Abstract
The transcription factor csal1 is an important molecule that plays a critical regulatory function in ovarian follicle development, as confirmed by our previous data. However, the candidate genes of csal1 and its regulatory mechanism remain poorly understood in the granulosa cells (GCs) of chicken prehierarchical follicles (PFs). Six transcriptomes of csal1 and empty vector were analyzed in Chinese Dagu hens by RNA sequencing. Six cDNA libraries were constructed, with more than 42 million clean reads and 16,779 unigenes. Of these 16,779 unigenes, 2,762 differentially expressed genes (DEGs) were found in GCs, including 1,605 upregulated and 1,157 downregulated unigenes. Fourteen genes, including BMP5, TACR2, AMH, PLAG1, MYOD1, BOP1, SIPA1, NOTCH1, BCL2L1, SOX9, ADGRA2, WNT5A, SLC7A11, and GATAD2B, were related to GC proliferation and differentiation, hormone production, ovarian follicular development, regulation of reproductive processes, and signaling pathways in the PFs. Further analysis demonstrated the DEGs in GCs of ovarian follicles were enriched in neuroactive ligand–receptor interaction, cell adhesion molecules, and pathways related to cytochrome P450, indicating a critical function for csal1 in the generation of egg-laying features by controlling ovarian follicle development. For the first time, the current study represents the transcriptome analysis with ectopic csal1 expression. These findings provide significant evidence for investigating the molecular mechanism by which csal1 controls PF development in the hen ovary.
Key words: Chinese Dagu hens, csal1, differentially expressed gene, ovarian follicle, transcriptomics
INTRODUCTION
Ovarian follicle development in hens represents a greatly complex event that involves multiple endocrine, paracrine, and autocrine processes spatially and temporally. It features substantial alterations in the course of follicle selection (Woodruff and Shea, 2007). The problem of follicular development can result in reduced reproductivity, especially low egg-laying rates, which constitute a major economic feature in poultry industry worldwide. A strict control of follicular hierarchy occurs in hens (Chen et al., 2021). Only a single dominant follicle is usually recruited into the preovulatory hierarchy in approximately one day, which is selected from 6 to 8-mm diameter prehierarchical follicles (PFs), reaches maturity, and undergoes ovulation. Therefore, PF development is very important. Diverse biological processes, cellular components, molecular functions, or some signaling pathways may affect proliferation, differentiation, hormone production, secretion of granulosa, regulation of reproductive processes, and even growth in PFs (Xu et al., 2018). In the ovary, granulosa cells (GCs) constitute a critical site of estrogen biosynthesis for local utilization and provide endocrine signals to other tissues (Nelson and Bulun, 2001).
In recent years, RNA sequencing (RNA-seq) has been instrumental in functional genomics. The information about all the transcripts of an organism in a particular state could be generated in a comprehensive and quick manner to solve complex biological problems (Miao et al., 2016; Wu et al., 2020; Wang et al., 2021b). Several studies also revealed many mRNA transcripts in ovarian follicles (Yang et al., 2008; Wang and Ma, 2019), which are associated with genetic improvement in the domestic chicken breed.
Chinese Dagu chicken is an important animal resource, which has been bred for more than 200 yr. It is widely distributed in Liaoning Province of China, mainly produced in Zhuanghe City of Liaoning Province, so it is also called Zhuanghe chicken. Chinese Dagu chicken is a dual-purpose local chicken breed characterized by large eggs. The chicken is large and solid, with strong foraging ability, quality and quantity eggs, thick and solid eggshell, fresh and tender meat, and strong disease resistance. Our study had confirmed the correlation between csal1 and laying traits of Chinese Dagu chicken (Zhu et al., 2018). However, the commercial production of Chinese Dagu chicken lacks systematic research on their laying performance. So, RNA-seq was carried out for identifying DEGs between overexpression and normal expression of csal1 in GCs of hen ovarian prehierarchical follicles to comprehensively characterize the regulatory role of csal1 in PF development.
Chick orthologue of the SALL1 gene, csal1, is a member of chicken spalt family which was characterized by multiple C2H2-type zinc-finger motifs (ZFs). To data, three members of the spalt family have been identified including csal1, csal3, and csal4. As a transcriptional repressor, the expression of csal1 is predominantly detected in the developing heart, mesoderm, ectoderm, and neural tube of the early embryo (Sweetman et al., 2005). Our previous study, which was the first related report, showed that csal1 suppresses the GC proliferation and steroidogenesis to regulate development and growth of ovarian follicles (Zhu et al., 2019).
Here, we aimed to obtain downstream target genes of csal1 in GCs from PFs through high-throughput transcriptome technology, which were the key candidate genes contributing to ovarian function control, thus influencing egg-laying potential. The study also focused on the regulatory role of csal1 in the biological processes and pathways associated with egg-laying in Chinese Dagu chicken. Additionally, RNA-seq data were validated by quantitative real-time polymerase chain reaction (RT-qPCR). This study provided important evidence for exploring the mechanisms underpinning egg-laying in poultry, and then provide molecular genetic markers for better egg production in the indigenous Chinese chicken breeding.
MATERIALS AND METHODS
Chicken and Samples
Chinese Dagu hens, provided by the Farm of Jinzhou Medical University, were euthanized at the age of 21 wk (n = 10), and their PFs (6 to 8-mm diameter) (Gilbert et al., 1983) were collected for experiments. GCs from PFs (6 to 8-mm diameter) underwent culture as reported in a previous study (Zhu et al., 2019). All the procedures involving animals followed the guidelines formulated by the Ministry of Agriculture of the People's Republic of China, with approval from the ethics committee of Jinzhou Medical University (201805024, Jinzhou, China).
Cell Culture and Cell Transfection
GCs were obtained as described in our previous report (Zhu et al., 2019). The specificity of GCs was detected by fluorescent and H&E staining procedures (Lyu et al., 2016). Simply, the pYr-adshuttle-4-csal1 plasmid and the corresponding empty vector were transfected into GCs with Lipofectamine 2000 (Invitrogen, Carlsbad, CA ). The generation of pYr-adshuttle-4-csal1 has been described previously (Zhu et al., 2019). Cells (105/well in 24-well plates) were cultured in basal medium with 1 μL/mL polybrene (Sigma, St. Louis, MO) at 37°C in a humid environment containing 5% CO2 (Xu et al., 2018) for 24 h.
Library Construction and Illumina Sequencing
Total RNA extraction utilized TRIzol Reagent (Invitrogen). RNA purity and amounts were assessed on a NanoDrop 8000 (Thermo Fisher Scientific, Waltham, MA ) and a Bioanalyzer 2100 system (Agilent, Santa Clara, CA). RNA (3 µg) was utilized for RNA-seq. The remaining RNA was kept at –80°C for validation assays. cDNA library construction utilized TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA), as directed by the manufacturer. The mRNA purification from total RNA utilized poly-T oligo-linked magnetic beads. 300-bp cDNA fragments were screened, AMPure XP system (Beckman Coulter, Fullerton, CA) was used for library fragment purification. DNA fragments containing adaptors at both ends underwent selective enrichment with the Illumina PCR Primer Cocktail by PCR amplification (15 cycles). Finally, PCR products underwent purification (AMPure XP system), and the quality of libraries was evaluated on a Bioanalyzer 2100 system.
RNA-Seq Data Analysis
The prepared libraries underwent sequencing by next-generation sequencing on Illumina HiSeq 2500. Raw reads (FASTQ format) were refined by removing low-quality reads as well as adaptors with in-house perl scripts to yield clean reads. Meanwhile, the contents of Q20, Q30, and N (fuzzy base) in raw data were determined. Clean reads were used in all downstream analyses, with alignment with the reference genome carried out with HISAT 2.1.0 (Pertea et al., 2016; Kim et al., 2019).
The reads mapped to various genes were separately counted using the HTSeq v0.6.1 software (https://htseq.readthedocs.io/en/master/). Gene expression levels were reflected by the amounts of fragments per kilobase of transcript per million fragments (FPKM) after normalization. The expression patterns of DEGs were assessed with the Multi-Experiment Viewer software v4.9.0 (https://mybiosoftware.com/mev-4-6-2-multiple-experiment-viewer.html) according to the normalized FPKM + 1 value between the case and control groups after normalization. The Benjamini and Hochberg's method was utilized to control the false discovery rate and derive the adjusted P value (Padj). The degree of gene expression was expressed as |log2FoldChange| (Log2FC). Genes with Padj < 0.05 and Log2FC>1 were identified with DESeq. Volcano plots for visualizing significantly regulated genes were obtained with the R package “ggplot2” (v3.5.0) (Wang et al., 2010). The R program was utilized for data analysis, including principle component analysis (PCA).
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Analyses of DEGs
Gene Ontology (GO; http://www.geneontology.org/) enrichment analysis was used for annotating and analyzing DEGs (Ashburner et al., 2000). GO terms include cell component (CC), molecular function (MF) and biological process (BP), in which DEGs were enriched. Bonferroni corrected P < 0.05 was used as a criterion for GO annotation. Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) was used to assess pathway enrichment. KEGG is a comprehensive database resource for analyzing DEGs through KEGG pathway enrichment. The results with P < 0.05 and an FDR close to 0 were considered significantly enriched by DEGs.
Protein Interaction Analysis
DEG–protein interactions were analyzed with STRING (http://string-db.org/). DEG–protein interaction network data files were visually edited using Cytoscape v3.5.1 (http://www.cytoscape.org/).
RNA-Seq Validation by RT-qPCR
Fourteen candidate genes were screened to validate RNA-seq data by RT-qPCR. Total RNA was isolated from csal1-transfected cells at 1 × 106/well using an RNA extraction kit (TaKaRa, China), and the first strand cDNA was synthesized from 1 μg of total RNA using PrimeScript RT reagent Kit (TaKaRa, RR047A) according to the manufacturer's instructions. Csal1 and 18S rRNA amounts were examined by RT-qPCR with TB Green (TaKaRa, RR820A) on a 7500 Fast Real-Time PCR System (Applied Biosystems, Grand Island, NY). The 2−ΔΔCt method was used for determining relative mRNA amounts, normalized to 18S rRNA expression. Table 1 showed all RT-qPCR primers (TaKaRa). Three independent triplicate assays were carried out.
Table 1.
The sequences of genes for RT-qPCR.
| Gene | Primer | Sequence (5′-3′) | PCR products |
|---|---|---|---|
| 18S | Forward | TAGTTGGTGGAGCGATTTGTCT | 169bp |
| Reverse | CGGACATCTAAGGGCATCACA | ||
| csal1 | Forward | CCCAGGAAGCAAGGTGTA | 167bp |
| Reverse | CGTTGGCATGTCCGTATT | ||
| BMP5 | Forward | GTGAGCAAAAGCAAGCAT | 260bp |
| Reverse | ATCAAAGTAGAGCACGGAG | ||
| TACR2 | Forward | CAGATTTGCTGATGTCCG | 310bp |
| Reverse | CACTTTGTTGTCCCGTTG | ||
| AMH | Forward | GGAGGAGATGGGACTTGGAG | 261bp |
| Reverse | AAGGCACTTTCATAGCGGG | ||
| PLAG1 | Forward | ATGATGGGTCTGGGGAGGT | 311bp |
| Reverse | TCGTGGTAGGGTGGTGGTT | ||
| SLC7A11 | Forward | GAAAAAAACATACCCCTCG | 214bp |
| Reverse | CACCATTCATAGACCCAAA | ||
| MYOD1 | Forward | CACGGGAACCCACACGAGGA | 352bp |
| Reverse | AGCGAGGGCTGGAGGCATCT | ||
| BOP1 | Forward | CAGAGGAAGATGAGGGTGAACG | 173bp |
| Reverse | TCGGAGCCTGATGCCAAC | ||
| SIPA1 | Forward | GCCTCGGCTACATCCCCAAT | 168bp |
| Reverse | GACCCCACCGCTCATCTTCA | ||
| NOTCH1 | Forward | CTGGGGACACCACCTACGA | 195bp |
| Reverse | GCTGGCACTCATCCACATC | ||
| BCL2L1 | Forward | CTTTCAGCGACCTCACCTCC | 192bp |
| Reverse | ACACAATGCGTCCCACCA | ||
| SOX9 | Forward | AATACCTGCCCCCCAACG | 382bp |
| Reverse | TACTGCGAGCGGGTGATG | ||
| ADGRA2 | Forward | TACACCAACGACATCACCCG | 146bp |
| Reverse | GCCACATAGAGGACATCAACCA | ||
| WNT5A | Forward | CTTCAACGCACCCACCAT | 151bp |
| Reverse | GCAGCACATCAGTTCACAGC | ||
| GATAD2B | Forward | GCTGGTGCTGCTGAAGAAG | 105bp |
| Reverse | GGAGATGGCTGGACGATAGA |
RESULTS
RNA-Seq Data Quality
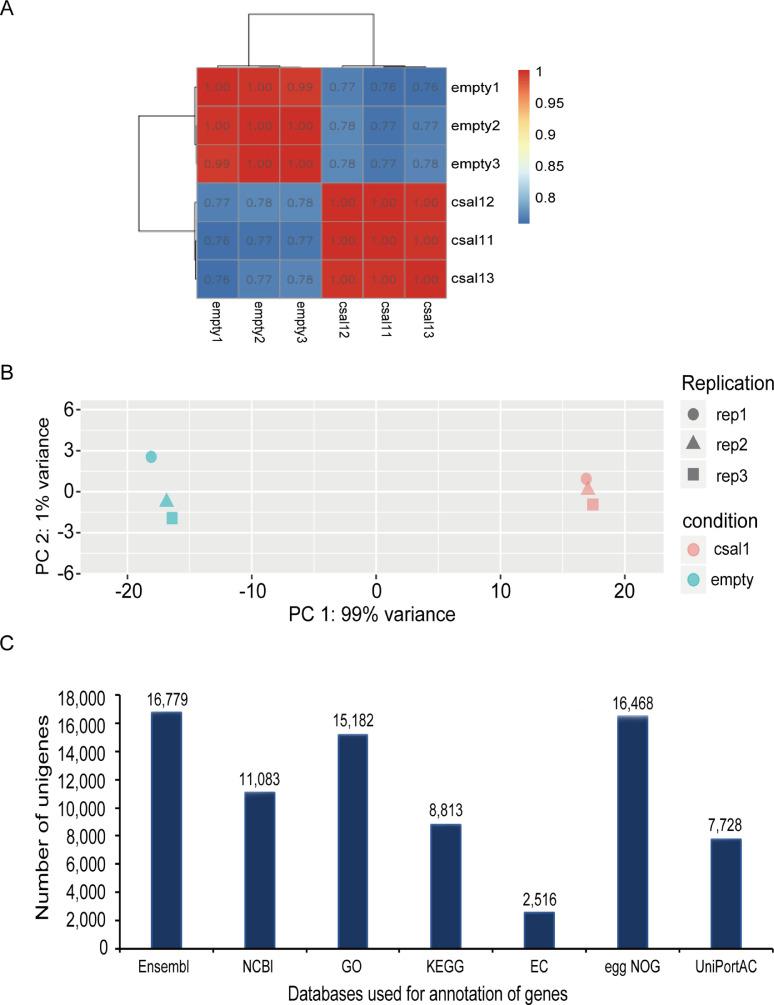
Six cDNA libraries were constructed for csal1 overexpressing and control GCs from 6 to 8-mm diameter PFs using the Illumina HiSeq 2500 platform. RNA-seq raw sequences in each library ranged from 45,486,652 to 52,635,202 reads. Of these raw reads, >97.26% and 92.73% showed quality scores at the Q20 and Q30 levels, respectively. The highest N (base) frequency in raw data was 0.001% (Supplementary Table 1). After filtering, >41 million clean reads per library were obtained. High-quality clean reads and bases (bp) of clean data were 90.93 to 92.98% of all sequences of raw reads (Supplementary Table 2). The percentage of all mapped sequences/reads ranged from 92.43 to 94.17% of the clean reads. Mapped reads included 5.29 to 5.70% of clean reads that were multiple mapped reads, and 94.30 to 94.71% of clean reads were uniquely mapped reads (URs) (Supplementary Table 3). Of all RNA-seq mapped events, read distribution was determined. All reads mapped to genes (MGs) covered 85.51 to 86.68% of the identified URs, and exons accounted for 73.67 to 89.03% of reads from MGs (Supplementary Table 4). FPKM were quantified to assess gene expression, and Pearson's correlation coefficients of paired specimens with their biological replicates were >0.99, suggesting the general reproducibility of sequencing data, which was close to 0.8 (Figure 1A, Supplementary Figure 1). The values of PC1 and PC2 were, respectively, 99% and 1% from the PCA, and an overview of transcriptomic variation was obtained (Figure 1B). Genes with greatest variance (PC1) were further examined by GO analysis. The fastq files have been submitted to Sequence Read Archive database (SRA accession PRJNA854338).
Figure 1.
Features of sequencing data. (A) Pairwise Pearson's correlation analysis of sequencing data in triplicate for 2 specimens. (B) Principal component analysis (PCA) of transcriptomic variations. Shapes, distinct replication patterns of the collected cells; colored dots, distinct specimens. (C) Annotations of gene numbers according to various databases.
The reference genome of Ensembl of sequencing unigenes was assessed and annotated NCBI and 5 additional databases. Among these, 16,779 unigenes underwent annotation from the 6 cDNA libraries. In the aforementioned database, the annotation numbers and percentages of genes are shown in Figure 1C.
DGE Analysis
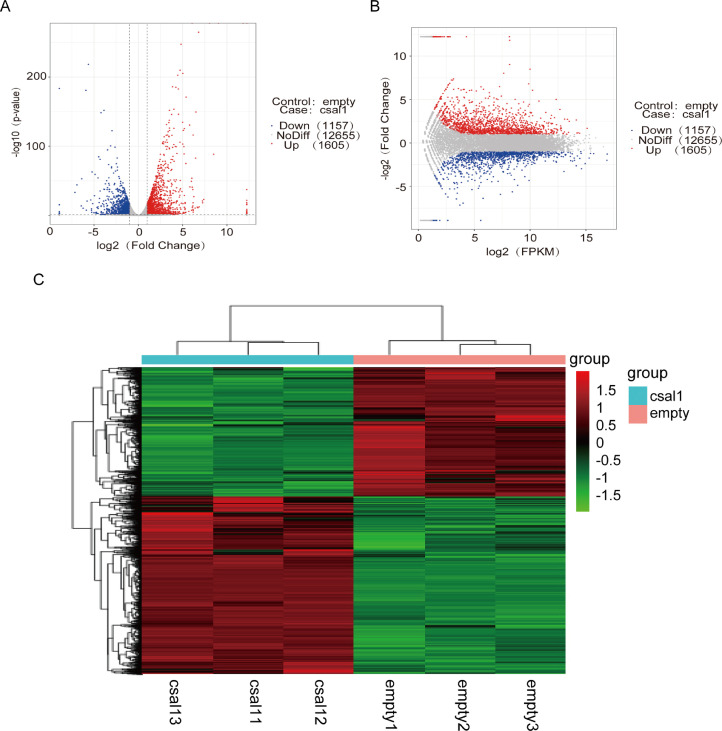
Of the 16,779 unigenes, 2,762 DEGs from GCs of ovarian follicles (6 to 8-mm diameter; SYF) were determined in the 6 cDNA libraries, with 1,605 significantly upregulated and 1,157 markedly downregulated unigenes (|log2FoldChange| > 1, P < 0.05; Figures 2A and 2B). The FPKM hierarchical clustering map (Figure 2C) visually represents gene expression patterns in the specimens and highlighted data repeatability and accuracy.
Figure 2.
Detection and analysis of DEGs between csal1 overexpressing and nontransfected granulosa cells from hen ovarian prehierarchical follicles. (A) Volcano plot of DEGs. The X-axis is log2foldchange; the Y-axis is - log10 (P-value). The two vertical dashed lines show the two-fold change threshold; the horizontal dashed line shows a P-value of 0.05. Red, blue, and gray indicate significantly upregulated, downregulated, and non-differentially expressed genes, respectively. (B) MA of DEGs. The X-axis is log2 Fragments per Kilobase of transcript per Million mapped reads (FPKM); the Y-axis is log2foldchange. Red, blue and gray dots represent significantly upregulated, downregulated, and non-differentially expressed genes, respectively. (C) Clustering of DEGs. Genes and samples are placed horizontally and as column, respectively. Red and green indicate high- and low-expression genes, respectively.
GO Analysis of DEGs
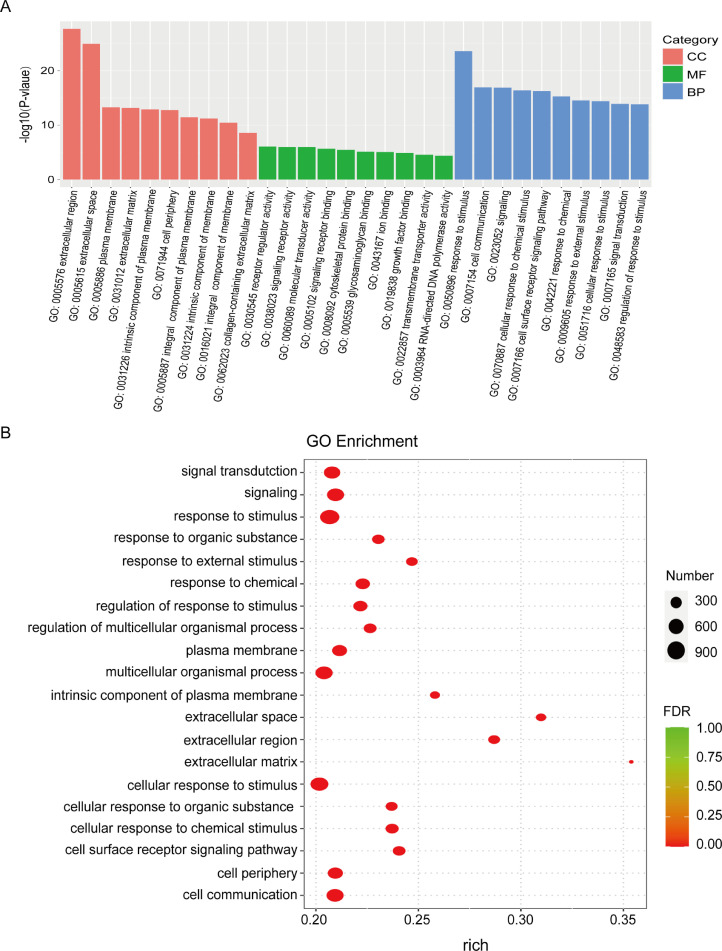
Based on the comparison of GCs from ovarian PFs in the csal1 overexpression and control groups, 2,762 DEGs were enriched in 12,184 distinct GO terms. A total of 9,157, 1,013, and 2,014 DEGs were assigned to the BP, CC, and MF GO categories, respectively. The top 10 GO terms with significance for BP, CC and MF are shown in Figure 3A (P < 0.05). This finding also indicated that the main function of csal1 involved biological processes in ovarian follicle development. Based on GO enrichment data, enrichment degree was assessed by the richness factor, FDR and the amounts of genes enriched in a given GO term. The higher the richness factor, the higher the degree of enrichment, the more significant the enrichment. The top 20 GO terms with the lowest FDRs are displayed in Figure 3B. Moreover, the top 10 GO terms for BP, MF, and CC are shown in the GO DAG structure (Supplementary Figure 2).
Figure 3.
GO enrichment analysis of DEGs between csal1 overexpressing and nontransfected granulosa cells from hen ovarian prehierarchical follicles. (A) The bar plot of Go enrichment analysis. X-axis, GO terms: biological process, cellular component, and molecular function. Y-axis, - log10 (P-value) of each term. (B) Bubble diagram of GO enrichment analysis. Circles of different sizes represent quantities, and different colors represent distinct FDR values.
KEGG Pathways Involving the DEGs
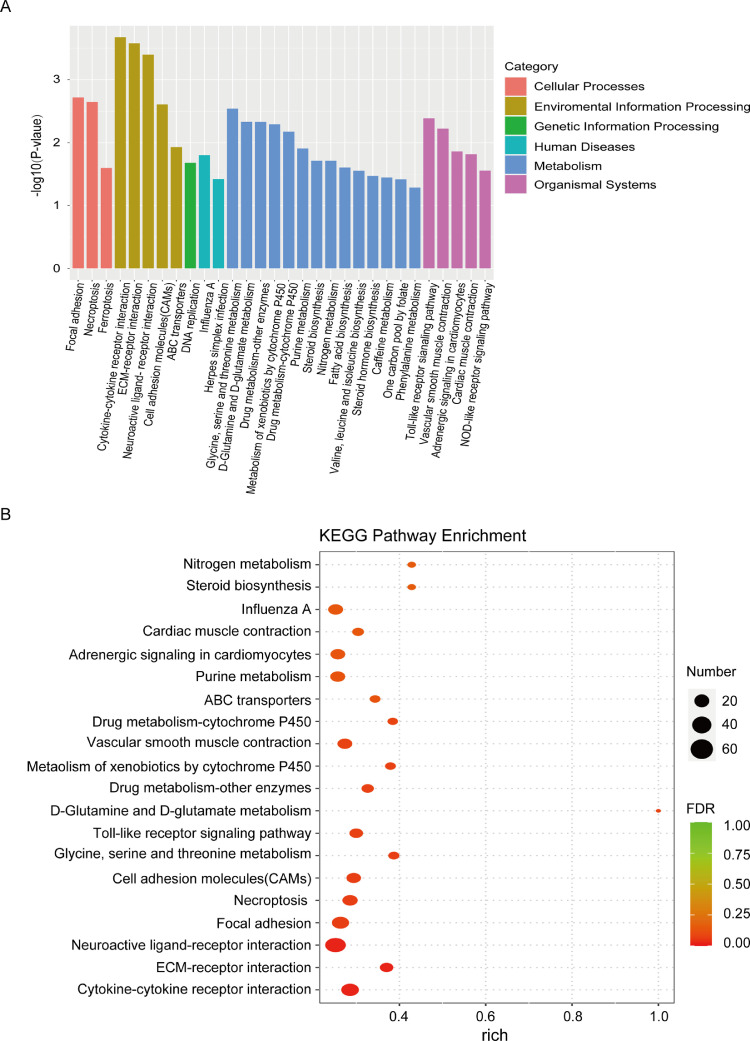
KEGG pathway enrichment analysis was carried out to assess csal1 overexpressing and nontransfected GCs from ovarian follicles of 6 to 8-mm diameter (SYF) to further reveal the most relevant signaling pathways associated with ovarian follicle development and identify GO-annotated DEGs involved in the transducing events. Among these, 30 most relevant pathways with remarkable enrichment are shown in Figure 4A. To summarize, 534 DEGs from these 20 significantly KEGG pathways, including 342 upregulated and 192 downregulated, were enriched (Figure 4B).
Figure 4.
KEGG pathway enrichment analysis of DEGs between csal1 overexpressing and non-transfected granulosa cells from hen ovarian prehierarchical follicles. (A) The bar plot of KEGG pathway enrichment analysis. X-axis, GO terms: cellular processes, environmental information processing, genetic information processing, human diseases, metabolism, organismal systems. Y-axis, - log10 (P-value) of every term. (B) Bubble diagram of KEGG pathway enrichment analysis. Circles of different sizes represent quantities, and different colors represent distinct FDR values.
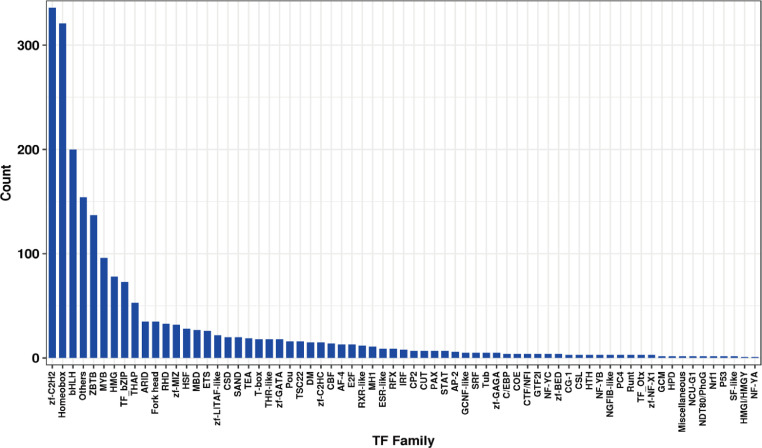
Transcription Factor Family Analysis
The transcription factors and their respective families were predicted and obtained through the comparison with the animal transcription factor database. About 2,073 transcription factors, which belonged to 69 transcription factor families, were obtained (Figure 5). The transcription factor family with the largest number of members was zf-C2H2, successively followed by homeobox and bHLH. They all had more than 200 members, and zf-C2H2 had 316 members.
Figure 5.
Statistical map of transcription factor family. X-axis, different transcription factor families. Y-axis, number of genes falling into a given transcription factor family.
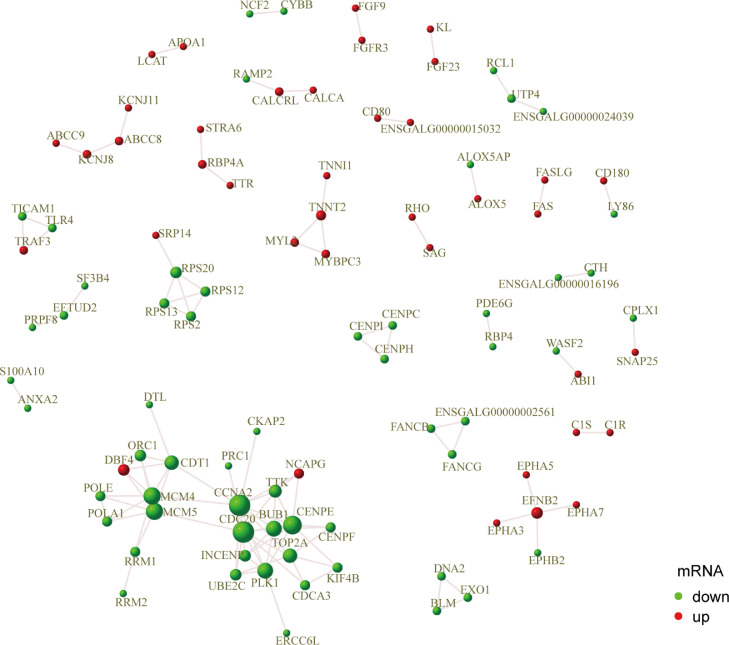
Protein Interaction Network
The associations among the protein products of the identified DEGs were analyzed for further clarifying the potential genetic mechanism of ovarian follicle development in chickens. The parameters were set to score >0.95. The protein-protein interaction network is depicted in Figure 6. In PFs, interactions among 253 proteins were found. PLK1, TTK, CDC20, CENPC, INCENP, KIF4B, MCM, NCAPG, FGF, FN1, ORC1, RAC2, and VEGFC, which are mainly involved in mitosis, embryonic development and cell proliferation and differentiation, might be in central effectors for these interactions.
Figure 6.
Predicted protein-protein interaction network. The points in the figure are genes (corresponding proteins). Red and green indicate upregulated and downregulated genes, respectively.
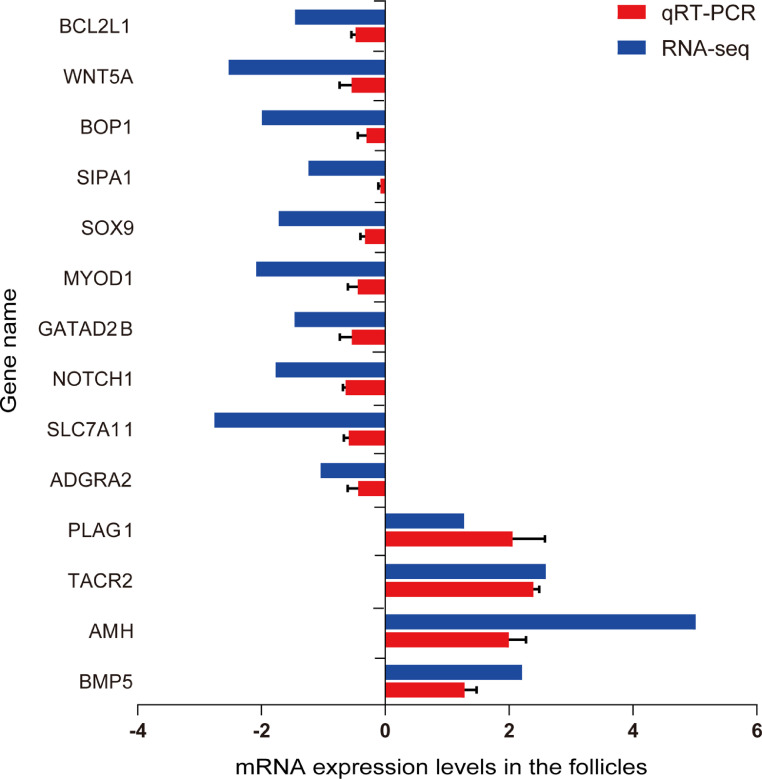
Validation of the Selected DGEs by RT-qPCR
Fourteen candidate DGEs were chosen for RT-qPCR validation (Table 2), including 4 relevant upregulated genes (BMP5, TACR2, AMH, and PLAG1) and 10 related downregulated genes (MYOD1, BOP1, SIPA1, NOTCH1, BCL2L1, SOX9, ADGRA2, WNT5A, SLC7A11, and GATAD2B). As shown in Figure 7, all 4 DGEs upregulated in RNA-seq were upregulated in GCs with csal1 overexpression from ovarian PFs. The 10 tested downregulated DGEs were also downregulated. These results showed that differences in the expression levels of these candidate DGEs detected by RT-qPCR were consistent with RNA-seq data. The observed expression trends confirmed the RNA-seq results, implying that the above RNA-seq results were reliable.
Table 2.
List of partially representative DEGs.
| Genes | Different expression type | Description | log2FoldChange | pval | KEGG | UniProtAC | eggNOG |
|---|---|---|---|---|---|---|---|
| BMP5 | Upregulated | bone morphogenetic protein 5 | 2.207191257 | 2.84E-17 | K04663 | P87373 | veNOG00413 |
| TACR2 | Upregulated | tachykinin receptor 2 | 2.592664911 | 0.027384 | K04223 | - | veNOG07359 |
| AMH | Upregulated | anti-Mullerian hormone | 5.012564505 | 6.5E-206 | K04665 | Q788U7 | veNOG16048 |
| PLAG1 | Upregulated | PLAG1 zinc finger | 1.273295028 | 6.69E-09 | K19484 | Q58NQ5 | veNOG02243 |
| MYOD1 | Downregulated | myogenic differentiation 1 | −2.084990972 | 0.006683 | K09064 | P16075 | opiNOG36948;veNOG00749 |
| BOP1 | Downregulated | block of proliferation 1 | −1.992773591 | 1.14E-27 | - | - | veNOG15316;fiNOG04039 |
| SIPA1 | Downregulated | signal-induced proliferation-associated 1-like protein 3-like | −1.242977568 | 0.000297 | - | - | veNOG04528 |
| NOTCH1 | Downregulated | notch 1 | −1.77078885 | 1.46E-24 | K02599 | F1NZ70 | veNOG02575 |
| BCL2L1 | Downregulated | BCL2 like 1 | −1.458977008 | 2.23E-10 | K04570 | Q07816 | veNOG10445 |
| SOX9 | Downregulated | SRY (sex determining region Y)-box 9 | −1.722821315 | 1.12E-12 | K18435 | P48434 | veNOG11187 |
| ADGRA2 | Downregulated | adhesion G protein-coupled receptor A2 | −1.045490949 | 8.4E-05 | K08461 | - | veNOG03321 |
| WNT5A | Down-regulated | Wnt family member 5A | −2.532140807 | 0.000399 | K00444 | Q9YGX6 | veNOG08444 |
| SLC7A11 | Downregulated | solute carrier family 7 member 11 | −2.760443999 | 1.57E-28 | K13869 | - | veNOG05087 |
| GATAD2B | Downregulated | GATA zinc finger domain containing 2B | −1.463635505 | 1.85E-16 | - | - | veNOG13402 |
Figure 7.
Gene expression levels (Log2FC) of 14 candidate genes assessed by RNA-Seq and RT-qPCR. Relative expression levels for various genes were obtained by the 2−ΔΔCt method with 18S rRNA as a reference gene.
DISCUSSION
The egg production trait, which is a critical feature in the global poultry industry, is affected by follicular development. Follicles at distinct developmental stages have different molecular features and functions in ovary growth and development. In particular, the genetic regulation of 6 to 8-mm diameter follicles contributes to follicle selection (Johnson and Woods, 2009; Johnson, 2015) and may have a unique effect on the hierarchy of undifferentiated PFs. Therefore, SYF (6 to 8-mm diameter) was selected in this study. Multiple reports identified factors controlling follicular development and the selection of undifferentiated PFs into the differentiated preovulatory follicle hierarchy (Johnson, 2015; Xu et al., 2018). Among these, our previous study revealed that csal1 suppresses the GC proliferation and steroidogenesis (Zhu et al., 2019). However, the exact regulatory effect of csal1 in PF development remains undefined.
The high-throughput sequencing technology, for example, RNA-seq, has been widely used in animal breeding research. RNA-Seq could assess the expression levels, structures, and modulatory profiles of genes by examining the transcriptomes of different cells or tissues. The recent findings provide novel insights into reproduction in chickens and identify effective biomarkers for genetic improvement in the chicken breed (Wang and Ma, 2019; Zhang et al., 2019; Mishra et al., 2020; Chen, et al., 2021).
Here, transcriptome analysis indicated that the main function of csal1 is to regulate BP in ovarian follicle development. Of which, 14 DEGs were screened, which were related to GC proliferation, differentiation, hormone production, ovarian follicular development, regulation of reproductive processes, and signaling pathways in the PFs of hen ovary included BMP5, TACR2, AMH, PLAG1, MYOD1, BOP1, SIPA1, NOTCH1, BCL2L1, SOX9, ADGRA2, WNT5A, SLC7A11, and GATAD2B. In addition, the 14 DEGs were characterized by KEGG pathway enrichment analysis, 14 DEGs were enriched in 13 KEGG pathways, including cytokine–cytokine receptor interaction, neuroactive ligand–receptor interaction, progesterone-mediated oocyte maturation, Wnt signaling pathway, and TGF-β signaling pathway, which were significantly enriched and are involved in signaling molecules and interaction, endocrine system, amino acid biosynthesis and metabolism, signal transduction, and cell growth and death. The identified DEGs were tightly associated with multiple physiological events of follicle growth and development, indicating their possible involvement in hen egg-laying potential.
The DEGs identified in this study included many receptor genes, such as follicle-stimulating hormone receptor (FSHR), estrogen receptor 1 (ESR1), nerve growth factor receptor (NGFR), G protein–coupled receptor (GPCR), gamma-aminobutyric acid type A receptor (GABRP), interleukin-1 receptor (IL1RL1), and so on. Of these genes, follicle-stimulating hormone (FSH) represents a key player in germ cell formation (Wang et al., 2021b). It participates in GC proliferation, antrum formation in secondary ovarian follicles, antral follicle development and estradiol biosynthesis, ultimately promoting folliculogenesis, oogenesis, oocyte meiotic maturation, and oocyte competence (Padmanabhan and Cardoso, 2020). FSH secretion relies on the HPG axis. FSH represents a glycoprotein with α and β subunits bound by non-covalent interactions (Das and Kumar, 2018). FSHβ affects target cells (GCs) when connected to its FSHR in the cell membrane (Szyma´nska et al., 2018). FSHR is a member of the G protein–coupled receptor (GPCR) family, which activates diverse pathways such as cAMP/PKA, PKC/MAPK and Ca+/CaMKII pathways (Szyma´nska et al., 2018). In addition, recent findings showed that FSH has new functions, such as regulation of bone formation, fat metabolism, energy homeostasis, cholesterol biosynthesis, and cardiovascular pathologies by interacting with its receptor, FSHR (Recchia et al., 2021).
Anti-Mullerian hormone (AMH) is secreted by the GCs of primary and preantral follicles, affecting folliculogenesis mainly by suppressing preantral follicles’ sensitivity to FSH and downregulating aromatase, inducing dominant follicle selection. In addition, AMH suppresses the growth of primordial follicles in the ovarian reserve, thereby suppressing follicular development (Dewailly et al., 2016). The AMH effect was further demonstrated by FSH treatment of pre-pubertal AMH (−/−) mice, promoting follicle growth. Similar findings have been obtained in a human study examining AMH added to culture GCs (Durlinger et al., 2001; Pellatt et al., 2011). AMH amounts are elevated during puberty because of the elevated oocyte maturation rate, and low in menopause, which was consistent with this study. Not only their systemic regulation by hormones (gonadotropins) but also their intraovarian regulation by gonadal steroids, growth factors, cytokines, and intracellular proteins, such as B cell lymphoma/leukemia 2 (BCL2) family members which are widely expressed in embryonic tissues (Matsuda et al. 2012). As one of the key transcription factors governing early gonad's differentiation, SOX9 was abnormally expressed in exposed ovary, while the formation of follicle was partially restored by SOX9 knockdown (Qiqi et al. 2022). The expression of Sox9 is regulated by members of the FGF, transforming growth factor-b (TGF-b), Wnt families, and bone morphogenetic protein (BMP) (Quintana et al. 2009). And BMP-SMAD1/5/8 signaling is critical for follicular activation and development, as well as GC proliferation, atresia and luteinization. In addition, Notch signaling is also induced by gonadotrophins, with a critical role in oocyte development (Jozkowiak et al., 2020).
Among the DEGs, solute carrier family 7 member 11 (SLC7A11) was significantly downregulated after the overexpression of csal1. Although the physiological function of the SLC7A family has been widely explored in mammals, such studies are scarce in poultry. SLC7A11 (or xCT), a cystine and glutamate antiporter, imports cystine for glutathione synthesis and antioxidation (Xu et al., 2021). The SLC7A family comprises sodium-independent amino acid transporters inducing the transport of multiple amino acids, including glycine, serine, alanine and cysteine. As neuronal transporters, they primarily regulate excitatory glutamatergic neurotransmission (Sawada et al., 2002). Amino acids regulate the reproductive, cardiovascular, and immune systems, implying their metabolism in the pituitary gland may be critical in modulating reproduction in chickens. Therefore, we speculated SLC7A11 downregulation after csal1 overexpression controlling amino acid synthetic, transport, and metabolic pathways might result in altered amino acid use, which might influence the development and functions of downstream target organs via the endocrine vasculature, finally affecting reproductive physiology and egg-laying potential in hens.
The spalt members, featuring many C2H2-type zinc finger (ZF) motifs, are transregulators of genes in growth and developmental events. Of these, SALL1 can interact with β-catenin and synergistically activate a reporter construct that responds to Wnt signaling by recruiting remodeling factors to heterochromatin (Zhu et al., 2019). In this study, DEGs related to the ZF structure and Wnt/β-catenin signaling pathway were also screened, such as ovo like ZF 2 (OVOL2) and pleomorphic adenoma gene 1 (PLAG1), Wnt family member 5A (Wnt5A), Dickkopf Wnt signaling pathway inhibitor 3 (Liu et al., 2021), and frizzled related protein (Tuerlings et al., 2022).
Previous studies reported PLAG1 encodes a ZF transcription factor. PLAG1 contributes to HGMA2–PLAG1–IGF2 signaling that after disruption causes Silver–Russell syndrome, which is manifested by severe intrauterine growth restriction. PLAG1 is abundant during development in the mouse brain (Tran et al., 2021). It constitutes cell proliferation via direct regulation of multiple target genes, for example, many growth factors, including IGF2 (Van Dyck et al., 2007). Also, PLAG1-deficient mice show growth retardation, which is presumed to be associated with IGF2 regulation by PLAG1 to some extent (Hensen et al., 2004). Further, it was shown PLAG1 controls milk production, reproductive potential, muscle generation, and body height in livestock (Fink et al., 2017). This suggested that PLAG1 affects growth and embryonic development in domesticated animals, possibly in follicles, which corroborated our results. As one of the subunits forming the nucleosome remodeling and histone deacetylation complex, GATA ZF domain containing 2B (GATAD2B) is critical in chromatin alteration and transcriptional modulation (Wang et al., 2021a). This structure and function are highly consistent with those of csal1. Wnt5A represents a Wnt family member transcriptionally regulated by Wnt signaling, greatly affecting cell proliferation, differentiation, migration and movement. Wnt5A also contributes to development, homeostasis and disease (Astudillo, 2022). The growth of hair follicles is controlled by diverse pathways, for example, Wnt, Bmp, Notch, and others. Of these, Wnt signaling is critically important. In addition, overexpressed Wnt5a promotes β-catenin degradation, inhibits the classical Wnt/β-catenin pathway and suppresses hair follicle growth (Wei et al., 2022). Generally speaking, these results corroborated the aforementioned findings.
Here, the screened DEGs contributed to controlling GC proliferation, differentiation and apoptosis, oocyte meiosis and maturation, follicular differentiation and atresia, and gonadotrophin-release hormone secretion through crosstalk or interactions with multiple pathways. These findings provide compelling evidence of the significant differences in gene expression levels in the whole genome during the follicular development in Chinese Dagu chickens, which confirmed the role of csal1 in follicular development. Taken together, the present data provide new insights into the regulation of follicular development in chickens.
CONCLUSIONS
In summary, this study was the first to reveal the transcriptome analysis of GCs from ovarian PFs (SYF) between those with overexpression of csal1 and normal ones. The current data suggested that the screened DGEs could constitute a basis for in-depth exploration of the mechanism by which csal1 controls genes that contribute to PF development and growth in the hen ovary from Chinese Dagu chickens in vivo. This study also explored the biological events and pathways associated with egg-laying and follicle development in chickens. An ongoing study by our team aims to precisely examine the regulatory role of the csal1 gene in PF development, which could provide major insights into the methods for improving egg production in poultry. Finally, identifying valuable biomarkers of the egg-laying trait could improve breeding programs to enhance egg production by Chinese Dagu chickens.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China [grant number 32002154]; the Natural Science Foundation of Liaoning Province [grant number 2021-BS-262]; Basic scientific research projects of Liaoning Education Department [grant number LJKMZ20221240]; and the Training Programs of Innovation and Entrepreneurship for Undergraduates of Jinzhou Medical University [grant number 201824]
DISCLOSURES
The authors declare no conflicts of interest.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2022.102310.
Appendix. Supplementary materials
REFERENCES
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astudillo P. An emergent Wnt5a/YAP/TAZ regulatory circuit and its possible role in cancer. Semin. Cell Dev. Biol. 2022;125:45–54. doi: 10.1016/j.semcdb.2021.10.001. [DOI] [PubMed] [Google Scholar]
- Chen X., Sun X., Chimbaka I.M., Qin N., Xu X., Liswaniso S., Xu R., Gonzalez J.M. Transcriptome analysis of ovarian follicles reveals potential pivotal genes associated with increased and decreased rates of chicken egg production. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.622751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das N., Kumar T.R. Molecular regulation of follicle-stimulating hormone synthesis, secretion and action. J. Mol. Endocrinol. 2018;60:R131–R155. doi: 10.1530/JME-17-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewailly D., Robin G., Peigne M., Decanter C., Pigny P., Catteau-Jonard S. Interactions between androgens, FSH, anti-Mullerian hormone and estradiol during folliculogenesis in the human normal and polycystic ovary. Hum. Reprod. Update. 2016;22:709–724. doi: 10.1093/humupd/dmw027. [DOI] [PubMed] [Google Scholar]
- Durlinger A.L., Gruijters M.J., Kramer P., Karels B., Kumar T.R., Matzuk M.M., Rose U.M., de Jong F.H., Uilenbroek J.T., Grootegoed J.A., Themmen A.P. Anti-Mullerian hormone attenuates the effects of FSH on follicle development in the mouse ovary. Endocrinology. 2001;142:4891–4899. doi: 10.1210/endo.142.11.8486. [DOI] [PubMed] [Google Scholar]
- Fink T., Tiplady K., Lopdell T., Johnson T., Snell R.G., Spelman R.J., Davis S.R., Littlejohn M.D. Functional confirmation of PLAG1 as the candidate causative gene underlying major pleiotropic effects on body weight and milk characteristics. Sci. Rep. 2017;7:44793. doi: 10.1038/srep44793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert A.B., Perry M.M., Waddington D., Hardie M.A. Role of atresia in establishing the follicular hierarchy in the ovary of the domestic hen (Gallus domesticus) J. Reprod. Fertil. 1983;69:221–227. doi: 10.1530/jrf.0.0690221. [DOI] [PubMed] [Google Scholar]
- Hensen K., Braem C., Declercq J., Van Dyck F., Dewerchin M., Fiette L., Denef C., Van de Ven W.J. Targeted disruption of the murine Plag1 proto-oncogene causes growth retardation and reduced fertility. Dev. Growth Differ. 2004;46:459–470. doi: 10.1111/j.1440-169x.2004.00762.x. [DOI] [PubMed] [Google Scholar]
- Johnson A.L. Ovarian follicle selection and granulosa cell differentiation. Poult. Sci. 2015;94:781–785. doi: 10.3382/ps/peu008. [DOI] [PubMed] [Google Scholar]
- Johnson A.L., Woods D.C. Dynamics of avian ovarian follicle development: cellular mechanisms of granulosa cell differentiation. Gen. Comp. Endocrinol. 2009;163:12–17. doi: 10.1016/j.ygcen.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Jozkowiak M., Hutchings G., Jankowski M., Kulcenty K., Mozdziak P., Kempisty B., Spaczynski R.Z., Piotrowska-Kempisty H. The stemness of human ovarian granulosa cells and the role of resveratrol in the differentiation of MSCs–a review based on cellular and molecular knowledge. Cells. 2020;9:1418. doi: 10.3390/cells9061418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Ma L., Zhang J. MicroRNA-934 promotes colorectal cancer cell proliferation by directly targeting Dickkopf-related protein 2. Exp. Ther. Med. 2021;22:1041. doi: 10.3892/etm.2021.10473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu Z., Qin N., Tyasi T.L., Zhu H., Liu D., Yuan S., Xu R. The Hippo/MST pathway member SAV1 plays a suppressive role in development of the prehierarchical follicles in hen ovary. PLoS One. 2016;11 doi: 10.1371/journal.pone.0160896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F., Inoue N., Manabe N., Ohkura S. Follicular growth and atresia in mammalian ovaries: regulation by survival and death of granulosa cells. J. Reprod. Dev. 2012;58:44–50. doi: 10.1262/jrd.2011-012. [DOI] [PubMed] [Google Scholar]
- Miao X., Luo Q., Qin X. Genome-wide transcriptome analysis in the ovaries of two goats identifies differentially expressed genes related to fecundity. Gene. 2016;582:69–76. doi: 10.1016/j.gene.2016.01.047. [DOI] [PubMed] [Google Scholar]
- Mishra S.K., Chen B., Zhu Q., Xu Z., Ning C., Yin H., Wang Y., Zhao X., Fan X., Yang M., Yang D., Ni Q., Li Y., Zhang M., Li D. Transcriptome analysis reveals differentially expressed genes associated with high rates of egg production in chicken hypothalamic-pituitary-ovarian axis. Sci. Rep. 2020;10:5976. doi: 10.1038/s41598-020-62886-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson L.R., Bulun S.E. Estrogen production and action. J. Am. Acad. Dermatol. 2001;45:S116–S124. doi: 10.1067/mjd.2001.117432. [DOI] [PubMed] [Google Scholar]
- Padmanabhan V., Cardoso R.C. Neuroendocrine, autocrine, and paracrine control of follicle-stimulating hormone secretion. Mol. Cell Endocrinol. 2020;500 doi: 10.1016/j.mce.2019.110632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellatt L., Rice S., Dilaver N., Heshri A., Galea R., Brincat M., Brown K., Simpson E.R., Mason H.D. Anti-Mullerian hormone reduces follicle sensitivity to follicle-stimulating hormone in human granulosa cells. Fertil. Steril. 2011;96 doi: 10.1016/j.fertnstert.2011.08.015. 1246-51e1. [DOI] [PubMed] [Google Scholar]
- Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiqi L., Junlin H., Xuemei C., Yi H., Fangfang L., Yanqing G., Yan Z., Lamptey J., Zhuxiu C., Fangfei L., Yingxiong W., Xinyi M. Fetal exposure of Aristolochic Acid I undermines ovarian reserve by disturbing primordial folliculogenesis. Ecotoxicol. Environ. Saf. 2022;236 doi: 10.1016/j.ecoenv.2022.113480. [DOI] [PubMed] [Google Scholar]
- Quintana L., zur Nieden N.I., Semino C.E. Morphogenetic and regulatory mechanisms during developmental chondrogenesis: new paradigms for cartilage tissue engineering. Tissue Eng. Part B Rev. 2009;15:29–41. doi: 10.1089/ten.teb.2008.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recchia K., Jorge A.S., Pessoa L.V.F., Botigelli R.C., Zugaib V.C., de Souza A.F., Martins D.D.S., Ambrosio C.E., Bressan F.F., Pieri N.C.G. Actions and roles of FSH in germinative cells. Int. J. Mol. Sci. 2021;22:10110. doi: 10.3390/ijms221810110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada H., Takahashi Y., Fujino J., Flores S.Y., Yokosawa H. Localization and roles in fertilization of sperm proteasomes in the ascidian Halocynthia roretzi. Mol. Reprod. Dev. 2002;62:271–276. doi: 10.1002/mrd.10089. [DOI] [PubMed] [Google Scholar]
- Sweetman D., Smith T.G., Farrell E.R., Munsterberg A. Expression of csal1 in pre limb-bud chick embryos. Int. J. Dev. Biol. 2005;49:427–430. doi: 10.1387/ijdb.051985ds. [DOI] [PubMed] [Google Scholar]
- Szymanska K., Kalafut J., Przybyszewska A., Paziewska B., Adamczuk G., Kielbus M., Rivero-Muller A. FSHR trans-activation and oligomerization. Front. Endocrinol (Lausanne) 2018;9:760. doi: 10.3389/fendo.2018.00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran S.C., Jaehne E.J., Dye L.E., Wong J., Bakas J.S., Gasperoni J.G., Hale M.W., van den Buuse M., Dworkin S., Grommen S.V.H., De Groef B. Effect of pleomorphic adenoma gene 1 deficiency on selected behaviours in adult mice. Neuroscience. 2021;455:30–38. doi: 10.1016/j.neuroscience.2020.12.003. [DOI] [PubMed] [Google Scholar]
- Tuerlings M., van Hoolwerff M., van Bokkum J.M., Suchiman H.E.D., Lakenberg N., Broekhuis D., Nelissen R., Ramos Y.F.M., Mei H., Cats D., Coutinho de Almeida R., Meulenbelt I. Long non-coding RNA expression profiling of subchondral bone reveals AC005165.1 modifying FRZB expression during osteoarthritis. Rheumatology (Oxford) 2022;61:3023–3032. doi: 10.1093/rheumatology/keab826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyck F., Declercq J., Braem C.V., Van de Ven W.J. PLAG1, the prototype of the PLAG gene family: versatility in tumour development (review) Int. J. Oncol. 2007;30:765–774. [PubMed] [Google Scholar]
- Wang B., Zhu S., Deng Y., Sai X., Chen Z., Liu J., Li G., Liu N., Chen J., Yu C., Sun T., Zhu P. Establishment of a CRISPR/Cas9-mediated GATAD2B homozygous knockout human embryonic stem cell line. Stem Cell Res. 2021;57 doi: 10.1016/j.scr.2021.102590. [DOI] [PubMed] [Google Scholar]
- Wang C., Ma W. Hypothalamic and pituitary transcriptome profiling using RNA-sequencing in high-yielding and low-yielding laying hens. Sci. Rep. 2019;9:10285. doi: 10.1038/s41598-019-46807-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.Q., Zhang W.D., Yuan B., Zhang J.B. Advances in the regulation of mammalian follicle-stimulating hormone secretion. Animals (Basel) 2021;11:1134. doi: 10.3390/ani11041134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Feng Z., Wang X., Wang X., Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Wei H., Xu X., Yang S., Liu C., Li Q., Jin P. The potential role of hsa_circ_0001079 in androgenetic alopecia via sponging hsa-miR-136-5p. J. Clin. Lab. Anal. 2022;36:e24021. doi: 10.1002/jcla.24021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff T.K., Shea L.D. The role of the extracellular matrix in ovarian follicle development. Reprod. Sci. 2007;14:6–10. doi: 10.1177/1933719107309818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhao X., Chen L., Wang J., Duan Y., Li H., Lu L. Transcriptomic analyses of the hypothalamic-pituitary-gonadal axis identify candidate genes related to egg production in Xinjiang Yili Geese. Animals (Basel) 2020;10:90. doi: 10.3390/ani10010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Guan Y., Xue L., Zhang P., Li M., Gao M., Chong T. The roles of ferroptosis regulatory gene SLC7A11 in renal cell carcinoma: a multi-omics study. Cancer Med. 2021;10:9078–9096. doi: 10.1002/cam4.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Qin N., Xu X., Sun X., Chen X., Zhao J. Inhibitory effect of SLIT2 on granulosa cell proliferation mediated by the CDC42-PAKs-ERK1/2 MAPK pathway in the prehierarchical follicles of the chicken ovary. Sci. Rep. 2018;8:9168. doi: 10.1038/s41598-018-27601-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K.T., Lin C.Y., Huang H.L., Liou J.S., Chien C.Y., Wu C.P., Huang C.W., Ou B.R., Chen C.F., Lee Y.P., Lin E.C., Tang P.C., Lee W.C., Ding S.T., Cheng W.T., Huang M.C. Expressed transcripts associated with high rates of egg production in chicken ovarian follicles. Mol. Cell. Probes. 2008;22:47–54. doi: 10.1016/j.mcp.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Zhang T., Chen L., Han K., Zhang X., Zhang G., Dai G., Wang J., Xie K. Transcriptome analysis of ovary in relatively greater and lesser egg producing Jinghai Yellow Chicken. Anim. Reprod. Sci. 2019;208 doi: 10.1016/j.anireprosci.2019.106114. [DOI] [PubMed] [Google Scholar]
- Zhu H., Qin N., Tyasi T.L., Jing Y., Liu D., Yuan S., Xu R. Genetic effects of the transcription factors-sal-like 1 and spalt-like transcription factor 3 on egg production-related traits in Chinese Dagu hens. J. Exp. Zool. A Ecol. Integr. Physiol. 2018;329:23–28. doi: 10.1002/jez.2156. [DOI] [PubMed] [Google Scholar]
- Zhu H., Qin N., Xu X., Sun X., Chen X., Zhao J., Xu R., Mishra B. Synergistic inhibition of csal1 and csal3 in granulosa cell proliferation and steroidogenesis of hen ovarian prehierarchical developmentdagger. Biol. Reprod. 2019;101:986–1000. doi: 10.1093/biolre/ioz137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.