Abstract
Plants have evolved unique morphological and developmental adaptations to cope with the abiotic stresses imposed by (hyper)arid environments. Such adaptations include the formation of rhizosheath–root system in which mutualistic plant–soil microbiome associations are established: the plant provides a nutrient-rich and shielded environment to microorganisms, which in return improve plant-fitness through plant growth promoting services. We hypothesized that the rhizosheath–root systems represent refuge niches and resource islands for the desert edaphic microbial communities. As a corollary, we posited that microorganisms compete intensively to colonize such “oasis” and only those beneficial microorganisms improving host fitness are preferentially selected by plant. Our results show that the belowground rhizosheath–root micro-environment is largely more hospitable than the surrounding gravel plain soil with higher nutrient and humidity contents, and cooler temperatures. By combining metabarcoding and shotgun metagenomics, we demonstrated that edaphic microbial biomass and community stability increased from the non-vegetated soils to the rhizosheath–root system. Concomitantly, non-vegetated soil communities favored autotrophy lifestyle while those associated with the plant niches were mainly heterotrophs and enriched in microbial plant growth promoting capacities. An intense inter-taxon microbial competition is involved in the colonization and homeostasis of the rhizosheath zone, as documented by significant enrichment of antibiotic resistance genes and CRISPR-Cas motifs. Altogether, our results demonstrate that rhizosheath–root systems are “edaphic mini-oases” and microbial diversity hotspots in hyperarid deserts. However, to colonize such refuge niches, the desert soil microorganisms compete intensively and are therefore prepared to outcompete potential rivals.
Subject terms: Microbial ecology, Environmental microbiology
Introduction
Deserts are heterogeneous habitats that cover approximately a third of the global land surface [1] and which are expanding with global climate change [2, 3]. In addition to aridity, numerous abiotic stresses are imposed on hot desert indigenous (micro)biota, including oligotrophy, elevated daily temperatures and radiation, high salinity, eolian erosion and environmental-physical instability [1, 4, 5]. Consequently, deserts are characterized by a lower biodiversity than more temperate ecosystems [3, 6], and are populated by macro- and microorganisms adapted to poly-extreme conditions [7–9].
Xerophytic plants are desert specialists that play key roles in desert ecosystem functioning. These plants have evolved both the aerial (stem and leaf) and subterranean (root system) organs to prevent water loss, improve water storage and optimize water and nutrient uptake [4, 10–12]. Thus, xerophytic plants represent ecological and fertility islands [13] where animals and microorganisms can find shelter, nutrients and thermal protection [10, 14]. Among these, desert speargrass species of the Poaceae family represent highly successful examples found in most deserts [4, 15]. The majority of speargrasses growing in arid soils develop a rhizosheath (RS)–root system constituted by soil particles (sand and other small mineral particles) that physically adhere to the surface of the entire root system [15–18]. Root hairs, fungal hyphae and adhesive agents, such as microbial- and plant-derived mucilages, are responsible for the aggregation of soil particles along the RS structure and for water/nutrient retention and uptake [17, 19–21]. Therefore, RS–root systems ultimately improve the overall fitness of plants under stressful environmental conditions [22–24].
The higher moisture and nutrient contents of RS (compared to non-vegetated (NV) soil) create favorable niches for colonization by edaphic microorganisms [15, 18, 25, 26]. In return, the RS microbiome provides beneficial plant growth promoting (PGP) services by improving the plant’s nutrient (e.g., P, N and Fe) status, growth, and/or resistance to abiotic and biotic stresses, such as drought, salinity and phytopathogens [27–32].
Inevitably, the process of colonizing and subsequently residing in the favorable RS niche must involve complex microbe–microbe competition dynamics [33, 34]. By using amplicon and shotgun metagenomic sequencing, we aimed to evaluate the microbial competition–colonization course associated with the RS–root system in xerophytic desert plants. We used the perennial Stipagrostis ciliata (Desf.) De Winter var. capensis (Trin. & Rupr.) De Winter (Poaceae family)—locally known as “Tall bushman-grass”—as the model plant in the study. This species grows extensively in the Namib Desert gravel plains [35, 36] and has shown the capacity to rapidly establish RS structure and biotic interactions with the surrounding soil microbiota after limited rain events (10–20 mm precipitation [35]). We hypothesize that (1) the compartmentalization of the RS–root structures of S. ciliata (root tissues (RT), RS, rhizosphere (RH)) will lead to deterministic processes dominating the assembly of its associated microbial communities [15] and that (2) the communities from the less extreme niches (the cooler and more humid RS and RH) would be enriched in microbial cells and stable than those from the most extreme NV soils [15, 37]. Furthermore, by adapting the Darwinian “Survival of the fittest” theory [38] to the microbial world, we expect that (3) desert soil microorganisms strongly compete to colonize the protected favorable RS–root system niches. Consequently, we anticipate that the microbial communities colonizing this niche will present significantly more markers indicative of competitive interactions (e.g., antibiotic production and resistance) to remain in the refuge than those inhabiting the NV soil. Similarly, we predict that (4) the microbial communities associated with plant will exhibit significantly more PGP traits (e.g., biopromotion and biofertilization) to favor the host and their own survival in such extreme environment.
Material and methods
Rhizosheath–root system sampling and processing
To cope with the hyperarid and oligotrophic conditions of the desert habitat, S. ciliata rapidly responds to rare and limited moisture events, adopting an amphiphytic lifestyle; i.e., either facultatively annual from seed growth or perennial by re-sprouting from existing live grass clumps [35]. In April 2017, in an area of 1 km2 of the Namib Desert gravel plains (S 23°32’53”; E 15°08’11”; Supplementary Fig. S1a), a total of ten S. ciliata plants of similar size were randomly selected to collect the RS–root systems. All selected plants had green leaves growing from the base of the clump, as evidence of active growth (Fig. 1a and Supplementary Fig. S1b, c). The entire plants were carefully exhumed from the rocky soil to preserve their RS–root systems (Fig. 1b). Intact portions of the RS–root system were subsequently excised from each plant using sterile scissors and tweezers and transferred into sterile plastic tubes (example of excised RS portion in Supplementary Fig. S1d). In addition, control NV soil samples were collected (0–10 cm depth; n = 10) at approximately one meter from each plant. All samples were collected under the research/collecting permit number 2248/2017, issued by the Namibian Ministry of Environment and Tourism.
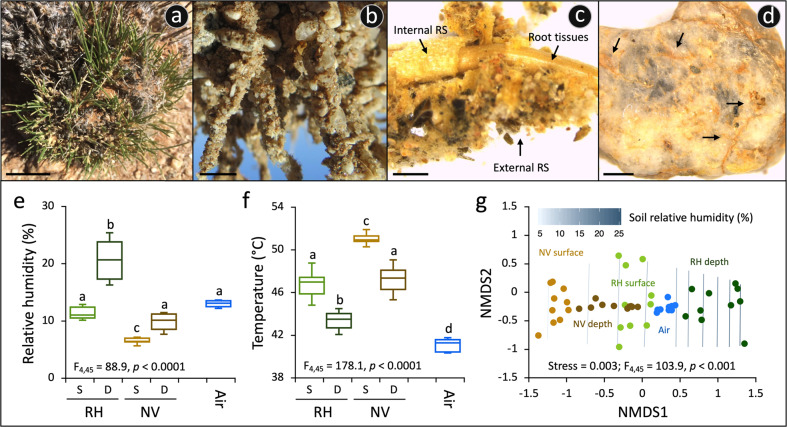
Fig. 1. Rhizosheath–root system niche of S. ciliata in Namib Desert gravel plain.
a The speargrass S. ciliata growing in the gravel plain as shown by the new green leaves developing from the basal portion of the plant (bar, 2 cm). b Close-up photograph of the S. ciliata rhizosheath–root system extracted from the soil (bar, 1 cm). The rhizosheath (RS) is composed of sand grains physically attached to the root, along with trapped stones and sand grains. c Stereomicroscope image of the rhizosheath–root system structure shows the external RS layer of the matrix with long root hairs developing from the epidermis (internal layer of RS; i.e., outermost cells of the root) that entrap sand grains and stones, as well as the central root tissue of vascular plants (bar, 1 mm). d Magnification of a stone detached from the RS; biological mineral weathering is indicated by black arrows (bar, 1 mm). e Relative humidity (%) and f temperature (°C) measured (n = 10; ±standard deviation) in NV soils (surface and in-depth) and soils under S. ciliata plants (surface and in-depth RH); S, surface, and D, in-depth. Values from air are also reported. Results of the ANOVA main test are indicated, along with lowercase letters referring to results of the post-hoc multiple comparison Tukey’s tests. g Non-parametric multidimensional scaling (NMDS) ordination plot showing the relative distribution of humidity and temperature measured; the relative humidity trend is plotted onto the ordination space. The result of the PERMANOVA main test is reported.
Soil and air temperature and relative humidity were measured in situ using handheld Ebro Electronic™ Hygrothermometer (TFH 620 with TPH 100 Air Probe) during five consecutive days (April 24th–28th; n = 10). Measurements were conducted between 12:00 and 13:00 p.m. as these were the hottest hours of the day in the central Namib Desert gravel plains [39]. The edaphic measurements were recorded at the soil surface (first 5 cm) and shallow sub-surface (from 5 to 10 cm deep) from two contrasting niches: NV soils and within S. ciliata plant clumps in the vicinity of their RS–root system. Data were analyzed by performing non-metric multidimensional scaling on the Euclidean distance matrix in R [40, 41]; permutational multivariate analysis of variance (PERMANOVA) was performed in Primer v.6.1 [42] using sample categories as explanatory variables (five levels: under plant sub-surface or surface, NV soil sub-surface or surface, and air).
In the laboratory, portions of the RS–root system were visualized with a stereomicroscope (Leica S8 APO) to define and divide the different compartments (i.e., RT, RS and RH) and measure the average sizes of RT and RS from a portion of RS–root from each plant. The RH was characterized as loosely adherent sand, detachable from the RS by gentle shaking. The RS (root coating containing sand and mineral particles physically trapped by the root hairs along the entire length) and RT (internal tissues) were separated with sterile scalpels [15].
The NV soil (n = 3), RH (n = 3) and RS (n = 3) samples were used for physicochemical analyses (2–3 g per sample); water content, pH, conductivity, salinity, organic matter (OM), carbon (TC, POC, PIC) nitrogen (TN, PON, PIN, nitrite, nitrate and ammonium), phosphate (PO43−), silicate, potassium (K), calcium (Ca), magnesium (Mg), and sulfur (S) contents were analyzed by the commercial company GEOMAR (Wischhofstraße, Germany) using standard protocols. Data were analyzed by performing PERMANOVA in Primer v.6.1 [42] on the Euclidean distance matrix, using sample categories as explanatory variables (three levels: NV soil, RH and RS). NV soil structure (percentages of rock, sand, silt, and clay) was also characterized at the Soil Science Laboratory of the University of Pretoria (South Africa).
DNA extraction
The surfaces of RT obtained from the RS–root system were sterilized as previously described [43]. Briefly, the RT were soaked in 70% ethanol for 3 min, followed by sodium hypochlorite 2.5% for 5 min and 70% ethanol for 30 s. Several washes with sterile water were performed to remove any trace of the chemicals used for surface sterilization. Subsequently, all samples (sterile RT, RS, RH and NV soil) were separately homogenized in liquid nitrogen with sterile mortars and pestles. Total DNA from the homogenized edaphic samples (NV soil, RH and RS) was extracted using 0.7 ± 0.05 g of sample and the PowerSoil® DNA Isolation Kit (MoBio Inc., USA), while from RT total DNA was extracted from 1 ± 0.1 g of sample and the DNeasy Plant Maxi Kit (Qiagen, Germany). The total DNA extracted was quantified on a Qubit Fluorometer using the high sensitivity dsDNA assay kit. The concentration of total DNA (ng/g of soil) extracted from RH and NV soils was further used as biomass proxies [44] and t-test was used to evaluate differences among the two compartments. Total DNA concentrations of the RT and of the RS compartments were not included in this comparison as large amounts of plant DNA were co-extracted.
Amplicon library preparation for phylogenetic analysis of the prokaryotic 16S rRNA genes and microeukaryotic ITS regions
The V3–V4 hypervariable regions of the prokaryotic 16S rRNA gene were amplified using the universal primers 341f and 785f, and the microeukaryotic ITS2 region amplified using the primers ITS3f and ITS4r [15]. PCR reactions mixture of 30 μl were performed for each sample using 1 U of Platinum® Taq DNA Polymerase, High Fidelity (Invitrogen) with 1× High Fidelity Buffer, 1.5 mM of MgSO4, 0.3 mM dNTPs mix, 0.3 µM each of forward and reverse primers, and ca. 10 ng of template DNA. The reaction conditions were as follows: denaturation at 95 °C for 5 min, followed by 25 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 68 °C for 45 s, final extension at 68 °C for 5 min. Total DNA extracted from sterile water and PCR mix (reagent without DNA) was used as an additional control in the amplicon PCRs; no amplification was detected by running the PCR product on 1% agarose gel. All the amplicon products obtained were used to incorporate the sequencing adapters by using the 96 Nextera XT Index Kit (Illumina). All tagged samples were pooled together, concentrated in a CentriVap DNA Concentrator (Labconco) and sequenced with the Illumina MiSeq platform at the Bioscience Core Lab, King Abdullah University of Science and Technology (Saudi Arabia). Raw forward and reverse reads for each sample were assembled into paired-end reads (minimum overlap of 30 nucleotides and maximum of one mismatch within the region) using the fastq-join algorithm (https://code.google.com/p/ea-utils/wiki/FastqJoin) and analyzed using the Qiime 1.9 pipeline. After quality filtering, trimming, dereplication, and paired-end merging of the sequences, a total of 1,830,127 (average length of 405 bases) and 3,669,396 (average length of 310 bases) sequences were obtained for prokaryotic and microeukaryotic components, respectively. Operational taxonomic units (OTUs) were clustered at 97% sequence similarity. Prokaryotic representative sequences of each OTU97 were searched against the SILVA 138 database [45], using uclust command, while microeukaryotes OTU97 representative were searched against the UNITE database [46], using the blast command. OTUs not identified as prokaryotes (i.e., chloroplast, mitochondria and unclassified or microeukaryotes (i.e., non-fungi and unclassified), and OTUs present in PCR and DNA blank controls were removed from the dataset (number of reads and OTUs removed per compartment are reported in Supplementary Table S2). OTUs showing low relative abundances (<0.01% for the prokaryotic 16S rRNA gene and <0.001% for the microeukaryotic ITS region datasets) were also removed (Supplementary Table S2). Rarefaction curves are shown in Supplementary Fig. S2.
Alpha- and beta-diversity, taxonomic distribution, and statistical analyses
Compositional similarity matrices (Bray–Curtis (BC) of the log-transformed OTU tables) were calculated in PRIMER v.6.1 [42, 47, 48] and homogeneity of multivariate dispersions (PERMDISP) tested to evaluate the dispersion of samples for each compartment. The specific roles of RS–root system compartmentalization in explaining the variation of microbial communities were quantified using the PERMANOVA function adonis2 in the vegan package in R [49]. Principal coordinates analysis (PCoA) and multivariate generalized linear models (many GLM, main and multiple comparison tests, using negative binomial family errors; Supplementary Fig. S3) were performed, with the factor “compartment” (of four levels: RT, RS, RH, and NV soil) as explanatory variables and by using PRIMER v.6.1 and the R package mvabund [50], respectively. Components of beta-diversity (similarity, richness difference and replacement) in the microbial communities associated with the root-system compartments and the NV soil were also quantified using the function beta.div.comp of the R package adespatial [51, 52]. The occurrence of distance-decay patterns in the RS–root system and NV soil was tested using the linear regression (GraphPad Prism 7 software, La Jolla California, USA) between the similarity of bacterial/microeukaryotic communities (BC) and the distance between the different compartments. Alpha diversity indices (richness and evenness) were calculated using the PAST software [53]. Shared and exclusive bacterial and microeukaryotic OTUs (fungi and algae) in the different compartments were visualized using Venn-diagrams. The Kruskal–Wallis test (FDR p correction) was used to detect significant differences (p < 0.05) among taxonomic groups in RS–root system compartments.
Co-occurrence network analysis
To identify ecological clusters of strongly associated microorganisms, a co-occurrence network was generated for each RS–root system compartment and for the NV soils using the CoNet plugin of Cytoscape 3.4 [54, 55] and Gephi 0.9.1 [56] for computation and visualization, respectively. We used the compartment-OTU tables described above for the analysis and merged the bacterial and microeukaryotic datasets using their relative abundance. We considered all the OTUs present in a compartment (i.e., RT, RS, RH and NV soil) to identify co-occurring OTUs within each network, possibly indicating functional/physical interactions among them in the different compartments. A combination of BC and Kullback–Leiber dissimilarity indices, along with the Pearson and Spearman correlation coefficients were used to build the networks. Edge-specific permutation and bootstrap score distributions with 2000 iterations were performed. For each measure and edge, 100 permutations and bootstrap scores were generated. The obtained data were normalized to detect statistically significant non-random events of co-occurrences, i.e., co-presences and mutual exclusions. The p values were computed by z-scoring the permuted null and bootstrap confidence intervals using pooled variance [57]. The most important statistical network descriptors were calculated [58], along with the normalized degree (number of node connections standardized by the total number of connections [59]), node betweenness centrality, and frequency of edges connecting the three microbial components; i.e., bacteria–bacteria, fungi–fungi, bacteria–fungi, and algae–algae/bacteria/fungi. Hubs (nodes with degree >75th percentile among all network nodes) and keystone taxa (nodes with betweenness centrality >75th percentile among hubs) were identified in the final networks.
Metagenome library preparation, sequencing and analyses
A total of nine samples (three RS, three RH and three NV soil) were selected for metagenomic analyses. Metagenomic libraries were prepared at the Bioscience Core Lab, King Abdullah University of Science and Technology (Saudi Arabia). First, 100 ng of total DNA were diluted in 52.5 ml of Nuclease-Free water (Ambion), in Covaris snap cap microtube (PN 520045, Covaris) and it was further fragmented by sonication with Covaris M220 to target DNA fragments of 300 bp, following this sonication protocol: treatment time 70 s, cycle per bust 200, duty factor 20%, peak incident power 50 W. The 300 bp fragmented DNA was used as input to prepare metagenomic libraries by using Illumina TruSeq Nano DNA Library Prep kit according to manufacturer’s instructions. Individual libraries were pooled, and the quality and quantity checked using a BioAnalyzer (Agilent) and the KAPA Library Quantification Kit (Roche). Pooled libraries were loaded on Illumina NovaSeq 6000 using 150 bp × 2 paired-end sequencing with an S1 flow cell following the NovaSeq XP workflow at the Biological Core Lab, King Abdullah University for Science and Technology (Saudi Arabia). Raw read sequences were quality filtered and trimmed using Trimmomatic v0.32 [60] to remove adapter sequences and leading and trailing bases with a quality score below 20 and reads with an average per base quality of 20 over a 4-bp window. The metagenomes were quality checked and filtered using FastQC v0.11.9 [61] and PRINSEQ-lite v0.20.4 to remove short sequences and those with ambiguous bases [62]. The sequencing depth of each metagenome was assessed using Nonpareil 3 [63]. All metagenomic reads were taxonomically classified using Kraken 2 [64] against the standard NCBI RefSeq database (accessed April 2022). Kraken 2 uses a k-mer based lowest common ancestor method with whole genome references to estimate read taxonomy to the deepest possible taxonomic resolution. High-quality reads were then assembled into contigs using metaSPAdes v3.12.0 [65]. The assembled metagenomes were quality checked using MetaQUAST v5.0.2 [66] after which we identified open reading (ORFs) using Prodigal v2.6.3 [67] through Prokka v1.12 [68]. These data were searched for genes related to plant growth-promotion (PGP, i.e., biofertilisation, biopromotion and bioprotection), nutrient and energy acquisition (N and P cycling, cellular metabolism, and energy acquisition) and competition (biotic competition and trophic interaction); the list of genes analyzed is given in Supplementary Table S3. Ribosomal genes were identified and taxonomically annotated from the unassembled sequence data using SingleM from which an OTU table of unique sequences was constructed. Secondary metabolic biosynthetic gene clusters (BGCs) were identified from contigs longer than 5 kb using antiSMASH v5.0 [69] under strict settings which only detects well-defined clusters containing all required parts. Carbohydrate-active enzymes were analyzed against the dbCAN2 database [70] whereby hits in two of the following databases were required to be accepted: HMMER (E value <1e−15, coverage >0.35), DIAMOND [71] (E value <1e−102) and Hotpep (Frequency > 2.6, Hits >6). Using the standards reported in Supplementary Methods S1, metagenome-assembled genomes (MAGs) were assembled with high-quality (>90% complete, <5% contamination) and medium-quality (>50% complete, <10% contamination) drafts.
Results
Rhizosheath–root systems of S. ciliata create favorable resource islands
The NV soils of the central Namib Desert gravel plain were extremely oligotrophic (0.33 ± 0.01 µg/mg of organic carbon (POC) and 0.046 ± 0.01 µg/mg of total nitrogen (TN)) and were structurally constituted by fine/very-fine sand particles and stones (Table 1 and Supplementary Table S4). In these soils, S. cilata is the dominant plant species [35] and often constitutes a monospecific vegetation cover after rain events exceeding 20 mm (Supplementary Fig. S1a). All the S. ciliata plant clumps tested (Fig. 1a and Supplementary Fig. S1b, c) exhibited a well-developed RS–root system (Fig. 1b). The RS matrix (RS and RH) appeared as thick and compact sandy cylinder, covering the entire length of each root (Fig. 1b, c and Supplementary Fig. S1d) and showed significantly different physicochemical properties from the surrounding NV soils (PERMANOVA: F2,6 = 37.53, p = 0.001; pairwise comparison of RS/RH vs. NV soil: t = 8.45, p = 0.001 and t = 2.61, p = 0.017, respectively). In general, RH and RS were less oligotrophic than NV soils with significantly higher concentrations of total carbon (TC; organic [OM, POC]) and total nitrogen (TN; organic [PON] and ammonium [NH4+]; Table 1 and Supplementary Table S4); the inorganic C/N forms [PIC and PIN] were only enriched in the RH. The RS soils had also the highest concentrations of phosphate [PO43−], calcium [Ca], magnesium [Mg] and sulfur [S] (Supplementary Table S4). Notably, the rocks associated with the RS showed signs of biological minerals weathering, possibly mediated by microorganisms and S. ciliata roots and root hairs (Fig. 1d and Supplementary Fig. S1e–g).
Table 1.
Characterization of carbon and nitrogen contents of the gravel plain non-vegetated (NV) soil, speargrasses rhizosheath (RS) and rhizosphere (RH); values are expressed as mean of three replicates ± standard deviation.
| Variable | Unit | ANOVA (p) | NV | RH | RS |
|---|---|---|---|---|---|
| Organic matter | % | 0.0008 | 0.066 ± 0.002 (a) | 2.869 ± 0.723 (b) | 0.140 ± 0.076 (a) |
| Total C | µg/mg | <0.0001 | 2.030 ± 0429 (a) | 17.185 ± 2.962 (b) | 2.199 ± 0.886 (a) |
| POC | µg/mg | 0.0008 | 0.332 ± 0.010 (a) | 14.344 ± 3.616 (b) | 0.698 ± 0.382 (a) |
| PIC | µg/mg | 0.0197 | 1.698 ± 0.421 (a) | 2.842 ± 2.130 (b) | 1.502 ± 0.622 (ab) |
| Total N | µg/mg | 0.0021 | 0.038 ± 0.100 (a) | 0.418 ± 0.100 (b) | 0.056 ± 0.021 (a) |
| PON | µg/mg | 0.0074 | 0.043 ± 0.002 (a) | 0.357 ± 0.099 (b) | 0.056 ± 0.018 (a) |
| PIN | µg/mg | <0.0001 | 0.005 ± 0.004 (a) | 0.0611 ± 0.008 (b) | 0.0006 ± 0.002 (a) |
Results of ANOVA tests are reported (p value); different letters indicate the significant difference across samples (Tukey’s multiple comparisons test, p < 0.05). The other physicochemical parameters measured are listed in Supplementary Table S4.
POC particulate organic carbon, PIC particulate inorganic carbon, PON particulate organic nitrogen, PIN particulate inorganic nitrogen, NV non vegetated, RH rhizosphere, RS rhizosheath.
The relative humidity of surface and sub-surface NV soils was significantly lower than the soils under the plants (RH, Fig. 1e). The niche that showed the highest relative humidity values was the sub-surface RH (ranging from 17 to 25%), while those with the lowest values was the surface of NV soil (6% ± 0.5%). Both these variables varied significantly with the distance between S. ciliata individuals and the corresponding NV soils (relative humidity: p < 0.0001, R2 = 0.81, r = −0.89; temperature: p < 0.0001, R2 = 0.78, r = 0.88). Consequently, day ambient temperatures, which ranged from 40 °C to 52 °C, were significantly lower in the RH of speargrass clumps than in NV soils at equivalent depths (4.2 °C ± 1.3 °C and 3.8 °C ± 1.2 °C cooler for surface and sub-surface horizons, respectively; Fig. 1f). The field parameters measured showed the presence of an environmental gradient, with the more favorable conditions in the deep S. ciliata RH and the harshest conditions in the surface NV soils (Fig. 1g). Altogether, these results and the fact that the most recent rain event recorded occurred over a month prior to sampling (Supplementary Table S1) show the enhanced water retention capacity of the RS–root systems.
Niche-partitioning dominates the assembly of S. ciliata rhizosheath–root microbial communities’ structure
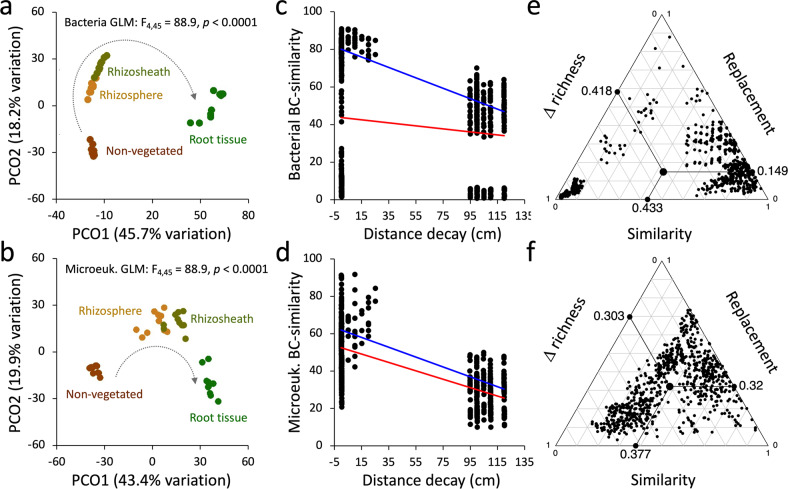
The “horseshoe shape” distribution of samples in the PCoA ordination plots reveals that both bacterial and microeukaryotic communities were assembled via niche-partitioning processes along the RS–root system compartments (manyGLM, F3,36 = 64815, p = 0.001 and F3,36 = 7196, p = 0.001, respectively; Fig. 2a, b and Supplementary Tables S5 and S6). Even though the NV soils was the microbial-source of the vast majority of the plant-associated bacterial and microeukaryotic OTUs (91% of OTUs (96% of relative abundance) and 56% of OTUs (97% of relative abundance), respectively; Supplementary Fig. S5), the selective process exerted by the RS–root system defined a non-random distribution of OTUs across the compartments with a significant relationship between microbial occurrence and abundance (Supplementary Fig. S6). The niche partitioning was further confirmed by the significant decline in compositional BC similarities with increasing distance; i.e., the edaphic microbial communities associated with close compartments (RH and RS) were more similar than those from the distant NV soils for both bacteria and microeukaryotes (p = 0.0003, R2 = 0.02 and p < 0.0001, R2 = 0.45; red regression lines in Fig. 2c, d). While the dissimilarities among bacterial communities were determined by similarity and richness differences, for microeukaryotes it was equally driven by the three components (i.e., similarity, richness differences and replacement; Fig. 2e, f and Supplementary Result S1). Despite such difference, the significant and positive relationship between bacterial and microeukaryotic BC similarity matrices (F1,778 = 523.9, p < 0.001, R2 = 0.4; considering only edaphic compartments: F1,433 = 1464, p < 0.001, R2 = 0.77) indicated commonalities in the processes driving the assembly, diversity and composition of the two microbial communities’ components at the RS–root system micro-scale.
Fig. 2. Diversity and dynamics of microbial communities associated with the S. ciliata rhizosheath–root system.
Principal coordinate analysis (PCoA) of (a) bacterial and (b) microeukaryotic communities associated with the rhizosheath–root system (root tissue (RT), rhizosheath (RS) and rhizosphere (RH)) and non-vegetated (NV) soil. Arrows indicate the “horseshoes” shape distribution of microbial communities associated to different compartments, starting from the RT, and ending into the NV soils. Decay relationships among microbial communities’ similarities (BC: Bray–Curtis) and compartment relative distance (cm) for (c) bacteria and (d) microeukaryotes; red regression lines indicate the significant correlation among BC’ similarity and distance considering all the compartments. We note that when excluding the root tissues from the analyses, the correlation coefficients increased; blue regression lines, bacteria: p < 0.0001, R2 = 0.81; microeukaryotes: p < 0.0001, R2 = 0.73. Ternary plot presenting the variation in species composition among sites (beta-diversity) as result of its three components: similarity, replacement and richness difference [51]; the indices decomposing beta-diversity are visualized for the (e) bacterial and (f) microeukaryotic communities. Each point represents a pair of samples, and its position is determined by a triplet of values from the similarity, replacement, and richness difference. In each ternary plot, the large central dots where the lines start are the centroid of the points for each beta-diversity component; the lines represent their mean values.
The impact of niche partitioning was also reflected in the microbial community compositions of each compartment. Significant compartment-specific differences for bacterial and microeukaryotic richness (F3,36 = 834.3, p < 0.0001 and F3,36 = 41.8, p < 0.0001, respectively) and evenness (F3,36 = 126, p < 0.0001 and F3,36 = 10.4, p < 0.0001, respectively) were detected. Specifically, lower species richness and evenness metrics were detected in RT and increased outwards from the plant in the RS up to reach the highest values in the RH and NV soils (Supplementary Table S7). However, the concentrations of the total DNA extracted, which can be used as a proxy for microbial biomass in desert soils [44], showed significantly higher values in the RH than in the NV soils (357 ± 137 vs. 85 ± 33 ng DNA/g soil, respectively; t1,18 = 6.097, p < 0.0001), revealing a local enrichment of microbial cells in the vicinity of the plant.
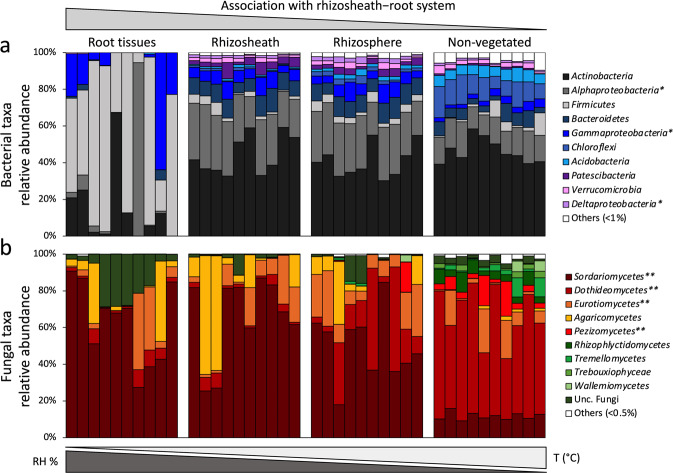
The edaphic bacterial communities were consistently dominated by Actinobacteria (45%, 41% and 47% of the RS, RH and NV soil communities, respectively), Alphaproteobacteria (27, 26 and 15%) and Bacteroidetes (9, 9 and 6%), while RT bacterial communities were dominated by Firmicutes (59%) and Gammaproteobacteria (14%) (Fig. 3a; Supplementary Table S8 and Supplementary Data S1). The microeukaryotic communities were less diverse; Dothideomycetes, Pezizomycetes, Tremellomycetes, and Rhizophylyctidomycetes classes dominated the NV soils, while Sordariomycetes and unclassified fungi were significantly enriched in all compartments of the speargrass RS–root systems (RT, RS and RH; Fig. 3b; Supplementary Table S8 and Supplementary Data S1).
Fig. 3. Taxonomical composition of microbial communities associated with the S. ciliata rhizosheath–root system and non-vegetated soil.
Relative abundance of (a) bacterial and (b) microeukaryotic phyla/classes associated with rhizosheath–root system compartments (root tissue (RT), rhizosheath (RS) and rhizosphere (RH)) and non-vegetated (NV) soil. Relative abundances are expressed as percentages; star (*) indicates classes belonging to the Proteobacteria phylum; stars (**) indicate classes belonging to the Ascomycota phylum. Gradients of relative humidity and temperature are also schematically reported on the top.
Rhizosheath–root system niches enhance microbial connectivity and network stability
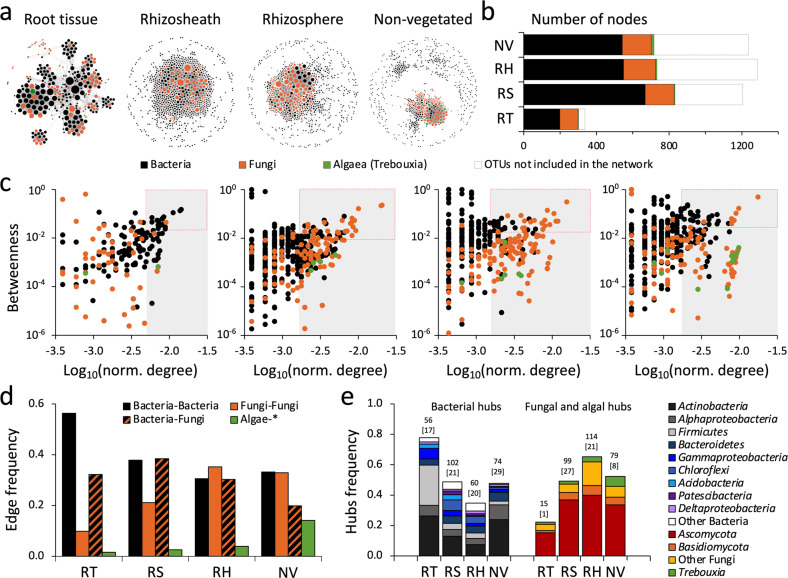
A total of 66% (±15%) of all microbial OTUs significantly co-occurred with one another (Fig. 4a, b and Table 2). Networks based on these co-occurrences were bacteria-dominated (66–80% of nodes). Fungal OTUs represented the remaining portion (19–33% of nodes), along with algal OTUs (<2% of nodes). A reduction in heterogeneity and modularity, together with an increase in connectedness and centrality of nodes, was observed from the NV soils to the RT (Fig. 4a and Table 2). The analysis of the node connectivity (i.e., normalized degrees per node) revealed a significantly higher level of connection in the RT network when compared to the edaphic networks (RS, RH and NV; ANOVA: F3,2579 = 125.7, p < 0.0001; Supplementary Fig. S7a). The contribution of each microbial group was compartment dependent: in RT the bacterial nodes showed the higher level of connections, while in all the edaphic compartments those of fungi were the most connected (Fig. 4c; Supplementary Result S2 and Supplementary Fig. S7b). In the RT network, the key connectors were the Firmicutes (including the well-known plant-associated Bacillus and Paenibacillus genera [72]), Actinobacteria (Nocardioides, Isoptericola and Streptomyces), and Gammaproteobacteria (Acinetobacter, Ramlibacter and Pseudoxanthomonas), as well as fungal PGP taxa such as Preussia (Ascomycota [73]), Rhodotorula (Basidiomycota [74]) and Mortierella (Zigomycota [75]; (Fig. 4e; Supplementary Data S2 and Supplementary Result S2). The edaphic RS, RH and NV soil networks showed significantly more fungal hubs and keystones species than the RT (Fig. 4e). Hub fungal taxa were affiliated to Ascomycota (among others, Acremonium, Aspergillus, Bipolaris, Chaetomium, Chrysosporium, Curvularia, Eurotium, Monosporascus, Pseudospiromastix and Thermomyces) and Basidiomycota (Coprinopsis, Filobasidium, Rhodotorula and Wallemia; Supplementary Data S2). The edaphic bacterial hubs/keystone species mostly belonged to the Actinobacteria and Alphaproteobacteria (Fig. 4e and Supplementary Data S2). It is also noteworthy that the photoautotrophic unicellular alga Trebouxia was ubiquitously identified as a network hub in the edaphic compartments (Fig. 4e).
Fig. 4. Bacterial and fungal co-occurrence network in S. ciliata rhizosheath–root system compartments.
a Significant interactions (edge, p < 0.05) between bacterial and fungal OTUs in root tissue (RT), rhizosheath (RS) and rhizosphere (RH) of S. ciliate and non-vegetated (NV) soil were visualized by co-occurrence network. Circles (nodes) represent individual OTUs (bacteria, fungi, and algae); size of circles indicates the number of connections of such node (degree); nodes were colored according to their taxonomic affiliation: black, orange and green indicate bacteria, fungi and algae, respectively. For co-occurrence networks’ properties refer to Table 2. b Proportion of bacterial, fungal and algae OTUs included in co-occurrence networks. Portion of OTUs not included in the network is also reported (white portion of bars). c Relationship between node-normalized degree (log10) and betweenness centrality in networks of rhizosheath–root system and non-vegetated soil compartments. Colors indicate the taxonomic affiliation of nodes (bacterial, fungal and algal OTUs); gray-box indicate the nodes with high degrees defined as hubs; red-dashed boxes delineate the keystone species of each network (high degree and high betweenness). d Frequency of edges (connections) in the rhizosheath–root system compartments’ networks. Connections among the three components of the network are showed: bacteria–bacteria, fungi–fungi, bacteria–fungi, and algae with all the others (* = bacteria, fungi and algae). e Taxonomic affiliation of bacterial and fungal hubs detected for each network (c) expressed as normalized frequency; total number of bacterial and fungal hubs are reported on each bar, along with the number of keystone species accounted among them (numbers in square brackets). Details regarding taxonomy are reported in Supplementary Data S2.
Table 2.
Co-occurrence network topology indices reported for the three rhizosheath–root system compartments (root tissue (RT), rhizosheath (RS) and rhizosphere (RH)) and the non-vegetated (NV) soil.
| Network parameter | RT | RS | RH | NV |
|---|---|---|---|---|
| Node | 301 | 832 | 733 | 717 |
| N. bacteria (%) | 199 (66) | 669 (80) | 549 (75) | 542 (76) |
| N. fungi (%) | 100 (33) | 159 (19) | 176 (24) | 159 (22) |
| N. algae (%) | 2 (1) | 4 (1) | 8 (1) | 16 (2) |
| Interaction | 2548 | 2550 | 2320 | 2660 |
| N. positive (%) | 2133 (84) | 1484 (58) | 1568 (68) | 2107 (79) |
| N. negative (%) | 415 (16) | 1066 (42) | 752 (32) | 553 (21) |
| Degreea | 17 | 6 | 6 | 7 |
| Betweennessa | 608 | 2354 | 2116 | 952 |
| Betweenness centralitya | 0.010 | 0.016 | 0.025 | 0.028 |
| Cluster coefficient | 0.719 | 0.279 | 0.278 | 0.236 |
| Centralization | 0.188 | 0.118 | 0.089 | 0.069 |
| Average path length | 3.167 | 4.678 | 5.52 | 5.939 |
| Average neighbors | 16.93 | 6.129 | 6.33 | 5.419 |
| Density | 0.056 | 0.007 | 0.009 | 0.001 |
| Heterogeneity | 0.704 | 1.433 | 1.444 | 1.513 |
| Modularity | 0.137 | 0.199 | 0.322 | 0.647 |
RT root tissue, RS rhizosheath, RH rhizosphere, NV non vegetated.
aMedian values are reported for each compartment.
Microbial communities in the root–rhizosheath–rhizosphere continuum favor a heterotrophic lifestyle
The shotgun metagenomes from the edaphic compartments (RS, RH and NV soils) supported the taxonomic results obtained by amplicon sequencing, i.e., each compartment showed taxonomically distinct communities (PERMANOVA; R2 = 0.73, adjusted p < 0.005; Supplementary Fig. S8; Supplementary Results S3 and Supplementary Data S3). Evaluation of indicator genes for the potential-acquisition of carbon, nitrogen and phosphorous showed evidence of different lifestyles across the edaphic niches (PERMANOVA; R2 = 0.72, adjusted p < 0.003). For example, compared to NV soil metagenomes, those in RS and RH were significantly enriched in genes involved in the metabolism of simple carbon substrates, such as sugars, amino acids and organic acids, that are typically present as root exudates [18, 76], as well as in carbohydrate-active enzymes, such as glycosyl hydrolases and glycoside transferases; p < 0.05) and cellulases (celE; p < 0.04). Similarly, N and P microbial uptake genes suggest that the RH and RS microbial communities relied on readily assimilable and/or abundant substrates (Table 1), including ammonia and nitrate (NO3–; ammonia monooxygenase subunits (amoABC; p < 0.001) and assimilatory nitrate reductases (nas; p < 0.02)), ammonia/ammonium (NH3/NH4+; nitrate reductase (nrfA; p < 0.03)), inorganic phosphate (low-affinity phosphate importers, pit (p < 0.002) and glycerol-3-phosphate ABC transporters ugpABCE (all p < 0.02)) and phytic acid (a plant-derived molecule; acid phosphatases (acpp; p < 0.05) and 3-phytases phyA (p < 0.03)). In the RS we also observed significant overrepresentation of transporter genes (hsrA, pbuE, sugar efflux transporter C, hmuU, gsiA), suggesting a high capacity for metabolite uptake, such as for photosynthates released by the speargrass root system.
In contrast, the NV soil communities were significantly enriched in aerobic carbon fixation capacity (prkB, rbcLS, RuBisCO; p < 1.52 e−3), assimilatory hydrogenases ((NiFe) hydrogenases; p < 0.015), the Wood-Ljungdahl pathway for anaerobic carbon fixation (acdABCSD; p < 0.03) and aerobic respiration (cytochrome C oxidases; p < 9.38 e−3), indicating a prioritization of energy acquisition strategies based on gasses (i.e., CO2 and H2). A preference for autotrophic metabolism was consistent with the higher proportion of biological N fixation (nifH) genes (p < 0.0004) in the NV soil metagenomes, as well as with the higher presence of genes encoding for the acquisition of inorganic phosphate (PO43–) directly from the environment through alkaline phosphatases (phoA, p < 0.0001), the Pho regulon (phoBR, p < 0.02) and the high-affinity phosphate transport system (pstSCAB; p < 0.0001). Along with these, in the NV soil microbial communities we observed a significant enrichment of genes involved in abiotic stress mitigation, including DNA repair mechanisms (radA, recNO, mutHLS; p < 0.02), breakdown of reactive oxygen species (superoxide dismutase; p < 0.015) and UV-damage repair (uvrABC; p < 0.03).
Rhizosheath–root system microbiome is enriched in plant-beneficial traits
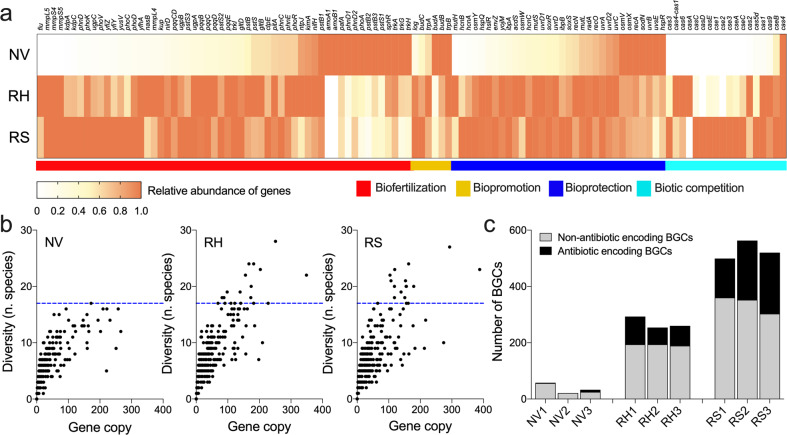
We explored the ecological services potentially provided to the plant by the RS and RH microbiomes, focusing on three distinct plant-beneficial functions: biofertilization (solubilization of nutrients for the enhancement of plant nutrition), biopromotion (stimulation of plant growth mediated by microbial-derived phytohormones and volatile compounds), and bioprotection (mitigation of plant abiotic and biotic stresses; list provided in Supplementary Table S3). Overall, plant-associated RS and RH microbial communities were significantly enriched in PGP traits from all the three categories, compared to those associated with NV soil communities (PERMANOVA; R2 = 0.85, p < 0.05; Fig. 5a, b). For instance, the RS and the RH metagenomes had significantly higher abundances of biofertilization markers involved in siderophore production (catecholate siderophore receptor (fiu; p < 0.05), siderophore-binding lipoprotein (yfiY; p < 0.0001) and heme uptake protein (mmpLS, p < 0.0001)), ammonification (nitrite reductase (nirD, p = 0.002)), potassium metabolism (potassium transporters (kimA; p < 0.002, and kdpC; p < 0.0001)) and phosphate solubilization (pqqBCDE, p = 0.04) compared to the NV soil metagenomes (Fig. 5a and Supplementary Data S3). The selection of beneficial microorganisms by S. ciliata was also indicated by the significant enrichment in the RS and the RH metagenomes of microbial genes encoding for cytokinin (cytokinins riboside 5’-monophosphate phosphoribohydrolase (LOG9; p = 0.04)), ACC deaminase (acdS; p = 0.04), auxin (indole-3-glycerol phosphate synthase, idpC; p = 0.003) and exopolysaccharide synthesis (epsF; p = 0.002). Genes involved in pathogen inhibition mechanisms, such as hydrogen cyanide production (hcnABC; p = 0.04), chitinase B (p < 0.0001) and ABC transporters (ytfQ, yphF, uup, ramA, modF, natB and natA [77]; all p < 0.05) were more common in the RS and RH metagenomes than those of NV soils (Fig. 5a).
Fig. 5. Functional potential of microbial communities from root-associated edaphic niches of the Namib Desert.
a Heatmap showing the relative normalized abundances of metabolic marker genes from the metagenome assemblies in rhizosheath (RS), rhizosphere (RH) and non-vegetated (NV) soil. Relative abundances are calculated by first normalizing by sequencing depth and then scaling against the highest proportion for each marker gene; genes within the PGP categories biofertilization, biopromotion and bioprotection (abiotic and biotic stresses), along with genes involved in biotic competition, are used in this analysis. b Relationship between the copies of PGP/biotic competition genes (x-axis) and the number of microbial species presenting these genes (y-axis) is visualized in NV, RH and RS. c Number of biosynthetic gene clusters (BGCs) detected across RS, RH and NV soil metagenomes; antibiotic-encoding BGCs are indicate in black, while the remaining BGCs in gray.
Analysis of PGP traits and taxonomic diversity revealed that the RH and RS microbial communities had a higher functional redundancy than NV soil communities (i.e., coexistence of multiple distinct taxa capable of performing a given PGP function [78]; Fig. 5b). Considering all the PGP genes analyzed, we found that RS, RH and NV had 5.6, 5.2 and 3.7 microbial species per PGP gene, respectively, and the relationship between gene copies and diversity (number of species) was significantly different among the three compartments (GLM: χ21,2 = 14.35, p < 0.0007). RH and RS also showed a higher number of occurrences for gene encoding PGP traits compared to NV soils (average count of genes: 35.7, 33.1 and 24.3, respectively). Functional genetic PGP potential of the 102 medium- and high-quality MAG confirmed the prevalence of multiple PGP traits in RS and RH (Supplementary Data S4).
Microbial competition biomarkers are abundant in rhizosheath–root system compartments
BGCs encoding for secondary metabolite production, such as antibiotics, pigments and sunscreens [79], were analyzed (Fig. 5c and Supplementary Data S3). The vast majority of BGCs belonged to bacteria, especially Actinobacteria, with only 252 BGCs assigned to fungal guilds, possibly due to the low percentage of reads assigned to the latest (Supplementary Results S3). The bacterial BGCs significantly decreased in abundance from the RS to the NV soil metagenomes (527 ± 33 BGCs in RS, 269 ± 21 in RH and 37 ± 19 in NV; p < 0.0001; Fig. 5c), suggesting that the RS and RH microbiomes are subject to higher levels of inter-taxa competition than NV soil microbiomes. Beside their limited number, also the fungal BGCs become more prominent in proximity of the plant (5 fungal BGCs in NV, 103 in RH and 144 in RS). Many BGCs were found to encode for antibiotic production (RS = 36% ± 7%, RH = 28% ± 5% and NV = 9% ± 13%; Fig. 5c and Supplementary Data S3) but the RH and RS metagenomes were also significantly enriched in antibiotic resistance genes (p < 0.001). The number of phage-related contigs showed the same trend with 75 ± 15, 34 ± 16 and 2 ± 3 viral contigs in the RS, RH and NV soil metagenomes, respectively (Supplementary Result S3). Ninety three percent of these phage contigs were unclassified using protein-sharing networks. The classified viral contigs belonged to Siphoviridae (Caudovirales, dsDNA tailed phage; Supplementary Data S3), possibly infecting the dominant actinobacterial genera, such as Streptomyces, Actinoplanes and Arthrobacter. Similarly, we observed a significantly higher proportion of CRISPR-Cas systems, which are proxies for previous viral infection events and bacterial immunity acquisition processes [80], in the RS and RH metagenomes than in the NV soil metagenomes (p < 0.05; Supplementary Data S3).
Discussion
In hyperarid desert ecosystems, resource (i.e., water and nutrients) availability is a key challenge for macro- and microorganisms. In this context, xerophytic plants and their root systems serve as important resource hotspots that affect both the structure and function of the surrounding edaphic microbial communities [15, 18, 81]. Here, we investigated the effects of the RS–root system of S. ciliata on the surrounding edaphic microbial community in the extremely oligotrophic gravel plain soils of the hyperarid central Namib Desert. S. ciliata is an amphiphytic speargrass which is abundant on rock-sandy gravel plains of the Namib Desert after spatio–temporally isolated rainfall events exceeding 20 mm [35]. A conspicuous adaptive trait evolved by Stipagrostis plants, and other xerophytic plants within the Poaceae family, is the development of a RS–root system [15], a cylindrical sheath of sand particles surrounding all elements of the root system. The RS is rich in exopolymers and mucigels produced by the root and its associated microorganisms [18, 22, 82] to efficiently retain water by reducing evapotranspiration [21, 83], as confirmed by the fact that the S. ciliata RS studied retained water more than a month after a light (<7 mm) rainfall.
An additional key adaptive trait of S. ciliata is the recruitment of specific microbial taxa and their functional capacity in the RS–root system. Indeed, the RS and RH edaphic niches have been described as “resource islands” and “diversity hotspots” in (hyper)oligotrophic and (hyper)arid soils [15, 18, 84, 85]. As already observed for Namib Desert dune speargrass species [15], S. ciliata recruits and assembles the microbial community in the RS–root system via strong deterministic niche-partitioning processes. While such communities have a reduced (RS) or similar (RH) compositional diversity than NV soil in term of richness, they present high functional redundancy (i.e., a same function can potentially be carried out by different microorganisms) with multiple taxa carrying a given PGP trait, including auxin and EPS production and provision of essential nutrients, e.g., P, N and Fe [15, 86–91]. Such functional redundancy suggests that the RS and RH compartments represent “functional shields” to support/ensure the fitness of the holobiont during extended dry periods, as well as “functional reservoirs” from which the plant selects its intimately associated endophytic communities.
RS and RH microbial communities of S. ciliata form more cohesive co-occurrence networks than those of NV soils, supporting the concept that both water and nutrient availability are determinant factors in the assembly and stability of desert edaphic microbial communities [37, 59, 92, 93]. Key components of the co-occurrence networks are microorganisms known to possess PGP traits that improve plant-fitness, such as Firmicutes of Bacillus and Paenibacillus genera. Members of these genera are often recognized as PGP taxa resistant/tolerant to drought [27, 72, 94–96] and have previously been found as connectors in RS-RH microbial networks of other Namib Desert speargrass species, namely S. sabulicola, S. seelyae and Cladoraphis spinosa [15]. This suggests that members of these bacterial genera have developed an intimate relationship, and possible co-evolution [97], with Namib Desert xerophytic plants forming RS–root structures. Similarly, the root fungal colonizers Mortierella sp., Auxarthron sp. and Xylaria sp. [98, 99] were also identified as important connectors in the RS and RH co-occurrence networks. These fungi support plant nutrient uptake [75, 100] and plant protection from herbivores and phytopathogens [101]. Their hyphae can further promote soil aggregation and stabilization, and therefore participate in the development of the RS structure surrounding plant roots [15], ultimately increasing soil water-holding capacity [102–105] and connectivity between edaphic microorganisms [90, 106, 107].
The inter-taxon interactions established between the microbial community members can either be positive (mutualism) or negative (competition) [15, 39, 90, 108]. While positive co-occurrences dominate in the oligotrophic NV soil networks, those negative increase along the RS and RH compartments (Table 2). This strongly suggests that desert edaphic microorganisms associated with the “limited niche-space” of the RS oasis compete more with each other than those inhabiting the unhospitable NV soil to reduce the invasion of possible competitors. The fact that a larger number of genes involved in antibiotic production, export and resistance was observed in the RS and RH strongly supports this view [93, 109]. As well, the presence of CRISPR-Cas proteins in these plant niches hints at defense mechanisms against the numerose and diverse edaphic phages [110] that might act as moderating agents within the community by controling population density [111].
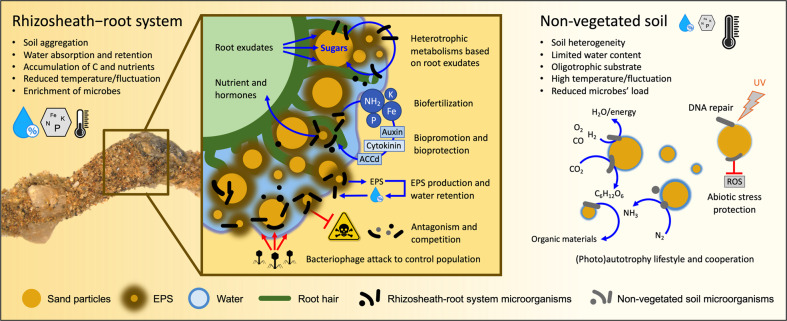
Altogether, our study supports the contention that plant RS–root systems represent resource islands and microbial density and competition hotspots. We therefore propose a dual selection process as a model of plant–microbe interaction in the colonization and occupation of the RS–root system niches (Fig. 6): to colonize the plant-associated niches desert edaphic microorganisms must (1) possess the capacity to improve the fitness and survival of the plant (plant selection) and be able (2) to compete successfully against other microorganisms (microbial competition); i.e., that both long-term plant–microbe co-evolution [112] and a microbial “arms race” [113, 114], respectively, co-occur. In contrast, NV soil desert microorganisms are less competitive but better equipped with self-sustaining processes to cope with the extreme/oligotrophic conditions to which they are exposed to. By some ecological analogies with the RS–root system, we speculate that similar processes should occur in other desert microbial refuge niches, such as lithic environments and biological soil crusts [8].
Fig. 6. Ecological race and interactions of microbial partners attracted by the rich root–rhizosphere niche under limited available resources in the hyperarid desert.
Schematic illustration of microbial communities associated with S. ciliata rhizosheath–root system (RS and RH) and NV soil and their contribute to niche-ecosystem functioning. Environmental, ecological, and functional characteristics of microbial communities associated with S. ciliata rhizosheath–root system and NV soil are reported considering both plant/soil–microbe and microbe–microbe interactions. Trophic modes and functions from metagenomes are resumed. More detailed explanation is reported in the “Discussion” section of the manuscript.
Conclusions
The RS–root system of S. ciliata is an “oasis” for hyperarid desert edaphic microorganisms as it provides more favorable abiotic conditions (with more nutrients and water) in otherwise poly-extreme and highly oligotrophic gravel plains. Consequently, these refuge niches and resource islands are hotspots for which edaphic microorganisms compete intensively to colonize. This is particularly emphasized by the fact that, despite presenting similar diversity in term of species number, the RH communities presented a significantly higher microbial density than the NV soil communities. This is in agreement with our initial hypothesis that the RS–root system of S. ciliata acts as hotspot for microbial colonization. The microorganisms able to colonize and remain in association with the RS–root system are (1) functionally equipped to support the host (and their own) growth and survival through biopromotion and biofertilization activities, that is they have been selected for their plant growth promoting capabilities, and (2) they have been selected to compete against the other microorganisms (e.g., through antibiotic production and viral pathogen resistance). Indeed, while presenting more favorable abiotic conditions, the biotic pressure endured by the resource island microbial colonizers is also an important factor to be considered to disentangle the mechanisms regulating the colonization of such “resource islands” in poly-extreme deserts. By applying Darwin’s evolutionary theory [37], we further postulate that such density/competitive niches may also represent evolutionary hotspots that can enhance the resilience and success of the RS–root microbial communities and their host, i.e., the plant holobiont in the extremely harsh and inconsistent environmental conditions of deserts and those arising from climate change.
Supplementary information
Acknowledgements
The authors thank the staff of the Gobabeb—Namib Research Institute for excellent assistance, fruitful discussions, and plant species identification; Taskeen Begum for her indispensable help in lab organization; Biological Core Lab team, Luke E. Esau and Emanuele Palescandolo for the metagenome libraries preparation, Novaseq sequencing, and MiSeq assistance.
Author contributions
R.M. and D.D. contributed to the conceptual development and experimental design of the study. R.M., M.F., J.-B.R., and D.D. collected samples. G.M.K. contributed to the plant identification. R.M. completed all lab work. R.M., J.-B.R., M.W.V.G., and M.F. performed all the analyses and interpreted the data. R.M. wrote the first draft of the manuscript. J.-B.R., M.W.V.G., M.F., D.A.C., K.S., G.M.K., and D.D. contributed to the manuscript editing. All authors read and approved the final manuscript.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors and it was financially supported (baseline fund to DD) by King Abdullah University of Science and Technology (KAUST). J-BR and DAC were supported by the South African National Research Foundation.
Data availability
The datasets generated during the current study are available in the NCBI SRA repository under the BioProject ID PRJNA628615. The raw, unassembled metagenomes are available in the NCBI SRA repository under the BioProject ID PRJNA731454.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ramona Marasco, Email: ramona.marasco@kaust.edu.sa.
Daniele Daffonchio, Email: daniele.daffonchio@kaust.edu.sa.
Supplementary information
The online version contains supplementary material available at 10.1038/s43705-022-00130-7.
References
- 1.Laity JJ. Deserts and desert environments. John Wiley & Sons; UK, 2009.
- 2.Huang J, Yu H, Guan X, Wang G, Guo R. Accelerated dryland expansion under climate change. Nat Clim Chang. 2015;6:166–71. doi: 10.1038/nclimate2837. [DOI] [Google Scholar]
- 3.Berdugo M, Delgado-Baquerizo M, Soliveres S, Hernández-Clemente R, Zhao Y, Gaitán JJ, et al. Global ecosystem thresholds driven by aridity. Science. 2020;367:787–90. doi: 10.1126/science.aay5958. [DOI] [PubMed] [Google Scholar]
- 4.Danin A. Plant adaptations to environmental stresses in desert dunes. In: Cloudsley-Thompson J, Punzo F, editors. Adaptations of desert organisms. Plant of desert dunes. Springer; Verlag Berlin Heidelberg, 1996.
- 5.Makhalanyane TP, Valverde A, Gunnigle E, Frossard A, Ramond J-B, Cowan DA. Microbial ecology of hot desert edaphic systems. FEMS Microbiol Rev. 2015;39:203–21. doi: 10.1093/femsre/fuu011. [DOI] [PubMed] [Google Scholar]
- 6.Fierer N, Leff JWJ, Adams BJ, Nielsen UN, Bates ST, Lauber CL, et al. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci USA. 2012;109:21390–5. doi: 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ronca S, Ramond J-BB, Jones BE, Seely M, Cowan DA. Namib Desert dune/interdune transects exhibit habitat-specific edaphic bacterial communities. Front Microbiol. 2015;6:1–12. doi: 10.3389/fmicb.2015.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pointing SB, Belnap J. Microbial colonization and controls in dryland systems. Nat Rev Microbiol. 2012;10:551–62. doi: 10.1038/nrmicro2831. [DOI] [PubMed] [Google Scholar]
- 9.Noy-Meir I. Desert ecosystems: higher trophic levels. Annu Rev Ecol Syst. 1974;5:195–214. doi: 10.1146/annurev.es.05.110174.001211. [DOI] [Google Scholar]
- 10.Danin A. Plants of desert dunes. In: Cloudsley-Thompson J, editor. Adaptations of desert organisms. Springer; Verlag Berlin Heidelberg, 2000.
- 11.Roth-Nebelsick A, Ebner M, Miranda T, Gottschalk V, Voigt D, Gorb S, et al. Leaf surface structures enable the endemic Namib Desert grass Stipagrostis sabulicola to irrigate itself with fog water. J R Soc Interface. 2012;9:1965–74. doi: 10.1098/rsif.2011.0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebner M, Miranda T, Roth-Nebelsick A. Efficient fog harvesting by Stipagrostis sabulicola (Namib dune bushman grass) J Arid Environ. 2011;75:524–31. doi: 10.1016/j.jaridenv.2011.01.004. [DOI] [Google Scholar]
- 13.Cartwright J. Ecological islands: conserving biodiversity hotspots in a changing climate. Front Ecol Environ. 2019;17:fee.2058. doi: 10.1002/fee.2058. [DOI] [Google Scholar]
- 14.André HM, Noti MI, Jacobson KM. The soil microarthropods of the Namib Desert: a patchy mosaic. J African Zool. 1997;111:499–517. [Google Scholar]
- 15.Marasco R, Mosqueira MJ, Fusi M, Ramond J, Merlino G, Booth JM, et al. Rhizosheath microbial community assembly of sympatric desert speargrasses is independent of the plant host. Microbiome. 2018;6:215. doi: 10.1186/s40168-018-0597-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown LK, George TS, Neugebauer K, White PJ. The rhizosheath—a potential trait for future agricultural sustainability occurs in orders throughout the angiosperms. Plant Soil. 2017;418:115–28. doi: 10.1007/s11104-017-3220-2. [DOI] [Google Scholar]
- 17.Pang J, Ryan MH, Siddique KHMM, Simpson RJ. Unwrapping the rhizosheath. Plant Soil. 2017;418:129–39. doi: 10.1007/s11104-017-3358-y. [DOI] [Google Scholar]
- 18.Marasco R, Fusi M, Mosqueira M, Booth JM, Rossi F, Cardinale M, et al. Rhizosheath–root system changes exopolysaccharide content but stabilizes bacterial community across contrasting seasons in a desert environment. Environ Microbiome. 2022;17:14. doi: 10.1186/s40793-022-00407-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreno-Espíndola IP, Rivera-Becerril F, de Jesús Ferrara-Guerrero M, De León-González F. Role of root-hairs and hyphae in adhesion of sand particles. Soil Biol Biochem. 2007;39:2520–6. doi: 10.1016/j.soilbio.2007.04.021. [DOI] [Google Scholar]
- 20.Wullstein LHH, Pratt SAA. Scanning electron microscopy of rhizosheaths of Oryzopsis hymenoides. Am J Bot. 1981;68:408–19. doi: 10.1002/j.1537-2197.1981.tb06379.x. [DOI] [Google Scholar]
- 21.Young IM. Variation in moisture contents between bulk soil and the rhizosheath of wheat (Triticum aestivum L. cv. Wembley) New Phytol. 1995;130:135–9. doi: 10.1111/j.1469-8137.1995.tb01823.x. [DOI] [Google Scholar]
- 22.Ashraf M, Hasnain S, Berge O, Campus Q. Effect of exo-polysaccharides producing bacterial inoculation on growth of roots of wheat (Triticum aestivum L.) plants grown in a salt-affected soil. Int J Environ Sci Technol. 2006;3:45–53. doi: 10.1007/BF03325906. [DOI] [Google Scholar]
- 23.George TS, Brown LK, Ramsay L, White PJ, Newton AC, Bengough AG, et al. Understanding the genetic control and physiological traits associated with rhizosheath production by barley (Hordeum vulgare) New Phytol. 2014;203:195–205. doi: 10.1111/nph.12786. [DOI] [PubMed] [Google Scholar]
- 24.Ndour PMS, Heulin T, Achouak W, Laplaze L, Cournac L. The rhizosheath: from desert plants adaptation to crop breeding. Plant Soil. 2020;456:1–13. doi: 10.1007/s11104-020-04700-3. [DOI] [Google Scholar]
- 25.Othman AA, Amer WM, Fayez M, Monib M, Hegazi NA. Biodiversity of diazotrophs associated to the plant cover of north sinai deserts. Arch Agron Soil Sci. 2003;49:683–705. doi: 10.1080/03650340310001615110. [DOI] [Google Scholar]
- 26.Bergmann D, Zehfus M, Zierer L, Smith B, Gabel M. Grass rhizosheaths: associated bacterial communities and potential for nitrogen fixation. West North Am Nat. 2009;69:105–14. doi: 10.3398/064.069.0102. [DOI] [Google Scholar]
- 27.Marasco R, Rolli E, Ettoumi B, Vigani G, Mapelli F, Borin S, et al. A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS ONE. 2012;7:e48479. doi: 10.1371/journal.pone.0048479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marasco R, Mapelli F, Rolli E, Mosqueira MJ, Fusi M, Bariselli P, et al. Salicornia strobilacea (synonym of Halocnemum strobilaceum) grown under different tidal regimes selects rhizosphere bacteria capable of promoting plant growth. Front Microbiol. 2016;7:1–11. doi: 10.3389/fmicb.2016.01286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rolli E, Marasco R, Vigani G, Ettoumi B, Mapelli F, Deangelis ML, et al. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ Microbiol. 2015;17:316–31. doi: 10.1111/1462-2920.12439. [DOI] [PubMed] [Google Scholar]
- 30.Alsharif W, Saad MM, Hirt H. Desert microbes for boosting sustainable agriculture in extreme environments. Front Microbiol. 2020;11:1666. [DOI] [PMC free article] [PubMed]
- 31.Zhang Y, Du H, Xu F, Ding Y, Gui Y, Zhang J, et al. Root-bacteria associations boost rhizosheath formation in moderately dry soil through ethylene responses. Plant Physiol. 2020;183:780–92. doi: 10.1104/pp.19.01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soussi A, Ferjani R, Marasco R, Guesmi A, Cherif H, Rolli E, et al. Plant-associated microbiomes in arid lands: diversity, ecology and biotechnological potential. Plant Soil. 2016;405:357–70. doi: 10.1007/s11104-015-2650-y. [DOI] [Google Scholar]
- 33.Livingston G, Matias M, Calcagno V, Barbera C, Combe M, Leibold MA, et al. Competition-colonization dynamics in experimental bacterial metacommunities. Nat Commun. 2012;3:1–8. doi: 10.1038/ncomms2239. [DOI] [PubMed] [Google Scholar]
- 34.Smith GR, Steidinger BS, Bruns TD, Peay KG. Competition–colonization tradeoffs structure fungal diversity. ISME J. 2018;12:1758–67. doi: 10.1038/s41396-018-0086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seely MK. The Namib dune desert: an unusual ecosystem. J Arid Environ. 1978;1:117–28. doi: 10.1016/S0140-1963(18)31736-1. [DOI] [Google Scholar]
- 36.Klaassen E, Craven P. Checklist of grasses in Namibia. SABONET; Pretoria & Windhoek, 2014. (Produced by National Botanical Research Institute Private Bag 13184).
- 37.Neilson JW, Califf K, Cardona C, Copeland A, van Treuren W, Josephson KL, et al. Significant impacts of increasing aridity on the arid soil microbiome. mSystems. 2017;2:1–15. doi: 10.1128/mSystems.00195-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darwin C. On the origin of species. London: Routledge; 1859.
- 39.Gunnigle E, Frossard A, Ramond J-B, Guerrero L, Seely M, Cowan DA. Diel-scale temporal dynamics recorded for bacterial groups in Namib Desert soil. Sci Rep. 2017;7:40189. doi: 10.1038/srep40189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wickham H. ggplot2: Elegant graphics for data analysis. Media. Springer; New York, NY 2016.
- 41.RC-Team. R: A language and environment for statistical computing (Version 3.5. 2, R foundation for statistical computing, Vienna, Austria, 2018). R Foundation for Statistical Computing; 2019.
- 42.Anderson MMJJ, Gorley RNRN, Clarke KRR. PERMANOVA + for PRIMER: guide to software and statistical methods; PRIMER-E. Plymouth, UK: PRIMER-E Ltd.; 2008.
- 43.Cherif H, Marasco R, Rolli E, Ferjani R, Fusi M, Soussi A, et al. Oasis desert farming selects environment-specific date palm root endophytic communities and cultivable bacteria that promote resistance to drought. Environ Microbiol Rep. 2015;7:668–78. doi: 10.1111/1758-2229.12304. [DOI] [PubMed] [Google Scholar]
- 44.Lee KC, Caruso T, Archer SDJ, Gillman LN, Lau MCY, Craig Cary S, et al. Stochastic and deterministic effects of a moisture gradient on soil microbial communities in the McMurdo dry valleys of Antarctica. Front Microbiol. 2018;9:1–12. doi: 10.3389/fmicb.2018.02619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2014;22:5271–7. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 47.Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol. 2007;62:142–60. doi: 10.1111/j.1574-6941.2007.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clarke KR, Gorley RN. PRIMER v7: user manual/tutorial. Plymouth, UK: PRIMER-E; 2015.
- 49.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara B, et al. The vegan R package: community ecology. 2013:0–291
- 50.Wang Y, Naumann U, Wright ST, Warton DI. mvabund—an R package for model-based analysis of multivariate abundance data. Methods Ecol Evol. 2012;3:471–4. doi: 10.1111/j.2041-210X.2012.00190.x. [DOI] [Google Scholar]
- 51.Legendre P. Interpreting the replacement and richness difference components of beta diversity. Glob Ecol Biogeogr. 2014;23:1324–34. doi: 10.1111/geb.12207. [DOI] [Google Scholar]
- 52.Dray S, Blanchet G, Borcard D, Guenard G, Jombart T, Larocque G, et al. Package ‘adespatial’. R package version. 2018.
- 53.Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:1–9. [Google Scholar]
- 54.Weiss S, Van Treuren W, Lozupone C, Faust K, Friedman J, Deng Y, et al. Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J. 2016;10:1669–81. doi: 10.1038/ismej.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10:538–50. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 56.Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. Third International AAAI Conference on Weblogs and Social Media. 2009;8:361–2.
- 57.Barberán A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012;6:343–51. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech Theory Exp. 2008;2008:P10008. doi: 10.1088/1742-5468/2008/10/P10008. [DOI] [Google Scholar]
- 59.de Vries FT, Griffiths RI, Bailey M, Craig H, Girlanda M, Gweon HS, et al. Soil bacterial networks are less stable under drought than fungal networks. Nat Commun. 2018;9:3033. doi: 10.1038/s41467-018-05516-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andrews S. FastQC: a quality control tool for high throughput sequence data. Cambridge, United Kingdom: Babraham Bioinformatics, Babraham Institute; 2010.
- 62.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–4. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodriguez-R LM, Gunturu S, Tiedje JM, Cole JR, Konstantinidis KT. Nonpareil 3: fast estimation of metagenomic coverage and sequence diversity. mSystems. 2018;3:1–9. doi: 10.1128/mSystems.00039-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27:824–34. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mikheenko A, Saveliev V, Gurevich A. MetaQUAST: evaluation of metagenome assemblies. Bioinformatics. 2016;32:1088–90. doi: 10.1093/bioinformatics/btv697. [DOI] [PubMed] [Google Scholar]
- 67.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 69.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81–7. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2014;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 72.Vigani G, Rolli E, Marasco R, Dell’Orto M, Michoud G, Soussi A, et al. Root bacterial endophytes confer drought resistance and enhance expression and activity of a vacuolar H+-pumping pyrophosphatase in pepper plants. Environ Microbiol. 2019;21:3212–28. doi: 10.1111/1462-2920.14272. [DOI] [PubMed] [Google Scholar]
- 73.Al-Hosni K, Shahzad R, Khan AL, Muhammad Imran Q, Al Harrasi A, Al Rawahi A, et al. Preussia sp. BSL-10 producing nitric oxide, gibberellins, and indole acetic acid and improving rice plant growth. J Plant Interact. 2018;13:112–8. doi: 10.1080/17429145.2018.1432773. [DOI] [Google Scholar]
- 74.Sen D, Paul K, Saha C, Mukherjee G, Nag M, Ghosh S, et al. A unique life-strategy of an endophytic yeast Rhodotorula mucilaginosa JGTA-S1—a comparative genomics viewpoint. DNA Res. 2019;26:131–46. doi: 10.1093/dnares/dsy044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johnson JM, Ludwig A, Furch ACU, Mithöfer A, Scholz S, Reichelt M, et al. The beneficial root-colonizing fungus Mortierella hyalina promotes the aerial growth of Arabidopsis and activates calcium-dependent responses that restrict Alternaria brassicae–induced disease development in roots. Mol Plant-Microbe Interact. 2019;32:351–63. doi: 10.1094/MPMI-05-18-0115-R. [DOI] [PubMed] [Google Scholar]
- 76.van Dam NM, Bouwmeester HJ. Metabolomics in the rhizosphere: tapping into belowground chemical communication. Trends Plant Sci. 2016;21:256–65. doi: 10.1016/j.tplants.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 77.Zeng Y, Charkowski AO. The role of ATP-binding cassette transporters in bacterial phytopathogenesis. Phytopathology®. 2021;111:600–10. doi: 10.1094/PHYTO-06-20-0212-RVW. [DOI] [PubMed] [Google Scholar]
- 78.Louca S, Polz MF, Mazel F, Albright MBN, Huber JA, O’Connor MI, et al. Function and functional redundancy in microbial systems. Nat Ecol Evol. 2018;2:936–43. doi: 10.1038/s41559-018-0519-1. [DOI] [PubMed] [Google Scholar]
- 79.Balskus EP, Walsh CT. The genetic and molecular basis for sunscreen biosynthesis in cyanobacteria. Science. 2010;329:1653–6. doi: 10.1126/science.1193637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smith VH. Effects of resource supplies on the structure and function of microbial communities. Antonie Van Leeuwenhoek. 2002;81:99–106. doi: 10.1023/A:1020533727307. [DOI] [PubMed] [Google Scholar]
- 82.Albalasmeh AA, Ghezzehei TA. Interplay between soil drying and root exudation in rhizosheath development. Plant Soil. 2014;374:739–51. doi: 10.1007/s11104-013-1910-y. [DOI] [Google Scholar]
- 83.Devitt DA, Smith SD. Root channel macropores enhance downward movement of water in a Mojave Desert ecosystem. J Arid Environ. 2002;50:99–108. doi: 10.1006/jare.2001.0853. [DOI] [Google Scholar]
- 84.Othman AA, Amer WM, Fayez M, Hegazi NA. Rhizosheath of sinai desert plants is a potential repository for associative diazotrophs. Microbiol Res. 2004;159:285–93. doi: 10.1016/j.micres.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 85.Naseem H, Ahsan M, Shahid MA, Khan N. Exopolysaccharides producing rhizobacteria and their role in plant growth and drought tolerance. J Basic Microbiol. 2018;58:1009–22. doi: 10.1002/jobm.201800309. [DOI] [PubMed] [Google Scholar]
- 86.Toju H, Peay KG, Yamamichi M, Narisawa K, Hiruma K, Naito K, et al. Core microbiomes for sustainable agroecosystems. Nat Plants. 2018;4:247–57. doi: 10.1038/s41477-018-0139-4. [DOI] [PubMed] [Google Scholar]
- 87.Banerjee S, Schlaeppi K, van der Heijden MGAA. Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol. 2018;16:567–76. doi: 10.1038/s41579-018-0024-1. [DOI] [PubMed] [Google Scholar]
- 88.Agler MT, Ruhe J, Kroll S, Morhenn C, Kim S-TT, Weigel D, et al. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016;14:1–31. doi: 10.1371/journal.pbio.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Delgado-Baquerizo M, Reich PB, Trivedi C, Eldridge DJ, Abades S, Alfaro FD, et al. Multiple elements of soil biodiversity drive ecosystem functions across biomes. Nat Ecol Evol. 2020;4:210–20. doi: 10.1038/s41559-019-1084-y. [DOI] [PubMed] [Google Scholar]
- 90.Hassani MA, Durán P, Hacquard S. Microbial interactions within the plant holobiont. Microbiome. 2018;6:58. doi: 10.1186/s40168-018-0445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lopez BR, Bacilio M. Weathering and soil formation in hot, dry environments mediated by plant–microbe interactions. Biol Fertil Soils. 2020;56:447–59. doi: 10.1007/s00374-020-01456-x. [DOI] [Google Scholar]
- 92.Hernandez DJ, David AS, Menges ES, Searcy CA, Afkhami ME. Environmental stress destabilizes microbial networks. ISME J. 2021;15:1722–34. doi: 10.1038/s41396-020-00882-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yuan MM, Guo X, Wu L, Zhang Y, Xiao N, Ning D, et al. Climate warming enhances microbial network complexity and stability. Nat Clim Chang. 2021;11:343–8. doi: 10.1038/s41558-021-00989-9. [DOI] [Google Scholar]
- 94.Safronova VI, Kuznetsova IG, Sazanova AL, Belimov AA, Andronov EE, Chirak ER, et al. Microvirga ossetica sp. nov., a species of rhizobia isolated from root nodules of the legume species Vicia alpestris Steven. Int J Syst Evol Microbiol. 2017;67:94–100. doi: 10.1099/ijsem.0.001577. [DOI] [PubMed] [Google Scholar]
- 95.Jiménez-Gómez A, Saati-Santamaría Z, Igual J, Rivas R, Mateos P, García-Fraile P. Genome insights into the novel species Microvirga brassicacearum, a rapeseed endophyte with biotechnological potential. Microorganisms. 2019;7:354. doi: 10.3390/microorganisms7090354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu T, Ye N, Wang X, Das D, Tan Y, You X, et al. Drought stress and plant ecotype drive microbiome recruitment in switchgrass rhizosheath. J Integr Plant Biol. 2021;63:1753–74. [DOI] [PubMed]
- 97.Blouin M. Chemical communication: an evidence for co-evolution between plants and soil organisms. Appl Soil Ecol. 2018;123:409–15. doi: 10.1016/j.apsoil.2017.10.028. [DOI] [Google Scholar]
- 98.Sarrocco S, Diquattro S, Baroncelli R, Cimmino A, Evidente A, Vannacci G, et al. A polyphasic contribution to the knowledge of Auxarthron (Onygenaceae). Mycol Prog. 2015;14:112.
- 99.Macías-Rubalcava ML, Sánchez-Fernández RE. Secondary metabolites of endophytic Xylaria species with potential applications in medicine and agriculture. World J Microbiol Biotechnol. 2017;33:15. [DOI] [PubMed]
- 100.Zhang K, Bonito G, Hsu C, Hameed K, Vilgalys R, Liao H-L. Mortierella elongata increases plant biomass among non-leguminous crop species. Agronomy. 2020;10:754. doi: 10.3390/agronomy10050754. [DOI] [Google Scholar]
- 101.Kobayashi DY, Crouch JA. Bacterial/fungal interactions: from pathogens to mutualistic endosymbionts. Annu Rev Phytopathol. 2009;47:63–82. doi: 10.1146/annurev-phyto-080508-081729. [DOI] [PubMed] [Google Scholar]
- 102.Asmelash F, Bekele T, Birhane E. The potential role of arbuscular mycorrhizal fungi in the restoration of degraded lands. Front Microbiol. 2016;7:1–15. doi: 10.3389/fmicb.2016.01095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kohlmeier S, Smits THM, Ford RM, Keel C, Harms H, Wick LY. Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ Sci Technol. 2005;39:4640–6. doi: 10.1021/es047979z. [DOI] [PubMed] [Google Scholar]
- 104.Warmink JA, Nazir R, Corten B, van Elsas JD. Hitchhikers on the fungal highway: the helper effect for bacterial migration via fungal hyphae. Soil Biol Biochem. 2011;43:760–5. doi: 10.1016/j.soilbio.2010.12.009. [DOI] [Google Scholar]
- 105.Booth JM, Fusi M, Marasco R, Michoud G, Fodelianakis S, Merlino G, et al. The role of fungi in heterogeneous sediment microbial networks. Sci Rep. 2019;9:7537. doi: 10.1038/s41598-019-43980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Deveau A, Bonito G, Uehling J, Paoletti M, Becker M, Bindschedler S, et al. Bacterial-fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol Rev. 2018;42:335–52. doi: 10.1093/femsre/fuy008. [DOI] [PubMed] [Google Scholar]
- 107.Simon A, Hervé V, Al-Dourobi A, Verrecchia E, Junier P. An in situ inventory of fungi and their associated migrating bacteria in forest soils using fungal highway columns. FEMS Microbiol Ecol. 2017;93:fiw217. doi: 10.1093/femsec/fiw217. [DOI] [PubMed] [Google Scholar]
- 108.Faust K, Raes J. CoNet app: inference of biological association networks using Cytoscape. F1000Research. 2016;5:1519. doi: 10.12688/f1000research.9050.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012;6:1007–17. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zablocki O, Adriaenssens EM, Cowan D. Diversity and ecology of viruses in hyperarid desert soils. Appl Environ Microbiol. 2016;82:770–7. doi: 10.1128/AEM.02651-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Van Goethem MW, Swenson TL, Trubl G, Roux S, Northen TR. Characteristics of wetting-induced bacteriophage blooms in biological soil crust. MBio. 2019;10:e02287–19. [DOI] [PMC free article] [PubMed]
- 112.Lambers H, Mougel C, Jaillard B, Hinsinger P. Plant-microbe-soil interactions in the rhizosphere: an evolutionary perspective. Plant Soil. 2009;321:83–115. doi: 10.1007/s11104-009-0042-x. [DOI] [Google Scholar]
- 113.Ghoul M, Mitri S. The ecology and evolution of microbial competition. Trends Microbiol. 2016;24:833–45. doi: 10.1016/j.tim.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 114.Schlatter DC, Kinkel LL. Antibiotics: conflict and communication in microbial communities. Microbe Mag. 2014;9:282–8. doi: 10.1128/microbe.9.282.1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available in the NCBI SRA repository under the BioProject ID PRJNA628615. The raw, unassembled metagenomes are available in the NCBI SRA repository under the BioProject ID PRJNA731454.