Abstract
Large-scale nanoarrays of single biomolecules enable high-throughput assays while unmasking the underlying heterogeneity within ensemble populations. Until recently, creating such grids which combine the advantages of microarrays and single-molecule experiments (SMEs) has been particularly challenging due to the mismatch between the size of these molecules and the resolution of top-down fabrication techniques. DNA origami placement (DOP) combines two powerful techniques to address this issue: (i) DNA origami, which provides a ~100 nm self-assembled template for single-molecule organization with 5 nm resolution and (ii) top-down lithography, which patterns these DNA nanostructures, transforming them into functional nanodevices via large-scale integration with arbitrary substrates. Presently, this technique relies on state-of-the-art infrastructure and highly trained personnel, making it prohibitively expensive for researchers. Here, we introduce a cleanroom-free, $1 benchtop technique to create meso-to-macro-scale DNA origami nanoarrays using self-assembled colloidal nanoparticles, thereby circumventing the need for top-down fabrication. We report a maximum yield of 74%, 2-fold higher than the statistical limit of 37% imposed on non-specific molecular loading alternatives. Furthermore, we provide a proof-of-principle for the ability of this nanoarray platform to transform traditionally low-throughput, stochastic, single-molecule assays into high-throughput, deterministic ones, without compromising data quality. Our approach has the potential to democratize single-molecule nanoarrays and demonstrates their utility as a tool for biophysical assays and diagnostics.
Keywords: DNA nanotechnology, DNA origami placement, self-assembly, nanosphere lithography, single molecule experiments, nanoarray, Poisson statistics
Graphical Abstract
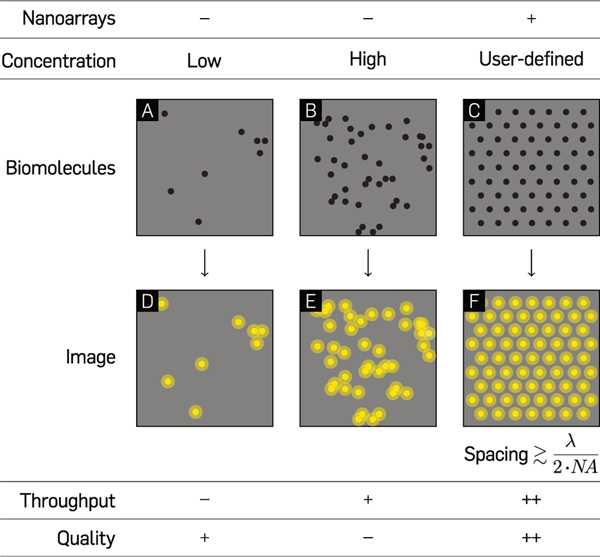
Bulk measurements yield little information about the heterogeneity prevalent at the single-molecule level.1,2 The interest in gaining quantitative and mechanistic insight into these molecular processes spurred the development of novel biophysical and analytical single-molecule methods is over the past few decades.2–4 Since the introduction of total internal reflection fluorescence (TIRF) microscopy,5,6 single-molecule experiments of biomolecular kinetics, conformational fluctuations, and folding mechanisms have become common-place in biophysics laboratories.7,8 Classical single-molecule experiments such as these are stochastic in nature,7–14 with biophysicists lacking the ability to control where individual molecules bind on surfaces. This leads to the possibility that two or more molecules may occupy the same diffraction-limited spot, often leading to confounding data (Figure 1A and B). Reducing the concentration of molecules to overcome this issue has the caveat of lowering experimental throughput. This concentration versus throughput conundrum is a major limitation of conventional single-molecule studies.1 Maximizing throughput while controlling the positions of molecules-of-interest for optimal data quality on a substrate would require close-packing (Figure 1C), ideally at the diffraction limit of light ≃λ/2NA, where λ is the wavelength of excitation light, and NA the numerical aperture of the objective lens. However, any deterministic positioning of molecules requires precise positional control on experimentally relevant substrates, and the size of most single molecules is well below the resolution of current micro-to-nano-manipulation techniques.
Figure 1.
Comparison between experiments with unpatterned molecules and single-molecule nanoarrays. Hypothetical microscopy samples of low (A and D) and high (B and E) concentrations of randomly immobilized single molecules on optical substrates and their corresponding images. (C and F) Single molecule arrays on a patterned glass substrate at an intermolecular distance marginally larger than the diffraction limit of light microscopy.
DNA origami15 is regarded as a molecular breadboard and bridge between the bottom-up world of biochemistry and the top-down world of lithography.16 DNA origami nanotechnology is modular and spatially programmable;17–22 an assembled origami unit is capable of carrying up to 200 individually addressable molecules-of-interest.23–26 In the past decade, origami nanostructures have been utilized for a myriad of applications ranging from electronic27,28 and optical devices,14,26,29,30 to single-molecule biophysics,9–11,31,32 biosensing,33–35 and nanofabrication.36–40 Being synthesized in solution, spatial stochasticity is intrinsically linked with the deposition of planar origami and their payload on glass substrates for optical experiments. A 2D DNA origami nanostructure (~100 nm), however, is more than an order of magnitude larger than other molecules, which makes it amenable to lithographic manipulation and deterministic positioning.
Electron beam lithography-based DNA origami placement (DOP)36–38 leverages the ability of origami nanostructures—through their electrostatic or covalent coupling to mica, glass, silicon, and silicon nitride—to interface biomolecular functional moieties with the outside world for visual probing. A recent application of this method by Gopinath et al.36 demonstrates the large-scale integration of functionalized DNA origami through placement on ~100 nm binding sites with >90% single-binding efficiency for hybrid nanodevice fabrication. Such a composite nano-to-micro-manipulation technique enables bilevel control—first, through the arbitrary decoration of molecules with a resolution of 5 nm on origami nanostructures and, second, by positioning the origami themselves on lithographically patterned sites on a desired substrate. The major drawback of lithographic techniques for origami placement is their high cost owing to the manufacturing complexity of top-down fabrication. The wide-scale utilization of such processes is therefore impractical for scientific research such as biophysics, which traditionally does not use sophisticated top-down nanofabrication.
Bottom-up, self-assembly based approaches have the potential to provide a framework for parallel fabrication of structures from components either too diminutive or innumerable to be handled robotically.41 Such processes were predicted to be a cornerstone of the field of nanotechnology during its nascent stages.42 Self-assembly techniques like nanosphere lithography (NSL), while limited in terms of their ability to create arbitrary shapes, offer a variety of advantages—they are cheap, facilitate fast, parallel-processing, and a variety of crystallization techniques exist for covering arbitrarily large surface topologies.43,44 In NSL, a flat, hydrophilic substrate is coated with a monodisperse colloidal suspension of spheres, and upon drying, a hexagonal-close-packed (HCP) layer called a colloidal crystal mask is formed. Attractive capillary forces and convective nanosphere transport are the dominant factors in the self-assembly process.43 The order and quality of the assembled arrays are substantially affected by the rates of solvent evaporation.45,46 Control over the temperature and the humidity of the system on a slightly tilted substrate can yield colloidal monolayers.47 Methods such as spin-coating,48 Langmuir–Blodgett deposition,49 and controlled evaporation50 have been used to assemble large-scale monolayers of colloidal suspensions.
Here, we present the application of NSL to the controlled placement of DNA origami nanostructures on glass substrates as a framework for the fabrication of large-scale single-molecule nanoarrays. This method for benchtop, cleanroom-free, DNA origami placement in meso-to-macro-scale grids utilizes tunable colloidal nanosphere masks44,51–54 and surface chemistry. This technique is similar to previous work55 which patterned gold nanoparticle arrays, but here we place the emphasis on maximizing single-molecule occupancy. A recent study39 reported DNA origami adsorption in nanohole arrays created using NSL, but critical process steps were reliant on a cleanroom environment, extremely long incubation periods were utilized, and single molecule occupancy was limited to approximately 50%. In the study reported here, we identify the optimal binding site diameter for circular origami and subsequently characterize the single origami binding. We report a maximum efficiency of 74%, 2-fold higher than the Poisson limit of 37% achievable with conventional, stochastic loading of single molecules.56,57 We provide evidence for the utility of our technique by demonstrating data quality comparable with classical, stochastic super-resolution DNA-points accumulation for imaging in nanoscale topography (DNA-PAINT)9–11,13,31 experiments, but with up to an order of magnitude higher throughput. This self-assembly based technique enables the highest 2D packing efficiency and approaches the single-molecule binding yield of top-down electron-beam lithography (EBL) based patterning at ~50× lesser cost and significantly lower complexity. It has the potential to address the concentration vs throughput conundrum in SMEs (Figure 1), and function as a robust platform for deterministic, high-throughput biophysical studies, thereby making DOP more feasible and accessible to the scientific community at large.
RESULTS AND DISCUSSION
Nanosphere Lithography and Surface Chemistry Transform Resource-Intensive DOP into a Facile Bench-top Process.
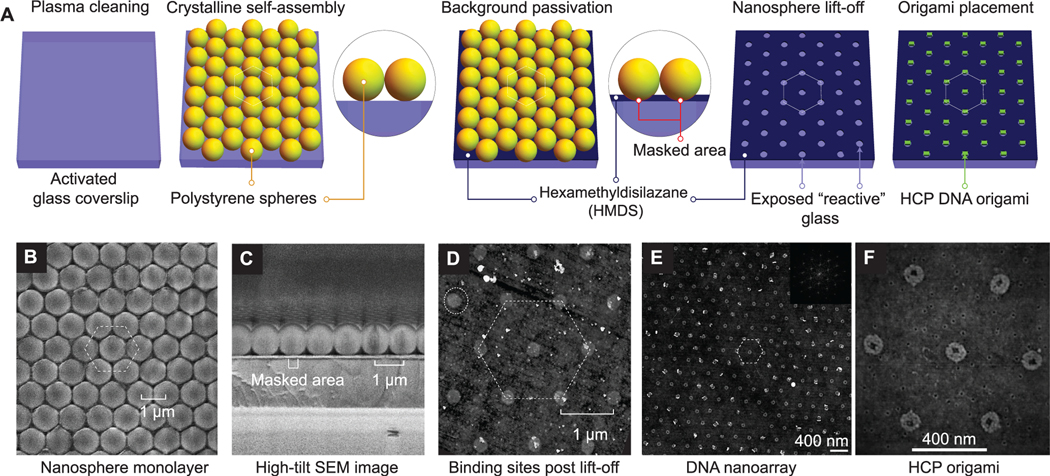
To achieve the highest theoretical packing density, we position DNA origami nanostructures through DOP onto a hexagonal array with a defined spacing that is marginally larger than the wavelength of visible light (Figure 2A). We create binding sites for origami nanostructures through a self-assembly based NSL technique (Figure 2B–F). In a typical experiment, upon drying of polystyrene nanospheres in a solvent-based aqueous solution, a close-packed crystalline layer of colloidal nanoparticles is observed on a slightly tilted (~45°), 1 cm2 hydrophilic glass surface (Figure 2B and C). Cross-sectional scanning electron microscope (SEM) images reveal contact areas between individual nanospheres and the glass substrate that can be utilized as “masks” for bulk vapor-phase passivation with hexamethyldisilazane (HMDS) (Figure 2C), similar to passivation in top-down DOP.37 Subsequent nanosphere “lift-off” by sonication in water results in the creation of nanosphere-dependent binding sites in these masked areas. Finally, controlled origami placement is achieved by tuning pH, Mg2+ concentration, origami concentration, and incubation time as previously reported by Gopinath and Rothemund.37 Results presented hereafter are from experiments performed at optimal values for these parameters. For the close-packed nanoparticles, closer visual inspection using (top-view) SEM images revealed continuous crystalline domains of up to 0.05 mm2 for 1 μm particle diameters (Figure S1). We found that the coverage and uniformity of crystal domains improves with a reduction in particle diameters. Additionally, we find instances of multilayer deposition in the close-packing process. Due to the gaseous nature and minuscule size of HMDS, the additional layers of nanospheres do not affect bulk surface modification.
Figure 2.
Bench-top DNA origami nanoarray fabrication. (A) Schematic illustration of the DNA origami patterning process through 2D nanosphere close-packing, selective passivation, lift-off, and finally, Mg2+-mediated origami placement. (B, C) SEM images of nanosphere close-packing (top view and cross-section), respectively. (D) AFM images of binding sites. (E, F) AFM images of microscale origami placement. (inset) 2D FFT demonstrating close-packing. Experimental results demonstrate data analogous to schematic depiction (A) of process steps.
Nanosphere Diameter Determines the Spacing s, the Binding Site Size a, and the Single DNA Origami Occupancy.
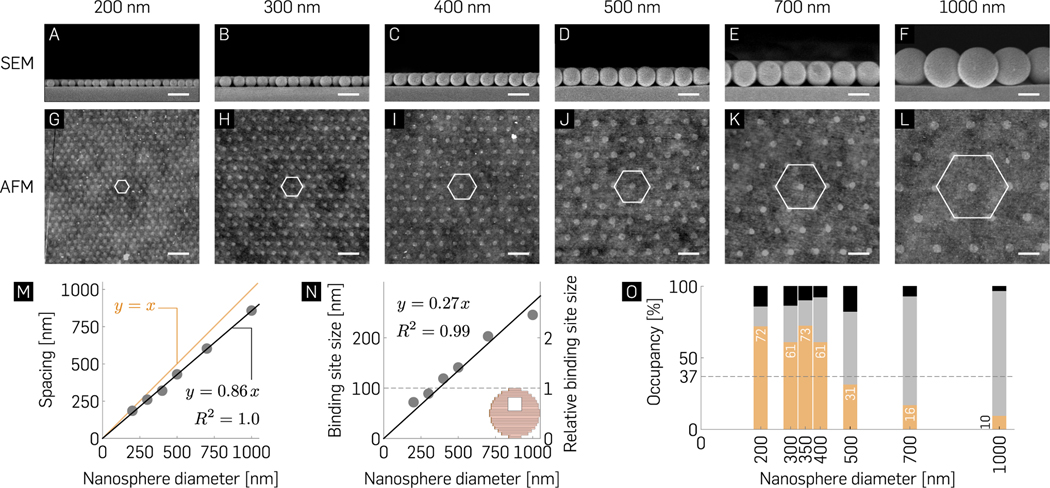
To characterize the size of the binding site as a function of nanosphere size, we imaged the binding site formation for nanospheres with diameters ranging from 200 to 1000 nm using SEM (Figure 3A–F) and atomic force microscopy (AFM, Figure 3G–L). Within our experimental range, the spacing between neighboring binding sites s increases linearly with nanosphere diameter dns (s = 0.86dns; R2 = 1.0; Figure 3M). In an HCP arrangement, each nanosphere makes mechanical contact with its six neighbors. These intermolecular interactions induce deformation and give rise to the less-than 1:1 s/dns ratio. Origami nanostructures align themselves on binding sites to maximize the number of silanol-Mg2+-origami bridges (Figure S2), presumably by a process of 2D diffusion once they land on the surface.37 A size match between the binding site and origami geometry is therefore paramount to maximizing single origami occupancy per binding site through steric occlusion37 (Figure 3N and O). Each nanosphere-glass contact point deforms both bodies, with the nanosphere acting as a mask for subsequent vapor deposition of HMDS (Figure S3). The nanosphere diameter dns and binding site diameter a relationship is defined by a Hertzian contact equation58 below:
Where, Eg and Ens are the elastic moduli and νg and νns are the Poisson’s ratios associated with glass and nanospheres, respectively, and P is the applied intermolecular pressure. The binding site diameter a increases linearly with the nanosphere diameter dns and is given by a = 0.27dns (R2 = 0.99). The linear fit suggests that within the experimental range, the applied pressure P and the mechanical properties of the nanospheres (νns and Ens) are independent of the nanosphere diameter. We find that 100 nm binding sites can be routinely fabricated using nanosphere diameters of 300–400 nm. Therefore, benchtop NSL enables controlled fabrication of arrays of binding sites for the placement of single origami molecules on glass substrates.
Figure 3.
Nanosphere diameter-dependent occupancy statistics. (A–F) Cross-sectional EM images of hexagonal packing with indicated nanospheres reveal their mechanical contact with the glass surface and (G–L) their corresponding origami binding sites observed post lift-off via AFM imaging. Scale bars are 500 nm. (M, N) Spacing and binding site size of nanoarray patterning as a function of indicated nanosphere diameters. (inset) 100 nm circular origami for comparison. (O) Mean percentage binding of zero (black), exactly one (orange), and two or more origami (gray) as a function of nanosphere diameter (N ≥ 600) demonstrating non-Poisson statistics for single molecule binding with maximal 72.4 ± 2.14% and 72 ± 6.84% single origami binding for 350 and 200 nm nanospheres, respectively. The >70% measured probability for single origami binding is ~2× higher than the Poisson limit (horizontal dashed line and Figure S4). Refer to Table S4 for the ± SD of the mean percentage bindings.
Single Origami Binding Statistics—~2× Better than Poisson Statistics and ~50× Cheaper than Top-Down DOP.
Circular origami with a square hole59 (section S1 and Table S2), 100 nm across, were utilized for experiments owing to their geometric similarity with binding sites. The hole served to guide the orientation of DNA origami in previously published DOP work by Gopinath et al.59 Our experimental observations indicate a maximal, 74% single origami occupancy when the origami are 350 nm apart from each other, i.e. at the limit of diffraction for a light microscope. Incubation conditions such as time and origami concentration were altered based on nanosphere diameter used; smaller nanospheres produce a larger number of binding sites and therefore require higher values of both of these parameters. The pH (8.3–8.4) and Mg2+ (40 mM) concentration remained constant for all experiments reported herein. All of the micrographs presented here were obtained via imaging on (ethanol-) dehydrated substrates (section S2). While the efficiency of single-molecule occupancy reported here is lower than the >95% previously reported using EBL, we argue that the ~20% occupancy difference is offset by the technique’s simplicity and ~50X lower cost (Table S3). Results reported here corroborate our prediction that the highest single origami occupancy values would be observed around the 300–400 nm nanosphere diameter range owing to the origami and binding site geometries being almost identical to each other. Further, we find ~100% occupancy of all binding sites under optimal incubation conditions. Similar to the study by Gopinath and Rothemund,37 our measurement statistics likely underestimate the number of single and multiple bindings of origami on the binding sites and are, in fact, a more comprehensive reflection of the fabrication process quality. Comparable to this previous study, we found only a fractional drop in single binding efficiency in slightly undersized sites as a result of using 200 nm nanospheres. The characterization process suggests a trade-off involved in the selection of appropriate nanosphere diameters with respect to the following parameters: throughput, single origami binding efficiency, and diffraction-limited experimental observation. We explored origami placement at a lower Mg2+ (15 mM) concentration, and preliminary results (Figure S5) indicate that origami binding efficiency can be optimized by rationally tuning the primary global parameters. Variability associated with placement results can be attributed, in part, to manual washing steps prior to drying and AFM characterization. An automated washing process was implemented, and preliminary experiments demonstrate 66% single origami occupancy with the automatic wash setup. The setup comprises a peristaltic pump and 3D printed tube holder (Figure S6) for positional alignment between runs without manual intervention.
Nanoarray Platform is Robust at Low Divalent Cation Concentrations.
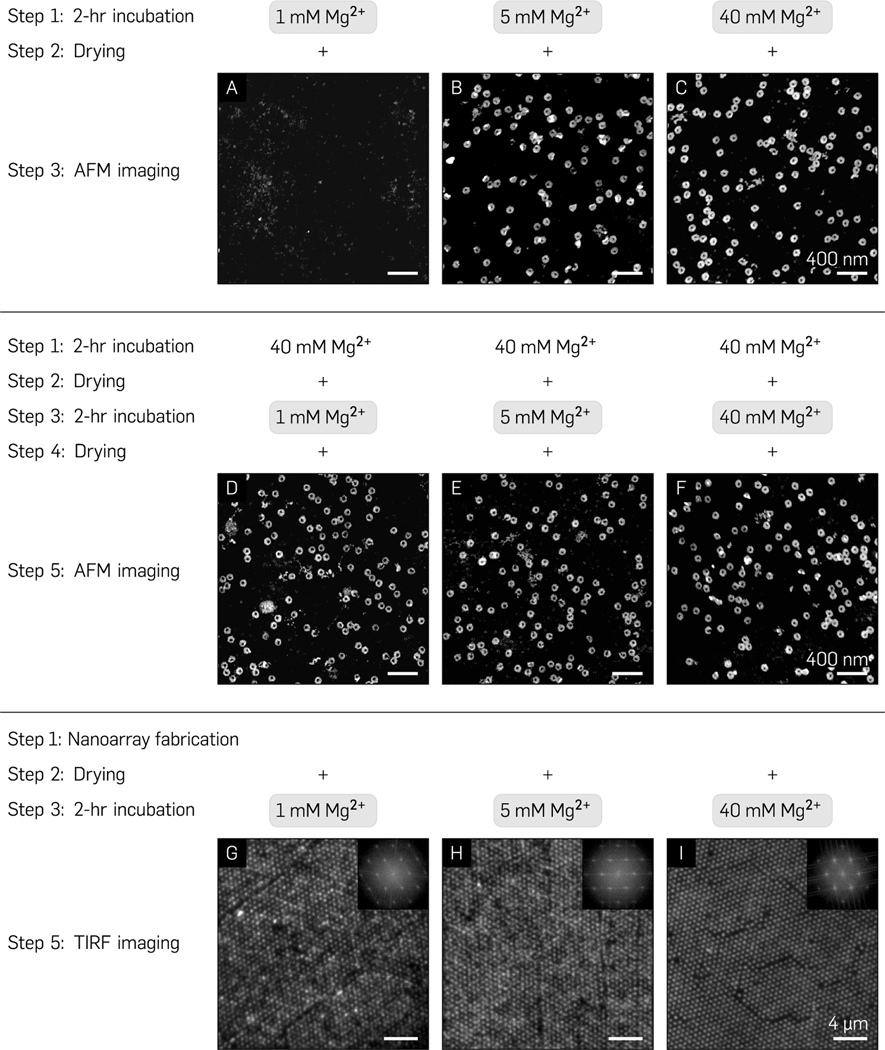
Single-molecule experiments for studying dynamic events are generally performed at less than 10 mM divalent cation concentrations as they are physiologically relevant and minimize the formation of biomolecular aggregates. On the contrary, DNA origami nanostructures are traditionally synthesized and stored in >10 mM Mg2+ buffers for electrostatic screening.60 Therefore, we explored the robustness of the nanoarray platform to assess its relevance for biophysical experiments conducted at low divalent cation concentrations. We first performed control experiments on multiple glass substrates to ascertain the quality of random immobilization of circular origami (250 pM, 30 min incubation) suspended in varying Mg2+ concentrations (1–40 mM) and observed that a minimum of 5 mM Mg2+ was required to stabilize origami on an activated glass surface post-rehydration (Figure 4A–C). Next, we randomly deposited origami suspended in placement buffer (40 mM Mg2+, pH 8.3), (ethanol-) dehydrated the surface for AFM characterization, rehydrated the origami in buffer with 1–40 mM Mg2+ for 2 h, dehydrated once more for imaging, and observed little-to-no apparent change in the quality of origami immobilization under AFM (Figure 4D–F). Finally, to confirm that this process translated effectively to programmatic placement, we patterned origami on a 700 nm “grid” (40 mM Mg2+, pH 8.3, 300 pM), dehydrated the surface, rehydrated for 2 h in 1–40 mM Mg2+ concentrations, and dehydrated in ethanol once more. We observed high-quality grids via fluorescence micrographs as assessed by their 2D Fourier transforms (Figure 4G–I), which confirmed the conservation of spatial conformation over time. These results demonstrate the robustness of this platform at low salt concentrations and validate its use for physiologically relevant single-molecule biophysics experiments. Prior to each dehydration step, the substrate was washed for 1 min in 1× TAE (12.5 mM Mg2+) buffer to remove any non-specifically bound origami.
Figure 4.
Nanoarray platform sensitivity to biologically relevant multivalent cation concentrations. (A-C) AFM images of origami immobilization post ethanol drying and rehydration on an activated glass substrate through direct incubation in 1, 5, and 40 mM Mg2+ buffer, respectively. (D-F) Incubation in 40 mM Mg2+ followed by ethanol drying and 2 h rehydration in 1, 5, and 40 mM Mg2+ and consequent drying. (G-I) TIRF images and their corresponding FFTs (insets) of Mg2+-mediated immobilization on the nanoarray platform with 700 nm pitch at 40 mM Mg2+ followed by a 2 h rehydration in 1, 5, and 40 mM Mg2+ buffers.
Previous DOP studies have demonstrated robustness at low salt conditions through multiple surface chemistry remodeling steps, as well as covalent linkage of origami molecules onto already modified substrates-of-interest.36,37 It is evident that surface chemistry forms the backbone of interactions between origami nanostructures and the substrate for DOP-based biophysical experiments. For simplicity and reproducibility, we optimized all experimental parameters for commercially available glass substrates routinely utilized for single-molecule biophysics experiments. With respect to the sensitivity of the nanoarray to low divalent concentrations, we suspect that the ethanol drying process sequesters and stabilizes Mg2+ bridges between the origami and the silanol groups on the binding sites such that origami are conserved in an entropically favorable energy state, mitigating dissociation or structural disintegration upon rehydration. Subsequent resuspension in lower divalent salt concentration has little-to-no adverse effects on the immobilized origami. To understand the physics underlying the drying process, additional experiments may be necessary. Most SMEs tend to last on the order of a few minutes to tens of minutes rather than a few hours, and they would benefit from the robust nature of this platform. Further proof of functional and structural robustness is found in its long shelf life of several months post-drying at room temperature without the need for sophisticated storage (Figure S7).
Nanoarray Platform Facilitates Optimal Quality, High Throughput, and Deterministic Single-Molecule Biophysics Experiments.
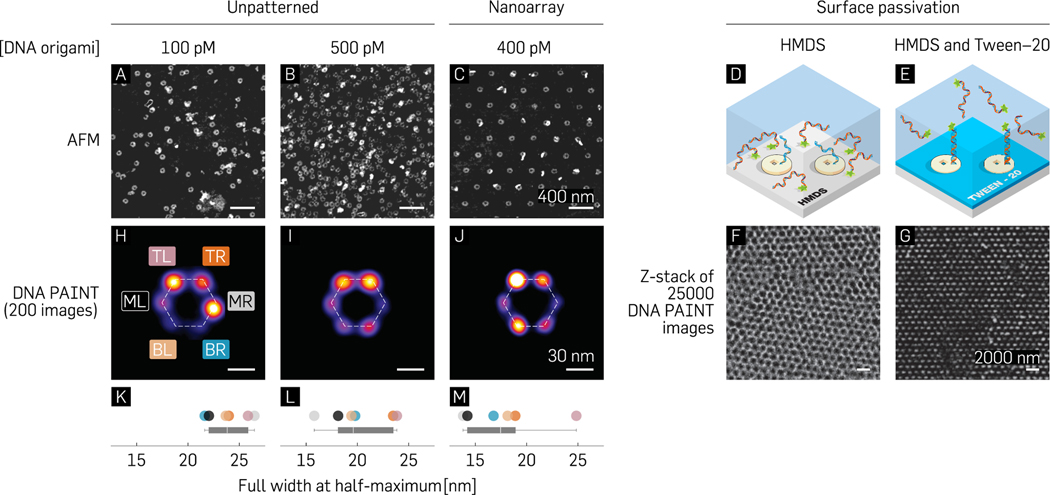
In order to quantify the efficiency of single-molecule incorporation prior to performing biophysics experiments on the nanoarray platform, we designed each origami baseplate to attach six fluorophore-labeled strands (in a hexagonal arrangement). We measured the incorporation efficiency of the designed strands to be 56% (Figure S8 and section S7). Following the photobleaching experiment, we used the optical characterization technique of DNA-PAINT to benchmark the accessibility of biomolecules on the nanoarray platform (Figure 1C). NSL-based placement positions a single origami nanostructure in a diffraction-limited area (Figure S9) 74% of the time as opposed to traditional DNA-PAINT experiments that rely on randomly deposited DNA origami.61 We used particle averaging, previously utilized in DNA-PAINT studies,9,11 as a means to improve image resolution and compare control experiments (stochastic immobilization) against nanoarray-based experiments (deterministic immobilization). We first provide AFM images as evidence that even at low concentrations of origami (100 pM; Figure 5A), it is likely that 2 or more structures could colocalize in a diffraction-limited spot. An increase in concentration to improve throughput results in a higher fraction of structures overlapping each other (500 pM; Figure 5B). However, when patterned on a glass substrate by a distance slightly greater than the diffraction-limit (400 pM, Figure 5C), up to 74% of origami molecules singly occupy individual binding sites (Figure 3O). We arranged three “docking” strands per vertex of a hexagon (Figure S10) to counteract the low conjugation efficiency and transiently bind fluorescently labeled “imager” strands in solution. Control experiments with randomly dispersed origami were first performed to justify conducting DNA-PAINT on origami immobilized through Mg2+-bridges on activated and/or dehydrated glass coverslips. In addition to the HMDS layer (Figure 5D), which is intrinsically part of the nanoarray fabrication process, we passivated the glass surface against nonspecific interactions of fluorescent, ssDNA via a 0.05% (v/v) Tween-20 detergent62 in the 40 mM Mg2+, Tris-HCl “placement” buffer, pH 8.3 (Figure 5E). In the absence of Tween-20 passivation, a honeycomb lattice corresponding to single-stranded imager strands interacting non-specifically with the background was observed (Figure 5F). Therefore, by facilitating specific interactions with the probe strands on origami nanostructures in the binding sites, Tween-20 passivation aided in improving the signal-to-noise ratio (SNR). This technique of passivation enabled DNA-PAINT imaging quality on the nanoarray platform comparable to that routinely reported with PAINT studies using standard imaging and data processing protocols11 (Figure 5G and Movie S1). High-density PAINT experiments were subsequently performed on patterned substrates with inter-origami pitches of 350–400 nm to provide mostly a single origami per binding site and maintain diffraction-limited resolvability of grids.
Figure 5.
(A–C) AFM images contrasting stochastic single-molecule immobilization for low (100 pM) and high (500 pM) origami concentrations with origami deterministically patterned at the diffraction limit (400 pM). (D–G) Schematic representation and experimental results from DNA-PAINT studies without (D and F) and with (E and G) Tween-20 treatment. (H–J) Averaged images of 200 manually picked structures for low and high concentrations of stochastically immobilized origami (H and I) and patterned origami (J) and (K–M) their respective full widths at half-maximum.
Individual structures were averaged using the image processing software, Picasso11 (Figure 5H–J), and their full width at half-maximum (fwhm) measured as a metric to characterize the point spread functions (PSFs) for the “sum” image. A standard analysis pipeline in Picasso comprises drift correction followed by manual or automatic single particle selection/picking and, finally, particle averaging.11 We performed PSF comparisons (Figure 5K–M) between the low (100 pM) and high concentrations (500 pM) of randomly immobilized origami with patterned origami (400 pM) for manually picked structures (Figure 5H–M) and automatically picked structures (Figure S11). We note that all origami used for these experiments broke up–down symmetry (20-T staple strands as previously reported by Gopinath et al.59) and were therefore expected to have specific interactions with the imager strands. We also present a fluorescence micrograph of an exemplary patterned PAINT data set of 11 000 frames (350 nm) collapsed along the Z-axis prior to analysis (Figure S9). To generate an averaged image, Picasso allows manual picking of structures (Figure 5H–J) or automatically picks structures similar to an initial user input of 5–10 structures (Figure S11). The averaging process of multiple structures involves aligning using their center of mass and finally through translational and rotational alignment over multiple iterations.
We picked 200 structures with ≥4 vertices present for the averaged image in each data set (Figure 5H–J). While the manually picked structures from the randomly immobilized origami had PSFs (point spread functions; mean ± SEM) of 23.94 ± 1.69 nm (low) and 20.07 ± 1.42 nm (high), the patterned data exhibited a slightly lower average value of 17.78 ± 1.26 nm (Figure 5K–M). For automatic picking-based averaging, it is plausible that multiple overlapping structures in the high concentration case confound the software’s ability to accurately localize individual origami structures and their associated docking strands, ultimately leading to a degradation in averaged image quality (Figure S11). We hypothesize that the bright points distinctly visible in both automatically as well as manually picked structures could be a combination of two factors: location/sequence-dependent strand conjugation efficiency, as well as random noise. We expect random noise to be a factor especially in the case of automatically picked structures with its source being non-specific interactions with the surface, deformed origami, multiple overlapping origami, or gold nanoparticles used as fiducial markers for drift correction. For the manually picked structures, we expect that the symmetry of the hexagonal pattern contributes to the software localizing certain vertices more brightly than others. This may be due to uncontrollable parameters (scoring function, alignment precision) in the analysis pipeline as well as the low occurrence of structures containing all six vertices. As a quality control check to ascertain that the bright vertices were not solely a random function of the software analysis and might indicate a probability of certain strands being conjugated/accessible more than others, we excluded the “docking” strands for two vertices (Figure S10). We observed that there was a low occurrence of at least one of the locations along the horizontal axis of the origami. Another possible reason for this could be the rotational symmetry along the vertical axis biasing the software rotational alignment to make one vertex brighter than the other. Regardless, our observations provide evidence that the nanoarray platform could serve as a potential solution to the concentration vs throughput conundrum without compromising data quality. Furthermore, due to its intrinsically deterministic nature, it is amenable to software automation for simpler data analysis paradigms. These advantages of the platform could be leveraged for the benefit of a myriad of SMEs, such as single-molecule FRET.7
CONCLUSION
In summary, we have developed a cleanroom-free, DNA origami placement technique which surpasses the single-molecule binding efficiency imposed by Poisson statistics on traditional single-molecule deposition methods. The technique circumvents the need for sophisticated equipment and training previously required for fabricating single-molecule nanoarrays on the meso-to-macro-scale; all at ~$1 per chip. We characterized binding site sizes concomitant with various nanosphere diameters via AFM and EM. This provides a framework for the programmed placement of appropriately sized 2D or 3D DNA nanostructures for various single-molecule applications on addressable glass substrates. We report that a nanosphere diameter of ~350 nm is essential to optimize the binding, and diffraction-limited imaging of single, circular DNA origami nanostructures (~75%) and their associated payloads on high-density grids. We validate the robustness of this platform for in vitro single-molecule experiments under low divalent salt concentrations and demonstrate a shelf life of up to 10 months. We demonstrate the high-throughput and deterministic single-molecule experiments such as super-resolution, traditionally stochastic, DNA-PAINT without compromising data quality. We envision that the platform will be of great utility to biophysics, protein biochemistry, and digital diagnostics owing to its ability to democratize maximum throughput single-molecule experiments with benchtop fabrication in any conventional laboratory setting.
MATERIALS AND METHODS
Origami Formation and Purification.
A circular origami with a square hole was designed using caDNAno http://cadnano.org/ as detailed by Gopinath et al.59 To control the side of the origami that binds to the binding site, all staple ends were positioned on the same face of the origami so that single-stranded 20T extensions to 5′ staple ends would all project from the same face of the origami. Staple strands (640 nM each in water, Integrated DNA Technolgies, Iowa) and the scaffold strand (single-stranded p8064, 100 nM from Tilibit Nano-systems, Germany) were mixed together to target concentrations of 100 nM (each staple) and 20 nM, respectively (a 5:1 staple:scaffold ratio) in 40 mM Tris, 20 mM Acetate, and 1 mM ethylenediaminetetraacetic acid (EDTA) with a typical pH around 8.6 and 12.5 mM magnesium chloride (MgCl2) [1× TAE (12.5 mM Mg2+)]. 100 μL volumes of staple/scaffold mixture were heated to 90 °C for 5 min and annealed from 90 to 25 °C at 0.1 °C/min in a PCR machine. A high concentration of excess staples will compete with DNA origami and inhibit DNA origami placement. Thus, origami were purified away from excess staples using 100 kDa molecular weight cut-offfilters (MWCO) spin filters (Amicon Ultra-0.5 Centrifugal Filter Units with Ultracel-100 membranes, Millipore, UFC510024). Once purified, the origami were stored in 0.5 mL DNA LoBind tubes (Eppendorf) to minimize loss of origami to the sides of the tube.
Fabrication of Origami Binding Sites.
A 1 cm2 glass chip was first washed with isopropanol (IPA) for 2 min. The chip was then blow dried with nitrogen and exposed to a 10 min air plasma cleaning in Harrick plasma cleaner at ~18 W (“High” setting). In an Eppendorf tube, 10 drops (~360 μL suspension) of 1 μm/700 nm/500 nm/400 nm PS nanospheres were poured and gently vortexed before use. The tube was spun at 8000–10 000 rpm for 5 min (faster and/or longer spinning for smaller nanosphere sizes). The supernatant was removed, and 360 μL of ultrapure water was added to resuspend the pellet. The spin process at 8000–10 000 rpm for 5 min was repeated. Finally, the supernatant was removed, and the pellet resuspended in 25% ethanol and 75% water (~3.5× more concentrated, i.e. 100 μL). The tube was vortexed aggressively to resuspend all particles (~6.5e10 particles/mL for 1 μm nanospheres at 1% w/w solids). The nanospheres were drop-cast onto an activated chip surface and dried at a ~45° angle at R.T (resting against a glass stirrer or similar object). Covering the entire surface generally requires 5–10 μL for a 1 cm2 chip. Once dried, a diffraction pattern (crystalline structure) can be observed—confirming the existence of a close-packed monolayer/multilayer of nanospheres. The chip was then heated at 60 °C for 5 min to remove any moisture. A 2 min “descum” plasma in air at ~18 W in Harrick plasma cleaner was used to remove any particulate matter. In a desiccator, 8–10 drops of HMDS were added to a glass cuvette and deposited onto the chip surface under vacuum seal for 20 min. The PS nanospheres were then lifted off the surface with water sonication in a Branson ultrasonic bath for 30–60 s to create origami binding sites. In the absence of an ultrasonic bath, continuous stirring in water for a longer period of time was adequate. The nanospheres visibly came off the surface at the end of this process. Finally, the surface was blown dry with a nitrogen “gun” and baked at 120 °C for 5 min to stabilize the HMDS on the surface.
Placement Protocol.
Chips with appropriately sized binding sites were incubated with ~100–200 pM origami (nominal concentration for 1 μm pitch, concentration inversely proportional to nanosphere size) in ~40 mM Mg, Tris-HCl (40 mM Tris) buffer (pH 8.3) for 60 min. They were subsequently washed in ~40 mM Mg, Tris-HCl (40 mM Tris) buffer (pH 8.3) for 5 min either manually or automatically using a peristaltic pump or shaker in a Petri dish. The chips were then transferred to a buffer solution of ~40 mM Mg, Tris-HCl (40 mM Tris) buffer (pH 8.3) + 0.07% Tween-20 and washed for 5 min. A final 5 min wash step was performed in ~35 mM Mg, 10 mM Tris (pH-8.9) to hydrolyze HMDS and lift off origami nonspecifically bound to the background. For AFM characterization, the chips were transferred to an ethanol drying series: 10 s in 50% ethanol, 20 s in 75% ethanol, and 2 min in 85% ethanol. They were then air-dried, followed by AFM/fluorescence verification of patterning.
Binding Efficiency Characterization.
All AFM images were acquired using a Dimension FastScan Bio (Bruker) using the “short and fat” or “long and thin” ScanAsyst-IN AIR or ScanAsyst-FLUID+ cantilever (“sharp nitride lever” (SNL), 2 nm tip radius, Bruker) in ScanAsyst Air or Fluid mode. All samples were ethanol dried prior to imaging. Single and multiple binding events for placed origami were hand-annotated for origami occupancy statistics and image averaging of arrays (imageJ) was used to determine binding site size. The mean spacing between neighboring binding sites in Figure 3M was measured from the fast Fourier transforms (FFTs) of the corresponding AFM images (Figure 3G–L) using imageJ.
Images of close-packed nanosphere crystals, as well as individual nanosphere cross sections, were obtained using a Hitachi S-4700 field emission scanning electron microscope (ASU Nanofab, Center for Solid State Electronics Research, Tempe, AZ) at 1–5 keV, and the stage (or electron beam) was manipulated as required. In order to prevent charging effects and distortion of the image collected, a sputter coater (Denton Vacuum Desk II, New Jersey) was used to coat the specimen (glass with nanospheres) with gold—palladium (Au-Pd), and carbon tape was used to provide a conduction path from the glass surface to the SEM stub (ground). For the cross-sectional images specifically, the glass coverslip was broken in half post sputter-coating and wedged inside a standard cross-sectional SEM sample holder such that the electron beam impinged directly on the flat edge of the glass coverslip to visualize the contact areas between the nanospheres and the glass surface. Measurements from high-resolution images were made manually using imageJ.
DNA-PAINT and Photobleaching Experiments.
All TIRF experiments were conducted on a benchtop super-resolution Oxford Nanoimager (Oxford, UK). For control DNA-PAINT experiments, a glass chip was activated for 10 min, followed by the creation of a “flow chamber” (using double sticky Kapton polyimide adhesive tape; Amazon) and 30 min incubation of 400 pM DNA origami at 40 mM Mg2+. Nonspecifically bound origami were washed off using several rounds of wicking the incubation buffer through the chamber. Next, a 0.05% Tween-20 (cat no. P1379, Sigma-Aldrich) v/v in 40 mM Mg2+ placement buffer was flown through several times before incubating the solution for 5 min. This prevented nonspecific ssDNA binding during the experiment (Figure 5E and G). Subsequent washing in Tween buffer and placement buffer was followed by the introduction of up to 5 nM P1-imager solution in placement-Tween buffer, 10× dilution of 40 nm gold nanoparticles (fiducials for drift correction, Sigma-Aldrich 741981), and an oxygen scavenging system (2×, 3×, 5× concentrations of PCA, PCD, and Trolox-Quinone, respectively). To ensure the gold nanoparticles settle on the bottom chip, it was taped to a 96-well plate holder in a centrifuge and spun at 150 g for 5 min, ensuring that the inlet and outlet of the flow chamber were sealed prior to spinning. Experiments with patterned chips were conducted by sticking the 10 mm × 10 mm coverslip onto a double-sided sticky Kapton tape and repeating the procedure as outlined above starting with incubation with 0.05% Tween-20 in placement buffer. DNA-PAINT data were analysed using Picasso.11
For photobleaching experiments, intensity traces of origami molecules in response to laser excitation were recorded and steps corresponding to independent stochastic fluorophore quenching events were analyzed. Based on the consequent histogram of the number of fluorophores experimentally found to be incorporated per origami baseplate, the conjugation efficiency was calculated. Photobleaching experiments were performed immediately after grid formation in imaging buffer and oxygen scavenging similar to DNA-PAINT. Fluorophore-labeled strands were added at a concentration between 10 and 100 nM (i.e., 10–100× excess). Laser intensity was adjusted in order to have a slow gradient of fluorophore intensity bleaching to make step-counting easier. Steps were quantified using two methods: imageJ63 and iSMS64 (http://inano.au.dk/about/research-groups/single-molecule-biophotonics-group-victoria-birkedal/software/). For the latter, the field-of-view was cropped, and the two ROIs were aligned to distinctly count the number of steps.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Kennedy and E. Le for support with data collection and H. Sasaki, A. Auer, and R. Jungmann for helpful discussions on DNA-PAINT. This work was supported by a National Institutes of Health Director’s New Innovator Award (1DP2AI144247, to R.F.H.); the Arizona Biomedical Research Consortium (ADHS17–00007401, to R.F.H.); the Office of Naval Research (N00014-17-1-2610 and N00014-18-1-2649, to P.W.K.R.), and the National Science Foundation (CCF-1317694 and CMMI-1636364, to P.W.K.R. and MCB-2027165, to A.G.). AFM data were collected in the lab of H. Yan at Arizona State University. SEM images were acquired at the Center for Solid State and Electronics Research at Arizona State University.
All data supporting the findings of this study are available within the paper and its Supporting Information. All RAW data are available upon request. All sequences of the DNA origami design are included as Supporting Information File 2 and Table S2.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.1c01150.
Detailed materials and methods provide details about origami design, preparation, and purification; experimental protocols and step-by-step workflow for the benchtop fabrication of DNA origami nanoarrays; SEM and AFM characterization protocols; DNA-PAINT and photobleaching experiment protocols; and a guide to troubleshooting the various steps of the fabrication processes (supplementary sections S1–S7). Supplementary Figures S1–S11 provide additional information relevant to the results described in the main text. Supplementary Tables S2–S4 provide details about DNA sequences, process cost, and origami binding statistics (PDF)
Supplementary Movie S1 provides a representative data set for DNA-PAINT imaging on the nanoarray platform.All sequences of the DNA origami design, a staple map, and the caDNAno file are also included (ZIP)
Additional data pertaining to Figure 3, specifically, nanosphere diameter vs. binding site diameter and origami binding statistics are also included (ZIP)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsnano.1c01150
The authors declare the following competing financial interest(s): A US patent application (WO2019108954A1) has been filed based on this work.
Contributor Information
Rishabh M. Shetty, Biodesign Center for Molecular Design and Biomimetics (at the Biodesign Institute) at Arizona State University, Tempe, Arizona 85287, United States; School of Biological and Health Systems Engineering, Arizona State University, Tempe, Arizona 85287, United States Department of Mechanical Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, United States.
Sarah R. Brady, Biodesign Center for Molecular Design and Biomimetics (at the Biodesign Institute) at Arizona State University, Tempe, Arizona 85287, United States
Paul W. K. Rothemund, Department of Bioengineering, Computational and Mathematical Sciences, and Computation and Neural Systems, California Institute of Technology, Pasadena, California 91125, United States
Rizal F. Hariadi, Biodesign Center for Molecular Design and Biomimetics (at the Biodesign Institute) at Arizona State University, Tempe, Arizona 85287, United States; Department of Physics, Arizona State University, Tempe, Arizona 85287, United States.
Ashwin Gopinath, Department of Bioengineering, Computational and Mathematical Sciences, and Computation and Neural Systems, California Institute of Technology, Pasadena, California 91125, United States; Department of Mechanical Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, United States.
REFERENCES
- (1).Leake MC The Physics of Life: One Molecule at a Time. Philos. Trans. R. Soc., B 2013, 368, 20120248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Coelho M; Maghelli N; Tolic-Nørrelykke IM Single-Molecule Imaging in Vivo: The Dancing Building Blocks of the Cell. Integr. Biol 2013, 5 (5), 748–758. [DOI] [PubMed] [Google Scholar]
- (3).Dulin D; Lipfert J; Moolman MC; Dekker NH Studying Genomic Processes at the Single-Molecule Level: Introducing the Tools and Applications. Nat. Rev. Genet 2013, 14 (1), 9–22. [DOI] [PubMed] [Google Scholar]
- (4).Yu Ji. Single-Molecule Studies in Live Cells. Annu. Rev. Phys. Chem 2016, 67, 565–585. [DOI] [PubMed] [Google Scholar]
- (5).Axelrod D. Total Internal Reflection Fluorescence Microscopy in Cell Biology. Traffic 2001, 2 (11), 764–774. [DOI] [PubMed] [Google Scholar]
- (6).Axelrod D; Burghardt TP; Thompson NL Total Internal Reflection Fluorescence. Annu. Rev. Biophys. Bioeng 1984, 13 (1), 247–268. [DOI] [PubMed] [Google Scholar]
- (7).Ha T. Single-Molecule Fluorescence Resonance Energy Transfer. Methods 2001, 25 (1), 78–86. [DOI] [PubMed] [Google Scholar]
- (8).Roy R; Hohng S; Ha T. A Practical Guide to Single-Molecule FRET. Nat. Methods 2008, 5 (6), 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Dai M; Jungmann R; Yin P. Optical Imaging of Individual Biomolecules in Densely Packed Clusters. Nat. Nanotechnol 2016, 11 (9), 798–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jungmann R; Avendaño MS; Dai M; Woehrstein JB; Agasti SS; Feiger Z; Rodal A; Yin P. Quantitative Super-Resolution Imaging with qPAINT. Nat. Methods 2016, 13 (5), 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Schnitzbauer J; Strauss MT; Schlichthaerle T; Schueder F; Jungmann R. Super-Resolution Microscopy with DNA-PAINT. Nat. Protoc 2017, 12 (6), 1198–1228. [DOI] [PubMed] [Google Scholar]
- (12).Stehr F; Stein J; Schueder F; Schwille P; Jungmann R. Flat-Top TIRF Illumination Boosts DNA-PAINT Imaging and Quantification. Nat. Commun 2019, 10 (1), 1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Strauss MT; Schueder F; Haas D; Nickels PC; Jungmann R. Quantifying Absolute Addressability in DNA Origami with Molecular Resolution. Nat. Commun 2018, 9 (1), 1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Acuna GP; Möller FM; Holzmeister P; Beater S; Lalkens B; Tinnefeld P. Fluorescence Enhancement at Docking Sites of DNA-Directed Self-Assembled Nanoantennas. Science 2012, 338 (6106), 506–510. [DOI] [PubMed] [Google Scholar]
- (15).Rothemund PWK Folding DNA to Create Nanoscale Shapes and Patterns. Nature 2006, 440 (7082), 297–302. [DOI] [PubMed] [Google Scholar]
- (16).Xu A; Harb JN; Kostiainen MA; Hughes WL; Woolley AT; Liu H; Gopinath A. DNA Origami: The Bridge from Bottom to Top. MRS Bull. 2017, 42 (12), 943–950. [Google Scholar]
- (17).Pinheiro AV; Han D; Shih WM; Yan H. Challenges and Opportunities for Structural DNA Nanotechnology. Nat. Nanotechnol 2011, 6 (12), 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Basu AS Digital Assays Part II: Digital Protein and Cell Assays. SLAS Technol. 2017, 22 (4), 387–405. [DOI] [PubMed] [Google Scholar]
- (19).Bald I; Keller A. Molecular Processes Studied at a Single-Molecule Level Using DNA Origami Nanostructures and Atomic Force Microscopy. Molecules 2014, 19 (9), 13803–13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Heucke SF; Baumann F; Acuna GP; Severin PMD; Stahl SW; Strackharn M; Stein IH; Altpeter P; Tinnefeld P; Gaub HE Placing Individual Molecules in the Center of Nanoapertures. Nano Lett. 2014, 14 (2), 391–395. [DOI] [PubMed] [Google Scholar]
- (21).Pibiri E; Holzmeister P; Lalkens B; Acuna GP; Tinnefeld P. Single-Molecule Positioning in Zeromode Waveguides by DNA Origami Nanoadapters. Nano Lett. 2014, 14 (6), 3499–3503. [DOI] [PubMed] [Google Scholar]
- (22).Bui H; Onodera C; Kidwell C; Tan Y; Graugnard E; Kuang W; Lee J; Knowlton WB; Yurke B; Hughes WL Programmable Periodicity of Quantum Dot Arrays with DNA Origami Nanotubes. Nano Lett. 2010, 10 (9), 3367–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Hariadi RF; Sommese RF; Sivaramakrishnan S. Tuning Myosin-Driven Sorting on Cellular Actin Networks. eLife 2015, 4, No. e05472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ko SH; Du K; Liddle JA Quantum-Dot Fluorescence Lifetime Engineering with DNA Origami Constructs. Angew. Chem., Int. Ed 2013, 52 (4), 1193–1197. [DOI] [PubMed] [Google Scholar]
- (25).Kuhler P; Roller E-M; Schreiber R; Liedl T; Lohmuller T; Feldmann J. Plasmonic DNA-Origami Nanoantennas for Surface-Enhanced Raman Spectroscopy. Nano Lett. 2014, 14 (5), 2914–2919. [DOI] [PubMed] [Google Scholar]
- (26).Pal S; Deng Z; Ding B; Yan H; Liu Y. DNA-Origami-Directed Self-Assembly of Discrete Silver-Nanoparticle Architectures. Angew. Chem., Int. Ed 2010, 49 (15), 2700–2704. [DOI] [PubMed] [Google Scholar]
- (27).Geng C; Wei T; Wang X; Shen D; Hao Z; Yan Q. Enhancement of Light Output Power from LEDs Based on Monolayer Colloidal Crystal. Small 2014, 10 (9), 1668–1686. [DOI] [PubMed] [Google Scholar]
- (28).Ding B; Deng Z; Yan H; Cabrini S; Zuckermann RN; Bokor J. Gold Nanoparticle Self-Similar Chain Structure Organized by DNA Origami. J. Am. Chem. Soc 2010, 132 (10), 3248–3249. [DOI] [PubMed] [Google Scholar]
- (29).Dutta PK; Varghese R; Nangreave J; Lin S; Yan H; Liu Y. DNA-Directed Artificial Light-Harvesting Antenna. J. Am. Chem. Soc 2011, 133 (31), 11985–11993. [DOI] [PubMed] [Google Scholar]
- (30).Kuzyk A; Schreiber R; Fan Z; Pardatscher G; Roller E-M; Hogele A; Simmel FC; Govorov AO; Liedl T. DNA-Based SelfAssembly of Chiral Plasmonic Nanostructures with Tailored Optical Response. Nature 2012, 483 (7389), 311–314. [DOI] [PubMed] [Google Scholar]
- (31).Heydarian H; Schueder F; Strauss MT; van Werkhoven B; Fazel M; Lidke KA; Jungmann R; Stallinga S; Rieger B. Template-Free 2D Particle Fusion in Localization Microscopy. Nat. Methods 2018, 15 (10), 781–784. [DOI] [PubMed] [Google Scholar]
- (32).Shrestha P; Emura T; Koirala D; Cui Y; Hidaka K; Maximuck WJ; Endo M; Sugiyama H; Mao H. Mechanical Properties of DNA Origami Nanoassemblies Are Determined by Holliday Junction Mechanophores. Nucleic Acids Res. 2016, 44 (14), 6574–6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Godonoga M; Lin T-Y; Oshima A; Sumitomo K; Tang MSL; Cheung Y-W; Kinghorn AB; Dirkzwager RM; Zhou C; Kuzuya A; Tanner JA; Heddle JG A DNA Aptamer Recognising a Malaria Protein Biomarker can Function as Part of a DNA Origami Assembly. Sci. Rep 2016, 6, 21266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ochmann SE; Vietz C; Trofymchuk K; Acuna GP; Lalkens B; Tinnefeld P. Optical Nanoantenna for Single Molecule-Based Detection of Zika Virus Nucleic Acids without Molecular Multiplication. Anal. Chem 2017, 89 (23), 13000–13007. [DOI] [PubMed] [Google Scholar]
- (35).Voigt NV; Tørring T; Rotaru A; Jacobsen MF; Ravnsbæk JB; Subramani R; Mamdouh W; Kjems J; Mokhir A; Besenbacher F; Gothelf KV Single-Molecule Chemical Reactions on DNA Origami. Nat. Nanotechnol 2010, 5 (3), 200–203. [DOI] [PubMed] [Google Scholar]
- (36).Gopinath A; Miyazono E; Faraon A; Rothemund PWK Engineering and Mapping Nanocavity Emission via Precision Placement of DNA Origami. Nature 2016, 535 (7612), 401–405. [DOI] [PubMed] [Google Scholar]
- (37).Gopinath A; Rothemund PWK Optimized Assembly and Covalent Coupling of Single-Molecule DNA Origami Nanoarrays. ACS Nano 2014, 8 (12), 12030–12040. [DOI] [PubMed] [Google Scholar]
- (38).Kershner RJ; Bozano LD; Micheel CM; Hung AM; Fornof AR; Cha JN; Rettner CT; Bersani M; Frommer J; Rothemund PWK; Wallraff GM Placement and Orientation of Individual DNA Shapes on Lithographically Patterned Surfaces. Nat. Nanotechnol 2009, 4 (9), 557–561. [DOI] [PubMed] [Google Scholar]
- (39).Brassat K; Ramakrishnan S; Bürger J; Hanke M; Doostdar M; Lindner JKN; Grundmeier G; Keller A. On the Adsorption of DNA Origami Nanostructures in Nanohole Arrays. Langmuir 2018, 34 (49), 14757–14765. [DOI] [PubMed] [Google Scholar]
- (40).Takabayashi S; Kotani S; Flores-Estrada J; Spears E; Padilla JE; Godwin LC; Graugnard E; Kuang W; Sills S; Hughes WL Boron-Implanted Silicon Substrates for Physical Adsorption of DNA Origami. Int. J. Mol. Sci 2018, 19 (9), 2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Whitesides GM Self-Assembly at All Scales. Science 2002, 295 (5564), 2418–2421. [DOI] [PubMed] [Google Scholar]
- (42).Whitesides G; Mathias J; Seto CT Molecular Self-Assembly and Nanochemistry: A Chemical Strategy for the Synthesis of Nanostructures. Science 1991, 254 (5036), 1312–1319. [DOI] [PubMed] [Google Scholar]
- (43).Vogel N; Weiss CK; Landfester K. From Soft to Hard: The Generation of Functional and Complex Colloidal Monolayers for Nanolithography. Soft Matter 2012, 8 (15), 4044–4061. [Google Scholar]
- (44).Vogel N; Retsch M; Fustin C-A; del Campo A; Jonas U. Advances in Colloidal Assembly: The Design of Structure and Hierarchy in Two and Three Dimensions. Chem. Rev 2015, 115 (13), 6265–6311. [DOI] [PubMed] [Google Scholar]
- (45).Kralchevsky PA; Denkov ND Capillary Forces and Structuring in Layers of Colloid Particles. Curr. Opin. Colloid Interface Sci 2001, 6 (4), 383–401. [Google Scholar]
- (46).Denkov N; Velev O; Kralchevski P; Ivanov I; Yoshimura H; Nagayama K. Mechanism of Formation of Two-Dimensional Crystals from Latex Particles on Substrates. Langmuir 1992, 8 (12), 3183–3190. [Google Scholar]
- (47).Micheletto R; Fukuda H; Ohtsu M. A Simple Method for the Production of a Two-Dimensional, Ordered Array of Small Latex Particles. Langmuir 1995, 11 (9), 3333–3336. [Google Scholar]
- (48).Colson P; Cloots R; Henrist C. Experimental Design Applied to Spin Coating of 2D Colloidal Crystal Masks: A Relevant Method? Langmuir 2011, 27 (21), 12800–12806. [DOI] [PubMed] [Google Scholar]
- (49).Grandidier J; Weitekamp RA; Deceglie MG; Callahan DM; Battaglia C; Bukowsky CR; Ballif C; Grubbs RH; Atwater HA Solar Cell Efficiency Enhancement via Light Trapping in Printable Resonant Dielectric Nanosphere Arrays. Phys. Status Solidi A 2013, 210 (2), 255–260. [Google Scholar]
- (50).Zhang L; Xiong Y. Rapid Self-Assembly of Submicrospheres at Liquid Surface by Controlling Evaporation and Its Mechanism. J. Colloid Interface Sci 2007, 306 (2), 428–432. [DOI] [PubMed] [Google Scholar]
- (51).Colson P; Henrist C; Cloots R. Nanosphere Lithography: A Powerful Method for the Controlled Manufacturing of Nanomaterials. J. Nanomater 2013, 21, 1. [Google Scholar]
- (52).Deckman HW; Dunsmuir JH Applications of Surface Textures Produced with Natural Lithography. J. Vac. Sci. Technol., B: Microelectron. Process. Phenom 1983, 1 (4), 1109–1112. [Google Scholar]
- (53).Wu Y; Zhang C; Yuan Y; Wang Z; Shao W; Wang H; Xu X. Fabrication of Wafer-Size Monolayer Close-Packed Colloidal Crystals via Slope Self-Assembly and Thermal Treatment. Langmuir 2013, 29 (46), 14017–14023. [DOI] [PubMed] [Google Scholar]
- (54).Ye X; Qi L. Two-Dimensionally Patterned Nanostructures Based on Monolayer Colloidal Crystals: Controllable Fabrication, Assembly, and Applications. Nano Today 2011, 6 (6), 608–631. [Google Scholar]
- (55).Li J-R; Lusker KL; Yu J-J; Garno JC Engineering the Spatial Selectivity of Surfaces at the Nanoscale Using Particle Lithography Combined with Vapor Deposition of Organosilanes. ACS Nano 2009, 3 (7), 2023–2035. [DOI] [PubMed] [Google Scholar]
- (56).Korlach J; Marks PJ; Cicero RL; Gray JJ; Murphy DL; Roitman DB; Pham TT; Otto GA; Foquet M; Turner SW Selective Aluminum Passivation for Targeted Immobilization of Single DNA Polymerase Molecules in Zero-Mode Waveguide Nanostructures. Proc. Natl. Acad. Sci. U. S. A 2008, 105 (4), 1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Beer NR; Hindson BJ; Wheeler EK; Hall SB; Rose KA; Kennedy IM; Colston BW On-Chip, Real-Time, Single-Copy Polymerase Chain Reaction in Picoliter Droplets. Anal. Chem 2007, 79 (22), 8471–8475. [DOI] [PubMed] [Google Scholar]
- (58).Hertz HR Ueber Die Beruehrung Elastischer Koerper (On Contact between Elastic Bodies). Gesammelte werke (Collected works) 1882, 156. [Google Scholar]
- (59).Gopinath A; Thachuk C; Mitskovets A; Atwater HA; Kirkpatrick D; Rothemund PWK Absolute and Arbitrary Orientation of Single-Molecule Shapes. Science 2021, 371 (6531), eabd6179. [DOI] [PubMed] [Google Scholar]
- (60).Kielar C; Xin Y; Shen B; Kostiainen MA; Grundmeier G; Linko V; Keller A. On the Stability of DNA Origami Nanostructures in Low-Magnesium Buffers. Angew. Chem., Int. Ed 2018, 57 (30), 9470–9474. [DOI] [PubMed] [Google Scholar]
- (61).Jungmann R; Avendaño MS; Woehrstein JB; Dai M; Shih WM; Yin P. Multiplexed 3d Cellular Super-Resolution Imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 2014, 11 (3), 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Pan H; Xia Y; Qin M; Cao Y; Wang W. A Simple Procedure to Improve the Surface Passivation for Single Molecule Fluorescence Studies. Phys. Biol 2015, 12 (4), 045006. [DOI] [PubMed] [Google Scholar]
- (63).Schindelin J; Arganda-Carreras I; Frise E; Kaynig V; Longair M; Pietzsch T; Preibisch S; Rueden C; Saalfeld S; Schmid B; Tinevez J-Y; White DJ; Hartenstein V; Eliceiri K; Tomancak P; Cardona A. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9 (7), 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Preus S; Noer SL; Hildebrandt LL; Gudnason D; Birkedal V. iSMS: Single-Molecule FRET Microscopy Software. Nat. Methods 2015, 12 (7), 593–594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.