Abstract
DEK, a chromatin-remodeling phosphoprotein, is associated with various functions and biological pathways in the periphery, including inflammation, oncogenesis, DNA repair, and transcriptional regulation. We recently identified an association between DEK loss and central nervous system diseases, such as Alzheimer’s. To understand DEK’s potential role in disease, it is critical to characterize DEK in healthy human brain to distinguish between neural DEK expression and function in healthy versus diseased states like dementia. We utilized two public databases, BrainCloud and Human Brain Transcriptome, and analyzed DEK mRNA expression across the lifespan in learning and memory relevant brain regions. Since DEK loss induces phenotypes associated with brain aging (e.g., DNA damage, apoptosis), we hypothesized that neural DEK expression may be highest during fetal development and lower in elderly individuals. In agreement with this hypothesis, DEK was most prominently expressed during fetal development in all queried forebrain areas, relative to other ages. Consistent with its roles in the periphery, pathways related to DEK in the brain were associated with cellular proliferation, DNA replication and repair, apoptosis, and inflammation. We also found novel neural development-relevant pathways (e.g., synaptic transmission, neurite outgrowth, myelination) to be enriched from genes correlated with DEK expression. These findings suggest that DEK is important for human brain development. Overall, we highlight age-related changes in neural DEK expression across the human lifespan and illuminate novel biological pathways associated with DEK that are distinct from normal brain aging. These findings may further our understanding of how DEK impacts brain function and disease susceptibility.
Graphical Abstract
Our lab recently identified a link between DEK loss and Alzheimer’s disease. It is difficult to determine the impact of DEK loss in neurodegenerative diseases without characterizing its expression in healthy human brain. We report that DEK expression is highest in several queried forebrain regions during the prenatal vs. postnatal period. Pathway analyses suggest DEK is critical for embryonic brain development. This study provides a framework to further exploit DEK’s role in brain function and disease.
Introduction
Our group was the first to associate DEK, a chromatin-associated phosphoprotein, with cognitive-related disorders, such as Alzheimer’s disease, using human cells (Greene et al. 2020) and postmortem brain tissue (Ghisays et al. 2018, O’Donovan et al. 2018). Specifically, a gene ontology analysis revealed that DEK loss is associated with age-related dementias and Alzheimer’s disease. Furthermore, we demonstrated that DEK loss in a neuronal cell model resulted in phenotypes associated with dementias and Alzheimer’s disease, including apoptosis and hyperphosphorylated Tau. Notably, these findings mirror what is observed in peripheral tissues, with DEK loss leading to DNA damage (Smith et al. 2017), cellular senescence (Wise-Draper et al. 2005), and cell death (Kavanaugh et al. 2011, Smith et al. 2017). In 2018 we reported that DEK is expressed throughout the murine brain, including in areas important for learning and memory, such as the hippocampus, amygdala, and prefrontal cortex (Ghisays et al. 2018). Here, we expand upon our previous work by characterizing DEK expression in the healthy human brain across the lifespan.
DEK has previously been established as a proto-oncoprotein that is overexpressed in a majority of solid tumors (Sanchez-Carbayo et al. 2003, Grasemann et al. 2005, Wu et al. 2008, Khodadoust et al. 2009, Liu et al. 2012, Privette Vinnedge et al. 2015). Classically, DEK is known to promote cellular proliferation and DNA repair and inhibit apoptosis (Waldmann et al. 2002, Wise-Draper et al. 2006, Khodadoust et al. 2009, Kavanaugh et al. 2011, Privette Vinnedge et al. 2011, Broxmeyer et al. 2012, Koleva et al. 2012, Waidmann et al. 2014, Privette Vinnedge et al. 2015, Smith et al. 2017). DEK is multifaceted and can impact immune function, as well. For example, DEK secreted from immune cells can act as a chemoattractant (Mor-Vaknin et al. 2006), bind to DEK antibodies in autoimmune disease (Sierakowska et al. 1993, Dong et al. 2000, Mor-Vaknin et al. 2011), and facilitate the formation of neutrophil extracellular traps (NETs; (Mor-Vaknin et al. 2017). Thus, we sought to determine whether DEK is also closely associated with these biological pathways in the brain.
Based on previous studies in the periphery, we posit that DEK may be regulated by steroid hormones in the brain. DEK is an estrogen receptor alpha (ERα) target gene and is associated with positive hormone receptor status in breast cancer (Privette Vinnedge et al. 2012). The link between DEK and female-biased diseases, such as Alzheimer’s disease, allows us to postulate that there could be a sex difference in DEK protein expression across the human lifespan, which may be brain region dependent. Through our characterization of DEK expression in wild-type C57/Bl6 mice, we observed a sex difference in the number of DEK-positive cells in the CA1 region of the hippocampus; specifically, female mice had more DEK-positive cells in this area than male mice (Ghisays et al. 2018). The CA1 is highly implicated in spatial and contextual learning and memory (Pittenger et al. 2002, Ji and Maren 2008, Bartsch et al. 2011, Jeong et al. 2018). Using human postmortem brain tissue, we found a decrease in DEK expression in the anterior cingulate cortex in women with severe dementia, but not in men (O’Donovan et al. 2018). However, no sex difference in DEK protein expression levels was observed in individuals without cognitive impairment. It is possible, then, that there is a sex difference in neural DEK expression that is limited to a diseased state. Understanding how disease impacts DEK expression or how dysfunction in DEK may contribute to disease susceptibility is important, but there is a need to characterize DEK expression in the healthy human brain in order to ultimately understand DEK’s potential role in diseases of the central nervous system. In line with this goal, we sought to determine whether biological factors (i.e., sex and age) affect DEK expression in the brain and to present potential biological processes that DEK could be involved in through pathway enrichment analysis of DEK-correlated genes.
We hypothesize that DEK expression will be highest in the human brain during early development, and will decline with age, because normal-to-high DEK expression is associated with cell growth and proliferation. Since DEK is an ERα target gene, we also postulated that DEK expression will be higher in pre-menopausal females, which have the highest levels of circulating estrogen. We hypothesize that DEK expression will be associated with the expression of development-, immune system-, and steroid hormone-related genes, as it is in the periphery. Last, we posit that DEK will be associated with biological pathways relevant to neural development during the fetal stage and will be related to different processes, such as hormone signaling, in later stages of life. Given that our previous findings suggest a prominent role for DEK in learning and memory, we focused our attention on learning and memory relevant forebrain regions including the dorsolateral prefrontal cortex (dlPFC), medial prefrontal cortex (mPFC), ventrolateral prefrontal cortex (vlPFC), hippocampus, and amygdala. The cerebellum was included as a control hindbrain region that is associated with learning and memory. The inclusion of this hindbrain region allows us to determine if any changes across DEK across the lifespan is limited to distinct areas of the brain.
Methods
Human data
To confirm the expression of DEK in multiple cell types and regions in the brain, we accessed the Brain RNA-Seq and Human Protein Atlas online tools. The former dataset contains RNA-seq of cell types isolated from mouse and human brain (Zhang et al. 2016), https://www.brainrnaseq.org/). The Human Protein Atlas, specifically the brain atlas, contains mRNA expression data of human genes in various areas of the brain (Sjöstedt et al. (2020), https://www.proteinatlas.org/humanproteome/brain), drawing from the FANTOM5 Forrest et al. (2014), https://fantom.gsc.riken.jp/5/) and GTex (Genotype-Tissue Expression Project, https://gtexportal.org/home/, dbGaP accession number phs000424.vN.pN) datasets.
Human postmortem gene expression data from brain tissue was obtained from the BrainCloud (Colantuoni et al. 2011) and Human Brain Transcriptome (HBT; (Johnson et al. 2009, Kang et al. 2011) databases. In the BrainCloud dataset, RNA was isolated from the dorsolateral prefrontal cortex grey matter tissue in 269 individuals with no neuropathological or neuropsychiatric diagnosis. According to Colantuoni et al., total RNA was extracted and amplified, then hybridized to microarrays. Once normalized, log2 intensity ratios were adjusted using surrogate variable analysis to reduce the impact of noise on gene expression levels. For more details on sample collection and data preparation, see Colantuoni et al. (2011).
The HBT dataset provides mRNA expression data from a multitude of brain regions, including forebrain areas such as prefrontal cortices, amygdala, and hippocampus, as well as hindbrain (cerebellum). RNA was collected postmortem from 57 cognitively healthy donors. Authors of the HBT dataset used Partek Genomics Suite version 6.5 (Partek Incorporated, St. Louis, MO, USA) to perform RMA background correction, quantile normalization, mean probe set summarization, and log2 transformation (Kang et al. 2011). For gene set enrichment pathway analysis (GSEA), genes from HBT were filtered using the criteria of a log2-transformed signal intensity of greater than or equal to 6 in at least one sample/subject.
Statistical analysis
To analyze DEK expression across the lifespan, two-way ANCOVAs were performed using SPSS Statistics 18.0.0. Because the post-mortem interval (PMI; the time between death and sample collection) of the human samples significantly predicted DEK expression via linear regression, PMI was included as a covariate in this analysis. Age and Sex were main factors in the two-way ANCOVAs for each brain region. Sidak post-hoc tests were run after finding a significant interaction or main effect of age and/or sex. Results were plotted using the means and standard errors from the model adjusted for the covariate, PMI, in GraphPad Prism 8.
To determine what biological pathways are related to DEK in the brain, the Pearson correlation coefficient with DEK was calculated for each gene that met expression criteria. Pearson correlations were calculated within each age group separately as well as using all samples/subjects from all age groups together. GSEA pathway analysis was performed using the Pearson correlation coefficient as a rank for each gene. To further determine what pathways are uniquely associated with DEK in specific age groups, genes with a correlation coefficient greater than or equal to 0.6 were compared across age groups such that lists of genes expressed uniquely in a particular age group were generated using ToppGene to be used for the comparison of DEK-associated pathways in the hippocampus vs. cerebellum (Table 4). To consolidate gene ontology (GO) terms to be visually represented in figures and tables, GO IDs and Odds Ratios were retrieved using Enrichr (Chen et al. 2013, Kuleshov et al. 2016, Xie et al. 2021). The top 500 GO term IDs, ranked by p-value, were input into REVIGO to remove obsolete and redundant GO terms (Supek et al. 2011). Subjects were not separated by sex; we found no overall difference in DEK expression between males and females (p=0.074; data not shown). GSEA and ToppGene analyses yielded results for DEK-associated molecular functions, cellular components, biological processes, and human phenotypes, but only biological processes are discussed here to simplify and consolidate the results. The REVIGO interactive graphs were exported into Cytoscape (version 3.9.1) and used to create bubble maps in Figures 3–5.
Table 4.
A comparison of DEK-associated gene ontologies uniquely enriched in adulthood in the forebrain (hippocampus) vs. hindbrain (cerebellum). Enriched pathways from genes positively associated with DEK expression are labeled in red; negatively enriched pathways are in blue. These results were rendered with ToppGene to determine genes correlated to DEK only in distinct age groups and Enrichr to retrieve the biological processes associated with these genes. REVIGO was used to consolidate ontology terms.
| Association | Hippocampus | Cerebellum | ||||
|---|---|---|---|---|---|---|
| GO term: Biological Process | P-value | Odds Ratio | GO term: Biological Process | P-value | Odds Ratio | |
| Positive | mitochondrion organization | 0.0000138 | 4.073500561 | neuron projection maintenance | 0.00026539 | 18.45337159 |
| mitochondrial transport | 4.03639E-05 | 7.171081678 | dephosphorylation | 0.000321767 | 3.036031183 | |
| nucleotide metabolic process | 0.001742217 | 8.992283773 | protein ubiquitination | 0.000467178 | 1.952604146 | |
| ribosomal large subunit biogenesis | 0.001949861 | 5.038914027 | post-translational protein modification | 0.000525664 | 2.193394046 | |
| regulation of mitochondrial membrane potential | 0.002408125 | 5.940241228 | neuron cell-cell adhesion | 0.001944462 | 9.223816356 | |
| negative regulation of lipid transport | 0.005062474 | 28.37472767 | neuron projection organization | 0.001944462 | 9.223816356 | |
| cellular response to oxidative stress | 0.008456816 | 2.931567329 | cellular response to reactive oxygen species | 0.006362487 | 3.465958213 | |
| regulation of organelle assembly | 0.008679242 | 3.61578703 | neuron migration | 0.007771372 | 3.777959451 | |
| protein stabilization | 0.008765581 | 2.541361078 | transport across blood-brain barrier | 0.01052808 | 2.844710845 | |
| intermembrane lipid transfer | 0.010310514 | 17.02309368 | gamma-aminobutyric acid metabolic process | 0.011432611 | 18.40343348 | |
| membrane lipid biosynthetic process | 0.010712995 | 4.031363787 | sensory perception of sour taste | 0.011432611 | 18.40343348 | |
| ribosome disassembly | 0.013539029 | 14.18518519 | chemical synaptic transmission | 0.022996688 | 1.73965959 | |
| energy derivation by oxidation of organic compounds | 0.019206567 | 5.81093688 | positive regulation of neuron death | 0.023779258 | 3.293821839 | |
| myelin maintenance | 0.021106579 | 10.63779956 | synapse organization | 0.033296143 | 2.132281014 | |
| regulation of integrin-mediated signaling pathway | 0.04017501 | 7.090413943 | calcium-mediated signaling using intracellular calcium source | 0.035643088 | 4.603868195 | |
| steroid hormone mediated signaling pathway | 0.045662986 | 6.544662309 | associative learning | 0.037501141 | 7.885550787 | |
| modulation of chemical synaptic transmission | 0.037818355 | 2.194276571 | ||||
| MAPK cascade | 0.039065538 | 1.652256369 | ||||
| glutamate receptor signaling pathway | 0.039607783 | 3.350463023 | ||||
| Negative | antibacterial humoral response | 0.007909055 | 4.41613153 | neurotransmitter biosynthetic process | 0.007094776 | 6.503626943 |
| positive regulation of synaptic transmission, GABAergic | 0.01537952 | 13.72932862 | DNA replication checkpoint signaling | 0.014680245 | 5.001992826 | |
| negative regulation of cell projection organization | 0.049969331 | 3.055476753 | regulation of chromosome segregation | 0.01802418 | 4.644460893 | |
| cell cycle checkpoint signaling | 0.028997209 | 5.415300546 | ||||
| positive regulation of low-density lipoprotein receptor activity | 0.029983069 | 10.82471264 | ||||
| mitotic DNA integrity checkpoint signaling | 0.03610366 | 4.873511648 | ||||
| extracellular structure organization | 0.047244335 | 1.572933459 | ||||
Figure 3.
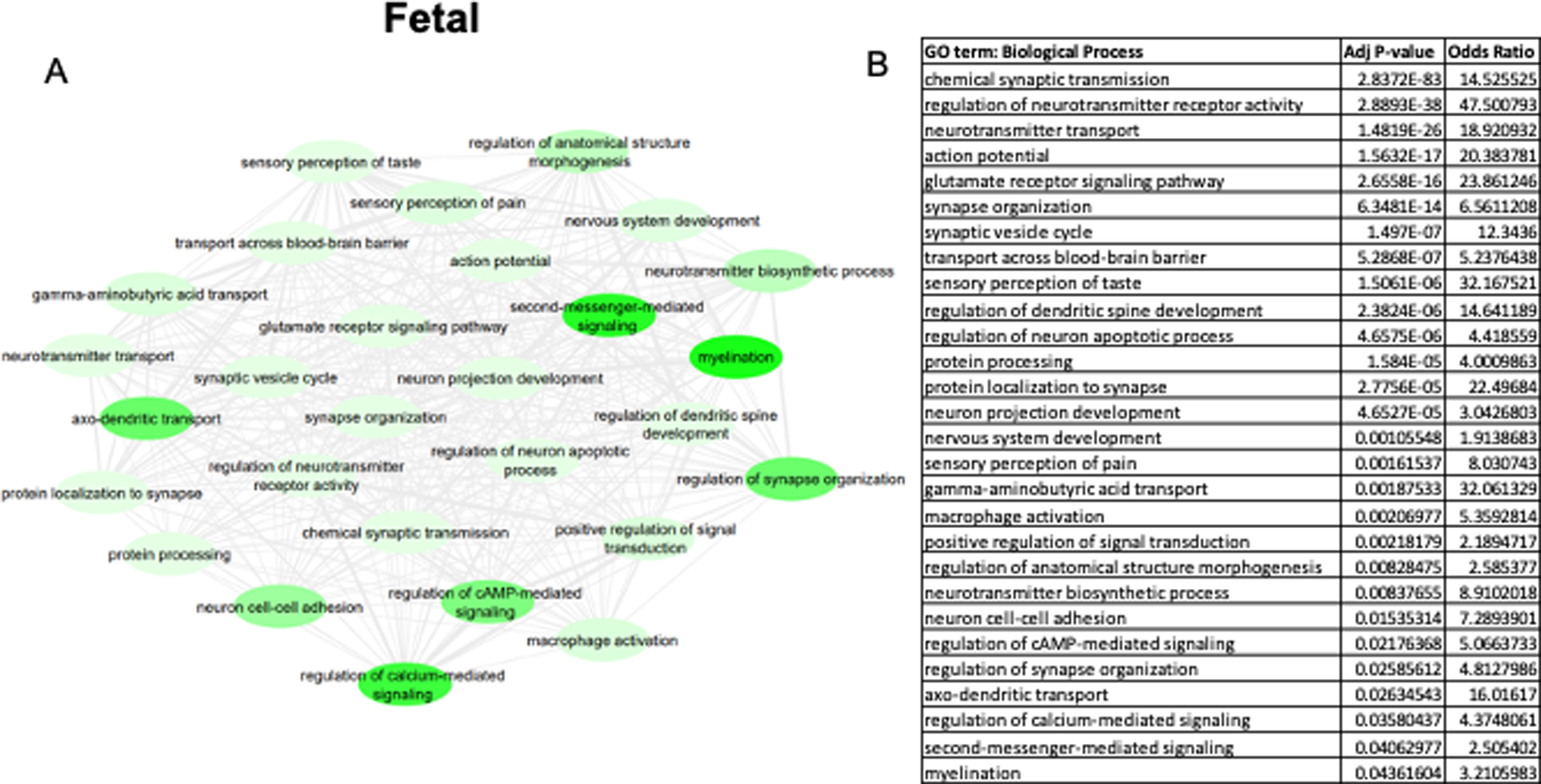
DEK-associated gene ontologies in all brain regions during fetal development (6–38 post-conceptional weeks). The most significantly enriched pathways from genes negatively correlated with DEK are represented in the map (A) and the corresponding table (B). A lighter green bubble color is indicative of a lower p-value. REVIGO was used to consolidate ontology terms and bubble map was generated in Cytoscape.
Figure 5.
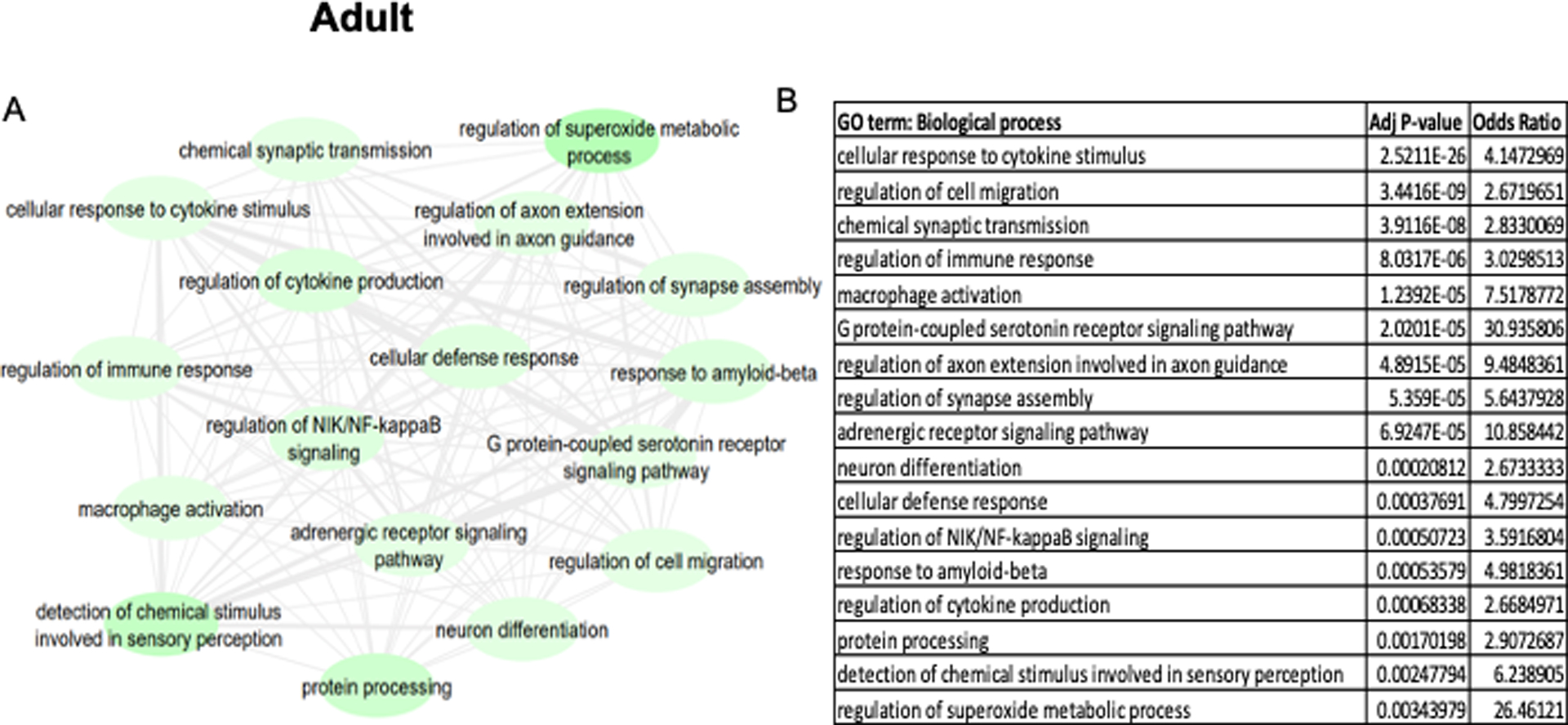
DEK-associated gene ontologies in all brain regions during adulthood (20+ years old). The most significantly enriched pathways from genes negatively correlated with DEK are represented in the map (A) and the corresponding table (B). A lighter green bubble color is indicative of a lower p-value. REVIGO was used to consolidate ontology terms and bubble map was generated in Cytoscape.
To identify age-associated pathways that are independent of DEK, a differential gene expression analysis was conducted for each age group versus the remaining age groups using Limma package v3.42.2 in R 3.6.1. Gene expression was compared at each age vs the other two ages (i.e., group 3 vs. Group 1&2). The gene lists were then filtered based on fold change >1.5 or <0.5 (log2FC >0.58 or <−0.58) and p values <0.05. The resulting filtered gene lists were then analyzed for enriched gene ontologies using the ToppFun function of ToppGene (https://toppgene.cchmc.org/enrichment.jsp) with FDR and p<0.05. Gene ontologies were validated, and odds ratios were retrieved using Enrichr. The top 100 ontologies from biological processes were then summarized and filtered for redundant gene ontology (GO) terms using REVIGO (http://revigo.irb.hr) (Tables 5–8).
Table 5.
Top 10 positively enriched age-associated gene ontologies in the hippocampus and five select gene ontologies of interest. Gene ontologies are listed in order by lowest p-value, to display the most highly enriched ones at the top of each list. These results were rendered with the Limma package in R to determine genes that are differentially expressed in distinct age groups. The ToppFun function in the ToppGene suite and Enrichr were used to identify the 100 top gene ontologies of biological processes of the differentially expressed genes. REVIGO was used to consolidate ontology terms.
| Hippocampus Upregulated Gene Ontologies | ||||||||
|---|---|---|---|---|---|---|---|---|
| Fetal | Pre-adult | Adult | ||||||
| GO term | p value | Odds Ratio | GO term | p value | Odds Ratio | GO term | p value | Odds Ratio |
| chromosome organization | 2.01E-58 | 0.41542118 | chemical synaptic transmission | 1.16E-43 | 0.674345326 | chemical synaptic transmission | 3.00E-32 | 0.592972918 |
| cell cycle process | 2.76E-42 | 0.203456613 | modulation of chemical synaptic transmission | 4.46E-29 | 0.470511559 | ion transmembrane transport | 2.74E-26 | 0.395176726 |
| cell division | 8.09E-37 | 0.747710754 | regulation of membrane potential | 2.28E-21 | 3.158672799 | modulation of chemical synaptic transmission | 1.41E-21 | 0.92270779 |
| negative regulation of cellular macromolecule biosynthetic process | 4.31E-29 | 0.588053036 | neuron projection development | 3.08E-21 | 0.602625883 | regulation of ion transport | 2.65E-20 | 0.348431597 |
| DNA repair | 1.88E-28 | 0.106522018 | ion transmembrane transport | 9.31E-20 | 0.742212259 | regulation of membrane potential | 3.82E-20 | 1.681851048 |
| chromosome segregation | 1.89E-24 | 1.518573922 | regulation of ion transport | 6.65E-19 | 0.654853042 | gliogenesis | 4.34E-17 | 4.147826087 |
| head development | 3.40E-19 | 0.885592399 | neurotransmitter transport | 3.08E-16 | 1.684012265 | locomotory behavior | 1.54E-16 | 0.447963365 |
| positive regulation of transcription, DNA-templated | 6.20E-18 | 0.633676804 | import into cell | 6.90E-16 | 2.165918268 | response to metal ion | 9.46E-14 | 0.191571119 |
| central nervous system development | 9.09E-18 | 1.772992814 | regulation of neuron projection development | 2.33E-14 | 1.815103671 | regulation of transporter activity | 2.55E-12 | 1.12099301 |
| protein localization to chromosome | 1.40E-15 | 0.321663405 | learning or memory | 6.46E-14 | 1.805451128 | cell junction organization | 5.24E-12 | 0.583918396 |
| signal transduction by p53 class mediator | 2.36E-13 | 1.900458142 | regulation of nervous system process | 2.26E-13 | 1.837548387 | learning or memory | 5.70E-11 | 2.696301668 |
| regulation of chromosome segregation | 7.01E-12 | 1.518573922 | regulation of neurogenesis | 1.72E-11 | 1.284852951 | blood circulation | 2.15E-10 | 0.395176726 |
| response to ionizing radiation | 1.35E-12 | 0.225675492 | central nervous system myelination | 4.96E-11 | 0.536556043 | regulation of neurotransmitter transport | 2.72E-10 | 0.896649795 |
| peptidyl-lysine modification | 8.68E-12 | 0.308344312 | brain development | 4.66E-10 | 2.419898064 | regulation of neuron projection development | 3.89E-10 | 2.198580166 |
| histone modification | 2.01E-11 | 0.589005575 | action potential | 6.31E-10 | 2.198512586 | oligodendrocyte differentiation | 8.01E-10 | 0.707729193 |
Table 8.
Top 10 negatively enriched age-associated gene ontologies in the cerebellum and five select gene ontologies of interest. Gene ontologies are listed in order by lowest p-value, to display the most highly enriched ones at the top of each list. These results were rendered with the Limma package in R to determine genes that are differentially expressed in distinct age groups. The ToppFun function in the ToppGene suite and Enrichr were used to identify the 100 top gene ontologies of biological processes of the differentially expressed genes. REVIGO was used to consolidate ontology terms.
| Cerebellum Downregulated Gene Ontologies | ||||||||
|---|---|---|---|---|---|---|---|---|
| Fetal | Pre-adult | Adult | ||||||
| GO term | p value | Odds Ratio | GO term | p value | Odds Ratio | GO term | p value | Odds Ratio |
| chemical synaptic transmission | 4.06E-33 | 2.653812317 | neuron projection development | 1.76E-21 | 1.75425923 | cell morphogenesis | 6.28E-37 | 1.953977195 |
| modulation of chemical synaptic transmission | 2.15E-21 | 3.516443154 | central nervous system development | 7.14E-20 | 2.288827043 | regulation of nervous system development | 2.47E-26 | 2.556508604 |
| neurotransmitter transport | 1.03E-20 | 3.136246236 | cell division | 1.97E-13 | 2.895959265 | cell adhesion | 2.32E-24 | 2.236123452 |
| regulation of neurotransmitter levels | 4.35E-19 | 3.564158055 | chemotaxis | 8.62E-11 | 2.729971989 | regulation of plasma membrane bounded cell projection organization | 6.17E-24 | 0.698254915 |
| neuron projection development | 3.92E-17 | 1.647426744 | mitotic cell cycle process | 4.56E-11 | 1.182282565 | cell-cell adhesion via plasma-membrane adhesion molecules | 1.49E-21 | 4.663199388 |
| regulation of ion transport | 1.98E-16 | 4.057524087 | cell-cell adhesion via plasma-membrane adhesion molecules | 2.64E-10 | 4.355540563 | synapse organization | 3.86E-20 | 4.056598985 |
| secretion | 3.06E-13 | 0.85369715 | organelle fission | 1.37E-09 | 2.559086134 | regulation of synapse structure or activity | 8.52E-20 | 4.056598985 |
| response to metal ion | 5.44E-13 | 1.824531295 | synaptic signaling | 1.06E-08 | 1.201627821 | cell junction organization | 1.90E-19 | 3.427677925 |
| regulation of system process | 5.64E-13 | 2.775877724 | animal organ morphogenesis | 1.76E-08 | 0.949685535 | chemotaxis | 1.94E-19 | 2.701182168 |
| regulation of transporter activity | 5.98E-13 | 2.220581114 | growth | 7.39E-08 | 1.682682048 | growth | 9.58E-18 | 1.620351077 |
| cellular homeostasis | 4.63E-12 | 1.811767044 | protein localization to chromosome | 9.36E-08 | 7.4526356 | cell division | 9.59E-17 | 2.228699288 |
| protein localization to synapse | 1.58E-11 | 5.560642814 | microtubule-based process | 9.49E-08 | 2.408304498 | mitotic cell cycle process | 3.36E-12 | 1.367827087 |
| amino acid transport | 8.07E-11 | 2.119554205 | chromosome segregation | 1.00E-07 | 5.12237395 | stem cell development | 3.37E-12 | 4.050725669 |
| myelination | 1.47E-10 | 6.721834862 | small GTPase mediated signal transduction | 1.05E-07 | 3.373686496 | Ras protein signal transduction | 1.37E-11 | 2.417567633 |
| gliogenesis | 6.15E-10 | 1.448537549 | hindbrain development | 2.97E-07 | 7.4526356 | regulation of locomotion | 1.39E-11 | 0.62833002 |
Results
DEK expression in neural cell types
DEK is expressed in most major cell types in the brain, including astrocytes, microglia, oligodendrocytes, endothelial cells, and neurons (Fig 1A). Fetal astrocytes, oligodendrocytes, and microglia had higher levels of DEK expression compared to other cell types. In addition, it was confirmed that DEK is expressed ubiquitously throughout the human brain (Fig 1B).
Figure 1.
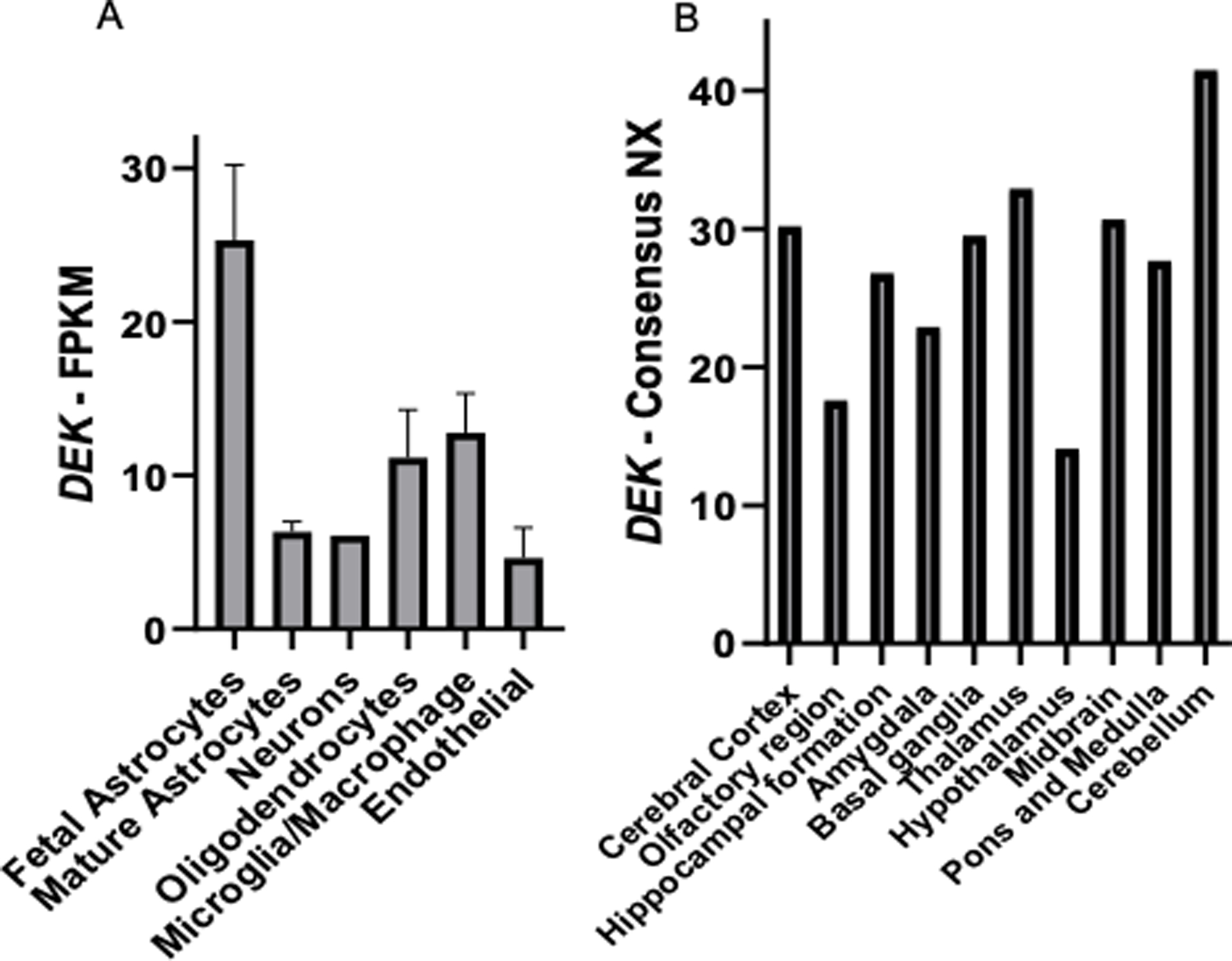
DEK is expressed in multiple cell types and regions in the human brain. A) Data from the Brain RNA-Seq database demonstrates that DEK is expressed in multiple cell types in the human brain. FPKM = Fragments Per Kilobase of transcript per Million mapped reads. Original data and graph can be found at https://www.brainrnaseq.org/. B) Normalized expression (NX) of DEK RNA expression across human brain regions in adults. NX values were determined by combining data from the GTEx Human brain RNA-Seq dataset and FANTOM5 Human brain CAGE dataset. Original data and graphs can be found at https://www.proteinatlas.org/ENSG00000124795-DEK/brain.
DEK expression across the lifespan
Two-way ANCOVA revealed an interaction between the effects of age and sex on DEK expression in the dorsolateral prefrontal cortex ( dlPFC )(HBT; F (2, 45) = 5.104, p = 0.01), medial prefrontal cortex (mPFC) (F (2, 45) = 7.259, p = 0.002), and vlPFC (F (2, 45) = 3.915, p = 0.027). A main effect of age on DEK expression was observed in all brain regions queried, including the dlPFC (BrainCloud, Fig. 2A, F (5, 256) = 34.141, p < 0.001; HBT, Fig 2B, F (2, 45) = 12.882, p < 0.001), the mPFC (Fig. 2C, F (2, 46) = 9.634, p < 0.001), the vlPFC (Fig. 2D, F (2, 44) = 10.382, p < 0.001), the hippocampus (Fig. 2E, F (2, 44) = 6.499, p = 0.003), the amygdala (Fig. 2F, F (2, 41) = 24.916, p < 0.001), and cerebellum (Fig. 2G, F (2, 40) = 3.469, p = 0.041). Specifically, in all forebrain regions, DEK expression was highest during fetal development stages and sharply declined during early childhood through adulthood (dlPFC; BC, p < 0.001; HBT, p < 0.001; mPFC, p ≤ 0.001; vlPFC, p ≤ 0.001; Hippocampus, Fetal v. Pre-adult p = 0.014, Fetal v. Adult p = 0.003; Amygdala, p < 0.001). In the cerebellum, DEK expression was lower in adulthood (Pre-adult v. Adult p = 0.037). A main effect of sex was found in the dlPFC with data from BrainCloud (F (1, 256) = 7.569, p = 0.006). In this region, males had significantly higher DEK expression than females throughout life (p = 0.006). However, DEK expression in the prefrontal cortices, using data from HBT, was highest in the brains of male fetuses but this was reversed in adulthood, such that DEK levels were higher in adult females compared to males (dlPFC, Fig. 2B, F (2, 45) = 5.104, p = 0.01; mPFC, Fig. 2C, F (2, 46) = 7.259, p = 0.002; vlPFC, Fig. 2D, F (2, 44) = 3.915, p = 0.027).
Figure 2.
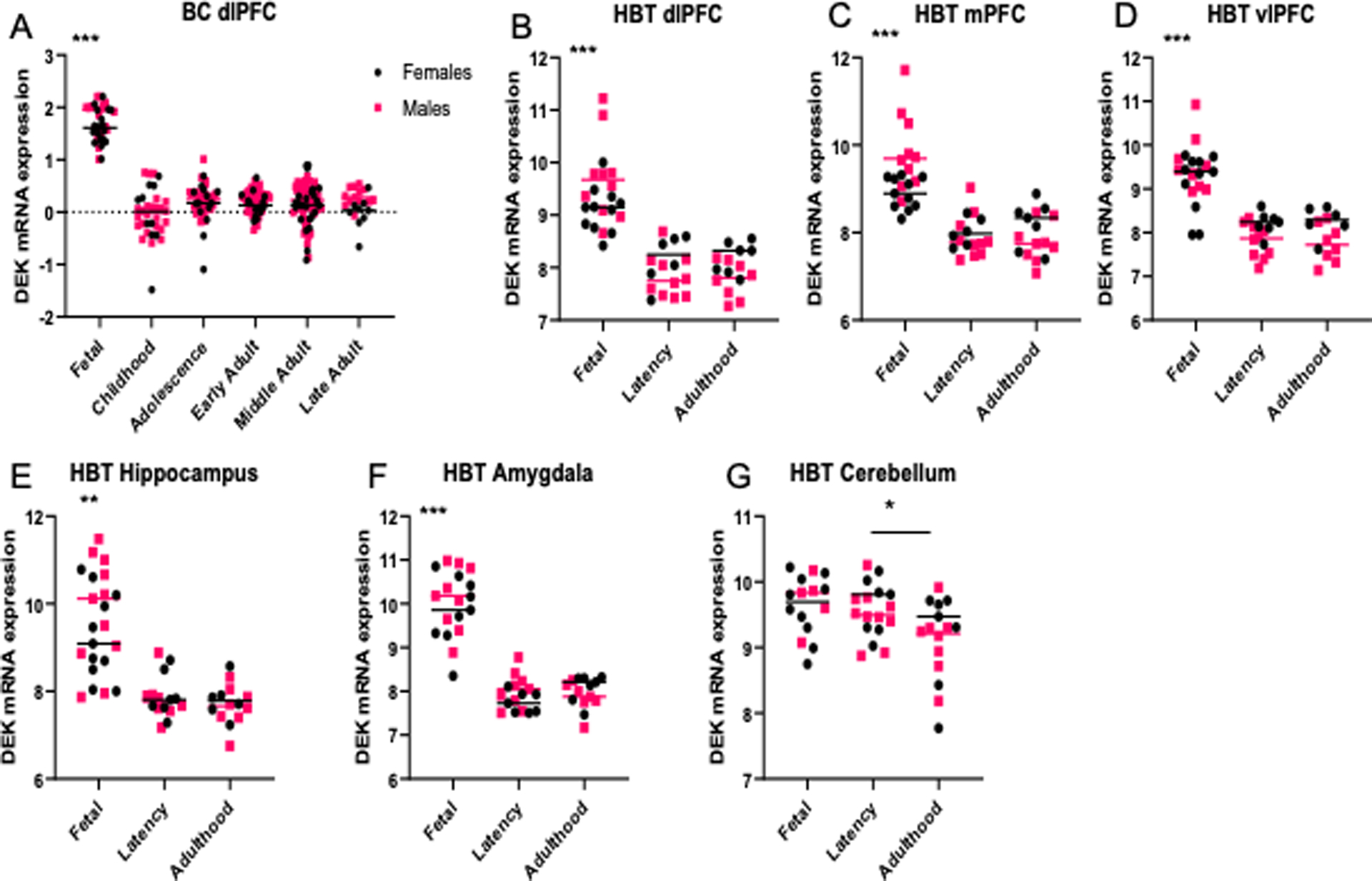
DEK expression across the lifespan. A) DEK expression is significantly greater during fetal development (6–38 weeks post-conception; n=38) than in childhood (0–10 years old; n=34), adolescence (10–18 years old; n=45), early adulthood (18–39 years old; n=57), middle adulthood (40–59 years old; n=73), and late adulthood (60+ years old; n=22); ***p<0.001. Data from BrainCloud in dorsolateral prefrontal cortex (dlPFC). B-D) DEK expression is significantly greater during fetal development (6–38 weeks post-conception; n=15–22) than in pre-adults (0–20 years old; n=14–16) or in adulthood (20+ years old; n=14–15) in the dlPFC, medial prefrontal cortex (mPFC), and ventrolateral prefrontal cortex (vlPFC); ***p<0.001. In addition, there is an interaction effect between age and sex on DEK expression in these regions (dlPFC, p=0.01; mPFC, p=0.002; vlPFC, p=0.027). Data from Human Brain Transcriptome (HBT). E-F) DEK expression is significantly greater during fetal development in the hippocampus (**p<0.01) and amygdala (***p<0.001). Data from HBT. G) A significant main effect of age on DEK expression was found in the cerebellum (p=0.041). Post-hoc analysis revealed that DEK expression in adulthood is significantly decreased compared to pre-adults (*p<0.05). Data from HBT.
DEK-associated pathway analyses
Across all age groups and brain regions, biological pathways enriched from genes positively correlated with DEK expression (Table 1, in red) include DNA replication and repair, RNA processing, cell cycle, gene expression regulation, and ubiquitin activity. Other positively enriched pathways include ribosome biogenesis and metabolism, as well as hormone secretion (not displayed in the table; p=0.0034; OR=4.82). Biological pathways enriched from genes negatively correlated with DEK expression (Table 1, in blue) include synaptic signaling and neurotransmitter transport, macrophage activation, glutamate receptor signaling, regulation of neuronal death, memory, and steroid hormone signaling (not displayed in the table; p=0013; OR=7.56).
Table 1.
DEK-associated gene ontologies in all brain regions and age groups. Enriched pathways from genes positively negatively associated with DEK expression are labeled in red; negatively enriched pathways are in blue. Pathways with the lowest p-value begin at the top of the list for each color/association. Normalized enrichment score (NES) reflects the degree to which a gene set is overrepresented at the top or bottom of a ranked list of genes, accounting for the difference in gene set sizes. Odds ratio represents the relative abundance of DEK-associated genes in each pathway vs. genes not in the pathway.
| Association | GO term: Biological Process | Adj. P-value | NES | Odds Ratio |
|---|---|---|---|---|
| Positive | ribosome biogenesis | 3.2218E-68 | 3.810316 | 18.009808 |
| DNA replication | 1.6042E-64 | 3.818609 | 37.949158 | |
| cellular response to DNA damage stimulus | 2.627E-63 | 3.292782 | 9.4652765 | |
| regulation of mRNA splicing, via spliceosome | 1.1487E-32 | 2.928372 | 17.84078 | |
| regulation of gene silencing by RNA | 5.7028E-28 | 3.017145 | 28.026503 | |
| DNA damage response, signal transduction by p53 class mediator | 3.3227E-24 | 2.869532 | 15.624064 | |
| chromosome organization | 2.1607E-19 | 3.587029 | 8.9705635 | |
| DNA synthesis involved in DNA repair | 1E-17 | 2.820032 | 20.367827 | |
| regulation of ATP metabolic process | 1E-17 | 1.808558 | 14.691581 | |
| protein localization to chromosome | 7.7E-16 | 2.816571 | 23.621524 | |
| nucleocytoplasmic transport | 1.1227E-11 | 2.284504 | 13.203869 | |
| regulation of chromosome segregation | 2.7247E-11 | 3.590709 | 37.799067 | |
| histone modification | 1.3067E-09 | 2.556048 | 5.001056 | |
| regulation of cell cycle | 1.2124E-08 | 3.296365 | 2.928732 | |
| regulation of ubiquitin protein ligase activity | 1.3124E-07 | 2.169337 | 15.962811 | |
| Negative | chemical synaptic transmission | 7.54E-72 | −2.22474 | 14.455558 |
| neurotransmitter transport | 4.85E-27 | −2.85102 | 21.703556 | |
| action potential | 1.0314E-17 | −2.40512 | 22.98916 | |
| regulation of neuronal synaptic plasticity | 1.9162E-14 | −2.70312 | 30.954329 | |
| glutamate receptor signaling pathway | 5.3858E-10 | −2.32602 | 14.856009 | |
| response to amyloid-beta | 7.9402E-10 | −1.31689 | 12.461061 | |
| long-term memory | 1.9565E-09 | −2.18819 | 29.845313 | |
| synapse organization | 1.6659E-07 | −1.87446 | 4.889468 | |
| macrophage activation | 5.3244E-07 | −2.18501 | 10.856086 | |
| regulation of neuron death | 3.3848E-06 | −1.63992 | 5.3672935 | |
| sensory perception of taste | 7.664E-05 | −2.34644 | 21.585311 | |
| regulation of dendrite extension | 0.00091983 | −1.78438 | 10.789266 | |
| receptor metabolic process | 0.00200576 | −1.89467 | 4.5074253 | |
| retrograde axonal transport | 0.00413632 | −2.1452 | 9.7978658 | |
| gamma-aminobutyric acid metabolic process | 0.00622536 | −2.03437 | 32.275338 | |
| regulation of MAPK cascade | 0.00872801 | −1.42452 | 2.4750818 | |
| regulation of calcium-mediated signaling | 0.00885153 | −1.93225 | 5.8819723 |
As seen in Table 2, the top genes harboring a positive association with DEK expression include USP1, SRBD1, RMI1, RBL1, and NUP107, which have functions in ubiquitin processing, RNA binding, DNA repair, cell cycle regulation, and nuclear transport, respectively (Stelzer et al. 2016). The top genes with a negative correlation to DEK expression include GRIN1, IGSF8, PPP2R2C, IQSEC2, and MROH1, which function in synaptic plasticity via NMDA receptors, response to viral infection, cell growth and division, synaptic organization, and binding, respectively (Stelzer et al. 2016).
Table 2.
Top 100 genes correlated with DEK expression in all brain regions and age groups. Genes with the strongest correlation to DEK expression start at the top of each list (negatively or positively correlated with DEK).
| Positive | Negative | ||
|---|---|---|---|
| Gene | Correlation Coefficient | Gene | Correlation Coefficient |
| USP1 | 0.94 | GRIN1 | −0.85 |
| SRBD1 | 0.92 | KCNMA1 | −0.84 |
| RMI1 | 0.92 | IGSF8 | −0.83 |
| LIN9 | 0.92 | PTPRN | −0.82 |
| RBL1 | 0.91 | PDE4A | −0.82 |
| NUP107 | 0.91 | PPP2R2C | −0.82 |
| TMPO | 0.91 | RAPGEFL1 | −0.82 |
| KlF20B | 0.91 | IQSEC2 | −0.82 |
| NUP54 | 0.91 | ARHGEF4 | −0.82 |
| NUP205 | 0.91 | BIN1 | −0.82 |
| RRM1 | 0.91 | MROH1 | −0.82 |
| PRPF40A | 0.91 | CEP170B | −0.81 |
| MCM6 | 0.9 | NDRG4 | −0.81 |
| POLE2 | 0.9 | MINK1 | −0.81 |
| ERI1 | 0.9 | ABLIM2 | −0.81 |
| SMC4 | 0.9 | CASKIN1 | −0.81 |
| MSH6 | 0.9 | TUBG2 | −0.8 |
| HAUS6 | 0.9 | PITPNM2 | −0.8 |
| SRSF10 | 0.9 | TMEM59L | −0.8 |
| WDR76 | 0.89 | DNM1 | −0.8 |
| POLA1 | 0.89 | CAMTA2 | −0.8 |
| UGDH | 0.89 | UNC13A | −0.8 |
| STAG1 | 0.89 | FBXO44 | −0.8 |
| CPSF3 | 0.89 | ARF3 | −0.8 |
| NUP43 | 0.89 | LYPD5 | −0.8 |
| STIL | 0.89 | TPPP | −0.8 |
| ACTL6A | 0.89 | CPNE6 | −0.79 |
| DNA2 | 0.89 | SYP | −0.79 |
| C18orf54 | 0.89 | RAB11FlP4 | −0.79 |
| SUZ12 | 0.89 | KIAA0513 | −0.79 |
| KNTC1 | 0.89 | MAPK8IP3 | −0.79 |
| STAG2 | 0.88 | THY1 | −0.79 |
| GABPB1 | 0.88 | JPH3 | −0.79 |
| DHX15 | 0.88 | FBXL18 | −0.79 |
| CASP8AP2 | 0.88 | DMTN | −0.79 |
| PCNA | 0.88 | HSPA12A | −0.79 |
| CNOT9 | 0.88 | TOM1L2 | −0.79 |
| SPATA5 | 0.88 | CYP46A1 | −0.78 |
| CDKAL1 | 0.88 | CAMK2B | −0.78 |
| CHD1 | 0.88 | TNK2 | −0.78 |
| PHF6 | 0.88 | ABHD12 | −0.78 |
| SASS6 | 0.88 | SNPH | −0.78 |
| RFC4 | 0.88 | ZBTB47 | −0.78 |
| ATAD5 | 0.88 | AGAP2 | −0.78 |
| ZMYM1 | 0.88 | RAB40B | −0.78 |
| WDHD1 | 0.88 | AZIN2 | −0.78 |
| SPDL1 | 0.87 | PLEKHA6 | −0.78 |
| CNTLN | 0.87 | TTBK1 | −0.78 |
| NDC1 | 0.87 | GPR61 | −0.78 |
| SLC25A24 | 0.87 | CACNA1D | −0.78 |
When DEK-associated biological pathways are separated by age group, distinctions can be made about DEK’s possible functions during different age periods. To do this, we explored enriched biological processes associated with DEK during different age groups. Table 3 lists the top 20 biological processes positively enriched from DEK-correlated genes in each age group, while Figures 3–4 display the negatively enriched pathways. The pathways from genes negatively correlated with DEK tend to offer more novel functions for DEK in the central nervous system, as seen in Table 1, so more focus was placed on the negatively enriched pathways. Positively associated pathways remain fairly stable across age groups; DEK is associated with functions such as mRNA processing, DNA repair, ribosome biogenesis, and gene expression throughout life (Table 3). Differently, in adulthood, DEK appears to be involved in additional pathways related to translation (Table 3). Pathways enriched from genes negatively correlated with DEK expression only during fetal development (6–38 post-conceptional weeks) include neuron projection development, neurotransmitter biosynthesis, and myelination (Figure 3). After birth, specifically in pre-adulthood (0–20 years old), DEK is negatively associated with glial cell proliferation, hippocampal neuron apoptosis, and reproductive system processes (Figure 4). One pathway negatively associated with DEK only in adulthood (20+ years old), seen in Figure 5, is superoxide metabolism, a function important in aging (Sasaki et al. 2008). Interestingly, there are several biological processes associated with DEK after birth that are not highly enriched in fetal development. For example, immune system and inflammatory response, serotonin and adrenergic signaling, response to amyloid-beta, neuron differentiation, and cell migration are all associated with DEK in pre-adulthood and adulthood but not during fetal development.
Table 3.
Top 20 positively enriched DEK-associated gene ontologies within each age groups in all brain regions. Pathways are listed in order by lowest adjusted p-value, then by highest odds ratio to display the most highly enriched pathways at the top of each list. Enrichr was used to retrieve the gene ontologies (GO terms) enriched from positively correlated genes and the associated statistics.
| Fetal Development | Pre-adulthood | Adulthood | ||||||
|---|---|---|---|---|---|---|---|---|
| GO term: Biological Process | Adj P-value | Odds Ratio | GO term: Biological Process | Adj P-value | Odds Ratio | GO term: Biological Process | Adj P-value | Odds Ratio |
| mRNA processing | 2.567E-146 | 28.258433 | mRNA processing | 3.281E-106 | 18.589207 | mRNA processing | 1.723E-131 | 23.384215 |
| mRNA splicing, via spliceosome | 1.318E-141 | 30.671828 | mRNA splicing, via spliceosome | 2.0004E-95 | 18.05645 | mRNA splicing, via spliceosome | 1.783E-128 | 25.610369 |
| RNA splicing | 3.664E-131 | 30.97823 | RNA splicing | 5.7125E-90 | 18.547828 | RNA splicing | 3.193E-117 | 25.269163 |
| DNA repair | 8.2448E-90 | 15.078876 | gene expression | 1.7439E-87 | 12.601939 | gene expression | 1.0481E-94 | 12.852176 |
| DNA metabolic process | 2.2702E-87 | 15.765298 | RNA processing | 3.2451E-68 | 19.624996 | ribosome biogenesis | 7.8587E-72 | 18.327958 |
| double-strand break repair | 2.4508E-70 | 22.47355 | nuclear-transcribed mRNA catabolic process | 6.697E-62 | 18.379107 | ncRNA processing | 9.8052E-68 | 16.146435 |
| mitotic spindle organization | 1.3356E-63 | 20.769036 | DNA metabolic process | 1.6458E-53 | 9.9259487 | rRNA processing | 7.2205E-66 | 18.635317 |
| RNA processing | 8.6536E-63 | 17.410882 | ncRNA processing | 1.1198E-52 | 12.978199 | RNA processing | 3.8599E-65 | 17.60567 |
| cellular response to DNA damage stimulus | 1.1702E-62 | 9.2185648 | DNA repair | 7.6505E-51 | 8.9556035 | cellular macromolecule biosynthesis | 1.1534E-62 | 9.6612988 |
| DNA replication | 2.3212E-61 | 34.476404 | ribosome biogenesis | 2.3765E-49 | 12.690657 | translation | 1.8412E-58 | 12.808307 |
| gene expression | 3.1434E-54 | 8.0800181 | cellular macromolecule biosynthesis | 3.1708E-49 | 8.3734627 | translational elongation | 2.1686E-57 | 32.216584 |
| ncRNA processing | 2.1412E-53 | 12.882187 | regulation of translation | 3.3982E-47 | 13.035326 | mitochondrial translation | 1.8936E-55 | 29.770635 |
| microtubule cytoskeleton organization | 4.5666E-53 | 21.193448 | mRNA export from nucleus | 6.1379E-46 | 22.447106 | rRNA metabolic process | 6.5297E-55 | 16.056575 |
| mRNA export from nucleus | 4.59E-53 | 27.328143 | rRNA processing | 4.0714E-45 | 12.795513 | translational termination | 1.6863E-52 | 31.588519 |
| ribosome biogenesis | 2.9806E-51 | 12.90128 | RNA metabolic process | 5.5813E-44 | 16.307516 | mitochondrial translational elongation | 1.4217E-51 | 35.160785 |
| RNA export from nucleus | 1.065E-50 | 25.807262 | mRNA-containing ribonucleoprotein complex export from nucleus | 3.3218E-42 | 22.15944 | DNA metabolic process | 7.9233E-51 | 8.9019139 |
| mRNA transport | 3.8339E-50 | 26.096154 | mRNA transport | 8.5464E-42 | 20.498498 | mitochondrial translational termination | 4.24E-50 | 33.209937 |
| mRNA-containing ribonucleoprotein complex export from nucleus | 1.9574E-49 | 27.512852 | cellular response to DNA damage stimulus | 1.3949E-41 | 6.8062948 | transcription by RNA polymerase II | 9.6503E-49 | 7.6908903 |
| rRNA processing | 2.1589E-49 | 13.74466 | RNA splicing | 2.6719E-41 | 21.766331 | transcription, DNA-templated | 1.4255E-48 | 10.332887 |
Figure 4.
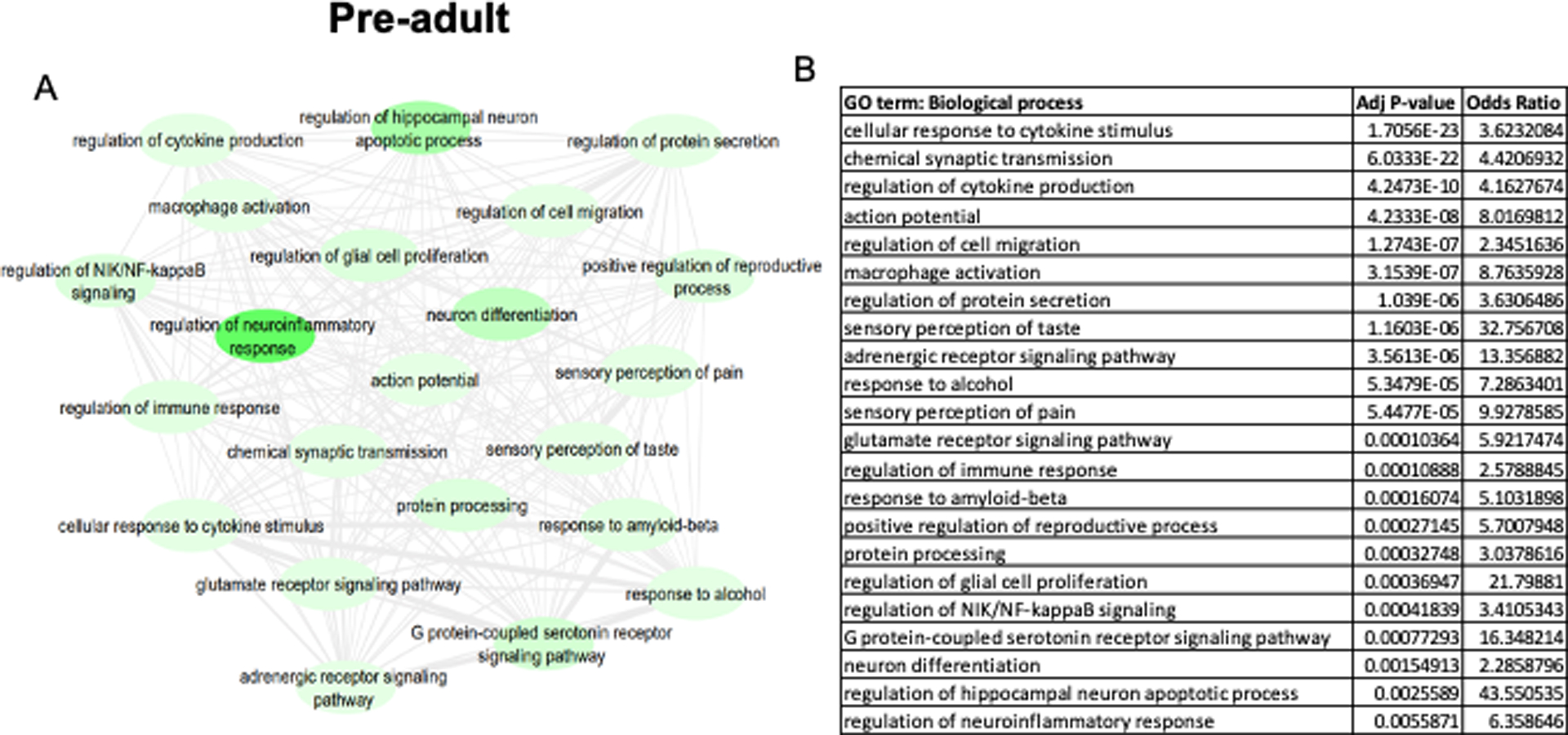
DEK-associated gene ontologies in all brain regions during pre-adulthood (0–20 years old). The most significantly enriched pathways from genes negatively correlated with DEK are represented in the map (A) and the corresponding table (B). A lighter green bubble color is indicative of a lower p-value. REVIGO was used to consolidate ontology terms and bubble map was generated in Cytoscape.
Because of the different expression pattern of DEK in the forebrain vs. the hindbrain (cerebellum), we investigated the difference in potential biological functions of DEK in these regions across life. We also chose to differentiate between pathways related to DEK in the cerebellum vs. hippocampus because these regions are important for learning and memory. In addition, we know that DEK is highly expressed in the hippocampus in the murine brain (Ghisays et al. 2018). We evaluated the difference in DEK-associated pathways in these regions only during adulthood to begin to understand potential variations in DEK function during this age group in which we know DEK expression is important (O’Donovan et al. 2018). In the hippocampus, many pathways positively related to DEK expression represent mitochondrial processes, and signaling pathways such as steroid hormone signaling (Table 4, left column). In contrast, DEK expression in the cerebellum is associated with synaptic transmission, protein modifications, neuron death, neurotransmitter metabolism, and sensory perception (Table 4, right column). In both the hippocampus and cerebellum, DEK expression in adulthood could be important for the cellular response to oxidative stress and maintaining neuronal projections.
Age-associated pathway analyses
In order to determine if DEK associated expression in genes and gene ontologies (categories) are primarily related to typical changes that occur in the aging brain, we compared the results of the correlation analysis vs pathway analysis using differential gene expression analysis. When we queried age-related pathways in the hippocampus, the fetal stage of development is primarily characterized by an upregulation of genes that are associated with DNA repair, chromosome organization and segregation, and histone modification which are not seen in the preadult and adult stages (Table 5). The pre-adult and adult stages had several overlapping biological pathways that were upregulated including those associated with neuron projection development, neurotransmitter function, chemical synaptic transmission, mitotic cell cycle process, regulation of membrane potential/action potential, and learning and memory. (Table 5). In contrast, many of the gene ontologies that were upregulated in the fetal stage of development were downregulated in the preadult and adult stages including chromosome organization and segregation, DNA repair and replication, and brain and head development (Table 6). In this vein, the fetal stage is associated with downregulation of gene ontologies that are typically highly expressed in the pre-adult and adult stage including chemical synaptic transmission, regulation of membrane potential, regulation of neuron projection development, and behavior (Table 6).
Table 6.
Top 10 negatively enriched age-associated gene ontologies in the hippocampus and five select gene ontologies of interest. Gene ontologies are listed in order by lowest p-value, to display the most highly enriched ones at the top of each list. These results were rendered with the Limma package in R to determine genes that are differentially expressed in distinct age groups. The ToppFun function in the ToppGene suite and Enrichr were used to identify the 100 top gene ontologies of biological processes of the differentially expressed genes. REVIGO was used to consolidate ontology terms.
| Hippocampus Downregulated Gene Ontologies | ||||||||
|---|---|---|---|---|---|---|---|---|
| Fetal | Pre-adult | Adult | ||||||
| GO term | p value | Odds Ratio | GO term | p value | Odds Ratio | GO term | p value | Odds Ratio |
| cation transport | 4.12E-38 | 2.612946958 | mitotic cell cycle | 4.43E-46 | 2.349890359 | chromosome organization | 2.65E-44 | 2.230512366 |
| modulation of chemical synaptic transmission | 5.16E-37 | 3.901537552 | chromosome organization | 7.46E-46 | 4.225031996 | mitotic cell cycle process | 9.41E-38 | 1.519985359 |
| chemical synaptic transmission | 4.06E-33 | 3.243656908 | cell division | 2.68E-27 | 2.17324263 | organelle fission | 2.39E-35 | 0.840448625 |
| regulation of membrane potential | 5.87E-33 | 3.692982062 | chromosome segregation | 2.93E-23 | 5.065349144 | cell division | 1.53E-27 | 2.792411981 |
| regulation of ion transport | 3.10E-32 | 3.883237604 | regulation of cell cycle | 1.19E-19 | 2.700191142 | chromatin assembly | 3.27E-23 | 3.808146096 |
| behavior | 2.15E-30 | 1.180983776 | negative regulation of transcription, DNA-templated | 6.36E-15 | 1.766189266 | chromosome segregation | 8.64E-20 | 5.188542422 |
| neurotransmitter transport | 8.91E-28 | 3.77004812 | cell cycle checkpoint signaling | 1.49E-14 | 12.68469657 | Head (and brain) development | 3.78E-19 | 2.333058003 |
| vesicle-mediated transport in synapse | 2.18E-23 | 2.467956251 | chromatin remodeling | 2.72E-14 | 2.137986877 | negative regulation of transcription, DNA-templated | 3.47E-18 | 1.612272621 |
| regulation of system process | 1.01E-21 | 2.657940663 | protein localization to chromosome, centromeric region | 3.78E-14 | 13.87229437 | DNA replication | 6.44E-17 | 5.0219347 |
| cell junction organization | 2.59E-21 | 1.145134492 | cellular response to DNA damage stimulus (DNA repair) | 4.01E-13 | 1.974011064 | regulation of cell cycle | 1.05E-16 | 1.528470364 |
| import into cell | 8.03E-19 | 1.477486233 | microtubule-based process | 1.60E-10 | 1.486377709 | neuron projection development | 3.97E-15 | 1.201478451 |
| regulation of transporter activity | 1.75E-18 | 1.933432109 | signal transduction by p53 class mediator | 1.38E-09 | 1.630475382 | DNA repair | 1.23E-14 | 2.117709487 |
| organelle fusion | 7.22E-18 | 0.505803485 | megakaryocyte differentiation | 4.17E-09 | 2.296858201 | axon guidance | 6.66E-13 | 2.456917444 |
| response to metal ion | 5.44E-13 | 1.74662065 | brain development | 4.24E-09 | 2.419898064 | neural precursor cell proliferation (regulation of neurogenesis) | 4.70E-11 | 2.837070938 |
| regulation of neuron projection development | 3.77E-10 | 2.586608702 | growth | 7.05E-09 | 1.404720023 | microtubule-based process | 7.36E-10 | 1.223290106 |
The cerebellum is characterized by an upregulation of pathways that are associated with hindbrain development, developmental growth, chromosome organization, microtubule-based process that are unique to the fetal stage (Table 7). The pre-adult and adult stages share several upregulated gene ontologies including chemical synaptic transmission, gliogenesis, and myelination. (Table 7). Similar to the hippocampus, many of the biological processes (i.e., chemical synaptic transmission, neurotransmitter transport, regulation of ion transport, myelination, and gliogenesis) that are highly expressed during the pre-adult and adult stages are downregulated in the fetal stage in the cerebellum (Table 8). Within the cerebellum, the preadult and adult periods are characterized by some overlapping pathways including those related to chemotaxis, growth, synaptic organization or signaling, cell-cell adhesion via plasma membrane adhesion molecules, and nervous system development (Table 8). However, there are some distinct pathways in the cerebellum between preadult (small GTPase mediated signal transduction) and adult (Ras protein signal transduction) stages (Table 8). Overall, in both the hippocampus and cerebellum, genes and gene ontologies that are highly expressed in the fetal stage are downregulated in the pre-adult and adult stages. For example, in the cerebellum, gene ontologies that collectively control organismal/developmental growth or growth factor responses are upregulated in the fetal stage and downregulated in the pre-adult or adult stages.
Table 7.
Top 10 positively enriched age-associated gene ontologies in the cerebellum and five select gene ontologies of interest. Gene ontologies are listed in order by lowest p-value, to display the most highly enriched ones at the top of each list. These results were rendered with the Limma package in R to determine genes that are differentially expressed in distinct age groups. The ToppFun function in the ToppGene suite and Enrichr were used to identify the 100 top gene ontologies of biological processes of the differentially expressed genes. REVIGO was used to consolidate ontology terms.
| Cerebellum Upregulated Gene Ontologies | ||||||||
|---|---|---|---|---|---|---|---|---|
| Fetal | Pre-adult | Adult | ||||||
| GO term | p value | Odds Ratio | GO term | p value | Odds Ratio | GO term | p value | Odds Ratio |
| cell morphogenesis | 3.21E-39 | 0.357650551 | chemical synaptic transmission | 3.22E-21 | 2.296701 | chemical synaptic transmission | 1.02E-22 | 1.325412176 |
| neuron projection development | 3.25E-41 | 1.647426744 | modulation of chemical synaptic transmission | 3.10E-14 | 1.156861966 | inorganic ion transmembrane transport | 2.76E-18 | 1.093730539 |
| synapse organization | 3.49E-23 | 1.169569261 | behavior | 4.66E-14 | 4.545306313 | regulation of membrane potential | 1.09E-14 | 2.439391503 |
| developmental growth | 1.27E-21 | 1.189545455 | ion homeostasis | 1.13E-13 | 1.409166486 | modulation of chemical synaptic transmission | 4.93E-14 | 1.914227531 |
| cell division | 1.29E-21 | 0.615693013 | cation transport | 7.89E-13 | 1.464437046 | neurotransmitter transport | 1.27E-13 | 1.388766584 |
| mitotic cell cycle process | 2.17E-21 | 0.452033283 | regulation of system process | 9.98E-11 | 2.405352078 | regulation of ion transport | 3.12E-12 | 1.005942173 |
| homophilic cell adhesion via plasma membrane adhesion molecules | 1.24E-19 | 1.467915358 | regulation of synaptic plasticity | 1.87E-10 | 30.81789474 | central nervous system development | 3.59E-12 | 2.419729407 |
| regulation of synapse organization | 3.96E-17 | 0.443631961 | myelination | 7.48E-09 | 1.77895506 | synaptic vesicle cycle | 5.13E-11 | 0.650030849 |
| cell adhesion | 5.22E-17 | 1.859751122 | neuron death | 3.76E-08 | 1.479188332 | behavior | 9.11E-11 | 2.096752816 |
| chemotaxis | 9.53E-16 | 1.14846395 | oligodendrocyte development | 9.72E-08 | 2.271604938 | neuron development | 1.31E-09 | 1.422312049 |
| hindbrain development | 1.70E-15 | 5.560642814 | cell junction organization | 1.89E-07 | 3.747303544 | myelination | 2.22E-09 | 2.199090192 |
| central nervous system neuron differentiation | 2.14E-15 | 2.383424408 | actin filament-based process | 4.12E-07 | 5.09512275 | regulation of transporter activity | 1.62E-08 | 1.572316103 |
| microtubule-based process | 1.22E-13 | 0.652474436 | regulation of Ras protein signal transduction | 1.54E-06 | 2.53712839 | gliogenesis | 5.71E-08 | 5.822532403 |
| chromosome organization | 2.16E-13 | 0.433893388 | gliogenesis | 1.63E-06 | 2.55581761 | cell junction organization | 8.25E-08 | 3.427677925 |
| neural precursor cell proliferation | 8.68E-13 | 0.693515436 | response to metal ion | 3.31E-06 | 0.582569632 | regulation of cell projection organization | 6.86E-07 | 4.443024312 |
Discussion
The goal of the current study was to characterize DEK expression in the healthy human brain. To accomplish this goal, we analyzed DEK expression in multiple learning and memory-relevant brain regions across the lifespan using the BrainCloud and Human Brain Transcriptome (HBT) databases. Next, in order to further our understanding of the potential role of DEK in human brain, we evaluated the biological pathways enriched from genes correlated with DEK expression in various brain regions. We also compared the results across age groups to discover whether DEK-related functions change throughout the human lifespan.
All forebrain areas analyzed (prefrontal cortex, hippocampus, amygdala) have significantly higher DEK expression during fetal development which declined after birth, while DEK expression in the hindbrain target region of the cerebellum remained relatively stable until adulthood where it begins to decline. This presents an interesting difference in the potential roles of DEK in the forebrain versus the hindbrain. We hypothesized that there would be a sex difference in DEK mRNA expression, with females having higher DEK mRNA expression relative to males in at least some of the queried brain regions. This hypothesis was based on previous data indicating that DEK is an ERα target gene (Privette Vinnedge et al. 2012) and our previous report of increased DEK expression in the hippocampus in female mice compared with male mice (Ghisays et al. 2018). However, the findings from the BrainCloud database demonstrated that males had higher DEK expression in the dlPFC throughout life relative to females. Even though data from HBT in the dlPFC showed the same age-related effect on DEK expression as seen in BrainCloud, we did not observe a similar sex effect as there was no difference in DEK expression in males and females. This could be due to differences in sample collection methods, sample size, RNA extraction techniques, post mortem intervals of the donors, or data normalization methods between the two databases. Interestingly, there was an interaction between age and sex in the dlPFC, mPFC, and vlPFC from HBT, with male DEK expression being higher during fetal development, and females having higher DEK levels after birth. While males have an overall total brain volume that is typically greater than females, females have larger volumes of prefrontal cortices than males and reach their peak volume at an earlier age after puberty (Lenroot and Giedd 2010, Liu et al. 2020). DEK is likely important for neural development, which could account for its expression differences between males and females in the frontal cortices during stages when neural development occurs at a faster rate, i.e., before birth in males and after birth in females. Future in vitro and in vivo (rodent) studies may investigate the potential significance of these sex differences in neural DEK with regards to brain function, physiology, and behavior.
To evaluate possible functions that DEK may have in the brain, we took a non-targeted approach by completing pathway analysis of genes whose expression are correlated with that of DEK. Pathways enriched from a set of genes with a positive correlation to DEK expression across age groups and brain regions include many functions that DEK is known to carry out in the periphery, such as DNA replication and repair (Kavanaugh et al. 2011, Deutzmann et al. 2015, Smith et al. 2017), RNA processing (McGarvey et al. 2000), cell cycle (Nakashima et al. 2017), gene expression regulation (Fu et al. 1997, Hollenbach et al. 2002, Campillos et al. 2003, Koleva et al. 2012), hormone signaling (Privette Vinnedge et al. 2012), and metabolism (Matrka et al. 2017). In contrast, many of the pathways enriched from genes negatively correlated to DEK expression are novel to what is known about DEK’s functions. For example, DEK has never before been linked to synaptic structure and transmission, action potential, neurotransmitter transport, or sensory perception. These results help to inform future directions of study of neural DEK by highlighting key biological functions that DEK may be involved in. By understanding DEK’s functions in the healthy brain, we can use this to inform and compare to DEK’s role in the diseased brain.
To take a closer look at the specific genes that are most strongly correlated with DEK throughout the brain, the complete lists of significantly positively and negatively associated genes were sorted by correlation coefficient. The top 50 genes from each list are named in Table 1. As expected, some genes positively correlated with DEK are associated with cancer and immunodeficiency, such as RRM1, POLE2, SMC4, MSH6, and SRSF10 (Stelzer et al. 2016). In the healthy brain, many of these genes are involved in processes such as DNA replication and repair, chromatin remodeling, and RNA splicing (Stelzer et al. 2016). In contrast, some of the genes negatively correlated with DEK expression are potential tumor suppressors, such as KCNMA1, BIN1, and NDRG4 (Stelzer et al. 2016). Given DEK’s history as an oncogene, it follows that DEK would be negatively associated with tumor suppressive genes. Other negatively correlated genes with DEK, including MINK1, SYP, SNPH, DNM1, JPH3, and TPPP, are related to learning/memory processes and cognition-related disorders, such as AD, Huntington’s disease, epileptic encephalopathy, and intellectual disability (Stelzer et al. 2016). It is notable that our research group and others have identified neurodegenerative diseases, including AD and Huntington’s, as potentially related to changes in DEK expression, particularly the loss of DEK expression (Christodoulou et al. 2020, Greene et al. 2020, Miao et al. 2020). Again, this highlights the interesting difference between positively and negatively correlated genes and pathways to DEK. However, there are exceptions to each of these observations, such as tumor suppressor genes’ expression being positively correlated with DEK, or other positively correlated genes to DEK being associated with intellectual disability. Overall, though, there is a general pattern for genes positively correlated with DEK to be related to functions previously associated with DEK in the periphery, while negatively correlated genes tend to be related to novel functions for DEK, many of which are linked with learning and memory. Because the effects of posttranslational modifications of DEK on its function are largely unknown, it is difficult at this time to differentiate between the functional significance of DEK expression being positively or negatively correlated with particular pathways. For the purpose of focusing on the unique roles of DEK by age and brain region, the pathways enriched from positively and negatively correlated genes will be discussed jointly here after.
To explore the potential age-specific functions of DEK in the brain, we compared DEK-associated biological processes across age groups, focusing on negatively enriched pathways. During fetal development, the most significantly enriched pathways related to DEK include processes such as nervous system development, apoptosis, biosynthesis, synaptic transmission, and cellular organization. Cellular communication and signaling are crucial for healthy brain development (Basson 2012, Perrimon et al. 2012, Navarro Quiroz et al. 2018) and regulation of cell death is also important to ensure that healthy cells are allowed to proceed with differentiation (Blaschke et al. 1996). DEK is known to regulate apoptosis and cell death in the periphery (Wise-Draper et al. 2005, Wise-Draper et al. 2006), and we now find that this function of DEK may be important during neural development in utero. Enriched DEK-associated pathways during fetal development also include microtubule-based processes such as neuron projection and dendritic spine development. We have previously observed a link between DEK loss and Tau hyperphosphorylation and development of neurite processes (Greene et al. 2020). This pathway analysis corroborates our hypothesis that DEK may be important for microtubule stability and neurite outgrowth.
In early life, i.e., both fetal development and pre-adulthood (before 20 years old), DEK-related pathways include immune system development processes and glial cell proliferation. DEK’s relationship with immune system functions may shift as humans age, from immune system development to inflammatory responses. After birth, in both pre-adulthood and adulthood, DEK-associated pathways include cytokine production, NF-κB signaling, and neuroinflammation. In general, DEK seems to be important for the cellular response to various stimuli, whether it is a virus, DNA damage catalyst, or sensory stimulus. DEK’s roles in DNA damage and immune responses are recognized outside of the brain (Kavanaugh et al. 2011, Pease et al. 2015, Smith et al. 2017, Pease et al. 2020), but we now have reason to believe that DEK can have these same functions in the central nervous system.
DEK is an estrogen receptor alpha (ERα) target gene and its expression is upregulated in vitro in response to estrogen, progesterone, and androgen administration (Privette Vinnedge et al. 2012). Here, we also see that DEK may be related to sex steroid hormone signaling pathways, as pathways enriched from genes correlated with DEK during pre-adulthood and adulthood include reproductive processes and steroid hormone signaling. Neurotransmission is another type of cell communication we found to be associated with DEK expression. For example, adrenergic signaling and serotonin receptor signaling were enriched from genes correlated with DEK after birth. The adrenergic system is important for cardiac function (Rengo 2014); many enriched pathways from genes correlated with DEK were relevant to cardiac function but we did not include them in order to focus on the central nervous system in the current manuscript. Serotonin can have many physiological actions, from regulating sleep/wake cycles, to cardiac function and digestion (Berger et al. 2009). It is difficult to say at this time how DEK is specifically linked to the serotonin system. Other DEK-associated pathways found in our current analysis include neurotransmitter biosynthesis, metabolism, and transport. Future studies may investigate the precise role of DEK in neurotransmission and regulating neurotransmitter levels, such as serotonin. Previous research indicates that altered DEK expression could reprogram cellular metabolism (Matrka et al. 2017), but it is not yet known if DEK is involved specifically in neurotransmitter metabolism.
Aside from age-specific roles for DEK in the brain, it is important to establish if there is a difference in DEK-associated pathways in various brain regions. To begin to answer this question, we compared DEK expression and DEK-associated biological pathways in the forebrain and hindbrain. We chose the hippocampus and cerebellum to represent these areas because DEK is highly expressed in these structures and they are known to be important for learning and memory (Thompson and Kim 1996). First, there are several distinct differences in DEK expression and related pathways between forebrain regions, including the hippocampus, mPFC, and amygdala, and the cerebellum. DEK expression remains relatively high throughout life in the cerebellum, while DEK levels sharply decline in the forebrain after birth. Although the lower postnatal mRNA expression levels of DEK in the forebrain do not take away from its potential functional significance, it is possible that the higher transcriptional levels of DEK in the cerebellum hint at an important function for DEK throughout life in this brain region that may be different than its role(s) in the forebrain. For example, sensory perception is associated with DEK in the cerebellum during adulthood, but not in the hippocampus. There is increasing evidence to suggest that the cerebellum plays a role in perceptual processes, such as nociception, visual and auditory processing, and attentional adjustment of perceptual discrimination (Rondi-Reig et al. 2014, Baumann et al. 2015, Breska and Ivry 2021). It is even proposed that the white matter integrity of the cerebellum can affect sensory processing (Narayan et al. 2021). Given DEK’s potential involvement in myelination, it is possible that DEK expression could be important for maintaining white matter integrity in various brain regions, including the cerebellum. In addition, DNA replication and cell cycle, which are related to DEK in the periphery and are positively associated with DEK expression in the whole brain throughout life, are among the pathways negatively enriched in the cerebellum during adulthood. Interestingly, these processes are not associated with DEK in the adult hippocampus, again suggesting that DEK may have unique roles in the forebrain versus hindbrain which could account for differences in DEK expression between these regions across age groups.
An additional goal of this study was to parse out DEK-related pathways across the lifespan from “normal” age-related changes in brain. In doing so, we identified genes and gene ontologies that were more prominently expressed when DEK was considered as a factor vs. when it was not (age-only). For example, biological pathways associated with ribosome biogenesis, mRNA splicing, mRNA transport, DNA metabolic processes, double strand break repair, cellular responses to DNA damage, and mitochondrial function were more prominently associated with DEK. Again, these findings in brain are consistent with its reported role in the periphery. Not surprisingly, there were some shared gene ontologies between DEK dependent vs DEK independent (age-only) pathways including DNA repair and microtubule organization. These findings suggest that changes in DEK expression in brain over the lifespan yields a unique pattern of gene expression that is not simply due to brain aging alone.
Not only can DEK have functions across multiple biological pathways in various brain regions, but it is expressed in different cell types. Because we have previously observed that DEK is co-expressed with neurons, microglia, and astrocytes in the murine brain (Ghisays et al. 2018), we postulate that DEK may have roles in multiple neural cell types in the human brain. Consistent with this hypothesis, in the human brain, DEK is expressed in numerous cell types including fetal and mature astrocytes, neurons, microglia, oligodendrocytes, and endothelial cells (represented in Figure 1; http://www.brainrnaseq.org/; Zhang et al. (2016)). Thus, there is congruency between the localization of DEK in major cell types between the murine and human brain. Notably, its expression pattern in brain is increased in astrocytic tumors in glioblastoma (Feng et al. 2017), which is consistent with its role in the periphery as a proto-oncoprotein. These findings suggest that the expression of DEK in distinct neural cell types in the brain may differ in healthy vs diseased states. Future studies will investigate DEK’s roles in different cell types in the brain to better understand its role in the healthy aging vs. aging associated with neuropathological conditions (e.g., Alzheimer’s disease).
There are several limitations to this study. First, it is correlative in nature and while the findings suggest potential roles for DEK in the human brain, we cannot fully determine the functional significance of the results. Second, while the pattern of DEK expression across ages is similar in the dlPFC across the two databases we used, there was a main effect of sex on DEK using BrainCloud data, but from the HBT data we found an interaction between age and sex. This could be due to differences between the two databases in sample size, tissue collection, RNA extraction, and data normalization. Third, the HBT sample size was smaller than BrainCloud; therefore, we could not stratify the samples from HBT into as many age groups. Thus, we combined age groups that would not normally be analyzed within one group. As a consequence, we could have missed subtleties in the effect of age on DEK expression. For example, both early childhood and adolescence were included in the pre-adult age group, but those are very distinct developmental periods. Nonetheless, it is clear that DEK expression in the selected forebrain regions (e.g., prefrontal cortex, amygdala, hippocampus) and hindbrain structure is highest during the fetal stage of development (e.g., 6–38 weeks post-conception) and in the forebrain its expression declines after birth, no matter the specific age group. For example, pre-adults (individuals between 0 and 20 years old) and aged individuals (20+ years old) do not have significantly different DEK mRNA levels. It is important to note, though, that the relatively lower levels of DEK mRNA expression in older individuals vs. fetal development is not necessarily indicative of the fact that DEK is not important in the aged brain. It is worth noting that in the healthy adult human brain, expression of the DEK protein is still easily detectable (O’Donovan et al. 2018). Finally, we focused on a limited number of brain regions in this study. The databases we drew from did not include midbrain structures, so we cannot say if the age-related change in neural DEK expression is limited to forebrain areas. However, DEK is expressed in midbrain regions (represented in Figure 2; https://www.proteinatlas.org/; Sjöstedt et al. (2020)), therefore this is a discrepancy that could be addressed in future studies.
Further, the gene set enrichment analysis method used here is limited in its nature by analyzing each pathway independently for their enrichment of DEK-associated genes. Because genes overlap between pathways, while one relevant pathway may be enriched in its association to DEK expression, other pathways may also be significantly enriched because of the overlapping genes, resulting in potential false positives. Similarly, if a small number of genes highly correlated to DEK expression are common to many pathways, there could be many irrelevant pathways, i.e., background noise, that are enriched due to the broad functions of those genes. Additionally, genes are assigned to pathways based on their known functions from previous research, and genes can have different levels of experimental evidence from the literature to corroborate their annotation. Therefore, it may be beneficial in future studies to use multiple gene expression pathway analysis methods and multiple databases to determine what pathways overlap between analyses and are indeed likely associated with DEK in the brain.
This is the first study to characterize DEK in the human brain across the lifespan. We have established that DEK is highly expressed in the forebrain during fetal development and confirmed that neural DEK is associated with biological pathways that it is known to be related to in the periphery. In addition, we report for the first time that DEK may be important for neural development, synaptic transmission, and sensory perception. This is a necessary first step into understanding DEK’s roles in the human brain. We know that DEK loss in the aged brain has implications for cognitive impairment and could result in phenotypes of Alzheimer’s disease and other neurodegenerative disorders, such as Huntington’s disease (Ghisays et al. 2018, O’Donovan et al. 2018, Greene et al. 2020, Miao et al. 2020, Xiang et al. 2020). Taken together, the data suggest an interesting association of DEK expression with neural development and neurodegeneration. Therefore, it could be the case that DEK is important for brain development, but that its precipitous decline during aging in certain individuals (e.g., genetically vulnerable) may uniquely predispose them to certain neurodegenerative diseases. Clearly, future studies should employ a more thorough analysis of brain developmental stages (e.g., before and after the onset of neurogenesis) or neuronal differentiation models to better inform us of the contribution of DEK in various processes at play in the embryonic brain. In addition, future studies will determine whether there are differences in DEK-associated gene pathways in the aged brain of healthy patients and those with neuropathological conditions to further our understanding of its function in the central nervous system.
Acknowledgements
This work was supported by the Local Initiative for Excellence (L.I.F.E.) Foundation and the Cincinnati Children’s Hospital Medical Center Mind Brain Behavior Research, Innovation, and Pilot (RIP) funding program. Research support was provided to ANG through a NIH T32 award to the University of Cincinnati Neuroscience Graduate Program (NIH-T32-NS007453).
List of Abbreviations
- ANCOVA
Analysis of covariance
- BIN1
Bridging Integrator 1
- CA1
cornu ammonis
- dbGaP
database of Genotypes and Phenotypes
- DNA
Deoxyribonucleic acid
- POLE2
DNA Polymerase Epsilon 2, Accessory Subunit
- dlPFC
dorsolateral prefrontal
- ERα
Estrogen Receptor alpha
- GO
gene ontology
- GSEA
gene set enrichment pathway analysis
- GTex
Genotype-Tissue Expression Project
- GRIN1
Glutamate Ionotropic Receptor NMDA Type Subunit 1
- HBT
Human Brain Transcriptome
- IGSF8
Immunoglobulin superfamily member 8
- IQSEC2
IQ Motif And Sec7 Domain ArfGEF 2
- MROH1
Maestro Heat Like Repeat Family Member 1
- mPFC
medial prefrontal cortex
- MSH6
mutS homolog 6
- NMDA
N-methyl-D-aspartate
- NDRG4
N-Myc Downstream Regulated Gene 4
- NF-κB
Nuclear factor-κB
- NUP107
nucleoporin 107
- PMI
post-mortem interval
- KCNMA1
Potassium Calcium-Activated Channel Subfamily M Alpha 1
- PPP2R2C
Protein Phosphatase 2 Regulatory Subunit Bgamma
- RBL1
RB Transcriptional Corepressor Like 1
- RMI1
RecQ Mediated Genome Instability 1
- RNA
Ribonucleic acid
- RRM1
Ribonucleotide Reductase Catalytic Subunit M1
- RMA
Robust Multichip Average
- SRBD1
S1 RNA Binding Domain 1
- SRSF10
Serine And Arginine Rich Splicing Factor 10
- SMC4
Structural Maintenance Of Chromosomes 4
- USP1
ubiquitin-specific protease 1
- vlPFC
ventrolateral prefrontal cortex
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Data Availability Statement
The data that support the findings of this study are openly available in the Human Brain Transcriptome database at https://hbatlas.org/, and by request from BrainCloud at https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000417.v2.p1.
References
- Bartsch T, Döhring J, Rohr A, Jansen O and Deuschl G (2011). “CA1 neurons in the human hippocampus are critical for autobiographical memory, mental time travel, and autonoetic consciousness.” Proc Natl Acad Sci U S A 108(42): 17562–17567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson MA (2012). “Signaling in cell differentiation and morphogenesis.” Cold Spring Harbor perspectives in biology 4(6): a008151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann O, Borra RJ, Bower JM, Cullen KE, Habas C, Ivry RB, Leggio M, Mattingley JB, Molinari M, Moulton EA, Paulin MG, Pavlova MA, Schmahmann JD and Sokolov AA (2015). “Consensus paper: the role of the cerebellum in perceptual processes.” Cerebellum 14(2): 197–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M, Gray JA and Roth BL (2009). “The expanded biology of serotonin.” Annu Rev Med 60: 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke AJ, Staley K and Chun J (1996). “Widespread programmed cell death in proliferative and postmitotic regions of the fetal cerebral cortex.” Development 122(4): 1165–1174. [DOI] [PubMed] [Google Scholar]
- Breska A and Ivry RB (2021). “The human cerebellum is essential for modulating perceptual sensitivity based on temporal expectations.” eLife 10: e66743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxmeyer HE, Kappes F, Mor-Vaknin N, Legendre M, Kinzfogl J, Cooper S, Hangoc G and Markovitz DM (2012). “DEK regulates hematopoietic stem engraftment and progenitor cell proliferation.” Stem Cells Dev 21(9): 1449–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campillos M, García MA, Valdivieso F and Vázquez J (2003). “Transcriptional activation by AP-2alpha is modulated by the oncogene DEK.” Nucleic acids research 31(5): 1571–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR and Ma’ayan A (2013). “Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool.” BMC Bioinformatics 14: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou CC, Zachariou M, Tomazou M, Karatzas E, Demetriou CA, Zamba-Papanicolaou E and Spyrou GM (2020). “Investigating the Transition of Pre-Symptomatic to Symptomatic Huntington’s Disease Status Based on Omics Data.” International Journal of Molecular Sciences 21(19): 7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR and Kleinman JE (2011). “Temporal dynamics and genetic control of transcription in the human prefrontal cortex.” Nature 478(7370): 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutzmann A, Ganz M, Schönenberger F, Vervoorts J, Kappes F and Ferrando-May E (2015). “The human oncoprotein and chromatin architectural factor DEK counteracts DNA replication stress.” Oncogene 34(32): 4270–4277. [DOI] [PubMed] [Google Scholar]
- Dong X, Wang J, Kabir FN, Shaw M, Reed AM, Stein L, Andrade LEC, Trevisani VFM, Miller ML, Fujii T, Akizuki M, Pachman LM, Satoh M and Reeves WH (2000). “Autoantibodies to DEK oncoprotein in human inflammatory disease.” Arthritis & Rheumatism 43(1): 85–93. [DOI] [PubMed] [Google Scholar]
- Feng T, Liu Y, Li C, Li Z and Cai H (2017). “DEK proto-oncogene is highly expressed in astrocytic tumors and regulates glioblastoma cell proliferation and apoptosis.” Tumor Biology 39(7): 1010428317716248. [DOI] [PubMed] [Google Scholar]
- Forrest ARR, Kawaji H, Rehli M, Kenneth Baillie J, de Hoon MJL, Haberle V, Lassmann T, Kulakovskiy IV, Lizio M, Itoh M, Andersson R, Mungall CJ, Meehan TF, Schmeier S, Bertin N, Jørgensen M, Dimont E, Arner E, Schmidl C, Schaefer U, Medvedeva YA, Plessy C, Vitezic M, Severin J, Semple CA, Ishizu Y, Young RS, Francescatto M, Alam I, Albanese D, Altschuler GM, Arakawa T, Archer JAC, Arner P, Babina M, Rennie S, Balwierz PJ, Beckhouse AG, Pradhan-Bhatt S, Blake JA, Blumenthal A, Bodega B, Bonetti A, Briggs J, Brombacher F, Maxwell Burroughs A, Califano A, Cannistraci CV, Carbajo D, Chen Y, Chierici M, Ciani Y, Clevers HC, Dalla E, Davis CA, Detmar M, Diehl AD, Dohi T, Drabløs F, Edge ASB, Edinger M, Ekwall K, Endoh M, Enomoto H, Fagiolini M, Fairbairn L, Fang H, Farach-Carson MC, Faulkner GJ, Favorov AV, Fisher ME, Frith MC, Fujita R, Fukuda S, Furlanello C, Furuno M, Furusawa J.-i., Geijtenbeek TB, Gibson AP, Gingeras T, Goldowitz D, Gough J, Guhl S, Guler R, Gustincich S, Ha TJ, Hamaguchi M, Hara M, Harbers M, Harshbarger J, Hasegawa A, Hasegawa Y, Hashimoto T, Herlyn M, Hitchens KJ, Ho Sui SJ, Hofmann OM, Hoof I, Hori F, Huminiecki L, Iida K, Ikawa T, Jankovic BR, Jia H, Joshi A, Jurman G, Kaczkowski B, Kai C, Kaida K, Kaiho A, Kajiyama K, Kanamori-Katayama M, Kasianov AS, Kasukawa T, Katayama S, Kato S, Kawaguchi S, Kawamoto H, Kawamura YI, Kawashima T, Kempfle JS, Kenna TJ, Kere J, Khachigian LM, Kitamura T, Peter Klinken S, Knox AJ, Kojima M, Kojima S, Kondo N, Koseki H, Koyasu S, Krampitz S, Kubosaki A, Kwon AT, Laros JFJ, Lee W, Lennartsson A, Li K, Lilje B, Lipovich L, Mackay-sim A, Manabe R.-i., Mar JC, Marchand B, Mathelier A, Mejhert N, Meynert A, Mizuno Y, de Lima Morais DA, Morikawa H, Morimoto M, Moro K, Motakis E, Motohashi H, Mummery CL, Murata M, Nagao-Sato S, Nakachi Y, Nakahara F, Nakamura T, Nakamura Y, Nakazato K, van Nimwegen E, Ninomiya N, Nishiyori H, Noma S, Nozaki T, Ogishima S, Ohkura N, Ohmiya H, Ohno H, Ohshima M, Okada-Hatakeyama M, Okazaki Y, Orlando V, Ovchinnikov DA, Pain A, Passier R, Patrikakis M, Persson H, Piazza S, Prendergast JGD, Rackham OJL, Ramilowski JA, Rashid M, Ravasi T, Rizzu P, Roncador M, Roy S, Rye MB, Saijyo E, Sajantila A, Saka A, Sakaguchi S, Sakai M, Sato H, Satoh H, Savvi S, Saxena A, Schneider C, Schultes EA, Schulze-Tanzil GG, Schwegmann A, Sengstag T, Sheng G, Shimoji H, Shimoni Y, Shin JW, Simon C, Sugiyama D, Sugiyama T, Suzuki M, Suzuki N, Swoboda RK, ‘t Hoen PAC, Tagami M, Takahashi N, Takai J, Tanaka H, Tatsukawa H, Tatum Z, Thompson M, Toyoda H, Toyoda T, Valen E, van de Wetering M, van den Berg LM, Verardo R, Vijayan D, Vorontsov IE, Wasserman WW, Watanabe S, Wells CA, Winteringham LN, Wolvetang E, Wood EJ, Yamaguchi Y, Yamamoto M, Yoneda M, Yonekura Y, Yoshida S, Zabierowski SE, Zhang PG, Zhao X, Zucchelli S, Summers KM, Suzuki H, Daub CO, Kawai J, Heutink P, Hide W, Freeman TC, Lenhard B, Bajic VB, Taylor MS, Makeev VJ, Sandelin A, Hume DA, Carninci P, Hayashizaki Y, F. C. The, R. P. the and Clst (2014). “A promoter-level mammalian expression atlas.” Nature 507(7493): 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu GK, Grosveld G and Markovitz DM (1997). “DEK, an autoantigen involved in a chromosomal translocation in acute myelogenous leukemia, binds to the HIV-2 enhancer.” Proc Natl Acad Sci U S A 94(5): 1811–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisays V, Nguyen ET, Streicher J, Pease NA, Fitzgerald M, Estrada CM, Franco-Villanueva A, Privette Vinnedge L and Solomon MB (2018). “Neuroanatomical Distribution of DEK Protein in Corticolimbic Circuits Associated with Learning and Memory in Adult Male and Female Mice.” Neuroscience 371: 254–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasemann C, Gratias S, Stephan H, Schüler A, Schramm A, Klein-Hitpass L, Rieder H, Schneider S, Kappes F, Eggert A and Lohmann DR (2005). “Gains and overexpression identify DEK and E2F3 as targets of chromosome 6p gains in retinoblastoma.” Oncogene 24(42): 6441–6449. [DOI] [PubMed] [Google Scholar]
- Greene AN, Parks LG, Solomon MB and Privette Vinnedge LM (2020). “Loss of DEK Expression Induces Alzheimer’s Disease Phenotypes in Differentiated SH-SY5Y Cells.” Front Mol Neurosci 13: 594319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R and Grosveld G (2002). “Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek.” J Cell Sci 115(Pt 16): 3319–3330. [DOI] [PubMed] [Google Scholar]
- Jeong Y, Huh N, Lee J, Yun I, Lee JW, Lee I and Jung MW (2018). “Role of the hippocampal CA1 region in incremental value learning.” Sci Rep 8(1): 9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J and Maren S (2008). “Differential roles for hippocampal areas CA1 and CA3 in the contextual encoding and retrieval of extinguished fear.” Learning & memory (Cold Spring Harbor, N.Y.) 15(4): 244–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanović D, Geschwind DH, Mane SM, State MW and Sestan N (2009). “Functional and evolutionary insights into human brain development through global transcriptome analysis.” Neuron 62(4): 494–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, Guennel T, Shin Y, Johnson MB, Krsnik Z, Mayer S, Fertuzinhos S, Umlauf S, Lisgo SN, Vortmeyer A, Weinberger DR, Mane S, Hyde TM, Huttner A, Reimers M, Kleinman JE and Sestan N (2011). “Spatio-temporal transcriptome of the human brain.” Nature 478(7370): 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh GM, Wise-Draper TM, Morreale RJ, Morrison MA, Gole B, Schwemberger S, Tichy ED, Lu L, Babcock GF, Wells JM, Drissi R, Bissler JJ, Stambrook PJ, Andreassen PR, Wiesmüller L and Wells SI (2011). “The human DEK oncogene regulates DNA damage response signaling and repair.” Nucleic Acids Res 39(17): 7465–7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodadoust MS, Verhaegen M, Kappes F, Riveiro-Falkenbach E, Cigudosa JC, Kim DS, Chinnaiyan AM, Markovitz DM and Soengas MS (2009). “Melanoma proliferation and chemoresistance controlled by the DEK oncogene.” Cancer Res 69(16): 6405–6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleva RI, Ficarro SB, Radomska HS, Carrasco-Alfonso MJ, Alberta JA, Webber JT, Luckey CJ, Marcucci G, Tenen DG and Marto JA (2012). “C/EBPα and DEK coordinately regulate myeloid differentiation.” Blood 119(21): 4878–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW and Ma’ayan A (2016). “Enrichr: a comprehensive gene set enrichment analysis web server 2016 update.” Nucleic Acids Res 44(W1): W90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenroot RK and Giedd JN (2010). “Sex differences in the adolescent brain.” Brain Cogn 72(1): 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Seidlitz J, Blumenthal JD, Clasen LS and Raznahan A (2020). “Integrative structural, functional, and transcriptomic analyses of sex-biased brain organization in humans.” Proc Natl Acad Sci U S A 117(31): 18788–18798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wang X, Sun F, Kong J, Li Z and Lin Z (2012). “DEK overexpression is correlated with the clinical features of breast cancer.” Pathology International 62(3): 176–181. [DOI] [PubMed] [Google Scholar]
- Matrka MC, Watanabe M, Muraleedharan R, Lambert PF, Lane AN, Romick-Rosendale LE and Wells SI (2017). “Overexpression of the human DEK oncogene reprograms cellular metabolism and promotes glycolysis.” PLoS One 12(5): e0177952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey T, Rosonina E, McCracken S, Li Q, Arnaout R, Mientjes E, Nickerson JA, Awrey D, Greenblatt J, Grosveld G and Blencowe BJ (2000). “The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes.” J Cell Biol 150(2): 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Jing J, Shao Y and Sun H (2020). “MicroRNA-138 promotes neuroblastoma SH-SY5Y cell apoptosis by directly targeting DEK in Alzheimer’s disease cell model.” BMC Neurosci 21(1): 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor-Vaknin N, Kappes F, Dick AE, Legendre M, Damoc C, Teitz-Tennenbaum S, Kwok R, Ferrando-May E, Adams BS and Markovitz DM (2011). “DEK in the synovium of patients with juvenile idiopathic arthritis: characterization of DEK antibodies and posttranslational modification of the DEK autoantigen.” Arthritis Rheum 63(2): 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor-Vaknin N, Punturieri A, Sitwala K, Faulkner N, Legendre M, Khodadoust MS, Kappes F, Ruth JH, Koch A, Glass D, Petruzzelli L, Adams BS and Markovitz DM (2006). “The DEK nuclear autoantigen is a secreted chemotactic factor.” Mol Cell Biol 26(24): 9484–9496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor-Vaknin N, Saha A, Legendre M, Carmona-Rivera C, Amin MA, Rabquer BJ, Gonzales-Hernandez MJ, Jorns J, Mohan S, Yalavarthi S, Pai DA, Angevine K, Almburg SJ, Knight JS, Adams BS, Koch AE, Fox DA, Engelke DR, Kaplan MJ and Markovitz DM (2017). “DEK-targeting DNA aptamers as therapeutics for inflammatory arthritis.” Nature communications 8: 14252–14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima T, Tomita H, Hirata A, Ishida K, Hisamatsu K, Hatano Y, Kanayama T, Niwa A, Noguchi K, Kato K, Miyazaki T, Tanaka T, Shibata T and Hara A (2017). “Promotion of cell proliferation by the proto-oncogene DEK enhances oral squamous cell carcinogenesis through field cancerization.” Cancer medicine 6(10): 2424–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan A, Rowe MA, Palacios EM, Wren-Jarvis J, Bourla I, Gerdes M, Brandes-Aitken A, Desai SS, Marco EJ and Mukherjee P (2021). “Altered Cerebellar White Matter in Sensory Processing Dysfunction Is Associated With Impaired Multisensory Integration and Attention.” Frontiers in Psychology 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro Quiroz E, Navarro Quiroz R, Ahmad M, Gomez Escorcia L, Villarreal JL, Fernandez Ponce C and Aroca Martinez G (2018). “Cell Signaling in Neuronal Stem Cells.” Cells 7(7): 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan SM, Franco-Villanueva A, Ghisays V, Caldwell JL, Haroutunian V, Privette Vinnedge LM, McCullumsmith RE and Solomon MB (2018). “Sex differences in DEK expression in the anterior cingulate cortex and its association with dementia severity in schizophrenia.” Schizophr Res 202: 188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease NA, Shephard MS, Sertorio M, Waltz SE and Vinnedge LMP (2020). “DEK Expression in Breast Cancer Cells Leads to the Alternative Activation of Tumor Associated Macrophages.” Cancers 12(7): 1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease NA, Wise-Draper T and Privette Vinnedge L (2015). “Dissecting the Potential Interplay of DEK Functions in Inflammation and Cancer.” J Oncol 2015: 106517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N, Pitsouli C and Shilo B-Z (2012). “Signaling mechanisms controlling cell fate and embryonic patterning.” Cold Spring Harbor perspectives in biology 4(8): a005975–a005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger C, Huang YY, Paletzki RF, Bourtchouladze R, Scanlin H, Vronskaya S and Kandel ER (2002). “Reversible inhibition of CREB/ATF transcription factors in region CA1 of the dorsal hippocampus disrupts hippocampus-dependent spatial memory.” Neuron 34(3): 447–462. [DOI] [PubMed] [Google Scholar]
- Privette Vinnedge LM, Benight NM, Wagh PK, Pease NA, Nashu MA, Serrano-Lopez J, Adams AK, Cancelas JA, Waltz SE and Wells SI (2015). “The DEK oncogene promotes cellular proliferation through paracrine Wnt signaling in Ron receptor-positive breast cancers.” Oncogene 34(18): 2325–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privette Vinnedge LM, Ho S-M, Wikenheiser-Brokamp KA and Wells SI (2012). “The DEK oncogene is a target of steroid hormone receptor signaling in breast cancer.” PloS one 7(10): e46985–e46985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privette Vinnedge LM, McClaine R, Wagh PK, Wikenheiser-Brokamp KA, Waltz SE and Wells SI (2011). “The human DEK oncogene stimulates β-catenin signaling, invasion and mammosphere formation in breast cancer.” Oncogene 30(24): 2741–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengo G (2014). “The adrenergic system in cardiovascular pathophysiology: a translational science point of view.” Frontiers in Physiology 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondi-Reig L, Paradis A-L, Lefort JM, Babayan BM and Tobin C (2014). “How the cerebellum may monitor sensory information for spatial representation.” Frontiers in Systems Neuroscience 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Carbayo M, Socci ND, Lozano JJ, Li W, Charytonowicz E, Belbin TJ, Prystowsky MB, Ortiz AR, Childs G and Cordon-Cardo C (2003). “Gene discovery in bladder cancer progression using cDNA microarrays.” Am J Pathol 163(2): 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Unno K, Tahara S, Shimada A, Chiba Y, Hoshino M and Kaneko T (2008). “Age-related increase of superoxide generation in the brains of mammals and birds.” Aging Cell 7(4): 459–469. [DOI] [PubMed] [Google Scholar]
- Sierakowska H, Williams KR, Szer IS and Szer W (1993). “The putative oncoprotein DEK, part of a chimera protein associated with acute myeloid leukaemia, is an autoantigen in juvenile rheumatoid arthritis.” Clin Exp Immunol 94(3): 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöstedt E, Zhong W, Fagerberg L, Karlsson M, Mitsios N, Adori C, Oksvold P, Edfors F, Limiszewska A, Hikmet F, Huang J, Du Y, Lin L, Dong Z, Yang L, Liu X, Jiang H, Xu X, Wang J, Yang H, Bolund L, Mardinoglu A, Zhang C, von Feilitzen K, Lindskog C, Pontén F, Luo Y, Hökfelt T, Uhlén M and Mulder J (2020). “An atlas of the protein-coding genes in the human, pig, and mouse brain.” Science 367(6482). [DOI] [PubMed] [Google Scholar]
- Smith EA, Gole B, Willis NA, Soria R, Starnes LM, Krumpelbeck EF, Jegga AG, Ali AM, Guo H, Meetei AR, Andreassen PR, Kappes F, Vinnedge LMP, Daniel JA, Scully R, Wiesmüller L and Wells SI (2017). “DEK is required for homologous recombination repair of DNA breaks.” Scientific Reports 7: 44662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M and Lancet D (2016). “The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses.” Current Protocols in Bioinformatics 54(1): 1.30.31–31.30.33. [DOI] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N and Šmuc T (2011). “REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms.” PLOS ONE 6(7): e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RF and Kim JJ (1996). “Memory systems in the brain and localization of a memory.” Proceedings of the National Academy of Sciences 93(24): 13438–13444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waidmann S, Kusenda B, Mayerhofer J, Mechtler K and Jonak C (2014). “A DEK Domain-Containing Protein Modulates Chromatin Structure and Function in Arabidopsis.” The Plant Cell 26(11): 4328–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann T, Eckerich C, Baack M and Gruss C (2002). “The ubiquitous chromatin protein DEK alters the structure of DNA by introducing positive supercoils.” J Biol Chem 277(28): 24988–24994. [DOI] [PubMed] [Google Scholar]
- Wise-Draper TM, Allen HV, Jones EE, Habash KB, Matsuo H and Wells SI (2006). “Apoptosis Inhibition by the Human DEK Oncoprotein Involves Interference with p53 Functions.” 26(20): 7506–7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise-Draper TM, Allen HV, Thobe MN, Jones EE, Habash KB, Münger K and Wells SI (2005). “The Human DEK Proto-Oncogene Is a Senescence Inhibitor and an Upregulated Target of High-Risk Human Papillomavirus E7.” Journal of Virology 79(22): 14309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Li Z, Lin H, Han L, Liu S and Lin Z (2008). “DEK overexpression in uterine cervical cancers.” Pathology International 58(6): 378–382. [DOI] [PubMed] [Google Scholar]
- Xiang C, Cong S, Liang B and Cong S (2020). “Bioinformatic gene analysis for potential therapeutic targets of Huntington’s disease in pre-symptomatic and symptomatic stage.” Journal of Translational Medicine 18(1): 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, Jeon M and Ma’ayan A (2021). “Gene Set Knowledge Discovery with Enrichr.” Current Protocols 1(3): e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sloan Steven A., Clarke Laura E., Caneda C, Plaza Colton A., Blumenthal Paul D., Vogel H, Steinberg Gary K., Edwards Michael S. B., Li G, Duncan John A. III, Cheshier Samuel H., Shuer Lawrence M., Chang Edward F., Grant Gerald A., Gephart Melanie G. H. and Barres Ben A. (2016). “Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse.” Neuron 89(1): 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in the Human Brain Transcriptome database at https://hbatlas.org/, and by request from BrainCloud at https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000417.v2.p1.


