Abstract
Developing functional machine learning (ML)-based models to address unmet clinical needs requires unique considerations for optimal clinical utility. Recent debates about the rigours, transparency, explainability, and reproducibility of ML models, terms which are defined in this article, have raised concerns about their clinical utility and suitability for integration in current evidence-based practice paradigms. This featured article focuses on increasing the literacy of ML among clinicians by providing them with the knowledge and tools needed to understand and critically appraise clinical studies focused on ML. A checklist is provided for evaluating the rigour and reproducibility of the four ML building blocks: data curation, feature engineering, model development, and clinical deployment. Checklists like this are important for quality assurance and to ensure that ML studies are rigourously and confidently reviewed by clinicians and are guided by domain knowledge of the setting in which the findings will be applied. Bridging the gap between clinicians, healthcare scientists, and ML engineers can address many shortcomings and pitfalls of ML-based solutions and their potential deployment at the bedside.
Keywords: Machine learning, Healthcare, Critical appraisal, Bias, Guidelines, Quality
Introduction
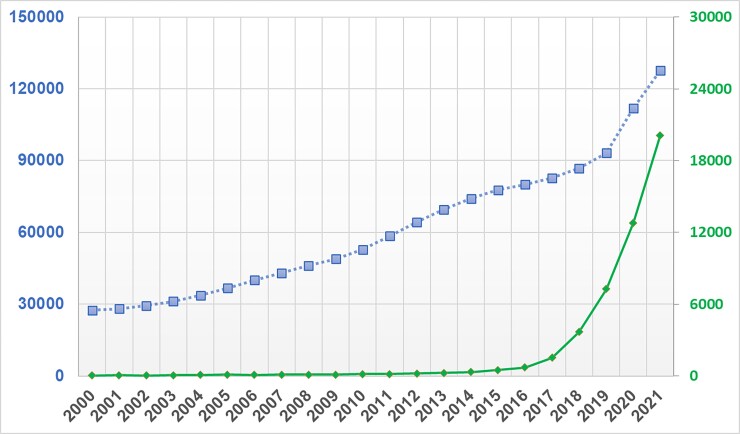
Machine learning (ML) is a field that lies at the intersection of mathematics and computer science, integrating principles from computing, optimization, and statistics. Successive advances in this field over the past few decades have brought a suite of very powerful mathematical algorithms able to learn hidden patterns from large quantities of data. From a data science-oriented perspective, ML is simply a collection of mathematical theories and statistical techniques that enable machines to improve at undertaking a given task with experience (e.g. recognition, prediction, prescription). Given that recognition and prediction are the backbone of clinical practice, many of these ML algorithms have proven efficient in addressing some longstanding challenges frequently encountered in analysing high dimensional, complex clinical data.1–6 These promising potentials have led to a rapid expansion in the number of articles published in clinical journals that focus on ML. Figure 1 shows the number of clinical diagnostic accuracy studies published in PubMed between 2000 and 2021 and the sub-portion of these studies that use ML methods. These trends translate to an annual growth rate of 8% compared with 39%, respectively. It is reported that nearly 25% of all diagnostic accuracy studies submitted to leading journals focus on the performance of ML algorithms.7 This is one in four papers in any given field to which an average clinician could be exposed.
Figure 1.
Temporal trends in machine learning-centered articles published in PubMed between 2000 and 2021. This figure shows the results of a simple search on PubMed for clinical diagnostic studies focused on ‘diagnosis’ between 2000 and 2021 (line with square markers) and the sub-portion of those studies that reference ‘machine learning’, ‘artificial intelligence’, or ‘deep learning’ in the title or abstract (line with diamond markers).
This exponential growth in the use of ML techniques to address unmet clinical needs has not been effectively translated to the bedside due to many scientific, technical, and logistical challenges, dampening the enthusiasm by many healthcare providers. First, as target end-users at the bedside, many clinicians are not familiar with the concepts of ML and whether its applications in healthcare can be trusted. This could create a barrier to change and limit the potential deployment at the bedside. Many reviewers or editors of medical journals are also not very familiar with such ML methodologies. In an interesting experiment at a ML-focused conference (NeurIPS), double-blinded reviewers failed to reach consensus on more than 57% of submitted papers at 22.5% acceptance rate.8 Such a large margin of disagreement among reviewers might deter journals from accepting high-quality ML papers, or more worrisome, publish poorly performed or flawed ones. Second, because of this gap in common language between end-users and developers, clinicians are rarely integral members of data science teams, potentially diminishing the clinical relevance, model explainability, and workflow compatibility of many ML-centered solutions.9 Finally, the field of ML itself continues to suffer from shortcomings that have led to hot debates about its usefulness in recent years, including the black box label as well as gender and racial bias to mention a few.10,11 These concerns, coupled with the lack of a clear regulatory pathway12 and poor access to large and high-quality datasets, have limited the availability of approved ML-based medical devices in the USA and Europe, creating a clinical paradigm with sceptical stakeholders and growing mistrust between clinicians and ML applications.
Understanding the complexity (and subjectiveness) of modelling decisions involved in building a functional ML application is central to critically appraising clinical ML studies. Figure 2 showcases the kind of ‘decisions’ that each data scientist typically considers throughout the various steps of designing a ML pipeline (i.e. from data to decisions). This featured article focuses on increasing literacy of ML among clinicians by providing them with the knowledge and tools needed to understand and critically appraise clinical studies focused on ML. Clinical applications of ML in cardiovascular disease and cardiac imaging have been described in detail elsewhere.3 Herein, we provide a succinct review of commonly used ML techniques and best practices and considerations for building and translating effective ML models. We also highlight the most crucial design flaws and serious red flags for clinicians to consider while critically appraising ML-centered articles in healthcare.
Figure 2.
The complexity of decisions considered when designing a machine learning model. This figure showcases the complexity and subjectiveness in the questions/decisions the data scientist may consider when building a machine learning algorithm. Ideally, these important decisions need to be made in conjunction with collaborating clinicians to enhance clinical relevance and utility. EDA, exploratory data analysis.
Basic definitions and terminologies
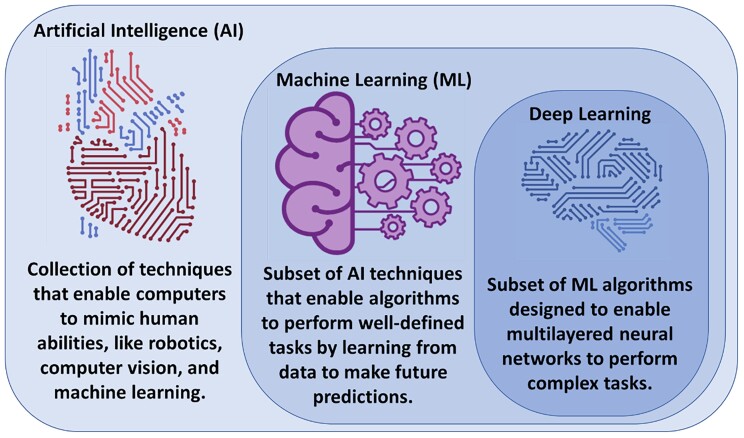
Artificial intelligence (AI) is the machine’s ability to mimic humans in learning and behaviour with automatic improvement and without explicit programming. Artificial intelligence encompasses multiple fields, including computer vision, robotics, and ML.13 Machine learning as a subfield of AI entails a collection of computer algorithms developed to automate regular processes and services or predict and forecast events of interest in a certain domain quickly and accurately using previously recorded historical data of that event. In healthcare, ML has been extensively used for clinical diagnostics, early prediction of outcomes, and disease phenotyping.14 Deep learning (DL), on the other hand, is a subclass of ML algorithms (i.e. multi-layered neural networks) that are designed to learn complex tasks from massive amounts of structured and unstructured data types like video, voice, image, or text.13 Figure 3 visualizes the relationship between AI, ML, and DL as interrelated and overlapping fields.
Figure 3.
Relationship between artificial intelligence, machine learning, and deep learning. Artificial intelligence is the general umbrella that encompasses machine learning with other domains, whereas deep learning is a subclass of machine learning algorithms (Credit: Salah Al-Zaiti. Created with BioRender.com).
Artificial intelligence was born back in the 1950s when a group of computer scientists started exploring whether computers could be designed to ‘think’ like humans. Early chess programmes are a good example of scientists’ first attempts of making computers act like humans. In this case, the programme involved hardcoded rules without any interactive learning. This approach, at that time, was called symbolic AI and dominated the field from the 1950s to the 1980s. Although symbolic AI proved to be a good approach to solve well-defined and logical problems, it turned out to be ill-suited to solving more complex and fuzzy problems such as image classification, speech recognition, and language translation. In late 1980s, ML arose to replace symbolic AI in automating intellectual tasks normally performed by human intelligence.
The concept behind ML algorithms is distinct from the rule-based logic seen in symbolic AI. Rather than programming a set of conditional statements derived from domain knowledge to guide decision making, scientists feed data rather than rules to train the model, refine its parameters, and test its performance for a given task (i.e. interactive learning). This model is then built in production. A key difference here is that rule-based algorithms use current knowledge already adopted by clinicians when making a decision (e.g. diagnosis), whereas a ML approach might use different rules or ‘features’ that it discovered during the data-driven process of model development. If we consider the task of diagnosing acute myocardial infarction (MI) using electrocardiogram (ECG) data as an example, a rule-based automated interpretation system would follow programmed rules like if ST amplitude ≥ 0.1 mV, then print ‘>>>Acute MI<<<’. On the contrary, a ML-based model would ‘learn’ the features and data-driven decision rules for classifying ‘>>>Acute MI<<<’ from an existing and labelled ECG dataset without any preexisting domain knowledge. The performance of this model is then tested on new ECG tracings not included in the original datasets. This is analogous to a cardiology fellow who is learning to read ECGs by reviewing few thousand examples under supervision until mastering his/her own unique approach; before interpreting ECGs in a real-world setting on his own.
The question then remains how ML is different from statistics used in the general medical literature. The main task of both is to find a mathematical representation in a multidimensional probability distribution, yet the focus and degree of formal development between both are different. Statistics is a branch of applied probabilities with well-outlined theoretical concepts focusing on drawing population inferences from sample data to understand the causal relationship between the predictors and the outcome variable (i.e. knowledge discovery to increase our understanding of a given phenomenon).15 For example, a statistician might explore the relationship between the presence of comorbidities and incidental cardiovascular disease using logistic regression. Here, the interest is inferring a function that can explain the most variability in incidental cardiovascular disease using a parsimonious subset of comorbidities, which would contribute to our understanding of disease pathogenesis (i.e. reduced dimensionality for knowledge discovery).16 On the other hand, ML uses general-purpose mathematical learning algorithms (beyond probabilities) to find generalizable patterns in high-dimensional data space without requiring prior assumptions about either the population or the residuals. In the previous example, a data scientist would explore a suite of learning models (which might include logistic regression) to build a model that yields the highest classification performance (i.e. sensitivity and specificity), with little attention to the impact of data properties on population density functions (i.e. no inferences). Here the interest is to find the model that best learns generalizable rules when applied to new unseen data (i.e. prediction rather than comprehension).15,17 Nevertheless, statistics and ML share numerous principles and techniques, and the overlap between both sometimes might be difficult to delineate. In fact, it has been shown that adapting statistical inferential techniques in learning algorithms (i.e. causal ML) dramatically boosts ML model performance.18
Subtypes of machine learning models
Machine learning models can be classified into four subtypes based on the degree of human supervision applied on data: supervised, unsupervised, semi-supervised, and reinforcement learning (RL). In supervised learning, a set of input variables is used to predict an outcome of interest that has been labelled by experts. Supervised techniques can be broken down into two subtypes according to the level of measurement of the outcome variable: regression or classification. In regression, the outcome is measured as a continuous numeric variable such as blood pressure, BMI, height, or weight. A regular prediction algorithm, such as linear regression or regression trees, is used when input data are collected at one time. If input data are longitudinal (i.e. time-series), then forecasting algorithms such Auto-Regressive Integrated Moving Average or exponential smoothing models could be used. In classification, the outcome of interest is measured as a categorical variable at either two levels (binary) or more (ordinal or nominal).
Table 1 summarizes the commonly used regression and classification techniques in supervised ML. This table also summarizes the adjustable free parameters needed to optimize each model performance (i.e. hyperparameters). Hyperparameters refer to the configurations of a model’s architecture that cannot be inferred from the data and need to be arbitrarily decided by the data scientist. For instance, when building a simple decision tree, the data scientist needs to decide on the tree depth (number of levels) and number of nodes (decision splits) before a tree can be built. The selection of these hyperparameters is arbitrary and can be done by searching all potential combinations of hyperparameters that perform best on the historical data. Finding the best hyperparameters plays an important role in optimizing the goodness-of-fit on the sample data, ideally leading to good performance when generalized to new unseen data.
Table 1.
Common supervised machine learning techniques used in healthcare
| Algorithm | Class | Technique description | Hyperparameters |
|---|---|---|---|
| Linear regression | Regression | Estimates the coefficients of a set of predictors (x1, x2, …) that would yield the best approximation of y with the least error in prediction. | None |
| Ridge regression | Regression | Similar to linear regression but applies stricter rules (penalty) to shrink the estimated coefficients to further improve the prediction (called L2 regularization). | Shrinkage value (penalty term) |
| LASSO regression | Regression or classification | Similar to ridge regression but assigns a larger penalty term to the estimated coefficients, which shrinks some coefficients to zero, reducing the number of features (called L1 regularization). | Shrinkage value (penalty term) |
| Elastic−net regression | Regression | A method based on weighted combination of both ridge regression and LASSO. | Shrinkage, weight between Ridge and LASSO |
| Logistic regression | Classification | An extension to linear regression replacing the linear slope needed to predict values of y with a step function needed to split classes and predict y as a binary outcome. | None |
| Linear discriminant analysis (LDA) | Classification | Estimates n-dimensional hyperplane that separates two classes by maximizing the ratio of between-groups to within-groups variance of distributions. | None |
| Support vector machine (SVM) | Classification | Estimates n-dimensional hyperplane that separates two classes based on maximizing the margin between data points in the decision boundary and the hyperplane projection. | Cost, curvature of the decision boundaries |
| Naïve Bayes (NB) | Classification | Uses Bayes rule to compute the conditional probability of the outcome assuming that features in that class are independent of each other. | Prior, smooth |
| K-nearest neighbours (KNN) | Regression or classification | Predicts new class or value based on the majority vote of closest k samples in the dataset. | Number of neighbours |
| Trees | Regression or classification | Rule-based branching tree representation that maps decisions at every split choice and their possible consequences in a way that maximizes data purity at each split. | Tree depth, number of nodes, number per leaf |
| Ensemble techniques | Regression or classification | Combining hundreds of base classifiers (e.g. trees) and bootstrapping to learn the ‘wisdom of the crowd’. Fusing base classifiers can happen in a parallel fashion by counting a majority vote (e.g. random forest) or in a sequential fashion by learning from one classifier at a time (e.g. gradient- or X-boosting). | Number of trees, interaction depth, shrinkage, observations per terminal nodes, number of candidate parameters at each split |
LASSO, least absolute shrinkage and selection operator.
The second subtype of ML models is unsupervised learning where an algorithm autonomously draws associations from unlabelled data without any a priori knowledge of true labels. With the emergence of big data, data sources became massive, making labelling labour-intensive, time-consuming, and very costly. With the wide availability of such massive quantities of unlabelled training data, unsupervised learning techniques become essential. Table 2 summarizes the four most common unsupervised techniques along with their potential applications in healthcare. Clustering entails grouping together patients with approximately similar characteristics given a set of features. The emerging clusters can potentially identify unique phenotypes of a given disease. A clustering example would be grouping patients with chest pain based on their age, sex, risk factors, symptoms, lab tests, medications, and angiographic findings. The resulting clusters can then be used to identify specific phenotypes like Type 1 vs. Type 2 MI. Anomaly detection entails learning the baseline pattern of how features typically aggregate in order to identify potential deviations from this normal state. An example of anomaly detection would be learning certain clinical contexts when medication X is prescribed to a given patient so any new deviations from this norm can be flagged as a potential medication error. Dimensionality reduction entails the mathematical summarization of data across features using orthogonal (independent) vectors for simplified modelling and visualization. For instance, principal component analysis has been historically used to summarize ECG waveform data from 192 body surface potential maps (BSPM) to yield only 12 independent waveforms that capture >90% of prognostic information in the data, significantly enhancing the clinical utility of BSPM for bedside care. Finally, association rule mining entails approaches to learn strong rules with interesting relationships between-groups of variables in large datasets. For example, implementing an apriori algorithm on electronic health record data in an emergency department might yield the rule (chest pain, ECG) → (troponin), indicating that a chief complaint of chest pain with documented ECG record are pre-requisites to the availability of troponin results in the medical charts.
Table 2.
Common unsupervised machine learning techniques and their potential use in healthcare
| Technique | Learning algorithms | Healthcare applications |
|---|---|---|
| Clustering | K-means, hierarchical clustering, DBSCAN | Disease phenotyping, genetic pathways, drug discovery, etc. |
| Anomaly detection | One-class SVM, isolation forest, neural networks | Waveform segmentation, physiological signal denoising, medications error warning, etc. |
| Visualization and dimensionality reduction | Principal component analysis (PCA), linear or stochastic neighbour embedding, TDA | Genetic data, ECG waveform analysis, inflammatory pathways, etc. |
| Association rule mining | Apriori, Eclat | EHR data mining, text mining, etc. |
DBSCAN, density-based spatial clustering of applications with noise; SVM, support vector machine; EHR, electronic health records; ECLAT, Equivalence Class Clustering and bottom-up Lattice Traversal; TDA, topological data analysis.
A third subtype of ML models is semi-supervised learning. Despite the scarcity of labelled data in many fields (e.g. medical images, ECG signals), many datasets contain a mixture of both labelled and unlabelled data, which created an opportunity for leveraging techniques from both supervised and unsupervised learning. These new semi-supervised learning techniques are broadly grouped under two general umbrellas: self-learning algorithms and co-learning algorithms.19 In self-learning, a single classifier is trained on a small subset of ‘seed’ data then used to classify unlabelled data. Data points classified with high confidence are added to the ‘seed’ subset and the base classifier is re-trained. This iterative process continues until all data points are labelled. This technique has been successfully implemented in image classification.20 In co-learning, a similar approach is used to train three classifiers, rather than one, on the ‘seed’ subset, and then using the consensus of the three algorithms in order to add new data points to the ‘seed’ set before re-training. This technique has been successfully used for classifying the level of activity of a patient using motion data from cameras and sensors to monitor and determine when assistance is needed (e.g. after falling).21 Unlike self-learning and co-learning, a more novel approach is to train a single classifier on a ‘seed’ set, then use this model to select the data points with the lowest confidence in prediction and thereafter ask users to manually label these informative examples before adding them to the ‘seed’ set and re-training the classifier. This latter approach is called active learning and has been successfully used in ECG beat classification, image classification, gene expression, and artefact detection.22,23
A fourth subtype of learning models is RL, which constitutes a totally different paradigm in terms of how the model learns. In general, there will be an agent which observes and learns the best policy (e.g. decision rules) by weighing actions (e.g. available treatment options) against subsequent rewards (e.g. short-term patient outcomes) where the policy can be adapted over time. Reinforcement learning is widely used in robotics, gaming, computer vision, and autonomous control.24 In healthcare, applications of RL are limited because its techniques warrant due diligence. Some of the successful applications include HIV therapy optimization, seizure control, and sepsis management.25 Reinforcement learning is a complex topic and has been explained in detail elsewhere.26
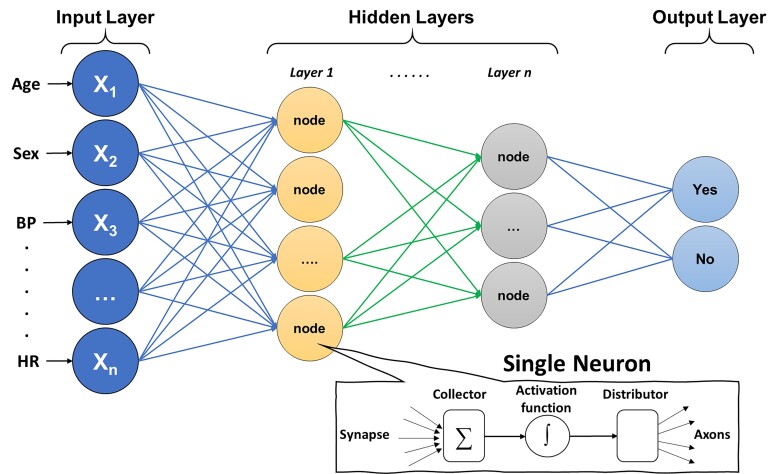
Finally, although not a distinct subtype of learning approaches, it is worth noting that DL has wide applications across supervised regression and classification, unsupervised, and semi-supervised learning, as well as RL. As illustrated in Figure 4, the architecture of DL is based on a neural network composed of an input layer (features), hidden layers (function nodes), and an output layer (prediction). Input features can either be structured data elements or unstructured data (e.g. pixels). The functional unit at each ‘synaptic connection’ is called a neuron and includes a summation and activation functions. A ML engineer will first need to define the network architecture and hyperparameters (e.g. number of hidden layers, number of nodes per layer, type of activation function, connection topologies, etc.). The next and most computationally exhaustive task is optimizing the synaptic weights in each neuron, which is frequently achieved using an optimization approach called gradient descent. Calculating the weights that optimize gradient descent is done using a first order iterative technique called backpropagation. Deep learning is a complex topic and has been explained in detail elsewhere,27 but Table 3 provides a simple summary of the common DL algorithms generally used in medical literature.
Figure 4.
Basic architecture of a deep neural network. The figure shows the basic architecture of a deep neural network, which is composed of an input layer (features), hidden layers (function nodes), and an output layer (prediction). The functional unit at each ‘synaptic connection’ is called a neuron and includes a summation and activation functions.
Table 3.
Common deep learning techniques with exemplary applications in healthcare
| Approach | Description |
|---|---|
| Artificial neural network (ANN) | This technique follows the architecture presented in Figure 4 with a feed-forward approach where inputs are only processed in a forward direction from one layer to the next until reaching the output layer which is the prediction/classification. |
| Recurrent neural network (RNN) | This is similar to ANN but with feed backward connections. Rather than feeding the output of neurons from one layer to the next, the outputs are re-fed to the previous layer (or to the neurons in the same layer). The synaptic weights are then recalibrated using the new information, making the optimization technique very exhaustive and time-consuming. |
| Convolutional neural network (CNN) | This is a multi-layer network specifically designed for processing unstructured data like images. Input features are usually values of pixels and hidden layers are designed to convolute these values to extract high-level features. Extracted features are then pooled and fed into an ANN. |
| Generative adversarial network (GAN) | A technique to design two competing neural networks, a generative one to learn producing data outputs and another to discriminate artificial data. GANs have been applied in assessing gender and racial algorithmic bias or in altering medical images or signals and augmenting data to train ML models. |
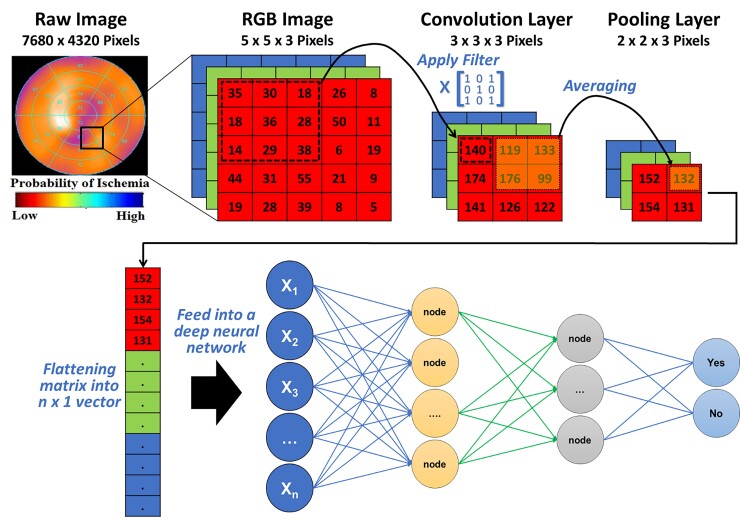
A DL technique that is widely used in cardiovascular literature is convolutional neural networks (CNN). This technique is specifically useful for image and signal processing where a series of convolution and pooling layers are used to extract numeric features from the unstructured images and use these extracted features in a multi-layer ANN. Figure 5 shows a simple illustration of a CNN model with one convolution layer and one pooling layer. Briefly, an image is usually stored as n-dimensional byte array with a colour value of each pixel for each of the three main colour channels: red, green, and blue (i.e. RGB image). First, a Kaufman filter (i.e. Kernel) of a given matrix size (e.g. 3 × 3) hovers over each colour channel and the resulting multiplication of pixel values and kernel values is stored in a new byte array. This step is called ‘convolution’ and is used to extract the low-level features of the image (e.g. edges, gradient). Next, to reduce the spatial dimensionality of the byte arrays, adjacent pixel values in a given size (e.g. 2 × 2) are summarized together by using either the mean or the max value (i.e. average pooling or max pooling, respectively). This step is called ‘pooling’ and is useful for extracting dominant features. After a series of iterations between convolution and pooling layers, the resulting byte array can be flattened into n × 1 array and fed into a classical multi-layer ANN to train the model on classifying the images based on known outcomes of interest.
Figure 5.
Basic architecture of a convolutional neural network. This figure illustrates how features can be extracted from a raw image (e.g. single photon emission computed tomography myocardial perfusion scan) for use in a neural network. The pixel values in each colour channel are multiplied by a kernel filter to extract low-level image features (convolution layer). Next, adjacent pixel values are grouped together using mean or max value to reduce spatial dimensionality (pooling layer). After repeated iterations of these two layers, the final byte array matrix is flattened and fed into a classical neural network to make predictions.
Understanding model development
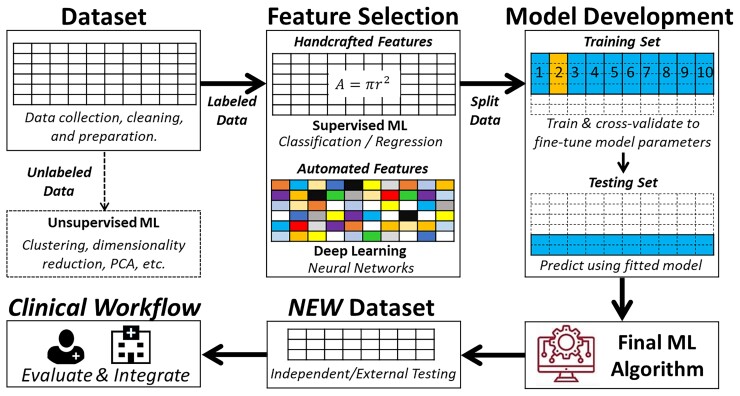
Learning from labelled cases (data) is the central premise of the field of ML. However, the process is far from simply dumping a dataset into a number-crunching machine and then wishing the machine to magically produce a useful model. Instead, both computational and clinical experts need to collaborate to guide the process, make key decisions along the way, and assess learned models to ensure that the data are properly used, and the learned model meets the needed performance. It is also possible that the learning process is an iterative one. Additional data collection and curation effort and/or algorithm improvement may need to happen between iterations to achieve the ultimate goal. Therefore, it is obvious that there are additional steps and considerations beyond invoking a programmed learning algorithm to process a dataset and obtain a ML model. Collectively, we consider in this section the whole process of transforming a dataset into a classifier or a regressor during model development. Figure 6 summarizes the four main building blocks of a ML pipeline.
Figure 6.
The iterative steps for developing a functional machine learning model. This is a simplified depiction of the actual iterative steps in building a machine learning pipeline. The first step in model development is data preprocessing followed by either supervised machine learning or unsupervised machine learning based on availability of labelled outcome data. Then, input features are pre-computed (i.e. handcrafted) or raw non-tabular data (i.e. image, waveform, etc.) is used for model development. Next, the dataset is partitioned into a training set and a testing set (usually 2:1). The training subset is further partitioned into k-folds to iteratively derive and update model hyperparameters, and the other testing subset is used to fine-tune and select the outperforming classifier or regressor. The best-performing model is then externally validated on new unseen data to determine generalizability before integration in the clinical workflow. Reproduced with permission from Helman et al.28
The first ML building block is data curation and preprocessing. Data quality requirements for ML models are steep, and the quality and quantity of data used to train a model determine its subsequent performance. Similar to any other clinical investigation, unbiased sampling techniques, valid and reliable measurement methods, and accurate outcome adjudication and ascertainment are essential pre-requisites to valid ML models. Once these pre-requisites are met, additional indicators of poor data quality include data ‘missingness’, incompleteness, inconsistency, inaccuracy, duplication, outliers, and crowdsourcing (i.e. irrelevant data).29 Thus, ML engineers should invest extensive amount of time for handling missing data, noise, and outliers; dealing with duplicates; and feature engineering. These steps entail univariate and bivariate data exploration, visualization, and proper data reduction or clustering before any model development.
The second ML building block is feature engineering. The word ‘feature’ is typically interchangeable with ‘variable’ or ‘independent variable’. Model performance depends on the appropriate selection and inclusion of input features. An important consideration is the number of features in relation to the size of training subset where a larger number of features requires exponentially larger sample size (i.e. called ‘curse of dimensionality’). Thus, it is important to use appropriate feature selection techniques to remove irrelevant features that might simply introduce noise into predictions while ensuring that relatively important features are not omitted. Table 4 summarizes the commonly used feature selection techniques in ML. While most techniques for feature selection are data driven, it is worth noting that ML engineers should seek domain experts’ advice for identifying subsets of features that are mechanistically linked to predicting the outcome at hand,30,31 an approach that has been shown to significantly improve model performance.32
Table 4.
Common feature selection techniques in machine learning
| Technique | Description |
|---|---|
| Recursive feature elimination (RFE) | Iteratively removing features that do not contribute to model performance based on predetermined accuracy metric. Machine learning engineers can specify desired number of features for inclusion in final model. |
| LASSO | In this approach, a penalty term that shrinks marginal coefficients toward zero. Features with estimates equal to zero will be dropped from the regression model; indirectly selecting features while optimizing the least squares estimator. |
| Principal component analysis (PCA) | PCA uses linear equations to create informative combinations between features in the dataset. The new combinations are weighted in a way that the first few vectors explain most variability in features in the dataset (principal components or eigenvalues). These principal components are independent and can be used in subsequent analysis rather than using the original features in the dataset. This compresses the dimensionality in the data by reducing the number of features and trading accuracy for simplicity. |
The third and most critical ML building block is model development. The starting point of this process is typically a partition of the dataset into a training subset and a testing subset. The ratio of this split is arbitrary and is frequently based on sample size. Common partitioning splits seen in clinical research include 70%/30%, 80%/20%, or 90%/10%. These two parts are disjoint with regard to a chosen variable (e.g. subject identifier, time of observations, etc.). The most important consideration is to make sure data from the same subject do not end up in both training and testing subsets (i.e. data leakage bias). When possible, the partitioning is done in a random fashion and at least preserves the prevalence of data samples per each class in the testing subset in comparison with the target population for which the model will be used. However, many data scientists elect to artificially keeping the prevalence of disease at ∼50% in the training dataset by oversampling the positive class (cases) or under-sampling the negative class (controls). This dataset balancing technique is meant to counteract false predictions caused by the algorithm’s over-reliance on the probabilistic distribution of the dominant class. For instance, in an unbalanced dataset with disease prevalence of only 10%, the algorithm would learn to predict ‘no disease’ whenever the uncertainty is high because there is a 90% chance this prediction would be true. Thus, balancing the training dataset can improve the classification performance during model development, although this can adversely affect generalizability to real-world data.
After the partitioning, the testing subset is set aside and not touched till ‘lockdown’ models have been obtained. To obtain lockdown models, training data are used. Using the whole training dataset for this task is impractical because a single training set is insufficient to find optimal hyperparameters or to assess variability in performance or risk of overfitting. Thus, to build more reliable lockdown models, the training subset is further partitioned into k-folds to use as cross-validation (CV) folds. These folds need to be created with proper stratification to maintain approximately equal ratios between positive and negative classes. The most used number of folds is 10 (i.e. 10-fold CV), which would allow adequate statistical comparison between variations in performance induced by different choices of parameters across these 10 folds. This is done in a round-robin way where the model is trained with nine folds (or k-1) and tested on the remaining fold. In each of the 10 iterative rounds, a different combination of hyperparameters is used (i.e. grid search). A performance metric [e.g. area under receiver operating characteristic (ROC) curve] is computed for each of the 10 folds and one-way ANOVA is used to determine whether different combinations of parameters have a statistically significant impact on performance. Next, the model (or models) that produces the highest average performance rank across the 10 CV folds is chosen and a final lockdown model is trained on the full training subset under the selected hyperparameters.
It is worth noting that if the training subset is not large enough to yield reliable distribution ratios in k-folds, the above procedure is modified by using an alternative CV approach referred to as leave one out cross-validation (i.e. LOOCV). In this approach, the grid search for selecting the hyperparameters for the lockdown is iteratively developed on n-1 training subsets and keeping that last subject in each iteration for parameter fine tuning. In either approach (k-fold CV or LOOCV), the final lockdown model is validated on the hold-out testing subset to assess how learned prediction rules generalize to new data and obtain final performance metrics. Table 5 summarizes the standardized performance assessment metrics used in predictive analytics.
Table 5.
Performance metrics for assessing machine learning models
| Metric | Description |
|---|---|
| Regression-based metrics | |
| Root mean square error (RMSE) | A measure for the deviation of the predicted values from the actual ones. The lower RMSE the more accurate the model. |
| Mean absolute error (MAE) | A measure of the absolute values for the difference between the predicted and the actual values. |
| R2 or adjusted R2 | The proportion of variance in the outcome that can be explained by the predictors. Adjusted R2 is more robust when new variables are added to the model. |
| Akaike’s information criterion (AIC) | Provides an indication of the model performance that accounts for model complexity. |
| Bayesian information criterion (BIC) | A measure similar to AIC but using Bayesian approach. It performs better for positive findings. |
| Classification-based metrics | |
| Area under ROC curve | ROC visually plots a curve between the true positive rate (recall) and the false positive rate with a total possible area under the curve of 1. The larger this area the more precise the classification performance. |
| Precision | The accuracy of the positive predictions (i.e. positive predictive value). Precision = TP/(TP + FP). |
| Recall (sensitivity) | The ratio of positive instances that are correctly detected by the classifier (i.e. sensitivity). Recall = TP/(TP + FN) |
| F1 score | The harmonic mean between precision and recall, which is a measure that works well with imbalanced data. F1 score = 2 × [(precision × recall)/(precision + recall)]. |
| Precision-recall curve | Precision-recall curve visually plots a curve between true positive rate (precision) and sensitivity (recall). Precision-recall curve is typically used in real-time alerting systems where high false positive rate is problematic (e.g. alarm fatigue). |
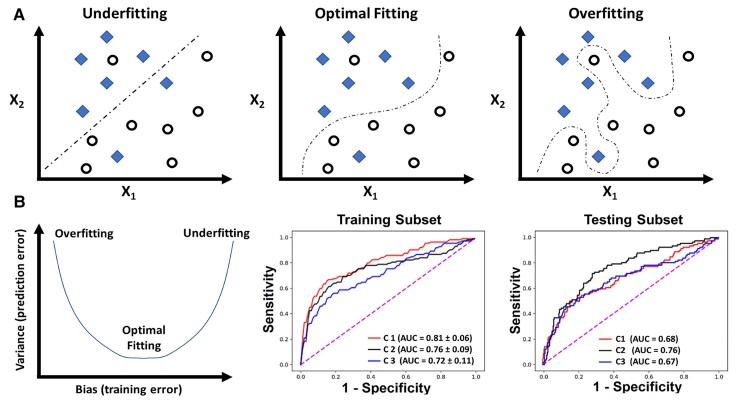
The above procedure for partitioning the dataset during model development is used to assess for overfitting; the biggest challenge in building a valid and generalizable ML model. Overfitting implies that the algorithm has captured patterns in the data irrelevant to outcome of interest (e.g. confounders, redundancy, missingness, outliers, etc.) rather than the real associations between variables. Such ‘noise’ causes the model to overfit the training data, so it generalizes poorly to new data (Figure 7A). To assess for overfitting, one first must estimate the overall bias in model predictions during training and compare it to the associated variance during testing. Bias refers to the overall model accuracy on historical data (e.g. root mean square error for regression, area under ROC curve for classification), whereas variance refers to the consistency in performance on future data. There is a tradeoff between bias and variance (Figure 7B). This means that very accurate models during training could yield large prediction error on new data (low bias – high variance), whereas less accurate ones during training could generalize well on new data without loss in performance (high bias − low variance). These challenges emphasize the need for evaluating a suite of regressors or classifiers during model development before selecting the model that best fits the data (i.e. optimizes bias − variance tradeoff).
Figure 7.
Overfitting and bias-variance tradeoff in machine learning model development. (A) The simple classification case of a binary outcome (denoted by diamonds and circles) using two variables X1 and X2. Unlike the first two classifiers that focused on capturing a real association between X1, X2, and the outcome, the last classifier seemed to capture patterns in the data irrelevant to outcome of interest (e.g. confounding, redundancy, missingness, outliers, etc.), thus ‘overfitting’ the model to training data. In (B), the plot to the left demonstrates three dynamic phases of the tradeoff between bias (training error) and variance (testing error): low bias − high variance (overfitting), low bias – low variance (optimal fitting), and high bias – high variance (underfitting). The two plots to the right show the area under receiver operating characteristic curve of three classifiers (C1, C2, and C3) fitted on a training cohort of n = 745 and testing cohort of n = 499.32 C1 shows the lowest bias (high area under receiver operating characteristic) during training but high variance (low area under receiver operating characteristic) during testing (i.e. overfitting), whereas C3 shows the highest bias (low area under receiver operating characteristic) during training and highest variance (low area under receiver operating characteristic) during testing (i.e. underfitting).
When training a deep neural network with stochastic gradient descent, the risk of overfitting is high. Therefore, at each round of training, it is typically necessary to further reserve a certain amount of data (10% of training data used in this round) as a validation dataset and to stop further gradient descent when a chosen performance metric or simply the value of the loss function stops improving after a set number of training epochs. Another challenge in training a deep neural network is the high cost in terms of computational power and therefore a random grid-based search of hyperparameters cannot be done with very fine resolution using an affordable amount of resources. Therefore, more recent approaches around AutoML need to be adopted where more effective ways of exploring the hyperparameter space can be achieved via algorithms such as genetic algorithm or Bayesian model optimization.
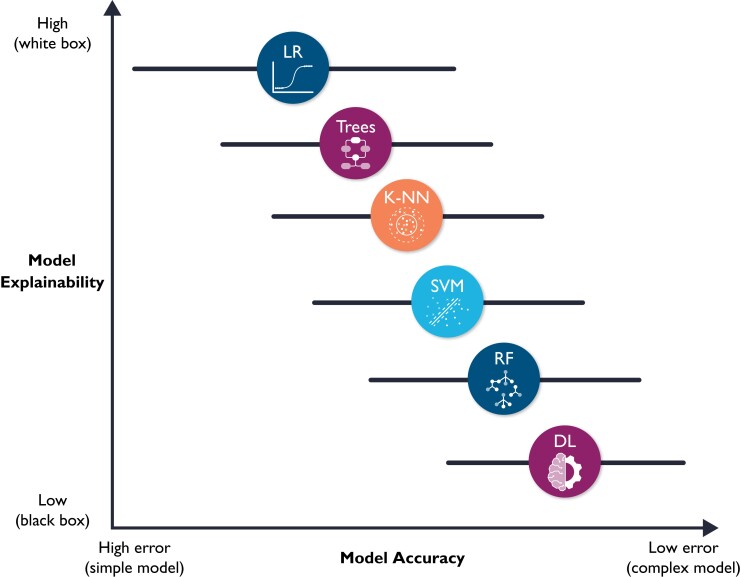
The fourth and final ML building block is prospective validation and integration into clinical workflow, which are not trivial tasks. Implementation into clinical workflow requires system training, performance engineering, monitoring, and system updating.33 More importantly, before any implementation, ML models require unique considerations for evaluation for optimal clinical utility, including prospective validation in representative clinical settings, as well as establishing benchmarks against reference standards. Unfortunately, almost 80% of DL-based models are based on open-source datasets.34 Nearly half of studies carried out in representative clinical settings show no incremental diagnostic gain over existing clinical decision support tools.35 These observations suggest a need for more sophisticated models to warrant a change in clinical practice. However, it is worth noting that the improved accuracy which accompanies increased model complexity also accompanies diminished model explainability. Figure 8 shows a hypothetical tradeoff between model accuracy and mode explainability. Explainability here refers to a human’s ability to understand the algorithm’s ‘logic’ (e.g. how and why a certain class was assigned). Explainable ML models are essential in healthcare not only because of their compatibility with clinical workflow and bedside care, but also for improving rigour and reproducibility of clinical investigation. There are currently numerous ‘under-the-hood’ techniques to improve ML model explainability, including partial dependence plots, rule extraction (e.g. fuzzy rules), feature importance (e.g. local interpretable model-agnostic explanations or LIME), decision trees, sensitivity analysis, layer-wise relevance propagation, and attention mechanisms (e.g. heatmaps).36–38
Figure 8.
Hypothetical tradeoff between model accuracy and model explainability. This figure shows a hypothetical relationship between model accuracy (computational cost) and model explainability. It is worth noting that this is an over-simplistic view of the relationship between these two constructs, and that the relationship between the selected classifiers is not linear. Yet, this figure emphasizes that predictive modelling is ‘mission critical’; explainable (simple) models are preferred because they will be trusted more, and thus used more.11 These models might also be more accurate than complex ones (note the horizontal error bars for accuracy). DL, deep learning; RF, random forest; SVM, support vector machine; K-NN, nearest neighbours; LR, logistic regression.
Methodological considerations for design rigour
A main challenge in any scientific inquiry is ensuring that conclusions are valid and reliable. This implies that published results must be reproducible.8 Unfortunately, more than 70% of scientists failed in replicating others’ findings and nearly half failed in reproducing their very own findings,39 making bias, or systematic errors, an unprecedented threat to evidence-based practice.40 Using robust experimental workflows to reduce unintentional errors is at the heart of scientific inquiry principles. There are numerous types of bias specific to ML literature. Table 6 summarizes numerous sources of bias relevant to ML research as previously identified by Mehrabi et al.41 In this table, we also relate the magnitude of threat of each source of bias to prediction accuracy of the ML model and its generalizability to new data.
Table 6.
Summary of common sources of bias relevant to machine learning literature41
| Type of bias | Description | Threat to prediction accuracy | Threat to generalizability to new data |
|---|---|---|---|
| 1. Bias related to data quality and curation | |||
| Sampling bias | Systematic error due to non-random sampling of subgroups. | + | +++ |
| Measurement bias | Systematic error due to invalid or unreliable tools to quantify a particular feature. | +++ | + |
| Omitted feature bias | Systematic error due to absence of important features | +++ | + |
| Representation bias | Systematic error due to lack of diversity in measured features. | + | +++ |
| Aggregation bias | Systematic error due to analysing heterogenous subgroups together. | +++ | + |
| Longitudinal data fallacy | Systematic error due analysing temporally diverse cohorts into a single time point. | +++ | + |
| 2. Modelling bias | |||
| Algorithmic bias | Systematic error introduced by algorithm design choices. | +++ | +++ |
| Presentation bias | Systematic error due to how information is presented. | +++ | + |
| Evaluation bias | Systematic error due to using inappropriate benchmarks during model design and selection. | +++ | +++ |
| 3. Model deployment bias | |||
| Historical bias | Systematic error due to learning implicit bias in data itself (e.g. clinician annotation behaviour). | ++ | ++ |
| Population bias | Systematic error due to mismatch between training data and target population (i.e. intent of use). | + | +++ |
| Temporal bias | Systematic error due changes in practice over time. | + | +++ |
Herein we highlight few sources of ML bias that are closely related to healthcare research. The first important methodological consideration is the impact of study design on ML models. Most ML models are developed on historical data from observational research (cohort and case–control designs). Each of these designs bring inherited methodological limitations on data quality and potential clinical use. While cohort studies are methodologically suitable for disease prognosis (forecasting) and diagnostics (predictive analytics), case–control studies are less reliable for designing predictive analytics. Specifically, predictive models developed on case–control studies were poorly calibrated and had less discriminative power when compared with cohort studies for modelling the same outcome.42 This poor performance can be attributed to poor control over known and unknown confounders during the selection of samples in case–control designs, emphasizing that careful construction of a case–control design can lead to comparable discriminative performance as a cohort design. Other limitations for the poor model calibration in case–control designs are (i) the over-representation of the outcome class, (ii) the poor representation of data available during the time of diagnosis, and (iii) the overemphasis on features closer to the outcome of interest (temporal bias).43 Thus, a ML model based on case–control studies must be prospectively validated using a cohort design for fair evaluation and recalibration before any clinical deployment.42
A second important consideration for ML applications in healthcare is sampling bias. Although most ML engineers focus on the adequacy of training samples and the numeric distribution of input features, little attention is paid to the sampling techniques used to collect the historical data in the first place. If biased sampling techniques were used, then the probabilistic distribution of features in training data might be different than that in representative clinical settings.44 This might produce models that are not only poorly generalizable to unseen data, but also lack physiological plausibility. Another important sampling concern is using multiple samples from the same patient across training and testing subsets. Unless these multiple samples are physiologically distinct, then the modelling dataset would be flawed. An example would be using each heartbeat within a 10-s ECG rhythm strip as a training sample. This essentially violates the Gaussian distribution principles where input features are expected to be independent and identically distributed. This frequently yields unrealistic and exaggerated performance metrics during model training, which would again poorly generalize to new unseen data.
Another critical consideration in ML research is the rigour of the ascertainment of the outcome variable. The conclusion validity of any supervised ML model depends heavily on the quality of labels on which the model was trained. High-quality labels need to be based on a good reference standard (e.g. gold standard) or a majority panel vote (e.g. consensus adjudication) where reviewers are blinded to model predictions.45 Poorly labelled outcome data will negatively affect model building twice, once during model training and once during model evaluation, leading to high bias − high variance tradeoff. Moreover, given that sophisticated ML models require large amount of data, many studies rely on ‘silver’ labels. Outcome ascertainment using silver labels can be based on with electronic health records (EHR)-phenotyping (i.e. rule-based queries) or semi-supervised labelling (i.e. predictions by active learning), each of which comes with its own limitations. For example, EHR-based phenotyping has been shown to miss up to 21% of acute MI events.46 Thus, assessing the quality and robustness of outcome ascertainment is important before evaluating model performance claims.
Quality assessment checklist
Ensuring rigours and reproducibility in ML research is essential for designing functional algorithms for clinical use, and there are currently numerous reporting guidelines and user guides for designing such ML models.1,8,33,45,47–52 However, evaluating the quality of a published ML paper requires a comprehensive understanding of the available ML techniques and approaches; the main building blocks of a functional ML pipeline; and potential design flaws and methodological biases involved. Table 7 provides a summary checklist for systematically evaluating published ML studies. This checklist can guide clinicians while evaluating the rigour of a published ML study by asking the most relevant questions and searching for potential pitfalls and red flags.
Table 7.
A checklist for Ruling Out Bias Using Standard Tools in Machine Learning
| Quality items | Important red flags to observe |
|---|---|
| 1. General design considerations | |
| 1.1. Was the study design and data collection methods appropriate? | Poorly designed case–control study, over-represented outcome class, etc. |
| 1.2. Was there any evidence for sampling bias? | Misrepresentation of features; using dependent or duplicate samples, etc. |
| 1.3. Is the experiment reproducible? | Insufficient details on data provenance of ML processes; paper does not follow reporting guidelines; failure to adhere to code or data availability policies, etc. |
| 2. Data quality considerations | |
| 2.1. Was the dataset size commensurate with the learning task and model complexity? | Large number of features relative to dataset size, large number of prediction classes relative to dataset size, <5 positive labels per input feature, DL on only few hundred examples, etc. |
| 2.2. Was data of high quality? | Data missingness, incompleteness, inconsistency, inaccuracy, duplication, outliers, noise, crowdsourcing, etc. |
| 2.3. Was missingness in the data characterized and properly handled? | >5% missing data, data not missing at random, excessive data imputation, etc. |
| 2.4. Were data properly visualized and exploratory analyses performed? | Lack of exploratory data analysis (tables or visual graphs of important features), data representation bias, etc. |
| 2.5. Were raw data collected as per accepted clinical standards, protocols, and techniques (valid and reliable measurement tools)? | Erroneous raw data, high interrater variability, inaccurate feature computation, etc. |
| 2.6. Were criteria and procedures that were used to assign the labels robust and acceptable? | Using only a single reviewer, lack of consensus, high interrater reliability, using surrogate outcomes or ‘soft’ silver labels, etc. |
| 3. Feature engineering considerations | |
| 3.1. Was the number of features commensurate to size of dataset? | Features to sample size ratio is ∼1:1 (without dimensionality reduction), etc. |
| 3.2. Were features selection techniques implemented? | Inclusion of all features without exploring feature selection, inclusion of features not typically available to algorithm in the intended target use, no feature ranking, or review by domain experts, etc. |
| 3.3. Was there any feature omission bias? | Important predictors missing, relevant confounders not controlled, etc. |
| 3.4. Was there any consultation with clinical domain experts on feature selection appropriateness? | Data-driven features not reviewed by domain experts, unintentional removal of features essential in clinical decision making, etc. |
| 3.5. Were irrelevant features removed? | Inadequate feature selection in highly dimensional datasets, etc. |
| 4. Model development considerations | |
| 4.1. Was dataset partitioning appropriate? | Inadequate training samples, duplicate observation in training and testing sets, etc. |
| 4.2. Was cross-validation (CV) done properly? | Cross-validation not used, mismatch between number of folds and sample size, using k-fold CV instead of LOOCV with very small sample sizes, not reporting confidence interval for the performance of CV, etc. |
| 4.3. Were parametric assumptions satisfied whenever indicated? | Skewed data with no transformations, high data collinearity, etc. |
| 4.4. Was there any data leakage (information from outside the training dataset is used to create the model)? | Scaling features before dataset partitioning, unblinding of labels, duplicate samples in both training and testing subsets, etc. |
| 4.5. Was special attention paid when assessing models with imbalanced classes? | Improper evaluation metrics when positive labels are rare, balancing the test subset (prevalence higher than expected), etc. |
| 4.6. Was the selection of best fitting algorithm appropriate? | Using complex models to simple tasks, comparing <3–4 predictive models, poor optimization techniques, etc. |
| 4.7. Was bias-variance tradeoff adequately assessed (underfitting vs. overfitting)? | Only results on testing set are presented, omitting confidence intervals, etc. |
| 4.8. Was there any evaluation bias (systematic error in predictions)? | Using only a single performance assessment metric, lack of adequate benchmarks, no interrogation of sources of error in false positives, and false negatives etc. |
| 4.9. Was there any algorithmic bias (underperformance in population subgroups like females or minorities)? | No post hoc sensitivity analyses, no AI fairness assessment, etc. |
| 4.10. Were there any unexpected results? | Very heterogenous samples (aggregation bias), analysing repeated measures cross-sectionally (longitudinal data fallacy), data based on old practices (temporal bias) |
| 5. Considerations for clinical utility | |
| 5.1. Did the dataset match the target clinical setting in which the model will be used? | Inappropriate use of an open-source dataset, data based on outdated practices (temporal bias), population bias, etc. |
| 5.2. Were results interpretable? | Lack of results on model explainability, false predictions are not interrogated, no actual case studies presented to demonstrate true or false predictions/classification, etc. |
| 5.3. Was the model assessed for gender- and racial bias? | No use of AI fairness assessment tools, historical bias, no subgroup sensitivity analyses, etc. |
| 5.4. Was model compared with a clinical benchmark to establish incremental gain? | No comparison against a reference standard (no comparison against existing algorithms that are in use today or comparison with experts with no significance testing), unacceptable reference standard, etc. |
| 5.5. Was the model externally validated on data from a different setting? | No independent validation set or test set from a different hospital or region, etc. |
| 5.6. Was technology acceptability empirically assessed? | Inadequate domain expertise, no collaborating clinicians, etc. |
Summary
Developing functional ML-based models to address unmet clinical needs requires unique considerations to reach the stage of potential clinical utility. This review summarized the main ML building blocks and identified important red flags clinicians should observe while critically appraising ML applications in healthcare. Bridging the gap between clinicians, healthcare scientists, and ML engineers can address many shortcomings and pitfalls of ML-based solutions and their potential deployment at the bedside. It is important for clinical ML studies to be reviewed by clinicians, and a checklist such as the one provided herein may serve as an aid. It is worth noting, though, that this checklist requires subsequent revisions using a formal Delphi consensus process so that it can be listed on the EQUATOR Network as a formal quality assessment tool.
Acknowledgments
A.P. and R.B. are supported by the European Union’s INTERREG VA programme, managed by the Special EU Programmes Body (SEUPB). Their work is associated with the project—‘Centre for Personalised Medicine—Clinical Decision Making and Patient Safety’. The views and opinions expressed in this study do not necessarily reflect those of the European Commission or the Special EU Programmes Body (SEUPB).
Funding: None.
Conflicts of interest: None declared.
Contributor Information
Salah S Al-Zaiti, Department of Acute and Tertiary Care, Department of Emergency Medicine, and Division of Cardiology, University of Pittsburgh, Pittsburgh PA, USA.
Alaa A Alghwiri, Data Science Core, The Provost Office, University of Pittsburgh, Pittsburgh PA, USA.
Xiao Hu, Center for Data Science, Emory University, Atlanta, GA, USA.
Gilles Clermont, Departments of Critical Care Medicine, Mathematics, Clinical and Translational Science, and Industrial Engineering, University of Pittsburgh, Pittsburgh, PA, USA.
Aaron Peace, The Clinical Translational Research and Innovation Centre, Northern Ireland, UK.
Peter Macfarlane, Institute of Health and Wellbeing, Electrocardiology Section, University of Glasgow, Glasgow, UK.
Raymond Bond, School of Computing, Ulster University, Ulster, UK.
Data availability
No new data were generated or analysed in support of this research.
References
- 1. Leisman DE, Harhay MO, Lederer DJ, Abramson M, Adjei AA, Bakker J, Ballas ZK, Barreiro E, Bell SC, Bellomo R, Bernstein JA, Branson RD, Brusasco V, Chalmers JD, Chokroverty S, Citerio G, Collop NA, Cooke CR, Crapo JD, Donaldson G, Fitzgerald DA, Grainger E, Hale L, Herth FJ, Kochanek PM, Marks G, Moorman JR, Ost DE, Schatz M, Sheikh A, Smyth AR, Stewart I, Stewart PW, Swenson ER, Szymusiak R, Teboul J-L, Vincent J-L, Wedzicha JA, Maslove DM. Development and reporting of prediction models: guidance for authors from editors of respiratory, sleep, and critical care journals. Crit Care Med 2020;48:623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rajkomar A, Dean J, Kohane I. Machine Learning in Medicine. N Engl J Med 2019;380:1347–1358. [DOI] [PubMed] [Google Scholar]
- 3. Al’Aref SJ, Anchouche K, Singh G, Slomka PJ, Kolli KK, Kumar A, Pandey M, Maliakal G, Van Rosendael AR, Beecy AN, Berman DS, Leipsic J, Nieman K, Andreini D, Pontone G, Schoepf UJ, Shaw LJ, Chang H-J, Narula J, Bax JJ, Guan Y, Min JK. Clinical applications of machine learning in cardiovascular disease and its relevance to cardiac imaging. Eur Heart J 2019;40:1975–1986. [DOI] [PubMed] [Google Scholar]
- 4. He J, Baxter SL, Xu J, Xu J, Zhou X, Zhang K. The practical implementation of artificial intelligence technologies in medicine. Nat Med 2019;25:30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kagiyama N, Shrestha S, Farjo PD, Sengupta PP. Artificial intelligence: practical primer for clinical research in cardiovascular disease. J Am Heart Assoc 2019;8:e012788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thottakkara P, Ozrazgat-Baslanti T, Hupf BB, Rashidi P, Pardalos P, Momcilovic P, Bihorac A. Application of machine learning techniques to high-dimensional clinical data to forecast postoperative complications. PLoS One 2016;11:e0155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bluemke DA, Moy L, Bredella MA, Ertl-Wagner BB, Fowler KJ, Goh VJ, Halpern EF, Hess CP, Schiebler ML, Weiss CR. Assessing radiology research on artificial intelligence: a brief guide for authors, reviewers, and readers—from the radiology editorial board. Radiology 2020;294:487–489. [DOI] [PubMed] [Google Scholar]
- 8. Pineau J, Vincent-Lamarre P, Sinha K, Larivière V, Beygelzimer A, d’Alché-Buc F, Fox E, Larochelle H. Improving reproducibility in machine learning research: a report from the NeurIPS 2019 reproducibility program. J Mach Learn Res 2021;22:1–20. [Google Scholar]
- 9. Helman S, Terry MA, Pellathy T, Williams A, Dubrawski A, Clermont G, Pinsky MR, Al-Zaiti S, Hravnak M. Engaging clinicians early during the development of a graphical user display of an intelligent alerting system at the bedside. Int J Med Inform 2021;159:104643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McCradden MD, Joshi S, Mazwi M, Anderson JA. Ethical limitations of algorithmic fairness solutions in health care machine learning. Lancet Digital Health 2020;2:e221–e223. [DOI] [PubMed] [Google Scholar]
- 11. Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intell 2019;1:206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Muehlematter UJ, Daniore P, Vokinger KN. Approval of artificial intelligence and machine learning-based medical devices in the USA and Europe (2015–20): a comparative analysis. Lancet Digital Health 2021;3:e195–e203. [DOI] [PubMed] [Google Scholar]
- 13. Yarali A. Applications of Artificial Intelligence, ML, and DL Intelligent Connectivity: AI, IoT, and 5G: IEEE; 2022: 279–297. [Google Scholar]
- 14. Saria S, Butte A, Sheikh A. Better medicine through machine learning: what’s real, and what’s artificial? PLoS Med 2019;15:e1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bzdok D, Altman N, Krzywinski M. Statistics versus machine learning. Nat Methods 2018;15:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fan J, Li R. Statistical challenges with high dimensionality: feature selection in knowledge discovery. arXiv preprint math/0602133. 2006. [Google Scholar]
- 17. Friedman JH. On bias, variance, 0/1—loss, and the curse-of-dimensionality. Data Min Knowl Discov 1997;1:55–77. [Google Scholar]
- 18. Richens JG, Lee CM, Johri S. Improving the accuracy of medical diagnosis with causal machine learning. Nat Commun 2020;11:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Longstaff B, Reddy S, Estrin D. Improving activity classification for health applications on mobile devices using active and semi-supervised learning. 2010 4th International Conference on Pervasive Computing Technologies for Healthcare. 2010:1–7. [Google Scholar]
- 20. Li L-J, Fei-Fei L. Optimol: automatic online picture collection via incremental model learning. Int J Comput Vis 2010;88:147–168. [Google Scholar]
- 21. Guan D, Yuan W, Lee Y-K, Gavrilov A, Lee S. Activity recognition based on semi-supervised learning. 13th IEEE International Conference on Embedded and Real-Time Computing Systems and Applications (RTCSA 2007). 2007:469–475. [Google Scholar]
- 22. Chai H, Liang Y, Wang S, Shen H-W. A novel logistic regression model combining semi-supervised learning and active learning for disease classification. Sci Rep 2018;8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xia Y, Xie Y. A novel wearable electrocardiogram classification system using convolutional neural networks and active learning. IEEE Access 2019;7:7989–8001. [Google Scholar]
- 24. Naeem M, Rizvi STH, Coronato A. A gentle introduction to reinforcement learning and its application in different fields. IEEE Access 2020;8:209320–209344. [Google Scholar]
- 25. Gottesman O, Johansson F, Komorowski M, Faisal A, Sontag D, Doshi-Velez F, Celi LA. Guidelines for reinforcement learning in healthcare. Nat Med 2019;25:16–18. [DOI] [PubMed] [Google Scholar]
- 26. Liu S, See KC, Ngiam KY, Celi LA, Sun X, Feng M. Reinforcement learning for clinical decision support in critical care: comprehensive review. J Med Internet Res 2020;22:e18477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, Cui C, Corrado G, Thrun S, Dean J. A guide to deep learning in healthcare. Nat Med 2019;25:24–29. [DOI] [PubMed] [Google Scholar]
- 28. Helman SM, Herrup EA, Christopher AB, Al-Zaiti SS. The role of machine learning applications in diagnosing and assessing critical and non-critical CHD: a scoping review. Cardiol Young 2021;31:1770–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gudivada V, Apon A, Ding J. Data quality considerations for big data and machine learning: going beyond data cleaning and transformations. Int J Adv Softw 2017;10:1–20. [Google Scholar]
- 30. Bond R, Finlay D, Al-Zaiti SS, Macfarlane P. Machine learning with electrocardiograms: a call for guidelines and best practices for ‘stress testing’ algorithms. J Electrocardiol 2021;69:1–6. [DOI] [PubMed] [Google Scholar]
- 31. Al-Zaiti S, Besomi L, Bouzid Z, Faramand Z, Frisch S, Martin-Gill C, Gregg R, Saba S, Callaway C, Sejdić E. Machine learning-based prediction of acute coronary syndrome using only the pre-hospital 12-lead electrocardiogram. Nat Commun 2020;11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bouzid Z, Faramand Z, Gregg Richard E, Frisch Stephanie O, Martin-Gill C, Saba S, Callaway C, Sejdić E, Al-Zaiti S. In search of an optimal subset of ECG features to augment the diagnosis of acute coronary syndrome at the emergency department. J Am Heart Assoc 2021;10:e017871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pencina MJ, Goldstein BA, D’Agostino RB. Prediction models-development, evaluation, and clinical application. N Engl J Med 2020;382:1583–1586. [DOI] [PubMed] [Google Scholar]
- 34. Hong S, Zhou Y, Shang J, Xiao C, Sun J. Opportunities and challenges of deep learning methods for electrocardiogram data: a systematic review. Comput Biol Med 2020;122:103801. [DOI] [PubMed] [Google Scholar]
- 35. Vasey B, Ursprung S, Beddoe B, Taylor EH, Marlow N, Bilbro N, Watkinson P, McCulloch P. Association of clinician diagnostic performance with machine learning-based decision support systems: a systematic review. JAMA Netw Open 2021;4:e211276–e211276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hicks SA, Isaksen JL, Thambawita V, Ghouse J, Ahlberg G, Linneberg A, Grarup N, Strümke I, Ellervik C, Olesen MS, Hansen T, Graff C, Holstein-Rathlou N-H, Halvorsen P, Maleckar MM, Riegler MA, Kanters JK. Explaining deep neural networks for knowledge discovery in electrocardiogram analysis. Sci Rep 2021;11:10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Payrovnaziri SN, Chen Z, Rengifo-Moreno P, Miller T, Bian J, Chen JH, Liu X, He Z. Explainable artificial intelligence models using real-world electronic health record data: a systematic scoping review. J Am Med Inform Assoc 2020;27:1173–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lauritsen SM, Kristensen M, Olsen MV, Larsen MS, Lauritsen KM, Jørgensen MJ, Lange J, Thiesson B. Explainable artificial intelligence model to predict acute critical illness from electronic health records. Nat Commun 2020;11:3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baker M. 1,500 scientists lift the lid on reproducibility. Nature 2016;533:452–454. [DOI] [PubMed] [Google Scholar]
- 40. Sarewitz D. Beware the creeping cracks of bias. Nature 2012;485:149–149. [DOI] [PubMed] [Google Scholar]
- 41. Mehrabi N, Morstatter F, Saxena N, Lerman K, Galstyan A. A survey on bias and fairness in machine learning. ACM Comput Surv (CSUR) 2021;54:1–35. [Google Scholar]
- 42. Reps J, Ryan PB, Rijnbeek PR, Schuemie MJ. Design matters in patient-level prediction: evaluation of a cohort vs. case–control design when developing predictive models in observational healthcare datasets. J Big Data 2021;8:1–8.33425651 [Google Scholar]
- 43. Yuan W, Beaulieu-Jones BK, Yu K-H, Lipnick SL, Palmer N, Loscalzo J, Cai T, Kohane IS. Temporal bias in case–control design: preventing reliable predictions of the future. Nat Commun 2021;12:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kukull WA, Ganguli M. Generalizability: the trees, the forest, and the low-hanging fruit. Neurology 2012;78:1886–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu Y, Chen P-HC, Krause J, Peng L. How to read articles that use machine learning: users’ guides to the medical literature. JAMA 2019;322:1806–1816. [DOI] [PubMed] [Google Scholar]
- 46. Mentz RJ, Newby LK, Neely B, Lucas JE, Pokorney SD, Rao MP, Jackson LR, Grau-Sepulveda MV, Smerek MM, Barth P, Nelson CL, Pencina MJ, Shah BR. Assessment of administrative data to identify acute myocardial infarction in electronic health records. J Am Coll Cardiol 2016;67:2441–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Collins GS, Dhiman P, Navarro CLA, Ma J, Hooft L, Reitsma JB, Logullo P, Beam AL, Peng L, Van Calster B. Protocol for development of a reporting guideline (TRIPOD-AI) and risk of bias tool (PROBAST-AI) for diagnostic and prognostic prediction model studies based on artificial intelligence. BMJ Open 2021;11:e048008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Faes L, Liu X, Wagner SK, Fu DJ, Balaskas K, Sim DA, Bachmann LM, Keane PA, Denniston AK. A clinician's guide to artificial intelligence: how to critically appraise machine learning studies. Transl Vis Sci Technol 2020;9:7–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scott I, Carter S, Coiera E. Clinician checklist for assessing suitability of machine learning applications in healthcare. BMJ Health Care Inform 2021;28:e100251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sounderajah V, Ashrafian H, Aggarwal R, De Fauw J, Denniston AK, Greaves F, Karthikesalingam A, King D, Liu X, Markar SR, McInnes MDF, Panch T, Pearson-Stuttard J, Ting DSW, Golub RM, Moher D, Bossuyt PM., Darzi A. Developing specific reporting guidelines for diagnostic accuracy studies assessing AI interventions: the STARD-AI Steering Group. Nat Med 2020;26:807–808. [DOI] [PubMed] [Google Scholar]
- 51. Sounderajah V, Ashrafian H, Rose S, Shah NH, Ghassemi M, Golub R, Kahn CE, Esteva A, Karthikesalingam A, Mateen B, Webster Dale, Milea Dan, Ting Daniel, Treanor Darren, Cushnan D, King D, McPherson D, Glocker B, Greaves F, Harling L, Ordish J, Cohen JF, Deeks Jon, Leeflang M, Diamond M, McInnes MDF, McCradden M, Abràmoff MD, Normahani P, Markar SR, Chang S, Liu X, Mallett S, Shetty S, Denniston A, Collins GS, Moher D, Whiting P, Bossuyt PM, Darzi A. A quality assessment tool for artificial intelligence-centered diagnostic test accuracy studies: QUADAS-AI. Nat Med 2021;27:1663–1665. [DOI] [PubMed] [Google Scholar]
- 52. Luo W, Phung D, Tran T, Gupta S, Rana S, Karmakar C, Shilton A, Yearwood J, Dimitrova N, Ho TB, Venkatesh S, Berk M. Guidelines for developing and reporting machine learning predictive models in biomedical research: a multidisciplinary view. J Med Internet Res 2016;18:e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analysed in support of this research.