Abstract
Colonization of the gastric mucosa by Helicobacter pylori is the major cause of gastroduodenal pathologies in humans. Studying the outcome of the humoral immune response directed against this gastric pathogen may contribute substantially to vaccine development and to the improvement of diagnostic techniques based on serology. By using two-dimensional gel electrophoresis, 29 proteins from H. pylori G27 were identified which strongly react with sera derived from H. pylori-infected patients suffering from different gastroduodenal pathologies. These antigens were characterized by mass spectrometry and proved to correspond to products of open reading frames predicted by the H. pylori genome sequence. The comparison of the antigenic patterns recognized by these sera revealed no association of specific H. pylori antigens with antibodies in patients with particular gastroduodenal pathologies.
Helicobacter pylori is a spiral-shaped, microaerophilic, gram-negative microorganism which colonizes human gastric epithelial cell surfaces and the overlying mucous layer. Infection with H. pylori, which affects approximately 50% of the world's population, causes chronic gastric inflammation, which in most cases remains asymptomatic. However, 10% of the H. pylori carriers develop severe gastric illness such as gastric or duodenal ulcer, atrophic gastritis, antral adenocarcinoma, or mucosa-associated lymphoid tissue (MALT) lymphoma. Therefore, infection by H. pylori causes a major health problem worldwide, especially in developing countries, where infection rates of >90% are encountered (13).
Several factors associated with the pathogenesis of H. pylori have been characterized so far, including flagella (18, 32); urease, which probably enables H. pylori to survive in the acidic environment of the stomach (9); an adhesin binding to the Lewis b blood group antigen (22); and the vacuolating cytotoxin VacA (3). In vitro VacA induces the formation of large acidic vacuoles in a number of eukaryotic cells (19). Furthermore, a 40-kb pathogenicity island (PAI) named cag has been identified in a subset of strains (1, 6). Based on the presence of the cag PAI, the H. pylori isolates are subdivided into two types. Type I strains, containing the cag PAI, exhibit increased virulence, since they are predominantly associated with severe gastric disease, while type II strains, lacking the cag PAI, are more frequently isolated from asymptomatic carriers. It has been demonstrated that some of the proteins encoded by the cag PAI trigger severe inflammatory responses in the host (6). However, the precise function of the gene products of the cag PAI and their role in virulence remain to be elucidated.
Pharmaceutical therapy to treat the H. pylori infection involves expensive combinations of various antibiotics, proton pump inhibitors, and bismuth compounds but shows only a limited efficacy (of approximately 80 to 90%) and does not prevent reinfection after successful eradication. In addition, H. pylori strains resistant to the most potent antibiotics used in the treatment of H. pylori infections, metronidazole and clarithromycin, are emerging rapidly (5). Considering further that the number of infected people worldwide requiring treatment is far beyond the reach of the antibiotic triple therapy, development of a vaccine seems to be the only suitable approach for the global control of H. pylori infection. It has been shown by various researchers that in animal models of infection protective immunity can be achieved by the coadministration of an appropriate mucosal adjuvant and various H. pylori antigens, either separately or in combination, via the orogastric route. The protective antigens identified include the urease; VacA; CagA, the immunodominant marker protein for the presence of the cag PAI; catalase; and HspA and HspB, the H. pylori homologs of the heat shock proteins GroES and GroEL (14, 24, 28, 30). In particular, the H. pylori urease gave rise to a high degree of protective immunity in vaccinated animals, and it was reported that 100% protection in H. pylori-challenged mice could be achieved by the administration of urease via a live carrier Salmonella strain expressing recombinant H. pylori subunits A and B (17). Furthermore, it has been demonstrated that therapeutic vaccination with recombinant VacA and CagA eradicates a chronic H. pylori infection in mice, demonstrating that the inability of the natural immune response to clear H. pylori infection can be overcome (16).
Considering the advantage of an efficacious vaccine, it is important to identify the H. pylori proteins which elicit a strong immune response in humans in order to analyze their capability to confer protective immunity. Furthermore, the identification and characterization of immunodominant proteins will contribute to the improvement of serological tests for detecting and monitoring H. pylori infections. Another important question is whether there exists a correlation between the presence of antibodies directed against specific H. pylori antigens and the particular H. pylori-associated gastroduodenal pathology from which a patient is suffering. In the present study, we used the proteome technology to identify common patterns of H. pylori antigens which are recognized by sera from patients showing various gastroduodenal pathologies.
Identification of immunogenic proteins of H. pylori by the proteome technology.
H. pylori G27 (36) was grown on Columbia agar plates containing 5% horse blood and 0.2% cyclodextrin as described previously (4). The bacteria were harvested from the plates, washed with phosphate-buffered saline, and lysed by incubation in lysis buffer (35 mM Tris, 9 M urea, 65 mM dithiothreitol, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS]) for 10 min at room temperature. Two-dimensional (2D) gel electrophoresis was performed by the method of O'Farrell (27), modified by Hochstrasser et al. (20, 21). Protein samples containing up to 200 μg of protein were subjected to isoelectric focusing (IEF) in a pH gradient ranging from pH 4 to pH 8. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed with pairs of identical IEF samples, and the gels were further processed in parallel by silver staining or immunoblotting using either control sera derived from five individuals identified H. pylori negative by serological tests (23) or the sera from 16 H. pylori-infected persons suffering from different gastroduodenal pathologies. These sera were collected at the Universitätsklinik Würzburg, Würzburg, Germany, the Hospital Calderon Guardia, San Jose, Costa Rica, and the Hospital Max Peralta, Cartago, Costa Rica, and were taken from patients identified as H. pylori positive by endoscopy and suffering from gastritis (AR, KE, BB, EE, MM, and SR), gastric or duodenal ulcer (CR3/4, CR6/10, CR7, CR9/11, and CR5/8), gastric cancer (18129, CR15, CR16, and CR19) or MALT lymphoma (H6031). Immunogenic H. pylori proteins identified in this way were eluted from preparative gels stained with colloidal Coomassie blue (26) and analyzed by digestion with trypsin followed by LC-mass spectrometry (MS). Briefly, the Coomassie blue-stained protein bands were precisely excised from the acrylamide gel, cut into small cubes, and rinsed several times with water (100 μl) for 15 to 30 min each. The gel pieces were washed three times with 100 μl of acetonitrile-water (1:1) for 10 to 20 min. To shrink the gel and extract residual water, pure acetonitrile was added for 10 min. The acetonitrile was removed, and 30 to 50 μl of digestion buffer (50 mM N-methylmorpholine [pH 8.1]) as well as trypsin (0.5 μg) were added. Digestion was performed at 37°C for 6 to 12 h. The supernatant containing the resulting peptides was recovered, and the gel pieces were extracted twice with 0.1% trifluoroacetic acid (20 to 30 min). The volume of the combined extracts was reduced to 5 μl in a Speed-Vac concentrator. LC-MS and collision-induced fragmentation (CID) spectra were recorded on a Finnigan LCQ ion trap mass spectrometer equipped with an electrospray ionization source. Grouping of fragment ion (CID) spectra originated from the same precursor ion, and cross-correlation analysis of the data was performed by using the Sequest program (10). The Sequest algorithm compares the measured fragment ion spectra of all selected peptides to the predicted spectra of tryptic peptides contained in the database and exhibiting the same molecular weight. Identification of multiple peptides derived from the same protein and evaluation of their cross-correlation scores results in unambiguous identification of the protein. The 2,591 database entries for peptide searches were created by selecting all annotated and predicted H. pylori proteins from the composite OWL protein sequence database (version 30.2).
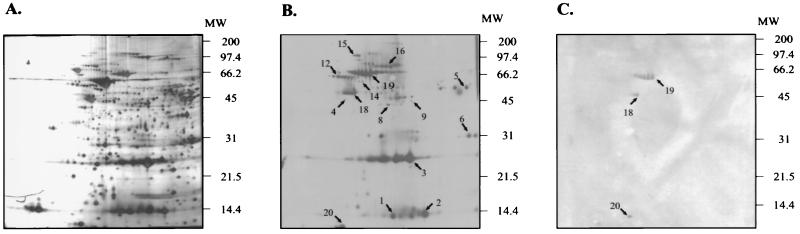
Figure 1 shows a typical 2D map of H. pylori proteins as well as the immunoblots obtained with an H. pylori-negative control serum sample (N1) and a serum sample taken from a H. pylori carrier with chronic gastritis (AR). In total, five control sera were tested individually. These sera showed a low level of cross-reactivity with H. pylori proteins, and the most prominent hybridizing spots were identified as HspB, the H. pylori homolog of the heat shock protein GroEL (spot 19, present in two out of five sera); the translation elongation factor EF-Tu (spot 18, present in two out of five sera); the ribosomal protein L7/L12 (spot 20, present in three out of five sera); the outer membrane protein 18 (spot 17, present in two out of five sera); flagellin A (spot 12, present in three out of five sera); and the urease subunit B (spot 16, present in one out of five sera). Spots 17, 18, 19, and 20 were also detected when an antiserum directed against whole-cell lysates of Escherichia coli was used in the immunoblot analysis (data not shown). Using the sera from 16 H. pylori carriers in individual experiments, a total of 120 hybridizing spots were detected, 29 of which could be unambiguously correlated to proteins in the 2D maps obtained by silver staining. These proteins were analyzed by LC-MS, and the peptide mass peaks obtained in all cases matched with the products of open reading frames (ORFs) predicted by the H. pylori genome sequence (33). The identified proteins are listed in Table 1, and their position in the H. pylori 2D protein map is shown in Fig. 2. Several proteins associated with the ability of H. pylori to colonize the gastric mucosa were detected, like the urease B subunit; the major flagellin, FlaA; and a flagellar hook-associated protein, FliD. However, most of the immunogenic proteins proved to be housekeeping enzymes involved in energy metabolism (pyruvate ferredoxin oxidoreductase, isocitrate dehydrogenase, ATP synthase F1, 3-oxoadipate coenzyme A transferase, aconitase B, and hydrogenase expression/formation protein), amino acid biosynthesis (aspartate ammonia-lyase and glutamine synthase), fatty acid and phospholipid metabolism (biotin carboxyl carrier protein), general cellular processes (bacterioferritin, trigger factor, alkyl hydroperoxide reductase, cochaperone HspA, HspB, the 70-kDa chaperone DnaK, hemolysin secretion protein, and the ATP-dependent protease binding subunit), DNA recombination and repair (RecA protein) and translation (elongation factors EF-P, EF-Tu, and EF-Ts and ribosomal protein L7/L12). A single antigen associated with the cell envelope, peptidoglycan-associated lipoprotein, could be detected. The peptide mass peaks of two proteins matched with ORF products HP0318 and HP1037, which are annotated as hypothetical proteins of unknown function in the genomic sequence of H. pylori strain 26695 (33) and which show the typical features of cytoplasmic proteins. It should be noted that the ORF product (JHP387) corresponding to HP1037 in the recently published genome sequence of the unrelated H. pylori isolate J99 was annotated as proline peptidase (2). Therefore, with the exception of the essential colonization factors UreB, FlaA, and FliD, no antigenic proteins which obviously could contribute to the pathogenic potential of H. pylori could be detected. It has been reported that purified NapA is able to activate the increased expression of CD11b/CD18 in human neutrophils (11); however, it is unclear whether this observation has in vivo relevance, since NapA was recently shown to belong to the class of bacterioferritins (12). The fact that the detected immunodominant proteins represent mainly housekeeping functions may indicate a general limitation of our approach, since only proteins which are abundant in the cell could be detected. Furthermore, the scope of antigens detectable by the proteome technology is restricted by the pH gradient applied in IEF, which ranged from pH 4 to 8 in our experiments, as well as by the resolution of the 2D gels. For example, antibodies to the CagA protein, which was shown previously to be the immunodominant marker protein for the presence of the cag PAI (7, 15), could not be detected for that reason, since its pI (9.6) is beyond the pH range of our IEF gels. Comparable observations were made in a similar study by McAtee et al. (25), who reported the characterization of 22 immunogenic proteins by hybridization of H. pylori whole-cell lysates with a serum pool from 14 H. pylori-positive individuals, 9 of which are identical to antigens detected in this study. However, these limitations might partially be overcome, if whole-cell lysates are size fractionated prior to their application to IEF gels in order to increase the total amount of protein to be analyzed, and if immobilized pH gradients are used in IEF to provide a broader range of resolution, particularly at higher pH. Of the 29 identified immunogenic proteins, only 1, peptidoglycan-associated lipoprotein precursor (Omp18, HP1125), is a component of the outer membrane and therefore meets the theoretical demands for a vaccine antigen; however, it showed cross-reactivity with two of the five H. pylori-negative control sera as well as with the anti-E. coli antiserum (data not shown). However, it is believed that urease and HspA, which are generally considered cytoplasmic proteins but confer protective immunity in animal models of H. pylori infection, become associated with the bacterial surface after lysis of a subpopulation of H. pylori cells (29); therefore, this might also be the case for other cytoplasmic H. pylori antigens, which could then be considered vaccine candidates.
FIG. 1.
2D map (pH 4 to 8) of a whole-cell lysate of H. pylori G27 and identification of H. pylori antigens by immunoblot analysis. A 100-μg portion of a whole-cell lysate of H. pylori G27 was loaded onto the IEF gels. Identified proteins are indicated by the spot numbers given in Table 1. The positions of molecular weight (MW) standards are indicated on the right. (A) Silver stain of a typical 2D gel. (B) Western blot of a duplicate 2D gel hybridized with serum AR, which is derived from an H. pylori-infected individual suffering from gastritis. Western blots were developed using the enhanced chemiluminescence detection system (Amersham). (C) Western blot of a duplicate 2D gel hybridized with a control serum from an H. pylori-negative individual (N1).
TABLE 1.
Identification of immunogenic H. pylori proteins by immunoblotting and LC-MSa
| Spot no. | Antigen | HP no. | MW (103) | pI |
|---|---|---|---|---|
| 1 | Neutrophil-activating protein (NapA) (bacterioferritin) | HP0243 | 16 | 5.9 |
| 2 | Cochaperone HspA (GroES) | HP0011 | 13 | 6.6 |
| 3 | Alkyl hydroperoxide reductase | HP1563 | 22 | 6.3 |
| 4 | DNA-directed RNA polymerase, α subunit | HP1293 | 38 | 4.8 |
| 5 | Isocitrate dehydrogenase | HP0027 | 47 | 7.9 |
| 6 | Conserved hypothetical protein | HP0318 | 28 | 7.7 |
| 7 | Translation elongation factor EF-P | HP0177 | 20 | 5.4 |
| 8 | RecA protein | HP0153 | 38 | 5.8 |
| 9 | Translation elongation factor EF-Ts | HP1555 | 39 | 6.5 |
| 10 | Pyruvate ferredoxin oxidoreductase, α subunit | HP1110 | 45 | 5.8 |
| 11 | Aspartate ammonia-lyase | HP0649 | 51 | 6.8 |
| 12 | Flagellin A (FlaA) | HP0601 | 53 | 6.3 |
| 13 | 70-kDa chaperone (DnaK) | HP0109 | 68 | 4.8 |
| 14 | Trigger factor | HP0795 | 49 | 5.2 |
| 15 | Flagellar hook-associated protein 2 (FliD) | HP0752 | 74 | 5.1 |
| 16 | Urease B subunit (UreB) | HP0072 | 63 | 5.9 |
| 17 | Peptidoglycan-associated lipoprotein precursor (Omp18) | HP1125 | 20 | 5.3 |
| 18 | Translation elongation factor EF-Tu | HP1205 | 44 | 5.0 |
| 19 | Chaperone and heat shock protein HspB (GroEL) | HP0010 | 60 | 5.6 |
| 20 | Ribosomal protein L7/L12 | HP1199 | 14 | 5.0 |
| 21 | ATP synthase F1, subunit β | HP1132 | 52 | 5.2 |
| 22 | Biotin carboxyl carrier protein | HP0371 | 17 | 5.4 |
| 23 | Hydrogenase expression/formation protein | HP0900 | 27 | 5.3 |
| 24 | Glutamine synthetase | HP0512 | 53 | 6.1 |
| 25 | Hemolysin secretion protein precursor | HP0599 | 48 | 6.2 |
| 26 | Hypothetical protein | HP1037 | 39 | 6.1 |
| 27 | 3-oxoadipate coenzyme A-transferase subunit A | HP0691 | 25 | 5.9 |
| 28 | Aconitase B | HP0779 | 94 | 6.5 |
| 29 | ATP-dependent protease binding subunit (ClpB) | HP0264 | 94 | 6.3 |
Theoretical molecular weight and isoelectric point were calculated from the ORFs annotated in the genomic sequence of H. pylori 26695 (33). Note that ORF products corresponding to HP0599 and HP1037 were annotated as methyl-accepting chemotaxis protein (JHP546; 97% pairwise protein identity to HP0599) and proline peptidase (JHP387; 96.4% pairwise protein identity to HP1037) in the genomic sequence of the unrelated H. pylori strain J99 (2).
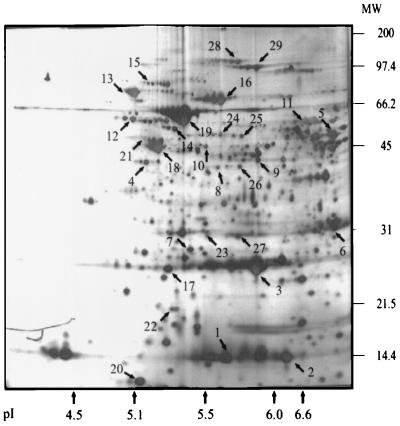
FIG. 2.
Silver-stained 2D map of a whole-cell lysate of H. pylori G27 showing the positions of the 29 identified immunogenic proteins listed in Table 1. The positions of molecular weight (MW) standards are indicated on the right. Isoelectric points are indicated at the bottom of the figure and were obtained by running a 2D sodium dodecyl sulfate-polyacrylamide gel electrophoresis standard under conditions identical to those applied for sample separation.
Comparison of antigenic patterns of H. pylori lysates obtained with different patient sera.
The H. pylori-positive sera used in this study were taken from patients suffering from chronic gastritis (six sera), peptic ulcer (five sera), gastric cancer (four sera), or MALT lymphoma (one serum). The antigenic patterns which were obtained when the different sera were hybridized to whole-cell protein lysates of H. pylori G27 in individual immunoblot experiments were compared with each other for the presence of a subset of 20 of the previously analyzed 29 spots which could be easily identified according to the electrophoretic mobility of the corresponding proteins. The results of this comparison are listed in Table 2 and demonstrate a highly variable humoral immune response in the 16 individuals under investigation. A single antigen, the flagellin A antigen, was recognized by all the sera tested, while antibodies against a second component of the flagella, flagellar hook-associated protein 2, and the chaperone GroEL were present in 14 sera. Several other antigens, i.e., urease subunit B, trigger factor, neutrophil-activating protein, alkyl hydroperoxide reductase, pyruvate ferredoxin oxidoreductase, EF-Tu, ribosomal protein L7/L12, Omp18, and the DnaK protein, reacted with about two-thirds of the analyzed sera, while EF-Ts, isocitrate dehydrogenase, and aspartate ammonia-lyase were recognized less frequently. However, it should be noted that FlaA, UreB, GroEL, EF-Tu, ribosomal protein L7/L12, and Omp18 also reacted with a low frequency with sera from H. pylori-negative individuals (Table 2).
TABLE 2.
Comparison of the antigenic patterns obtained with sera from 16 H. pylori-infected individuals based on a subset of 20 selected antigensa
| G27 spot no. | Antigenic pattern of sera from patients with:
|
Frequencyb | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No infection (control)
|
Gastritis
|
Gastric or duodenal ulcer
|
Gastric cancer
|
MALT (H6031) | ||||||||||||||||||
| N1 | N2 | N3 | N4 | N5 | AR | BB | EE | KE | MM | SR | CR 3/4 | CR 5/8 | CR 6/10 | CR 7 | CR 9/11 | 18129 | CR 15 | CR 16 | CR 19 | |||
| 1 | − | − | − | − | − | + | + | + | + | + | (+) | + | + | − | − | − | + | − | − | − | + | 10 |
| 2 | − | − | − | − | − | + | − | + | + | + | − | + | − | − | − | + | − | − | − | − | + | 7 |
| 3 | − | − | − | − | − | + | − | + | (+) | + | − | − | + | − | + | − | − | (+) | + | + | − | 9 |
| 4 | − | − | − | − | − | + | − | − | + | − | − | + | + | − | − | − | + | (+) | − | − | (+) | 7 |
| 5 | − | − | − | − | − | + | − | − | + | − | − | − | − | − | − | + | − | − | − | − | − | 3 |
| 6 | − | − | − | − | − | + | − | − | + | + | − | + | − | − | − | + | + | + | − | − | + | 8 |
| 7 | − | − | − | − | − | − | − | − | + | + | − | − | − | (+) | − | − | + | − | + | − | − | 5 |
| 8 | − | − | − | − | − | + | − | − | − | + | + | + | + | − | − | − | (+) | − | − | − | − | 6 |
| 9 | − | − | − | − | − | + | − | − | − | − | − | − | − | + | − | − | − | − | − | − | + | 3 |
| 10 | − | − | − | − | − | + | (+) | + | + | − | − | + | + | − | − | + | + | − | − | − | + | 9 |
| 11 | − | − | − | − | − | − | + | − | + | − | − | + | + | − | − | − | − | − | − | − | − | 4 |
| 12 | − | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 16 |
| 13 | − | − | − | − | − | + | + | + | − | − | + | + | + | + | − | + | − | − | − | − | + | 9 |
| 14 | − | − | − | − | − | + | + | − | + | + | − | + | + | − | + | + | + | − | − | (+) | + | 11 |
| 15 | − | − | − | − | − | + | + | + | + | + | − | + | + | + | + | + | + | − | + | (+) | + | 14 |
| 16 | − | − | − | + | − | + | + | + | + | + | + | + | − | + | − | (+) | − | + | (+) | − | − | 11 |
| 17 | − | + | − | − | + | − | + | − | − | + | − | + | − | + | + | + | + | + | − | − | + | 9 |
| 18 | + | − | − | + | − | + | − | − | − | + | + | − | + | − | + | + | + | + | − | − | + | 9 |
| 19 | + | − | + | − | − | + | + | + | + | + | + | + | + | − | + | + | + | − | + | + | + | 14 |
| 20 | + | − | − | + | + | + | + | − | + | + | − | + | − | − | − | + | + | + | − | − | + | 9 |
Sera were obtained from patients suffering from gastritis (sera AR, BB, EE, KE, MM, and SR), gastric or duodenal ulcer (sera CR3/4, CR5/8, CR6/10, CR7, and CR9/11), gastric cancer (sera 18129, CR15, CR16, and CR19) or MALT lymphoma (serum H6031) and from five H. pylori-negative individuals (N1 to N5). Spot numbers of antigens are as given in Table 1. Parentheses indicate a low degree of reactivity.
Detection frequency of a given antigen by the 16 H. pylori-positive sera under investigation.
The identification of highly immunogenic H. pylori proteins is a prerequisite for the development of serological test kits based on recombinant antigens. Along with the urea breath test, serology is a noninvasive method for the rapid diagnosis of H. pylori infection, and several serological test kits are commercially available which are based mainly on pooled H. pylori antigens. The defined composition of such a test kit should greatly increase both its sensitivity and its specificity. Although the humoral immune response to H. pylori seems to be quite variable, combinations of frequently recognized antigens could prove useful for diagnostic purposes. The metabolic enzymes identified here seem to be good candidates to be included into a serological test kit, since they (i) do not cross-react with sera from noninfected individuals and (ii) should, according to their housekeeping function, be highly conserved between different H. pylori strains irrespective of their classification as type I or type II. In contrast, antigens associated with H. pylori virulence, such as CagA or VacA, show quite a high degree of sequence variability (8, 34, 35, 37) or might be subject to phase variation as a consequence of slipped-strand mispairing, as suggested for several H. pylori outer membrane proteins (31, 33).
The comparison of the antigenic patterns of whole-cell lysates of H. pylori G27 obtained by hybridization with sera from patients suffering from different gastroduodenal pathologies revealed no correlation between the presence of a particular antibody and the state of disease, at least on the basis of the subset of 20 antigens under investigation (Table 2). This is not surprising because, as mentioned above, with the exception of UreB and FlaA, none of the immunogenic proteins identified in this study is clearly correlated to H. pylori virulence and could therefore account for different clinical outcomes of infection. Although the proteome technique proved to be a useful approach for the identification of immunogenic proteins, the detection of marker antigens correlated with a particular gastroduodenal pathology could have failed for several reasons. (i) Putative virulence factors such as the gene products of the cag PAI, which are expressed in the type I strain G27, are obviously produced in amounts which are below the detection limit of the applied method. (ii) Factors contributing to virulence may not be expressed under in vitro culture conditions, but their expression may depend on certain in vivo stimuli, for example cell-to-cell contact. (iii) It is generally assumed that genetic factors of both the bacterium and the host contribute to the clinical outcome of H. pylori infection. H. pylori G27 is a type I isolate which is adapted to laboratory growth conditions and might therefore have lost virulence traits due to prolonged in vitro passages. Furthermore, nothing is known about the way in which the course of the chronic H. pylori infection influences the immune response. Therefore, to profoundly address whether marker antigens correlated with specific states of H. pylori-associated disease do exist, a large number of human sera together with the infecting strain have to be analyzed. Currently, our study is being extended to the analysis of the antigenic patterns of fresh clinical H. pylori isolates obtained with the sera of their respective human hosts.
Acknowledgments
We thank Fernando Garcia, Uwe Gross, Albert Haas, and Wolfgang Scheppach for providing human sera. Vincenzo Scarlato is acknowledged for critical reading of the manuscript.
D.B. is a recipient of a postdoctoral fellowship from the Deutsche Krebsforschungszentrum. This study was supported by a grant from the BMBF (BMBF 01 KI9608 TP2) to W.G. and R.G.
REFERENCES
- 1.Akopyants N S, Clifton S W, Kersulyte D, Crabtree J E, Youree B E, Reece C A, Bukanov N O, Drazek E S, Roe B A, Berg D E. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol. 1998;28:37–53. doi: 10.1046/j.1365-2958.1998.00770.x. [DOI] [PubMed] [Google Scholar]
- 2.Alm R A, Ling L-S L, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 3.Atherton J C, Cao P, Peek R M, Jr, Tummuru M K R, Blaser M J, Cover T L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori: association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 4.Beier D, Spohn G, Rappuoli R, Scarlato V. Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J Bacteriol. 1997;179:4676–4683. doi: 10.1128/jb.179.15.4676-4683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckley M J M, Deltenre M. Therapy of H. pylori infection. Curr Opin Gastroenterol. 1997;13:56–62. [Google Scholar]
- 6.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, Rappuoli R. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cover T L, Tummuru M K R, Cao P, Thompson S A, Blaser M J. Divergence of genetic sequences for vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- 9.Cussac V, Ferrero R L, Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J Bacteriol. 1992;174:2466–2473. doi: 10.1128/jb.174.8.2466-2473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eng J K, McCormak A L, Yates J R., Jr An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 11.Evans D J, Jr, Evans D G, Takemura T, Nakano H, Lampert H C, Graham D Y, Granger D N, Kvietys P R. Characterization of a Helicobacter pylori neutrophil-activating protein. Infect Immun. 1995;63:2213–2220. doi: 10.1128/iai.63.6.2213-2220.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans D J, Jr, Evans D G, Lampert H C, Nakano H. Identification of four new prokaryotic bacterioferritins, from Helicobacter pylori, Anabaena variabilis, Bacillus subtilis and Treponema pallidum, by analysis of gene sequences. Gene. 1995;153:123–127. doi: 10.1016/0378-1119(94)00774-m. [DOI] [PubMed] [Google Scholar]
- 13.Feldman R A, Eccersley A J, Hardie J M. Epidemiology of Helicobacter pylori: acquisition, transmission, population prevalence and disease-to-infection ratio. Br Med Bull. 1998;54:39–53. doi: 10.1093/oxfordjournals.bmb.a011678. [DOI] [PubMed] [Google Scholar]
- 14.Ferrero R L, Thiberge J-M, Kansau I, Wuscher N, Huerre M, Labigne A. The GroES homolog of Helicobacter pylori confers protective immunity against mucosal infection in mice. Proc Natl Acad Sci USA. 1995;92:6499–6503. doi: 10.1073/pnas.92.14.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerstenecker B, Eschweiler B, Vogele H, Koch H K, Hellerich U, Kist M. Serodiagnosis of Helicobacter pylori infections with an enzyme immunoassay using the chromatographically purified 120 kilodalton protein. Eur J Clin Microbiol Infect Dis. 1992;11:595–601. doi: 10.1007/BF01961665. [DOI] [PubMed] [Google Scholar]
- 16.Ghiara P, Rossi M, Marchetti M, Di Tommaso A, Vindigni C, Ciampolini F, Covacci A, Telford J L, De Magistris M T, Pizza M, Rappuoli R, Del Giudice G. Therapeutic intragastric vaccination against Helicobacter pylori in mice eradicates an otherwise chronic infection and confers protection against reinfection. Infect Immun. 1997;65:4996–5002. doi: 10.1128/iai.65.12.4996-5002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez-Duarte O G, Lucas B, Yan Z-X, Panthel K, Haas R, Meyer T F. Protection of mice against gastric colonization by Helicobacter pylori by single oral dose immunization with attenuated Salmonella typhimurium producing urease subunits A and B. Vaccine. 1998;16:460–471. doi: 10.1016/s0264-410x(97)00247-8. [DOI] [PubMed] [Google Scholar]
- 18.Haas R, Meyer T F, van Putten J P M. Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using shuttle mutagenesis. Mol Microbiol. 1993;8:753–760. doi: 10.1111/j.1365-2958.1993.tb01618.x. [DOI] [PubMed] [Google Scholar]
- 19.Harris P R, Cover T L, Crowe D R, Orenstein J M, Graham M F, Blaser M J, Smith P D. Helicobacter pylori cytotoxin induces vacuolation of primary human mucosal epithelial cells. Infect Immun. 1996;64:4867–4871. doi: 10.1128/iai.64.11.4867-4871.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochstrasser D F, Augsburger V, Funk M, Appel R, Pelegrini C, Müller A F. Immobilized pH gradients in capillary tubes and two-dimensional gel electrophoresis. Electrophoresis. 1986;7:505–511. [Google Scholar]
- 21.Hochstrasser D F, Harrington M G, Hochstrasser A C, Miller M J, Merril C R. Methods for increasing the resolution of two-dimensional protein electrophoresis. Anal Biochem. 1988;173:424–435. doi: 10.1016/0003-2697(88)90209-6. [DOI] [PubMed] [Google Scholar]
- 22.Ilver D, Arnqvist A, Ögren J, Frick I-M, Kersulyte D, Incecik E T, Berg D E, Covacci A, Engstrand L, Boren T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 23.Karvar S, Karch H, Frosch M, Burghardt W, Gross U. Use of serum-specific immunoglobulins A and G for detection of Helicobacter pylori infection in patients with chronic gastritis by immunoblot analysis. J Clin Microbiol. 1997;35:3058–3061. doi: 10.1128/jcm.35.12.3058-3061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchetti M, Rossi M, Gianelli V, Giuliani M M, Pizza M, Censini S, Covacci A, Massari P, Pagliaccia C, Manetti R, Telford J L, Douce G, Dougan G, Rappuoli R, Ghiara P. Protection against Helicobacter pylori infection in mice by intragastric vaccination with H. pylori antigens is achieved using a non-toxic mutant of E. coli heat-labile enterotoxin (LT) as adjuvant. Vaccine. 1998;16:33–37. doi: 10.1016/s0264-410x(97)00153-9. [DOI] [PubMed] [Google Scholar]
- 25.McAtee C P, Lim M Y, Fung K, Velligan M, Fry K, Chow T, Berg D E. Identification of potential diagnostic and vaccine candidates of Helicobacter pylori by two-dimensional gel electrophoresis, sequence analysis, and serum profiling. Clin Diagn Lab Immunol. 1998;5:537–542. doi: 10.1128/cdli.5.4.537-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 27.O'Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 28.Pappo J, Thomas W D, Jr, Kabok Z, Taylor N, Murphy J C, Fox J G. Effect of oral immunization with recombinant urease on murine Helicobacter felis gastritis. Infect Immun. 1995;63:1246–1252. doi: 10.1128/iai.63.4.1246-1252.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phadnis S H, Parlow M H, Levy M, Ilver D, Caulkins C M, Connors J B, Dunn B E. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect Immun. 1996;64:905–912. doi: 10.1128/iai.64.3.905-912.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radcliff F J, Hazell S L, Kolesnikow T, Doidge C, Lee A. Catalase, a novel antigen for Helicobacter pylori vaccination. Infect Immun. 1997;65:4668–4674. doi: 10.1128/iai.65.11.4668-4674.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saunders N J, Peden J F, Hood D W, Moxon E R. Simple sequence repeats in the Helicobacter pylori genome. Mol Microbiol. 1998;27:1091–1098. doi: 10.1046/j.1365-2958.1998.00768.x. [DOI] [PubMed] [Google Scholar]
- 32.Suerbaum S, Josenhans C, Labigne A. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J Bacteriol. 1993;175:3278–3288. doi: 10.1128/jb.175.11.3278-3288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 34.van der Ende A, Pan Z-J, Bart A, van der Hulst R W M, Feller M, Xiao S-D, Tytgat G N J, Dankert J. cagA-positive Helicobacter pylori populations in China and The Netherlands are distinct. Infect Immun. 1998;66:1822–1826. doi: 10.1128/iai.66.5.1822-1826.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Doorn L J, Figueiredo C, Rossau R, Jannes G, van Asbroeck M, Sousa J C, Carneiro F, Quint W G V. Typing of Helicobacter pylori vacA gene and detection of cagA gene by PCR and reverse hybridization. J Clin Microbiol. 1998;36:1271–1276. doi: 10.1128/jcm.36.5.1271-1276.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang Z, Censini S, Bayelli P F, Telford J L, Figura N, Rappuoli R, Covacci A. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect Immun. 1995;63:94–98. doi: 10.1128/iai.63.1.94-98.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaoka Y, Kodama T, Kashima K, Graham D Y, Sepulveda A R. Variants of the 3′ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J Clin Microbiol. 1998;36:2258–2263. doi: 10.1128/jcm.36.8.2258-2263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]