Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) protein binds angiotensin-converting enzyme 2 as its primary infection mechanism. Interactions between S and endogenous proteins occur after infection but are not well understood. We profiled binding of S against >9000 human proteins and found an interaction between S and human estrogen receptor α (ERα). Using bioinformatics, supercomputing, and experimental assays, we identified a highly conserved and functional nuclear receptor coregulator (NRC) LXD-like motif on the S2 subunit. In cultured cells, S DNA transfection increased ERα cytoplasmic accumulation, and S treatment induced ER-dependent biological effects. Non-invasive imaging in SARS-CoV-2–infected hamsters localized lung pathology with increased ERα lung levels. Postmortem lung experiments from infected hamsters and humans confirmed an increase in cytoplasmic ERα and its colocalization with S in alveolar macrophages. These findings describe the discovery of a S-ERα interaction, imply a role for S as an NRC, and advance knowledge of SARS-CoV-2 biology and coronavirus disease 2019 pathology.
Interactions identified between the SARS-CoV-2 Spike protein and estrogen receptors in humans and laboratory animals.
INTRODUCTION
Coronavirus disease 2019 (COVID-19) is an infectious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The most frequent symptom of severe COVID-19 is pneumonia, accompanied by fever, cough, and dyspnea commonly associated with cytokine storm, systemic inflammatory response, and coagulopathy (1, 2). The elderly and those with underlying comorbidities are more likely to develop severe illness and mortality (3, 4).
SARS-CoV-2 is characterized by four structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins (5). Currently, most COVID-19 vaccines use S as the target antigen, as it is an important determinant capable of inducing a robust protective immune response (6). Furthermore, it is a critical component for cell infection via direct interaction with angiotensin-converting enzyme 2 (ACE2) (7, 8). S is composed of 1273 amino acids. It consists of a signal peptide located at the N terminus (amino acids 1 to 13), the S1 subunit (14 to 685 residues), and the S2 subunit (686 to 1273 residues). The S1 subunit contains the ACE2 receptor binding domain (RBD), whereas the S2 subunit is responsible for viral and host cell membrane fusion (9, 10) that requires other proteins (8, 11–14). Cells are still susceptible to infection and show S-dependent biological responses independent of ACE2 (15–19), suggesting that S may promote pathology independent from its capacity to bind ACE2. Given the multitude and complex array of systemic symptoms associated with COVID-19, it is possible that other molecular targets of S may exist. Identification of additional S targets would be critical for advancing our understanding of SARS-CoV-2 infection and COVID-19 pathobiology.
RESULTS
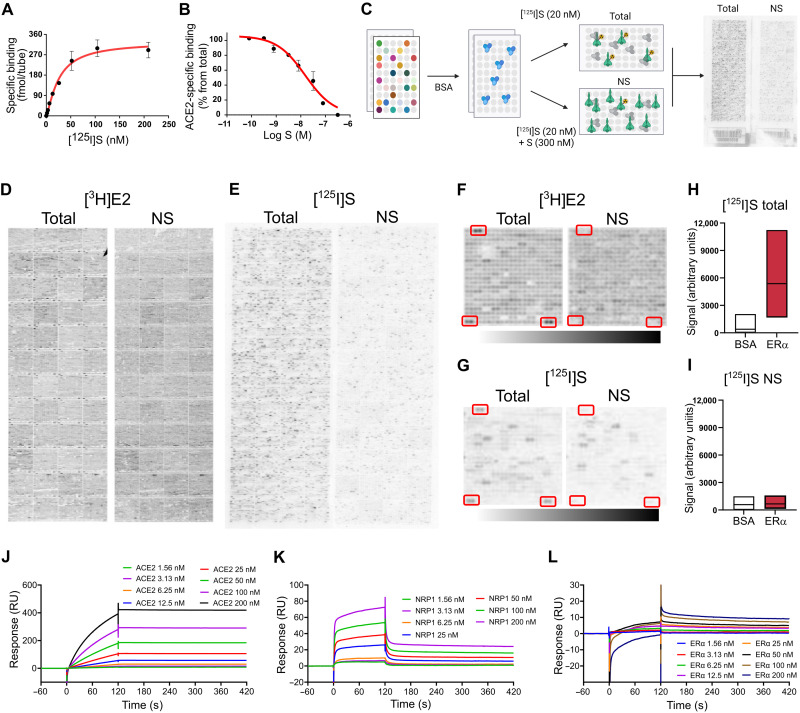
S binds estrogen receptor α with high affinity
First, we radiolabeled full-length recombinant S with 125I and confirmed its binding to human ACE2 [Equilibrium dissociation constant (KD) = 27.8 ± 5.0 nM; Fig. 1, A and B] (13, 20). Next, we used ProtoArray protein array slides to screen for [125I]S binding against >9000 human proteins (Fig. 1C). As per ProtoArray protocol, we also tested the binding of [3H]estradiol (E2) (1 nM), which serves as a positive control. As expected, incubation with [3H]E2 labeled the full-length estrogen receptor α (ERα; Fig. 1, D and F). Other arrays were incubated with [125I]S (20 nM) with or without [3H]E2 (1 nM) or in the presence of nonradioactive S (300 nM) to control for nonspecific binding (Fig. 1E). We detected a specific [125I]S signal at seven proteins on the array, including neuropilin 1 (NRP1; table S1), a known S target protein (12). Unexpectedly, we also detected a specific and reproducible [125I]S signal at the multiple ERα sites (Fig. 1, G to I). We then confirmed the S-NRP1 and S-ERα interactions in secondary assays by immobilizing S and performing surface plasmon resonance (SPR) kinetic analyses with recombinant ACE2 (KD = 0.58 nM), NRP1 (KD = 89.4 nM), and ERα (KD = 9.7 nM) (Fig. 1, J to L).
Fig. 1. S binds ERα with high affinity.
(A) [125I]S saturation and (B) competition binding to recombinant ACE2. (C) Schematic of ProtoArray experimental design. (D) Positive control ProtoArray autoradiograms showing total and nonspecific (NS) binding of [3H]E2. (E) ProtoArray autoradiograms (experiment 1 of 3) showing total and nonspecific binding of [125I]S. (F and G) Representative array blocks showing total and nonspecific [3H]E2 and [125I]S binding. Red rectangles show location of ERα proteins. (H and I) Quantification of total and nonspecific [125I]S binding at ERα and bovine serum albumin (BSA) (control). Data are representative of three independent experiments. (J to L) Representative SPR sensorgrams showing kinetic and equilibrium binding analyses of immobilized S exposed to increasing concentrations of ACE2, NRP1, and ERα protein (Kon = 2.03 × 105, Koff = 1.96 × 10−3, and KD = 9.7 nM). In (A) and (B), data are represented as means ± SEM. In (H) and (I), data are presented as median ± min and max limits.
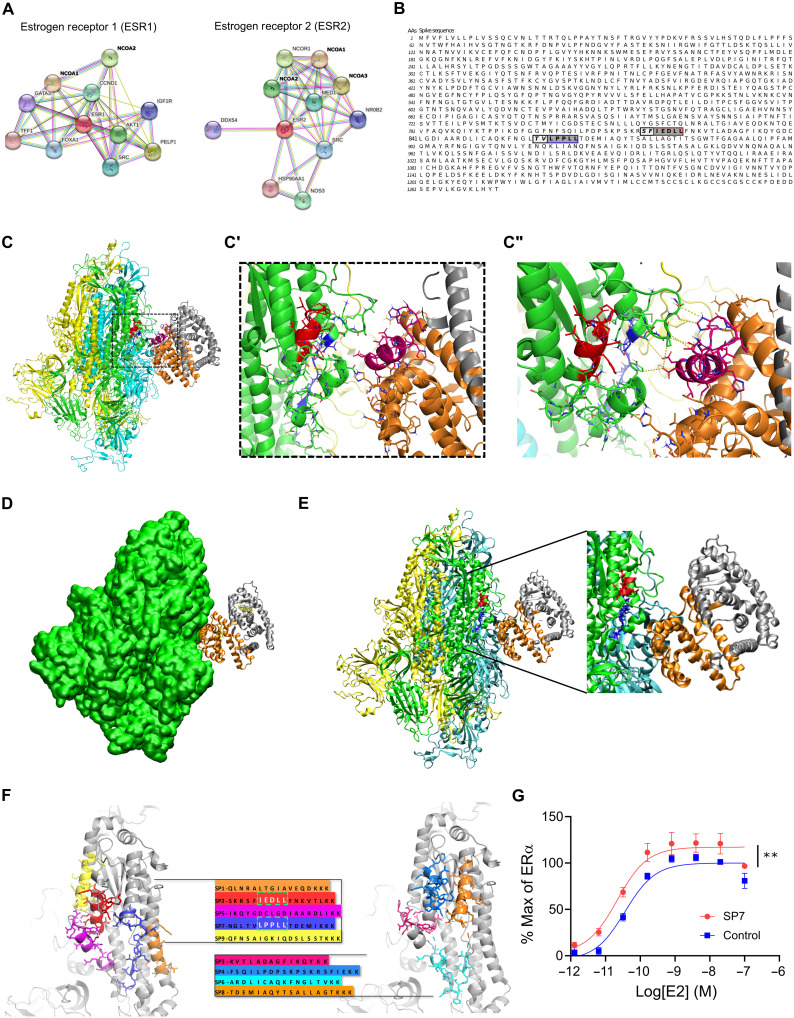
S and ER interact at conserved LXD-like nuclear receptor coregulator motifs
To identify discrete structural domains involved in S-ER interactions, we used bioinformatics and the EXaSCale smArt pLatform Against paThogEns (EXSCALATE) supercomputing platform (21). First, a network analysis confirmed prominent interactions between ERα, ERβ, and other proteins (Fig. 2A) including known interactions with NR coactivators 1, 2, and 3 (NCOA1, NCOA2, and NCOA3; table S2). NCOAs bind to the activation function 2 (AF-2) region on ERs to modulate ligand-mediated activation of ER transcription via a region called the NR box that includes an LXD motif, known as the LXXLL core consensus sequence (where L is leucine and X is any amino acid; fig. S1) (22). This motif is necessary and sufficient for nuclear receptor coregulator (NRC) binding to ligand-bound ERs and for ER function. Using this information and the EXSCALATE platform, we identified two ER-interacting LXD-like motifs in the S sequence (Fig. 2B). We then analyzed and compared the sequences of other coronavirus S proteins, including SARS-CoV, MERS-CoV (Middle East respiratory syndrome coronavirus), HCoV (human coronavirus), and MHV (murine coronavirus) (table S3) to search for conserved LDX-like motifs. We found discrete shared amino acid patterns across species (fig. S2), suggesting a conserved functional role of these regions. We then verified the conservation of the two LDX-like motifs and found the LPPLL pattern at residues 861 to 865 conserved among SARS-CoV-2, SARS-CoV, HCoV, and MERS-CoV (Fig. 2C and fig. S2), while the LXD-like pattern IEDLL at residues 818 to 822 is also conserved among the same viruses. Note that a standard LEDLL pattern is found in HCoV-HKU1 in the same position. Notably, this LXD-like region, which is solvent-exposed in the S experimental structures, retains well-defined three-dimensional (3D) structural characteristics (α helix–12 folding; magenta in Fig. 2C) found in the ER-NCOA complexes 3UUD (23) and 3OLL (24). On the contrary, the LXXLL motif, less solvent-exposed, is unstructured (blue in Fig. 2C). It is well known, however, that the motif region may assume the α helix folding only after the binding with ER (25), implying a conformational rearrangement of the two molecular partners.
Fig. 2. S and ER interact at conserved LXD NRC motifs.
(A) ER interaction network showing known and predicted protein associations. (B) LXD-like patterns in the S sequence. The LXXLL motif and a homologous region are highlighted in blue and red boxes, respectively, with dark gray background. Positions (−1 and −2) are reported in italic and light gray background, respectively. AAs, amino acids. (C and C′) The LPPLL and IEDLL residues of the two motifs are shown in the 3D x-ray S structure [Protein Data Bank (PDB) ID 6VYB] with blue and red colors, respectively. The ER dimer is in orange and gray, while the helix-12 is reported in magenta. (C″) The image shows favorable interactions between ER and the S’s regions containing the LXXLL motifs. The interacting residues and the predicted interactions are reported in stick and yellow dots, respectively. (D) S-ER motif–oriented docking. The best 3D docking hypothesis is shown. The ER dimer is in orange and gray, and S is green. (E) Alignment between the best-pose and the 3OLL model tied with NCOA1. The region occupied by S’s α helix interacts in the area where the NCOA fragment was crystallized. (F) S protein peptides and their location with respect to the S 3D structure. (G) The SP7 peptide containing the LPPLL motif significantly increased ERα activation [F(1,48) = 30.38, **P < 0.01, two-way analysis of variance (ANOVA); peptide treatment main effect]. Data are shown as means ± SEM.
We then performed in silico molecular docking simulations to identify a putative S-ER binding mode. An S-ERα 3D model was built on the basis of Protein Data Bank (PDB) 6VYB in its wild-type (WT) and fully glycosylated form and PDB 3OLL, which contained both E2 and NCOA1 (24). For protein-protein docking, S in glycosylated form and ERα/β were used as receptor and ligand, respectively. The top 100 predicted complex structures were selected, and the 10 best hypotheses were visually inspected to confirm the reliability of the calculation (26). Since the revised LXD-like motifs identified were located outside the S RBD, the ability of ER to interact outside this region was evaluated by means of blind docking (26). The best binding hypothesis included evidence of a high-affinity ER interaction toward the lateral region of S, which includes the so-called “fusion peptide portion” (fig. S3). The structural information that ER residues are recognized by NCOA was then used to guide S-ER docking studies by optimizing protein-protein interactions. The best binding hypothesis obtained highlighted the binding of ER to the S region containing the two described LDX-like motifs (Fig. 2, D and E). Several molecular dynamics (MD) simulations of the best docking complexes were then carried out. MD results showed the formation of a strong interaction between ER and S even in the first phase of the recognition (fig. S4). We then extracted nine peptide sequences (SP1 to SP9) based on their proximity to the putative S-ER binding region and their LXD-like domain sequence similarities (Fig. 2F), synthesized each peptide, and examined their effects on ERα-mediated transcriptional activation (GeneBLAzer ERα/β-UAS-bla GripTite cells). One peptide (SP7), which solely contained the LPPLL motif, significantly increased the potency of E2 in stimulating ERα transcriptional activation (Fig. 2G and fig. S5).
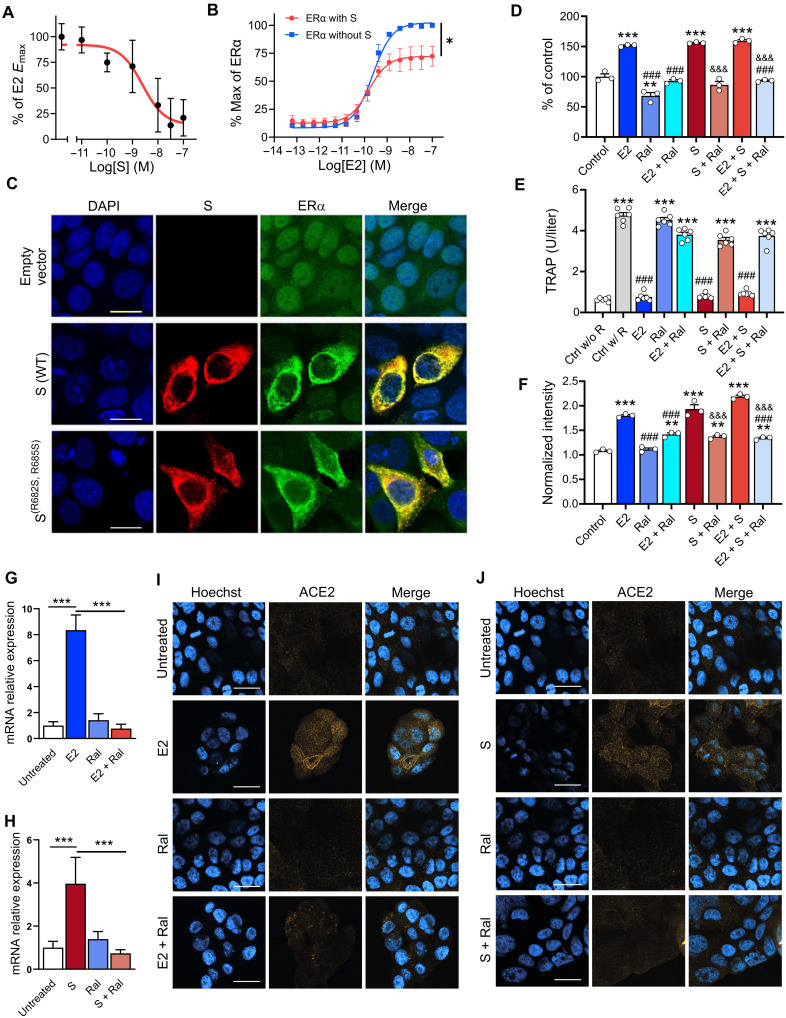
S modulates ER-dependent biological functions
We used MCF-7 nuclear extracts and the TransAM ER assay (27) to measure E2-stimulated ERα DNA binding. We found that full-length S [median inhibitory concentration (IC50) = 2.4 ± 1.5 nM] and S trimer (IC50 = 72 ± 2.6 nM), but not S-RBD, inhibited E2-stimulated ERα DNA binding (Fig. 3A and fig. S6, A and B). We also assessed whether S affected ER-mediated transcriptional activation using ERα-Ligand binding domain (LBD) and ERβ-LBD reporter cell lines (GeneBLAzer ERα/β-UAS-bla GripTite). S overexpression significantly decreased the Emax of E2-stimulated ERα (EmaxS = 72 ± 8.8% and EmaxControl = 100%) and ERβ (EmaxS = 85 ± 2.3% and EmaxControl = 100%) transcriptional activation without significantly affecting their median effective concentration (EC50) (ERα, EC50S = 0.15 ± 0.04 nM and EC50Control = 0.23 ± 0.05 nM; ERβ, EC50S = 0.51 ± 0.03 nM and EC50Control = 0.70 ± 0.06 nM; Fig. 3B and fig. S6C), suggesting a selective partial antagonism of the ER-induced transcriptional effect.
Fig. 3. S modulates ER-dependent biological functions.
S inhibits (A) E2-induced ERα DNA binding in MCF-7 nuclear extracts and (B) transcriptional activation in an ERα reporter cell line [F(1,28) = 21.73, *P = 0.01, two-way ANOVA; S treatment × E2 concentration interaction effect]. (C) Immunofluorescent staining of S and endogenous ERα in MCF-7 cells transfected with empty vector, WT, or the furin cleavage site mutant S(R682S, R685S). Scale bars, 16 mm. (D) S increases MCF-7 cell proliferation in an ER-dependent manner (**P < 0.01 and ***P < 0.001 versus control; ###P < 0.001 versus E2; &&&P < 0.001 versus S; one-way ANOVA with Tukey post hoc test). (E) S decreases osteoclast differentiation in an ER-dependent manner (***P < 0.001 versus control without RANKL; ###P < 0.001 versus control with RANKL; one-way ANOVA with Tukey post hoc test). (F) S and E2 increase ACE2 protein levels in MCF-7 cells in an ER-dependent manner (***P < 0.001 versus control; ###P < 0.001 versus E2; &&&P < 0.001 versus S; one-way ANOVA with Tukey post hoc test). (G and H) E2 and S increase ACE2 mRNA (***P < 0.001, one-way ANOVA with Tukey post hoc test) and (I and J) protein in the Calu-3 lung cell line in an ER-dependent manner. Scale bars, 30 mm. All data are shown as means ± SEM.
To visualize the cellular distribution of S and ERα, we transfected MCF-7 cells with the WT S or a mutant S(R682S, R685S) stabilized at the furin cleavage site and performed immunocytochemistry. Cells transfected with the empty pcDNA3.1 vector showed the expected ERα nuclear-enriched distribution pattern and no S signal (Fig. 3C). In contrast, overexpression of either WT or mutant S increased ERα cytoplasmic labeling (Fig. 3C), indicating that S, either with or without an intact furin cleavage site, leads to an increase in ERα and its redistribution from the nucleus to the cytoplasm.
E2 increases MCF-7 cell proliferation, whereas raloxifene, a potent selective ER modulator, blocks MCF-7 cell proliferation (28). As expected, E2 treatment increased MCF-7 cell proliferation, and this effect was blocked by raloxifene (2 μM; Fig. 3D). S (10 ng/ml) itself also increased MCF-7 cell proliferation, and this effect was also blocked by raloxifene (2 μM) and fulvestrant (ICI 182,780; 1 μM) (fig. S7A), indicating that it was ER dependent. Notably, exposure of MCF-7 cells to E2 and S did not lead to an additive proliferation response, and neither E2 nor S induced proliferation in an ER-lacking cell line (MDA-MB-231; fig. S7B).
E2 inhibition of osteoclast differentiation is an ERα-dependent effect linked to its therapeutic use (21, 29, 30). RAW264.7, a murine macrophage cell line that expresses ERα, was induced to differentiate into osteoclasts by receptor activator of nuclear factor κB ligand (RANKL) treatment in the presence or absence of E2 (1 nM), S (10 ng/ml), or their combination. E2 or S, as well as their combination, abolished RANKL-induced osteoclast differentiation (Fig. 3E), and these effects were completely blocked by raloxifene (2 μM), indicating that they were ER dependent.
To assess the relevance of S and ER signaling to SARS-CoV-2 cell entry mechanisms, we first assessed the effect of E2 (1 nM) and S (10 ng/ml) on ACE2 levels in MCF-7 cells via enzyme-linked immunosorbent assay (ELISA). E2 or S, as well as their combination, significantly increased ACE2 levels, and in both cases, these effects were blocked by raloxifene (2 μM; Fig. 3F), indicating that the ACE2 increases were ER dependent. We also tested the effect of S and E2 on ACE2 expression in Calu-3 cells, a human airway epithelial cell line used to study SARS-CoV-2 infection. Both E2 (200 nM) and S (10 ng/ml) increased ACE2 mRNA (Fig. 3, G and H) and ACE2 membrane protein expression (Fig. 3, I and J). In both cases, raloxifene (20 μM) reverted these effects, indicating that they were ER dependent.
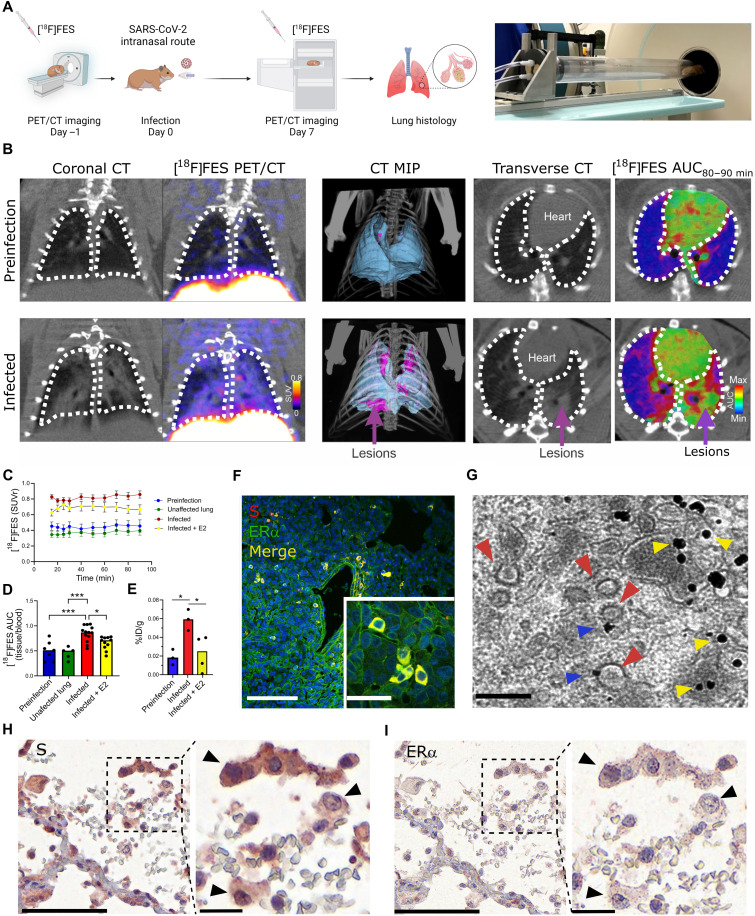
SARS-CoV-2 infection increases cytoplasmic ERα accumulation and S-ERα colocalization in pulmonary macrophages
To extend the relevance of the above findings to COVID-19, we performed in vivo SARS-CoV-2 infection experiments in Syrian golden hamsters. A SARS-CoV-2/USA-WA1/2020 strain (BEI Resources) was propagated with one passage in cell culture in a biosafety level-3 (BSL-3) laboratory. Syrian golden hamsters (male, 6 to 8 weeks old; Envigo, Indianapolis, IN) were exposed to a 1.5 × 105 median tissue culture infective dose (TCID50) in 100 μl of Dulbecco’s modified Eagle’s medium (DMEM) by the intranasal route as previously described (31). Male hamsters were imaged longitudinally inside in-house developed and sealed BSL-3–compliant biocontainment devices (31) at 1 day before (day −1) and at 7 days (day 7) after infection using the positron emission tomography (PET) radiopharmaceutical [18F]fluoroestradiol [[18F]FES; 20 megabecquerels (MBq) per animal, n = 5] and computed tomography (CT; Fig. 4A). A 90-min dynamic PET acquisition was performed immediately after intravenous [18F]FES injection to visualize the hamster body from the eyes to thighs (starting at the skull vertex). Following PET, a CT scan was immediately performed as previously described (31). SARS-CoV-2 infection in hamsters produced marked pathology in the lung (as detected by CT and an established image algorithm and analysis pipeline) (31) at day 7 after infection compared to day −1 (before infection) (Fig. 4, B to D). No distinguishable [18F]FES uptake was present in the lungs at day −1 (Fig. 4B). In contrast, the pattern of lung lesions detected via CT overlapped with the lung [18F]FES uptake at day 7 (Fig. 4B). Specifically, lung [18F]FES uptake at day 7 was significantly higher in infected lung regions compared to these same sites at day −1 and at unaffected areas at day 7 (Fig. 4, C and D). Furthermore, [18F]FES lung uptake at day 7 was significantly decreased after pretreatment with a pharmacological dose (1 mg/kg, intravenously) of E2, indicating that it reflected specific ERα binding (Fig. 4, C and D). To further corroborate these findings, we performed ex vivo biodistribution studies using [18F]FES. At 120 min after [18F]FES dosing, hamsters were euthanized, and the lungs were harvested and counted for radioactivity. In line with the PET data, SARS-CoV-2–infected hamsters had significantly greater lung [18F]FES uptake compared to both uninfected hamsters and SARS-CoV-2–infected hamsters pretreated with E2 (Fig. 4E).
Fig. 4. SARS-CoV-2 infection increases cytoplasmic ERα accumulation and S-ERα colocalization in pulmonary macrophages.
(A) Schematic showing experimental design of SARS-CoV-2 hamster studies and BSL-3 imaging compartment (Created with BioRender). (B) CT, [18F]FES PET, and area under the curve (AUC) heatmap overlay images from hamsters at preinfection (day −1) and infection (day 7). MIP, maximum intensity projection. (C) Time activity curves showing standard uptake value ratio (SUVr; tissue/blood [18F]FES content) in each experimental group. n = 4 to 5 per group. ***P < 0.001 versus preinfection (D) [18F]FES uptake expressed as area under the curve ratio (AUCr; tissue/blood [18F]FES content). F(3,32) = 12.15, ***P < 0.001 and *P < 0.05. (E) [18F]FES uptake expressed as % injected dose (%ID)/gram of body weight in postmortem hamster lung (harvested 110 min after injection; n = 3 to 4 per group). F(2,7) = 7.161, *P < 0.05. (F) Hamster lung IHC showing colocalization of S and ERα immunoreactivity (experiment 1 of 2). (G) Immunogold electron microscopy showing SARS-CoV-2 particles (red arrowheads) and ERα-bound gold nanoparticles (blue arrowheads) in a hamster alveolar macrophage. Scale bars, 200 nm. Yellow arrowheads correspond to cytoplasmic ERα accumulation. (H) S and (I) ERα immunostaining in SARS-CoV-2–infected human lung showing S-ERα colocalization in macrophages (black arrowheads). Scale bars, 100 nm (low mag) and 25 nm (high mag). All data are shown as means ± SEM.
We exposed additional cohorts of male hamsters to SARS-CoV-2 as above, then euthanized them at day 7 after infection along with uninfected controls, and collected their lungs to perform fluorescent immunohistochemistry (IHC) with anti-S and anti-ERα antibodies. As expected, we observed no S or ERα signal in uninfected hamster tissue (fig. S9). In contrast, the vast majority of cells from infected hamsters that were positive for S exhibited ERα immunoreactivity (Fig. 4F and fig. S8). In infected hamsters, S-positive cells accounted for 14 ± 5%, while ERα-positive cells accounted for 13 ± 5% of lung cells. Moreover, ERα in these cells showed cytoplasmic accumulation in a pattern as we observed in MCF-7 cells transfected with S DNA (Fig. 3C).
To examine the subcellular expression of ERα, we performed immunoelectron microscopy using gold nanoparticles targeting anti-ERα antibodies in lung tissue from uninfected and SARS-CoV-2–infected hamsters. In agreement with the fluorescent IHC results, we found high levels of gold nanoparticle ERα labeling in various cytoplasmic compartments in lung cells from infected hamsters, whereas uninfected hamsters showed low ERα labeling (fig. S8). The vast majority of cells from infected hamsters with ERα labeling constituted alveolar macrophages (fig. S8), and in these cells, we specifically observed gold nanoparticle accumulation at the surface of SARS-CoV-2 virions (Fig. 4G), confirming that S-ERα interact in vivo.
Last, to extend these findings to humans, we performed S and ERα IHC in postmortem lung tissue derived from four human COVID-19 autopsies. We observed S labeling in only one of the four cases and in that sample S staining colocalized with granular cytoplasmic labeling of ERα in cells favored to be macrophage lineages (alveolar > interstitial; Fig. 4, H and I, and fig. S9). The other three cases did not show ERα staining. The granular cytoplasmic ERα cytoplasmic staining pattern in the S-labeled cells markedly differed from the nuclear pattern of ERα normally found in breast cancer tissue (fig. S9) and was similar to the pattern observed in MCF-7 cells transfected with S DNA and in alveolar macrophage cells in SARS-CoV-2–infected hamsters, supporting the notion that S and ERα colocalize in the cytoplasm of SARS-CoV-2–infected human alveolar macrophage lung cells.
DISCUSSION
E2 is the most potent endogenous estrogen and is highly selective for ER. In the absence of E2, ERs exist within target cells in a transcriptionally inactive form. Upon ligand activation, ERs undergo homodimerization and binding to discrete DNA regions present at enhancers of specific target genes. Gene regulation occurs when the ER homodimer builds a transcriptional complex with NRC proteins, which can either activate or inactivate transcriptional activity (32, 33). Our results, together with prior observations (34), suggest that S-ERα interactions are involved in SARS-CoV-2 infection and COVID-19 pathology via modulation of ERα signaling, transcriptional regulation of ACE2, and potentially of other genes with roles in inflammation and immunity. Our collective findings indicate that S exhibits structural and functional properties consistent with a role as an NRC at ERα, and it is plausible that this function may extend to other NRs as well. Furthermore, given its conserved LXD motif, it is possible that these properties may also extend to S proteins from other coronavirus strains.
Alveolar macrophages are abundant in the lungs where they play a central role as a first-line defense against various pathogens (35) including SARS-CoV-2 (36, 37). Estrogens are not only responsible for the maturation and proper functioning of the female reproductive system but also play important roles in immunity (32, 38–41). In particular, ERα signaling in alveolar macrophages is considered a key component of the immune response to infection (42–44). We observed ER-dependent biological effects of S in the RAW264.7 macrophage cell line. Whereas ERα is mainly localized to the cell nucleus in MCF-7 cells (45), we found that ERα showed cytoplasmic localization in MCF-7 cells transfected with S DNA.
We observed a similar ectopic localization pattern in lung cells, especially in alveolar macrophages from SARS-CoV-2–infected hamsters and humans. More specifically, we found that cytoplasmic ERα colocalized at the surface of SARS-CoV-2 virions within alveolar macrophages, confirming that direct S-ERα interactions occur in the context of SARS-CoV-2 infection in this cell type. Our results, together with prior findings, suggest that S-ERα interactions in alveolar macrophages may play a critical role in SARS-CoV-2 infection and COVID-19 pathology.
We observed an apparent discrepancy between the effect of S on ERα cytosolic localization and E2-mediated biological effects such as E2-induced ER transcription, MCF-7 cell proliferation, and osteoclast differentiation. ER signaling is complex and can have contrasting biological effects across different cell types. For this reason, we interpret here the effect of S-ERα cytosolic colocalization as a modulator and not necessarily an inhibitor of ER signaling. In regard to the effects of S on MCF-7 cell proliferation specifically, we believe that the most plausible explanation for this effect is that S-ERα cytosolic localization may result in a potentiation of membrane-bound ERα signaling either by a direct S/membrane-bound ERα or, more likely, as an indirect consequence of an imbalance in the finely tuned ER-mediated hormone response. In line with this hypothesis, although membrane ERs have lost the capacity to bind to specific response elements or transcription initiation complexes onto DNA, the rapid membrane activation of these receptors stimulates DNA synthesis and cell proliferation (46). In addition, the membrane-bound ER structures maintain the AF-2 transcriptional activation domain (47) that we proposed to be the S binding site on ERs. Last, membrane ER activation is inhibited by the same synthetic ER antagonists that block transcriptional activation by the classically described ERs (48).
One of the most frequently reported COVID-19 epidemiologic findings is sex-related mortality and specifically male-related susceptibility. The evidence to date supports a higher predominance of men in several countries; thus, the male sex has been considered a poor prognostic factor (49). In line with these reports, male laboratory animals are more susceptible to SARS-CoV and SARS-CoV-2 infection and related pathology as compared to females (31, 50, 51). ER signaling contributes to these sex differences (45, 51), and the potential protective effects of estrogens in COVID-19 have been widely debated in the literature (52), although a recent study showed that E2 treatment did not alleviate lung complications in SARS-CoV-2–infected male hamsters (51). Accordingly, here, we focused our efforts on male hamsters due to their known higher susceptibility to SARS-CoV-2 infection compared to females. Nevertheless, future studies will aim to address this limitation by examining whether SARS-CoV-2 infection leads to similar effects in female hamsters as those observed here in males. Notably, sex-based differences have been reported in various chronic inflammatory responses associated with lung disease (53) and, specifically, as a function of ER signaling in activated macrophages (42). Whereas circulating estrogens play a protective role by regulating both the innate and adaptive immune response to infection (54), it may be possible that the modulation of ER signaling in SARS-CoV-2–infected lung tissue may stimulate proinflammatory signals leading to hypertrophy, vasoconstriction, and vessel obstruction. As compared to female patients, hyperactivation of ER signaling in pulmonary tissue in males has been associated with lower frequency but more severe progression of vascular obliteration in pulmonary arterial hypertension (53). This model could also potentially explain the widely discussed effect of ER modulation in SARS-CoV-2 infection and the reported protective effect of antiestrogenic treatment on COVID-19 prevalence in women with ovarian and breast cancer (4).
In conclusion, we report novel interactions between the SARS-CoV-2 S and ERα that may have important therapeutic implications for COVID-19. Our results also highlight the use of multimodal PET/CT imaging and the Food and Drug Administration–approved [18F]FES radiopharmaceutical as a translational approach and biomarker for the longitudinal assessment of COVID-19 lung pathology.
MATERIALS AND METHODS
SARS-CoV-2 S radiolabeling
To an Eppendorf vial was added 0.25 M phosphate buffer (PB; 80 μl) (pH 7.5), SARS-CoV-2 S(R683A, R685A), His-Tag (20 μg; ACROBiosystems, #SPN-C52H4) in water, lactoperoxidase (2 μg), Na125I (0.7 mCi), and H2O2 (0.4 × 103%). After incubation for 60 min at 35°C, the reaction was quenched with ascorbic acid (0.1 mg). The mixture was allowed to stand for 10 min, and then bovine serum albumin (BSA; 3 mg) was added. The mixture was then applied to a G-25 desalting column (GE Healthcare) to separate the radioiodinated S from unreacted radioiodine. Approximately 0.2 mCi of product was obtained. The purified radiolabeled protein was formulated with 1% BSA and 10% sucrose, divided into aliquots, and stored at −20°C.
[125I]S radioligand binding assays
For saturation assays, the radioligand specific activity was adjusted with unlabeled peptide to enable a radioligand concentration range appropriate for the KD of the receptor. For plate preparation, Protein A–coated plates (Thermo Fisher Scientific, catalog no. 15130) were washed with wash buffer [50 mM tris, 5 mM MgCl2, and 0.1 mM EDTA (pH 7.4)] and incubated with human ACE2 Fc Tag (0.2 μg per well; ACROBiosystems, #AC2-H5257) in incubation buffer [50 mM tris, 5 mM MgCl2, 0.1 mM EDTA, and 2% BSA (pH 7.4)] for 60 min with gentle shaking. The plates were then washed with wash buffer (4 × 0.3 ml) followed by high salt wash buffer [50 mM tris, 5 mM MgCl2, 0.1 mM EDTA, and 125 mM NaCl (pH 7.4)] and used directly for the binding assay. For incubation and filtration, 75 ml of buffer, 25 ml of the unlabeled protein (for competition assays) or buffer, and 25 ml of radioligand solution in binding buffer were added to each well. The plate was incubated at room temperature (RT) for 60 min with gentle agitation. The incubation was stopped by washing the wells with (i) incubation buffer (1 × 0.3 ml, ice cold), (ii) wash buffer (three washes, ice cold), and (iii) high-salt wash buffer (one wash, ice cold). Following washing, NaOH (0.1 M) was added to each well, and the plates were incubated at 40°C for 1 hour to digest the protein. Following digestion, the radioactivity was transferred to a counting plate, neutralized and scintillation cocktail (BetaPlate Scint, PerkinElmer) was added, and the radioactivity was counted in a Wallac TriLux 1450 MicroBeta counter. For each concentration of radioligand, nonspecific binding was subtracted from total binding to give specific binding. Nonspecific binding was determined using wells incubated without ACE2. For saturation assays, the bound radioactivity (in counts per minute per well) was converted to molar amounts (in femtomoles per well) from the specific activity of the radioligand. A counting efficiency for 125I of 71% was used for counts per minute to disintegrations per minute calculations. Data were fitted using the nonlinear curve fitting routines in Prism (GraphPad Software Inc.).
ProtoArray
The Invitrogen ProtoArray Human Protein Arrays (Thermo Fisher Scientific) are high-density microarrays that contain more than 9000 unique human proteins individually purified and arrayed onto a nitrocellulose-coated slide. We followed the manufacturer’s instructions to probe the arrays for small tritiated molecules. Briefly, protein microarrays were blocked for 30 to 40 min in blocking buffer [50 Hepes, 250 NaCl, 20 glutathione, 1 dithiothreitol, 1% (or 2%) BSA, and 0.1% Tween 20]. The blocking buffer was then gently aspirated off and replaced with incubation buffer [phosphate-buffered saline (PBS), 0.1% Tween 20, 1% (or 2%) BSA, and with or without 1 nM 3H-E2] containing the radioligand (20 nM [125I]S). To determine nonspecific binding, 300 nM S was added to the incubation mix. Every condition was tested in duplicate. After incubation, slides were washed three times in ice-cold washing buffer (PBS and 0.1% Tween 20) and rinsed with ice-cold distilled water. Slides were then air-dried and placed into a Hypercassette and covered by a tritium-sensitive phosphor screen (GE Healthcare), exposed for 1 day, and then scanned on a PerkinElmer Cyclone scanner. The digitized images were also analyzed using ProtoArray Prospector v5.2, and potential hits were identified using the software’s algorithm.
SPR using immobilized SARS-CoV-2 S
SPR measurements were performed using a Biacore apparatus (Biacore) using CM5 sensor chips. To find out the optimal pH for S (ACROBiosystems) immobilization, we conducted pH scouting. The S was prepared in 10 mM sodium acetate buffer at pH 4.0 to 5.5. The best pH for immobilization was 4.0 (fig. S10A). After covalent immobilization, there was approximately 8500 Response units (RUs) of S on the sensor surface (fig. S10B). Increasing concentrations of full-length ERα (Invitrogen), ACE2 (ACROBiosystems), and NRP1 (ACROBiosystems) from 1.56 to 200 nM were injected. Protein binding responses were analyzed using BIAeval software. All curves were globally fitted to a single-site binding model to determine an approximate fit. The chi-square value was noted to indicate the goodness of fit. The remaining data were refitted to the single-site binding model and the improvement in fit (reduction in chi-square value) noted. Kon, Koff, and KD were reported by the global fit model. These data were also fitted to a two-site binding model to determine whether this gave a better fit (indicated by a lower chi-square value than the single-site model). For the equilibrium model, binding response amplitudes were fitted to a saturation binding curve, from which the Rmax and KD (concentration at half Rmax) were determined.
Interactome analysis
The STRING database (55), which integrates all known and predicted associations between proteins, including both physical interactions and functional associations, has been used to analyze functional associations between biomolecules. Each protein-protein interaction is annotated with a “score.” This score does not indicate the strength or the specificity of the interaction but the confidence. All scores rank from 0 to 1, with 1 being the highest possible confidence.
3D model selection and MD simulation protocol
S 3D model was built on the basis of PDB 6VYB returned to its WT form and fully glycosylated (56). An asymmetric glycosylation of the three protomers has been derived by glycoanalyitic data for the N-glycans and O-glycans according to the work of Tagliamonte et al. (57) and Casalino et al. (58). For the ER, the x-ray PDB model with code 3UUD was used, containing ERα and NCOA223, and code 3OLL, containing ERβ and NCOA124. The proteins were modeled using Amber14SB force field (59) and the carbohydrate moieties by the GLYCAM06j-1 version of GLYCAM06 force field (60), and the general amber force field (61) was used for the E2 bound to ER receptor. The so-prepared structure was used as starting point for MD simulations. Protein was inserted in a clinic box, extending up to 10 Å from the solute, and immersed in TIP3P water molecules (62). Counterions were added to neutralize the overall charge with the genion GROMACS tool. After energy minimizations, the system was relaxed for 5 ns by applying positional restraints of 1000 kJ/mol per square nanometer to the protein atoms. Following this step, unrestrained MD simulation was carried out with a time step of 2 fs, using GROMACS 2020.2 simulation package (supercomputer Marconi-100, Cineca, Bologna, Italy) (63). V-rescale temperature coupling was used to keep the temperature constant at 300 K (64). The particle-mesh Ewald method was used for the treatment of the long-range electrostatic interactions (65). The first 5 ns of each trajectory were excluded from the analysis. The trajectory obtained after 1-ms MD simulation has been clustered to obtain representative structures. In particular, the structure used for the docking studies is the first centroid of the first cluster extracted from the MD experiment. For the ER, the x-ray PDB model with code 3OLL was used, containing E2 and NCOA1 (24).
Protein-protein docking procedure
The input of two individual proteins, one for receptor and the other for ligand, was provided. In particular, the S and ER were used as receptor and ligand, respectively. Then, the HDOCK tool will perform docking to sample putative binding modes through a fast Fourier transform–based search method and then scoring the protein-protein interactions. Last, the top 100 predicted complex structures are provided, and the best 10 hypotheses were visually inspected to confirm the reliability of the calculation. The entire workflow is well described in the work published by Yan et al. (26).
ERα reporter gene assays
Peptide transfection
GeneBLAzer ERα-UAS-bla GripTite (human embryonic kidney 293 ERα-bla, Invitrogen, Carlsbad, CA, USA) cells comprise a mammalian one-hybrid system stably expressing a β-lactamase reporter gene under the control of the GAL4 DNA binding site and a fusion protein consisting of the human ERα ligand-binding domain and the GAL4 DNA binding domain. The cells were plated at 8000 cells per well in black-wall/clear-bottom plate. After cultured overnight, cells were treated with peptide delivery reagent, 20 mM Hepes containing peptides, and PULSin reagent (Polyplus-transfection, NY USA), for 4 hours in DMEM supplemented with 1 μM nonessential amino acid (NEAA), penicillin (100 U/ml), and streptomycin (100 μg/ml). Then, the medium was replaced with phenol red–free DMEM supplemented with 2% charcoal-stripped fetal bovine serum (FBS), 1 μM NEAA, penicillin (100 U/ml), streptomycin (100 μg/ml), and 10 μM sodium pyruvate.
S transfection
One million cells were plated into each well of six-well plates in 2 ml of DMEM supplemented with 10% FBS, 1 μM NEAA, penicillin (100 U/ml), and streptomycin (100 μg/ml). On the next day, the medium was replaced with 2 ml of DMEM supplemented with 10% FBS. Then, the cells in each well were transfected with 2.5 μg of different S plasmids with Lipofectamine 3000. After 24 hours of incubation, cells were detached from six-well plate using tryptophan and plated at 8000 cells per well in black-wall/clear-bottom 384-well plate. Transfected cells were treated with E2 for 18 hours. The next day, 8 μl of LiveBLAzer (Life Technologies, Madison, WI) detection mixture was added to each well, and the plates were incubated at RT in the dark for 2 hours. Fluorescence intensity at 460- and 530-nm emission and 405-nm excitation was measured by an PHERAstar plate reader (BMG LABTECH, Cary, NC). Data were represented as the ratio of the emission wavelengths (460 nm/530 nm).
Human ERα transcriptional activation assays
The ERα activity was determined by the ERα transcription factor activation assay kit (Abcam, ab207203) according to the manufacturer’s directions. Briefly, MCF-7 nuclear extracts (5 μg; Abcam, ab14860) were treated with S (0.01 to 300 nM; ACROBiosystems), S-RBD (1 to 100 nM; ACROBiosystems), S-trimer (1 to 100 nM; provided by M. Borgnia), and/or E2 (100 nM: Tocris). Extracts were added to each well coated with the ER consensus binding site (5′-GGTCACAGTGACC-3′). The wells were washed and then incubated with rabbit anti-ERα (1:2000; 1 hour at RT) and horseradish peroxidase (HRP)–conjugated secondary antibody (1:2000; 1 hour at RT) that were provided with the kit. Colorimetric reaction was measured by spectrophotometry at a wavelength of 450 nm. Values were given as percentage of the effect induced by E2.
Immunofluorescence
About 80,000 MCF-7 cells were placed in each chamber of a four-well chamber slide (Thermo Fisher Scientific, catalog no. 177399) containing 500 μl of DMEM + 10% FBS and cultured overnight at 37°C in a 5% CO2 incubator. The next day, cells in each well were transfected with 1.5 μl of ViaFect reagent (Promega, catalog no. E498A) and 0.5 μg of empty pcDNA3.1 vector or an expression vector for the WT SARS-CoV2 S with a C-terminal hemagglutinin (HA) epitope tag (pBOB-CAG-SARS-CoV2-S-HA) or the double-mutant (R682S, R685S) SARS-CoV2 S with a C-terminal flag epitope tag (pCAGGS-SARS2-S-FKO). pBOB-CAG-SARS-CoV2-S-HA was a gift from G. Pao (Addgene plasmid #141347; http://n2t.net/addgene:141347; RRID: Addgene 141347). pCAGGS-SARS2-S-FKO (C-flag) was a gift from H. Choe and M. Farzan (Addgene plasmid #159364; http://n2t.net/addgene:159364; RRID: Addgene 159364). After 48 hours, the cells were fixed in 4% formaldehyde for 20 min at RT, rinsed with 1× PBS, permeabilized with 0.1% Triton X-100 for 25 min, rinsed with PBS, and incubated for 2 hours in blocking buffer (Rockland Immunochemicals Inc., catalog no. MB-070). The cells were then incubated at 4°C overnight with 2 μg/ml each of anti-ERα (H222) rat immunoglobulin G1 (IgG1) monoclonal antibody (mAb; Santa Cruz Biotechnology, sc-5349; 1:100) and HA-probe (F-7) mouse IgG2a mAb (Santa Cruz Biotechnology, sc-7392X; 1:1000), OctA-probe (anti-flag) mouse IgG1 mAb (Santa Cruz Biotechnology, sc-51590; 1:50), or normal mouse IgG (Santa Cruz Biotechnology, catalog no. sc-2025; 1:200) as a negative control. Afterward, the cells were washed four times with PBS + 0.1% Tween 20 (PBS-T) for 5 min and incubated at RT for 1 hour in the dark with a fluorescent secondary antibody mixture contaning mouse IgGκ BP-CFL594 (Santa Cruz Biotechnology, sc-516178; 1:100) and anti-rat IgG Alexa Fluor 488 (Thermo Fisher Scientific, catalog no. A-11006; 1:500). The cells were then washed four times with PBS-T for 5 min in the dark and rinsed with PBS. Each slide was carefully detached from its gasket and immediately mounted with a 1.5-T glass coverslip using EverBrite Hardset mounting medium with 4′,6-diamidino-2-phenylindole (DAPI; Biotium, catalog no. 23004). The mounted slides were allowed to cure for 24 hours in the dark at RT and stored in a slide box at 4°C. The slides were imaged at 63× magnification using a Zeiss LSM 880 Airyscan inverted confocal microscope (Max Planck Florida Institute for Neuroscience). Each image represents the average of 16 scans. Images were prepared for presentation using ImageJ v.1.53c software [National Institutes of Health (NIH)].
Proliferation assays
MCF-7 and MDA-MB-23 cells were obtained from American Type Culture Collection (ATCC) and growth in DMEM without phenol red, supplemented with 10% FBS and penicillin/streptomycin at 37°C in a 5% CO2 and 95% humidified atmosphere. For each assay, cells were seeded at the density of 104 cells/cm2. Before treatments, to reduce estrogen levels in FBS and avoiding any interference, cells were cultured for 24 hours in medium containing 5% dextran-coated charcoal-treated serum. Then, cells were treated for 24 hours with E2 (Sigma-Aldrich, catalog no. E1024, batch: SLCC8875), S (R&D Systems, catalog no. 1059-CV, batch: DODR0220111), raloxifene (Sigma-Aldrich, catalog no. R1402, batch: MKCJ7180), S + raloxifene, E2 + S, and E2 + raloxifene + S. In particular, the concentration tested for S was 10 ng/ml, that for raloxifene was 2 μM, that for E2 was 1 nM, and that for fulvestrant (ICI 182,780) was 1 μM.
Cell proliferation was measured using a 5-bromo-2-deoxyuridine (BrdU) labeling and a proliferation ELISA kit (Abcam, ab126556) following the manufacturer’s instructions. Briefly, BrdU was added to wells for 24 hours, and then cells were fixed using fixing solution. Then, cells were washed and incubated with detector anti-BrdU antibody for 1 hour at RT. After the incubation, cells were washed and incubated with the HRP-conjugated goat anti-mouse antibody for 30 min at RT. For the detection, the chromogenic substrate tetramethylbenzidine was added, and the colored product has been detected using a spectrophotometer (450/550 nm). Values were given as percentage of cells grown only in serum-free medium. At least two independent assays were performed with eight duplicates each.
Tartrate-resistant acid phosphatase activity by ELISA assay in RAW264.7-osteoclasts
RAW264.7 (murine macrophages, ATCC, USA) were cultured as per the manufacturer’s protocol and grown in DMEM without phenol red, supplemented with 10% FBS and penicillin/streptomycin at 37°C in a 5% CO2 and 95% humidified atmosphere. Then, 1.5× 105 cells/cm2 in 24-well dishes were seeded, and mouse RANKL (Miltenyi Biotec, Germany) was added at the final concentration of 35 ng/ml to initiate osteoclasts development (day 0) as previously described (66). At day 3, cells were examined under the microscope and refed with fresh medium containing RANKL. At day 6, RAW264.7-osteoclast population was prevalent and ready for treatments and then biochemical studies. Before treatments, to reduce estrogen levels in FBS and avoiding any interference, cells were cultured for 24 hours in medium containing 5% dextran-coated charcoal-treated serum. Cells were treated with E2 (1 nM), S (10 ng/ml), and raloxifene (2 μM) and the combination of them for 24 hours. After 24 hours of treatment, we quickly collected the cells by sterile tubes and resuspended the cells using PBS (pH 7.4) to dilute cell suspension to the concentration of approximately 1 million per milliliter. Then, cells were subjected to repeated freeze-thaw cycles to let out the inside components. In the meantime, the reagents of the kit were brought to RT.
Tartrate-resistant acid phosphatase (TRAP) activity was performed using an ELISA kit from MyBioSource (MBS1601167). The standard curve, reagents, and samples were prepared following the manufacturer’s protocol. Briefly, 50 μl of standard was added to standard wells and 40 μl of sample to sample wells and then added 10 μl of anti-TRAP antibody to sample wells and 50 μl of streptavidin-HRP to sample wells and standard wells. The plate was incubated 1 hour at 37°C. The plate was washed five times with wash buffer, and 50 μl of substrate solution A was added to each well plus 50 μl of substrate solution B and incubated 10 min at 37°C in the dark. Last, 50 μl of stop solution to each well was added, and the optical density was immediately determined using a microplate reader set at 450 nm.
Fluorescence-based assay for ACE2 in MCF-7 cells
Cells were obtained from ATCC and grew in DMEM without phenol red, supplemented with 10% FBS and penicillin/streptomycin at 37°C in a 5% CO2 and 95% humidified atmosphere. For each assay, cells were seeded at the density of 104 cells/cm2. Before treatments, to reduce estrogen levels in FBS and avoiding any interference, cells were cultured for 24 hours in medium containing 5% dextran-coated charcoal-treated serum. Then, cells were treated for 24 hours with E2, S, raloxifene, S + raloxifene, E2 + S, and E2 + raloxifene + S. In particular, the concentration tested for S was 10 ng/ml, that for raloxifene was 2 μM, and that for E2 was 1 nM.
Cells were cultured in a 96-well plate. Cells were washed in ice-cold PBS and then fixed with 2% formalin solution in PBS for 15 min. After further washes, to prevent the nonspecific binding, cells were blocked with a 10% BSA solution in PBS for 20 min and then incubated with the Alexa Fluor 647–conjugated antibody for human ACE-2 (R&D Systems). After several washes, the plate was read at the fluorescence intensity of 668 nm using a microplate reader (Spark, Tecan). Then, to normalize the results, DAPI was added, and the fluorescence intensity at 461 nm was evaluated.
ACE2 expression in Calu-3 cells
Calu-3 cell line was obtained from ATCC and maintained in Eagle’s minimum essential medium (Lonza) supplemented with 10% FBS, 1% l-glutamine, and 1% penicillin/streptomycin solution at 37°C in a humidified atmosphere of 5% CO2.
RNA extraction and quantitative reverse transcription polymerase chain reaction
Cells were seeded in six-well plates and, after incubation, were treated according to the experimental protocol with E2 (200 nM), raloxifene (20 μM), and S (10 ng/ml). Total RNA was extracted from cell lines using RNeasy Plus Mini Kit (QIAGEN). cDNA was made using GenePro thermal cycler (Bioer). Reverse transcription polymerase chain reaction (PCR) analysis was performed on an Applied Biosystem QuantStudio 5 Real-Time PCR system (Thermo Fisher Scientific) using Itaq Universal SYBR (Bio-Rad) gene expressions assays. Primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH; forward primer, AATCCCATCACCATCTTCCA; reverse primer, TGGACTCCACGACGTACTCA) and ACE2 (forward primer, AAAGTGGTGGGAGATGAAGC; reverse primer, GAGATGCGCGGTCACAGTAT) were used. Samples were assayed in runs that were composed of three stages: hold stage at 95°C for 20 min; PCR stage at 60°C for 25 min; and melt curve stage at 95°C for 1 min, 60°C for 20 min, and 95°C for 1 min again. Gene expressions were normalized by GAPDH levels using the 2−ΔΔCt method.
Immunocytochemistry assay
Cells were seeded on glass coverslips precoated with collagen in 24-well plates. After incubation at 37°C, cells were treated according to the experimental protocol with E2 (200 nM), raloxifene (20μM), and S (10 ng/ml). After 72 hours, cells were washed, fixed with 4% formaldehyde, permeabilized with 0.1% Triton X-100 in PBS, and stained overnight at 4°C with ACE2 protein–specific antibody (Abcam, Ab15348). Cells were then incubated with anti-rabbit secondary antibody (Alexa Fluor 536 anti-rabbit, Invitrogen Life Technologies) for 1 hour at 37°C. Nuclei were labeled with Hoechst 33342 (Thermo Fisher Scientific) for nuclear staining for 20 min. Cells were mounted with Fluor mount (Sigma-Aldrich, St. Louis, MO, USA), and images were acquired through confocal microscope LSM 800, 60× magnification, software ZEN 2.1 blue edition (Carl Zeiss, Jenza, Germany) and analyzed with ImageJ software.
PET imaging
Animal studies
All animal protocols were approved by the Johns Hopkins University Biosafety, Radiation Safety, and Animal Care and Use Committees. Male golden Syrian hamsters (7 to 8 weeks of age) were purchased from Envigo (Haslett, MI). Animals were housed under standard housing conditions in positive/negative control cages (Alentown, NJ) in animal biological safety level 3 (ABSL-3) facility at the Johns Hopkins University–Koch Cancer Research Building. After 1 to 2 weeks of acclimation, animals were inoculated with 1.5 × 105 TCID50 of SARS-CoV-2 USA-WA1/2020 in 100 μl of DMEM (50 μl per nostril) through the intranasal route under ketamine (60 to 80 mg/kg) and xylazine (4 to 5 mg/kg) anesthesia administered intraperitoneally, as previously described (51). Control animals received an equivalent volume of DMEM.
Imaging
Hamsters were imaged inside in-house developed, sealed biocontainment devices compliant with ABSL-3. A cohort of male noncastrated hamsters was imaged longitudinally 1 day before SARS-CoV-2 infection and 7 days after infection using [18F]FES (n = 4 to 5). A second cohort of SARS-CoV-2–infected male hamsters was intravenously coinjected with [18F]FES and E2 (0.3 mg/kg; 3% dimethyl sulfoxide; Sigma-Aldrich) on day 7 after infection (n = 4). Each animal was injected 16.1 ± 1.5 MBq of [18F]FES intravenously via the penile vein. A 90-min PET acquisition and subsequent CT were performed using the nanoScan PET/CT (Mediso, Arlington, VA). For each animal, 8 to 13 volumes of interest (VOIs) were manually selected using CT as a guide and applied to the PET dataset using VivoQuantTM 2020 (Invicro, Boston, MA) for visualization and quantification. A VOI was placed on the left ventricle of the heart to measure the blood uptake. [18F]FES PET activity was calculated for each hamster (n = 4 to 5 hamsters per group) as the average activity of all VOIs normalized by the mean standardized uptake value (SUVmean). All animals were euthanized 110 min after injection, and the lung was harvested to quantify associated radioactivity using an automated γ-counter. Heatmap overlays were implemented using RStudio version 1.2.1335 (R Foundation) as previously described (67). Multiple comparisons were performed using two-way repeated-measures analysis of variance, followed by Bonferroni’s multiple-comparison test.
Hamster histology
In hamster lung sample preparation for confocal and electron microscopy, we perfused uninfected and infected hamsters with heparin solution (1000 U/ml), followed by fixative solution [4% paraformaldehyde (PFA), 0.15% glutaraldehyde, and 15% picric acid solution in 0.1 M PB (pH 7.4)] or fixative solution [4% PFA and 15% picric acid solution in 0.1 M PB (pH 7.4)]. After perfusion, we kept the lungs in the same fixative solution at 4°C for another 2 hours. We placed the lungs in 2% PFA fixative solution and postfixed the lungs at 4°C overnight. After rinsing in 0.1 M PB, serial sections (50 μm) were cut with a vibratome (VT1000S, Leica Microsystems Inc).
IHC staining in hamster tissue
The vibratome lung sections were rinsed and incubated for 1 hour in 0.1 M PB supplemented with 4% BSA and 0.3% Triton X-100. Sections were then incubated with cocktails of primary antibodies: rabbit anti-SARS-CoV-2 S protein (1:100; Invitrogen, #MA5-36087) + rat anti-ERα H222 (1:100; Santa Cruz Biotechnology, #sc53492) overnight at 4°C. After rinsing 3 × 10 min in PB, sections were incubated in a cocktail of the corresponding fluorescence secondary antibodies: Alexa Fluor 594–donkey anti-rabbit (Jackson ImmunoResearch Laboratories, 711-585-152) + Alexa Fluor 488–donkey anti-rat (Jackson ImmunoResearch Laboratories, 712-545-153) for 2 hours at RT. After rinsing, sections were mounted on slides. Fluorescent images were collected with a Zeiss LSM 880 with Cy7.5 Confocal System (Zeiss). Images were taken sequentially with different lasers with 20× objectives. Images were quantified with the “cell counter” plug-in in ImageJ (NIH).
Electron microscopy
The vibratome lung sections were rinsed and incubated with 1% sodium borohydride to inactivate free aldehyde groups, rinsed, and then incubated with blocking solution. Sections were then incubated with the primary antibodies rat anti-ERα H222 (1:100; Santa Cruz Biotechnology, #sc53492), diluted in 1% normal goat serum, 4% BSA, and 0.02% saponin in PB at 4°C overnight. Sections were rinsed and incubated overnight at 4°C in the secondary antibody Nanogold-Fab′ goat anti-rat-IgG (1:100; Nanoprobes, #2008) for ERα protein detection. Sections were rinsed in PB, and then sections were postfixed with 1.5% glutaraldehyde for 10 min and rinsed in PB and double-distilled water, followed by silver enhancement of the gold particles with the Nanoprobe Silver Kit (Nanoprobes, 2012) for 7 min at RT. Sections were rinsed with PB and fixed with 0.5% osmium tetroxide in PB for 25 min, washed in PB, followed by double-distilled water, and then contrasted in freshly prepared 1% uranyl acetate for 35 min. Sections were dehydrated through a series of graded alcohols and with propylene oxide. Afterward, they were flat embedded in Durcupan ACM epoxy resin (Electron Microscopy Sciences, 14040). Resin-embedded sections were polymerized at 60°C for 2 days. Sections of 60 nm were cut from the outer surface of the tissue with an ultramicrotome UC7 (Leica Microsystems) using a diamond knife (Diatome). The sections were collected on formvar-coated single-slot grids and counterstained with Reynold’s lead citrate. Sections were examined and photographed using a Tecnai G2 12 transmission electron microscope (Thermo Fisher Scientific) equipped with the OneView digital micrograph camera (Gatan). Alveolar macrophages were identified by three known morphological characteristics from electron microscopy experiments: (i) a large single-lobe nucleus, (ii) phagolysosomes inside the cytoplasm, and (iii) pseudo-podium stretched out from cell membrane (68).
Serial IHC staining in human tissue
Formalin-fixed and paraffin-embedded human breast and human COVID-19 lung tissues were achieved from Pathology Department of Johns Hopkins Medicine under an Institutional Review Board–approved protocol. The tissue slides were deparaffinized with xylene and rehydrated with gradient concentrations of ethanol and boiled in a high-pressure cooker with a citrate buffer (Bio SB, catalog no. BSB 0032) for 15-min retrieval. Then, slides were subjected to the serial IHC. On day 1, slides were blocked with a peroxidase blocker (Bio SB, catalog no. BSB 0054), washed with an immunoDNA washer buffer (Bio SB, catalog no. BSB 0150), and then incubated with anti-SARS-CoV-2 S glycoprotein antibody (0.2 μg/ml; Abcam, catalog no. ab272504) for 1 hour. After three washes, the Mouse/Rabbit PolyDetector Plus link & HRP label (Bio SB, catalog no. BSB 0270) was applied. The AEC-red chromogen (Vector Laboratories, catalog no. SK-4205) and a hematoxylin solution were used for color development and countered stain. The slides were mounted with a VectaMount AQ Aqueous Mounting medium (Vector Laboratories, catalog no. H-5501) and scan at 20× and 40× magnification using a MoticEasyScan Pro 6 (Meyer Instruments Inc., Houston TX). After scan, the slides were incubated in a PBS buffer for coverslip detachment. On day 2, the slides of detached coverslip were dehydrated in 70% ethanol, decolorized in 90% ethanol for 10 min, and rehydrated with 70% ethanol and distilled H2O. The decolorized slides were stripped with an antibody elution buffer [0.2% SDS, 62.5 mM tris-HCI (pH 6.8), 5% glycerol, and 0.08% β-mercaptoethanol] within a 130°C oven for 20 min (until boiling). The slides were washed with distilled H2O three times, each 15 min; washed with wash buffer twice, each 10 min; and then incubated in PBS buffer for 10 min. After antibody elution, an estrogen-α antibody (Abcam, catalog no. ab108398) at 1:250 work solution was applied for 90 min and then followed previous procedure for stain development. Last, the slides were mounted and scanned for data collection. Macrophages were identified using four known morphological characteristics from immunohistochemical experiments: (i) They comprised discrete cells, (ii) with large pale nuclei with nucleoli and (iii) a “foaminess” to the cytoplasm, and (iv) they were localized within alveolar walls in the lungs.
Acknowledgments
We thank N. Urban (Max Planck Institute for Neuroscience), M. Borgnia (National Institute on Environmental Health Sciences), L. Shi (National Institute on Drug Abuse), S. Coletti (Chelonia Applied Science), M. Hall (National Center for Advancing Translational Sciences), and A. Grottesi (Department HPC, CINECA, via dei Tizii 6, 00185 Roma, Italy) for sharing reagents, technical advice, and discussions.
Funding: This work was supported by the National Institute on Drug Abuse (ZIA000069) to M.M.; the R01-AI153349, R01-AI161829-A1, and the Center for Infection and Inflammation, Johns Hopkins University to S.K.J.; the National Institute of Biomedical Imaging and Bioengineering P41 (EB024495) to M.G.P.; and the Intramural Research Programs of National Center for Advancing Translational Sciences, National Institutes of Health.
Author contributions: Conceptualization: M.A. and M.Mi. Methodology: O.S., A.R.B., D.I., C.T., C.A.R.-B., J.C.N., A.C., V.Ca., R.B., M.Mon., V.Co., S.B., I.G.P., K.F., M.B., A.N., G.C., M.O., X.Y., S.Z., J.B., M.G.P., J.E.H., M.X., W.Z., M.Mor., A.Z.R., K.W.N., S.K.J., and M.A. Investigation: O.S., A.R.B., D.I., C.T., C.A.R.-B., J.C.N., A.C., V.Ca., R.B., M.Mon., V.Co., S.B., I.G.P., K.F., M.B.. A.N., G.C., M.O., X.Y., S.Z., J.B., M.G.P., and J.E.H. Visualization: O.S., A.R.B., D.I., C.T., C.A.R.-B., and J.C.N. Supervision: M.X., W.Z., M.Mor., A.Z.R., K.W.N., S.K.J., M.A., and M.Mi. Writing—original draft: O.S., M.A., and M.Mi. Writing—review and editing: All co-authors.
Competing interests: M.M. has received research funding from AstraZeneca, Redpin Therapeutics, and Attune Neurosciences. All other co-authors declare that they have no competing interests.
Data and materials availability: All data are available in the main text or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Tables S1 to S3
REFERENCES AND NOTES
- 1.Zhou X., Cheng Z., Luo L., Zhu Y., Lin W., Ming Z., Chen W., Hu Y., Incidence and impact of disseminated intravascular coagulation in COVID-19 a systematic review and meta-analysis. Thromb. Res. 201, 23–29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitrovic M., Sabljic N., Cvetkovic Z., Pantic N., Zivkovic Dakic A., Bukumiric Z., Libek V., Savic N., Milenkovic B., Virijevic M., Vucinic V., Milosevic I., Pravdic Z., Suvajdzic N., Fareed J., Antic D., Rotational thromboelastometry (ROTEM) profiling of COVID-19 patients. Platelets 32, 690–696 (2021). [DOI] [PubMed] [Google Scholar]
- 3.La Vignera S., Cannarella R., Condorelli R. A., Torre F., Aversa A., Calogero A. E., Sex-specific SARS-CoV-2 mortality: Among hormone-modulated ACE2 expression, risk of venous thromboembolism and hypovitaminosis D. Int. J. Mol. Sci. 21, 2978 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montopoli M., Zorzi M., Cocetta V., Prayer-Galetti T., Guzzinati S., Bovo E., Rugge M., Calcinotto A., Clinical outcome of SARS-CoV-2 infection in breast and ovarian cancer patients who underwent antiestrogenic therapy. Ann. Oncol. 32, 676–677 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y., Liu Q., Guo D., Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 92, 418–423 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z., Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 11, 1620 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y., Wang Q., Zhou H., Yan J., Qi J., Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181, 894–904.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Müller M. A., Drosten C., Pöhlmann S., SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Letko M., Marzi A., Munster V., Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5, 562–569 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y., Yang C., Xu X.-F., Xu W., Liu S.-W., Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 41, 1141–1149 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F., Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 117, 11727–11734 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daly J. L., Simonetti B., Klein K., Chen K.-E., Williamson M. K., Antón-Plágaro C., Shoemark D. K., Simón-Gracia L., Bauer M., Hollandi R., Greber U. F., Horvath P., Sessions R. B., Helenius A., Hiscox J. A., Teesalu T., Matthews D. A., Davidson A. D., Collins B. M., Cullen P. J., Yamauchi Y., Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370, 861–865 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walls A. C., Park Y.-J., Tortorici M. A., Wall A., McGuire A. T., Veesler D., Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y., Zhang Z., Yang L., Lian X., Xie Y., Li S., Xin S., Cao P., Lu J., The MERS-CoV receptor DPP4 as a candidate binding target of the SARS-CoV-2 spike. iScience 23, 101160 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amendola A., Garoffolo G., Songia P., Nardacci R., Ferrari S., Bernava G., Canzano P., Myasoedova V., Colavita F., Castilletti C., Sberna G., Capobianchi M. R., Piacentini M., Agrifoglio M., Colombo G. I., Poggio P., Pesce M., Human cardiosphere-derived stromal cells exposed to SARS-CoV-2 evolve into hyper-inflammatory/pro-fibrotic phenotype and produce infective viral particles depending on the levels of ACE2 receptor expression. Cardiovasc. Res. 117, 1557–1566 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu J., Korteweg C., Pathology and pathogenesis of severe acute respiratory syndrome. Am. J. Pathol. 170, 1136–1147 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki Y. J., Nikolaienko S. I., Dibrova V. A., Dibrova Y. V., Vasylyk V. M., Novikov M. Y., Shults N. V., Gychka S. G., SARS-CoV-2 spike protein-mediated cell signaling in lung vascular cells. Vascul. Pharmacol. 137, 106823 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dosch S. F., Mahajan S. D., Collins A. R., SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-kappaB pathway in human monocyte macrophages in vitro. Virus Res. 142, 19–27 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shirato K., Kizaki T., SARS-CoV-2 spike protein S1 subunit induces pro-inflammatory responses via toll-like receptor 4 signaling in murine and human macrophages. Heliyon 7, e06187 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., Graham B. S., McLellan J. S., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allegretti M., Cesta M. C., Zippoli M., Beccari A., Talarico C., Mantelli F., Bucci E. M., Scorzolini L., Nicastri E., Repurposing the estrogen receptor modulator raloxifene to treat SARS-CoV-2 infection. Cell Death Differ. 29, 156–166 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McInerney E. M., Rose D. W., Flynn S. E., Westin S., Mullen T. M., Krones A., Inostroza J., Torchia J., Nolte R. T., Assa-Munt N., Milburn M. V., Glass C. K., Rosenfeld M. G., Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12, 3357–3368 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delfosse V., Grimaldi M., Pons J.-L., Boulahtouf A., le Maire A., Cavailles V., Labesse G., Bourguet W., Balaguer P., Structural and mechanistic insights into bisphenols action provide guidelines for risk assessment and discovery of bisphenol A substitutes. Proc. Natl. Acad. Sci. U.S.A. 109, 14930–14935 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Möcklinghoff S., Rose R., Carraz M., Visser A., Ottmann C., Brunsveld L., Synthesis and crystal structure of a phosphorylated estrogen receptor ligand binding domain. Chembiochem 11, 2251–2254 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Plevin M. J., Mills M. M., Ikura M., The LxxLL motif: A multifunctional binding sequence in transcriptional regulation. Trends Biochem. Sci. 30, 66–69 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Yan Y., Tao H., He J., Huang S.-Y., The HDOCK server for integrated protein-protein docking. Nat. Protoc. 15, 1829–1852 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Sadler T. M., Gavriil M., Annable T., Frost P., Greenberger L. M., Zhang Y., Combination therapy for treating breast cancer using antiestrogen, ERA-923, and the mammalian target of rapamycin inhibitor, temsirolimus. Endocr. Relat. Cancer 13, 863–873 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Fryar-Tita E. B., Das J. R., Davis J. H., Desoto J. A., Green S., Southerland W. M., Bowen D., Raloxifene and selective cell cycle specific agents: A case for the inclusion of raloxifene in current breast cancer treatment therapies. Anticancer Res 27, 1393–1399 (2007). [PubMed] [Google Scholar]
- 29.Hong S., Chang J., Jeong K., Lee W., Raloxifene as a treatment option for viral infections. J. Microbiol. 59, 124–131 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shevde N. K., Bendixen A. C., Dienger K. M., Pike J. W., Estrogens suppress RANK ligand-induced osteoclast differentiation via a stromal cell independent mechanism involving c-Jun repression. Proc. Natl. Acad. Sci. U.S.A. 97, 7829–7834 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz-Bedoya C. A., Mota F., Ordonez A. A., Foss C. A., Singh A. K., Praharaj M., Mahmud F. J., Ghayoor A., Flavahan K., De Jesus P., Bahr M., Dhakal S., Zhou R., Solis C. V., Mulka K. R., Bishai W. R., Pekosz A., Mankowski J. L., Villano J., Klein S. L., Jain S. K., 124I-Iodo-DPA-713 positron emission tomography in a hamster model of SARS-CoV-2 infection. Mol. Imaging Biol. 24, 135–143 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel H. K., Bihani T., Selective estrogen receptor modulators (SERMs) and selective estrogen receptor degraders (SERDs) in cancer treatment. Pharmacol. Ther. 186, 1–24 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Zella L. A., Chang C.-Y., McDonnell D. P., Pike J. W., The vitamin D receptor interacts preferentially with DRIP205-like LxxLL motifs. Arch. Biochem. Biophys. 460, 206–212 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aguilar-Pineda J. A., Albaghdadi M., Jiang W., Vera-Lopez K. J., Nieto-Montesinos R., Alvarez K. L. F., Davila Del-Carpio G., Gómez B., Lindsay M. E., Malhotra R., Lino Cardenas C. L., Structural and functional analysis of female sex hormones against SARS-CoV-2 cell entry. Int. J. Mol. Sci. 22, 11508 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joshi N., Walter J. M., Misharin A. V., Alveolar macrophages. Cell. Immunol. 330, 86–90 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Abassi Z., Knaney Y., Karram T., Heyman S. N., The lung macrophage in SARS-CoV-2 infection: A friend or a foe? Front. Immunol. 11, 1312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shieh W.-J., Hsiao C.-H., Paddock C. D., Guarner J., Goldsmith C. S., Tatti K., Packard M., Mueller L., Wu M.-Z., Rollin P., Su I.-J., Zaki S. R., Immunohistochemical, in situ hybridization, and ultrastructural localization of SARS-associated coronavirus in lung of a fatal case of severe acute respiratory syndrome in Taiwan. Hum. Pathol. 36, 303–309 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levenson A. S., Wolf D. M., Catherino W. H., Takei H., Jordan V. C., Understanding the antiestrogenic actions of raloxifene and a mechanism of drug resistance to tamoxifen. Breast Cancer 5, 99–106 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Pearce S. T., Jordan V. C., The biological role of estrogen receptors α and β in cancer. Crit. Rev. Oncol. Hematol. 50, 3–22 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Heldring N., Pike A., Andersson S., Matthews J., Cheng G., Hartman J., Tujague M., Ström A., Treuter E., Warner M., Gustafsson J.-A., Estrogen receptors: How do they signal and what are their targets. Physiol. Rev. 87, 905–931 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Taneja V., Sex hormones determine immune response. Front. Immunol. 9, 1931 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vegeto E., Cuzzocrea S., Crisafulli C., Mazzon E., Sala A., Krust A., Maggi A., Estrogen receptor-alpha as a drug target candidate for preventing lung inflammation. Endocrinology 151, 174–184 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuentes N., Pesantez J., Nicoleau M., Wal R. V., Chroneos Z., Silveyra P., Role of estrogen on alveolar macrophage polarization in response to particulate matter exposure. FASEB J. 33, 735.2 (2019). [Google Scholar]
- 44.Keselman A., Fang X., White P. B., Heller N. M., Estrogen signaling contributes to sex differences in macrophage polarization during asthma. J. Immunol. 199, 1573–1583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kocanova S., Mazaheri M., Caze-Subra S., Bystricky K., Ligands specify estrogen receptor alpha nuclear localization and degradation. BMC Cell Biol. 11, 98 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz N., Verma A., Bivens C. B., Schwartz Z., Boyan B. D., Rapid steroid hormone actions via membrane receptors. Biochim. Biophys. Acta 1863, 2289–2298 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Flouriot G., Brand H., Denger S., Metivier R., Kos M., Reid G., Sonntag-Buck V., Gannon F., Identification of a new isoform of the human estrogen receptor-alpha (hER-alpha) that is encoded by distinct transcripts and that is able to repress hER-alpha activation function 1. EMBO J. 19, 4688–4700 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russell K. S., Haynes M. P., Sinha D., Clerisme E., Bender J. R., Human vascular endothelial cells contain membrane binding sites for estradiol, which mediate rapid intracellular signaling. Proc. Natl. Acad. Sci. U.S.A. 97, 5930–5935 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mo P., Xing Y., Xiao Y., Deng L., Zhao Q., Wang H., Xiong Y., Cheng Z., Gao S., Liang K., Luo M., Chen T., Song S., Ma Z., Chen X., Zheng R., Cao Q., Wang F., Zhang Y., Clinical characteristics of refractory coronavirus disease 2019 in Wuhan, China. Clin. Infect. Dis. 73, e4208–e4213 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Channappanavar R., Fett C., Mack M., Ten Eyck P. P., Meyerholz D. K., Perlman S., Sex-based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. J. Immunol. 198, 4046–4053 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dhakal S., Ruiz-Bedoya C. A., Zhou R., Creisher P. S., Villano J. S., Littlefield K., Ruelas Castillo J., Marinho P., Jedlicka A. E., Ordonez A. A., Bahr M., Majewska N., Betenbaugh M. J., Flavahan K., Mueller A. R. L., Looney M. M., Quijada D., Mota F., Beck S. E., Brockhurst J.; Johns Hopkins COVID-19 Hamster Study Group , Sex differences in lung imaging and SARS-CoV-2 antibody responses in a COVID-19 golden syrian hamster model. MBio 12, e0097421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Al-Kuraishy H. M., Al-Gareeb A. I., Faidah H., Al-Maiahy T. J., Cruz-Martins N., Batiha G. E.-S., The looming effects of estrogen in covid-19: A rocky rollout. Front. Nutr. 8, 649128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ambhore N. S., Kalidhindi R. S. R., Sathish V., Sex-steroid signaling in lung diseases and inflammation. Adv. Exp. Med. Biol. 1303, 243–273 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nadkarni S., McArthur S., Oestrogen and immunomodulation: New mechanisms that impact on peripheral and central immunity. Curr. Opin. Pharmacol. 13, 576–581 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Szklarczyk D., Gable A. L., Nastou K. C., Lyon D., Kirsch R., Pyysalo S., Doncheva N. T., Legeay M., Fang T., Bork P., Jensen L. J., von Mering C., The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borocci S., Cerchia C., Grottesi A., Sanna N., Prandi I. G., Abid N., Beccari A. R., Chillemi G., Talarico C., Altered local interactions and long-range communications in UK variant (B.1.1.7) spike glycoprotein. Int. J. Mol. Sci. 22, 5464 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tagliamonte M. S., Abid N., Borocci S., Sangiovanni E., Ostrov D. A., Kosakovsky Pond S. L., Salemi M., Chillemi G., Mavian C., Multiple recombination events and strong purifying selection at the origin of SARS-CoV-2 spike glycoprotein increased correlated dynamic movements. Int. J. Mol. Sci. 22, 80 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Casalino L., Gaieb Z., Goldsmith J. A., Hjorth C. K., Dommer A. C., Harbison A. M., Fogarty C. A., Barros E. P., Taylor B. C., McLellan J. S., Fadda E., Amaro R. E., Beyond shielding: The roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 6, 1722–1734 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maier J. A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K. E., Simmerling C., ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kirschner K. N., Yongye A. B., Tschampel S. M., González-Outeiriño J., Daniels C. R., Foley B. L., Woods R. J., GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 29, 622–655 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J., Wolf R. M., Caldwell J. W., Kollman P. A., Case D. A., Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 63.Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., Lindahl E., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 (2015). [Google Scholar]
- 64.Bussi G., Donadio D., Parrinello M., Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Darden T., York D., Pedersen L., Particle mesh Ewald: AnN·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
- 66.Collin-Osdoby P., Osdoby P., RANKL-mediated osteoclast formation from murine RAW 264.7 cells. Methods Mol. Biol. 816, 187–202 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Ordonez A. A., Wang H., Magombedze G., Ruiz-Bedoya C. A., Srivastava S., Chen A., Tucker E. W., Urbanowski M. E., Pieterse L., Fabian Cardozo E., Lodge M. A., Shah M. R., Holt D. P., Mathews W. B., Dannals R. F., Gobburu J. V. S., Peloquin C. A., Rowe S. P., Gumbo T., Ivaturi V. D., Jain S. K., Dynamic imaging in patients with tuberculosis reveals heterogeneous drug exposures in pulmonary lesions. Nat. Med. 26, 529–534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aitken E. H., Negri E. M., Barboza R., Lima M. R. I., Álvarez J. M., Marinho C. R. F., Caldini E. G., Epiphanio S., Ultrastructure of the lung in a murine model of malaria-associated acute lung injury/acute respiratory distress syndrome. Malar. J. 13, 230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Tables S1 to S3