Abstract
Animals are shaped through the movement of large cellular collectives. Such morphogenetic processes require cadherin-based cell adhesion to maintain tissue cohesion and planar cell polarity to coordinate movement. Despite a vast literature surrounding cadherin-based adhesion and planar cell polarity, it is unclear how these molecular networks interface. Here we investigate the relationship between cadherins and planar cell polarity during gastrulation cell movements in Xenopus laevis. We first assessed bulk cadherin localization and found that cadherins were enriched at a specific subset of morphogenetically active cell–cell junctions. We then found that cadherin and actin had coupled temporal dynamics and that disruption of planar cell polarity uncoupled these dynamics. Next, using superresolution time-lapse microscopy and quantitative image analysis, we were able to measure the lifespan and size of individual cadherin clusters. Finally, we show that planar cell polarity not only controls the size of cadherin clusters but, more interestingly, regulates cluster stability. These results reveal an intriguing link between two essential cellular properties, adhesion and planar polarity, and provide insight into the molecular control of morphogenetic cell movements.
INTRODUCTION
Embryonic development requires the stereotyped movement of cellular collectives. An animal’s anterior–posterior (head-to-tail) axis is established in part through a specific form of collective cell movements termed convergent extension (CE) in which cells converge along one axis, resulting in extension along the perpendicular axis (Keller and Sutherland, 2020). CE is a deeply conserved and essential cellular process and has been described in organisms ranging from nematodes to mammals (Huebner and Wallingford, 2018), and failure of CE is linked to human birth defects (Wallingford et al., 2013). Thus, cultivating a deeper knowledge of CE not only will inform our understanding of this essential developmental process but will also provide insight into the etiology of human disease.
Vertebrate CE is patterned through the asymmetric localization of planar cell polarity (PCP) proteins, which allows cells to sense directionality along the anterior–posterior axis (Butler and Wallingford, 2017). CE also requires cell–cell adhesion to maintain tissue integrity and to transduce forces between neighboring cells (Lecuit and Yap, 2015). Such adhesion is mediated by cadherins, homophilic cell adhesion molecules that are required for a multitude of collective cell behaviors (Arslan et al., 2021), including vertebrate CE (Lee and Gumbiner, 1995). While PCP and cadherin-based cell adhesions have been studied in great depth, relatively little is known about how these cellular activities interface during CE.
PCP signaling has been shown to function upstream of cadherin-mediated cell adhesion in Xenopus, Drosophila, zebrafish, mice, and cell culture (Mirkovic et al., 2011; Kraft et al., 2012; Dohn et al., 2013; Tatin et al., 2013; Nagaoka et al., 2014), and multiple mechanisms have been proposed for the underlying interaction. One such mechanism is that PCP controls the ability of cadherins to form higher ordered clusters that modulate adhesion (Kraft et al., 2012). Recently we found that perturbation of PCP disrupted cis-clustering of Xenopus Cdh3 (aka C-cadherin, aka mammalian P-cadherin) (Huebner et al., 2021). This result led us to perform a deeper investigation into the relationship between PCP and cadherin during axis elongation in Xenopus.
Here, we show that Cdh3 is enriched specifically at shortening cell–cell junctions during Xenopus CE. Next, we determine that PCP was required for the temporal coordination of cadherin and actin dynamics. We then developed a new quantitative imaging approach to measure the size and, importantly, the lifespan of individual cadherin clusters, revealing that clusters are highly dynamic and heterogeneous in both size and lifespan. Finally, we demonstrate that perturbation of PCP reduced cluster size and, more interestingly, reduced cluster lifespan. These results confirm a relationship between PCP and cadherin clustering and specifically show that PCP is required for stabilizing clusters. These data not only improve our understanding of two essential molecular networks, PCP and cadherin-based adhesions, but also deepen our understanding of the molecular control of a fundamental developmental process, convergent extension.
RESULTS AND DISCUSSION
Cadherins are enriched at shortening cell–cell junctions during CE
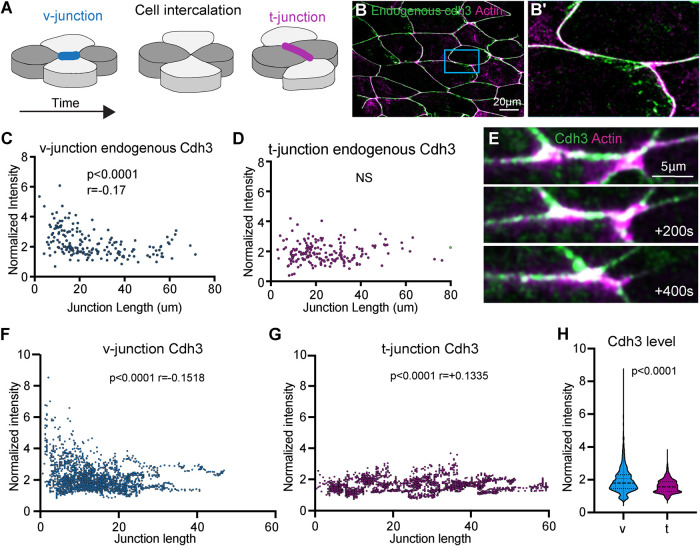
Xenopus CE results from cell intercalation, during which cells come together along the mediolateral axis and are pushed apart along the anterior–posterior direction (Shih and Keller, 1992) (Figure 1A). During this process, mediolaterally oriented junctions (termed “v-junctions” by convention) shorten and are replaced by anterior–posterior-oriented junctions (termed “t-junctions”) (Bertet et al., 2004; Blankenship et al., 2006) (Figure 1A). Cadherins are planar polarized to t-junctions during CE in the Drosophila germ band (Blankenship et al., 2006), so we began our study by asking whether cadherins are similarly polarized during CE in Xenopus.
FIGURE 1:
Cdh3 is enriched at shortening v-junctions. (A) Schematic depicting the cell intercalation movements that drive convergent extension. (B) Image of Xenopus mesodermal cells immunostained for Cdh3 (green) and labeled for actin (magenta). (B′) Zoomed image of a single v-junction from the inset in B. This image highlights that endogenous Cdh3 is present at v-junctions. (C) Plot showing endogenous Cdh3 intensity vs. cell–cell junction length at v-junctions. A Spearman correlation was used to assess the relationship between Cdh3 and junction length. (D) Plot of endogenous Cdh3 intensity vs. t-junction length. The relationships between intensity and junction length were compared using a Spearman correlation. (E) Frames from a time-lapse movie of a shortening v-junction labeled with Cdh3-GFP (green) and the actin marker lifeact-RFP (magenta). (F) Graph comparing mean Cdh3 intensity to v-junction length from time-lapse movies of actively shortening v-junctions. The relationship between Cdh3 intensity and v-junction length was assessed by Spearman correlation. (G) Comparison of Cdh3 intensity to t-junction length from time-lapse movies. Spearman correlation was used to assess the relationship between Cdh3 intensity and t-junction length. (H) Graph of the mean intensity at shortening v-junctions or at t-junctions. Intensities were collected from time-lapse movies and were normalized to account for bleaching. Conditions were statistically compared using a Mann–Whitney test.
To assess Cdh3 polarity we used immunostaining to observe the endogenous Cdh3 localization at v- and t-junctions. Visual inspection showed that Cdh3 was present at v-junctions (Figure 1, B and B′). We next measured endogenous Cdh3 fluorescence intensities at v- and t-junctions. Interestingly we observed a slight but significant negative correlation between Cdh3 intensity and v-junction length, such that Cdh3 was enriched at the shortest v-junctions (Figure 1C). No such correlation was observed at t-junctions (Figure 1D). This result led us to ask how Cdh3 levels change specifically at shortening v-junctions.
To address this, we used a well-characterized, functional Cdh3-GFP fusion protein (Pfister et al., 2016; Huebner et al., 2021) to visualize and quantify Cdh3 protein localization in real time at shortening v-junctions (Figure 1E). Quantification of mean Cdh3-GFP intensity at shortening v-junctions showed a clear enrichment of Cdh3 during junction shortening, such that the shortest v-junctions had the highest Cdh3 intensity (Figure 1F). No such correlation was observed between junction length and Cdh3 intensity at t-junctions (Figure 1G). Further, when comparing Cdh3 intensity at shortening v-junctions to t-junctions, there was a significant enrichment for Cdh3 at the shortening v-junctions (Figure 1H). Our interpretation of these data is that Cdh3 is not necessarily polarized during Xenopus CE but instead becomes increasingly enriched at shortening v-junctions.
These results are intriguing because cadherins in Xenopus CE are not planar polarized as observed in Drosophila, which has been the paradigm for studying cadherin function during CE (Blankenship et al., 2006). These results are further surprising because the patterns of actomyosin localization are largely the same in these two systems (Bertet et al., 2004; Blankenship et al., 2006; Shindo and Wallingford, 2014; Butler and Wallingford, 2018). Thus, it seemsthat there is a fundamental difference in Cdh3 function during Xenopus mesenchymal CE compared with cadherins during Drosophila germ band extension CE.
PCP is required for synchronized cadherin and actin oscillations
The actomyosin cytoskeleton provides the force to move cellular collectives, and it has become increasingly clear that actin and myosin function through oscillatory periods of activity and inactivity (Coravos et al., 2017). In Xenopus, actomyosin oscillations are observed at v-junctions and proper tuning of the frequency and amplitude of oscillations is required for efficient cell intercalation (Shindo et al., 2019; Huebner et al., 2022). Further, Cdh3 has also been observed to oscillate at v-junctions in Xenopus, and oscillatory changes in cadherin cluster size correlate with v-junction shortening events (Huebner et al., 2021). These results led us to ask whether there is a temporal relationship between Cdh3 and actin oscillations.
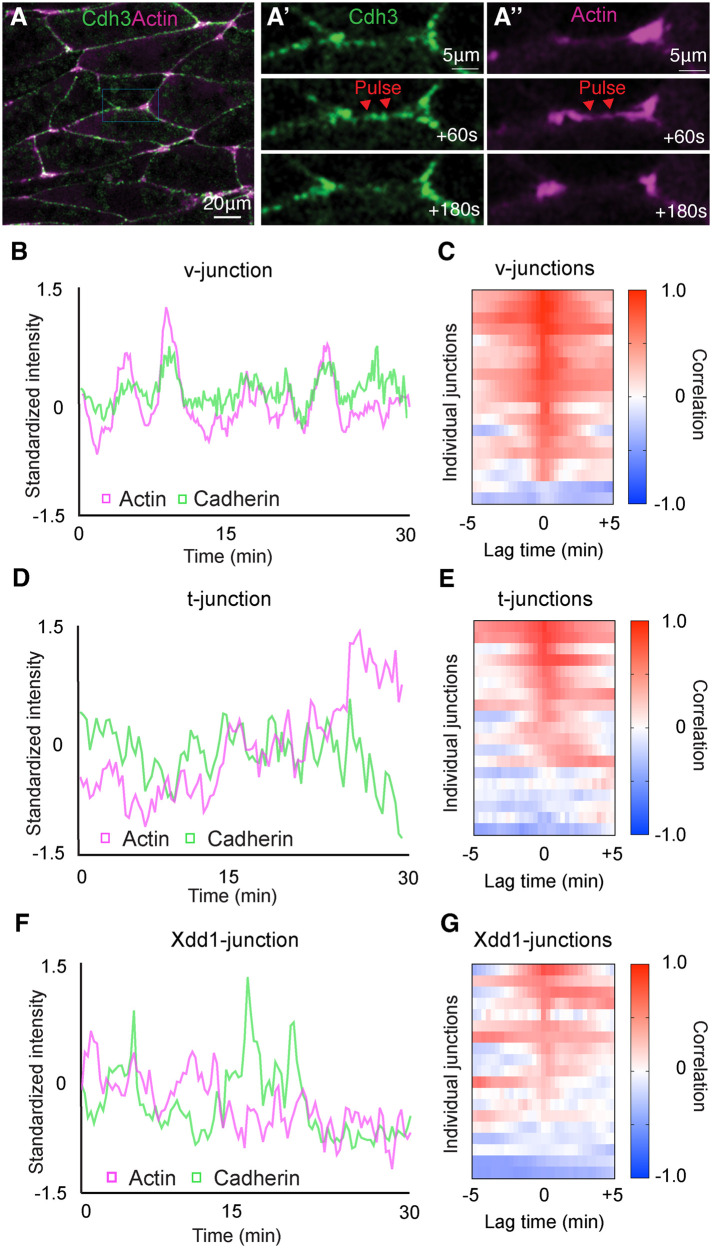
To this end, we collected time-lapse movies of Cdh3 and actin at shortening v-junctions. We observed oscillatory pulses of Cdh3 and actin, and these were clearly correlated in time (Figure 2, A–A′′ and B). Moreover, cross-correlation analysis confirmed a strong positive correlation between actin and Cdh3 oscillations (Figure 2C). Interestingly, such coordinated behavior was specific to shortening v-junctions, as Cdh3 and actin intensity were poorly coordinated at t-junctions when compared with v-junctions (Figure 2, D and E). These data show a clear coordination of Cdh3 and actin oscillations specifically at shortening v-junctions, which led us to ask whether PCP was required for this asymmetric oscillatory behavior.
FIGURE 2:
PCP is required for synchronized cadherin and actin dynamics. (A) Frame from a time-lapse movie of intercalating cells labeled for Cdh3 in green and actin in magenta. (A′) Time series of Cdh3 from the inset in A. Here we are highlighting a shortening v-junction and showing that Cdh3 undergoes oscillatory pulses at said junctions. (A′′) Time series of actin from the inset in A. In this time series there is an actin pulse that occurs concurrently with the Cdh3 pulse shown in A′. (B) Graph plotting Cdh3 and actin intensity over time at a shortening v-junction. Here we observe concurrent peaks of Cdh3 and actin. Intensities were normalized to account for bleaching and then standardized to emphasize the overlap in the two signals over time. (C) Heatmap of the cross-correlation analysis for 19 shortening v-junctions. Each line on the y-axis of this heatmap represents the cross-correlation for a single shortening v-junction. The value of the correlation is represented in color code with red being positive correlation and blue being negative correlation. The x-axis shows the lag time, and the fact that we observe a high positive correlation at lag time 0 indicates that Cdh3 and actin pulses occur concurrently. (D) Graph plotting Cdh3 and actin intensities at t-junctions. (E) Heatmap of the cross-correlation analysis for 19 t-junctions. (F) Graph of Cdh3 and actin over time measured at junctions where Xdd1 was present in the background. (G) Heatmap of the cross-correlation analysis for 19 junctions with Xdd1 in the background.
Next, we used a dominant negative dishevelled-2 (Xdd1), which specifically disrupts PCP, to test whether PCP was required for the coordinated Cdh3 and actin oscillations (Sokol, 1996; Wallingford et al., 2000). We collected time-lapse movies of Cdh3 and actin with Xdd1 in the background, and here we specifically chose junctions oriented as v-junctions. We found that Cdh3 and actin were poorly cross-correlated after PCP perturbation (Figure 2, F and G). These data suggest that PCP has a role in coupling Cdh3 and actin dynamics and overall, these results show that Cdh3 and actin undergo PCP-dependent coordinated oscillations specifically at v-junctions.
Planar cell parity is not required for cadherin turnover
Having determined that PCP was required for the bulk localization and temporal dynamics of Cdh3, we next asked how PCP controls these features. Cadherin protein levels and dynamics can be modulated by altering the protein turnover at cell junctions (Cavey et al., 2008). One method to assess bulk protein turnover is fluorescence recovery after photobleaching (FRAP) (Reits and Neefjes, 2001). We therefore performed FRAP on Cdh3 (Supplemental Figure 1A) in the presence or absence of Xdd1. Surprisingly, we observed no difference in the FRAP curves, the recovery half-time, or the recovery mobile fraction (Supplemental Figure 1, B–D). These results show that PCP does not govern bulk Cdh3 turnover at cell–cell junctions and instead must regulate some other aspect of Cdh3 behavior.
Cadherins clusters are dynamic and heterogeneous during Xenopus CE
Cadherins form intercellular and intracellular interactions with fellow cadherins to generate adhesion complexes, termed clusters, that are observable from the nano to the micron scale (Yap et al., 2015). We have recently shown that cadherin clustering is required for CE and that perturbation of PCP altered cadherin cluster sizes (Huebner et al., 2021). However, our previous work was averaged over large data sets and did not allow us to study the behavior of individual clusters. We therefore sought to develop a method to measure the size and, importantly, the lifespan of individual Cdh3 clusters. It is of note that we are particularly interested in cluster lifespan as temporal clustering dynamics correlates with v-junction shortening (Huebner et al., 2021) and because, to our knowledge, there are few or no current methods to measure cluster lifespan.
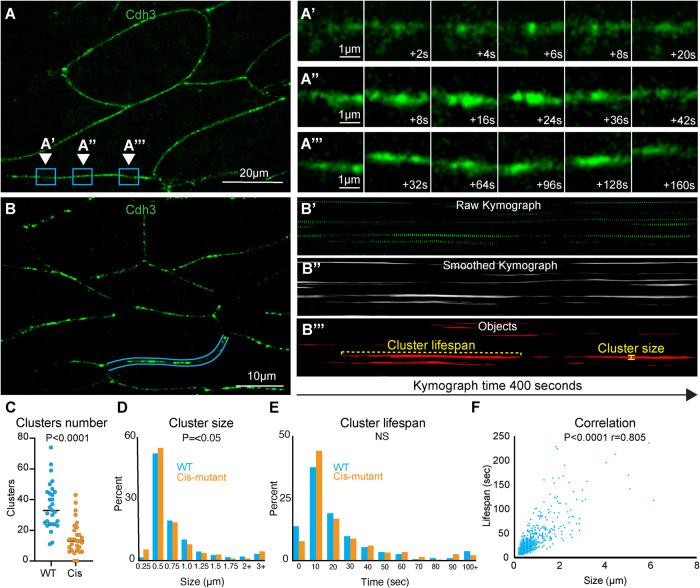
Here we collected superresolution time-lapse movies of the classical cadherin Cdh3 during CE. We were able to clearly observe the formation and dissipation of submicron-scale cadherin clusters (Figure 3, A–A′′′). From these movies it was apparent that cluster size and lifespan were heterogeneous (Figure 3, A–A′′′). To measure cluster size and lifespan, we first made kymographs across the length of individual cell–cell junctions (Figure 3, B and B′). Kymographs were then thresholded to remove background and smoothed along the time axis to make clusters a continuous line in the image (Figure 3B′′). A binary mask was then applied to the kymographs that allowed us to identify each cluster as an individual object, and objects touching the edges of the kymograph were discarded (Figure 3B′′′). Using this method, we were able to measure the size, lifespan, and number of clusters observed at a cell junction (Figure 3, B′′′–E). Interestingly, we observed a tight correlation between cluster size and lifespan, indicating that these features are not independent (Figure 3F). Importantly, the mean cluster size observed with this new method matched well with the previously published mean cluster size observed in our large averaged data sets (Huebner et al., 2021), providing evidence that our new approach is effective.
FIGURE 3:
Cadherin clusters display heterogeneous spatiotemporal dynamics during CE. (A) Still frame from a superresolution time-lapse movie of Cdh3. Three different regions of a single junction are highlighted; each region is the position where a cluster formed and dissipated. These regions were chosen to highlife the heterogeneity in cluster size and lifespan. (A′) Frames from the time-lapse movie shown in A with a zoom on the inset labeled A′. These images show a small, short lifespan Cdh3 cluster. (A′′) Frames from the time-lapse movie shown in A with a zoom on the inset labeled A′′. Images show a large cluster with an intermediate lifespan. (A′′′) Frames from the time-lapse movie shown in A with a zoom on the inset labeled A′′′. Images show a particularly long-lived Cdh3 cluster. (B) Still frame from a time-lapse superresolution movie of Cdh3. Here the blue lines highlight the cell junction that was used to make the kymographs in B′–B′′′. (B′) The raw kymograph of the cell junction highlighted in B. This kymograph shows Cdh3 over a 400 s time period beginning at the left side of the image. (B′′) Smoothed image of the kymograph shown in B′. This kymograph was smoothed along the time axis to connect the Cdh3 spots into uninterrupted clusters. (B′′′) This image shows a thresholded version of the kymograph in B′′. Here the kymograph was thresholded and converted to a binary image that allowed us to identify clusters as individual objects. Objects that touched the edges of the kymograph were removed so that we evaluated only complete clusters. The cluster lifespan was then determined as the cluster length, and the cluster size was determined by the width of each cluster. (C) Graph showing the number of clusters per junction for control and Cdh3-cis-mutant junctions. Each spot represents a single junction, and all junctions were measured over a 400 s time frame. Conditions were statistically compared using a Mann–Whitney test. (D) Histogram displaying the size distribution of control and Cdh3-cis-mutant clusters. Conditions were statistically compared using a Kolmogorov–Smirnov test. (E) Histogram displaying the Cdh3 cluster lifespan distribution for control and Cdh3-cis-mutant clusters. Conditions were statistically compared using a Kolmogorov–Smirnov test. (F) Graph plotting Cdh3 cluster size vs. cluster lifespan from control embryos. Each dot represents a single cluster, and the relationship between size and lifespan was assessed with a Spearman correlation.
We next tested the validity of this new method by applying it to two negative controls, a membrane marker that does not form clusters and a well-characterized cadherin mutant with disrupted clustering (Cdh3-cis-mutant) (Harrison et al., 2011; Huebner et al., 2021). As expected, we observed almost no clustering with the membrane marker (Supplemental Figure 2, A–A′′′). To assess clustering in the Cdh3-cis-mutant, we used a previously developed method for knockdown of the wild-type Cdh3 and replacement with the Cdh3-cis-mutant (Huebner et al., 2021). As expected, expression of the Cdh3-cis-mutant resulted in a significant reduction in the number of clusters observed at each cell junction using the new method (Figure 3C; Supplemental Figure 2, B–B′′′). Interestingly, however, when we compared the distributions of Cdh3 cluster size and lifespan with those of the mutant, we found only a modest difference in size and no difference in lifespan (Figure 3, D and E). These data suggest that the mutant inhibits the ability of clusters to form but has little effect on the dynamics of clusters that do form. Importantly, these results show that we have developed an effective method to measure individual Cdh3 cluster dynamics.
PCP controls individual cadherin cluster size and stability
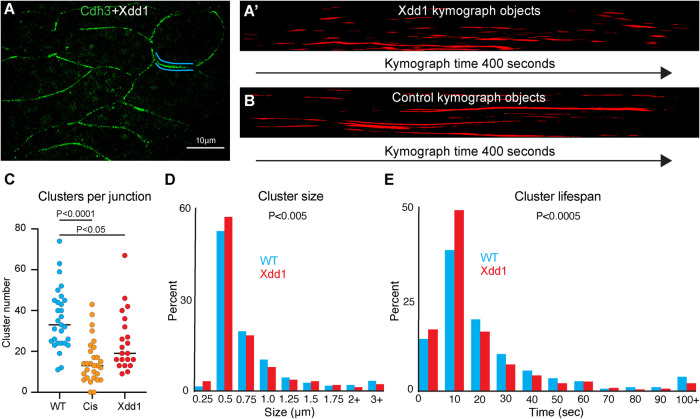
With this new cluster analysis method, we sought to develop a more refined understanding of how PCP regulates Cdh3 clustering. Here we overexpressed Xdd1 and collected superresolution time-lapse movies of Cdh3 (Figure 4A). When visualizing cell junction kymographs of Xdd1-expressing cells, we immediately noticed that the clusters appeared to have much shorter lifespans when compared with control kymographs (Figure 4, A′ and B). Measurement of the number of clusters per junction showed a significant reduction in clusters in the presence of Xdd1, consistent with our observations of the Cdh3-cis-mutant (Figure 4C). We also observed a reduction in cluster size when comparing with Xdd1 (Figure 4D). However, the most striking result was a reduction in cluster lifespan in the presence of Xdd1 (Figure 4E). This change in cluster lifespan was specific to Xdd1 as it was not observed in the Cdh3-cis-mutant. These results indicate that PCP has a role in stabilizing Cdh3 clusters and provides an exciting new link between PCP and cadherin-based cell adhesions.
FIGURE 4:
PCP regulates cadherin cluster stability. (A) Still frame of cells labeled for Cdh3 in green with Xdd1 in the background. Blue lines highlight the junction shown in the kymograph in A′. (A′) Kymograph of the cell junction highlighted in A. This kymograph has been smoothed, thresholded, and converted to objects. (B) Kymograph of a control cell junction for comparison to the Xdd1 junction. This kymograph has also been smoothed, thresholded, and converted to objects. (C) Graph of the number of Cdh3 clusters per junction for control, Cdh3-cis-mutant, and Xdd1. Each dot represents one junction over a 400 s measurement interval. Conditions were compared using an analysis of variance. (D) Histogram displaying the size distribution of control clusters and clusters with Xdd1 in the background. Conditions were statistically compared using a Kolmogorov–Smirnov test. (E) Histogram displaying the cadherin cluster lifespan distribution of control and Xdd1 clusters. Conditions were statistically compared using a Kolmogorov–Smirnov test.
This work is an investigation into the relationship between PCP and cadherin-based cell adhesions during Xenopus gastrulation. Our initial question asked whether Cdh3 was planar polarized to t-junctions during Xenopus CE, as would be expected based on results from Drosophila germband CE (Blankenship et al., 2006). However, we found that Cdh3 became increasingly enriched at shortening v-junctions. This result hints at a critical difference in the behavior of cadherins during Xenopus CE compared with that of Drosophila. Next, we found that PCP was required for coordinated oscillations of Cdh3 and actin, suggesting that PCP controls the coupling of cadherin and actin dynamics. This result was also of interest because previous perturbations of actin or cadherin oscillations altered the frequency (Shindo et al., 2019) or amplitude (Huebner et al., 2022) of oscillations. However, Xdd1 controlled the coupling of the actin and cadherin dynamics, which is a new phenotype relating to oscillations.
Next, we wanted to investigate the relationship between PCP and cadherin clustering as two previous reports linked PCP to clusters in Xenopus (Kraft et al., 2012; Huebner et al., 2021). To this end we developed a method to measure the size, number, and lifespan of individual cadherin clusters. Previous research has shown cadherins physically interacting with the PCP proteins Vangl and Wnt-11/Frizzled-7, and in both cases the interaction with PCP components is thought to sequester cadherins (Kraft et al., 2012; Nagaoka et al., 2014). Here we perturbed the core PCP protein Dishevelled and found that this perturbation resulted in smaller and shorter-lived cadherin clusters. This result provides a new connection between the core PCP protein Dishevelled and the classic cadherin Cdh3. However, it must be noted that our data do not indicate a direct physical interaction between Dishevelled and Cdh3.
We prefer a model in which Dishevelled modulates the actomyosin cytoskeleton and this in turn alters Cdh3 clustering. First, we have published Cdh3 affinity-purification mass spectrometry data from the Xenopus dorsal mesoderm and found no direct interactions with core PCP proteins (Huebner et al., 2022). Second, core PCP proteins, Dishevelled in particular, are known to regulate actomyosin dynamics (Butler and Wallingford, 2017). Third, there is clear evidence that the actomyosin cytoskeleton regulates cluster size (Yap et al., 2015). Finally, we report that actomyosin and cadherin dynamics become uncoupled in the presence of Xdd1. We predict that perturbation of Dishevelled modulates the actomyosin cytoskeleton, which results in poor coupling of actomyosin to cadherins and destabilization of cadherin clusters. While we believe this model accounts for the current data further investigation is required to determine the exact mechanistic link between Dishevelled and Cdh3.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Xenopus embryo manipulations
Adult female Xenopus animals were induced to ovulate by injection of 600 U of human chorionic gonadotropin and kept at 16°C overnight. The following day the ovulating females were gently squeezed to stimulate egg laying and then eggs were fertilized in vitro. Approximately 1.5 h after fertilization, embryos were dejellied with 3% cysteine (pH 8) for 10 min and then washed in 1/3× Marc’s modified Ringer’s (MMR) solution. Before microinjection, embryos were placed in 2% Ficoll in 1/3× MMR, and following microinjection embryos were reared in 1/3× MMR. Embryos were injected using a Parker’s Picospritizer III with an MK1 Manipulator. Embryos were injected at the four-cell stage in the dorsal blastomeres, targeting the presumptive dorsal marginal zone. Embryos were dissected at stage 10.25 in Steinberg’s solution to isolate Keller explants (Keller et al., 1992).
Plasmids, antibody, and morpholino
Cdh3-GFP (Pfister et al., 2016), Cdh3-cis-mutant (Huebner et al., 2021), lifeact-RFP, and membrane-BFP plasmids were made in pCS105, and Xdd1 (Sokol, 1996) was made in CS2myc. The Cdh3 antibody was from the Developmental Studies Hybridoma Bank (catalogue number 6B6). The Cdh3 morpholino was ordered from Gene Tools and had been previously characterized (Ninomiya et al., 2012).
mRNA microinjections
Capped mRNAs were generated using the Thermo Fisher SP6 mMessage mMachine kit (catalogue number AM1340). mRNAs were injected at the indicated concentrations per blastomere: Cdh3-GFP (50 pg), Cdh3-cis-mutant (300 pg), lifeact-RFP (100 pg), membrane-BFP (100 pg), and Xdd1 (1 ng). Cdh3 morpholino was injected at a concertation of 10 ng per blastomere.
Imaging Xenopus explants
Following dissection, explants were incubated at room temperature for 4 h or 16°C overnight before imaging. Explants were maintained in either Steinberg’s solution or Danilchik’s for Amy solution during imaging and were mounted on fibronectin-coated glass coverslips. Superresolution images were acquired with a BioVision Technologies instantaneous structured illumination microscope. Standard resolution confocal images were acquired with either a Zeiss LSM 700 or a Nikon A1R. Superresolution movies were collected with a 2 s time interval, and confocal movies were acquired with a 20 s time interval. All images were acquired at an approximate depth of 5 μm into the tissue.
Measurement of protein intensities at cell–cell junctions
The open-source image analysis software Fiji (Schindelin et al., 2012) was used for image processing and quantification. First, to better visualize the cadherin clusters, images were processed with a 50-pixel rolling ball radius background subtraction and smoothed with a 3 × 3 averaging filter. The segmented line tool was then used to set a line of interest (LOI) across the length of the cell–cell junction with a line width set to the thickness of the junction (16 pixels). The measure tool was then used to measure the mean intensity values. For time-lapse movies, we used the Fiji time-lapse plug-in line interpolator tool to make successive measurements for each time point. Here a LOI was drawn every 10–30 frames and the line interpolator tool was used to fill in the LOIs between the manually drawn LOIs. The measure tool was then used to extract mean intensities at each time point.
Measurement of cadherin cluster sizes and lifespans
Here we used the Fiji time-lapse line interpolator tool to draw LOIs (as described above) across the length of a cell junction for 200 frames (400 s) of a movie. Then the time-lapse line interpolator tool was used to generate a kymograph of all the LOIs. Kymographs were thresholded at two times the mean intensity of the kymograph. We then used a 40-pixel smooth along the time axis to connect the LOIs. The Fiji analyze particle tool was used to identify individual clusters, and clusters that hit the edge of the image were excluded so that we analyzed only complete clusters. We next used the measure bounding rectangle tool to identify the width (cluster size) and length (cluster lifespan) of each cluster. Finally, clusters of 1–2 pixels in width were filtered out as noise.
FRAP
Bleaching experiments were acquired on a Zeiss 700 confocal with a 4 s time interval, and images were acquired for 4 min postbleaching. Regions of interest were bleached using 405- and 488-nm-wavelength lasers set at 35% power. A second region of interest was from a neighboring nonbleached cell was used for bleach correction. Bleach correction and curve-fitting were carried out using a Python script (modified from https://imagej.net/tutorials/analyze-frap-movies).
Cdh3 immunostaining
Explants were prepared as described above and then fixed in 1× MEMFA (MEM formaldehyde) for 1 h at room temperature. Samples were then washed three times in phosphate-buffered saline (PBS) to remove fixative and permeabilized with 0.05% Triton X-100 in PBS for 30 min. Next, samples were blocked in 1% normal goat serum (NGS) in PBS for 2 h at room temperature. The primary antibody was diluted 1:100 in 1% NGS/PBS, and samples were incubated with primary antibody at 4°C overnight. We then performed a second incubation in blocking solution for 1 h at room temperature. Secondary antibody (goat anti-mouse 488, #A32723) was diluted 1:500, and samples were incubated with secondary antibody for 1 h at room temperature. Finally, samples were washed three times in 1× PBS and imaged.
Supplementary Material
Acknowledgments
We would like to acknowledge Shinuo Weng and Austin Baldwin for critical reading of this manuscript. This work was supported by the NICHD (RO1HD099191).
Abbreviations used:
- Cdh3
cadherin3
- CE
convergent extension
- FRAP
fluorescence recovery after photobleaching
- PCP
planar cell polarity
- Xdd1
dominant negative dishevelled-2.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-06-0194) on October 12, 2022.
REFERENCES
- Arslan FN, Eckert J, Schmidt T, Heisenberg C-P (2021). Holding it together: when cadherin meets cadherin. Biophys J 120, 4182–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertet C, Sulak L, Lecuit T (2004). Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature 429, 667–671. [DOI] [PubMed] [Google Scholar]
- Blankenship JT, Backovic ST, Sanny JS, Weitz O, Zallen JA (2006). Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev Cell 11, 459–470. [DOI] [PubMed] [Google Scholar]
- Butler MT, Wallingford JB (2017). Planar cell polarity in development and disease. Nat Rev Mol Cell Biol 18, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MT, Wallingford JB (2018). Spatial and temporal analysis of PCP protein dynamics during neural tube closure. eLife 7, e36456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavey M, Rauzi M, Lenne P-F, Lecuit T (2008). A two-tiered mechanism for stabilization and immobilization of E-cadherin. Nature 453, 751. [DOI] [PubMed] [Google Scholar]
- Coravos JS, Mason FM, Martin AC (2017). Actomyosin pulsing in tissue integrity maintenance during morphogenesis. Trends Cell Biol 27, 276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohn MR, Mundell NA, Sawyer LM, Dunlap JA, Jessen JR (2013). Planar cell polarity proteins differentially regulate extracellular matrix organization and assembly during zebrafish gastrulation. Dev Biol 383, 39–51. [DOI] [PubMed] [Google Scholar]
- Harrison OJ, Jin X, Hong S, Bahna F, Ahlsen G, Brasch J, Wu Y, Vendome J, Felsovalyi K, Hampton CM, et al. (2011). The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure 19, 244–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner RJ, Malmi-Kakkada AN, Sarıkaya S, Weng S, Thirumalai D, Wallingford JB (2021). Mechanical heterogeneity along single cell-cell junctions is driven by lateral clustering of cadherins during vertebrate axis elongation. eLife 10, e65390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner RJ, Wallingford JB (2018). Coming to consensus: a unifying model emerges for convergent extension. Dev Cell 46, 389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner RJ, Weng S, Lee C, Sarıkaya S, Papoulas O, Cox RM, Marcotte EM, Wallingford JB (2022). ARVCF catenin controls force production during vertebrate convergent extension. Dev Cell 57, 1119–1131.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R, Shih J, Sater A (1992). The cellular basis of the convergence and extension of the Xenopus neural plate. Dev Dyn 193, 199–217. [DOI] [PubMed] [Google Scholar]
- Keller R, Sutherland A (2020). Convergent extension in the amphibian, Xenopus laevis. Curr Top Dev Biol 136, 271–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft B, Berger CD, Wallkamm V, Steinbeisser H, Wedlich D (2012). Wnt-11 and Fz7 reduce cell adhesion in convergent extension by sequestration of PAPC and C-cadherin. J Cell Biol 198, 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T, Yap AS (2015). E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat Cell Biol 17, 533. [DOI] [PubMed] [Google Scholar]
- Lee C-H, Gumbiner BM (1995). Disruption of gastrulation movements in Xenopus by a dominant-negative mutant for C-cadherin. Dev Biol 171, 363–373. [DOI] [PubMed] [Google Scholar]
- Mirkovic I, Gault WJ, Rahnama M, Jenny A, Gaengel K, Bessette D, Gottardi CJ, Verheyen EM, Mlodzik M (2011). Nemo kinase phosphorylates β-catenin to promote ommatidial rotation and connects core PCP factors to E-cadherin–β-catenin. Nat Struct Mol Biol 18, 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka T, Inutsuka A, Begum K, Hafiz KMB, Kishi M (2014). Vangl2 regulates E-cadherin in epithelial cells. Sci Rep 4, 6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya H, David R, Damm EW, Fagotto F, Niessen CM, Winklbauer R (2012). Cadherin-dependent differential cell adhesion in Xenopus causes cell sorting in vitro but not in the embryo. J Cell Sci 125, 1877–1883. [DOI] [PubMed] [Google Scholar]
- Pfister K, Shook DR, Chang C, Keller R, Skoglund P (2016). Molecular model for force production and transmission during vertebrate gastrulation. Development 143, 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reits EAJ, Neefjes JJ (2001). From fixed to FRAP: measuring protein mobility and activity in living cells. Nat Cell Biol 3, E145–E147. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih J, Keller R (1992). Cell motility driving mediolateral intercalation in explants of Xenopus laevis. Development 116, 901. [DOI] [PubMed] [Google Scholar]
- Shindo A, Inoue Y, Kinoshita M, Wallingford JB (2019). PCP-dependent transcellular regulation of actomyosin oscillation facilitates convergent extension of vertebrate tissue. Dev Biol 446, 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo A, Wallingford JB (2014). PCP and septins compartmentalize cortical actomyosin to direct collective cell movement. Science 343, 649–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol SY (1996). Analysis of Dishevelled signalling pathways during Xenopus development. Curr Biol 6, 1456–1467. [DOI] [PubMed] [Google Scholar]
- Tatin F, Taddei A, Weston A, Fuchs E, Devenport D, Tissir F, Makinen T (2013). Planar cell polarity protein Celsr1 regulates endothelial adherens junctions and directed cell rearrangements during valve morphogenesis. Dev Cell 26, 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB, Niswander LA, Shaw GM, Finnell RH (2013). The continuing challenge of understanding, preventing, and treating neural tube defects. Science 339, 1222002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB, Rowning BA, Vogeli KM, Rothbacher U, Fraser SE, Harland RM (2000). Dishevelled controls cell polarity during Xenopus gastrulation. Nature 405, 81–85. [DOI] [PubMed] [Google Scholar]
- Yap AS, Gomez GA, Parton RG (2015). Adherens junctions revisualized: organizing cadherins as nanoassemblies. Dev Cell 35, 12–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.