Abstract
All flaviviruses, including Zika virus, produce noncoding subgenomic flaviviral RNA (sfRNA), which plays an important role in viral pathogenesis. However, the exact mechanism of how sfRNA enables viral evasion of antiviral response is not well defined. Here, we show that sfRNA is required for transplacental virus dissemination in pregnant mice and subsequent fetal brain infection. We also show that sfRNA promotes apoptosis of neural progenitor cells in human brain organoids, leading to their disintegration. In infected human placental cells, sfRNA inhibits multiple antiviral pathways and promotes apoptosis, with signal transducer and activator of transcription 1 (STAT1) identified as a key shared factor. We further show that the production of sfRNA leads to reduced phosphorylation and nuclear translocation of STAT1 via a mechanism that involves sfRNA binding to and stabilizing viral protein NS5. Our results suggest the cooperation between viral noncoding RNA and a viral protein as a novel strategy for counteracting antiviral responses.
A mechanism by which Zika virus noncoding RNA enables viral evasion of antiviral response in vertebrates.
INTRODUCTION
Flaviviruses are small enveloped viruses with single-stranded positive sense RNA genomes (1). A unique feature of flavivirus infection is the production of noncoding RNA derived from viral 3′ untranslated regions (3′UTRs), which accumulates in infected cells in high abundance (2, 3). Generation of this RNA, termed subgenomic flaviviral RNA (sfRNA), is determined by highly conserved RNA elements in the 3′UTRs that resist degradation by the cellular 5′ → 3′ exoribonuclease XRN-1 (4, 5). Previously, sfRNA was shown to facilitate replication and pathogenesis of West Nile virus (WNV) (6) and dengue virus (DENV) (7–9). However, the molecular mechanism that underpins this activity is currently not well established (10). Furthermore, the role of sfRNA in the pathogenesis of other flaviviruses, including Zika virus (ZIKV), remains poorly defined.
ZIKV is a mosquito-borne pathogenic flavivirus capable of causing reoccurring severe outbreaks in human populations (11). Among flaviviruses, ZIKV is rather unique in its ability to disseminate through the placenta and establish infection in the fetal brain, resulting in neurodevelopmental abnormalities (12). Infected mothers subsequently have a high risk of giving birth to infants with microcephaly and other neurological disorders known collectively as congenital Zika syndrome (13). The 3′UTR of ZIKV contains two XRN-1–resistant RNA elements (xrRNAs) and is processed into two sfRNAs that differ in length (14). To determine the role of these RNAs in ZIKV infection, we recently generated sfRNA-deficient mutant viruses and used them to show that sfRNA facilitates ZIKV transmission by mosquitoes with a mechanism involving the inhibition of apoptosis in mosquito tissues, leading to increased virus dissemination and secretion into saliva (15). Here, we used this loss-of-function system to investigate the role of sfRNA during ZIKV infection of the vertebrate host.
Using a combination of in vivo, ex vivo, and in cellulo approaches, we demonstrated that sfRNA facilitates replication and pathogenesis of ZIKV and is required for virus-induced cytopathic effects (CPEs). We also found that sfRNA is crucial for viral dissemination into the fetal brain and apoptosis of infected neural progenitors. Furthermore, using transcriptome profiling and pathway enrichment analysis, we found that ZIKV sfRNA inhibits multiple pathways in the infected placental cells and identified signal transducer and activator of transcription 1 (STAT1) as a critical shared component. Further experiments revealed that the production of sfRNA caused a marked reduction in STAT1 phosphorylation and thus suppressed antiviral signaling by type I, II, and III interferons (IFNs). Mechanistically, we showed that inhibition of STAT1 phosphorylation by sfRNA required cooperation between sfRNA and the viral protein nonstructural protein 5 (NS5), whereby sfRNA bound to and stabilized NS5, leading to the accumulation of NS5 in the amounts required for efficient inhibition of STAT1 phosphorylation. Hence, we concluded that stabilization of NS5 by sfRNA is the key mechanism by which sfRNA exerts its functions in immune evasion and pathogenesis of ZIKV.
RESULTS
ZIKV sfRNA facilitates virus replication, determines CPE, and promotes viral pathogenesis
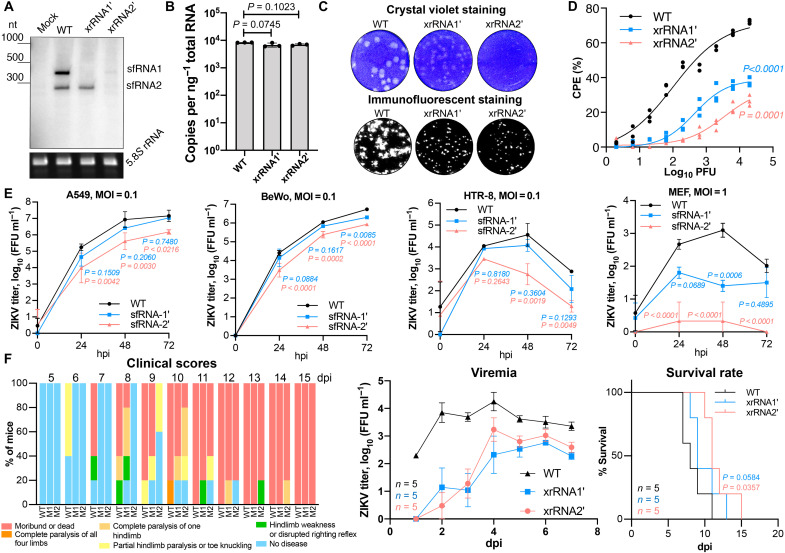
To elucidate the functions of ZIKV sfRNA, we previously introduced point mutations (xrRNA1′ and xrRNA2′) into each and both xrRNAs within the 3′UTR of the African ZIKV strain, ZIKVMR766 (15). In mosquito cells, individual mutations in xrRNA1 and xrRNA2 abrogated the production of sfRNA-1 and sfRNA-2, respectively, while virus containing mutations in both xrRNAs was not viable (15). To determine the sfRNA production phenotypes of xrRNA1′ and xrRNA2′ mutants in a vertebrate host, Vero cells were infected with wild-type (WT) ZIKV and the two viral mutants. The IFN-deficient Vero cells (16) were used in this experiment because they support the replication of all three viruses at comparable levels (15). We found that, unlike in insect cells (15), the mutation in xrRNA2 not only abolished the production of sfRNA-2 but also markedly reduced the accumulation of sfRNA-1 in mammalian cells (Fig. 1A), although cells infected with WT and mutant viruses contained similar levels of viral genomic RNA (Fig. 1B). These results are consistent with a previous report showing the cooperation between two xrRNAs of DENV and the impairment of the production of both sfRNAs when xrRNA1 is disrupted (17). Considering that mutation in xrRNA2 of ZIKV impaired the production of both sfRNAs in mammalian but not insect cells (15), we can assume that cooperation between ZIKV xrRNAs is host specific. As the mutation in xrRNA2 virtually abolishes the production of both sfRNAs in vertebrate cells without compromising virus viability, the xrRNA2′ ZIKV mutant can be used to assess the effect of nearly complete sfRNA deficiency on virus replication and pathogenesis. In turn, the xrRNA1′ mutant, which is deficient in sfRNA-1 only, can be used to study the impact of partial sfRNA deficiency.
Fig. 1. ZIKV sfRNA facilitates viral replication, cytotoxicity, and pathogenesis in the mammalian host.
(A) Northern blotting showing the production of ZIKV sfRNA in Vero cells infected with WT, xrRNA1′, and xrRNA2′ ZIKV. Bottom displays ribosomal RNA (rRNA) visualized by ethidium brimide staining as a loading control. nt, nucleotide. (B) The quantitative reverse transcription polymerase chain reaction (qRT-PCR) quantification of viral RNA in samples used in (A). (C) ZIKV plaque morphology on a monolayer of Vero cells at 72 hours postinfection (hpi). Bottom: Virus replication foci visualized by immunostaining of Vero cells inoculated with the same virus samples. (D) Cytotoxicity of WT and sfRNA-deficient ZIKV mutants determined using Viral ToxGlo Assay. Vero cells were infected at the indicated multiplicities of infection (MOIs), and CPE was measured at 72 hpi. % CPE is calculated with reference to uninfected cells. (E) Viral growth kinetics in human lung (A549), human placental (BeWo and HTR-8), and mouse embryonic fibroblast (MEF) cells infected at MOI = 0.1. (F) Replication of WT and sfRNA-deficient ZIKV in AG129 mice inoculated with 104 focus-forming units (FFU) per mouse. Animals were monitored for disease symptoms for 15 days, and blood was collected daily via tail bleeding. Images in (A) and (C) are representative of three independent experiments. Values in (B), (E), and (F) (viremia) are the means from three biological replicates ± SD. Statistical analysis is by Student’s t test in (B), regression analyses in (D) with the P values indicating the differences in means, one-way analysis of variance (ANOVA) in (E), area under the curve method for viremia in (F), and Gehan-Breslow-Wilcoxon test for survival rates in (F). Titers in (E) and (F) were determined by a foci-forming immunoassay of C6/36 cells. dpi, days postinfection.
We previously demonstrated that sfRNA facilitated the replication of WNV and was required for virus-induced CPE (14). To elucidate whether sfRNA of ZIKV has similar functions, we first assessed the plaque-forming properties of WT and sfRNA-deficient ZIKV. We found that WT ZIKV formed large and clear plaques on Vero cells, whereas plaques formed by xrRNA1′ mutant had reduced size and were less clear (Fig. 1C, top row). No clearly defined plaques were visible in cell monolayers infected with xrRNA2′ mutant (Fig. 1C, top row), while the immunoassay clearly showed virally infected cell foci (Fig. 1C, bottom row), indicative of productive infection. The quantitative CPE assay revealed significantly higher levels of CPE in Vero cells infected with WT virus compared to the sfRNA-deficient mutants (Fig. 1D and table S2). Together, these results demonstrated that sfRNAs are required for the CPE induced by ZIKV infection in vertebrate cells. Given that Vero cells infected with WT and mutant viruses contained similar levels of viral RNA (Fig. 1B), the reduced cytopathic properties of sfRNA-deficient ZIKV cannot be attributed to the lower viral load. Therefore, the sfRNA likely has a specific function in promoting cell death.
In IFN-competent human cell lines A549, BeWo, and HTR-8 infected at multiplicity of infection (MOI) = 0.1, the xrRNA1′ ZIKV replicated with efficiency comparable to WT virus, whereas the replication of xrRNA2′ mutant was highly attenuated (Fig. 1E). In WT mouse embryonic fibroblasts (MEFs), both mutants were incapable of productive infection after virus inoculation at MOI = 0.1 (fig. S1). At MOI = 1, the xrRNA1′ mutant established infection, although at a lower level than WT virus, whereas replication of xrRNA2′ was still barely detectable (Fig. 1E). These results demonstrated that the production of sfRNAs is required for efficient replication of ZIKV in IFN response–competent mammalian cells.
To determine whether sfRNAs contribute to the pathogenesis of ZIKV infection in vivo, AG129 mice were used as an infection model. These mice are deficient in receptors for IFN-α/β and IFN-γ while still competent in IFN-λ signaling. Unlike WT mice, they support the replication of ZIKV and develop disease (18). Mice infected with sfRNA-deficient mutants showed a later onset of disease manifestations, lower viremia, and a delay in mortality compared to mice infected with WT virus (Fig. 1F, fig. S2, and table S1). This illustrated that the production of sfRNAs also promotes the replication and pathogenesis of ZIKV in vivo. In summary, sfRNAs are required for optimal virus replication in vertebrate cells and in vivo, thereby increasing CPE and disease, respectively.
Production of sfRNAs is required for ZIKV infection in the placenta and dissemination into the fetal brain
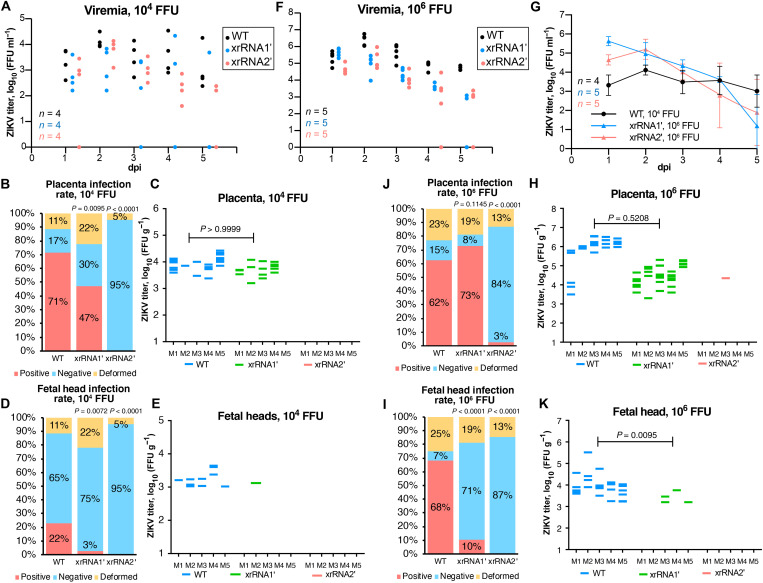
To determine whether the production of sfRNAs contributes to ZIKV pathogenesis during pregnancy, an established animal pregnancy model (19, 20) with mice deficient in the IFN-α/β receptor (IFNAR−/−) was used. IFNAR−/− mice usually survive infection with Brazilian (Asian lineage) virus strains but die after infection with African lineage viruses (18). The xrRNA1′ and xrRNA2′ mutants of the Brazilian ZIKV strain Natal (ZIKVNatal) were thus constructed (fig. S3A), and pregnant dams were inoculated with either WT or the sfRNA-deficient ZIKVNatal viruses [104 focus-forming units (FFU) per mouse] at embryonic day 12.5 (E12.5). All three viruses established viremias in IFNAR−/− dams (Fig. 2A), with mutant viruses only showing significant differences at 2 days postinfection (dpi) for WT versus xrRNA1′ and at 4 dpi for WT versus xrRNA2′ (Fig. 2A and table S3). The mutant and WT viruses had no significant effect on the placental or fetal weights (fig. S3B) or on the proportion of deformed fetuses (Fig. 2B). Notably, WT and xrRNA1′ ZIKV established productive infections in the placenta, whereas no virus was detected in the placentas of dams infected with the xrRNA2′ mutant (Fig. 2, B and C) despite similar viral titers in blood to those seen for the xrRNA1′ mutant (Fig. 2A). This indicated that the production of sfRNAs is crucial for ZIKV infection of the placenta. Moreover, despite the presence of virus in the placenta at levels comparable to WT, the xrRNA1′ mutant was detected in only 1 fetal head (3% infection rate; Fig. 2, D and E), whereas WT ZIKV was detected in 11 fetal heads (22% infection rate; Fig. 2, D and E). This indicated that production of sfRNAs is important for dissemination of ZIKV into fetal brain. All fetal heads were negative in dams infected with the xrRNA2′ mutant (Fig. 2B), as might be expected since placentas were also not infected (Fig. 2, B and C).
Fig. 2. ZIKV sfRNA is required for placental infection and viral dissemination into the fetal brain.
(A, F, and G) Serum viremia in ZIKV-infected dams. Pregnant IFNAR−/− mice were inoculated with 104 FFU (A) and 106 FFU (F) of WT or sfRNA-deficient ZIKV Natal via subcutaneous injection. (G) Side-by-side comparison of viremia curves in mice infected with WT ZIKV at 104 FFU and xrRNA2′ ZIKV at 106 FFU. Values in (G) are means ± SD. (B to E) Infection rate (B and D) and viral titers (C and E) in fetal tissues collected from mice infected at 104 FFU. (H to K) Infection rate (I and J) and viral titers (H and K) in fetal tissues collected from mice infected at 106 FFU. Fetal material was collected at 5 dpi and weighed. Placentas and fetal heads were separated, homogenized, and used for virus titration. Deformed fetuses (fetal tissue masses) were not used for virus titration, as placentas and heads could not be separated. Viral titers were normalized to the tissue weight. The only virus-positive placenta sample for xrRNA2′ mutant in (E) contains reverse mutation to the WT sequence. Statistical analyses for differences in viremias in (A) and (F) are shown in tables S2 and S3. Statistical analyses for differences in viral loads in (G) are shown in tables S4 and S5. Statistical analyses for (B), (D), (J), and (I) were undertaken using Fisher’s exact tests and by Mann-Whitney U tests for (C), (E), (H), and (K); P values are in comparison to the WT-infected group. All titers were determined by a foci-forming immunoassay on C6/36 cells.
To determine whether inefficient placental infection by sfRNA-deficient ZIKV mutants was simply the consequence of lower viremia in the maternal blood compared to WT virus, the effect of a higher viral inoculation dose was assessed. Dams were infected with a 100 times higher dose of each virus (106 FFU), which increased the viremia levels for all the viruses by ≈1 to 2 logs compared to the 104 FFU infectious dose (fig. S3C). Although the serum titers for the mutant viruses were significantly lower than those for WT at multiple time points after infection with 106 FFU (Fig. 2F and table S4), when 104 FFU of WT was compared with 106 FFU of xrRNA1′ virus, the overall viral load (area under the curve) was significantly higher for xrRNA1′ (Fig. 2G, fig. S3C, and table S5). There was no significant difference in viremia between animals inoculated with 104 FFU of WT ZIKV and 106 FFU of xrRNA2′ virus (Fig. 2G, fig. S3D, and table S6). While higher viremias observed after inoculation with 106 FFU increased the placental infection for the xrRNA1′ mutant from 47 to 73% compared to 104 FFU, the placental infection for the xrRNA2′ mutant virus only rose from 0 to 3% (Fig. 2, J and H). The latter represented a single placenta, with the follow-up sequencing of the viral RNA identifying reversion of mutations to the WT sequence. The 22% dissemination into fetal heads for the WT virus after infection with 104 FFU (Fig. 2D) rose to 68% after infection with a 100-fold higher viral dose (Fig. 2, I and K), and for the xrRNA1′ mutant, this proportion increased from 3 to 10%. However, the infection for the xrRNA2′ mutant virus remained zero (Fig. 2I). Considering that the 100-fold increase in the viremia in xrRNA2′ mutant virus–infected dams achieved with the 106 FFU infection dose (which is higher than WT viremia after 104 FFU infection) did not restore placental infection and dissemination into fetal brain of this sfRNA-deficient virus, we can confidently conclude that this phenotype cannot be explained simply by the amount of virus in maternal blood. This indicates that at least one isoform of sfRNA (sfRNA1 or sfRNA2) is required to permit ZIKV replication in the placenta and points toward the functional significance of sfRNAs for viral infection of the placental tissue. Collectively, the results in pregnant animals demonstrated that the sfRNAs of ZIKV are required for efficient viral infection of the placenta, transplacental virus migration, and fetal brain infection.
sfRNA facilitates ZIKV-induced apoptosis of neural progenitor cells in the developing human brain tissue
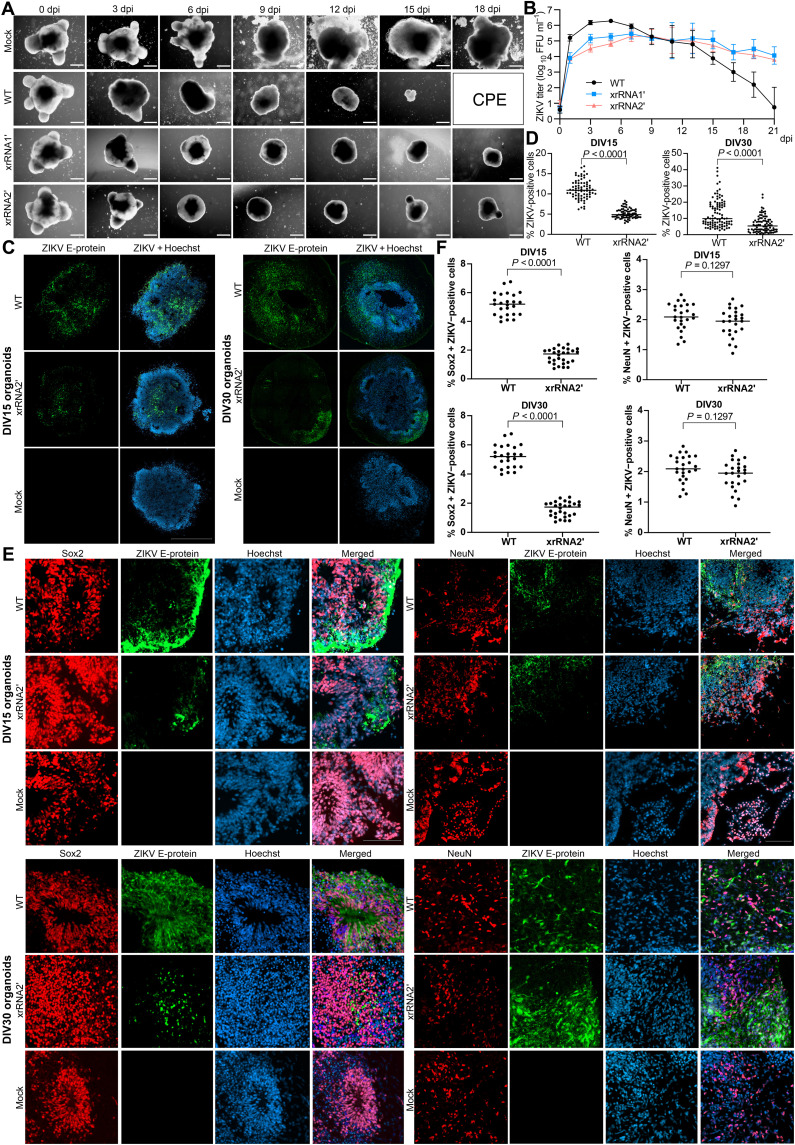
To gain insights into the effects of ZIKV sfRNAs on fetal neurovirulence and neuropathogenesis in humans, we used induced pluripotent stem cell (iPSC)–derived human cerebral organoids (hCOs). This well-established model of human brain development has been used extensively to study microcephaly and ZIKV infection in the developing brain tissue [reviewed in (21)]. Typically, 1- to 5-week-old cerebral organoids are used for ZIKV infections, as they closely mimic the early first trimester of gestational human brain age, the period when the fetus is most vulnerable to the virus (22, 23). Neural progenitor cells (NPCs) have been previously identified as the primary sites of ZIKV replication in the fetal brain (24). Accordingly, hCOs at 9 to 15 days in vitro (DIV9 to DIV15), consisting primarily of NPCs, are most permissive to ZIKV infection and associated cytotoxicity (20). Consistent with previous reports, we observed overt shrinkage of DIV15 hCOs and loss of their typical structure by 6 days after infection with WT and mutant ZIKVMR766 (Fig. 3A). However, only WT virus caused CPE and complete disintegration of hCOs by 15 to 18 dpi. In contrast, organoids infected with either of the sfRNA-deficient mutants survived through the course of the infection, although remaining smaller than uninfected organoids (Fig. 3A). In DIV15 organoids, viral titers at early time points after infection with the WT virus were higher than those after infection with the mutant viruses (Fig. 3B and table S7). In organoids infected at DIV30, xrRNA2′ mutant exhibited significantly reduced replication through the entire time course and became undetectable at 7 dpi, while xrRNA1′ mutant showed only slight attenuation compared to the WT ZIKV (fig. S4A and table S8). No CPE was detected in DIV30 organoids throughout the course of infection with WT or mutant viruses. Consistent with differences in viral titers, at 3 dpi, WT virus–infected DIV15 and DIV30 organoids also had significantly more infected cells than organoids infected with the mutant viruses (Fig. 3, C and D). Immunostaining of the histological section prepared from DIV15 and DIV30 organoids showed that WT virus and both mutants primarily infected sex determining region Y-box 2 (Sox2)–positive NPC with very limited infection in neuron-specific nuclear protein (NeuN)–positive mature neurons (Fig. 3, E and F). This is consistent with a previously described tropism of ZIKV in neural tissues (24).
Fig. 3. The sfRNA facilitates ZIKV replication in hCOs and viral infection of neural progenitors.
(A) Morphology of hCOs infected with WT and sfRNA-deficient ZIKV at DIV15. Scale bars, 500 μm. (B) Viral titers in culture supernatants from hCOs infected with WT and sfRNA-deficient ZIKV are shown in (A). The 14- to 15-day-old organoids were infected with 104 FFU of ZIKV. Viral titers were determined by a foci-forming assay. Values are the means from three independent organoid differentiation experiments, each containing five to six organoids per group ±SD. Statistical analyses are shown in table S7. (C and D) Infection efficiency of WT and sfRNA-deficient ZIKV in hCOs. (E and F) Localization of ZIKV replication in cells expressing markers of neural progenitors (Sox2) and mature neurons (NeuN). Cerebral organoids at DIV15 and DIV30 were infected with WT or xrRNA2′ ZIKV and sectioned at 3 dpi. The images in (C) and (E) are representative from three independent experiments (independent organoid differentiation) that all showed similar results. Mann-Whitney U tests were used in (D) and (F). All tests are two-sided.
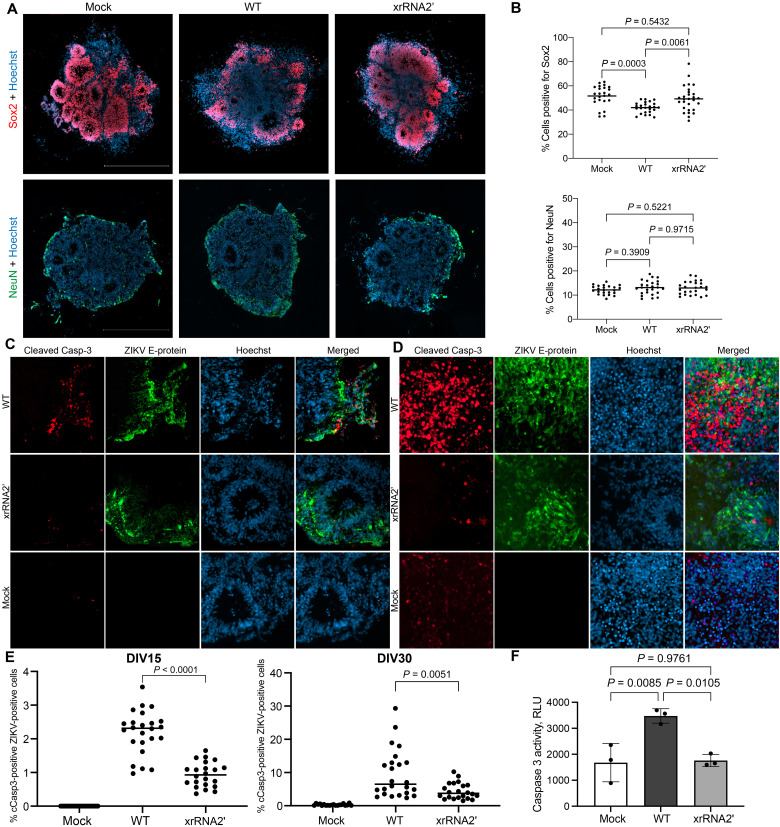
In addition, the analysis of ZIKV-infected hCOs stained for the NPC marker Sox2 demonstrated that WT ZIKV caused a significant decrease in the number of Sox2-positive cells at 3 dpi, while hCOs infected with sfRNA-deficient xrRNA2′ ZIKV (which will be further referred to as a sfRNA-deficient mutant) had a similar number of Sox2-positive cells to uninfected control (Fig. 4, A and B, and fig. S4B). Premature neuron differentiation and apoptosis of immature neurons and neural progenitors have previously been identified as the reasons for ZIKV-induced microcephaly (25). Here, we found no difference in the quantity of NeuN-positive cells between hCOs infected with WT and xrRNA2′ ZIKV or between infected organoids and mock (Fig. 4, A and B, and fig. S4B). This eliminates premature differentiation as the possible reason for the observed disappearance of Sox2-positive cells in organoids infected by WT virus. Subsequently, immunohistochemical detection of cleaved caspase 3 (cCasp3) was used to elucidate the effect of sfRNAs on the induction of apoptosis in hCOs. Sections of DIV15 and DIV30 organoids fixed at 3 days after infection with WT or the sfRNA-deficient ZIKV mutant demonstrated that WT virus induced caspase cleavage in a significantly higher proportion of infected cells when compared to sfRNA-deficient mutant virus (Fig. 4, C to E). In agreement with these data, a significant increase in the caspase 3/7 activity in response to infection with WT but not sfRNA-deficient mutant viruses was observed in a homogeneous monolayer culture of the human neural progenitor ReNcell at 72 hours postinfection (hpi; Fig. 4F), although the cells infected with either virus had similar viral loads at this time point (fig. S4C). This provides further evidence for the specific function of sfRNAs in virus-induced apoptosis of NPC.
Fig. 4. Production of ZIKV sfRNA promotes apoptosis of infected NPCs.
(A) Neural progenitors (Sox2-positive cells) and mature neurons (NeuN-positive cells) in hCOs infected with WT and sfRNA-deficient ZIKV. (B) Quantification of cells expressing Sox2 and NeuN in ZIKV-infected hCOs. (C and D) Effect of sfRNA production on caspase 3 activation in ZIKV-infected human brain organoids at DIV15 (C) and DIV30 (D). ZIKV E-protein and cCasp3 were visualized by immunofluorescent staining. (E) Image quantification of (C) and (D). Cells positive for ZIKV E-protein and cCasp3 were counted across sections of multiple organoids from three independent experiments. (F) Caspase 3/7 activity in immortalized human NPC ReNCell infected with WT and sfRNA-deficient ZIKV. Cells were infected at MOI = 1, and caspase 3/7 activity was assessed at 72 hpi. Values are the means from three biological replicates ± SD. Statistical analysis is one-way ANOVA in (B) and (F) and Mann-Whitney U test in (E). All tests are two-sided. RLU, relative luciferase units.
Collectively, the experiments performed in hCOs identified sfRNAs as an important neurovirulence factor of ZIKV. The sfRNAs were found to facilitate ZIKV replication in the developing human brain tissue and to be required for the neuropathic effect of the virus. Production of sfRNAs was found to be a prerequisite for the elimination of ZIKV-infected neural progenitors via apoptotic cell death.
ZIKV sfRNA inhibits signaling pathways that converge at STAT1
We previously demonstrated that the sfRNA of WNV acts by inhibiting type I IFN response (6). The data for ZIKV presented in Figs. 1F and 2 argue that sfRNAs facilitate ZIKV pathogenesis in AG129 mice deficient in receptors for IFN-α/β/γ and are required for the infection of the placenta in mice deficient in IFNAR. This suggests that sfRNAs may affect other type I/II IFN response–independent antiviral pathways. To identify all pathways and processes affected by ZIKV sfRNA, we performed the transcriptome-wide gene expression profiling of cells infected with WT and sfRNA-deficient ZIKV. The human placental cell line BeWo was used for this experiment, as it is known to support ZIKV replication; is capable of responding to all three types of IFNs, IFN-α/β, IFN-γ, and IFN-λ (26); and has a placental origin (and thus relevant to ZIKV tropism).
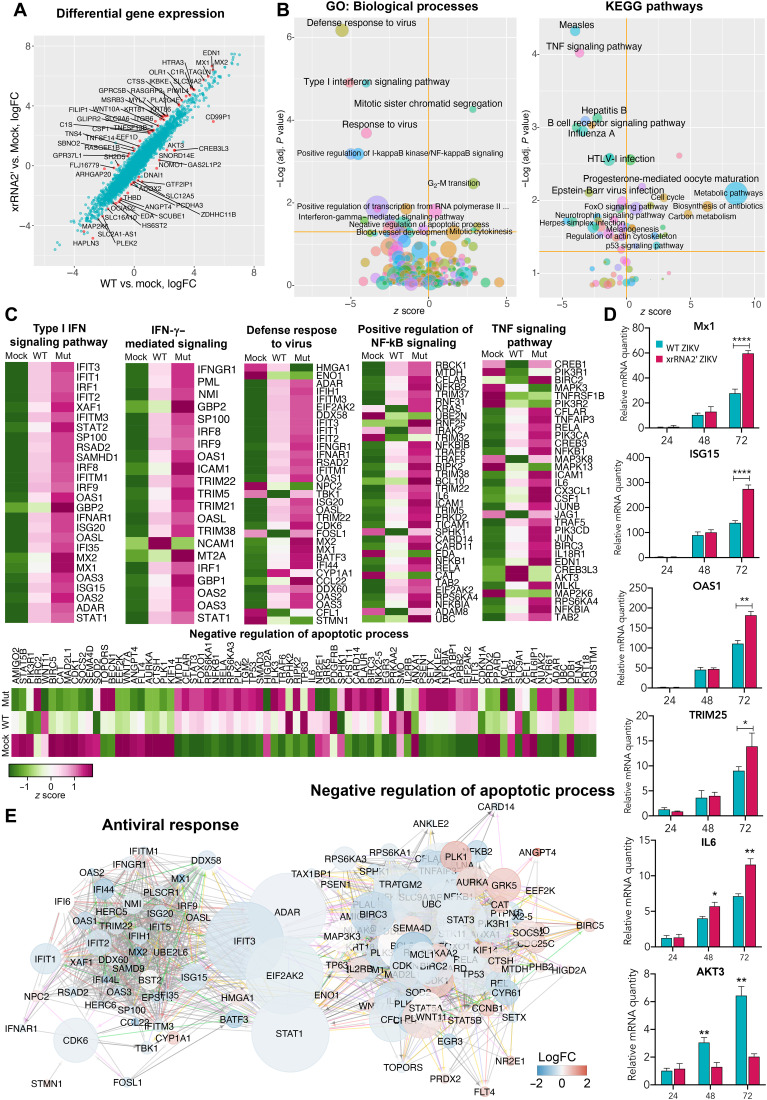
Cells were inoculated with WT ZIKV or xrRNA2′ mutant viruses at MOI = 1. At 72 hpi, total RNA was isolated from the cells and used for RNA sequencing (RNA-seq). Notably, both viruses produced a similar amount of intracellular viral RNA at this time point (fig. S5A). Principal components analysis of the library size-normalized RNA-seq counts showed a clear separation of samples infected with WT and xrRNA2′ ZIKV (fig. S5B). This indicates the presence of a gene expression component specifically associated with sfRNA production. Differential gene expression analysis demonstrated that multiple genes, including known IFN-stimulated genes (ISGs) such as Mx1, Mx2, and ISG15, showed stronger induction in response to xrRNA2′ mutant virus compared to WT virus (Fig. 5A and table S9). Subsequently, the gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses identified type I IFN signaling as the most enriched biological process inhibited by sfRNA in these cells (Fig. 5B). In addition, the enrichment analyses revealed that ZIKV sfRNAs inhibited IFN-γ, nuclear factor κB (NF-κB), and tumor necrosis factor (TNF) signaling (Fig. 5B). While IFN-γ was not produced by these cells, STAT2 degradation by ZIKV (27) is known to skew signaling toward the STAT1/STAT1 pathway, which is the primary factor activated by IFN-γ signaling. As a result, a prominent signature associated with stimulation of IFN-γ signaling emerges (Fig. 5C). Moreover, sfRNAs appeared to decrease the activation of “negative regulation of apoptotic processes” (Fig. 5B, left) and slightly, albeit significantly, induce p53 signaling (Fig. 5B, right). This provides additional evidence for the proapoptotic function of sfRNAs. Notably, the sfRNA-affected genes related to IFN responses and antiviral defense were represented by ISGs, receptors for IFN-α/β and IFN-γ, and the components of IFN-α/β/γ signaling, whereas IFNs themselves were not identified as sfRNA-affected genes at this time point (Fig. 5C and table S9). This suggests that ZIKV sfRNAs act downstream of IFN-α/β/γ receptors while not affecting IFN-α/β production. The expression of ISGs was significantly higher in cells infected with xrRNA2′ ZIKV than WT ZIKV. A similar pattern was also observed in the expression of proinflammatory (TNF signaling pathway) and antiapoptotic genes. Collectively, the results of gene expression profiling demonstrated that ZIKV sfRNAs inhibit type I and II IFN signaling and TNF signaling and promote apoptosis. Potentially, they also inhibit type III IFN response, which shares gene expression signatures with the response to type I IFNs. Therefore, it cannot be distinguished from type I IFN pathway using current bioinformatic annotations. To validate the results of RNA-seq, we performed quantitative reverse transcription polymerase chain reaction (qRT-PCR) for selected differentially expressed genes in infected BeWo cells at different time points after infection. Consistent with the transcriptomic data, we observed the inhibitory effect of sfRNAs on the expression of ISGs and the antiapoptotic cytokine interleukin-6 (IL-6) later in infection (72 hpi), while the expression of the proapoptotic AKT3 gene was elevated at this time point (Fig. 5D).
Fig. 5. Effect of sfRNA production on expression of host genes in infected human placental cells.
(A) Differential gene expression in BeWo cells infected with WT and xrRNA2′ ZIKV. Red dots show genes that responded differently to infection with WT and mutant viruses (false discovery rate–adjusted P < 0.05). LogFC, log fold change. (B) Pathways and biological processes affected by the production of ZIKV sfRNA for genes identified in (A). (C) Expression of genes associated with biological processes affected by the production of sfRNA in BeWo cells. Values are z scores and are the means from three biological replicates. (D) Expression of selected ISGs and apoptosis-regulating genes in BeWo cells infected with MOI = 1 of WT (magenta bars) and xrRNA2′ (cyan bars) ZIKV and determined by qRT-PCR. Relative mRNA quantity was determined using the ΔΔCt method and is relative to mock with normalization to GAPDH. Values are the means ± SD, n = 3. Statistical analysis is by t test. (E) Network of interactions between the sfRNA-affected genes involved in antiviral response and apoptosis. Two individual networks were reconstructed from the genes identified in (C) using the GeneMANIA Cytoscape plug-in and then merged. The resulting network was subjected to Cytoscape network analysis to calculate the betweenness centrality values as a measure for the weight of each node in the combined network. Size of the nodes indicates betweenness centrality, and the color of the nodes indicates the difference in gene expression between the cells infected with WT and xrRNA2′ mutant ZIKV. Positions of the nodes are relative to their connectivity within and between the two subnetworks. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To further dissect the molecular processes affected by ZIKV sfRNAs, we queried whether the regulatory networks associated with antiviral response and apoptosis intersect. To this end, we determined the relative weight of each individual gene in these networks to indicate their importance in the respective signaling. The network analysis identified STAT1, adenosine deaminase RNA specific (ADAR), interferon induced protein with tetratricopeptide repeats 3 (IFIT3), and eukaryotic translation initiation factor 2 alpha kinase 2 (EIF2AK2) also known as protein kinase R (PKR) as shared components of the antiviral response and negative regulation of apoptosis (Fig. 5E). Moreover, these genes also have the biggest weight in the joint network as indicated by their betweenness centrality, the measure that indicates which node has the biggest control over the network (Fig. 5E). The involvement of these genes in antiviral response and negative regulation of apoptosis suggests that their inhibition should simultaneously impair antiviral response and promote apoptosis. Therefore, we concluded that these genes might represent molecular targets of sfRNAs. Among these proteins, ADAR, IFIT3, and PKR are the effectors and the most downstream components of the type I and III IFN signaling (28) and were therefore deemed unlikely candidates for mediating the effect of sfRNAs on the expression of multiple ISGs. STAT1, however, is a key regulatory component in all types of IFN signaling. It is also known to antagonize apoptosis in cells exposed to IFN-γ (29). In addition, analysis of pathways differentially regulated by WT and sfRNA-deficient virus infections using Ingenuity Pathway Analysis software identified STAT1 as the most significant direct upstream regulator of the differentially expressed genes affected by sfRNA (table S10), indicating the key role of ZIKV sfRNAs in inhibiting STAT1-regulated pathways. On the basis of these results, we hypothesized that STAT1 represents the key target of ZIKV sfRNAs, which allows them to modulate multiple host response pathways.
ZIKV sfRNAs inhibit IFN signaling but not IFN production
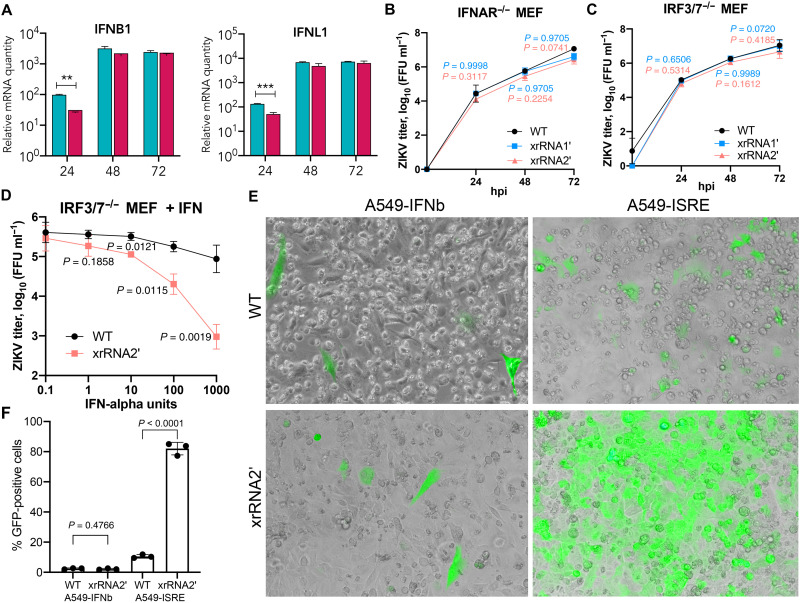
The sfRNA of DENV2 has been reported to inhibit pattern recognition receptor (PRR) signaling and IFN expression (8, 9). However, this was not the case for other flaviviruses, including other strains of DENV (7, 30), and our RNA-seq analysis showed no effect of ZIKV sfRNAs on the expression of IFNs, suggesting that sfRNA of ZIKV does not target PRRs and IFN production. To validate these findings, we first assessed the expression of IFNB1 and IFNL1 in BeWo cells at different time points after infection with WT and sfRNA-deficient ZIKV using qRT-PCR. Consistent with the results of RNA-seq, we observed no inhibitory effect of ZIKV sfRNAs on the expression of these IFN genes at any time point (Fig. 6A). Instead, the expression of IFNB1 and IFNL1 early in infection (24 hpi) was higher in cells infected with WT ZIKV compared to cells infected with xrRNA2′ mutant virus (Fig. 6A). In addition, replication of sfRNA-deficient ZIKV mutants was restored in the IFNAR-knockout MEF (Fig. 6B) compared to the WT MEF (Fig. 1E). This provides additional evidence that ZIKV sfRNA acts by inhibiting IFN signaling and not IFN expression. Moreover, we found that in IRF3/7−/− MEF deficient in intrinsic IFN production, xrRNA2′ mutant ZIKV replicated at comparable levels to the WT virus (Fig. 6C) but exhibited significantly higher sensitivity to the treatment with exogenous IFN-α (Fig. 6D). This further indicates that compared to WT ZIKV, the sfRNA-deficient mutant virus has decreased ability to counteract IFN-α/β signaling. Furthermore, the reporter assay with A549 cells expressing green fluorescent protein (GFP) under the control of either IFN-β promoter (A549-IFNb) or IFN-stimulated response element (A549-ISRE) demonstrated that infection with WT or sfRNA-deficient ZIKV induced IFNB1 gene promoter at comparable levels, while sfRNA-deficient ZIKV caused significantly stronger activation of the ISRE promoter than WT virus (Fig. 6E). Therefore, we can conclude that ZIKV sfRNAs inhibit downstream signaling from IFN receptors but not the expression of IFNs.
Fig. 6. ZIKV sfRNA inhibits IFN signaling but not PRR signaling and IFN production.
(A) Expression of IFNs in human placental BeWo cells infected with WT (cyan) and sfRNA-deficient (magenta) ZIKV. Cells were infected at MOI = 1, and total RNA was isolated at the indicated time points and used for qRT-PCR. Relative mRNA quantity was determined using the ΔΔCt method relative to mock with normalization to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (B and C) Replication of WT and sfRNA-deficient ZIKV mutants in IFNAR−/− and IRF3/7−/− MEF. Cells were infected at MOI = 0.1. (D) Effect of IFN treatment on the replication of WT and sfRNA-deficient ZIKV in IRF3/7−/− MEF incapable of endogenous IFN production. Cells were pretreated with IFN-α, infected at MOI = 1, and titers were determined at 48 hpi. (E) Effect of ZIKV sfRNA on the activity of IFNb and ISRE promoters. IFNb-GFP and ISRE-GFP reporter A549 cells were infected with WT or sfRNA-deficient ZIKV at MOI = 0.1, and GFP fluorescence was documented at 48 hpi. The images are merged bright-field and epifluorescent microphotographs of live cells and are representative of three experiments that showed similar results. (F) Quantification of GFP-positive cells in (E). Values in (A) to (D) and (F) are the means of three biological replicates ± SD. Statistical analysis was performed using one-way ANOVA with Dunnett’s correction (A to D) and Student’s t test (F). **P < 0.01, ***P < 0.001.
Production of ZIKV sfRNAs decreases phosphorylation and nuclear translocation of STAT1 in infected cells
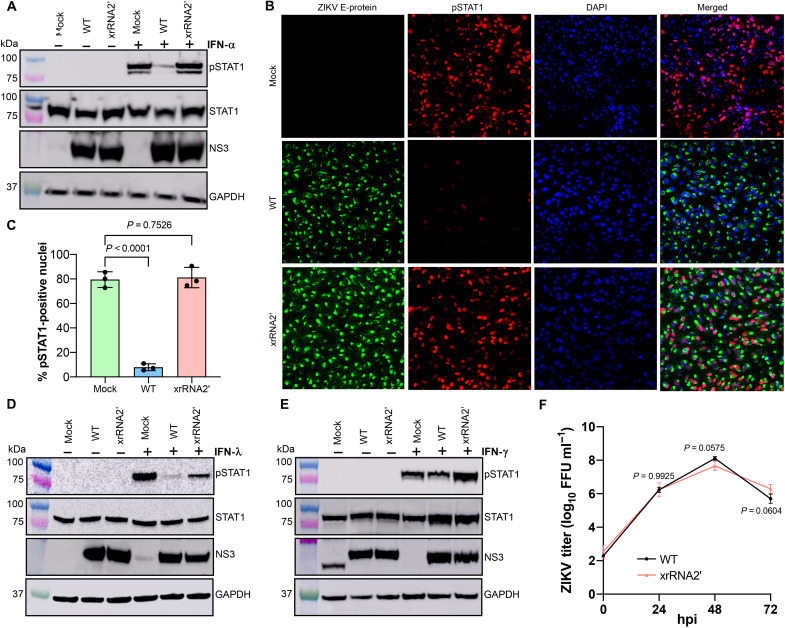
The network reconstruction analysis identified STAT1 as a potential key target of ZIKV sfRNA (Fig. 5E). STAT1 plays a central role in signal transduction from all three types of IFNs. In response to IFNs, it becomes phosphorylated, dimerizes with STAT2 or another STAT1 subunit, translocates into the nucleus, and activates the expression of antiviral genes. To elucidate the effect of sfRNAs on STAT1 activation, the levels of total and phosphorylated STAT1 were determined in Vero cells infected with WT or xrRNA2′ ZIKV after treatment with IFN-α. We found that IFN-α induced substantial accumulation of Tyr701-phosphorylated STAT1 (pSTAT1) in mock-infected cells and in cells infected with the sfRNA-deficient ZIKV but not in cells infected with the WT virus (Fig. 7A). Equal amounts of viral NS3 protein were detected in both infections, which is consistent with a previous observation that sfRNA deficiency does not affect viral replication in IFN-deficient Vero cells (15). The total STAT1 levels were not affected by the infections with both viruses (Fig. 7A), indicating that sfRNAs inhibited STAT1 phosphorylation rather than affecting STAT1 expression. In addition, immunofluorescent microscopy revealed that infection with WT, but not with xrRNA2′ ZIKV, efficiently prevented the accumulation of pSTAT1 in the nuclei of infected cells in response to IFN-α treatment (Fig. 7, B and C), which suggests that transcriptional activation of STAT1-regulated ISGs would be inhibited by sfRNAs.
Fig. 7. ZIKV sfRNA inhibits type I and III IFN signaling by suppressing phosphorylation and nuclear translocation of STAT1.
(A) Effect of ZIKV sfRNA on the phosphorylation of STAT1 in response to type I IFN. (B) Immunofluorescent detection of Tyr701-phosphorylated STAT1 in ZIKV-infected Vero cells treated with IFN-α1. The image is representative of three independent experiments that showed similar results. DAPI, 4′,6-diamidino-2-phenylindole. (C) Image quantification for (B). The pSTAT1-positive nuclei were quantified in ZIKV-positive cells in three independent experiments. (D and E) Effect of ZIKV sfRNA on the phosphorylation of STAT1 in response to type II (E) and type III (D) IFN. The band in mock after probing for NS3 in (E) represents nonspecific antibody binding and has different NS3 molecular weight. (F) Replication of WT and sfRNA-deficient ZIKV in STAT1-deficient U3A cells. Cells were infected at MOI = 0.1; titers were determined by a foci-forming assay of C6/36 cells. In (A) to (E), Vero (A to C and E) or HTR-8 cells (D) were infected with WT or xrRNA2′ ZIKV or left uninfected (mock). At 48 hpi, cells were treated with human IFN-α1 (A to C), IFN-λ1 (D), or IFN-γ (E) for 20 (A to C and E) or 60 min (D). Levels of Tyr701-phosphorylated STAT1 (pSTAT1) and total STAT1 indicate phosphorylation and expression of STAT1, respectively. Levels of ZIKV NS3 indicate viral loads in the infected cells. GAPDH levels indicate the total protein input. In (C) and (F), the values are the means from three independent experiments ± SD. The statistical analysis is by Student’s t test in (C) and one-way ANOVA in (F). All tests are two-sided.
We next tested the ability of sfRNAs to inhibit STAT1 phosphorylation in response to IFN-λ in human placental HTR-8 cells that have functional IFN-λ signaling (26). To achieve comparable viral loads in cells infected with WT and sfRNA-deficient ZIKV, a high MOI (MOI = 5) was used in this experiment (fig. S7). The results demonstrated that sfRNAs decreased IFN-λ–induced phosphorylation of STAT1 while having no effect on the total STAT1 levels (Fig. 7D). ZIKV sfRNA thus also disrupts type III IFN signaling by inhibiting the phosphorylation of STAT1.
Type II IFN signaling is exclusively regulated by STAT1; hence, the effect of sfRNA on IFN-γ signaling was also examined. Vero cells capable of responding to IFN-γ were used for this experiment. The results showed that although the total STAT1 level rose after IFN-γ treatment in cells containing either virus, the level of phosphorylated STAT1 was lower in cells infected with WT virus than in cells infected with xrRNA2′ mutant ZIKV (Fig. 7E), illustrating inhibition of IFN-γ signaling by sfRNA.
Hence, we demonstrated that sfRNAs play a key role in the inhibition of STAT1 phosphorylation in response to all three types of IFNs. In addition, replication of the xrRNA2′ mutant ZIKV was similar to that of the WT virus in STAT1-deficient U3A cells (Fig. 7F). The difference in CPE between WT and sfRNA-deficient viruses was also diminished in STAT1−/− cells (fig. S8). These results identify STAT1 as the host factor responsible for the attenuated phenotype of sfRNA-deficient ZIKV mutants and demonstrate the functional significance of sfRNA-mediated inhibition of STAT1 signaling in ZIKV-host interactions.
ZIKV sfRNA interacts with viral protein NS5 to inhibit STAT1 phosphorylation
To further characterize the inhibitory activity of ZIKV sfRNAs on STAT1 phosphorylation and nuclear translocation, Vero cells were transfected with in vitro generated ZIKV sfRNA or GFP RNA fragment (control; fig. S9A), followed by treatment with IFN-α. The sfRNA-transfected cells showed substantial accumulation of pSTAT1 in the nuclei, similar to the levels in cells transfected with control RNA (fig. S9B). As only phosphorylated STAT1 translocates to the nuclei following IFN stimulation, these results imply that sfRNA requires other viral or virus-induced host factors to execute this activity.
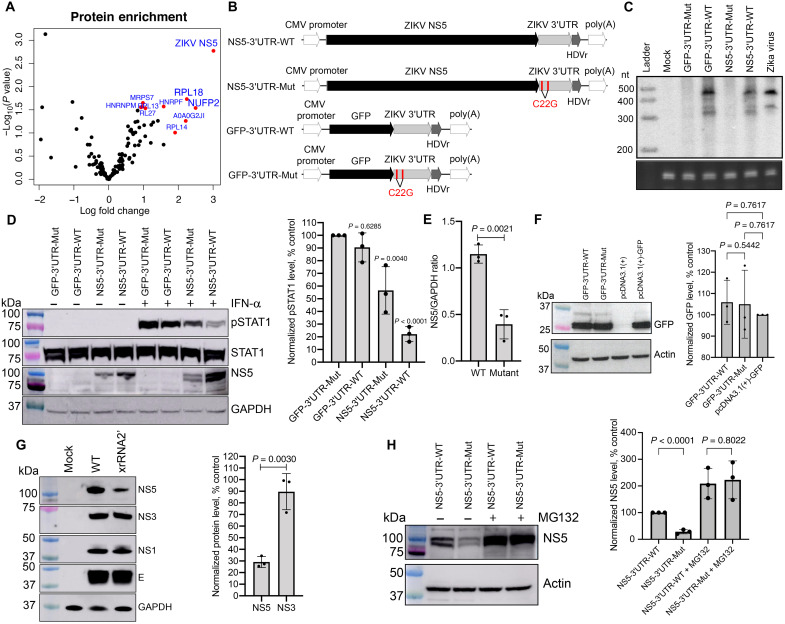
The activities of sfRNA in flaviviruses have previously been associated with interactions between sfRNA and host proteins (8, 9, 30). These interactions are usually identified by an RNA affinity pulldown experiment in which lysates of uninfected cells are used with in vitro transcribed sfRNA (31, 32). However, given the lack of inhibitory activity of sfRNA alone on STAT1 phosphorylation and nuclear translocation in uninfected cells (fig. S9B), functional interactions of sfRNAs affecting STAT1 phosphorylation need to be studied in the context of virus infection. Because in vitro transcribed sfRNA used as a bait in pulldowns would compete with sfRNAs produced in cells infected with the WT virus, lysates of cells infected with the sfRNA-deficient virus were used for sfRNA affinity pulldown assays. Streptavidin-binding aptamer-tagged ZIKV sfRNA and control GFP RNA (fig. S10) were generated by in vitro transcription and used in RNA affinity pulldowns with the lysate of cells infected with xrRNA2′ mutant virus to identify sfRNA-interacting proteins. Label-free quantitative sequential window acquisition of all theoretical fragment ion spectra mass spectrometry (SWATH-MS) was then used to identify proteins enriched in sfRNA pulldowns compared to GFP RNA pulldown. This approach revealed ZIKV NS5 as the most significantly enriched sfRNA-interacting protein (Fig. 8A). NS5 of WNV (33) and Japanese encephalitis virus (JEV) (34) were previously shown to inhibit STAT1 phosphorylation. However, NS5 ectopically expressed from plasmids was unable to inhibit STAT1 phosphorylation to the same extent as NS5 produced during infection, suggesting the requirement of another viral factor for this activity (35). Considering the identified sfRNA-NS5 interaction, we hypothesized that sfRNA is the viral factor that acts in conjunction with NS5 to provide efficient inhibition of STAT1 phosphorylation.
Fig. 8. ZIKV sfRNA inhibits STAT1 phosphorylation by stabilizing NS5 via direct binding.
(A) The sfRNAZIKV-binding proteins identified by RNA affinity pulldown. Top 10 most significant sfRNA-interacting proteins are highlighted. The values are the means from three independent experiments. (B) Schematics of the reporter constructs for the production of ZIKV sfRNA and NS5 alone and in combination. The mutations leading to sfRNA deficiency are shown in red. CMV, cytomegalovirus; poly(A), polyadenylate. (C) Production of sfRNA in HEK293T cells transfected with the constructs shown in (B). Bottom: 5.8S rRNA. (D) Detection of pSTAT1, total STAT1, NS5, and GAPDH (loading control) in HEK293T cells transfected with the plasmids shown in (B) and treated with IFN at 48 hours posttransfection (hpt). (E) Quantification of NS5 levels in (D) by densitometry. (F) Western blot for GFP expression in HEK293T cells transfected with the plasmids expressing GFP with and without sfRNAZIKV. (G) Detection of structural (E-protein) and nonstructural (NS5, NS3, and NS1) proteins in Vero cells infected with WT and xrRNA2′ ZIKV. (H) Effect of the proteasome inhibitor MG132 on the accumulation of ZIKV NS5 in HEK293T cells 24 hours after transfection with indicated constructs. Right panels in (D) and (F) to (H) shows image densitometry for the respective blots based on three independent experiments. Values are normalized to GAPDH or actin and expressed as a percentage relative to control. The blots in (C), (D), and (F) to (H) are representative of three independent experiments. Error bars in (D) to (H) represent SD. Statistical analysis was performed by one-way ANOVA (E) or t test (E to H).
To test this hypothesis, expression plasmids producing ZIKV NS5 and sfRNA (NS5-3′UTR-WT), ZIKV NS5 alone (NS5-3′UTR-Mut), GFP and ZIKV sfRNA (GFP-3′UTR-WT), and GFP alone (GFP-3′UTR-Mut) were generated (Fig. 8B). To recapitulate the biogenesis of viral protein and sfRNA in infection, they were designed to encode a single transcript consisting of NS5 open reading frame (ORF) (or GFP in control) followed by ZIKV 3′UTR. These transcripts were expected to be translated and undergo decapping, thus becoming the accessible substrate for XRN-1 for further conversion into sfRNA in a similar manner to viral RNA during infection. Hepatitis virus delta ribozyme (HDVR) was added immediately downstream of the 3′ end of the encoded mRNA to remove the polyadenylate tail and produce an authentic 3′-OH terminus present in viral RNA and sfRNAs. The mutations were then introduced into both xrRNAs to create constructs deficient in sfRNA production yet encoding respective proteins (Fig. 8B). The constructs were confirmed to either produce (3′UTR-WT constructs) or not produce (3′UTR-MUT constructs) both sfRNAs in cells transfected with plasmid DNA using Northern blotting (Fig. 8C). In addition to the expected sfRNA bands, the band with a slightly higher electrophoretic mobility than sfRNA-2 (appeared as lower–molecular weight band) was evident in the samples from transfected cells (Fig. 8C), which can be explained by incomplete denaturation of highly structured sfRNA. Human embryonic kidney (HEK) 293T cells were then transfected with each plasmid, treated with IFN-α, and analyzed by Western blot for pSTAT1 (Fig. 8D). Consistent with the results of sfRNA transfection (fig. S9), similarly robust levels of pSTAT1 were observed in cells expressing only sfRNA (GFP-3′UTR-WT) and in cells transfected with a control construct producing neither sfRNA nor NS5 (GFP-3′UTR-mut; Fig. 8D). Cells expressing NS5 without sfRNA (NS5-3′UTR-Mut) exhibited slightly decreased accumulation of pSTAT1 (Fig. 8D), which illustrated that NS5 alone could inhibit STAT1 phosphorylation only to a limited extent. In contrast, accumulation of pSTAT1 was markedly reduced in cells transfected with the construct expressing both ZIKV NS5 and sfRNA (NS5-3′UTR-WT; Fig. 8D). These results show that the cooperative action of NS5 and sfRNA is required to inhibit STAT1 phosphorylation efficiently.
ZIKV sfRNA stabilizes NS5 protein
While analyzing the levels of phosphorylated and total STAT1 in cells transfected with the expression constructs, we also assessed intracellular levels of NS5 and GFP. Notably, cells coexpressing NS5 and ZIKV sfRNA had significantly higher levels of NS5 compared to cells expressing NS5 alone (Fig. 8, D and E), while expression of GFP was not affected by sfRNA production (Fig. 8F). This indicates that sfRNA specifically facilitates the accumulation of NS5. The lack of difference in GFP expression also suggests that the introduction of viral 3′UTR with or without xrRNA1′2′ mutations into mRNAs does not affect the transcription or translation of mRNAs produced from these constructs. Instead, this demonstrates a specific posttranslational effect of ZIKV sfRNA on NS5, which we hypothesized is likely due to protein stabilization.
To further elucidate the effect of ZIKV sfRNA on NS5 accumulation, the levels of nonstructural (NS5, NS3, and NS1) and structural (E) ZIKV proteins were assessed in Vero cells infected with WT and sfRNA-deficient ZIKV mutant xrRNA2′. Cells infected with the sfRNA-deficient ZIKV contained less NS5 than cells infected with the WT virus (Fig. 8G) while containing comparable levels of other viral proteins (Fig. 8G). Considering that all viral proteins are produced by the proteolytic cleavage of a single polyprotein precursor, this further indicates that sfRNA selectively stabilizes NS5 protein and does not affect transcription or translation. To validate this hypothesis, the levels of NS5 were assessed in HEK293 cells transfected with plasmid constructs expressing NS5 with sfRNA (NS5-3′UTR-WT) or without sfRNA (NS5-3′UTR-Mut) followed by the treatment with the inhibitor of proteasomal protein degradation N-carbobenzyloxy-l-leucyl-l-leucyl-l-leucinal (MG132). Expectedly, MG132 increased NS5 levels in cells transfected with both constructs, indicating for the efficient inhibition of proteasomal activity (Fig. 8H). Upon suppression of protein degradation by the drug, the difference in the levels of NS5 in cells transfected with NS5-3′UTR-WT and NS5-3′UTR-Mut constructs observed in untreated cells was no longer present (Fig. 8H). Together, the data in Fig. 8 demonstrate that sfRNA binds to and stabilizes NS5 protein, which results in enhanced inhibition of STAT1 phosphorylation.
ZIKV NS5 was previously shown to induce STAT2 degradation (27), and a recent study demonstrated that sfRNA-deficient ZIKV mutant was also capable of this activity (36), thereby excluding a role for ZIKV sfRNA in STAT2 degradation. Consistent with these results, we also showed that deficiency in sfRNA production and associated decrease in NS5 level did not affect STAT2 degradation in cells either infected with viruses or transfected with expression constructs (fig. S11). This suggests that STAT2 degradation can be efficiently induced by a relatively small amount of NS5, while inhibition of STAT1 phosphorylation likely requires higher amounts of NS5 achieved via stabilization by sfRNA.
DISCUSSION
The production of sfRNA is an evolutionarily conserved feature of all flaviviruses. The sfRNA is so important for virus propagation that flaviviruses have evolved functional redundancy in their 3′UTRs to enable robust sfRNA generation and protect it from adverse mutations (37). Multiple studies by us and others have established the importance of sfRNA for the replication of diverse flaviviruses in insect and vertebrate hosts [reviewed in (10)]. Until recently, WNV and DENV were the main focus of these studies. The sfRNA of WNV was previously shown to facilitate viral replication in mammalian (4, 6, 14) and insect (4, 14, 38, 39) cells and to be required for WNV pathogenesis in mice (4, 14), CPE in cell culture (4, 14) and virus transmission by mosquitoes (39). We previously found that in the mammalian host, sfRNA of WNV acts by inhibiting signal transduction from IFNAR but not the production of type I IFN, although the molecular mechanism of this function remains to be identified (6). On the contrary, the sfRNA of some but not all DENV2 strains was shown to inhibit PRR signaling and IFN production by binding to the proteins Carpin and guanylate Binding Protein 3 (G3BP) (9) or tripartite motif-containing protein 25 (TRIM25) (8). Hence, it is currently unclear whether the molecular function of sfRNA is conserved between different flaviviruses, is virus specific, or is a combination of both. The phenotypical and biochemical analyses of sfRNA functions in other members of the genus Flavivirus are therefore required to resolve this matter.
In this study, we focused on the functions of sfRNA of the latest pandemic flavivirus, ZIKV. The sfRNA of ZIKV has been previously shown to promote virus replication in the insect host and is required for viral transmission (15, 32). In the mammalian host, deletion of the entire stem-loop elements from dengue viral 3′UTR inhibited viral replication (17). However, as this deletion causes major structural disruption in viral 3′UTR, which is also required for viral replication (40), this model does not allow separation of the effects caused by sfRNA deficiency from the effect of compromised genome integrity. Another study used partially sfRNA-deficient ZIKV (still producing sfRNA2, equivalent to our xrRNA1′ mutant) and found that the mutation did not affect viral replication in cell lines while causing reduced maternal blood viremia and placental viral loads in a mouse pregnancy model (41). However, this study could not assess the effect of sfRNA on transplacental virus dissemination and fetal brain infection, as even WT ZIKV in these experiments was not detected in fetal heads (41).
Here, we used a ZIKV mutant with nearly complete sfRNA deficiency (lacking both sfRNA1 and sfRNA2) and a combination of experimental models and approaches to overcome the limitations of the previous studies and identify the biological functions of ZIKV sfRNAs at the molecular level. We demonstrated that sfRNAs are required for efficient replication of ZIKV and that deficiency in the production of sfRNAs results in significant attenuation of virus replication in human cells and in a mouse model in vivo. We found that deficiency in sfRNAs prevented the virus from infecting the mouse placenta and disseminating into the fetal brain. We also demonstrated that ZIKV sfRNAs promote virus-induced CPE in cultured cells and apoptosis of neural progenitors in the infected developing human brain tissue. We found that sfRNAs exert these functions by inhibiting the phosphorylation of STAT1 enabled by sfRNA binding to and stabilizing the viral IFN antagonist, NS5 protein. This provides a fundamental understanding of the role of sfRNAs in the subversion of host responses during ZIKV infection.
Mammalian cells produce three types of IFNs, of which type I IFN signaling is the most ubiquitous antiviral mechanism active against flaviviruses (42). Type III IFNs are also involved in defense against flaviviruses but function in a tissue-specific manner. In particular, IFN-λ pathway is active in the placenta and the female reproductive tract and plays a crucial role in protecting these tissues from ZIKV infection (26). Type II IFN is produced by natural killer cells as part of the innate immune response and subsequently by T cells as adaptive immunity develops (43). Many cells, including placental trophoblasts, express IFN-γ receptors and mount antiviral responses after exposure to IFN-γ (44). To date, IFN-γ has been demonstrated to act against WNV and DENV to prevent systemic virus spread (45, 46). In addition, treatment with IFN-γ was shown to reduce ZIKV replication in human cells (47). Here, we showed that ZIKV sfRNAs play a significant role in inhibiting the responses to IFN-γ, IFN-α, and IFN-λ by preventing the phosphorylation of STAT1.
Signal transduction from type I and III IFNs relies on Tyr701 phosphorylation of STAT1, which then associates with phosphorylated STAT2 and interferon regulatory factor 9 (IRF9) to form the transcription complex ISGF3, which translocates into the nucleus and up-regulates the expression of ISGs (48). In addition, phosphorylated STAT1 can form homodimers that mediate signal transduction from type II IFN and act as a noncanonical signaling pathway in response to type I IFN (49). Moreover, STAT1 plays a rather complex role in the regulation of apoptosis, being able to exert pro- and antiapoptotic functions depending on the type of phosphorylation and cellular context. In particular, STAT1 protects cells exposed to IFN-γ from apoptosis by activating the transcription of genes that encode the suppressors of apoptosis (29). Here, using transcriptomic data analysis of human placental cells infected with WT and sfRNA-deficient viruses, we identified STAT1 as a common link between major antiviral pathways affected by sfRNAs, IFN signaling and the negative regulation of apoptosis.
Inhibition of STAT1 phosphorylation in flavivirus-infected cells was previously linked to the activity of the viral protein NS5 (33). However, this activity of ectopically expressed NS5 in the absence of infection was found to be significantly lower than in infected cells, suggesting a requirement for another viral factor (35). Here, we identified sfRNA as this viral factor by showing that it binds to and stabilizes NS5, which results in NS5 accumulation to the levels required for inhibition of STAT1 phosphorylation. To the best of our knowledge, this is the first report of a noncoding viral RNA binding to a viral protein to inhibit host antiviral responses. ZIKV NS5 is also known to trigger proteolytic cleavage of STAT2 (27). Consistent with a recent report (36), we demonstrate that sfRNA is not required for this activity and that the small amounts of NS5 present in cells infected with a sfRNA-deficient virus were sufficient to degrade STAT2.
The concentration-dependent requirement for NS5 in the inhibition of STAT1 phosphorylation and concentration-independent requirement for NS5 in STAT2 degradation can be mechanistically explained on the basis of our data and existing knowledge (fig. S12). WNV and ZIKV NS5 were previously shown to inhibit STAT1 phosphorylation. They were also found to bind heat shock protein 90 (Hsp90), which is required to fold tyrosine kinase 2 (Tyk2), the upstream kinase that phosphorylates STAT1. Therefore, it was suggested that suppression of STAT1 phosphorylation by flavivirus NS5 could be due to this direct protein-protein interaction (35). It was proposed that binding of NS5 causes inactivation of Hsp90, resulting in incorrect folding and subsequent proteasomal degradation of Tyk2, thereby preventing downstream STAT1 phosphorylation in response to IFN receptor signaling (35). Assuming that NS5-Hsp90 binding is a reversible reaction, high levels of NS5 would be required to suppress the activity of abundantly expressed Hsp90. This is likely achieved by stabilization of NS5 by sfRNA, as we showed here, which would then allow efficient inhibition of STAT1 phosphorylation. In contrast, the effect of NS5 on STAT2 is irreversible, as it triggers the energy-dependent ubiquitination of STAT2, followed by its proteasomal degradation with NS5 being recycled. NS5 thereby acts as a catalyst (27) with small amounts of NS5 sufficient for STAT2 degradation.
A previous study indicated that NS5-induced STAT2 degradation switches signaling from STAT1/STAT2-driven responses to STAT1/STAT1-driven responses (50) in ZIKV-infected cells. Here, we propose that ZIKV counteracts type I and III IFN responses by using a small amount of available NS5 to induce STAT2 degradation and prevent the formation of STAT1/STAT2 heterodimers. Removal of STAT2 switches signaling to the STAT1/STAT1 homodimer pathway, which is used primarily by type II IFN response. sfRNA inhibits this pathway by binding to and stabilizing NS5, thus enabling NS5 to counteract STAT1 phosphorylation. Thus, the sfRNA-mediated inhibition of all three types of IFN response allows ZIKV to spread systemically, reach and replicate in the placenta, and eventually reach the fetal brain, where it induces apoptosis and neuropathogenicity.
MATERIALS AND METHODS
Cells
Female African green monkey (Cercopithecus aethiops) kidney fibroblast cells [Vero, American Type Culture Collection (ATCC), CCL-81], male human alveolar carcinoma A549 cells (ATCC, CCL-185), male human placental trophoblast BeWo cells (ATCC, CCL-98), human placental trophoblast HTR-8 cells (ATCC, CRL-3271), HEK293T cells (ATCC, CRL-3216), and Aedes albopictus larvae C6/36 cells (ATCC, CRL-1660) were obtained from ATCC. WT, IFNAR−/−, and IRF3/7−/− MEFs have been generated previously (6). U3A STAT1-deficient 2fTGH cells (51) were obtained from CellBank Australia. ReNcell human NPCs were provided by B. Day and U. Baumgartner from QIMR Berghofer Medical Research Institute (QIMRB), QLD, Australia. Reporter cell lines A459-IFNb and A549-ISRE were provided by R. Randall from the University of St Andrews, UK and described previously (52). Vero, MEF, HEK293T, and HTR-8 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS). A459 and BeWo cells were maintained in Ham’s F-12K (Kaighn’s) medium supplemented with 10% (v/v) FBS. ReNcells were cultured in KnockOut DMEM/F-12 supplemented with 2% StemPro Neural Supplement, 10 μg of epidermal growth factor recombinant human protein, 5 μg of fibroblast growth factor-basic (FGFb) recombinant human protein, and 1% GlutaMAX (Gibco) in flasks coated with Matrigel matrix basement membrane [1:100 dilution in phosphate-buffered saline (PBS); Corning]. C6/36 cells were cultured in RPMI 1640 supplemented with 10% (v/v) FBS and 10 mM Hepes (pH 7.4). All vertebrate cells were incubated at 37°C with 5% CO2. Insect C6/36 cells were cultured at 28°C in sealed containers. All cell culture media and reagents were from Gibco, USA unless specified otherwise.
Viruses and infection of cells
ZIKV strain MR766 was obtained from the Victorian Infectious Diseases Reference Laboratory. Natal strain of ZIKV was previously assembled from synthetic DNA fragments (53). The viruses were passaged once in C6/36 cells, and viral titers were determined by a foci-forming assay on Vero76 cells. The viral genomes were sequenced and confirmed to match GeneBank reference sequences MK105975 for ZIKVMR766 and KU955594 for ZIKVNatal. All infections were performed at the indicated MOI by incubating cells with 50 μl of inoculum per square centimeter of growth area for 1 hour at 37°C. Inoculated cells were then maintained in the growth medium containing a reduced amount of FBS (2%) to prevent overgrowth.
RNA isolation
Viral RNA was isolated from cell culture fluids using the NucleoSpin RNA Virus Kit (Macherey-Nagel, Germany). Total RNA from cells was isolated using TRI Reagent (Sigma-Aldrich, USA). RNA from pulldown samples was isolated using TRI Reagent LS (Sigma-Aldrich, USA). All RNA isolation procedures were conducted according to the manufacturer’s instructions.
Generation of sfRNA-deficient ZIKV mutants
The sfRNA-deficient mutants of ZIKVMR766 xrRNA1′ and xrRNA2′ were generated previously (15). To generate xrRNA1′ and xrRNA2′ mutants of ZIKVNatal, the pUC19 plasmid containing the fragment of ZIKV cDNA with full-length 3′UTR (54) was used as a template for PCR-directed site-specific mutagenesis. The primers ZIKV_xrRNA1′F and ZIKV_xrRNA1′R or ZIKV-xrRNA2′F and xrRNA2′R (table S12) were used to introduce C to G substitution into the position of each of the xrRNAs critical for XRN-1 resistance (5, 15). Mutagenesis was performed using the Q5 Site-Directed Mutagenesis Kit (NEB, USA) according to the manufacturer’s instructions. Fragments containing the mutations were then amplified from the plasmid using primers Natal 7F and Natal 7R (table S11) and used to assemble infectious cDNA via circular polymerase extension reaction, which was performed as described previously (55).
Reverse transcription polymerase chain reaction
To confirm the retention of mutations in P0 and P1 viruses, viral RNA was isolated from culture fluid samples, and ZIKV 3′UTR was amplified using primers Natal 3′UTR F and Natal 3′UTR R (table S11). RT-PCR was performed using the SuperScript III One-Step RT-PCR Kit with Platinum Taq (Invitrogen, USA) according to the manufacturer’s recommendations and the following cycling conditions: 60°C for 15 min, 94°C for 2 min followed by 30 cycles of 94°C for 15 s, 55°C for 30 s, and 68°C for 1 min with a final extension at 68°C for 5 min.
Foci-forming immunoassay
Viral titers were determined by a foci-forming assay on C6/36 cells as described previously (15).
Plaque assay
Ten-fold serial dilutions of virus samples were prepared, and 200 μl of each was used to inoculate Vero cells grown in six-well plates by incubation for 1 hour. Cells were then overlaid with DMEM containing 0.5% low–melting point agarose (Bio-Rad, USA) and 2% FBS. At 72 hpi, cells were fixed with 4% PBS-buffered formaldehyde for 1 hour, stained with 0.1% crystal violet for 30 min, and washed with water.
Northern blotting
Detection of sfRNA was performed by Northern blotting as described previously (15, 37).
Viral cytotoxicity assay
Vero cells were seeded into 96-well plates at a density of 104 cells per well. At 24 hours after seeding, serial three-fold dilutions of virus samples were prepared, and 100 μl of each dilution was used to infect the cells. Eight wells on each plate were left uninfected (mock) to be used as a control. Infection was performed by incubation with the inocula for 24 hours, which were then replaced with 100 μl of DMEM supplemented with 2% FBS. At 3 dpi, CPEs were determined on the basis of cellular adenosine triphosphate levels using Viral ToxGlo Assay (Promega, USA) according to the manufacturer’s instructions. Luminescence was measured on a DTX880 multimode detector (Beckman Coulter). Percentage survival was determined as the percentage of luciferase activity (luminescence value) in infected cells compared to uninfected control, and the CPE was calculated as 100% − (% survival). The experiment was performed in duplicate. Data were fitted into sigmoidal curves using GraphPad Prism v8.0.
Virus growth kinetics
Cells were seeded at 106 cells per well in six-well plates and inoculated with WT or mutated ZIKV at indicated MOIs by incubating for 1 hour with 200 μl of virus inoculum. Incubations were performed at 37°C; then, inoculum was removed, and cells were washed three times with PBS and overlaid with 2 ml of their relevant culture medium supplemented with 2% FBS. At time point zero, 100 μl of culture medium was immediately collected from the wells, and infected cells were then incubated for 3 days. Culture fluid samples (100 μl) were then harvested at 24, 48, and 72 hpi and subjected to a focus-forming assay to determine the virus titers from which growth curves were plotted.
Animal experiments
AG129 mice, which lack receptors for IFN-α/β and IFN-γ, were obtained from the animal facility at the Australian Institute for Bioengineering and Nanotechnology of The University of Queensland. The 5- to 6-week-old AG129 mice (mix gender) were injected via intraperitoneal route with 103 FFU per mouse of WT or sfRNA-deficient ZIKVMR766. Signs of encephalitis were monitored and scored as previously described (56). Mice were monitored two times a day for signs of neurological disease. All mice with a clinical score of three or more were immediately euthanized within 30 min by CO2 suffocation and cervical dislocation. Blood specimens were collected from the caudal vein at indicated days after infection, incubated at room temperature for 30 min, and centrifuged at 10,000g at 4°C for 20 min to separate the serum. For the pregnancy experiment, approximately 10-week-old pregnant IFNAR1−/− dams [C57BL/6J background (53)] were infected via subcutaneous injection with WT, xrRNA1′, or xrRNA2′ mutant of ZIKVNatal at the dose of 104 or 106 FFU per mouse via subcutaneous injection. Pregnant IFNAR−/− dams were infected at E12.5 as described. Mice were then monitored for 5 days, and blood samples were collected daily as described above. At 5 days after infection (E17.5), dams were euthanized, and fetuses were weighed and photographed. Fetal heads and placenta were then processed for the determination of tissue titers. One-half of each tissue sample was homogenized in 1 ml of DMEM containing 2% FBS supplemented with penicillin-streptomycin (Gibco, USA) for 5 min at 30 Hz using a TissueLyser II (QIAGEN, USA). Homogenates were then centrifuged at 10,000g for 5 min at 4°C, and supernatants were collected for virus titration. Viral loads in mouse serum and tissue homogenates were determined by a focus-forming assay on C6/36 cells using a human recombinant monoclonal antibody against ZIKV E-protein (hZ67).
Human embryonic stem cell culture and generation of iPSC-derived human brain organoids
Human embryonic stem cells, GENEA022 (provided by Genea Biocells), were cultured on extracellular matrix gel from Engelbreth-Holm-Swarm murine sarcoma (Sigma-Aldrich Pty Ltd., catalog no. E1270-5X10ML) in mTeSR medium (STEMCELL Technologies, catalog no. 85851). Cortical organoids were generated by an optimized protocol (57) where patterned embryoid bodies were expanded for 4 days in N2 medium: DMEM/F12 (Gibco, catalog no. 11320-33), 1% N-2 supplement (Gibco, catalog no. 17502-048), 2% B-27 supplement (Gibco, catalog no. 17504044), 1% MEM nonessential amino acids (Gibco, catalog no. 11140-050), 1% penicillin-streptomycin (Gibco, catalog no. 15140148), and 0.1% β-mercaptoethanol (Gibco, catalog no. 21985-023) with daily supplementation of basic fibroblast growth factor (20 ng/ml; R&D, catalog no. 233-FB-01M). Embryoid bodies were then embedded in 15 μl of Matrigel (STEMCELL Technologies, catalog no. 354277), and the media were changed to a 1:1 mixture of Neurobasal medium (Invitrogen, catalog no. 21103049) and DMEM/F12 medium supplemented with 1:200 MEM-NEAA, 1:100 GlutaMAX (Invitrogen, catalog no. 35050-038), 1:100 N2 supplement, 1:50 B-27 supplement, 1% penicillin-streptomycin, 50 μM 2-mercaptoethanol, and 0.25% insulin solution (Sigma-Aldrich, catalog no. I9278). Fresh medium was replaced thrice a week.
Infection of iPSC-derived human brain organoids
Cerebral organoids on day 15 were used for viral infection. A virus inoculum titer of 104 FFU per 50 μl of each virus was added to a single organoid-containing well of a round-bottom, ultralow-attachment 96-well plate and incubated at 37°C overnight. Each ZIKV-infected organoid was then transferred to a single well of a 24-well plate containing 500 μl of ND medium. To determine viral growth kinetics in infected organoids, 160 μl of culture supernatant was harvested from each well at the indicated time points and then replaced by the same amount of fresh culture medium. Harvested culture fluids were titered by a focus-forming assay. Three biologically independent organoids per virus were used. Infected organoids were imaged by dark-field microscopy with ×4 magnification using a Nikon Eclipse TE200 inverted microscope.
Immunohistological analysis of iPSC-derived human brain organoids
Organoids were fixed in 4% paraformaldehyde for 1 hour at room temperature and washed three times with 1× PBS at room temperature. Fixed organoids were then immersed in 30% sucrose in PBS at 4°C. Organoids were allowed to sink before embedding in a solution containing a 3:2 ratio of optimal cutting temperature compound and 30% sucrose. Embedded organoids were sectioned at a thickness of 12 μm with a Thermo Scientific CryoStar NX70 cryostat. Organoid sections were air-dried and washed three times for 10 min at room temperature followed by blocking and permeabilizing for 60 min with a solution containing 3% bovine serum albumin (Sigma-Aldrich, catalog no. A9418-50G) and 0.1% Triton X-100 in 1× PBS. Afterward, sections were incubated with primary antibodies overnight at 4°C and washed three times with 1× PBS for 10 min each at room temperature. Primary antibodies were anti-ZIKV E-protein 4G2 antibody used at 1:100 dilution, anti-NeuN (1:500; Millipore, ABN78), anti-cCasp3 (1:500; Cell Signaling Technology, 9661), and anti-Sox2 (1:500; Cell Signaling Technology, D9B8N). Secondary fluorophore-conjugated antibodies were then added for 1 hour at room temperature. All sections were incubated with Hoechst 33342 (Invitrogen, catalog no. H3570) for nuclei detection. Images were acquired using a Zeiss Axioscan Z1 based in the School of Biomedical Sciences Imaging Facilities at The University of Queensland. The number of positive cells per organoid for the indicated markers was analyzed by the imaging software CellProfiler using the same pipeline for each sample in the same experiment.
Caspase 3/7 activity assay
ReNcell human NPCs were seeded at the density of 5 × 105 cells per well in six-well plates coated with Matrigel matrix basement membrane (Corning, USA) diluted 1:100 in PBS. The next day, cells were infected at MOI = 1. At 72 hpi, culture medium was removed, and cells were washed twice with PBS and dislodged by incubation with 200 μl per well of Accutase Cell Detachment Solution (Innovative Cell Technologies, USA). Incubation was performed for 5 min at 37°C. Then, digestion was terminated by the addition of 200 μl of 0.1% trypsin inhibitor solution (Sigma-Aldrich, USA), and cells were resuspended in 1 ml of PBS and then pelleted by centrifugation at 400g for 5 min. Cells were resuspended in 500 μl of PBS, and 50 μl of suspension was transferred into an opaque 96-well plate for caspase assay. Caspase 3/7 activity was then assessed using the Caspase-Glo 3/7 Assay System (Promega, USA) according to the manufacturer’s recommendations. Luminescence was measured using a CLARIOstar Plus microplate reader (BMG Labtech, Germany).
Next-generation sequencing and bioinformatic analysis
For the analysis of BeWo cells, RNA enrichment and library preparation were performed using the TruSeq 2 Library Preparation Kit (Illumina, USA). All sample groups used for the analysis contained biological triplicates. Libraries were sequenced on an Illumina NextSeq 500 instrument using NextSeq 500/550 High Output Kit v2.5, for 1 × 75 cycles (Illumina, USA). Image analysis was performed in real time by the NextSeq Control Software and Real Time Analysis. Quality control of raw sequencing data was performed using FastQC software v.0.72. Data were then trimmed to remove PCR primers, adapters, and short reads using TRIMMOMATIC v.0.36.6 (with the following settings: ILLUMINACLIP: TruSeq2-SE:2:30:10 LEADING:32 TRAILING:32 SLIDINGWINDOW:4:20 MINLEN:25) and subjected to another quality analysis with FastQC. Trimmed reads were mapped to the human genome assembly hg38 using HISAT2 v.2.1.0, allowing one mismatch. Feature counting was performed using featureCounts v1.6.4 with counting mode set to “union” and strand to “unstranded”; feature type was “exon,” and ID attribute was Gene_ID. Genome FASTA and GFF3 files were obtained from the National Center for Biotechnology Information GenBank. Differential gene expression analysis was performed using edgeR v.3.24.0. Low-abundance reads (<1 cpm) were removed from the dataset, and data were normalized to library sizes and composition bias using the trimmed mean of M values method. Normalized data were subjected to the multidimensional scaling analysis and used to build a quasi-likelihood negative binomial generalized log-linear model. The quasi-likelihood F test (glmQLFTest) was then applied to the contrasts WT-Mock, xrRNA2′-Mock, and (WT-Mock)-(xrRNA2′-Mock). Genes were considered differentially expressed if false discovery rate–corrected P values were <0.05. Gene expression data were plotted using ggplot2 v.3.3.2. GO and KEGG pathway enrichment analyses were performed using Database for Annotation, Visualization and Integrated Discovery v6.8. Enrichment data were then combined with expression values, and z scores were calculated using the R package GOplot v.1.0.2 and plotted using ggplot2 v.3.3.2. Heatmaps were generated using heatmap.2 function of R package gplots v3.1.1. Gene interaction networks were reconstructed using Cytoscape v3.8.0. Genetic interactions were identified using the GeneMANIA Cytoscape plug-in. Betweenness centrality values represent the numbers of the shortest paths that pass through each node in the network and were calculated using the “analyze network” function of Cytoscape.
Quantitative RT-PCR
First-strand cDNA synthesis was performed using qScript cDNA SuperMix (Quantabio, USA) according to the manufacturer’s instructions. Quantitative PCR was then conducted using QuantStudio 6 Flex Real Time PCR Instrument (Applied Biosystems, USA) according to the manufacturer’s instructions. Gene expression levels were determined by ΔΔCt method relative to mock and normalized to TBP. Viral genomic RNA levels were determined using a standard curve approach by comparing the Ct values of the samples to the Ct values observed in the amplification of serial dilutions (102 to 108 copies per reaction) of a PCR-amplified and purified ZIKV genomic fragment. For each experiment, RNA from three biological replicates was used, and PCR amplification of each cDNA sample was performed in triplicate. Negative controls were included for each set of primers.
IFN treatment
To examine STAT1 phosphorylation in infected cells, Vero or HTR-8 cells were seeded in 24-well plates at a density of 2 × 105 cells per well. The next day, cells were infected with WT or the xrRNA2′ mutant of ZIKVMR766 at MOI = 5. At 48 hours posttransfection (hpt), cell culture medium was replaced with 1 ml of fresh medium containing 104 U of human IFN-α2 (Abcam, UK) or human IFN-λ1 (100 ng/ml; PeproTech, USA). Vero cells were incubated with IFN-α2 for 20 min, and HTR-8 cells were incubated with IFN-λ1 for 1 hour. Immediately after incubations, cells were lysed for Western blotting or fixed for immunofluorescent protein detection. For IFN resistance assay, IRF3/7−/− MEFs were seeded in 24-well plates and pretreated with mouse IFN-α (#HC1040A, Hycult Biotech, USA) at indicated concentrations for 6 hours. Then, cells were infected at MOI = 0.1 by incubation with inoculum for 1 hour. After the inoculum was removed, cells were washed with PBS and maintained in the media containing the same concentrations of IFN. Culture fluids were collected for analysis at 44 hpi.
Western blotting
Cells were lysed in 100 μl per well of a 24-well plate of Bolt LDS sample buffer (Thermo Fisher Scientific, USA) supplemented with Bolt sample reducing agent (Thermo Fisher Scientific, USA) and protease inhibitor cocktail (Sigma-Aldrich, USA). Lysates were sonicated using a Branson Digital Sonifier 450 (Marshall Scientific) at 10% output for 15 s with a pulse on for 3 s and off for 1 s. Lysates were then incubated at 95°C for 5 min, cooled on ice, and loaded into the Bolt 4 to 2% bis-tris gels (Thermo Fisher Scientific, USA). Electrophoresis was performed for 1 hour at 170 V in Bolt MES SDS running buffer (Thermo Fisher Scientific, USA). Proteins were then electroblotted onto a nitrocellulose membrane using iBlot2 Dry Blotting System (Thermo Fisher Scientific, USA) and iBlot 2 Transfer Stack (Thermo Fisher Scientific, USA). Membranes were blocked in clear milk blocking buffer (Pierce, USA) for 1 hour, incubated with primary antibodies overnight at 4°C, then washed four times for 5 min with Tris-buffered saline pH=7.4 with 0.05% Tween 20 (TBS-T), incubated with horseradish peroxidase (HRP)–conjugated secondary antibodies for 1 hour at room temperature, and washed four times for 5 min with TBS-T. Detection of HRP activity was performed using SuperSignal West Pico PLUS chemiluminescence substrate (Pierce, USA), and signal was visualized using Amersham Imager 600. Blot densitometry was conducted using ImageJ Fiji v.2.1.0. The primary and secondary antibodies and their dilutions are listed in table S12.
Immunofluorescence assay
Cells for immunofluorescence assay (IFA) were seeded on glass coverslips contained in a 24-well plate. After IFN treatment, cells were washed once with PBS and fixed with ice-cold 100% methanol (500 μl per well) at −20°C for 20 min. Fixed cells were rinsed twice with 500 μl of TBS and incubated with 500 μl of 50 mM glycine for 30 min at room temperature to quench autofluorescence. Cells were then washed three times for 5 min with 500 μl of TBS, blocked for 1 hour at room temperature with 500 μl of 1% bovine serum albumin in TBS, and incubated at 37°C for 1 hour with 250 μl of primary antibodies diluted in blocking solution. The mixture of antibodies contained rabbit monoclonal antibody against phosphoTyr701-STAT1 (#9167, Cell Signaling Technology, USA) diluted 1:200 and mouse monoclonal antibody 4G2 against flavivirus E-protein diluted 1:50. Cells were then washed three times for 5 min with TBS (500 μl per well) and incubated at 37°C for 1 hour with 250 μl of a mixture containing goat anti-rabbit immunoglobulin G (IgG) (H+L) highly cross-adsorbed secondary antibody conjugated with Alexa Fluor plus 488 (Invitrogen, USA) and goat anti-mouse IgG (H+L) highly cross-adsorbed secondary antibody conjugated with Alexa Fluor 594 (Invitrogen, USA), each diluted 1:500 in blocking solution. After the incubation, cells were washed three times for 5 min with 500 μl of TBS per well, and coverslips were mounted using ProLong Gold Antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen, USA). Imaging was performed using a Zeiss LSM710 confocal microscope.
In vitro transcription and transfection of RNA
For in vitro transcription, plasmids containing the sequence of interest under the control of the T7 promoter were linearized by restriction digest, and 1 μg of purified digested DNA was used as a template for the reaction performed using the MEGAscript T7 Transcription Kit (Ambion, USA) as specified by the manufacturer and purified by LiCl precipitation. For the generation of ZIKV sfRNA and GFP RNA fragment, fluorescein-labeled RNA was prepared in parallel using Fluorescein RNA Labeling Mix (Roche, Switzerland) and mixed with unlabeled RNA in 1:10 ratio to serve as a tracer. RNA was then incubated with 10 U of RppH (NEB, USA) and 1 U of XRN-1 (NEB, USA) in NEB3 buffer for 1 hour at 37°C. RNA was then purified by phenol:chloroform extraction. All in vitro transcribed RNAs were analyzed by electrophoresis in 1.5% formaldehyde agarose gel, and RNA concentration was determined on a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, USA). RNA was transfected into cells using an in-suspension protocol (58).
RNA affinity pulldown
To generate the plasmids for in vitro transcription of sfRNA and negative control RNA fused with four streptavidin affinity tags (4xS1m), the aptamer sequences followed by ZIKV sfRNA sequence or GFP gene fragment were inserted under the T7 promoter into the pUC19 vector. The DNA fragments containing the T7 promoter with 2xS1m and 2xS1m alone (T7_2xS1m_SacI_BamHI_F/R and 2xS1m_BamHI_XbaI_F/R in table S12) were obtained as two sets of complementary single-stranded Ultramer DNA oligos (IDT, USA). These oligos were designed to form terminal overhangs compatible with restriction enzyme cleavage sites within the pUC19 vector upon annealing. Before cloning, 2 nmol of each complementary oligonucleotide was annealed by incubation at 95°C for 5 min, followed by gradual cooling to room temperature in 50 μl of Duplex buffer (IDT, USA). Annealed oligos were gel-purified from 2% agarose gel, and T7_2xS1m_SacI_BamHI DNA fragment was ligated with Sac I + Bam HI–digested pUC19 plasmid using T4 DNA ligase (NEB, USA). The resultant plasmid was digested with Bam HI and Xba I and ligated with 2xS1m_BamHI_XbaI dsDNA oligo to generate pUC19-T7-4xS1m construct. The ZIKV sfRNA sequence and GFP gene fragment were PCR-amplified from pUC19-ZIKV-F4 (15) and pMYC-GFP (Addgene, #42142) plasmids, respectively, using primers HindIII_ZIKV_sfRNA and XbaI_GFP (table S11) and PrimeSTAR GXL polymerase (Takara, Japan). Cycling conditions were as follows: 35 cycles of 98°C for 10 s, 55°C for 30 s, and 68°C for 3 min and 30 s, followed by a final extension at 68°C for 5 min. PCR products were gel-purified, digested with Xba I and Hind III (NEB, USA), and ligated into pUC19_4xS1m plasmid digested with the same enzymes. The resulting constructs pUC19_sfRNA_4xS1m and pUC19_GFP_4xS1m and all intermediate plasmids were Sanger sequenced and shown to conform to the desired design and sequence.
RNA affinity purification was performed using optimized S1m streptavidin RNA aptamer (59) and the previously described protocol with minor modifications (32). A549 cells were seeded in 10× T175 flasks at a density of 1 × 107 cells per flask and infected with ZIKVMR766-xrRNA2′ at MOI = 0.5. At 48 hpi, the cells were washed once with ice-cold diethyl pyrocarbonate (DEPC)–treated PBS, harvested by scraping in 10 ml of DEPC-PBS per flask, pelleted by centrifugation at 300g for 5 min, and washed twice in DEPC-PBS.Cells were then lysed by incubation on ice for 30 min in 2 ml of SA-RNA lysis buffer [20 mM tris-HCl (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 2 mM dithiothreitol (DTT), and 2 mM vanadyl ribonucleoside complex ribonuclease inhibitor; NEB, USA], supplemented with 1× protease inhibitor cocktail (Sigma-Aldrich), 1× PhosphoSTOP phosphatase inhibitor cocktail (Sigma-Aldrich, USA), and 1% NP-40 (Sigma-Aldrich, USA). The cell lysates were then sonicated using Branson Digital Sonifier 450 (Marshall Scientific) at 10% output for 15 s with a pulse on for 3 s and off for 1 s. Lysates were cleared by centrifugation at 16,000g for 15 min at 4°C and split into halves for incubation with sfRNA bait and GFP bait.
The in vitro transcribed, aptamer-containing RNA was mixed with SA-RNA lysis buffer to make up a final volume of 50 μl. RNA was renatured by incubation at 56°C for 5 min, 37°C for 10 min, and room temperature for 2 min and then chilled on ice. For RNA-to-bead coupling, 100 μl of Streptavidin Sepharose High-Performance bead slurry (GE Healthcare, USA) was first equilibrated by washing three times with 500 μl of SA-RNP lysis buffer and collected by centrifugation at 100g for 2 min, then resuspended in 100 μl of SA-RNP lysis buffer supplemented with 80 U RNasin (Promega, USA), and incubated with 30 μg of renatured RNA with overhead rotation at 4°C for 2.5 hours. The A549 cell lysates were precleared by incubation with 100 μl of equilibrated SA beads for 2.5 hours at 4°C with overhead rotation. Beads were then removed by centrifugation at 16,000g for 1 min. Next, lysates were supplemented with 40 U of RNasin (Promega, USA) and incubated with sfRNA_4xS1m or GFP_4xS1m–coupled SA beads at 4°C for 3.5 hours with overhead rotation. Then, the protein-bound SA beads were collected by centrifugation at 100g for 2 min and washed five times with SA wash buffer [20 mM tris-HCl (pH 7.5), 300 mM NaCl, 5 mM MgCl2, 2 mM DTT, 1× VRC, and 1× protease inhibitor cocktail; Sigma-Aldrich], followed by a final wash with SA wash buffer supplemented without supplements. Bound proteins were eluted from the beads by incubation with 100 μl of 8 M urea/50 mM ammonium bicarbonate (ABC) for 15 min with overhead rotation. Beads were removed by centrifugation at 3800g for 1 min, and the elutes were collected.
Mass spectrometry
Pulldown elutes were centrifuged at 16,000g for 10 min to remove insoluble materials, and protein concentrations were determined using the Pierce BCA Protein Assay Kit (Pierce, USA). Samples containing 10 μg of protein were transferred to an Amicon Ultra 10k 0.5-ml centrifugal filter column (Amicon, USA) and centrifuged at 14,000g for 40 min. The column was washed with 500 μl of wash solution (8 M urea and 50 mM ABC) and centrifuged at 14,000g for 4 min. The on-column reduction of the proteins was then performed in a 200-μl wash solution containing 5 mM DTT at 56°C for 30 min. Then, iodoacetamide was added to the solution to a final concentration of 25 mM, and alkylation was performed at room temperature in the dark for 30 min. The reaction was terminated by adding DTT to a final concentration of 10 mM, and the reaction solution was removed by centrifugation at 14,000g. Proteins were then resuspended in 100 μl of 50 mM ABC and digested overnight at 37°C using 0.2 μg of trypsin per 10 μg of protein. To collect the peptides, the column was transferred to another collection tube and centrifuged at 14,000g at room temperature for 40 min. Fifty microliters of 0.5 M NaCl was added to the column and centrifuged to ensure the complete elution of the peptides. The elutes were combined, and the peptides were further purified using C18 ZipTip Pipette Tips (Millipore, USA) as per the manufacturer’s instructions.
Peptides were separated using reversed-phase chromatography on a Shimadzu Prominence NanoLC system. Using a flow rate of 30 μl/min, samples were desalted on an Agilent C18 trap (0.3 mm by 5 mm, 5 μm) for 3 min, followed by separation on a Vydac Everest C18 (300 A, 5 μm, 150 mm by 150 μm) column at a flow rate of 1 μl/min. A gradient of 10 to 60% buffer B over 45 min, where buffer A = 1% ACN and 0.1% FA and buffer B = 80% ACN and 0.1% FA, was used to separate peptides. Eluted peptides were directly analyzed on a TripleTOF 5600 instrument (AB Sciex) using a NanoSpray III interface. Gas and voltage settings were adjusted as required. Time-of-flight mass spectrometry scan across mass/charge ratio (m/z) of 350 to 1800 was performed for 0.5 s, followed by the information-dependent acquisition of the top 20 peptides across m/z of 40 to 1800 (0.05 s per spectra).
Reference protein sequences for human and ZIKVMR766 proteins were obtained from Swiss-Prot database. Mass spectrometry data were converted to mgf format and searched using Paragon method and ProteinPilot software (Sciex, USA). Search settings were as follows: Trypsin was set as the enzyme; the instrument was set to TripleTOF 5600, and cys modifications were set to iodoacetamide. Peak areas were quantified in PeakView (Sciex, USA) using the SWATH Acquisition MicroApp (Sciex, USA). Protein enrichment analysis was performed on peak area data using Limma v3.46.0 and R v3.6.3.
Generation of expression plasmids
To generate the expression plasmids that produce ZIKV NS5 with sfRNA and GFP with ZIKV sfRNA, DNA fragments containing Kozak sequence followed by the corresponding ORF, ZIKV 3′UTR, and HDVR were cloned into pcDNA3.1(+) expression vector. The fragments were obtained as synthetic custom genes cloned in the pUCIDT vector (IDT, USA). To introduce the restriction sites compatible with pcDNA3.1(+), fragments were PCR-amplified from the plasmids using PrimeSTAR GXL polymerase (Takara, Japan) and Nhel_IDT_F and EcoRI_ITD_R primers (table S11), which are complementary to the pUCIDT vector backbone immediately upstream and downstream of the insert. The cycling conditions were as follows: 98°C for 2 min, 35 cycles of 98°C for 10 s, 55°C for 30 s, and 68°C for 3 min and 30 s, followed by a 5-min extension at 68°C. The PCR amplicons were gel-purified, digested with Nhe I and Eco RI, and ligated with pcDNA3.1(+) vector cut with the same enzymes.
To generate the plasmids pcDNA3.1(+)-ZIKV_NS5_3′UTR-Mut and pcDNA3.1(+)-GFP_ZIKV_3′UTR that respectively produce ZIKV NS5 without sfRNA and neither NS5 nor sfRNA (negative control), PCR-directed site-specific mutagenesis was used to introduce point mutations into two xrRNA regions of 3′UTRs within pcDNA3.1(+)-ZIKV_NS5_3′UTR and pcDNA3.1(+)-GFP_ZIKV_3′UTR plasmids. Mutagenesis was performed in two steps using a Q5 site-directed mutagenesis kit (NEB, USA) as per the manufacturer’s instructions. The first round of PCR mutagenesis was conducted using xrRNA1_C22G primers (table S11). The cycling conditions were as follows: 98°C for 30 s, 35 cycles of 98°C for 10 s, 55°C for 30 s, and 72°C for 4 min and 30 s, followed by a 2-min extension at 72°C. The PCR-amplified plasmids were circularized, propagated in Escherichia coli, and used as templates for the second round of PCR mutagenesis, under the same cycling conditions with xrRNA2_C22G primers. All expression plasmids were deep-sequenced using the Oxford Nanopore platform.
Nanopore sequencing
Nanopore sequencing of amplicons was performed as described previously (60). The regions of the expression plasmids containing the entire inserts were PCR-amplified using PrimeSTAR GXL Polymerase (Takara, Japan) and Nanopore_pCMV_F and Nanopore_BGH_R primers (table S11). The amplicons were gel-purified and subjected to the preparation of barcoded libraries using the PCR Barcoding Expansion Kit (EXP-PBC001, Oxford Nanopore Technologies, UK). Sequencing runs were conducted using the MinION Flongle flow cell using MinKNOW software (Oxford Nanopore Technologies, UK). Base calling and adapter trimming of the fast5 files containing sequencing reads were conducted using guppy_basecaller v 3.3.0+ef22818 (Oxford Nanopore Technologies, UK). The fastq files containing the reads were used for reads mapping against the reference plasmid sequence using the Bowtie2 tool (Galaxy Australia). The binary alignment file was visualized using Integrative Genomics Viewer 2.8 (Broad Institute, USA).
Plasmid transfection
HEK293T cells were seeded in a 24-well plate at a density of 1.5 × 105 cells per well and transfected with 0.14 pmol of plasmid DNA using Lipofectamine 2000 (2.5 μl per well; Invitrogen, USA) according to the manufacturer’s instructions. At 48 hpt, cells in each well were treated with 50 U of human IFN-α2 (Abcam, UK) for 15 min or left untreated as a control, then washed with PBS, and lysed for Western blotting.
MG132 treatment
HEK293 cells were transfected with the plasmids as described above. At 24 hpt, the cell culture medium was supplemented with 50 μM MG132, and cells were incubated with the drug for 24 hours and then lysed for Western blot analysis.
Statistical analysis
Statistical analysis was performed using Graph Pad Prism software version 8, R Studio version 0.99.893, and IBM SPSS Statistics v23. Specific statistical treatments are described in the figure legends and supplementary tables.
Research standards
All mouse work was conducted in accordance with the “Australian code for the care and use of animals for scientific purposes” as defined by the National Health and Medical Research Council of Australia. Mouse work was approved by the QIMR Berghofer Medical Research Institute animal ethics committee (P2195) and The University of Queensland animal ethics committee (AE31401).
All experiments with human stem cells were carried out in accordance with the ethical guidelines of The University of Queensland and with the approval by The University of Queensland Human Research Ethics Committee (approval no. 2019000159). A commercially available human embryonic stem cell (hESC) line was used. The manufacturer reported that karyotyped embryo cells were fully consented to the development of stem cells by all responsible people through an informed consent process (a signed deidentified consent form can be provided upon request). Donors have received no payment or other benefits for their donation. Donated embryos were originally created by assisted reproduction technology for the purpose of procreation. Embryos were identified as unsuitable for implantation, biopsy, or freezing because of abnormal development. Embryonic outgrowths were developed for consented clinical investigation studies.
Acknowledgments
We acknowledge the excellent support of A. Nouwens and P. Josh from the School of Chemistry and Molecular Biosciences (The University of Queensland) for mass spectrometry and proteomics data analysis. We thank M. Kasherman and S. Walters from the School of Biomedical Sciences Imaging and Analytical facilities (The University of Queensland) for technical support. We are thankful to R. Hall (The University of Queensland), A. Merits and E. Žusinaite (University of Tartu), and B. Day and U. Baumgartner (QIMR Berghofer Medical Research Institute) for providing antibodies and cell lines. Next-generation sequencing was performed at ACE Sequencing (The University of Queensland). Sanger sequencing was performed by the Australian Genomics Research Facility.
Funding: This work was supported by National Health and Medical Research Council (NHMRC) Ideas grants APP1127916 and APP1059794 (to A.A.K.); NHMRC Investigator grant APP1173880 (to A.Su.); NHMRC Ideas grants APP1138795, APP1127976, APP1144806, and APP1130168 (to E.W.); Australian Research Council (ARC) Discovery grant DP210103401 (to E.W.); NHMRC Ideas grant APP2001408 (to J.A.); The University of Queensland Early Career Researcher Grant UQECR2058457 (to J.A.); and a Jérôme Lejeune Postdoctoral Fellowship and Brisbane Children’s Hospital Foundation grant 50308 (to J.A.). E.W. was supported by the BrAshA-T Foundation and the Perry Cross Spinal Research Foundation.
Author contributions: Conceptualization: A.Sl. and A.A.K. Methodology: A.Sl. and X.W. Investigation: A.Sl., X.W., H.C., J.A., M.E.F., K.Y., F.J.T., A.A.A., R.B., Y.X.S., J.D.J.S., D.W., and N.P. Software: A.Sl. Visualization: A.Sl. and A.Su. Supervision: A.A.K., A.Su., E.W., and A.Sl. Writing (original draft): A.Sl. and A.A.K. Writing (review and editing): A.Sl., A.Su., and A.A.K.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq datasets are deposited in the NCBI Gene Expression Omnibus database with accession number GSE171648.
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S12
REFERENCES AND NOTES
- 1.Mazeaud C., Freppel W., Chatel-Chaix L., The multiples fates of the flavivirus RNA genome during pathogenesis. Front. Genet. 9, 595 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clarke B. D., Roby J. A., Slonchak A., Khromykh A. A., Functional non-coding RNAs derived from the flavivirus 3′ untranslated region. Virus Res. 206, 53–61 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Scherbik S. V., Paranjape J. M., Stockman M., Silverman R. H., Brinton M. A., Stockman B. M., Silverman R. H., Brinton M. A., RNase L plays a role in the antiviral response to West Nile virus. J. Virol. 80, 2987–2999 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Funk A., Truong K., Nagasaki T., Torres S., Floden N., Balmori Melian E., Edmonds J., Dong H., Shi P.-Y., Khromykh A. A., RNA structures required for production of subgenomic flavivirus RNA. J. Virol. 84, 11407–11417 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akiyama B. M., Laurence H. M., Massey A. R., Costantino D. A., Xie X., Yang Y., Shi P.-Y. Y., Nix J. C., Beckham J. D., Kieft J. S., Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science 354, 1148–1152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuessler A., Funk A., Lazear H. M., Cooper D. A., Torres S., Daffis S., Jha B. K., Kumagai Y., Takeuchi O., Hertzog P., Silverman R., Akira S., Barton D. J., Diamond M. S., Khromykh A. A., West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J. Virol. 86, 5708–5718 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filomatori C. V., Carballeda J. M., Villordo S. M., Aguirre S., Pallarés H. M., Maestre A. M., Sánchez-Vargas I., Blair C. D., Fabri C., Morales M. A., Fernandez-Sesma A., Gamarnik A. V., Dengue virus genomic variation associated with mosquito adaptation defines the pattern of viral non-coding RNAs and fitness in human cells. PLOS Pathog. 13, e1006265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manokaran G., Finol E., Wang C., Gunaratne J., Bahl J., Ong E. Z., Tan H. C., Sessions O. M., Ward A. M., Gubler D. J., Harris E., Garcia-Blanco M. A., Ooi E. E., Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350, 217–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bidet K., Dadlani D., Garcia-Blanco M. A., G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLOS Pathog. 10, e1004242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slonchak A., Khromykh A. A., Subgenomic flaviviral RNAs: What do we know after the first decade of research. Antiviral Res. 159, 13–25 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Ali S., Gugliemini O., Harber S., Harrison A., Houle L., Ivory J., Kersten S., Khan R., Kim J., LeBoa C., Nez-Whitfield E., O’Marr J., Rothenberg E., Segnitz R. M., Sila S., Verwillow A., Vogt M., Yang A., Mordecai E. A., Environmental and social change drive the explosive emergence of zika virus in the Americas. PLOS Negl. Trop. Dis. 11, e0005135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mlakar J., Korva M., Tul N., Popović M., Poljšak-Prijatelj M., Mraz J., Kolenc M., Rus K. R., Vipotnik T. V., Vodušek V. F., Vizjak A., Pižem J., Petrovec M., Zupanc T. A., Zika virus associated with microcephaly. N. Engl. J. Med. 374, 951–958 (2016). [DOI] [PubMed] [Google Scholar]
- 13.de Araújo T. V. B., de A. Ximenes R. A., de B. Miranda-Filho D., Souza W. V., Montarroyos U. R., de Melo A. P. L., Valongueiro S., de F. P. M. de Albuquerque M., Braga C., Filho S. P. B., Cordeiro M. T., Vazquez E., di C. S. Cruz D., Henriques C. M. P., Bezerra L. C. A., da S. Castanha P. M., Dhalia R., Marques-Júnior E. T. A., Martelli C. M. T., Rodrigues L. C., Dhalia C., Santos M., Cortes F., de Oliveira W. K., Coelho G. E., Cortez-Escalante J. J., Campelo de Albuquerque de Melo C. F., Ramon-Pardo P., Aldighieri S., Mendez-Rico J., Espinal M., Torres L., Hazin A. N., Van der Linden A., Coentro M., Dimech G. S., Siqueira de Assuncao R., Ismael de Carvalho P., Oliveira V. F., Association between microcephaly, Zika virus infection, and other risk factors in Brazil: Final report of a case-control study. Lancet Infect. Dis. 18, 328–336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pijlman G. P., Funk A., Kondratieva N., Leung J., Torres S., van der Aa L., Liu W. J., Palmenberg A. C., Shi P.-Y. Y., Hall R. A., Khromykh A. A., A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 4, 579–591 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Slonchak A., Hugo L. E., Freney M. E., Hall-Mendelin S., Amarilla A. A., Torres F. J., Setoh Y. X., Peng N. Y. G., Sng J. D. J., Hall R. A., van den Hurk A. F., Devine G. J., Khromykh A. A., Zika virus noncoding RNA suppresses apoptosis and is required for virus transmission by mosquitoes. Nat. Commun. 11, 2205 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emeny J. M., Morgan M. J., Regulation of the interferon system: Evidence that vero cells have a genetic defect in interferon production. J. Gen. Virol. 43, 247–252 (1979). [DOI] [PubMed] [Google Scholar]
- 17.De Borba L., Villordo S. M., Marsico F. L., Carballeda J. M., Filomatori C. V., Gebhard L. G., Pallarés H. M., Lequime S., Lambrechts L., Vargas I. S., Blair C. D., Gamarnik A. V., RNA structure duplication in the dengue virus 3′ UTR: Redundancy or host specificity? mBio. 10, e02506-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradley M. P., Nagamine C. M., Animal models of Zika virus. Comp. Med. 67, 242–252 (2017). [PMC free article] [PubMed] [Google Scholar]
- 19.Prow N. A., Liu L., Nakayama E., Cooper T. H., Yan K., Eldi P., Hazlewood J. E., Tang B., Le T. T., Setoh Y. X., Khromykh A. A., Hobson-Peters J., Diener K. R., Howley P. M., Hayball J. D., Suhrbier A., A vaccinia-based single vector construct multi-pathogen vaccine protects against both Zika and chikungunya viruses. Nat. Commun. 9, 1230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hobson-Peters J., Harrison J. J., Watterson D., Hazlewood J. E., Vet L. J., Newton N. D., Warrilow D., Colmant A. M. G., Taylor C., Huang B., Piyasena T. B. H., Chow W. K., Setoh Y. X., Tang B., Nakayama E., Yan K., Amarilla A. A., Wheatley S., Moore P. R., Finger M., Kurucz N., Modhiran N., Young P. R., Khromykh A. A., Bielefeldt-Ohmann H., Suhrbier A., Hall R. A., A recombinant platform for flavivirus vaccines and diagnostics using chimeras of a new insect-specific virus. Sci. Transl. Med. 11, eaax7888 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Sutarjono B., Can we better understand how zika leads to microcephaly? A systematic review of the effects of the zika virus on human brain organoids. J Infect Dis 219, 734–745 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Watanabe M., Buth J. E., Vishlaghi N., de la Torre-Ubieta L., Taxidis J., Khakh B. S., Coppola G., Pearson C. A., Yamauchi K., Gong D., Dai X., Damoiseaux R., Aliyari R., Liebscher S., Schenke-Layland K., Caneda C., Huang E. J., Zhang Y., Cheng G., Geschwind D. H., Golshani P., Sun R., Novitch B. G., Self-organized cerebral organoids with human-specific features predict effective drugs to combat Zika virus infection. Cell Rep. 21, 517–532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian X., Nguyen H. N., Song M. M., Hadiono C., Ogden S. C., Hammack C., Yao B., Hamersky G. R., Jacob F., Zhong C., Yoon K. J., Jeang W., Lin L., Li Y., Thakor J., Berg D. A., Zhang C., Kang E., Chickering M., Nauen D., Ho C. Y., Wen Z., Christian K. M., Shi P. Y., Maher B. J., Wu H., Jin P., Tang H., Song H., Ming G. L., Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238–1254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferraris P., Cochet M., Hamel R., Gladwyn-Ng I., Alfano C., Diop F., Garcia D., Talignani L., Montero-Menei C. N., Nougairède A., Yssel H., Nguyen L., Coulpier M., Missé D., Zika virus differentially infects human neural progenitor cells according to their state of differentiation and dysregulates neurogenesis through the Notch pathway. Emerg. Microbes Infect. 8, 1003–1016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang N., Zhang N., Qin C. F., Liu X., Shi L., Xu Z., Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell 19, 120–126 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Bayer A., Lennemann N. J., Ouyang Y., Bramley J. C., Morosky S., Marques E. T. D. A., Cherry S., Sadovsky Y., Coyne C. B., Sirohi D., Kuhn R. J., Bayer A., Lennemann N. J., Ouyang Y., Bramley J. C., Morosky S., Marques E. T. D. A., Cherry S., Sadovsky Y., Coyne C. B., Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host Microbe 19, 705–712 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant A., Ponia S. S., Tripathi S., Balasubramaniam V., Miorin L., Sourisseau M., Schwarz M. C., Sánchez-Seco M. P., Evans M. J., Best S. M., García-Sastre A., Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19, 882–890 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Platanias L. C., Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Lostao L., Briones J., Forné I., Martinez-Gallo M., Ferrer B., Sierra J., Rodriguez-Sanchez J. L., Juarez C., Role of the STAT1 pathway in apoptosis induced by fludarabine and JAK kinase inhibitors in B-cell chronic lymphocytic leukemia. Leuk. Lymphoma 46, 435–442 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Soto-Acosta R., Xie X., Shan C., Baker C. K., Shi P.-Y., Rossi S. L., Garcia-Blanco M. A., Bradrick S., Fragile X mental retardation protein is a Zika virus restriction factor that is antagonized by subgenomic flaviviral RNA. eLife 7, e39023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillips S. L., Soderblom E. J., Bradrick S. S., Garcia-blanco M. A., Identification of proteins bound to dengue viral RNA in vivoreveals new host proteins important for virus replication. MBio 7, e01865–e01815 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Göertz G. P., Van Bree J. W. M., Hiralal A., Fernhout B. M., Steffens C., Boeren S., van Bree J. W. M., Hiralal A., Fernhout B. M., Steffens C., Boeren S., Visser T. M., Vogels C. B. F., Abbo S. R., Fros J. J., Koenraadt C. J. M., van Oers M. M., Pijlman G. P., Subgenomic flavivirus RNA binds the mosquito DEAD/H-box helicase ME31B and determines Zika virus transmission by Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 116, 19136–19144 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W. J., Wang X. J., Wang X. J., Mokhonov V. V., Shi P.-Y. P., Randall R., Khromykh A. A., Inhibition of interferon signaling by the New York 99 strain and kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 79, 1934–1942 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin R.-J., Chang B.-L., Yu H.-P., Liao C.-L., Lin Y.-L., Blocking of interferon-induced jak-stat signaling by japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 80, 5908–5918 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roby J. A., Esser-nobis K., Dewey-verstelle E. C., Fairgrieve M. R., Schwerk J., Lu A. Y., Soveg F. W., Hemann E. A., Hatfield L. D., Keller B. C., Shapiro A., Forero A., Stencel-baerenwald J. E., Savan R., Gale M. Jr., Flavivirus nonstructural protein NS5 dysregulates HSP90 to broadly inhibit JAK/STAT Signaling. Cell 9, 899 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pallarés H. M., Soledad G., Navarro C., Villordo S. M., Merwaiss F., De Borba L., Gonzalez M. M., Ledesma L., Ojeda D. S., Henrion-Lacritick A., Morales M. A., Fabri C., María C., Saleh C., Gamarnik A. V., Zika virus subgenomic flavivirus RNA generation requires cooperativity between duplicated RNA structures that are essential for productive infection in human cells. J. Virol. 94, 343–363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slonchak A., Parry R., Pullinger B., Sng J. D. J., Wang X., Buck T. F., Torres F. J., Harrison J. J., Colmant A. M. G., Hobson-Peters J., Hall R. A., Tuplin A., Khromykh A. A., Structural analysis of 3’UTRs in insect flaviviruses reveals novel determinants of sfRNA biogenesis and provides new insights into flavivirus evolution. Nat. Commun. 13, 1279 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnettler E., Sterken M. G., Leung J. Y., Metz S. W., Geertsema C., Goldbach R. W., Vlak J. M., Kohl A., Khromykh A. A., Pijlman G. P., Non-coding flavivirus RNA displays RNAi suppressor activity in insect and mammalian cells. J. Virol. 86, 13486–13500 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Göertz G. P., Fros J. J., Miesen P., Vogels C. B. F., van der Bent M. L., Geertsema C., Koenraadt C. J. M., van Rij R. P., van Oers M. M., Pijlman G. P., Noncoding subgenomic flavivirus RNA is processed by the mosquito rna interference machinery and determines West Nile virus transmission by culex pipiens mosquitoes. J. Virol. 90, 10145–10159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.J. A. Roby, A. Funk, A. A. Khromykh, Flavivirus Replication and Assembly, in Molecular Virology and Control of Flaviviruses, I. Shipy, Ed. (Caister Academic Press, 2012), pp. 21–49. [Google Scholar]
- 41.Sparks H., Monogue B., Akiyama B., Kieft J., Beckham J. D., Disruption of Zika virus xrRNA1-dependent sfRNA1 production results in tissue-specific attenuated viral replication. Viruses 12, 1177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lazear H. M., Schoggins J. W., Diamond M. S., Shared and distinct functions of type I and type III interferons. Immunity 50, 907–923 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Randall R. E., Goodbourn S., Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89, 1–47 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Banerjee S., Smallwood A., Moorhead J., Chambers A. E., Papageorghiou A., Campbell S., Nicolaides K., Placental expression of interferon-γ (IFN-γ) and its receptor IFN-γR2 fail to switch from early hypoxic to late normotensive development in preeclampsia. J. Clin. Endocrinol. Metab. 90, 944–952 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Shrestha B., Wang T., Samuel M. A., Whitby K., Craft J., Fikrig E., Diamond M. S., Gamma interferon plays a crucial early antiviral role in protection against West Nile virus infection. J. Virol. 80, 5338–5348 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prestwood T. R., Morar M. M., Zellweger R. M., Miller R., May M. M., Yauch L. E., Lada S. M., Shresta S., Gamma interferon (IFN-γ) receptor restricts systemic dengue virus replication and prevents paralysis in IFN-α/β receptor-deficient mice. J. Virol. 86, 12561–12570 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamel R., Dejarnac O., Wichit S., Ekchariyawat P., Neyret A., Luplertlop N., Perera-Lecoin M., Surasombatpattana P., Talignani L., Thomas F., Cao-Lormeau V.-M., Choumet V., Briant L., Desprès P., Amara A., Yssel H., Missé D., Biology of Zika virus infection in human skin cells. J. Virol. 89, 8880–8896 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mogensen T. H., IRF and STAT transcription factors—From basic biology to roles in infection, protective immunity, and primary immunodeficiencies. Front. Immunol. 9, 3047 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramana C. V., Chatterjee-Kishore M., Nguyen H., Stark G. R., Complex roles of Stat1 in regulating gene expression. Oncogene 19, 2619–2627 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Chaudhary V., Yuen K.-S., Chan J. F., Chan C.-P., Wang P.-H., Cai J.-P., Zhang S., Liang M., Kok K.-H., Chan C.-P., Yuen K.-Y., Jin D.-Y., Selective activation of type II interferon signaling by Zika virus NS5 protein. J. Virol. 91, e00163-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKendry R., John J., Flavell D., Muller M., Kerr I. M., Stark G. R., High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc. Natl. Acad. Sci. U.S.A. 88, 11455–11459 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S., Short J. A. L., Young D. F., Killip M. J., Schneider M., Goodbourn S., Randall R. E., Heterocellular induction of interferon by negative-sense RNA viruses. Virology 407, 247–255 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Setoh Y. X., Prow N. A., Peng N., Hugo L. E., Devine G., Hazlewood J. E., Suhrbier A., Khromykh A. A., De novo generation and characterization of new Zika virus isolate using sequence data from a microcephaly case. mSphere 2, e00190-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Setoh Y. X., Amarilla A. A., Peng N. Y. G., Griffiths R. E., Carrera J., Freney M. E., Nakayama E., Ogawa S., Watterson D., Modhiran N., Nanyonga F. E., Torres F. J., Slonchak A., Periasamy P., Prow N. A., Tang B., Harrison J., Hobson-Peters J., Cuddihy T., Cooper-White J., Hall R. A., Young P. R., Mackenzie J. M., Wolvetang E., Bloom J. D., Suhrbier A., Khromykh A. A., Determinants of Zika virus host tropism uncovered by deep mutational scanning. Nat. Microbiol. 4, 876–887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piyasena T. B. H., Setoh Y. X., Hobson-Peters J., Newton N. D., Bielefeldt-Ohmann H., McLean B. J., Vet L. J., Khromykh A. A., Hall R. A., Infectious DNAs derived from insect-specific flavivirus genomes enable identification of pre- and post-entry host restrictions in vertebrate cells. Sci. Rep. 7, 2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lazear H. M., Govero J., Smith A. M., Platt D. J., Fernandez E., Miner J. J., Diamond M. S., A mouse model of Zika virus pathogenesis. Cell Host Microbe 19, 720–730 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lancaster M. A., Renner M., Martin C. A., Wenzel D., Bicknell L. S., Hurles M. E., Homfray T., Penninger J. M., Jackson A. P., Knoblich J. A., Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slonchak A., Shannon R. P., Pali G., Khromykh A. A., Human microRNA miR-532-5p exhibits antiviral activity against West Nile virus via suppression of host genes SESTD1 and TAB3 required for virus replication. J. Virol. 90, 2388–2402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leppek K., Stoecklin G., An optimized streptavidin-binding RNA aptamer for purification of ribonucleoprotein complexes identifies novel ARE-binding proteins. Nucleic Acids Res. 42, e13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rawle D. J., Nguyen W., Dumenil T., Parry R., Warrilow D., Tang B., Le T. T., Slonchak A., Khromykh A. A., Lutzky V. P., Yan K., Suhrbier A., Sequencing of historical isolates, k-mer mining and high serological cross-reactivity with ross river virus argue against the presence of getah virus in australia. Pathogens 9, 848 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Tables S1 to S12