Abstract
Objectives
By deploying a novel combination of machine learning approaches, we aim to investigate the contributions of each local and systemic risk factors in multi-etiology of knee osteoarthritis (KOA) to disease onset and deterioration.
Methods
A machine-learning-based KOA progression prediction model is developed using the data from the National Institute of Health Osteoarthritis Biomarkers Consortium. According to Kellgren-Lawrence (KL) grade of plain radiographs at baseline, the subjects are divided into either KOA onset or deterioration study groups. The disease progression is defined as the changes in both joint space width (JSW) and WOMAC pain score. In addition to radiographic and symptomatic data, the anthropological particulars, history of the knee injury and surgery, metabolic syndrome and living habits were deployed in a multi-layer perceptron (MLP) to predict disease progression in each study group. The relative contributions of each risk factors were weighted via DeepLIFT gradient. Additionally, statistical interactions among risk factors were identified compared.
Results
Our model achieved AUC of 0.843 (95% CI 0.824, 0.862) and 0.765 (95% CI 0.756, 0.774) in prediction of KOA onset and deterioration, respectively. For KOA onset prediction, history of injury has attained the highest DeepLIFT gradient except medial joint space narrowing; while for KOA deterioration prediction, diabetes and habit of smoking obtained second and third highest gradients respectively aside from medial joint space narrowing, surpassing the impact of the injury.
Conclusion
We developed a machine learning workflow which effectively dissects the risk factors’ contributions and their mutual interactions for onset and deterioration of KOA respectively.
Keywords: Knee osteoarthritis, Machine learning, Neural network, t-SNE
1. Introduction
Knee osteoarthritis (KOA) is a multi-factorial disease, subject to the interplay between local and systemic risk factors. KOA was traditionally conceived as a local problem of mechanics, leading to wear and tear of articular cartilage that cushions the joint during the movement [1]. The abnormal mechanical loading, such as joint instability after ligament or meniscal injury, may also induce the mechanoflammation in the pathogenesis of KOA [2,3], ultimately resulting in cartilage matrix degradation. Meanwhile, despite ageing and obesity once being conceived as the major systemic risk factors to trigger KOA, mounting evidence suggests that they are inadequate to explain the sharp growth of the diseased population [4]. Recently, there emerged a growing interest in metabolic syndromes, such as diabetes and hypertension to characterize the disease from a systemic perspective and improve KOA disease management [[5], [6], [7], [8], [9], [10]].
Given the fact that KOA was attributed to a plethora of local and systemic risk factors, to our best knowledge, there is a lack of information regarding their relative contributions to the disease onset and deterioration. The apprehension of the underlying predominant risk factors and their interrelationship would be instrumental in developing an effective preventive and therapeutic plan for KOA.
The medical community has been attaching much importance to big data analysis by its remarkable capability to uncover meaningful patterns from large amounts of data without a prior hypothesis, which becomes an indispensable tool to elucidate the effect of different risk factors to KOA development [11], such as the work by Du et al. who applied neural networks for automatic classification of KOA severity based on the knee cartilage damage index [12]. Recent attempts have leveraged this technique for KOA phenotyping and progression prediction [11,13,14]. Lamentably, only a narrow scope of risk factors was investigated, failing to give a comprehensive insight into KOA’s disease nature. Moreover, another limitation stemmed from the use of traditional statistical models, namely Logistic Regression and tree-based models [13,14]. The former one may fail to model the potential non-linearity in the clinical dataset and implicit statistical interactions between risk factors while the latter requires laborious feature engineering based on specific domain knowledge to ensure promising prediction performance [15,16].
In this study, we aim to address the long-standing question regarding the relative contribution of the local and systemic factors towards the onset and deterioration of KOA by applying a novel deep learning approach. Thus, a neural network-based KOA progression prediction model was constructed with the database from Osteoarthritis Initiative (OAI) in the Foundation for the National Institutes of Health Osteoarthritis (FNIH OA) Biomarkers Consortium. To strive for a more holistic analysis and robust prediction model than the previous studies, three major measures have been implemented. First, individuals with or without KOA history at baseline were further categorized into disease onset and deterioration study groups respectively [17]. Second, a more stringent KOA progression definition was employed based on the joint progression in symptomatic and radiographic perspectives, and over 20 well-received KOA risk factors in the realms of symptomatic information, demographic particulars, radiographic information, living habits, metabolic syndrome, and mechanical factors, were recruited. Third, our model was built on the multi-layer perceptron (MLP) deep neural network, which could outperform the aforementioned statistical frameworks for its capability in modelling non-linearity, and implicit statistical interaction among the input risk factors without intensive prior feature engineering [18]. On top of the carefully trained model, recently developed deep learning algorithms were also applied to decipher the relative contributions of each risk factors and the patterns of their statistical interactions in the onset and deterioration of KOA.
2. Methodology
2.1. Acquisition of dataset
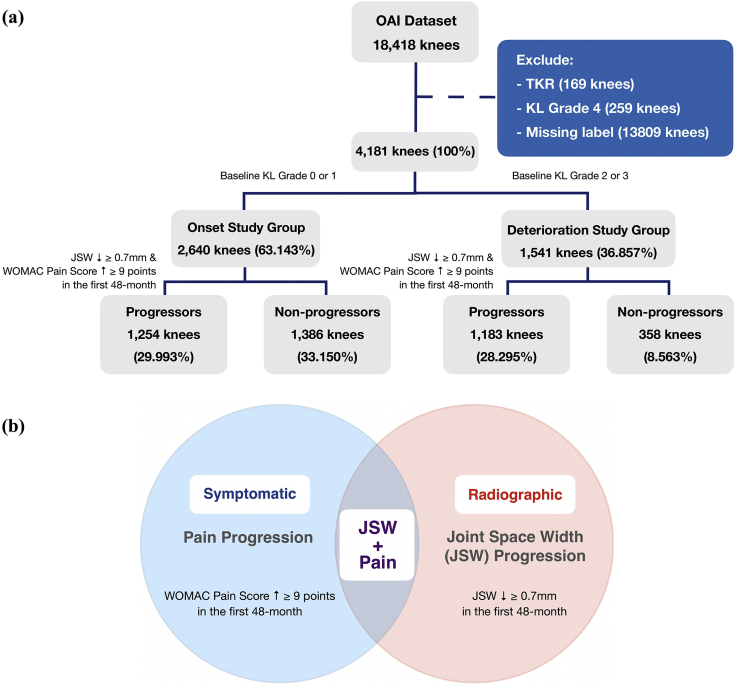
This project used the dataset from a publicly available source from OAI in the FNIH OA Biomarkers Consortium (https://data-archive.nimh.nih.gov/oai). The dataset comprised features, i.e. clinical and radiographic measurable properties, from 18,418 subjects, with each subject constituting to a data point for analysis. In our study, we only consider the knees with no, doubtful and moderate OA defined by Kellgren Lawrence Grade (KL-grade 0, 1, 2 and 3) at baseline (first-time visit). As a result, the entries with baseline KL-grade 4 and those having received total knee replacement (TKR) surgery were first excluded. Moreover, samples that showed no progression and dropped out of the study before the 48-month follow-up were view as data with missing labels and were subsequently ruled out. After the selection procedure, we employed the remaining 4181 knees for our following investigations. Later, to allow for the prediction of disease onset and deterioration separately, the dataset was further divided to categorize the subjects with and without KOA history by examining their KL-grade at baseline. Specifically, subjects with KL-grade 0 or 1, which signify normal and doubtful cases, were classified as historically unaffected by KOA (onset study group), comprising 2640 knees. While those with KL-grade 2 or above at baseline were regarded as pre-existing confirmed cases that form the deterioration study group, containing 1541 knees. (Fig. 1a).
Fig. 1.
(a) The progression definition adopted by this paper. (b) The data selection flow diagram.
2.2. Class labelling
After obtaining the data from OAI, its data points must be labelled as different classes to give a framework for the supervised learning algorithm to be trained. KOA is usually divided into symptomatic and radiographic as these two subtypes are not always overlapped [19]. There are two definitions for KOA development: one for pain, and one for radiographic progression. Pain progression refers to a persistent increase of at least 9 points under a normalized scale from 0 to 100 of McMaster Universities Osteoarthritis Index (WOMAC) Pain Score [20,21]. While radiographic progression is indicated by the loss in the medial knee joint space width (JSW) of at least 0.7mm.
Here, we adopted the definition of KOA based on both radiographic and symptomatic KOA changes, which has been demonstrated to clearly cluster the progressors and non-progressors in the OAI dataset [11](Fig. 1b). Progressor class consisted of subjects who demonstrated progression in the perspectives of both JSW and Pain simultaneously within the first 48-month period after the first clinical record while the rest would fall under the category of non-progressor class. Altogether, in the KOA onset study group, there are 1254 progressive and 1386 non-progressive knees. Whereas the disease deterioration study group comprises 1,183and 358 progressive and non-progressive knees respectively (Fig. 1a).
2.3. Missing data handling
In the database, there were over 60% of samples having missing entries for at least one risk factor. In order to fully utilize the entire dataset and maintain an adequate sample size for subsequent training of the supervised learning model, instead of dropping those incomplete samples, the Multivariate Imputation by Chained Equation (MICE) algorithm was employed. The algorithm focuses on one input variable at a time, and leverage all other variables to predict its missing values based on a regression model, such operation is performed recurrently for each variable with missing entries [22] to complete the dataset. However, for subjects with any missing labels to determine their progression class, their entire entry would be excluded from the study.
2.4. Class imbalance handling
As shown in Fig. 1a, a noticeable class imbalance could be observed among KOA Progressors and Non-progressors. In particular, the number of data points in the Non-progressor class was extremely small, such class imbalance could result in a highly biased model. To mitigate this problem, Synthetic Minority Over-sampling Technique (SMOTE) was implemented during the training of both MLP and Logistic Regression Models to statistically synthesise more data in the minority class [23], thereby balancing the class distribution which ensures a better convergence of the models. To be specific, SMOTE first selects a minority class sample X randomly and finds its k-nearest minority class neighbours. The synthetic sample is subsequently created by choosing one of the k nearest neighbours Y at random, followed by connecting X and Y to form a line segment in the high dimensional feature space. The synthetic instances are generated as a convex combination of the two chosen instances X and Y [24]. This approach is effective as new synthetic samples from the minority class were created in a plausible manner conforming to the geometrical distribution of the original data from the minority class. Compared to other imbalanced data handling approaches, such as under-sampling of the majority class or the naive over-sampling minority class method by randomly duplicating the existing minority class data, SMOTE ameliorates the data imbalance situation without loss of training data, and prevents overfitting stemming from repeatedly learning on duplicated samples [23].
2.5. Machine learning model
Two Multi-layer Perceptron (MLP) models, one for onset progression prediction and one for deterioration prediction, were built as a binary classifier to discern Progressors and Non-progressors. The MLP architectures, i.e. the number of hidden layers and hidden units in each layer, were tuned to produce optimal classification results. For the KOA onset prediction, the most optimised architecture was found to be consisting of four hidden layers, each with 100, 100, 50 and 10 hidden units respectively. Whereas the MLP for deterioration prediction contains 3 hidden layers each has 80, 80 and 30 hidden units respectively. Both neural networks have an output layer containing sigmoid function which generates binary classification outputs. Rectified linear unit (ReLU) was chosen to be the activation function in each neuron. In particular, preceded by normalization with L2 norm of the input variables, the model was trained using PyTorch 1.4.0 library in Python 3.6 environment with the Binary Cross-Entropy (BCE) as loss function and adaptive moment estimation (Adam) as optimisation algorithm [25]. The learning rate and weight decay were set to be 0.01 and 0.0005 respectively. Additionally, to avoid overfitting, dropout = 0.5 was applied in every hidden-layer during the training process. The aforementioned hyperparameters, including the number of hidden layers, number of hidden units, dropout probability, weight decay and number of training epochs were obtained using random search [26] with 5-fold cross-validation. The tuning ranges of the hyperparameters are listed in (Supplementary Table 2).
In order to demonstrate the added value of using MLP over the traditional statistical models used in previous literature [13,14], utilizing Scikit-learn 0.23.1 package, we additionally trained a Logistic Regression with the same progression definition and input risk factors as the MLP for both KOA onset and deterioration cases. Concurrently, a decision tree was also implemented with an aim for elucidating whether MLP is more advantageous over other non-linear classifiers in our dataset. For all models, 60% of the data were allocated as the training set and 20% for validation, while the remaining 20% of the samples were recruited as the test set (see Table 1).
Table 1.
Summary of statistics of risk factors employed in the study.
| Risk Factor Category | Risk Factor | Onset Study Group | Deterioration Study Group | |
|---|---|---|---|---|
| Demographic Information | Age | Range | 45–79 | 45–79 |
| Mean | 59.39 | 62.26 | ||
| Standard Dev. | 8.88 | 8.64 | ||
| Weight | Range | 42.6–132.8 kg | 48.3–135.5 kg | |
| Mean | 79.59 kg | 85.14 kg | ||
| Standard Dev. | 15.89 kg | 15.54 kg | ||
| Sex | Male | 42.93% | 40.12% | |
| Female | 57.07% | 59.88% | ||
| Race | White | 86.47% | 74.57% | |
| Black | 11.38% | 23.18% | ||
| Asian | 1.36% | 0.71% | ||
| Other | 0.79% | 1.54% | ||
| Mechanical Factors | History of Injury | Yes | 19.22% | 32.01% |
| No | 79.68% | 67.99% | ||
| History of Surgery | Yes | 4.67% | 18.35% | |
| No | 95.32 | 81.58% | ||
| Radiographic Information | Joint Space Width (Medial) | Range | 2.67–7.30 mm | 0.48–8.49 mm |
| Mean | 4.71 mm | 3.93 mm | ||
| Standard Dev. | 0.86 mm | 1.35 mm | ||
| OARSI Joint Space Narrowing (Medial) | Range | 0-1 (out of 3) | 0-2 (out of 3) | |
| Mean | 0.1576 | 1.000 | ||
| Standard Dev. | 0.3644 | 0.8150 | ||
| OARSI Joint Space Narrowing (Lateral) | Range | 0-1 (out of 3) | 0-2 (out of 3) | |
| Mean | 0.02 | 0.17 | ||
| Standard Dev. | 0.13 | 0.50 | ||
| OARSI Osteophyte (Medial Femoral + Medial Tibial) | Range | 0-2 (out of 3) | 0-6 (out of 6) | |
| Mean | 0.13 | 2.19 | ||
| Standard Dev. | 0.35 | 1.55 | ||
| OARSI Osteophyte (Lateral Femoral + Lateral Tibial) | Range | 0-2 (out of 3) | 0-6 (out of 6) | |
| Mean | 0.09 | 1.47 | ||
| Standard Dev. | 0.31 | 1.57 | ||
| Symptomatic Information | WOMAC Total Score | Range | 0-62 (out of 96) | 0-96 (out of 96) |
| Mean | 7.43 | 15.60 | ||
| Standard Dev. | 10.88 | 16.68 | ||
| WOMAC Pain Score | Range | 0-15 (out of 20) | 0-20 (out of 20) | |
| Mean | 1.46 | 3.14 | ||
| Standard Dev. | 2.39 | 3.68 | ||
| WOMAC Stiffness Score | Range | 0-8 (out of 8) | 0-8 (out of 8) | |
| Mean | 1.06 | 1.82 | ||
| Standard Dev. | 1.33 | 1.73 | ||
| WOMAC Disability Score | Range | 0-43 (out of 68) | 0-68 (out of 68) | |
| Mean | 4.91 | 10.64 | ||
| Standard Dev. | 7.86 | 12.05 | ||
| Metabolic Syndromes | Body Mass Index (BMI) | Range | 17.70–43.72 | 18.51–48.73 |
| Mean | 27.93 | 30.08 | ||
| Standard Dev. | 4.48 | 4.81 | ||
| Systolic Blood Pressure | Range | 86–190 | 90–200 | |
| Mean | 121.95 | 126.67 | ||
| Standard Dev. | 15.83 | 16.16 | ||
| Diastolic Blood Pressure | Range | 44–110 | 48–120 | |
| Mean | 75.09 | 76.71 | ||
| Standard Dev. | 9.64 | 9.98 | ||
| Mean Arterial Pressure | Range | 18–98 | 18–106 | |
| Mean | 46.86 | 49.96 | ||
| Standard Dev. | 13.25 | 13.48 | ||
| Diabetes | Yes | 5.84% | 8.56% | |
| No | 91.67% | 90.01% | ||
| Missing | 2.49% | 1.43% | ||
| Living Habits | Alcohol Intake Habit | Range | 0–7 | 0–7 |
| Definition | 0: None; 1: less than once/week; 2: 1–3 drinks/week; 3: 4–7 drinks/week; 4: 8–14 drinks/week; 5: 15–21 drinks/week; 6: 22–27 drinks/week; 7: 28+ drinks/week |
|||
| Mean | 1.81 | 1.76 | ||
| Standard Dev. | 1.44 | 1.52 | ||
| Beer Intake Habit | Range | 1–9 | 1–9 | |
| Definition | 1: Never; 2: A few times per year; 3: Once/month; 4: 2–3 times/month; 5: Once/week; 6: Twice/week; 7: 3–4 times/week; 8: 5–6 times/week; 9: Every day |
|||
| Mean | 2.74 | 2.66 | ||
| Standard Dev. | 2.02 | 2.11 | ||
| Milk Intake Habit | Range | 1–9 | 1–9 | |
| Definition | 1: Never; 2: A few times per year; 3: Once/month; 4: 2–3 times/month; 5: Once/week; 6: Twice/week; 7: 3–4 times/week; 8: 5–6 times/week; 9: Every day |
|||
| Mean | 1.54 | 1.64 | ||
| Standard Dev. | 1.36 | 1.61 | ||
| Smoking Habit | Yes | 23.37% | 21.96% | |
| No | 76.63% | 77.46% | ||
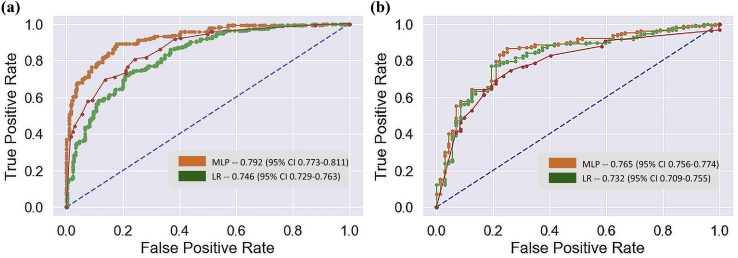
All the models were assessed by five performance metrics, namely accuracy, precision, recall, F1-score and area under receiver operating characteristic curve (AUC) to give a more holistic view and comparison of the models’ performance (Table 2). To compute performance metrics on the test set, stratified bootstrapping with 10 iterations was employed, which enabled us to reliably assess the confidence intervals of each score. In addition to the scores, ROC curves (Fig. 2) were also plotted for model comparison in both onset and deterioration predictions of KOA.
Table 2.
Detailed summary of performance of the statistical learning models on the test set. LR – Logistic Regression. DT – Decision Tree. MLP – Multilayer Perceptron.
| Model | Progression Status | Accuracy | Precision | Recall | F1 | AUROC |
|---|---|---|---|---|---|---|
| LR | Onset | 0.746 (0.725–0.767) | 0.728 (0.709–0.747) | 0.745 (0.725–0.765) | 0.736 (0.721–0.751) | 0.746 (0.729–0.763) |
| DT | 0.771 (0.752–0.790) | 0.846 (0.826–0.866) | 0.633 (0.612–0.654) | 0.724 (0.705–0.743) | 0.764 (0.742–0.786) | |
| MLP | 0.843 (0.823–0.863) | 0.826 (0.809–0.843) | 0.849 (0.830–0.868) | 0.837 (0.816–0.858) | 0.843 (0.824–0.862) | |
| LR | Deterioration | 0.692 (0.669–0.715) | 0.917 (0.900–0.934) | 0.658 (0.638–0.678) | 0.766 (0.748–0.784) | 0.742 (0.719–0.765) |
| DT | 0.680 (0.656–0.704) | 0.920 (0.894–0.946) | 0.637 (0.615–0.659) | 0.753 (0.728–0.778) | 0.710 (0.690–0.730) | |
| MLP | 0.744 (0.724–0.764) | 0.943 (0.920–0.966) | 0.709 (0.690–0.728) | 0.810 (0.790–0.83) | 0.765 (0.756–0.774) |
Fig. 2.
Comparison of the statistical models’ performance with ROC curves. Blue dashed lines signify the performance of a random classifier. The legends in the subplots indicates the AUC of the statistical models with 95% confidence intervals. (a) Demonstrates ROC curves of the Multi-layer Perceptron, Decision Tree and Logistic Regression models for prediction of KOA onset and (b) for deterioration of KOA.
2.6. Clustering
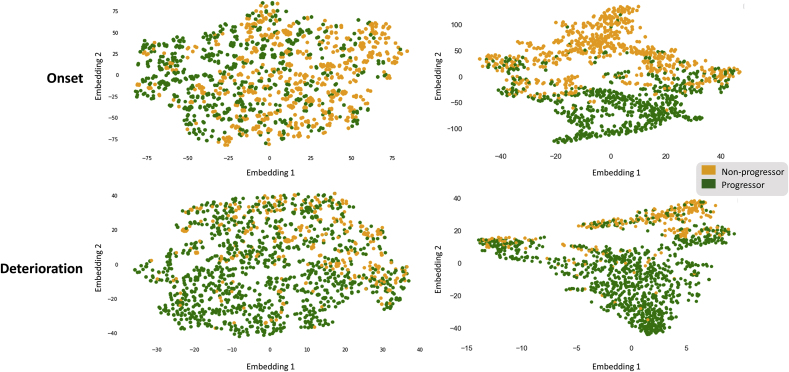
As a visualization of separation between the Progressor and Non-progressor Classes, the t-distributed Stochastic Neighbor Embedding (t-SNE) was employed. This dimensionality reduction method is capable of embedding high-dimensional data into low-dimensional representation, leading to the easier realization of segregation of classes within a complex data structure [27]. Here, t-SNE was employed to provide a visual comparison between the raw data as well as the data projected by the first two hidden-layers of the MLPs to demonstrate the class segregation efficiency of the deep neural network model (Fig. 3). Additionally, we leverage the clustering technique to visualize the class distribution of raw data under different progression definitions, namely JSW, Pain and JSW-Pain progressions. (Supplementary Figure 2).
Fig. 3.
The t-SNE clustering visualization. The upper panel corresponds to the onset KOA progression, the lower panel shows the data distribution of the pre-existing KOA progression. The two plots on the left reveal the distribution of the data before being processed by the MLP, whereas the plots on the right-hand side visualize the distribution of data after projection by the MLP.
2.7. Feature contribution and statistical interaction
Since the neural network is considered as a black box, in which the complex decision process is unknown [28]. In an attempt to visualize its decision-making rationale, a recently developed approach in the deep learning community, namely DeepLIFT, was employed to the trained MLP model [29,30]. In this analysis, the contribution of each risk factor to the model prediction would be quantified by calculating their relative backpropagated gradients, with respect to the model prediction output. More specifically, the algorithm aims to explain the difference in the output from some reference output in terms of the difference of the input from some reference input where the reference is chosen in a problem-specific manner, usually a numerical value which is absent in the range of the input data. In our case, as all the input values are greater than 0, so reference = −1 was chosen. In a nutshell, the greater the difference a unit change of an input feature exerts onto the model output, the greater the contribution of it towards the prediction [31]. As a result, the algorithm effectively dissects the trained neural network to readily interpret the importance of each risk factor. We implemented DeepLIFT algorithm using Captum library in Python 3.6 environment.
Statistical interaction describes a situation in which the effect of one causal input variable on the outcome depends on a second causal variable, or equivalently, the effects of the two interacting variables towards the model prediction are non-additive. Unravelling the interaction patterns within the dataset allows identification of the synergistic and joint involvements among the risk factors [32,33]. Unlike regular additive statistical models, neural networks inherently process the feature interactions through their non-linear activation functions and densely connected hidden units [18] in a way that the interaction information is encoded in the weight matrices [34]. To this end, we employed the Neural Interaction Detection (NID) method [18] to uncover the statistical interactions between risk factors captured by the MLP model. Through the acquisition of trained weights from the hidden layers in the neural network, the pairwise statistically interacting candidates were discovered with a special coefficient signifying the strength of the detected interaction, the stronger it is, the larger the value. We subsequently constructed a feature interaction network from the interaction strength coefficients using NetworkX 2.5 library in Python. Here, the nodes and edges represent the input risk factors and the detected pairwise statistical interactions respectively, whereas greater interaction strengths are highlighted by thicker and darker edges.
3. Results
Three different machine learning models have been built, including logistic regression, decision tree and a multilayer perceptron (MLP) to predict the onset and deterioration of KOA respectively. Their performance was compared in Table 2 and Fig. 2. The MLP model outperforms the logistic regression models in both disease onset and deterioration prediction. Such a phenomenon could be attributed to the complex non-linearity lying in the data structure. Thus, linear classification model may fall short in handling the complexity of the data point distribution in this dataset when compared with its non-linear counterpart. Coupled with that, we also compared our MLP model with a decision tree, which is also a non-linear model adopted by Lazzarini et al. [14]. It is worth noticing that despite the non-linear nature of the tree-based model, it is still outperformed by the MLP in most performance metrics. The result is in line with our hypothesis that the dataset is not only non-linear by nature but also contains confounded and interacting risk factors which could be well-captured by the MLP.
The clustering visualization with t-SNE gave an intuitive perception as to how are the data correspond to the progressor and non-progressor classes distributed by projecting them on an arbitrary two-dimensional plane. As shown in Fig. 3, compared to the distribution of the raw data, two distinct clusters representing Non-progressors and Progressors respectively, emerge in the data projected by the first two layers of the MLP with much less overlapping region among the two classes, especially in the onset prediction; in the KOA deterioration prediction, although the improvement of the clustering is not as obvious, one could still observe that after the projection, the non-progressors are becoming more densely localised in the upper right corner compared to the raw data. This indicates the MLP’s capability in projecting the data onto latent space where clearer decision boundaries can be found between two groups of data in both KOA onset and deterioration predictions, thereby better model performance could be achieved, which also conformed with the performance scores shown in Table 2. Besides, distributions of data under different progression definitions were also demonstrated, as shown from Supplementary Figure 2, the Progressors and Non-progressors are largely overlapped under the JSW and Pain progression definitions in both onset and deterioration conditions. Despite opaque decision boundaries, notable reductions in the overlapping region between opposite classes can be observed under the definition of JSW-Pain joint progression. This result may suggest the combination of both symptomatic and radiographic information as OA progression definition would allow better initial segregation of the two classes, thereby benefitting the subsequent classification by the statistical learning models, Consistent with this notion, as reflected by our data documented in Supplementary Table 3, the MLP model trained based on the definition of JSW-Pain progression significantly outperforms those using only single perspective (i.e. either JSW or Pain) as progression definitions, in all performance metrics when considering onset and deterioration predictions of the disease.
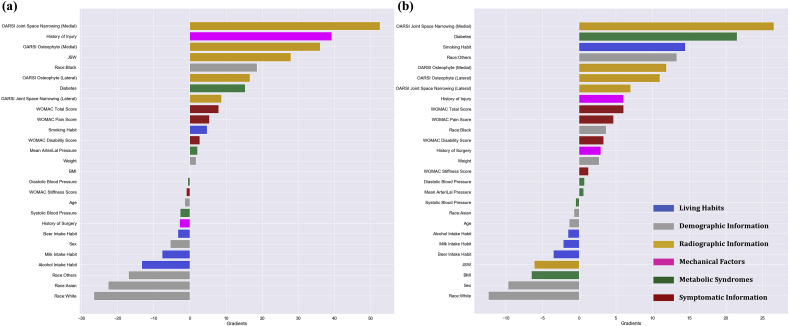
Fig. 4 displayed the gradients of every feature from the two MLP models respectively. Considering the onset scenario, the greatest contribution to non-progression (negative gradients), was seen for the habits of milk and alcohol intake, races of white, Asian and other ethnic minorities in the United States. On the flip, the increase in OARSI joint space narrowing and osteophyte grades in the medial compartment, joint space width, history of mechanical injury are found conducive to the progression. In the perspective of KOA deterioration, the increase in OARSI joint space narrowing score, diabetes and habits of smoking are inducive factors towards progression. While white race, female sex, increase in BMI and joint space width are identified as major contributors to non-progression.
Fig. 4.
DeepLIFT gradient plots of the input risk factors. The risk factor categories are indicated by different colors, blue for living habits, grey for demographic information of the subjects, orange for radiographic information, pink for mechanical factors, green for metabolic syndromes and brown for symptomatic information. (a) Demonstrates the gradient of each risk factor in the KOA onset prediction and (b) displays gradients in the prediction of KOA deterioration. Positive gradient would indicate that as the value of the feature increases, the prediction of positive class (i.e. progressors) is favored. Contrariwise, a negative gradient implies increasing the feature value would result in a higher tendency of prediction towards the negative class (i.e. non-progressors). Meanwhile, the magnitude of the gradient correlates to the extent of influence and contribution in the model’s prediction.
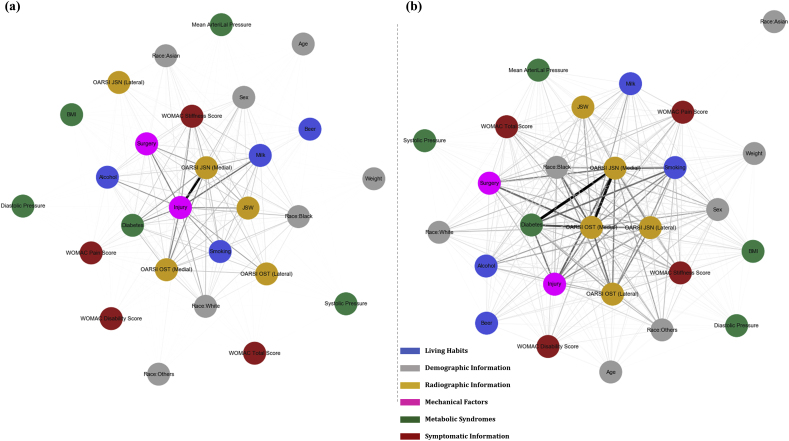
Further efforts have been made to identify the statistical interactions captured by the MLP. From the network in Fig. 5, it is observed that, for onset progression, history of injury is strongly interacting with the medial OARSI joint space narrowing score. Other risk factors, such as the history of surgery, diabetes, habits of milk and alcohol intake as well as medial OARSI osteophyte score also show noticeable statistical interaction with each other. On the other hand, in the case of disease deterioration, diabetes, medial OARSI joint space narrowing and osteophyte scores forged the most significant interaction with each other, meaning a high degree of association could be found between these factors in the context of disease deterioration prediction. It is noteworthy that, unlike the onset scenario, lateral OARSI joint space narrowing and osteophyte scores were also found in the subset of risk factors having conspicuous interactions with the medial OARSI features. Besides, distinguished interactions were identified among other factors, such as smoking habit and history of injury and surgery. In a greater perspective, more complex statistical interaction structure could be observed, in which a larger subset of risk factors was found to statistically interacting with one another in the network of KOA deterioration than that of disease onset.
Fig. 5.
Statistical feature interaction network. The network visualizes the non-linear interaction detected in our MLP models by the Neural Interaction Detection algorithm. The colors signify different categories of the risk factors, blue for living habits, grey for demographic information of the subjects, orange for radiographic information, pink for mechanical injury information and green for metabolic syndromes and brown for symptomatic information. (a) Reveals statistical interactions in the KOA onset prediction and (b) displays statistical interactions in the prediction of KOA deterioration.
4. Discussion
In this study, through the application of an advanced data-driven approach to the FNIH cohort database, a machine-learning-based KOA progression prediction model was developed where a multitude of risk factors, including symptomatic information, demographic particulars, radiographic information, living habits, metabolic syndrome, and mechanical factors, were recruited simultaneously for a holistic analysis. Deep feedforward neural network (or inter-changeably MLP) was developed to specifically capture the non-linearity in the dataset and confounded interactions among the input risk factors. The model has attained high performance of 0.843 (95% CI 0.824, 0.862) and 0.765 (95% CI 0.756, 0.774) AUC in the prediction of KOA onset and deterioration respectively, which is well-exhibited by a clear separation of progressor and non-progressor classes in the t-SNE plot after the data were processed by the MLPs. In addition, by benchmarking with the models, such as logistic regression and decision tree which were employed in previous pieces of literature [13,14], we further demonstrated the superiority of our method over the traditional statistical learning approaches.
It is worth noticing that in this study, following Nelson et al.’s approach [11], a joint progression definition based on both symptomatic and radiographic perspectives were adopted. While most of the existing machine learning models only rely on single-perspective KOA progression for model development [13,14,35]; through the visualization of the data distribution, it was however revealed that either symptomatic or radiographic perspective is insufficient to define a clear decision boundary for KOA progression prediction (Supplementary Figure 2). On the flip side, when considering both distinct perspectives simultaneously, two clusters correspond to Progressors and Non-progressors class becomes more conspicuous. On top of that, the MLP models developed based on JSW-Pain joint progression outperformed the ones trained on single-perspective KOA progressions by a significant degree (Supplementary Table 3) in both onset and deterioration predictions. These results provide a strong indication that recruiting both KOA progression types, JSW and pain progressions would establish a more holistic and stringent classification basis, hence providing a multiple-perspective analysis for the prediction model to learn from.
Later, via the application of a recently developed technique from the deep learning community, DeepLIFT on our trained MLP, we successfully identified the relative contribution of each risk factor, where it was revealed that the onset of KOA is highly attributable to the mechanical injury. This is in agreement with previous studies which identified mechanical injury as a major risk factor predisposing the development of KOA [[36], [37], [38], [39], [40]]. More than that, our result further indicates mechanical injury’s particular influence in the disease’s onset, where the early joint injury was recently argued to trigger an inflammatory response or abnormal mechanics, resulting in cartilage degradation. The repercussion of such damage in the knee joint would be the commencement of KOA in the end [2,3]. As one of the few modifiable or preventable risk factors, this finding may suggest future prevention programme in reducing the risk of knee OA onset.
On the other hand, our findings also suggest the predominant role of metabolic syndromes, especially diabetes in triggering the deterioration of the disease. Recent understanding of OA has been suggesting its correspondence to not merely local mechanical features, but also a broader spectrum of systemic factors. Particularly, mounting evidence has propounded a significant association between diabetes and OA [[6], [7], [8]]. As one of the most instrumental risk factors identified in our study, we show further evidence that diabetes may play a pivotal role specifically in the deterioration of KOA. Such a result may as well be evident to a recent hypothesis that connects diabetes and the worsening of KOA: Firstly, the peripheral nervous system may be altered by diabetic neuropathy in the OA patients, leading to muscle weakness and joint laxity and as a result causing deterioration of KOA. Besides, hyperglycemia-induced low-grade systemic inflammatory response, which is potentially associated with the loss in cartilage and delayed tendon healing after injury [41], would also induce further progression of OA.
Second to diabetes, smoking was found to be another highly contributory feature conduces to KOA deterioration, which however seemingly contradicts to a number of findings contending smoking’s protective role towards OA [42,43], as explained by the upregulation of glycosaminoglycan and collagen synthetic activity of articularchondrocytes by nicotine found in cigarettes [44]. On the flip side, nicotine has been reported to excite corresponding receptors on neuronal cells which could ultimately engender musculoskeletal pain [45]. Meanwhile, increased carbon monoxide levels in arterial blood upon cigarette smoking could contribute to tissue hypoxia, which may, in turn, hinder cartilage repair in smokers [46]. As such, smoking might bear contradictory effects in radiographic and symptomatic perspectives of OA on account of its involvement in a multitude of pathways pertains to the disease’s etiology [45]. As a corollary, in light of the JSW-Pain joint progression definition, our result regarding smoking is attributed to the summation of effects from both radiographic and symptomatic pathways of the disease. The identification of diabetes and smoking as influential predictors to the worsening of KOA could bring deeper hindsight to disease management by intervening in the sufferers’ living style, for instance, diet planning and cessation of smoking.
Finally, the interactions between risk factors were also, to our best knowledge for the first time, being recovered in both KOA onset and deterioration scenarios using the “Neural Interaction Detection” (NID) algorithm. The detected pairwise statistical interactions were visualised as network, in which the interacting risk factors would impose non-additive effects on the prediction output owing to the confounded high-order associations among them [47]. In the prediction of KOA onset, history of mechanical injury and medial OARSI joint space narrowing grade forms the most significant interaction; considering the disease deterioration, in place of history of injury, diabetes becomes the most profoundly interacting factor with not only the medial but also lateral OARSI osteophyte and joint space narrowing grades, indicating the whole joint involvement and possible coalition with systemic factors upon KOA deterioration. Generally speaking, the interactions among local and systemic risk factors in the deterioration cases was much more complex and complicated than in the onset cases.
5. Conclusion
We have applied a novel deep learning framework to decipher the multi-etiology of knee osteoarthritis. Our model, which is capable of modelling nonlinearity and interactions between input factors, has achieved high performance in predicting the predisposal of disease’s onset and deterioration. More notably, our approach has demonstrated the discrepancy in the contribution of a variety of local and systemic risk factors and statistical interaction patterns in the onset and deterioration of KOA. To be concise, local risk factor such as the history of injury is predominant in the onset of disease; whereas systemic risk factor, diabetes and smoking, in particular, comes to our notice regarding their dominant role in disease deterioration. Besides, with more complex statistical interaction patterns, the worsening process of KOA may involve the interplay between a larger subset of risk factors than the disease incidence. Our findings highlight the importance of injury prevention and cessation of smoking in the management of KOA. Management of the co-morbidities in KOA patients, such as diabetes, is also essential to halt disease progression. In a broader sense, it is anticipated that the approach presented herein may as well inspire further development of self-efficiency and self-management programs for early detection of KOA.
Author contributions
HL, LC, PC and CW conceived the study. HL and LC collected data. All authors contributed to the writing of the manuscript and approved the final version.
Role of funding source
This work was supported by Research Grants Council of Hong Kong Early Career Scheme (PolyU 251008/18 M), PROCORE-France/Hong Kong Joint Research Scheme (F-PolyU504/18) and also Health and Medical Research Fund Scheme (01150087#, 15161391#, 16172691#).
Declaration of competing interest
The authors have no relevant competing interests to disclose.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ocarto.2020.100135.
Contributor Information
L.C. Chan, Email: lc-justin.chan@connect.polyu.hk.
H.H.T. Li, Email: toby.li@connect.polyu.hk.
P.K. Chan, Email: cpk464@yahoo.com.hk.
C. Wen, Email: chunyi.wen@polyu.edu.hk.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wen C., Lu W.W., Chiu K.Y. Importance of subchondral bone in the pathogenesis and management of osteoarthritis from bench to bed. Journal of Orthopaedic Translation. 2014;2(1):16–25. [Google Scholar]
- 2.Felson D.T. Osteoarthritis as a disease of mechanics. Osteoarthritis Cartilage. 2013;21(1):10–15. doi: 10.1016/j.joca.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincent T.L. vol. 2019. Elsevier; 2019. Mechanoflammation in osteoarthritis pathogenesis; pp. S36–S38. (Seminars in Arthritis and Rheumatism). [DOI] [PubMed] [Google Scholar]
- 4.Wallace I.J., Worthington S., Felson D.T., Jurmain R.D., Wren K.T., Maijanen H., et al. Knee osteoarthritis has doubled in prevalence since the mid-20th century. Proc. Natl. Acad. Sci. Unit. States Am. 2017;114(35):9332–9336. doi: 10.1073/pnas.1703856114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhuo Q., Yang W., Chen J., Wang Y. Metabolic syndrome meets osteoarthritis. Nat. Rev. Rheumatol. 2012;8(12):729. doi: 10.1038/nrrheum.2012.135. [DOI] [PubMed] [Google Scholar]
- 6.Eymard F., Parsons C., Edwards M., Petit-Dop F., Reginster J.-Y., Bruyère O., et al. Diabetes is a risk factor for knee osteoarthritis progression. Osteoarthritis Cartilage. 2015;23(6):851–859. doi: 10.1016/j.joca.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Nieves-Plaza M., Castro-Santana L.E., Font Y.M., Mayor A.M., Vilá L.M. Association of hand or knee osteoarthritis with diabetes mellitus in a population of Hispanics from Puerto Rico. J. Clin. Rheumatol.: practical reports on rheumatic & musculoskeletal diseases. 2013;19(1) doi: 10.1097/RHU.0b013e31827cd578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Louati K., Vidal C., Berenbaum F., Sellam J. Association between diabetes mellitus and osteoarthritis: systematic literature review and meta-analysis. RMD open. 2015;1(1) doi: 10.1136/rmdopen-2015-000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veronese N., Stubbs B., Solmi M., Smith T.O., Noale M., Schofield P., et al. Knee osteoarthritis and risk of hypertension: a longitudinal cohort study. Rejuvenation Res. 2018;21(1):15–21. doi: 10.1089/rej.2017.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wen C., Chen Y., Tang H., Yan C., Lu W., Chiu K. Bone loss at subchondral plate in knee osteoarthritis patients with hypertension and type 2 diabetes mellitus. Osteoarthritis Cartilage. 2013;21(11):1716–1723. doi: 10.1016/j.joca.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 11.Nelson A.E., Fang F., Arbeeva L., Cleveland R.J., Schwartz T.A., Callahan L.F., et al. A machine learning approach to knee osteoarthritis phenotyping: data from the FNIH Biomarkers Consortium. Osteoarthritis Cartilage. 2019;27(7):994–1001. doi: 10.1016/j.joca.2018.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du Y., Shan J., Almajalid R., Zhang M. 2018 IEEE/ACM International Conference on Connected Health: Applications, Systems and Engineering Technologies (CHASE); 2018. IEEE; 2018. Knee osteoarthritis severity level classification using whole knee cartilage damage Index and ANN; pp. 19–21. [Google Scholar]
- 13.Tiulpin A., Klein S., Bierma-Zeinstra S.M.A., Thevenot J., Rahtu E., Meurs Jv, et al. Multimodal machine learning-based knee osteoarthritis progression prediction from plain radiographs and clinical data. Sci. Rep. 2019;9(1):20038. doi: 10.1038/s41598-019-56527-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazzarini N., Runhaar J., Bay-Jensen A., Thudium C., Bierma-Zeinstra S., Henrotin Y., et al. A machine learning approach for the identification of new biomarkers for knee osteoarthritis development in overweight and obese women. Osteoarthritis Cartilage. 2017;25(12):2014–2021. doi: 10.1016/j.joca.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Hastie T., Tibshirani R., Friedman J. second ed. ed. 2009. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. New York, NY. [Google Scholar]
- 16.Rendle S. vol. 2010. IEEE; 2010. Factorization machines; pp. 995–1000. (2010 IEEE International Conference on Data Mining). [Google Scholar]
- 17.Koster I.M., Oei E.H., Hensen J.-H.J., Boks S.S., Koes B.W., Vroegindeweij D., et al. Predictive factors for new onset or progression of knee osteoarthritis one year after trauma: MRI follow-up in general practice. Eur. Radiol. 2011;21(7):1509–1516. doi: 10.1007/s00330-011-2089-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsang M., Cheng D., Liu Y. 2017. Detecting Statistical Interactions from Neural Network Weights. arXiv preprint arXiv:170504977. [Google Scholar]
- 19.Conaghan P.G., Vanharanta H., Dieppe P.A. Is progressive osteoarthritis an atheromatous vascular disease? Ann. Rheum. Dis. 2005;64(11):1539–1541. doi: 10.1136/ard.2005.039263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roos M.K., Lohmander L.S., Womac E.M. Osteoarthritis Index: reliability, validity, and responsiveness in patients with arthroscopically assessed osteoarthritis. Scand. J. Rheumatol. 1999;28(4):210–215. doi: 10.1080/03009749950155562. [DOI] [PubMed] [Google Scholar]
- 21.Angst F., Aeschlimann A., Michel B.A., Stucki G. Minimal clinically important rehabilitation effects in patients with osteoarthritis of the lower extremities. J. Rheumatol. 2002;29(1):131–138. [PubMed] [Google Scholar]
- 22.Buuren Sv, Groothuis-Oudshoorn K. mice: Multivariate imputation by chained equations in R. J. Stat. Software. 2010:1–68. [Google Scholar]
- 23.Chawla N.V., Bowyer K.W., Hall L.O., Kegelmeyer W.P. SMOTE: synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002;16:321–357. [Google Scholar]
- 24.He H., Ma Y. John Wiley & Sons; 2013. Imbalanced Learning: Foundations, Algorithms, and Applications. [Google Scholar]
- 25.Kingma D.P., Ba J. 2014. Adam: A Method for Stochastic Optimization. arXiv preprint arXiv:14126980. [Google Scholar]
- 26.Bergstra J., Bengio Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012;13(1):281–305. [Google Scholar]
- 27.van der Maaten L.J.P., Hinton G.E. Visualizing high-dimensional data using t-SNE. J. Mach. Learn. Res. 2008;9(nov):2579–2605. [Google Scholar]
- 28.Fong R.C., Vedaldi A. vol. 2017. 2017. Interpretable explanations of black boxes by meaningful perturbation; pp. 3429–3437. (Proceedings of the IEEE International Conference on Computer Vision). [Google Scholar]
- 29.Shrikumar A., Greenside P., Kundaje A. 2017. Learning Important Features through Propagating Activation Differences. arXiv preprint arXiv:170402685. [Google Scholar]
- 30.Sundararajan M., Taly A., Yan Q. 2017. Axiomatic Attribution for Deep Networks. arXiv preprint arXiv:170301365. [Google Scholar]
- 31.Ancona M., Ceolini E., Öztireli C., Gross M. 2017. Towards Better Understanding of Gradient-Based Attribution Methods for Deep Neural Networks. arXiv preprint arXiv:171106104. [Google Scholar]
- 32.De Gonzalez A.B., Cox D.R. Interpretation of interaction: a review. Ann. Appl. Stat. 2007;1(2):371–385. [Google Scholar]
- 33.Jaccard J., Wan C.K., Turrisi R. The detection and interpretation of interaction effects between continuous variables in multiple regression. Multivariate Behav. Res. 1990;25(4):467–478. doi: 10.1207/s15327906mbr2504_4. [DOI] [PubMed] [Google Scholar]
- 34.Tsang M., Liu H., Purushotham S., Murali P., Liu Y. Neural interaction transparency (nit): disentangling learned interactions for improved interpretability. Adv. Neural Inf. Process. Syst. 2018;2018:5804–5813. [Google Scholar]
- 35.Kerkhof H., Bierma-Zeinstra S., Arden N., Metrustry S., Castano-Betancourt M., Hart D., et al. Prediction model for knee osteoarthritis incidence, including clinical, genetic and biochemical risk factors. Ann. Rheum. Dis. 2014;73(12):2116–2121. doi: 10.1136/annrheumdis-2013-203620. [DOI] [PubMed] [Google Scholar]
- 36.Muthuri S., McWilliams D., Doherty M., Zhang W. History of knee injuries and knee osteoarthritis: a meta-analysis of observational studies. Osteoarthritis Cartilage. 2011;19(11):1286–1293. doi: 10.1016/j.joca.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 37.Driban J.B., Eaton C.B., Lo G.H., Ward R.J., Lu B., McAlindon T.E. Association of knee injuries with accelerated knee osteoarthritis progression: data from the Osteoarthritis Initiative. Arthritis Care Res. 2014;66(11):1673–1679. doi: 10.1002/acr.22359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roos E.M. Joint injury causes knee osteoarthritis in young adults. Curr. Opin. Rheumatol. 2005;17(2):195–200. doi: 10.1097/01.bor.0000151406.64393.00. [DOI] [PubMed] [Google Scholar]
- 39.Lohmander L., Östenberg A., Englund M., Roos H. High prevalence of knee osteoarthritis, pain, and functional limitations in female soccer players twelve years after anterior cruciate ligament injury. Arthritis Rheum.: Official Journal of the American College of Rheumatology. 2004;50(10):3145–3152. doi: 10.1002/art.20589. [DOI] [PubMed] [Google Scholar]
- 40.Øiestad B.E., Engebretsen L., Storheim K., Risberg M.A. Winner of the 2008 systematic review competition: knee osteoarthritis after anterior cruciate ligament injury. Am. J. Sports Med. 2009;37(7):1434–1443. doi: 10.1177/0363546509338827. [DOI] [PubMed] [Google Scholar]
- 41.King K., Rosenthal A. The adverse effects of diabetes on osteoarthritis: update on clinical evidence and molecular mechanisms. Osteoarthritis Cartilage. 2015;23(6):841–850. doi: 10.1016/j.joca.2015.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berenbaum F. Diabetes-induced osteoarthritis: from a new paradigm to a new phenotype. Postgrad. Med. 2012;88(1038):240–242. doi: 10.1136/pgmj.2010.146399rep. [DOI] [PubMed] [Google Scholar]
- 43.Järvholm B., Lewold S., Malchau H., Vingård E. Age, bodyweight, smoking habits and the risk of severe osteoarthritis in the hip and knee in men. Eur. J. Epidemiol. 2005;20(6):537–542. doi: 10.1007/s10654-005-4263-x. [DOI] [PubMed] [Google Scholar]
- 44.Gullahorn L., Lippiello L., Karpman R. Smoking and osteoarthritis: differential effect of nicotine on human chondrocyte glycosaminoglycan and collagen synthesis. Osteoarthritis Cartilage. 2005;13(10):942–943. doi: 10.1016/j.joca.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Felson D., Zhang Y. Elsevier; 2015. Smoking and Osteoarthritis: a Review of the Evidence and its Implications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amin S., Niu J., Guermazi A., Grigoryan M., Hunter D.J., Clancy M., et al. Cigarette smoking and the risk for cartilage loss and knee pain in men with knee osteoarthritis. Ann. Rheum. Dis. 2007;66(1):18–22. doi: 10.1136/ard.2006.056697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao J., Ye H., He X., Zhang H., Wu F., Chua T.-S. 2017. Attentional Factorization Machines: Learning the Weight of Feature Interactions via Attention Networks. arXiv preprint arXiv:170804617. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.