Abstract
The rhizosheath is a belowground area that acts as a communication hub at the root–soil interface to promote water and nutrient acquisition. Certain crops, such as white lupin (Lupinus albus), acquire large amounts of phosphorus (P), owing partially to exudation of acid phosphatases (APases). Plant growth-promoting rhizobacteria also increase soil P availability. However, potential synergistic effects of root APases and rhizosheath-associated microbiota on P acquisition require further research. In this study, we investigated the roles of root purple APases (PAPs) and plant growth-promoting rhizobacteria in rhizosheath formation and P acquisition under conditions of soil drying (SD) and P treatment (+P: soil with P fertilizer; –P: soil without fertilizer). We expressed purple acid phosphatase12 (LaPAP12) in white lupin and rice (Oryza sativa) plants and analyzed the rhizosheath-associated microbiome. Increased or heterologous LaPAP12 expression promoted APase activity and rhizosheath formation, resulting in increased P acquisition mainly under SD–P conditions. It also increased the abundance of members of the genus Bacillus in the rhizosheath-associated microbial communities of white lupin and rice. We isolated a phosphate-solubilizing, auxin-producing Bacillus megaterium strain from the rhizosheath of white lupin and used this to inoculate white lupin and rice plants. Inoculation promoted rhizosheath formation and P acquisition, especially in plants with increased LaPAP12 expression and under SD–P conditions, suggesting a functional role of the bacteria in alleviating P deficit stress via rhizosheath formation. Together, our results suggest a synergistic enhancing effect of LaPAP12 and plant growth-promoting rhizobacteria on rhizosheath formation and P acquisition under SD–P conditions.
Phosphorus-responsive PURPLE ACID PHOSPHATASE12 and auxin-producing Bacillus spp. promote rhizosheath formation in Lupinus albus under soil drying conditions, enhancing phosphorus acquisition.
Introduction
Plants grow under diverse environmental conditions, with varying levels of soil nutrients and water availability (Castrillo et al., 2017). Phosphorus (P) is present in soils in different forms, including phosphoesters, phytates, and inorganic orthophosphate (Pi). P is required for both plant growth and microbial proliferation, and Pi is a plant-growth-limiting factor affecting crop productivity (Lynch, 2007; Rose et al., 2013; Zhu et al., 2016). To avoid soil P deficiency, phosphate fertilizer is commonly applied in agricultural cropping systems. However, only up to 80% of the bound P (as iron and aluminum oxides and hydroxides in acid soils, and calcium oxide and hydroxide in alkaline soils) (Richardson et al., 2001; Richardson and Simpson, 2011) in soil might be available for plant uptake. Plants have thus evolved several soil Pi-acquisition strategies (Honvault et al., 2021), such as structural modifications of their roots, organic acid and phosphatase exudation, and beneficial associations with microbes (Lambers et al., 2008; Harrison, 2012; Hacker et al., 2015; Lambers et al., 2015; Hiruma et al., 2016; Sasse et al., 2018).
White lupin (Lupinus albus) is an emerging model crop that responds to P status via structural and functional root modifications, in the form of cluster roots (CRs) (Xu et al., 2020; Aslam et al., 2021). CR are bottlebrush-like structures composed of hundreds of small rootlets specializing in the mobilization and uptake of Pi and other nutrients in impoverished soils (Pueyo et al., 2021), such as by the release of large amounts of organic acids and phosphatases under low P (Wasaki et al., 2009). CR formation is also induced by Fe deficiency and Al toxicity, with the exudates of these roots playing an important role in toxic metal immobilization both in the root and rhizosphere (Quiñones et al., 2021, 2022). Plants with CR are much more efficient in terms of Pi absorption than non-CR plants (Uhde-Stone, 2017). Acid phosphatases (APases) are ubiquitous enzymes (Bhadouria and Giri, 2021) in root exudates that are able to hydrolyze organic phosphate (Po) to release Pi (Tran et al., 2010b; Plaxton and Tran, 2011). Intracellular and extracellular APases can remobilize, recycle, and hydrolyze Po (Ticconi and Abel, 2004; Richardson et al., 2009). Purple acid phosphatases (PAPs) are members of the APase family and their ability to increase P use efficiency under P starvation conditions has been reported (Xie and Shang, 2018). Emerging evidence suggests that the PAPs of many plant species are induced by Pi deprivation (Tran et al., 2010a; Wang et al., 2014; Deng et al., 2020). Genetic manipulation of APase-encoding genes enhanced Pi acquisition from soil Po sources (Ticconi and Abel, 2004; Xiao et al., 2006b; Li et al., 2011). The heterologous expression of MtPAP1 in Arabidopsis (Arabidopsis thaliana) resulted in 4.6- to 9.9-fold higher APase activity, which substantially improved organic P acquisition and plant biomass (Xiao et al., 2006a). This is consistent with the enhanced P-acquisition efficiency associated with AtPAP15 expression in soybean (Glycine max) (Wang et al., 2009). Tran et al. (2010a) found that AtPAP12 and AtPAP26 are predominantly root-secreted APases that scavenge Pi from extracellular phosphoesters, with AtPAP26 also acting as a dual-targeted intracellular APase involved in vacuolar Pi recycling in P-starved A. thaliana. Similarly, the overexpression of PvPAP3 substantially enhanced Pi uptake in common bean (Phaseolus vulgaris), indicating a role for PAPs in enhancing P utilization efficiency (Liang et al., 2010). Collectively, the data show that –P promotes the secretion of APases from roots, which is imperative for enhancing Pi acquisition from soil Po sources.
Root exudates are also key for the establishment and modulation of the associations between roots and the microbial community (Sweeney et al., 2020; Honvault et al., 2021; Tian et al., 2021). The rhizosheath is a dynamic, subterranean root environment that acts as a communication hub between roots and microorganisms at the root–soil interface. It is thus essential for nutrient exchange, particularly under conditions of P scarcity (Haling et al., 2013; James et al., 2016), and is a key drought tolerance-associated trait; for instance, ethylene-dependent root–microbe associations trigger rhizosheath formation, particularly under conditions of moderate soil drying (SD) (Zhang et al., 2020b). The rhizosheath also plays a pivotal role in other abiotic stresses, including nutrient deficiency and soil acidity (Lynch and de Leij, 2012; Rabbi et al., 2018; Zhang et al., 2020b).
Kalayu (2019) reported that up to 50% of rhizosheath-associated bacteria are P-solubilizers that strongly promote Pi acquisition (Menezes-Blackburn et al., 2018), thereby improving plant health. Honvault et al. (2021) reported that malate and malonate exudation from roots corresponded positively with altered plant P content and rhizosheath P concentrations. Similarly, carboxylate exudation is thought to play a key role in root–rhizosheath–microbe interactions, by scavenging P from the soil surrounding the roots. Several species of root-colonizing rhizosphere bacteria, including members of Azospirillum, Bacillus, Pseudomonas, and Rhizobium, have a similar mechanism of auxin production; their root colonization stimulates plant root growth and branching, thereby enhancing nutrient uptake (Spaepen and Vanderleyden, 2011; Talboys et al., 2014; Liu et al., 2016). For instance, Bacillus methylotrophicus extracted from the maize (Zea mays) rhizosphere produces bioactive volatiles that increase auxin synthesis and reprogram root architecture (Pérez-Flores et al., 2017). We recently provided evidence that, under moderate SD, Piriformospora indica and Bacillus cereus promote rice (Oryza sativa) rhizosheath formation through root auxin modulation (Xu et al., 2021). The co-inoculation of Bacillus megaterium and Rhizobium tropici substantially increased plant biomass and P uptake in P. vulgaris (Korir et al., 2017), as also demonstrated in Z. mays (Vinci et al., 2018) and cucumber (Cucumis sativus) (Liu et al., 2016) inoculated with Bacillus amyloliquefaciens. These findings indicate a major contribution of root-associated bacteria to auxin biosynthesis and root growth.
In earlier work, we showed that APases are important for Pi acquisition in white lupin (Xu et al., 2020). Here, we investigated the involvement of APases and the bacterial community in the rhizosheath in P acquisition under SD and –P conditions, by heterologously expressing LaPAP12 (encoding a white lupin PAPase) in white lupin and rice. We also analyzed the composition of the rhizosheath bacterial community to identify bacteria associated with Pi acquisition. The effects of inoculation with a rhizosheath-associated B. megaterium strain on root growth, rhizosheath formation, and P acquisition were also determined. Our results indicate an important role of enhanced APase secretion in rhizosheath formation and bacterial attraction, and that both APases and rhizosheath bacteria promote P acquisition under SD–P conditions.
Results
Characterization of the white lupin PAP gene family reveals strong induction of LaPAP12 expression under P deficiency
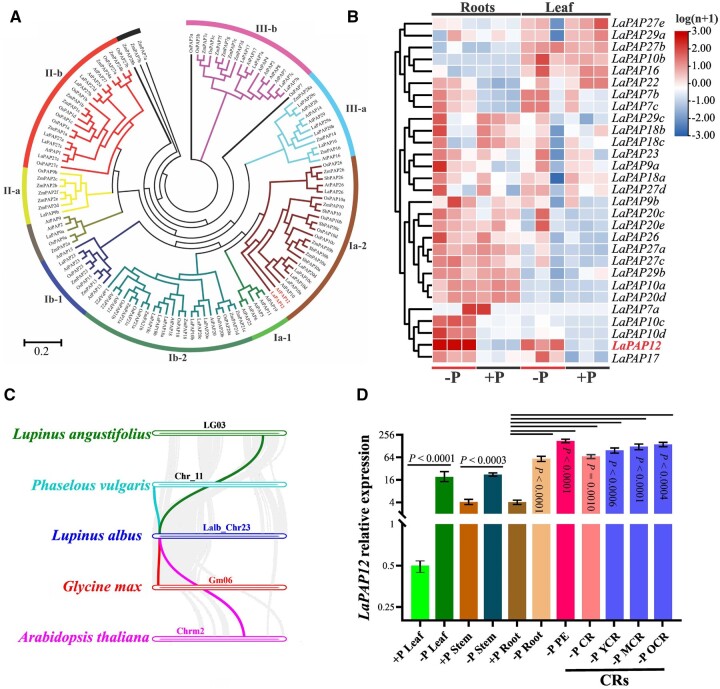
In a previous study (Xu et al., 2020), we showed that PAPs play an important role in plant Pi absorption. In the present study, A. thaliana PAP protein sequences served as the query in a search against the whole white lupin genome, which revealed 29 putative LaPAP genes. In accordance with a previous classification of A. thaliana PAP genes (Li et al., 2002), the 29 LaPAPs were categorized into three distinct groups (I–III) and eight subgroups (Ia-1, Ia-2, Ib-1, Ib-2, IIa, IIb, IIIa, and IIIb). A phylogenetic tree including L. albus, A. thaliana, O. sativa, sorghum (Sorghum bicolor), and Z. mays PAPs is presented in Figure 1A. LaPAPs were named based on their closest A. thaliana orthologs (protein sequence similarity 68.59%), and the genes within each clade were assigned lowercase letters (a–d). The LaPAP12 gene belongs to the Ia-2 subgroup, which also includes LaPAP10 and LaPAP26, and has high sequence identity to AtPAP12. The tissue-specific expression patterns of the 29 LaPAPs in roots and leaves were investigated under –P and +P conditions by RT-qPCR. The results revealed substantially higher expression of LaPAP12 in both roots and leaves under –P than +P conditions (Figure 1B). Moreover, a comparative syntenic analysis showed that LaPAP12 has a common origin with three related legume species (Lupinus angustifolius, G. max, and P. vulgaris) and one outgroup species (A. thaliana) (Figure 1C). Analysis of the transcriptional response of LaPAP12 in different tissues showed that expression was significantly higher in the CR pre-emergence zone, and in old CRs (OCR), than in other plant tissues under –P conditions (Figure 1D). Taken together, these finding suggested that LaPAP12 is a candidate gene to enhance P use efficiency.
Figure 1.
Identification and characterization of white lupin PAPs. A, Phylogenetic analysis of PAPs in L. albus, A. thaliana, Z. mays, and O. sativa. The PAPs were aligned to generate an unrooted maximum-likelihood phylogenetic tree using IQ-TREE software, with 1,000 bootstrap replicates. B, Heatmap of the expression of white lupin PAPs in leaf and root grown under +P (soil with P fertilizer) and –P (soil without P fertilizer) conditions, based on the results of RT-qPCR. C, Results of a co-evolutionary analysis of PAP12 genes in L. albus, L. angustifolius, P. vulgaris, G. max, and A. thaliana, shown by the colored lines. D, Tissue/organ-specific expression of LaPAP12 in the leaf, stem, root, CR pre-emergence zone (PE), CRs, young CR (YCR), mature CR (MCR), and OCR of lupin grown under +P and–P hydroponic conditions. For two soil-P treatments: +P is 142.6 ppm (100 ppm P from P fertilizer + 42.6 ppm soil Olsen-P) and −P is 42.6 ppm (soil Olsen-P).
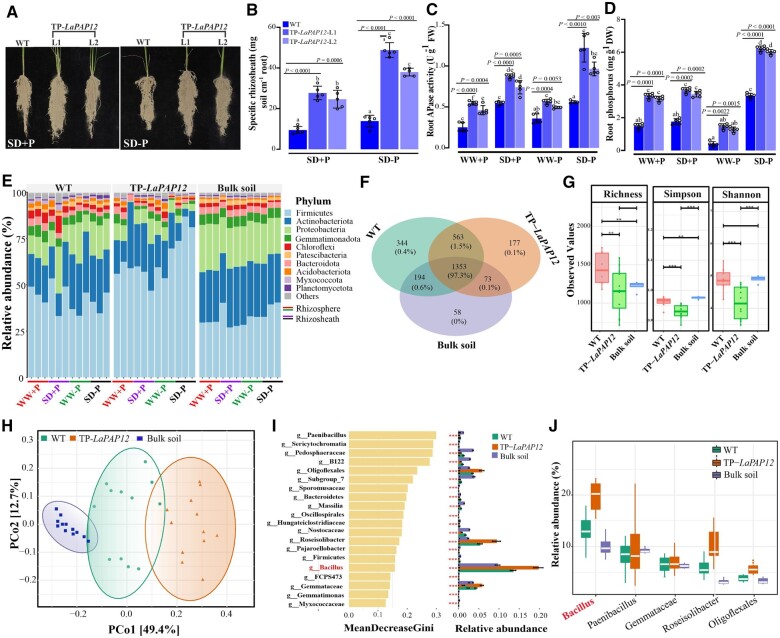
Bacillus is the dominant bacterial genus associated with the white lupin rhizosheath
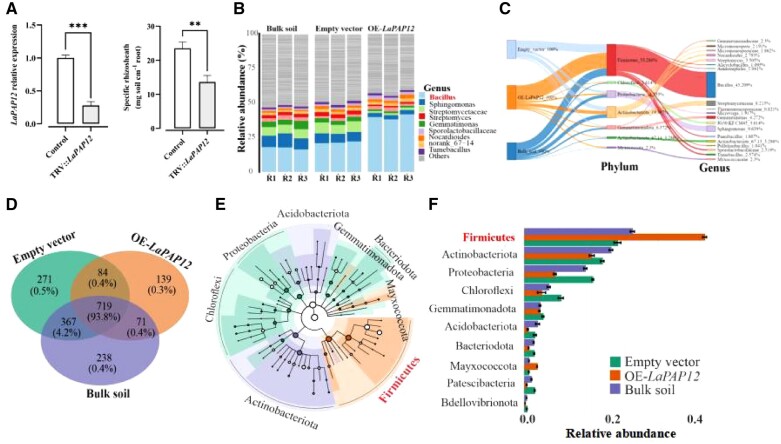
To investigate the role of LaPAP12, knockdown plants of LaPAP12 were generated with virus-induced gene silencing (VIGS). The knockdown plants of LaPAP12 (TRV::LaPAP12), which show reduced PAP12 gene expression (Figure 2A) and root APase activity (Supplemental Figure S1a), have decreased rhizosheath formation (Supplemental Figure S2, a and b) and root P concentration (Supplemental Figure S1b) when compared with vector control. Moreover, white lupin transgenic hairy roots overexpressing LaPAP12 (OE-LaPAP12) and transgenic rice plants heterologously expressing LaPAP12 were also generated. When transferred into soil, hydroponically grown transgenic hairy roots OE-LaPAP12 (Supplemental Figure S2a) exhibited improved root growth, expressed as total root length (Supplemental Figure S3b), and increased rhizosheath formation (Supplemental Figure S3c), compared with hairy roots transformed with empty vector (EV). In addition, root APase activity and P content were significantly higher in OE-LaPAP12 than EV roots (Supplemental Figures S3, d and e). The composition of the bacterial community associated with rhizosheath formation in white lupin transgenic hairy root OE-LaPAP12 was analyzed by subjecting the rhizosheath and bulk soil to a 16S microbial sequencing analysis.
Figure 2.
Composition of the bacterial community associated with rhizosheath formation in white lupin transgenic hairy root (OE-LaPAP12). A, LaPAP12 expression and Rhizosheath formation in TRV::LaPAP12 and the EV control. Data are means ± standard error (n = 3), the significant differences are indicated **P < 0.01 and ***P < 0.001, unpaired two-tailed Student’s t test. B, Relative abundance (%) of bacterial taxa in the rhizosheath soil of OE-LaPAP12 and EV, and in bulk soil. R1–R3 represent three replicates in each group. C, Sankey diagram of the proportion of bacteria, from the phylum to the genus level. D, Venn diagram of bacterial OTUs in the rhizosheath soil of OE-LaPAP12 and EV, and bulk soil. E, Cladogram showing the differentially expressed taxa (small circles) as different shades (according to LEfSe) in the rhizosheath soil of OE-LaPAP12, EV and in bulk soil. Three replicates of OE-LaPAP12-L1 were subjected to 16S microbiome sequencing analysis. F, Relative abundance of different bacteria in OE-LaPAP12, EV rhizosheath, or bulk soil.
Bacillus was common to all soils and was the dominant genus, accounting for >80% of all bacterial sequences. The relative abundance of Bacillus was higher in the rhizosheath of OE-LaPAP12 roots than in the rhizosheath of EV roots or bulk soil. Other well-represented taxa included Sphingomonas and Streptomycetaceae, which were less abundant in OE-LaPAP12 than in EV rhizosheath (Figure 2B). An interactive Sankey diagram at the phylum to genus levels showed that, in the OE-LaPAP12 rhizosheath, Firmicutes was most abundant at the phylum level (53%) and Bacillus at the genus level (43%) (Figure 2C). To determine common and distinct bacterial operational taxonomic units (OTUs), a Venn diagram was generated, which revealed 719 core OTUs (139, 217, and 238 unique OTUs in OE-LaPAP12 rhizosheath, EV rhizosheath, and bulk soil, respectively). In addition, EV rhizosheath shared 84 OTUs with OE-LaPAP12 rhizosheath and 367 with bulk soil, while OE-LaPAP12 rhizosheath shared 71 OTUs with bulk soil (Figure 2D). A cladogram further illustrated the prevalence of the Firmicutes phylum and Bacillus genus in OE-LaPAP12 rhizosheath, while different phyla and genera were associated with EV rhizosheath and bulk soil (Figure 2E). A similar trend can be seen in Figure 2F, with Firmicutes being more abundant, and Actinobacteria and Proteobacteria less abundant, in OE-LaPAP12 than EV rhizosheath, or bulk soil. In summary, the relative abundance of the associated bacterial communities was clearly different for each soil.
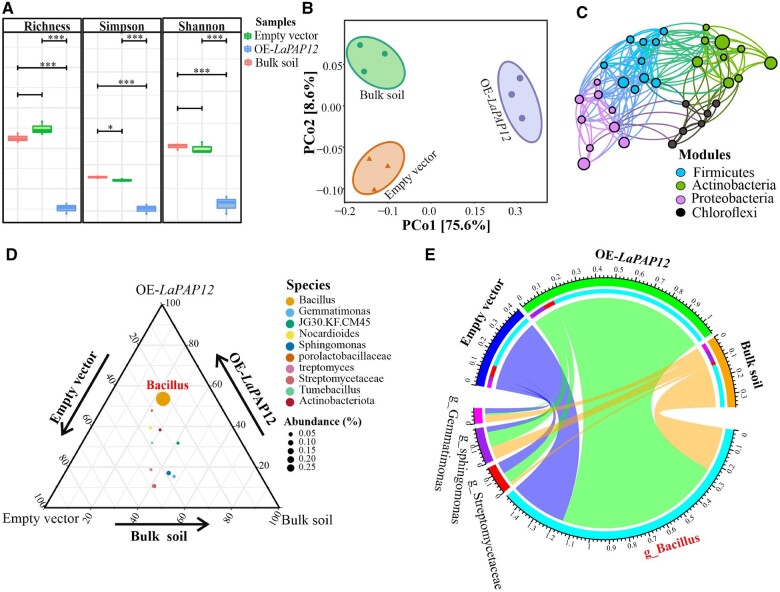
Alpha-bacterial diversity, analyzed using the richness, Simpson, and Shannon indices, differed significantly in the EV rhizosheath and bulk soil compared with the OE-LaPAP12 rhizosheath (Figure 3A). A principal coordinate analysis (PCoA) distinguished the bacterial communities of the three different soils. Based on unweighted and weighted UniFrac matrices, the bacterial community compositions of OE-LaPAP12, EV, and bulk soils differed substantially from each other (Figure 3B). Analysis of the distribution network of highly abundant phyla showed a higher proportion of OTUs in bulk soils (Figure 3C). Ternary analysis revealed that, compared with both EV and bulk soil, OE-LaPAP12 soil harbored a distinctive bacterial community, in which Bacillus was the most abundant genus (Figure 3D). By contrast, EV and bulk soil consisted of other mixed bacterial communities with lower relative abundances than Bacillus. In a Circos plot of the top four bacterial genera, Bacillus predominated over the other three genera in OE-LaPAP12 soil (Figure 3E). Taken together, these results show that Bacillus was the predominant bacterial genus in all three soils, but was substantially more abundant in OE-LaPAP12 hairy root rhizosheath.
Figure 3.
Bacillus is a highly dominant bacterial community in white lupin OE-LaPAP12-L1 hairy root rhizosheath. A, Alpha-diversity indices. B, PCoA showing similarities and dissimilarities among groups. C, Cooccurrence-based network analysis of the top four bacterial taxa at the phylum level. D, Ternary analysis of the bacterial community in the rhizosheath soil of EV and OE-LaPAP12, and in bulk soil. E, Circos plot of the differentially abundant taxa in the rhizosheath soil of EV, OE-LaPAP12 and in bulk soil. Three replicates of each sample were used. OE-LaPAP12-L1 replicates were subjected to 16S microbiome sequencing analysis.
Inoculation of white lupin with a rhizosheath-associated B. megaterium strain promotes rhizosheath formation and P acquisition
To examine the role of B. megaterium in rhizosheath formation and P acquisition, a phosphate-solubilizing strain was isolated from white lupin rhizosheath. When incubated on Pikovskaya’s medium supplemented with Ca3(PO4)2 as the sole source of phosphate, this strain produced a clear halo zone, indicating its phosphate-solubilizing activity (Supplemental Figure S4 and Supplemental Table S1). In addition, the B. megaterium strain was able to produce indole-3-acetic acid (IAA) in LB medium alone, with the highest production level occurring in LB medium supplemented with l-tryptophan (Supplemental Table S2). The IAA concentration declined after 96 h, perhaps due to the intracellular storage or degradation of IAA by bacterially secreted IAA oxidase (Ji et al., 2012).
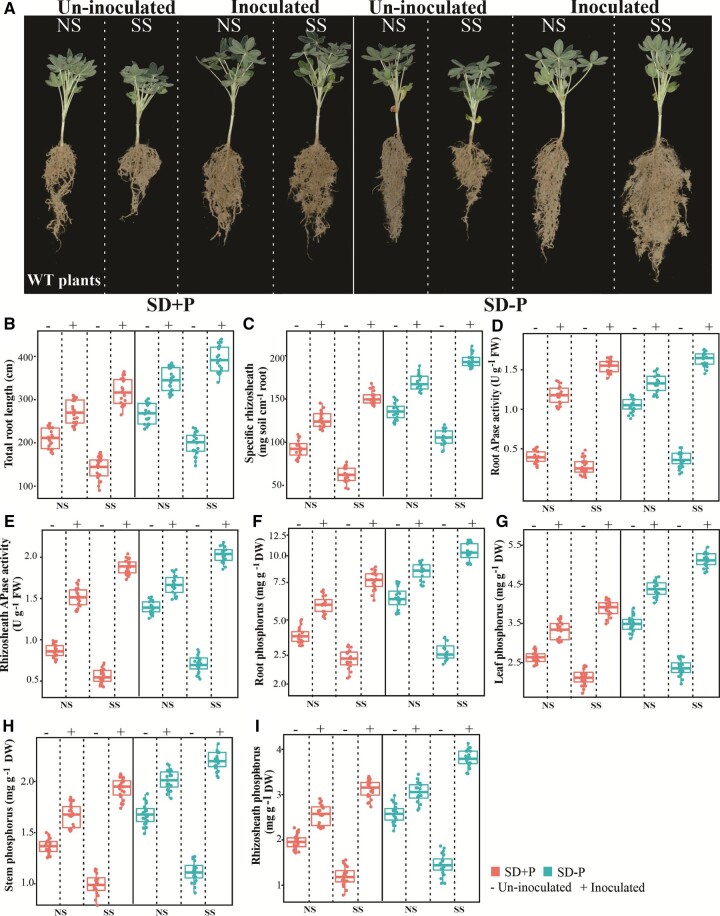
The inoculation of white lupin with the isolated B. megaterium strain had profound effects on plant growth under both SD+P and SD–P conditions, as root growth and rhizosheath formation were substantially higher than in uninoculated plants in both natural soil (NS) and sterilized soil (SS) (Figure 4A). Inoculation with B. megaterium increased total root length (Figure 4B) and specific rhizosheath formation (Figure 4C) in NS, and especially in SS. In both soils, white lupin had higher APase activity in roots and rhizosheath soil under SD–P than SD+P conditions (Figure 4, D and E). Root APase and rhizosheath APase activities were higher in inoculated plants, and the increases were more marked in plants grown in SS than NS (Figure 4, D and E). Compared with uninoculated plants, the P contents in roots, leaves, and stems were higher in B. megaterium = inoculated plants (Figure 4, G–H), and more P accumulated in the rhizosheath (Figure 4I) under both SD+P and SD–P conditions, and in both soils.
Figure 4.
Effect of Bacillus inoculation on rhizosheath formation in WT white lupin, and P acquisition under SD. A, Phenotypic characteristics of WT white lupin grown under SD+P and SD–P. B, Total root length (cm). C, Specific rhizosheath soil DW (mg soil cm−1 root). D, Root APase activity (U g−1 FW). E, APase activity in rhizosheath soil (U g−1 FW). F, Root P concentration (mg g−1 DW). G, Rhizosheath P concentration (mg g−1 DW). H, Leaf P content (mg g−1 DW). I, Stem P content (mg g−1 DW). Data are mean ± standard error (n = 20 replicates). Uninoculated, soil without inoculated B. megaterium; inoculated, soil inoculated with B. megaterium.
APase-mediated rhizosheath formation enhances P acquisition in rice
Insertion of the LaPAP12 into the rice genome did not result in phenotypic differences among the EV, wild-type (WT), and transgenic (TP) lines (Supplemental Figure S5a). The transcript level of LaPAP12 was significantly increased in TP lines (Supplemental Figure S5b). In rice, rhizosheath formation was observed only under SD conditions (Figure 5A). The relative expression of LaPAP12 in TP-LaPAP12 lines was significantly upregulated (Supplemental Figure S5c) under SD conditions, with more significant increases seen under SD–P than well-watered (WW)+P or WW–P conditions.
Figure 5.
Rhizosheath APase activity and Bacillus, as the dominant bacterial community in the rhizosheath of the rice line TP-LaPAP12-L1, facilitate P acquisition. A, Phenotypes of rice plants heterologously expressing LaPAP12 under SD+P and SD–P conditions. B, Specific rhizosheath soil DW (mg soil cm−1 root) under SD+P and SD–P. C, Root APase activity. D, Root P concentration in response to WW and SD conditions, in the presence or absence of sufficient P (+P/–P) in WT rice and lines expressing LaPAP12. E, Relative abundance (%) of the bacterial community, determined at the phylum level. Three replicates of each group were subjected to 16S microbiome sequencing. F, Venn diagrams showing the number of common and unique OTUs in WT and TP-LaPAP12, and in bulk soil. G, Alpha-diversity, measured by the richness, Simpson, and Shannon diversity indices, in the rhizosheath soil of WT and LaPAP12, and in bulk soil. H, PCoA plot showing the microbiota compositions in the rhizosheath soil of WT and LaPAP12, and in bulk soil. I, Mean decrease in the Gini index in rhizosheath soil of WT and LaPAP12, and in bulk soil. The abscissa on the left is the mean decrease in Gini index, and the bar plot on the right is the relative abundance (***P < 0.001). J, Box plot showing the relative abundances of the top five genera. Measurements were made in five independent biological replicates; error bars represent the standard error. Different letters at the top of the bars indicate significant differences among LaPAP12 overexpression lines and WT under different treatments. WW+P, WW with sufficient P; SD+P, SD with P fertilizer; WW–P, WW without fertilizer; SD–P, SD without fertilizer; and TP-LaPAP12-L1, LaPAP12-overexpressing line 1.
Under SD+P and SD–P conditions, rhizosheath development was higher in transgenic TP-LaPAP12 rice plants than WT plants, and was highest under SD–P (Figure 5B). Root and rhizosheath APase activities in the TP-LaPAP12 lines were significantly higher than in WT plants under all treatments. Additionally, root APase and rhizosheath activities were enhanced in SD+P/SD–P compared with WW+P/WW–P conditions, with the highest activities occurring in the TP-LaPAP12 lines under SD+P conditions (Figure 5C and Supplemental Figure S6b). Root P content was also highest in transgenic plants under SD–P conditions (Figure 5D), which might be a result of enhanced Pi uptake by increased rhizosheath formation and root APase activity (Figure 5, B and C). The same pattern was observed for shoot and rhizosheath P contents, and for the growth parameters root length and total plant dry weight (DW) (Supplemental Figure S6, a–e).
Bacillus is the predominant genus in the rhizosheath-associated bacterial community of TP-LaPAP12 rice
The composition of the bacterial community associated with rhizosheath formation in TP-LaPAP12 rice was also analyzed, based on 1,321,572 sequences. The average length of the sequence reads was 419 bp. The sequences clustered into 855 bacterial OTUs, corresponding to 338 genera. In total, 22 phyla, 58 classes, 124 orders, 201 families, 338 genera, and 506 species were obtained. In total, 2,762 OTUs were identified from the WT, TP-LaPAP12, and bulk soil samples (Supplemental Figure S7). The relatively high number of OTUs indicates high species richness in soil. Overall, the rhizosheath of WT and TP-LaPAP12 plants, and the bulk soil, were dominated by three bacterial phyla, Firmicutes, Actinobacteria, and Proteobacteria, whose relative abundances varied under SD−P conditions. Among the top 10 bacterial phyla, Firmicutes was more abundant in the rhizosheath of TP-LaPAP12 than in that of WT (80%, t test, P = 0.001). The relative abundance of Firmicutes was greater under SD–P (80%) than SD+P (55%) conditions (Figure 5E). A Venn diagram showed that 1,353 core OTUs (97.3%) were shared by the rhizosheaths of WT, TP-LaPAP12, and bulk soil, which possessed 344 (0.4%), 177 (0.1%), and 58 (0%) unique OTUs, respectively. WT and TP-LaPAP12 rhizosheaths shared 563 (1.5%) OTUs, WT rhizosheaths and bulk soil shared 194 (0.6%) OTUs, and TP-LaPAP12 rhizosheaths and bulk soil 73 (0.1%) shared OTUs (Figure 5F).
Alpha-diversity was highest in the bacterial community of the WT soil, followed by that of bulk soil and then OE-LaPAP12 soil (Figure 5G). The richness diversity index of bulk soil was higher than that of TP-LaPAP12 soil, although the difference was not statistically significant (P > 0.05). The Shannon indexes of bulk soil and WT rhizosheath/rhizosphere were similar, and significantly higher than that of TP-LaPAP12 rhizosheath (Figure 5G). The PCoA revealed that the composition of the bacterial community in TP-LaPAP12 soil was distinct from that in WT and bulk soil, indicating that LaPAP12 expression, and thus LaPAP12 presence/activity in the root and rhizosheath, had a substantial effect on bacterial community composition. The first two PCoA components accounted for 49.4% and 12.7% of the variance (Figure 5H). TP-LaPAP12 soil was significantly richer in g_Bacillus than in WT and bulk soils, while g_Pedosphaeraceae, g_Massilia, and g_Nostocaceae were abundant in bulk soil only (Figure 5I). Among the top five genera, Bacillus predominated in all soils, but especially in TP-LaPAP12 soil compared with WT and bulk soils (P < 0.001, Figure 5J).
The Sankey plot revealed a distinct difference in bacterial community composition at the genus level among the WT, TP-LaPAP12, and bulk soils. The TP-LaPAP12 soil was enriched in bacteria of the Bacillaceae family, which includes the Bacillus genus (Supplemental Figure S8a). A phylum-based network analysis of the five most abundant phyla (Firmicutes, Proteobacteria, Actinobacteria, Gemmatimonadota, and Chloroflexi) showed that they were strongly positively correlated (Supplemental Figure S8b). The first two axes of the redundancy analysis together explained 94.9% of the total variance in microbial distribution. The rhizosheath of TP-LaPAP12 accounted for the largest amount of variance in the soil microbial community, followed by that of WT and then bulk soils (Supplemental Figure S8c). The proportion of common bacterial taxa among the rhizosheaths of both rice genotypes and bulk soil was highly variable. A ternary analysis revealed substantial variation in bacterial taxa at the phylum level for both rice genotypes (WT and TP-LaPAP12) (Supplemental Figure S8d).
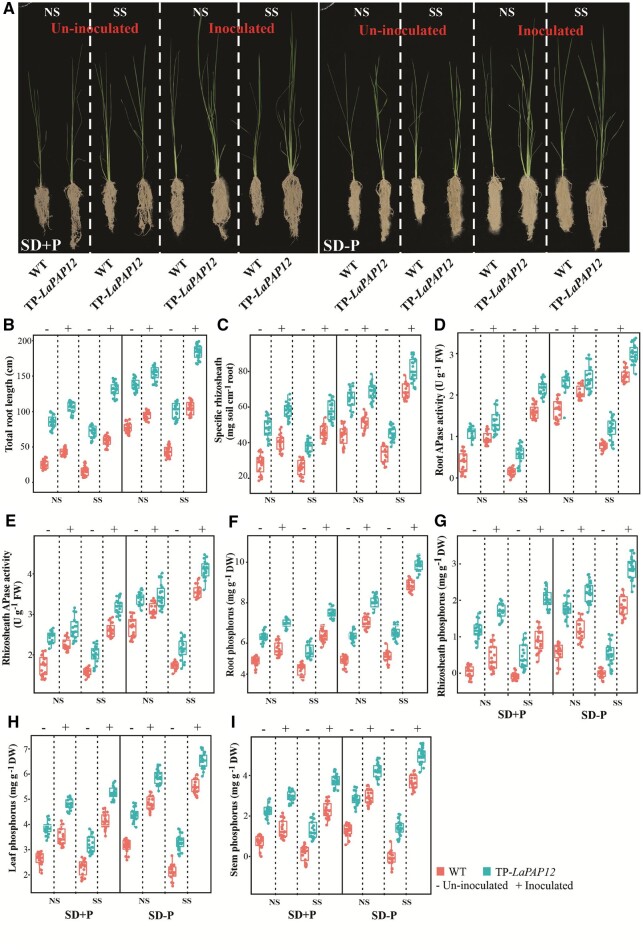
Inoculation of rice with B. megaterium promotes rhizosheath formation and P acquisition under SD and P-deficiency conditions
To investigate whether inoculation with B. megaterium affects rhizosheath formation and P acquisition in rice, WT and TP-LaPAP12 rice were inoculated with the selected strain in both NS and SS soils under SD+P and SD–P conditions. Inoculation substantially promoted rhizosheath formation in TP-LaPAP12 plants, particularly under SD–P conditions (Figure 6A). TP-LaPAP12 roots were longer than WT roots regardless of inoculation and soil treatments. The effects of the P level on total root length were substantial, with a substantially larger increase seen under SD–P than SD+P conditions in both NS and SS. In addition, B. megaterium inoculation of WT and TP-LaPAP12, under both SD treatments and in both soils, substantially increased total root length, especially in SS (Figure 6B). Similarly, the specific rhizosheath weight increased upon inoculation of TP-LaPAP12 rice with B. megaterium in NS, with an even larger increase seen in SS (Figure 6C). Similar trends were observed for related parameters, including APase activities in roots and rhizosheaths (Figure 6, D and E), root P content (Figure 6F), rhizosheath P concentration (Figure 6G), and leaf and stem P contents (Figure 6, H and I).
Figure 6.
Bacillus inoculation increases rice rhizosheath formation in association with P acquisition under SD−P conditions. A, Phenotypic characteristics of WT and TP-LaPAP12 rice. B, Total root length (cm). C, Specific rhizosheath weight (mg soil cm−1 root). D, Root APase activity (U g−1 FW). E, Rhizosheath APase activity (U g−1 FW). F, Root P content (mg g−1 DW). G, Rhizosheath P concentration (mg g−1 DW). H, Leaf P content (mg g−1 DW). I, Stem P content (mg g−1 DW). Data are mean ± standard error (n = 20). Uninoculated, soil without inoculated B. megaterium; inoculated, soil inoculated with B. megaterium.
Discussion
White lupin PAP LaPAP12 is important for rhizosheath formation and P acquisition
Plants deploy an array of adaptive strategies to cope with P deprivation, including root architecture modification (Wang et al., 2010) and APases secretion (Baldwin et al., 2001). PAPs are key players in P acquisition, which is achieved through the hydrolysis of phytate, a major source of Po (Tran et al., 2010b; Robinson et al., 2012). Low P triggers extracellular APase secretion, which converts P0 into the Pi required for plant growth and development (Tran et al., 2010b). The role of root-secreted PAPs has not yet been characterized in white lupin under SD and –P conditions. In this work, we identified a white lupin PAP gene, LaPAP12, which was substantially induced under –P, suggesting a role in increasing Pi availability. LaPAP12 overexpression promoted root APase activity and secretion, increased root growth, and substantially enhanced rhizosheath formation under SD conditions, thus assisting P acquisition.
LaPAP12, a white lupin homolog of Arabidopsis AtPAP12, is part of a large family of PAPs that include several subfamilies. LaPAP12 belongs to subgroup-Ia, which is a major secretory group (Lu et al., 2016; Mehra et al., 2017). PAPs in this group play a critical role in the mobilization of extracellular P from Po sources (Lu et al., 2016). Rice OsPAP10c, another member of subgroup-Ia, is also strongly induced by Pi deficiency, suggesting its importance in external Po utilization (Lu et al., 2016). Secretory PAPs hydrolyze Po, which leads to improved plant growth. Both SD and root exudation facilitate rhizosheath formation (Albalasmeh and Ghezzehei, 2014), thus contributing to water and nutrient availability (Aslam et al., 2021; Karanja et al., 2021). In a previous study, we showed that white lupin develops a substantial rhizosheath under SD conditions (Aslam et al., 2021), which can be considered an adaptive trait of the plant to withstand water deficit. Here, we found that white lupin transgenic hairy roots OE-LaPAP12 increased rhizosheath formation under SD conditions, by increasing total root length and root APase activity. In these plants, P acquisition was notably enhanced under –P. Few studies have examined the genetics of rhizosheath formation (George et al., 2014). Several traits are related to rhizosheath formation, including root length, root hairs (Carminati et al., 2017; Kohli et al., 2021), CR (Aslam et al., 2021), root exudation (Albalasmeh and Ghezzehei, 2014; Carminati et al., 2017), microbial activities (Hanna et al., 2013; Zhang et al., 2020b), and soil texture (Haling et al., 2010). We recently reported that enhanced rhizosheath formation in white lupin is associated with increased P acquisition (Aslam et al., 2021), concomitant with CR formation under SD–P conditions. To further investigate the role of white lupin LaPAP12 in rhizosheath formation, we chose rice as an outgroup species due to its ability to develop a rhizosheath under moderate SD (Zhang et al., 2020a). The overexpression of LaPAP12 in rice remarkably increased root APase activity and promoted rhizosheath formation under SD–P conditions. These findings suggest that enhanced P acquisition in LaPAP12-overexpressing lines is associated with increased rhizosheath formation, which in part can be attributed to increased APase activity and root growth.
Our results also showed that the expression levels of the LaPAP12 gene in TP rice lines were significantly induced under SD–P (Supplemental Figure S5c), suggesting the involvement of this gene in –P responses under SD, as well as in the regulation of P acquisition and solubilization. Our analysis showed that LaPAP12 was more highly expressed in TP lines under SD than WW conditions. This result is consistent with previous studies demonstrating the need for root-secreted APases for plant growth and development in several crop species (Richardson, 2009; Tran et al., 2010a, 2010b; Plaxton and Tran, 2011; Wang et al., 2011; Robinson et al., 2012; Wang and Liu, 2018). For instance, overexpression of GmPAP4, a PAP gene with high phytase activity, resulted in enhanced APase activity and considerable translocation of root P to shoots in A. thaliana (Kong et al., 2014).
The overexpression of OsPAP21b leads to increased APase activity and P accumulation in transgenic rice, both in hydroponic culture and in soil supplemented with an organic P source (Mehra et al., 2017). Based on previous work, we hypothesized that the formation of a larger rhizosheath is associated with greater root APase activity and significantly enhances P acquisition in both white lupin and rice. Consistent with our previous study (Aslam et al., 2021), here we showed that, under SD–P conditions, rhizosheath formation is substantially enhanced in transgenic rice heterologously expressing LaPAP12 compared with WT plants, with the former being characterized by enhanced root and shoot P contents, as well as a higher rhizosheath P concentration. The direct link between plant P status and rhizosheath mass suggests the importance of rhizosheath formation in P acquisition.
LaPAP12 overexpression and B. megaterium inoculation synergistically enhance rhizosheath formation and P acquisition
Plants release a complex mixture of root exudates to attract beneficial soil bacteria, which in turn use root exudates as their energy source (el Zahar Haichar et al., 2008; Ndour et al., 2017). Approximately 20% of photosynthetic carbon is released into the soil, where it fuels the plant–bacterial interactions that support plant growth by increasing nutrient availability (Bais et al., 2006); this allows for plant survival in response to environmental stresses (Paterson and Mwafulirwa, 2021). Studying the role of LaPAP12 in APase secretion in association with soil bacterial communities can provide insight into possible synergic effects, that is, the stimulation of rhizosheath formation and P acquisition under SD conditions. Previous studies showed that several species of bacteria in the Bacillus genus, including B. megaterium, are plant growth-promoting rhizobacteria having a positive effect on root growth and enhancing tolerance to several abiotic stresses (Chinnaswamy et al., 2018; Wei et al., 2019). Beneficial bacteria, such as Bacillus, Paenibacillus, Staphylococcus, and Brevibacterium, exhibit plant growth-promoting activities, including phosphate solubilization and IAA production (Yahaghi et al., 2018). Chen and Liu (2019) reported that Pantoea sp. S32, isolated from the rhizosphere of alfalfa, exhibits high P-solubilization activity under both Pi and Po conditions, and thus improves plant growth. Similarly, high P solubilization and IAA production rates were demonstrated in Pseudomonas putida (BHUJY23) (Lavakush and Verma, 2012). These plant growth-promoting attributes of microorganisms show promise for improving the overall growth of plants. In this study, B. megaterium cultured in LB medium supplemented with l-tryptophan displayed a clear halo zone around the colony on Pikovskaya’s agar medium, with IAA levels reaching a maximum after 48 h (Supplemental Table S2). Therefore, the B. megaterium strain isolated in this study is a promising candidate to improve P acquisition from soil. The relative abundance of Bacillus was highly enriched in the rhizosheath of white lupin transgenic hairy roots, and in rice roots OE-LaPAP12 under SD–P conditions. In both cases, this increase was probably due to APase release from the root, which increases the availability of P, a nutrient promoting bacterial growth and proliferation. The higher relative abundance of rhizosheath-associated Bacillus under SD–P also suggests that these bacteria can improve soil P availability. Positive effects of soil bacterial communities on plant rhizosheath formation were reported previously (Marasco et al., 2018). u Rahman et al. (2021) demonstrated that the availability of soil P corresponded positively with the abundance of Bacillus, Pseudomonas, and Trichoderma spp. Our demonstration of the positive effects of APase exudation on rhizosheath formation build on the findings of Liu et al. (2019), who showed that root exudates, particularly sugars and amino acids, are mainly responsible for rhizosheath formation and microbial abundance in soil (Liu et al., 2017), and on other studies showing that these compounds enhance soil P solubilization and acquisition (Divjot et al., 2021; Kang et al., 2021).
In our study, Bacillus inoculation of white lupin and rice enhanced P content in the root, rhizosheath, leaf, and stem. Recently, Kang et al. (2021) reported that P-solubilizing Enterobacter ludwigii (AFFR02) and B. megaterium (Mj1212) improved alfalfa growth under drought. Similarly, co-inoculation with B. megaterium considerably improved P and potassium accumulation in C. sativus (Zhao et al., 2021). To explore the synergistic mediating effects of root APase secretion and the associated bacterial on P acquisition, B. megaterium was inoculated into NS and SS soils, where it increased total root length, root APase activity, and rhizosheath formation under SD conditions in white lupin and rice plants. These effects were greater in plants that overexpressed LaPAP12. Wang et al. (2021) similarly demonstrated that A. thaliana inoculated with B. megaterium substantially promoted lateral root initiation via auxin influx, suggesting a role of Bacillus in auxin-mediated root growth. A very recent study by our group demonstrated increased IAA production in rice inoculated with B. cereus, which favored root growth and rhizosheath formation under moderate SD conditions (Xu et al., 2021). Thus, Bacillus may contribute to rhizosheath formation through auxin modulation, leading to increased root growth. These results suggest a synergistic interaction of APases with native soil bacteria-promoting rhizosheath formation under SD. The combination of high APase activity and inoculation with rhizosheath-associated plant growth-promoting rhizobacteria is a promising approach to enhance P acquisition and plant growth through rhizosheath formation.
In conclusion, our study demonstrated that the overexpression of LaPAP12 enhances APase secretion; by attracting Bacillus bacteria, this in turn promotes root growth and rhizosheath formation. Under SD and –P conditions, this association leads to improved P acquisition in both white lupin and rice plants. These findings improve our understanding of how APase secretion and the bacterial community together contribute to the development of P-efficient agricultural crops.
Materials and methods
Identification of white lupin PAPs
The protein sequences of LaPAP12 (Lalb_Chr23g0266391) and its orthologs in Narrow-leaf lupin (L. angustifolius), A. thaliana, G. max, P. vulgaris, Barrel clover (Medicago truncatula), Mung Bean (Vigna radiata), Adzuki Bean (Vigna angularis), Ipomoea triloba, red clover (Trifolium pratense), and Black Cottonwood (Populus trichocarpa) were retrieved and aligned in ClustalW (https://www.genome.jp/tools-bin/clustalw). The aligned sequences were then used to construct an unrooted maximum-likelihood phylogenetic tree with IQ-TREE (http://www.iqtree.org/) and 1,000 bootstrap values. The conserved amino acid regions of the PAP12 protein in the selected plant species were analyzed using Jalview software.
Plant material and soil treatments
Lupinus albus (white lupin) seeds and O. sativa (rice) WT and transgenic (TP-LaPAP12) seeds were surface-sterilized with 75% (v/v) ethanol for 1 min and then rinsed three times with sterile water. The seeds were then sandwiched between sheets of wet moist filter paper and kept at 37°C for 4 days to allow their germination. The seedlings were grown in miniature hydroponics, consisting of 1-mL unlidded tip boxes filled with sterile water, for 3–5 days (until seedling length reached 3 cm) under controlled conditions: 14/10-h light/dark cycle, light intensity of 300 μmol photons m−2 s−1, and 40% relative humidity at 26°C. The soil was collected from paddy rice fields (at 20 cm depth) in Huayang, Jiangxi Province, China (Aslam et al., 2021); its chemical properties were as follows: 0.65 g total P kg−1; 27.7 g K kg−1; 92.0 mg exchangeable K kg−1; 1.75 g N kg−1; 20.5 g organic C kg−1; and 42.6 mg Olsen-P kg−1. To ensure soil homogeneity, the soil was air-dried for at least 3 days under sunlight before being sieved through a 4-mm mesh; it was then stored until use. SS was autoclaved three times and then heat-incubated until complete dehydration (Xu et al., 2021); NS was used as the control. Separate pots for white lupin (8-cm diameter/20-cm height; 1.0 kg soil) and rice (14 cm diameter/10 cm height, 1.8 kg soil) were prepared. The P treatments were +P (100 mg P kg soil−1) and –P (0 mg P kg soil−1), with KH2PO4 used as the P source. For soil P treatment, +P treatment is 142.6 mg P kg soil−1 (100 + 42.6), −P treatment is 42.6 mg P kg soil−1 (soil Olsen-P). The soils were supplemented with nutrient solution containing 0.5 mM Ca(NO3)2, 0.025 μM (NH4)6Mo7O24, and 1.75-mM K2SO4 as the N and K sources. Four treatment were applied: WW with sufficient P (WW+P), WW with deficient P (WW−P), SD with sufficient P (SD+P), and SD with deficient P (SD–P). For the WW treatment, the plants received 80 mL of water for white lupin and 140 mL of water for rice, every 2 days. SD white lupin and rice plants were irrigated with 80 and 140 mL of water every 6 days, respectively, as done previously (Zhang et al., 2020a) but with slight modifications. The pots were weighed daily to estimate the soil water content. All experiments were conducted with at least five biological replicates.
Construction of plant transformation vectors and plant transformation
White lupin LaPAP12-overexpressing hairy roots were generated as described previously (Uhde-Stone et al., 2005), with slight modifications. The open-reading frame of LaPAP12 (Lalb_Chr23g0266391) was amplified using a gene-specific primer (overhang with BamHI and SmaI) (Supplemental Table S3) followed by ligation of the amplified product into plasmid pFGC5941 under the control of the CaMV 35S promoter. The resulting expression vector, containing p35S::LaPAP12, was then transferred using the heat shock method to Agrobacterium rhizogenes (K599 strain). White lupin sterilized seeds were germinated on moist filter paper until a 20-mm radicle emerged, and then transferred to deionized water for 3 days of incubation in the dark. The primary root apex (5–10 mm) was excised, infected with A. rhizogenes (K599 strain) carrying the vector containing the p35S::LaPAP12 construct or EV (control), and co-cultured on 0.5 MS medium (100 µM acetosyringone) in the dark for 3 days at room temperature. Seedlings bearing roots transformed with OE-LaPAP12 or EV were grown hydroponically with or without P for 2 weeks, transferred to soil, and subjected to SD treatment. For rhizosheath collection, the plants were harvested after 2 weeks. White lupin genome insertion with the targeted gene was confirmed by PCR amplification of the bar gene and further validated by quantifying the relative expression in transgenic OE-LaPAP12 and EV hairy roots.
pTRV1 and pTRV2 VIGS vectors were obtained from YouBio Biotechnology Company (Changsha, China). A 186-bp LaPAP12 fragment was amplified (using primers described in Supplemental Table S3), from white lupin root cDNA and cloned into pTRV2 VIGS vector. Plants infected with tobacco rattle virus (TRV) were considered as virus vector control. The Agrobacterium tumefaciens (strain GV3101) containing pTRV1 or pTRV2 derivatives were grown at 28°C in YEB medium containing appropriate antibiotics. The cells were harvested from overnight grown cultures and re-suspended in the infiltration buffer (10 mM 2-(N-morpholino) ethanesulfonic acid; 0.1 mM acetosyringone, 10 mM MgCl2) to a final OD600 = 0.5 and incubated for 2 h at room temperature. For leaf infiltration, each A. tumefaciens (strain GV3101) containing pTRV1 and pTRV2 derivatives were mixed in 1:1 ratio and used for inoculation. Agrobacterium tumefaciens (strain GV3101) carrying pTRV1 and pTRV2 were delivered into plants by inoculation of Agrobacterium cultures carrying VIGS vector into plant roots (three times in a 5-day interval). Fourteen days after the last inoculation, the plants were harvested.
For the transformation of rice, LaPAP12 was cloned into the pBWA(V)HS overexpression vector driven by a constitutive CaMV 35S promoter using the In-Fusion HD seamless cloning kit (GBI, Suzhou, China) following the manufacturer’s instructions, and then transformed into A. tumefaciens (strain GV3101). Agrobacterium tumefaciens-mediated transformation was used to generate rice lines heterologously expressing LaPAP12 according to a previously described protocol (Chen et al., 2003). Hygromycin resistance was used as a selection marker to screen for positive rice plants. Relative expression in transformed lines was then analyzed.
Soil sample collection and analysis of root morphological traits
For rhizosheath collection, the pots were carefully disassembled at week 6 of plant growth. The plants were shaken to remove loosely attached soil, leaving the rhizosheath only, after which the roots were detached from the shoots. Soil firmly bound to the root was collected as rhizosheath and washed with sterile water in a clean tray. To assess absolute rhizosheath DW, the collected rhizosheaths were dried at 105°C for 3 days, while specific rhizosheath weight (mg cm−1) was calculated as the ratio of dry rhizosheath weight to total root length (cm) for each plant. For WW treatment, soil attached to the roots was collected as the rhizosphere. Lastly, bulk soil (both WW and SD) was collected from pots without plants.
Root length was measured using winRHIZO software (Regent Instruments, Quebec, Canada). For root hair length measurement, five independent root segments (1 mm) were excised from a plant root, placed in ethanol (50% v/v), and photographed under a stereomicroscope (SMZ18; Olympus, Tokyo, Japan) equipped with a DS-U3 camera (Nikon, Tokyo, Japan). Root hair length was measured using ImageJ software (National Institutes of Health, Bethesda, Maryland, USA) as described by George et al. (2014). The roots and shoots of each plant were dried at 60°C for 3 days and their DW was then recorded. Five independent biological replicates were used.
APase activity and P content
APase activity in the collected soil and plant samples was analyzed using a commercial APase kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) following the manufacturer’s instructions. Activity (U g−1 fresh weight [FW]) was measured using a spectrophotometer (SpectraMax; Molecular Devices, San Jose, California, USA) at 410 nm, following the protocol of Xu et al. (2020). For P determination, the dried plant tissues and soil samples were digested in concentrated H2SO4 and the P concentration was then measured using the ammonium molybdate/vanadate method and a spectrometer at 600 nm, as described previously (Aslam et al., 2021). Measurements were taken from three to five independent replicates.
Soil sampling and DNA extraction
To investigate the bacterial composition and diversity of the rhizosheath (SD), rhizosphere (WW), and bulk soil (SD and WW), samples were collected from three replicates and pooled. The pooled sample was considered a biological replicate. Samples were immediately frozen in liquid nitrogen and preserved at −80°C until further treatment. Three biological replicates were collected for each soil. In an independent experiment, ∼1.0 g of soil sample was ground and homogenized to extract soil DNA using the Soil DNA Isolation Plus kit (cat. 64000-NB). DNA quality was determined by agarose gel electrophoresis (1% w/v) and the DNA was quantified using the Nanodrop ONE instrument (Thermo Scientific, Waltham, Massachusetts, USA). The purified, high-quality soil DNA samples were stored at −80°C until further analysis.
Illumina sequencing of 16S rRNA gene amplicons and data processing
The V4 region of the 16S rRNA gene was amplified using F388 and R806 primers (Supplemental Table S3) with a 6-bp long barcode and Illumina adapter sequences. A 50-µL PCR mixture was prepared, consisting of 25 µL of Phusion High Fidelity PCR Master Mix along with HF buffer (New England Biolabs, Ipswich, Massachusetts, USA), 3 µL of each primer (10 µM), 3 µL of DMSO, 6 µL of ddH2O, and 10 µL of DNA template. The PCR samples were placed in a thermocycler and processed using the following parameters: initial denaturation at 98°C for 15 s, annealing at 58°C for 15 s, elongation at 72°C for 5 s, and final elongation at 72°C for 60 s. The PCR products were then purified using Agencourt AMPure XP beads (Beckman Coulter, Brea, California, USA) to eliminate nonspecific products, quantified using the PicoGreen dsDNA assay kit (Invitrogen, Carlsbad, California, USA), and pooled in equimolar concentrations. Paired-end (2 × 150 bp) sequencing was accomplished using the HiSeq4000 platform (Illumina, San Diego, California, USA). The QIIME (Quantitative Insight into Microbial Ecology) tool was used to analyze 16S microbial sequences, following the protocol of Zhang et al. (2020b).
B. megaterium isolation and inoculation
The effects of B. megaterium inoculation on rhizosheath formation and P acquisition were investigated in white lupin (WT) plants and rice (WT and TP-LaPAP12 lines) grown in autoclaved SS and natural, non-autoclaved soil (NS; used as control) under SD conditions (SD+P and SD–P). Bacillus megaterium was isolated from fresh rhizosheath and cultured on V8 juice agar medium following a previously described protocol (Wei et al., 2019) with slight modifications. The B. megaterium species was determined by PCR using the primers listed in Supplemental Table S3, with sequence validation through sequencing and an NCBI search. A single colony was recultured in liquid nutrient broth (N611; PhytoTech Labs, Lenexa, Kansas, USA) at 28°C with constant shaking (150 rpm) for 2 days, at a pH of 6.0. Cultured bacteria were centrifuged at 4,032 g for 12 min; the bacterial pellet was washed twice with NaCl and then resuspended in sterilized water. Both white lupin and rice seeds were inoculated with the B. megaterium suspension (108 cells g−1 soil) and then transplanted into SS or NS. Un-inoculated seedlings were treated with sterilized double-distilled water as described previously (Rolli et al., 2015; Xu et al., 2021). Specific rhizosheath weight, root length, root hair length, phosphorous content, and APase activity were measured as described above. Twenty white lupin (WT) and rice (WT and TP-LaPAP12) plants were grown for each SD treatment, and the experiment was repeated twice.
Phosphate solubilization assay
The phosphate solubilization ability of the B. megaterium strain was analyzed using Pikovskaya’s agar medium supplemented with tricalcium phosphate (0.5% w/v), as described previously (Yahaghi et al., 2018). First, 5 µL of bacterial culture in the center of an agar plate was inoculated and then incubated in the dark at 28°C for 7 days. The formation of a clear halo zone around the colony was interpreted as phosphate solubilization. Four plates were used as biological replicates, each containing one colony.
IAA production assay
Bacillus megaterium was cultured in three different flasks containing 30 mL of LB medium, with or without 0.5 mg of l-tryptophan/mL, and kept at 30°C on an orbital shaker at 150 rpm. After incubation for 24, 48, and 96 h, 1 mL of bacterial culture from each flask was centrifuged at 7,104 g for 10 min and vigorously mixed with 2 mL of Salkowski’s reagent (1 mL 0.5 M FeCl3 mixed with 49 mL of HClO4 [35% v/v]). A pink color developed after 30 min of incubation and absorbance was measured at 530 nm, with the IAA concentration determined using a calibration curve of pure IAA (Supplemental Table S2). Three flasks (n = 3, biological replicates) were measured three times.
RNA extraction and RT-qPCR analysis
Fresh leaf samples of WT lupin and WT and TP-LaPAP12 rice were crushed using a bead tissue lyser and homogenized to extract total RNA using TRIzol reagent (Nanjing Vazyme Biotech Co., Ltd, Nanjing, China) following the manufacturer’s instructions. RNA quantity and quality were measured using the Agilent Bio-analyzer 2100 system (Agilent Technologies, Santa Clara, California, USA) and 2% (w/v) agarose gel electrophoresis, respectively. To perform the RT-qPCR analyses, isolated RNA was reverse-transcribed into cDNA using the PrimeScript reagent kit with gDNA Eraser (Takara, Dalian, China), following the manufacturer’s instructions. Each reaction system contained a total volume of 10 µL with 1 µL of diluted first-strand cDNA, 5 µL of SYBR Green PCR Master Mix (Applied Biosystems, Waltham, Massachusetts, USA), and 10 µM of forward and reverse primers. The PCR samples were processed using the Applied Biosystems 7500 real-time PCR system. Gene expression was normalized using the ACTIN gene and calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). The gene-specific primers were designed using Primer Express Software (Applied Biosystems) and are listed in Supplemental Table S3.
Statistical analyses
All experiments were performed with five independent biological replicates, each of which comprised five technical replicates. All statistical analyses were carried out using R software (version 4.0.2; R Development Core Team, Vienna, Austria). Two-way analysis of variance (ANOVA) was used to determine the effects of soil treatments, genotypes, and their interaction on all parameters. Means were compared using the least significant difference test at P < 0.05. For analysis of the RT-qPCR data, Student’s t test (P ≤ 0.05) was applied. Data normality was checked using the Shapiro–Wilk test.
Accession numbers
Sequence data for the LaPAP12 from this article can be found in the White Lupin Genome data libraries (https://www.whitelupin.fr/index.html) under accession number: Lalb_Chr23g0266391.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Root-secreted APase activity and root P content in knock-down plant of LaPAP12 (TRV::LaPAP12) and control.
Supplemental Figure S2. Rhizosheath formation in white lupin OE-LaPAP12, knock-down plant of LaPAP12 (TRV::LaPAP12), and the EV.
Supplemental Figure S3. White lupin transgenic hairy root initiation and other root-related traits under P treatment and SD conditions.
Supplemental Figure S4. Ca3(PO4)2 solubilization by B. megaterium strain.
Supplemental Figure S5. Phenotypic observation and expression analysis of transgenic overexpressing TP-LaPAP12 and WT rice plants.
Supplemental Figure S6. Physiological and biochemical traits displayed by rice plants overexpressing TP-LaPAP12 under four different treatments.
Supplemental Figure S7. The total number of bacterial OTUs identified in WT and LaPAP12-overexpressing rice lines.
Supplemental Figure S8. Bacillus enriched rice microbial community of TP-LaPAP12-L1 rhizosheath and rhizosphere soils.
Supplemental Table S1. Solubility index for Ca3(PO4)2 solubilization by B. megaterium strain.
Supplemental Table S2. Estimation of IAA by the B. megaterium after 24, 48, and 96 h of incubation using spectrophotometry.
Supplemental Table S3. List of primers used in this study.
Supplementary Material
Acknowledgments
We would like to thank Prof. Song from Haixia Institute of Fujian Agriculture and Forestry University for providing us with the transformation vector.
Funding
This work was supported by the National Natural Science Foundation of China (31761130073 and 31872169) and the Agencia Estatal de Investigación, Spain (AGL2017-88381-R and PID2021-125371OB-I00).
Conflict of interest statement. None declared.
Contributor Information
Mehtab Muhammad Aslam, Joint International Research Laboratory of Water and Nutrient in Crops, Haixia Institute of Ecology and Environmental Engineering, College of Resource and Environment, Fujian Agriculture and Forestry University, Fuzhou 350002, China; College of Agriculture, Yangzhou University, Yangzhou 225009, China; Department of Biology, Hong Kong Baptist University, Hong Kong; State Key Laboratory of Agrobiotechnology, Chinese University of Hong Kong, Hong Kong.
José J Pueyo, Institute of Agricultural Sciences, ICA-CSIC, Madrid 28006, Spain.
Jiayin Pang, School of Agriculture and Environment, UWA Institute of Agriculture, University of Western Australia, Perth, Western Australia 6009, Australia.
Jinyong Yang, Joint International Research Laboratory of Water and Nutrient in Crops, Haixia Institute of Ecology and Environmental Engineering, College of Resource and Environment, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Weiguo Chen, Joint International Research Laboratory of Water and Nutrient in Crops, Haixia Institute of Ecology and Environmental Engineering, College of Resource and Environment, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Hao Chen, Joint International Research Laboratory of Water and Nutrient in Crops, Haixia Institute of Ecology and Environmental Engineering, College of Resource and Environment, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Muhammad Waseem, College of Horticulture, South China Agricultural University, Guangzhou 510642, China.
Ying Li, College of Agriculture, Yangzhou University, Yangzhou 225009, China.
Jianhua Zhang, Department of Biology, Hong Kong Baptist University, Hong Kong; State Key Laboratory of Agrobiotechnology, Chinese University of Hong Kong, Hong Kong.
Weifeng Xu, Joint International Research Laboratory of Water and Nutrient in Crops, Haixia Institute of Ecology and Environmental Engineering, College of Resource and Environment, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
W.X. and M.M.A. conceived the study design. W.X. supervised the project. M.M.A., W.C., H.C., and J.Y. carried out all the experiments. M.M.A., W.C., H.C., and J.Y. performed all data analyses, designed the figures, and wrote the manuscript. J.P. critically edited and revised the manuscript. W.X., J.P., M.W., J.J.P., Y.L., and J.Z. reviewed the manuscript. All authors discussed the results and contributed to the final manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Weifeng Xu (wfxu@fafu.edu.cn).
References
- Albalasmeh AA, Ghezzehei TA (. 2014) Interplay between soil drying and root exudation in rhizosheath development. Plant Soil 374: 739–751 [Google Scholar]
- Aslam MM, Karanja JK, Yuan W, Zhang Q, Zhang J, Xu W (. 2021) Phosphorus uptake is associated with the rhizosheath formation of mature cluster roots in white lupin under soil drying and phosphorus deficiency. Plant Physiol Biochem 166: 531–539 [DOI] [PubMed] [Google Scholar]
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM (. 2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57: 233–266 [DOI] [PubMed] [Google Scholar]
- Baldwin JC, Karthikeyan AS, Raghothama KG (. 2001) LEPS2, a phosphorus starvation-induced novel acid phosphatase from tomato. Plant Physiol 125: 728–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadouria J, Giri J (. 2021) Purple acid phosphatases: roles in phosphate utilization and new emerging functions. Plant Cell Rep 1–19 [DOI] [PubMed] [Google Scholar]
- Carminati A, Benard P, Ahmed MA, Zarebanadkouki M (. 2017) Liquid bridges at the root–soil interface. Plant Soil 417: 1–15 [Google Scholar]
- Castrillo G, Teixeira PJPL, Paredes SH, Law TF, de Lorenzo L, Feltcher ME, Finkel OM, Breakfield NW, Mieczkowski P, Jones CD (. 2017) Root microbiota drive direct integration of phosphate stress and immunity. Nature 543: 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Liu S (. 2019) Identification and characterization of the phosphate-solubilizing bacterium Pantoea sp. S32 in reclamation soil in Shanxi, China. Front Microbiol 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Jin W, Wang M, Zhang F, Zhou J, Jia Q, Wu Y, Liu F, Wu P (. 2003) Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J 36: 105–113 [DOI] [PubMed] [Google Scholar]
- Chinnaswamy A, Coba de la Peña T, Stoll A, de la Peña Rojo D, Bravo J, Rincón A, Lucas MM, Pueyo JJ (. 2018) A nodule endophytic Bacillus megaterium strain isolated from Medicago polymorpha enhances growth, promotes nodulation by Ensifer medicae and alleviates salt stress in alfalfa plants. Ann Appl Biol 172: 295–308 [Google Scholar]
- Deng S, Lu L, Li J, Du Z, Liu T, Li W, Xu F, Shi L, Shou H, Wang C (. 2020) Purple acid phosphatase 10c (OsPAP10c) encodes a major acid phosphatase and regulates the plant growth under phosphate deficient condition in rice. J Exp Bot 71: 4321–4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divjot K, Rana KL, Tanvir K, Yadav N, Yadav AN, Kumar M, Kumar V, Dhaliwal HS, Saxena AK (. 2021) Biodiversity, current developments and potential biotechnological applications of phosphorus-solubilizing and-mobilizing microbes: a review. Pedosphere 31: 43–75 [Google Scholar]
- el Zahar Haichar F, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent JM, Heulin T, Achouak W (. 2008) Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2: 1221–1230 [DOI] [PubMed] [Google Scholar]
- George TS, Brown LK, Ramsay L, White PJ, Newton AC, Bengough AG, Russell J, Thomas WTB (. 2014) Understanding the genetic control and physiological traits associated with rhizosheath production by barley (Hordeum vulgare). New Phytol 203: 195–205 [DOI] [PubMed] [Google Scholar]
- Hacker N, Ebeling A, Gessler A, Gleixner G, González Macé O, de Kroon H, Lange M, Mommer L, Eisenhauer N, Ravenek J (. 2015) Plant diversity shapes microbe–rhizosphere effects on P mobilisation from organic matter in soil. Ecol Lett 18: 1356–1365 [DOI] [PubMed] [Google Scholar]
- Haling RE, Brown LK, Bengough AG, Young IM, Hallett PD, White PJ, George TS (. 2013) Root hairs improve root penetration, root–soil contact, and phosphorus acquisition in soils of different strength. J Exp Bot 64: 3711–3721 [DOI] [PubMed] [Google Scholar]
- Haling RE, Richardson AE, Culvenor RA, Lambers H, Simpson RJ (. 2010) Root morphology, root-hair development and rhizosheath formation on perennial grass seedlings is influenced by soil acidity. Plant Soil 335: 457–468 [Google Scholar]
- Hanna AL, Youssef HH, Amer WM, Monib M, Fayez M, Hegazi NA (. 2013) Diversity of bacteria nesting the plant cover of north Sinai deserts, Egypt. J Adv Res 4: 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ ( 2012) Cellular programs for arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol 15: 691–698 [DOI] [PubMed] [Google Scholar]
- Hiruma K, Gerlach N, Sacristán S, Nakano RT, Hacquard S, Kracher B, Neumann U, Ramírez D, Bucher M, O’Connell RJ (. 2016) Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 165: 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honvault N, Houben D, Firmin S, Meglouli H, Laruelle F, Fontaine J, Lounès‐Hadj Sahraoui A, Coutu A, Lambers H, Faucon MP (. 2021) Interactions between belowground traits and rhizosheath fungal and bacterial communities for phosphorus acquisition. Funct Ecol 35: 1603–1619 [Google Scholar]
- James RA, Weligama C, Verbyla K, Ryan PR, Rebetzke GJ, Rattey A, Richardson AE, Delhaize E (. 2016) Rhizosheaths on wheat grown in acid soils: phosphorus acquisition efficiency and genetic control. J Exp Bot 67: 3709–3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji LY, Zhang WW, Yu D, Cao YR, Xu H (. 2012) Effect of heavy metal-solubilizing microorganisms on zinc and cadmium extractions from heavy metal contaminated soil with Tricholoma lobynsis. World J Microbiol Biotechnol 28: 293–301 [DOI] [PubMed] [Google Scholar]
- Kalayu G ( 2019) Phosphate solubilizing microorganisms: promising approach as biofertilizers. Int J Agron 2019 [Google Scholar]
- Kang S-M, Khan M-A, Hamayun M, Kim L-R, Kwon E-H, Kang Y-S, Kim K-Y, Park J-J, Lee I-J (. 2021) Phosphate-solubilizing Enterobacter ludwigii AFFR02 and Bacillus megaterium Mj1212 rescues alfalfa’s growth under post-drought stress. Agriculture 11: 485 [Google Scholar]
- Karanja JK, Aslam MM, Qian Z, Yankey R, Dodd IC, Weifeng X. (. 2021) Abscisic acid mediates drought-enhanced rhizosheath formation in tomato. Front Plant Sci 12: 658787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli PS, Maurya K, Thakur JK, Bhosale R, Giri J (. 2021) Significance of root hairs in developing stress‐resilient plants for sustainable crop production. Plant Cell Environ 45: 677–694 [DOI] [PubMed] [Google Scholar]
- Kong Y, Li X, Ma J, Li W, Yan G, Zhang C (. 2014) GmPAP4, a novel purple acid phosphatase gene isolated from soybean (Glycine max), enhanced extracellular phytate utilization in Arabidopsis thaliana. Plant Cell Rep 33: 655–667 [DOI] [PubMed] [Google Scholar]
- Korir H, Mungai NW, Thuita M, Hamba Y, Masso C (. 2017) Co-inoculation effect of rhizobia and plant growth promoting rhizobacteria on common bean growth in a low phosphorus soil. Front Plant Sci 8: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H, Martinoia E, Renton M (. 2015) Plant adaptations to severely phosphorus-impoverished soils. Curr Opin Plant Biol 25: 23–31 [DOI] [PubMed] [Google Scholar]
- Lambers H, Raven JA, Shaver GR, Smith SE (. 2008) Plant nutrient-acquisition strategies change with soil age. Trends Ecol Evol 23: 95–103 [DOI] [PubMed] [Google Scholar]
- Lavakush JY, Verma JP (. 2012) Isolation and characterization of effective plant growth promoting rhizobacteria from rice rhizosphere of Indian soil. Asian J Biol Sci 5: 294–303 [Google Scholar]
- Li D, Zhu H, Liu K, Liu X, Leggewie G, Udvardi M, Wang D (. 2002) Purple acid phosphatases of Arabidopsis thaliana: comparative analysis and differential regulation by phosphate deprivation. J Biol Chem 277: 27772–27781 [DOI] [PubMed] [Google Scholar]
- Li Y-S, Gao Y, Tian Q-Y, Shi F-L, Li L-H, Zhang W-H (. 2011) Stimulation of root acid phosphatase by phosphorus deficiency is regulated by ethylene in Medicago falcata. Environ Exp Bot 71: 114–120 [Google Scholar]
- Liang C, Tian J, Lam H-M, Lim BL, Yan X, Liao H (. 2010) Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiol 152: 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Carvalhais LC, Crawford M, Singh E, Dennis PG, Pieterse CMJ, Schenk PM (. 2017) Inner plant values: diversity, colonization and benefits from endophytic bacteria. Front Microbiol 8: 2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T-Y, Chen M-X, Zhang Y, Zhu F-Y, Liu Y-G, Tian Y, Fernie AR, Ye N, Zhang J (. 2019) Comparative metabolite profiling of two switchgrass ecotypes reveals differences in drought stress responses and rhizosheath weight. Planta 250: 1355–1369 [DOI] [PubMed] [Google Scholar]
- Liu Y, Chen L, Zhang N, Li Z, Zhang G, Xu Y, Shen Q, Zhang R (. 2016) Plant–microbe communication enhances auxin biosynthesis by a root-associated bacterium, Bacillus amyloliquefaciens SQR9. Mol Plant Microbe Interact 29: 324–330 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (. 2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu L, Qiu W, Gao W, Tyerman SD, Shou H, Wang C (. 2016) OsPAP10c, a novel secreted acid phosphatase in rice, plays an important role in the utilization of external organic phosphorus. Plant Cell Environ 39: 2247–2259 [DOI] [PubMed] [Google Scholar]
- Lynch JM, de Leij F (. 2012) Rhizosphere. eLS 1–10Oxford University Press [Google Scholar]
- Lynch JP ( 2007) Roots of the second green revolution. Aust J Bot 55: 493–512 [Google Scholar]
- Marasco R, Mosqueira MJ, Fusi M, Ramond J-B, Merlino G, Booth JM, Maggs-Kölling G, Cowan DA, Daffonchio D (. 2018) Rhizosheath microbial community assembly of sympatric desert speargrasses is independent of the plant host. Microbiome 6: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra P, Pandey BK, Giri J (. 2017) Improvement in phosphate acquisition and utilization by a secretory purple acid phosphatase (OsPAP21b) in rice. Plant Biotechnol J 15: 1054–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes-Blackburn D, Giles C, Darch T, George TS, Blackwell M, Stutter M, Shand C, Lumsdon D, Cooper P, Wendler R (. 2018) Opportunities for mobilizing recalcitrant phosphorus from agricultural soils: a review. Plant Soil 427: 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndour P, Gueye M, Barakat M, Ortet P, Bertrand-Huleux M, Pablo A-L, Dezette D, Chapuis-Lardy L, Assigbetsé K, Kane NA (. 2017) Pearl millet genetic traits shape rhizobacterial diversity and modulate rhizosphere aggregation. Front Plant Sci 8: 1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson E, Mwafulirwa L (. 2021) Root–soil–microbe interactions mediating nutrient fluxes in the rhizosphere. In VVSR Gupta, AK Sharma, eds, Rhizosphere Biology: Interactions Between Microbes and Plants. Springer, pp 75–91 [Google Scholar]
- Pérez-Flores P, Valencia-Cantero E, Altamirano-Hernández J, Pelagio-Flores R, López-Bucio J, García-Juárez P, Macías-Rodríguez L (. 2017) Bacillus methylotrophicus M4-96 isolated from maize (Zea mays) rhizoplane increases growth and auxin content in Arabidopsis thaliana via emission of volatiles. Protoplasma 254: 2201–2213 [DOI] [PubMed] [Google Scholar]
- Plaxton WC, Tran HT (. 2011) Metabolic adaptations of phosphate-starved plants. Plant Physiol 156: 1006–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pueyo JJ, Quiñones MA, Coba de la Peña T, Fedorova EE, Lucas MM (. 2021) Nitrogen and phosphorus interplay in lupin root nodules and cluster roots. Front Plant Sci 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñones MA, Fajardo S, Fernández-Pascual M, Lucas MM, Pueyo JJ (. 2021) Nodulated white lupin plants growing in contaminated soils accumulate unusually high mercury concentrations in their nodules, roots and especially cluster roots. Horticulturae 7: 302 [Google Scholar]
- Quiñones MA, Lucas MM, Pueyo JJ (. 2022) Adaptive mechanisms make lupin a choice crop for acidic soils affected by aluminum toxicity. Front Plant Sci 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbi SMF, Tighe MK, Flavel RJ, Kaiser BN, Guppy CN, Zhang X, Young IM (. 2018) Plant roots redesign the rhizosphere to alter the three‐dimensional physical architecture and water dynamics. New Phytol 219: 542–550 [DOI] [PubMed] [Google Scholar]
- Richardson AE ( 2009) Regulating the phosphorus nutrition of plants: molecular biology meeting agronomic needs. Plant Soil 322: 17–24 [Google Scholar]
- Richardson AE, Hadobas PA, Hayes JE, O’Hara CP, Simpson RJ (. 2001) Utilization of phosphorus by pasture plants supplied with myo-inositol hexaphosphate is enhanced by the presence of soil micro-organisms. Plant Soil 229: 47–56 [Google Scholar]
- Richardson AE, Hocking PJ, Simpson RJ, George TS (. 2009) Plant mechanisms to optimise access to soil phosphorus. Crop Pasture Sci 60: 124–143 [Google Scholar]
- Richardson AE, Simpson RJ (. 2011) Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiol 156: 989–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WD, Park J, Tran HT, Del Vecchio HA, Ying S, Zins JL, Patel K, McKnight TD, Plaxton WC (. 2012) The secreted purple acid phosphatase isozymes AtPAP12 and AtPAP26 play a pivotal role in extracellular phosphate-scavenging by Arabidopsis thaliana. J Exp Bot 63: 6531–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolli E, Marasco R, Vigani G, Ettoumi B, Mapelli F, Deangelis ML, Gandolfi C, Casati E, Previtali F, Gerbino R (. 2015) Improved plant resistance to drought is promoted by the root‐associated microbiome as a water stress‐dependent trait. Environ Microbiol 17: 316–331 [DOI] [PubMed] [Google Scholar]
- Rose TJ, Impa SM, Rose MT, Pariasca-Tanaka J, Mori A, Heuer S, Johnson-Beebout SE, Wissuwa M (. 2013) Enhancing phosphorus and zinc acquisition efficiency in rice: a critical review of root traits and their potential utility in rice breeding. Ann Bot 112: 331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse J, Martinoia E, Northen T (. 2018) Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci 23: 25–41 [DOI] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J (. 2011) Auxin and plant–microbe interactions. Cold Spring Harb Perspect Biol 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney CJ, De Vries FT, Van Dongen BE, Bardgett RD (2020) Root traits explain rhizosphere fungal community composition among temperate grassland plant species. New Phytol 0–2 [DOI] [PubMed] [Google Scholar]
- Talboys PJ, Owen DW, Healey JR, Withers PJA, Jones DL (. 2014) Auxin secretion by Bacillus amyloliquefaciens FZB42 both stimulates root exudation and limits phosphorus uptake in Triticum aestivium. BMC Plant Biol 14: 51–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Pei Y, Huang W, Ding J, Siemann E (. 2021) Increasing flavonoid concentrations in root exudates enhance associations between arbuscular mycorrhizal fungi and an invasive plant. ISME J 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticconi CA, Abel S (. 2004) Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci 9: 548–555 [DOI] [PubMed] [Google Scholar]
- Tran HT, Hurley BA, Plaxton WC (. 2010a) Feeding hungry plants: the role of purple acid phosphatases in phosphate nutrition. Plant Sci 179: 14–27 [Google Scholar]
- Tran HT, Qian W, Hurley BA, She YM, Wang D, Plaxton WC (. 2010b) Biochemical and molecular characterization of AtPAP12 and AtPAP26: the predominant purple acid phosphatase isozymes secreted by phosphate‐starved Arabidopsis thaliana. Plant Cell Environ 33: 1789–1803 [DOI] [PubMed] [Google Scholar]
- u Rahman MK, Wang X, Gao D, Zhou X, Wu F (. 2021) Root exudates increase phosphorus availability in the tomato/potato onion intercropping system. Plant Soil 1–18.34720209 [Google Scholar]
- Uhde-Stone C ( 2017) White lupin: a model system for understanding plant adaptation to low phosphorus availability. In S Sulieman, LSP Tran, eds, Legume Nitrogen Fixation in Soils with Low Phosphorus Availability. Springer, pp 243–280 [Google Scholar]
- Uhde-Stone C, Liu J, Zinn KE, Allan DL, Vance CP (. 2005) Transgenic proteoid roots of white lupin: a vehicle for characterizing and silencing root genes involved in adaptation to P stress. Plant J 44: 840–853 [DOI] [PubMed] [Google Scholar]
- Vinci G, Cozzolino V, Mazzei P, Monda H, Savy D, Drosos M, Piccolo A (. 2018) Effects of Bacillus amyloliquefaciens and different phosphorus sources on Maize plants as revealed by NMR and GC-MS based metabolomics. Plant Soil 429: 437–450 [Google Scholar]
- Wang BL, Tang XY, Cheng LY, Zhang AZ, Zhang WH, Zhang FS, Liu JQ, Cao Y, Allan DL, Vance CP (. 2010) Nitric oxide is involved in phosphorus deficiency‐induced cluster‐root development and citrate exudation in white lupin. New Phytol 187: 1112–1123 [DOI] [PubMed] [Google Scholar]
- Wang L, Li Z, Qian W, Guo W, Gao X, Huang L, Wang H, Zhu H, Wu J-W, Wang D (. 2011) The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol 157: 1283–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu D (. 2018) Functions and regulation of phosphate starvation-induced secreted acid phosphatases in higher plants. Plant Sci 271: 108–116 [DOI] [PubMed] [Google Scholar]
- Wang L, Lu S, Zhang Y, Li Z, Du X, Liu D (. 2014) Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J Integr Plant Biol 56: 299–314 [DOI] [PubMed] [Google Scholar]
- Wang S, Na X, Yang L, Liang C, He L, Jin J, Liu Z, Qin J, Li J, Wang X, et al. (2021) Bacillus megaterium strain WW1211 promotes plant growth and lateral root initiation via regulation of auxin biosynthesis and redistribution. Plant Soil 466: 491–504 [Google Scholar]
- Wang X, Wang Y, Tian J, Lim BL, Yan X, Liao H (. 2009) Overexpressing AtPAP15 enhances phosphorus efficiency in soybean. Plant Physiol 151: 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasaki J, Maruyama H, Tanaka M, Yamamura T, Dateki H, Shinano T, Ito S, Osaki M (. 2009) Overexpression of the LASAP2 gene for secretory acid phosphatase in white lupin improves the phosphorus uptake and growth of tobacco plants. Soil Sci Plant Nutr 55: 107–113 [Google Scholar]
- Wei Z, Gu Y, Friman V-P, Kowalchuk GA, Xu Y, Shen Q, Jousset A (. 2019) Initial soil microbiome composition and functioning predetermine future plant health. Sci Adv 5: eaaw0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Katagi H, Harrison M, Wang Z-Y (. 2006a) Improved phosphorus acquisition and biomass production in Arabidopsis by transgenic expression of a purple acid phosphatase gene from M. truncatula. Plant Sci 170: 191–202 [Google Scholar]
- Xiao K, Zhang JH, Harrison M, Wang ZY (. 2006b) Ectopic expression of a phytase gene from Medicago truncatula barrel medic enhances phosphorus absorption in plants. J Integr Plant Biol 48: 35–43 [Google Scholar]
- Xie L, Shang Q (. 2018) Genome-wide analysis of purple acid phosphatase structure and expression in ten vegetable species. BMC Genomics 19: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Liao H, Zhang Y, Yao M, Liu J, Sun L, Zhang X, Yang J, Wang K, Wang X, et al. (2021) Coordination of root auxin with the fungus Piriformospora indica and bacterium Bacillus cereus enhances rice rhizosheath formation under soil drying. ISME J 16: 801–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Zhang Q, Yuan W, Xu F, Aslam MM, Miao R, Li Y, Wang Q, Li X, Zhang X (. 2020) The genome evolution and low-phosphorus adaptation in white lupin. Nat Commun 11: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahaghi Z, Shirvani M, Nourbakhsh F, de la Peña TC, Pueyo JJ, Talebi M (. 2018) Isolation and characterization of Pb-solubilizing bacteria and their effects on Pb uptake by Brassica juncea: implications for microbe-assisted phytoremediation. J Microbiol Biotechnol 28: 1156–1167 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Du H, Gui Y, Xu F, Liu J, Zhang J, Xu W (. 2020a) Moderate water stress in rice induces rhizosheath formation associated with abscisic acid and auxin responses. J Exp Bot 71: 2740–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Du H, Xu F, Ding Y, Gui Y, Zhang J, Xu W (. 2020b) Root–bacteria associations boost rhizosheath formation in moderately dry soil through ethylene responses. Plant Physiol 183: 780–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Mao X, Zhang M, Yang W, Di HJ, Ma L, Liu W, Li B (. 2021) The application of Bacillus megaterium alters soil microbial community composition, bioavailability of soil phosphorus and potassium, and cucumber growth in the plastic shed system of North China. Agric Ecosyst Environ 307: 107236 [Google Scholar]
- Zhu Q, Riley WJ, Tang J, Koven CD (. 2016) Multiple soil nutrient competition between plants, microbes, and mineral surfaces: model development, parameterization, and example applications in several tropical forests. Biogeosciences 13: 341–363 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.