Abstract
Breast cancer is the most commonly diagnosed cancer and second-leading cause of cancer deaths in women. Breast cancer stem cells (BCSCs) promote metastasis and therapeutic resistance contributing to tumor relapse. Through activating genes important for BCSCs, transcription factors contribute to breast cancer metastasis and therapeutic resistance, including the signal transducer and activator of transcription (STAT) family of transcription factors. The STAT family consists of six major isoforms, STAT1, STAT2, STAT3, STAT4, STAT5, and STAT6. Canonical STAT signaling is activated by the binding of an extracellular ligand to a cell-surface receptor followed by STAT phosphorylation, leading to STAT nuclear translocation and transactivation of target genes. It is important to note that STAT transcription factors exhibit diverse effects in breast cancer; some are either pro- or anti-tumorigenic while others maintain dual, context-dependent roles. Among the STAT transcription factors, STAT3 is the most widely studied STAT protein in breast cancer for its critical roles in promoting BCSCs, breast cancer cell proliferation, invasion, angiogenesis, metastasis, and immune evasion. Consequently, there have been substantial efforts in developing cancer therapeutics to target breast cancer with dysregulated STAT3 signaling. In this comprehensive review, we will summarize the diverse roles that each STAT family member plays in breast cancer pathobiology, as well as, the opportunities and challenges in pharmacologically targeting STAT proteins and their upstream activators in the context of breast cancer treatment.
Keywords: Breast cancer, Breast cancer stem cells (BCSCs), Signal transducer and activator of transcription (STAT), STAT inhibitors, Breast cancer therapeutics
1. Introduction
Breast cancer is the most frequently diagnosed cancer in women; an estimated 287,000 will be diagnosed with breast cancer in 2022 [1]. Despite advances in early detection and breast cancer therapeutics, breast cancer patients that present with distant metastases have poor prognoses and high probabilities of drug resistance resulting in tumor relapse [2–4]. Metastasis to distant organs is responsible for the majority of breast cancer-related deaths [5], underscoring the importance of identifying mechanisms or cell populations that drive and facilitate breast cancer metastasis.
Breast cancer stem cells (BCSCs) are a small percentage of cells within breast tumors that maintain the ability to self-renew and regenerate the heterogeneous tumor lesions, known as tumor relapse [2,6,7]. These BCSCs are few in number, often quiescent, express high levels of ATP-binding cassette transporters, maintain upregulated DNA-repair capacity, and retain resistance to high levels of reactive oxygen species (ROS) contributing to therapeutic resistance and poor patient prognoses [7–9]. Given that BCSCs are responsible for metastasis and contribute to therapeutic resistance, this supports the clinical need to study and therapeutically target these cell populations [6,7].
Transcription factors play essential roles in eukaryotic gene expression by binding specific DNA sites and regulating transcription of almost every gene in a cell’s genome [10–12]. It is estimated that there are more than 1600 transcription factors in the human genome, nearly 20% have been associated with different disease phenotypes [10,13]. Multiple transcription factors have been identified as molecular markers and/or mediators of BCSCs that are correlated with breast cancer disease stage [14–18]. Since some transcription factors have been established to play important roles in promoting cancer progression [19], they often represent valid therapeutic opportunities.
Signal transducer and activator of transcription (STAT) family of transcription factors were originally discovered as ligand-induced transcription factors in interferon (IFN)-treated cells that mediate cytokine signaling pathways [20–22]. STATs play important roles in the regulation of cell proliferation, differentiation, apoptosis, and modulate the immune cell landscape [23,24]. In breast cancer, some members of the STAT family, particularly STAT3 and STAT5, are frequently hyperactivated by multiple cytokines leading to the enrichment of BCSCs and other malignant phenotypes of more aggressive breast cancer [25]. Thus, a deeper understanding of how these transcription factors modulate breast cancer progression remains essential in the development of novel breast cancer therapeutics in order to improve patient prognoses and reduce tumor relapse and metastasis.
2. Methods
Databases including open access journals, PubMed (Central), Scopus, Medline, Web of Science, and Google Scholar were utilized to obtain articles related to specific topics of interest. Key terms used during data collection include “STAT” with “breast cancer”, “treatment”, “inhibitors”, and “therapeutics”. Boolean logic (AND, OR) applied to connect the terms when searching databases. The comprehensive review included the most relevant or original articles for Sections 1–4. Section 5 (STAT proteins in breast cancer) includes all references that described individual STAT proteins in breast cancer and any tested STAT inhibitors in breast cancer.
3. Breast cancer, BCSCs, and transcription factors
3.1. Breast cancer
Breast cancer represents 31% of all diagnosed cancer cases in women and is the second leading cause of cancer-related deaths in women [1]. Though a large percentage of breast cancer cases are treatable, metastatic breast cancer patients have a dismal 5-year survival of 22%, with metastasis to distant organs causing the majority of breast cancer deaths [4,5]. Breast cancers can be categorized by intrinsic subtypes (PAM50), which are broadly named Luminal A, Luminal B, human epidermal growth factor receptor 2 (HER2)-enriched, and triple-negative breast cancer (TNBC), which encompasses claudin-low and basal-like TNBCs [26,27]. To make the breast cancer classifications more clinically relevant, the surrogate intrinsic subtypes are based on molecular and histological characteristics that are more widely used: Luminal A-like, Luminal B-like HER2-negative, Luminal B-like HER2-positive, HER2-enriched (non-luminal), and triple-negative [26]. Luminal A-like tumors are the least proliferative and tend to express high levels of specific hormone receptors: estrogen receptor (ER) and progesterone receptor (PR) [28]. Luminal B-like HER2-negative tumors express ER and PR, but at levels lower than in Luminal A-like and lack HER2 expression. Luminal B-like HER2-positive tumors are significantly more proliferative (indicated by a higher Ki-67 index) than Luminal A-like and Luminal B-like HER2-negative, which is partly attributed to expression of HER2. HER2-enriched (non-luminal) tumors lack expression of both hormone receptors and are considered one of the most aggressive breast cancer subtypes. While many HER2-enriched breast tumors respond to HER2-targeted therapies, it is not uncommon that these tumors become refractory to HER2-targeted therapies after 1–3 years [26,29]. TNBCs are characterized by the lack of ER, PR, and HER2 receptors excluding these patients from hormone- or HER2-targeted therapies. Due to the lack of these important receptors and incomplete knowledge of the mechanisms that drive TNBC, prognoses for TNBC patients are poor. Moreover, TNBC and HER2-enriched breast cancers also maintain the highest propensity to metastasize to distant organs [5,30]. The most common sites of metastasis for breast cancer patients include: bone, lungs, brain, and liver [4]. Despite advances in breast cancer therapeutics [29,31,32], there are limited FDA-approved therapies for TNBC patients and few options for HER2-enriched breast cancer patients who exhibit tumor relapse underscoring the clinical importance for further investigating the molecular mechanisms that drive metastatic breast cancer.
3.2. The discovery of cancer stem cells (CSCs) and BCSCs
Stem cell properties and the manipulation of “stemness” by cancer has been widely accepted for years [33]. The first “cancer stem cell” was discovered in the 1930 s when researchers identified a single cell from a mouse tumor initiates a new tumor in a recipient mouse [34]. Nearly 30 years later, single cells isolated from malignant teratocarcinomas have the ability to differentiate into multiple cell lineages and differentiate into non-tumorigenic tumor types [35]. Multiple findings led to the first definition of a cancer stem cell (CSC) that describes cancer cells as those that mimic tissue renewal, retain stemness properties, and contain malignant stem cells that maintain the ability to proliferate, but have limited ability to differentiate [33,36]. In 1995, multiple subtypes of acute myeloid leukemia were consistently engrafted into immunodeficient mice; engraftment was only successful when initiated from CD34-positive CD38-negative populations [37].
The first BCSCs were identified in 2003 when researchers established that breast cancer cells within a single breast tumor were incredibly heterogeneous [6]. Furthermore, as few as 100 CD44high/CD24low cells could initiate tumors in a mouse xenograft assay. In contrast, thousands of cells with combinations of markers other than CD44high/CD24low were unable to establish tumors in immunodeficient mice. Notably, the subpopulation of tumorigenic cells could be serially passaged and at each passage, the CD44high/CD24low subpopulation could give rise to both CD44high/CD24low cells, as well as, non-tumorigenic cells of differing phenotypes. Though specific subsets of markers have been identified to be expressed heterogeneously in numerous cancer types, combinations of markers are variable and remain to be fully understood as there are still conceptual and technological challenges within the CSC field [33,38]. However, since the discovery and isolation of BCSCs [6], an abundance of exciting BCSC research has flourished in the last two decades leading to the discovery of important factors known as transcription factors.
3.3. Transcription factors and breast cancer
Transcription factors are an essential class of proteins that control expression of nearly the entire cell genome, and have become a focus of cancer research and therapeutics following the discovery of oncogenes [19,39,40]. The main function of transcription factors is to bind specific sites on DNA and recruit transcriptional machinery to regulate gene expression [13]. Transcription factors typically act as the nuclear effectors of signal transduction pathways that starts with the binding of an extracellular ligand to a cell-surface receptor with an extracellular domain for that ligand [13,19]. This interaction may cause conformational, chemical, or biological changes that activate intracellular signaling. Here, latent transcription factors are activated by a host of modifications including phosphorylation or interactions with other factors. The activated transcription factors then translocate to the nucleus where they modulate the cell’s transcriptome. Given the critical regulatory control that transcription factors exhibit within cells, dysregulation of these gene expression networks often leads to cancer [41].
In breast cancer, there are numerous transcription factor families known to promote tumor malignancy including the STAT family of proteins. Since their discovery in 1994 [20–23], significant advances have been made in elucidating the roles of and mechanisms for the STAT transcription factors in facilitating or suppressing breast cancer. In recent years, substantial efforts have been invested in developing STAT-targeted therapeutics with promising outcomes and challenges. This review will focus on the STAT family of transcription factors, their important roles in breast cancer, and the advancements made in STAT-targeted breast cancer therapeutics.
4. STAT family of transcription factors
4.1. STAT transcription factors and structure
The discovery of STAT proteins resulted from the examination of IFN-related pathways, which led to the identification of a previously unrecognized signal transduction pathway [20–22,42]. Since the discovery of the STAT family, STATs play essential roles in the regulation of a myriad of physiological and biological processes in cells [23]. Given that STATs regulate proliferation, metastasis, and chemoresistance, STAT function is often hyperactivated or dysregulated in many cancer types [24,43]. STAT transcription factors maintain the ability to transduce signals resulting in STAT translocation to the nucleus and subsequent gene expression modulation [23]. The STAT family consists of six major isoforms including STAT1, STAT2, STAT3, STAT4, STAT5, and STAT6, which have been identified at these chromosome locations within the human genome 2q32.2, 12q13.3, 17q21.2, 2q32.2, 17q21.2, 17q21.2, and 12q13.3, respectively [23,44,45]. Of note, multiple isoforms for each STAT protein have been identified, so full-length isoforms are denoted with α and the following isoforms named with β-δ [46].
STAT family members are ~740–900 amino acid residues in length and are mostly structurally conserved. In general, the canonical STAT members contain these common domains/regions including 1) the N-terminal domain (NTD; NH2), 2) the coiled-coil domain (CCD), 3) the DNA-binding domain (DBD), 4) the linker domain (LD), 5) the SRC homology 2 domain (SH2), 6) the tyrosine-phosphorylation site (pY), and 7) the C-terminal transactivation domain (TAD) [23,46–53] (Fig. 1).
Fig. 1. STAT structure.

The signal transducer and activator of transcription (STAT) family members are ~740–900 amino acids in length and maintain relatively conserved regions in their structures. Each STAT protein contains these functionally conserved domains: 1) N-terminal domain (NTD) capped with NH2, 2) coiled-coiled domain (CCD), 3) DNA-binding domain (DBD), 4) linker domain (LD), 5) SRC homology 2 domain (SH2), 6) tyrosine phosphorylation site (pY), and 7) C-terminal transactivation domain (TAD) containing COOH. STAT4 is the shortest STAT protein (748), while STAT2 is the longest (851). Additionally, there is another conserved phospho-serine residue in the TAD of STAT1, STAT3, STAT5a, STAT5b, and STAT6.
The NTD is a hook-like structure made up of several alpha-helices [51,54]. Importantly, the NTD mediates critical protein-protein interactions between the STAT family members leading to dimerization and cooperative DNA-binding [47,51]. The CCD also consists of multiple alpha-helices, this time forming a rope-like structure [46]. In contrast to the NTD, the CCD facilitates STAT protein binding to other transcription factors or co-activators [46,55]. For example, STAT1 and STAT3-STAT6 interact with N-Myc interactor (Nmi), which is stimulated by the ligands interleukin-2 (IL-2) and IFNɣ [55]. In another example, downstream of IFNα, the STAT1-STAT2 heterodimer recruits and interacts with p48 to mediate STAT-driven transcription [56]. In addition, STAT5a and STAT5b interact with silencing mediator for retinoic acid receptor and thyroid hormone receptor, which suppresses STAT5-mediated transcription, demonstrating that the CCD also mediates interactions with co-repressors [57]. The CCD also contributes to nuclear translocation as the nuclear localization signal (NLS) interacts with importins to facilitate nuclear translocation [46,58,59]. The DBD contains an immunoglobulin fold that mediates recognition, binding to specific DNA sequences, and stabilization of DNA interacting elements [46,47,50]. The LD is an essential contact point that provides structural stability for the STAT-driven complex [46,56]. The SH2 domain structure enables the protein the ability to bind specific phosphotyrosine-containing motifs, also referred to as tyrosine-phosphorylation sites (pY) [60–62]. This region is highly conserved in a variety of signaling molecules in order to mediate protein-protein interactions [61]. More specifically, the SH2 domains of STAT proteins can bind both pYs on receptor complexes and pYs on the same STAT protein leading to subsequent dimerization [46, 63]. Moreover, tyrosine phosphorylation is necessary for activation of a myriad of transcription factors including those in the STAT family [21, 44,64,65]. Finally, the C-terminal TAD facilitates STAT interactions with transcriptional co-factors [66,67]. For example, TAD enables STAT2 to bind p300/CREB-binding protein [68] and Brahma-related gene 1 in response to IFNα signaling [69]. Additionally, STAT6 can directly interact with p300/CBP and the nuclear coactivator 1 via the TAD to enhance transcriptional activation [70].
4.2. Canonical STAT functions
The canonical STAT pathway is evolutionarily conserved and activated by ligands, such as IFNs or ILs, binding to cell surface receptors [47,53]. Activation by ligand binding to its respective surface receptor initiates a cascade of signaling events leading to STAT translocation to the nucleus and regulation of downstream target genes (Fig. 2). Inactive (i.e. unphosphorylated) STAT proteins reside in the cytoplasm prior to ligand binding [47,53]. Once an extracellular ligand binds a cell surface receptor, it undergoes conformational changes leading to the recruitment of Janus kinases (JAKs), non-receptor tyrosine kinase family, and subsequent autophosphorylation of tyrosine residues within the receptor complex [21,44,53,71]. Activated JAKs now provide docking sites for a diverse number of signaling molecules with SH2 domains, such as STATs [72]. Given that multiple ligands can activate this JAK/STAT signal cascade to modulate many cell phenotypes, the pY-SH2 interaction is highly specific as a way of regulating STAT activation. When receptor-mediated STATs are recruited to the activated JAK complex, the JAKs phosphorylate a specific pY on the unactivated STAT molecule resulting in reciprocal binding of the pY on one phosphorylated STAT (pSTAT) to the SH2 domain of the pSTAT associated with the second receptor. This reciprocal binding leads to STAT homo- or heterodimerization releasing the complex from the extracellular receptor-JAK complex and allows the pSTAT dimer to translocate the nucleus, which is facilitated by importins and other helper proteins [73–75]. Unactivated STATs are also phosphorylated by tyrosine kinases that retain intrinsic kinase activity, which do not require JAKs (Fig. 2). In this mechanism, a growth factor or ligand binds its respective receptor tyrosine kinase, which phosphorylates STAT proteins, allows for STAT dimerization and translocation to the nucleus [43,76]. Though the physical STAT functions within the nucleus are relatively conserved, nuclear import of each STAT varies representing an important aspect of STAT regulation. However, the mechanistic findings of how STATs 1–6 are imported and exported from the nucleus go beyond the scope of this review [73,77–79].
Fig. 2.
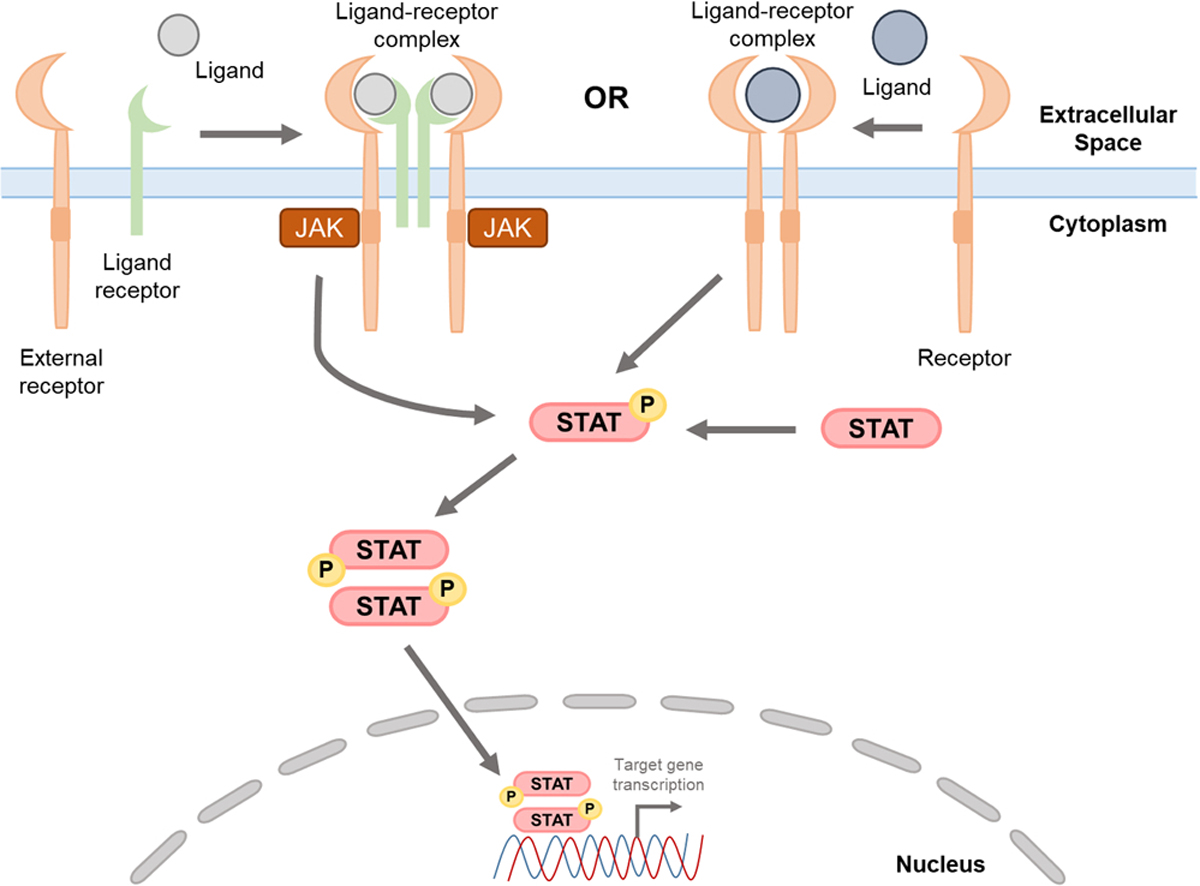
Canonical STAT pathway activation. STAT proteins are activated by a myriad of cytokines or growth factors (i.e. ligands), which bind to extracellular domains on cell-surface receptors (i.e. receptors) to form ligand-receptor complexes. The canonical IL-6/IL-6Ralpha/STAT3 pathway (left), for example, involves the IL-6 cytokine (ligand) binding to the IL-6Ralpha, which activates gp130 association with IL-6/IL-6Ralpha. Two trimeric IL-6/IL-6Ralpha/STAT3 complexes interact, recruit JAKs, phosphorylate the intracellular domains of gp130, activate STAT3 via phosphorylation, pSTAT3 dimers then translocate to the nucleus where they modulate gene expression of downstream target genes. Additional receptors, such as EGF or FGF receptors (right), retain intrinsic kinase activities and can directly phosphorylate STAT proteins leading to STAT phosphorylation, dimerization, and subsequent translocation to the nucleus. Though there are reports of alternative STAT activation including methods where STAT proteins form dimers without phosphorylation, this pathway describes the most common STAT pathway activation.
Within the nucleus, the pSTAT dimer binds specific DNA sequences within promoter or regulatory regions of genes, often referred to as DNA regulatory elements, leading to robust transcriptional changes that alter expression of a multitude of target genes [44,47,80–82]. Though canonical STAT signaling involves STATs homo- or heterodimerizing following phosphorylation by JAKs, multiple studies report non-canonical STAT signaling. For example, STAT1 and STAT3 can form homodimers prior to phosphorylation [83–86]. However, these non-phosphorylated STAT dimers are unable to translocate to the nucleus [84]. In another report, the majority of unactivated STAT3 (as well as other STATs) is detected in multimeric complexes, referred to as “statosomes”, indicating an alternate method of STAT3 activation that is retained in the cytoplasm [87]. In contrast to previous reports, unphosphorylated STAT1 translocates to the nucleus and upregulates immune regulatory genes in the absence of IFN stimulation [88]. Interestingly, STAT5 forms tetramers, which mediate IL-2-dependent transcriptional activation of a specific subset of genes involved in immune function that canonical STAT5 dimers are unable to activate [89, 90]. Alternative STAT activation also occurs through other proteins, such as non-receptor tyrosine kinases or G-protein coupled receptors (GPCRs) that facilitate STAT recruitment to JAKs as STAT proteins are unable to phosphorylate themselves or other proteins since they do not maintain any tyrosine kinase activity [23,82,91,92]. Though these reports shed some light on alternative STAT signaling, non-canonical mechanisms of STAT signaling remain largely unexplored.
There are main regulators of STAT signaling including suppressors of cytokine signaling (SOCS), which are expressed at the start of STAT signaling [93]. SOCS have the ability to bind and inhibit both STAT proteins and/or JAK kinases as they maintain an SH2 domain and kinase inhibitory regions to inhibit each, respectively [93,94]. For example, SOCS3 is an important JAK-STAT pathway inhibitor that specifically prevents STAT3 expression and pSTAT5 in breast cancer [95,96]. Moreover, treatment with isolinderalactone increases SOCS3 and subsequently suppresses pSTAT3 and apoptosis-inducing factors in TNBC [97]. Additionally, SOCS2, SOCS3, SOCS5, SOCS6, and SOCS7 are highly, constitutively expressed in breast cancer cells compared to normal breast epithelial cells [98]. Phosphatases are another category of proteins that can negatively regulate STAT pathway activation mediated by dephosphorylation of tyrosine residues [99]. Though regulation of STATs are important for understanding STAT pathways, further detail into the exact mechanisms of the negative regulators of STATs go beyond the scope of this review [53,93].
It is widely understood that STAT proteins play critical roles in many signal transduction pathways that are important in human physiology and biology. Furthermore, over 50 cytokines and growth factors can activate a combination of STAT signal cascades [21,44], suggesting that dysregulation of the extracellular activators (i.e. cytokines or growth factors), the intrinsic players (i.e. STATs or associated factors and kinases), the negative regulators of STAT proteins, or any combination of these events may lead to cancer [53]. More specifically, STAT3 contributes to breast cancer progression and chemoresistance with many of these findings leading to the development of natural compounds and drug inhibitors that target STAT3 for breast cancer treatment. While STAT proteins besides STAT3 are not as well understood in breast cancer, there are promising findings investigating all STAT family members in breast cancer. This review will summarize functions for all STAT proteins in breast cancer, discuss STAT inhibitors reported in breast cancer, and their clinical implications.
5. STAT proteins in breast cancer
5.1. STAT1
STAT1 signaling modulates pathways involved in cell growth, differentiation, homeostasis, immune signaling, and immune response [100]. Similar to other STAT family members, STAT1 is activated by both types of IFNs (I and II) [80,101]. Numerous reports demonstrate that STAT1 displays tumor suppressive functions in many cancer cell types, including breast cancer, with loss of STAT1 expression or activation leading to cancer progression [23,102,103]. Moreover, higher STAT1 activation, as indicated by both tyrosine phosphorylation and DNA binding, is associated with longer relapse-free survival and overall survival for breast cancer patients [104]. Furthermore, high STAT1 mRNA expression using a STAT1-immune-related gene signature correlates with longer distant metastasis-free survival (DMFS) in ER-negative/PR-negative and TNBC patients [105]. Additionally, chemotherapy-induced IFN target genes are associated with STAT1 phosphorylation and IFNɣ secretion in treatment-sensitive breast cancers, suggesting that IFN/STAT1 activation predicts response of ER-negative breast cancers [106]. PR also suppresses IFN-activated pSTAT1 decreasing immune surveillance and increasing immune evasion in PR-positive breast tumors [107]. STAT1 inhibition decreases breast cancer cell proliferation, ERα protein levels and target gene expression [108]. Further investigation reveals that STAT1 directly binds the promoter region of ERα, suggesting an important role for STAT1 in ER activity [108].
Utilizing diverse transgenic and alternative mouse models, STAT1 exhibits anti-tumor function including inhibition of mammary gland tumorigenesis and promotion of tumor immune surveillance in vivo [102,109–113]. For example, STAT1 suppresses ErbB2/Neu (or Her2/Neu)-induced tumorigenesis in transgenic mice [110]. In another study, human breast tumors had little to no expression of STAT1; normal breast epithelial cells express significantly elevated levels of STAT1 compared to adjacent breast tumor tissue [102]. From the same study, STAT1-deficient transgenic mice develop spontaneous ER-positive/PR-positive mammary tumors whereas none of the wild-type mice developed tumors [102]. Moreover, STAT1 antagonizes the JAK2-STAT3/5 A/5B pathway, induced by the prolactin receptor, to inhibit ER-positive tumorigenesis [114]. STAT1 also downregulates NAD(P)H quinone dehydrogenase 1 resulting in increased oxidative stress in sensitized patient-derived xenografts for both HER2-positive and TNBCs [115].
In contrast to multiple studies demonstrating a tumor-suppressive role for STAT1, high STAT1 activity also correlates with poorer patient prognoses [116]. STAT1 is activated by CD95 (Fas), a receptor that mediates apoptosis by activating a caspase cascade, which promotes STAT1-dependent mammospheres [117]. Inhibition of DNA methyltransferase 3 beta by microRNA (miR)− 29c results in upregulated TIMP3/STAT1/FOXO1 signaling, which promotes breast cancer progression [118]. Phospholipid scramblase 1 directly binds and increases STAT3 interactions with the STAT1 promoter, leading to enhanced transactivation and breast cancer cell proliferation and invasion [119]. Overexpression of STAT1 also promotes myeloid-derived suppressor cell (MDSC) migration and suppresses both CD4-positive and CD8-positive T cells in mouse tumors [116]. STAT1 knockdown in cancer-associated fibroblasts extends breast cancer progression and combination treatment of a STAT1 inhibitor with doxorubicin suppresses tumor progression in mouse mammary tumors [120]. STAT1 also interacts with ERα and STAT1 and ERα directly interact with the promoter region of interferon-induced transmembrane protein 1 (IFITM1) resulting in upregulated IFN-mediated gene expression to promote breast cancer cell survival [121]. In another example, STAT1 activation, in cooperation with mucin 1, associates with shortened recurrence-free survival and overall survival in breast cancer patients [122], suggesting that STAT1 functions in breast cancer still remain to be fully understood.
Given that BCSCs are major drivers of therapeutic resistance, STAT1’s role in modulating BCSCs may be important in understanding how to utilize STAT1 in breast cancer treatment. Low levels of surface-CD24, concurrently with high levels of surface CD44, indicate cancer-initiating cells or BCSCs [6]. CD24-mediated inhibition of the Sonic Hedgehog (SHH) pathway and STAT1 activity suppresses breast cancer cell proliferation and invasion [123]. Similarly, sphingosine kinase 1 promotes breast cancer cell proliferation and mammosphere formation, while suppressing apoptosis, in part by downregulating STAT1 activation [124]. Following irradiation, an increase in STAT1 is attributed to a significant decrease in apoptosis of mammospheres [125]. In contrast, overexpressed and hyperactive STAT1 decreases CD44-expressing cells in breast cancer cells [126]. STAT1’s role in modulating BCSCs and potential contribution to radioresistance highlight the need to further examine STAT1 mechanisms in breast cancer.
STAT1 and STAT3 have been reported to functionally interact, antagonize, and cooperate in cancer cells [127,128], suggesting these two STAT family members have complex networks. STAT1 is enriched by nuclear epidermal growth factor receptor (EGFR) and STAT3 binds to the STAT1 promoter to increase STAT1 expression in breast cancer cells [127]. EGFR is an important transcriptional regulator in breast cancer and nuclear EGFR staining is a predictive marker for patient prognosis [129]. Moreover, inhibition of EGFR and subsequent suppression of STAT3 activation with tannic acid promotes cell cycle arrest and apoptosis in breast cancer cells [130]. Further investigation reveals that tannic acid also increases pSTAT1, leading to increased expression of p21, a cyclin-dependent kinase inhibitor, leading to cell cycle arrest and apoptosis [130]. When comparing STAT expression in lymph nodes and primary breast tumors using a tissue microarray, the lymph nodes exhibit increased cytoplasmic STAT1, pSTAT3, and STAT5 and nuclear pSTAT3 [131].
In another study using breast cancer specimens, high STAT1 and STAT3 activation correlate with decreased tumor grade; high pSTAT1 associates with increased immune infiltration (inflammation-induced) whereas increased pSTAT3 associates with suppressed CD4-positive T-cell infiltrate [132]. The Shc1 scaffold activates STAT3 and inhibits STAT1 activation to promote immunosuppression while inhibiting anti-tumor immune surveillance in breast cancer cells, respectively [128]. Loss of Shc1-tyrosine kinase signaling increases IFNɣ, while WT-Shc1-tyrosine kinase signaling represses IFNɣ secretion [128]. Further investigation into the relationship between STAT1 and STAT3 in anti-tumor immunity reveals that STAT1 loss in high STAT3-activated mammary tumors suppresses tumor onset in mice. Interestingly, STAT1 loss in low STAT3-activated mammary tumors significantly promotes tumor growth, suggesting that STAT3 activity may be critical in determining STAT1’s pro- or anti-tumor roles [128].
The programmed cell death 1 (PD-1) and programmed death-ligand 1 (PD-L1) interaction, which inhibits hyperactivation of immune cells to balance normal immune homeostasis, is a common strategy for tumor cells to inhibit and evade anti-tumor immune cells [133,134]. IFNɣ induces PD-L1 expression through the JAK1/2-STAT1 pathway in TNBC cells [135,136]. PD-L1 and pSTAT1 are positively correlated in breast tumor specimens [137]. Furthermore, upregulated PD-L1 in CSCs promotes immune evasion or suppression [138]. One study elucidating the mechanisms of PD-L1 in breast cancer utilizes dual inhibition of both STAT1 and STAT3, and found that combined inhibition abrogates expression of PD-L1; pSTAT1 and pSTAT3 form a heterodimer in the cytosol, translocate to the nucleus, and directly bind the promoter region of PD-L1 [139]. Taken together, STAT1 plays critical roles in modulating breast cancer cells, BCSCs, and the pro- and anti-tumor immune system. However, the major regulators or modulators that determine whether STAT1 will promote or antagonize breast cancer remain unclear, emphasizing the importance of continuing to investigate STAT1 in breast cancer.
Since the tumor suppressive role of STAT1 is highly reported, there are few STAT1 inhibitors currently used for the treatment of breast cancer (Table 1). Epigallocatechin gallate (EGCG), the main active component in green tea and a known STAT1 inhibitor, reduces cell proliferation and induces apoptosis in luminal A breast cancer cells [131,140]. Furthermore, EGCG decreases tumor volume and increases PARP expression to inhibit tumor growth in vivo [140]. EGCG also exhibits a dual impact by suppressing proliferation and migration and by modulating vascular endothelial growth factor (VEGF) in TNBC, suggesting an important role for angiogenesis in TNBC [141]. Another STAT1 inhibitor, Fludarabine [142,143], suppresses STAT1 expression, pSTAT1, and PD-L1 expression in breast cancer cells [139]. In conclusion, the pro- or anti-tumor roles may be dependent on the status of additional factors (i.e. STAT3) or other aspects in the tumor microenvironment, which warrants further investigation.
Table 1.
Preclinical STAT inhibitors in breast cancer.
| STAT | Drug/Therapeutic Inhibitor | Main Function | Cancer and/or Cell Type | Ref. |
|---|---|---|---|---|
|
| ||||
| STAT1 | EGCG | Suppresses cell proliferation of breast cancer cells | Breast cancer (MCF-7) cells | [131] |
| STAT1 | Fludarabine | Suppresses STAT1, pSTAT1, and PD-L1 expression in breast cancer cells | Breast cancer (MCF-7, MDA-MB-231 and MDA-MB-468) cells | [139] |
| STAT1 | Fludarabine and doxorubicin | Decreased STAT1-positive cells and suppressed mammary tumor growth of murine mammary tumor cells | Breast cancer (MDA-MB-231 and PNA.Met1) cells; PNA.Met1 (murine mammary tumor line derived from spontaneous mammary tumors from MMTV-PyMT model) | [120] |
| STAT3 | 10,11-dehydrocurvularin (DCV) | Macrolide, derived from marine fungus, inhibits pSTAT3 without impacting upstream JAK1 or JAK2, leading to decreased breast cancer cell proliferation, migration, and invasion; DCV suppressed tumor growth through inhibited pSTAT3 | Breast cancer (MDA-MB-231 and MDA-MB-468) and mammary epithelial (MCF-10A) cells; MDA-MB-231 cells in mouse xenografts | [366] |
| STAT3 | 17o | Decreases pSTAT3 in breast cancer cells | Breast cancer (MDA-MB-468) and multiple myeloma cells | [367] |
| STAT3 | 6Br-6a | Induces cell cycle arrest and apoptosis by blocking activation of STAT3; decreases tumor growth and size in mouse xenografts | Breast cancer (MDA-MB-231 and MCF-7) cells; MDA-MB-231 in right flank of nude mice | [368] |
| STAT3 | 7β-(3-Ethyl-cis-crotonoyloxy)– 1α-(2-methylbutyryloxy)– 3,14-dehydro-Z-notonipetranone (ECN) | A sesquiterpenoid (from Farfarae Flos, traditional medicine used to treat inflammation) suppresses pSTAT3, pJAK1/2, pSrc and induces apoptosis in breast cancer cells | Breast cancer (MDA-MB-231) cells | [369] |
| STAT3 | Alantolactone | A sesquiterpene lactone induces apoptosis and ROS levels; activates caspases and decreases mitochondrial membrane potential and pSTAT3 in breast cancer cells | Breast cancer (MDA-MB-231 and MCF-7) cells | [370] |
| STAT3 | Bazedoxifene | Suppresses pSTAT3, cell proliferation, migration and invasion of breast cancer cells; induces apoptosis in breast cancer cells | Breast cancer (SUM159, MDA-MB-231, and MDA-MB-468) cells | [371] |
| STAT3 | Betulinic acid | Inhibits proliferation, migration, and invasion of breast cancer cells; decreases tumor growth, volume, and lung metastasis in vivo | Breast cancer (MCF-7, 4T1, and MDA-MB-231) cells; subcutaneous 4T1 cells in BALB/C mice | [372] |
| STAT3 | Bisindole-PBD (5b) | Suppresses angiogenesis by suppressing expression of VEGF and STAT3 in breast cancer cells | Breast cancer (MDA-MB-231 and MCF-7) and human endothelial cells | [373] |
| STAT3 | BP-1–102 | Analog of S31–201.1066; blocks pSTAT3 and STAT3 activation, suppresses growth, migration, and invasion of tumor cells and suppresses expression of STAT3-target genes; inhibits growth of human breast tumor xenografts in mice | Breast cancer (MDA-MB-231), prostate, non-small cell lung cancer, and pancreatic cancer cells; MDA-MB-231 xenografts in mice | [374] |
| STAT3 | Cantharidin | Decreases cell growth in breast cancer cells, inhibited adhesion of breast cancer cells to platelets | Breast cancer (MCF-7) cells | [375] |
| STAT3 | Cantharidin | Inhibits cell proliferation, migration, and invasion and induces cell cycle arrest in TNBC cells; inhibits tumor growth in xenograft mouse model | Breast cancer (MDA-MB-231) cells; MDA-MB-231 cells in breast xenograft mouse model | [376] |
| STAT3 | Cantharidin | Inhibits cell proliferation and autophagy and induced apoptosis of TNBC cells; inhibited growth of TNBC xenografts in nude mice | Breast cancer (MDA-MB-231 and MDA-MB-468) cells; MDA-MB-231 and MDA-MB-468 cells in nude mice | [377] |
| STAT3 | Caffeic acid p-nitro-phenethyl ester (CAPE-pNO2) | CAPE-pNO2 decreases pEGFR, pSTAT3, pAkt, MMP2, MMP9, and VEGFA; suppresses breast cancer cell proliferation, migration, and invasion; suppresses tumor growth and induces apoptosis in xenograft mouse model | Breast cancer (MDA-MB-231) cells; subcutaneous injection of MDA-MB-231 cells in nude mice | [378] |
| STAT3 | Carfilzomib | Decreases proliferation and mitosis in breast cancer cell lines; decreases serum IL-6 in mice with MDA-MB-231 tumor | Breast cancer (MDA-MB-231 and MDA-MB-468) cells; MDA-MB-231 cells in nude mice | [379] |
| STAT3 | Catechol | Inhibits proliferation and mammosphere formation (and CD44high/CD24low subpopulation) of breast cancer cells | Breast cancer (MDA-MB-231 and MCF-7) cells | [380] |
| STAT3 | CDDO-Methyl Ester (CDDO-ME) | Inhibits IL-6-induced and constitutive JAK1 activity, and inhibits IL-6-induced STAT3 activation and dimerization | Breast cancer (MDA-MB-468) and cervical cancer cells | [381] |
| STAT3 | Compound 6b | Curcumin-benzo[b]thiophene 1, 1-dioxide (Curcumin-BTP) hybrids as STAT3 inhibitors; Compound 6B suppressed pSTAT3, nuclear translocation, and DNA-binding activity in breast cancer cells; Compound 6B induces apoptosis in breast cancer cells, but not normal breast epithelial cells; inhibits IL-6-mediated pSTAT3; suppresses tumor growth of breast cancer cells in mice with reduced pSTAT3 in tumors | Breast cancer (MCF-7), normal breast epithelial (MCF-10A), and normal liver cells; MCF-7 cells subcutaneously injected in BALB/C mice | [382] |
| STAT3 | Coumarin-benzo[b]thiophene 1, 1-dioxide | Decreases pSTAT3 and cell proliferation and induced apoptosis and ROS generation of breast cancer cells; decreased tumor growth and pSTAT3 in vivo | Breast cancer (MDA-MB-231, MCF-7, and 4T1), mammary epithelial (MCF-10A), hepatocellular carcinoma, and colonic carcinoma cells; 4T1 mouse xenograft model | [383] |
| STAT3 | CPA-1, CPA-7 | Inhibition of constitutive STAT3 activation and decrease in pSTAT3 and cell viability and induces apoptosis in breast cancer cells | Breast cancer (MDA-MB-231, MDA-MB-435, MDA-MB-453, MCF-7, and MDA-MB-468), melanoma, prostate cancer, lung carcinoma and colon cancer cells | [384] |
| STAT3 | Cucurbitacin E | (CuE) inhibits growth and pSTAT3 and induces apoptosis and cell cycle arrest of breast cancer cells | Breast cancer (MDA-MB-231 and Bcap-37) cells | [385] |
| STAT3 | Curcumin alone and curcumin with EGCG | Curcumin and epigallocatechin gallate (EGCG) reduced pSTAT3, STAT3 interaction with NFkB, CD44-positive cells, and mammospheres | Breast cancer (MDA-MB-231 and MCF-7) cells | [267, 386] |
| STAT3 | Cyanidin-3-glucoside (C3G) | C3G, an anthocyanin in many fruits, reduced angiogenesis by inhibiting VEGF expression and secretion in breast cancer cells | Breast cancer (MDA-MB-231 and Hs-578 T) cells | [387] |
| STAT3 | Deguelin | Decreases pSTAT3 and expression of EGFR-downstream target proteins and cell proliferation of breast cancer cells; inhibits growth of MDA-MB-231 cells in dorsal flank of nude mice | Breast cancer (MDA-MB-231, MDA-MB-468, BT-20 and BT-549) cells; MDA-MB-231 cells in dorsal flank of nude mice | [388] |
| STAT3 | Dihydrotanshinone (DHTS) | Decreases mammosphere formation of breast cancer cells; inhibits tumor growth in mouse xenograft model | Breast cancer (MDA-MB-231 and MCF-7) cells; MCF-7 in tumor xenografts in mice | [389] |
| STAT3 | DT-13 | Inhibits migration via suppression of pSTAT3 and PLOD2 expression in breast cancer cells; DT-13 had little effect on tumor growth, but decreased lung and liver metastasis burden in mice | Breast cancer (MDA-MB-231 and MD-MB-468) and pre-adipocyte cells; MDA-MB-231 in orthotopic xenograft tumor model | [390] |
| STAT3 | Esculentoside A | Derived from root of Phytolacca esculenta; inhibits proliferation and mammospheres and induces apoptosis in breast cancer cells; suppresses breast CSC initiated tumor growth in mice | Breast cancer (EMT6, MCF-7), normal mammary epithelial (MCF10A), and normal liver cells | [271] |
| STAT3 | Eupalinolide J | Suppresses pSTAT3, proliferation and induces apoptosis and cell cycle arrest of TNBC cells; suppresses tumor growth in vivo | Breast cancer (MDA-MB-231 and MDA-MB-468) and mammary epithelial (MCF-10A) cells; MDA-MB-231 cells in nude mouse xenograft model | [391] |
| STAT3 | Evista (Raloxifene-HCl) | (Raloxifene-HCl) Inhibits pSTAT3 and IL-6-induced pSTAT3, decreases cell viability, and increases caspase-3 activity in breast cancer cells | Breast cancer (MCF-7, MDA-MB-231, and SUM159), colon cancer, multiple myeloma, and Hela cells | [392] |
| STAT3 | FLLL31/FLLL32 | Derived from curcumin (main compound in turmeric); designed to bind JAK2 and STAT3 SH2 domain; downregulates pSTAT3, DNA-binding activity, cell viability, and invasion of breast cancer cells; induces apoptosis of breast cancer cells; FLLL32 suppresses tumor growth and vascularization in mouse xenografts | Breast cancer (MDA-MB-231, SK-BR-3, MDA-MB-468, MDA-MB-453, and SUM159), pancreatic cancer, and human embryonic kidney cells; MDA-MB-231 mouse xenografts | [268] |
| STAT3 | Flubendazole | Inhibits pSTAT3 and cell viability and induces apoptosis in TNBC cells; suppresses BCSC-enriched TNBC tumor growth, angiogenesis, and metastasis in vivo | Breast cancer (MDA-MB-231, Hs578T, BT-549, and 4T1) cells; 4T1 cells in MFP of BALB/C mice | [393] |
| STAT3 | Galiellalactone, SG-1709, and SG-1721 | Galiellalactone and two analogues (SG-1709 and SG-1721) block pSTAT3 and suppress cell viability and proliferation in TNBC cells; SG-1721 induces cell cycle arrest and apoptosis; SG-1721 inhibits tumor growth of TNBC cells in vivo | Breast cancer (BT-549, BT-20, MDA-MB-468, MCF-7, T47D, SK-BR-3, MDA-MB-453) and mammary epithelial (MCF-10A) cells; TNBC cells (exact cell line not stated) subcutaneous implantation in nude mice | [394] |
| STAT3 | Ganoderic acid A | Decreases pJAK2, pSTAT3 and suppresses invasion and induces apoptosis of breast cancer cells | Breast cancer (MDA-MB-231) cells | [395] |
| STAT3 | Genistein | Phytotherapeutic Genistein suppresses breast cancer cell proliferation | Breast cancer (MDA-MB-453) cells | [396] |
| STAT3 | GO-Y030 | Suppresses pSTAT3, STAT3 transcriptional activity, cell viability and induces apoptosis in breast cancer cells | Breast cancer (MDA-MB-231) and pancreatic cancer cells | [397] |
| STAT3 | HJC0123 | Inhibits pSTAT3 and STAT3-mediated luciferase reporter activity and induced apoptosis in TNBC cells; inhibits tumor growth in TNBC xenograft mouse model | Breast cancer (MCF-7 and MDA-MB-231) and pancreatic cancer cells; MDA-MB-231 cells in xenograft mouse model | [398] |
| STAT3 | HJC0416 | (5-chloro-N-(1,1-dioxo-1 H-1λ6-benzo[b]thiophen-6-yl)–2-hydroxybenzamide) HJC0416 inhibits cell cycle progression and promotes apoptosis in breast cancer cells; HJC0416 reduces tumor volume in breast cancer murine xenografts in vivo | Breast cancer (MDA-MB-231) cells; MDA-MB-231 tumor xenografts in mice | [399] |
| STAT3 | Hydrazinocurcumin | (Analog of curcumin) inhibits pSTAT3, downstream STAT3 target genes, cell migration and invasion and induces apoptosis in breast cancer cells | Breast cancer (MDA-MB-231 and MCF-7) cells | [400] |
| STAT3 | Ilamycin C | Ilamycins are cyclic peptides produced by (isolated from) Streptomyces; Increased toxicity against TNBC cells vs. LumA cells; suppresses pSTAT3, proliferation, migration and invasion and induces apoptosis in TNBC cells | Breast cancer (MDA-MB-231, BT-549, and MCF-7) and normal breast (MCF10A) cells | [180] |
| STAT3 | IS3 295 | Decreased cell proliferation and constitutive STAT3 activation in breast cancer cells; induced apoptosis and promoted cell cycle arrest in breast cancer cells | Breast cancer (MDA-MB-231, MDA-MB-435, MDA-MB-453, and MDA-MB-468), prostate cancer, melanoma, pancreatice cancer, and fibroblast cells | [401] |
| STAT3 | Isoharringtonine (IHT) | IHT decreases pSTAT3 and suppresses proliferation, migration, and BCSCs in breast cancer cells | Breast cancer (HCC1937, HCC1806 and MCF-7) cells | [402] |
| STAT3 | LLL12 | Inhibits STAT3 phosphorylation, DNA binding activity, STAT3-dependent luciferase activity, cell viability and migration in breast cancer cells; decreased tumor volume in mouse tumor xenografts | Breast cancer (MDA-MB-231, MDA-MB-453, and SK-BR-3), pancreatic cancer, glioblastoma, human hepatocytes, and human lung fibroblast cells; MDA-MB-231 tumors in mice | [403] |
| STAT3 | LLY17 | Inhibits pSTAT3 in breast cancer cells; suppresses cell viability and migration and induces apoptosis in TNBC cells, not Luminal A breast cancer cells; suppresses tumor growth of 4T1 cells in vivo | Breast cancer (MDA-MB-468, MDA-MB-231, SUM159, BT-549, and 4T1) cells; 4T1 in MFP mouse model in BALB/C mice | [404] |
| STAT3 | LYR71 | 6-methyl-2-propylimino-6, 7-dihydro-5 H-benzo [1,3]-oxathiol4-one (LYR71), a derivative of trimeric resveratrol, suppresses pSTAT3, MMP9 expression and activity, migration and invasion of breast cancer cells; inhibits growth of MDA-MB-231 tumors in mice | Breast cancer (MDA-MB-231) cells; MDA-MB-231 subcutaneously injected in flank of nude mice | [405] |
| STAT3 | Napabucasin | (BBI608) Decreases pSTAT3, cell viability, and sternness of breast cancer cells | Breast cancer (MCF-7) cells | [406] |
| STAT3 | Naringenin | Decreases pSTAT3, cell viability and induces apoptosis of breast cancer cells; increases caspase 3 and 9 activity | Breast cancer (MDA-MB-231) cells | [407] |
| STAT3 | Niclosamide | Inhibits proliferation, migration, and invasion and induces apoptosis in breast cancer cells; decreased IL-6 and pSTAT3 in breast cancer cells | Breast cancer (MDA-MB-468 and MCF-7) cells | [408] |
| STAT3 | Nifuroxazide | Reduces pSTAT3 and expression of MMP2 and MMP9, decreases cell viability, migration and invasion and induces apoptosis of breast cancer cells; suppressed tumor growth of 4T1 cells in vivo | Breast cancer (MDA-MB-231, MCF-7, and 4T1) cells; 4T1 cells in BALB/C mice | [409] |
| STAT3 | Pectolinarigenin | Suppresses proliferation, migration and invasion and induces apoptosis of breast cancer cells; had no effect on tumor growth of subcutaneously injected 4T1 cells, but decreased spontaneous lung metastases; increased CD8 + T cells in peripheral blood and in lung metastatic tissue | Breast cancer (4T1, MDA-MB-231, and MCF-7) cells; subcutaneous 4T1 cells in mice | [410] |
| STAT3 | Picrasidine G | Decreases pSTAT3 and STAT3-regulated gene expression and induced apoptosis in TNBC cells | Breast cancer (MCF-7, T47D, MDA-MB-361, SKBR3, Hs578T, MDA-MB-231 and MDA-MB-468) and mammary epithelial (MCF10A) cells | [411] |
| STAT3 | Pimozide | Decreases pSTAT3, suppresses migration and invasion and increases apoptosis in TNBC cells | Breast cancer (BT-549, MDA-MB-231, MDA-MB-468, and HCC1806) cells | [260] |
| STAT3 | Piperlongumine (PL) | PL, a natural alkaloid, decreases pSTAT3 and pJAK2, suppresses breast cancer cell proliferation, colony formation, and induces apoptosis | Breast cancer (MDA-MB-231 and MDA-MB-453) cells | [412] |
| STAT3 | PM-73 G | Phosphopeptidomimetic prodrug blocks constitutive pSTAT3, but not Akt or Tyr861 phosphorylation in MDA-MB-468 cells (first ref). Treatment inhibits orthotopic MDA-MB-468 human breast tumor xenograft tumor proliferation in nude mice (second ref) | Breast cancer (MDA-MB-468) cells; MDA-MB-468 human breast tumor xenografts in nude mice | [413, 414] |
| STAT3 | Pyrimethamine | Decreases pSTAT3 in breast cancer cells and mouse tumors; decreases proliferation and invasion of breast cancer cells and decreases T reg cells in mice | Breast cancer (TUBO) cells | [415] |
| STAT3 | Resveratrol (RES) | (aka RES, aka trans-3,4′,5-trihydroxystilbene) Blocks Src tyorisine kinase activity, thereby inhibiting constitutive STAT3 activation (pSTAT3), cell proliferation, cell cycle progression and induces apoptosis in breast cancer cells | Breast cancer (MDA-MB-231 and MDA-MB-468), pancreatic cancer, prostate cancer, and mouse fibroblast cells | [416, 417] |
| STAT3 | rPP-C8 | Recombinant STAT3-specific inhibitor (rPP-C8) suppresses STAT3 target gene expression and proliferation, and induced apoptosis of breast cancer cells | Breast cancer (SKBR3, MDA-MB-468, MCF-7, T47D, and 4T1), glioblastoma, melanoma, and prostate cancer cells | [418] |
| STAT3 | S3I-1757 | Disrupts STAT3-STAT3 dimerization, which is required for nuclear translocation; inhibits pSTAT3, nuclear accumulation of P-Y705-STAT3, STAT3 DNA-binding and suppresses expression of STAT3 target genes (BCL2Ll, BIRC5, CCND1, and MMP9); suppresses migration and invasion of breast cancer cells | Breast cancer (MDA-MB-468, MDA-MB-231, and MDA-MB-453), lung cancer, mammary epithelial (MCF10A), and embryonic kidney cells | [419] |
| STAT3 | S3I-201 | (NSC #74859) Chemical probe inhibitor of STAT3 activity; binds to STAT3 phosphotyrosine peptide derived from the x-ray crystal structure of the STAT3beta homodimer; inhibits growth and induces apoptosis preferentially in tumor cells; inhibits expression of stat3-regulated genes (cyclin D1, Bcl-xl, and survivin) and inhibits growth of human breast tumors in vivo | Breast cancer (MDA-MB-231, MDA-MB-435, and MDA-MB-468) and other (NIH 3T3/v-Src) cells; MDA-MB-231 breast tumor xenografts in mice | [256] |
| STAT3 | Sabutoclax | Pan-active BCL-2 protein antagonist; induces apoptosis and caspase activity, and decreases pSTAT3 and CD44high/CD24low subpopulation in breast cancer cells; increased apoptosis, decreases CD44 and ALDH1 cells and decreases pSTAT3 in fresh human breast tumors | Breast cancer (MCF-7 and Cal51) cells; human breast tumor samples | [420] |
| STAT3 | Saikosaponin b2 (SSb2) | Saikosaponin b2 (SSb2) decreases pSTAT3, VASP, MMP2, and MMP9; suppresses breast cancer cell proliferation and migration; limited to no liver and kidney toxicity in mice | Breast cancer (MCF-7) cells | [270] |
| STAT3 | Satraplatin | A tetravalent platinum derivative decreases pSTAT3 and increases apoptosis of breast cancer cells | Breast cancer (MCF-7 and MDA-MB-231) cells | [249] |
| STAT3 | Schisandrin A | Inhibits P-gp mRNA and protein expression in breast cancer cells; decreased pSTAT3 in breast cancer cells | Breast cancer (MCF-7) and DOX-resistant (BEL-7402 and K-562) cells | [421] |
| STAT3 | SH48 | SH48 inhibits STAT3 dimerization and translocation into the nucleus; induces autophagy of mammary epithelial cells transformed by H-ras oncogene | Mammary epithelial cells transformed by H-ras oncogene (MCF10A-ras), prostate cancer, and cervical cancer cells | [422] |
| STAT3 | SH5–07, SH4–54 | Hydroxamic acid (SH5–07) and benzoic acid (SH4–54) analogues inhibit STAT3 activation and DNA binding activity; suppressed breast cancer cell proliferation to greater extent when the cells exhibit constitutive STAT3 activity; both compounds (individually) suppressed growth of breast cancer cells and decreased expression of STAT3-target genes in vivo | Breast cancer (MDA-MB-231 and MCF7), pancreatic, prostate, and normal mouse fibroblast cells; MDA-MB-231 cells injected into left flank athymic mice | [259] |
| STAT3 | PMMB-187 | PMMB-187 (a shikonin derivative) inhibits constitutive STAT3 activation, nuclear translocation, DNA binding activity, and subsequent target gene expression in breast cancer cells; induces breast cancer cell apoptosis; inhibits breast tumor growth in mouse xenografts | Breast cancer (MCF-7, MDA-MB-231 and MDA-MB-468) and non-tumor breast epithelial (MCF-10A) cells; MDA-MB-231 cells in mouse xenografts | [423] |
| STAT3 | Silibinin | (Legasil), Made from flavonoid extracts from milk thistle seeds; blocks STAT3 signaling in reactive astrocytes resulting in decreased brain metastases from multiple cancers (including breast cancer) in mice | Brain-tropic breast cancer (MDA-MB-231-BrM2) and melanoma cells | [263] |
| STAT3 | SPI | Potent and selective inhibitor of the STAT3 SH2 domain; blocks constitutive pSTAT3, DNA binding and activity, and transcriptional function in multiple cancers | Breast cancer (MDA-MB-231, MDA-MB-435, MCF-7), pancreatic cancer, prostate cancer, non-small cell lung cancer, and normal mouse fibroblast cells | [424] |
| STAT3 | STA-21 | (NSC #628869) STA-21 inhibits STAT3 DNA binding activity, dimerization, and STAT3-dependent luciferase activity; inhibits survival of breast cancer cells with constitutive STAT3 signaling | Breast cancer (MDA-MB-231, MDA-MB-435S, MDA-MB-453, MDA-MB-468, and MCF-7), human ovarian carcinoma, and human skin fibroblast cells | [425] |
| STAT3 | Stattic | (6-nitro-benzo[b]thiophene-1,1-dioxide 1) Retains STAT3 in cytosol by inhibiting binding of a phosphotyrosine-containing peptide derived from the gp130 receptor to the STAT3 SH2 domain | Breast cancer (MDA-MB-231 and MDA-MB-435S) and HepG2 cells | [258] |
| STAT3 | STX-0119 | Inhibited STAT3 promoter binding and pSTAT3 in breast cancer cells | Breast cancer (MDA-MB-468) cells | [426] |
| STAT3 | WP1066 | (modified structure of AG490) WP1066 decreases pSTAT3 in breast cancer cells; decreases breast cancer brain metastases in intracardiac mouse model; reduces proliferation, MM9 expression, and VEGF expression in brain metastatic cells | Breast cancer (MDA-MB-231BR and BT-474BR) cells; MDA-MB-231BR brain metastases in mice | [261, 262] |
| STAT3 | XZH-5 | Decreases pSTAT3, STAT3 downstream target genes, colony formation and migration and induces apoptosis in breast cancer cells, inhibits IL-6-induced pSTAT3 and nuclear accumulation in breast cancer cells | Breast cancer (MDA-MB-231, SUM159 and MCF-7) and pancreatic cancer cells | [427] |
| STAT3 and STAT5 | SH-4–54 | Suppressed breast cancer cell proliferation; decreases TNBC cell plasma membrane antiporter system (xC) and cystine import, both of which are essential for cancer cells to adapt to increased levels of ROS | Breast cancer (MDA-MB-231 and T47D) cells | [428] |
| STAT3 and STAT5 | Withacnistin | Blocks EGF- or IL-6-stimulated STAT3 and STAT5 binding to EGFR and gp130; suppresses pSTAT3 nuclear nuclear translocation and DNA binding activity resulting in decreased transactivation and STAT3-target gene expression and induces apoptosis in breast cancer cells; treatment suppresses breast tumor growth | Breast cancer (MDA-MB-468), lung cancer, and mouse fibroblast cells | [429] |
| STAT5 | CAS 285986–31-4 | Nonpeptidic nicotinoyl hydrazine compound STAT5 inhibitor; decreases pSTAT5 and proliferation in breast cancer cells | Breast cancer (T47D) cells | [314] |
| STAT5a | Pimozide | Pimozide sensitives DOX-resistant cells through the suppression of STAT5a; decreases tumor growth in subcutaneous xenograft mouse model | Breast cancer (MCF-7) cells | [322, 328] |
| STAT6 | AS1517499 | Inhibits pSTAT6 and M2 macrophage differentiation of mouse macrophages; suppresses tumor growth and liver metastasis of orthotopic 4T1 breast cancer mouse model | Breast cancer (4T1) and mouse macrophage cells; 4T1 cells in orthotopic mouse model | [353] |
5.2. STAT2
STAT2 was discovered through investigation of IFN signaling pathways and relays important immunomodulatory and anti-viral functions of IFN-I [144–146]. STAT2 is the longest STAT family protein and is physically and functionally conserved; IFNɣ signaling is restored with mouse STAT2 in STAT2-deficient human cells [147,148]. Canonical STAT2 signaling involves IFN-I or –III activation of the heterotrimeric interferon-stimulated gene factor 3 (ISGF3) complex, which contains STAT1, STAT2, and interferon regulatory factor 9 (IRF9) [42,144]. The ISGF3 complex is unique to STAT2 activation as pSTAT2 molecules do not form homodimers despite the fact that alternative homo- and heterodimers of the other STAT proteins are identified to mediate IFN signaling [148]. Notably, the STAT2-mediated ISGF-3 complex can upregulate IL-6 gene expression leading to increased IL-6/STAT3 signaling [149,150], which is commonly dysregulated and hyperactivated in breast cancer [24].
PR suppresses IFN signaling in breast cancer cells by promoting an immunosuppressive microenvironment [151]. Further investigation reveals that STAT2 is critical for IFN-I pathway activation and that PR-mediated suppression of IFN signaling occurs by increasing ubiquitination and subsequent degradation of STAT2 in breast cancer cells [151]. STAT2-deficiency suppresses breast cancer cell proliferation, migration, and invasion, while STAT2 overexpression upregulates expression of IFITM1, contributing to aggressive inflammatory breast cancer phenotypes [152]. Covalent addition of ISG15 ubiquitin like modifier (ISG15), a ubiquitin-like protein tag, to STAT2 promotes secretion of chemokine ligands leading to a subsequent increase in CD8 + T cells and suppression of breast cancer growth and metastasis [153]. Analysis of breast cancer patient data reveals that high STAT2 mRNA expression correlates with worse post-progression survival, but better relapse-free survival [154,155]. Given that STAT2 is not as commonly studied, there are currently no published natural compounds or pharmacological inhibitors for STAT2 in breast cancer.
5.3. STAT3
In 1994, STAT3 was first described as a DNA-binding protein activated by IL-6 in hepatocytes [156,157]. A year later, the Src oncoprotein was reported to activate STAT3, making this publication the first to implicate STAT3 in cancer [158]. Nearly 20 years later, STAT3 is now the most widely studied STAT family member in breast cancer as it regulates networks of genes involved in oncogenesis [159], cancer cell proliferation [43], cell cycle progression [43], angiogenesis [160,161], metastasis, and evasion of apoptosis [162–164]. Moreover, STAT3 is the only STAT protein that is essential for embryonic development as homozygous genetic deletion is lethal in mice [165]. STAT3 activation is triggered by the largest and most diverse number of cytokines of all the STAT family members, underscoring its vast influence on many physiological pathways [53, 166, 167]. These ligands include the IL-6 cytokine family, IL-10 cytokine family, additional interleukins (i.e. IL-21), multiple IFNs, EGF, FGF, IGF, leptin, and granulocyte-colony stimulating factor (G-CSF) [21, 44, 53, 168, 169]. Additionally, there are multiple isoforms of STAT3, though most publications studying STAT3 in cancer refer to the full-length isoform (STAT3α) [170].
Though in normal conditions STAT3 is regulated by many molecular factors [23], STAT3 is dysregulated in multiple cancer types including breast cancer [171,172]. The IL-6 family of cytokines are the most predominant STAT3 activators, in particular, IL-6 is a major mediator of breast cancer cell growth, angiogenesis, tumor growth, metastasis, and immune evasion or modulation [24, 43, 169, 173–175]. Increased serum IL-6 upregulates STAT3 activity, which increases IL-6 expression through STAT3 directly binding and transactivating the IL-6 promoter region, further promoting this IL-6/JAK/STAT3 positive-feedback loop [173,174]. Hyperactivation of STAT3 enriches expression of genes involved in cancer stemness (CD44) [176], cell cycle (cyclin D1) [159, 177], apoptosis or cell survival (Bcl-2, Bcl-xL, and Mcl-1) [178–180], invasion and migration (matrix metalloproteases or MMPs, ERRα) [181, 182], angiogenesis (VEGF and HIF1α) [183,184], and immunosuppression (IL-10 and TGFβ) [174, 183, 185, 186]. Furthermore, pSTAT3 correlates with poor prognosis for breast cancer patients [187]. Though STAT3-mediated gene signatures are identified in most breast cancer subtypes, STAT3-mediated gene signatures are upregulated in basal-like compared to luminal subtypes, suggesting STAT3 activity is more likely to be hyperactivated in TNBC subtypes of breast cancer [188].
Oncostatin M (OSM) upregulates STAT3-dependent expression of IL-6 and high OSM associates with worse patient survival [189,190]. Increased IL-6 secretion and pSTAT3 promote breast cancer progression resulting in shortened patient survival [189]. OSM activates STAT3/SMAD3 signaling in breast cancer cells, which leads to increased Snail and epithelial-to-mesenchymal transition (EMT) [191]. IFNβ antagonizes this OSM/STAT3/OSM pathway, thus inhibiting BCSC phenotypes and increasing STAT1 and pSTAT1 in breast cancer cells [191]. Increased IL-8 and growth-regulated oncogene in inflammatory breast cancer promotes BCSCs, which is enhanced by co-culture with macrophages [192]. On the other hand, IL-17 suppresses STAT3 activation [193].
In addition to cytokines and chemokines, proteins or other factors also activate STAT3 signaling in breast cancer. Long noncoding RNA (lncRNA) MAFG-antisense 1 (MAFG-AS1), whose expression is elevated in breast tumors compared to normal breast tissue, increases pJAK2 and pSTAT3; knockdown of MAFG-AS1 suppresses breast cancer cell proliferation and decreases tumor growth in mice [194]. Enhancer of zeste homolog 2 methylates STAT3 to increase nuclear localization of STAT3 and promote breast cancer progression [195]. Endogenous breast tumor kinase (Brk) leads to upregulated pSTAT3 and STAT3 transcriptional activation to promote breast cancer oncogenesis [196]. Furthermore, STAP-2 enhances STAT3 activation by directly binding and interacting with STAT3 and indirectly through Brk; increased STAT3 activation promotes breast cancer cell proliferation [197–199].
EGFR is highly upregulated and often constitutively activated in breast cancer [200–202]. Importantly, EGFR activation leads to JAK autophosphorylation and subsequent STAT activation [203,204]. EGFR and pSTAT3 promote breast cancer cell proliferation and invasion and are upregulated in TNBC tissues [203]. Growth factor receptor-binding protein 2 (Grb2), the adaptor protein that directly binds EGFR, downregulates EGF-mediated activation of STAT3, which decreases STAT3-mediated gene transcription [205].
Tropomyosin receptor tyrosine kinases (Trk), a family of tyrosine kinase receptors, bind neurotrophins and other ligands to regulate multiple cellular processes [206]. TrkA and phosphorylated-TrkA (pTrkA) levels are elevated in breast tumors compared to normal breast tissue; overexpression promotes breast cancer cell proliferation, migration, and invasion [207]. Our lab found that TrkA and JAK2/STAT3 are co-overexpressed and activated in HER2-enriched and TNBC [208]. TrkA and JAK2/STAT3 also physically and functionally interact in HER2-enriched and TNBC [208]. Moreover, TrkA interaction with STAT3 promotes STAT3 phosphorylation resulting in STAT3 translocation to the nucleus and increases STAT3 transcriptional activity. Additionally, co-activation of the TrkA-STAT3 pathway promotes BCSCs and correlates with poor patient prognosis [208].
Interestingly, STAT3 also physically and functionally interacts with truncated glioma-associated oncogene homolog 1 (tGLI1), a gain-of-function isoform of the transcription factor GLI1, increasing mammosphere forming ability of breast cancer cells [171,209]. tGLI1, originally discovered in glioblastoma [210], is tumor-specific [210,211], and can regulate GLI1 target genes as well as eight novel target genes, which include VEGF-A, VEGF-C, VEGFR2, TEM7, HPSE, CD24, CD44, and OCT4 [171, 212–218]. tGLI1 has also been shown to promote breast cancer brain metastasis (BCBM) by enriching BCSCs [214]. More recently we found that tGLI1-positive breast cancer cells upregulate extracellular vesicle-derived miR-1290 and miR-1246 to activate astrocytes to promote the progression of brain metastases; astrocytes overexpressing miR-1290 promoted the growth of co-implanted breast cancer cells in the brain in vivo [219].
There are also multiple negative regulators of STAT3 reported in breast cancer. Double PHD fingers 3 (DPF3 or CERD4) suppresses breast cancer cell proliferation and higher expression correlates with better patient prognoses; downregulation of DPF3 activates the JAK2/STAT3 pathway to promote breast cancer proliferation and migration [220]. WW domain-containing oxidoreductase (Wwox) is lost in TNBC compared to luminal breast cancer cells, while overexpression of Wwox suppresses cell proliferation and metastasis in TNBC [221]. Further investigation reveals Wwox inhibits pJAK2, pSTAT3, and STAT3 association with the IL-6 promoter region, implicating Wwox as an important negative regulator of STAT3 activity [221]. Gametogenetin-binding protein suppresses breast cancer cell proliferation, migration, invasion, BCSCs, and induces apoptosis partly through the inhibition of the IL-6/JAK/STAT3 pathway [222]. GRAM domain-containing protein 1B decreases breast cancer cell migration by modulating cell morphology and decreasing STAT3 signaling [223].
MicroRNAs (miRNAs), small noncoding RNAs that regulate gene expression by targeting mRNAs, also modulate STAT3 activity in breast cancer [30, 224, 225]. Overexpression of miR-124 decreases mRNA and protein levels of STAT3; miR-124 directly binds to STAT3 mRNA leading to decreased breast cancer cell growth and invasion [226]. Moreover, lncRNA nuclear enriched abundant transcript 1, which is increased in breast cancer compared to normal breast tissues, promotes proliferation and cell cycle progression of breast cancer cells, potentially by downregulating the STAT3-inhibitor miR-124 [227]. miR-125a and miRNA let-7e directly target the 3’- untranslated regions of IL-6 receptor (IL-6R) and STAT3 to mediate breast cancer cell proliferation, chemosensitivity, and endothelial cell adhesion [228]. The cytokine resistin increases expression of LIN28A, which suppresses let-7a in breast cancer cells [229]. Furthermore, downregulation of let-7a increases expression of target genes STAT3 and IL-6 and overexpression of let-7a suppresses resistin-mediated breast cancer cell proliferation and mammosphere formation [229]. miR-93–5p also suppresses cell proliferation by downregulating STAT3 in breast cancer [230]. miR-519d negatively regulates STAT3 to suppress breast cancer cell proliferation and invasion while inducing apoptosis [231]. Similarly, miR-520c suppresses breast cancer cell migration, invasion, and EMT-associated markers by downregulating STAT3 [232]. miR-204 suppresses expression and subsequent activity of JAK2, decreasing pSTAT3 and breast cancer cell proliferation [233]. Similarly, miR-375 suppresses BCSCs and Adriamycin resistance by directly targeting JAK2 and subsequent STAT3 activation [234]. In contrast, miR-18a overexpression correlates with a decrease in protein inhibitor of activated signal transducer and activator of transcription 3 expression to counteract STAT3 downregulation [235].
Immune modulation is another way STAT3 promotes a pro-tumorigenic microenvironment for breast cancer. Macrophages play important roles in immune surveillance and the anti-tumor immune response. However, macrophages can be transformed into tumor-promoting cells (referred to as tumor-associated macrophages, TAMS) that facilitate cancer cell evasion of the immune response as well as promote tumor cell growth [236–239]. Breast cancer cell-derived chemokine C-C motif ligand 5 promotes the M2 macrophage phenotype through activating STAT3 from receptor C-C chemokine receptor type 5 [240]. TAM-secreted IL-6 also induces BCSCs by activating STAT3 signaling to promote migration and angiogenesis [241]. In contrast, overexpression of Kruppel-like family of transcription factor-14 (KLF14), which is downregulated in breast cancer, inhibits M2 macrophage polarization as well as suppresses breast cancer cell invasion in vitro and tumor growth in vivo [242]. Further investigation reveals that KLF14 reduces invasion of breast cancer cells by activating expression of SOCS3, leading to the suppression of RhoA/Rock/STAT3 signaling in breast cancer cells [242].
STAT3 signaling also upregulates immune evasion to promote breast cancer progression. STAT3-deficient mice developed early lesions, suggesting that STAT3 is not required for tumor initiation. However, the lesions regressed over time, which is attributed to an increase in immune infiltration including increased CD8 + T cells and macrophages resulting in tumor clearance [243]. Interestingly, STAT3-wild-type mice developed a high number of lung metastases, while STAT3-deficient mice did not develop any lung metastases [243]. These findings are consistent with additional reports of high STAT3 activity in BCSC-enriched TNBC cells increasing metastatic potential [244]. In addition, IL-35, secreted from breast cancer cells, suppresses canonical T cell proliferation and induces differentiation of regulatory T cells [245]. These induced-regulatory T cells activate both STAT1 and STAT3 activity to promote breast cancer cell proliferation and T cell-specific immune evasion [245].
STAT3 can also promote breast cancer progression through mediating resistance to breast cancer treatments. SIRT4 decreases pSTAT3 in breast cancer cells and increases sensitivity to tamoxifen [246]. Exosomal miR-378a-3p and miR-378d are highly secreted by breast cancer cells treated with doxorubicin or paclitaxel and increase drug resistance by upregulating Wnt and Notch pathways while downregulating Dickkopf WNT Signaling Pathway Inhibitor 3 and NUMB Endocytic Adaptor Protein [247]. Furthermore, STAT3 promotes a drug resistant phenotype by binding the promoter regions of both miR-378a-3p and miR-378d in the breast cancer cells. miR-124, previously reported to downregulate STAT3 [226], sensitizes doxorubicin-resistant BCSCs by downregulating STAT3 and hypoxia-inducible factor-1 (HIF-1) signaling pathways in breast cancer [248]. Consistent with these findings, pharmacological inhibition of STAT3 also sensitizes breast cancer cells to doxorubicin [249]. Higher expression of Bcl-2, a regulator of apoptosis, and STAT3 activity is elevated in metastatic breast cancer cell lines compared to their parental lines and additional functional assays reveal that constitutively active STAT3 upregulates Bcl-2 in a mechanism that promotes chemoresistance of metastatic breast cancer cells [250]. Inhibition of HER2 and subsequent STAT3 activity sensitizes HER2-positive breast cancer cells to radiotherapy [251]. JAK/STAT3 signaling also regulates fatty acid beta-oxidation (FAO) by activating CPT1B and JAK inhibition suppresses FAO-mediated BCSCs and chemoresistance in breast cancer cells [252]. Overexpression of leukemia inhibitory factor receptor (LIFR)-mediated STAT3 activation promotes resistance to trastuzumab-emtansine (T-DM1), while STAT3 inhibition sensitizes T-DM1-resistant breast cancer cells [253]. Given that STAT3 promotes the progression of breast cancer by upregulating pro-tumorigenic pathways, STAT3 is a very favorable target for breast cancer therapeutics both pre-clinically and clinically [254].
As previously mentioned, STAT3 is the most widely studied STAT protein in breast cancer. Given STAT3’s various roles contributing to breast cancer progression, there are over 60 STAT3 inhibitors published in breast cancer to date (Table 1). Since the early 2000 s, STAT3 inhibitors ranging from pharmacological small-molecule inhibitors to natural plant derivatives have been a major focus for breast cancer therapeutics. It is important to note that many synthetic or natural compound STAT3 inhibitors are not fully characterized in breast cancer as not every publication shows direct and specific STAT3 inhibition. This section will discuss STAT3 inhibitors for breast cancer listed in Table 1.
For example, novel platinum compounds, CPA-1/7, inhibit STAT3 activation and DNA-binding, resulting in a decrease in cell proliferation and increase in apoptosis in breast cancer cells [255]. S31–201 inhibits activated-STAT3 dimerization to suppress breast cancer cell proliferation and induce apoptosis [256]. An analog of S3I-201, referred to as BP-1–102, inhibits STAT3 activation, which results in a decrease in breast cancer cell proliferation, migration, invasion, and increase apoptosis [257]. Furthermore, BP-1–102 intravenously injected into mice bearing breast tumor xenografts suppresses tumor growth, tumoral pSTAT3, and additional STAT3-mediated gene expression [257]. Stattic (or 6-nitro-benzo[b]thiophene-1,1-dioxide 1), an important STAT3 inhibitor, acts by selectively inhibiting STAT3 activation by blocking the GP130 receptor and STAT3-SH2 domain interaction, STAT3 dimerization, and nuclear translocation [258]. In addition, Stattic induces apoptosis in highly activated STAT3 breast cancer cell lines. Though Stattic is highly selective for STAT3, Stattic can bind STAT1 and STAT5b, but with significantly reduced binding affinities [258]. Hydroamic and benzoic acid analogues SH5–07 or SH4–54 inhibit pSTAT3 and DNA-binding and both effectively suppress breast tumor growth in vivo [259]. Furthermore, pimozide, an antipsychotic drug originally synthesized to treat schizophrenia, suppresses pSTAT3, migration and invasion and induces apoptosis in multiple TNBC cell lines [260].
WP1066, originally discovered to inhibit STAT3 in human glioma cells [261], inhibits STAT3 activation and reduces macrometastases of brain-tropic breast cancer cells in the brain of nude mice [262]. Moreover, WP1066 suppresses breast cancer cell invasion and MMP9 expression and angiogenesis, implicating WP1066 as a potential therapeutic for BCBM [262]. Interestingly, STAT3 activation distinguishes a subpopulation of reactive astrocytes that promote BCBM; loss of STAT3 in reactive astrocytes abrogates brain metastases in mice, in part, by modulating both innate and adaptive immunity in the brain [263]. Silibinin (Legasil), a STAT3 inhibitor derived from a nutraceutical product extract from milk thistle seeds, crosses the blood-brain barrier [264, 265], and suppresses brain metastases in both mouse models and in patients [263].
In addition to silibinin, there are numerous natural compounds that exhibit anticancer effects in breast cancer. Curcumin, for example, is an important phytochemical derived from turmeric that is implicated in prevention and treatment of a multitude of diseases including cancer [266]. Furthermore, curcumin alone and with EGCG suppressed mammosphere formation, pSTAT3, and CD44-positive cells of TNBC and HER2-enriched breast cancer cells [267]. In another example, FLLL31/FLLL32, STAT3-specific small molecule inhibitors designed to biochemically mimic curcumin inhibit pSTAT3, DNA-binding, and subsequent transactivation in breast cancer cells; FLLL32 suppresses tumor growth in mouse xenografts [268]. Saikosaponin b2 (SSb2) is extracted from the root of Bupleurum plants and is traditionally used to treat inflammation [269]. SSb2 decreases pSTAT3 and suppresses proliferation and migration of breast cancer cells [270]. Additionally, Esculentoside A (EsA) is extracted from Phytolacca esculenta roots; the triterpene saponin suppresses cell proliferation and mammosphere formation of BCSCs as well as induces apoptosis [271]. Furthermore, treatment with EsA decreases tumor growth of mouse breast tumors in vivo [271].
Medicinal mushrooms can also exhibit pharmacological properties, including antitumor and immunotherapeutic effects [272,273]. Hericium erinaceus (H. erinaceus) water extract, a medicinal mushroom demonstrated to exhibit anticancer properties, inhibits cell viability, and induces apoptosis and cell cycle arrest in breast cancer cells [274]. Moreover, whole genome and transcriptome analysis revealed that the JAK-STAT pathway is significantly enriched in response to H. erinaceus treatment based on differential gene expression results [274]. Furthermore, Micotherapy U-care, which contains medicinal mushroom extracts from Agaricus blazei, Ophiocordyceps sinensis, Ganoderma lucidum, Grifola frondosa, and Lentinula edodes, has been shown to decrease pulmonary metastases and reduce NOS, COX2, and IL-6 expression in a TNBC mouse model [275]. Reduced IL-6 expression in metastatic and bronchial epithelial cells suggested that Micotherapy U-care could suppress STAT3 activation in TNBC.
There are over 60 STAT3 inhibitors published in breast cancer and more in clinical trials for the treatment of numerous cancers [276,277]. However, there are currently no FDA-approved STAT3 inhibitors for the treatment of breast cancer underscoring the clinical importance of these preclinical studies in breast cancer.
5.4. STAT4
The crystal structure of the NTD of multiple STATs and the specific characterization of the secondary structure of this NTD for STAT4 was discovered in 1998 [51]. STAT4 is not as commonly studied as other STAT proteins, which may be due to the fact that very few cytokines activate the STAT4-mediated immune pathways [23]. IL-12 is the main ligand that triggers JAK2-Tyk2 phosphorylation, STAT4 homodimerization, translocation to the nucleus, and subsequent modulation of STAT4-mediated genes [278,279]. IL-12 increases production of IFNɣ, differentiation of T helper cells, and facilitates innate and adaptive immunity in a mainly tumor-suppressive manner [279–282]. Though few cytokines trigger STAT4 pathways, STAT4 has the ability to bind multiple target gene promoter regions including MYD88, IFNɣ, TNF, IL18R1, Furin, and IL18RAP [280,283]. Additionally, STAT4-deficient mice have underdeveloped immunity against both parasitic and bacterial infections [278]. Though IL-12 is the master regulator of STAT4 activation, IL-23, IFNα, IL-2, IL-27, and IL-35 also trigger STAT4 signaling [284–286].
STAT4’s functions in breast cancer are not well understood. Immunohistochemical analysis of STAT4 expression reveals higher STAT4 expression in non-cancer breast tissues compared to the breast cancer tissue samples [287]. High STAT4 expression correlates with better overall survival and relapse-free survival in breast cancer patients [154]. High expression of STAT4 and IL12B1/2 also correlates with better survival in breast cancer patients [288]. Furthermore, pSTAT4 is upregulated by cryptotanshinone in CD4-positive T cells, which suppresses breast cancer cell growth in vivo [289]. STAT4 co-expressed genes are also enriched in adaptive immune responses, specifically T cell activation and signaling as well as cytokine regulation [290]. The role of STAT4 in breast cancer or specifically in the breast-immune microenvironment warrants further investigation. Given that there are limited studies and that most of breast cancer studies implicate STAT4 in a tumor suppressive role, there currently no published STAT4 inhibitors for breast cancer.
5.5. STAT5
STAT5 often refers to two proteins: STAT5a and STAT5b [291]. Originally described as mammary gland factor (MGF), STAT5 was later renamed as it shared amino acid sequence homology with STAT1 and STAT2 [292]. Further investigation reveals that there are two STAT5 proteins, STAT5a and STAT5b, as the proteins share 94% structural homology, but originate from separate genes [293–295]. Structurally, the CCD, DBD, LD, and SH2 domain remain conserved throughout the STAT family; however, the NTD and TAD exhibit the most diversity and allow for different signaling activities [295]. STAT5 proteins are mainly activated by the IL-2 family of cytokines, which are characterized by signaling through the IL-2 receptor γ and includes IL-2, IL-4, IL-7, IL-9, and IL-15 [296–299]. The IL-2 family and receptors are critical for normal immune cell development as well as mediate lymphocyte activation and expansion during immune responses [291]. Importantly, STAT5a and STAT5b dimers bind to similar core consensus sequences, suggesting these STAT5 isoforms can regulate many of the same pathways and genes [291]. Moving forward, “STAT5” will be used to describe both STAT5 isoforms unless otherwise specified.
In normal mammary development, STAT5 transcription factors modulate genes involved in mammary tissue differentiation, development of alveolar progenitors, and lactation associated with pregnancy [300–303]. Dysregulated activation of STAT5 results in over-aggressive alveolar development, impaired mammary gland remodeling, and mammary tumor formation through upregulating Akt/phosphatidylinositol 3-kinase (PI3K) signaling components [304]. Constitutively active STAT5 also significantly promotes mammary tumor formation in mice with loss of phosphatase and tensin homolog, an important tumor suppressor in the PI3K pathway [304]. STAT5a and Nmi are downregulated in 70% of metastatic breast tissues compared to primary tumors, suggesting that these two cooperatively regulate normal breast tissue and that dysregulation may contribute to breast cancer metastasis [305]. N-α-Acetyltransferase 10 protein (Naa10p) decreases migration and invasion of breast cancer cells [306]. Furthermore, Naa10p physically and functionally interacts with STAT5a and antagonizes STAT5a-JAK2 activity leading to suppression of breast cancer metastasis in vitro and in vivo [306].
Activation of STAT5 signaling has been reported to contribute to breast cancer. For example, 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase 4 (PFKFB4) promotes angiogenesis by upregulating IL-6 in breast cancer cells, which activates pSTAT5 in endothelial cells [307]. Pharmacological inhibition of PFKFB4 suppresses breast cancer tumor growth and decreases angiogenesis in mouse xenografts [307]. Fibroblast growth factor 2 (FGF2) activates STAT5, ERK, and AKT to increase breast cancer cell proliferation [308]. Additionally, breast cancer cells with constitutively active FGFR-2, the receptor for FGF that co-localizes in the nucleus with STAT5, promotes breast tumor growth in an orthotopic mouse model [308]. Signal-transducing adaptor protein-2 (STAP-2) promotes STAT5 activation through Brk phosphorylation of STAT5; knockdown of STAP-2 suppresses breast cancer cell proliferation [309]. In another study, parathyroid hormone-related protein overexpressed in mammary tissue promotes tumor growth in transgenic mice, in part, through the activation of STAT5 and subsequent downstream target genes [310]. FYN, a Src tyrosine kinase family member, promotes breast cancer cell migration and invasion through the activation of STAT5 and subsequent upregulation of Jagged-1 and Delta Like Canonical Notch Ligand 4 [300]. Consistent with in vitro findings, knockdown of FYN suppresses breast cancer lung metastases in mice [300]. ABL non-receptor tyrosine kinases upregulate pSTAT5 to promote STAT5-target gene-mediated osteolysis of breast cancer cells [311]. ABL kinase knockdown, and subsequent STAT5 downregulation, suppresses breast cancer bone metastasis in vivo [311].
Prolactin (PRL) signaling is implicated in breast cancer progression [312]. PRL-induced pSTAT5 directly upregulates heat shock protein-90α to promote breast cancer cell survival by evading apoptosis [313]. Further investigation into the mechanism by which PRL-induces STAT5 activation reveals that PRL promotes the dissociation of linker histone H1, which negatively regulates STAT5 signaling by limiting chromatin accessibility, thereby increasing breast cancer cell proliferation [314]. Moderate extracellular acidosis suppresses PRL-STAT5 signaling in breast cancer, but did not impact signaling triggered by other ligands, such as OSM [315]. Additionally, protein tyrosine phosphatase 1B (PTP1B) negatively regulates PRL-induced pSTAT5 by inhibiting JAK2 phosphorylation in breast cancer cells [316]. Given that PTP1B and pSTAT3 negatively correlate in breast cancer patient datasets [316], further investigation into PTP1B and additional modulators of the PRL-STAT5 signaling axis may inform breast cancer treatments.
On the other hand, STAT5 activation, not protein expression, is decreased or lost in breast cancers, especially those with lymph-node positive or metastasized breast cancers [317]. Moreover, PRL-STAT5 suppresses invasive phenotypes, such as migration, invasion, MMP secretion, and regulation of E-cadherin in breast cancer cells [318]. Stabilized and upregulated E-cadherin is recapitulated in breast cancer xenografts with STAT5 activation [318].
Breast cancer cells can modulate immune cells in the tumor microenvironment to create a pro-tumorigenic niche. In response, anti-tumor immune cells may activate pathways associated with tumor clearance. For example, STAT5 activation by breast cancer cells secreting GM-CSF promotes expression of immune-related genes and loss of STAT5 in macrophages promotes breast cancer cell migration in vitro and breast cancer lung metastasis in vivo [319].
STAT5a exhibits dual roles in luminal breast cancer depending on which tyrosine residue is phosphorylated [320]. Where phosphorylation at one residue decreases cell proliferation, the other suppresses clonogenicity, suggesting STAT5a may be able to switch phosphorylation-dependent phenotypes. Prognostic assessment of STAT mRNA expression reveals that high STAT5a and high STAT5b expression associates with better overall survival and relapse-free survival in breast cancer patients [154,155]. Interestingly, an endogenous dominant-negative STAT5 isoform, identified in luminal A breast cancer cells, inhibits transcriptional activity of STAT5a, STAT5b, and both estrogen receptors [321]. The dominant-negative STAT5 also induces apoptosis in some luminal breast cancer cells, suggesting the full mechanism of action of this STAT5 isoform has yet to be elucidated.
Despite the abundance of studies focused on STAT5 in breast cancer, there are limited studies describing the role of STAT5 in therapeutic resistance. STAT5a expression is increased in chemoresistant patient breast tumors compared to chemosensitive breast tumors [322]. STAT5a also upregulates ABCB1 expression, which mediates chemoresistance in breast cancer cells [323].
STAT3 and STAT5 are activated in 29% of breast tumors stained [324,325]. STAT3 and STAT5 are upregulated by STAP-2 in breast cancer cells and knockdown of STAP-2 or STAT5 decreases proliferation of breast cancer cells [197,309]. Interestingly, STAT3 and STAT5 can exhibit antagonistic functions in breast cancer. For example, LIF-mediated STAT3 activation induces apoptosis in mammary epithelial cells whereas STAT5 activation inhibits apoptosis [326,327]. STAT5, with STAT3 or alone, plays important roles in breast cancer emphasizing the importance of studying STAT5 inhibitors for the treatment of breast cancer.
Despite STAT5’s numerous roles in breast cancer, there are only a few STAT5 inhibitors published for the treatment of breast cancer (Table 1). A nicotinoyl hydrazine compound, CAS 285986–31–4, decreases pSTAT5 to reduce breast cancer cell proliferation [314]. Pimozide, an antipsychotic drug previously mentioned to target STAT3, inhibits cell proliferation and promotes cell cycle arrest and DNA damage in breast cancer cells [328,329]. Consistent with these findings, pimozide decreases pSTAT5 and ABCB1 and sensitizes doxorubicin-resistant luminal A breast cancer cells to doxorubicin treatment [322]. Interestingly, pharmacological inhibition of JAK2 and subsequent inactivation of STAT5 in combination with PI3K/mTOR inhibitors exhibits synergism. Combination treatment decreases cell proliferation and suppresses breast tumor growth and metastasis in an orthotopic mouse model [330]. This JAK2/STAT5 signaling pathway is induced by IL-8 suggesting that IL-8 mediated activation of JAK2/STAT5 signaling promotes breast cancer progression and metastasis and should be investigated further. Though few STAT5 inhibitors are reported in breast cancer, inhibition of STAT5-mediated pathways reveals promising results for breast cancer therapeutics.
5.6. STAT6
STAT6 was first discovered in B cells when researchers identified a transcription factor whose activation was upregulated by IL-4 [331, 332]. Further investigation reveals that IL-4 activation of STAT6 allows STAT6 to translocate to the nucleus and influence gene expression [333–335]. IL-13, which is structurally and functionally similar to IL-4 and shares the same receptor, also triggers canonical STAT6 signaling [332, 336, 337]. STAT6 activation promotes differentiation of macrophages and expansion and proliferation of B and T cells [332, 338, 339]. Though STAT6 is implicated in multiple cancer and disease types, the crystal structure was not solved until 2016, where researchers confirmed that only the activated STAT6 homodimer exhibits DNA-binding activity as opposed to the non-phosphorylated form [340].
STAT6 regulates G1/S cell cycle progression as STAT6 knockdown increases proliferation and decreases p21 and p27, G1 cyclin-dependent kinase (CDK) inhibitors, in breast cancer cells [341]. Furthermore, STAT6 interacts with transcription factor Sp1 to transcriptionally regulate p21 and p27 expression [341]. Interestingly, TNBC cells expressing low levels of STAT6 modulate expression of genes associated with decreased metastatic potential and apoptotic resistance as compared to the luminal A, STAT6-high expressing breast cancer cells [342]. STAT6 increases breast cancer cell resistance to apoptosis [342, 343]. STAT6-deficient mice challenged with 4T1 cells exhibit reduced primary breast tumor growth and decreased metastases, suggesting that loss of STAT6 may promote anti-tumor immunity in vivo [344].
Interestingly, reduction of IL-13 receptor subunit alpha 2 (IL13RA2) results in elevated pSTAT6 and suppresses migration of breast cancer cells [345]. Silencing IL13RA2 with IL-13 treatment increases expression of tumor protein 63, a tumor suppressor protein, and suppresses STAT6-mediated breast cancer lung metastasis [345]. Additionally, overexpression of IL-4 and subsequent STAT6 activation suppresses cell proliferation and increases apoptosis of breast cancer cells [346]. Suppression of STAT6 by miR-1207–5p decreases breast cancer cell proliferation and promotes cell cycle arrest [347]. Trastuzumab-resistant breast cancer cells express significantly decreased levels of STAT6 compared to trastuzumab-sensitive breast cancer cells [323]. Loss of STAT6 increases resistance to trastuzumab in HER2-positive breast cancer cells [348]. Moreover, STAT6 knockout modulates expression of genes associated with EMT, promotes mammosphere formation, and increases anchorage-independence of breast cancer cells [323,348]. In addition, loss of STAT6 transformed a non-tumorigenic breast cancer cell line to promote tumor formation in mice [348]. High STAT6 mRNA expression correlates with favorable overall survival relapse-free survival in breast cancer patients [154].
IL-4 and IL-13, STAT6 activators, also promote macrophage polarization [349,350]. In contrast to previous findings, STAT6 upregulates pro-tumorigenic, M2 phenotype associated genes [351,352], suggesting that inhibition of STAT6 may prevent TAM differentiation. M2 macrophages exhibit high levels of pSTAT6, while STAT6 knockdown or inhibition prevents M2 phenotype differentiation of mouse macrophages [353]. Furthermore, pharmacological inhibition of STAT6 suppresses tumor growth of 4T1 tumors and liver metastasis in mice [353]. Similarly, inhibition of both IL-4- and IL-13-induced pSTAT6 suppresses M2 macrophages in breast cancer [354]. Though STAT6 is not as widely studied in breast cancer compared to its other STAT family members, STAT6-mediated pathways could provide important therapeutic targets for breast cancer treatment.
Though STAT6 inhibitors are used in multiple cancer types, very few are published for breast cancer (Table 1) [254]. Notably, AS1517499, a potent STAT6 inhibitor originally synthesized to treat asthma and other allergic reaction-associated diseases [355,356], inhibits M2 macrophage polarization in mouse macrophages to suppress breast cancer tumor growth and liver metastasis [353]. To assess the clinical relevance of this STAT6 inhibitor in breast cancer, AS1517499 was used in a 4T1-orthotopic mouse model and found to suppress tumor growth and liver metastasis in vivo [353]. These findings suggest that STAT6-mediated signaling pathways may contain important therapeutic targets and warrant further investigation.
6. Clinical trials targeting STAT family members in breast cancer
All of the STAT protein family members are implicated in breast cancer, either in pro-tumorigenic or anti-cancer roles. As previously mentioned, STAT3 is the most commonly studied STAT protein in breast cancer and contributes to breast cancer by promoting breast cancer cell and tumor growth, metastasis, and immune evasion [43]. TTI-101, an orally available STAT3 inhibitor, is currently in Phase I clinical trials (NCT03195699) for patients with advanced cancers. These include breast cancer, head and neck squamous cell carcinoma, non-small cell lung cancer, hepatocellular cancer, colorectal cancer, gastric adenocarcinoma, and melanoma. This Phase I clinical trial, which started recruiting as early as 2017, aims to determine compound administration safety, patient response (if any), and will focus on pharmacodynamics by measuring pSTAT3, PD1 and PD-L1 expression. Though there are additional clinical trials also directly targeting STAT3, many do not include breast cancer patients or are no longer recruiting patients due to adverse effects.
Though inhibition of STAT proteins has been demonstrated to be effective in suppressing breast cancer in preclinical models, there are numerous challenges to targeting transcription factors in the clinic. Transcription factors are reported to be “intrinsically disordered”, which not only effects the structure of potential binding pockets, but also means that the rate, frequency, and affinity for protein-protein or protein-nucleic acid interactions is highly variable [13,357]. Therefore, to target STAT3, JAK1/2 inhibitors are currently being utilized in clinical trials instead [24,358]. Ruxolitinib is a JAK1/2 receptor tyrosine kinase inhibitor and is currently approved to treat patients with myelofibrosis or polycythemia [359–361]. Ruxolitinib was evaluated in a non-randomized phase II clinical trial for patients with metastatic TNBC (NCT01562873). Patients were enrolled on the basis of tumoral-pSTAT3 IHC staining, and while pharmacodynamic analysis of ruxolitinib indicates that the proportion of pSTAT3-positive cells decreased, the study was terminated as other objective responses were not met [359]. In a randomized, phase II study designed for patients with advanced HER2-negative breast cancer (NCT02120417), ruxolitinib or placebo was evaluated in combination with capecitabine, a fluorouracil prodrug that is approved for metastatic breast cancer [362, 363]. Patients were enrolled based on prior chemotherapy treatments for hormone receptor positivity, advanced, or metastatic disease and patient prognosis (overall survival and progression-free survival) was the main objective [363]. Though the overall response rate to the combination treatment was increased, the primary endpoint (overall survival) did not improve compared to capecitabine and placebo so the study was terminated. Combination of ruxolitinib and paclitaxel (NCT02041429), a chemotherapeutic approved to treat node-positive breast cancer, was evaluated in HER2-negative breast cancer patients and demonstrates promise as the combination treatment was well tolerated [364]. Similarly, the same combination was evaluated for TNBC patients with inflammatory Brca, though those results are still ongoing (NCT02041429, NCT02876302). A phase I/II clinical trial investigating the safety and efficacy of ruxolitinib in combination with trastuzumab (NCT02066532), a monoclonal antibody approved for the treatment of HER2-positive breast cancer, yielded less than successful results as there was no improvement in progression-free survival and hematologic-related adverse events were observed [365].
To improve upon current, FDA-approved treatments, ruxolitinib in combination with pembrolizumab, an anti-PD-1 immunotherapy, is being evaluated in a phase I study for patients with advanced or metastatic TNBC (NCT03012230). This study, which is still recruiting patients, aims to determine the safety and efficacy and examine PD-1, PD-L1, JAK2, and pSTAT3 tumoral expression. Interestingly, there is also a randomized phase II study evaluating ruxolitinib in the treatment of premalignant breast cancer cells with objectives focused on apoptosis and pSTAT5 levels (NCT02928978). Given that numerous clinical trials utilizing ruxolitinib alone or in combination have been terminated or completed, an open-label study for patients with breast cancer, colorectal cancer, pancreatic cancer or lung cancer is currently recruiting with the purpose of continuing ruxolitinib treatment alone or with a previously administered chemotherapy (NCT02955940). In summary, targeting STAT3 activity by directly or indirectly targeting STAT3 activity, such as JAK1/2 inhibitors, is a major focus for breast cancer treatment.
7. Conclusion
Since the discovery of the first STAT protein over 20 years ago, all of the STAT family members are considered mediators of multiple cellular processes leading to the progression or suppression of breast cancer. In this review, we discussed the STAT family of transcription factors, their many roles in breast cancer, and therapeutic strategies to target them for breast cancer treatment. Each STAT signaling pathway is activated by a set of cytokines or ligands and that dysregulation of the ligands, cell-surface receptors, STATs, negative regulators, or downstream target genes can lead to breast cancer. Further investigation reveals that understanding how one STAT member acts in a specific setting may not be enough to determine a patient’s outcome, but that focusing on the balance between two or more STATs is incredibly beneficial. Though most studies focus on the role of STATs within breast cancer cells, emphasis on STAT signaling in tumor microenvironmental cells and the immune cell populations is gaining more attention. Insight into the communication between breast cancer cells and immune cells or stromal cells has led to clinical trials utilizing combination therapies yielding promising results.
Despite these promising findings, many of the mechanisms reported to date are still incomplete as STAT signaling is clearly complex in breast cancer. Furthermore, breast cancer is an incredibly heterogeneous malignancy and current understanding of breast cancer subtypes are constantly being modified. A better understanding of STAT expression and activation in different subtypes of breast cancer is critical in developing more effective STAT inhibitors for breast cancer treatment. Even with hundreds of published articles describing STAT mechanisms and dozens of STAT3 inhibitors in breast cancer, there are still no FDA-approved STAT inhibitors for the treatment of breast cancer. Moreover, the limited number of STAT inhibitors in clinical trials further underscores the need to continue to study STAT proteins in breast cancer to advance treatment for breast cancer patients. Though individual STAT proteins correlate with different patient outcomes, it is clear that a better understanding of the balance between the STAT family members is critical for breast cancer diagnoses and treatments. Important future tasks may include 1) the development or manipulation of current diagnostic tools to detect multiple STAT proteins simultaneously, 2) development of reliable, predictive biomarkers to determine STAT activity, 3) utilization of multiple methods to stratify patients for treatment, and 4) counteract or resolve issues in current STAT-targeted or JAK1/2-targeted treatments that are responsible for adverse events.
Acknowledgments
This research was funded by NIH grants 1R01CA228137–01A1 (HWL), F31CA261027–01A1 (DD), P30CA012197 (WB), as well as, DoD grants, W81XWH-17–1–0044 (HWL), W81XWH-19–1–0072 (HWL), W81XWH-19–1–0753 (HWL), and W81XWH-20–1–0044 (HWL). The authors would like to thank the Carpenter Library at Wake Forest University School of Medicine for open-access literature support.
Abbreviations:
- BCSCs
breast cancer stem cells
- CCD
coiled-coil domain
- CDK
cyclin-dependent kinase
- CSC
cancer stem cell
- DBD
DNA-binding domain
- DMFS
distant metastasis-free survival
- DPF3
double PHD fingers 3
- EGCG
epigallocatechin gallate
- EGFR
epidermal growth factor receptor
- EMT
epithelial-to-mesenchymal transition
- ER
estrogen receptor
- EsA
Esculentoside A
- FAO
fatty acid beta-oxidation
- FGF2
fibroblast growth factor 2
- G-CSF
granulocyte-colony stimulating factor
- GPCRs
G-protein coupled receptors
- Grb2
growth factor receptor-binding protein 2
- HER2
human epidermal growth factor receptor 2
- HIF-1
hypoxia-inducible factor-1
- IFITM1
interferon-induced transmembrane protein 1
- IFN
interferon
- IL13RA2
interleukin 13 receptor subunit alpha 2
- IL-2
interleukin-2
- IRF9
interferon regulatory factor 9
- ISG15
ISG15 ubiquitin like modifier
- ISGF3
interferon-stimulated gene factor 3
- JAK
Janus kinase
- KLF14
Kruppel-like family of transcription factor-14
- LIF
leukemia inhibitory factor
- LIFR
leukemia inhibitory factor receptor
- MAFG-AS1
MAFG-antisense 1
- MDSC
myeloid-derived suppressor cell
- MGF
mammary gland factor
- Naa10p
N-α-Acetyltransferase 10 protein
- NLS
nuclear localization signal
- NMI
N-Myc and STAT interactor
- NTD
N-terminal domain
- OSM
oncostatin M
- PD-1
programmed cell death 1
- PD-L1
programmed death-ligand 1
- PFKFB4
6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase 4
- PI3K
phosphatidylinositol 3-kinase
- PR
progesterone receptor
- PRL
prolactin
- pSTAT
phosphorylated STAT
- PTP1B
protein tyrosine phosphatase 1B
- pY
tyrosine-phosphorylation site
- ROS
reactive oxygen species
- SHH
Sonic Hedgehog
- SIRT4
sirtuin 4
- SOCS
suppressors of cytokine signaling
- SSb2
saikosaponin b2
- STAP-2
signal-transducing adaptor protein 2
- STAT
signal transducer and activator of transcription
- TAD
transactivation domain
- TAMS
tumor-associated macrophages
- T-DM1
trastuzumab-emtansine
- TNBC
triple-negative breast cancer
- Trk
Tropomyosin receptor tyrosine kinases
- VEGF
vascular endothelial growth factor
- Wwox
WW domain-containing oxidoreductase
Footnotes
Conflict of interest
Authors declare no conflict of interest.
CRediT authorship contribution statement
Grace L. Wong: Conceptualized the manuscript, Selected the reviewed literature, Compiled the review tables and figures, Wrote the manuscript. Sara G. Manore: Contributed to manuscript editing. Daniel L. Doheny: Contributed to manuscript editing. Hui-Wen Lo: Supervised the design and writing of the manuscript, Edited the figures and manuscript, Obtained the funding for this publication.
Data availability
No data was used for the research described in the article.
References
- [1].Siegel RL, Miller KD, Fuchs HE, Jemal A, Cancer Stat. 72 (1) (2022) 7–33. [DOI] [PubMed] [Google Scholar]
- [2].Zhang T, Zhou H, Wang K, Wang X, Wang M, Zhao W, Xi X, Li Y, Cai M, Zhao W, Xu Y, Shao R, Role, molecular mechanism and the potential target of breast cancer stem cells in breast cancer development, Biomed. Pharmacother. = Biomed. Pharmacother. 147 (2022), 112616. [DOI] [PubMed] [Google Scholar]
- [3].Ghislain I, Zikos E, Coens C, Quinten C, Balta V, Tryfonidis K, Piccart M, Zardavas D, Nagele E, Bjelic-Radisic V, Cardoso F, Sprangers MAG, Velikova G, Bottomley A, Health-related quality of life in locally advanced and metastatic breast cancer: methodological and clinical issues in randomised controlled trials, Lancet Oncol. 17 (7) (2016) e294–e304. [DOI] [PubMed] [Google Scholar]
- [4].Chen MT, Sun HF, Zhao Y, Fu WY, Yang LP, Gao SP, Li LD, Jiang HL, Jin W, Comparison of patterns and prognosis among distant metastatic breast cancer patients by age groups: a SEER population-based analysis, Sci. Rep. 7 (1) (2017) 9254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wu Q, Li J, Zhu S, Wu J, Chen C, Liu Q, Wei W, Zhang Y, Sun S, Breast cancer subtypes predict the preferential site of distant metastases: a SEER based study, Oncotarget 8 (17) (2017) 27990–27996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF, Prospect. Identif. Tumor Breast Cancer Cells 100 (7) (2003) 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Batlle E, Clevers H, Cancer stem cells revisited, Nat. Med. 23 (10) (2017) 1124–1134. [DOI] [PubMed] [Google Scholar]
- [8].Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC, Wong H, Rosen J, Chang JC, Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy, J. Natl. Cancer Inst. 100 (9) (2008) 672–679. [DOI] [PubMed] [Google Scholar]
- [9].Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, Joshua B, Kaplan MJ, Wapnir I, Dirbas FM, Somlo G, Garberoglio C, Paz B, Shen J, Lau SK, Quake SR, Brown JM, Weissman IL, Clarke MF, Association of reactive oxygen species levels and radioresistance in cancer stem cells, Nature 458 (7239) (2009) 780–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT, The Human Transcription Factors, Cell 172 (4) (2018) 650–665. [DOI] [PubMed] [Google Scholar]
- [11].Vernimmen D, Bickmore WA, The hierarchy of transcriptional activation: from enhancer to promoter, Trends Genet: TIG 31 (12) (2015) 696–708. [DOI] [PubMed] [Google Scholar]
- [12].Lee TI, Young RA, Transcriptional regulation and its misregulation in disease, Cell 152 (6) (2013) 1237–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Henley MJ, Koehler AN, Advances in targeting ‘undruggable’ transcription factors with small molecules, Nat. Rev. Drug Discov. 20 (9) (2021) 669–688. [DOI] [PubMed] [Google Scholar]
- [14].Apostolou P, Toloudi M, Chatziioannou M, Ioannou E, Papasotiriou I, Cancer stem cells stemness transcription factors expression correlates with breast cancer disease stage, Curr. Stem Cell Res. Ther. 7 (6) (2012) 415–419. [DOI] [PubMed] [Google Scholar]
- [15].Lu X, Mazur SJ, Lin T, Appella E, Xu Y, The pluripotency factor nanog promotes breast cancer tumorigenesis and metastasis, Oncogene 33 (20) (2014) 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cho Y, Kang HG, Kim S-J, Lee S, Jee S, Ahn SG, Kang MJ, Song JS, Chung J-Y, Yi EC, Chun K-H, Post-translational modification of OCT4 in breast cancer tumorigenesis, Cell Death Differ. 25 (10) (2018) 1781–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu P, Tang H, Song C, Wang J, Chen B, Huang X, Pei X, Liu L, SOX2 Promotes Cell Proliferation and Metastasis in Triple Negative Breast Cancer, Front Pharm. 9 (2018), 942–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi MJ, Hernandez-Garcia S, Elorriaga K, Pandiella A, Rezola R, Martin AG, Sox2 expression in breast tumours and activation in breast cancer stem cells, Oncogene 31 (11) (2012) 1354–1365. [DOI] [PubMed] [Google Scholar]
- [19].Darnell JE, Transcription factors as targets for cancer therapy, Nat. Rev. Cancer 2 (10) (2002) 740–749. [DOI] [PubMed] [Google Scholar]
- [20].Sadowski HB, Shuai K, Darnell JE Jr., Gilman MZ, A common nuclear signal transduction pathway activated by growth factor and cytokine receptors, Sci. (N. Y., N. Y. ) 261 (5129) (1993) 1739–1744. [DOI] [PubMed] [Google Scholar]
- [21].Darnell JE Jr., Kerr IM, Stark GR, Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins, Sci. (N. Y., N. Y. ) 264 (5164) (1994) 1415–1421. [DOI] [PubMed] [Google Scholar]
- [22].Wegenka UM, Lütticken C, Buschmann J, Yuan J, Lottspeich F, Müller-Esterl W, Schindler C, Roeb E, Heinrich PC, Horn F, The interleukin-6-activated acute-phase response factor is antigenically and functionally related to members of the signal transducer and activator of transcription (STAT) family, Mol. Cell. Biol. 14 (5) (1994) 3186–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Verhoeven Y, Tilborghs S, Jacobs J, De Waele J, Quatannens D, Deben C, Prenen H, Pauwels P, Trinh XB, Wouters A, Smits ELJ, Lardon F, van Dam PA, The potential and controversy of targeting STAT family members in cancer, Semin. Cancer Biol. 60 (2020) 41–56. [DOI] [PubMed] [Google Scholar]
- [24].Manore SG, Doheny DL, Wong GL, Lo HW, IL-6/JAK/STAT3 signaling in breast cancer metastasis: biology and treatment, Front. Oncol. 12 (2022), 866014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chung SS, Giehl N, Wu Y, Vadgama JV, STAT3 activation in HER2-overexpressing breast cancer promotes epithelial-mesenchymal transition and cancer stem cell traits, Int. J. Oncol. 44 (2) (2014) 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, Ruddy K, Tsang J, Cardoso F, Breast cancer, Nat. Rev. Dis. Prim. 5 (1) (2019) 66. [DOI] [PubMed] [Google Scholar]
- [27].Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge Ø, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale A-L, Brown PO, Botstein D, Molecular portraits of human breast tumours, Nature 406 (6797) (2000) 747–752. [DOI] [PubMed] [Google Scholar]
- [28].Cheang MC, Martin M, Nielsen TO, Prat A, Voduc D, Rodriguez-Lescure A, Ruiz A, Chia S, Shepherd L, Ruiz-Borrego M, Calvo L, Alba E, Carrasco E, Caballero R, Tu D, Pritchard KI, Levine MN, Bramwell VH, Parker J, Bernard PS, Ellis MJ, Perou CM, Di Leo A, Carey LA, Defining breast cancer intrinsic subtypes by quantitative receptor expression, oncologist 20 (5) (2015) 474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Oh D-Y, Bang Y-J, HER2-targeted therapies — a role beyond breast cancer, Nat. Rev. Clin. Oncol. 17 (1) (2020) 33–48. [DOI] [PubMed] [Google Scholar]
- [30].Wong GL, Abu Jalboush S, Lo H-W, Exosomal Micro Organo Breast Cancer Metastas-.−. 12 (7) (2020) 1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li Y, Zhan Z, Yin X, Fu S, Deng X, Targeted therapeutic strategies for triple-negative breast cancer, Front. Oncol. 11 (2021), 731535–731535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yin L, Duan J-J, Bian X-W, Yu S-C, Triple-negative breast cancer molecular subtyping and treatment progress, Breast Cancer Res. 22 (1) (2020) 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Clevers H, The cancer stem cell: premises, promises and challenges, Nat. Med 17 (3) (2011) 313–319. [DOI] [PubMed] [Google Scholar]
- [34].Furth J, Kahn MC, Breedis C, The Transmission of Leukemia of Mice with a Single Cell1, Am. J. Cancer 31 (2) (1937) 276–282. [Google Scholar]
- [35].Kleinsmith LJ, Pierce GB Jr., Multipotentiality of single embryonal carcinoma cells, Cancer Res. 24 (1964) 1544–1551. [PubMed] [Google Scholar]
- [36].Pierce GB, Speers WC, Tumors as caricatures of the process of tissue renewal: prospects for therapy by directing differentiation, Cancer Res. 48 (8) (1988) 1996–2004. [PubMed] [Google Scholar]
- [37].Uckun FM, Sather H, Reaman G, Shuster J, Land V, Trigg M, Gunther R, Chelstrom L, Bleyer A, Gaynon P, et al. , Leukemic cell growth in SCID mice as a predictor of relapse in high-risk B-lineage acute lymphoblastic leukemia, Blood 85 (4) (1995) 873–878. [PubMed] [Google Scholar]
- [38].Wang C, Xu K, Wang R, Han X, Tang J, Guan X, Heterogeneity of BCSCs contributes to the metastatic organotropism of breast cancer, J. Exp. Clin. Cancer Res. 40 (1) (2021) 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Varmus HE, Oncogenes and transcriptional, Control 238 (4832) (1987) 1337–1339. [DOI] [PubMed] [Google Scholar]
- [40].Lis JT, A 50 year history of technologies that drove discovery in eukaryotic transcription regulation, Nat. Struct. Mol. Biol. 26 (9) (2019) 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bushweller JH, Targeting transcription factors in cancer — from undruggable to reality, Nat. Rev. Cancer 19 (11) (2019) 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fu XY, Kessler DS, Veals SA, Levy DE, Darnell JE Jr., ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains, Proc. Natl. Acad. Sci. USA 87 (21) (1990) 8555–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ma JH, Qin L, Li X, Role of STAT3 signaling pathway in breast cancer, Cell Commun. Signal.: CCS 18 (1) (2020) 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Darnell JE Jr., STATs and gene regulation, Sci. (N. Y., N. Y. ) 277 (5332) (1997) 1630–1635. [DOI] [PubMed] [Google Scholar]
- [45].Clevenger CV, Roles and regulation of stat family transcription factors in human breast cancer, Am. J. Pathol. 165 (5) (2004) 1449–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lim CP, Cao X, Structure, function, and regulation of STAT proteins, Mol. Biosyst. 2 (11) (2006) 536–550. [DOI] [PubMed] [Google Scholar]
- [47].Awasthi N, Liongue C, Ward AC, STAT proteins: a kaleidoscope of canonical and non-canonical functions in immunity and cancer, J. Hematol. Oncol. 14 (1) (2021) 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Copeland NG, Gilbert DJ, Schindler C, Zhong Z, Wen Z, Darnell JE, Mui ALF, Miyajima A, Quelle FW, Ihle JN, Jenkins NA, Distribution of the Mammalian Stat Gene Family in Mouse Chromosomes, Genomics 29 (1) (1995) 225–228. [DOI] [PubMed] [Google Scholar]
- [49].Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW, Signaling through the JAK/STAT pathway, recent advances and future challenges, Gene 285 (1) (2002) 1–24. [DOI] [PubMed] [Google Scholar]
- [50].Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE, Kuriyan J, Crystal Structure of a Tyrosine Phosphorylated STAT-1 Dimer Bound to DNA, Cell 93 (5) (1998) 827–839. [DOI] [PubMed] [Google Scholar]
- [51].Vinkemeier U, Moarefi I, Darnell JE, Kuriyan J, Structure of the Amino-Terminal Protein Interaction Domain of STAT-4, Sci. (N. Y., N. Y. ) 279 (5353) (1998) 1048–1052. [DOI] [PubMed] [Google Scholar]
- [52].Becker S, Groner B, Müller CW, Three-dimensional structure of the Stat3β homodimer bound to DNA, Nature 394 (6689) (1998) 145–151. [DOI] [PubMed] [Google Scholar]
- [53].Hu X, Li J, Fu M, Zhao X, Wang W, The JAK/STAT signaling pathway: from bench to clinic, Signal Transduct. Target. Ther. 6 (1) (2021) 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Strehlow I, Schindler C, Amino-terminal signal transducer and activator of transcription (STAT) domains regulate nuclear translocation and STAT deactivation, J. Biol. Chem. 273 (43) (1998) 28049–28056. [DOI] [PubMed] [Google Scholar]
- [55].Zhu M, John S, Berg M, Leonard WJ, Functional association of Nmi with Stat5 and Stat1 in IL-2- and IFNgamma-mediated signaling, Cell 96 (1) (1999) 121–130. [DOI] [PubMed] [Google Scholar]
- [56].Yang E, Wen Z, Haspel RL, Zhang JJ, Darnell JE Jr., The linker domain of Stat1 is required for gamma interferon-driven transcription, Mol. Cell. Biol. 19 (7) (1999) 5106–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Nakajima H, Brindle PK, Handa M, Ihle JN, Functional interaction of STAT5 and nuclear receptor co-repressor SMRT: implications in negative regulation of STAT5-dependent transcription, EMBO J. 20 (23) (2001) 6836–6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Brown S, Hu N, Hombría JC, Novel level of signalling control in the JAK/STAT pathway revealed by in situ visualisation of protein-protein interaction during Drosophila development, Dev. (Camb., Engl. ) 130 (14) (2003) 3077–3084. [DOI] [PubMed] [Google Scholar]
- [59].Ma J, Zhang T, Novotny-Diermayr V, Tan AL, Cao X, A novel sequence in the coiled-coil domain of Stat3 essential for its nuclear translocation, J. Biol. Chem. 278 (31) (2003) 29252–29260. [DOI] [PubMed] [Google Scholar]
- [60].Moran MF, Koch CA, Anderson D, Ellis C, England L, Martin GS, Pawson T, Src homology region 2 domains direct protein-protein interactions in signal transduction, Proc. Natl. Acad. Sci. USA 87 (21) (1990) 8622–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hidaka M, Homma Y, Takenawa T, Highly conserved eight amino acid sequence in SH2 is important for recognition of phosphotyrosine site, Biochem. Biophys. Res. Commun. 180 (3) (1991) 1490–1497. [DOI] [PubMed] [Google Scholar]
- [62].Lu W, Gong D, Bar-Sagi D, Cole PA, Site-specific incorporation of a phosphotyrosine mimetic reveals a role for tyrosine phosphorylation of SHP-2 in cell signaling, Mol. Cell 8 (4) (2001) 759–769. [DOI] [PubMed] [Google Scholar]
- [63].Shuai K, Stark GR, Kerr IM, Darnell JE Jr., A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma, Sci. (N. Y., N. Y. ) 261 (5129) (1993) 1744–1746. [DOI] [PubMed] [Google Scholar]
- [64].Fu XY, A transcription factor with SH2 and SH3 domains is directly activated by an interferon alpha-induced cytoplasmic protein tyrosine kinase(s), Cell 70 (2) (1992) 323–335. [DOI] [PubMed] [Google Scholar]
- [65].Schindler C, Darnell JE Jr., Transcriptional responses to polypeptide ligands: the JAK-STAT pathway, Annu. Rev. Biochem. 64 (1995) 621–651. [DOI] [PubMed] [Google Scholar]
- [66].Parrini M, Meissl K, Ola MJ, Lederer T, Puga A, Wienerroither S, Kovarik P, Decker T, Müller M, Strobl B, The C-Terminal Transactivation Domain of STAT1 Has a Gene-Specific Role in Transactivation and Cofactor Recruitment, Front. Immunol. 9 (2018) 2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Moriggl R, Berchtold S, Friedrich K, Standke GJ, Kammer W, Heim M, Wissler M, Stӧcklin E, Gouilleux F, Groner B, Comparison of the transactivation domains of Stat5 and Stat6 in lymphoid cells and mammary epithelial cells, Mol. Cell. Biol. 17 (7) (1997) 3663–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston DM, Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha, Nature 383 (6598) (1996) 344–347. [DOI] [PubMed] [Google Scholar]
- [69].Huang M, Qian F, Hu Y, Ang C, Li Z, Wen Z, Chromatin-remodelling factor BRG1 selectively activates a subset of interferon-alpha-inducible genes, Nat. Cell Biol. 4 (10) (2002) 774–781. [DOI] [PubMed] [Google Scholar]
- [70].Razeto A, Ramakrishnan V, Litterst CM, Giller K, Griesinger C, Carlomagno T, Lakomek N, Heimburg T, Lodrini M, Pfitzner E, Becker S, Structure of the NCoA-1/SRC-1 PAS-B Domain Bound to the LXXLL Motif of the STAT6 Transactivation Domain, J. Mol. Biol. 336 (2) (2004) 319–329. [DOI] [PubMed] [Google Scholar]
- [71].Babon JJ, Lucet IS, Murphy JM, Nicola NA, Varghese LN, The molecular regulation of Janus kinase (JAK) activation, Biochem. J. 462 (1) (2014) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Liu KD, Gaffen SL, Goldsmith MA, JAK/STAT signaling by cytokine receptors, Curr. Opin. Immunol. 10 (3) (1998) 271–278. [DOI] [PubMed] [Google Scholar]
- [73].Reich NC, STATs get their move on, Jak. -Stat. 2 (4) (2013), e27080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kӧster M, Hauser H, Dynamic redistribution of STAT1 protein in IFN signaling visualized by GFP fusion proteins, Eur. J. Biochem. 260 (1) (1999) 137–144. [DOI] [PubMed] [Google Scholar]
- [75].Kawashima T, Bao YC, Nomura Y, Moon Y, Tonozuka Y, Minoshima Y, Hatori T, Tsuchiya A, Kiyono M, Nosaka T, Nakajima H, Williams DA, Kitamura T, Rac1 and a GTPase-activating protein, MgcRacGAP, are required for nuclear translocation of STAT transcription factors, J. Cell Biol. 175 (6) (2006) 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wang Y, van Boxel-Dezaire AH, Cheon H, Yang J, Stark GR, STAT3 activation in response to IL-6 is prolonged by the binding of IL-6 receptor to EGF receptor, Proc. Natl. Acad. Sci. USA 110 (42) (2013) 16975–16980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Meyer T, Vinkemeier U, STAT nuclear translocation: potential for pharmacological intervention, Expert Opin. Ther. Targets 11 (10) (2007) 1355–1365. [DOI] [PubMed] [Google Scholar]
- [78].Banninger G, Reich NC, STAT2 nuclear trafficking, J. Biol. Chem. 279 (38) (2004) 39199–39206. [DOI] [PubMed] [Google Scholar]
- [79].McBride KM, Reich NC, The ins and outs of STAT1 nuclear transport, Sci. ‘S. STKE: Signal Transduct. Knowl. Environ. 2003 (195) (2003) Re13. [DOI] [PubMed] [Google Scholar]
- [80].Levy DE, Darnell JE, STATs: transcriptional control and biological impact, Nat. Rev. Mol. Cell Biol. 3 (9) (2002) 651–662. [DOI] [PubMed] [Google Scholar]
- [81].Aaronson DS, Horvath CM, A road map for those who don’t know JAK-STAT, Sci. (N. Y., N. Y. ) 296 (5573) (2002) 1653–1655. [DOI] [PubMed] [Google Scholar]
- [82].Decker T, Kovarik P, Serine phosphorylation of STATs, Oncogene 19 (21) (2000) 2628–2637. [DOI] [PubMed] [Google Scholar]
- [83].Braunstein J, Brutsaert S, Olson R, Schindler C, STATs Dimerize in the Absence of Phosphorylation*, J. Biol. Chem. 278 (36) (2003) 34133–34140. [DOI] [PubMed] [Google Scholar]
- [84].Haan S, Kortylewski M, Behrmann I, Müller-Esterl W, Heinrich PC, Schaper F, Cytoplasmic STAT proteins associate prior to activation, The, Biochem. J. 345 Pt 3 (Pt 3) (2000) 417–421. [PMC free article] [PubMed] [Google Scholar]
- [85].Novak U, Ji H, Kanagasundaram V, Simpson R, Paradiso L, STAT3 forms stable homodimers in the presence of divalent cations prior to activation, Biochem. Biophys. Res. Commun. 247 (3) (1998) 558–563. [DOI] [PubMed] [Google Scholar]
- [86].Stancato LF, David M, Carter-Su C, Larner AC, Pratt WB, Preassociation of STAT1 with STAT2 and STAT3 in Separate Signalling Complexes Prior to Cytokine Stimulation (*), J. Biol. Chem. 271 (8) (1996) 4134–4137. [DOI] [PubMed] [Google Scholar]
- [87].Ndubuisi MI, Guo GG, Fried VA, Etlinger JD, Sehgal PB, Cellular physiology of STAT3: Where’s the cytoplasmic monomer? J. Biol. Chem. 274 (36) (1999) 25499–25509. [DOI] [PubMed] [Google Scholar]
- [88].Cheon H, Stark GR, Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes, Proc. Natl. Acad. Sci. USA 106 (23) (2009) 9373–9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].John S, Vinkemeier U, Soldaini E, Darnell JE Jr., W.J. Leonard, The significance of tetramerization in promoter recruitment by Stat5, Mol. Cell. Biol. 19 (3) (1999) 1910–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Lin JX, Li P, Liu D, Jin HT, He J, Ata Ur Rasheed M, Rochman Y, Wang L, Cui K, Liu C, Kelsall BL, Ahmed R, Leonard WJ, Critical Role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function, Immunity 36 (4) (2012) 586–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Brooks AJ, Dai W, O’Mara ML, Abankwa D, Chhabra Y, Pelekanos RA, Gardon O, Tunny KA, Blucher KM, Morton CJ, Parker MW, Sierecki E, Gambin Y, Gomez GA, Alexandrov K, Wilson IA, Doxastakis M, Mark AE, Waters MJ, Mech. Act. Protein Kinase JAK2 Growth Horm. Recept. 344 (6185) (2014) 1249783. [DOI] [PubMed] [Google Scholar]
- [92].Park OK, Schaefer TS, Nathans D, In vitro activation of Stat3 by epidermal growth factor receptor kinase, 93(24), 1996: 13704–13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Owen KL, Brockwell NK, Parker BS, JAK-STAT Signaling: A Double-Edged Sword of Immune Regulation and Cancer Progression, 11(12), 2019: 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola NA, Hilton DJ, A family of cytokine-inducible inhibitors of signalling, Nature 387 (6636) (1997) 917–921. [DOI] [PubMed] [Google Scholar]
- [95].Santillán-Benítez JG, Mendieta-Zerón H, Gómez-Oliván LM, Ordóñez Quiroz A, Torres-Juárez JJ, González-Bañales JM, JAK2, STAT3 and SOCS3 gene expression in women with and without breast cancer, Gene 547 (1) (2014) 70–76. [DOI] [PubMed] [Google Scholar]
- [96].Barclay JL, Anderson ST, Waters MJ, Curlewis JD, SOCS3 as a tumor suppressor in breast cancer cells, and its regulation by PRL, Int. J. Cancer 124 (8) (2009) 1756–1766. [DOI] [PubMed] [Google Scholar]
- [97].Yen MC, Shih YC, Hsu YL, Lin ES, Lin YS, Tsai EM, Ho YW, Hou MF, Kuo PL, Isolinderalactone enhances the inhibition of SOCS3 on STAT3 activity by decreasing miR-30c in breast cancer, Oncol. Rep. 35 (3) (2016) 1356–1364. [DOI] [PubMed] [Google Scholar]
- [98].Evans MK, Yu CR, Lohani A, Mahdi RM, Liu X, Trzeciak AR, Egwuagu CE, Expression of SOCS1 and SOCS3 genes is differentially regulated in breast cancer cells in response to proinflammatory cytokine and growth factor signals, Oncogene 26 (13) (2007) 1941–1948. [DOI] [PubMed] [Google Scholar]
- [99].Villarino AV, Kanno Y, O’Shea JJ, Mechanisms and consequences of Jak-STAT signaling in the immune system, Nat. Immunol. 18 (4) (2017) 374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ramana CV, Chatterjee-Kishore M, Nguyen H, Stark GR, Complex roles of Stat1 in regulating gene expression, Oncogene 19 (21) (2000) 2619–2627. [DOI] [PubMed] [Google Scholar]
- [101].Schindler C, Levy DE, Decker T, JAK-STAT signaling: from interferons to cytokines, J. Biol. Chem. 282 (28) (2007) 20059–20063. [DOI] [PubMed] [Google Scholar]
- [102].Chan SR, Vermi W, Luo J, Lucini L, Rickert C, Fowler AM, Lonardi S, Arthur C, Young LJT, Levy DE, Welch MJ, Cardiff RD, Schreiber RD, STAT1-deficient mice spontaneously develop estrogen receptor α-positive luminal mammary carcinomas, Breast Cancer Res. 14 (1) (2012) R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Zhang Y, Molavi O, Su M, Lai R, The clinical and biological significance of STAT1 in esophageal squamous cell carcinoma, BMC Cancer 14 (1) (2014) 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Widschwendter A, Tonko-Geymayer S, Welte T, Daxenbichler G, Marth C, Doppler W, Prognostic significance of signal transducer and activator of transcription 1 activation in breast cancer, Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 8 (10) (2002) 3065–3074. [PubMed] [Google Scholar]
- [105].Yau C, Esserman L, Moore DH, Waldman F, Sninsky J, Benz CC, A multigene predictor of metastatic outcome in early stage hormone receptor-negative and triple-negative breast cancer, Breast Cancer Res.: BCR 12 (5) (2010) R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Legrier M-E, Bièche I, Gaston J, Beurdeley A, Yvonnet V, Déas O, Thuleau A, Château-Joubert S, Servely J-L, Vacher S, Lassalle M, Depil S, Tucker GC, Fontaine J-J, Poupon M-F, Roman-Roman S, Judde J-G, Decaudin D, Cairo S, Marangoni E, Activation of IFN/STAT1 signalling predicts response to chemotherapy in oestrogen receptor-negative breast cancer, Br. J. Cancer 114 (2) (2016) 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Goodman ML, Trinca GM, Walter KR, Papachristou EK, D’Santos CS, Li T, Liu Q, Lai Z, Chalise P, Madan R, Fan F, Markiewicz MA, Jin VX, Carroll JS, Hagan CR, Progesterone Receptor Attenuates STAT1-Mediated IFN Signaling in Breast Cancer, J. Immunol. (Baltim., Md.: 1950) 202 (10) (2019) 3076–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Hou Y, Li X, Li Q, Xu J, Yang H, Xue M, Niu G, Zhuo S, Mu K, Wu G, Li X, Wang H, Zhu J, Zhuang T, STAT1 facilitates oestrogen receptor α transcription and stimulates breast cancer cell proliferation, 22(12), 2018: 6077–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Koromilas AE, Sexl V, The tumor suppressor function of STAT1 in breast cancer, Jak. -Stat. 2 (2) (2013), e23353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Raven JF, Williams V, Wang S, Tremblay ML, Muller WJ, Durbin JE, Koromilas AE, Stat1 is a suppressor of ErbB2/Neu-mediated cellular transformation and mouse mammary gland tumor formation, Cell Cycle (Georget., Tex. ) 10 (5) (2011) 794–804. [DOI] [PubMed] [Google Scholar]
- [111].Klover PJ, Muller WJ, Robinson GW, Pfeiffer RM, Yamaji D, Hennighausen L, Loss of STAT1 from mouse mammary epithelium results in an increased Neu-induced tumor burden, Neoplasia (N. Y., N. Y. ) 12 (11) (2010) 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Schneckenleithner C, Bago-Horvath Z, Dolznig H, Neugebauer N, Kollmann K, Kolbe T, Decker T, Kerjaschki D, Wagner KU, Müller M, Stoiber D, Sexl V, Putting the brakes on mammary tumorigenesis: loss of STAT1 predisposes to intraepithelial neoplasias, Oncotarget 2 (12) (2011) 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Mori H, Chen JQ, Cardiff RD, Pénzváltó Z, Hubbard NE, Schuetter L, Hovey RC, Trott JF, Borowsky AD, Pathobiology of the 129:Stat1−/− mouse model of human age-related ER-positive breast cancer with an immune infiltrate-excluded phenotype, Breast Cancer Res. 19 (1) (2017) 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Chan SR, Rickert CG, Vermi W, Sheehan KC, Arthur C, Allen JA, White JM, Archambault J, Lonardi S, McDevitt TM, Bhattacharya D, Lorenzi MV, Allred DC, Schreiber RD, Dysregulated STAT1-SOCS1 control of JAK2 promotes mammary luminal progenitor cell survival and drives ERα(+) tumorigenesis, Cell death Differ. 21 (2) (2014) 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Totten SP, Im YK, Cepeda Cañedo E, Najyb O, Nguyen A, Hébert S, Ahn R, Lewis K, Lebeau B, La Selva R, Sabourin V, Martínez C, Savage P, Kuasne H, Avizonis D, Santos Martínez N, Chabot C, Aguilar-Mahecha A, Goulet M-L, Dankner M, Witcher M, Petrecca K, Basik M, Pollak M, Topisirovic I, Lin R, Siegel PM, Kleinman CL, Park M, St-Pierre J, Ursini-Siegel J, STAT1 potentiates oxidative stress revealing a targetable vulnerability that increases phenformin efficacy in breast cancer, Nat. Commun. 12 (1) (2021) 3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Hix LM, Karavitis J, Khan MW, Shi YH, Khazaie K, Zhang M, Tumor STAT1 transcription factor activity enhances breast tumor growth and immune suppression mediated by myeloid-derived suppressor cells, J. Biol. Chem. 288 (17) (2013) 11676–11688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Qadir AS, Stults AM, Murmann AE, Peter ME, The mechanism of how CD95/Fas activates the Type I IFN/STAT1 axis, driving cancer stemness in breast cancer, Sci. Rep. 10 (1) (2020) 1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Li W, Yi J, Zheng X, Liu S, Fu W, Ren L, Li L, Hoon DSB, Wang J, Du G, miR-29c plays a suppressive role in breast cancer by targeting the TIMP3/STAT1/FOXO1 pathway, Clin. Epigenet. 10 (1) (2018) 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Huang P, Liao R, Chen X, Wu X, Li X, Wang Y, Cao Q, Dong C, Nuclear translocation of PLSCR1 activates STAT1 signaling in basal-like breast cancer, Theranostics 10 (10) (2020) 4644–4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Zellmer VR, Schnepp PM, Fracci SL, Tan X, Howe EN, Zhang S, Tumor-induced Stromal STAT1 Accelerates Breast Cancer via Deregulating Tissue Homeostasis, Mol. Cancer Res.: MCR 15 (5) (2017) 585–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Escher TE, Dandawate P, Sayed A, Hagan CR, Anant S, Lewis-Wambi J, Enhanced IFNα Signaling Promotes Ligand-Independent Activation of ERα to Promote Aromatase Inhibitor Resistance in Breast Cancer, Cancers 13 (20) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Khodarev N, Ahmad R, Rajabi H, Pitroda S, Kufe T, McClary C, Joshi MD, MacDermed D, Weichselbaum R, Kufe D, Cooperativity of the MUC1 oncoprotein and STAT1 pathway in poor prognosis human breast cancer, Oncogene 29 (6) (2010) 920–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Suyama K, Onishi H, Imaizumi A, Shinkai K, Umebayashi M, Kubo M, Mizuuchi Y, Oda Y, Tanaka M, Nakamura M, Katano M, CD24 suppresses malignant phenotype by downregulation of SHH transcription through STAT1 inhibition in breast cancer cells, Cancer Lett. 374 (1) (2016) 44–53. [DOI] [PubMed] [Google Scholar]
- [124].Hii L-W, Chung FF-L, Mai CW, Yee ZY, Chan HH, Raja VJ, Dephoure NE, Pyne NJ, Pyne S, Leong C-O, Sphingosine Kinase 1 Regulates the Survival of Breast Cancer Stem Cells and Non-stem Breast Cancer Cells by Suppression of STAT1, 9(4), 2020: 886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Zhan JF, Chen LH, Yuan YW, Xie GZ, Sun AM, Liu Y, Chen ZX, STAT1 promotes radioresistance of CD44(+)/CD24(-/low) cells in breast cancer, Exp. Biol. Med. (Maywood, N. J. ) 236 (4) (2011) 418–422. [DOI] [PubMed] [Google Scholar]
- [126].Bonuccelli G, Castello-Cros R, Capozza F, Martinez-Outschoorn UE, Lin Z, Tsirigos A, Xuanmao J, Whitaker-Menezes D, Howell A, Lisanti MP, Sotgia F, The milk protein α-casein functions as a tumor suppressor via activation of STAT1 signaling, effectively preventing breast cancer tumor growth and metastasis, Cell Cycle (Georget., Tex. ) 11 (21) (2012) 3972–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Han W, Carpenter RL, Cao X, Lo H-W, STAT1 gene expression is enhanced by nuclear EGFR and HER2 via cooperation With STAT3, 52(12), 2013: 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Ahn R, Sabourin V, Bolt AM, Hébert S, Totten S, De Jay N, Festa MC, Young YK, Im YK, Pawson T, Koromilas AE, Muller WJ, Mann KK, Kleinman CL, Ursini-Siegel J, The Shc1 adaptor simultaneously balances Stat1 and Stat3 activity to promote breast cancer immune suppression, Nat. Commun. 8 (1) (2017) 14638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Lo HW, Xia W, Wei Y, Ali-Seyed M, Huang SF, Hung MC, Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer, Cancer Res. 65 (1) (2005) 338–348. [PubMed] [Google Scholar]
- [130].Darvin P, Joung YH, Kang DY, Sp N, Byun HJ, Hwang TS, Sasidharakurup H, Lee CH, Cho KH, Park KD, Lee HK, Yang YM, Tann. Acid. Inhib. EGFR/STAT1/3 Enhanc. p38/STAT1 Signal. axis Breast Cancer Cells 21 (4) (2017) 720–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Huang R, Faratian D, Sims AH, Wilson D, Thomas JS, Harrison DJ, Langdon SP, Increased STAT1 signaling in endocrine-resistant breast cancer, PloS One 9 (4) (2014), e94226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Gujam FJ, McMillan DC, Edwards J, The relationship between total and phosphorylated STAT1 and STAT3 tumour cell expression, components of tumour microenvironment and survival in patients with invasive ductal breast cancer, Oncotarget 7 (47) (2016) 77607–77621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Yi M, Niu M, Xu L, Luo S, Wu K, Regulation of PD-L1 expression in the tumor microenvironment, J. Hematol. Oncol. 14 (1) (2021) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Han Y, Liu D, Li L, PD-1/PD-L1 pathway: current researches in cancer, Am. J. Cancer Res. 10 (3) (2020) 727–742. [PMC free article] [PubMed] [Google Scholar]
- [135].Chen M, Pockaj B, Andreozzi M, Barrett MT, Krishna S, Eaton S, Niu R, Anderson KS, JAK2 and PD-L1 Amplification Enhance the Dynamic Expression of PD-L1 in Triple-negative Breast Cancer, Clin. Breast Cancer 18 (5) (2018) e1205–e1215. [DOI] [PubMed] [Google Scholar]
- [136].Gilad Y, Eliaz Y, Yu Y, Han SJ, O’Malley BW, Lonard DM, Drug-induced PD-L1 expression and cell stress response in breast cancer cells can be balanced by drug combination, Sci. Rep. 9 (1) (2019) 15099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Nakayama Y, Mimura K, Tamaki T, Shiraishi K, Kua LF, Koh V, Ohmori M, Kimura A, Inoue S, Okayama H, Suzuki Y, Nakazawa T, Ichikawa D, Kono K, Phospho-STAT1 expression as a potential biomarker for anti-PD-1/anti-PD-L1 immunotherapy for breast cancer, Int. J. Oncol. 54 (6) (2019) 2030–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Darvin P, Sasidharan Nair V, Elkord E, PD-L1 Expression in Human Breast Cancer Stem Cells Is Epigenetically Regulated through Posttranslational Histone Modifications, J. Oncol. 2019 (2019) 3958908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Sasidharan Nair V, Toor SM, Ali BR, Elkord E, Dual inhibition of STAT1 and STAT3 activation downregulates expression of PD-L1 in human breast cancer cells, Expert Opin. Ther. Targets 22 (6) (2018) 547–557. [DOI] [PubMed] [Google Scholar]
- [140].Zan L, Chen Q, Zhang L, Li X, Epigallocatechin gallate (EGCG) suppresses growth and tumorigenicity in breast cancer cells by downregulation of miR-25, Bioengineered 10 (1) (2019) 374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Braicu C, Gherman CD, Irimie A, Berindan-Neagoe I, Epigallocatechin-3-Gallate (EGCG) inhibits cell proliferation and migratory behaviour of triple negative breast cancer cells, J. Nanosci. Nanotechnol. 13 (1) (2013) 632–637. [DOI] [PubMed] [Google Scholar]
- [142].Frank DA, Mahajan S, Ritz J, Fludarabine-induced immunosuppression is associated with inhibition of STAT1 signaling, Nat. Med. 5 (4) (1999) 444–447. [DOI] [PubMed] [Google Scholar]
- [143].Plunkett W, Gandhi V, Huang P, Robertson LE, Yang LY, Gregoire V, Estey E, Keating MJ, Fludarabine: pharmacokinetics, mechanisms of action, and rationales for combination therapies, Semin. Oncol. 20 (5 Suppl 7) (1993) 2–12. [PubMed] [Google Scholar]
- [144].Fu XY, Schindler C, Improta T, Aebersold R, Darnell JE Jr., The proteins of ISGF-3, the interferon alpha-induced transcriptional activator, define a gene family involved in signal transduction, Proc. Natl. Acad. Sci. USA 89 (16) (1992) 7840–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE Jr., Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha, Proc. Natl. Acad. Sci. USA 89 (16) (1992) 7836–7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Park C, Li S, Cha E, Schindler C, Immune response in Stat2 knockout mice, Immunity 13 (6) (2000) 795–804. [DOI] [PubMed] [Google Scholar]
- [147].Paulson M, Pisharody S, Pan L, Guadagno S, Mui AL, Levy DE, Stat protein transactivation domains recruit p300/CBP through widely divergent sequences, J. Biol. Chem. 274 (36) (1999) 25343–25349. [DOI] [PubMed] [Google Scholar]
- [148].Lee CJ, An HJ, Cho ES, Kang HC, Lee JY, Lee HS, Cho YY, Stat2 stability regulation: an intersection between immunity and carcinogenesis, Exp. Mol. Med. 52 (9) (2020) 1526–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Blaszczyk K, Olejnik A, Nowicka H, Ozgyin L, Chen YL, Chmielewski S, Kostyrko K, Wesoly J, Balint BL, Lee CK, Bluyssen HA, STAT2/IRF9 directs a prolonged ISGF3-like transcriptional response and antiviral activity in the absence of STAT1, Biochem. J. 466 (3) (2015) 511–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Nan J, Wang Y, Yang J, Stark GR, IRF9 and unphosphorylated STAT2 cooperate with NF-κB to drive IL6 expression, 115(15), 2018: 3906–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Walter KR, Balko JM, Hagan CR, Progesterone receptor promotes degradation of STAT2 to inhibit the interferon response in breast cancer, OncoImmunology 9 (1) (2020) 1758547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Ogony J, Choi HJ, Lui A, Cristofanilli M, Lewis-Wambi J, Interferon-induced transmembrane protein 1 (IFITM1) overexpression enhances the aggressive phenotype of SUM149 inflammatory breast cancer cells in a signal transducer and activator of transcription 2 (STAT2)-dependent manner, Breast Cancer Res. 18 (1) (2016) 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Fan J-B, Miyauchi S, Xu H-Z, Liu D, Kim LJY, Burkart C, Cheng H, Arimoto K-I, Yan M, Zhou Y, Győrffy B, Knobeloch K-P, Rich JN, Cang H, Fu X-D, Zhang D-E, Type I Interferon Regulates a Coordinated Gene Network to Enhance Cytotoxic T Cell–Mediated Tumor Killing, Cancer Discov. 10 (3) (2020) 382–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Wang S, Yu L, Shi W, Li X, Yu L, Prognostic roles of signal transducers and activators of transcription family in human breast cancer, Biosci. Rep. 38 (6) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Wu H-T, Liu J, Li G-W, Shen J-X, Huang Y-T, The transcriptional STAT3 is a potential target, whereas transcriptional STAT5A/5B/6 are new biomarkers for prognosis in human breast carcinoma, Oncotarget 8 (22) (2017) 36279–36288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Akira S, Nishio Y, Inoue M, Wang XJ, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T, Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway, Cell 77 (1) (1994) 63–71. [DOI] [PubMed] [Google Scholar]
- [157].Lütticken C, Wegenka UM, Yuan J, Buschmann J, Schindler C, Ziemiecki A, Harpur AG, Wilks AF, Yasukawa K, Taga T, et al. , Association of transcription factor APRF and protein kinase Jak1 with the interleukin-6 signal transducer gp130, Sci. (N. Y., N. Y. ) 263 (5143) (1994) 89–92. [DOI] [PubMed] [Google Scholar]
- [158].Yu CL, Meyer DJ, Campbell GS, Larner AC, Carter-Su C, Schwartz J, Jove R, Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein, Sci. (N. Y., N. Y. ) 269 (5220) (1995) 81–83. [DOI] [PubMed] [Google Scholar]
- [159].Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE Jr., Stat3 as an oncogene, Cell 98 (3) (1999) 295–303. [DOI] [PubMed] [Google Scholar]
- [160].Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, Zhang S, Wang T, Sinibaldi D, Coppola D, Heller R, Ellis LM, Karras J, Bromberg J, Pardoll D, Jove R, Yu H, Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis, Oncogene 21 (13) (2002) 2000–2008. [DOI] [PubMed] [Google Scholar]
- [161].Wei D, Le X, Zheng L, Wang L, Frey JA, Gao AC, Peng Z, Huang S, Xiong HQ, Abbruzzese JL, Xie K, Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis, Oncogene 22 (3) (2003) 319–329. [DOI] [PubMed] [Google Scholar]
- [162].Lo H-W, Hsu S-C, Ali-Seyed M, Gunduz M, Xia W, Wei Y, Bartholomeusz G, Shih J-Y, Hung M-C, Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway, Cancer Cell 7 (6) (2005) 575–589. [DOI] [PubMed] [Google Scholar]
- [163].Lo H-W, Hsu S-C, Xia W, Cao X, Shih J-Y, Wei Y, Abbruzzese JL, Hortobagyi GN, Hung M-C, Epidermal Growth Factor Receptor Cooperates with Signal Transducer and Activator of Transcription 3 to Induce Epithelial-Mesenchymal Transition in Cancer Cells via Up-regulation of TWIST Gene Expression, Cancer Res. 67 (19) (2007) 9066–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Lo HW, Cao X, Zhu H, Ali-Osman F, Constitutively activated STAT3 frequently coexpresses with epidermal growth factor receptor in high-grade gliomas and targeting STAT3 sensitizes them to Iressa and alkylators, Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 14 (19) (2008) 6042–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S, Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality, Proc. Natl. Acad. Sci. USA 94 (8) (1997) 3801–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Erdogan F, Radu TB, Orlova A, Qadree AK, de Araujo ED, Israelian J, Valent P, Mustjoki SM, Herling M, Moriggl R, Gunning PT, JAK-STAT core Cancer Pathw.: Integr. Cancer Inter. Anal. 26 (7) (2022) 2049–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Garg M, Shanmugam MK, Bhardwaj V, Goel A, Gupta R, Sharma A, Baligar P, Kumar AP, Goh BC, Wang L, Sethi G, The pleiotropic role of transcription factor STAT3 in oncogenesis and its targeting through natural products for cancer prevention and therapy, Med. Res. Rev (2020). [DOI] [PubMed] [Google Scholar]
- [168].Zhong Z, Wen Z, Darnell JE Jr., Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6, Sci. (N. Y., N. Y. ) 264 (5155) (1994) 95–98. [DOI] [PubMed] [Google Scholar]
- [169].Jones SA, Jenkins BJ, Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer, Nat. Rev. Immunol. 18 (12) (2018) 773–789. [DOI] [PubMed] [Google Scholar]
- [170].Aigner P, Just V, Stoiber D, STAT3 isoforms: Alternative fates in cancer? Cytokine 118 (2019) 27–34. [DOI] [PubMed] [Google Scholar]
- [171].Sirkisoon SR, Carpenter RL, Rimkus T, Anderson A, Harrison A, Lange AM, Jin G, Watabe K, Lo H-W, Interaction between STAT3 and GLI1/tGLI1 oncogenic transcription factors promotes the aggressiveness of triple-negative breast cancers and HER2-enriched breast cancer, Oncogene 37 (19) (2018) 2502–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Wang H-Q, Man Q-W, Huo F-Y, Gao X, Lin H, Li S-R, Wang J, Su F-C, Cai Lulu, Shi Y, Liu Bing, Bu L-L, STAT3 Pathw. Cancer.:, Present, Future 3 (2) (2022), e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Chang Q, Bournazou E, Sansone P, Berishaj M, Gao SP, Daly L, Wels J, Theilen T, Granitto S, Zhang X, Cotari J, Alpaugh ML, de Stanchina E, Manova K, Li M, Bonafe M, Ceccarelli C, Taffurelli M, Santini D, Altan-Bonnet G, Kaplan R, Norton L, Nishimoto N, Huszar D, Lyden D, Bromberg J, The IL-6/JAK/Stat3 feed-forward loop drives tumorigenesis and metastasis, Neoplasia (N. Y., N. Y. ) 15 (7) (2013) 848–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Johnson DE, O’Keefe RA, Grandis JR, Targeting the IL-6/JAK/STAT3 signalling axis in cancer, Nature Reviews, Clin. Oncol. 15 (4) (2018) 234–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].To SQ, Dmello RS, Richards AK, Ernst M, Chand AL, STAT3 Signal. Breast Cancer.: Multicell. Actions Ther. Potential 14 (2) (2022) 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Chung SS, Aroh C, Vadgama JV, Constitutive activation of STAT3 signaling regulates hTERT and promotes stem cell-like traits in human breast cancer cells, PloS One 8 (12) (2013), e83971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Lee J-L, Wang M-J, Chen J-Y, Acetylation and activation of STAT3 mediated by nuclear translocation of CD44, J. Cell Biol. 185 (6) (2009) 949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Bhattacharya S, Ray RM, Johnson LR, STAT3-mediated transcription of Bcl-2, Mcl-1 and c-IAP2 prevents apoptosis in polyamine-depleted cells, Biochem. J. 392 (Pt 2) (2005) 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Zaanan A, Okamoto K, Kawakami H, Khazaie K, Huang S, Sinicrope FA, The Mutant KRAS Gene Up-regulates BCL-XL Protein via STAT3 to Confer Apoptosis Resistance That Is Reversed by BIM Protein Induction and BCL-XL Antagonism, J. Biol. Chem. 290 (39) (2015) 23838–23849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Xie Q, Yang Z, Huang X, Zhang Z, Li J, Ju J, Zhang H, Ma J, Ilamycin C induces apoptosis and inhibits migration and invasion in triple-negative breast cancer by suppressing IL-6/STAT3 pathway, J. Hematol. Oncol. 12 (1) (2019) 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].x Xie T, Wei D, Liu M, Gao AC, Ali-Osman F, Sawaya R, Huang S, Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis, Oncogene 23 (20) (2004) 3550–3560. [DOI] [PubMed] [Google Scholar]
- [182].Ma JH, Qi J, Lin SQ, Zhang CY, Liu FY, Xie WD, Li X, S.T.A.T.3 Targets, ERR-α to Promote Epithelial-Mesenchymal Transition, Migration, and Invasion in Triple-Negative Breast Cancer Cells, Mol. Cancer Res.: MCR 17 (11) (2019) 2184–2195. [DOI] [PubMed] [Google Scholar]
- [183].Yu H, Kortylewski M, Pardoll D, Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment, Nat. Rev. Immunol. 7 (1) (2007) 41–51. [DOI] [PubMed] [Google Scholar]
- [184].Kujawski M, Kortylewski M, Lee H, Herrmann A, Kay H, Yu H, Stat3 mediates myeloid cell-dependent tumor angiogenesis in mice, J. Clin. Investig. 118 (10) (2008) 3367–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Yu H, Pardoll D, Jove R, STATs in cancer inflammation and immunity: a leading role for STAT3, Nat. Rev. Cancer 9 (11) (2009) 798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Carpenter RL, Lo HW, STAT3 Target Genes Relevant to Human Cancers, Cancers 6 (2) (2014) 897–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Nakagawa T, Oda G, Kawachi H, Ishikawa T, Okamoto K, Uetake H, Nuclear Expression of p-STAT3 Is Associated with Poor Prognosis in ER(−) Breast Cancer, Clin. Pract. 12 (2) (2022) 157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Tell RW, Horvath CM, Bioinformatic analysis reveals a pattern of STAT3-associated gene expression specific to basal-like breast cancers in human tumors, 111(35), 2014: 12787–12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Tawara K, Scott H, Emathinger J, Wolf C, LaJoie D, Hedeen D, Bond L, Montgomery P, Jorcyk C, HIGH expression of OSM and IL-6 are associated with decreased breast cancer survival: synergistic induction of IL-6 secretion by OSM and IL-1β, Oncotarget 10 (21) (2019) 2068–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Tawara K, Scott H, Emathinger J, Ide A, Fox R, Greiner D, LaJoie D, Hedeen D, Nandakumar M, Oler AJ, Holzer R, Jorcyk C, Co-Expression of VEGF and IL-6 Family Cytokines is Associated with Decreased Survival in HER2 Negative Breast Cancer Patients: Subtype-Specific IL-6 Family Cytokine-Mediated VEGF Secretion, Transl. Oncol. 12 (2) (2019) 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Doherty MR, Parvani JG, Tamagno I, Junk DJ, Bryson BL, Cheon HJ, Stark GR, Jackson MW, The opposing effects of interferon-beta and oncostatin-M as regulators of cancer stem cell plasticity in triple-negative breast cancer, Breast Cancer Res. 21 (1) (2019) 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Valeta-Magara A, Gadi A, Volta V, Walters B, Arju R, Giashuddin S, Zhong H, Schneider RJ, Inflammatory Breast Cancer Promotes Development of M2 Tumor-Associated Macrophages and Cancer Mesenchymal Cells through a Complex Chemokine Network, Cancer Res. 79 (13) (2019) 3360–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Ma M, Huang W, Kong D, IL-17 inhibits the accumulation of myeloid-derived suppressor cells in breast cancer via activating STAT3, Int. Immunopharmacol. 59 (2018) 148–156. [DOI] [PubMed] [Google Scholar]
- [194].Ding M, Fu Y, Guo F, Chen H, Fu X, Tan W, Zhang H, Long non-coding RNA MAFG-AS1 knockdown blocks malignant progression in breast cancer cells by inactivating JAK2/STAT3 signaling pathway via MAFG-AS1/miR-3196/TFAP2A axis, Int J. Clin. Exp. Pathol. 13 (10) (2020) 2455–2473. [PMC free article] [PubMed] [Google Scholar]
- [195].Zhao Y, Hu Z, Li J, Hu T, EZH2 Exacerbates Breast Cancer by Methylating and Activating STAT3 Directly, J. Cancer 12 (17) (2021) 5220–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Liu L, Gao Y, Qiu H, Miller WT, Poli V, Reich NC, Identification of STAT3 as a specific substrate of breast tumor kinase, Oncogene 25 (35) (2006) 4904–4912. [DOI] [PubMed] [Google Scholar]
- [197].Ikeda O, Miyasaka Y, Sekine Y, Mizushima A, Muromoto R, Nanbo A, Yoshimura A, Matsuda T, STAP-2 is phosphorylated at tyrosine-250 by Brk and modulates Brk-mediated STAT3 activation, Biochem. Biophys. Res. Commun. 384 (1) (2009) 71–75. [DOI] [PubMed] [Google Scholar]
- [198].Minoguchi M, Minoguchi S, Aki D, Joo A, Yamamoto T, Yumioka T, Matsuda T, Yoshimura A, STAP-2/BKS, an Adaptor/Docking Protein, Modulates STAT3 Activation in Acute-phase Response through Its YXXQ Motif*, J. Biol. Chem. 278 (13) (2003) 11182–11189. [DOI] [PubMed] [Google Scholar]
- [199].Ikeda O, Sekine Y, Mizushima A, Nakasuji M, Miyasaka Y, Yamamoto C, Muromoto R, Nanbo A, Oritani K, Yoshimura A, Matsuda T, Interactions of STAP-2 with Brk and STAT3 participate in cell growth of human breast cancer cells, J. Biol. Chem. 285 (49) (2010) 38093–38103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Williams CB, Phelps-Polirer K, Dingle IP, Williams CJ, Rhett MJ, Eblen ST, Armeson K, Hill EG, Yeh ES, HUNK phosphorylates EGFR to regulate breast cancer metastasis, Oncogene 39 (5) (2020) 1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Sainsbury JR, Farndon JR, Needham GK, Malcolm AJ, Harris AL, Epidermal-growth-factor receptor status as predictor of early recurrence of and death from breast cancer, Lancet (Lond., Engl. ) 1 (8547) (1987) 1398–1402. [DOI] [PubMed] [Google Scholar]
- [202].Sirkisoon SR, Carpenter RL, Rimkus T, Miller L, Metheny-Barlow L, Lo H-W, EGFR and HER2 signaling in breast cancer brain metastasis, Front Biosci. (Elite Ed. ) 8 (2) (2016) 245–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Song X, Liu Z, Yu Z, EGFR Promotes the Development of Triple Negative Breast Cancer Through JAK/STAT3 Signaling, Cancer Manag Res 12 (2020) 703–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Lo HW, Hsu SC, Ali-Seyed M, Gunduz M, Xia W, Wei Y, Bartholomeusz G, Shih JY, Hung MC, Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway, Cancer Cell 7 (6) (2005) 575–589. [DOI] [PubMed] [Google Scholar]
- [205].Zhang T, Ma J, Cao X, Grb2 regulates Stat3 activation negatively in epidermal growth factor signalling, Biochem. J. 376 (Pt 2) (2003) 457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Lange AM, Lo H-W, Inhib. TRK Proteins Clin. Cancer Ther. 10 (4) (2018) 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Lagadec C, Meignan S, Adriaenssens E, Foveau B, Vanhecke E, Romon R, Toillon RA, Oxombre B, Hondermarck H, Le X, Bourhis, TrkA overexpression enhances growth and metastasis of breast cancer cells, Oncogene 28 (18) (2009) 1960–1970. [DOI] [PubMed] [Google Scholar]
- [208].Regua AT, Aguayo NR, Jalboush SA, Doheny DL, Manore SG, Zhu D, Wong GL, Arrigo A, Wagner CJ, Yu Y, Thomas A, Chan MD, Ruiz J, Jin G, Strowd R, Sun P, Lin J, Lo HW, TrkA Interacts with and Phosphorylates STAT3 to Enhance Gene Transcription and Promote Breast Cancer Stem Cells in Triple-Negative and HER2-Enriched Breast Cancers, Cancers 13 (10) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Doheny D, Manore SG, Wong GL, Lo H-W, Hedgehog Signaling and Truncated GLI1 in Cancer, Cells 9 (9) (2020) 2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Lo HW, Zhu H, Cao X, Aldrich A, Ali-Osman F, A novel splice variant of GLI1 that promotes glioblastoma cell migration and invasion, Cancer Res. 69 (17) (2009) 6790–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Fan YH, Ding J, Nguyen S, Liu XJ, Xu G, Zhou HY, Duan NN, Yang SM, Zern MA, Wu J, Aberrant hedgehog signaling is responsible for the highly invasive behavior of a subpopulation of hepatoma cells, Oncogene 35 (1) (2016) 116–124. [DOI] [PubMed] [Google Scholar]
- [212].Cao X, Geradts J, Dewhirst MW, Lo HW, Upregulation of VEGF-A and CD24 gene expression by the tGLI1 transcription factor contributes to the aggressive behavior of breast cancer cells, Oncogene 31 (1) (2012) 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Rimkus TK, Carpenter RL, Sirkisoon S, Zhu D, Pasche BC, Chan MD, Lesser GJ, Tatter SB, Watabe K, Debinski W, Lo H-W, Truncated Glioma-Associated Oncogene Homolog 1 (tGLI1) Mediates Mesenchymal Glioblastoma via Transcriptional Activation of CD44, 78(10), 2018: 2589–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Sirkisoon SR, Carpenter RL, Rimkus T, Doheny D, Zhu D, Aguayo NR, Xing F, Chan M, Ruiz J, Metheny-Barlow LJ, Strowd R, Lin J, Regua AT, Arrigo A, Anguelov M, Pasche B, Debinski W, Watabe K, Lo H-W, TGLI1 transcription factor mediates breast cancer brain metastasis via activating metastasis-initiating cancer stem cells and astrocytes in the tumor microenvironment, Oncogene 39 (1) (2020) 64–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Carpenter RL, Paw I, Zhu H, Sirkisoon S, Xing F, Watabe K, Debinski W, Lo HW, The gain-of-function GLI1 transcription factor TGLI1 enhances expression of VEGF-C and TEM7 to promote glioblastoma angiogenesis, Oncotarget 6 (26) (2015) 22653–22665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Di Mauro C, Rosa R, D’Amato V, Ciciola P, Servetto A, Marciano R, Orsini RC, Formisano L, De Falco S, Cicatiello V, Di Bonito M, Cantile M, Collina F, Chambery A, Veneziani BM, De Placido S, Bianco R, Hedgehog signalling pathway orchestrates angiogenesis in triple-negative breast cancers, Br. J. Cancer 116 (11) (2017) 1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Zhu H, Carpenter RL, Han W, Lo HW, The GLI1 splice variant TGLI1 promotes glioblastoma angiogenesis and growth, Cancer Lett. 343 (1) (2014) 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Doheny D, Sirkisoon S, Carpenter RL, Aguayo NR, Regua AT, Anguelov M, Manore SG, Arrigo A, Jalboush SA, Wong GL, Yu Y, Wagner CJ, Chan M, Ruiz J, Thomas A, Strowd R, Lin J, Lo H-W, Combined inhibition of JAK2-STAT3 and SMO-GLI1/tGLI1 pathways suppresses breast cancer stem cells, tumor growth, and metastasis, Oncogene 39 (42) (2020) 6589–6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Sirkisoon SR, Wong GL, Aguayo NR, Doheny DL, Zhu D, Regua AT, Arrigo A, Manore SG, Wagner C, Thomas A, Singh R, Xing F, Jin G, Watabe K, Lo H-W, Breast cancer extracellular vesicles-derived miR-1290 activates astrocytes in the brain metastatic microenvironment via the FOXA2→CNTF axis to promote progression of brain metastases, Cancer Lett. 540 (2022), 215726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Lin WH, Dai WG, Xu XD, Yu QH, Zhang B, Li J, Li HP, Downregulation of DPF3 promotes the proliferation and motility of breast cancer cells through activating JAK2/STAT3 signaling, Biochem. Biophys. Res. Commun. 514 (3) (2019) 639–644. [DOI] [PubMed] [Google Scholar]
- [221].Chang R, Song L, Xu Y, Wu Y, Dai C, Wang X, Sun X, Hou Y, Li W, Zhan X, Zhan L, Loss of Wwox drives metastasis in triple-negative breast cancer by JAK2/STAT3 axis, Nat. Commun. 9 (1) (2018) 3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Liu J, Liu L, Yagüe E, Yang Q, Pan T, Zhao H, Hu Y, Zhang J, GGNBP2 suppresses triple-negative breast cancer aggressiveness through inhibition of IL-6/STAT3 signaling activation, Breast Cancer Res. Treat. 174 (1) (2019) 65–78. [DOI] [PubMed] [Google Scholar]
- [223].Khanna P, Lee JS, Sereemaspun A, Lee H, Baeg GH, GRAMD1B regulates cell migration in breast cancer cells through JAK/STAT and Akt signaling, Sci. Rep. 8 (1) (2018) 9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Gebert LFR, MacRae IJ, Regulation of microRNA function in animals, Nat. Rev. Mol. Cell Biol. 20 (1) (2019) 21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Cao Q, Li YY, He WF, Zhang ZZ, Zhou Q, Liu X, Shen Y, Huang TT, Interplay between microRNAs and the STAT3 signaling pathway in human cancers, Physiol. Genom. 45 (24) (2013) 1206–1214. [DOI] [PubMed] [Google Scholar]
- [226].Shi P, Chen C, Li X, Wei Z, Liu Z, Liu Y, MicroRNA-124 suppresses cell proliferation and invasion of triple negative breast cancer cells by targeting STAT3, Mol. Med Rep. 19 (5) (2019) 3667–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Pang Y, Wu J, Li X, Wang C, Wang M, Liu J, Yang G, NEAT1/miR-124/STAT3 feedback loop promotes breast cancer progression, Int. J. Oncol. 55 (3) (2019) 745–754. [DOI] [PubMed] [Google Scholar]
- [228].Park Y, Kim J, Regulation of IL-6 signaling by miR-125a and let-7e in endothelial cells controls vasculogenic mimicry formation of breast cancer cells, BMB Rep. 52 (3) (2019) 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Deshmukh SK, Srivastava SK, Zubair H, Khan MA, Singh AP, Singh S, Resistin Induces LIN28A-Mediated Let-7a Repression in Breast Cancer Cells Leading to IL-6 and STAT3 Upregulation, 13(18), 2021: 4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Li JP, Xiang Y, Fan LJ, Yao A, Li H, Liao XH, Long noncoding RNA H19 competitively binds miR-93–5p to regulate STAT3 expression in breast cancer, J. Cell. Biochem. 120 (3) (2019) 3137–3148. [DOI] [PubMed] [Google Scholar]
- [231].Deng XIN, Zhao YI, Wang B, miR-519d-mediated downregulation of STAT3 suppresses breast cancer progression, Oncol. Rep. 34 (4) (2015) 2188. [DOI] [PubMed] [Google Scholar]
- [232].Wang N, Wei L, Huang Y, Wu Y, Su M, Pang X, Ji F, Zhong C, Chen T, Li B, miR520c blocks EMT progression of human breast cancer cells by repressing STAT3, Oncol. Rep. 37 (3) (2017) 1537. [DOI] [PubMed] [Google Scholar]
- [233].Wang X, Qiu W, Zhang G, Xu S, Gao Q, Yang Z, MicroRNA-204 targets JAK2 in breast cancer and induces cell apoptosis through the STAT3/BCl-2/survivin pathway, Int J. Clin. Exp. Pathol. 8 (5) (2015) 5017–5025. [PMC free article] [PubMed] [Google Scholar]
- [234].Zhao Q, Liu Y, Wang T, Yang Y, Ni H, Liu H, Guo Q, Xi T, Zheng L, MiR-375 inhibits the stemness of breast cancer cells by blocking the JAK2/STAT3 signaling, Eur. J. Pharmacol. 884 (2020), 173359. [DOI] [PubMed] [Google Scholar]
- [235].Nomair AM, Ahmed SS, Nomeir HM, El Mansy H, Mohammed AF, The role of protein inhibitor of activated STAT3 and miRNA-18a expressions in breast cancer, Egypt. J. Med. Hum. Genet. 20 (1) (2019) 15. [Google Scholar]
- [236].Deligne C, Midwood KS, Macrophages and Extracellular Matrix in Breast Cancer: Partners in Crime or Protective Allies?, 11, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Akashi K, Traver D, Miyamoto T, Weissman IL, A clonogenic common myeloid progenitor that gives rise to all myeloid lineages, Nature 404 (6774) (2000) 193–197. [DOI] [PubMed] [Google Scholar]
- [238].Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, Littman DR, Cumano A, Geissmann F, A clonogenic bone marrow progenitor specific for macrophages and dendritic cells, Sci. (N. Y., N. Y. ) 311 (5757) (2006) 83–87. [DOI] [PubMed] [Google Scholar]
- [239].Wu S-Y, Watabe K, The roles of microglia/macrophages in tumor progression of brain cancer and metastatic disease, Front Biosci. (Landmark Ed. ) 22 (10) (2017) 1805–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].y Zhu Y, c Zhao Y, Chen C, Xie M, CCL5 secreted by luminal B breast cancer cells induces polarization of M2 macrophages through activation of MEK/STAT3 signaling pathway via CCR5, Gene 812 (2022), 146100. [DOI] [PubMed] [Google Scholar]
- [241].Radharani NNV, Yadav AS, Nimma R, Kumar TVS, Bulbule A, Chanukuppa V, Kumar D, Patnaik S, Rapole S, Kundu GC, Tumor-associated macrophage derived IL-6 enriches cancer stem cell population and promotes breast tumor progression via Stat-3 pathway, Cancer Cell Int. 22 (1) (2022) 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Chu J, Hu XC, Li CC, Li TY, Fan HW, Jiang GQ, KLF14 alleviated breast cancer invasion and M2 macrophages polarization through modulating SOCS3/RhoA/Rock/STAT3 signaling, Cell. Signal. 92 (2022), 110242. [DOI] [PubMed] [Google Scholar]
- [243].Jones LM, Broz ML, Ranger JJ, Ozcelik J, Ahn R, Zuo D, Ursini-Siegel J, Hallett MT, Krummel M, Muller WJ, STAT3 Establishes an Immunosuppressive Microenvironment during the Early Stages of Breast Carcinogenesis to Promote Tumor Growth and Metastasis, Cancer Res. 76 (6) (2016) 1416–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Oh E, Kim Y-J, An H, Sung D, Cho T-M, Farrand L, Jang S, Seo JH, Kim JY, Flubendazole elicits anti-metastatic effects in triple-negative breast cancer via STAT3 inhibition, 143(8), 2018: 1978–1993. [DOI] [PubMed] [Google Scholar]
- [245].Hao S, Chen X, Wang F, Shao Q, Liu J, Zhao H, Yuan C, Ren H, Mao H, Breast cancer cell-derived IL-35 promotes tumor progression via induction of IL-35-producing induced regulatory T cells, Carcinogenesis 39 (12) (2018) 1488–1496. [DOI] [PubMed] [Google Scholar]
- [246].Xing J, Li J, Fu L, Gai J, Guan J, Li Q, SIRT4 enhances the sensitivity of ER-positive breast cancer to tamoxifen by inhibiting the IL-6/STAT3 signal pathway, 8(16), 2019: 7086–7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Yang Q, Zhao S, Shi Z, Cao L, Liu J, Pan T, Zhou D, Zhang J, Chemotherapy-elicited exosomal miR-378a-3p and miR-378d promote breast cancer stemness and chemoresistance via the activation of EZH2/STAT3 signaling, J. Exp. Clin. Cancer Res. 40 (1) (2021) 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Liu C, Xing H, Guo C, Yang Z, Wang Y, Wang Y, MiR-124 reversed the doxorubicin resistance of breast cancer stem cells through STAT3/HIF-1 signaling pathways, Cell Cycle (Georget., Tex. ) 18 (18) (2019) 2215–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Gariboldi MB, Ravizza R, Molteni R, Osella D, Gabano E, Monti E, Inhibition of Stat3 increases doxorubicin sensitivity in a human metastatic breast cancer cell line, Cancer Lett. 258 (2) (2007) 181–188. [DOI] [PubMed] [Google Scholar]
- [250].Real PJ, Sierra A, De Juan A, Segovia JC, Lopez-Vega JM, Fernandez-Luna JL, Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells, Oncogene 21 (50) (2002) 7611–7618. [DOI] [PubMed] [Google Scholar]
- [251].Kim J-S, Kim H-A, Seong M-K, Seol H, Oh JS, Kim E-K, Chang JW, Hwang S-G, Noh WC, STAT3-survivin signaling mediates a poor response to radiotherapy in HER2-positive breast cancers, Oncotarget 7 (6) (2016) 7055–7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Wang T, Fahrmann JF, Lee H, Li Y-J, Tripathi SC, Yue C, Zhang C, Lifshitz V, Song J, Yuan Y, Somlo G, Jandial R, Ann D, Hanash S, Jove R, Yu H, JAK/STAT3-Regulated Fatty Acid β-Oxidation Is Critical for Breast Cancer Stem Cell Self-Renewal and Chemoresistance, Cell Metab. 27 (1) (2018) 136–150. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Wang L, Wang Q, Gao M, Fu L, Li Y, Quan H, Lou L, STAT3 activation confers trastuzumab-emtansine (T-DM1) resistance in HER2-positive breast cancer, Cancer Sci. 109 (10) (2018) 3305–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Miklossy G, Hilliard TS, Turkson J, Therapeutic modulators of STAT signalling for human diseases, Nat. Rev. Drug Discov. 12 (8) (2013) 611–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Turkson J, Zhang S, Palmer J, Kay H, Stanko J, Mora LB, Sebti S, Yu H, Jove R, Inhibition of constitutive signal transducer and activator of transcription 3 activation by novel platinum complexes with potent antitumor activity, Mol. Cancer Ther. 3 (12) (2004) 1533–1542. [PubMed] [Google Scholar]
- [256].Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, Lawrence HR, Yip MLR, Jove R, McLaughlin MM, Lawrence NJ, Sebti SM, Turkson J, Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity, 104(18), 2007: 7391–7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Zhang X, Yue P, Page BDG, Li T, Zhao W, Namanja AT, Paladino D, Zhao J, Chen Y, Gunning PT, Turkson J, Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts, 109 (24), 2012: 9623–9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Schust J, Sperl B, Hollis A, Mayer TU, Berg T, Stattic: A Small-Molecule Inhibitor of STAT3 Activation and Dimerization, Chem. Biol. 13 (11) (2006) 1235–1242. [DOI] [PubMed] [Google Scholar]
- [259].Yue P, Lopez-Tapia F, Paladino D, Li Y, Chen C-H, Namanja AT, Hilliard T, Chen Y, Tius MA, Turkson J, Hydroxamic Acid and Benzoic Acid–Based STAT3 Inhibitors Suppress Human Glioma and Breast Cancer Phenotypes In Vitro and In Vivo, Cancer Res. 76 (3) (2016) 652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Dees S, Pontiggia L, Jasmin J-F, Mercier I, Phosphorylated STAT3 (Tyr705) as a biomarker of response to pimozide treatment in triple-negative breast cancer, Cancer Biol. Ther. 21 (6) (2020) 506–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Iwamaru A, Szymanski S, Iwado E, Aoki H, Yokoyama T, Fokt I, Hess K, Conrad C, Madden T, Sawaya R, Kondo S, Priebe W, Kondo Y, A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo, Oncogene 26 (17) (2007) 2435–2444. [DOI] [PubMed] [Google Scholar]
- [262].Lee HT, Xue J, Chou PC, Zhou A, Yang P, Conrad CA, Aldape KD, Priebe W, Patterson C, Sawaya R, Xie K, Huang S, Stat3 orchestrates interaction between endothelial and tumor cells and inhibition of Stat3 suppresses brain metastasis of breast cancer cells, Oncotarget 6 (12) (2015) 10016–10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Priego N, Zhu L, Monteiro C, Mulders M, Wasilewski D, Bindeman W, Doglio L, Martínez L, Martínez-Saez E, Ramón y Cajal S, Megías D, Hernández-Encinas E, Blanco-Aparicio C, Martínez L, Zarzuela E, Muñoz J, Fustero-Torre C, Piñeiro-Yáñez E, Hernández-Laín A, Bertero L, Poli V, Sanchez-Martinez M, Menendez JA, Soffietti R, Bosch-Barrera J, Valiente M, STAT3 labels a subpopulation of reactive astrocytes required for brain metastasis, Nat. Med. 24 (7) (2018) 1024–1035. [DOI] [PubMed] [Google Scholar]
- [264].Lee Y, Park HR, Chun HJ, Lee J, Silibinin prevents dopaminergic neuronal loss in a mouse model of Parkinson’s disease via mitochondrial stabilization, 93(5), 2015: 755–765. [DOI] [PubMed] [Google Scholar]
- [265].Bosch-Barrera J, Sais E, Cañete N, Marruecos J, Cuyàs E, Izquierdo A, Porta R, Haro M, Brunet J, Pedraza S, Menendez JA, Response of brain metastasis from lung cancer patients to an oral nutraceutical product containing silibinin, Oncotarget 7 (22) (2016) 32006–32014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Prajapati KS, Gupta S, Kumar S, Targeting Breast Cancer-Derived Stem Cells by Dietary Phytochemicals: A Strategy for Cancer Prevention and Treatment, 14(12), 2022: 2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Chung SS, Vadgama JV, Curcumin and epigallocatechin gallate inhibit the cancer stem cell phenotype via down-regulation of STAT3-NFκB signaling, Anticancer Res. 35 (1) (2015) 39–46. [PMC free article] [PubMed] [Google Scholar]
- [268].Lin L, Hutzen B, Zuo M, Ball S, Deangelis S, Foust E, Pandit B, Ihnat MA, Shenoy SS, Kulp S, Li PK, Li C, Fuchs J, Lin J, Novel STAT3 phosphorylation inhibitors exhibit potent growth-suppressive activity in pancreatic and breast cancer cells, Cancer Res. 70 (6) (2010) 2445–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Cui X, Wang Y, Kokudo N, Fang D, Tang W, Traditional Chinese medicine and related active compounds against hepatitis B virus infection, Biosci. Trends 4 (2) (2010) 39–47. [PubMed] [Google Scholar]
- [270].Ma Q, Gao FF, He X, Li K, Gao Y, Xu XL, Jiang NH, Ding L, Song WJ, He YQ, Pan WT, Wei L, Zhang JW, Antitumor effects of saikosaponin b2 on breast cancer cell proliferation and migration, Mol. Med Rep. 20 (2) (2019) 1943–1951. [DOI] [PubMed] [Google Scholar]
- [271].Liu C, Dong L, Sun Z, Wang L, Wang Q, Li H, Zhang J, Wang X, Esculentoside A suppresses breast cancer stem cell growth through stemness attenuation and apoptosis induction by blocking IL-6/STAT3 signaling pathway, Phytother. Res.: PTR 32 (11) (2018) 2299–2311. [DOI] [PubMed] [Google Scholar]
- [272].Jakopovic B, Oršolić N, Jakopovich I, Prote Res. Antitumor Prop. Med. Mushrooms 26 (21) (2021) 6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Neergheen VS, Hip Kam A, Pem Y, Ramsaha S, Bahorun T, Regulation of cancer cell signaling pathways as key events for therapeutic relevance of edible and medicinal mushrooms, Semin. Cancer Biol. 80 (2022) 145–156. [DOI] [PubMed] [Google Scholar]
- [274].Atay S, Ak H, Kalmis E, Kayalar H, Aydin HH, Transcriptome-Wide Analysis Reveals the Molecular Mechanism of Tumoricidal Effects of Lion’s Mane Medicinal Mushroom, Hericium erinaceus (Agaricomycetes), on MCF-7 Breast Cancer Cells, Int. J. Med. Mushrooms 23 (1) (2021) 91–106. [Google Scholar]
- [275].Roda E, De Luca F, Di Iorio C, Ratto D, Siciliani S, Ferrari B, Cobelli F, Borsci G, Priori EC, Chinosi S, Ronchi A, Franco R, Di Francia R, Berretta M, Locatelli CA, Gregori A, Savino E, Bottone MG, Rossi P, Novel Medicinal Mushroom Blend as a Promising Supplement in Integrative Oncology: A Multi-Tiered Study using 4T1 Triple-Negative Mouse Breast Cancer Model, 21(10), 2020: 3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Zou S, Tong Q, Liu B, Huang W, Tian Y, Fu X, Targeting STAT3 in Cancer Immunotherapy, Mol. Cancer 19 (1) (2020) 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Qin J-J, Yan L, Zhang J, Zhang W-D, STAT3 as a potential therapeutic target in triple negative breast cancer: a systematic review, J. Exp. Clin. Cancer Res 38 (1) (2019), 195–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Kaplan MH, STAT4: a critical regulator of inflammation in vivo, Immunol. Res. 31 (3) (2005) 231–242. [DOI] [PubMed] [Google Scholar]
- [279].Trinchieri G, Interleukin-12 and the regulation of innate resistance and adaptive immunity, Nat. Rev. Immunol. 3 (2) (2003) 133–146. [DOI] [PubMed] [Google Scholar]
- [280].Lawless VA, Zhang S, Ozes ON, Bruns HA, Oldham I, Hoey T, Grusby MJ, Kaplan MH, Stat4 regulates multiple components of IFN-gamma-inducing signaling pathways, J. Immunol. (Baltim., Md.: 1950) 165 (12) (2000) 6803–6808. [DOI] [PubMed] [Google Scholar]
- [281].Tugues S, Burkhard SH, Ohs I, Vrohlings M, Nussbaum K, Vom Berg J, Kulig P, Becher B, New insights into IL-12-mediated tumor suppression, Cell death Differ. 22 (2) (2015) 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Morinobu A, Gadina M, Strober W, Visconti R, Fornace A, Montagna C, Feldman GM, Nishikomori R, O’Shea JJ, STAT4 serine phosphorylation is critical for IL-12-induced IFN-gamma production but not for cell proliferation, Proc. Natl. Acad. Sci. USA 99 (19) (2002) 12281–12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [283].Good SR, Thieu VT, Mathur AN, Yu Q, Stritesky GL, Yeh N, O’Malley JT, Perumal NB, Kaplan MH, Temporal induction pattern of STAT4 target genes defines potential for Th1 lineage-specific programming, J. Immunol. (Baltim., Md.: 1950) 183 (6) (2009) 3839–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [284].Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, Gadina M, O’Shea JJ, Biron CA, Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection, Sci. (N. Y., N. Y. ) 297 (5589) (2002) 2063–2066. [DOI] [PubMed] [Google Scholar]
- [285].Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, Vega F, Yu N, Wang J, Singh K, Zonin F, Vaisberg E, Churakova T, Liu M, Gorman D, Wagner J, Zurawski S, Liu Y, Abrams JS, Moore KW, Rennick D, de Waal-Malefyt R, Hannum C, Bazan JF, Kastelein RA, Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12, Immunity 13 (5) (2000) 715–725. [DOI] [PubMed] [Google Scholar]
- [286].Yang C, Mai H, Peng J, Zhou B, Hou J, Jiang D, STAT4: an immunoregulator contributing to diverse human diseases, Int. J. Biol. Sci. 16 (9) (2020) 1575–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [287].He R, Chen H, Feng Z, Dang Y, Gan T, Chen G, Yang LJIJCEM, High. Lev. STAT4 Expr. Is. Assoc. deterioration Breast Cancer 9 (6) (2016) 11612–11618. [Google Scholar]
- [288].Núñez-Marrero A, Assessing the Role of the Interleukin-12/STAT4 Axis in Breast Cancer by a Bioinformatics Approach, Int. J. Sci., Basic Appl. Res. 48 (2) (2019) 38–52. [PMC free article] [PubMed] [Google Scholar]
- [289].Zhou J, Xu XZ, Hu YR, Hu AR, Zhu CL, Gao GS, Cryptotanshinone induces inhibition of breast tumor growth by cytotoxic CD4+ T cells through the JAK2/STAT4/ perforin pathway, Asian Pac. J. Cancer Prev.: APJCP 15 (6) (2014) 2439–2445. [DOI] [PubMed] [Google Scholar]
- [290].Zhou Y, Jiang S, Yu S, Zhu L, Liu Y, Li S, Hao N, Ren YJGS, Mining the prognostic significance and immune infiltration of STAT family members in human breast cancer by bioinformatics analysis, 2022 11(4), 2022: 720–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [291].Lin J-X, Leonard WJ, The role of Stat5a and Stat5b in signaling by IL-2 family cytokines, Oncogene 19 (21) (2000) 2566–2576. [DOI] [PubMed] [Google Scholar]
- [292].Wakao H, Gouilleux F, Groner B, Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response, EMBO J. 13 (9) (1994) 2182–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [293].Hou J, Schindler U, Henzel WJ, Wong SC, McKnight SL, Identification and purification of human Stat proteins activated in response to interleukin-2, Immunity 2 (4) (1995) 321–329. [DOI] [PubMed] [Google Scholar]
- [294].Liu X, Robinson GW, Gouilleux F, Groner B, Hennighausen L, Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue, Proc. Natl. Acad. Sci. USA 92 (19) (1995) 8831–8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Lin JX, Mietz J, Modi WS, John S, Leonard WJ, Cloning of human Stat5B. Reconstitution of interleukin-2-induced Stat5A and Stat5B DNA binding activity in COS-7 cells, J. Biol. Chem. 271 (18) (1996) 10738–10744. [PubMed] [Google Scholar]
- [296].Giri JG, Ahdieh M, Eisenman J, Shanebeck K, Grabstein K, Kumaki S, Namen A, Park LS, Cosman D, Anderson D, Utilization of the beta and gamma chains of the IL-2 receptor by the novel cytokine IL-15, EMBO J. 13 (12) (1994) 2822–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Russell SM, Keegan AD, Harada N, Nakamura Y, Noguchi M, Leland P, Friedmann MC, Miyajima A, Puri RK, Paul WE, Leonard WJ, Interleukin-2 Receptor γ Chain: A Functional Component of the Interleukin-4 Receptor, Sci. (N. Y., N. Y. ) 262 (5141) (1993) 1880–1883. [DOI] [PubMed] [Google Scholar]
- [298].Kondo M, Takeshita T, Ishii N, Nakamura M, Watanabe S, Arai K, Sugamura K, Sharing of the interleukin-2 (IL-2) receptor gamma chain between receptors for IL-2 and IL-4, Sci. (N. Y., N. Y. ) 262 (5141) (1993) 1874–1877. [DOI] [PubMed] [Google Scholar]
- [299].Noguchi M, Nakamura Y, Russell SM, Ziegler SF, Tsang M, Cao X, Leonard WJ, Interleukin-2 receptor gamma chain: a functional component of the interleukin-7 receptor, Sci. (N. Y., N. Y. ) 262 (5141) (1993) 1877–1880. [DOI] [PubMed] [Google Scholar]
- [300].Lee G-H, Yoo K-C, An Y, Lee H-J, Lee M, Uddin N, Kim M-J, Kim I-G, Suh Y, Lee S-J, FYN promotes mesenchymal phenotypes of basal type breast cancer cells through STAT5/NOTCH2 signaling node, Oncogene 37 (14) (2018) 1857–1868. [DOI] [PubMed] [Google Scholar]
- [301].Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L, Stat5a is mandatory for adult mammary gland development and lactogenesis, Genes Dev. 11 (2) (1997) 179–186. [DOI] [PubMed] [Google Scholar]
- [302].Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng C-X, Robinson GW, Hennighausen L, Inactivation of Stat5 in Mouse Mammary Epithelium during Pregnancy Reveals Distinct Functions in Cell Proliferation, Surviv., Differ 24 (18) (2004) 8037–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [303].Wagner K-U, Rui H, Jak2/Stat5 Signaling in Mammogenesis, Breast Cancer Initiation and Progression, J. Mammary Gland Biol. Neoplasia 13 (1) (2008) 93–103. [DOI] [PubMed] [Google Scholar]
- [304].Schmidt JW, Wehde BL, Sakamoto K, Triplett AA, Anderson SM, Tsichlis PN, Leone G, Wagner K-U, Stat5 Regul. Phosphatidylinositol 3-Kinase/Akt1 Pathw. Mammary Gland Dev. Tumor 34 (7) (2014) 1363–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [305].Alsheikh HAM, Metge BJ, Pruitt HC, Kammerud SC, Chen D, Wei S, Shevde LA, Samant RS, Disruption of STAT5A and NMI signaling axis leads to ISG20-driven metastatic mammary tumors, Oncogenesis 10 (6) (2021) 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [306].Zeng Y, Min L, Han Y, Meng L, Liu C, Xie Y, Dong B, Wang L, Jiang B, Xu H, Zhuang Q, Zhao C, Qu L, Shou C, Inhibition of STAT5a by Naa10p contributes to decreased breast cancer metastasis, Carcinogenesis 35 (10) (2014) 2244–2253. [DOI] [PubMed] [Google Scholar]
- [307].Li D, Tang J, Gao R, Lan J, Shen W, Liu Y, Chen Y, Sun H, Yan J, Nie Y, Luo N, PFKFB4 promotes angiogenesis via IL-6/STAT5A/P-STAT5 signaling in breast cancer, J. Cancer 13 (1) (2022) 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [308].Cerliani JP, Guillardoy T, Giulianelli S, Vaque JP, Gutkind JS, Vanzulli SI, Martins R, Zeitlin E, Lamb CA, Lanari C, Interaction between FGFR-2, STAT5, and Progesterone Receptors in Breast Cancer, Cancer Res. 71 (10) (2011) 3720–3731. [DOI] [PubMed] [Google Scholar]
- [309].Ikeda O, Mizushima A, Sekine Y, Yamamoto C, Muromoto R, Nanbo A, Oritani K, Yoshimura A, Matsuda T, Involv. STAP-2 Brk-Mediat. Phosphorylation Act. STAT5 Breast Cancer Cells 102 (4) (2011) 756–761. [DOI] [PubMed] [Google Scholar]
- [310].Grinman DY, Boras-Granic K, Takyar FM, Dann P, Hens JR, Marmol C, Lee J, Choi J, Chodosh LA, Sola MEG, Wysolmerski JJ, PTHrP induces STAT5 activation, secretory differentiation and accelerates mammary tumor development, Breast Cancer Res. 24 (1) (2022) 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [311].Wang J, Rouse C, Jasper JS, Pendergast AM, ABL kinases promote breast cancer osteolytic metastasis by modulating tumor-bone interactions through TAZ and STAT5 signaling, 9(413), 2016: ra12–ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [312].Tworoger SS, Eliassen AH, Rosner B, Sluss P, Hankinson SE, Plasma prolactin concentrations and risk of postmenopausal breast cancer, Cancer Res. 64 (18) (2004) 6814–6819. [DOI] [PubMed] [Google Scholar]
- [313].Perotti C, Liu R, Parusel CT, Bӧcher N, Schultz J, Bork P, Pfitzner E, Groner B, Shemanko CS, Heat shock protein-90-alpha, a prolactin-STAT5 target gene identified in breast cancer cells, is involved in apoptosis regulation, Breast Cancer Res. 10 (6) (2008) R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].Schauwecker SM, Kim JJ, Licht JD, Clevenger CV, Histone H1 and Chromosomal Protein HMGN2 Regulate Prolactin-induced STAT5 Transcription Factor Recruitment and Function in Breast Cancer Cells*, J. Biol. Chem. 292 (6) (2017) 2237–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Yang N, Liu C, Peck AR, Girondo MA, Yanac AF, Tran TH, Utama FE, Tanaka T, Freydin B, Chervoneva I, Hyslop T, Kovatich AJ, Hooke JA, Shriver CD, Rui H, Prolactin-Stat5 signaling in breast cancer is potently disrupted by acidosis within the tumor microenvironment, Breast Cancer Res. 15 (5) (2013) R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Johnson KJ, Peck AR, Liu C, Tran TH, Utama FE, Sjolund AB, Schaber JD, Witkiewicz AK, Rui H, PTP1B suppresses prolactin activation of Stat5 in breast cancer cells, Am. J. Pathol. 177 (6) (2010) 2971–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [317].Nevalainen MT, Xie J, Torhorst J, Bubendorf L, Haas P, Kononen J, Sauter G, Rui H, Signal transducer and activator of transcription-5 activation and breast cancer prognosis, J. Clin. Oncol.: Off. J. Am. Soc. Clin. Oncol. 22 (11) (2004) 2053–2060. [DOI] [PubMed] [Google Scholar]
- [318].Sultan AS, Xie J, LeBaron MJ, Ealley EL, Nevalainen MT, Rui H, Stat5 promotes homotypic adhesion and inhibits invasive characteristics of human breast cancer cells, Oncogene 24 (5) (2005) 746–760. [DOI] [PubMed] [Google Scholar]
- [319].Jesser EA, Brady NJ, Huggins DN, Witschen PM, O’Connor CH, Schwertfeger KL, STAT5 is activated in macrophages by breast cancer cell-derived factors and regulates macrophage function in the tumor microenvironment, Breast Cancer Res. 23 (1) (2021) 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [320].Woock AE, Grible JM, Olex AL, Harrell JC, Zot P, Idowu M, Clevenger CV, Serine residues 726 and 780 have nonredundant roles regulating STAT5a activity in luminal breast cancer, Sci. Rep. 11 (1) (2021) 13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [321].Yamashita H, Iwase H, Toyama T, Fujii Y, Naturally occurring dominant-negative Stat5 suppresses transcriptional activity of estrogen receptors and induces apoptosis in T47D breast cancer cells, Oncogene 22 (11) (2003) 1638–1652. [DOI] [PubMed] [Google Scholar]
- [322].Li Z, Chen C, Chen L, Hu D, Yang X, Zhuo W, Chen Y, Yang J, Zhou Y, Mao M, Zhang X, Xu L, Ju S, Shen J, Wang Q, Dong M, Xie S, Wei Q, Jia Y, Zhou J, Wang L, STAT5a Confers Doxorubicin Resistance to Breast Cancer by Regulating ABCB1, Front. Oncol. 11 (2021), 697950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [323].DiScala M, Najor MS, Yung T, Morgan D, Abukhdeir AM, Cobleigh MA, Loss of STAT6 leads to anchorage-independent growth and trastuzumab resistance in HER2+ breast cancer cells, PloS One 15 (6) (2020), e0234146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [324].Walker SR, Nelson EA, Zou L, Chaudhury M, Signoretti S, Richardson A, Frank DA, Reciprocal effects of STAT5 and STAT3 in breast cancer, Mol. Cancer Res.: MCR 7 (6) (2009) 966–976. [DOI] [PubMed] [Google Scholar]
- [325].Halim CE, Deng S, Ong MS, Yap CT, Involvement of STAT5 in Oncogenesis, Biomedicines 8 (9) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [326].Walker SR, Xiang M, Frank DA, Distinct roles of STAT3 and STAT5 in the pathogenesis and targeted therapy of breast cancer, Mol. Cell. Endocrinol. 382 (1) (2014) 616–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [327].Clarkson RWE, Boland MP, Kritikou EA, Lee JM, Freeman TC, Tiffen PG, Watson CJ, The Genes Induced by Signal Transducer and Activators of Transcription (STAT)3 and STAT5 in Mammary Epithelial Cells Define the Roles of these STATs in Mammary Development, Mol. Endocrinol. 20 (3) (2006) 675–685. [DOI] [PubMed] [Google Scholar]
- [328].Dakir el H, Pickard A, Srivastava K, McCrudden CM, Gross SR, Lloyd S, Zhang SD, Margariti A, Morgan R, Rudland PS, El-Tanani M, The anti-psychotic drug pimozide is a novel chemotherapeutic for breast cancer, Oncotarget 9 (79) (2018) 34889–34910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [329].Strobl JS, Kirkwood KL, Lantz TK, Lewine MA, Peterson VA, Worley JF 3rd, Inhibition of human breast cancer cell proliferation in tissue culture by the neuroleptic agents pimozide and thioridazine, Cancer Res. 50 (17) (1990) 5399–5405. [PubMed] [Google Scholar]
- [330].Britschgi A, Andraos R, Brinkhaus H, Klebba I, Romanet V, Müller U, Murakami M, Radimerski T, Bentires-Alj M, JAK2/STAT5 Inhibition Circumvents Resistance to PI3K/mTOR Blockade: A Rationale for Cotargeting These Pathways in Metastatic Breast Cancer, Cancer Cell 22 (6) (2012) 796–811. [DOI] [PubMed] [Google Scholar]
- [331].Boothby M, Gravallese E, Liou H-C, Glimcher LH, DNA A, Binding Protein Regulated by IL-4 and by Differentiation in B Cells, Sci. (N. Y., N. Y. ) 242 (4885) (1988) 1559–1562. [DOI] [PubMed] [Google Scholar]
- [332].Hebenstreit D, Wirnsberger G, Horejs-Hoeck J, Duschl A, Signaling mechanisms, interaction partners, and target genes of STAT6, Cytokine Growth Factor Rev. 17 (3) (2006) 173–188. [DOI] [PubMed] [Google Scholar]
- [333].Kotanides H, Reich NC, Requirement of Tyrosine Phosphorylation for Rapid Activation of a DNA Binding Factor by IL-4, Sci. (N. Y., N. Y. ) 262 (5137) (1993) 1265–1267. [DOI] [PubMed] [Google Scholar]
- [334].Kӧhler I, Rieber EP, Allergy-associated Iϵ and Fcϵ receptor II (CD23b) genes activated via binding of an interleukin-4-induced transcription factor to a novel responsive element, 23(12), 1993: 3066–3071. [DOI] [PubMed] [Google Scholar]
- [335].Hou J, Schindler U, Henzel WJ, Ho TC, Brasseur M, McKnight SL, Inter. −4- Induc. Transcr. Factor.: IL-4 Stat. 265 (5179) (1994) 1701–1706. [DOI] [PubMed] [Google Scholar]
- [336].Brown KD, Zurawski SM, Mosmann TR, Zurawski G, A family of small inducible proteins secreted by leukocytes are members of a new superfamily that includes leukocyte and fibroblast-derived inflammatory agents, growth factors, and indicators of various activation processes, J. Immunol. (Baltim., Md.: 1950) 142 (2) (1989) 679–687. [PubMed] [Google Scholar]
- [337].Hershey GK, IL-13 receptors and signaling pathways: an evolving web, J. Allergy Clin. Immunol. 111 (4) (2003) 677–690. [DOI] [PubMed] [Google Scholar]
- [338].Gordon S, Alternative activation of macrophages, Nat. Rev. Immunol. 3 (1) (2003) 23–35. [DOI] [PubMed] [Google Scholar]
- [339].Ansel KM, Djuretic I, Tanasa B, Rao A, REGULATION OF TH2 DIFFERENTIATION AND Il4 LOCUS ACCESSIBILITY, 24(1) (2006) 607–656. [DOI] [PubMed] [Google Scholar]
- [340].Li J, Rodriguez JP, Niu F, Pu M, Wang J, Hung L-W, Shao Q, Zhu Y, Ding W, Liu Y, Da Y, Yao Z, Yang J, Zhao Y, Wei G-H, Cheng G, Liu Z-J, Ouyang S, Struct. basis DNA Recognit. STAT6 113 (46) (2016) 13015–13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [341].Wei M, Liu B, Gu Q, Su L, Yu Y, Zhu Z, Stat6 cooperates with Sp1 in controlling breast cancer cell proliferation by modulating the expression of p21 (Cip1/WAF1) and p27 (Kip1), Cell. Oncol. (Dordr. ) 36 (1) (2013) 79–93. [DOI] [PubMed] [Google Scholar]
- [342].Zhang WJ, Li BH, Yang XZ, Li PD, Yuan Q, Liu XH, Xu SB, Zhang Y, Yuan J, Gerhard GS, Masker KK, Dong C, Koltun WA, Chorney MJ, IL-4-induced Stat6 activities affect apoptosis and gene expression in breast cancer cells, Cytokine 42 (1) (2008) 39–47. [DOI] [PubMed] [Google Scholar]
- [343].Porter HA, Perry A, Kingsley C, Tran NL, Keegan AD, IRS1 is highly expressed in localized breast tumors and regulates the sensitivity of breast cancer cells to chemotherapy, while IRS2 is highly expressed in invasive breast tumors, Cancer Lett. 338 (2) (2013) 239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [344].Ostrand-Rosenberg S, Grusby MJ, Clements VK, Cutting edge: STAT6-deficient mice have enhanced tumor immunity to primary and metastatic mammary carcinoma, J. Immunol. (Baltim., Md.: 1950) 165 (11) (2000) 6015–6019. [DOI] [PubMed] [Google Scholar]
- [345].Papageorgis P, Ozturk S, Lambert AW, Neophytou CM, Tzatsos A, Wong CK, Thiagalingam S, Constantinou AI, Targeting IL13Ralpha2 activates STAT6-TP63 pathway to suppress breast cancer lung metastasis, Breast Cancer Res. 17 (1) (2015) 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [346].Gooch JL, Christy B, Yee D, STAT6 mediates interleukin-4 growth inhibition in human breast cancer cells, Neoplasia (N. Y., N. Y. ) 4 (4) (2002) 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [347].Yan C, Chen Y, Kong W, Fu L, Liu Y, Yao Q, Yuan Y, PVT1-derived miR-1207–5p promotes breast cancer cell growth by targeting STAT6, 108(5), 2017: 868–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [348].Cobleigh MA, DiScala M, Najor MS, Yung T, Abukhdeir AM, STAT6 expression and trastuzumab resistance in HER2 + breast cancer, 38(15_suppl), 2020: e13006–e13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [349].Wang N, Liang H, Zen K, Molecular mechanisms that influence the macrophage m1-m2 polarization balance, Front. Immunol. 5 (2014) 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [350].Yu T, Gan S, Zhu Q, Dai D, Li N, Wang H, Chen X, Hou D, Wang Y, Pan Q, Xu J, Zhang X, Liu J, Pei S, Peng C, Wu P, Romano S, Mao C, Huang M, Zhu X, Shen K, Qin J, Xiao Y, Modulation of M2 macrophage polarization by the crosstalk between Stat6 and Trim24, Nat. Commun. 10 (1) (2019) 4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [351].Sica A, Mantovani A, Macrophage plasticity and polarization: in vivo veritas, J. Clin. Investig. 122 (3) (2012) 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [352].Pauleau AL, Rutschman R, Lang R, Pernis A, Watowich SS, Murray PJ, Enhancer-mediated control of macrophage-specific arginase I expression, J. Immunol. (Baltim., Md.: 1950) 172 (12) (2004) 7565–7573. [DOI] [PubMed] [Google Scholar]
- [353].Binnemars-Postma K, Bansal R, Storm G, Prakash J, Targeting the Stat6 pathway in tumor-associated macrophages reduces tumor growth and metastatic niche formation in breast cancer, 32(2), 2018: 969–978. [DOI] [PubMed] [Google Scholar]
- [354].Rahal OM, Wolfe AR, Mandal PK, Larson R, Tin S, Jimenez C, Zhang D, Horton J, Reuben JM, McMurray JS, Woodward WA, Blocking Interleukin (IL)4- and IL13-Mediated Phosphorylation of STAT6 (Tyr641) Decreases M2 Polarization of Macrophages and Protects Against Macrophage-Mediated Radioresistance of Inflammatory Breast Cancer, Int. J. Radiat. Oncol., Biol., Phys. 100 (4) (2018) 1034–1043. [DOI] [PubMed] [Google Scholar]
- [355].Nagashima S, Yokota M, Nakai E, Kuromitsu S, Ohga K, Takeuchi M, Tsukamoto S, Ohta M, Synthesis and evaluation of 2-{[2-(4-hydroxyphenyl)-ethyl]amino}pyrimidine-5-carboxamide derivatives as novel STAT6 inhibitors, Bioorg. Med. Chem. 15 (2) (2007) 1044–1055. [DOI] [PubMed] [Google Scholar]
- [356].Nagashima S, Nagata H, Iwata M, Yokota M, Moritomo H, Orita M, Kuromitsu S, Koakutsu A, Ohga K, Takeuchi M, Ohta M, Tsukamoto S, Identification of 4-benzylamino-2-[(4-morpholin-4-ylphenyl)amino]pyrimidine-5-carboxamide derivatives as potent and orally bioavailable STAT6 inhibitors, Bioorg. Med. Chem. 16 (13) (2008) 6509–6521. [DOI] [PubMed] [Google Scholar]
- [357].Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK, Intrinsic disorder in transcription factors, Biochemistry 45 (22) (2006) 6873–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [358].Ruiu R, Di Lorenzo A, Cavallo F, Conti L, Are Cancer Stem Cells a Suitable Target Breast Cancer Immunother.? 12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [359].Stover DG, Gil Del Alcazar CR, Brock J, Guo H, Overmoyer B, Balko J, Xu Q, Bardia A, Tolaney SM, Gelman R, Lloyd M, Wang Y, Xu Y, Michor F, Wang V, Winer EP, Polyak K, Lin NU, Phase II study of ruxolitinib, a selective JAK1/2 inhibitor, in patients with metastatic triple-negative breast cancer, NPJ Breast Cancer 4 (2018) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [360].Verstovsek S, Mesa RA, Gotlib J, Levy RS, Gupta V, DiPersio JF, Catalano JV, Deininger M, Miller C, Silver RT, Talpaz M, Winton EF, Harvey JH Jr., Arcasoy MO, Hexner E, Lyons RM, Paquette R, Raza A, Vaddi K, Erickson-Viitanen S, Koumenis IL, Sun W, Sandor V, Kantarjian HM, A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis, N. Engl. J. Med. 366 (9) (2012) 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [361].Harrison C, Kiladjian JJ, Al-Ali HK, Gisslinger H, Waltzman R, Stalbovskaya V, McQuitty M, Hunter DS, Levy R, Knoops L, Cervantes F, Vannucchi AM, Barbui T, Barosi G, JAK inhibition with ruxolitinib versus best available therapy for myelofibrosis, N. Engl. J. Med. 366 (9) (2012) 787–798. [DOI] [PubMed] [Google Scholar]
- [362].Huo X, Li J, Zhao F, Ren D, Ahmad R, Yuan X, Du F, Zhao J, The role of capecitabine-based neoadjuvant and adjuvant chemotherapy in early-stage triple-negative breast cancer: a systematic review and meta-analysis, BMC Cancer 21 (1) (2021) 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [363].O’Shaughnessy J, DeMichele A, Ma CX, Richards P, Yardley DA, Wright GS, Kalinsky K, Steis R, Diab S, Kennealey G, Geschwindt R, Jiang W, Rugo HS, A randomized, double-blind, phase 2 study of ruxolitinib or placebo in combination with capecitabine in patients with advanced HER2-negative breast cancer and elevated C-reactive protein, a marker of systemic inflammation, Breast Cancer Res Treat. 170 (3) (2018) 547–557. [DOI] [PubMed] [Google Scholar]
- [364].Lynce F, Williams JT, Regan MM, Bunnell CA, Freedman RA, Tolaney SM, Chen WY, Mayer EL, Partridge AH, Winer EP, Overmoyer B, Phase I study of JAK1/2 inhibitor ruxolitinib with weekly paclitaxel for the treatment of HER2-negative metastatic breast cancer, Cancer Chemother. Pharmacol. 87 (5) (2021) 673–679. [DOI] [PubMed] [Google Scholar]
- [365].Kearney M, Franks L, Lee S, Tiersten A, Makower DF, Cigler T, Mundi P, Chi D-C, Goel A, Klein P, Andreopoulou E, Sparano J, Trivedi M, Accordino M, Califano A, Hershman DL, Silva J, Kalinsky K, Phase I/II trial of ruxolitinib in combination with trastuzumab in metastatic HER2 positive breast cancer, Breast Cancer Res. Treat. 189 (1) (2021) 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [366].Zhao Q, Bi Y, Zhong J, Li X, Guo J, Liu YX, Pan LR, Tan Y, Deng ZS, Yu XJ, 10,11-dehydrocurvularin exerts antitumor effect against human breast cancer by suppressing STAT3 activation, Acta Pharmacol. Sin. 42 (5) (2021) 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [367].Page BD, Fletcher S, Yue P, Li Z, Zhang X, Sharmeen S, Datti A, Wrana JL, Trudel S, Schimmer AD, Turkson J, Gunning PT, Identification of a non-phosphorylated, cell permeable, small molecule ligand for the Stat3 SH2 domain, Bioorg. Med. Chem. Lett. 21 (18) (2011) 5605–5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [368].Liu Z, Ge X, Gu Y, Huang Y, Liu H, Yu M, Liu Y, Small molecule STAT3 inhibitor, 6Br-6a suppresses breast cancer growth in vitro and in vivo, Biomed. Pharmacother. = Biomedecine Pharmacother. 121 (2020), 109502. [DOI] [PubMed] [Google Scholar]
- [369].Jang H, Ko H, Song K, Kim YS, Sesquiterpenoid A from Farfarae Flos Induces Apoptosis of MDA-MB-231 Human Breast Cancer Cells through Inhibition of JAK–STAT3 Signaling, 9(7), 2019: 278.A. Sesquiterpenoid Farfarae Flos Induces Apoptosis MDA-MB-231 Hum. Breast Cancer Cells Inhib. JAK–STAT3 Signal. 9 7 2019. 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [370].Cui L, Bu W, Song J, Feng L, Xu T, Liu D, Ding W, Wang J, Li C, Ma B, Luo Y, Jiang Z, Wang C, Chen J, Hou J, Yan H, Yang L, Jia X, Apoptosis induction by alantolactone in breast cancer MDA-MB-231 cells through reactive oxygen species-mediated mitochondrion-dependent pathway, Arch. pharmacal Res. 41 (3) (2018) 299–313. [DOI] [PubMed] [Google Scholar]
- [371].Tian J, Chen X, Fu S, Zhang R, Pan L, Cao Y, Wu X, Xiao H, Lin HJ, Lo HW, Zhang Y, Lin J, Bazedoxifene is a novel IL-6/GP130 inhibitor for treating triple-negative breast cancer, Breast Cancer Res Treat. 175 (3) (2019) 553–566. [DOI] [PubMed] [Google Scholar]
- [372].Zeng AQ, Yu Y, Yao YQ, Yang FF, Liao M, Song LJ, Li YL, Yu Y, Li YJ, Deng YL, Yang SP, Zeng CJ, Liu P, Xie YM, Yang JL, Zhang YW, Ye TH, Wei YQ, Betulinic acid impairs metastasis and reduces immunosuppressive cells in breast cancer models, Oncotarget 9 (3) (2018) 3794–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [373].Sarma P, Ramaiah MJ, Pal D, Bhadra U, Pal M, Bhadra A novel bisindole-PBD conjugate inhibits angiogenesis by regulating STAT3 and VEGF in breast cancer cells, Life Sci. 151 (2016) 264–276. [DOI] [PubMed] [Google Scholar]
- [374].Zhang X, Yue P, Page BD, Li T, Zhao W, Namanja AT, Paladino D, Zhao J, Chen Y, Gunning PT, Turkson J, Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts, Proc. Natl. Acad. Sci. USA 109 (24) (2012) 9623–9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [375].Shou LM, Zhang QY, Li W, Xie X, Chen K, Lian L, Li ZY, Gong FR, Dai KS, Mao YX, Tao M, Cantharidin and norcantharidin inhibit the ability of MCF-7 cells to adhere to platelets via protein kinase C pathway-dependent downregulation of α2 integrin, Oncol. Rep. 30 (3) (2013) 1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [376].Gu XD, Xu LL, Zhao H, Gu JZ, Xie XH, Cantharidin suppressed breast cancer MDA-MB-231 cell growth and migration by inhibiting MAPK signaling pathway, Braz. J. Med. Biol. Res. = Rev. Bras. De. Pesqui. Med. e Biol. 50 (7) (2017), e5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [377].Li HC, Xia ZH, Chen YF, Yang F, Feng W, Cai H, Mei Y, Jiang YM, Xu K, Feng DX, Cantharidin Inhibits the Growth of Triple-Negative Breast Cancer Cells by Suppressing Autophagy and Inducing Apoptosis in Vitro and in Vivo, Cell. Physiol. Biochem.: Int. J. Exp. Cell. Physiol., Biochem., Pharmacol. 43 (5) (2017) 1829–1840. [DOI] [PubMed] [Google Scholar]
- [378].Huang Q, Li S, Zhang L, Qiao X, Zhang Y, Zhao X, Xiao G, Li Z, CAPE-pNO2 Inhibited the Growth and Metastasis of Triple-Negative Breast Cancer via the EGFR/STAT3/Akt/E-Cadherin Signaling Pathway, 9, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [379].Vyas D, Lopez-Hisijos N, Shah P, Deshpande KS, Basson MD, Vyas A, Chaturvedi LS, Second-Generation A, Proteasome Inhibitor and Doxorubicin Modulates IL-6, pSTAT-3 and NF-kB Activity in MDA-MB-231 Breast Cancer Cells, J. Nanosci. Nanotechnol. 17 (1) (2017) 175–185. [DOI] [PubMed] [Google Scholar]
- [380].Choi HS, Kim JH, Kim SL, Deng HY, Lee D, Kim CS, Yun BS, Lee DS, Catechol derived from aronia juice through lactic acid bacteria fermentation inhibits breast cancer stem cell formation via modulation Stat3/IL-6 signaling pathway, Mol. Carcinog. 57 (11) (2018) 1467–1479. [DOI] [PubMed] [Google Scholar]
- [381].Ahmad R, Raina D, Meyer C, Kufe D, Triterpenoid CDDO-methyl ester inhibits the Janus-activated kinase-1 (JAK1)–>signal transducer and activator of transcription-3 (STAT3) pathway by direct inhibition of JAK1 and STAT3, Cancer Res. 68 (8) (2008) 2920–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [382].Zhang W, Guo J, Li S, Ma T, Xu D, Han C, Liu F, Yu W, Kong L, Discovery of monocarbonyl curcumin-BTP hybrids as STAT3 inhibitors for drug-sensitive and drug-resistant breast cancer therapy, Sci. Rep. 7 (2017) 46352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [383].Cai G, Yu W, Song D, Zhang W, Guo J, Zhu J, Ren Y, Kong L, Discovery of fluorescent coumarin-benzo[b]thiophene 1, 1-dioxide conjugates as mitochondria-targeting antitumor STAT3 inhibitors, Eur. J. Med. Chem. 174 (2019) 236–251. [DOI] [PubMed] [Google Scholar]
- [384].Turkson J, Zhang S, Palmer J, Kay H, Stanko J, Mora LB, Sebti S, Yu H, Jove R, Inhibition of constitutive signal transducer and activator of transcription 3 activation by novel platinum complexes with potent antitumor activity, Mol. Cancer Ther. 3 (12) (2005) 1533–1542. [PubMed] [Google Scholar]
- [385].Lan T, Wang L, Xu Q, Liu W, Jin H, Mao W, Wang X, Wang X, Growth inhibitory effect of Cucurbitacin E on breast cancer cells, Int J. Clin. Exp. Pathol. 6 (9) (2013) 1799–1805. [PMC free article] [PubMed] [Google Scholar]
- [386].Banik U, Parasuraman S, Adhikary AK, Othman NH, Curcumin: the spicy modulator of breast carcinogenesis, J. Exp. Clin. Cancer Res 36 (1) (2017) 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [387].Ma X, Ning S, Cyanidin-3-glucoside attenuates the angiogenesis of breast cancer via inhibiting STAT3/VEGF pathway, 33(1), 2019: 81–89. [DOI] [PubMed] [Google Scholar]
- [388].Mehta R, Katta H, Alimirah F, Patel R, Murillo G, Peng X, Muzzio M, Mehta RG, Deguelin action involves c-Met and EGFR signaling pathways in triple negative breast cancer cells, PloS One 8 (6) (2013), e65113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [389].Kim SL, Choi HS, Kim JH, Jeong DK, Kim KS, Lee DS, Dihydrotanshinone-Induced NOX5 Activation Inhibits Breast Cancer Stem Cell through the ROS/Stat3 Signaling Pathway, Oxid. Med. Cell. Longev. 2019 (2019) 9296439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [390].He J, Wei X, Li S, Quan X, Li R, Du H, Yuan S, Sun L, DT-13 suppresses breast cancer metastasis by modulating PLOD2 in the adipocytes microenvironment, Phytomedicine: Int. J. Phytother. Phytopharm. 59 (2019), 152778. [DOI] [PubMed] [Google Scholar]
- [391].Lou C, Chen Y, Zhang J, Yang B, Zhao H, Eupalinolide J Suppresses the Growth of Triple-Negative Breast Cancer Cells via Targeting STAT3 Signaling Pathway, 10, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [392].Shi W, Yan D, Zhao C, Xiao M, Wang Y, Ma H, Liu T, Qin H, Zhang C, Li C, Lin J, Li S, Lv J, Lin L, Inhibition of IL-6/STAT3 signaling in human cancer cells using Evista, Biochem. Biophys. Res. Commun. 491 (1) (2017) 159–165. [DOI] [PubMed] [Google Scholar]
- [393].Oh E, Kim YJ, An H, Sung D, Cho TM, Farrand L, Jang S, Seo JH, Kim JY, Flubendazole elicits anti-metastatic effects in triple-negative breast cancer via STAT3 inhibition, Int. J. Cancer 143 (8) (2018) 1978–1993. [DOI] [PubMed] [Google Scholar]
- [394].Ko H, Lee JH, Kim HS, Kim T, Han YT, Suh YG, Chun J, Kim YS, Ahn KS, Novel Galiellalactone Analogues Can Target STAT3 Phosphorylation and Cause Apoptosis in Triple-Negative Breast Cancer, Biomolecules 9 (5) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [395].Yang Y, Zhou H, Liu W, Wu J, Yue X, Wang J, Quan L, Liu H, Guo L, Wang Z, Lian X, Zhang Q, Ganoderic acid A exerts antitumor activity against MDA-MB-231 human breast cancer cells by inhibiting the Janus kinase 2/signal transducer and activator of transcription 3 signaling pathway, Oncol. Lett. 16 (5) (2018) 6515–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [396].Sharma S, Malhotra L, Yadav P, Mishra V, Sharma RS, Abdul E, Samath, Genistein: A novel inhibitor of IL-6/IL-6R interface of the Interleukin-6–mediated STAT3 dependent pathway of carcinogenesis, J. Mol. Struct. 1258 (2022), 132668. [Google Scholar]
- [397].Hutzen B, Friedman L, Sobo M, Lin L, Cen L, De Angelis S, Yamakoshi H, Shibata H, Iwabuchi Y, Lin J, Curcumin analogue GO-Y030 inhibits STAT3 activity and cell growth in breast and pancreatic carcinomas, Int. J. Oncol. 35 (4) (2009) 867–872. [DOI] [PubMed] [Google Scholar]
- [398].Chen H, Yang Z, Ding C, Chu L, Zhang Y, Terry K, Liu H, Shen Q, Zhou J, Fragment-based drug design and identification of HJC0123, a novel orally bioavailable STAT3 inhibitor for cancer therapy, Eur. J. Med. Chem. 62 (2013) 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [399].Chen H, Yang Z, Ding C, Xiong A, Wild C, Wang L, Ye N, Cai G, Flores RM, Ding Y, Shen Q, Zhou J, Discovery of potent anticancer agent HJC0416, an orally bioavailable small molecule inhibitor of signal transducer and activator of transcription 3 (STAT3), Eur. J. Med. Chem. 82 (2014) 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [400].Wang X, Zhang Y, Zhang X, Tian W, Feng W, Chen T, The curcumin analogue hydrazinocurcumin exhibits potent suppressive activity on carcinogenicity of breast cancer cells via STAT3 inhibition, Int. J. Oncol. 40 (4) (2012) 1189–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [401].Turkson J, Zhang S, Mora LB, Burns A, Sebti S, Jove R, A novel platinum compound inhibits constitutive Stat3 signaling and induces cell cycle arrest and apoptosis of malignant cells, J. Biol. Chem. 280 (38) (2005) 32979–32988. [DOI] [PubMed] [Google Scholar]
- [402].Chen W, Wang H, Cheng M, Ni L, Zou L, Yang Q, Cai X, Jiao B, Isoharringtonine inhibits breast cancer stem-like properties and STAT3 signaling, Biomed. Pharmacother. 103 (2018) 435–442. [DOI] [PubMed] [Google Scholar]
- [403].Lin L, Hutzen B, Li PK, Ball S, Zuo M, DeAngelis S, Foust E, Sobo M, Friedman L, Bhasin D, Cen L, Li C, Lin J, A novel small molecule, LLL12, inhibits STAT3 phosphorylation and activities and exhibits potent growth-suppressive activity in human cancer cells, Neoplasia (N. Y., N. Y. ) 12 (1) (2010) 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [404].Pan L, Chen X, Fu S, Yu W, Li C, Wang T, Lo HW, Lin J, LLY17, a novel small molecule STAT3 inhibitor induces apoptosis and suppresses cell migration and tumor growth in triple-negative breast cancer, Breast Cancer Res Treat. 181 (1) (2020) 31–41. [DOI] [PubMed] [Google Scholar]
- [405].Kim JE, Kim HS, Shin Y-J, Lee CS, Won C, Lee S-A, Lee JW, Kim Y, Kang J-S, Ye S-K, Chung M-H, LYR71, a derivative of trimeric resveratrol, inhibits tumorigenesis by blocking STAT3-mediated matrix metalloproteinase 9 expression, Exp. Mol. Med. 40 (5) (2008) 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [406].Liu X, Huang J, Xie Y, Zhou Y, Wang R, Lou J, Napabucasin Attenuates Resistance of Breast Cancer Cells to Tamoxifen by Reducing Stem Cell-Like Properties, Med. Sci. Monit.: Int. Med. J. Exp. Clin. Res. 25 (2019) 8905–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [407].Noori S, Rezaei Tavirani M, Deravi N, Mahboobi Rabbani MI, Zarghi A, Naringenin Enhances the Anti-Cancer Effect of Cyclophosphamide against MDA-MB-231 Breast Cancer Cells Via Targeting the STAT3 Signaling Pathway, Iran. J. Pharm. Res.: IJPR 19 (3) (2020) 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [408].Gyamfi J, Lee YH, Min BS, Choi J, Niclosamide reverses adipocyte induced epithelial-mesenchymal transition in breast cancer cells via suppression of the interleukin-6/STAT3 signalling axis, Sci. Rep. 9 (1) (2019) 11336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [409].Yang F, Hu M, Lei Q, Xia Y, Zhu Y, Song X, Li Y, Jie H, Liu C, Xiong Y, Zuo Z, Zeng A, Li Y, Yu L, Shen G, Wang D, Xie Y, Ye T, Wei Y, Nifuroxazide induces apoptosis and impairs pulmonary metastasis in breast cancer model, Cell death Dis. 6 (3) (2015), e1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [410].Li Y, Gan C, Zhang Y, Yu Y, Fan C, Deng Y, Zhang Q, Yu X, Zhang Y, Wang L, He F, Xie Y, Ye T, Yin W, Inhibition of Stat3 Signaling Pathway by Natural Product Pectolinarigenin Attenuates Breast Cancer Metastasis, Front Pharm. 10 (2019) 1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [411].Yamashita N, Kondo M, Zhao S, Li W, Koike K, Nemoto K, Kanno Y, Picrasidine G decreases viability of MDA-MB 468 EGFR-overexpressing triple-negative breast cancer cells through inhibition of EGFR/STAT3 signaling pathway, Bioorg. Med. Chem. Lett. 27 (11) (2017) 2608–2612. [DOI] [PubMed] [Google Scholar]
- [412].Chen D, Ma Y, Li P, Liu M, Fang Y, Zhang J, Zhang B, Hui Y, Yin Y, Piperlongumine Induces Apoptosis and Synergizes with Doxorubicin by Inhibiting the JAK2-STAT3 Pathway in Triple-Negative Breast Cancer, 24(12), 2019: 2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [413].Mandal PK, Gao F, Lu Z, Ren Z, Ramesh R, Birtwistle JS, Kaluarachchi KK, Chen X, Bast RC Jr., Liao WS, McMurray JS, Potent and selective phosphopeptide mimetic prodrugs targeted to the Src homology 2 (SH2) domain of signal transducer and activator of transcription 3, J. Med. Chem. 54 (10) (2011) 3549–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [414].Auzenne EJ, Klostergaard J, Mandal PK, Liao WS, Lu Z, Gao F, Bast RC Jr., F.M. Robertson, J.S. McMurray, A phosphopeptide mimetic prodrug targeting the SH2 domain of Stat3 inhibits tumor growth and angiogenesis, J. Exp. Ther. Oncol. 10 (2) (2012) 155–162. [PMC free article] [PubMed] [Google Scholar]
- [415].Khan MW, Saadalla A, Ewida AH, Al-Katranji K, Al-Saoudi G, Giaccone ZT, Gounari F, Zhang M, Frank DA, Khazaie K, The STAT3 inhibitor pyrimethamine displays anti-cancer and immune stimulatory effects in murine models of breast cancer, Cancer Immunol., Immunother.: CII 67 (1) (2018) 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [416].Kotha A, Sekharam M, Cilenti L, Siddiquee K, Khaled A, Zervos AS, Carter B, Turkson J, Jove R, Resveratrol inhibits Src and Stat3 signaling and induces the apoptosis of malignant cells containing activated Stat3 protein, Mol. Cancer Ther. 5 (3) (2006) 621–629. [DOI] [PubMed] [Google Scholar]
- [417].Kohandel Z, Farkhondeh T, Aschner M, Pourbagher-Shahri AM, Samarghandian S, STAT3 pathway as a molecular target for resveratrol in breast cancer treatment, Cancer Cell Int. 21 (1) (2021) 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [418].Borghouts C, Tittmann H, Delis N, Kirchenbauer M, Brill B, Groner B, The intracellular delivery of a recombinant peptide derived from the acidic domain of PIAS3 inhibits STAT3 transactivation and induces tumor cell death, Mol. Cancer Res.: MCR 8 (4) (2010) 539–553. [DOI] [PubMed] [Google Scholar]
- [419].Zhang X, Sun Y, Pireddu R, Yang H, Urlam MK, Lawrence HR, Guida WC, Lawrence NJ, Sebti SM, A novel inhibitor of STAT3 homodimerization selectively suppresses STAT3 activity and malignant transformation, Cancer Res. 73 (6) (2013) 1922–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [420].Hu Y, Yagüe E, Zhao J, Wang L, Bai J, Yang Q, Pan T, Zhao H, Liu J, Zhang J, Sabutoclax, pan-active BCL-2 protein family antagonist, overcomes drug resistance and eliminates cancer stem cells in breast cancer, Cancer Lett. 423 (2018) 47–59. [DOI] [PubMed] [Google Scholar]
- [421].Zhang ZL, Jiang QC, Wang SR, Schisandrin A reverses doxorubicin-resistant human breast cancer cell line by the inhibition of P65 and Stat3 phosphorylation, Breast Cancer (Tokyo, Jpn. ) 25 (2) (2018) 233–242. [DOI] [PubMed] [Google Scholar]
- [422].Hong SJ, Kim JT, Kim SJ, Cho NC, Kim K, Lee S, Suh YG, Cho KC, Kim KP, Surh YJ, An Electrophilic Deguelin Analogue Inhibits STAT3 Signaling in H-Ras-Transformed Human Mammary Epithelial Cells: The Cysteine 259 Residue as a Potential Target, Biomedicines 8 (10) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [423].Qiu HY, Fu JY, Yang MK, Han HW, Wang PF, Zhang YH, Lin HY, Tang CY, Qi JL, Yang RW, Wang XM, Zhu HL, Yang YH, Identification of new shikonin derivatives as STAT3 inhibitors, Biochem. Pharmacol. 146 (2017) 74–86. [DOI] [PubMed] [Google Scholar]
- [424].Zhao W, Jaganathan S, Turkson J, A cell-permeable Stat3 SH2 domain mimetic inhibits Stat3 activation and induces antitumor cell effects in vitro, J. Biol. Chem. 285 (46) (2010) 35855–35865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [425].Song H, Wang R, Wang S, Lin J, A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells, 102(13), 2005: 4700–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [426].Matsuno K, Masuda Y, Uehara Y, Sato H, Muroya A, Takahashi O, Yokotagawa T, Furuya T, Okawara T, Otsuka M, Ogo N, Ashizawa T, Oshita C, Tai S, Ishii H, Akiyama Y, Asai A, Identification of a New Series of STAT3 Inhibitors by Virtual Screening, ACS Med. Chem. Lett. 1 (8) (2010) 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [427].Liu A, Liu Y, Jin Z, Hu Q, Lin L, Jou D, Yang J, Xu Z, Wang H, Li C, Lin J, XZH-5 Inhibits STAT3 Phosphorylation and Enhances the Cytotoxicity of Chemotherapeutic Drugs in Human Breast and Pancreatic Cancer Cells, PloS One 7 (10) (2012), e46624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [428].Linher-Melville K, Nashed MG, Ungard RG, Haftchenary S, Rosa DA, Gunning PT, Singh G, Chronic Inhibition of STAT3/STAT5 in Treatment-Resistant Human Breast Cancer Cell Subtypes: Convergence on the ROS/SUMO Pathway and Its Effects on xCT Expression and System xc- Activity, PloS One 11 (8) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [429].Zhang X, Blaskovich MA, Forinash KD, Sebti SM, Withacnistin inhibits recruitment of STAT3 and STAT5 to growth factor and cytokine receptors and induces regression of breast tumours, Br. J. Cancer 111 (5) (2014) 894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.


