Abstract
Background: Mupirocin resistance of methicillin-resistant Staphylococcus aureus (MRSA) was frequently reported, but heterogeneous mupirocin resistance in Staphylococcus aureus (S. aureus) was rarely recognized. This study aims to investigate the prevalence of mupirocin heteroresistance among clinical S. aureus isolates and its possible molecular mechanism. Methods: Disk diffusion and agar dilution were used to detect the resistance features of mupirocin resistant S. aureus isolates collected form a tertiary teaching hospital in China. Population analysis profiling was used to identify the mupirocin heteroresistant isolates. Multi locus sequence typing and Staphylococcus protein A gene molecular typing were used to discriminate the genetic features of the heteroresistant isolates. Mutations in the isoleucyl tRNA synthetase (ileS) gene of S. aureus isolates were detected by gene sequencing technique. Results: Mupirocin heteroresistant isolates were identified in 27.67% (83/300) strains. The dominant clones with mupirocin heteroresistance were ST239-t030 MRSAs (25.30%, 21/83). Mutations of G1762T and A637G in ileS gene could be detected in the mupirocin resistant and heteroresistant isolates. The resistance of resistant subpopulations with mutation of G1762T in ileS gene could stabilize for at least 25 passages. Conclusions: This study first revealed a higher prevalence of mupirocin heteroresistance in S. aureus. The mutation of G1762T in ileS gene is closely correlated with both mupirocin resistant and heteroresistant S. aureus isolates, supportingo ileS as a potential marker for fast identification of mupirocin resistant S. aureus.
Keywords: Heteroresistance, mupirocin resistance, Staphylococcus aureus
Introduction
Staphylococcus aureus (S. aureus) is a major human pathogen contributing to multiple community and hospital acquired infections, such as skin and soft tissue infections, abscess, sepsis and blood stream infection [1]. Mupirocin is widely used for the prevention and treatment of local skin and soft tissue S. aureus infections [2] and is routinely prescribed as a de-colonization agent for methicillin-resistant S. aureus (MRSA) colonizers [3-5]. As a topical antimicrobial agent, mupirocin can competitively bind to isoleucine t-RNA synthetase (ileS) and inhibit bacterial growth correspondingly. Recently, mupirocin resistance was frequently reported, and the mechanism of resistance have been explored [6-8].
It is well known that mupirocin resistance was categorized into two kinds of resistance phenotypes, low-level mupirocin resistance (LLMR) and high-level mupirocin resistance (HLMR), and the resistance mechanisms differed greatly [9]. However, to our knowledge, heterogeneous mupirocin resistance in S. aureus is rarely reported, and its potential clinical significance was not elucidated. The characterization of heterogeneous resistance was defined as only a small number of highly resistant subpopulations presented in the cultures from a single-cell inocula, while the majority of cells showed low or moderate level of resistance [10]. Heteroresistance is a common phenomenon in S. aureus toward multiple antimicrobial agents, such as β-lactam antibiotics, macrolide and vancomycin [11-14]. Upon exposure to sub-inhibitory concentrations of antibiotics, clones with heterogeneous resistance could develop into homogeneous and highly resistant ones [15], which may lead to anti-infection failure or high-level of homogeneous resistance. However, the definition of heterogeneous resistance is varied for different bacteria, and there is no clear definition to mupirocin in S. aureus. Hence, the present study defined the heterogeneous mupirocin resistance in S. aureus as the isolates with heteroresistant subpopulation at a frequency (≥10-9), with an MIC ≥8 μg/mL to mupirocin of the resistant subpopulations [16], and carrying well-known mutations in ileS gene correlated with LLMR or mupA gene contributing to HLMR in S. aureus.
In this study, we try to investigate the actual prevalence of mupirocin heteroresistance among methicillin-susceptible S. aureus (MSSA) and MRSA isolates. Meanwhile, its possible resistance mechanism was explored.
Methods
Isolates identification
S. aureus strains used in this study were isolated from different patients admitted to a 3000-bed teaching hospital in China from March 2011 to February 2020, and the strains were isolated from routine clinical samples, identified with VITEK-2 compact fully automated microbiology analysis system (BioMérieux, France) and then stored at -80°C. The isolates were obtained from different clinical samples, including sputum (41%, 123/300), secretion samples (26.6%, 79/300), wound samples (10.3%, 31/300), whole blood (6.9%, 21/300), nasopharyngeal swabs (6.9%, 21/300), urine (3%, 9/300), pus (5%, 15/300) and bronchoalveolar lavage fluid (0.3%, 1/300). During this study, the isolates were again subcultivated at blood agar plates and verified again by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (microTyper MS, Tianrui, China) and 16S-rRNA gene sequencing [17].
Mupirocin susceptibility testing
Disc diffusion was used to screen LLMR and HLMR isolates using 5 μg and 120 μg mupirocin discs [18,19]. The MICs of S. aureus to mupirocin was further detected using agar dilution [20]. The concentration of mupirocin was set from 0.015 μg/mL to 512 μg/mL with two-fold dilution. Strains with mupirocin MICs between 8 and 256 µg/mL were considered as LLMR, and the ones with an MIC ≥512 µg/mL were considered to be HLMR.
Identification of mupirocin heteroresistant S. aureus isolates
Classical population analysis profiling (PAP) was used to identify mupirocin heteroresistant S. aureus isolates. The assay was performed according to the procedure suggested by Tomasz et al. [21] with minor modifications. Generally, approximately 109 CFU of the cultures at stationary-phase were plated onto a series of tryptone soya agar (TSA) plates containing serial concentrations of mupirocin (4 µg/mL to 512 µg/mL with two-fold dilution), and the plates were incubated at 35°C for 48 h. The number of colonies grew on each plate were recorded, and suspected resistant colonies were subcultivated and identified using MALDI-TOF MS technique. The frequency of heteroresistant subpopulations obtained from each isolate was calculated. To verify the stability of resistance, the subpopulations were subcultured for 50 passages on TSA plates without antibiotics.
Molecular typing
Multi-locus sequence typing (MLST) was performed according to the procedure previously described [22]. The total genome was extracted using Qiagen bacterial total DNA extracting kit (Qiagen Ltd, Germany). Seven respective PCR assays were conducted to amplify seven housekeeping genes for S. aureus, including carbamatek inase (arcC), shikimate dehydrogenase (aroE), glycerol kinase (glpF), guanylate kinase (gmk), phosphate acetyltransferase (pta), triosephosphate isomerase (tpi) and acetyle coenzyme A acetyltransferase (yqi). The PCR products were sequenced, and the sequences were compared with the known alleles in the MLST databases (http://saureus.mlst.net). The variable repeat region of staphylococcus protein A gene (spa) was amplified based on the procedure previously described [23]. Then, the PCR products were sequenced, and the sequences were analyzed using the Ridom web server (http://spaserver.ridom.de).
PCR amplification and sequencing of the ileS gene
To detect the possible mutation of ileS gene of the mupirocin resistant subpopulations, the whole sequence of ileS gene was amplified using a pair of primers, ileS-F: TACCGCGAGCAATCGTCCCT, ileS-R: TGTTGGCATCGTGGGCATAG. The PCR products were purified with QIA quick PCR purification kit (Qiagen, GmbH, Germany) and sequenced with the following primers, ileS-WF: CAATCCAGTGCTTCTGCTAC, ileS-WR: AGACTTTGGGTAAGTAGTACG. The DNA sequences for each isolate was compared with S. aureus ileS gene (GenBank accession number X74219) to identify potential mutation points.
Detection of HLMR encoding genes
MupA and mupB genes, contributing to HLMR of S. aureus, were amplified by PCR using the following primers, mupA-F: TATATTATGCGATGGAAGGTTGG, mupA-R: AATAAAATCAGCTGGAAAGTGTTG, mupB-F: CTAGAAGTCGATTTTGGAGTAG, and mupB-R: AGTGTCTAAAATGATAAGACGATC. The amplification conditions were set according the protocol previously described [24]. The positive PCR products were verified by gene sequencing.
Stability of mupirocin resistant subpopulations
The resistant subpopulations originated from mupirocin heteroresistant S. aureus strains were subcultured on TSA plates without antibiotics for 50 passages. The resistance levels to mupirocin were detected with agar dilution, and mutations in ileS gene were detected using the method described above.
Time-kill kinetics assay
S. aureus ATCC25923 and one isolate with mupirocin heteroresistance were selected for time-kill kinetics assay. The culture tubes with each isolate were incubated at 35°C with shaking at 200 rpm, and 106 cfu/mL TSB broth culture at logarithmic-phase of each isolate was prepared. Then, mupirocin was added to yield concentrations of 0, 0.5, 1, 2, 4, 8, 16, 32 and 64 × MIC of the tested isolate. At 0 min, 20 min, 40 min, 60 min, 2 h, 6 h and 24 h after mupirocin addition, the samples of each broth culture were collected, and appropriate dilution were performed. Thereafter, the diluted bacterial suspensions (10 µL) were spirally plated on TSA plates. After 24 h cultivation at 35°C, the viable colonies were counted and the time-kill curves were plotted.
Results
Mupirocin resistance features
The MIC50 and MIC90 of 300 S. aureus strains to mupirocin were 0.25 μg/mL and 0.5 μg/mL, respectively. As shown in Table 1, comparing with those of MSSA isolates analyzed in this study, both MIC50 and MIC90 of the MRSA isolates to mupirocin increased to 2 and 4 folds, respectively. By both disk diffusion and agar microdilution, the LLMR rates for MSSA and MRSA were 4.7% (7/150) and 0.6% (1/150), respectively. No HLMR isolate was identified.
Table 1.
Mupirocin resistance features of MSSA and MRSA isolates
| Strains | MIC50 (μg/mL) | MIC90 (μg/mL) |
|---|---|---|
| MSSA (n=150) | 0.125 | 0.25 |
| MRSA (n=150) | 0.25 | 1.00 |
MSSA: Methicillin-Susceptible Staphylococcus Aureus; MRSA: Methicillin-Resistant Staphylococcus Aureus.
Epidemiological characteristics of mupirocin heteroresistant isolates
In total, 27.7% (83/300) mupirocin sensitive strains showed significant heterogeneous resistance, and two different heterogeneous resistance patterns were identified by PAP, including heteroresistant-LLMR mode (r-LLMR) and heteroresistant-HLMR mode (r-HLMR) (Supplementary Figure 1), which accounted for 27.3% (82/300) and 3.3% (1/300), respectively. The distribution of the samples positive with mupirocin heteroresistant S. aureus isolates were shown in Table 2. The majority of the mupirocin heteroresistant S. aureus isolates were isolated from wound secretion (39.8%, 33/83), followed by sputum (31.3%, 26/83), whole blood (10.8%, 9/83) and pus (8.4%, 7/83). The isolates from wound secretion were mainly composed by MSSA (26.5%, 22/83), while those from sputum were mainly composed by MRSA isolates (20.5%, 17/83). Based on the definition of heterogeneous resistance in our study, the frequency of heteroresistant subpopulations among 40 strains of MRSA and 43 strains of MSSA were between 10-9 to 10-8.
Table 2.
Distribution of clinical samples with mupirocin heteroresistant S. aureus isolates
| Samples | Percentages (n, %) | MRSA (n, %) | MSSA (n, %) |
|---|---|---|---|
| Wound secretion | 33, 39.8% | 11, 13.3% | 22, 26.5% |
| Sputum | 26, 31.3% | 17, 20.5% | 9, 10.8% |
| Whole blood | 9, 10.8% | 3, 3.6% | 6, 7.2% |
| Nasopharyngeal swab | 4,4.8% | 1, 1.2% | 3, 3.6% |
| Pus | 7, 8.4% | 5, 6% | 2, 2.4% |
| Puncture fluid | 3, 3.6% | 2, 2.4% | 1, 1.2% |
| Urine | 1, 1.2% | - | 1, 1.2% |
MSSA: Methicillin-Susceptible Staphylococcus Aureus; MRSA: Methicillin-Resistant Staphylococcus Aureus.
Results of PAP assay showed that mupirocin resistant subpopulations generated from the heterogeneous resistant isolates were presenting at a lower frequency, ranging from 10-9 to 10-8, and the resistance levels of these highly resistant subpopulations could reach up to 256 μg/mL or 512 μg/mL. For only one r-HLMR isolate identified in this study, 25 highly resistant subpopulations (MIC ≥512 μg/mL) were obtained. Meanwhile, the resistance to mupirocin of the subpopulations originated from 82 r-LLMR isolates varied from strain to strain (8-256 μg/mL).
Molecular typing results
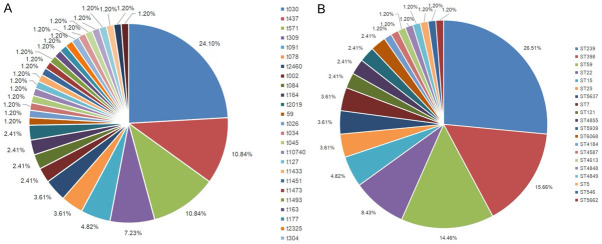
Among 83 heterogeneous mupirocin resistant S. aureus, ST-239 (26.51%, 22/83) was the most prevalent, followed by ST-398 (15.66%, 13/83), ST-59 (14.46%, 12/83), ST-22 (8.43%, 7/83), ST15 (4.82%, 4/83), ST-7 (3.61%, 3/83), ST-25 (3.6%, 3/83), ST-5637 (3.6%, 3/83), ST-121 (2.41%, 2/83), ST-4855 (2.4%, 2/83), ST-5939 (2.41%, 2/83), ST-6068 (2.41%, 2/83), ST-5 (1.2%, 1/83), ST-546 (1.2%, 1/83), ST-4587 (1.2%, 1/83), ST-4613 (1.2%, 1/83), ST-4848 (1.2%, 1/83), ST-4849 (1.2%, 1/83), ST-4855 (1.2%, 1/83) and ST-5662 (1.2%, 1/83). By spa typing, 32 spa types were found. The most prevalent type was t030 (25.30%, 21/83), followed by t437 (10.84%, 9/83), t571 (10.84%, 9/83), t309 (7.23%, 6/83), t091 (4.82%, 4/83), t078 (3.61%, 3/83), t2460 (2.41%, 2/83), t164 (2.41%, 2/83), t002 (2.41%, 2/83) and t084 (2.41%, 2/83) (Figure 1). Combination of STs with spa types, the most predominant clone was ST239-t030 (25.30%, 21/83), followed by ST59-t437 (10.84%, 9/83), ST398-t571 (10.84%, 9/83) and ST22-t309 (7.23%, 6/83). The 21 strains of ST239-t030 (100%, 21/21) and 8 isolates of ST59-t437 (88.89%, 8/9) were mainly composed by MRSA, while 8 strains of ST398-t571 (88.89%, 8/9) and 5 isolates of ST22-t309 (83.33%, 5/6) were mainly composed by MSSA.
Figure 1.
Distribution of STs and spa types of heterogeneous mupirocin resistant MRSA and MSSA. Note: A. Distribution of different spa types; B. Distribution of different ST types. MSSA: Methicillin-Susceptible Staphylococcus Aureus; MRSA: Methicillin-Resistant Staphylococcus Aureus; SPA: Staphylococcus Protein A Gene.
IleS gene mutation, mupA and mupB gene detection
Among 115 mupirocin sensitive S. aureus isolates, 83 mupirocin heteroresistant S. aureus parent isolates and 83 randomly selected resistant subpopulations originated from 83 mupirocin heteroresistant S. aureus isolates, neither mupA or mupB gene was detected. On the contrary, multiple point mutations (G1762T, A637G, A1263T, G937T, A1312G, C1468T) in ileS gene were identified and varied greatly among different isolates, and mutation of A637G and G1762T were the most frequent (Table 3). Only mutation of G1762T was observed in mupirocin heteroresistant isolates and their resistant subpopulations. No significant differences in types of mutations in ileS gene among resistant subpopulations originated from mupirocin heteroresistant MRSA and MSSA isolates was observed (Table 4).
Table 3.
Distribution of point mutations of ileS gene in mupirocin sensitive and mupirocin heteroresistant strains
| Sense Mutations in ileS gene | Mupirocin sensitive strains (n=115) | Heterogeneous mupirocin resistant strains | ||
|---|---|---|---|---|
|
| ||||
| Parent heteroresistant isolates (n=83) | LLMR subpopulations (n=82) | HLMR subpopulations (n=1) | ||
| A637G | 39 | 1 | 74 | 1 |
| G937T | 5 | 13 | 11 | 0 |
| A1263T | 5 | 13 | 13 | 0 |
| A1312G | 18 | 10 | 11 | 0 |
| C1468T | 14 | 14 | 11 | 1 |
| G1762T | 0 | 0 | 63 | 1 |
Note: Resistant subpopulations were randomly selected. LLMR: Low-Level Mupirocin Resistance; HLMR: High-Level Mupirocin Resistance.
Table 4.
Major mutations in ileS gene among randomly selected mupirocin resistant subpopulations originated from S. aureus isolates with mupirocin heteroresistance
| Mutation points | Resistant subpopulations originated from r-LLMR | Resistant subpopulations originated from r-HLMR | ||
|---|---|---|---|---|
|
|
|
|||
| MRSA (n=40) | MSSA (n=42) | MRSA (n=0) | MSSA (n=1) | |
| A637G (Asn-Asp) | 36 | 38 | 0 | 1 |
| G1762T (Val-Phe) | 28 | 38 | 0 | 1 |
Note: Resistant subpopulations were randomly selected. MSSA: Methicillin-Susceptible Staphylococcus Aureus; MRSA: Methicillin-Resistant Staphylococcus Aureus; LLMR: Low-Level Mupirocin Resistance; HLMR: High-Level Mupirocin Resistance.
Stability of mupirocin resistant subpopulations
Twenty-seven mupirocin resistant subpopulations originated from 9 mupirocin heteroresistant S. aureus isolates were randomly selected to assess their stability of resistance to mupirocin during successive subcultivation for 50 generations. The diversity of mutation in ileS gene of mupirocin resistant subpopulations was observed in the same isolate, for either MRSA or MSSA isolates. As shown in Supplementary Table 1, among 12 subpopulations with mutation of G1762T in ileS gene, the resistance of 75.0% (9/12) subpopulations to mupirocin was stable after 50th successive passages, and the stability of resistance to mupirocin of 25% (3/12) could maintain for at least 25th generations of subcultivation on TSA plates without antibiotics. On the contrary, the stability of resistance to mupirocin of 53.3% (8/15) subpopulations without mutation of G1762T in ileS gene reverted to sensitive within 15th generations of subcultivation on TSA plates without antibiotics. Furthermore, mutations in ileS gene of the different subpopulations originated from the same isolate were not unique, such as subpopulations from r-314 and r-724 isolates.
Time-kill feature of mupirocin heteroresistant strains
Mupirocin sensitive strain (S. aureus, ATCC25923) and mupirocin heteroresistant isolate (r-724) showed different growth features in TSB broth with gradient concentrations of mupirocin. The growth of ATCC25923 at different concentrations of mupirocin were all inhibited until 24 hours (Figure 2). The growth of r-724 isolate was inhibited when the concentrations of mupirocin ≥0.5 μg/mL, while the isolate at 0.25 μg/mL showed delayed growth at 24 hours, and the subpopulations were further confirmed to be LLMR.
Figure 2.
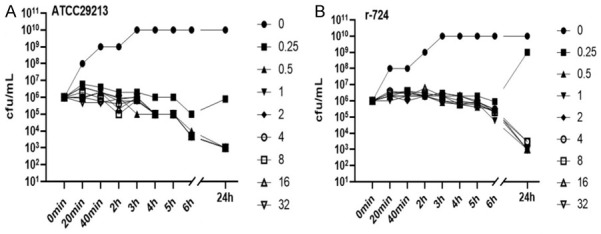
Time-kill features of ATCC25923 and mupirocin heteroresistant isolate (r-724). Note: A. Staphylococcus aureus ATCC29213; B. Mupirocin heteroresistant Staphylococcus aureus r-724.
Discussion
In this study, the prevalence rate of mupirocin resistant S. aureus was close to that reported by Yu et al. [25], who revealed that only 4.7% and 0.6% LLMR isolates were detected from MRSA and MSSA isolates, and no HLMR isolate was found. Comparing with that of MSSA isolates, the mupirocin MIC90 of MRSA isolates increased from 0.25 μg/mL to 1 μg/mL in this study. Because the number of isolates investigated in this study is small, the actual prevalence of mupirocin resistance among MRSA and MSSA needs to be further ascertained. Surprisingly, the prevalence of heterogeneous resistance of MSSA and MRSA to mupirocin reached 28.0% (42/150) and 26.7% (40/150), respectively. It is known that, oxacillin sensitive MRSAs commonly show typical heterogeneous resistance and can easily convert to highly resistant populations under sub-inhibitory concentrations of oxacillin and/or mupirocin, which could lead to failure of anti-infection treatment when they are incorrectly treated as susceptible ones [13]. How to accurately detect the resistance of mupirocin in S. aureus and the effectiveness of routine mupirocin based MRSA de-colonization protocol should be reevaluated when the patients have been confirmed to be colonized with heterogeneous mupirocin resistant MRSA. Also, the effectiveness of routine usage of mupirocin ointment agents should be carefully considered in treatment of patients with suspect S. aureus infection.
Since high prevalence of LLMR among MRSA and MSSA strains was found in this study, we try to further investigate the molecular characteristics of mupirocin heteroresistant S. aureus strains. Most of the strains were isolated from sputum, secretion and blood, et al. It is revealed that ST239-t030, ST59-t437, ST398-t571 and ST22-t309 clones were dominant ones with heterogeneous resistance. ST239-t030 and ST59-t437 are two known hospital acquired MRSA and community acquired MRSA clones prevalent in Asia [26,27]. ST239-t030 isolates in this study were fully composed of MRSA isolates, while ST398-t571 and ST22-t309 clones were mainly composed of MSSA isolates. The mupirocin heteroresistant strains were isolated most frequently from wound secretion samples (39.8%) in this study, which suggested that mupirocin heteroresistance should be taken into account when mupirocin was used in treatment of wound infections by S. aureus.
To rapidly screen mupirocin resistant S. aureus, disc diffusion with 5 μg or 120 μg mupirocin was recommended for LLMR or HLMR detection [18,19]. In this study, excellent agreement between disc diffusion and agar dilution was observed. Considering the wide application of automated microbial identification and antimicrobial detection equipment in clinical laboratories, the consistency between disc diffusion and BD PhoenixTM PMIC/ID panel on detection of mupirocin resistance detection was further compared, and it was confirmed that BD PhoenixTM PMIC/ID panel could accurately identify LLMR strains (data not shown). Despite that disc diffusion was routinely applied in many laboratories and could easily discriminate heteroresistance by observing clonal growth within inhibition zone [16], no literature reported its efficiency on detect mupirocin heteroresistance in S. aureus yet. No heteroresistant isolate was identified by disk diffusion with 5 μg mupirocin in this study. Lower frequency of resistant subpopulations (10-9-10-8) and low density of bacterial cells inoculated in the plates maybe the major reasons leading to low detection efficiency of disk diffusion during the identification of mupirocin heteroresistant clones [16]. Also, no significant correlation was observed between mupirocin MICs of the primary isolates with mupirocin heteroresistance and the frequencies of resistant subpopulations. Hence, it is not practical to predict mupirocin heteroresistance and possible failure of mupirocin treatment based on the mupirocin MICs of the primary isolates [28]. Although PAP is tedious and expensive to perform routinely in clinical laboratories, it is still the best method to identify mupirocin heteroresistance based on experience in this study. A simplified PAP with single concentration (4 μg/mL mupirocin) or double concentrations (4 μg/mL and 256 μg/mL mupirocin) maybe an alternative method in clinical microbiological laboratories. Moreover, other techniques with more detection sensitivity may provide more choices to detect heteroresistance, such as droplet digital PCR [29] and plasmonic colloidosomes coupled MALDI-TOF MS technique [30].
To further explore the resistance mechanism of mupirocin heteroresistant S. aureus, some well-known mutations within ileS gene responsible for LLMR were detected. It was found that G1762T, a point mutation, appeared at a high frequency in mupirocin resistant subpopulations. G1762T is a mutation within ileS gene correlated with LLMR in MRSA. The substitution can lead to an amino acid change (V588F) at codon 588 from valine to phenylalanine, and the Rossman Fold of Isoleucyl-tRNA Synthetase can be affected [31]. However, another well-known mutation (V631F) was not detected in the mupirocin resistant subpopulations in this study. In contrast, another point mutation of A637G appeared at a high frequency at mupirocin resistant subpopulations. However, the correlation between mutation of A637G in ileS gene of S. aureus isolates and the resistance of LLMR couldn’t be confirmed, since this mutation also appeared in 39 mupirocin sensitive strains in this study. Furthermore, the resistance to mupirocin of the subpopulations with G1762T mutation was stable after at least 25 successive passages. However, the resistance to mupirocin of the resistant subpopulations with mutation in A637G of ileS gene was not always stable, because 40% (6/14) mupirocin resistant subpopulations returned to mupirocin sensitive after 15 successive passages on TSA plates without antibiotics. Hence, based on the results of this study, it is presumable that mutation of G1762T in ileS gene is a key mechanism contributing to mupirocin heteroresistance in S. aureus. To define mupirocin heteroresistance of S. aureus, appearance of G1762T mutation in ileS gene or not should be considered as an important index. Also, detection of mutation of G1762T with suitable molecular methods may be a potential target in rapid discrimination of LLMR or heteroresistant S. aureus isolates.
The limitations of this study include following two aspects. Firstly, owing to limitation of expenditure, not all of the possible molecular mechanisms contributing to heteroresistance found in other bacterial pathogens were investigated in this study, such as gene amplification-driven heteroresistance [32] or overexpression of genes encoding proteins involved in efflux [33]. Secondly, possible new mechanisms contributing to heteroresistance is not investigated during this study. Whole genome sequencing or other new techniques should be adopted to explore other possible molecular mechanisms related to heteroresistance in S. aureus.
Conclusions
As far as we know, high prevalence and stability of mupirocin heteroresistance in S. aureus was firstly reported in this study, which indicates that accurate and rapid identification of mupirocin heteroresistant S. aureus seems to be an important issue for rational application of mupirocin agents in de-colonization of MRSA and treatment of skin and soft tissue infections resulted by S. aureus. ST239-t030 MRSA, ST59-t437 MRSA, ST398-t571 MSSA as well as ST22-t309 MSSA clones are easier to present with mupirocin heteroresistance and should be paid more attention. Also, mutation of G1762T in ileS gene seems to be an important molecular target in rapid screening of mupirocin resistant or heteroresistant S. aureus using PCR or other molecular methods.
Acknowledgements
We thank for all of the staff of laboratory medicine for assisting with isolate storage. The work is supported by Natural science foundation of Inner Mongolia Autonomous Region of China (2016MS0829).
This work is exempt from formal ethical approval and informed consent according to the local ethical guidelines, and was approved by Ethics Committee of Affiliated hospital of Inner Mongolian medical university (Reference number KY2020028).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Woodford N, Livermore DM. Infections caused by Gram-positive bacteria: a review of the global challenge. J Infect. 2009;59(Suppl 1):S4–S16. doi: 10.1016/S0163-4453(09)60003-7. [DOI] [PubMed] [Google Scholar]
- 2.Forcade NA, Parchman ML, Jorgensen JH, Du LC, Nyren NR, Trevino LB, Pena J, Mann MW, Munoz A, Trevino SB, Mortensen EM, Wickes BL, Pollock BH, Frei CR. Prevalence, severity, and treatment of community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) skin and soft tissue infections in 10 medical clinics in Texas: a South Texas ambulatory research network (STARNet) study. J Am Board Fam Med. 2011;24:543–550. doi: 10.3122/jabfm.2011.05.110073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang SS, Singh R, Mckinnell JA, Park S, Gombosev A, Eells SJ, Gillen DL, Kim D, Rashid S, Macias-Gil R, Bolaris MA, Tjoa T, Cao C, Hong SS, Lequieu J, Cui E, Chang J, He J, Evans K, Peterson E, Simpson G, Robinson P, Choi C, Bailey CJ, Leo JD, Amin A, Goldmann D, Jernigan JA, Platt R, Septimus E, Weinstein RA, Hayden MK, Miller LG. Decolonization to reduce postdischarge infection risk among MRSA carriers. N Engl J Med. 2019;380:638–650. doi: 10.1056/NEJMoa1716771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiefer A, Bogdan C, Melichar VO. Successful eradication of newly acquired MRSA in six of seven patients with cystic fibrosis applying a short-term local and systemic antibiotic scheme. BMC Pulm Med. 2018;18:20. doi: 10.1186/s12890-018-0588-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellis MW, Griffith ME, Dooley DP, Mclean JC, Jorgensen JH, Patterson JE, Davis KA, Hawley JS, Regules JA, Rivard RG, Gray PJ, Ceremuga JM, Dejoseph MA, Hospenthal DR. Targeted intranasal mupirocin to prevent colonization and infection by community-associated methicillin-resistant Staphylococcus aureus strains in soldiers: a cluster randomized controlled trial. Antimicrob Agents Chemother. 2007;51:3591–3598. doi: 10.1128/AAC.01086-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khoshnood S, Heidary M, Asadi A, Soleimani S, Motahar M, Savari M, Saki M, Abdi M. A review on mechanism of action, resistance, synergism, and clinical implications of mupirocin against Staphylococcus aureus. Biomed Pharmacother. 2019;109:1809–1818. doi: 10.1016/j.biopha.2018.10.131. [DOI] [PubMed] [Google Scholar]
- 7.Seah C, Alexander DC, Louie L, Simor A, Low DE, Longtin J, Melano RG. MupB, a new high-level mupirocin resistance mechanism in Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56:1916–1920. doi: 10.1128/AAC.05325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dadashi M, Hajikhani B, Darban-Sarokhalil D, van Belkum A, Goudarzi M. Mupirocin resistance in Staphylococcus aureus: a systematic review and meta-analysis. J Glob Antimicrob Resist. 2020;20:238–247. doi: 10.1016/j.jgar.2019.07.032. [DOI] [PubMed] [Google Scholar]
- 9.Hodgson JE, Curnock SP, Dyke KG, Morris R, Sylvester DR, Gross MS. Molecular characterization of the gene encoding high-level mupirocin resistance in Staphylococcus aureus J2870. Antimicrob Agents Chemother. 1994;38:1205–1208. doi: 10.1128/aac.38.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Halfawy OM, Valvano MA. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev. 2015;28:191–207. doi: 10.1128/CMR.00058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saravolatz SN, Martin H, Pawlak J, Johnson LB, Saravolatz LD. Ceftaroline-heteroresistant Staphylococcus aureus. Antimicrob Agents Chemother. 2014;58:3133–3136. doi: 10.1128/AAC.02685-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomes DM, Ward KE, Laplante KL. Clinical implications of vancomycin heteroresistant and intermediately susceptible Staphylococcus aureus. Pharmacotherapy. 2015;35:424–432. doi: 10.1002/phar.1577. [DOI] [PubMed] [Google Scholar]
- 13.Chung M, Kim CK, Conceicao T, Aires-De-Sousa M, De Lencastre H, Tomasz A. Heterogeneous oxacillin-resistant phenotypes and production of PBP2A by oxacillin-susceptible/mecA-positive MRSA strains from Africa. J Antimicrob Chemother. 2016;71:2804–2809. doi: 10.1093/jac/dkw209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen DK, Lai HY, Yang DM, Xu HT. Mechanism of hetero-erythromycin resistant Staphylococcus aureus and a comparison of detection methods. Zhonghua Yi Xue Za Zhi. 2013;93:3867–3871. [PubMed] [Google Scholar]
- 15.Lee AS, Gizard Y, Empel J, Bonetti EJ, Harbarth S, Francois P. Mupirocin-induced mutations in ileS in various genetic backgrounds of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2014;52:3749–3754. doi: 10.1128/JCM.01010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersson DI, Nicoloff H, Hjort K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat Rev Microbiol. 2019;17:479–496. doi: 10.1038/s41579-019-0218-1. [DOI] [PubMed] [Google Scholar]
- 17.Monday SR, Bohach GA. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J Clin Microbiol. 1999;37:3411–3414. doi: 10.1128/jcm.37.10.3411-3414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finlay JE, Miller LA, Poupard JA. Interpretive criteria for testing susceptibility of staphylococci to mupirocin. Antimicrob Agents Chemother. 1997;41:1137–1139. doi: 10.1128/aac.41.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Thirtieth Informational Supplement. M100-S30. Wayne, PA: Clinical and Laboratory Standards Institute; 2020. [Google Scholar]
- 20.European committee for antimicrobial susceptibility testing (EUCAST) of the European society of clinical microbiology and infectious diseases (ESCMID) EUCAST definitive document E.DEF 3.1, June 2000: determination of minimum inhibitory concentrations (MICs) of antibacterial agents by agar dilutions. Clin Microbiol Infect. 2000;6:509–515. doi: 10.1046/j.1469-0691.2000.00142.x. [DOI] [PubMed] [Google Scholar]
- 21.Tomasz A, Nachman S, Leaf H. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob Agents Chemother. 1991;35:124–129. doi: 10.1128/aac.35.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellmann A, Friedrich AW, Rosenkotter N, Rothganger J, Karch H, Reintjes R, Harmsen D. Automated DNA sequence-based early warning system for the detection of methicillin-resistant Staphylococcus aureus outbreaks. PLoS Med. 2006;3:e33. doi: 10.1371/journal.pmed.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahmoudi S, Mamishi S, Mohammadi M, Banar M, Ashtiani M, Mahzari M, Bahador A, Pourakbari B. Phenotypic and genotypic determinants of mupirocin resistance among Staphylococcus aureus isolates recovered from clinical samples of children: an Iranian hospital-based study. Infect Drug Resist. 2019;12:137–143. doi: 10.2147/IDR.S185610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Jin Y, Lin C, Hao Z, Duan J, Guo Y, Wang S, Hu L, Wang L, Yu F. Low prevalence of mupirocin resistance among Staphylococcus aureus clinical isolates from a Chinese tertiary hospital. J Med Microbiol. 2019;68:201–205. doi: 10.1099/jmm.0.000911. [DOI] [PubMed] [Google Scholar]
- 26.Cheng H, Yuan W, Zeng F, Hu Q, Shang W, Tang D, Xue W, Fu J, Liu J, Liu N, Zhu J, Yang J, Hu Z, Yuan J, Zhang X, Li S, Chen Z, Hu X, Rao X. Molecular and phenotypic evidence for the spread of three major methicillin-resistant Staphylococcus aureus clones associated with two characteristic antimicrobial resistance profiles in China. J Antimicrob Chemother. 2013;68:2453–2457. doi: 10.1093/jac/dkt213. [DOI] [PubMed] [Google Scholar]
- 27.Song JH, Hsueh PR, Chung DR, Ko KS, Kang CI, Peck KR, Yeom JS, Kim SW, Chang HH, Kim YS, Jung SI, Son JS, So TM, Lalitha MK, Yang Y, Huang SG, Wang H, Lu Q, Carlos CC, Perera JA, Chiu CH, Liu JW, Chongthaleong A, Thamlikitkul V, Van PH. Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. J Antimicrob Chemother. 2011;66:1061–1069. doi: 10.1093/jac/dkr024. [DOI] [PubMed] [Google Scholar]
- 28.Nicoloff H, Hjort K, Levin BR, Andersson DI. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat Microbiol. 2019;4:504–514. doi: 10.1038/s41564-018-0342-0. [DOI] [PubMed] [Google Scholar]
- 29.Sun L, Talarico S, Yao L, He L, Self S, You Y, Zhang H, Zhang Y, Guo Y, Liu G, Salama NR, Zhang J. Droplet digital PCR-based detection of clarithromycin resistance in helicobacter pylori isolates reveals frequent heteroresistance. J Clin Microbiol. 2018;56:e00019–18. doi: 10.1128/JCM.00019-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai YC, Li CY, Yi J, Qin Q, Liu BH, Qiao L. Plasmonic colloidosome-coupled MALDI-TOF MS for bacterial heteroresistance study at single-cell level. Anal Chem. 2020;92:8051–8057. doi: 10.1021/acs.analchem.0c00494. [DOI] [PubMed] [Google Scholar]
- 31.Antonio M, Mcferran N, Pallen MJ. Mutations affecting the Rossman fold of isoleucyl-tRNA synthetase are correlated with low-level mupirocin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:438–442. doi: 10.1128/AAC.46.2.438-442.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hjort K, Nicoloff H, Andersson DI. Unstable tandem gene amplification generates heteroresistance (variation in resistance within a population) to colistin in Salmonella enterica. Mol Microbiol. 2016;102:274–289. doi: 10.1111/mmi.13459. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, Hu DX, Zhang QJ, Liao XP, Liu YH, Sun J. Efflux pump overexpression contributes to tigecycline heteroresistance in Salmonella enterica serovar Typhimurium. Front Cell Infect Microbiol. 2017;7:37. doi: 10.3389/fcimb.2017.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.