Abstract
SARS-CoV-2 viral load and detection of infectious virus in the respiratory tract are the two key parameters for estimating infectiousness. As shedding of infectious virus is required for onward transmission, understanding shedding characteristics is relevant for public health interventions. Viral shedding is influenced by biological characteristics of the virus, host factors and pre-existing immunity (previous infection or vaccination) of the infected individual. Although the process of human-to-human transmission is multifactorial, viral load substantially contributed to human-to-human transmission, with higher viral load posing a greater risk for onward transmission. Emerging SARS-CoV-2 variants of concern have further complicated the picture of virus shedding. As underlying immunity in the population through previous infection, vaccination or a combination of both has rapidly increased on a global scale after almost 3 years of the pandemic, viral shedding patterns have become more distinct from those of ancestral SARS-CoV-2. Understanding the factors and mechanisms that influence infectious virus shedding and the period during which individuals infected with SARS-CoV-2 are contagious is crucial to guide public health measures and limit transmission. Furthermore, diagnostic tools to demonstrate the presence of infectious virus from routine diagnostic specimens are needed.
Subject terms: SARS-CoV-2, Viral infection, Clinical microbiology
A better understanding of the transmission of SARS-CoV-2 is essential to inform public health measures. In this Review, Puhach, Meyer and Eckerle explore insights into what influences SARS-CoV-2 shedding, how this drives transmission and the tools available to measure this and determine infectiousness.
Introduction
At the end of 2019, a novel coronavirus emerged, later termed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is the causative agent of coronavirus disease 2019 (COVID-19). SARS-CoV-2 primarily targets multiciliated cells in the upper respiratory tract (URT), but was also reported to infect cells outside the URT1. It can spread to the lower respiratory tract (LRT), where it infects alveoli, leading to reduced gas exchange, inflammation and pulmonary pathologies that are typical of COVID-19 (ref.2). Individuals who are infected shed the virus through the URT, with emission of infectious virus leading to secondary transmission and thus further spread of the virus.
Because of their nonspecific clinical presentation, precise diagnostic tools are needed to identify SARS-CoV-2 infections. Specific real-time PCR (RT-PCR) assays were quickly available after the emergence of the virus, later followed by antigen-detecting (rapid) diagnostic tests (Ag-RDTs) and serological assays. Although detection of viral RNA in respiratory specimens by RT-PCR is highly sensitive and specific, it does not distinguish between replication-competent virus and residual RNA. In the absence of a diagnostic test, infectiousness is often established using one of two proxies: the presence of viral RNA above a defined cycle threshold (Ct) value, or a positive Ag-RDT. RT-PCR is a useful tool for initial diagnosis, whereas Ag-RDTs can serve as an indicator for ending the isolation period. This is because viral RNA (which would be picked up by RT-PCR) remains detectable in the absence of infectious virus, whereas positivity of Ag-RDTs better correlates with the presence of infectious virus.
Aside from the respiratory tract, SARS-CoV-2 RNA has been detected in peripheral blood, stool, urine and ocular secretions3–7. Virus isolation from non-respiratory specimens was unsuccessful in most studies4,8,9, with very few reported cases of infectious virus presence in non-respiratory specimens10–13. Furthermore, viral loads from respiratory tract samples were found to be much higher than from other materials, the latter often with RNA viral loads that are incompatible with the presence of infectious virus. Such specimens are not considered relevant for transmission and therefore, we concentrate on SARS-CoV-2 virus shedding only through the respiratory tract.
Here, we elucidate the relationship between SARS-CoV-2 viral load and infectious virus presence, the biological and host factors that determine infectious virus shedding, measurement of infectious virus and the role diagnostics can have as a proxy for infectious virus shedding.
Measuring SARS-CoV-2 viral load
The gold standard for laboratory diagnosis of a respiratory tract infection is demonstration of viral RNA with a virus-specific (semi-)quantitative RT-PCR from material collected from the respiratory tract. The most commonly used materials are swab specimens from the nasopharynx or oropharynx, but swabs of the nasal cavity, saliva or gargled liquid solution have also been suggested as alternative materials, with the advantage of being a less uncomfortable procedure for the participant. Viral load as determined by RT-PCR is either expressed as the number of viral RNA copies per millilitre of viral transport medium or per swab, or by the arbitrary test-specific Ct value. By contrast, infectiousness is determined by qualitative or quantitative assessment of infectious virus in a clinical specimen by replication of virus in cell culture. The limitations to measuring viral shedding are described in Box 1. In this Review, we refer to viral particles that can cause infection as infectious virus, and to viral RNA levels (which are widely used as surrogates for infectious virus) as viral load.
Box 1 Limitations to measuring viral load.
Specimen selection site
The anatomical site chosen to collect the swab specimen for detection of SARS-CoV-2 might influence viral load detection. Higher RNA viral load was reported from nasopharyngeal than oropharyngeal swabs28,124,181. As a result, nasopharyngeal samples show the highest diagnostic accuracy compared with other upper respiratory tract samples182. Similarly, higher virus isolation success was reported from nasopharyngeal swabs than from saliva, nasal or sublingual swabs124. However, another study found higher RNA viral loads in the throat and sputum than from nasal swabs30. Two studies indicate that virus can be detected earlier in the throat29 or saliva33, but reaches significantly higher levels and remains detectable for longer in the nose29,33. A meta-analysis, which evaluated different clinical sampling methods using nasopharyngeal swab as a reference, demonstrated that pooled nasal and throat swabs showed the best diagnostic performance183. Notably, this analysis revealed higher heterogeneity of results in studies using nasal or saliva specimens than using pooled nasal and throat swabs183.
The effect of the swabbing method (self-administered or performed by trained person) on measured viral loads cannot be overlooked: the sensitivity of antigen-detecting (rapid) diagnostic tests achieved by health-care professionals was higher than for self-tests57,184.
Impact of individual infection kinetics
To date, there is a limited number of studies that describe the longitudinal dynamics of SARS-CoV-2 shedding29,33. Most of the studies used only a single time point to collect respiratory swabs from individuals who were infected for measurement of viral load. As a result, different times from symptom onset can be a confounding factor when comparing viral load between different patients, which might also explain the variation of available data on viral load.
Influence of epidemic period
RNA viral loads across the specimens collected at single time points were found to indicate the trajectory of the epidemic, as a high proportion of individuals who were recently infected with low cycle threshold values correlates with a higher reproduction number, indicative of a growing epidemic185. Similarly, the rise and fall of RNA viral load correlated with the number of COVID-19 cases and hospital admissions across the population as it was identified using SARS-CoV-2 RNA measurement in wastewater samples186. Moreover, variation in estimates of the mean incubation period was shorter before the epidemic peak in China than after the peak187. Therefore, sampling at single time points can be biased by the epidemic period and might reflect more epidemiological dynamics than individual shedding kinetics.
Influence of SARS-CoV-2 variant of concern
Available data on SARS-CoV-2 variants of concern demonstrated that, although the overall pattern of viral load dynamics is conserved between the variants, infection with different SARS-CoV-2 variants of concern led to highly distinct infectious virus amounts and RNA viral loads25,86,87,90,92,94 and variations in SARS-CoV-2 incubation period113. Therefore, extrapolation of our understanding from shedding of current or earlier SARS-CoV-2 variants to newly emerged variants may be of only limited value.
Detection of infectious virus
The gold standard for determining the presence of infectious (that is, replication competent) virus in respiratory specimens is the recovery of virus in cell culture, a procedure that is commonly termed virus isolation (Fig. 1).
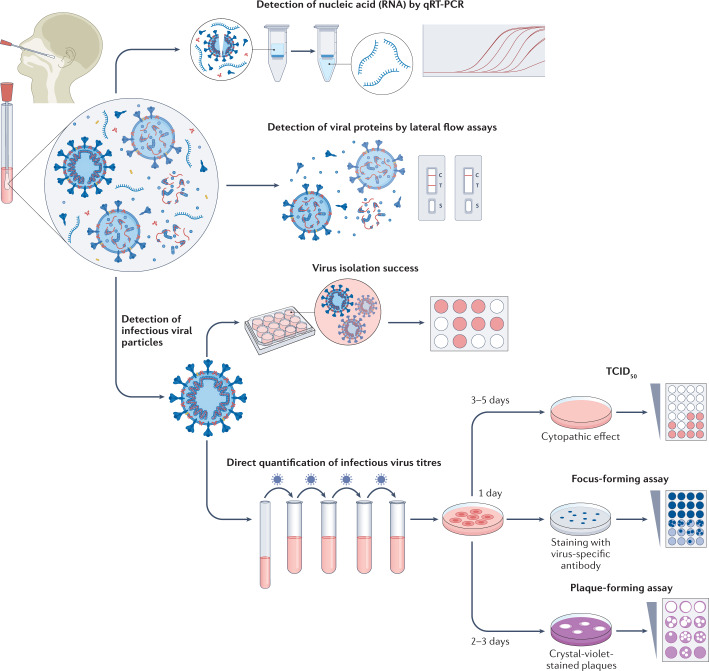
Fig. 1. Methods to measure infectious virus and RNA viral load.
Swab specimens from the nasopharynx or oropharynx are used for detection of SARS-CoV-2 viral loads. Detection of viral nucleic acids (RNA) is performed by quantitative real-time PCR (qRT-PCR). Viral RNA is extracted from lysed virus, reverse transcribed and amplified by qPCR using primers specific for one or more target regions in the viral genome. The amplification cycle at which samples cross the threshold (cycle threshold) defines the amount of viral RNA. RNA viral load can be expressed as the number of viral RNA copies per millilitre, or by the arbitrary test-specific cycle threshold value. Lateral flow assays detect the presence of specific viral proteins in the lysed viral particles. SARS-CoV-2 nucleocapsid is used in most antigen-detecting (rapid) diagnostic tests. The presence of infectious (replication-competent) virus in respiratory specimens can only be determined by the recovery of virus in cell culture by isolation or by quantification of infectious virus titres using 50% tissue culture infectious dose (TCID50), focus-forming assays or plaque-forming assays. Virus isolation is performed by applying infectious medium on the monolayer of cells; isolation success is determined by the presence of a cytopathic effect approximately 3–5 days post-infection. White colour indicates the presence of a cytopathic effect in cells. For quantification of infectious virus titres, serial dilutions of respiratory samples are performed and used for inoculation on the monolayer of cells. In TCID50, 3–5 days post-infection, viral-induced cytopathic effect is classically defined using microscopy. In focus-forming assays, cells are fixed 1 day post-infection and immunostaining with virus-specific antibodies is performed to detect groups of infected cells (foci). The foci, indicating the presence of infectious virus, are displayed in blue. In plaque-forming assays, plates are fixed 2–3 days post-infection and stained with crystal violet; wells with individual plaques are used to determine viral titres. The plaques, indicating the presence of infectious virus, are displayed in white.
In the case of SARS-CoV-2, various cell lines and primary cells can be used for virus isolation, including those that express angiotensin-converting enzyme 2 (ACE2; the receptor required for virus entry) or transmembrane protease 2 (TMPRSS2; which is also important for virus entry)14. A cell line derived from African green monkey kidney cells, Vero E6, is commonly used for virus isolation, propagation and titration15. Other human cell lines that have been successfully used for SARS-CoV-2 isolation are a colorectal adenocarcinoma cell line (Caco-2), a lung adenocarcinoma cell line (Calu-3), a lung adenocarcinoma cell line ectopically overexpressing ACE2 (A549) and a human hepatocellular carcinoma cell line (Huh7)16,17.
The presence of infectious virus in the cell culture is qualitatively assessed using light microscopy, which can be used to identify cells undergoing the cytopathic effects (and death) caused by SARS-CoV-2 infection, consisting of syncytium formation, cell rounding, detachment and degeneration17. Infection is usually confirmed by a second method, either by a specific RT-PCR for viral RNA from the supernatant of infected cells, indicating virus replication by an increase of viral load over time in comparison to the baseline sample, or by immunostaining for viral proteins15,18.
This qualitative measurement of virus presence cannot, however, quantify the infectious virions in the inoculated specimens, although samples with lower viral load commonly show delayed development of a cytopathic effect19. Instead, methods such as plaque assays, focus-forming assays or 50% tissue culture infectious dose (TCID50) can be used to quantify infectious virus in a patient sample.
The above methodologies are reliable tools to detect infectious virus in clinical specimens of individuals who are infected with SARS-CoV-2, although there are limitations. Detection of viable virus particles is highly influenced by the quality of the sample, and infectious viral particles can quickly lose their infectiousness in unsuitable storage conditions. To preserve infectious virus in specimens, swab samples from patients infected with SARS-CoV-2 should be immediately submerged in a viral transport medium suitable for cell culture and stored at −80 °C as early as possible after collection. Prolonged exposure to higher temperatures or repeated freeze–thaw cycles can drastically influence the quality of the sample, leading to potentially complete loss of infectious viral particles. Therefore, many factors can influence the reproducibility of the results between different laboratories. Furthermore, cell lines used for isolation can show a high variability between laboratories even when they are presumably the same. Consumables used during cell culture, such as culture medium or additives such as fetal bovine serum and antibiotics, could potentially also impact virus isolation success. In human primary airway epithelial cells, which mimic the primary site of entry in the human respiratory tract, the probability of isolating infectious virus was reduced compared with that of Vero E6 cells, indicating that infectious virus determined using Vero E6 cells might be overestimated for assessing transmission risks in vivo20.
Importantly, all cell culture work with SARS-CoV-2 is done under biosafety level 3 conditions, so only specially trained personnel in laboratories with advanced infrastructure can perform these experiments. Thus, detection of viable virus through virus isolation is not suitable for diagnostics and is restricted to research only.
Detection of RNA viral load
Techniques for detecting viral RNA by RT-PCR were quickly established at the beginning of the pandemic21,22 (Fig. 1). The high specificity and sensitivity of RT-PCR make it the gold standard for diagnosing SARS-CoV-2 infections. Quantitative RT-PCR assays provide a Ct value, which is inversely correlated with the concentration of the target viral RNA in the clinical sample (that is, the higher the value, the lower the target RNA in the sample). By using an external standard with a defined number of RNA copies, Ct values can be transformed into absolute viral RNA copy numbers or international units per millilitre of viral transport medium or per total swab.
Although RT-PCR cannot directly determine infectiousness owing to its inability to differentiate between replication-competent (infectious) virus and residual (non-infectious) viral RNA, a correlation between RNA viral load and the presence of infectious virus has been sought. Several studies have attempted to correlate the quantity of viral RNA with infectiousness by isolating virus across a range of Ct values. Indeed, there was a stepwise decrease in the probability of virus isolation with increasing Ct values in samples collected during the first 8 days post-onset of symptoms (dpos)18,23. However, other studies have found that the correlation between infectious virus and RNA viral load was low and that viral load (or Ct values as a proxy) is only a weak predictor of infectious virus presence in the first 5 dpos4,20,24,25. Furthermore, when taking a certain Ct value or RNA copy number as a threshold, it is not possible to determine whether the RNA viral load is increasing or already decreasing; therefore, a low viral load could be measured at the end of infection or in the early (pre-)symptomatic phase before reaching peak viral load.
In a routine diagnostic context, analytical sensitivity and limits of detection may vary between the tests and laboratories where they are applied. An analytical performance comparison between different RT-PCR assays showed variation between the measured Ct values and the detection rate26. Therefore, application of RNA standards and calculation of RNA genome copy number based on a standard curve can improve comparability between laboratories and assays. To facilitate easier calibration and control of nucleic acid amplification techniques, an international standard with assigned potency in the form of an inactivated SARS-CoV-2 isolate was introduced by the World Health Organization (WHO)27.
As with the detection of infectious virus, several other parameters can influence whether viral load can be detected. The site of specimen collection can impact the findings on viral load; although some studies report higher RNA viral load in nasal or nasopharyngeal swabs28,29, others show higher RNA viral load in throat samples30. Moreover, the transport media used for the sample, storage condition and quality of the sample may further influence the detection of viral RNA and their usefulness and limitations when extrapolating to potential infectiousness.
Although new variants have impacted some gene targets, in most instances, they did not have a major effect on molecular diagnostics, owing to the use of dual-target assays (in which at least two viral genes are detected simultaneously)31.
Antigen-detecting rapid diagnostic tests
Most lateral flow tests are designed to detect SARS-CoV-2 nucleocapsid protein, as a proxy for infectious virus, in nasal or nasopharyngeal swabs32–36 (Fig. 1). Indeed, most studies on Ag-RDT detection show good concordance with RT-PCR positivity when Ct values are below 25–30, a viral load compatible with the presence of infectious virus, whereas higher Ct values give less reliable results34,37–41.
Early time points during infection often give negative results with Ag-RDT in individuals who have tested positive by PCR29,42. On average, the first positive Ag-RDT results are obtained about 1–2 days later than positive PCR results37, whereas the highest sensitivity in patients was shown during the first 7 dpos in the studies with ancestral SARS-CoV-2 (refs.42–44). Antigen tests show highest sensitivity for specimens containing infectious virus and with Ct values below 25 (refs.45–49), and their positivity highly correlates with the presence of infectious virus34,45,47,50. By contrast, Ag-RDTs are less sensitive to low RNA viral loads (which have higher Ct values)51. Several studies have demonstrated a strong correlation between Ag-RDT positivity and the period in which infectious virus can be detected, indicating that Ag-RDTs can add an additional safety layer for deciding when to end isolation29,39.
However, some inconsistencies between studies and tests have been noted. For instance, there have been reports (across a range of studies and Ag-RDTs) of failure to detect viral antigens in specimens with a low Ct value and/or containing infectious virus (beyond the early acute phase)46,50. Moreover, there are seldom reports of Ag-RDTs remaining positive after more than 10 dpos42,46,50. As most studies failed to isolate infectious virus after more than 10 dpos, it remains unclear whether Ag-RDT positivity beyond 10 dpos correlates with infectious virus shedding. One study showed that antigen tests predict infectiousness more accurately at 1–5 dpos, than at 6–11 dpos52. Notably, there was a good correlation between Ag-RDT positivity and infectious virus isolation within the first 11 dpos52.
Conflicting results were found for sensitivity and specificity of Ag-RDTs for detection of SARS-CoV-2 variants, with large variations between manufacturers, the type of setting in which the Ag-RDTs were used (self-tests versus tests collected by a health-care professional) and the type of sample used for detection (nasal versus oral)53–57. With increasing hybrid immunity and the presence of mucosal antibodies, Ag-RDTs may further lose sensitivity58.
Viral load and shedding dynamics
Viral loads are used as a proxy to characterize infectious viral shedding. The exact time for which individuals remain infectious is laborious to estimate and is likely to vary between patients. Viral factors, such as viral variant, and host factors, such as patient age and sex and immune status, influence shedding dynamics.
Viral load as a key determinant of viral shedding
After the emergence of SARS-CoV-2 in late 2019, the first details on viral load and infectious virus shedding were measured in a cluster of infections that occurred in January 2020 in Germany, assessing nine immunocompetent individuals with a mild course of disease4. Peak RNA viral loads were reached in the early symptomatic period at 5 dpos, a finding that was confirmed by other studies reporting peak viral loads at the time of symptom onset or even shortly before4,7,28,59. RNA viral loads gradually declined over the course of the disease in the nasopharyngeal and throat swabs, reaching low or undetectable levels 2 weeks after symptom onset4,23,59,60 (Fig. 2). Declining RNA viral load is associated with resolution of clinical symptoms and gradual increase in antibody titres, for both binding and neutralizing antibodies18,23. However, ongoing detection of viral RNA has been described for prolonged periods up to 28 dpos in otherwise healthy individuals61, and some studies have reported low-level detection of RNA by RT-PCR even for months62. Participants who continue to shed viral RNA for more than 4 weeks after initial detection by RT-PCR represent a minority of non-severe cases, estimated to be around 3%63, 14%64 or less than 20%65.
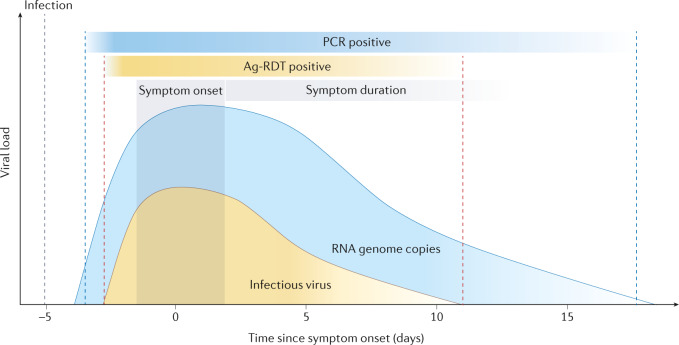
Fig. 2. Kinetics of RNA viral loads and infectious virus for ancestral SARS-CoV-2 in patients with mild-to-moderate disease.
According to different studies, the incubation period for ancestral SARS-CoV-2 was estimated to lie between 4.6 and 6.4 days. On average, symptoms continue to persist for 10 days. RNA can already be detected before the onset of symptoms; RNA levels peak around the onset of symptoms and then gradually decline. Median clearance for RNA viral load is 16 days post-onset of symptoms. Infectious virus titres are highest around symptom onset, and infectious virus can be isolated up to 8 or 10 days post-onset of symptoms. RNA can be detected for prolonged periods by real-time PCR, when infectious virus is no longer detectable, whereas virus detection by antigen-detecting (rapid) diagnostic tests (Ag-RDTs) was shown to be a better correlate for infectiousness. Gradients reflect variability between individuals (lighter shade towards the end of infection shows that viral loads continue to be detected in some but not all individuals). The grey dashed line marks the initial infection, the blue dashed lines mark the PCR-positive period and the red dashed lines mark Ag-RDT positivity. Details of the underlying studies used to generate Fig. 2 can be found in Supplementary Table 1.
Infectious virus shedding of the ancestral SARS-CoV-2 strain, as determined by virus isolation in cell culture, was reported to correlate with high RNA viral load in the early acute phase after symptom onset23. Importantly, daily longitudinal sampling of respiratory specimens from individuals with mild disease or asymptomatic infection revealed that infectious virus can already be detected before the onset of symptoms33. Successful infectious virus isolation was reported within the first 8–10 dpos, but culture probability after this time period rapidly declined4,7,23,29,66,67. Studies that assessed infectious virus quantitatively found that infectious virus titres declined over the first 10 dpos25,29. In addition, a reduced chance of virus isolation coincided with the time of seroconversion in hospitalized patients and, as a result, infectious virus could no longer be isolated from seroconverted patients with detectable antibody titres18,68,69. Although similar seroconversion studies performed on mildly symptomatic patients are missing, the number of immunologically naive individuals is declining and this broadly existing underlying immunity makes such an assessment more complex.
Most studies on infectious virus shedding in the acute symptomatic period were on immunocompetent patients that had mild-to-moderate disease, representing the majority of COVID-19 cases in the community. Therefore, the assessment of the presence of infectious virus in the URT from those studies was used to define the duration of the period of infectiousness and contributed to best public health practices for isolation and quarantine62,70. Although the pattern of infection is broadly similar in patients with mild and severe disease, key differences do exist. The first week of illness is comparable in terms of RNA viral load between patients with mild and severe disease. However, patients with severe disease have elevated RNA viral loads in the second week of illness, and RNA was detected for prolonged periods71. Moreover, infectious virus was recovered from hospitalized patients for prolonged periods of up to 32 dpos18,72,73; however, the median time from symptom onset to viral clearance in culture was similar to that of patients with mild or moderate disease18,73. Severe COVID-19 is also characterized by high and persistent RNA viral load in the LRT, whereas non-severe cases have similar viral loads in the URT and LRT74.
Prolonged detection of viral RNA was also reported in immunocompromised patients; for example, 224 days after the beginning of the infection, virus was still detected in a man infected with HIV, including the detection of subgenomic RNA (sgRNA) indicating active viral replication75. Also, infectious virus was recovered up to 61 dpos in nasopharyngeal swabs collected from immunocompromised patients76, and low RNA viral loads were still detected at 60 dpos in another study77. Infectious virus was isolated from bronchoalveolar fluids from patients receiving chimeric antigen receptor (CAR) T cell therapy up to 28 days after admission to an intensive care unit78. A case report on an immunocompromised patient showed isolation of infectious virus up to 78 dpos79. The reports of infectious virus isolation from severely ill or immunocompromised patients are limited (owing to the low number of patients), so it is difficult to define the proportion of cases with prolonged shedding.
The characteristics of viral shedding of other respiratory viruses are outlined in Box 2.
Box 2 Shedding of respiratory viruses.
The dynamics of viral shedding differs between respiratory viruses, which influences their transmission and has an effect on diagnostics and measures applied to contain the outbreaks.
SARS-CoV
The epidemic of severe acute respiratory syndrome coronavirus (SARS-CoV) started in November 2002 in the Guangdong province of China and rapidly spread outside China. The virus was airborne and could also be spread via droplets of saliva, but is only moderately transmissible among humans188. Only low viral loads were detected in the early symptomatic period, generally peaking in the upper respiratory tract (URT) around 10–14 days post-onset of symptoms (dpos)189,190, and then dropping to low levels at 3–4 weeks post-infection191. In patients infected with SARS-CoV, viral RNA was detectable for a maximum of 8 weeks in samples collected from the URT191 and for 52 days in sputum samples192, whereas infectious virus was isolated up to 28 dpos from stool and respiratory specimens and up to 36 dpos from urine samples191,193. SARS-CoV replicated less efficiently at low temperatures; thus, virus replication was more efficient in the lower respiratory tract (LRT) than in the URT194. Notably, asymptomatic or pre-symptomatic viral shedding and transmission were not recorded for SARS-CoV190,195; the peaks of transmission occurred around 2 and 10 dpos195. As a result, outbreaks were successfully contained through isolation of symptomatic patients infected with SARS-CoV, which reduced onward transmission196.
MERS-CoV
Middle East respiratory coronavirus (MERS-CoV) was isolated from a patient with pneumonia in Saudi Arabia in 2012 and was shown to be the causative agent of a cluster of severe respiratory tract infections in the Middle East197. The disease caused by MERS-CoV is characterized by a wide range of clinical severities and by predominantly respiratory symptoms, such as acute viral pneumonia, with a high case fatality ratio198. The virus is capable of airborne transmission and has low transmissibility among humans, with a maximum estimated reproduction number below 1 (ref.198). Higher RNA viral loads were detected in the LRT than in the URT. Estimated mean shedding duration is 15.3 days in the URT and 16.3 days in the LRT62. Prolonged PCR positivity and higher RNA viral loads in the URT and LRT were associated with increased disease severity62,199. Viral RNA was also detected in the urine, stool and serum200. One study reported detection of viral RNA in the blood for 34 days and showed that presence of viral RNA in the blood is associated with higher mortality201; however, another study failed to isolate virus from PCR-positive serum samples200.
Influenza virus
In symptomatic patients, RNA viral loads start to be detectable by real-time PCR 2 days before the onset of symptoms and peak at 1 dpos202. Human challenge trials with influenza A viruses show that viral loads already sharply increase at 1 day post-inoculation, reach a peak at 2 days post-inoculation and become undetectable at 8 days post-inoculation. The mean duration of viral shedding for influenza viruses is 4.8 days, and the maximum duration is between 6 and 7 days203,204. Kinetics of infectious viral titres were similar to the viral load trends detected by real-time PCR for different strains of influenza205. Lower RNA viral loads and shorter infectious viral shedding were noted in asymptomatic patients202.
Human respiratory syncytial virus
This virus is the most frequent causative agent of LRT infections, leading to morbidity and mortality particularly in young children and older adults206. The virus is transmitted by contact with nasal secretions or large aerosols. Viral loads and symptoms increased simultaneously, reaching a peak at 5.4 days207. In human challenge trials, respiratory syncytial virus titres were detectable for an average of 4.6 days. Viral RNA could be still detected up to 9 dpos, whereas infectious virus titres could be detected from 1 to 8 dpos in adults208 and up to 9 dpos in children209.
Viral shedding of SARS-CoV-2 variants
Viral evolution of SARS-CoV-2 over time has led to the emergence of numerous variants. Combined with increasing population immunity due to vaccination or natural infection, this has led to a need to reassess our knowledge of viral shedding patterns.
The WHO designated variants as variants of concern (VOCs) if they were associated with one or more of the following: elevated transmissibility or a detrimental change in COVID-19 epidemiology; increased virulence or a change in clinical disease presentation; or decreased effectiveness of public health measures or available diagnostics, vaccines or therapeutics80. To date, five VOCs are recognized: Alpha, Beta, Gamma, Delta and Omicron. In contrast to ancestral SARS-CoV-2, VOCs display some differences in evasion from immunity, viral loads, shedding period or even incubation period, resulting in drastically different levels of transmission81–85 (Fig. 3).
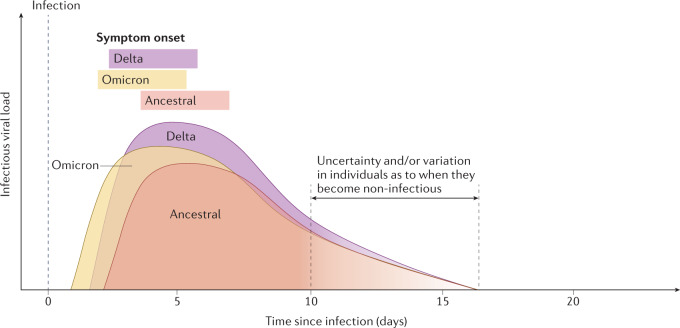
Fig. 3. Infectious viral load and symptom onset in SARS-CoV-2 Delta and Omicron BA.1 variants of concern.
Overall patterns of shedding dynamics are conserved between SARS-CoV-2 variants. In comparison to ancestral SARS-CoV-2, Delta and Omicron BA.1 have shorter incubation periods, estimated as approximately 3.7–4 days for Delta and approximately 3–3.4 days for Omicron BA.1. Higher infectious viral loads were detected in patients infected with Delta than in patients infected with Omicron BA.1 or ancestral SARS-CoV-2. Only a limited number of studies have determined when virus shedding for Delta and Omicron BA.1 ends, so this time point is not well defined. Owing to the low number of studies comparing the end of the infectious period between different SARS-CoV-2 variants of concern, the end point of infectivity is not well defined (shown as a colour gradient). Details of the underlying studies used to generate Fig. 3 can be found in Supplementary Table 2.
All VOCs have shown changes in viral load compared with ancestral SARS-CoV-2. One study reported that infection with Alpha leads to approximately tenfold higher RNA viral load and an increased probability of cell culture isolation compared with the ancestral virus86. However, another study did not find a substantial difference in the infectious virus titre between Alpha and ancestral SARS-CoV-2 (ref.33). Delta reportedly led to an even higher increase in RNA viral load: one study reported a 1,000× increase relative to the ancestral virus87, and other studies reported 1.7× (ref.88) or 6.2× higher89 viral load than Alpha. Furthermore, Delta demonstrated elevated probability of cell culture isolation90 and higher infectious virus titres than Alpha91. Although Omicron was shown to be highly transmissible, lower RNA viral loads92, lower cell culture isolation probability93 and lower infectious virus titres25 were observed in patients infected with Omicron BA.1 than in those infected with Delta. Even within the Omicron clade, there are differences between sub-lineages, with infection with Omicron BA.2 leading to higher levels of RNA viral loads and longer time to viral clearance than with Omicron BA.1 (refs.94–96).
Similarly, VOCs have shown differences in the duration of viral shedding. Analysis of Ct values in respiratory specimens found that Delta showed longer persistence of viral RNA than ancestral SARS-CoV-2 (ref.97). Another study demonstrated that there was not significant difference in the mean duration of viral RNA presence in Delta and Omicron BA.1 infections92. The duration of infectious virus shedding appears to be similar to that observed with ancestral SARS-CoV-2, with culturable virus obtained at 5 dpos85 and no replication-competent virus isolated beyond 10 dpos in patients infected with Delta and Omicron BA.1 (refs.84,98). It is important to note that pre-existing immunity to SARS-CoV-2, either from infection or vaccination, might influence the duration of infectious virus shedding (alongside immune status and disease severity, as discussed above), which may have driven some of these differences during the course of the pandemic.
Influence of age and sex on viral shedding
There is some evidence that age-associated and sex-associated differences in innate and adaptive immunity, as well as higher ACE2 expression in adults than in children, result in an increased risk for severe disease in older male patients99–101. Moreover, a few studies have found that age and sex influence viral loads and shedding dynamics. In cases of infection with ancestral SARS-CoV-2, resolution of RNA shedding was faster in participants <18 years of age and slower in participants >50 years of age61. According to one study, viral RNA can be detected for longer times in male patients infected with ancestral SARS-CoV-2 (ref.102), and RNA viral loads were elevated in male patients infected with either Alpha or Delta variants compared with female patients88. However, a possible association of viral load dynamics with age or sex is highly debated, as other studies demonstrated that they have no influence on infectious virus25 or RNA viral loads59.
Early studies in ancestral SARS-CoV-2 did not find a difference in virus isolation success103 or RNA viral loads between children and adults104–106, but sample sizes were small. Slightly lower RNA viral loads and a more rapid clearance of viral RNA was observed in children than in adults when analysing much larger cohorts, whereas the patterns of shedding curves over time were similar between children and adults107. Furthermore, large-scale analysis of viral loads across different age groups showed no differences of distribution of RNA viral load between children and adults108 or only slightly lower viral loads (<0.5 log10 units) in children <5 years of age86.
Symptoms as a correlate for shedding
One of the key epidemiological parameters for SARS-CoV-2 transmission is the incubation period, defined as the time from exposure or infection to the onset of symptoms. Studies on ancestral SARS-CoV-2 have estimated that the incubation period on average is between 4.6 and 6.4 days59,109–111 (Fig. 2). A human challenge trial with ancestral SARS-CoV-2 demonstrated that symptoms start to appear 2–4 days after inoculation, and RNA viral loads reach their peak 4–5 days after inoculation29. Thus, artificial inoculation of the virus confirmed the timing of peak viral loads observed in naturally infected individuals, whereas onset of symptoms was faster in the human challenge cases. In contrast to natural infection, in artificial inoculation, virus-containing drops with high viral load are directly applied in the nose and therefore reach the nasal epithelium more quickly, which might lead to the more rapid appearance of symptoms. For Delta, the estimated incubation period was between 3.7 and 4 days81–83,97, whereas infection with Omicron BA.1 was characterized by an even shorter incubation period of 3–3.4 days83,112,113 (Fig. 3). However, as the time point of infection is rarely known outside of human challenge trials, dpos is most commonly used when analysing viral load and infectious virus.
Considering that high viral loads can be detected in the URT of infected individuals regardless of their clinical manifestations, the presence of symptoms is an unreliable indicator of infectiousness. Notably, individuals infected with SARS-CoV-2 can be infectious before the onset of symptoms59, and it was estimated that about half of secondary transmissions take place in the pre-symptomatic phase59,114. Moreover, according to population surveys, asymptomatic cases represent around 40% of all SARS-CoV-2 infections with ancestral SARS-CoV-2 (refs.115–117), and tracing of close contacts of confirmed cases of SARS-CoV-2 found that up to 23% of infections were asymptomatic118.
There are conflicting findings regarding viral shedding differences in symptomatic and asymptomatic patients. Comparison of viral loads between symptomatic and asymptomatic patients remains challenging, as the time of exposure cannot be clearly identified in asymptomatic individuals, and dpos cannot be used when comparing viral loads with symptomatic individuals. Furthermore, individuals who do not show clinical symptoms at the time of testing can represent either true asymptomatic individuals or pre-symptomatic individuals who will develop symptoms later. Thus, only well-controlled studies with a follow-up of assessed individuals can make a clear distinction between pre-symptomatic and asymptomatic individuals. A study on ancestral SARS-CoV-2, which followed COVID-19 confirmed cases hospitalized for isolation and recorded symptoms daily, found similar initial Ct values between asymptomatic and symptomatic individuals119. Similarly, no significant difference in RNA viral loads between symptomatic and asymptomatic patients was found in other studies in which patients were followed longitudinally and the presence of symptoms was either monitored by health-care professionals120 or was self-reported115. By contrast, other studies, in which symptoms were also recorded by clinicians, reported lower RNA viral loads in asymptomatic participants121,122. In addition, one study found a faster clearance of viral RNA in asymptomatic than in symptomatic individuals123, and another recorded a longer median duration of viral RNA shedding among asymptomatic patients119.
There are limited data regarding the presence of infectious virus in asymptomatic patients. One study showed lower virus isolation success from asymptomatic patients124, but only a small number of patients were included. Therefore, more studies evaluating infectious virus in asymptomatic patients would help to elucidate the differences in their infectivity compared with symptomatic patients.
SARS-CoV-2 transmission
Viral loads have a key role in the SARS-CoV-2 transmission. As previously discussed, host (role of vaccination or previous infection) and viral factors (SARS-CoV-2 variants) greatly influence viral load dynamics and therefore further influence viral transmission.
Influence of viral load on transmission
SARS-CoV-2 can be transmitted via larger droplets and aerosols produced when breathing, speaking, sneezing or coughing and to a lesser extend also by contaminated surfaces. As an infection can only be induced by infectious viral particles and not by remnant RNA or protein alone, the presence of infectious SARS-CoV-2 is required for secondary transmission. Although transmission is a multifactorial process that is also influenced, for example, by environmental and behavioural factors (such as humidity, air quality, exposure time or closeness of contact), the viral load of SARS-CoV-2 in the URT is considered to be a proxy for transmission risk.
An epidemiological study that included viral load analysis found that viral load of an index case strongly correlates with onward transmission, with higher viral loads for ancestral SARS-CoV-2 presenting a greater secondary attack rate risk125. In this study, viral load was identified as the main driver of transmission, with a more pronounced effect in household settings than in non-household settings (hospitals and nursing homes, among others). Transmission probability peaks around symptom onset, when infectious virus titres are estimated to be the highest during the course of infection. As viral load decreases with time, the probability of transmission also gradually declines in cases of infection with ancestral SARS-CoV-2 (ref.126). On this note, a study of health-care workers infected with ancestral virus documented no transmission from index cases later than 6 dpos, which is in line with findings showing reduced virus isolation success towards the end of week 1 of symptomatic disease127.
However, there are limitations when using viral load of an index case as a proxy for transmission. To date, the infectious dose of SARS-CoV-2 required to lead to a secondary transmission is not yet known, and the association between presence of infectious virus in the respiratory tract and infectiousness of the same individuals is poorly understood. In the only available human challenge trial that was conducted with ancestral SARS-CoV-2, an initial infectious dose of 10 TCID50 did not lead to an infection in 16 of 36 participants29. Other factors, such as symptoms, type of contact, protective measures, vaccination status and other host factors may have an additionally strong effect on transmission128–133.
Viral load can markedly vary between individuals (as a result of individual susceptibility and of immunity from previous infections or vaccination), which leads to differences in their propensity to transmit the virus. Indeed, differences have been observed in the duration of infectious virus detection and in nasal and oral viral loads for both ancestral SARS-CoV-2 and Alpha33. Inter-individual variability was suggested to have a role in the observed heterogeneity of viral load dynamics, as some early immune signatures were significantly associated with higher oropharyngeal RNA viral loads in patients134. Therefore, observed heterogeneity between individuals has an important role in ongoing viral transmission33.
Such differences can lead to heterogeneity in virus transmission. Modelling with ancestral SARS-CoV-2 and Alpha estimated that individuals who are highly infectious, known as superspreaders, shed 57-fold more virus over the course of infection than those with lowest infectiousness33. By contrast, most patients with COVID-19 do not infect other individuals as they expel few to no viral particles from their airways135. Indeed, only a minority (about 8%) of patients positive for SARS-CoV-2 infected with ancestral SARS-CoV-2 or Alpha have significantly higher infectious virus titres than the rest of the population (as shown in a study measuring virus isolation probability in a large cohort of patients)86. Moreover, only 15%114 to 19%136 of individuals that were infected led to 80% of secondary transmissions of ancestral SARS-CoV-2. Similar trends were confirmed for Omicron BA.1 and BA.2, for which only 9%137 to 20%138 of the infectious contacts were responsible for 80% of all transmissions.
Superspreading events are therefore characterized by infectious individuals having close contact with a high number of susceptible individuals and by a higher probability of transmission per contact. Aside from biological factors influencing these events, sociobehavioural and environmental factors contribute to the likelihood of superspreading (for example, large indoor gatherings with poor ventilation and no other infection prevention measures). Moreover, particular locations can represent a higher risk of transmission (for example, many superspreading events take place in crowded indoor settings, such as cruise ships, family gatherings, parties, elderly care centres and hospitals)139.
The role of pre-existing immunity on viral shedding and transmission
All currently licensed SARS-CoV-2 vaccines are administered intramuscularly, leading to a rise in serum antibodies and protection from severe disease and death due to COVID-19, but not to long-term protection from infection140–142. The levels of circulating antibodies generated following vaccination decline over time, but can be elevated by a booster dose143,144. Furthermore, currently available vaccines were developed against the ancestral SARS-CoV-2 strain using the spike protein of the first sequenced virus, and the degree of protection from severe disease against other genetic variants was shown to vary145. Moreover, vaccination leads to limited induction of neutralizing antibodies on mucosal surfaces, which may have a role in mitigating virus replication and prevention of more pronounced disease146,147. For instance, secretory component antibodies, which are specific to mucosal surfaces, were detected in the saliva in 58% of participants 2 weeks post-vaccination with mRNA vaccines in one study, but the levels were significantly lower than in convalescent participants, and their neutralizing capacity significantly decayed 6 months post-vaccination148. A study on a small group of individuals uninfected or infected with Delta demonstrated that mucosal antibody responses induced by vaccination were low or undetectable, but breakthrough infections led to substantial increases of antibody titres in saliva149. However, the role of pre-existing mucosal immunity on infectious virus shedding and the possible correlation between the mucosal antibodies and viral loads in humans has not been elucidated.
As a result of waning antibodies and the emergence of VOCs with immune-evading properties, breakthrough infections have been increasingly reported among vaccinated individuals, mainly since the emergence of the Delta and Omicron VOCs. It has been debated whether vaccination with current SARS-CoV-2 vaccines impacts viral load (and therefore shedding) in breakthrough infections. The effect of vaccination on viral load and shedding is therefore of interest as it would mean that vaccination not only protects the vaccinee but can also help to mitigate virus spread by reducing infectious virus titres or shortening infectious shedding periods, thus having an impact beyond protection of the individual.
Overall, vaccination has been found to lead to reduced viral load (Fig. 4), although this decreases with time. Vaccination with ChAdOx1 vaccine (the Oxford–AstraZeneca vaccine) or BNT162b2 (the Pfizer/BioNTech vaccine) leads to lower RNA viral loads in individuals infected with Alpha, but the effect was weaker for breakthrough infections with Delta150,151. Immunization with BNT162b2 led to reduced RNA viral loads in Delta breakthrough infections, although this effect declined 2 months after vaccination and ultimately faded 6 months after vaccination152. Immunization with ChAdOx1 vaccine also led to a reduction of RNA viral load in breakthrough infections with Alpha VOC153. Faster clearance of RNA viral loads was detected in the group of vaccinated patients who mostly received mRNA vaccines154,155, and lower probability of isolation of infectious virus from patients vaccinated with mRNA or adenoviral vector vaccines was observed156,157. Even though not all studies could demonstrate a reduction of RNA viral loads in Delta breakthrough infections150,154, infectious virus titres were reported to be lower in individuals vaccinated with mRNA or adenoviral vector vaccines despite similar levels of viral RNA25,93,157. Vaccination was also found to influence infectious virus isolation. Viable virus in cell culture was detected for significantly longer median time periods in unvaccinated patients infected with Delta than in vaccinated patients infected with Delta155,158. However, no significant differences in RNA viral loads were found between unvaccinated, fully vaccinated or boosted patients infected with Omicron BA.1 or BA.2 (refs.93,159), whereas infectious virus titres, measured quantitively at 5 dpos, were lower in Omicron BA.1 breakthrough infections only after a booster dose25. Other studies showed that vaccination status did not influence infectious virus isolation success93 or the time from initial positive PCR assay to culture conversion in patients infected with Omicron BA.1 (ref.85). These studies indicate that triple vaccination reduces infectious viral load but not the time period during which infectious virus can be isolated from Omicron breakthrough infections.
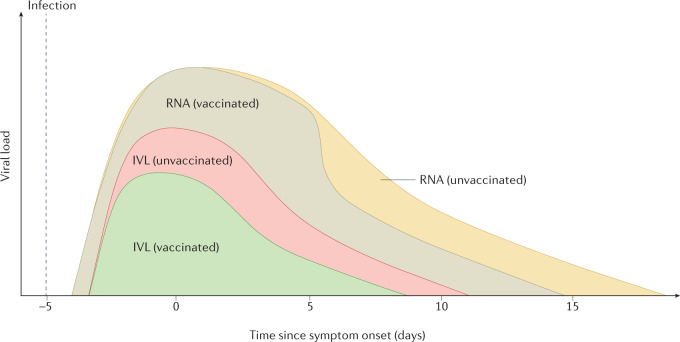
Fig. 4. Influence of vaccination on viral load.
Similar RNA viral loads were detected in vaccinated and unvaccinated patients infected with the Delta variant of concern during the first 5 days post-onset of symptoms. However, faster clearance of viral RNA was shown in vaccinated patients. Infectious viral loads (IVLs) were significantly lower in vaccinated individuals and declined faster than in unvaccinated individuals infected with Delta. Dynamics of viral loads in vaccinated individuals may vary widely in case of infection with another variant. Details of the underlying studies used to generate Fig. 4 can be found in Supplementary Table 3.
There are limited data on the effect of previous infection on viral shedding. A study performed on ancestral SARS-CoV-2 demonstrated lower RNA viral loads among seropositive individuals than among seronegative individuals160. Although higher levels of reinfection with Omicron BA.1 were demonstrated among unvaccinated patients previously infected with other SARS-CoV-2 variants161, there are no relevant data on the effect of previous infections on viral load dynamics.
Together, these findings suggest that vaccinated individuals are less infectious than unvaccinated individuals, although the duration of this effect has not been studied systematically. Nevertheless, there are some conflicting data on the effect of vaccination on onward transmission. An epidemiological study performed in the UK found that, despite RNA viral load declining faster among fully vaccinated than unvaccinated patients infected with Delta, the peak RNA viral loads were similar, and the secondary attack rate among household contacts exposed to fully vaccinated or unvaccinated index cases did not differ151. By contrast, data from Israel showed that less Delta transmission took place in households with vaccinated participants than with unvaccinated participants130. Another study from the UK showed that both BNT162b2 and ChAdOx1 vaccines led to the reduction of onward transmission from vaccinated index patients, although a stronger reduction was detected for Alpha than for Delta129, probably owing to the higher viral loads in the case of infection with Delta, as shown previously88,89,129. Finally, another study found that vaccination was associated with reduced onward transmission of Delta breakthrough infection due to shorter duration of viable virus shedding158.
Overall, even though the currently used vaccines are still based on the ancestral virus spike protein and elicit mainly a systemic rather than a mucosal immune response, some effect on viral load, infectious virus shedding and transmission has been observed129,130,162. Furthermore, with increasing rates of breakthrough infections in the Omicron waves since the end of 2021, many individuals display hybrid immunity consisting of vaccination combined with one or more natural infections before or after vaccination163,164. It is thought that such hybrid immunity may provide better control of virus replication in the mucosa149,163,165.
With the constant emergence of novel variants that can evade existing immunity, our understanding of the effect of vaccination on viral shedding should be constantly updated166. Better understanding of the role of mucosal immunity, and potentially vaccines that elicit local rather than systemic immune responses, are needed to aim for viral load reduction as a means to control SARS-CoV-2 circulation167–169.
Influence of SARS-CoV-2 VOCs on transmission
There are several possible underlying causes of increased transmissibility of newly emerging variants, which allow VOCs to quickly outcompete previously circulating strains, including increased viral loads, a lower infectious dose required to establish infection and prolonged period of infectiousness170. Furthermore, the immune-evading properties of new variants lead to higher susceptibility of infection for vaccinated and previously infected individuals and result in higher transmissibility, as was observed with Omicron166,171.
The rapid emergence of SARS-CoV-2 variants with altered biological properties has shown that knowledge on viral loads, viral kinetics and infectious virus shedding is variant specific, and each emerging variant requires a reassessment. Although understanding of mutational profiles and associated phenotypes of SARS-CoV-2 variants has improved, reasons for enhanced transmissibility are manifold and not all understood yet. To date, shedding characteristics and transmission properties cannot be easily predicted based on sequences. Unlike immune-evasion mechanisms, shedding dynamics, such as kinetics of infectious virus titres or incubation periods of the SARS-CoV-2 variants, cannot be predicted from specific mutation patterns. With a still highly dynamic situation in terms of viral evolution of SARS-CoV-2, understanding viral kinetics and their effect on transmission remains of high public health interest.
SARS-CoV-2 diagnostics in public health
Our ability to define the presence of infectious virus is key to guiding public health measures, as it will enable the isolation of infectious individuals to limit secondary transmission. Unfortunately, no point-of-care diagnostic test currently exists to determine infectious SARS-CoV-2 in a patient sample172, and virus culture as described above is not suited for diagnostic purposes. Thus, a range of approaches have been suggested to find a proxy for infectiousness to guide isolation periods.
One example is the detection of sgRNA transcripts, which are generated during virus replication, and specifically the synthesis of negative-strand RNA. Although sgRNAs are transcribed in infected cells, they are not packaged in the virions and can therefore serve as an indicator of active replication and thus of infectious virus. Specific RT-PCR assays were developed to detect sgRNAs in addition to the diagnostic detection of genomic SARS-CoV-2 RNA, but such assays have not made their way into routine diagnostic use owing to their lower sensitivity than conventional RT-PCR assays. Some studies found that detection of sgRNA correlates with detection of infectious virus4,173,174, and that sgRNA was rarely detectable 8 dpos67. However, sgRNA was detected in diagnostic samples up to 17 days after initial detection of infection175 or in culture-negative samples176, probably owing to the stability and nuclease resistance of double-membrane vesicles containing sgRNAs. Thus, although the absence of sgRNA would indicate absence of viral replication, the presence of sgRNA does not necessarily indicate infectiousness19.
Ct values have also been used as a proxy for infectiousness, as described above. However, as already discussed, low-quality specimens resulting from technical mistakes during the collection process can falsely indicate an absence of infectious virus. Furthermore, owing to the quick increase of RNA viral load at the beginning of the infection, a low viral load, especially in the absence of symptoms or in the early symptomatic period, does not preclude that an individual will not soon enter the infectious period with the highest transmission risk. At such a period, viral loads reach their peak levels, causing the majority of transmission events59,126.
Even though Ag-RDTs are less sensitive than RT-PCR, they are less expensive, can be performed outside of laboratory settings and give faster results, and so are useful tools to guide isolation and limit transmission177. RT-PCR tests have a limit of detection of 102–103 genome copies per millilitre, whereas Ag-RDTs have a limit of detection corresponding to 104–106 genome copies per millilitre177–180. Infectious individuals typically have RNA viral loads of >106 genome copies per millilitre, which corresponds largely with a Ct of 25 in most RT-PCR assays4, indicating that Ag-RDT is a good proxy for infectiousness177. However, the obvious limitations of Ag-RDT, such as lower sensitivity of infectious virus detection towards the end of infection47,52, should not be neglected. Ag-RDTs have also shown variation in their sensitivity and specificity for detection of SARS-CoV-2 VOCs53,54, which is a challenge as new variants emerge.
Overall, all of the currently available diagnostic methods have certain limitations for detection of infectious virus. However, even if these tests serve only as imperfect tools when used as proxies for infectiousness, their implementation as part of a public health strategy is not intended to prevent every single infection, but rather to reduce the number of infectious people in the community and thus to decrease the number of secondary transmissions.
Conclusions
Entering the third year of the pandemic, much knowledge on SARS-CoV-2 viral loads, infectious virus shedding and windows of infectiousness has been gained, although emerging SARS-CoV-2 variants and an increasing population immunity add more complexity to the situation.
Although much progress has been made during the pandemic in the field of diagnostics, to date, no diagnostic tests exist that reliably determine the presence of infectious virus. Continuing evaluation of viral-shedding characteristics under these changing circumstances and understanding the biological properties of novel SARS-CoV-2 variants when it comes to viral shedding remain of importance to guide public health practices.
Supplementary information
Acknowledgements
The authors thank E. Boehm for help with literature search and proofreading. The work was funded by the COVID-19 National Research Program (grant number 198412) of the Swiss National Science Foundation.
Glossary
- Chimeric antigen receptor (CAR) T cell therapy
A way to treat cancer by using T cells expressing genetically engineered receptors to target cancer cells.
- Cycle threshold (Ct) value
The number of amplifications required for a target gene to cross the threshold determined by real-time PCR. Arbitrary test-specific Ct values inversely correlate with viral load.
- Focus-forming assays
Assays that count the number of ‘foci’, defined as a cluster of adjacent cells expressing viral antigen stained by a specific antibody.
- Immunostaining
A method for the detection of specific proteins in individual cells or tissues using antibodies. In the case of SARS-CoV-2, anti-nucleocapsid antibodies are used to detect virus in infected cells.
- Index case
The infected individual who is triggering an outbreak or a cluster by transmitting an infectious agent to others. There might be multiple index cases in an outbreak or epidemiological study.
- Plaque assays
Assays that quantify the number of infectious virions by counting plaques in a cell monolayer that correspond to single infectious particles.
- Secondary attack rate
The probability that an infection spreads from an index case to susceptible people in a specific setting (usually, a household or close contacts). The term is used to evaluate the risk of onward transmission of pathogen within a population.
- Seroconversion
The development of specific antibodies in the serum as a consequence of immunization by natural infection or vaccination.
- sgRNA
Subgenomic RNA fragments that occur during viral replication.
- TCID50
A measurement of the presence of cytopathic effects in cells upon infection with serial dilutions of virus specimens, which indicates the dose needed to induce a cytopathic effect in 50% of the inoculated wells.
Author contributions
O.P. and I.E. wrote the manuscript. B.M. created the figures. All authors contributed to the discussion of the content, and reviewed and edited the manuscript before submission.
Peer review
Peer review information
Nature Reviews Microbiology thanks Nancy Leung, Quanyi Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41579-022-00822-w.
References
- 1.Puelles VG, et al. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamers MM, Haagmans BL. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 2022;20:270–284. doi: 10.1038/s41579-022-00713-0. [DOI] [PubMed] [Google Scholar]
- 3.Peng L, et al. SARS-CoV-2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J. Med. Virol. 2020;92:1676–1680. doi: 10.1002/jmv.25936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wölfel R, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pérez-Bartolomé F, Sánchez-Quirós J. Ocular manifestations of SARS-CoV-2: literature review. Arch. Soc. Esp. Oftalmol. 2021;96:32–40. doi: 10.1016/j.oftal.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vetter P, et al. Daily viral kinetics and innate and adaptive immune response assessment in COVID-19: a case series. mSphere. 2020 doi: 10.1128/mSphere.00827-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeong HW, et al. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin. Microbiol. Infect. 2020;26:1520–1524. doi: 10.1016/j.cmi.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerrada-Romero C, et al. Excretion and viability of SARS-CoV-2 in feces and its association with the clinical outcome of COVID-19. Sci. Rep. 2022;12:7397. doi: 10.1038/s41598-022-11439-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dergham J, Delerce J. Isolation of viable SARS-CoV-2 virus from feces of an immunocompromised patient suggesting a possible fecal mode of transmission. J. Clin. Med. 2021 doi: 10.3390/jcm10122696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao F, et al. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J, et al. Isolation of infectious SARS-CoV-2 from urine of a COVID-19 patient. Emerg. Microbes Infect. 2020;9:991–993. doi: 10.1080/22221751.2020.1760144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colavita F, et al. SARS-CoV-2 isolation from ocular secretions of a patient with COVID-19 in Italy with prolonged viral RNA detection. Ann. Intern. Med. 2020;173:242–243. doi: 10.7326/M20-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuyama S, et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl Acad. Sci. USA. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Case JB, Bailey AL, Kim AS, Chen RE, Diamond MS. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology. 2020;548:39–48. doi: 10.1016/j.virol.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baggen J, Vanstreels E, Jansen S, Daelemans D. Cellular host factors for SARS-CoV-2 infection. Nat. Microbiol. 2021;6:1219–1232. doi: 10.1038/s41564-021-00958-0. [DOI] [PubMed] [Google Scholar]
- 17.Chu H, et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Kampen JJA, et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19) Nat. Commun. 2021;12:267. doi: 10.1038/s41467-020-20568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruce EA, et al. Predicting infectivity: comparing four PCR-based assays to detect culturable SARS-CoV-2 in clinical samples. EMBO Mol. Med. 2022;14:e15290. doi: 10.15252/emmm.202115290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Essaidi-Laziosi M, Perez Rodriguez FJ. Estimating clinical SARS-CoV-2 infectiousness in Vero E6 and primary airway epithelial cells. Lancet Microbe. 2021;2:e571. doi: 10.1016/S2666-5247(21)00216-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu R, et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin. Chim. Acta. 2020;505:172–175. doi: 10.1016/j.cca.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corman VM, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eur. Surveill. 2020 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bullard J, et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin. Infect. Dis. 2020;71:2663–2666. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jefferson T, Spencer EA, Brassey J, Heneghan C. Viral cultures for coronavirus disease 2019 infectivity assessment: a systematic review. Clin. Infect. Dis. 2021;73:e3884–e3899. doi: 10.1093/cid/ciaa1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puhach O, et al. Infectious viral load in unvaccinated and vaccinated individuals infected with ancestral, Delta or Omicron SARS-CoV-2. Nat. Med. 2022;28:1491–1500. doi: 10.1038/s41591-022-01816-0. [DOI] [PubMed] [Google Scholar]
- 26.van Kasteren PB, et al. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020;128:104412. doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bentley, E. et al. Collaborative study for the establishment of a WHO international standard for SARS-CoV-2 RNA (WHO, 2020).
- 28.Zou L, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Killingley B, et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat. Med. 2022;28:1031–1041. doi: 10.1038/s41591-022-01780-9. [DOI] [PubMed] [Google Scholar]
- 30.Yu F, et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin. Infect. Dis. 2020;71:793–798. doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.European Centre for Disease Prevention and Control & World Health Organization Regional Office for Europe. Methods for the detection and identification of SARS-CoV-2 variants: second update, August 2022 (WHO, 2022).
- 32.Ke R, et al. Longitudinal analysis of SARS-CoV-2 vaccine breakthrough infections reveals limited infectious virus shedding and restricted tissue distribution. Open Forum Infect. Dis. 2022;9:ofac192. doi: 10.1093/ofid/ofac192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ke R, et al. Daily longitudinal sampling of SARS-CoV-2 infection reveals substantial heterogeneity in infectiousness. Nat. Microbiol. 2022;7:640–652. doi: 10.1038/s41564-022-01105-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pekosz A, et al. Antigen-based testing but not real-time polymerase chain reaction correlates with severe acute respiratory syndrome coronavirus 2 viral culture. Clin. Infect. Dis. 2021;73:e2861–e2866. doi: 10.1093/cid/ciaa1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monel B, et al. Release of infectious virus and cytokines in nasopharyngeal swabs from individuals infected with non-Alpha or Alpha SARS-CoV-2 variants: an observational retrospective study. EBioMedicine. 2021;73:103637. doi: 10.1016/j.ebiom.2021.103637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirby JE, et al. SARS-CoV-2 antigen tests predict infectivity based on viral culture: comparison of antigen, PCR viral load, and viral culture testing on a large sample cohort. Clin. Microbiol. Infect. 2022 doi: 10.1016/j.cmi.2022.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pickering S, et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests and association with detection of infectious virus in clinical specimens: a single-centre laboratory evaluation study. Lancet Microbe. 2021;2:e461–e471. doi: 10.1016/S2666-5247(21)00143-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tariq M, et al. Viable severe acute respiratory syndrome coronavirus 2 isolates exhibit higher correlation with rapid antigen assays than subgenomic RNA or genomic RNA. Front. Microbiol. 2021;12:718497. doi: 10.3389/fmicb.2021.718497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu VT, et al. Comparison of home antigen testing with RT-PCR and viral culture during the course of SARS-CoV-2 infection. JAMA Intern. Med. 2022;182:701–709. doi: 10.1001/jamainternmed.2022.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albert E, et al. Field evaluation of a rapid antigen test (Panbio™ COVID-19 Ag rapid test device) for COVID-19 diagnosis in primary healthcare centres. Clin. Microbiol. Infect. 2021;27:472.e7–472.e10. doi: 10.1016/j.cmi.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ford L, et al. Epidemiologic characteristics associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigen-based test results, real-time reverse transcription polymerase chain reaction (rRT-PCR) cycle threshold values, subgenomic RNA, and viral culture results from university testing. Clin. Infect. Dis. 2021;73:e1348–e1355. doi: 10.1093/cid/ciab303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berger A, et al. Diagnostic accuracy of two commercial SARS-CoV-2 antigen-detecting rapid tests at the point of care in community-based testing centers. PLoS ONE. 2021;16:e0248921. doi: 10.1371/journal.pone.0248921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brümmer LE, et al. Accuracy of novel antigen rapid diagnostics for SARS-CoV-2: a living systematic review and meta-analysis. PLoS Med. 2021;18:e1003735. doi: 10.1371/journal.pmed.1003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ngo Nsoga MT, et al. Diagnostic accuracy of Panbio rapid antigen tests on oropharyngeal swabs for detection of SARS-CoV-2. PLoS ONE. 2021;16:e0253321. doi: 10.1371/journal.pone.0253321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korenkov M, et al. Evaluation of a rapid antigen test to detect SARS-CoV-2 infection and identify potentially infectious individuals. J. Clin. Microbiol. 2021;59:e0089621. doi: 10.1128/JCM.00896-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamayoshi S, et al. Comparison of rapid antigen tests for COVID-19. Viruses. 2020;12:1420. doi: 10.3390/v12121420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKay SL, et al. Performance evaluation of serial SARS-CoV-2 rapid antigen testing during a nursing home outbreak. Ann. Intern. Med. 2021;174:945–951. doi: 10.7326/M21-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nordgren J, et al. SARS-CoV-2 rapid antigen test: high sensitivity to detect infectious virus. J. Clin. Virol. 2021;140:104846. doi: 10.1016/j.jcv.2021.104846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandez-Montero A, Argemi J. Validation of a rapid antigen test as a screening tool for SARS-CoV-2 infection in asymptomatic populations. Sensitivity, specificity and predictive values. EClinicalMedicine. 2021;37:100954. doi: 10.1016/j.eclinm.2021.100954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Currie DW, et al. Relationship of SARS-CoV-2 antigen and reverse transcription PCR positivity for viral cultures. Emerg. Infect. Dis. 2022;28:717–720. doi: 10.3201/eid2803.211747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corman VM, et al. Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: a single-centre laboratory evaluation study. Lancet Microbe. 2021;2:e311–e319. doi: 10.1016/S2666-5247(21)00056-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopera TJ, Alzate-Ángel JC. The usefulness of antigen testing in predicting contagiousness in COVID-19. Microbiol. Spectr. 2022;10:e0196221. doi: 10.1128/spectrum.01962-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Osterman A, et al. Impaired detection of Omicron by SARS-CoV-2 rapid antigen tests. Med. Microbiol. Immunol. 2022;211:105–117. doi: 10.1007/s00430-022-00730-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galliez RM, et al. Evaluation of the Panbio COVID-19 antigen rapid diagnostic test in subjects infected with Omicron using different specimens. Microbiol. Spectr. 2022;10:e0125022. doi: 10.1128/spectrum.01250-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raïch-Regué D, et al. Performance of SARS-CoV-2 antigen-detecting rapid diagnostic tests for Omicron and other variants of concern. Front. Microbiol. 2022;13:810576. doi: 10.3389/fmicb.2022.810576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Ogtrop ML, van de Laar TJW, Eggink D, Vanhommerig JW, van der Reijden WA. Comparison of the performance of the Panbio COVID-19 antigen test in SARS-CoV-2 B.1.1.7 (Alpha) variants versus non-B.1.1.7 variants. Microbiol. Spectr. 2021;9:e0088421. doi: 10.1128/Spectrum.00884-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lindner AK, et al. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with self-collected nasal swab versus professional-collected nasopharyngeal swab. Eur. Respir. J. 2021;57:2003961. doi: 10.1183/13993003.03961-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meiners, L. & Horn, J. SARS-CoV-2 rapid antigen test sensitivity and viral load in freshly symptomatic hospital employees, December 2020 to February 2022. Preprint at 10.2139/ssrn.4099425 (2022). [DOI] [PubMed]
- 59.He X, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 60.Néant N, et al. Modeling SARS-CoV-2 viral kinetics and association with mortality in hospitalized patients from the French COVID cohort. Proc. Natl Acad. Sci. USA. 2021;118:e2017962118. doi: 10.1073/pnas.2017962118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Owusu D, et al. Persistent SARS-CoV-2 RNA shedding without evidence of infectiousness: a cohort study of individuals with COVID-19. J. Infect. Dis. 2021;224:1362–1371. doi: 10.1093/infdis/jiab107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cevik M, et al. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen X, et al. Associations of clinical characteristics and treatment regimens with the duration of viral RNA shedding in patients with COVID-19. Int. J. Infect. Dis. 2020;98:252–260. doi: 10.1016/j.ijid.2020.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim SM, Hwang YJ. Prolonged SARS-CoV-2 detection and reversed RT-PCR results in mild or asymptomatic patients. Infect. Dis. 2021;53:31–37. doi: 10.1080/23744235.2020.1820076. [DOI] [PubMed] [Google Scholar]
- 65.Talmy T, Tsur A. Duration of SARS-CoV-2 detection in Israel Defense Forces soldiers with mild COVID-19. J. Med. Virol. 2021;93:608–610. doi: 10.1002/jmv.26374. [DOI] [PubMed] [Google Scholar]
- 66.Singanayagam, A. et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 10.2807/1560-7917.ES.2020.25.32.2001483 (2020). [DOI] [PMC free article] [PubMed]
- 67.Perera R, et al. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg. Infect. Dis. 2020;26:2701–2704. doi: 10.3201/eid2611.203219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Killerby ME, et al. Shedding of culturable virus, seroconversion, and 6-month follow-up antibody responses in the first 14 confirmed cases of coronavirus disease 2019 in the United States. J. Infect. Dis. 2021;224:771–776. doi: 10.1093/infdis/jiab125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Glans H, et al. Shedding of infectious SARS-CoV-2 by hospitalized COVID-19 patients in relation to serum antibody responses. BMC Infect. Dis. 2021;21:494. doi: 10.1186/s12879-021-06202-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Badu K, et al. SARS-CoV-2 viral shedding and transmission dynamics: implications of WHO COVID-19 discharge guidelines. Front. Med. 2021;8:648660. doi: 10.3389/fmed.2021.648660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Munker D, et al. Dynamics of SARS-CoV-2 shedding in the respiratory tract depends on the severity of disease in COVID-19 patients. Eur. Respir. J. 2021;58,:2002724. doi: 10.1183/13993003.02724-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Folgueira MD, Luczkowiak J. Prolonged SARS-CoV-2 cell culture replication in respiratory samples from patients with severe COVID-19. Clin. Microbiol. Infect. 2021;27:886–891. doi: 10.1016/j.cmi.2021.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim MC, et al. Duration of culturable SARS-CoV-2 in hospitalized patients with Covid-19. N. Engl. J. Med. 2021;384:671–673. doi: 10.1056/NEJMc2027040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen PZ, et al. SARS-CoV-2 shedding dynamics across the respiratory tract, sex, and disease severity for adult and pediatric COVID-19. eLife. 2021 doi: 10.7554/eLife. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cunha MDP, et al. Atypical prolonged viral shedding with intra-host SARS-CoV-2 evolution in a mildly affected symptomatic patient. Front. Med. 2021;8:760170. doi: 10.3389/fmed.2021.760170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aydillo T, et al. Shedding of viable SARS-CoV-2 after immunosuppressive therapy for cancer. N. Engl. J. Med. 2020;383:2586–2588. doi: 10.1056/NEJMc2031670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Caillard S, Benotmane I, Gautier Vargas G, Perrin P, Fafi-Kremer S. SARS-CoV-2 viral dynamics in immunocompromised patients. Am. J. Transpl. 2021;21:1667–1669. doi: 10.1111/ajt.16353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roedl K, et al. Viral dynamics of SARS-CoV-2 in critically ill allogeneic hematopoietic stem cell transplant recipients and immunocompetent patients with COVID-19. Am. J. Respir. Crit. Care Med. 2021;203:242–245. doi: 10.1164/rccm.202009-3386LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leung WF, et al. COVID-19 in an immunocompromised host: persistent shedding of viable SARS-CoV-2 and emergence of multiple mutations: a case report. Int. J. Infect. Dis. 2022;114:178–182. doi: 10.1016/j.ijid.2021.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.European Centre for Disease Prevention and Control. Rapid risk assessment: assessing SARS-CoV-2 circulation, variants of concern, non-pharmaceutical interventions and vaccine rollout in the EU/EEA, 15th update (European Centre for Disease Prevention and Control, 2021).
- 81.Grant R, et al. Impact of SARS-CoV-2 Delta variant on incubation, transmission settings and vaccine effectiveness: results from a nationwide case–control study in France. Lancet Reg. Health Eur. 2022;13:100278. doi: 10.1016/j.lanepe.2021.100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ogata T, Tanaka H. Shorter incubation period among unvaccinated Delta variant coronavirus disease 2019 patients in Japan. Int. J. Environ. Res. Public Health. 2022 doi: 10.3390/ijerph19031127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Backer JA, et al. Shorter serial intervals in SARS-CoV-2 cases with Omicron BA.1 variant compared with Delta variant, the Netherlands, 13 to 26 December 2021. Eur. Surveill. 2022 doi: 10.2807/1560-7917.ES.2022.27.6.2200042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takahashi K, et al. Duration of infectious virus shedding by SARS-CoV-2 Omicron variant-infected vaccinees. Emerg. Infect. Dis. 2022;28:998–1001. doi: 10.3201/eid2805.220197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boucau J, et al. Duration of shedding of culturable virus in SARS-CoV-2 Omicron (BA.1) infection. N. Engl. J. Med. 2022;387:275–277. doi: 10.1056/NEJMc2202092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jones TC, et al. Estimating infectiousness throughout SARS-CoV-2 infection course. Science. 2021 doi: 10.1126/science.abi5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li B, et al. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant. Nat. Commun. 2022;13:460. doi: 10.1038/s41467-022-28089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bolze A, et al. SARS-CoV-2 variant Delta rapidly displaced variant Alpha in the United States and led to higher viral loads. Cell Rep. Med. 2022;3:100564. doi: 10.1016/j.xcrm.2022.100564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Earnest R, et al. Comparative transmissibility of SARS-CoV-2 variants Delta and Alpha in New England, USA. Cell Rep. Med. 2022;3:100583. doi: 10.1016/j.xcrm.2022.100583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Luo CH, et al. Infection with the SARS-CoV-2 Delta variant is associated with higher recovery of infectious virus compared to the Alpha variant in both unvaccinated and vaccinated individuals. Clin. Infect. Dis. 2021;75:e715–e725. doi: 10.1093/cid/ciab986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Despres HW, et al. Measuring infectious SARS-CoV-2 in clinical samples reveals a higher viral titer: RNA ratio for Delta and Epsilon vs. Alpha variants. Proc. Natl Acad. Sci. USA. 2022 doi: 10.1073/pnas.2116518119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hay JA, et al. Quantifying the impact of immune history and variant on SARS-CoV-2 viral kinetics and infection rebound: a retrospective cohort study. eLife. 2022;11:e81849. doi: 10.7554/eLife.81849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fall A, et al. The displacement of the SARS-CoV-2 variant Delta with Omicron: an investigation of hospital admissions and upper respiratory viral loads. EbioMedicine. 2022 doi: 10.1016/j.ebiom.2022.104008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lentini A, Pereira A. Monitoring of the SARS-CoV-2 Omicron BA.1/BA.2 lineage transition in the Swedish population reveals increased viral RNA levels in BA.2 cases. Med. 2022;3:636–664. doi: 10.1016/j.medj.2022.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Qassim SH, et al. Effects of BA.1/BA.2 subvariant, vaccination, and prior infection on infectiousness of SARS-CoV-2 Omicron infections. J. Travel Med. 2022;29:taac068. doi: 10.1093/jtm/taac068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marking U, et al. Correlates of protection, viral load trajectories and symptoms in BA.1, BA.1.1 and BA.2 breakthrough infections in triple vaccinated healthcare workers. medRxiv. 2022 doi: 10.1101/2022.04.02.22273333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Y, et al. Transmission, viral kinetics and clinical characteristics of the emergent SARS-CoV-2 Delta VOC in Guangzhou, China. EClinicalMedicine. 2021;40:101129. doi: 10.1016/j.eclinm.2021.101129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Siedner MJ, et al. Duration of viral shedding and culture positivity with postvaccination SARS-CoV-2 Delta variant infections. JCI Insight. 2022 doi: 10.1172/jci.insight.155483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pierce CA, et al. Immune responses to SARS-CoV-2 infection in hospitalized pediatric and adult patients. Sci. Transl Med. 2020;12:eabd5487. doi: 10.1126/scitranslmed.abd5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takahashi T, et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588:315–320. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bunyavanich S, Do A, Vicencio A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA. 2020;323:2427–2429. doi: 10.1001/jama.2020.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zheng S, et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January–March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.L’Huillier AG, Torriani G. Culture-competent SARS-CoV-2 in nasopharynx of symptomatic neonates, children, and adolescents. Emerg. Infect. Dis. 2020;26:2494–2497. doi: 10.3201/eid2610.202403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Baggio S, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral load in the upper respiratory tract of children and adults with early acute coronavirus disease 2019 (COVID-19) Clin. Infect. Dis. 2021;73:148–150. doi: 10.1093/cid/ciaa1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chung E, et al. Comparison of symptoms and RNA levels in children and adults with SARS-CoV-2 infection in the community setting. JAMA Pediatr. 2021;175:e212025. doi: 10.1001/jamapediatrics.2021.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Han MS, et al. Clinical characteristics and viral RNA detection in children with coronavirus disease 2019 in the Republic of Korea. JAMA Pediatr. 2021;175:73–80. doi: 10.1001/jamapediatrics.2020.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bellon M, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral load kinetics in symptomatic children, adolescents, and adults. Clin. Infect. Dis. 2021;73:e1384–e1386. doi: 10.1093/cid/ciab396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Madera S, et al. Nasopharyngeal SARS-CoV-2 viral loads in young children do not differ significantly from those in older children and adults. Sci. Rep. 2021;11:3044. doi: 10.1038/s41598-021-81934-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lauer SA, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Elias C, Sekri A, Leblanc P, Cucherat M, Vanhems P. The incubation period of COVID-19: a meta-analysis. Int. J. Infect. Dis. 2021;104:708–710. doi: 10.1016/j.ijid.2021.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dhouib W, et al. The incubation period during the pandemic of COVID-19: a systematic review and meta-analysis. Syst. Rev. 2021;10:101. doi: 10.1186/s13643-021-01648-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brandal LT, et al. Outbreak caused by the SARS-CoV-2 Omicron variant in Norway, November to December 2021. Eur. Surveill. 2021;26:2101147. doi: 10.2807/1560-7917.ES.2021.26.50.2101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu Y, et al. Incubation period of COVID-19 caused by unique SARS-CoV-2 strains: a systematic review and meta-analysis. JAMA Netw. Open. 2022;5:e2228008. doi: 10.1001/jamanetworkopen.2022.28008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sun K, et al. Transmission heterogeneities, kinetics, and controllability of SARS-CoV-2. Science. 2021;371:eabe2424. doi: 10.1126/science.abe2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lavezzo E, et al. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo’. Nature. 2020;584:425–429. doi: 10.1038/s41586-020-2488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gudbjartsson DF, et al. Spread of SARS-CoV-2 in the Icelandic population. N. Engl. J. Med. 2020;382:2302–2315. doi: 10.1056/NEJMoa2006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Glenet M, et al. Asymptomatic COVID-19 adult outpatients identified as significant viable SARS-CoV-2 shedders. Sci. Rep. 2021;11:20615. doi: 10.1038/s41598-021-00142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang Y, et al. Characterization of an asymptomatic cohort of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infected individuals outside of Wuhan, China. Clin. Infect. Dis. 2020;71:2132–2138. doi: 10.1093/cid/ciaa629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Long QX, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 120.Lee S, et al. Clinical course and molecular viral shedding among asymptomatic and symptomatic patients with SARS-CoV-2 infection in a community treatment center in the Republic of Korea. JAMA Intern. Med. 2020;180:1447–1452. doi: 10.1001/jamainternmed.2020.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou R, et al. Viral dynamics in asymptomatic patients with COVID-19. Int. J. Infect. Dis. 2020;96:288–290. doi: 10.1016/j.ijid.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hall SM, et al. Comparison of anterior nares CT values in asymptomatic and symptomatic individuals diagnosed with SARS-CoV-2 in a university screening program. PLoS ONE. 2022;17:e0270694. doi: 10.1371/journal.pone.0270694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kissler SM, et al. Viral dynamics of acute SARS-CoV-2 infection and applications to diagnostic and public health strategies. PLoS Biol. 2021;19:e3001333. doi: 10.1371/journal.pbio.3001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tallmadge RL, et al. Viral RNA load and infectivity of SARS-CoV-2 in paired respiratory and oral specimens from symptomatic, asymptomatic, or postsymptomatic individuals. Microbiol. Spectr. 2022;10:e0226421. doi: 10.1128/spectrum.02264-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marks M, et al. Transmission of COVID-19 in 282 clusters in Catalonia, Spain: a cohort study. Lancet Infect. Dis. 2021;21:629–636. doi: 10.1016/S1473-3099(20)30985-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Marc A, et al. Quantifying the relationship between SARS-CoV-2 viral load and infectiousness. eLife. 2021 doi: 10.7554/eLife.69302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cheng HY, et al. Contact tracing assessment of COVID-19 transmission dynamics in Taiwan and risk at different exposure periods before and after symptom onset. JAMA Intern. Med. 2020;180:1156–1163. doi: 10.1001/jamainternmed.2020.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mugglestone MA, et al. Presymptomatic, asymptomatic and post-symptomatic transmission of SARS-CoV-2: joint British Infection Association (BIA), Healthcare Infection Society (HIS), Infection Prevention Society (IPS) and Royal College of Pathologists (RCPath) guidance. BMC Infect. Dis. 2022;22:453. doi: 10.1186/s12879-022-07440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Eyre DW, et al. Effect of Covid-19 vaccination on transmission of Alpha and Delta variants. N. Engl. J. Med. 2022;386:744–756. doi: 10.1056/NEJMoa2116597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Prunas O, et al. Vaccination with BNT162b2 reduces transmission of SARS-CoV-2 to household contacts in Israel. Science. 2022;375:1151–1154. doi: 10.1126/science.abl4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lyngse FP, et al. Household transmission of SARS-CoV-2 Omicron variant of concern subvariants BA.1 and BA.2 in Denmark. Nat. Commun. 2022;13:5760. doi: 10.1038/s41467-022-33498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Leung NHL. Transmissibility and transmission of respiratory viruses. Nat. Rev. Microbiol. 2021;19:528–545. doi: 10.1038/s41579-021-00535-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Alihsan B, et al. The efficacy of facemasks in the prevention of COVID-19: a systematic review. medRxiv. 2022 doi: 10.1101/2022.07.28.22278153. [DOI] [Google Scholar]
- 134.Hu Z, et al. Early immune markers of clinical, virological, and immunological outcomes in patients with COVID-19: a multi-omics study. Elife. 2022;11:e77943. doi: 10.7554/eLife.77943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen PZ, et al. Heterogeneity in transmissibility and shedding SARS-CoV-2 via droplets and aerosols. eLife. 2021 doi: 10.7554/eLife.65774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Adam DC, et al. Clustering and superspreading potential of SARS-CoV-2 infections in Hong Kong. Nat. Med. 2020;26:1714–1719. doi: 10.1038/s41591-020-1092-0. [DOI] [PubMed] [Google Scholar]
- 137.Guo Z, et al. Superspreading potential of infection seeded by the SARS-CoV-2 Omicron BA.1 variant in South Korea. J. Infect. 2022;85:e77–e79. doi: 10.1016/j.jinf.2022.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Guo Z, et al. Superspreading potential of COVID-19 outbreak seeded by Omicron variants of SARS-CoV-2 in Hong Kong. J. Travel Med. 2022;29:taac049. doi: 10.1093/jtm/taac049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Althouse BM, et al. Superspreading events in the transmission dynamics of SARS-CoV-2: opportunities for interventions and control. PLoS Biol. 2020;18:e3000897. doi: 10.1371/journal.pbio.3000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Baden LR, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lopez Bernal J, et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N. Engl. J. Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Feikin DR, et al. Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: results of a systematic review and meta-regression. Lancet. 2022;399:924–944. doi: 10.1016/S0140-6736(22)00152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Belik M, et al. Comparative analysis of COVID-19 vaccine responses and third booster dose-induced neutralizing antibodies against Delta and Omicron variants. Nat. Commun. 2022;13:2476. doi: 10.1038/s41467-022-30162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pegu A, et al. Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science. 2021;373:1372–1377. doi: 10.1126/science.abj4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lipsitch M, Krammer F, Regev-Yochay G, Lustig Y, Balicer RD. SARS-CoV-2 breakthrough infections in vaccinated individuals: measurement, causes and impact. Nat. Rev. Immunol. 2022;22:57–65. doi: 10.1038/s41577-021-00662-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mostaghimi D, Valdez CN, Larson HT, Kalinich CC, Iwasaki A. Prevention of host-to-host transmission by SARS-CoV-2 vaccines. Lancet Infect. Dis. 2021 doi: 10.1016/S1473-3099(21)00472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Russell MW, Moldoveanu Z, Ogra PL, Mestecky J. Mucosal immunity in COVID-19: a neglected but critical aspect of SARS-CoV-2 infection. Front. Immunol. 2020;11:611337. doi: 10.3389/fimmu.2020.611337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sheikh-Mohamed S, et al. Systemic and mucosal IgA responses are variably induced in response to SARS-CoV-2 mRNA vaccination and are associated with protection against subsequent infection. Mucosal Immunol. 2022;15:799–808. doi: 10.1038/s41385-022-00511-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Collier AY, et al. Characterization of immune responses in fully vaccinated individuals after breakthrough infection with the SARS-CoV-2 Delta variant. Sci. Transl Med. 2022;14:eabn6150. doi: 10.1126/scitranslmed.abn6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pouwels KB, et al. Effect of Delta variant on viral burden and vaccine effectiveness against new SARS-CoV-2 infections in the UK. Nat. Med. 2021;27:2127–2135. doi: 10.1038/s41591-021-01548-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Singanayagam A, et al. Community transmission and viral load kinetics of the SARS-CoV-2 Delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: a prospective, longitudinal, cohort study. Lancet Infect. Dis. 2021;22:183–195. doi: 10.1016/S1473-3099(21)00648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Levine-Tiefenbrun M, et al. Viral loads of Delta-variant SARS-CoV-2 breakthrough infections after vaccination and booster with BNT162b2. Nat. Med. 2021;27:2108–2110. doi: 10.1038/s41591-021-01575-4. [DOI] [PubMed] [Google Scholar]
- 153.Emary KRW, et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet. 2021;397:1351–1362. doi: 10.1016/S0140-6736(21)00628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chia PY, et al. Virological and serological kinetics of SARS-CoV-2 Delta variant vaccine breakthrough infections: a multicentre cohort study. Clin. Microbiol. Infect. 2021;28:612.e1–612.e7. doi: 10.1016/j.cmi.2021.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Garcia-Knight M, et al. Infectious viral shedding of SARS-CoV-2 Delta following vaccination: a longitudinal cohort study. PLoS Pathog. 2022;18:e1010802. doi: 10.1371/journal.ppat.1010802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Peña-Hernández MA, et al. Comparison of infectious SARS-CoV-2 from the nasopharynx of vaccinated and unvaccinated individuals. medRxiv. 2022 doi: 10.1101/2021.12.28.21268460. [DOI] [Google Scholar]
- 157.Shamier MC, et al. Virological characteristics of SARS-CoV-2 vaccine breakthrough infections in health care workers. medRxiv. 2021 doi: 10.1101/2021.08.20.21262158. [DOI] [Google Scholar]
- 158.Jung J, et al. Transmission and infectious SARS-CoV-2 shedding kinetics in vaccinated and unvaccinated individuals. JAMA Netw. Open. 2022;5:e2213606. doi: 10.1001/jamanetworkopen.2022.13606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hirotsu Y, et al. Similar viral loads in Omicron infections regardless of vaccination status. medRxiv. 2022 doi: 10.1101/2022.04.19.22274005. [DOI] [Google Scholar]
- 160.Letizia AG, et al. SARS-CoV-2 seropositivity and subsequent infection risk in healthy young adults: a prospective cohort study. Lancet Respir. Med. 2021;9:712–720. doi: 10.1016/S2213-2600(21)00158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pulliam JRC, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science. 2022;376:eabn4947. doi: 10.1126/science.abn4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Harris RJ, et al. Effect of vaccination on household transmission of SARS-CoV-2 in England. N. Engl. J. Med. 2021;385:759–760. doi: 10.1056/NEJMc2107717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bates TA, et al. Vaccination before or after SARS-CoV-2 infection leads to robust humoral response and antibodies that effectively neutralize variants. Sci. Immunol. 2022;7:eabn8014. doi: 10.1126/sciimmunol.abn8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wratil PR, et al. Three exposures to the spike protein of SARS-CoV-2 by either infection or vaccination elicit superior neutralizing immunity to all variants of concern. Nat. Med. 2022;28:496–503. doi: 10.1038/s41591-022-01715-4. [DOI] [PubMed] [Google Scholar]
- 165.Malato J, et al. Risk of BA.5 infection among persons exposed to previous SARS-CoV-2 variants. N. Engl. J. Med. 2022;387:953–954. doi: 10.1056/NEJMc2209479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cao Y, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hassan AO, et al. A single-dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell. 2020;183:169–184. doi: 10.1016/j.cell.2020.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lapuente D, et al. Protective mucosal immunity against SARS-CoV-2 after heterologous systemic prime-mucosal boost immunization. Nat. Commun. 2021;12:6871. doi: 10.1038/s41467-021-27063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Afkhami S, et al. Respiratory mucosal delivery of next-generation COVID-19 vaccine provides robust protection against both ancestral and variant strains of SARS-CoV-2. Cell. 2022;185:896–915. doi: 10.1016/j.cell.2022.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Grubaugh ND, Hodcroft EB. Public health actions to control new SARS-CoV-2 variants. Cell. 2021;184:1127–1132. doi: 10.1016/j.cell.2021.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Carreño JM, et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- 172.Ashcroft P, Lehtinen S, Bonhoeffer S. Test-trace-isolate-quarantine (TTIQ) intervention strategies after symptomatic COVID-19 case identification. PLoS ONE. 2022;17:e0263597. doi: 10.1371/journal.pone.0263597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Santos Bravo M, et al. Viral culture confirmed SARS-CoV-2 subgenomic RNA value as a good surrogate marker of infectivity. J. Clin. Microbiol. 2022;60:e0160921. doi: 10.1128/JCM.01609-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kim JY, et al. Diagnostic usefulness of subgenomic RNA detection of viable SARS-CoV-2 in patients with COVID-19. Clin. Microbiol. Infect. 2022;28:101–106. doi: 10.1016/j.cmi.2021.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Alexandersen S, Chamings A. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun. 2020;11:6059. doi: 10.1038/s41467-020-19883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bonenfant G, et al. Surveillance and correlation of SARS-CoV-2 viral RNA, antigen, virus isolation, and self-reported symptoms in a longitudinal study with daily sampling. Clin. Infect. Dis. 2022 doi: 10.1093/cid/ciac282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Peeling RW, Heymann DL, Teo YY, Garcia PJ. Diagnostics for COVID-19: moving from pandemic response to control. Lancet. 2022;399:757–768. doi: 10.1016/S0140-6736(21)02346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pilarowski G, et al. Performance characteristics of a rapid severe acute respiratory syndrome coronavirus 2 antigen detection assay at a public plaza testing site in San Francisco. J. Infect. Dis. 2021;223:1139–1144. doi: 10.1093/infdis/jiaa802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Leber W, et al. Comparing the diagnostic accuracy of point-of-care lateral flow antigen testing for SARS-CoV-2 with RT-PCR in primary care (REAP-2) EClinicalMedicine. 2021;38:101011. doi: 10.1016/j.eclinm.2021.101011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kohmer N, et al. The comparative clinical performance of four SARS-CoV-2 rapid antigen tests and their correlation to infectivity in vitro. J. Clin. Med. 2021 doi: 10.3390/jcm10020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wang W, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lee, R. A., Herigon, J. C., Benedetti, A., Pollock, N. R. & Denkinger, C. M. Performance of saliva, oropharyngeal swabs, and nasal swabs for SARS-CoV-2 molecular detection: a systematic review and meta-analysis. J. Clin. Microbiol. 59, e02881-20 (2021). [DOI] [PMC free article] [PubMed]
- 183.Tsang NNY, et al. Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: a systematic review and meta-analysis. Lancet Infect. Dis. 2021;21:1233–1245. doi: 10.1016/S1473-3099(21)00146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Peto T. COVID-19: rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine. 2021;36:100924. doi: 10.1016/j.eclinm.2021.100924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hay JA, et al. Estimating epidemiologic dynamics from cross-sectional viral load distributions. Science. 2021;373,:eabh0635. doi: 10.1126/science.abh0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Peccia J, et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Xin H, et al. The incubation period distribution of coronavirus disease 2019: a systematic review and meta-analysis. Clin. Infect. Dis. 2021;73:2344–2352. doi: 10.1093/cid/ciab501. [DOI] [PubMed] [Google Scholar]
- 188.Yu IT, et al. Evidence of airborne transmission of the severe acute respiratory syndrome virus. N. Engl. J. Med. 2004;350:1731–1739. doi: 10.1056/NEJMoa032867. [DOI] [PubMed] [Google Scholar]
- 189.Cheng PK, et al. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet. 2004;363:1699–1700. doi: 10.1016/S0140-6736(04)16255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Peiris JS, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chan PKS, et al. Laboratory diagnosis of SARS. Emerg. Infect. Dis. 2004;10:825–831. doi: 10.3201/eid1005.030682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Liu W, et al. Long-term SARS coronavirus excretion from patient cohort, China. Emerg. Infect. Dis. 2004;10:1841–1843. doi: 10.3201/eid1010.040297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Xu D, et al. Persistent shedding of viable SARS-CoV in urine and stool of SARS patients during the convalescent phase. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24:165–171. doi: 10.1007/s10096-005-1299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.V’Kovski P, et al. Disparate temperature-dependent virus–host dynamics for SARS-CoV-2 and SARS-CoV in the human respiratory epithelium. PLoS Biol. 2021;19:e3001158. doi: 10.1371/journal.pbio.3001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Pitzer VE, Leung GM, Lipsitch M. Estimating variability in the transmission of severe acute respiratory syndrome to household contacts in Hong Kong, China. Am. J. Epidemiol. 2007;166:355–363. doi: 10.1093/aje/kwm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Riley S, et al. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science. 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- 197.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 198.Breban R, Riou J, Fontanet A. Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet. 2013;382:694–699. doi: 10.1016/S0140-6736(13)61492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Oh MD, et al. Viral load kinetics of MERS coronavirus infection. N. Engl. J. Med. 2016;375:1303–1305. doi: 10.1056/NEJMc1511695. [DOI] [PubMed] [Google Scholar]
- 200.Corman VM, et al. Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus Infection. Clin. Infect. Dis. 2016;62:477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Min C-K, et al. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci. Rep. 2016;6:25359. doi: 10.1038/srep25359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ip DK, et al. Viral shedding and transmission potential of asymptomatic and paucisymptomatic influenza virus infections in the community. Clin. Infect. Dis. 2017;64:736–742. doi: 10.1093/cid/ciw841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Carrat F, et al. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am. J. Epidemiol. 2008;167:775–785. doi: 10.1093/aje/kwm375. [DOI] [PubMed] [Google Scholar]
- 204.Pawelek KA, et al. Modeling within-host dynamics of influenza virus infection including immune responses. PLoS Comput. Biol. 2012;8:e1002588. doi: 10.1371/journal.pcbi.1002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ip DKM, et al. The dynamic relationship between clinical symptomatology and viral shedding in naturally acquired seasonal and pandemic influenza virus infections. Clin. Infect. Dis. 2016;62:431–437. doi: 10.1093/cid/civ909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Nair H, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Bagga B, et al. Comparing influenza and RSV viral and disease dynamics in experimentally infected adults predicts clinical effectiveness of RSV antivirals. Antivir. Ther. 2013;18:785–791. doi: 10.3851/IMP2629. [DOI] [PubMed] [Google Scholar]
- 208.DeVincenzo JP, et al. Viral load drives disease in humans experimentally infected with respiratory syncytial virus. Am. J. Respir. Crit. Care Med. 2010;182:1305–1314. doi: 10.1164/rccm.201002-0221OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kutter JS, et al. Small quantities of respiratory syncytial virus RNA only in large droplets around infants hospitalized with acute respiratory infections. Antimicrob. Resist. Infect. Control. 2021;10:100. doi: 10.1186/s13756-021-00968-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.