Abstract
Bacillus amyloliquefaciens is an important plant disease-preventing and growth-promoting microorganism. B. amyloliquefaciens WS-8 can stimulate plant growth and has strong antifungal properties. In this study, we sequenced the complete genome of B. amyloliquefaciens WS-8 by Pacific Biosciences RSII (PacBio) Single Molecule Real-Time (SMRT) sequencing. The genome consists of one chromosome (3,929,787 bp) and no additional plasmids. The main bacteriostatic substances were determined by genome, transcriptome, and mass spectrometry data. We thereby laid a theoretical foundation for the utilization of the strain. By genomic analysis, we identified 19 putative biosynthetic gene clusters for secondary metabolites, most of which are potentially involved in the biosynthesis of numerous bioactive metabolites, including difficidin, fengycin, and surfactin. Furthermore, a potential class II lanthipeptide biosynthetic gene cluster and genes that are involved in auxin biosynthesis were found. Through the analysis of transcriptome data, we found that the key bacteriostatic genes, as predicted in the genome, exhibited different levels of mRNA expression. Through metabolite isolation, purification, and exposure experiments, we found that a variety of metabolites of WS-8 exert an inhibitory effect on the necrotrophic fungus Botrytis cinerea, which causes gray mold; by mass spectrometry, we found that the main substances are mainly iturins and fengycins. Therefore, this strain has the potential to be utilized as an antifungal agent in agriculture.
Keywords: Bacillus amyloliquefaciens WS-8, genome sequence, biosynthetic gene cluster, antifungal, plant growth promoting
Introduction
Pathogenic microorganisms affecting plant health are a major and chronic threat to food production and ecosystem stability all over the world [1]. At present, chemical control is the main approach to control plant diseases [2]. However, the long-term use of chemical fungicides has resulted in serious and multiple resistance of pathogenic microorganisms. At the same time, pesticide residues have polluted the environment, endangered human health, and destroyed the ecological balance. Biological control has attracted more and more attention and played an increasingly important role in the world because of its advantages to the environment, ecology, and human health.
Bacillus spp. are an important source of plant disease-preventing and growth-promoting microorganisms. The prevention of plant disease is mainly achieved by secreting various antimicrobial substances [3]. Johnson et al. [4] discovered that Bacillus subtilis can produce antimicrobial substances, many of which, including polypeptides, lipopeptides and antimicrobial proteins, have been isolated from different Bacillus species. Bacillus amyloliquefaciens, a non-pathogenic bacterial species that is widespread in nature, is known to produce bioactive compounds and is capable to promote plant growth. It has therefore been used as a biological control agent in agriculture [1].
The whole genome of B. amyloliquefaciens FZB42 [5], published in 2007, has provided a deeper understanding of B. amyloliquefaciens and its capacity to produce multiple secondary metabolites, promoting its biological utilization. The major active substances of B. amyloliquefaciens are antifungal peptides. B. amyloliquefaciens can produce antifungal peptides by two main pathways, i.e., the ribosome pathway and the non-ribosome pathway. As an ever-increasing number of genomes of different B. amyloliquefaciens strains are sequenced, marked differences have been found between gene clusters of secondary metabolites in different strains [6], and the active substances of some strains may be totally different.
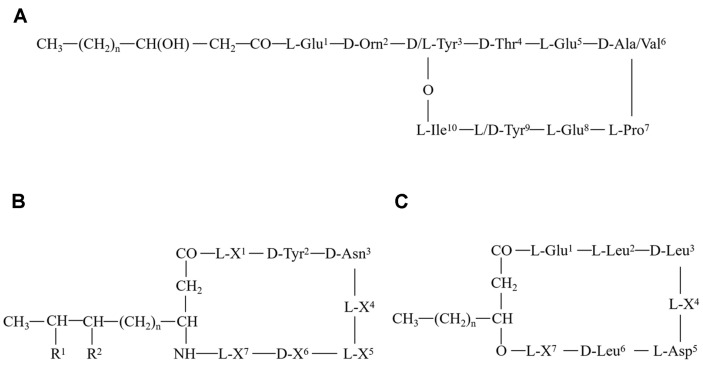
Fengycin, iturin, surfactin [7], difficidin, bacilysin [8], and macrolactin [9] are among the main antifungal peptides synthesized by the non-ribosomal pathway of B. amyloliquefaciens. The fengycins are a family of cyclic octapeptide-containing decapeptides (amino acid sequences are usually suggested as L-Glu-D-Orn-[D or L]-Tyr-D-Thr-L-Glu-D-[Ala or Val]-L-Pro-L-G1u-[L or D]-Tyr-L-Ile), which linked to a β-hydroxy fatty acid chain between C12 and C19 (Fig. 1A) [7]. Iturins and surfactins both are heptapeptides. The iturins are a family of cyclic heptapeptides (the 2nd and 3rd amino acids being always D-Tyr-D-Asn), which linked to a β-amino fatty acid chain between C15 and C18 (Fig. 1B) [10]. The surfactins are another family of cyclic heptapeptides (L-Glu-L-Leu-D-Leu-L-X4-L-Asp-D-Leu-L-X7, two of which are variable) [11], which linked to a β-amino fatty acid chain between C13 and C17 (Fig. 1C).
Fig. 1.
The chemical structure of fengycins (A), iturins (B), surfactins (C).
Although many strains have been used to control plant diseases, more efficient strains are still needed in agriculture. Before the use of a new strain as a biocontrol agent, it is recommendable to identify its main active substances. B. amyloliquefaciens WS-8, which can stimulate plant growth, has strong antifungal properties. Our preliminary study showed that the WS-8 strain exerts strong antagonistic activity against Fulvia fulva (Cooke) Cif and can promote growth of Lycopersicon esculentum Mill [12]. In order to determine the antifungal mechanism of B. amyloliquefaciens WS-8, we sequenced its complete genome. To elucidate the expression levels of core genes of secondary metabolic gene clusters, we also sequenced the transcriptome in the late logarithmic phase. Moreover, the anti-gray-mold compounds were isolated, purified, and characterized through chromatography and liquid chromatography-tandem mass spectrometry (LC-MS/MS). These results improve our understanding of the antifungal mechanisms of the WS-8 strain, and they will enable us to make better use of this strain as a fungicidal agent in agriculture.
Materials and Methods
Microorganisms and Culture Conditions
B. amyloliquefaciens WS-8 was isolated from hillside soil from Langya Mountain in Hebei, China, and stored at the China General Microbiology Culture Collection Centre (CGMCC: 11787). Luria Bertani (LB) broth medium (containing 10 g/l tryptone, 5 g/l yeast extract, and 5 g/l NaCl in distilled water) was used as growth medium. Bacteria were cultured at 32°C for 32 h with continuous shaking at 200 rpm.
DNA Isolation, Genome Sequencing and Assembly
Genomic DNA of B. amyloliquefaciens WS-8 was extracted using the SDS method. A library with a 10-kb insert size was constructed for sequencing. The genome was sequenced using a PacBio RS II sequencing platform by Beijing Novogene Bioinformatics Technology Co., Ltd. The reads were assembled using the SMRT portal [13]. The complete genome sequence of B. amyloliquefaciens WS-8 has been submitted to GenBank under the accession number CP018200.
Gene Prediction and Identification of Secondary Metabolite Clusters
Gene prediction was performed on the WS-8 genome assembly by GeneMarkS [14] with an integrated model which combines the GeneMarkS generated (native) and heuristic model parameters. Gene annotation was added by the NCBI Prokaryotic Genome Annotation Pipeline [15]. The genome comparison was carried out by RAST version 2.0 [16]. Genes potentially involved in the biosynthesis of antibiotics and secondary metabolites were identified using antiSMASH3.0 [17]. Genome overview was created by CGView Server to show the annotation information of WS-8 [18].
Transcriptomic Analysis Using RNA-Seq
For RNA extraction, cells were cultivated for 32 h and harvested by centrifugation at 10,000 ×g for 5 min at 4°C. The pellets were immediately frozen in liquid nitrogen. Total RNA was extracted using an RNAprep Pure Cell/Bacteria Kit (TIANGEN BIOTECH, China) according to the manufacturer’s instructions. RNA integrity was assessed using the RNA Nano 6000 Assay Kit and a Bioanalyzer 2100 system (Agilent Technologies, USA). Sequencing libraries were generated using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 4000 platform and paired-end reads were generated.
Raw data (raw reads, fastq files) were first processed using in-house Perl scripts from Beijing Novogene Bioinformatics Technology Co., Ltd. In this step, clean data (clean reads) were obtained by removing reads containing adapters, reads containing poly-N tails, and low-quality reads from the raw data. Q20, Q30, and GC content of the clean data were calculated. All of the downstream analyses were based on the high-quality clean data. The remaining clear reads were mapped to the reference genome of WS-8 using Bowtie2 software based on the local alignment algorithm [19]. HTSeq v0.6.1 was used to count the reads mapped to each gene. The fragments per kilobase of transcripts per million mapped reads (FPKM) of each gene was calculated based on the length of the gene and read count mapped to this gene [20]. The transcriptomic data of B. amyloliquefaciens WS-8 has been submitted to GenBank under the accession number SRX6564092.
Purification of Anti-Gray-Mold Compounds
The strain WS-8 was grown in nutrient broth (NB) at 32°C for 32 h. After centrifugation (10,000 rpm, 30 min, and 4°C), cells were removed and the cell-free supernatant (4 liters) was shaken with 200 g of Amberlite XAD-7HP (Sigma, USA) for 12 h at 18°C. The antimicrobial crude extract (CE) was obtained following the protocol of Xin [21]. Further purification was carried out by HPLC (SHIMADZU LC-20A, Japan) with a C18 column (250 × 4.6 mm, 5 µm; WONDASIL, Japan) at room temperature. The mobile phase consisted of acetonitrile and HPLC-grade water (with 0.1%trifluoroacetic acid [TFA]). A linear gradient was used for elution at a flow rate of 1 ml/min as follows: 0-60 min, from 10% to 80%acetonitrile (linear gradient); 60-65 min, from 80% to 90% acetonitrile (linear gradient); 65-75 min, 90% acetonitrile (isocratic); 75-80 min, from 90% to 10% acetonitrile (linear gradient); 80-90 min, 10%acetonitrile (isocratic). Elution was monitored by determining absorbance at 214 nm, and fractions were manually collected each minute. Using the agar well diffusion method [22, 23], fractions with anti-gray-mold (Botrytis cinerea) activity were detected and selected for LC-MS/MS analysis.
LC-MS/MS Analysis
Liquid chromatograph-mass spectrometry (LC-MS) was performed by the Thermo Fisher UltiMate 3000 UPLC/Q-Exactive Orbitrap MS system. The UHPLC conditions were as follows: Thermo GOLD HYPERSIL column (C18, 50 × 2.1 mm, 1.9 µm; Thermo), eluent A was H2O/0.1% formic acid, eluent B was CH3OH/0.1% formic acid, flow rate was 300 µl/min, elution gradient was 70%A/30%B to 10%A/90%B, 20 min. The MS operating conditions were as follows: the temperature was 300°C, a sheath gas rate of 35 l/min, auxiliary gas rate of 35 l/min, electrospray voltage of 3.0 kV for positive full scan mode, and an m/z range of 100 - 3000. Detailed sequence information for antimicrobial peptides was obtained using the targeted MS/MS mode. The target ion was isolated and fragmented by adding a voltage of 35 V.
Results
General Genome Features of B. amyloliquefaciens WS-8
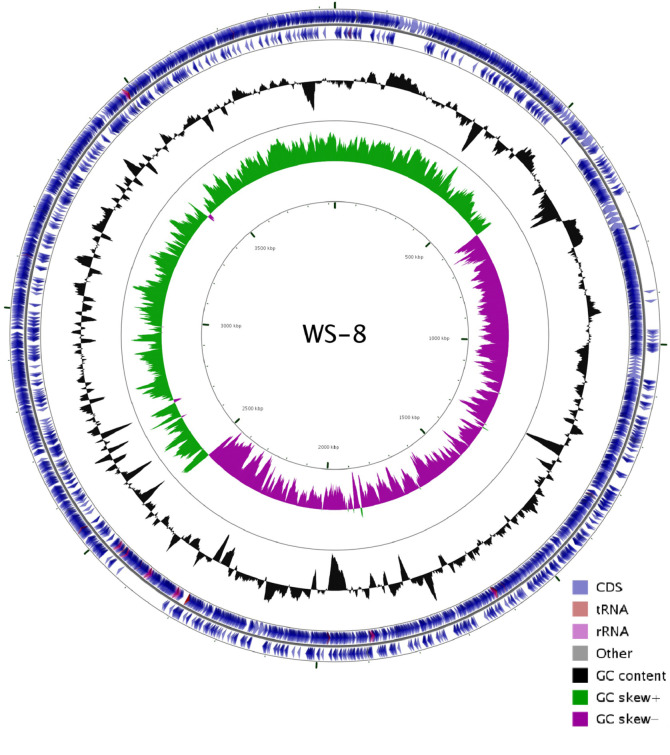
A total of 90,776 reads, with approximately 311-fold coverage (approximately 1.227 G), were obtained. Compared to the type strain B. amyloliquefaciens FZB42, they constructed a library with fragments of 1.5 to 3.0 kbp, and abtained a total of 40,000 sequences [5]. Our sequencing coverage is at least 6 times more than the type stain FZB42. With the increase of sequencing coverage, the error of sequencing can be corrected, and the error rate of the whole genome can be reduced. The reads were assembled using the SMRT portal. The complete genome of WS-8 is composed of one gapless circular chromosome of 3,929,787 bp, almost the same as the genome of B. amyloliquefaciens FZB42 (3,918,589 bp) [5]. It shows a G+C content of 46.5%, and no plasmid was found. The genome contains 3895 predicted genes, 3777 protein coding genes, 107 pseudogenes, 86 tRNA genes, 27 rRNA genes, and 5 ncRNA genes (Table 1 and Fig. 2). Meanwhile, 32 long tandem repeats, 28 transposons, 32 long interspersed nuclear elements, 7 short interspersed nuclear elements, and 152 tandem repeat sequences were also identified in the genome. Among the identified genes, 2,895, 2,727, and 2,307 genes were classified to functional categories based on clusters of, orthologous genes of proteins (COG) [24], Kyoto Encyclopedia of Genes and Genomes designation (KEGG) [25], and gene ontology (GO) [26], respectively.
Table 1.
General genome features of B. amyloliquefaciens WS-8.
| Feature | Value |
|---|---|
| Genome size (bp) | 3929787 |
| GC content [%] | 45.6 |
| Predicted genes | 3895 |
| Protein coding genes (CDSs) | 3777 |
| Pseudo genes | 107 |
| tRNA genes | 86 |
| rRNA genes | 27 |
| ncRNAs genes | 5 |
Fig. 2.
Circular genome map of B. amyloliquefaciens WS-8. The circular map consists of 5 circles. From the outermost circle inwards, circle (1) and circle (2) show the coding gene distributions in the forward strand and the backward strand, respectively, including tRNA (brown), rRNA (pink), and other (gray); and circle (3) shows GC content; circle (4) and circle (5) show GC skew+ and GC skew-, respectively.
Using RAST version 2.0 under the Compare Metabolic Reconstruction model, a total of 2,428 functioning parts genes were found, 170 and 108 unique genes were identified from B. amyloliquefaciens WS-8 and B. amyloliquefaciens FZB42 respectively.
Functional Gene Annotation
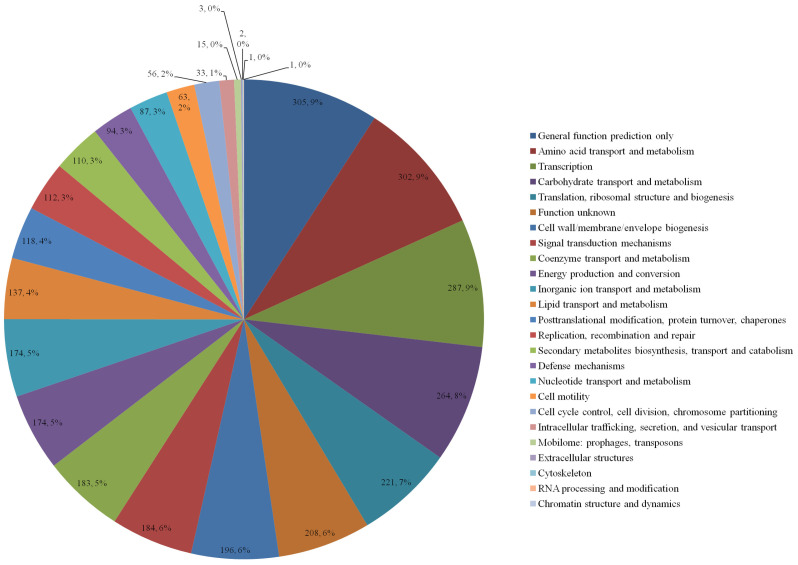
Function and classification of COG. The COG functional categories of the complete genome sequence of B. amyloliquefaciens WS-8 are shown in Fig. 3. Among the 3,895 genes, 2,895 genes were classified into COG categories. The major categories of B. amyloliquefaciens WS-8 were general function prediction only, amino acid transport and metabolism, transcription, carbohydrate transport and metabolism, translation, ribosomal structure and biogenesis, function unknown, and cell wall/membrane/envelope biogenesis. Additionally, the genome contains genes probably involved in the promotion of plant growth, i.e., 31 genes for nitrogen metabolism, 40 genes for sulfur metabolism, 33 genes for phosphorus metabolism, 10 genes for potassium metabolism, and 4 genes for auxin biosynthesis.
Fig. 3.
COG categories of B. amyloliquefaciens WS-8.
Identification of secondary metabolite clusters with antiSMASH. After annotation, we identified genes and gene clusters related to the biosynthesis of interesting secondary metabolites using antiSMASH. We found 19 putative biosynthetic gene clusters for secondary metabolites (Table 2), i.e., seven microcins, four non-ribosomal peptides, four polyketides, two terpenes, one lantipeptide, and one other metabolite. Seven putative gene clusters showed high similarity (> 70% of genes showing similarity) to reported difficidin, fengycin, bacillaene, macrolactin, surfactin, bacilysin, and bacillibactin biosynthesis gene clusters. Cluster 9 showed 7% similarity to butirosin, and the other gene clusters were not similar to any known cluster. Additionally, we identified a potential class II lanthipeptide biosynthetic pathway. In this gene cluster, we found three LanA-like (precursor peptide) genes, two LanM-like (modification enzyme) genes, one LanT-like (transporter, with a peptidase domain) gene, one LanI-like (immunity protein) gene, and five regulatory element genes [27, 28].
Table 2.
Secondary metabolite clusters in B. amyloliquefaciens WS-8 identified by antiSMASH 3.0.
| Cluster | Type | From | To | Most similar known biosynthetic gene cluster (percent of similarity) | MIBiG BGC-ID |
|---|---|---|---|---|---|
| 1 | Polyketide | 106469 | 206922 | Difficidin (93% ) | BGC0000176_c1 |
| 2 | Polyketide | 321899 | 363008 | NA | NA |
| 3 | Terpene | 426326 | 448209 | NA | NA |
| 4 | Nonribosomal peptide | 473436 | 611237 | Fengycin (93%) | BGC0001095_c1 |
| 5 | Nonribosomal peptide | 675868 | 778542 | Bacillaene (92%) | BGC0001089_c1 |
| 6 | Polyketide | 1004652 | 1090557 | Macrolactin (90%) | BGC0000181_c1 |
| 7 | Lantipeptide | 1259447 | 1288335 | NA | NA |
| 8 | Terpene | 1408827 | 1429567 | NA | NA |
| 9 | Polyketide | 1511611 | 1552855 | Butirosin (7%) | BGC0000693_c1 |
| 10 | Microcin | 1609461 | 1629609 | NA | NA |
| 11 | Microcin | 1878888 | 1899036 | NA | NA |
| 12 | Nonribosomal peptide | 2089305 | 2154712 | Surfactin (78%) | BGC0000433_c1 |
| 13 | Microcin | 2298675 | 2324765 | NA | NA |
| 14 | Microcin | 2369207 | 2395180 | NA | NA |
| 15 | Microcin | 2435066 | 2455214 | NA | NA |
| 16 | Microcin | 2456802 | 2476950 | NA | NA |
| 17 | Other | 2776305 | 2817723 | Bacilysin (85%) | BGC0001184_c1 |
| 18 | Nonribosomal peptide | 3354032 | 3405823 | Bacillibactin (92%) | BGC0000309_c1 |
| 19 | Microcin | 3470058 | 3490206 | NA | NA |
Transcriptome Analysis of Secondary Metabolite Clusters
The transcriptome of stationary phase of the strain WS-8 was sequenced with the HiSeq4000 sequencing platform. A total of 9,006,772 clean Reads and 1.35 G clean data were obtained. The Q20 and Q30 of clean data were 98.07% and 94.69%, respectively. The GC content of the transcriptome was 48.08%. About 97.97% of the clean reads were mapped to the WS-8 genome. Using the annotation of antiSMASH for mapping and FPKM values to indicate the expression levels, the mapped genes were classified into four groups (Table 3). Using a threshold of FPKM > 1 to define potential gene expression [29], 3,540 expressed genes were sequenced. As shown in Table 3, more than 93% of the genes were expressed in the stationary phase. Most genes were expressed at a medium level, and more than 13% of the genes were expressed at a high level. In addition, there were 277 non-expressed genes.
Table 3.
Transcriptomic features of B. amyloliquefaciens WS-8.
| FPKM Interval | Gene counts | Percentage |
|---|---|---|
| 0~1 | 277 | 60.2% |
| 1~10 | 0 | 0.00% |
| 10~500 | 3,045 | 80.83% |
| >500 | 495 | 13.14% |
The transcripts were assessed based on FPKM values: high expression (FPKM ≥ 500), medium expression (10 ≤ FPKM< 500), low expression (1 ≤ FPKM < 10), and no expression (FPKM < 1).
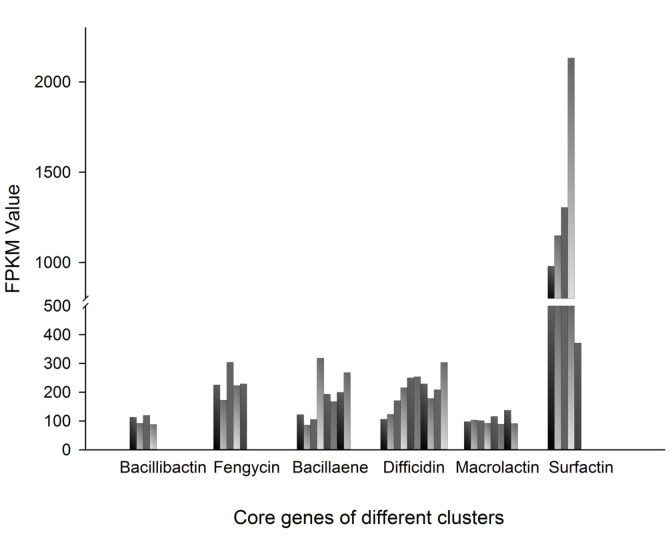
Using the FPKM value, the expression levels of core genes in gene clusters related to antibacterial substances were analyzed. We found that core genes of six gene clusters, which are homologous to bacillibactin, fengycin, bacillaene, difficidin, macrolactin, and surfactin biosynthetic gene cluster were all expressed (Fig. 4). All of the bacillibactin genes (bsuF, bsuB, bsuE, and bsuA), fengycin genes (fenA, fenB, fenC, fenD, and fenE), bacillaene genes (baeE, baeD, baeC, baeR, baeN, baeM, baeL, and baeJ), difficidin genes (difA, difD, difE, difF, difG, difH, difI, difJ, difK, and difL), and macrolactin genes (mlnA, mlnB, mlnC, mlnD, mlnE, mlnF, mlnG, and mlnH) were expressed at a medium level. In the surfactin gene cluster, there were four highly expressed genes (srf'AA, srf'AB, srf'AC, srf'AD), and one medium expressed gene (sfp). These data suggest that the products of these gene clusters may exist in WS-8. For the lantipeptide gene cluster, all of the precursor peptide genes, synthetase genes, and transport genes were also expressed.
Fig. 4.
FPKM values of core genes of different gene clusters.
Purification and UPLC-MS Analysis of Antibacterial Substances from WS-8
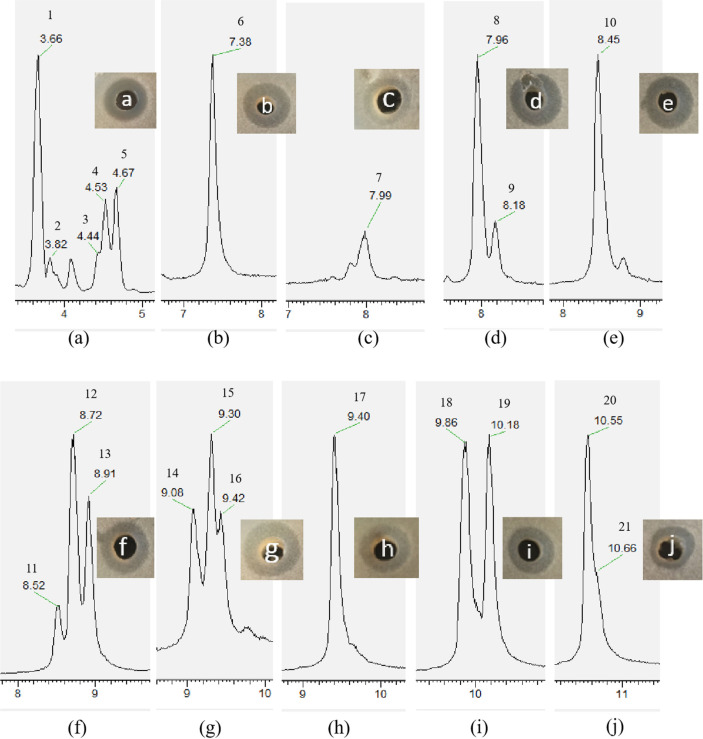
The antimicrobial compounds were enriched by Amberlite XAD-7HP from the cell-free supernatant and isolated by HPLC. Using gray mold as indicator bacteria, we tested all of the HPLC fractions. We found 25 fractions with anti-gray-mold activity. The Thermo Fisher UltiMate 3000 UPLC/Q-Exactive Orbitrap MS system was employed to analyze the above 25 fractions, and we found many fractions containing the same compound. Ten of these fractions (a, b, c, d, e, f, g, h, i, and j) containing non-repeating substances for further analysis were chosen, and 21 compounds showing anti-gray-mold activity were identified (Fig. 5).
Fig. 5.
UPLC and anti-gray-mold activity analysis of the 10 fractions from B. amyloliquefaciens WS-8.
Using MS to elucidate the exact molecular weight of these 21 compounds, we identified 14 lipopeptides belonging to two main types: iturin and fengycin (Table 4). Many of the components have similar charge-to-mass ratios (m/z), so we analyzed their secondary mass spectra one by one. All of the compounds in fraction (a) were identified as iturins. Compound 1 (m/z ratio of the protonated molecules [M+H]+ molecule is 1043.55), with the characteristic fragment ions of which have m/z ratios of 212.10, 392.16, 638.39, 801.44, 915.50, and 932.51 (Fig. S1), was identified as C14 iturin A by comparison with Xu’s data [7]. The fragment patterns of compounds 2 and 3 (Fig. S2), both with an m/z ratio of the protonated [M+H]+ molecule of 1044.53, show that their characteristic fragment ions are basically identical and they would be identified as the same molecule by mass spectrometry. However, due to the different retention time of the liquid phase, it can be concluded that these two compounds may be derivatives of C14 iturin B. Compounds 4 and 5, both with an m/z ratio of the protonated [M+H]+ molecule of 1057.56, also have similar fragment patterns (Fig. S3), indicating that these two compounds may be derivatives of C15 iturin A.
Table 4.
The m/z value of active substance detected by Q-Exactive Orbitrap MS.
| Fraction no. | Compound no. | m/z | m/z | Characteristic fragment ions | Retention time (min) | Identification |
|---|---|---|---|---|---|---|
| [M+H]+ | [M+Na]+ | |||||
| a | 1 | 1043.5474 | 1065.5284 | - | 3.66 | C14Iturin A |
| 2 | 1044.5328 | 1066.5145 | - | 3.82 | C14 Iturin B | |
| 3 | 1044.5334 | 1066.5149 | - | 4.44 | C14 Iturin B | |
| 4 | 1057.5643 | 1079.5454 | - | 4.53 | C15Iturin A | |
| 5 | 1057.5636 | 1079.5449 | - | 4.67 | C15Iturin A | |
| b | 6 | 1435.766 | 1457.7474 | 540.77 | 7.38 | C14 Fengycin A |
| c | 7 | 1449.7848 | 1471.7635 | 540.77 | 7.99 | C15 Fengycin A |
| d | 8 | 1463.7975 | 1485.7784 | 554.79 | 7.96 | C14Fengycin B |
| 9 | 1449.7836 | 1471.7632 | 540.77 | 8.18 | C15 Fengycin A | |
| e | 10 | 1449.7816 | 1471.7626 | 540.77 | 8.45 | C15 Fengycin A |
| f | 11 | 1477.8138 | 1499.7944 | 554.79 | 8.52 | C15 Fengycin B |
| 12 | 1463.7969 | 1485.7777 | 540.77 | 8.72 | C16 Fengycin A | |
| 13 | 1463.7974 | 1485.7789 | 540.77 | 8.91 | C16 Fengycin A | |
| g | 14 | 1477.8138 | 1499.7888 | 540.77 | 9.08 | C17 Fengycin A |
| 15 | 1477.814 | 1499.7932 | 540.77 | 9.3 | C17 Fengycin A | |
| 16 | 1477.8157 | 1499.7963 | 540.77 | 9.42 | C17 Fengycin A | |
| h | 17 | 1491.8285 | 1513.8091 | 554.79 | 9.4 | C16 Fengycin B |
| i | 18 | 1505.8439 | 1527.825 | 554.79 | 9.86 | C17 Fengycin B |
| 19 | 1447.8036 | 1469.7852 | 540.77 | 10.18 | C15:1 Fengycin A | |
| j | 20 | 1461.8197 | 1483.8 | 540.77 | 10.55 | C16:1 Fengycin A |
| 21 | 1475.8328 | 1497.8135 | 554.79 | 10.66 | C15:1 Fengycin B |
The remaining 16 compounds were identified as fengycins. Fengycin A and fengycin B are different in their sixth amino acids of cyclic octapeptide (Ala and Val respectively). So, in the process of mass spectrometry, the daughter ion of MS peaks are different. Therefore, ion fragments (m/z 1080) and ion fragments (m/z 1108) are usually used as characteristic fragment ions to distinguish fengycin A and fengycin B. In this research, their molecular ion peaks mainly exist in the form of doubly charged [M+2H]2+ and singly charged [M+H]+ ions; most are doubly charged ions. Therefore, their fragment patterns of doubly charged ions were used in the following analysis.
Compounds 6, 17, 18, 19, 20, and 21 have unique m/z ratios. After analysis of their secondary ion mass spectra (Fig. S4), they were identified as C14 fengycin A, C16 fengycin B, C17 fengycin B, C15:1 fengycin A, C16:1 fengycin A, and C15:1 fengycin B, respectively. The carbon chains of compounds 19, 20, and 21 all contain a double bond.
Besides, three compounds (7, 9, and 10), with an m/z ratio of the protonated [M+H]+ molecule of 1449.78, were identified as C15 fengycin A. Upon analisis of secondary ion mass spectra of the doubly charged ions, we found that these three compounds had similar fragments (Fig. S5). Therefore, these three compounds may be derivatives of C15 fengycin A.
Compounds 8, 12, and 13 have similar molecular ion peaks (m/z ratio of the protonated [M+H]+ molecules is 1463.79), but the characteristic ion peaks in their secondary mass spectra (Fig. S6) are quite different. Compound 8, containing doubly charged fragment ions of m/z 554.79, equal to the characteristics fragment ions of fengycin B, was identified as C14 fengycin B. Compounds 12 and 13, containing doubly charged fragment ions of m/z 540.77, equal to the characteristics fragment ions of fengycin A, were identified as C16 fengycin A.
The last four compounds (11, 14, 15, and 16) show a similar molecular ion peak of the protonated [M+H]+ fragment at 1477.81, but their characteristic fraction ions (Fig. S7) are also different. Compound 11, containing doubly charged fragment ions of m/z 554.79, equal to the characteristics fragment ions of fengycin B, was identified as C15 fengycin B. Compounds 14, 15, and 16, containing doubly charged fragment ions of m/z 540.77, equal to the characteristic fragment ions of fengycin A, were identified as C17 fengycin A.
Discussion
Plant disease has always been an important problem in agriculture. B. cinerea, an airborne plant pathogenic fungus that causes gray mold disease, affecting more than 200 crop species worldwide, is one of the most important problems in agricultural production [30]. The problem is most serious in vegetables and fruits, such as tomato, cucumber, and strawberry, causing huge economic losses [31]. The application of chemical pesticides has for a long time been the primary strategy for the prevention and control of plant diseases, but the use of chemical pesticides brings a series of problems, such as environmental pollution, so biological control has gradually become an appealing alternative. Many strains of Bacillus spp. are considered as safe microorganisms in agriculture [32]. They have the potential to produce more than two dozen antibiotics with an amazing variety of structures, and many of these substances have been successfully used in agricultural and industrial applications [33, 34].
B. amyloliquefaciens can inhibit a variety of plant pathogens and is widely used in agriculture. Since B. amyloliquefaciens FZB42 [5] was sequenced, more than 60 strains of B. amyloliquefaciens have been sequenced (including the genome reported in this study). Belbahri et al. analyzed the genomic information of 48 strains of B. amyloliquefaciens. They found that these strains have various secondary metabolite synthesis gene clusters and that obvious differences exist between strains [6]. The genome of this study was also discussed in their analysis. Through antiSMASH analysis, we found that the WS-8 genome contains seven gene clusters similar to known antibacterial biosynthetic gene clusters, which indicates that WS-8 bacteria may produce these antibacterial agents or their homologues. By using the FPKM values of the transcriptome, we found that core genes of six gene clusters, which are homologous to bacillibactin, fengycin, bacillaene, difficidin, macrolactin, and surfactin biosynthetic gene clusters, were all expressed. This indicates that the secondary metabolites of B. amyloliquefaciens WS-8 may include many bioactive substances.
Ten fractions (a, b, c, d, e, f, g, h, i, j, and k) containing 21 compounds with strong anti-gray-mold activity were fractionated from the cell-free supernatants of B. amyloliquefaciens WS-8 culture by a combination of Amberlite XAD-7HP resin and reversed phase chromatography. Compared with published LC-MS/MS data, we found that these active compounds are identified as iturins and fengycins. The iturin compounds identified in this study were C14 iturin A, C14 iturin B and C15 iturin A; two derivatives of C14 iturin B and C15 iturin A were found. Iturin compounds have been found in many strains of B. amyloliquefaciens and their applications in biological control have also been reported [32, 35]. In this study, 11 fengycin compounds were identified, i.e., C14 fengycin A, C15 fengycin A, C16 fengycin A, C17 fengycin A, C15:1 fengycin A, C16:1 fengycin A, C14 fengycin B, C15 fengycin B, C16 fengycin B, C17 fengycin B, and C15:1 fengycin B. Among these, derivatives of C15 fengycin A, C16 fengycin A, and C17 fengycin A were found. In addition, three fengycins with a double bond were found, i.e., C15:1 fengycin A, C16:1 fengycin A, and C15:1 fengycin B. Xu et al. analyzed the metabolites of B. siamensis JFL15, and they reported that fengycins were the main active substances [7]. The above results suggest that the strain WS-8 produces many bioactive derivatives.
Surfactin does not have antifungal activity, but it can enhance the antifungal activity of other lipopeptide, especially iturin [36, 37]. In this research, although core genes of surfactin biosynthesis cluster were under high expression level, but no surfatin was detected. Because of no anti-graymold activity, the fractions containing surfactin probably were abandoned after the active test.
In conclusion, the genome of B. amyloliquefaciens WS-8 was sequenced and annotated; based on the genome data, the active substances of this strain were isolated and identified. A total of 21 known metabolites were identified, five of which were iturins and 16 were fengycins, according to MS/MS spectra data. Our results indicate that B. amyloliquefaciens WS-8 can be used as an effective biocontrol agent in agriculture. Future studies will be necessary to further clarify the effects of this strain on pathogenic microorganisms in different crops.
Supplemental Materials
Supplementary data for this paper are available on-line only at http://jmb.or.kr.
Acknowledgments
This study was supported by grants from the Science and technology planning project of Hebei academy of sciences (19304), High-level talents funding projects of Hebei province (B2018003019), and High-level talents training and funding projects of Hebei academy of sciences (2018G01). We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest to declare.
REFERENCES
- 1.Compant S, Duffy B, Nowak J, Clement C, Barka EA. Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 2005;71:4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirooka T, Ishii H. Chemical control of plant diseases. J. General Plant Pathol. 2013;79:390–401. doi: 10.1007/s10327-013-0470-6. [DOI] [Google Scholar]
- 3.Fickers P. Antibiotic compounds from Bacillus: Why are they so Amazing? Am. J. Biochem. Biotechnol. 2012;8:38–43. [Google Scholar]
- 4.Johnson BA, Anker H, Meleney FL. Bacitracin: A New antibiotic produced by a member of the B. Subtilis Group. Science. 1945;102:376–377. doi: 10.1126/science.102.2650.376. [DOI] [PubMed] [Google Scholar]
- 5.Chen XH, Koumoutsi A, Scholz R, Eisenreich A, Schneider K, Heinemeyer I, et al. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 2007;25:1007–1014. doi: 10.1038/nbt1325. [DOI] [PubMed] [Google Scholar]
- 6.Belbahri L, Chenari Bouket A, Rekik I, Alenezi FN, Vallat A, Luptakova L, et al. Comparative genomics of Bacillus amyloliquefaciens strains reveals a core genome with traits for habitat adaptation and a secondary metabolites rich accessory genome. Front. Microbiol. 2017;8:1438. doi: 10.3389/fmicb.2017.01438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu BH, Lu YQ, Ye ZW, Zheng QW, Wei T, Lin JF, et al. Genomics-guided discovery and structure identification of cyclic lipopeptides from the Bacillus siamensis JFL15. PLoS One. 2018;13:e0202893. doi: 10.1371/journal.pone.0202893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen XH, Scholz R, Borriss M, Junge H, Mogel G, Kunz S, et al. Difficidin and bacilysin produced by plant-associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J. Biotechnol. 2009;140:38–44. doi: 10.1016/j.jbiotec.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 9.Schneider K, Chen XH, Vater J, Franke P, Nicholson G, Borriss R, et al. Macrolactin is the polyketide biosynthesis product of the pks2 cluster of Bacillus amyloliquefaciens FZB42. J. Nat. Prod. 2007;70:1417–1423. doi: 10.1021/np070070k. [DOI] [PubMed] [Google Scholar]
- 10.Schneider J, Taraz K, Budzikiewicz H, Deleu M, Thonart P, Jacques P. The structure of two fengycins from Bacillus subtilis S499. Z. Naturforsch. C. 1999;54:859–866. doi: 10.1515/znc-1999-1102. [DOI] [PubMed] [Google Scholar]
- 11.Peypoux F, Bonmatin JM, Labbe H, Grangemard I, Das BC, Ptak M, Wallach J, et al. [Ala4]surfactin, a novel isoform from Bacillus subtilis studied by mass and NMR spectroscopies. Eur. J. Biochem. 1994;224:89–96. doi: 10.1111/j.1432-1033.1994.tb19998.x. [DOI] [PubMed] [Google Scholar]
- 12.Cui LT, Liu HW, Cheng HC, Wang YN, Wang Q, Li QY, et al. Cloning and bioinformatics analysis of LanM gene in Bacillus amyloliquefaciens WS-8. Genomics Appl. Biol. 2019;38:644–649. [Google Scholar]
- 13.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 14.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angiuoli SV, Gussman A, Klimke W, Cochrane G, Field D, Garrity G, et al. Toward an online repository of Standard Operating Procedures (SOPs) for (meta) genomic annotation. Omics. 2008;12:137–141. doi: 10.1089/omi.2008.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant JR, StothardP The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frazee AC, Pertea G, Jaffe AE, Langmead B, Salzberg SL, Leek JT. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 2015;33:243–246. doi: 10.1038/nbt.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xin B, Zheng J, Xu Z, Li C, Ruan L, Peng D, et al. Three novel lantibiotics, ticins A1, A3, and A4, have extremely stable properties and are promising food biopreservatives. Appl. Environ. Microbiol. 2015;81:6964–6972. doi: 10.1128/AEM.01851-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xin B, Zheng J, Xu Z, Song X, Ruan L, Peng D, et al. The Bacillus cereus group is an excellent reservoir of novel lanthipeptides. Appl. Environ. Microbiol. 2015;81:1765–1774. doi: 10.1128/AEM.03758-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devillers J, Steiman R, Seigle-Murandi F. The usefulness of the agar-well diffusion method for assessing chemical toxicity to bacteria and fungi. Chemosphere. 1989;19:1693–1700. doi: 10.1016/0045-6535(89)90512-2. [DOI] [Google Scholar]
- 24.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie L, Miller LM, Chatterjee C, Averin O, Kelleher NL, van der Donk WA. Lacticin 481: in vitro reconstitution of lantibiotic synthetase activity. Science. 2004;303:679–681. doi: 10.1126/science.1092600. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Ge X, Zhang L, Teng K, Zhong J. One-pot synthesis of class II lanthipeptide bovicin HJ50 via an engineered lanthipeptide synthetase. Sci. Rep. 2016;6:38630. doi: 10.1038/srep38630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhi Y, Wu Q, Xu Y. Genome and transcriptome analysis of surfactin biosynthesis in Bacillus amyloliquefaciens MT45. Sci. Rep. 2017;7:40976. doi: 10.1038/srep40976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williamson B, Tudzynski B, Tudzynski P, Van Kan JAL. Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 2007;8:561–580. doi: 10.1111/j.1364-3703.2007.00417.x. [DOI] [PubMed] [Google Scholar]
- 31.ten Have A, Mulder W, Visser J, van Kan JAL. The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol. Plant Microbe Interact. 1998;11:1009–1016. doi: 10.1094/MPMI.1998.11.10.1009. [DOI] [PubMed] [Google Scholar]
- 32.Yu GY, Sinclair JB, Hartman GL, Bertagnolli BL. Production of iturin A by Bacillus amyloliquefaciens suppressing Rhizoctonia solani. Soil Biol. Biochem. 2002;34:955–963. doi: 10.1016/S0038-0717(02)00027-5. [DOI] [Google Scholar]
- 33.Stein T. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 2005;56:845–857. doi: 10.1111/j.1365-2958.2005.04587.x. [DOI] [PubMed] [Google Scholar]
- 34.Beric T, Kojic M, Stankovic S, Topisirovic L, Degrassi G, Myers M, et al. Antimicrobial activity of Bacillus sp natural isolates and their potential use in the biocontrol of phytopathogenic bacteria. Food Technol. Biotechnol. 2012;50:25–31. [Google Scholar]
- 35.Arrebola E, Jacobs R, Korsten L. Iturin A is the principal inhibitor in the biocontrol activity of Bacillus amyloliquefaciens PPCB004 against postharvest fungal pathogens. J. Appl. Microbiol. 2010;108:386–395. doi: 10.1111/j.1365-2672.2009.04438.x. [DOI] [PubMed] [Google Scholar]
- 36.Kim PI, Bai H, Bai D, Chae H, Chung S, Kim Y, et al. Purification and characterization of a lipopeptide produced by Bacillus thuringiensis CMB26. J. Appl. Microbiol. 2004;97:942–949. doi: 10.1111/j.1365-2672.2004.02356.x. [DOI] [PubMed] [Google Scholar]
- 37.Maget-Dana R, Thimon L, Peypoux F, Ptak M. Surfactin/iturin A interactions may explain the synergistic effect of surfactin on the biological properties of iturin A. Biochimie. 1992;74:1047–1051. doi: 10.1016/0300-9084(92)90002-V. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.