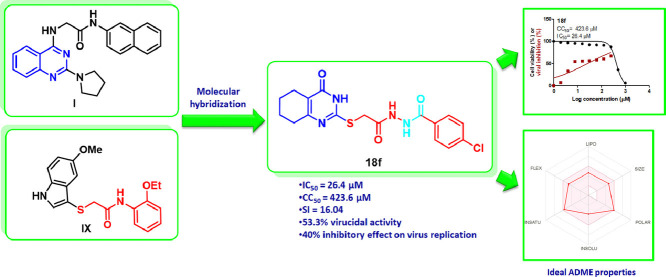
Abstract
In the current investigation, two novel series of (tetrahydro)thioquinazoline-N-arylacetamides and (tetrahydro)thioquinazoline-N-arylacetohydrazides were designed, synthesized and investigated for their antiviral activity against SARS-CoV-2. The thioquinazoline-N-arylacetamide 17g as well as the tetrahydrothioquinazoline-N-arylacetohydrazides 18c and 18f showed potent antiviral activity with IC50 of 21.4, 38.45 and 26.4 µM, respectively. In addition, 18c and 18f demonstrated potential selectivity toward the SARS-CoV-2 over the host cells with SI of 10.67 and 16.04, respectively. Further evaluation of the mechanism of action of the three derivatives 17g, 18c, and 18f displayed that they can inhibit the virus at the adsorption as well as at the replication stages, in addition to their virucidal properties. In addition, 17g, 18c, and 18f demonstrated satisfactory physicochemical properties as well as drug-likeness properties to be further optimized for the discovery of novel antiviral agents. The docking simulation on Mpro binding site predicted the binding pattern of the target compounds rationalizing their differential activity based on their hydrophobic interaction and fitting in the hydrophobic S2 subsite of the binding site
Keywords: Thioquinazoline, SARS-CoV-2, Molecular docking studies
Graphical abstract
1. Introduction
Recently, coronavirus disease 2019 (COVID-19) has been identified as a global pandemic disease that affects the survival of population all over the world [1]. COVID-19 is a respiratory disease that causes upper and lower respiratory tract infection which can be further progressed further into respiratory failure by complex mechanisms and may end up with premature mortality [1,2]. It was reported that COVID-19 is caused by a novel zoonotic member of betacoronaviruses called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) which is a single stranded RNA virus of the Coronaviridae family [3,4].
The huge problem, which is currently facing the world, is the remarkable ability of SARS-CoV-2 to mutate over a short period of time [5]. Hence, a plethora of studies have been performed to get some insightful knowledge about SARS-CoV-2 virus [6]. Successfully, some information has been reported including its molecular structure, life cycle, and its interactions with the host cells. This enabled the development various vaccines with different mechanisms of action to be used by humans [7].
Up till now, the effective and specific antiviral agents for the treatment of SARS-CoV-2 infection are limited or rare [8]. Hence, numerous investigations have been carried out to identify new targets to control this pathogen without affecting the host cells. Recently, some structural elements that can act as potential therapeutic targets have been recognized. Spike glycoprotein was identified as a promising target that is present on the virus surface, and it is responsible for the virus binding to the host cell [9]. RNA-dependent RNA polymerase (RdRp) is another attractive target participating in the replication of RNA from an RNA template [10]. Meanwhile, the 3-C-like protease (3CLpro or Mpro) and papain-like protease (PLpro) were pointed out to be the most important targets for the design of promising antiviral agents against SARS-CoV-2 as they play a key role in the life cycle of SARS-CoV-2 virus with no homologues proteins in human cells [11], [12], [13], [14]. Hence, inhibition of Mpro and/or PLpro can result in a selective antiviral activity without side effects on humans [12].
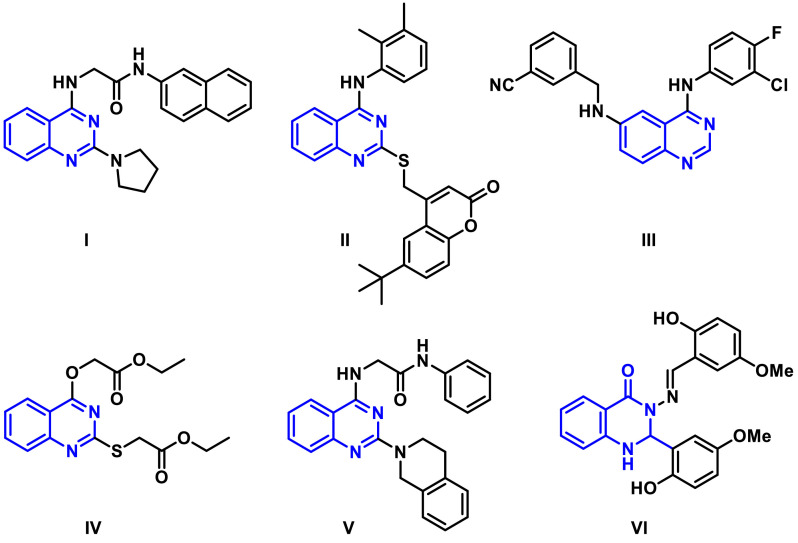
In this context, quinazolines are regarded as one of the most interesting scaffolds for designing antiviral agents [15,16]. Quinazolines were reported to possess potential antiviral activity against diverse types of RNA viruses including respiratory syncytial virus (RSV) [17], influenza A virus (IAV) [18] as well as hepatitis C virus (HCV) [19]. For instance, Zhang and co-workers [18] reported the design of a series of 2,4-disubstituted quinazoline derivatives incorporating S-acetamide and NH-acetamide moieties at position 4. Compound I, a representative of this series, was found to have a potent antiviral activity against IAV. Moreover, Hwu et al., [19] described the potent antiviral activity of a class of quinazoline-coumarin conjugates, for example compound II showed potent antiviral activity against hepatitis C virus and chikungunya virus. Meanwhile, Lee et al. [20] reported the synthesis of a series of 4-anilino-6-aminoquinazolines as anti-MERS-CoV inhibitors, compound III, a representative example of the synthesized series, showed IC50 = 0.157 µM, CC50 = 3.59 µM and SI = 25. Recently, some studies reported the activity of quinazoline derivatives against SARS‑CoV‑2 [21,22]. For example, Zhao et al. [21] reported the promising activity of a series of quinolone and quinazoline derivatives in inhibiting RNA synthesis driven by SARS-CoV-2 RdRp. For instance, compounds IV and V revealed 58.43% and 58.83% inhibition on SARS-CoV-2 RdRp at 10 µM concentration. Additionally, Rothan and Teoh [22] reported the expected interesting potential of quinazoline derivatives, for example compound VI, in inhibiting SARS‑CoV‑2 Main Protease (Mpro) in a high throughput virtual screening campaign (Fig. 1 ).
Fig. 1.
Examples of antiviral agents I-VI incorporating quinazoline scaffold.
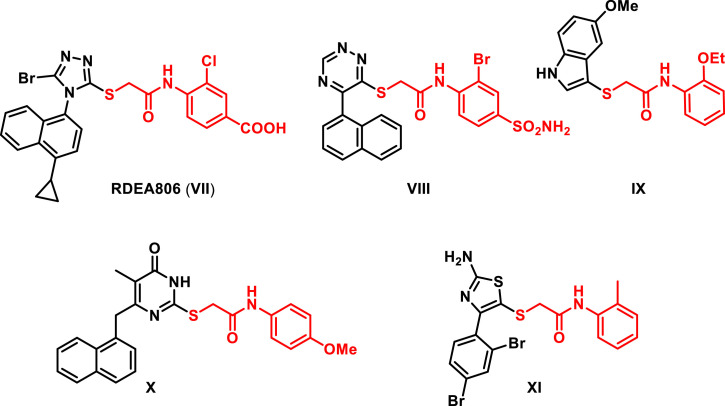
On the other hand, several studies reported the promising antiviral properties of various heterocycles substituted with N-aryl-2-(thio)acetamide moieties. For instance, RDEA806 (VII) was reported to possess a potential HIV-1 reverse transcriptase (RT) inhibitory activity against wild-type (WT) as well as some non-nucleoside reverse transcriptase (NNRT) resistant viruses [23]. Moreover, Zhan and co-workers [24] reported the synthesis and the interesting antiviral properties of novel 1,2,4-triazin-6-yl-thioacetamide derivatives as potent HIV-1 NNRTIs. Compound VIII was an example of this series displaying nanomolar activity against HIV-1(WT) as well as a moderate potency against the double mutant strain RES056. Furthermore, Zhang et al. [25] reported the discovery of some indol-3-yl-thio-N-phenyl-acetamides with potent antiviral activity. For instance, compound IX revealed a dual potency against RSV and IAV [25]. Moreover, Yu et al. [26] reported the potent anti-influenza activity of different pyrimidines substituted at 2 position with a N-aryl-2-(thio)acetamide moiety. For example, compound X demonstrated a broad activity against IAV and IBV. In addition, Zhan and co-workers [27] reported the anti-influenza properties of some thiazolyl-N-aryl-2-(thio)acetamides, for example, the derivative XI displayed a potent activity on influenza A/H1N1 virus (Fig. 2 ).
Fig. 2.
Structures of antiviral agents VII-XI incorporating N-aryl-2-(thio)acetamide moiety.
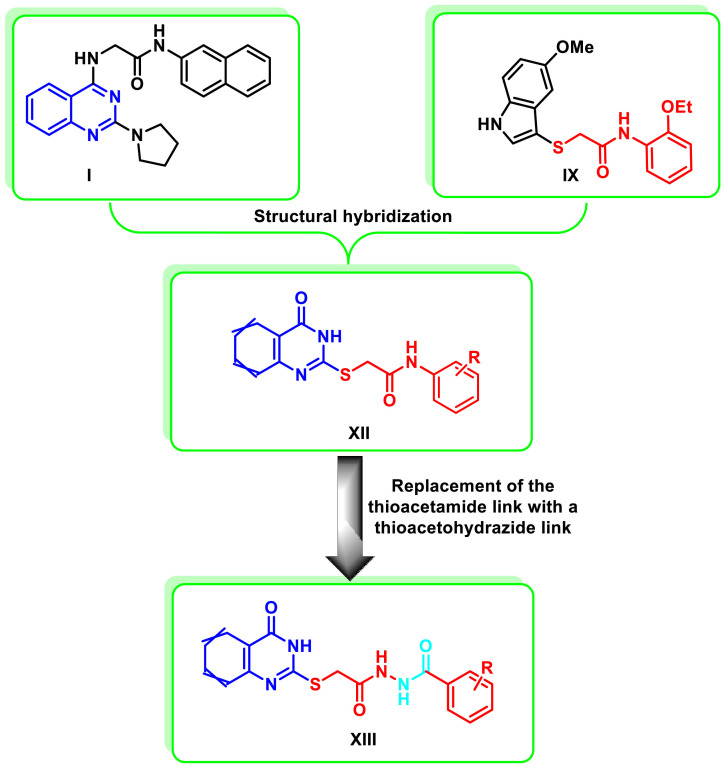
Rationale design of thioquinazoline-N-aryl-acetamide/N-arylacetohydrazide hybrids
Encouraged by the previous findings, and since most of the antiviral drugs in the clinical use were not designed specifically for SARS-CoV-2, we were interested in the current study in designing a new series of anti-SARS-CoV-2 through the application of the molecular hybridization strategy between the quinazoline ring and the N-aryl-2-(thio)acetamide moiety to design a novel series of (tetrahydro)thioquinazoline-N-arylacetamide hybrids XII (Fig. 3 ). For further structure-activity relationship investigation, further elongation of the N-aryl-2-(thio)acetamido linkage was carried out by its replacement with a 2-(thio)acetohydrazide linker to afford scaffold XIII (Fig. 3). Derivatives from the designed scaffolds were synthesized utilizing the conventional methods of organic chemistry. Subsequently, the synthesized compounds were assayed for their antiviral activity against SARS-CoV-2. Promising hits were further examined for their expected mode of antiviral activity. Additionally, their physicochemical as well as pharmacokinetic properties were predicted using the SwissADME free webtool. Concurrently, in silico molecular docking studies were performed in the SARS‑CoV‑2 Main Protease (Mpro) binding site to study their binding mode in order to rationalize their promising antiviral activity.
Fig. 3.
The design strategy of the target (tetrahydro)thioquinazoline-N-arylacetamides XII and (tetrahydro)thioquinazoline-N-arylacetohydarzides XIII as anti-SARS-CoV-2 agents.
2. Results and discussion
2.1. Chemistry
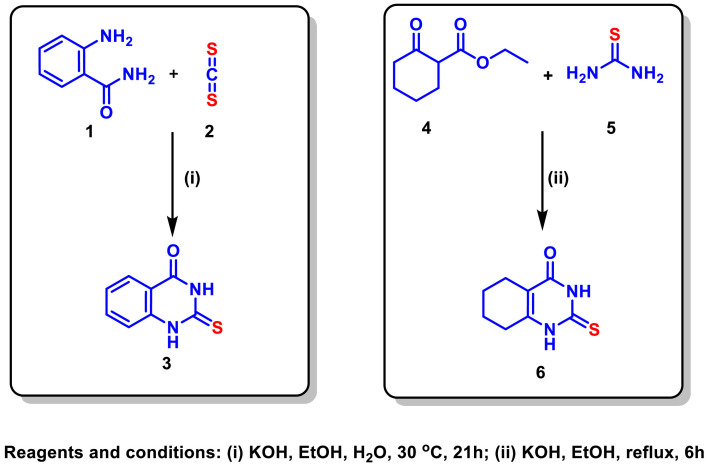
Initially, the starting quinazoline 3 was synthesized by the reaction of 1 with CS2 (2) in the presence of KOH at 30 oC [28,29]. Meanwhile, the starting material 6 was synthesized by the condensation of 4 with thiourea (5) under basic conditions [30,31] (Scheme 1 ).
Scheme 1.
Synthesis of the starting quinazolines 3 and 6.
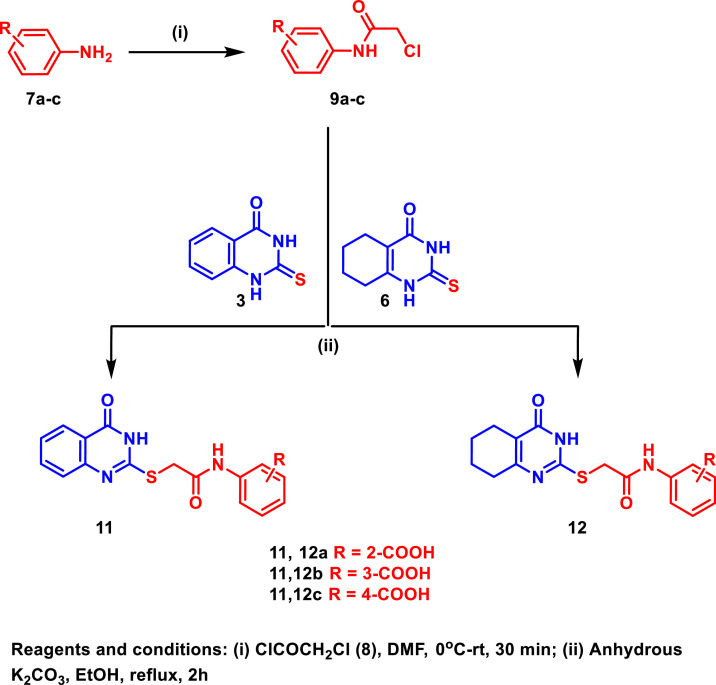
For the synthesis of the target compounds 11a-c and 12a-c; 2-aminobenzoic acid (7a), 3-aminobenzoic acid (7b), and 4-aminobenozic acid (7c) were reacted with chloroacetyl chloride (8) in DMF to afford the intermediates 9a-c, respectively, which were subsequently reacted with the starting materials 3 and 6 under basic conditions to afford the target compounds 11a-c and 12a-c, respectively, in excellent yields (Scheme 2 ).
Scheme 2.
Synthesis of the target compounds 11 and 12.
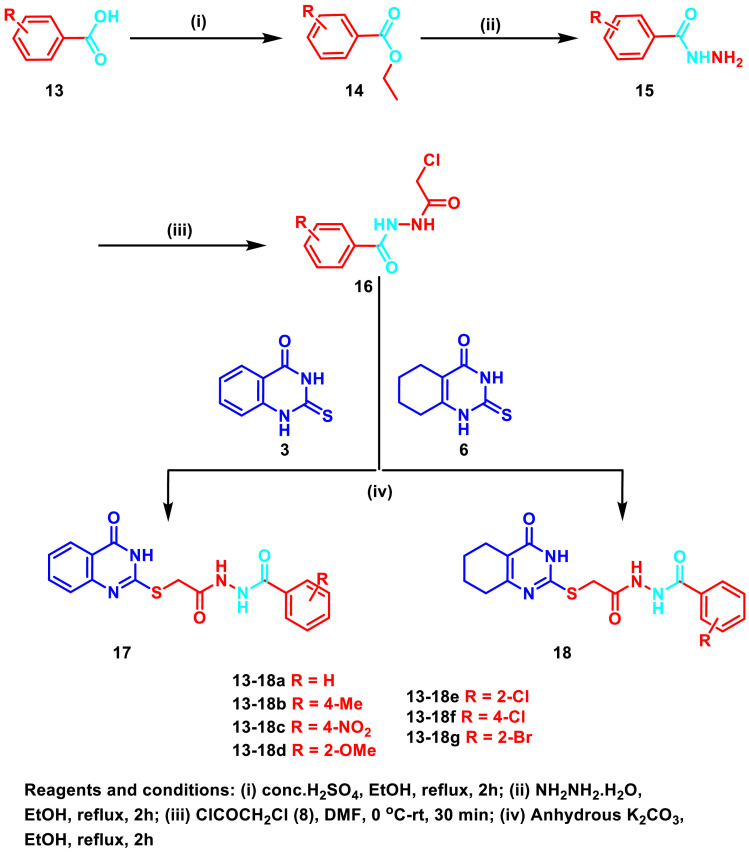
For the synthesis of the target quinazolines 17 and 18, different benzoic acid derivatives 13 were first esterified to give the corresponding esters 14 which were subsequently reacted with hydrazine hydrate to afford the corresponding acid hydrazides 15 (Scheme 3 ). Further reaction of 15 with chloroacetyl chloride (8) was performed to give the intermediates 16 which were reacted with the thioquinazolines 3 and 6 to yield the target compounds 17 and 18, respectively, in excellent yields.
Scheme 3.
Synthesis of the target compounds 17 and 18.
2.2. Biological evaluation
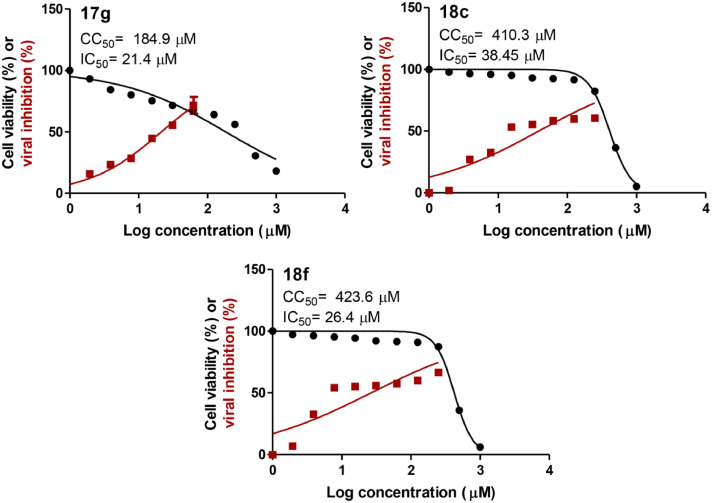
All the target compounds 11a-c, 12a-c, 17a-g and 18a-g were evaluated for their antiviral activity against NRC-03-nhCoV as well as for their cytotoxic activity on Vero-E6 cells employing MTT assay [32]. The CC50 (concentration necessary for 50% growth inhibition of normal cell line compared to the control experiment) of the target compounds on Vero-E6 cells and IC50 (concentration necessary for 50% reduction of virus-induced cytopathic effect (CPE) compared to the virus control experiment) of the target compounds against NRC-03-nhCoV virus in Vero-E6 cells are presented in Table 1 . The selectivity indices which are the ratio of CC50 relative to IC50 of the tested compounds were calculated and depicted in Table 1. In addition, Fig. 4 presents the inhibition curves of the most potent compounds 17g, 18c, and 18f (For the rest of the inhibition curves see the SI).
Table 1.
IC50 against NRC-03-nhCoV, CC50 on Vero-E6, and the selectivity index (SI) [CC50/IC50] of the target compounds.
| Compound ID | R | IC50a (μM) | CC50b (μM) | SIc |
|---|---|---|---|---|
| 11a | 2-COOH | 104 | 450.5 | 4.3 |
| 11b | 3-COOH | 79.7 | 394 | 4.9 |
| 11c | 4-COOH | 79.2 | 442.6 | 5.6 |
| 12a | 2-COOH | 173.6 | 431.4 | 2.5 |
| 12b | 3-COOH | 531.3 | 524.3 | 0.98 |
| 12c | 4-COOH | 530.4 | 333.5 | 0.63 |
| 17a | H | 317.5 | 186.4 | 0.59 |
| 17b | 4-Me | 126.7 | 271.4 | 2.14 |
| 17c | 4-NO2 | 121.1 | 316.1 | 2.61 |
| 17d | 2-OMe | 258.8 | 101.2 | 0.4 |
| 17e | 2-Cl | 457.8 | 114.2 | 0.24 |
| 17f | 4-Cl | 74.3 | 319 | 4.3 |
| 17g | 2-Br | 21.4 | 184.9 | 8.64 |
| 18a | H | 140.2 | 235 | 1.68 |
| 18b | 4-Me | 113.7 | 503.8 | 4.43 |
| 18c | 4-NO2 | 38.45 | 410.3 | 10.67 |
| 18d | 2-OMe | 107 | 315.5 | 2.95 |
| 18e | 2-Cl | 459.8 | 524.5 | 1.14 |
| 18f | 4-Cl | 26.4 | 423.6 | 16.04 |
| 18g | 2-Br | 93.4 | 488.8 | 5.23 |
aIC50 (half maximal inhibitory concentration); bCC50 (half maximal cytotoxic concentration); cSI (Selectivity index).
Fig. 4.
Dose-inhibition curves of 17g, 18c, and 18f against NRC-03-nhCoV [33] and Vero-E6 cells; IC50 and CC50 values were calculated using nonlinear regression analysis of GraphPad Prism software (version 5.01) by plotting log inhibitor versus normalized response (variable slope).
The presented study showed promising activity in comparison to the results reported in the literature [20,21]. From the IC50 results presented in Table 1, it is obvious that the synthesized thioquinazolines and tetrahydrothioquinazolines displayed moderate to potent antiviral activity against NRC-03-nhCoV. Compounds 17g, 18c and 18f showed the most potent inhibitory activity with IC50 of 21.4, 38.45 and 26.4 µM, respectively, on NRC-03-nhCoV. Close investigation of the antiviral results revealed that the thioquinazoline-N-aryl-acetamide hybrids 11a-c demonstrated moderate antiviral properties with an IC50 range of 79.2 to 104 µM. Compounds 11b and 11c incorporating 3 and 4 carboxylic groups, respectively, showed higher potencies in comparison to the 2-carboxylic acid derivative 11a with IC50 of 79.7, 79.2, and 104 µM, respectively, as well as higher SIs. Replacing the thioquinazoline moiety in series 11a-c with a tertahydrothioquinazoline moiety in series 12a-c decreased the antiviral activity (IC50 range of 173.6 to 531.3 µM). Compound 12a, with a carboxylic acid moiety at the 2 position, showed an IC50 of 173.6 µM and a 2.5-fold higher selectivity toward the virus over the host cells (CC50 = 431.4 µM). Shifting the COOH moiety to the 3 and 4 positions in 12b and 12c, respectively, resulted in more than a 2-fold decrease in the antiviral potency with no preferential selectivity (SI = 0.98 and 0.63, respectively).
Replacing the thio-N-aryl-acetamide moiety of series 11a-c with thioacetohydrazide moiety in series 17a-g, resulted in a different pattern of potency as well as specificity. The thioquinazoline-N-aryl-acetohydrazide derivative 17a with an unsubstituted terminal phenyl moiety displayed a weak antiviral activity (IC50 = 317.5 µM) and high cytotoxic activity (CC50 = 186.4 µM, SI = 0.59). Introduction of 4-Me and 4-NO2 groups at the terminal phenyl in 17b and 17c increased the antiviral properties with IC50 of 126.7 and 121.1 µM, respectively and SI of 2.14 and 2.61 µM, respectively. Moreover, the 2-OMe 17d and 2-chloro 17e derivatives demonstrated a decrease in the antiviral activity with IC50 of 258.8 and 457.8 µM, respectively, with concomitant high cytotoxic (CC50 = 101.2 and 114.2 µM, respectivley). Introduction of 4-chloro group at the terminal phenyl group in 17f resulted in a more than 4-fold increase in the antiviral activity (IC50 = 74.3 µM) in comparison to 17a (IC50 = 317.5 µM) with reduced cytotoxic activity (CC50 = 319 µM and SI = 4.30). Furthermore, the 2-bromo congener 17g demonstrated the highest antiviral activity (IC50 = 21.4 µM) with 8.64 times higher selectivity towards the virus over the host cells (CC50 =184.9 µM).
Hydrogenation of the fused phenyl group of the thioquinazoline ring in series 17a-g to afford the tetrahydrothioquinazoline series 18a-g increased the antiviral activity in all cases with the exception of compounds 18e and 18g. Compound 18a with unsubstituted phenyl group displayed a moderate antiviral activity (IC50 = 140.2 µM) with a low SI of 1.68 (CC50 = 235 µM). Introduction of 4-NO2 and 4-Cl groups at the terminal phenyl moiety to yield 18c and 18f, respectively, resulted in a more than three-fold increase in potency in comparison to 18a with IC50 of 38.45 and 26.4 µM, respectively, in addition to a more than 10-fold higher selectivity toward the virus over the host cell (SI = 10.67 and 16.04, respectively). On the other hand, introduction of 4-Me, 2-OMe, 2-Br groups at the terminal phenyl moiety to give 18b, 18d, and 18g showed a decrease in the antiviral activity with IC50 = 93.40 to 113.7 µM. Slight decrease in the antiviral activity was found for 18e (IC50 = 459.8 µM) in comparison to 17e (IC50 = 457.8 µM) and more than four-fold reduction was obsereved for 2-bromo derivative 18g (IC50 = 93.40 µM) in copmarison to 17g (IC50 = 21.4 µM).
In summary, although compound 17g displayed the highest antiviral activity with IC50 of 21.4 µM, it was less favored because of its cytotoxic properties (CC50 = 184.9 µM and SI = 8.64). On the contrary, 18f and 18c were regarded to be the most promising derivatives in terms of their potency (IC50 = 26.4 and 38.45 µM) as well as their selectively and safety (SI = 16.04 and 10.67).
2.3. Mechanism of Anti-SARS-CoV-2 Activity
For further investigation of the mechanism of virus inhibition of the most promising derivatives 17g, 18c and 18f, a plaque infectivity reduction assay was performed [34]. This assay studied whether the promising derivatives affected the virus at the adsorption stage and/or replication stage and/or due to their direct virucidal effect. The inhibition results are depicted in Table 2 . Interestingly, the three derivatives were found to have multiple inhibitory effects to different extents in the three stages. Nevertheless, the replication stage was found to be the most affected following treatment with the tested compounds (40-50% viral inhibition at 125 µM and 38.9-40% viral inhibition at 62.5 µM).
Table 2.
Mechanisms of action of 17g, 18c and 18f against SARS-CoV-2.
| Compound ID |
Conc (µM) |
Mode of action Virus inhibition% |
||
|---|---|---|---|---|
| Adsorption | Replication | Virucidal | ||
|
17g |
125 | 29.6 ± 8.3 | 50.0 ± 3.3 | 47.8 ± 5.1 |
| 62.5 | 26.7 ± 6.65 | 45.1 ± 1.71 | 37.3 ± 6.43 | |
| 31.25 | 16.7 ± 3.35 | 38.2 ± 4.29 | 25.6 ± 5.1 | |
|
18c |
125 | 44.4 ± 3.87 | 43.3 ± 3.35 | 38.9 ± 2.31 |
| 62.5 | 28.9 ± 10.18 | 38.9 ± 1.91 | 33.6 ± 6.03 | |
| 31.25 | 20.0 ± 3.3 | 30.0 ± 3.3 | 22.2 ± 5.1 | |
| 18f | 125 | 20.0 ± 3.3 | 40.0 ± 10 | 53.3 ± 3.35 |
| 62.5 | 13.3 ± 3.35 | 40.0 ± 3.3 | 34.4 ± 5.1 | |
| 31.25 | 6.7 ± 6.65 | 31.6 ± 4.27 | 15.6 ± 13.89 | |
As can be seen in Table 2, the thioquinazoline-N-aryl acetamide derivative 17g showed at 125 µM a moderate inhibition of the virus at the adsorption stage 29.6%, whereas it showed a potent inhibition at the replication stage (50%) as well as a potent virucidal effect (47.8%). The tetrahydrothioquinazoline-N-arylacetohydrazide 18c displayed moderate inhibitory activity on the three tested mechanisms. At 125 µM, it revealed inhibition% of 44.4 and 43.3% by applying adsorption and replication mechanisms, respectively, and a 38.9% virucidal effect was noticed. Moreover, at 125 µM, compound 18f was found to have 53% virucidal activity as well as a 40% inhibitory effect on virus replication and only 20% inhibition of the viral adsorption mechanism.
2.4. Molecular modeling
Protease enzyme plays a critical role in viral protein maturation by cleaning proproteins after their translation into the host cell cytosol. As a result, viral proteases are considered potential antiviral drug targets [35]. The inhibition of a viral protease can reduce the assembly of mature viral particles. Therefore, SARS-CoV-2 main protease (Mpro) could be a plausible target for the newly synthesized compounds, especially with its reported pyrimidinedione inhibitors which are analogues to our designed antiviral derivatives [36], thus, molecular docking simulations have been carried out to study the binding pattern of the target compounds 11a-c, 12a-c, 17a-g, and 18a-g in the active site of SARS-CoV-2 main protease (Mpro). For this end, the X-ray crystallographic structure of SARS-CoV-2 main protease (Mpro) co-crystalized with a pyrimidine-2,4-dione inhibitor (YD1) (PDB ID: 7LTJ) was retrieved from the protein data bank (https://www.rcsb.org/) [36]. The molecular docking protocol was first validated by self-docking of the co-crystallized ligand (YD1) in the vicinity of the enzyme active site. The self-docking step reproduced the co-crystalized ligand pose efficiently with a docking score (S) of −13.26 kcal/mol and a root mean square deviation (RMSD) of 1.538 Å. Moreover, the docking protocol reproduced all the key interactions with the active site amino acids (Figs. 5 and 6 ). Using the validated molecular docking protocol, the target compounds 11a-c, 12a-c, 17a-g, and 18a-g were docked in the SARS-CoV-2 Mpro active site.
Fig. 5.
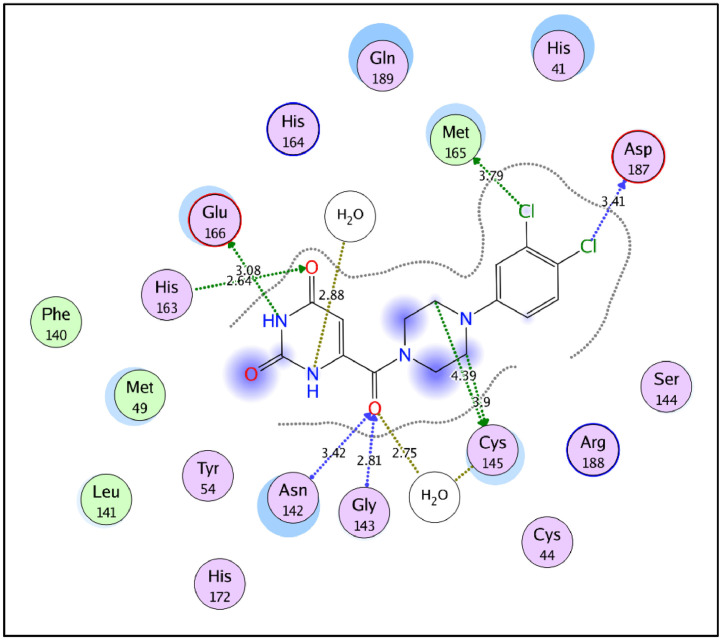
2D interaction diagram of the co-crystalized inhibitor YD1 in Mpro active site (PDB ID: 7LTJ).
Fig. 6.
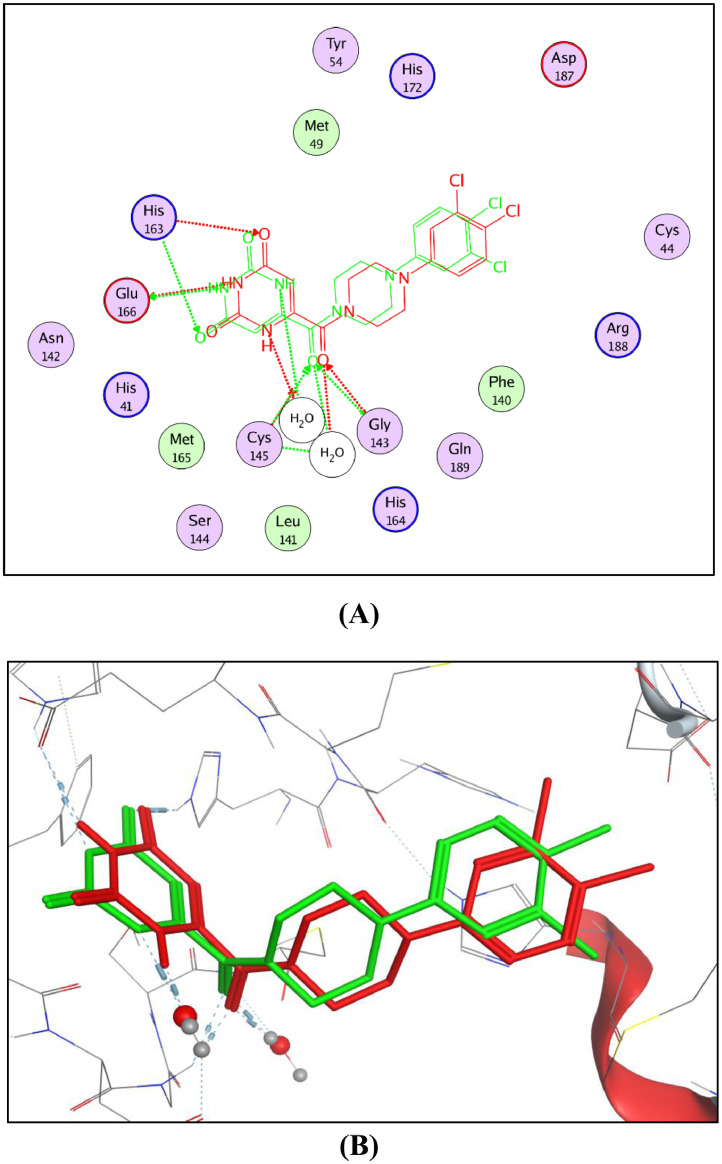
2D diagram (A) and 3D representation (B) of the superimposition of the co-crystallized (red) and the docking pose (green) of YD1 in the active site of Mpro (PDB ID: 7LTJ).
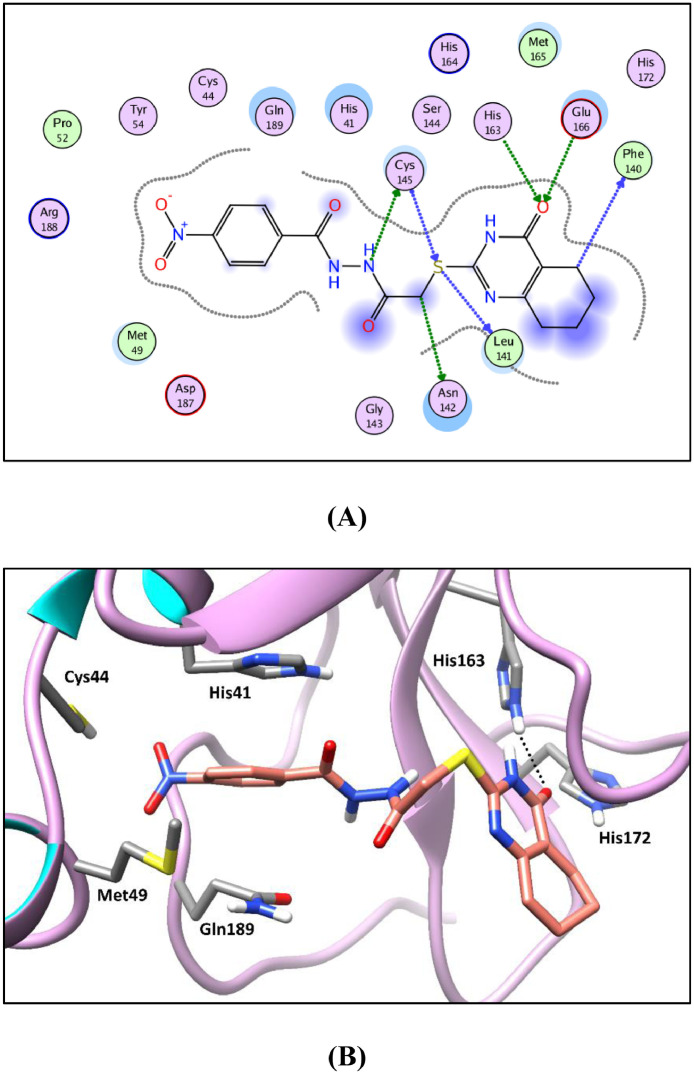
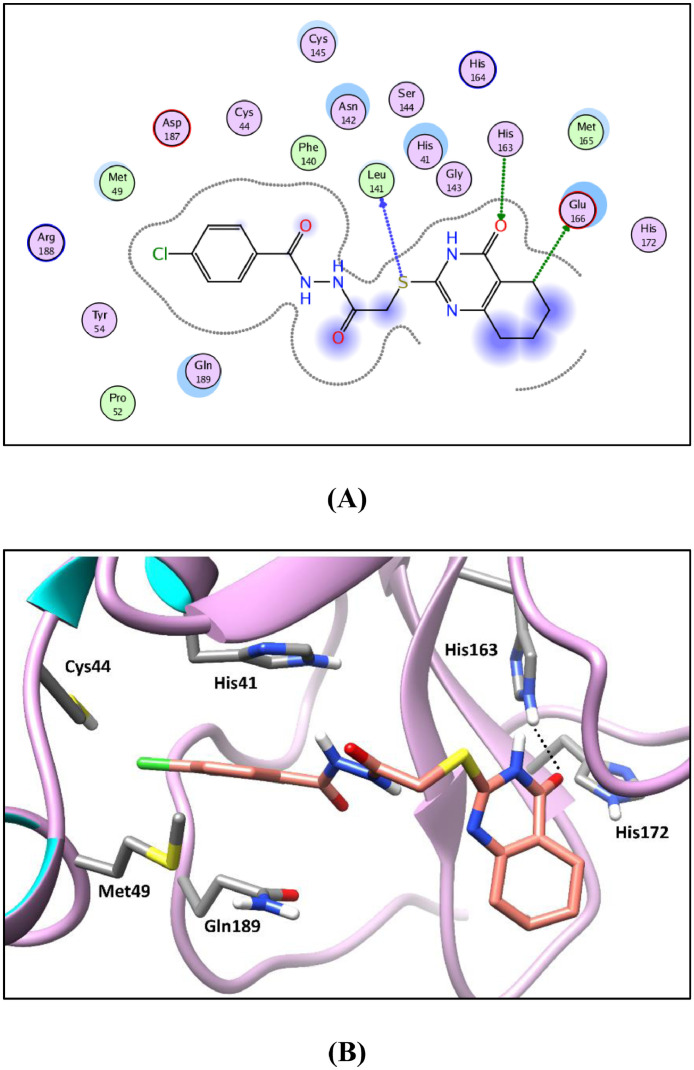
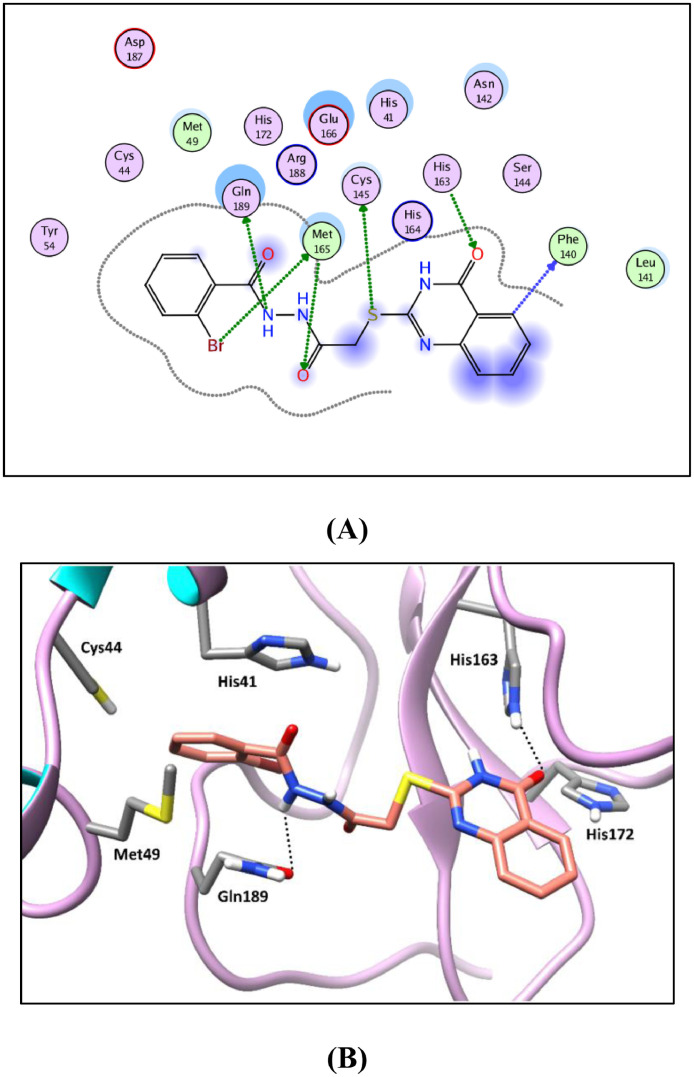
Generally, the target compounds showed a common binding pattern in the target enzyme (Mpro) active site accommodated in subsites S1 and S2 (Fig. 7, Fig. 5, Fig. 6 ). The (tetrahydro)quinazolinone moiety occupies YD1 uracil binding subsite S1 driven by polar contacts interacting by its carbonyl group with the key amino acid His163 through hydrogen bonding. On the other side, the substituted phenyl group occupies the largely hydrophobic S2 subsite occupied by YD1 dichlorophenyl moiety. The target compounds’ substituted phenyl group is stabilized through π-π stacking with the imidazole side chain of the key catalytic amino acid His41. The substituted phenyl group is sandwiched between the side chains of the amino acids His41 and Gln189. The linker in-between interacts through multiple H-bond interactions with the surrounding key amino acids Asn142, Gly143, Cys145, and His164. In compounds 11a-c and 12a-c, the peripheral carboxylate moiety on the distal phenyl group is involved in extra hydrogen bond interactions with the amino acids His41, Cys44, Met49, Pro52, and/or Met165 (For further details see supporting materials).
Fig. 8.
2D diagram (A) and 3D representation (B) of compound 18c showing its interaction in Mpro active site (PDB ID: 7LTJ).
Fig. 9.
2D diagram (A) and 3D representation (B) of compound 18f showing its interaction in Mpro active site (PDB ID: 7LTJ).
Fig. 7.
2D diagram (A) and 3D representation (B) of compound 17g showing its interaction in Mpro active site (PDB ID: 7LTJ).
Table 3 shows the docking score of the target compounds and the co-crystalized ligand (YD1). The newly synthesized compounds show a predicted docking score range of −13.99 to −11.73 kcal/mol, whereas the co-crystalized ligand YD1 showed a predicted docking score of −13.26 kcal/mol. As for the most promising compounds 17g, 18c, and 18f, compound 18c showed the most negative docking score −13.99 kcal/mol more negative than that of YD1. Whereas compounds 17g and 18f exhibited comparable docking score (−12.45 and −12.07 kcal/mol, respectively) which is less negative than that of the co-crystalized ligand (YD1) (−13.26 kcal/mol) indicating their less predicted binding affinity than YD1. Compounds 17d, 18a, 17a, and 17e showed the least negative predicted docking scores (−11.54, −11.47, −11.38, and −11.37 kcal/mol) which agree with their poor experimental activity (258.8, 140.2, 317.5, and 457.8 μM).
Table 3.
Docking energy scores (S) in kcal/mol for the target compounds and the co-crystalized compound (YD1) in Mpro active site (PDB ID: 7LTJ).
| Compound | Energy score (S) kcal/mol | IC50 (μM) |
|---|---|---|
| 11a | −11.60 | 104 |
| 11b | −11.58 | 79.7 |
| 11c | −11.92 | 79.2 |
| 12a | −11.98 | 173.6 |
| 12b | −11.96 | 531.3 |
| 12c | −12.39 | 530.4 |
| 17a | −11.38 | 317.5 |
| 17b | −12.16 | 126.7 |
| 17c | −12.99 | 121.1 |
| 17d | −11.54 | 258.8 |
| 17e | −11.37 | 457.8 |
| 17f | −11.98 | 74.3 |
| 17g | −12.45 | 21.4 |
| 18a | −11.47 | 140.2 |
| 18b | −13.36 | 113.7 |
| 18c | −13.99 | 38.45 |
| 18d | −11.97 | 107 |
| 18e | −11.78 | 459.8 |
| 18f | −12.07 | 26.4 |
| 18g | −13.18 | 93.4 |
| YD1 | −13.26 | 4.2 [36] |
The predicted binding pattern of the target compounds could rationalize their differential activity based on their hydrophobic interaction and fitting in the hydrophobic S2 subsite of the binding site. Compounds 11a-c and 12a-c showed a relatively less predicted binding affinity due to their ionic polar carboxylate substitution on their distal phenyl group which decreases the probable hydrophobic interactions with the hydrophobic S2 subsite, however, this loss of proper hydrophobic interactions is somehow compensated by the carboxylate involvement in multiple extra hydrogen bond interactions with the amino acids His41, Cys44, Met49, Pro52, and/or Met165 like in case of compound 11b. On the other hand, compounds 17a-g and 18a-g show better predicted binding affinity because of their hydrophobic substitutions on the distal phenyl group and the longer thioacetohydrazide linker which makes the (substituted)phenyl group better fitted in the hydrophobic S2 subsite. Compounds achieving higher hydrophobic interaction with S2 subsite and better fit of their (substituted)phenyl group show strong predicted binding affinity as reflected in their promising experimental activity e.g., compounds 17g, 18c, and 18f with o-bromo, p-nitro, and p-chloro substitution, respectively. Alternatively, compounds with unsubstituted distal phenyl ring 17a and 18a show less predicted binding affinity relative to their substituted congeners due to their less possible hydrophobic interactions with S2 subsite. Compounds with less hydrophobic substituents (OMe) e.g., compounds 17d or that are not well fitted in the S2 subsite e.g., compounds 17e and 18e show weaker predicted binding affinity which is reflected in their weak experimental activity (IC50 = 258.8, 457.8, and 459.8 μM).
2.5. Estimation of physicochemical, pharmacokinetic and ADME properties
Encouraged by the promising antiviral properties of 17g, 18c and 18f, they were further selected to predict their physicochemical and ADME properties using SwissADME free web tool [37]. Table 4 presents some selected results of 17g, 18c and 18g.
Table 4.
Selected calculated physicochemical and pharmacokinetic properties of 17g, 18c and 18f from SwissADME free webtool [37].
| Product | 17g | 18c | 18f |
|---|---|---|---|
| MW | 433.28 | 403.41 | 392.86 |
| Rotatable bonds | 7 | 8 | 7 |
| H-bond acceptors | 4 | 6 | 4 |
| H-bond donors | 3 | 3 | 3 |
| MR | 102.11 | 103.16 | 99.35 |
| TPSA | 129.25 | 175.07 | 129.25 |
| iLogP | 1.77 | 1.60 | 2.62 |
| GI absorption | High | Low | High |
| BBB permeant | No | No | No |
| P-gp substrate | No | Yes | Yes |
Analysis of the obtained results (Table 4) revealed that the target thioquinazoline-N-aryl acetamide 17g and tetrahydrothioquinazoline-N-aryl acetohydrazides 18c and 18f express acceptable levels of physicochemical properties. The molecular weight of 17g, 18c and 18f is spanning between 392.86 to 433.28 g/mol. They incorporate acceptable numbers of hydrogen bond donors (less than 5) and acceptors (less than 10). Additionally, the topological polar surface area (TPSA) for 17g and 18f is 129.25 Å, while it is slightly high in case of the 18c TPSA = 175.07 Å and the ilogP (octanol–water partition coefficient) is ranging from 1.60 to 2.62 [38].
The target (tetrahydro)thioquinazoline-N-arylacetamides 17g and 18f are predicted to be well absorbed from the GIT while 18c is predicted to have low GIT absorption. The three target compounds 17g, 18c and 18f have no predicted ability to penetrate the blood brain barrier which decreases their probable adverse effect at the central level. Compounds 18c and 18f were found to be substrates for P-glycoprotein (P-gp), which plays a significant role in the removal of strange substances outside the cells. Meanwhile, 17g is not a substrate for P-glycoprotein (P-gp) [39].
Fig. 10 presents the bioavailability radar chart of compounds 17g, 18c and 18f provided by the SwissADME web tool [37]. Generally, the bioavailability radar presented by SwissADME demonstrates a pink-colored area that identifies the optimum space of six physicochemical parameters for oral bioavailability. These six properties are size, polarity, lipophilicity, solubility, flexibility, and saturation. Close investigation of the presented radar charts showed that compound 18f occupies the ideal space of the six physicochemical properties for oral bioavailability, whereas the derivatives 17g and 18c are nearly fully located in the pink area, only the degree of unsaturation slightly deviates from the optimum in 17g while the polarity is slightly deviating from the ideal for 18c (Fig. 10).
Fig. 10.
Bioavailability radar plot from SwissADME online web tool for 17g, 18c and 18f.
Moreover, it is attractive that 17g and 18f follow all rules of drug-likeness, they do not violate Lipinski's rule [40], Veber rule [41], Ghose-filter [42], Egan [43] or Muegge´s filter [44]. Because of the high TPSA of compound 18c, it satisfies only Lipinski's rule. In addition, it worth pointing out that the three derivatives do not incorporate in their structures Pan Assay Interference (PAINS) fragments [45]. Hence, in addition to the promising antiviral activity of the target compounds, their promising drug-likeness parameters suggests their potential to be subjected for future optimization for the discovery of chemotherapeutic agents.
3. Conclusion
The presented study involves the design, synthesis, and anti-SARS-CoV-2 activity evaluation of two novel series of (tetrahydro)thioquinazoline-N-arylacetamides and (tetrahydro)thioquinazoline-N-arylacetohydrazides. The thioquinazoline-N-arylacetamide 17g beside the tetrahydrothioquinazoline-N-arylacetohydrazides 18c and 18f showed potent antiviral activity with IC50 = 21.4, 38.45, and 26.4 µM, respectively. The derivatives 18c and 18f exhibited SI = 10.67 and 16.04, respectively, towards the virus over the host cells. In addition, the three derivatives 17g, 18c, and 18f showed promising virucidal properties beside their ability to inhibit the virus at the adsorption as well as at the replication stages. Moreover, besides their promising antiviral activity, compounds 17g, 18c and 18f displayed acceptable physicochemical and pharmacokinetic properties for further optimization as antiviral agents.
4. Experimental
4.1. Chemistry
4.1.1. General remarks
Chemicals along with solvents used for chemical reaction were obtained from commercial companies. Follow up of the reactions were carried out using analytical thin layer chromatography (TLC). Uncorrected melting points were recorded on a Stuart SMP30 melting point apparatus. Elemental analyses of the synthesized hybrids were recorded in the micro analytical labs, National Research Centre, Cairo, Egypt. IR spectra (4000–400 cm−1) were recorded on Jasco FT/IR 300 E Fourier transform infrared spectrophotometer. 1H NMR as well as 13C NMR spectra were measured in DMSO-d 6 as a solvent at 500 (125) MHz and 400 (100) MHz on Bruker instruments.
4.1.2. General procedure I for the synthesis of the target compounds 11a-c, 12a-c, 17a-g and 18a-g
A mixture of 3 or 6 and anhydrous K2CO3 was stirred for 30 min at room temperature then 3a-c or 16a-g was added and the mixture was heated under reflux for 2 h. The reaction mixture was then cooled to room temperature poured on ice and neutralized with few drops of 2N HCl and the precipitated product was filtered, dried and purified by crystallization from MeOH/ DCM 1:1 mixture to give the corresponding target products 11a-c, 12a-c, 17a-g, 18a-g in analytical pure form.
2-(2-((4-Oxo-3,4-dihydroquinazolin-2-yl)thio)acetamido)benzoic acid (11a)
Off white powder; yield = 95%; mp 250-252 °C; 1H-NMR (500 MHz; DMSO-d 6) δ H 4.15 (s, 2H), 7.13 (t, 3 J = 7.5 Hz, 1H), 7.38 (t, 3 J = 7.5 Hz, 1H), 7.46 (d, 3 J = 8.0 Hz, 1H), 7.56 (t, 3 J = 7.5 Hz, 1H), 7.69 (t, 3 J = 7.5 Hz, 1H), 7.95 (d, 3 J = 7.5 Hz, 1H), 8.00 (d, 3 J = 7.5 Hz, 1H), 8.49 (d, 3 J = 8.5 Hz, 1H), 11.77 (br., 1H), 12.47 (br., 1H), 12.76 ppm (br., 1H); 13C-NMR (125 MHz; DMSO-d 6) δ C 35.08, 115.85, 116.70, 119.94, 122.92, 124.32, 125.79, 126.02, 131.13, 133.97, 134.56, 140.47, 148.22, 154.62, 161.17, 166.65, 169.34 ppm; Anal. Calcd for C17H13N3O4S: C, 57.46; H, 3.69; N, 11.82. Found: C, 57.73; H, 3.93; N, 11.65.
3-(2-((4-Oxo-3,4-dihydroquinazolin-2-yl)thio)acetamido)benzoic acid (11b)
Off white powder; yield = 87%; mp 252-254 °C; 1H-NMR (400 MHz; DMSO-d 6) δ H 4.19 (s, 2H), 7.40 (d, 3 J = 7.2 Hz, 1H), 7.43 (t, 3 J = 9.2 Hz, 1H), 7.63 (d, 3 J = 7.6 Hz, 1H), 7.72 (dt, 3 J = 7.8 Hz, 4 J = 1.2 Hz, 1H), 7.80 (dd, 3 J = 8.6 Hz, 4 J = 1.2 Hz, 1H), 8.02 (d, 3 J = 8.0 Hz, 4 J = 1.2 Hz, 1H), 8.25 (s, 1H), 10.55 (s, 1H), 12.68 (br., 2H), 12.95 ppm (br., 1H); 13C-NMR (100 MHz; DMSO-d 6) δ C 35.13, 115.86, 116.21, 119.91, 123.27, 124.28, 125.76, 126.10, 129.12, 131.39, 134.67, 139.18, 148.21, 155.26, 161.14, 166.24, 167.11 ppm; Anal. Calcd for C17H13N3O4S: C, 57.46; H, 3.69; N, 11.82. Found: C, 57.15; H, 3.87; N, 12.03.
4-(2-((4-Oxo-3,4-dihydroquinazolin-2-yl)thio)acetamido)benzoic acid (11c)
Pale yellow powder; yield = 83%; mp 249-251 °C; 1H-NMR (400 MHz; DMSO-d 6) δ H 4.11 (s, 2H), 7.29-7.40 (m, 2H), 7.68-7.69 (m, 2H), 7.88-7.89 (m, 2H), 7.99-8.00 (m, 2H), 11.06 (br., 2H), 12.45 ppm (br., 1H); Anal. Calcd for C17H13N3O4S: C, 57.46; H, 3.69; N, 11.82. Found: C, 57.64; H, 3.90; N, 11.66.
2-(2-((4-Oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)thio)acetamido)benzoic acid (12a)
White powder; yield = 90%; mp 227-229 °C; 1H-NMR (500 MHz; DMSO-d 6) δ H 1.58-1.60 (m, 4H), 2.14-2.24 (m, 2H), 2.35-2.36 (m, 2H), 4.01 (s, 2H), 7.14-7.15 (m, 1H), 7.56-7.57 (m, 1H), 7.95-7.96 (m, 1H), 8.47 (d, 3 J = 6.5 Hz, 1H), 11.61 (s, 1H), 12.71 ppm (br., 2H); 13C-NMR (125 MHz; DMSO-d 6) δ C 21.35, 21.38, 21.67, 30.86, 34.97, 116.59, 116.67, 119.97, 122.88, 131.08, 133.96, 140.44, 155.90, 162.72, 166.71, 169.23 ppm; Anal. Calcd for C17H17N3O4S: C, 56.81; H, 4.77; N, 11.69. Found: C, 56.57; H, 4.45; N, 11.48.
3-(2-((4-Oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)thio)acetamido)benzoic acid (12b)
White powder; yield = 81%; mp 263-265 °C; 1H-NMR (400 MHz; DMSO-d 6) δ H 1.61-1.62 (m, 4H), 2.24-2.25 (m, 2H), 2.39-2.40 (m, 2H), 4.04 (s, 2H), 7.41 (t, 3 J = 8.0 Hz, 1H), 7.62 (d, 3 J = 7.6 Hz, 1H), 7.76 (d, 3 J = 8.0 Hz, 1H), 8.20 (s, 1H), 10.45 (s, 1H), 12.53 ppm (br., 1H); 13C-NMR (100 MHz; DMSO-d 6) δ C 21.40, 21.72, 30.83, 34.95, 116.53, 119.92, 123.14, 124.24, 129.03, 131.72, 139.08, 156.49, 159.42, 162.79, 166.28, 167.28 ppm; Anal. Calcd for C17H17N3O4S: C, 56.81; H, 4.77; N, 11.69. Found: C, 56.62; H, 4.95; N, 11.47.
4-(2-((4-Oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)thio)acetamido)benzoic acid (12c)
White powder; yield = 87%; mp 237-239 °C; 1H-NMR (500 MHz; DMSO-d 6) δ H 1.61-1.63 (m, 4H), 2.24-2.25 (m, 2H), 2.37-2.38 (m, 2H), 4.04 (s, 2H), 7.66 (d, 3 J = 8.0 Hz, 2H), 7.88 (d, 3 J = 8.0 Hz, 2H), 10.65 (s, 1H), 12.58 ppm (br., 2H); 13C-NMR (125 MHz; DMSO-d 6) δ C 21.95, 22.25, 31.30, 35.58, 116.98, 118.94, 126.38, 130.93, 143.27, 157.23, 163.56, 167.15, 167.71, 167.81 ppm; Anal. Calcd for C17H17N3O4S: C, 56.81; H, 4.77; N, 11.69. Found: C, 56.98; H, 4.53; N, 11.81.
N'-(2-((4-Oxo-3,4-dihydroquinazolin-2-yl)thio)acetyl)benzohydrazide (17a)
White powder; yield = 85%; mp 249-251 °C; 1H-NMR (500 MHz; DMSO-d 6) δ H 4.11 (s, 2H), 7.40-7.43 (m, 1H), 7.47-7.50 (m, 2H), 7.54-7.60 (m, 2H), 7.75-7.76 (m, 1H), 7.84-7.85 (m, 2H), 8.01-8.03 (m, 1H), 10.37 (br., 1H), 10.49 (br., 1H), 12.62 ppm (br., 1H); Anal. Calcd for C17H14N4O3S: C, 57.62; H, 3.98; N, 15.81. Found: C, 57.32; H, 4.15; N, 15.54.
4-Methyl-N'-(2-((4-oxo-3,4-dihydroquinazolin-2-yl)thio)acetyl)benzohydrazide (17b)
Pale yellow powder; yield = 91%; mp 243-245 °C;1H-NMR (500 MHz; DMSO-d 6) δ H 2.34 (s, 3H), 4.11 (s, 2H), 7.27-7.28 (m, 2H), 7.42-7.43 (m, 2H), 7.60-7.61 (m, 1H), 7.76-7.77 (m, 2H), 8.02-8.04 (m, 1H), 10.31 (s, IH), 10.41 (s, 1H), 12.69 ppm (br., 1H); 13C-NMR (125 MHz; DMSO-d 6) δ C 20.95, 32.10, 119.97, 125.75, 125.96, 126.20, 127.46, 128.92, 129.54, 134.56, 141.81, 148.26, 154.78, 161.13, 165.25, 166.56 ppm; Anal. Calcd for C18H16N4O3S: C, 58.68; H, 4.38; N, 15.21. Found: C, 58.45; H, 4.68; N, 15.57.
4-Nitro-N'-(2-((4-oxo-3,4-dihydroquinazolin-2-yl)thio)acetyl)benzohydrazide (17c)
Pale yellow powder; yield = 96%; mp 256-258 °C;1H-NMR (500 MHz; DMSO-d 6) δ H 4.13 (s, 2H), 7.39-7.44 (m, 1H), 7.60 (t, 3 J = 6.5 Hz, 1H), 7.76-7.78 (m, 1H), 8.02 (t, 3 J = 6.5 Hz, 1H), 8.07-8.09 (m, 2H), 8.33-8.34 (m, 2H), 10.50 (s, IH), 10.87 (s, 1H), 12.68 ppm (br., 1H); 13C-NMR (125 MHz; DMSO-d 6) δ C 32.05, 119.93, 123.64, 125.74, 125.97, 128.99, 134.53, 137.96, 148.21, 149.37, 154.87, 161.20, 163.78, 166.50 ppm; Anal. Calcd for C17H13N5O5S: C, 51.13; H, 3.28; N, 17.54. Found: C, 51.46; H, 3.55; N, 17.16.
2-Methoxy-N'-(2-((4-oxo-3,4-dihydroquinazolin-2-yl)thio)acetyl)benzohydrazide (17d)
White solid; yield = 85%; mp 224-226 °C; 1H-NMR (500 MHz; DMSO-d 6) δ H 3.86 (s, 3H), 4.10 (s, 2H), 7.05 (t, 3 J = 7.5 Hz, 1H), 7.14 (d, 3 J = 8.5 Hz, 1H), 7.42 (t, 3 J = 7.5 Hz, 1H), 7.50 (dt, 3 J = 8.0 Hz, 4 J = 1.5 Hz, 1H), 7.58 (d, 3 J = 8.0 Hz, 1H), 7.71 (dd, 3 J = 7.8 Hz, 4 J = 1.5 Hz, 1H), 7.76 (dt, 3 J = 7.5 Hz, 4 J = 1.5 Hz, 1H), 8.03 (dd, 3 J = 7.5 Hz, 4 J = 1.0 Hz, 1H), 10.08 (s, 1H), 10.65 (br., 1H), 12.71 ppm (br., 1H); 13C-NMR (125 MHz; DMSO-d 6) δ C 31.97, 55.93, 112.11, 119.94, 120.55, 121.14, 125.70, 125.97, 130.39, 132.80, 134.52, 148.15, 155.07, 156.99, 161.28, 163.39, 165.52 ppm; Anal. Calcd for C18H16N4O4S: C, 56.24; H, 4.20; N, 14.58. Found: C, 56.49; H, 4.53; N, 14.81.
2-Chloro-N'-(2-((4-oxo-3,4-dihydroquinazolin-2-yl)thio)acetyl)benzohydrazide (17e)
White solid; yield = 74%; mp 246-248 °C; 1H-NMR (500 MHz; DMSO-d 6) δ H 4.09 (s, 2H), 7.40-7.43 (m, 2H), 7.45-7.49 (m, 2H), 7.52 (d, 3 J = 7.5 Hz, 1H), 7.59 (d, 3 J = 8.0 Hz, 1H), 7.75 (t, 3 J = 7.5 Hz, 1H), 8.02 (d, 3 J = 7.5 Hz, 1H), 10.47 (s, 1H), 10.50 (br., 1H), 12.70 ppm (br., 1H); 13C-NMR (125 MHz; DMSO-d 6) δ C 32.04, 119.95, 125.74, 125.94, 126.21, 127.07, 129.28, 129.78, 130.39, 131.42, 134.54, 140.40, 148.25, 154.77, 161.14, 165.05, 166.22 ppm; Anal. Calcd for C17H13ClN4O3S: C, 52.51; H, 3.37; N, 14.41. Found: C, 52.21; H, 3.66; N, 14.74.
4-Chloro-N'-(2-((4-oxo-3,4-dihydroquinazolin-2-yl)thio)acetyl)benzohydrazide (17f)
White solid; yield = 86%; mp 257-259 °C; 1H-NMR (500 MHz; DMSO-d 6) δ H 4.11 (s, 2H), 7.42 (t, 3 J = 8.0 Hz, 1H), 7.57 (d, 3 J = 8.5 Hz, 2H), 7.60 (d, 3 J = 8.0 Hz, 1H), 7.77 (t, 3 J = 8.5 Hz, 1H), 7.87 (d, 3 J = 8.5 Hz, 2H), 8.03 (d, 3 J = 7.5 Hz, 1H), 10.39 (s, IH), 10.60 (s, 1H), 12.70 ppm (br., 1H); 13C-NMR (125 MHz; DMSO-d 6) δ C 32.08, 119.94, 125.76, 125.97, 126.21, 128.57, 129.38, 131.08, 134.57, 136.70, 148.26, 154.75, 161.13, 164.37, 166.56 ppm; Anal. Calcd for C17H13ClN4O3S: C, 52.51; H, 3.37; N, 14.41. Found: C, 52.77; H, 3.05; N, 14.73.
2-Bromo-N'-(2-((4-oxo-3,4-dihydroquinazolin-2-yl)thio)acetyl)benzohydrazide (17g)
White solid; yield = 93%; mp 233-235 °C; 1H-NMR (500 MHz; DMSO-d 6) δ H 4.10 (s, 2H), 7.41-7.42 (m, 4H), 7.58 (t, 3 J = 8.0 Hz, 1H), 7.66 (t, 3 J = 8.0 Hz, 1H), 7.73-7.75 (m, 1H), 8.01 (t, 3 J = 8.0 Hz, 1H), 10.46 (s, 1H), 10.50 (br., 1H), 12.69 ppm (br., 1H); 13C-NMR (125 MHz; DMSO-d 6) δ C 32.03, 119.27, 119.94, 125.73, 125.93, 126.20, 127.52, 129.30, 131.52, 132.91, 134.53, 136.56, 148.24, 154.78, 161.11, 165.85, 166.19 ppm; Anal. Calcd for C17H13BrN4O3S: C, 47.13; H, 3.02; N, 12.93. Found: C, 47.38; H, 3.32; N, 12.66.
N'-(2-((4-oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)thio)acetyl)benzohydrazide (18a)
White powder; yield = 90%; mp 246-248 °C; 1H-NMR (500 MHz; DMSO-d 6) δ H 1.60-1.65 (m, 4H), 2.24-2.25 (m, 2H), 2.49 (ov., 2H), 3.96 (s, 2H), 7.44-7.47 (m, 2H), 7.54 (t, 3 J = 7.5 Hz, 1H), 7.83 (d, 3 J = 7.5 Hz, 2H), 10.23 (s, IH), 10.44 (s, 1H), 12.52 ppm (br., 1H); 13C-NMR (125 MHz; DMSO-d 6) δ C 21.42, 21.77, 30.99, 31.92, 116.68, 127.43, 128.41, 131.81, 132.34, 156.28, 160.44, 162.73, 165.36, 166.66 ppm; Anal. Calcd for C17H18N4O3S: C, 56.97; H, 5.06; N, 15.63. Found: C, 56.67; H, 5.29; N, 15.92.
4-Methyl-N'-(2-((4-oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)thio)acetyl)benzohydrazide (18b)
Off white powder; yield = 91%; mp 245-247 °C; 1H-NMR (500 MHz; DMSO-d 6) δ H 1.61-1.66 (m, 4H), 2.24-2.25 (br., 2H), 2.32 (s, 3H), 2.49 (br. ov, 2H), 3.95 (s, 2H), 7.26 (d, 3 J = 7.0 Hz, 2H), 7.74 (d, 3 J = 6.5 Hz, 2H), 10.19 (s, 1H), 10.35 (s, 1H), 12.52 ppm (br., 1H); 13C-NMR (125 MHz; DMSO-d 6) δ C 20.95, 21.41, 21.78, 30.96, 31.92, 116.84, 127.44, 128.91, 129.52, 141.79, 155.53, 160.59, 162.58, 165.21, 166.62 ppm; Anal. Calcd for C18H20N4O3S: C, 58.05; H, 5.41; N, 15.04. Found: C, 58.27; H, 5.23; N, 15.38.
4-Nitro-N'-(2-((4-oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)thio)acetyl)benzohydrazide (18c)
Pale yellow powder; yield = 92%; mp 251-253 °C; 1H-NMR (500 MHz; DMSO-d 6) δ H 1.61-1.66 (m, 4H), 2.25 (br., 2H), 2.49 (br., 2H), 3.96 (s, 2H), 8.05 (d, 3 J = 8.5 Hz, 2H), 8.31 (d, 3 J = 8.5 Hz, 2H), 10.35 (s, 1H), 10.84 (br., 1H), 12.44 ppm (br., 1H); 13C-NMR (125 MHz; DMSO-d 6) δ C 21.43, 21.78, 30.98, 31.88, 116.60, 123.64, 129.01, 137.97, 149.38, 156.12, 161.02, 163.80, 166.59 ppm; Anal. Calcd for C17H17N5O5S: C, 50.61; H, 4.25; N, 17.36. Found: C, 50.89; H, 4.05; N, 17.63.
2-Methoxy-N'-(2-((4-oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)thio)acetyl)benzohydrazide (18d)
Off white powder; yield = 93%; mp 212-214 °C; 1H-NMR (500 MHz; DMSO-d 6) δ H 1.60-1.66 (m, 4H), 2.25-2.26 (m, 2H), 2.49 (ov. br., 2H), 3.85 (s, 3H), 3.95 (s, 2H), 7.04 (t, 3 J = 7.5 Hz, 1H), 7.14 (d, 3 J = 8.5 Hz, 1H), 7.49 (t, 3 J = 7.0 Hz, 1H), 7.69 (d, 3 J = 7.5 Hz, 1H), 10.04 (s, 1H), 10.52 (br., 1H), 12.57 ppm (br., 1H); 13C-NMR (125 MHz; DMSO-d 6) δ C 21.41, 21.78, 30.97, 31.79, 55.95, 112.12, 116.53, 120.57, 121.12, 130.16, 130.41, 132.83, 157.01, 160.14, 162.47, 163.36, 165.59 ppm; Anal. Calcd for C18H20N4O4S: C, 55.66; H, 5.19; N, 14.42. Found: C, 55.45; H, 5.47; N, 14.08.
2-Chloro-N'-(2-((4-oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)thio)acetyl)benzohydrazide (18e)
Off white powder; yield = 95%; mp 238-240 °C; 1H-NMR (500 MHz; DMSO-d 6) δ H1.61-1.65 (m, 4H), 2.26 (br. ov, 2H), 2.49 (br., 2H), 3.94 (s, 2H), 7.42-7.50 (m, 4H), 10.38 (s, 1H), 10.41 (s, 1H), 12.54 ppm (br., 1H); 13C-NMR (125 MHz; DMSO-d 6) 21.40, 21.77, 30.92, 31.85, 116.59, 127.05, 127.99, 129.27, 129.77, 130.38, 131.41, 134.45, 160.36, 162.50, 165.01, 166.28 ppm; Anal. Calcd for C17H17ClN4O3S: C, 51.97; H, 4.36; N, 14.26. Found: C, 51.73; H, 4.62; N, 14.54.
4-Chloro-N'-(2-((4-oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)thio)acetyl)benzohydrazide (18f)
white powder; yield = 95%; mp 252-254 °C; 1H-NMR (500 MHz; DMSO-d 6) δ H1.61-1.66 (m, 4H), 2.19-2.26 (br., 2H), 2.34-2.49 (m, 2H), 3.95 (s, 2H), 7.54 (d, 3 J = 8.0 Hz, 2H), 7.85 (d, 3 J = 8.5 Hz, 2H), 10.23 (s, 1H), 10.52 (s, 1H), 12.49 ppm (br., 1H); 13C-NMR (125 MHz; DMSO-d 6) δ C 21.40, 21.77, 30.95, 31.88, 116.34, 126.43, 128.54, 129.35, 131.06, 136.68, 160.13, 162.32, 164.31, 166.60 ppm; Anal. Calcd for C17H17ClN4O3S: C, 51.97; H, 4.36; N, 14.26. Found: C, 51.77; H, 4.72; N, 14.48.
2-Bromo-N'-(2-((4-oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)thio)acetyl)benzohydrazide (18g)
White powder; yield = 94%; mp 248-250 °C; 1H-NMR (500 MHz; DMSO-d 6) δ H 1.61-1.65 (m, 4H), 2.25-2.26 (m, 2H), 2.49 (br. ov, 2H), 3.94 (s, 2H), 7.38-7.45 (m, 3H), 7.65-7.67 (m, 1H), 10.39 (s, 2H), 12.55 ppm (br., 1H); 13C-NMR (125 MHz; DMSO-d 6) δ C 21.43, 21.80, 30.96, 31.88, 119.30, 127.57, 129.34, 131.58, 132.95, 132.96, 136.59, 156.27, 160.34, 162.55, 165.91, 166.35 ppm; Anal. Calcd for C17H17BrN4O3S: C, 46.69; H, 3.92; N, 12.81. Found: C, 46.45; H, 3.78; N, 12.57.
4.2. Biological evaluation
4.2.1. In vitro bioassay of cytotoxicity and antiviral activity
4.2.1.1. MTT cytotoxicity assay
The cytotoxic activity of the synthesized compounds were determined employing MTT assay as previously described [32]
4.2.1.2. Inhibitory concentration 50 (IC50) determination
The values of IC50 for the target quinazolines were determined as reported [34].
4.2.1.3. Mechanism of action(s)
To investigate whether the most potent candidates affect the (a) viral adsorption, (b) viral replication, or (c) has a virucidal effect, the plaque infectivity reduction assay was performed according to the reported procedure [34].
4.3. Molecular modeling
Molecular docking studies were carried out using Molecular Operating Environment (MOE, 2020.0901) software. All minimizations were performed with MOE until an RMS gradient of 0.05 kcal∙mol−1Å− 2 with MMFF94x force field and the partial charges were automatically calculated. The X-ray crystallographic structure of SARS-CoV-2 main protease (Mpro) co-crystalized with a pyrimidine-2,4-dione inhibitor (YD1) (PDB ID: 7LTJ) was downloaded from the protein data bank [36]. Water molecules and ligands which are not involved in the binding were first removed. Next, the protein structure was prepared for the molecular docking study using QuickPrep protocol in MOE with the default options. YD1 exhibits several binding interactions with Mpro active site with the amino acids Asn142, Gly143, Cys145, His163, Met165, Glu166, and Asp187 either directly or through water mediated interactions (Fig. 5). To perform the molecular docking study, the co-crystalized ligand (YD1) was used to define the active site and Triangle Matcher placement method and London dG scoring function were used.
The molecular docking protocol was first validated by self-docking of the co-crystallized ligand (YD1) in the vicinity of the enzyme active site. The self-docking step reproduced the co-crystalized ligand pose efficiently with docking score (S) of −13.26 kcal/mol and a root mean square deviation (RMSD) of 1.538 Å, moreover, the docking protocol reproduced all the key interactions with the active site amino acids indicating the suitability of the adopted molecular docking protocol for the intended molecular docking study (Fig. 6).
4.4. Estimation of physicochemical, pharmacokinetic and ADME properties
The open SwissADME web tool available from the Swiss Institute of Bioinformatics (SIB) was used for the calculation of the physicochemical descriptors as well as to predict the ADME parameters, and pharmacokinetic properties of the most potent compounds [37]. The compounds’ structures were drawn on the web user interface, converted to SMILES notations, then submitted to the online server for calculation.
Author statement
Heba T. Abdel-Mohsen: Participated in the development of the idea of the project, organic synthesis of the target compounds, structure elucidation of the target compounds, analysis of the biological results, writing, revising and finalizing the manuscript.
Mohamed A. Omar: Participated in organic synthesis of the target compounds and revising the interpretation of NMR results.
Omnia Kutkat: Participated in analysis of the synthesized compounds for their antiviral activity.
Ahmed M. El Kerdawy: Run the computational studies to the target compounds, analysed the obtained results and revised the manuscript.
Alaa A. Osman: Participated in the in silico analyses of the target compounds.
Mohamed GabAllah: Participated in analysis of the synthesized compounds for their antiviral activity.
Ahmed Mostafa: Participated in the development of the idea of the project, analysis of the synthesized compounds for their antiviral activity and revising the biological part of the manuscript.
Mohamed A. Ali: Participated in the development of the idea of the project and revising the biological part of the manuscript.
Hoda I. El Diwani: Participated in the development of the idea of the project and revising the manuscript.
Declaration of Competing Interest
The authors have no conflict of interest to declare.
Acknowledgments
Special thanks to National Research Centre (Egypt) for their fund through the project ID 12060119.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2022.134690.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- 1.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet (London, England) 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen T., Wu D., Chen H., Yan W., Yang D., Chen G., Ma K., Xu D., Yu H., Wang H., Wang T., Guo W., Chen J., Ding C., Zhang X., Huang J., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Consortium C.-G.U., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saied E.M., El-Maradny Y.A., Osman A.A., Darwish A.M.G., Abo Nahas H.H., Niedbala G., Piekutowska M., Abdel-Rahman M.A., Balbool B.A., Abdel-Azeem A.M. A comprehensive review about the molecular structure of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): insights into natural products against COVID-19. Pharmaceutics. 2021;13(11) doi: 10.3390/pharmaceutics13111759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J., Zhao S., Ou J., Zhang J., Lan W., Guan W., Wu X., Yan Y., Zhao W., Wu J., Chodosh J., Zhang Q. COVID-19: coronavirus vaccine development updates. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.602256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Artese A., Svicher V., Costa G., Salpini R., Di Maio V.C., Alkhatib M., Ambrosio F.A., Santoro M.M., Assaraf Y.G., Alcaro S., Ceccherini-Silberstein F. Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses. Drug Resist. Updat. 2020;53 doi: 10.1016/j.drup.2020.100721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elfiky A.A. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in silico perspective. J. Biomol. Struct. Dyn. 2021;39(9):3204–3212. doi: 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 12.Osipiuk J., Azizi S.A., Dvorkin S., Endres M., Jedrzejczak R., Jones K.A., Kang S., Kathayat R.S., Kim Y., Lisnyak V.G., Maki S.L., Nicolaescu V., Taylor C.A., Tesar C., Zhang Y.A., Zhou Z., Randall G., Michalska K., Snyder S.A., Dickinson B.C., Joachimiak A. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021;12(1):743. doi: 10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huff S., Kummetha I.R., Tiwari S.K., Huante M.B., Clark A.E., Wang S., Bray W., Smith D., Carlin A.F., Endsley M., Rana T.M. Discovery and mechanism of SARS-CoV-2 main protease inhibitors. J. Medicin. Chem. 2021 doi: 10.1021/acs.jmedchem.1c00566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cannalire R., Cerchia C., Beccari A.R., Di Leva F.S., Summa V. Targeting SARS-CoV-2 proteases and polymerase for COVID-19 treatment: state of the art and future opportunities. J. Medicin. Chem. 2020 doi: 10.1021/acs.jmedchem.0c01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bansal R., Malhotra A. Therapeutic progression of quinazolines as targeted chemotherapeutic agents. Eur. J. Med. Chem. 2021;211 doi: 10.1016/j.ejmech.2020.113016. [DOI] [PubMed] [Google Scholar]
- 16.Das R., Mehta D.K., Dhanawat M. Bestowal of quinazoline scaffold in anticancer drug discovery. Anticancer Agent. Med. Chem. 2021;21(11):1350–1368. doi: 10.2174/1871520620666200627205321. [DOI] [PubMed] [Google Scholar]
- 17.Matharu D.S., Flaherty D.P., Simpson D.S., Schroeder C.E., Chung D., Yan D., Noah J.W., Jonsson C.B., White E.L., Aube J., Plemper R.K., Severson W.E., Golden J.E. Optimization of potent and selective quinazolinediones: inhibitors of respiratory syncytial virus that block RNA-dependent RNA-polymerase complex activity. J. Medicin. Chem. 2014;57(24):10314–10328. doi: 10.1021/jm500902x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang G., Wang M., Zhao J., Wang Y., Zhu M., Wang J., Cen S., Wang Y. Design, synthesis and in vitro anti-influenza A virus evaluation of novel quinazoline derivatives containing S-acetamide and NH-acetamide moieties at C-4. Eur. J. Med. Chem. 2020;206 doi: 10.1016/j.ejmech.2020.112706. [DOI] [PubMed] [Google Scholar]
- 19.Hwu J.R., Kapoor M., Gupta N.K., Tsay S.C., Huang W.C., Tan K.T., Hu Y.C., Lyssen P., Neyts J. Synthesis and antiviral activities of quinazolinamine-coumarin conjugates toward chikungunya and hepatitis C viruses. Eur. J. Med. Chem. 2022;232 doi: 10.1016/j.ejmech.2022.114164. [DOI] [PubMed] [Google Scholar]
- 20.Lee J.Y., Shin Y.S., Lee J., Kwon S., Jin Y.H., Jang M.S., Kim S., Song J.H., Kim H.R., Park C.M. Identification of 4-anilino-6-aminoquinazoline derivatives as potential MERS-CoV inhibitors. Bioorg. Med. Chem. Lett. 2020;30(20) doi: 10.1016/j.bmcl.2020.127472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J., Zhang Y., Wang M., Liu Q., Lei X., Wu M., Guo S., Yi D., Li Q., Ma L., Liu Z., Guo F., Wang J., Li X., Wang Y., Cen S. Quinoline and quinazoline derivatives inhibit viral RNA synthesis by SARS-CoV-2 RdRp. ACS Infect. Dis. 2021;7(6):1535–1544. doi: 10.1021/acsinfecdis.1c00083. [DOI] [PubMed] [Google Scholar]
- 22.Rothan H.A., Teoh T.C. Cell-based high-throughput screening protocol for discovering antiviral inhibitors against SARS-COV-2 main protease (3CLpro) Mol. Biotechnol. 2021;63(3):240–248. doi: 10.1007/s12033-021-00299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moyle G., Boffito M., Stoehr A., Rieger A., Shen Z., Manhard K., Sheedy B., Hingorani V., Raney A., Nguyen M., Nguyen T., Ong V., Yeh L.T., Quart B. Phase 2a randomized controlled trial of short-term activity, safety, and pharmacokinetics of a novel nonnucleoside reverse transcriptase inhibitor, RDEA806, in HIV-1-positive, antiretroviral-naive subjects. Antimicrob. Agents Chemother. 2010;54(8):3170–3178. doi: 10.1128/AAC.00268-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhan P., Li X., Li Z., Chen X., Tian Y., Chen W., Liu X., Pannecouque C., De Clercq E. Structure-based bioisosterism design, synthesis and biological evaluation of novel 1,2,4-triazin-6-ylthioacetamides as potent HIV-1 NNRTIs. Bioorg. Med. Chem. Lett. 2012;22(23):7155–7162. doi: 10.1016/j.bmcl.2012.09.062. [DOI] [PubMed] [Google Scholar]
- 25.Zhang G.N., Li Q., Zhao J., Zhang X., Xu Z., Wang Y., Fu Y., Shan Q., Zheng Y., Wang J., Zhu M., Li Z., Cen S., He J., Wang Y. Design and synthesis of 2-((1H-indol-3-yl)thio)-N-phenyl-acetamides as novel dual inhibitors of respiratory syncytial virus and influenza virus A. Eur. J. Med. Chem. 2020;186 doi: 10.1016/j.ejmech.2019.111861. [DOI] [PubMed] [Google Scholar]
- 26.Yu M., Liu A., Du G., Naesens L., Vanderlinden E., De Clercq E., Liu X. Discovery of dihydro-alkyloxy-benzyl-oxopyrimidines as promising anti-influenza virus agents. Chem. Biol. Drug Des. 2011;78(4):596–602. doi: 10.1111/j.1747-0285.2011.01180.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhan P., Wang L., Liu H., Chen X., Li X., Jiang X., Zhang Q., Liu X., Pannecouque C., Naesens L., De Clercq E., Liu A., Du G. Arylazolyl(azinyl)thioacetanilide. Part 9: Synthesis and biological investigation of thiazolylthioacetamides derivatives as a novel class of potential antiviral agents. Arch. Pharm. Res. 2012;35(6):975–986. doi: 10.1007/s12272-012-0604-y. [DOI] [PubMed] [Google Scholar]
- 28.Chou S.-Y., Yin W.-K., Chung Y.-S., Chang L.-S., Liu C.-W., Chen S.-F., Shih K.-S. Kilogram-scale synthesis of a highly selective α 1-adrenoceptor antagonist (DL-028A) Org. Process Res. Dev. 2002;6(3):273–278. [Google Scholar]
- 29.Abdel-Mohsen H.T., Omar M.A., Petreni A., Supuran C.T. Novel 2-substituted thioquinazoline-benzenesulfonamide derivatives as carbonic anhydrase inhibitors with potential anticancer activity. Arch. Pharm. (Weinheim) 2022 doi: 10.1002/ardp.202200180. [DOI] [PubMed] [Google Scholar]
- 30.Abdel-Mohsen H.T., Conrad J., Harms K., Nohr D., Beifuss U. Laccase-catalyzed green synthesis and cytotoxic activity of novel pyrimidobenzothiazoles and catechol thioethers. RSC Adv. 2017;7(28):17427–17441. [Google Scholar]
- 31.Abdel-Mohsen H.T., Petreni A., Supuran C.T. Investigation of the carbonic anhydrase inhibitory activity of benzenesulfonamides incorporating substituted fused-pyrimidine tails. Arch. Pharm. (Weinheim) 2022;355(11) doi: 10.1002/ardp.202200274. [DOI] [PubMed] [Google Scholar]
- 32.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunolog. Method. 1983;65(1):55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 33.Kandeil A., Mostafa A., El-Shesheny R., Shehata M., Roshdy W.H., Ahmed S.S., Gomaa M., Taweel A.E., Kayed A.E., Mahmoud S.H., Moatasim Y., Kutkat O., Kamel M.N., Mahrous N., Sayes M.E., Guindy N.M.E., Naguib A., Ali M.A. Coding-complete genome sequences of two SARS-CoV-2 isolates from Egypt. Microbiol. Resour. Announc. 2020;9(22) doi: 10.1128/MRA.00489-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mostafa A., Kandeil A., Y A.M.M.E., Kutkat O., Moatasim Y., Rashad A.A., Shehata M., Gomaa M.R., Mahrous N., Mahmoud S.H., GabAllah M., Abbas H., Taweel A.E., Kayed A.E., Kamel M.N., Sayes M.E., Mahmoud D.B., El-Shesheny R., Kayali G., Ali M.A. FDA-approved drugs with potent in vitro antiviral activity against severe acute respiratory syndrome Coronavirus 2. Pharmaceuticals (Basel) 2020;13(12) doi: 10.3390/ph13120443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mengist H.M., Dilnessa T., Jin T. Structural Basis of Potential Inhibitors Targeting SARS-CoV-2 Main Protease. Front Chem. 2021;9 doi: 10.3389/fchem.2021.622898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clyde A., Galanie S., Kneller D.W., Ma H., Babuji Y., Blaiszik B., Brace A., Brettin T., Chard K., Chard R., Coates L., Foster I., Hauner D., Kertesz V., Kumar N., Lee H., Li Z., Merzky A., Schmidt J.G., Tan L., Titov M., Trifan A., Turilli M., Van Dam H., Chennubhotla S.C., Jha S., Kovalevsky A., Ramanathan A., Head M.S., Stevens R. High-throughput virtual screening and validation of a SARS-CoV-2 main protease noncovalent inhibitor. J. Chem. Inform. Model. 2022;62(1):116–128. doi: 10.1021/acs.jcim.1c00851. [DOI] [PubMed] [Google Scholar]
- 37.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daina A., Michielin O., Zoete V. iLOGP: a simple, robust, and efficient description of n-octanol/water partition coefficient for drug design using the GB/SA approach. J. Chem. Inform. Model. 2014;54(12):3284–3301. doi: 10.1021/ci500467k. [DOI] [PubMed] [Google Scholar]
- 39.Amin M.L. P-glycoprotein inhibition for optimal drug delivery. Drug Target Insight. 2013;7:27–34. doi: 10.4137/DTI.S12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug. Deliv. Rev. 2001;46(1-3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 41.Veber D.F., Johnson S.R., Cheng H.Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Medicin. Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 42.Ghose A.K., Viswanadhan V.N., Wendoloski J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999;1(1):55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- 43.Egan W.J., Merz K.M., Jr., Baldwin J.J. Prediction of drug absorption using multivariate statistics. J. Medicin. Chem. 2000;43(21):3867–3877. doi: 10.1021/jm000292e. [DOI] [PubMed] [Google Scholar]
- 44.Muegge I., Heald S.L., Brittelli D. Simple selection criteria for drug-like chemical matter. J. Medicin. Chem. 2001;44(12):1841–1846. doi: 10.1021/jm015507e. [DOI] [PubMed] [Google Scholar]
- 45.Baell J.B., Holloway G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Medicin. Chem. 2010;53(7):2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.