Abstract
Malassezia is the most abundant genus in the fungal microflora found on human skin, and it is associated with various skin diseases. Among the 18 different species of Malassezia that have been identified to date, M. restricta and M. globosa are the most predominant fungal species found on human skin. Several studies have suggested a possible link between Malassezia and skin disorders. However, our knowledge on the physiology and pathogenesis of Malassezia in human body is still limited. Malassezia is unable to synthesize fatty acids; hence, it uptakes external fatty acids as a nutrient source for survival, a characteristic compensated by the secretion of lipases and degradation of sebum to produce and uptake external fatty acids. Although it has been reported that the activity of secreted lipases may contribute to pathogenesis of Malassezia, majority of the data were indirect evidences; therefore, enzymes’ role in the pathogenesis of Malassezia infections is still largely unknown. This review focuses on the recent advances on Malassezia in the context of an emerging interest for lipases and summarizes the existing knowledge on Malassezia, diseases associated with the fungus, and the role of the reported lipases in its physiology and pathogenesis.
Keywords: Lipase, Malassezia, M. globose, M. restricta, skin microflora
Introduction
Malassezia, an abundant genus of the fungal microflora found on human skin, is recognized as an opportunistic fungus owing to its association with various skin diseases [1-3]. The phylum, subphylum, class, order, and family of Malassezia are Basidiomycota, Ustilaginomycotina, Malasseziomycetes, Malasseziales, and Malasseziaceae, respectively [4]. Fourteen well-established species of Malassezia and four additional species have been identified to date. Out of the 18 species, 10 species (M. restricta, M. globosa, M. arunalokei, M. sympodialis, M. dermatis, M. slooffiae, M. furfur, M. obtusa, M. japonica, and M. yamatoensis) were isolated mainly from human skin, whereas the others are normally isolated from animal skin [5]. In the case of zoophilic species, M. pachydermatis is one of the most dominant fungal species isolated from companion animals, such as dogs [6]. M. restricta and M. globosa have been identified as the most predominant fungal species on human skin by several studies involving early culture-based, targeted polymerase chain reaction (PCR)-based, or recent skin mycobiome analysis-based studies [1, 7-9].
Malassezia seems to have evolved in niche specific manner and is well-adapted to the skin environment, wherein carbohydrates are restricted but lipids are abundant, a condition that favors the lipophilic characteristics of the fungus. Malassezia possesses multiple genes encoding lipases; hence, a number of studies have suggested that the secreted lipases might play an important role in not only the survival but also in the pathogenesis of the fungus in the hosts [10-12]. In this review article, we focused on the recent advances on Malassezia in the context of an emerging interest for lipases. We present the background information for the skin diseases associated with Malassezia and summarize the current knowledge on the fungal lipases.
Malassezia-Associated Skin Diseases in Humans
Numerous studies that analyzed the fungal communities on human skin suggested Malassezia as the predominant fugal genus. Mycobiome analyses of samples collected from 14 different body sites of 10 healthy adults revealed that M. restricta and M. globosa were commonly isolated from the glabella, external auditory canal, and retroauricular crease; and the occiput, back, and inguinal crease; respectively [1]. Comparison of the fungal communities inhabiting skin between healthy adults and children revealed that M. restricta dominates at the sebaceous sites in adults rather than in children [9]. Furthermore, it was shown that the occiput, back, and manubrium exhibited M. globosa predominance, whereas M. restricta was predominant in the external auditory canal, retroauricular crease, and forehead [9]. Since Malassezia lives on the skin surface, it is usually associated with numerous skin diseases, such as seborrheic dermatitis/dandruff, atopic dermatitis, and pityriasis versicolor. Although various studies suggested that skin disorders and Malassezia are linked, our knowledge regarding the physiology and pathogenesis of Malassezia in skin diseases is still largely limited.
Seborrheic dermatitis and dandruff are the most common skin diseases worldwide. Seborrheic dermatitis, a chronic and inflammatory skin disease that occurs on the areas with a rich supply of sebaceous glands, including face, scalp, chest, and upper trunk, is characterized by scaling and redness. Dandruff is a mild form of seborrheic dermatitis and its pathological skin condition can be defined as abnormal scalp flakes accompanied by itching without inflammation [13]. The causal relationship of seborrheic dermatitis and dandruff with Malassezia has been shown: (1) effective treatment of the disorder following the use of antifungal drugs, (2) improvement of symptoms accompanied by quantitative reduction of Malassezia, and (3) recurrence of dandruff induced by Malassezia metabolite [14-17]. Besides, a number of large-scale sequencing analyses showed positive correlations between seborrheic dermatitis, dandruff, and Malassezia suggesting the fungus to be a potential disease causing agent.
Among the different species of Malassezia, M. restricta may play a critical role in the pathogenesis of seborrheic dermatitis and dandruff. Analysis of the fungal community in the scalp of the French population showed M. restricta as the most abundant fungal species on scalps with dandruff and scalps that were healthy. However, M. restricta was more abundant in the scalps with dandruff than that in healthy scalps and in the dandruff rich areas than in the non-dandruff areas [18]. Similar results were observed for the mycobiome analysis of the scalp of Chinese and Koreans subjects with and without dandruff, thereby verifying the disequilibrium of a balanced microbiome and the correlation between increased M. restricta population and dandruff [2, 19, 20]. A recent mycobiome and metagenome analyses involving the Indian female population revealed a significantly increased predominance of M. restricta over M. globosa in the swab samples from the scalps with dandruff compared to that in the healthy scalp, thus reiterating the critical role of M. restricta in dandruff [3]. Metagenome analysis showed enrichment of the N-glycan biosynthesis pathway of M. restricta in the scalp with dandruff compared to that in the healthy scalp. Since glycoprotein of the fungal cells is involved with adhesion to the host tissue [21], this pathway may be related to colonization of M. restricta on the scalp surface, thus further demonstrating the critical role of the fungus in dandruff [3]. Moreover, a mycobiome study on healthy scalps and scalps with dandruff revealed two different operational taxonomic unit levels of M. restricta, suggesting that distinct subspecies within the same species might play a different role in the disease [22].
Atopic dermatitis (AD) is a complex inflammatory disease caused by abnormal skin barrier function and immune response [23]. Owing to the defective epidermal barrier, skin with AD gets easily infected by the residential microflora, triggering the host immune responses. Among the resident microflora of the skin, Malassezia is thought to possess an antigenic property that induces type I hypersensitivity reaction in the skin. One of the hallmarks of AD is the elevated level of immunoglobulin E (IgE) in total serum of most patients, leading to IgE-mediated allergic reaction [24]. Several studies have reported hypersensitization to Malassezia-specific IgE in patients with AD. For example, a high proportion of AD patients were sensitized to M. sympodialis extract and recombinant M. sympodialis allergens, whereas patients with seborrheic dermatitis and healthy individuals exhibited no such sensitization [25]. Serum IgE from AD patients was more sensitized to Malassezia antigens [26]. A study reported positive correlation between Malassezia-specific IgE levels and disease severity in adult AD patients [27]. A study showed the increased levels of specific IgE against M. restricta compared to the other species of Malassezia in the sera of AD patients and suggested that M. restricta might also play an important role in AD [28]. In addition, a study that focused on the link between the allergens from Malassezia and AD showed that M. globosa allergen (MGL_1304), identified in sweat, induces the release of histamine from the basophils in AD patients but not in healthy individuals [29]. Moreover, the levels of MGL_1304-specific IgE in the sera of AD patients were significantly increased compared to those of the healthy individuals [30], and the activity of histamine release by MGL_1304 and its homolog Mala r 8 from M. restricta were higher than that of Mala s 8 produced by M. sympodialis [31]. These results imply that IgE-mediated sensitization against Malassezia and its allergens can act as a triggering factor for AD.
Pityriasis versicolor, characterized by hypo- or hyper-pigmentation in the seborrheic lesions of skin without inflammation, is one of the most common skin diseases caused by Malassezia infection of the stratum corneum. In general, pityriasis versicolor occurs frequently in adolescents and young adults with highly active sebaceous glands. Abundance of Malassezia in the lesions of patients with pityriasis versicolor is significantly increased compared to those in the unaffected skin sites of the patients or in healthy individuals, which suggests the etiological correlations between the disorder and the fungus [32, 33]. Although the exact role of Malassezia in pityriasis versicolor is not yet clear, the role of the metabolites produced by Malassezia in the pathogenesis has been demonstrated. Examples of these metabolites include indoles and indole derivatives derived from tryptophan by Malassezia, M. furfur in particular. Indirubin, malassezin, pityriacitrin, 6-formylindolo[3,2-b]carbazole, and indolo[3,2-b]carbazole generated by M. furfur serves as ligands of the aryl hydrocarbon receptor, which is a nuclear receptor belonging to the basic helix-loop-helix family of the transcription factor with implications in the association of the metabolites with pityriasis versicolor [34-36].
Malassezia is not only a part of the residential microflora of human skin, but it is also found in various other organs. Mycobiome analysis using stool samples from healthy individuals showed a high prevalence of Malassezia, especially M. restricta [37-40]. Furthermore, high prevalence of M. restricta in the water-lavage samples obtained from non-inflamed intestinal regions of patients with Crohn’s disease undergoing screening colonic endoscopy suggested the association of the fungal species with the disease [41]. A recent study also showed increased abundance of Malassezia in fecal samples from patients with pancreatic or colorectal cancer compared to those in the healthy control groups implying that the fungus might be linked to cancer in organs, as well as the diseases related to skin [42, 43]. Furthermore, a respiratory mycobiome study revealed the association between Malassezia and cystic fibrosis lung disease [44]. These findings imply that Malassezia plays a critical role in the human body than what is believed.
The Potential Role of Lipases in the Pathogenesis of Malassezia
Skin is the largest organ of the human body, which form the first barrier against external environment. Lipids, the main constituents of the stratum corneum produced by the sebaceous glands, contribute significantly to the barrier function of skin. The lipid mixture within the stratum corneum primarily comprises ceramides, cholesterol, and fatty acids. Human sebum contains triglycerides, wax monoesters, squalene, and a small amount of cholesterol and cholesterol esters [45]. Given that Malassezia requires lipids for growth, the lipid-rich skin environment serves as a suitable habitat for the fungus. A cell culture-based study showed increased Malassezia population in adult than in children [46]; sebum level of the skin of the former was significantly high than that on the latter population [47]. Although culture-independent real-time PCR-based analysis and a mycobiome study revealed Malassezia as the predominant fungal genus on the adult skin, fungal communities were more diverse on the skin of children [8, 9].
Since species of Malassezia are unable to synthesize fatty acids, they must uptake external fatty acids as a nutrient source for survival. Genome analyses of multiple Malassezia species revealed the absence of the gene encoding fatty acid synthase [10, 12]. This lack of fatty acid synthesis ability is responsible for the lipid dependency of Malassezia, a characteristic which is compensated by the secretion of various lipases and degradation of sebum to produce and uptake external fatty acids [10]. Indeed, genome analyses of several Malassezia species showed significantly higher number of lipase encoding genes than that in other fungi [10-12].
Pathogenic microorganisms secrete enzymes with lipolytic activities to survive within the host. Among the different lipolytic enzymes, extracellular lipase, which primarily catalyzes the hydrolysis of triglycerides, is considered an important virulence factor [48, 49]. In this regard, several studies have suggested the important roles of lipases in microbial pathogenesis. For example, almost all clinically isolated bacterial pathogens, including Staphylococcus aureus, exhibited lipolytic activities [50, 51], and inhibition of chemotaxis and reduction of granulocytes mediated phagocytic killing of S. aureus cells by purified lipase from the bacteria [52, 53]. Moreover, a recent study showed that Geh lipase inhibits activation of innate immune cells, thereby verifying the critical roles of lipases in the pathogenesis of S. aureus [54].
Lipases also play an important role in fungal pathogenesis. For example, Candida albicans possesses at least 10 genes (CaLIP1–CaLIP10) encoding lipases. All the CaLIP genes, except CaLIP7, contain a signal peptide, implying that they produce extracellular lipases. Expression of CaLIP5, CaLIP6, CaLIP8, and CaLIP9 during systemic candidiasis suggested their association with the pathogenesis of C. albicans [55]. Despite the temporal and stage-specific expression of CaLIP genes in general, CaLIP5 and CaLIP8 were predominantly expressed in the liver and kidney of infected mice, and CaLIP4, CaLIP5, and CaLIP8 were expressed in at least 50% of the saliva specimens from patients with oral candidiasis [56]. Moreover, a CaLIP8 deficient mutant with significantly reduced virulence in the murine intravenous infection model confirmed the importance of lipases in the pathogenesis of C. albicans [57]. Contribution of lipases in the fungal virulence was also suggested in other species of Candida such as C. parapsilosis, which possesses CpLIP1 and CpLIP2 genes encoding lipases. These lipases were required for intracellular survival of C. parapsilosis within the macrophages and reduced production of inflammatory cytokines in the host [58, 59].
Given the lipid-dependent characteristics of Malassezia and the crucial role of lipases in fungal survival on the skin, a large number of studies have been carried out to characterize the lipases in Malassezia (Table 1). Malassezia left the unsaturated fatty acids, oleic acid in particular, on the skin after consuming the saturated fatty acids in sebum, and it triggered dandruff formation in the dandruff susceptible individuals, the finding of which supports the role of lipases in the pathogenesis of Malassezia [60].
Table 1.
Lipases in different species of Malassezia that are commonly found on human skin.
| Species | Predicted function from Pfam analysis (ID/accession) | Gene ID | Signal peptide | Optimal condition | Substrate | Reference |
|---|---|---|---|---|---|---|
| M. restricta KCTC 27527 | Class 3 (PF01764) | MRET_0019 (MrLIP1) | O | pH 5, 34°C | Mono- and diglyceride | [12, 71-73] |
| MRET_1032 | × | [12, 71] | ||||
| MRET_2826 (MrLIP4) | O | [12, 71] | ||||
| MRET_3772 | O | [12, 71] | ||||
| MRET_4032 (MrLIP2) | O | Mono- and diglyceride | [12, 71, 72] | |||
| MRET_4356 | × | [12, 71] | ||||
| LIP family (PF03583) | MRET_0930 (MrLIP5) | O | pH 7 | [12, 71, 74] | ||
| MRET_1179 (MrLIP3) | O | Mono-, di-, and triglyceride | [12, 71, 72] | |||
| MRET_4098 | O | [12, 71] | ||||
| MRET_4099 | O | [12, 71] | ||||
| Partial alpha/beta-hydrolase lipase region (PF04083) | MRET_1507 | × | [12, 71] | |||
| MRET_4144 | × | [12, 71] | ||||
| Putative serine esterase, Lipase-like (PF05057) | MRET_3765 | × | [12, 71] | |||
| GDSL-like Lipase/Acylhydrolase (PF00657) | MRET_3994 | O | [12, 71] | |||
| M. globosaCBS 7966 | Class 3 (PF01764) | MGL_0279 | × | [11] | ||
| MGL_0797 (MgLIP1) | O | Mono- and diglyceride | [11, 70] | |||
| MGL_0798 (MgMDL3) | O | Mono- and diglyceride | [11, 70] | |||
| MGL_0799 (MgMDL2) | O | pH 6, 15°C | Mono- and diglyceride | [11, 68, 70] | ||
| MGL_0800 (MgMDL4) | O | Mono- and diglyceride | [11, 70] | |||
| MGL_1311 (MgMDL5) | O | Mono- and diglyceride | [11, 70] | |||
| MGL_1769 | × | [11] | ||||
| MGL_3878 (MgMDL6) | O | Mono- and diglyceride | [11, 70] | |||
| LIP family (PF03583) | MGL_1311 (MgLIP4) | O | Mono-, di-, and tri-glyceride | [11, 70] | ||
| MGL_3507 (MgLIP6) | O | Diglyceride | [11, 70] | |||
| MGL_4051 (MgLIP7) | O | Mono-, di-, and triglyceride | [11, 70] | |||
| MGL_4052 (MgLIP5) | O | Mono-, di-, and triglyceride | [11, 70] | |||
| MGL_4054 (MgLIP2) | O | pH 5, 30°C | Mono-, di-, and triglyceride | [11, 67, 70] | ||
| MGL_4197 (MgLIP3) | O | Mono-, di-, and triglyceride | [11, 70] | |||
| Partial alpha/beta-hydrolase lipase region (PF04083) | MGL_1975 | × | [11] | |||
| MGL_2531 | × | [11] | ||||
| Putative serine esterase, Lipase-like (PF05057) | MGL_4063 | × | [11] | |||
| GDSL-like Lipase/Acylhydrolase (PF00657) | MGL_1366 | O | [11] | |||
| M. sympodialis ATCC 42132 | Class 3 (PF01764) | MSYG_1326 | O | [81] | ||
| MSYG_2002 | × | [81] | ||||
| MSYG_2462 | O | [81] | ||||
| MSYG_2628 | O | [81] | ||||
| MSYG_3125 | O | [81] | ||||
| MSYG_3126 | O | [81] | ||||
| MSYG_4275 | × | [81] | ||||
| LIP family (PF03583) | MSYG_2467 | O | [81] | |||
| MSYG_2468 | O | [81] | ||||
| MSYG_2630 | O | [81] | ||||
| MSYG_3257 | O | [81] | ||||
| Partial alpha/beta-hydrolase lipase region (PF04083) | MSYG_0158 | × | [81] | |||
| MSYG_2613 | × | [81] | ||||
| Putative serine esterase, Lipase-like (PF05057) | MSYG_3580 | × | [81] | |||
| GDSL-like Lipase/Acylhydrolase (PF00657) | MSYG_1283 | × | [81] |
The lipase activity was first described in M. furfur [61, 62]. MfLIP1, encoding extracellular lipase, was also first identified and characterized in M. furfur. Recombinant MfLip1 enzyme expressed in Pichia pastoris exhibited hydrolytic activity against monoglycerides and showed highest activity at 40°C and pH 5.8 [63]. Elevated activity of an extracellular lipase in M. furfur with increased pH suggested that the environmental conditions critically influence the activity of the enzyme [64]. During the yeast to hyphal dimorphic transition of M. furfur, extracellular lipase activity was reported to be higher in hypha form than in the yeast from [65]. The same lipase activity was also observed in M. globosa. MgLip1 hydrolyzed mono- and diglycerides, but not triglycerides. Expression of MgLIP1 on the human scalp demonstrated using reverse transcription-PCR suggested that lipase may play an important role in the survival of M. globosa on the host skin surface [66]. Another lipase, MgLip2 exhibited its optimal activity at 30°C and pH 5.0 [67]. A putative diacylglycerol-like lipase, MgMdl2, was also identified and characterized. Its recombinant form showed optimal activity at 15°C and pH 6.0. Similar to MgLip1, recombinant MgMdl2 also utilized mono- and diglycerides, but not triglycerides [68]. Interestingly, a structural study showed that amino acid substitutions, such as Phe278Ala and Glu282Ala converted the substrate specificity of MgMdl2 from mono- and diglycerides to triglycerides, suggesting the critical role of these two amino acid residues in substrate recognition [69]. Subsequently, genome analysis of M. globosa revealed that the fungus possesses at least 14 lipase encoding genes [11]. Thirteen of them were expressed in P. pastoris and the hydrolysis activity of each enzyme was analyzed against various substrates. Results revealed that MgLip1, MgMdl2, MgMdl3, MgMdl4, MgMdl5, and MgMdl6 are the Class 3 family lipases with specificity for mono- and diglycerides, whereas MgLip2, MgLip3, MgLip4, MgLip5, and MgLip7 are the LIP family lipases that hydrolyze all substrates including triglycerides, one of the key components in human sebum [70].
In M. restricta, initially 12 lipase genes were identified in the genome of the strain KCTC 27527; however, a recently resequenced genome analysis of the same strain revealed two additional lipase genes [12, 71]. Among the 14 lipases identified in M. restricta, three secretory lipases (MrLip1, MrLip2, and MrLip3) were purified and characterized. MrLip1 and MrLip2 were members of Class 3 family, and MrLip3 belonged to the LIP family of lipases. Similar to MgLip1 in M. globosa, MrLip1 and MrLip2 specifically degrade mono- and diglycerides. In contrast, MrLip3 hydrolyzes triglycerides as well as mono- and diglycerides [72]. MrLip1 exhibited optimal enzyme activity at 34°C and pH 5.0; conditions similar to those found on normal healthy skin surface. Furthermore, expression patterns of MrLip1 were well correlated with the optimal pH and temperature of the enzyme activity [73]. Our previous study showed that MrLIP5, which encodes the LIP family lipase, was expressed on the scalp of 53 out of 56 patients with severe dandruff, suggesting a possible association of MrLip5 with dandruff and pathogenesis of Malassezia [12]. Subsequent purification of MrLip5, and analysis of the gene expression levels and enzyme activity under different pH conditions revealed highest expression level and optimal enzyme activity at pH 7.0–8.0 [74]. Given that the pH increases in the diseased skin surface [75-77], our observation suggested that MrLip5 might play a major role in the pathogenesis of Malassezia on the diseased skin surface.
In addition to the possible pathological roles of secreted lipases in Malassezia, association between secreted lipases and mechanism of the action of an antifungal agent was suggested. Example includes treatment of Malassezia with the antifungal drug zinc pyrithione (ZPT). Clinical isolates of M. restricta treated with ZPT showed significantly reduced expression levels of several secreted lipases, including MrLip1, MrLip3, and MrLip5. This finding indicated that inhibition of lipase functions is one of the key mechanisms of action of ZPT; hence, the lipases secreted from Malassezia could be a potential target for antifungal drug [78].
Concluding Remarks
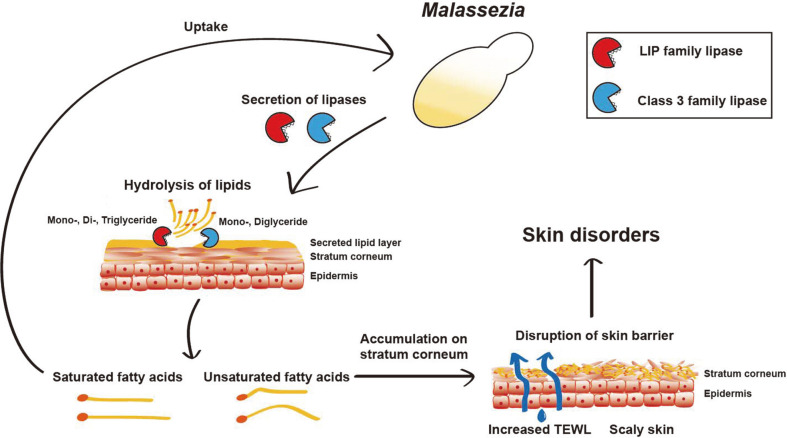
Research on Malassezia has gained more attention owing to its association with not only diseases on the skin but also its presence in other organs of the human body. However, the roles of Malassezia in human body are still largely unknown. Existing studies suggest that lipases may play critical role in the survival and pathogenesis of Malassezia on the human skin surface. The lipases decompose sebum to free fatty acids. Saturated fatty acids are primarily consumed by Malassezia, whereas the unsaturated fatty acid, oleic acid in particular, remains on the stratum corneum and likely triggers disruption of the skin barrier function (Fig. 1) [17].
Fig. 1. The potential role of lipases secreted by Malassezia in skin diseases.
Lipases secreted by Malassezia decompose the human skin sebum-derived lipids, such as mono-, di-, and triglycerides, into saturated and unsaturated fatty acids. The saturated fatty acids are consumed by Malassezia for survival, whereas the unsaturated fatty acids accumulate on the stratum corneum. This accumulation might interfere with the permeability of the skin barrier, thereby leading to various skin disorders. TEWL: Transepidermal water loss.
Similar to other pathogenic microorganisms, secreted lipases in Malassezia may also modulate or interfere with the host immune cell responses. However, to date, information on the roles of lipase in Malassezia is restricted to in vivo observation of lipase gene transcripts and in vitro analysis of purified enzyme activity. Genetic experiments using a mutant lacking the gene encoding lipase are certainly required to understand the role of lipases in physiology and pathogenesis of Malassezia. The gene knockout method utilizing Agrobacterium tumefaciens-mediated transformation is available for few Malassezia species, including M. furfur, M. sympodialis, and M. pachydermatis [79, 80]. However, a similar gene manipulation technique is yet to be developed for M. restricta and M. globosa, the two most dominant species of Malassezia in human body. Therefore, future studies are warranted to develop molecular genetic tools for increasing our understanding of the physiology and pathogenesis of M. restricta and M. globosa and to elucidate the role of lipases in Malassezia infections.
Acknowledgments
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT and Future Planning 2019R1F1A1061930 and 2019R1A4A1024764.
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest to declare.
REFERENCES
- 1.Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang LL, Clavaud C, Bar-Hen A, Cui M, Gao J, Liu YY, et al. Characterization of the major bacterial-fungal populations colonizing dandruff scalps in Shanghai, China, shows microbial disequilibrium. Exp. Dermatol. 2015;24:398–400. doi: 10.1111/exd.12684. [DOI] [PubMed] [Google Scholar]
- 3.Saxena R, Mittal P, Clavaud C, Dhakan DB, Hegde P, Veeranagaiah MM, et al. Comparison of healthy and dandruff scalp microbiome reveals the role of commensals in scalp health. Front. Cell Infect. Microbiol. 2018;8:346. doi: 10.3389/fcimb.2018.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q-M, Theelen B, Groenewald M, Bai F-Y, Boekhout T. Moniliellomycetes and Malasseziomycetes, two new classes in Ustilaginomycotina. Persoonia. 2014;33:41–47. doi: 10.3767/003158514X682313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Theelen B, Cafarchia C, Gaitanis G, Bassukas ID, Boekhout T, Dawson TL., Jr Malassezia ecology, pathophysiology, and treatment. Med. Mycol. 2018;56:S10–S25. doi: 10.1093/mmy/myx134. [DOI] [PubMed] [Google Scholar]
- 6.Tang S, Prem A, Tjokrosurjo J, Sary M, Van Bel MA, Rodrigues-Hoffmann A, et al. The canine skin and ear microbiome: A comprehensive survey of pathogens implicated in canine skin and ear infections using a novel next-generation-sequencing-based assay. Vet. Microbiol. 2020;247:108764. doi: 10.1016/j.vetmic.2020.108764. [DOI] [PubMed] [Google Scholar]
- 7.Gupta AK, Kohli Y. Prevalence of Malassezia species on various body sites in clinically healthy subjects representing different age groups. Med. Mycol. 2004;42:35–42. doi: 10.1080/13693780310001610056. [DOI] [PubMed] [Google Scholar]
- 8.Sugita T, Suzuki M, Goto S, Nishikawa A, Hiruma M, Yamazaki T, et al. Quantitative analysis of the cutaneous Malassezia microbiota in 770 healthy Japanese by age and gender using a real-time PCR assay. Med. Mycol. 2010;48:229–233. doi: 10.3109/13693780902977976. [DOI] [PubMed] [Google Scholar]
- 9.Jo J-H, Deming C, Kennedy EA, Conlan S, Polley EC, Ng W-I, et al. Diverse human skin fungal communities in children converge in adulthood. J. Invest. Dermatol. 2016;136:2356–2363. doi: 10.1016/j.jid.2016.05.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu G, Zhao H, Li C, Rajapakse MP, Wong WC, Xu J, et al. Genus-wide comparative genomics of Malassezia delineates its phylogeny, physiology, and niche adaptation on human skin. PLoS Genet. 2015;11:e1005614. doi: 10.1371/journal.pgen.1005614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J, Saunders CW, Hu P, Grant RA, Boekhout T, Kuramae EE, et al. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc. Natl. Acad. Sci. USA. 2007;104:18730–18735. doi: 10.1073/pnas.0706756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park M, Cho YJ, Lee YW, Jung WH. Whole genome sequencing analysis of the cutaneous pathogenic yeast Malassezia restricta and identification of the major lipase expressed on the scalp of patients with dandruff. Mycoses. 2017;60:188–197. doi: 10.1111/myc.12586. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz JR, Messenger AG, Tosti A, Todd G, Hordinsky M, Hay RJ, et al. A comprehensive pathophysiology of dandruff and seborrheic dermatitis - towards a more precise definition of scalp health. Acta Derm. Venereol. 2013;93:131–137. doi: 10.2340/00015555-1382. [DOI] [PubMed] [Google Scholar]
- 14.Heng MC, Henderson CL, Barker DC, Haberfelde G. Correlation of Pityosporum ovale density with clinical severity of seborrheic dermatitis as assessed by a simplified technique. J. Am. Acad. Dermatol. 1990;23:82–86. doi: 10.1016/0190-9622(90)70191-J. [DOI] [PubMed] [Google Scholar]
- 15.Peter RU, Richarz-Barthauer U. Successful treatment and prophylaxis of scalp seborrhoeic dermatitis and dandruff with 2% ketoconazole shampoo: results of a multicentre, double-blind, placebo-controlled trial. Br. J. Dermatol. 1995;132:441–445. doi: 10.1111/j.1365-2133.1995.tb08680.x. [DOI] [PubMed] [Google Scholar]
- 16.Pierard GE, Arrese JE, Pierard-Franchimont C P DED, author. Prolonged effects of antidandruff shampoos - time to recurrence of Malassezia ovalis colonization of skin. Int. J. Cosmet. Sci. 1997;19:111–117. doi: 10.1046/j.1467-2494.1997.171706.x. [DOI] [PubMed] [Google Scholar]
- 17.DeAngelis YM, Gemmer CM, Kaczvinsky JR, Kenneally DC, Schwartz JR, Dawson TL., Jr Three etiologic facets of dandruff and seborrheic dermatitis: Malassezia fungi, sebaceous lipids, and individual sensitivity. J. Investig. Dermatol. Symp. Proc. 2005;10:295–297. doi: 10.1111/j.1087-0024.2005.10119.x. [DOI] [PubMed] [Google Scholar]
- 18.Clavaud C, Jourdain R, Bar-Hen A, Tichit M, Bouchier C, Pouradier F, et al. Dandruff is associated with disequilibrium in the proportion of the major bacterial and fungal populations colonizing the scalp. PLoS One. 2013;8:e58203. doi: 10.1371/journal.pone.0058203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park T, Kim HJ, Myeong NR, Lee HG, Kwack I, Lee J, et al. Collapse of human scalp microbiome network in dandruff and seborrhoeic dermatitis. Exp. Dermatol. 2017;26:835–838. doi: 10.1111/exd.13293. [DOI] [PubMed] [Google Scholar]
- 20.Lin Q, Panchamukhi A, Li P, Shan W, Zhou H, Hou L, et al. Malassezia and Staphylococcus dominate scalp microbiome for seborrheic dermatitis. Bioprocess Biosyst Eng. 2020 doi: 10.1007/s00449-020-02333-5. https://doi.org/10.1007/s00449-020-02333-5. [DOI] [PubMed] [Google Scholar]
- 21.de Groot PW, Bader O, de Boer AD, Weig M, Chauhan N. Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryot. Cell. 2013;12:470–481. doi: 10.1128/EC.00364-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Z, Wang Z, Yuan C, Liu X, Yang F, Wang T, et al. Dandruff is associated with the conjoined interactions between host and microorganisms. Sci. Rep. 2016;6:24877. doi: 10.1038/srep24877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de la O-Escamilla NO, Sidbury R. Atopic dermatitis: Update on pathogenesis and therapy. Pediatric. Ann. 2020;49:e140–e146. doi: 10.3928/19382359-20200217-01. [DOI] [PubMed] [Google Scholar]
- 24.Kasperkiewicz M, Schmidt E, Ludwig RJ, Zillikens D. Targeting IgE antibodies by immunoadsorption in atopic dermatitis. Front. Immunol. 2018;9:254. doi: 10.3389/fimmu.2018.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansson C, Sandström M, Bartosik J, Särnhult T, Christiansen J, Zargari A, et al. Atopy patch test reactions to Malassezia allergens differentiate subgroups of atopic dermatitis patients. Br. J. Dermatol. 2003;148:479–488. doi: 10.1046/j.1365-2133.2003.05093.x. [DOI] [PubMed] [Google Scholar]
- 26.Brodská P, Panzner P, Pizinger K, Schmid-Grendelmeier P. IgE-mediated sensitization to Malassezia in atopic dermatitis: more common in male patients and in head and neck type. Dermatitis. 2014;25:120–126. doi: 10.1097/DER.0000000000000040. [DOI] [PubMed] [Google Scholar]
- 27.Glatz M, BuCHNER M, Von Bartenwerffer W, Schmid-Grendelmeier P, Worm M, Hedderich J, et al. Malassezia spp.-specific immunoglobulin E level is a marker for severity of atopic dermatitis in adults. Acta Derm. Venereol. 2015;95:191–196. doi: 10.2340/00015555-1864. [DOI] [PubMed] [Google Scholar]
- 28.Kato H, Sugita T, Ishibashi Y, Nishikawa A. Detection and quantification of specific IgE antibodies against eight Malassezia species in sera of patients with atopic dermatitis by using an enzyme-linked immunosorbent assay. Microbiol. Immunol. 2006;50:851–856. doi: 10.1111/j.1348-0421.2006.tb03860.x. [DOI] [PubMed] [Google Scholar]
- 29.Hiragun T, Ishii K, Hiragun M, Suzuki H, Kan T, Mihara S, et al. Fungal protein MGL_1304 in sweat is an allergen for atopic dermatitis patients. J. Allergy Clin. Immunol. 2013;132:608–615.:e604. doi: 10.1016/j.jaci.2013.03.047. [DOI] [PubMed] [Google Scholar]
- 30.Hiragun M, Hiragun T, Ishii K, Suzuki H, Tanaka A, Yanase Y, et al. Elevated serum IgE against MGL_1304 in patients with atopic dermatitis and cholinergic urticaria. Allergol. Int. 2014;63:83–93. doi: 10.2332/allergolint.13-OA-0611. [DOI] [PubMed] [Google Scholar]
- 31.Kohsaka T, Hiragun T, Ishii K, Hiragun M, Kamegashira A, Hide M. Different hypersensitivities against homologous proteins of MGL_1304 in patients with atopic dermatitis. Allergol. Int. 2018;67:103–108. doi: 10.1016/j.alit.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Honnavar P, Dogra S, Handa S, Chakrabarti A, Rudramurthy SM. Molecular identification and quantification of Malassezia species isolated from pityriasis versicolor. Indian Dermatol. Online J. 2020;11:167–170. doi: 10.4103/idoj.IDOJ_142_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elshabrawy WO, Saudy N, Sallam M. Molecular and phenotypic identification and speciation of Malassezia yeasts isolated from Egyptian patients with pityriasis versicolor. J. Clin. Diagn Res. 2017;11:DC12–DC17. doi: 10.7860/JCDR/2017/27747.10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vlachos C, Schulte BM, Magiatis P, Adema GJ, Gaitanis G. Malassezia-derived indoles activate the aryl hydrocarbon receptor and inhibit Toll-like receptor-induced maturation in monocyte-derived dendritic cells. Br. J. Dermatol. 2012;167:496–505. doi: 10.1111/j.1365-2133.2012.11014.x. [DOI] [PubMed] [Google Scholar]
- 35.Magiatis P, Pappas P, Gaitanis G, Mexia N, Melliou E, Galanou M, et al. Malassezia yeasts produce a collection of exceptionally potent activators of the Ah (dioxin) receptor detected in diseased human skin. J. Invest. Dermatol. 2013;133:2023–2030. doi: 10.1038/jid.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esser C, Bargen I, Weighardt H, Haarmann-Stemmann T, Krutmann J. Functions of the aryl hydrocarbon receptor in the skin. Semin. Immunopathol. 2013;35:677–691. doi: 10.1007/s00281-013-0394-4. [DOI] [PubMed] [Google Scholar]
- 37.Suhr MJ, Banjara N Hallen‐Adams HE, author. Sequence‐based methods for detecting and evaluating the human gut mycobiome. Lett. Appl. Microbiol. 2016;62:209–215. doi: 10.1111/lam.12539. [DOI] [PubMed] [Google Scholar]
- 38.Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome. 2017;5:153. doi: 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Auchtung TA, Fofanova TY, Stewart CJ, Nash AK, Wong MC, Gesell JR, et al. Investigating colonization of the healthy adult gastrointestinal tract by fungi. MSphere. 2018;3:e00092–18. doi: 10.1128/mSphere.00092-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raimondi S, Amaretti A, Gozzoli C, Simone M, Righini L, Candeliere F, et al. Longitudinal survey of fungi in the human gut: ITS profiling, phenotyping and colonization. Front. Microbiol. 2019;10:1575. doi: 10.3389/fmicb.2019.01575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, et al. Malassezia is associated with Crohn's disease and exacerbates colitis in mouse models. Cell Host Microbe. 2019;25:377–388. doi: 10.1016/j.chom.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019;574:264–267. doi: 10.1038/s41586-019-1608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut. 2019;68:654–662. doi: 10.1136/gutjnl-2018-317178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soret P, Vandenborght L-E, Francis F, Coron N, Enaud R, Avalos M, et al. Respiratory mycobiome and suggestion of interkingdom network during acute pulmonary exacerbation in cystic fibrosis. Sci. Rep. 2020;10:3589. doi: 10.1038/s41598-020-60015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wertz PW. Lipids and the permeability and antimicrobial barriers of the skin. J. Lipids. 2018;2018:5954034. doi: 10.1155/2018/5954034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta AK, Kohli Y. Prevalence of Malassezia species on various body sites in clinically healthy subjects representing different age groups. Med. Mycol. 2004;42:35–42. doi: 10.1080/13693780310001610056. [DOI] [PubMed] [Google Scholar]
- 47.Oranges T, Dini V, Romanelli M. Skin physiology of the neonate and infant: clinical implications. Adv. Wound Care (New Rochelle). 2015;4:587–595. doi: 10.1089/wound.2015.0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stehr F, Kretschmar M, Kröger C, Hube B, Schäfer WJJomcBe. Microbial lipases as virulence factors. J. Mol. Catal. B Enzym. 2003;22:347–355. doi: 10.1016/S1381-1177(03)00049-3. [DOI] [Google Scholar]
- 49.Park M, Do E, Jung WH. Lipolytic enzymes involved in the virulence of human pathogenic fungi. Mycobiology. 2013;41:67–72. doi: 10.5941/MYCO.2013.41.2.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Priya R, Mythili A, Singh YR, Sreekumar H, Manikandan P, Panneerselvam K, et al. Virulence, speciation and antibiotic susceptibility of ocular coagualase negative Staphylococci (CoNS) J. Clin. Diagn. Res. 2014;8:DC33–37. doi: 10.7860/JCDR/2014/7867.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Al-Wali WI, Elvin SJ, Mason CM, Clark A, Tranter HS. Comparative phenotypic characteristics of Staphylococcus aureus isolates from line and non-line associated septicaemia, CAPD peritonitis, bone/joint infections and healthy nasal carriers. J. Med. Microbiol. 1998;47:265–274. doi: 10.1099/00222615-47-3-265. [DOI] [PubMed] [Google Scholar]
- 52.Rollof J, Braconier JH, Soderstrom C, Nilsson-Ehle P. Interference of Staphylococcus aureus lipase with human granulocyte function. Eur. J. Clin. Microbiol. Infect. Dis. 1988;7:505–510. doi: 10.1007/BF01962601. [DOI] [PubMed] [Google Scholar]
- 53.Braconier JH, Rollof J. Influence by staphylococcal lipase on granulocyte metabolism and killing of bacteria. Zentralbl. Bakteriol. 1991;276:68–72. doi: 10.1016/S0934-8840(11)80220-3. [DOI] [PubMed] [Google Scholar]
- 54.Chen X, Alonzo F., 3rd Bacterial lipolysis of immune-activating ligands promotes evasion of innate defenses. Proc. Natl. Acad. Sci. USA. 2019;116:3764–3773. doi: 10.1073/pnas.1817248116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hube B, Stehr F, Bossenz M, Mazur A, Kretschmar M, Schafer W. Secreted lipases of Candida albicans: cloning, characterisation and expression analysis of a new gene family with at least ten members. Arch. Microbiol. 2000;174:362–374. doi: 10.1007/s002030000218. [DOI] [PubMed] [Google Scholar]
- 56.Stehr F, Felk A, Gacser A, Kretschmar M, Mahnss B, Neuber K, et al. Expression analysis of the Candida albicans lipase gene family during experimental infections and in patient samples. FEMS Yeast Res. 2004;4:401–408. doi: 10.1016/S1567-1356(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 57.Gacser A, Stehr F, Kroger C, Kredics L, Schafer W, Nosanchuk JD. Lipase 8 affects the pathogenesis of Candida albicans. Infect. Immun. 2007;75:4710–4718. doi: 10.1128/IAI.00372-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neugnot V, Moulin G, Dubreucq E, Bigey F. The lipase/acyltransferase from Candida parapsilosis: Molecular cloning and characterization of purified recombinant enzymes. Eur. J. Biochem. 2002;269:1734–1745. doi: 10.1046/j.1432-1327.2002.02828.x. [DOI] [PubMed] [Google Scholar]
- 59.Tóth A, Németh T, Csonka K, Horváth P, Vágvölgyi C, Vizler C, et al. Secreted Candida parapsilosis lipase modulates the immune response of primary human macrophages. Virulence. 2014;5:555–62. doi: 10.4161/viru.28509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ro BI, Dawson TL. The role of sebaceous gland activity and scalp microfloral metabolism in the etiology of seborrheic dermatitis and dandruff. J. Investig. Dermatol. Symp. Proc. 2005;10:194–197. doi: 10.1111/j.1087-0024.2005.10104.x. [DOI] [PubMed] [Google Scholar]
- 61.Ran Y, Yoshiike T, Ogawa H. Lipase of Malassezia furfur: Some properties and their relationship to cell growth. J. Med. Vet. Mycol. 1993;31:77–85. doi: 10.1080/02681219380000081. [DOI] [PubMed] [Google Scholar]
- 62.Plotkin LI, Squiquera L, Mathov I, Galimberti R, Leoni J. Characterization of the lipase activity of Malassezia furfur. J. Med. Vet. Mycol. 1996;34:43–48. doi: 10.1080/02681219680000071. [DOI] [PubMed] [Google Scholar]
- 63.Brunke S, Hube B. MfLIP1, a gene encoding an extracellular lipase of the lipid-dependent fungus Malassezia furfur. Microbiology. 2006;152:547–554. doi: 10.1099/mic.0.28501-0. [DOI] [PubMed] [Google Scholar]
- 64.Juntachai W, Chaichompoo A, Chanarat S. Ambient pH regulates secretion of lipases in Malassezia furfur. Microbiology. 2020;166:288–295. doi: 10.1099/mic.0.000879. [DOI] [PubMed] [Google Scholar]
- 65.Juntachai W, Kajiwara S. Differential expression of extracellular lipase and protease activities of mycelial and yeast forms in Malassezia furfur. Mycopathologia. 2015;180:143–151. doi: 10.1007/s11046-015-9900-7. [DOI] [PubMed] [Google Scholar]
- 66.DeAngelis YM, Saunders CW, Johnstone KR, Reeder NL, Coleman CG, Kaczvinsky JR, Jr, et al. Isolation and expression of a Malassezia globosa lipase gene, LIP1. J. Invest. Dermatol. 2007;127:2138–2146. doi: 10.1038/sj.jid.5700844. [DOI] [PubMed] [Google Scholar]
- 67.Juntachai W, Oura T, Kajiwara S. Purification and characterization of a secretory lipolytic enzyme, MgLIP2, from Malassezia globosa. Microbiology. 2011;157:3492–3499. doi: 10.1099/mic.0.054528-0. [DOI] [PubMed] [Google Scholar]
- 68.Xu H, Lan D, Yang B, Wang Y. Biochemical properties and structure analysis of a DAG-Like lipase from Malassezia globosa. Int. J. Mol. Sci. 2015;16:4865–4879. doi: 10.3390/ijms16034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lan D, Xu H, Xu J, Dubin G, Liu J, Iqbal Khan F, et al. Malassezia globosa MgMDL2 lipase: Crystal structure and rational modification of substrate specificity. Biochem. Biophys. Res. Commun. 2017;488:259–265. doi: 10.1016/j.bbrc.2017.04.103. [DOI] [PubMed] [Google Scholar]
- 70.Sommer B, Overy DP, Haltli B, Kerr RG. Secreted lipases from Malassezia globosa: recombinant expression and determination of their substrate specificities. Microbiology. 2016;162:1069–1079. doi: 10.1099/mic.0.000299. [DOI] [PubMed] [Google Scholar]
- 71.Cho Y-J, Park M, Jung WHJMra. Resequencing the genome of Malassezia restricta strain KCTC 27527. Microbiol. Resour. Announc. 2019;8:e00213–19. doi: 10.1128/MRA.00213-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sommer B, Overy DP, Kerr RG. Identification and characterization of lipases from Malassezia restricta, a causative agent of dandruff. FEMS Yeast Res. 2015;15:fov078. doi: 10.1093/femsyr/fov078. [DOI] [PubMed] [Google Scholar]
- 73.Park M, Jung WH, Han SH, Lee YH, Lee YW. Characterisation and expression analysis of MrLip1, a class 3 family lipase of Malassezia restricta. Mycoses. 2015;58:671–678. doi: 10.1111/myc.12412. [DOI] [PubMed] [Google Scholar]
- 74.Park M, Lee JS, Jung WH, Lee YW. pH-dependent expression, stability, and activity of Malassezia restricta MrLip5 Lipase. Ann. Dermatol. 2020;32:473–480. doi: 10.5021/ad.2020.32.6.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eberlein-Konig B, Schafer T, Huss-Marp J, Darsow U, Mohrenschlager M, Herbert O, et al. Skin surface pH, stratum corneum hydration, trans-epidermal water loss and skin roughness related to atopic eczema and skin dryness in a population of primary school children. Acta Derm. Venereol. 2000;80:188–191. doi: 10.1080/000155500750042943. [DOI] [PubMed] [Google Scholar]
- 76.Moalli R, Meyers RS, Ultsch GR, Jackson DC. Acid-base balance and temperature in a predominantly skin-breathing salamander, Cryptobranchus alleganiensis. Respir. Physiol. 1981;43:1–11. doi: 10.1016/0034-5687(81)90083-9. [DOI] [PubMed] [Google Scholar]
- 77.Jung YC, Kim EJ, Cho JC, Suh KD, Nam GW. Effect of skin pH for wrinkle formation on Asian: Korean, Vietnamese and Singaporean. J. Eur. Acad. Dermatol. Venereol. 2013;27:e328–332. doi: 10.1111/j.1468-3083.2012.04660.x. [DOI] [PubMed] [Google Scholar]
- 78.Park M, Cho YJ, Lee YW, Jung WH. Understanding the mechanism of action of the anti-dandruff agent zinc pyrithione against Malassezia restricta. Sci. Rep. 2018;8:12086. doi: 10.1038/s41598-018-30588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ianiri G, Averette AF, Kingsbury JM, Heitman J, Idnurm A. Gene function analysis in the ubiquitous human commensal and pathogen Malassezia genus. mBio. 2016;7:e01853–16. doi: 10.1128/mBio.01853-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Celis AM, Vos AM, Triana S, Medina CA, Escobar N, Restrepo S, et al. Highly efficient transformation system for Malassezia furfur and Malassezia pachydermatis using Agrobacterium tumefaciens-mediated transformation. J. Microbiol. Methods. 2017;134:1–6. doi: 10.1016/j.mimet.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 81.Gioti A, Nystedt B, Li W, Xu J, Andersson A, Averette AF, et al. Genomic insights into the atopic eczema-associated skin commensal yeast Malassezia sympodialis. mBio. 2013;4:e00572–00512. doi: 10.1128/mBio.00572-12. [DOI] [PMC free article] [PubMed] [Google Scholar]