Abstract
Carcass traits play important roles in the broiler industry and single nucleotide polymorphism (SNP) can be efficient molecular markers for marker-assisted breeding of chicken carcass traits. Based on our previous RNA-seq data (accession number GSE58755), cysteine rich with epidermal growth factor like domains 1 (CRELD1) and DnaJ heat shock protein family member C30 (DNAJC30) are differentially expressed in breast muscle between white recessive rock chicken (WRR) and Xinghua chicken (XH). In this study, we further characterize the potential function and SNP mutation of CRELD1 and DNAJC30 in chicken for the first time. According to protein interaction network and enrichment analysis, CRELD1 and DNAJC30 may play some roles in chicken muscle development and fat deposition. In WRR and XH, the results of the relative tissue expression pattern demonstrated that CRELD1 and DNAJC30 are not only differentially expressed in breast muscle but also leg muscle and abdominal fat. Therefore, we identified 5 SNP sites of CRELD1 and 7 SNP sites of DNAJC30 and genotyped them in an F2 chicken population. There are 4 sites of CRELD1 and 3 sites of DNAJC30 are associated with chicken carcass traits like breast muscle weight, body weight, dressed weight, leg weight percentage, eviscerated weight with giblet percentage, intermuscular adipose width, shank length, and girth. These results suggest that the SNP sites of CRELD1 and DNAJC30 can be potential molecular markers to improve the chicken carcass traits and lay the foundation for marker-assisted selection.
Key words: broiler, CRELD1, DNAJC30, carcass trait
INTRODUCTION
Domestic chicken (Gallus gallus domesticus) has become the most extensively distributed poultry in the world since their domestication (Scanes, 2007; Wang et al., 2020). Chicken also plays an irreplaceable role in the human diet nowadays and the production of chicken meat and eggs has been increasing quickly in recent years (Connerton et al., 2018; Cartoni Mancinelli et al., 2022). However, poultry is the terminal host of various diseases like avian influenza (Fusaro et al., 2019; Escalera-Zamudio et al., 2020). For this reason, the European Union has already placed a ban on live bird imports to prevent the spread of avian influenza, and the trade controls and import restrictions of live poultry can control the epidemic effectively (Reino et al., 2017; Chen et al., 2020).
Based on the above background, slaughtering poultry and storing it on ice for transportation to sale have gradually replaced the live poultry market, which makes carcass trait become one of the most important traits of poultry, especially in chicken. Carcass traits are one kind of quantitative trait and they can't be measured without slaughter, which becomes a major challenge to the further development of the poultry industry (Uemoto et al., 2009). Skeletal muscle development and fat deposition have a significant impact on poultry carcass traits (Zerehdaran et al., 2004; Lotfi et al., 2011; Li et al., 2021).
Xinhua chicken (XH) and white recessive rock chicken (WRR) are kinds of leaner and hypertrophic broiler respectively (Hu et al., 2013; Teng et al., 2019). WRR demonstrates significantly faster muscle growth and lower fat deposition compared to XH in chicken carcass traits due to the complex gene regulatory network (Qiu et al., 2006; Ouyang et al., 2015; Li et al., 2019). In our previous study, RNA sequencing was performed to construct a LncRNA-microRNA-gene interaction network between WRR and XH's breast muscle tissues (Li et al., 2019). Among all differentially expressed genes, CRELD1 and DnaJ heat shock protein family member C30 (DNAJC30) are the top 50 significantly upregulated genes in WRR's breast muscle compared to XH, which may play some roles in chicken carcass traits.
Cysteine rich with epidermal growth factor like domains 1 (CRELD1) is a multifunctional gene that mostly plays important roles in myocardial development (Beckert et al., 2021) and immune system homeostasis (Bonaguro et al., 2020). Most human fetal and adult tissues express CRELD1 and the gene expression in skeletal muscle and adult heart has a higher level (Rupp et al., 2002). Besides, CRELD1 is identified as a key gene required for the synaptic expression of ionotropic acetylcholine receptors in C2C12 myoblasts (D'Alessandro et al., 2018). Studies revealed that CRELD1 highly expresses and has an important biological function in skeletal muscle, but its relationship with chicken carcass traits are not yet investigated.
Heat shock proteins family (HSP) is one kind of molecular chaperone family that plays critical roles in protein folding and assembly (Gomez-Pastor et al., 2018). Many studies have shown that HSP play an important role in skeletal muscle development (Du SJ et al., 2008; Sin et al., 2019). DNAJC30 is one of the HSP and it has been reported as an auxiliary component of ATP-synthase machinery in mitochondria (Tebbenkamp et al., 2018). In addition, DNAJC30 is also the protein chaperone required for the exchange of damaged complex I subunits in mitochondria (Stenton et al., 2021). ATP-synthase and mitochondrial complex I subunits are highly correlated with the metabolism and development of muscle and fat, which suggested that DNAJC30 may play some roles in skeletal muscle and fat metabolism (Hong et al., 2014; Sanchez-Gonzalez et al., 2022). However, there are no reports about their function in skeletal muscle development and their relationship with the chicken carcass.
Single nucleotide polymorphism (SNP) is an important genetic factor that is usually used for marker-assisted breeding of economic traits in agriculture (Sun et al., 2020). Studies have shown SNP in functional genes can be significantly associated with chicken carcass traits (Lei et al., 2005; Lu et al., 2012). In this study, we first examined the relative tissue expression pattern of CRELD1 and DNAJC30 and found that these 2 genes were significantly differentially expressed in WRR and XH's breast muscle, leg muscle, and abdominal fat. After that, SNP sites in CRELD1 and DNAJC30 were detected and significantly associated with chicken carcass traits. Our study demonstrated that the SNP sites of CRELD1 and DNAJC30 were potential molecular markers in the breeding of carcass traits.
MATERIALS AND METHODS
Experimental Animals and Determination of Their Carcass Traits
The F2 generation of Chinese indigenous chicken was the object of this study. All tissue samples and carcass trait data were collected from KwangFeng Industrial Co., Ltd (Guangzhou, Guangdong, China). The animal experiment performed in this study satisfied strict the requirements of the Institutional Animal Care and Use Committee at the South China Agricultural University (approval ID: 2021-f074).
Chinese indigenous spotted chicken line was crossed with Chinese indigenous yellow chicken to produce F1 individuals. 15 pairs of F1 individuals were used to construct full-sib families to produce the F2 individual. The F2 individuals were raised in floor pens and fed with commercial corn-soybean-based diets to 80 d of age. The 410 F2 individuals were slaughtered according to standard procedures. Blood samples were collected and stored at −20℃ before slaughter. The measured carcass traits included subcutaneous fat thickness, intermuscular adipose width, eviscerated weight (EW), EW with giblet, abdominal visceral fat weight (AVFW), legs weigh (LW), breast muscle weight (BMW), body weight (BW), shank length (SL), shank girth (SG), dressed weight (DW), BMW percentage, LW percentage, AVFW percentage, EW with giblet percentage (EWGP), EW percentage and slaughter percentage.
Genomic DNA Extraction, PCR Amplification, and SNP Detection
The genomic DNA of the chicken blood sample was extracted using NRBC Blood DNA Kit (Omega, Georgia, CA) according to the manufacturer's protocol. The primers were designed using Primer 5. The genomic DNA of each chicken sample was subjected to PCR amplification using 2×Taq MasterMix (CWBIO, Jiangsu, China) according to the manufacturer's protocol. The annealing temperature of both genes was 58℃, and the cycle number was 32. The PCR products were sequenced using ABI-3730XL by Tianyi Biotech Co., Ltd (Guangzhou, China). The information on primers used for PCR amplification and Sanger sequencing was listed in Table S1. Names for SNP sites follow the nomenclature standards approved by the HGVS (Human Genome Variation Society).
RNA Extraction, cDNA Synthesis, and qPCR Assay
The total RNA of 12 tissues was extracted using RNAiso Plus (Takara, Kyoto, Japan) and the HiPure Universal RNA Mini Kit (Magen, Guangzhou, China) following the manufacturer's protocol. cDNA was synthesized using MonScript 5×RTIII All−in−One Mix Kits (Monad, Shanghai, China) for reverse transcription. Primers were designed in Primer 5 software. cDNA samples were subjected to ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) following the manufacturer's protocol. The 2 −ΔΔCt method and internal normalization were used to analyze quantification results. The information on primers used for qPCR amplification was listed in Table S1.
Protein Interaction Network, GO, and KEGG Analysis
STRING online database (https://cn.string-db.org/, accessed on 10 September 2021) was employed to analyze the protein interaction network between CRELD1 and DNCJC30 (von Mering et al., 2005; Szklarczyk et al., 2017). Proteins enriched in the network were clustered into 3 clusters using the kmeans method. GO enrichment and KEGG pathway enrichment of these proteins were performed and visualized using the g: Profiler tool (https://biit.cs.ut.ee/gprofiler/gost, accessed on 10 September 2021) (Raudvere et al., 2019).
Statistical Analysis
SNP Genotyping was performed using DNASTAR software (DNASTAR, Inc. Madison, WI), Linkage disequilibrium (LD) and haplotype analysis were performed using SHEsis online tool (http://analysis.bio-x.cn/, accessed on 30 September 2021) (Shi and He, 2005; Li et al., 2009).
Microsoft Excel (Microsoft Corp., Redmond, WA) was used to calculate the genotype frequency allele frequency, and diversity parameters. Diversity parameters contained Hardy–Weinberg p-value (Pw), observed heterozygosity (Ho), expected heterozygosity (He), and the polymorphism information content (PIC).
Correlation among traits was considered by calculating the inter-trait Pearson's correlation for all pairwise combinations of traits within each class of traits. SPSS 25 software (SPSS Inc., Chicago, IL).
Association analysis of SNP sites and carcass traits was performed with a generalized linear model as the following model:
Yij is the observed value of different carcass traits, µ is the overall population mean, Gi is the effect of each genotype, and eij is the random error. For each carcass trait, the analysis of variance was carried out using SAS 9.4 (SAS Institute Inc, Cary, NC), and the significance was determined by Duncan's multiple tests. Significance between SNP sites is represented by letters where different letters indicate a significant difference (P < 0.05).
RESULTS
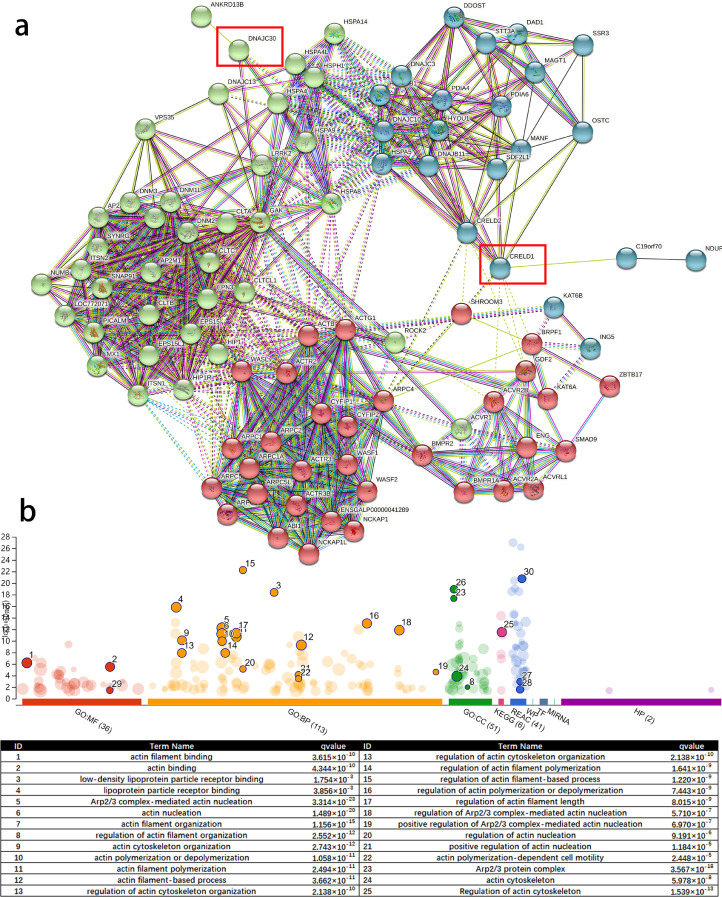
The Interaction Network of CRELD1 and DNAJC30
STRING database was employed to construct the protein interaction network of these 2 genes (Figure 1A). Ninety genes were determined in the interaction network and Kmeans clustering was run to generate 3 clusters, including a muscle-related cluster (red cluster), an endocytosis-related cluster (green cluster), and an N-glycan-related cluster (blue cluster). CRELD1 and DNAJC30 were contained in the green cluster and blue cluster respectively, and these 2 clusters interacted through the muscle-related cluster. It indicated that CRELD1 and DNAJC30 probably play some role in muscle development. GO and KEGG enrichment analysis also suggested that the interaction network of CRELD1 and DNAJC30 enriched 23 muscle-related GO terms and KEGG pathway, which further confirmed the potential muscle-regulatory function of CRELD1 and DNAJC30 (Figure 1B). Besides, there are also 2 lipid-related GO terms, which indicated that CRELD1 and DNAJC30 may be also the candidate genes for lipid metabolism. Overall, these results showed that CRELD1 and DNAJC30 were associated with muscle and fat development and metabolism to some extent.
Figure 1.
The protein interaction network and enrichment analysis. (A) 92 genes were determined in the interaction network and Kmeans clustering was run to generate 3 clusters (red, blue, and green). (B) Muscle and fat-related terms were enriched in the interaction network between CRELD1 and DNCJC30. Abbreviations: CRELD1, cysteine rich with epidermal growth factor like domains 1; DNAJC30, DnaJ heat shock protein family member C30.
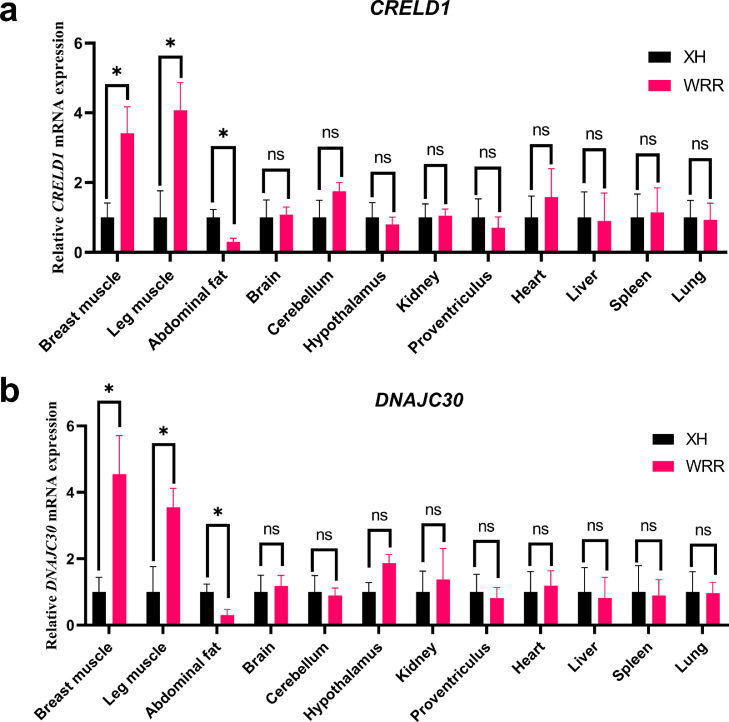
Relative mRNA Expression Level of CRELD1 and DNAJC30 in Tissue of WRR and XH
To further predict the potential function of CRELD1 and DNAJC30 in chicken, the relative mRNA expression level of these 2 genes was measured in XH and WRR using qPCR. The results showed that mRNA expression of CRELD1 and DNAJC30 between WRR and XH were significantly different in breast muscle, leg muscle, and abdominal fat (P < 0.05) but showed no significant differences in other 9 tissue (brain, cerebellum, hypothalamus, kidney, proventriculus, heart, liver, spleen, and lung) (Figure 2). In skeletal muscle tissues (breast and leg muscle), the mRNA expression of CRELD1 in WRR was significantly higher than XH, with the expression level being the highest in the leg muscle (P < 0.05; Figure 2A). Interestingly, the expression level of DNAJC30 in WRR was also significantly higher in skeletal muscle tissues but with the highest expression level in breast muscle (Figure 2B; P < 0.05). It is also interesting to note that both mRNA expression of CRELD1 and DNAJC30 in WRR was the significantly lowest expression in abdominal fat than XH (P < 0.05; Figures 2A and 2B). The expression of CRELD1 and DNAJC30 in WRR were higher in skeletal muscle and lower in abdominal fat than XH, emphasizing their essential roles in the carcass trait of chicken.
Figure 2.
The relative tissue expression pattern of CRELD1 and DNAJC30 in Xinhua chicken (XH) and White Recessive Rock chicken (WRR). (A) Relative CRELD1 expression of different tissues between WRR and XH. (B) Relative DNAJC30 expression of different tissues between WRR and XH. Symbols “*” and “**” indicate a significant difference at P < 0.05 and P < 0.01, respectively. Abbreviations: CRELD1, cysteine rich with epidermal growth factor like domains 1; DNAJC30, DnaJ heat shock protein family member C30
The Polymorphism of CRELD1 and DNAJC30 Genes
The PCR-amplified DNA sequences of 410 individuals were confirmed by Sanger sequencing and genotyped by DNASTAR. 5 SNP sites of CRELD1 and 7 SNP sites of DNAJC30 were obtained. In CRELD1, 2 SNP sites were located on exon and the other 3 sites were located on intron. These 2 sites in exon were synonymous mutations. Besides, In DNAJC30, 2 sites were in the exon and the other 5 sites were in the intron and the 2 exon-located sites were also synonymous mutations. The allele that was consistent with the NCBI reference genome (bGalGal1.mat.broiler. GRCg7b Primary Assembly) was defined as wild type. Otherwise, the allele was defined as mutant type (Table 1).
Table 1.
The information on SNPs in CRELD1 and DNAJC30 in this study.
| Gene | SNP ID | Abbreviation | Location | Amino change | Wild type | Heterozygote | Mutant type |
|---|---|---|---|---|---|---|---|
| CRELD1 | NC_052543.1: g.11683326 G>A | C-SNP1 | Intron 5 | NO | GG (n = 115) |
GA (n = 196) |
AA (n = 98) |
| NC_052543.1: g.11683378 G>A | C-SNP2 | Exon 6 | NO | GG (n = 206) |
GA (n = 167) |
AA (n = 36) |
|
| NC_052543.1: g.11683497 T>C | C-SNP3 | Intron 6 | NO | TT (n = 38) |
TC (n = 164) |
CC (n = 207) |
|
| NC_052543.1: g.11683588 A>G | C-SNP4 | Exon 7 | NO | AA (n = 36) |
AG (n = 165) |
GG (n = 208) |
|
| NC_052543.1: g.11683643 G>C | C-SNP5 | Intron 7 | NO | GG (n = 231) |
GC (n = 147) |
CC (n = 31) |
|
| DNAJC30 | NC_052550.1: g.618125 A>G | D-SNP1 | Exon | NO | AA (n = 89) |
AG (n = 104) |
GG (n = 213) |
| NC_052550.1: g.618020 T>C | D-SNP2 | Exon | NO | TT (n = 118) |
TC (n = 123) |
CC (n = 165) |
|
| NC_052550.1: g.617962 C>T | D-SNP3 | Intron | NO | CC (n = 261) |
CT (n = 88) |
TT (n = 57) |
|
| NC_052550.1: g.617664 T>C | D-SNP4 | Intron | NO | TT (n = 33) |
TC (n = 93) |
CC (n = 280) |
|
| NC_052550.1: g.617659 C>T | D-SNP5 | Intron | NO | CC (n = 36) |
CT (n = 88) |
TT (n = 282) |
|
| NC_052550.1: g.617631 G>T | D-SNP6 | Intron | NO | GG (n = 294) |
GT (n = 74) |
TT (n = 38) |
|
| NC_052550.1: g.617467 T>C | D-SNP7 | Intron | NO | TT (n = 267) |
TC (n = 70) |
CC (n = 69) |
Abbreviations: CRELD1, cysteine rich with epidermal growth factor like domains 1; DNAJC30, DnaJ heat shock protein family member C30; SNP, single nucleotide polymorphism.
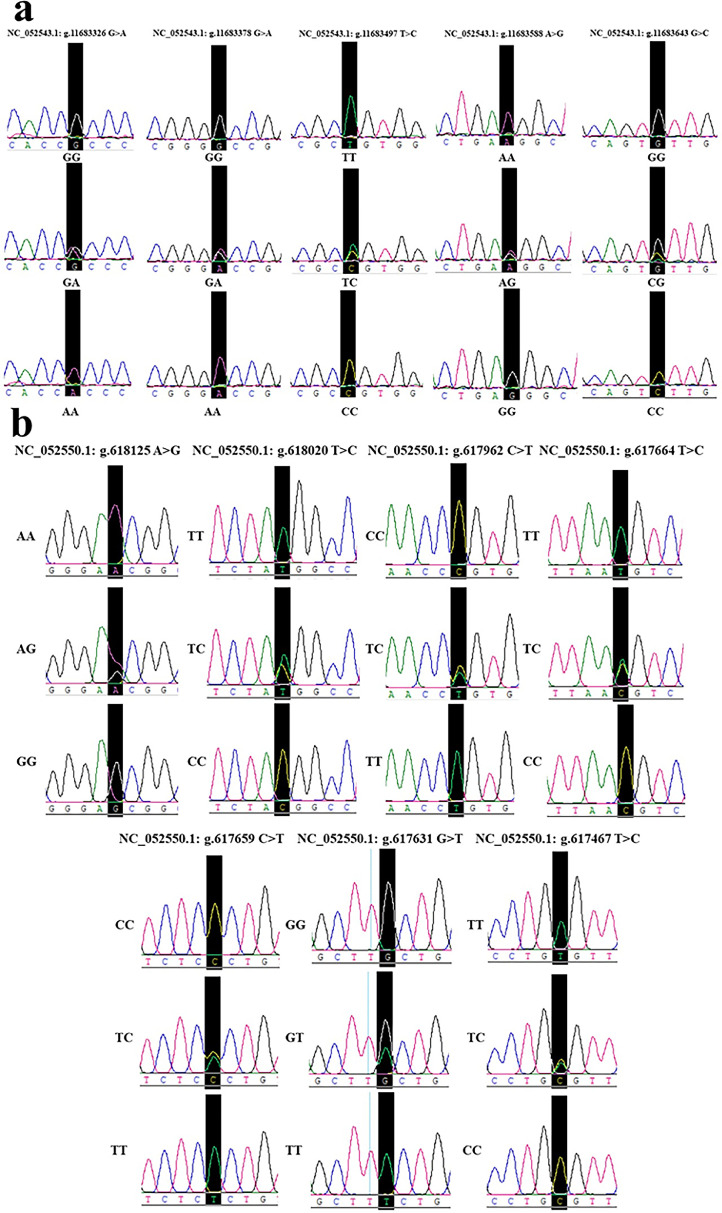
The 5 SNP sites of CRELD1 detected in this study were NC_051543.1: g.11683326 (C-SNP1), NC_051543.1: g.11683378 (C-SNP2), NC_051543.1: g.11683497 (C-SNP3), NC_051543.1: g.11683588 (C-SNP4) and NC_051543.1: g.11683643 (C-SNP5) The 7 SNP sites of DNAJC30 detected in this study were NC_052550.1: g.618125 (D-SNP1), NC_052550.1: g.618020 (D-SNP2), NC_052550.1: g.617962 (D-SNP3), NC_052550.1: g.617664 (D-SNP4), NC_052550.1: g.617659 (D-SNP5), NC_052550.1: g.617631 (D-SNP6), NC_052550.1: g.617467 (D-SNP7). The peak maps of all 3 genotypes of SNP sites mentioned above were shown in Figure 3.
Figure 3.
The peak maps of all 3 genotypes of SNP sites in CRELD1 and DNAJC30. (A) SNP sites of CRELD1 in this study. (B) SNP sites of DNAJC30 in this study. Abbreviations: CRELD1, cysteine rich with epidermal growth factor like domains 1; DNAJC30, DnaJ heat shock protein family member C30; SNP, single nucleotide polymorphism.
After genotyping, the genotype frequency, allele frequency, and diversity parameters of SNP sites in CRELD1 and DNAJC30 were calculated and listed in Table 2. Interestingly, all 5 SNP sites in CRELD1 were consistent with Hardy-Weinberg Equilibrium (Pw > 0.05) while all 7 SNP sites in DNAJC30 deviated from Hardy-Weinberg Equilibrium (Pw < 0.05). These variations of DNAJC30 may be due to a strong artificial selection. The PIC of SNP sites was calculated and revealed that all sites of CRELD1 and DNAJC30 were moderate polymorphism (0.25 < PIC < 0.5).
Table 2.
Genotype and allele frequency, and diversity parameters of SNPs in CRELD1 and DNAJC30.
| Gene | SNPs1 ID (Abbreviation) | Genotype frequency |
Allele frequency |
Pw2 | Ho3 | He4 | PIC5 | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | Heterozygote | Mutant type | Wild type | Mutant type | ||||||
| CRELD1 |
NC_052543.1: g.11683326 G>A (C-SNP1) |
0.28 | 0.48 | 0.24 | 0.52 | 0.48 | 0.72 | 0.50 | 0.50 | 0.37 |
|
NC_052543.1: g.11683378 G>A (C-SNP2) |
0.50 | 0.41 | 0.09 | 0.71 | 0.29 | 0.97 | 0.59 | 0.41 | 0.33 | |
|
NC_052543.1: g.11683497 T>C (C-SNP3) |
0.09 | 0.40 | 0.51 | 0.29 | 0.71 | 0.80 | 0.59 | 0.41 | 0.33 | |
|
NC_052543.1: g.11683588 A>G (C-SNP4) |
0.09 | 0.40 | 0.51 | 0.29 | 0.71 | 0.92 | 0.59 | 0.41 | 0.33 | |
|
NC_052543.1: g.11683643 G>C (C-SNP5) |
0.56 | 0.36 | 0.08 | 0.74 | 0.26 | 0.54 | 0.62 | 0.38 | 0.31 | |
| DNAJC30 |
NC_052550.1: g.618125 A>G (D-SNP1) |
0.22 | 0.26 | 0.52 | 0.35 | 0.65 | 0.00 | 0.55 | 0.45 | 0.35 |
|
NC_052550.1: g.618020 T>C (D-SNP2) |
0.29 | 0.30 | 0.41 | 0.44 | 0.56 | 0.00 | 0.51 | 0.49 | 0.37 | |
|
NC_052550.1: g.617962 C>T (D-SNP3) |
0.64 | 0.22 | 0.14 | 0.75 | 0.25 | 0.00 | 0.63 | 0.37 | 0.30 | |
|
NC_052550.1: g.617664 T>C (D-SNP4) |
0.08 | 0.23 | 0.69 | 0.20 | 0.80 | 0.00 | 0.69 | 0.31 | 0.27 | |
|
NC_052550.1: g.617659 C>T (D-SNP5) |
0.09 | 0.22 | 0.69 | 0.20 | 0.80 | 0.00 | 0.68 | 0.32 | 0.27 | |
|
NC_052550.1: g.617631 G>T (D-SNP6) |
0.72 | 0.18 | 0.09 | 0.82 | 0.18 | 0.00 | 0.70 | 0.30 | 0.26 | |
|
NC_052550.1: g.617467 T>C (D-SNP7) |
0.66 | 0.17 | 0.17 | 0.74 | 0.26 | 0.00 | 0.62 | 0.38 | 0.31 | |
SNPs: single nucleotide polymorphism sites.
Pw= the χ 2 test of Hardy–Weinberg equilibrium. Pw < 0.05 means Hardy-Weinberg disequilibrium (HWD) and Pw > 0.05 means Hardy–Weinberg equilibrium.
Ho: observed heterozygosity.
He: expected heterozygosity.
PIC: polymorphism information content. Suggested PIC value classes are as follows: PIC>0.5 means high polymorphism, 0.25<PIC< 0.5 means moderate polymorphism, and PIC <0.25 means low polymorphism.
Abbreviations: CRELD1, cysteine rich with epidermal growth factor like domains 1; DNAJC30, DnaJ heat shock protein family member C30; SNP, single nucleotide polymorphism.
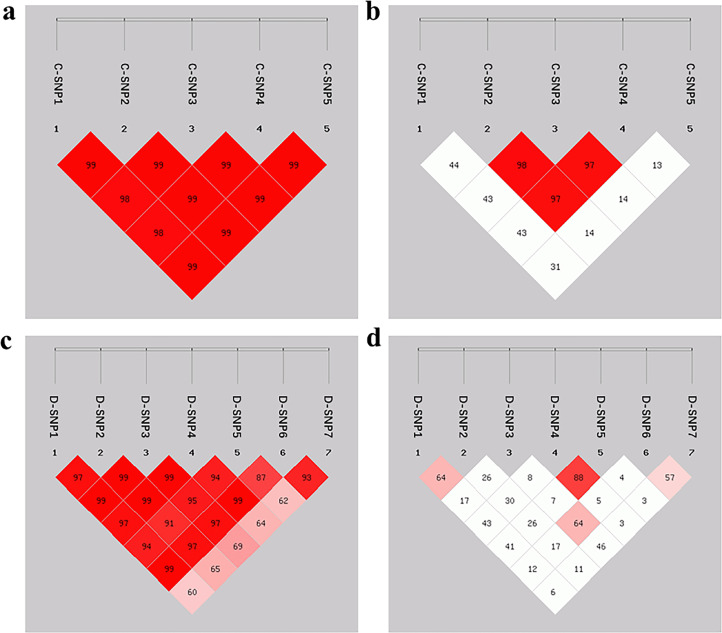
To further explore the relationship among the SNP sites in CRELD1 and DNAJC30 repetitively, LD and haplotype analyses were performed. LD analysis was performed by calculating D′ and r2 values and the results were shown in Figure 4. SNP sites in CRELD1 and DNAJC30 were at high LD level according to the D′ and r2 values (Table S2 and S3). Besides, 11 haplotypes (4 in CRELD1 and 7 in DNAJC30) were found and the information on haplotypes was listed in Table S4 and Table S5.
Figure 4.
Genetic analysis of linkage equilibrium on SNP sites in CRELD1 and DNAJC30 genes. (A) D' of SNP sites in CRELD1; (B) r2 of SNP sites in CRELD1; (C) D' of SNP sites in DNAJC30; (D) r2 of SNP sites in DNAJC30. Abbreviations: CRELD1, cysteine rich with epidermal growth factor like domains 1; DNAJC30, DnaJ heat shock protein family member C30; SNP, single nucleotide polymorphism.
Correlation Analysis of Chicken Carcass Traits
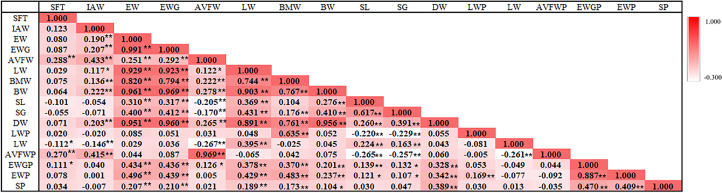
To further understand the detailed information of the 410 individual chicken population, the Pearson correlation coefficient was calculated among these 17 carcass traits. The values of pairwise correlation between chicken carcass traits were shown in Figure 5 and the descriptive statistics of 17 carcass traits in the study population were shown in Table S6. Interestingly, we found that SL and SG were significantly correlated with muscle development and fat deposition-related carcass traits AVFW, eviscerated weight with giblet percentage, LW, BW, BMW and so on (P < 0.05).
Figure 5.
Pearson's correlation analysis of carcass traits. The result was shown as a heat map. Symbol “*” and “**” indicate a significant difference at P < 0.05 and P < 0.01 (2-tailed), respectively.
Association Analysis of CRELD1 and DNAJC30 SNPs With Chicken Carcass Traits
To explore the relationship between obtained SNP sites and chicken carcass traits, association analysis was performed and the association results were shown in Table 3. In CRELD1, all SNP sites were associated with chicken carcass traits except C-SNP5. C-SNP1 was significantly associated with 5 carcass traits like BMW, BW, DW, SL, and SG (P < 0.05). C-SNP2, C-SNP3, and C-SNP4 were all associated with EWGP, and C-SNP4 was also associated with SG. It is interesting to note that the heterozygote of all obtained SNP sites in CRELD1 was significantly higher than its wild type or mutant type in associated carcass traits. In DNAJC30, a total of 7 SNP sites were found but only 3 of them were associated with chicken carcass traits (P < 0.05). Both D-SNP1 and D-SNP2 were associated with EWGP, and D-SNP1 and D-SNP2 were also associated with SG and LW percentage, respectively (P < 0.05). D-SNP6 was associated with intermuscular adipose width and the mutant type was significantly higher than the heterozygote (P < 0.05). SL and SG were correlated with muscle development and fat deposition-related carcass traits according to correlation analysis results (Figure 5). Thus, the above results indicated that SNPs of CRELD1 and DNAJC30 can be potential molecular markers for marker-assist breeding of the chicken carcass traits.
Table 3.
Association analysis of SNPs in CRELD1 and DNAJC30 with chicken carcass traits (MEANS ± SD).
| Gene | SNPs abbreviation | Carcass trait | Genotype |
Available Sample | P-value | ||
|---|---|---|---|---|---|---|---|
| Wild type | Heterozygote | Mutant type | |||||
| CRELD1 | C-SNP1 G>A | BMW (g) | 104.51 ± 17.87b (n = 108) |
111.08 ± 16.59a (n = 173) |
107.88 ± 18.79ab (n = 90) |
371 | 0.0094 |
| BW (g) | 1834.86 ± 229.65b (n = 108) |
1922.08 ± 211.63a (n = 177) |
1901.80 ± 227.52a (n = 91) |
376 | 0.0051 | ||
| DW (g) | 1621.50 ± 218.01b (n = 108) |
1700.90 ± 197.55a (n = 175) |
1672.34 ± 224.16ab (n = 90) |
373 | 0.0090 | ||
| SL (cm) | 7.49 ± 0.83ab (n = 108) |
7.62 ± 0.55a (n = 177) |
7.32 ± 1.01b (n = 91) |
376 | 0.0104 | ||
| SG (cm) | 11.90 ± 1.11ab (n = 108) |
12.02 ± 0.90a (n = 177) |
11.66 ± 1.02b (n = 91) |
376 | 0.0188 | ||
| C-SNP2 G>A | EWGP (%) | 79.71 ± 2.65a (n = 182) |
80.01 ± 1.84a (n = 137) |
78.71 ± 3.39b (n = 34) |
353 | 0.0217 | |
| C-SNP3 T>C | EWGP (%) | 78.78 ± 3.30b (n = 36) |
80.01 ± 1.86a (n = 134) |
79.71 ± 2.65a (n = 183) |
353 | 0.0287 | |
| C-SNP4 A>G | EWGP (%) | 78.71 ± 3.39b (n = 34) |
80.01 ± 1.85a (n = 136) |
79.70 ± 2.65a (n = 183) |
353 | 0.0222 | |
| SG (cm) | 11.48 ± 0.94b (n = 35) |
11.94 ± 0.98a (n = 152) |
11.94 ± 1.01a (n = 189) |
376 | 0.0316 | ||
| DNAJC30 | D-SNP1 A>G | BMWP (%) | 8.73 ± 0.85a (n = 81) |
8.61 ± 0.78ab (n = 97) |
8.44 ± 0.79b (n = 193) |
371 | 0.0167 |
| SG (cm) | 11.63 ± 1.03b (n = 82) |
11.98 ± 1.03a (n = 95) |
11.98 ± 0.96a (n = 196) |
373 | 0.0200 | ||
| D-SNP2 T>C | BMWP (%) | 8.70 ± 0.83a (n = 106) |
8.56 ± 0.79ab (n = 114) |
8.44 ± 0.78b (n = 151) |
371 | 0.0425 | |
| LWP (%) | 17.45 ± 0.86b (n = 106) |
17.73 ± 0.89a (n = 114) |
17.50 ± 0.87b (n = 151) |
371 | 0.0344 | ||
| D-SNP6 G>T | IAW (mm) | 27.09 ± 9.83ab (n = 267) |
23.93 ± 10.66b (n = 70) |
29.02 ± 9.67a (n = 34) |
371 | 0.0228 | |
The different letters stand for significant differences (P < 0.05), while the same letter indicates no difference (P > 0.05).
Abbreviations: BMW, breast muscle weight; BMWP, breast muscle weight percentage; BW, body weight; CRELD1, cysteine rich with epidermal growth factor like domains 1; DNAJC30, DnaJ heat shock protein family member C30; DW, dressed weight; EWGP, eviscerated weight with giblet percentage; IAW, intermuscular adipose width; LWP, leg weight percentage; SG, shank girth; SL, shank length; SNP, single nucleotide polymorphism.
DISCUSSION
Chicken consumption has become one of the most widespread meat in the world (Zhao et al., 2019). Carcass traits are one kind of the most important traits that have a great impact on the development of the domestic chicken industry (Santos et al., 2021). Promoting muscle development and reducing fat deposition are the top priority in chicken breeding (Guo et al., 2019). Therefore, many studies have been devoted to exploring the potential candidate genes, regulatory network, and molecular mechanism to improve chicken carcass traits (Li et al., 2015; Allais et al., 2019; Fu et al., 2020; Zhang et al., 2020; Wang et al., 2021; Li et al., 2022; Yang et al., 2022). However, it still needs further research to unravel the underlying mechanisms of chicken carcass traits.
Previous studies have reported that CRELD1 is highly expressed in muscle tissue including skeletal muscle and myocardial tissue (Rupp et al., 2002; Beckert et al., 2021). Importantly, CRELD1 plays an important role in controlling the abundance of ionotropic acetylcholine receptors in C2C12 myoblasts (D'Alessandro et al., 2018). Mitochondrial function is important for basic cell metabolism including muscle cells and adipocytes (Jokinen et al., 2017; Migliavacca et al., 2019). As a member of HSP, DNAJC30 is a key gene in mitochondrial function which is required for damaged mitochondrial complex I subunits exchange and ATP synthase complex function (Tebbenkamp et al., 2018; Stenton et al., 2021). These 2 genes have gradually become research hotspots in human disease phenotypes but there is no research in poultry science.
Interestingly, we found that both CRELD1 and DNAJC30 are the differentially expressed genes in the muscle tissue between XH chicken and WRR chicken based on our previous work. It indicated that CRELD1 and DNAJC30 may play some role in chicken muscle development. Besides, the interaction network between CRELD1 and DNAJC30 is highly associated with muscle development according to the enrichment analysis. Interestingly, some lipid deposition-related terms are also enriched in addition to muscle-related terms, which further suggests that CRELD1 and DNAJC30 are likely to be associated with chicken carcass traits.
The bioinformatics analysis results are consistent with the relative gene expression pattern in different tissues between WRR and XH. We found that CRELD1 and DNAJC30 are expressed in almost all chicken tissues, but these 2 genes are differentially expressed in breast muscle, leg muscle, and abdominal fat between XH and WRR. In WRR, CRELD1 and DNAJC30 show a higher expression level of breast muscle and leg muscle but show a lower expression level of abdominal fat compared to XH. XH is one kind of Chinese indigenous chicken, which has a slower muscle growth rate and more fat deposition compared with commercial chicken breeds like WRR (Tian et al., 2021). Based on these results, we demonstrate that CRELD1 and DNAJC30 may play some role in chicken muscle development and fat deposition, which may have an impact on chicken carcass traits.
To further confirm the relationship between CRELD1 and DNAJC30 genes and chicken carcass traits, we detect SNP mutation in CRELD1 and DNAJC30 and performed association analysis with chicken carcass traits. Our results show that a total of 4 of 5 SNP sites in CRELD1 and 3 of 7 SNP sites in DNAJC30 are significantly associated with carcass traits. These SNP sites could serve as candidate sites for marker-assisted breeding in chicken carcass traits and confirm the underlying functions of CRELD1 and DNAJC30 for regulatory muscle development and fat deposition. Notably, C-SNP1 located in intron 5 of CRELD1 is associated with most carcass traits including BMW, BW, DW, SL, and SG. SL and SG are correlated with other muscle and fat-related carcass traits according to our correlation analysis results of chicken carcass traits. Multiple studies have reported that shank traits may be related to fat deposition and muscle development (Ye et al., 2014; Luo et al., 2016; Chen et al., 2021). Other SNP sites in CRELD1 are only associated with 1 or 2 carcass traits. The SNP sites of DNAJC30 are also only associated with 1 or 2 carcass traits. It may be due to the smaller number of individual chickens in this population. Nevertheless, we first proved that CRELD1 and DNAJC30 have the potential to become candidate genes for muscle development and fat deposition to affect chicken carcass traits. However, there are still many unanswered questions about the underlying molecular mechanism of the function of CRELD1 and DNAJC30. Future studies on the current topic are therefore recommended.
CONCLUSIONS
In summary, this is the first investigation to analyze the function of CRELD1 and DNAJC30 in chicken. Our results demonstrated that CRELD1 and DNAJC30 are related to muscle development and fat deposition in chicken according to bioinformatic analysis and tissue gene expression profiles. SNP mutation of CRELD1 and DNAJC30 are significantly associated with chicken carcass traits. Our study identifies several SNP sites in CRELD1 and DNAJC30 for molecular-marker-assist breeding to improve chicken carcass traits.
ACKNOWLEDGMENTS
This work was supported by the Science and Technology Program of Guangzhou, China (202103000084), China Agriculture Research System (CARS-41-G03), the Construction Project of Modern Agricultural Science and Technology Innovation Alliance in Guangdong Province (2021KJ128), and the Science and Technology Program of Guangdong province, China (2020B1212060060).
DISCLOSURES
The authors declare no conflicts of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2022.102324.
Appendix. Supplementary materials
REFERENCES
- Allais S., Hennequet-Antier C., Berri C., Salles L., Demeure O., Le Bihan-Duval E. Mapping of QTL for chicken body weight, carcass composition, and meat quality traits in a slow-growing line. Poult. Sci. 2019;98:1960–1967. doi: 10.3382/ps/pey549. [DOI] [PubMed] [Google Scholar]
- Beckert V., Rassmann S., Kayvanjoo A.H., Klausen C., Bonaguro L., Botermann D.S., Krause M., Moreth K., Spielmann N., Da Silva-Buttkus P., Fuchs H., Gailus-Durner V., de Angelis M.H., Händler K., Ulas T., Aschenbrenner A.C., Mass E., Wachten D. CRELD1 regulates myocardial development and function. J. Mol. Cell. Cardiol. 2021;156:45–56. doi: 10.1016/j.yjmcc.2021.03.008. [DOI] [PubMed] [Google Scholar]
- Bonaguro L., Köhne M., Schmidleithner L., Schulte-Schrepping J., Warnat-Herresthal S., Horne A., Kern P., Günther P., ter Horst R., Jaeger M., Rahmouni S., Georges M., Falk C.S., Li Y., Mass E., Beyer M., Joosten L.A.B., Netea M.G., Ulas T., Schultze J.L., Aschenbrenner A.C. CRELD1 modulates homeostasis of the immune system in mice and humans. Nat. Immunol. 2020;21:1517–1527. doi: 10.1038/s41590-020-00811-2. [DOI] [PubMed] [Google Scholar]
- Cartoni Mancinelli A., Mattioli S., Twining C., Dal Bosco A., Donoghue A.M., Arsi K., Angelucci E., Chiattelli D., Castellini C. Poultry meat and eggs as an alternative source of n-3 long-chain polyunsaturated fatty acids for human Nutrition. Nutrients. 2022;14:1969. doi: 10.3390/nu14091969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Liu Z., Liu L., Wang J., Jin Q. Enhanced potency of a broad H7N9-neutralizing antibody HNIgGA6 through structure-based design. Front. Microbiol. 2020;11:1313. doi: 10.3389/fmicb.2020.01313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Chen J., Wu J., Ren X., Li L., Lu S., Cheng T., Tan L., Liu M., Luo Q., Liang S., Nie Q., Zhang X., Luo W. Integrative analyses of mRNA expression profile reveal SOCS2 and CISH play important roles in GHR mutation-induced excessive abdominal fat deposition in the sex-linked dwarf chicken. Front. Genet. 2021;11 doi: 10.3389/fgene.2020.610605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerton P.L., Richards P.J., Lafontaine G.M., O'Kane P.M., Ghaffar N., Cummings N.J., Smith D.L., Fish N.M., Connerton I.F. The effect of the timing of exposure to Campylobacter jejuni on the gut microbiome and inflammatory responses of broiler chickens. Microbiome. 2018;6:88. doi: 10.1186/s40168-018-0477-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessandro M., Richard M., Stigloher C., Gache V., Boulin T., Richmond J.E., Bessereau J. CRELD1 is an evolutionarily-conserved maturational enhancer of ionotropic acetylcholine receptors. Elife. 2018;7 doi: 10.7554/eLife.39649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S.J, Li H., Bian Y., Zhong Y. Heat-shock protein 90alpha1 is required for organized myofibril assembly in skeletal muscles of zebrafish embryos. Proc. Natl. Acad. Sci. U S A. 2008;105:554–559. doi: 10.1073/pnas.0707330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalera-Zamudio M., Golden M., Gutierrez B., Theze J., Keown J.R., Carrique L., Bowden T.A., Pybus O.G. Parallel evolution in the emergence of highly pathogenic avian influenza A viruses. Nat. Commun. 2020;11:5511. doi: 10.1038/s41467-020-19364-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu R., Ren T., Li W., Liang J., Mo G., Luo W., He D., Liang S., Zhang X. A novel 65-bp indel in the GOLGB1 gene is associated with chicken growth and carcass traits. Animals (Basel) 2020;10:475. doi: 10.3390/ani10030475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro A., Zecchin B., Vrancken B., Abolnik C., Ademun R., Alassane A., Arafa A., Awuni J.A., Couacy-Hymann E., Coulibaly M., Gaidet N., Go-Maro E., Joannis T., Jumbo S.D., Minoungou G., Meseko C., Souley M.M., Ndumu D.B., Shittu I., Twabela A., Wade A., Wiersma L., Akpeli Y.P., Zamperin G., Milani A., Lemey P., Monne I. Disentangling the role of Africa in the global spread of H5 highly pathogenic avian influenza. Nat. Commun. 2019;10:5310. doi: 10.1038/s41467-019-13287-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pastor R., Burchfiel E.T., Thiele D.J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell. Biol. 2018;19:4–19. doi: 10.1038/nrm.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Wang J., Chen H., Su H., Wang Z., Wan Y., Huang Y., Jiang R. Effects of exercise on carcass composition, meat quality, and mRNA expression profiles in breast muscle of a Chinese indigenous chicken breed. Poult. Sci. 2019;98:5241–5246. doi: 10.3382/ps/pez415. [DOI] [PubMed] [Google Scholar]
- Hong J., Kim B.W., Choo H.J., Park J.J., Yi J.S., Yu D.M., Lee H., Yoon G.S., Lee J.S., Ko Y.G. Mitochondrial complex I deficiency enhances skeletal myogenesis but impairs insulin signaling through SIRT1 inactivation. J. Biol. Chem. 2014;289:20012–20025. doi: 10.1074/jbc.M114.560078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Xu H., Li Z., Zheng X., Jia X., Nie Q., Zhang X. Comparison of the genome-wide DNA methylation profiles between fast-growing and slow-growing broilers. PLoS One. 2013;8:e56411. doi: 10.1371/journal.pone.0056411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokinen R., Pirnes-Karhu S., Pietilainen K.H., Pirinen E. Adipose tissue NAD (+)-homeostasis, sirtuins and poly (ADP-ribose) polymerases -important players in mitochondrial metabolism and metabolic health. Redox Biol. 2017;12:246–263. doi: 10.1016/j.redox.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M.M., Nie Q.H., Peng X., Zhang D.X., Zhang X.Q. Single nucleotide polymorphisms of the chicken insulin-like factor binding protein 2 gene associated with chicken growth and carcass traits. Poult. Sci. 2005;84:1191–1198. doi: 10.1093/ps/84.8.1191. [DOI] [PubMed] [Google Scholar]
- Li H., Wang S., Yan F., Liu X., Jiang R., Han R., Li Z., Li G., Tian Y., Kang X., Sun G. Effect of polymorphism within miRNA-1606 gene on growth and carcass traits in chicken. Gene. 2015;566:8–12. doi: 10.1016/j.gene.2015.03.037. [DOI] [PubMed] [Google Scholar]
- Li K., Huang W., Wang Z., Chen Y., Cai D., Nie Q. circTAF8 regulates myoblast development and associated carcass traits in chicken. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.743757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Huang W., Wang Z., Chen Y., Cai D., Nie Q. circTAF8 regulates myoblast development and associated carcass traits in chicken. Front. Genet. 2022;12 doi: 10.3389/fgene.2021.743757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Cai B., Abdalla B.A., Zhu X., Zheng M., Han P., Nie Q., Zhang X. LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway. J. Cachexia Sarcopenia Muscle. 2019;10:391–410. doi: 10.1002/jcsm.12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang Z., He Z., Tang W., Li T., Zeng Z., He L., Shi Y. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn) Cell. Res. 2009;19:519–523. doi: 10.1038/cr.2009.33. [DOI] [PubMed] [Google Scholar]
- Lotfi E., Zerehdaran S., Ahani A.M. Genetic evaluation of carcass composition and fat deposition in Japanese quail. Poult. Sci. 2011;90:2202–2208. doi: 10.3382/ps.2011-01570. [DOI] [PubMed] [Google Scholar]
- Lu Y., Chen S.R., Liu W.B., Hou Z.C., Xu G.Y., Yang N. Polymorphisms in Wnt signaling pathway genes are significantly associated with chicken carcass traits. Poult. Sci. 2012;91:1299–1307. doi: 10.3382/ps.2012-02157. [DOI] [PubMed] [Google Scholar]
- Luo W., Lin S., Li G., Nie Q., Zhang X. Integrative analyses of miRNA-mRNA interactions reveal let-7b, miR-128 and MAPK pathway involvement in muscle mass loss in sex-linked dwarf chickens. Int. J. Mol. Sci. 2016;17:276. doi: 10.3390/ijms17030276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliavacca E., Tay S., Patel H.P., Sonntag T., Civiletto G., McFarlane C., Forrester T., Barton S.J., Leow M.K., Antoun E., Charpagne A., Seng C.Y., Descombes P., Feng L., Francis-Emmanuel P., Garratt E.S., Giner M.P., Green C.O., Karaz S., Kothandaraman N., Marquis J., Metairon S., Moco S., Nelson G., Ngo S., Pleasants T., Raymond F., Sayer A.A., Ming S.C., Slater-Jefferies J., Syddall H.E., Fang T.P., Titcombe P., Vaz C., Westbury L.D., Wong G., Yonghui W., Cooper C., Sheppard A., Godfrey K.M., Lillycrop K.A., Karnani N., Feige J.N. Mitochondrial oxidative capacity and NAD (+) biosynthesis are reduced in human sarcopenia across ethnicities. Nat. Commun. 2019;10:5808. doi: 10.1038/s41467-019-13694-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang H., He X., Li G., Xu H., Jia X., Nie Q., Zhang X. Deep sequencing analysis of miRNA expression in breast muscle of fast-growing and slow-growing broilers. Int. J. Mol. Sci. 2015;16:16242–16262. doi: 10.3390/ijms160716242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu F.F., Nie Q.H., Luo C.L., Zhang D.X., Lin S.M., Zhang X.Q. Association of single nucleotide polymorphisms of the insulin gene with chicken early growth and fat deposition. Poult. Sci. 2006;85:980–985. doi: 10.1093/ps/85.6.980. [DOI] [PubMed] [Google Scholar]
- Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J. g: Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reino L., Figueira R., Beja P., Araujo M.B., Capinha C., Strubbe D. Networks of global bird invasion altered by regional trade ban. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1700783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp P.A., Fouad G.T., Egelston C.A., Reifsteck C.A., Olson S.B., Knosp W.M., Glanville R.W., Thornburg K.L., Robinson S.W., Maslen C.L. Identification, genomic organization and mRNA expression of CRELD1, the founding member of a unique family of matricellular proteins. Gene. 2002;293:47–57. doi: 10.1016/s0378-1119(02)00696-0. [DOI] [PubMed] [Google Scholar]
- Sanchez-Gonzalez C., Herrero M.J., Salegi A.B., Nunez D.A.C., Stancic B., Pereira M.P., Contreras L., Cuezva J.M., Formentini L. Chronic inhibition of the mitochondrial ATP synthase in skeletal muscle triggers sarcoplasmic reticulum distress and tubular aggregates. Cell. Death Dis. 2022;13:561. doi: 10.1038/s41419-022-05016-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M.N., Rothschild D., Widowski T.M., Barbut S., Kiarie E.G., Mandell I., Guerin M.T., Edwards A.M., Torrey S. In pursuit of a better broiler: carcass traits and muscle myopathies in conventional and slower-growing strains of broiler chickens. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanes C.G. The Global Importance of Poultry. Poult. Sci. 2007;86:1057–1058. doi: 10.1093/ps/86.6.1057. [DOI] [PubMed] [Google Scholar]
- Shi Y.Y., He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell. Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- Sin T.K., Zhang G., Zhang Z., Gao S., Li M., Li Y.P. Cancer takes a toll on skeletal muscle by releasing heat shock proteins-an emerging mechanism of cancer-induced cachexia. Cancers (Basel) 2019;11 doi: 10.3390/cancers11091272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenton S.L., Sheremet N.L., Catarino C.B., Andreeva N.A., Assouline Z., Barboni P., Barel O., Berutti R., Bychkov I., Caporali L., Capristo M., Carbonelli M., Cascavilla M.L., Charbel Issa P., Freisinger P., Gerber S., Ghezzi D., Graf E., Heidler J., Hempel M., Heon E., Itkis Y.S., Javasky E., Kaplan J., Kopajtich R., Kornblum C., Kovacs-Nagy R., Krylova T.D., Kunz W.S., La Morgia C., Lamperti C., Ludwig C., Malacarne P.F., Maresca A., Mayr J.A., Meisterknecht J., Nevinitsyna T.A., Palombo F., Pode-Shakked B., Shmelkova M.S., Strom T.M., Tagliavini F., Tzadok M., van der Ven A.T., Vignal-Clermont C., Wagner M., Zakharova E.Y., Zhorzholadze N.V., Rozet J., Carelli V., Tsygankova P.G., Klopstock T., Wittig I., Prokisch H. Impaired complex I repair causes recessive Leber’s hereditary optic neuropathy. J. Clin. Invest. 2021;131 doi: 10.1172/JCI138267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Dong Z., Zhao L., Ren Y., Zhang N., Chen F. The Wheat 660K SNP array demonstrates great potential for marker-assisted selection in polyploid wheat. Plant Biotechnol. J. 2020;18:1354–1360. doi: 10.1111/pbi.13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., Jensen L.J., von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebbenkamp A.T.N., Varela L., Choi J., Paredes M.I., Giani A.M., Song J.E., Sestan-Pesa M., Franjic D., Sousa A.M.M., Liu Z., Li M., Bichsel C., Koch M., Szigeti-Buck K., Liu F., Li Z., Kawasawa Y.I., Paspalas C.D., Mineur Y.S., Prontera P., Merla G., Picciotto M.R., Arnsten A.F.T., Horvath T.L., Sestan N. The 7q11.23 protein DNAJC30 interacts with ATP synthase and links mitochondria to brain development. Cell. 2018;175:1088–1104. doi: 10.1016/j.cell.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng J., Gao N., Zhang H., Li X., Li J., Zhang H., Zhang X., Zhang Z. Performance of whole genome prediction for growth traits in a crossbred chicken population. Poult. Sci. 2019;98:1968–1975. doi: 10.3382/ps/pey604. [DOI] [PubMed] [Google Scholar]
- Tian W., Wang Z., Wang D., Zhi Y., Dong J., Jiang R., Han R., Li Z., Kang X., Li H., Liu X. Chromatin interaction responds to breast muscle development and intramuscular fat deposition between Chinese indigenous chicken and fast-growing broiler. Front. Cell. Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.782268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemoto Y., Sato S., Odawara S., Nokata H., Oyamada Y., Taguchi Y., Yanai S., Sasaki O., Takahashi H., Nirasawa K., Kobayashi E. Genetic mapping of quantitative trait loci affecting growth and carcass traits in F2 intercross chickens. Poult. Sci.. 2009;88:477–482. doi: 10.3382/ps.2008-00296. [DOI] [PubMed] [Google Scholar]
- von Mering C., Jensen L.J., Snel B., Hooper S.D., Krupp M., Foglierini M., Jouffre N., Huynen M.A., Bork P. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Hu H., Tian Y., Li J., Scheben A., Zhang C., Li Y., Wu J., Yang L., Fan X., Sun G., Li D., Zhang Y., Han R., Jiang R., Huang H., Yan F., Wang Y., Li Z., Li G., Liu X., Li W., Edwards D., Kang X., Wittkopp P. The chicken pan-genome reveals gene content variation and a promoter region deletion in IGF2BP1 affecting body size. Mol. Biol. Evol. 2021;38:5066–5081. doi: 10.1093/molbev/msab231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.S., Thakur M., Peng M.S., Jiang Y., Frantz L., Li M., Zhang J.J., Wang S., Peters J., Otecko N.O., Suwannapoom C., Guo X., Zheng Z.Q., Esmailizadeh A., Hirimuthugoda N.Y., Ashari H., Suladari S., Zein M., Kusza S., Sohrabi S., Kharrati-Koopaee H., Shen Q.K., Zeng L., Yang M.M., Wu Y.J., Yang X.Y., Lu X.M., Jia X.Z., Nie Q.H., Lamont S.J., Lasagna E., Ceccobelli S., Gunwardana H., Senasige T.M., Feng S.H., Si J.F., Zhang H., Jin J.Q., Li M.L., Liu Y.H., Chen H.M., Ma C., Dai S.S., Bhuiyan A., Khan M.S., Silva G., Le T.T, Mwai O.A., Ibrahim M., Supple M., Shapiro B., Hanotte O., Zhang G., Larson G., Han J.L., Wu D.D., Zhang Y.P. 863 genomes reveal the origin and domestication of chicken. Cell. Res. 2020;30:693–701. doi: 10.1038/s41422-020-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Xian Y., Li Z., Wang Z., Nie Q. G0S2 gene polymorphism and its relationship with carcass traits in chicken. Animals (Basel) 2022;12:916. doi: 10.3390/ani12070916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Lin S., Mu H., Tang X., Ou Y., Chen J., Ma Y., Li Y. Analysis of differentially expressed genes and signaling pathways related to intramuscular fat deposition in skeletal muscle of sex-linked dwarf chickens. Biomed. Res. Int. 2014;2014:1–7. doi: 10.1155/2014/724274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerehdaran S., Vereijken A.L., van Arendonk J.A., van der Waaijt E.H. Estimation of genetic parameters for fat deposition and carcass traits in broilers. Poult. Sci. 2004;83:521–525. doi: 10.1093/ps/83.4.521. [DOI] [PubMed] [Google Scholar]
- Zhang H., Shen L., Xu Z., Kramer L.M., Yu J., Zhang X., Na W., Yang L., Cao Z., Luan P., Reecy J.M., Li H. Haplotype-based genome-wide association studies for carcass and growth traits in chicken. Poult. Sci. 2020;99:2349–2361. doi: 10.1016/j.psj.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Song S., Ma Y., Xu X., Zhou G., Li C. Short-term feeding of dietary casein increases abundance of lactococcus lactis and upregulates gene expression involving obesity prevention in cecum of young rats compared with dietary chicken protein. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.02411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.