengenaert et al., 2022). Introduction of scopoletin into lignin theoretically introduces ethers that make it more cleavable than WT lignin. Introduction of the metabolic route to scopoletin results in the successful incorporation of scopoletin into lignin and improved saccharification after alkaline pretreatment, making this a promising strategy for translation into trees (Hoengenaert et al., 2022).
Specific laccases may be required to allow the coupling of certain exotic monomers into the lignin polymer. It has been shown that laccase ChLAC4 catalyzes the oxidation of coniferyl alcohol and its subsequent coupling into lignin in the seed coat of Cleome hassleriana and that ChLAC4 does not efficiently oxidize caffeyl alcohol (Zhuo et al., 2022). Instead, ChLAC8 is responsible for oxidation of caffeyl alcohol and its coupling into the lignin of C. hassleriana seed coats (Zhuo et al., 2022). These findings suggest that ChLAC8 could be essential in lignin-engineering strategies aimed at introducing caffeyl alcohol into the lignin of trees and also that laccases might be engineered to enable or facilitate polymerization of specific exotic monomers.
Lessons learned from over 20 years of field trials with lignin-engineered trees
For practical and regulatory reasons, most studies on lignin engineering have been performed with greenhouse-grown trees (see above and Table 1). However, unlike greenhouse-grown trees, field-grown trees undergo annual seasonal cycles of growth and dormancy and interact with a range of biotic and abiotic environmental factors, such as field location, wind, drought, and pathogens. These factors greatly influence the cell-wall composition and growth of plants (as illustrated in Table 2 for WT trees and described below for lignin-engineered trees) (Cesarino, 2019). Because field trials also allow the trees to be evaluated under relevant agricultural practices (such as short-rotation coppice culture), they are a crucial step in translating fundamental knowledge generated in the greenhouse to conditions closer to industrial exploitation (Pilate et al., 2015).
Table 2.
Biomass, lignin amount, and lignin composition of WT P. trichocarpa trees grown in the greenhouse and at 3 different field locations in North Carolina (USA).
| Biomass (g) | Lignin amount (%) | Lignin composition (S/G ratio) | |
|---|---|---|---|
| WT (greenhouse) | n.a. | 22.0 ± n.a. | 2.5 ± n.a. |
| WT (field, Piedmont site) | 93.8 ± 9.2 | 26.4 ± 0.4 | 1.7 ± 0.1 |
| WT (field, coastal plain site) | 136.1 ± 14.0 | 25.5 ± 0.5 | 1.9 ± 0.1 |
| WT (field, mountain site) | 1088.9 ± 88.5 | 23.4 ± 0.3 | 2.0 ± 0.0 |
In the field, the trees were planted in April 2009, coppiced for the first time in January 2010, and coppiced for the second time in January 2011. Values (means ± standard error) are given for plant material derived from the greenhouse or the second coppice in the field. n.a., not available.
Source: Stout et al. (2014).
Table 3 summarizes reports on all field trials performed with lignin-engineered trees. To date, eucalyptus trees downregulated in C3′H or C4H and poplar trees downregulated in 4CL, CSE, CCoAOMT, CCR, COMT, CAD, or LTF1 have been evaluated in the field. Of particular interest are studies that examined the performance of the very same transgenic line in both the greenhouse and the field, preferably planted in multiple contrasting locations and harvested over multiple rotations. Data from these studies lead to the following general observations:
-
(1)
The lignin reduction and growth phenotype of trees engineered to produce less lignin are highly dependent on the growth environment (genotype × environment interactions). Indeed, the very same transgenic line can behave significantly differently depending on whether it is grown in the greenhouse or the field. For example, the reduction in lignin in 4CL-, CCoAOMT-, CCR-, or CAD-downregulated poplars was greater when the trees were grown in the greenhouse than when they were grown in the field (Wang et al., 2012; Stout et al., 2014; Van Acker et al., 2014, 2017; Xiang et al., 2017; De Meester et al., 2022a). Moreover, although CCR-, CAD-, and vessel-specific 4CL-downregulated poplars grew normally in the greenhouse, the same lines displayed a yield penalty when grown in the field (Leplé et al., 2007; Van Acker et al., 2017; Cao et al., 2020; De Meester et al., 2022a). Lignin reduction is also highly dependent on biotic and abiotic environmental factors determined by the field location. For example, the reduction in lignin in 4CL-downregulated poplars grown for 2 years in the field was different depending on whether the lines were grown in mountain, coastal plain, or Piedmont regions of North Carolina (Stout et al., 2014). Similarly, a 2-year-old CAD-downregulated line exhibited different levels of lignin reduction depending on whether it was grown in a field in the UK or in a field in France (Pilate et al., 2002).
-
(2)
The lignin reduction and growth phenotype of trees engineered to produce less lignin are highly dependent on tree age. Indeed, consistent with the first conclusion, the lignin reduction in field-grown trees engineered to produce less lignin often becomes less pronounced throughout development and is generally smaller than in their greenhouse-grown clonal ramets. For example, 4CL-downregulated lines grown in the greenhouse had severely reduced lignin amounts (up to 47% less lignin than the WT control), whereas the same transgenic lines grown on a mountain site field had mildly reduced lignin amounts (up to 8%) after 2 years and lignin amounts similar to that of the WT after 3 years of growth (Xiang et al., 2017). Similarly, greenhouse-grown CAD-downregulated poplars had 10% less lignin (Van Acker et al., 2017), whereas the same transgenic lines grown in the field displayed 10% less lignin after 1 year and had lignin amounts equal to that of the WT after 3 years of growth (De Meester et al., 2022a). The lignin content in 1-year-old greenhouse-grown 4CL- and CCoAOMT-downregulated poplars was reduced by 16%–42%, whereas that in 5-year-old field-grown clonal ramets was reduced by only 6%–10% (Wang et al., 2012). This trend might be explained by the fact that greenhouse-grown trees continuously grow and develop new xylem because of the long-daylight regime typically used in greenhouses, but trees grown in the field in a temperate climate zone cease growth in late summer every year and may continue to lignify their cell walls—aided by the good neighbor cells—until they enter dormancy (Van Acker et al., 2014; De Meester et al., 2022a). Alternatively, these observations might be explained by the enhanced exposure to (multiple cycles of) environmental stresses in the field, such as drought, leading to enhanced lignin biosynthesis (Jubany-Marí et al., 2009; Lee et al., 2012; Cesarino, 2019; Liu et al., 2021b). They might also be explained by fluctuations in the level of gene downregulation. Indeed, the extent of RNA interference (RNAi)-induced downregulation of a specific gene is dependent not only on the specific transfer DNA (T-DNA) integration event and thus the specific transgenic line but also on the individual plant and the specific environment. For example, substantial variation in the level of downregulation was observed among clonal ramets of the same CCR2-downregulated line, as judged from variation in the reddish-brown xylem phenotype associated with CCR2 downregulation (Van Acker et al., 2014). To exclusively evaluate the effects of environment and age on lignin content reduction (and to exclude possible interactions between the environment/tree age and the level of gene silencing), CRISPR-Cas9-generated lignin-engineered trees, whose gene knockdowns or knockouts are stable by definition, will need to be tested in different locations over multiple years (Zhou et al., 2015; De Meester et al., 2020; de Vries et al., 2021a).
Table 3.
Biomass, lignin amount, and lignin composition of lignin-engineered trees grown in the field compared with greenhouse conditions.
| Species | Gene | Method | Tested in GH? | Location | # Lines | Field size | Field duration | Age of the stems at harvest | Effect on total lignin | Effect on lignin composition | Saccharification efficiency | Pulping efficiency | Biomass yield | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. urophylla × E. grandis | C4H | RNAi | no | FL, USA | 9 | n.a. | 2 years | 2 years | ↓40% to ↓30% | ↓S/G | ↑ | n.d. | ↓ | Sykes et al. (2015) |
| E. urophylla × E. grandis | C3′H | RNAi | no | FL, USA | 6 | n.a. | 2 years | 2 years | ↓28% to ↓25% | ↓S/G | ↑ | n.d. | ↓ | Sykes et al. (2015) |
| P. tremula × P. alba | 4CL | antisense | no | OR, USA | 14 | 10–15 plants per line | 2 years | 2 years | ↓10% to WT | ↓S/G to ↑S/G | WT | n.d. | ↓ | Voelker et al. (2010) |
| P. tomentosa Carr. | 4CL | sense and antisense | no | Hebei, China | 5 | 60 plants per line | 6 years | 1 year | ↓28% to ↓5% | ↑S/G | n.d. | n.d. | ↑ | Tian et al. (2013) |
| P. nigra × Populus maximowiczii | 4CL | antisense | yes | mountain, NC, USA | 3 | n.a. | 3 years | 2 years | ↓8% to WT [↓47% to ↓8%] | ↑S/V [↑S/V] | ↑ [n.d.] | n.d. [n.d.] | n.d. [n.d.] | Xiang et al. (2017) [Xiang et al., 2017] |
| P. nigra × P. maximowiczii | 4CL | antisense | yes | mountain, NC, USA | 3 | n.a. | 3 years | 3 years | WT [↓47% to ↓8%] | ↓S/V to ↑S/V [↑S/V] | ↑ [n.d.] | n.d. [n.d.] | n.d. [n.d.] | Xiang et al. (2017) [Xiang et al., 2017] |
| P. nigra × P. maximowiczii | 4CL | antisense | yes | coastal plain, NC, USA | 3 | n.a. | 3 years | 2 years | ↓7% to WT [↓47% to ↓8%] | ↓S/V to ↑S/V [↑S/V] | WT [n.d.] | n.d. [n.d.] | n.d. [n.d.] | Xiang et al. (2017) [Xiang et al., 2017] |
| P. nigra × P. maximowiczii | 4CL | antisense | yes | coastal plain, NC, USA | 3 | n.a. | 3 years | 3 years | ↓9% to WT [↓47% to ↓8%] | ↑S/V [↑S/V] | ↑ [n.d.] | n.d. [n.d.] | n.d. [n.d.] | Xiang et al. (2017) [Xiang et al., 2017] |
| P. trichocarpa | 4CL | antisense | yes | coastal plain, NC, USA | 12 | 149 plants per line | 2 years | 1 year (2 y/o roots) | ↓37% to WT [↓46% to ↓5%] | ↓S/G to ↑S/G [↓S/G and ↑S/G] | n.d. [n.d.] | n.d. [n.d.] | ↓90% to ↑83% [n.d.] | Stout et al. (2014) [Stout et al., 2014] |
| P. trichocarpa | 4CL | antisense | yes | Piedmont, NC, USA | 12 | 149 plants per line | 2 years | 1 year (2 y/o roots) | ↓33% to WT [↓46% to ↓5%] | ↓S/G to ↑S/G [↓S/G and ↑S/G] | n.d. [n.d.] | n.d. [n.d.] | ↓92% to ↓14% [n.d.] | Stout et al. (2014) [Stout et al., 2014] |
| P. trichocarpa | 4CL | antisense | yes | mountain, NC, USA | 12 | 78 plants per line | 2 years | 1 year (2 y/o roots) | ↓24% to WT [↓46% to ↓5%] | ↓S/G to ↑S/G [↓S/G and ↑S/G] | n.d. [n.d.] | n.d. [n.d.] | ↓92% to ↑24% [n.d.] | Stout et al. (2014) [Stout et al., 2014] |
| P. tomentosa | 4CL | antisense | yes | Beijing, China | 2 | n.a. | 5 years | 5 years | ↓8% to ↓6% [↓42% to ↓26%] | n.d. [n.d.] | WT [n.d.] | n.d. [n.d.] | WT [WT] | Wang et al. (2012) [Jia et al., 2004] |
| P. deltoides × P. euramericana | 4CL | fiber-specific suppression | yes | Shanghai, China | 3 | 30 plants per line | 1 year | 1 year | ↓∼25% [↓21%] | n.d. [n.d.] | n.d. [n.d.] | n.d. [n.d.] | WT [WT] | Cao et al. (2020) [Cao et al., 2020] |
| P. deltoides × P. euramericana | 4CL | vessel-specific suppression | yes | Shanghai, China | 3 | 30 plants per line | 1 year | 1 year | ↓∼20% [↓26%] | n.d. [n.d.] | n.d. [n.d.] | n.d. [n.d.] | ↓ [WT] | Cao et al. (2020) [Cao et al., 2020] |
| P. alba × P. glandulosa | CSE1 | CRISPR-Cas9 | yes | Republic of Korea | 1 or 3a | n.a. | 1 year | 8 months or 1 yearb | ↓ ∼24% [↓ up to 16%] | n.d. [n.d.] | ↑ [n.d.] | n.d. [n.d.] | WT [WT] | Jang et al. (2021) [Jang et al., 2021] |
| P. alba × P. glandulosa | CSE2 | CRISPR-Cas9 | yes | Republic of Korea | 1 or 3a | n.a. | 1 year | 8 months or 1 yearb | ↓29% [↓ up to 16%] | n.d. [n.d.] | ↑ [n.d.] | n.d. [n.d.] | WT [WT] | Jang et al. (2021) [Jang et al., 2021] |
| P. tomentosa | CCoAOMT | antisense | yes | Beijing, China | 2 | n.a. | 5 years | 5 years | ↓10% to ↓9% [↓16%] | n.d. [n.d.] | ↑ [n.d.] | n.d. [n.d.] | WT [WT] | Wang et al. (2012) [Jing et al., 2004] |
| P. tremula × P. alba | CCoAOMT | antisense | no | n.a. | 1 | n.a. | 3 years | 3 years | ↓13% | ↑S/G | n.d. | ↑ | WT | Wei et al. (2008) |
| P. tremula × P. alba | CCR | sense and antisense | yes | Orléans,France | 4 | 10 plants per line | 8 years | 2, 4, and 5 yearsc | ↓47% to WT [↓30% to ↓22%] | ↓S/G, incorporation of FA [↓S/G, incorporation of FA] | n.d. [n.d.] | ↑ [n.d.] | ↓ [WT] | Leplé et al. (2007) [Leplé et al., 2007] |
| P. tremula × P. alba | CCR | sense | yes | East Flanders, Belgium | 2 | 120 plants per line | 5 years | 10 months | ↓8% to ↓5% [↓30% to ↓22%] | WT S/G, incorporation of FA [↓S/G, incorporation of FA] | ↑ [↑] | n.d. [n.d.] | ↓ [WT] | Van Acker et al. (2014) [Leplé et al., 2007] |
| P. tremula × P. alba | CCR | sense and antisense | yes | Orléans, France | 2 | 120 plants per line | 5 years | 2 years | ↓24% to ↓9% [↓30% to ↓22%] | ↓S/G to WT, incorporation of FA [↓S/G, incorporation of FA] | ↑ [↑] | n.d. [n.d.] | ↓ [WT] | Van Acker et al. (2014) [Leplé et al., 2007] |
| P. tremula × P. alba | COMT | antisense | yes | Orléans, France | 2 | 10 plants per line | 5 years | 2 years | WT [WT] | ↓S/G, incorporation of 5-OHG [↓S/G, incorporation of 5-OHG] | n.d. [n.d.] | n.d. [↓] | WT [WT] | Pilate et al. (2002) [Lapierre et al., 1999; Van Doorsselaere et al., 1995] |
| P. tremula × P. alba | COMT | antisense | yes | Orléans, France | 2 | 10 plants per line | 5 years | 2 years (4 y/o roots) | WT [WT] | ↓S/G, incorporation of 5-OHG [↓S/G, incorporation of 5-OHG] | n.d. [n.d.] | ↓ [↓] | WT [WT] | Pilate et al. (2002) [Lapierre et al., 1999; Van Doorsselaere et al., 1995] |
| P. tremula × P. alba | COMT | antisense | yes | Berkshire, England | 2 | 12 plants per line | 4 years | 4 years | WT [WT] | ↓S/G, incorporation of 5-OHG [↓S/G, incorporation of 5-OHG] | n.d. [n.d.] | ↓ [↓] | WT [WT] | Pilate et al. (2002) [Lapierre et al., 1999; Van Doorsselaere et al., 1995] |
| P. tremula × P. alba | COMT | antisense | yes | n.a. | 2 | n.a. | 2 years | 2 years | WT [WT] | ↓S/G, incorporation of 5-OHG [↓S/G, incorporation of 5-OHG] | n.d. [n.d.] | ↓ [↓] | WT [WT] | Lapierre et al. (1999) [Lapierre et al., 1999; Van Doorsselaere et al., 1995] |
| P. tremula × P. alba | CAD | sense and antisense | yes | n.a. | 3 | n.a. | 2 years | 2 years | ↓4% to WT [↓4% to WT] | WT S/G, ↑ free phenolic G and S units [WT S/G] | n.d. [n.d.] | ↑ [↑] | WT [WT] | Lapierre et al. (1999) [Lapierre et al., 1999] |
| P. tremula × P. alba | CAD | antisense | yes | Orléans, France | 2 | 10 plants per line | 5 years | 2 years | Slightly ↓ [WT] | WT S/G, ↑ free phenolic G and S units [WT S/G] | n.d. [n.d.] | n.d. [↑] | WT [WT] | Pilate et al. (2002) [Baucher et al., 1996] |
| P. tremula × P. alba | CAD | antisense | yes | Orléans, France | 2 | 10 plants per line | 5 years | 2 years (4 y/o roots) | Slightly ↓ [WT] | WT S/G, ↑ free phenolic G and S units [WT S/G] | n.d. [n.d.] | ↑ [↑] | WT [WT] | Pilate et al. (2002) [Baucher et al., 1996] |
| P. tremula × P. alba | CAD | antisense | yes | Berkshire, England | 2 | 12 plants per line | 4 years | 4 years | Slightly ↓ [WT] | WT S/G, ↑ free phenolic G and S units [WT S/G] | n.d. [n.d.] | ↑ [↑] | WT [WT] | Pilate et al. (2002) [Baucher et al., 1996] |
| Morus alba | CAD | natural mutantd | no | Tsukuba, Japan | 1 | n.a. | n.a. | 1 year | Lighter color in Mäule staining | n.d. | ↑ | ↑ | n.d. | Ikeda et al. (2021) |
| Morus alba | CAD | natural mutantd | no | Tsukuba, Japan | 1 | n.a. | n.a. | n.a. | ↓33% | ↓S/G | n.d. | n.d. | n.d., lodging phenotype | Yamamoto et al. (2020) |
| P. tremula × P. alba | CAD | RNAi | yes | East Flanders, Belgium | 3 | 240 plants per line | 4 years | 1 year | ↓10% to ↓8% [↓10% to ↓6%] | ↓S/G [↓S/G] | ↑ [↑] | n.d. [n.d.] | ↓ [WT] | De Meester et al. (2022b) [Van Acker et al., 2017] |
| P. tremula × P. alba | CAD | RNAi | yes | East Flanders, Belgium | 3 | 240 plants per line | 4 years | 3 years (4 y/o roots) | WT [↓10% to ↓6%] | WT [↓S/G] | ↑ [↑] | n.d. [n.d.] | ↓ [WT] | De Meester et al. (2022b) [Van Acker et al., 2017] |
| P. deltoides × P. euramericana | LTF1 | fiber-specific suppression | yes | Shanghai, China | 2 | n.a. | 1 year | 1 year | ↓ [↓43%] | n.d. [↓S/G] | ↑ [n.d.] | n.d. [n.d.] | WT [WT] | Gui et al. (2020) [Gui et al., 2020] |
| P. deltoides × P. euramericana | LTF1 | vessel-specific suppression | yes | Shanghai, China | 2 | n.a. | 1 year | 1 year | ↓ [↓16%] | n.d. [↑S/G ] | ↓ [n.d.] | n.d. [n.d.] | ↓ [↓] | Gui et al. (2020) [Gui et al., 2020] |
# lines, number of lines with the intended transgenic construct or mutant allele (thus not including control lines); age of stem at harvest, if not explicitly stated otherwise, the age of the root is equal to that of the stem; n.d., not determined; n.a., not available; y/o, year-old; S/G, syringyl/guaiacyl ratio; S/V, syringaldehyde/vanillin ratio; FA, ferulic acid; 5-OHG, 5-hydroxyguaiacyl. Text in square brackets [ ] refers to data from greenhouse-grown trees of the same transgenic line, if available. For saccharification efficiency, see the corresponding reference for specific information about biomass pretreatments. Effect on total lignin is given as a percentage of the total lignin analyzed via different methods; see the corresponding reference for specific information.
One line for lignin amount determination, three lines for biomass determination.
Eight-month-old trees for total lignin determination, 12-month-old trees for biomass determination.
Two-year-old trees for total lignin and lignin composition determination, 4-year-old trees for biomass yield determination, 5-year-old trees for chemical pulping experiments.
The data presented are for the natural mutant Sekizaisou in comparison with other mulberry lines.
In many cases, the enhanced wood-processing efficiencies typically found for greenhouse-grown lignin-engineered trees are also observed for field-grown trees, albeit sometimes to a lesser extent and/or accompanied by a yield penalty (Tables 1 and 3). Although there may be ways to circumvent the frequently observed yield penalty in lignin-engineered trees (see below), the observation that the lignin content of trees RNAi-engineered to produce less lignin often gradually increases with age, generally implying that the processing efficiency gradually decreases, may call for a significant revision of research strategies. If similar observations are made for CRISPR-Cas9-generated lignin-engineered trees, a better strategy to deploy lignin-engineered trees for applications might be to grow the trees in very short-rotation coppice (e.g., harvesting every year or 2 years). Alternatively, engineering the lignin composition instead of the lignin amount or reducing the relative amounts of lignin in the cell wall by increasing cellulose or matrix polysaccharides (e.g., by making use of transcription factors) are valuable strategies.
Origins of the yield penalty often observed in lignin-engineered trees
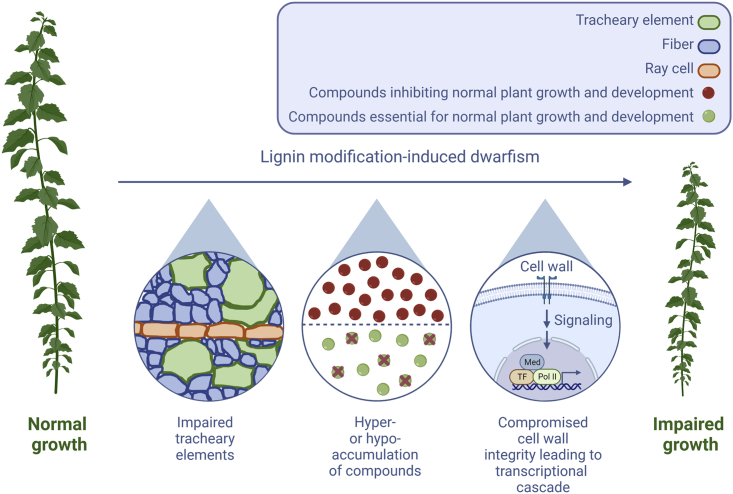
Lignin-modified plants that show the greatest improvement in processing efficiency often also exhibit biomass yield penalties (also termed lignin modification-induced dwarfism [LMID]), thereby limiting the benefits of their enhanced processing efficiency (Chen and Dixon, 2007; Bonawitz and Chapple, 2013; Van Acker et al., 2014; Muro-Villanueva et al., 2019). Because lignin provides rigidity and hydrophobicity to plant cell walls (Turner and Somerville, 1997), one might expect this growth reduction to be directly correlated with the level of lignin reduction. However, the literature shows that some transgenic poplars with severely reduced amounts of lignin had biomass yields similar to the WT, whereas poplars with unaltered or mildly reduced lignin amounts displayed severe yield penalties. For example, 4CL-downregulated poplars had up to 40%–45% less lignin and normal or increased growth (Hu et al., 1999; Li et al., 2003; Jia et al., 2004), whereas C4H-downregulated poplars had 13% less lignin and reduced growth (Kim et al., 2020). Similarly, PAL-downregulated lines (i.e., lines i7-10 and i7-2) had severely reduced amounts of lignin (−57% to −42%) and mildly reduced heights (−17% to −7%), whereas HCT-downregulated trees (i.e., line i19-4) had moderately reduced lignin amounts (−21% to −17%) and severely reduced heights (−65% to −43%) (Wang et al., 2018). Wang et al. (2018) also showed that the growth of lignin-engineered trees is not associated with a particular lignin subunit composition or specific linkages. If not changes in lignin content and composition, what mechanisms give rise to the impaired growth observed in many lignin-engineered trees (and other plants)? Several hypotheses explaining LMID in lignin-engineered plants have been postulated (Bonawitz and Chapple, 2013; Muro-Villanueva et al., 2019; Ha et al., 2021). Below, we summarize these hypotheses (which were based mainly upon observations in Arabidopsis) and discuss whether experimental support for each model also exists in trees (Figure 3).
Figure 3.
Hypotheses explaining lignin modification-induced dwarfism (LMID) in forest trees.
First, LMID might be caused by impaired functioning of TEs, resulting in perturbed water and nutrient transport in the stem. Second, a block in the phenylpropanoid pathway could lead to the accumulation of growth-inhibitory compounds or prevent the biosynthesis of compounds essential for growth. Third, alterations in the cell wall might be detected by cell-wall integrity sensors that activate a transcriptional response requiring Mediator (Med). TF, transcription factor; Pol II, RNA polymerase II.
First, LMID in lignin-modified trees could be caused by impaired functioning of the tracheary elements (TEs; vessels in hardwood, tracheids in softwood) as a result of (i) vascular collapse (Leplé et al., 2007; Coleman et al., 2008; Wagner et al., 2009; Zhou et al., 2018; De Meester et al., 2020; de Vries et al., 2021a), (ii) blockage of TEs with tyloses and phenolic deposits (Kitin et al., 2010), or (iii) enlarged vessels (which generally enhance xylem vulnerability to embolism) combined with reductions in relative water content and free water available to support the hydraulic network (De Meester et al., 2022b). Consequently, the stem sap flow and/or hydraulic conductance may be perturbed, potentially leading to enhanced susceptibility to drought stress and/or impaired tree growth (Cochard et al., 2004; Coleman et al., 2008; Cao et al., 2020; White, 2021; De Meester et al., 2022b). Impaired TEs have been observed in growth-impaired angiosperm trees with reduced expression of HCT (Zhou et al., 2018), C3′H (Coleman et al., 2008; Zhou et al., 2018), 4CL (Kitin et al., 2010; Voelker et al., 2010, 2011), CSE (de Vries et al., 2021a), CCR (Leplé et al., 2007; De Meester et al., 2020), and CAD (De Meester et al., 2022b), as well as growth-impaired gymnosperm trees with reduced expression of 4CL (Wagner et al., 2009). Similar to findings in Arabidopsis (Turner and Somerville, 1997; Brown et al., 2007; Peña et al., 2007), collapsed TEs were also found in (growth-impaired) cellulose- and hemicellulose-deficient trees, highlighting that not only lignin but also other cell-wall components are important for vascular integrity in trees (Joshi et al., 2011; Lee et al., 2011; Yu et al., 2013). As in Arabidopsis (Yang et al., 2013; Vargas et al., 2016; De Meester et al., 2018), the impaired TE hypothesis is supported by the observation that LMID in trees can be (largely) avoided by targeting the lignin reduction specifically to fibers while still allowing sufficient lignin to be deposited in TEs (see also below) (Cao et al., 2020; Gui et al., 2020; De Meester et al., 2021).
Second, LMID might be caused by hyper- or hypoaccumulation of pathway intermediates and/or derivates. When a certain enzymatic conversion in the lignin biosynthesis pathway is perturbed, compounds located upstream of this conversion or their derivatives may accumulate, and the abundance of compounds located downstream of this conversion will decrease. In addition, a transcriptional cascade aimed at increasing flux through the pathway is activated to compensate for the loss of lignin building blocks (e.g., by the activation of shikimate and phenylpropanoid pathways), resulting in the (additional) toxic buildup of compounds because the pathway remains blocked (Vanholme et al., 2012). Hyper- or hypoaccumulation of phenylpropanoid-related compounds can have a strong effect on the stress- or defense-related transcriptome (Tsai et al., 2020) and, hence, on the abundance of other growth-related compounds, potentially leading or at least contributing to the observed yield penalties. For example, the Arabidopsis ref3 mutant accumulates high levels of cinnamate-derived compounds owing to its reduced C4H activity. Cinnamate hyperaccumulation in ref3 mutants impairs auxin signaling, which might underlie the observed yield penalty (Schilmiller et al., 2009; Steenackers et al., 2019; El Houari et al., 2021). Arabidopsis and Medicago HCT-deficient plants accumulate high levels of the stress hormone salicylic acid (SA), which negatively influences stem height in the transgenic plants (Gallego-Giraldo et al., 2011a). Reducing SA levels in these plants restored plant growth. Furthermore, overexpression of LAC4 or LAC17 in high-monolignol glucoside-containing lines (MYB58-OE or MYB63-OE) decreases levels of monolignol glucosides and rescues impaired plant growth (Perkins et al., 2020). The growth-inhibitory effect of specific hyperaccumulating phenylpropanoids and phenylpropanoid-derived compounds might explain why, e.g., HCT-downregulated poplars show severe growth defects, whereas PAL-downregulated lines show no/very mild growth defects (Wang et al., 2018). Based on current insights into the phenylpropanoid pathway (Figure 1), HCT-downregulated lines are expected to accumulate the HCT substrate p-coumaroyl-CoA and its derivates that remain soluble and potentially interfere with cellular processes, whereas PAL-downregulated lines accumulate the substrate phenylalanine, which can be further metabolized in primary processes such as protein synthesis. Following this rationale, valuable lignin-engineering strategies might focus on targeting enzymes that work upstream of C4H, resulting in a buildup of substrates that could be used in primary metabolism (thus avoiding the accumulation of useless and potentially toxic carbon sinks). For example, the transcription factors PtrHSFB3-1 and PtrMYB092 have been shown to activate transcription of multiple genes encoding enzymes in the phenylpropanoid and monolignol-specific pathways, including PAL family enzymes and most likely also enzymes upstream of PAL (Liu et al., 2021a). Poplar knockout mutants in either PtrHSFB3-1 or PtrMYB092 display a low lignin content (up to −17% or −27%, respectively) without adverse effects on biomass yield (Liu et al., 2021a). Notably, although several lignin-engineered trees have been shown to hyper- or hypoaccumulate a plethora of compounds (e.g., Leplé et al., 2007; Saleme et al., 2017; De Meester et al., 2020; de Vries et al., 2021a), to date, no support for a link between the abundance of a specific compound and growth has been found in trees. This is due at least in part to the fact that the metabolic response to pathway perturbations is complex, involving many unknown metabolites.
Third, LMID could be triggered by a cell-wall integrity system that senses defects in the cell wall and consequently activates a cascade of transcriptional changes to overcome such defects. This reallocation of resources toward the constitutive production of cell-wall components and/or defense compounds might come at the expense of plant growth. Indeed, judging from their increased expression of pathogenesis-related genes, HCT-deficient alfalfa and Arabidopsis plants exhibit constitutive activation of defense responses (Gallego-Giraldo et al., 2011a, 2011b). More recently, a study with HCT- and CCR1-deficient Arabidopsis showed that defense gene induction and release of elicitors result from the ectopic expression of ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 1 (ADPG1), which encodes a pectin-degrading enzyme (Gallego-Giraldo et al., 2020). The role of transcriptional changes in LMID has been demonstrated by the (partial) restoration of biomass yield upon mutation of Med5A/5B subunits of the transcriptional coregulator Mediator in Arabidopsis c3′h1 mutants (Bonawitz et al., 2014). Nevertheless, the role of Mediator in LMID of lignin-engineered trees has not been studied to date.
It should be noted that the occurrence, cause, and severity of LMID depend on the position of the perturbation in the pathway (as described above for PAL- versus HCT-downregulated poplars). For example, although the growth of dwarfed HCT-deficient Arabidopsis is restored by reducing SA levels (Gallego-Giraldo et al., 2011a), other phenylpropanoid mutants exhibit dwarfed phenotypes and decreased SA levels (Huang et al., 2010) or grow normally and have increased SA levels (Bonawitz et al., 2014). Although the growth of c3′h1 Arabidopsis mutants is largely restored by mutating the med5a/5b subunits of the Mediator complex (Bonawitz et al., 2014), the stunted growth of c4h and high-monolignol MYB63-OE Arabidopsis mutants is not recovered by disrupting Med5A/5B (Bonawitz et al., 2014; Perkins et al., 2020). Finally, the species, the specific genotype, and the environment play roles in the expression of the phenotype, as illustrated by 4CL-downregulated poplar; the growth of 4CL-deficient Populus tomentosa in a field in China was increased by 8% (Tian et al., 2013), whereas 4CL-deficient P. tremula × P. alba grown in a field in Oregon displayed yield penalties. These differences may be due to different genetic backgrounds, different levels of 4CL downregulation, or different environments.
Overcoming LMID in lignin-engineered trees
Despite the fact that our understanding of the potential reasons for LMID remains fragmentary, strategies can be designed to eliminate or at least mitigate LMID by, e.g., avoiding impaired TEs, avoiding the (excessive) buildup or shortage of certain compounds, or avoiding the (excessive) triggering of the cell-wall integrity-monitoring pathway. Moreover, these potential causes might be interconnected; relieving vascular collapse might also relieve the triggering of the cell-wall-monitoring pathway and the buildup of (toxic) compounds and vice versa.
To avoid LMID, the level of target gene downregulation is of major importance. For example, CSE-downregulated P. tremula × P. alba trees generated by RNAi had 25% less lignin and normal growth (Saleme et al., 2017), whereas CRISPR-Cas9-generated CSE-knockout P. tremula × P. alba had 35% less lignin and perturbed growth (de Vries et al., 2021a). In addition to RNAi, gene editing or screening of natural populations can also be used to obtain a range of lines with various levels of target enzyme activity. For example, CCR2-knockout P. tremula × P. alba generated using CRISPR-Cas9 were extremely dwarfed and could barely survive when grown in soil (De Meester et al., 2020). By contrast, P. tremula × P. alba harboring one knockout CCR2 allele and one weak CCR2 allele (with a 3-bp deletion, resulting in a 114I115A-to-114T conversion in the CCR2 protein), had 10% less lignin, grew normally, and showed improved wood-processing efficiency (De Meester et al., 2020). In a second example, HCT-downregulated poplars made with RNAi had severe growth reductions (Wang et al., 2018), whereas a naturally occurring poplar HCT mutant (homozygous for a point mutation leading to a truncated HCT protein lacking 73 amino acids at the C-terminal end) had a significantly altered lignin composition but showed normal growth (Vanholme et al., 2013a).
Next to residual enzyme activity, the spatial distribution of the (remaining active) lignin biosynthesis enzyme also seems to be critical for avoiding LMID. In Arabidopsis, reintroduction of lignin biosynthesis specifically in the TEs (and not the fibers) of c4h, cse, and ccr1 mutants (largely) restores the collapsed TEs and stunted growth (Yang et al., 2013; Vargas et al., 2016; De Meester et al., 2018). However, because of the presence of “good neighbors for lignification” monolignols synthesized in TE cells not only lignify the TE cell wall but also contribute to cell-wall lignification of neighboring cells (Pesquet et al., 2013; Smith et al., 2013, 2017; De Meester et al., 2018). For example, in Arabidopsis ccr1 mutants expressing CCR1 under the control of a vessel-specific promoter (ccr1 ProSNBE:AtCCR1), the production of monolignols is restricted to TE cells, but xylary fibers (and not interfascicular fibers) also show enhanced lignin deposition. In poplar, expression of ProSNBE:AtCCR1 in CRISPR-Cas9-generated ccr2 mutants results in TE- and ray-specific monolignol biosynthesis and restores vascular integrity and biomass yield (De Meester et al., 2021). In transgenic lines in which monolignols are abundantly produced by TEs and ray cells, these monolignols migrate across multiple cell layers into adjoining and non-adjoining fibers, resulting in a restoration of lignin amount in all xylem cells. In lines whose supply of monolignols is limited, monolignols are incorporated only into the cell walls of the TEs and rays producing them and their adjoining cells, resulting in the desired fiber hypolignification. Alternatively, fiber hypolignification can be achieved by repressing lignin biosynthesis in fiber cells of poplar by, e.g., fiber-specific downregulation of 4CL1 or fiber-specific expression of the constitutive repressor LTF1AA (Cao et al., 2020; Gui et al., 2020). In both cases, the transgenic poplars displayed hypolignified fibers but normal TEs and growth. In the 4CL1-engineered poplars, monolignol-producing TEs act as good neighbors for lignification of adjoining fibers but not fibers located further away (Cao et al., 2020). However, no (or very little) lignification of both adjoining and non-adjoining fibers was observed in LTF1AA poplars, probably because all genes involved in lignin biosynthesis in the fibers were suppressed, including those encoding cell-wall-localized laccases and peroxidases involved in dehydrogenation of the lignin monomers (Gui et al., 2020). By contrast, when lignin biosynthesis was specifically allowed in fiber cells, leaving the TEs hypolignified, the transgenic poplars had collapsed TEs and severe growth perturbations (Cao et al., 2020; Gui et al., 2020). These observations show that the presence of sufficient lignin in the TEs is crucial for avoiding LMID in trees, whereas hypolignification of the fibers does not appear to affect growth.
Breeding lignin traits in forest trees
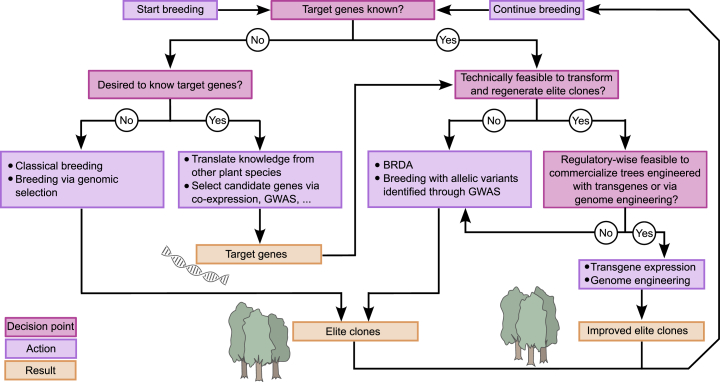
Unlike fruit trees that have been domesticated for thousands of years, forest trees were not actively subjected to domestication until the 1950s (Lebedev et al., 2020). Nevertheless, the use of traditional breeding to improve lignin traits in forest trees has great potential. Large variations in lignin amount (15.7%–27.9% and 24.4%–32.1%) and S/G ratio (1.0–3.0 and 1.8–4.2) are present in undomesticated Populus trichocarpa (Studer et al., 2011) and Eucalyptus grandis × Eucalyptus urophylla F2 full-sib families (Marco de Lima et al., 2019), respectively. Thus, substantial improvements in lignin content and composition of forest trees are possible by traditional breeding alone (Studer et al., 2011). As traditional breeding is time consuming owing to long generation times and the fact that wood quality traits are typically best evaluated at rotation age (Ahmar et al., 2021), tree breeding can benefit from the use of genomic information (Figure 4). For instance, genomic selection has substantially accelerated the speed of forest tree breeding (Grattapaglia et al., 2018; Howe et al., 2020; Lebedev et al., 2020; Ahmar et al., 2021; Grattapaglia, 2022). This technique enables the prediction of multiple traits based on thousands of molecular markers and therefore depends on low-cost, high-throughput sequencing techniques (Grattapaglia and Resende, 2011; Rambolarimanana et al., 2018; Grattapaglia, 2022). The underlying genes and alleles that cause the desired trait—let alone the biochemical mechanism behind it—usually remain unknown. Genomic selection is currently used to optimize lignin traits in eucalyptus (Rambolarimanana et al., 2018; Marco de Lima et al., 2019; Bouvet et al., 2020).
Figure 4.
Simplified flowchart reflecting the integration of various breeding strategies for lignin engineering in forest trees.
In general, the most efficient breeding strategy uses a combination of techniques as follows: first, breed via classical breeding and genomic selection for a series of traits such as wood quality, disease resistance, and growth architecture, thus producing elite clones. Next, use transgene or genome engineering to further improve or introduce the desired lignin traits, thereby avoiding the loss of genetic constitution in elite lines. BRDA, breeding with rare defective alleles; GWAS, genome-wide association studies.
Complementary approaches to improve lignin properties in trees are breeding techniques that target specific genes known to be involved in lignin biosynthesis (Figure 4). As described above, many genes involved in lignification are already known (Figure 1). These have been discovered over the past decades through fundamental research on both model organisms and tree species, e.g., by forward genetic approaches and after the selection of candidate genes based on co-expression analysis followed by reverse genetic approaches. Genome-wide association studies (GWAS) are of particular interest because GWAS can be used to identify genes/alleles that are causal for a specific trait in a population (Fahrenkrog et al., 2017). For instance, association genetics in different poplar species has revealed associations between lignin amount or composition and genes encoding β-TUBULIN 15, CCoAOMT1, an FAD-binding domain-containing protein, ADP-ribosylation factor (ARF) GTPase-activating protein (GAP) domain 11, the KANADI transcription factor, and EPSPS (Guerra et al., 2013; Porth et al., 2013; Muchero et al., 2015; Xie et al., 2018, 2020; Yao et al., 2021). The roles of CCoAOMT1 and EPSPS in lignification have been validated by reverse genetics (Meyermans et al., 2000; Zhong et al., 2000; Xie et al., 2020). Further reverse genetics support is needed to validate the roles of β-TUBULIN 15, the FAD-binding domain-containing protein, ARF-GAP domain 11, and KANADI in lignin biosynthesis.
If the causal genes for a given trait are known, traditional tree breeding can also be accelerated by screening large populations of trees by next-generation sequencing for individuals with rare alleles of target genes and by introducing these genes into breeding programs (Marroni et al., 2011; Vanholme et al., 2013a), a strategy termed breeding with rare defective alleles (BRDA). The identification of a natural mutant allele of HCT1 that increased the frequency of H units in Populus nigra served as a proof of concept for the use of this strategy to alter lignin traits in poplar (Vanholme et al., 2013a). The potential of using natural allelic variation was also illustrated by the discovery of a natural cad null mutant in Pinus taeda L. (loblolly pine) (MacKay et al., 1997). Lignin was lowered by about 10% and was composed of 15% coniferaldehyde and 30% dihydroconiferyl alcohol in the mutant, whereas these values were 7% and 3%, respectively, in control pine (MacKay et al., 1997; Ralph et al., 1997). In addition, the Morus alba (mulberry) cultivar Sekizaisou was revealed to be a natural cad null mutant (Yamamoto et al., 2020). The tree showed reduced lignin levels with increased incorporation of hydroxycinnamaldehydes, improved wood delignification efficiency, and increased pulp yield under alkaline pulping conditions compared with a set of reference genotypes that lacked the cad mutation (Ikeda et al., 2021). Use of such naturally occurring rare defective alleles in breeding programs can be useful if they are present in the breeding germplasm, but modern breeding techniques enable engineering of the desired alleles directly into elite genotypes.
Modern breeding techniques, such as genetic engineering via transgene expression and gene editing, have opened up new possibilities in forest tree breeding (Chanoca et al., 2019; Min et al., 2022). Once elite genotypes have been generated by classical breeding, use of these modern breeding techniques enables them to be further improved without breaking up their genetic constitution. Moreover, these techniques enable the introduction of traits that are not present in the species and the generation of allelic variants that are absent or rare in the population (Goralogia et al., 2021). Use of transgenes enables alteration of target gene expression levels and introduction of genes from pathways that are not found in natural populations. Lignin modifications that rely on the biosynthesis of alternative monomers, such as coniferyl ferulate and sinapyl-p-coumarate, can therefore be accomplished via transgenic approaches (Wilkerson et al., 2014; Lapierre et al., 2021). Although small-scale field trials have shown the benefits of lignin engineering (Table 3) (Lapierre et al., 1999; Pilate et al., 2002; Wei et al., 2008; Wang et al., 2012; Tian et al., 2013; Cao et al., 2020; Gui et al., 2020), to our knowledge, no transgenic elite lines with altered lignin are currently grown in commercial forestry applications because of the expensive regulatory burden that was installed for genetically modified plants decades ago and is no longer consistent with current scientific data (Strauss et al., 2015, 2019; Custers et al., 2016).
Genetically improved trees obtained via gene editing have also not yet made it into commercial plantations. Nevertheless, the potential of gene editing to engineer cell wall traits in forest trees is great. From a regulatory standpoint, gene-editing techniques have the advantage over genetic engineering, as there is relatively swift regulation in many regions of the world, including major forestry countries such as Canada and Brazil, for bringing gene-edited trees to the market (Ellens et al., 2019; Nepomuceno et al., 2020; Jenkins et al., 2021). However, at least for now, in the European Union (EU), organisms obtained by gene editing are subject to the same regulatory obligations as those obtained by genetic engineering (Court of Justice of the European Union, 2018). Also, from a technological perspective, gene editing has a number of advantages. Gene editing enables, by definition, more stable engineering than RNAi-based gene downregulation (Van Acker et al., 2013; De Meester et al., 2022a). In addition, individual genes from a gene family can be altered without affecting close gene family members. For instance, when 4CL1 was mutated via CRISPR-Cas9 in poplar, 4CL5 (which shows 89% nucleotide identity with 4CL1) remained unaltered (Zhou et al., 2015), and CSE1 and CSE2 have been independently mutated via CRISPR-Cas9 in poplar despite their 88% nucleotide identity (de Vries et al., 2021a; Jang et al., 2021). Gene editing also enables the stable engineering of weak alleles. For instance, a poplar line with one ccr2 knockout allele and one ccr2 weak allele has a stably reduced lignin content and improved wood processing efficiency (De Meester et al., 2021). Furthermore, genome editing allows multiple genes to be edited simultaneously through multiplex editing; e.g., to edit multiple genes of the same pathway or to stack lignin traits with yield and disease-resistance traits.
However, an important limitation of the use of CRISPR-Cas in trees is that the T-DNA with the CRISPR-Cas sequence, which is currently stably integrated into the genome, must be eliminated before unconfined field release in order to take advantage of the low regulatory burden and swift regulation of gene-edited crops (Dalla Costa et al., 2020). Elimination of the T-DNA via crossing and Mendelian segregation is not preferred because it is time consuming and would break up the genetic constitution of highly heterozygous commercially relevant elite lines, leading to a loss of desired traits (Norelli et al., 2017). More research and development are needed to obtain efficient protocols for generating transgene-free gene-edited trees, as has already been successfully achieved for many other plant species (reviewed in He et al., 2022).
Concluding remarks and perspectives
Modification of lignin to improve wood processing has been successfully achieved in several forest tree species. However, commercialization of trees with reduced lignin levels or modified lignin is hindered by a number of factors. First, lignin engineering often results in lower biomass yields. Future efforts should therefore be made to address this issue, e.g., by tuning enzyme activity to obtain more subtle shifts in lignin levels, by reducing lignin content only in fibers, and by mutating genes located upstream of C4H (thus avoiding the accumulation of phenylpropanoids to potentially toxic levels) or genes encoding transcription factors that affect the entire lignin biosynthetic pathway. A second limitation is the lack of data on field-grown lignin-modified trees. Field trials of the most promising lignin-engineered lines are essential for the translation of basic research into applications. Importantly, in multi-year field trials, gradual fading of the reduction in wood lignin content of trees RNAi-engineered to produce less lignin has often been observed. Field trials of CRISPR-Cas-engineered trees are needed to further investigate whether this is a general phenomenon or whether confounding factors, such as variations in the level of downregulation caused by RNAi, are at play. A third hurdle is the lack of data on lignin engineering in elite lines. Here, establishing transformation protocols for elite lines would be helpful. In addition, protocols for gene editing that avoid the integration of the CRISPR-Cas-encoding DNA would further facilitate field studies and commercialization of new varieties. Furthermore, lignin engineering would benefit from evaluating the effects of stacking different lignin traits and stacking lignin traits with other desirable forestry traits (e.g., yield, wood density, pest resistance). Therefore, multiplex gene-editing strategies in which edits in different genes are stacked in a single elite line, followed by high-throughput screening of the engineered lines for combinations of edits, would further accelerate the development of lignin-engineered commercial forest trees.
Funding
We thank Annick Bleys for preparing this manuscript for submission. B.D.M. is indebted to Research Foundation Flanders (FWO; grant G020618N) and the Energy Transition Fund (ETF) projects AD-LIBIO and ADV_BIO for a postdoctoral fellowship. T.M. is indebted to the ETF project AD-LIBIO for a postdoctoral fellowship. W.B. is additionally indebted to the interuniversity iBOF project Next-BioRef.
Author contributions
B.D.M., T.M., and R.V. wrote this review article. W.B. conceived and supervised the study.
Acknowledgments
Figure 3 was created with BioRender.com. No conflict of interest is declared.
Published: October 27, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
References
- Abu-Omar M.M., Barta K., Beckham G.T., Luterbacher J.S., Ralph J., Rinaldi R., Román-Leshkov Y., Samec J.S.M., Sels B.F., Wang F. Guidelines for performing lignin-first biorefining. Energy Environ. Sci. 2021;14:262–292. [Google Scholar]
- Ahmar S., Ballesta P., Ali M., Mora-Poblete F. Achievements and challenges of genomics-assisted breeding in forest trees: from marker-assisted selection to genome editing. Int. J. Mol. Sci. 2021;22:10583. doi: 10.3390/ijms221910583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki D., Hanaya Y., Akita T., Matsushita Y., Yoshida M., Kuroda K., Yagami S., Takama R., Fukushima K. Distribution of coniferin in freeze-fixed stem of Ginkgo biloba L. by cryo-TOF-SIMS/SEM. Sci. Rep. 2016;6:31525. doi: 10.1038/srep31525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher M., Chabbert B., Pilate G., Van Doorsselaere J., Tollier M.T., Petit-Conil M., Cornu D., Monties B., Van Montagu M., Inzé D., et al. Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in poplar. Plant Physiol. 1996;112:1479–1490. doi: 10.1104/pp.112.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher M., Halpin C., Petit-Conil M., Boerjan W. Lignin: genetic engineering and impact on pulping. Crit. Rev. Biochem. Mol. Biol. 2003;38:305–350. doi: 10.1080/10409230391036757. [DOI] [PubMed] [Google Scholar]
- Berthet S., Demont-Caulet N., Pollet B., Bidzinski P., Cézard L., Le Bris P., Borrega N., Hervé J., Blondet E., Balzergue S., et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell. 2011;23:1124–1137. doi: 10.1105/tpc.110.082792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet S., Thevenin J., Baratiny D., Demont-Caulet N., Debeaujon I., Bidzinski P., Leple J.-C., Huis R., Hawkins S., Gomez L.-D., et al. Role of plant laccases in lignin polymerization. Adv. Bot. Res. 2012;61:145–172. [Google Scholar]
- Blaschek L., Champagne A., Dimotakis C., Nuoendagula Decou R., Decou R., Hishiyama S., Kratzer S., Kajita S., Pesquet E. Cellular and genetic regulation of coniferaldehyde incorporation in lignin of herbaceous and woody plants by quantitative Wiesner staining. Front. Plant Sci. 2020;11:109. doi: 10.3389/fpls.2020.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- Bonawitz N.D., Chapple C. Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr. Opin. Biotechnol. 2013;24:336–343. doi: 10.1016/j.copbio.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Bonawitz N.D., Kim J.I., Tobimatsu Y., Ciesielski P.N., Anderson N.A., Ximenes E., Maeda J., Ralph J., Donohoe B.S., Ladisch M., et al. Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature. 2014;509:376–380. doi: 10.1038/nature13084. [DOI] [PubMed] [Google Scholar]
- Bouvet J.-M., Makouanzi Ekomono C.G., Brendel O., Laclau J.-P., Bouillet J.-P., Epron D. Selecting for water use efficiency, wood chemical traits and biomass with genomic selection in a Eucalyptus breeding program. For. Ecol. Manage. 2020;465:118092. [Google Scholar]
- Bozell J.J., Holladay J.E., Johnson D., White J.F. Top value-added chemicals from biomass - volume II—results of screening for potential candidates from biorefinery lignin. 2007. https://www.pnnl.gov/main/publications/external/technical_reports/PNNL-16983.pdf
- Brown D.M., Goubet F., Wong V.W., Goodacre R., Stephens E., Dupree P., Turner S.R. Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J. 2007;52:1154–1168. doi: 10.1111/j.1365-313X.2007.03307.x. [DOI] [PubMed] [Google Scholar]
- Bryant N.D., Pu Y., Tschaplinski T.J., Tuskan G.A., Muchero W., Kalluri U.C., Yoo C.G., Ragauskas A.J. Transgenic poplar designed for biofuels. Trends Plant Sci. 2020;25:881–896. doi: 10.1016/j.tplants.2020.03.008. [DOI] [PubMed] [Google Scholar]
- Cai Y., Zhang K., Kim H., Hou G., Zhang X., Yang H., Feng H., Miller L., Ralph J., Liu C.-J. Enhancing digestibility and ethanol yield of Populus wood via expression of an engineered monolignol 4-O-methyltransferase. Nat. Commun. 2016;7:11989. doi: 10.1038/ncomms11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Huang C., Luo L., Zheng S., Zhong Y., Sun J., Gui J., Li L. Cell-specific suppression of 4-coumarate-CoA ligase gene reveals differential effect of lignin on cell physiological function in Populus. Front. Plant Sci. 2020;11:589729. doi: 10.3389/fpls.2020.589729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarino I. Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 2019;56:209–214. doi: 10.1016/j.copbio.2018.12.012. [DOI] [PubMed] [Google Scholar]
- Chanoca A., de Vries L., Boerjan W. Lignin engineering in forest trees. Front. Plant Sci. 2019;10:912. doi: 10.3389/fpls.2019.00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Dixon R.A. Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 2007;25:759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- Chen H.-C., Li Q., Shuford C.M., Liu J., Muddiman D.C., Sederoff R.R., Chiang V.L. Membrane protein complexes catalyze both 4-and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc. Natl. Acad. Sci. USA. 2011;108:21253–21258. doi: 10.1073/pnas.1116416109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L., He X., Shu W., Wang L., Tang F. Knockdown of miR393 promotes the growth and biomass production in poplar. Front. Plant Sci. 2021;12:714907. doi: 10.3389/fpls.2021.714907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochard H., Froux F., Mayr S., Coutand C. Xylem wall collapse in water-stressed pine needles. Plant Physiol. 2004;134:401–408. doi: 10.1104/pp.103.028357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman H.D., Samuels A.L., Guy R.D., Mansfield S.D. Perturbed lignification impacts tree growth in hybrid poplar - a function of sink strength, vascular integrity, and photosynthetic assimilation. Plant Physiol. 2008;148:1229–1237. doi: 10.1104/pp.108.125500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corma A., Iborra S., Velty A. Chemical routes for the transformation of biomass into chemicals. Chem. Rev. 2007;107:2411–2502. doi: 10.1021/cr050989d. [DOI] [PubMed] [Google Scholar]
- Court of Justice of the European Union . Court of Justice of the European Union; Luxembourg: 2018. Organisms obtained by mutagenesis are GMOs and are, in principle, subject to the obligations laid down by the GMO Directive.https://curia.europa.eu/jcms/upload/docs/application/pdf/2018-07/cp180111en.pdf [Google Scholar]
- Custers R., Bartsch D., Fladung M., Nilsson O., Pilate G., Sweet J., Boerjan W. EU regulations impede market introduction of GM forest trees. Trends Plant Sci. 2016;21:283–285. doi: 10.1016/j.tplants.2016.01.015. [DOI] [PubMed] [Google Scholar]
- Dalla Costa L., Piazza S., Pompili V., Salvagnin U., Cestaro A., Moffa L., Vittani L., Moser C., Malnoy M. Strategies to produce T-DNA free CRISPRed fruit trees via Agrobacterium tumefaciens stable gene transfer. Sci. Rep. 2020;10:20155. doi: 10.1038/s41598-020-77110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meester B., de Vries L., Özparpucu M., Gierlinger N., Corneillie S., Pallidis A., Goeminne G., Morreel K., De Bruyne M., De Rycke R., et al. Vessel-specific reintroduction of CINNAMOYL-COA REDUCTASE1 (CCR1) in dwarfed ccr1 mutants restores vessel and xylary fiber integrity and increases biomass. Plant Physiol. 2018;176:611–633. doi: 10.1104/pp.17.01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meester B., Madariaga Calderón B., de Vries L., Pollier J., Goeminne G., Van Doorsselaere J., Chen M., Ralph J., Vanholme R., Boerjan W. Tailoring poplar lignin without yield penalty by combining a null and haploinsufficient CINNAMOYL-CoA REDUCTASE2 allele. Nat. Commun. 2020;11:5020. doi: 10.1038/s41467-020-18822-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meester B., Oyarce P., Vanholme R., Van Acker R., Tsuji Y., Vangeel T., Van den Bosch S., Van Doorsselaere J., Sels B., Ralph J., et al. Engineering curcumin biosynthesis in poplar affects lignification and biomass yield. Front. Plant Sci. 2022;13:943349. doi: 10.3389/fpls.2022.943349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meester B., Van Acker R., Wouters M., Traversari S., Steenackers M., Neukermans J., Van Breusegem F., Déjardin A., Pilate G., Boerjan W. Field and saccharification performances of poplars severely downregulated in CAD1. New Phytol. 2022 doi: 10.1111/nph.18366. [DOI] [PubMed] [Google Scholar]
- De Meester B., Vanholme R., de Vries L., Wouters M., Van Doorsselaere J., Boerjan W. Vessel- and ray-specific monolignol biosynthesis as an approach to engineer fiber-hypolignification and enhanced saccharification in poplar. Plant J. 2021;108:752–765. doi: 10.1111/tpj.15468. [DOI] [PubMed] [Google Scholar]
- de Vries L., Brouckaert M., Chanoca A., Kim H., Regner M.R., Timokhin V.I., Sun Y., De Meester B., Van Doorsselaere J., Goeminne G., et al. CRISPR-Cas9 editing of CAFFEOYL SHIKIMATE ESTERASE 1 and 2 shows their importance and partial redundancy in lignification in Populus tremula × P. alba. Plant Biotechnol. J. 2021;19:2221–2234. doi: 10.1111/pbi.13651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries L., Guevara-Rozo S., Cho M., Liu L.-Y., Renneckar S., Mansfield S.D. Tailoring renewable materials via plant biotechnology. Biotechnol. Biofuels. 2021;14:167. doi: 10.1186/s13068-021-02010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries L., MacKay H.A., Smith R.A., Mottiar Y., Karlen S.D., Unda F., Muirragui E., Bingman C., Vander Meulen K., Beebe E.T., et al. pHBMT1, a BAHD-family monolignol acyltransferase, mediates lignin acylation in poplar. Plant Physiol. 2022;188:1014–1027. doi: 10.1093/plphys/kiab546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Río J.C., Rencoret J., Gutiérrez A., Elder T., Kim H., Ralph J. Lignin monomers from beyond the canonical monolignol biosynthetic pathway: another brick in the wall. ACS Sustain. Chem. Eng. 2020;8:4997–5012. [Google Scholar]
- del Río J.C., Rencoret J., Gutiérrez A., Kim H., Ralph J. Unconventional lignin monomers—extension of the lignin paradigm. Adv. Bot. Res. 2022;104:1–39. [Google Scholar]
- Derbyshire P., Ménard D., Green P., Saalbach G., Buschmann H., Lloyd C.W., Pesquet E. Proteomic analysis of microtubule interacting proteins over the course of xylem tracheary element formation in Arabidopsis. Plant Cell. 2015;27:2709–2726. doi: 10.1105/tpc.15.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Houari I., Boerjan W., Vanholme B. Behind the scenes: the impact of bioactive phenylpropanoids on the growth phenotypes of Arabidopsis lignin mutants. Front. Plant Sci. 2021;12:734070. doi: 10.3389/fpls.2021.734070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellens K.W., Levac D., Pearson C., Savoie A., Strand N., Louter J., Tibelius C. Canadian regulatory aspects of gene editing technologies. Transgenic Res. 2019;28:165–168. doi: 10.1007/s11248-019-00153-2. [DOI] [PubMed] [Google Scholar]
- Eudes A., Pereira J.H., Yogiswara S., Wang G., Teixeira Benites V., Baidoo E.E.K., Lee T.S., Adams P.D., Keasling J.D., Loqué D. Exploiting the substrate promiscuity of hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase to reduce lignin. Plant Cell Physiol. 2016;57:568–579. doi: 10.1093/pcp/pcw016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog A.M., Neves L.G., Resende M.F.R., Jr., Vazquez A.I., de los Campos G., Dervinis C., Sykes R., Davis M., Davenport R., Barbazuk W.B., et al. Genome-wide association study reveals putative regulators of bioenergy traits in Populus deltoides. New Phytol. 2017;213:799–811. doi: 10.1111/nph.14154. [DOI] [PubMed] [Google Scholar]
- Fan D., Li C., Fan C., Hu J., Li J., Yao S., Lu W., Yan Y., Luo K. MicroRNA6443-mediated regulation of FERULATE 5-HYDROXYLASE gene alters lignin composition and enhances saccharification in Populus tomentosa. New Phytol. 2020;226:410–425. doi: 10.1111/nph.16379. [DOI] [PubMed] [Google Scholar]
- Fournand D., Cathala B., Lapierre C. Initial steps of the peroxidase-catalyzed polymerization of coniferyl alcohol and/or sinapyl aldehyde: capillary zone electrophoresis study of pH effect. Phytochemistry. 2003;62:139–146. doi: 10.1016/s0031-9422(02)00573-3. [DOI] [PubMed] [Google Scholar]
- Franke R., McMichael C.M., Meyer K., Shirley A.M., Cusumano J.C., Chapple C. Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J. 2000;22:223–234. doi: 10.1046/j.1365-313x.2000.00727.x. [DOI] [PubMed] [Google Scholar]
- Freudenberg K. Biosynthesis and constitution of lignin. Nature. 1959;183:1152–1155. doi: 10.1038/1831152a0. [DOI] [PubMed] [Google Scholar]
- Galán-Martín Á., Tulus V., Díaz I., Pozo C., Pérez-Ramírez J., Guillén-Gosálbez G. Sustainability footprints of a renewable carbon transition for the petrochemical sector within planetary boundaries. One Earth. 2021;4:565–583. [Google Scholar]
- Gallego-Giraldo L., Escamilla-Trevino L., Jackson L.A., Dixon R.A. Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc. Natl. Acad. Sci. USA. 2011;108:20814–20819. doi: 10.1073/pnas.1117873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Giraldo L., Jikumaru Y., Kamiya Y., Tang Y., Dixon R.A. Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.) New Phytol. 2011;190:627–639. doi: 10.1111/j.1469-8137.2010.03621.x. [DOI] [PubMed] [Google Scholar]
- Gallego-Giraldo L., Liu C., Pose-Albacete S., Pattathil S., Peralta A.G., Young J., Westpheling J., Hahn M.G., Rao X., Knox J.P., et al. ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 1 (ADPG1) releases latent defense signals in stems with reduced lignin content. Proc. Natl. Acad. Sci. USA. 2020;117:3281–3290. doi: 10.1073/pnas.1914422117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goralogia G.S., Redick T.P., Strauss S.H. Gene editing in tree and clonal crops: progress and challenges. Vitro Cell Dev. Biol. Plant. 2021;57:683–699. [Google Scholar]
- Gou M., Ran X., Martin D.W., Liu C.-J. The scaffold proteins of lignin biosynthetic cytochrome P450 enzymes. Native Plants. 2018;4:299–310. doi: 10.1038/s41477-018-0142-9. [DOI] [PubMed] [Google Scholar]
- Grabber J.H., Quideau S., Ralph J. p-Coumaroylated syringyl units in maize lignin: implications for β-ether cleavage by thioacidolysis. Phytochemistry. 1996;43:1189–1194. [Google Scholar]
- Grattapaglia D., Resende M.D.V. Genomic selection in forest tree breeding. Tree Genet. Genomes. 2011;7:241–255. [Google Scholar]
- Grattapaglia D., Silva-Junior O.B., Resende R.T., Cappa E.P., Müller B.S.F., Tan B., Isik F., Ratcliffe B., El-Kassaby Y.A. Quantitative genetics and genomics converge to accelerate forest tree breeding. Front. Plant Sci. 2018;9:1693. doi: 10.3389/fpls.2018.01693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grattapaglia D. Twelve years into genomic selection in forest trees: climbing the slope of enlightenment of marker assisted tree breeding. Forests. 2022;13:1554. [Google Scholar]
- Guan M., Li C., Shan X., Chen F., Wang S., Dixon R.A., Zhao Q. Dual mechanisms of coniferyl alcohol in phenylpropanoid pathway regulation. Front. Plant Sci. 2022;13:896540. doi: 10.3389/fpls.2022.896540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra F.P., Wegrzyn J.L., Sykes R., Davis M.F., Stanton B.J., Neale D.B. Association genetics of chemical wood properties in black poplar (Populus nigra) New Phytol. 2013;197:162–176. doi: 10.1111/nph.12003. [DOI] [PubMed] [Google Scholar]
- Gui J., Lam P.Y., Tobimatsu Y., Sun J., Huang C., Cao S., Zhong Y., Umezawa T., Li L. Fibre-specific regulation of lignin biosynthesis improves biomass quality in Populus. New Phytol. 2020;226:1074–1087. doi: 10.1111/nph.16411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J., Luo L., Zhong Y., Sun J., Umezawa T., Li L. Phosphorylation of LTF1, an MYB transcription factor in Populus, acts as a sensory switch regulating lignin biosynthesis in wood cells. Mol. Plant. 2019;12:1325–1337. doi: 10.1016/j.molp.2019.05.008. [DOI] [PubMed] [Google Scholar]
- Guo L., Wang P., Jaini R., Dudareva N., Chapple C., Morgan J.A. Dynamic modeling of subcellular phenylpropanoid metabolism in Arabidopsis lignifying cells. Metab. Eng. 2018;49:36–46. doi: 10.1016/j.ymben.2018.07.003. [DOI] [PubMed] [Google Scholar]
- Ha C.M., Escamilla-Trevino L., Yarce J.C.S., Kim H., Ralph J., Chen F., Dixon R.A. An essential role of caffeoyl shikimate esterase in monolignol biosynthesis in Medicago truncatula. Plant J. 2016;86:363–375. doi: 10.1111/tpj.13177. [DOI] [PubMed] [Google Scholar]
- Ha C.M., Rao X., Saxena G., Dixon R.A. Growth-defense trade-offs and yield loss in plants with engineered cell walls. New Phytol. 2021;231:60–74. doi: 10.1111/nph.17383. [DOI] [PubMed] [Google Scholar]
- He Y., Mudgett M., Zhao Y. Advances in gene editing without residual transgenes in plants. Plant Physiol. 2022;188:1757–1768. doi: 10.1093/plphys/kiab574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoengenaert L., Wouters M., Kim H., De Meester B., Morreel K., Vandersyppe S., Pollier J., Desmet S., Goeminne G., Ralph J., et al. Overexpression of the scopoletin biosynthetic pathway enhances lignocellulosic biomass processin. Sci. Adv. 2022;8:eabo5738. doi: 10.1126/sciadv.abo5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa M., Suzuki S., Umezawa T., Sato Y. Progress of lignification mediated by intercellular transportation of monolignols during tracheary element differentiation of isolated Zinnia mesophyll cells. Plant Cell Physiol. 2001;42:959–968. doi: 10.1093/pcp/pce124. [DOI] [PubMed] [Google Scholar]
- Howe G.T., Jayawickrama K., Kolpak S.E., Kling J., Trappe M., Hipkins V., Ye T., Guida S., Cronn R., Cushman S.A., et al. An Axiom SNP genotyping array for Douglas-fir. BMC Genom. 2020;21:9. doi: 10.1186/s12864-019-6383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Kamimura N., Sakamoto S., Nagano S., Takata N., Liu S., Goeminne G., Vanholme R., Uesugi M., Yamamoto M., et al. Rerouting of the lignin biosynthetic pathway by inhibition of cytosolic shikimate recycling in transgenic hybrid aspen. Plant J. 2022;110:358–376. doi: 10.1111/tpj.15674. [DOI] [PubMed] [Google Scholar]
- Hu W.J., Harding S.A., Lung J., Popko J.L., Ralph J., Stokke D.D., Tsai C.J., Chiang V.L. Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat. Biotechnol. 1999;17:808–812. doi: 10.1038/11758. [DOI] [PubMed] [Google Scholar]
- Huang J., Gu M., Lai Z., Fan B., Shi K., Zhou Y.-H., Yu J.-Q., Chen Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010;153:1526–1538. doi: 10.1104/pp.110.157370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Peng X., Kong L., Wu W., Chen Y., Maravelias C.T. Greenhouse gas emission mitigation potential of chemicals produced from biomass. ACS Sustain. Chem. Eng. 2021;9:14480–14487. [Google Scholar]
- Ikeda T., Takata N., Sakamoto S., Hu S., Hishiyama S., Hishiyama S., Mitsuda N., Boerjan W., Ralph J., Kajita S. Improved chemical pulping and saccharification of a natural mulberry mutant deficient in cinnamyl alcohol dehydrogenase. Holzforschung. 2021;75:968–977. [Google Scholar]
- Isikgor F.H., Becer C.R. Lignocellulosic biomass: a sustainable platform for the production of bio-based chemicals and polymers. Polym. Chem. 2015;6:4497–4559. [Google Scholar]
- Jang H.-A., Bae E.-K., Kim M.-H., Park S.-J., Choi N.-Y., Pyo S.-W., Lee C., Jeong H.-Y., Lee H., Choi Y.-I., et al. CRISPR-knockout of CSE gene improves saccharification efficiency by reducing lignin content in hybrid poplar. Int. J. Mol. Sci. 2021;22:9750. doi: 10.3390/ijms22189750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins D., Dobert R., Atanassova A., Pavely C. Impacts of the regulatory environment for gene editing on delivering beneficial products. In Vitro Cell Dev. Biol. Plant. 2021;57:609–626. doi: 10.1007/s11627-021-10201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia C., Zhao H., Wang H., Xing Z., Du K., Song Y., Wei J. Obtaining the transgenic poplars with low lignin content through down-regulation of 4CL. Chin. Sci. Bull. 2004;49:905–909. [Google Scholar]
- Lu J., Zhao H., Wei J., He Y., Shi C., Wang H., Song Y. Lignin reduction in transgenic poplars by expressing antisense CCoAOMT gene. Prog. Nat. Sci. 2004;14:1060–1063. [Google Scholar]
- Joshi C.P., Thammannagowda S., Fujino T., Gou J.-Q., Avci U., Haigler C.H., McDonnell L.M., Mansfield S.D., Mengesha B., Carpita N.C., et al. Perturbation of wood cellulose synthesis causes pleiotropic effects in transgenic aspen. Mol. Plant. 2011;4:331–345. doi: 10.1093/mp/ssq081. [DOI] [PubMed] [Google Scholar]
- Jouanin L., Goujon T., de Nadaï V., Martin M.-T., Mila I., Vallet C., Pollet B., Yoshinaga A., Chabbert B., Petit-Conil M., et al. Lignification in transgenic poplars with extremely reduced caffeic acid O-methyltransferase activity. Plant Physiol. 2000;123:1363–1374. doi: 10.1104/pp.123.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubany-Marí T., Munné-Bosch S., López-Carbonell M., Alegre L. Hydrogen peroxide is involved in the acclimation of the Mediterranean shrub, Cistus albidus L., to summer drought. J. Exp. Bot. 2009;60:107–120. doi: 10.1093/jxb/ern274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlen S.D., Zhang C., Peck M.L., Smith R.A., Padmakshan D., Helmich K.E., Free H.C.A., Lee S., Smith B.G., Lu F., et al. Monolignol ferulate conjugates are naturally incorporated into plant lignins. Sci. Adv. 2016;2:e1600393. doi: 10.1126/sciadv.1600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Li Q., Karlen S.D., Smith R.A., Shi R., Liu J., Yang C., Tunlaya-Anukit S., Wang J.P., Chang H.-M., et al. Monolignol benzoates incorporate into the lignin of transgenic Populus trichocarpa depleted in C3H and C4H. ACS Sustain. Chem. Eng. 2020;8:3644–3654. [Google Scholar]
- Kim M.H., Cho J.S., Bae E.K., Choi Y.I., Eom S.H., Lim Y.J., Lee H., Park E.J., Ko J.H. PtrMYB120 functions as a positive regulator of both anthocyanin and lignin biosynthetic pathway in a hybrid poplar. Tree Physiol. 2021;41:2409–2423. doi: 10.1093/treephys/tpab082. [DOI] [PubMed] [Google Scholar]
- Kitin P., Voelker S.L., Meinzer F.C., Beeckman H., Strauss S.H., Lachenbruch B. Tyloses and phenolic deposits in xylem vessels impede water transport in low-lignin transgenic poplars: a study by cryo-fluorescence microscopy. Plant Physiol. 2010;154:887–898. doi: 10.1104/pp.110.156224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Morreel K., Lu F., Rencoret J., Carlos Del Río J., Voorend W., Vermerris W., Boerjan W., Ralph J. Maize tricin-oligolignol metabolites and their implications for monocot lignification. Plant Physiol. 2016;171:810–820. doi: 10.1104/pp.16.02012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre C., Pilate G., Pollet B., Mila I., Leplé J.C., Jouanin L., Kim H., Ralph J. Signatures of cinnamyl alcohol dehydrogenase deficiency in poplar lignins. Phytochemistry. 2004;65:313–321. doi: 10.1016/j.phytochem.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Lapierre C., Pollet B., Petit-Conil M., Toval G., Romero J., Pilate G., Leplé J.-C., Boerjan W., Ferret V., De Nadai V., et al. Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have an opposite impact on the efficiency of industrial kraft pulping. Plant Physiol. 1999;119:153–164. doi: 10.1104/pp.119.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre C., Sibout R., Laurans F., Lesage-Descauses M.-C., Déjardin A., Pilate G. p-Coumaroylation of poplar lignins impacts lignin structure and improves wood saccharification. Plant Physiol. 2021;187:1374–1386. doi: 10.1093/plphys/kiab359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedev V.G., Lebedeva T.N., Chernodubov A.I., Shestibratov K.A. Genomic selection for forest tree improvement: methods, achievements and perspectives. Forests. 2020;11:1190. [Google Scholar]
- Lee B.-R., Muneer S., Jung W.-J., Avice J.-C., Ourry A., Kim T.-H. Mycorrhizal colonization alleviates drought-induced oxidative damage and lignification in the leaves of drought-stressed perennial ryegrass (Lolium perenne) Physiol. Plantarum. 2012;145:440–449. doi: 10.1111/j.1399-3054.2012.01586.x. [DOI] [PubMed] [Google Scholar]
- Lee C., Teng Q., Zhong R., Ye Z.-H. Molecular dissection of xylan biosynthesis during wood formation in poplar. Mol. Plant. 2011;4:730–747. doi: 10.1093/mp/ssr035. [DOI] [PubMed] [Google Scholar]
- Lee S., Mo H., Kim J.I., Chapple C. Genetic engineering of Arabidopsis to overproduce disinapoyl esters, potential lignin modification molecules. Biotechnol. Biofuels. 2017;10:40. doi: 10.1186/s13068-017-0725-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplé J.C., Dauwe R., Morreel K., Storme V., Lapierre C., Pollet B., Naumann A., Kang K.-Y., Kim H., Ruel K., et al. Downregulation of cinnamoyl-coenzyme A reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell. 2007;19:3669–3691. doi: 10.1105/tpc.107.054148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhou Y., Cheng X., Sun J., Marita J.M., Ralph J., Chiang V.L. Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc. Natl. Acad. Sci. USA. 2003;100:4939–4944. doi: 10.1073/pnas.0831166100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Kim J.I., Pysh L., Chapple C. Four isoforms of Arabidopsis 4-coumarate:CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 2015;169:2409–2421. doi: 10.1104/pp.15.00838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Koelewijn S.F., Van den Bossche G., Van Aelst J., Van den Bosch S., Renders T., Navare K., Nicolaï T., Van Aelst K., Maesen M., et al. A sustainable wood biorefinery for low-carbon footprint chemicals production. Science. 2020;367:1385–1390. doi: 10.1126/science.aau1567. [DOI] [PubMed] [Google Scholar]
- Liu B., Liu J., Yu J., Wang Z., Sun Y., Li S., Lin Y.-C.J., Chiang V.L., Li W., Wang J.P. Transcriptional reprogramming of xylem cell wall biosynthesis in tension wood. Plant Physiol. 2021;186:250–269. doi: 10.1093/plphys/kiab038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Jiang Y., Jin Y., Wang C., Yang J., Qi H. Drought-induced ABA, H2O2 and JA positively regulate CmCAD genes and lignin synthesis in melon stems. BMC Plant Biol. 2021;21:83. doi: 10.1186/s12870-021-02869-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., Ralph J., Morreel K., Messens E., Boerjan W. Preparation and relevance of a cross-coupling product between sinapyl alcohol and sinapyl p-hydroxybenzoate. Org. Biomol. Chem. 2004;2:2888–2890. doi: 10.1039/B411428K. [DOI] [PubMed] [Google Scholar]
- Lu S., Li Q., Wei H., Chang M.-J., Tunlaya-Anukit S., Kim H., Liu J., Song J., Sun Y.-H., Yuan L., et al. Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. USA. 2013;110:10848–10853. doi: 10.1073/pnas.1308936110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay J.J., O’Malley D.M., Presnell T., Booker F.L., Campbell M.M., Whetten R.W., Sederoff R.R. Inheritance, gene expression, and lignin characterization in a mutant pine deficient in cinnamyl alcohol dehydrogenase. Proc. Natl. Acad. Sci. USA. 1997;94:8255–8260. doi: 10.1073/pnas.94.15.8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon E.L., de Vries L., Jang S.-K., Middar S., Kim H., Unda F., Ralph J., Mansfield S.D. Exogenous chalcone synthase expression in developing poplar xylem incorporates naringenin into lignins. Plant Physiol. 2022;188:984–996. doi: 10.1093/plphys/kiab499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon E.L., Mansfield S.D. Tailor-made trees: engineering lignin for ease of processing and tomorrow's bioeconomy. Curr. Opin. Biotechnol. 2019;56:147–155. doi: 10.1016/j.copbio.2018.10.014. [DOI] [PubMed] [Google Scholar]
- Mansfield S.D., Kang K.-Y., Chapple C. Designed for deconstruction - poplar trees altered in cell wall lignification improve the efficacy of bioethanol production. New Phytol. 2012;194:91–101. doi: 10.1111/j.1469-8137.2011.04031.x. [DOI] [PubMed] [Google Scholar]
- Marco de Lima B., Cappa E.P., Silva-Junior O.B., Garcia C., Mansfield S.D., Grattapaglia D. Quantitative genetic parameters for growth and wood properties in Eucalyptus “urograndis” hybrid using near-infrared phenotyping and genome-wide SNP-based relationships. PLoS One. 2019;14:e0218747. doi: 10.1371/journal.pone.0218747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroni F., Pinosio S., Di Centa E., Jurman I., Boerjan W., Felice N., Cattonaro F., Morgante M. Large-scale detection of rare variants via pooled multiplexed next-generation sequencing: towards next-generation Ecotilling. Plant J. 2011;67:736–745. doi: 10.1111/j.1365-313X.2011.04627.x. [DOI] [PubMed] [Google Scholar]
- Meyermans H., Morreel K., Lapierre C., Pollet B., De Bruyn A., Busson R., Herdewijn P., Devreese B., Van Beeumen J., Marita J.M., et al. Modifications in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J. Biol. Chem. 2000;275:36899–36909. doi: 10.1074/jbc.M006915200. [DOI] [PubMed] [Google Scholar]
- Min T., Hwarari D., Li D., Movahedi A., Yang L. CRISPR-based genome editing and its applications in woody plants. Int. J. Mol. Sci. 2022;23:10175. doi: 10.3390/ijms231710175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K., Ralph J., Kim H., Lu F., Goeminne G., Ralph S., Messens E., Boerjan W. Profiling of oligolignols reveals monolignol coupling conditions in lignifying poplar xylem. Plant Physiol. 2004;136:3537–3549. doi: 10.1104/pp.104.049304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K., Ralph J., Lu F., Goeminne G., Busson R., Herdewijn P., Goeman J.L., Van der Eycken J., Boerjan W., Messens E. Phenolic profiling of caffeic acid O-methyltransferase-deficient poplar reveals novel benzodioxane oligolignols. Plant Physiol. 2004;136:4023–4036. doi: 10.1104/pp.104.049312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottiar Y., Vanholme R., Boerjan W., Ralph J., Mansfield S.D. Designer lignins: harnessing the plasticity of lignification. Curr. Opin. Biotechnol. 2016;37:190–200. doi: 10.1016/j.copbio.2015.10.009. [DOI] [PubMed] [Google Scholar]
- Mottiar Y., Karlen S.D., Goacher R.E., Ralph J., Mansfield S.D. Metabolic engineering of p-hydroxybenzoate in poplar lignin. Plant Biotechnol. J. 2022 doi: 10.1111/pbi.13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchero W., Guo J., DiFazio S.P., Chen J.-G., Ranjan P., Slavov G.T., Gunter L.E., Jawdy S., Bryan A.C., Sykes R., et al. High-resolution genetic mapping of allelic variants associated with cell wall chemistry in Populus. BMC Genom. 2015;16:24. doi: 10.1186/s12864-015-1215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro-Villanueva F., Mao X., Chapple C. Linking phenylpropanoid metabolism, lignin deposition, and plant growth inhibition. Curr. Opin. Biotechnol. 2019;56:202–208. doi: 10.1016/j.copbio.2018.12.008. [DOI] [PubMed] [Google Scholar]
- Nepomuceno A.L., Fuganti-Pagliarini R., Felipe M.S.S., Molinari H.B.C., Velini E.D., Pinto E.R.d.C., Dagli M.L.Z., Andrade Filho G., Fernandes P.M.B. Brazilian biosafety law and the new breeding technologies. Front. Agric. Sci. Eng. 2020;7:204–210. [Google Scholar]
- Norelli J.L., Wisniewski M., Fazio G., Burchard E., Gutierrez B., Levin E., Droby S. Genotyping-by-sequencing markers facilitate the identification of quantitative trait loci controlling resistance to Penicillium expansum in Malus sieversii. PLoS One. 2017;12:e0172949. doi: 10.1371/journal.pone.0172949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani M., Demura T. The quest for transcriptional hubs of lignin biosynthesis: beyond the NAC-MYB-gene regulatory network model. Curr. Opin. Biotechnol. 2019;56:82–87. doi: 10.1016/j.copbio.2018.10.002. [DOI] [PubMed] [Google Scholar]
- Oyarce P., De Meester B., Fonseca F., de Vries L., Goeminne G., Pallidis A., De Rycke R., Tsuji Y., Li Y., Van den Bosch S., et al. Introducing curcumin biosynthesis in Arabidopsis enhances lignocellulosic biomass processing. Native Plants. 2019;5:225–237. doi: 10.1038/s41477-018-0350-3. [DOI] [PubMed] [Google Scholar]
- Peña M.J., Zhong R., Zhou G.-K., Richardson E.A., O'Neill M.A., Darvill A.G., York W.S., Ye Z.-H. Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell. 2007;19:549–563. doi: 10.1105/tpc.106.049320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins M., Smith R.A., Samuels L. The transport of monomers during lignification in plants: anything goes but how? Curr. Opin. Biotechnol. 2019;56:69–74. doi: 10.1016/j.copbio.2018.09.011. [DOI] [PubMed] [Google Scholar]
- Perkins M.L., Schuetz M., Unda F., Chen K.T., Bally M.B., Kulkarni J.A., Yan Y., Pico J., Castellarin S.D., Mansfield S.D., et al. Monolignol export by diffusion down a polymerization-induced concentration gradient. Plant Cell. 2022;34:2080–2095. doi: 10.1093/plcell/koac051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins M.L., Schuetz M., Unda F., Smith R.A., Sibout R., Hoffmann N.J., Wong D.C.J., Castellarin S.D., Mansfield S.D., Samuels L. Dwarfism of high-monolignol Arabidopsis plants is rescued by ectopic LACCASE overexpression. Plant Direct. 2020;4:e00265. doi: 10.1002/pld3.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesquet E., Korolev A.V., Calder G., Lloyd C.W. The microtubule-assosiated protein AtMAP70-5 regulates secondary wall patterning in Arabidopsis wood cells. Curr. Biol. 2010;20:744–749. doi: 10.1016/j.cub.2010.02.057. [DOI] [PubMed] [Google Scholar]
- Pesquet E., Zhang B., Gorzsás A., Puhakainen T., Serk H., Escamez S., Barbier O., Gerber L., Courtois-Moreau C., Alatalo E., et al. Non-cell-autonomous postmortem lignification of tracheary elements in Zinnia elegans. Plant Cell. 2013;25:1314–1328. doi: 10.1105/tpc.113.110593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilate G., Allona I., Boerjan W., Dejardin A., Fladung M., Gallardo F., Häggman H., Jansson S., Van Acker R., Halpin C. IUFRO Tree Biotechnology Conference 2015; Treebiotech2015 Florence, Italy. hal-02741586. 2015. Field trials with genetically engineered forest trees: past experiences and future prospects. [Google Scholar]
- Pilate G., Guiney E., Holt K., Petit-Conil M., Lapierre C., Leplé J.C., Pollet B., Mila I., Webster E.A., Marstorp H.G., et al. Field and pulping performances of transgenic trees with altered lignification. Nat. Biotechnol. 2002;20:607–612. doi: 10.1038/nbt0602-607. [DOI] [PubMed] [Google Scholar]
- Porth I., Klapšte J., Skyba O., Hannemann J., McKown A.D., Guy R.D., DiFazio S.P., Muchero W., Ranjan P., Tuskan G.A., et al. Genome-wide association mapping for wood characteristics in Populus identifies an array of candidate single nucleotide polymorphisms. New Phytol. 2013;200:710–726. doi: 10.1111/nph.12422. [DOI] [PubMed] [Google Scholar]
- Qin S., Fan C., Li X., Li Y., Hu J., Li C., Luo K. LACCASE14 is required for the deposition of guaiacyl lignin and affects cell wall digestibility in poplar. Biotechnol. Biofuels. 2020;13:197. doi: 10.1186/s13068-020-01843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J. In: The Science and Lore of the Plant Cell Wall Biosynthesis, Structure and Function. Hayashi T., editor. Universal Publishers (Brown Walker Press); Boca Raton, FL: 2006. What makes a good monolignol substitute? pp. 285–293. [Google Scholar]
- Ralph J., Kim H., Lu F., Grabber J.H., Leplé J.C., Berrio-Sierra J., Derikvand M.M., Jouanin L., Boerjan W., Lapierre C. Identification of the structure and origin of a thioacidolysis marker compound for ferulic acid incorporation into angiosperm lignins (and an indicator for cinnamoyl CoA reductase deficiency) Plant J. 2008;53:368–379. doi: 10.1111/j.1365-313X.2007.03345.x. [DOI] [PubMed] [Google Scholar]
- Ralph J., Lapierre C., Boerjan W. Lignin structure and its engineering. Curr. Opin. Biotechnol. 2019;56:240–249. doi: 10.1016/j.copbio.2019.02.019. [DOI] [PubMed] [Google Scholar]
- Ralph J., Lundquist K., Brunow G., Lu F., Kim H., Schatz P.F., Marita J.M., Hatfield R.D., Ralph S.A., Christensen J.H., et al. Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochemistry Rev. 2004;3:29–60. [Google Scholar]
- Ralph J., MacKay J.J., Hatfield R.D., O'Malley D.M., Whetten R.W., Sederoff R.R. Abnormal lignin in a loblolly pine mutant. Science. 1997;277:235–239. doi: 10.1126/science.277.5323.235. [DOI] [PubMed] [Google Scholar]
- Rambolarimanana T., Ramamonjisoa L., Verhaegen D., Leong Pock Tsy J.-M., Jacquin L., Cao-Hamadou T.-V., Makouanzi G., Bouvet J.-M. Performance of multi-trait genomic selection for Eucalyptus robusta breeding program. Tree Genet. Genomes. 2018;14:71. [Google Scholar]
- Regner M., Bartuce A., Padmakshan D., Ralph J., Karlen S.D. Reductive cleavage method for quantitation of monolignols and low-abundance monolignol conjugates. ChemSusChem. 2018;11:1600–1605. doi: 10.1002/cssc.201800617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S., Kamimura N., Tokue Y., Nakata M.T., Yamamoto M., Hu S., Masai E., Mitsuda N., Kajita S. Identification of enzymatic genes with the potential to reduce biomass recalcitrance through lignin manipulation in Arabidopsis. Biotechnol. Biofuels. 2020;13:97. doi: 10.1186/s13068-020-01736-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleme M.d.L.S., Cesarino I., Vargas L., Kim H., Vanholme R., Goeminne G., Van Acker R., Fonseca F.C.d.A., Pallidis A., Voorend W., et al. Silencing CAFFEOYL SHIKIMATE ESTERASE affects lignification and improves saccharification in poplar. Plant Physiol. 2017;175:1040–1057. doi: 10.1104/pp.17.00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels A.L., Rensing K.H., Douglas C.J., Mansfield S.D., Dharmawardhana D.P., Ellis B.E. Cellular machinery of wood production: differentiation of secondary xylem in Pinus contorta var. latifolia. Planta. 2002;216:72–82. doi: 10.1007/s00425-002-0884-4. [DOI] [PubMed] [Google Scholar]
- Sawada D., Kalluri U.C., O’Neill H., Urban V., Langan P., Davison B., Pingali S.V. Tension wood structure and morphology conducive for better enzymatic digestion. Biotechnol. Biofuels. 2018;11:44. doi: 10.1186/s13068-018-1043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A.L., Stout J., Weng J.-K., Humphreys J., Ruegger M.O., Chapple C. Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 2009;60:771–782. doi: 10.1111/j.1365-313X.2009.03996.x. [DOI] [PubMed] [Google Scholar]
- Serk H., Gorzsás A., Tuominen H., Pesquet E. Cooperative lignification of xylem tracheary elements. Plant Signal. Behav. 2015;10:e1003753. doi: 10.1080/15592324.2014.1003753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.A., Schuetz M., Karlen S.D., Bird D., Tokunaga N., Sato Y., Mansfield S.D., Ralph J., Samuels A.L. Defining the diverse cell populations contributing to lignification in Arabidopsis stems. Plant Physiol. 2017;174:1028–1036. doi: 10.1104/pp.17.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.A., Schuetz M., Roach M., Mansfield S.D., Ellis B., Samuels L. Neighboring parenchyma cells contribute to Arabidopsis xylem lignification, while lignification of interfascicular fibers is cell autonomous. Plant Cell. 2013;25:3988–3999. doi: 10.1105/tpc.113.117176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenackers W., El Houari I., Baekelandt A., Witvrouw K., Dhondt S., Leroux O., Gonzalez N., Corneillie S., Cesarino I., Inzé D., et al. cis-Cinnamic acid is a natural plant growth-promoting compound. J. Exp. Bot. 2019;70:6293–6304. doi: 10.1093/jxb/erz392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J.J., Akiyama T., Chapple C., Ralph J., Mansfield S.D. The effects on lignin structure of overexpression of ferulate 5-hydroxylase in hybrid poplar. Plant Physiol. 2009;150:621–635. doi: 10.1104/pp.109.137059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout A.T., Davis A.A., Domec J.-C., Yang C., Shi R., King J.S. Growth under field conditions affects lignin content and productivity in transgenic Populus trichocarpa with altered lignin biosynthesis. Biomass Bioenergy. 2014;68:228–239. [Google Scholar]
- Strauss S.H., Boerjan W., Chiang V., Costanza A., Coleman H., Davis J.M., Lu M.Z., Mansfield S.D., Merkle S., Myburg A., et al. Certification for gene-edited forests. Science. 2019;365:767–768. doi: 10.1126/science.aay6165. [DOI] [PubMed] [Google Scholar]
- Strauss S.H., Costanza A., Séguin A. BIOTECHNOLOGY. Genetically engineered trees: paralysis from good intentions. Science. 2015;349:794–795. doi: 10.1126/science.aab0493. [DOI] [PubMed] [Google Scholar]
- Studer M.H., DeMartini J.D., Davis M.F., Sykes R.W., Davison B., Keller M., Tuskan G.A., Wyman C.E. Lignin content in natural Populus variants affects sugar release. Proc. Natl. Acad. Sci. USA. 2011;108:6300–6305. doi: 10.1073/pnas.1009252108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M., Liu Y., Lyu J., Zhao S., Wang Y. Chemical and structural responses to downregulated p-hydroxycinnamoyl-coenzyme A: quinate/shikimate p-hydroxycinnamoyltransferase in poplar cell walls. Front. Plant Sci. 2021;12:679230. doi: 10.3389/fpls.2021.679230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes R.W., Gjersing E.L., Foutz K., Rottmann W.H., Kuhn S.A., Foster C.E., Ziebell A., Turner G.B., Decker S.R., Hinchee M.A.W., et al. Down-regulation of p-coumaroyl quinate/shikimate 3'-hydroxylase (C3'H) and cinnamate 4-hydroxylase (C4H) genes in the lignin biosynthetic pathway of Eucalyptus urophylla x E. grandis leads to improved sugar release. Biotechnol. Biofuels. 2015;8:128. doi: 10.1186/s13068-015-0316-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H.M., Liu S., Hill-Skinner S., Wu W., Reed D., Yeh C.-T., Nettleton D., Schnable P.S. The maize brown midrib2 (bm2) gene encodes a methylenetetrahydrofolate reductase that contributes to lignin accumulation. Plant J. 2014;77:380–392. doi: 10.1111/tpj.12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Xie J., Zhao Y., Lu H., Liu S., Qu L., Li J., Gai Y., Jiang X. Sense-antisense- and RNAi-4CL1 regulate soluble phenolic acids, cell wall components and growth in transgenic Populus tomentosa Carr. Plant Physiol. Biochem. 2013;65:111–119. doi: 10.1016/j.plaphy.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Tobimatsu Y., Schuetz M. Lignin polymerization: how do plants manage the chemistry so well? Curr. Opin. Biotechnol. 2019;56:75–81. doi: 10.1016/j.copbio.2018.10.001. [DOI] [PubMed] [Google Scholar]
- Tokunaga N., Sakakibara N., Umezawa T., Ito Y., Fukuda H., Sato Y. Involvement of extracellular dilignols in lignification during tracheary element differentiation of isolated Zinnia mesophyll cells. Plant Cell Physiol. 2005;46:224–232. doi: 10.1093/pcp/pci017. [DOI] [PubMed] [Google Scholar]
- Tsai C.J., Popko J.L., Mielke M.R., Hu W.-J., Podila G.K., Chiang V.L. Suppression of O-methyltransferase gene by homologous sense transgene in quaking aspen causes red-brown wood phenotypes. Plant Physiol. 1998;117:101–112. doi: 10.1104/pp.117.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.J., Xu P., Xue L.J., Hu H., Nyamdari B., Naran R., Zhou X., Goeminne G., Gao R., Gjersing E., et al. Compensatory guaiacyl lignin biosynthesis at the expense of syringyl lignin in 4CL1-knockout poplar. Plant Physiol. 2020;183:123–136. doi: 10.1104/pp.19.01550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuyama T., Kawai R., Shitan N., Matoh T., Sugiyama J., Yoshinaga A., Takabe K., Fujita M., Yazaki K. Proton-dependent coniferin transport, a common major transport event in differentiating xylem tissue of woody plants. Plant Physiol. 2013;162:918–926. doi: 10.1104/pp.113.214957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S.R., Somerville C.R. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unda F., Mottiar Y., Mahon E.L., Karlen S.D., Kim K.H., Loqué D., Eudes A., Ralph J., Mansfield S.D. A new approach to zip-lignin? 3, 4-Dihydroxybenzoate is compatible with lignification. New Phytol. 2022;235:234–246. doi: 10.1111/nph.18136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Väisänen E., Takahashi J., Obudulu O., Bygdell J., Karhunen P., Blokhina O., Laitinen T., Teeri T.H., Wingsle G., Fagerstedt K.V., et al. Hunting monolignol transporters: membrane proteomics and biochemical transport assays with membrane vesicles of Norway spruce. J. Exp. Bot. 2020;71:6379–6395. doi: 10.1093/jxb/eraa368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker R., Déjardin A., Desmet S., Hoengenaert L., Vanholme R., Morreel K., Laurans F., Kim H., Santoro N., Foster C., et al. Different routes for conifer- and sinapaldehyde and higher saccharification upon deficiency in the dehydrogenase CAD1. Plant Physiol. 2017;175:1018–1039. doi: 10.1104/pp.17.00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker R., Leplé J.C., Aerts D., Storme V., Goeminne G., Ivens B., Légée F., Lapierre C., Piens K., Van Montagu M.C.E., et al. Improved saccharification and ethanol yield from field-grown transgenic poplar deficient in cinnamoyl-CoA reductase. Proc. Natl. Acad. Sci. USA. 2014;111:845–850. doi: 10.1073/pnas.1321673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker R., Vanholme R., Storme V., Mortimer J.C., Dupree P., Boerjan W. Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol. Biofuels. 2013;6:46. doi: 10.1186/1754-6834-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bosch S., Schutyser W., Vanholme R., Driessen T., Koelewijn S.-F., Renders T., De Meester B., Huijgen W.J.J., Dehaen W., Courtin C.M., et al. Reductive lignocellulose fractionation into soluble lignin-derived phenolic monomers and dimers and processable carbohydrate pulps. Energy Environ. Sci. 2015;8:1748–1763. [Google Scholar]
- Doorsselaere J., Baucher M., Chognot E., Chabbert B., Tollier M.-T., Petit-Conil M., Leplé J.C., Pilate G., Cornu D., Monties B., et al. A novel lignin in poplar trees with a reduced caffeic acid 5-hydroxyferulic acid O-methyltransferase activity. Plant J. 1995;8:855–864. [Google Scholar]
- Vanholme B., Cesarino I., Goeminne G., Kim H., Marroni F., Van Acker R., Vanholme R., Morreel K., Ivens B., Pinosio S., et al. Breeding with rare defective alleles (BRDA): a natural Populus nigra HCT mutant with modified lignin as a case study. New Phytol. 2013;198:765–776. doi: 10.1111/nph.12179. [DOI] [PubMed] [Google Scholar]
- Vanholme B., Desmet T., Ronsse F., Rabaey K., Van Breusegem F., De Mey M., Soetaert W., Boerjan W. Towards a carbon-negative sustainable bio-based economy. Front. Plant Sci. 2013;4:174. doi: 10.3389/fpls.2013.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R., Cesarino I., Rataj K., Xiao Y., Sundin L., Goeminne G., Kim H., Cross J., Morreel K., Araujo P., et al. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science. 2013;341:1103–1106. doi: 10.1126/science.1241602. [DOI] [PubMed] [Google Scholar]
- Vanholme R., De Meester B., Ralph J., Boerjan W. Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 2019;56:230–239. doi: 10.1016/j.copbio.2019.02.018. [DOI] [PubMed] [Google Scholar]
- Vanholme R., Storme V., Vanholme B., Sundin L., Christensen J.H., Goeminne G., Halpin C., Rohde A., Morreel K., Boerjan W. A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell. 2012;24:3506–3529. doi: 10.1105/tpc.112.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas L., Cesarino I., Vanholme R., Voorend W., de Lyra Soriano Saleme M., Morreel K., Boerjan W. Improving total saccharification yield of Arabidopsis plants by vessel-specific complementation of caffeoyl shikimate esterase (cse) mutants. Biotechnol. Biofuels. 2016;9:139. doi: 10.1186/s13068-016-0551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaas J.V., Dixon R.A., Chen F., Mansfield S.D., Boerjan W., Ralph J., Crowley M.F., Beckham G.T. Passive membrane transport of lignin-related compounds. Proc. Natl. Acad. Sci. USA. 2019;116:23117–23123. doi: 10.1073/pnas.1904643116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker S.L., Lachenbruch B., Meinzer F.C., Jourdes M., Ki C., Patten A.M., Davin L.B., Lewis N.G., Tuskan G.A., Gunter L., et al. Antisense down-regulation of 4CL expression alters lignification, tree growth, and saccharification potential of field-grown poplar. Plant Physiol. 2010;154:874–886. doi: 10.1104/pp.110.159269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker S.L., Lachenbruch B., Meinzer F.C., Kitin P., Strauss S.H. Transgenic poplars with reduced lignin show impaired xylem conductivity, growth efficiency and survival. Plant Cell Environ. 2011;34:655–668. doi: 10.1111/j.1365-3040.2010.02270.x. [DOI] [PubMed] [Google Scholar]
- Wagner A., Donaldson L., Kim H., Phillips L., Flint H., Steward D., Torr K., Koch G., Schmitt U., Ralph J. Suppression of 4-coumarate-CoA ligase in the coniferous gymnosperm Pinus radiata. Plant Physiol. 2009;149:370–383. doi: 10.1104/pp.108.125765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Xue Y., Chen Y., Li R., Wei J. Lignin modification improves the biofuel production potential in transgenic Populus tomentosa. Ind. Crop. Prod. 2012;37:170–177. [Google Scholar]
- Wang J.P., Chuang L., Loziuk P.L., Chen H., Lin Y.-C., Shi R., Qu G.-Z., Muddiman D.C., Sederoff R.R., Chiang V.L. Phosphorylation is an on/off switch for 5-hydroxyconiferaldehyde O-methyltransferase activity in poplar monolignol biosynthesis. Proc. Natl. Acad. Sci. USA. 2015;112:8481–8486. doi: 10.1073/pnas.1510473112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.P., Matthews M.L., Williams C.M., Shi R., Yang C., Tunlaya-Anukit S., Chen H.-C., Li Q., Liu J., Lin C.-Y., et al. Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat. Commun. 2018;9:1579. doi: 10.1038/s41467-018-03863-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.P., Naik P.P., Chen H.-C., Shi R., Lin C.-Y., Liu J., Shuford C.M., Li Q., Sun Y.-H., Tunlaya-Anukit S., et al. Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin in Populus trichocarpa. Plant Cell. 2014;26:894–914. doi: 10.1105/tpc.113.120881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chao N., Zhang A., Kang J., Jiang X., Gai Y. Systematic analysis and biochemical characterization of the caffeoyl shikimate esterase gene family in poplar. Int. J. Mol. Sci. 2021;22:13366. doi: 10.3390/ijms222413366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yao S., Htet W.P.P.M., Yue Y., Zhang Z., Sun K., Chen S., Luo K., Fan D. MicroRNA828 negatively regulates lignin biosynthesis in stem of Populus tomentosa through MYB targets. Tree Physiol. 2022;42:1646–1661. doi: 10.1093/treephys/tpac023. [DOI] [PubMed] [Google Scholar]
- Wei J., Wang Y., Wang H., Li R., Lin N., Ma R., Qu L., Song Y. Pulping performance of transgenic poplar with depressed caffeoyl-CoA O-methyltransferase. Sci. Bull. 2008;53:3553–3558. [Google Scholar]
- White F.M. McGraw-Hill Higher Education; New York, NY: 2021. Fluid Mechanics. [Google Scholar]
- Widhalm J.R., Gutensohn M., Yoo H., Adebesin F., Qian Y., Guo L., Jaini R., Lynch J.H., McCoy R.M., Shreve J.T., et al. Identification of a plastidial phenylalanine exporter that influences flux distribution through the phenylalanine biosynthetic network. Nat. Commun. 2015;6:8142. doi: 10.1038/ncomms9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson C.G., Mansfield S.D., Lu F., Withers S., Park J.-Y., Karlen S.D., Gonzales-Vigil E., Padmakshan D., Unda F., Rencoret J., et al. Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science. 2014;344:90–93. doi: 10.1126/science.1250161. [DOI] [PubMed] [Google Scholar]
- Xiang Z., Sen S.K., Min D., Savithri D., Lu F., Jameel H., Chiang V., Chang H.-m. Field-grown transgenic hybrid poplar with modified lignin biosynthesis to improve enzymatic saccharification efficiency. ACS Sustain. Chem. Eng. 2017;5:2407–2414. [Google Scholar]
- Xie M., Muchero W., Bryan A.C., Yee K., Guo H.-B., Zhang J., Tschaplinski T.J., Singan V.R., Lindquist E., Payyavula R.S., et al. A 5-enolpyruvylshikimate 3-phosphate synthase functions as a transcriptional repressor in Populus. Plant Cell. 2018;30:1645–1660. doi: 10.1105/tpc.18.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M., Zhang J., Singan V.R., McGranahan M.J., LaFayette P.R., Jawdy S.S., Engle N., Doeppke C., Tschaplinski T.J., Davis M.F., et al. Identification of functional single nucleotide polymorphism of Populus trichocarpa PtrEPSP-TF and determination of its transcriptional effect. Plant Direct. 2020;4:e00178. doi: 10.1002/pld3.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Tomiyama H., Koyama A., Okuizumi H., Liu S., Vanholme R., Goeminne G., Hirai Y., Shi H., Nuoendagula and Takata N., et al. A century-old mystery unveiled: Sekizaisou is a natural lignin mutant. Plant Physiol. 2020;182:1821–1828. doi: 10.1104/pp.19.01467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Liu J., Kim H., Liu B., Huang X., Yang Z., Lin Y.J., Chen H., Yang C., Wang J.P., et al. CAD1 and CCR2 protein complex formation in monolignol biosynthesis in Populus trichocarpa. New Phytol. 2019;222:244–260. doi: 10.1111/nph.15505. [DOI] [PubMed] [Google Scholar]
- Yang F., Mitra P., Zhang L., Prak L., Verhertbruggen Y., Kim J.-S., Sun L., Zheng K., Tang K., Auer M., et al. Engineering secondary cell wall deposition in plants. Plant Biotechnol. J. 2013;11:325–335. doi: 10.1111/pbi.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wang X.-C., Dai M., Chen B., Qiao Y., Deng H., Zhang D., Zhang Y., Villas Bôas de Almeida C.M., Chiu A.S., et al. Shifting from fossil-based economy to bio-based economy: status quo, challenges, and prospects. Energy. 2021;228:120533. [Google Scholar]
- Yao T., Feng K., Xie M., Barros J., Tschaplinski T.J., Tuskan G.A., Muchero W., Chen J.-G. Phylogenetic occurrence of the phenylpropanoid pathway and lignin biosynthesis in plants. Front. Plant Sci. 2021;12:704697. doi: 10.3389/fpls.2021.704697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R., de Oliveira M.V.V., Kleven B., Maeda H.A. The entry reaction of the plant shikimate pathway is subjected to highly complex metabolite-mediated regulation. Plant Cell. 2021;33:671–696. doi: 10.1093/plcell/koaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Sun J., Li L. PtrCel9A6, an endo-1, 4-β-glucanase, is required for cell wall formation during xylem differentiation in Populus. Mol. Plant. 2013;6:1904–1917. doi: 10.1093/mp/sst104. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Yu X.-H., Liu C.-J. The inducible accumulation of cell wall-bound p-hydroxybenzoates is involved in the regulation of gravitropic response of poplar. Front. Plant Sci. 2021;12:755576. doi: 10.3389/fpls.2021.755576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yu X., Lam P.-Y., Zhang K., Tobimatsu Y., Liu C.-J. Monolignol acyltransferase for lignin p-hydroxybenzoylation in Populus. Native Plants. 2021;7:1288–1300. doi: 10.1038/s41477-021-00975-1. [DOI] [PubMed] [Google Scholar]
- Zhong R., Lee C., Ye Z.-H. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 2010;15:625–632. doi: 10.1016/j.tplants.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Zhong R., Morrison W.H., III, Himmelsbach D.S., Poole F.L., Ye Z.H., Ye Z.-H. Essential role of caffeoyl coenzyme A O-methyltransferase in lignin biosynthesis in woody poplar plants. Plant Physiol. 2000;124:563–578. doi: 10.1104/pp.124.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Jacobs T.B., Xue L.J., Harding S.A., Tsai C.J. Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4-coumarate:CoA ligase specificity and redundancy. New Phytol. 2015;208:298–301. doi: 10.1111/nph.13470. [DOI] [PubMed] [Google Scholar]
- Zhou X., Ren S., Lu M., Zhao S., Chen Z., Zhao R., Lv J. Preliminary study of cell wall structure and its mechanical properties of C3H and HCT RNAi transgenic poplar sapling. Sci. Rep. 2018;8:10508. doi: 10.1038/s41598-018-28675-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo C., Wang X., Docampo-Palacios M., Sanders B.C., Engle N.L., Tschaplinski T.J., Hendry J.I., Maranas C.D., Chen F., Dixon R.A. Developmental changes in lignin composition are driven by both monolignol supply and laccase specificity. Sci. Adv. 2022;8:eabm8145. doi: 10.1126/sciadv.abm8145. [DOI] [PMC free article] [PubMed] [Google Scholar]