Abstract
RNA G-quadruplexes (rG4s) are RNA secondary structures, which are formed by guanine-rich sequences and have important cellular functions. Existing computational tools for rG4 prediction rely on specific sequence features and/or were trained on small datasets, without considering rG4 stability information, and are therefore sub-optimal. Here, we developed rG4detector, a convolutional neural network to identify potential rG4s in transcriptomics data. rG4detector outperforms existing methods in both predicting rG4 stability and in detecting rG4-forming sequences. To demonstrate the biological-relevance of rG4detector, we employed it to study RNAs that are bound by the RNA-binding protein G3BP1. G3BP1 is central to the induction of stress granules (SGs), which are cytoplasmic biomolecular condensates that form in response to a variety of cellular stresses. Unexpectedly, rG4detector revealed a dynamic enrichment of rG4s bound by G3BP1 in response to cellular stress. In addition, we experimentally characterized G3BP1 cross-talk with rG4s, demonstrating that G3BP1 is a bona fide rG4-binding protein and that endogenous rG4s are enriched within SGs. Furthermore, we found that reduced rG4 availability impairs SG formation. Hence, we conclude that rG4s play a direct role in SG biology via their interactions with RNA-binding proteins and that rG4detector is a novel useful tool for rG4 transcriptomics data analyses.
Graphical Abstract
Graphical Abstract.
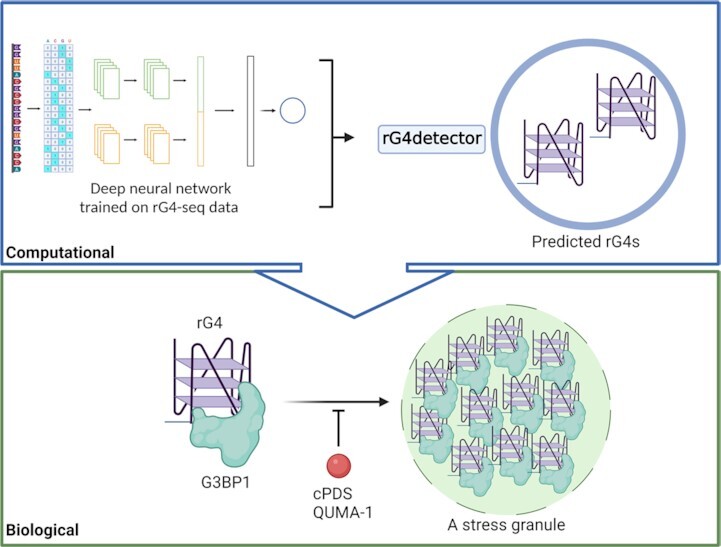
In this study, we developed rG4detector, a novel method based on deep neural networks to predict RNA G-quadruplexes from an RNA sequence, and used it to reveal that G3BP1 interacts with rG4s in a stress-dependent manner and that rG4–protein interactions contribute to stress granules formation.
INTRODUCTION
RNA G-quadruplexes (rG4s) are non-canonical higher-order RNA secondary structures, which fold from guanine (G)-rich RNA strands due to the propensity of Gs to self-assemble in a plane and form G-quartets via Hoogsteen hydrogen bonds (1–3). The formation and subsequent self-stacking of G-quartets provide rG4s with a high thermodynamic stability, which is further regulated by the binding of monovalent cations (e.g. K+) in between the G-quartet planes and also by steric factors such as loop length (the number of intervening nucleobases between G stretches) and the number of G-quartets (dictated by the number of Gs within each stretch) (3).
rG4-forming sequences (rG4FSs) in the human transcriptome have been detected by various experimental assays based on RNA next-generation sequencing (NGS), including rG4-seq (with >13 000 rG4FSs (4) in vitro) and G4RP-seq (with >6000 rG4FSs in vivo) (5,6). Several bioinformatic tools (7), including G4RNA screener (8), indicate that ∼60% of the human transcripts have at least one rG4FS. This prevalence is strongly indicative of rG4s’ functional relevance. Accordingly, numerous studies have shown that rG4s play regulatory roles in key cellular processes such as transcription, RNA splicing, translation, and stress response (1,3,9,10).
A main feature of the cellular stress response is formation of stress granules (SGs), which are cytoplasmic biomolecular condensates that assemble in response to a variety of cellular stresses. SGs are composed of untranslated mRNA and RNA-binding proteins (RBPs) (11,12) and regulate different aspects of RNA metabolism, including translation, sequestration of RBPs and mRNA molecules and signaling cascades (12–16). Aberrant SG dynamics have been implicated in several human neurodegenerative disorders, including amyotrophic lateral sclerosis (ALS) (12,17–21). This highlights the importance of SGs in cells’ function but also of the critical need to better understand SG biology in mechanisms underlying pathologies.
Several lines of evidence indirectly implicate rG4 function in SG biology. Notable changes in SG formation have been observed in response to addition of exogenous G4-forming repeated RNA sequences (22), or as a result of perturbations of DHX36 expression, a main rG4-helicase (23). In addition, RAS GTPase-activating binding protein 1 (G3BP1), a SG core protein, has been suggested to bind rG4s (24,25). Taken together, rG4FSs appear to be key players in SG biology. However, to date, firm evidence demonstrating direct involvement and roles of rG4s in SG biology is still lacking.
Several computational methods were developed to predict rG4 formation in the human transcriptome (7). QGRS was developed to identify sequences that contain four G-tracts (26), while cGcC-scoring (27) and G4Hunter (28) are based on a scheme that attributes a high score for any G- or C-tract. Hence, these methods for the discovery of rG4s rely on simple scoring schemes that mostly identify canonical G4 motifs. In contrast, G4NN is a neural network trained to distinguish between experimentally validated rG4 sequences and non-rG4 sequences (8). G4NN is thus a powerful approach but its accuracy is still suboptimal as it was trained on a limited number of rG4FSs (<500) and does not include quantitative data of rG4 stability. Therefore, the need for an accurate rG4 prediction method is not fully met.
rG4-seq assesses the prevalence of rG4s in vitro on the basis of their ability to act as roadblocks to reverse transcriptase (RT). Hence, rG4 sites are ascribed to RT stalling (RTS) sites, which is measured both in rG4-defavoring conditions (Li+-rich buffer, as a control) and rG4-promoting conditions (K+-rich buffer and with the presence of pyridostatin (PDS), a well-known G4-stabilizer (29)). An improved processing of available high-throughput datasets was achieved by rG4-seeker (30), which was developed to process the rG4-seq dataset (4). rG4-seeker calculates the ratio of stalled reads (RSR), which is correlated with the stability of the rG4 upstream of the RSR (4), to improve the detection of significant RTS sites (especially dedicated to the identification of non-canonical rG4FS). These rich and accurate quantitative data were still not utilized for developing machine-learning-based methods for rG4 prediction.
In this work, we tested the hypothesis that a machine-learning-based detector can improve rG4 prediction and lead to new biological discoveries. We developed rG4detector, a convolutional neural network for predicting rG4 folding of any given sequence based on rG4-seq data. rG4detector assigns an rG4 propensity score for any RNA sequence, and outperforms G4NN, G4Hunter, and cGcC-scoring. In addition, we interrogated rG4detector's biological relevance and discovered both known and novel biological principles behind rG4 folding. To demonstrate that rG4detector effectively predicts rG4FSs and can set forward testable biological hypotheses, we characterized the dynamic enrichment of rG4s bound by G3BP1 in SGs. On this basis, we demonstrate that endogenous rG4s play a direct role in SG formation through rG4 interactions with RBPs.
MATERIALS AND METHODS
Computational materials
Datasets
In this work, we used the raw data produced by the rG4-seq protocol on RNA from human HeLa cells (4) (accession code GSE77282) to calculate RSR scores across the human transcriptome (Supplementary Figure S1A). We extracted reads using SRA Toolkit, trimmed adapters with Cutadapt (31), aligned them to the hg38 reference genome using STAR-2.7 (32), and merged all output files using SAMtools (33). Then, we used rG4-seeker, an improved statistical pipeline for processing rG4-seq data, to extract read coverage and stalling-events counts per position. Using these counts, we calculated the RSR scores in single-nucleotide resolution. For adjacent nucleotides we summed their number of stalling events and the total read coverage, and then calculated their combined RSR (as described in rG4-seeker (30)). We then calculated an RSR-ratio score, which we defined as a proxy to rG4 stability, as follows:
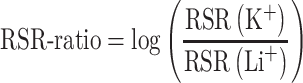 |
To attain high-quality RSR-ratio scores, we filtered out RSR-ratio scores based on <1000 total read count. We were left with 53 991 high-quality RSR-ratio scores (Supplementary Figure S1B).
We used several independent datasets to validate the prediction performance of rG4detector. First, we used Guo and Bartel rG4 experimental dataset (34). This dataset, which contains 11 606 sequences, includes RTS frequencies over the mouse transcriptome. RTS was measured using the fold-enrichment value, i.e. the ratio of the number of reads stalled at a given position over the background read density of the read-stalled-position nucleotide within the same transcript. We used as labels the log of the ratio of fold-enrichment under K+ over fold-enrichment under Li+. Second, we used rG4-seq data on Arabidopsis thaliana transcriptome (35), which contains 178 sequences with RTS measurements. Third, we applied rG4detector to classify rG4s from the G4RNA dataset. We obtained all unique G4RNA sequences from the G4RNA platform (http://scottgroup.med.usherbrooke.ca/G4RNA) and merged overlapping sequences, which had the same label, leaving 128 sequences, where 103 are experimentally verified rG4s and 25 are non-rG4s. Last, we used the dataset from Zhang et al. (36) low-throughput experiment, which examined the dependence of rG4 thermodynamic stability on its loop length. In this experiment, 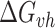 values were measured for 29 canonical rG4s with variable loop lengths (27 sequences with all combinations of loop lengths from 1 to 3, and 2 sequences of loop lengths of 4 and 5). In this dataset, loop nucleotide content is any nucleotide except for guanine in the adjacent positions to the G-tracts.
values were measured for 29 canonical rG4s with variable loop lengths (27 sequences with all combinations of loop lengths from 1 to 3, and 2 sequences of loop lengths of 4 and 5). In this dataset, loop nucleotide content is any nucleotide except for guanine in the adjacent positions to the G-tracts.
Sequence extraction
To extract the sequences corresponding to the RSR-ratio measurements, we followed the guidelines proposed by Uhl et al. (37). We mapped each RSR-ratio position to a single transcript, which has the highest transcript support level, i.e. is correlated with the most prominent transcript isoform. For each position associated with an RSR-ratio score, we extracted sequences of length 30nt upstream of the RTS location, and since previous studies demonstrated that G4-flanking sequences are informative of G4-folding potential (38), we appended each sequence we assigned an RSR-ratio for by its 25nt-long upstream and downstream flanks, obtaining a sequence in total length of 80nt. In case of sequences crossing transcript boundaries, we used zero padding before the 5’ end or past the 3’ end.
Computational methods
rG4detector architecture
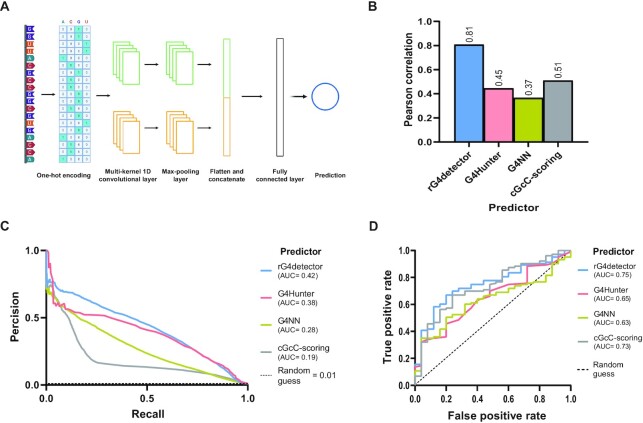
rG4detetor is a random initialization ensemble of multi-kernel convolutional neural networks, based on an architecture previously proposed by Zhang et al. (39) (Figure 1A). Convolutional neural networks are very popular in genomics for their ability to capture specific sequence patterns. But, using a kernel of fixed size might not be beneficial for identifying potential rG4 sequence features because of their variable lengths. Therefore, we implemented in rG4detector two parallel one-dimensional convolution layers with 128 kernels each of sizes of 10 and 17, respectively. rG4detector input is a 80nt-long one-hot-encoded RNA sequence. We replaced N positions in the RNA sequence with a uniform vector of 0.25. The one-hot-encoded matrix is first processed by the convolutional layer. The output of each one of the convolutional kernels goes through a max-pooling layer with pool sizes of 2. The max-pooling output matrices are flattened and concatenated to form a single numerical vector. This vector is the input to a fully connected layer with 64 nodes with ReLU activation function. Finally, the output of the fully connected layer is the input to a single neuron, which outputs the network prediction. To prevent overfitting, we used in the convolution layers kernel regularizers (regularizer weight of 0.0005) and dropout (probability 0.2). Dropout was also used following the fully connected layer.
Figure 1.
rG4detector accurately predicts rG4s in various datasets and tasks. (A) A diagram depicting rG4detector's convolutional neural network architecture. The RNA sequence is first one-hot encoded and then processed by a one-dimensional multi-convolutional layer. The outputs of each kernel go through a max-pooling layer. All max-pooling outputs are concatenated and passed into a fully connected layer followed by a single output neuron, which outputs the network RSR-ratio prediction. (B) Prediction performance gauged by Pearson correlation for rG4 propensity by rG4detector and G4Hunter (28), G4NN (8), and cGcC-scoring (27) on a human rG4 dataset (4). (C) Precision–recall curves of rG4detector, G4Hunter, G4NN and cGcC-scoring for rG4 detection on human transcripts. Area under the curve (AUC) value is indicated. (D) Receiver operating characteristic (ROC) curves of rG4detector, G4Hunter, G4NN, and cGcC-scoring in binary classification over the G4RNA dataset.
To train the model and select optimal hyper-parameters, we excluded the data of chromosome 2 from the training dataset and used it as a validation set. We chose the hyper-parameters values using a search over 600 random hyper-parameter combinations with a pre-defined range for each parameter (Supplementary Data S1, Table S1). Once the hyper-parameters were set, we trained 50 models with different initial weights and evaluated their performance on the validation set. We tested 15 ensemble models based on 1–15 of the best-performing models, and observed that an ensemble of 11 models performs the best in this setting (Supplementary Figure S1C). rG4detector final prediction is the average of all 11 models’ outputs. We held out the data of chromosome 1 to use it as a test set. We implemented rG4detector using Keras library with Tensorflow backend (version 2.9.1). The models were trained over five training epochs, with batch sizes of 128 and were optimized using Adam optimizer with learning rate of 0.005 and default β1 and β2 values (0.9 and 0.999, respectively).
Running existing methods
We generated rG4 predictions of G4Hunter (28), G4NN (8) and cGcC-scoring (27), by locally running G4RNA screener (gitlabscottgroup.med.usherbrooke.ca/J-Michel/g4rna_screener) (8), a tool that runs the three methods. We ran G4RNA screener with three window sizes: 60 (G4RNA screener default), 25 (G4Hunter chosen window size) and 80 (rG4detector input size), and with step size of 1, preferring high prediction resolution over computing time. We assigned the maximum prediction per sequence to be the final prediction. At each task, we report the results on the window size that enabled each method to achieve best performance (performances over all window sizes are in Supplementary Data S1, Table S2).
Detection of rG4FSs
We utilized the predictions of rG4detector and existing methods to identify rG4FSs in a given transcript. To test rG4 detection ability, we retrieved all human transcripts which contain an rG4 by the rG4-seq experiment in the held-out test set (chromosome 1). For each transcript, we assigned a binary label to each nucleotide: 1 for a nucleotide belonging to an rG4FS, and 0 otherwise. We scanned each transcript to obtain predictions with window size of 80nt and step size 1nt. Since rG4detector input includes both the potential rG4FSs and its flanking sequences, whose size varies according to the rG4FSs size, we used a Gaussian weighted average to combine each position's predictions into one final score. By doing so, we assigned more weight for predictions where the given nucleotide is closer to the center of rG4detector input as follows:
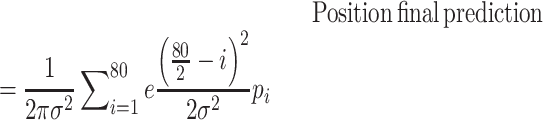 |
(1) |
with  , which led to best performance on the validation set over
, which led to best performance on the validation set over  .
.
We performed the same process using G4NN and cGcC-scoring with window sizes of 25, 60, and 80. For G4Hunter, we implemented and applied the transcriptome-wide detection algorithm as previously described by the developers of G4Hunter (28) with window sizes of 25, 60, and 80. We gauged detection performance by the precision–recall curve.
Interpretability of rG4detector
To deduce insights behind the molecular mechanism of rG4 formation, we interrogated the rG4detector-trained model. We first visualized the impact of loop lengths on predicted RSR-ratio scores. We predicted RSR-ratio scores for a canonical rG4 with variable loop lengths while varying the length of each loop separately between 1 to 12. The loops and flanks were encoded such that each nucleotide at those positions was assigned an equal uniform probability, i.e. 0.25.
Next, we further validated the effect of loop length on RSR-ratio predictions. We composed a dataset of canonical rG4s with multiple combinations of loops lengths as previously defined (36). We generated 27 sequences with all possible combinations of loop lengths from 1 to 3, and two sequences with constant loop length of 4 and 5. Following the experimental protocol, we set loop nucleotide content to equal uniform probability for any nucleotide, except at positions adjacent to the G-tracts, which we set to have equal probability for A, C, and U only, i.e. 1/3 for each.
To examine the effect of G-tracts length on RSR-ratio predictions, we predicted the RSR-ratio scores for a canonical rG4 sequence with a constant length of G-tracts, which we varied between 1 and 8, with loop lengths between 1 to 4. We set positions outside the G-tracts to equal probability distribution for any nucleotide, i.e. 0.25.
In addition, we visualized the importance of each position and nucleotide in a given RNA sequence on its RSR-ratio prediction using a mutation heatmap and a sequence logo. For this visualization, we chose one of the rG4 sequences that were identified in the human rG4-seq experiment, and predicted the RSR-ratio score of all possible single-nucleotide mutated sequences. We then calculated their difference from the original sequence prediction and visualized the differences in a heatmap. In addition, we used the integrated gradients (IG) approach to identify key features within the given sequence (40). The IG approach attributes a differentiable model's prediction to features of the input relative to a neutral baseline and assigns an importance score to each feature. The method computes the path integral of the gradients along a straight path between the input and the baseline. We used an all-0.25 matrix baseline representing equal probability for each nucleotide in each position of the sequence. To visualize the preferences learned by rG4detector by IG we generated a sequence logo using logomaker (41).
Last, we explored the impact of mutations in the G-tracts on predicted RSR-ratio scores. For this aim, we defined the wild type rG4 as GGGNGGGNGGGNGGG. We predicted an RSR-ratio score for each rG4 variant with a mutation in one of the Gs in the G-tracts to all possible nucleotides. The impact of the mutation was measured as the difference between the predicted score of the wild-type rG4 and the mutated rG4.
MEME analysis
To evaluate the most common motif within G3BP1-bound RNAs, we ran Multiple Expectation maximization for Motif Elicitation analysis (MEME) online (MEME suite version 5.4.1; https://meme-suite.org/meme/index.html) by using the parameters: classic motif discovery mode, distribution of any number of repetition, search for ten motifs with minimum width of 6nt, and maximum width of 20nt.
Experimental materials
Reagents
We used the next stock reagents in this study, specifically for in vitro experiments: MgCl2 (powder; Mallinckrodt CHEMICALS, 6066-04), 100% glycerol (Sigma-Aldrich, G5516), RNase/DNase-free UPW (Invitrogen, 10977-035), 1 M Tris–HCl pH 8.0 (Invitrogen, 15568-025), 1 M Tris–HCl pH 7.5 (Invitrogen, 15567-027), 0.5 M EDTA (Invitrogen, AM9261), 1 M DTT (Sigma-Aldrich, 43816), TBE ×10 (Fisher BioReagents, BP13334). Stock solutions of 3 M KCl (powder; MERCKGaA, 104936) or 3 M LiCl (powder; J.T. Baker, 2370-01), were prepared by dissolving the powders in UPW and treated with DEPC (Sigma-Aldrich, D5758) before use.
RNA oligos
We chemically synthesized 6FAM-labeled RNA sequences (Supplementary data S1, Table S3) from Sigma-Aldrich/MERCK or from Integrated DNA Technologies (IDT). We dissolved the oligos in RNase-free TEx1 buffer (10 mM Tris–HCl pH 7.5 and 1 mM EDTA) for stock concentration of 100 uM and stored them at –80°C in aliquotes to avoid from thaw-freeze cycles.
Cell culture
We cultured G3BP1-GFP expressing U2OS cells (Human Bone Osteosarcoma Epithelial Cells) in growth media consisting of Dulbecco's modified Eagle's medium (DMEM, Biological Industries, 01-050-1A) supplemented with 1% penicillin-streptomycin (Sartorius, 03-031-1B), and 10% fetal bovine serum (FBS, Sartorius, 04-007-1A), at 37°C, with 5% CO2.
Experimental methods
RNA G4 preparation
We diluted the FAM-labeled RNAs to desired concentration in a TEx1 buffer with or without 150 mM DEPC-treated KCl or 150 mM DEPC-treated LiCl. Then, using a PCR machine we annealed the diluted RNAs to form secondary structures by heating to 90°C for 5 min and then lowering the temperature to 25°C in 5°C intervals (from 95°C to 50°C and from 30°C to 25°C) or in 10°C intervals (between 50°C to 30°C) as follow: 85–70°C for 5 min each, 65–50°C for 15 min each, 40–30°C for 30 min each and 25°C for 2 h. After that, we stored the RNAs at 4°C.
Circular dichroism (CD) spectroscopy
We annealed 10uM FAM-labeled RNA oligos in TEx1 buffer with or without 150 mM DEPC-treated KCl or LiCl, as mentioned above. We performed CD experiments at 25°C using Chirascan™-plus ACD spectropolarimeter with a quartz cuvette with a 1 mm path length. We collected CD spectra from 320 to 210 nm. The bandwidth was 1 nm, and the response time was 1s. We baseline-corrected all CD spectra for the signal by the buffer and represented the average of 3–5 runs.
Thermal difference absorbance spectrum (TDS)
We annealed 10 uM FAM-labeled RNA oligos in TEx1 buffer with or without 150 mM DEPC-treated KCl or LiCl, as mentioned above. We obtained the TDS spectra from the difference between the absorbance recorded at high temperature (90°C) and low temperature (25°C) under the same conditions. For technical details of the runs, see the above CD section.
Electro-mobility shift assays (EMSA)
We prepared 20 ul reaction mixtures, which contained 160 nM 5’ 6FAM-labeled RNA oligos of both rG4FSs and non-rG4FSs (Supplementary data S1, Table S3), and binding buffer consisting of 10 mM Tris–HCl pH 8.0, 150 mM KCl, 1 mM EDTA, 2 mM MgCl2, 10% glycerol, 2 mM DTT, 0.1 mg/ml Ultrapure-BSA (Invitrogen, AM2616) and Ribolock (1:40; Thermo Scientific, E00381), with or without recombinant human G3BP1 protein (Prospec, Enz-048). We incubated the binding reactions at 37°C for 1 h and then loaded them onto a 5% native non-denaturing polyacrylamide (acrylamide:bis-acrylamide 29:1 (30%); Bio-Lab, UN3426) gel consisting of (for 12.5 ml) 9 ml DEPC-ddW, 1.25 ml TBE ×10, 2.075 ml 30% polyacrylamide, 125 ul 10% ammonium persulfate (APS; Bio-Rad, 1610700) and 12.5 ul TEMED (Bio-Rad, 1610801). We performed gel electrophoresis at 100 V for 50 min in TBEx1 buffer on ice and in the dark. After 50 min, we performed gel scanning using ImageQuant LAS4000 (GE Healthcare) gel imager at cy2 channel (488 nm). For the EMSA experiments with the oligo r(AGG)5 we then electro-transferred the RNA-protein complex from the gel to a nitrocellulose membrane (Whatmann; 10401383) and used primary anti-G3BP1 antibody (Santa cruz; sc-365338) to detect the recombinant hG3BP1 within the lanes.
Staining procedure in fixed cells
For staining of BioTASQ (produced as described previously (5)), we seeded 50K G3BP1-GFP expressing U2OS cells per well 24 h prior to the stress. After the stress induction, we fixed the cells and permeabilized them with ice-cold 100% methanol (Bio-Lab, UN1230) for 10 min at RT, and washed them with RNase-free PBS ×1 (Gibco, 14200-067) once for 5 min. Next, we incubated the cells with 25 uM BioTASQ for 1 h at RT. Then, we washed the cells with RNase-free PBS ×1 three times for 5 min and blocked by Cas-block reagent (Life Technologies, 008120) for 10 min at RT, and incubated them with Streptavidin-TexasRed antibody (1:200; Invitrogen, S872) for 1 h at RT in the dark, and washed them with RNase-free PBS ×1 three times for 5 min each, and shortly dried and mounted them with DAPI (FleuroShield with DAPI; Sigma-Aldrich, F6057).
For QUMA-1 (Sigma-Aldrich, SCT056), we seeded 20K G3BP1-GFP expressing U2OS cells per well 24 h prior to the stress. After the stress induction, we fixed the cells with 4% PFA (Alfa Aesar, 43368) for 10 min and washed them with RNase-free PBS ×1 three times. Then, we incubated the cells with 2 uM QUMA-1 and Hoechst 33342 (dilution of 1:8000; Sigma-Aldrich, B2261) for 15 min at 37°C. We saved the plate in the dark from this point. Next, we washed the cells with RNase-free PBSx1 three times 5 min each. We treated the cells with UPW or DMSO as controls (no stress conditions). For SG induction, we incubated the cells with several stressors and conditions as follows: NaAsO2 (400 μM for 30 min, Sigma-Aldrich, 71287), Puromycin (200 ug/ml for 4 h, Invivogen, ANT-PR), MG-132 (100 uM for 1hr, Sigma-Aldrich, C2211), and Thapsigargin (1 uM for 1 h, Sigma-Aldrich, T9033). We acquired the fixed cells (in the procedures) via a Zeiss LSM800 laser scanning confocal microscopy system equipped with a Zeiss Axiovert microscope and using a 63× 1.4 NA oil immersion lens.
Live-cell imaging
We seeded 12K-15K G3BP1-GFP expressing U2OS cells per well 24 or 48 h prior to the experiment in a 96-well plate (Brooks, MGB096-1-2-LG-L). We incubated the cells with 10 uM cPDS (carboxypyridostatin trifluoroacetate salt; Sigma-Aldrich, SML1176) for 24 h or with 1 uM QUMA-1 for 3 h before the experiment. Then, we replaced the medium with a 150 uM NaAsO2-added medium and immediately took them to the microscope to monitor SG formation. We took SG live imaging by a PCO-Edge sCMOS camera controlled by VisView installed on a VisiScope Confocal Cell Explorer system (Yokogawa spinning disk scanning unit; CSU-W1) and an inverted Olympus microscope (60× oil objective; excitation wavelength: GFP: 488 nm). We analyzed SG and cell areas using surface features in Imaris software 9.5.1.
Cell lysis and western-blotting
We seeded 500K stable G3BP1-GFP expressing U2OS cells in 6-well plates in triplicates for the experiment. We incubated the cells with varying concentrations of cPDS for 24 h or of QUMA-1 for 3 h. In parallel, as a control, we exposed the cells to 400 uM sodium arsenate stress for 30 min without small rG4-binding molecules immediately before lysis for western blot (WB) analysis. The cells were lysed in RIPA buffer supplemented with cOmplete Protease Inhibitor Cocktail (Roche; 4693116001) and PhosSTOP (Roche; 4906837001), and incubated for 10 min on ice with vortexing each for 2 min. Then, the samples were cleared by centrifugation at 14 000 × g for 5 min at 4 °C, and the supernatant was transferred to new Eppendorf tubes. We quantified the protein concentrations with Protein Assay Dye Reagent (Bio-Rad; 500-0006), and we resolved the protein at 50 μg of total protein per well by 10% SDS–PAGE at 100 for 10 min and at 120 V for the rest time up to 80 min. After gel electrophoresis, we transferred the proteins to nitrocellulose membranes (Whatmann; 10401383) at 250 mA for 70 min. We blocked the membranes for 1 h at room temperature with 3% bovine albumin fraction V (MPBio; 160069) in PBS containing 0.05% Tween-20 (PBST) and then we incubated the membranes with primary antibodies for p-eIF2alpha (Santa Cruz Biotechnologie; sc-101670s), and Lamin A/C (Santa Cruz Biotechnologies; sc-20681) as control overnight at 4 °C with rocking in antibody solution (5% albumin, 0.02% sodium azide and five drops of phenol red in 0.05% PBST). Following primary antibody incubation, we washed the membranes three times for 5 min at room temperature with 0.05% PBST and they were incubated for 1h at room temperature with horseradish peroxidase-conjugated species-specific secondary antibodies. Then, we washed them three times for 5 min each in 0.05% PBST at room temperature and we visualized them using EZ-ECL Chemiluminescence (Biological Industries, 20500-120) by ImageQuant LAS 4000 (GE Healthcare Life Sciences). We performed densitometric analysis using FiJi software (NIH) and representative bands are presented. For WB analysis after EMSA we used an antibody against human G3BP1 (Santa Cruz Biotechnologies; sc-365338).
Statistical analysis
We performed statistics with Prism software 9.3.1 or with R (version 4.0.5) (42) apart from the hypergeometric tests of the intersections between SG-transcriptome and rG4 datasets, which were calculated online: http://nemates.org/MA/progs/overlap_stats.html. We used geom_density function of the ggplot2 package in R to analyze overlapping binned data of the SG-transcriptome and rG4 datasets. We used unpaired t-test or Mann-Whithney test for pairwise comparisons. We used Pearson's Chi-squared test with Yates’ continuity correction for the pairwise comparison of rG4-positive sequences fraction under stress and basal conditions. We analyzed multiple-group comparisons using one-way ANOVA with Bonferroni correction. For analysis of live-imaging experiments, we used repeated-measures two-way ANOVA test. We tested the normal distribution of the data (by histograms) and used the Levene test to compare variances between the treatments within the data. Statistical tests were considered significant if P-values or FDR corrected q-values ≤0.05. We show data as means ± SEM or SD or plotted using boxplots as noted in the text.
Figures’ design
We placed and organized all the figures by using Illustrator software. We generated all the plots by Prism software. We generated Figure 1A and Figure 6C by BioRender.com. The all raw gels for EMSA and WB experiments are found at Zenodo website (https://zenodo.org/record/7225796#.Y1AX7nZByUk).
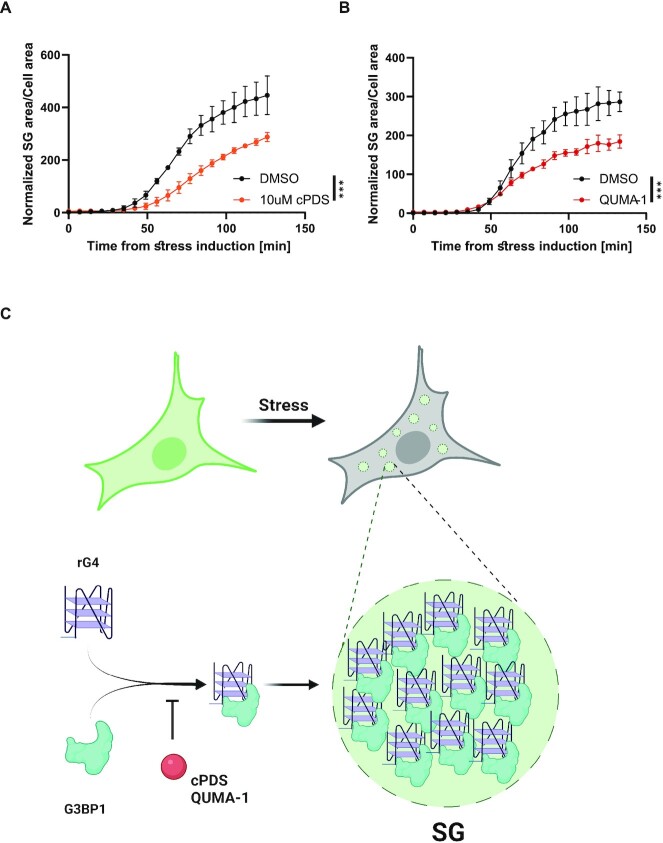
Figure 6.
rG4 availability regulates stress granule formation. Live imaging and quantification of SG formation by assessing G3BP1-GFP in U2OS cells incubated with (A) 1 uM QUMA-1 for 3 h or (B) 10 uM cPDS for 24 h prior to stress induction (150 uM of sodium arsenate). SG area/cell area normalized to DMSO treatment. Four sites per well, 3–4 wells per condition. *** P < 0.001, **** P < 0.0001, two-way ANOVA repeated measure. (C) A model for the regulatory role of rG4s in SG formation. rG4-G3BP1 interactions promote SG formation, while limited rG4 availability diminishes SG condensation.
RESULTS
rG4detector improves rG4 stability prediction compared to existing methods
We first trained and validated rG4detector (schematic architecture in Figure 1A) on the rG4-seq dataset from human transcripts (4) and compared rG4detector performance on a held-out subset of the data to G4Hunter (28), G4NN (8) and cGcC-scoring (27). To evaluate all methods in RSR-ratio prediction, we calculated the Pearson correlation coefficient of predicted and measured RSR-ratio scores on the held-out data. The Pearson correlation is an appropriate metric for skewed distributions, such as RSR-ratio scores where most values are centered around 0.5 (as this metric is based on capturing the variance in the data; Supplementary Figure S1B). The performance of rG4detector (r = 0.81; Supplementary Figure S2A) was superior to G4Hunter (r = 0.45), G4NN (r = 0.37) and cGcC-scoring (r = 0.51) (Figure 1B). Similarly, improved rG4 prediction performance was obtained on the mouse (34) and plant transcriptomes (35), albeit with worse prediction performance of all methods compared to the human transcriptome (Supplementary Figure S2B).
rG4detector outperforms existing methods in rG4 detection
Next, we assessed the ability of rG4detector to identify rG4FSs in discrete transcripts. To this end, we retrieved all transcripts from the held-out test set (chromosome 1) containing an rG4 detected by rG4-seq. For each transcript, we calculated the Gaussian-weighted average of predictions for each nucleic-acid position in an 80nt-long window. The precision–recall curves confirm that the prediction performance of rG4detector is superior to G4Hunter, G4NN, and cGcC-scoring with an improvement in precision consistent over almost all recall values (area under the precision–recall curve (AUPR) of 0.42 versus 0.38, 0.28, and 0.19, respectively; Figure 1C).
We further used the area under the receiver operating characteristic curve (AUROC) metric to evaluate rG4detector classification performance in a binary rG4 classification task based on the G4RNA dataset (43). We padded all sequences with their genomic flanks, and predicted all overlapping windows of 80nt. we assigned the maximum value to be the final sequence prediction. We found rG4detector AUROC to be better (0.75) than existing methods (0.65, 0.63, and 0.73 for G4Hunter, G4NN, and cGcC-scoring, respectively; Figure 1D).
G4-folding principles learned by rG4detector
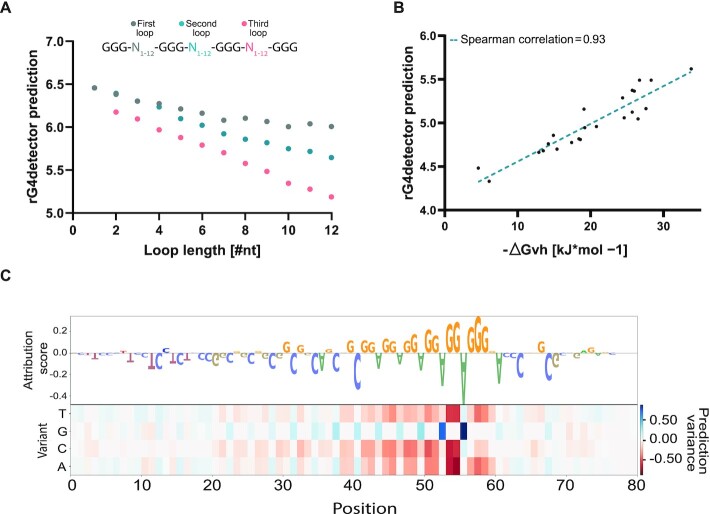
To validate that rG4detector learned meaningful biochemical principles, we defined canonical rG4FSs as GGGN1–12GGGN1–12GGGN1–12GGG (N for any intervening nucleotide with loop lengths between 1–12nt). We found that increased loop lengths decrease the predicted RSR-ratio score (Figure 2A). The most substantial decrease was observed for the third loop, at the 3' end, while the least substantial effect was observed for the first loop, at the 5' end. This inverse association between loop length and rG4 stability is reassuring since it was previously observed in a low-throughput assay (36). In addition, we predicted multiple combinations of loop lengths as was previously experimentally tested in (36). rG4detector predicted the sequence's stability with high Spearman correlation of 0.93 (Figure 2B).
Figure 2.
rG4detector identifies key sequence features to predict rG4 propensity. (A) Loop length inversely correlates with rG4detector prediction on canonical rG4 (GGGNGGGNGGGNGGG) sequences with variable loop lengths. (B) rG4detector prediction correlates with experimentally tested thermodynamic stability of rG4s with variable loop lengths. (C) Heatmap of rG4detector sensitivity to mutations and the corresponding attribution logo, which visualizes nucleotide importance to the model's prediction for the sequence r(AGG)5.
We next examined the effect of G-tract length on rG4 stability. By varying the G-tract length between 1 and 4 over multiple loop lengths, we observed an increase in rG4 stability predictions with G4-tract length (Supplementary Figure S3A). This observation is in agreement with previous studies, which confirmed that rG4s with three or four G-quartets are more stable than those with only two G-quartets (44,45). For longer G-tract length, we observed variable trends in rG4 stability predictions depending on the loop length. Shorter loops led to a decrease in rG4 stability prediction with G-tract length, while longer loops led to an increase. We speculate that this observation is due to the fact that increasing the G-tracts length in sequences with longer loops increases the number of potential combinations of G-quartets compared to sequences with shorter loops. To conclude, rG4detector learned the known rG4 stability dependency on G-tract length up to length 4, and discovered novel dependencies for longer G-tracts.
Lastly, we used a mutation map and sequence logo to visualize the key features identified by rG4detector in a given sequence. The attribution scores indicate that rG4detector assigns high importance to G-rich sequences (Figure 2C). According to the model, the relevant guanines reside in continuous stretches, while cytosines, especially in the loops within the rG4FSs, are disfavored for rG4 propensity. The highest attribution scores are assigned to G in the 3' end of the sequence hinting on a possible edge effect in the data (Supplementary Figure S3B).
rG4detector reveals increased enrichment of potential rG4-containing sequences bound by G3BP1 under stress
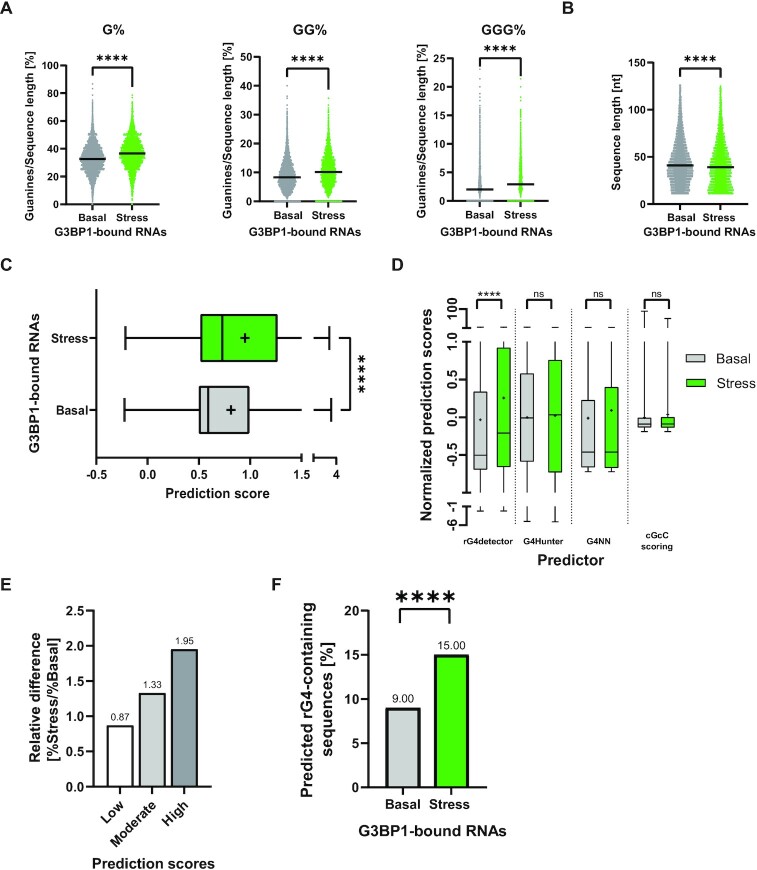
To demonstrate the value of rG4detector, we used it to generate new biological hypotheses. G3BP1, a central protein in the stress response (46,47), has been recently identified as an rG4-binding protein (24,25). Therefore, we hypothesized that G3BP1 displays stress-dependent preference towards binding of rG4s. We tested this hypothesis on a G3BP1-bound RNA data from an enhanced crosslinking and immuno-precipitation (eCLIP) study of Markmiller et al. (48). We discovered a significantly higher percentage of G-rich sequences under stress conditions relative to basal (non-stressed) conditions (G%: 36.84 versus 32.81, GG%: 10.49 versus 8.73 and GGG%: 3.33 versus 2.44, respectively; Figure 3A and Supplementary data S1, Tables S4 and S5). Then, for each G3BP1-bound RNA sequence, we retrieved the sub-sequence with the maximum rG4detector score over all 80nt-long sub-sequences. We tested if there is a length effect to rule out that longer sequences were biased toward higher predictions due to more predictions per sequence. Indeed, a slightly longer average length of G3BP1-bound RNAs under basal conditions was observed (Figure 3B and Supplementary data S1, Tables S4 and S5). However, the predicted propensity of RNAs to form rG4s was higher under stress compared to basal conditions (Figure 3C and Supplementary data S1, Table S6). Notably, rG4detector identifies a unique behavior (as average rG4 prediction scores) for G3BP1-bound RNAs under stress versus basal conditions, which was overlooked by existing prediction methods (Figure 3D and Supplementary data S1, Table S7). Altogether, rG4detector uncovered that under stress G3BP1 binds RNAs that are more likely to form rG4 structures.
Figure 3.
rG4detector reveals G3BP1 association with rG4-forming sequences is enhanced under stress. Analysis of G3BP1 eCLIP data from Markmiller et al. (48) reveals (A) stress-dependent enrichment in binding of single, double, or triple guanine-rich sequences by G3BP1 relative to basal conditions. (B) Comparison of the sequence length in basal and stress groups. (C) Comparison of the distribution of rG4 propensity scores under stress and basal conditions. (D) Distribution of predicted rG4 propensity scores under stress and basal conditions by rG4detector, G4Hunter, G4NN, and cGcC-scoring. Data standardized to obtain a mean of zero and standard deviation of 1. Line – median, cross – mean. (E) Categorical rG4 propensity scores to bins by RSR-ratio values: ‘low’ (0 to 1), ‘moderate’ (1 to 2) and ‘high’ (2+). (F) The fraction of rG4-containing sequences under stress and basal conditions. Pairwise Mann–Whitney test ****P < 0.0001, ns – non significant. (A–D); Pearson's chi-squared test with Yates’ continuity correction (F).
To further understand the difference in rG4FS prevalence between the two conditions, we binned G3BP1-bound RNAs into three categories of ‘low’ (0–1), ‘moderate’ (1–2) and ‘high’ (2+) predicted RSR-ratio scores. Intriguingly, ‘moderate’ and ‘high’ bins are more prevalent under stress conditions compared to basal conditions (relative difference of 1.33 and 1.95 folds, respectively), supporting that G3BP1-bound RNAs under stress are more likely to form rG4 structures (Figure 3E and Supplementary data S1, Table S6). Finally, we classified G3BP1-bound RNAs as rG4FSs and non-rG4FSs based on a calculated threshold (threshold of 1.56; Supplementary Figure S4). We revealed a higher percentage of rG4FSs under stress conditions compared to basal conditions (15% compared to 9%; Figure 3F and Supplementary data S1, Table S6). Consistently, similar analyses on and comparisons between basal and stress conditions with non-overlapping G3BP1-bound RNAs, which were uniquely found in either stress or basal conditions, provided similar results (Supplementary Figure S5 and Supplementary data S1, Tables S8–S11). Together, rG4detector revealed a broad and stress-sensitive binding of rG4FSs by G3BP1.
G3BP1 binds to its endogenous binding motif r(AGG)5 in a competitive manner
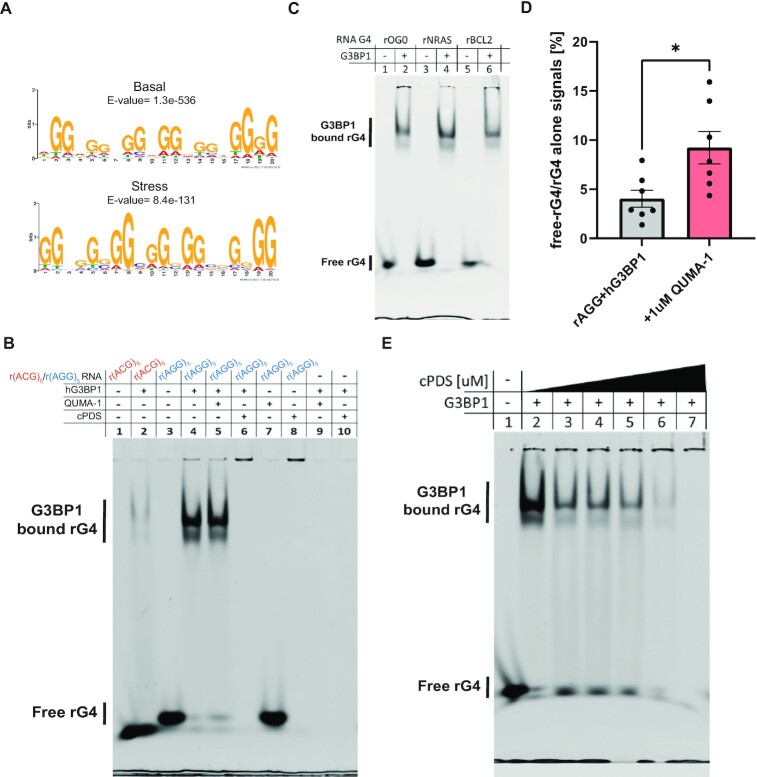
By using MEME analysis (49) to study G3BP1-bound RNAs (48), we found that r(AGG)5 is the most probable motif for the sequences (with only slight differences between the conditions: AGGAGGAGG AGGAGGTGGGG, E-value = 1.3e–536 (basal) or GGAGGAGGAGGTGGATGAGG, E-value = 8.4e–131 (stress); Figure 4A). This observation is in line with a recent report (25), but it also implies that r(AGG)5 RNA is prone to fold into an rG4 structure, which was not directly tested so far.
Figure 4.
G3BP1 interaction with endogenous rG4-forming sequences depends on their availability. (A) MEME analysis of G3BP1 eCLIP data under both basal (top logo) and stress (bottom logo) conditions (48) reveals potential rG4-forming motifs, which share the G3BP1-bound sequence r(AGG)5. (B) EMSA of rG4-forming sequence r(AGG)5 or mutated form r(ACG)5 bound to recombinant hG3BP1 without or with rG4-binders QUMA-1 (1 uM) or cPDS (10 uM). (C) EMSA of a variety of rG4-forming sequences reveal binding to recombinant hG3BP1. rG4-forming oligos are rOG0 (24), rNRAS (25), and rBCL2 (24). (D) Quantification of free rG4 without or with QUMA-1. *P < 0.05, unpaired two-tailed t-test. (E) EMSA of r(AGG)5 bound to recombinant hG3BP1 with different concentrations of cPDS (0, 0.1, 0.5, 1, 5, 10 uM). EMSA were in 5% native PAGE. The binding reaction contains 160 nM 6FAM-labeled oligo without or with 1.25 ug recombinant hG3BP1 per lane. RNA is labeled with 6FAM and detected at a wavelength of 488 nm. ≥3 experimental repeats.
We demonstrated that r(AGG)5 RNA is folding into a parallel rG4 structure in vitro by a CD assay and by measuring TDS profiles (Supplementary Figure S6) of the sequence with KCl or LiCl buffer or without addition of cations. Next, via an EMSA, we demonstrated in vitro that a human G3BP1 recombinant protein (hG3BP1) is able to bind to the rG4-r(AGG)5 but negligibly binds to its ‘mutated’ version (r(ACG)5), which does not form an rG4 structure (Figure 4B and Supplementary Figures S7A and S8). To substantiate the evidence that hG3BP1 binds rG4s, we validated through EMSA experiments that hG3BP1 binds to additional hG3BP1-bound rG4FSs, which were previously reported by others: rOG0, rNRAS, and rBCL2 (24,25) (Figure 4C and Supplementary Figures S7B–D). Therefore, hG3BP1 is an rG4-binding protein with preference for binding rG4-(AGG)5 motifs.
The rG4-binding molecules QUMA-1 (50) and carboxy pyridostatin (cPDS) (51) are competitive binders of rG4 sequences (52). We sought to test if QUMA-1 or cPDS can compete with the binding of G3BP1 to rG4-r(AGG)5. We performed EMSA experiments of r(AGG)5 and hG3BP1 without or with QUMA-1 or cPDS. The binding of QUMA1 or cPDS to the rG4 inhibited hG3BP1 binding to rG4-(AGG)5 (Figure 4B, D–E). The competition by cPDS delays the rG4-r(AGG)5 electrophoresis although the migration of hG3BP1 was not affected (Figure 4B and Supplementary Figure S8). To substantiate the evidence for cPDS-dependent competition, we further performed titrations with different cPDS concentrations, which resulted in progresive reduction in the amount of hG3BP1-bound rG4-(AGG)5 (Figure 4E). The effect of cPDS on the hG3BP1-rG4 complex abundance is consistent with data about rG4-OG0 (Supplementary Figure S9) and supports the interpretation that cPDS competes for available rG4-(AGG)5. The different rG4 sequestration patterns observed can be explained by different molecular size of cPDS (706.71 g/mol) and QUMA-1 (628.17 g/mol) or potential aggregation of cPDS with rG4s that may inhibited its migration through the elephoetic field.
To exclude the possibility that QUMA-1 or cPDS interact with hG3BP1 directly, or affect the protein's electrophoresis, we performed additional EMSA experiments. Incubating QUMA-1 or cPDS with rG4-r(AGG)5 alone, without hG3BP1, resulted in a band mobility similar to the one which includes hG3BP1 (Figure 4B, Supplementary Figure S8). Furthermore, the hG3BP1 band migrates to the same level through the gel suggesting that it is not retarded by biding to QUMA-1 or to cPDS (Supplementary Figure S8). These results demonstrate that cPDS and QUMA-1 bind to the rG4 independently of hG3BP1 and do not influence hG3BP1’s gel mobility. We conclude that small rG4-binding molecules sequester rG4s. By doing so, they affect rG4 availability and binding to G3BP1 and perhaps other rG4BPs.
SGs are colocalized with endogenous rG4s that regulate their formation
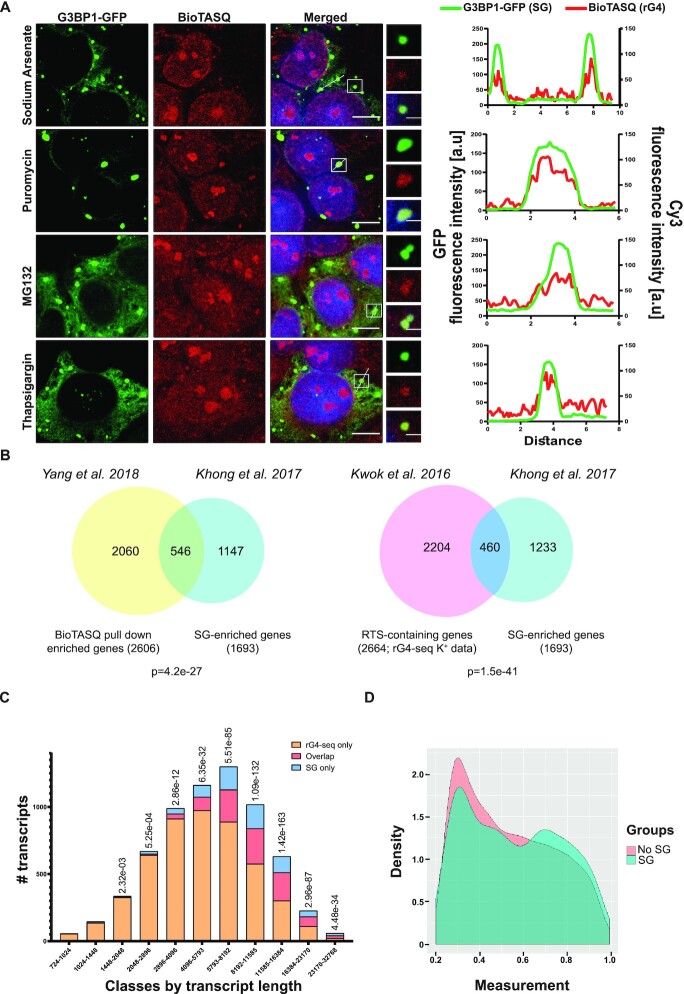
Because of the impact of rG4 ligands on G3BP1 in vitro, we hypothesized that small rG4-binding molecules might impact G3BP1-dependent SG formation. To better understand rG4 roles in SG biology, we visualized endogenous rG4s in U2OS cells using the biotinylated small rG4-binding molecule BioTASQ (5,53). Under basal (no-stress) conditions, the BioTASQ and G3BP1-GFP signals were dispersed. However, BioTASQ fluorescence was enhanced within cytoplasmic condensation compared to surrounding cytoplasmic signal under a variety of stress conditions (Figure 5A and Supplementary Figure S10A). We verified that these cytoplasmic condensates are genuine SGs by the expression of a G3BP1-GFP reporter that enabled straightforward rG4/G3BP1 colocalization. This observation was confirmed by similar experiments performed with QUMA-1 (Supplementary Figure S10B). Hence, endogenous rG4s are enriched in SGs regardless of the stress type.
Figure 5.
Enrichment of endogenous rG4s in stress granules. (A) Confocal micrographs of bioTASQ-detected rG4 enrichment in stress granules of U2OS cells under a variety of stressors. rG4 (Cy3, red), SGs (G3BP1-GFP, green), nuclei (DAPI, blue). ×63 lens. Scale bar: 10 um. Inset scale bar: 2 um. Intensity profiles for colocalization analysis between SGs (GFP; green) and rG4s (BioTASQ; red) were quantified for representative granule(s) in each of the stress conditions using the FiJi software. (B) Venn diagrams of rG4-containing transcripts (Kwok et al. 2016; 2664 transcripts (4) or Yang et al. 2018; 2060 transcripts (5)), intersected with 1693 SG-enriched transcripts (Khong et al. 2017 (54)). 460 or 546 transcripts are shared significantly more than is expected at random (hypergeometric test, given 12 300, 11 944 and 15 340 sequenced RNAs, respectively). (C) Bar plot of overlap between SG-enriched and rG4-containing transcripts based on rG4-seq data in discrete transcript length bins. Overlapping SG-enriched transcripts with rG4-containing transcripts (red), rG4-containing transcripts, which are not enriched in SG (orange), and SG transcripts, which do not contain rG4s (blue). We removed less than 0.1% of the data to improve figure clarity. (D) The distribution of reverse transcriptase stalling (RTS) values (measured from rG4-seq data) of stress granule-associated transcripts (blue), or transcripts that are not associated with stress granules (pink), normalized using kernel density estimation statistics in R. Data for analysis in (C, D) from Khong et al. 2017 (54) and Kwok et al. 2016 (4).
To evaluate the prevalence of rG4-containing transcripts in the SG-transcriptome, we next compared reported rG4-containing transcripts (4,5) and SG-associated transcripts (54). We found that rG4-containing transcripts from both G4RP-seq (5) and rG4-seq (4) datasets are significantly enriched in SGs, compared to transcripts that were not associated with SGs (460 or 546 mRNAs, respectively, among 1693 SG transcripts, Figure 5B). Although the enrichment over the rG4-seq dataset in the SG transcriptome is statistically significant, we also found that the enrichment is length dependent. The observed bias for longer transcript length (>1400nt) is reasonable because SG transcriptome is associated with longer transcripts (54). Furthermore, the degree of association varies with transcript length, but the maximum is not at the longest transcripts suggesting that other, unknown, parameters come into play (Figure 5C). Ranking of SG-associated transcripts (54) according to their reported RTS score (4), indicated higher rG4 potential relative to transcripts that were not associated with SGs (Figure 5D). Finally, to investigate endogenous rG4 roles in SG formation, we incubated U2OS cells with small rG4-binding molecules, cPDS (51) or QUMA-1 (50). Live-cell imaging revealed that SG formation was hindered by the presence of small rG4-binding molecules (in concentrations of 10 uM for 24 h or 1 uM for 3 h, respectively. Figure 6A, B). As indicated by our in vitro experiments (Figure 4C, D), this probably reflects sequestration of rG4s from SGs. Importantly, none of the small rG4-binding molecules we used induced phosphorylation of eIF2 alpha, indicating that they do not affect the cellular stress response at the concentrations we used (Supplementary Figure S11). Altogether, we conclude that rG4s are enriched in SGs and regulate SG formation (Figure 6C).
DISCUSSION
Over the past years, a series of computational methods have been developed to assess the transcriptome-wide prevalence of rG4FSs (7). A significant leap has been recently taken with the introduction of machine-learning-based models, but this emerging trend needed to be strengthened upon training on NGS-based high-throughput datasets, which are now available.
Here, we developed a new machine-learning model named rG4detector, which is based on a convolutional neural network and is trained on rG4-seq data. rG4detector displays improved performance relative to existing rG4 predictors and can predict rG4 stability. In addition, as rG4detector model input is the RNA sequence one-hot-encoded matrix, no feature engineering is required, and as a result biased assumptions on rG4 formation factors are minimized.
Although rG4detector outperforms existing methods in predicting RTS measurements in three different species, the correlation of rG4detector predictions on the mouse and plant transcriptomes were substantially lower than in the human transcriptome (Figure 1B and Supplementary Figure S2). We assume that this is due to the use of different calculations for RTS experimental scores. Extracting the RSR-ratio scores from the raw data of the mouse and plant experiments will provide a more accurate comparison between rG4detector predictions and the RTS measurements of these different datasets.
However, rG4detector is not bias-free. A primary example is the higher sensitivity for positions near the 3’ end of rG4FSs compared to the 5’ end, which is likely a result of the RTS sites that occur at the 3’ end of the sequences (due to the processivity of the enzyme). Consequently, RSR-ratio predictions are biased toward the 3’ end. This issue may be addressed by training rG4detector on a combination of rG4-seq with newer datasets, which are produced by alternative NGS-based approaches, such as DMS-seq over the human transcriptome (10) that would diminish the 3’-end biases. In addition, training rG4detector on upcoming high-throughput datasets, such as rG4-seq 2.0 (55), can improve prediction performance. Moreover, rG4detector paves the way to future deep-learning models for predicting complex types of RNA structures, including intermolecular and DNA/RNA hybrid G4s, which currently remain unexplored.
By applying rG4detector to eCLIP data of G3BP1-bound RNAs under stress versus basal conditions, which demonstrated a dynamic emergence of sequences bound to G3BP1 between the conditions (48), we unexpectedly found that G3BP1-bound RNAs contain more potential rG4s under stress compared to basal conditions. This suggests that under stress G3BP1 binds RNAs that are more likely to form rG4 structures. Nevertheless, MEME analysis found a highly similar binding motif of G3BP1, r(AGG)5, under both basal and stress conditions. We reason that the differences in G3BP1 binding preferences are too subtle for MEME to detect and thus are not reflected in the most probable binding motif. From a molecular and biochemical perspective, it is likely that the linear consensus sequence remains similar, but more RNAs are prone to form rG4 structures under stress compared to a basal condition as recently observed by a preprint from the Ivanov lab (10).
We confirmed rG4-G3BP1 interaction in vitro, including with its endogenous binding motif r(AGG)5, demonstrating that endogenous rG4s are enriched in SGs and that RBP-rG4 interactions are necessary for SG formation. Our results along with others’ (10) contribute to the emerging focus on rG4 and stress biology. rG4s might potentially promote SG formation via anchoring RBPs and through inter- and intra-molecular RNA–RNA interactions (56–60). In this context, past studies suggested that G3BP1 displays preference to unfolded double-stranded RNAs (61,62), but our results and others’ (25) show that G3BP1 elicits high affinity for rG4 motifs. A way to reconcile these conclusions was proposed recently by suggesting that unstructured and structured RNAs serve different functions in condensations (63).
The G3BP1-binding motif r(AGG)5 is not a common and conventional sequence for forming rG4s (4) suggesting that there is room for additional structural and biochemical studies of non-canonical rG4s. As rG4s are also known to be involved with regulation of paraspeckles (64), research about rG4s in biomolecular condensates may be an emerging field of interest. Finally, rG4-prediction methods, such as our novel rG4detector, can pave the way to robust unbiased research about rG4 regulation and further advance this field.
DATA AVAILABILITY
rG4detector code, trained models, and processed datasets are publicly available via https://github.com/OrensteinLab/rG4detector.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Shira Albeck and Tamar Unger (Weizmann Institute of Science), as well as Shira Roth (Bar-Ilan University) for technical advice. We thank Dr Yosef Scolnik for CD training. We thank members of Orenstein, Hornstein and Monchaud groups for helpful critiques, discussions, advice and protocols.
Contributor Information
Maor Turner, School of Electrical and Computer Engineering, Ben-Gurion University of the Negev, Be’er-Sheva 8410501, Israel.
Yehuda M Danino, Department of Molecular Genetics, Weizmann Institute of Science, Rehovot 7610001, Israel; Department of Molecular Neuroscience, Weizmann Institute of Science, Rehovot 7610001, Israel.
Mira Barshai, School of Electrical and Computer Engineering, Ben-Gurion University of the Negev, Be’er-Sheva 8410501, Israel.
Nancy S Yacovzada, Department of Molecular Genetics, Weizmann Institute of Science, Rehovot 7610001, Israel; Department of Molecular Neuroscience, Weizmann Institute of Science, Rehovot 7610001, Israel.
Yahel Cohen, Department of Molecular Genetics, Weizmann Institute of Science, Rehovot 7610001, Israel; Department of Molecular Neuroscience, Weizmann Institute of Science, Rehovot 7610001, Israel.
Tsviya Olender, Department of Molecular Genetics, Weizmann Institute of Science, Rehovot 7610001, Israel.
Ron Rotkopf, Bioinformatics Unit, Life Sciences Core Facilities, Weizmann Institute of Science, Rehovot 7610001, Israel.
David Monchaud, Institut de Chimie Moleculaire, ICMUB CNRS UMR 6302, UBFC Dijon, France.
Eran Hornstein, Department of Molecular Genetics, Weizmann Institute of Science, Rehovot 7610001, Israel; Department of Molecular Neuroscience, Weizmann Institute of Science, Rehovot 7610001, Israel.
Yaron Orenstein, School of Electrical and Computer Engineering, Ben-Gurion University of the Negev, Be’er-Sheva 8410501, Israel; Department of Computer Science, Bar-Ilan University, Ramat-Gan 5290002, Israel; The Mina & Everard Goodman Faculty of Life Sciences, Bar-Ilan University, Ramat-Gan 5290002, Israel.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research in the Orenstein lab was supported by the Israel Cancer Association [20221519]; Israel Science Foundation [358/21]; Israeli Council for Higher Education (CHE) via Data Science Research Center, Ben-Gurion University of the Negev, Israel; E.H. is the Mondry Family Professorial Chair at Weizmann Institute of Science; Research in the Hornstein lab is supported by Andrea L. and Lawrence A. Wolfe Family Center for Research on Neuroimmunology and Neuromodulation; CReATe consortium and ALSA (program: ‘Prognostic Value of miRNAs in Biofluids From ALS Patients’); RADALA Foundation; AFM Telethon (20576); Weizmann–Brazil Center for Research on Neurodegeneration at Weizmann Institute of Science; Minerva Foundation, with funding from the Federal German Ministry for Education and Research; ISF Legacy Heritage Fund [828/17]; Israel Science Foundation [135/16, 3497/21, 424/22, 425/22]; Target ALS [118945]; Thierry Latran Foundation for ALS Research; European Research Council under the European Union's Seventh Framework Program [[FP7/2007–2013]/ERC grant agreement number 617351]; United States–Israel Binational Science Foundation [2021181]; ERA-Net for Research Programs on Rare Diseases [eRARE FP7] via the Israel Ministry of Health; Dr Sydney Brenner and friends; Edward and Janie Moravitz; A. Alfred Taubman through IsrALS; Yeda-Sela; Yeda-CEO; Israel Ministry of Trade and Industry; Y. Leon Benoziyo Institute for Molecular Medicine; the Nella and Leon Benoziyo Center for Neurological Diseases; Kekst Family Institute for Medical Genetics; David and Fela Shapell Family Center for Genetic Disorders Research; Crown Human Genome Center; Nathan, Shirley, Philip and Charlene Vener New Scientist Fund; Julius and Ray Charlestein Foundation; Fraida Foundation; the Wolfson Family Charitable Trust; Adelis Foundation; Merck (UK); M. Halphen; estates of F. Sherr, L. Asseof and L. Fulop; Goldhirsh-Yellin Foundation; Redhill Foundation–Sam and Jean Rothberg Charitable Trust; Dr Dvora and Haim Teitelbaum Endowment Fund; Anita James Rosen Foundation; Y.M.D. is funded by a fellowship from CNRS-WIS center for research of RNA secondary structures. Funding for open access charge: Same as funding grants.
Conflict of interest statement. None declared.
REFERENCES
- 1. Fay M.M., Lyons S.M., Ivanov P.. RNA G-quadruplexes in biology: principles and molecular mechanisms. J. Mol. Biol. 2017; 429:2127–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kwok C.K., Marsico G., Balasubramanian S.. Detecting RNA G-quadruplexes (rG4s) in the transcriptome. Cold Spring Harb. Perspect. Biol. 2018; 10:a032284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Varshney D., Spiegel J., Zyner K., Tannahill D., Balasubramanian S.. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020; 21:459–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kwok C.K., Marsico G., Sahakyan A.B., Chambers V.S., Balasubramanian S.. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods. 2016; 13:841–844. [DOI] [PubMed] [Google Scholar]
- 5. Yang S.Y., Lejault P., Chevrier S., Boidot R., Gordon Robertson A., Wong J.M.Y., Monchaud D.. Transcriptome-wide identification of transient RNA G-quadruplexes in human cells. Nat. Commun. 2018; 9:4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang S.Y., Monchaud D., Wong J.M.Y.. Global mapping of RNA G-quadruplexes (G4-RNAs) using G4RP-seq. Nat. Protoc. 2022; 17:870–889. [DOI] [PubMed] [Google Scholar]
- 7. Puig Lombardi E., Londoño-Vallejo A.. A guide to computational methods for G-quadruplex prediction. Nucleic Acids Res. 2020; 48:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garant J.-M., Perreault J.-P., Scott M.S.. Motif independent identification of potential RNA G-quadruplexes by G4RNA screener. Bioinformatics. 2017; 33:3532–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dumas L., Herviou P., Dassi E., Cammas A., Millevoi S.. G-Quadruplexes in RNA biology: recent advances and future directions. Trends Biochem. Sci. 2021; 46:270–283. [DOI] [PubMed] [Google Scholar]
- 10. Kharel P., Fay M., Manasova E.V., Anderson P.J., Kurkin A.V., Guo J.U., Ivanov P.. Stress promotes RNA G-quadruplex folding in human cells. 2022; bioRxiv doi:03 March 2022, preprint: not peer reviewed 10.1101/2022.03.03.482884. [DOI] [PMC free article] [PubMed]
- 11. Alberti S., Carra S.. Quality control of membraneless organelles. J. Mol. Biol. 2018; 430:4711–4729. [DOI] [PubMed] [Google Scholar]
- 12. Protter D.S.W., Parker R.. Principles and properties of stress granules. Trends Cell Biol. 2016; 26:668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mahboubi H., Stochaj U.. Cytoplasmic stress granules: dynamic modulators of cell signaling and disease. Biochim. Biophys. Acta (BBA) - Mol. Basis Di. 2017; 1863:884–895. [DOI] [PubMed] [Google Scholar]
- 14. Kedersha N., Ivanov P., Anderson P.. Stress granules and cell signaling: more than just a passing phase?. Trends Biochem. Sci. 2013; 38:494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ivanov P., Kedersha N., Anderson P.. Stress granules and processing bodies in translational control. Cold Spring Harb. Perspect. Biol. 2019; 11:a032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anderson P., Kedersha N.. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 2009; 10:430–436. [DOI] [PubMed] [Google Scholar]
- 17. Lee K.-H., Zhang P., Kim H.J., Mitrea D.M., Sarkar M., Freibaum B.D., Cika J., Coughlin M., Messing J., Molliex A.et al.. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016; 167:774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maharjan N., Künzli C., Buthey K., Saxena S.. C9ORF72 regulates stress granule formation and its deficiency impairs stress granule assembly, hypersensitizing cells to stress. Mol. Neurobiol. 2017; 54:3062–3077. [DOI] [PubMed] [Google Scholar]
- 19. Boeynaems S., Bogaert E., Kovacs D., Konijnenberg A., Timmerman E., Volkov A., Guharoy M., De Decker M., Jaspers T., Ryan V.H.et al.. Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell. 2017; 65:1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chew J., Cook C., Gendron T.F., Jansen-West K., Del Rosso G., Daughrity L.M., Castanedes-Casey M., Kurti A., Stankowski J.N., Disney M.D.et al.. Aberrant deposition of stress granule-resident proteins linked to C9orf72-associated TDP-43 proteinopathy. Mol. Neurodegener. 2019; 14:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Y.R., King O.D., Shorter J., Gitler A.D.. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 2013; 201:361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fay M.M., Anderson P.J., Ivanov P.. ALS/FTD-Associated C9ORF72 repeat RNA promotes phase transitions in vitro and in cells. Cell Rep. 2017; 21:3573–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sauer M., Juranek S.A., Marks J., De Magis A., Kazemier H.G., Hilbig D., Benhalevy D., Wang X., Hafner M., Paeschke K.. DHX36 prevents the accumulation of translationally inactive mRNAs with G4-structures in untranslated regions. Nat. Commun. 2019; 10:2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Su H., Xu J., Chen Y., Wang Q., Lu Z., Chen Y., Chen K., Han S., Fang Z., Wang P.et al.. Photoactive G-Quadruplex ligand identifies multiple G-Quadruplex-Related proteins with extensive sequence tolerance in the cellular environment. J. Am. Chem. Soc. 2021; 143:1917–1923. [DOI] [PubMed] [Google Scholar]
- 25. He X., Yuan J., Wang Y.. G3BP1 binds to guanine quadruplexes in mRNAs to modulate their stabilities. Nucleic Acids Res. 2021; 49:11323–11336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kikin O., D’Antonio L., Bagga P.S. QGRS mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006; 34:W676–W82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beaudoin J.-D., Jodoin R., Perreault J.-P.. New scoring system to identify RNA G-quadruplex folding. Nucleic Acids Res. 2014; 42:1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bedrat A., Lacroix L., Mergny J.-L.. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016; 44:1746–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rodriguez R., Müller S., Yeoman J.A., Trentesaux C., Riou J.-F., Balasubramanian S.. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J. Am. Chem. Soc. 2008; 130:15758–15759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chow E.Y.-C., Lyu K., Kwok C.K., Chan T.-F.. rG4-seeker enables high-confidence identification of novel and non-canonical rG4 motifs from rG4-seq experiments. RNA Biol. 2020; 17:903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:10–12. [Google Scholar]
- 32. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. 1000 Genome Project Data Processing Subgroup Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guo J.U., Bartel D.P.. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016; 353:aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang X., Cheema J., Zhang Y., Deng H., Duncan S., Umar M.I., Zhao J., Liu Q., Cao X., Kwok C.K.et al.. RNA G-quadruplex structures exist and function in vivo in plants. Genome Biol. 2020; 21:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang A.Y.Q., Bugaut A., Balasubramanian S.. A sequence-independent analysis of the loop length dependence of intramolecular RNA G-quadruplex stability and topology. Biochemistry. 2011; 50:7251–7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Uhl M., Tran V.D., Backofen R.. Improving CLIP-seq data analysis by incorporating transcript information. BMC Genomics. 2020; 21:894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sahakyan A.B., Chambers V.S., Marsico G., Santner T., Di Antonio M., Balasubramanian S.. Machine learning model for sequence-driven DNA G-quadruplex formation. Sci. Rep. 2017; 7:14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Q., Zhu L., Huang D.-S.. High-Order convolutional neural network architecture for predicting DNA-Protein binding sites. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019; 16:1184–1192. [DOI] [PubMed] [Google Scholar]
- 40. Sundararajan M., Taly A., Yan Q.. Precup D., Teh Y.W.. Axiomatic attribution for deep networks. Proceedings of the 34th International Conference on Machine Learning, Proceedings of Machine Learning Research. 2017; 70:PMLR; 3319–3328. [Google Scholar]
- 41. Tareen A., Kinney J.B.. Logomaker: beautiful sequence logos in python. Bioinformatics. 2020; 36:2272–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Team R.C. R development core team. RA Lang. Environ. Stat. Comput. 2013; 55:275–286. [Google Scholar]
- 43. Garant J.-M., Luce M.J., Scott M.S., Perreault J.-P.. G4RNA: an RNA G-quadruplex database. Database. 2015; 2015:bav059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pandey S., Agarwala P., Maiti S.. Effect of loops and G-quartets on the stability of RNA G-quadruplexes. J. Phys. Chem. B. 2013; 117:6896–6905. [DOI] [PubMed] [Google Scholar]
- 45. Matsumoto S., Tateishi-Karimata H., Takahashi S., Ohyama T., Sugimoto N.. Effect of molecular crowding on the stability of RNA G-Quadruplexes with various numbers of quartets and lengths of loops. Biochemistry. 2020; 59:2640–2649. [DOI] [PubMed] [Google Scholar]
- 46. Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., Tazi J.. The rasgap-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003; 160:823–831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47. Ge Y., Jin J., Li J., Ye M., Jin X.. The roles of G3BP1 in human diseases. Gene. 2022; 821:146294. [DOI] [PubMed] [Google Scholar]
- 48. Markmiller S., Sathe S., Server K.L., Nguyen T.B., Fulzele A., Cody N., Javaherian A., Broski S., Finkbeiner S., Bennett E.J.et al.. Persistent mRNA localization defects and cell death in ALS neurons caused by transient cellular stress. Cell Rep. 2021; 36:109685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bailey T.L., Elkan C.. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994; 2:28–36. [PubMed] [Google Scholar]
- 50. Chen X.-C., Chen S.-B., Dai J., Yuan J.-H., Ou T.-M., Huang Z.-S., Tan J.-H.. Tracking the dynamic folding and unfolding of RNA G-quadruplexes in live cells. Angew. Chem. Weinheim Bergstr. Ger. 2018; 130:4792–4796. [DOI] [PubMed] [Google Scholar]
- 51. Biffi G., Di Antonio M., Tannahill D., Balasubramanian S.. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014; 6:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Umar M.I., Kwok C.K.. Specific suppression of D-RNA G-quadruplex–protein interaction with an L-RNA aptamer. Nucleic Acids Res. 2020; 48:10125–10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Renard I., Grandmougin M., Roux A., Yang S.Y., Lejault P., Pirrotta M., Wong J.M.Y., Monchaud D. Small-molecule affinity capture of DNA/RNA quadruplexes and their identification in vitro and in vivo through the G4RP protocol. Nucleic Acids Res. 2019; 47:5502–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Khong A., Matheny T., Jain S., Mitchell S.F., Wheeler J.R., Parker R.. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell. 2017; 68:808–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhao J., Chow E.Y.-C., Yeung P.Y., Zhang Q.C., Chan T.-F., Kwok C.K.. rG4-seq 2.0: enhanced transcriptome-wide RNA G-quadruplex structure sequencing for low RNA input samples. 2022; bioRxiv doi:10 February 2022, preprint: not peer reviewed 10.1101/2022.02.10.479665. [DOI] [PMC free article] [PubMed]
- 56. Decker C.J., Burke J.M., Mulvaney P.K., Parker R.. RNA is required for the integrity of multiple nuclear and cytoplasmic membrane-less RNP granules. EMBO J. 2022; 41:e110137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Van Treeck B., Parker R.. Emerging roles for intermolecular RNA–RNA interactions in RNP assemblies. Cell. 2018; 174:791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Van Treeck B., Protter D.S.W., Matheny T., Khong A., Link C.D., Parker R.. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:2734–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tauber D., Tauber G., Khong A., Van Treeck B., Pelletier J., Parker R.. Modulation of RNA condensation by the DEAD-Box protein eIF4A. Cell. 2020; 180:411–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sanders D.W., Kedersha N., Lee D.S.W., Strom A.R., Drake V., Riback J.A., Bracha D., Eeftens J.M., Iwanicki A., Wang A.et al.. Competing Protein-RNA interaction networks control multiphase intracellular organization. Cell. 2020; 181:306–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang P., Mathieu C., Kolaitis R.-M., Zhang P., Messing J., Yurtsever U., Yang Z., Wu J., Li Y., Pan Q.et al.. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell. 2020; 181:325–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Guillén-Boixet J., Kopach A., Holehouse A.S., Wittmann S., Jahnel M., Schlüßler R., Kim K., Trussina I.R.E.A., Wang J., Mateju D.et al.. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell. 2020; 181:346–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mann J.R., Donnelly C.J.. RNA modulates physiological and neuropathological protein phase transitions. Neuron. 2021; 109:2663–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Simko E.A.J., Liu H., Zhang T., Velasquez A., Teli S., Haeusler A.R., Wang J.. G-quadruplexes offer a conserved structural motif for NONO recruitment to NEAT1 architectural lncRNA. Nucleic Acids Res. 2020; 48:7421–7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
rG4detector code, trained models, and processed datasets are publicly available via https://github.com/OrensteinLab/rG4detector.