Abstract
Objective
Exercise is known to induce beneficial effects in synovial joints. However, the mechanisms underlying these are unclear. Synovial joints experience repeated mechanical loading during exercise. These mechanical stimuli are transduced into biological responses through cellular mechanotransduction. Mechanotransduction in synovial joints is typically studied in tissues. However, synovial fluid directly contacts all components of the joint, and thus may produce a whole-joint picture of the mechanotransduction response to loading. The objective of this study was to determine if metabolic phenotypes are present in the synovial fluid after acute exercise as a first step to understanding the beneficial effects of exercise on the joint.
Material and methods
Mice underwent a single night of voluntary wheel running or standard housing and synovial fluid was harvested for global metabolomic profiling by LC-MS. Hierarchical unsupervised clustering, partial least squares discriminant, and pathway analysis provided insight into exercise-induced mechanotransduction.
Results
Acute exercise produced a distinct metabolic phenotype in synovial fluid. Mechanosensitive metabolites included coenzyme A derivatives, prostaglandin derivatives, phospholipid species, tryptophan, methionine, vitamin D3, fatty acids, and thiocholesterol. Enrichment analysis identified several pathways previously linked to exercise including amino acid metabolism, inflammatory pathways, citrulline-nitric oxide cycle, catecholamine biosynthesis, ubiquinol biosynthesis, and phospholipid metabolism.
Conclusion
To our knowledge, this is the first study to investigate metabolomic profiles of synovial fluid during in vivo mechanotransduction. These profiles indicate that exercise induced stress-response processes including both pro- and anti-inflammatory pathways. Further research will expand these results and define the relationship between the synovial fluid and the serum.
1. Introduction
Synovial joints allow force transmission and joint rotation between long bones. Synovial joints, such as the knee, hip, hands, and shoulder, are common sites of osteoarthritis (OA). Because synovial joints experience frequent mechanical loading during joint motion, the biomechanical properties and mechanobiological responses of tissues from these joints have been subject to extensive research.
Mechanotransduction is the conversion of a mechanical stimulus into a biological response. Within the synovial joint, the articular cartilage and periarticular bone have been most studied in the context of mechanotransduction. Chondrocytes, the highly specialized resident cell type in articular cartilage, produce and maintain the cartilage matrix by responding to the mechanical environment in the joint [1]. Studies show that physiological dynamic loading in the joint maintains the cartilage matrix by promoting anabolic processes in chondrocytes [2]. In contrast, cartilage matrix synthesis is reduced and breakdown is increased when loading is absent or exceeds the physiologic range, as occurs with high impact loads and tissue strains resulting from injury or overuse [3,4]. When chondrocytes fail to maintain homeostasis of these anabolic and catabolic processes, osteoarthritis (OA) may develop [5]. Physiological loading is also necessary to maintain bone homeostasis. A lack of physiological loading results in bone resorption, whereas enhanced physiological loading stimulates bone formation [6]. Osteoblasts and osteocytes are the main mechanosensitive cells in the bone that respond to mechanical stimuli by promoting bone growth, remodeling, and repair to maintain homeostasis of the bone [6].
Synovial fluid (SF) has received less focus in joint mechanotransduction studies despite it being a mechanosensitive fluid with non-Newtonian flow characteristics that bathes the articular cartilage and periarticular bone surfaces [7,8]. SF is a plasma dialysate that is secreted by the synovial membrane. It contains a pool of molecules originating in both the blood plasma and the joint. Numerous cells throughout the joint may secrete factors into the SF, including synoviocytes, chondrocytes, osteoblasts, osteoclasts, osteocytes, fibroblasts, resident and infiltrating immune cells, and cells from other connective joint tissues including ligaments, tendons, and menisci [8]. Consequently, SF is likely to reflect the overall local state of the joint. Although mechanotransduction studies have focused on the lubricating properties of SF, a metabolic analysis of SF may provide a whole-joint perspective on mechanotransduction in synovial joints.
Metabolomics, or the study of small molecules in a biological system, is a promising method to characterize changes in joint biology [9]. Changes in the metabolome occur on a much faster timescale than in the proteome or genome, and thus more sensitively reflect the physiological state of the system [10]. Global metabolomic profiling, specifically, is a large-scale approach that detects all possible metabolites in a biological system, as opposed to targeted approaches that focus on a specific predetermined subset [11]. Thus, a metabolomic analysis of the SF provides a unique approach to evaluate mechanosensitive pathways that are stimulated during joint loading.
Mechanotransduction in synovial joints has been of particular interest in regard to OA. OA is the most common degenerative joint and is most associated with the deterioration of articular cartilage, and clinical studies demonstrate that exercise and muscle strengthening can reduce symptoms of OA [[12], [13], [14], [15], [16]]. Despite these empirical data, the mechanisms underlying the beneficial effects of exercise on joint health remain unclear.
While there is evidence that exercise has beneficial effects in the management of OA, no study to date has evaluated the effect of exercise on global joint metabolism. To take the first steps in understanding how exercise affects the joint metabolome, we examined metabolomic perturbations in SF following a single bout of nightly voluntary wheel running in young adult healthy male mice. Control sedentary animals were housed under standard cage conditions. Global metabolomic profiling of SF was conducted by untargeted LC-MS analysis. We aimed to identify specific mechanosensitive metabolites and mechanosensitive pathways in the SF to provide insight into the overall joint response to acute exercise. For this manuscript we use a broad definition of mechanosensitive to include metabolites induced either by direct mechanical loading of relevant joint tissues or indirectly through the systemic effects of exercise. We hypothesized that acute exercise would decrease inflammatory pathways and increase anabolic pathways associated with joint homeostasis.
2. Methods
2.1. Experimental design
Four separate litters of 14-week-old male C57BL/6J mice were obtained from a breeding colony at the Oklahoma Medical Research Foundation (OMRF). Mice from each litter were randomly assigned to the control or exercise groups. The control group (control: n = 6) was single-housed for one night in standard caging with a hut and bedding but no wheel. The exercised group (ex: n = 8) was single-housed in a standard cage that included bedding and a stainless-steel mouse running wheel (diameter = 11.5 cm, width = 5.2 cm, Mini Mitter, Bend, OR). Wheel running data were automatically collected via magnetic reed switch sensors that count the number of wheel revolutions per minute as previously described [17]. Mice were placed in their respective cages between 4:30–4:45PM and removed between 7:30–8:00AM the following day. All experiments were conducted within the OMRF vivarium, which provides temperature-controlled rooms maintained at 22 ± 3 °C on 14/10-h light/dark cycles (8:00PM off/6:00AM on) with ad libitum access to food and water. All litters of animals were tested within a 3-week period, and each litter contributed 1–2 animals per experimental group. Litters were tested on separate days so that no more than 4 animals were tested at a given time. Animals were euthanized between 8:00–9:30AM, alternating between control and exercise groups. Animals were euthanized under isoflurane anesthesia by cardiac puncture and exsanguination. All animal procedures were conducted in accordance with a protocol approved by the Institutional Animal Care and Use Committee at OMRF.
2.2. Synovial fluid collection
Immediately after euthanasia, SF was recovered from the knee joint using a calcium sodium alginate compound (CSAC) as previously described [18]. The patellar tendon was transected superior to the patella and slightly retracted distally to allow a 2 mm diameter biopsy punch of Melgisorb (Tendra, REF 250600; Goteborg, Sweden) wound dressing to be inserted into the joint space using fine forceps. The joint was manually articulated for 10 s to absorb the SF. The wound dressing was then carefully removed and placed in an Eppendorf tube. 35 μL of Alginate Lyase in H2O (1 unit/mL concentration; derived from Flavobacterium, Sigma-Aldrich A1603-100 MG) was added to the tube, vortexed, and digested at 34 °C for 30 min. The viscosity was lowered by adding 15 μL of 1.0 M sodium citrate (C6H5Na3O7) to chelate the Ca2+ ions. The samples were measured for total volume and then frozen at −80 °C. Frozen samples were shipped on dry ice to the June Lab at Montana State University and maintained at −80 °C until metabolite extraction and analysis.
2.3. Metabolite extraction
Metabolites were extracted from SF as previously described with slight modification [19,20]. Thawed SF samples were centrifuged at 4 °C at 500×g for 5 min to remove cells and debris. The supernatant was collected and vacuum concentrated for ∼2 h. The dried pellet was re-suspended in mass spectrometry grade 50:50 water: acetonitrile at −20 °C for 30 min to extract metabolites. The sample was then vortexed for 3 min and centrifuged at 16,100×g for 5 min at 4 °C. Proteins were precipitated by adding 250 μL of acetone, followed by 3 min of shaking and overnight refrigeration at 4 °C. The mixture was then centrifuged for 5 min at 16,100×g for 5 min and supernatant was collected and vacuum concentrated. The dried pellet was re-suspended in 50:50 water:acetonitrile.
2.4. Analysis by HPLC-MS
Metabolite extracts were analyzed for metabolomic profiling using an Agilent 1290 UPLC system (Agilent, Santa Clara, CA) coupled to an Agilent 6538 Q-TOF mass spectrometer (Agilent Santa Clara, CA) in positive mode with a resolution of ∼20,000 and accuracy of ∼5 ppm. Samples were run on a Cogent Diamond Hydride HILIC 150 × 2.1 mm column (MicroSolv, Eatontown, NJ) in normal phase using previously optimized gradient elution methods [19]. Total run time was 15 min including a 2-min wash prior to each sample.
3. Data processing
Raw data were converted from Agilent's proprietary file format (.d files) to mzXML files using Agilent MSConvert prior to being imported to MZMine 2.14 for data processing [21]. A noise level threshold of 1000 was applied. Retention times and mass-to-charge ratios (m/z) were normalized across spectra with a retention time tolerance of 0.25 min and m/z tolerance of 0.01 m/z or 30 ppm and peaks were aligned. Peaks were detected with a minimum time span of 0.1 min, minimum height of 1000, and m/z tolerance of 0.1 m/z or 30 ppm. Metabolite features with median intensity values of zero in both control and exercised groups were eliminated and any remaining zeroes were replaced with one-half the minimum peak intensity for further analysis [22]. For analysis, metabolite features were identified by their experimentally measured mass-to-charge (m/z) ratios. Common adducts formed during electrospray ionization include Na+ and H+ adducts. Mass-to-charge ratios were matched to putative metabolite identities using the metabolite database, METLIN (adducts: H+ and Na+; mass tolerance: 15 ppm; removed drugs and toxins) [23,24].
3.1. Metabolomic profiling and statistical analyses
Statistical analyses were performed in MetaboAnalyst [22] and MATLAB (Mathworks, Inc.). Raw data were normalized by the median, log transformed, standardized (mean-centered and divided by standard deviation), and filtered by standard deviation. Normalized data were used for all analyses unless otherwise noted, and data were checked to meet the relevant assumptions of the specific statistical tests. Kolmogorov-Smirnov (KS) tests were used to compare the median metabolite distributions of control and exercised datasets (a priori pks<0.05). To identify differentially expressed metabolite features, fold changes (FC) were calculated (on data prior to normalization to compare the absolute differences, which can be skewed by normalization). To preserve the magnitude of the differences, fold changes were calculated based on the raw mean metabolite intensities between the control and exercised groups. Data were then normalized to the median on a per-metabolite basis and Student's T-tests and volcano plot analysis were applied. Enrichment analysis of differentially expressed metabolite features was performed using the MS Peaks to Pathways mummichog application in MetaboAnalyst [22,25]. Mummichog matches all detected metabolite features to potential metabolites (mass tolerance: 0.1 ppm) in the global network of pathways and searches for local pathway enrichment to identify the pathway activity and most likely metabolite identifications. Mummichog reports the activity network as pathways and calculates an FDR-corrected p-value to reveal the most likely enriched pathways (FDR-corrected p-value<0.05).
Hierarchical clustering analysis (HCA), clustergrams, and partial least squares-discriminant analysis (PLS-DA) were used to visualize both sample-to-sample variation and overall metabolomic profiles of control and exercised SF. PLS-DA score plots include 95% confidence ellipses to assess separation between the metabolomic profiles of control and exercised mice based on the first 2 components [26]. Mechanosensitive metabolites were identified using the variable of importance projection (VIP) scores from PLS-DA, correlation coefficients, and volcano plot analysis. VIP scores indicated which metabolite features contributed the most to the variation between exercised and sedentary groups.
To identify strongly correlated metabolite features we calculated pointwise biserial correlation coefficients, rpb. These were calculated on a per-metabolite basis between the intensities of the control and exercise groups. These correlations are a measure of the effect size of the median metabolite intensities between controls and exercised [27]: correlation values greater than zero represent metabolites with higher intensities in the exercised group and correlation values less than zero represent metabolites with higher intensity in the control group. We visualized these correlations by plotting the empirical cumulative distribution.
Lastly, significantly up- and down-regulated metabolite features revealed by volcano plot analysis (greater than twofold change and p-value<0.05) were considered mechanosensitive metabolites. Mechanosensitive metabolites were putatively identified using the metabolite database, METLIN (adducts: H+ and Na+; mass tolerance: 15 ppm; removed drugs and toxins) [23,24].
4. Results
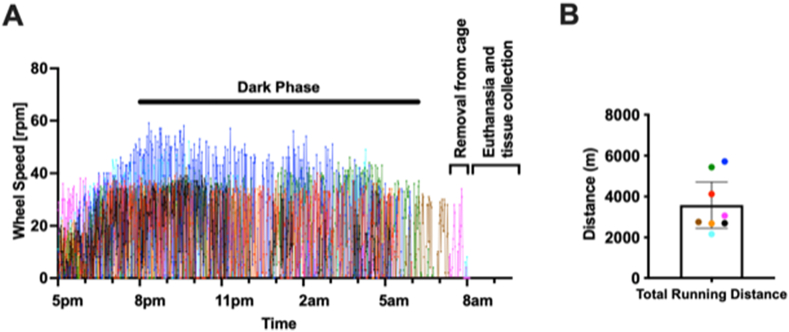
The objective of this study was to determine the whole-joint metabolic response to acute exercise by profiling changes in the synovial fluid. On average, Ex animals voluntarily ran 3,581 m between 5pm and 8am [95%CI: 2450–4,712 m], prior to SF collection (Fig. 1). Most animals greatly reduced their running activity when the room lights came on at 6am, although some animals continued low levels of running until removal (Fig. 1).
Fig. 1.
Acute exercise in mice. Mice selected for the exercise group (n = 8) were individually housed in cages equipped with a stainless-steal running wheel (diameter = 11.5 cm) between approximately 4:30pm to 7:30am the following morning. (A) Wheel speed in revolutions per minute (rpm) plotted against time. Each color denotes a separate animal, and dark phase refers to the time during which the room lights were turned off. (B) Total running distance in meters was calculated as the sum of the total revolutions from 5pm until removal from the cage multiplied by the wheel circumference. Each colored symbol refers to an individual animal as indicated in panel A. Value equals mean ± 95% CI.
4.1. Acute exercise altered global metabolomic profiles
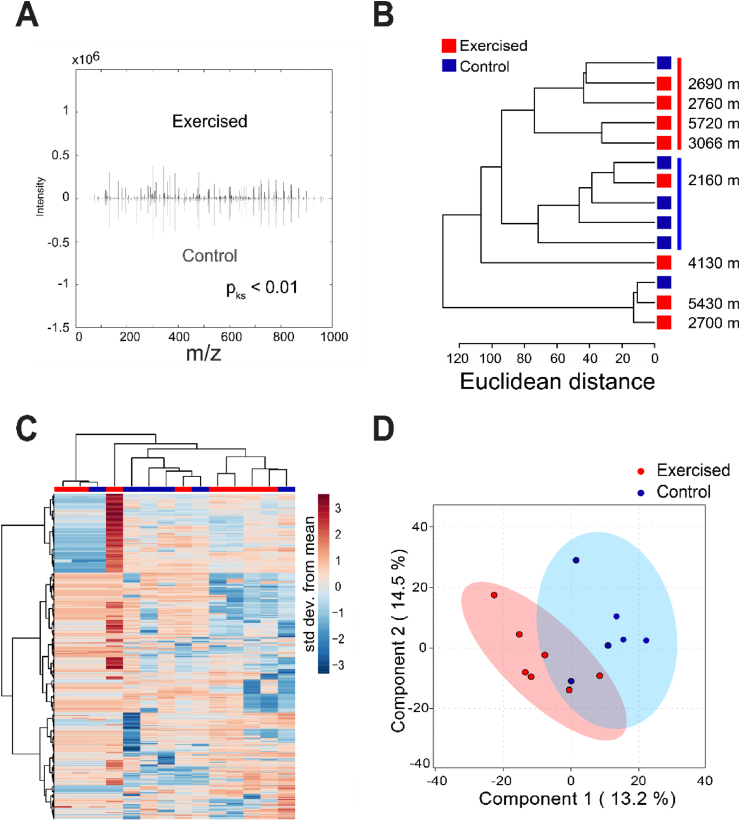
A total of 2812 metabolite features were detected in mouse SF in this study. These features were used to generate global metabolomic profiles for exercised and sedentary cohorts. These analyses revealed that acute exercised induced substantial metabolomic changes in the SF. The global metabolomic profile of acute exercised SF was significantly different than sedentary SF (pks<0.01; Fig. 2A). Unsupervised HCA assessed grouping of samples and calculated distances between clusters. This distinguished the majority of exercised samples from controls (Fig. 2B). However, HCA did not perfectly distinguish control from exercised samples with one sample of exercised SF falling within a cluster of four control samples. This exercised sample was from the mouse that ran the least of all mice in this study (Fig. 2B). Clustergram analysis found groups of co-regulated metabolites and inter-sample variation that likely contributes to the overall clustering. The distinct metabolomic profiles between exercised and sedentary controls further supports the SF as a mechanosensitive fluid, with a single bout of physical activity inducing metabolic changes in the SF.
Fig. 2.
Acute exercise results in distinct metabolomic profiles. (A) Acute exercise alters the cumulative metabolite distributions. Mirrored metabolite feature distributions of m/z values and corresponding intensities were plotted to illustrate any differences in distributions. KS test showed a statistically significant (pks<0.01) change in metabolite distributions comparing control to acute exercised SF. (B) Unsupervised hierarchical cluster analysis illustrates unsupervised clustering of most exercised (red line) and control samples (blue line). Tree distances are represented as Euclidean distances. Distances run by exercised mice shown on the right. Tree distances are represented as Euclidean distances. Red represents exercised SF and blue represents control SF. (C) Clustergram representation of all metabolite features detected shows overall metabolomic profiles of each sample. Clustering of samples is illustrated by the dendrogram on the top, and clustering of co-metabolites is illustrated by the dendrogram on the side. (D) Supervised PLSA-DA clearly discriminated between acute exercised and control samples. PLS-DA score plot illustrated separation between of exercised and control samples, with PC1 containing 13.2% of the variation and PC2 containing 14.5% of the variation.
A supervised clustering method, PLS-DA, was used to further examine the metabolomic profiles of acute exercised and control SF. PLS-DA clearly separated control from acute exercised SF samples with the first 2 components containing 27.7% of the variation between datasets (Fig. 2D). The 95% confidence ellipses show separation between exercised and control cohorts, with some overlap between cohort clusters (Fig. 2D). Taken together, KS-test, HCA, and PLS-DA suggest that acute exercise induced metabolic changes in the SF in comparison to unexercised controls, further supporting the SF as a fluid that captures changes associated with exercise.
4.2. Mechanosensitive metabolites
We investigated specific metabolite features associated with acute exercise in SF to gain insight into in vivo mechanotransduction in the joint. Variable Importance in Projection (VIP) in PLS-DA, correlation coefficient analysis, and volcano plots analysis were used to reveal mechanosensitive metabolites.
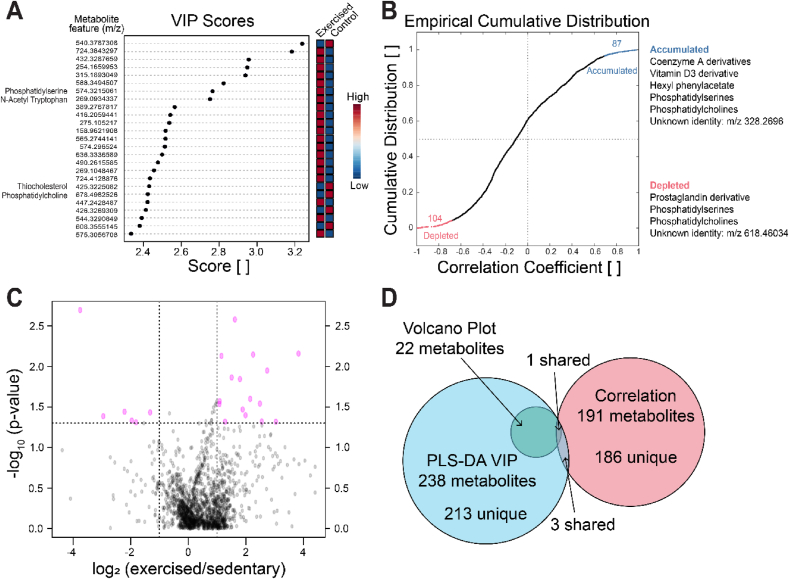
VIP in PLS-DA found metabolites that contribute the most to the separation between exercised and control cohorts in the PLS-DA score plot (Fig. 2D; Fig. 3A). Metabolite features with the highest scores have the greatest ability to classify samples into their respective cohorts. Phosphatidylcholine (PC), phosphatidylserine (PS), thiocholesterol, and tryptophan derivatives were some of the putatively identified mechanosensitive metabolites (Table 1, Supplemental Table 4). Interestingly, 20 of 25 metabolites with the highest VIP scores had metabolite intensities greater in exercised than control SF (Fig. 3A, Table 1) Supplemental Table 4. This indicates that the differences between exercised and sedentary control SF are mainly attributed to exercised-induced increases in metabolite expression as opposed to decreases.
Fig. 3.
Mechanosensitive metabolites in mouse synovial fluid. (A) PLS-DA VIP scores determined the top 25 mechanosensitive metabolite features plotted by their m/z value that contributed the most to the variation between datasets. The majority of metabolite features with high VIP scores were higher in exercised than controls. These features are matched to metabolite identities in Table 1. Select mechanosensitive metabolite features higher in exercised than control SF matched with metabolite identities N-acetyl tryptophan and phosphatidylserine. (B) Cumulative distribution of pointwise biserial correlations for metabolite features correlated to acute exercise. Features with a correlation coefficient greater than 0.7 or less than −0.7 were considered accumulated (blue) or depleted (red), respectively. The top 10 accumulated and top 10 depleted metabolite features were considered mechanosensitive metabolites and matched with metabolite identities in Table 2 (C) Volcano plot analysis shows metabolite features significantly upregulated or downregulated by exercise. Metabolite features in the upper right and left regions (dashed lines) have a p-value<0.05 and greater than twofold change. The significant metabolite features are matched to metabolite identities in Table 3 (D) Venn Diagram showing metabolites used in fold-change analysis. There was substantial overlap of metabolites from the volcano plot and VIP analysis. However, there was limited overlap between the metabolites used from the correlation analysis. Note m/z ratios (mass-to-charge) may match multiple metabolite identities depending on adduct formation (H+ or Na+) occurring during ionization.
Table 1.
Mechanosensitive metabolites identified by PLS-DA VIP scores. VIP scores in PLS-DA identified metabolite features that contributed the most to the variation between control and exercised datasets. The top 25 metabolite features were matched with potential metabolite identities using the metabolite database METLIN (adducts: H+, Na+; mass tolerance: 15 ppm). False positives and drugs were removed from compound matches. An uncondensed list of compound matches is shown in Supplemental Table 4. Mass-to-charge ratios may match multiple metabolite identities because of either isoptomers or differential adduct formation (H+ or Na+) during ionization. Columns: Metabolite identity-putative metabolite match between experimental data and Metlin database. Mass-mass of putative metabolite. PPM—difference between database and experimental metabolite in parts per million. Adduct—ion adduct of experimental metabolite feature. Feature—mass of experimental metabolite feature. VIP score—variable importance projection score for PLS-DA analysis. High/Low—arrows indicate if the expression increased or decreased with exercise.
| MECHANOSENSITIVE METABOLITE FEATURES | ||||||
|---|---|---|---|---|---|---|
| PLS-DA VIP | ||||||
| Metabolite Identity | Mass | PPM | Adduct | Feature | VIP score | High (↑)/Low (↓) with Ex |
| 540.3787308 | 3.2385 | ↓ | ||||
| 724.3843297 | 3.1837 | ↑ | ||||
| 432.3287659 | 2.9562 | ↑ | ||||
| 254.1659953 | 2.9487 | ↑ | ||||
| alpha-licanic acid | 292.2038 | 11 | Na | 315.1893049 | 2.9388 | ↑ |
| Oxo-phytodienoic Acid | 292.2038 | 11 | ||||
| Colnelenic acid | 292.2038 | 11 | ||||
| Octadecatrienoic acid derivatives | 292.2038 | 11 | ||||
| Etherolenic acid | 292.2038 | 11 | ||||
| Na | 588.3494507 | 2.8234 | ↑ | |||
| Phosphatidylserine | 573.3067 | 13 | H | 574.3215061 | 2.7651 | ↑ |
| Phenyalanyl-cysteine | 268.0882 | 7 | H | 269.0934337 | 2.7517 | ↑ |
| benzopyrene derivatives | 268.0888 | 9 | ||||
| 3-hydroxyphenoytoin | 268.0848 | 5 | ||||
| 5-(4-hydroxyphenyl-5-phenylhydantoin | 268.0848 | 5 | ||||
| 2,4-Imidazolidinedione | 268.0848 | 5 | ||||
| N-acetyl-d-tryptophan | 246.1004 | 14 | Na | |||
| Methionyl-prolnie/prolyl-methionine | 246.1038 | 1 | ||||
| 389.2767817 | 2.5654 | ↑ | ||||
| 416.2059441 | 2.5421 | ↑ | ||||
| (1xi,3S)-1,2,3,4-Tetrahydro-1-methyl-beta-carboline-1,3-dicarboxylic acid | 274.0954 | 9 | H | 275.105217 | 2.5367 | ↑ |
| p-(3,4-Dihydro-6-methoxy-2-naphthyl)phenol | 252.115 | 3 | Na | |||
| (1Z,4Z)-1,5-bis(4-hydroxyphenyl)-1,4-pentadiene | 252.115 | 3 | ||||
| 2-Phenylethyl 3-phenyl-2-propenoate | 252.115 | 3 | ||||
| Cinnamyl phenylacetate | 252.115 | 3 | ||||
| 158.9621908 | 2.5173 | ↑ | ||||
| Chaetoglobosin N | 542.2781 | 12 | Na | 565.2744141 | 2.5167 | ↑ |
| 574.296524 | 2.5135 | ↑ | ||||
| 636.3336589 | 2.4987 | ↑ | ||||
| 490.2615585 | 2.4765 | ↑ | ||||
| Idebenone metabolite (QS-4) | 268.0947 | 10 | H | 269.1048467 | 2.4558 | ↑ |
| diethyl 2,6-dimethyl-4-oxo-4h-pyran-3,5-dicarboxylate | 268.0947 | 10 | ||||
| 3,3′-diindolylmethane | 246.1157 | 0 | Na | |||
| Phosphatidylserine | 701.4268 | 4 | Na | 724.4128876 | 2.4345 | ↑ |
| Unoprostone isopropyl ester | 424.3189 | 8 | H | 425.3225082 | 2.4303 | ↓ |
| 4,4′-Diaponeurosporene | 402.3287 | 10 | Na | |||
| Vitamin D3 derivatives | 402.3298 | 8 | ||||
| Thiocholesterol | 402.332 | 2 | ||||
| Phosphatidylcholine | 677.4996 | 12 | H | 678.4982526 | 2.4231 | ↓ |
| clavulone derivatives | 446.2305 | 11 | H | 447.2428487 | 2.4219 | ↑ |
| 6B-Hydroxybudesonide | 446.2305 | 11 | ||||
| ethyl-1-(2,4,6-trihydroxy-3-isobutyrylphenyl)butyl]-4-cyclohexene-1,3-dione | 446.2304 | 11 | ||||
| 1-heptadecanoyl-sn-glycerol 3-phosphate | 424.259 | 11 | Na | |||
| Phosphatidic acid | 424.259 | 11 | ||||
| 426.3269309 | 2.4128 | ↓ | ||||
| 544.3290849 | 2.391 | ↑ | ||||
| 608.3555145 | 2.3803 | ↓ | ||||
| 2,3-diacetoxy-7,8-epoxy-24,29-dinor-1,3,5-friedelatriene-20-carboxylic acid | 13 | 13 | Na | 575.3056708 | 2.3351 | ↑ |
We expanded our analysis of PLS-DA VIP scores from the top 25 metabolite features to all features with a VIP score greater than 1.5 to investigate pathways differences. Discriminative metabolite features were involved in corticosteroid, prostaglandin, and ubiquinol biosynthesis as determined by enrichment analysis (Supplemental Table 3).
Mechanosensitive metabolites that had the greatest accumulation or depletion with acute exercise were identified by correlation coefficient analysis. Correlation analysis examined the effects of acute exercise on metabolite intensities in comparison to controls. There were 191 statistically significant metabolite features that correlated with acute exercise, 87 of which were positively correlated (accumulated) and 104 were negatively correlated (depleted, Fig. 3B). The strongest positively and negatively correlated metabolites were considered mechanosensitive metabolites (Table 2, Supplemental Table 5). Of those metabolite features identified as mechanosensitive, putative metabolite identity matches included coenzyme A derivatives, a vitamin D3 derivative, a prostaglandin derivative, PS, and PC (Fig. 3B, Table 2, Supplemental Table 5).
Table 2.
Mechanosensitive metabolites identified by pairwise biserial correlation coefficient analysis. Correlation coefficient analysis identified metabolite features that were accumulated or depleted after acute exercise. Strongly correlated features were matched with metabolite identities using METLIN (adducts: H+, Na+; mass tolerance: 15 ppm). False positives and drugs were removed from compound matches. An uncondensed list of compound matches is shown in Supplemental Table 5. Mass-to-charge ratios may match multiple metabolite identities because of either isoptomers or differential adduct formation (H+ or Na+) during ionization. Columns: Metabolite identity-putative metabolite match between experimental data and Metlin database. Mass-mass of putative metabolite. PPM—difference between database and experimental metabolite in parts per million. Adduct—ion adduct of experimental metabolite feature. Feature—mass of experimental metabolite feature. Coefficient—correlation coefficient. Accumulated/Depleted—indicates if the metabolite increased or decreased with exercise.
| MECHANOSENSITIVE METABOLITE FEATURES | ||||||
|---|---|---|---|---|---|---|
| Correlation Coefficient Analysis | ||||||
| Metabolite Identity | Mass | PPM | Adduct | Feature | rpb | P-value |
| 491.4027405 | −0.992 | 0.008 | ||||
| 618.4603464 | −0.981 | 0.019 | ||||
| Phosphatidylcholine | 591.39 | 9 | Na | 614.3850952 | −0.969 | 0.006 |
| 755.0263367 | −0.967 | 0.033 | ||||
| Phosphatidylserine | 511.3274 | 11 | Na | 534.3226522 | −0.963 | 0.002 |
| 928.7206607 | −0.960 | 0.040 | ||||
| 6-Monoacetyl morphine-3-O-glucuronide | 373.2406 | 0 | H | 374.2474714 | −0.959 | 0.002 |
| Androsta-5,16-dienol [17,16-b] quinolin-3beta-ol | 373.2406 | 0 | ||||
| Dodecylphosphocholine | 351.2538 | 11 | Na | |||
| 2,3-dinor-6-keto Prostaglandin F1α-d9 | 351.2607 | 6 | ||||
| 141.9570389 | −0.956 | 0.011 | ||||
| 491.8885193 | −0.954 | 0.046 | ||||
| 686.3826643 | −0.952 | 0.003 | ||||
| 250.1348 | 0.929 | 0.023 | ||||
| 342.2328 | 0.934 | 0.020 | ||||
| Vitamin D3 derivative | 522.3764 | 1 | H | 545.3847 | 0.944 | 0.056 |
| 697.9982 | 0.948 | 0.052 | ||||
| 328.2565 | 0.949 | 0.004 | ||||
| Phenylglyoxylyl-CoA | 899.1363 | 11 | H | 900.1329 | 0.961 | 0.039 |
| 3,4-didehydroadipyl-CoA semialdehyde | 877.152 | 9 | Na | |||
| 328.2696 | 0.966 | 0.007 | ||||
| Phosphatidyserine | 899.6615 | 9 | H | 900.6772 | 0.973 | 0.027 |
| Phosphatidylcholine | 877.6924 | 4 | Na | |||
| 359.2435 | 0.973 | 0.005 | ||||
| Hydroxypentobarbital | 242.1267 | 2 | H | 243.133316 | 0.976 | 0.024 |
| 2,6-di-tert-butylbenzoquinone | 220.1463 | 9 | Na | |||
| 3-Phenylpropyl isovalerate | 220.1463 | 9 | ||||
| 13-Nor-6-eremophilene-8,11-dione | 220.1463 | 9 | ||||
| Hexyl phenylacetate | 220.1463 | 9 | ||||
| 2-Phenylethyl hexanoate | 220.1463 | 9 | ||||
| Tetradecene-1,3-diyne-diol derivatives | 220.1463 | 9 | ||||
| 2-Methyl-1-phenyl-2-propanyl butyrate | 220.1463 | 9 | ||||
Volcano plot analysis showed mechanosensitive metabolite features significantly upregulated (accumulated) and downregulated (depleted) based on p-value (before FDR-correction) and magnitude of fold change (Fig. 3C). 16 metabolite features were significantly upregulated and 6 were significantly downregulated (p-value<0.05, greater than twofold change; Fig. 3C). These mechanosensitive metabolite features were matched to putative metabolite identities shown in Table 3 (Supplemental Table 6). Select mechanosensitive features significantly upregulated by exercise included fatty acids, cysteinyl-phenylalanine, and tryptophan. Select mechanosensitive features significantly downregulated by exercise included vitamin D3 derivatives and thiocholesterol (Fig. 3C, Table 3, Supplemental Table 6).
Table 3.
Mechanosensitive metabolites identified by volcano plot analysis. Mechanosensitive features upregulated or downregulated with exercise were identified by volcano plot analysis. Features with a greater than twofold change and p-value (prior to FDR correction) less than 0.05 were considered mechanosensitive metabolites. Metabolite identities were assigned via METLIN (adducts H+, Na+; mass tolerance: 15 ppm). False positives and drugs were removed from compound matches. An uncondensed list of compound matches is shown in Supplemental Table 6. Mass-to-charge ratios may match multiple metabolite identities because of either isoptomers or differential adduct formation (H+ or Na+) during ionization. Columns: Metabolite identity-putative metabolite match between experimental data and Metlin database. Mass-mass of putative metabolite. PPM—difference between database and experimental metabolite in parts per million. Adduct—ion adduct of experimental metabolite feature. Feature—mass of experimental metabolite feature. Fold Change—fold change of metabolite between exercise and control group. P-value—FDR-corrected p-value for comparing between exercise and control. Accumulated/Depleted—indicate if expression increased or decreased with exercise.
| MECHANOSENSITIVE METABOLITE FEATURES | |||||||
|---|---|---|---|---|---|---|---|
| Volcano Plot Analysis | |||||||
| Metabolite Identity | Mass | PPM | Adduct | Feature | Fold Change | P-value | Accumulated (↑)/Depleted (↓) |
| 540.3787 | 0.07398 | 0.002015 | ↓ | ||||
| 724.3843 | 3.0682 | 0.002631 | ↑ | ||||
| 432.3288 | 14.191 | 0.006931 | ↑ | ||||
| 254.1660 | 4.7594 | 0.007132 | ↑ | ||||
| alpha-licanic acid | 292.2038 | 11 | Na | 315.1893 | 2.2216 | 0.007407 | ↑ |
| Colnelenic acid | 292.2038 | 11 | |||||
| Etherolenic acid | 292.2038 | 11 | |||||
| Octadecatrienoic acid derivatives | 292.2038 | 11 | |||||
| Oxo-phytodienoic acid derviatives | 292.2038 | 11 | |||||
| 588.3495 | 6.6728 | 0.01123 | ↑ | ||||
| Phosphatidylserine | 573.3067 | 13 | H | 574.3215 | 2.8375 | 0.01366 | ↑ |
| 3-hydroxyphenytoin | 268.0848 | 5 | H | 269.0934 | 3.4668 | 0.01427 | ↑ |
| 2,4-Imidazolidinedione, 5-(7-oxabicyclo [4.1.0]-hepta-2,4-dien-3-yl)-5-phenyl- | 268.0848 | 5 | |||||
| 2,3-Dihydroxycarbamazepine | 268.0848 | 5 | |||||
| Benzo [a]pyrene derivatives | 268.0888 | 9 | |||||
| Cysteinyl-phenylalanine | 268.0882 | 7 | |||||
| Phenylalanyl-cysteine | 268.0882 | 7 | |||||
| N-acetyl-tryptophan | 246.1004 | 14 | Na | ||||
| methionyl-proline | 246.1038 | 1 | |||||
| prolyl-methionine | 246.1038 | 1 | |||||
| 389.2768 | 4.4307 | 0.02512 | ↑ | ||||
| 416.2059 | 2.1272 | 0.02681 | ↑ | ||||
| 158.9622 | 5.6077 | 0.02872 | ↑ | ||||
| 574.2965 | 2.1147 | 0.02902 | ↑ | ||||
| idebenone metabolite (QS-4) | 268.0947 | 10 | H | 269.1048 | 3.6993 | 0.03387 | ↑ |
| diethyl 2,6-dimethyl-4-oxo-4h-pyran-3,5-dicarboxylate | 268.0947 | 10 | |||||
| 3,3′-diindolylmethane | 246.1157 | 0 | Na | ||||
| Unoprostone isopropyl ester | 424.3189 | 8 | H | 425.3225 | 0.2154 | 0.03620 | ↓ |
| 4,4′-Diaponeurosporene | 402.3287 | 10 | Na | ||||
| fluorovitamin D3/fluorocholecalciferol derivatives | 402.3298 | 8 | |||||
| 25-Fluorovitamin D3 | 402.3298 | 8 | |||||
| Thiocholesterol | 402.3298 | 8 | |||||
| Phosphatidylcholine | 677.4996 | 12 | H | 678.4983 | 0.3988 | 0.03687 | ↓ |
| Phosphatidylethanolamine | 677.4995 | 12 | |||||
| 544.3291 | 3.9614 | 0.04001 | ↑ | ||||
| 608.3555 | 0.1292 | 0.04109 | ↓ | ||||
| 598.4739 | 0.2565 | 0.04653 | ↓ | ||||
| 291.2023 | 5.8616 | 0.04775 | ↑ | ||||
| 716.9248 | 2.4243 | 0.04794 | ↑ | ||||
| 782.4581 | 8.2303 | 0.04807 | ↑ | ||||
| Phosphatidylcholine | 523.4002 | 6 | Na | 546.3927 | 0.2842 | 0.04888 | ↓ |
| 2-o-methyl PAF C18 | 523.4002 | 6 | |||||
Many mechanosensitive metabolite features overlapped among PLS-DA and volcano plot analyses, but none of these overlapped with the metabolites detected in the correlation analyses (Fig. 3D) A select few include phosphatidylserines, phosphatidylcholines, and vitamin D3 derivatives, which were identified in all three analyses. Thiocholesterol and a variety of fatty acids (i.e. octadecatrienoic acid derivatives) were identified by PLS-DA and volcano plot analysis. Taken together, these results suggest that phospholipids, vitamin D derivatives, and fatty acids are prominent mechanosensitive metabolites in the SF in response to acute exercise.
4.3. Mechanosensitive pathways
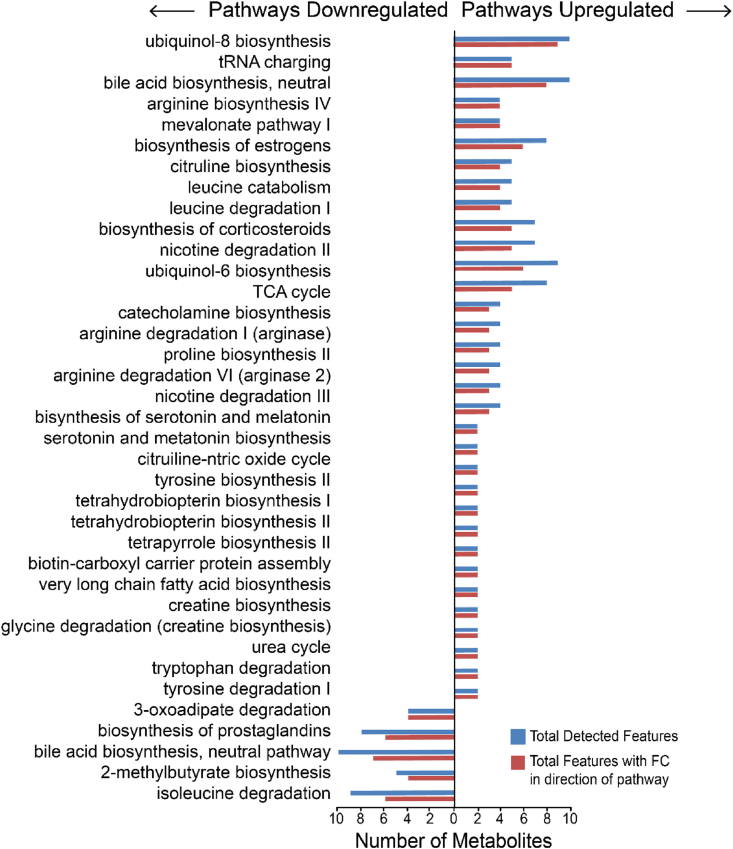
To generate a metabolomic phenotype of SF following acute exercise, the computational algorithm, mummichog, was employed to predict functional pathway activity from the detected metabolite features [25]. Pathway analysis of metabolite features with higher intensities (FC > 1) after acute exercise mapped to 33 pathways (FDR-corrected p-value<0.05) including ubiquinol (antioxidant) biosynthesis, tRNA charging, amino acid biosynthesis (arginine, proline, leucine, and tyrosine), citrulline and nitric oxide metabolism, the urea cycle, creatine biosynthesis, catecholamine biosynthesis, and fatty acid biosynthesis. Metabolite features with lower intensities after exercise (FC < 1) mapped to 5 significant pathways (FDR-corrected p-value<0.05) including 3-oxoadipate degradation, biosynthesis of prostaglandins (inflammatory mediator), bile acid biosynthesis, 2-methylbutyrate biosynthesis, isoleucine degradation, and ketone oxidation (Fig. 4, Supplemental Tables 1-2). Taken together, these pathways show that the synovial fluid of exercised mice captures metabolites related to amino acid synthesis, as well as both pro- and anti-inflammatory pathways that indicate a complex response to exercise.
Fig. 4.
Acute exercise alters metabolic pathways in mouse synovial fluid. Enrichment analysis of metabolite features upregulated (right side) or downregulated (left side) following acute exercise identified significantly different biochemical pathways (FDR-corrected p-value<0.05). Blue bars represent all metabolite features detected in the dataset. Red bars represent metabolite features that were upregulated or downregulated corresponding to the directionality of the pathway (i.e. Leucine catabolim: a total of 9 metabolites in this pathway were detected but only 6 had a fold change less than one corresponding to the directionality of this pathway—downregulated)). Supplemental Tables 1 and 2 show the full lists of perturbed pathways.
5. Discussion
To our knowledge, this is the first description of global metabolomic changes measured in SF after acute exercise. We measured changes in joint metabolism following acute exercise using global metabolomic profiling of SF from mice undergoing one night of voluntary wheel running. We detected a total of 2812 metabolite features in mouse SF. Multiple analyses (i.e. KS test, PLS-DA, and HCA) showed distinct metabolomic profiles between exercised and control SF samples. These results demonstrate that SF captures the response to exercise and associated mechanical loads. In the SF, changes in joint metabolism occur within hours of a diurnal bout of increased physical activity. Importantly, the SF likely captures changes in metabolites produced by synovial joint mechanosensitive cells as well as systemic exercise responses that diffuse into the SF.
Mechanosensitive metabolites were identified using VIP scores from PLS-DA, correlation coefficient analysis, and volcano plot analysis. Mechanosensitive metabolites included coenzyme A derivatives, prostaglandin derivatives, phospholipid species, tryptophan, methionine, vitamin D3, fatty acids, and thiocholesterol. Enrichment analysis identified many pathways previously linked to exercise including amino acid metabolism, inflammatory pathways, the citrulline-nitric oxide cycle, catecholamine biosynthesis, ubiquinol biosynthesis, and phospholipid metabolism.
Global metabolomic profiling detects small-molecules including substrates, co-factors, and other cytosolic molecules to generate a high-dimensional phenotype of a biological system. SF metabolomic profiles describe a comprehensive whole-joint phenotype generated by molecules potentially originating from multiple cell types across the joint [8]. The present data provide a biochemical profile of mechanosensitive metabolites involved in the overall joint response to a single night of wheel-running exercise in mice. These data provide an initial view of how exercise promotes healthy joint metabolism and how exercise therapy may function mechanistically as an intervention for OA joints to alleviate OA symptoms. It is important to recognize, however, that the results from a single exercise bout may not reflect the changes that occur following chronic, repeated bouts of exercise.
The metabolic phenotype generated in this study sheds light on the effects of acute exercise and its contribution to maintaining overall joint health. Acute exercise upregulated amino acid metabolism (arginine, proline, leucine, and tyrosine) in mouse SF. The SF is in direct contact with the articular cartilage—a known mechanosensitive tissue that relies on mechanical stimuli to maintain a healthy matrix [1,2]. The articular cartilage is an avascular tissue so chondrocytes rely on the SF for nutrients. Previous studies have found increased amino acid metabolism in primary chondrocytes in response to short-duration mechanical stimuli [[28], [29], [30]]. The results herein suggest that the SF may support the maintenance of the cartilage extracellular matrix by replenishing amino acids to the chondrocytes in response to mechanical stimuli.
Acute exercise, defined here as one night of voluntary wheel running, stimulated both inflammatory and anti-inflammatory pathways in mouse SF. Prostaglandin biosynthesis was downregulated after exercise, and a prostaglandin derivative was identified as a significantly depleted mechanosensitive metabolite. Given the pro-inflammatory role of prostaglandins, the observed decrease in the intensity of metabolites involved in prostaglandin biosynthesis may be an anti-inflammatory effect of acute exercise. Other inflammation-related pathways altered with acute exercise included citrulline-nitric oxide cycle and catecholamine biosynthesis. Nitric oxide regulates blood flow and plays an anti-inflammatory role under physiologically normal conditions, and catecholamines can both elicit and attenuate inflammatory responses during exercise [31,32]. These results suggest that acute exercise may stimulate both inflammatory and immunosuppressive effects, consistent with previous findings [31,33]. The emerging paradigm of OA research incorporates several biological processes beyond mechanical wear including inflammation in the synovium, articular cartilage, and SF [34,35]. Inflammation is also associated with OA risk factors including joint trauma, age, and obesity [36]. This may partially explain the importance of exercise in managing inflammation-associated OA pain and damage in the joint.
Ubiquinol, an endogenous antioxidant, was significantly upregulated after acute exercise. Recent evidence suggests that oxidative stress in the joint damages the surrounding tissue and can lead to increased OA severity [37,38]. Regulation of oxidative stress in chondrocytes has been of particular interest for therapeutic targets to slow OA progression [38]. Further studies are needed using an OA mouse model to determine if acute exercise reduces oxidative stress-associated damage to the joint by upregulating antioxidants, such as ubiquinol. This may provide a mechanistic explanation for the use of exercise as a therapeutic intervention to manage OA.
Acute exercise significantly altered a variety of phospholipids. PC, PS, and PA were identified as mechanosensitive metabolites, and fatty acid biosynthesis was significantly upregulated after acute exercise. These data are from metabolite features detected in the synovial fluid, so cytosolic responses could differ. The mechanical function of SF is to lubricate articular cartilage surfaces. The major components of SF that contribute to its boundary lubrication function include lubricin, hyaluronan, and membrane phospholipids [39]. Previous studies show that altered composition and concentration of lubricating agents in SF are associated with damaged articular cartilage surfaces in both OA and RA [40]. Specifically, the composition of phospholipid species and their concentrations are altered in OA [41,42]. Exercise may play a role in maintaining the composition and concentration of SF phospholipids to support the SF's function as a boundary lubricant. Prior targeted studies have examined the plasma and serum metabolomic response to exercise [[43], [44], [45]]. While there are many similarities between these studies, one differences is that Nayor et al. find evidence for exercise-induced lipolysis in the circulation [43]. In contrast, this study found evidence for fatty acid biosynthesis within the synovial fluid after exercise. Exercise is known to induce substantial changes in lipid metabolism [45], and further studies involving different durations and intensities of exercise, as well as paired serum and synovial fluid analysis, are needed to determine if the observed fatty acid biosynthesis is unique to synovial fluid.
While this study illustrates important metabolomic responses to acute exercise, some limitations apply. It is likely that some of the identified mechanosensitive metabolites are involved in the systemic effects of exercise derived from the circulation. Future studies involving paired serum and SF analyses are required to discriminate these two categories of metabolites. Further, the selection of metabolites using arbitrary thresholds (fold change >2, VIP scores >1.5, correlation >0.7 or < 0.7) may affect the pathway analysis outcomes. PLS-DA includes the risk of overfitting which may also affect these results. Global metabolomic profiling by LC-MS is beneficial for quantifying the physiological state of a biological system, but it cannot identify all metabolite features due to the lack of comprehensive metabolite databases and standards libraries. Future studies will employ additional targeted metabolomics methods to confirm putative identities of metabolites in pathways of interest. Conducting this study in mice provides an additional limitation as the small size of the knee joint provides a limited volume of synovial fluid that is difficult to collect and accurately quantify. This difficulty likely increases the variance of metabolite measurements. Additionally, despite the physiologic benefits of using voluntary wheel running as an exercise stimulus, the dose and intensity of exercise was variable among animals, which also likely contributed to increased variability. Finally, because the mice of this study were young adult aged (14 weeks), these results may not apply to osteoarthritic joints in older patients.
Despite these limitations, this study holds important findings for joint mechanobiology. Global metabolomic profiling can discriminate between acute exercised and control SF, thereby identifying mechanosensitive metabolites and pathways. The results of this study provide several avenues to further explore the beneficial effects of acute exercise on joint metabolism. Specifically, this study suggests that acute exercise stimulates both pro-inflammatory and anti-inflammatory metabolic pathways. Chronic exercise conditions may reveal more beneficial effects on joint metabolism and inflammation. These findings also suggest that exercise may promote joint homeostasis by facilitating amino acid metabolism for protein synthesis and increasing the production of antioxidants to reduce oxidative-stress in joint tissues. This study provides a metabolic phenotype for comparison for future studies that investigate the impact of acute exercise in an OA joint.
Acknowledgements
The authors acknowledge funding provided by NSF (CMMI 1554708), NIH (R01AR073964, P30GM114731, R03AR066828), the M.J. Murdock Charitable Trust (FSU-2017207), and the Department of Veterans Affairs (I01BX004666, I01BX004882). We would like to acknowledge the Mass Spectrometry CORE Facility at Montana State University for their assistance with mass spectrometry and metabolomics. Funding for the Mass Spectrometry Facility used in this publication was made possible in part by the MJ Murdock Charitable Trust, the National Institute of General Medical Sciences of the National Institutes of Health under Award Number P20GM103474, and the Office of the VPREDGE at MSU. The funding sources were not involved in this study. Finally, we would also like to acknowledge Dr. Erik Adams his feedback on clinical relevance, and Dr. Mark Greenwood for his input regarding statistical analyses.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ocarto.2021.100228.
Contributor Information
Timothy M. Griffin, Email: Tim-Griffin@omrf.org.
Ronald K. June, Email: rjune@montana.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sophia Fox A.J., Bedi A., Rodeo S.A. The basic science of articular cartilage: structure, composition, and function. Sport Health. 2009;1:461–468. doi: 10.1177/1941738109350438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buckwalter J.A., Mankin H.J. Articular cartilage: tissue design and chondrocyte-matrix interactions. Instr. Course Lect. 1998;47:477–486. [PubMed] [Google Scholar]
- 3.Kurz B., Jin M., Patwari P., Cheng D.M., Lark M.W., Grodzinsky A.J. Biosynthetic response and mechanical properties of articular cartilage after injurious compression. J. Orthop. Res. 2001;19:1140–1146. doi: 10.1016/S0736-0266(01)00033-X. [DOI] [PubMed] [Google Scholar]
- 4.Patwari P., Fay J., Cook M.N., Badger A.M., Kerin A.J., Lark M.W., et al. In vitro models for investigation of the effects of acute mechanical injury on cartilage. Clin. Orthop. Relat. Res. 2001:S61–S71. doi: 10.1097/00003086-200110001-00007. [DOI] [PubMed] [Google Scholar]
- 5.Torzilli P.A., Grigiene R., Borrelli J., Jr., Helfet D.L. Effect of impact load on articular cartilage: cell metabolism and viability, and matrix water content. J. Biomech. Eng. 1999;121:433–441. doi: 10.1115/1.2835070. [DOI] [PubMed] [Google Scholar]
- 6.Liedert A, Kaspar D, Augat P, Ignatius A, Claes L. Mechanobiology of bone tissue and bone cells. In: Mechanosensitivity in Cells and Tissues, Kamkin A, Kiseleva I Eds. Moscow2005. [PubMed]
- 7.Cowman M.K., Schmidt T.A., Raghavan P., Stecco A. Viscoelastic properties of hyaluronan in physiological conditions. F1000Res. 2015;4:622. doi: 10.12688/f1000research.6885.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamer T.M. Hyaluronan and synovial joint: function, distribution and healing. Interdiscipl. Toxicol. 2013;6:111–125. doi: 10.2478/intox-2013-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams S.B., Jr., Setton L.A., Nettles D.L. The role of metabolomics in osteoarthritis research. J. Am. Acad. Orthop. Surg. 2013;21:63–64. doi: 10.5435/JAAOS-21-01-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollywood K., Brison D.R., Goodacre R. Metabolomics: current technologies and future trends. Proteomics. 2006;6:4716–4723. doi: 10.1002/pmic.200600106. [DOI] [PubMed] [Google Scholar]
- 11.Roessner U., Bowne J. What is metabolomics all about? Biotechniques. 2009;46:363–365. doi: 10.2144/000113133. [DOI] [PubMed] [Google Scholar]
- 12.Bennell K.L., Hinman R.S., Metcalf B.R., Buchbinder R., McConnell J., McColl G., et al. Efficacy of physiotherapy management of knee joint osteoarthritis: a randomised, double blind, placebo controlled trial. Ann. Rheum. Dis. 2005;64:906–912. doi: 10.1136/ard.2004.026526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ettinger W.H., Jr., Burns R., Messier S.P., Applegate W., Rejeski W.J., Morgan T., et al. A randomized trial comparing aerobic exercise and resistance exercise with a health education program in older adults with knee osteoarthritis. The Fitness Arthritis and Seniors Trial (FAST) J. Am. Med. Assoc. 1997;277:25–31. [PubMed] [Google Scholar]
- 14.Griffin T.M., Huebner J.L., Kraus V.B., Yan Z., Guilak F. Induction of osteoarthritis and metabolic inflammation by a very high-fat diet in mice: effects of short-term exercise. Arthritis Rheum. 2012;64:443–453. doi: 10.1002/art.33332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter D.J., Eckstein F. Exercise and osteoarthritis. J. Anat. 2009;214:197–207. doi: 10.1111/j.1469-7580.2008.01013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Runhaar J., Beavers D.P., Miller G.D., Nicklas B.J., Loeser R.F., Bierma-Zeinstra S., et al. Inflammatory cytokines mediate the effects of diet and exercise on pain and function in knee osteoarthritis independent of BMI. Osteoarthritis Cartilage. 2019;27:1118–1123. doi: 10.1016/j.joca.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Griffin T.M., Batushansky A., Hudson J., Lopes E.B.P. Correlation network analysis shows divergent effects of a long-term, high-fat diet and exercise on early stage osteoarthritis phenotypes in mice. J Sport Health Sci. 2020;9:119–131. doi: 10.1016/j.jshs.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seifer D.R., Furman B.D., Guilak F., Olson S.A., Brooks S.C., 3rd, Kraus V.B. Novel synovial fluid recovery method allows for quantification of a marker of arthritis in mice. Osteoarthritis Cartilage. 2008;16:1532–1538. doi: 10.1016/j.joca.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlson A.K., Rawle R.A., Adams E., Greenwood M.C., Bothner B., June R.K. Application of global metabolomic profiling of synovial fluid for osteoarthritis biomarkers. Biochem. Biophys. Res. Commun. 2018;499(2):182–188. doi: 10.1016/j.bbrc.2018.03.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S., Hwang J., Xuan J., Jung Y.H., Cha H.S., Kim K.H. Global metabolite profiling of synovial fluid for the specific diagnosis of rheumatoid arthritis from other inflammatory arthritis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pluskal T., Castillo S., Villar-Briones A., Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinf. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia J., Wishart D.S. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinformatics. 2016;55(14 10 11) doi: 10.1002/cpbi.11. 14 10 91. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Z.J., Schultz A.W., Wang J., Johnson C.H., Yannone S.M., Patti G.J., et al. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Protoc. 2013;8:451–460. doi: 10.1038/nprot.2013.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guijas C., Montenegro-Burke J.R., Domingo-Almenara X., Palermo A., Warth B., Hermann G., et al. METLIN: a technology platform for identifying knowns and unknowns. Anal. Chem. 2018;90:3156–3164. doi: 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S., Park Y., Duraisingham S., Strobel F.H., Khan N., Soltow Q.A., et al. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Worley B., Halouska S., Powers R. Utilities for quantifying separation in PCA/PLS-DA scores plots. Anal. Biochem. 2013;433:102–104. doi: 10.1016/j.ab.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornbrot D. Point biserial correlation. In: Wiley StatsRef: Statistics Reference Online.
- 28.McCutchen C.N., Zignego D.L., June R.K. Metabolic responses induced by compression of chondrocytes in variable-stiffness microenvironments. J. Biomech. 2017;64:49–58. doi: 10.1016/j.jbiomech.2017.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salinas D., Minor C.A., Carlson R.P., McCutchen C.N., Mumey B.M., June R.K. Combining targeted metabolomic data with a model of glucose metabolism: toward progress in chondrocyte mechanotransduction. PLoS One. 2017;12 doi: 10.1371/journal.pone.0168326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zignego D.L., Hilmer J.K., June R.K. Mechanotransduction in primary human osteoarthritic chondrocytes is mediated by metabolism of energy, lipids, and amino acids. J. Biomech. 2015;48:4253–4261. doi: 10.1016/j.jbiomech.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woods J.A., Wilund K.R., Martin S.A., Kistler B.M. Exercise, inflammation and aging. Aging Dis. 2012;3:130–140. [PMC free article] [PubMed] [Google Scholar]
- 32.Zouhal H., Jacob C., Delamarche P., Gratas-Delamarche A. Catecholamines and the effects of exercise, training and gender. Sports Med. 2008;38:401–423. doi: 10.2165/00007256-200838050-00004. [DOI] [PubMed] [Google Scholar]
- 33.Pedersen B.K., Hoffman-Goetz L. Exercise and the immune system: regulation, integration, and adaptation. Physiol. Rev. 2000;80:1055–1081. doi: 10.1152/physrev.2000.80.3.1055. [DOI] [PubMed] [Google Scholar]
- 34.Griffin T.M., Guilak F. The role of mechanical loading in the onset and progression of osteoarthritis. Exerc. Sport Sci. Rev. 2005;33:195–200. doi: 10.1097/00003677-200510000-00008. [DOI] [PubMed] [Google Scholar]
- 35.Sohn D.H., Sokolove J., Sharpe O., Erhart J.C., Chandra P.E., Lahey L.J., et al. Plasma proteins present in osteoarthritic synovial fluid can stimulate cytokine production via Toll-like receptor 4. Arthritis Res. Ther. 2012;14:R7. doi: 10.1186/ar3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldring M.B., Otero M. Inflammation in osteoarthritis. Curr. Opin. Rheumatol. 2011;23:471–478. doi: 10.1097/BOR.0b013e328349c2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maneesh M., Jayalekshmi H., Suma T., Chatterjee S., Chakrabarti A., Singh T.A. Evidence for oxidative stress in osteoarthritis. Indian J. Clin. Biochem. 2005;20:129–130. doi: 10.1007/BF02893057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poulet B., Beier F. Targeting oxidative stress to reduce osteoarthritis. Arthritis Res. Ther. 2016;18:32. doi: 10.1186/s13075-015-0908-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kosinska M.K., Liebisch G., Lochnit G., Wilhelm J., Klein H., Kaesser U., et al. Sphingolipids in human synovial fluid - a lipidomic study. PLoS One. 2014;9 doi: 10.1371/journal.pone.0091769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elsaid K.A., Jay G.D., Warman M.L., Rhee D.K., Chichester C.O. Association of articular cartilage degradation and loss of boundary-lubricating ability of synovial fluid following injury and inflammatory arthritis. Arthritis Rheum. 2005;52:1746–1755. doi: 10.1002/art.21038. [DOI] [PubMed] [Google Scholar]
- 41.Hills B.A. Surface-active phospholipid: a Pandora's box of clinical applications. Part II. Barrier and lubricating properties. Intern. Med. J. 2002;32:242–251. doi: 10.1046/j.1445-5994.2002.00201.x. [DOI] [PubMed] [Google Scholar]
- 42.Hills B.A., Crawford R.W. Normal and prosthetic synovial joints are lubricated by surface-active phospholipid: a hypothesis. J. Arthroplasty. 2003;18:499–505. doi: 10.1016/s0883-5403(03)00072-x. [DOI] [PubMed] [Google Scholar]
- 43.Nayor M., Shah R.V., Miller P.E., Blodgett J.B., Tanguay M., Pico A.R., et al. Metabolic architecture of acute exercise response in middle-aged adults in the community. Circulation. 2020;142:1905–1924. doi: 10.1161/CIRCULATIONAHA.120.050281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alzharani M.A., Alshuwaier G.O., Aljaloud K.S., Al-Tannak N.F., Watson D.G. Metabolomics profiling of plasma, urine and saliva after short term training in young professional football players in Saudi Arabia. Sci. Rep. 2020;10:19759. doi: 10.1038/s41598-020-75755-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanford J.A., Nogiec C.D., Lindholm M.E., Adkins J.N., Amar D., Dasari S., et al. Molecular transducers of physical activity consortium (MoTrPAC): mapping the dynamic responses to exercise. Cell. 2020;181:1464–1474. doi: 10.1016/j.cell.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.