Abstract
Scrub typhus, caused by Orientia tsutsugamushi infection, is characterized by local as well as systemic inflammatory manifestations. Inflammation is initiated by O. tsutsugamushi-infected macrophages and endothelial cells in the dermis. We investigated the regulation of chemokine induction in macrophage cell line J774A.1 in response to O. tsutsugamushi infection. The mRNAs for macrophage inflammatory proteins 1α/β (MIP-1α/β), MIP-2, and macrophage chemoattractant protein 1 were induced within 30 min, and their levels showed a transitory peak for 3 to 12 h. However, the lymphotactin, eotaxin, gamma interferon-inducible protein 10, and T-cell activation gene 3 mRNAs were not detected by RNase protection assays. Heat-killed O. tsutsugamushi induced a similar extent of chemokine responses. Induction of the chemokine genes was not blocked by the eukaryotic protein synthesis inhibitor cycloheximide, suggesting that de novo synthesis of host cell protein is not required for these transcriptional responses. The induction of chemokine mRNAs by O. tsutsugamushi was blocked by the inhibitors of NF-κB activation. Furthermore, O. tsutsugamushi induced the nuclear translocation and activation of NF-κB. These results demonstrate that heat-stable molecules of O. tsutsugamushi induce a subset of chemokine genes and that induction involves activation of the transcription factor NF-κB.
Orientia tsutsugamushi, an obligate intracellular bacterium, is the causative agent of scrub typhus (20). The disease is characterized by fever, rash, eschar, pneumonitis, menigitis, and disseminated intravascular coagulation, which leads to severe multiorgan failure (1, 11, 62). O. tsutsugamushi causes local inflammations accompanying eschars at the site of infection, which then spread systemically (6). O. tsutsugamush infects a variety of cells in vitro and in vivo, including macrophages, polymorphonuclear leukocytes (PMN), lymphocytes, and endothelial cells (26, 38, 42, 47).
Analysis of early immunologic responses to O. tsutsugamushi infection in mice showed that macrophage-mediated cellular immunity is essential for resolution of this infection (8, 39). Resistance to the lethal effects of acute rickettsia infection is under unigenic dominant control by the Ric locus (21). Macrophages infiltrate both susceptible (Rics) and resistant (Ricr) mouse strains in response to O. tsutsugamushi infection (25, 39). A slight increase occurs in the number of infiltrating cells recovered from resistant mice. Although susceptible mice experienced slower cellular infiltration, the number of infiltrating macrophages was larger than that in resistant mice (39). The resistant strain of mice was reported to have less PMN response to O. tsutsugamushi than a susceptible strain did (26). Induction of nonspecific inflammation leading to the recruitment of PMN rendered resistant mice susceptible to rickettsia infection (26). As a result, susceptible mice died within 2 weeks of infection. By contrast, Ricr strains showed a minimal level of infection over 2 weeks and survived the infection (27, 39). Mononuclear cells such as lymphocytes and macrophages as well as PMN were observed in eschars and rashes caused by scrub typhus (1). It is notable that in a patient who died after 2 days, infiltration of considerable numbers of PMN was observed around some of the blood vessels (1). Early host inflammatory responses seem to play a key role in determining the fate of the host infected with O. tsutsugamushi (39, 53). For these reasons, the regulatory components that determine the quality and magnitude of the cellular influx to the site of the rickettsia infection should be analyzed. Proinflammatory mediators and chemokines play an important role in these processes (4, 24). The expression of chemokines and their kinetics, however, have not been elucidated in the disease caused by O. tsutsugamushi.
Chemokines are the key players in the processes of leukocyte recruitment into inflammatory tissues. The interaction of different chemokines with their receptors on leukocytes allows selective activation and chemotaxis of neutrophils, lymphocytes, or monocytes necessary for migration to the sites of evolving inflammation. It has been shown that during infection, infected macrophages produce a subset of chemokines (17, 46, 56). The cellular influx into inflamed tissue is provoked by gradients of chemokines that contribute to the adhesion of leukocytes to endothelium, transendothelial migration, and movement through the extracellular matrix (24).
Activated monocytes and macrophages are the major chemokine-secreting cells (4). Various sets of chemokines are produced by monocytes and macrophages infected with different pathogenic microorganisms (7, 17, 45, 46, 56). The kinetics of chemokine gene expression also varies according to the microorganism studied. The regulation of chemokine gene expression, as a defense mechanism against pathogenic microorganism, seems to be related to the clinical courses of the infected host (33, 34, 37, 44).
The transcription factor NF-κB is known to play an important role in the regulation of inflammatory mediators, such as cytokines, acute-phase proteins, and adhesion molecules (5, 19). Since many of the chemokine genes are also regulated by NF-κB (61), it is possible that O. tsutsugamushi induces the chemokine genes through activation of NF-κB. In this study, we analyzed the transcriptional activation of a subset of chemokine genes in a murine macrophage cell line during O. tsutsugamushi infection. The activation of transcription factor NF-κB was also shown to be involved in the induction of chemokine genes by O. tsutsugamushi.
MATERIALS AND METHODS
Cell culture.
J774A.1 cells were obtained from the American Type Culture Collection, Rockville, Md., and cultured in Dulbecco's modified Eagle's medium (Gibco BRL, Grand Island, N.Y.) containing 10% (vol/vol) heat-inactivated fetal bovine serum (FBS) (Gibco BRL), 100 μg of streptomycin per ml, 100 U of penicillin per ml, and 2 mM l-glutamine (DMEM-10) in a humidified 5% CO2 atmosphere at 37°C. The cells were seeded onto six-well plates (Becton Dickinson Labware, Franklin Lakes, N.J.) for the preparation of mRNA or onto 100-mm dishes (Becton Dickinson Labware) for the preparation of nuclear extract. The prototype strain, O. tsutsugamushi Karp (American Type Culture Collection) was propagated in monolayers of L-929 cells as described previously (32, 51). When more than 90% of the cells were infected, as determined by an indirect immunofluorescent-antibody technique (9), the cells were collected, homogenized with a glass Dounce homogenizer (Wheaton Inc., Millville, N.J.), and centrifuged at 500 × g for 5 min. The supernatant was centrifuged at 10,000 × g for 10 min, and the rickettsia pellet was resuspended in DMEM-10 and stored in liquid nitrogen until use. The infectivity titer of the inoculum was determined as described previously with modification (31, 57). Briefly, fivefold serially diluted rickettsia samples were inoculated onto L-929 cell layers on 24-well tissue culture plates. After 3 days of incubation, the cells were collected, fixed, and stained as described previously (31). The ratio of infected cells to the counted number of cells was determined microscopically, and infected-cell counting units (ICU) of the rickettsia sample were calculated as follows (57): ICU = (total number of cells used in infection) × (percentage of infected cells) × (dilution rate of the rickettsiae suspension)/100.
A total of 2.8 × 106 ICU of O. tsutsugamushi was used to infect J774A.1 cells cultured in six-well plates for the preparation of total RNA, and 1.4 × 107 ICU was used in 100-mm dishes for the preparation of nuclear extract. Infection was confirmed by an immunofluorescent-antibody assay 2 h after infection (5 to 10 bacteria were found per cell). The L929 cell lysate was prepared as described above and was used in infection of the macrophage cell line for the control experiments. Lipopolysaccharide (LPS) derived from Escherichia coli (Sigma Chemical Co., St. Louis, Mo.), which is known to induce the production of chemokines in murine and human monocytes/macrophages (61), was used as a positive control for each experiment. In the inhibition assays, J774A.1 cells were preincubated with 25 μM pyrrolidinedithiocarbamate (PDTC; Sigma), 50 μM N-tosyl-l-phenylalanine chloromethyl ketone (TPCK; Sigma), or 10 μg of cycloheximide (CHX; Sigma) per ml for 1 h before O. tsutsugamushi was inoculated. Inhibitors were maintained during the course of inhibition assays. To exclude the possible LPS contamination in the medium or in the inoculum, 30 μg of polymyxin B sulfate (Sigma) per ml was added to the cell culture to neutralize the LPS. The concentration of polymyxin B used was the maximum concentration that did not cause toxic effects to mammalian cells (59). Polystyrene beads (Polyscience Inc., Warrington, Pa.), 1 μm in diameter, were used in phagocytosis assay. Heat-inactivated inoculum was obtained by heating O. tsutsugamushi at 100°C for 10 min.
RNase protection assay.
Total RNA was prepared with RNeasy kit (Qiagen GmbH, Hilden, Germany) as specified by the manufacturer and was quantified spectrophotometrically. Detection and semiquantification of various murine chemokine mRNAs were performed with the multiprobe RNase protection assay system from Pharmingen (San Diego, Calif.). In brief, a mixture of [32P]CTP-labeled antisense riboprobes was generated from chemokine template DNAs including lymphotactin (Ltn), RANTES, eotaxin, macrophage inflammatory protein 1α (MIP-1α), MIP-1β, MIP-2, gamma interferon-inducible protein 10 (IP-10), macrophage chemoattractant protein-1 (MCP-1), and T-cell activation gene 3 (TCA-3). The template DNAs for the murine housekeeping genes encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and a murine ribosomal protein, L32, was also included to ensure equal loading of total RNA onto the gels. Total RNAs from each sample (10 μg each) were hybridized overnight at 56°C with 2 × 105 cpm of the 32P-labeled antisense riboprobe mixture. After hybridization, the samples were digested with a mixture of RNases A and T1. Nuclease-protected RNA fragments were precipitated with ethanol. The samples were resolved on a 5% polyacrylamide sequencing gel (52). The bands were observed after autoradiography. The specific chemokine bands were identified on the basis of their individual mobilities compared with labeled standard probes. The band intensities shown in autoradiography were digitized by scanning the images and analyzed with TINA software (Raytest Isotopenmeßgeräte GmbH, Straubenhardt, Germany). The densitometric intensity was normalized with respect to the intensities of the band for the housekeeping genes, GAPDH and L32.
Semiquantitative RT-PCR.
Total RNA extracted from each sample (2 to 5 μg per sample) was subjected to first-strand cDNA synthesis at 42°C for 1 h in a 40-μl reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 5 mM MgCl2, 1 mM deoxynucleoside triphosphate mixture, 1 U of RNasin per μl, 2.5 μM oligo(dT) primer, and 100 U of murine leukemia virus reverse transcriptase (RT) (all from Perkin-Elmer, Branchburg, N.J.). The cDNA was heated at 94°C for 5 min and diluted with water. The cDNA amounts equivalent to 100 ng of total RNA were subjected to PCR amplification in a 20-μl reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 0.5 mM deoxynucleoside triphosphate mixture, 1 μM each primer, and 0.2 U of AmpliTaq DNA polymerase (Perkin-Elmer) in a Gene Cycler (Bio-Rad Laboratories Inc., Hercules, Calif.). The reaction mixture was prepared as a master mixture to minimize reaction variation. One PCR cycle consisted of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min. The PCR products (5-μl samples) were electrophoresed in a 1.5% agarose gel containing 0.5 μg of ethidium bromide per ml. If not otherwise specified, a 123-bp DNA ladder (Gibco BRL) was used at 1 μg/lane as molecular size markers to provide bands from 4,182 to 123 bp. The amplified DNA fragments in the gels were identified according to their size predicted by cDNA sequences reported previously (2, 15, 22, 30, 54, 58). The densities of the bands were analyzed as described previously (52). The densitometric intensity was normalized by comparing the ratio of chemokine bands with that of β-actin. PCR was performed for the following number of cycles for each set of primers to ensure that the assay was in the linear range according to the amount of template (data not shown): RANTES, 30; MIP-1α, 20; MIP-1β, 25; MIP-2, 25; MCP-1, 25; β-actin, 25. The 5′ and 3′ sequences of the primers and the size of PCR products are as follows: RANTES (215 bp), 5′-CCT CAC CAT CAT CCT CAC TGC A-3′, 5′-TCT TCT CTG GGT TGG CAC ACA C-3′; MIP-1β (390 bp), 5′-AAC CCC GAG CAA CAC CAT GAA G-3′, 5′-TGA ACG TGA GGA GCA AGG ACG C-3′; MIP-1α (357 bp) 5′-GGT CTC CAC CAC TGC CCT TGC-3′, 5′-GGT GGC AGG AAT GTT CGG CTC-3′; MIP-2 (536 bp), 5′-AGT TTG CCT TGA CCC TGA AGC C-3′, 5′-CCA TGA AAG CCA TCC GAC TGC A-3′; MCP-1 (582 bp), 5′-TCT CTT CCT CCA CCA CCA TGC AG-3′, 5′-GGA AAA ATG GAT CCA CAC CTT GC-3′; β-actin (349 bp), 5′-TGG AAT CCT GTG GGA TCC ATG AAA C-3′, 5′-TAA AAC GCA GCT CAG TAA CAG TCC G-3′.
EMSA.
Nuclear extraction and electrophoretic mobility shift assay (EMSA) were performed as described previously with some modifications (10). Following infection with O. tsutsugamushi, J774A.1 cells were washed with cold phosphate-buffered saline (PBS) and collected by centrifugation (500 × g for 5 min). The cells were resuspended in 100 μl of buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride), vigorously vortexed for 15 s, and allowed to stand in ice for 10 min. The nuclei were pelleted by centrifugation (400 × g for 2 min) and resuspended for 20 min on ice in 50 μl of cold buffer containing 20 mM HEPES (pH 7.9), 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, and 0.2 mM phenylmethylsulfonyl fluoride. Nuclear debris were removed by centrifugation (13,000 × g for 5 min) at 4°C, and nuclear extracts were collected. Protein concentrations were determined with the bicinchoninic acid protein assay reagent (Pierce Chemical Co. Rockford, Ill.). Aliquots of the supernatant were frozen in liquid nitrogen and stored at −70°C until use. Equal amounts of nuclear extracts (10 μg of protein) from each sample were incubated for 30 min at 25°C in 30 μl of binding buffer (10 mM Tris-HCl [pH 7.5], 75 mM KCl, 1 mM dithiothreitol, 1 mM EDTA, 4% Ficoll) containing 2 μg of sonicated salmon sperm DNA and 30,000 cpm of an NF-κB-specific oligonucleotide probe that was radiolabeled with [γ-32P]ATP (Amersham Ltd., Little Chalfont, England). The sequence of the NF-κB-specific probe was 5′-AGT TGA GGG GAC TTT CCC AGG C-3′. To ascertain the specific binding of nuclear extracts with NF-κB probe, a competition assay was performed with a 50-fold molar excess of unlabeled oligonucleotides. Nuclear translocation of NF-κB heterodimer was analyzed by a supershift assay with anti-p65 antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.). The nuclear extract proteins were mixed with 4 μg of anti-p65 antibody and were hybridized with the NF-κB-specific oligonucleotide probe. The supershifted bands were analyzed after separation on 5% nondenaturing polyacrylamide gels and autoradiography.
RESULTS
Induction of chemokine gene expression.
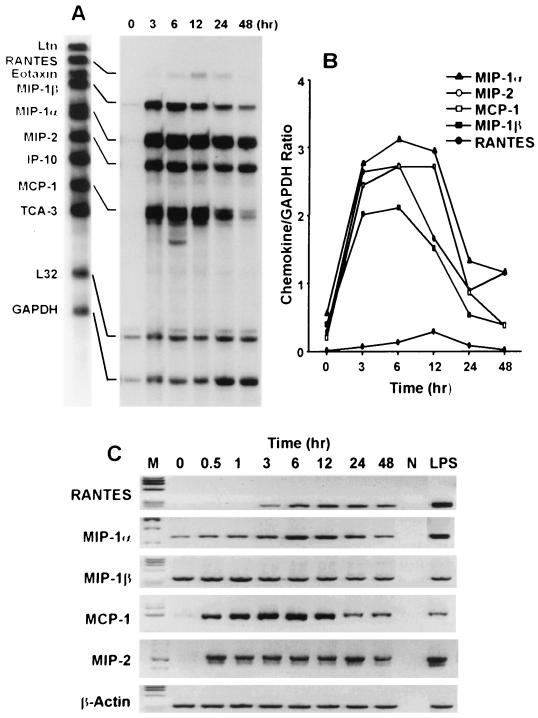
Before and after exposure of J774A.1 cells to O. tsutsugamushi, the levels of chemokine transcripts were assayed at each time point by an RNase protection assay and semiquantitative RT-PCR (Fig. 1). Although there were some variations in the ratio of RNA transcripts for the control and the test groups between the sets of RT-PCR experiments, the changes within a set of experiments were reproducible throughout this study. The absence of contamination of RNA with genomic DNA was monitored by the size of PCR products from the pair of primers whose binding sites are located in different exons. The mRNAs of the CC chemokines MIP-1α, MIP-1β, and MCP-1 were constitutively expressed at low levels in noninfected J774A.1 cells (Fig. 1). Basal levels of these chemokines are also expressed constitutively in monocytes and macrophages (17, 56). The mRNAs for MIP-1α, MIP-1β, MCP-1, and MIP-2 were up-regulated and detected as early as 30 min after infection, peaked at 6 h, and began to decrease from 6 to 12 h after infection. While the MIP-1α and MIP-2 mRNAs persisted after incubation for 48 h, the levels of transcripts for MIP-1β and MCP-1 were reduced to the levels in uninfected cells by 48 h. The transcript for RANTES was also detectable as early as 3 h after infection. However, the level of this transcript was significantly lower than those of other induced chemokines. Expression of RANTES mRNA was characterized by slower kinetics compared to those of other induced chemokine mRNAs. The peak response for RANTES was observed 12 h after infection and decreased to the level of uninfected cells by 48 h. Similar kinetics of mRNA expression were detected when the levels of chemokine mRNAs were analyzed by either the RNase protection assay or semiquantitative RT-PCR (Fig. 1). No mRNA for Ltn, eotaxin, IP-10, and TCA-3 was detected during infection by RNase protection.
FIG. 1.
Time course of O. tsutsugamushi-stimulated chemokine induction by the J774A.1 cell line. (A) Before and after incubation of J774A.1 cells with O. tsutsugamushi. The levels of chemokine mRNAs at each time point were assayed by the RNase protection assay. (B) Normalized expression level of each chemokine mRNA. (C) mRNA levels of chemokine genes induced by the infection of O. tsutsugamushi, analyzed by semiquantitative RT-PCR at each time point. M, φX174 DNA digested with HaeIII; N, negative control (reactions performed without cDNA).
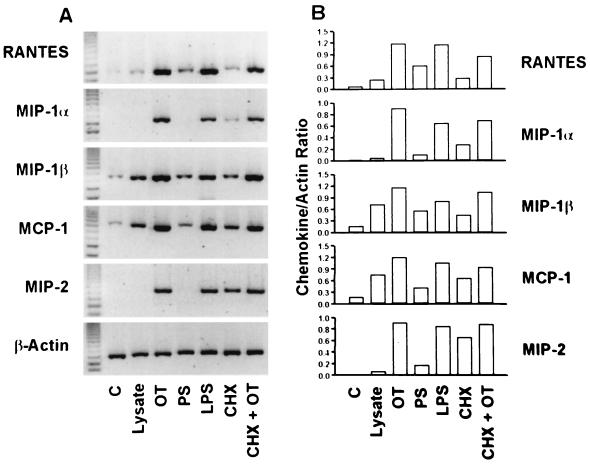
Figure 2 shows the profile of chemokine mRNA expression analyzed by semiquantitative RT-PCR. Cells treated with E. coli LPS were included as a positive control for each experiment. In the cells stimulated with 1 μg of LPS per ml for 6 h, comparable but slightly smaller amounts of chemokine mRNAs were detected compared with those in O. tsutsugamushi-infected cells. Cells treated with medium alone or L-929 lysates were used as negative controls. When the cells were treated with lysate of uninfected L-929 cell lysate, the mRNA levels of all chemokines were slightly increased. The mRNA levels of MIP-1β and MCP-1 showed an approximately fivefold increase in their optical densities. Although the RANTES, MIP-1β, and MCP-1 mRNAs were detected in cells treated with medium or L-929 cell lysate, the levels in cells incubated with O. tsutsugamushi increased by approximately two- to fivefold as measured by their optical densities. Compared to control groups, cells incubated for 6 h with O. tsutsugamushi resulted in higher levels of mRNAs of all the chemokines tested (Fig. 2). To determine whether the chemokine induction was a specific response to O. tsutsugamushi infection, we investigated whether the phagocytosis of polystyrene beads similar in size to O. tsutsugamushi would provide a stimulus for chemokine gene expression. Although the levels of mRNA of RANTES, MIP-1β, and MCP-1 were increased by incubation with polystyrene beads, they were similar to those of the cells treated with an L-929 cell lysate. The mRNA levels in the cells infected with O. tsutsugamushi were approximately 2- to 10-fold higher as measured by their optical densities than were those in the cells treated with polystyrene beads. The chemokine genes are induced by proinflammatory cytokines such as interleukin-1 (IL-1) and tumor necrosis factor alpha (61). To investigate whether the chemokine induction was a consequence of the host cytokine expression, cells were incubated for 1 h with CHX, a eukaryotic protein synthesis inhibitor, and then infected with O. tsutsugamushi. Although the chemokine genes were induced when the cells were treated only with CHX (16, 61), higher levels of chemokine mRNAs were observed when CHX-treated cells were infected with O. tsutsugamushi (Fig. 2).
FIG. 2.
(A) Determination of chemokine mRNA induction in J774A.1 cells treated with polystyrene beads or CHX, by semiquantitative RT-PCR. (B) The band intensities were determined with TINA software, and the level of each chemokine mRNA expression was normalized with mRNA level of β-actin. J774A.1 cells stimulated for 6 h with medium alone (C), L-929 cell lysate (Lysate), O. tsutsugamushi (OT), polystyrene beads (PS), LPS derived from E. coli (LPS), cycloheximide (CHX), or cycloheximide and O. tsutsugamushi (CHX + OT).
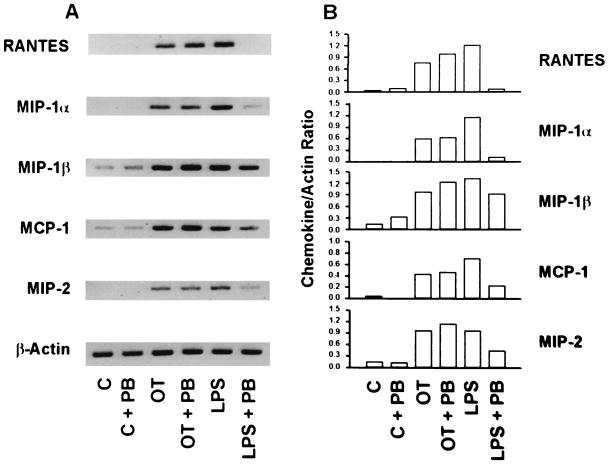
Cells treated with medium alone or medium and polymyxin B contained little or no detectable mRNA (Fig. 3). In the presence of polymyxin B, LPS-mediated chemokine induction was significantly reduced. In addition to the mRNAs of RANTES, MIP-1β, and MCP-1, the levels of the mRNAs of MIP-1α and MIP-2 were dramatically reduced to those similar to the levels in the control group by polymyxin B treatment. In contrast, in cells treated with polymyxin B and O. tsutsugamushi, mRNA levels for all the chemokines tested did not differ significantly from those induced by stimulation with O. tsutsugamushi. These results show that possible exogenous sources of LPS are not responsible for the induction of the chemokine genes.
FIG. 3.
(A) Semiquantitative RT-PCR to determine the effect of polymyxin B on the levels of O. tsutsugamushi-induced chemokine mRNAs in J774A.1 cells. (B) The band intensities were determined and normalized as for the experiment in Fig. 2. J774A.1 cells were stimulated for 6 h with medium (C), O. tsutsugamushi (OT), and LPS derived from E. coli (LPS) in the absence or presence of polymyxin B (PB).
Chemokine expression by NF-κB activation.
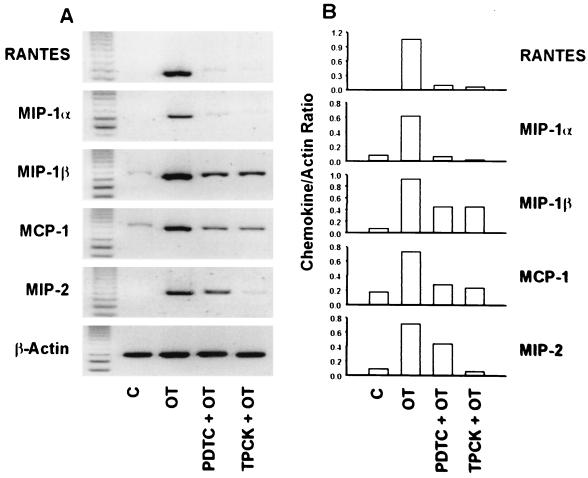
To examine whether NF-κB activation is involved in the chemokine induction of O. tsutsugamushi-exposed J774A.1 cells, we used two inhibitors of NF-κB activation, the antioxidant PDTC (50) and the proteasome inhibitor TPCK (36). When the cells were incubated with O. tsutsugamushi in the presence of TPCK, induction of RANTES, MIP-1α, and MIP-2 was inhibited completely (Fig. 4). Although induction of MIP-1β and MCP-1 was not completely blocked by the inhibitor, the levels of their transcripts were reduced by one-half and one-third, respectively, compared to those in the cells treated with O. tsutsugamushi alone. Expression of MIP-1β, MIP-2, and MCP-1 genes was also inhibited in the presence of PDTC by approximately one-half to one-third as judged by measurement of their optical densities, while induction of RANTES and MIP-1α was also completely blocked in O. tsutsugamushi-infected cells (Fig. 4).
FIG. 4.
Effect of PDTC and TPCK on the levels of O. tsutsugamushi-induced chemokine mRNAs in J774A.1 cells. (A) Levels of each chemokine mRNA were analyzed in total RNA samples prepared from uninfected cells (C), O. tsutsugamushi-infected cells (OT), and infected cells in the presence of PDTC (PDTC + OT) or TPCK (TPCK + OT) by RT-PCR analysis. (B) The intensities of bands were determined and normalized as specified for Fig. 2.
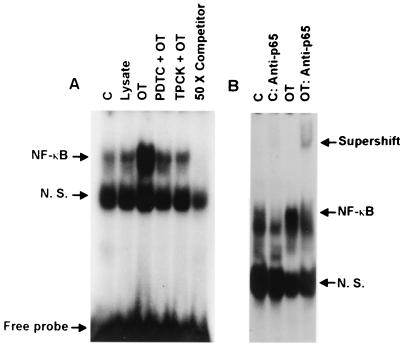
NF-κB activation by O. tsutsugamushi was directly evaluated by nuclear translocation of NF-κB and EMSA (Fig. 5). At 2 h after infection of macrophages with O. tsutsugamushi, we could detect a mobility-shifted complex which was competed off by an unlabeled probe (50-fold excess of competitor) corresponding to the κB binding domain of the murine kappa light-chain gene enhancer. The bands at the front of NF-κB complex observed in all lanes might be probe binding to nonspecific proteins (36). Although the basal levels of NF-κB complexes were detected in the cells treated with medium or L-929 cell lysate, the activation and nuclear translocation of NF-κB were remarkably increased when stimulated with O. tsutsugamushi. When the levels of NF-κB activation were normalized to nonspecific bands, the level of activation in the O. tsutsugamushi-infected cells was increased two- or threefold compared to that in cells treated with L-929 cell lysate or medium only. In the presence of NF-κB activation inhibitors, however, the levels of NF-κB complex were decreased and comparable to those of control groups. The proteasome inhibitor TPCK was more effective in inhibiting NF-κB activation than was PDTC. The p50/p65 heterodimeric form of NF-κB is the prototypical and transcriptionally active complex, while the p50/p50 homodimeric form is constitutively present and is thought to be an inactive or repressive complex (5, 19). The p50/p65 heterodimeric form of the NF-κB complex was confirmed by a supershift assay with a p65-specific antibody (Fig. 5B). The NF-κB complexes shown in Fig. 5A were more widely separated, and, furthermore, incubation with anti-p65 antibody resulted in the loss of a band (Fig. 5B). The remaining lower complex of NF-κB might represent the p50/p50 homodimeric form of NF-κB (16), although we did not identify the homodimeric complex by using a p50-specific antibody. The basal level of the heterodimeric complex of NF-κB was also detected in the cells treated with medium only. The level of the heterodimeric form in the O. tsutsugamushi-infected cells was approximately three times higher, as measured by optical density, than in the control group after nonspecific bands were normalized for. The supershifted complex was detectable only in the O. tsutsugamushi-stimulated cells.
FIG. 5.
Activation of the transcription factor NF-κB by O. tsutsugamushi and effect of PDTC and TPCK on O. tsutsugamushi-induced activation of NF-κB. (A) NF-κB activation was analyzed by EMSA for nuclear extracts prepared from J774A.1 cells treated for 2 h with medium (C), L-929 cell lysate (Lysate), and O. tsutsugamushi (OT). The nuclear extracts from the cells pretreated with PDTC (PDTC + OT) or TPCK (TPCK + OT) for 1 h before infection with O. tsutsugamushi were also analyzed. A competitive inhibition assay was performed on nuclear extracts preincubated with the unlabeled NF-κB consensus oligonucleotide (50 × Competitor). (B) A supershift assay was also performed. Nuclear extract was preincubated with antibodies against the p65 subunit of NF-κB. N. S., nonspecific.
Heat stability of the stimulating molecule.
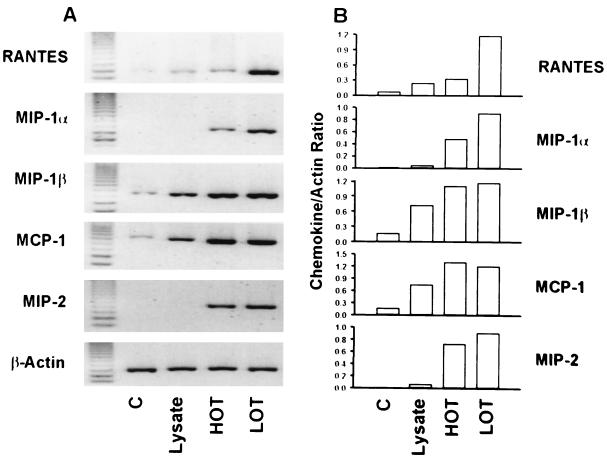
To evaluate whether active rickettsia replication was required for chemokine induction, we exposed macrophages to heat-inactivated O. tsutsugamushi for 6 h. As shown in Fig. 6, mRNA levels of MIP-1β, MCP-1, and MIP-2 in cells treated with heat-inactivated O. tsutsugamushi were comparable to those in cells treated with the live microorganism. The kinetics of chemokine expression in cells stimulated with heat-inactivated O. tsutsugamushi were also similar to those of expression in the cells treated with live O. tsutsugamushi (data not shown). However, in cells treated with heat-inactivated O. tsutsugamushi, the levels of RANTES and MIP-1α were reduced in optical density by 30 and 50%, respectively, compared to those in cells treated with active O. tsutsugamushi.
FIG. 6.
Chemokine responses to inactivated or active O. tsutsugamushi. (A) The levels of chemokine mRNAs were compared by semiquantitative RT-PCR after incubation of the J774A.1 cells for 6 h with medium (C), L-929 cell lysate (Lysate), or heat-inactivated (HOT) or live (LOT) O. tsutsugamushi. (B) The intensities of bands were determined and normalized as specified for Fig. 2.
DISCUSSION
It has been well documented that macrophages play a pivotal role in early immune responses to O. tsutsugamushi infection (27, 39–41). Although the inactive tissue macrophages could support the growth of O. tsutsugamushi at the site of infection, subsequent cellular influxes, especially of activated macrophages and lymphocytes, have been suggested to be important in protection against O. tsutsugamushi infection (25, 26). Early PMN responses seem to provide a cellular population for rickettsia replication instead of providing antirickettsial activity in vivo (26). The cellular recruitment is controlled largely by chemokines which are secreted by stimulated cells such as macrophages at the site of primary infection (34, 61).
In this study, we showed that the murine macrophage cell line J774A.1 induced the expression of MIP-1α, MIP-1β, RANTES, MCP-1, and MIP-2 in response to O. tsutsugamushi infection. With the exception of RANTES, the induction of the chemokine genes occurred within 30 min and peaked transiently between 3 and 12 h. The inducibility and the kinetics of these chemokines are different from those of murine macrophages infected with other pathogenic microorganisms (45, 46). The differences in the patterns of early chemokine responses to various pathogens are likely to be related to disease manifestations (49). Although we did not perform assays to confirm the secretion of active chemokine protein following gene induction, several recent studies have shown a correlation between mRNA expression and chemokine protein secretion (7, 17, 46, 56).
The chemokine genes were induced specifically in response to O. tsutsugamushi infection. Ingestion of polystyrene beads by macrophages resulted in little or no induction of the chemokine genes tested. Phagocytosis of inert particles such as latex beads by murine macrophages did not affect the basal levels of cytokines and chemokines (45). Contamination by LPS during the preparation of O. tsutsugamushi was also examined. It has been previously reported that the cell wall component in O. tsutsugamushi is deficient in LPS (3). Blocking LPS with polymyxin B did not decrease chemokine responses in macrophages infected with O. tsutsugamushi. These results suggest that O. tsutsugamushi-mediated induction of chemokine genes requires certain signals which are not generated by nonspecific phagocytosis of macrophages and, in addition, are not mediated by LPS. This finding is intriguing in light of the fact that throughout the entire course of the O. tsutsugamushi infection, the macrophage is one of the main target cells for rickettsia parasitism. The ability of O. tsutsugamushi to selectively induce the expression of a subset of chemokines in vitro represents the earliest host response to infection and could play a role in early manifestations following skin infection in vivo.
Previous studies had already demonstrated that various cytokines stimulate chemokine expression in vitro. We therefore examined the role of newly synthesized proteins in chemokine gene expression (61). When CHX, the eukaryotic protein synthesis inhibitor, was included in macrophage cultures, the levels of the chemokine mRNAs were similar to those in cells infected with O. tsutsugamushi alone. This indicated that chemokine induction was not an indirect effect due to prior induction of tumor necrosis factor alpha or IL-1, which are known to induce chemokine production in macrophages (4).
The transcription factor NF-κB/Rel family plays a central role in the regulation of a variety of genes involved in host innate immunity, including various chemokines (19). For all the chemokine genes tested in this study, regulation by NF-κB either has been demonstrated or is suggested by the presence of the NF-κB consensus motif in the promoter (14, 18, 61, 63). We have found that O. tsutsugamushi induces an increase in the levels of active NF-κB in the nucleus, particularly of the p65/p50 heterodimer. The induction of the chemokine mRNAs by O. tsutsugamushi is completely or partially blocked by inhibitors of NF-κB activation. PDTC, an antioxidant, inhibits the phosphorylation of IκB (50), a prerequisite for its subsequent proteolytic degradation. TPCK, an inhibitor of chymotryptic activity associated with the proteasome, blocks activation of NF-κB by inhibiting proteasome-dependent degradation of the inhibitory peptides (36). These chemically unrelated compounds reduce O. tsutsugamushi-induced RANTES and MIP-1α mRNA levels, implicating NF-κB as the main transcription factor in the expression of these chemokines. The mRNA expression of MIP-1β and MCP-1 was partially blocked by treatment with PDTC and TPCK. These findings strongly suggest that O. tsutsugamushi induces gene expression of the chemokines in J774A.1 cells via proteasome-sensitive and reactive oxygen intermediate-sensitive pathways that have been implicated in the activation of NF-κB (5, 19). Our data indicates that TPCK is likely to be more effective in inhibiting the expression of the chemokine genes, especially for MIP-2, and in activating NF-κB. Although NF-κB is essential for the transcription of the chemokine genes, a number of other transcription factors form activating complexes capable of up-regulating chemokine gene expression. Various transcriptional regulatory elements apart from NF-κB are required for the expression of MCP-1 (18, 60, 61). In addition, various potential cis-regulatory elements have been identified in the upstream region of the MIP-1β gene (63). For these reasons, it appears that O. tsutsugamushi activates signal transduction pathways leading to activation of those transcription factors as well as to activation of NF-κB. Although direct evidence was not provided, this data suggests that the induction of chemokines in J774A.1 cells and the activation of NF-κB are physiologically relevant.
A recent report has suggested that NF-κB activation by rickettsia infection is related to the inhibition of apoptosis in endothelial cells and fibroblasts. This provides a possible mechanism to enable host cells to remain as a site for rickettsiae replication (13). However, it has also been reported that apoptotic death of macrophages and lymphocytes occurs in the spleen and lymph nodes of O. tsutsugamushi-infected mice (29). Furthermore, apoptosis was also observed in J774A.1 cells within 12 h of O. tsutsugamushi infection (12). Further study of apoptosis modulation in rickettsia-infected cells by NF-κB activation is needed.
In addition, we tried to investigate rickettsia molecules eliciting chemokine responses of macrophages. The physicochemical characteristic of the molecule was analyzed after O. tsutsugamushi was subjected to heat treatment. The expression of chemokine mRNAs was unchanged whether the cells were treated with heat-killed or living O. tsutsugamushi. This suggests that heat-stable rickettsia molecules may be involved in activating transcription factors and that proliferation of O. tsutsugamushi within infected macrophages is not a prerequisite for expression of those chemokines. Further studies on stimulatory components of O. tsutsugamushi and signal transduction pathways in host cells during rickettsia infection will provide valuable insights into the mechanisms controlling the inflammatory responses during O. tsutsugamushi infection.
Protective immunity against O. tsutsugamushi is largely due to cell-mediated immune responses, particularly those provided by macrophages and T cells (28, 40, 51). The explanation for a susceptible/resistant mouse phenotype to O. tsutsugamushi infection was provided by the analysis of the early T-lymphocyte activation 1 (Eta-1)/osteopontine (Op) gene, which maps to the Ric locus (21, 43). Eta-1/Op has been thought to enhance resistance to rickettsia infection by affecting the ability of macrophages to migrate to sites of infection and/or to express bactericidal activity (43). However, the infiltration of T lymphocytes and their secretion of Eta-1/Op in the early stage of infection should be preceded by activation of macrophages and their chemokine secretions, which recruit specific and nonspecific immune cells. In other studies, genetic susceptibility to infectious disease has been shown to be associated with the expression of different cytokine profiles (23). Members of the CC chemokine subfamily, which include RANTES, MIP-1α, MIP-1β, and MCP-1, preferentially attract monocytes and lymphocytes. Those of the CXC chemokine subfamily, such as IL-8 and MIP-2, are potent neutrophil attractants (4). Furthermore, a correlation between chemokines and a subset of T-cell responses has been described (35, 48, 55). While the CC chemokines MIP-1α, MIP-1β, and RANTES were found to be efficient chemoattractants for Th1 cells, Th2 cells were not attracted by these chemokines (55). Stimulation of T cells in the presence of MIP-1α enhanced gamma interferon production by Th1 cells, while stimulation of T cells in the presence of MCP-1 led to an increase IL-4 production (35). Based on these studies, we hypothesize that a delicate balance of chemokines exists between the induction of a resistant and a susceptible immune response to rickettsia infection. Further study is required to determine whether qualitative and quantitative differences in the production of chemokines can be correlated with the resistant or susceptible mouse phenotype.
ACKNOWLEDGMENT
This work was supported by the Korea Research Foundation of the Republic of Korea (grant 97110814).
REFERENCES
- 1.Allen A C, Spitz S. A comparative study of the pathology of scrub typhus (tsutsugamushi disease) and other rickettsial disease. Am J Pathol. 1945;21:603–681. [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso S, Minty A, Bourlet Y, Buckingham M. Comparison of three actin-coding sequences in the mouse: evolutionary relationships between the actin genes of warm-blooded vertebrates. J Mol Evol. 1986;23:11–22. doi: 10.1007/BF02100994. [DOI] [PubMed] [Google Scholar]
- 3.Amano K, Tamura A, Ohashi N, Urakami H, Kaya S, Fukushi K. Deficiency of peptidoglycan and lipopolysaccharide components in Rickettsia tsutsugamushi. Infect Immun. 1987;55:2290–2292. doi: 10.1128/iai.55.9.2290-2292.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baggiolini M, Dewald B, Moser B. Interleukin-8 and related chemotactic cytokines—CXC and CC chemokines. Adv Immunol. 1994;55:97–179. [PubMed] [Google Scholar]
- 5.Baldwin A S., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 6.Burnett J W. Rickettsioses: a review for the dermatologist. J Am Acad Dermatol. 1980;2:359–373. doi: 10.1016/s0190-9622(80)80355-0. [DOI] [PubMed] [Google Scholar]
- 7.Bussfeld D, Kaufmann A, Meyer R G, Gemsa D, Sprenger H. Differential mononuclear leukocyte attracting chemokine production after stimulation with active and inactivated influenza A virus. Cell Immunol. 1998;186:1–7. doi: 10.1006/cimm.1998.1295. [DOI] [PubMed] [Google Scholar]
- 8.Catanzaro P J, Shirai A, Hilderbrandt P K, Osterman J V. Host defenses in experimental scrub typhus: histopathological correlates. Infect Immun. 1976;13:861–875. doi: 10.1128/iai.13.3.861-875.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang W H, Kang J S, Lee W K, Choi M S, Lee J H. Serological classification by monoclonal antibodies of Rickettsia tsutsugamushi isolated in Korea. J Clin Microbiol. 1990;28:685–68. doi: 10.1128/jcm.28.4.685-688.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen B C, Chou C F, Lin W W. Pyrimidinoceptor-mediated potentiation of inducible nitric-oxide synthase induction in J774 macrophages. Role of intracellular calcium. J Biol Chem. 1998;273:29754–29763. doi: 10.1074/jbc.273.45.29754. [DOI] [PubMed] [Google Scholar]
- 11.Chi W C, Huang J J, Sung J M, Lan R R, Ko W C, Chen F F. Scrub typhus associated with multiorgan failure: a case report. Scand J Infect Dis. 1997;29:634–635. doi: 10.3109/00365549709035911. [DOI] [PubMed] [Google Scholar]
- 12.Choi N J, Kim M K, Park H J, Lim B U, Kang J S. Apoptosis of murine macrophage-like cells infected with Orientia tsutsugamushi. J Korean Soc Microbiol. 1998;33:399–406. [Google Scholar]
- 13.Clifton D R, Goss R A, Sahni S K, van Antwerp D, Baggs R B, Marder V J, Silverman D J, Sporn L A. NF-kappa B-dependent inhibition of apoptosis is essential for host cell survival during Rickettsia rickettsii infection. Proc Natl Acad Sci USA. 1998;95:4646–4651. doi: 10.1073/pnas.95.8.4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danoff T M, Lalley P A, Chang Y S, Heeger P S, Neilson E G. Cloning, genomic organization, and chromosomal localization of the Scya5 gene encoding the murine chemokine RANTES. J Immunol. 1994;152:1182–1189. [PubMed] [Google Scholar]
- 15.Davatelis G, Tekamp-Olson P, Wolpe S D, Hermsen K, Luedke C, Gallegos C, Coit D, Merryweather J, Cerami A. Cloning and characterization of a cDNA for murine macrophage inflammatory protein (MIP), a novel monokine with inflammatory and chemokinetic properties. J Exp Med. 1988;167:1939–1944. doi: 10.1084/jem.167.6.1939. . (Errarum, 170:2189, 1989.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebnet K, Brown K D, Siebenlist U K, Simon M M, Shaw S. Borrelia burgdorferi activates nuclear factor-kappa B and is a potent inducer of chemokine and adhesion molecule gene expression in endothelial cells and fibroblasts. J Immunol. 1997;158:3285–3292. [PubMed] [Google Scholar]
- 17.Flesch I E, Barsig J, Kaufmann S H. Differential chemokine response of murine macrophages stimulated with cytokines and infected with Listeria monocytogenes. Int Immunol. 1998;10:757–765. doi: 10.1093/intimm/10.6.757. [DOI] [PubMed] [Google Scholar]
- 18.Freter R R, Alberta J A, Hwang G Y, Wrentmore A L, Stiles C D. Platelet-derived growth factor induction of the immediate-early gene MCP-1 is mediated by NF-kappaB and a 90-kDa phosphoprotein coactivator. J Biol Chem. 1996;271:17417–17424. doi: 10.1074/jbc.271.29.17417. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh S, May M J, Kopp E B. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert D N, Moore W L, Jr, Hedberg C L, Sanford J P. Potential medical problems in personnel returning from Vietnam. Ann Intern Med. 1968;68:662–678. doi: 10.7326/0003-4819-68-3-662. [DOI] [PubMed] [Google Scholar]
- 21.Groves M G, Rosenstreich D L, Taylor B A, Osterman J V. Host defenses in experimental scrub typhus: mapping the gene that controls natural resistance in mice. J Immunol. 1980;125:1395–1399. [PubMed] [Google Scholar]
- 22.Heeger P, Wolf G, Meyers C, Sun M J, O'Farrell S C, Krensky A M, Neilson E G. Isolation and characterization of cDNA from renal tubular epithelium encoding murine Rantes. Kidney Int. 1992;41:220–225. doi: 10.1038/ki.1992.31. [DOI] [PubMed] [Google Scholar]
- 23.Heinzel F P, Rerko R M, Ahmed F, Pearlman E. Endogenous IL-12 is required for control of Th2 cytokine responses capable of exacerbating leishmaniasis in normally resistant mice. J Immunol. 1995;155:730–739. [PubMed] [Google Scholar]
- 24.Imhof B A, Dunon D. Leukocyte migration and adhesion. Adv Immunol. 1995;58:345–416. doi: 10.1016/s0065-2776(08)60623-9. [DOI] [PubMed] [Google Scholar]
- 25.Jerrells T R. Association of an inflammatory I region-associated antigen-positive macrophage influx and genetic resistance of inbred mice to Rickettsia tsutsugamushi. Infect Immun. 1983;42:549–557. doi: 10.1128/iai.42.2.549-557.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jerrells T R, Osterman J V. Host defenses in experimental scrub typhus: inflammatory response of congenic C3H mice differing at the Ric gene. Infect Immun. 1981;31:1014–1022. doi: 10.1128/iai.31.3.1014-1022.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jerrells T R, Osterman J V. Role of macrophages in innate and acquired host resistance to experimental scrub typhus infection of inbred mice. Infect Immun. 1982;37:1066–1073. doi: 10.1128/iai.37.3.1066-1073.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jerrells T R, Osterman J V. Host defenses in experimental scrub typhus: delayed-type hypersensitivity responses of inbred mice. Infect Immun. 1982;35:117–123. doi: 10.1128/iai.35.1.117-123.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasuya S, Nagano I, Ikeda T, Goto C, Shimokawa K, Takahashi Y. Apoptosis of lymphocytes in mice induced by infection with Rickettsia tsutsugamushi. Infect Immun. 1996;64:3937–3941. doi: 10.1128/iai.64.9.3937-3941.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawahara R S, Deuel T F. Platelet-derived growth factor-inducible gene JE is a member of a family of small inducible genes related to platelet factor 4. J Biol Chem. 1989;264:679–682. [PubMed] [Google Scholar]
- 31.Kee S H, Choi I H, Choi M S, Kim I S, Chang W H. Detection of Rickettsia tsutsugamushi in experimentally infected mice by PCR. J Clin Microbiol. 1994;32:1435–1439. doi: 10.1128/jcm.32.6.1435-1439.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim I S, Seong S Y, Woo S G, Choi M S, Chang W H. High-level expression of a 56-kilodalton protein gene (bor56) of Rickettsia tsutsugamushi Boryong and its application to enzyme-linked immunosorbent assays. J Clin Microbiol. 1993;31:598–605. doi: 10.1128/jcm.31.3.598-605.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kusugami K, Ando T, Ohsuga M, Imada A, Shinoda M, Konagaya T, Ina K, Kasuga N, Fukatsu A, Ichiyama S, Nada T, Ohta M. Mucosal chemokine activity in Helicobacter pylori infection. J Clin Gastroenterol. 1997;25(Suppl. 1):S203–S210. doi: 10.1097/00004836-199700001-00032. [DOI] [PubMed] [Google Scholar]
- 34.Locati M, Murphy P M. Chemokines and chemokine receptors: biology and clinical relevance in inflammation and AIDS. Annu Rev Med. 1999;50:425–440. doi: 10.1146/annurev.med.50.1.425. [DOI] [PubMed] [Google Scholar]
- 35.Lukacs N W, Chensue S W, Karpus W J, Lincoln P, Keefer C, Strieter R M, Kunkel S L. C-C chemokines differentially alter interleukin-4 production from lymphocytes. Am J Pathol. 1997;150:1861–1868. [PMC free article] [PubMed] [Google Scholar]
- 36.Mackman N. Protease inhibitors block lipopolysaccharide induction of tissue factor gene expression in human monocytic cells by preventing activation of c-Rel/p65 heterodimers. J Biol Chem. 1994;269:26363–26367. [PubMed] [Google Scholar]
- 37.Mastroianni C M, Lancella L, Mengoni F, Lichtner M, Santopadre P, D'Agostino C, Ticca F, Vullo V. Chemokine profiles in the cerebrospinal fluid (CSF) during the course of pyogenic and tuberculous meningitis. Clin Exp Immunol. 1998;114:210–214. doi: 10.1046/j.1365-2249.1998.00698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murata M, Sudo K, Suzuki K, Aoyama Y, Nogami S, Tanaka H, Kawamura A., Jr Proliferating sites of Rickettsia tsutsugamushi in mice by different routes of inoculation evidenced with immunofluorescence. Jpn J Exp Med. 1985;55:193–199. [PubMed] [Google Scholar]
- 39.Nacy C A, Groves M G. Macrophages in resistance to rickettsial infections: early host defense mechanisms in experimental scrub typhus. Infect Immun. 1981;31:1239–1250. doi: 10.1128/iai.31.3.1239-1250.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nacy C A, Meltzer M S. Macrophages in resistance to rickettsial infection: macrophage activation in vitro for killing of Rickettsia tsutsugamushi. J Immunol. 1979;123:2544–2549. [PubMed] [Google Scholar]
- 41.Nacy C A, Meltzer M S. Macrophages in resistance to rickettsial infections: protection against lethal Rickettsia tsutsugamushi infections by treatment of mice with macrophage-activating agents. J Leukoc Biol. 1984;35:385–396. doi: 10.1002/jlb.35.4.385. [DOI] [PubMed] [Google Scholar]
- 42.Ng F K, Oaks S C, Jr, Lee M, Groves M G, Lewis G E., Jr A scanning and transmission electron microscopic examination of Rickettsia tsutsugamushi-infected human endothelial, MRC-5, and L-929 cells. Jpn J Med Sci Biol. 1985;38:125–139. doi: 10.7883/yoken1952.38.125. [DOI] [PubMed] [Google Scholar]
- 43.Patarca R, Freeman G J, Singh R P, Wei F Y, Durfee T, Blattner F, Regnier D C, Kozak C A, Mock B A, Morse H C. Structural and functional studies of the early T lymphocyte activation 1 (Eta-1) gene. Definition of a novel T cell-dependent response associated with genetic resistance to bacterial infection. J Exp Med. 1989;170:145–161. doi: 10.1084/jem.170.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polo S, Veglia F, Malnati M S, Gobbi C, Farci P, Raiteri R, Sinicco A, Lusso P. Longitudinal analysis of serum chemokine levels in the course of HIV-1 infection. AIDS. 1999;13:447–454. doi: 10.1097/00002030-199903110-00002. [DOI] [PubMed] [Google Scholar]
- 45.Racoosin E L, Beverley S M. Leishmania major: promastigotes induce expression of a subset of chemokine genes in murine macrophages. Exp Parasitol. 1997;85:283–295. doi: 10.1006/expr.1996.4139. [DOI] [PubMed] [Google Scholar]
- 46.Rhoades E R, Cooper A M, Orme I M. Chemokine response in mice infected with Mycobacterium tuberculosis. Infect Immun. 1995;63:3871–3877. doi: 10.1128/iai.63.10.3871-3877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rikihisa Y, Ito S. Intracellular localization of Rickettsia tsutsugamushi in polymorphonuclear leukocytes. J Exp Med. 1979;150:703–708. doi: 10.1084/jem.150.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sallusto F, Lanzavecchia A, Mackay C R. Chemokines and chemokine receptors in T-cell priming and Th1/Th2-mediated responses. Immunol Today. 1998;19:568–574. doi: 10.1016/s0167-5699(98)01346-2. [DOI] [PubMed] [Google Scholar]
- 49.Schluger N W, Rom W N. Early responses to infection: chemokines as mediators of inflammation. Curr Opin Immunol. 1997;9:504–508. doi: 10.1016/s0952-7915(97)80102-1. [DOI] [PubMed] [Google Scholar]
- 50.Schmidt K N, Traenckner E B, Meier B, Baeuerle P A. Induction of oxidative stress by okadaic acid is required for activation of transcription factor NF-kappa B. J Biol Chem. 1995;270:27136–27142. doi: 10.1074/jbc.270.45.27136. [DOI] [PubMed] [Google Scholar]
- 51.Seong S Y, Huh M S, Jang W J, Park S G, Kim J G, Woo S G, Choi M S, Kim I S, Chang W H. Induction of homologous immune response to Rickettsia tsutsugamushi Boryong with a partial 56-kilodalton recombinant antigen fused with the maltose-binding protein MBP-Bor56. Infect Immun. 1997;65:1541–1545. doi: 10.1128/iai.65.4.1541-1545.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seong S Y, Park S G, Huh M S, Jang W J, Choi M S, Chang W H, Kim I S. T-track PCR fingerprinting for the rapid detection of genetic polymorphism. FEMS Microbiol Lett. 1997;152:37–44. doi: 10.1111/j.1574-6968.1997.tb10406.x. [DOI] [PubMed] [Google Scholar]
- 53.Seong S Y, Park S G, Kim H R, Han T H, Kang J S, Choi M S, Kim I S, Chang W H. Isolation of a new Orientia tsutsugamushi serotype. Microbiol Immunol. 1997;41:437–443. doi: 10.1111/j.1348-0421.1997.tb01876.x. [DOI] [PubMed] [Google Scholar]
- 54.Sherry B, Tekamp-Olson P, Gallegos C, Bauer D, Davatelis G, Wolpe S D, Masiarz F, Coit D, Cerami A. Resolution of the two components of macrophage inflammatory protein 1, and cloning and characterization of one of those components, macrophage inflammatory protein 1 beta. J Exp Med. 1988;168:2251–2259. doi: 10.1084/jem.168.6.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siveke J T, Hamann A. T helper 1 and T helper 2 cells respond differentially to chemokines. J Immunol. 1998;160:550–554. [PubMed] [Google Scholar]
- 56.Sprenger H, Krause A, Kaufmann A, Priem S, Fabian D, Burmester G R, Gemsa D, Rittig M G. Borrelia burgdorferi induces chemokines in human monocytes. Infect Immun. 1997;65:4384–4388. doi: 10.1128/iai.65.11.4384-4388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamura A, Urakami H. Easy method for infectivity titration of Rickettsia tsutsugamushi by infected cell counting. Nippon Saikingaku Zasshi. 1981;36:783–785. . (In Japanese.) [PubMed] [Google Scholar]
- 58.Tekamp-Olson P, Gallegos C, Bauer D, McClain J, Sherry B, Fabre M, van Deventer S, Cerami A. Cloning and characterization of cDNAs for murine macrophage inflammatory protein 2 and its human homologues. J Exp Med. 1990;172:911–919. doi: 10.1084/jem.172.3.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thelestam M, Mollby R. Cultured human fibroblasts as a model for evaluation of potential in vivo toxicity of membrane damaging antibiotics. Chem Biol Interact. 1980;29:315–325. doi: 10.1016/0009-2797(80)90150-7. [DOI] [PubMed] [Google Scholar]
- 60.Ueda A, Okuda K, Ohno S, Shirai A, Igarashi T, Matsunaga K, Fukushima J, Kawamoto S, Ishigatsubo Y, Okubo T. NF-kappa B and Sp1 regulate transcription of the human monocyte chemoattractant protein-1 gene. J Immunol. 1994;153:2052–2063. [PubMed] [Google Scholar]
- 61.Vaddi K, Keller M, Newton R C. The chemokine FactsBook. New York, N.Y: Academic Press, Inc.; 1997. [Google Scholar]
- 62.Watt G, Strickman D. Life-threatening scrub typhus in a traveler returning from Thailand. Clin Infect Dis. 1994;18:624–626. doi: 10.1093/clinids/18.4.624. [DOI] [PubMed] [Google Scholar]
- 63.Widmer U, Manogue K R, Cerami A, Sherry B. Genomic cloning and promoter analysis of macrophage inflammatory protein (MIP)-2, MIP-1 alpha, and MIP-1 beta, members of the chemokine superfamily of proinflammatory cytokines. J Immunol. 1993;150:4996–5012. [PubMed] [Google Scholar]