The omicron (B.1.1.529) variant of SARS-CoV-2 evolved into several sublineages, three of which (BA.1, BA.2, and BA.5) became globally dominant. Currently, the prevalence of omicron subvariants BQ.1 (a subvariant of BA.5), its sublineage BQ.1.1, and XBB (a recombinant of two different BA.2 subvariants) is increasing rapidly in the USA, France, Singapore, India, and elsewhere. BQ.1.1 and XBB possess substitutions relative to BA.5 and BA.2, respectively, in the receptor-binding domain of their spike protein (appendix p 4), which is the major target for vaccines and therapeutic monoclonal antibodies (mAbs) for COVID-19. Both variants have the substitution R346T, which confers resistance to certain therapeutic antibodies,1 raising concerns that mAbs or vaccines might be less effective against BQ.1.1 and XBB than against other omicron strains. We showed that BQ.1.1 and XBB have enhanced immune evasion capabilities compared with earlier omicron variants, including BA.5 and BA.2, by evaluating the efficacy of therapeutic mAbs against BQ.1.1 and XBB.2 However, the neutralising ability of plasma from convalescent individuals and COVID-19 vaccinees against BQ.1.1 and XBB clinical isolates remained unknown.
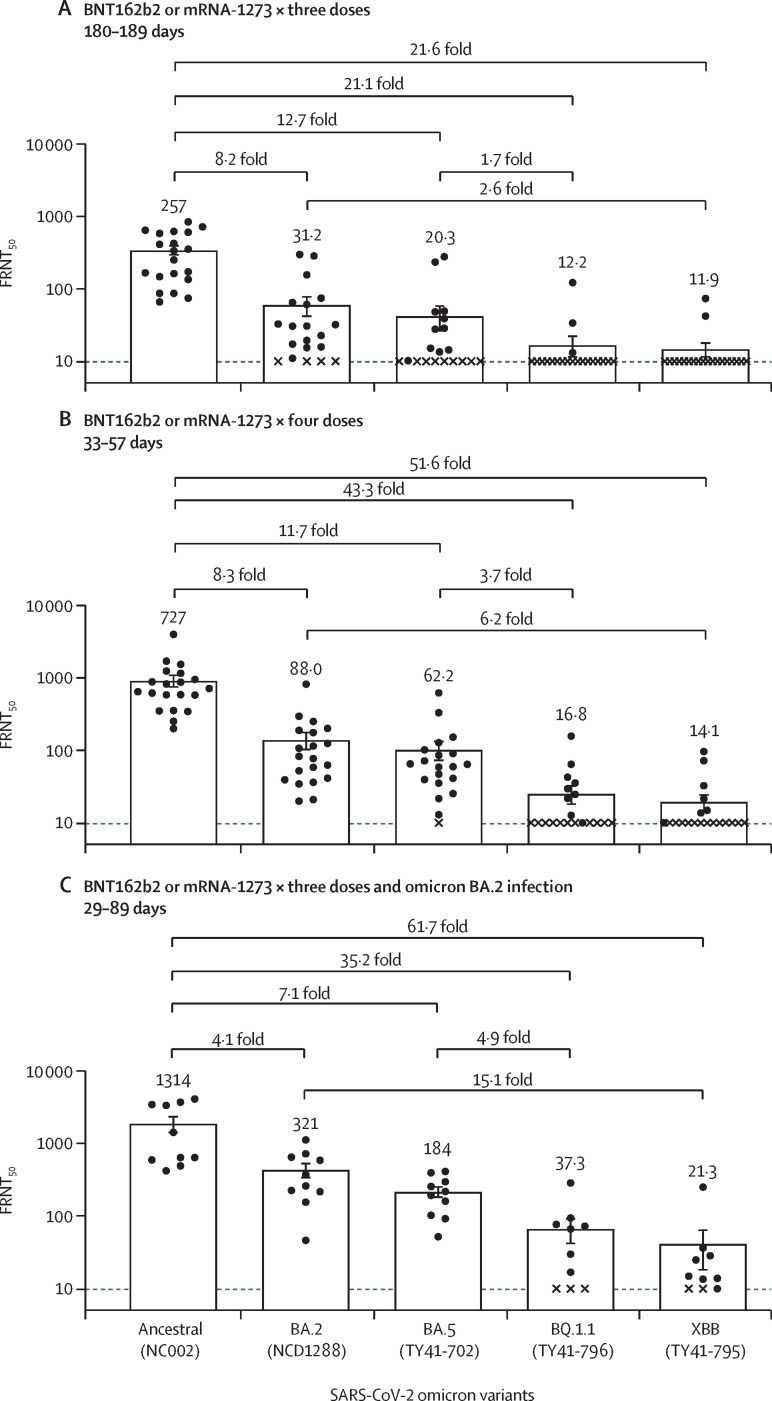
Accordingly, we evaluated the neutralising ability of antibodies in plasma from three different groups against BQ.1.1 and XBB clinical isolates: individuals (180–189 days after the third dose; n=20) who received three doses of the monovalent mRNA vaccine BNT162b2 (Pfizer–BioNTech) or mRNA-1273 (Moderna), or both; individuals (33–57 days after the fourth dose; n=20) who received four doses of the monovalent mRNA vaccine BNT162b2 or mRNA-1273, or both; and individuals (29–89 days after the infection; n=10) who received three doses of monovalent BNT162b2 or mRNA-1273 before the BA.2 breakthrough infection. Using a live-virus neutralisation assay, we determined the 50% focus reduction neutralisation titre (FRNT50) of the plasma samples against BA.2 (hCoV-19/Japan/UT-NCD1288-2N/2022), BA.5 (hCoV-19/Japan/TY41-702/2022), BQ.1.1 (hCoV-19/Japan/TY41-796/2022), and XBB (hCoV-19/Japan/TY41-795/2022). For plasma from individuals who received a third dose of the mRNA vaccine, 17 (85%) of 20 samples or 18 (90%) of 20 samples had FRNT50 values that were below the limit of detection (<10-fold dilution) against BQ.1.1 or XBB, respectively. To calculate the geometric mean titre of each group, we assigned samples that were under the limit of detection of an FRNT50 value of ten. The FRNT50 geometric mean titres against BQ.1.1 and XBB were 21·1-fold and 21·6-fold lower, respectively, than those against the ancestral strain (SARS-CoV-2/UT-NC002-1T/Human/2020/Tokyo) (figure A , appendix p 5). In addition, the geometric mean titres against BQ.1.1 and XBB were 1·7-fold and 2·6-fold lower, respectively, than those against BA.5 and BA.2. Similar results were obtained with samples from individuals who received four doses of mRNA vaccine (figure B); the FRNT50 geometric mean titres against BQ.1.1 and XBB were 43·3-fold and 51·6-fold lower, respectively, than those against the ancestral strain, and were 3·7-fold and 6·2-fold lower than those against BA.5 and BA.2, respectively (figure B, appendix p 6). In contrast, most of the samples from vaccinees with BA.2 breakthrough infection neutralised BQ.1.1 and XBB; however, the FRNT50 geometric mean titres against BQ.1.1 and XBB were 35·2-fold and 61·7-fold lower, respectively, than those against the ancestral strain, and were 4·9-fold and 15·1-fold lower than those against BA.5 and BA.2, respectively (figure C, appendix p 7).
Figure.
Antibody responses to SARS-CoV-2 omicron variants
(A) Neutralising antibody titres of human plasma obtained from individuals immunised with a third dose of BNT162b2 or mRNA-1273 vaccine. Samples were collected 180–189 days after the third immunisation (n=20). (B) Neutralising antibody titres of human plasma obtained from individuals immunised with four doses of BNT162b2 or mRNA-1273 vaccine. Samples were collected 33–57 days after the fourth immunisation (n=20). (C) Neutralising antibody titres of human plasma obtained from individuals who were infected with omicron BA.2 after three doses of BNT162b2 or mRNA-1273 vaccine. Samples were collected 29–89 days after symptom onset (n=10). Each dot represents data from one individual. The lower limit of detection (value=10) is indicated by the horizontal dashed line. Samples under the detection limit (<10-fold dilution) were assigned an FRNT50 value of 10 and are represented by X. Geometric mean titres are shown. FRNT50=50% focus reduction neutralisation titre.
Our data suggest that the omicron sublineages BQ.1.1 and XBB effectively evade current humoral immunity induced by mRNA vaccines or natural infection. A previous study using pseudotyped viruses reported that BQ.1.1 and XBB were less well recognised than BA.2 and BA.4/5 by plasma from convalescent individuals and mRNA vaccinees.3 These findings show that BQ.1.1 and XBB clinical isolates have higher immune evasion abilities than earlier omicron variants, including BA.5 and BA.2.
YK is supported by grants from the Center for Research on Influenza Pathogenesis (HHSN272201400008C) and from the Center for Research on Influenza Pathogenesis and Transmission (75N93021C00014), funded by the National Institute of Allergy and Infectious Disease; and by a Research Program on Emerging and Reemerging Infectious Diseases (JP21fk0108552 and JP21fk0108615), a Project Promoting Support for Drug Discovery (JP21nf0101632), the Japan Program for Infectious Diseases Research and Infrastructure (JP22wm0125002), and a grant (JP223fa627001) from the Japan Agency for Medical Research and Development. All other authors declare no competing interests. RU, MI, and YF contributed equally.
Supplementary Material
References
- 1.Miller NL, Clark T, Raman R, Sasisekharan R. Insights on the mutational landscape of the SARS-CoV-2 omicron variant receptor-binding domain. Cell Rep Med. 2022;3:100527. doi: 10.1016/j.xcrm.2022.100527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imai M, Ito M, Kiso M, et al. Efficacy of antiviral agents against omicron BQ.1.1 and XBB subvariants. N Engl J Med (in press). [DOI] [PMC free article] [PubMed]
- 3.Cao Y, Jian F, Wang J, et al. Imprinted SARS-CoV-2 humoral immunity induces convergent omicron RBD evolution. bioRxiv. 2022 doi: 10.1101/2022.09.15.507787. published online Sept 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.