Abstract
Amyotrophic lateral sclerosis (ALS), a devastating progressive neurodegenerative disease, has no effective treatment. Recent evidence supports a strong metabolic component in ALS pathogenesis. Indeed, metabolic abnormalities in ALS correlate to disease susceptibility and progression, raising additional therapeutic targets against ALS. Ozone (O3), a natural bioactive molecule, has been shown to elicit beneficial effects to reduce metabolic disturbances and improved motor behavior in TDP-43A315T mice. However, it is fundamental to determine the mechanism through which O3 acts in ALS. To characterize the association between O3 exposure and disease-associated weight loss in ALS, we assessed the mRNA and protein expression profile of molecular pathways with a main role in the regulation of the metabolic homeostasis on the hypothalamus and the brown adipose tissue (BAT) at the disease end-stage, in TDP-43A315T mice compared to age-matched WT littermates. In addition, the impact of O3 exposure on the faecal bacterial community diversity, by Illumina sequencing, and on the neuromuscular junctions (NMJs), by confocal imaging, were analysed. Our findings suggest the effectiveness of O3 exposure to induce metabolic effects in the hypothalamus and BAT of TDP-43A315T mice and could be a new complementary non-pharmacological approach for ALS therapy.
Subject terms: Molecular biology, Neuroscience
Introduction
Amyotrophic Lateral Sclerosis (ALS) is considered the third most common neurodegenerative disease worldwide1, and is becoming a disease with a significant impact and much community awareness for many countries across the globe. Over 60% of patients die within 3–5 years of diagnosis. There is no cure for ALS. Although much effort has been made in the past two decades to understand the complexity and heterogeneity of ALS, the mechanisms by which progressive degeneration and death of motor neurons occur have not yet been elucidated. Indeed, both genetic and risk factors from gene-environment interactions, including metabolic alterations, contribute to the disease progression and pathogenesis.
Not until a couple of years ago that both clinical and epidemiological studies provided for the first time evidence that ozone (O3), a highly oxidative gas, may be a potential novel therapeutic strategy in the treatment of degenerative disorders2–5. In this context, clinical trials evidenced the effectiveness of O3 therapy in the treatment of multiple sclerosis2,3,5, due to its potential to induce controlled oxidative stress6, through a mechanism of action involving its interaction with the nuclear factor erythroid-derived 2–like 2 (Nrf2)3, a key regulator of inducible antioxidant responses7. Additionally, clinical data indicated the immunomodulatory effect of O3 therapy8, and animal work demonstrated the effectiveness of this gas to activate the hypothalamus–pituitary-adrenal (HPA) axis9, which has the potential to alter the composition of the gut microbiota and increase gastrointestinal permeability10,11, suggesting the potential role of O3 to induce controlled metabolic effects.
We previously reported that repeated exposure to O3 significantly reduce metabolic disturbances in the mouse model of ALS based on TDP-43 proteinopathy (TDP-43A315T mice)12. However, the underlying molecular changes that link O3 exposures to disease-associated weight loss in this transgenic (Tg) line were not determined. In the current study, we have examined the relationships between O3 exposure and induced metabolic effects in TDP-43A315T mice, by mean of the study of the molecular pathways with a main role in the regulation of the metabolic homeostasis in TDP-43A315T mice. To our knowledge, this work represents the first insight regarding the potential use of this gas as a potential complementary approach to improve metabolic and endocrine response in ALS.
Methods
Animals
Cohorts of male Tg Prp-TDP43A315T mice (Strain No. 010700, United States)12 and age-matched WT littermates (non-Tg C57Bl6/J mice) were used in this study. This Tg mouse model of ALS13 expresses human transactive response DNA-binding protein 43 (TDP-43) with an A315T mutation (hTDP-43A315T)12. Animals expressing the hTDP-43 transgene were confirmed via PCR following the protocol described14. Animals were group-housed under standard housing conditions with a 12 h light–dark cycle, and food and water ad libitum. The maintenance and use of mice and all experimental procedures were approved by the Animal Ethics Committee of the Hospital Nacional de Parapléjicos (Approval no. 26/OH 2018) accordingly with the Spanish Guidelines for the Care and Use of Animals for Scientific Purposes in compliance with ARRIVE guidelines (https://arriveguidelines.org). All analyses were conducted by personnel blinded to the animal genotype.
Experimental design and O3 exposure
Mice at 42 days of age (asymptomatic stage of disease) underwent two consecutive weeks of O3 vs. FA (Filtered Air) exposure following the protocol described13,15. Each genotype of mice was divided into two subgroups (n = 3- 7 TDP-43A315T mice/subgroup and n = 5–8 WT mice/subgroup) according to the exposure. O3 was generated from pure O2 with a BTM 802 N generator and distributed in a Plexiglas chamber (50 × 35 × 35 cm) together with zero air at a total flow of 15 L/min. The O3 concentration of the ambient air in the chamber was kept constant at 0.25 ppm and was continuously monitored by an Environment O342M analyser (Envea, France). This concentration is higher than both EPA National Ambient Air Quality Standard (NAAQS) (0.075 ppm) and the recommended OMS guideline value (0.05 ppm), but mice are routinely recognized as less susceptible to O3 exposure than humans due to obligate nose breathing and other intrinsic factors16. FA was obtained by filtering regular air through activated charcoal to reduce O3 concentration to a minimum of (< 0.02 ppm). During the 15 consecutive days of exposure (4 h/day), mice had food and water ad libitum, and their general health were monitored closely in terms of their mobility or level of activity. No difference in body weight gain between groups (O3 group vs. FA group) was observed. Then, to monitor disease onset (defined as the last day of individual peak body weight before a gradual loss occurs) all mice were weighed and assessed three times per week, after which they were checked daily until the end-stage of disease, defined as when weight is 20% below the initial weight on three consecutive days (~ 95–100 ± 2 days). Then the mice were euthanized.
Fecal collection, DNA extraction and Illumina sequencing and analysis
Two to three freshly evacuated fecal pellets were collected from each mouse in the morning, prior to the end-stage of disease. Briefly, mice temporarily were placed individually into empty autoclaved cages and allowed to defecate. Fecal pellets were collected aseptically from each mouse and placed into a sterile tube. Faecal samples were stored at − 80 °C prior to DNA extraction.
Genomic DNA was extracted with a Purelink Microbiome DNA purification kit (Invitrogen) according to the manufacturer’s instructions, and subjected to high throughput sequencing. DNA yields were determined through fluorometry (Qubit, Life Technologies, Carlsbad, CA) by using a reagent kit (Quant-iT BR dsDNA Kit, Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Amplicons purification and Illumina sequencing were performed commercially by Macrogen, Inc., Korea (www.macrogen.com).
A total of 3,484,598 reads were obtained with an average GC content of (54.5 ± 0.6)%, whereas the ratio of bases that have pared quality score of over 20 (Q20) and 30 (Q30) were (93.4 ± 0.5)% and (86.1 ± 0.7)%, respectively. Raw sequences were processed using CUTADAPT17 to remove primers. Then, forward and reverse reads were trimmed at 250 and 210 bp, respectively, using DADA218. After being denoised and dereplicated, the remaining forward and reverse reads were assembled. Chimeras were removed using the ‘removeBiomeraDenova’ function in the DADA2 pipeline, which resulted in the final taxon table based on 1,053,851 reads. Taxonomic annotation was performed using the SILVA 132 database19. Analyses and graphical representations were performed in R version 4.1.0 (R Core Team., 2013)20 using vegan21, phyloseq22, Microbiome23, DESeq224, MASS25, ggplot226, ampvis227 and VennDiagram28 packages. Sample metadata, taxonomic units as amplicon sequencing variants (ASV) matrix and taxa information were imported into a phyloseq object. Then, alpha diversity measures were calculated (i.e. ASV observed, Chao1, Shannon, Gini-Simpson and Pielou’s Eveness indexes). Bray–Curtis distances were used for calculating beta diversity and non-metric multidimensional scaling (NMDS). Permutational multivariate analysis of variance (PerMANOVA) was carried out based on the Bray–Curtis dissimilarity matrix by using permutest and adonis functions from vegan package.
Tissue collection and sample preparation
Animals were euthanized with sodium pentobarbitone (140 mg/kg) injected intraperitoneally, and transcardially perfused with 0.01 M phosphate buffered saline (PBS; pH 7.4). For molecular biology experiments, tissue samples of hypothalamus and brown adipose tissue (BAT) were processed independently to extract both mRNA and proteins, for real time PCR or Western-blotting analysis following the protocol described29. Hypothalamus and BAT were immediately frozen on dry ice and stored at − 80 °C for later analysis. For IHC analysis, TA samples were dissected, post-fixed overnight in 4% paraformaldehyde (PFA in 0.01 M PBS), and then transferred to 18% and then 30% sucrose solutions overnight30. Serial cryosections (50 µm thick) were cut on a cryostat (Leica CM 1850).
RNA isolation and RT-qPCR
Total RNA was isolated from hypothalamus and BAT, respectively, using the RNeasy Lipid Tissue Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Complementary DNA (cDNA) (0.5 µg of total RNA for hypothalamus and 1 µg for BAT, respectively) and the relative quantification of each gene was performed as described previously31. The 18S rRNA was used as a control to normalize gene expression32. The reactions were run on an ABI PRISM 7900 Fast Sequence Detection System instrument and software (Applied Biosystem) according to the manufacturer's protocol. Primers were designed using NCBI/Primer-BLAST software (Tables 1, 2). Relative quantification for each gene was performed by the ∆∆Ct method33.
Table 1.
Primers used for quantitative real time reverse transcription-polymerase chain reaction (RT-qPCR) of genes implicated in regulation of energy homeostasis in the hypothalamus.
| Gene name | Forward primer | Reverse primer |
|---|---|---|
| POMC | 5′-AGAACGCCATCATCAAGAAC-3′ | 5′-AAGAGGCTAGAGGTCATCAG-3′ |
| AgRP | 5′-TGGCCTCAAGAAGACAACTG-3′ | 5′-CATTGGCTAGGTGCGACTAC-3′ |
Table 2.
Primers used for RT-qPCR of genes involved in the metabolism of BAT.
| Gene name | Forward primer | Reverse primer |
|---|---|---|
| Prdm16 | 5′-ATGGGCTTTGACCATACCCG-3′ | 5′-TCGGCTCCAAAGCTAACAGG-3′ |
| PGC1α | 5′-GCACGCAGCCCTATTCATTG-3′ | 5′-TGAGTCTCGACACGGAGAGT-3′ |
| PPARγ | 5′-TGAAAGAAGCGGTGAACCACTG-3′ | 5′-TGGCATCTCTGTGTCAACCATG-3′ |
| AdipoQ | 5′-TCCCAATGTACCCATTCGCT-3′ | 5′-AGAGTCCCGGAATGTTGCAG-3′ |
| Fabp4 | 5′-CCTTCAAACTGGGCGTGGAA-3′ | 5′-CCCCGCCATCTAGGGTTATG-3′ |
| GLUT4 | 5′-GCTCTGACCGATGGGGAACC-3′ | 5′-AAACTGAAGGGAGCCAAGCA-3′ |
| UCP1 | 5′-AGTACCCAAGCGTACCAAGC-3′ | 5′-GACCCGAGTCGCAGAAAGA-3′ |
| Leptin | 5′-TTCACACACGCAGTCGGTAT-3′ | 5′-GAAGTCCAAGCCAGTGACCC-3′ |
| Ob-Rb | 5′-CCCCCACTGAAAGACAGCTT-3′ | 5′-TTCACATCCCCGAAGACTGC-3′ |
Protein extraction and western blot analysis
Proteins from the hypothalamus were extracted using RIPA buffer (Sigma Aldrich) containing a cocktail of protease inhibitors (Roche) as described previously29. Denatured protein samples (30 µg) from each group were electrophoresed into Bolt® Bis–Tris Plus gels (Invitrogen), transferred to PVDF membranes (BioRad) and incubated with primary antibodies [rabbit anti-long-form of the leptin receptor (Ob-Rb) (1:500; Abcam), mouse anti-Akt (1:250; Santa Cruz), rabbit anti-phospho Akt (Ser473) (1:500; Cell Signaling), mouse anti-STAT3 (1:250; Santa Cruz Biotechnology), rabbit anti-phospho STAT3 (Tyr705) (1:500; Santa Cruz Biotechnology), rabbit anti-supressor of cytokine signaling 3 (SOCS-3) (1:500; Cell Signaling), and rabbit anti-neuropeptide Y (NPY) (1:200; Abcam)] overnight. Subsequently, a corresponding anti-rabbit or anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody (Vector Laboratories) were used as described previously29. Mouse anti-GAPDH (1:5000, Millipore) and/or mouse anti-actin (1:1000; Cell Signaling) were used as a loading control and band intensity was measured as the integrated intensity using ImageJ software (v1.4; NIH). All data were normalized to control values on each membrane.
Muscle histology and analysis of NMJs
Tibialis anterior (TA) samples were processed as is previously described. The stain of NMJs were conducted following the protocol described50, with slight modifications. Briefly, slides were brought up to RT and incubated for 1 h with alpha(α)-bungarotoxin (α-BTX) (Invitrogen) at 1:500 in TBS. Then, slides were incubated in blocking solution (Dako) for 1 h. Primary antibodies against beta(β)-III-tubulin (Promega) and synaptophysin (SYN) (GeneTex) were applied at 1:150 and 1:250, respectively, in blocking solution, and incubated at 4 °C overnight. Sections were then incubated with their corresponding mouse Alexa488 and rabbit Alexa647-conjugated secondary antibodies (Thermo Fisher Scientific), both diluted 1:1000 in blocking solution, for 1 h at room temperature (RT). Finally, the slides were coverslipped in “Immumount” (Thermo Scientific) and confocal images were collected at RT with × 20 objective, and LAS-AF software (Leica).
NMJs from flattened z stacks of TA were analysed (ImageJ software v1.4; NIH) by personnel blinded to the animal genotype. Brightness and contrast thresholds were set to optimize the signal-to-noise ratio of the presynaptic staining (β-III-tubulin and SYN). Innervated NMJs were defined as having observed overlap of staining for pre- and post-synaptic elements. Partial denervated NMJs were defined as α-BTX signal in the presence of incomplete presynaptic staining. Denervated NMJs were defined as α-BTX signal in the absence of presynaptic staining50–53.
Statistical analysis
For molecular biology experiments and IHC analysis, respectively, two-way ANOVA was used followed by Dunett’s post hoc test, to compare all groups with control WT-FA mice, while Tukey’s post hoc test were used for multiple comparisons between all groups. Statistical analysis was performed using GraphPad Prism software (version 8.3.1) with p < 0.05 (CI 95%) considered significant. Values were reported as means ± standard error of the mean (SEM).
Results
O3 exposure significantly altered the expression levels of Akt pathway in the hypothalamus of TDP-43A315T mice
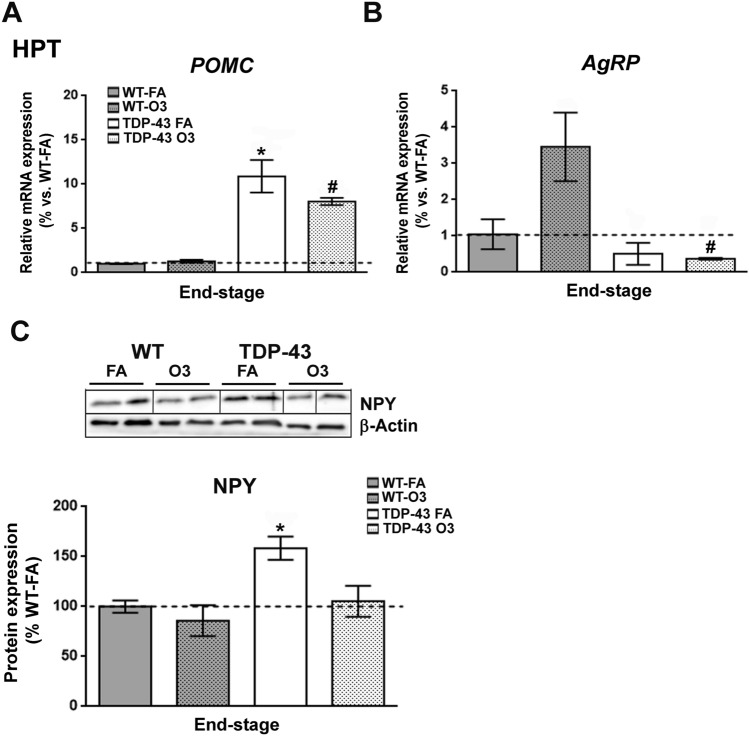
Since metabolic abnormalities have been reported in both ALS patients34 and mouse models of ALS29,35–37, and as we previously reported a progressive decline in body weight in TDP-43A315T mice13, we evaluated if genes involved in metabolism were affected in the hypothalamus of TDP-43A315T mice compared to WT mice in responses to FA or O3 exposure (Fig. 1). RT-qPCR analysis demonstrated that there was a significant effect of genotype on the expression profile of POMC (p < 0.0001) and AgRP (p = 0.01) transcripts in the hypothalamus (Fig. 1A,B). Tukey’s post hoc test demonstrated a significant increase in POMC mRNA expression in both TDP-43A315T mice FA and O3-exposed compared to WT controls in response to FA and O3 (p = 0.0003 and p = 0.003, respectively; Fig. 1A). Conversely, Tukey’s post hoc test demonstrated a significant downregulation of AgRP mRNA levels in the hypothalamus of TDP-43A315T mice O3-exposed compared to WT controls in response to O3 (p = 0.01; Fig. 1B). Additionally, immunoblotting analysis demonstrated that there was a significant effect of genotype on the protein expression levels profile of NPY (p = 0.01) in the hypothalamus (Fig. 1C; Supplementary Fig. S1). Tukey’s post hoc test demonstrated a significant hypothalamic upregulation of NPY protein levels in TDP-43A315T mice FA-exposed relative to WT controls in response to FA (p = 0.04; Fig. 1C; Supplementary Fig. S1).
Figure 1.
Alterations in anorexigenic and orexigenic neuropeptides in the hypothalamus of TDP-43A315T mice in response to O3. (A) mRNA expression of POMC and (B) AgRP neuropeptides was assessed by RT-qPCR in the hypothalamus of TDP-43A315T mice exposed to FA (n = 3 mice) or O3 (n = 4 mice) compared to WT controls exposed to FA (n = 5 mice) or O3 (n = 5 mice), at the end-stage of disease. (C) Representative β-Actin-normalized immunoblot images and quantification of NPY protein in the hypothalamus of TDP-43A315T mice compared to age-matched WT littermate controls. The white lines in the immunoblot images represent the lanes that were run on the same gel but were non-contiguous. Values are expressed as the mean ± SEM for the different groups. Comparison between groups was performed by two-way ANOVA followed by Dunnett’s post hoc test to compare all groups with WT-FA, while Tukey’s post hoc test was used for multiple comparisons between all groups, where * p < 0.05 vs. WT mice FA-exposed; # p < 0.05 vs. WT mice O3-exposed; ** p < 0.05 vs. TDP-43A315T FA-exposed. In the immunoblot images, representative bands were run on the same gel but were non-continuous. Abbreviations: WT, Wild-type mice; TDP-43,TDP-43A315T mice; FA, filtered air; O3, ozone; HPT, hypothalamus.
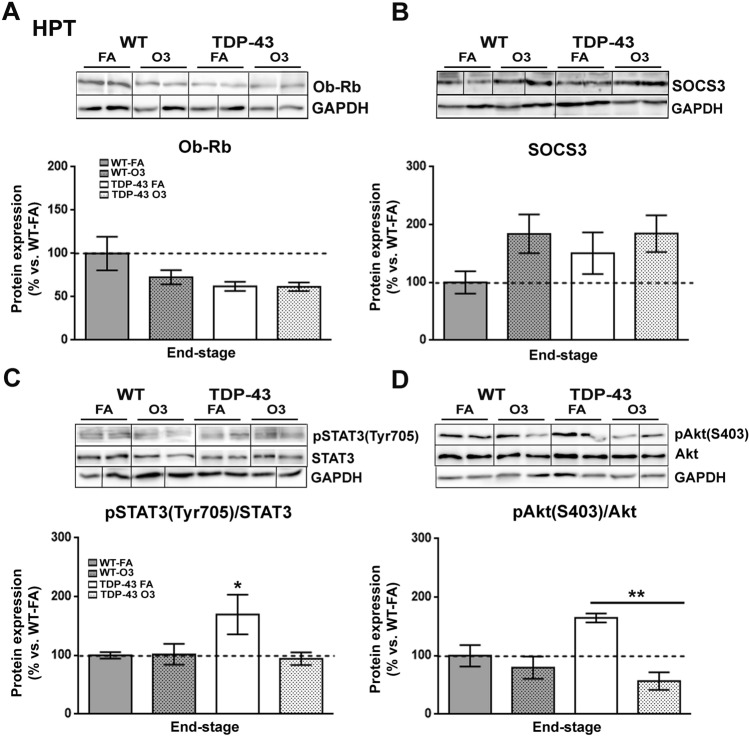
We next investigated the protein levels of Ob-Rb, the long isoform of leptin receptor, SOCS3, a main inhibitor of the leptin signaling, and the phosphorylation levels of Akt (pSer473-Akt) and STAT3 (pTyr705-STAT3) proteins (Fig. 2), as they are important targets in the regulation of glucose and energy metabolism38. Immunoblotting analysis demonstrated no significant effect of either genotype or O3 exposure on Ob-Rb and SOCS3 protein expression levels (Fig. 2A,B; Supplementary Fig. S2). With respect of leptin signaling pathways, immunoblotting analysis demonstrated a significant increase on the phosphorylation levels of STAT3 protein in the hypothalamus of TDP-43A315T mice FA-exposed compared to WT controls in response to FA (p = 0.04) (Fig. 2C; Supplementary Fig. S2). However, there was a significant effect of the O3 exposure (p = 0.0035) on the phosphorylation levels of Akt protein in the hypothalamus of TDP-43A315T mice (Fig. 2D). Tukey’s post hoc test demonstrated a statistically significant hypothalamic decrease on the phosphorylation levels of Akt protein in TDP-43A315T mice O3-exposed compared to TDP-43A315T mice (p = 0.008) in responses to FA (Fig. 2D; Supplementary Fig. S2).
Figure 2.
Alterations in serine phosphorylation of Akt in the hypothalamus of TDP-43A315T mice in response to O3 exposure. (A) Representative β-Actin-normalized immunoblot images and quantitation of Ob-Rb receptor, (B) SOCS3, (C) pSTAT3 (pTyr705-STAT3) protein, (D) pAkt (pSer473-Akt) protein, respectively, in the hypothalamus of TDP-43A315T mice exposed to FA (n = 3 mice) or O3 (n = 4 mice) compared to WT controls exposed to FA (n = 5 mice) or O3 (n = 5 mice) at the end-stage of disease. The white lines in the immunoblot images represent the lanes that were run on the same gel but were non-contiguous. Values are expressed as the mean ± SEM for the different groups. Comparison between groups was performed by two-way ANOVA followed by Dunnett’s post hoc test to compare all groups with WT-FA, while Tukey’s post hoc test was used for multiple comparisons between all groups, where * p < 0.05 vs. WT mice FA-exposed; # p < 0.05 vs. WT mice O3-exposed; ** p < 0.05 vs. TDP-43A315T FA-exposed. In the immunoblot images, representative bands were run on the same gel but were non-continuous. Abbreviations: HPT, hypothalamus; Ob-Rb, long form of leptin receptor; SOCS3, suppressor of cytokine signaling 3; STAT3, signal transducer and activator of transcription 3; Akt, serine/threonine kinase.
O3 exposure decreased the expression profile of genes involved in metabolism and thermogenesis in the BAT of TDP-43A315T mice
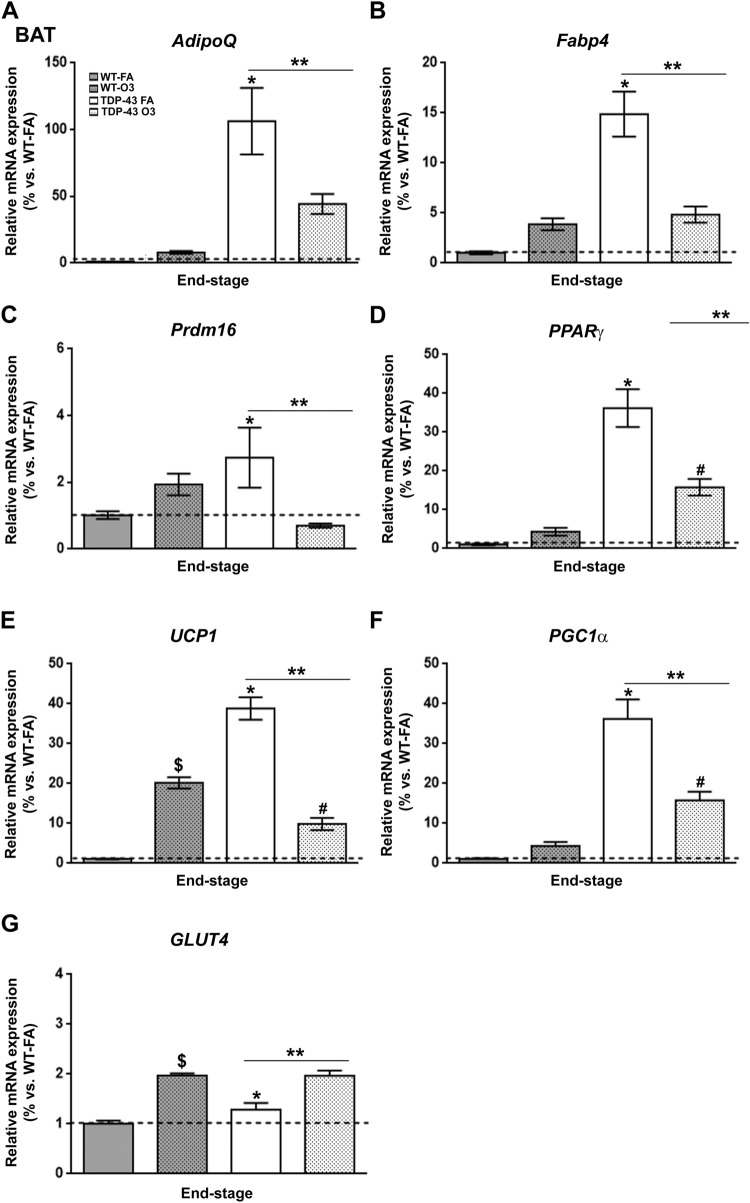
BAT is a thermogenic organ with an important role in controlling energy expenditure and the regulation of body weight39. Interestingly, animal work has shown that acute O3 exposure causes endocrine and metabolic changes, increasing food intake and body fat mass40. In this context, to determine the metabolic impact of O3 exposure on altering BAT thermogenic activity in TDP-43A315T mice compared to WT mice in responses to FA or O3 exposure, we first conducted RT-qPCR analysis to examine the expression profile of genes involved in brown adipocyte differentiation (Fig. 3). Our results indicated a significant effect of genotype and exposure (p < 0.05) on the expression profile of AdipoQ, Fabp4, Prdm16 and PPARγ transcripts in BAT across groups (Fig. 3A–D). In TDP-43A315T mice FA-exposed, AdipoQ, Fabp4, Prdm16 and PPARγ mRNA levels were significantly upregulated compared to WT controls in responses to FA (p = 0.0002, p < 0.0001, p = 0.04 and p < 0.0001, respectively). Additionally, Tukey’s post hoc test demonstrated an overall statistically significant downregulation of all genes (AdipoQ: p = 0.01, Fig. 3A; Fabp4: p < 0.0001, Fig. 3B; Prdm16: p = 0.01, Fig. 3C; and PPARγ: p = 0.0001, Fig. 3D) in the BAT of TDP-43A315T mice O3-exposed compared to TDP-43A315T mice in responses to FA.
Figure 3.
Alterations in the expression of genes involved in brown adipocyte differentiation in the BAT of TDP-43A315T mice in response to O3 exposure. (A) AdipoQ, (B) Fabp4, (C) Prdm16, (D) PPARγ, (E) UCP1, (F) PGC1α and (G) GLUT4 mRNA expression was assessed by RT-qPCR in the BAT of TDP-43A315T mice exposed to FA (n = 5 mice) or O3 (n = 7 mice) compared to WT controls exposed to FA (n = 6 mice) or O3 (n = 8 mice) at the end-stage of disease. Values are expressed as the mean ± SEM for the different groups (n = 3–8 per genotype/exposure). Comparison between groups was performed by two-way ANOVA followed by Dunnett’s post hoc test to compare all groups with WT-FA, while Tukey’s post hoc test was used for multiple comparisons between all groups, where $ p < 0.05 WT O3-exposed vs. WT mice FA-exposed; * p < 0.05 vs. WT mice FA-exposed; # p < 0.05 vs. WT mice O3-exposed; ** p < 0.05 vs. TDP-43A315T FA-exposed. Abbreviations: WT, Wild-type mice; TDP-43, TDP-43A315T mice; FA, filtered ai; O3, ozone; BAT, brown adipose tissue.
We next examined the expression of genes involved in BAT-mediated thermogenesis, including UCP1 and PGC1α (Fig. 3E,F). There were significant effects of genotype (p < 0.0001) and exposure (p = 0.008 and p = 0.009, respectively) on the expression profile of both genes in the BAT. In TDP-43A315T mice FA-exposed, UCP1 and PGC1α mRNA levels were significantly upregulated compared to WT controls in responses to FA (p = 0.003 and p = 0.0001, respectively), while a different pattern of expression was observed in TDP-43A315T mice O3-exposed compared to WT controls in responses to O3 (Fig. 3E,F). Additionally, RT-qPCR analysis demonstrated that O3 exposure modified UCP1 and PGC1α mRNA levels in TDP-43A315T mice, with a statistically significant downregulation on their expression compared to TDP-43A315T mice in responses to FA (p < 0.0001, Fig. 3E,F).
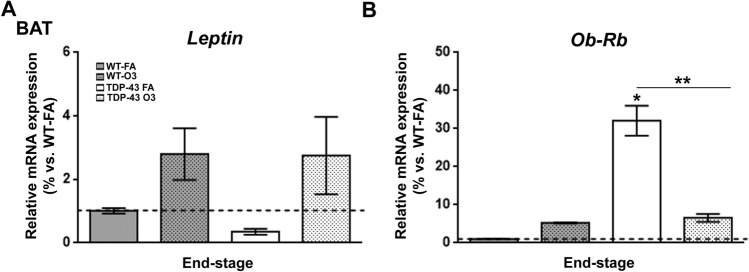
Finally, to further examine the effect of O3 exposure on BAT, we analysed the expression profile of genes known to mediate glucose uptake, particularly GLUT4, which is significantly expressed in BAT41 (Fig. 3G). Remarkably, as leptin modulates body weight gain and energy expenditure42,43, at least in part, by modulating thermogenesis44, and, as it has been provided that leptin treatment increase body temperature in leptin deficient mice (ob/ob mice)45, we also asked how Leptin and Ob-Rb transcripts were affected in the BAT of TDP-43A315T mice in response to O3 exposure (Fig. 4). RT-qPCR analysis demonstrated a statistically significant upregulation of GLUTt4 mRNA levels in the BAT of TDP-43A315T mice in responses to O3 exposure (p < 0.0015, Fig. 3G). There was no effect of genotype on the expression profile of Leptin transcript in the BAT (Fig. 4A). In contrast, RT-qPCR analysis demonstrated a statistically significant BAT upregulation of Ob-Rb mRNA levels in TDP-43A315T mice FA-exposed compared to WT controls in responses to FA exposure (p < 0.0001, Fig. 4B). Tukey’s post hoc test demonstrated a statistically significant downregulation of Ob-Rb mRNA levels (p < 0.0001, Fig. 4B) in the BAT of O3-exposed TDP-43A315T mice compared to TDP-43A315T mice in responses to FA.
Figure 4.
Alterations in Ob-Rb in the BAT of TDP-43A315T mice in response to O3 exposure. (A) Leptin and (B) Ob-Rb mRNA expression was assessed by RT-qPCR in the BAT of TDP-43A315T mice exposed to FA (n = 5 mice) or O3 (n = 7 mice) compared to WT controls exposed to FA (n = 6 mice) or O3 (n = 8 mice) at the end-stage of disease. Values are expressed as the mean ± SEM for the different groups. Comparison between groups was performed by two-way ANOVA followed by Dunnett´s post hoc test to compare all groups with WT-FA, while Tukey’s post hoc test was used for multiple comparisons between all groups, where * p < 0.05 vs. WT mice FA-exposed; # p < 0.05 vs. WT mice O3-exposed; ** p < 0.05 vs. TDP-43A315T FA-exposed. Abbreviations: WT, Wwild-type mice; TDP-43, TDP-43A315T mice; FA, filtered air; O3 , ozone; BAT, brown adipose tissue.
O3 exposure did not induce fecal microbiota compositional changes in TDP-43A315T mice
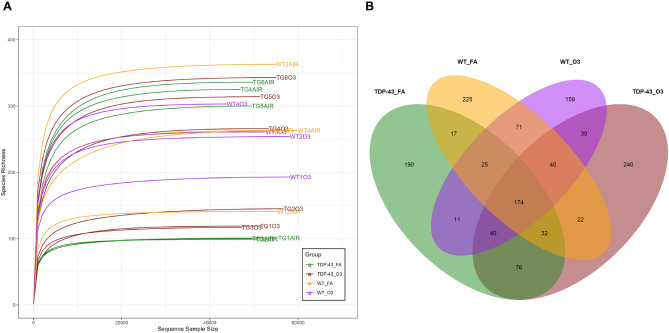
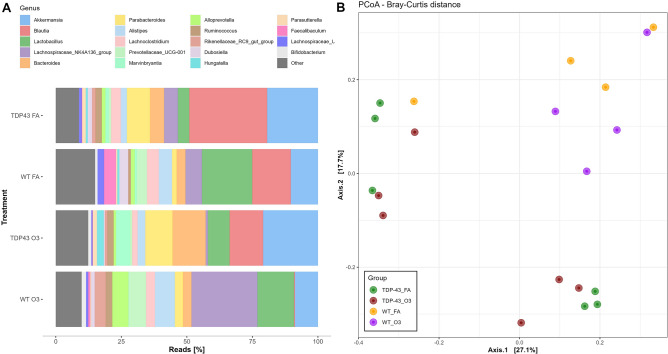
As it has been reported that the HPA axis closely interact with the gut microbiota46, we next compared fecal gut microbiome composition in TDP-43A315T mice and age-matched WT littermate controls, in responses to FA or O3 exposure. Microbial-mediated effects on ALS neuropathology were assessed through 16S rRNA amplicon sequencing in fecal samples from TDP-43A315T and WT controls prior to the end-stage of disease. The rarefaction curve suggested that ASV richness is not affected by either genotype or exposure, since any trend is observed as indicated by the mix higher curves under the same sequencing depth (Fig. 5A). Additionally, all the curves tend to be smooth and reach the plateau, which indicates that the amount of sequencing data was sufficient and reasonable, and that more data would only produce a few new ASVs. The total number of observed genera (alpha diversity) was similar in both TDP-43A315T mice and WT controls whether they were exposed to FA or O3. However, evenness was higher in WT mice than in TDP-43A315T mice (p = 0.03), suggesting that a more even distribution of taxa can be found in WT controls compared to TDP-43A315T mice. The Venn diagram exhibited the exact number of ASVs common for the four groups (174). This number was higher than the number of unique ASVs in WT controls exposed to O3, but lower than the unique ASVs observed in the other groups (Fig. 5B). Actually, the gut microbiome composition of TDP-43A315T mice and WT controls yielded significantly different compositions (Fig. 6). TDP-43A315T mice showed a significant increase in ASVs within Proteobacteria phyla like Parasutterella and Escherichia/Shigella genus. In addition, several ASVs within specific genera were observed in TDP-43A315T mice but not in WT controls (e.g. Ruminococcus, Parabacteroides, Marvinbryantia, Blautia) (Supplementary Fig. S3).
Figure 5.
WT controls and TDP-43A315T mice fecal microbiota diversity. (A) Rarefaction curves for the samples analysed within the four groups (TDP-43-FA, TDP-43-O3, WT-FA and WT-O3). (B) Venn diagram of four groups (TDP-43-FA, TDP-43-O3, WT-FA and WT-O3) and their intersections. The numbers correspond to the number of ASVs in each subset and intersection. Abbreviations: WT, Wild-type mice; TDP-43, TDP-43A315T mice; FA, filtered air; O3, ozone.
Figure 6.
TDP-43 A315T mice develop gut microbiome compositional changes. (A) Relative abundance at genus level obtained by 16S rRNA amplicon sequencing. (B) Bray–Curtis PCoA at the end-stage of disease. Abbreviations: WT, Wild-type mice; TG, TDP-43A315T mice; FA, filtered air; O3, ozone.
O3 exposure did not impair the NMJs in the skeletal muscle of TDP-43A315T mice
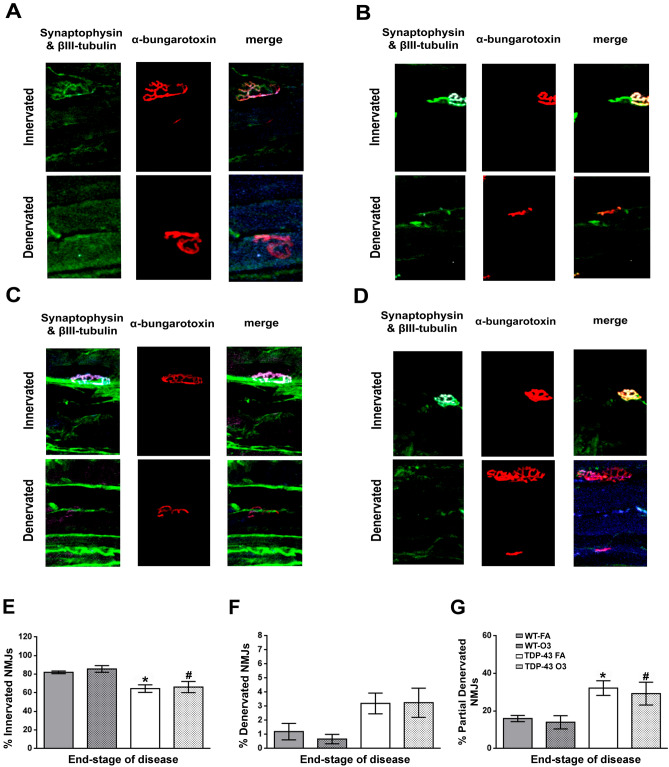
We next analised alterations in NMJs in TDP-43A315T mice compared to age-matched WT littermate controls in responses to FA or O3 exposure, as the hypothalamus regulates skeletal muscle metabolism47, and NMJ pathology is well-known in mouse models of ALS48. To this aim we examined the TA muscle by IHC analysis (Fig. 7). Densitometric analysis demonstrated a significant effect of genotype on the percentage of innervated and denervated (p < 0.0001, respectively) NMJs in the TA muscle of TDP-43A315T mice compared to WT (Fig. 7E–G). Tukey’s post hoc test demonstrated a significant decrease on the percentage of innervated NMJs in the TA muscle of TDP-43A315T mice exposed to FA and O3 relative to WT mice in response to FA (p = 0.0013 and p = 0.007, respectively) and O3 exposure (p = 0.0007 and p = 0.003, respectively) (Fig. 7E). Similarly, IHC analysis showed a significant decrease on the percentage of denervated NMJs in the TA muscle of TDP-43A315T mice exposed to FA and O3 compared to WT in response to FA and O3 exposure (p = 0.002 in all cases; Fig. 7F). There was a slight decrease in partially denervation of NMJs in the TA muscle of TDP-43A315T mice O3-exposed compared to FA-exposed, however, the differences were not statistically significant (Fig. 7G).
Figure 7.
O3 exposure did not alter the NMJs denervation state in the TA muscle in TDP-43A315T mice at the end-stage of the disease. Representative examples of NMJ immunostaining in TA muscle of WT mice exposed to FA (n = 6 mice) (A) or O3 (n = 8 mice) (B), and TDP-43A315T mice exposed to FA (n = 5 mice) (C) or O3 (n = 7 mice) (D). Quantification of innervation (E), denervation (F) and partial denervation (G) at the end-stage of disease. Values are expressed as the mean ± SEM for the different groups. Comparison between groups was performed by two-way ANOVA followed by Dunnett’s post hoc test to compare all groups with WT-FA, while Tukey’s post hoc test was used for multiple comparisons between all groups, where * p < 0.05 vs. WT mice FA-exposed; # p < 0.05 vs. WT mice O3-exposed. Abbreviations: WT, Wild-type mic; TDP-43, TDP-43A315T mice; FA, filtered air; O3, ozone.
Discussion
Although the cellular basis for motor neuron degeneration in ALS is not yet fully understood, it is increasingly recognized that energy homeostasis (the balance between energy intake and expenditure) appears to be compromised in ALS49. Indeed, ALS patients develop prominent changes in weight and eating behavior that result from and mediate the underlying neurodegenerative process. In this context, emerging research suggests that these alterations may be mediated through changes in the hypothalamic function50.
Hypothalamic neuropeptides derived from POMC provide a strong anorexigenic effect (i.e. decreases food intake), while NPY/AgRP neurons have a potent orexigenic effect (i.e. increase food intake). Here, we report a downregulation of AgRP mRNA levels and a significantly upregulation of POMC transcripts in TDP-43A315T mice compared to WT, which is in accordance with previously data reporting a progressive loss of body weight in TDP-43A315T mice13,51–54. In addition, although no statistical difference was obtained, our results show a reduction of NPY protein expression levels in TDP-43A315T mice O3-exposed relative to FA-exposed. It is conceivable that changes in the expression of NPY in the hypothalamus of TDP-43A315T mice could potentially be due to modifications in food intake. Indeed, in a situation of negative energy balance, such as the malnutrition occurring in the majority of ALS patients55,56, the expression of NPY would be normally increased57,58. However, future experiments with a larger sample size should try to corroborate this hypothesis.
Besides sensitive genes involved in metabolism, we also examined leptin signaling in the hypothalamus of TDP-43A315T mice, as central hypothalamic leptin signaling has a critical role in promoting energy homeostasis via modulation of food intake and energy expenditure59. Although the precise dynamics of Ob-Rb regulation in the hypothalamus is not completely understood, no differences in Ob-Rb protein expression levels between groups were obtained. However, a significant decrease in serine phosphorylation of Akt was determined in the hypothalamus of TDP-43A315T mice in response to O3 exposures. The alterations in Akt levels could partly explain the hypoglycemic state observed in TDP-43A315T mice13, as Akt is an important target in the regulation of glucose and energy metabolism38, which is in accordance with previous data from our group showing how O3 exposure resulted in higher plasma glucose levels at later time points13. In addition, this data might reflect disruption of insulin signaling in the hypothalamus of TDP-43A315T mice, as Akt signaling pathway is part of the insulin cascade60. Indeed, insulin signaling in the hypothalamus plays a role in maintaining body weight61, and insulin resistance is related to disease severity in ALS62. However, future experiments should try to corroborate this hypothesis.
Considering that BAT has profound effects on body weight and metabolism in rodents63, it is conceivable that the reported improvement on loss of body weight in TDP-43A315T mice in response to O3 exposure, could be due to the effect of this gas to modify its thermogenic function. Indeed, the ability of this adipose tissue to expend energy has increased interest in stimulating thermogenesis to treat metabolic diseases such as obesity and diabetes type II64. In this context, our results confirm a significant upregulation of genes involved in brown adipocyte differentiation, including AdipoQ, Fabp4, Prdm16 and PPARγ mRNAs in BAT of TDP-43A315T mice in response to FA compared with O3 exposure. We also found significant effects of O3 exposure on the expression profile of genes involved in BAT-mediated thermogenesis such as UCP1 and PGC1α. These data are of interest because evidence supports that the decrease in thermogenesis is likely associated with a greater predisposition to body weight gain65, which is in accordance with previous data from our group showing a less severe decline disease-associated weight loss in TDP-43A315T mice exposed to O3. Indeed, we also found a significant upregulation of GLUT4 mRNA levels in the BAT of TDP-43A315T mice in responses to O3 exposure, which is an interesting data as we previously reported plasma glucose levels were highest at the end-stage of disease after O3 exposure in TDP-43A315T mice13. Finally, concomitantly to the decrease in the expression profile of AdipoQ, Fabp4, Prdm16, PPARγ, UCP1 and PGC1α in the BAT of TDP-43A315T mice, our results confirm a significant downregulation of Ob-Rb mRNA expression levels in responses to O3 exposure, which is in accordance with experimental data showing how obese mice that lack of Ob-Rb receptor showed a decreased BAT thermogenic capacity66. As a whole, these transcriptional modifications in TDP-43A315T mice exposed to O3 might reflect the physiological response of the hypothalamus to this gas to overcome adipose atrophy and loss of body weight. Indeed, growing experimental research suggests the importance of the hypothalamus and the role of hypothalamic peptides and neurons in the control of BAT thermogenesis67. However, future experiments should try to corroborate this hypothesis.
In attempt to have a better knowledge on the potential major role of O3 exposure to induce controlled metabolic effects, we examined fecal microbiome composition in TDP-43A315T mice compared to WT. Gut microbiome has been implicated in ALS development68, and thus, it is conceivable that the metabolic changes determined in TDP-43A315T mice in responses to O3 exposure could be also due to a compensatory effect of O3 on gut dysbiosis (an imbalance in the gut microbiota community). Indeed, in ALS disease gut dysbiosis affects the central nervous system via pro-inflammatory mediators (i.e. cytokines and hormone-like molecules), thus, impacting gut-brain communications69. In this context, our data showed how the gut microbiome composition of TDP-43A315T mice yielded significantly different composition compared with WT controls. Although no statistical changes were found on gut bacterial communities in TDP-43A315T mice in response to O3 compared with FA exposure, an increase in the relative abundance of genera like Bacteroides, Parabacteroides or Marvinbryantia was observed as a result of such exposure (Supplementary Fig. S1). In addition, gut microbiome composition of WT mice showed a significant increase in Lachnospiraceae NK4A136 group as a result of O3 exposure. This data is of interest because this group is a potential butyrate-producer associated with probiotic activity in mice70. Indeed, it has been found that butyrate is one of the main short-chain fatty acids (SCFAs) produced by microbiota helping to maintain gut barrier integrity and inhibit inflammation71. These results might indicate the capacity of this O3 to restore physiological homeostasis and potentially improve dysbiosis.
Finally, as metabolic alterations in ALS are associated with the progression of disease pathology, we examined potential changes in NMJ innervation in TA muscles of TDP-43A315T mice, as degenerative processes in the skeletal muscle, particularly involving NMJs, are observed throughout disease progression in ALS72,73. Our densitometric analysis showed no significant changes on NMJs in TA muscles in TDP-43A315T mice in response to O3 exposure compared with FA exposure, although a significant impairment of NMJs in TDP-43A315T mice compared to WT controls was found, which is in agreement with previous data reported in mutant TDP-43 mice (i.e. Tg NEFH-hTDP-43ΔNLS or TDP-43 rNLS bigenic mice) showing a progressive NMJs denervation followed by spinal cord motor neuron loss74.
In summary, our study provides the first experimental evidence underlying the potential effect of O3 exposure on hypothalamic function in TDP-43A315T mice, which might result in modifications on neural signaling, affecting thermogenesis and energy homeostasis in ALS. Molecular biology analysis has demonstrated that O3 modified the expression profile of hypothalamic neuropeptides, and significantly altering phosphorylation levels of Akt, concomitantly to decrease the expression of genes involved in metabolism and thermogenesis in the BAT of TDP-43A315T mice O3-exposed. Composition of fecal gut microbiome of O3-exposed TDP-43A315T mice varied significantly compared to WT controls, and densitometric analysis of NMJs, indicated that O3 does not impair the progression of disease in the skeletal muscle. However, further functional studies are necessary to determine the mechanism of actions of O3 which may provide a new avenue for therapeutic development for this fatal condition.
Supplementary Information
Acknowledgements
The authors would like to gratefully acknowledge Agueda Ferrer-Donato and Surgery Unit of the Hospital Nacional de Parapléjicos for their excellent technical support. Sara Rodríguez-Sánchez is supported by a PhD Fellowship from the UCLM research plan (2019-PREDUCLM-10801).
Author contributions
Conceptualization: C.M.F.-M.; Data curation: S.R., N.V., S.S., M.C.-P.; Formal analysis: S.R., N.V., S.S., M.C.-P., A.R., C.M.F.-M.; Software: S.R., N.V., S.S., C.M.F.-M.; Funding acquisition: C.M.F.-M.; Investigation: S.R., N.V., S.S., C.M.F.-M.; Methodology: S.R., N.V., S.S., M.C.-P., A.R., C.M.F.-M.; Project administration: C.M.F.-M. Resources: C.M.F.-M; Supervision: A.A., L.L.P., C.M.F.-M. Validation: A.A., L.L.P., C.M.F.-M. Writing—original draft: S.R., N.V., S.S., C.M.F.-M. Writing—review & editing: S.R., N.V., S.S., A.R., A.A., L.L.P., C.M.F.-M.
Funding
This work was supported by funding from the Consejería de Educación, Cultura y Deportes, Fondo Europeo de Desarrollo Regional (FEDER), Junta de Comunidades de Castilla-La Mancha (JCCM) (SBPLY/17/180501/000303).
Data availability
The datasets generated and/or analysed during the current study are available upon reasonable request to the corresponding author, Dr Carmen M. Fernandez-Martos (cmfernandezm@sescam.jccm.es; Carmen.fernandez-martos@utas.edu.au), and the raw demultiplexed sequence data (amplicon reads) have been uploaded to the Sequence Read Archive (SRA) with BioProject accession number PRJNA886986 (https://www.ncbi.nlm.nih.gov/bioproject/886986). Additional material including source data is available online.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Nicolas Valiente and Susana Seseña.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-25033-4.
References
- 1.Pfrieger FW. Neurodegenerative diseases and cholesterol: Seeing the field through the players. Front. Aging Neurosci. 2021;13:766587. doi: 10.3389/fnagi.2021.766587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Izadi M, et al. Changes in Th17 cells frequency and function after ozone therapy used to treat multiple sclerosis patients. Mult. Scler. Relat. Disord. 2020;46:102466. doi: 10.1016/j.msard.2020.102466. [DOI] [PubMed] [Google Scholar]
- 3.Scassellati C, Galoforo AC, Bonvicini C, Esposito C, Ricevuti G. Ozone: A natural bioactive molecule with antioxidant property as potential new strategy in aging and in neurodegenerative disorders. Ageing Res. Rev. 2020;63:101138. doi: 10.1016/j.arr.2020.101138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mašán J, Sramka M, Rabarova D. The possibilities of using the effects of ozone therapy in neurology. Neuro Endocrinol. Lett. 2021;42:13–21. [PubMed] [Google Scholar]
- 5.Delgado-Roche L, et al. Medical ozone promotes Nrf2 phosphorylation reducing oxidative stress and pro-inflammatory cytokines in multiple sclerosis patients. Eur. J. Pharmacol. 2017;811:148–154. doi: 10.1016/j.ejphar.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 6.Braidy N, et al. Therapeutic relevance of ozone therapy in degenerative diseases: Focus on diabetes and spinal pain. J. Cell. Physiol. 2018;233:2705–2714. doi: 10.1002/jcp.26044. [DOI] [PubMed] [Google Scholar]
- 7.Gupte AA, Lyon CJ, Hsueh WA. Nuclear factor (erythroid-derived 2)-like-2 factor (Nrf2), a key regulator of the antioxidant response to protect against atherosclerosis and nonalcoholic steatohepatitis. Curr. Diab. Rep. 2013;13:362–371. doi: 10.1007/s11892-013-0372-1. [DOI] [PubMed] [Google Scholar]
- 8.Diaz-Luis J, Menéndez-Cepero S, Macias-Abrahan C. Ozone therapy immunomodulatory effect in the selective immunoglobulin A deficiency. J. Ozone Ther. 2015 doi: 10.7203/jo3t.1.1.2015.12161. [DOI] [Google Scholar]
- 9.Xia Y, et al. Personal ozone exposure and stress hormones in the hypothalamus-pituitary-adrenal and sympathetic-adrenal-medullary axes. Environ. Int. 2022;159:107050. doi: 10.1016/j.envint.2021.107050. [DOI] [PubMed] [Google Scholar]
- 10.Tashiro H, Shore SA. The gut microbiome and ozone-induced airway hyperresponsiveness. Mechanisms and therapeutic prospects. Am. J. Respir. Cell Mol. Biol. 2021;64:283–291. doi: 10.1165/rcmb.2020-0288TR. [DOI] [PubMed] [Google Scholar]
- 11.Alderete T, et al. Exposure to ambient air pollutants is associated with the composition of the gut microbiome in adolescents from Southern California. Environ. Epidemiol. 2019;3:6–7. doi: 10.1097/01.EE9.0000605664.50459.e7. [DOI] [Google Scholar]
- 12.Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18809–18814. doi: 10.1073/pnas.0908767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez A, et al. Repeated exposure to ozone produces changes in metabolic disturbances present in the TDP-43A315T transgenic model of amyotrophic lateral sclerosis. Diagn. Pathol. Open Access. 2021;S6:001. doi: 10.1101/2021.02.12.430915. [DOI] [Google Scholar]
- 14.Alvaro-Alonso C, Ferrer-Donato A, Fernandez-Torres E, Carballo-Villa M, Fernandez-Martos CM. Methodology Aspects of Colony Maintain for a Murine Model of Amyotrophic Lateral Sclerosis (ALS) TDP-43 Proteinopathy. Animals. 2020;10:2329. doi: 10.3390/ani10122329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bello-Medina PC, Prado-Alcala RA, Rivas-Arancibia S. Effect of ozone exposure on dendritic spines of CA1 pyramidal neurons of the dorsal hippocampus and on object-place recognition memory in rats. Neuroscience. 2019;402:1–10. doi: 10.1016/j.neuroscience.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 16.Chuang GC, et al. Pulmonary ozone exposure induces vascular dysfunction, mitochondrial damage and atherogenesis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2009;297:L209–216. doi: 10.1152/ajplung.00102.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:2011. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 18.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2013. [Google Scholar]
- 21.Oksanen, J. et al. Vegan: Community ecology package [online]. R Package version 2.0–5 (2011).
- 22.McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lahti L, S. S. et al. Tools for microbiome analysis in R. Microbiome package version 1.15.1. Bioconductor, 2017. http://microbiome.github.com/microbiome/ (2017). Accessed 4 Aug 2022.
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ripley, B. e. a. Package “mass”. Cran r. 538, 113–120 (2013).
- 26.Wickham H. ggplot2 WIREs. Comput. Stat. 2011;3:180–185. doi: 10.1002/wics.147. [DOI] [Google Scholar]
- 27.Andersen KS, Kirkegaard RH, Karst SM, Albertsen M. Ampvis2: An R package to analyse and visualise 16S rRNA amplicon data. Biorxiv. 2018 doi: 10.1101/299537. [DOI] [Google Scholar]
- 28.Dusa, A. CRAN—package venndiagram. https://cran.r-project.org/web/packages/VennDiagram/ (2021). Accessed 4 Aug 2022.
- 29.Ferrer-Donato A, Contreras A, Frago LM, Chowen JA, Fernandez-Martos CM. Alterations in leptin signaling in amyotrophic lateral sclerosis (ALS) Int. J. Mol. Sci. 2021;22:10305. doi: 10.3390/ijms221910305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, et al. Cytoskeletal changes during development and aging in the cortex of neurofilament light protein knockout mice. J. Comp. Neurol. 2013;521:1817–1827. doi: 10.1002/cne.23261. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez CM, et al. The expression of rat resistin isoforms is differentially regulated in visceral adipose tissues: Effects of aging and food restriction. Metabolism. 2009;58:204–211. doi: 10.1016/j.metabol.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 32.Fernandez-Martos CM, et al. Differential expression of Wnts after spinal cord contusion injury in adult rats. PLoS ONE. 2011;6:e27000. doi: 10.1371/journal.pone.0027000PONE-D-11-06684[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.D'Amico E, et al. Metabolic abnormalities, dietary risk factors and nutritional management in amyotrophic lateral sclerosis. Nutrients. 2021;13:2273. doi: 10.3390/nu13072273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim MA, et al. Genetically altering organismal metabolism by leptin-deficiency benefits a mouse model of amyotrophic lateral sclerosis. Hum. Mol. Genet. 2014;23:4995–5008. doi: 10.1093/hmg/ddu214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shan X, Chiang PM, Price DL, Wong PC. Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 2010;107:16325–16330. doi: 10.1073/pnas.1003459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang W, et al. The ALS disease-associated mutant TDP-43 impairs mitochondrial dynamics and function in motor neurons. Hum. Mol. Genet. 2013;22:4706–4719. doi: 10.1093/hmg/ddt319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varela L, Horvath TL. Leptin and insulin pathways in POMC and AgRP neurons that modulate energy balance and glucose homeostasis. EMBO Rep. 2012;13:1079–1086. doi: 10.1038/embor.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scotney H, et al. Glucocorticoids modulate human brown adipose tissue thermogenesis in vivo. Metabolism. 2017;70:125–132. doi: 10.1016/j.metabol.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nappi F, et al. Endocrine aspects of environmental “Obesogen” pollutants. Int. J. Environ. Res. Public Health. 2016 doi: 10.3390/ijerph13080765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu Y, et al. Increased expression of glucose transporter GLUT-4 in brown adipose tissue of fasted rats after cold exposure. Am. J. Physiol. 1993;264:E890–895. doi: 10.1152/ajpendo.1993.264.6.E890. [DOI] [PubMed] [Google Scholar]
- 42.Pan WW, Myers MG., Jr Leptin and the maintenance of elevated body weight. Nat. Rev. Neurosci. 2018;19:95–105. doi: 10.1038/nrn.2017.168. [DOI] [PubMed] [Google Scholar]
- 43.Pandit R, Beerens S, Adan RAH. Role of leptin in energy expenditure: The hypothalamic perspective. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2017;312:R938–R947. doi: 10.1152/ajpregu.00045.2016. [DOI] [PubMed] [Google Scholar]
- 44.Barrios V, et al. Leptin modulates the response of brown adipose tissue to negative energy balance: Implication of the GH/IGF-I axis. Int. J. Mol. Sci. 2021 doi: 10.3390/ijms22062827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischer AW, et al. Leptin raises defended body temperature without activating thermogenesis. Cell Rep. 2016;14:1621–1631. doi: 10.1016/j.celrep.2016.01.041. [DOI] [PubMed] [Google Scholar]
- 46.Carabotti M, Scirocco A, Maselli MA, Severi C. The gut-brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015;28:203–209. [PMC free article] [PubMed] [Google Scholar]
- 47.Braun TP, Marks DL. Hypothalamic regulation of muscle metabolism. Curr. Opin. Clin. Nutr. Metab. Care. 2011;14:237–242. doi: 10.1097/MCO.0b013e328345bbcd. [DOI] [PubMed] [Google Scholar]
- 48.Mejia Maza A, et al. NMJ-Analyser identifies subtle early changes in mouse models of neuromuscular disease. Sci. Rep. 2021;11:12251. doi: 10.1038/s41598-021-91094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dupuis L, Pradat PF, Ludolph AC, Loeffler JP. Energy metabolism in amyotrophic lateral sclerosis. Lancet Neurol. 2011;10:75–82. doi: 10.1016/S1474-4422(10)70224-6. [DOI] [PubMed] [Google Scholar]
- 50.Vercruysse P, Vieau D, Blum D, Petersen A, Dupuis L. Hypothalamic alterations in neurodegenerative diseases and their relation to abnormal energy metabolism. Front. Mol. Neurosci. 2018;11:2. doi: 10.3389/fnmol.2018.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Esmaeili MA, Panahi M, Yadav S, Hennings L, Kiaei M. Premature death of TDP-43 (A315T) transgenic mice due to gastrointestinal complications prior to development of full neurological symptoms of amyotrophic lateral sclerosis. Int. J. Exp. Pathol. 2013;94:56–64. doi: 10.1111/iep.12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo Y, et al. HO-1 induction in motor cortex and intestinal dysfunction in TDP-43 A315T transgenic mice. Brain Res. 2012;1460:88–95. doi: 10.1016/j.brainres.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 53.Hatzipetros T, et al. C57BL/6J congenic Prp-TDP43A315T mice develop progressive neurodegeneration in the myenteric plexus of the colon without exhibiting key features of ALS. Brain Res. 2014;1584:59–72. doi: 10.1016/j.brainres.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 54.Medina DX, Orr ME, Oddo S. Accumulation of C-terminal fragments of transactive response DNA-binding protein 43 leads to synaptic loss and cognitive deficits in human TDP-43 transgenic mice. Neurobiol. Aging. 2014;35:79–87. doi: 10.1016/j.neurobiolaging.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 55.Ludolph AC, et al. Effect of high-caloric nutrition on survival in amyotrophic lateral sclerosis. Ann. Neurol. 2020;87:206–216. doi: 10.1002/ana.25661. [DOI] [PubMed] [Google Scholar]
- 56.Lopez-Gomez JJ, et al. Malnutrition at diagnosis in amyotrophic lateral sclerosis (als) and its influence on survival: Using glim criteria. Clin. Nutr. 2021;40:237–244. doi: 10.1016/j.clnu.2020.05.014. [DOI] [PubMed] [Google Scholar]
- 57.Caron A, et al. POMC neurons expressing leptin receptors coordinate metabolic responses to fasting via suppression of leptin levels. Elife. 2018 doi: 10.7554/eLife.33710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pedroso JA, et al. Changes in leptin signaling by SOCS3 modulate fasting-induced hyperphagia and weight regain in mice. Endocrinology. 2016;157:3901–3914. doi: 10.1210/en.2016-1038. [DOI] [PubMed] [Google Scholar]
- 59.Munzberg H, Singh P, Heymsfield SB, Yu S, Morrison CD. Recent advances in understanding the role of leptin in energy homeostasis. F1000Res. 2020 doi: 10.12688/f1000research.24260.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Asano T, et al. Insulin receptor substrate is a mediator of phosphoinositide 3-kinase activation in quiescent pancreatic cancer cells. Cancer Res. 2005;65:9164–9168. doi: 10.1158/0008-5472.CAN-05-0779. [DOI] [PubMed] [Google Scholar]
- 61.Ono H. Molecular mechanisms of hypothalamic insulin resistance. Int. J. Mol. Sci. 2019;20:1317. doi: 10.3390/ijms20061317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muddapu VR, Dharshini SAP, Chakravarthy VS, Gromiha MM. Neurodegenerative diseases—Is metabolic deficiency the root cause? Front. Neurosci. 2020;14:213. doi: 10.3389/fnins.2020.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Svensson PA, et al. Gene expression in human brown adipose tissue. Int. J. Mol. Med. 2011;27:227–232. doi: 10.3892/ijmm.2010.566. [DOI] [PubMed] [Google Scholar]
- 64.Jung SM, Sanchez-Gurmaches J, Guertin DA. Brown adipose tissue development and metabolism. Handb. Exp. Pharmacol. 2019;251:3–36. doi: 10.1007/164_2018_168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tremblay A, Royer MM, Chaput JP, Doucet E. Adaptive thermogenesis can make a difference in the ability of obese individuals to lose body weight. Int. J. Obes. 2013;37:759–764. doi: 10.1038/ijo.2012.124. [DOI] [PubMed] [Google Scholar]
- 66.Trayhurn P, Thurlby PL, James WP. A defective response to cold in the obese (obob) mouse and the obese Zucker (fafa) rat [proceedings] Proc. Nutr. Soc. 1976;35:133A. [PubMed] [Google Scholar]
- 67.Zhang W, Bi S. Hypothalamic regulation of brown adipose tissue thermogenesis and energy homeostasis. Front. Endocrinol. 2015;6:136. doi: 10.3389/fendo.2015.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boddy SL, et al. The gut microbiome: A key player in the complexity of amyotrophic lateral sclerosis (ALS) BMC Med. 2021;19:13. doi: 10.1186/s12916-020-01885-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rowin J, Xia Y, Jung B, Sun J. Gut inflammation and dysbiosis in human motor neuron disease. Physiol. Rep. 2017;5:e13443. doi: 10.14814/phy2.13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu, M.-R., Chou, T.-S., Huang, C.-Y. & Hsiao, J.-K. A potential probiotic- Lachnospiraceae NK4A136 group: Evidence from the restoration of the dietary pattern from a high-fat diet. Res. Sq. [Preprint]. 10.21203/rs.3.rs-48913/v1. (2020).
- 71.Ma L, et al. Spermidine improves gut barrier integrity and gut microbiota function in diet-induced obese mice. Gut Microbes. 2020;12:1–19. doi: 10.1080/19490976.2020.1832857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dupuis L, Loeffler JP. Neuromuscular junction destruction during amyotrophic lateral sclerosis: Insights from transgenic models. Curr. Opin. Pharmacol. 2009;9:341–346. doi: 10.1016/j.coph.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 73.Dadon-Nachum M, Melamed E, Offen D. The “dying-back” phenomenon of motor neurons in ALS. J. Mol. Neurosci. 2011;43:470–477. doi: 10.1007/s12031-010-9467-1. [DOI] [PubMed] [Google Scholar]
- 74.Walker AK, et al. Functional recovery in new mouse models of ALS/FTLD after clearance of pathological cytoplasmic TDP-43. Acta Neuropathol. 2015;130:643–660. doi: 10.1007/s00401-015-1460-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available upon reasonable request to the corresponding author, Dr Carmen M. Fernandez-Martos (cmfernandezm@sescam.jccm.es; Carmen.fernandez-martos@utas.edu.au), and the raw demultiplexed sequence data (amplicon reads) have been uploaded to the Sequence Read Archive (SRA) with BioProject accession number PRJNA886986 (https://www.ncbi.nlm.nih.gov/bioproject/886986). Additional material including source data is available online.