Abstract
The abusive use of antimicrobial compounds and the associated appearance of antimicrobial resistant strains are a major threat to human health. An improved antimicrobial administration involves a faster diagnosis and detection of resistances. Antimicrobial susceptibility testing (AST) are the reference techniques for this purpose, relying mainly in the use of culture techniques. The long time required for analysis and the lack of reproducibility of these techniques have fostered the development of high-throughput AST methods, including electrochemical biosensors. In this review, recent electrochemical methods used in AST have been revised, with particular attention on those used for the evaluation of new drug candidates. The role of nanomaterials in these biosensing platforms has also been questioned, inferring that it is of minor importance compared to other applications.
Graphical Abstract

Keywords: Electrochemical biosensors, Antimicrobial compounds, Antimicrobial susceptibility testing (AST), Nanomaterials, Screening methods
Introduction
The abuse in the use of antibiotic treatments both in the livestock and agricultural sectors and in human healthcare, together with a deficient treatment of antibiotic waste, has enhanced the problem of antimicrobial resistance (AMR) [1, 2]. This worldwide challenge translates into an increase in the minimal inhibitory concentrations (MIC) bacteria could tolerate, leading to an enlarged mortality and morbidity due to the prevalence of AMR superbugs. According to the European Centre for Disease Prevention and Control (ECDC), each year, more than 670,000 infections are initiated by AMR bacteria, leading to the death of over 33,000 person each year just in the European Union/European Economic Area [3] and around 700,000 worldwide. Without action, this value is expected to increase to over 10 million deaths per year by 2050 [4].
The current pandemic situation due to the SARS-CoV-2 (COVID-19 disease) has required the use of antibiotics as co-adjuvant treatments to stop more severe symptoms of this disease, what has just aggravated the AMR problem [5–8].
Resistance of bacteria against traditional antibiotic treatments (e.g., broad-spectrum β-lactam antibiotics) is generated by two main genetic mechanisms: gene mutations and horizontal gene transfer [9]. These mechanisms lead to the production of enzymes or modification in the bacteria characteristics that do not allow drug penetration, modify the antimicrobial target, or generate global changes in metabolic pathways. Hydrolytic extensive spectrum β-lactamase enzyme is an example of these resistance mechanisms. By hydrolyzing the β-lactam ring of the broad-spectrum β-lactam antibiotics, bacteria inhibit antibiotic effect by avoiding the incorporation of the β-lactam ring to bacteria cell walls through penicillin-binding proteins (PBPs), thus stopping the mechanism of action of these drugs, avoiding bacteria growth [10].
Developing resistance mechanisms is the expected consequence of the remarkable genetic plasticity of bacteria and the co-evolution of bacteria and antimicrobial compounds present in nature. However, the misuse of clinical antibiotic treatments since their discovery has favored the scaling up of acquired resistance.
But, although bacteria adaptability to antibiotics is an aspect we could not modulate, there are two features on antibiotic usage in which mankind could definitively make things better. Antibiotic release to the environment is one of them [11]. Through human and domestic animal excretion, production or excess drug waste handling, or direct rivers and sea contamination, antibiotics reach the environment, favoring environmental selection [12–16]. A proper identification of contaminated environments by antibiotics is fundamental to address this problem, and in this sense, biosensing approaches, specially electrochemical biosensors, have been deeply studied as useful tools for point-of-care, rapid, and low-cost detection of antibiotics in complex samples [17–19].
However, even antibiotic detection in complex environmental samples is helpful, what it is really a challenge is to provide valuable knowledge on both the effect and the dose required of antimicrobial compounds to fight infection. Antibiotic screening approaches are used in this sense, and in this field of research, electrochemical biosensors have also played their role. However, their use specially for screening new antibiotic compounds is far to be completely exploited.
In this review, a revision onto the electrochemically based antibiotic screening methods has been done, focusing on personalize medicine and those that have potential as screening platforms for new antibiotic development. A special consideration will have electrochemical sensors incorporating nanomaterials, which purpose and need will be critically evaluated.
Antibiotic screening in the clinical practice
Antibiotic screening is divided in two purposes, the selection of suitable treatments for antimicrobial resistant organisms and the identification of new antibiotic compounds. The first one has extended application in the clinical practice and relies on the use of antimicrobial susceptibility testing (AST). AST is defined as the identification of the susceptibility of a microorganism against a determined drug to search for resistance and provide information on the suitability of a treatment [20]. The use of AST is intended to warrant that antibiotics are prescribed properly and to construct patterned roadmaps of the antimicrobial resistant organisms present in a local area [21]. AST methods are standardized by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the Clinical Laboratory Standards Institute (CLSI), being necessary for a new AST method to be tested against standardized ones to be validated [22].
Main AST techniques are based on bacterial growth in solid or liquid media and rely on the diffusion or dilution of antibiotics across the media [23]. From diffusion test, disk diffusion method is the gold standard technique used in clinical microbiology laboratories. This test consists in placing a permeable disk impregnated with antibiotics on top of an agar plate seeded with the bacteria wanted to be tested [24]. After overnight inoculation, a bright clear ring is formed around the disk if the antibiotic is effective, being the diameter directly related to the bacteria susceptibility against the compound. However, a direct quantification of the MIC is just suitable for certain types of bacteria and antibiotics and implies the use of complex algorithms [25]. Differentiation between bactericidal (kills the bacteria) and bacteriostatic (suppresses the growth) effect is not possible also, as only growth inhibition is recorded [26]. Although this method is simple and cost-effective, the time required and the lack of information make it clearly improvable. However, it is still the reference technique not only in a clinical scenario but also in drug discovery [27–29].
Dilution methods in contrast are the more reliable techniques for MIC determination. Both in agar and in broth, the method consists in adding increasing and known concentrations of antibiotic to a fix concentration of inoculum, thus considering as the MIC the lowest concentration at which growth is completely inhibited by naked eye [30]. Although the simplicity of the method allows its use as antimicrobial screening platform, the irreproducibility of this quantification approach has led to a deep research on visual and colorimetric techniques that allow a more accurate MIC quantification, especially in broth dilution methods [31–34].
A combination of agar dilution and disk diffusion known as the antimicrobial gradient method is commercially available under the name Etest® and allows to determine the MIC of a compound using diffusion techniques. By impregnating a strip with different concentrations of an antimicrobial compound, the MIC could be extrapolated, in an easy-to-use approach that combines the simplicity of disk diffusion and the quantification of agar dilution. However, the cost per strip (between $2 and 3) makes their use as screening platform less cost-effective [23, 35].
Another drawback of the currently available AST methods is the lack of appropriateness for complex human samples such as blood. In this sense, EUCAST has recently moved research closer to a solution by developing a rapid antimicrobial susceptibility testing (RAST) able to detect infections in the bloodstream [36].
Moreover, four automatized equipment for AST are also approved by the FDA and available in the market: VITEK2 (bioMérieux), MicroScan WalkAway (Siemens Healthcare Diagnostics), BD Phoenix (BD Diagnostics), and Sensititre ARIS 2 × (Trek Diagnostic Systems). All of them require between 3.5 and 16 h for giving a result, apart from the time needed for pre-incubating the samples, which could last between 24 and 48 h [37].
But the use of AST methods is not limited to the identification of resistance and suitable treatments in a clinical scenario. They are also powerful tools to help in the development of alternative antibiotic compounds to substitute traditional ones, a real challenge that has been scarcely dealt. With only 43 new antibiotic compounds in development, a value that is comparably reduced considering the more than 4000 immuno-oncology drugs being researched [38], antibiotic development is the problem that pharmaceutical industry is trying to avoid. The high cost of development and reduced revenue of antibiotic commercialization has made big pharmaceutical industries leave the antibiotic development race, with only 4 top companies still in the lead.
Approximately, 45% of the costs associated to antibiotic development are expended in preclinical stages, also the more risky ones [39]. Novel approximations to faster antimicrobial development pipeline have emerged to help reduce the cost associated to these initial steps. This is the case of artificial intelligence (AI), a resource deeply needed for going through the more than 1030 drug-like compounds that it is estimated that still could be discovered [40, 41]. But once synthetically discovered, the efficacy of the compounds must be evaluated, for what AST are the preferred choice. However, although there is a deep investigation on new natural and synthetic antimicrobial compounds, the need to modify the characteristic of AST assays (e.g., inoculum size, medium, and growth conditions) to achieve a proper result hinders comparison between different research works [23].
Antibiotic screening through electrochemical means
The limitations of the traditional AST methods, such as the long time required or the standardization, have prompted the development of alternative screening methods that allow not only to identify resistant bacteria but also to serve as platform for new drug screening.
Sensors and biosensors have stood out due to their specificity, rapid response, easy to use, portability, low cost, and suitability as point-of-care (POC) devices [42]. Biosensors are formed by two main components: a versatile recognition element (e.g., antibodies and aptamers) and the transducer that detects the recognition event and converts it in a measurable signal (e.g., electrochemical, magnetic, surface plasmon resonance, and optical) [43]. Electrochemical transduction systems have gained attention due to their simplicity, affordability, and portability, bringing POC testing to a reality [44].
Their usefulness as simple platforms have been applied to several applications including AST, ranging from the use of different types of electrodes to a variety of electrochemical techniques (Table 1). The choice of a direct or indirect detection approach for bacteria cell viability testing is one of the points that differentiate the electrochemical AST methods developed until this moment. The evaluation of AST methods in complex samples has been pointed out in those works intended to be used for resistance detection, although many works meant to be used as antibiotic screening platforms do not consider this parameter.
Table 1.
Current electrochemistry methods for AST detection according to the electrochemical technique used. Bacteria used, antibiotic detected, and time required are summarized in this table as the most relevant parameters in an AST sensor
| Electrochemical technique | Bacteria | Antibiotic | Concentrations tested | Detection time | Reference |
|---|---|---|---|---|---|
| LSV | E. coli | Penicillin and streptomycin | Not specified | 2 h | [45] |
| DPV | E. coli and K. pneumoniae | Ampicillin, kanamycin, and tetracycline | 10 μg/mL | 1 h | [46] |
| DPV | E. coli | Gentamicin sulfate | 1.55 μM | 90 min | [47] |
| CV | E. coli | Ampicillin and kanamycin | 0–16 μg/mL and 0–64 μg/mL, respectively | 1 h | [48] |
| Amperometric oxygen sensor | E. coli, E. adecarboxylata, C. acidovorans, C. glutamicum, and S. epidermidis | Tetracycline, ampicillin, and chloramphenicol | 1 μg/mL, 5 μg/mL, and 5 μg/mL, respectively | 8 h | [49] |
| SWV | E. coli | Erythromycin, amikacin, ampicillin, and cefepime | 13.6 μM, 0.852 mM, 1.43 mM, and 1.04 mM, respectively | 2–5 h | [50] |
| EIS and DPV | S. aureus and methicillin-resistant S. aureus | Amoxicillin and oxacillin | 8 μg/mL | < 45 min | [51] |
| EIS | E. coli | Streptomycin | 4 μg/mL | 2.5 h | [52] |
| Impedance | E. coli | Ampicillin | 10 mg/L | 1–2 h | [53] |
| EIS | S. aureus | Flucloxacillin | 300 mg/mL | 2 h | [54] |
| Impedance | E. coli, S. aureus, and P. aeruginosa | Ampicillin, chloramphenicol, gentamicin, and amikacin | 0–128 mg/mL | 4 h | [55] |
| Impedance | E. coli and methicillin-resistant S. aureus | Ampicillin, ciprofloxacin, erythromycin, daptomycin, gentamicin, and methicillin | 0.1–100 μg/mL | < 90 min | [56] |
| Capacitance | E. coli and S. aureus | Gentamicin, tetracycline, and ampicillin | 0–50 μg/mL, 2 μg/mL, and 8 μg/mL, respectively | Not specified | [57] |
| Electrical resistance | E. coli, K. pneumoniae, and S. saprophyticus | Ampicillin and nalidixic acid | 10 mg/L and 20 mg/L, respectively | 2 h | [58] |
CV, cyclic voltammetry; DPV, differential pulse voltammetry; EIS, electrochemical impedance spectroscopy; LSV, linear sweep voltammetry; SWV, square wave voltammetry
Electrochemical AST methods using redox dye labels
The use of redox active dye labels to provide an indirect analytical signal has been extensive in electrochemical sensing, including, unsurprisingly, AST sensors.
Using a conventional three-electrode setup, fluorescein diacetate was used as a label [45] to evaluate the susceptibility of E. coli against penicillin and streptomycin. In the presence of active bacteria, fluorescein diacetate is hydrolysed by the enzymes secreted by E. coli. The oxidation of this product is monitored through voltammetric scans, being an increase in the current associated to an increase in the bacteria growth.
Using a conventional three-electrode setup, Mishra et al. [46] presented a fast-screening method to evaluate the response of bacteria to antibiotics. Using a Pt modified glass substrate as working electrode, they measured bacterial cell metabolic activity via the use of resazurin. Resazurin is a blue-colored, electroactive redox dye [59] able to penetrate bacteria cell walls and be oxidized by bacteria metabolic enzymes into a pink fluorescence resorufin product [60]. The methodology of this work is based on the incubation separately of the dye with bacteria Klebsiella pneumoniae and E. coli, reducing the dye and consequently lowering the current peak recorded through differential pulse voltammetry (DPV). However, when antibiotics such as ampicillin, kanamycin, and tetracycline are present, the reduction is inhibited due to the bacterial cell death.
The use of resazurin as dye in AST methodologies is extensive. Combining resazurin with screen-printed electrodes, a platform for the electrochemical antibiotic susceptibility testing of E. coli against gentamicin sulfate was developed [47]. DPV was also used allowing the determination of antibiotic susceptibility after 90 min, including the steps of inoculation, pre-incubation, and testing. This is a considerable reduction of the assay time required compared to previous resazurin-based biosensors, and a costless and easy-to-use AST methodology that can be easily implemented in a clinical scenario. As the developed biosensor was intended to be used for the detection of urinary tract infections, artificial urine samples were used during antibiotic testing; however, no thorough matrix effects and selectivity experiments were undergone.
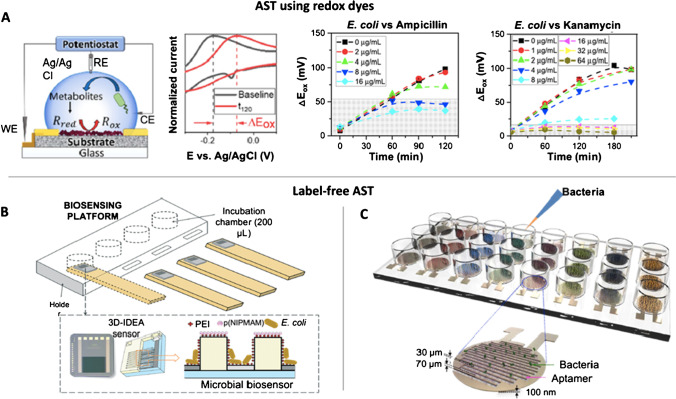
Metabolic activity of bacteria has been used in combination with dyes attending to different parameters. A recent work by Bolotsky et al. [48] proposed a different approach for a rapid antibiotic susceptibility testing using bacteria pH changes as signal generator (Fig. 1A). Electrodeposition was performed to create a redox-active organic crystalline layer (RZx) on pyrolytic graphite sheets (PGS), used to monitor the bacterial metabolic activity. The sensors were proved with E. coli K-12 incubated with two different antibiotics: ampicillin and kanamycin. The principle relies in the fact that the RZx creates responses to the bacterial metabolism by monitoring the pH change with the cell proliferation. One of the main advantages of the sensor is that it is highly stable, being viable after 60 days storage, which can be of great interest for the point-of-care antibiotic susceptibility testing. Moreover, detection of bacterial viability has been demonstrated in spiked human blood and low-fat milk as complex solutions, with no matrix effects observed. Additionally, measurement in complex samples was compared to traditional AST methods as optical density at 600 nm measurement, for which the opacity of complex samples makes measurement impossible.
Fig. 1.
AST electrochemical sensors using redox dyes (A) and label-free (B, C). A Test of ampicillin and kanamycin against E. coli using a redox-active crystalline layer on pyrolytic graphite sheets (RZx-PGS) electrode. Reprinted from [48] with permission of Elsevier. B Biosensing platform with a PDMS impedimetric transducer functionalized with polyethyleneimine (PEI), polyIJN-isopropylmethacrylamide) (pNIPMAM), and E. coli for the susceptibility testing of ampicillin. Used with permission of the Royal Society of Chemistry from [53]; permission conveyed through Copyright Clearance Center, Inc. C Capacitance AST array with aptamers as recognition elements immobilized in between electrodes. Bacteria are recognized by aptamers increasing the capacitance recorded. In the presence of an antibiotic, this capacitance is decreased. Reprinted from [57] with permission of Elsevier
Label-free AST methods
Although labels are widely used in biosensing applications, there is an increasing interest in developing label-free biosensors. These devices are more point-of-care orientated as they involve less steps and reagents during measurement process [61].
Equally to label-based sensors, some label-free ones also rely on bacteria metabolic changes as signal generators. As example, Karasinski et al. [49] proposed the use of amperometric signals with a multi-array dissolved oxygen electrochemical sensor to monitor bacteria susceptibility. The effects of different antibiotics on the bacterial growth and their respiratory activity over time were studied. The approach is based on the addition of small concentrations of antibiotics to the bacterial medium. The presence of increasing antibiotic concentrations is directly related to the consumption of oxygen performed by the bacteria, creating a unique fingerprint for each species. Tetracycline, ampicillin, and chloramphenicol were tested by measuring the level change of oxygen against five species of bacteria: E. coli, E. adecarboxylata, Comamonas acidovorans, Corynebacterium glutamicum, and Staphylococcus epidermidis. A pattern recognition was applied using a principal component analysis (PCA), generating a template that can be used to select specific combinations and concentrations of cell/antibiotics.
The direct electron transfer capacity that bacteria intrinsically present has also been exploited for cell growth viability testing in AST sensors [62]. Disposable screen-printed electrodes, modified with membranous didodecylmethylammonium bromide (DDAB), were used to estimate the susceptibility of Gram-negative Escherichia coli JM109 against well-known antibiotics such as erythromycin, amikacin, ampicillin, and cefepime [50]. The dynamic direct electron transfer capacities of E. coli were used in this work as a label-free way to monitor cell growth through cyclic voltammetry and square wave voltammetry. DDAB/E. coli biofilms, deposited on the surface of the electrode, promoted conductivity without affecting bacteria viability. In less than 5 h, they were able to confirm that cefepime, amikacin, and ampicillin inhibited cell growth while erythromycin had any effect.
Alternatively, Hannah et al. [51] modified screen-printed gold electrodes with a hydrogel of agarose for bacterial growth monitoring through electrochemical impedance spectroscopy (EIS) and DPV. Drug-resistant Staphylococcus aureus was deposited on top of the hydrogel to monitor the influence of the antibiotic’s amoxicillin and oxacillin in less than 45 min, taking measurements in periods of 5 min. The electrodes showed a good resolution, being able to differentiate between bacteria growth in absence of antibiotic and under low antibiotic concentrations. In a later work [52], the same methodology was used to monitor the electrochemical growth profiles of E. coli against streptomycin, throwing differences in approximately 2.5 h. The main difference between these two protocols is the enlarged growing of bacteria among time obtained with the second method due to the prolonged integrity of the gel-modified electrode compared to the hydrogel used in the previous work. The bacterial growth profiles were monitored by EIS.
EIS was also used by Brosel-Uliu et al. [53] by the immobilization of the bacteria E. coli in a three-dimensional interdigitated electrode array (3D-IDEA) modified with microgels to prevent bacteria deposition and increase reproducibility and sensitivity (Fig. 1B). This microbial sensor allows to monitor bacterial response to ampicillin by monitoring impedance fluctuations for 24 h. The 3D-IDEA sensor showed a large decrease in Rs in the first 2 h, while after the fourth hour, it remained quite stable.
Abeyrathne et al. [54] used also EIS technique in a non-faradic approach for detecting the antibiotic susceptibility of S. aureus against flucloxacillin in less than 2 h using interdigitated electrodes as platforms. Electrodes were SiO2 passivated and functionalized with antibodies as recognition element. Through EIS measurements, they were able to differentiate live and dead bacteria cells while they were exposed to antibiotics due to the change of the medium conductivity as a result of the bacteria metabolic process. As next step, the modifying of the sensor for the detection of S. aureus in whole blood must be studied. For that purpose, addition of a filter paper to the proposed sensor that remove erythrocytes and neutrophils (bigger diameters respect to the bacteria) and a subsequent wash step to remove unbound cells will be investigated.
Puttaswamy et al. [55] presented a rapid electrical method to determine the effect of antibiotics on bacteria using a different type of “impedance microbiology.” The bacterial metabolism shows the conductance/impedance at a single frequency since this method uses measurements at 500 different frequencies to estimate the electric charge that is stored due to the charge polarization at cell membranes of the living bacteria. It allows to track the number of living bacteria every 1 h that the measure is taken. So, the decrease in the number of bacteria that are proliferating in presence of antibiotic can be determined. For the study, different strains such as E. coli and S. aureus were tested against ampicillin and chloramphenicol while P. aeruginosa was exposed against gentamicin and amikacin in about 4 h.
By taking advantage of the use of printed electrodes and impedance electrochemical measurements, Safavieh et al. [56] reported a biosensor that allows the detection of pathogens, the identification of the correct antibiotics through antibiotic susceptibility testing, and the monitoring of mutations that help to adjust the correct therapy. The biosensor presents a rapid (< 90 min), label-free, and real-time analysis via capturing the target bacteria on flexible plastic-based microchips using electrodes modified with antibodies and monitoring the impedimetric response in the presence and absence of the antibiotics after incubation for 1 h. The microchip was evaluated with E. coli and methicillin-resistant Staphylococcus aureus (MRSA) for different antibiotics such as ampicillin, erythromycin, daptomycin, ciprofloxacin, daptomycin, methicillin, and gentamicin. Also, the ability of the microchip was demonstrated with MRSA-spiked whole blood with different clinically relevant concentrations of bacteria, being able to be used in urine samples too.
Another common recognition element used in biosensing to increase selectivity are aptamers [63]. The use of aptamers in AST was reported by Jo et al. [57] in a functionalized capacitance sensor that allows the monitoring of antibiotic susceptibility and the bacterial growth in real time (Fig. 1C). Due to the intrinsic high selectivity of aptamers, the bacteria can be identified withing 1 h using this sensor. The combination in the use of aptamers and electrical sensors allows the direct and rapid identification of the bacteria and the evaluation of their resistance against different antibiotics with high accuracy, sensitivity, and selectivity. In this sensor, bacteria are bounded over the sensor surface via aptamers between electrodes, acting as capacitors that are connected between parallel electrodes. For the antimicrobial susceptibility tests, different antibiotics such as gentamicin, tetracycline, and ampicillin were tested. For the culture of E. coli and S. aureus, the bacteria death is monitored by the change of capacitance when treated with antibiotics demonstrating the applicability of this sensor for rapid AST.
The use of microfluidics to perform rapid drug testing has also been exploited in AST. Their feasibility and use as organ-on-a-chip platforms has facilitated their implementation in AST screening [64]. Using microfluidics, Yang et al. [58] described an ultrasensitive all-electrical measurement constituted by a set of microfluidic channels that allows the flow of a liquid bacteria sample in the device for the subsequent incubation with different antibiotics. The signal measured is the electrical resistance of the microchannels that changes in proportion to the cell viability, allowing a rapid AST within 2 h. In addition, the constant fluctuations due to the antibiotics in the electrical resistance can be related to morphological changes of the bacteria. Ampicillin and nalidixic acid, antibiotics with different action mechanisms, were evaluated against E. coli, K. pneumoniae, and S. saprophyticus. Interestingly, the electrical measurement developed in this work is suitable for a multiplexed analysis desirable for a large antibiotic screening testing. As a drawback, application in clinical test could be limited in patients with complex urine matrices, not being able to be tested in the proposed device.
AST in drug screening
Although AST techniques are also used for drug screening purposes, the electrochemical AST sensors developed have been predominantly focused on addressing antimicrobial resistance. However, there are just a few examples that exploit the potential of electrochemical sensors for a faster and simple screening of new drug candidates (Table 2).
Table 2.
Potential electrochemical AST sensors for screening new drug candidates classified according to their most relevant parameters: electrochemical technique used, bacteria detected, antibiotic tested (both type and concentration), and time required for performing the measurement
| Electrochemical technique | Bacteria | Antibiotic | Concentrations tested | Detection time | Ref |
|---|---|---|---|---|---|
| SWV | P. aeruginosa | Colistin sulfate | 4, 16, and 100 mg/L | 45 h | [65] |
| SWV | P. aeruginosa | Antimicrobial peptides | 5–50 μM | 300 s | [66] |
| SWV | P. aeruginosa | RA13 | 5–50 μM | 300 s | [67] |
RA13, reverse amide 2-aminoimidazole derivative; SWV, square wave voltammetry
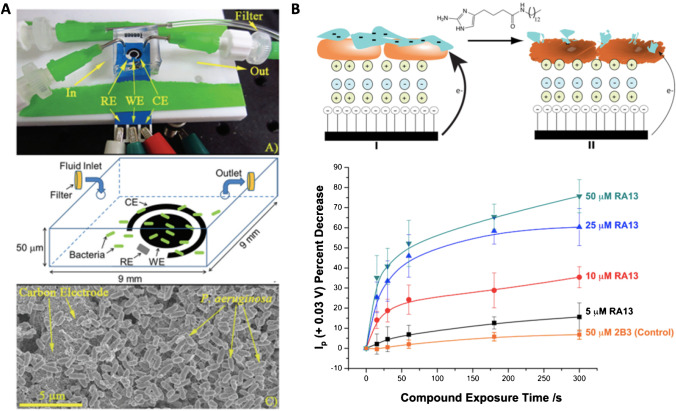
Combining the use of a disposable screen-printed electrode coupled to a microfluidic chamber, Webster et al. [65] monitored the antibiotic susceptibility of P. aeruginosa via electrochemical detection of the virulence factor pyocyanin (Fig. 2A). Bacteria was exposed to different concentrations of colistin sulfate as antibiotic. For the electrochemical measurement, square wave voltammetry was used as technique due to its increased sensitivity and ability to monitor the reduction peak corresponding to pyocyanin, directly correlated with a decrease in cell growth. The main disadvantage of this methodology is that the time required for completing measurements is of 45 h, comparable with the standard techniques currently used.
Fig. 2.
AST methods for the screening of new drug candidates. A Screen-printed electrode-based sensor covered with a microfluidic chamber with bacteria trapped on the inside. Reproduced from [65] with permission from the Royal Society of Chemistry. B Electrode based on the formation of cationic (green) and anionic (blue) polymers and P. aeruginosa biofilm (orange). After anti-biofilm exposure, the structure is compromised reducing the current recorded due to the electroactive phenazines produced by P. aeruginosa. Reprinted from [67] with permission of Elsevier
Antibiotic compounds are not the only therapeutic target in antimicrobial research. Anti-biofilms have been subject of study for many pathological microorganisms as the formation of biofilms enhances multidrug tolerance. Besides, the incorporation of electrochemical scaffolds together with anti-biofilm compounds has also been confirmed to have a co-adjuvant effect in antimicrobial treatment [68].
Evaluation of new anti-biofilm forming compounds has also been done using an electrochemical sensor using ferricyanide as redox indicator. In this case, alginate, one of the main components of mucoid P. aeruginosa strain biofilms [69], was immobilized on top of a pyrolytic graphite electrode modified layer-by-layer with poly(diallyldimethylammonium) chloride and polystyrene sulfonate. Both polymers have opposite charge, which allows the retaining of alginate in the upper electrode layer [66]. By the addition of antimicrobial peptides, the alginate was broken and ferricyanide was able to go through the layers and reach the electrode, providing an increase in the oxidation current recorded. In a later work, the same group used the same type of electrodes but in this case with the immobilization of P. aeruginosa on the upper layer (Fig. 2B). Direct electrochemical reduction of phenazine compounds produced by P. aeruginosa was recorded through SWV in a label-free sensor. With this sensor, the establishment of the half maximal inhibition concentration (IC50) and half maximal effective concentration (EC50) of an anti-biofilm compound was possible in just a few minutes [67].
Nanomaterials in the screening of antibiotic compounds: are they really needed?
Nanomaterials are defined as those materials that present at least one dimension in the range of 1–100 nm. Their small size provides them with outstanding properties not presented by their counterpart bulk materials [70].
In the last decades, nanomaterials have gained significant importance in multiple applications, going from medical imaging, drug delivery, food technology, cosmetics, and biomolecular electronic devices [71]. Nanomaterials have been incorporated in biosensing as they present enhanced electrical conductivity, improved biological sensing accuracy, and biocompatibility, what allows to increase sensitivities and decrease detection limits. Besides this, their high surface area also facilitates the immobilization of increased amounts of different bioreceptors [72] through an extended range of chemical reactions [73].
Nanomaterials are typically incorporated in electrochemical biosensing either as electrode modifiers or as detection labels [74]. The incorporation of nanomaterials, such as carbon-based nanomaterials, quantum dots, or metallic nanoparticles, has helped to overcome the slack electrode surface kinetics, acting as electrocatalysts or transduction systems [75]. However, the immobilization of these nanomaterials on the electrode surface is still a challenge to overcome [76].
As detection labels, nanomaterials have been explored as substitutes to traditional enzymatic labels that, although sensitive, require harsh conditions and present a low thermal stability, hindering their integration into commercial devices. To overcome these problems, nanomaterials have emerged as alternative, both alone or in combination with enzymes [77]. Again, metallic nanoparticles such as silver nanoparticles, gold nanoparticles, bimetallic nanoparticles, or zinc or cerium oxide nanoparticles stand out for their redox properties and electrocatalytic activities [78–80].
Their superior qualities have also made them ideal candidates as promising antimicrobial drugs [81, 82]. With silver nanoparticles (AgNPs) showing up for their intrinsic antimicrobial properties against known bacterial strains such as E. coli, S. aureus, or P. aeruginosa, multimetallic nanoparticles have also seem to be effective [83].
Although their use is extended in electrochemical biosensors, even for the detection of antibiotic residues [17], their application on AST is scant, being introduced in a reduced number of recent works (Table 3). The toxicity of some of these nanomaterials against microorganisms could be one of the reasons why their use is not as spread compared to other fields of research [74, 77]. However, their potential deserves a wider look onto their implementation in AST electrochemical sensing.
Table 3.
Nanomaterial-based electrochemical biosensors for AST classified according to the nanomaterial used and their function, the electrochemical technique used, bacteria detected, antibiotic tested and their concentration, and the time required for performing the analysis
| Nanomaterial | Nanomaterial function | Electrochemical technique | Bacteria | Antibiotics | Concentrations tested | Detection time | Ref |
|---|---|---|---|---|---|---|---|
| ʟ-CeONP/ITO | Working electrode | CV | Bacillus subtilis, Escherichia coli | Ciprofloxacin, cefixime, and amoxycillin | 2 μg/μL | 15 min | [84] |
| MWCNTs and AuNPs | Enhance the sensitivity of SPCEs | DPV | Salmonella gallinarum | Ofloxacin and penicillin | 0.0625–256 μg/mL | 4 h | [85] |
| Silicon nano transistors | Sensor design | SiNWFETs | E. coli, S. saprophyticus, and S. aureus | Ampicillin, cefotaxime, and ciprofloxacin | 100 mg/L, 20 mg/L, and 1–4 mg/L, respectively | 30 min | [86] |
| CDs | Bacterial growth-monitoring sensor | CV | E. coli and ampicillin resistant E. coli | Ampicillin | 100 μg/mL | 20 min | [87] |
| AgNPs-invertase complexes | Inhibition of enzymatic activity | PGM | E. coli | Colistin, spectinomycin, streptomycin, and tetracycline | 0–65 μg/mL | 4 h | [88] |
| Nanochannels | Sensing platforms | DPV | S. aureus | RIP, YSPWTNF-NH2 | 50 μg/mL | 24 h | [89] |
Nanomaterials: AgNPs-invertase complexes, silver nanoparticles-invertase complexes; CDs, carbon nanodots; ʟ-CeONP/ITO, ʟ-lysine-functionalized cerium oxide nanoparticle coated indium tin oxide; MWNCTs and AuNPs, multiwalled carbon nanotubes and gold nanoparticles; RIP, RNAIII-inhibiting peptide. Electrochemical methods: CV, cyclic voltammetry; DPV, differential pulse voltammetry; EIS, electrochemical impedance spectroscopy; LSV, linear sweep voltammetry; SWV, square wave voltammetry
As example, cerium oxide nanoparticles (CeNPs) have been used in an electrochemical AST as ITO electrode modifiers [84] allowing the monitoring within 15 min by time-lapse microscopy video and electrochemistry of the susceptibility of Gram-positive Bacillus subtilis and Gram-negative E. coli against the antibiotics ciprofloxacin, cefixime, and amoxycillin. In this work, the toxicity of CeNPs against E. coli has also been considered and explored as a relevant parameter in the antimicrobial screening biosensor developed. For that purpose, bulk CeNPs, CeNPs functionalized with L-lysine (L-CeNPs), and with pluronic acid (P-CeNPs) were compared, observing that L-lysine presents a protective effect against the antimicrobial activity, allowing their use in an AST sensor. In terms of conductivity, the modification of the ITO electrode with L-CeNPs significantly increased the conductivity obtained.
Carbon nanotubes (MWCNTs) and gold nanoparticles (AuNPs) have also been used as electrode modifiers in combination with resazurin as label [85]. The incorporation of these two nanomaterials combined into screen-printed carbon electrodes (SPCEs) increased the peak current recorded, what it is in correlation with the enhanced conductivity and electropolymerization that these nanomaterials present. Ofloxacin and penicillin antibiotics were tested against Salmonella gallinarum isolates by mixing bacteria, resazurin, and different concentrations of the antibiotics. DPV was selected for the detection of viable bacteria, been able to detect them above 102 CFU/mL after 1 h of incubation. The methodology was also applied in egg liquid sample obtaining a decrease of the signal sensor due to the change of the resistance of the medium, although the absolute change of current was maintained.
Taking advantage of the acidification properties of bacteria while growing, an ion-selective silicon nanowire field-effect transistor (SiNWFET) sensor [86] was developed. E. coli and other pathogen species such as S. aureus and S. saprophyticus were tested in presence and absence of different antibiotics in less than 30 min. SiNWFET used in this work contained a H+-selective sensing oxide layer for a specific detection of pH changes even in media with a high ionic concentration that increase background signal. Moreover, the use of this technology also minimizes variation between sensors, what it is desirable both to perform parallel AST screening and for research transference purposes. Largely, the use of this SiNWFET allowed the detection of tiny changes on the pH even under high ionic background concentrations, providing a better sensitivity. In addition, the proposed sensor could be modified to add a pre-filtering system that allows the detection of real samples without any preparation step as pre-cultivation.
In a different approach, but also considering bacteria acidification, carbon nanodots (CDs) have been used as electrochemical labels in an AST method [87] able to detect antibiotic susceptibility in 20 min (Fig. 3A). CDs have many advantages such as small size, good conductivity, low toxicity, and high solubility. Also, the economical and one-step synthesis of CDs makes them interesting materials for applications in biosensing. The sensor consisted in the encapsulation of CDs in alginate microspheres, together with E. coli (non-resistant) and E. coli + pET32 (resistant) and antibiotic concentrations. The presence of the alginate microspheres allows an enhanced 3D growing of the bacteria, better mimicking a real-case scenario. The change in the redox potential over time of the CDs due to the pH changes generated by the bacteria metabolism was recorded by cyclic voltammetry at 0 and 20 min, showing a discrimination between low bacterial counts of < 103 CFU/mL. The biocompatibility of the used microspheres with the bacteria tested was also evaluated, confirming that the bacteria growing rate was not affected by the presence of these nanomaterials in the microenvironment.
Fig. 3.
Nanomaterial-based AST sensors. A Using carbon nanodots as pH sensitive labels encapsulated in alginate microspheres for a better 3D bacterium growing. Reprinted from [87] with permission from Elsevier. B Using a nanochannel-based immunosensor for the detection of hyaluronidase, a virulence factor of Gram-positive bacteria, and the evaluation of a quorum sensing inhibitor (RIP). Adapted with permission from [89].
Copyright 2022 American Chemical Society
Nanoparticles can also be used as encapsulating agents as they improve availability of active compounds [90]. In this line, Laibao et al. [88] proposed the use of a modified personal glucometer as a biosensor for rapid (withing 4 h) and reliable antimicrobial susceptibility testing using polyethyleneimine AgNPs (PEI-AgNPs) to encapsulate invertase complexes. Cationic PEI-AgNPs could reversibly bind the anionic enzyme invertase, inhibiting the catalytic activity by forming an electrostatic interaction between both of them. In the presence of bacteria, invertase is released from the complex, as the cationic PEI-AgNPs bind to the anionic surface of bacteria, thus releasing invertase. The enzyme is then active to convert sucrose into glucose, a change that is recorded by the glucometer. The AST of E. coli was tested with four different antibiotics (colistin, spectinomycin, streptomycin, and tetracycline) within 5 h. The PEI-AgNPs used in this case act as a detection mechanism for bacteria identification. However, a main drawback of the work is the lack of specificity of the proposed biosensor, as it responds to different bacteria which can be an issue for application in real samples.
Nanopore/nanochannel-based materials have also been shown as outstanding tools for electrochemical biosensing [91, 92]. Regarding antibiotic screening, an innovative approach to monitor the effect of new quorum sensing inhibitors against S. aureus using a nanochannel-based electrochemical immunosensor has been recently reported [89]. The sensor was able to differentiate between S. aureus and P. aeruginosa by monitoring hyaluronidase detection, a virulence factor primarily secreted by Gram-positive bacteria. Additionally, the effect of anti-infective RNAIII-inhibiting peptide (RIP, YSPWTNF-NH2), a quorum sensing inhibitor, as suppressor of bacterial growth and virulence was evaluated using the developed sensor, by monitoring the decrease in the hyaluronidase secreted levels (Fig. 3B). The use of nanochannel membranes in this work facilitates the identification of biomarkers in complex samples due to their intrinsic filtering properties. Moreover, nanochannel membranes serve as platform for the immobilization of biorecognition elements, what leads to a label-free identification of the analytes of interest.
In general terms, the nanomaterials used in AST methods covered both the implementation such as electrode modifiers, labels, or even encapsulating agents. And although their use is relevant in terms of the bacteria concentration that could be detected, with a low CFU/mL counting in many of the works revised, this is not translated in lower antibiotic concentrations that could be tested or even the time required for obtaining a result. It is worthy to mention that the potential toxicity of these materials and how it could affect bacterial growth is not considered in many of the works, being a parameter of paramount importance for AST methods.
All considered, although the use of nanomaterials seems promising for increasing conductivity and lowering limits of detection, it is not translated into improved results for antibiotic resistance identification. However, the intrinsic characteristics of nanomaterials could be more interestingly exploited in the screening of new antimicrobial compounds and in the identification of MIC concentrations, for which reaching a lower CFU/mL count is more relevant.
Commercial potential of electrochemical AST methods: barriers to overcome
Antimicrobial resistance is a health challenge that research, industry, and policymakers should stop ignoring. With an increasing tendency of superbugs becoming more deadly than cancer, the need to find solutions to this problem is increasingly imminent. And these solutions go through both a control administration of existent antimicrobial drugs and the development of new ones. But the lack of fast, accurate, and easy-to-use diagnosis devices complicates them. Thus, the development of specific and rapid point-of-care devices can assist in managing this global health crisis, preventing unnecessary antibiotic administration [93]. Although some rapid infection testing are out on the market, they still lack from integration and require the extraction of invasive samples [94].
For that reason, the use of electrochemical biosensors both for the identification of antimicrobial resistance and for the development of new drugs seems to be a promising approach, and it has been seen like that by the research community. But still, the commercialization of these type of devices is far to become a reality.
Technical parameters such as long-term stability of the sensors developed, matrix effects, cost, or feasibility of their use by non-specialized personnel are scarcely evaluated by research works on electrochemical sensors [63] and are also lacking in the works revised in this review. Although LOD, sensitivity, and specificity have been clearly adopted as parameters that should be optimized in biosensing, it is also important to keep in mind the above-mentioned characteristics. Moreover, cost-effectivity is also an important criterion to consider, being the low profit rate the main reason for the reduced investment of pharmaceutical industry in antibiotic development [39].
Thus, the inclusion of a deeper evaluation of these and other parameter should be promoted in research works if we want to be able to transmute laboratory results into real clinically relevant devices.
Moreover, the development of multiplexed systems able to identify or evaluate more than one superbug is also desirable, as these complex infections are worsened by the presence of polymicrobial interactions [95]. This is especially relevant in the development of new antimicrobial compounds as polymicrobial infections have been directly related with an enhanced antimicrobial resistance [96].
Despite some promising leads, the way electrochemical AST methods have been developed needs to be revisited to really address market needs.
Outlook
The implementation of new AST methods is constantly evolving given the increasingly worrying incidence of antimicrobial resistances. However, traditional AST methods still do not solve this issue, which leads to increased research in alternative high-throughput screening strategies such as biosensors. Biosensing field has proved to be relevant in reducing antimicrobial resistance through environmental monitoring of antibiotics, with many biosensors developed in this field. The reduced complexity of environmental samples could be a fundamental factor to explain this difference. Detection of antimicrobial resistance through electrochemical means in human samples is a different matter. Although promising, as they help to reduce time and increase sensitivity compared to traditional AST methods, they still not accomplish all what it is claimed to AST methods. Cost of the final assay is not even considered in research works revised and is one of the main arguments that must be revised to point out biosensors as alternative to AST methods. This cost reduction is fundamental to improve antimicrobial screening research, being the main obstacle to overcome for pharmaceutical industry. It is also noticeable the scarcity of bacteria tested with these sensors, using E. coli as preferred choice as it is an easy-to-use bacterial model. But the reality is that genders of bacteria such as Klebsiella pneumoniae, Acinetobacter baumannii, and MRSA are predominantly associated to nosocomial infections and represent a larger burden for healthcare systems [97]. Bactericidal and bacteriostatic effects are lacked also in these works, and it is one of the main demands done to traditional AST methods. This aspect is less relevant in the identification of antibiotic resistance, but it is definitively a concern in the screening of new drug candidates. In this last field, it is surprising that the low number of electrochemical biosensors was developed. Although they have a greater potential, antimicrobial screening electrochemical sensors focused on the activity testing of new antimicrobial compounds are scarce. The lack of standardization of these methods compared to the AST techniques approved by the EUCAST and CLSI could be one of the reasons. Potentiating communication between electrochemical groups and microbiology research groups in charge of discovering new antibiotic compounds is a suitable way to promote this synergy and favor antimicrobial compounds development.
Regarding the use of nanomaterials, although promising, it is still not relevant enough to make a significant difference compared to non-nanomaterial–based biosensors. However, the low CFU/mL that these methodologies can reach, open the path to their use for MIC determination. That is why their use is not unjustified, but the type of analysis to be carried out with these instruments must be modified.
As we have outlined in this review, electrochemical biosensors are promising AST methods to both detect resistance in the clinical setting and to serve as screening platforms for new drug candidates, but further improvements are still required to be relevant in the combat against antimicrobial resistance.
Acknowledgements
C. Toyos-Rodríguez and D. Valero-Calvo thank the Spanish Ministry of Science and Innovation (MICINN) for the award of the FPI Grants PRE2018-084953 and PRE2021-097567, respectively. A. de la Escosura-Muñiz also acknowledges the MICINN for the “Ramón y Cajal” Research Fellow (RyC-2016-20299).
Funding
This work has been supported by the MCI-21-PID2020-115204RBI00 project from the Spanish Ministry of Science and Innovation (MICINN) and the SV-PA-21-AYUD/2021/51323 project from the Asturias Regional Government.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Published in the topical collection Electrochemical Biosensors – Driving Personalized Medicine with guest editors Susana Campuzano Ruiz and Maria Jesus Lobo-Castañón.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Miethke M, Pieroni M, Weber T, Brönstrup M, Hammann P, Halby L, Arimondo PB, Glaser P, Aigle B, Bode HB, Moreira R, Li Y, Luzhetskyy A, Medema MH, Pernodet J-L, Stadler M, Tormo JR, Genilloud O, Truman AW, Weissman KJ, Takano E, Sabatini S, Stegmann E, Brötz-Oesterhelt H, Wohlleben W, Seemann M, Empting M, Hirsch AKH, Loretz B, Lehr C-M, Titz A, Herrmann J, Jaeger T, Alt S, Hesterkamp T, Winterhalter M, Schiefer A, Pfarr K, Hoerauf A, Graz H, Graz M, Lindvall M, Ramurthy S, Karlén A, van Dongen M, Petkovic H, Keller A, Peyrane F, Donadio S, Fraisse L, Piddock LJV, Gilbert IH, Moser HE, Müller R. Towards the sustainable discovery and development of new antibiotics. Nat Rev Chem. 2021;5:726–749. doi: 10.1038/s41570-021-00313-1. [DOI] [PubMed] [Google Scholar]
- 2.Ventola CL. The antibiotic resistance crisis P&T. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 3.WHO Regional Office for Europe/European Centre for Disease Prevention and Control (2022) Antimicrobial resistance surveillance in Europe 2022–2020 data. In: Copenhagen: WHO Regional Office for Europe; 2022. https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2022-2020-data#:~:text=Antimicrobial%20resistance%20(AMR)%20remains%20a,people%20die%20as%20a%20direct. Accessed 29 Aug 2022.
- 4.O'Neill J. Tackling drug-resistant infections globally: final report and recommendations. 2016. https://amrreview.org/sites/default/files/160525_Final%20paper_with%20cover.pdf. Accessed 29 Aug 2022.
- 5.Vaughn VM, Gandhi TN, Petty LA, Patel PK, Prescott HC, Malani AN, Ratz D, McLaughlin E, Chopra V, Flanders SA. Empiric antibacterial therapy and community-onset bacterial coinfection in patients hospitalized with coronavirus disease 2019 (COVID-19): a multi-hospital cohort study. Clin Infect Dis. 2021;72:e533–e541. doi: 10.1093/cid/ciaa1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharifipour E, Shams S, Esmkhani M, Khodadadi J, Fotouhi-Ardakani R, Koohpaei A, Doosti Z, Golzari EJ, S, Evaluation of bacterial co-infections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect Dis. 2020;20:646. doi: 10.1186/s12879-020-05374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nori P, Szymczak W, Puius Y, Sharma A, Cowman K, Gialanella P, Fleischner Z, Corpuz M, Torres-Isasiga J, Bartash R, Felsen U, Chen V, Guo Y. Emerging co-pathogens: New Delhi metallo-beta-lactamase producing Enterobacterales infections in New York City COVID-19 patients. Int J Antimicrob Agents. 2020;56:106179. doi: 10.1016/j.ijantimicag.2020.106179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Getahun H, Smith I, Trivedi K, Paulin S, Balkhy HH. Tackling antimicrobial resistance in the COVID-19 pandemic. Bull World Health Organ. 2020;98:442–442A. doi: 10.2471/BLT.20.268573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munita JM, Arias CA (2016) Mechanisms of antibiotic resistance. Microbiol Spectr 4:4.2.15. 10.1128/microbiolspec.VMBF-0016-2015 [DOI] [PMC free article] [PubMed]
- 10.Rice LB. Mechanisms of resistance and clinical relevance of resistance to β-lactams, glycopeptides, and fluoroquinolones. Mayo Clin Proc. 2012;87:198–208. doi: 10.1016/j.mayocp.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsson DGJ, Flach C-F. Antibiotic resistance in the environment. Nat Rev Microbiol. 2022;20:257–269. doi: 10.1038/s41579-021-00649-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milaković M, Vestergaard G, González-Plaza JJ, Petrić I, Šimatović A, Senta I, Kublik S, Schloter M, Smalla K, Udiković-Kolić N. Pollution from azithromycin-manufacturing promotes macrolide-resistance gene propagation and induces spatial and seasonal bacterial community shifts in receiving river sediments. Environ Int. 2019;123:501–511. doi: 10.1016/j.envint.2018.12.050. [DOI] [PubMed] [Google Scholar]
- 13.Taylor P, Reeder R. Antibiotic use on crops in low and middle-income countries based on recommendations made by agricultural advisors. CABI Agric Biosci. 2020;1:1. doi: 10.1186/s43170-020-00001-y. [DOI] [Google Scholar]
- 14.Cabello FC, Godfrey HP, Buschmann AH, Dölz HJ. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect Dis. 2016;16:e127–e133. doi: 10.1016/S1473-3099(16)00100-6. [DOI] [PubMed] [Google Scholar]
- 15.Anwar M, Iqbal Q, Saleem F. Improper disposal of unused antibiotics: an often overlooked driver of antimicrobial resistance. ERATCK. 2020;18:697–699. doi: 10.1080/14787210.2020.1754797. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Chu L, Wojnárovits L, Takács E. Occurrence and fate of antibiotics, antibiotic resistant genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant: an overview. Sci Total Environ. 2020;744:140997. doi: 10.1016/j.scitotenv.2020.140997. [DOI] [PubMed] [Google Scholar]
- 17.Sun Y, Zhao J, Liang L. Recent development of antibiotic detection in food and environment: the combination of sensors and nanomaterials. Microchim Acta. 2021;188:21. doi: 10.1007/s00604-020-04671-3. [DOI] [PubMed] [Google Scholar]
- 18.Joshi A, Kim K-H. Recent advances in nanomaterial-based electrochemical detection of antibiotics: challenges and future perspectives. Biosens Bioelectron. 2020;153:112046. doi: 10.1016/j.bios.2020.112046. [DOI] [PubMed] [Google Scholar]
- 19.Hermouche L, Bendany M, Abbi K, El Hamdouni Y, Labjar N, El Mahi M, Lotfi E mostapha, Dalimi M, Dhiba D, El Hajjaji S (2021) Electrochemical sensors for tetracycline antibiotics detection based on carbon electrode materials modified by biological and chemical compounds: a review. J Environ Anal Chem 1–23 10.1080/03067319.2021.1946525
- 20.Giske CG, Turnidge J, Cantón R, Kahlmeter G. Update from the European Committee on Antimicrobial Susceptibility Testing (EUCAST) JCM. 2022;60:e00276–e321. doi: 10.1128/jcm.00276-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Belkum A, Bachmann TT, Lüdke G, Lisby JG, Kahlmeter G, Mohess A, Becker K, Hays JP, Woodford N, Mitsakakis K, Moran-Gilad J, Vila J, Peter H, Rex JH, WmM D. Developmental roadmap for antimicrobial susceptibility testing systems. Nat Rev Microbiol. 2019;17:51–62. doi: 10.1038/s41579-018-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasala A, Hytönen VP, Laitinen OH. Modern tools for rapid diagnostics of antimicrobial resistance. Front Cell Infect Microbiol. 2020;10:308. doi: 10.3389/fcimb.2020.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: a review. J Pharm Anal. 2016;6:71–79. doi: 10.1016/j.jpha.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorgensen JH, Ferraro MJ. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. CLIN INFECT DIS. 2009;49:1749–1755. doi: 10.1086/647952. [DOI] [PubMed] [Google Scholar]
- 25.Nijs A, Cartuyvels R, Mewis A, Peeters V, Rummens JL, Magerman K. Comparison and evaluation of Osiris and Sirscan 2000 antimicrobial susceptibility systems in the clinical microbiology laboratory. J Clin Microbiol. 2003;41:3627–3630. doi: 10.1128/JCM.41.8.3627-3630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernatová S, Samek O, Pilát Z, Šerý M, Ježek J, Jákl P, Šiler M, Krzyžánek V, Zemánek P, Holá V, Dvořáčková M, Růžička F. Following the mechanisms of bacteriostatic versus bactericidal action using Raman spectroscopy. Molecules. 2013;18:13188–13199. doi: 10.3390/molecules181113188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das K, Tiwari RKS, Shrivastava DK. Techniques for evaluation of medicinal plant products as antimicrobial agent: current methods and future trends. J Med Plant Res. 2010;4:104–111. doi: 10.5897/JMPR09.030. [DOI] [Google Scholar]
- 28.Lage O, Ramos M, Calisto R, Almeida E, Vasconcelos V, Vicente F. Current screening methodologies in drug discovery for selected human diseases. Mar Drugs. 2018;16:279. doi: 10.3390/md16080279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sani I, Ukwuani-Kwaja AN, Abdulkadir D (2022) Antibacterial activities of plant-derived metallic nanoparticles on some selected multidrug-resistant clinical isolates. Asian J Biol Sci 15:15–26. 10.17311/ajbs.2022.15.26
- 30.Humphries RM, Ambler J, Mitchell SL, Castanheira M, Dingle T, Hindler JA, Koeth L, Sei K, Hardy D, Zimmer B, Butler-Wu S, Dien Bard J, Brasso B, Shawar R, Dingle T, Humphries R, Sei K, Koeth L. CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J Clin Microbiol. 2018;56:e01934–e2017. doi: 10.1128/JCM.01934-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanegas D, Abril-Novillo A, Khachatryan A, Jerves-Andrade L, Peñaherrera E, Cuzco N, Wilches I, Calle J, León-Tamariz F. Validation of a method of broth microdilution for the determination of antibacterial activity of essential oils. BMC Res Notes. 2021;14:439. doi: 10.1186/s13104-021-05838-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gun MA, TanriverdiCayci Y, Durupinar B, Coban AY. A new colorimetric method for rapid detection of antibiotic resistance in Escherichia coli isolates. Jundishapur J Microbiol. 2022;14:e119858. doi: 10.5812/jjm.119858. [DOI] [Google Scholar]
- 33.Philips S, Van Hoecke F, De Laere E, Vervaeke S, De Smedt R, Boelens J, De Geyter D, Piérard D, Lagrou K. Comparison of two commercial colorimetric broth microdilution tests for Candida susceptibility testing: Sensititre YeastOne versus MICRONAUT-AM. JoF. 2021;7:356. doi: 10.3390/jof7050356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gutiérrez-del-Río I, López-Ibáñez S, Magadán-Corpas P, Fernández-Calleja L, Pérez-Valero Á, Tuñón-Granda M, Miguélez EM, Villar CJ, Lombó F. Terpenoids and polyphenols as natural antioxidant agents in food preservation. Antioxidants. 2021;10:1264. doi: 10.3390/antiox10081264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffner SE, Klintz L, Olsson-Liljequist B, Bolmström A. Evaluation of Etest for rapid susceptibility testing of Mycobacterium chelonae and M. fortuitum. J Clin Microbiol. 1994;32:1846–1849. doi: 10.1128/jcm.32.8.1846-1849.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Åkerlund A, Jonasson E, Matuschek E, Serrander L, Sundqvist M, Kahlmeter G, the RAST Study Group, Dzajic E, Hansen DS, Agergaard harlotte N, Pätäri-Sampo A, Manninen R, Grönroos JO, Rasigade J-P, Salka W, Boyer PH, Lebessi E, Zapaniotis N, Petinaki E, Spiliopoulou I, Kolonitsiou F, Helgason KO, Brazil J, Riccobono E, Lo Cascio G, Maccacaro L, Kolstad H, Haukeland TS, Kellokumpu P-L, Fossum Mjøen A, Tofteland S, Harbak B, Hartzen SH, Haug Hänsgen S, Gammelsrud KW, Skolbekken U, Michalsen N, Brekken AL, Pedersen B, Guennigsman B, Lia A, Berg AK, Marco F, Pitart C, Egea P, Cortes-Cuevas JL, Machuca J, Wietzke M, Dammström M, Granström R, Corneliusson M, Skarstedt M, Frykfeldt K, Ivarsson CL, Sergejev A, Hagström S, Lidén U, Rydberg J, Ramström H, Fröding I, Petropoulos EA, Ininbergs K, Jalal S, Persson A-LS, Kamenska N, Granlund K, Smekal A-K, Hill A, Rådberg G, Heyman G, Rodriguez L, Vennberg L, Hazırolan G, Akyar I, Gülşen Altınkanat G, Kaygisiz ANS, Dzajic E, Hansen DS, Agergaard harlotte N, Pätäri-Sampo A, Manninen R, Grönroos JO, Rasigade J-P, Salka W, Boyer PH, Lebessi E, Zapaniotis N, Petinaki E, Spiliopoulou I, Kolonitsiou F, Helgason KO, Brazil J, Riccobono E, Lo Cascio G, Maccacaro L, Kolstad H, Haukeland TS, Kellokumpu P-L, Fossum Mjøen A, Tofteland S, Harbak B, Hartzen SH, Haug Hänsgen S, Gammelsrud KW, Skolbekken U, Michalsen N, Brekken AL, Pedersen B, Guennigsman B, Lia A, Berg AK, Marco F, Pitart C, Egea P, Cortes-Cuevas JL, Machuca J, Wietzke M, Dammström M, Granström R, Corneliusson M, Skarstedt M, Frykfeldt K, Ivarsson CL, Sergejev A, Hagström S, Lidén U, Rydberg J, Ramström H, Fröding I, Petropoulos EA, Ininbergs K, Jalal S, Persson A-LS, Kamenska N, Granlund K, Smekal A-K, Hill A, Rådberg G, Heyman G, Rodriguez L, Vennberg L, Hazırolan G, Akyar I, Gülşen Altınkanat G, Kaygisiz ANS (2020) EUCAST rapid antimicrobial susceptibility testing (RAST) in blood cultures: validation in 55 European laboratories. J Antimicrob Chemother 75:3230–3238 10.1093/jac/dkaa333 [DOI] [PMC free article] [PubMed]
- 37.van Belkum A, Burnham C-AD, Rossen JWA, Mallard F, Rochas O, Dunne WM. Innovative and rapid antimicrobial susceptibility testing systems. Nat Rev Microbiol. 2020;18:299–311. doi: 10.1038/s41579-020-0327-x. [DOI] [PubMed] [Google Scholar]
- 38.The Pew Charitable Trusts. (2021) Tracking the global pipeline of antibiotics in development, March 2021. https://www.pewtrusts.org/en/research-and-analysis/issue-briefs/2021/03/tracking-the-global-pipeline-of-antibiotics-in-development. Accessed 2 Sept 2022.
- 39.Plackett B. Why big pharma has abandoned antibiotics. Nature. 2020;586:S50–S50. doi: 10.1038/d41586-020-02884-3. [DOI] [Google Scholar]
- 40.Melo MCR, Maasch JRMA, de la Fuente-Nunez C. Accelerating antibiotic discovery through artificial intelligence. Commun Biol. 2021;4:1050. doi: 10.1038/s42003-021-02586-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider G. Automating drug discovery. Nat Rev Drug Discov. 2018;17:97–113. doi: 10.1038/nrd.2017.232. [DOI] [PubMed] [Google Scholar]
- 42.Anik Ü (2017) Electrochemical medical biosensors for POC applications. In: RJ Narayan (ed.), Medical biosensors for point of care (POC) applications. Duxford: Woodhead Publishing, pp 275–292.
- 43.Ahmadian E, Samiei M, Hasanzadeh A, Kavetskyy T, Jafari S, Alipour M, Salatin S, Rameshrad M, Sharifi S, Eftekhari A, Hasanzadeh M. Monitoring of drug resistance towards reducing the toxicity of pharmaceutical compounds: past, present and future. J Pharm Biomed. 2020;186:113265. doi: 10.1016/j.jpba.2020.113265. [DOI] [PubMed] [Google Scholar]
- 44.Hayat A, Catanante G, Marty JL. Current trends in nanomaterial-based amperometric biosensors. Sensors (Switzerland) 2014;14:23439–23461. doi: 10.3390/s141223439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menon S, Vishnu N, Panchapakesan SSS, Kumar AS, Sankaran K, Unrau P, Parameswaran MA. Electrochemical sensing methodology for antibiogram assays. J Electrochem Soc. 2014;161:B3061–B3063. doi: 10.1149/2.011402jes. [DOI] [Google Scholar]
- 46.Mishra P, Singh D, Mishra KP, Kaur G, Dhull N, Tomar M, Gupta V, Kumar B, Ganju L. Rapid antibiotic susceptibility testing by resazurin using thin film platinum as a bio-electrode. J Microbiol Methods. 2019;162:69–76. doi: 10.1016/j.mimet.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Crane B, Hughes JP, Rowley Neale SJ, Rashid M, Linton PE, Banks CE, Shaw KJ. Rapid antibiotic susceptibility testing using resazurin bulk modified screen-printed electrochemical sensing platforms. Analyst. 2021;146:5574–5583. doi: 10.1039/d1an00850a. [DOI] [PubMed] [Google Scholar]
- 48.Bolotsky A, Muralidharan R, Butler D, Root K, Murray W, Liu Z, Ebrahimi A. Organic redox-active crystalline layers for reagent-free electrochemical antibiotic susceptibility testing (ORACLE-AST) Biosens Bioelectron. 2021;172:112615. doi: 10.1016/j.bios.2020.112615. [DOI] [PubMed] [Google Scholar]
- 49.Karasinski J, White L, Zhang Y, Wang E, Andreescu S, Sadik OA, Lavine BK, Vora M. Detection and identification of bacteria using antibiotic susceptibility and a multi-array electrochemical sensor with pattern recognition. Biosens Bioelectron. 2007;22:2643–2649. doi: 10.1016/j.bios.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 50.Chalenko Y, Shumyantseva V, Ermolaeva S, Archakov A. Electrochemistry of Escherichia coli JM109: direct electron transfer and antibiotic resistance. Biosens Bioelectron. 2012;32:219–223. doi: 10.1016/j.bios.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 51.Hannah S, Addington E, Alcorn D, Shu W, Hoskisson PA, Corrigan DK. Rapid antibiotic susceptibility testing using low-cost, commercially available screen-printed electrodes. Biosens Bioelectron. 2019;145:111696. doi: 10.1016/j.bios.2019.111696. [DOI] [PubMed] [Google Scholar]
- 52.Hannah S, Dobrea A, Lasserre P, Blair EO, Alcorn D, Hoskisson PA, Corrigan DK. Development of a rapid, antimicrobial susceptibility test for E. coli based on low-cost, screen-printed electrodes. Biosensors. 2020;10:153. doi: 10.3390/bios10110153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brosel-Oliu S, Mergel O, Uria N, Abramova N, van Rijn P, Bratov A. 3D impedimetric sensors as a tool for monitoring bacterial response to antibiotics. Lab Chip. 2019;19:1436–1447. doi: 10.1039/C8LC01220B. [DOI] [PubMed] [Google Scholar]
- 54.Abeyrathne C, Huynh D, Mcintire T, Nguyen T, Nasr B, Zantomio D, Chana G, Abbot I, Choong P, Catton M, Skafidas E. Lab on a chip sensor for rapid detection and antibiotic resistance determination of Staphylococcus aureus. Analyst. 2016;21:1922–1929. doi: 10.1039/c5an02301g. [DOI] [PubMed] [Google Scholar]
- 55.Puttaswamy S (2013) Novel electrical method for the rapid determination of minimum inhibitory concentration (MIC) and assay of bactericidal/bacteriostatic activity. J Biosens Bioelectron 2 10.4172/2155-6210.S2-003
- 56.Safavieh M, Pandya HJ, Venkataraman M, Thirumalaraju P, Kanakasabapathy MK, Singh A, Prabhakar D, Chug MK, Shafiee H. Rapid real-time antimicrobial susceptibility testing with electrical sensing on plastic microchips with printed electrodes. ACS Appl Mater Interfaces. 2017;9:12832–12840. doi: 10.1021/acsami.6b16571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jo N, Kim B, Lee S-M, Oh J, Park IH, Jin Lim K, Shin J-S, Yoo K-H. Aptamer-functionalized capacitance sensors for real-time monitoring of bacterial growth and antibiotic susceptibility. Biosens Bioelectron. 2018;102:164–170. doi: 10.1016/j.bios.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 58.Yang Y, Gupta K, Ekinci KL. All-electrical monitoring of bacterial antibiotic susceptibility in a microfluidic device. Proc Natl Acad Sci USA. 2020;117:10639–10644. doi: 10.1073/pnas.1922172117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rampersad SN. Multiple applications of Alamar Blue as an indicator of metabolic function and cellular health in cell viability bioassays. Sensors. 2012;12:12347–12360. doi: 10.3390/s120912347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McGaw LJ, Elgorashi EE, Eloff JN (2014) Cytotoxicity of African medicinal plants against normal animal and human cells. In: Toxicological survey of African medicinal plants. Elsevier, pp 181–233.
- 61.Kanagavalli P, Andrew C, Veerapandian M, Jayakumar M. In-situ redox-active hybrid graphene platform for label-free electrochemical biosensor: insights from electrodeposition and electroless deposition. TrAC. 2021;143:116413. doi: 10.1016/j.trac.2021.116413. [DOI] [Google Scholar]
- 62.Yang Y, Xu M, Guo J, Sun G. Bacterial extracellular electron transfer in bioelectrochemical systems. Process Biochem. 2012;47:1707–1714. doi: 10.1016/j.procbio.2012.07.032. [DOI] [Google Scholar]
- 63.Al Mamun M, Wahab YA, Hossain MAM, Hashem A, Johan MR. Electrochemical biosensors with aptamer recognition layer for the diagnosis of pathogenic bacteria: barriers to commercialization and remediation. TrAC. 2021;145:116458. doi: 10.1016/j.trac.2021.116458. [DOI] [Google Scholar]
- 64.Dai J, Hamon M, Jambovane S. Microfluidics for antibiotic susceptibility and toxicity testing. Bioengineering. 2016;3:25. doi: 10.3390/bioengineering3040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Webster TA, Sismaet HJ, Chan IJ, Goluch ED. Electrochemically monitoring the antibiotic susceptibility of Pseudomonas aeruginosa biofilms. Analyst. 2015;140:7195–7201. doi: 10.1039/C5AN01358E. [DOI] [PubMed] [Google Scholar]
- 66.Vinogradov SM, Satterwhite-Warden JE, Hicks RP, Anderson E, Hvastkovs EG. Electrochemical detection of alginate penetration in immobilized layer-by-layer films by unnatural amino acid containing antimicrobial peptides. Electrochim Acta. 2015;186:245–252. doi: 10.1016/j.electacta.2015.10.051. [DOI] [Google Scholar]
- 67.Robb AJ, Vinogradov S, Danell AS, Anderson E, Blackledge MS, Melander C, Hvastkovs EG. Electrochemical detection of small molecule induced Pseudomonas aeruginosa biofilm dispersion. Electrochim Acta. 2018;268:276–282. doi: 10.1016/j.electacta.2018.02.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sultana ST, Call DR, Beyenal H. Eradication of Pseudomonas aeruginosa biofilms and persister cells using an electrochemical scaffold and enhanced antibiotic susceptibility. NPJ Biofilms Microbiomes. 2016;2:2. doi: 10.1038/s41522-016-0003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramsey DM, Wozniak DJ. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis: P. aeruginosa pathogenesis. Mol Microbiol. 2005;56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- 70.Malhotra BD, Ali MA. Nanomaterials in biosensors: fundamentals and applications. Nanomaterials for Biosensors. 2018;1:1–74. 10.1016/B978-0-323-44923-6.00001-7.
- 71.Anjum S, Ishaque S, Fatima H, Farooq W, Hano C, Abbasi BH, Anjum I. Emerging applications of nanotechnology in healthcare systems: grand challenges and perspectives. Pharmaceuticals. 2021;14:707. doi: 10.3390/ph14080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holzinger M, Le Goff A, Cosnier S (2014) Nanomaterials for biosensing applications: a review. Front Chem 2. 10.3389/fchem.2014.00063 [DOI] [PMC free article] [PubMed]
- 73.Malik P, Gupta R, Malik V, Ameta RK. Emerging nanomaterials for improved biosensing. Meas Sensors. 2021;16:100050. doi: 10.1016/j.measen.2021.100050. [DOI] [Google Scholar]
- 74.Toyos-Rodríguez C, García-Alonso FJ, de la Escosura-Muñiz A. Electrochemical biosensors based on nanomaterials for early detection of Alzheimer’s disease. Sensors. 2020;20:4748. doi: 10.3390/s20174748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baig N, Sajid M, Saleh TA. Recent trends in nanomaterial-modified electrodes for electroanalytical applications. TrAC. 2019;111:47–61. doi: 10.1016/j.trac.2018.11.044. [DOI] [Google Scholar]
- 76.Ahmad R, Wolfbeis OS, Hahn Y-B, Alshareef HN, Torsi L, Salama KN. Deposition of nanomaterials: a crucial step in biosensor fabrication. Materials Today Communications. 2018;17:289–321. doi: 10.1016/j.mtcomm.2018.09.024. [DOI] [Google Scholar]
- 77.Iglesias-Mayor A, Amor-Gutiérrez O, Costa-García A, de la Escosura-Muñiz A. Nanoparticles as emerging labels in electrochemical immunosensors. Sensors. 2019;19:5137. doi: 10.3390/s19235137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Malathi S, Pakrudheen I, Kalkura SN, Webster TJ, Balasubramanian S. Disposable biosensors based on metal nanoparticles. Sensors International. 2022;3:100169. doi: 10.1016/j.sintl.2022.100169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kaushal S, Nanda SS, Samal S, Yi DK. Strategies for the development of metallic-nanoparticle-based label-free biosensors and their biomedical applications. ChemBioChem. 2020;21:576–600. doi: 10.1002/cbic.201900566. [DOI] [PubMed] [Google Scholar]
- 80.Toyos-Rodríguez C, Adawy A, García-Alonso FJ, de la Escosura-Muñiz A. Enhancing the electrocatalytic activity of palladium nanocluster tags by selective introduction of gold atoms: application for a wound infection biomarker detection. Biosens Bioelectron. 2022;200:113926. doi: 10.1016/j.bios.2021.113926. [DOI] [PubMed] [Google Scholar]
- 81.Makabenta JMV, Nabawy A, Li C-H, Schmidt-Malan S, Patel R, Rotello VM. Nanomaterial-based therapeutics for antibiotic-resistant bacterial infections. Nat Rev Microbiol. 2021;19:23–36. doi: 10.1038/s41579-020-0420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morena AG, Bassegoda A, Natan M, Jacobi G, Banin E, Tzanov T. Antibacterial properties and mechanisms of action of sonoenzymatically synthesized lignin-based nanoparticles. ACS Appl Mater Interfaces. 2022;14:37270–37279. doi: 10.1021/acsami.2c05443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Basavegowda N, Baek K-H. Multimetallic nanoparticles as alternative antimicrobial agents: challenges and perspectives. Molecules. 2021;26:912. doi: 10.3390/molecules26040912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rao RP, Sharma S, Mehrotra T, Das R, Kumar R, Singh R, Roy I, Basu T. Rapid electrochemical monitoring of bacterial respiration for Gram-positive and Gram-negative microbes: potential application in antimicrobial susceptibility testing. Anal Chem. 2020;92:4266–4274. doi: 10.1021/acs.analchem.9b04810. [DOI] [PubMed] [Google Scholar]
- 85.Ren Y, Ji J, Sun J, Pi F, Zhang Y, Sun X. Rapid detection of antibiotic resistance in Salmonella with screen printed carbon electrodes. J Solid State Electrochem. 2020;24:1539–1549. doi: 10.1007/s10008-020-04645-8. [DOI] [Google Scholar]
- 86.Xu X, Chen S, Yu Y, Virtanen P, Wu J, Hu Q, Koskiniemi S, Zhang Z. All-electrical antibiotic susceptibility testing within 30 min using silicon nano transistors. Sens Actuators, B: Chemical. 2022;357:131458. doi: 10.1016/j.snb.2022.131458. [DOI] [Google Scholar]
- 87.Das R, Singh N. Exploring electrochemistry of carbon nanodots and its application in noninvasive bacterial growth monitoring. Biosens Bioelectron. 2019;144:111640. doi: 10.1016/j.bios.2019.111640. [DOI] [PubMed] [Google Scholar]
- 88.Zheng L, Shen Y, Dong W, Zheng C, Zhou R, Lou Y-L. Rapid detection and antimicrobial susceptibility testing of pathogens using AgNPs-invertase complexes and the personal glucose meter. Front Bioeng Biotechnol. 2022;9:795415. doi: 10.3389/fbioe.2021.795415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de la Escosura-Muñiz A, de la, Ivanova K, Tzanov T, Electrical evaluation of bacterial virulence factors using nanopores. ACS Appl Mater Interfaces. 2019;11:13140–13146. doi: 10.1021/acsami.9b02382. [DOI] [PubMed] [Google Scholar]
- 90.Olivieri F, Castaldo R, Cocca M, Gentile G, Lavorgna M. Mesoporous silica nanoparticles as carriers of active agents for smart anticorrosive organic coatings: a critical review. Nanoscale. 2021;13:9091–9111. doi: 10.1039/D1NR01899J. [DOI] [PubMed] [Google Scholar]
- 91.de la Escosura-Muñiz A, Merkoçi A. Nanochannels for electrical biosensing TrAC. 2016;79:134–150. doi: 10.1016/j.trac.2015.12.003. [DOI] [Google Scholar]
- 92.de la Escosura-Muñiz A, Merkoçi A. Nanochannels preparation and application in biosensing. ACS Nano. 2012;6:7556–7583. doi: 10.1021/nn301368z. [DOI] [PubMed] [Google Scholar]
- 93.Burnham C-AD, Leeds J, Nordmann P, O’Grady J, Patel J. Diagnosing antimicrobial resistance. Nat Rev Microbiol. 2017;15:697–703. doi: 10.1038/nrmicro.2017.103. [DOI] [PubMed] [Google Scholar]
- 94.Lebovitz EE, Burbelo PD. Commercial multiplex technologies for the microbiological diagnosis of sepsis. Mol Diagn Ther. 2013;17:221–231. doi: 10.1007/s40291-013-0037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Azimi S, Lewin GR, Whiteley M (2022) The biogeography of infection revisited. Nat Rev Microbiol. 10.1038/s41579-022-00683-3 [DOI] [PMC free article] [PubMed]
- 96.Yung DBY, Sircombe KJ, Pletzer D. Friends or enemies? The complicated relationship between Pseudomonas aeruginosa and Staphylococcus aureus. Mol Microbiol. 2021;116:1–15. doi: 10.1111/mmi.14699. [DOI] [PubMed] [Google Scholar]
- 97.Christaki E, Marcou M, Tofarides A. Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J Mol Evol. 2020;88:26–40. doi: 10.1007/s00239-019-09914-3. [DOI] [PubMed] [Google Scholar]