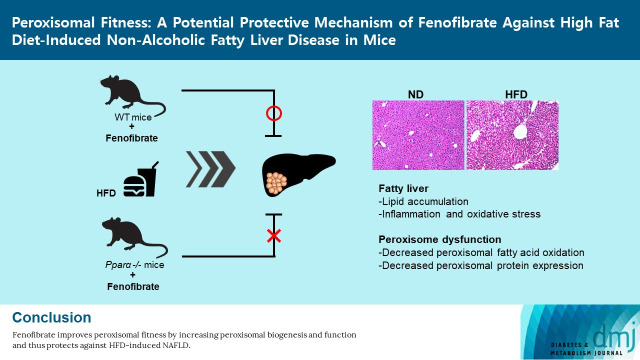
Abstract
Background
Non-alcoholic fatty liver disease (NAFLD) has been increasing in association with the epidemic of obesity and diabetes. Peroxisomes are single membrane-enclosed organelles that play a role in the metabolism of lipid and reactive oxygen species. The present study examined the role of peroxisomes in high-fat diet (HFD)-induced NAFLD using fenofibrate, a peroxisome proliferator-activated receptor α (PPARα) agonist.
Methods
Eight-week-old male C57BL/6J mice were fed either a normal diet or HFD for 12 weeks, and fenofibrate (50 mg/kg/day) was orally administered along with the initiation of HFD.
Results
HFD-induced liver injury as measured by increased alanine aminotransferase, inflammation, oxidative stress, and lipid accumulation was effectively prevented by fenofibrate. Fenofibrate significantly increased the expression of peroxisomal genes and proteins involved in peroxisomal biogenesis and function. HFD-induced attenuation of peroxisomal fatty acid oxidation was also significantly restored by fenofibrate, demonstrating the functional significance of peroxisomal fatty acid oxidation. In Ppara deficient mice, fenofibrate failed to maintain peroxisomal biogenesis and function in HFD-induced liver injury.
Conclusion
The present data highlight the importance of PPARα-mediated peroxisomal fitness in the protective effect of fenofibrate against NAFLD.
Keywords: Fenofibrate, Non-alcoholic fatty liver disease, Peroxisomal disorders, PPAR alpha
Graphical abstract
INTRODUCTION
Non-alcoholic fatty liver disease (NAFLD) is the most common chronic liver disease with a worldwide prevalence of 20% to 30% [1], and its prevalence is even higher in high caloric intake and obese populations [2]. Though NAFLD is commonly benign, it may develop into inflammation, fibrosis, cirrhosis (non-alcoholic steatohepatitis [NASH]), and eventually cancer of the liver [3]. NAFLD is a metabolic disorder caused by the accumulation of fat in the liver leading to liver dysfunction [4]. Unfortunately, specific and potent treatment options for NAFLD have not been recognized yet. Thus, there is an urgent need for exploring strategies for the proper treatment of NAFLD.
A number of studies have reported a pathogenic role of oxidative stress in NAFLD [5]. Markers of oxidative stress such as lipid peroxidation and reactive oxygen species (ROS) are increased in the liver of NAFLD patients [6]. The accumulation of triglycerides (TG) in the cytoplasm of hepatocytes is associated with NAFLD, and exposure to hydrogen peroxide (H2O2) increases the cytoplasmic TG level in HepG2 cells, a human hepatocyte carcinoma cell line [7]. Besides, various antioxidants, including curcumin, resveratrol, quercetin, and lycopene, decrease oxidative stress and the features of NAFLD [8]. Thus, inhibition of oxidative stress may play a key role in inhibiting NAFLD progression.
Peroxisomes are single membrane-bound organelles that rapidly assemble, multiply, and are degraded in response to metabolic needs. Peroxisomal biogenesis is regulated by de novo biogenesis, the growth and division of pre-existing peroxisomes, and pexophagy [9]. de novo biogenesis requires the fusion of two pre-peroxisomal vesicles, one from the endoplasmic reticulum (ER) and the other from mitochondria [10]. Proteins that are involved in the peroxisomal biogenesis process are called peroxins (peroxisomal biogenesis factor [PEX]). Peroxisomes regulate many metabolic functions, such as the β-oxidation of fatty acids (FA) as well as ROS homeostasis [11]. Peroxisomal β-oxidation of very-long-chain fatty acids (VLCFA) occurs through ATP binding cassette subfamily D member 1 (ABCD1), while α-oxidation of branched-chain fatty acids (BCFA), synthetic ether-chain phospholipids and bile acids occurs through ABCD3 (also named as PMP70) [12]. Pex2 deficiency increases cholesterol synthesis in the liver of newborn mice [13]. Pex11b deficiency increases neuronal apoptosis and causes defects in peroxisomal FA β-oxidation and peroxisomal ether lipid biosynthesis in Zellweger syndrome mice [14]. Also, Pex11b deficiency results in developmental delay of kidneys and livers [14].
The peroxisome has a dense crystalline core within a large number of antioxidant enzymes, such as catalase, peroxiredoxin (PRX) 1, 5, and 6, copper- and zinc-containing superoxide dismutase (Cu/ZnSOD), and epoxide hydrolase, which may play important roles in ROS metabolism [12]. Endogenous catalase has protective effects on the kidney from diabetic stress through maintaining peroxisomal fitness [15]. Also, redox imbalance in peroxisomes of catalase knockout mice accelerates NAFLD in mice [16]. Impaired peroxisomal fitness may enhance oxidative stress and inflammation in white adipose tissue leading to obesity [17]. Thus, any alteration in peroxisomal function can potentially exacerbate the oxidative stress leading to tissue injury [15-18]. Accordingly, the loss of peroxisomes and impaired peroxisomal functions have been demonstrated to occur in inflammatory conditions, including NASH and NAFLD [16,18].
Fenofibrate 2-[4-(4-chlorobenzoyl)phenoxy]-2-methylpropanoic acid, 1-methylethyl ester), a peroxisome proliferator-activated receptor α (PPARα) agonist, is widely used in the clinic as a lipid-lowering agent against mixed dyslipidemia or primary hypercholesterolemia [19], while PPARα is the key regulator of fatty acid oxidation (FAO) [20]. Fenofibrate reduces the activity of acetyl coenzyme A carboxylase (ACC) and fatty acid synthase (FAS), thus inhibiting the de novo synthesis of FAs. Fenofibrate may reduce the risk of cardiovascular disease and diabetic retinopathy in diabetic patients [21,22]. Fenofibrate protects mice against high-fat diet (HFD)-induced kidney injury [23]. Also, fenofibrate induces the expression of Pex11a in the kidney, which may increase the elongation and number of peroxisomes [24]. Fenofibrate prevents fasting-refeeding process-induced abnormal liver function by increasing peroxisomal FAO and peroxisome biogenesis [25]. On the other hand, a PPARα independent action of fenofibrate also has been reported in various tissues and cells [23,26,27]. Thus, it is important to understand the underlying molecular mechanisms of fenofibrate-mediated peroxisomal fitness in the liver.
The purpose of this study was (1) to evaluate the role of the peroxisome and (2) to determine the involvement of PPARα in fenofibrate-induced peroxisomal fitness against HFD-induced NAFLD in mice.
METHODS
Materials
Chemicals and immunoblotting antibodies were obtained from Sigma-Aldrich Company (St. Louis, MO, USA), Nunc (Rochester, NY, USA), and Cell Signaling Technology (Danvers, MA, USA), respectively, unless otherwise stated.
Animals
In series I, 8-week-old male C57BL/6J wild-type (WT) mice were used. In series II, 8-week-old male Ppara-deficient (Ppara-/-) [28] mice were used. The mice were housed in a temperature-controlled room on a 12-hour light-dark cycle. They were fed a normal diet (ND; 10% fat, Research Diets D12450) or a HFD (60% fat, Research Diets D12492) for 12 weeks [17]. Fenofibrate (F6020, Sigma-Aldrich) was prepared at 0.5% dissolved in carboxymethyl cellulose (CMC) and administered to ND, HFD, and HFD Ppara-/- mice daily at a dose of 50 mg/kg (200 to 400 μL/mice) by oral gavage for 12 weeks. ND and HFD mice not treated by fenofibrate were administered with an equal volume of CMC. After 12 weeks, all mice were sacrificed. Blood plasma and liver tissues were collected for further analysis. All animal studies were approved by the Institutional Animal Care and Use Committee of Ewha Womans University (No.15-062 and No.18-054).
Measurement of blood parameters
Blood samples were collected with a heparinized syringe and centrifuged at 3,000 rpm for 20 minutes at 4°C to collect the plasma. Plasma alanine aminotransferase (ALT) levels were measured using an EnzyChrom L-Alanine Assay Kit (EALA-100, BioAssay Systems, Hayward, CA, USA).
Real-time polymerase chain reaction
Tissue samples were subjected to real-time polymerase chain reaction (qPCR) analysis as previously described [15]. Briefly, the mRNA levels were measured by qPCR using a SYBR Green PCR Master Mix kit (Applied Biosystems, Foster City, CA, USA) with the StepOne Real-Time PCR System (Applied Biosystems). The relative quantitation of each gene was calculated after normalization to 18S rRNA levels. The primer sequences are listed in Supplementary Table 1.
Histology and immunohistochemistry staining
Tissue samples were subjected to immunohistochemistry (IHC) analysis as previously described [15]. Briefly, liver tissue was fixed in 4% paraformaldehyde-lysine-periodate, dehydrated, and embedded in paraffin. To examine the liver histology and morphology, 5 μm liver sections were stained with hematoxylin and eosin (H&E). For IHC staining, anti-4-hydroxynonena (4-HNE; 1:200, MHN-100P, CiteAb, New Bond St, UK), anti-F4/80 (1:200; Santa Cruz Biotechnology Inc., Dallas, TX, USA), anti-nitrotyrosine (NT, 1:400, sc-32757, Santa Cruz Biotechnology Inc.), anti-8-hydroxyguanine (8-oxo-dG; 1:200; 4354-MC-050, Trevigen, Gaithersburg, MD, USA), and anti-collagen 1 (COL1; 1:200, 1310-01, Southern Biotech, Birmingham, AL, USA) antibodies were used and incubated with the tissue sections overnight at 4°C. Images were captured using a Zeiss microscope equipped with an Axio Cam HRC digital camera and Axio Cam software (Carl Zeiss, Thornwood, NY, USA). The staining intensities were quantified using Image-Pro Plus 4.5 software (Cybernetics, Silver Spring, MD, USA).
Immunofluorescence staining
Liver sections were incubated with the indicated primary antibodies, such as anti-ABCD3 (1:200, ab85550, Abcam, Cambridge, UK), anti-catalase (1:200, sc271803, Santa Cruz Biotechnology Inc.), and anti-adipose differentiation-related protein (ADFP, 1:200, ab52356, Abcam). After incubation with the primary antibodies, the liver sections were subsequently incubated with Alexa 488-conjugated goat anti-mouse (1:1,000, A11018, Invitrogen, Carlsbad, CA, USA) and Alexa 568-conjugated goat anti-rabbit (1:1,000, A11070, Invitrogen). 4´,6-Diamidino-2-phenylindole dihydrochloride (DAPI, 1:1,000, 62248, Thermo Fisher Scientific, Waltham, MA, USA) was used for cell nuclei staining.
Immunoblotting analysis
Tissue samples were subjected to immunoblotting analysis as described previously [15]. Briefly, liver tissue was homogenized in lysis buffer and centrifuged at 13,000 rpm, 4°C for 15 minutes. The total concentration of protein was measured using the Bradford protein assay dye (Bio-Rad Laboratories, Hercules, CA, USA). The whole lysate was mixed with 5× sample buffer and denatured at 95°C for 6 minutes. The total proteins were separated by SDS-PAGE gel electrophoresis and transferred onto a polyvinylidene fluoride membrane (GE Healthcare BioSciences Co., Piscataway, NJ, USA). After protein blocking, the membranes were incubated with anti-phospho-nuclear factor kappa B (p-NF-κB, 1:1,000, #3033, Cell Signaling Technology), anti-total-nuclear factor kappa B (t-NF-κB, 1:1,000, #8243, Cell Signaling Technology), anti-acyl-CoA oxidase 1 (ACOX1; 1:1,000, sc-98499, Santa Cruz Biotechnology Inc.), and anti-β-actin (1:3,000, A5326, Sigma-Aldrich) antibodies. The blots were reacted with peroxidase-conjugated secondary antibodies (Vector Laboratories Inc., Burlingame, CA, USA), and the positive immunoreactive protein bands were detected using LAS-3000 film (FUJIFILM Corporation, Tokyo, Japan). All protein levels were normalized to β-actin.
Measurement of peroxisomal FAO
Liver FAO levels were measured as previously described [17]. Liver tissue (50 mg) was placed in reaction buffer containing 0.2 mM palmitate (NEC075H250UC, 14C-palmitate at 1.25 μCi/mL, NEN Life Science, Boston, MA, USA). Homogenized liver samples were incubated on an orbital shaker-incubator (Vision, Daejeon, Korea) at 30°C. The FAO reaction produced 14CO2 was trapped with 1 N NaOH solution. After 2 hours of incubation, the reaction was stopped by the addition of 4 N sulfuric acid. The trapped 14CO2 solution was mixed with a liquid scintillation cocktail (Ultima Gold, PerkinElmer, Waltham, MA, USA) and measured using a Packard TopCount NXT Luminescence and Scintillation Counter (Packard, San Diego, CA, USA). Peroxisomal FAO was measured in the reaction buffer in the presence of 100 µM antimycin A and 12.5 µM rotenone.
Statistical analysis
All results are expressed as mean±standard error (SE). Using Statview 5.0 software, the results were analyzed by one-way analysis of variance (ANOVA) among multiple groups. A P<0.05 was considered significant.
RESULTS
Fenofibrate ameliorates HFD-induced liver injury in mice
HFD feeding for 12 weeks accelerated the gain of body weight, and body weight at the time of sacrifice were 25.5±0.5 and 33.7±0.5 g in ND- and HFD-fed mice (P<0.05), respectively. Fenofibrate effectively prevented HFD-induced weight gain, and body weight of treated HFD mice were 30.1±0.4 g (P<0.05 compared to HFD-fed mice). The protective effects of fenofibrate against HFD-induced liver injury have been established [25,29,30]. Consistently, immunofluorescence (IF) staining of ADFP (also called as perilipin-2), a marker of lipid droplets, was increased in HFD-fed mice liver and inhibited by fenofibrate (Fig. 1A) in the present study. Macrophage infiltration in the fatty liver was elevated as indicated by an increase in F4/80-positive staining area, which was significantly decreased by fenofibrate treatment (Fig. 1A and B). 8-oxo-dG, NT, and 4-HNE immunostaining were used to determine the state of oxidative stress in HFD mice after fenofibrate treatment. HFD significantly increased 8-oxo-dG, NT, and 4-HNE accumulation, which were inhibited by fenofibrate (Fig. 1A, C, D, and E). HFD-induced Il1b, Il6, F4/80, and monocyte chemoattractant protein 1 (Mcp1) mRNA levels were also inhibited by fenofibrate (Fig. 1F). Also, the protein levels of liver p-NF-κB were increased in HFD-fed mice, which were inhibited by fenofibrate (Fig. 1G and H). HFD-fed mice showed significantly increased plasma ALT levels, which were effectively decreased by fenofibrate (Fig. 1I).
Fig. 1.
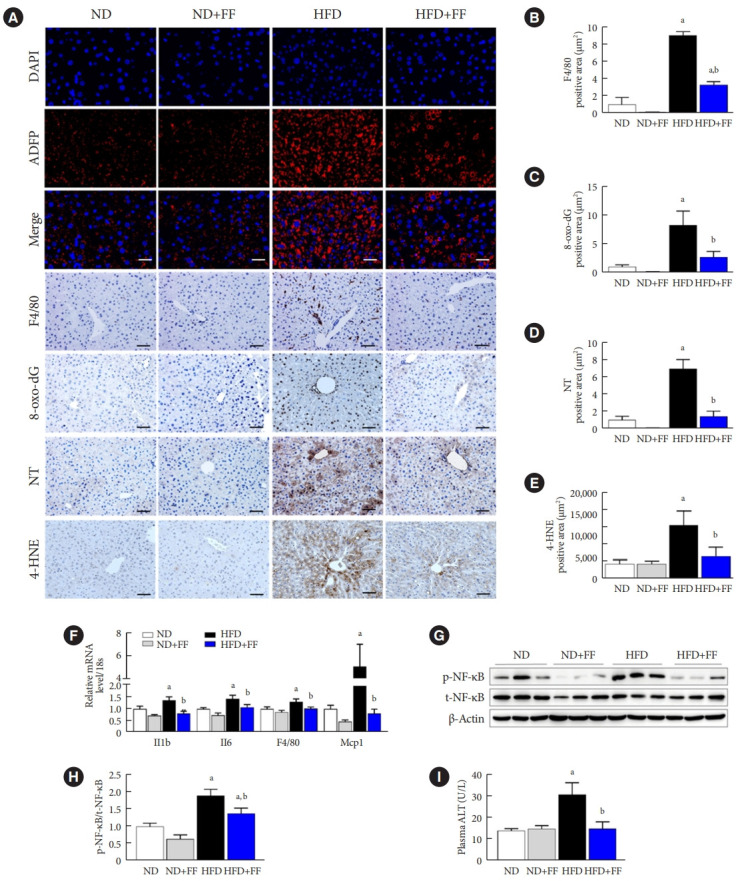
Fenofibrate (FF) ameliorates high-fat diet (HFD)-induced liver dysfunction in wild-type (WT) mice. (A) Liver sections were immunofluorescence (IF) stained for adipose differentiation-related protein (ADFP; red) and 4´,6-diamidino-2-phenylindole dihydrochloride (DAPI) nuclear counterstaining (blue). Original magnification, 200×; scale bar, 100 μm. (A, B, C, D, E) Liver sections were also stained with anti-F4/80, 8-hydroxyguanine (8-oxo-dG), nitrotyrosine (NT), and 4-hydroxynonena (4-HNE) antibodies and were quantified. Original magnification, 100×; scale bar, 200 μm, n=4. (F) Interleukin 1β (Il1b), Il6, F4/80, and monocyte chemoattractant protein 1 (Mcp1) were measured by real-time polymerase chain reaction, and the results were normalized to the 18S rRNA levels. (G, H) The protein levels of phospho-nuclear factor kappa B (p-NF-κB) and total-NF-κB (t-NF-κB) were measured by Western blotting. (I) Plasma alanine aminotransferase (ALT) levels were measured using an enzyme-linked immunosorbent assay (ELISA) kit. Data are expressed as the mean±standard error of 6 mice/group. ND, normal diet. aP<0.05 vs. ND mice, bP<0.05 vs. HFD mice.
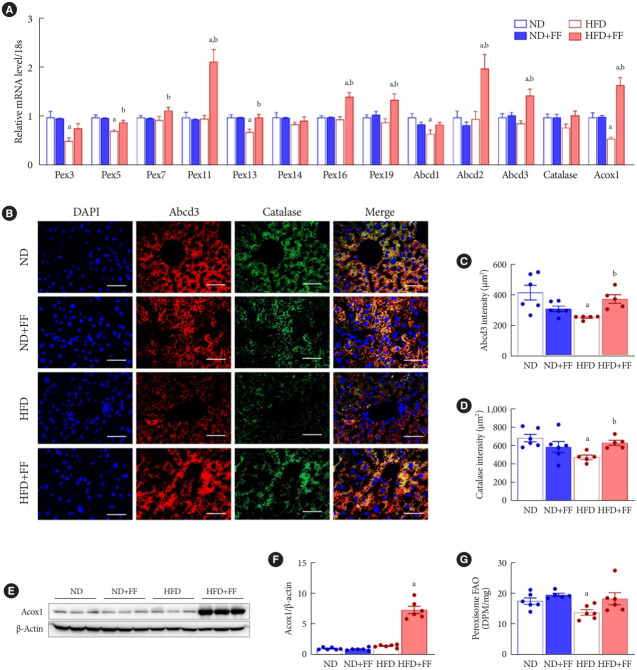
Fenofibrate increases liver peroxisomal biogenesis in HFD mice
HFD impairs liver peroxisomal biogenesis, resulting fatty liver in mice [16]. Fenofibrate restores fasting/refeeding-induced impairment of liver peroxisomal biogenesis in mice [25]. Interestingly, in the current study, expression of Pex3, Pex5, Pex13, Abcd1, and Acox1 mRNA was significantly decreased in HFD-fed mice liver (Fig. 2A). And nine of 13 analyzed genes (Pex5, Pex7, Pex11, Pex13, Pex16, Pex19, Abcd2, Abcd3, and Acox1) involved in peroxisomal fitness were upregulated in the liver of fenofibrate-treated HFD mice compared to each of HFD-fed mice. The administration of fenofibrate did not affect the expression of any gene in ND-fed mice. We, thus, measured mRNA expression of PPARα target genes such as cluster of differentiation 36 (Cd36), fatty-acid-binding protein (Fabp), and peroxisome proliferator-activated receptor co-activator-1α (Pgc1a). Fenofibrate upregulated the expression of Cd36 and Fabp, but not Pgc1a, mRNA in ND-fed mice (Supplementary Fig. 1). These data suggest that not all target genes are simultaneously regulated to the same extent.
Fig. 2.
Fenofibrate (FF) improves liver peroxisomal function in high-fat diet (HFD)-fed wild-type (WT) mice. (A) Peroxisome-related genes were analyzed by real-time polymerase chain reaction, and the results were normalized to 18S rRNA levels. (B, C, D) Liver sections were used for immunofluorescence (IF) staining of ATP binding cassette subfamily D member 3 (ABCD3; red), catalase (green), and 4´,6-diamidino-2-phenylindole dihydrochloride (DAPI) counterstaining (blue) and were quantified. Original magnification, 200×; scale bar, 200 μm. (E, F) Protein levels of acyl-CoA oxidase 1 (ACOX1) were measured by Western blotting, and the results were normalized to β-actin levels. (G) Peroxisomal fatty acid oxidation (FAO) was measured in liver tissue. Data are expressed as the mean±standard error of 6 mice/group. ND, normal diet; Pex, peroxisomal biogenesis factor; DPM, disintegration per minute. aP<0.05 vs. ND mice, bP<0.05 vs. HFD mice.
Fenofibrate improves liver peroxisomal function in HFD mice
Catalase is the most abundant peroxisomal antioxidant enzyme [31], while ABCD3, a major component of the peroxisomal membrane, is involved in metabolic transport of long-chain acyl-CoA [32]. To examine the effect of fenofibrate on peroxisomal function, we co-stained ABCD3 and catalase in the liver sections. HFD-fed mice showed decreased expression of ABCD3 and catalase, which were effectively inhibited by fenofibrate (Fig. 2B-D), suggesting that fenofibrate increased functional peroxisomes in the fatty liver. Fenofibrate increased the protein levels of ACOX1, a rate-limiting enzyme in the peroxisomal β-oxidation pathway, in HFD-fed mice without significant effect on ND-fed mice (Fig. 2E and F). Importantly, HFD feeding significantly reduced peroxisomal FAO in mice liver, which was also effectively increased by fenofibrate (Fig. 2G).
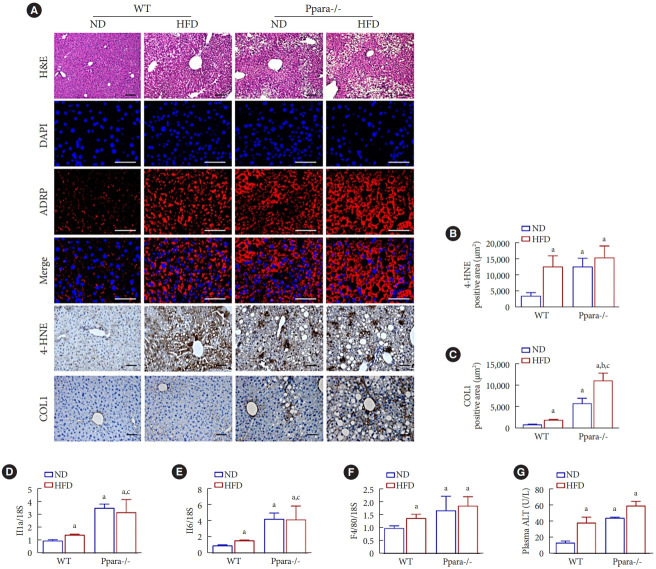
PPARα is important for maintaining liver homeostasis in mice
PPARα activation is implicated in improving steatosis, inflammation, and fibrosis in various pre-clinical models of NAFLD [33]. Hepatocyte-specific deletion of Ppara promotes NAFLD phenotypes even under ND in mice [34]. In silico analysis using Gene Expression Omnibus (GES83452) data of human liver biopsy of normal and NASH showed decreased PPARA expression in patients with NASH (Supplementary Fig. 2). Interestingly, increased lipid accumulation in Ppara-/- mice under ND was exacerbated by HFD (Fig. 3A). Deficiency of Ppara increased the levels of 4-HNE and COL1 even under ND, and HFD feeding further increased COL1 immunostaining in Ppara-/- mice (Fig. 3A-C). Basal expression of Il1b, Il6, and F4/80 mRNA were significantly increased in Ppara-/- mice (Fig. 3D-F). Consistently, a deficiency of Ppara increased plasma ALT levels in mice even under ND (Fig. 3G). Fenofibrate failed to reduce HFD-induced ALT in Ppara-/- mice (Supplementary Fig. 3).
Fig. 3.
Role of peroxisome proliferator-activated receptor α (PPARα) in maintaining liver homeostasis in Ppara-/- mice. (A, B, C) Liver morphology was detected by H&E staining. Original magnification, 100×; scale bar, 200 μm. Liver sections were immunofluorescence (IF) stained for adipose differentiation-related protein (ADFP; red) and 4´,6-diamidino-2-phenylindole dihydrochloride (DAPI) nuclear counterstaining (blue). Original magnification, 200×; scale bar, 200 μm. Liver sections were also stained with anti-4-hydroxynonena (4-HNE) and collagen 1 (COL1) antibody and the positive area were quantified. Original magnification, 100×; scale bar, 200 μm. (D, E, F) Interleukin 1β (Il1b), Il6, and F4/80 were measured by real-time polymerase chain reaction, and the results were normalized to the 18S rRNA levels. (G) Plasma alanine aminotransferase (ALT) levels were measured using an enzyme-linked immunosorbent assay (ELISA) kit. Data are expressed as the mean±standard error of 6 mice/group. ND, normal diet; WT, wild-type; HFD, high-fat diet; ADRP, adipose differentiation-related protein. aP<0.05 vs. WT mice with ND, bP<0.05 vs. Ppara-/- mice with ND, cP<0.05 vs. WT mice with HFD.
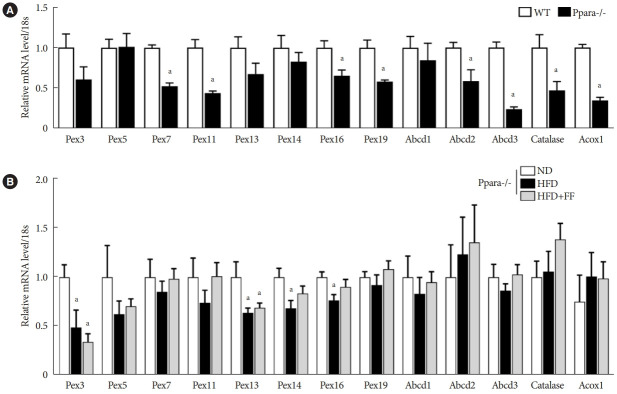
Fenofibrate fails to maintain peroxisomal biogenesis in Ppara-/- mice
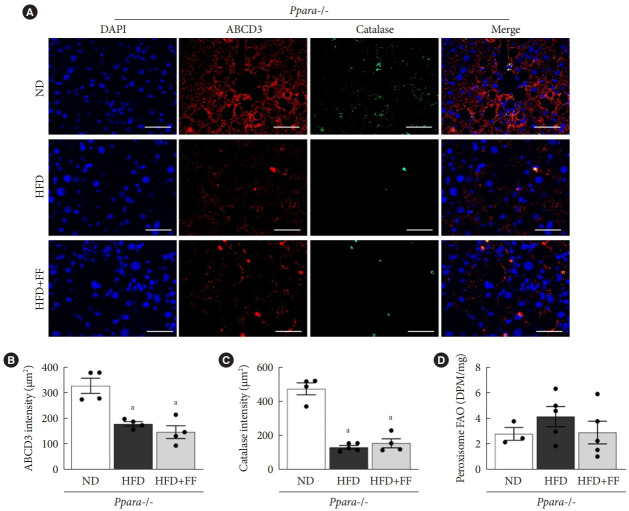
Basal mRNA expression of PPARα target genes such as Pex11, Abcd2, Abcd3, and Acox1 were significantly decreased in Ppara-/- mice liver (Fig. 4A). Basal expression of Pex7, Pex16, Pex19, and catalase mRNA were also significantly decreased in Ppara-/- mice liver (Fig. 4A). Functional peroxisome estimated by ABCD3 and catalase immunostaining were significantly decreased in Ppara-/- mice liver (Supplementary Fig. 4). HFD feeding decreased the mRNA levels of Pex3, Pex13, Pex14, and Pex16 in Ppara-/- mice liver, which was not affected by fenofibrate (Fig. 4B).
Fig. 4.
Fenofibrate fails to maintain peroxisomal biogenesis in Ppara-/- mice. (A) The expression of peroxisome-related genes was decreased in Ppara-/- mice compared to wild-type (WT) mice. (B) Peroxisome-related genes in Ppara-/- mice were analyzed by real-time polymerase chain reaction, and the results were normalized to 18S rRNA levels. Data are expressed as the mean±standard error of 6 mice/group. PPAR, peroxisome proliferator-activated receptor; Pex, peroxisomal biogenesis factor; Abcd, ATP binding cassette subfamily D member; Acox1, acyl-CoA oxidase 1; ND, normal diet; HFD, high-fat diet; FF, fenofibrate. aP<0.05 vs. WT mice or Ppara-/- mice with ND.
Fenofibrate fails to improve peroxisomal function in Ppara-/- mice
IF staining showed that ABCD3 and catalase expression were reduced in HFD-fed Ppara-/- mice compared to ND-fed Ppara-/- mice. As expected, there was little effect of fenofibrate on ABCD3 or catalase protein expression in HFD-fed Ppara-/- mice (Fig. 5A-C). Basal peroxisomal FAO in Ppara-/- mice liver was remarkably suppressed (Fig. 5D) compared to that of WT (Fig. 2G), and there was little difference in FAO among ND, HFD, and fenofibrate-treated HFD Ppara-/- mice (Fig. 5D). These data suggest that fenofibrate fails to maintain peroxisomal function in Ppara-/- mice.
Fig. 5.
Fenofibrate fails to improve peroxisomal function in Ppara-/- mice. (A, B, C) Liver sections were immunofluorescence stained for ATP binding cassette subfamily D member 3 (ABCD3; red), catalase (green), and 4´,6-diamidino-2-phenylindole dihydrochloride (DAPI) nuclear counterstaining (blue) and intensities were quantified. Original magnification, 200×; scale bar, 200 μm. (D) Peroxisomal fatty acid oxidation (FAO) was measured in liver tissue. Data are expressed as the mean±standard error of 6 mice/group. PPAR, peroxisome proliferator-activated receptor; ND, normal diet; HFD, high-fat diet; FF, fenofibrate; DPM, disintegration per minute. aP<0.05 vs. Ppara-/- mice with ND.
DISCUSSION
In this study, administration of fenofibrate to HFD-fed mice (1) effectively prevented liver steatosis and injury characterized by ALT, inflammation, oxidative stress, and lipid accumulation; (2) significantly increased the expression of liver peroxisomal antioxidant- and biogenesis-related markers; and (3) increased liver peroxisomal FAO. In Ppara deficient mice, fenofibrate failed to maintain peroxisomal biogenesis and FAO in HFD-induced liver injury. Taken together, the present data suggest that fenofibrate improves peroxisomal fitness by increasing peroxisomal biogenesis and function and thus protects against HFD-induced NAFLD.
Extensive studies have revealed that fenofibrate decreases HFD-induced plasma TG, ALT, and insulin levels in mice [27,35,36]. In addition, fenofibrate decreases HFD-induced lipid accumulation, inflammation (Il1b, Il6, Mcp1, and tumor necrosis factor-α), oxidative stress, and fibrosis (α-smooth muscle actin [α-SMA] and COL1) in liver [27,35,36]. In the liver of HFD-fed mice, fenofibrate suppresses lipogenesis makers (sterol regulatory element-binding protein 1 [SREBP1] and ACC) [37], increases lipolysis markers (adipose triglyceride lipase [ATGL]) [38] and β-oxidant marker (ACOX1) [36]. In line with these previous reports, the present study has shown that HFD-induced liver lipid drops and plasma ALT levels were decreased in response to fenofibrate treatment. In addition, HFD-induced liver inflammation, oxidative stress, and fibrosis were reduced by fenofibrate. We also confirmed that fenofibrate increased ACOX1 expression in the liver of HFD mice, suggesting increased FAO in response to fenofibrate. However, concerns on the effect of fenofibrate on liver lipid accumulation have been reported; mice under ND treated with fenofibrate for 10 days showed increased liver TG [39], and fenofibrate simultaneous induced FAO, FAS, and FA elongation in the liver of mice under ND [40]. It remains to be important to understand the underlying mechanism of this contradictory effect of fenofibrate on liver lipid accumulation in order to develop effective strategies against NAFLD.
In this context of fenofibrate’s contradicting effect, fenofibrate increases cell viability along with upregulation of nuclear factor erythroid 2–related factor 2 (NRF2) and antioxidant enzymes only under stress condition (high glucose or hypoxia-reoxygenation injury) not at basal in cultured cardiac myocytes [41]. There are differences in activation of PPARα and the hypolipidemic effects of fenofibrate in fish between HFD and ND [42], which could be a reference for other species.
Since fenofibrate is a ligand for PPARα, we have focused on the peroxisome. More than 14 proteins are involved in peroxisomal biogenesis [11]. PEX3, PEX16, and PEX19 play important roles in the early stages of peroxisomal biogenesis, membrane integrity, and protein transport across membranes [43-45]. Also, ABCD3 has been suggested to be involved in metabolic transport of long-chain acyl-CoA [32]. Analysis of Gene Expression Omnibus, a public database, showed that PEX16 and PEX19 were significantly reduced in patients with NASH (GES164760). PEX13, a peroxisome membrane protein that helps import proteins into the peroxisome, was also significantly reduced in patients with NASH (GSE17470). In addition, deficiency of Pex11a, a peroxin involved in peroxisomal division and proliferation [46], reduces peroxisomal biogenesis and FAO, contributing to increased lipid accumulation in the liver [47]. Newborn Pex2 knockout mice exhibit cholesterol synthesis in the liver [13]. In the present study, fenofibrate increased the expression of Pex5, Pex7, Pex11, Pex13, Pex16, Pex19, Abcd2, Abcd3, and Acox1 in the liver of HFD-fed mice, suggesting that fenofibrate increases peroxisomal biogenesis and function in the liver. Since peroxisomal biogenesis is regulated by de novo biogenesis, the growth and division of pre-existing peroxisomes, and pexophagy [9], it will be interesting to understand how these three pathways govern fenofibrate-induced peroxisomal biogenesis.
The main metabolic functions of peroxisomes in mammalian cells include β-oxidation of VLCFA and ROS metabolism [12]. Catalase is the most abundant peroxisomal antioxidant enzyme [31] and effectively removes H2O2 produced during peroxisomal β-oxidation, maintaining both the cellular and the peroxisomal redox homeostasis [48]. Recent studies have demonstrated that endogenous catalase exerts beneficial effects in protecting against liver injury, including lipid accumulation and inflammation, by maintaining the liver redox balance from the early stage of HFD-induced metabolic stress [49] and NAFLD [50]. Inhibition of catalase activity augmented mitochondrial ROS production and DNA damage and impaired cell growth in human diploid fibroblasts [51]. In addition, catalase deficiency enhanced diabetic kidney injury through peroxisomal dysfunction [52]. In the present study, catalase estimated by immunostaining was increased in the liver of HFD-fed mice by fenofibrate. Although we have not measured catalase activity in the present study, decreased oxidative stress in the face of increased peroxisomal FAO in fenofibrate-treated HFD mice suggests that fenofibrate increases functional peroxisomes in the liver under HFD stress. The present data were obtained at 12 weeks after HFD feeding with or without fenofibrate; it is necessary to determine the peroxisomal biogenesis and function including FAO over time, rather than a single instant, for a comprehensive understanding.
PPARα is highly expressed in the liver and brown adipose tissue and is a key regulator of FAO [20]. PPARα is associated with peroxisomal lipid oxidation and synthesis [53]. PPARα activation not only increases the expression of FAO genes but also molecules regulating peroxisomal biogenesis in the liver [54]. When PPARα is activated, it increases the expression of Pex11, which is involved in peroxisome biosynthesis by promoting the division of peroxisomes [55]. In addition, PPARα governs inflammatory process, mainly by trans-repression of proinflammatory genes [33,36], and lipid accumulation and inflammation are intertwined [56]. Pharmacological activation of PPARα also prevents intrahepatic inflammation and fibrosis by inhibiting activated macrophages and stellate cells and lowering the expression of fibrotic markers [33]. In the liver, PPARA gene expression was significantly decreased in patients with NASH compared to those without NASH (Supplementary Fig. 2). In our study, the deficiency of Ppara augmented liver injury by increasing plasma ALT, liver inflammation and fibrosis, and lipid accumulation even under ND. In addition, the deficiency of Ppara failed to maintain peroxisomal biogenesis and function. Thus, the present data suggest a potential role of PPARα in maintaining liver homeostasis and peroxisomal fitness. However, it should be noted that there is a PPARα-independent effect of fenofibrate [23,26,27].
Further investigations are necessary to support the current findings. To confirm the involvement of the peroxisome in NAFLD, knockdown of major genes related to peroxisomal biogenesis and function need to be performed. Although delayed treatment with fenofibrate protects against HFD-induced kidney injury [23], the therapeutic effects of fenofibrate on peroxisomal biogenesis and function against NAFLD need to be investigated. Although PEX13, 16, and 19 were significantly reduced in patients with NASH, data about the involvement of peroxisomes in NAFLD in humans are not available yet.
In conclusion, the present results confirm that fenofibrate protects against HFD-induced liver injury such as inflammation, oxidative stress, fibrosis, and lipid accumulation in mice. Importantly, fenofibrate increases peroxisome biogenesis and function via PPARα in the liver of HFD mice (Fig. 6). Thus, it is suggested that improved peroxisomal fitness induced by fenofibrate may play an important role in protecting against NAFLD.
Fig. 6.
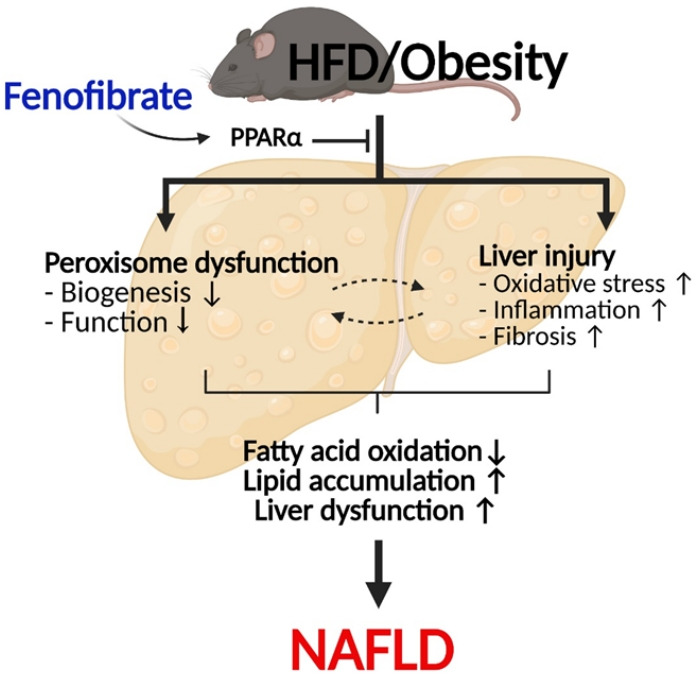
Suggested model of fenofibrate-mediated peroxisomal fitness against high-fat diet (HFD)-induced non-alcoholic fatty liver disease (NAFLD). HFD or obesity decreases peroxisomal biogenesis and function and increases liver injury, including oxidative stress, inflammation, and fibrosis, due to inhibition of the peroxisome proliferator-activated receptor α (PPARα) pathway. Subsequently, it results in decreased hepatic fatty acid oxidation, increased lipid accumulation, and the induction of liver dysfunction, which leads to the development of NAFLD. Fenofibrate maintains peroxisomal biogenesis and function through activation of the PPARα pathway. It also attenuates liver injury and increases hepatic fatty acid oxidation. Thus, fenofibrate may mediate protective effects against NAFLD by maintaining peroxisomal biogenesis and function.
Acknowledgments
We are immensely grateful to Frank J. Gonzalez (National Institutes of Health/ National Cancer Institute) for providing the Ppara-/- mice.
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
AUTHOR CONTRIBUTIONS
Conception or design: H.H.
Acquisition, analysis, or interpretation of data: S.J., X.Y., L.P., D.D.
Drafting the work or revising: S.J., M.J.U.
Final approval of the manuscript: S.J., M.J.U., X.Y., L.P., D.D., G.T.O., H.H.
FUNDING
This work was supported by National Research Foundation (No. 2020R1I1A1A01072879 and 2019R1A2C2002720), Brain Pool program funded by the Ministry of Science and ICT through the National Research Foundation (No. 2020H1D3A-2A02110924), and research grant from Ewha Womans University (1-2022-0767-001-1), Republic of Korea.
SUPPLEMENTARY MATERIALS
Supplementary materials related to this article can be found online at https://doi.org/10.4093/dmj.2021.0274.
Primer sequences used for RT-qPCR analysis
The expression of peroxisome proliferator-activated receptor α (PPARα) target genes in wild-type (WT) mice. (A) Cluster of differentiation 36 (Cd36), (B) fatty-acid-binding protein (Fabp), and (C) peroxisome proliferator-activated receptor co-activator-1α (Pgc1a) were measured by real-time polymerase chain reaction, and the results were normalized to the 18S rRNA levels. Data are expressed as the mean±standard error of 6 mice/group. FF, fenofibrate; HFD, high-fat diet; ND, normal diet. aP<0.05 vs. ND, bP<0.05 vs. HFD.
Differently expressed peroxisome proliferator-activated receptor α (PPARα) in patients with or without non-alcoholic steatohepatitis (NASH). Analysis of PPARA gene expression using Gene Expression Omnibus database (GSE83452) was performed. Data of GSE83452 were obtained for 224 subjects (98 no NASH and 126 NASH). aP<0.05 vs. no NASH.
Plasma alanine aminotransferase (ALT) levels in Ppara-/- mice. Plasma ALT levels were measured using an enzyme-linked immunosorbent assay (ELISA) kit. Data are expressed as the mean±standard error of 6 mice/group. ND, normal diet; HFD, high-fat diet; FF, fenofibrate. aP<0.05 vs. ND.
Peroxisome biogenesis and activity in wild-type (WT) and Ppara-/- mice. (A, B) ATP binding cassette subfamily D member 3 (ABCD3) and catalase levels were decreased in Ppara-/- mice. DAPI, 4´,6-diamidino-2-phenylindole dihydrochloride. aP<0.05 vs. WT.
REFERENCES
- 1.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2014;384:766–81. doi: 10.1016/S0140-6736(14)60460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tiniakos DG, Vos MB, Brunt EM. Nonalcoholic fatty liver disease: pathology and pathogenesis. Annu Rev Pathol. 2010;5:145–71. doi: 10.1146/annurev-pathol-121808-102132. [DOI] [PubMed] [Google Scholar]
- 4.Bril F, Barb D, Portillo-Sanchez P, Biernacki D, Lomonaco R, Suman A, et al. Metabolic and histological implications of intrahepatic triglyceride content in nonalcoholic fatty liver disease. Hepatology. 2017;65:1132–44. doi: 10.1002/hep.28985. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z, Tian R, She Z, Cai J, Li H. Role of oxidative stress in the pathogenesis of nonalcoholic fatty liver disease. Free Radic Biol Med. 2020;152:116–41. doi: 10.1016/j.freeradbiomed.2020.02.025. [DOI] [PubMed] [Google Scholar]
- 6.Zelber-Sagi S, Ivancovsky-Wajcman D, Fliss-Isakov N, Hahn M, Webb M, Shibolet O, et al. Serum malondialdehyde is associated with non-alcoholic fatty liver and related liver damage differentially in men and women. Antioxidants (Basel) 2020;9:578. doi: 10.3390/antiox9070578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekiya M, Hiraishi A, Touyama M, Sakamoto K. Oxidative stress induced lipid accumulation via SREBP1c activation in HepG2 cells. Biochem Biophys Res Commun. 2008;375:602–7. doi: 10.1016/j.bbrc.2008.08.068. [DOI] [PubMed] [Google Scholar]
- 8.Ferramosca A, Di Giacomo M, Zara V. Antioxidant dietary approach in treatment of fatty liver: new insights and updates. World J Gastroenterol. 2017;23:4146–57. doi: 10.3748/wjg.v23.i23.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Germain K, Kim PK. Pexophagy: a model for selective autophagy. Int J Mol Sci. 2020;21:578. doi: 10.3390/ijms21020578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugiura A, Mattie S, Prudent J, McBride HM. Newly born peroxisomes are a hybrid of mitochondrial and ER-derived preperoxisomes. Nature. 2017;542:251–4. doi: 10.1038/nature21375. [DOI] [PubMed] [Google Scholar]
- 11.Smith JJ, Aitchison JD. Peroxisomes take shape. Nat Rev Mol Cell Biol. 2013;14:803–17. doi: 10.1038/nrm3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lodhi IJ, Semenkovich CF. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 2014;19:380–92. doi: 10.1016/j.cmet.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovacs WJ, Charles KN, Walter KM, Shackelford JE, Wikander TM, Richards MJ, et al. Peroxisome deficiency-induced ER stress and SREBP-2 pathway activation in the liver of newborn Pex2 knock-out mice. Biochim Biophys Acta. 2012;1821:895–907. doi: 10.1016/j.bbalip.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Baumgart E, Morrell JC, Jimenez-Sanchez G, Valle D, Gould SJ. PEX11 beta deficiency is lethal and impairs neuronal migration but does not abrogate peroxisome function. Mol Cell Biol. 2002;22:4358–65. doi: 10.1128/MCB.22.12.4358-4365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang I, Lee J, Huh JY, Park J, Lee HB, Ho YS, et al. Catalase deficiency accelerates diabetic renal injury through peroxisomal dysfunction. Diabetes. 2012;61:728–38. doi: 10.2337/db11-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang I, Uddin MJ, Pak ES, Kang H, Jin EJ, Jo S, et al. The impaired redox balance in peroxisomes of catalase knockout mice accelerates nonalcoholic fatty liver disease through endoplasmic reticulum stress. Free Radic Biol Med. 2020;148:22–32. doi: 10.1016/j.freeradbiomed.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 17.Piao L, Dorotea D, Jiang S, Koh EH, Oh GT, Ha H. Impaired peroxisomal fitness in obese mice, a vicious cycle exacerbating adipocyte dysfunction via oxidative stress. Antioxid Redox Signal. 2019;31:1339–51. doi: 10.1089/ars.2018.7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Islam S, Won J, Khan M, Chavin KD, Singh I. Peroxisomal footprint in the pathogenesis of nonalcoholic steatohepatitis. Ann Hepatol. 2020;19:466–71. doi: 10.1016/j.aohep.2019.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Farnier M. Update on the clinical utility of fenofibrate in mixed dyslipidemias: mechanisms of action and rational prescribing. Vasc Health Risk Manag. 2008;4:991–1000. doi: 10.2147/vhrm.s3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montagner A, Polizzi A, Fouche E, Ducheix S, Lippi Y, Lasserre F, et al. Liver PPARα is crucial for whole-body fatty acid homeostasis and is protective against NAFLD. Gut. 2016;65:1202–14. doi: 10.1136/gutjnl-2015-310798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jo SH, Nam H, Lee J, Park S, Lee J, Kyoung DS. Fenofibrate use is associated with lower mortality and fewer cardiovascular events in patients with diabetes: results of 10,114 patients from the Korean National Health Insurance Service Cohort. Diabetes Care. 2021;44:1868–76. doi: 10.2337/dc20-1533. [DOI] [PubMed] [Google Scholar]
- 22.Elam MB, Ginsberg HN, Lovato LC, Corson M, Largay J, Leiter LA, et al. Association of fenofibrate therapy with long-term cardiovascular risk in statin-treated patients with type 2 diabetes. JAMA Cardiol. 2017;2:370–80. doi: 10.1001/jamacardio.2016.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sohn M, Kim K, Uddin MJ, Lee G, Hwang I, Kang H, et al. Delayed treatment with fenofibrate protects against high-fat dietinduced kidney injury in mice: the possible role of AMPK autophagy. Am J Physiol Renal Physiol. 2017;312:F323–34. doi: 10.1152/ajprenal.00596.2015. [DOI] [PubMed] [Google Scholar]
- 24.Weng H, Ji X, Endo K, Iwai N. Pex11a deficiency is associated with a reduced abundance of functional peroxisomes and aggravated renal interstitial lesions. Hypertension. 2014;64:1054–60. doi: 10.1161/HYPERTENSIONAHA.114.04094. [DOI] [PubMed] [Google Scholar]
- 25.Lee JN, Dutta RK, Kim SG, Lim JY, Kim SJ, Choe SK, et al. Fenofibrate, a peroxisome proliferator-activated receptor α ligand, prevents abnormal liver function induced by a fastingrefeeding process. Biochem Biophys Res Commun. 2013;442:22–7. doi: 10.1016/j.bbrc.2013.10.140. [DOI] [PubMed] [Google Scholar]
- 26.Araki H, Tamada Y, Imoto S, Dunmore B, Sanders D, Humphrey S, et al. Analysis of PPARalpha-dependent and PPARalpha-independent transcript regulation following fenofibrate treatment of human endothelial cells. Angiogenesis. 2009;12:221–9. doi: 10.1007/s10456-009-9142-8. [DOI] [PubMed] [Google Scholar]
- 27.Hua H, Yang J, Lin H, Xi Y, Dai M, Xu G, et al. PPARα-independent action against metabolic syndrome development by fibrates is mediated by inhibition of STAT3 signalling. J Pharm Pharmacol. 2018;70:1630–42. doi: 10.1111/jphp.13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akiyama TE, Nicol CJ, Fievet C, Staels B, Ward JM, Auwerx J, et al. Peroxisome proliferator-activated receptor-alpha regulates lipid homeostasis, but is not associated with obesity: studies with congenic mouse lines. J Biol Chem. 2001;276:39088–93. doi: 10.1074/jbc.M107073200. [DOI] [PubMed] [Google Scholar]
- 29.Kondo K, Sugioka T, Tsukada K, Aizawa M, Takizawa M, Shimizu K, et al. Fenofibrate, a peroxisome proliferator-activated receptor alpha agonist, improves hepatic microcirculatory patency and oxygen availability in a high-fat-diet-induced fatty liver in mice. Adv Exp Med Biol. 2010;662:77–82. doi: 10.1007/978-1-4419-1241-1_10. [DOI] [PubMed] [Google Scholar]
- 30.Kostapanos MS, Kei A, Elisaf MS. Current role of fenofibrate in the prevention and management of non-alcoholic fatty liver disease. World J Hepatol. 2013;5:470–8. doi: 10.4254/wjh.v5.i9.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walton PA, Brees C, Lismont C, Apanasets O, Fransen M. The peroxisomal import receptor PEX5 functions as a stress sensor, retaining catalase in the cytosol in times of oxidative stress. Biochim Biophys Acta Mol Cell Res. 2017;1864:1833–43. doi: 10.1016/j.bbamcr.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 32.Imanaka T, Aihara K, Suzuki Y, Yokota S, Osumi T. The 70- kDa peroxisomal membrane protein (PMP70), an ATP-binding cassette transporter. Cell Biochem Biophys. 2000;32:131–8. doi: 10.1385/cbb:32:1-3:131. [DOI] [PubMed] [Google Scholar]
- 33.Pawlak M, Lefebvre P, Staels B. Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J Hepatol. 2015;62:720–33. doi: 10.1016/j.jhep.2014.10.039. [DOI] [PubMed] [Google Scholar]
- 34.Regnier M, Polizzi A, Smati S, Lukowicz C, Fougerat A, Lippi Y, et al. Hepatocyte-specific deletion of Pparα promotes NAFLD in the context of obesity. Sci Rep. 2020;10:6489. doi: 10.1038/s41598-020-63579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jain MR, Giri SR, Bhoi B, Trivedi C, Rath A, Rathod R, et al. Dual PPARα/γ agonist saroglitazar improves liver histopathology and biochemistry in experimental NASH models. Liver Int. 2018;38:1084–94. doi: 10.1111/liv.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rakhshandehroo M, Knoch B, Muller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 2010;2010:612089. doi: 10.1155/2010/612089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng S, Liang S, Liu Q, Deng Z, Zhang Y, Du J, et al. Diosgenin prevents high-fat diet-induced rat non-alcoholic fatty liver disease through the AMPK and LXR signaling pathways. Int J Mol Med. 2018;41:1089–95. doi: 10.3892/ijmm.2017.3291. [DOI] [PubMed] [Google Scholar]
- 38.Chen WL, Chen YL, Chiang YM, Wang SG, Lee HM. Fenofibrate lowers lipid accumulation in myotubes by modulating the PPARα/AMPK/FoxO1/ATGL pathway. Biochem Pharmacol. 2012;84:522–31. doi: 10.1016/j.bcp.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Yan F, Wang Q, Xu C, Cao M, Zhou X, Wang T, et al. Peroxisome proliferator-activated receptor α activation induces hepatic steatosis, suggesting an adverse effect. PLoS One. 2014;9:e99245. doi: 10.1371/journal.pone.0099245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oosterveer MH, Grefhorst A, van Dijk TH, Havinga R, Staels B, Kuipers F, et al. Fenofibrate simultaneously induces hepatic fatty acid oxidation, synthesis, and elongation in mice. J Biol Chem. 2009;284:34036–44. doi: 10.1074/jbc.M109.051052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cortes-Lopez F, Sanchez-Mendoza A, Centurion D, CervantesPerez LG, Castrejon-Tellez V, Del Valle-Mondragon L, et al. Fenofibrate protects cardiomyocytes from hypoxia/reperfusion- and high glucose-induced detrimental effects. PPAR Res. 2021;2021:8895376. doi: 10.1155/2021/8895376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ning LJ, He AY, Lu DL, Li JM, Qiao F, Li DL, et al. Nutritional background changes the hypolipidemic effects of fenofibrate in Nile tilapia (Oreochromis niloticus) Sci Rep. 2017;7:41706. doi: 10.1038/srep41706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Honsho M, Tamura S, Shimozawa N, Suzuki Y, Kondo N, Fujiki Y. Mutation in PEX16 is causal in the peroxisome-deficient Zellweger syndrome of complementation group D. Am J Hum Genet. 1998;63:1622–30. doi: 10.1086/302161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sacksteder KA, Jones JM, South ST, Li X, Liu Y, Gould SJ. PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J Cell Biol. 2000;148:931–44. doi: 10.1083/jcb.148.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghaedi K, Tamura S, Okumoto K, Matsuzono Y, Fujiki Y. The peroxin pex3p initiates membrane assembly in peroxisome biogenesis. Mol Biol Cell. 2000;11:2085–102. doi: 10.1091/mbc.11.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delille HK, Dodt G, Schrader M. Pex11pβ-mediated maturation of peroxisomes. Commun Integr Biol. 2011;4:51–4. doi: 10.4161/cib.4.1.13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weng H, Ji X, Naito Y, Endo K, Ma X, Takahashi R, et al. Pex11α deficiency impairs peroxisome elongation and division and contributes to nonalcoholic fatty liver in mice. Am J Physiol Endocrinol Metab. 2013;304:E187–96. doi: 10.1152/ajpendo.00425.2012. [DOI] [PubMed] [Google Scholar]
- 48.Walker CL, Pomatto L, Tripathi DN, Davies K. Redox regulation of homeostasis and proteostasis in peroxisomes. Physiol Rev. 2018;98:89–115. doi: 10.1152/physrev.00033.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piao L, Choi J, Kwon G, Ha H. Endogenous catalase delays high-fat diet-induced liver injury in mice. Korean J Physiol Pharmacol. 2017;21:317–25. doi: 10.4196/kjpp.2017.21.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shin SK, Cho HW, Song SE, Bae JH, Im SS, Hwang I, et al. Ablation of catalase promotes non-alcoholic fatty liver via oxidative stress and mitochondrial dysfunction in diet-induced obese mice. Pflugers Arch. 2019;471:829–43. doi: 10.1007/s00424-018-02250-3. [DOI] [PubMed] [Google Scholar]
- 51.Koepke JI, Wood CS, Terlecky LJ, Walton PA, Terlecky SR. Progeric effects of catalase inactivation in human cells. Toxicol Appl Pharmacol. 2008;232:99–108. doi: 10.1016/j.taap.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 52.Hwang I, Uddin MJ, Lee G, Jiang S, Pak ES, Ha H. Peroxiredoxin 3 deficiency accelerates chronic kidney injury in mice through interactions between macrophages and tubular epithelial cells. Free Radic Biol Med. 2019;131:162–72. doi: 10.1016/j.freeradbiomed.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 53.Wang YX. PPARs: diverse regulators in energy metabolism and metabolic diseases. Cell Res. 2010;20:124–37. doi: 10.1038/cr.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schrader M, Grille S, Fahimi HD, Islinger M. Peroxisome interactions and cross-talk with other subcellular compartments in animal cells. Subcell Biochem. 2013;69:1–22. doi: 10.1007/978-94-007-6889-5_1. [DOI] [PubMed] [Google Scholar]
- 55.Li X, Gould SJ. PEX11 promotes peroxisome division independently of peroxisome metabolism. J Cell Biol. 2002;156:643–51. doi: 10.1083/jcb.200112028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Z, Yu R, Xiong Y, Du F, Zhu S. A vicious circle between insulin resistance and inflammation in nonalcoholic fatty liver disease. Lipids Health Dis. 2017;16:203. doi: 10.1186/s12944-017-0572-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences used for RT-qPCR analysis
The expression of peroxisome proliferator-activated receptor α (PPARα) target genes in wild-type (WT) mice. (A) Cluster of differentiation 36 (Cd36), (B) fatty-acid-binding protein (Fabp), and (C) peroxisome proliferator-activated receptor co-activator-1α (Pgc1a) were measured by real-time polymerase chain reaction, and the results were normalized to the 18S rRNA levels. Data are expressed as the mean±standard error of 6 mice/group. FF, fenofibrate; HFD, high-fat diet; ND, normal diet. aP<0.05 vs. ND, bP<0.05 vs. HFD.
Differently expressed peroxisome proliferator-activated receptor α (PPARα) in patients with or without non-alcoholic steatohepatitis (NASH). Analysis of PPARA gene expression using Gene Expression Omnibus database (GSE83452) was performed. Data of GSE83452 were obtained for 224 subjects (98 no NASH and 126 NASH). aP<0.05 vs. no NASH.
Plasma alanine aminotransferase (ALT) levels in Ppara-/- mice. Plasma ALT levels were measured using an enzyme-linked immunosorbent assay (ELISA) kit. Data are expressed as the mean±standard error of 6 mice/group. ND, normal diet; HFD, high-fat diet; FF, fenofibrate. aP<0.05 vs. ND.
Peroxisome biogenesis and activity in wild-type (WT) and Ppara-/- mice. (A, B) ATP binding cassette subfamily D member 3 (ABCD3) and catalase levels were decreased in Ppara-/- mice. DAPI, 4´,6-diamidino-2-phenylindole dihydrochloride. aP<0.05 vs. WT.