Abstract
Objective
Novel biomarkers related to main clinical hallmarks of Chronic obstructive pulmonary disease (COPD), a heterogeneous disorder with pulmonary and extra-pulmonary manifestations, were investigated by profiling the serum levels of 1305 proteins using Slow Off-rate Modified Aptamers (SOMA)scan technology.
Methods
Serum samples were collected from 241 COPD subjects in the multicenter French Cohort of Bronchial obstruction and Asthma to measure the expression of 1305 proteins using SOMAscan proteomic platform. Clustering of the proteomics was applied to identify disease subtypes and their functional annotation and association with key clinical parameters were examined. Cluster findings were revalidated during a follow-up visit, and compared to those obtained in a group of 47 COPD patients included in the Melbourne Longitudinal COPD Cohort.
Results
Unsupervised clustering identified two clusters within COPD subjects at inclusion. Cluster 1 showed elevated levels of factors contributing to tissue injury, whereas Cluster 2 had higher expression of proteins associated with enhanced immunity and host defense, cell fate, remodeling and repair and altered metabolism/mitochondrial functions. Patients in Cluster 2 had a lower incidence of exacerbations, unscheduled medical visits and prevalence of emphysema and diabetes. These protein expression patterns were conserved during a follow-up second visit, and substanciated, by a large part, in a limited series of COPD patients. Further analyses identified a signature of 15 proteins that accurately differentiated the two COPD clusters at the 2 visits.
Conclusions
This study provides insights into COPD heterogeneity and suggests that overexpression of factors involved in lung immunity/host defense, cell fate/repair/ remodelling and mitochondrial/metabolic activities contribute to better clinical outcomes. Hence, high throughput proteomic assay offers a powerful tool for identifying COPD endotypes and facilitating targeted therapies.
Introduction
Chronic obstructive pulmonary disease (COPD) is a difficult to treat disease, characterized by irreversible airflow obstruction and often associated with lung emphysema. These events result from persistent lung inflammation and tissue remodeling that leads to respiratory insufficiency and functional disability [1]. Cigarette smoke, but also genetic/epigenetic alterations leading to lung accelerated aging, have been shown to predispose individuals to COPD [2–4]. However, whether markers of these processes are detected in peripheral blood of patients and relate to clinical traits of the disease remains elusive.
COPD manifests in different clinical phenotypes according to the degree of airflow obstruction, frequency of acute exacerbations, emphysema and airway inflammation [5]. In addition, pulmonary and cardio-metabolic comorbidities may impact COPD prognosis and therapeutic management [6]. Although numerous studies have addressed analytical approaches for identifying novel COPD endotypes underlying these clinical phenotypes [7–14], those approaches did not include the measurement of biomarkers of lung injury/repair and of pulmonary, or extra-pulmonary co-morbidities, these factors being major contributors of COPD onset and progression [6].
Given the heterogeneity in COPD pathophysiology and clinical presentation, robust and wide analytic methodologies are required to characterize novel endotypes. To this end, High-throughput proteomic technology, Slow Off-rate Modified Aptamers (SOMA)scan, has been developed for quantitatively assessing hundreds of proteins specifically in serum and plasma samples with high sensitivity and specificity [15, 16]. This platform has previously proved useful in being able to measure simultaneously large numbers of proteins in different organ diseases, including lung [17, 18].
The current study was aimed at identifying novel biomarkers related to main clinical hallmarks of COPD, to its most frequent co-morbidities and to medication. This was performed by profiling the serum levels of 1305 proteins by SOMAscan in 241 patients included in the multicenter prospective French Cohort of Bronchial Obstruction and Asthma (COBRA) [19, 20]. Unsupervised clustering of differentially expressed proteins combined to gene ontology (GO) pathway analysis, classified COPD patients into clinical clusters. Findings were validated in another independent series of COPD patients enrolled in the Melbourne Longitudinal COPD Cohort (MLCC) [21]. Further analyses were carried out for determining a short protein fingerprint linked to GO biological processes, as a useful tool to discriminate patient clusters, for monitoring its stability at a follow-up and, ultimately, for identifying potential novel therapeutic targets to adapt clinical managements.
Methods
Study populations
Stable COPD patients (n = 241) were included in the COBRA cohort [19, 20] (CPP Ile-de-France I Ethics Committee, n° 09–11962) and written informed consent was obtained before inclusion (S1 Table in S1 File). Serum aliquots were collected for the measurement of the levels of hemoglobin and of C reactive protein (CRP) and for SOMAscan analyses. The evolution of the clinical outcomes and of proteomic profiles was assessed in 163 COPD patients out of the 241 having a follow up visit 7.5 ± 6.6 months (mean ± SD) after inclusion (S1 Table in S1 File). SOMAscan data were validated in a separate series of 47 COPD patients originating from the MLCC [21] (S2 Table in S1 File). Serum samples from n = 50 control healthy subjects were used for comparisons.
SOMAscan analysis
A total of 1305 analytes were quantified in patient serum using the SOMAscan high throughput proteomic assay (SomaLogic, Boulder, CO, USA) at National Jewish Health (Denver, Colorado, United States of America) [22]. The raw SOMAscan data were standardized by four steps: hybridization normalization, place scaling, median signal normalization and calibration, according to manufacturer’s instructions (http://somalogic.com/wp-content/uploads/2017/06/SSM-071-Rev-0-Technical-Note-SOMAscan-Data-Standardization.pdf). The normalized expression values were then log2 transformed for downstream analyses. Proteins were described in the manuscript using SOMAscan target names.
Data processing and bioinformatical approaches
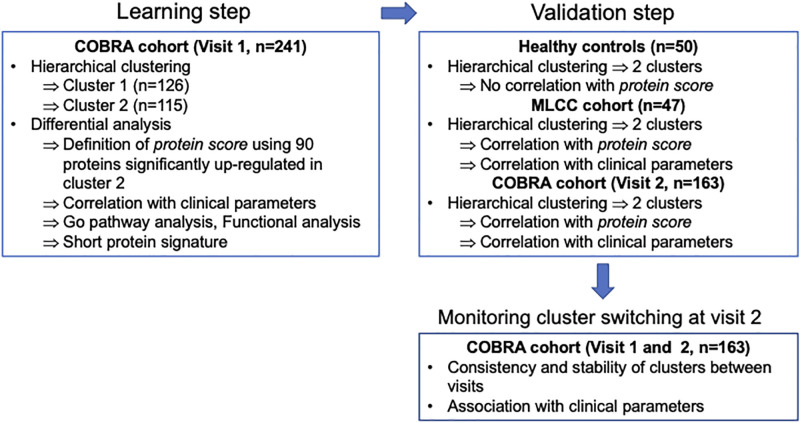
The bioinformatics approaches are summarized in Fig 1 and follow a typical machine learning approach with a learning step and a validation step. For each cohort consisting exclusively of either diseased or healthy patients, hierarchical clustering was performed using R, based on the top 10% proteins by expression variation, to first segment the population. Differential expression analyses between the detected clusters were performed using the Limma package [23]. Proteins with fold change >1.5 and false discovery rate (FDR) <0.05 were defined as significant. Functional analyses of the differentially expressed proteins were performed using the GO pathway database.
Fig 1. Study flow diagram.
Stable COPD patients (n = 241) were included in the COBRA cohort at visit 1 and serum SOMAscan analyses were performed. Two COPD clusters, Cluster 1 (n = 126) and Cluster 2 (n = 115) with distinct protein expression patterns were identified through unsupervised hierarchical clustering. The differentially expressed proteins in Cluster 2 were submitted to GO pathways analysis for further functional enrichment study, then median expression values from 90 proteins up-regulated in Cluster 2 (defined as “protein scores”) were generated for each COPD cluster. Association studies were performed between COPD clusters or corresponding protein and clinical parameters. The evolution of the clinical outcomes and of proteomic profiles was assessed in 163 COPD patients out of the 241 having a follow up visit (visit 2) after inclusion (visit 1). Expression patterns associated with Cluster 1 (n = 97) and Cluster 2 (n = 66) were verified in COBRA cohort at visit 2 on the basis of protein scores previously defined at visit 1. Protein signatures representative of each COPD cluster were identified to monitor clinical patient stability at visit 2. SOMAscan data obtained in the COBRA cohort were validated in a separate group of 47 COPD patients originating from the MLCC cohort. Serum samples from n = 50 control healthy subjects were included for comparisons.
During the learning step, hierarchical clustering was performed for COPD patients from the COBRA cohort at the time of inclusion (n = 241). Two COPD clusters: Cluster 1 (n = 126) and Cluster 2 (n = 115) with different protein expression patterns were identified. Using the 10% of proteins that differed most between the two clusters, the protein score for each patient was defined as the median expression of 90 proteins that were upregulated in Cluster 2. In addition, a short signature consisting of 15 proteins WAS associated with each cluster. Cluster 2-associated biomarkers were identified by sorting 10% of the most differentially expressed proteins representative of highly enriched pathways from GO pathway analysis and exhibiting the highest odd-ratios (OD) within their corresponding pathways; Cluster-1 associated biomarkers included top four of the 6 up-regulated biomarkers after exclusion of C3b that failed to show any differential expression in the MLCC cohort. Association studies between COPD clusters or corresponding proteins and clinical parameters were performed.
During the validation step, the reproducibility of the clustering patterns found in the learning step was assessed in 163 COPD patients of the 241 patients from the COBRA cohort who had a follow-up visit (visit 2) after inclusion, in 47 COPD patients from the MLCC cohort, and in n = 50 healthy subjects, included in the analysis as negative controls. Two main clusters were identified for each data set by hierarchical clustering. Importantly, clustering was based on all proteins (i.e., significant proteins found at visit 1 in the COBRA cohort were not considered) to ensure that our validation approach was agnostic to the results found at visit 1. We then tested whether clusters found in subsequent clusters were significantly different on the basis of the protein score defined in the learning step. In addition, we identified differentially expressed proteins and evaluated their enrichment with respect to significant proteins found in the learning step.
Finally, the short protein signature identified during the learning step was used to monitor the evolution of patients from the COBRA cohort between visit 1 and visit 2.
Data processing and statistical analyses are described in details in the S1 File.
Results
Subject analyzed
GOLD stage distribution, smoking habits, respiratory function, and medication were comparable in COPD patients included in the COBRA cohort, when comparing visit 1 and visit 2 (S1 Table in S1 File). In contrast, a significant reduction in the incidence of patients having exacerbations (p = 0.003) was observed at visit 2, as compared to visit 1 (S1 Table in S1 File). The remaining 78 COPD patients having only visit 1 had less severe disease than the 163 having 2 visits. This was attested by their higher distribution into GOLD 1 group, values of % predicted post-bronchodilator forced expiratory volume in one second (FEV1) and FEV1/forced vital capacity (FVC), transfer factor of the lung for carbon monoxide (DLCO), unscheduled medical visits, oral corticosteroid (OCS) use, but similar onset and number of exaberbations and comorbidities (S1 Table in S1 File).
The 47 COPD patients from the MLCC cohort were more severe and symptomatic than those included in the COBRA cohort, in terms of GOLD stage, airflow obstruction, incidence of cough, treatments with muscarinic antagonists, long-term inhaled steroids (ICS) and oxygen therapy (S1 and S2 Tables in S1 File).
Identification of two COPD subtypes in association with distinct biological hallmarks
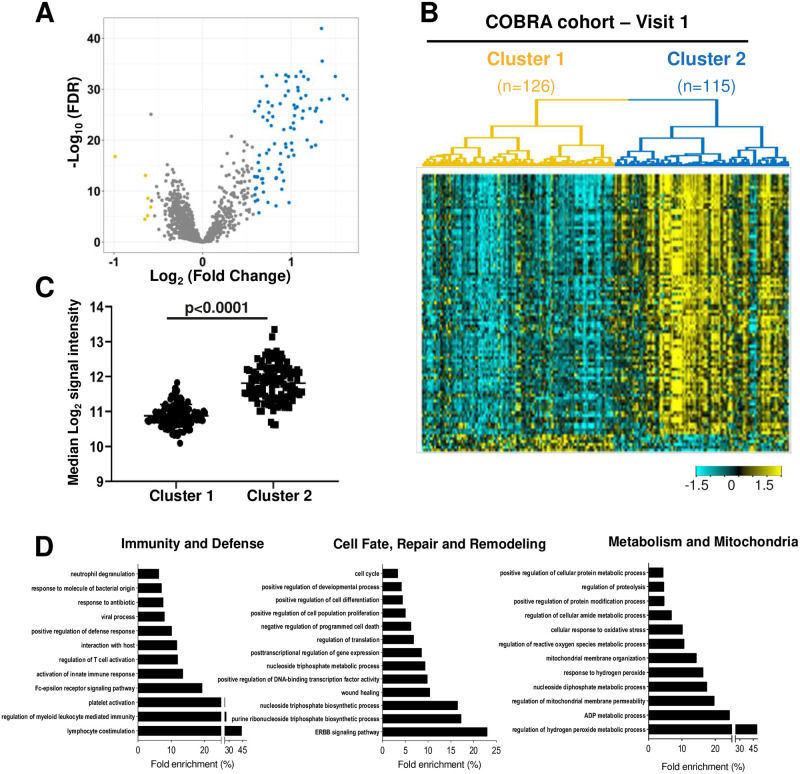
Hierarchical clustering, with the top 10% proteins by expression variation, revealed two COPD subtypes (126 and 115 patients in Clusters 1 and 2, respectively) within the 241 patients from the COBRA cohort at visit 1. A total of 96 proteins were differentially expressed between the two clusters (fold change >1.5 and FDR<0.05). Of these, 6 and 90 proteins were significantly up-regulated, in Clusters 1 and 2 respectively (Fig 2A and 2B, Table 1 and S3 Table in S1 File). Evaluation of the protein score (median expression) of the 90 proteins associated with Cluster 2, confirmed this significantly elevated levels (Fig 2C).
Fig 2. Two COPD patient Clusters show differentially expressed proteins enriched for distinct biological pathways.
(A) Volcano plot showing differentially expressed proteins between Cluster 1 and Cluster 2. X axis corresponds to log2 (fold change) and Y axis corresponds to -log10 (FDR). Yellow indicates proteins with significant downregulation and blue indicates those with significant upregulation (Fold change >1.5 and FDR <0.05 for both comparisons); (B) Heatmap showing the expression pattern of differentially expressed proteins between the two clusters. (C) Median expression of 90 proteins associated with Cluster 2; (D) Top GO pathways comparing Cluster 2 versus Cluster 1, regrouped in three core functions defined as “Immunity and defense”, “Cell fate, repair and remodeling”, and “Metabolism and mitochondria”.
Table 1. Biological classification of the top 90 proteins enriched in Cluster 2 COPD patients.
| Biological processes (originating from the GO database) | Proteins |
|---|---|
| • ERBB signaling pathway | AREG, GRB2 adapter protein, SRCN1, PDPK1, PKC-A, SHC1, MK01 |
| • Wound healing | PKC-B-II, VAV, SRCN1, LYN, SMAD2, LYNB, Caspase-3, PDPK1, RAC1, Haemoglobin, METAP1, PRKACA, annexin I, PKC-A, FYN, 14-3-3 protein ζ/δ, PTP-1C, GPVI, ERK-1, NCC27, MK01 |
| • Posttranscriptional regulation of gene expression | eIF-4H, 14-3-3 protein β/α, 14-3-3 protein ζ/δ, GAPDH, PKC-A, RPS6KA3, hnRNP A2/B1, ERK-1, MK01, eIF-5A-1 |
| • Negative regulation of programmed cell death | SRCN1, NDP kinase B, Caspase-3, PDPK1, CK2-A1:B, Sphingosine kinase 1, α-Synuclein, annexin I, PKC-A, RPS6KA3, UFM1, FYN, 14-3-3 protein ζ/δ, PPID, TCTP, PA2G4, SHC1, BAD, eIF-5A-1, CD40 ligand, Ubiquitin+1, Lactoferrin, Azurocidin, STAT3 |
| • Positive regulation of cell proliferation | AREG, FGF16, SHC1, CK2-A1:B, PTP-1C, BAD, MK01, eIF5A-1, STAT3 |
| • Positive regulation of cell differentiation | BTK, LYN, SMAD2, LYNB, NDP kinase B, RAC1, annexin I, PKC-A, RPS6KA3, FYN, PA2G4, BAD, CPNE1, NCC27, eIF-5A-1, Lactoferrin |
| • Positive regulation of developmental process | BTK, PKC-B-II, SRCN1, LYN, SMAD2, LYNB, NDP kinase B, PDPK1, RAC1, Sphingosine kinase 1, annexin I, PKC-A, RPS6KA3, FYN, HXK2, PA2G4, BAD, CPNE1, NCC27, eIF5A-1, Lactoferrin, STAT3 |
| • Cell cycle | FER, PKC-B-II, IMB1, SRCN1, NSF1C, SBDS, CK2-A1:B, IF4G2, PRKACA, PKC-A, RPS6KA3, RAN, 41, PTP-1C, PA2G4, UBC9, ERK-1, MK01 |
| • Regulation of myeloid leukocyte mediated immunity | BTK, FER, LYN, LYNB, PDPK1, ARGI1, Sorting nexin 4 |
| • Fce receptor signaling pathway | BTK, FER, VAV, GRB2 adapter protein, LYN, LYNB, PDPK1, RAC1, SHC1, ERK-1, MK01 |
| • Activation of innate immune response | BTK, SP-D, SRCN1, LYN, LYNB, PDPK1, UBE2N, PRKACA, RPS6KA3, FYN, Ubiquitin+1 |
| • Regulation of T cell activation | CD40 ligand, CSK, SRCN1, Caspase-3, PDPK1, PTP-1C, DUSP3 |
| • Positive regulation of defense response | BTK, SP-D, VAV, SRCN1, LYN, LYNB, PDPK1, UBE2N, α-Synuclein, PRKACA, RPS6KA3, FYN, ARGI1, ERK-1, Ubiquitin+1, Lactoferrin, Sorting nexin 4 |
| • Neutrophil degranulation | IMB1, NDP kinase B, CK2-A1:B, RAC1, Haemoglobin, IMDH1, PTP-1C, PA2G4, ARGI1, Cyclophilin A, CPNE1, BPI, MK01, Lactoferrin, Azurocidin |
| • ADP metabolic process | GAPDH, Myokinase, HXK2, Triosephosphate isomerase, BAD |
| • Mitochondrial membrane organisation and permeability | Cyclophilin F, α-Synuclein, 14-3-3 protein ζ/δ, HXK2, BAD, STAT3, 14-3-3 protein β/α |
| • Cellular response to oxidative stress | Carbonic anhydrase XIII, FER, SRCN1, NDP kinase B, α-Synuclein, annexin I, ARGI1, ERK-1, MK01 |
| • Positive regulation of protein modification process | AREG, CSK, SRCN1, LYN, LYNB, PDPK1, CK2-A1:B, RAC1, Sphingosine kinase 1, UBE2N, α-Synuclein, PRKACA, PKC-A, FYN, UBC9, EDAR, SHC1, ERK-1, MK01, CD40 ligand, Ubiquitin+1, Lactoferrin, Azurocidin, STAT3, Ubiquitin+1 |
| • Cellular response to environmental stimulus | H2A3, GRB2 adapter protein |
| • Actin filament organisation | Tropomyosin 4, GRB2 adapter protein, FER |
41: Protein 4.1; AREG: Amphiregulin; ARGI1: Arginase-1; BAD: Bcl2-associated agonist of cell death; BPI: Bactericidal permeability-increasing protein; BTK: Tyrosine-protein kinase BTK; CK2A1:B: Casein kinase II 2-alpha:2-beta haeterotetramer; CPNE1: Copine1; CSK: Tyrosine-protein kinase CSK; PPID: Peptidyl-prolyl cis-trans isomerase D or Cyclophilin D; Cyclophilin F; DUS3: Dual specificity protein phosphatase 3; EDAR: Tumor necrosis factor receptor superfamily member EDAR; eIF-4H: Eukaryotic translation initiation factor 4H; eIF-5A-1: Eukaryotic translation initiation factor 5A-1; ERK-1: Mitogen-activated protein kinase 3 (MAPK3); FER: Tyrosine-protein kinase Fer; FGF-16: Fibroblast growth factor 16; FYN: Tyrosine-protein kinase Fyn; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; GPVI: Platelet glycoprotein VI; GRB2 adapter protein: Growth factor receptor-bound protein 2; hnRNP A2/B1: Heterogeneous nuclear ribonucleoproteins A2/B1; HXK2: Hexokinase-2; IF4G2: Eukaryotic translation initiation factor 4 gamma 2; IMB1: Importin subunit β-1; IMDH1: Inosine-5’-monophosphate dehydrogenase 1; LYN: Tyrosine-protein kinase Lyn; LYNB: Tyrosine-protein kinase Lyn, isoform B; MK01: Mitogen-activated protein kinase 1; M2-PK: Pyruvate kinase PKM; METAP1: Methionine aminopeptidase 1; NCC27 or CLIC1: Chloride intracellular channel protein 1; NDP kinase B: Nucleoside diphosphate kinase B; NSF1C: NSFL1 cofactor p47; PA2G4: Proliferation-associated protein 2G4; PDPK1: 3-phosphoinositide-dependent protein kinase 1; PRKACA: cAMP-dependent protein kinase catalytic subunit α; PKC-A: Protein kinase C, α type; PKC-B-II: Protein kinase C, β type (splice variant β-II); PTP-1C or PTPN6: Tyrosine-protein phosphatase non-receptor type 6; RAC1: Ras-related C3 botulinum toxin substrate 1; RAN: GTP-binding nuclear protein Ran; RPS6KA3: Ribosomal protein S6 kinase α3; SBDS: Ribosome maturation protein SBDS; SHC1: SHC-transforming protein 1; SMAD2: Mothers against decapentaplegic homolog 2; SP-D: Pulmonary surfactant associated protein D; SRCN1 or SRC: Proto-oncogene tyrosine-protein kinase Src; STAT3: Signal transducer and activator of transcription 3; SUMO3: Small ubiquitin-related modifier 3; TCTP: Translationally-controlled tumour protein; UBC9: SUMO-conjugating enzyme UBC9; UBE2N: Ubiquitin-conjugating enzyme E2 N; UFM1: Ubiquitin-fold modifier 1; VAV: Proto-oncogene vav.
In bold are indicated the proteins belonging to the short signature.
To further ascertain the biological processes and molecular functions associated with each COPD subtype, we performed GO pathway analysis on the Top90 up-regulated proteins in Cluster 2 (FDR<0.0001). Our data highlighted the enrichement of this cluster in hallmarks of lung immunity/host defense, cell fate/repair/remodelling and mitochondrial/metabolic activities (Fig 2D, Table 1 and S3 Table in S1 File).
Clinical parameters in Cluster 1- and Cluster-2-related COPD patients
We then examined whether COPD patients from the two clusters showed distinct clinical characteristics by considering the parameters describing the COBRA cohort at visit 1 (Table 2). Cluster 2 patients had a lower incidence (p = 0.01) and number (p = 0.02) of exacerbations and of unscheduled medical visits (p = 0.0002) in the previous year, compared to those in Cluster 1 (Table 2). The proportion of COPD patients with emphysema, as assessed by CT scan and/or by the measurement DLCO, was also lower in Cluster 2 than in Cluster 1 (p<0.0001) (Table 2). Further, Cluster 2 patients had a lower prevalence of hypertension (p = 0.02), diabetes (p = 0.009) and obstructive sleep apnea (p = 0.04) (Table 2). In addition, the proportion of patients requiring long-acting β2-agonists (LABA) alone was significantly lower in Cluster 2 than in Cluster 1 (p = 0.007) and this was accompanied by a trend towards a lower rate of patients treated with LABA, in combination with long-lasting muscarinic antagonists (LAMA) (p = 0.06) (Table 2). In contrast, the incidence of ICS use, alone or in combination with LABA and LAMA, was not significantly different between the two clusters (p = 0.79) (Table 2).
Table 2. Differences in clinical characteristics between COPD patients of Cluster 1 and Cluster 2 at visit 1.
| Parameter | Cluster 1—number * | Cluster 2—number * | Values in Cluster 1 | Values in Cluster 2 | p value ** |
|---|---|---|---|---|---|
| Male sex—no. (%) | 126 | 115 | 82 (65) | 82 (71) | |
| Age (years) | 126 | 115 | 63.7 ± 9.7 | 62.5 ± 9.9 | 0.34 |
| Caucasian origin—no. (%) | 125 | 114 | 115 (91) | 101 (88) | 0.39 |
| GOLD stages | |||||
| GOLD I—no. (%) | 125 | 114 | 23 (18) | 31 (27) | 0.12 |
| GOLD II—no. (%) | 125 | 114 | 49 (39) | 37 (32) | 0.28 |
| GOLD III—no. (%) | 125 | 114 | 29 (23) | 24 (21) | 0.76 |
| GOLD IV—no. (%) | 125 | 114 | 23 (18) | 21 (18) | 1.00 |
| Body Mass Index (kg per m2) | 126 | 115 | 27.1 ± 6.8 | 25.8 ± 5.1 | 0.09 |
| Smoking history | |||||
| Never smokers—no. (%) | 126 | 115 | 1 (1) | 7 (6) | 0.02 |
| Active smokers—no. (%) | 126 | 115 | 46 (37) | 38 (33) | 0.59 |
| Biology | |||||
| Blood leukocytes (no. per mm3) | 97 | 68 | 7400 (6450–12500) | 7600 (6525–8875) | 0.42 |
| Blood eosinophils (no. per mm3) | 97 | 68 | 168 (118–228) | 158 (88–240) | 0.80 |
| With blood eosinophils ≥ 300 per mm3—no. (%) | 97 | 68 | 15 (15) | 12 (18) | 0.83 |
| Hemoglobin—g per deciliter | 84 | 31 | 14.6 (13.7–15.2) | 14.1 (13.2–15.7) | 0.74 |
| CRP—mg per Liter | 85 | 27 | 4.0 (3.0–8.9) | 5.0 (2.3–7.5) | 0.79 |
| With CRP ≥ 3 mg per Liter—no. (%) | 85 | 27 | 65 (76) | 23 (85) | 0.43 |
| Respiratory function | |||||
| Pre-bronchodilator FEV1 (% predicted) | 122 | 76 | 58.0 ± 21.1 | 61.3 ± 27.3 | 0.36 |
| Post-bronchodilator FEV1 (% predicted) | 111 | 95 | 60.8 ± 20.9 | 66.0 ± 28.9 | 0.22 |
| Pre-bronchodilator FEV1 / FVC (% predicted) | 122 | 80 | 52.4 ± 13.7 | 54.2 ± 16.9 | 0.51 |
| Post-bronchodilator FEV1 / FVC (% predicted) | 113 | 96 | 52.2 ± 14.1 | 55.3 ± 17.6 | 0.21 |
| FRC—% | 43 | 81 | 138.9 ± 49.3 | 137.2 ± 31.2 | 0.82 |
| RV—% | 49 | 86 | 160.4 ± 61.1 | 154.1 ± 48.1 | 0.51 |
| TLC—% | 52 | 85 | 114.0 ± 23.1 | 113.6 ± 16.5 | 0.92 |
| DLCO—% | 42 | 73 | 54.4 ± 22.6 | 61.7 ± 20.9 | 0.08 |
| Symptoms | |||||
| With emphysema no. (%) | 126 | 115 | 65 (52) | 27 (31) | < 0.0001 |
| With exacerbations in the previous 12 months—no. (%) | 126 | 115 | 77 (61) | 51 (44) | 0.01 |
| Number of exacerbations in the previous 12 months—no. | 126 | 115 | 1.61 ± 0.25 | 1.24 ± 0.20 | 0.02 |
| With unscheduled medical visits in the previous 12 months—no. (%) | 126 | 115 | 67 (53) | 38 (33) | 0.0002 |
| With hospitalizations for COPD in the previous 12 months—no. (%) | 126 | 115 | 31 (25) | 17 (15) | 0.08 |
| Comorbidities | |||||
| Cardiovascular—no. (%) | 126 | 115 | 61 (48) | 42 (37) | 0.07 |
| Hypertension—no. (%) | 126 | 115 | 49 (39) | 28 (24) | 0.02 |
| Diabetes—no. (%) | 126 | 115 | 21 (17) | 7 (6) | 0.009 |
| Obstructive sleep apnea—no. (%) Treatments | 126 | 115 | 17 (13) | 5 (4) | 0.04 |
| On SABA—no. (%) | 125 | 106 | 85 (68) | 59 (56) | 0.06 |
| On LABA alone—no. (%) | 125 | 108 | 20 (16) | 6 (6) | 0.02 |
| On LAMA alone—no. (%) | 125 | 108 | 9 (7) | 7 (6) | 0.83 |
| On ICS alone—no. (%) | 125 | 108 | 1 (1) | 3 (3) | 0.25 |
| Daily dose of ICS alone—μg of equivalents beclomethasone | 1 | 3 | 1000 | 517 ± 275 | 0.27 |
| On OCS—no. (%) | 125 | 108 | 4 (3) | 4 (4) | 1.00 |
| Daily dose of prednisone (mg) | 125 | 108 | 24.5 ± 10.4 | 16.3 ± 11.1 | 0.32 |
| On LABA + LAMA—no. (%) | 125 | 108 | 15 (12) | 7 (6) | 0.06 |
| On LABA + LAMA + ICS—no. (%) | 125 | 108 | 44 (35) | 34 (31) | 0.55 |
| On anti-hypertensive drugs—no. (%) | 124 | 108 | 52 (42) | 28 (26) | 0.01 |
| On statins—no. (%) | 124 | 108 | 37 (30) | 19 (18) | 0.03 |
| Other—no. (%) | 124 | 107 | 66 (53) | 36 (34) | 0.003 |
| Adherence to treatment—no. (%) | 124 | 100 | 107 (86) | 96 (96) | 0.02 |
Data are expressed as numbers (%) and as means ± SD
DLCO = transfer factor of the lung for carbon monoxide; ICS = inhaled corticosteroids; SABA = short-acting b2-agonists; LABA = long-acting b2-agonists; LAMA = long lasting muscarinic antagonists.
* indicates the number of patients with each available variable;
** Students’ t test or Fisher exact test 2-tailed
Finally, the proportion of COPD patients necessitating anti-hypertensive drugs (p = 0.01), statins (p = 0.03), or other therapies (p = 0.003) was lower in Cluster 2 than in Cluster 1 (Table 2). No difference was observed between the two clusters in terms of gender, age, ethnic origin, GOLD stages, smoking history, biology (including serum levels of CRP), respiratory function, symptoms and treatements (Table 2). Failure to observe differences in these parameters may be linked, at least in part, to the higher proportion of COPD patients belonging to GOLD I and II stages in each cluster. For example, when considering the distribution of values of pre-bronchodilator FEV1 across COPD severity, 56% and 61% patients belonged to GOLD I and II in Cluster 1 and 2, respectively (S1 Fig).
Correlations between cluster-associated proteins and key clinical parameters
Correlation analyses showed positive associations between lower incidence of exacerbations and/or prevalence of emphysema and circulating levels of different biomarkers implicated in the regulation of EGFR pathway (eg. SHC1, AREG, GRB2 adapter protein), of host defense and innate immune responses (BTK), oxidant stress (eg. cyclophilin F, α-Synclein and Carbonic anhydrase XIII, 14-3-3 protein β/α and ζ/δ, BAD), wound healing and cell survival (eg. Tropomyosin 4, 14-3-3 protein β/α and ζ/δ, eIF-5A-1, FGF-16, PA2G4, TCTP, BAD), as well as some components of epithelial-mesenchymal remodelling (eg. RAC1, GRB2 adapter protein, ARGI1, Prostatic binding protein, DRG-1, CPNE1). Positive correlations were also found between lower prevalence of emphysema and levels of biomarkers of T cell activation (CD40 ligand), metabolism (M2-PK and 6-phosphogluconate dehydrogenase) and proteostasis (eg. Ubiquitin+1, SUMO3, UBC9, Sorting nexin 4, SNAA, UFM1) markers (S4 Table in S1 File).
Finally, a negative association was found between serum levels of MMP-12 and renin, which were up-regulated in Cluster 1, and lower prevalence of emphysema (S4 Table in S1 File).
Examination of Clusters 1 and 2 in an independent COPD cohort and in healthy subjects
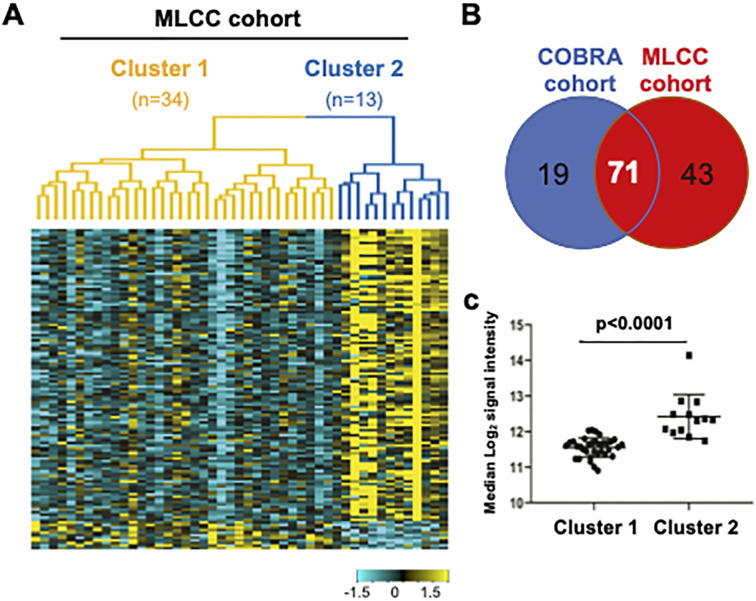
We then attempted to validate these clusters in another COPD population of 47 patients [21]. Unsupervised hierarchical clustering, with the top 10% proteins by expression variation, showed two clusters within these COPD subjects (34 and 13 subjects in Clusters 1 and 2, respectively) where 125 proteins were differentially expressed between Cluster 1 and Cluster 2 (Fold change>1.5 and FDR<0.05) (Fig 3A). While 114 proteins were significantly up-regulated in Cluster 2, 71 of these (62%), overlapped with the proteins that were higher in Cluster 2 of the COBRA cohort at visit 1 (Fig 3B). We also found no similarities between the 2 cohorts regarding the few up-regulated proteins in Cluster 1. We calculated the protein score (median expression of the 90 Cluster 2-associated proteins defined in the COBRA cohort), and showed that it was significantly up-regulated in Cluster 2, as compared to Cluster 1 in the MLCC cohort (p<0.0001) (Fig 3C).
Fig 3. COPD Clusters are observed in an independent patient cohort, but not in healthy controls.
(A) Heatmap showing levels of differentially expressed proteins in the MLCC cohort. Yellow indicates high expression and cyan indicates low expression. (B) Overlap of the Cluster 2-associated up-regulated proteins (Fold change >1.5 for Cluster 2 vs Cluster 1 and FDR <0.05) between COBRA and MLCC cohorts. (C) Protein score (defined in CORBA cohort) in Cluster 1- and 2 in the MLCC cohort.
Comparisons of the clinical parameters between the two clusters showed significant lower prevalence of emphysema and of incidence of hospitalizations for COPD during the last 12 months, as well as a diminished requirement of treatment with LABA, in Cluster 2 compared to Cluster 1 (S6 Table in S1 File). This was accompanied by a trend towards a lower onset of exacerbations and of cardio-metabolic co-morbidities, although the differences between the two clusters failed to achieve statistical significance (S5 Table in S1 File).
To determine whether the subtyping was specific of COPD, we performed the same analyses in serum samples from a group of 50 healthy donors. Although hierarchical clustering generated two subtypes within these subjects, the protein score, as defined in the COBRA cohort, failed to show significant difference between the 2 Clusters (median log2 signal intensity between Cluster 1 (n = 19 subjects) and Cluster 2 (n = 31 subjects) [5%-95%]: 10.6 [10.5–10.7] and 10.7 [10.6–10.8], respectively, p = 0.129).
Identification of a protein fingerprint associated with COPD endotypes
To establish a short protein signature reflecting changes in clinical outcomes in relation with lung biological processes, we generated a fingerprint of 15 biologically relevant biomarkers of the top 96 differentially expressed proteins that were mainly selected from the different GO biological processes previously identified (Fig 2B, S3 and S4 Tables in S1 File). These biomarkers included 11 and 4 proteins representative of Cluster 2 and 1, respectively (Table 3).
Table 3. Correlation analyses between the 15 differentially regulated proteins and main clinical parameters in COPD patients of the COBRA cohort.
| Biological processes | Target | With exacerbations (Y/N) | With emphysema (Y/N) | With unscheduled medical visits (Y/N) | |||
|---|---|---|---|---|---|---|---|
| OR (2.5–97.5%) | FDR | OR (2.5–97.5%) | FDR | OR (2.5–97.5%) | FDR | ||
| Cell fate, remodeling and repair | AREG | 0.78 (0.53–1.15) | 0.310 | 0.49 (0.3–0.8) | 0.004 | 0.69 (0.45–1.05) | 0.122 |
| FGF16 | 0.81 (0.55–1.19) | 0.347 | 0.54 (0.34–0.85) | 0.007 | 0.74 (0.5–1.1) | 0.164 | |
| SHC1 | 0.64 (0.4–1.03) | 0.102 | 0.33 (0.19–0.59) | 0.001 | 0.58 (0.36–0.95) | 0.057 | |
| 14-3-3 protein β/α | 0.51 (0.32–0.81) | 0.026 | 0.4 (0.24–0.68) | 0.002 | 0.46 (0.28–0.75) | 0.015 | |
| eIF-4H | 0.63 (0.46–0.86) | 0.026 | 0.58 (0.41–0.82) | 0.003 | 0.62 (0.45–0.86) | 0.015 | |
| Tropomyosin 4 | 0.78 (0.64–0.96) | 0.042 | 0.71 (0.57–0.89) | 0.004 | 0.78 (0.63–0.96) | 0.049 | |
| Metabolism and mitochondria | Cyclophilin F | 0.72 (0.57–0.91) | 0.026 | 0.65 (0.5–0.84) | 0.002 | 0.69 (0.54–0.88) | 0.015 |
| Carbonic anhydrase XIII | 0.80 (0.65–0.99) | 0.068 | 0.74 (0.59–0.94) | 0.013 | 0.77 (0.62–0.96) | 0.049 | |
| H2A3 | 0.87 (0.69–1.09) | 0.310 | 0.81 (0.63–1.04) | 0.099 | 0.87 (0.69–1.1) | 0.268 | |
| Immunity and defense | BTK | 0.72 (0.57–0.92) | 0.026 | 0.63 (0.49–0.82) | 0.002 | 0.73 (0.57–0.93) | 0.035 |
| CD40 ligand | 0.91 (0.64–1.3) | 0.598 | 0.54 (0.35–0.81) | 0.004 | 0.72 (0.5–1.05) | 0.127 | |
| Midkine | 1.4 (1.06–1.84) | 0.042 | 1.45 (1.08–1.94) | 0.012 | 1.32 (1–1.74) | 0.093 | |
| Lactadherin | 1.57 (1.07–2.3) | 0.042 | 1.97 (1.31–2.95) | 0.002 | 1.38 (0.94–2.01) | 0.131 | |
| Tissue injury | MMP-12 | 1.13 (0.82–1.55) | 0.484 | 1.45 (1.04–2.03) | 0.031 | 1.11 (0.81–1.53) | 0.561 |
| Renin | 1.10 (0.86–1.41) | 0.484 | 0.99 (0.77–1.27) | 0.933 | 1.07 (0.84–1.37) | 0.567 | |
Abbreviations: DLCO, transfer factor of the lung for carbon monoxide; OR, odds-ratio; FDR, false discovery rate; AREG, Amphiregulin; FGF16, Fibroblast growth factor 16; SHC, SHC-transforming protein 1; eIF-4H: Eukaryotic translation initiation factor 4H; BTK, Tyrosine-protein kinase BTK; MMP-12, metallopreoteinase-12.
OR was obtained through a logistic model.
Bold denotes statistical significance
Specifically, Cluster 2-associated Metabolism/Mitochondria markers, namely Cyclophilin F, Carbonic anhydrase XIII and H2A3, were sorted from GO pathways involved in mitochondrial membrane organization and permeability and cellular response to oxidative stress and to environmental stimulus, respectively, whereas immunity/defense markers, including BTK and CD40 ligand, were sorted from activation of innate immune response and T cell regulation, respectively (S4 Table in S1 File).
To cover the different aspects of cell fate/repair/remodeling processes, rational ranking of differentially expressed proteins led to the prioritisation of: 1) the representative transcription factor, eIF-4H, that is involved in the post-translational regulation of gene expression; 2) the two growth factors, AREG and FGF-16, that promote tissue regeneration through ERBB signaling pathway and that positively regulate cell proliferation; 3) the cytosolic protein regulating cell proliferation, SHC1, 4) the key regulator of apoptotic and nutrient-sensing signaling, 14-3-3 protein β/α; and 5) the tissue-remodeling mediator, Tropomypsin 4, that contributes to actin filament organization. Lastly, to predict Cluster 1-COPD endotype, a selection of 4 of the 6 up-regulated proteins in Cluster 1 (S3 Table in S1 File), was performed, after exclusion of C3b that failed to show any differential expression in the MLCC cohort. These 4 proteins included two hallmarks of tissue injury (MMP-12 and renin) and two modulators of lung immune responses (Midkine and Lactadherin).
We next investigated the association between each of the 15 proteins composing this short signature and main hallmarks of COPD, such as the prevalence of emphysema, the incidence of exacerbations and of unscheduled medical visits and the number of exacerbations and values of DLCO recorded during the previous 12 months (Table 3). We showed that 7 and 5 out of the 15 proteins were associated with the incidence of exacerbations and with that of unscheduled medical visits, respectively, and that 12 of the 15 proteins correlated with the prevalence of emphysema (Table 3). The proteins that displayed the most significant correlations with exacerbations and unscheduled medical visits were 14-3-3 protein β/α, eIF-4H, that regulate cell renewal by controlling apoptosis, and cyclophilin F, a marker of metabolism and of mitochondrial functions (FDR values between 0.015 and 0.026) (Table 3). The 12 proteins associated with the prevalence of emphysema fitted wth all biological processes and they showed FDR values ranging from 0.03 and 0.001 (Table 3). In contrast, we found no significant correlation between the expression of each of the 15 proteins and the number of exacerbations, or DLCO values (rho between 0.01 and 0.11, p values from 0.79 and 0.94, for exacerbations, and rho between 0.02 and 0.15, p values ranging from 0.90 and 0.38, for DLCO, Pearson correlation).
Monitoring cluster switching at a second visit
To determine whether the 2 protein clusters were maintained in the 163 subjects from COBRA cohort during the follow-up visit, we first performed unsupervised hierarchical clustering on these subjects. Consistent with the results at inclusion, the two distinct expression patterns were confirmed at visit 2 (97 subjects in Cluster 1 and 66 subjects in Cluster 2) (S1A and S1B Fig). The expression of 86 proteins was significantly different between the two clusters (Fold change>1.5 and FDR<0.05), with 83 and 3 up-regulated proteins in Cluster 2 and Cluster 1, respectively, when compared to their Cluster 1 and 2 counterparts. Seventy-five out of these 83 upregulated proteins (eg. 90%) were also enriched in Cluster 2 at visit 1, whereas only one of the 3 up-regulated proteins in Cluster 1 (eg. Lactadherin) was also elevated at visit 1. Furthermore, the protein score (median expression) of the 90 Cluster 2-associated proteins defined at visit 1 was significantly higher in Cluster 2 at visit 2, as compared to Cluster 1 at visit 1 (p<0.0001, S1C Fig).
Next, we demonstrated that clustering pattern was significantly consistent between the two visits, with 108 subjects among 163 showing the same cluster identify (68 for Cluster 1 and 40 for Cluster 2) (p<0.001, Fisher exact test). However, 55 subjects changed their cluster identity, since 29 patients belonging to Cluster 1 at visit 1 exhibited mixed profile combining Cluster 1 and 2 protein signatures at visit 2 and 26 switched from Cluster 2 at visit 1 to Cluster 1 at visit 2 (S6 Table in S1 File). COPD patient switching from to Cluster 1 to 2 displayed lower incidence of exacerbations (p = 0.02), of unscheduled medical visits (p = 0.04) and of hospitalizations for COPD (p = 0.02) in the previous year. COPD patients switching from Cluster 2 to 1 showed a significant higher incidence of hospitalizations for COPD (p = 0.05) and of LAMA use (p = 0.04) (S6 Table in S1 File).
Discussion
Using SOMAscan, we profiled the serum levels of 1305 proteins in 241 COPD patients from the COBRA cohort and identified two distinct subtypes throught unsupervised hierarchical clustering of 96 differential expressed proteins. These two clusters appeared to be clinically relevant since Cluster 2 showed lower number and incidence of exacerbations, unscheduled medical visits, hospitalizations for COPD within the previous year and reduced prevalence of emphysema than Cluster 1.
Lung function was similar between the 2 clusters, which could be explained by the higher proportion of GOLD I and II patients in the COBRA cohort, who manifested little or no airflow obstruction.
The occurrence of co-morbidities, particularly hypertension, diabetes and obstructive sleep apnea, the need of LABA of treatements for cardiovascular and metabolic comorbidities (i.e., anti-hypertensive drugs and statins), were all lower in Cluster 2 than in Cluster 1.
Despite the limited number of COPD patients included in the MLCC patient group, we were able to cross-validate, at least in part, our findings by showing a reduction in the prevalence of emphysema and on the incidence of hospitalizations for COPD during the last 12 months and a trend towards a lower occurrence of exacerbations and of cardio-metabolic co-morbidities months in Cluster 2 versus Cluster 1. Therefore, our results highlight the valuable use of SOMAscan technology as reproducible screening tool to identify specific serum biomarkers associated to COPD phenotypes.
To initiate mechanism understanding, we combined GO pathway analysis with mapping of the 96 differentially expressed proteins into biological processes. This approach led us to characterize the enrichment of Cluster 2 with hallmarks of lung immunity/host defense, cell fate/repair/remodelling and mitochondrial/metabolic activities. These findings reflect high glycolytic activity, mitochondrial functions and proteostasis that may maintain mucosal barrier by regulating metabolic energy in proliferating immune and structural cells upon tissue remodelling under oxidative stress in Cluster 2 [24]. Therefore, these results are consitent with proteomic profiling of the bronchoalveolar lavages from COPD and lung cancer patients [25].
To distinguish between the different COPD clusters, the top96 differentially expressed proteins were refined into a clinically feasible fingerprint composed of 15 biologically biomarkers, including 11 and 4 up-regulated proteins in Cluster 2 and Cluster 1, respectively. The 11 up-regulated biomarkers in Cluster 2 have been reported to promote tissue repair and they included the pro-survival protein, 14-3-3 protein β/α, the transcription factor-regulating stem cell renewal, eIF4H, and the growth factors-promoting stem cell activity, AREG, SHC1 and FGF-16. These findings suggest that Cluster 2-patients exhibit better lung regeneration than those in Cluster 1. Of note, the mesenchymal biomarker, Tropomyosin 4, and Carbonic anhydrase XIII, one of the main hallmarks of systemic and local oxidative stress, were also augmented in Cluster 2 patients. Previous studies have shown upregulation of Tropomyosin 4 expression in muscle fibres of diaphragm of COPD patients under enhanced resistance to fatigue [26] and elevated levels Carbonic anhydrase XIII in COPD skeletal muscles in relation with a gain of force [27]. Together, these results suggest that enrichement of Tropomyosin 4 and of Carbonic anhydrase XIII in Cluster 2 translated into a better efficiency of the antioxidant systems to improve muscle dysfunction in response to chronic exercise [28]. Representative hallmarks of proteostasis (eg. Cyclophilin F, Carbonic anhydrase XIII and H2A3), and of innate immune response, host defense and inflammasome activity (BTK and CD40 ligand) [29–31] were also higher in Cluster 2 than in Cluster 1. Given that Cyclophilin F, an ubiquitously expressed immunophilin involved in protein folding/trafficking and mitochondrial permeability [32], can be secreted under inflammatory stimuli and oxidative stress [33], its elevated expression, along with BTK and CD40 ligand, may indicate an improved immune/inflammatory responses upon lung injury and oxidative stress in Cluster 2.
Finally, among the 4 proteins of our fingerprint that were up-regulated in Cluster 1, Midkine and Lactadherin, were reported to be prominent during several chronic immune and metabolic disorders, such as atherosclerosis, cardiac, kidney and metabolic diseases [34–36]. Elevated levels of these proteins in Cluster 1 COPD patients are consistent with higher incidence of hypertension, diabetes and use of anti-hypertensive drugs and statins and they support the contribution of cardiometabolic co-morbidities to COPD severity [6, 37]. MMP-12 levels were also higher in Cluster 1 than in Cluster 2 and they were associated with greater prevalence of emphysema. This finding is consistent with MMP-12 involvement in lung injury secondary to the degradation of extracellular matrix in COPD [38, 39]. Likewise, elevated renin expression, an enzyme involved in the renin-angiotensin II-aldosterone axis regulating blood pressure [40], was associated with higher prevalence of hypertension in Cluster 1, as previously demonstrated [41].
Correlation analyses between each of the 15 proteins and major clinical determinants of COPD demonstrated that our signature reflected mainly the prevalence of emphysema, rather than the incidence of exacerbations and of unscheduled medical visits, although 5 to 7 proteins out of the 15 and related to cell fate, remodeling and repair (14-3-3 protein β/α, eIF-4H and tropomyosin 4) and to immunity and defense (BTK, midkine and lactadherin) were associated with these two latter clinical parameters.
Importantly, 12 proteins belonging to all classes of biological processes were highly-significantly associated with emphysema. These included AREG, FGF16, SHC1, 14-3-3 protein β/α, eIF-4H and tropomyosin 4 (cell fate remodeling and repair), cyclophilin F and carbonic anhydrase XIII (metabolism and mitochondria), BTK, midkine and lactadherin (immunity and defense) and MMP-12 (tissue injury). These data indicated that lung injury and renewal, oxidative stress, diaphragm muscle dysfuntions, immune responses and subcellular alterations, predominated in our patient cohort in association with emphysema.
Intriguingly, we found no association between the protein fingerprint and values of DLCO. However, in the COBRA cohort, emphysema was not monitored homogeneously across the hospital centers participating in patient inclusion, since either quantitative computed tomography (CT) scan, or DLCO was used. The latter being also a marker of altered alveolar-capillary permeability, its use to map emphysema may have confounded the analyses and explain this discrepancy.
Serum proteomic profiling at visit 2 showed that approximately 34% of COPD COBRA patients changed their clusters with no link to acute events (eg. exacerbations, pneumonia), or co-morbidities, all patients being in a stable state during the month preceding blood sampling. Patients exhibiting combined Cluster 1 and Cluster 2 protein profiles (18%) displayed significant lower incidence of exacerbations, of unscheduled medical visits and of hospitalizations for COPD, whereas switching from Cluster 2 to Cluster 1 (16% of the patients) was associated with an increase in the onset of hospitalizations for COPD and with a trend towards higher LAMA use.
Whereas emphysema status did not improve in patients with combined protein profiles, the increase in markers of host defense and lung repair after 6–12 months at visit 2, suggested a better recovering of the lung tissue, potentially as a result of the activation of repairing mechanisms. Whether the enhancement in lung repair can translate into a long-term improvement of lung function within this group of patients, requires further investigation.
Among the 15 protein fingerprint that correlated with the prevalence of emphysema and the incidence of exacerbations, FGF-16, a growth factor contributing to tissue regeneration [42, 43], may represent a starting point for the development of new therapeutics. Consistently, the airway administration of FGF-2, another member of FGF family promoting tissue repair, was reported to reduce emphysema and to enhance lung repair in cigarette smoke-exposed or elastase-induced COPD mouse models, possibly by attenuating inflammation and alveolar cell death [44]. In addition, a few randomized clinical trials performed in patients with periodontitis and osteoarthritis, two chronic inflammatory diseases with progressive tissue degeneration, demonstrated improved tissue repair upon the administration of recombinant human FGF-2 [45, 46]. Whether FGF-16 promotes similar beneficial effects in COPD patients deserves further investigation.
In an effort to profile COPD patients, specific markers have been previously established for identifying patients with high versus low rate of exacerbations [10], or by defining COPD-subtypes using CT imaging, namely, emphysema- and airway-dominant diseases [11, 12]. In an extention of these latter studies, CT-based phenotying was shown to be associated with gender signatures involving different types of leukocytes and of mitochondrial-related genes, as assessed in bronchial brushes [13]. These generated profiles, however, have not been validated across multiple cohorts. An additional integrative sputum microbiome analysis stratified COPD patients into two neutrophilic subgroups differing by the predominance of airway Haemophilus infection and by the interchangeability with eosinophilic inflammation [14], suggesting that different therapeutic strategies may succesfully target these phenotypes. Of significance, our study provides novel insights into COPD heterogeneity and suggests that overexpression of factors involved in lung immunity/host defense, cell fate/repair/remodelling and mitochondrial/metabolic activities contribute to better clinical outcomes. Despite the non-invasive nature of our COPD phenotyping through SOMAscan proteomic analysis applied to serum samples, this study has some limitations.
Firstly, SOMAscan can detect very low levels of proteins compared to immunoassays, but, as a discovery tool, this platform provides only relative quantification, rather than absolute concentrations. Although this technology is rapidly growing in popularity due to the quantification of large numbers of proteins efficiently and cost-effectively, comparisons to conventional immunoassays are required potentially due to lack of specificity for some aptamers, or differences in signal to noise ratios.
Secondly, the association studies between the different protein signatures and the COPD clusters as shown herein suggest that distinct molecular mechanisms of lung inflammation, injury/repair and oxidative stress operate differently in Cluster 1 and Cluster 2-COPD patients. Whether our cluster-specific protein signatures can predict disease pathophysiology and progression requires further investigations.
Thirdly, we were able to cross-validated our findings only in a small group of COPD patients belonging to the MLCC cohort, that included mainly frequent exacerbators. Therefore, it would be appropriate to further confirm the existence of the 2 COPD clusters currently described in a larger series of fully-clinically defined COPD patients. Also, additional tissue validation studies using patient biopsies, or primary lung cells, are required to confirm the presence of our 15-protein signature.
Fourthly, exacerbations were reported at 6- or 12-month intervals in the COBRA cohort and relied on self-reporting. This method could lead to underreporting of mild or moderate events that may manifest potentially in patients switching clusters in the COBRA cohort, but is unlikely to influence the identification of severe exacerbators, as those included in MLCC cohort.
Lastly, the nature of exacerbation triggers, such as viral, or bacterial respiratory cultures, air pollution and others, that may influence the cluster switch, is not detailed in either cohorts, COBRA and MLCC.
Conclusion
Overall, this study demonstrates that the SOMAscan technique offers a powerful and reproducible screening tool to identify specific serum proteins and pathways that allows a better understanding of the largely unknown heterogeneity of COPD and the characterization of novel endotypes. In addition, the currently described short protein fingerprint may lead to a better management of COPD patients in terms of treatment options and monitoring, with more frequent follow-up visits for the patients belonging to Cluster 1. Also, this signature may offer a valuable tool for selecting patients to be included in clinical trials and for identifying potential new therapeutic targets.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(TIFF)
(TIFF)
Acknowledgments
We are grateful to the patients who made this study possible and to the Investissement d’Avenir—Program ANR-11-IDEX-0005-02, Laboratoire d’Excellence, INFLAMEX. We also thank Dr. Russell Bowler and his team at National Jewish Health (Denver, Colorado, United Sates of America) for performing the SOMAscan proteomic assays.
The associated pulmonologists and research staff of the COBRA cohort are as follows: Michel Aubier, Bernard Aguilaniu, Noussaier Ben Salem, Patrick Berger, Philippe Bonniaud, Arnaud Bourdin, Laurent Boyer, Arnaud Chambellan, Pascal Chanez, Cécile Chenivesse, Frédéric De Blay, Gilles Devouassoux, Gaétan Deslée, Gilles Garcia, Jodelle Gerbier, Pierre-Olivier Girodet, Laurent Guilleminault, Marc Humbert, Sylvie Leroy, Cécile Londner, Antoine Magnan, Bernard Maitre, Charles-Hugo Marquette, Roger Marthan, Aurélie Metsu, Nicolas Molinari, Laurie Anne Pahus, Marina Pretolani, Christophe Pison, Nicolas Roche, Marine Servat, Thomas Similowski, Camille Taillé, Angelica Tiotiu, Anne Tsicopoulos, Isabelle Vachier, Eric Van Ganse.
Third parties contributing to this study include the Biological Ressources Centre of the Bichat Hospital in Paris (Dr. Sarah Tubiana) that stored and tracked the serum samples of the COPBA cohort and LaSer Analytica, Paris, France (http://www.la-ser.com), that elaborated and managed the COBRA clinical database.
Data Availability
We are not able to share publicly neither the clinical nor the biological raw data, even upon request. The reasons for this restriction are as follows: 1) Inserm is the owner of the clinical data originating from the COBRA cohort, 2) the informed consent signed by the patients of both cohorts does not mention that their data can be freely shared, even if they would be anonymized, and 3) this study was financially supported by AstraZeneca and in the legal contract with Inserm there is no agreement that allows sharing the data. These reasons are now listed in the appropriate box related to Data access. In this setting, it is not possible to publish the SOMAscan proteomic data as suggested by the PLOS ONE research data policy. To publish such data, even in anonymized profiles, would violate the written consent obtained from the participants and thus would not be in accordance with European General Data Protection Regulation. Due to these restrictions, the authors of this study will share only the used information guideline and a summary of all used data as main or supplemental material to this paper.
Funding Statement
The COBRA cohort was funded by Inserm, Legs Poix - Chancellerie des Universités, AstraZeneca, Chiesi, GlaxoSmithKline, MedImmune LLC, Novartis Pharma AG and Roche. Marina Pretolani’s institution has received funding from AstraZeneca. Rania Dagher, Chia-Chien Chiang, Jennifer Kearley and Daniel Muthas are employees and shareholders of AstraZeneca. Jingya Wang, Roland Kolbeck and Alison A. Humbles were previous employees and shareholders of AstraZeneca. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Vogelmeier CF, Criner GJ, Martinez FJ, Anzueto A, Barnes PJ, Bourbeau J, et al. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Lung Disease 2017 Report: GOLD Executive Summary. Eur Respir J. 2017. Apr;22(3):575–601. doi: 10.1111/resp.13012 Epub 2017 Mar 7. [DOI] [PubMed] [Google Scholar]
- 2.Aggarwal T, Wadhwa R, Thapliyal N, Sharma K, Rani V, Maurya PK. Oxidative, inflammatory, genetic, and epigenetic biomarkers associated with chronic obstructive pulmonary disorder. J Cell Physiol. 2019. Mar;234(3):2067–2082. doi: 10.1002/jcp.27181 Epub 2018 Sep 1. . [DOI] [PubMed] [Google Scholar]
- 3.Brandsma C-A, de Vries M, Costa R, Woldhuis RR, Königshoff M, Timens W. Lung ageing and COPD: is there a role for ageing in abnormal tissue repair? Eur Respir Rev. 2017. Dec 6;26(146):170073. doi: 10.1183/16000617.0073-2017 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes PJ. Senescence in COPD and Its Comorbidities. Annu Rev Physiol. 2017. Feb 10;79:517–539. doi: 10.1146/annurev-physiol-022516-034314 Epub 2016 Dec 9. [DOI] [PubMed] [Google Scholar]
- 5.Segal LN, Martinez FJ. Chronic obstructive pulmonary disease subpopulations and phenotyping. J. Allergy Clin Immunol. 2018. Jun;141(6):1961–1971. doi: 10.1016/j.jaci.2018.02.035 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown JP, Martinez CH. Chronic obstructive pulmonary disease comorbidities. Curr Opin Pulm Med. 2016. Mar;22(2):113–8. doi: 10.1097/MCP.0000000000000241 [DOI] [PubMed] [Google Scholar]
- 7.Agustí A, Celli B, Faner R. What does endotyping mean for treatment in chronic obstructive pulmonary disease? Lancet Lond Engl. 2017. Sep 2;390(10098):980–987. doi: 10.1016/S0140-6736(17)32136-0 . [DOI] [PubMed] [Google Scholar]
- 8.Sidhaye VK, Nishida K, Martinez FJ. Precision medicine in COPD: where are we and where do we need to go? Eur Respir Rev. 2018. Aug 1;27(149):180022. doi: 10.1183/16000617.0022-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen AB, Moseley PL, Oprea TI, Ellesøe SG, Eriksson R, Schmock H, et al. Temporal disease trajectories condensed from population-wide registry data covering 6.2 million patients. Nat Commun. 2014. Jun 24;5:4022. doi: 10.1038/ncomms5022 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou A, Zhou Z, Zhao Y, Chen P. The recent advances of phenotypes in acute exacerbations of COPD. Int J Chron Obstruct Pulmon Dis. 2017. Mar 27;12:1009–1018. doi: 10.2147/COPD.S128604 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramanian DR, Gupta S, Burggraf D, Vom Silberberg SJ, Heimbeck I, Heiss-Neumann MS, et al. Emphysema- and airway-dominant COPD phenotypes defined by standardised quantitative computed tomography. Eur Respir J. 2016. Jul;48(1):92–103. doi: 10.1183/13993003.01878-2015 Epub 2016 May 26. [DOI] [PubMed] [Google Scholar]
- 12.Keene JD, Jacobson S, Kechris K, Kinney GL, Foreman MG, Doerschuk CM, et al. COPDGene and SPIROMICS Investigators ‡. Biomarkers Predictive of Exacerbations in the SPIROMICS and COPDGene Cohorts. Am J Respir Crit Care Med. 2017. Feb 15;195(4):473–481. doi: 10.1164/rccm.201607-1330OC . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteve-Codina A, Hofer TP, Burggraf D, Heiss-Neumann MS, Gesierich W, Boland A, et al. Gender specific airway gene expression in COPD sub-phenotypes supports a role of mitochondria and of different types of leukocytes. Sci Rep. 2021. Jun 18;11(1):12848. doi: 10.1038/s41598-021-91742-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Locantore N, Haldar K, Ramsheh MY, Beech AS, Ma W, et al. Inflammatory Endotype-associated Airway Microbiome in Chronic Obstructive Pulmonary Disease Clinical Stability and Exacerbations: A Multicohort Longitudinal Analysis. Am J Respir Crit Care Med. 2021. Jun 15;203(12):1488–1502. doi: 10.1164/rccm.202009-3448OC . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PloS One 2010. Dec 7;5(12):e15004. doi: 10.1371/journal.pone.0015004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeBoer EM, Wagner BD, Popler J, Harris JK, Zemanick ET, Accurso FJ, et al. Novel Application of Aptamer Proteomic Analysis in Cystic Fibrosis Bronchoalveolar Lavage Fluid. Proteomics Clin Appl. 2019. May;13(3):e1800085. doi: 10.1002/prca.201800085 Epub 2019 Jan 3. [DOI] [PubMed] [Google Scholar]
- 17.Deterding RR, Wagner BD, Harris JK, DeBoer EM. Pulmonary Aptamer Signatures in Children’s Interstitial and Diffuse Lung Disease. Am J Respir Crit Care Med. 2019. Dec 15;200(12):1496–1504. doi: 10.1164/rccm.201903-0547OC . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Norman KC, O’Dwyer DN, Salisbury ML, DiLillo KM, Lama VN, Xia M, et al. Identification of a unique temporal signature in blood and BAL associated with IPF progression. Sci Rep. 2020. Jul 21;10(1):12049. doi: 10.1038/s41598-020-67956-w . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourdin A, Suehs CM, Marin G, Vachier I, Matzner-Lober E, Chanez P, et al. COBRA Consortium. Asthma, COPD, and overlap in a national cohort: ACO on a gradient. J Allergy Clin Immunol. 2018. Apr;141(4):1516–1518. doi: 10.1016/j.jaci.2017.11.049 Epub 2018 Jan 31. . [DOI] [PubMed] [Google Scholar]
- 20.Pretolani M, Soussan D, Poirier I, Thabut G, Aubier M, COBRA Study Group, COBRA cohort Study Group. Clinical and biological characteristics of the French COBRA cohort of adult subjects with asthma. Eur Respir J. 2017. Aug 24;50(2):1700019. doi: 10.1183/13993003.00019-2017 . [DOI] [PubMed] [Google Scholar]
- 21.Hutchinson AF, Ghimire AK, Thompson MA, Black JF, Brand CA, Lowe AJ, et al. A community-based, time-matched, case-control study of respiratory viruses and exacerbations of COPD. Respir Med. 2007. Dec;101(12):2472–81. doi: 10.1016/j.rmed.2007.07.015 Epub 2007 Sep 5. . [DOI] [PubMed] [Google Scholar]
- 22.Rohloff JC, Gelinas AD, Jarvis TC, Ochsner UA, Schneider DJ, Gold L, et al. Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents. Mol Ther Nucleic Acids 2014. Oct 7;3(10):e201. doi: 10.1038/mtna.2014.49 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015. Apr 20;43(7):e47. doi: 10.1093/nar/gkv007 Epub 2015 Jan 20. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelsen SG. The Unfolded Protein Response in Chronic Obstructive Pulmonary Disease. Ann Am Thorac Soc. 2016. Apr;13 Suppl 2(Suppl 2):S138–45. doi: 10.1513/AnnalsATS.201506-320KV . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pastor MD, Nogal A, Molina-Pinelo S, Meléndez R, Salinas A, González De la Peña M, et al. Identification of proteomic signatures associated with lung cancer and COPD. J Proteomics 2013. Aug 26;89:227–37. doi: 10.1016/j.jprot.2013.04.037 Epub 2013 May 9. . [DOI] [PubMed] [Google Scholar]
- 26.Levine S, Kaiser L, Leferovich J, Tikunov B. Cellular adaptations in the diaphragm in chronic obstructive pulmonary disease. N Engl J Med. 1997. Dec 18;337(25):1799–806. doi: 10.1056/NEJM199712183372503 . [DOI] [PubMed] [Google Scholar]
- 27.Jaitovich A, Barreiro E. Skeletal Muscle Dysfunction in Chronic Obstructive Pulmonary Disease. What We Know and Can Do for Our Patients. Am J Respir Crit Care Med. 2018. Jul 15;198(2):175–186. doi: 10.1164/rccm.201710-2140CI [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barreiro E, Gea J, Matar G, Hussain SNA. Expression and carbonylation of creatine kinase in the quadriceps femoris muscles of patients with chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2005. Dec;33(6):636–42. doi: 10.1165/rcmb.2005-0114OC Epub 2005 Sep 15. . [DOI] [PubMed] [Google Scholar]
- 29.Bittner ZA, Liu X, Mateo Tortola M, Tapia-Abellán A, Shankar S, Andreeva L, et al. BTK operates a phospho-tyrosine switch to regulate NLRP3 inflammasome activity. J Exp Med. 2021. Nov 1;218(11):e20201656. doi: 10.1084/jem.20201656 Epub 2021 Sep 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Florence JM, Krupa A, Booshehri LM, Gajewski AL, Kurdowska AK. Disrupting the Btk Pathway Suppresses COPD-Like Lung Alterations in Atherosclerosis Prone ApoE-/- Mice Following Regular Exposure to Cigarette Smoke. Int J Mol. Sci. 2018. Jan 24;19(2):343. doi: 10.3390/ijms19020343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacy M, Bürger C, Shami A, Ahmadsei M, Winkels H, Nitz K, et al. Cell-specific and divergent roles of the CD40L-CD40 axis in atherosclerotic vascular disease. Nat Commun. 2021. Jun 18;12(1):3754. doi: 10.1038/s41467-021-23909-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nigro P, Pompilio G, Capogrossi MC. Cyclophilin A: a key player for human disease. Cell Death Dis. 2013. Oct 31;4(10):e888. doi: 10.1038/cddis.2013.410 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M, Tang J, Yin J, Wang X, Feng X, Yang X, et al. The clinical implication of serum cyclophilin A in patients with chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 2018. Jan 19;13:357–363. doi: 10.2147/COPD.S152898 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Şalaru DL, Arsenescu-Georgescu C, Chatzikyrkou C, Karagiannis J, Fischer A, Mertens PR. Midkine, a heparin-binding growth factor, and its roles in atherogenesis and inflammatory kidney diseases. Nephrol Dial Transplant. 2016. Nov;31(11):1781–1787. doi: 10.1093/ndt/gfw083 Epub 2016 May 17. . [DOI] [PubMed] [Google Scholar]
- 35.Bădilă E, Daraban AM, Ţintea E, Bartoş D, Alexandru N, Georgescu A. Midkine proteins in cardio-vascular disease. Where do we come from and where are we heading to? Eur J Pharmacol. 2015. Sep 5;762:464–71. doi: 10.1016/j.ejphar.2015.06.040 Epub 2015 Jun 20. . [DOI] [PubMed] [Google Scholar]
- 36.Lee HA, Lim J, Joo HJ, Lee Y-S, Jung YK, Kim JH, et al. Serum milk fat globule-EGF factor 8 protein as a potential biomarker for metabolic syndrome. Clin Mol Hepatol. 2021. Jul;27(3):463–473. doi: 10.3350/cmh.2020.0351 Epub 2021 Feb 15. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cebron Lipovec N, Beijers RJHCG, van den Borst B, Doehner W, Lainscak M, Schols AMWJ. The Prevalence of Metabolic Syndrome In Chronic Obstructive Pulmonary Disease: A Systematic Review. COPD 2016. Jun;13(3):399–406. doi: 10.3109/15412555.2016.1140732 Epub 2016 Feb 25. . [DOI] [PubMed] [Google Scholar]
- 38.Shipley JM, Wesselschmidt RL, Kobayashi DK, Ley TJ, Shapiro SD. Metalloelastase is required for macrophage-mediated proteolysis and matrix invasion in mice. Proc Natl Acad Sci USA. 1996. Apr 30;93(9):3942–6. doi: 10.1073/pnas.93.9.3942 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hao W, Li M, Zhang Y, Zhang C, Xue Y. Expressions of MMP-12, TIMP-4, and Neutrophil Elastase in PBMCs and Exhaled Breath Condensate in Patients with COPD and Their Relationships with Disease Severity and Acute Exacerbations. J Immunol Res 2019; 2019 Apr 17;2019:7142438. doi: 10.1155/2019/7142438 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuba K, Imai Y, Penninger JM. Angiotensin-converting enzyme 2 in lung diseases. Curr Opin Pharmacol. 2006. Jun;6(3):271–6. doi: 10.1016/j.coph.2006.03.001 Epub 2006 Apr 3. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jan Danser AH. Renin and prorenin as biomarkers in hypertension. Curr Opin Nephrol Hypertens. 2012. Sep;21(5):508–14. doi: 10.1097/MNH.0b013e32835623aa . [DOI] [PubMed] [Google Scholar]
- 42.Davies DE, Polosa R, Puddicombe SM, Richter A, Holgate ST. The epidermal growth factor receptor and its ligand family: their potential role in repair and remodelling in asthma. Allergy 1999. Aug;54(8):771–83. . [PubMed] [Google Scholar]
- 43.Zheng Y, Zhang C, Croucher DR, Soliman MA, St-Denis N, Pasculescu A, et al. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature 2013. Jul 11;499(7457):166–71. doi: 10.1038/nature12308 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang L, Zhou F, Zheng D, Wang D, Li X, Zhao C, et al. FGF/FGFR signaling: From lung development to respiratory diseases. Cytokine Growth Factor Rev. 2021. Dec;62:94–104. doi: 10.1016/j.cytogfr.2021.09.002 Epub 2021 Sep 20. . [DOI] [PubMed] [Google Scholar]
- 45.Cochran DL, Oh T-J, Mills MP, Clem DS, McClain PK, Schallhorn RA, et al. A Randomized Clinical Trial Evaluating rh-FGF-2/β-TCP in Periodontal Defects. J Dent Res. 2016. May;95(5):523–30. doi: 10.1177/0022034516632497 Epub 2016 Feb 23. . [DOI] [PubMed] [Google Scholar]
- 46.Shah SS, Mithoefer K. Current Applications of Growth Factors for Knee Cartilage Repair and Osteoarthritis Treatment. Curr Rev Musculoskelet Med. 2020. Dec;13(6):641–650. doi: 10.1007/s12178-020-09664-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(TIFF)
(TIFF)
Data Availability Statement
We are not able to share publicly neither the clinical nor the biological raw data, even upon request. The reasons for this restriction are as follows: 1) Inserm is the owner of the clinical data originating from the COBRA cohort, 2) the informed consent signed by the patients of both cohorts does not mention that their data can be freely shared, even if they would be anonymized, and 3) this study was financially supported by AstraZeneca and in the legal contract with Inserm there is no agreement that allows sharing the data. These reasons are now listed in the appropriate box related to Data access. In this setting, it is not possible to publish the SOMAscan proteomic data as suggested by the PLOS ONE research data policy. To publish such data, even in anonymized profiles, would violate the written consent obtained from the participants and thus would not be in accordance with European General Data Protection Regulation. Due to these restrictions, the authors of this study will share only the used information guideline and a summary of all used data as main or supplemental material to this paper.