SUMMARY
Mammalian developmental and disease-associated genes concentrate large quantities of the transcriptional machinery by forming membrane-less compartments known as transcriptional condensates. However, it is unknown whether these structures are evolutionarily conserved or involved in 3D genome reorganization. Here, we identify inducible transcriptional condensates in the yeast heat shock response (HSR). HSR condensates are biophysically dynamic spatiotemporal clusters of the sequence-specific transcription factor Heat shock factor 1 (Hsf1) with Mediator and RNA Pol II. Uniquely, HSR condensates drive the coalescence of multiple Hsf1 target genes, even those located on different chromosomes. Binding of the chaperone Hsp70 to a site on Hsf1 represses clustering, while an intrinsically disordered region on Hsf1 promotes condensate formation and intergenic interactions. Mutation of both Hsf1 determinants reprograms HSR condensates to become constitutively active without intergenic coalescence, which comes at a fitness cost. These results suggest that transcriptional condensates are ancient and flexible compartments of eukaryotic gene control.
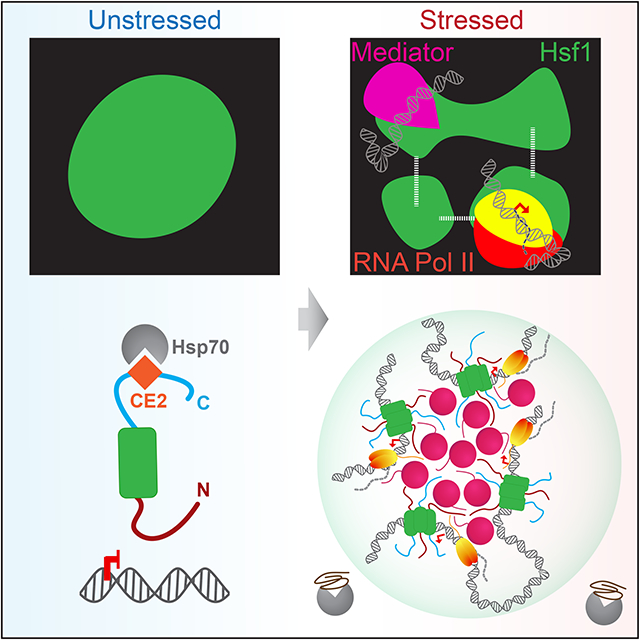
Graphical Abstract
eTOC blurb
Chowdhary et al. show that the transcription factor Hsf1, together with Mediator and Pol II, forms dynamic transcriptional condensates to drive 3D genome reorganization that promotes cellular fitness during stress. They demonstrate that transcriptional condensates can exist outside of the mammalian lineage and represent conserved elements of eukaryotic gene control
INTRODUCTION
Eukaryotes have evolved specialized mechanisms to ensure robust expression of critically important genes such as those activated to specify cell lineage and in response to stress. Mammalian cells utilize clusters of transcription factor binding sites, termed “super-enhancers” (SEs), to drive high-level gene expression by concentrating a large fraction of transcriptional activators and coactivators into extensive DNA looping networks (Hnisz et al., 2013; Whyte et al., 2013; Beagrie et al., 2017; Dowen et al., 2014; Huang et al., 2018). Recent studies suggest that SEs function through cooperative assembly of the transcriptional apparatus into “biomolecular condensates” (Boija et al., 2018; Guo et al., 2019; Sabari et al., 2018), defined as self-organized membrane-free compartments that concentrate specific biomolecules (Banani et al., 2017). Transcriptional condensates at SEs are hypothesized to drive transcriptional bursting of associated genes (Henninger et al., 2021; Hnisz et al., 2017; Sabari et al., 2018).
Biomolecular condensates – and transcriptional condensates in particular – are commonly established and maintained by multivalent interactions. In many cases, multivalent interactions are driven by intrinsically disordered regions (IDRs) (Banani et al., 2017; Shin and Brangwynne, 2017). Some IDRs can engage in weak and transient “fuzzy” interactions with other proteins and nucleic acids and promote the assembly of phase-separated condensates, (Boehning et al., 2018; Boija et al., 2018; Chong et al., 2018; Guo et al., 2019; Hnisz et al., 2017; Sabari et al., 2018; Sanborn et al., 2021; Shin and Brangwynne, 2017; Tuttle et al., 2021; Wei et al., 2020; Sanders et al., 2020; Lu et al., 2019; Lu et al., 2020; Nair et al., 2019; Zamudio et al., 2019). However, intermediate concentrations of transcriptional activators can also lead to formation of active hubs that are not necessarily phase separated (Chong et al., 2018; Baek et al., 2021), suggesting biophysical diversity among different transcriptional condensates.
IDRs are features of transcription factors across the eukaryotic lineage, but it is unknown whether transcriptional condensates are conserved beyond mammalian cells. Moreover, it is unclear how transcriptional condensates conduct gene control in three-dimensions (3D). Recent studies in budding yeast have revealed profound remodeling of the 3D chromatin architecture following activation of the heat shock response (HSR) by the transcription factor Hsf1 (Chowdhary et al., 2017; Chowdhary et al., 2019). HSR genes share several key features with mammalian SEs, such as recruitment of high levels of the transcriptional machinery and extensive chromatin looping (Kainth et al., 2021). However, unlike SE-regulated genes, HSR genes physically interact with other HSR genes separated by large distances and even chromosomes (Chowdhary et al., 2017; Chowdhary et al., 2019). Here we provide experimental evidence implicating biomolecular condensation as the underlying mechanism of 3D gene control during the yeast HSR. We show that Hsf1, together with Mediator and RNA Pol II, forms inducible and dynamic transcriptional condensates that promote intergenic interactions among HSR genes. We identify a C-terminal binding site for the chaperone Hsp70 and the N-terminal IDR of Hsf1 as molecular determinants that endow HSR condensates with inducibility and the capacity to drive intergenic interactions. Finally, we generate a separation-of-function mutant of Hsf1 that carries mutation of both determinants and lacks intergenic coalescence, which comes at a fitness cost in this system. Our work reveals that transcriptional condensates are conserved features of eukaryotic gene control that can reversibly reconfigure the 3D genome.
RESULTS
Inducible high-level occupancy of the transcriptional machinery at HSR genes
The yeast HSR genes – defined as genes under the control of Hsf1 – show prolific levels of transcription. To assess the extent to which HSR genes concentrate the transcriptional machinery, we reanalyzed chromatin immunoprecipitation sequencing (ChIP-seq) data of Hsf1, Med15 (subunit of the Mediator complex known to interact with Hsf1) and Rpb1 (subunit of RNA Pol II) (Albert et al., 2019; Anandhakumar et al., 2016; Kim and Gross, 2013; Pincus et al., 2018; Sarkar et al., 2022). We found strong enrichment of all three factors at HSR genes upon heat shock and observed that the Hsf1 and Med15 peaks colocalize at heat shock elements (HSEs), the known binding sites for Hsf1 (Figures 1A, S1A-D).
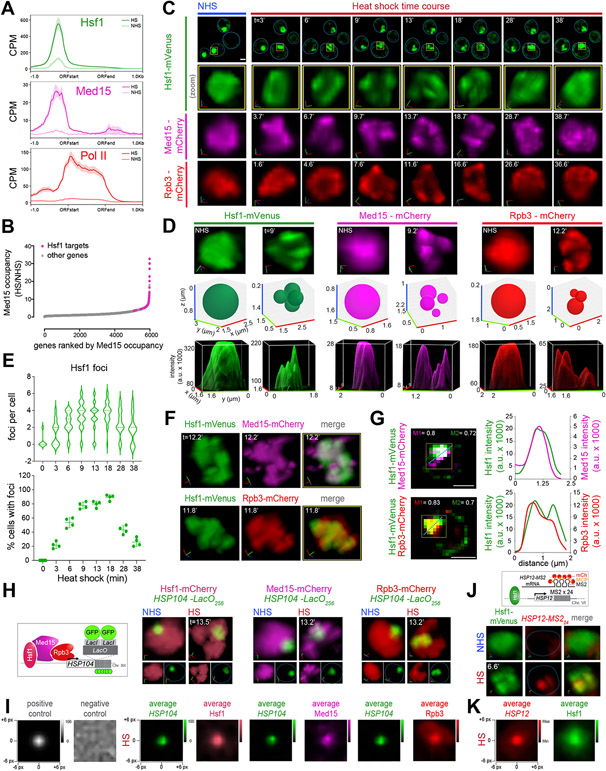
Figure 1. Hsf1, Mediator and RNA Pol II form transcriptionally active clusters at HSR genes upon heat shock.
A) Metagene plots of Hsf1 (top), Med15 (middle) and Pol II (Rpb1; bottom) normalized ChIP-seq reads of HSR genes. Shown are means +/− SE (shaded region). NHS, non-heat shock; HS, heat shock; CPM, counts per million reads.
B) Rank order of fold change in Med15 occupancy across Pol II-transcribed genes upon heat shock.
C) 4D imaging of single cells expressing Hsf1-mVenus, Med15-mCherry, or Rpb3-mCherry before (NHS, 24-26°C) and following HS (39°C) for the times (t) indicated. Row 1: images of cells expressing Hsf1-mVenus. Representative planes are shown. Blue line highlights cell boundary. Scale bar is 2 μm. Row 2: zoomed-in 3D volumetric rendering of the nucleus showing Hsf1-mVenus in one cell (yellow box in Row 1). x (red), y (green) and z (blue) axes are indicated. Rows 3 and 4: 3D volumetric renderings of the nuclei of a cell expressing either Med15-mCherry or Rpb3-mCherry.
D) Top row: 3D volumetric rendering as in C. Middle row: 3D bubble charts of Hsf1, Med15 and Rpb3 foci as detected by FindFoci for cells shown in the top row. Bottom row: 3D surface plots of signal intensity.
E) Top: number of Hsf1 foci per living cell quantified by automated analysis. Bottom: percentage of cells with >2 Hsf1 foci. 40-60 cells were evaluated per time point. Shown are means +/− SD.
F) Live imaging of cells co-expressing Hsf1-mVenus with Med15-mCherry (top) or Rpb3-mCherry (bottom).
G) Left: Quantification of colocalization of Hsf1 with Med15 and Rpb3 using Mander’s Overlay Coefficient. M1 indicates the fraction of mCherry that overlaps mVenus; M2, fraction overlap of mVenus with mCherry. Scale bar is 2 μm. Right: averaged intensity profiles along blue dotted lines as indicated in the images.
H) 3D rendered micrographs of Hsf1-mCherry, Med15-mCherry or Rpb3-mCherry and the HSP104-LacO256 gene locus. Blue dotted line highlights nuclear boundary.
I) Contour plots of Hsf1-mCherry, Med15-mCherry or Rpb3-mCherry showing averaged signal intensities centered at the HSP104-LacO256 gene locus 13 minutes following heat shock. Negative and positive controls were generated from simulation analysis of a random array or a 2D gaussian distribution, respectively.
J) Top: schematic of Hsf1 (green) and HSP12-MS224 mRNA (red). Bottom: 3D rendered micrographs of representative cells. Blue dotted line highlights nuclear boundary.
K) Contour plot of Hsf1-mVenus showing averaged intensity signal centered at HSP12-MS224 mRNA. Cells were imaged at 5 minutes of heat shock.
See also Figures S1-3.
Although HSR genes constitute <1% of the yeast genome, 30% of Med15 and 20% of Pol II ChIP-seq reads overlapped with Hsf1 reads upon heat shock (Figure S1C). Rank order analysis of protein-coding genes by change in Med15 occupancy during acute heat shock revealed that all HSR genes are in the top 20%, with the majority residing above the inflection point of the curve (Figure 1B). To determine if Hsf1 binding is sufficient to recruit high levels of Mediator upon heat shock, we performed Med15 ChIP at the non-HSR gene BUD3 in a wild type strain and a strain harboring a chromosomal integration of a high-affinity HSE at the BUD3 locus (Chowdhary et al., 2019). While neither Hsf1 nor Med15 were detectable at the wild type BUD3 locus, heat shock induced massive recruitment of Hsf1 and Med15 to BUD3 with the engineered HSE, suggesting that Mediator is robustly and selectively recruited by Hsf1 at its gene targets. (Figure S1E). Together, these data demonstrate that HSR genes disproportionately concentrate the transcriptional machinery upon activation.
Hsf1, Mediator and RNA Pol II colocalize in subnuclear clusters upon heat shock
We hypothesized that HSR genes engage a large fraction of transcriptional machinery by spatially concentrating it within the nucleus. Therefore, we investigated the spatiotemporal distributions of Hsf1, Mediator and RNA Pol II in live single cells upon heat shock (Figures 1C, D). In unstressed cells, endogenously tagged Hsf1-mVenus was predominantly nuclear and diffusely localized (Figure 1C, NHS). However, Hsf1 formed semi-discrete subnuclear clusters within 6 min of heat shock that persisted for 20-25 min, after which Hsf1 returned to its diffuse localization pattern (Figure 1C, heat shock). Hsf1 clusters were identified within 3D images of live cells using the automated FindFoci algorithm in ImageJ (Figure 1D, E). Here, we have interpreted inhomogeneities in signal distribution as clusters (or foci) of locally enriched target molecules. This analysis revealed the presence of Hsf1 clusters in >80% of cells during acute heat shock (Figure 1E). We typically observed 4 clusters of Hsf1 per individual nucleus, although the number varied between 2 and 6 (Figure 1C, D, E). These results are consistent for both the deconvolved images and those processed without deconvolution (Figure S2A, C).
To provide best representation of the total number of clusters and their nuclear positions, we rendered images in 3D and displayed foci assignments as 3D bubble plots. For displaying inhomogeneities, we visualized images as 3D surface plots (Figure 1C, D). To rule out processing or deconvolution-related artifacts in the image analyses, we compared inhomogeneities within images processed with and without deconvolution, and the images processed without deconvolution displayed the same number of foci and peaks as deconvolved images (Figure S2B). Analysis of diploid cells expressing Hsf1-GFP gave virtually identical results as haploid cells, indicating that the number of Hsf1 clusters does not correlate with ploidy (Figure S3C). To distinguish whether redistribution of Hsf1 into clusters is a physical consequence of elevated temperature or a regulatory consequence of Hsf1 activation, we imaged Hsf1-mVenus expressing cells following treatment with azetidine-2-carboxylic acid (AZC), a proline analog that incorporates into nascent proteins, impairs protein folding, and activates Hsf1 (Trotter et al., 2001; Zheng et al., 2018). Following AZC treatment, Hsf1 formed intranuclear clusters (Figure S3D). Therefore, Hsf1 clustering appears to be driven by activation rather than heat per se.
We next tested whether Mediator and RNA Pol II also formed intranuclear clusters during heat shock by imaging endogenously tagged Med15-mCherry or Rpb3-mCherry. In the absence of heat shock, Med15 and Rpb3 were diffusely localized within the nucleus (Figure 1C and D, NHS). However, like Hsf1, both formed an average of 4 subnuclear clusters in >80% of cells upon heat shock (Figures 1C, D and S3A, B). Clusters formed by both Med15 and Rpb3 co-localized with Hsf1 during heat shock with a fractional overlap of at least 70% (Figures 1F, G and S3E). These data provide evidence of spatiotemporal interactions between Hsf1, Mediator and RNA Pol II inside living cells following heat shock.
Hsf1 clusters activate transcription of HSR genes
To determine whether HSR genes associate with the Hsf1/Med15/Rpb3 clusters in yeast, we inserted arrays of 256 lacO-repeats adjacent to the HSR gene HSP104 (Figure S1D) and co-expressed LacI-GFP with either Hsf1-, Med15- or Rpb3-mCherry (Figure 1H). Under basal conditions, HSP104-lacO256 minimally overlapped with Hsf1-, Med15- and Rpb3-mCherry but co-localized with these factors during heat shock (Figure 1H and S3F). Across a population of cells, the average fluorescence intensities of Hsf1, Med15 and Rpb3 were enriched at the center of the HSP104-lacO256 focus (Figure 1I). These results indicate that the Hsf1, Mediator and Pol II clusters associate with a canonical HSR gene.
To test whether Hsf1 clusters are sites of active transcription, we employed the MS2 RNA imaging system to visualize nascent transcript production (Hocine et al., 2013; Lenstra and Larson, 2016). We inserted MS2 repeats into the 3' untranslated region of HSP12, another HSR target (Figure S1D), in a strain expressing the MS2 coat protein fused to mCherry (MCP-mCherry) and co-expressing Hsf1-mVenus. We observed no HSP12-MS24 fluorescence under non-heat shock conditions, indicating a lack of transcription of HSP12 mRNA, while heat shock induced the transient appearance of a concentrated focus of nascent HSP12-MS24 mRNA that colocalized with Hsf1-mVenus (Figures 1J and S3G). The average fluorescence intensity of Hsf1 was enriched at the center of the HSP12-MS224 mRNA focus across a population of heat-shocked cells (Figure 1K). These data demonstrate that, upon heat shock, Hsf1 forms subnuclear clusters with the transcriptional machinery that mark sites of nascent mRNA production.
Hsf1 clusters exhibit properties of biomolecular condensates
To investigate the dynamics of Hsf1 clusters in living cells, we employed stimulated emission depletion (STED) super-resolution imaging. We took time-series images of live cells expressing Hsf1-mVenus every minute and found that the Hsf1 clusters exhibited a stereotyped temporal signature along the course of heat shock: (1) an early demixing phase; (2) an intermediate intermixing phase; and (3) a late remixing phase (Figure 2A).
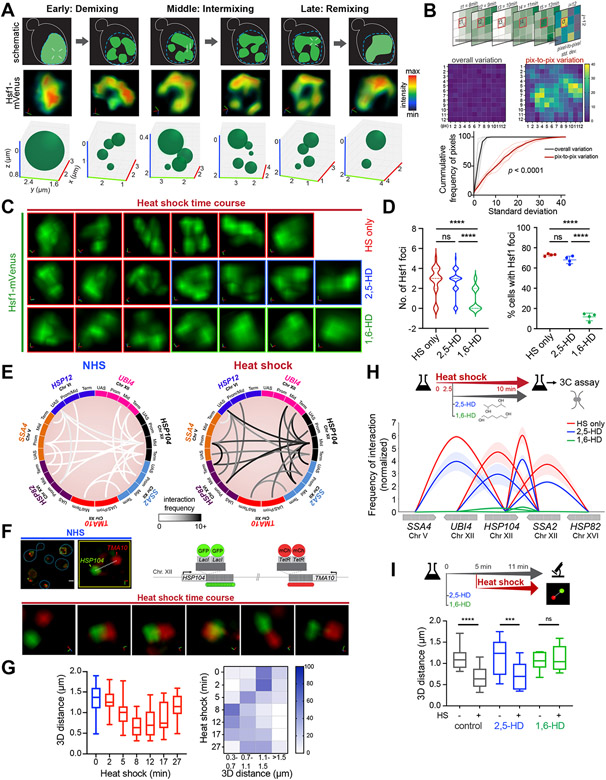
Figure 2. Hsf1 clusters are dynamic and associated with HSR gene coalescence.
A) Schematic (top), STED super-resolution imaging (middle) and 3D bubble plots (bottom) depicting Hsf1-mVenus foci dynamics at different stages of heat shock.
B) Top: schematic of the method used for computing variation of pixel intensity over time during the intermixing phase of heat shock. Middle: Heatmap of pixel variation in a representative nucleus against overall variation within the image. Bottom: Cumulative distribution of standard deviations for pixel-to-pixel (red) and overall (black) variation within six individual cells.
C) 3D rendered nuclei of representative cells subjected to heat shock alone or in the presence of either 2,5-HD or 1,6-HD. HD was added after 9 min of HS. Two different cells were imaged before and after HD treatment.
D) Number of Hsf1 foci per cell and the percentage of cells showing >2 Hsf1 foci in the conditions in C (n=110-140 cells/condition). Cells were imaged 3 minutes after adding HD. ****P <0.0001; ns (not significant), P >0.05. P values were calculated by ANOVA followed by Tukey’s post hoc analysis.
E) Circos plots depicting the two most frequent intergenic interactions between indicated gene pairs as determined by Taq I-3C derived from (Chowdhary et al., 2017; Chowdhary et al., 2019). Arc shades depict the frequencies of chromatin interactions between indicated regions.
F) 4D imaging of a diploid strain expressing HSP104-lacO256, TMA10-tetO200, LacI-GFP and TetR-mCherry (schematic, top right). Top left: micrographs of representative cells under NHS conditions (blue line highlights cell boundary) and an enlarged 3D view of the nuclear region indicated within the yellow box. Bottom: enlarged 3D micrographs of a representative cell subjected to heat shock. d, 3D distances measured between signal centroids.
G) Distribution of 3D distances between lacO-tagged HSP104 and tetO-tagged TMA10 gene loci in cells following heat shock (n=13 for NHS; n=20-30 for HS time points). Bottom: Heat map depicting percentage of cells with tagged gene loci within indicated 3D distances binned at intervals of 0.4 μm.
H) Intergenic contacts (solid arcs) between Hsf1 target gene pairs as determined by TaqI-3C in presence and absence of hexanediol. Genes are segmented as UAS, promoter, mid-ORF and terminator. Depicted are means +/− SD; n=2, qPCR=4.
I) Distribution of 3D distances between lacO-tagged HSP104 and tetO-tagged TMA10 under conditions as indicated. Cells were left untreated or pre-treated for 5 min with either 2,5- or 1,6-HD followed by no heat shock or heat shock for 6 min. n =13-22. ****P <0.0001; ***P <0.001; ns (not significant), P >0.05 (calculated using two-tail t test).
See also Figure S4.
The demixing phase is marked by formation of semi-discrete clusters of Hsf1 during the first 6-7 min of heat shock induction. In this phase, Hsf1 nuclear distribution transitions from a homogenous to a heterogenous regime. In the intermixing phase (8-27 min), Hsf1 clusters rearrange continuously within the nucleus, rapidly exchanging molecules. During the intermixing phase, Hsf1 signal intensity displayed high pixel variation over time – indicating rapid spatial dynamics – while overall Hsf1 signal in the nuclei remained nearly constant (Figure 2B). Also, the numbers, sizes and positions of Hsf1 foci all varied over time, indicating rapid rearrangements among clusters (Figure 2A, bottom). Finally, in the remixing phase (28-32 min), clusters disperse and Hsf1 reverts to its pre-heat shocked diffuse localization and uniformly distributed regime. These results demonstrate that Hsf1 clusters are remarkably dynamic: they form quickly, rearrange rapidly, and dissolve abruptly.
To further probe the material properties of Hsf1 clusters, we tested their sensitivity to an aliphatic alcohol, 1,6-hexanediol (1,6-HD). 1,6-HD has historically been used to disrupt liquid-like biomolecular condensates, including transcriptional condensates in mammalian cells. While the mechanism of action remains unclear, evidence suggests that it impacts hydrophobic and aromatic interactions more strongly than electrostatic effects (Cho et al., 2018; Kroschwald et al., 2015; Patel et al., 2015; Sabari et al., 2018). Extended treatment of cells with 1,6-HD can cause membrane blebbing, disrupt chromatin architecture, and inhibit kinase and phosphatase activity (Duster et al., 2021; Itoh et al., 2021; Kroschwald et al., 2017). To minimize aberrant effects of 1,6-HD, we optimized the concentration and duration of hexanediol treatment for two most widely studied nuclear condensates: the nucleolus and the nuclear pore complex (NPC) (Banani et al., 2017; Kato and McKnight, 2018). To this end, we applied 1,6-HD to living cells expressing endogenous Nop56-mRFP (nucleolar component) and Pom34-GFP (nuclear pore component) at three different concentrations (3, 5 and 10%) for 20-30 minutes (Figure S4A, B). Treatment with 3% 1,6-HD disintegrated the nucleolus and NPC after 15 minutes; 10% dissolved them in less than 4 minutes and caused membrane blebbing. Treatment with 5% 1,6-HD dissolved the nucleolus and NPC after 7 minutes with no signs of membrane blebbing until after 20 minutes (Figure S4B). We also scored the effects of 2,5-hexanediol (2,5-HD), a less hydrophobic alcohol with the same atomic composition as 1,6-HD. 2,5-HD had minimal effect on nucleolar and NPC integrity at 3 or 5%. However, at 10%, the effects of 2,5-HD were the same as observed with 1,6-HD (Figure S4A, B). Based on these observations, we identified 5% for less than 20 minutes as the optimum concentration and treatment duration for both hexanediols.
To determine the effect of hexanediols on Hsf1 clusters, we first heat shocked cells for 8 minutes to allow cluster formation and then treated cells expressing Hsf1-mVenus with either 5% 1,6-HD or 2,5-HD. Hsf1 clusters were dissolved within 4 minutes upon addition of 1,6-HD but were unaffected by 2,5-HD treatment and mimicked cells that were only heat-shocked (Figure 2C, D). We observed similar effects on Med15 clusters, except they were even more rapidly dissolved upon 1,6-HD treatment (Figure S4C, D). Together, these data indicate that Hsf1 clusters display hallmark properties of biomolecular condensates. We will hereon refer to them as HSR condensates.
HSR condensates drive intergenic interactions among HSR genes
HSR genes engage in selective intra- and inter-chromosomal interactions upon heat shock (Chowdhary et al., 2017; Chowdhary et al., 2019), and we hypothesized that these interactions are driven by condensate formation (Kainth et al., 2021). Using a highly sensitive chromosome conformation capture technique (TaqI-3C) (Chowdhary et al., 2020), we have demonstrated interactions among seven Hsf1-dependent genes spanning distinct chromosomes (Chowdhary et al., 2019) (Figure 2E). The intergenic interactions between Hsf1-dependent HSR genes occur with low frequency under non-heat shock conditions but increase many -fold upon acute heat shock.
To complement this molecular assay, we directly imaged intergenic coalescence of HSR genes in a strain with operator arrays marking two HSR genes, HSP104-lacO256 and TMA10-tetO200, that co-expresses LacI-GFP and TetR-mCherry (Chowdhary et al., 2019) (Figure 2F). Under non-heat shock conditions, HSP104-lacO256 and TMA10-tetO200 were spatially separated, as indicated by a large 3D distance between the two loci (d =1.6 μm), consistent with the respective locations of HSP104 and TMA10 on the far left and right arms of chromosome XII. However, upon heat shock, HSP104-lacO256 and TMA10-tetO200 came in close proximity within 5 minutes, coalescing into a partially-overlapping focus after 10 minutes that ultimately dissipated after 16 minutes of heat shock (Figure 2F). We obtained consistent results for images processed with or without deconvolution (Figure S2D). Across a population of cells, HSP104-lacO256 and TMA10-tetO200 were separated by much larger 3D distances under non-inducing and chronic heat shock conditions (17 min and later), while the distribution was skewed toward shorter distances in acutely heat-shocked cells (Figure 2G). By contrast, HSP104-lacO256 remained spatially separated from PGM2-lacO128, an Hsf1-independent, Msn2/4-regulated gene, despite strong activation of both genes in response to heat shock (Figure S4G, H; (Chowdhary et al., 2019)). Taken together, these observations indicate that coalescence of HSR genes is specific and follows the kinetics of Hsf1 cluster formation and dissolution.
To determine if HSR condensates are involved in intergenic coalescence, we perturbed Hsf1 clusters and assayed for gene coalescence. We first induced Hsf1 clusters by subjecting cells to brief heat shock treatment for 2.5 minutes before adding 1,6-HD or 2,5-HD and performed TaqI-3C (Figures 2H, S4E). Interactions between HSR genes were disrupted by 1,6-HD but not 2,5-HD. As determined by single cell imaging analysis, the coalescence of HSP104-lacO256 and TMA10-tetO200 was likewise abolished in cells pretreated with 1,6-HD but not 2,5-HD before heat shock (Figure 2I and Figure S4F). These results indicate that 1,6-HD dissolves Hsf1 clusters and disrupts intergenic interactions between HSR genes.
Hsp70 binding represses Hsf1 cluster formation
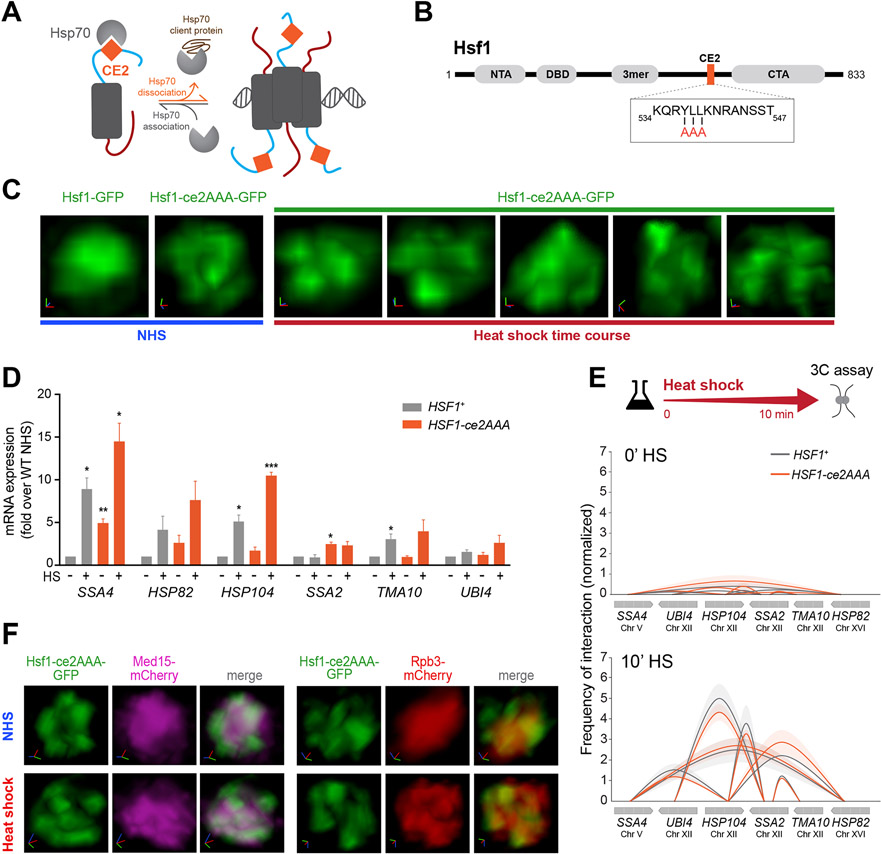
Hsf1 activity is repressed when it is bound by the chaperone Hsp70 (Figure 3A) (Krakowiak et al., 2018; Masser et al., 2019; Peffer et al., 2019; Zheng et al., 2016), so we wondered if Hsp70 binding would also repress HSR condensate formation. To test this, we disrupted the Hsp70 binding site known as “conserved element 2” (CE2) (Krakowiak et al., 2018; Peffer et al., 2019) (Figure 3A, B). We expressed GFP-tagged Hsf1-ce2AAA as the only copy of Hsf1 and imaged its localization. In contrast to the diffuse nuclear localization of wild type Hsf1, we observed constitutive clustering of Hsf1-ce2AAA in unstressed cells that persisted during heat shock (Figure 3C).
Figure 3. Hsp70 binding to Hsf1 represses cluster formation.
A) Schematic of the Hsp70 titration model of Hsf1 regulation. Hsp70 represses Hsf1 by binding to the CE2 domain. Sequestration of Hsp70 by its client proteins activates Hsf1.
B) Domain map of Hsf1. NTA, N-terminal activation domain; DBD, DNA binding domain; 3mer, trimerization domain; CE2, conserved element 2 domain (orange); CTA, C-terminal activation domain. Box: CE2 sequence and alanine substitutions in the Hsf1-ce2AAA mutant.
C) 3D rendered images of wild type and Hsf1-ce2AAA mutant cells.
D) mRNA expression measured by RT-qPCR of representative Hsf1-regulated genes in HSF1+ and HSF1-ce2AAA strains under NHS and 10 min-HS conditions. ***P <0.001; **P <0.01; *P <0.05 (calculated using multiple unpaired t tests).
E) Intergenic contacts between indicated Hsf1 target gene pairs determined by TaqI-3C.
F) Live imaging of cells co-expressing Hsf1 ce2AAA-GFP with Med15-mCherry or Rpb3-mCherry.
To determine whether the constitutive clusters formed by Hsf1-ce2AAA are transcriptionally active, we measured mRNA levels of representative HSR genes (Figure 3D). We detected a significant increase in the basal mRNA levels of some targets in Hsf1-ce2AAA cells compared to wild type. However, the basal levels of most target transcripts in the mutant were lower than their respective levels in wild type cells following heat shock, suggesting that the constitutive clusters formed by Hsf1-ce2AAA are less active than those formed by wild type Hsf1 upon activation. Further supporting a lack of full activation, both Med15 and Rpb3 were diffusely localized under basal conditions in Hsf1-ce2AAA cells and only formed clusters with Hsf1-ce2AAA upon heat shock (Figure 3F). Moreover, HSR genes showed no detectable intergenic interactions as measured by TaqI-3C under basal conditions in Hsf1-ce2AAA cells, but both intra- and intergenic interactions were induced upon heat shock to the same degree as in wild type cells (Figures 3E and S5D). Thus, mutation of the CE2 binding site for Hsp70 results in constitutive Hsf1 cluster formation, but this is not sufficient to drive intergenic interactions.
The N-terminal domain of Hsf1 promotes formation of HSR condensates
Hsf1 contains functional N-terminal and C-terminal activation domains referred to as NTA and CTA (Figure 4A). The NTA both represses Hsf1 under basal conditions – likely due to a second Hsp70 binding site – and plays a positive role in transactivation, while the CTA constitutes a strong transactivation domain (Kim and Gross, 2013; Krakowiak et al., 2018; Peffer et al., 2019; Sorger, 1990). The NTA and CTA are disordered and act synergistically to stably recruit Mediator to HSR gene promoters (Figure 4A, B; Figure S5G) (Kim and Gross, 2013). To address whether they play a role in intergenic interactions and HSR condensate formation, we separately removed both regions.
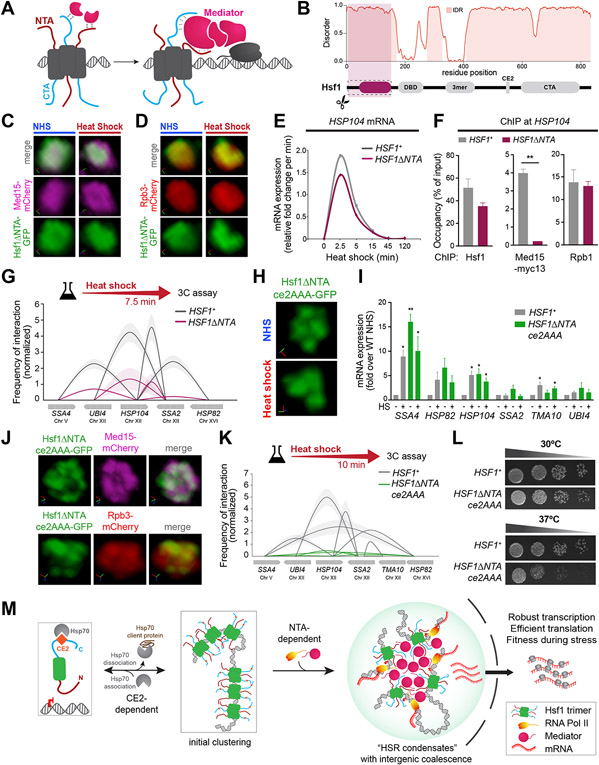
Figure 4. HSR condensates drive inducible intergenic coalescence by the combined actions of the CE2 and NTA regions of Hsf1.
A) Cartoon depiction of Mediator recruitment via dual Hsf1 activation domains.
B) Domain map and prediction of disorder in Hsf1 by Metapredict V2 (Emenecker et al., 2022).
C) Live imaging of Hsf1ΔNTA -GFP and Med15-mCherry.
D) Live imaging of Hsf1ΔNTA -GFP and Rpb3-mCherry.
E) Transcription rates of HSP104 in HSF1+ and HSF1-ΔNTA deduced from RT-qPCR.
F) Hsf1 and Med15-myc13 ChIP at the UASs and Rpb1 ChIP at the promoter region of HSP104 in wild type and Hsf1-ΔNTA strains heat-shocked for 7.5 min. Depicted are means + SD. **, P <0.01 (calculated using two-tail t-test).
G) Intergenic contacts between indicated Hsf1 target gene pairs in 7.5 min-HS conditions as determined by TaqI-3C.
H) Representative 3D rendering of Hsf1-GFP and Hsf1-ΔNTA-ce2AAA-GFP.
I) mRNA expression measured by RT-qPCR of representative Hsf1-regulated genes in HSF1+ and HSF1-ΔNTAce2AAA strains under NHS and 10 min-HS conditions. **P <0.01; *P <0.05 (calculated using multiple unpaired t tests)
J) Live imaging of cells co-expressing Hsf1ΔNTAce2AAA -GFP with Med15-mCherry or Rpb3-mCherry under NHS conditions.
K) Intergenic contacts) between indicated Hsf1 target gene pairs following 10 min-HS conditions as determined by TaqI-3C.
L) Spot dilution analysis of HSF1+ and HSF1-ΔNTAce2AAA cells. Plates were incubated at 30°C or 37°C for 30 h.
M) Schematic depiction of HSR condensate formation.
We observed that Hsf1-ΔNTA-GFP failed to form clusters even upon heat shock (Figure 4C, D). Moreover, neither Rpb3 nor Med15 clustered in the Hsf1-ΔNTA mutant (Figure 4C, D). While Hsf1-ΔNTA was able to robustly bind to the HSP104, HSP82, and SSA4 promoters and recruit RNA Pol II upon heat shock, it was unable to recruit high levels of Med15 (Figure 4F, S5B). Yet, the rate of inducible transcription of HSR genes and local chromatin looping were comparable in Hsf1-ΔNTA cells and wild type (Figures 4E, S5A, C). Since Mediator is indispensable for transcriptional activation (Anandhakumar et al., 2016), the lack of detectable Med15 in the ChIP assay suggests that Hsf1-ΔNTA is only able to transiently recruit but not stably interact with Mediator. Along with the lack of stable Med15 recruitment, Hsf1-ΔNTA cells showed a substantial decrease in intergenic interactions between HSP104 and other HSR genes compared to wild type (Figure 4G). Loss of the CTA likewise decreased Mediator recruitment and intergenic interactions among HSR genes, but it also impaired fitness under all growth conditions, rendering it too severe a mutation to interpret specifically (Figure S5H-J) (Krakowiak et al., 2018; Sorger, 1990). These data indicate that the NTA promotes the ability of Hsf1 to cluster with Mediator and Pol II and drive intergenic interactions among HSR genes.
Inducible HSR condensates promote cellular fitness under stress
Since loss of CE2 resulted in constitutive Hsf1 clustering, and loss of the NTA reduced intergenic interactions, we reasoned that combining these mutations could reprogram Hsf1 to be constitutively clustered but unable to drive intergenic interactions. To test this, we generated a double mutant, Hsf1-ΔNTAce2AAA, GFP-tagged it and expressed it as the only copy of Hsf1 from the endogenous HSF1 promoter. Indeed, Hsf1-ΔNTAce2AAA-GFP formed constitutive clusters (Figure 4H) and was fully activated, driving transcription of some target genes under basal conditions to levels exceeding their expression during heat shock in wild type cells (Figure 4I). In contrast to the constitutive clusters of Hsf1-ce2AAA single mutant, Hsf1-ΔNTAce2AAA-GFP colocalized in clusters with Med15- and Rpb3-mCherry under basal conditions (Figure 4J). Upon heat shock, the double mutant showed no further increase in transcription of HSR genes, and levels of several HSR targets even decreased (Figure S5E). While Hsf1-ΔNTAce2AAA-GFP clusters persisted during heat shock (Figure 4H), Med15- and Rpb3-mCherry clusters were diminished (Figure S5F). Despite its constitutive transcriptional activity and clustering, Hsf1-ΔNTAce2AAA was unable to drive intergenic interactions among HSR genes, even during heat shock (Figure 4K). Finally, Hsf1-ΔNTAce2AAA cells showed impaired growth at elevated temperature (Figure 4L), suggesting that the ability to form inducible transcriptional condensates capable of driving intergenic interactions promotes fitness during stress. Thus, the CE2 and NTA are determinants that endow HSR condensates with inducibility and the capacity to drive intergenic interactions, features necessary for cell survival under stress.
DISCUSSION
Our discovery of HSR condensates in yeast demonstrates that transcriptional condensates are likely ancient and conserved compartments of eukaryotic gene control. The distinguishing features of HSR condensates – their inducible kinetics and intergenic interactions – indicate that transcriptional condensates are capable of rapidly and reversibly remodeling the 3D genome in response to environmental cues. HSR condensates encode these unique properties within the primary sequence of Hsf1 via Hsp70 binding sites and the intrinsically disordered NTA. As such, genetic disruption of the CE2 combined with ablation of the NTA was sufficient to reprogram HSR condensates to be constitutive structures that lack intergenic coalescence. Thus, relatively simple genetic changes can have dramatic consequences in the regulatory properties of transcriptional condensates, endowing these structures with evolutionary plasticity.
Since deletion of the NTA resulted in loss of condensates and intergenic coalescence (Figure 4C, G), it may be that cooperative binding among the IDRs of Hsf1, Mediator and RNA Pol II (Figures 4B, S5K, L) serves to nucleate HSR condensates and bring HSR genes in 3D proximity (Figure 4M). The other possibility is that Hsf1 initiates the condensation itself via cooperative binding of Hsf1 trimers to multiple HSEs of gene targets (Erkine et al., 1999), with each activator molecule then tethering or engaging one or more polymerases and/or Mediator subunits (Baek et al., 2021). These local multivalent interactions between Hsf1 and Mediator at individual HSR genes could then intermix with other such local assemblies leading to intergenic coalescence.
The role of Hsp70 in repressing Hsf1 clustering represents a cell biological manifestation of the known role of Hsp70 in repressing Hsf1 transcriptional activity (Kmiecik et al., 2020; Krakowiak et al., 2018; Masser et al., 2019; Zheng et al., 2016). While we do not elucidate the biochemical basis of this repression in the current study, two mechanisms are possible: Hsp70 could passively block clustering by steric occlusion, and/or Hsp70 could mechanically pull Hsf1 out of the clusters. Precedents exist for both putative mechanisms in repression of condensates (Ruff et al., 2021; Yoo et al., 2022). Regardless of the specific mechanisms, Hsp70 may function as a “molecular stir bar” to antagonize condensates. Expanding on this notion, Hsf1 may have evolved to form condensates upon activation that mimic other condensates recognized by Hsp70 to peg its activity – and therefore the expression of the HSR regulon – to the availability of Hsp70. Other chaperones such as Hsp90, which is known to associate with nuclear hormone receptors that also condense with the transcriptional machinery (Frank et al., 2021; Nair et al., 2019; Picard et al., 1990), may also remodel transcriptional condensates.
Are HSR condensates likely to be conserved in mammalian cells? In human cells, HSF1 forms large stress-induced foci upon heat shock (Jolly et al., 1997). However, these large HSF1 foci localize to noncoding DNA regions and are anti-correlated with HSR gene expression (Gaglia et al., 2020; Jolly et al., 2004). In addition to these large HSF1 clusters, a recent study provides compelling evidence that smaller HSF1 foci represent transcriptionally active condensates at HSR genes (Zhang et al., 2022). Beyond HSF1, the mammalian Pol II-pausing factor NELF (negative elongation factor), which is associated with transcriptional dowregulation during stress, also forms condensates upon heat shock that are distinct from HSF1 foci (Rawat et al., 2021). NELF condensates may liberate transcriptional machinery from housekeeping genes that can then be recruited by HSF1 (Kainth et al., 2021; Mahat et al., 2016). It remains unclear whether mammalian HSF1 condensates play a role in 3D genome organization. Although the organization of mammalian genomes into topologically associated domains (TADs) (Dixon et al., 2012) would seem to preclude the ability of HSR genes to coalesce upon activation, multiple HSR genes are clustered adjacent to each other in the human genome, which may allow intergenic interactions among HSR genes (Kainth et al., 2021). Thus, key features of the HSR condensates may be conserved.
HSR condensates may provide several advantages to cells under stress (Figure 4M). First, condensation concentrates large quantities of the transcriptional machinery to enable robust induction of HSR genes critical to counteracting the stress. Second, the coalescence of multiple HSR genes separated by large distances could reduce search time for repeated transcriptional firing events and coordinate transcriptional bursting across the HSR regulon. Third, the localized transcription of multiple HSR genes could promote mRNA clustering into “transperons” that may bypass normal mRNA quality control and increase efficiency of post-transcriptional steps of gene expression such as nuclear export (Nair et al., 2021; Nair et al., 2022; Zander et al., 2016). Fourth, HSR condensates may enable target messages to avoid cytosolic stress granules and thereby promote their privileged translation (Zid and O'Shea, 2014), perhaps by recruiting translation factors or mRNA modifying enzymes. HSR condensates may thus promote cell survival under stress by controlling both transcriptional and post-transcriptional events. HSR condensates represent the founding example of inducible transcriptional clusters that drive intergenic interactions between stress-responsive genes. Given these attributes, it may be the case that analogous structures have evolved to execute other stimulus-regulated transcriptional programs within budding yeast and beyond.
Limitations of the study
The major limitation of this study is the spatiotemporal resolution of the live imaging data of HSR condensates in yeast cells. The principal challenge is the small size of the yeast nucleus (<3 μm3), which means that the HSR condensates are distributed in a very compact space. The low abundance of the components (on the order of 103 molecules of Hsf1 or Med15 per cell) and the rapid kinetics of formation and dissolution of HSR condensates presented further technical challenges. We relied on relatively long exposures, requiring 15 seconds to capture all z-stacks at each time point, and we were limited by photobleaching. Together, these challenges made it difficult to discern whether the HSR condensates in budding yeast are discrete foci like transcriptional condensates in mammalian cells. Although HSR condensates share components and properties with mammalian transcriptional condensates, we do not rule out the possibility that HSR condensates might be qualitatively distinct. Perhaps, HSR condensates are constantly ebbing and flowing, forming mobile regions of local enrichment of the transcriptional machinery that rapidly recruit and release target genes as they dynamically survey the genome.
In addition to the condensates themselves, the same constraints apply to the LacO and TetR arrays we used to image and quantify the distance between genomic loci. Perhaps due to the long acquisition times, these arrays – which typically appear as diffraction-limited spots in the literature – spanned multiple pixels in our deconvolved images, limiting the precision of our 3D distance measurements. Although we cannot precisely quantify distance with the arrays in single cells, our TaqI-3C data complement the microscopy-based analyses. Together, these orthogonal approaches support the model that Hsf1 target genes preferentially get closer to one another upon heat shock in a manner that requires Hsf1 to form condensates.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. David Pincus (pincus@uchicago.edu).
Materials availability
S. cerevisiae strains and plasmids generated in this study are available from the lead contact upon request.
Data and code availability
Raw images associated with the key figure were deposited on Mendeley at doi: 10.17632/cr972zx459.1. Other microscopy and non-imaging data reported in this paper will be shared by the lead contact upon request.
This paper does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All S. cerevisiae strains were grown in YPD (yeast extract-peptone-dextrose) or SDC (synthetic dextrose complete) media at 30°C as indicated below.
METHOD DETAILS
Yeast Strains
For tagging Hsf1, Med15 and Rpb3 with mCherry, PCR amplicons with mCherry coding sequence and homology to 3’-ends of either HSF1, MED15 and RPB3 were amplified from plasmid pFA6a-mCherry-hphMX6. The amplicons were transformed into the respective haploid strains for in-frame insertion of mCherry. Primers used in strain construction are listed in Table S6.
For Myc x 9 tagging of Med15, genomic DNA of a previously myc9-tagged MED15 strain (ASK201) (Anandhakumar et al., 2016) was used as template to amplify MYCx 9::TRP1 cassette flanked by DNA homologous to 3’-end of MED15. This amplicon was transformed in strains DSG144 and LRY003 to obtain strains ASK213 and ASK214, respectively. LRY003 is a derivative of previously described strain ASK804 (Chowdhary et al., 2019) in which TRP1 was deleted by replacing with KAN-MX. For Myc x 13 tagging of Med15, plasmid pFA6a-13Myc-His3MX was used as template to obtain MYC13-HIS3 amplicon with homology to 3’-end of MED15. This amplicon was transformed in strains DPY144, DPY417 and DPY418 to obtain strains ASK215, ASK216 and ASK217, respectively.
For MS2-MCP labelling of HSP12 mRNA, MCP-mCherry-URA3 cassette was amplified from plasmid pSH100 with primers Fw MCP-mCherry-Ura3 and Rv MCP-mCherry-Ura3. This cassette was inserted at the endogenous URA3 locus in strain DPY032 to obtain strain SCY008. Next, 24xMS2-loxP-KANMX6-loxP cassette was amplified from plasmid pDZ415 using primers Fw HSP12-MS2-loxp-KanMX6-loxp and Rv HSP12-MS2-loxp-KanMX6-loxp. This cassette was inserted in the 3’-UTR region of HSP12 (immediately after stop codon) of strain SCY008 to obtain strain SCY009. Finally, plasmid pSH69 was transformed to express Cre recombinase in strain SCY010 that led to the removal of KANMX in strain SCY011.
The diploid strain ASK741 was created by crossing a MATα derivative of strain DSG200 (Chowdhary et al., 2019) with MATa DSG200.
Plasmids pNH604-HSF1pr-HSF1-GFP and pNH604-HSF1pr-HSF1(147-833)-GFP were used as templates for quick change PCR (Primers, Fw subCE2-AAA and Rv subCE2-AAA) to create CE2->AAA mutation in WT and ΔNTA HSF1, respectively. These plasmids were linearized with Pme I (New England Biolabs) and transformed in strain DPY034 for integration at the TRP1 locus to obtain strains DPY1805 and DPY1818. Loss of parental HSF1 plasmid was confirmed by growth on 5-FOA media.
A complete list of strains is provided in Table S1. PCR primer sequences are provided in Table S6.
Culture Conditions
For microscopy, cells were grown at 30°C in SDC (synthetic dextrose complete) media to early-log density (A600 = 0.4-0.5).
For 3C, ChIP and RT-qPCR analyses, cells were grown at 30°C in YPD (yeast extract-peptone-dextrose) to a mid-log density (A600 = 0.65-0.8). A portion of the culture was maintained at 30°C as non-heat-shocked (NHS) sample while the remainder (heat-shocked (HS) sample) was subjected to an instantaneous 30°C to 39°C thermal upshift for the indicated duration.
For spot dilution analysis, cells were grown to stationary phase in YPD media. Master suspensions were prepared by diluting the saturated cultures to a uniform cell density (A600 = 0.3) and were transferred to a 96-well microtiter plate. These were then serially diluted (five-fold). 4 μl of each dilution were transferred onto solid YPDA plates. Cells were grown at either 30° or 37°C for 30 h.
Chromosome Conformation Capture
TaqI-3C was conducted essentially as previously described (Chowdhary et al., 2017; 2020; Chowdhary et al., 2019). Briefly, cells were cultured to a mid-log density (A600 = 0.8) at 30°C. They were either maintained at 30°C or heat-shocked at 39°C for 10 min (or as indicated), and then crosslinked with formaldehyde (1% final concentration). For 3C assay involving hexanediol treatment, cells were heat-shocked at 39°C for 2.5 min followed by treatment with either 2,5- or 1,6-hexanediol (5% final concentration), and then crosslinked with formaldehyde. Crosslinked cells were harvested and lysed in FA lysis buffer (50 mM HEPES pH 7.9, 140 mM NaCl, 1% Triton X-100, 0.1% Sodium deoxycholate, 1 mM EDTA, 1 mM PMSF) for two cycles (20 min each) of vortexing at 4°C. A 10% fraction of the chromatin lysate was digested using 200 U of Taq I (New England Biolabs) at 60°C for 7 h. Taq I was heat-inactivated at 80°C for 20 min. The digested chromatin fragments were centrifuged, and the pellet was resuspended in 100 μl of 10 mM Tris-HCl (pH 7.5). The Taq I-digested chromatin was diluted 7-fold to which 10,000 cohesive end units of Quick T4 DNA ligase (New England Biolabs) were added. Proximity ligation was performed at 25°C for 2h. The ligated sample was digested with RNase at 37°C for 20 min and then Proteinase K at 65°C for 12 h. The 3C DNA template was extracted using phenol-chloroform and then precipitated.
The interaction frequencies were quantified using qPCR. Quantitative PCR was performed on a CFX Real-Time PCR system (Bio-Rad) using Power SYBR Green PCR master mix (Fisher Scientific). Sequences of 3C primers used in this study are provided in Table S2. Normalization controls were used to account for the following: (i) variation in primer pair efficiencies; (ii) primer dimer background; (iii) variation in the recovery of 3C templates; and (iv) to ensure a ligation-dependent 3C signal. For detailed algorithms to calculate normalized 3C interaction frequencies, see below and (Chowdhary et al., 2020).
Chromatin Immunoprecipitation (ChIP)
ChIP was conducted essentially as previously described (Chowdhary et al., 2019). Briefly, the cells were heat-shocked at 39°C for 7.5 min (or maintained at 30°C) and crosslinked with 1% formaldehyde. Cells were then harvested and subjected to glass bead lysis in lysis buffer (50 mM HEPES pH 7.5, 140 mM NaCl, 1% Triton X-100, 0.1% Sodium deoxycholate, 1 mM EDTA, 2 mM PMSF, and 250 μg/ml cOmplete™, EDTA-free Protease Inhibitor Cocktail) for 30 min at 4°C. The chromatin lysate was sonicated to an average size of −250 bp using 40 cycles of sonication (30 sec on/off High-Power setting; Diagenode Biorupter Plus). A 20% fraction of the sonicated chromatin was incubated with one of the following antibodies: 1 μl of anti-Rpb1; 1 μl of anti-Hsf1 (Chowdhary et al., 2019) or 2.5 μl of anti-Myc (Santa Cruz Biotechnology) for 16 h at 4°C. Antibody-chromatin complexes were immobilized on Protein A-Sepharose beads (GE Healthcare) for 16 h at 4°C, then washed sequentially with lysis buffer, high salt buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 1% Triton X-100, 0.1% Sodium Deoxycholate, 1 mM EDTA), wash buffer (10 mM Tris pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% Sodium Deoxycholate, 1 mM EDTA) and finally 1x TE (10 mM Tris-HCl pH 8.0, 0.5 mM EDTA). Chromatin was eluted by incubating the beads in elution buffer (50 mM Tris pH 8.0, 1% SDS, 10 mM EDTA) at 65°C for 30 min. RNA and proteins were removed by DNase-free RNase (final concentration of 200 μg/ml; incubation at 37°C for 1 h) and Proteinase K (final concentration of 50 μg/ml; incubation at 60°C for 16 h). The ChIP template was extracted using phenol-chloroform and precipitated in presence of ethanol.
ChIP occupancy signals were quantified using qPCR. Sequences of ChIP primers used in this study are provided in Table S4. The ChIP DNA quantities were deduced from interpolation of a standard curve generated using genomic DNA template. The qPCR signal for each primer combination was normalized to the corresponding signal from the input DNA. The input DNA control was incorporated to correct for variation in the recovery of ChIP DNA templates.
Reverse Transcription-qPCR (RT-qPCR)
Cells were cultured to a mid-log density (A600 = 0.8) at 30°C, and were either maintained at 30°C or heat-shocked at 39°C for times indicated. 20 mM sodium azide was added at appropriate times to terminate transcription. Cells were then harvested and subjected to glass bead lysis in presence of TRIzol (Invitrogen) and chloroform for 10 min at 4°C. Total RNA was precipitated in ethanol. A fraction of total RNA (~20 μg) was treated with DNase I (RNase-free; New England Biolabs) at 37°C for 15 min. DNase I was heat-inactivated at 75°C for 10 min. RNA was purified using the RNA clean and concentrator kit (Zymo Research). 2-3 μg of the purified RNA template and random hexamers were used for preparing cDNA (Superscript IV first-strand synthesis system; Invitrogen).
The cDNA reaction mix was diluted 2-fold, and 2 μl of the diluted cDNA template was used for qPCR reaction. Sequences of primers used for RT-qPCR analysis in this study are provided in Table S5. Relative cDNA levels were quantified using the ΔΔCt method (see Chowdhary et al., 2017). qPCR signal from SCR1 Pol III transcript was used as a normalization control. This accounted for variation in the recovery of cDNA templates. Relative fold change per minute in mRNA expression was calculated by dividing mean mRNA levels (derived from two independent biological samples) for a given time point by previous time point, and then by the time elapsed in minutes.
Fluorescence microscopy
For live-cell imaging, cells were grown at 30°C to early log phase (A600 = 0.5) in synthetic dextrose complete (SDC) medium. An aliquot of living cells was immobilized onto concanavalin A-coated glass bottom dish. Fresh SDC medium was added in the dish before imaging. Images were taken on Leica TCS SP5 II STED-CW laser scanning confocal microscope (Leica Microsystems, Inc., Buffalo Grove, IL) equipped with GaAsP hybrid detector and STED mode turned off. Samples were subjected to heat shock at 39°C by heating the objective (HCX PL APOCS 63x/1.4 oil UV) with Bioptechs objective heater system and by controlling the temperature of air flowing through the incubator chamber enclosing the microscope. Argon laser was used at lines 488 and 514 nm for excitation of GFP and mVenus. The orange HeNe laser light source was used for excitation of mCherry at 594 nm. STED laser (592 nm) was turned off for all live imaging experiments, except during STED imaging in Figures 2A, B (see below for details). For experiments involving hexanediol treatment, cells were heat-shocked and imaged for ~8 min. Immediately after this, the SDC media in the dish was replaced with pre-warmed (39°C) SDC supplemented with either 2,5- or 1,6-hexanediol (5% final concentration), and cells were imaged at 39°C for the times indicated.
The images were acquired in xyz scan mode, covering 8 to 10 planes on the z-axis with an interplanar distance of 0.25 μm. For dual-color imaging of live cells, fluorophores were sequentially scanned in two channels with scanning modes switched on between lines. The sequential capture allowed rapid scanning of fluorophores in both channels while minimizing bleed-through or channel crosstalk. Post-acquisition, images were deconvolved by YacuDecu function that utilizes Richardson-Lucy algorithm for calculating theoretical Point Spread Functions (PSFs) (https://github.com/bnorthan) (Rueden et al., 2017). The PSFs were computed each time based on the set of microscopy parameters used in the imaging analyses. Custom plugins for Fiji (Schindelin et al., 2012) were used to colorize, split or merge channels, make composites and adjust brightness of the images. 3D rendering and visualization were performed using either ClearVolume (Royer et al., 2015) or arivis Vision 4D software v. 3.1 (render mode: maximum intensity; arivis AG, Rostok, Germany). For images processed without deconvolution (Figure S2A-D), images were acquired as above and custom plugins for Fiji were used to colorize, split or merge channels, subtract background and apply gaussian blur.
For colocalization analyses, the fractional overlap metric scores (Manders’ colocalization coefficients) were calculated using JACoP plugin implemented in ImageJ (Bolte and Cordelieres, 2006; Schneider et al., 2012). The average intensity plots were generated using the plot profile feature in ImageJ. Intensities of pixels were obtained along the line path (as indicated in Figure 1G) for each z-section. Intensities across nine individual z-sections were combined and the average was plotted for each channel.
For analysis of gene coalescence, nine z-planes with an interplanar distance of 0.5 μm, covering the entire depth of the nucleus, were inspected for location of tagged genomic loci. The relative nuclear positions of the tagged loci were assessed by measuring 3D distances between them. 3D distances were measured as 3D polyline lengths between the signal centroids of tagged gene loci. A cell was scored positive for coalescence if the 3D distance between the centroids was between 0.3 and 0.7 μm. 3D distance measurements, 3D rendering, and visualization were performed in arivis Vision4D.
Stimulated emission depletion (STED) super-resolution microscopy in live cells
Samples for STED imaging were prepared as described for live imaging, except live cells were subjected to heat shock at 39°C by heating the objective (STED rated HCX PL APO 100x/1.4 oil) and incubator chamber enclosing the microscope. High-resolution STED images of cells expressing Hsf1-mVenus were acquired in xyz scan mode (z=9; interplanar distance=0.25 μm) on Leica TCS SP5 II STED-CW laser scanning confocal microscope with STED mode turned on. The 514 nm line from argon ion laser light source was used to excite mVenus. Emission depletion was accomplished with 592 nm continuous wave STED laser. Images were deconvolved as above, then rendered (render mode: volumetric) and visualized in arivis Vision4D. STED microscopy was used for acquiring images analyzed and presented in Figure 2A, B.
QUANTIFICATION AND STATISTICAL ANALYSIS
Hsf1, Med15 and Rpb1 ChIP-seq data analysis
Hsf1 ChIP-seq data were obtained from GSE117653 (Pincus et al., 2018). Med15 data are from PRJNA657372 (Sarkar et al., 2022). Rpb1 ChIP-seq data were obtained from GSE125226 (Albert et al., 2019). Reads were aligned to the S. cerevisiae reference genome (SacCer3) using Bowtie 2 (Langmead and Salzberg, 2012). SAM files were converted to BAM format using SAMtools (Li et al., 2009). BAM files were then converted to bigWig and bedGraph format at bin size of 1 (−bs 1) and normalized to the library size (--normalizeUsing CPM) using bamCoverage function of deepTools2 (Ramirez et al., 2016). The bigWig files were used to obtain genome browser tracks in Integrative Genomics Viewer (IGV) browser (Robinson et al., 2011), and to make metagene plots. Metagene profiles were created using computeMatrix and plotProfile function tools in deepTools2. The plots are scaled +/−1 kb of ORFs of the Hsf1-dependent genes (Pincus et al., 2018). Occupancy of Hsf1, Med15 and Rpb1 (Pol II) were computed using the bedtools map function. The occupancy of Hsf1 and Med15 were obtained as sum of signals across 1 kb region upstream of ORFs of protein-coding genes. For occupancy of Pol II, sum of signals across ORFs were normalized to length of the ORFs.
For analysis of co-occupancy between Hsf1 and Med15, peaks were called using MACS2 algorithm (Zhang et al., 2008) with -g 1.2e7 --keep-dup auto flags. Peaks identified on repetitive regions of telomeres and rDNA were excluded from the analysis. The overlap of Med15 and Hsf1 peaks was computed using intersectBed -wa -u -a Med15_peaks.bed -b Hsf1_peaks.bed option.
For analysis of Pol II distribution at Hsf1 genes, CPKM of Pol II reads were obtained for all non-overlapping, verified ORFs archived in SGD (Cherry et al., 2012) using the bedtools map function. These normalized counts were then used to calculate % Pol II reads at Hsf1 gene targets in comparison to all other genes in the yeast genome.
Nascent transcript sequencing data analysis
NAC-seq data were obtained from GSE117653 (Pincus et al., 2018). Reads were aligned to the S. cerevisiae reference genome (SacCer3) using TopHat2 (Kim et al., 2013) with --segment-length 20 -I 2500 options. Wiggle files were generated by normalizing to the library size (--normalizeUsing CPM) using the bamCoverage function of deepTools2 and visualized in IGV genome browser.
Foci counting and characterization
Hsf1, Med15 and Rpb3 foci were detected and quantified within 3D live-cell images using the automated FindFoci plugin (Herbert et al., 2014) in ImageJ. For foci detection in 3D, a region of interest was marked around the nucleus of each cell and then foci within the marked regions were identified across all z planes (z=9). We used the optimizer function in the plugin submenu to train FindFoci algorithm and predict optimum parameter settings for foci assignment. The following set of optimized parameters were applied to all images in this study: (a) background, one standard deviation above mean; (b) search method, above background with an optimal value of 0.3; and (c) merge option, relative above background with peak parameter of 0.2 and minimum size as 1. Finally, we used the x, y, z position coordinates and sizes of peak intensity regions to present foci as 3D bubble charts. The 3D bubble charts were created by calling bubblechart3 function in MATLAB (MATLAB, 2021. version 9.11.0.1873467 (R2021b), Natick, MA: The MathWorks, Inc.).
For visualizing images as 3D surface plots, a square of 10 × 10 pixels was marked around the nucleus of each cell type in each condition (as indicated in Figure 1D). Representative planes of each cell were analyzed and displayed as 3D surface plots using the Interactive 3D Surface Plot plugin in ImageJ (contributing author: Kai Uwe Barthel, Germany). Appropriate parameter settings for scale, rotation, smoothing, and lightning were adapted for the display of 3D surface plots. Note that for each cell type, similar parameter settings were used in both non-heat shock and heat shock conditions.
Analysis of average relative localization of factors
Foci of LacO-tagged HSP104 gene locus or MS2-tagged HSP12 mRNA locus were manually identified in a specific z plane and centered within a square of 12 × 12 pixels. Distribution of signal from another channel, depicting nuclear distribution of the secondary factor, was gathered from corresponding 12 × 12 squares in the same z plane. A composite text image was created by computing average of centered images from roughly 30 cells per condition. The intensity of each pixel was scaled from 0 to 100 using a formula for uniform scaling: [(intensity-minimum) / (maximum-minimum)] × 100. For negative control, fifteen 12 × 12 arrays were generated with random numbers from 0 to 100. A composite text image was then created by computing an average of the corresponding values. For positive control, kernel density of a 2D Gaussian distribution was generated in an array and computed using the following formula: (e^(−z2/(2σ2)) / σ (2π)−1/2). Here, z is in range (−k, k+1) and σ = 1. This array was then used to create a text image with a scale ranging between 0 and 100. Finally, text images of controls or test samples were used to generate contour plots using the Plotly function (Plotly Technologies Inc., 2015) in R (R Core Team, 2020). The intensity minima and maxima were split into 800 steps for all contour plots.
Quantification of overall and pixel-to-pixel variation within STED-acquired images
For demonstration of internal dynamics of Hsf1 foci, the variation between corresponding pixels of Hsf1 foci was compared against overall variation in the images. For this analysis, we used STED-acquired images of cells expressing Hsf1-mVenus captured at every minute over the intermixing phase of heat shock (8 to 12 min-HS). We marked 12 (i) × 12 (j) squares bounding the nuclei of six individual cells imaged every minute between t1 (8min) and t5 (12min). Next, we obtained maximum intensity projections of the 3D images stacks for each individual nuclei at each of five heat shock time points. All pixel intensities were scaled from 0 to 100 to account for variation in image brightness by computing into the following formula: [(intensity-minimum) / (maximum-minimum)] × 100. Standard deviation (σ) was calculated for the min-max normalized pixels to determine pixel-to-pixel variation in Hsf1 levels at the given nuclear locations. Overall standard deviation was computed from average intensities of each image in the time course. The overall standard deviation was then used to populate a 12 × 12-pixel square for comparison to pixel-to-pixel variation of Hsf1 intensity and presented as heat maps. Six such distributions, each of overall variation and pixel-to-pixel variation, were used to plot cumulative frequency of pixels as a function of standard deviation.
Quantification of 3C
TaqI-3C data was quantified as described in (Chowdhary et al., 2020; Chowdhary et al., 2019). The percent digestion efficiency was determined by amplifying a region across Taq I restriction site using a pair of convergent primers (sequences provided in Table S3). The percent digestion efficiencies were determined for each primer combination and incorporated into the following formula:
Here, CtR is the cycle threshold (Ct) quantification of the digested only (DO) or undigested (UND) templates, and CtARS504 the cycle threshold quantification of the ARS504 locus (a region lacking a Taq I site).
For measurement of normalized frequency of intragenic or intergenic interactions, the Ct values for digested only (DO3C) and ligated (Lig3C) templates for crosslinked chromatin, and digested only and ligated genomic DNA (LiggDNA and DOgDNA, respectively) were incorporated into the following formula:
(i): ΔCt values were obtained by subtracting Ct (no-template) from those of Lig3C, DO3C, or gDNA templates.
(ii): 2−ΔCt/ARS504 are the fold-over signals normalized to ARS504 locus.
(iii): Ligation-dependent signals (LDS) are determined as ratio of fold-over normalized signals of Lig3C and DO3C templates (2−ΔCtLig3C/ARS504Lig3C)/(2ΔCtDO3C/ARS504DO3C); also applicable to the gDNA control.
(iv): LDS were corrected for variation in Taq I digestion efficiencies of sites 1 and 2 (as detailed above).
(v): Normalized Frequency of Interaction is defined as the ratio of ligation-dependent signals of 3C and gDNA control templates after correcting for differences in their digestion efficiencies.
Statistical tests used
Student’s t test (two-tailed) was used to calculate statistical significance between all pairwise comparisons (as assumptions of parametric distributions were fulfilled), except in Figure 2B and D, two sample Kolmogorov–Smirnov test and one-way ANOVA followed by Tukey’s post hoc analysis were used, respectively.
Each pairwise comparison is done with means of two independent biological samples (N=2) +SD. n.s., P>0.05; *, P<0.05; **, P<0.01; ***, P<0.001.
Supplementary Material
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Polyclonal anti-Hsf1 | Gross Lab | N A |
| Rabbit Polyclonal anti-Rpb1 | Gross Lab | N A |
| Mouse monoclonal anti-Myc (9E10) | Santa Cruz Biotechnology | Cat #sc-40; RRID:AB_627268 |
| Chemicals, peptides, and recombinant proteins | ||
| TaqI-v2 | New England BioLabs | Cat#R0149 |
| Quick T4 DNA Ligase | New England BioLabs | Cat#M2200L |
| 1,6-Hexanediol | Sigma-Aldrich | Cat#240117 |
| 2,5-Hexanediol | Sigma-Aldrich | Cat#H11904 |
| RNase (DNase-Free) | Sigma Aldrich | Cat#11579681001 |
| Proteinase K | Invitrogen | Cat#25530049 |
| Power SYBR Green PCR Master Mix | ThermoFisher Scientific | Cat#4367660 |
| Protein A Sepharose Beads | GE Life Sciences | Cat#17096303 |
| TRIzol Reagent | Invitrogen | Cat#15596026 |
| DNase I (RNase-free) | New England BioLabs | Cat#M0303 |
| Critical commercial assays | ||
| RNA Clean and Concentrator-25 | Zymo Research | Cat#R1018 |
| Superscript IV first-strand synthesis system | Invitrogen | Cat#18091050 |
| Deposited data | ||
| Hsf1 ChIP-seq | (Pincus et al., 2018) | GEO# GSE117653 |
| Med15 ChIP-seq | (Sarkar et al., 2020) | NCBI Short Read Archive Project# PRJNA657372 |
| Rpb1 ChIP-seq | (Albert et al., 2019) | GEO# GSE125226 |
| NAC-seq | (Pincus et al., 2018) | GEO# GSE117653 |
| Imaging data | This paper | doi: 10.17632/cr972zx459.1 |
| Experimental models: Organisms/strains | ||
|
Saccharomyces cerevisiae W303 and BY4741 strain backgrounds (Table S1) |
This paper | N A |
| Oligonucleotides | ||
| Primers for 3C analysis (Table S2 and S3) | This paper | N A |
| Primers for ChIP analysis (Table S4) | This paper | N A |
| Primers for RT-qPCR (Table S5) | This paper | N A |
| Primers for strain construction (Table S6) | This paper | N A |
| Recombinant DNA | ||
| pFA6a-mCherry-hphMX6 | (Wang et al., 2014) Jian-Qui Wu |
Addgene plasmid # 105156 RRID: Addgene_105156 |
| pSH100 (YCplac33 MET25pro MCP-mCherry | (Hocine et al., 2013) Robert Singer and Daniel Zenklusen |
Addgene plasmid # 45930 RRID: Addgene_45930 |
| pDZ415 (24MS2SL loxP-Kan-loxP) | (Hocine et al., 2013) Robert Singer and Daniel Zenklusen |
Addgene plasmid # 45162 RRID: Addgene_45162 |
| pFA6a-13Myc-His3MX | (Longtine et al., 1998) John Pringle |
Addgene plasmid # 41602 RRID: Addgene_41602 |
| pSH69 | (Hegemann and Heick, 2011) | Euroscarf (P30675) |
| pNH604-HSF1pr-HSF1-GFP | This paper | N A |
| pNH604-HSF1pr-HSF1(147-833)-GFP | This paper | N A |
| Software and algorithms | ||
| Bowtie 2 | (Langmead and Salzberg, 2012) | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| SAMtools | (Li et al., 2009) | http://samtools.sourceforge.net |
| deepTools2 | (Ramirez et al., 2016) | https://deeptools.readthedocs.io/en/develop/ |
| Integrative Genomics Viewer (IGV) browser | (Robinson et al., 2011) | https://software.broadinstitute.org/software/igv/ |
| MACS2 | Zhang et al., 2008 | https://pypi.org/project/MACS2/ |
| TopHat2 | (Kim et al., 2013) | https://ccb.jhu.edu/software/tophat/index.shtml |
| Plotly | (Plotly Technologies Inc., 2015) | N A |
| R | (R Core Team, 2020) | https://www.r-project.org |
| Leica Application Suite-X | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| ImageJ2 | (Rueden et al., 2017) | https://imagej.net/software/imagej2/ |
| YacuDecu | Brian Northan | https://github.com/bnorthan |
| Fiji | (Schindelin et al., 2012) | https://imagej.net/software/fiji/#publication |
| ClearVolume | (Royer et al., 2015) | https://clearvolume.github.io |
| arivis Vision4D | arivis AG, Rostok, Germany | https://imaging.arivis.com/en/imaging-science/imaging-science |
| FindFoci | Herbert et al., 2014 | http://www.sussex.ac.uk/gdsc/intranet/microscopy/UserSupport/AnalysisProtocol/imagej/findfoci |
| MATLAB | MATLAB, 2021. version 9.11.0.1873467 (R2021b), Natick, MA: The MathWorks, Inc. | https://www.mathworks.com/products/matlab.html |
| JACoP | (Bolte and Cordelieres, 2006) | https://imagej.nih.gov/ij/plugins/track/jacop.html |
| Other | ||
| Bioruptor Plus | Diagenode | Cat#B01020001 |
| MatTek glass bottom dish (35mm) | MatTek | Cat#P35G-1.5-14-C |
| Bioptechs Delta TC3 Culture Dish Micro-Observation Temperature Control System | Bioptechs | Model# Delta TC3 |
HIGHLIGHTS.
Yeast Hsf1 forms inducible transcriptional condensates with Mediator and RNA Pol II
HSR condensates promote clustering of HSR genes located on different chromosomes
Hsp70 chaperone represses Hsf1 clustering, while Hsf1 activation domain promotes it
Transcriptional condensates are conserved and flexible features of eukaryotic gene control
ACKNOWLEDGEMENTS
We would like to thank V. Bindokas and C. Labno at the University of Chicago Integrated Light Microscopy Core (RRID: SCR_019197) for imaging assistance. We thank R. Ranganathan and A. Ruthenburg for sharing reagents and equipment. We also thank L. Dyer for help with strain construction and Pincus and Gross lab members for helpful discussions. This work was supported by NIH grants R01 GM138689 to D.P. and R01 GM138988 and R15 GM128065 to D.S.G, and NSF QLCI QuBBE grant OMA-2121044 to D.P.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Albert B, Kos-Braun IC, Henras AK, Dez C, Rueda MP, Zhang X, Gadal O, Kos M, and Shore D (2019). A ribosome assembly stress response regulates transcription to maintain proteome homeostasis. Elife 8. 10.7554/eLife.45002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandhakumar J, Moustafa YW, Chowdhary S, Kainth AS, and Gross DS (2016). Evidence for Multiple Mediator Complexes in Yeast Independently Recruited by Activated Heat Shock Factor. Mol Cell Biol 36, 1943–1960. 10.1128/MCB.00005-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek I, Friedman LJ, Gelles J, and Buratowski S (2021). Single-molecule studies reveal branched pathways for activator-dependent assembly of RNA polymerase II pre-initiation complexes. Mol Cell 81, 3576–3588 e3576. 10.1016/j.molcel.2021.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, and Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18, 285–298. 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagrie RA, Scialdone A, Schueler M, Kraemer DC, Chotalia M, Xie SQ, Barbieri M, de Santiago I, Lavitas LM, Branco MR, et al. (2017). Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543, 519–524. 10.1038/nature21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehning M, Dugast-Darzacq C, Rankovic M, Hansen AS, Yu T, Marie-Nelly H, McSwiggen DT, Kokic G, Dailey GM, Cramer P, et al. (2018). RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat Struct Mol Biol 25, 833–840. 10.1038/s41594-018-0112-y. [DOI] [PubMed] [Google Scholar]
- Boija A, Klein IA, Sabari BR, Dall'Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. (2018). Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 175, 1842–1855 e1816. 10.1016/j.cell.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, and Cordelieres FP (2006). A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224, 213–232. 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. (2012). Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40, D700–705. 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, and Cisse II (2018). Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415. 10.1126/science.aar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, Heckert A, Banala S, Lavis L, Darzacq X, and Tjian R (2018). Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361. 10.1126/science.aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary S, Kainth AS, and Gross DS (2017). Heat Shock Protein Genes Undergo Dynamic Alteration in Their Three-Dimensional Structure and Genome Organization in Response to Thermal Stress. Mol Cell Biol 37. 10.1128/MCB.00292-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary S, Kainth AS, and Gross DS (2020). Chromosome conformation capture that detects novel cis- and trans-interactions in budding yeast. Methods 170, 4–16. 10.1016/j.ymeth.2019.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary S, Kainth AS, Pincus D, and Gross DS (2019). Heat Shock Factor 1 Drives Intergenic Association of Its Target Gene Loci upon Heat Shock. Cell Rep 26, 18–28 e15. 10.1016/j.celrep.2018.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, and Ren B (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztanyi Z, Csizmok V, Tompa P, and Simon I (2005). IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434. 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, and Young RA (2014). Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159, 374–387. 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duster R, Kaltheuner IH, Schmitz M, and Geyer M (2021). 1,6-Hexanediol, commonly used to dissolve liquid-liquid phase separated condensates, directly impairs kinase and phosphatase activities. J Biol Chem 296, 100260. 10.1016/j.jbc.2021.100260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emenecker RJ, Griffith D, Holehouse AS (2022). Metapredict V2: An update to metapredict, a fast, accurate, and easy-to-use predictor of consensus disorder and structure. bioRxiv, 2022.06.06.494887. 10.1101/2022.06.06.494887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkine AM, Magrogan SF, Sekinger EA, and Gross DS (1999). Cooperative binding of heat shock factor to the yeast HSP82 promoter in vivo and in vitro. Mol Cell Biol 19, 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Liu X, and Ortlund EA (2021). Glucocorticoid receptor condensates link DNA-dependent receptor dimerization and transcriptional transactivation. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2024685118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglia G, Rashid R, Yapp C, Joshi GN, Li CG, Lindquist SL, Sarosiek KA, Whitesell L, Sorger PK, and Santagata S (2020). HSF1 phase transition mediates stress adaptation and cell fate decisions. Nat Cell Biol 22, 151–158. 10.1038/s41556-019-0458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall'Agnese A, Hannett NM, Spille JH, Afeyan LK, Zamudio AV, Shrinivas K, et al. (2019). Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548. 10.1038/s41586-019-1464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann JH, and Heick SB (2011). Delete and repeat: a comprehensive toolkit for sequential gene knockout in the budding yeast Saccharomyces cerevisiae. Methods Mol Biol 765, 189–206. 10.1007/978-1-61779-197-0_12. [DOI] [PubMed] [Google Scholar]
- Henninger JE, Oksuz O, Shrinivas K, Sagi I, LeRoy G, Zheng MM, Andrews JO, Zamudio AV, Lazaris C, Hannett NM, et al. (2021). RNA-Mediated Feedback Control of Transcriptional Condensates. Cell 184, 207–225 e224. 10.1016/j.cell.2020.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert AD, Carr AM, and Hoffmann E (2014). FindFoci: a focus detection algorithm with automated parameter training that closely matches human assignments, reduces human inconsistencies and increases speed of analysis. PLoS One 9, e114749. 10.1371/journal.pone.0114749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, and Young RA (2013). Super-enhancers in the control of cell identity and disease. Cell 155, 934–947. 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, and Sharp PA (2017). A Phase Separation Model for Transcriptional Control. Cell 169, 13–23. 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocine S, Raymond P, Zenklusen D, Chao JA, and Singer RH (2013). Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat Methods 10, 119–121. 10.1038/nmeth.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Li K, Cai W, Liu X, Zhang Y, Orkin SH, Xu J, and Yuan GC (2018). Dissecting super-enhancer hierarchy based on chromatin interactions. Nat Commun 9, 943. 10.1038/s41467-018-03279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Iida S, Tamura S, Nagashima R, Shiraki K, Goto T, Hibino K, Ide S, and Maeshima K (2021). 1,6-hexanediol rapidly immobilizes and condenses chromatin in living human cells. Life Sci Alliance 4. 10.26508/lsa.202001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, and Vourc'h C (2004). Stress-induced transcription of satellite III repeats. J Cell Biol 164, 25–33. 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Morimoto R, Robert-Nicoud M, and Vourc'h C (1997). HSF1 transcription factor concentrates in nuclear foci during heat shock: relationship with transcription sites. J Cell Sci 110 ( Pt 23), 2935–2941. [DOI] [PubMed] [Google Scholar]
- Kainth AS, Chowdhary S, Pincus D, and Gross DS (2021). Primordial super-enhancers: heat shock-induced chromatin organization in yeast. Trends in cell biology 31, 801–813. 10.1016/j.tcb.2021.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, and McKnight SL (2018). A Solid-State Conceptualization of Information Transfer from Gene to Message to Protein. Annu Rev Biochem 87, 351–390. 10.1146/annurev-biochem-061516-044700. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36. 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, and Gross DS (2013). Mediator recruitment to heat shock genes requires dual Hsf1 activation domains and mediator tail subunits Med15 and Med16. J Biol Chem 288, 12197–12213. 10.1074/jbc.M112.449553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiecik SW, Le Breton L, and Mayer MP (2020). Feedback regulation of heat shock factor 1 (Hsf1) activity by Hsp70-mediated trimer unzipping and dissociation from DNA. The EMBO journal 39, e104096. 10.15252/embj.2019104096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakowiak J, Zheng X, Patel N, Feder ZA, Anandhakumar J, Valerius K, Gross DS, Khalil AS, and Pincus D (2018). Hsf1 and Hsp70 constitute a two-component feedback loop that regulates the yeast heat shock response. Elife 7. 10.7554/eLife.31668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nuske E, Poser I, Richter D, and Alberti S (2015). Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife 4, e06807. 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, and Simon A (2017). Hexanediol: a chemical probe to investigate the material properties of membrane-less compartments. [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra TL, and Larson DR (2016). Single-Molecule mRNA Detection in Live Yeast. Curr Protoc Mol Biol 113, 14 24 11–14 24 15. 10.1002/0471142727.mb1424s113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, and Pringle JR (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. . [DOI] [PubMed] [Google Scholar]
- Lu F, Portz B, and Gilmour DS (2019). The C-Terminal Domain of RNA Polymerase II Is a Multivalent Targeting Sequence that Supports Drosophila Development with Only Consensus Heptads. Mol Cell 73, 1232–1242 e1234. 10.1016/j.molcel.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wu T, Gutman O, Lu H, Zhou Q, Henis YI, and Luo K (2020). Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat Cell Biol 22, 453–464. 10.1038/s41556-020-0485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Salamanca HH, Duarte FM, Danko CG, and Lis JT (2016). Mammalian Heat Shock Response and Mechanisms Underlying Its Genome-wide Transcriptional Regulation. Mol. Cell 62, 63–78. 10.1016/j.molcel.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masser AE, Kang W, Roy J, Mohanakrishnan Kaimal J, Quintana-Cordero J, Friedlander MR, and Andreasson C (2019). Cytoplasmic protein misfolding titrates Hsp70 to activate nuclear Hsf1. Elife 8. 10.7554/eLife.47791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair RR, Pataki E, and Gerst JE (2022). Transperons: RNA operons as effectors of coordinated gene expression in eukaryotes. Trends Genet. 10.1016/j.tig.2022.07.005. [DOI] [PubMed] [Google Scholar]
- Nair RR, Zabezhinsky D, Gelin-Licht R, Haas BJ, Dyhr MC, Sperber HS, Nusbaum C, and Gerst JE (2021). Multiplexed mRNA assembly into ribonucleoprotein particles plays an operon-like role in the control of yeast cell physiology. Elife 10. 10.7554/eLife.66050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SJ, Yang L, Meluzzi D, Oh S, Yang F, Friedman MJ, Wang S, Suter T, Alshareedah I, Gamliel A, et al. (2019). Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat Struct Mol Biol 26, 193–203. 10.1038/s41594-019-0190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. (2015). A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–1077. 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Peffer S, Goncalves D, and Morano KA (2019). Regulation of the Hsf1-dependent transcriptome via conserved bipartite contacts with Hsp70 promotes survival in yeast. J Biol Chem 294, 12191–12202. 10.1074/jbc.RA119.008822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D, Khursheed B, Garabedian M, Fortin M, Lindquist S, and Yamamoto K (1990). Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature 348, 166–168. [DOI] [PubMed] [Google Scholar]
- Pincus D, Anandhakumar J, Thiru P, Guertin MJ, Erkine AM, and Gross DS (2018). Genetic and epigenetic determinants establish a continuum of Hsf1 occupancy and activity across the yeast genome. Mol Biol Cell 29, 3168–3182. 10.1091/mbc.E18-06-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotly Technologies Inc. (2015). Collaborative data science. Plotly Technologies Inc. [Google Scholar]
- R Core Team (2020). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–165. 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat P, Boehning M, Hummel B, Aprile-Garcia F, Pandit AS, Eisenhardt N, Khavaran A, Niskanen E, Vos SM, Palvimo JJ, et al. (2021). Stress-induced nuclear condensation of NELF drives transcriptional downregulation. Mol Cell 81, 1013–1026 e1011. 10.1016/j.molcel.2021.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat Biotechnol 29, 24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer LA, Weigert M, Gunther U, Maghelli N, Jug F, Sbalzarini IF, and Myers EW (2015). ClearVolume: open-source live 3D visualization for light-sheet microscopy. Nat Methods 12, 480–481. 10.1038/nmeth.3372. [DOI] [PubMed] [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, and Eliceiri KW (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529. 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff KM, Choi YH, Cox D, Ormsby AR, Myung Y, Ascher DB, Radford SE, Pappu RV, and Hatters DM (2021). Sequence grammar underlying unfolding and phase separation of globular proteins. bioRxiv, 2021.2008.2020.457073. 10.1101/2021.08.20.457073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. (2018). Coactivator condensation at super-enhancers links phase separation and gene control. Science 361. 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall'Agnese A, and Young RA (2020). Biomolecular Condensates in the Nucleus. Trends Biochem Sci 45, 961–977. 10.1016/j.tibs.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Yeh BT, Feigerle JT, Hao CV, Townshend RJ, Lieberman Aiden E, Dror RO, and Kornberg RD (2021). Simple biochemical features underlie transcriptional activation domain diversity and dynamic, fuzzy binding to Mediator. Elife 10. 10.7554/eLife.68068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A, et al. (2020). Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 181, 306–324 e328. 10.1016/j.cell.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D, Zhu ZI, Knoll ER, Paul E, Landsman D, and Morse RH (2022). Mediator dynamics during heat shock in budding yeast. Genome Res 32, 111–123. 10.1101/gr.275750.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, and Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science 357. 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Sorger PK (1990). Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell 62, 793–805. [DOI] [PubMed] [Google Scholar]
- Trotter EW, Berenfeld L, Krause SA, Petsko GA, and Gray JV (2001). Protein misfolding and temperature up-shift cause G1 arrest via a common mechanism dependent on heat shock factor in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 98, 7313–7318. 10.1073/pnas.121172998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle LM, Pacheco D, Warfield L, Wilburn DB, Hahn S, and Klevit RE (2021). Mediator subunit Med15 dictates the conserved "fuzzy" binding mechanism of yeast transcription activators Gal4 and Gcn4. Nat Commun 12, 2220. 10.1038/s41467-021-22441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Lo Presti L, Zhu YH, Kang M, Wu Z, Martin SG, and Wu JQ (2014). The novel proteins Rng8 and Rng9 regulate the myosin-V Myo51 during fission yeast cytokinesis. J Cell Biol 205, 357–375. 10.1083/jcb.201308146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei MT, Chang YC, Shimobayashi SF, Shin Y, Strom AR, and Brangwynne CP (2020). Nucleated transcriptional condensates amplify gene expression. Nat Cell Biol 22, 1187–1196. 10.1038/s41556-020-00578-6. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, and Young RA (2013). Master transcription factors and Mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319. S0092-8674(13)00392-9 [pii] 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H, Bard JAM, Pilipenko EV, and Drummond DA (2022). Chaperones directly and efficiently disperse stress-triggered biomolecular condensates. Mol Cell 82, 741–755 e711. 10.1016/j.molcel.2022.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio AV, Dall'Agnese A, Henninger JE, Manteiga JC, Afeyan LK, Hannett NM, Coffey EL, Li CH, Oksuz O, Sabari BR, et al. (2019). Mediator Condensates Localize Signaling Factors to Key Cell Identity Genes. Mol Cell 76, 753–766 e756. 10.1016/j.molcel.2019.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander G, Hackmann A, Bender L, Becker D, Lingner T, Salinas G, and Krebber H (2016). mRNA quality control is bypassed for immediate export of stress-responsive transcripts. Nature 540, 593–596. 10.1038/nature20572 [DOI] [PubMed] [Google Scholar]
- Zhang H, Shao S, Zeng Y, Wang X, Qin Y, Ren Q, Xiang S, Wang Y, Xiao J, and Sun Y (2022). Reversible phase separation of HSF1 is required for an acute transcriptional response during heat shock. Nat Cell Biol 24, 340–352. 10.1038/s41556-022-00846-7. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Beyzavi A, Krakowiak J, Patel N, Khalil AS, and Pincus D (2018). Hsf1 Phosphorylation Generates Cell-to-Cell Variation in Hsp90 Levels and Promotes Phenotypic Plasticity. Cell Rep 22, 3099–3106. 10.1016/j.celrep.2018.02.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Krakowiak J, Patel N, Beyzavi A, Ezike J, Khalil A, and Pincus D (2016). Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. Elife 5. 10.7554/eLife.18638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, and O'Shea EK (2014). Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature 514, 117–121. 10.1038/nature13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw images associated with the key figure were deposited on Mendeley at doi: 10.17632/cr972zx459.1. Other microscopy and non-imaging data reported in this paper will be shared by the lead contact upon request.
This paper does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.