Abstract
Over the past few years, the field of pediatric cancer has experienced a shift in momentum and this has led to new and exciting findings that have relevance beyond pediatric malignancies. Here we present the current status of key aspects of pediatric cancer research. We have focused on genetic and epigenetic drivers of disease, cellular origins of different pediatric cancers, disease models, the tumor microenvironment, and cellular immunotherapies.
Keywords: Pediatric Cancer, Genomics, Epigenetics, Microenvironment, Immunotherapy
INTRODUCTION:
The treatment of pediatric cancers has been a success story, with current overall survival of ~80% in the U.S. It is estimated that there are currently >500,000 survivors of pediatric cancer in the U.S. Nonetheless, this success has occurred at a significant price, as the prevalence of severe chronic health disorders among long-term survivors of pediatric cancer is 3-fold higher than in matched controls. In addition, there are still close to 13,000 pediatric cancer deaths per year in the U.S, and there are several tumors for which long-term survival remains poor. Recently, the adult cancer research community has seen major advances in detailing the molecular and cellular mechanisms of disease that have resulted in the generation of novel targeted therapies and immunotherapies, some of which are applicable to pediatric tumors. Here, we have reviewed several major topics that we believe have had a major impact already on fundamental understanding of the biology of pediatric cancers and will continue to impact the field going forward. To maintain the current relevance of this review, we have restricted most of our literature search to the last 5–6 years and have included a few older references for context. The following sections focus on new insights into the genetic and epigenetic underpinnings of these cancers, therapeutic breakthroughs, developmental and cellular origins of these tumors, model systems that have provided new perspectives, microenvironmental influences on tumor biology, and immunotherapy approaches to treatment. While a comprehensive review of the current status of pediatric cancer research is of course beyond the scope of this manuscript, we believe that each of the areas discussed will continue to generate new ideas and hypotheses in the near future that hopefully will lead to new and improved therapies that ultimately improve both short-term and long-term outcomes of childhood cancer.
GENOMIC AND EPIGENOMIC PROFILING STUDIES
Pediatric Pan-Cancer Profiling Studies:
Large scale genomic profiling of various hematologic and non-hematologic tumors, so called “pan cancer studies”, have been instrumental in revealing the overall landscape of adult malignancies allowing for the identification of genetic drivers of disease, discovery of mutational signatures and complex structural aberrations, and investigation of drug targets (1,2). Recently, several pediatric pan-cancer genomic studies have been completed and have reported that pediatric tumors contain fewer coding mutations than adult tumors, that TP53 remains the most frequently recurring mutation, and that mutations in epigenetic-associated genes were common events (3–6). Profiling close to 1,700 pediatric leukemias and solid tumors revealed 142 somatic driver gene mutations and found that copy number alterations and structural variants were the most prevalent types of alterations to impact these driver genes (3). Furthermore, this study discovered two new mutation signatures in addition to the existing signatures within the COSMIC (Catalogue Of Somatic Mutations In Cancer) database.
Analysis of DNA sequencing data obtained from 961 tumors across 24 different pediatric, adolescent, and young adult patient histologies identified 34 genomic loci recurrently impacted by copy number alterations (4). This study also demonstrated that of the 77 significantly mutated genes identified across the entire sample cohort, the majority of these mutations were exclusive to specific tumor types (4). Furthermore, the authors highlighted the differences between the genomic landscapes of adults and pediatrics showing that only 30% of the significantly mutated genes identified in pediatric tumors were present in adults. Of the 24 tumor types profiled in this study, osteosarcomas and adrenocortical carcinomas were the most genomically complex, hallmarked by an increased rate of hyperdiploidy and chromothripsis, a catastrophic shattering of chromosomes with subsequent error-prone repair (4,7). Whether the observed genomic complexity in adrenocortical carcinoma and osteosarcoma represents the culmination of persistent chromosomal instability or the product of a single early cataclysmic event is yet to be experimentally determined. Collectively, these pan-cancer studies further highlight the unique biology driving several pediatric cancers (Figure 1). Moreover, the data from these studies validate the seminal publication by Bert Vogelstein and colleagues illustrating the age-associated differences in the mutational landscapes across multiple cancer types (8).
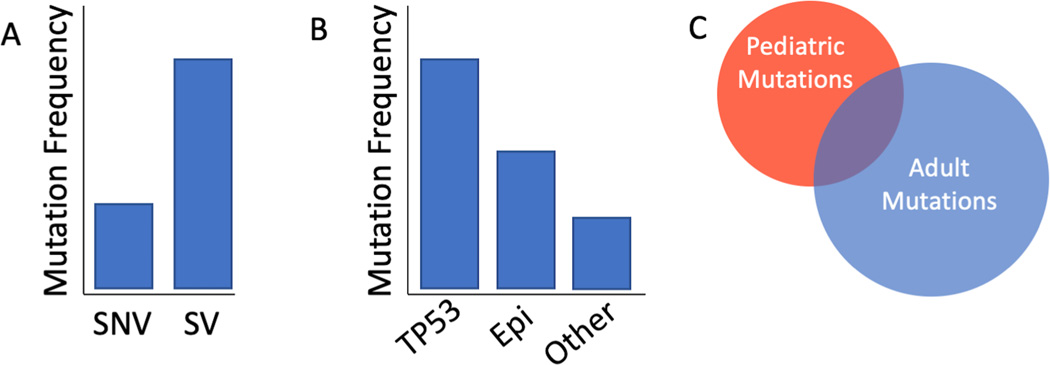
Figure 1: Features of pediatric tumors.
(A) SV’s are more common than SNVs. (B) Somatic mutations in TP53 are frequently recurrent and mutations in epigenetic genes are also common. (C) The mutation spectra between adult and pediatric tumors share little overlap. SNV, single nucleotide variant; SV, structural variant; Epi; epigenetic associated genes.
DNA Methylation Profiling of Brain Tumors
DNA methylation refers to the addition of methyl groups to chromosomal DNA to regulate gene transcription. Recently, numerous studies have emerged demonstrating the utility of DNA methylation analysis to subcategorize central nervous system (CNS) tumors. Genome-wide DNA methylation profiling of 3,093 meningiomas revealed that clear cell meningiomas, a histology predominantly observed in children and young adults, clustered independently of all other subtypes (9). DNA methylation arrays subdivided Group 3 and Group 4 medulloblastomas into 8 different molecular subgroups (10). DNA methylation analysis of pilocytic astrocytoma specimens showed that tumors arising in the infratentorial, midline, and cortical regions of the brain had distinct methylation profiles (11). A separate study comparing diffuse astrocytoma and pilocytic astrocytoma specimens showed that the pilocytic specimens were hypomethylated and had a different methylation profile than the diffuse astrocytoma specimens (12). Integrated epigenetic profiling of CNS atypical teratoid/rhabdoid tumors, identified three disease subgroups with distinct DNA methylation profiles and epigenetic landscapes (13,14). Together, these data illustrate the power and applicability of DNA methylation to subcategorize CNS tumors which has clinical implications pertaining to the refinement of ambiguous diagnoses (15,16).
Mutational Activation of RAS-MAPK Pathway
Upstream and downstream components of the RAS and MAPK pathways are recurrently mutated in various pediatric CNS and solid tumors (4,5,17). Analysis of the Foundation Medicine sequencing data generated from 1,215 different pediatric tumors showed MAPK mutations were common in hypermutant tumors driven by replication repair deficiency and that the RAS/MAPK pathway was indeed active in these tumors (5). By performing functional experiments using pediatric hypermutant glioma and pediatric hypermutant colorectal cancer patient-derived xenograft (PDX) models, the authors showed that these RAS-MAPK mutant replication repair deficient tumors were susceptible to MEK inhibition, highlighting a potential context for synthetic lethality (5).
Whole genome sequencing of matched primary-relapsed neuroblastoma specimens led to the discovery that 78% of relapsed specimens contained mutations in genes involved in RAS-MAPK signaling (18). Similarly, mutations in PHOX2B, CIC, and DMD activated RAS-MAPK signaling in neuroblastoma cell lines and tumor specimens (19). Importantly, both studies demonstrated that mutations in genes involved in RAS-MAPK signaling conferred increased sensitivity to pharmacologic MEK inhibition both in vitro and in vivo (5,19).
In the largest study of its kind, mutational analysis of DNA from 641 rhabdomyosarcoma patients indicated that mutations in the RAS pathway occurred at a frequency of 56% in fusion negative tumors that lack the pathogenomic FOXO1 fusion oncogenes (20). This study also showed that isoform-specific RAS mutations associated with age where HRAS mutations were enriched in infants, KRAS in toddlers, and NRAS in adolescents (20). This incredibly intriguing finding implies that the different RAS isoforms may impart specific age-restricted oncogenic programs.
Somatic mutations to the RAS-MAPK pathway have also been reported in aggressive pediatric hematological malignancies. Survivors of pediatric cancer are at increased risk for subsequent malignancies due to the late effects of their highly cytotoxic chemotherapy treatments (21,22). Genomic analysis revealed a high frequency of KRAS and NRAS mutations in pediatric cancer survivors who had developed therapy related myeloid neoplasms (23). A combination of exome and whole genome sequencing discovered that 44% of relapsed acute lymphoblastic leukemia (ALL) specimens had somatic mutations in NRAS, KRAS, or PTPN11 (24). Using isogeneic leukemia cell lines expressing either wild-type or the G12D KRAS mutant, this study showed that oncogenic KRAS conferred resistance to methotrexate and an unexpected sensitivity to vincristine. Although juvenile myelomonocytic leukemia has long been associated with oncogenic RAS signaling, integrated genomic and DNA methylation analysis revealed that mutations in NRAS, KRAS, and PTPN11 were differentially enriched in independent molecular subgroups (25). While each of the subgroups were associated with different clinical and molecular features, the extent to which the respective somatic RAS pathway mutations contributed to these subgroup-specific features was not reported.
Germline Variants Associated with Predisposition and Increased Disease Risk:
The importance of understanding the deleterious effects of germline mutations in pediatric cancer patients can be traced back to Knudson’s original 1971 publication describing the two-hit hypothesis of tumorigenesis in patients with retinoblastoma (26). Five decades later, the prevalence and impact of germline variants in patients with pediatric tumors is still an active area of research due, in large part, to the advances in genome sequencing technology. A comprehensive summary of the various germline variants associated with pediatric cancers and cancer predisposition syndromes is beyond the scope of this manuscript but have been thoroughly reviewed (27–31). Analysis of germline DNA sequencing data from 1,120 pediatric cancer patients diagnosed with leukemia, CNS tumors, neuroblastoma, retinoblastoma, osteosarcoma, Ewing sarcoma, rhabdomyosarcoma, adrenocortical carcinoma, and melanoma revealed that the overall rate of pathogenic or likely pathogenic germline variants was a striking 8.5% (32). Two additional studies estimate that the frequency of causative germline variants in pediatric cancer patients to be slightly lower at 6% and 6.9%, respectively (4,33). When hematologic malignancies were excluded, the percentage of pediatric patients diagnosed with solid tumors and CNS tumors that carried germline variants in known cancer predisposition genes increased to 12% (34). Strikingly, germline variants were frequently observed in the following DNA repair genes: TP53, BRCA2, PMS2, CHECK2, MSH2, and MSH6 (32,35).
The frequency of germline variants in TP53 varies by tumor type. Germline variants in TP53 increased the risk of developing sonic hedgehog (SHH) medulloblastomas and were associated with an increased prevalence of chromothripsis in these patients (36). This is consistent with the finding that germline variants in TP53 were associated with a higher number of somatic chromosomal structural variations (4). In adrenocortical carcinoma, germline TP53 variants were observed in approximately 50% of patients and rather than clustering in hotspots, these variants were distributed throughout the gene (37). Between 4–5.3% of patients with osteosarcoma harbored pathogenic or likely pathogenic germline TP53 variants, with almost half of these mutations occurring de novo (38,39). These osteosarcoma patients tended to be younger, have disease within the axial skeleton, were more likely to present with metastatic disease upon diagnosis, and had an inferior prognosis (38). Germline TP53 variants were observed at a frequency of 2% in pediatric B-cell ALL (40). According to this study, pathogenic TP53 variants were enriched in older children and those diagnosed with hypodiploid ALL, a more aggressive form of this disease. Moreover, these patients experienced inferior outcomes and were at a higher risk of developing a secondary malignancy (40).
As stated above, there are several germline variants in genes other than TP53 that have also been documented in pediatric cancers. Patients with osteosarcoma were found to have pathogenic or likely pathogenic germline variants in CDKN2A, MEN1, VHL, POT1, APC, MSH2, and ATRX (38). Amazingly, this study concluded that 28% of patients with osteosarcoma had a pathogenic or likely pathogenic germline variant in a cancer predisposition gene (38). Furthermore, the GLDC/IL33 locus has been identified as an osteosarcoma susceptibility locus and two single nucleotide polymorphisms (SNPs), rs3765555 and rs55933544, were associated with significantly decreased survival and decreased IL33 expression (41,42). Data from the Children’s Oncology Group identified germline variants in 15 genes at a frequency of 7.3% in children with rhabdomyosarcoma (43). Similar results were obtained in a separate rhabdomyosarcoma study showing that approximately 7% of patients had a germline variant in a known cancer predisposition gene (44). Interestingly, in both studies these germline variants were enriched in patients with FOXO1 fusion-negative disease. Separate genome wide association studies (GWAS) have identified new SNPs associated with Ewing sarcoma risk at the 6p25.1, 20p11.22, and 20p11.23 loci (45,46). In patients with Ewing sarcoma, pathogenic or likely pathogenic germline variants occurred at a frequency of 13% and were enriched in several genes associated with DNA damage repair (47). GWAS of 707 neuroblastoma patients revealed that the rs6720708 SNP at the BARD1 locus, a known high-risk neuroblastoma susceptibility locus, was significantly associated with the development of adrenal neuroblastoma versus developing thoracic disease (48).
A study consisting of 280 California patients with glioblastoma, anaplastic astrocytoma, and astrocytoma not otherwise specified reported that pathogenic germline variants were present in approximately 11% of the patients and that these variants were enriched in patients with glioblastoma (49). In medulloblastoma, germline variants were differentially enriched according to disease subgroups. The SHH subgroup of medulloblastoma had the highest rate of germline alterations affecting various genes including TP53, SUFU, PTCH1, PABL2, BRCA2, GPR161, and ELP1 (36,50,51). Germline variants in APC were enriched in the WNT subgroup whereas PALB2 and BRCA2 germline variants were observed in SHH, Group 3, and Group 4 medulloblastomas (36).
ETV6 germline variants were associated with increased risk of developing childhood ALL and patients with these predisposing germline variants were more likely to be older at the time of diagnosis and have hyperdiploid ALL, a leukemia subtype with a favorable prognosis (52). The rs3824662 SNP in GATA3 was recently discovered to confer increased risk of relapse in patients with ALL (53). New ALL risk loci were also identified at 5q31.1, 6p21.31, 9q21.31, and 17q21.32 and associated with specific ALL subtypes (54). Two SNPs rs886285 (at 5q31.1) and rs210143 (at 6p21.31) were associated with high-hyperdiploidy ALL, rs10853104 (at 17q21.32) with the ETV6-RUNX1 subtype, and rs76925697 (at 9q21.31) with B-cell ALL (54). Patients with Down syndrome have an extraordinarily high risk for developing various leukemias during childhood, including acute myeloid leukemia (AML), ALL, and megakaryoblastic leukemia (55). ALL patients with Down syndrome exhibited an increased frequency of a risk allele in CDKN2A, a previously established susceptibility locus (56). The IKZF1 SNP rs58923657, which maps to an active B-cell enhancer, was also enriched in patients with Down syndrome and the risk allele decreased the enhancer activity (56). Moreover, silencing IKZF1 preferentially increased the proliferation of lymphoblastoid cells from individuals with Down syndrome versus cells from those without Down syndrome (56). Together, these data suggest that presence of trisomy 21 modifies the phenotypic effect of the rs58923657 risk allele.
Telomeres are complexes of proteins bound to repetitive DNA sequences positioned at the end of each chromosome that shorten with each cell division, thus acting as a cellular clock recording each replicative event. Telomere length is disproportionately associated with cancer incidence where shorter telomeres seem to be protective while longer telomeres confer an increased risk (57–59). For instance, longer telomere length was associated with a higher risk of developing ependymoma with adolescent and adult onset (60). In osteosarcoma, a weighted polygenic risk score revealed that longer telomere length was a risk factor for developing disease in Hispanic, Asian/Pacific Islander, and African American children (61). The association between telomere length and osteosarcoma risk was recapitulated in a separate study where the data were not stratified by ethnicity (62). Additionally, this GWAS study also found that SNPs in known telomere maintenance genes increased the risk of developing neuroblastoma and ALL (62).
Given that pediatric tumors are rare, germline variants that moderately increase risk in adult disease may indeed have a larger affect size in certain pediatric tumors. That said, not all germline variants in known cancer predisposition genes are causative of disease but may impact disease severity and/or other phenotypic aspects of the tumor. Similarly, germline variants in genes not previously associated with adult diseases may have biological and/or clinical relevance in children and vice versa (63). However, the accessibility to large scale genomic databases combined with growing institutional genomic profiling efforts affords researchers a collaborative opportunity to perform clinically annotated genotyping studies to further investigate the impact of germline variants in pediatric cancers. For example, the MSK-IMPACT study (NCT01775072) is performing prospective clinical germline sequencing on pediatric solid tumor patients and using the resultant data to identify actionable targeted therapeutics and prioritize patients and families that may benefit from further screening/surveillance (64). These types of clinical studies can also inform the surveillance protocols for these patients. In 2017, the Pediatric Cancer Working Group of the American Association for Cancer Research convened a Childhood Cancer Predisposition Workshop where a set of screening and surveillance recommendations were established. This meeting included the discussions on the use and interpretation of whole-body MRI, the need for registries to enable natural history studies, the consultancy role of syndrome-specific centralized centers of excellence, and modifications to existing surveillance protocols (65–71).
In summary, the widespread adoption of advanced genomic technologies in both the clinic and the research laboratory have led to the discovery of numerous germline variants. Experimental investigations aimed at determining the functional impact of these and other reported variants in racially and ethnically diverse cohorts will further our understanding of the biological and clinical significance of germline genetics in pediatric malignancies. Nonetheless, the current data clearly indicate that subsets of pediatric cancer patients are genetically predisposed to developing disease and supports the observed inverse relationship between germline variants and age (37,38,72) (Figure 2).
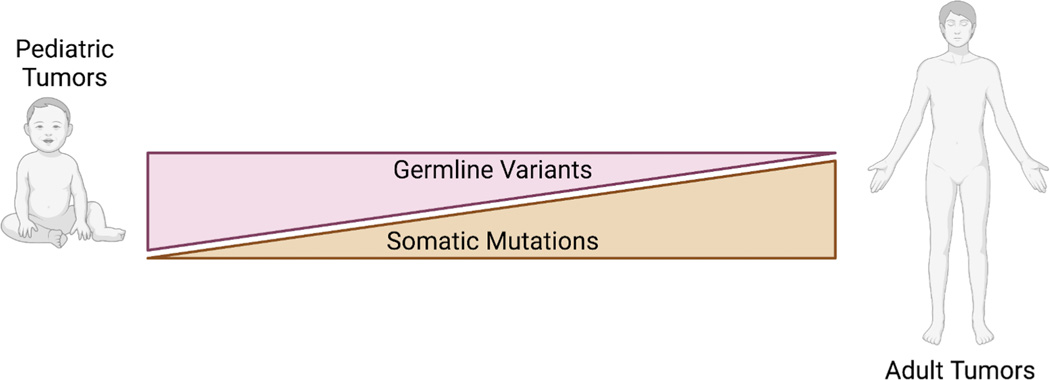
Figure 2: Comparison of germline and somatic mutations in pediatric versus adult cancers.
Germline variants are more frequently observed in pediatric cancer patients than in adult cancer patients whereas adult tumors have an increased somatic mutation burden than pediatric tumors.
Liquid Biopsies
Disease monitoring is an important aspect to the management of both pediatric and adult disease. However, unlike the adult setting where repeated biopsies are more readily obtained, performing serial biopsies in pediatric patients has traditionally been discouraged unless deemed medically necessary. This “one and done” approach constrains the ability to identify and/or develop reliable biomarkers for the longitudinal study of central nervous system (CNS) and solid tumors in children. Liquid biopsies are minimally invasive tests that utilize accessible body fluids (i.e.: blood, urine, saliva, and/or cerebral spinal fluid) to detect cellular and/or molecular biomarkers of a given disease such as cell free DNA (cfDNA), circulating tumor cells, microRNAs, proteins, and metabolites (73–75). Emerging data suggests that liquid biopsies may circumvent these sampling limitations and allow for longitudinal assessments of disease burden. Detection and quantification of cfDNA has been documented in various solid and CNS tumors of childhood, with the earlier studies largely relying on polymerase chain reaction-based approaches (76). These methods were reliable in detecting cfDNA from diseases in which known recurrent driver mutations existed, for example histone mutations in high grade glioma (HGG) (77,78).
More recently, next generation sequencing (NGS) based methods have been employed and have the potential to identify mutations in tumors a priori. NGS profiling studies of cfDNA from patients with neuroblastoma, Ewing sarcoma, Wilms tumor, osteosarcoma, alveolar rhabdomyosarcoma, medulloblastoma, and high-grade glioma have consistently reported positive associations between cfDNA abundance and disease burden (78–88). The anatomical site from which cfDNA is obtained can impact the cfDNA yield for subsequent downstream analyses. Studies have shown improved detection of cfDNA isolated from cerebral spinal fluid versus that of plasma in patients with medulloblastoma and brain stem glioma (84,88). For Wilms tumor, detecting mutations in cfDNA isolated from urine was not only achievable, but more mutations were detected in the urine versus plasma from these patients (83). Optimization of this urine-based Wilms tumor cfDNA profiling approach is particularly appealing as it presents zero risk to the patient, allows for serial collections in large quantities, and can be done at home. Moreover, this study provides rationale to investigate the applicability of cfDNA profiling in other childhood kidney tumors such as renal cell carcinoma, malignant rhabdoid tumors, and clear cell sarcoma of the kidney.
cfDNA has also been used to gain important biological insight. Longitudinal profiling of cfDNA from patients with neuroblastoma demonstrated the ability to identify clonal populations with distinct mutational patterns, model clonal evolution throughout the treatment regimen, and identify relapse specific clones (79). Analysis of gene-specific 5-hydroxymethylcytosine modified cfDNA from neuroblastoma patients identified unique profiles associated with disease burden (81). Further analysis revealed differentially enriched biological processes and pathways in patients with and without active disease. Methylation analysis of cfDNA isolated from the cerebral spinal fluid of patients with medulloblastoma was used to derive an epigenetic signature based on differentially methylated CpG sites that could specifically and sensitively identify medulloblastoma subtypes (85).
Several companies have gained, or are seeking, US Food and Drug Administration (FDA) approval for their respective adult oncology liquid biopsy assays including Guardant, FoundationOne, Grail, and Thrive (89–95). Currently, liquid biopsies have not yet been approved for use with pediatric patients but trials investigating the clinical utility of liquid biopsies to monitor disease burden and treatment response in various pediatric cancers are underway. It is likely that additional studies on the utility of liquid biopsies will increase in the coming years and ultimately integrate into standard of care for patients with certain tumor types.
Filling in the Knowledge Gaps
Despite the recent advances in exploring the genomic landscapes of various pediatric cancers, knowledge gaps remain. One such knowledge gap is that there are only a few studies that detail the genomic and transcriptomic profiles of specimens from patients with recurrent, refractory, and/or relapsed CNS and solid tumors as the majority of published genomic studies were performed using treatment naive specimens. The emerging data from the few studies performed on recurrent, refractory, and/or relapsed specimens from pediatric patients with CNS and solid tumors suggest a comparatively higher mutation burden compared to treatment naïve patient specimens (4,18,96–98). Additional descriptive and functional studies are needed to uncover the mechanisms associated with disease progression in each of these tumors and tumor subtypes.
Another knowledge gap is that most of the genomic profiling data has been generated using short-read sequencing methods. While the identification of single nucleotide variants from these data is well established, many short-read sequencing analytical tools struggle to resolve complex genomic aberrations, necessitating the implementation of sophisticated algorithms, each with their own biases and limitations (99,100). Long-read sequencing platforms routinely produce continuous sequencing read lengths of >10kb and have been demonstrated to resolve complex genomic rearrangements, including chromothripsis (101,102). As part of the Gabriella Miller Kids First (GMKF) Pediatric Research Program, long-read sequencing technologies will be used to uncover the genomic complexities associated with certain pediatric cancers and birth defects. It is anticipated that long-read sequencing technologies may supplement the existing data to better resolve the complex structural variants inherent to tumors such as osteosarcoma and adrenocortical carcinoma.
BREAKTHROUGHS IN TARGETED THERAPIES
Unlike adults, few efficacious targeted therapies exist for childhood cancers. Recently, larotrectinib and selumetinib received FDA and European Medicines Agency (EMA) approval, a significant therapeutic advance for the treatment pediatric cancer patients bearing driver mutations targeted by these kinase inhibitors. Larotrectinib targets TRK proteins and is indicated for cancers with oncogenic TRK-fusions such as those found in 2.22% of pediatric patients (103,104). The overall response rate for patients with TRK-fusion positive tumors treated with Larotrectinib was 79% with 16% having complete responses (105). In addition to its potent and durable antineoplastic effect, larotrectinib improved the quality of life for patients, the gold standard for any therapeutic agent (106).
Selumetinib is a MEK1/2 inhibitor indicated for patients with neurofibromatosis type 1 with inoperable plexiform neurofibromas (107–109). Of the patients treated with selumetinib, >70% exhibited a confirmed durable partial response of >20% reduction in tumor size (107,108). Impressively, all the patients who had received selumetinib had experienced some degree of tumor shrinkage. Selumetinib is also currently under clinical investigation for pediatric patients with BRAF mutant pilocytic astrocytoma and NF1-associated low-grade gliomas (110). The approval of selumetinib marks the first ever approved targeted therapy for patients with neurofibromatosis. Moreover, these two targeted agents validate the necessity of a pediatric drug development pipeline.
EPIGENETIC DRIVERS OF PEDIATRIC CANCER
Histone H3K27:
Since the first reports of somatic histone H3 mutations at lysine 27 (H3K27M) and glycine 34 (H3G34) in pediatric high-grade gliomas (HGG) in 2012, the interest in these “onco-histones” has sparked numerous investigations aimed at understanding their role in tumorigenesis (111,112). Functional studies show that the H3K27M mutation increased the self-renewal capacity of neurospheres in vitro whereas CRISPR-mediated correction of the H3K27M mutation in glioma cell lines decreased cell proliferation (113,114). In genetically engineered murine HGG models, the presence of the H3K27M mutation decreased tumor latency (113,115). Multiple studies have shown that the H3K27M mutation regulated genes involved in neurogenesis and differentiation (113,114,116–118). Bivalent promoters are chromatin domains that are marked by both repressive and activating histone modifications. Mutant H3K27M histones colocalized with the H3K27Ac mark at bivalent promoters and enhancers, diminished the deposition and spreading of H3K27me3 marks, and allowed for transcription of polycomb repressed genes (113,114,117,118). Interestingly, the H3K27M mutation did not sequester polycomb repressor complex 2 (PRC2) (117,118). Correction of the mutation restored the balance between H3K27me3 and H3K27Ac marked loci, reestablished polycomb mediated repression of developmental genes, and promoted glial lineage commitment (114,116). Therapeutically, H3K27M mutant cell lines were sensitive to pharmacologic inhibition of EZH2, the PRC2 histone methyltransferase, and did so in a p16INK4a-dependent manner and such interventions prolonged the survival of tumor bearing mice (115). Bromodomain and extra-terminal (BET) proteins are “chromatin readers” that bind to acetylated lysines to recruit chromatin remodelers that subsequently regulate transcription. The BET inhibitor JQ1 forced the differentiation of H3K27M mutant glioma cells in vitro and prolonged survival in vivo (118). Interestingly, an aggressive subset of pediatric posterior fossa ependymomas have been identified as having low levels of H3K27me3 and increased levels of H3K27Ac (119). These tumors exhibited DNA methylation profiles like that of H3K27M mutant gliomas as well as overlapping H3K27me3 ChIP-seq peaks. This signifies that histone mutations are not the only means by which to achieve the molecular and phenotypic characteristics attributed to the H3K27M mutation.
Enhancer Dysregulation:
Enhancers are regulatory chromatin domains that recruit transcriptional machinery to promote or repress gene expression from a specific locus. The accessibility and activity of enhancers are developmentally regulated and associated with lineage commitment but can be dysregulated to aberrantly drive oncogenic transcriptional profiles. In pediatric HGG, the presence of the H3G34 mutation arrested the cells in a developmental state where the chromatin conformation juxtaposed the GSX2 enhancer and PDGFRA locus and allowed for PDGFRA to hijack the enhancer of this developmentally regulated transcription factor to drive pathologic PGDFRA expression (120). Enhancer hijacking has also been observed in MYCN-amplified neuroblastoma. An integrative genomic approach identified two classes of MYCN amplicons (121). Class I amplicons were simple amplification events that contained both MYCN and its local enhancer whereas class II amplicons contained MYCN without the local enhancer. Further investigation revealed that these Class II amplicons were double minute chromosomes and circumvented the lack of the local MYCN enhancer by hijacking distal enhancers through complex chromosomal structural rearrangements (121).
Differential enhancer activity was observed in matched primary-metastatic osteosarcoma tumors and isogenic cell line pairs, revealing and that these metastatic-specific enhancers promoted transcriptional programs necessary for metastatic seeding (122). Further analysis indicated that an enhancer regulating the expression of coagulation factor 3 was positively selected for in the metastatic cell lines and that perturbation of this enhancer reduced pulmonary metastasis in vivo (122). The EWS-FLI1 chimeric oncoprotein is the product of the EWSR1-FLI1 translocation that defines 85% of Ewing sarcoma tumors. GGAA repeats are the known EWS-FLI1 binding motif in Ewing sarcoma (123). Recent data demonstrated that these GGAA repeats are enriched at super-enhancers, were bound by EWS-FLI1, and identified MEIS1, a developmentally regulated homeobox protein, as a super-enhancer driven oncogenic transcription factor (124). Additionally, MEIS1 and EWS-FLI1 co-localized at a subset of super-enhancers to further drive transcriptional dysregulation.
Recently both mesenchymal stem-like and adrenergic-like cells were identified in primary and established neuroblastoma cell lines, and each subpopulation of cells had unique super-enhancer landscapes that were responsible for maintaining their respective lineage identities (125). Additional functional analysis showed that ectopic expression of the mesenchymal stem-like super-enhancer associated transcription factor PRRX1 partially reprogramed the adrenergic-like neuroblastoma cells towards a more mesenchymal stem-like state as evidenced by global shifts in the super-enhancer landscape and transcriptional profiles (125).
The PAX3/7-FOXO1 translocations, the hallmark chromosomal aberrations in alveolar rhabdomyosarcoma, brings PAX3/7 gene transcription under the control of the FOXO1 super-enhancer (126). Mutations in the RAS-MAPK pathway drive a subset of fusion negative rhabdomyosarcomas tumors (127). In this context, RAS-mutant cells were developmentally arrested due to oncogenic MAPK signaling which imparted a RAS-dependent super-enhancer landscape (128). Inhibition of MAPK signaling by trametinib altered the chromatin accessibility and restored the myogenic super-enhancer landscape to drive differentiation of fusion negative rhabdomyosarcoma cells (128). SNAI2 was shown to compete with MYOD1, a master myogenic transcription factor, and inactivate myogenic enhancers to halt differentiation in fusion negative rhabdomyosarcoma (129). A separate study demonstrated that TWIST2 repressed myogenesis by both upregulating SNAI2 expression and competing with MYOD1 binding at target genes associated with muscle differentiation (130). The loss of H3K27Ac and deposition of H3K27me3 at TWIST2-bound myogenic loci indicated that TWIST is capable of recruiting PRC2 to inhibit differentiation in fusion negative rhabdomyosarcoma cells.
Pioneer Factors:
Pioneer factors are the first transcription factors to bind specific heterochromatic loci and alter the local chromatin accessibility by recruiting additional chromatin remodelers. OTX2 is a developmentally regulated transcription factor that was found to occupy several active enhancers in Group 3 medulloblastoma cells (131). Additionally, NEUROD1, an established neuronal pioneering factor, was shown to interact with OTX2 at enhancers (131,132). Ectopic expression of OTX2 induced a Group 3 medulloblastoma chromatin accessibility and enhancer signature in mesenchymal stem cells, suggesting that OTX2 functions as a pioneer factor (131). The EWS-FLI1 oncogenic fusion gene endowed Ewing sarcoma cells with a unique enhancer profile (133). Mechanistically, EWS-FLI1 bound to GGAA repeats to remodel chromatin and generate de novo enhancers and displaced ETS transcription factors to promote transcription of target genes (134). The PAX3-FOXO1 fusion was found to function as a pioneering factor in alveolar rhabdomyosarcoma and created de novo super-enhancers that drove oncogenic and myogenic transcription programs in a BRD4-dependent manner (135). BRD4 is a member of the BET family of “chromatin readers” and CHD4 is a chromatin remodeler. CHD4 interacted with BRD4 and colocalized with PAX3-FOXO1 at a subset of super-enhancers in alveolar rhabdomyosarcoma (136). These data also demonstrated that PAX3-FOXO1 binding was, at least in part, dependent on the presence of CHD4 chromatin remodeling (136).
Redirecting/Hijacking of Chromatin Remodelers:
BAF is the mammalian equivalent of the SWI/SNF chromatin remodeling complex, of which SS18 is a member. The SS18-SSX chimeric oncoprotein, the gene product of the translocation that defines synovial sarcoma, competed with SS18 to hijack the BAF complex (137). The SS18-SSX fusion antagonized PRC2-mediated gene repression by directing the BAF complex away from enhancers and towards bivalent genes to promote aberrant gene transcription (138,139). Additionally, the histone demethylase KDM2B, a member of the non-canonical PRC1.1 complex, has been shown to recruit BAF complexes containing the SS18-SSX fusion to repressed developmentally regulated genes, resulting in their transcription (140).
At active enhancers, the interaction between EWS-FLI1 and RING1B of the PRC1 complex was necessary for target gene expression (141). These data also showed that RING1B maintained its repressive function at non-EWS-FLI1 bound loci. Together, the data suggest that RING1B targets EWS-FLI1 to repressed enhancers to recruit additional chromatin modifiers and transcription factors to activate these enhancers and subsequent transcriptional programs (141). The prion domain present in wild type EWS is retained in the EWS-FLI1 fusion and this domain was deemed critical for recruiting the BAF complex to activate enhancers (142).
As previously mentioned, most pediatric tumors have a relatively low mutational burden. It is becoming clear that in contradistinction, many pediatric tumors have significantly reprogramed epigenomes, leading to widespread transcriptional dysregulation. Better mechanistic understanding of the means by which this epigenetic reprograming occurs may lead to new therapeutic “targeted” approaches in a variety of pediatric tumors.
DEVELOPMENTAL AND CELLULAR ORIGINS OF PEDIATRIC TUMORS
Despite decades of research and various hypotheses, the question regarding the cell of origin of childhood tumors has plagued the field of pediatric cancer research. Recent advances in single cell RNA sequencing (scRNA-seq) technologies combined with powerful informatic analyses have allowed investigators to more deeply probe this question. Since many pediatric tumors are “embryonal” in appearance, an integrated approach utilized across various studies from multiple groups has involved comparing the single cell transcriptional profiles of normal developing tissues with that of tumors derived from the same tissue. Comparative single cell analysis of transcriptomes from normal human fetal adrenal glands and dissociated neuroblastoma tumors suggested that, of the cells profiled, neuroblastoma cells most closely resembled developmentally arrested sympathoblasts (143). Strikingly, this comparison was consistent across the different neuroblastoma risk groups. A separate study used scRNA-seq data from embryonic and post-natal murine adrenal glands to demonstrate that neuroblastoma cells significantly correlated with neuroblasts, a cell type whose signature partially overlapped with that of sympathoblasts (144). This study also highlighted a subset of neuroblastoma cells that closely resembled Schwann cell precursors which were hallmarked by decreased MYCN and ALK expression (144). Another study mapped neuroblastoma cells to cells isolated from early human embryos and fetal adrenal glands and showed that neuroblastoma cells most closely resembled undifferentiated chromaffin cells (145). In these studies, chromaffin cells clustered adjacently to sympathoblasts, thus approximating the potential cellular origins of neuroblastoma (143–145). The creation of a single cell transcriptome atlas derived from healthy fetal, pediatric, adolescent, and adult kidneys and ureters identified ureteric bud cells and primitive vesicle cells of the developing nephron as the cells that most closely correlated with Wilms tumor cells (146).
Similar comparative scRNA-seq approaches have been used to identify the cellular origin in pediatric brain tumors. Comparing scRNA-seq data from human medulloblastoma cells to cells from the developing mouse brain revealed that WNT subgroup medulloblastoma cells were heterogenous and did not explicitly correlate with any cell type in the murine cerebellum but, in fact, resembled lower rhombic lip mossy-fiber neurons of the pons (147,148). Group 4 medulloblastoma cells mapped to unipolar brush cells and glutamatergic cerebellar nuclei (147,148). The SHH subgroup were enriched in granule neuron progenitors that varied in their differentiation status in accordance with age, highlighting the differences between adult and pediatric disease (147). scRNA-seq on both the developing brain and tumor cells from a mouse model of SHH medulloblastoma revealed significant correlations between cerebellar granule neuron progenitor cells and the murine tumor cells, thereby independently validating the conclusions of similar studies (147–149). Projection of bulk RNA-seq data onto a single cell atlas derived from scRNA-seq data of the developing human brain stem, murine pons, and murine forebrain identified neural progenitor cells as the potential cell of origin for H3K27M pontine gliomas (148). Similarly, the H3G34 mutation arrested HGG tumor cells in a developmental state most consistent with interneuron progenitor cells (120). Analysis of embryonal tumors with multilayered rosettes suggested that these tumors were likely derived from fetal radial glial cells (148). The cellular origins of atypical teratoid/rhabdoid tumors (ATRT) were more ambiguous and, while the precise cell of origin was not identified, the data suggested that these tumors likely arose from early progenitor cells of non-neuroectodermal origin (148).
EXPERIMENTAL MODEL SYSTEMS
Comprehensively Profiled Patient Derived Xenografts
In comparison to adult diseases, the rate of progress in the field of pediatric cancer research has been somewhat constrained due to the relative rarity of these diseases, which ultimately equates to a paucity of reliable models for experimentation. PDX models help to circumvent this issue as the implanted tumor cells reflect the natural history of the disease. However, assessing the fidelity of these models is of paramount importance in interpreting the resultant data, thus necessitating comprehensive characterization of these models. Recently, different groups have established large banks of clinically annotated and genomically profiled pediatric PDX models (98,150–154). Collectively, these PDX models comprised hematologic malignancies, solid tumors, CNS tumors, and rare histologies obtained from patients with primary, relapsed, and/or metastatic disease and have been made available to the research community (Table 1). A combination of genomic, transcriptomic, and epigenomic profiling concluded that these PDX models closely resembled the original tumor tissues from which they were derived.
Table 1:
PDX Repositories
| Childhood Cancer Repository | www.cccells.org/xenografts |
| EuroPDX Consortium | www.europdx.eu |
| PDX Development and Trial Centers Research Network | www.pdxnetwork.org |
| PDXFinder | www.pdxfinder.org |
| Pediatric Preclinical Testing Consortium | www.ncipptc.org |
| Childhood Solid Tumor Network at St Jude Children’s Research Hospital | www.stjude.org/research/why-st-jude/data-tools/childhood-solid-tumor-network.html |
Genomic data derived from molecularly characterized osteosarcoma PDX models were used to identify druggable targets and demonstrated the efficacy of genome-informed therapeutic approaches (150). Genomically characterized PDX models of high-risk rhabdomyosarcoma were used to conduct Phase II and Phase III pre-clinical trials and identified that the addition of a WEE1 inhibitor improved the therapeutic response to irinotecan and vincristine (151). A separate Pediatric Preclinical Testing Consortium study evaluating the efficacy of WEE1 inhibition in combination with irinotecan in neuroblastoma, osteosarcoma, and Wilms tumor xenografts yielded similar results (155). Importantly, these two independent studies provided compelling pre-clinical data resulting in the establishment of a Phase I/II clinical trial to investigate the combination of WEE1 inhibition with irinotecan in pediatric patients with relapse or refractory solid tumors (NCT02095132).
While these thoroughly characterized models are useful in facilitating the study of cell autonomous mechanisms of disease and preclinical identification/evaluation of therapeutic targets, they are not ideal for mechanistic immuno-oncology investigations, which require an intact immune system not present in PDX models. Expanding the use of these models into humanized mice can partially address this deficiency, albeit certain species incompatibility issues and other limitations remain (156). Furthermore, it is important to note that, although PDX models exhibit high fidelity with respect to the source material, sampling bias still exists, due to the variable extent of intratumoral heterogeneity present in CNS and solid tumors. Therefore, depending on the abundance and distribution of clonal and/or sub-clonal populations present in the tumor location from which the specimen was obtained, the PDX generated from this material may or may not be representative of the bulk tumor, but rather a regional sub-population. Nonetheless, PDX models are incredibly valuable in advancing the field of pediatric cancer research.
Patient Derived Organoids
Several factors impact the engraftment of tumor tissues and thus the rate of successful PDX generation varies markedly. The patient derived organoid is a newer model system that fills the gap between PDX models and patient derived cancer cell lines (157). Recently a pediatric renal tumor organoid biobank comprised of 54 organoids derived from tumors including Wilms tumor, metanephric adenoma, malignant rhabdoid tumors, renal cell carcinoma, congenital mesoblastic nephroma, and a hyperplastic intralobular nephrogenic rest was established (158). Corresponding normal tissue organoids were also developed. A hepatoblastoma tumor-normal organoid pair was also recently reported (159). In both studies, genomic and transcriptomic profiling revealed that the organoid models resembled the tumors from which they were derived and demonstrated the applicability of these tumor derived organoids for in vitro drug screening (158,159).
Somatic Genome Engineering of Oncogenic Translocations
Numerous pediatric solid tumors are defined by hallmark translocations that give rise to fusion oncoproteins that drive disease, most notably Ewing sarcoma (EWSR1-FLI1), desmoplastic small round cell tumors (EWSR1-WT1), and rhabdomyosarcoma (PAX3/7-FOXO1). Somatic genome engineering via CRISPR-Cas9 has been used to successfully generate these translocations. Functional EWSR1-FLI1 translocations have been engineered into HEK293 cells (160,161) as well as mesenchymal stem cells and induced pluripotent stem cells (162). Functional EWSR1-WT1 translocations have also been engineered using CRISPR-Cas9 (161,163). The PAX3-FOXO1 translocation was engineered into mouse myoblasts using a Cre-Lox strategy to invert the FOXO1 locus followed by CRISPR-Cas9 to create intronic double strand breaks in PAX3 and FOXO1 (164). Here, the rate of successful translocation formation varied between forelimb and hindlimb myoblasts and was attributed to the differences in the 3D genome organization and physical proximity of the PAX3 and FOXO1 loci.
All experimental models have inherent flaws, and their utility is dependent upon the specific questions being interrogated. However, a thorough comprehension of the benefits and limitations of each model will allow for strategic utilization of the most appropriate model(s) to generate useful data that will drive the field forward. It is likely that integrated studies that utilize scRNA-seq of tumor tissues and/or organoids to map the cellular origins of disease will be critical in the development of new models where disease-specific mutations are engineered into the presumed permissive cells of origin. If successful, this approach could provide new insights into diseases such as Ewing sarcoma, where despite immense effort put forth by international groups of researchers, murine models have yet to be successfully engineered (165).
IMMUNE MICROENVIRONMENT PROFILES OF PEDIATRIC TUMORS
The tumor microenvironment is comprised of neoplastic cells, immune cells, fibroblasts, endothelial cells, pericytes, various extracellular matrix components, growth factors, soluble stimulatory and inhibitory molecules, nutrient gradients, and variable oxygen tension that work in concert to sustain tumor growth (166). Over the last decade, the number of tumor microenvironment studies in adult cancers has increased dramatically. While still less studied, recent reports documenting the landscape of the tumor microenvironment in different pediatric cancers have increased. The use of computational tools to impute cell types from RNA sequencing data has significantly contributed to this increase by allowing for data mining and retrospective inquiry of established datasets (167–169). Functional studies are also beginning to emerge, albeit at a reduced frequency which is likely attributable to a scarcity of robust immunocompetent tumor models to study. In reviewing the available literature, one more consistent finding is that the molecular and cellular microenvironmental profiles of pediatric CNS and solid tumors is incredibly heterogeneous and are thus deserving of their own discussion.
Osteosarcoma:
scRNA-seq analysis of osteosarcoma specimens revealed marked cellular heterogeneity between primary, metastatic, and recurrent disease states (170). This study also reported a decreased abundance of CD4+ and CD8+ lymphocytes in the recurrent and metastatic specimens with CD8+ cells expressing markers of T-cell exhaustion (170). This data is consistent with reports that metastatic osteosarcoma specimens contained fewer CD8+ lymphocytes than non-metastatic specimens, expressed low cytotoxicity scores, and exhibited lymphocyte exclusion (171–173). Additionally, an integrated multi-omic analysis showed that osteosarcomas exhibited an ineffective immune response, and low neoantigen expression (174). This study also showed that copy number alterations inversely correlated with immune cell abundance. One of the most striking findings from this study was the data showing the correlation between patient age and tumor inflammation with tumor infiltrating lymphocytes being more abundant in adult specimens versus pediatric specimens (174). This data further highlights the differences between pediatric and adult disease.
Myeloid cells are an important feature of the osteosarcoma microenvironment. Analysis of osteosarcoma specimens demonstrated that myeloid lineage cells were the most abundant immune cells present in these tissues (170). Clustering analysis of this data identified 10 distinct subgroups of myeloid cells, highlighting the heterogeneity of myeloid lineage cells in osteosarcoma. Experimental data showed that M2 tumor associated macrophages (TAMs) promoted osteosarcoma metastasis and that treatment with all-trans retinoic acid reduced TAM-dependent metastasis in vivo (175). Myeloid derived suppressor cells (MDSCs) are highly suppressive immature myeloid cells with demonstrated ability to limit the efficacy of chimeric antigen receptor (CAR) T-cells in osteosarcoma models in vivo (176,177). All-trans retinoic acid was sufficient to eliminate monocytic MDSCs and relieve the suppressive phenotype of granulocytic MDSCs (177). These studies highlight the biological significance, phenotypic plasticity, and functional heterogeneity of myeloid lineage cells within the osteosarcoma microenvironment.
Ewing sarcoma:
Ewing sarcoma specimens exhibited the lowest lymphocyte abundance when compared to other pediatric solid tumors and the presence of CD8+ T-cells did not confer a survival benefit to patients (178,179). Examination of pregnancy-associated plasma protein-A (PAPPA) expression across various pediatric solid tumors illustrated that Ewing sarcoma had the highest expression levels (180). Functional analysis demonstrated that inhibition of PAPPA expression upregulated the expression of numerous genes associated with an active immune response, suggesting that PAPPA is immunosuppressive. Immunohistochemistry revealed that suppressive HLA-G+ lymphocytes outnumbered HLA-G- lymphocytes in Ewing sarcoma biopsy specimens and that HLA-G was upregulated on xenografted tumors cells in response to CAR T-cell treatment (181). Moreover, CCL21, an immunostimulatory cytokine, was shown downregulated in metastatic patient specimens, was inversely correlated with the CD4+/CD8+ T-cell ratio and was associated with a better prognosis (182). Together, these data highlight the immunosuppressive nature intrinsic to Ewing sarcoma. Exactly how the EWS-FLI1 fusion oncoprotein contributes to the immunosuppressive phenotype of Ewing sarcoma is not yet known.
Neuroblastoma:
Gene expression and immunohistochemical analysis of neuroblastoma specimens revealed that T-cells positively correlated with intratumoral dendritic cells (DC) and natural killer (NK) cells, both of which served as positive prognosticators of overall survival (183). When compared to other pediatric solid tumors, the gene expression level of CD200 was significantly higher in neuroblastoma specimens than in any of the other tumor types (184). Moreover, CD200-high neuroblastoma specimens contained fewer CD4+ and CD8+ T-cells and expressed lower levels of IFNγ and TNFα, associating CD200 with a dampened T-cell response. MYCN-amplified neuroblastoma tumors were shown to be less immunogenic than non-amplified tumors as evidenced by lower immune scores and decreased MHC class I gene expression (185,186). Interestingly, a separate study using RNA sequencing data from the TARGET (Therapeutically Applicable Research to Generate Effective Treatments) and GMKF cohorts demonstrated that the activation of downstream MYCN transcriptional programs, rather than amplification of MYCN itself was inversely correlated with T-cell abundance and tumor inflammation (187). This study also showed that increased T-cell infiltration and a higher neoantigen load were independently associated with improved overall survival in these patients (187). For high-risk MYCN non-amplified neuroblastoma specimens with a high MYCN gene signature, T-cell clonal expansion was associated with improved outcomes while a subgroup of these specimens demonstrated T-cell exhaustion (185). Increased abundance of CSF1R+ myeloid cells was associated with inferior relapse-free survival for patients with neuroblastoma and therapeutically targeting these cells both decreased tumor burden and sensitized animals to PD-1 inhibition (188).
Wilms Tumor
In Wilms tumor specimens, an inverse relationship between M1 macrophage abundance and tumor stage was observed, whereas the opposite was true for M2 macrophages (189). Functional studies show that loss of the tumor suppressor WT1 upregulated COX2 expression in normal kidneys and that Wilms tumors derived from the WT1-IGF2 murine model demonstrated robust COX2 expression, elucidating one possible mechanism by which Wilms tumor cells promote a suppressive microenvironment (190). This study also highlighted the immunosuppressive microenvironment of Wilms tumors as indicated by increased abundance of regulatory T-cells, the expression of suppressive cytokines, and a diminished Th1 response when compared to normal kidneys (190).
Rhabdomyosarcoma
While the abundance of infiltrating immune cells did not dramatically differ between embryonal and alveolar rhabdomyosarcoma, their distribution throughout the tumor tissue did indeed vary. The vast majority of both CD3+ and CD163+ cells localized within 15um of tumor blood vessels in alveolar specimens whereas the perivascular localization in embryonal specimens was more diffuse (191). Additionally, tertiary lymphoid structures were more frequently associated with the alveolar histology. Consistent across the histologies was that both embryonal and alveolar rhabdomyosarcoma contained higher numbers of TAMs than T-cells (191,192). Fibrocytes have been shown to promote an immunosuppressive microenvironment and undermine the efficacy of immune checkpoint inhibition in a syngeneic embryonal rhabdomyosarcoma model (193).
Medulloblastoma
Analysis of immune infiltrates across multiple CNS tumors revealed a consistent inverse correlation between tumor grade and immune cell content (194). In comparison to the other major brain tumor histologies, medulloblastomas were characterized as being immunologically “cold” (195). However, within medulloblastomas, the relative abundance of immune cells varied by subgroup with SHH and WNT tumors having higher proportion of CD8+ T-cells than Group 3 and Group 4 tumors (194). In low-risk Group 3 medulloblastoma, a decreased T-reg abundance was associated with inferior progression free survival (194). A murine model of SHH and Group 3 medulloblastoma also revealed subtype-specific differences where SHH medulloblastoma tumors contained higher numbers of infiltrating lymphocytes and myeloid-lineage cells than Group 3 tumors (196). Interestingly, a murine model of SHH medulloblastoma demonstrated that tumoricidal TAMs were recruited to the tumors in a CCL2/CCR2-dependent manner, suggesting a beneficial role of macrophages in the SHH medulloblastoma microenvironment (197). A separate study using a p53-mutant medulloblastoma model demonstrated that mutant p53 inhibited the presentation of MHC class-I by negatively regulating the expression of TAP1 and ERAP1, molecules needed for successful antigen loading (198). Importantly, activation of TNFR2 and LTβR signaling was sufficient to restore MHC class-I expression in the tumor cells in vitro.
Glioma
The microenvironmental heterogeneity of pediatric gliomas varied in accordance with grade, subgroup, and histone mutation status (194,199,200). Multiplexed immunofluorescent imaging showed that low grade gliomas (LGG) had a higher density of CD3+ cells than HGG and that, amongst the LGGs, pilocytic astrocytomas had the lowest T-cell density (200). This study also reported increased vascularity in recurrent versus primary pilocytic astrocytomas. Immunohistochemical evaluation of infiltrating immune cells demonstrated that there were no statistically significant differences in the abundance of natural killer cells or CD163+ myeloid cells between LGG and HGG (201). In a separate study, diffuse intrinsic pontine gliomas were shown to be “immunologically cold” tumors with little immune infiltrate (202,203). Proteomic and phospho-proteomic analysis revealed marked heterogeneity in the immune microenvironment of both LGG and HGG and showed that different subgroups of gliomas were classified as “hot” or “cold” irrespective of histology and/or diagnosis (195).
The pediatric tumor microenvironment field, while gaining interest, is still emerging and additional studies are needed to comprehend the dynamic interaction between tumor cells and the various components of their microenvironment. Studies directed at answering important questions pertaining to the microenvironmental mechanisms that promote immunosuppression and drive resistance to current immunotherapies are of critical clinical importance and will contribute to shaping the therapeutic landscape.
IMMUNOTHERAPEUTIC APPROACHES FOR PEDIATRIC TUMORS
The 2011 approval of ipilimumab by both FDA and EMA marked the start of the immunotherapy revolution (204,205). This rapidly evolving therapeutic landscape has dramatically changed the treatment paradigm for adult and select pediatric malignancies (Table 2). With the various late effects associated with certain prolonged cytotoxic chemotherapy and radiation therapy regimens, it is the hope that immunotherapy may increase the quality of life for pediatric and adolescent patients in addition to providing durable responses, but this will require long-term follow-up.
Table 2:
FDA approved immunotherapies for pediatric oncology patients
| Drug | Trade Name | Pediatric Indication | Target | Type of Immunotherapy |
|---|---|---|---|---|
| Ipilimumab | Yervoy | Colorectal Cancer, Melnaoma | CTLA-4 | Monoclonal Antibody |
| Nivolumab | Opdivo | Colorectal Cancer | PD-1 | Monoclonal Antibody |
| Pembrolizumab | Keytruda | Hodgkin Lymphoma, Merkel Cell Carcinoma, Non-Hodgkin Lymphoma, Solid Tumors | PD-1 | Monoclonal Antibody |
| Blinatumomab | Blincyto | Acute Lymphoblastic Leukemia | C19, CD3 | Bi-specific T-cell engager |
| Tisagenlecleucel | Kymriah | Acute Lymphoblastic Leukemia, | CD19 | Chimeric Antigen Receptor T-cell |
| Gemtuzumab Ozogamicin | Mylotarg | Acute Myeloid Leukemia | CD33 | Antibody-drug conjugate |
| Avelumab | Bavencio | Merkel Cell Carcinoma | PD-L1 | Monoclonal Antibody |
| Dinutuximab | Unituxin | Neuroblastoma | GD2 | Monoclonal Antibody |
| Naxitamab-gqgk | Danyelza | Neuroblastoma | GD2 | Monoclonal Antibody |
Table contents obtained from National Cancer Institute: cancer.gov/about-cancer/treatment/drugs/childhood-cancer-fda-approved-drugs
Immune Checkpoint Inhibition in CNS and Solid Tumors
Data shows that robust expression of PD-L1 is not a universally common finding in most pediatric tumors, although a subset of 11q-deleted neuroblastomas exhibited increased expression of PD-L1 (202,206–208). Furthermore, somatic copy number gains at loci encompassing genes that encode for PD-L1/PD-L2 has been shown in a subset of osteosarcoma specimens (209). Two patients with hypermutant glioblastoma driven by biallelic mismatch repair deficiency syndrome were successfully treated with single agent nivolumab and exhibited durable clinical and radiologic responses (210). In a separate study, a patient with a hypermutant glioma and constitutional mismatch repair deficiency was treated with pembrolizumab yet succumbed to their disease (64). These two studies support the findings that mismatch repair deficient tumors exhibit variable responses to checkpoint inhibition (211). Nonetheless, immune checkpoint inhibition targeting PD-1 or CTLA4 has shown limited therapeutic efficacy for most pediatric patients with CNS and solid tumors with some notable exception (206,212–214). Given the clinical inefficacy of currently available immune checkpoint inhibition, adoptive cell therapies have played a much larger role to date in the immunotherapy of pediatric tumors.
CD19 CAR T-Cell Therapy in Leukemia
One of the most remarkable advances in cancer research has been the development, clinical implementation, and efficacy of chimeric antigen receptor (CAR) T-cell therapy in high-risk pediatric B-cell ALL. Furthermore, the fact that the first FDA approved the use of CAR T-cell therapy, tisagenlecleucel, was for a pediatric and adolescent indication before being approved for adults was a ground-breaking milestone for field of pediatric oncology. Infusion of these autologous CD19-directed CAR T-cells into pediatric and young adult patients with relapsed/refractory B-cell ALL achieved a remission rate of >80% with no evidence of residual disease at 3 months post-infusion (215). Additionally, these patients demonstrated 1-year event free survival (EFS) and overall survival (OS) rates of 50% and 76%, respectively. Nonetheless, this therapy is not without side effects as a high incidence of cytokine release syndrome was observed with 40% of these patients experiencing neurologic events (215). CD19 CAR T-cell therapy has also demonstrated efficacy in pediatric patients with CNS B-cell ALL (216,217). In these patients, neurotoxicity was associated with an increased CNS disease burden prior to treatment (217). The acute effects of cytokine release syndrome can be managed but late effects may exist in children and longitudinal monitoring is needed in these patients (218).
Leukemic B-cells can escape killing by CAR T-cells by employing a variety of mechanisms, including downregulating the expression of CD19 (219). To address this, studies investigating sequential infusion of CD19 and CD22 CAR T-cells and preclinical efficacy of CD19/CD22 bivalent CAR T-cells are underway (220–222). Additionally, the use of low affinity CD19 CAR T-cells demonstrated superior in vitro and in vivo T-cell responses when compared to high affinity CAR T-cells (223). In patients, these lower affinity CAR T-cells were associated with increased persistence and expansion of the adoptively transferred cells, lower toxicity, a remission rate of 85%, a 1-year EFS rate of 64%, and a 1-year OS rate of 46% (223). With a median follow-up of 4.8 years, a recent study found that allogeneic hematopoietic stem cell transplant after CD19 CAR T-cell infusion significantly reduced the relapse rate in patients with recurrent B-ALL (224).
GD2-targeted CAR T-Cell Therapy
GD2 is a carbohydrate-containing sphingolipid that is expressed by many cell types throughout the body and highly expressed by neuroblastoma cells (225). GD2-targeted CAR T-cells engineered to co-express IL15 reduced T-cell exhaustion in vitro and enhanced the therapeutic efficacy in vivo (226). Interestingly, GD2-targeted CAR T-cells were effective at killing neuroblastoma xenografts but were ineffective in osteosarcoma and Ewing sarcoma xenografts, both of which expressed GD2 (177). Moreover, the expansion and immunosuppressive effects of MDSCs in the sarcoma microenvironment was responsible for the lack of therapeutic effect of the CAR T-cells (177).
Tumor cells can evade GD2 CAR T-cells. Upon encountering GD2-targeted CAR T-cells, osteosarcoma cells upregulated PD-L1 to induce exhaustion and apoptosis of the CAR T-cells (227). Systemic administration of GD2 CAR T-cells in orthotopic DIPG xenografts resulted in a durable reduction in tumor burden and improved survival, however a small subset of xenografted tumor cells lost antigen expression (228). Retinoblastoma cells that were initially responsive to GD2 CAR T-cell therapy escaped killing by downregulating GD2 expression and increasing the expression of PD-L1 and PD-1 on the surface of tumor cells and CAR T-cells, respectively (229,230). Sequential exposure of retinoblastoma cells to CAR T-cells against different targets (GD2 and CD171) enhanced tumor cell killing in vivo (230). In neuroblastoma xenograft models, engineering GD2 CAR T-cells to express IL15 improved the therapeutic efficacy and reduced the expression of PD-1 on the CAR T-cells (231).
In phase 1 studies of patients with relapsed/refractory neuroblastoma, lymphodepletion prior to infusion of GD2 CAR T-cells was deemed safe (232,233). In one study, despite demonstrated evidence of transient tumor regression in 25% of patients, the investigators did not observe an objective clinical response (233). The second study showed a significant CAR T-cell expansion in the patients who received prior lymphodepleting therapy versus those show did not and that inhibition of PD1 did not improve the performance of the CAR T-cells (232). This study also observed an expansion of CD11b+CD163+ myeloid cells after GD2 CAR T-cell infusion. While the results are promising, comprehensive investigations of the tumor-specific microenvironments needs to be performed in order to better understand the complexities of immune escape and optimize immunotherapeutic interventions.
NY-ESO-1 Engineered T Cell Receptors
NY-ESO-1 is a cancer testes antigen expressed in the majority of synovial sarcoma tumors and a subset of malignant peripheral nerve sheath tumors (234,235). Synovial sarcoma patients treated with autologous T-cells expressing a NY-ESO-1 engineered T cell receptor (NY-ESO-1 c259T-cells) resulted in an overall response rate of 50% (236). After infusion, NY-ESO-1 c259T-cells generated a memory T-cell response, did not exhibit signs of T-cell exhaustion, were clonally expanded, and persisted in vivo. A second study revealed that increased expression of NY-ESO-1 was likely associated with more robust and durable therapeutic responses (237). These studies provide a degree of optimism for a difficult to manage adolescent/young adult tumor.
Despite some successes, immunotherapy for pediatric cancer patients is still in the early phase of development. To realize the full potential of immunotherapy to provide efficacious and durable responses for pediatric patients, especially those with CNS and solid tumors, a better understanding of the interaction between the tumor, the microenvironment and the immune cells will be required to move this field forward. To address this, the Cancer Moonshot has created the Pediatric Immunotherapy Discovery and Development Network and the Cancer Immunotherapy Trials Network. The goal of these networks is to collaboratively advance the field of pediatric cancer immunotherapy by characterizing new targets, generating suitable experimental models for preclinical evaluation, and better understand pediatric tumor immunology.
CONCLUSIONS AND OUTLOOK
The last 5 years has witnessed significant advances in our understanding of the genetic and epigenetic underpinning of pediatric cancers as well as the treatments of some pediatric tumors. At the genetic level, it is clear that many genetic changes identified are distinct from adult cancers and will require unique interventions (FIGURE 2). Furthermore, it is also clear that pediatric tumors appear to have a high degree of epigenetic changes that lead to widespread alterations in gene expression secondary to these changes. This in turn may ultimately lead to novel therapeutic approaches in the future. Germline genetic variations associated with predisposition to cancer appears to occur at a higher frequency in pediatric patients with cancer compared to adult cancer patients, and these findings may ultimately allow both earlier intervention in identified high-risk children and hopefully prevention in the future. Finally, adoptive immunotherapy, particularly CAR-T cells have had a major impact on the treatment of childhood ALL, but immune checkpoint inhibitors have to date been of minimal use in pediatric tumors. Ongoing work identifying the impact of the tumor microenvironment on trafficking of immune cells necessary for ICI activity may help improve the effect of these agents in the future.
ACKNOWLEDGEMENTS
We are indebted to the patients and their families who have participated in the various studies, without which, progress would not have been made. We are both grateful and thankful to our scientific and clinical colleagues around the world that have dedicated their careers to investigating pediatric malignancies. Given the numerous recent developments and achievements in pediatric cancer research, we regret that we were not able to encompass all these studies into this review. Nonetheless, your collective accomplishments and efforts are noted and appreciated. T.A.M. is supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. The views and opinions contained within this article do not necessarily reflect those of the National Institutes of Health or the US Department of Health and Human Services. The mention of trade names and/or commercialized products does not indicate endorsement by the US government.
Footnotes
Conflict of Interest: The authors have no financial interests to declare as it pertains to this manuscript
REFERENCES
- 1.Cieslik M, Chinnaiyan AM. Global genomics project unravels cancer’s complexity at unprecedented scale. Nature 2020;578:39–40 [DOI] [PubMed] [Google Scholar]
- 2.Consortium ITP-CAoWG. Pan-cancer analysis of whole genomes. Nature 2020;578:82–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma X, Liu Y, Alexandrov LB, Edmonson MN, Gawad C, Zhou X, et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 2018;555:371–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grobner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, et al. The landscape of genomic alterations across childhood cancers. Nature 2018;555:321–7 [DOI] [PubMed] [Google Scholar]
- 5.Campbell BB, Galati MA, Stone SC, Riemenschneider AN, Edwards M, Sudhaman S, et al. Mutations in the RAS/MAPK pathway drive replication repair deficient hypermutated tumors and confer sensitivity to MEK inhibition. Cancer Discov 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong M, Mayoh C, Lau LMS, Khuong-Quang DA, Pinese M, Kumar A, et al. Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk pediatric cancer. Nat Med 2020;26:1742–53 [DOI] [PubMed] [Google Scholar]
- 7.Hatch EM, Hetzer MW. Chromothripsis. Curr Biol 2015;25:R397–9 [DOI] [PubMed] [Google Scholar]
- 8.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW. Cancer genome landscapes. Science 2013;339:1546–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sievers P, Sill M, Blume C, Tauziede-Espariat A, Schrimpf D, Stichel D, et al. Clear cell meningiomas are defined by a highly distinct DNA methylation profile and mutations in SMARCE1. Acta Neuropathol 2021;141:281–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Northcott PA, Buchhalter I, Morrissy AS, Hovestadt V, Weischenfeldt J, Ehrenberger T, et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017;547:311–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sexton-Oates A, Dodgshun A, Hovestadt V, Jones DTW, Ashley DM, Sullivan M, et al. Methylation profiling of paediatric pilocytic astrocytoma reveals variants specifically associated with tumour location and predictive of recurrence. Mol Oncol 2018;12:1219–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeyapalan JN, Doctor GT, Jones TA, Alberman SN, Tep A, Haria CM, et al. DNA methylation analysis of paediatric low-grade astrocytomas identifies a tumour-specific hypomethylation signature in pilocytic astrocytomas. Acta Neuropathol Commun 2016;4:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torchia J, Golbourn B, Feng S, Ho KC, Sin-Chan P, Vasiljevic A, et al. Integrated (epi)-Genomic Analyses Identify Subgroup-Specific Therapeutic Targets in CNS Rhabdoid Tumors. Cancer Cell 2016;30:891–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johann PD, Erkek S, Zapatka M, Kerl K, Buchhalter I, Hovestadt V, et al. Atypical Teratoid/Rhabdoid Tumors Are Comprised of Three Epigenetic Subgroups with Distinct Enhancer Landscapes. Cancer Cell 2016;29:379–93 [DOI] [PubMed] [Google Scholar]
- 15.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature 2018;555:469–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danielsson A, Nemes S, Tisell M, Lannering B, Nordborg C, Sabel M, et al. MethPed: a DNA methylation classifier tool for the identification of pediatric brain tumor subtypes. Clin Epigenetics 2015;7:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryall S, Zapotocky M, Fukuoka K, Nobre L, Guerreiro Stucklin A, Bennett J, et al. Integrated Molecular and Clinical Analysis of 1,000 Pediatric Low-Grade Gliomas. Cancer Cell 2020;37:569–83.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eleveld TF, Oldridge DA, Bernard V, Koster J, Colmet Daage L, Diskin SJ, et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet 2015;47:864–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eleveld TF, Schild L, Koster J, Zwijnenburg DA, Alles LK, Ebus ME, et al. RAS-MAPK Pathway-Driven Tumor Progression Is Associated with Loss of CIC and Other Genomic Aberrations in Neuroblastoma. Cancer Res 2018;78:6297–307 [DOI] [PubMed] [Google Scholar]
- 20.Shern JF, Selfe J, Izquierdo E, Patidar R, Chou HC, Song YK, et al. Genomic Classification and Clinical Outcome in Rhabdomyosarcoma: A Report From an International Consortium. J Clin Oncol 2021:JCO2003060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moke DJ, Song Z, Liu L, Hamilton AS, Deapen D, Freyer DR. A Population-Based Analysis of 30-Year Mortality among Five-Year Survivors of Adolescent and Young Adult Cancer: The Roles of Primary Cancer, Subsequent Malignancy, and Other Health Conditions. Cancers (Basel) 2021;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zahnreich S, Schmidberger H. Childhood Cancer: Occurrence, Treatment and Risk of Second Primary Malignancies. Cancers (Basel) 2021;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz JR, Ma J, Kamens J, Westover T, Walsh MP, Brady SW, et al. The acquisition of molecular drivers in pediatric therapy-related myeloid neoplasms. Nat Commun 2021;12:985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oshima K, Khiabanian H, da Silva-Almeida AC, Tzoneva G, Abate F, Ambesi-Impiombato A, et al. Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia. Proc Natl Acad Sci U S A 2016;113:11306–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipka DB, Witte T, Toth R, Yang J, Wiesenfarth M, Nöllke P, et al. RAS-pathway mutation patterns define epigenetic subclasses in juvenile myelomonocytic leukemia. Nat Commun 2017;8:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knudson AG. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A 1971;68:820–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller DB, Piccolo SR. Compound Heterozygous Variants in Pediatric Cancers: A Systematic Review. Front Genet 2020;11:493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Capasso M, Montella A, Tirelli M, Maiorino T, Cantalupo S, Iolascon A. Genetic Predisposition to Solid Pediatric Cancers. Front Oncol 2020;10:590033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kratz CP, Jongmans MC, Cavé H, Wimmer K, Behjati S, Guerrini-Rousseau L, et al. Predisposition to cancer in children and adolescents. Lancet Child Adolesc Health 2021;5:142–54 [DOI] [PubMed] [Google Scholar]
- 30.Plon SE, Lupo PJ. Genetic Predisposition to Childhood Cancer in the Genomic Era. Annu Rev Genomics Hum Genet 2019;20:241–63 [DOI] [PubMed] [Google Scholar]
- 31.Mitchell SG, Pencheva B, Porter CC. Germline Genetics and Childhood Cancer: Emerging Cancer Predisposition Syndromes and Psychosocial Impacts. Curr Oncol Rep 2019;21:85. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Walsh MF, Wu G, Edmonson MN, Gruber TA, Easton J, et al. Germline Mutations in Predisposition Genes in Pediatric Cancer. N Engl J Med 2015;373:2336–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagener R, Taeubner J, Walter C, Yasin L, Alzoubi D, Bartenhagen C, et al. Comprehensive germline-genomic and clinical profiling in 160 unselected children and adolescents with cancer. Eur J Hum Genet 2021;29:1301–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akhavanfard S, Padmanabhan R, Yehia L, Cheng F, Eng C. Comprehensive germline genomic profiles of children, adolescents and young adults with solid tumors. Nat Commun 2020;11:2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gröbner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, et al. The landscape of genomic alterations across childhood cancers. Nature 2018;555:321–7 [DOI] [PubMed] [Google Scholar]
- 36.Waszak SM, Northcott PA, Buchhalter I, Robinson GW, Sutter C, Groebner S, et al. Spectrum and prevalence of genetic predisposition in medulloblastoma: a retrospective genetic study and prospective validation in a clinical trial cohort. Lancet Oncol 2018;19:785–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wasserman JD, Novokmet A, Eichler-Jonsson C, Ribeiro RC, Rodriguez-Galindo C, Zambetti GP, et al. Prevalence and functional consequence of TP53 mutations in pediatric adrenocortical carcinoma: a children’s oncology group study. J Clin Oncol 2015;33:602–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirabello L, Zhu B, Koster R, Karlins E, Dean M, Yeager M, et al. Frequency of Pathogenic Germline Variants in Cancer-Susceptibility Genes in Patients With Osteosarcoma. JAMA Oncol 2020;6:724–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diessner BJ, Pankratz N, Hooten AJ, Mirabello L, Sarver AL, Mills LJ, et al. Nearly Half of TP53 Germline Variants Predicted To Be Pathogenic in Patients With Osteosarcoma Are De Novo: A Report From the Children’s Oncology Group. JCO Precis Oncol 2020;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian M, Cao X, Devidas M, Yang W, Cheng C, Dai Y, et al. TP53 Germline Variations Influence the Predisposition and Prognosis of B-Cell Acute Lymphoblastic Leukemia in Children. J Clin Oncol 2018;36:591–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koster R, Panagiotou OA, Wheeler WA, Karlins E, Gastier-Foster JM, Caminada de Toledo SR, et al. Genome-wide association study identifies the GLDC/IL33 locus associated with survival of osteosarcoma patients. Int J Cancer 2018;142:1594–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin Q, Han J, Sun Q, Wen L, Wang S. Functional variant of IL33 is associated with survival of osteosarcoma patients. J Bone Oncol 2020;20:100270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Sisoudiya SD, Martin-Giacalone BA, Khayat MM, Dugan-Perez S, Marquez-Do DA, et al. Germline Cancer-Predisposition Variants in Pediatric Rhabdomyosarcoma: A Report from the Children’s Oncology Group. J Natl Cancer Inst 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J, Light N, Subasri V, Young EL, Wegman-Ostrosky T, Barkauskas DA, et al. Pathogenic Germline Variants in Cancer Susceptibility Genes in Children and Young Adults With Rhabdomyosarcoma. JCO Precis Oncol 2021;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Machiela MJ, Grunewald TGP, Surdez D, Reynaud S, Mirabeau O, Karlins E, et al. Genome-wide association study identifies multiple new loci associated with Ewing sarcoma susceptibility. Nat Commun 2018;9:3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin SH, Sampson JN, Grunewald TGP, Surdez D, Reynaud S, Mirabeau O, et al. Low-frequency variation near common germline susceptibility loci are associated with risk of Ewing sarcoma. PLoS One 2020;15:e0237792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brohl AS, Patidar R, Turner CE, Wen X, Song YK, Wei JS, et al. Frequent inactivating germline mutations in DNA repair genes in patients with Ewing sarcoma. Genet Med 2017;19:955–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oldridge DA, Truong B, Russ D, DuBois SG, Vaksman Z, Mosse YP, et al. Differences in Genomic Profiles and Outcomes Between Thoracic and Adrenal Neuroblastoma. J Natl Cancer Inst 2019;111:1192–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muskens IS, de Smith AJ, Zhang C, Hansen HM, Morimoto L, Metayer C, et al. Germline cancer predisposition variants and pediatric glioma: a population-based study in California. Neuro Oncol 2020;22:864–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Begemann M, Waszak SM, Robinson GW, Jager N, Sharma T, Knopp C, et al. Germline GPR161 Mutations Predispose to Pediatric Medulloblastoma. J Clin Oncol 2020;38:43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waszak SM, Robinson GW, Gudenas BL, Smith KS, Forget A, Kojic M, et al. Germline Elongator mutations in Sonic Hedgehog medulloblastoma. Nature 2020;580:396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moriyama T, Metzger ML, Wu G, Nishii R, Qian M, Devidas M, et al. Germline genetic variation in ETV6 and risk of childhood acute lymphoblastic leukaemia: a systematic genetic study. Lancet Oncol 2015;16:1659–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, Liu AP, Devidas M, Lee SH, Cao X, Pei D, et al. Association of GATA3 Polymorphisms With Minimal Residual Disease and Relapse Risk in Childhood Acute Lymphoblastic Leukemia. J Natl Cancer Inst 2021;113:408–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vijayakrishnan J, Qian M, Studd JB, Yang W, Kinnersley B, Law PJ, et al. Identification of four novel associations for B-cell acute lymphoblastic leukaemia risk. Nat Commun 2019;10:5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marlow EC, Ducore J, Kwan ML, Cheng SY, Bowles EJA, Greenlee RT, et al. Leukemia Risk in a Cohort of 3.9 Million Children with and without Down Syndrome. J Pediatr 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown AL, de Smith AJ, Gant VU, Yang W, Scheurer ME, Walsh KM, et al. Inherited genetic susceptibility to acute lymphoblastic leukemia in Down syndrome. Blood 2019;134:1227–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McNally EJ, Luncsford PJ, Armanios M. Long telomeres and cancer risk: the price of cellular immortality. J Clin Invest 2019;129:3474–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ackermann S, Fischer M. Telomere Maintenance in Pediatric Cancer. Int J Mol Sci 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haycock PC, Burgess S, Nounu A, Zheng J, Okoli GN, Bowden J, et al. Association Between Telomere Length and Risk of Cancer and Non-Neoplastic Diseases: A Mendelian Randomization Study. JAMA Oncol 2017;3:636–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang C, Ostrom QT, Semmes EC, Ramaswamy V, Hansen HM, Morimoto L, et al. Genetic predisposition to longer telomere length and risk of childhood, adolescent and adult-onset ependymoma. Acta Neuropathol Commun 2020;8:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang C, Hansen HM, Semmes EC, Gonzalez-Maya J, Morimoto L, Wei Q, et al. Common genetic variation and risk of osteosarcoma in a multi-ethnic pediatric and adolescent population. Bone 2020;130:115070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walsh KM, Whitehead TP, de Smith AJ, Smirnov IV, Park M, Endicott AA, et al. Common genetic variants associated with telomere length confer risk for neuroblastoma and other childhood cancers. Carcinogenesis 2016;37:576–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang KL, Mashl RJ, Wu Y, Ritter DI, Wang J, Oh C, et al. Pathogenic Germline Variants in 10,389 Adult Cancers. Cell 2018;173:355–70.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fiala EM, Jayakumaran G, Mauguen A, Kennedy JA, Bouvier N, Kemel Y, et al. Prospective pan-cancer germline testing using MSK-IMPACT informs clinical translation in 751 patients with pediatric solid tumors. Nat Cancer 2021;2:357–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brodeur GM, Nichols KE, Plon SE, Schiffman JD, Malkin D. Pediatric Cancer Predisposition and Surveillance: An Overview, and a Tribute to Alfred G. Knudson Jr. Clin Cancer Res 2017;23:e1–e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Porter CC, Druley TE, Erez A, Kuiper RP, Onel K, Schiffman JD, et al. Recommendations for Surveillance for Children with Leukemia-Predisposing Conditions. Clin Cancer Res 2017;23:e14–e22 [DOI] [PubMed] [Google Scholar]
- 67.Walsh MF, Chang VY, Kohlmann WK, Scott HS, Cunniff C, Bourdeaut F, et al. Recommendations for Childhood Cancer Screening and Surveillance in DNA Repair Disorders. Clin Cancer Res 2017;23:e23–e31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greer MC, Voss SD, States LJ. Pediatric Cancer Predisposition Imaging: Focus on Whole-Body MRI. Clin Cancer Res 2017;23:e6–e13 [DOI] [PubMed] [Google Scholar]
- 69.Druker H, Zelley K, McGee RB, Scollon SR, Kohlmann WK, Schneider KA, et al. Genetic Counselor Recommendations for Cancer Predisposition Evaluation and Surveillance in the Pediatric Oncology Patient. Clin Cancer Res 2017;23:e91–e7 [DOI] [PubMed] [Google Scholar]
- 70.Kratz CP, Achatz MI, Brugières L, Frebourg T, Garber JE, Greer MC, et al. Cancer Screening Recommendations for Individuals with Li-Fraumeni Syndrome. Clin Cancer Res 2017;23:e38–e45 [DOI] [PubMed] [Google Scholar]
- 71.Tabori U, Hansford JR, Achatz MI, Kratz CP, Plon SE, Frebourg T, et al. Clinical Management and Tumor Surveillance Recommendations of Inherited Mismatch Repair Deficiency in Childhood. Clin Cancer Res 2017;23:e32–e7 [DOI] [PubMed] [Google Scholar]
- 72.Qing T, Mohsen H, Marczyk M, Ye Y, O’Meara T, Zhao H, et al. Germline variant burden in cancer genes correlates with age at diagnosis and somatic mutation burden. Nat Commun 2020;11:2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alix-Panabières C, Pantel K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov 2021;11:858–73 [DOI] [PubMed] [Google Scholar]
- 74.Kahana-Edwin S, Cain LE, Karpelowsky J. Roadmap to Liquid Biopsy Biobanking from Pediatric Cancers-Challenges and Opportunities. Biopreserv Biobank 2021;19:124–9 [DOI] [PubMed] [Google Scholar]
- 75.Trigg RM, Shaw JA, Turner SD. Opportunities and challenges of circulating biomarkers in neuroblastoma. Open Biol 2019;9:190056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Andersson D, Fagman H, Dalin MG, Stahlberg A. Circulating cell-free tumor DNA analysis in pediatric cancers. Mol Aspects Med 2020;72:100819 [DOI] [PubMed] [Google Scholar]
- 77.Stallard S, Savelieff MG, Wierzbicki K, Mullan B, Miklja Z, Bruzek A, et al. CSF H3F3A K27M circulating tumor DNA copy number quantifies tumor growth and in vitro treatment response. Acta Neuropathol Commun 2018;6:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang TY, Piunti A, Lulla RR, Qi J, Horbinski CM, Tomita T, et al. Detection of Histone H3 mutations in cerebrospinal fluid-derived tumor DNA from children with diffuse midline glioma. Acta Neuropathol Commun 2017;5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chicard M, Colmet-Daage L, Clement N, Danzon A, Bohec M, Bernard V, et al. Whole-Exome Sequencing of Cell-Free DNA Reveals Temporo-spatial Heterogeneity and Identifies Treatment-Resistant Clones in Neuroblastoma. Clin Cancer Res 2018;24:939–49 [DOI] [PubMed] [Google Scholar]
- 80.Trigg RM, Turner SD, Shaw JA, Jahangiri L. Diagnostic accuracy of circulating-free DNA for the determination of MYCN amplification status in advanced-stage neuroblastoma: a systematic review and meta-analysis. Br J Cancer 2020;122:1077–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Applebaum MA, Barr EK, Karpus J, West-Szymanski DC, Oliva M, Sokol EA, et al. 5-Hydroxymethylcytosine Profiles in Circulating Cell-Free DNA Associate with Disease Burden in Children with Neuroblastoma. Clin Cancer Res 2020;26:1309–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shulman DS, Klega K, Imamovic-Tuco A, Clapp A, Nag A, Thorner AR, et al. Detection of circulating tumour DNA is associated with inferior outcomes in Ewing sarcoma and osteosarcoma: a report from the Children’s Oncology Group. Br J Cancer 2018;119:615–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miguez ACK, Barros BDF, de Souza JES, da Costa CML, Cunha IW, Barbosa P, et al. Assessment of somatic mutations in urine and plasma of Wilms tumor patients. Cancer Med 2020;9:5948–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Escudero L, Llort A, Arias A, Diaz-Navarro A, Martinez-Ricarte F, Rubio-Perez C, et al. Circulating tumour DNA from the cerebrospinal fluid allows the characterisation and monitoring of medulloblastoma. Nat Commun 2020;11:5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li J, Zhao S, Lee M, Yin Y, Li J, Zhou Y, et al. Reliable tumor detection by whole-genome methylation sequencing of cell-free DNA in cerebrospinal fluid of pediatric medulloblastoma. Sci Adv 2020;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Klega K, Imamovic-Tuco A, Ha G, Clapp AN, Meyer S, Ward A, et al. Detection of Somatic Structural Variants Enables Quantification and Characterization of Circulating Tumor DNA in Children With Solid Tumors. JCO Precis Oncol 2018;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cimmino F, Lasorsa VA, Vetrella S, Iolascon A, Capasso M. A Targeted Gene Panel for Circulating Tumor DNA Sequencing in Neuroblastoma. Front Oncol 2020;10:596191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pan C, Diplas BH, Chen X, Wu Y, Xiao X, Jiang L, et al. Molecular profiling of tumors of the brainstem by sequencing of CSF-derived circulating tumor DNA. Acta Neuropathol 2019;137:297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Woodhouse R, Li M, Hughes J, Delfosse D, Skoletsky J, Ma P, et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One 2020;15:e0237802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li G, Pavlick D, Chung JH, Bauer T, Tan BA, Peguero J, et al. Genomic profiling of cell-free circulating tumor DNA in patients with colorectal cancer and its fidelity to the genomics of the tumor biopsy. J Gastrointest Oncol 2019;10:831–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leighl NB, Page RD, Raymond VM, Daniel DB, Divers SG, Reckamp KL, et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer. Clin Cancer Res 2019;25:4691–700 [DOI] [PubMed] [Google Scholar]
- 92.Strickler JH, Loree JM, Ahronian LG, Parikh AR, Niedzwiecki D, Pereira AAL, et al. Genomic Landscape of Cell-Free DNA in Patients with Colorectal Cancer. Cancer Discov 2018;8:164–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV, Consortium C. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol 2020;31:745–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lennon AM, Buchanan AH, Kinde I, Warren A, Honushefsky A, Cohain AT, et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science 2020;369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chang W, Brohl AS, Patidar R, Sindiri S, Shern JF, Wei JS, et al. MultiDimensional ClinOmics for Precision Therapy of Children and Adolescent Young Adults with Relapsed and Refractory Cancer: A Report from the Center for Cancer Research. Clin Cancer Res 2016;22:3810–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang Y, Chen F, Donehower LA, Scheurer ME, Creighton CJ. A pediatric brain tumor atlas of genes deregulated by somatic genomic rearrangement. Nat Commun 2021;12:937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rokita JL, Rathi KS, Cardenas MF, Upton KA, Jayaseelan J, Cross KL, et al. Genomic Profiling of Childhood Tumor Patient-Derived Xenograft Models to Enable Rational Clinical Trial Design. Cell Rep 2019;29:1675–89 e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cameron DL, Di Stefano L, Papenfuss AT. Comprehensive evaluation and characterisation of short read general-purpose structural variant calling software. Nat Commun 2019;10:3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van Belzen IAEM, Schönhuth A, Kemmeren P, Hehir-Kwa JY. Structural variant detection in cancer genomes: computational challenges and perspectives for precision oncology. NPJ Precis Oncol 2021;5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lei M, Liang D, Yang Y, Mitsuhashi S, Katoh K, Miyake N, et al. Long-read DNA sequencing fully characterized chromothripsis in a patient with Langer-Giedion syndrome and Cornelia de Lange syndrome-4. J Hum Genet 2020;65:667–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mitsuhashi S, Ohori S, Katoh K, Frith MC, Matsumoto N. A pipeline for complete characterization of complex germline rearrangements from long DNA reads. Genome Med 2020;12:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Federman N, McDermott R. Larotrectinib, a highly selective tropomyosin receptor kinase (TRK) inhibitor for the treatment of TRK fusion cancer. Expert Rev Clin Pharmacol 2019;12:931–9 [DOI] [PubMed] [Google Scholar]
- 104.Zhao X, Kotch C, Fox E, Surrey LF, Wertheim GB, Baloch ZW, et al. NTRK Fusions Identified in Pediatric Tumors: The Frequency, Fusion Partners, and Clinical Outcome. JCO Precis Oncol 2021;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Laetsch TW, DuBois SG, Mascarenhas L, Turpin B, Federman N, Albert CM, et al. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: phase 1 results from a multicentre, open-label, phase 1/2 study. Lancet Oncol 2018;19:705–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kummar S, Berlin J, Mascarenhas L, van Tilburg CM, Geoerger B, Lassen UN, et al. Quality of Life in Adult and Pediatric Patients with Tropomyosin Receptor Kinase Fusion Cancer Receiving Larotrectinib. Curr Probl Cancer 2021:100734 [DOI] [PubMed] [Google Scholar]
- 107.Dombi E, Baldwin A, Marcus LJ, Fisher MJ, Weiss B, Kim A, et al. Activity of Selumetinib in Neurofibromatosis Type 1-Related Plexiform Neurofibromas. N Engl J Med 2016;375:2550–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gross AM, Wolters PL, Dombi E, Baldwin A, Whitcomb P, Fisher MJ, et al. Selumetinib in Children with Inoperable Plexiform Neurofibromas. N Engl J Med 2020;382:1430–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jackson S, Baker EH, Gross AM, Whitcomb P, Baldwin A, Derdak J, et al. The MEK inhibitor selumetinib reduces spinal neurofibroma burden in patients with NF1 and plexiform neurofibromas. Neurooncol Adv 2020;2:vdaa095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fangusaro J, Onar-Thomas A, Young Poussaint T, Wu S, Ligon AH, Lindeman N, et al. Selumetinib in paediatric patients with BRAF-aberrant or neurofibromatosis type 1-associated recurrent, refractory, or progressive low-grade glioma: a multicentre, phase 2 trial. Lancet Oncol 2019;20:1011–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012;482:226–31 [DOI] [PubMed] [Google Scholar]
- 112.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 2012;44:251–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Larson JD, Kasper LH, Paugh BS, Jin H, Wu G, Kwon CH, et al. Histone H3.3 K27M Accelerates Spontaneous Brainstem Glioma and Drives Restricted Changes in Bivalent Gene Expression. Cancer Cell 2019;35:140–55 e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen KY, Bush K, Klein RH, Cervantes V, Lewis N, Naqvi A, et al. Reciprocal H3.3 gene editing identifies K27M and G34R mechanisms in pediatric glioma including NOTCH signaling. Commun Biol 2020;3:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mohammad F, Weissmann S, Leblanc B, Pandey DP, Hojfeldt JW, Comet I, et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med 2017;23:483–92 [DOI] [PubMed] [Google Scholar]
- 116.Silveira AB, Kasper LH, Fan Y, Jin H, Wu G, Shaw TI, et al. H3.3 K27M depletion increases differentiation and extends latency of diffuse intrinsic pontine glioma growth in vivo. Acta Neuropathol 2019;137:637–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Harutyunyan AS, Krug B, Chen H, Papillon-Cavanagh S, Zeinieh M, De Jay N, et al. H3K27M induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis. Nat Commun 2019;10:1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Piunti A, Hashizume R, Morgan MA, Bartom ET, Horbinski CM, Marshall SA, et al. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med 2017;23:493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bayliss J, Mukherjee P, Lu C, Jain SU, Chung C, Martinez D, et al. Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci Transl Med 2016;8:366ra161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen CCL, Deshmukh S, Jessa S, Hadjadj D, Lisi V, Andrade AF, et al. Histone H3.3G34-Mutant Interneuron Progenitors Co-opt PDGFRA for Gliomagenesis. Cell 2020;183:1617–33 e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Helmsauer K, Valieva ME, Ali S, Chamorro Gonzalez R, Schopflin R, Roefzaad C, et al. Enhancer hijacking determines extrachromosomal circular MYCN amplicon architecture in neuroblastoma. Nat Commun 2020;11:5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Morrow JJ, Bayles I, Funnell APW, Miller TE, Saiakhova A, Lizardo MM, et al. Positively selected enhancer elements endow osteosarcoma cells with metastatic competence. Nat Med 2018;24:176–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Guillon N, Tirode F, Boeva V, Zynovyev A, Barillot E, Delattre O. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS One 2009;4:e4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lin L, Huang M, Shi X, Mayakonda A, Hu K, Jiang YY, et al. Super-enhancer-associated MEIS1 promotes transcriptional dysregulation in Ewing sarcoma in co-operation with EWS-FLI1. Nucleic Acids Res 2019;47:1255–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.van Groningen T, Koster J, Valentijn LJ, Zwijnenburg DA, Akogul N, Hasselt NE, et al. Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat Genet 2017;49:1261–6 [DOI] [PubMed] [Google Scholar]
- 126.Gryder BE, Wachtel M, Chang K, El Demerdash O, Aboreden NG, Mohammed W, et al. Miswired Enhancer Logic Drives a Cancer of the Muscle Lineage. iScience 2020;23:101103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 2014;4:216–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yohe ME, Gryder BE, Shern JF, Song YK, Chou HC, Sindiri S, et al. MEK inhibition induces MYOG and remodels super-enhancers in RAS-driven rhabdomyosarcoma. Sci Transl Med 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pomella S, Sreenivas P, Gryder BE, Wang L, Milewski D, Cassandri M, et al. Interaction between SNAI2 and MYOD enhances oncogenesis and suppresses differentiation in Fusion Negative Rhabdomyosarcoma. Nat Commun 2021;12:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li S, Chen K, Zhang Y, Barnes SD, Jaichander P, Zheng Y, et al. Twist2 amplification in rhabdomyosarcoma represses myogenesis and promotes oncogenesis by redirecting MyoD DNA binding. Genes Dev 2019;33:626–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Boulay G, Awad ME, Riggi N, Archer TC, Iyer S, Boonseng WE, et al. OTX2 Activity at Distal Regulatory Elements Shapes the Chromatin Landscape of Group 3 Medulloblastoma . Cancer Discov 2017;7:288–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Matsuda T, Irie T, Katsurabayashi S, Hayashi Y, Nagai T, Hamazaki N, et al. Pioneer Factor NeuroD1 Rearranges Transcriptional and Epigenetic Profiles to Execute Microglia-Neuron Conversion. Neuron 2019;101:472–85 e7 [DOI] [PubMed] [Google Scholar]
- 133.Tomazou EM, Sheffield NC, Schmidl C, Schuster M, Schonegger A, Datlinger P, et al. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Rep 2015;10:1082–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suva ML, et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 2014;26:668–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gryder BE, Yohe ME, Chou HC, Zhang X, Marques J, Wachtel M, et al. PAX3-FOXO1 Establishes Myogenic Super Enhancers and Confers BET Bromodomain Vulnerability. Cancer Discov 2017;7:884–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Marques JG, Gryder BE, Pavlovic B, Chung Y, Ngo QA, Frommelt F, et al. NuRD subunit CHD4 regulates super-enhancer accessibility in rhabdomyosarcoma and represents a general tumor dependency. Elife 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 2013;153:71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Boulay G, Cironi L, Garcia SP, Rengarajan S, Xing YH, Lee L, et al. The chromatin landscape of primary synovial sarcoma organoids is linked to specific epigenetic mechanisms and dependencies. Life Sci Alliance 2021;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.McBride MJ, Pulice JL, Beird HC, Ingram DR, D’Avino AR, Shern JF, et al. The SS18-SSX Fusion Oncoprotein Hijacks BAF Complex Targeting and Function to Drive Synovial Sarcoma. Cancer Cell 2018;33:1128–41 e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Banito A, Li X, Laporte AN, Roe JS, Sanchez-Vega F, Huang CH, et al. The SS18-SSX Oncoprotein Hijacks KDM2B-PRC1.1 to Drive Synovial Sarcoma. Cancer Cell 2018;33:527–41 e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sanchez-Molina S, Figuerola-Bou E, Blanco E, Sanchez-Jimenez M, Taboas P, Gomez S, et al. RING1B recruits EWSR1-FLI1 and cooperates in the remodeling of chromatin necessary for Ewing sarcoma tumorigenesis. Sci Adv 2020;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell 2017;171:163–78 e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kildisiute G, Kholosy WM, Young MD, Roberts K, Elmentaite R, van Hooff SR, et al. Tumor to normal single-cell mRNA comparisons reveal a pan-neuroblastoma cancer cell. Sci Adv 2021;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hanemaaijer ES, Margaritis T, Sanders K, Bos FL, Candelli T, Al-Saati H, et al. Single-cell atlas of developing murine adrenal gland reveals relation of Schwann cell precursor signature to neuroblastoma phenotype. Proc Natl Acad Sci U S A 2021;118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dong R, Yang R, Zhan Y, Lai HD, Ye CJ, Yao XY, et al. Single-Cell Characterization of Malignant Phenotypes and Developmental Trajectories of Adrenal Neuroblastoma. Cancer Cell 2020;38:716–33 e6 [DOI] [PubMed] [Google Scholar]
- 146.Young MD, Mitchell TJ, Vieira Braga FA, Tran MGB, Stewart BJ, Ferdinand JR, et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 2018;361:594–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hovestadt V, Smith KS, Bihannic L, Filbin MG, Shaw ML, Baumgartner A, et al. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature 2019;572:74–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jessa S, Blanchet-Cohen A, Krug B, Vladoiu M, Coutelier M, Faury D, et al. Stalled developmental programs at the root of pediatric brain tumors. Nat Genet 2019;51:1702–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ocasio J, Babcock B, Malawsky D, Weir SJ, Loo L, Simon JM, et al. scRNA-seq in medulloblastoma shows cellular heterogeneity and lineage expansion support resistance to SHH inhibitor therapy. Nat Commun 2019;10:5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sayles LC, Breese MR, Koehne AL, Leung SG, Lee AG, Liu HY, et al. Genome-Informed Targeted Therapy for Osteosarcoma. Cancer Discov 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stewart E, Federico SM, Chen X, Shelat AA, Bradley C, Gordon B, et al. Orthotopic patient-derived xenografts of paediatric solid tumours. Nature 2017;549:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Smith KS, Xu K, Mercer KS, Boop F, Klimo P, DeCupyere M, et al. Patient-derived orthotopic xenografts of pediatric brain tumors: a St. Jude resource. Acta Neuropathol 2020;140:209–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Brabetz S, Leary SES, Grobner SN, Nakamoto MW, Seker-Cin H, Girard EJ, et al. A biobank of patient-derived pediatric brain tumor models. Nat Med 2018;24:1752–61 [DOI] [PubMed] [Google Scholar]
- 154.Richter-Pechanska P, Kunz JB, Bornhauser B, von Knebel Doeberitz C, Rausch T, Erarslan-Uysal B, et al. PDX models recapitulate the genetic and epigenetic landscape of pediatric T-cell leukemia. EMBO Mol Med 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kolb EA, Houghton PJ, Kurmasheva RT, Mosse YP, Maris JM, Erickson SW, et al. Preclinical evaluation of the combination of AZD1775 and irinotecan against selected pediatric solid tumors: A Pediatric Preclinical Testing Consortium report. Pediatr Blood Cancer 2020;67:e28098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Stripecke R, Munz C, Schuringa JJ, Bissig KD, Soper B, Meeham T, et al. Innovations, challenges, and minimal information for standardization of humanized mice. EMBO Mol Med 2020;12:e8662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer 2018;18:407–18 [DOI] [PubMed] [Google Scholar]
- 158.Calandrini C, Schutgens F, Oka R, Margaritis T, Candelli T, Mathijsen L, et al. An organoid biobank for childhood kidney cancers that captures disease and tissue heterogeneity. Nat Commun 2020;11:1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Saltsman JA, Hammond WJ, Narayan NJC, Requena D, Gehart H, Lalazar G, et al. A Human Organoid Model of Aggressive Hepatoblastoma for Disease Modeling and Drug Testing. Cancers (Basel) 2020;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Torres R, Martin MC, Garcia A, Cigudosa JC, Ramirez JC, Rodriguez-Perales S. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nat Commun 2014;5:3964. [DOI] [PubMed] [Google Scholar]
- 161.Spraggon L, Martelotto LG, Hmeljak J, Hitchman TD, Wang J, Wang L, et al. Generation of conditional oncogenic chromosomal translocations using CRISPR-Cas9 genomic editing and homology-directed repair. J Pathol 2017;242:102–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Torres-Ruiz R, Martinez-Lage M, Martin MC, Garcia A, Bueno C, Castano J, et al. Efficient Recreation of t(11;22) EWSR1-FLI1(+) in Human Stem Cells Using CRISPR/Cas9. Stem Cell Reports 2017;8:1408–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Vanoli F, Tomishima M, Feng W, Lamribet K, Babin L, Brunet E, et al. CRISPR-Cas9-guided oncogenic chromosomal translocations with conditional fusion protein expression in human mesenchymal cells. Proc Natl Acad Sci U S A 2017;114:3696–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lagutina IV, Valentine V, Picchione F, Harwood F, Valentine MB, Villarejo-Balcells B, et al. Modeling of the human alveolar rhabdomyosarcoma Pax3-Foxo1 chromosome translocation in mouse myoblasts using CRISPR-Cas9 nuclease. PLoS Genet 2015;11:e1004951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Minas TZ, Surdez D, Javaheri T, Tanaka M, Howarth M, Kang H-J, et al. Combined experience of six independent laboratories attempting to create an Ewing sarcoma mouse model. Oncotarget 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science 2015;348:74–80 [DOI] [PubMed] [Google Scholar]
- 167.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019;37:773–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhou Y, Yang D, Yang Q, Lv X, Huang W, Zhou Z, et al. Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat Commun 2020;11:6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gomez-Brouchet A, Illac C, Gilhodes J, Bouvier C, Aubert S, Guinebretiere JM, et al. CD163-positive tumor-associated macrophages and CD8-positive cytotoxic lymphocytes are powerful diagnostic markers for the therapeutic stratification of osteosarcoma patients: An immunohistochemical analysis of the biopsies fromthe French OS2006 phase 3 trial. Oncoimmunology 2017;6:e1331193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ligon JA, Choi W, Cojocaru G, Fu W, Hsiue EH, Oke TF, et al. Pathways of immune exclusion in metastatic osteosarcoma are associated with inferior patient outcomes. J Immunother Cancer 2021;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sorenson L, Fu Y, Hood T, Warren S, McEachron TA. Targeted transcriptional profiling of the tumor microenvironment reveals lymphocyte exclusion and vascular dysfunction in metastatic osteosarcoma. Oncoimmunology 2019;8:e1629779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wu CC, Beird HC, Andrew Livingston J, Advani S, Mitra A, Cao S, et al. Immuno-genomic landscape of osteosarcoma. Nat Commun 2020;11:1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhou Q, Xian M, Xiang S, Xiang D, Shao X, Wang J, et al. All-Trans Retinoic Acid Prevents Osteosarcoma Metastasis by Inhibiting M2 Polarization of Tumor-Associated Macrophages. Cancer Immunol Res 2017;5:547–59 [DOI] [PubMed] [Google Scholar]
- 176.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 2009;9:162–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Long AH, Highfill SL, Cui Y, Smith JP, Walker AJ, Ramakrishna S, et al. Reduction of MDSCs with All-trans Retinoic Acid Improves CAR Therapy Efficacy for Sarcomas. Cancer Immunol Res 2016;4:869–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Machado I, Lopez-Guerrero JA, Scotlandi K, Picci P, Llombart-Bosch A. Immunohistochemical analysis and prognostic significance of PD-L1, PD-1, and CD8+ tumor-infiltrating lymphocytes in Ewing’s sarcoma family of tumors (ESFT). Virchows Arch 2018;472:815–24 [DOI] [PubMed] [Google Scholar]
- 179.Mochizuki K, Kawana S, Yamada S, Muramatsu M, Sano H, Kobayashi S, et al. Various checkpoint molecules, and tumor-infiltrating lymphocytes in common pediatric solid tumors: Possibilities for novel immunotherapy. Pediatr Hematol Oncol 2019;36:17–27 [DOI] [PubMed] [Google Scholar]
- 180.Heitzeneder S, Sotillo E, Shern JF, Sindiri S, Xu P, Jones R, et al. Pregnancy-Associated Plasma Protein-A (PAPP-A) in Ewing Sarcoma: Role in Tumor Growth and Immune Evasion. J Natl Cancer Inst 2019;111:970–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Spurny C, Kailayangiri S, Altvater B, Jamitzky S, Hartmann W, Wardelmann E, et al. T cell infiltration into Ewing sarcomas is associated with local expression of immune-inhibitory HLA-G. Oncotarget 2018;9:6536–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sand LG, Berghuis D, Szuhai K, Hogendoorn PC. Expression of CCL21 in Ewing sarcoma shows an inverse correlation with metastases and is a candidate target for immunotherapy. Cancer Immunol Immunother 2016;65:995–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Melaiu O, Chierici M, Lucarini V, Jurman G, Conti LA, De Vito R, et al. Cellular and gene signatures of tumor-infiltrating dendritic cells and natural-killer cells predict prognosis of neuroblastoma. Nat Commun 2020;11:5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Xin C, Zhu J, Gu S, Yin M, Ma J, Pan C, et al. CD200 is overexpressed in neuroblastoma and regulates tumor immune microenvironment. Cancer Immunol Immunother 2020;69:2333–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wei JS, Kuznetsov IB, Zhang S, Song YK, Asgharzadeh S, Sindiri S, et al. Clinically Relevant Cytotoxic Immune Cell Signatures and Clonal Expansion of T-Cell Receptors in High-Risk MYCN-Not-Amplified Human Neuroblastoma. Clin Cancer Res 2018;24:5673–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Layer JP, Kronmuller MT, Quast T, van den Boorn-Konijnenberg D, Effern M, Hinze D, et al. Amplification of N-Myc is associated with a T-cell-poor microenvironment in metastatic neuroblastoma restraining interferon pathway activity and chemokine expression. Oncoimmunology 2017;6:e1320626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bao R, Spranger S, Hernandez K, Zha Y, Pytel P, Luke JJ, et al. Immunogenomic determinants of tumor microenvironment correlate with superior survival in high-risk neuroblastoma. J Immunother Cancer 2021;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Mao Y, Eissler N, Blanc KL, Johnsen JI, Kogner P, Kiessling R. Targeting Suppressive Myeloid Cells Potentiates Checkpoint Inhibitors to Control Spontaneous Neuroblastoma. Clin Cancer Res 2016;22:3849–59 [DOI] [PubMed] [Google Scholar]
- 189.Tian K, Wang X, Wu Y, Wu X, Du G, Liu W, et al. Relationship of tumour-associated macrophages with poor prognosis in Wilms’ tumour. J Pediatr Urol 2020;16:376 e1- e8 [DOI] [PubMed] [Google Scholar]
- 190.Maturu P, Jones D, Ruteshouser EC, Hu Q, Reynolds JM, Hicks J, et al. Role of Cyclooxygenase-2 Pathway in Creating an Immunosuppressive Microenvironment and in Initiation and Progression of Wilms’ Tumor. Neoplasia 2017;19:237–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chen L, Oke T, Siegel N, Cojocaru G, Tam AJ, Blosser RL, et al. The Immunosuppressive Niche of Soft-Tissue Sarcomas is Sustained by Tumor-Associated Macrophages and Characterized by Intratumoral Tertiary Lymphoid Structures. Clin Cancer Res 2020;26:4018–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kather JN, Horner C, Weis CA, Aung T, Vokuhl C, Weiss C, et al. CD163+ immune cell infiltrates and presence of CD54+ microvessels are prognostic markers for patients with embryonal rhabdomyosarcoma. Sci Rep 2019;9:9211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Highfill SL, Cui Y, Giles AJ, Smith JP, Zhang H, Morse E, et al. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci Transl Med 2014;6:237ra67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Grabovska Y, Mackay A, O’Hare P, Crosier S, Finetti M, Schwalbe EC, et al. Pediatric pan-central nervous system tumor analysis of immune-cell infiltration identifies correlates of antitumor immunity. Nat Commun 2020;11:4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Petralia F, Tignor N, Reva B, Koptyra M, Chowdhury S, Rykunov D, et al. Integrated Proteogenomic Characterization across Major Histological Types of Pediatric Brain Cancer. Cell 2020;183:1962–85 e31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Pham CD, Flores C, Yang C, Pinheiro EM, Yearley JH, Sayour EJ, et al. Differential Immune Microenvironments and Response to Immune Checkpoint Blockade among Molecular Subtypes of Murine Medulloblastoma. Clin Cancer Res 2016;22:582–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Maximov V, Chen Z, Wei Y, Robinson MH, Herting CJ, Shanmugam NS, et al. Tumour-associated macrophages exhibit anti-tumoural properties in Sonic Hedgehog medulloblastoma. Nat Commun 2019;10:2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Garancher A, Suzuki H, Haricharan S, Chau LQ, Masihi MB, Rusert JM, et al. Tumor necrosis factor overcomes immune evasion in p53-mutant medulloblastoma. Nat Neurosci 2020;23:842–53 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 199.Bockmayr M, Klauschen F, Maire CL, Rutkowski S, Westphal M, Lamszus K, et al. Immunologic Profiling of Mutational and Transcriptional Subgroups in Pediatric and Adult High-Grade Gliomas. Cancer Immunol Res 2019;7:1401–11 [DOI] [PubMed] [Google Scholar]
- 200.Robinson MH, Vasquez J, Kaushal A, MacDonald TJ, Velazquez Vega JE, Schniederjan M, et al. Subtype and grade-dependent spatial heterogeneity of T-cell infiltration in pediatric glioma. J Immunother Cancer 2020;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Haberthur K, Brennan K, Hoglund V, Balcaitis S, Chinn H, Davis A, et al. NKG2D ligand expression in pediatric brain tumors. Cancer Biol Ther 2016;17:1253–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lieberman NAP, DeGolier K, Kovar HM, Davis A, Hoglund V, Stevens J, et al. Characterization of the immune microenvironment of diffuse intrinsic pontine glioma: implications for development of immunotherapy. Neuro Oncol 2019;21:83–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lin GL, Nagaraja S, Filbin MG, Suva ML, Vogel H, Monje M. Non-inflammatory tumor microenvironment of diffuse intrinsic pontine glioma. Acta Neuropathol Commun 2018;6:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hanaizi Z, van Zwieten-Boot B, Calvo G, Lopez AS, van Dartel M, Camarero J, et al. The European Medicines Agency review of ipilimumab (Yervoy) for the treatment of advanced (unresectable or metastatic) melanoma in adults who have received prior therapy: summary of the scientific assessment of the Committee for Medicinal Products for Human Use. Eur J Cancer 2012;48:237–42 [DOI] [PubMed] [Google Scholar]
- 205.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Davis KL, Fox E, Merchant MS, Reid JM, Kudgus RA, Liu X, et al. Nivolumab in children and young adults with relapsed or refractory solid tumours or lymphoma (ADVL1412): a multicentre, open-label, single-arm, phase 1–2 trial. Lancet Oncol 2020;21:541–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Majzner RG, Simon JS, Grosso JF, Martinez D, Pawel BR, Santi M, et al. Assessment of programmed death-ligand 1 expression and tumor-associated immune cells in pediatric cancer tissues. Cancer 2017;123:3807–15 [DOI] [PubMed] [Google Scholar]
- 208.Coronado E, Yanez Y, Vidal E, Rubio L, Vera-Sempere F, Canada-Martinez AJ, et al. Intratumoral immunosuppression profiles in 11q-deleted neuroblastomas provide new potential therapeutic targets. Mol Oncol 2021;15:364–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.McEachron TA, Triche TJ, Sorenson L, Parham DM, Carpten JD. Profiling targetable immune checkpoints in osteosarcoma. Oncoimmunology 2018;7:e1475873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bouffet E, Larouche V, Campbell BB, Merico D, de Borja R, Aronson M, et al. Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. J Clin Oncol 2016;34:2206–11 [DOI] [PubMed] [Google Scholar]
- 211.Mandal R, Samstein RM, Lee KW, Havel JJ, Wang H, Krishna C, et al. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science 2019;364:485–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Le Cesne A, Marec-Berard P, Blay JY, Gaspar N, Bertucci F, Penel N, et al. Programmed cell death 1 (PD-1) targeting in patients with advanced osteosarcomas: results from the PEMBROSARC study. Eur J Cancer 2019;119:151–7 [DOI] [PubMed] [Google Scholar]
- 213.Geoerger B, Kang HJ, Yalon-Oren M, Marshall LV, Vezina C, Pappo A, et al. Pembrolizumab in paediatric patients with advanced melanoma or a PD-L1-positive, advanced, relapsed, or refractory solid tumour or lymphoma (KEYNOTE-051): interim analysis of an open-label, single-arm, phase 1–2 trial. Lancet Oncol 2020;21:121–33 [DOI] [PubMed] [Google Scholar]
- 214.Merchant MS, Wright M, Baird K, Wexler LH, Rodriguez-Galindo C, Bernstein D, et al. Phase I Clinical Trial of Ipilimumab in Pediatric Patients with Advanced Solid Tumors. Clin Cancer Res 2016;22:1364–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Maude SL, Laetsch TW, Buechner J, Rives S, Boyer M, Bittencourt H, et al. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N Engl J Med 2018;378:439–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhang L, Zuo Y, Lu A, Wu J, Jia Y, Wang Y, et al. Safety and Efficacy of Chimeric Antigen Receptor T-Cell Therapy in Children With Central Nervous System Leukemia. Clin Lymphoma Myeloma Leuk 2021;21:e410–e4 [DOI] [PubMed] [Google Scholar]
- 217.Tan Y, Pan J, Deng B, Ling Z, Song W, Xu J, et al. Toxicity and effectiveness of CD19 CAR T therapy in children with high-burden central nervous system refractory B-ALL. Cancer Immunol Immunother 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Shalabi H, Gust J, Taraseviciute A, Wolters PL, Leahy AB, Sandi C, et al. Beyond the storm - subacute toxicities and late effects in children receiving CAR T cells. Nat Rev Clin Oncol 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Li X, Chen W. Mechanisms of failure of chimeric antigen receptor T-cell therapy. Curr Opin Hematol 2019;26:427–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Pan J, Zuo S, Deng B, Xu X, Li C, Zheng Q, et al. Sequential CD19–22 CAR T therapy induces sustained remission in children with r/r B-ALL. Blood 2020;135:387–91 [DOI] [PubMed] [Google Scholar]
- 221.Hua J, Qian W, Wu X, Zhou L, Yu L, Chen S, et al. Sequential Infusion of Anti-CD22 and Anti-CD19 Chimeric Antigen Receptor T Cells for a Pediatric Ph-Like B-ALL Patient That Relapsed After CART-Cell and Haplo-HSCT Therapy: A Case Report and Review of Literature. Onco Targets Ther 2020;13:2311–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Qin H, Ramakrishna S, Nguyen S, Fountaine TJ, Ponduri A, Stetler-Stevenson M, et al. Preclinical Development of Bivalent Chimeric Antigen Receptors Targeting Both CD19 and CD22. Mol Ther Oncolytics 2018;11:127–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ghorashian S, Kramer AM, Onuoha S, Wright G, Bartram J, Richardson R, et al. Enhanced CAR T cell expansion and prolonged persistence in pediatric patients with ALL treated with a low-affinity CD19 CAR. Nat Med 2019;25:1408–14 [DOI] [PubMed] [Google Scholar]
- 224.Shah NN, Lee DW, Yates B, Yuan CM, Shalabi H, Martin S, et al. Long-Term Follow-Up of CD19-CAR T-Cell Therapy in Children and Young Adults With B-ALL. J Clin Oncol 2021:JCO2002262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Nazha B, Inal C, Owonikoko TK. Disialoganglioside GD2 Expression in Solid Tumors and Role as a Target for Cancer Therapy. Front Oncol 2020;10:1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Xu X, Huang W, Heczey A, Liu D, Guo L, Wood M, et al. NKT Cells Coexpressing a GD2-Specific Chimeric Antigen Receptor and IL15 Show Enhanced In Vivo Persistence and Antitumor Activity against Neuroblastoma. Clin Cancer Res 2019;25:7126–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Chulanetra M, Morchang A, Sayour E, Eldjerou L, Milner R, Lagmay J, et al. GD2 chimeric antigen receptor modified T cells in synergy with sub-toxic level of doxorubicin targeting osteosarcomas. Am J Cancer Res 2020;10:674–87 [PMC free article] [PubMed] [Google Scholar]
- 228.Mount CW, Majzner RG, Sundaresh S, Arnold EP, Kadapakkam M, Haile S, et al. Potent antitumor efficacy of anti-GD2 CAR T cells in H3-K27M(+) diffuse midline gliomas. Nat Med 2018;24:572–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sujjitjoon J, Sayour E, Tsao ST, Uiprasertkul M, Sanpakit K, Buaboonnam J, et al. GD2-specific chimeric antigen receptor-modified T cells targeting retinoblastoma - assessing tumor and T cell interaction. Transl Oncol 2021;14:100971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Andersch L, Radke J, Klaus A, Schwiebert S, Winkler A, Schumann E, et al. CD171- and GD2-specific CAR-T cells potently target retinoblastoma cells in preclinical in vitro testing. BMC Cancer 2019;19:895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Chen Y, Sun C, Landoni E, Metelitsa L, Dotti G, Savoldo B. Eradication of Neuroblastoma by T Cells Redirected with an Optimized GD2-Specific Chimeric Antigen Receptor and Interleukin-15. Clin Cancer Res 2019;25:2915–24 [DOI] [PubMed] [Google Scholar]
- 232.Heczey A, Louis CU, Savoldo B, Dakhova O, Durett A, Grilley B, et al. CAR T Cells Administered in Combination with Lymphodepletion and PD-1 Inhibition to Patients with Neuroblastoma. Mol Ther 2017;25:2214–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Straathof K, Flutter B, Wallace R, Jain N, Loka T, Depani S, et al. Antitumor activity without on-target off-tumor toxicity of GD2-chimeric antigen receptor T cells in patients with neuroblastoma. Sci Transl Med 2020;12. [DOI] [PubMed] [Google Scholar]
- 234.Shurell E, Vergara-Lluri ME, Li Y, Crompton JG, Singh A, Bernthal N, et al. Comprehensive adipocytic and neurogenic tissue microarray analysis of NY-ESO-1 expression - a promising immunotherapy target in malignant peripheral nerve sheath tumor and liposarcoma. Oncotarget 2016;7:72860–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Iura K, Maekawa A, Kohashi K, Ishii T, Bekki H, Otsuka H, et al. Cancer-testis antigen expression in synovial sarcoma: NY-ESO-1, PRAME, MAGEA4, and MAGEA1. Hum Pathol 2017;61:130–9 [DOI] [PubMed] [Google Scholar]
- 236.D’Angelo SP, Melchiori L, Merchant MS, Bernstein D, Glod J, Kaplan R, et al. Antitumor Activity Associated with Prolonged Persistence of Adoptively Transferred NY-ESO-1 (c259)T Cells in Synovial Sarcoma. Cancer Discov 2018;8:944–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ramachandran I, Lowther DE, Dryer-Minnerly R, Wang R, Fayngerts S, Nunez D, et al. Systemic and local immunity following adoptive transfer of NY-ESO-1 SPEAR T cells in synovial sarcoma. J Immunother Cancer 2019;7:276. [DOI] [PMC free article] [PubMed] [Google Scholar]