Abstract
Objectives
There is an unmet medical need for biomarkers in OA which can be applied in clinical drug development trials. The present study describes the development of a specific and robust assay measuring type II collagen degradation (T2CM) and discusses its potential as a noninvasive translational biomarker.
Methods
A type II collagen specific neoepitope (T2CM) was identified by mass spectrometry and monoclonal antibodies were raised towards the epitope, employed in a chemiluminescence immunoassay. T2CM was assessed in bovine cartilage explants with or without MMP-13 inhibitor, and explant supernatants were analyzed by Western blot. T2CM was measured in plasma samples from one study (n = 48 patients) where OA patients were referred to total knee replacement (TKR). Additionally, T2CM was quantified in serum from OA patients receiving salmon calcitonin treatment (sCT) (n = 50) compared to placebo (n = 57).
Results
The T2CM assay was technically robust (13/4 % inter/intra-variation) and specific for the type II collagen fragment cleaved by MMP-1 and -13. The MMP-13 inhibitor reduced the T2CM release from bovine cartilage explants receiving catabolic treatment. These results were confirmed by Western blot. In human end-stage OA patients (scheduled for TKR), the T2CM levels were elevated compared to moderate OA (p<0.004). The OA patients receiving sCT had lower levels of T2CM compared to placebo group after 1, 6, and 24 months of treatment (p = 0.0285, p = 0.0484, p = 0.0035).
Conclusions
To our knowledge, T2CM is the first technically robust serological biomarker assay which has shown biological relevance in ex vivo models and OA cohorts. This suggests that T2CM may have potential as a translational biomarker for cartilage degradation.
Keywords: Biomarker, Extracellular matrix, Cartilage, Type II collagen, T2CM
1. Introduction
Osteoarthritis (OA) is the most prevalent degenerative joint disorder which can advance to chronic disability and cause reduced quality of life in adults [1]. Although OA pathology involves tissue of the whole joint structure, the progressive destruction of articular cartilage extracellular matrix (ECM) is considered its hallmark [1,2]. During cartilage remodeling, type II collagen and aggrecan, which are the most abundant molecules of the cartilage ECM, are consecutively cleaved by matrix degrading enzymes [3]. Enzymes cleaving type II collagen include gelatinases and collagenases, which are members of the matrix metalloproteinase (MMP) family. In OA, the tissue homeostasis is disrupted with a predominance of catabolic activities leading to increased expression of collagenases cleaving the ECM. Especially MMP-13 is considered to be the main collagenase in OA and is known to actively cleave and degrade type II collagen [[4], [5], [6]]. When MMPs cleave type II collagen, it results in protein fragments called neoepitopes, which are released into circulation. These protein fragments can reflect a pathology-specific enzymatic modification and may serve as biomarkers indicating the degree of cartilage destruction [7]. For several years, many potential biomarkers of type II collagen have been investigated in OA [8]. One of the most studied potential biomarker in the OA field is the urinary C-terminal cross-linked telopeptide of type II collagen (CTX-II) which has demonstrated to be a valuable biochemical marker in terms of diagnosis, severity of disease, and prognosis [[9], [10], [11], [12]]. However, when CTX-II is quantified in serum, it has shown inadequate utility to reflect clinical relevance in OA cohort compared to urinary CTX-II [13,14]. Identifying a serological type II collagen degradation biomarker would be advantageous, since serum is relatively easily accessible and is largely present at sites where many of the body's metabolic processes are taking place. Consequently, it is likely that many of the metabolic changes occurring in OA may be represented in serum [15]. Besides identifying serological biomarkers, biomarkers reflecting MMP-13 activity would also be a valuable tool. Studies have indicated that MMP-13 plays a critical role in type II collagen degradation in articular cartilage in OA, and could therefore be an important treatment target during OA progression [16,17]. OA drug candidates fail in clinical trials and hence, no therapies that modify the onset or progression of structural damage in OA have been approved for use in patients. Applying objective biomarkers in combination with the current methods (clinical examinations, questionnaires, and radiographs) may advance the development of approved OA drugs. Biomarkers can be a valuable tool in the clinical drug development by facilitating patient selection and study design optimization. For years, the OA patient heterogeneity including patients with low disease progression have been hindering the clinical trials. However, identifying the patients who are fast progressors of OA and those who may respond to the therapy will be a paramount [18].
Identifying a serological type II collagen degradation biomarker which reflects MMP-13 activity may facilitate therapeutic advances and allow for strategies to reduce the costs of clinical trials. To our knowledge such biomarker assay has not been developed before and tested in OA cohorts.
In the presented work, we developed and validated a novel specific assay targeting a neoepitope fragment, called T2CM (Type II collagen MMP-derived), generated from MMP-1 and -13 cleavage in the C-terminal of the type II collagen helical domain. The potential as a translational biomarker and biological value of T2CM was evaluated in supernatant from a bovine full-depth cartilage explant (BEX) model with catabolic and MMP-13 inhibitory treatment compared to an untreated control group. In addition, the potential of T2CM as a noninvasive biomarker was investigated by quantifying its concentration in EDTA plasma samples from end-stage OA patients scheduled for total knee replacement (TKR). Furthermore, T2CM was measured in serum samples from OA patients receiving oral salmon calcitonin treatment (sCT) compared to the placebo group. Salmon calcitonin has previously shown to reduce OA pathogenesis including bone resorption and cartilage degradation [[19], [20], [21]]. Thus, an ideal treatment in OA cohort for testing the potential of T2CM as a pharmacodynamic biomarker.
2. Methods
All the reagents used were standard chemicals from Merck (Whitehouse Station, NJ, USA) and Sigma (St. Louis, MO, USA) unless stated otherwise. All synthetic peptides for antibody production and assay validation were purchased from Genscript (Piscataway, NJ, USA) (Table 1).
Table 1.
Amino acid sequences of the synthetic peptides used for monoclonal antibody production and CLIA validation.
| Peptide type | Sequence |
|---|---|
| Immunogenic peptide | MSAFAGLGPR-GGC-KLHa |
| Standard peptide | MSAFAGLGPR |
| Screening peptide | MSAFAGLGPRK-Biotin |
| Elongated peptide | DMSAFAGLGPR |
| Truncated peptide | SAFAGLGPR |
| Non-sense coating peptide | HDFSSDLENVK-Biotin |
Keyhole Limpet Hemocyanin
2.1. Identification and selection of neoepitope fragments
The neoepitope-containing fragment was discovered by mass spectrometry (MS) in conditioned media of a bovine full-depth cartilage explant (BEX) model, described previously [22]. Briefly, full-depth bovine femoral condyle articular cartilage explants were cultured in serum-free medium in the absence, or presence of pro-inflammatory cytokines, and conditioned media were harvested three times a week for three weeks. Conditioned media were harvested at selected timepoints from IL-17 [100 ng/mL], oncostatin M [10 ng/mL] combined with TNF-α [20 ng/mL], and control (untreated) explant groups. The harvested supernatants were analyzed by shotgun LC-MS/MS for identification and label-free quantification peptides. For endogenous cleavage sites discovery, in addition to MS analysis procedure of trypsin-digested samples described previously [22], conditioned media were also analyzed without trypsin-digestion: 100 μL of media from each timepoint were reduced and alkylated as previously described, mixed with equal volume of 1 M NaCl containing 1 % formic acid, ultrafiltered through a 30 kDa filter (PALL Life Sciences, Westborough, MA, USA) and the <30 kDa filtrate desalted with reversed-phase Vydac UltraMicro Spin C18 columns (Harvard Apparatus, cat#74–7206) according to the manufacturer's instructions. The MS analysis parameters for these samples were the same as for trypsin-digested samples, but with the following modifications: MS 1E06, scan 100 ms 200–1800 m/z, MS/MS 1E06, 60 ms.
Peptide identification was done with Proteome Discoverer 2.1 software (ThermoFisher Scientific). For tryptic digests, the processing workflow consisted of the following nodes: Spectrum Selector for spectra pre-processing (precursor mass range: 350–5000 Da; S/N Threshold: 1.5), Sequest-HT search engine (Protein Database: Bos Taurus proteome, UniProt (https://www.uniprot.org/) proteome ID UP000009136, n23868, downloaded June 08, 2015); Enzyme: Semi-trypsin; Max. missed cleavage sites: 2; Peptide length range 6–144 amino acids; Precursor mass tolerance: 10 ppm; Fragment mass tolerance: 0.02 Da; Static modification: cysteine carbamidomethylation; Dynamic modification: methionine oxidation, hydroxyproline and pyro-glutamic acid (N-terminal Glu to pyroglutamic acid), and Percolator for peptide validation (FDR <1 % based on peptide q-value). For ≤30 kDa filtrates, the processing workflow consisted of the following nodes: Spectrum Selector for spectra pre-processing (precursor mass range: 10–30000 Da; S/N Threshold: 1.5), Sequest-HT search engine (Protein Database: as before; Enzyme: No enzyme; Max. missed cleavage sites: 0; Peptide length range 6–144 amino acids; Precursor mass tolerance: 10 ppm; Fragment mass tolerance: 0.02 Da; and Percolator for peptide validation (FDR <1 % based on peptide q-value). Peptide intensities were quantified using a proprietary algorithm developed in Proteome Discoverer 2.1.
The fragment for monoclonal antibody development was selected based on PEP score (<0.05), q-value (<0.01), Xcorr>2 and the frequency of occurrence, and it was verified to be unique for type II collagen by BLAST using NPS@: Network Protein Sequence Analysis with the UniprotKB/Swiss-prot database [23]. The sequence was also aligned between human, mouse, bovine, and rat to see if it is conserved across species using Uniprot's Align tool (Clustal Omega algorithm) [24]. In-house bioinformatic tools were used to screen and shortlist epitopes of interest before further investigation.
2.2. Monoclonal antibody development
Five 6- to 7-week-old Balb/C female mice were immunized subcutaneously with 200 μL emulsified antigen and 50 μg immunogenic peptide (MSAFAGLGPR -GGC-KLH) using Stimune Immunogenic Adjuvant (SPECOL) (Invitrogen, Carlsbad, CA, US). The immunizations were performed at 2-week intervals until stable serum titer levels were reached. The mouse with the highest serum titer rested for a month and then was boosted intravenously with 50 μg immunogenic peptide in 100 μL 0.9 % NaCl solution 3 days before isolation of the spleen for cell fusion. The mouse spleen cells were fused with SP2/0 myeloma cells to produce hybridoma cells, method previously described by Gefter et al. (1977) [25]. The selected hybridoma clones were grown in 96-well microtiter plates using standard limiting dilution method to secure monoclonal growth.
2.3. Monoclonal antibody characterization
Native reactivity and peptide affinity of the generated monoclonal antibodies were evaluated by displacement using human serum samples and the standard peptide in a preliminary ELISA using 4 mg/mL coating peptide on a streptavidin-coated microtiter plate (Roche, Basel, Switzerland) and the supernatant (containing the antibodies) from the monoclonal hybridoma cells. Two hybridoma clones (NBH237B#33-14F2-1D5-2C8 and NBH237B#33-14F2-1D9-1B6) were selected based on their high reactivity to the native samples, including their reactivity with only the standard peptide, and not the truncated, elongated, or non-sense coating peptide. The immunoglobulins were purified from supernatant using HiTrap Protein G HP affinity columns according to manufacturer's instructions (GE Healthcare Life Science, Buckinghamshire, UK) and antibody isotype was determined using Rapid ELISA Mouse monoclonal antibody Isotyping Kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. An antibody of one hybridoma clone (NBH237B#33-14F2-1D5-2C8) was chosen for further assay development and labelled with horseradish peroxidase (HRP) using a Peroxidase Labelling Kit (Roche, Basel, Switzerland) following the manufacturer's instructions.
2.4. Assay protocol of T2CM CLIA
The development of the competitive chemiluminescence immunoassay (CLIA) included several preliminary optimizing experiments where the reagents, their concentrations, incubation-time and -temperature were analyzed by several checkerboards. The definitive T2CM CLIA protocol comprised of following: A 96-well streptavidin-coated white microplate (Greiner Bio-One, Kremsmünster, Austria) was coated with 2.5 ng/mL biotinylated synthetic peptide (MSAFAGLGPRK-Biotin) dissolved in assay buffer (50 mM phosphate buffered saline (PBS), 1 % bovine serum albumin, 0.1 % Tween-20, 0.36 % Bronidox, 8 % NaCl, adjusted to pH 7.4 at 20 °C) and incubated for 30 min at 20 ᵒC with constant shaking (300 rpm) in darkness. Next, 20 μL/well of standard peptide (100 ng/mL) and samples were added to the appropriate wells, followed by the addition of 100 μL/well of HRP-labelled antibody diluted in assay buffer to the concertation of 25 ng/mL and incubated for 1 h at 20 °C with constant shaking (300 rpm) in darkness. After each incubation step, wells were washed five times with standard washing buffer (20 mM Tris, 50 mM NaCl, pH 7.2). The chemiluminescence substrate (Roche, BM Chemiluminescence ELISA substrate (POD), Basel, Switzerland) working solutions were mixed 15 min before use and 100 μL/well were added to plate and incubated for 3 min at 20 °C with constant shaking (300 rpm) in darkness. The relative light units were measured at all wavelengths within 5 min on a microplate luminometer reader (SpectraMax M5, Molecular Devices, CA, USA). A standard curve was plotted using a 4-parameter logistic curve fit Y = (A − D)/(1 + (x/C)^B) + D, where R > 0.9. Data were analyzed using the SoftMax Pro version 7.0.3 software.
2.5. Technical evaluation of T2CM CLIA
Technical assay validation was completed according to international guidelines. The intra- and inter-assay variation was calculated by 10 independent runs including five quality controls and two internal control samples covering the detection range. The lower limit of detection (LLOD) was calculated as the mean + 3 standard deviations (3 SD) from 21 determinations of blank samples (i.e., assay buffer). The upper limit of detection (ULOD) was measured as the mean back-calibration calculation – 3 SD of 10 independent determinations of the highest standard concentration (standard A). The measurement range was defined as the range between lower limit of measurement range (LLMR) and the upper limit of measurement range (ULMR), which were determined from 10 independent runs with the standard peptide. Measurements below LLMR or above ULMR were assigned the value of LLMR/ULMR respectively. IC50 (half-maximal inhibition concentration) was determined from the standard curve. Dilution recovery assessment was performed from 2-fold dilutions of serum, plasma and ex vivo supernatant to calculate the linearity as recovery percentages of 100% with the undiluted sample as reference value. The peptide spiking recovery was assessed by comparing different concentrations of peptide spiked in buffer and human serum to identify the matrix effect of serum samples. The analyte stability was examined through temperature tests and repeated freeze-thaw cycles of serum samples. The temperature tests included different time point and temperatures where T2CM levels were measured in three human serum samples after 0, 2, 4, 24, and 48 h incubation at either 4 °C, 20 °C or 37 °C. The recovery was estimated with 0 h sample as a reference. Furthermore, the effect of four repeated freeze/thaw cycles of three serum samples was assessed where freeze/thaw recovery was calculated with the zero cycle samples as a reference. In addition, analytical interference was performed by adding a low/high content of hemoglobin (2.50/5 mg/mL), lipemia/lipids (1.50/5 mg/mL) and biotin (3/9 ng/mL) to a serum sample of known concentration. Recovery percentage was calculated with the normal serum sample as reference. The normal reference levels for hemoglobin, lipidemia/lipids and biotin were 0–10 mg/dl (0–0.00161 mmol/L), <150 mg/dl (<1.6935 mmol/L) and 0.221–3.004 ng/mL, respectively. Each sample was run in double determination.
2.6. Bovine cartilage explant model
The explants were isolated from the femoral condyles of stifle bovine joints, as described previously [26]. Explants were cultured for 21 days with media being changed every 2–3 days and addition with one of the following treatments: 1) medium without catabolic treatment (w/o), 2) oncostatin M [10 ng/mL] combined with TNF-α [40 ng/mL] (OSM + TNF), 3) OSM + TNF [10/20 ng/mL], 4) OSM + TNF [10/10 ng/mL], 5) OSM + TNF [20/40 ng/mL], 6) OSM + TNF [20/20 ng/mL], 7) OSM + TNF [20/10 ng/mL] in the presence or absence of MMP-13 inhibitor compound, MSC2392891A-3 (Merck, Darmstadt, Germany). The culture media were harvested every 2–3 days and stored until biochemical analysis. Chondrocyte metabolic activity was evaluated by the alamarBlue assay (Thermo Fisher Scientific, Waltham, MA, USA) once weekly until termination day.
2.7. In vitro protease cleavage
OA human articular cartilage from a total knee joint-replacement surgery was cut into small pieces of same weight using a scalpel. The cartilage was snap-frozen in liquid nitrogen and crushed with a metal mortar to increase the permeability of the tissue. Immediately, the smashed cartilage was placed in digestion buffer (50 mM Tris-HCL, 150 mM NaCl, 10 mM CaCl2, 10 μM ZnCl2, 0.05 % Brij35, Mili-Q water) for 2 h followed by gentle agitation to remove blood and contamination. Digestion buffer was removed and replaced with new digestion buffer and the recombinant enzymes MMP-1, -2, -9, -13 (Giotto Biotech, Sesto Fiorentino FI, Italy) were added and incubated with the cartilage for 3 days at 37 °C. The supernatant was harvested and EDTA (5 mM) was added to stop the digestion reaction. All supernatants were stored at −80 °C until measurement with the T2CM assay to identify which proteases that generated this specific neoepitope peptide. All MMP cleavages included a control group containing only digestion buffer and the MMP. Furthermore, all MMP reactions and controls were performed in two replicates.
2.8. Western blot of BEX supernatant
Supernatant from BEX treated with OSM + TNF [20/20 ng/mL], OSM + TNF [20/40 ng/mL], or untreated group (w/o) were harvested on day 7, 14, and 21 and electrophoresed on a NuPAGE 4–12 % Bis-Tris gel (Invitrogen, Carlsbad, CA, US) under reducing conditions using NuPAGE® MES SDS running buffer (Invitrogen, Carlsbad, CA, US). By using an iBlot® Dry blotting system (Life Technologies, Carlsbad, CA, US), the proteins from the polyacrylamide gel were transferred onto an iBlot® nitrocellulose membrane (Life Technologies, Bengaluru, India). Following, the membrane was blocked for 1 h with 5 % skim milk (Sigma–Aldrich, St. Louis, MO, USA) in TBST (Tris-Buffered Saline (TBS) with 0.1 %. Tween-20). The membrane was incubated overnight at 4 °C with monoclonal antibody NBH237B#33-14F2-1D5-2C8. Next, membrane was washed in TBST three times 10 min and incubated 1–5 min in SuperSignal west femto maximum sensitivity substrate (Thermo Fisher Scientific, Waltham, MA, USA). The bands were visualized through C-DiGit™ Blot Scanner (LI-COR Biosciences, Lincoln, NE, USA).
2.9. Clinical validation of T2CM
The clinical relevance of T2CM was evaluated in two OA cohorts. In one of the OA cohorts, T2CM was measured in EDTA plasma samples from 48 patients displaying symptomatic knee OA and underwent TKR, previously published [27]. OA patients were classified by the Kellgren-Lawrence (KL) score system. Additionally, T2CM was measured in serum samples from 50 OA patients receiving 0.8 mg oral sCT twice daily including a placebo group of 57 OA patients, previously published [28,29]. Serum samples were collected at baseline (before treatment), and after 1, 6, 12, 24 months of treatment. The TKR patients were compared to baseline T2CM levels in 54 patients with moderate OA (KL 2) from sCT study. Samples were collected after informed consent and approved by the local ethical committee in compliance with the Helsinki Declaration of 1975.
2.10. Statistical analysis
For the statistical calculations of T2CM levels in BEX model, two-way ANOVA with Tukey's post-hoc test was used to achieve the difference between the treatment groups at each timepoint. Characteristics of the OA patient cohort are presented as a percentage or as a mean SD unless otherwise stated (Table 3A–B). sCT patients with moderate OA (KL 2) were compared to the OA patients referred to TKR (KL 4), and the difference between groups were analyzed by Mann-Whitney test. Results are shown as Tukey boxplots. Furthermore, T2CM results from all OA patients in sCT study were analyzed by one-way analysis of covariance (ANCOVA) with Bonferroni post hoc test, adjusting for covariates (age, gender, and BMI) at each time point. For the covariates to be normally distributed, following natural logarithm transformation equation was used: [30]. Results are presented with percentage (%) difference from baseline. All values are presented as mean SD if not otherwise stated and were considered statistically significant for p < 0.05. Graphs and statistical analyses were performed using GraphPad Prism version 7.01 (GraphPad Software, Inc., CA, USA) except for the ANCOVA with adjustment of BMI, sex, and age by MedCalc Statistical Software version 18 (MedCalc Software, Ostend, Belgium).
Table 3.
A-B. A) Demographics and T2CM measurements of patients referred to TKR with severe OA (KL4)[27] combined with moderate OA patients (KL2) from the oral salmon calcitonin treated cohort at baseline [[28], [29]]. B) Demographics and T2CM measurements of the oral salmon calcitonin treated OA patients and placebo group from cohort 3 [[28], [29]]. Significant difference was observed in gender distribution between placebo and sCT (p < 0.0001). n, number of patients; TKR, Total knee replacement. sCT, salmon calcitonin treatment.
| A | Male/Female(%) |
Age Mean (SD) |
BMI Mean (SD) |
KL score |
T2CM concentration Median with upper/lower fences |
|---|---|---|---|---|---|
| Moderate OA sCT patients before treatment (n = 54) | 43/57 | 68.3 (4.0) | 29.3 (4.7) | 2 | 3.088, 14.45, 1.3 |
| TKR (n = 48) |
42/58 |
68.8 (9.8) |
29.9 (5.5) |
4 |
5.693, 12.95, 2.74 |
|
B |
Male/Female(%) |
Age Mean (SD) |
BMI Mean (SD) |
KL score(%) |
T2CM concentration Mean % difference from baseline with (SD)for 1,6,12,24 months |
| Placebo (n = 57) | 54/45 | 63.1 (6.1) | 27.9 (3.3) | 2: 86 3: 14 |
−33.18 (8.50), −41.46 (8.31), −38.27 (8.86), −22.19 (8.76) |
| sCT (n = 50) | 66/34 | 63.9 (7.0) | 30.2 (5.0) | 2: 88 3: 12 |
−61.49 (9.10), −66.35 (8.90), −59.55 (9.58), −61.73 (9.49) |
Ethical statement
All animals were treated according to the guidelines for animal welfare. Monoclonal antibody production in mice was approved by the Danish National Authority (The Animal Experiments Inspectorate) under approval number 2013-15-2934-00956. Bovine tissue use for explant studies was obtained from the Danish Veterinary and Food Administration under Ministry of Environment and Food of Denmark (J.nr.: 2015-12-711-03228/AMSE, DK-10-3-oth-005).
3. Results
3.1. Identification of the T2CM neoepitope by mass spectrometry and antibody characterization
The neoepitope fragment [D]↓1220MSAFAGLGPR1229 (where ↓ indicates the endogenous cleavage site and [D] is the amino acid preceding the cleavage site) was identified by MS analysis in conditioned media from OSM + TNF-treated bovine articular cartilage explants. It was verified to be unique for type II collagen by BLAST, to be conserved across human, mouse, bovine, and rat species by Clustal Omega multiple sequence alignment (Fig. 1) and was selected for monoclonal antibody development based on PEP score (<0.05), q-value (<0.01), and Sequest-HT Xcorr>2. The monoclonal antibody with the best native reactivity and peptide affinity in the assay setting was selected from the antibody producing clones for assay development. The antibody isotype was determined to be IgG2b, kappa.
Fig. 1.
Sequence alignment of the targeted T2CM sequence in human, mouse, bovine, and rat species (black box). The sequences were aligned using Clustal Omega algorithm.
3.2. Development and technical evaluation of the T2CM assay
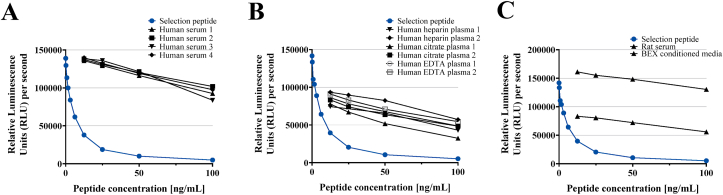
A full technical validation was performed to evaluate the novel T2CM assay. Based on 10 independent assay runs, the intra- and inter-variation was determined to be 4% and 13% respectively, which is within the acceptance criteria <10% for the intra-assay variation and <15% for the inter-assay variation. Linearity of human samples was observed from undiluted to a 1:8 dilution for human serum, human EDTA plasma, human citrate plasma, human heparin plasma, rat serum, mouse serum and BEX supernatant (Fig. 2A–C). The recommended dilution for human samples and BEX supernatant was 1:2 and the mean linearity was within 100 ± 20%. Hemoglobin, lipids and biotin did not interfere with T2CM measurements in serum. The measurement range (LLMR-ULMR) of the assay was determined to be 0.3–19.1.00 ng/mL. The analyte stability was acceptable at prolonged storage of human serum samples at 4 °C (92% after 24h), 20 °C (93% after 24h) and 37 °C (94% after 24h). Moreover, the mean analyte recovery in three human serum samples was 90 % during up to four freeze/thaw cycles (Table 2).
Fig. 2.
A-C. Standard curve and native reactivity to various matrices. Standard curves and native reactivity against A) human serum, B) human plasma (EDTA, Citrate and Heparin), and C) animal samples (rat serum, mouse serum and BEX supernatant), diluted from 1:1, 1:2, 1:4 and 1:8 for the T2CM assay. Signals are shown as relative luminescence units (all wavelengths) per second, as a function of standard peptide concentration.
Table 2.
T2CM assay technical validation summary.
| TECHNICAL VALIDATION | RESULTS (Accepted recovery: 100 ± 20 %) |
|---|---|
| IC50 | 5.63 ng/mL |
| Measurement range (LLMR-ULMR) | 0.3–19.1 ng/mL |
| Inter-assay variationa | 13% |
| Intra-assay variationa | 4% |
| Dilution recovery of human serum (n = 3)a | 92% |
| Dilution recovery of human EDTA plasma (n = 3)a | 94% |
| Dilution recovery of human plasma citrate (n = 3)a | 83% |
| Dilution recovery of human heparin (n = 3)a | 96% |
| Dilution recovery of BEX supernatant (n = 2)a | 114% |
| Dilution recovery of rat serum (n = 2)a | 94% |
| Spiking recovery (serum in serum) (n = 3)a | 95% |
| Analyte recovery 24h, 4ᵒC/20ᵒC/37ᵒCa | 92%/93%/94% |
| Analyte recovery, 4 freeze/thaw cyclesa | 90% |
| Hemoglobin recoverya | 102% |
| Lipemia recoverya | 91% |
| Biotin recoverya | 113% |
Percentages are reported as mean of n, number of measurements. LLMR, lower limit of measuring range; ULMR, upper limit of measuring range.
3.3. Assay specificity and characterization
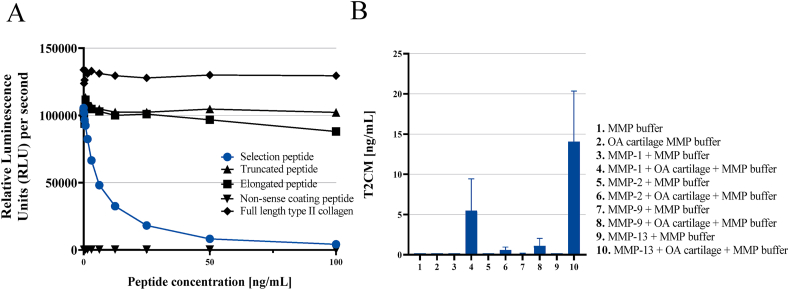
The antibody NBH237B #33 14F2-1D5-2C8 detected the standard peptide (MSAFAGLGPR) generating a standard curve following a 4-parameter logistic curve while no signal was observed when adding the elongated peptide (DMSAFAGLGPR), truncated peptide (SAFAGLGPR), or non-sense coating peptide (HDFSSDLENVK-Biotin) including the full-length type II collagen (Merck, Whitehouse Station, NJ, USA) (Fig. 3A). These data indicate that the antibody was specific towards the unique C-terminal site of the human target sequence ↓1220MSAFAGLGPR1229. The target was generated predominantly by collagenases MMP -1 and −13 in human cartilage (Fig. 3B).
Fig. 3.
A-B. A) High specificity of the T2CM assay. Reactivity towards the standard peptide (MSAFAGLGPR), truncated peptide (SAFAGLGPR), elongated peptide (DMSAFAGLGPR). No background signal was detected when using a non-sense screening (coating) peptide (HDFSSDLENVK-Biotin). Signals are shown as relative luminescence units (RLU) per second, as a function of standard peptide concentration. B) MMP-1 and -13 primarily mediate the T2CM neoepitope. Generation of T2CM fragment by MMP-mediated cleavage of human articular cartilage obtained from an OA patient undergoing total knee replacement surgery. Results are shown as bars with as mean ± SD.
3.4. Translational value and biological evaluation of T2CM in BEX supernatants and human OA samples
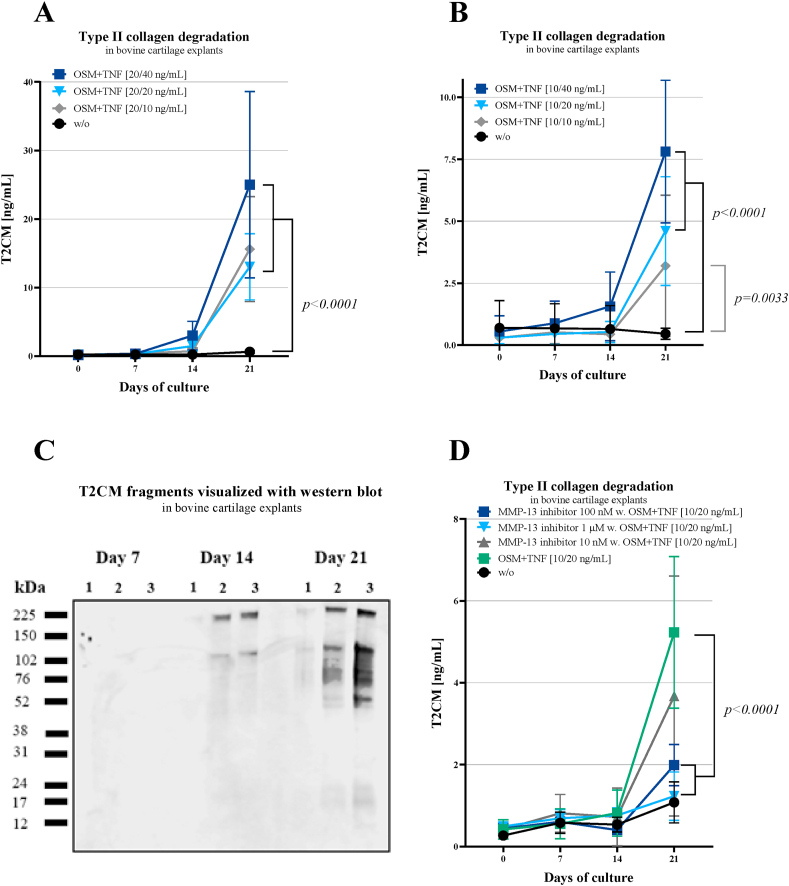
T2CM was measured in BEX supernatant collected on day 0, 7, 14, and 21. All OSM + TNF treated groups showed elevated T2CM levels compared to untreated group as a function of time (p<0.0001, p=0.0033, Fig. 4A–B). These results were further confirmed by Western blot analysis, where T2CM detected the strongest bands at day 21 (~225, ~110, ~76, ~52 and ~17 kDa bands), compared to day 7 (no bands) and day 14 (~225 and ~110 kDa bands) (Fig. 4C). Thus, the number of bands in OSM + TNF treated samples increased with the duration of tissue cultivation. Additionally, the OSM + TNF stimulation was combined with an MMP-13 inhibitor. Here, the T2CM levels were significantly decreased in groups receiving OSM + TNF combined with the two highest concentrations (100 nM and 1 μM) of the MMP-13 inhibitor compared to the OSM + TNF group on day 21 (p<0.0001, Fig. 4D).
Fig. 4.
A-D. A-B) T2CM was increased during prolonged stimulation with OSM + TNF in BEX supernatant. T2CM levels were measured in supernatant from the BEX model harvested on day 0, 7, 14, and 21. Explants were either without treatment (w/o) or treated with different concentrations of OSM + TNF-α (O + T). Group comparisons were done using two-way ANOVA with Tukey's multiple comparison at each timepoint, n = 6. Error bars are shown with mean ± SD. C) Western blot results confirmed the BEX results. T2CM fragments were visualized in explants conditioned media from day 7, 14, and 21, with strongest bands at day 21 which confirms the BEX results. 1) without treatment, 2) 20/20 ng/mL OSM + TNF-α, 3) 20/40 ng/mL OSM + TNF-α. D) MMP-13 inhibitor decreased the release of T2CM. T2CM release measured in BEX supernatant treated with MMP-13 inhibitor in combination with OSM + TNF-α (O + T). Supernatants were harvested on day 0, 7, 14, and 21. Group comparisons were done using two-way ANOVA with Tukey's multiple comparison, n = 6. Error bars are shown with mean ± SD.
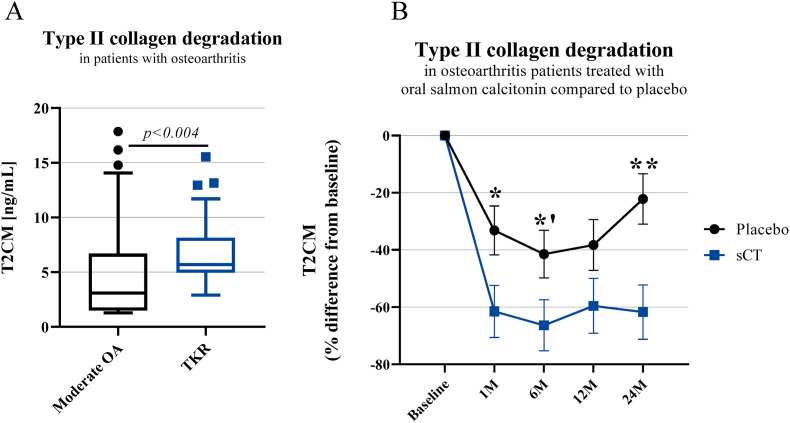
The T2CM levels were also evaluated in human EDTA plasma samples from OA patients directed to TKR (KL 4) and compared to moderate OA patients (KL 2). Average age of the TKR patients was 68.8 years (9.8) and 58 % were female while 68.3 years (4.0) and 57 % were female for the moderate OA patients (Table 3A). T2CM was significantly elevated in OA patients referred to TKR compared to the moderate OA patients (p<0.004) (Fig. 5A). The sCT patients were in average 63.9 years (7.0) and 34 % were female and placebo group in average 63.1 years (6.1) and 45 % were female. There was a statistical difference in the gender distribution observed between sCT patients and placebo (p<0.0001) (Table 3B). Additionally, T2CM was significantly decreased in OA patients receiving sCT compared to the placebo group after 1, 6, and 24 months of treatment (p=0.0285, p=0.0484, p=0.0035) (Fig. 5B).
Fig. 5.
A-B. A) T2CM were increased in patients undergoing total knee replacement. In EDTA plasma levels of T2CM were assessed in patients with moderate OA (n = 54) and patients referred to total knee replacement (TKR) (n = 48). Data were analyzed by Mann-Whitney test, and data are presented as a Tukey boxplot. B) T2CM was decreased in OA patients treated with oral salmon calcitonin compared to placebo. Serum levels of T2CM were assessed in oral salmon calcitonin treated (sCT) OA patients (n = 50) compared to placebo (n = 57). Data were analyzed by one-way analysis of covariance (ANCOVA) with Bonferroni post hoc test corrected for age, gender and BMI and presented as percentage (%) difference from baseline. All values are presented as mean ± SD. Significance difference is given by ∗ p=0.0285, ∗’ p = 0.0484, ∗∗p=0.0035. M: Month.
4. Discussion
The present study described the development and validation of a novel competitive CLIA assay for the detection of MMP-1 and -13 generated fragment of type II collagen, named T2CM. The main findings included: 1) A technically robust and specific assay towards the T2CM fragment 1220MSAFAGLGPR1229 primarily generated by MMP-1 and -13, 2) T2CM was measurable in human serum, human plasma, rat serum, mouse serum, and supernatant from the BEX model, 3) T2CM was elevated in BEX cultures treated with OSM + TNF after 14 days, 4) T2CM was decreased when OSM + TNF treatment was combined with MMP-13 inhibitor, 5) T2CM was upregulated in OA patients referred to TKR compared to patients with moderate OA, and 6) T2CM was downregulated in OA patients receiving sCT compared to the placebo group.
Destruction and fragmentation of articular cartilage and its components (particularly type II collagen and aggrecan) is the most prominent feature of OA [31]. This destruction is partly mediated by MMPs. For example, MMP-1 is upregulated in arthritic disease and degrades native triple-helical fibrillar collagens including type II collagen [32]. Furthermore, MMP-13 possesses robust ability to cleave type II collagen and is considered a major catabolic effector playing a critical role in OA induction, thus frequently targeted in OA studies [32,33]. Hence, MMP-13 inhibitors are thought to have the ability to halt cartilage erosion in early OA [32]. Although MMP-13 is considered to be one of the major collagenases in OA, it is also involved in other diseases characterized by destruction of the collagenous tissue architecture such as rheumatoid arthritis and atherosclerosis [34]. Consequently, increased levels of T2CM released into circulation may be a result of other diseases apart from OA where MMP-13 is also involved.
In the BEX model, we hypothesized that T2CM is released into the supernatant when explants are treated with a catabolic stimulus such as OSM and TNF. As previously reported, OSM + TNF treatment has been shown to upregulate MMP activity [22,35] and induce MMP-mediated type II collagen (C2) degradation measured by the C2M biomarker in the BEX model [22,36]. Indeed, the T2CM release was enhanced with increasing OSM + TNF treatment verified by both CLIA and Western blot, which confirms the MS results from the biomarker discovery phase. Since the neoepitope sequence is located close to the C-terminal of the triple helical region, it may correspond to a rather small band in the Western blot. However, collagen cross-links (on lysine residues) close to the neoepitope could potentially explain the larger sized immunoreactive bands observed in the Western blot [37]. As the T2CM fragment was generated by cartilage cleavage with MMP-1 and -13, and its release is decreased when treated with MMP-13 inhibitor, we hypothesized that this biomarker may have clinical relevance in OA or other diseases with altered type II collagen turnover. T2CM was found to be upregulated in OA patients referred to TKR compared to patients diagnosed with moderate OA. A broader range of the T2CM levels was observed in the moderate OA patients compared to TKR. Theoretically, the broader range of T2CM would be expected in the TKR group. However, it cannot be ruled out that some of the moderate OA patients may be fast disease progressors with more severe OA compared to their diagnosis. Despite the wide application of the KL classification, it suffers from subjectivity of a practitioner and is criticized for its application to disease progression and insensitivity to change. Furthermore, there is a lack of a clear cut between KL 3–4 in joint space narrowing.
Levels of T2CM were decreased in OA patients treated with oral salmon calcitonin compared to placebo group. Above findings highlight the potential of T2CM as a translational biomarker that can be applied in both preclinical compound screening and in clinical studies, either alone or as a part of a biomarker panel. As OA is a heterogeneous disease involving alterations of the entire joint tissue [5], combining T2CM (representing cartilage) with ECM markers of other joint tissues such as bone and synovial membrane may help explain the pathological alterations of the whole joint as an organ.
Various potential biomarker candidates of type II collagen have been investigated in OA. However, to our knowledge no similar serological type II collagen degradation biomarker reflecting MMP-1 and -13 activity has been developed. A biomarker fragment generated from MMP-13 cleavage of type II collagen could be advantages especially in terms of identifying new disease-modifying treatments for OA. MMP-13 has been proposed as a favorable therapeutic target to decelerate articular cartilage degradation. Besides cleaving type II collagen in cartilage, MMP-13 also degrades proteoglycan, types IV and type IX collagen, osteonectin and perlecan in cartilage [38]. Clinical investigation has shown that patients with articular cartilage deterioration have high MMP-13 expression, suggesting that enhanced MMP-13 may be associated with cartilage break down [39]. New waves of potential drug candidates inhibiting MMP-13 directly or indirectly have shown remarkably improved selectivity compared to the initially failed inhibitor compounds [17,40]. However, the road to clinical implementation of MMP-13 inhibitors is not straightforward. The development of MMP-inhibitor drugs is complex and can involve unfavorable clinical outcomes as seen in cancer with adverse effects [41]. To promote future development of MMP-13 inhibitor therapeutics in OA, an MMP-13 driven biomarker such as T2CM may play a crucial role in all stages of drug development.
Limitations of the study include the lack EDTA plasma samples from a healthy group with matching average age for comparison of T2CM levels to groups with moderate OA and TKR. The T2CM neoepitope fragment is measured in serum and EDTA plasma samples which include a pool of degraded type II collagen not exclusively from articular cartilage. Type II collagen is also present in hyaline cartilage located in the nose, ears, ends of the ribs and in parts of the respiratory system [42]. Hence not all T2CM fragments may originate from the synovial joint of the knee. The EDTA plasma and serum samples from the two OA cohorts including supernatant from the BEX studies were collected for another purpose which explains the limited sample sizes. Another limitation is the absence of MMP-1 inhibitor, since T2CM is generated from both MMP-1 and -13. Based on these limitations, future investigations should seek to explore and validate the diagnostic and prognostic value of T2CM in OA and other diseases involving MMP-1 and -13 breakdown of type II collagen.
5. Conclusion
In conclusion, a novel neoepitope-based biomarker, T2CM, measuring a MMP-1 and -13 generated fragment of type II collagen was developed and validated in a translational animal model to OA human clinical set-ups. T2CM was upregulated in a bovine ex vivo cartilage model and in patients with severe OA compared to moderate OA. In addition, T2CM was downregulated in OA patients treated with oral salmon calcitonin. Based on these results, T2CM may be considered as a potential biomarker for assessing cartilage degradation in joint degenerative diseases. In addition, the assay may have potential as a compound screening tool and/or as a translational noninvasive serological biomarker for OA, providing a surrogate measure of current degradation status of cartilage.
Authors contributions
Dovile Sinkeviciute and Patrik Önnerfjord performed the mass spectrometry and identified the biomarker sequence through in-house bioinformatic tools developed by Joseph Blair and Line Mærsk Staunstrup. Solveig Skovlund Groen and Signe Holm Nielsen developed the immunoassay assay targeting T2CM and did the technical and biological validation of the assay. Christian S. Thudium, Sven Lindemann, and Daniela Werkmann designed the preclinical BEX studies. Morten A. Karsdal, Anne-Christine Bay-Jensen, and Lars Arendt-Nielsen designed and coordinated the allocation of OA studies. Solveig Skovlund Groen did the data analysis and drafted the manuscript. All authors contributed to and approved the final manuscript. All authors edited the manuscript, with Simon Francis Thomsen providing meticulous revisions.
Role of the funding source
We would like to thank The Danish Council for Technology and Innovation and The Danish National Advanced Technology Foundation for the support to conduct the two OA human studies.
Declaration of competing interest
The study was supported by The Danish Council for Technology and Innovation and The Danish National Advanced Technology Foundation. Morten A. Karsdal, Anne-Christine Bay-Jensen, Christian S. Thudium and Signe Holm Nielsen, Joseph Blair, and Line Mærsk are employees of Nordic Bioscience. Morten A. Karsdal holds stock in Nordic Bioscience. Sven Lindemann and Daniela Werkmann are employees of Merck. Solveig Skovlund Groen, Simon Francis Thomsen, Dovile Sinkeviciute, Patrik Önnerfjord, and Lars Arendt-Nielsen have no competing interest.
Acknowledgements
We would like to acknowledge Tülay Dastan for her work in antibody production phase and Helene Sofie Hector for her help with bovine explant cultures. Furthermore, a special thanks to Ole Simonsen for his valuable and highly appreciated work in conducting the OA studies.
References
- 1.Shi Y., Hu X., Cheng J., et al. A small molecule promotes cartilage extracellular matrix generation and inhibits osteoarthritis development. Nat. Commun. 2019;10:1914. doi: 10.1038/s41467-019-09839-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen D., Shen J., Zhao W., et al. Osteoarthritis: toward a comprehensive understanding of pathological mechanism. Bone Res. 2017;5:16044. doi: 10.1038/boneres.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sophia Fox A.J., Bedi A., Rodeo S.A. The basic science of articular cartilage: structure, composition, and function. Sport Health. 2009;1:461–468. doi: 10.1177/1941738109350438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billinghurst R.C., Dahlberg L., Ionescu M., et al. Enhanced cleavage of type II collagen by collagenases in osteoarthritic articular cartilage. J. Clin. Invest. 1997;99:1534–1545. doi: 10.1172/JCI119316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loeser R.F., Goldring S.R., Scanzello C.R., Goldring M.B. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum. 2012;64:1697–1707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell P.G., Magna H.A., Reeves L.M., et al. Cloning, expression, and type II collagenolytic activity of matrix metalloproteinase-13 from human osteoarthritic cartilage. J. Clin. Invest. 1996;97:761–768. doi: 10.1172/JCI118475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karsdal M.A., Henriksen K., Leeming D.J., et al. Biochemical markers and the FDA critical Path: how biomarkers may contribute to the understanding of pathophysiology and provide unique and necessary tools for drug development. Biomarkers. 2009;14:181–202. doi: 10.1080/13547500902777608. [DOI] [PubMed] [Google Scholar]
- 8.Henrotin Y., Addison S., Kraus V., Deberg M. Type II collagen markers in osteoarthritis: what do they indicate? Curr. Opin. Rheumatol. 2007;19:444–450. doi: 10.1097/BOR.0b013e32829fb3b5. [DOI] [PubMed] [Google Scholar]
- 9.Lotz M., Martel-Pelletier J., Christiansen C., et al. Value of biomarkers in osteoarthritis: current status and perspectives. Ann. Rheum. Dis. 2013;90:171–178. doi: 10.1136/annrheumdis-2013-203726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Spil W.E., DeGroot J., Lems W.F., Oostveen J.C.M., Lafeber F.P.J.G. Serum and urinary biochemical markers for knee and hip-osteoarthritis: a systematic review applying the consensus BIPED criteria. Osteoarthritis Cartilage. 2010;18:605–612. doi: 10.1016/j.joca.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Hao H.Q., Zhang J.F., He Q.Q., Wang Z. Cartilage oligomeric matrix protein, C-terminal cross-linking telopeptide of type II collagen, and matrix metalloproteinase-3 as biomarkers for knee and hip osteoarthritis (OA) diagnosis: a systematic review and meta-analysis. Osteoarthritis Cartilage. 2019;27:726–736. doi: 10.1016/j.joca.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Kraus V.B., Collins J.E., Hargrove D., et al. Predictive validity of biochemical biomarkers in knee osteoarthritis: data from the FNIH OA Biomarkers Consortium. Ann. Rheum. Dis. 2017;76:186–195. doi: 10.1136/annrheumdis-2016-209252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oestergaard S., Chouinard L., Doyle N., et al. The utility of measuring C-terminal telopeptides of collagen type II (CTX-II) in serum and synovial fluid samples for estimation of articular cartilage status in experimental models of destructive joint diseases. Osteoarthritis Cartilage. 2006;14:670–679. doi: 10.1016/j.joca.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Luo Y., He Y., Karsdal M., Bay-Jensen A.-C. Serological CTX-II does not measure the same as urinary CTX-II. Osteoarthr Cartil Open. 2020;2:100081. doi: 10.1016/j.ocarto.2020.100082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munjal A., Bapat S., Hubbard D., Hunter M., Kolhe R., Fulzele S. Advances in Molecular biomarker for early diagnosis of Osteoarthritis. Biomol. Concepts. 2019;10:111–119. doi: 10.1515/bmc-2019-0014. [DOI] [PubMed] [Google Scholar]
- 16.Ruan G., Xu J., Wang K., et al. Associations between knee structural measures, circulating inflammatory factors and MMP13 in patients with knee osteoarthritis. Osteoarthritis Cartilage. 2018;26:1063–1069. doi: 10.1016/j.joca.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Li H., Wang D., Yuan Y., Min J. New insights on the MMP-13 regulatory network in the pathogenesis of early osteoarthritis. Arthritis Res. Ther. 2017;19:248. doi: 10.1186/s13075-017-1454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter D.J., Nevitt M., Losina E., Kraus V. Biomarkers for osteoarthritis: current position and steps towards further validation. Best Pract. Res. Clin. Rheumatol. 2014;28:61–71. doi: 10.1016/j.berh.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen R.H., Bay-Jensen A.C., Byrjalsen I., Karsdal M.A. Oral salmon calcitonin reduces cartilage and bone pathology in an osteoarthritis rat model with increased subchondral bone turnover. Osteoarthritis Cartilage. 2011;19:466–473. doi: 10.1016/j.joca.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Karsdal M.A., Byrjalsen I., Henriksen K., et al. The effect of oral salmon calcitonin delivered with 5-CNAC on bone and cartilage degradation in osteoarthritic patients: a 14-day randomized study. Osteoarthritis Cartilage. 2010;18:150–159. doi: 10.1016/j.joca.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 21.Sondergaard B.C., Madsen S.H., Segovia-Silvestre T., et al. Investigation of the direct effects of salmon calcitonin on human osteoarthritic chondrocytes. BMC Muscoskel. Disord. 2010;11:62. doi: 10.1186/1471-2474-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinkeviciute D., Aspberg A., He Y., Bay-Jensen A.-C., Önnerfjord P. Characterization of the interleukin-17 effect on articular cartilage in a translational model: an explorative study. BMC Rheumatol. 2020;4:30. doi: 10.1186/s41927-020-00122-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Combet C., Blanchet C., Geourjon C., Deléage G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 24.Pundir S., Martin M.J., O'Donovan C. UniProt tools. Curr Protoc Bioinforma. 2016;53 doi: 10.1002/0471250953.bi0129s53. 1.29.1-1.29.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gefter M.L., Margulies D.H., Scharff M.D. A simple method for polyethylene glycol-promoted hybridization of mouse myeloma cells. Somat. Cell Genet. 1977;3:231–236. doi: 10.1007/BF01551818. [DOI] [PubMed] [Google Scholar]
- 26.Gudmann N.S., Wang J., Hoielt S., et al. Cartilage turnover reflected by metabolic processing of type II collagen: a novel marker of anabolic function in chondrocytes. Int. J. Mol. Sci. 2014;15:18789–18803. doi: 10.3390/ijms151018789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siebuhr A.S., Petersen K.K., Arendt-Nielsen L., et al. Identification and characterisation of osteoarthritis patients with inflammation derived tissue turnover. Osteoarthritis Cartilage. 2014;22:44–50. doi: 10.1016/j.joca.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 28.Karsdal M.A., Bihlet A., Byrjalsen I., et al. OA phenotypes, rather than disease stage, drive structural progression - identification of structural progressors from 2 phase III randomized clinical studies with symptomatic knee OA. Osteoarthritis Cartilage. 2015;23:550–558. doi: 10.1016/j.joca.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 29.Karsdal M.A., Byrjalsen I., Alexandersen P., et al. Treatment of symptomatic knee osteoarthritis with oral salmon calcitonin: results from two phase 3 trials. Osteoarthritis Cartilage. 2015;23:532–543. doi: 10.1016/j.joca.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 30.Cole T.J., Altman D.G. Statistics Notes: percentage differences, symmetry, and natural logarithms. BMJ. 2017;358:j3683. doi: 10.1136/bmj.j3683. [DOI] [PubMed] [Google Scholar]
- 31.Zheng S., Hunter D.J., Xu J., Ding C. Monoclonal antibodies for the treatment of osteoarthritis. Expet Opin. Biol. Ther. 2016;16:1529–1540. doi: 10.1080/14712598.2016.1229774. [DOI] [PubMed] [Google Scholar]
- 32.Mehana E.S.E., Khafaga A.F., El-Blehi S.S. The role of matrix metalloproteinases in osteoarthritis pathogenesis: an updated review. Life Sci. 2019;234:116786. doi: 10.1016/j.lfs.2019.116786. [DOI] [PubMed] [Google Scholar]
- 33.Rose B.J., Kooyman D.L. A tale of two joints: the role of matrix metalloproteases in cartilage biology. Dis. Markers. 2016;2016:4895050. doi: 10.1155/2016/4895050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto K., Okano H., Miyagawa W., et al. MMP-13 is constitutively produced in human chondrocytes and co-endocytosed with ADAMTS-5 and TIMP-3 by the endocytic receptor LRP1. Matrix Biol. 2016;56:57–73. doi: 10.1016/j.matbio.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sondergaard B.C., Wulf H., Henriksen K., et al. Calcitonin directly attenuates collagen type II degradation by inhibition of matrix metalloproteinase expression and activity in articular chondrocytes. Osteoarthritis Cartilage. 2006;14:759–768. doi: 10.1016/j.joca.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Kjelgaard-Petersen C.F., Platt A., Braddock M., et al. Translational biomarkers and ex vivo models of joint tissues as a tool for drug development in rheumatoid arthritis. Arthritis Rheum. 2018;70:1419–1428. doi: 10.1002/art.40527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eyre D.R., Weis M.A., Wu J.J. Advances in collagen cross-link analysis. Methods. 2008;45:65–74. doi: 10.1016/j.ymeth.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiomi T., Lemaître V., D'Armiento J., Okada Y. Matrix metalloproteinases, a disintegrin and metalloproteinases, and a disintegrin and metalloproteinases with thrombospondin motifs in non-neoplastic diseases: review Article. Pathol. Int. 2010;60:477–496. doi: 10.1111/j.1440-1827.2010.02547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roach H.I., Yamada N., Cheung K.S.C., et al. Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis Rheum. 2005;52:3110–3124. doi: 10.1002/art.21300. [DOI] [PubMed] [Google Scholar]
- 40.Young D.A., Barter M.J., Wilkinson D.J. Recent advances in understanding the regulation of metalloproteinases. F1000Research. 2019;8:195. doi: 10.12688/f1000research.17471.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roy R., Yang J., Moses M.A. Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J. Clin. Oncol. 2009;27:5287–5297. doi: 10.1200/JCO.2009.23.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watkins J. Biomechanics of musculoskeletal adaptation. Comprehensive Biomedical Physics. 2014;10:1–37. [Google Scholar]