Abstract
The pressure to optimize the enzymatic rate acceleration for adenylate kinase (AK)-catalyzed phosphoryl transfer has led to the evolution of an induced-fit mechanism, where the binding energy from interactions between the protein and substrate adenosyl group is utilized to drive a protein conformational change that activates the enzyme for catalysis. The adenine group of adenosine contributes 11.8 kcal mol–1 to the total ≥14.7 kcal mol–1 adenosine stabilization of the transition state for AK-catalyzed phosphoryl transfer to AMP. The relative third-order rate constants for activation of adenylate kinase, by the C-5 truncated adenosine 1-(β-d-erythrofuranosyl)adenine (EA), for catalysis of phosphoryl transfer from ATP to phosphite dianion (HP, kcat/KHPKAct = 260 M–2 s–1), fluorophosphate (47 M–2 s–1), and phosphate (9.6 M–2 s–1), show that substitution of −F for −H and of −OH for −H at HP results, respectively, in decreases in the reactivity of AK for catalysis of phosphoryl transfer due to polar and steric effects of the −F and −OH substituents. The addition of a 5′-CH2OH to the EA activator results in a 3.0 kcal mol–1 destabilization of the transition state for AK-activated phosphoryl transfer to HP due to a steric effect. This is smaller than the 8.3 kcal mol–1 steric effect of the 5′-CH2OH substituent at OMP on HP-activated OMPDC-catalyzed decarboxylation of 1-(β-d-erythrofuranosyl)orotate. The 2′-OH ribosyl substituent shows significant interactions with the transition states for AK-catalyzed phosphoryl transfer from ATP to AMP and for adenosine-activated AK-catalyzed phosphoryl transfer from ATP to HP.
Introduction
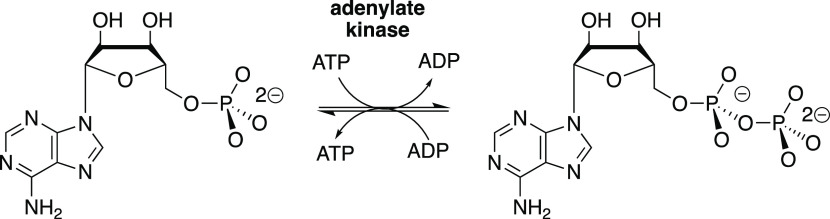
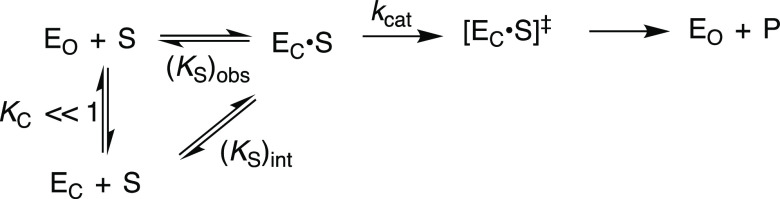
Adenylate kinase (AK) catalyzes reversible phosphoryl group transfer between two molecules of adenosine 5′-diphosphate (ADP) to form adenosine 5′-triphosphate (ATP) and adenosine 5′-monophosphate (AMP, Scheme 1). The enzyme is present at high levels in mammalian tissues, where it acts to maintain an equilibrium between the adenine nucleotides ATP, ADP, and AMP. The enzyme functions over short periods of stress, when there is a requirement for high energy output, to convert ADP from myosin-catalyzed hydrolysis of ATP to AMP and ATP so that both high-energy phosphates of ATP contribute to muscle motion.1,2 This role in the organism’s “fight or flight” response places adenylate kinase under intense evolutionary pressure to optimize catalytic activity. The exertion of this pressure has led to a distinctive and highly efficient catalytic motif,3,4 where the AK protein–substrate binding interactions are utilized to drive a change in enzyme conformation, from an inactive open form EO to a closed complex EC, which traps the substrate in a protein cage (Scheme 2).5 This substrate-driven conformational change organizes the AK protein side chains at positions that optimize their stabilizing interactions with the enzyme-bound transition state.6,7
Scheme 1. Adenylate Kinase-Catalyzed Phosphoryl Transfer from ATP to AMP.
Scheme 2. Induced Fit Mechanism for the Utilization of Intrinsic Substrate Binding Energy (ΔGint) to Drive an Enzyme-Activating Conformational Change from EO to EC.
The induced-fit mechanism was proposed more than 60 years ago to rationalize enzyme specificity in the catalysis of reactions of substrates that provide binding energy to drive a protein conformational change (Scheme 2).8,9 We have shown that such substrate-driven conformational changes are the defining property of many enzymes that follow this mechanism,10−12 where the use of substrate binding energy in the formation of the Michaelis complex ensures that the enzymatic transition state binds with a higher affinity than substrate.
Enzymes that catalyze a diverse set of proton transfer,13,14 hydride transfer,14,15 and decarboxylation14,16 reactions of phosphorylated substrates use the intrinsic phosphodianion binding energy to drive thermodynamically unfavorable protein conformational changes.10,11,17 The dianion binding energy so utilized is not expressed at the Michaelis complex, but is only expressed at the transition state and provides for specificity in binding of enzymatic transition states with a higher affinity than substrate.10−12 This is often required to avoid tight and effectively irreversible substrate binding.18 Two of these enzymes, orotidine monophosphate decarboxylase (OMPDC) and triosephosphate isomerase (TIM), have received prominent attention in mechanistic studies: OMPDC because of the enzyme’s extraordinary rate acceleration19,20 and TIM because the fierce evolutionary pressure to optimize the enzymatic rate acceleration has resulted in an enzyme that has achieved close to perfection in catalytic efficiency.21,22 The similar induced-fit mechanisms for these enzymes are consistent with the notion that this mechanism is enforced for metabolic enzymes that show the highest catalytic proficiency.
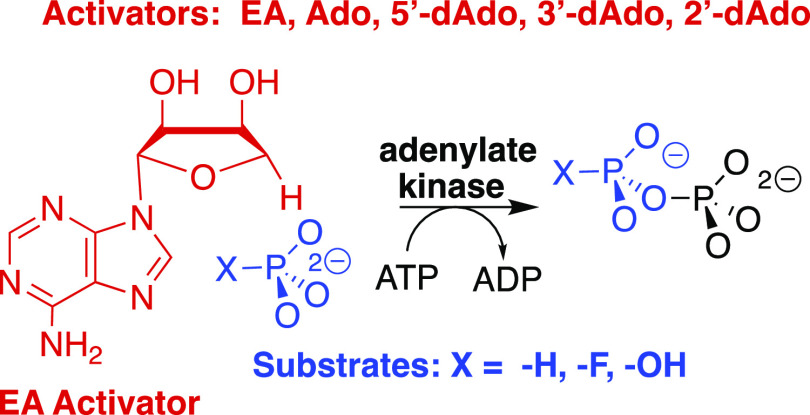
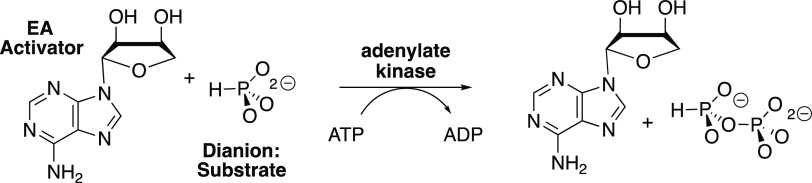
With this in mind, we targeted adenylate kinase for study, to test our hypothesis that the pressure to optimize the enzymatic rate acceleration led to the utilization of protein–adenosyl group binding interactions to drive a protein conformational change, which activates the enzyme for catalysis of phosphoryl transfer.10 This hypothesis was supported by our recent report that: (1) the binding interactions between human adenylate kinase isozyme 1 (HAdK1) and the adenosyl group of substrate AMP provide a ≥14.7 kcal mol–1 stabilization of the transition state for phosphoryl transfer from ATP to AMP (Scheme 1).23 (2) Greater than 8.7 kcal mol–1 of this transition state stabilization is recovered, after severing the covalent connection between the adenosyl and phosphate substrate fragments of AMP, as activation of HAdK1-catalyzed phosphoryl transfer from ATP to the substrate piece phosphite dianion by the second nonreacting piece 1-(β-d-erythrofuranosyl)adenine (EA) (Scheme 3).23
Scheme 3. Adenylate Kinase-Catalyzed Phosphoryl Transfer from ATP to Phosphite Dianion That Is Activated by 1-(β-d-Erythrofuranosyl)adenine (EA)23.
The substrate binding pockets for enzymes that catalyze the reactions of phosphate monoester substrates may be divided into catalytic and dianion activation sites. The specificity of the dianion binding site for activation by five inorganic tetrahedral dianions was characterized in an earlier study on the reactions catalyzed by TIM, OMPDC, and glycerol 3-phosphate dehydrogenase.24,25 Our experimental results on adenylate kinase likewise show that the AMP-binding pocket is divided into a catalytic site for dianion substrates and an activation site for the adenosyl group that uses the ligand binding energy to activate the enzyme for catalysis at the second site. We report here experiments to characterize the specificity of HAdK1 for catalysis of phosphoryl transfer from ATP to several tetrahedral dianions and the enzyme specificity for activation by adenosine and several deoxyadenosines.
Experimental Section
Materials
The following reagent-grade chemicals were obtained from Sigma-Aldrich: triethanolamine (TEA) hydrochloride, potassium chloride, imidazole, pyruvate (sodium salt), phosphoenolpyruvate (PEP, monosodium salt hydrate), d-ribose 5′-phosphate (disodium salt) (R5P), adenosine 5′-monophosphate (AMP, monosodium salt), adenosine 5′-diphosphate (ADP, monosodium salt), adenosine 5′-triphosphate (ATP, disodium salt), the reduced form of β-nicotinamide adenine dinucleotide (NADH, disodium salt dihydrate), adenosine (Ado), 5′-deoxyadenosine (5′-dAdo), 2′-deoxyadenosine monohydrate (2′-dAdo), and isopropyl β-d-1-thiogalactopyranoside (IPTG). Cordycepin (3′-deoxyadenosine, 3′-dAdo) was obtained from the Cayman Chemical Company. Sodium chloride, magnesium chloride hexahydrate, dithiothreitol, and tris(hydroxymethyl)aminomethane (Tris, free base) were from Fisher. Sodium phosphite pentahydrate was from Riedel–de Haën. Sodium fluorophosphate was from Alfa Aesar. Sodium hydrogen phosphate was from Fluka. Luria–Bertani broth was from IBI Scientific. Ampicillin (sodium salt) and chloramphenicol were from VWR. Bovine serum albumin (BSA) and bovine pancreas DNAse I were from Roche Diagnostics. Rabbit muscle adenylate kinase was purchased from Creative Enzymes. Rabbit muscle pyruvate kinase (PK) and porcine heart lactate dehydrogenase (LDH) were from Calzyme. Pierce protease inhibitor tablets were obtained from Thermo Scientific. Escherichia coli BL21 (DE3) competent cells were from Lucigen. 1-(β-d-Erythrofuranosyl)adenine (EA) was purchased from Apex Molecular.
Methods
The protocol for the following experiments is described in an earlier publication:23 (i) The preparation of HAdK1. (ii) The assays for the activity of LDH and PK. (iii) The coupled enzyme assay to monitor AK-catalyzed phosphoryl transfer from 1 mM ATP to AMP. (iv) The coupled enzyme assay to monitor EA-activated AK-catalyzed phosphoryl transfer from 1 mM ATP to phosphite dianion, where the velocity of conversion of NADH to NAD is equal to the velocity of conversion of ATP to ADP. This assay was also used to monitor the conversion of ATP to ADP in the nucleoside-activated AK-catalyzed phosphoryl transfer from 1 mM ATP to fluorophosphate and phosphate dianion. It was shown in a control experiment for a standard pyruvate kinase assay that there is no detectable activation of pyruvate kinase-catalyzed conversion of PEP to pyruvate by 4 mM EA plus 10 mM phosphite dianion. The reactions were monitored at 25 °C by following the change in absorbance at 340 nm using either a temperature-controlled Cary 3500 Multicell Peltier UV–vis spectrophotometer or a Cary 3E spectrophotometer equipped with a temperature-controlled Peltier block multicell changer. The concentrations of phosphite and phosphate dianion at pH 7.5 were calculated from the total concentrations of the mono- and dianion, using a pKa of 6.4 for HPO32–,26 and 7.2 for HOPO32–.27
All enzyme assays were carried out in solutions of ionic strength of 0.15 (I = 0.15) that were prepared by the addition of measured amounts of NaCl to the assay mixture. Assays for HAdK1-catalyzed phosphoryl transfer from 1 mM ATP to phosphite, fluorophosphate, or phosphate dianion in the presence of increasing activator concentrations were in 1.0 mL solutions that contain 25 mM TEA at pH 7.5 (I = 0.15, NaCl), 30 mM KCl, 3 mM MgCl2, 0.5 mM PEP, 1 mM ATP, 0.1 mg/mL BSA, 0.2 mM NADH, 5–25 mM total monoanion + dianion substrate, 2–10 mM of the specified activator, one unit each of LDH and PK, and the following concentration of adenylate kinase for experiments with the given dianion and activator: rabbit muscle AdK (RAdK), EA + phosphite (1–4 μM RAdK); RAdK, EA + fluorophosphate (1 μM RAdK); HAdK1, EA + phosphite (2 μM HAdK1); HAdK1, EA + fluorophosphate (4 μM HAdK1); HAdK1, EA + phosphate (70 μM HAdK1); HAdK1, adenosine + phosphite (30 μM HAdK1); HAdK1, 5′-deoxyadenosine + phosphite (12 μM HAdK1); HAdK1, 3′-deoxyadenosine + phosphite (40 μM HAdK1); HAdK1, 2′-deoxyadenosine + phosphite (40 μM HAdK1); HAdK1, ribose 5-phosphate (20 μM HAdK1); HAdK1, adenine + ribose 5-phosphate (20 μM HAdK1). The observed initial reaction velocity, vobs, for oxidation of NADH in this coupled enzyme assay was determined by monitoring at 340 nm the disappearance of <5% of the limiting reagent NADH for a period of 30 min. All experimental points were performed in triplicate. The kinetic parameters reported in the Tables were obtained from the linear or nonlinear least-squares fits of the experimental data to the appropriate kinetic equation determined using Prism 8 for MacOS from GraphPad Software.
Results
The initial velocity, vobs, at 25 °C for AK-catalyzed phosphoryl transfer from 1.0 mM ATP (Km = 0.06 mM for the enzyme from rabbit muscle)28 to different acceptors to form ADP and phosphorylated product was determined by coupling the formation of ADP to PK-catalyzed phosphoryl transfer from PEP to ADP to form pyruvate and ATP, followed by LDH-catalyzed reduction of pyruvate by NADH. The reaction was monitored by following the decrease in absorbance at 340 nm. Figure S1 in the Supporting Information (SI) shows the Michaelis–Menten plot of vobs/[E] against [AMP] for commercial rabbit muscle adenylate kinase (RAdK)-catalyzed phosphoryl transfer from 1.0 mM ATP to AMP; and, Figure S2 shows the plot of vobs/[E] against [2′-dAMP] for HAdK1-catalyzed phosphoryl transfer from 1 mM ATP to 2′-dAMP. The solid lines for Figures S1 and S2 show the nonlinear least-squares fits of these data to the Michaelis–Menten equation using the kinetic parameters reported in Table 1. The table also lists kinetic parameters for HAdK1-catalyzed phosphoryl transfer from 1.0 mM ATP to AMP, and an upper limit for kcat/Km for phosphoryl transfer from 1.0 mM ATP to phosphite dianion determined in earlier work.23
Table 1. Kinetic Parameters for Phosphoryl Transfer from 1 mM ATP to Different Phosphoryl Acceptors, Determined at pH 7.5 and 25 °C in 25 mM TEA Buffer and I = 0.15 (NaCl).
| enzyme | phosphoryl acceptor | kcat (s–1)a | Km (M)a | kcat/Km (M–1s–1) |
|---|---|---|---|---|
| RAdK | AMPb | 100 ± 2 | (8.6 ± 0.6) × 10–5 | 1.2 × 106 |
| HAdK1 | AMPc | 475 | 6.8 × 10–5 | 7.0 × 106 |
| 2′-dAMPd | 92 | (1.05 ± 0.06) × 10–3 | 8.8 × 104 | |
| HPc | no reaction detected | ≤1.0 × 10–4 | ||
| R5Pe | not saturated | 0.014 ± 0.01 | ||
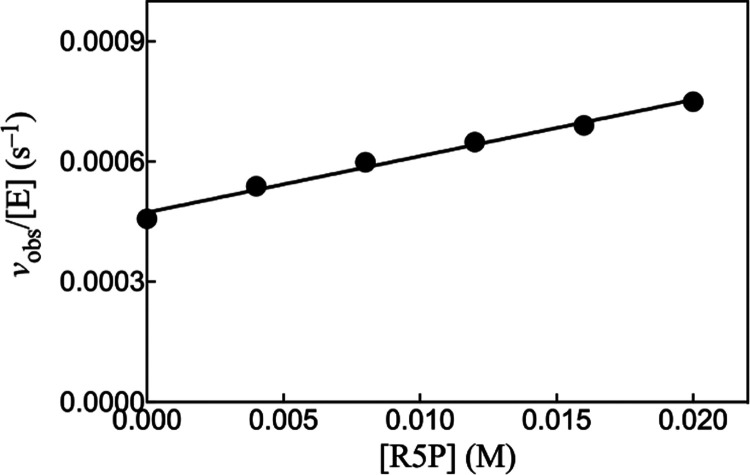
Figure 1 shows the linear increase, with increasing [R5P], in vobs/[E] for HAdK1-catalyzed phosphoryl transfer from 1 mM ATP to ribose 5′-phosphate. The solid line in Figure 1 shows the fit of the experimental data to eq 1 to give kcat/Km = 0.014 M–1 s–1 (Table 1). There is no detectable activation of the reaction of the substrate piece R5P by the addition of the missing fragment, as shown by the identical values of vobs/[E] determined for the HAdK1-catalyzed reaction of R5P in the presence and absence of 5 mM adenine.
| 1 |
Figure 1.
Increase in vobs/[E] with increasing concentrations of ribose 5-phosphate for HAdK1-catalyzed (20 μM) reactions of 1 mM ATP at pH 7.5, I = 0.15 (NaCl), and 25 °C.
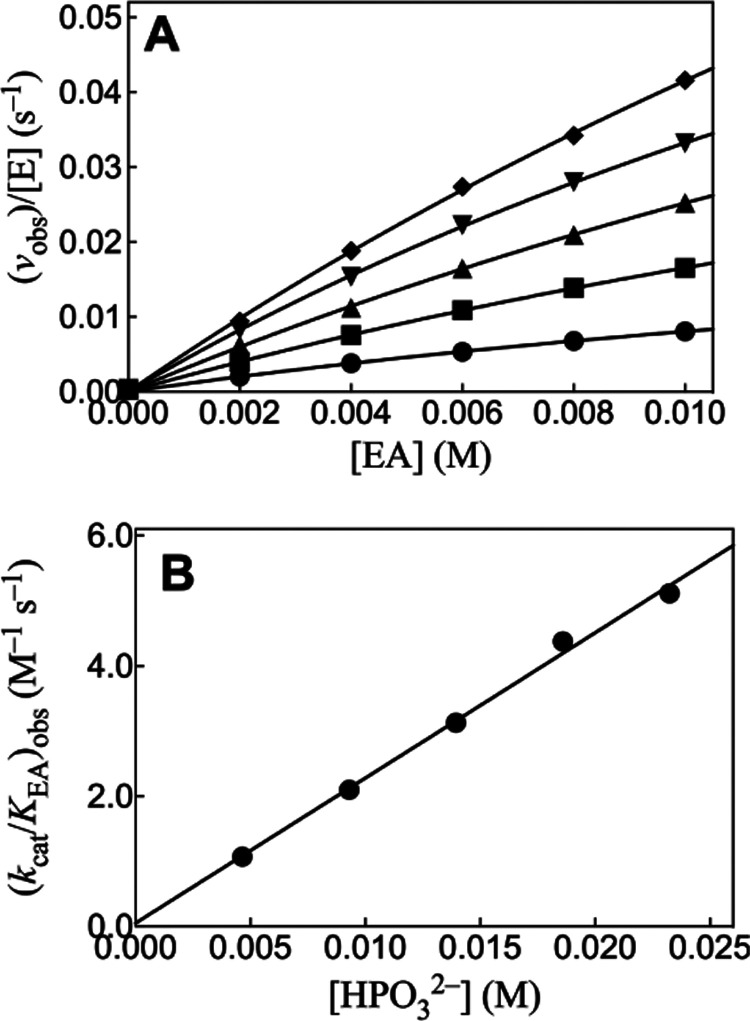
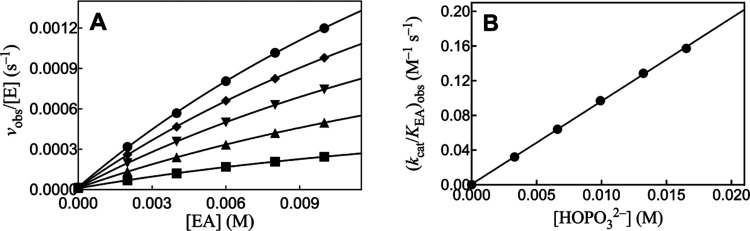
We previously reported kinetic parameters for EA-activated HAdK1-catalyzed phosphoryl transfer from ATP to phosphite dianion (Table 2) and presented 31P-NMR spectral data which confirm the structure of the isohypophosphate reaction product.23Figure 2A shows the effect of increasing [EA] on vobs/[E] for rabbit muscle adenylate kinase-catalyzed (RAdK) reactions of 1 mM ATP with different fixed [HPO32–]. The solid lines for Figure 2A show the nonlinear least-squares fits of these kinetic data to eq 2, derived for Scheme 4 (KHP ≫ [HP]). The slight curvature with increasing [EA] for the individual plots of Figure 2A is consistent with the weak partial saturation of RAdK by high [EA]. Values of the apparent second-order rate constants (kcat/KAct)obs for reactions at different fixed [HPO32–] were determined as the slopes of the linear correlations at low [EA] ([Act] ≪ KAct, eq 2). Figure 2B shows the linear plot of (kcat/KAct)obs determined for Figure 2A, against [HPO32–]. The fit of this linear plot to eq 3 gives the value of (kcat)XP·Act/KXPKact = 220 ± 10 M–2 s–1 reported in Table 2.
| 2 |
| 3 |
Table 2. Third-Order Rate Constants (Scheme 4) for EA Activation of RAdK- and HAdK1-Catalyzed Phosphoryl Transfer from ATP to Dianionsa.
| enzyme | dianion substrate | (kcat)XP·Act/KXPKAct (M–2 s–1) |
|---|---|---|
| Rabbit Muscle (kcat/Km)AMP = 1.2 × 106 M–1 s–1b | HPO32–d | 220 ± 10 |
| FPO32–e | 55 ± 3 | |
| Human (kcat/Km)AMP = 7.0 × 106 M–1 s–1c | HPO32–c | 260 ± 7 |
| FPO32–f | 47 ± 0.5 | |
| HOPO32–g | 9.6 ± 1.0 |
Determined at pH 7.5 and 25 °C in 25 mM TEA buffer at I = 0.15 (NaCl). The uncertainty in the kinetic parameters is the standard error from the least-squares fits of the kinetic data to eq 3.
Published data.23
Determined for data from Figure S3A,B.
Figure 2.
(A) Increase in vobs/[E] with increasing [EA] for RAdK-catalyzed reactions of 1 mM ATP with phosphite dianion at pH 7.5, I = 0.15 (NaCl), and 25 °C. Key: ◆ 23 mM [HPO32–] and 2 μM RAdK; ▼ 19 mM [HPO32–] and 2 μM RAdK; ▲ 14 mM [HPO32–] and 2 μM RAdK; ■ 9.2 mM [HPO32–] and 1 μM RAdK; ● 4.6 mM [HPO32–] and 1 μM RAdK. (B) Effect of increasing [HPO32–] on the values of (kcat/KAct)obs determined for (A).
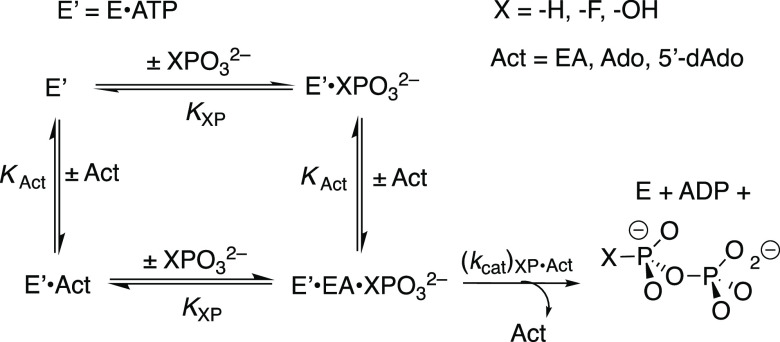
Scheme 4. Kinetic Mechanism for the Activation of HAdK1-Catalyzed Phosphoryl Transfer from ATP to Dianions XPO32– by EA, Ado, and 5′-dAdo.
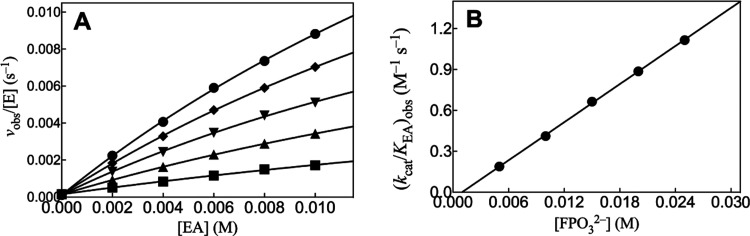
Figures 3A and 4A show, respectively, the effect of increasing [EA] on vobs/[E] for HAdK1-catalyzed reactions of 1 mM ATP at increasing fixed [FPO32–] or [HOPO32–]. The nonlinear least-squares fits of the experimental data to eq 2, derived for Scheme 4, (KXP ≫ [XP]) gave values of (kcat/KAct)obs for reactions at the different [XPO32–]. Figures 3B and 4B show, respectively, linear plots of values from Figures 3A or 4A (eq 3), against [FPO32–] and [HOPO32–]. The kinetic parameter (kcat)XP·Act/KXPKAct for HAdK reported in Table 2 were determined as the slopes of these linear plots. Figure S3A shows the effect of increasing [EA] on vobs/[E] for RAdK1-catalyzed reactions of 1 mM ATP at increasing fixed [FPO32–]. The nonlinear least-squares fits of the experimental data to eq 2, derived for Scheme 4, (KXP ≫ [XP]) gave values of (kcat/KAct)obs for reactions at the different [FPO32–]. Figure S3B shows the linear plot of values from Figure S3A (eq 3) against [FPO32–]. The kinetic parameter (kcat)XP·Act/KXPKact for RAdK reported in Table 2 were determined as the slopes of this plot.
Figure 3.
(A) Increase in vobs/[E] with increasing [EA] for HAdK1-catalyzed (4 μM) reactions of 1 mM ATP with FPO32– at pH 7.5, I = 0.15 (NaCl), and 25 °C. Key: ● 25 mM [FPO32–]; ◆ 20 mM [FPO32–]; ▼ 15 mM [FPO32–]; ▲ 10 mM [FPO32–]; ■ 5 mM [FPO32–]. (B) Effect of increasing [FPO32–] on the values of (kcat/KAct)obs determined for (A).
Figure 4.
(A) Increase in vobs/[E] with increasing [EA] for HAdK1-catalyzed (70 μM) reactions of 1 mM ATP with HOPO32– at pH 7.5, I = 0.15 (NaCl), and 25 °C. Key: ● 17 mM [HOPO32–]; ◆ 13 mM [HOPO32–]; ▼ 10 mM [HOPO32–]; ▲ 6.6 mM [HOPO32–]; ■ 3.3 mM [HOPO32–]. (B) Effect of increasing [HOPO32–] on the values of (kcat/KAct)obs determined for (A).
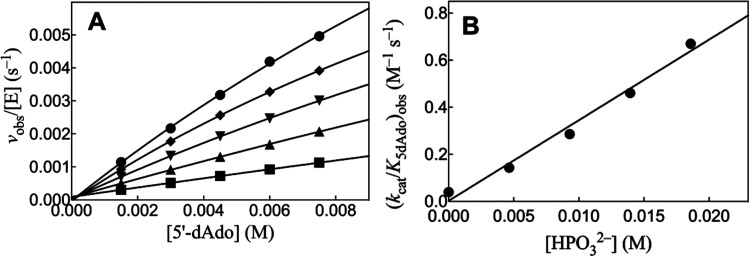
Figure 5A shows the effect of increasing [5′-dAdo] on vobs/[E] for HAdK1-catalyzed reactions of 1 mM ATP with different fixed [HPO32–]. The nonlinear least-squares fits of the experimental data to eq 2, derived for Scheme 4 (KXP ≫ [XP]), gave values of (kcat/K5′dAdo)obs for reactions at the different [XPO32–]. Figure 5B shows the linear plot of values of (kcat/K5′dAdo)obs from Figure 5A, against [HPO32–]. The fit of this linear plot to eq 3 gives the value of (kcat)XP·Act/KXPKact reported in Table 3. Figure S4A,B shows similar kinetic data for adenosine-activated HAdK1-catalyzed phosphoryl transfer from ATP to phosphite. The fit of the linear plot of (kcat/KAdo)obs against [Ado] to eq 3 gives the value of (kcat)XP·Act/KXPKact reported in Table 3. Figures S5 and S6 show, respectively, the effect of increasing [2′-dAdo] and [3′-dAdo] on vobs/[E] for HAdK1-catalyzed reactions of 1 mM ATP with 23 mM [HPO32–] at pH 7.5, I = 0.15 (NaCl) and 25 °C. There is no significant activation of phosphoryl transfer from ATP to phosphite dianion by 2′-Ado and 3′-Ado, but rather a ca. 10% decrease in vobs/[E] is observed as the nucleoside concentration is increased from 0.0 to 10 mM (Table 3).
Figure 5.
(A) Increase in vobs/[E] with increasing [5′-dAdo] for HAdK1-catalyzed (12 μM) reactions of 1 mM ATP with phosphite dianion at pH 7.5, I = 0.15 (NaCl), and 25 °C. Key: ● 23 mM [HPO32–]; ◆ 19 mM [HPO32–]; ▼ 14 mM [HPO32–]; ▲ 9.2 mM [HPO32–]; ■ 4.6 mM [HPO32–]. (B) Effect of increasing [HPO32–] on the values of (kcat/K5′dAdo)obs determined for (A).
Table 3. Third-Order Rate Constants (Scheme 4) for Activation of HAdK1-Catalyzed Phosphoryl Transfer Reactions by EA, Adenosine, and Deoxyadenosinesa.
Discussion
We have chosen HAdK1 as the focus for these studies because this is the major cytosolic adenylate kinase in humans. HAdK1 is present in most human tissues, with the highest expression levels in skeletal muscle, brain, and erythrocytes.29,30 We present a brief comparison of results obtained for HAdK1 with results for the more widely studied, commercially available, adenylate kinase from rabbit muscle. The similar third-order rate constants for EA-activated reactions of phosphite and fluorophosphate determined for catalysis by HAdK1 and RAdK show that this activation is a general phenomenon for mammalian adenylate kinases. The observation that substrate binding drives large conformational changes for adenylate kinases from bacteria3,31−35 suggests that EA-activated AK-catalyzed phosphoryl transfer to phosphite dianion will be observed for most or all adenylate kinases
It is interesting that there is no detectable saturation of HAdK1 or RAdK (Figure 2) by 23 mM of the substrate piece phosphite dianion and only weak saturation by 10 mM of the activator piece EA. The tighter binding of the whole substrate AMP (Km = 6.8 × 10–5 M, Table 1) reflects the entropic advantage to the binding of a single whole substrate compared to the substrate in pieces.36 This has been noted in other studies on enzyme-catalyzed reactions of substrate pieces.14,37
The nine known isozymes of human AK show a large variation in their kinetic parameters and substrate specificity.30 The mechanistic significance of these variations is unclear but might be probed through studies on the reaction of the substrate pieces. For example, HAdK1 shows a high specificity for the catalysis of phosphoryl transfer to AMP, but a much smaller specificity for the catalysis of the reaction of different NTP donors.30 This suggests that the high specificity of HAdK1 for phosphoryl transfer to AMP is due to the many specific protein-adenine transition state contacts that give rise to the ≥14.7 kcal mol–1 intrinsic binding energy for the adenosine group of AMP.23 We predict that AK isozymes with a broader specificity for phosphoryl transfer to different NMPs will show a smaller intrinsic binding energy for the nucleoside at the NMP site. We are also interested in determining the intrinsic binding energy for the adenine ring at the ATP site but have not yet developed an assay for the reaction of ATP in pieces.
Intrinsic Binding Energies for Nucleoside Monophosphates
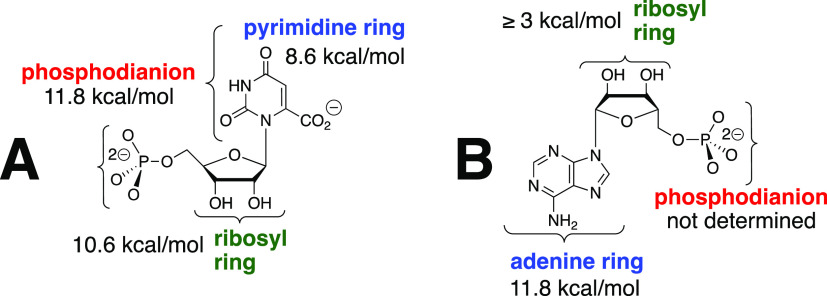
It is informative to compare the large intrinsic binding energies for nucleoside monophosphate substrates in the stabilization of transition states for OMPDC-catalyzed decarboxylation of OMP, and for HAdK1-catalyzed phosphoryl transfer to AMP. The 31 kcal mol–1 transition state stabilization for OMPDC-catalyzed decarboxylation of OMP is obtained from ca. 10 kcal mol–1 interactions with the phosphodianion, pyrimidine ring, and ribosyl ring fragments of OMP (Figure 6A).16,38 By comparison, the transition state binding energy of AMP in HAdK1-catalyzed phosphoryl transfer may be partitioned as follows (Figure 6B): (i) the 11.8 kcal mol–1 stabilization by interactions with the adenine ring, calculated from the ratio of the values of kcat/Km for the HAdK1-catalyzed reaction of AMP (7 × 106 M–1 s–1) and the truncated substrate ribose 5-phosphate (0.014 M–1 s–1). (ii) The transition state stabilization by the entire adenosine moiety is ≥14.7 kcal mol–1.23 This gives a lower limit of (14.7–11.8) ≥ 3 kcal mol–1 for transition state stabilization from interactions with the ribosyl ring, which probably sharply underestimates these interactions. (iii) The stabilization from the interaction between the protein catalyst and the AMP phosphodianion has not been determined because there is no simple protocol for estimating these protein–ligand interactions at the pentavalent transition state for concerted phosphoryl transfer from ATP to AMP.39
Figure 6.
(A) Partitioning of the intrinsic transition state binding energy of substrate OMP for OMPDC-catalyzed decarboxylation of OMP into interactions with the three substrate fragments.16,38 (B) Partial partitioning of the transition state binding energy of substrate AMP for adenylate kinase-catalyzed phosphoryl transfer into interactions with the corresponding substrate fragments.
The difference between the small observed binding energy for the substrates OMP and AMP, respectively, of reactions catalyzed by OMPDC and adenylate kinase; and, their much larger intrinsic transition state binding energies represents, in part, the substrate binding energy utilized to drive unfavorable enzyme conformational changes (ΔGC, Scheme 2).10 It is important to emphasize that this binding energy may also be used to induce destabilizing electrostatic stress between protein side chains and OMP bound to OMPDC12,18,40 or between the terminal phosphate dianions of ATP and AMP bound to adenylate kinase that is relieved at the enzymatic transition states.18 There is evidence that the former destabilizing interactions are small for OMPDC-catalyzed decarboxylation,41,42 but the binding energy utilized to introduce electrostatic stress at the ternary Michaelis complex for adenylate kinase has not been evaluated.
The activation barrier to turnover of OMP bound at the caged complex to OMPDC can be calculated from the experimental rate constant kcat. This barrier has been successfully modeled in computational studies.40,43,44 By comparison, we know of no reports of successful computational studies to model the observed substrate binding energy at the Michaelis complex to OMPDC, presumably because of the difficulty associated with modeling the barrier to the enzyme conformational change that accompanies substrate binding (Scheme 2). Computational studies on adenylate kinase have focused on modeling the complex conformational transitions associated with ligand binding.31−33,45 To the best of our knowledge, there are no reports of successful computational studies to model the values of either kcat or Km for adenylate kinase-catalyzed phosphoryl transfer reactions.
Adenylate Kinase-Catalyzed Reactions of Substrate Pieces
Catalytic Site
We previously showed that EA, an AMP fragment, activated adenylate kinase for catalysis of phosphoryl transfer from ATP to phosphite dianion (Scheme 3), and identified the isohypophosphate reaction product by 31P-NMR.23 The kinetic data shown in Figures 3 and 4, respectively, provide evidence that EA activates adenylate kinase for catalysis of phosphoryl transfer from ATP to FPO32– and HOPO32–. We were unable to identify the products of these putative phosphoryl transfer reactions. We assume, by analogy to the reaction of HPO32– (Scheme 3), that the reactions give, respectively, fluoropyrophosphate and pyrophosphate.
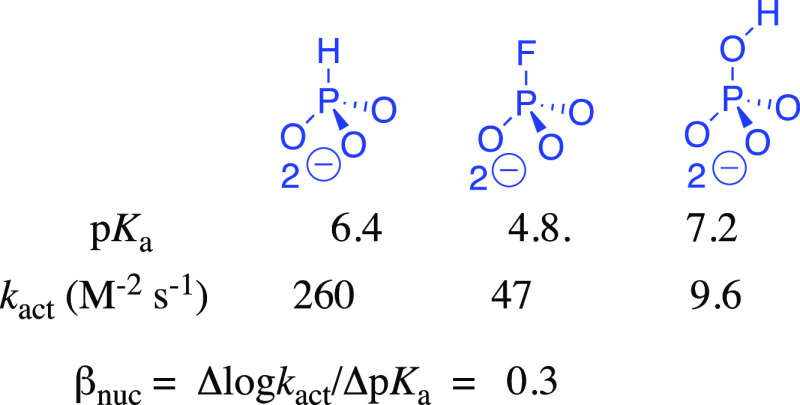
The 5.5-fold larger value of kact = (kcat)XP·Act/KXPKAct (Scheme 4) for phosphoryl transfer from ATP to phosphite dianion (pKa = 6.4)26 compared to fluorophosphate dianion (pKa = 4.8)27 gives a value of βnuc = 0.3 for a two-point Brønsted plot (Figure 7). The Brønsted parameter of 0.3 will overestimate the bond order of the partial transition state bond between the dianion nucleophile and the transferred γ-phosphoryl group of ATP,46 if there is an unfavorable steric interaction between HAdK1 and the fluorine of fluorophosphate. In any case, the results are consistent with an open reaction transition state.39 By comparison, phosphate dianion (pKa = 7.2)27 is more basic than phosphite dianion, but much less reactive toward HAdK1-catalyzed phosphoryl transfer (kact = 9.6 M–2 s–1). We suggest that this falloff in activity is due to an unfavorable interaction between the −OH at phosphate dianion and the EA activator. Similarly, the sulfur for oxygen substitution at AMP to give AMPS results in a 100-fold falloff in kcat/Km for adenylate kinase-catalyzed phosphoryl transfer to the pro S oxygen of AMPS to form SP-adenosine 5′-O-(1-thiodiphosphate) ((SP) ADPαS),47 but only a 1.4-unit decrease in the pKa of the reacting functional group.48 We propose that the falloff in the activity of AMPS compared with AMP is likewise partly due to the steric effect of sulfur, which is larger than oxygen, on adenylate kinase-catalyzed phosphoryl transfer.
Figure 7.
Third-order rate constants kact and substrate pKas for activation of HAdK1-catalyzed phosphoryl transfer by EA. Substitution of −F for −H at HP results in a 6-fold falloff in kact and reduction in substrate basicity that is consistent with βnuc = 0.3 on a two-point Brønsted plot for phosphoryl transfer. Substitution of −OH for −H at HP causes an increase in dianion basicity that should enhance nucleophilic reactivity; the large 27-fold falloff in kact is consistent with a large steric effect of the −OH on enzyme activation.
Activation Site
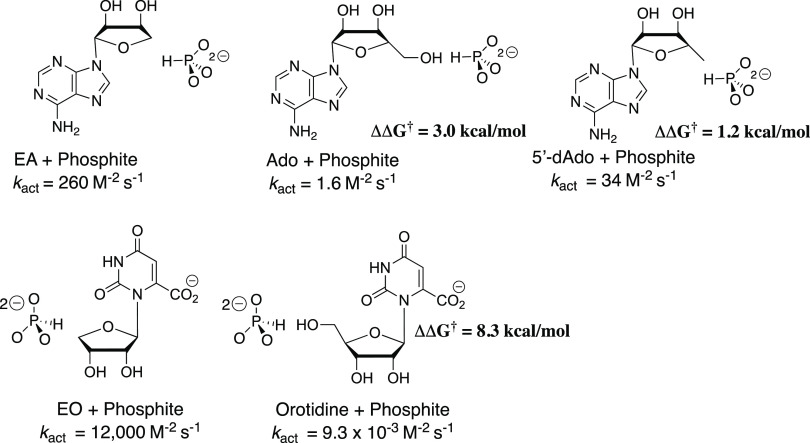
Chart 1 shows that the −CH2OH substituent at C-4′ of EA and 1-(β-D-erythrofuranosyl)orotate (EO), respectively, results in substantial decreases in kact (M–2 s–1) for activation of HAdK1- and OMPDC-catalyzed reactions, where EA and EO serve different functions as activator and substrate for these enzyme-catalyzed reactions. The -CH2OH substituent effects correspond to a 3.0 and 8.3 kcal mol–1 destabilization, respectively, of the transition states for the two enzyme-catalyzed reactions. This is consistent with a more severe steric crowding between the nucleoside 4′-CH2OH and phosphite dianion at the transition state for the OMPDC-catalyzed decarboxylation compared with the HAdK1-catalyzed phosphoryl transfer reaction. The small 1.2 kcal mol–1 effect of the nucleoside 5-CH3 group on the stability of the transition state for EA-activated phosphoryl transfer from ATP to phosphite dianion (Chart 1) is consistent with only weak steric interaction between the 5′-CH3 group and the phosphite dianion at the transition state for the HAdK1-activated reaction.
Chart 1. Third-Order Rate Constants, kact, for Activation of HAdK1- and OMPDC-Catalyzed Reactions of Truncated Substratesa.
a The activator is shown on the left and the substrate on the right. The values of ΔΔG† give the effect of the change in activator or substrate on ΔG† for the optimal reactions of EA or EO.
There is no detectable activation of HAdk1-catalyzed phosphoryl transfer from ATP to phosphite dianion by either 2′-dAdo (Figure S5) or 3′-dAdo (Figure S6). By comparison, the value of kcat/Km = 7.0 × 106 M–1 s–1 for HAdK1-catalyzed phosphoryl transfer from 1 mM ATP to AMP is nearly 80-fold larger than kcat/Km = 8.8 × 104 M–1 s–1 for phosphoryl transfer to 2′-dAMP. The value of kact = 0.02 M–1 s–1 calculated assuming similar 2′-OH substituent effects on kcat/Km = 7.0 × 106 M–1 s–1 for HAdK1-catalyzed phosphoryl transfer to AMP and on kact = 1.6 M–1 s–1 for activation by adenosine (Chart 1) would have been too small to measure by our experimental methods. We conclude that the interactions between HAdK1 and the 2′- and 3′-OH of Ado are used to activate the enzyme for catalysis of phosphoryl transfer from ATP to phosphite dianion, presumably by stabilizing the active closed form of HAdK1 (Scheme 2). We have likewise shown that binding interactions between OMPDC and the 2′- and 3′-OH of OMP are utilized to activate OMPDC for catalysis of decarboxylation of OMP.38,49
Intrinsic Adenine Binding Energy
There is an 11.8 kcal mol–1 stabilization of the transition state for enzyme-catalyzed phosphorylation of AMP by ATP by the adenine ring (Figure 6), but there is no detectable activation of HAdK1-catalyzed phosphoryl transfer from ATP to the truncated substrate R5P by 5 mM adenine. We propose that steric interactions between the HAdK1-bound adenine and R5P substrate pieces inhibit the binding of adenine in the conformation that shows strong interactions with the closed form of HAdK1.
Summary
The binding interactions between the phosphodianion of OMP and OMPDC provide a 12 kcal mol–1 stabilization of the transition state for decarboxylation at the orotate ring,24,37 while the binding interactions between the adenosine of AMP and HAdK1 provide a ≥14.7 kcal mol–1 stabilization of the transition state for phosphoryl transfer from ATP to AMP (Table 1). A substantial fraction of this stabilizing binding energy is observed for both OMPDC24,37,50 and HAdK1 (Table 2) in the absence of a covalent connection between the activator and reacting substrate pieces. The dianion activation site at OMPDC24 and the dianion substrate site at HAdK1 (Table 2) each accept several tetrahedral dianions as activators or substrates.
The similarity in the results of studies on the reactions of substrate pieces catalyzed by enzymes that stabilize the transition states for distinct decarboxylation and phosphoryl transfer reactions of nucleoside monophosphate substrates is a consequence of a striking similarity in the enzyme architecture,3,4,10,11,51 which enables utilization of the binding energy of nonreacting substrate fragments to drive large protein conformational changes.10 A significant difference is the effect of the addition of a 4′-CH2OH substituent at EO and EA on the stability of the transition state for reactions of the substrate pieces (Chart 1). The much larger 4′-CH2OH substituent effect on the OMPDC- compared with the HAdK1-catalyzed reaction is consistent with the notion that the closed-caged complex of substrate to OMPDC is tighter and more confined than for HAdK1 so that the 4′-CH2OH introduces more severe steric crowding at the complex to OMPDC.
The binding interactions between the phosphodianion of OMP and the Q215, Y217, and R235 side chains of OMPDC are used to drive closure of a phosphodianion gripper loop over enzyme-bound orotate ring.51 This conformational change is activating because the interactions between the gripper side chains and the phosphodianion-truncated substrate EO have essentially no effect on the stability of the transition state for the decarboxylation of the orotate ring.52,53 We have concluded that the phosphodianion-driven loop closure at OMPDC has the effect of placing the protein side chains at the orotate ring at positions that provide optimal stabilization of the decarboxylation transition state.6,7,10,11
The 11.8 kcal mol–1 adenine ring binding energy (Figure 6) is utilized to drive an enzyme conformational change,3,4 which places the protein side chains at positions around the bound ATP and AMP substrates that provide for optimal stabilization of the transition state for enzyme-catalyzed phosphoryl transfer. The side chains and other structural elements that function to drive this conformational change have not yet been identified.6,7 Studies on the effect of site-directed substitutions at adenylate kinase on enzyme activity for catalysis of phosphoryl transfer reactions of whole and truncated substrates, that follow protocols described in studies on TIM,54−56 OMPDC,16,49,57 and glycerol 3-phosphate dehydrogenase15,58,59 have the potential to provide original insight into the origin of the catalytic rate acceleration for HAdK1.
Conclusions
This work builds upon the results from earlier studies that highlight certain limitations for calculations directed toward the de novo design of active protein catalysts. These calculations assume that the binding energy from nonreacting substrate fragments is used exclusively for the stabilization of the Michaelis complex to substrate. They have been useful in guiding the design of active catalysts that provide large and, by some criteria, enzyme-like, transition state stabilization.60−63 However, these catalysts are primitive in comparison to enzymes that catalyze rapid and specific metabolic reactions. We have shown that the most proficient enzyme catalysts of metabolic reactions, such as AK, TIM, and OMPDC, use binding interactions with nonreacting fragments to construct the active catalyst “on the fly” by driving the conversion of inactive open proteins to active closed forms (Scheme 2). We suggest that an important goal for protein engineers, who are working to mimic Nature’s most proficient catalysts, should be to develop procedures to identify proteins that utilize substrate binding energy to drive conformational changes to form catalytically active protein–substrate cages, which provide a large stabilization of enzymatic transition states.
Glossary
Abbreviations
- Ado
adenosine
- 2′-dAdo
2′-deoxyadenosine
- 3′-dAdo
3′-deoxyadenosine
- 5′-dAdo
5′-deoxyadenosine
- AdK
adenylate kinase
- HAdK1
Homo sapiens adenylate kinase 1
- RAdK1
rabbit muscle adenylate kinase
- AMP
adenosine 5′-monophosphate
- ADP
adenosine 5′-diphosphate
- ATP
adenosine 5′-triphosphate
- BSA
bovine serum albumin
- EA
1-(β-d-erythrofuranosyl)adenine
- HP
phosphite dianion
- HPPi
isohypophosphate
- LDH
lactate dehydrogenase
- OMP
orotidine 5′-monophosphate
- OMPDC
orotidine 5′-monophosphate decarboxylase
- PK
pyruvate kinase
- R5P
ribose 5-phosphate
- SI
Supporting Information
- TEA
triethanolamine
- TIM
triosephosphate isomerase
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.2c00531.
Michaelis–Menten plot of vobs/[E] against [AMP] for RAdK-catalyzed phosphoryl transfer from ATP (Figure S1); Michaelis–Menten plot of vobs/[E] against [2′-dAMP] for HAdK1-catalyzed phosphoryl transfer from ATP (Figure S2); kinetic data for EA-activated RAdK-catalyzed phosphoryl transfer from ATP to FPO32– (Figure S3A,B); kinetic data for adenosine-activated HAdK-catalyzed phosphoryl transfer from ATP to HPO32– (Figure S4A,B); and the effect of increasing [2′-dAdo] (Figure S5) and [3′-dAdo] (Figure S6) on vobs/[E] for HAdK1-catalyzed phosphoryl transfer from ATP to phosphite dianion (PDF)
Accession Codes
Author Contributions
All authors have given approval to the final version of the manuscript.
The authors acknowledge the National Institutes of Health Grant GM134881 for support of this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Noda L. Adenosine Triphosphate-Adenosine Monophosphate Transphosphorylase III. Kinetic Studies. J. Biol. Chem. 1958, 232, 237–250. 10.1016/S0021-9258(18)70391-8. [DOI] [PubMed] [Google Scholar]
- Zeleznikar R. J.; Heyman R. A.; Graeff R. M.; Walseth T. F.; Dawis S. M.; Butz E. A.; Goldberg N. D. Evidence for compartmentalized adenylate kinase catalysis serving a high energy phosphoryl transfer function in rat skeletal muscle. J. Biol. Chem. 1990, 265, 300–311. 10.1016/S0021-9258(19)40230-5. [DOI] [PubMed] [Google Scholar]
- Kerns S. J.; Agafonov R. V.; Cho Y.-J.; Pontiggia F.; Otten R.; Pachov D. V.; Kutter S.; Phung L. A.; Murphy P. N.; Thai V.; Alber T.; Hagan M. F.; Kern D. The energy landscape of adenylate kinase during catalysis. Nat. Struct. Mol. Biol 2015, 22, 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzler-Wildman K. A.; Lei M.; Thai V.; Kerns S. J.; Karplus M.; Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature 2007, 450, 913–916. 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- Richard J. P.; Amyes T. L.; Goryanova B.; Zhai X. Enzyme architecture: on the importance of being in a protein cage. Curr. Opin. Chem. Biol. 2014, 21, 1–10. 10.1016/j.cbpa.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshel A.; Sharma P. K.; Kato M.; Xiang Y.; Liu H.; Olsson M. H. M. Electrostatic basis for enzyme catalysis. Chem. Rev. 2006, 106, 3210–3235. 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- Warshel A. Electrostatic Origin of the Catalytic Power of Enzymes and the Role of Preorganized Active Sites. J. Biol. Chem. 1998, 273, 27035–27038. 10.1074/jbc.273.42.27035. [DOI] [PubMed] [Google Scholar]
- Thomas J. A.; Koshland D. E. Jr. Competitive Inhibition by Substrate during Enzyme Action. Evidence for the Induced-fit Theory. J. Am. Chem. Soc. 1960, 82, 3329–3333. 10.1021/ja01498a025. [DOI] [Google Scholar]
- Koshland D. E. Jr. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proc. Natl. Acad. Sci. 1958, 44, 98–104. 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard J. P. Enabling Role of Ligand-Driven Conformational Changes in Enzyme Evolution. Biochemistry 2022, 61, 1533–1542. 10.1021/acs.biochem.2c00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard J. P. Protein Flexibility and Stiffness Enable Efficient Enzymatic Catalysis. J. Am. Chem. Soc 2019, 141, 3320–3331. 10.1021/jacs.8b10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amyes T. L.; Richard J. P. Specificity in transition state binding: The Pauling model revisited. Biochemistry 2013, 52, 2021–2035. 10.1021/bi301491r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard J. P. A Paradigm for Enzyme-Catalyzed Proton Transfer at Carbon: Triosephosphate Isomerase. Biochemistry 2012, 51, 2652–2661. 10.1021/bi300195b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez P. L.; Nagorski R. W.; Cristobal J. R.; Amyes T. L.; Richard J. P. Phosphodianion Activation of Enzymes for Catalysis of Central Metabolic Reactions. J. Am. Chem. Soc. 2021, 143, 2694–2698. 10.1021/jacs.0c13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristobal J. R.; Richard J. P. Glycerol-3-Phosphate Dehydrogenase: The K120 and K204 Side Chains Define an Oxyanion Hole at the Enzyme Active Site. Biochemistry 2022, 61, 856–867. 10.1021/acs.biochem.2c00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard J. P.; Amyes T. L.; Reyes A. C. Orotidine 5′-Monophosphate Decarboxylase: Probing the Limits of the Possible for Enzyme Catalysis. Acc. Chem. Res. 2018, 51, 960–969. 10.1021/acs.accounts.8b00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amyes T. L.; Malabanan M. M.; Zhai X.; Reyes A. C.; Richard J. P. Enzyme Activation Through the Utilization of Intrinsic dianion binding energy. Prot. Eng., Des. Sel. 2017, 30, 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jencks W. P.Binding energy, specificity, and enzymic catalysis: the Circe effect, Adv. Enzymol. Relat. Areas Mol. Biol. 1975, 43, 219–410, Adv Enzymol Relat Areas Mol Biol. [DOI] [PubMed] [Google Scholar]
- Wolfenden R.; Snider M. J. The Depth of Chemical Time and the Power of Enzymes as Catalysts. Acc. Chem. Res. 2001, 34, 938–945. 10.1021/ar000058i. [DOI] [PubMed] [Google Scholar]
- Radzicka A.; Wolfenden R. A proficient enzyme. Science 1995, 267, 90–93. 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- Knowles J. R.; Albery W. J. Perfection in enzyme catalysis: the energetics of triosephosphate isomerase. Acc. Chem. Res. 1977, 10, 105–111. 10.1021/ar50112a001. [DOI] [Google Scholar]
- Albery W. J.; Knowles J. R. Evolution of enzyme function and the development of catalytic efficiency. Biochemistry 1976, 15, 5631–5640. 10.1021/bi00670a032. [DOI] [PubMed] [Google Scholar]
- Fernandez P. L.; Richard J. P. Adenylate Kinase-Catalyzed Reaction of AMP in Pieces: Enzyme Activation for Phosphoryl Transfer to Phosphite Dianion. Biochemistry 2021, 60, 2672–2676. 10.1021/acs.biochem.1c00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A. C.; Zhai X.; Morgan K. T.; Reinhardt C. J.; Amyes T. L.; Richard J. P. The Activating Oxydianion Binding Domain for Enzyme-Catalyzed Proton Transfer, Hydride Transfer and Decarboxylation: Specificity and Enzyme Architecture. J. Am. Chem. Soc. 2015, 137, 1372–1382. 10.1021/ja5123842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spong K.; Amyes T. L.; Richard J. P. Enzyme Architecture: The Activating Oxydianion Binding Domain for Orotidine 5′-Monophophate Decarboxylase. J. Am. Chem. Soc. 2013, 135, 18343–18346. 10.1021/ja4107513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang W.-Y.; Amyes T. L.; Richard J. P. A Substrate in Pieces: Allosteric Activation of Glycerol 3-Phosphate Dehydrogenase (NAD+) by Phosphite Dianion. Biochemistry 2008, 47, 4575–4582. 10.1021/bi8001743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jencks W. P.; Regenstein J.. Ionization Constants of Acids and Bases. In Handbook of Biochemistry and Molecular Biology (Physical and Chemical Data), 3rd ed.; Fasman G. D., Ed.; CRC Press: Cleveland, OH, 1976; pp 305–351. [Google Scholar]
- Sheng X. R.; Li X.; Pan X. M. An Iso-random Bi Bi Mechanism for Adenylate Kinase. J. Biol. Chem. 1999, 274, 22238–22242. 10.1074/jbc.274.32.22238. [DOI] [PubMed] [Google Scholar]
- Khoo J. C.; Russell P. J. Isoenzymes of adenylate kinase in human tissue. Biochim. Biophys. Acta, Enzymol. 1972, 268, 98–101. 10.1016/0005-2744(72)90202-1. [DOI] [PubMed] [Google Scholar]
- Panayiotou C.; Solaroli N.; Karlsson A. The many isoforms of human adenylate kinases. Int. J. Biochem. Cell Biol. 2014, 49, 75–83. 10.1016/j.biocel.2014.01.014. [DOI] [PubMed] [Google Scholar]
- Adkar B. V.; Jana B.; Bagchi B. Role of Water in the Enzymatic Catalysis: Study of ATP + AMP → 2ADP Conversion by Adenylate Kinase. J. Phys. Chem. A 2011, 115, 3691–3697. 10.1021/jp104787s. [DOI] [PubMed] [Google Scholar]
- Arora K.; Brooks C. L. Large-scale allosteric conformational transitions of adenylate kinase appear to involve a population-shift mechanism. Proc. Natl. Acad. Sci. 2007, 104, 18496–18501. 10.1073/pnas.0706443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.; Liu M. S.; Ji B. Mapping the Dynamics Landscape of Conformational Transitions in Enzyme: The Adenylate Kinase Case. Biophys. J. 2015, 109, 647–660. 10.1016/j.bpj.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellinzoni M.; Haouz A.; Graña M.; Munier-Lehmann H.; Shepard W.; Alzari P. M. The crystal structure of Mycobacterium tuberculosisadenylate kinase in complex with two molecules of ADP and Mg2+ supports an associative mechanism for phosphoryl transfer. Protein Sci. 2006, 15, 1489–1493. 10.1110/ps.062163406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M. B.; Phillips G. N. Jr. Crystal structures of Bacillus stearothermophilus adenylate kinase with bound Ap5A, Mg2+ Ap5A, and Mn2+ Ap5A reveal an intermediate lid position and six coordinate octahedral geometry for bound Mg2+ and Mn2+. Proteins: Struct., Funct., Genet. 1998, 32, 276–288. . [DOI] [PubMed] [Google Scholar]
- Jencks W. P. On the attribution and additivity of binding energies. Proc. Natl. Acad. Sci. 1981, 78, 4046–4050. 10.1073/pnas.78.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amyes T. L.; Richard J. P.; Tait J. J. Activation of orotidine 5′-monophosphate decarboxylase by phosphite dianion: The whole substrate is the sum of two parts. J. Am. Chem. Soc. 2005, 127, 15708–15709. 10.1021/ja055493s. [DOI] [PubMed] [Google Scholar]
- Reyes A. C.; Amyes T. L.; Richard J. P. Enzyme Architecture: Erection of Active Orotidine 5′-Monophosphate Decarboxylase by Substrate-Induced Conformational Changes. J. Am. Chem. Soc 2017, 139, 16048–16051. 10.1021/jacs.7b08897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassila J. K.; Zalatan J. G.; Herschlag D. Biological Phosphoryl-Transfer Reactions: Understanding Mechanism and Catalysis. Annu. Rev. Biochem. 2011, 80, 669–702. 10.1146/annurev-biochem-060409-092741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N.; Mo Y.; Gao J.; Pai E. F. Electrostatic stress in catalysis: structure and mechanism of the enzyme orotidine monophosphate decarboxylase. Proc. Natl. Acad. Sci. 2000, 97, 2017–2022. 10.1073/pnas.050417797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rindfleisch S.; Krull M.; Uranga J.; Schmidt T.; Rabe von Pappenheim F.; Kirck L. L.; Balouri A.; Schneider T.; Chari A.; Kluger R.; Bourenkov G.; Diederichsen U.; Mata R. A.; Tittmann K. Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5′-monophosphate decarboxylase catalysis. Nat. Catal. 2022, 5, 332–341. 10.1038/s41929-022-00771-w. [DOI] [Google Scholar]
- Goryanova B.; Amyes T. L.; Richard J. P. Role of the Carboxylate in Enzyme-Catalyzed Decarboxylation of Orotidine 5′-Monophosphate: Transition State Stabilization Dominates Over Ground State Destabilization. J. Am. Chem. Soc. 2019, 141, 13468–13478. 10.1021/jacs.9b04823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardi-Kilshtain A.; Doron D.; Major D. T. Quantum and Classical Simulations of Orotidine Monophosphate Decarboxylase: Support for a Direct Decarboxylation Mechanism. Biochemistry 2013, 52, 4382–4390. 10.1021/bi400190v. [DOI] [PubMed] [Google Scholar]
- Warshel A.; Strajbl M.; Villa J.; Florian J. Remarkable Rate Enhancement of Orotidine 5′-Monophosphate Decarboxylase Is Due to Transition-State Stabilization Rather Than to Ground-State Destabilization. Biochemistry 2000, 39, 14728–14738. 10.1021/bi000987h. [DOI] [PubMed] [Google Scholar]
- Matsunaga Y.; Fujisaki H.; Terada T.; Furuta T.; Moritsugu K.; Kidera A. Minimum Free Energy Path of Ligand-Induced Transition in Adenylate Kinase. PLoS Comp. Biol. 2012, 8, e1002555 10.1371/journal.pcbi.1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jencks W. P. A primer for the Bema Hapothle. An empirical approach to the characterization of changing transition-state structures. Chem. Rev. 1985, 85, 511–527. 10.1021/cr00070a001. [DOI] [Google Scholar]
- Sheu K.-F. R.; Frey P. A. Enzymatic and 31P Nuclear Magnetic Resonance Study of Adenylate Kinase-catalyzed Stereospecific Phosphorylation of Adenosine 5′-Phosphorothioate. J. Biol. Chem. 1977, 252, 4445–4448. [PubMed] [Google Scholar]
- Song B.; Sigel R. K. O.; Sigel H. Acid-Base Properties of Adenosine 5′-O-Thiomonophosphate in Aqueous Solution. Chem - Eur. J. 1997, 3, 29–33. 10.1002/chem.19970030106. [DOI] [Google Scholar]
- Cristobal J. R.; Brandão T. A. S.; Reyes A. C.; Richard J. P. Protein–Ribofuranosyl Interactions Activate Orotidine 5′-Monophosphate Decarboxylase for Catalysis. Biochemistry 2021, 60, 3362–3373. 10.1021/acs.biochem.1c00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryanova B.; Amyes T. L.; Gerlt J. A.; Richard J. P. OMP Decarboxylase: Phosphodianion Binding Energy Is Used To Stabilize a Vinyl Carbanion Intermediate. J. Am. Chem. Soc. 2011, 133, 6545–6548. 10.1021/ja201734z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B. G.; Hassell A. M.; Wolfenden R.; Milburn M. V.; Short S. A. Anatomy of a proficient enzyme: the structure of orotidine 5′-monophosphate decarboxylase in the presence and absence of a potential transition state analog. Proc. Natl. Acad. Sci. 2000, 97, 2011–2016. 10.1073/pnas.030409797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amyes T. L.; Ming S. A.; Goldman L. M.; Wood B. M.; Desai B. J.; Gerlt J. A.; Richard J. P. Orotidine 5′-monophosphate decarboxylase: Transition state stabilization from remote protein-phosphodianion interactions. Biochemistry 2012, 51, 4630–4632. 10.1021/bi300585e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman L. M.; Amyes T. L.; Goryanova B.; Gerlt J. A.; Richard J. P. Enzyme Architecture: Deconstruction of the Enzyme-Activating Phosphodianion Interactions of Orotidine 5′-Monophosphate Decarboxylase. J. Am. Chem. Soc. 2014, 136, 10156–10165. 10.1021/ja505037v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai X.; Reinhardt C. J.; Malabanan M. M.; Amyes T. L.; Richard J. P. Enzyme Architecture: Amino Acid Side-Chains That Function To Optimize the Basicity of the Active Site Glutamate of Triosephosphate Isomerase. J. Am. Chem. Soc. 2018, 140, 8277–8286. 10.1021/jacs.8b04367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai X.; Amyes T. L.; Richard J. P. Role of Loop-Clamping Side Chains in Catalysis by Triosephosphate Isomerase. J. Am. Chem. Soc. 2015, 137, 15185–15197. 10.1021/jacs.5b09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai X.; Amyes T. L.; Richard J. P. Enzyme Architecture: Remarkably Similar Transition States for Triosephosphate Isomerase-Catalyzed Reactions of the Whole Substrate and the Substrate in Pieces. J. Am. Chem. Soc. 2014, 136, 4145–4148. 10.1021/ja501103b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S.; Nguyen B. T.; Richard J. P.; Kluger R.; Gao J. Origin of Free Energy Barriers of Decarboxylation and the Reverse Process of CO2 Capture in Dimethylformamide and in Water. J. Am. Chem. Soc. 2021, 143, 137–141. 10.1021/jacs.0c12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R.; Cristobal J. R.; Gong N. J.; Richard J. P. Hydride Transfer Catalyzed by Glycerol Phosphate Dehydrogenase: Recruitment of an Acidic Amino Acid Side Chain to Rescue a Damaged Enzyme. Biochemistry 2020, 59, 4856–4863. 10.1021/acs.biochem.0c00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristobal J. R.; Reyes A. C.; Richard J. P. The Organization of Active Site Side Chains of Glycerol-3-phosphate Dehydrogenase Promotes Efficient Enzyme Catalysis and Rescue of Variant Enzymes. Biochemistry 2020, 59, 1582–1591. 10.1021/acs.biochem.0c00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovelock S. L.; Crawshaw R.; Basler S.; Levy C.; Baker D.; Hilvert D.; Green A. P. The road to fully programmable protein catalysis. Nature 2022, 606, 49–58. 10.1038/s41586-022-04456-z. [DOI] [PubMed] [Google Scholar]
- Weitzner B. D.; Kipnis Y.; Daniel A. G.; Hilvert D.; Baker D. A computational method for design of connected catalytic networks in proteins. Protein Sci. 2019, 28, 2036–2041. 10.1002/pro.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kries H.; Blomberg R.; Hilvert D. De novo enzymes by computational design. Curr. Opin. Chem. Biol. 2013, 17, 221–228. 10.1016/j.cbpa.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Privett H. K.; Kiss G.; Lee T. M.; Blomberg R.; Chica R. A.; Thomas L. M.; Hilvert D.; Houk K. N.; Mayo S. L. Iterative approach to computational enzyme design. Proc. Natl. Acad. Sci. 2012, 109, 3790–3795. 10.1073/pnas.1118082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.