Abstract
Anemia affects over 25% of the world’s population with the heaviest burden borne by women and children. Genetic hemoglobin (Hb) variants, such as sickle cell disease, are among the major causes of anemia. Anemia and Hb variant are pathologically interrelated and have an overlapping geographical distribution. We present the first point-of-care (POC) platform to perform both anemia detection and Hb variant identification, using a single paper-based electrophoresis test. Feasibility of this new integrated diagnostic approach is demonstrated via testing individuals with anemia and/or sickle cell disease. Hemoglobin level determination is performed by an artificial neural network (ANN) based machine learning algorithm, which achieves a mean absolute error of 0.55 g/dL and a bias of −0.10 g/dL against the gold standard (95% limits of agreement: 1.5 g/dL) from Bland-Altman analysis on the test set. Resultant anemia detection is achieved with 100% sensitivity and 92.3% specificity. With the same tests, subjects with sickle cell disease were identified with 100% sensitivity and specificity. Overall, the presented platform enabled, for the first time, integrated anemia detection and hemoglobin variant identification using a single point-of-care test.
Keywords: Anemia, blood hemoglobin level, genetic hemoglobin disorders, sickle cell anemia, machine learning, neural network, point-of-care diagnostics
A point-of-care diagnosticx technology and approach is presented to perform both anemia detection and hemoglobin variant identification in a single test using paper-based microchip electrophoresis.
INTRODUCTION
Anemia, characterized by a low blood hemoglobin (Hb) level, is among the world’s most common and serious health conditions 1. Anemia has high prevalence affecting a third of the world’s population or about 1.62 billion people with the heaviest morbidity and mortality among women and children living in low-and-middle-income countries 2–4. In sub-Saharan Africa, the prevalence of anemia among preschool children is extremely high and has been reported to be as high as 91% in West Africa 5. Anemia can cause severe consequences including impaired cognitive and behavioral development in children6, and poor birth outcomes 7, 8, and, decreased work productivity in adults 9.
Genetic Hb disorders, such as sickle cell disease (SCD), are among the major causes for anemia globally 1, 10. Inherited Hb disorders afflict nearly 7% of the world’s population, with most structural Hb variants having the recessive β-globin gene mutations, βS (Hb S) and βC (Hb C) 11–13. SCD arises when these mutations are inherited homozygously (Hb SS, SCD-SS) or paired with another β-globin gene mutation, such as hemoglobin C (Hb SC or SCD-SC).
Anemia and Hb variant are pathologically interrelated and have an overlapping geographical distribution. Under deoxygenation, sickle hemoglobin polymerizes, which induces the sickling of red blood cells (RBCs). Sickled RBCs have increased rigidity and fragility and are prone to intravascular hemolysis, leading to chronic hemolytic anemia, vasculopathy, and increased morbidity and mortality. Anemia and SCD have an overlapping pathophysiology and geographical distribution, causing high morbidity and mortality predominantly in resource-constrained regions of sub-Saharan Africa and south-east Asia14–16.
Anemia and SCD-related complications can be mitigated by early diagnosis followed by timely intervention 17–21. Anemia treatment depends on the accurate characterization of the cause, such as inherited Hb disorders 22, 23. Meanwhile, Hb disorders such as SCD benefit from close monitoring of blood Hb levels during treatment21. As a result, it is uniquely valuable to integrate analyses of blood Hb levels, i.e., anemia status, and of Hb variants, especially in sub-Saharan Africa and India where anemia and inherited Hb disorders are the most prevalent 24. Blood Hb levels (in g/dL) is used as the indicator of anemia 25, while the presence of Hb variants in blood is the primary indicator of an inherited disorder 26. In resource-rich countries, standard clinical laboratory tests, including complete blood counts (CBCs) and high-performance liquid chromatography (HPLC), are typically used in the diagnosis of anemia and inherited Hb disorders 25, 26. However, advanced laboratory techniques require trained personnel and state-of-the-art facilities, which are lacking in the very countries in which both anemia and Hb disorders are most prevalent 14, 19. Therefore, there is a need for affordable, portable, easy-to-use, accurate point-of-care tests that facilitate decentralized anemia detection and Hb variant identification 14, 19, 23, 27.
In a 2019 report, the World Health Organization (WHO) listed both anemia and sickle cell as the most essential in vitro diagnostic (IVD) tests needed for primary care use in low and middle income countries 28. Point-of-care (POC) assays including HemoCue® 29, AnemoCheckTM 30, and smartphone based technologies31, 32 have been developed for anemia testing. Lateral flow assays including Sickle SCAN 33 and HemoTypeSC 34 have been developed for Hb variant testing. However, these POC assays only enables individual detection of Hb level or Hb variant. Therefore, measurement of these two pathologically interconnected and geographically overlapped clinical endpoints require using of two distinct platforms 35.
Hb electrophoresis has recently been added to the WHO essential list of IVDs for diagnosing SCD and sickle cell trait 36. We have previously developed and extensively validated an affordable paper-based Hb electrophoresis platform, Hb Variant (HbV) to conduct semi-quantitatively Hb variant detection 37. HbV relied on osmotic blood lysing, manual sample loading, and measured Hb variant band intensity within a pre-defined region of interest in single captured image at pre-determined time point. As a result, HbV enabled semi-quantitative determination of relative percentage (in %) for identified Hb variants 37.
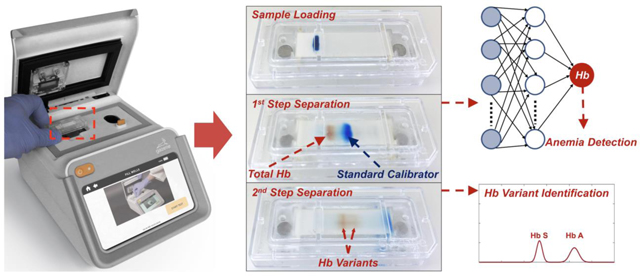
Accurate and reproducible anemia detection depends on accurate determination of absolute Hb levels. Here, we developed an integrated point-of-care quantitative anemia and Hb variant test, the Hb Variant-Anemia (HbVA) test 38. HbVA enables determination of absolute Hb level (in g/dL) by further engineering our previously reported HbV system through: 1) using chemical-assisted blood lysing, 2) using customized sample loading system (Fig.1), and 3) implementing an artificial neural network (ANN) based machine learning algorithm. The chemical-assisted blood lysing ensures release of all Hb molecules. The customized sample loading system provides consistent Hb and standard calibrator bands with controlled dimension and position. The ANN takes minimally processed intensity time series extracted from the raw image videos generated by the microchip electrophoresis POC platform, identifies salient features from the spectral pattern, and predicts a corresponding Hb level for the sample. We show the feasibility of this new diagnostic approach via a comprehensive test set comprising of 46 subjects, including individuals with or without anemia, and individuals with or without sickle cell disease (homozygous or heterozygous). We demonstrate that HbVA reproducibly and accurately determines blood Hb concentration with a mean absolute error of 0.55 g/dL and a bias of −0.10 g/dL (95% limits of agreement: 1.5 g/dL) against the gold standard according to Bland-Altman analysis. Resultant anemia determination is achieved with 100% sensitivity and 92.3% specificity with a receiver operating characteristic area under the curve (AUC) of 0.99. Further, following from our original study, Hb variant determination is performed using the same method, leading to identification of subjects with sickle cell disease with 100% sensitivity and specificity. This also validates our prior published device design and analysis. Overall, we show that HbVA platform enables, for the first time, reproducible, accurate, and integrated blood Hb level determination, anemia detection, and Hb variant identification in a single integrated POC test.
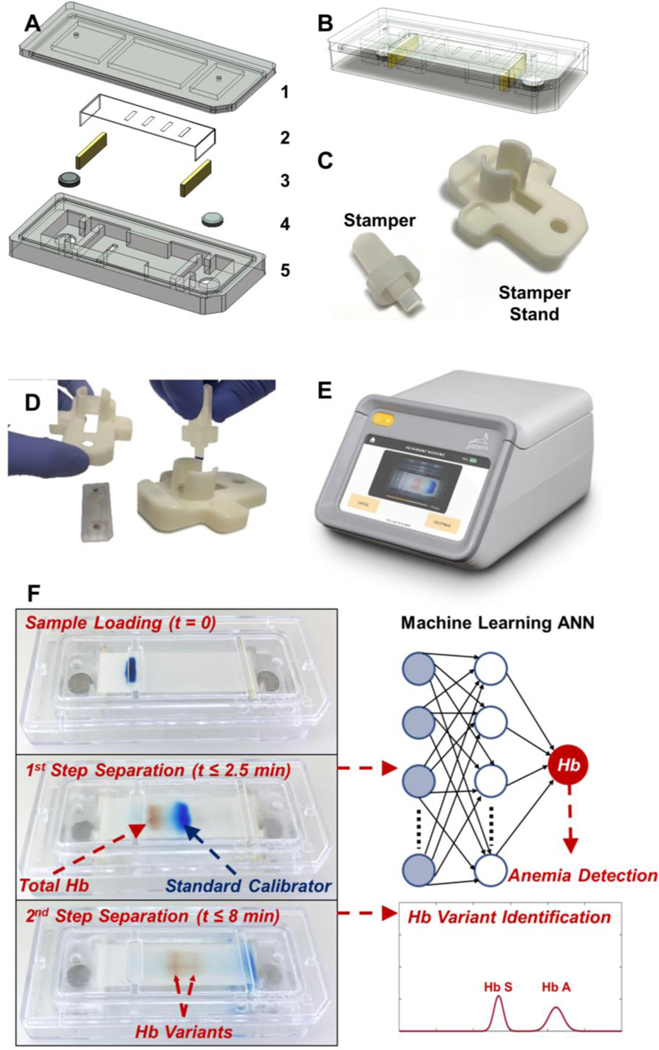
Figure. 1. POC microchip electrophoresis for integrated anemia and hemoglobin variant testing.
(A) HbVA cartridge consists of: (1) a plastic top cover, (2) a strip of cellulose acetate (CA) paper, (3) a pair of blotting pads, (4) embedded stainless-steel round electrodes, and (5) a plastic bottom part. Top and bottom plastic parts were injection molded. (B) HbVA cartridge design alloweds high volume manufacturing and assembly for affordability and single use. (C) A custom designed Stamper and Stamper Stand pair is utilized to achieve reproducible blood sample application into the HbVA cartridge. (D) HbVA cartridge was placed under the Stamper Stand and the blood sample was applied to the cartridge using the Stamper in a single step. (E) HbVA portable battery-operated reader unit accommodated the cartridge and ran the built software and the image analysis algorithm that tracked and analyzed the electrophoresis process in real-time. (F) A schematic representation of the HbVA test process is shown. First, a drop of Blood (red) was mixed with Standard Calibrator (xylene cyanol, blue) and applied on the CA paper in the cartridge (t=0). Within the first 2.5 minutes (t ≤ 2.5 min), the total Hb (red) and standard calibrator (blue) were electrophoretically separated, at which time blood Hb level (g/dL) and anemia status was determined by the algorithm. Next, Hbn variant separation occurred (t ≤ 8 min), which is then analyzed to determine the presence of major hemoglobin variants and types in the blood sample (i.e., Hb A, F, S, and C). The entire electrophoresis process was tracked in real-time and the captured data was analyzed by the machine learning artificial neural network (ANN) algorithm for integrated blood Hb level determination, anemia detection, and Hb variant identification in a single test.
RESULTS
HbVA performs 2-step electrophoresis in a single test tracked by real-time imaging. The fundamental principle behind the HbVA technology is of Hb electrophoresis, in which different (bio)molecules including total hemoglobin, standard calibrator, and hemoglobin variants can be separated based on their charge-to-mass ratio when exposed to an electric field in the presence of a carrier substrate38. HbVA is single-use and cartridge-based, which can be mass-produced at low-cost (Fig. 1A, B). The HbVA test works with a standard finger-prick or a heel-prick blood sample that is collected according to the World Health Organization (WHO) guidelines for drawing blood, which typically yields about 25 μL per drop39. The sampled blood is mixed and lysed with a customized standard calibrator solution. A customized sample stamper set including stamper and stamper stand (Fig. 1C) is used to transfer the mixture containing lysed blood and standard calibrator into the cartridge for electrophoresis (Fig. 1D). Tris/Borate/EDTA (TBE) buffer is used to provide the necessary ions for electrical conductivity at a pH of 8.4 in the cellulose acetate paper 40, 41. The pH induced net negative charges of Hb and of standard calibrator molecules, causing them to travel from the negative to the positive electrode when placed in an electric field (Fig. 1F). HbVA performs a 2-step separation based on processing time (Fig. 1F & 2A). During the first step, the major mobility difference between total Hb and standard calibrator allows the total Hb including tested Hb variants to separates from the standard calibrator within a short period of time (<2.5 minutes) (Fig. 1F, middle-left & Fig. 2A t0 to t150). During the second step, the finer mobility differences among Hb variants allow them to separate (Figure 1F, middle-right & Fig. 2A t150 to t480 & C, inset). The natural red visible hemoglobin, and naturally blue, visible standard calibrator (Xylene Cyanol), combined with optically clear HbVA cartridge in transmission mode within the reader’s imaging chamber, obviate the need for picrosirius staining, which is otherwise utilized in benchtop cellulose acetate hemoglobin electrophoresis 40, 41.
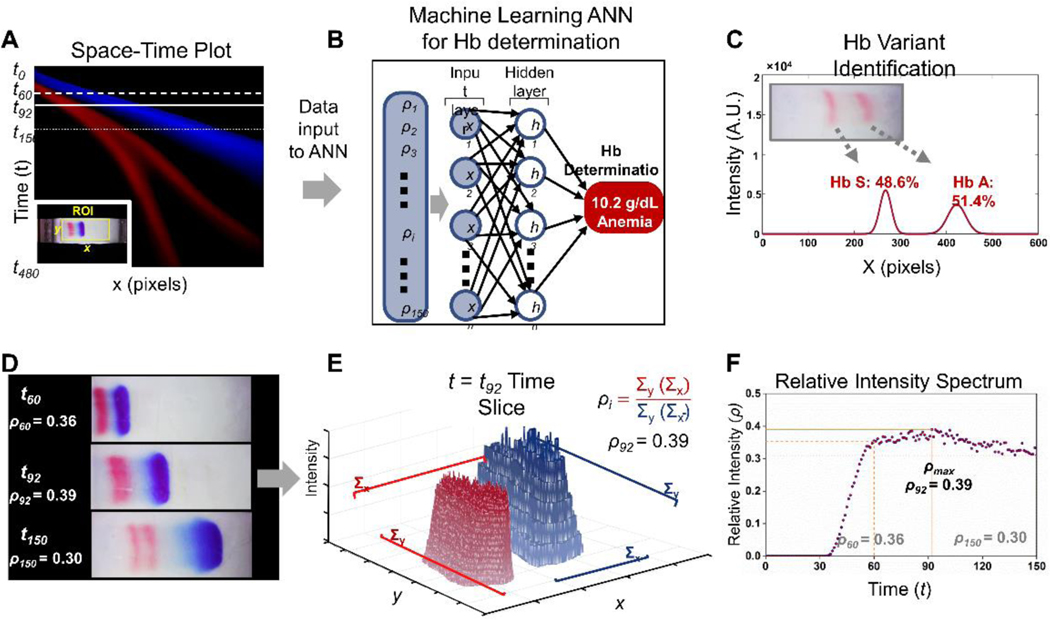
Figure. 2. Overview of HbVA integrated hemoglobin level determination, anemia detection, and hemoglobin variant identification.
(A) 2D space-time plot represents HbVA test band trajectory in time (y-axis) and space (x axis), visually illustrating the full electrophoretic band separation process across a whole video in a single image. Each pixel row of the image corresponds to a single one second frame, with time increasing from top (0 s) to bottom (480 s). For each point on the x axis we plotted the total intensities for the two-color bands (red = Hb and blue = standard calibrator), summed across the y range of the region of interest (ROI). The ROI was illustrated in the inset for a representative video frame. (B) The processing algorithm extracted the relative Hb and calibrator band information within the ROI by generating a relative intensity time series vector for t=0 to 150 s, as input for the trained ANN. Process of computing demonstrated in (E) and (F) and also described in main text. (C) Hb variant was identified based on the final location Hb variant band at the end of the test (t=480 s). (D) Individual frames within the ROI at 3 representative time points during HbVA test. At 60 s, detectable separation initiated between Hb band and standard calibrator band due to their major mobility differences while the Hb band remain unseparated (Top frame, ). At 92 s, Hb band and standard calibrator band further separates thus increasing band separation resolution (Middle frame ). At 150 s, total hemoglobin starts to separate into hemoglobin variants due to their minor mobility differences (Bottom frame, ). (E) 3D intensity profile of Hextracted from image acquired from frame at time slice t = 92 s (solid horizontal line in A and middle image in C). (F) An example pattern of time series vector including to recognized by the trained ANN.
Integrated Hb level determination, anemia detection, and Hb variant identification in one single HbVA test
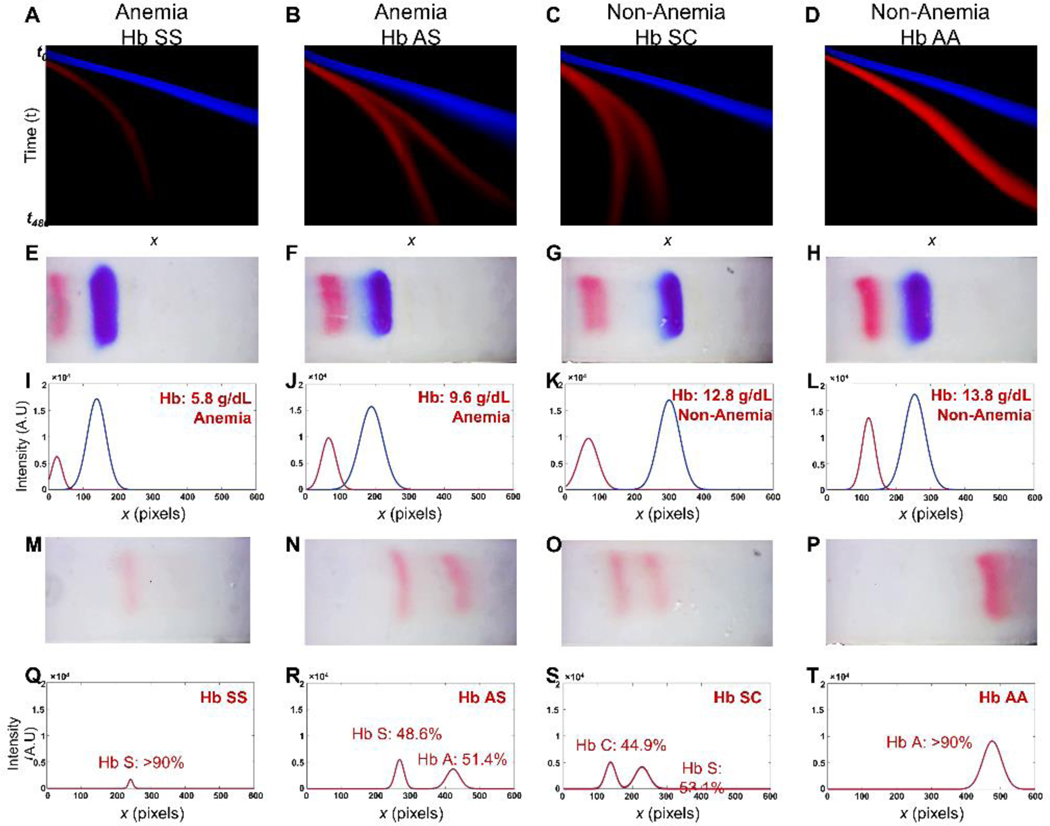
The HbVA 2-step electrophoresis separation is tracked in real-time (Fig. 2A-C), and analyzed using a two-step process according to the stage of separation. Briefly, for the first step (time t0 → t150), the algorithm tracks the relative Hb and calibrator band movements (Fig. 2A) within the ROI (Fig. 2A Inset). This is done by generating a time series of the relative intensity between Hb band and the standard calibrator band, , as input for a pre-trained ANN (Fig. 2B). The vector encoding the relatively intensity, , is obtained from the information within ROI during the first 150 s in HbVA test (Fig. 2D). It is evaluated as the relative intensity ratio of the total Hb band intensity over total standard calibrator band intensity for each frame (Fig. 2E&F). The trained ANN extracts salient features from input , and performs regression analysis by associating these features with a Hb level in g/dL and corresponding anemia status detection (Fig. 2B). The ANN has been pre trained on a training set comprising samples spanning a wide range of Hb levels, enabling anemia detection over a comparable range (Fig. 3A-L). This is followed by the second step of Hb variant identification. From the same test, late time (t150 → t480) data is used to identify Hb variants using our previously published algorithm (Fig. 2C, Fig. 3M-T) 37. The HbVA algorithm tracks Hb variant band migration and identifies Hb variants based on their final locations at the end of test (Fig. 2C, t ≤ 8 min). In case of a single Hb variant detected, such as Hb SS (Fig. 3M&Q) vs. Hb AA (Fig. 3P&T), the HbVA algorithm reports a percentage value greater of >90%, which agrees with the results reported by the reference standard method (HPLC). If there is more than one peak identified, then the areas under each of the peaks are calculated and the relative percentages are reported, for example in the cases of Hb AS (Fig. 3N&R) and Hb SC (Fig. 3O&S).
Figure. 3. Integrated Hb level determination, anemia detection, and Hb variant identification in 4 representative tests on clinical samples with different Hb levels and Hb variants.
(A-D) The first row includes 2D representation of HbVA test band trajectories. (E-H) The second row illustrates a representative frame for each test from the image arrays used to generate relative intensity ratio time series vectors , which are then utilized by the artificial neural network (ANN) to determine the Hb levels following the procedure outlined in Fig. 2. (I-L) The third row demonstrates the electropherogram corresponding to the image frames in the second row generated from the intensity profile envelopes. The Hb levels determined by HbVA (red) are compared against the reference method complete blood count (CBC) reported results. (M-P) The fourth row demonstrate the frames utilized to identify Hb variants. (Q-T) The fifth row demonstrates the electropherogram generated according to the band information in the fourth row. Each column represents HbVA test result for each patient. First column: HbVA test result for patient at Hb level of 6.0 g/dL and with homozygous HbSS (sickle cell disease, SCD patient); Second column: HbVA test result for patient at Hb level of 10.3 g/dL and with heterozygous HbAS (SCD patient undergoing transfusion therapy); Third column: HbVA test result for patient at Hb level of 12.7 g/dL and with heterozygous Hb SC disease (hemoglobin C disease); Fourth column: HbVA test results for patient at Hb level of 14.5 g/dL and with homozygous HbAA (healthy subject). The HbVA Hb level determination and anemia detection results are compared against the reference method complete blood count (CBC) reported results. The Hb variant identified by HbVA are compared against the reference method high performance liquid chromatography (HPLC) reported results. HbVA demonstrated agreement in Hb level determination, anemia detection and Hb variant identification with reference standard methods CBC and HPLC. (Patient 1: HbVA: 5.8 g/dL, Anemia, Hb SS vs. CBC&HPLC: 6.0 g/dL, Anemia, Hb SS; Patient 2: HbVA: 9.6 g/dL, Anemia, Hb AS vs. CBC&HPLC: 10.3 g/dL, Anemia, Hb AS; Patient 3: HbVA: 12.8 g/dL, Anemia, Hb SC vs. CBC&HPLC: 12.7 g/dL, Anemia, Hb SC; Patient 4: HbVA: 13.8 g/dL, Non-anemia, Hb AA vs. CBC&HPLC: 14.5 g/dL, Non-anemia, Hb AA). These results demonstrate HbVA’s integrated blood Hb level determination and Hb variant identification.
Efficacy of ANN based Hb level determination
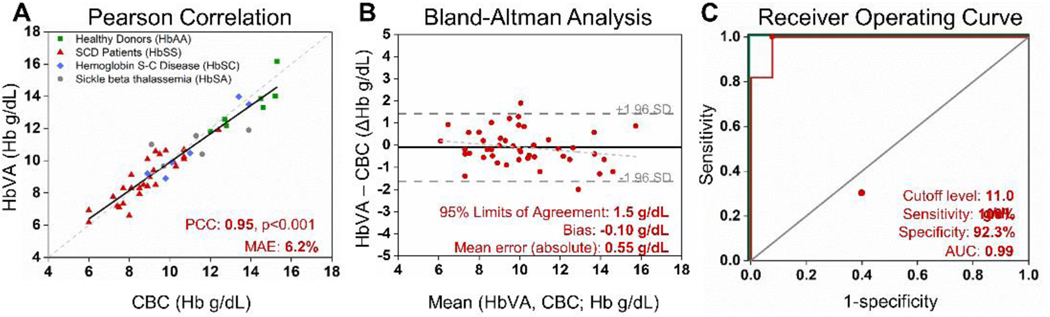
Hb level determination from HbVA video data was performed using a machine learning approach utilizing an ANN Machine learning implementation details can be found in the Methods section. The efficacy of our network’s performance has been validated using a standard repeated sub sampling validation routine. Details and results have been quoted in the Supplementary Information. The final optimally trained network achieved 93.8% mean relative error (MRE) against the gold standard and Pearson correlation coefficient 0.95 on a test set of 46 samples.
HbVA tests are repeatable and agnostic to user variance
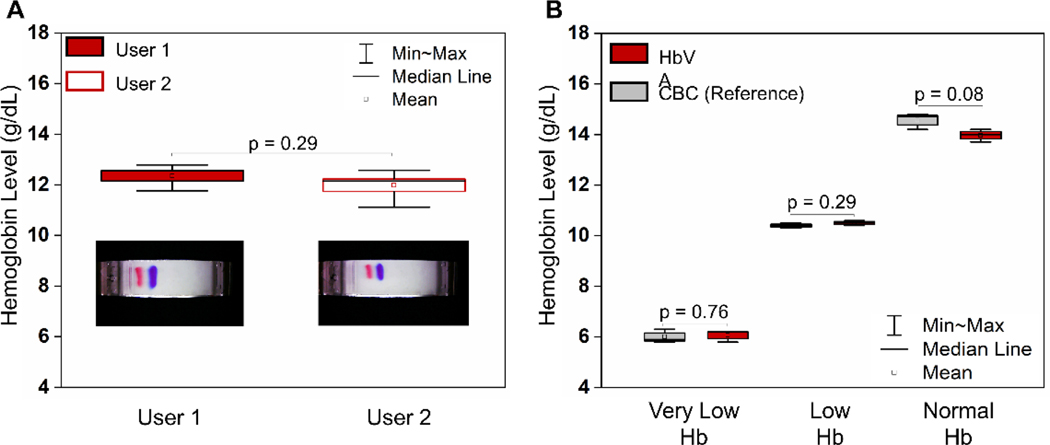
HbVA utilizes a blue standard calibrator with consistent charge-to-mass ratio at the test pH of 8.4. Utilization of relative Hb intensity over standard calibrator intensity as ANN input for Hb level determination compensates for user variance and enhances test repeatability (Fig. 4A, user 1 vs user 2). Among the 10 repeated tests (Fig. 4A), HbVA determined consistent Hb levels by 2 users (Fig. 5A, Mean ± SD (User 1 vs User 2), = 11.99 ± 0.55 vs. 12.35 ± 0.45, coefficient of variance (COV) = 4.2%). Additionally, all 10 tests demonstrated agreement within ±1.0 g/dL with the 12.7 g/dL Hb level reported by reference standard of complete blood count (CBC). These results indicate that HbVA determines Hb level and detects anemia repeatable and are agnostic to user variance.
Figure. 4. Repeatability and Reproducibility of Hemoglobin Variant/Anemia (HbVA) test:
(A) Repeatability test of HbVA Hb level determination was tested with 10 repeated tests using the same sample, comparing variances between 2 users (demonstrated in the inset figures). The determined Hb levels between 2 users demonstrate strong repeatability (Mean ± SD (User 1 vs User 2), = 11.99 ± 0.55 vs. 12.35 ± 0.45, coefficient of variance (COV) = 4.2%). All 10 repeated tests demonstrated agreement within ±1.0 g/dL with the 12.7 g/dL Hb level reported by reference standard of complete blood count (CBC). (B) Reproducibility test of HbVA Hb level determination were tested using 3 samples with very low, low and normal Hb levels reported from CBC. Each sample was tested 3 times by both HbVA and CBC. The standard deviation of HbVA determined Hb levels were within 4% COV across very low, low and normal Hb levels (Mean ± SD and COV for low, middle and high Hb levels: 6.1 ± 0.2 g/dL, 3.8%; 10.5 ± 0.1 g/dL, 1.0%; and 14.0 ± 0.3 g/dL, 2.1%, respectively, n=3 for each sample). HbVA determined Hb levels also demonstrated good accuracy within ±0.6 g/dL and ±5.0% with the CBC reported comparing to CBC reported data.
Figure. 5. Hemoglobin Variant/Anemia (HbVA) artificial neural network (ANN) based machine learning algorithm accurately determines Hb levels and anemia status.
(A) HbVA measures blood Hb levels were strongly associated with CBC measured results (PCC=0.95, p<0.001). The dashed line represents the ideal result where HbVA Hb level is equal to the CBC Hb level whereas solid line represents the actual data fit. (B) Bland-Altman analysis revealed HbVA determines blood Hb levels to within ±0.55 of the Hb level (absolute mean error) with minimal experimental bias with −0.1 g/dL, indicating that Hb determination has very small bias. The dashed light grey line indicated the relationship between the residual and the average Hb level measurements obtained from the CBC and HBVA (r = −0.07). The dashed dark grey line represented 95% limits of agreement (±1.5 g/dL). (C) The receiver-operating characteristic (ROC) analysis graphically illustrates HbVA’s performance against a random chance diagnosis (grey line), with an area under the curve of 0.99, and a perfect diagnostic (green lines), with an area under the curve of 1. The area under the curve of 0.99 suggested HbVA’s viable diagnostic performance. n=46
HbVA reproducibly determines Hb level for anemia detection
We performed reproducibility testing using 3 samples at different Hb ranges (Sample 1: 6.0 g/dL (≤9.0 g/dL, very low Hb), Sample 2: 10.4 g/dL (within 9.1–12.0 g/dL, low Hb level) and Sample 3: 14.6 g/dL (within 12.1–17.0 g/dL, normal Hb level) 42. Each sample was tested 3 times using both HbVA and reference standard method CBC. HbVA determined Hb level consistently over all 3 tested samples (Fig. 4B). The standard deviations among individual tests were ±0.2 g/dL, ±0.1 g/dL and ±0.3 g/dL (coefficient of variation (COV) = 3.8%, 1.0% and 1.8%, respectively) for samples at Hb level of Sample 1, 2, and 3, respectively (Fig. 4B). In addition, HbVA determined Hb levels were within ±0.6 g/dL (<5%) when compared with CBC results demonstrating consistent agreement with the reference standard over the inspected Hb level range. These results indicate HbVA determines Hb levels reproducibly over very low, low and normal Hb levels.
HbVA determines Hb level accurately
The scattered plot includes the ANN determined Hb levels by HbVA (y axis) versus the Hb levels reported by the standard reference (CBC) within 46 tested samples with a variety of Hb variants mixed with healthy subjects (Fig. 5A). Our results demonstrate a Pearson correlation coefficient (PCC) of 0.95, p<0.001 revealing strong association between HbVA determined Hb levels and CBC reported Hb levels (Fig. 5A). Bland-Altman analysis showed HbVA determines blood Hb levels with a mean absolute error of 0.55 g/dL and a bias of −0.10 g/dL (95% limits of agreement: 1.5 g/dL) in 46 patients (Fig. 5B). The receiver-operating characteristic analysis revealed that HbVA achieves a strong performance with an area under the curve of 0.99 (Fig. 5C) and highlights the accuracy of HbVA through the entire range of tested Hb levels (6.0 – 15.3 g/dL). Hb levels and HbVA measured residual (r=−0.07) showed that HbVA performance remained consistent throughout the range of tested Hb level (Fig. 5B), This degree of accuracy is on par with reported accuracy values in POC settings of other clinically used single-function anemia detection devices 29, 43.
HbVA performs integrated anemia detection and Hb variant identification with high sensitivity and specificity
We investigated the sensitivity and specificity of HbVA anemia detection from ANN determined Hb levels, using single Hb cutoff level of 11.0 g/dL to differentiate anemia and non-anemia subjects. Hb levels at <11.0 g/dL is a well-established threshold for defining anemia44. The sensitivity and specificity of HBVA to detect anemia was 100% and 92.3% (95% CI, 84.6% – 100%), respectively (Table 1). The high sensitivity and specificity indicate the potential for this test to serve as an integrated assessment tool for anemia. We investigated sensitivity and specificity of HbVA Hb variant identification by comparing HbVA identified Hb variant with standard reference method HPLC reported results. Overall, HbVA demonstrated 100% sensitivity and specificity on identifying Hb variants including HbSS, HbAS, HbSC and HbAA among the tested 46 samples using the results from the same tests for Hb level measurement and anemia detection (Table 2).
Table 1.
Sensitivity and specificity of Hb Variant/Anemia on determining anemia severity
| Anemia vs. Non-Anemia (Cut-off: 11.0 g/dL) | |
|---|---|
|
| |
| True Positive, TP | 33 |
| True Negative, TN | 12 |
| False Positive, FP | 1* |
| False Negative, FN | 0 |
| Sensitivity, TP/(TP+FN) | 100% |
| Specificity, TN/(TN+FP) | 92.30% |
False Positive: CBC reported Hb level = 11.6 g/dL while HbVA determined Hb level = 10.4 g/dL
Table 2.
Sensitivity and specificity of Hb Variant/Anemia on Hb variant identification
| Hb SS vs. others | Hb SC vs. others | Hb AA vs. others | |
|---|---|---|---|
|
| |||
| True Positive, TP | 9 | 6 | 7 |
| True Negative, TN | 37 | 40 | 39 |
| False Positive, FP | 0 | 0 | 0 |
| False Negative, FN | 0 | 0 | 0 |
| Sensitivity, TP/(TP+FN) | 100% | 100% | 100% |
| Specificity, TN/(TN+FP) | 100% | 100% | 100% |
Diagnostic profile of HbVA integrated Hb level determination, anemia detection and Hb variant identification
Overall, 7 healthy subjects (HbAA), 6 subjects with Hb SC, 5 sickle beta+ thalassemia (HbSBeta+) patients, and 28 SCD (HbSS) patients were tested (Fig. 5A). It should be noted that 21 of the 28 SCD patients tested in this study were on exchange transfusion therapy thus their Hb variants were identified as a mix of HbA and HbS by both the HbVA and standard reference method HPLC. Among the 46 tested patients, HbVA demonstrated high accuracy in Hb level determination as well as high sensitivity and specificity in both anemia detection as well as in Hb variant identification, compared with the reference CBC and HPLC.
DISCUSSION
Timely and effective treatments are available for anemia upon early detection and the treatment methods are highly dependent on the causes of anemia, including but not limited to nutritional deficiencies, malaria, and Hb disorders. Therefore, anemia detection is often carried out along with identification/screening procedures of other diseases. For example, a policy in Kenya states that women should be tested for anemia, HIV, syphilis, and malaria at first antenatal care 45.
Iron deficiency and SCD are among the most prevalent causes for anemia in low resource settings. Iron deficiency anemia can be treated with iron supplements or blood transfusion 46. Current treatment and emerging therapies for SCD include blood transfusion, hydroxyurea, Crizanlizumab, and Voxelotor47, 48, and are becoming more available for low-resource settings. However, high intracellular iron in patients with SCD are associated with markers of inflammation and mortality 49, thus therapeutic or supplemental iron may have adverse clinical consequences in patients with SCD and unnecessary transfusions therefore are strongly suggested to be minimized 50, 51. The goal of therapeutic interventions for patients with SCD is to maintain the HbS% below a target threshold to reduce SCD-related complications 52. As a result, application of transfusion therapy for patients with SCD requires careful consideration of multiple biomarkers including Hb level and percentage of hemoglobin S (HbS%) 52, 53. Furthermore, both Hb level and HbS% are also critical clinical endpoints for evaluating of the efficacy of hydroxyurea in treating SCD 54. Together, early, accurate and simultaneous diagnosis, as well as consistent monitoring of both anemia and Hb disorders is essential to facilitate timely lifesaving interventions, especially in low resource settings, where anemia and Hb disorders are most prevalent. In fact, a recent study has been performed in Tanzania to detect anemia and SCD using two mono-functional POC devices for detecting individual disease 35.
Many commercially available point-of-care (POC) assays including the WHO Haemoglobin Colour Scale, HemoCue®, DiaSpectTM, Mission® Hemoglobin Plus, and AnemoCheck™, Haemospect®, and NBM-200, as well as various emerging POC assays and smartphone based technologies are in the horizon for performing individual diagnosis of anemia (Table 3) 31, 55–63. Several POC assays including hemoglobin solubility test, density test, and lateral flow assays including Sickle SCAN, and HemoTypeSC are available for individual diagnosis of SCD (Table 3)64–70. These POC assays provide POC solutions for individual diagnosis of anemia and SCD, and have been tested and validated as summarized in our previous review articles 27, 71. However, these POC assays so far only focus on detection of single disease, while none of them is capable of integrated detection of both anemia and SCD using a single assay.
Table 3.
Comparison of available anemia diagnostic technologies with Hb Variant/Anemia
| Technology | Anemia | Hb Variant | Low cost | Accuracy* | Reproducibility | Ref |
|---|---|---|---|---|---|---|
|
| ||||||
| Complete blood count (Standard Reference) |
✓ | ☓□ | ☓□ | ✓ | ✓ | 25 |
| HemoCue | ✓ | ☓□ | ✓ | ✓ | ✓ | 29 |
| WHO color scale | ✓ | ☓□ | ✓ | ☓□ | ✓ | 78 |
| Non-invasive methods (including smart phone-based technologies) |
✓ | ☓□ | ✓ | ☓□ | ☓□ | 29 |
| HPLC (Standard Reference) |
☓□ | ✓ | ☓ □ | ✓ | ✓ | 79 |
| Paper-based hemoglobin solubility | ☓□ | □✓* | ✓ | ✓ | ✓ | 64 |
| Density-based HbS detection | ☓□ | □✓* | ✓ | ✓ | ✓ | 80 |
| Sickle SCAN | ☓□ | ✓ | ✓ | ✓ | ✓ | 33 |
| HemoTypeSC | ☓□ | ✓ | ✓ | ✓ | ✓ | 34 |
|
Hb Variant/Anemia
(HbVA) |
✓ | ✓ | ✓ | ✓ | ✓ | This paper |
Only distinguishes Hb S from Hb A, does not distinguish other Hb variants such as Hb C.
Previously, we leveraged the WHO recommended Hb electrophoresis test to develop an affordable paper-based Hb electrophoresis platform, HbV, for single function Hb variant identification 37. HbV’s Hb variant identification capability has been extensively validated in multiple locations using >700 clinical patient samples and has demonstrated its capability to identify and quantify the relative percentage (%) of Hb variants, with the goal of establishing a clinical standard test for Hb variant screening and diagnosis worldwide 37.
In this work, we improved the paper-based electrophoresis platform from various engineering perspectives including hardware, reagent, and software algorithm. These advances (in Lysing, Stamper set, dynamic LOI determination, machine learning recognition of pattern) enabled accurate quantification of absolute level of total Hb (in g/dL) and anemia detection, in addition to the previous validated Hb variant identification. To our knowledge, HbVA is the first POC device providing integrated Hb level determination, anemia detection and Hb variant identification.
Here, we determined Hb level by comparing Hb band intensity to the intensity of the standard calibrator. This is established on the foundation that the standard calibrator has consistent and significantly different electric mobility from Hbs, at the test pH of 8.4. This mobility difference allows the total Hb band to be separated from the standard calibrator band within a short period of time (≤2.5 min), before the Hb variants start separating.
Compared to relative percentage of Hb variant, determination of absolute level of total Hb level requires complete lysing of RBCs in whole blood. The customized standard calibration solution utilized here demonstrated >99.99% lysing of RBCs. This lysing efficiency of the improved solution is significantly higher than the lysing efficiency achieved previously, without inducing any noticeable alteration on Hb molecule electrophoretic property.
Reproducible and accurate determination of total Hb level depends on consistently generated quantifiable Hb band and standard calibrator band for samples at a range of Hb levels. Compared to the previous Hb quantification system, the sample applicator has reduced width and sample volume holding capacity. This improved sample applicator design constrained the loaded sample to maintain >2.5 mm margin from the edge of CA paper. This margin reduces the impact of band diffusion and CA paper saturation on Hb and standard calibrator bands quantification. Additionally, the customized sample stamper and stamper stand control the total volume and position of the loaded sample, providing efficient tool for consistent sample loading which can potentially reduce both test variance and user variance, especially in rapid clinical test at the point of care.
The ANN based machine learning algorithm leveraged in this study is essential for real-time tracking of the electrophoresis band separation process and extracting salient features from the data for accurate determination of Hb level and anemia detection. The algorithm extracts the dynamics of relative Hb to calibrator band position within the ROI. The band information is tracked in real-time to evaluate the relative intensity ratio of the total Hb band intensity over total standard calibrator to generate a time series vector , as input for the trained ANN The ANN examines this relative intensity time series spectrum to extract the dynamics of relative Hb to calibrator band position within the ROI. Additionally, the learning-oriented approach implemented in HbVA holds the perquisite of being flexible to ANN re-training and algorithm upgrade. Future increased test population will augment the current data set enabling ANN re-training and further tuning and enhancement of network performance.
Clinical testing of HbVA demonstrated accurately determined blood Hb concentration with low error and bias, according to Bland-Altman analysis. Anemia determination was achieved with high sensitivity and specificity with a receiver operating characteristic area under the curve (AUC) of 0.99. In addition to the new feature of Hb level determination and anemia detection, HbVA maintained its performance on Hb variant identification demonstrating 100% sensitivity and specificity identifying SCD.
A potential limitation of this study is that tested population was focused on patients with inherited hemoglobin disorders, who are typically anemic. Future studies will expand the test population by including higher number of healthy subjects with higher Hb levels. This system also shares the common limitation as other electrophoresis techniques for the function of Hb variant identification. Some Hb types appear in the same electrophoretic window, as they exhibit similar charge to mass ratio thus electrophoretic mobility in a given condition 72, 73, which may be improved using curve fitting methods. For example, Hb G and Hb D are difficult to separate because they have identical migration in gel electrophoresis, in capillary electrophoresis, and they have overlapping elution times in HPLC. This sharing of detection window or peak overlapping is also a challenge for the reference standard method, HPLC as well as its alternatives 72, 74, 75.
In summary, we show that merging engineering innovation and machine learning algorithms can be used to quantitatively measure Hb levels in paper-based electrophoresis, enabling, for the first time, reproducible, accurate, and integrated anemia detection and Hb variant identification in a single test at the point-of-care. HbVA is a versatile and mass-producible microchip electrophoresis platform technology that may address unmet needs in biology and medicine, when rapid, decentralized Hb or protein analyses are needed.
METHODS
Fully integrated microchip electrophoresis cartridge
The machine learning assisted HbVA cellulose acetate paper-based microchip electrophoresis system (Fig. 1) facilities, integrated Hb level determination, anemia detection and Hb variant identification (Video S1). The HbVA cartridge is manufactured using two injection molded plastic parts made of Optix® CA-41 Polymethyl Methacrylate Acrylic (Fig.1A-1&A-5). The injection molded HbVA cartridge embodies a pair of round corrosion-resistant, biomedical grade electrodes made of 316 stainless steel 316. The electrode pair is in direct contact to the electrophoresis buffer solution connected to the cellulose acetate paper strip in the buffer pools housed in the cartridge (Fig. 1A&B). One corner of the HemeChip cartridge is chamfered to facilitate correct orientation during use (Fig. 1A&B).
Repeatability and reproducibility of HbVA
We first tested HbVA’s repeatability by performing 10 repeated tests using same sample. The 10 tests were performed by 2 users with 5 tests each and compared for user variance. In addition, we tested HbVA’s reproducibility over very low, low and normal Hb level ranges. We performed reproducibility test using 3 samples at different Hb levels. Each sample was tested 3 times using both HbVA and reference standard method CBC of 6.0 g/dL (≤9.0 g/dL, very low Hb level), 10.4 g/dL (within 9.1–12.0 g/dL, low Hb level) and 14.6 g/dL (within 12.1–17.0 g/dL, normal Hb level).
Materials and Reagents
1x Tris/Borate/EDTA (TBE) buffer at pH 8.4 was used as background electrolyte for electrophoresis, reconstituted from 10x TBE buffer (ThermoFisher Scientific, Waltham, MA). Whole blood lysing buffer was prepared using ultrapure water (ThermoFisher Scientific, Waltham, MA) and customized standard calibrator (a fiducial marker that is negatively charged at pH 8.4) with 0.01% saponin and was used to perform whole blood lysing to release Hb from red blood cells.
HbVA Test for Integrated Hb Level Determination, Anemia Detection and Hb Variant Identification
The HbVA test can be performed by minimally trained personnel to produce fast, accurate, and reproducible results. The complete procedure consists of three steps: (i) chip preparation, (ii) sample preparation, (iii) Hb separation (10 minutes), followed by machine learning ANN algorithm-based data analysis. Briefly, Cellulose acetate paper within HbVA cartridge is wetted by introducing 40 μL background electrolyte. The cartridge is positioned onto the customized stamper. Sample is prepared by diluting whole blood sample into customized standard calibrator solution at 2:1. The calibrator solution is composed of electrophoresis marker xylene cyanol (molecular weight: 538.61 g/mol, Sigma Aldrich). The mixed sample is then loaded into wetted cellulose acetate paper using the customized stamper toolset. Finally, 200 μL background electrolyte is injected into the buffer ports at each end of the cartridge to provide sufficient contact between electrode-electrolyte-paper to complete circuit connection and provide stable current for electrophoresis. The prepared cartridge is then housed into the HbVA reader with preset voltage applied to initiate Hb separation.
HbVA integrated tests involve 2 steps separation in progression with time. The step 1 separation takes place within 3 minutes after initiation of separation process. During the short separation time, step 1 separates total Hb (red) from lysed blood from the applied standard calibrator (blue) (Fig. 1F, middle-left&2A) due to the major mass-to-charge ratio between Hb and standard calibrator. Region of interest (ROI) is defined by the algorithm. Within the ROI, the relative intensity ratio of the total Hb band intensity over total standard calibrator band intensity is evaluated by = 0.39 for demonstrated frame in Fig. 2B at t = 92 s). The electrophoresis separation process within ROI is tracked for the first 150 frames (Fig. 2C) and the relative intensity ratio is then calculated for the first 150 frames after HbVA test initiation (Fig. 2D) tracking the relative Hb band intensity to generate a time series vector (Fig. 2E). The obtained is then input to a trained artificial neural network (ANN) which performs regression by examining underlying time series data to determine the Hb level. The step 2 separation takes place between 3 – 10 minutes during HbVA process (Fig. 2F). As separation time progresses, step 2 separates total Hb into individual Hb subtypes according to their finer mass-to-charge ration among various Hb subtypes. After 10 minutes, the test is stopped automatically by the reader software.
Clinical testing of HbVA
Clinical study design, study participants, sample size calculation, and details on test methods are designed according to the Reference and Selected Procedures for the Quantitative Determination of Hb in Blood; Approved Standard – Third Edition (H15-A3) published by the CLSI 42, 76. We tested 46 samples including 37 clinical patient samples and 9 healthy subject samples. Since the focus of HbVA is to perform integrated anemia detection (reflected by low Hb level) and Hb variant identification, we mainly examined samples from SCD patients most of whom were anemic. Among 46 patients, 17 tested samples (37% of the total tested samples) were within the very low Hb range of ≤9.0 g/dL; 19 tested samples (41% of the total tested samples) were within the low Hb range of 9.1–12.0 g/dL; and 11 tested samples (22% of the total tested samples) are within Hb level range of 12.1–17.0 g/dL. Institutional Review Board (IRB) approved protocols included the following common objectives: to validate HbVA technology as a point-of-care platform for Hb testing, to compare the screening results obtained from HbVA with that obtained from laboratory CBC and HPLC as the standard reference methods, to determine the diagnostic accuracy of the test including sensitivity and the specificity, and to determine the feasibility of using HbVA as a point-of-care testing platform in low and middle income countries. All experiments were performed in compliance with the study protocol approved by the IRB committee at University Hospitals Cleveland Medical Center (UHCMC IRB# 04–17-15). Study participants were fully informed regarding the purposes of the study and consent was obtained.
Statistical Methods
Statistical significance (p < 0.05) was determined via two-tailed Student’s t-test assuming unequal variance. All statistical tests (calculation of regression correlation coefficients and Student’s t-tests were conducted using Minitab 17 Statistical Software. 95% confidence intervals for sensitivity and specificity are calculated according to the efficient-score method. The Bland-Altman method was used to determine the repeatability of HbVA Hb determination using residual analysis by comparison to the standard reference method. The coefficient of repeatability was set as 1.96 times the standard deviations of the differences between the two measurements. Sensitivity was determined as the ratio of true positive results divided by the summation of true positive results and false negative results. Specificity was determined as the ratio of true negative results divided by the summation of true negative results and false positive results.
Machine learning for Hb level determination: ANN implementation
The machine learning processing pipeline was established using the open source Keras machine learning library on top of a TensorFlow backend 77. The workflow can be deconstructed into the following steps, also illustrated for a representative HbVA test in Fig. 2 and SI video (insert video reference) : 1) Region of interest (ROI), is carefully defined to capture all relevant pixel information for both Hb band (red) and standard calibrator band (blue). Each frame from the video is sequentially split into its constituent red and blue channels. 2)The spatial summed relative intensity between red channel and blue channels (ρi = (ΣY ΣX )Red /(ΣY ΣX )Blue, Fig. 2E) is calculated for each frame from time 0 to 150 s, monitoring the relative abundancy of total hemoglobin and standard calibrator within that time period, to construct a relative intensity time series vector (Fig. 2F). is fed as the input feature vector to a trained artificial neural network (ANN) that examines the intensity ratio pattern and reports the corresponding Hb level in g/dL (Fig. 2B). The result is used to determine the anemia status of the test subject according to the standard.
The ANN in our workflow has been developed for a Python environment. For choice of ANN performing this regression problem, we used a vanilla feed forward network, also known as a multi-layer perceptron (MLP). The constructed ANN has three densely connected layers- an input layer, a hidden dense layer, and an output layer. The input and hidden layers each have 32 nodes with rectified linear unit (ReLU) activations. The ANN takes the pre-processed relative intensity ratio time series vector as input feature vector. For the size of our choice of input vector , this corresponds to5921 trainable parameters in the network. The network was trained and tested on a comprehensive data set of 68 HbVA tests. The training set consisted of 27 samples, out of which 4 were further split into a validation set (i.e 15% of training set). The remaining 41 samples were kept aside for the test set, and later augmented by a further set of 5 samples to make up a combined test set of 46 samples. Training was run on an NVIDIA GeForce RTX™ 2060 GPU. Assuming the error in the input data CBC responses to be normally distributed, we chose the mean squared logarithmic error (MSLE) as our choice of loss function to minimize over the training process. To prevent overfitting- along with allocating 15% of our training set into a holdout validation set, we stopped training when the validation loss performance stopped changing over a set number of epochs. The optimal network reached an MSLE loss of 0.9% for training and 1.1% for validation. Further details about the efficacy testing of the ANN are shown in the Supplementary Materials.
Supplementary Material
ACKNOWLEDGEMENTS
Authors acknowledge National Heart Lung and Blood Institute Small Business Innovation Research Program (R44HL140739, R41HL151015), National Heart Lung and Blood and the National Center for Complementary & Integrative Health (NCCIH) U54HL143541, National Institute of Diabetes and Digestive and Kidney Diseases Small Business Innovation Research Program (R41DK119048), and National Heart Lung and Blood Institute R01HL133574 and T32HL134622. This article’s contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
DECLARATION OF INTEREST STATEMENT
RA, CP, JAL, UAG, and Case Western Reserve University have financial interests in Hemex Health Inc. JAL, EK, UAG, and Case Western Reserve University have financial interests in BioChip Labs Inc. UAG and Case Western Reserve University have financial interests in Xatek Inc. UAG has financial interests in DxNow Inc. Financial interests include licensed intellectual property, stock ownership, research funding, employment, and consulting. Hemex Health Inc. offers point-of-care diagnostics for hemoglobin disorders, anemia, and malaria. BioChip Labs Inc. offers commercial clinical microfluidic biomarker assays for inherited or acquired blood disorders. Xatek Inc. offers point-of-care global assays to evaluate the hemostatic process. DxNow Inc. offers microfluidic and bio-imaging technologies for in vitro fertilization, forensics, and diagnostics.
Footnotes
Competing interests of Case Western Reserve University employees are overseen and managed by the Conflict of Interests Committee according to a Conflict-of-Interest Management Plan.
DATA AND MATERIALS AVAILABILITY
All reasonable requests for materials and data will be fulfilled by the corresponding author of this publication.
REFERENCES
- 1.Kassebaum NJ, Jasrasaria R, Naghavi M, Wulf SK, Johns N, Lozano R, Regan M, Weatherall D, Chou DP, Eisele TP, Flaxman SR, Pullan RL, Brooker SJ and Murray CJ, Blood, 2014, 123, 615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaparro CM and Suchdev PS, Ann N Y Acad Sci, 2019, 1450, 15–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.T. W. H. Organization.
- 4.W. H. Organization, WHO Global Database on Anaemia, 2008.
- 5.Soares Magalhães RJ and Clements ACA, Bull World Health Organ, 2011, 89, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker SP, Wachs TD, Gardner JM, Lozoff B, Wasserman GA, Pollitt E. and Carter JA, Lancet, 2007, 369, 145–157. [DOI] [PubMed] [Google Scholar]
- 7.Haider BA, Olofin I, Wang M, Spiegelman D, Ezzati M. and Fawzi WW, BMJ : British Medical Journal, 2013, 346, f3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasmussen K, J Nutr, 2001, 131, 590S–601S; discussion 601S-603S. [DOI] [PubMed] [Google Scholar]
- 9.Haas JD and T. t. Brownlie, J Nutr, 2001, 131, 676S–688S; discussion 688S-690S. [DOI] [PubMed] [Google Scholar]
- 10.Barnett R, Lancet, 2017, 389, 998–998. [Google Scholar]
- 11.Modell B. and Darlison M, Bull World Health Organ, 2008, 86, 480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jamison DT, Bank W. and Project DCP, Disease Control Priorities in Developing Countries, Oxford University Press, 2006. [PubMed] [Google Scholar]
- 13.Weatherall DJ and Clegg JB, Bull World Health Organ, 2001, 79, 704–712. [PMC free article] [PubMed] [Google Scholar]
- 14.Weatherall DJ, Blood, 2010, 115, 4331–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato GJ, Piel FB, Reid CD, Gaston MH, Ohene-Frempong K, Krishnamurti L, Smith WR, Panepinto JA, Weatherall DJ, Costa FF and Vichinsky EP, Nature Reviews Disease Primers, 2018, 4, 18010. [DOI] [PubMed] [Google Scholar]
- 16.Mburu J. and Odame I, Int. J. Lab. Hematol, 2019, 41, 82–88. [DOI] [PubMed] [Google Scholar]
- 17.Vichinsky E, Hurst D, Earles A, Kleman K. and Lubin B, Pediatrics, 1988, 81, 749–755. [PubMed] [Google Scholar]
- 18.Frempong T. and Pearson HA, Conn Med, 2007, 71, 9–12. [PubMed] [Google Scholar]
- 19.Mburu J. and Odame I, International Journal of Laboratory Hematology, 2019, 41, 82–88. [DOI] [PubMed] [Google Scholar]
- 20.Quarmyne MO, Dong W, Theodore R, Anand S, Barry V, Adisa O, Buchanan ID, Bost J, Brown RC, Joiner CH and Lane PA, Am J Hematol, 2017, 92, 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tshilolo L, Tomlinson G, Williams TN, Santos B, Olupot-Olupot P, Lane A, Aygun B, Stuber SE, Latham TS, McGann PT and Ware RE, The New England journal of medicine, 2019, 380, 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.G. W. H. Organization, 2017.
- 23.G. W. H. Organization, Licence: CC BY-NC-SA 3.0 IGO, 2017.
- 24.Howard J. and Thein SL, Hematology, 2019, 2019, 505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Northrop-Clewes CAand Thurnham DI, J Blood Med, 2013, 4, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.C. f. D. C. a. Prevention, 2015.
- 27.Alapan Y, Fraiwan A, Kucukal E, Hasan MN, Ung R, Kim M, Odame I, Little JA and Gurkan UA, Expert Rev Med Devices, 2016, 13, 1073–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.G. W. H. Organization, 2019. ((WHO Technical Report Series, No. 1017)).
- 29.Lamhaut L, Apriotesei R, Combes X, Lejay M, Carli P. and Vivien B, Anesthesiology, 2011, 115, 548–554. [DOI] [PubMed] [Google Scholar]
- 30.Perez-Plazola MS, Tyburski EA, Smart LR, Howard TA, Pfeiffer A, Ware RE, Lam WA and McGann PT, BMC Medicine, 2020, 18, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mannino RG, Myers DR, Tyburski EA, Caruso C, Boudreaux J, Leong T, Clifford GD and Lam WA, Nature Communications, 2018, 9, 4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dimauro G, De Ruvo S, Di Terlizzi F, Ruggieri A, Volpe V, Colizzi L. and Girardi F, Electronics, 2020, 9.
- 33.Kanter J, Telen MJ, Hoppe C, Roberts CL, Kim JS and Yang X, BMC Medicine, 2015, 13, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steele C, Sinski A, Asibey J, Hardy-Dessources M-D, Elana G, Brennan C, Odame I, Hoppe C, Geisberg M, Serrao E. and Quinn CT, American Journal of Hematology, 2019, 94, 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smart LR, Ambrose EE, Raphael KC, Hokororo A, Kamugisha E, Tyburski EA, Lam WA, Ware RE and McGann PT, Annals of Hematology, 2018, 97, 239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.W. H. O. (WHO), 2019.
- 37.Hasan MN, Fraiwan A, An R, Alapan Y, Ung R, Akkus A, Xu JZ, Rezac AJ, Kocmich NJ, Creary MS, Oginni T, Olanipekun GM, Hassan-Hanga F, Jibir BW, Gambo S, Verma AK, Bharti PK, Riolueang S, Ngimhung T, Suksangpleng T, Thota P, Werner G, Shanmugam R, Das A, Viprakasit V, Piccone CM, Little JA, Obaro SK and Gurkan UA, Analyst, 2020, 145, 2525–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An R, Man Y, Iram S, Kucukal E, Hasan MN, Solis-Fuentes A, Bode A, Hill A, Cheng K, Huang Y, Ahuja S, Little JA, Hinczewski M. and Gurkan UA, Blood, 2020, 136, 46–47. [Google Scholar]
- 39.Organization GWH, Available from: https://www.ncbi.nlm.nih.gov/books/NBK138650/, 2020.
- 40.Bain BJ, Haemoglobinopathy Diagnosis, Wiley, 2005. [Google Scholar]
- 41.Bain BJ, Wild B, Stephens A. and Phelan L, Variant Haemoglobins: A Guide to Identification, Wiley, 2011. [Google Scholar]
- 42.N. C. f. C. L. Standards, 2000.
- 43.Marn H. and Critchley JA, Lancet Glob Health, 2016, 4, e251–265. [DOI] [PubMed] [Google Scholar]
- 44.Thaver IH and Baig L, J Pak Med Assoc, 1994, 44, 282–284. [PubMed] [Google Scholar]
- 45.Young N, Taetgmeyer M, Zulaika G, Aol G, Desai M, Ter Kuile F. and Langley I, BMC Public Health, 2019, 19, 1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jimenez K, Kulnigg-Dabsch S. and Gasche C, Gastroenterol Hepatol (N Y), 2015, 11, 241–250. [PMC free article] [PubMed] [Google Scholar]
- 47.Neumayr LD, Hoppe CC and Brown C, Am J Manag Care, 2019, 25, S335–s343. [PubMed] [Google Scholar]
- 48.Vichinsky E, Hoppe CC, Ataga KI, Ware RE, Nduba V, El-Beshlawy A, Hassab H, Achebe MM, Alkindi S, Brown RC, Diuguid DL, Telfer P, Tsitsikas DA, Elghandour A, Gordeuk VR, Kanter J, Abboud MR, Lehrer-Graiwer J, Tonda M, Intondi A, Tong B. and Howard J, N. Engl. J. Med, 2019, 381, 509–519. [DOI] [PubMed] [Google Scholar]
- 49.van Beers EJ, Yang Y, Raghavachari N, Tian X, Allen DT, Nichols JS, Mendelsohn L, Nekhai S, Gordeuk VR, Taylor JG and Kato GJ, Circulation Research, 2015, 116, 298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koduri PR, Am J Hematol, 2003, 73, 59–63. [DOI] [PubMed] [Google Scholar]
- 51.Raghupathy R, Manwani D. and Little JA, Advances in Hematology, 2010, 2010, 272940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chou ST, Alsawas M, Fasano RM, Field JJ, Hendrickson JE, Howard J, Kameka M, Kwiatkowski JL, Pirenne F, Shi PA, Stowell SR, Thein SL, Westhoff CM, Wong TE and Akl EA, Blood Advances, 2020, 4, 327–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uter S, An HH, Linder GE, Kadauke S, Sesok-Pizzini D, Kim HC, Friedman DF and Chou ST, Blood Advances, 2021, 5, 2586–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.John CC, Opoka RO, Latham TS, Hume HA, Nabaggala C, Kasirye P, Ndugwa CM, Lane A. and Ware RE, N. Engl. J. Med, 2020, 382, 2524–2533. [DOI] [PubMed] [Google Scholar]
- 55.Tyburski EA, Gillespie SE, Stoy WA, Mannino RG, Weiss AJ, Siu AF, Bulloch RH, Thota K, Cardenas A, Session W, Khoury HJ, O’Connor S, Bunting ST, Boudreaux J, Forest CR, Gaddh M, Leong T, Lyon LA and Lam WA, J Clin Invest, 2014, 124, 4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGann PT, Tyburski EA, de Oliveira V, Santos B, Ware RE and Lam WA, Am J Hematol, 2015, 90, 1122–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee J, Song J, Choi J-H, Kim S, Kim U, Nguyen V-T, Lee J-S and Joo C, Scientific Reports, 2020, 10, 8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanchis-Gomar F, Cortell-Ballester J, Pareja-Galeano H, Banfi G. and Lippi G, J Lab Autom, 2013, 18, 198–205. [DOI] [PubMed] [Google Scholar]
- 59.Darshana LGT and Uluwaduge DI, Anemia, 2014, 2014, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hiscock R, Kumar D. and Simmons SW, Anaesthesia and Intensive Care, 2015, 43, 341–350. [DOI] [PubMed] [Google Scholar]
- 61.Ahmad NA, Awaluddin SM, Samad R, Kasim NM, Yusof M, Abd Razak MA, Ying CY and Sahril N, Int. J. Biomed, 2015, 5, 91–94. [Google Scholar]
- 62.Ahankari AS, Fogarty AW, Tata LJ, Dixit JV and Myles PR, BMJ Innovations, 2016, 2, 70. [Google Scholar]
- 63.Punter-Villagrasa J, Cid J, Páez-Avilés C, Rodríguez-Villarreal I, Juanola-Feliu E, Colomer-Farrarons J. and Miribel-Català PL, Sensors (Basel), 2015, 15, 4564–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang X, Kanter J, Piety NZ, Benton MS, Vignes SM and Shevkoplyas SS, Lab Chip, 2013, 13, 1464–1467. [DOI] [PubMed] [Google Scholar]
- 65.Quinn CT, Paniagua MC, DiNello RK, Panchal A. and Geisberg M, Br J Haematol, 2016, 175, 724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McGann PT, Schaefer BA, Paniagua M, Howard TA and Ware RE, Am J Hematol, 2016, 91, 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar AA, Patton MR, Hennek JW, Lee SY, D’Alesio-Spina G, Yang X, Kanter J, Shevkoplyas SS, Brugnara C. and Whitesides GM, Proc Natl Acad Sci U S A, 2014, 111, 14864–14869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumar AA, Chunda-Liyoka C, Hennek JW, Mantina H, Lee SYR, Patton MR, Sambo P, Sinyangwe S, Kankasa C, Chintu C, Brugnara C, Stossel TP and Whitesides GM, PLOS ONE, 2014, 9, e114540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Knowlton SM, Sencan I, Aytar Y, Khoory J, Heeney MM, Ghiran IC and Tasoglu S, Scientific Reports, 2015, 5, 15022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanter J, Telen MJ, Hoppe C, Roberts CL, Kim JS and Yang X, BMC Med, 2015, 13, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.An R, Huang Y, Man Y, Valentine RW, Kucukal E, Goreke U, Sekyonda Z, Piccone C, Owusu-Ansah A, Ahuja S, Little JA and Gurkan UA, Lab on a Chip, 2021, 21, 1843–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Keren DF, Hedstrom D, Gulbranson R, Ou CN and Bak R, Am J Clin Pathol, 2008, 130, 824–831. [DOI] [PubMed] [Google Scholar]
- 73.Borbely N, Phelan L, Szydlo R. and Bain B, Journal of clinical pathology, 2013, 66, 29–39. [DOI] [PubMed] [Google Scholar]
- 74.Nusrat M, Moiz B, Nasir A. and Rasool Hashmi M, BMC Research Notes, 2011, 4, 103–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sharma P. and Das R, World Journal of Methodology, 2016, 6, 20–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.N. C. f. C. L. Standards, 2018.
- 77.Shan G, Li X. and Huang W, The Innovation, 2020, 1, 100031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marn H. and Critchley JA, The Lancet Global Health, 2016, 4, e251–e265. [DOI] [PubMed] [Google Scholar]
- 79.Arishi WA, Alhadrami HA and Zourob M, Micromachines (Basel), 2021, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumar AA, Patton MR, Hennek JW, Lee SYR, D’Alesio-Spina G, Yang X, Kanter J, Shevkoplyas SS, Brugnara C. and Whitesides GM, Proceedings of the National Academy of Sciences, 2014, 111, 14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All reasonable requests for materials and data will be fulfilled by the corresponding author of this publication.