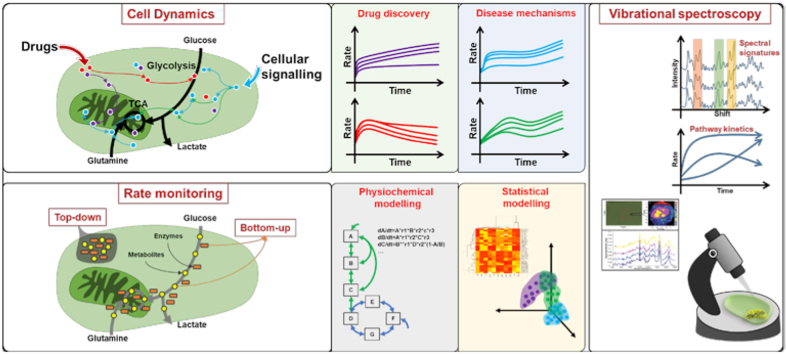
Abstract
Background
The dynamics of the cellular glycolysis pathway underpin cellular function and dysfunction, and therefore ultimately health, disease, diagnostic and therapeutic strategies. Evolving our understanding of this fundamental process and its dynamics remains critical.
Scope of review
This paper reviews the medical relevance of glycolytic pathway in depth and explores the current state of the art for monitoring and modelling the dynamics of the process. The future perspectives of label free, vibrational microspectroscopic techniques to overcome the limitations of the current approaches are considered.
Major conclusions
Vibrational microspectroscopic techniques can potentially operate in the niche area of limitations of other omics technologies for non-destructive, real-time, in vivo label-free monitoring of glycolysis dynamics at a cellular and subcellular level.
Keywords: Glycolysis, Cellular metabolism, Systems biology, Dynamics, Modelling, Vibrational spectroscopy, Chemometrics
Graphical abstract
1. Introduction
Most of the metabolic networks for energy production/nutrient utilisation were elucidated during the ‘golden age of biochemistry’ (roughly 1920s–1960s) [1], resulting in around fifteen Noble Prizes related to energy balance or core metabolic pathways [1]. Glycolysis, an anaerobic mechanism, is believed to be the first ATP production pathway to evolve, as it is common to both prokaryotes and eukaryotes [2,3]. It is involved in aerobic and anaerobic energy production, responsible for production of precursors for other metabolic pathways and serves as the primary energy source for cells without mitochondria [4]. The glycolysis process was first explored by Louis Pasteur in the 1800s, and it took almost 100 years to fully elucidate the complete pathway, proposed by Embden, Meyerhof and Parnas in the 1940s [5]. Glycolysis is intricately linked with several human disease mechanisms and drug toxicities [1,6,7]. It is involved in a multitude of epidemic diseases such as cancer [8], neurological disorders [9], diabetes [10], etc., and has attracted substantial attention from scientific community for disease diagnostics [11], therapy [12,13], drug targeting [14], etc. Almost 8 decades after the complete elucidation of the pathway, new insights continue to emerge, and reconsideration of the original dogma of the pathway has even been proposed [15].
Technologies to monitor glucose metabolism with increasing efficiencies continue to emerge, taking more and more holistic approaches to map the inter- and intra-cellular interactions. Omics approaches (metabolomics, proteomics, lipidomics, transcriptomics, genomics, etc.), employing high-throughput technologies, have opened up new opportunities in systems-biology, such that different genotypic/phenotypic levels of intracellular metabolic activities can be captured in a snapshot with high-precision and efficiency [16]. The massive amount of data generated requires intensive statistical analysis to resolve the underlying processes and, additionally, this data can be computationally-modelled and simulated to enrich the experimental observations, guide experimental strategies, and predict pathway kinetics to hypothesise the behaviour of the system and its components [17,18]. Knowledge and understanding of cellular metabolic dynamics are essential for differentiation between healthy and diseased cells [19], identifying target pathways and drug discovery for disease therapy [12,[20], [21], [22]], etc., which opens up scope for development of novel strategies for dynamic, non-destructive, single-cell metabolic analysis. Dynamic analysis of the systemic interactions remain a challenge, given the multi-omics analysis techniques are destructive to cell/tissue samples, and demand multiple experimental setups. Furthermore, omics technologies are restricted by the minimum sample detection limit which makes single-cell analysis complicated, as due to the cellular heterogeneity some features might be missed in analysis of cell populations [23]. Cellomics, also referred to as high-content analysis/screening, involves labelling, imaging, analysis, and visualisation of the biological system to provide a better alternative for decoding single-cell dynamics. However, labelling techniques are, by definition, limited to visualisation of what has been labelled and assume an a priori knowledge of the process to be monitored. As an alternative, vibrational spectroscopy provides a label-free, holistic representation of the biomolecular content of the sample, in real-time [24]. It is a relatively well established analytical approach, compared to modern omics approaches, but is being increasingly investigated for clinical and pharmaceutical applications, given the development of ever more sophisticated microscopic instrumentation enabling cellular and subcellular analysis, as well as tools to datamine and analyse the complex datasets [[25], [26], [27]]. Although many approaches to date have focussed on analysis of tissues, and biofluids for disease diagnostics [28,29], the potential of vibrational spectroscopy for cellular and subcellular analysis of dynamic processes is increasingly being explored [[30], [31], [32], [33], [34]], including for in vitro drug screening and toxicology analyses of cell lines [27,[35], [36], [37]].
In this paper, we review the medical relevance of the glycolytic pathway in detail and briefly explore the current state of art for its monitoring and modelling. The future perspectives specifically focuses on the potential of vibrational microspectroscopic techniques, which are non-destructive, and can be harnessed to gain insights into the cellular metabolism in a label-free, dynamic manner. Additionally, the high resolution of microspectroscopy can be used for analysis at organelle level in a single-cell.
2. The glycolysis pathway: current understanding
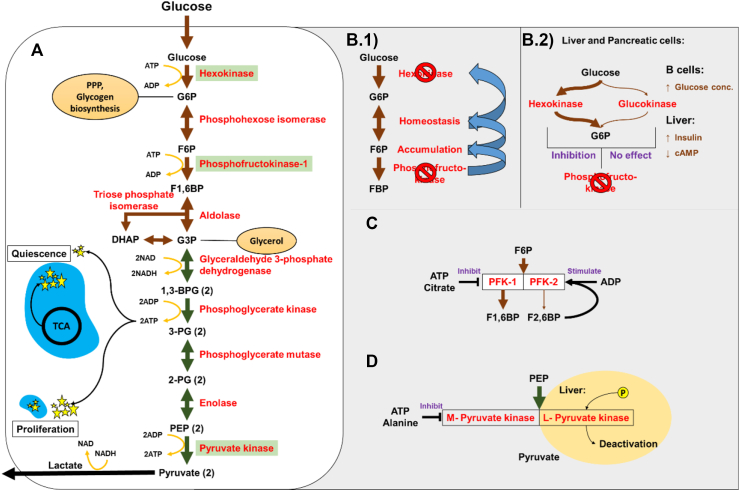
Glycolysis is a ten-step anaerobic process (Figure 1_A) divided into two phases. The first five steps are collectively referred to as the preparatory/investment phase, in which the six-carbon glucose molecule is broken down into two, three-carbon carbon molecules by investing energy, while the proceeding five steps are termed the payoff phase, in which energy is produced.
Figure 1.
Schematic representation of Glycolysis pathway and the regulatory enzymes. (A) 10 steps of glycolytic pathway. Brown and green arrows represent the preparatory phase and payoff phase respectively. Yellow curved arrows represent the energy consumption and production nodes. The enzymes involved in each node are written in red and the regulatory enzymes are highlighted with green background. The energy production during quiescence and proliferation from cytoplasm (glycolysis) and mitochondria (blue ovals with TCA cycle) are represented with yellow stars. Circles emerging from different nodes of the pathway represent the precursor molecules which end up in different pathways. (B.1) The inhibition of PFK leads to accumulation of F6P which goes into homeostasis with G6P. The excessive accumulation of G6P leads to inhibition of hexokinase. (B.2) The isozyme of hexokinase, glucokinase with low affinity to glucose is represented with a thin arrow. Both the enzymes phosphorylate glucose to G6P. The effect of PFK inhibition on respective enzyme is indicated in purple. The regulation of the enzymes in beta cells and liver is represented by the regulatory molecule and its effect is represented by an arrow. (C) Two PFK enzymes in the glycolytic pathway are represented and the effect of the regulatory molecules is indicated in purple. The different rates of activity of both the enzymes is represented by the thicknesses of the arrows. (D) Final regulatory steps of the pathway with two isoforms of the pyruvate kinase are represented. The phosphorylation of L isoform leads to deactivation of the enzyme and the effect of the regulatory molecules is represented in purple.
The first, irreversible, regulatory investment step in glycolysis is phosphorylation of glucose to glucose-6-phosphate (G6P), catalysed by hexokinase (cofactors Mg++, Mn++). Consuming one adenosine triphosphate (ATP) molecule ensures maximum intake of glucose as the added charge prevents it diffusing out of the cell [38]. Glucokinase is a differentially regulated isozyme of hexokinase, present only in the liver and beta cells along with hexokinase [39]. Glucokinase is stimulated by insulin and supressed by cyclic adenosine monophosphate in the liver, and activated by increased glucose concentration in the pancreas [40,41]. Glucokinase has significantly lower affinity to glucose, lowering its uptake in liver cells during starvation to spare glucose for more important organs. Glucokinase remains unaffected by the accumulation of G6P and continues to phosphorylate glucose (Figure 1_B.2). Hence, the presence of an extra isozyme helps liver cells regulate the glucose concentration in the body.
The second irreversible, regulatory investment step is the phosphorylation of fructose-6-phosphate (F6P) to fructose-1,6-bisphosphate (F1,6BP), catalysed by phosphofructokinase (PFK, cofactor Mg++), consuming one ATP molecule. It is the commitment step of glycolysis, as, if the reaction proceeds beyond this point, it must proceed through the entire pathway. Deactivation of PFK leads to F6P accumulation which is freely converted back to G6P, as per Le Chatelier's principle [42], and inhibits hexokinase (Figure 1_B.1). The two types of PFK; PFK 1 and PFK 2 in humans have the same substrate but yield F1,6BP and F2,6BP, respectively. PFK2 has additional phosphatase activity along with the kinase activity. The PFK enzyme is allosterically inhibited by ATP, citrate and upregulated by AMP and F2,6BP (Figure 1_C). The feed forward stimulation in liver promotes the activity of PFK using F2,6 P.
The first energy production step (step 6 in the pathway, payoff phase) is the conversion of glyceraldehyde 3-phosphate (G3P) to 1,3-bisphosphoglycerate (1,3BPG), which is catalysed by glyceraldehyde-3-phosphate dehydrogenase (GAPDH), an oxidoreductase enzyme. This reaction converts a Nicotinamide adenine dinucleotide (NAD) molecule to NADH, which can be further utilised in electron transport chain to produce ATP. Further, 1,3BPG is converted to 3-phosphoglycerate (step 7 in pathway) which produces an ATP molecule from adenosine diphosphate (ADP). This reaction is catalysed by phosphoglycerate kinase (cofactors Mg++, Mn++).
The final step of glycolysis, conversion of phosphoenol pyruvate to pyruvate (step 10 in the pathway) is catalysed by the pyruvate kinase (PK) (cofactors Mg++, K++) enzyme. This is an irreversible regulatory reaction. Elevated levels of ATP and alanine create negative feedback to inhibit PK. Fructose 1,6 bisphosphate stimulates this enzyme in the absence of ATP. In liver cells, an additional L isozyme of PK is present along with the M isozyme (L-liver, M-muscle) but the L form is predominant. The L form is controlled by phosphorylation, which deactivates the molecule (Figure 1_D). During starvation, this enzyme isoform phosphorylates to avoid taking in glucose.
The net energy produced by glycolysis is two ATP molecules (four produced, two consumed) and two reduced NAD molecules. In addition to glucose catabolism and energy production, glycolysis produces several relevant molecules essential for cell sustenance. G6P can enter the pentose phosphate pathway, producing several pentose sugars and NADPH for cholesterol and fatty acid synthesis. G6P is a starting point for glycogen synthesis. G3P produces glycerol, essential for production of triglycerides and phospholipids. Glycolysis is directly and indirectly involved in the biosynthesis of several amino acids, which are the building blocks of proteins.
In the quiescent state, the glycolytic rate is at the basal level (Pasteur's effect) [43] and the majority of the ATP produced is by mitochondrial oxidative phosphorylation pathway (OxPP). However, during cell proliferation, the OxPP rate drops, and high glycolytic rate is observed. This leads to drop in NAD/NADH ratio which is compensated by converting pyruvate to lactate, oxidising NADH to NAD (Figure 1_A) [43].
3. Medical relevance and therapies
Glucose is generally considered the main carbon source, and the glycolytic pathway significantly impacts cellular metabolism and proliferation [44]. Glycolysis dysfunction plays a key role in diabetes and obesity [10], cancer [45], neurodegenerative diseases such as amyotrophic lateral sclerosis, Alzheimer's and Parkinson's disease [46,47].
Diabetes is caused by the improper functioning of the glycolysis, due to compromised cellular signalling. Insulin secretion is regulated by glycolysis [10], and hepatic glucose production (HGP) was shown to be an effective way to maintain euglycemia [[48], [49], [50]]. Type-1 diabetes insulin insufficiency leads to a decrease in glycolysis rate, whereas type-2 diabetes hyperinsulinemia leads to an increased rate of glycolysis in liver, pancreatic beta cells and adipose tissue [10]. Hexokinase-2 (HK2) influenced by insulin [51,52] plays an essential role in diabetes [52,53] and was reduced specifically (28 ± 0.5% and 31 ± 4% diabetic, 40 ± 0.5% and 47 ± 7% control at 0.11 and 11.0 mM/L of glucose, respectively [54]) in the skeletal muscles of type-2 diabetic patients [54,55]. Under hyperinsulinemic conditions (450 pmol m−2. min−1 for 3 h [55]; 40 and 240 mU.m−2. min−1 [56]), upregulation of HK2 activity in muscles and adipose tissues was observed in obese and normal patients (93 ± 20 control 1, 194 ± 37 control 2, 127 ± 35 obese [55] and 5 ± 0.08 control, 4.33 ± 0.66 obese (averages of soluble/particulate and both hyperinsulinemic flow rates) [56]), but was reduced in type-2 diabetic patients (9 ± 18 [55]; 3.10 ± 0.10 [56]). Genetic mutation, leading to loss of function of glucokinase [[57], [58], [59]], decreased glucose phosphorylation and decreased insulin secretion [60], was observed in ‘maturity onset diabetes of young’ patients. Glucokinase activity was reduced in obese type-2 patients by about 50% [61] which were later reported to exhibit active glucokinase mutation leading to hypoglycaemia [62,63]. An independent risk factor for the development of type-2 diabetes can be the elevated levels of lactate in plasma (27% aetiologic fraction) [64]. The pyruvate and lactate interconversion rates (diabetic 46 ± 9 to 108 ± 31 and control 21 ± 3 to 50 ± 13 μmol/min/kg) were observed to be significantly elevated, probably due to impaired glucose oxidative pathway in type-2 diabetes [65]. A single gene or protein cannot explain the complete pathophysiology of diabetes, hence insights into the dynamics of glucose catabolism rate could provide an extra layer of detail for categorising the disease features which may aid in its therapy [39].
Glucose is the only source of energy to the brain, which performs differential aerobic respiration to satisfy its rapid energy demands, undertaking intermittent biosynthesis of metabolites and maintain the redox states [66]. Aerobic glycolysis fulfils the energy requirement of the membrane bound ATP dependent processes, such as the pumping action of NA+/K + ATPase [67,68]. This process is not restricted to brain and was observed in human red blood cell membranes [69], skeletal muscle [70], vascular smooth muscle [71], and neurons [72]. One primary reason for opting such an inefficient process generating only two over thirty ATP molecules is the rate of production. The higher rate of glycolysis over OxPP accommodates the small, rapidly changing energy requirements by the cell/organ [73]. Brain glucose-hypometabolism [74], glucose-accumulation [75] and reduced glycolytic pathway flux [75] was observed in Alzheimer's disease (AD) patients [47,76] and the well documented link between AD and diabetes can also be plausibly explained by brain hyperglycemia [47], higher levels of the apolipoprotein E gene (ApoE2) making the brain more resistant to AD due to increased glycolytic robustness [47]. Glycolytic dysfunction in peripheral cells was also observed in other neurodegenerative diseases (Huntington's disease, Parkinson's disease and amyotrophic lateral sclerosis) indicating that the pathology of the disease is not limited to nervous cells and strengthening the notion of glycolytic dysfunction as a common pathway leading to neurodegeneration [9,47]. A dynamic sense of glycolytic rate to differentiate the normal from the abnormal could benefit in disease prevention, anti-ageing strategies and to unravel the mystery of the most essential organ, the brain!
Mature erythrocytes lack nuclei and mitochondria, making glycolysis the sole pathway for ATP production [77]. Although rare, the most common glycolytic abnormality in erythrocytes is PK deficiency [78] resulting in lack of energy, abnormal membrane function, potassium and water leakage from the cell, increase of calcium concentration and ultimately to cell rigidity, loss of flexibility, early susceptibility to splenic sequestration which collectively leads to haemolysis [78]. Other enzyme abnormalities linked to hexokinase, phosphoglycerate kinase, phosphoglucose isomerase, phosphofructokinase, aldolase, triosephosphate isomerase, etc., have also been identified but are scarce [79].
The role of glycolysis is significant in cancer research, as tumour cells perform glycolysis at a rate which is ∼ten times higher than normal cells. A comprehensive understanding of glycolysis can foster cancer detection, identification, classification, and cure by identifying drug targets. In the 1920s, Warburg observed that tumour cells consume surprisingly large amounts of glucose, compared to normal cells [43]. Multiple mechanisms leading to this phenomenon such as mitochondrial DNA mutation, nuclear DNA mutation, oncogenic transformation and the influence of the microenvironment around a tumour are reported [[80], [81], [82]]. The phenomenon is being exploited using fluorodeoxyglucose positron emission tomography (FDG-PET) for the detection of tumours [83]. Studies on proliferating lymphocyte cells demonstrated that around 90% of the pyruvate was being converted to lactate, concluding that the Warburg effect is not specific to tumour cells [[84], [85], [86], [87]]. Increased activity of hexokinase (79% breast cancer patients [88]), lactate dehydrogenase A (in 61.8% mixed stage gastric carcinoma patients with 56.3% survival rate compared to 78.4% survival rate in low activity patients [89]) and glyceraldehyde-3-phosphate (2.5 fold under hypoxia for 24 h in MFC-7 cells [90]) in tumours and cancer cell-lines was reported [[88], [89], [90]]. Inhibiting cancer cell proliferation, apoptosis induction and reversing multidrug resistance can be achieved by silencing the overexpressed enzymes such as lactate dehydrogenase A and PK [[91], [92], [93]]. However, these enzymes are multifunctional. For instance, hexokinase and enolase are critical in transcription regulation [94,95] and glucose-6-phosphate isomerase might affect cell motility [96]. Hence, identification of novel tumour features and drug targets which are distinct from normal cells is crucial for the disease therapy. The glycolysis pathway dynamics can be used for classification and drug discovery for different cancer phenotypes.
The Warburg effect and subsequent rewiring of cellular metabolism was also observed during atherosclerosis [97], whereby vulnerable human atherosclerotic lesions exhibit an enhanced expression of glycolytic markers compared to stable plaques [98]. The main risk factors for atherosclerosis include vascular endothelial cell (VEC) injury, lipid deposition, inflammatory and immune dysfunction, the former being considered one of the main triggers driving the occurrence and development of subclinical atherosclerosis [99,100]. Several seminal studies have provided compelling evidence that VECs are highly glycolytic, in both healthy and dysfunctional activated states [101]. VECs in healthy vessels are quiescent and actively maintain blood flow, vascular tone, and transport across the vessel wall. However, in response to extracellular events, VECs can become activated and induce angiogenic or pro-inflammatory signals [101]. Activation is accompanied by changes in cellular metabolism that provide energy and metabolic intermediates that fuel important biological processes including angiogenesis, inflammation and barrier function [102]. VECs can become adhesive and adopt a prothrombotic phenotype that orchestrates a vascular inflammatory response, including leukocyte recruitment and increased vascular permeability [99]. In these pathological conditions, the glycolytic process is compromised, leading to metabolic processes being activated to compensate for ATP shortage and to increased oxidative stress, cell dysfunction, as well as cell death [103,104]. Upon activation, VEC metabolism becomes disordered, represented by increased glycolysis and expression level and activity of fatty acid synthase [103]. These enhance the proliferation, migration, and inflammation of VECs, leading to VEC dysfunction and vascular disease.
Similar studies of the metabolism of vascular smooth muscle cells (vSMCs) have revealed an important role for glycolysis in atherosclerosis [105]. During the development of an atherosclerotic plaque, the accumulation of SMC-like cells following de-differentiation of medial vSMCs and/or partial myogenic differentiation of resident vascular stem cell progenitors is accompanied by enhanced aerobic glycolysis [106]. Thus, modulating VEC and vSMC metabolism via antiglycolytic therapies may be a potential therapeutic target for atherosclerosis.
4. Glycolysis monitoring techniques
For an excellent general review of methodologies employed in monitoring metabolic processes, in vitro and in vivo, the reader is referred to the work of Duraj et al. [107]. Techniques to monitor the glycolysis process either as a static snapshot or dynamic process, can be categorised as either top-down or bottom-up approaches [108]. The bottom-up approach tries to understand individual components or a smaller network of the system, whereas the top-down approach holistically targets the entire system using high-throughput techniques to obtain a large dataset [108]. Both approaches rely on advanced computational methods to model the behaviour of the system, which will be discussed in section_5. For a mode extensive and detailed discussion of the techniques, the reader is directed to excellent reviews of statistical/physiochemical modelling [109] and modelling from top-down and bottom-up approaches [108].
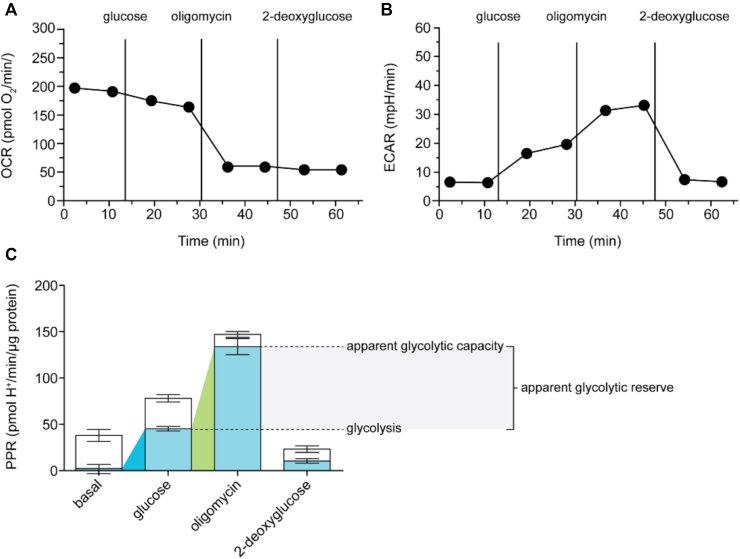
In what is considered a bottom-up approach, monitoring the extracellular acidification rate (ECAR) in live cells using simple biochemical assays, in a kinetic spectrophotometric mode can be used to decode the dynamics of glycolysis pathway [110]. The ECAR assay can be coupled to the O2 consumption assay in the same experiment to quantitatively derive and subtract the CO2 acidification as the detection elements in both assays fluoresce at different wavelengths [111]. This assay is non-destructive and provides a dynamic profile of glucose metabolism which can be used for statistical and physiochemical modelling [112,113]. (Figure 2) represents the data obtained from one of such dynamic experiments obtained from Agilent Seahorse instrument [114]. The glycolysis rate is at basal level when no glucose is added, while the cell relies on oxidative phosphorylation for ATP demand [114]. Upon addition of glucose, the cell reduces its oxygen consumption and the glycolytic rate increases. If the mitochondrial ATP production is blocked (oligomycin) the cell relies solely on glycolysis for ATP demand and the glycolysis rate increases to its full capacity [114]. The glycolytic rate drops to basal level if a competitive inhibitor, 2-deoxyglucose is added. Mitochondrial respiration also contributes to the extracellular acidification to some extent, which is represented in (Figure 2_C) [114].
Figure 2.
Representative figure for the dynamic monitoring the OCR and ECAR in C2C12 myoblast cells using Agilent Seahorse instrument. The OCR (A) and ECAR (B) were monitored dynamically using Agilent Seahorse instrument in C2C12 cells in the basal condition (without any glucose) and the sequential addition of 10 mM glucose, 2 μg/ml oligomycin (ATP synthase inhibitor/glycolysis stimulator) and 10 mM 2DG (competitive inhibitor of hexokinase and phosphoglucoisomerase). (C) Respiratory (blank column) and glycolytic (blue column) proton production rate (PPR) form the ‘A’ and ‘B’ experiment. Coloured wedges indicate glycolysis in basal condition (blue) and glycolytic capacity (green) with their difference as glycolytic reserve. The data is an average of 6 replicates. Reproduced with permission from [114].
Alternatively, targeting specific enzymes/metabolites in the pathway provide more specific details of the pathway intermediates [115]. Radioactive ((2-deoxy-d-[1,2–3H]-glucose); (2-deoxy-d-[1–14C]-glucose); (2-deoxy-2-(18F)-fluoro-D-glucose)) or fluorescent (2-[N-(7-nitrobenz-2-oxa-1,3-diaxol-4-yl)amino]-2-deoxyglucose); glucose analogues can provide a reliable quantitative estimate of glucose entering the glycolytic pathway in a cell [115]. Alternately, the activity of rate limiting glycolytic enzymes can be determined by quantifying their catalytic products in vivo [115]. The simplicity and the fact that these assays can be coupled with several activators/inhibitors make them extremely useful in pharmaceutical research and development [111]. The data from these assays is suitable for physiochemical and to some extent statistical modelling [116]. Unfortunately, the cell must be lysed to quantify/harvest the metabolites/enzymes at fixed time-points to obtain in situ pathway dynamics. Furthermore, the biochemical assays are not sensitive enough for single-cell analysis.
Positron emission tomography (PET) uses radiolabelled tracers for static and dynamic imaging but its use has been largely limited to drug development and clinical research [117]. Sequential and simultaneous PET and magnetic resonance imaging (MRI) have emerged in recent years as kinetic imaging strategies with high clinical importance [117]. In this technique, a positron-emitting isotopic compound is non-invasively injected intravenously in trace quantities and its biodistribution is used to infer the physiological and biochemical processes such as glycolysis [117]. The potential benefit of kinetic analysis has been demonstrated for a number of tracers such as 18-F-labelled 2-deoxy-2-d-glucose (18F-FDG; tumour detection, response monitoring, tumour detection estimates) [[118], [119], [120]], 18F-DOPA (tumour-grade differentiation) [121], 18F-FMISO (tumour hypoxia response to therapy) [122], 18F-FLT (brain tumour progression, glioma therapy response modelling) [123,124]. This technique can potentially be used for in situ kinetic analysis in humans for diagnostics using statistical and physiochemical modelling with a downside of requiring radiolabels.
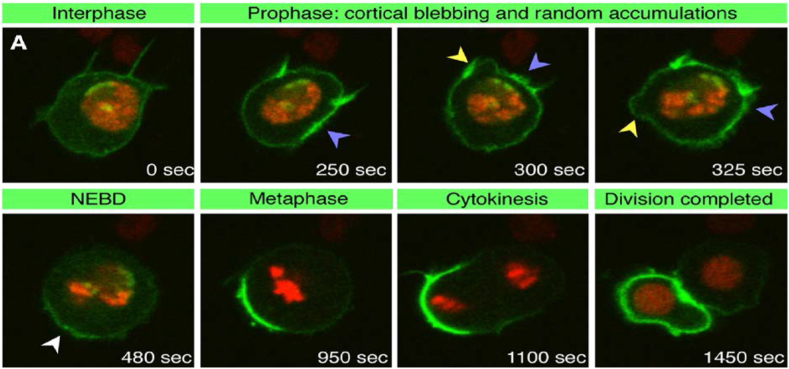
Green fluorescent proteins (GFP) are often used to study protein localisation, dynamics and interactions in single living cells [125,126]. They can be tagged to virtually any protein to form a fluorophore [125,127], and advancement in GFP biology has aided in optimised expression for a wide range of cell types [128], several variants of which have already been developed to tag and monitor different proteins simultaneously [129,130]. Microscopic techniques such as fluorescence recovery after photobleaching (FRAP), fluorescence resonance energy transfer (FRET) and fluorescence correlation spectroscopy (FCS) have also evolved in laser scanning microscopy [128] and dynamic imaging can be performed to obtain large kinetic data sets for statistical as well as physiochemical modelling. A representation of confocal microscopy and FRAP dynamic imaging with pon and histone2B proteins, tagged with GFP and RFP, respectively, in mitotically dividing drosophila sensory organ precursor cell is shown in (Figure 3) [131]. Although it can be monitored dynamically, the technique is limited by the fact that it only visualises what has been labelled, which assumes an a priori knowledge of the process to be monitored, and/or multiple probes for a more systemic understanding.
Figure 3.
Fluorescence microscopy of GFP-Pon protein (green) and Histone2B-RFP protein (red) in live drosophila sensory organ precursor cell to monitor the localisation of pon protein during mitotic cell division. Yellow arrowheads indicate cortical blebbing, blue arrowheads indicate random accumulations of GFP-Pon, and the white arrowhead indicates early GFP-Pon crescent; NEBD stands for nuclear-envelope breakdown. Reproduced with permission from [131].
More systemic approaches, which can be considered top-down, employ different omics techniques, including genomics [132,133], transcriptomics [[134], [135], [136]], proteomics [137,138] and metabolomics [139], by which the entire genome, transcriptome, proteome, and metabolome of the system can be analysed as a snapshot of the quenched system using high-throughput techniques [[140], [141], [142]]. These techniques are usually used in a targeted manner, focusing on specific aspects of a systemic response, such that they can also be considered bottom-up approaches [[143], [144], [145]].
Next generation sequencing has emerged as a powerful tool in scientific research and diagnostics, making use of fragmented nucleotide sequences to cost effectively construct a sequencing library of millions of nucleotide strands simultaneously in a short span of time with high accuracy [146,147]. The small sequenced fragments are then aligned using complex computer simulations to obtain complete or targeted genome/transcriptome sequences [146,147]. Genomic data can be used to accurately draw the pathway map (physiochemical modelling) for the whole cell while the quantitative transcriptomic data can be used for statistical modelling [148]. Predicting the phenotype from genotype is complex and the correlation is not yet clear. Schwannhäusser et al. established that the mRNA expression level was the best predictor of protein levels and could explain 40% of protein level variability in mouse fibroblast cells, while protein stability had a minor role [149]. However, this may not be universally the case, as another study in embryonic stem cells reported that changes in protein levels are not accompanied by changes in corresponding mRNA expression level [150]. Schwannhäusser et al. further indicated that, under different conditions, rate constants may vary and the protein stability might have a bigger role at single cell level or as a result of any perturbation [149] as protein degradation is involved in regulation of many cellular processes [151]. Although, the cellular genome does not change much during the lifetime of the organism, the cellular transcriptome is highly dynamic and depends on the state of the system. Several approaches for a wider range of systems at single and multi-cellular levels are already developed for dynamic transcriptome analysis [[152], [153], [154], [155]], but by the nature of the analytical technology, these approaches are destructive and rely on quenching the systemic information at pre-fixed states.
Single-cell genomics rely on the numerous approaches [[156], [157], [158]] for genome amplification so there is enough for sequencing, to get insights into the genome evolution, e.g. in cases such as cancer, and to understand the genomic basis of a specific phenotype, which is missed in bulk sequencing of a population [159,160]. Similarly, single-cell transcriptomics requires addition of an extra step of converting the transcripts into cDNA using reverse transcriptase which can be amplified and sequenced [160]. Alternatively, RNA hybridisation probes attached with fluorescent compounds can be used for specific sequence identification and use of different RNA probes can aid in building comprehensive transcriptome [161,162]. Single-cell transcriptomics is preferred over single-cell proteomics due to the ease of amplification over the difficulties associated with protein amplification and finds applications in studying gene dynamics, RNA splicing and cell typing [160].
The cellular proteome/metabolome defines the cellular phenotype. The proteomics and metabolomics approaches employ high-throughput analytical tools such as mass spectrometry (MS) or nuclear magnetic resonance spectroscopy (NMR) [163]. Quantitative detection of molecules/metabolites aid in statistical/physiochemical modelling and to determine the relative activity of the pathway. The proteomic data can be complicated for pathway flux analysis, due to the different layers of enzymatic regulation (post-translational modifications, allosteric control, etc.) and the lack of in vivo kinetic parameters for many enzymes [164]. Modern metabolomics approaches utilise stable isotopic tracers of hydrogen, carbon, nitrogen, oxygen, and sulphur incorporated in the primary carbon source. As pathways have unique carbon transitions, analysing the location of the stable isotope in the amino acids produced can reveal the chosen pathway. Despite these developments, there remain physical limitations, as only a small subset of metabolites can be quantified [165,166]. These limitations can be metabolite related (low abundance, low stability), instrumental (identical retention in chromatography and MS) [166] and experimental metabolism quenching, as high metabolome turnover rates are on the sub-second timescales [167] and the quenching methods induce leakage in cells [[168], [169], [170]]. Metabolic flux analysis describes the true flux through the pathway at a given state, but to get a dynamic view of the system which would describe the systems transition through different states at a molecular level would require a wide array of experimentation [171] which would again encounter the above mentioned challenges. Visualising the dynamics of the single-cell metabolome and proteome is possible using time-lapse microscopy or using time-resolved omics experiments [[172], [173], [174]].
For single-cell proteomics, several strategies are applied which entail either tagging the proteins of interest with a specific antibody attached to a distinctive fluorophore [175,176], quantum dot [177], etc., or tagging the genome with a fluorescent protein, to be analysed using fluorescence microscopy [178,179]. Alternatively, the proteins can be tagged with rare metal isotopes for mass cytometry [180,181]. Major steps for single cell proteomics by MS (SCoPE-MS) based analysis are sample preparation, separation and detection. To deal with the protein amplification issue, several groups focus on oocytes or very early cleavage cell stage, due to their large size and abundance of proteins [[182], [183], [184]] while some working on mammalian cells (10–15 μm diameter) combine carrier-cells and single-cell barcoding [185,186]. The second generation of SCoPE-MS [187], SCoPE2 [188,189] further increase throughput of sample preparation over 100 fold by miniaturising sample preparation step such as MAMS (Micro-Arrays for MS) which achieves high speed aliquoting by exploiting differences in wettability among recipient sites and surrounding areas [190]. Sample preparation can involve several steps to purify the proteins to some extent and then further separate them via different chromatographic techniques, before detection by MS/MS–MS. Although transcriptomics and proteomics have the same purpose, transcriptomics does not consider post-translational regulation.
Several fluorescence microscopy-based strategies can be used for single-cell metabolomics studies, such as usage of fluorescent proteins which illuminate when bound to the metabolite [191]. Not requiring fluorescent proteins for metabolites of interest and the detection sensitivity in the femtomole range, MS has been most frequently used for single-cell metabolomics [192]. Single-cell metabolomics using mass spectroscopy is progressing rapidly as the challenges are accepted and creatively explored by an increasing community and the continuous effort are moving the field towards more sensitive techniques with higher throughput, quantitative abilities and decreased technical variability [193,194]. The future development is directed towards techniques with high-throughput, high-sensitivity for molecules with low abundance/low ionisation efficiency, good replicability for single-cell metabolite detection [193]. The metabolomics profile illustrates how the cell adapts to environmental stresses at a molecular level, providing a dynamic understanding of the cell [191].
Given the complex nature of cells and the interactions within the different levels of central dogma (geno-pheno-envirotype interactions), combinatorial/multi-omics/pan-omics have emerged which can extend the scope of harvested information to elucidate the interactions between biological entities for precision medicine and biomarker discovery [195,196]. Proteogenomics harness the advantages of both genotype and phenotype by identifying/quantifying translated nucleotide sequences to generate candidate proteins to discover single amino acid variants (SAAV), splice proteoforms and additionally genome re-annotation to discover novel coding sequences which have found significant applications in precision medicine [197,198]. Epigenetics, on the other hand, deals with changes in the biochemistry, not by modifications in the genomic sequence, but by methylation, acetylation, phosphorylation, ubiquitylation, sumolyation, chromatin modifications, etc., which are responsible for several illnesses, behaviours, and other health indicators [199]. A wide array of multi-omics platforms are now available as databases and data processing toolboxes [200].
Although these sophisticated systemic strategies have come a long way, further development is required to establish in situ, non-destructive, label free approaches which can aid in dynamic sub-cellular analysis.
5. In silico modelling
I. silico modelling approaches describe a system in terms of simple mathematical equations to mimic the behaviour of the system [201] as described by the top down and bottom up approaches described in section_4 [108].
Modelling from the bottom up requires detailed datasets covering the full dynamic range of each reaction to define the behaviour of the system [108]. Homeostatic control in vivo does not allow the gathering of such detailed datasets, and hence the bottom-up approach relies on in vitro with cell and cell-free experiments to fit the parameters [108]. Often, models based on in vitro cell-free enzyme kinetics analysis draw from repositories of kinetic data to fulfil the data requirement [108], but trying to understand the in vivo metabolism dynamics in terms of in vitro cell-free kinetics of the constituent enzymes can generate erroneous results due to the discrepancy between the in vitro cell free rates and in vivo fluxes [202].
The semi-quantitative indices analysis using dynamic PET and MRI imaging can be used for kinetic modelling [117]. Dynamic imaging of GFP generates large, kinetically complex data sets which can be analysed with computational models to quantify biophysical properties of molecules and even processes [203]. This approach of computational cell biology can be used to kinetically monitor the essential pathways, mechanisms and controls of biological processes in real-time [203].
The systems biology approach of ‘metabolic flux analysis’ (MFA), also known as ‘fluxomics’, is a physiological counterpart of different omics techniques. MFA is an end result of the multiomics interplay which holistically describes the operation of the entire system by inferring the complex model from the available in vivo data [108]. Largely based on 13C tracers [164], MFA has developed over the last three decades from a black box balance approach of a few reactions [204] to simultaneous measurement of hundreds of metabolic fluxes [205], including reversible and parallel reactions [206]. Flux balance analysis (FBA), 13C-fluxomics and 13C constraint based FBA are different approaches of MFA and can be used in measuring dynamic, stationary and semi-stationary metabolite fluxes [164]. In principle, given the mathematical complexity of kinetic models, these are more challenging to construct and rely on approximate kinetic formalisms for parameter fitting [[207], [208], [209]]. This approach uses simplified equations which often result in loss of kinetic details, thermodynamic consistency and prevent identification of novel regulatory mechanisms [108].
The emergence of open source resources such as Cell Designer [210], which enable biochemical pathway models to be simply drawn, but also kinetically modelled by the user, have made the approaches much more accessible. Model/Graphical representation by Systems Biology Markup Language (SBML) [211] and Systems Biology Graphical Notation (SBGN) [212] means that the user defined models can be interfaced with many other databases, for pathway enrichment [213], a technique becoming increasingly popular in predictive toxicology [214]. Once validated with experimental data under different conditions, the models can accurately predict cellular metabolism at different steady states [215,216]. Numerous tools for steady state kinetic modelling are freely available [[217], [218], [219], [220], [221], [222]].
Modelling approaches can be differentiated as statistical and physiochemical modelling. Statistical modelling is employed when difficult to manage, abundant data is available, but the insight into the structure of system is sparse i.e., components of the system, their interaction and regulation. As no assumption can be made about the underlying mechanism of the system due to the lack of structure, the objective of modelling is to identify relevant variables or dimensions in the system [109]. This type of modelling employs statistical tools to connect different layer of data and gap filling. These models, however, rely on the predictability of the dimension considered to be significant e.g. a principal component analysis. Genome scale modelling falls under this method. Several statistical tools such as KEGG mapper, Metaboanalyst, etc. Are freely available for statistical modelling [217,222,223]. Berndt et al. modelled murine liver cancer cells using shotgun proteomics approach by LC-MS/MS [224]. They found significant upregulation of PFK (L isoform) and GAPDH enzymes. A twofold increase of the isoforms A and C of fructose-bisphosphate aldolase which preferentially contribute to glycolytic rate unlike the isoform B which contributes to gluconeogenic rate were observed [224]. Kelly et al. developed a kinetic model of hepatic glycolysis to observe the effect of the extracellular environment on the cytosolic free NAD/NADH in a cell culture [225]. From the data obtained from the model, they concluded that cytosolic the NAD/NADH ratio is maintained by two key enzymes GADPH and lactate dehydrogenase (LDH) [225].
Physicochemical models can be viewed as the translation of a pathway map into a mathematical form. These models predict specific biomolecular transformations such as covalent modifications, intermolecular associations and localisation as mathematical equations based on a prior knowledge of pathways for which components and connectivity are well established [[226], [227], [228], [229], [230]]. The equations describe the identifiable processes such as catalysis and assembly with physical interpretations of parameters such as concentration, binding affinity and reaction rate [231]. Modelling begins with identifying the elements in the model and generation of a schematic describing the interactions between them. These interactions are further translated into equations which require certain assumptions to be made for visualising the dynamics of the equations. Although the models assume how the interactions take place, they also aid in rapid hypothesis testing and model validation [109]. Ordinary and partial differential equation can be used for both deterministic and stochastic modelling. ODEs are most common for deriving simple mechanistic or empirical equations. Mass action kinetics, rate law (Michaelis Menten models, Hill kinetics), power law, lin-log approximations and thermokinetic considerations can be used to represent the effect of involved variables [109]. Maier et al. described a dynamic model for HepG2 cells by quantifying 25 extra and intra-cellular metabolites by 13C labelling and intermittent metabolite quantification from parallel cultures using high performance liquid chromatography (HPLC), liquid chromatography-mass spectroscopy-mass spectroscopy (LC-MS-MS) and gas chromatography–mass spectroscopy (GC–MS) [232]. The negative control of GAPDH and positive control of OxPP over glycolytic flux [232] supported the hypothesis of GAPDH and Warburg effect as potential targets for tumour treatment [232]. Yarmush et al. modelled steatotic HepG2 cells during defatting for applications in fatty liver transplant [233] by measuring 28 extracellular metabolites and intracellular triglycerides using HPLC and commercially available assay kits [233]. They observed that the glycolytic flux was high while the extracellular glucose consumption was very small compared to glycogenolysis, indicating that glycolysis and fatty acid co-oxidation might co-exist in steatotic HepG2 cells [233].
6. Future perspectives: label free microspectroscopic analysis
Vibrational spectroscopic microscopy has emerged as a label-free alternative to currently employed techniques, which can provide molecular specific signatures of biological processes and function at a cellular and subcellular level. The spectrum comprises contributions from each molecular bond and is a “signature” or “fingerprint” which is characteristic of a material, or changes associated with a physical or chemical process. In complex samples, notably biological cells or tissue, the spectroscopic signature incorporates characteristics of all constituent functional groups of lipids, proteins, nucleic acids and other specific biomolecules and metabolites [24,234] (Figure 4 [235]). Infrared (IR) absorption and Raman scattering are the most common forms of vibrational spectroscopy, and both provide a molecule fingerprint, although through different fundamental physical principles. IR absorption is the result of electric dipole transition between vibrational states, whereas Raman spectroscopy is based on an inelastic scattering process caused by the polarisability change of a molecule [24]. The OH group has a particular strong IR absorption, whereas it is a relatively weak Raman scatterer, and therefore the latter is often promoted for in vivo applications, or live cell imaging [28]. Raman is often conducted at visible or near IR wavelengths, and therefore, in microscopic mode, can have spatial resolution of <1 μm, whereas the longer wavelengths of the IR (5–20 μm) limit its spatial resolution. Additionally, certain molecules such as amino acid residues, S–S disulphide bridges, C–S linkages from proteins and nucleic acids are better highlighted in Raman spectroscopy compared to IR spectroscopy [24].
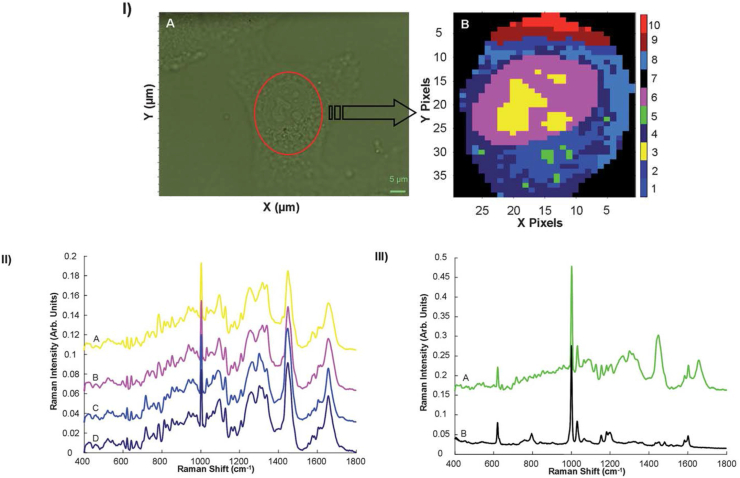
Figure 4.
: I) (A) Microscopic image of an A549 lung cancer cell, showing the reduced area identified for spectral mapping. (B) K-Means cluster map of the Raman profile of the same reduced area. II) K-means spectra of clusters 3 (A—representing nucleoli), 6 (B—representing nucleus), 1 and 4 (C and D, both from the cytoplasm). Spectra are offset for clarity. III) K-means spectrum of Cluster 5 (A), compared to the Raman spectrum polystyrene nanoparticles (B). Spectra are offset for clarity. (Reproduced with permission from [248]).
Using Raman and/or IR microspectroscopy, quantitative spectroscopic changes attributable to the biochemical processes occurring during cell culture and mitosis [236,237], proliferation [238], differentiation and activation [[239], [240], [241]], adhesion [242], death [243] and invasion [244] have previously been documented. Confocal Raman microspectroscopy has been demonstrated to provide detail at a subcellular level, of fixed or live cells, in 2D and/or 3D culture environments [[245], [246], [247]], as shown for the example of a Raman microspectroscopic map of an A549 human lung cancer cell, analysed by K-means clustering (Figure 4_I), which differentiates the nucleolar (yellow) regions of the nucleus (purple), which is in turn differentiated from regions of the cytoplasm (blue), according to their characteristic mean spectra (Figure 4_II) [248]. The analysis also unambiguously identified the presence of polystyrene nanoparticles (green) in the cytoplasm of the cell, identified by its characteristic spectrum (Figure 4_III). Similar studies have demonstrated the detection of cellular mitochondrial distribution [249] and phagosomes [250]. Label free analysis can identify signatures of subcellular phenomena not evident in a labelled approach, as for example the mechanistic role of accumulation of the chemotherapeutic agent doxorubicin in the nucleolus of cells [251], and potentially guide the identification of new biomarkers of cellular events. Thepphilpou et al. have reviewed the applications of different modes of vibrational spectroscopy for cellular analysis [252], and the reader is also referred to the graphical summary of the different methodologies in the work of Paraskevaidi et al. [253].
Critically, however, the analysis and interpretation of the spectral responses remains a challenge, even in the hands of “specialists”. By their very nature, label free techniques register all species within the sampling area, and identification of specific responses requires multivariate analytical techniques to data-mine the differential signatures, which are characteristic of cell injury events or response pathways. The characteristic spectral response can have (additive) contributions of increased or decreased concentrations of any or all constituent biomolecules, but also more complex contributions due to conformational, environmental (pH etc.) changes, and it is apparent that a novel combinatorial approach based on identification of the (differential) spectroscopic signatures rather than specific bands associated with specific biomolecules, may be more appropriate. The spectroscopic signature is, in itself, characteristic of the key cellular event, and can be used to identify and quantify it, in a “spectralomics” approach [36]. The evolution of the characteristic signatures may therefore be used to characterise the cellular pathway. The reported reproducibility of subcellular signatures of drug uptake, mechanisms of action and cellular response [36,251,[254], [255], [256], [257]], as well as nanoparticle trafficking and toxicity [37,[258], [259], [260], [261]], in multiple cell lines suggests that such an approach to label free characterisation of cellular processes according to characteristic spectroscopic signatures is feasible. Munck introduced similar terminology of the “spectral phenome”, in the discussion of the near IR spectroscopic analysis of a barley endosperm mutant model, evaluated by chemometrics [204], although metabolic pathways were not explicitly explored.
Although it cannot be considered truly label free, several studies have demonstrated that stable isotope labelling can be utilised in Raman spectroscopy for in situ monitoring of a kinetic process. Xu et al. used deuterated glucose and deuterated naphthalene as sole carbon sources for Escherichia coli and Pseudomonas sp. Cultures, respectively, and demonstrated that the Carbon-Deuterium vibration at ∼2700 cm−1 could be used to semi quantitatively and sensitively monitor the metabolism of the carbon sources, although the kinetics of the process were not explicitly monitored spectroscopically [262]. Zhang et al. used a similar technique based on simulated Raman scattering (SRS), which images the metabolic dynamics of newly synthesised macromolecules in a cell according to the signatures of C-D bonds derived from a deuterated glucose precursor [263]. They described the utility of this technique to visualise temporally separated glucose populations by using spectral differences of glucose isotopologues [263]. With a detection limit of 10 mM of C-D bonds, this technique is effective for tracing functional utilisation of glucose derived anabolic products over the conventional glucose imaging techniques which are limited to monitoring glucose uptake or catabolism activity [263]. Noothalpati and Shigeto explored ergosterol biosynthesis as a model system in single living fission yeast cells using stable isotope labelled Raman microspectroscopy to establish the technique for in vivo metabolic analysis [264]. Their study revealed the intrinsic spectra and relative abundances of all isotopomers of ergosterol, partially or fully substituted with 13C, when the spatially resolved Raman spectra was analysed by multivariate spectral data analysis [264]. Li et al. showed the application of stable isotope labelling and single cell Raman spectroscopy to link the food chain from 13C-glucose carbon substrate to E. coli microbe up to Caenorhabditis elegans predator in a quantitative and non-destructive manner [265]. Similarly, spatiotemporal and functional relationships between proteins and lipid droplets was dynamically visualised by 13C incorporation and in vivo time-lapse Raman imaging to illustrate the potential in proteome visualisation at intervals of 1 h over time periods of up to 40 h, without fluorophore tagging in single fission yeast cells [266]. Weber et al. explored the utility of Raman microspectroscopy coupled with stable isotope probing for monitoring the growth rates of heterologous bacteria at single-cell resolution, based on the 13C- and 12C-isotopologues of the amino acid phenylalanine [267], identifiable by the strong phenylalanine ring breathing modes 966 and 1,002 cm−1). Using a robust curve fitting process for precise deconvolution of partially overlapping peaks, the identified peaks provided an accurate reporter probe for quantification of cellular isotope fractional abundances (fcell) for complex 13C-labelled carbon source assimilation in E. coli in time-course experiments. The fcell and bacterial growth rates were validated using isotope ratio mass-spectroscopy and optical density measurements respectively.
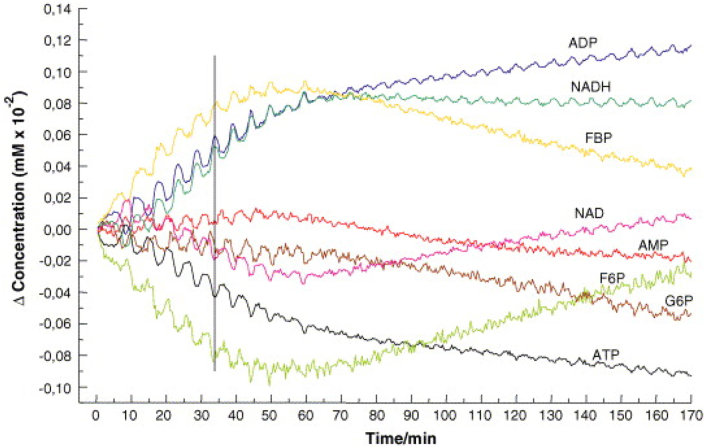
Analysis of the evolution of spectral profiles in cells has not been common, although kinetic, mechanistic approaches to analysis of organisms and organs have become well accepted in pharmaco kinetics/dynamics [268] and systems biology [269,270]. Mukherjee et al. used Raman mirospectroscopy to monitor effects of culture conditions in bacterial growth, although the spectral analysis was large based on peak ratios [271]. Mair et al. demonstrated that oscillatory kinetics of many glycolytic intermediates of yeast can be analysed by time resolved FT-IR spectroscopy [272]. Consecutive FT-IR-spectra of Saccharomyces carlsbergensis yeast extract were taken over a period of 1–2 h. The yeast spectra at all time points were least squares fitted with linear combinations of spectra of standard solutions of glycolytic intermediates (G6P, F6P, F1, 6P2, PEP, Pyr) and related compounds (ATP, ADP, AMP, NADH, NAD, trehalose, glycogen, ethanol, glycerol, phosphate), such that the kinetics of glycolytic intermediates could be extracted from the reconstructed spectra (Figure 5).
Figure 5.
Extraction of the kinetics of glycolytic intermediates with low concentration from the reconstructed spectra (Reproduced with permission from [272]).
Poonprasartporn and Chan conducted an FTIR spectroscopic study of live HEP2G cells, grown in either normal glucose (3.8 mM) or high glucose (25 mM) medium and compared their spectroscopic profiles after incubation times of 24, 48 and 72 h [273]. In the normal glucose treated cells, increases in the absorbance of bands associated with nucleic acid PO2− phosphate stretching and symmetric stretching of COO−, from fatty acids, amino acids and carboxylate metabolites. In contrast, the high glucose treated cells exhibited a clear reduction of the PO2− stretching mode band after 24 h, and further spectral changes in the region ∼1000–1200 cm−1 after 48 and 72 h, linked to the glycogen and ATP:ADP ratio. The study supports the use of the technique for monitoring metabolic processes in cells, and a more recent study from the same authors compared the effect of two anti-diabetic drugs on insulin resistant HEPG2 cells [274]. The analysis mainly focused on the glycogen peaks at 1150, 1080 and 1020 cm−1, which served as spectroscopic markers of cellular response to insulin. They discovered that the spectral changes were highly specific to the drug applied and were able to differentiate between the spectral profiles of normal and insulin resistant cells, the lack of response to insulin and the restoration of insulin resistance upon drug treatment in the resistant cells.
Pleitez et al. demonstrated the potential of mid-infrared optoacoustic microscopy (MiROM) for label-free, bond-selective, live-cell metabolic imaging for spatiotemporal dynamic monitoring of lipids, proteins and carbohydrates [275]. MiROM, based on mid-IR vibrational excitation and positive-contrast detection, offers greater sensitivity in the fingerprint spectral regions as compared to modern vibrational spectroscopic approaches which measure a reduction of transmission or reflected light intensity [[276], [277], [278], [279]], as evident from the lower limit of detection (LOD) (2.5 mM for DMSO and 1.5 μM for albumin) using laser powers in hundreds of microwatts (330 μW for DMSO and 500 μW for albumin), thereby significantly lowering the risk of phototoxicity to the cells [280]. They demonstrated MiROM could efficiently differentiate lipids (2,853 cm−1), proteins (sum of amide I (1,700 and 1,600 cm−1) and II (1,550 cm−1)), and carbohydrates (sum of 1,081 and 1,084 cm−1) to generate MiROM micrographs [275]. They further explored its potential for dynamic cellular analysis and used the optoacoustic contrasts to generate box plots to quantify the phenome at fixed time points to observe its dynamics [275]. Further, lipid and protein dynamics during isoproterenol (ISO) induced lipolysis in white and brown adipocytes was also showcased [275]. Lipid (2,857 cm−1) and protein (1,550 cm−1) were recorded every 5 min before and after addition of isoproterenol [275]. The lipid content slowly increased indicating lipogenesis until the lipogenesis was induced which induced nearly linear decrease in lipid content [275]. The study is significant for single-cell analysis, as differential lipolysis rates were observed in both white and brown adipocytes while some cells even remained unaffected [275].
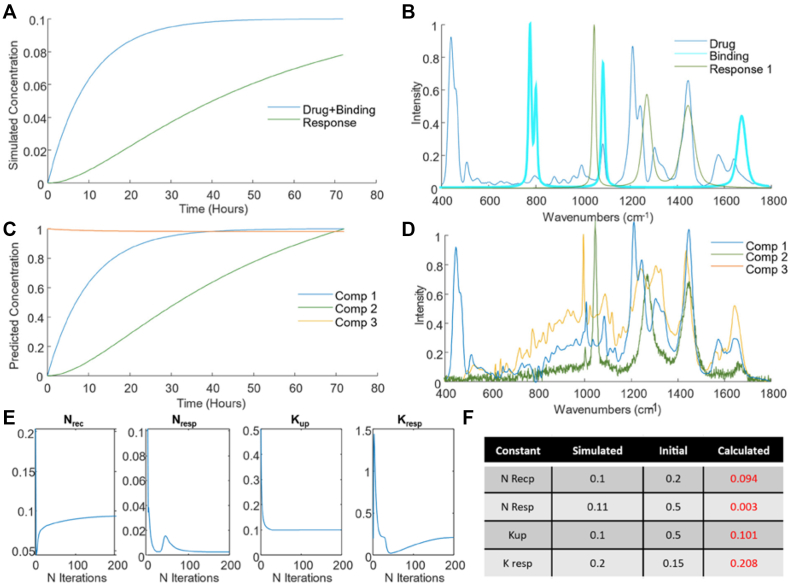
Accurately data mining the characteristic signatures of the subcellular interactions and processes represents a challenge. Multivariate curve resolution-alternating least squares (MCR-ALS) has been proposed as a more flexible approach to study the evolution of cellular processes [281]. MCR-ALS models the temporal evolution of the spectral dataset by considering a set of components whose concentrations evolve at different rates and have characteristic spectral features. MCR-ALS iteratively fits the target spectrum by least squares fitting multiple components, either provided a priori, based on previous or external information of the system, or estimated using techniques such as Singular Value Decomposition (SVD) [282]. Then, an initial estimation of concentration matrix or spectra is made by using information about the chemical components involved or using other methods such as Evolving Factor Analysis (EFA) [283]. Perez-Guaita et al. have previously demonstrated the application of MCR-ALS to gain insight into the pharmacodynamics and biochemical changes associated with drug exposure in an in vitro cellular model, as measured using Raman microspectroscopy [284], and a multimodal combination of Raman and Infrared microspectroscopies [285]. MCR-ALS uses external information and constraints to guide the iterative process, which can include initial estimates of the spectra of the starting point and intermediate constituents of the pathway, which can each be constrained to ensure that they contribute non negatively, and collectively constrained to ensure “closure”, or mass balance, in the hope of reducing the ambiguity of the result and leading to chemically interpretable solutions. One such constraint involves the introduction of kinetic equations to fit the evolution of the components to known chemical relations within the system, in a so-called hard-soft modelling approach. The approach has previously been used to model the evolution of spectra of very simple reactions (e.g., A- > B) [286], but more recently Perez-Guaita et al. have demonstrated that it is possible to constrain the least squares fitting procedure by a set of ordinary differential equations [30]. The approach was validated using the example of cellular uptake and binding of the chemotherapeutic drug doxorubicin, in vitro cell, accurately reproducing the spectroscopic profiles and uptake rates in simulated models (Figure 6).
Figure 6.
(A) Kinetic evolution of simulated drug uptake and binding (dark blue), and cellular response (green), (B) Raman spectrum of the drug, doxorubicin (dark blue), simulated spectral signature of drug binding (cyan) and of the subsequent cellular response (green), (C) predicted kinetic evolution of the MCR-ALS components, (D) MCR-ALS components, extracted after 50 iterations, (E) evolution of the kinetic constraint constants over 200 iterations of the MCR-ALS algorithm, (F) initial and final (after 200 iterations) constants employed in the MCR-ALS model. (Reproduced with permission from [30]).
The hard-soft MCR-ALS approach opens up the possibility of data mining the label free Raman microspectroscopic responses of a cellular metabolic process described by a phenomenological rate equation, or systems biology approach. Such a framework of label free, subcellular Raman microspectroscopic analysis, combined with a kinetic, mechanistic modelling approach, to underpin chemometric analysis protocols, provides a basis for the unambiguous interpretation of the evolution of the characteristic spectroscopic signatures. Note however, that these spectroscopic signatures, for example of the interaction of an intercalating agent in the nucleus of a cell, may be constituted by contributions of chemical moieties from multiple different biomolecules, or changes to them, and is a characteristic signature of the cellular event or process, rather than of any specific biomarker(s). The approach potentially lays the foundation for a spectralomics paradigm of label free high content spectroscopic analysis of cellular function, providing a holistic view of the cellular processes to augment conventional labelled and omics approaches. The combinatorial approach could guide identification of key biomarker targets for more in depth labelled and omics approaches, and, in its own right, could find future applications in preclinical drug screening and toxicology among others. However, it should be noted that the exploration of the techniques for such complex evolution of biochemically complex systems is very much in its infancy. The continuous evolution of novel and hybrid label free spectroscopic approaches promises increased capacity to rapidly screen cellular and subcellular processes in real time. Coherent Raman spectroscopic techniques promise large area screening at up to video rates [279,287], while the development of quantum cascade laser sources which are tunable across the mid IR region of the spectrum have given rise to hybrid techniques such as atomic force microscopy detected IR spectroscopy (AFM-IR) [288,289], as well as photoacoustic [290], and photothermal techniques, the most recent incarnations of which enable simultaneous measurement of IR and Raman on the same spot [291]. Nevertheless, it is crucial to demonstrate that techniques such as MCR-ALS and other multivariate datamining technique can reliably extract the signatures of interest, and that they can be interpreted in terms of their biochemical origin.
Author contributions
Nitin Patil: Conceptualization, Writing - Original Draft, Orla Howe: Writing - Original Draft, Paul Cahill: Writing - Original Draft. Hugh J. Byrne: Conceptualization, Writing - Original Draft, Supervision, Funding acquisition.
Funding acknowledgement
NP is funded by Science Foundation Ireland (SFI) Frontiers for the Future Award 20/FFPP/8517
Conflict of interest
None declared.
Data availability
No data was used for the research described in the article.
References
- 1.Deberardinis R.J., Thompson C.B. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148:1132–1144. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandel N.S. Glycolysis. Cold Spring Harbor Perspect Biol. 2021;13:a040535. doi: 10.1101/CSHPERSPECT.A040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romano A.H., Conway T. Evolution of carbohydrate metabolic pathways. Res Microbiol. 1996;147:448–455. doi: 10.1016/0923-2508(96)83998-2. [DOI] [PubMed] [Google Scholar]
- 4.Harris R.A. Encycl Biol Chem Second; 2013. Glycolysis overview; pp. 443–447. [DOI] [Google Scholar]
- 5.Barnett J.A. A history of research on yeasts 5: the fermentation pathway. Yeast. 2003;20:509–543. doi: 10.1002/YEA.986. [DOI] [PubMed] [Google Scholar]
- 6.Bugrim A., Nikolskaya T., Nikolsky Y. Early prediction of drug metabolism and toxicity: systems biology approach and modeling. Drug Discov Today. 2004;9:127–135. doi: 10.1016/S1359-6446(03)02971-4. [DOI] [PubMed] [Google Scholar]
- 7.Kell D.B. Systems biology, metabolic modelling and metabolomics in drug discovery and development. Drug Discov Today. 2006;11:1085–1092. doi: 10.1016/j.drudis.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Yu L., Chen X., Sun X., Wang L., Chen S. The glycolytic switch in tumors: how many players are involved? J Cancer. 2017;8:3430–3440. doi: 10.7150/jca.21125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell S.M., Burgess T., Lee J., Blackburn D.J., Allen S.P., Mortiboys H. Peripheral glycolysis in neurodegenerative diseases. Int J Mol Sci. 2020;21:1–19. doi: 10.3390/IJMS21238924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo X., Li H., Xu H., Woo S., Dong H., Lu F., et al. Glycolysis in the control of blood glucose homeostasis. Acta Pharm Sin B. 2012;2:358–367. doi: 10.1016/J.APSB.2012.06.002. [DOI] [Google Scholar]
- 11.Ussher J.R., Elmariah S., Gerszten R.E., Dyck J.R.B. The emerging role of metabolomics in the diagnosis and prognosis of cardiovascular disease. J Am Coll Cardiol. 2016;68:2850–2870. doi: 10.1016/J.JACC.2016.09.972. [DOI] [PubMed] [Google Scholar]
- 12.Ganapathy-Kanniappan S., Geschwind J.F.H. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer. 2013;12:1–11. doi: 10.1186/1476-4598-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abboud G., Choi S.C., Kanda N., Zeumer-Spataro L., Roopenian D.C., Morel L. Inhibition of glycolysis reduces disease severity in an autoimmune model of rheumatoid arthritis. Front Immunol. 2018;9:1973. doi: 10.3389/fimmu.2018.01973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z., Wang N., Chen J., Shen J. Emerging glycolysis targeting and drug discovery from Chinese medicine in cancer therapy. Evid Based Complement Alternat Med. 2012;2012:13. doi: 10.1155/2012/873175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schurr A., Gozal E. Glycolysis at 75: is it time to tweak the first elucidated metabolic pathway in history? Front Neurosci. 2015;9:170. doi: 10.3389/FNINS.2015.00170/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuang H.Y., Hofree M., Ideker T. A decade of systems biology. Annu Rev Cell Dev Biol. 2010;26:721–744. doi: 10.1146/annurev-cellbio-100109-104122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braun R. Systems analysis of high–throughput data. Adv Exp Med Biol. 2014;844:153. doi: 10.1007/978-1-4939-2095-2_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Machado D., Costa R.S., Rocha M., Ferreira E.C., Tidor B., Rocha I. Modeling formalisms in systems biology. Amb Express. 2011;1:1–14. doi: 10.1186/2191-0855-1-45/TABLES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberti M.V., Locasale J.W. The warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41:211. doi: 10.1016/J.TIBS.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jezewski A.J., Lin Y.-H., Reisz J.A., Culp-Hill R., Barekatain Y., Yan V.C., et al. Targeting host glycolysis as a strategy for antimalarial development. bioRxiv. 2021 doi: 10.1101/2020.10.09.331728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanzey M., Abdul Rahim S.A., Oudin A., Dirkse A., Kaoma T., Vallar L., et al. Comprehensive analysis of glycolytic enzymes as therapeutic targets in the treatment of glioblastoma. PLoS One. 2015;10 doi: 10.1371/JOURNAL.PONE.0123544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verlinde C.L.M.J., Hannaert V., Blonski C., Willson M., Périé J.J., Fothergill-Gilmore L.A., et al. Glycolysis as a target for the design of new anti-trypanosome drugs. Drug Resist Updates. 2001;4:50–65. doi: 10.1054/DRUP.2000.0177. [DOI] [PubMed] [Google Scholar]
- 23.Altschuler S.J., Wu L.F. Cellular heterogeneity: do differences make a difference? Cell. 2010;141:559–563. doi: 10.1016/J.CELL.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byrne H.J., Sockalingum G.D., Stone N. In: RSC anal. Spectrosc. Ser. Moss D., editor. Royal Society of Chemistry; 2011. Raman microscopy: complement or competitor? pp. 105–143. [DOI] [Google Scholar]
- 25.Byrne H.J., Ostrowska K.M., Nawaz H., Dorney J., Meade A.D., Bonnier F., et al. Vibrational spectroscopy: disease diagnostics and beyond. Challenges Adv. Comput. Chem. Phys. 2014;14:355–399. doi: 10.1007/978-94-007-7832-0_13. [DOI] [Google Scholar]
- 26.Old O.J., Fullwood L.M., Scott R., Lloyd G.R., Almond L.M., Shepherd N.A., et al. Vibrational spectroscopy for cancer diagnostics. Anal Methods. 2014;6:3901–3917. doi: 10.1039/C3AY42235F. [DOI] [Google Scholar]
- 27.Jamieson L.E., Byrne H.J. Vibrational spectroscopy as a tool for studying drug-cell interaction: could high throughput vibrational spectroscopic screening improve drug development? Vib Spectrosc. 2017;91:16–30. doi: 10.1016/J.VIBSPEC.2016.09.003. [DOI] [Google Scholar]
- 28.Baker M.J., Byrne H.J., Chalmers J., Gardner P., Goodacre R., Henderson A., et al. Clinical applications of infrared and Raman spectroscopy: state of play and future challenges. Analyst. 2018;143:1735–1757. doi: 10.1039/c7an01871a. [DOI] [PubMed] [Google Scholar]
- 29.Paraskevaidi M., Matthew B.J., Holly B.J., Hugh B.J., Thulya C.P.V., Loren C., et al. Clinical applications of infrared and Raman spectroscopy in the fields of cancer and infectious diseases. Appl Spectrosc Rev. 2021;56:804–868. doi: 10.1080/05704928.2021.1946076. [DOI] [Google Scholar]
- 30.Pérez-Guaita D., Quintás G., Farhane Z., Tauler R., Byrne H.J. Combining pharmacokinetics and vibrational spectroscopy: MCR-ALS hard-and-soft modelling of drug uptake in vitro using tailored kinetic constraints. Cells 2022. 2022;11:1555. doi: 10.3390/CELLS11091555. 11, Page 1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z., Parida S., Prasad R., Pandey R., Sharma D., Barman I. Vibrational spectroscopy for decoding cancer microbiota interactions: current evidence and future perspective. Semin Cancer Biol. 2021 doi: 10.1016/J.SEMCANCER.2021.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Richards O., Jenkins C., Griffiths H., Paczkowska E., Dunstan P.R., Jones S., et al. Vibrational spectroscopy: a valuable screening and diagnostic tool for obstetric disorders? Front Glob Women’s Heal. 2021;1 doi: 10.3389/fgwh.2020.610582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pence I., Mahadevan-Jansen A. Clinical instrumentation and applications of Raman spectroscopy. Chem Soc Rev. 2016;45:1958–1979. doi: 10.1039/c5cs00581g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Byrne H.J., Bonnier F., Casey A., Maher M., McIntyre J., Efeoglu E., et al. Advancing Raman microspectroscopy for cellular and subcellular analysis: towards in vitro high-content spectralomic analysis. Appl Opt. 2018;57:E11. doi: 10.1364/ao.57.000e11. [DOI] [PubMed] [Google Scholar]
- 35.Tomellini S.A., Finn J.W. Encycl Anal Chem; 2000. Vibrational spectroscopy in drug discovery, development and production. [DOI] [Google Scholar]
- 36.Farhane Z., Nawaz H., Bonnier F., Byrne H.J. In vitro label-free screening of chemotherapeutic drugs using Raman microspectroscopy: towards a new paradigm of spectralomics. J Biophot. 2018;11 doi: 10.1002/jbio.201700258. [DOI] [PubMed] [Google Scholar]
- 37.Efeoglu E., Maher M.A., Casey A., Byrne H.J. Label-free, high content screening using Raman microspectroscopy: the toxicological response of different cell lines to amine-modified polystyrene nanoparticles (PS-NH2) Analyst. 2017;142:3500–3513. doi: 10.1039/c7an00461c. [DOI] [PubMed] [Google Scholar]
- 38.Rajas F., Gautier-Stein A., Mithieux G. Glucose-6 phosphate, a central hub for liver carbohydrate metabolism. Metabolites. 2019;9 doi: 10.3390/METABO9120282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouché C., Serdy S., Kahn C.R., Goldfine A.B. The cellular fate of glucose and its relevance in type 2 diabetes. Endocr Rev. 2004;25:807–830. doi: 10.1210/ER.2003-0026. [DOI] [PubMed] [Google Scholar]
- 40.Magnuson M.A. Glucokinase gene structure. Functional implications of molecular genetic studies. Diabetes. 1990;39:523–527. doi: 10.2337/diab.39.5.523. [DOI] [PubMed] [Google Scholar]
- 41.Bedoya F.J., Matschinsky F.M., Shimizu T., O'Neil J.J., Appel M.C. Differential regulation of glucokinase activity in pancreatic islets and liver of the rat. J Biol Chem. 1986;261:10760–10764. doi: 10.1016/s0021-9258(18)67451-4. [DOI] [PubMed] [Google Scholar]
- 42.Peters B. 2017. Chemical equilibrium. React. Rate theory rare events simulations; pp. 19–37. [DOI] [Google Scholar]
- 43.DeBerardinis R.J., Lum J.J., Hatzivassiliou G., Thompson C.B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metabol. 2008;7:11–20. doi: 10.1016/J.CMET.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 44.Teuwen L.A., Geldhof V., Carmeliet P. How glucose, glutamine and fatty acid metabolism shape blood and lymph vessel development. Dev Biol. 2019;447:90–102. doi: 10.1016/j.ydbio.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 45.Alfarouk K.O., Verduzco D., Rauch C., Muddathir A.K., Bashir A.H.H., Elhassan G.O., et al. Glycolysis, tumor metabolism, cancer growth and dissemination. A new pH-based etiopathogenic perspective and therapeutic approach to an old cancer question. Oncoscience. 2014;1:777–802. doi: 10.18632/oncoscience.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bell S.M., Burgess T., Lee J., Blackburn D.J., Allen S.P., Mortiboys H. Peripheral glycolysis in neurodegenerative diseases. Int J Mol Sci. 2020;21:1–19. doi: 10.3390/ijms21238924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X., Alshakhshir N., Zhao L. Glycolytic metabolism, brain resilience, and Alzheimer's disease. Front Neurosci. 2021;15:476. doi: 10.3389/FNINS.2021.662242/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morral N. Novel targets and therapeutic strategies for type 2 diabetes. Trends Endocrinol Metabol. 2003;14:169–175. doi: 10.1016/S1043-2760(03)00031-6. [DOI] [PubMed] [Google Scholar]
- 49.Kurukulasuriya R., Link J., Madar D., Pei Z., Richards S., Rohde J., et al. Potential drug targets and progress towards pharmacologic inhibition of hepatic glucose production. Curr Med Chem. 2012;10:123–153. doi: 10.2174/0929867033368556. [DOI] [PubMed] [Google Scholar]
- 50.Saltiel A.R. New perspectives into the molecular pathogenesis and treatment of type 2 diabetes. Cell. 2001;104:517–529. doi: 10.1016/S0092-8674(01)00239-2. [DOI] [PubMed] [Google Scholar]
- 51.Vogt C., Ardehali H., Iozzo P., Yki-Jarvinen H., Koval J., Maezono K., et al. Regulation of hexokinase II expression in human skeletal muscle in vivo. Metabolism. 2000;49:814–818. doi: 10.1053/meta.2000.6245. [DOI] [PubMed] [Google Scholar]
- 52.Printz R.L., Koch S., Potter L.R., O'Doherty R.M., Tiesinga J.J., Moritz S., et al. Hexokinase II mRNA and gene structure, regulation by insulin, and evolution. J Biol Chem. 1993;268:5209–5219. doi: 10.1016/S0021-9258(18)53521-3. [DOI] [PubMed] [Google Scholar]
- 53.Chang P.Y., Jensen J., Printz R.L., Granner D.K., Ivy J.L., Moller D.E. Overexpression of hexokinase II in transgenic mice: evidence that increased phosphorylation augments muscle glucose uptake. J Biol Chem. 1996;271:14834–14839. doi: 10.1074/jbc.271.25.14834. [DOI] [PubMed] [Google Scholar]
- 54.Vestergaard H., Bjørbæk C., Hansen T., Larsen F.S., Granner D.K., Pedersen O. Impaired activity and gene expression of hexokinase II in muscle from non-insulin-dependent diabetes mellitus patients. J Clin Invest. 1995;96:2639–2645. doi: 10.1172/JCI118329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laville M., Bernard C., Lyon U., Vega N., Vidal H. Regulation by insulin of gene expression in human skeletal muscle and adipose tissue: evidence for specific defects in type 2 diabetes. Am Diabetes Assoc. 2001 doi: 10.2337/diabetes.50.5.1134. [DOI] [PubMed] [Google Scholar]
- 56.Pendergrass M., Koval J., Vogt C., Diabetes H.Y.J. Insulin-induced hexokinase II expression is reduced in obesity and NIDDM. Am Diabetes Assoc. 1998 doi: 10.2337/diabetes.47.3.387. 1998 undefined. [DOI] [PubMed] [Google Scholar]
- 57.Hattersley A.T., Turner R.C., Patel P., O'Rahilly S., Hattersley A.T., Patel P., et al. Linkage of type 2 diabetes to the glucokinase gene. Lancet. 1992;339:1307–1310. doi: 10.1016/0140-6736(92)91958-B. [DOI] [PubMed] [Google Scholar]
- 58.Froguel P., Vaxillaire M., Sun F., Velho G., Zouali H., Butel M.O., et al. Erratum: close linkage of glucokinase locus on chromosome 7p to early-onset non-insulin-dependent diabetes mellitus (Nature (1992) 356 (162-164)) Nature. 1992;357:607. doi: 10.1038/357607c0. [DOI] [PubMed] [Google Scholar]
- 59.Froguel P., Zouali H., Vionnet N., Velho G., Vaxillaire M., Sun F., et al. Familial hyperglycemia due to mutations in glucokinase -- definition of a subtype of diabetes mellitus. N Engl J Med. 1993;328:697–702. doi: 10.1056/NEJM199303113281005. [DOI] [PubMed] [Google Scholar]
- 60.Matschinsky F.M. A lesson in metabolic regulation inspired by the glucokinase glucose sensor paradigm. Diabetes. 1996;45:223–241. doi: 10.2337/diab.45.2.223. [DOI] [PubMed] [Google Scholar]
- 61.Caro J.F., Triester S., Patel V.K., Tapscott E.B., Frazier N.L., Dohm G.L. Liver glucokinase: decreased activity in patients with type II diabetes. Horm Metab Res. 1995;27:19–22. doi: 10.1055/S-2007-979899. [DOI] [PubMed] [Google Scholar]
- 62.Christesen H.B.T., Jacobsen B.B., Odili S., Buettger C., Cuesta-Munoz A., Hansen T., et al. The second activating glucokinase mutation (A456V): implications for glucose homeostasis and diabetes therapy. Diabetes. 2002;51:1240–1246. doi: 10.2337/diabetes.51.4.1240. [DOI] [PubMed] [Google Scholar]
- 63.Glaser B., Kesavan P., Heyman M., Davis E., Cuesta A., Buchs A., et al. Familial hyperinsulinism caused by an activating glucokinase mutation. N Engl J Med. 1998;338:226–230. doi: 10.1056/nejm199801223380404. [DOI] [PubMed] [Google Scholar]
- 64.Ohlson L.O., Larsson B., Björntorp P., Eriksson H., Svärdsudd K., Welin L., et al. Risk factors for Type 2 (non-insulin-dependent) diabetes mellitus. Thirteen and one-half years of follow-up of the participants in a study of Swedish men born in 1913. Diabetologia. 1988;31:798–805. doi: 10.1007/BF00277480. [DOI] [PubMed] [Google Scholar]
- 65.Avogaro A., Toffolo G., Miola M., Valerio A., Tiengo A., Cobelli C., et al. Intracellular lactate- and pyruvate-interconversion rates are increased in muscle tissue of non-insulin-dependent diabetic individuals. J Clin Invest. 1996;98:108–115. doi: 10.1172/JCI118754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vaishnavi S.N., Vlassenko A.G., Rundle M.M., Snyder A.Z., Mintun M.A., Raichle M.E. Regional aerobic glycolysis in the human brain. Proc Natl Acad Sci U S A. 2010;107:17757–17762. doi: 10.1073/pnas.1010459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Magistretti P.J., Chatton J.Y. Relationship between L-glutamate-regulated intracellular Na+ dynamics and ATP hydrolysis in astrocytes. J Neural Transm. 2005;112:77–85. doi: 10.1007/s00702-004-0171-6. [DOI] [PubMed] [Google Scholar]
- 68.Pellerin L., Magistretti P.J. Excitatory amino acids stimulate aerobic glycolysis in astrocytes via an activation of the na+/k+ atpase. Dev Neurosci. 1996;18:336–342. doi: 10.1159/000111426. [DOI] [PubMed] [Google Scholar]
- 69.Mercer R.W., Dunham P.B. Membrane-bound ATP fuels the Na/K pump: studies on membrane-bound glycolytic enzymes on inside-out vesicles from human red cell membranes. J Gen Physiol. 1981;78:547–568. doi: 10.1085/jgp.78.5.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Okamoto K.E.N., Wang W., Rounds J.A.N., Chambers E.A., Jacobs D.O. ATP from glycolysis is required for normal sodium homeostasis in resting fast-twitch rodent skeletal muscle. Am J Physiol Endocrinol Metab. 2001;281 doi: 10.1152/AJPENDO.2001.281.3.E479. [DOI] [PubMed] [Google Scholar]
- 71.Campbell J.D., Paul R.J. The nature of fuel provision for the Na+,K(+)-ATPase in porcine vascular smooth muscle. J Physiol. 1992;447:67–82. doi: 10.1113/jphysiol.1992.sp018991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu K., Aoki C., Elste A., Rogalski-Wilk A.A., Siekevitz P. The synthesis of ATP by glycolytic enzymes in the postsynaptic density and the effect of endogenously generated nitric oxide. Proc Natl Acad Sci U S A. 1997;94:13273–13278. doi: 10.1073/pnas.94.24.13273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tager H.S. Biochemistry: a functional approach. JAMA, J Am Med Assoc. 1984;251:1095. doi: 10.1001/jama.1984.03340320071034. [DOI] [Google Scholar]
- 74.Mosconi L., Pupi A., De Leon M.J. Brain glucose hypometabolism and oxidative stress in preclinical Alzheimer's disease. Ann N Y Acad Sci. 2008;1147:180–195. doi: 10.1196/ANNALS.1427.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.An Y., Varma V.R., Varma S., Casanova R., Dammer E., Pletnikova O., et al. Evidence for brain glucose dysregulation in Alzheimer's disease. Alzheimer's Dementia. 2018;14:318–329. doi: 10.1016/J.JALZ.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vlassenko A.G., Gordon B.A., Goyal M.S., Su Y., Blazey T.M., Durbin T.J., et al. Aerobic glycolysis and tau deposition in preclinical Alzheimer's disease. Neurobiol Aging. 2018;67:95–98. doi: 10.1016/J.NEUROBIOLAGING.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van Wijk R., Van Solinge W.W. The energy-less red blood cell is lost: erythrocyte enzyme abnormalities of glycolysis. Blood. 2005;106:4034–4042. doi: 10.1182/BLOOD-2005-04-1622. [DOI] [PubMed] [Google Scholar]
- 78.Climent F., Roset F., Repiso A., de la Ossa P. Red cell glycolytic enzyme disorders caused by mutations: an update. Cardiovasc Hematol Disord: Drug Targets. 2009;9:95–106. doi: 10.2174/187152909788488636. [DOI] [PubMed] [Google Scholar]
- 79.Valentine W.N., Tanaka K.R., Paglia D.E. Hemolytic anemias and erythrocyte enzymopathies. Ann Intern Med. 1985;103:245–257. doi: 10.7326/0003-4819-103-2-245. [DOI] [PubMed] [Google Scholar]
- 80.Heiden M.G.V., Cantley L.C., Thompson C.B. Understanding the warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/SCIENCE.1160809. 80- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stubbs M., Griffiths J.R. The altered metabolism of tumors: HIF-1 and its role in the Warburg effect. Adv Enzym Regul. 2010;50:44–55. doi: 10.1016/j.advenzreg.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 82.Bayley J.P., Devilee P. The Warburg effect in 2012. Curr Opin Oncol. 2012;24:62–67. doi: 10.1097/cco.0b013e32834deb9e. [DOI] [PubMed] [Google Scholar]
- 83.Shanmugam M., McBrayer S.K., Rosen S.T. Targeting the Warburg effect in hematological malignancies: from PET to therapy. Curr Opin Oncol. 2009;21:531–536. doi: 10.1097/CCO.0b013e32832f57ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang T., Marquardt C., Foker J. Aerobic glycolysis during lymphocyte proliferation. Natalia. 1976;261:702–705. doi: 10.1038/261702a0. 1976 2615562. [DOI] [PubMed] [Google Scholar]
- 85.Roos D., Loos J.A. Changes in the carbohydrate metabolism of mitogenically stimulated human peripheral lymphocytes: II. Relative importance of glycolysis and oxidative phosphorylation on phytohaemagglutinin stimulation. Exp Cell Res. 1973;77:127–135. doi: 10.1016/0014-4827(73)90561-2. [DOI] [PubMed] [Google Scholar]
- 86.Hedeskov C.J. Early effects of phytohaemagglutinin on glucose metabolism of normal human lymphocytes. Biochem J. 1968;110:373–380. doi: 10.1042/BJ1100373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brand K. Glutamine and glucose metabolism during thymocyte proliferation. Pathways of glutamine and glutamate metabolism. Biochem J. 1985;228:353–361. doi: 10.1042/BJ2280353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brown R.S., Goodman T.M., Zasadny K.R., Greenson J.K., Wahl R.L. Expression of hexokinase II and Glut-1 in untreated human breast cancer. Nucl Med Biol. 2002;29:443–453. doi: 10.1016/S0969-8051(02)00288-3. [DOI] [PubMed] [Google Scholar]
- 89.Kolev Y., Uetake H., Takagi Y., Sugihara K. Lactate dehydrogenase-5 (LDH-5) expression in human gastric cancer: association with hypoxia-inducible factor (HIF-1α) pathway, angiogenic factors production and poor prognosis. Ann Surg Oncol. 2008;15:2336–2344. doi: 10.1245/S10434-008-9955-5. [DOI] [PubMed] [Google Scholar]
- 90.Higashimura Y., Nakajima Y., Yamaji R., Harada N., Shibasaki F., Nakano Y., et al. Up-regulation of glyceraldehyde-3-phosphate dehydrogenase gene expression by HIF-1 activity depending on Sp1 in hypoxic breast cancer cells. Arch Biochem Biophys. 2011;509:1–8. doi: 10.1016/j.abb.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 91.Zhou M., Zhao Y., Ding Y., Liu H., Liu Z., Fodstad O., et al. Warburg effect in chemosensitivity: targeting lactate dehydrogenase-A re-sensitizes Taxol-resistant cancer cells to Taxol. Mol Cancer. 2010;9 doi: 10.1186/1476-4598-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shi H.-S., Li D., Zhang J., Wang Y.-S., Yang L., Zhang H.-L., et al. Silencing of pkm2 increases the efficacy of docetaxel in human lung cancer xenografts in mice. Wiley Online Libr. 2010;101:1447–1453. doi: 10.1111/j.1349-7006.2010.01562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xie H., Valera V.A., Merino M.J., Amato A.M., Signoretti S., Linehan W.M., et al. LDH-A inhibition, a therapeutic strategy for treatment of hereditary leiomyomatosis and renal cell cancer. Mol Cancer Therapeut. 2009;8:626–635. doi: 10.1158/1535-7163.MCT-08-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pastorino J., Hoek J. Hexokinase II: the integration of energy metabolism and control of apoptosis. Curr Med Chem. 2005;10:1535–1551. doi: 10.2174/0929867033457269. [DOI] [PubMed] [Google Scholar]
- 95.Ogino T., Yamadera T., Mizumoto K., Nonaka T., Imajoh-Ohmi S. Enolase, a cellular glycolytic enzyme, is required for efficient transcription of Sendai virus genome. Biochem Biophys Res Commun. 2001;285:447–455. doi: 10.1006/bbrc.2001.5160. [DOI] [PubMed] [Google Scholar]
- 96.Dobashi Y., Watanabe H., Matsubara M., Yanagawa T., Raz A., Shimamiya T., et al. Autocrine motility factor/glucose-6-phosphate isomerase is a possible predictor of metastasis in bone and soft tissue tumours. Wiley Online Libr. 2006;208:44–53. doi: 10.1002/path.1878. [DOI] [PubMed] [Google Scholar]
- 97.Yvan-Charvet L., Ivanov S. Metabolic reprogramming of macrophages in atherosclerosis: is it all about cholesterol? J Lipid Atheroscler. 2020;9:231–242. doi: 10.12997/JLA.2020.9.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Poels K., Schnitzler J.G., Waissi F., Levels J.H.M., Stroes E.S.G., Daemen M.J.A.P., et al. Inhibition of PFKFB3 hampers the progression of atherosclerosis and promotes plaque stability. Front Cell Dev Biol. 2020;8 doi: 10.3389/FCELL.2020.581641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gimbrone M.A., García-Cardeña G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res. 2016;118:620–636. doi: 10.1161/CIRCRESAHA.115.306301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rohlenova K., Veys K., Miranda-Santos I., De Bock K., Carmeliet P. Endothelial cell metabolism in health and disease. Trends Cell Biol. 2018;28:224–236. doi: 10.1016/J.TCB.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 101.Li X., Sun X., Carmeliet P. Hallmarks of endothelial cell metabolism in health and disease. Cell Metabol. 2019;30:414–433. doi: 10.1016/J.CMET.2019.08.011. [DOI] [PubMed] [Google Scholar]
- 102.Draoui N., De Zeeuw P., Carmeliet P. Angiogenesis revisited from a metabolic perspective: role and therapeutic implications of endothelial cell metabolism. Open Biol. 2017;7 doi: 10.1098/RSOB.170219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Leung S.W.S., Shi Y. The glycolytic process in endothelial cells and its implications. Acta Pharmacol Sin. 2021 432 2021;43:251–259. doi: 10.1038/s41401-021-00647-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Magnuson D.K., Maier R.V., Pohlman T.H. Protein kinase C: a potential pathway of endothelial cell activation by endotoxin, tumor necrosis factor, and interleukin-1. Surgery. 1989;106:216–223. doi: 10.5555/URI:PII:0039606089902110. [DOI] [PubMed] [Google Scholar]
- 105.Shi J., Yang Y., Cheng A., Xu G., He F. Metabolism of vascular smooth muscle cells in vascular diseases. Am J Physiol Heart Circ Physiol. 2020;319:H613–H631. doi: 10.1152/ajpheart.00220.2020. [DOI] [PubMed] [Google Scholar]
- 106.Chen Z., Liu M., Li L., Chen L. Involvement of the Warburg effect in non-tumor diseases processes. J Cell Physiol. 2018;233:2839–2849. doi: 10.1002/JCP.25998. [DOI] [PubMed] [Google Scholar]
- 107.Duraj T., Carrión-Navarro J., Seyfried T.N., García-Romero N., Ayuso-Sacido A. Metabolic therapy and bioenergetic analysis: the missing piece of the puzzle. Mol Metabol. 2021;54 doi: 10.1016/J.MOLMET.2021.101389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Saa P.A., Nielsen L.K. Formulation, construction and analysis of kinetic models of metabolism: a review of modelling frameworks. Biotechnol Adv. 2017;35:981–1003. doi: 10.1016/J.BIOTECHADV.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 109.Gerdtzen Z.P. Modeling metabolic networks for mammalian cell systems: general considerations, modeling strategies, and available tools. Adv Biochem Eng Biotechnol. 2012;127:71–108. doi: 10.1007/10_2011_120. [DOI] [PubMed] [Google Scholar]
- 110.Schwalfenberg M, Carey C, Hynes J, Technologies A. Metabolic profiling of cells in 3D cultures using MitoXpress xtra and pH-xtra assays. [n.d.]
- 111.Mcgarrigle R, Carey C, Hynes J. Assessing the impact of drug treatment on cardiomyocyte function [n.d].
- 112.Xintaropoulou C., Ward C., Wise A., Marston H., Turnbull A., Langdon S.P. A comparative analysis of inhibitors of the glycolysis pathway in breast and ovarian cancer cell line models. Oncotarget. 2015;6 doi: 10.18632/ONCOTARGET.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mookerjee S.A., Goncalves R.L.S., Gerencser A.A., Nicholls D.G., Brand M.D. The contributions of respiration and glycolysis to extracellular acid production. Biochim Biophys Acta Bioenerg. 2015;1847:171–181. doi: 10.1016/J.BBABIO.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 114.Mookerjee S.A., Nicholls D.G., Brand M.D. Determining maximum glycolytic capacity using extracellular flux measurements. PLoS One. 2016;11 doi: 10.1371/JOURNAL.PONE.0152016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Teslaa T., Teitell M.A. Techniques to monitor glycolysis. Methods Enzymol. 2014;542:91. doi: 10.1016/B978-0-12-416618-9.00005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barend Van Dyk J. 2020. Mathematical modelling of glycolysis in skeletal muscle cells. [Google Scholar]
- 117.Kotasidis F.A., Tsoumpas C., Rahmim A. Advanced kinetic modelling strategies: towards adoption in clinical PET imaging. Clin Transl Imaging. 2014;2:219–237. doi: 10.1007/s40336-014-0069-8. [DOI] [Google Scholar]
- 118.Kawai N., Nishiyama Y., Miyake K., Tamiya T., Nagao S. Evaluation of tumor FDG transport and metabolism in primary central nervous system lymphoma using [ 18F]fluorodeoxyglucose (FDG) positron emission tomography (PET) kinetic analysis. Ann Nucl Med. 2005;19:685–690. doi: 10.1007/BF02985117. [DOI] [PubMed] [Google Scholar]
- 119.Nishiyama Y., Yamamoto Y., Monden T., Sasakawa Y., Kawai N., Satoh K., et al. Diagnostic value of kinetic analysis using dynamic FDG PET in immunocompetent patients with primary CNS lymphoma. Eur J Nucl Med Mol Imag. 2007;34:78–86. doi: 10.1007/S00259-006-0153-Z. [DOI] [PubMed] [Google Scholar]
- 120.Anzai Y., Minoshima S., Wolf G.T., Wahl R.L. Head and neck cancer: detection of recurrence with three-dimensional principal components analysis at dynamic FDG PET. Radiology. 1999;212:285–290. doi: 10.1148/RADIOLOGY.212.1.R99JL02285. [DOI] [PubMed] [Google Scholar]
- 121.Schiepers C., Chen W., Cloughesy T., Dahlbom M., Huang S.-C. 18F-FDOPA kinetics in brain tumors. Soc Nucl Med. 2007;48:1651–1661. doi: 10.2967/jnumed.106.039321. [DOI] [PubMed] [Google Scholar]
- 122.Thorwarth D., Eschmann S.M., Paulsen F., Alber M. A kinetic model for dynamic [18F]-Fmiso PET data to analyse tumour hypoxia. Phys Med Biol. 2005;50:2209–2224. doi: 10.1088/0031-9155/50/10/002. [DOI] [PubMed] [Google Scholar]
- 123.Schiepers C., Chen W., Dahlbom M., Cloughesy T., Hoh C.K., Huang S.C. 18F-fluorothymidine kinetics of malignant brain tumors. Eur J Nucl Med Mol Imag. 2007;34:1003–1011. doi: 10.1007/S00259-006-0354-5. [DOI] [PubMed] [Google Scholar]
- 124.Wardak M., Schiepers C., Dahlbom M., Cloughesy T., Chen W., Satyamurthy N., et al. Discriminant analysis of 18F-fluorothymidine kinetic parameters to predict survival in patients with recurrent high-grade glioma. Clin Cancer Res. 2011;17:6553–6562. doi: 10.1158/1078-0432.CCR-10-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tsien R.Y. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/ANNUREV.BIOCHEM.67.1.509. [DOI] [PubMed] [Google Scholar]
- 126.Matz M.V., Fradkov A.F., Labas Y.A., Savitsky A.P., Zaraisky A.G., Markelov M.L., et al. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 127.Heim R., Prasher D.C., Tsien R.Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci U S A. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lippincott-Schwartz J., Snapp E., Kemvorthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. 2001 26. [DOI] [PubMed] [Google Scholar]
- 129.Pepperkok R., Squire A., Geley S., Bastiaens P.I.H. Simultaneous detection of multiple green fluorescent proteins in live cells by fluorescence lifetime imaging microscopy. Curr Biol. 1999;9:269–274. doi: 10.1016/S0960-9822(99)80117-1. [DOI] [PubMed] [Google Scholar]
- 130.Luo W.X., Cheng T., Guan B.Q., Li S.W., Miao J., Zhang J., et al. Variants of green fluorescent protein GFPxm. Mar Biotechnol. 2006;8:560–566. doi: 10.1007/s10126-006-6006-8. [DOI] [PubMed] [Google Scholar]
- 131.Mayer B., Emery G., Berdnik D., Wirtz-Peitz F., Knoblich J.A. Quantitative analysis of protein dynamics during asymmetric cell division. Curr Biol. 2005;15:1847–1854. doi: 10.1016/j.cub.2005.08.067. [DOI] [PubMed] [Google Scholar]
- 132.Ståhl P.L., Lundeberg J. Toward the single-hour high-quality genome. Annu Rev Biochem. 2012;81:359–378. doi: 10.1146/annurev-biochem-060410-094158. [DOI] [PubMed] [Google Scholar]
- 133.Harrison R.J. Understanding genetic variation and function- the applications of next generation sequencing. Semin Cell Dev Biol. 2012;23:230–236. doi: 10.1016/j.semcdb.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 134.Van Vliet A.H.M. Next generation sequencing of microbial transcriptomes: challenges and opportunities. FEMS Microbiol Lett. 2010;302:1–7. doi: 10.1111/j.1574-6968.2009.01767.x. [DOI] [PubMed] [Google Scholar]
- 135.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Slonim D.K., Yanai I. Getting started in gene expression microarray analysis. PLoS Comput Biol. 2009;5 doi: 10.1371/journal.pcbi.1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Thelen J.J., Miernyk J.A. The proteomic future: where mass spectrometry should be taking us. Biochem J. 2012;444:169–181. doi: 10.1042/BJ20110363. [DOI] [PubMed] [Google Scholar]
- 138.Angel T.E., Aryal U.K., Hengel S.M., Baker E.S., Kelly R.T., Robinson E.W., et al. Mass spectrometry-based proteomics: existing capabilities and future directions. Chem Soc Rev. 2012;41:3912–3928. doi: 10.1039/c2cs15331a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Reaves M.L., Rabinowitz J.D. Metabolomics in systems microbiology. Curr Opin Biotechnol. 2011;22:17–25. doi: 10.1016/j.copbio.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fondi M., Liò P. Multi -omics and metabolic modelling pipelines: challenges and tools for systems microbiology. Microbiol Res. 2015;171:52–64. doi: 10.1016/J.MICRES.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 141.Pavel A.B., Sonkin D., Reddy A. Integrative modeling of multi-omics data to identify cancer drivers and infer patient-specific gene activity. BMC Syst Biol. 2016;10:1–14. doi: 10.1186/S12918-016-0260-9/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Reuter J.A., Spacek D.V., Snyder M.P. High-throughput sequencing technologies. Mol Cell. 2015;58:586–597. doi: 10.1016/J.MOLCEL.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li P., Su M., Chatterjee M., Lämmerhofer M. Targeted analysis of sugar phosphates from glycolysis pathway by phosphate methylation with liquid chromatography coupled to tandem mass spectrometry. Anal Chim Acta. 2022;1221 doi: 10.1016/J.ACA.2022.340099. [DOI] [PubMed] [Google Scholar]
- 144.Gao Y., Fillmore T.L., Munoz N., Bentley G.J., Johnson C.W., Kim J., et al. High-throughput large-scale targeted proteomics assays for quantifying pathway proteins in Pseudomonas putida KT2440. Front Bioeng Biotechnol. 2020;8:1383. doi: 10.3389/FBIOE.2020.603488/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lichtenstein D., Mentz A., Sprenger H., Schmidt F.F., Albaum S.P., Kalinowski J., et al. A targeted transcriptomics approach for the determination of mixture effects of pesticides. Toxicology. 2021:460. doi: 10.1016/J.TOX.2021.152892. [DOI] [PubMed] [Google Scholar]
- 146.Thomas Brenn, Alexander J Lazar, McKee's pathology of the skin, 2 volume set E-book - J. Eduardo calonje, Steven Billings - Google Books n.d. https://books.google.ie/books?hl=en&lr=&id=pMN1DwAAQBAJ&oi=fnd&pg=PP1&dq=McKee%27s+Pathology+of+the+Skin+2020&ots=OF1IfQ_fSi&sig=iGTlURgocz00l3HB9GmDkow_8mE&redir_esc=y#v=onepage&q=next generation sequencing&f=false.
- 147.Next generation sequencing - an overview | ScienceDirect Topics n.d. https://www.sciencedirect.com/topics/medicine-and-dentistry/next-generation-sequencing.
- 148.Salzman J., Jiang H., Wong W.H. Statistical modeling of RNA-seq data. Stat Sci. 2011;26:62–83. doi: 10.1214/10-STS343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schwanhüusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/NATURE10098. [DOI] [PubMed] [Google Scholar]
- 150.Lu R., Markowetz F., Unwin R.D., Leek J.T., Airoldi E.M., MacArthur B.D., et al. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462:358–362. doi: 10.1038/NATURE08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kirkpatrick D.S., Denison C., Gygi S.P. Weighing in on ubiquitin: the expanding role of mass spectrometry-based proteomics. Nat Cell Biol. 2005;7:750. doi: 10.1038/NCB0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yang Y., Zhou R., Li W., Liu Y., Zhang Y., Ao H., et al. Dynamic transcriptome analysis reveals potential long non-coding RNAs governing postnatal pineal development in pig. Front Genet. 2019;10:409. doi: 10.3389/FGENE.2019.00409/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liu Z., Yu X., Qin A., Zhao Z., Liu Y., Sun S., et al. Research strategies for single-cell transcriptome analysis in plant leaves. Plant J. 2022 doi: 10.1111/TPJ.15927. [DOI] [PubMed] [Google Scholar]
- 154.Xu G.F., Gong C.C., Lyu H., Deng H.M., Zheng S.C. Dynamic transcriptome analysis of Bombyx mori embryonic development. Insect Sci. 2022;29:344–362. doi: 10.1111/1744-7917.12934. [DOI] [PubMed] [Google Scholar]
- 155.Hayashizaki Y., Kanamori M. Dynamic transcriptome of mice. Trends Biotechnol. 2004;22:161–167. doi: 10.1016/J.TIBTECH.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 156.Börgstrom E., Paterlini M., Mold J.E., Frisen J., Lundeberg J. Comparison of whole genome amplification techniques for human single cell exome sequencing. PLoS One. 2017;12 doi: 10.1371/JOURNAL.PONE.0171566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Binder V., Bartenhagen C., Okpanyi V., Gombert M., Moehlendick B., Behrens B., et al. A new workflow for whole-genome sequencing of single human cells. Hum Mutat. 2014;35:1260–1270. doi: 10.1002/HUMU.22625. [DOI] [PubMed] [Google Scholar]
- 158.Babayan A., Alawi M., Gormley M., Müller V., Wikman H., McMullin R.P., et al. Comparative study of whole genome amplification and next generation sequencing performance of single cancer cells. Oncotarget. 2016;8:56066–56080. doi: 10.18632/ONCOTARGET.10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kalisky T., Quake S.R. Single-cell genomics. Nat Methods. 2011 84 2011;8:311–314. doi: 10.1038/nmeth0411-311. [DOI] [PubMed] [Google Scholar]
- 160.Wang D., Bodovitz S. Single cell analysis: the new frontier in “omics. Trends Biotechnol. 2010;28:281–290. doi: 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lubeck E., Coskun A.F., Zhiyentayev T., Ahmad M., Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014 114 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen K.H., Boettiger A.N., Moffitt J.R., Wang S., Zhuang X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015:348. doi: 10.1126/science.aaa6090. 80- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Amberg A., Riefke B., Schlotterbeck G., Ross A., Senn H., Dieterle F., et al. NMR and MS methods for metabolomics. Methods Mol Biol. 2017;1641:229–258. doi: 10.1007/978-1-4939-7172-5_13. [DOI] [PubMed] [Google Scholar]
- 164.Winter G., Krömer J.O. Fluxomics - connecting ’omics analysis and phenotypes. Environ Microbiol. 2013;15:1901–1916. doi: 10.1111/1462-2920.12064. [DOI] [PubMed] [Google Scholar]
- 165.Van Dam J.C., Eman M.R., Frank J., Lange H.C., Van Dedem G.W.K., Heijnen S.J. Analysis of glycolytic intermediates in Saccharomyces cerevisiae using anion exchange chromatography and electrospray ionization with tandem mass spectrometric detection. Anal Chim Acta. 2002;460:209–218. doi: 10.1016/S0003-2670(02)00240-4. [DOI] [Google Scholar]
- 166.Luo B., Groenke K., Takors R., Wandrey C., Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J Chromatogr A. 2007;1147:153–164. doi: 10.1016/j.chroma.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 167.Koning W de, Dam K van. A method for the determination of changes of glycolytic metabolites in yeast on a subsecond time scale using extraction at neutral pH. Anal Biochem. 1992;204:118–123. doi: 10.1016/0003-2697(92)90149-2. [DOI] [PubMed] [Google Scholar]
- 168.Wittmann C., Krömer J.O., Kiefer P., Binz T., Heinzle E. Impact of the cold shock phenomenon on quantification of intracellular metabolites in bacteria. Anal Biochem. 2004;327:135–139. doi: 10.1016/j.ab.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 169.Bolten C.J., Kiefer P., Letisse F., Portais J.C., Wittmann C. Sampling for metabolome analysis of microorganisms. Anal Chem. 2007;79:3843–3849. doi: 10.1021/AC0623888. [DOI] [PubMed] [Google Scholar]
- 170.Dietmair S., Timmins N.E., Gray P.P., Nielsen L.K., Krömer J.O. Towards quantitative metabolomics of mammalian cells: development of a metabolite extraction protocol. Anal Biochem. 2010;404:155–164. doi: 10.1016/j.ab.2010.04.031. [DOI] [PubMed] [Google Scholar]
- 171.Antoniewicz M.R. Dynamic metabolic flux analysis — tools for probing transient states of metabolic networks. Curr Opin Biotechnol. 2013;24:973–978. doi: 10.1016/J.COPBIO.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 172.Sah S., Ma X., Botros A., Gaul D.A., Yun S.R., Park E.Y., et al. Space- and time-resolved metabolomics of a high-grade serous ovarian cancer mouse model. Cancers 2022. 2022;14:2262. doi: 10.3390/CANCERS14092262. 14, Page 2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Fessenden M. Metabolomics: small molecules, single cells. Natalia. 2016;540:153–155. doi: 10.1038/540153a. 2016 5407631. [DOI] [PubMed] [Google Scholar]
- 174.Papagiannakis A., Niebel B., Wit E.C., Heinemann M. Autonomous metabolic oscillations robustly gate the early and late cell cycle. Mol Cell. 2017;65:285–295. doi: 10.1016/J.MOLCEL.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 175.Giedt R.J., Pathania D., Carlson J.C.T., McFarland P.J., del Castillo A.F., Juric D., et al. Single-cell barcode analysis provides a rapid readout of cellular signaling pathways in clinical specimens. Nat Commun. 2018;9:1–10. doi: 10.1038/s41467-018-07002-6. 2018 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lin J.R., Izar B., Wang S., Yapp C., Mei S., Shah P.M., et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. Elife. 2018;7 doi: 10.7554/ELIFE.31657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zrazhevskiy P., True L.D., Gao X. Multicolor multicycle molecular profiling with quantum dots for single-cell analysis. Nat Protoc. 2013;8:1852–1869. doi: 10.1038/nprot.2013.112. 2013 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Slavov N. Unpicking the proteome in single cells. Science. 2020;367:512–513. doi: 10.1126/science.aaz6695. 80- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Levy E., Slavov N. Single cell protein analysis for systems biology. Essays Biochem. 2018;62:595–605. doi: 10.1042/EBC20180014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nair N., Mei H.E., Chen S.Y., Hale M., Nolan G.P., Maecker H.T., et al. Mass cytometry as a platform for the discovery of cellular biomarkers to guide effective rheumatic disease therapy. Arthritis Res Ther. 2015;17 doi: 10.1186/S13075-015-0644-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Spitzer M.H., Nolan G.P. Mass cytometry: single cells, many features. Cell. 2016;165:780–791. doi: 10.1016/J.CELL.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Virant-Klun I., Leicht S., Hughes C., Krijgsveld J. Identification of maturation-specific proteins by single-cell proteomics of human oocytes. Mol Cell Proteomics. 2016;15:2616–2627. doi: 10.1074/MCP.M115.056887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sun L., Dubiak K.M., Peuchen E.H., Zhang Z., Zhu G., Huber P.W., et al. Single cell proteomics using frog (Xenopus laevis) blastomeres isolated from early stage embryos, which form a geometric progression in protein content. Anal Chem. 2016;88:6653–6657. doi: 10.1021/acs.analchem.6b01921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lombard-Banek C., Reddy S., Moody S.A., Nemes P. Label-free quantification of proteins in single embryonic cells with neural fate in the cleavage-stage frog (Xenopus laevis) embryo using capillary electrophoresis electrospray ionization high-resolution mass spectrometry (CE-ESI-HRMS) Mol Cell Proteomics. 2016;15:2756–2768. doi: 10.1074/MCP.M115.057760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Specht H., Harmange G., Perlman D.H., Emmott E., Niziolek Z., Budnik B., et al. Automated sample preparation for high-throughput single-cell proteomics. bioRxiv. 2018 doi: 10.1101/399774. [DOI] [Google Scholar]
- 186.Budnik B., Levy E., Harmange G., Slavov N. Mass-spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. bioRxiv. 2018 doi: 10.1101/102681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Budnik B., Levy E., Harmange G., Slavov N. SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol. 2018;19:1–12. doi: 10.1186/S13059-018-1547-5/FIGURES/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Specht H., Emmott E., Petelski A.A., Huffman R.G., Perlman D.H., Serra M., et al. Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity using SCoPE2. Genome Biol. 2021;22:1–27. doi: 10.1186/S13059-021-02267-5/TABLES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Specht H., Emmott E., Petelski A.A., Huffman R.G., Perlman D.H., Serra M., et al. Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity. bioRxiv. 2020 doi: 10.1101/665307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Urban P.L., Jefimovs K., Amantonico A., Fagerer S.R., Schmid T., Mädler S., et al. High-density micro-arrays for mass spectrometry. Lab Chip. 2010;10:3206–3209. doi: 10.1039/C0LC00211A. [DOI] [PubMed] [Google Scholar]
- 191.Zenobi R. Single-cell metabolomics: analytical and biological perspectives. Science. 2013:342. doi: 10.1126/science.1243259. 80- [DOI] [PubMed] [Google Scholar]
- 192.Zhang L., Vertes A. Single-cell mass spectrometry approaches to explore cellular heterogeneity. Angew Chem Int Ed. 2018;57:4466–4477. doi: 10.1002/ANIE.201709719. [DOI] [PubMed] [Google Scholar]
- 193.Duncan K.D., Fyrestam J., Lanekoff I. Advances in mass spectrometry based single-cell metabolomics. Analyst. 2019;144:782–793. doi: 10.1039/C8AN01581C. [DOI] [PubMed] [Google Scholar]
- 194.Lanekoff I., Sharma V.V., Marques C. Single-cell metabolomics: where are we and where are we going? Curr Opin Biotechnol. 2022;75 doi: 10.1016/J.COPBIO.2022.102693. [DOI] [PubMed] [Google Scholar]
- 195.Metwaly A., Haller D. Multi-omics in IBD biomarker discovery: the missing links. Nat Rev Gastroenterol Hepatol. 2019;16:587–588. doi: 10.1038/S41575-019-0188-9. [DOI] [PubMed] [Google Scholar]
- 196.Olivier M., Asmis R., Hawkins G.A., Howard T.D., Cox L.A. The need for multi-omics biomarker signatures in precision medicine. Int J Mol Sci 2019. 2019;20:4781. doi: 10.3390/IJMS20194781. 20, Page 4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ang M.Y., Low T.Y., Lee P.Y., Wan Mohamad, Nazarie W.F., Guryev V., Jamal R. Proteogenomics: from next-generation sequencing (NGS) and mass spectrometry-based proteomics to precision medicine. Clin Chim Acta. 2019;498:38–46. doi: 10.1016/J.CCA.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 198.Sheynkman G.M., Shortreed M.R., Cesnik A.J., Smith L.M. Proteogenomics: integrating next-generation sequencing and mass spectrometry to characterize human proteomic variation. Annu Rev Anal Chem. 2016;9:521–545. doi: 10.1146/ANNUREV-ANCHEM-071015-041722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Weinhold B. Epigenetics: the science of change. Environ Health Perspect. 2006;114:A160. doi: 10.1289/EHP.114-A160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Subramanian I., Verma S., Kumar S., Jere A., Anamika K. Multi-omics data integration, interpretation, and its application. Bioinf Biol Insights. 2020;14 doi: 10.1177/1177932219899051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Colquitt R.B., Colquhoun D.A., Thiele R.H. In silico modelling of physiologic systems. Best Pract Res Clin Anaesthesiol. 2011;25:499–510. doi: 10.1016/J.BPA.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 202.Teusink B., Passarge J., Reijenga C.A., Esgalhado E., Van Der Weijden C.C., Schepper M., et al. Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur J Biochem. 2000;267:5313–5329. doi: 10.1046/J.1432-1327.2000.01527.X. [DOI] [PubMed] [Google Scholar]
- 203.Phair R.D., Misteli T. Kinetic modelling approaches to in vivo imaging. Nat Rev Mol Cell Biol. 2001;2:898–907. doi: 10.1038/35103000. 2001 212. [DOI] [PubMed] [Google Scholar]
- 204.Papoutsakis E.T. Equations and calculations for fermentations of butyric acid bacteria. Biotechnol Bioeng. 2000;67:813–826. doi: 10.1002/(sici)1097-0290. 20000320. 67:6<813::aid-bit17>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 205.Price N.D., Reed J.L., Palsson B. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat Rev Microbiol. 2004;2:886–897. doi: 10.1038/nrmicro1023. [DOI] [PubMed] [Google Scholar]
- 206.Wiechert W. 13C metabolic flux analysis. Metab Eng. 2001;3:195–206. doi: 10.1006/mben.2001.0187. [DOI] [PubMed] [Google Scholar]
- 207.Visser D., Van Der Heijden R., Mauch K., Reuss M., Heijnen S. Tendency modeling: a new approach to obtain simplified kinetic models of metabolism applied to Saccharomyces cerevisiae. Metab Eng. 2000;2:252–275. doi: 10.1006/mben.2000.0150. [DOI] [PubMed] [Google Scholar]
- 208.Neves A.R., Ventura R., Mansour N., Shearman C., Gasson M.J., Maycock C., et al. Is the glycolytic flux in Lactococcus lactis primarily controlled by the redox charge? Kinetics of NAD+ and NADH pools determined in vivo by 13C NMR. J Biol Chem. 2002;277:28088–28098. doi: 10.1074/jbc.M202573200. [DOI] [PubMed] [Google Scholar]
- 209.Curto R., Voit E.O., Sorribas A., Cascante M. Validation and steady-state analysis of a power-law model of purine metabolism in man. Biochem J. 1997;324:761–775. doi: 10.1042/bj3240761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Funahashi A., Matsuoka Y., Jouraku A., Morohashi M., Kikuchi N., Kitano H. CellDesigner 3.5: a versatile modeling tool for biochemical networks. Proc IEEE. 2008;96:1254–1265. doi: 10.1109/JPROC.2008.925458. [DOI] [Google Scholar]
- 211.Hucka M., Finney A., Sauro H.M., Bolouri H., Doyle J.C., Kitano H., et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 212.Moodie S., Le Novère N., Demir E., Mi H., Villéger A. Systems biology graphical notation: process description language level 1 version 1.3. J Integr Bioinform. 2015;12:263. doi: 10.2390/biecoll-jib-2015-263. [DOI] [PubMed] [Google Scholar]
- 213.Van Hemert J.L., Dickerson J.A., Valencia A. PathwayAccess: CellDesigner plugins for pathway databases. Bioinformatics. 2011;27:2345–2346. doi: 10.1093/bioinformatics/btq423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Oki N.O., Farcal L., Abdelaziz A., Florean O., Doktorova T.Y., Exner T., et al. Integrated analysis of in vitro data and the adverse outcome pathway framework for prioritization and regulatory applications: an exploratory case study using publicly available data on piperonyl butoxide and liver models. Toxicol Vitro. 2019;54:23–32. doi: 10.1016/j.tiv.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 215.Marín-Hernández A., Gallardo-Pérez J.C., Rodríguez-Enríquez S., Encalada R., Moreno-Sánchez R., Saavedra E. Modeling cancer glycolysis. Biochim Biophys Acta Bioenerg. 2011;1807:755–767. doi: 10.1016/J.BBABIO.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 216.Bruck J., Liebermeister W., Klipp E. Exploring the effect of variable enzyme concentrations in a kinetic model of yeast glycolysis. Genome Inform. 2008;20:1–14. doi: 10.1142/9781848163003_0001. [DOI] [PubMed] [Google Scholar]
- 217.Kanehisa M., Sato Y., Kawashima M. KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. 2022;31:47–53. doi: 10.1002/PRO.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Olivier B.G., Snoep J.L. Web-based kinetic modelling using JWS Online. Bioinformatics. 2004;20:2143–2144. doi: 10.1093/BIOINFORMATICS/BTH200. [DOI] [PubMed] [Google Scholar]
- 219.Chelliah V., Juty N., Ajmera I., Ali R., Dumousseau M., Glont M., et al. BioModels: ten-year anniversary. Nucleic Acids Res. 2015;43:D542–D548. doi: 10.1093/NAR/GKU1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Costa R.S., Veríssimo A., Vinga S. KiMoSys: a web-based repository of experimental data for KInetic MOdels of biological SYStems. BMC Syst Biol. 2014;8 doi: 10.1186/S12918-014-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Placzek S., Schomburg I., Chang A., Jeske L., Ulbrich M., Tillack J., et al. BRENDA in 2017: new perspectives and new tools in BRENDA. Nucleic Acids Res. 2017;45:D380–D388. doi: 10.1093/NAR/GKW952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Pang Z., Chong J., Zhou G., De Lima Morais D.A., Chang L., Barrette M., et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49:W388–W396. doi: 10.1093/NAR/GKAB382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kanehisa M., Sato Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020;29:28–35. doi: 10.1002/pro.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Berndt N., Egners A., Mastrobuoni G., Vvedenskaya O., Fragoulis A., Dugourd A., et al. Kinetic modelling of quantitative proteome data predicts metabolic reprogramming of liver cancer. Br J Cancer. 2019 1222 2019;122:233–244. doi: 10.1038/s41416-019-0659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kelly R.A., Leedale J., Harrell A., Beard D.A., Randle L.E., Chadwick A.E., et al. Modelling the impact of changes in the extracellular environment on the cytosolic free NAD+/NADH ratio during cell culture. PLoS One. 2018;13 doi: 10.1371/JOURNAL.PONE.0207803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Schoeberl B., Eichler-Jonsson C., Gilles E.D., Muüller G. Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat Biotechnol. 2002;20:370–375. doi: 10.1038/NBT0402-370. [DOI] [PubMed] [Google Scholar]
- 227.Markevich N.I., Hoek J.B., Kholodenko B.N. Signaling switches and bistability arising from multisite phosphorylation in protein kinase cascades. J Cell Biol. 2004;164:353–359. doi: 10.1083/JCB.200308060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Huang C.Y.F., Ferrell J.E. Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci U S A. 1996;93:10078–10083. doi: 10.1073/PNAS.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Hoffmann A., Levchenko A., Scott M.L., Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/SCIENCE.1071914. [DOI] [PubMed] [Google Scholar]
- 230.Bhalla U.S., Ram P.T., Iyengar R. MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science. 2002;297:1018–1023. doi: 10.1126/SCIENCE.1068873. [DOI] [PubMed] [Google Scholar]
- 231.Aldridge B.B., Burke J.M., Lauffenburger D.A., Sorger P.K. Physicochemical modelling of cell signalling pathways. Nat Cell Biol. 2006;8:1195–1203. doi: 10.1038/NCB1497. [DOI] [PubMed] [Google Scholar]
- 232.Maier K., Hofmann U., Reuss M., Mauch K. Dynamics and control of the central carbon metabolism in hepatoma cells. BMC Syst Biol. 2010;4:1–28. doi: 10.1186/1752-0509-4-54/TABLES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Yarmush G., Santos L., Yarmush J., Koundinyan S., Saleem M., Nativ N.I., et al. Metabolic flux distribution during defatting of steatotic human hepatoma (HepG2) cells. Metabolism 2016. 2016;6:1. doi: 10.3390/METABO6010001. Vol 6, Page 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.De Gelder J., De Gussem K., Vandenabeele P., Moens L. Reference database of Raman spectra of biological molecules. J Raman Spectrosc. 2007;38:1133–1147. doi: 10.1002/JRS.1734. [DOI] [Google Scholar]
- 235.Bonnier F., Knief P., Lim B., Meade A.D., Dorney J., Bhattacharya K., et al. Imaging live cells grown on a three dimensional collagen matrix using Raman microspectroscopy. Analyst. 2010;135:3169–3177. doi: 10.1039/c0an00539h. [DOI] [PubMed] [Google Scholar]
- 236.Boydston-White S., Romeo M., Chernenko T., Regina A., Miljković M., Diem M. Cell-cycle-dependent variations in FTIR micro-spectra of single proliferating HeLa cells: principal component and artificial neural network analysis. Biochim Biophys Acta Biomembr. 2006;1758:908–914. doi: 10.1016/j.bbamem.2006.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Matthäus C., Boydston-White S., Miljković M., Romeo M., Diem M. Raman and infrared microspectral imaging of mitotic cells. Appl Spectrosc. 2006;60:1–8. doi: 10.1366/000370206775382758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Short K.W., Carpenter S., Freyer J.P., Mourant J.R. Raman spectroscopy detects biochemical changes due to proliferation in mammalian cell cultures. Biophys J. 2005;88:4274–4288. doi: 10.1529/biophysj.103.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ami D., Neri T., Natalello A., Mereghetti P., Doglia S.M., Zanoni M., et al. Embryonic stem cell differentiation studied by FT-IR spectroscopy. Biochim Biophys Acta Mol Cell Res. 2008;1783:98–106. doi: 10.1016/j.bbamcr.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 240.Notingher I., Bisson I., Bishop A.E., Randle W.L., Polak J.M.P., Hench L.L. In situ spectral monitoring of mRNA translation in embryonic stem cells during differentiation in vitro. Anal Chem. 2004;76:3185–3193. doi: 10.1021/ac0498720. [DOI] [PubMed] [Google Scholar]
- 241.Pavillon N., Hobro A.J., Akira S., Smith N.I. Noninvasive detection of macrophage activation with single-cell resolution through machine learning. Proc Natl Acad Sci U S A. 2018;115:E2676–E2685. doi: 10.1073/pnas.1711872115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Meade A.D., Lyng F.M., Knief P., Byrne H.J. Growth substrate induced functional changes elucidated by FTIR and Raman spectroscopy in in-vitro cultured human keratinocytes. Anal Bioanal Chem. 2007;387:1717–1728. doi: 10.1007/s00216-006-0876-5. [DOI] [PubMed] [Google Scholar]
- 243.Gasparri F., Muzio M. Monitoring of apoptosis of HL60 cells by fourier-transform infrared spectroscopy. Biochem J. 2003;369:239–248. doi: 10.1042/BJ20021021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Liu K.Z., Jia L., Kelsey S.M., Newland A.C., Mantsch H.H. Quantitative determination of apoptosis on leukemia cells by infrared spectroscopy. Apoptosis. 2001;6:269–278. doi: 10.1023/A:1011383408381. [DOI] [PubMed] [Google Scholar]
- 245.Gargotti M., Efeoglu E., Byrne H.J., Casey A. Raman spectroscopy detects biochemical changes due to different cell culture environments in live cells in vitro. Anal Bioanal Chem. 2018;410:7537–7550. doi: 10.1007/s00216-018-1371-5. [DOI] [PubMed] [Google Scholar]
- 246.Bonnier F., Knief P., Meade A.D., Dorney J., Bhattacharya K., Lyng F.M., et al. Collagen matrices as an improved model for in vitro study of live cells using Raman microspectroscopy. Clin. Biomed. Spectrosc. Imaging II, 24th May. 2011;8087 doi: 10.1117/12.889872. 80870F-80870F – 10. [DOI] [Google Scholar]
- 247.Meade A.D., Clarke C., Draux F., Sockalingum G.D., Manfait M., Lyng F.M., et al. Studies of chemical fixation effects in human cell lines using Raman microspectroscopy. Anal Bioanal Chem. 2010;396:1781–1791. doi: 10.1007/S00216-009-3411-7/TABLES/2. [DOI] [PubMed] [Google Scholar]
- 248.Dorney J., Bonnier F., Garcia A., Casey A., Chambers G., Byrne H.J. Identifying and localizing intracellular nanoparticles using Raman spectroscopy. Analyst. 2012;137:1111–1119. doi: 10.1039/c2an15977e. [DOI] [PubMed] [Google Scholar]
- 249.Matthäus C., Chernenko T., Newmark J.A., Warner C.M., Diem M. Label-free detection of mitochondrial distribution in cells by nonresonant Raman microspectroscopy. Biophys J. 2007;93:668–673. doi: 10.1529/biophysj.106.102061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.van Manen H.-J., Kraan Y.M., Roos D., Otto C. Single-cell Raman and fluorescence microscopy reveal the association of lipid bodies with phagosomes in leukocytes. Proc Natl Acad Sci U S A. 2005;102:10159–10164. doi: 10.1073/pnas.0502746102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Farhane Z., Bonnier F., Casey A., Byrne H.J. Raman micro spectroscopy for in vitro drug screening: subcellular localisation and interactions of doxorubicin. Analyst. 2015;140:4212–4223. doi: 10.1039/c5an00256g. [DOI] [PubMed] [Google Scholar]
- 252.Theophilou G., Paraskevaidi M., Lima K.M., Kyrgiou M., Martin-Hirsch P.L., Martin F.L. Extracting biomarkers of commitment to cancer development: potential role of vibrational spectroscopy in systems biology. Expert Rev Mol Diagn. 2015;15:693–713. doi: 10.1586/14737159.2015.1028372. [DOI] [PubMed] [Google Scholar]
- 253.Paraskevaidi M., Matthew B.J., Holly B.J., Hugh B.J., Thulya C.P.V., Loren C., et al. Clinical applications of infrared and Raman spectroscopy in the fields of cancer and infectious diseases. Appl Spectrosc Rev. 2021;56:804–868. doi: 10.1080/05704928.2021.1946076. [DOI] [Google Scholar]
- 254.Szafraniec E., Majzner K., Farhane Z., Byrne H.J., Lukawska M., Oszczapowicz I., et al. Spectroscopic studies of anthracyclines: structural characterization and in vitro tracking. Spectrochim Acta Part A Mol Biomol Spectrosc. 2016;169:152–160. doi: 10.1016/j.saa.2016.06.035. [DOI] [PubMed] [Google Scholar]
- 255.Farhane Z., Bonnier F., Byrne H.J. Monitoring doxorubicin cellular uptake and trafficking using in vitro Raman microspectroscopy: short and long time exposure effects on lung cancer cell lines. Anal Bioanal Chem. 2017;409:1333–1346. doi: 10.1007/s00216-016-0065-0. [DOI] [PubMed] [Google Scholar]
- 256.Farhane Z., Bonnier F., Byrne H.J. An in vitro study of the interaction of the chemotherapeutic drug Actinomycin D with lung cancer cell lines using Raman micro-spectroscopy. J Biophot. 2018;11 doi: 10.1002/jbio.201700112. [DOI] [PubMed] [Google Scholar]
- 257.Farhane Z., Bonnier F., Howe O., Casey A., Byrne H.J. Doxorubicin kinetics and effects on lung cancer cell lines using in vitro Raman micro-spectroscopy: binding signatures, drug resistance and DNA repair. J Biophot. 2018;11 doi: 10.1002/jbio.201700060. [DOI] [PubMed] [Google Scholar]
- 258.Efeoglu E., Keating M., McIntyre J., Casey A., Byrne H.J. Determination of nanoparticle localisation within subcellular organelles in vitro using Raman spectroscopy. Anal Methods. 2015;7:10000–10017. doi: 10.1039/c5ay02661j. [DOI] [Google Scholar]
- 259.Efeoglu E., Casey A., Byrne H.J. In vitro monitoring of time and dose dependent cytotoxicity of aminated nanoparticles using Raman spectroscopy. Analyst. 2016;141:5417–5431. doi: 10.1039/c6an01199c. [DOI] [PubMed] [Google Scholar]
- 260.Efeoglu E., Casey A., Byrne H.J. Determination of spectral markers of cytotoxicity and genotoxicity using in vitro Raman microspectroscopy: cellular responses to polyamidoamine dendrimer exposure. Analyst. 2017;142:3848–3856. doi: 10.1039/c7an00969k. [DOI] [PubMed] [Google Scholar]
- 261.Efeoglu E., Maher M.A., Casey A., Byrne H.J. Toxicological assessment of nanomaterials: the role of in vitro Raman microspectroscopic analysis. Anal Bioanal Chem. 2018;410:1631–1646. doi: 10.1007/s00216-017-0812-x. [DOI] [PubMed] [Google Scholar]
- 262.Xu J., Zhu D., Ibrahim A.D., Allen C.C.R., Gibson C.M., Fowler P.W., et al. Raman deuterium isotope probing reveals microbial metabolism at the single-cell level. Anal Chem. 2017;89:13305–13312. doi: 10.1021/acs.analchem.7b03461. [DOI] [PubMed] [Google Scholar]
- 263.Zhang L., Shi L., Shen Y., Miao Y., Wei M., Qian N., et al. Spectral tracing of deuterium for imaging glucose metabolism. Nat Biomed Eng. 2019;3:402. doi: 10.1038/S41551-019-0393-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Noothalapati H., Shigeto S. Exploring metabolic pathways in vivo by a combined approach of mixed stable isotope-labeled Raman microspectroscopy and multivariate curve resolution analysis. Anal Chem. 2014;86:7828–7834. doi: 10.1021/AC501735C/SUPPL_FILE/AC501735C_SI_001.PDF. [DOI] [PubMed] [Google Scholar]
- 265.Li M., Huang W.E., Gibson C.M., Fowler P.W., Jousset A. Stable isotope probing and Raman spectroscopy for monitoring carbon flow in a food chain and revealing metabolic pathway. Anal Chem. 2013;85:1642–1649. doi: 10.1021/AC302910X/SUPPL_FILE/AC302910X_SI_003.GIF. [DOI] [PubMed] [Google Scholar]
- 266.Noothalapati Venkata H.N., Shigeto S. Stable isotope-labeled Raman imaging reveals dynamic proteome localization to lipid droplets in single fission yeast cells. Chem Biol. 2012;19:1373–1380. doi: 10.1016/J.CHEMBIOL.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 267.Weber F., Zaliznyak T., Edgcomb V.P., Taylor G.T. Using stable isotope probing and Raman microspectroscopy to measure growth rates of heterotrophic bacteria. Appl Environ Microbiol. 2021;87 doi: 10.1128/AEM.01460-21/SUPPL_FILE/AEM.01460-21-S0001.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ryan S.M., Frías J.M., Wang X., Sayers C.T., Haddleton D.M., Brayden D.J. PK/PD modelling of comb-shaped PEGylated salmon calcitonin conjugates of differing molecular weights. J Contr Release. 2011;149:126–132. doi: 10.1016/j.jconrel.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 269.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. 80- [DOI] [PubMed] [Google Scholar]
- 270.Steuer R., Junker B.H. Computational models of metabolism: stability and regulation in metabolic networks. Adv Chem Phys. 2008;142:105. doi: 10.1002/9780470475935.ch3. 251. [DOI] [Google Scholar]
- 271.Mukherjee R., Verma T., Nandi D., Umapathy S. Understanding the effects of culture conditions in bacterial growth: a biochemical perspective using Raman microscopy. J Biophot. 2020;13 doi: 10.1002/JBIO.201900233. [DOI] [PubMed] [Google Scholar]
- 272.Mair T., Zimányi L., Khoroshyy P., Müller A., Müller S.C. Analysis of the oscillatory kinetics of glycolytic intermediates in a yeast extract by FT-IR spectroscopy. Biosystems. 2006;83:188–194. doi: 10.1016/J.BIOSYSTEMS.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 273.Poonprasartporn A., Chan K.L.A. Live-cell ATR-FTIR spectroscopy as a novel bioanalytical tool for cell glucose metabolism research. Biochim Biophys Acta Mol Cell Res. 2021;1868 doi: 10.1016/j.bbamcr.2021.119024. [DOI] [PubMed] [Google Scholar]
- 274.Poonprasartporn A., Chan K.L.A. Label-free study of intracellular glycogen level in metformin and resveratrol-treated insulin-resistant HepG2 by live-cell FTIR spectroscopy. Biosens Bioelectron. 2022;212 doi: 10.1016/J.BIOS.2022.114416. [DOI] [PubMed] [Google Scholar]
- 275.Pleitez M.A., Khan A.A., Soldà A., Chmyrov A., Reber J., Gasparin F., et al. Label-free metabolic imaging by mid-infrared optoacoustic microscopy in living cells. Nat Biotechnol. 2020;38:293–296. doi: 10.1038/s41587-019-0359-9. [DOI] [PubMed] [Google Scholar]
- 276.Cheng J.X., Xie X.S. Vibrational spectroscopic imaging of living systems: an emerging platform for biology and medicine. Science. 2015:350. doi: 10.1126/SCIENCE.AAA8870. 80- [DOI] [PubMed] [Google Scholar]
- 277.Liao C.S., Huang K.C., Hong W., Chen J., Caroline K., Eakins G., et al. Stimulated Raman spectroscopic imaging by microsecond delay-line tuning. Opt. InfoBase Conf. Pap. 2014 doi: 10.1364/ACPC.2016.ATH2K.5. [DOI] [Google Scholar]
- 278.Hong W., Liao C.-S., Zhao H., Younis W., Zhang Y., Seleem M.N., et al. In situ detection of a single bacterium in complex environment by hyperspectral CARS imaging. Wiley Online Libr. 2016;1:513–517. doi: 10.1002/slct.201600166. [DOI] [Google Scholar]
- 279.Zhang C., Cheng J.X. Perspective: coherent Raman scattering microscopy, the future is bright. APL Photonics. 2018;3 doi: 10.1063/1.5040101. [DOI] [Google Scholar]
- 280.Minamikawa T., Murakami Y., Matsumura N., Niioka H., Fukushima S., Araki T., et al. Photo-induced cell damage analysis for single-and multifocus coherent anti-Stokes Raman scattering microscopy. J Spectrosc. 2017;2017 doi: 10.1155/2017/5725340. [DOI] [Google Scholar]
- 281.Jaumot J., Gargallo R., De Juan A., Tauler R. A graphical user-friendly interface for MCR-ALS: a new tool for multivariate curve resolution in MATLAB. Chemometr Intell Lab Syst. 2005;76:101–110. doi: 10.1016/j.chemolab.2004.12.007. [DOI] [Google Scholar]
- 282.Singular Value decomposition - MATLAB & simulink - MathWorks United Kingdom n.d. https://uk.mathworks.com/help/symbolic/singular-value-decomposition.html.
- 283.Maleš P., Brkljača Z., Crnolatac I., Bakarić D. Application of MCR-ALS with EFA on FT-IR spectra of lipid bilayers in the assessment of phase transition temperatures: potential for discernment of coupled events. Colloids Surf B Biointerfaces. 2021;201 doi: 10.1016/j.colsurfb.2021.111645. [DOI] [PubMed] [Google Scholar]
- 284.Perez-Guaita D., Quintas G., Farhane Z., Tauler R., Byrne H.J. Data mining Raman microspectroscopic responses of cells to drugs in vitro using multivariate curve resolution-alternating least squares. Talanta. 2020;208 doi: 10.1016/j.talanta.2019.120386. 0–21. [DOI] [PubMed] [Google Scholar]
- 285.Perez-Guaita D., Chrabaszcz K., Malek K., Byrne H.J. Multimodal vibrational studies of drug uptake in vitro: is the whole greater than the sum of their parts? J Biophot. 2020;13 doi: 10.1002/jbio.202000264. [DOI] [PubMed] [Google Scholar]
- 286.Vernooij R.R., Joshi T., Horbury M.D., Graham B., Izgorodina E.I., Stavros V.G., et al. Spectroscopic studies on photoinduced reactions of the anticancer prodrug, trans,trans,trans-[Pt(N3)2(OH)2(py)2] Chem Eur J. 2018;24:5790–5803. doi: 10.1002/chem.201705349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Zhang C., Zhang D., Cheng J.X. vol. 17. 2015. (Coherent Raman scattering microscopy in biology and medicine). Https://DoiOrg/101146/Annurev-Bioeng-071114-040554. 415–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Dazzi A., Policar C. AFM-IR: photothermal infrared nanospectroscopy: application to cellular imaging. Biointerface charact. By adv. IR spectrosc. Elsevier. 2011:245–278. doi: 10.1016/B978-0-444-53558-0.00009-6. [DOI] [Google Scholar]
- 289.Mathurin J., Deniset-Besseau A., Bazin D., Dartois E., Wagner M., Dazzi A. Photothermal AFM-IR spectroscopy and imaging: status, challenges, and trends. J Appl Phys. 2022;131 doi: 10.1063/5.0063902. [DOI] [Google Scholar]
- 290.Wissmeyer G., Pleitez M.A., Rosenthal A., Ntziachristos V. Looking at sound: optoacoustics with all-optical ultrasound detection. Light Sci Appl. 2018;7 doi: 10.1038/S41377-018-0036-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Kansiz M., Prater C., Dillon E., Lo M., Anderson J., Marcott C., et al. Optical photothermal infrared microspectroscopy with simultaneous Raman – a new non-contact failure analysis technique for identification of <10 μm organic contamination in the hard drive and other electronics industries. Micros Today. 2020;28:26. doi: 10.1017/S1551929520000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.