Abstract
The trace metal iron (Fe) controls the diversity and activity of phytoplankton across the surface oceans, a paradigm established through decades of in situ and mesocosm experimental studies. Despite widespread Fe-limitation within high-nutrient, low chlorophyll (HNLC) waters, significant contributions of the cyanobacterium Synechococcus to the phytoplankton stock can be found. Correlations among differing strains of Synechococcus across different Fe-regimes have suggested the existence of Fe-adapted ecotypes. However, experimental evidence of high- versus low-Fe adapted strains of Synechococcus is lacking, and so we investigated the transcriptional responses of microbial communities inhabiting the HNLC, sub-Antarctic region of the Southern Ocean during the Spring of 2018. Analysis of metatranscriptomes generated from on-deck incubation experiments reflecting a gradient of Fe-availabilities reveal transcriptomic signatures indicative of co-occurring Synechococcus ecotypes adapted to differing Fe-regimes. Functional analyses comparing low-Fe and high-Fe conditions point to various Fe-acquisition mechanisms that may allow persistence of low-Fe adapted Synechococcus under Fe-limitation. Comparison of in situ surface conditions to the Fe-titrations indicate ecological relevance of these mechanisms as well as persistence of both putative ecotypes within this region. This Fe-titration approach, combined with transcriptomics, highlights the short-term responses of the in situ phytoplankton community to Fe-availability that are often overlooked by examining genomic content or bulk physiological responses alone. These findings expand our knowledge about how phytoplankton in HNLC Southern Ocean waters adapt and respond to changing Fe supply.
Subject terms: Water microbiology, Transcriptomics
Introduction
Nutrient availability exerts a fundamental influence on microbial communities inhabiting surface oceans, constraining both oceanic and atmospheric biogeochemical cycling [1]. Specifically, the trace metal iron (Fe) influences carbon fixation (via primary productivity) and carbon export to the deep ocean [2, 3]. Fe-limitation of phytoplankton has been studied for decades, resulting in the discovery of high nutrient, low chlorophyll (HNLC) regions that account for ~40% of the world’s surface oceans [4]. Most field studies capture the community-response of Fe addition on the physiology of phytoplankton such as diatoms: these observations form our foundation of knowledge for how Fe impacts photosynthetic organisms [5, 6]. Indeed, the discovery of low-Fe adapted diatoms provided insight into the ability of specific phytoplankton to cope with chronic Fe-stress. Specifically, it highlighted how they acquire Fe, decrease cellular Fe requirements [7], and substitute enzymes that require large amounts of Fe with more parsimonious ones [8–10]. However, there is evidence that prokaryotic phytoplankton, the cyanobacteria [11], also exhibit adaptations to Fe-limitation. Despite widespread Fe-limitation in the surface ocean, Prochlorococcus and Synechococcus are abundant and contribute significantly to global primary production [12, 13]. These picophytoplankton have adapted to nutrient-poor conditions through genomic streamlining [14] and by maintaining small cell sizes, which (for spherical cells) increases their surface-to-volume ratios [15]. Some cyanobacteria also alter their cellular and photosynthetic structures, lose genes encoding high Fe proteins [16] and in some cases may use sophisticated Fe acquisition mechanisms [17, 18] to adapt to Fe-limited conditions.
There is a rich history of research into factors that select for different phytoplankton species in aquatic systems [19]. Genomic analyses have pointed to Synechococcus (and Prochlorococcus) ecotypes that are correlated with regions of differing Fe availabilities [20–23]. However, when comparing genomes to distinguish low-Fe (HNLC) and high-Fe (coastal) ecotypes of Synechococcus, observations of genome-encoded functions specific to adaptations to differing Fe-regimes do not fully correspond to the respective niches they occupy [21]. The hypothesis that local conditions shape microbial communities at a more complex scale than simply high-Fe versus low-Fe needs to be investigated to generate accurate predictions of how oceanic microbial communities respond to nutrient limitation. Furthermore, given the contributions of cyanobacteria to global carbon cycles and the paucity of field-based studies investigating gene-level responses to Fe availability, it is important to disentangle how this group of phytoplankton respond to Fe availability. Most importantly, it is critical to test the response(s) of putative Fe-ecotypes beyond correlative observations.
The Southern Ocean Time Series (SOTS) is located in the Sub-Antarctic Zone (SAZ) [24, 25], a region of the global ocean that plays a critical role in CO2 uptake and its subsequent export to the deep ocean [26]. While primary productivity in this region is typically limited by Fe availability seasonally [2, 27], the region encounters periodic inputs of trace metals and these processes are expected to change with new climate trajectories [28]. Thus, SOTS represents an ideal location for the assessment of phytoplankton community response to changes in Fe availability.
We examined the response of phytoplankton to manipulated Fe availability—both increasing and decreasing - within short-term, on-deck bottle-incubations during a Spring (March 2018) expedition at SOTS. High-throughput sequencing of the total microbial mRNA pool (metatranscriptomics) was done for the entire microbial community but presented here with a focus on marine Synechococcus spp. We initially hypothesized that Fe additions would stimulate total community productivity, and that Fe removal would lead to an Fe-stress response by all microbial members. During an examination of Synechococcus spp. transcripts, we observed a more complex response, suggesting the presence of both high and low-Fe adapted strains residing within the SOTS surface waters. This study also establishes an experimental framework to employ metatranscriptomics to characterize the entire phytoplankton community across a gradient of Fe availabilities in a manner that allows for a clearer interpretation of in situ population-level physiology and dynamics.
Methods
Cruise details, sample collection and iron incubation
Iron amendments and in situ water column sampling were conducted at SOTS onboard the R/V Investigator during March 2018 using trace-metal-clean conditions [29]. In situ samples were collected prior to sunrise from 15 m depth using 12 L Niskin bottles on a CTD rosette at 47° 00’ 01.6“S, 142° 01’ 14.7“E (March 5th), 46° 59’ 52.6“S, 141°59'57.6“E (March 7th), and 47° 00’ 01.8“S, 142° 00’ 07.9“E (March 9th). Concentrations of nitrate (NO3), phosphate (PO4), silicate (Si), nitrite (NO2), and ammonium (NH4) were determined for unfiltered samples using a Seal AA3 segmented flow system following previous procedures [30]. Two Fe incubation experiments were performed: one on March 5th, 2018 (GRW1) and one on March 14th, 2018 (GRW2). Homogenized seawater collected from a trace-metal clean pump at 5 m was allocated into 2 L Nalgene™ bottles. For GRW1, either 0.25 nM, 0.5 nM, 1.0 nM, or 2.5 nM of either desferrioxamine-B (DFB) or Fe chloride (FeCl3) was added to chelate Fe or increase Fe, respectively. For GRW2, the procedure was repeated except bottles were amended with either 1.25 nM, 5.0 nM, or 12.5 nM DFB, or 2.5 nM FeCl3 as Fe-limitation conditions (based on in situ chlorophyll a fluorescence and Fv/Fm) appeared to wane [31]. An control was included for each experiment and T = 0 h sample was also collected (in situ, 5 m). Bottles were incubated at ~33% incident irradiance and ambient surface temperature (~11o C). After 72 h, bottles were sampled for chlorophyll a, photosynthetic health (Fv/Fm), and total RNA. See the Supplemental Material for further details.
RNA extraction, preprocessing, and sequencing
RNA was extracted with a phenol-chloroform based approach using a publicly available protocol [32]. Residual DNA was removed using the Turbo DNA-free™ kit (Ambion®). Libraries were prepared and sequenced by the DOE Joint Genome Institute (JGI) using the low-input protocol for total RNA. Briefly, ribosomal RNA was depleted using the Illumina® Ribo-Zero rRNA Removal Kit (Bacteria) and sequenced using paired-151 nt reads on the Illumina Novaseq S4 platform. Biological duplicates for all treatments were sequenced for incubations except for one control bottle from both GRW1 and GRW2 due to insufficient RNA.
Bioinformatic analyses
Raw read filtering and trimming was done using BBDuk v38.67 and BBMap v38.84 from the BBtools suite of packages [33]. Trimmed reads from all 18 incubation samples for GRW1, including the T = 0 h in situ sample, were co-assembled using MEGAHIT v1.2.9 [34]. Open reading frames (ORFs) were called using MetaGeneMark v3.38 [35] and reads mapped to the assembly with BBMap v38.84 were tabulated using featureCounts v2.0.0 [36]. For direct comparison of the in situ samples, these trimmed reads were also mapped to the GRW1 assembly. Read counts were normalized using the transcripts-per-million (TPM) approach [37]. Data visualization was performed using ggplot2 in the R statistical platform [38, 39]. The Pheatmap R-package was also used for hierarchical clustering and heatmap generation of functional genes [40]. Raw and processed data used in this study are publicly available through the JGI Data Portal (https://data.jgi.doe.gov) under the Project ID numbers 1260738 and 1260735.
Taxonomic composition was probed using the DNA-directed RNA polymerase beta subunit (Rpb1/RpoB) marker gene [41]. Briefly, GRW1 proteins were queried against a reference Rpb1/RpoB database using DIAMOND BLASTp [42]. Hits were clustered at 95% aa similarity using CD-HIT [43] and queried against the NCBI non-redundant database using DIAMOND BLASTp to retain only Rpb1/RpoB. Hits ≥1200 aa were aligned to reference Rpb1/RpoB proteins (Supplementary Table 1) in MEGA7 [44] using ClustalW [45]. For further exploration of cyanobacterial RpoB phylogenies, unclustered protein sequences were used to construct a separate tree containing all eukaryotic and prokaryotic references, with additional Synechococcus RpoB reference sequences (Supplementary Table 2). The separate maximum-likelihood trees were constructed in PhyML [46], and hits <1200 aa were placed on the tree using pplacer [47] and visualized/annotated using iTOL v.4 [48].
ORFs were functionally annotated using eggNOG-mapper v2.0.1 [49] and FeGenie [50]. Taxonomic annotation of ORFs was done using the “best LAST hit” procedure by the IMG pipeline [51, 52]. Only genes annotated as Synechococcus using this tool and confirmed to be of cyanobacterial origin via eggNOG-mapper’s Best Tax Level, were referred to as “Synechococcus-like transcripts”. To investigate the numerous hits (763) annotated as “oprB-like” porins (Supplementary Table 3), translated sequences were aligned separately to Slr1908 protein sequences identified as “Fe-uptake porins” in cyanobacteria [53] or the NCBI non-redundant database (downloaded January 2022) using DIAMOND BLASTp [42], with an e-value threshold of 1e-10 retaining 5 hits per query (Supplementary Table 4). The ferritin phylogenetic tree was constructed using the same approach outlined for RpoB/RPB1 using Synechococcus isolate ferritin sequences downloaded from NCBI (June 2021). Putative Synechococcus ferritin sequences >190 aa were used within the base tree (Supplementary Table 5).
Competitive read recruitment to two representative Synechococcus genomes was done to expand upon the observations from metatranscriptome assemblies, and to better resolve differences in transcript abundance patterns that may be driven by differences in gene copy numbers. Coding sequences from sequenced genomes of Synechococcus sp. CC9311 (CP000435.1) and Synechococcus sp. BL107 (NZ_DS022298.1) were downloaded from NCBI on June 12th, 2021, chosen based on high similarity of rpoB and ferritin-like sequences within the assembly (Supplementary Figs. 2 and 5) representing isolates from divergent trophic environments (coastal vs open-ocean). Trimmed reads from the Fe-incubations and in situ metatranscriptomes were mapped to both genomes simultaneously with 85% similarity fraction over 90% read length in CLC Genomics Workbench v.21.0.4 (Qiagen), retaining non-redundantly mapped reads. See the Supplemental Material for further details on bioinformatic analysis.
Statistical analyses
Differential transcript abundance analysis (i.e., “Differential Expression”) and principal components analyses (PCA) were computed on the ORFs from the combined assembly using DeSeq2 v1.28.1 within R [39, 54]. For differential transcript analysis, fold change and adjusted p values (padj) were calculated for the following comparisons within the GRW1 incubation: 2.5 nM Fe/2.5 nM DFB, 2.5 nM Fe/Control, and 2.5 nM DFB/ Control. To compare transcript abundances of genes of interest between DFB-added incubations, Fe-added incubations, and surface in situ samples, ordinary one-way ANOVAs or Kruskal-Wallis tests were performed. Post-hoc multiple comparisons were adjusted with Tukey’s HSD (ANOVA post-hoc) or Dunn’s test (Kruskal-Wallis post-hoc). For comparisons between normalized transcripts of genes of interest grouped under DFB-added or Fe-added treatments, either unpaired two-tailed t tests, unpaired t tests with Welch’s correction, or Mann–Whitney tests were performed when appropriate. See Supplementary Methods for details on statistical analyses.
Results
Physicochemical status
The Southern Ocean Time Series (SOTS) is located along the northern edge of the sub-Antarctic zone close to the subtropical front (STF). Surface water temperature ranged between 11 and 13 °C, salinity between 34.5 and 34.9, NO3 between 8.35 and 12.7 µmol L−1 and Si between 0.6 and 0.8 µmol L−1, over an 11 day sampling period, consistent with northern sub-Antarctic zone water properties [53]. Dissolved Fe concentrations in the upper 50 m ranged between 0.15 and 0.35 nmol L−1. The variability in dissolved Fe concentrations mirrored salinity and temperature in the upper water column. Below the surface, Sub-Antarctic Mode Water (SAMW) and Antarctic Intermediate Water (AAIW) were present between about 200–400 m, and 600–1200 m, respectively.
Community physiological and transcriptional response to Fe availability
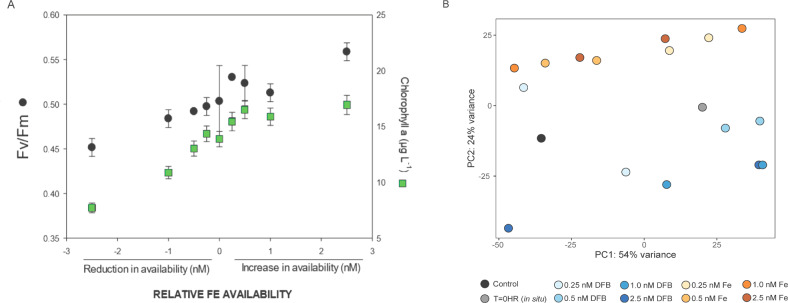
The changes in relative Fe availability generated in our experiments were positively correlated with measures of photosynthetic biomass (chlorophyll a) and photosystem II photochemical efficiency (Fv/Fm, Fig. 1A). Relative to the control, phytoplankton biomass increased with added FeCl3 and decreased with added DFB (Fig. 1B). For the second incubation (GRW2), Fe-addition stimulated increases in Fv/Fm while DFB addition reduced bulk chlorophyll a and Fv/Fm, although these values were similar between the 12.5 nM DFB and 5.0 nM DFB-added treatments (Supplementary Fig. 1). For GRW1 specifically, the effects of these treatments on gene transcript abundance between +Fe and +DFB resulted in distinct patterns, except for one 0.25 nM DFB treatment (Fig. 1B). Despite inherent bottle variability present, the genetic signatures driving the separation of +DFB and +Fe were apparent and thus further investigated.
Fig. 1. Bulk community response to on-deck incubations either reducing available Fe (DFB-added) or increasing available Fe (FeCl3-added) after 72 h.
A Total phytoplankton biomass estimates (chlorophyll a concentrations, green boxes) and photosynthetic efficiency (Fv/Fm, ratio of variable fluorescence to maximal fluorescence, black circles) as a function of relative Fe availability. Results are shown as the means ± error of technical duplicates, averaged between duplicate bottles. B Principal component analysis (PCA) displaying PC1 against PC2 of total community gene transcript abundance patterns, color coded by treatment (blue = DFB-added, orange = Fe added). The black circle is the control bottle and gray circle is the T = 0 h in situ sample. Each bottle duplicate is plotted, with the exception of the control.
Taxonomically-resolved community response to Fe
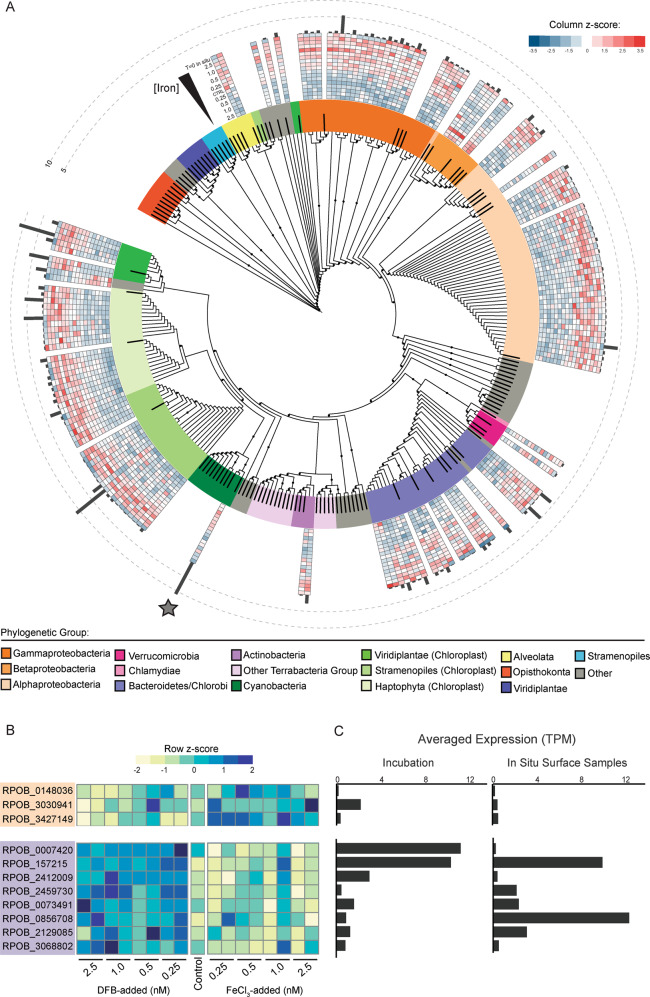
The DNA-directed RNA polymerase beta subunit genes (rpoB/rpb1), used previously as a housekeeping transcript for eukaryotes and prokaryotes within metatranscriptomes [41, 55, 56], was targeted to resolve taxonomic structure. Phylogenetic placement of candidate rpoB/rpb1 translated sequences (clustered at 95% amino acid similarity) responsive to Fe treatments show assignment to groups ranging from heterotrophic bacteria and photosynthetic eukaryotes (Fig. 2A). Cyanobacterial RpoB was most closely related to Synechococcus and Prochlorococcus references (Fig. 2A and Supplementary Fig. 2). These candidates showed high relative transcript abundance across the in situ surface samples and higher representation within +DFB treatments relative to +Fe (Fig. 2A). However, closer examination of un-clustered, putative Synechococcus RpoB (Supplementary Fig. 2) sequences show the presence of individual RpoB in both high-Fe and low-Fe treatments (Fig. 2B). When phylogenetically resolved, the high-Fe RpoB were most similar to Synechococcus sp. CC9311 reference, and low-Fe responsive RpoB similar to Synechococcus spp. HB1133, BL107, and CC9902 references (Supplementary Fig. 2). Analyses of chlorophyll biosynthetic genes for algal eukaryotes and prokaryotes (Supplementary Fig. 3, Supplementary Table 6) demonstrated Synechococcus sp. CC9902-, BL107-, and WH8016-like transcripts were higher in low-Fe treatments and in situ (Supplementary Fig. 3). Conversely, transcripts annotated as chlorophyll biosynthetic genes from Chlorophyta, Streptophyta, Synechococcus CC9311 and Prochlorococcus were higher under high-Fe conditions (Supplementary Fig. 3).
Fig. 2. Taxonomically resolved community activity in response to Fe availability using rpoB/rpb1 as a marker gene.
A Phylogenetic placement of translated candidate rpoB/rpb1 clustered at 95% amino acid identity across eukaryotes (blue branches) and prokaryotes (red branches), color coded by broad phylogenetic group. Reference proteins used to construct the tree are represented as black bars, and black dots are bootstrap values >0.5. Normalized transcript abundance (transcripts-per-million, TPM) levels are shown in the heatmap on the outer rings and are scaled (z-score, [Observed TPM – mean TPM]/standard deviation) for each tree candidate, going from low Fe to high Fe from the inner to outer ring. Black bars on the outermost ring represent in situ [T = 0 h, March 5th (5 m), March 5th (15 m), March 7th (15 m), March 9th (15 m)] averaged TPMs. The gray star represents the placement of a candidate in the phylum Cyanobacteria. B Un-clustered cyanobacterial-like rpoB transcript abundance (TPM, z-score) against Fe availability within the incubations. Orange block = sequences phylogenetically clustered with the Synechococcus sp. CC9311 reference RpoB. Purple block = sequences phylogenetically clustered with the Synechococcus spp. HB1133, BL107, and CC9902. The full phylogeny is shown in Supplementary Fig. 1C Averaged transcript abundance values (TPM) within the Fe incubation and the in situ samples, corresponding to each candidate rpoB row.
Functional response of Synechococcus to Fe availability
Differential transcript abundances of Fe cycling genes
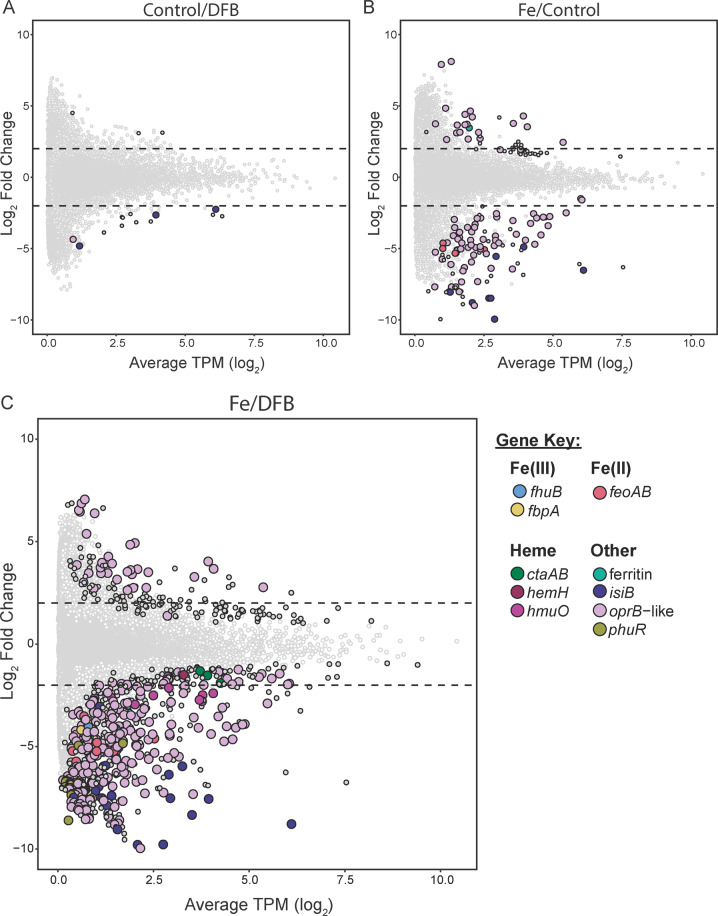
To investigate functional responses to Fe availability, differential transcript abundance analyses were performed with Synechococcus-like genes within the assembly between high-Fe (2.5 nM Fe), low-Fe (2.5 nM DFB), and control incubations (Fig. 3A–C, Supplementary Table 7). Results for all genes (including non-Synechococcus-like) are in Supplementary Table 8 as well. Comparing high-Fe/low-Fe treatments generated the most genes with significantly different transcript abundances (padj ≤ 0.1), whereas the control/low-Fe analysis had the fewest (Fig. 3A–C). Because most genes were associated with Fe metabolism, we focused on this functional group, although genes within nitrogen/carbon metabolism and photosynthesis also had significantly different transcript abundances (Supplementary Table 7).
Fig. 3. Synechococcus Fe-responsive genes with significantly different transcript abundances under high Fe versus low Fe conditions.
Genes with significantly different transcript abundances (padj < 0.1) are outlined in black. The smaller, gray circles are genes in the “other” category (not Fe-related). The dashed lines show -2 and +2 log2 fold change thresholds. A Control versus 2.5 nM DFB B 2.5 nM FeCl3 versus Control C 2.5 nM FeCl3 versus 2.5 nM DFB.
Transcripts for Fe(III), Fe(II), and putative heme-cycling genes were significantly decreased when Fe was added (Fig. 3B and C). It is possible that phuR, a “Heme/hemoglobin uptake outer membrane receptor” [57], targets non-heme sources, and so it has been denoted as an “organic iron complex“ transporter in the “Other” category (Fig. 3). Notably, 268 genes annotated as oprB-like Fe/carbon uptake porins were identified in the high-Fe/low-Fe analysis (Fig. 3C). Further analyses of all Synechococcus assigned oprB-like genes indicate 763 unique sequences with high sequence similarity to experimentally confirmed Fe-selective porins (Supplementary Table 9) in Synechocystis [53]. Comparison to all proteins within the NCBI non-redundant database also indicate numerous hits to “iron uptake porin” and/or proteins with the cl41527 conserved domain (“por_somb Superfamily/iron uptake porin”, Supplementary Table 4). Other top hits included “OprB Superfamily/Carbohydrate-selective porin” (Supplementary Table 4). The gene encoding the Fe-stress marker protein Flavodoxin, isiB, was also significantly decreased with Fe-addition (Fig. 3A–C).
Estimating Synechococcus Fe-status in situ
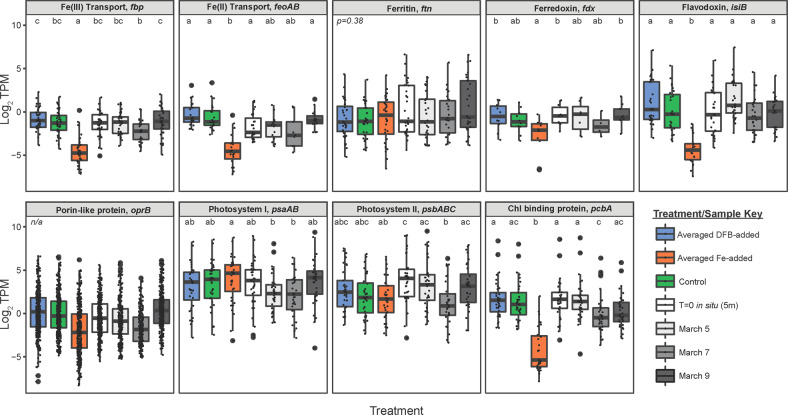
To assess the Fe-status of putative Synechococcus members in situ, genes indicative of Fe stress were queried and within 4 metatranscriptomes from surface samples (Fig. 4, Supplementary Table 10). Transcript abundances of genes assigned to a ferric binding protein (fbp), involved in ABC-transport of Fe(III) species [58], was significantly elevated in the +DFB, control, and in situ samples compared to +Fe (Fig. 4, Supplementary Table 11). Likewise, genes assigned an Fe(II) transport system, feoAB [58], and flavodoxin (isiB), an Fe-stress indicator [10], showed statistically significant similar trends (Fig. 4, Supplementary Table 11). Interestingly, ferredoxin (fdx), known to be replaced with flavodoxin under Fe-limitated conditions [10, 59–61], was significantly higher within the +DFB conditions compared to +Fe (padj = 0.0313) (Fig. 4, Supplementary Table 11). Furthermore, individual Synecoccocus fdx harbored on different contigs shows a separation between those elevated under DFB-added conditions (p = 0.0003), and those elevated under Fe-added conditions (p = 0.0426, Supplementary Fig. 4). Transcripts for genes assigned to the putative Fe-storage protein, ferritin [58, 62], displayed no statistically significant trends across conditions (Fig. 4, Supplementary Table 11). Transcripts for genes involved in electron transport of photosystem I (“PSI”, psaAB) were not significantly different between bottle incubations, but were significantly reduced in the March 5th and March 7th in situ samples compared to +Fe conditions (Fig. 4, Supplementary Table 11). Similarly, photosystem II (“PSII”, psbABC) genes did not differ significantly between treatments or between the in situ samples compared to the incubations (Fig. 4, Supplementary Table 11). The gene encoding the chlorophyll a/b binding antennae protein shown to be induced upon Fe-deficiency in cyanobacteria [63], pcbA, had significantly elevated transcript abundances under low-Fe conditions and the in situ samples compared to the +Fe conditions (Fig. 4, Supplementary Table 11).
Fig. 4. Interpretation of the in situ Fe status of Synechococcus communities using the incubations as reference.
A–I Comparison of the transcript abundance of different Fe cycling marker genes across low Fe (averaged DFB-added), high Fe (averaged Fe-added) and in situ surface samples (5–15 m depth). Transcript abundance values are in normalized TPM (log2).
Analysis of other Fe-responsive Synechococcus genes
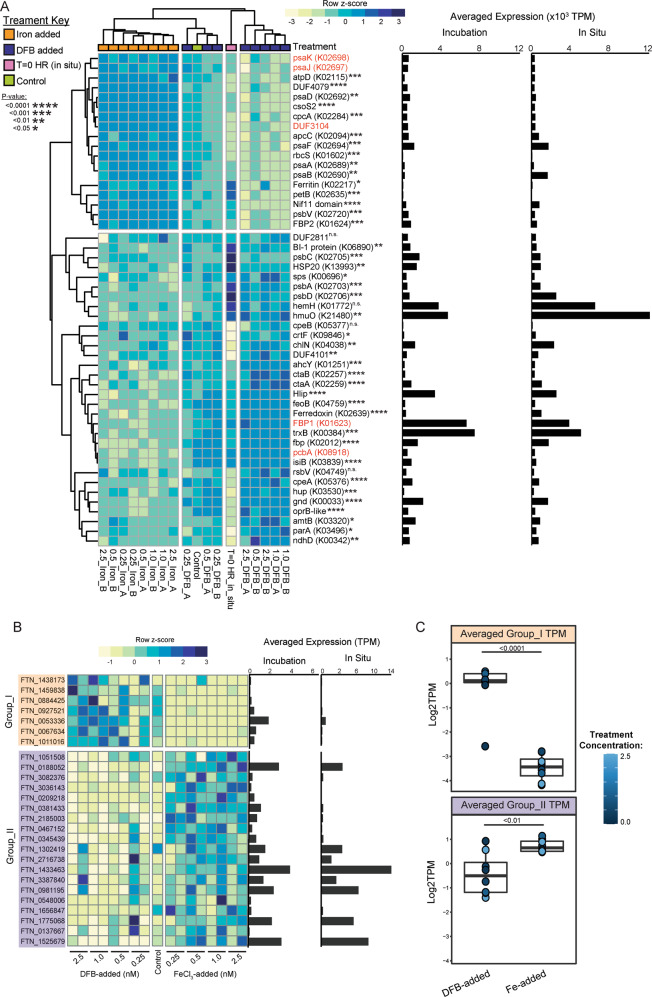
Genes with the highest variability in transcript abundances between the +DFB and +Fe incubations, deduced from ranking eigenvalues of each variable across PC2 (Fig. 1B), were queried. Here, the top 50 most abundant Synechococcus-like genes ranged in assigned function from Photosynthesis/Energy generation to Fe- and nitrogen-cycling (Supplementary Table 12). Genes encoding PSI components (psaABDFJK), F-type H + /Na + -transporting ATPase (atpD), carboxysome shell peptide (csoS2), phycocyanin alpha chain (cpcA), ribulose bisphosphate carboxylase (rbcS), fructose-bisphosphate aldolase class-I (FBP2), ferritin, Nif11 domain-like protein, cytochrome b6 (petB), and the phycobilisome core linker protein (apcC) had significantly elevated transcript abundance across the +Fe incubations compared to the +DFB incubations (Fig. 5A, Supplementary Table 13). Genes involved in PSII function (psbACD), phycoerythrin synthesis (cpeAB), light-independent protochlorophyllide reductase (chlN), ferredoxin, flavodoxin (isiB), chlorophyll a/b binding antennae protein (pcbA), thioredoxin reductase (trxB), a high light inducible protein (Hlip), NAD(P)H-quinone oxidoreductase chain 4 (ndhD), fructose-bisphosphate aldolase class-I (FBP1), SAM dependent carboxyl methyltransferase (crtF), heat shock protein 20 (HSP20), and ammonium transport (amtB) were elevated under DFB-added treatments. Fe cycling genes such as the heme utilization gene (hmuO), heme a- and o-synthase (ctaA and ctaB), Fe(II) transport (feoB), Fe(III) transport (fbp) had significantly higher transcript abundance across the +DFB treatments compared the to +Fe treatments (Fig. 5A, Supplementary Table 13). The heme biosynthesis ferrochelatase gene (hemH) trended toward having higher values within the +DFB conditions, but did not have statistically significantly higher transcript abundance in the +DFB relative to the +Fe conditions (Fig. 5A, Supplementary Table 13). Ferritin-like genes in this subset showed on-average higher representation under high-Fe conditions (Fig. 5A, Supplementary Table 13). However, analysis of individual Synechococcus ferritin-like transcripts revealed ferritins with higher abundances in the low-Fe treatments and ferritins with higher abundances in the high-Fe treatments (Fig. 5B). These sequences with contrasting patterns are phylogenetically distinct (Supplementary Fig. 5), with the low-Fe ferritin clustering closely (bootstrap > 0.5) with a Synechococcus sp. CC9311 reference (“Group_I”) and high-Fe ferritin clustering closely (bootstrap > 0.5) with Synechococcus sp. WH8020 and BL107 reference (“Group_II”, Supplementary Fig. 5). Comparison of averaged transcript abundances within each group of ferritin between +DFB and +Fe-added incubations show these patterns are statisticaly significant (Fig. 5C, Supplementary Table 13).
Fig. 5. Transcript abundance patterns of genes assigned to Synechococcus across Fe availability.
A The top 50 most abundant genes with the highest variance across Fe-added and DFB-added incubations. Each column is a different bottle from the GRW1 incubation, where “A” and “B” show the result for each biological replicate per treatment. Each value shows the averaged log2 normalized transcripts (TPM) across all genes identified as the annotation shown next to each row within the heatmap, scaled by row (z-score, [Observed TPM – mean TPM]/standard deviation) and clustered by sample and gene using a Euclidian distance metric. Full gene names are shown in Supplementary Table 9. P values next to each gene were calculated by performing appropriate unpaired t tests between log2(TPM) values from the +DFB-added incubations versus the +Fe-added incubations. Gene names in red were not statistically analyzed due to having a non-normal distribution and heteroskedasticity. The averaged transcript abundance values for each gene are shown alongside within the Fe incubations (left) and across in situ surface samples (right). B Heatmap of individual (un-averaged) Synechococcus ferritin separated between low (orange block, “Group I”) and high-Fe (purple block, “Group II”) expressed genes. Averaged TPM values across the incubation experiments and in situ surface samples for each are shown alongside. Phylogenetic placement of each gene is shown in Supplementary Fig. 4. C Comparison of the averaged values within each ferritin “Group” heatmap between DFB-added and Fe-added incubations. Each dot represents the averaged value within each incubation bottle, color coded by the level of either DFB or Fe added. P-values were calculated using unpaired t-test with Welch’s correction for Group_I values, and the Mann-Whitney non-parametric test for Group_II values.
Read recruitment to genomes of Synechococcus isolates
To expand our observations of ecotype-specific Synechococcus responses to Fe within the metatranscriptome assembly, and to build linkages to laboratory studies, we competitively mapped to genomes of Synechococcus sp. CC9311 (“CC9311”) and Synechococcus sp. BL107 (“BL107”). Around 0.40–0.62% of transcripts mapped to CC9311 and 1.13–2.07% of transcripts mapped to BL107 (Supplementary Table 14). Genes of interest from the assembly method were targeted for examination. The analysis revealed 5 ferritins and 3 ferredoxins with differential patterns of representation influenced by Fe availability within the CC9311 genome, whereas BL107 only had 1 ferritin and 1 ferredoxin responsive to Fe (Supplementary Fig. 6 and Supplementary Table 15). Additionaly, rpoB transcripts followed similar trends as seen for CC9311-like and BL107-like RpoB (Fig. 1B, Supplementary Fig. 2). In both genomes, there were multiple oprB-like porins responsive to Fe. Interestingly, CC9311 had reads mapped to feoAB and multiple ABC-transporters, whereas BL107 had few ABC transport components with recruitments and mostly had Fe-uptake porins responsive to Fe availability (Supplementary Fig. 6A). Most genes within the BL107 genome had higher transcript abundances than genes in the CC9311 genome across both the incubations and in situ surface samples (Supplementary Fig. 6B). To confirm the patterns seen here within the GRW1, we also mapped metatranscriptomic reads from GRW2 to these genomes. Read mapping to CC9311 and BL107 genomes from GRW2 metatranscriptomes resulted in 0.48–0.68% and 0.54–0.90% reads mapped, respectively (Supplementary Table 14). Genes of interest (rpoB, ferritin, ferredoxins, porins, ABC-type transporters, flavodoxin) within both CC9311 and BL107 genomes showed patterns similar to GRW1 for +Fe versus + DFB conditions (Supplementary Fig. 7 and Supplementary Table 15).
Discussion
Since the seminal work of Martin and colleagues (e.g., [64, 65]), it is clear that Fe constrains marine productivity in many surface oceans [4]. Moreover, subsequent studies have demonstrated the effects of Fe on microbial community diversity—especially for planktonic phototrophs [6, 66, 67]. Using current generation molecular techniques, correlative examinations present evidence that specific populations, with particular genetic capabilities, are distributed across the surface oceans that is in part constrained by Fe-availability [16, 21]. Building on these foundations, the present study provides an experimental approach to diagnose the specific responses of phytoplankton to changes across a gradient of Fe availabilities. Our results demonstrate how the titration of available Fe can be used to both discern community-level Fe-status in the surface-ocean, and to tease apart sub-populations, revealing the persistence and genetic potential of phytoplankton ecotypes. However useful this titration approach was when viewing “bulk” community photosynthetic parameters such as chlorophyll a and photosystem II health (Fv/Fm), it did not capture the responses of different microbial community members at high resolution. The employment of metatranscriptomics, however, allowed experimental demonstration of the potential for different Synechococcus ecotypes to respond across a gradient of available Fe, illuminating the strategies that different potential ecotypes employ.
We show that bulk photosynthetic biomass and health of the community increased linearly with Fe availability at SOTS, with a pronounced decrease in both photosynthetic biomass and health when DFB was added. This approach has been used previously in field studies to assess the microbial responses to Fe limitation, albeit with coarser (e.g., bulk pigment) metrics [68–70]. Analyses of photosynthetic health and flow cytometric cell counts using this approach in the subtropical Pacific Ocean (HNLC region) showed that most cell types (large eukaryotes versus cyanobacteria) increased in abundance with increased Fe availability, although cyanobacteria displayed less severe Fe limitation [68]. In the equatorial Pacific Ocean, Prochlorococcus exhibited a minimal response to Fe additions whereas large phytoplankton and Synechococcus increased in growth [71], leading to the hypothesis of its low-Fe adaptation, which is reflected in its genomic content [16]. Furthermore, culture-based transcriptome responses of Prochlorococcus light ecotypes to different Fe availabilities hint towards various mechanisms encoded within the genomes of strains (e.g., Fe transport systems, photosynthetic antennae proteins, Fe-deficiency-induced response genes) that can grow under low Fe concentrations compared to those that cannot [72]. Here, we see similar trends with Synechococcus, with an added layer being the presence of putative low- and high-Fe adapted ‘ecotypes’ at SOTS. An expedition in the HNLC, sub-Antarctic Southern Ocean (‘FeCycle’) measured high contributions of cyanobacteria to biotic Fe pools [73], and Synechococcus has been shown to be the primary genus (~70%) of cyanobacteria within the SOTS phytoplankton community [74]. It is possible that the seasonal Fe-limitation in the SAZ, combined with periodic inputs of Fe [28] and frequent injections of subtropical water across this region result in a diverse community of Synechococcus adapted to high and low Fe conditions. This transcriptomics approach demonstrated the co-occurrence of different Fe-phytoplankton within this region, overlooked by genomic analyses alone, and reflected overlapping niche space derived from episodic exposure, to and/or fluctuations in, Fe availability. That such variability in conditions results in variability in the phytoplankton community has been a central tenet of ocean sciences and microbiology for more than 60 years [19].
Functional analyses further indicated that variable Synechococcus types were present and adapted to differing Fe-conditions. Genes involved in Fe-cycling and acquisition were the most responsive to changes in Fe-availability, as many such pathways are controlled by the ferric uptake regulator protein, Fur [75]. Differential transcript abundance analysis of high- and low-Fe treatments highlight a potentially important Fe acquisition mechanism in cyanobacteria that was recently shown to involve a Fe-specific porin [53]. This gene was annotated as an oprB-like porin gene with significant sequence similarity to the porin recently identified in Qiu et al. 2021. The transcript abundance of this porin-like transporter under both high- and low-Fe conditions, as well as in situ, may provide an advantage to some cyanobacteria, as the passive diffusion through a substrate-specific porin may be an alternative Fe-uptake pathway aside from energy-costly active transport [76, 77]. However, it remains unclear which chemical forms of Fe are transported through the porin, or if the porin may transport other small molecules, as OprB-like proteins mediate sugar uptake in Pseudomonas aeruginosa [78] and the cyanobacterium Nostoc punctiforme [79]. Moreover, it is thought that a broad spectrum of compounds might make up the organic ligands known to bind Fe in seawater [80] and to be from sources ranging from active production [81] to predation and lysis byproducts [11, 82]. Unsurprisingly, we saw transcripts for genes encoding Fe(III) and Fe(II) active transport systems fbp and feoA also overrepresented under Fe-limiting conditions. Finally, heme has also been recognized as a potentially significant source of Fe for phytoplankton in the ocean when other forms of Fe are limited [83, 84], and our analysis indicate multiple genes involved in putative heme biosynthesis (ctaAB, hemH) and utilization (hmuO) processes with transcript abundances overrepresented under Fe-limited incubations and in situ. The Heme/hemoglobin/transferrin/lactoferrin TonB-dependent outer membrane receptor, phuR [57], had elevated transcript abundances under Fe-limited conditions as well, however it is possible that this gene could be involved in the uptake of other organic Fe sources. Thus, it is possible that these genes may not be directly involved the uptake and utilization of heme as an Fe-source under Fe-limiting conditions, but rather the management of intracellular heme pools and other cellular processes [83], warranting better characterization of the function of heme-cycling genes in cyanobacteria as a response to Fe-limitation. The availability of more resolved Fe-specific pathways from cultured isolate work in the future should allow for a better resolution of the specific Fe-sources these cells are using. While we cannot fully distinguish all forms of Fe preferred by Synechococcus, the analyses highlight the diverse mechanisms in which cyanobacteria can acquire Fe at SOTS.
Although we observed more low-Fe adapted Synechococcus representation under low-Fe conditions, metatranscriptomes suggested these cells were still exhibiting signatures of Fe-stress. Transcripts for common genetic markers of Fe-stress found to be transcribed under Fe-limiting conditions, namely the chlorophyll binding gene pcbA [63], and flavodoxin isiB [85–87] were elevated under low-Fe treatments and prevalent in situ (Fig. 3 & 4). Paradoxically, ferredoxin, which is known to be replaced by flavodoxin when the cell is Fe-stressed [61, 88, 89], was elevated under low-Fe conditions (Figs. 4F and 5A). A transcriptome study of genome-wide response to Fe-deficiency in Synechocystis sp. PCC6803 also found one out of 6 ferredoxin genes 3- to 4-fold up-regulated under Fe-limitation in contrast to the 5 down-regulated ferredoxins [90]. It was found that in Synechocystis sp. PCC6803, distinct ferredoxin paralogs played a role in the tolerance to oxidative and metal stress [91], and a unique ferredoxin (Fed2), not involved in photosynthetic electron transport, was shown to be involved in Fe-perception under low-Fe conditions [92]. Further, different ferredoxin paralogs across Synechococcus genomes had ecotype-specific patterns across high- and low-Fe oceanic regions [21], suggesting ecotype-specific purposes / repurposing for ferredoxin. Finer scale analysis of each Synechococcus-like ferredoxin detected in our analyses showed a similar trend, where a subset of ferredoxin genes had higher representation under high Fe conditions and another subset with higher representation under low Fe conditions, including those most confidently associated with Synechococcus CC9311 (Supplementary Fig. 4).
Ferritin is thought to serve as an Fe storage compound in eukaryotes [93] and cyanobacteria [94] when Fe is replete. It has been shown to also be involved in diel regulation of Fe-uptake and recycling and the maintenance of cellular Fe-homeostasis for Ostreococcus [95]. In the coastal strain Synechococcus sp. CC9311 [96, 97] and Synechocystis [98] it is thought to play a role in coping with oxidative stress. Ferritin paralogs also show different abundances across Fe regimes [21], hinting towards ferritins role in multiple cellular processes aside from Fe storage under Fe-replete conditions, as has been shown in diatoms [99]. Furthermore, different Prochlorococcus light-ecotypes with varying growth and transcriptome responses under low-Fe concentrations had strain-specific expression of ferritin-like genes, and it was hypothesized that ferritins may serve different functions in different strains depending on their response to Fe-limitation [72]. Here, we see a taxon-specific representation of ferritin-like transcripts, where most are represented under high-Fe conditions but some, clustering phylogenetically with Synechococcus sp. CC9311, are more represented under low Fe conditions (Fig. 5B and Supplementary Fig. 5). Indeed, competitive recruitment of the metatranscriptomes to Synechococcus sp. CC9311 (a high-nutrient adapted strain) and Synechococcus BL107 (an open-ocean/oligotrophic strain) revealed 5 ferritins and 3 ferredoxins with different transcript abundance patterns against Fe-availability from the CC9311 genome whereas BL107 had only 1 ferritin and 1 flavodoxin responsive to Fe (Supplementary Fig. 6). It is unexpected to see a higher representation of ferritin-like genes under low Fe conditions: we posit that CC9311 ferritins may play a role in an overall cellular response to stress for high-Fe adapted strains coping with Fe limitation. These analyses highlight the complex response of high-nutrient adapted CC9311-like strains in comparison to the low-nutrient adapted BL107-like strains with changes in Fe availability: CC9311-like strains use multiple Fe acquisition systems to obtain Fe under both conditions and expresses ferritin even under Fe-limited states to either attempt to store any Fe it takes in or to cope with cellular stress. In contrast, the more streamlined, low-Fe adapted strains, such as BL107 may persist better in low-Fe waters [16]. However, this strategy may place these strains at a disadvantage under short-term Fe increases. Although we cannot fully characterize the Synechococcus in our dataset to the strain level, these complex responses to variable Fe conditions point towards different strategies to cope with Fe-limitation, with evidence for both strategies in the surface waters at SOTS.
Overall, by leveraging metatranscriptomes across Fe titrations to interpret in situ samples, we captured adaptations to changes in Fe availability of an important group of phytoplankton at SOTS composed of different ecotypes that would have been overlooked in the “bulk” phytoplankton physiological responses. Indeed, altering Fe availability by adding DFB or FeCl3 stimulated net chlorophyll a and photosystem II health within the bulk community. Since this region is seasonally Fe-limited and encounters episodic inputs of Fe [28], it is possible that high-nutrient adapted “opportunists” respond quickly to these transient pulses, allowing their persistence in this region. Such local adaptation may drive the evolution of clade-specific paralogs of Fe acquisition genes, putative Fe storage genes (ferritins), ferredoxins, and flavodoxins, which have different abundances across varying Fe regimes [21] and could be transcriptionally activated under differing Fe concentrations as suggested by our data. These results warrant better characterization of potential Synechococcus Fe-ecotypes and their complex responses to episodic Fe availability in HNLC regions.
Supplementary information
Acknowledgements
We are grateful to the officers, crew, and research staff of the Marine National Facility and the R.V. Investigator for their help with sample collection and generation of hydrochemistry data. Dr. Shane Hogle and an anonymous reviewer greatly improved the manuscript with their comments. This research was financially supported under Australian Research Council’s Discovery program (DP170102108; DP130100679) and ship time from Australia’s Marine National Facility. This work was supported by a National Science Foundation grant (OCE- 1829641) to SWW. The work (proposal 504140, doi:10.46936/10.25585/60001188) conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Author contributions
Conceptualisation: NEG, GRL, SWW; Data curation: NEG, RFS; Formal analysis: NEG, GRL, SWW; Funding acquisition: PWB, MJE, SWW; Investigation: NEG, RFS, BST, GRL, MJE, PWB; Methodology: NEG, GRL, SWW, SR; Project administration: SWW, PWB, MJE; Resources: SWW, CP, PWB; Supervision: SWW, CP; Validation: NEG, GRL; Visualisation: NEG; Writing—original draft: NEG, SWW; Writing—review & editing: all authors.
Data availability
Raw and processed data used in this study are publicly available through the JGI Data Portal (https://data.jgi.doe.gov) under the Project ID numbers 1260738 and 1260735.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s43705-022-00132-5.
References
- 1.Moore CM, Mills MM, Arrigo KR, Berman-Frank I, Bopp L, Boyd PW, et al. Processes and patterns of oceanic nutrient limitation. Nat Geosci. 2013;6:701–10. doi: 10.1038/ngeo1765. [DOI] [Google Scholar]
- 2.Martin JH, Gordon RM, Fitzwater SE. The case for iron. Limnol Oceanogr. 1991;36:1793–802. doi: 10.4319/lo.1991.36.8.1793. [DOI] [Google Scholar]
- 3.Tagliabue A, Bowie AR, Boyd PW, Buck KN, Johnson KS, Saito MA. The integral role of iron in ocean biogeochemistry. Nature. 2017;543:51–9. doi: 10.1038/nature21058. [DOI] [PubMed] [Google Scholar]
- 4.Boyd PW, Jickells T, Law CS, Blain S, Boyle EA, Buesseler KO, et al. Mesoscale iron enrichment experiments 1993–2005: synthesis and future directions. Science. 2007;315:612–7. doi: 10.1126/science.1131669. [DOI] [PubMed] [Google Scholar]
- 5.Marchetti A, Maldonado MT, Lane ES, Harrison PJ. Iron requirements of the pennate diatom Pseudo-nitzschia: Comparison of oceanic (high-nitrate, low-chlorophyll waters) and coastal species. Limnol Oceanogr. 2006;51:2092–101. doi: 10.4319/lo.2006.51.5.2092. [DOI] [Google Scholar]
- 6.Marchetti A, Maldonado MT Iron. In: Borowitzka MA, Beardall J, Raven JA, editors. The Physiology of Microalgae. Cham: Springer International Publishing; 2016. p. 233-79.
- 7.Strzepek RF, Harrison PJ. Photosynthetic architecture differs in coastal and oceanic diatoms. Nature. 2004;431:689–92. doi: 10.1038/nature02954. [DOI] [PubMed] [Google Scholar]
- 8.Sunda WG, Swift DG, Huntsman SA. Low iron requirement for growth in oceanic phytoplankton. Nature. 1991;351:55–7. doi: 10.1038/351055a0. [DOI] [Google Scholar]
- 9.Peers G, Price NM. Copper-containing plastocyanin used for electron transport by an oceanic diatom. Nature. 2006;441:341–4. doi: 10.1038/nature04630. [DOI] [PubMed] [Google Scholar]
- 10.La Roche J, Boyd PW, McKay RML, Geider RJ. Flavodoxin as an in situ marker for iron stress in phytoplankton. Nature. 1996;382:802–5. doi: 10.1038/382802a0. [DOI] [Google Scholar]
- 11.Strzepek RF, Maldonado MT, Higgins JL, Hall J, Safi K, Wilhelm SW, et al. Spinning the “Ferrous Wheel”: the importance of the microbial community in an iron budget during the FeCycle experiment. Global Biogeochemical Cycles. 2005;19:GB4S26. doi: 10.1029/2005GB002490. [DOI] [Google Scholar]
- 12.Flombaum P, Gallegos JL, Gordillo RA, Rincón J, Zabala LL, Jiao N, et al. Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus. PNAS. 2013;110:9824–9. doi: 10.1073/pnas.1307701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li WKW. Primary production of prochlorophytes, cyanobacteria and eukaryotic ultraplankton: measurements from flow cytometric sorting. Limnol Oceanogr. 1994;39:169–75. doi: 10.4319/lo.1994.39.1.0169. [DOI] [Google Scholar]
- 14.Partensky F, Garczarek L. Prochlorococcus: advantages and limits of minimalism. Annu Rev Mar. Science. 2010;2:305–31. doi: 10.1146/annurev-marine-120308-081034. [DOI] [PubMed] [Google Scholar]
- 15.Partensky F, Blanchot J, Vaulot D Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review. Bulletin-Institut Oceanographique Monaco-Numero Special-. 1999:457-76.
- 16.Rusch DB, Martiny AC, Dupont CL, Halpern AL, Venter JC. Characterization of Prochlorococcus clades from iron-depleted oceanic regions. PNAS. 2010;107:16184–9. doi: 10.1073/pnas.1009513107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilhelm SW. Ecology of iron-limited cyanobacteria: A review of physiological responses and implications for aquatic systems. Aquat Microb Ecol. 1995;9:295–303. doi: 10.3354/ame009295. [DOI] [Google Scholar]
- 18.Wilhelm SW, Trick CG. Iron-limited growth of cyanobacteria: multiple siderophore production is a common response. Limnol Oceanogr. 1994;39:1979–84. doi: 10.4319/lo.1994.39.8.1979. [DOI] [Google Scholar]
- 19.Hutchinson GE. The paradox of the plankton. The American Naturalist. 1961;95:137–45. doi: 10.1086/282171. [DOI] [Google Scholar]
- 20.Lee MD, Ahlgren NA, Kling JD, Walworth NG, Rocap G, Saito MA, et al. Marine Synechococcus isolates representing globally abundant genomic lineages demonstrate a unique evolutionary path of genome reduction without a decrease in GC content. Environ Microbiol. 2019;21:1677–86. doi: 10.1111/1462-2920.14552. [DOI] [PubMed] [Google Scholar]
- 21.Ahlgren NA, Belisle BS, Lee MD. Genomic mosaicism underlies the adaptation of marine Synechococcus ecotypes to distinct oceanic iron niches. Environ Microbiol. 2020;22:1801–15. doi: 10.1111/1462-2920.14893. [DOI] [PubMed] [Google Scholar]
- 22.Sohm JA, Ahlgren NA, Thomson ZJ, Williams C, Moffett JW, Saito MA, et al. Co-occurring Synechococcus ecotypes occupy four major oceanic regimes defined by temperature, macronutrients and iron. The. ISME Journal. 2016;10:333–45. doi: 10.1038/ismej.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farrant GK, Doré H, Cornejo-Castillo FM, Partensky F, Ratin M, Ostrowski M, et al. Delineating ecologically significant taxonomic units from global patterns of marine picocyanobacteria. PNAS. 2016;113:E3365–E74. doi: 10.1073/pnas.1524865113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shadwick E, Trull T, Tilbrook B, Sutton A, Schulz E, Sabine C. Seasonality of biological and physical controls on surface ocean CO2 from hourly observations at the Southern Ocean Time Series site south of Australia. Global Biogeochem Cycles. 2015;29:223–38. doi: 10.1002/2014GB004906. [DOI] [Google Scholar]
- 25.Trull T, Schulz E, Bray S, Pender L, McLaughlan D, Tilbrook B, et al., editors. The Australian integrated marine observing system Southern Ocean time series facility. OCEANS 2010 IEEE-Sydney; 2010: IEEE.
- 26.Sabine CL, Feely RA, Gruber N, Key RM, Lee K, Bullister JL, et al. The oceanic sink for anthropogenic CO2. Science. 2004;305:367–71. doi: 10.1126/science.1097403. [DOI] [PubMed] [Google Scholar]
- 27.Blain S, Queguiner B, Armand L, Belviso S, Bombled B, Bopp L, et al. Effect of natural iron fertilization on carbon sequestration in the Southern Ocean. Nature. 2007;446:1070–U1. doi: 10.1038/nature05700. [DOI] [PubMed] [Google Scholar]
- 28.Henley SF, Cavan EL, Fawcett SE, Kerr R, Monteiro T, Sherrell RM, et al. Changing biogeochemistry of the Southern Ocean and Its ecosystem implications. Front Mar Sci. 2020;7:581. doi: 10.3389/fmars.2020.00581. [DOI] [Google Scholar]
- 29.Bruland KW, Franks RP, Knauer GA, Martin JH. Sampling and analytical methods for the determination of copper, cadmium, zinc, and nickel at the nanogram per liter level in sea water. Anal Chim Acta. 1979;105:233–45. doi: 10.1016/S0003-2670(01)83754-5. [DOI] [Google Scholar]
- 30.Rees C, Pender L, Sherrin K, Schwanger C, Hughes P, Tibben S, et al. Methods for reproducible shipboard SFA nutrient measurement using RMNS and automated data processing. Limnology and Oceanography: Methods. 2019;17:25–41. [Google Scholar]
- 31.Schallenberg C, Strzepek RF, Schuback N, Clementson LA, Boyd PW, Trull TW. Diel quenching of Southern Ocean phytoplankton fluorescence is related to iron limitation. Biogeosciences. 2020;17:793–812. doi: 10.5194/bg-17-793-2020. [DOI] [Google Scholar]
- 32.Gilbert N, Pound H, Wilhelm SW RNA extraction from Sterivex using phenol:chloroform - cutting off the filter from the Sterivex unit for low biomass samples. Protocols.io. 2020. 10.17504/protocols.io.bh75j9q6. Accessed March 24, 2022.
- 33.Bushnell B BBMap: a fast, accurate, splice-aware aligner. Lawrence Berkeley National Lab.(LBNL), Berkeley, CA (United States); 2014.
- 34.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–6. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 35.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132–e. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 37.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory in Biosciences. 2012;131:281–5. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 38.Wickham H. ggplot2. Wiley Interdiscip Rev Comput Stat. 2011;3:180–5. doi: 10.1002/wics.147. [DOI] [Google Scholar]
- 39.Team RC. R: A language and environment for statistical computing. 2013.
- 40.Kolde R, Kolde M Package ‘pheatmap’. Bioconductor 1–6. 2012.
- 41.Pound HL, Gann ER, Tang X, Krausfeldt LE, Huff M, Staton ME, et al. The “neglected viruses” of Taihu: abundant transcripts for viruses infecting eukaryotes and their potential role in phytoplankton succession. Front Microbiol. 2020;11:338. doi: 10.3389/fmicb.2020.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 43.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–2. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson JD, Gibson TJ, Higgins DG Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinform. 2003:2.3. 1-2.3. 22. [DOI] [PubMed]
- 46.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 47.Matsen FA, Kodner RB, Armbrust EV. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics. 2010;11:1–16. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucl Acids Res. 2019;47:W256–W9. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, Von Mering C, et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 2017;34:2115–22. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garber AI, Nealson KH, Okamoto A, McAllister SM, Chan CS, Barco RA, et al. FeGenie: a comprehensive tool for the identification of iron genes and iron gene neighborhoods in genome and metagenome assemblies. Front Microbiol. 2020;11:37. doi: 10.3389/fmicb.2020.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–93. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clum A, Huntemann M, Bushnell B, Foster B, Foster B, Roux S, et al. DOE JGI metagenome workflow. mSystems. 2021;6:e00804–20. doi: 10.1128/mSystems.00804-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu GW, Jiang HB, Lis H, Li ZK, Deng B, Shang JL, et al. A unique porin meditates iron‐selective transport through cyanobacterial outer membranes. Environ Microbiol. 2021;23:376–90. doi: 10.1111/1462-2920.15324. [DOI] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moniruzzaman M, Wurch LL, Alexander H, Dyhrman ST, Gobler CJ, Wilhelm SW. Virus-host relationships of marine single-celled eukaryotes resolved from metatranscriptomics. Nat Commun. 2017;8:16054. doi: 10.1038/ncomms16054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stough JMA, Kolton M, Kostka JE, Weston DJ, Pelletier DA, Wilhelm SW. Diversity of active viral infections within the Sphagnum microbiome. Appl Environ Microbiol. 2018;84e001124-18. [DOI] [PMC free article] [PubMed]
- 57.Smith AD, Wilks A. Differential contributions of the outer membrane receptors PhuR and HasR to heme acquisition in Pseudomonas aeruginosa. J Biol Chem. 2015;290:7756–66. doi: 10.1074/jbc.M114.633495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andrews SC, Robinson AK, Rodríguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbiology Reviews. 2003;27:215–37. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 59.Geider RJ, La Roche J. The role of iron in phytoplankton photosynthesis, and the potential for iron-limitation of primary productivity in the sea. Photosynth Res. 1994;39:275–301. doi: 10.1007/BF00014588. [DOI] [PubMed] [Google Scholar]
- 60.McKay RML, Geider RJ, LaRoche J. Physiological and biochemical response of the photosynthetic apparatus of two marine diatoms to Fe stress. Plant Physiol. 1997;114:615–22. doi: 10.1104/pp.114.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKay RML, LaRoche J, Yakunin AF, Durnford DG, Geider RJ. Accumulation of ferredoxin and flavodoxin in a marine diatom in response to Fe. J Phycol. 1999;35:510–9. doi: 10.1046/j.1529-8817.1999.3530510.x. [DOI] [Google Scholar]
- 62.Andrews SC. Iron storage in bacteria. Adv Microb Physiol. 1998;40:281–351. doi: 10.1016/S0065-2911(08)60134-4. [DOI] [PubMed] [Google Scholar]
- 63.Chen M, Bibby TS, Nield J, Larkum A, Barber J. Iron deficiency induces a chlorophyll d-binding Pcb antenna system around Photosystem I in Acaryochloris marina. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2005;1708:367–74. doi: 10.1016/j.bbabio.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 64.Martin JH, Coale KH, Johnson KS, Fitzwater SE, Gordon RM, Tanner SJ, et al. Testing the iron hypothesis in ecosystems of the equatorial pacific ocean. Nature. 1994;371:123–9. doi: 10.1038/371123a0. [DOI] [Google Scholar]
- 65.Martin JH, Fitzwater SE. Iron deficiency limits phytoplankton growth in the north-east pacific subarctic. Nature. 1988;331:41–43. doi: 10.1038/331341a0. [DOI] [Google Scholar]
- 66.Hutchins DA. Iron and the marine phytoplankton community. Progress in Phycological Research. 1995;11:1–48. [Google Scholar]
- 67.Martin JH, Fitzwater SE. Iron deficiency limits phytoplankton growth in the north-east Pacific subarctic. Nature. 1988;331:341–3. doi: 10.1038/331341a0. [DOI] [Google Scholar]
- 68.Eldridge ML, Trick CG, Alm MB, DiTullio GR, Rue EL, Bruland KW, et al. Phytoplankton community response to a manipulation of bioavailable iron in HNLC waters of the subtropical Pacific Ocean. Aquat Microb Ecol. 2004;35:79–91. doi: 10.3354/ame035079. [DOI] [Google Scholar]
- 69.Wells ML. Manipulating iron availability in nearshore waters. Limnol Oceanogr. 1999;44:1002–8. doi: 10.4319/lo.1999.44.4.1002. [DOI] [Google Scholar]
- 70.Wilhelm SW, King AL, Twining BS, LeCleir GR, DeBruyn JM, Strzepek RF, et al. Elemental quotas and physiology of a southwestern Pacific Ocean plankton community as a function of iron availability. Aquat Microb Ecol. 2013;68:185–94. doi: 10.3354/ame01611. [DOI] [Google Scholar]
- 71.Cavender-Bares KK, Mann EL, Chisholm SW, Ondrusek ME, Bidigare RR. Differential response of equatorial Pacific phytoplankton to iron fertilization. Limnol Oceanogr. 1999;44:237–46. doi: 10.4319/lo.1999.44.2.0237. [DOI] [Google Scholar]
- 72.Thompson AW, Huang K, Saito MA, Chisholm SW. Transcriptome response of high- and low-light-adapted Prochlorococcus strains to changing iron availability. ISME J. 2011;5:1580–94. doi: 10.1038/ismej.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boyd PW, Strzepek R, Ellwood M, Hutchins D, Nodder SD, Twining B, et al. Why are biotic iron pools uniform across high‐and low‐iron pelagic ecosystems? Global Biogeochem Cycles. 2015;29:1028–43. doi: 10.1002/2014GB005014. [DOI] [Google Scholar]
- 74.Fourquez M, Bressac M, Deppeler SL, Ellwood M, Obernosterer I, Trull TW, et al. Microbial competition in the subpolar southern ocean: an Fe–C Co-limitation experiment. Front Mar Sci. 2020;6:776. doi: 10.3389/fmars.2019.00776. [DOI] [Google Scholar]
- 75.González A, Sevilla E, Bes MT, Peleato ML, Fillat MF Chapter Five - Pivotal Role of iron in the regulation of cyanobacterial electron transport. In: Poole RK, editor. Advances in Microbial Physiology. 68: Academic Press; 2016. p. 169-217. [DOI] [PubMed]
- 76.Sutak R, Camadro J-M, Lesuisse E. Iron uptake mechanisms in marine phytoplankton. Front Microbiol. 2020;11:566691. [DOI] [PMC free article] [PubMed]
- 77.Qiu G-W, Lou W-J, Sun C-Y, Yang N, Li Z-K, Li D-L, et al. Outer membrane iron uptake pathways in the model cyanobacterium Synechocystis sp. strain PCC 6803. Appl Environ Microbiol. 2018;84:e01512–18. doi: 10.1128/AEM.01512-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van den Berg B. Structural basis for outer membrane sugar uptake in pseudomonads. J Biol Chem. 2012;287:41044–52. doi: 10.1074/jbc.M112.408518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ekman M, Picossi S, Campbell EL, Meeks JC, Flores EA. Nostoc punctiforme sugar transporter necessary to establish a cyanobacterium-plant symbiosis. Plant Physiol. 2013;161:1984–92. doi: 10.1104/pp.112.213116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rue EL, Bruland KW. Complexation of iron(III) by natural organic ligands in the Central North Pacific as determined by a new competitive ligand equilibration/adsorptive cathodic stripping voltammetric method. Marine Chem. 1995;50:117–38. doi: 10.1016/0304-4203(95)00031-L. [DOI] [Google Scholar]
- 81.Martinez JS, Carter-Franklin JN, Mann EL, Martin JD, Haygood MG. Structure and membrane affinity of a suite of amphiphilic siderophores produced by a marine bacterium. PNAS. 2003;100:3754–9. doi: 10.1073/pnas.0637444100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Poorvin L, Rinta-Kanto JM, Hutchins DA, Wilhelm SW. Viral release of iron and its bioavailability to marine plankton. Limnology Oceanogr. 2004;49:1734–41. doi: 10.4319/lo.2004.49.5.1734. [DOI] [Google Scholar]
- 83.Hogle SL, Barbeau KA, Gledhill M. Heme in the marine environment: from cells to the iron cycle. Metallomics. 2014;6:1107–20. doi: 10.1039/C4MT00031E. [DOI] [PubMed] [Google Scholar]
- 84.Hogle SL, Brahamsha B, Barbeau KA. Direct heme uptake by phytoplankton-associated Roseobacter bacteria. mSystems. 2017;2:e00124–16. doi: 10.1128/mSystems.00124-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ludwig M, Bryant D. Acclimation of the global transcriptome of the cyanobacterium Synechococcus sp. strain PCC 7002 to nutrient limitations and different nitrogen sources. Front Microbiol. 2012;3:145. [DOI] [PMC free article] [PubMed]
- 86.Hernández-Prieto MA, Schön V, Georg J, Barreira L, Varela J, Hess WR, et al. Iron deprivation in Synechocystis: inference of pathways, non-coding RNAs, and regulatory elements from comprehensive expression profiling. G3 Genes|Genomes|Genetics. 2012;2:1475–95. doi: 10.1534/g3.112.003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nodop A, Pietsch D, Höcker R, Becker A, Pistorius EK, Forchhammer K, et al. Transcript profiling reveals vew insights into the acclimation of the mesophilic fresh-water cyanobacterium Synechococcus elongatus PCC 7942 to iron starvation. Plant Physiol. 2008;147:747–63. doi: 10.1104/pp.107.114058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Straus NA, Bryant DA Iron deprivation: physiology and gene regulation. The molecular biology of cyanobacteria. Dordrecht: Springer; 1994. p. 731-50.
- 89.Erdner DL, Anderson DM. Ferredoxin and flavodoxin as biochemical indicators of iron limitation. Limnol Oceanogr. 1999;44:1609–15. doi: 10.4319/lo.1999.44.7.1609. [DOI] [Google Scholar]
- 90.Singh AK, McIntyre LM, Sherman LA. Microarray analysis of the genome-wide response to iron deficiency and iron reconstitution in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2003;132:1825–39. doi: 10.1104/pp.103.024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cassier-Chauvat C, Chauvat F. Function and regulation of ferredoxins in the cyanobacterium, Synechocystis PCC6803: Recent Advances. Life. 2014;4:666–80. doi: 10.3390/life4040666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schorsch M, Kramer M, Goss T, Eisenhut M, Robinson N, Osman D, et al. A unique ferredoxin acts as a player in the low-iron response of photosynthetic organisms. PNAS. 2018;115:E12111–E20. doi: 10.1073/pnas.1810379115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marchetti A, Parker MS, Moccia LP, Lin EO, Arrieta AL, Ribalet F, et al. Ferritin is used for iron storage in bloom-forming marine pennate diatoms. Nature. 2009;457:467–70. doi: 10.1038/nature07539. [DOI] [PubMed] [Google Scholar]
- 94.Keren N, Aurora R, Pakrasi HB. Critical roles of bacterioferritins in iron storage and proliferation of cyanobacteria. Plant Physiol. 2004;135:1666–73. doi: 10.1104/pp.104.042770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Botebol H, Lesuisse E, Šuták R, Six C, Lozano J-C, Schatt P, et al. Central role for ferritin in the day/night regulation of iron homeostasis in marine phytoplankton. PNAS. 2015;112:14652–7. doi: 10.1073/pnas.1506074112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bradley JM, Svistunenko DA, Pullin J, Hill N, Stuart RK, Palenik B, et al. Reaction of O2 with a diiron protein generates a mixed-valent Fe2+/Fe3+ center and peroxide. PNAS. 2019;116:2058–67. doi: 10.1073/pnas.1809913116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bradley JM, Svistunenko DA, Wilson MT, Hemmings AM, Moore GR, Le Brun NE. Bacterial iron detoxification at the molecular level. J Biol Chem. 2020;295:17602–23. doi: 10.1074/jbc.REV120.007746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shcolnick S, Summerfield TC, Reytman L, Sherman LA, Keren N. The Mechanism of iron homeostasis in the unicellular cyanobacterium Synechocystis sp. PCC 6803 and its relationship to oxidative stress. Plant Physiol. 2009;150:2045–56. doi: 10.1104/pp.109.141853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cohen NR, Mann E, Stemple B, Moreno CM, Rauschenberg S, Jacquot JE, et al. Iron storage capacities and associated ferritin gene expression among marine diatoms. Limnol Oceanogr. 2018;63:1677–91. doi: 10.1002/lno.10800. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data used in this study are publicly available through the JGI Data Portal (https://data.jgi.doe.gov) under the Project ID numbers 1260738 and 1260735.