Abstract
Myocardial infarction causes a massive loss of cardiomyocytes (CMs), which can lead to heart failure accompanied by fibrosis, stiffening of the heart, and loss of function. Heart failure causes high mortality rates and is a huge socioeconomic burden, which, based on diets and lifestyle in the developed world, is expected to increase further in the next years. At present, the only curative treatment for heart failure is heart transplantation associated with a number of limitations such as donor organ availability and transplant rejection among others. Thus, the development of cellular reprogramming and defined differentiation protocols provide exciting new possibilities for cell therapy approaches and which opened up a new era in regenerative medicine. Consequently, tremendous research efforts were undertaken to gain a detailed molecular understanding of the reprogramming processes and the in vitro differentiation of pluripotent stem cells into functional CMs for transplantation into the patient’s injured heart. In the last decade, non-coding RNAs, particularly microRNAs, long non-coding RNAs, and circular RNAs emerged as critical regulators of gene expression that were shown to fine-tune cellular processes both on the transcriptional and the post-transcriptional level. Unsurprisingly, also cellular reprogramming, pluripotency, and cardiac differentiation and maturation are regulated by non-coding RNAs. In here, we review the current knowledge on non-coding RNAs in these processes and highlight how their modulation may enhance the quality and quantity of stem cells and their derivatives for safe and efficient clinical application in patients with heart failure. In addition, we summarize the clinical cell therapy efforts undertaken thus far.
Keywords: Non-coding RNA, Cell-therapy, Pluripotency, Reprogramming, Cardiac therapy, Regeneration
1. Introduction
Cardiovascular diseases (CVDs) leading to heart failure are the most common cause of death worldwide, which is particularly attributed to the lack of sufficient regenerative potential of the heart. The high mortality and morbidity rates of CVDs contribute to a vast socioeconomic burden. The Western lifestyle and demographic changes are expected to account for drastically increasing patient numbers in the next years.1 According to the 2015 American Heart Association CVD Burden Report, 41.5% of the US population was diagnosed with at least one type of CVD including high blood pressure, coronary heart disease, stroke, congestive heart failure, and atrial fibrillation. Unsurprisingly, CVDs are by far the costliest chronic diseases.2 Despite advances in biomedical research in the last decades, pharmacological interventions primarily focus on symptom reduction, while the only curative treatment for heart failure available to date is heart transplantation, which is accompanied by long waiting time for a donor heart and immune suppression after transplantation.
Contrary to the long-standing assumption, the heart is not a post-mitotic organ and proliferation of cardiomyocytes (CMs) occurs throughout life, although at an extremely low level.3 This endogenous regeneration, however, does not balance the huge loss of CMs during injury such as myocardial infarction and thus, subsequent maladaptive remodelling can lead to heart failure.4 The knowledge of endogenous CM turnover and the emergence of cell therapies have led to two different regenerative strategies for CVD: first, promoting the limited intrinsic proliferation of CMs and secondly, the replacement of lost CMs by transplantation of pluripotent stem cells (PSCs), PSC-derived cardiac progenitors or CMs (PSC-CMs).
PSCs have the ability to self-renew and differentiate into all cell types derived from the three germ layers emphasizing their remarkable regenerative potential in versatile diseases. Initially, embryonic stem cells (ESCs) were utilized in regenerative medicine and research, but ethical and legal limitations hamper their clinical application.5 Therefore, the discovery of cellular reprogramming with the so-called Yamanaka factors and the establishment of induced pluripotent stem cells (iPSCs) in 2006 revolutionized the stem cell field and heralded a new era in regenerative medicine.6 To generate iPSCs, virtually any somatic cell may be reprogrammed by the introduction of four transcription factors Oct4, Klf4, Sox2, and c-Myc. Since the initial discovery, several transcription factors and combinations introduced as genes, proteins, or mRNAs, and the delivery by various integrating and non-integrating vectors were tested successfully.7 Nonetheless, several problems emerged using the iPSC technology and scientists around the globe constantly seek to find solutions. For instance, the accessibility of different genetic regions in somatic cells varies, explaining the impact on reprogramming efficiency and maturation of iPSCs and in this regard, the advantages of their counterpart, the ESCs. In addition to their potential in regenerative medicine, iPSCs are also heavily studied as patient-specific platforms for disease modelling in 2D and 3D cell culture systems.8 This contributes to a wider and more detailed understanding of disease phenotypes and their underlying mechanisms, and finally helps to discover new therapeutic strategies and targets. Moreover, those platforms can also be used for drug screenings either for personalized medicine or for specific patient cohorts that are not responding to traditional treatments. In CM cultures also cardiotoxic side effects, which frequently occur during anti-cancer treatments,9,10 can be studied as this is often the reason for market withdrawal of novel drugs.8
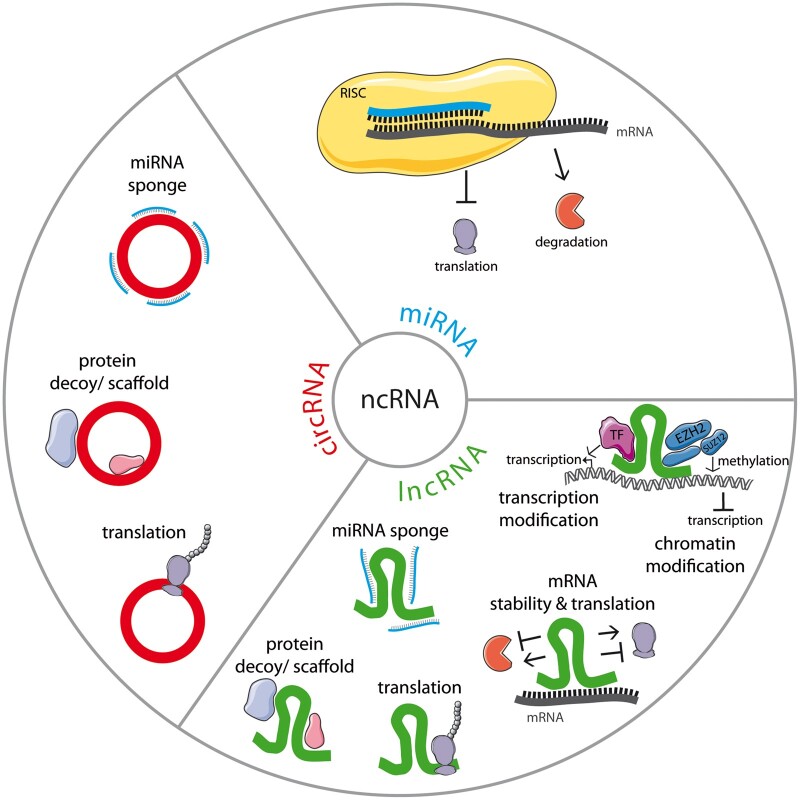
The human ENCODE project revealed that only 2% of the human transcriptome is protein-coding,11 whereas a large proportion accounts for non-coding RNA (ncRNA) transcripts, which are not translated into proteins. Those ncRNAs are emerging as crucial regulators of physiological and pathophysiological processes. Besides the well-described transfer RNAs and ribosomal RNAs, particularly microRNAs (miRNAs), long ncRNAs (lncRNAs) and circular RNAs (circRNAs) gained a lot of attention during the past two decades. While miRNAs are short oligo-ribonucleotides with a size of about 21–23 nucleotides, lncRNAs are defined by a length of more than 200 nucleotides.12 CircRNAs are the most recently described class of ncRNAs that are characterized by a ‘covalent closure’ forming a circRNA molecule.13 Functionally, miRNAs post-transcriptionally regulate gene expression by forming a miRNA-induced silencing complex with Ago proteins. The complex induces sequence-specific mRNA degradation or stalls mRNA translation12 (Figure 1). The modes of action of lncRNAs and circRNAs are much more heterogeneous. Both can regulate gene expression on the transcriptional and post-transcriptional level: in the nucleus, they can act as a scaffold for transcription factors and epigenetic modifiers, as protein decoys or as transcriptional enhancers. In the cytoplasm, they can influence mRNA stability, have a sponge effect on miRNAs or RNA-binding proteins, or serve as scaffolds for protein complexes.12,14 A small number of lncRNAs and circRNAs were even shown to code for micropeptides.15 Due to the relatively recent discovery of ncRNAs that make up the major part of our genome as well as the continuously growing number of newly described transcripts, ncRNA research is still in its infancy but has a promising potential for the development of novel regenerative strategies and for the treatment of many diseases. Additionally, all three classes of ncRNAs are found in extracellular fluids and can therefore serve as biomarkers.16
Figure 1.
NcRNAs fine-tune gene expression via versatile mechanisms. Cellular functions of ncRNAs are summarized here with miRNA in blue, lncRNA in green, and circRNA in red.
In this review, we focus on the exciting opportunities offered by ncRNAs as regulators of PSC-CMs for cardiac cell therapy. NcRNAs are promising targets to enhance or perform reprogramming of somatic cells, to improve the pluripotent state of PSCs, and to facilitate the differentiation into CMs in order to increase the quality and quantity of PSC-CMs for safe and efficient clinical application.
2. ncRNAs in cellular reprogramming
Cellular reprogramming is based on and accompanied by wide-ranging changes of the transcriptional and epigenetic landscape. Particularly, pluripotency-related genes are activated while genes crucial for cellular specification are silenced through chromatin remodelling processes. Since the discovery of reprogramming of murine fibroblasts into iPSCs with retroviruses containing Oct4, Klf4, Sox2, and c-Myc, alternative molecules and delivery strategies were identified to reprogram not only fibroblast but also other somatic cells successfully. As cellular reprogramming is a very inefficient process, different strategies were tested to enhance the generation of iPSCs through modulation, for example of telomerase and SV40 large T17 or p5318,19 in addition to the four reprogramming factors. Also the replacement of c-Myc with small molecules such as valproic acid (a histone deacetylase inhibitor)20 has been proven to be an effective strategy. Historically, murine embryonic fibroblasts had been the first described cell type that was reprogrammed,6 subsequently enabling the direct transfer of this fundamental discovery to human dermal fibroblasts.17 To date, successful generation of iPSCs from a wide range of cell types like keratinocytes, cord blood, or peripheral blood cells has been demonstrated.7
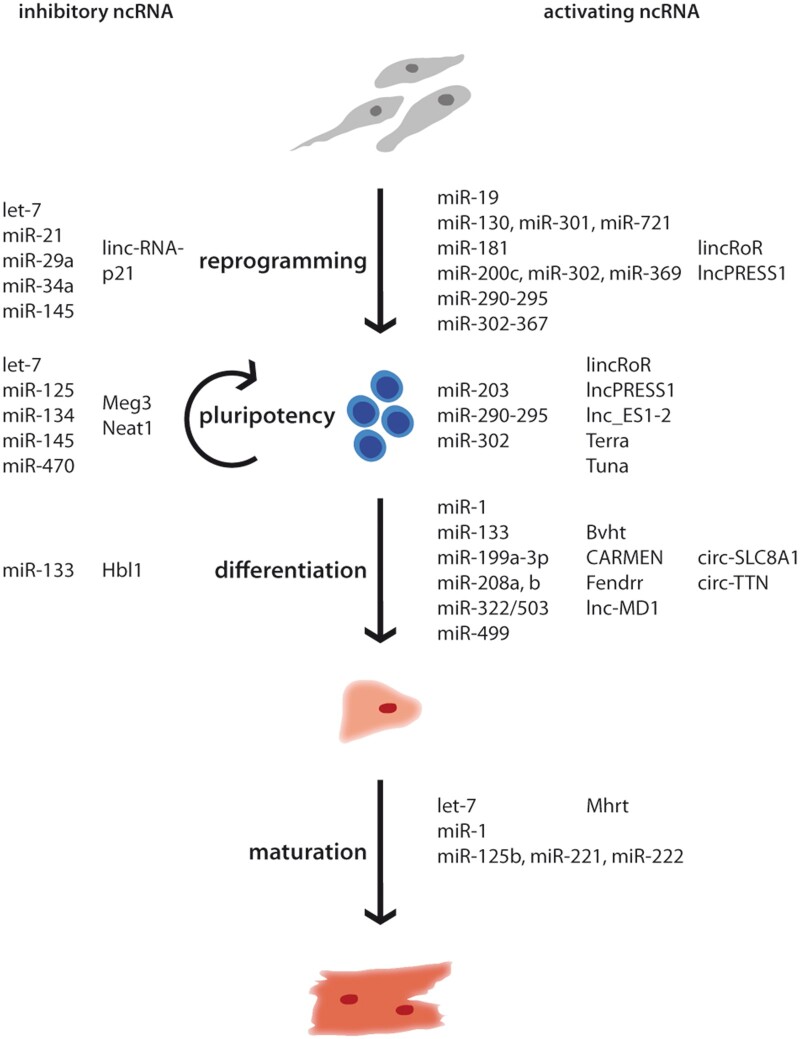
Not surprisingly, as master regulators of gene expression, ncRNAs were described to influence stem cell properties with some of them being sufficient to reprogram somatic cells alone (Figure 2). The miR-302–367 cluster was found to reprogram human cancer cell lines21 as well as murine and human fibroblasts22,23 even more efficient than Oct4, Klf4, Sox2, and c-Myc. miR-200c, miR-302, and miR-369 were sufficient for the reprogramming of adipose stromal cells and dermal fibroblasts, but exhibited a lower efficiency.24 A major advantage of synthetic and mature miRNA mimics as a reprogramming strategy is the non-integrative nature of those molecules, thus, no transgenes remain in the generated iPSCs. Importantly, if delivered by multiple consecutive transfections, miRNAs have a similar reprogramming efficiency compared to commonly used retroviruses.7
Figure 2.
NcRNA influence reprogramming of iPSCs, pluripotency, differentiation to CMs, and their maturation. NcRNAs regulating the process positively are summarized on the right side, inhibitory ncRNAs on the left.
miRNAs were also used in addition to or instead of one of the classical reprogramming factors. For example, the miR-290–295 cluster, known as ESC-specific cell cycle-regulating miRNAs, regulates the transition from G1 to S phase.25 From this family, miR-291, miR-294, and miR-295 also play a role in activating the stemness properties as these are under the transcriptional control of c-Myc and thus, each of them can substitute c-Myc to enhance reprogramming together with Oct4, Klf4, and Sox2.26 From this cluster, miR-294 seems to be the most important, because it interferes with super ordinated pathways such as Akt, Wnt, and TGFβ signalling.27 Also the other family members have further regulatory duties such as miR-291, which suppresses the expression of p65 and concurrently NF-κB formation.28 Interestingly, like miR-294, miR-181 also acts on Wnt and TGFβ pathways initiating reprogramming without having synergistic effect, suggesting that both miRNAs influence additional mRNAs, which are connected to reprogramming.27
Moreover, the expression of the core transcription factors Oct4, Sox2, and Nanog is controlled by miRNAs. miR-34a degrades the transcripts of Nanog, Sox2, and c-Myc. Consequently, reprogramming of miR-34a knockout murine fibroblasts is more efficient than their wild-type counterparts.29 Also the knockdown of miR-145 leads to a higher efficiency as this miRNA represses the translation of Oct4, Sox2, and Klf4.30 The inhibition of let-7 in combination with overexpression of Oct4, Sox2, and Klf4 results in a similar efficiency compared to reprogramming including c-Myc as let-7 inhibits LIN-41, which is another essential transcription factor for reprogramming.31 Also miR-130, miR-301, and miR-721 enhance iPSC generation by inhibiting the translation of mesenchyme homeobox 2 (Meox2), a homeobox transcription factor and negative regulator of reprogramming.32
Reprogramming can lead to genome destabilization and therefore activate tumour suppressors, which can be based on or enhanced by the reprogramming vector integration. In turn, miRNAs involved in those processes can be exploited to increase reprogramming efficiency. NcRNAs add an additional level of regulation as the control of such crucial processes has to be stringent, otherwise cells can transform into a cancerous state. For example, miR-19 represses phosphatase and tensin homolog (PTEN) and can be used as a substitute for c-Myc,33 while inhibition of miR-29a and miR-21 indirectly leads to depletion of p53, which is known to greatly facilitate reprogramming.34
Besides miRNAs, lncRNAs also play crucial roles during reprogramming (Figure 2). The first described lncRNA in reprogramming is regulator of reprogramming (lincRoR), which acts as a sponge for several miRNAs such as miR-145, which represses the expression of the core pluripotency factors.35,36 LncRNAs not only influence gene expression post-transcriptionally but also interact with chromatin modifiers. For example, lncPRESS1 acts as a decoy for the histone deacetylase SIRT6 maintaining the acetylation of pluripotency-related genes in pluripotency, while it is repressed by p53 during differentiation.37 In contrast, lincRNA-p21 is induced by p53 and prevents reprogramming by building a scaffold for histone and DNA methyltransferases at pluripotency genes.38
To conclude, ncRNAs influence the expression of genes regulating stem cell features on different levels while interfering in various processes during reprogramming. Due to the later discovery, less lncRNAs and barely any circRNAs are described in reprogramming,39 but this will certainly change within the next years. In particular, synthetic miRNAs or miRNA inhibitors can be used as a cocktail to induce or enhance ‘traceless’ reprogramming of somatic cells into iPSCs which is of great interest for the application of patient-specific iPSCs in medical sciences.
3. ncRNAs in control of pluripotency
Pluripotency is a complex process and tightly regulated on multiple levels. The pluripotency gene regulatory network orchestrates the pluripotent state of a cell via core transcription factors and, additionally, via chromatin-mediated and RNA-based processes, which fine-tune the regulation and increase the variety of translated proteins. The super-ordinated role of ncRNAs, in particular miRNAs, has been highlighted in a knockout model in PSCs, where core proteins of the riboprotein complex involved in miRNA processing, namely Dicer and DGCR8, were absent. As a consequence, the proliferative capacity and differentiation of PSCs into specialized cell types was severely impaired.40,41 Several miRNAs are described as regulators of pluripotency with a more pronounced effect on differentiation (Figure 2). Pluripotency is regulated on versatile levels by miRNAs, for example, miR-302, described above as a reprogramming factor, also inhibits the translation of the transcription factor NR2F2, which initiates neural development.42 The miR-290–295 cluster regulates cell cycle progression of PSCs and therefore plays not only an important role during reprogramming but also in maintaining self-renewal.25 Culture conditions of PSCs can be improved by miR-203 and lead to a higher differentiation capacity by repressing DNA methyltransferases.43 The differentiation of ESCs is promoted by miR-125, which inhibits Cbx7, a chromatin modifier, and enhances the maturation of the pro-differentiative miRNA let-7.44 The core pluripotency factors Nanog, Sox2, and Oct4 are negatively regulated by many miRNAs, for example miR-145, miR-470, miR-134, and miR-296, which inhibit their translation.30,45
LncRNAs often exhibit regulatory features specifically in distinct cell types, so it is already conceivable that they also modulate pluripotency (Figure 2). For example, lincRoR as mentioned above sponges miR-145, which derepresses the translation of Oct4, Sox2, and Nanog.36 The multifaceted modes of action of lncRNAs can be illustrated by the following lncRNAs. LncRNA Tuna (megamind) activates the transcription of Nanog, Sox2, and Fgf4 by recruiting RNA-binding proteins polypyrimidine tract-binding protein 1 (PTBP1), heterogeneous nuclear ribonucleoprotein K (hnRNP-K), and nucleolin (NCL) to their promoter region.46 Nanog was also assumed to be targeted by the lncRNA Cyrano,47 but could not be validated in a recent study that used several knockout and knockdown techniques to eliminate Cyrano in ESCs and iPSCs without any effect on pluripotency.48
LncRNAs are commonly involved in the epigenetic control of specific processes maintaining or inducing a pluripotent state. For instance, lnc_ES1 and lnc_ES2 regulate on the one hand the expression of Sox2 and on the other hand are involved in chromatin remodelling via targeting SUZ12, which is part of the histone-modifying complex PRC2.49 Furthermore, also Meg3 and lncPRESS1 influence pluripotency by distinct chromatin modifications, which are described above. Meg3 serves as a scaffold for PRC2 at promoters of pluripotency-associated genes.50
Pluripotency is also regulated by lncRNAs, which are well known in a stem cell-independent context. For example, Terra, a particular lncRNA that is transcribed from several sub-telomeric regions into the telomere, inhibits the transcription of the pluripotency repressor TCF3.51
Taken together, ncRNAs bear the potential to optimize culture conditions and keep PSCs in an optimal pluripotent state. It is therefore conceivable that panels of ncRNAs may even be exploited to assess the quality of PSCs before using them in subsequent manufacturing steps and applying them to clinics.
4. ncRNAs in cardiac differentiation
Many patients with CVD suffer from cardiac damage characterized by massive loss of CMs. The general idea is that these patients could benefit from a cardiac cell therapy that has already been explored in a large number of pre-clinical and clinical studies. One example is the clinical trial ESCORT (Transplantation of Human Embryonic Stem Cell-derived Progenitors in Severe Heart Failure) where 5–10 million ESC-derived cardiac progenitors were required per patient highlighting a major challenge for cardiac cell therapies (Table 1).52 To fulfil this high demand for cells, the manufacturing processes have to be scaled up in an effective and cost-efficient way while maintaining quality and integrity of the cells. Indeed, such upscaling has been the focus of research in recent years and substantial progress can be reported. Large-scale adherent monolayer platforms and three-dimensional suspension cultures in stirred tank reactors were established for cardiac differentiation.53 From a differentiation in monolayer, for example 7.2 × 108 iPSCs were differentiated to 6.2–7.0 × 108 CMs after purification (with a purity of 99%), whereas in a Good Manufacturing Practice (GMP) compliant process in a suspension culture 1 × 106/mL ESCs were differentiated in carrier-free aggregates resulting in 1.5–2 × 106/mL CMs (90% purity on Day 25 of differentiation).53 In addition to the high demand of needed cells, the cells were applied by different approaches in the clinical trials. Besides intracoronary infusions or epicardial injections, cells were also transplanted as patches. For the latter, as in the ESCORT trial, the cells have to undergo a tissue engineering process to embed the cells in a fibrin scaffold adding another step of complexity to the manufacturing process.
Table 1.
Overview of registered cell therapy trials for the treatment of CVD
| Study, year, phasea | Type of cells, number of cells, time point of intervention, and delivery | Results | Clinical trial number |
|---|---|---|---|
| Bone marrow (-derived) cells | |||
| TOPCARE-AMI, 2001, IIa | Bone marrow-derived progenitor cells or circulating progenitor cells (CD34/CD45-positive), 1.6–9.4 million cells, 3–7 days after AMI by intracoronary infusion |
|
Not available |
| BOOST, 2002, I | Autologous bone marrow cells, 2500 million cells, 4–8 days after PCI by intracoronary transfer |
|
NCT00224536 |
| ASTAMI, 2003, II | Autologous mononuclear bone marrow cells, 54–130 million cells, 4–7 days after PCI by intracoronary injection |
|
NCT00199823 |
| LEUVEN-AMI, 2003, II | Autologous bone marrow-derived stem cells, 176–432 million nucleated cells and 100–244 million mononucleated cells, by intracoronary injection |
|
NCT00264316 |
| STEMI, 2003, II | Autologous bone marrow-derived stem cells, 304 million cells, 1 day after reperfusion by intracoronary infusion |
|
NCT00264316 |
| REGENT, 2004 | Autologous bone marrow-derived unselected mononuclear cells or CD34+-CXCR4+ cells, 178 million BMNCs and 1.9 million CD34–CXCR4 cells (median), 3–12 days after PCI by intracoronary infusion |
|
NCT00316381 |
| BONAMI, 2004, II | Autologous bone marrow mononuclear cells, 89.6–107 million cells, 7–10 days after PCI by intracoronary injection | LV function↑, myocardial viability↑ | NCT00200707 |
| REPAIR-AMI, 2004, III | Autologous bone marrow-derived progenitor cells, 50 mL bone marrow aspirated, 3–7 days after reperfusion therapy by intracoronary infusion |
|
NCT00279175 |
| SCAMI, 2005, II | Autologous bone marrow cells, mean 381 million cells, mean 7 days after AMI by intracoronary administration with over-the-wire balloon catheter |
|
NCT00669227 |
| HEBE, 2005 | Autologous mononuclear bone marrow cells or peripheral mononuclear blood cells, 132–460 or 150–424 million cells, respectively, 3–8 days after AMI by intracoronary infusion | 4 months: ↔, clinical events↔ | NTR166 (Netherlands Trial Register) |
| TIME, 2008, II | Autologous bone marrow mononucleated stem cells, 150 million nucleated cells, 3/7 days after MI by intracoronary infusion |
|
NCT00684021 |
| LateTIME, 2008, II | Autologous bone marrow mononucleated stem cells, 150 million nucleated cells, 2–3 weeks after MI by intracoronary infusion | 6 months: ↔ | NCT00684060 |
| C-CURE, 2008, II/III | Bone marrow-derived cardiopoetic cells, 605–1168 million cells, MI/revascularization max. 2 months ago by endocardial injection |
|
NCT00810238 |
| BAMI, 2013, III | Autologous bone marrow-derived mononuclear cells, 2–8 days after reperfusion by percutaneous intracoronary intervention with over-the-wire balloon | Results not published yet, follow-up ended October 2019 | NCT01569178 |
| Mesenchymal stem cells | |||
| Prochymal, 2005, I | Allogeneic bone marrow-derived MSCs, 0.5/1.6/5 million cells per kilogram, by intravenous infusion |
|
NCT00114452 |
| SEED-MSC, 2007, II/III | Autologous bone marrow-derived MSCs, 1 million cells per kilogram body weight, mean 4 weeks after PCI by intracoronary injection |
|
NCT01392105 |
| TAC-HFT, 2008, I/II | Autologous mesenchymal and bone marrow cells, 100/200 million cells, by transendocardial injection during cardiac catheterization |
|
NCT00768066 |
| POSEIDON-Pilot, 2010, I/II | Autologous/allogeneic bone marrow-derived MSCs, 20/100/200 million cells, by transendocardial injection during cardiac catheterization |
|
NCT01087996 |
| WJ-MSC-AMI, 2011, II | Umbical Wharton’s Jelly-derived MSCs, 6 million cells, 4–7 days after reperfusion by intracoronary infusion |
|
NCT01291329 |
| CHART-1, 2012, III | Autologous bone marrow-derived mesenchymal cardiopoetic cells, 600 million cells, by intramyocardial injection |
|
NCT01768702 |
| TRIDENT, 2014, II | Allogenic adult MSCs, 20/100 million cells, by transendocardial injection |
|
NCT02013674 |
| Adipose tissue-derived regenerative cells | |||
| APOLLO, 2007, I | 20 million cells, 1 day after PCI by intracoronary infusion |
|
NCT00442806 |
| PRECISE, 2007, I | 0.4/0.8/1.2 million cells per kilogram body weight, by transendocardial injection |
|
NCT00426868 |
| ADVANCE, 2012, II | Autologous, via intracoronary route |
|
NCT01216995 |
| ATHENA, 2012/2014, II | Autologous, 0.4/0.8 million cells per kilogram body weight (max. 40/80 million cells), by intramyocardial injection |
|
NCT01556022 NCT02052427 |
| Cardiosphere-derived stem cells | |||
| CADUCEUS, 2009, I | Autologous, 12.5/25 million cells, 1.5–3 months after MI by intracoronary infusion |
|
NCT00893360 |
| ALLSTAR, 2012, I/II | Allogenic, 25 million cells, within 1 year after MI by intracoronary infusion |
|
NCT01458405 |
| Cardiac stem cells | |||
|
1 million cells, 3–5 months after surgery by intracoronary infusion |
|
NCT00474461 |
| CAREMI, 2014, I/II | Allogenic, 10/20/35 million cells, 5–7 days after reperfusion by intracoronary infusion |
|
NCT02439398 |
| Embryonic stem cell-derived progenitor cells | |||
| ESCORT, 2013, I | ESC-derived cardiac progenitors (SSEA-1+ Isl-1+), 5–10 million in a fibrin patch, MI min. 6 months ago, patch was fixed by ‘Kangaroo’ procedure |
|
NCT02057900 |
| Induced pluripotent stem cell-derived cardiomyocytes | |||
| 2018, I | Allogenic, 100 million cells in 4–5 cm 0.1 mm thick sheets | Ongoing | jRCT2053190081 (Japan Registry of Clinical Trials) |
| HEAL-CHF, 2019 | Allogenic, 100 million cells, 5, epicardial injection at coronary artery bypass surgery | Ongoing | NCT03763136 |
Study start according to clinicaltrial.gov
EED, early enhancement defect; LVEF, left ventricular ejection fraction; LVESV, left ventricular end-systolic volume; LVEDV, left ventricular end-diastolic volume; MR, magnetic resonance; NYHA, New York Heart Association; PCI, percutaneous coronary intervention; PET, positron emission tomography.
In vitro cardiac differentiation mimics the embryonic development of the heart. Several crucial signalling pathways including bone morphogenetic protein, Wnt/β-catenin, Notch or fibroblast growth factor (FGF) signalling are found to induce cardiomyocyte-specific gene programmes in an accurately coordinated way. For in vitro generation of CMs, exogenous regulation, which is accomplished by specific compounds, needs to be strictly timed as the differentiation process is extremely delicate.54 NcRNAs are also involved in cardiac differentiation (Figure 2), for example miR-1 promotes differentiation to the mesoderm by increasing the expression of transcription factors associated with cardiogenesis and the sarcomeric proteins in cooperation with miR-133.55 In addition to these regulatory effects during the early time points of differentiation, miR-1 inhibits Wnt and FGF signalling in cardiac progenitors leading to the differentiation of CMs,56 whereas miR-133 has an antagonistic role in those subsequent processes.55 The miRNAs miR-199a-3p and miR-483-3p are enriched in mesodermal progenitor cells and regulate their corresponding target genes ACVR2A and PGAM1, which play roles in the Nodal/TGFβ signalling and glycolysis pathway.57 The so-called myomiRs, which are CM exclusively expressed, miR-208a, miR-208b, and miR-499 are transcribed from the introns of myosin heavy chain (Myh) 6, 7, and 7b and regulate the expression of their host genes, therefore influencing the differentiation and progression to more adult CMs.58 miR-322/503 increases the cardiomyocyte yield by driving PSCs to the cardiac fate and inhibiting neural lineages.59
A well-known lncRNA in cardiac development is Braveheart, which initiates transcription of early cardiac genes like Mesp1, Nkx2.5, Tbx5, and Gata4 by interacting with Suz12 in mice, so far no human homologue was identified.60 LncRNAs are often poorly conserved in their primary structure, but many lncRNAs may have functional homologous across different species. In human cells, several lncRNAs are known to influence cardiac commitment. For example, lncRNA Fendrr modulates gene expression via recruiting methyltransferase mixed lineage leukemia (MLL) to the promoter of forkhead box F1 (Foxf1) and paired like homeodomain 2 (Pitx2), two important transcription factors of early embryogenesis (lateral plate mesoderm), where the methylation leads to increased transcription. Additionally Fendrr binds to PRC2 reducing gene expression.61 Initiating and maintaining cardiomyocyte identity is also dependent on the cardiac mesoderm enhancer-associated non-coding RNA (CARMEN). CARMEN is a super enhancer-associated lncRNA that interacts with SUZ12 and EZH2, which is enzymatically active domain of PRC2 and thereby influences the differentiation of cardiac progenitors.62 Lnc-MD1 sequesters miR-133 and miR-135, constructing a regulatory network of ncRNAs, preventing the repression of transcription factors needed for cardiomyocyte differentiation.63 Similarly, heart break lncRNA1 binds to miR-1 thus inhibiting differentiation and is in turn regulated by SOX2.64 Accordingly, lncRNAs mainly operate via repression of distinct regulatory pathways rather than activating desired pathways.65
In contrast to lncRNAs, circRNAs are enriched in the later stages of cardiac differentiation and show temporal expression patterns. Since circRNA research is still in its infancy, only few mechanistic studies were performed so far. First insights were gained for a circRNA from the titin (TTN) locus, circ-TTN, which is highly expressed in CMs and described to bind miR-432 competitively and therefore inhibiting the PI3K/Akt pathway.66,67 Additionally, circ-SLC8A1 reveals an enrichment in CMs suggesting a role in cardiac differentiation.66
Besides high demands on iPSC-CM quality and quantity for clinical application, which may be improved by modulation of ncRNAs, in particular the maturation of engineered CMs remains a critical issue. PSC-CMs differ from adult CMs in their transcriptome, cytoskeleton structure, metabolism, and electrophysiology.68 For the transplantation of cells, the electrical and mechanical capacity of the produced CMs should resemble the physiological parameters of the myocardium69; otherwise, as reported in animal studies, the transplantation of immature CMs can cause arrhythmias.70,71 Several strategies and concepts were tested to mature CMs in culture with a focus on a prolonged culturing time, stiff substrates resembling the collagen deposition during embryogenesis, cardiac engineering techniques, mechanical loading, electrical stimulation, or neurohormonal factors.69 Recent approaches include the modulation of ncRNAs as these are also involved in the maturation of CMs. The most prominent example is miR-1 where the overexpression leads to the functional maturation of electrophysiological properties of CMs by influencing the action potential.72 Let-7 inhibits the PI3K/AKT/insulin signalling cascade leading to the metabolic change from glucose to fatty acids typically observed during CMs maturation.73 Overexpression of miR-125b, miR-199a, miR-221, and miR-222 resulted in more mature PSC-CMs as observed by co-cultivation with endothelial cells.74 Furthermore, lncRNA Myheart inhibits the chromatin remodelling factor Brg1 and therefore impairs the expression of its target genes MYH6 and MYH7, which is accompanied by a decrease of the MYH7/6 ratio. This might be an important factor and could additionally influence maturation of CMs.75
5. Cardiac cell therapies in clinical trials
Curing cardiac injury by transplanting lost CMs with in vitro generated PSC-CMs is one of the scopes of regenerative medicine in CVD. The concept of cardiac cell therapeutic approaches has been fuelled by seminal discoveries including iPSC technology, large-scale cell production, defined cardiac cell specification, and cardiac tissue engineering over the past two decades. Pre-clinical animal models have been instrumental in the translation of these technologies into clinics. For example, transplantation of PSC-CMs after myocardial infarction ameliorated cardiac function in small animal models by engraftment and electrical coupling of the injected cells with the myocardium.76–81 Subsequently, applying cardiac cell therapies in large animal models like non-human primates (macaque)71,82 or pigs83 paved the way for translation into a clinical context. A recurring issue is the occurrence of arrhythmias after cell therapy. Noteworthy, these complications were especially detected in large animals, probably due to higher amount of transplanted PSC-CMs. Additionally, changes in the heart rate of small animals might be masked by their fast heart rate.71 Although in some studies no arrhythmic effects of ESC-CMs have been observed, arrhythmogenesis has to be studied carefully prior to clinical application of promising cell therapies.78
For ischaemic heart disease, several clinical trials using multipotent stem cells were initiated in the past decades (Table 1). Different cell types, e.g. bone marrow cells, mesenchymal stem cells (MSCs), cardiac stem cells, or cardiosphere-derived cells, served as a cell source. In general, the cell number, route of application, and time point after myocardial infarction had a considerable influence on the respective outcome. In some studies, a slight beneficial effect of stem cell therapies was documented, whereas others did not report any effects. The clinical trials revealed so far no safety issue, but whether cardiac cell therapies are the solution for the increasing number of patients with heart failure and their high burden remains unclear. The advantage of transplanting early progenitors is their high plasticity and adaption to the injured tissue environment, whereas more mature PSC-CMs have a higher potential to improve cardiac function by replacing the lost CMs. So far, most clinical trials were conducted with multipotent cells, PSC-CMs have only been used a couple of times. One clinical trial was based on ESC-derived cardiac progenitor cells (ESCORT, NCT02057900) and after the first year, no tumours or arrhythmias were detected in the study group consisting of five patients. The systolic motion of the heart area that received a fibrin patch with cells was improved.52 In 2018, the first clinical application of iPSC-derived CMs was approved in Japan and included 10 patients (Japan Registry of Clinical Trials jRCT2053190081). The first patient received a cell sheet of 100 million iPSC-CMs in January 2020. Also in 2018, a Chinese clinical trial was registered injecting iPSC-CMs to patients with chronic heart failure systemically, but did not recruit any patients so far (IDCVTCHF, NCT03759405). In 2019, the HEAL-CHF trial (NCT03763136) was enrolled to acquire the efficiency of injection iPSC-CM epicardially. A major shortcoming of many cell therapies is the washout of the infused or injected cells and the low engraftment rates after therapy. In two of the cardiac cell therapy studies, the PSC-CMs were transplanted as a cardiac patch. With this tissue-engineering approach, 3D structures of PSC-CMs are first created in vitro and then transplanted to foster retention of the cells with scaffolds such as natural hydrogels and synthetic polymers. Also natural, organ-based scaffolds like decellularized hearts are investigated.54
In an animal study, different application routes of bone marrow cells after myocardial infarction were compared: intramyocardial injection outcompeted infusions with 7% of cell retention vs. 1% and injecting the cells in a fibrin scaffold led to even higher retention rates of 17% 3 days after infarction.84 As the cell retention increases with engineering approaches, the cell number needed for transplantation reduces, which also make a huge positive impact on the financial expenditure for the cell production process.
In humans, after a myocardial infarction, the scar might be huge in relation to laboratory manufacturing scales, whereas the transplanted patch needs to have a similar size to rescue the heart function. With an increasing size of the engineered tissue, an adequate nutrition, which relies on diffusion processes, becomes problematic. Therefore, vascularization of cell sheets is investigated intensively, which can be achieved by embedding endothelial cells and distinct exogenous stimuli.85
Although transplanted cells do not persist in the heart for long, especially after intramyocardial injection or infusion, studies reported a beneficial effect beyond the persistence of the cells. Especially for MSC therapies, functional improvements based on remuscularization is improbable. This regenerative potential is assumed to be based on the secretome of the transplanted cells including cytokines, growth factors, and others often delivered to cells within the microenvironment via extracellular vesicles.86,87 Extracellular vesicles, which among other biomolecules contain miRNAs, improved the cardiac function when injected into the heart after purification from the producing cells.88 Besides paracrine effects, cell transplantation also generated an acute sterile immune response that improved cardiac function by the infiltration of macrophages89 by reducing fibroblast activity and the amount of extracellular matrix in the border zone. The beneficial effects after systemic application of cells for cardiac therapy, which rarely end up in the heart, support the paracrine hypothesis and suggest repeated dosing for long-lasting effects.90 Therefore, a longer retention of the cells would also lead to a prolonged paracrine effect combining the best of two worlds.
Stem cell-derived products bear several safety concerns like the risk of tumourigenesis by not terminally differentiated cells remaining in the cell product upon transplantation. iPSCs are commonly generated with integrating vectors and oncogenic factors.91 During long-term culture, the probability and number of chromosomal abnormalities raises, leading to a more serious risk for transforming events. After iPSC-CMs transplantation in animals, tumour and teratoma formation was reported by some groups, but the percentage was low in the large number of available studies and especially immune-deficient animals were more prone to tumour formation.92 Also the clinical trials with stem cells and stem cell-derived early progenitors did not show any increased tumour incidence in the patients. Still there is the potential of tumourigenic long-term effects of transplanted stem cells; therefore, several methods are tested to eliminate remaining PSCs in PSC-CMs productions. The confidence in scientists and their research in cardiac cell therapies suffered a major set-back after the wide-ranging retraction of papers connected to cardiac stem cell therapy including the SCIPIO trial (NCT00474461). Not only the use of cardiac progenitor cells but the whole field focusing on the development of new therapies in CVD with the help of applying cells was critically interrogated. Even though the initial studies on c-kit+ cells and the clinical trial were retracted, independent laboratories could show in different pre-clinical models a beneficial effect of those cardiac progenitor cells, which most likely rely on paracrine effects (discussed above).93
Efficient cardiac differentiation and advanced purification protocols are investigated, as well as the additional selection with suicide genes,94–97 cytotoxic antibodies,98,99 or miRNA switches.100,101 Regarding the latter, a construct with a constitutively expressed fluorescence reporter is generated with an upstream binding site of a miRNA, which is specifically expressed in mature CMs. Therefore, cells expressing the fluorophore can subsequently be eliminated by cell sorting as these are identified as non-mature CMs, because mature CMs express the miRNA that leads to the degradation of the fluorophore. For a purification independent of cell sorting, apoptosis-inducing genes can be cloned downstream of the miRNA-binding sites leading to autonomous induction of apoptosis in non-CMs.100
In summary, PSCs and derived CMs bear a huge potential in regenerative medicine but safety and efficacy have to be evaluated carefully before they can be applied as a standard clinical therapy.
6. Conclusions and future perspectives
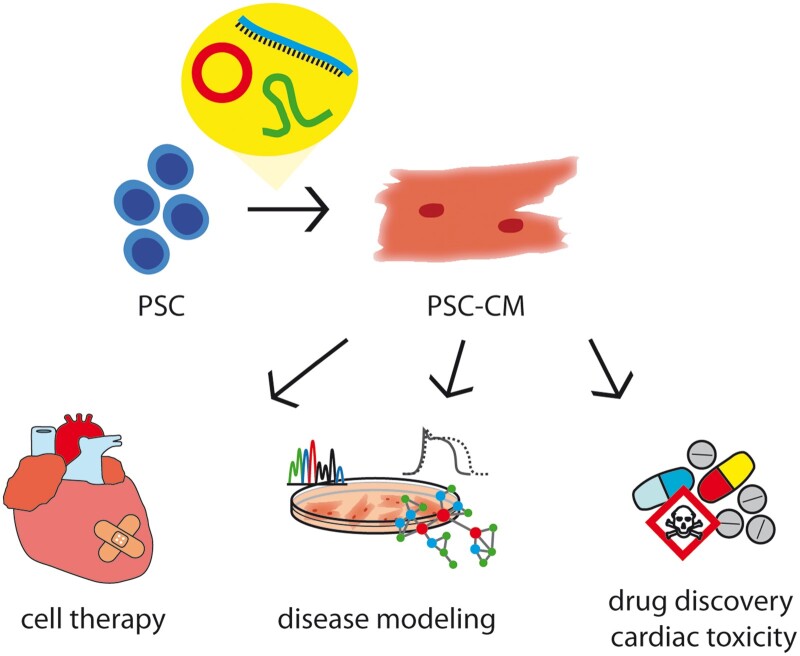
Besides tremendous efforts, cardiac cell therapies based on PSC-CMs have not entered clinical routine so far. Results of the ongoing clinical trials are awaited to earn more knowledge of safety and efficacy of PSC-CMs in humans. To solve the concerns regarding safety of the cell products, various strategies are tested including the exploitation of ncRNAs. By regulating cellular processes, ncRNAs can be used to fine-tune the different steps of PSC-CMs generation for clinical application: for reprogramming to iPSCs in a non-integrative manner, for optimized culture conditions of PSCs, more efficient cardiac differentiation and maturation (Figure 3). Cocktails of specific ncRNAs for the different steps of the manufacturing process are imaginable.
Figure 3.
NcRNAs might bridge the hurdles of PSC-CMs implementation in clinics and applied research.
Until application is clinically approved, disease modelling and drug screening on iPSC-based platforms will also benefit from further research and improvement of culture systems. When thinking of personalized medicine, a fast and efficient generation of patient-specific iPSCs and differentiation to the desired cell type has a huge impact on the health of patients. Also for the identification of novel drugs, an optimal screening platform is needed where the quality of PSC-CMs has drastic effects on the screening results and whether the discovery of novel therapeutic strategies is translatable from the dish to patients.
NcRNAs also bear the potential to design a panel for quality control to assess pluripotency, cardiac purity, and maturation of the cells. Especially lncRNAs and circRNAs reveal cell type-specific expression patterns and might serve as a suitable tool to examine the quality of the cells before using them in clinics.102
Among the many different strategies aiming to improve PSC-CMs for cardiac cell therapy, ncRNAs have an emerging role as gene regulators and chromatin modifiers, thus regulating the different manufacturing steps of iPSC-CMs. Further detailed studies are needed to identify novel ncRNAs and characterize their mode of action. Considerable progress can be expected by novel high-throughput loss-of-function approaches and technologies such as single-cell sequencing to identify new ncRNAs especially with a dynamically regulated expression.
Contributor Information
Hannah J Hunkler, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, 30625 Hannover, Germany.
Sonja Groß, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, 30625 Hannover, Germany.
Thomas Thum, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, 30625 Hannover, Germany; REBIRTH Center for Translational Regenerative Medicine, Hannover Medical School, Carl-Neuberg-Str. 1, 30625 Hannover, Germany; Fraunhofer Institute for Toxicology and Experimental Medicine, Nikolai-Fuchs-Str. 1, 30625 Hannover, Germany.
Christian Bär, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, 30625 Hannover, Germany; REBIRTH Center for Translational Regenerative Medicine, Hannover Medical School, Carl-Neuberg-Str. 1, 30625 Hannover, Germany.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (SFB/Transregio TRR267 to C.B. and T.T.), the Federal Ministry of Education and Research (BMBF, Germany, research grant ERA-CVD JTC2018 INNOVATION, 01KL1903, to C.B.), and a REBIRTH Synergy grant (to C.B.).
References
- 1. Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, Chiuve SE, Cushman M, Delling FN, Deo R, de Ferranti SD, Ferguson JF, Fornage M, Gillespie C, Isasi CR, Jiménez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Lutsey PL, Mackey JS, Matchar DB, Matsushita K, Mussolino ME, Nasir K, O’Flaherty M, Palaniappan LP, Pandey A, Pandey DK, Reeves MJ, Ritchey MD, Rodriguez CJ, Roth GA, Rosamond WD, Sampson UKA, Satou GM, Shah SH, Spartano NL, Tirschwell DL, Tsao CW, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS, Muntner P; American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee . Heart disease and stroke statistics-2018 update: a report from the American Heart Association. Circulation 2018;137:e67–e492. [DOI] [PubMed] [Google Scholar]
- 2. Khavjou O, Phelps D, Leib A.. Projections of Cardiovascular Disease Prevalence and Costs: 2015-2035. RTI International; 2016. p1–54. https://www.heart.org/-/media/files/get-involved/advocacy/burden-report-technical-report.pdf?la=en. [Google Scholar]
- 3. Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabé-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisén J.. Evidence for cardiomyocyte renewal in humans. Science 2009;324:98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sutton J, St MG, Sharpe N.. Left ventricular remodeling after myocardial infarction: pathophysiology and therapy. Circulation 2000;101:2981–2988. [DOI] [PubMed] [Google Scholar]
- 5. Yamanaka S. A fresh look at iPS cells. Cell 2009;137:13–17. [DOI] [PubMed] [Google Scholar]
- 6. Takahashi K, Yamanaka S.. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663–676. [DOI] [PubMed] [Google Scholar]
- 7. González F, Boué S, Belmonte JCI.. Methods for making induced pluripotent stem cells: reprogramming à la carte. Nat Rev Genet 2011;12:231–242. [DOI] [PubMed] [Google Scholar]
- 8. Matsa E, Burridge PW, Wu JC.. Human stem cells for modeling heart disease and for drug discovery. Sci Transl Med 2014;6:239ps6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chatterjee S, Gupta SK, Bär C, Thum T.. Noncoding RNAs: potential regulators in cardioncology. Am J PhysiolHeart Circ Physiol 2019;316:H160–H168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chatterjee S, Hofer T, Costa A, Lu D, Batkai S, Gupta SK, Bolesani E, Zweigerdt R, Megias D, Streckfuss-Bömeke K, Brandenberger C, Thum T, Bär C.. Telomerase therapy attenuates cardiotoxic effects of doxorubicin. Mol Ther 2021;29:1395–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. ENCODE Project Consortium . An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beermann J, Piccoli MT, Viereck J, Thum T.. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev 2016;96:1297–1325. [DOI] [PubMed] [Google Scholar]
- 13. Santer L, Bär C, Thum T.. Circular RNAs: a novel class of functional RNA molecules with a therapeutic perspective. Mol Ther 2019;27:1350–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bar C, Chatterjee S, Thum T.. Long noncoding RNAs in cardiovascular pathology, diagnosis, and therapy. Circulation 2016;134:1484–1499. [DOI] [PubMed] [Google Scholar]
- 15. Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, Olson EN.. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015;160:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de G-CD, Vea A, Bär C, Fiedler J, Couch LS, Brotons C, Llorente-Cortes V, Thum T.. Circulating non-coding RNAs in biomarker-guided cardiovascular therapy: a novel tool for personalized medicine? Eur Heart J 2019;40:1643–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ.. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 2008;451:141–146. [DOI] [PubMed] [Google Scholar]
- 18. Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, Song Z, Liu Y, Yong J, Zhang P, Cai J, Liu M, Li H, Li Y, Qu X, Cui K, Zhang W, Xiang T, Wu Y, Zhao Y, Liu C, Yu C, Yuan K, Lou J, Ding M, Deng H.. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell 2008;3:475–479. [DOI] [PubMed] [Google Scholar]
- 19. Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JCI.. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 2009;460:1140–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA.. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol 2008;26:795–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DTS, Chen DT, Ying SY.. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA 2008;14:2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE.. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 2011;8:376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kogut I, McCarthy SM, Pavlova M, Astling DP, Chen X, Jakimenko A, Jones KL, Getahun A, Cambier JC, Pasmooij AMG, Jonkman MF, Roop DR, Bilousova G.. High-efficiency RNA-based reprogramming of human primary fibroblasts. Nat Commun 2018;9:745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, Saito T, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, Mori M.. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 2011;8:633–638. [DOI] [PubMed] [Google Scholar]
- 25. Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R.. Embryonic stem cell specific microRNAs regulate the G1/S transition and promote rapid proliferation. Nat Genet 2008;40:1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Judson RL, Babiarz J, Venere M, Blelloch R.. Embryonic stem cell specific microRNAs promote induced pluripotency. Nat Biotechnol 2009;27:459–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Judson RL, Greve TS, Parchem RJ, Blelloch R.. MicroRNA-based discovery of barriers to dedifferentiation of fibroblasts to pluripotent stem cells. Nat Struct Mol Biol 2013;20:1227–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lüningschrör P, Stöcker B, Kaltschmidt B, Kaltschmidt C.. miR-290 cluster modulates pluripotency by repressing canonical NF-κB signaling. Stem Cells 2012;30:655–664. [DOI] [PubMed] [Google Scholar]
- 29. Choi YJ, Lin C-P, Ho JJ, He X, Okada N, Bu P, Zhong Y, Kim SY, Bennett MJ, Chen C, Ozturk A, Hicks GG, Hannon GJ, He L.. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol 2011;13:1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS.. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 2009;137:647–658. [DOI] [PubMed] [Google Scholar]
- 31. Worringer KA, Rand TA, Hayashi Y, Sami S, Takahashi K, Tanabe K, Narita M, Srivastava D, Yamanaka S.. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of pro-differentiation genes. Cell Stem Cell 2014;14:40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pfaff N, Fiedler J, Holzmann A, Schambach A, Moritz T, Cantz T, Thum T.. MiRNA screening reveals a new miRNA family stimulating iPS cell generation via regulation of Meox2. EMBO Rep 2011;12:1153–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He X, Cao Y, Wang L, Han Y, Zhong X, Zhou G, Cai Y, Zhang H, Gao P.. Human fibroblast reprogramming to pluripotent stem cells regulated by the miR19a/b-PTEN axis. PLoS One 2014;9:e95213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang C, Li Z, Rana TM.. microRNAs modulate iPS cell generation. RNA 2011;17:1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, Manos PD, Datta S, Lander ES, Schlaeger TM, Daley GQ, Rinn JL.. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 2010;42:1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X, Liu H.. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell 2013;25:69–80. [DOI] [PubMed] [Google Scholar]
- 37. Jain AK, Xi Y, McCarthy R, Allton K, Akdemir KC, Patel LR, Aronow B, Lin C, Li W, Yang L, Barton MC.. LncPRESS1 is a p53-regulated LncRNA that safeguards pluripotency by disrupting SIRT6-mediated de-acetylation of histone H3K56. Mol Cell 2016;64:967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bao X, Wu H, Zhu X, Guo X, Hutchins AP, Luo Z, Song H, Chen Y, Lai K, Yin M, Xu L, Zhou L, Chen J, Wang D, Qin B, Frampton J, Tse HF, Pei D, Wang H, Zhang B, Esteban MA.. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res 2015;25:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang X, Zhang J, Zheng K, Zhang H, Pei X, Yin Z, Wen D, Kong Q.. Long noncoding RNAs sustain high expression levels of exogenous octamer-binding protein 4 by sponging regulatory microRNAs during cellular reprogramming. J Biol Chem 2019;294:17863–17874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K.. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 2005;19:489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ.. Characterization of Dicer-deficient murine embryonic stem cells. PNAS 2005;102:12135–12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rosa A, Brivanlou AH.. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J 2011;30:237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Salazar-Roa M, Trakala M, Álvarez-Fernández M, Valdés-Mora F, Zhong C, Muñoz J, Yu Y, Peters TJ, Graña-Castro O, Serrano R, Zapatero-Solana E, Abad M, Bueno MJ, De Cedrón MG, Fernández-Piqueras J, Serrano M, Blasco MA, Wang D-Z, Clark SJ, Izpisua-Belmonte JC, Ortega S, Malumbres M.. Transient exposure to miR-203 enhances the differentiation capacity of established pluripotent stem cells. The EMBO Journal 2020;39:e104324. doi: 10.15252/embj.2019104324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O’Loghlen A, Muñoz-Cabello AM, Gaspar-Maia A, Wu H-A, Banito A, Kunowska N, Racek T, Pemberton HN, Beolchi P, Lavial F, Masui O, Vermeulen M, Carroll T, Graumann J, Heard E, Dillon N, Azuara V, Snijders AP, Peters G, Bernstein E, Gil J.. MicroRNA regulation of Cbx7 mediates a switch of polycomb orthologs during ESC differentiation. Cell Stem Cell 2012;10:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I.. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 2008;455:1124–1128. [DOI] [PubMed] [Google Scholar]
- 46. Lin N, Chang K-Y, Li Z, Gates K, Rana ZA, Dang J, Zhang D, Han T, Yang C-S, Cunningham TJ, Head SR, Duester G, Dong PDS, Rana TM.. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. 2014;53:1005–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smith KN, Starmer J, Miller SC, Sethupathy P, Magnuson T.. Long noncoding RNA moderates microRNA activity to maintain self-renewal in embryonic stem cells. Stem Cell Reports 2017;9:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hunkler HJ, Hoepfner J, Huang C-K, Chatterjee S, Jara-Avaca M, Gruh I, Bolesani E, Zweigerdt R, Thum T, Bär C.. The long non-coding RNA cyrano is dispensable for pluripotency of murine and human pluripotent stem cells. Stem Cell Reports 2020;15:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ng SY, Johnson R, Stanton LW.. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J 2012;31:522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kaneko S, Bonasio R, Saldaña-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D.. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell 2014;53:290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu X, Guo M, Zhang N, Ye S.. Telomeric noncoding RNA promotes mouse embryonic stem cell self-renewal through inhibition of TCF3 activity. Am J Physiol Cell Physiol 2018;314:C712–C720. [DOI] [PubMed] [Google Scholar]
- 52. Menasché P, Vanneaux V, Hagège A, Bel A, Cholley B, Parouchev A, Cacciapuoti I, Al-Daccak R, Benhamouda N, Blons H, Agbulut O, Tosca L, Trouvin J-H, Fabreguettes J-R, Bellamy V, Charron D, Tartour E, Tachdjian G, Desnos M, Larghero J.. Transplantation of human embryonic stem cell-derived cardiovascular progenitors for severe ischemic left ventricular dysfunction. J Am Coll Cardiol 2018;71:429–438. [DOI] [PubMed] [Google Scholar]
- 53. Le MNT, Hasegawa K.. Expansion culture of human pluripotent stem cells and production of cardiomyocytes. Bioengineering 2019;6:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liau B, Zhang D, Bursac N.. Functional cardiac tissue engineering. Regen Med 2012;7:187–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS, Srivastava D.. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell 2008;2:219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lu TY, Lin B, Li Y, Arora A, Han L, Cui C, Coronnello C, Sheng Y, Benos PV, Yang L.. Overexpression of microRNA-1 promotes cardiomyocyte commitment from human cardiovascular progenitors via suppressing WNT and FGF signaling pathways. J Mol Cell Cardiol 2013;63:146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ishikawa D, Diekmann U, Fiedler J, Just A, Thum T, Lenzen S, Naujok O.. miRNome profiling of purified endoderm and mesoderm differentiated from hESCs reveals functions of miR-483-3p and miR-1263 for cell-fate decisions. Stem Cell Reports 2017;9:1588–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu N, Olson EN.. MicroRNA regulatory networks in cardiovascular development. Dev Cell 2010;18:510–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shen X, Soibam B, Benham A, Xu X, Chopra M, Peng X, Yu W, Bao W, Liang R, Azares A, Liu P, Gunaratne PH, Mercola M, Cooney AJ, Schwartz RJ, Liu Y.. miR-322/-503 cluster is expressed in the earliest cardiac progenitor cells and drives cardiomyocyte specification. Proc Natl Acad Sci U S A 2016;113:9551–9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, Boyer LA.. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 2013;152:570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M, Herrmann BG.. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 2013;24:206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ounzain S, Micheletti R, Arnan C, Plaisance I, Cecchi D, Schroen B, Reverter F, Alexanian M, Gonzales C, Ng SY, Bussotti G, Pezzuto I, Notredame C, Heymans S, Guigó R, Johnson R, Pedrazzini T.. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J Mol Cell Cardiol 2015;89:98–112. [DOI] [PubMed] [Google Scholar]
- 63. Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I.. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011;147:358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu J, Li Y, Lin B, Sheng Y, Yang L.. HBL1 is a human long noncoding RNA that modulates cardiomyocyte development from pluripotent stem cells by counteracting miR-1. Dev Cell 2017;42:333–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hu W, Alvarez ‐Dominguez JR, Lodish HF.. Regulation of mammalian cell differentiation by long non‐coding RNAs. EMBO Rep 2012;13:971–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li Y, Zhang J, Huo C, Ding N, Li J, Xiao J, Lin X, Cai B, Zhang Y, Xu J.. Dynamic organization of lncRNA and circular RNA regulators collectively controlled cardiac differentiation in humans. EBioMedicine 2017;24:137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang X, Cao X, Dong D, Shen X, Cheng J, Jiang R, Yang Z, Peng S, Huang Y, Lan X, Elnour IE, Lei C, Chen H.. Circular RNA TTN acts as a miR-432 sponge to facilitate proliferation and differentiation of myoblasts via the IGF2/PI3K/AKT signaling pathway. Mol Ther Nucleic Acids 2019;18:966–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sayed N, Liu C, Wu JC.. Translation of human-induced pluripotent stem cells: from clinical trial in a dish to precision medicine. J Am Coll Cardiol 2016;67:2161–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yang X, Pabon L, Murry CE.. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res 2014;114:511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liao SY, Liu Y, Siu CW, Zhang Y, Lai WH, Au KW, Lee YK, Chan YC, Yip PMC, Wu EX, Wu Y, Lau CP, Li RA, Tse HF.. Proarrhythmic risk of embryonic stem cellderived cardiomyocyte transplantation in infarcted myocardium. Hear Rhythm 2010;7:1852–1859. [DOI] [PubMed] [Google Scholar]
- 71. Chong JJH, Yang X, Don CW, Minami E, Liu YW, Weyers JJ, Mahoney WM, Van Biber B, Cook SM, Palpant NJ, Gantz JA, Fugate JA, Muskheli V, Gough GM, Vogel KW, Astley CA, Hotchkiss CE, Baldessari A, Pabon L, Reinecke H, Gill EA, Nelson V, Kiem HP, Laflamme MA, Murry CE.. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature 2014;510:273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fu JD, Rushing SN, Lieu DK, Chan CW, Kong CW, Geng L, Wilson KD, Chiamvimonvat N, Boheler KR, Wu JC, Keller G, Hajjar RJ, Li RA.. Distinct roles of microRNA-1 and -499 in ventricular specification and functional maturation of human embryonic stem Cell-Derived cardiomyocytes. PLoS One 2011;6:e27417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kuppusamy KT, Jones DC, Sperber H, Madan A, Fischer KA, Rodriguez ML, Pabon L, Zhu W-Z, Tulloch NL, Yang X, Sniadecki NJ, Laflamme MA, Ruzzo WL, Murry CE, Ruohola-Baker H.. Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc Natl Acad Sci U S A 2015;112:E2785–E2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lee DS, Chen JH, Lundy DJ, Liu CH, Hwang SM, Pabon L, Shieh RC, Chen CC, Wu SN, Yan YT, Lee ST, Chiang PM, Chien S, Murry CE, Hsieh PCH.. Defined microRNAs induce aspects of maturation in mouse and human embryonic-stem-cell-derived cardiomyocytes. Cell Rep 2015;12:1960–1967. [DOI] [PubMed] [Google Scholar]
- 75. Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, Xiong Y, Chien H, Zhou B, Ashley E, Bernstein D, Chen PS, Chen HV, Quertermous T, Chang CP.. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014;514:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O'Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE.. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol 2007;25:1015–1024. [DOI] [PubMed] [Google Scholar]
- 77. Caspi O, Huber I, Kehat I, Habib M, Arbel G, Gepstein A, Yankelson L, Aronson D, Beyar R, Gepstein L.. Transplantation of human embryonic stem cell-derived cardiomyocytes improves myocardial performance in infarcted rat hearts. J Am Coll Cardiol 2007;50:1884–1893. [DOI] [PubMed] [Google Scholar]
- 78. Shiba Y, Fernandes S, Zhu WZ, Filice D, Muskheli V, Kim J, Palpant NJ, Gantz J, Moyes KW, Reinecke H, Van Biber B, Dardas T, Mignone JL, Izawa A, Hanna R, Viswanathan M, Gold JD, Kotlikoff MI, Sarvazyan N, Kay MW, Murry CE, Laflamme MA.. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature 2012;489:322–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kawamura M, Miyagawa S, Miki K, Saito A, Fukushima S, Higuchi T, Kawamura T, Kuratani T, Daimon T, Shimizu T, Okano T, Sawa Y.. Feasibility, safety, and therapeutic efficacy of human induced pluripotent stem cell-derived cardiomyocyte sheets in a porcine ischemic cardiomyopathy model. Circulation 2012;126:S29–S37. [DOI] [PubMed] [Google Scholar]
- 80. Masumoto H, Matsuo T, Yamamizu K, Uosaki H, Narazaki G, Katayama S, Marui A, Shimizu T, Ikeda T, Okano T, Sakata R, Yamashita JK.. Pluripotent stem cell-engineered cell sheets reassembled with defined cardiovascular populations ameliorate reduction in infarct heart function through cardiomyocyte-mediated neovascularization. Stem Cells 2012;30:1196–1205. [DOI] [PubMed] [Google Scholar]
- 81. Kawamura M, Miyagawa S, Fukushima S, Saito A, Miki K, Ito E, Sougawa N, Kawamura T, Daimon T, Shimizu T, Okano T, Toda K, Sawa Y.. Enhanced survival of transplanted human induced pluripotent stem cell-derived cardiomyocytes by the combination of cell sheets with the pedicled omental flap technique in a porcine heart. Circulation 2013;128:87–94. [DOI] [PubMed] [Google Scholar]
- 82. Shiba Y, Gomibuchi T, Seto T, Wada Y, Ichimura H, Tanaka Y, Ogasawara T, Okada K, Shiba N, Sakamoto K, Ido D, Shiina T, Ohkura M, Nakai J, Uno N, Kazuki Y, Oshimura M, Minami I, Ikeda U.. Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature 2016;538:388–391. [DOI] [PubMed] [Google Scholar]
- 83. Ye L, Chang YH, Xiong Q, Zhang P, Zhang L, Somasundaram P, Lepley M, Swingen C, Su L, Wendel JS, Guo J, Jang A, Rosenbush D, Greder L, Dutton JR, Zhang J, Kamp TJ, Kaufman DS, Ge Y, Zhang J.. Cardiac repair in a porcine model of acute myocardial infarction with human induced pluripotent stem cell-derived cardiovascular cells. Cell Stem Cell 2014;15:750–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nakamuta JS, Danoviz ME, Marques FLN, dos SL, Becker C, Gonçalves GA, Vassallo PF, Schettert IT, Tucci PJF, Krieger JE.. Cell therapy attenuates cardiac dysfunction post myocardial infarction: effect of timing, routes of injection and a fibrin scaffold. PLoS One 2009;4:e6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Oikonomopoulos A, Kitani T, Wu JC.. Pluripotent stem cell-derived cardiomyocytes as a platform for cell therapy applications: progress and hurdles for clinical translation. Mol Ther 2018;26:1624–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ong SG, Huber BC, Lee WH, Kodo K, Ebert AD, Ma Y, Nguyen PK, Diecke S, Chen WY, Wu JC.. Microfluidic single-cell analysis of transplanted human induced pluripotent stem cell-derived cardiomyocytes after acute myocardial infarction. Circulation 2015;132:762–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tachibana A, Santoso MR, Mahmoudi M, Shukla P, Wang L, Bennett M, Goldstone AB, Wang M, Fukushi M, Ebert AD, Woo YJ, Rulifson E, Yang PC.. Paracrine effects of the pluripotent stem cell-derived cardiac myocytes salvage the injured myocardium. Circ Res 2017;121:e22–e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Santoso MR, Ikeda G, Tada Y, Jung JH, Vaskova E, Sierra RG, Gati C, Goldstone AB, von BD, Shukla P, Wu JC, Wakatsuki S, Woo YJ, Yang PC.. Exosomes from induced pluripotent stem cell-derived cardiomyocytes promote autophagy for myocardial repair. J Am Heart Assoc 2020;9:e014345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Vagnozzi RJ, Maillet M, Sargent MA, Khalil H, Johansen AK, Schwanekamp JA, York AJ, Huang V, Nahrendorf M, Sadayappan S, Molkentin JD.. An acute immune response underlies the benefit of cardiac stem cell therapy. Nature 2020;577:405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wysoczynski M, Khan A, Bolli R.. New paradigms in cell therapy: repeated dosing, intravenous delivery, immunomodulatory actions, and new cell types. Circ Res 2018;123:138–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lu X, Zhao T.. Clinical therapy using iPSCs: hopes and challenges. Genomics Proteomics Bioinformatics 2013;11:294–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lalit PA, Hei DJ, Raval AN, Kamp TJ.. Induced pluripotent stem cells for post-myocardial infarction repair: remarkable opportunities and challenges. Circ Res 2014;114:1328–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Davis DR. Cardiac stem cells in the post-Anversa era. Eur Heart J 2019;40:1039–1041. [DOI] [PubMed] [Google Scholar]
- 94. Cao F, Lin S, Xie X, Ray P, Patel M, Zhang X, Drukker M, Dylla SJ, Connolly AJ, Chen X, Weissman IL, Gambhir SS, Wu JC.. In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation 2006;113:1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Cao F, Drukker M, Lin S, Sheikh AY, Xie X, Li Z, Connolly AJ, Weissman IL, Wu JC.. Molecular imaging of embryonic stem cell misbehavior and suicide gene ablation. Cloning Stem Cells 2007;9:107–117. [DOI] [PubMed] [Google Scholar]
- 96. Yagyu S, Hoyos V, Del Bufalo F, Brenner MK.. An inducible caspase-9 suicide gene to improve the safety of therapy using human induced pluripotent stem cells. Mol Ther 2015;23:1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Cho SJ, Kim SY, Jeong HC, Cheong H, Kim D, Park SJ, Choi JJ, Kim H, Chung HM, Moon SH, Cha HJ.. Repair of ischemic injury by pluripotent stem cell based cell therapy without teratoma through selective photosensitivity. Stem Cell Reports 2015;5:1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Tang C, Lee AS, Volkmer JP, Sahoo D, Nag D, Mosley AR, Inlay MA, Ardehali R, Chavez SL, Pera RR, Behr B, Wu JC, Weissman IL, Drukker MA.. Antibody against SSEA-5 glycan on human pluripotent stem cells enables removal of teratoma-forming cells. Nat Biotechnol 2011;29:829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ben-David U, Nudel N, Benvenisty N.. Immunologic and chemical targeting of the tight-junction protein Claudin-6 eliminates tumorigenic human pluripotent stem cells. Nat Commun 2013;4:1992. [DOI] [PubMed] [Google Scholar]
- 100. Miki K, Endo K, Takahashi S, Funakoshi S, Takei I, Katayama S, Toyoda T, Kotaka M, Takaki T, Umeda M, Okubo C, Nishikawa M, Oishi A, Narita M, Miyashita I, Asano K, Hayashi K, Osafune K, Yamanaka S, Saito H, Yoshida Y.. Efficient detection and purification of cell populations using synthetic microRNA switches. Cell Stem Cell 2015;16:699–711. [DOI] [PubMed] [Google Scholar]
- 101. Parr CJC, Katayama S, Miki K, Kuang Y, Yoshida Y, Morizane A, Takahashi J, Yamanaka S, Saito H.. MicroRNA-302 switch to identify and eliminate undifferentiated human pluripotent stem cells. Sci Rep 2016;6:32532–32514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Fico A, Fiorenzano A, Pascale E, Patriarca EJ, Minchiotti G.. Long non-coding RNA in stem cell pluripotency and lineage commitment: functions and evolutionary conservation. Cell Mol Life Sci 2019;76:1459–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]