Abstract
Circular RNAs are non-coding RNAs that widely exist in eukaryotes. The research progress of its generation mechanism and biological function show that circular RNAs may be used in the development of tumors, neurological diseases, cardiovascular diseases. They play an important role in the occurrence and development of diseases and has a potential to be used as a disease marker. Oral squamous cell carcinoma is one of the most common malignant tumors in oral surgery. It is difficult to treat, easy to metastasize, and has a poor prognosis. Due to its unclear mechanism, blocking oral squamous cell carcinoma at the genetic level cannot be achieved. The research progress of circular RNA in the field of oral squamous cell carcinoma will bring new ideas for the biological treatment of oral squamous cell carcinoma. This review summarizes the circRNAs mechanism, the biological function and the research progress in the development of tumors, especially oral squamous cell carcinoma.
Keywords: Non-coding RNAs, Circular RNAs, Biology function, Oral squamous cell carcinoma, Biomarker
1. Introduction
Circular RNAs (circRNAs) are a new member of the long non-coding RNAs (lncRNAs) family [1]. Unlike linear RNA, this RNA does not have the characteristics of 5′methylguanosine cap and 3′polyadenylation tail, but a covalent closed-loop structure, so circular RNA is not effective for ribonucleases (Rnases), what makes they more sensitive and more stable than linear RNA [2]. According to different sources, circular RNA can be divided into exonic circular RNA (exonic circRNA), intronic circular RNA (intronic circRNA), exon-intron circular RNA (exon-intron circRNA), and tRNA-derived circular RNA (TriRNA) [[3], [4], [5], [6]]. Research has shown that circular RNAs are widely involved in the pathogenesis of various diseases, and their complex and diverse functions, unique abundance, breadth, stability and tissue specificity play an important role in the pathological process and become a biological star in the field of science [7].
Oral squamous cell carcinoma (OSCC) is the 11th most common cancer in the world [8]. OSCC has the characteristics of local invasiveness, high recurrence, and easy metastasis. The 5-year survival rate is lower than performed in other solid tumors, which is only 50–60% [9]. However, the molecular mechanism of OSCC is still unclear, and there are no highly sensitive and specific biomarkers as monitoring indicators for the early diagnosis, treatment and prognosis of OSCC. Previous literature has shown that circRNAs are associated with the malignant progression of OSCC, suggesting that circRNAs have potential functions as markers for early diagnosis and biological treatment of OSCC. This article briefly introduces the characteristics and the generation process of circular RNAs, and discusses their research progress in tumors, especially OSCC, from their biological functions.
2. The mechanism of circular RNAs formation
In eukaryotes, canonical splicing is formed by the spliceosome machinery, which removes introns and joins exons into one linear RNA transcript [10]. Most circular RNAs are derived from exons. The whole process can be divided into two parts: 1) the upstream intron of one or more exon pairs is connected with the downstream intron; 2) the 2′-hydroxyl of the upstream intron is connected with the 5′-hydroxyl of the downstream intron The phosphate group reacts, and then the 3′-hydroxyl group of the 3′-exon reacts with the 5′-phosphate of the 5′-exon, finally forming a circular RNA [11]. ALU sequences in introns can also interact, promoting back splicing [12]. Some RNA-binding proteins act as regulators in this process. For example, the RNA-binding protein MBL joins proximal introns and promotes the production of circular RNAs; while the RNA editing enzyme ADAR1 inhibits the production of circular RNAs [13,14]. In addition, a lasso-driven circularization is another mechanism for the generation of circular RNAs, upstream splice acceptors and downstream donors are brought close to each other due to the presence of RNA lasso, while exons are formed by 5′-3′ phosphodimers ester linkage [15].
3. Biological functions of circular RNAs
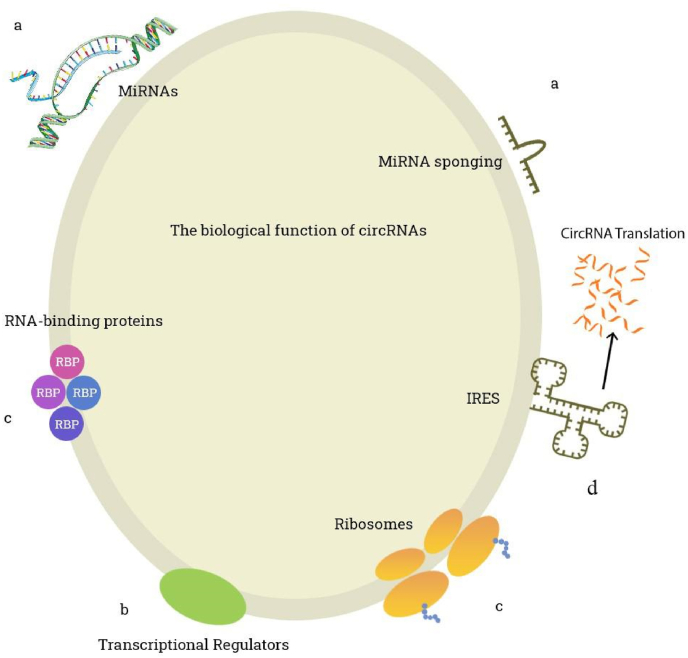
Circular RNAs have diverse functions as non-coding RNAs, which are mainly divided into the following four aspects:
-
a)
Sponging of miRNAs. miRNAs are considered to be endogenous non-coding RNAs, which can inhibit downstream gene expression, thereby inhibiting protein synthesis. More and more studies have found that circular RNAs with complementary sequences can sponge-like adsorb miRNAs and inhibit their downstream gene expression [7]. For example, cirs-7, also known as CDR1as, is a circular RNA containing more than 70 miR-7 binding sites, which can bind to miR-7 and act on its downstream mRNA [5,16]. This molecular pathway axis is widely expressed in diseases such as astrocytoma and lung cancer (Fig. 1) [17].
-
b)
Regulation of linear RNA transcription. Circular RNA can promote or inhibit the transcription of linear RNA and is a key regulator of alternative splicing or transcription. Circular RNAs formed by exons constitute a vast majority of known circular RNAs, and their generation process has a competitive impact on conventional splicing.
-
c)
Sponging on proteins. Circular RNAs can also adsorb RNA binding proteins (RNA binding proteins, RBPs) to regulate protein levels. For example, circ-MBL found in flies and humans can bind to MBL proteins at multiple binding sites [7]. An increasing number of studies have shown that some circular RNAs are also able to interact with proteins, thereby affecting the behavior of cells [18]. For example, circFOXO3, a circular RNA that has received the most extensive attention, has been shown to be an adaptor linking p21 and cyclindependent kinases 2 (CDK2). circFOXO3 promotes the release of CDK2 from p21, which can phosphorylate cyclin A and cyclin E, thereby promoting cell division and proliferation. High expression of circFOXO3 can also promote the interaction between p53 and MDM2, thereby accelerating the degradation of p53 [19].
-
d)
Regulation of protein translation. The translation is performed by ribosomes and involves initiation, elongation, termination and ribosome recycling [20]. The Initiation on eukaryotic mRNAs involves scanning by 43S preinitiation complexes from the 5′ cap-proximal point of attachment to the initiation codon, followed by ribosomal subunit joining and factor displacement [21]. Lacking the 5′-cap and 3′-tail, circRNA can only adopt cap-independent manners. In addition to previously described m6A-mediated translation [22], artificial and endogenous circRNAs containing an internal ribosome entry site (IRES) that directly recruits ribosomes [23], can also be translated. These two approaches may be coupled with each other. For example, m6A improves the efficiency of IRES-mediated translation of circZNF609 [24]. Additionally, circRNA with an infinite ORF undergoes rolling circle amplification in an IRES-independent manner, leading to a hundred-fold higher productivity than linear transcript [25]. Peptides encoded by circRNAs are generally truncated and their functions are mostly analogous to the full-length protein counterparts (circFBXW7-185aa [26]). However, some proteins originating from circRNAs exert functions are independent of or even opposed to those of their host gene products (circFNDC3B-218aa) [27]. These results broaden the range of human proteome. However, the regulatory mechanisms of circRNA translation and the processes of elongation and termination are still not completely understood.
Fig. 1.
The biological functions of circRNAs.
4. Circular RNAs and tumors
Studies have shown that circular RNAs play an important role in the occurrence and development of solid tumors and hematological malignancies, especially through their biological functions as miRNA sponges. Circular RNAs are involved in several characteristic processes of tumorigenesis and development, such as the evasion of growth suppressors, maintenance of proliferative signals, the evasion of cells’ death and senescence, and the ability to promote angiogenesis and activate the invasion and metastasis. In addition, the abnormal expression, the tissue specificity, the diversity and the stability of circRNAs in tumor cells make them have a great potential as tumor markers. In liver cancer, researchers have found that the expression of hsa_circ_0005075 in 60 groups of liver cancer tissues was significantly different from normal tissues, indicating that hsa_circ_0005075 is a potential biomarker [28]. In addition, the expression of hsa_circ_0001649 is also different, and is closely related to the size of HCC tumor and the occurrence of tumor thrombus, while circZFR, circFUT8 and circIPO11 have been proved to be useful for the identification of HCC specimens [29,30]. For lung cancer, both clinical cohort studies and cell-level studies have found that circRNAITCH plays an inhibitory role in lung cancer. Abnormal regulation of circRNA-ITCH can enhance the expression of its parent tumor suppressor gene ITCH by sponging miR-7 and miR-214, thereby regulating the proliferation of cancer cells [31]. circ-001569 can sponge-like adsorb miR-145 and upregulate the expression of its downstream genes such as E2F5, BAG4 and FMN12, resulting in an increase in G2/M phase cells and a decrease in tumor cell apoptosis [32]. In addition, in non-solid tumors, Salzman et al. serendipitously found genes producing hundreds of circular RNAs in acute myeloid leukemia, and these circular RNAs were also detected in HeLa cells [33]. Another study has confirmed that chemotherapy can inhibit the abnormal expression of hsa_circ_0004277 in patients with acute myeloid leukemia [34]. In addition to the above tumors, studies have also found that some circRNAs are associated with gastric cancer, breast cancer, colon cancer, bladder cancer, ovarian tumors, and skin squamous cells [[35], [36], [37], [38], [39], [40], [41], [42]]. It is closely related to various cancers such as epithelial carcinoma [43].
5. Circular RNAs and OSCC
5.1. Research status of circular RNAs in OSCC
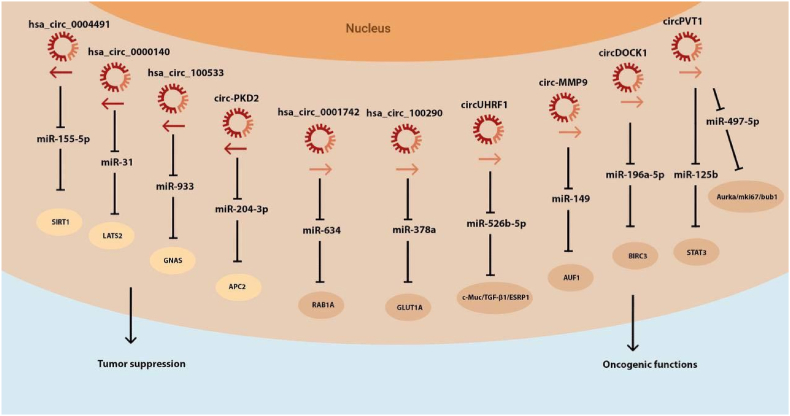
The application of high-throughput sequencing has allowed more and more OSCC-specifically expressed circular RNAs to be screened. Studies have screened circRNAs with significant differences between OSCC tissue and paracancerous tissue by circRNA chip technology, including 155 circRNAs with a relative expression ratio of more than 1.5 times. Circular RNA hsa_circ_0001874 was shown to be closely related to a tumor's clinical stage and a degree of differentiation, and its expression in low differentiated OSCC tissues has been significantly higher than in moderately and highly differentiated OSCC tissues [44]. Through the prediction of circular RNA target gene analysis software, it is inferred that miR-103A-3P, miR-107, miR-593–5p, miR-661 and miR-662 may be the sponge targets of hsa_circ_0001874, but the specific functions are still unknown. Similar studies have also included exploring the expression profiles of circRNAs in tongue squamous cell carcinoma tissues and normal paracancerous tissues. High-throughput sequencing was performed in 4 cases of cancer foci and 4 cases of adjacent tissues. A total of 17 171 circRNAs with differences were screened, among which 15 circRNAs had a 50-fold difference [45]. The upper circular RNA hsa-circ-0033 967 with a differential fold of 116.31 is predicted to have 163 potential binding sites for hsa-miR-608, and hsamiR-608 has been confirmed to play a tumor suppressor role in liver cancer glioma in recent years [46]. This finding suggested that hsa-circ-0033 967 could block the tumor suppressor effect of hsa-miR-608 through a sponge action, thereby promoting the occurrence and the development of tongue cancer. Some circular RNAs have been shown to have unique signaling pathways in the malignant process of OSCC, and have clinical application value. Cyclin-dependent kinases 6 (CDK6) promote tumorigenesis at some specific stages by regulating transcriptional responses. Wang et al. established a cell apoptosis model based on three oral squamous cell carcinoma cell lines CAL-7, SCC-9, and SCC-25 [47]. Compared with the control group, it was found that there were differences in the expression of 628 circRNAs, among which the circRNA DOCK1. It is one of the circular RNAs whose expression is significantly decreased in the apoptosis model. Real-time quantitative polymerase chain reaction found that this circular RNA is also highly expressed in OSCC cell lines and tissues. In this low-expression model of circular RNA, the expression level of miR-193a-5p, which plays an important role in various apoptosis pathways, increased, and at the same time, the apoptosis rate increased to varying degrees. Further experiments proved that miR-193a-5p could bind to the mRNA of baculoviral IAP repeat containing (BIRC) to reduce the content of BIRC in cells and increase the apoptosis rate. Chen X. et al. have found that circ_0014 359 was highly expressed in the OSCC tissues and cell lines compared to the normal controls and that expression was associated with the survival of patients. For the OSCC cell lines, circ_0014 359 knock down induced apoptosis and inhibited migration, invasion, and epithelial-mesenchymal transition of OSCC cells. In vivo, silencing the circ_0014 359 blocked the growth of OSCC tumors. The circ_0014 359 can directly interact with the micro-RNA-149 (miR-149) [48]. The existence of −196a-5p/BIRC3 signaling pathway-circRNA DOCK1 can indirectly affect the apoptosis of OSCC by regulating the expression of the protein BIRC, which reflects the potential of circRNA DOCK1 as a diagnostic biomarker and therapeutic target for OSCC. Another study has found that another circular RNA may also assist in the diagnosis of OSCC [49]. Cui L. et al. have found that CircCDR1as acted as an oncogene in OSCC progression through elevating SLC7A11 by targeting miR-876–5p [50]. The expression of hsa_circ_1001242 was significantly increased in 4 oral squamous cell carcinoma cell lines SCC-9, SCC-15, SCC25, and CAL-27, and the expression of hsa_circ_1001242 was significantly increased. Levels and tumor size were negatively correlated with T stage. Similarly, the expressions of hsa_circ_0001874 and hsa_circ_0001971 in saliva were significantly increased in OSCC samples, and they have the potential to be used as OSCC tumor markers to judge the degree of tumor development, because their contents are related to the TNM stage of OSCC, and the former is related to the tumor grade. The hsa_circ_0001874 had a higher expression level in the more malignant samples. Not only that, OSCC and oral leukoplakia can also be distinguished by comparing the differences in the expression levels of these two circular RNAs, and surgery will also reduce the content of the two circular RNAs in saliva [51]. Not all circRNAs in OSCC are reported below. The only circRNA forming the circRNA-mRNA regulatory axis is described, along with a description of the role in other forms of cancer to understand the diversity of function. Table 1 provides a succinct description of the regulatory network and expression levels in OSCC tissues.
Table 1.
A Short display of the above described circRNA-miRNA pathway regulatory axis.
| circRNA | Expression IN OSCC | Sponged miRNA | Regulatory Axis | Reference |
|---|---|---|---|---|
| ciRS-7 | Upregulated | miR-671–5p | ciRS-7/miR-671–5p/CDR1/AKT/ERK1/2/mTOR/ROS | 47 |
| circPVT1 | Upregulated | miRNA-125b | circPVT1/miRNA-125b/STAT3 | 52 |
| circHIPK3 | Upregulated | miR-124 | circHIPK3/miR-124/ITGB1 | 53 |
| circMDM2 | Upregulated | miR-532–3p | circMDM2/miR-532–3p/HK2 | 54 |
| circPKD2 | Downregulated | miR-204–3p | circPKD2/miR-204–3p/APC2/WNT/β-catenin/p-AKT/p-ERK1/2 | 55 |
| circDOCK1 | Upregulated | miR-196a-5p | circDOCK1/miR-196a-5p/BIRC3 | 56 |
| circATRNL1 | Downregulated | miR-23a-3p | circALTR1/miR-23a-3p/PTEN/AKT/P13K/ATM/ATR/P53 | 57 |
| hsa_circRNA_100 533 | Downregulated | miR-933 | hsa_circRNA_100533/miR-933/GNAS | 58 |
| hsa_circRNA_100 290 | Upregulated | miR-378a | hsa_circRNA_100 290/miR-378a/GLUT1 | 59 |
| hsa_circ_000140 | Downregulated | miR-31 | hsa_circ_000140/miR-31/LATS2 | 60 |
| hsa_circ_0001971 | Upregulated | miR-104, miR-204 | hsa_circ_0001971/miR-104/miR-204/P13K/AKT/FoxO3a | 61 |
| hsa_circ_0008309 | NC: Variation but significantly showed downregulation | miR-136–5P, miR-382–5P | hsa_circ_0008309/miR-136–5P/miR-382–5P/ATX1 | 62 |
5.2. Application value of circular RNAs in OSCC
Nowadays, some progress has been made in early diagnosis, surgical methods, and radiotherapy and chemotherapy for OSCC. However, the early symptoms of OSCC are not typical, and tissue biopsy is often required for final diagnosis. There is still a lack of rapid, accurate and non-invasive early diagnosis in clinical work. Circular RNAs are abundant, widespread, conserved, and can be stably expressed in saliva, blood, urine, and exocrine vesicles, and their expression in tissues and organs is stage-specific. Since OSCC occurs in the mouth, the test of saliva samples may become an early screening and a diagnosis method for OSCC high-risk markers, and the operation is simple and does not require invasive procedures to obtain samples. Several studies have shown that microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) in saliva can be used as early diagnostic markers for OSCC [63,64] [Fig. 2]. Compared with long non-coding RNAs, circular RNAs are less sensitive to RNases due to their closed loop structures, and the half-life of circular RNAs is often higher than 48 h, while the half-life of microRNAs is only 10 h on average. Therefore, circular RNAs are more suitable as markers [65]. Literature has shown that there are OSCC-specific circular RNAs in saliva. As mentioned above, the content of has-circ-0001874 and has-circ-0001971 in the saliva of OSCC patients are significantly increased. Specific circular RNAs are used as the strong evidence for the diagnosis of OSCC [51]. Similarly, circRNAs can also be used as prognostic markers for OSCC cyclic, and these OSCC-specific circRNAs can be used as monitoring indicators for regular follow-up of patients after surgery or medical treatment [66]. Research shows that some circRNAs can regulate the biological behavior of tumor cells through intracellular signaling pathways, so circRNAs are considered to be one of the future molecular biology therapeutic targets for OSCC in case circRNAs can be interfered by drugs and inhibit the proliferation of OSCC cells and promote apoptosis [67]. Nowadays, the most important method to interfere with the expression of circRNAs is to construct a targeted small interfering RNA (siRNA) of a specific type of circRNA through a database query, and to introduce it into OSCC cells, which can reach the level equivalent to the gene encoding the circRNA, thereby hindering the further development of cancer cells or cancerous tissue by the knockout effect. Research found that the introduction of circRNA-100290 targeting siRNA into OSCC cell lines inhibited the proliferation of OSCC cell lines in vitro and in vivo by regulating the circRNA-miR29 family-CDK6 pathway [25]. Similarly, Wang et al. constructed a low-expression model of this circRNA by introducing siRNA of circRNA DOCK1 and found that the apoptosis rate of CAL-7, SCC-9, and SCC-25 3 OSCC cell lines increased [68]. Using molecular biological methods to maintain the low expression state of circRNAs that promote the malignant development of OSCC has the potential to be used in clinical treatment.
Fig. 2.
Schematic diagram of the circRNA-miRNA networks in OSCC.
On the contrary, the overexpression of certain circular RNAs has a certain inhibitory effect on the proliferation and development of cancer cells. It has become possible to synthesize circular RNAs with specific benign effects by artificial means and to transfect them into cells to achieve therapeutic effects. Some researchers have successfully synthesized a circular RNA-scRNA21 that can significantly inhibit the proliferation of cancer cells in vitro [69]. This circular RNA can bind miR-21 through sponge action when introduced into cells, thereby up-regulating the target inhibition of miR-21. The expression of tumor suppressor gene Daxx induces the apoptosis of gastric cancer cells. By increasing the intracellular content of circRNAs and then affecting the biological behavior of OSCC cells, it provides a new idea for the treatment of OSCC.
6. Conclusions
Although circular RNAs have high advantages as biomarkers for diagnosis, prognosis and treatment of OSCC, the current research results still have great limitations. For example, when analyzing the differentially expressed circRNAs in OSCC patients, the size of a sample and a selection of the samples will affect the final data, and the current research results cannot represent the situation of all OSCC patients. In summary, as a molecule that has just been discovered to have special biological functions, circular RNA has good specificity and stability, and its potential as a tumor marker has been demonstrated in gastric cancer, colon cancer, breast cancer and other cancers [[29], [30], [31], [32], [33], [34]]. OSCC is a cancer that is prone to metastasis and has a poor prognosis, and its mechanism of occurrence and development is still unclear. Revealing the occurrence and development of OSCC at the molecular level will certainly provide more information on the early diagnosis, treatment and prognosis of OSCC and become a more efficient and accurate method. Studies on the role of circRNAs in OSCC have also appeared one after another, providing new ideas for tumor diagnosis and prognosis. Although more and more circRNAs that are differentially expressed between tumor tissues and normal tissues have been discovered, only a small number of circRNAs have been elucidated for their precise roles in cellular and molecular mechanisms, and the mechanism of action of the vast majority of circRNAs is still uncertain, and there is still much unknown to be explored [[70], [71], [72], [73], [74], [75], [76]]. Moreover, before the role of circRNA as a tumor diagnostic marker transitions from a theory to a clinical application and becomes a simple, efficient and accurate laboratory examination method, a plenty of research is needed as a basis.
Funding
This work was supported by the Bashkir State Medical University Strategic Academic Leadership Program (PRIORITY-2030).
Author contributions
Albert Sufianov and Sema Begliarzade conceptualized and designed the study. All authors have participated in the acquisition, analysis and interpretation of the data. Valentin Kudriashov has drafted the manuscript. Tatiana Ilyasova, Yanchao Liang contributed to the critical revisions of the manuscript. All authors agreed on the journal to which the article would be submitted, gave the final approval for the version to be published, and agreed to be accountable for all aspects of the work.
Declaration of competing interest
The authors declare they have no conflict of interest.
Contributor Information
Albert Sufianov, Email: sufianov@gmail.com.
Sema Begliarzade, Email: semanagiyeva@yandex.ru.
Valentin Kudriashov, Email: vkudryashov.uro@gmail.com.
Aferin Beilerli, Email: abeilerli@mail.ru.
Tatiana Ilyasova, Email: Iltanya67@yandex.ru.
Yanchao Liang, Email: liangyanchao@hrbmu.edu.cn.
Ozal Beylerli, Email: obeylerli@mail.ru.
References
- 1.Beilerli A., Gareev I., Beylerli O., Yang G., Pavlov V., Aliev G., Ahmad A. Circular RNAs as biomarkers and therapeutic targets in cancer. Semin. Cancer Biol. 2022 Aug;83:242–252. doi: 10.1016/j.semcancer.2020.12.026. [DOI] [PubMed] [Google Scholar]
- 2.Li J., Yang J., Zhou P., et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5(2):472–480. [PMC free article] [PubMed] [Google Scholar]
- 3.Jeck W.R., Sorrentino J.A., Wang K., et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talhouarne G.J., Gall J.G. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. RNA. 2014;20(9):1476–1487. doi: 10.1261/rna.045781.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meng X., Li X., Zhang P., et al. Circular RNA: an emerging key player in RNA world. Briefings Bioinf. 2017;18(4):547–557. doi: 10.1093/bib/bbw045. [DOI] [PubMed] [Google Scholar]
- 6.Lu Z., Filonov G.S., Noto J.J., et al. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA. 2015;21(9):1554–1565. doi: 10.1261/rna.052944.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J., Liu T., Wang X., et al. Circles reshaping the RNA world: from waste to treasure. Mol. Cancer. 2017;16(1):58. doi: 10.1186/s12943-017-0630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzmaurice C., Dicker D., Pain A., et al. The global burden of cancer 2013. JAMA Oncol. 2015;1(4):505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brocklehurst P.R., Baker S.R., Speight P.M. Oral cancer screening: what have we learnt and what is there still to achieve. Future Oncol. 2010;6(2):299–304. doi: 10.2217/fon.09.163. [DOI] [PubMed] [Google Scholar]
- 10.Demetrick D.J., Inoue M., Lester W.M., et al. Human papillomavirus type 16 associated with oral squamous carcinoma in a cardiac transplant recipient. Cancer. 1990;66(8):1726–1731. doi: 10.1002/1097-0142(19901015)66:8<1726::aid-cncr2820660814>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 11.Williams H.K. Molecular pathogenesis of oral squamous carcinoma. Mol. Pathol. 2000;53(4):165–172. doi: 10.1136/mp.53.4.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L.L., Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocquerelle C., Mascrez B., Hétuin D., et al. Mis-splicing yields circular RNA molecules. Faseb. J. 1993;7(1):155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 14.Meng S., Zhou H., Feng Z., et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer. 2017;16(1):94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lasda E., Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaphiropoulos P.G. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc. Natl. Acad. Sci. U. S. A. 1996;93(13):6536–6541. doi: 10.1073/pnas.93.13.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32(5):309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du W.W., Yang W., Liu E., et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du W.W., Fang L., Yang W., et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuller A.P., Green R. Roadblocks and resolutions in eukaryotic translation. Nat. Rev. Mol. Cell Biol. 2018;19(8):526–541. doi: 10.1038/s41580-018-0011-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson R.J., Hellen C.U., Pestova T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao J., Lee E.E., Kim J., Yang R., Chamseddin B., Ni C., et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun. 2019;10(1):2300. doi: 10.1038/s41467-019-10246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macejak D.G., Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353(6339):90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 24.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., et al. Circ-ZNF609 is a circular RNA that can be translated and functions in Myogenesis. Mol. Cell. 2017;66(1):22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abe N., Hiroshima M., Maruyama H., Nakashima Y., Nakano Y., Matsuda A., et al. Rolling circle amplification in a prokaryotic translation system using small circular RNA. Angew Chem. Int. Ed. Engl. 2013;52(27):7004–7008. doi: 10.1002/anie.201302044. [DOI] [PubMed] [Google Scholar]
- 26.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., et al. Novel role of FBXW7 circular RNA in repressing Glioma tumorigenesis. J. Natl. Cancer Inst. 2018;110(3):304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Z., Cai J., Lin J., Zhou H., Peng J., Liang J., et al. A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating snail in colon cancer. Mol. Cancer. 2020;19(1):71. doi: 10.1186/s12943-020-01179-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shang X., Li G., Liu H., et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine (Baltim.) 2016;95(22) doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X., Xiong Q., Wu Y., et al. Quantitative proteomics reveals the regulatory networks of circular RNA CDR1as in hepatocellular carcinoma cells[J] J. Proteome Res. 2017;16(10):3891–3902. doi: 10.1021/acs.jproteome.7b00519. [DOI] [PubMed] [Google Scholar]
- 30.Ren S., Xin Z., Xu Y., et al. Construction and analysis of circular RNA molecular regulatory networks in liver cancer. J] Cell Cycle. 2017;16(22):2204–2211. doi: 10.1080/15384101.2017.1346754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pei W., Tao L., Zhang L.W., et al. Circular RNA profiles in mouse lung tissue induced by radon[J] Environ. Health Prev. Med. 2017;22(1):36. doi: 10.1186/s12199-017-0627-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie H., Ren X., Xin S., et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salzman J., Gawad C., Wang P.L., et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W., Zhong C., Jiao J., et al. Characterization of hsa_circ_0004277 as a new biomarker for acute myeloid leukemia via circular RNA profile and bioinformatics analysis. Int. J. Mol. Sci. 2017;18(3):597. doi: 10.3390/ijms18030597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Liu H., Li W., et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY) 2017;9(6):1585–1594. doi: 10.18632/aging.101254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang M., He Y.R., Liang L.C., et al. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer[J] World. J. Gastroenterol. 2017;23(34):6330–6338. doi: 10.3748/wjg.v23.i34.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J., Li Y., Zheng Q., et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 38.Liang H.F., Zhang X.Z., Liu B.G., et al. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7(7):1566–1576. [PMC free article] [PubMed] [Google Scholar]
- 39.Lu L., Sun J., Shi P., et al. Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget. 2017;8(27):44096–44107. doi: 10.18632/oncotarget.17307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bachmayr-Heyda A., Reiner A.T., Auer K., et al. Correlation of circular RNA abundance with proliferation-exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci. Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong Z., Lü M., Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci. Rep. 2016;6 doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmed I., Karedath T., Andrews S.S., et al. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget. 2016;7(24):36366–36381. doi: 10.18632/oncotarget.8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sand M., Bechara F.G., Gambichler T., et al. Circular RNA expression in cutaneous squamous cell carcinoma. J. Dermatol. Sci. 2016;83(3):210–218. doi: 10.1016/j.jdermsci.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L., Wang M., Ren W., Li S., Zhi K., Gao L., Zheng J. Prognostic significance of CircRNAs expression in oral squamous cell carcinoma. Oral Dis. 2022 Mar 14 doi: 10.1111/odi.14188. [DOI] [PubMed] [Google Scholar]
- 45.Fan H.Y., Jiang J., Tang Y.J., Liang X.H., Tang Y.L. CircRNAs: a new chapter in oral squamous cell carcinoma biology. OncoTargets Ther. 2020 Sep 11;13:9071–9083. doi: 10.2147/OTT.S263655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang K., Liang Q., Wei L., et al. MicroRNA-608 acts as a prognostic marker and inhibits the cell proliferation in hepatocellular carcinoma by macrophage migration inhibitory factor. Tumour Biol. 2016;37(3):3823–3830. doi: 10.1007/s13277-015-4213-5. [DOI] [PubMed] [Google Scholar]
- 47.Wang L., Wei Y., Yan Y., et al. CircDOCK1 suppresses cell apoptosis via inhibition of miR-196a-5p by targeting BIRC3 in OSCC. Oncol. Rep. 2018;39(3):951–966. doi: 10.3892/or.2017.6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X., Chen J., Fu L., Chen J., Chen Y., Liu F. Circ_0014359 promotes oral squamous cell carcinoma progression by sponging miR-149. Acta Biochim. Pol. 2022 May 18;69(2):315–320. doi: 10.18388/abp.2020_5745. [DOI] [PubMed] [Google Scholar]
- 49.Sun S., Li B., Wang Y., et al. Clinical significance of the decreased expression of hsa_circ_001242 in oral squamous cell carcinoma. Dis. Markers. 2018;2018 doi: 10.1155/2018/6514795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cui L., Huang C., Zhou D. Overexpression of circCDR1as drives oral squamous cell carcinoma progression. Oral Dis. 2021 Nov 24 doi: 10.1111/odi.14085. [DOI] [PubMed] [Google Scholar]
- 51.Zhao S.Y., Wang J., Ouyang S.B., et al. Salivary circular RNAs Hsa_Circ_0001874 and Hsa_Circ_0001971 as novel biomarkers for the diagnosis of oral squamous cell carcinoma. Cell. Physiol. Biochem. 2018;47(6):2511–2521. doi: 10.1159/000491624. [DOI] [PubMed] [Google Scholar]
- 52.Xia B., Hong T., He X., Hu X., Gao Y. A circular RNA derived from MMP9 facilitates oral squamous cell carcinoma metastasis through regulation of MMP9 mRNA stability. Cell Transplant. 2019;28:1614–1623. doi: 10.1177/0963689719875409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng L., Yuan X.Q., Li G.C. The emerging landscape of circular RNA ciRS-7 in cancer (review) Oncol. For. Rep. 2015;33:2669–2674. doi: 10.3892/or.2015.3904. [DOI] [PubMed] [Google Scholar]
- 54.Verduci L., Ferraiuolo M., Sacconi A., Ganci F., Vitale J., Colombo T., Paci P., Strano S., Macino G., Rajewsky N., Blandino G. The oncogenic role of circPVT1 in head and neck squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD transcription-competent complex. Genome Biol. 2017;18:237. doi: 10.1186/s13059-017-1368-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wen J., Liao J., Liang J., Chen X.P., Zhang B., Chu L. Circular RNA HIPK3: a key circular RNA in a variety of human cancers. Front. Oncol. 2020;10:773. doi: 10.3389/fonc.2020.00773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patra K.C., Wang Q., Bhaskar P.T., Miller L., Wang Z., Wheaton W., Chandel N., Laakso M., Muller W.J., Allen E.L. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell. 2013;24:213–228. doi: 10.1016/j.ccr.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng C., So H.I., Yin S., Su X., Xu Q., Wang S., Duan W., Zhang E., Sun C., Xu Z. MicroRNA-532-3p suppresses malignant behaviors of tongue squamous cell carcinoma via regulating CCR7. Front. Pharmacol. 2019;10:940. doi: 10.3389/fphar.2019.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mohamed N.-E., Hay T., Reed K.R., Smalley M.J., Clarke A.R. APC2 is critical for ovarian WNT signalling control, fertility and tumour suppression. BMC Cancer. 2019;19:677. doi: 10.1186/s12885-019-5867-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu P., Li X., Guo X., Chen J., Li C., Chen M., Liu L., Zhang X., Zu X. Circular RNA DOCK1 promotes bladder carcinoma progression via modulating circDOCK1/hsa-miR-132-3p/Sox5 signalling pathway. Cell Prolif. 2019;52 doi: 10.1111/cpr.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li J., Sun D., Pu W., Wang J., Peng Y. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6:319–336. doi: 10.1016/j.trecan.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 61.Zambrano A., Molt M., Uribe E., Salas M. Glut 1 in cancer cells and the inhibitory action of resveratrol as a potential therapeutic strategy. Int. J. Mol. Sci. 2019;20:3374. doi: 10.3390/ijms20133374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chi H. miR-194 regulated AGK and inhibited cell proliferation of oral squamous cell carcinoma by reducing PI3K-Akt-FoxO3a signaling. Biomed. Pharmacother. 2015;71:53–57. doi: 10.1016/j.biopha.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 63.Dumache R. Early diagnosis of oral squamous cell carcinoma by salivary microRNAs. Clin. Lab. 2017;63(11):1771–1776. doi: 10.7754/Clin.Lab.2017.170607. [DOI] [PubMed] [Google Scholar]
- 64.Tang H., Wu Z., Zhang J., et al. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol. Med. Rep. 2013;7(3):761–766. doi: 10.3892/mmr.2012.1254. [DOI] [PubMed] [Google Scholar]
- 65.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen B., Wei W., Huang X., et al. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics. 2018;8(14):4003–4015. doi: 10.7150/thno.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Song L., Xiao Y. Downregulation of hsa_circ_0007534 suppresses breast cancer cell proliferation and invasion by targeting miR-593/MUC19 signal pathway. Biochem. Biophys. Res. Commun. 2018;503(4):2603–2610. doi: 10.1016/j.bbrc.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 68.Wang L., Wei Y., Yan Y., et al. CircDOCK1 suppresses cell apoptosis via inhibition of miR-196a-5p by targeting BIRC3 in OSCC. Oncol. Rep. 2018;39(3):951–966. doi: 10.3892/or.2017.6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu X., Abraham J.M., Cheng Y., et al. Synthetic circular RNA functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Mol. Ther. Nucleic Acids. 2018;13:312–321. doi: 10.1016/j.omtn.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gareev I., Beylerli O., Liang Y., Xiang H., Liu C., Xu X., Yuan C., Ahmad A., Yang G. The role of MicroRNAs in therapeutic resistance of malignant primary brain tumors. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.740303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun J., Sun Z., Gareev I., Yan T., Chen X., Ahmad A., Zhang D., Zhao B., Beylerli O., Yang G., Zhao S. Exosomal miR-2276-5p in plasma is a potential diagnostic and prognostic biomarker in glioma. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.671202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beylerli O., Gareev I., Sufianov A., Ilyasova T., Zhang F. The role of microRNA in the pathogenesis of glial brain tumors. Noncoding RNA Res. 2022;7(2):71–76. doi: 10.1016/j.ncrna.2022.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gareev I., Gileva Y., Dzidzaria A., Beylerli O., Pavlov V., Agaverdiev M., Mazorov B., Biganyakov I., Vardikyan A., Jin M., Ahmad A. Long non-coding RNAs in oncourology. Noncoding RNA Res. 2021 Aug 26;6(3):139–145. doi: 10.1016/j.ncrna.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beylerli O., Gareev I., Sufianov A., Ilyasova T., Guang Y. Long noncoding RNAs as promising biomarkers in cancer. Noncoding RNA Res. 2022 Feb 25;7(2):66–70. doi: 10.1016/j.ncrna.2022.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sufianov A., Begliarzade S., Ilyasova T., Liang Y., Beylerli O. MicroRNAs as prognostic markers and therapeutic targets in gliomas. Noncoding RNA Res. 2022 Jul 6;7(3):171–177. doi: 10.1016/j.ncrna.2022.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beylerli O., Khasanov D., Gareev I., Valitov E., Sokhatskii A., Wang C., Pavlov V., Khasanova G., Ahmad A. Differential non-coding RNAs expression profiles of invasive and non-invasive pituitary adenomas. Noncoding RNA Res. 2021 Jun 30;6(3):115–122. doi: 10.1016/j.ncrna.2021.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]