Abstract
Malus spectabilis ‘Duojiao’ is a spontaneous delayed-green leaf color mutant of M. spectabilis ‘Riversii’ and has chloroplasts with irregularly arranged vesicles and indistinct stromal lamellae. The yellow leaves of mutant have less chlorophyll (Chl), carotenoids, and flavonoids. Measurement of photosynthetic gas exchange indicated that the mutant has lower photosynthetic activity than ‘Riversii’ plants. Transcriptome sequencing with the Illumina platform was used to characterize differences in gene expression between the leaves of plants with yellow and green colors and elucidate the molecular mechanisms responsible for variation in leaf color in ornamental crabapple. In the comparison group of mutant yellow leaves and the maternal green leaves, 1848 differentially significant expressed genes (DEGs) were annotated by transcriptomic analysis. Many DEGs and transcription factors were identified related to chloroplast development, Chl synthesis and degradation, photosynthesis, carotenoid biosynthesis, flavonoid biosynthesis and other pathways related to plant leaf color formation. Among these, the Chl biosynthesis-related coproporphyrinogen gene, oxidative decarboxylase gene, and Chl a oxygenase gene were down-regulated, indicating that Chl biosynthesis was blocked. GLK1, which regulates chloroplast development, was down-regulated in yellow leaves. Parallel experiments showed that the content of the Chl synthesis precursors, protoporphyrinogen IX, chlorophyllide a, and chlorophyllide b and the activity of chlorophyllogen III oxidase and chlorophyllide a oxygenase in the yellow leaves of ‘Duojiao’ were lower than those in the green leaves of ‘Riversii’. Thus, leaf color formation is greatly affected by Chl metabolism and chloroplast development. The reliability of the RNA-sequencing data was confirmed by quantitative real-time PCR analysis with 12 selected DEGs.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-022-01248-7.
Keywords: Ornamental crabapple , Delayed-green leaf color mutant , Pigment , Photosynthesis , Chloroplast ultrastructure , RNA-seq
Introduction
Leaf color is a phenotypic trait of plants that is affected by internal genetic factors and external environmental conditions and plays an important role in enhancing the aesthetics of urban landscapes (Zhao et al. 2020). Leaf color mutants in particular have received increased research attention. Some leaf color mutants have special excellent characteristics and can be used as a marker to simplify the breeding of improved varieties and the production of hybrid varieties. (Walles 1971; Yan et al. 2019; Zhao et al. 2010). Some leaf color mutants have advantageous properties that provide high-quality germplasm resources for genetic breeding (Gan and Amasino 1995); these mutants can also be used to analyze and characterize gene function (Hansson et al. 1999) and interactions (López-Juez et al. 1998), as well as for breeding (Reyes-Arribas et al. 2010). These mutants have thus been a major focus of plant epigenetics research and have received much attention from plant breeders (Sinkkonen et al. 2012). The new variety ‘Duojiao’ is a rare germplasm resource. The study of its leaf color characteristics has theoretical and practical significance, which can not only enrich the landscape hierarchy and diversity in China, but also provide high-quality resources for cultivating new varieties with independent property rights (Zhang et al. 2020).
Ornamental crabapples (Rosaceae: Malus Mill.) are the most popular small trees used as ornamentals in the temperate zone (Zhou et al. 2019). Applications of crabapple germplasm are still limited to traditional cultivars; there is thus a need to enhance the domestication of wild resources and the selection and breeding of new varieties (Wu et al. 2015). The new variety of delayed-green-leaved ‘Duojiao’ is a rare germplasm resource. Exploration of its leaf color traits has theoretical and practical significance, as such studies could help enrich landscapes and provide high-quality resources that could be used to cultivate new varieties.
Transcriptome sequencing (RNA-Seq) is an efficient, comprehensive and quick research method in molecular biology (Wang et al. 2009a, b). It can accurately and quickly obtain almost all the transcripts of the samples to be tested, provide comprehensive transcriptome information, and understand the gene function and gene structure of plants at a specific stage from the overall level. It can also study non-model plants lacking genome sequences at the omics level and find unknown transcripts (Kristiansson et al. 2009; Li et al. 2018); RNA-seq is also used to study chlorophyll (Chl) biosynthesis in plants such as Populus deltoides Marsh (Zhang et al. 2019b), hot peppers (Ma et al. 2016), and wheat (Wu et al. 2018).
Here, we conducted an experiment that combined cytological and physiological measurements and transcriptome sequencing to clarify the mechanism underlying the leaf coloration of a delayed-green leaf crabapple mutant (‘Duojiao’). The results of this study provide new insight that could aid the future use of this cultivar; the valuable molecular data provided by our study could also aid future studies of the coloring mechanism in other plant species. Understanding the mechanism of leaf coloration will help horticulturists cultivate ornamental trees better.
Materials and methods
Experimental materials
The delayed-green leaf mutant ‘Duojiao’ was obtained through a bud mutation in 2014 that occurred on the branch of ‘Riversii’ with green leaves showing normal growth at Zhendong Nursery (Tai’an, Shandong Province, China, 36° 15′ N, 117° 16′ E; temperate monsoon climate). Two-year-old plants of the cultivar Malus spectabilis ‘Riversii’ and its mutant M. spectabilis ‘Duojiao’ were used to clarify the mechanism underlying the yellow leaf coloration of ‘Duojiao’.
A total of 100 plants of ‘Duojiao’ and ‘Riversii’ with similar growth and tree strength were selected in the nursery in June 7th, 2019, and a large number of leaves on the new shoots were randomly selected and mixed, including the yellow leaves of ‘Duojiao’, yellow leaves with green spots of ‘Duojiao’, green leaves of ‘Duojiao’, and green leaves of ‘Riversii’. Three biological replicates were used when sampling the leaves for chloroplast ultrastructure observations and transcriptome sequencing. After harvest, partial samples were immediately frozen in liquid nitrogen and stored at − 80 °C for transcriptome sequencing, another part of the samples were cut into 1 cm×1 cm pieces and fixed overnight with 2.5% (v/v) glutaraldehyde (Solarbio).
The leaf color of the mutant delayed-green leaf crabapple changes throughout the growing season (Fig. 1); in our experiments, we examined changes in the color of delayed-green crabapple leaves at different time points in 2020. A total of 100 grafted seedlings were sampled at fixed leaf positions, and one new shoot was identified at the periphery of each tree canopy in four directions. The tenth leaves from the lower part of the new shoots of ‘Duojiao’ and ‘Riversii’ were used to investigate changes in the content of Chl, flavonoids, and Chl precursors and the activity of enzymes in the Chl biosynthesis pathway of the delayed-green leaf mutant; there were three biological replicates for each assay. The leaves of ‘Duojiao’ were yellow in May, yellow with green spots in June, light green in early July, and all green in August, September, and October. The leaves of ‘Riversii’ were a healthy green color, and biological characters were normal over the entire growth period. After harvest, samples were immediately frozen in liquid nitrogen and stored at − 80 °C.
Fig. 1.
Phenotypes of mutant leaves from the same shoot of ‘Duojao’ in June
Determination of chl, carotenoid, and flavonoid content
According to the method of Lichtenthaler (1983), fresh leaves of ‘Duojiao’ and ‘Riversii’ were collected in the growing season and used to measure the content of various pigments. Chl and carotenoids (Car) were extracted in the dark with 96% ethanol at room temperature for 24 h and then were calculated from an absorbance of 665 nm, 649 nm, and 470 nm in a visible-ultraviolet spectrophotometer (UV2600, SHIMADZU, Japan). The content of Chl a (mg/g), Chl b (mg/g), and Car (mg/g) was calculated using the equation of Gang (2019). The content of coproporphyrinogen III, protoporphyrinogen IX, chlorophyllide a, and chlorophyllide b and the activity of the key enzymes coproporphyrinogen oxidative decarboxylase and chlorophyllide a oxygenase were measured according to Wen et al. (2019). Specifically, 50 µl of standards of different concentration gradients and 50 µl of samples diluted 5 times were added with 100 µl of HRP-conjugate detection antibody, incubated at 37 °C for 60 min, then discarded and added 50 µl each of chromogen solution A and chromogen solution B. The samples were incubated at 37 °C for 15 min, protected from light, then 50 µl of termination solution was added and the absorbance values were measured at 450 nm within 15 min. The sample concentration was calculated according to the standard curve.
The flavonoid content was determined using an UV-2600 spectrophotometer at 510 nm according to the method described by Jia (1999). Briefly, 0.5 g of leaves were cut (removing the main veins of leaves) and ground in liquid nitrogen. Five ml of 65% pre-cooled ethanol was then added; extraction took place for 4 h at 4 °C in the dark, followed by centrifugation at 12,000 rpm for 20 min. Next, 0.5 ml of supernatant was placed in a test tube. After adding 0.5 ml of 5% NaNO3, 0.5 ml of 10% Al(NO3)3, and 2 ml of 4% NaOH, the solution was left to stand for 15 min; 65% ethanol was used as the blank control, and the absorbance was measured by Shimadzu UV-2600 ultraviolet spectrophotometer spectrophotometer at 510 nm. The mean from three measurements was used in analyses.
Ultrastructural observation of chloroplast
The chloroplasts ultrastructure of ‘Duojiao’ leaves and ‘Riversii’ leaves was observed by transmission electron microscopy (TEM) following the modified methods of Li (2016).
Tissue samples were cut into small pieces of 1 cm×1 cm, immersed in 2% (v/v) paraformaldehyde 2.5% (v/v) glutaraldehyde fixative (Solarbio) and pumped so that the leaf tissues were completely soaked by the fixative and fixed overnight at 4 °C. Then the pre-fixed samples were washed with 0.1 M phosphate buffer (pH 7.0) for three times for 15 min each, fixed with 1% (v/v) OsO4 fixative for 1–1.5 h, transferred to 0.1 M phosphate buffer (pH 7.0) and rinsed three times for 15 min each. The fixed samples were dehydrated in a series of concentration gradients (50%, 70%, 90%, 100%, v/v) of acetone solution for 15 min each time and finally in pure acetone (100%) three times for 15–20 min each time. At room temperature, the dehydrated samples were placed in the mixture of pure acetone and Epon812 resin (2:1, v/v) for 0.5 h, then placed in the mixture of pure acetone and Epon812 resin (1:2, v/v) for 1.5 h, and finally placed in the pure Epon812 resin for 24 h at 37 °C, 45 and 60 °C respectively. Sections were cut using a Reichert-Jung ULTRACUT Eultrathin sectioning machine (Austria). Uranyl acetate staining for 15 min and lead citrate for 15 min were done. The sections were observed and photographed with an electronic JEM 1200 electron microscope (JEOL Ltd., Tokyo, Japan).
Measurement of photosynthetic characteristics
Photosynthetic parameters were measured from the yellow and green leaves of ‘Duojiao’ and green leaves of ‘Riversii’ on July 16, August 14, and September 9, 2019, using a CIRAS-3 portable photosynthesizer (PP System, USA). Using the built-in LED light source, the light intensity was set to 1000–1200 µmol m−2 s−1, and the intercellular CO2 concentration (Ci), stomatal conductance (Gs), net photosynthetic rate (A) and transpiration rate (E) were read after the instrument was stable. All measurements were carried out between 9 and 11 am on a clear day. The means from the five measurements were used in analyses.
RNA isolation, transcriptome sequencing, and qRT-PCR analysis
Total RNA was extracted by using the FastPure® Plant Total RNA Isolation Kit (Polysaccharides and Polyphenolics-rich) (Vazyme Biotech Co., Ltd.) from tissues of ‘Duojiao’ and ‘Riversii’. The integrity, concentration, and purity of extracted RNA samples were examined using an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA). The construction and sequencing of cDNA library were completed by Annoroad Gene Technology Co. Ltd. (Beijing, China). The raw reads obtained by Illumina sequencing (Illumina HiSeq™ 2500 sequencer, San Diego, CA, USA) were filtered to obtain clean reads, and the subsequent analyses were based on clean reads. The process of sequencing information analysis is mainly divided into three main parts: sequencing data quality control, comparative data analysis, and transcriptome analysis (Kim et al. 2015). Differentially expressed genes (DEGs) were determined using the following criteria: |log2 Foldchange|≥1 and q < 0.05. Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) pathway enrichment analyses and Gene Ontology (GO) analysis (http://geneontology.org/) were then conducted to determine the biological functions of the DEGs.
The qRT-PCR unigene-specific primers (Table S1) for testing gene expression were designed using IDT (https://sg.idtdna.com/PrimerQuest) based on the nucleotide database of the National Center for Biotechnology Information. HiScript® III 1st Strand cDNA Synthesis Kit (Vazyme Biotech Co., Ltd.) was used to reverse-transcribe RNA into cDNA. The amplification reactions were run in a Gene9600 real-time PCR detection system (Bioer) with the following cycling parameters: 94 °C for 30 s, 94 °C for 40 cycles for 10 s, and 72 °C for 30 s. Actin was selected as the internal reference genefor the gene relative expression level determined using the 2−△△Ct method (Livak and Schmittgen 2002).
Statistical analysis
Statistical analyses were performed using SPSS 25.0 Windows software (SPSS Inc., Chicago, IL, USA). Independent-sample t-test and variance analysis were conducted after the normal distribution and variance homogeneity test of the data. The variables were log-transformed when necessary to ensure that residuals were normal. GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA, USA) and Microsoft Excel 97-2003 (Microsoft Corporation, Redmond, WA, USA) were to draw plots and tables.
Results
Phenotypic characteristics
The delayed-green leaf mutant of crabapple was derived from a bud mutation that occurred on the branch of a plant with green leaves showing normal growth. New, unfolded leaves of ‘Duojiao’ were initially reddish-yellow and then gradually lightened to a bright yellow with a high degree of glossiness. The tender leaves changed from yellow to yellowish-green, with irregular green spots in the center of the veins, and finally matured into dark green (Fig. 1). The characteristics of these foliage remained stable and consistent over six years of observation (Fig. 2).
Fig. 2.

Growth of M. spectabilis ‘Duojiao’
Content of different pigments in chl-deficient mutants
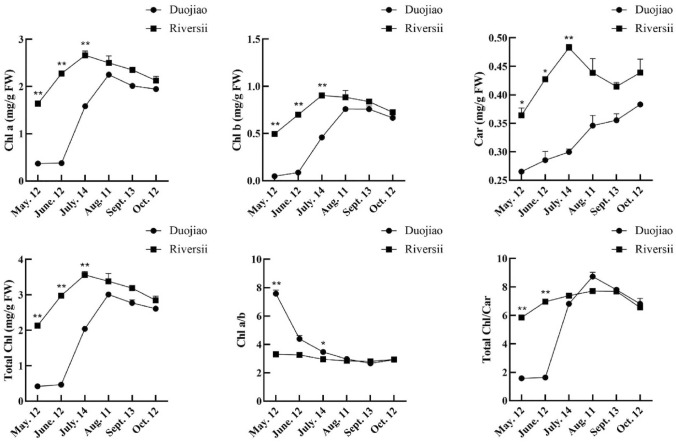
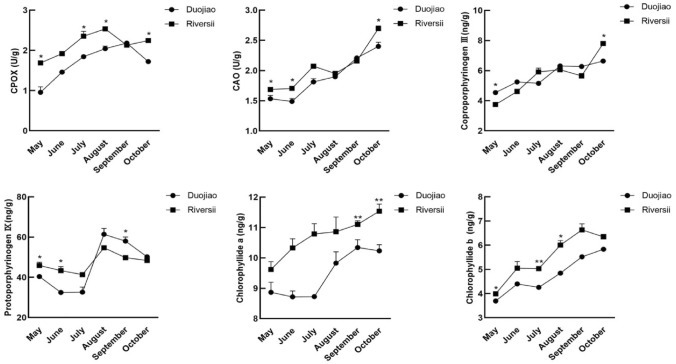
Next, we measured the content of various pigments of ‘Duojiao’ and ‘Riversii’. Specifically, we measured the Chl, Car, and flavonoid content from May to October. From May to August, the pigment content of Chl a, Chl b, and Car in the leaves of ‘Duojiao’, was increased (Fig. 3). However, all pigment content in the leaves of ‘Duojiao’ was lower than in the leaves of ‘Riversii’. In May, Chl a, Chl b, and carotenoid contents were 77%, 90%, and 27% lower in the yellow leaves of ‘Duojiao’ than in those of ‘Riversii’, respectively. Similarly, Chl a/b in the yellow leaves of ‘Duojiao’ was higher in May, June, and July compared with those of ‘Riversii’. The Chl/Car ratio in yellow leaves was markedly decreased. Parallel experiments showed that the contents of Chl synthesis precursors protoporphyrinogen IX, chlorophyllide a, and chlorophyllide b and the activity of chlorophyllogen III oxidase (CPOX) and chlorophyllide a oxygenase (CAO) were lower in the yellow leaf growth period of ‘Duojiao’ compared with the green leaves of ‘Riversii’. The content of coproporphyrinogen III was higher in ‘Duojiao’ than in ‘Riversii’ in May and June; differences in protoporphyrinogen IX, chlorophyllide a, and chlorophyllide b were also observed between ‘Duojiao’ and ‘Riversii’ (Fig. 4). Additionally, the total flavonoid content in ‘Duojiao’ was lower than that in ‘Riversii’ during the entire growth period (Fig. 5).
Fig. 3.
Determination of the pigment content in ‘Duojiao’ and ‘Riversii’ leaves. **, * indicate significant differences at P value < 0.01 and P value < 0.05, respectively, as determined by t-test
Fig. 4.
The content of chlorophyll biosynthetic precursors in the leaves of M. spectabilis ‘Riversii’ and M. spectabilis ‘Duojiao’. * indicate significant differences: *P value < 0.05, **P value < 0.01
Fig. 5.
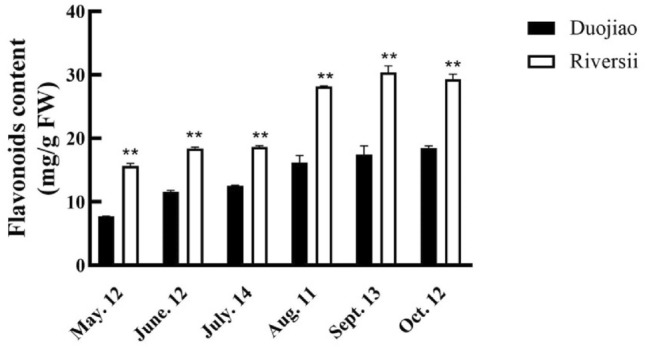
Determination of the flavonoid content in ‘Duojiao’ and ‘Riversii’ leaves. * indicate significant differences: *P value < 0.05, **P value < 0.01
Anatomical and photosynthetic changes in mutant leaves
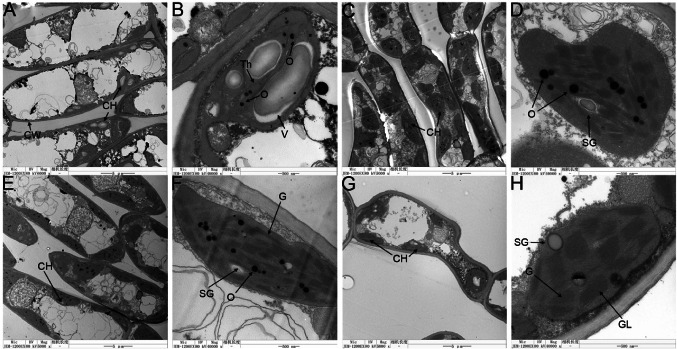
Next, we compared the ultrastructure of chloroplasts of ‘Riversii’ and ‘Duojiao’; as chloroplasts are the sites of photosynthesis, carbon fixation, and energy conversion (Douzery et al. 2004; Li et al. 2009; Jiang et al. 2012), changes in the anatomical structure of chloroplasts can affect photosynthetic and physiological activity. In the mesophyll cells of normal ‘Riversii’ green leaves, chloroplasts are full ellipsoidal or spherical, scattered in cells and arranged close to the cell membrane, with uniform grana distribution, well-developed thylakoids, orderly stacking and close arrangement. Their arrangement direction is parallel to the long axis of chloroplasts, and they contain starch without osmiophilic granules (Fig. 6G, H). In contrast, chloroplasts in etiolated leaves were clustered, scattered, and irregularly shaped; they also contained irregularly arranged vesicles and a greater number of osmiophilic granules. The thylakoid layer structure was destroyed (Fig. 6A, B), which was related to the reduction of Chl Zhu et al. (2014), Yang et al. (2015b), Zhang et al. (2017). The chloroplasts in the yellow leaves with green spots of ‘Duojiao’ were irregular in shape, contained more osmiophilic granules, had fewer and smaller starch granules, and had scattered grana (Fig. 6C, D). In the green leaves of ‘Duojiao’, chloroplasts were mostly arranged along the direction of the cell membrane and were regular in shape (mostly long ovals or spindle-shaped); however, they were not uniform in size, the grana lamellae of chloroplasts were neatly arranged but not highly stacked, and they were arranged parallel to the long axis of chloroplasts, with more osmiophilic granules and fewer, smaller starch granules (Fig. 6E, F).
Fig. 6.
Chloroplast ultrastructure of ‘Duojiao’ leaves and ‘Riversii’ leaves. A, B Yellow leaf of ‘Duojiao’. C, D Yellow leaf with green spots of ‘Duojiao’. E, F Green leaf of ‘Duojiao’. G, H Green leaf of ‘Riversii’. CH Chloroplast, CW Cell wall, G Grana, GL Grana lamellae, O Osmiophilic granules, SG Starch granule, Th Thylakoid, V Vesicles. Bars = 5 μm (A, C, E, G); Bars = 500 nm (B, D, F, H)
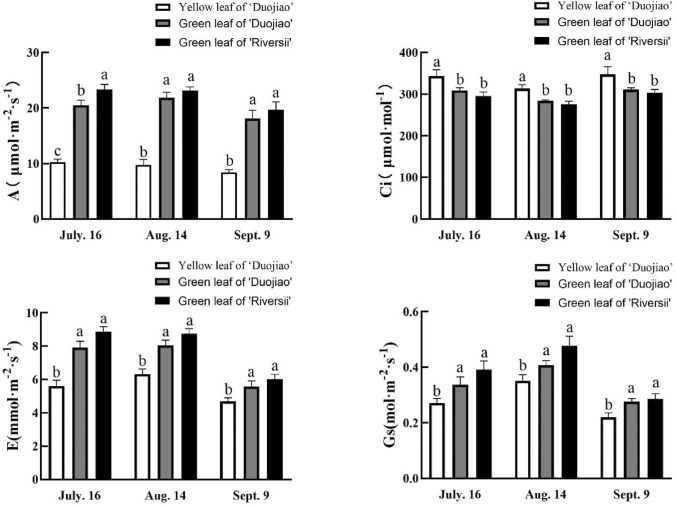
To study the effect of the low pigments content on photosynthesis of mutant, we measured the photosynthetic rate of yellow leaves and green leaves of the mutant ‘Duojiao’ and the green leaves of ‘Riversii’. The A, Gs, and E were decreased, but the Ci was increased in the mutant yellow leaves (Fig. 7). Abnormal chloroplast structure impaired photosynthetic efficiency.
Fig. 7.
Photosynthetic characteristics of the yellow leaf mutant M. spectabilis ‘Duojiao’ and M. spectabilis ‘Riversii’. Different lowercase letters above the bars indicate significant differences among groups at each time point (P < 0.05) according to Duncan’s multiple range test. A, net photosynthetic rate; Ci, intercellular CO2 concentration; Gs, stomatal conductance; E, transpiration rate
Transcriptome sequencing
Sequencing quality evaluation
Based on the above chloroplasts ultrastructure and physiological characteristics, we hypothesized that the expression model of genes responsible for chloroplast development and pigment biosynthesis had changed in the yellow leaves of ‘Duojiao’ (DS). We performed Illumina sequencing using the yellow and green leaves (DX) of the mutant and green leaves of ‘Riversii’ (XS, XX) and the apple genome (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/148/765/GCF_000148765.1_MalDomGD1.0/GCF_000148765.1_MalDomGD1.0_genomic.fna.gz) as the reference genome to clarify the molecular mechanisms of mutants delayed-green leaves formation.
The twelve samples were divided into four groups: DS, DX, XS and XX. A total of 37.56 Gb of clean data were produced. The lowest Q30 values obtained for each group were 95.25% (DS), 94.84% (DX), 95.26% (XS) and 94.78% (XX), indicating high sequencing quality. The ratio of sample to the reference genome sequence was between 82.89% and 86.53% (Table S2). These results show that the quality of obtained RNA-Seq data were high enough to proceed with subsequent analyses.
qRT-PCR verification
To investigate the reliability of the RNA-seq data, 12 genes with important functions and representativeness were selected, which were verified by qRT-PCR, including three up-regulated genes and nine down-regulated genes. The results of qRT-PCR and transcriptome sequencing showed that the expression patterns of these genes were similar. The correlations between the qPCR and RNA-seq data were as high as 99% (Fig. 8), which indicated that the transcriptome data were accurate.
Fig. 8.
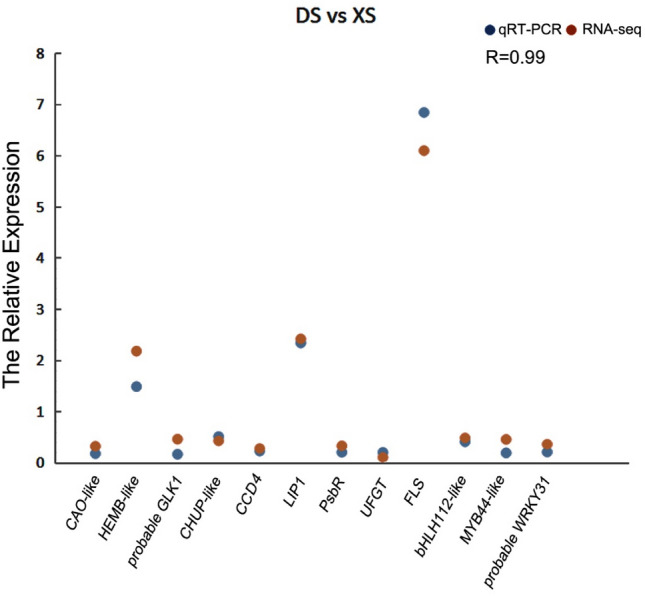
Comparison of qRT-PCR and RNA-seq to verify the accuracy of transcriptome data
DEG identification and GO and KEGG enrichment analyses
The criteria |log2(FoldChange)|>1 and q < 0.05 were used to identify DEGs (Figure S1A). Comparison of the DS with those of XS revealed 1,848 DEGs (983 up-regulated, 865 down-regulated). Hierarchical clustering of DEGs was carried out, and the abundances of DEGs was visually described by FPKM and color combination (Figure S1B). Most of the DEGs were up-regulated.
To characterize the primary biological functions of the mutant unigenes of yellow leaves, GO and KEGG analyses were conducted. In the GO functional classification, 1,848 DEGs were annotated into three GO categories: cellular component, molecular function and biological process (Figure S1C). Most DEGs were enriched in cellular process (864 DEGs, 46.75%) in biological process. In cellular component, DEGs were mostly enriched in cell part (1,108, 59.96%) and organelle (573, 31.01%). In molecular function, DEGs were mostly enriched in binding (862, 46.65%) and catalytic activity (858, 46.43%), and these DEGs may be closely tied to the differences in leaf color between ‘Duojiao’ and ‘Riversii’.
All DEGs were assigned to 102 KEGG pathways, and most of them were involved in metabolic pathways and the secondary metabolites biosynthesis. The main enriched pathways were ‘Isoquinoline alkaloid biosynthesis,’ ‘Zeatin biosynthesis’, and ‘Phenylpropanoid biosynthesis’ (Figure S1D). In mutant delayed-green leaves, many down-regulated DEGs were enriched in ‘Secondary metabolites biosynthesis’, ‘Porphyrin and chlorophyll metabolism’, and ‘Carotenoids biosynthesis’ pathways, which were likely involved in determining the leaf coloration of crabapple.
Identification and expression analysis of genes involved in chl, carotenoid, and flavonoid metabolism
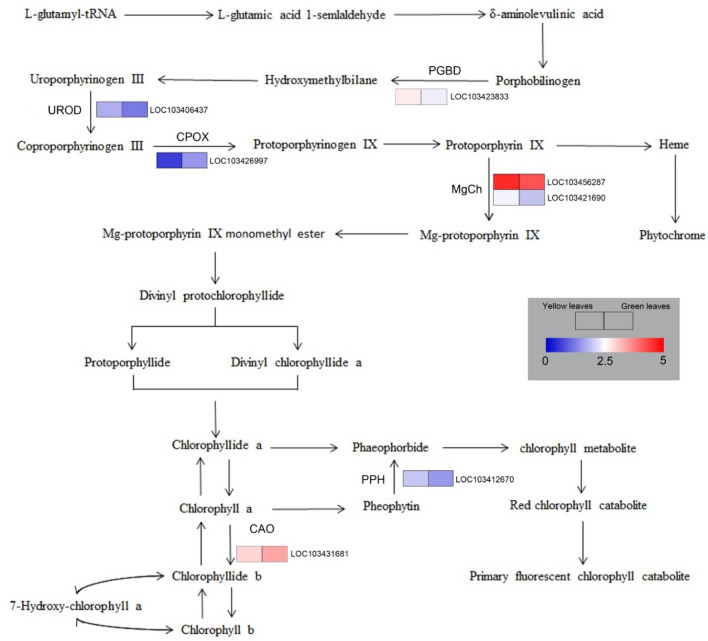
Color of plant leaves are ultimately determined by their pigment composition. Changes in leaf color are often also caused by Chl mutations, which directly or indirectly affect Chl metabolic pathway, leading to the changes in Chl content in plants and eventually lead to a yellowing phenotype. Six unigenes related to Chl biosynthesis and one unigene associated with Chl degradation were detected, and their expression levels were visualized by hierarchical cluster analysis (Fig. 9). The expression levels of DEGs involved in the transformation of Coprogen III and Protogen IX were lower in ‘DS’ than in ‘XS’, indicating that the conversion between Coprogen III and Protogen IX was lower in ‘DS’. In addition, the expression levels of CAO were significantly decreased in ‘DS’, indicating that the rate of biosynthesis of Chl b was lower in ‘DS’; this might reduce the efficiency of Chl biosynthesis. The up-regulation of the expression of Chl degradation-related PPH in mutant yellow leaves indicated that the Chl degradation was more rapid in ‘DS’ than in ‘XS’. After ‘Duojiao’ was turned green (DX) compared with ‘XX’, the expression of all differentially expressed genes that were significantly expressed in yellow leaves in the porphyrin and chlorophyll metabolism pathways, except for SUMT, returned to normal in the mutant turned green leaves (Table S3).
Fig. 9.
Expression profiles of DEGs associated with pigment metabolism between yellow leaves in ‘Duojiao’ (DS) and normal green leaves in ‘Riversii’ (XS)
Four DEGs related to carotenoid metabolism were identified. Compared with normal green leaves, the expression levels of CCD4 (probable carotenoid cleavage dioxygenase 4) and ZEP (zeaxanthin epoxidase) in the mutant were lower, while the expression levels of BCH (beta-carotene hydroxylase 2) and PLD (protein LUTEIN DEFICIENT 5, chloroplastic) in the mutant yellow leaves were higher compared with normal green leaves of ‘Riversii’ (Table S4). The expression of genes involved in the carotenoid metabolism pathway of ‘Duojiao’ after turning green (DX) returned to normal, and there was no significant difference in expression compared with the genes in the green leaves of ‘Riverssii’ (XX) (Table S5).
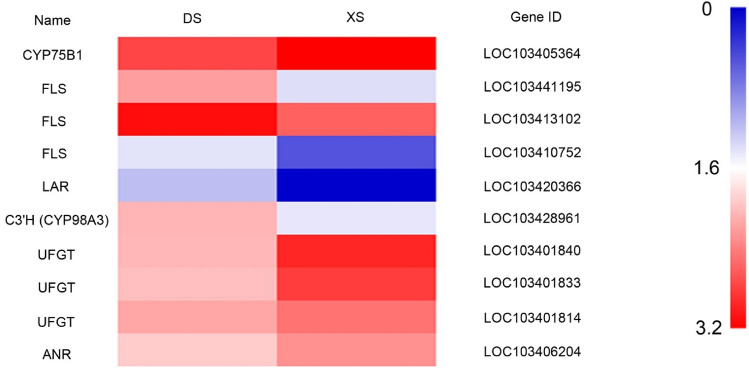
According to the KEGG analysis and GO annotations, 10 unigenes associated with flavonoid metabolism were detected (Fig. 10). Compared with the XS, one DEG (ANR) in the flavonoid synthesis pathway in DS was down-regulated. The expression levels of four DEGs (e.g., UFGT and CYP75B1) associated with flavonoid metabolism were significantly down-regulated in the mutant type.
Fig. 10.
Expression profiles of DEGs involved in flavonoid biosynthesis
DEGs related to chloroplast development and photosynthesis
Next, we explored the pathways of chloroplast development and photosynthesis-antenna proteins in more detail. Compared with ‘Riversii’, the expression levels of GLK (chloroplast development), PGT (plastidic glucose transporter), and CHUP1 (chloroplast unusual positioning 1) in the mutant yellow leaves were down-regulated (Table S6), indicating that the decrease of these genes may be related to the yellow leaves in mutant. After the yellow leaves of the mutant returned to green, the differential gene expression related to chloroplast development returned to normal level (Table S7).
Five DEGs associated with photosynthesis biosynthesis were annotated in this database (Table S8). Four of these DEGs were significantly down-regulated in ‘DS’. The down-regulated DEGs were PsbR (photosystem II, 10 kDa polypeptide), GBSS (granule-bound starch synthase 1), PPase DSP4 (phosphoglucan phosphatase DSP4), and FtsH6 (ATP-dependent zinc metalloprotease); ELIP (early light-induced protein) was significantly up-regulated.
Discussion
The yellow leaf phenotype is closely related to chl metabolism
The change in the type and proportion of pigments was the direct cause of the change in the color of the leaves of the mutants (Zhao et al. 2020). The delayed greening mechanisms of leaf mutants had been systematically studied, including physiological properties, absorption spectra, genetic characteristics, fluorescence, and microstructures (Runge et al. 1995; Falbel et al. 1996). Different from green leaf varieties, cultivars with yellow leaves had lower Chl concentrations, higher Chl a/b and Car/Chl ratios, and their chloroplasts lacked grana and stroma (Fu et al. 2013).
In our study, the young leaves of the mutant had a yellow phenotype, and older leaves turned green with the accumulation of Chl during the mature stage. The Chl a, Chl b, and Car contents in the yellow leaves of mutant plants were significantly lower than those of the green leaves of wild-type plants. The Chl a content in yellow leaves was approximately two-ninths of that in ‘Riversii’, the Chlb content was approximately one-eighth of that in ‘Riversii’, and the Car content was approximately three-fourths of that in ‘Riversii’. Many chlorotic forms of leaf color mutants show reduced Chl content ( Wang et al. 2016; Wu et al. 2018). As the mutant leaves turned green, the difference of pigments content between the mutant and the green leaves decreased; when the mutant leaves were green, the pigments content of the mutant was slightly lower than that of ‘Riversii’. The total Chl/Car of crabapple leaf color mutant increased significantly at first and then decreased with leaf development, indicating that the proportion of carotenoids in pigment composition gradually decreased with leaf development, and then increased (the leaves have turned green), indicating that in the yellow leaf stage, the decrease of leaf chlorophyll is much greater than that of carotenoids, resulting in the increase of relative content of carotenoid content and affecting leaf coloration. The Chl a/b ratio of the mutant (7.58) was highest at the yellow-leaf stage, most likely because Chl b synthesis was more likely to decrease than Chl a synthesis, which indicated damageto the light-collecting antenna complexes and photosystems in yellow leaves (Zhang et al. 2017). The leaves then turned green, and the Chl a/b ratio of the mutant decreased until it reached the level of the green leaves of ‘Riversii’. A novel rice (Oryza sativa) mutant was identified as a low Chl b mutant with a Chl a/b ratio of 4.7 (Chen et al. 2007). Most of the currently known mutants are deficient in Chl b. these mutants can be classified into two major groups according to the Chl a/b ratio (Falbel et al. 1996). One is a mutant without Chl b, such as the Arabidopsis thaliana mutant and the barley (Hordeum vulgare L.) mutant chlorina ƒ2(Bellemare et al. 1982; Murray and Kohorn 1991), which fails to accumulate sufficient amounts of light-harvesting Chl a/b-binding proteins under any growth conditions compared with the wild type (Harrison et al. 1993); in addition, the stacking of the thylakoid membrane is altered (Staehelin 1986). Another group of mutants shows a significant reduction in Chl b content, but the total Chl content and Chl a/b ratio are altered by changes in light intensity and temperature in the growth environment (Yang et al. 1990). An appropriate Chl a/b ratio is critical for photosynthetic antenna size and maintaining pigment function).
The Chl content in plants depends on the balance between Chl synthesis and degradation (Masuda and Fujita 2008). Some studies revealed that Chl-deficient mutants are mainly caused by the deficiency of enzymes during Chl synthesis process (Cui et al. 2001; Wu et al. 2007; Zhao et al. 2016). The transcriptome sequencing of the ‘DS’ and the ‘XS’ was conducted in order to explore the basic molecular mechanism underlying the unique phenotype of ‘Duojiao’. One DEG related to coproporphyrinogen III oxidase (CPOX, LOC103426997) was identified based on KEGG analysis, and the expression level of LOC103426997 was significantly reduced in DS compared with XS. This suggests that the Protogen IX stage of Chl biosynthesis was blocked, and this blockage might reduce the efficiency of Chl a biosynthesis. The expression of CAO (LOC103431681) was significantly down-regulated in DS; this gene encodes chlorophyllide a oxygenase to oxidize methyl formyl group of C7 side chain of Chl a to convert Chl b (Wang et al. 2009a, b). Biochemical and genetic analyses have shown that CAO is the only enzyme that converts Chl a to Chl b (Oster et al. 2000; Rüdiger 2002). CAO plays an important role in the the regulation of Chl biosynthesis (Sakuraba et al. 2007; Li et al. 2014) conducted genetic identification on rice 507ys yellow-green leaf mutant obtained by treatment with chemical mutagen ethyl methylsulfonate (EMS), and found that OsCAO1 (LOC_Os10g41780), which codes for chlorophyllin a oxygenase, had a single-site base mutation in the coding region, resulting in inactivation of CAO enzyme, blocked synthesis of Chl b, and yellow-green leaves. This finding indicate that CAO is essential for Chl biosynthesis and regulates leaf color formation. In our study, the decrease of Chl a and b further confirmed this result.
Leaf color mutants may also arise if the Chl degradation process is accelerated (Li et al. 2018). The expression of CHL2 and RCCR, which encode Chl-degrading enzymes, is significantly higher in Murray mutants than in wild-type plants, which results in accelerated Chl degradation and reduced Chl content and may explain the yellow mutation in Cymbidium sinense leaves (Zhu et al. 2015). The gene encoding Chl b reductase (NOL) is up-regulated in the light green stripe mutant in Oncidium ‘Milliongolds’, which increases the accumulation and activity of Chl b reductase (NOL), accelerates Chl b degradation, reduces the Chl b content in the mutant, and eventually leads to leaf chlorosis (Tian 2016). In our study, a gene involved in the regulation of demagnetizing chlorophyllase (PPH) was significantly up-regulated in DS_XS, which promoted Chl degradation in mutant yellow leaves.
Any step of Chl biosynthesis that is blocked can affect the Chl content (Li et al. 2018). The blockage of Chl biosynthesis in the rice leaf color mutants Chlorina-1 and Chlorina-9 occurred at the step involving the conversion of protoporphyrinogen IX to magnesium protoporphyrin (Zhang et al. 2006). A previous study examining the differences between green and white leaves in pineapple found that the step involving the conversion of porphobilinogen to uroporphyrinogen III is blocked, and this results in changes in leaf color (Li et al. 2017). All of these above research shows that as long as Chl biosynthesis is blocked in one step, the content of the products before blocking step will increase, and the content of all products after blocking step will decrease; this leads to impaired Chl biosynthesis and decreased leaf greenness (Cui et al. 2001). During the yellow leaf stage of the mutant, coprogen III accumulated; the content of protogen IX, chlorophyllide a, and chlorophyllide b was significantly lower in ‘Duojiao’ than in ‘Riversii’, and the activity of CPOX and CAO was significantly lower in ‘Duojiao’ leaves than in ‘Riversii’ green leaves, suggesting that the step of Chl biosynthesis was blocked in the mutant might be the conversion of coprogen III to protogen IX.
The content of Car decreased significantly in the yellow leaves of the mutant compared with the green leaves of ‘Riversii’. In previous studies, the Car content of yellow leaf mutant pylm and the yellow-green winter wheat mutant Ygm was significantly reduced (Zhang et al. 2017; Wu et al. 2018). The ratio of Car to Chl underlies yellow leaf coloration (Zhang et al. 2019b). In addition, the ratio of Car-to-Chl increased in both the Populus deltoides Marsh mutant and pylm mutant (Zhang et al. 2017, 2019b). Consistent with these findings, in our study the ratio of Car to Chl was 3.73 times higher in yellow leaves than in green leaves. Therefore, the increase in the ratio of Car to Chl was related to the yellow phenotype of M. spectabilis ‘Duojiao’.
Compared with ‘Riversii’, the photosynthetic pigments content in the mutant yellow leaves decreased significantly. The content of Chl b in ‘Riversii’ green leaves was 10.18 times higher than in yellow leaves of ‘Duojiao’. These results showed that the yellow foliage phenotype was the result of a deficiency of photosynthetic pigments, which alterd the ratios of Chl a/b and Car/Chl.
The yellow leaf phenotype is closely related to chloroplast development
The biosynthesis of Chl was related to the formation of thylakoid membrane and coordinated with the development of chloroplast (Yu et al. 2007). Through literature review, it was found that chloroplasts in mesophyll cells of most leaf-colored mutants were poorly developed and have abnormal ultrastructure. Compared with the control, chloroplasts of leaf-colored mutants usually contained fewer, thinner and irregularly arranged grana lamellae, and thylakoids usually expanded at different levels (Yoo et al. 2009; Yang et al. 2015b). The normal development of chloroplasts in higher plants required the coordination of nuclear genes and chloroplast genes (Li et al. 2018). Changes in transcription and expression levels of any gene type may affect the biogenesis of normal chloroplasts, and the resulting destruction of Chl metabolism and chloroplast assembly will lead to abnormal leaf color (Li et al. 2015; Yang et al. 2015a).
We examined the anatomical characteristics of ‘Duojiao’ and ‘Riversii’ and identified marked differences in leaf structure. The chloroplasts of the yellow leaves of the mutant have a high-density vesicular structure that is severely disintegrated internally; small, clustered osmiophilic granules; no grana lamellae; and no starch granules, indicating that the development of chloroplasts is abnormal in this mutant. By consulting the literature, it was found that chloroplasts in most leaf color mutant chloroplasts were poorly developed and had abnormal ultrastructure, and compared to controls, chloroplasts in leaf color mutants usually contained fewer, thinner and irregularly arranged grana lamellae, and thylakoids were usually expanded at different levels For example, the ultrastructure of the chloroplasts in mutant Lagerstroemia indica leaves is characterized by ruptured thylakoid membranes and indistinct or absent stromal lamellae (Li et al. 2015); the ultrastructure of the chloroplasts in mutant Capsicum annuum L. contain fewer thylakoid grana, irregular and unclear thylakoid grana, and more osmiophilic globules (Ma et al. 2016); the chloroplast ultrastructure of a novel rice mutant (Oryza sativa L. var. Zhenhui 249Y) had fewer grana lamellae and slightly swollen thylakoids (Chen et al. 2007). Similar results were obtained for the Arabidopsis Chl b-deficient mutant: the chloroplasts were smaller in size and had fewer grana lamellae early during leaf growth. TOC33, a component of the protein transport machine on the chloroplast outer envelope, was not expressed; thus, chloroplast protein transport was blocked, which ultimately blocked chloroplast development (Jarvis et al. 1998). These structural changes might lead to the decrease in Chl content, abnormal Chl a/b ratios, and delayed greening of leaves (Liu et al. 2016). These results were consistent with the observed changes in transcript abundance. As the ‘Duojiao’ leaves developed, the leaves turned green, the structure of the chloroplasts was gradually restored, and the thylakoids began to stack and form their characteristic grana structure; these findings were in line with the increases in the Chl content.
Studies in rice (Fitter et al. 2002) and tomato (Powell et al. 2012) have shown that GLK genes affect chloroplast development and photosynthetic gene expression. GLK, originally identified in maize, encodes an important TF that regulates chloroplast development (Hall et al. 1998). In a study of the mechanism underlying leaf color variation of the new species Acer buergerianum Miq. ‘Golden Qilu’, the expression of 17 GLK transcription-regulated genes in the mutant was reduced, the structure of the thylakoid membrane in the leaves of ‘Golden Qilu’ was broken or missing, and the stroma and grana lamellae were loose (Chu 2020). A transcriptome study of Ginkgo biloba variegata leaves and normal green leaves revealed that in addition to the down-regulated expression of GbGLK in Ginkgo biloba variegata yellow leaves, the structural development of the chloroplast was altered; this observation leads to the prediction that down-regulation of the expression of GbGLK inhibits normal chloroplast development in the leaves (Li et al. 2018). GLK1 (LOC103444434) in the yellow leaves of the ornamental crabapple mutant ‘Duojiao’ also showed significantly down-regulated expression, which was confirmed by qRT-PCR. The thylakoid structure of the yellow leaves of ‘Duojiao’ was not well developed, the grana lamellae were not stacked, and the chloroplasts were filled with vesicles. The down-regulated expression of GLK1 in the yellow leaves of the crabapple mutant may have inhibited the normal development of chloroplasts in the leaves and resulted in structural differences compared with normal green leaves.
In chloroplasts, starch was the major energy storage compound; it is mainly stored in the form of granules to store excess carbohydrates produced during photosynthesis (Liu et al. 2016). We identified two down-regulated DEGs associated with starch metabolism that differed in abundance, including granule-bound starch synthase (GBSS, LOC103439436) and phosphoglucan phosphatase DSP4 (PPase DSP4, LOC103401524). GBSS is a SS and a key enzyme for amylose formation in starches (Visser et al. 1991). The decreased of the abundance of GBSS and PPase DSP4 might indicate that the energy storage level in the yellow leaves of ‘Duojiao’ is low. The ultrastructure of chloroplast supports this conclusion: compared with ‘Riversii’, starch grain accumulation was rare in ‘Duojiao’ (Fig. 4A–D).
CHUP1 is involved in the chloroplast movement signaling process, which is necessary for chloroplast localization; it also plays an important role in avoiding light damage and improving photosynthetic efficiency in plants (Kodama et al. 2008; Oikawa et al. 2008). The significantly down-regulated CHUP1 gene was annotated in DS_XS, and electron microscopy showed that chloroplasts in mutant crabapple yellow leaves were disordered, suggesting that the down-regulation of CHUP1 affected chloroplast movement. Chloroplasts develop from protoplasts. The plastid undifferentiated (PUN) mutant in maize inhibited the biogenesis of plastid in mesophyll cells and vascular bundle sheath, resulting in the decrease of proteins encoded by chloroplast (Roth et al. 2001). Thus, variation in leaf color is related to the abnormal development and distribution of chloroplasts.
Leaf color changes in mutants may be caused by the abnormal development of plastids in mutated leaves (Li et al. 2018). The gene expression pattern shows that the DEGs involved in determining chloroplasts structure affected chloroplast development and decreased the content of photosynthetic pigments, which resulted in the development of a yellow phenotype.
Conclusion
The major mechanisms underlying leaf color variation in the mutant crabapple ‘Duojiao’ were studied by physiological characterization and transcriptome sequence analysis. The contents of Chl a, Chl b, and Car in the yellow leaves of ‘Duojiao’ were significantly lower than in the green leaves of ‘Riversii’. The Car/Total Chl ratio was high in the mutant yellow leaves, and the content of Chl was reduced more in the mutant yellow leaves; thus, the relative content of carotenoids in yellow leaves was high. The Chl a/b ratio was high in the mutant yellow leaves, and the content of Chl b was reduced more in the mutant yellow leaves; thus, the delayed-green leaf mutant ‘Duojiao’ is a Chl b-deficient mutant. The disruption of thylakoid stacking in the yellow leaf mutant hindered pigment synthesis, changed the proportion of pigment, which explained the change in leaf color.
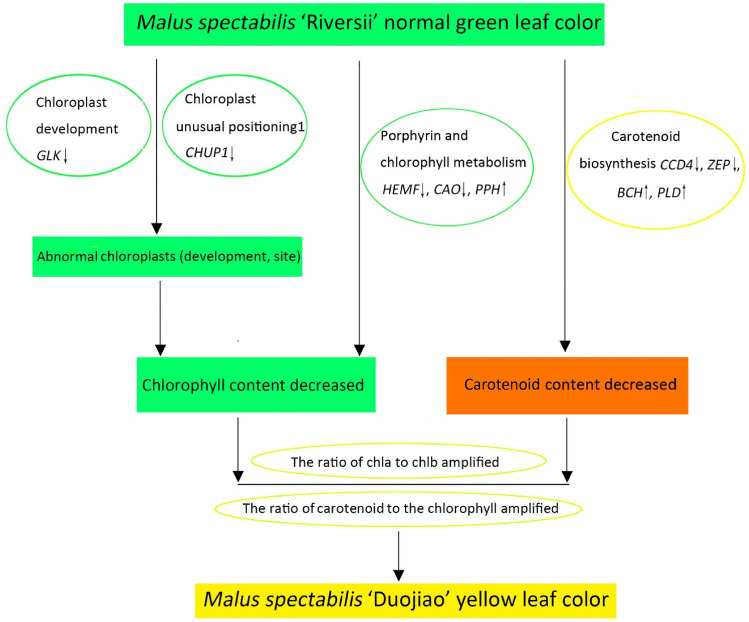
Three key candidate genes involved in pigment synthesis, chloroplast development, and degradation were identified by transcriptome sequencing, including genes involved in the synthesis of CPOX enzyme metabolism (LOC103426997); the gene encoding chlorophyllide a oxygenase, which catalyzes the conversion of Chl a to Chl b; and the gene GLK1, which affects chloroplast development. The down-regulation of chloroplast degeneration and the Chl biosynthesis-related unigenes together may contribute to the yellowing phenotype of ‘Duojiao’. We proposed a possible mechanism by which the yellow leaf phenotype in ‘Duojiao’ is formed in Fig. 11.
Fig. 11.
The proposed pathway by which the mutant leaf coloration in ‘Duojiao’ is formed
These findings provide new insight into the formation mechanism of the yellow leaves of ‘Duojiao’ and will facilitate improvements in the selective breeding of leaf color varieties. In addition, the leaf transcriptome data increases the existing data set in the databases of crabapple transcriptome and could contribute to ornamental crabapple breeding in the future.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Annoroad Gene Technology Co. Ltd. (Beijing, China) for help with the RNA-Seq data analysis and Canhong Zhang (Tai’an Zhendong Nursery, Tai’an, Shandong, China) for help with sampling.
Author contributions
LZ, JZ, and XS conceived and designed the experiments. LZ performed the experiments. LZ, JZ, YY, and YM analyzed the data. LZ, JZ, YM and YY contributed reagents/materials/analysis tools. LZ and JZ contributed to the writing of the manuscript. All authors approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [Grant Number 32,072,520]; the Natural Science Foundation of Shandong Province [Grant Number ZR2020MC132]; the Fruit Innovation Team of Modern Agricultural Industry Technology System in Shandong Province, China [Grant Number SDAIT-06-07]; the Key Technology Research and Development Program of Shandong, China [Grant Number 2018GNC113019]; and the Industrialization Project of Improved Varieties in Shandong Province, China [Grant Nnumber 2019LZGC007].
Declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bellemare G, Bartlett SG, Chua NH. Biosynthesis of chlorophyll a/b-binding polypeptides in wild type and the chlorina f2 mutant of barley. J Biol Chem. 1982;257:7762–7767. doi: 10.1016/S0021-9258(18)34446-6. [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang W, Xie YJ, Lu W, Zhang RX. Comparative proteomics of thylakoid membrane from a chlorophyll b-less rice mutant and its wild type. Plant Sci. 2007;173:397–407. doi: 10.1016/j.plantsci.2007.06.012. [DOI] [Google Scholar]
- Chu JT (2020) Study on Leaf Color Variation Mechanism of a New Cultivar ‘Golden Qilu’ in Acer buergerianum Miq., Dissertation for the Degree of Shandong Agricultural university. https://kns.cnki.net/KCMS/detail/detail.aspx?dbname=CMFD202002&filename=1020043462.nh
- Cui HR, Xia YW, Gao MW. Effects of temperature on leaf color and chlorophyll biosynthesis of rice mutant W1. He Nong Xue Bao. 2001;15:269–273. [Google Scholar]
- Douzery EJP, Snell EA, Bapteste E, Delsuc F, Philippe H. The timing of eukaryotic evolution: Does a relaxed molecular clock reconcile proteins and fossils? Proc Natl Acad Sci USA. 2004;101:15386. doi: 10.1073/pnas.0403984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falbel TG, Meehl JB, Staehelin LA. Severity of mutant phenotype in a series of chlorophyll-deficient wheat mutants depends on light intensity and the severity of the block in chlorophyll synthesis. Plant Physiol. 1996;112:821–832. doi: 10.1104/pp.112.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. GLK gene pairs regulate chloroplast development in diverse plant species. PlJ. 2002;31:713–727. doi: 10.1046/j.1365-313X.2002.01390.x. [DOI] [PubMed] [Google Scholar]
- Fu X, Zhou L, Huang J, Mo W, Zhang J, Li J, Wang H, Huang X. Relating photosynthetic performance to leaf greenness in litchi: a comparison among genotypes. Sci Hortic. 2013;152:16–25. doi: 10.1016/j.scienta.2013.01.001. [DOI] [Google Scholar]
- Gan S, Amasino RM. Inhibition of leaf senescence by autoregulated production of cytokinin. Science. 1995;270:1986–1988. doi: 10.1126/science.270.5244.1986. [DOI] [PubMed] [Google Scholar]
- Gang HX, Liu GF, Chen S, Jiang J. Physiological and transcriptome analysis of a yellow-green leaf mutant in birch (betula platyphylla × b. Pendula) Forests. 2019 doi: 10.3390/f10020120. [DOI] [Google Scholar]
- Hall LN, Rossini L, Cribb L, Langdale JA. GOLDEN 2: a novel transcriptional regulator of cellular differentiation in the Maize Leaf. Plant Cell. 1998;10:925–936. doi: 10.2307/3870679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson A, Kannangara CG, Wettstein DV, Hansson M. Molecular basis for semidominance of missense mutations in the XANTHA-H (42-kDa) subunit of magnesium chelatase. Proc Natl Acad Sci USA. 1999;96:1744–1749. doi: 10.1073/pnas.96.4.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MA, Nemson JA, Melis A. Assembly and composition of the chlorophyll a-b light-harvesting complex of barley (Hordeum vulgare L.): Immunochemical analysis of chlorophyll b-less and chlorophyll b-deficient mutants. Photosynth Res. 1993;38:141–151. doi: 10.1007/BF00146413. [DOI] [PubMed] [Google Scholar]
- Jarvis P, Chen LJ, Li H, Peto CA, Fankhauser C, Chory J. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science. 1998;282:100–103. doi: 10.1126/science.282.5386.100. [DOI] [PubMed] [Google Scholar]
- Jia ZS, Tang MC, Wu JM. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999;64:555–559. doi: 10.1016/S0308-8146(98)00102-2. [DOI] [Google Scholar]
- Jiang S, Zhang X, Zhang F, Xu Z, Chen W, Li Y. Identification and fine mapping of qCTH4, a quantitative trait loci controlling the chlorophyll content from tillering to heading in rice (Oryza sativa L.) J Hered. 2012;103:720–726. doi: 10.1093/jhered/ess041. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y, Tsuboi H, Kagawa T, Wada M. Low temperature-induced chloroplast relocation mediated by a blue light receptor, phototropin 2, in fern gametophytes. J Plant Res. 2008;121:441–448. doi: 10.1007/s10265-008-0165-9. [DOI] [PubMed] [Google Scholar]
- Kristiansson E, Asker N, Larsson DG. Characterization of the Zoarces viviparus liver transcriptome using massively parallel pyrosequencing. BMC Genomics. 2009;10:345. doi: 10.1186/1471-2164-10-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li NN, Yang YP, Ye JH, Lu JL, Zheng XQ, Liang YR. Effects of sunlight on gene expression and chemical composition of light-sensitive albino tea plant. Plant Growth Regul. 2016;78:253–262. doi: 10.1007/s10725-015-0090-6. [DOI] [Google Scholar]
- Li WX, Yang SB, Lu ZG, He ZC, Ye YL, Zhao BB, Wang L, Jin B. Cytological, physiological, and transcriptomic analyses of golden leaf coloration in Ginkgo biloba L. Hortic Res. 2018;5:12. doi: 10.1038/s41438-018-0015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kanakala S, He Y, Zhong X, Yu S, Li R, Sun L, Ma J. Physiological characterization and comparative transcriptome analysis of white and green leaves of Ananas comosus var. bracteatus. PLoS ONE. 2017;12:e0169838. doi: 10.1371/journal.pone.0169838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang ZY, Wang P, Wang SA, Ma LL, Li LF, Yang RT, Ma YZ, Wang Q. Comprehensive transcriptome analysis discovers novel candidate genes related to leaf color in a Lagerstroemia indica yellow leaf mutant. Genes Genomics. 2015;37:851–863. doi: 10.1007/s13258-015-0317-y. [DOI] [Google Scholar]
- Li YQ, Pu X, Li CM, Zhong P, Sun CH, Li XL, Deng XJ, Wang PR. Genetic identification and candidate gene analysis of yellow-green leaf mutant 507ys in Rice. Scientia Agricultura Sinica. 2014;47(2):221–229. doi: 10.3864/j.issn.0578-1752.2014.02.002. [DOI] [Google Scholar]
- Li Z, Wakao S, Fischer BB, Niyogi KK. Sensing and responding to excess light. Annu Rev Plant Biol. 2009;60:239–260. doi: 10.1146/annurev.arplant.58.032806.103844. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Analysis. 1983;11:591–592. doi: 10.1042/bst0110591. [DOI] [Google Scholar]
- Liu XL, Yu WW, Wang GB, Cao FL, Cai JF, Wang HL. Comparative proteomic and physiological analysis reveals the variation mechanisms of leaf coloration and carbon fixation in a xantha mutant of Ginkgo biloba L. Int J Mol Sci. 2016;17:1794. doi: 10.3390/ijms17111794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR. Methods. 2002;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- López-Juez E, Jarvis RP, Takeuchi A, Chory PJ. New arabidopsis cue mutants suggest a close connection between plastid- and phytochrome regulation of nuclear gene expression. Plant Physiol. 1998;118:803–815. doi: 10.1104/pp.118.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma ZH, Sun GS, Zhang CW, Wang Q, Dai ZL, Sun CQ, Pan YP. Chlorophyll content, chloroplast ultrastructure and transcriptome analysis in wild-type and yellow-bud-mutant hot peppers. J Agric Sci Technol. 2016;18:1065–1078. [Google Scholar]
- Masuda T, Fujita Y, Oikawa K, Yamasato A, Kong S-G, Kasahara M, Nakai M, Takahashi F, Ogura Y, Kagawa T, Wada M. Regulation and evolution of chlorophyll metabolism. Photochem Photobiol Sci. 2008;7:1131–1149. doi: 10.1039/b807210h. [DOI] [PubMed] [Google Scholar]
- Murray DL, Kohorn BD. Chloroplasts of Arabidopsis thaliana homozygous for the ch-1 locus lack chlorophyll b, lack stable LHCPII and have stacked thylakoids. Plant Mol Biol. 1991;16:71–79. doi: 10.1007/BF00017918. [DOI] [PubMed] [Google Scholar]
- Oikawa K, Yamasato A, Kong S-G, Kasahara M, Nakai M, Takahashi F, Ogura Y, Kagawa T, Wada M. Chloroplast outer envelope protein CHUP1 is essential for chloroplast anchorage to the plasma membrane and chloroplast movement. Plant Physiol. 2008;148:829–842. doi: 10.1104/pp.108.123075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster U, Tanaka R, Tanaka A, Rüdiger W. Cloning and functional expression of the gene encoding the key enzyme for chlorophyll b biosynthesis (CAO) from Arabidopsis thaliana. PlJ. 2000;21:305–310. doi: 10.1046/j.1365-313x.2000.00672.x. [DOI] [PubMed] [Google Scholar]
- Powell ALT, Nguyen CV, Hill T, Cheng KL, Figueroa-Balderas R, Aktas H, Ashrafi H, Pons C, Fernandez-Munoz R, Vicente A, Lopez-Baltazar J, Barry CS, Liu YS, Chetelat R, Granell A, Van Deynze A, Giovannoni JJ, Bennett AB. Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science. 2012;336:1711–1715. doi: 10.1126/science.1222218. [DOI] [PubMed] [Google Scholar]
- Reyes-Arribas T, Barrett JE, Huber DJ, Nell TA, Clark DG. Leaf senescence in a non-yellowing cultivar of chrysanthemum (Dendranthema grandiflora) Physiol Plant. 2010;111:540–544. doi: 10.1034/j.1399-3054.2001.1110415.x. [DOI] [PubMed] [Google Scholar]
- Roth R, Sawers RJ, Munn HL, Langdale JA. Plastids undifferentiated, a nuclear mutation that disrupts plastid differentiation in Zea mays L. Planta. 2001;213:647–658. doi: 10.1007/s004250100537. [DOI] [PubMed] [Google Scholar]
- Rüdiger W. Biosynthesis of chlorophyll b and the chlorophyll cycle. Photosynth Res. 2002;74:187–193. doi: 10.1023/A:1020959610952. [DOI] [PubMed] [Google Scholar]
- Runge S, Van Cleve B, Lebedev N, Armstrong G, Apel K. Isolation and classification of chlorophyll-deficient xantha mutants of Arabidopsis thaliana. Planta. 1995;197:490–500. doi: 10.1007/BF00196671. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Yamasato A, Tanaka R, Tanaka A. Functional analysis of N-terminal domains of Arabidopsis chlorophyllide a oxygenase. Plant Physiol Biochem. 2007;45:740–749. doi: 10.1016/j.plaphy.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Sinkkonen A, Somerkoski E, Paaso U, Holopainen JK, Rousi M, Mikola J. Genotypic variation in yellow autumn leaf colours explains aphid load in silver birch. New Phytol. 2012;195:461–469. doi: 10.1111/j.1469-8137.2012.04156.x. [DOI] [PubMed] [Google Scholar]
- Staehelin LA. Chloroplast structure and supramolecular organization of photosynthetic membranes. In: Staehelin LA, Arntzen CJ, editors. Photosynthesis III: photosynthetic membranes and light harvesting systems. Berlin: Springer; 1986. pp. 1–84. [Google Scholar]
- Tian WW (2016) Physiological and molecular basic resecrah of light green stripe mutant in Oncidium ‘Milliongolds’. Dissertation for the Degree of Chinese Academy of Forestry. https://kns.cnki.net/KCMS/detail/detail.aspx?dbname=CMFD201701&filename=1016251397.nh
- Visser RGF, Somhorst I, Kuipers GJ, Ruys NJ, Feenstra WJ, Jacobsen E. Inhibition of the expression of the gene for granule-bound starch synthase in potato by antisense constructs. Mol Gen Genet. 1991;225:289–296. doi: 10.1007/BF00269861. [DOI] [PubMed] [Google Scholar]
- Wang PR, Zhang FT, Gao JX, Sun XQ, Deng XJ. An overview of chlorophyll biosynthesis in higher plants. Acta Bot Boreal Occident Sin. 2009;29(3):629–636. doi: 10.3321/j.issn:1000-4025.2009.03.032. [DOI] [Google Scholar]
- Walles B. Chromoplast development in a carotenoid mutant of maize. Protoplasma. 1971;73:159–175. doi: 10.1007/bf01275592. [DOI] [Google Scholar]
- Wang YK, He YJ, Yang M, He JB, Xu P, Shao MQ, Chu P, Guan RZ. Fine mapping of a dominant gene conferring chlorophyll-deficiency in Brassica napus. Sci Rep. 2016;6:31419. doi: 10.1038/srep31419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen BB, Li C, Fu XL, Li DM, Li L, Chen XD, Wu HY, Cui XW, Zhang XH, Shen HY, Zhang WQ, Xiao W, Gao DS. Effects of nitrate deficiency on nitrate assimilation and chlorophyll synthesis of detached apple leaves. Plant Physiol Bioch. 2019;142:363–371. doi: 10.1016/j.plaphy.2019.07.007. [DOI] [PubMed] [Google Scholar]
- Wu HY, Shi NR, An XY, Liu C, Fu HF, Cao L, Feng Y, Sun DJ, Zhang LL. Candidate genes for yellow leaf color in common wheat (Triticum aestivum L.) and major related metabolic pathways according to transcriptome profiling. Int J Mol Sci. 2018;19:1594. doi: 10.3390/ijms19061594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XX, Liu FL, Fang YF, Jiang NN, Jiang LH, Song GF. A comprehensive evaluation on application value of 36 Euro-American ornamental crabapples. J Nanjing For Univ Natl Sci Ed. 2015;39:93–98. doi: 10.3969/j.issn.1000-2006.2015.01.017. [DOI] [Google Scholar]
- Wu ZM, Zhang X, He B, Diao LQ, Sheng SL, Wang JL, Guo XQ, Su N, Wang LF, Jiang L, Wang CM, Zhai HQ, Wan JM. A Chlorophyll-Deficient Rice Mutant with Impaired Chlorophyllide Esteriflcation in Chlorophyll Biosynthesis. Plant Physiol. 2007;145:29–40. doi: 10.1104/pp.107.100321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Yang SH, Wei JJ, Long Y, Jia RD, Zhao X, Ge H. Chloroplast ultrastructure, metabolite contents and gene expression involved in the pathway of chlorophyll biosynthesis of Rosa beggeriana‘Aurea’. Acta Horticulturae Sinica. 2019;46(11):2188–2200. doi: 10.16420/j.issn.0513-353x.2019-0032. [DOI] [Google Scholar]
- Yang C-M, Osterman JC, Markwell J. Temperature sensitivity as a general phenomenon in a collection of chlorophyll-deficient mutants of sweetclover (Melilotus alba) Biochem Genet. 1990;28:31–40. doi: 10.1007/BF00554819. [DOI] [PubMed] [Google Scholar]
- Yang HY, Xia XW, Fang W, Fu Y, An MM, Zhou MB. Identification of genes involved in spontaneous leaf color variation in Pseudosasa japonica. Genet Mol Res. 2015;14:11827–11840. doi: 10.4238/2015.October.2.16. [DOI] [PubMed] [Google Scholar]
- Yang Y, Chen X, Xu B, Li Y, Ma Y, Wang G. Phenotype and transcriptome analysis reveals chloroplast development and pigment biosynthesis together influenced the leaf color formation in mutants of Anthurium andraeanum ‘Sonate’. Front Plant Sci. 2015;6:139. doi: 10.3389/fpls.2015.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SC, Cho SH, Sugimoto H, Li J, Kusumi K, Koh HJ, Iba K, Paek NC. Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol. 2009;150:388–401. doi: 10.1104/pp.109.136648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Fu A, Aluru M, Park S, Xu Y, Liu H, Liu X, Foudree A, Nambogga M, Rodermel S. Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ. 2007;30:350–365. doi: 10.1111/j.1365-3040.2006.01630.x. [DOI] [PubMed] [Google Scholar]
- Zhang HT, Li JJ, Yoo J-H, Yoo S-C, Cho S-H, Koh H-J, Seo HS, Paek N-C. Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol Biol. 2006;62:325–337. doi: 10.1007/s11103-006-9024-z. [DOI] [PubMed] [Google Scholar]
- Zhang K, Liu ZY, Shan XF, Li CY, Tang XY, Chi MY, Feng H. Physiological properties and chlorophyll biosynthesis in a Pak-choi (Brassica rapa L. ssp. chinensis) yellow leaf mutant, pylm. Acta Physiol Plant. 2017;39:1–10. doi: 10.1007/s11738-016-2321-5. [DOI] [Google Scholar]
- Zhang LL, Mao YF, Wang YY, Yang L, Yin YJ, Shen X, Zhang CH, Zhang DJ. Malus spectabilis ‘Duojiao’: a new yellow-leaf cultivar. HortScience. 2020;55:1155–1158. doi: 10.21273/hortsci14865-20. [DOI] [Google Scholar]
- Zhang SZ, Wu XL, Cui J, Zhang F, Wan XQ, Liu QL, Zhong Y, Lin TT. Physiological and transcriptomic analysis of yellow leaf coloration in Populus deltoides Marsh. PLoS ONE. 2019;14:17. doi: 10.1371/journal.pone.0216879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MH, Li X, Zhang XX, Zhang H, Zhao XY. Mutation mechanism of leaf color in plants: a review. Forests. 2020 doi: 10.3390/f11080851. [DOI] [Google Scholar]
- Zhao SL, Long WH, Wang YH, Liu LL, Wang YL, Niu M, Zheng M, Wang D, Wan JM. A rice White-stripe leaf3 (wsl3) mutant lacking an HD domain-containing protein affects chlorophyll biosynthesis and chloroplast development. J Plant Biol. 2016;59:282–292. doi: 10.1007/s12374-016-0459-8. [DOI] [Google Scholar]
- Zhao Y, Wang ML, Zhang YZ, Du LF, Pan T. A chlorophyll-reduced seedling mutant in oilseed rape, Brassica napus, for utilization in F1 hybrid production. Plant Breed. 2010;119:131–135. doi: 10.1046/j.1439-0523.2000.00453.x. [DOI] [Google Scholar]
- Zhou T, Jiang H, Zhang D, Fan J, Zhang L, Wang G, Zhang W, Cao F. ‘Fen Balei’ Crabapple. HortScience. 2019;54:1433–1434. doi: 10.21273/hortsci14063-19. [DOI] [Google Scholar]
- Zhu GF, Yang FX, Shi SS, Li DM, Wang Z, Liu HL, Huang D, Wang CY. Transcriptome characterization of cymbidium sinense ‘Dharma’ using 454 pyrosequencing and its application in the identification of genes associated with leaf color variation. PLoS ONE. 2015;10:1–17. doi: 10.1371/journal.pone.0128592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LX, Zeng XH, Chen YL, Yang ZH, Qi LP, Pu YY, Yi B, Wen J, Ma CZ, Shen JX, Tu JX, Fu TD. Genetic characterisation and fine mapping of a chlorophyll-deficient mutant (BnaC.ygl) in Brassica napus. Mol Breed. 2014;34:603–614. doi: 10.1007/s11032-014-0060-0. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.