Abstract
Background
Management of advanced chordomas remains delicate considering their insensitivity to chemotherapy. Homozygous deletion of the regulatory gene CDKN2A has been described as the most frequent genetic alteration in chordomas and may be considered as a potential theranostic marker. Here, we evaluated the tumor efficacy of the CDK4/6 inhibitor palbociclib, as well as the PLK1 inhibitor volasertib, in three chordoma patient-derived xenograft (PDX) models to validate and identify novel therapeutic approaches.
Methods
From our chordoma xenograft panel, we selected three models, two of them harboring a homozygous deletion of CDKN2A/2B genes, and the last one a PBRM1 pathogenic variant (as control). For each model, we tested the palbociclib and volasertib drugs with pharmacodynamic studies together with RT-PCR and RNAseq analyses.
Results
For palbociclib, we observed a significant tumor response for one of two models harboring the deletion of CDKN2A/2B (p = 0.02), and no significant tumor response in the PBRM1-mutated PDX; for volasertib, we did not observe any response in the three tested models. RT-PCR and RNAseq analyses showed a correlation between cell cycle markers and responses to palbociclib; finally, RNAseq analyses showed a natural enrichment of the oxidative phosphorylation genes (OxPhos) in the palbociclib-resistant PDX (p = 0.02).
Conclusion
CDK4/6 inhibition appears as a promising strategy to manage advanced chordomas harboring a loss of CDKN2A/2B. However, further preclinical studies are strongly requested to confirm it and to understand acquired or de novo resistance to palbociclib, in the peculiar view of a targeting of the oxidative phosphorylation genes.
Keywords: CDKN2A/2B deletion, chordoma patient’s derived xenograft, CDK4/6 inhibitor, palbocicib, PLK1 inhibitor, volasertib
Introduction
Chordomas are rare bone tumors of the skull-base and spine which originate from remnants of the notochord, explaining their common location along the neuraxis. They are characterized by a locally invasive extension despite slow growth. Considering their chemoradioresistance, a radical resection associated with a high-dose radiation is the gold-standard therapeutic strategy (1). However, despite this aggressive treatment, the disease-free survival is generally short (2, 3) due to the fact that locoregional recurrence is a common event following the initial treatment (4). The management of these tumor relapses or progression after surgery and proton beam therapy is a clinical challenge, especially for patients with metastases and inoperable recurrence disease. Difficulties in managing advanced chordomas explain the poor long-term prognosis of this cancer with an overall survival evaluated at ~30% at 10 years (3). There is, therefore, an unmet need for systemic therapies, which are especially difficult to raise given the lack of data (5–15) concerning the tumor biology (genetic driver events), and that chordomas have a relatively low mutation burden with few therapeutically actionable alterations (5, 6). Over the years, a wider use of new targeted therapies, such as tyrosine kinase inhibitors (TKI), has been developed. Unfortunately, the proportion of objective responses to all these agents was very low in chordomas (16–24). The limited number of patients included in these studies, as well as the lack of follow-up, interfere with the preliminary proof of effectiveness of these drugs, that encourages the development of several pre-clinical models (25–34).
Recently, we established and well-characterized a large panel of 12 chordoma patient-derived xenografts (PDXs) (35), including all relevant clinical features, their immunohistochemical and genomic profiles, serving as a support for preclinical drug testing. In this way, the Next Sequencing Generation (NGS) genomic characterization of our models showed a preponderance (58%) of homozygous deletions of CDKN2A/2B and mutations affecting the mammalian SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling complexes (such as PBRM1, SMARCB1, ARID1A pathogenic mutations), as described in a majority of chordomas (5, 6, 14). Mutations in the SWI/SNF complexes were also mutually exclusive with homozygous deletions of CDKN2A/2B in our panel. Genes encoding subunits of SWI/SNF complexes are collectively altered in over 20% of human malignancies (36) and became an interesting target for new therapies as enhancer of zeste homolog 2 (EZH2) inhibitors (35, 37, 38).
Cyclin-dependent kinase (CDK) inhibitor 2A (CDKN2A) is a tumor suppressor gene encoding the p16ink4a protein which plays an important role in cell-cycle regulation through the inhibition of cyclin-dependent kinases (CDK) 4/6 that protects the p53 protein from degradation (39). CDKN2A loss or mutation is found in a wide range of malignancies and may lead to an increased CDK activity, bypassing a critical senescent signal (40). In chordomas, the regulatory gene CDKN2A is also frequently lost (5, 6), and thus, CDK4/6 may be in an over-activated state (41), although multiple upstream seem to be the input in the dysregulation of CDK4/6 activity (42). Palbociclib (PD 0332991), a selective inhibitor of CDK4/6, approved for the treatment of hormone-receptor positive breast cancer (43), has been recently tested in in vitro cell-line chordoma models harboring the loss of CDKN2A (41), suggesting a potential in vivo action of anti-CDK4/6 in chordomas harboring this genomic alteration.
The Polo-like kinase (PLK) family, particularly PLK1, has recently become an attractive oncogenic target considering its regular roles in the G2/M cell cycle checkpoint, via its effects on chromosome segregation, spindle assembly, and cytokinesis (44). Volasertib (BI 2536), a selective PLK1-inhibitor, showed a promising antitumor activity in other cancer preclinical models (45–47). Considering that several genomic analyses of chordoma samples reveal widespread cell cycle dysregulation (48), testing the inhibition of PLK1 in chordomas makes sense.
In this study, we therefore focused our pharmacological expertise on the CDK4/6 inhibitor palbociclib and the PLK1-inhibitor volasertib in two xenograft models harboring homozygous deletions of CDKN2A/2B, and in a third PBRM1-mutated model (control), in order to evaluate the CDKN2A/2B loss as a theranostic biomarker in chordomas.
Materials and methods
Chordoma patient-derived xenografts
Three previously established and characterized chordoma PDXs were used for in vivo experiments: cluster of differentiation (CD) 3, CD7, and CD39 (35). The main clinical, histological, and genomic features of our xenografts panel have been previously described (35). PDX models of chordomas were obtained by engrafting a primary tumor sample into nude mice (Charles River Laboratories). All in vivo experimental procedures, animal care, and housing were performed in accordance with the recommendations of the European Community (2010/63/UE) for the care and use of laboratory animals. Experimental procedures were specifically approved by the ethics committee of the Institut Curie CEEA-IC #118 (Authorization APAFiS# 25870-2020060410487032-v1 given by National Authority) in compliance with the international guidelines.
Next-generation sequencing
DNA was extracted from frozen tumor samples using a standard phenol/chloroform procedure. RNA was extracted from microdissected areas using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA).
DNA from xenografts and a part of patients’ tumors were then analyzed for gene mutations by targeted next-generation sequencing (NGS). The in-house NGS panel includes 571 genes of interest in oncology for diagnosis, prognosis, and theranostics including chordoma genes of interest such as PIK3CA, PTEN, CDKN2A/2B, PBRM1, SETD2, and ARID1A. The library preparation was performed using the Agilent Sureselect XT HS kit, and sequencing was completed on an Illumina NovaSeq 6000 sequencer. All variants, called using Varscan2 (v2.4.3-0), that passed the following thresholds were validated: allelic ratio above 5% and population frequency lower than 0.1% in 1000 g, ESP, or gnomAD. This large targeted NGS panel also allowed molecular analysis of tumors for CNV (copy number variation) status including CDKN2A homozygous deletion detection. In case of doubt concerning the originating link between the patient’s tumors and PDXs, an identity monitoring was performed based on polymorphisms.
Antitumor efficacy of targeted therapy drugs
Considering the results of the molecular analysis of our chordoma xenograft panel, two drugs on three PDXs (CD3, CD7, and CD39) were considered in our pharmacological program, i.e. the CDK4/6 inhibitor palbociclib (Ibrance®, Pfizer) and the PLK1 inhibitor volasertib (Boehringer Ingelheim). This last targeted treatment was tested in the field of our huge expertise developed in various types of human cancers, such as non-small cell lung cancers (46) and breast cancers (47). Palbociclib was administered orally at a dose of 75 mg/kg/day (tween 80 0.5% + methylcellulose 0.5% + water 99%), once daily, 5 days per week; volasertib was administered orally at the dose of 10 mg/kg/day (methylcellulose 0.5% + tween 80 0.5% + DMSO 2% + water 97%), once daily, 4 days per week. The treatment was administered from day 1 to mouse sacrifice. Drugs were sourced from the oncological department of Institut Curie, Paris, France.
For in vivo therapeutic studies, a 15 mm3 tumor fragment was grafted into 30 female immunodeficiency nude mice. Mice bearing growing tumors with a volume of 60–150 mm3 were randomly assigned to control or treatment groups. Animals with tumor volumes outside this range were excluded. Mice were weighted and tumors were measured once a week. Considering the slow growth of these tumors, xenografted mice were sacrificed when tumor volume reached a mean volume of 500 mm3.
Tumor volumes were calculated using two perpendicular diameters with calipers as follows: V (volume) = (a×b)2/2 where a and b are the largest and smallest perpendicular tumor diameters (in mm). Relative tumor volumes (RTV) were calculated from the following formula: RTV = (V x /V1), where V x is the tumor volume on day x and V1 is the tumor volume at initiation of therapy (day 1). Antitumor activity was evaluated according to tumor growth inhibition (TGI), calculated according to the following formula: percent GI = 100 − (RTVt/RTVc ×100), where RTVt is the median RTV of treated mice and RTVc is the median RTV of controls, both at a given time point when the antitumor effect was optimal. A meaningful biological effect was defined as a TGI of at least 50% (49). The statistical significance of differences observed between the individual RTVs corresponding to the treated mice and control groups was calculated using the two-tailed Mann–Whitney U test.
Moreover, to evaluate the response to treatments observed in the three models according to individual mouse variability, we decided to consider each mouse as one tumor-bearing entity. Hence, in all in vivo experiments, a relative tumor variation (RTVV)) was calculated for each treated mouse as follows: [(RTVt/mRTVc)], where RTVt is the relative tumor volume of the treated mouse and mRTVc is the median relative tumor volume of the corresponding control group at the end of treatment. We then calculated an overall response rate (ORR) for each treated mouse as follows: ORR = [(RTVV) - 1]. A tumor was considered to be responding to treatment, if the ORR was below -0.5. The statistical significance of the ORR between tested treatments was determined using Fisher’s Exact test. Finally, we evaluated the impact of treatments on tumor progression, by evaluating all progression-free survival probabilities taking tumor doubling time (RTV × 2) and time for RTV × 4 into account. A log-rank (Mantel–Cox) test was used to perform treatment comparisons.
Pharmacodynamics study by RT-PCR analysis and RNAseq after in vivo pharmacological experiments
A RT-PCR study was performed after the administration of palbociclib and volasertib drugs on the two models CD3 and CD7. The RT-PCR technique has been previously described by our group (50). Results, expressed as N-fold differences in target gene expressions relative to the murine and human TBP genes (both the murine and the human TBP transcripts), and termed Ntarget, were determined as Ntarget = 2ΔCtsample, where the ΔCt value of the sample was determined by subtracting the Ct value of the specific target gene from the Ct value of the TBP. The Ntarget values of the samples were subsequently normalized so that the value for “basal mRNA level” (smallest amount of quantifiable target gene mRNA, Ct = 35) was 1 for selected genes. The expression of the following cancer genes potentially implied in palbociclib resistance (51, 52) was studied: CCNE1, CDK4, PTEN, RB1, and E2F1. The PLK1 gene was also studied, as the target of PLK1 inhibitors. Finally, we studied some genes implied in the proliferation, or overexpressed in chordomas: MKI67, MYC, TBXT and EGFR ( 53). The nucleotide sequences of the gene primers used are available on request.
For RNAseq analyses, RNA was extracted from microdissected areas using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Library preparation was performed using the QuantSeq 3′ mRNA-Seq reverse (REV) Library Prep Kit (Lexogen, Vienna, Austria) using 150 ng of total RNA, and according to the manufacturer’s instruction. The pool was sequenced on a NovaSeq 6000 SP 2x75bp flow cell (Illumina Inc., San Diego, CA, USA). RNA sequencing data were analyzed using the BlueBee Genomics Platform (Lexogen, Vienna, Austria). Gene Ontology Enrichment Analysis was performed using the ShinyGO (http://bioinformatics.sdstate.edu/go/) and gProfiler (https://biit.cs.ut.ee/gprofiler/gost) Web sites.
Statistical analyses
Statistical analyses were performed using the Prism v9.0 software (GraphPad Software, Inc., La Jolla, CA, USA). Statistical characteristics were used to describe all variables. Numerical variables were expressed as the median or mean and standard deviation, as appropriate. Categorical variables were expressed as the count and percentage. Variables were tested by the Mann-Whitney U test, Fisher’s Exact test, or chi-squared test, as appropriate. Survival distributions were determined using the Kaplan–Meier method and the log-rank (Mantel–Cox) test was used to compare groups. Every statistical test used in this manuscript was two-tailed, and p-values less than 0.05 were considered as significant.
Results
Patient-derived xenograft selection
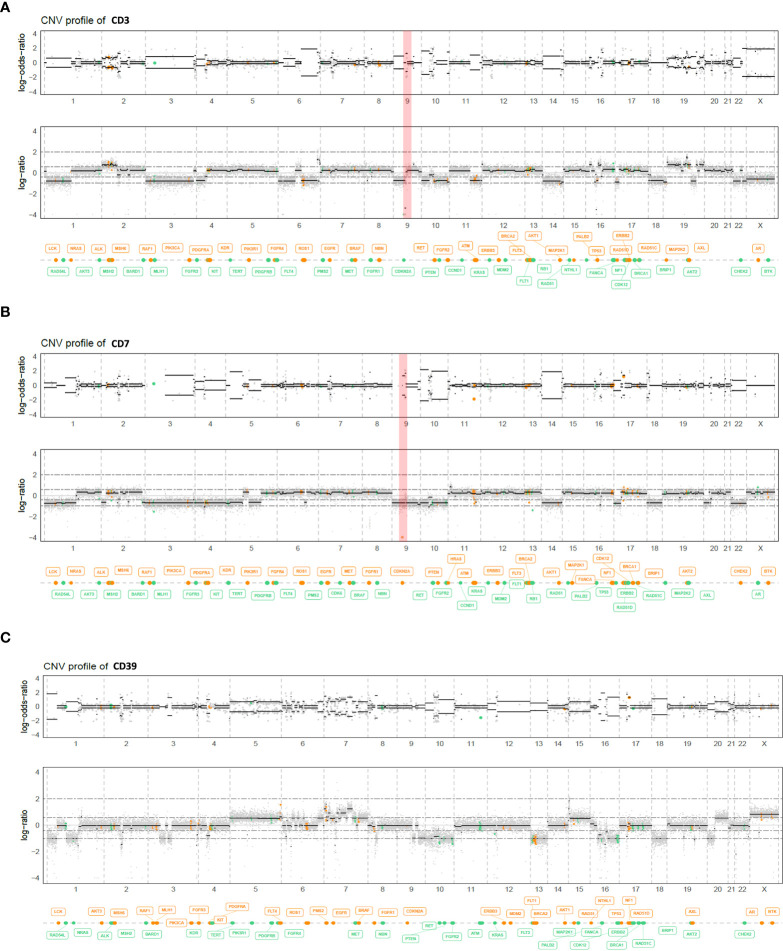
The histological and genomic features of the three PDXs are described in Table 1 . CD3 was obtained from a patient’s sacral primary tumor previously operated on and displaying a homozygous deletion of CDKN2A/2B; CD7 was obtained from a skull base primary tumor previously operated on and irradiated and displaying a homozygous deletion of CDKN2A/2B; CD39 was obtained from a skull base primary tumor previously operated on and presenting a pathogenic PBRM1 variant (c.599C>G), without CDKN2A/2B deletion ( Figure 1 ). The CD39 model which does not harbor any loss-of-function genomic alteration of CDKN2A or CDKN2B but a PBRM1-variant was chosen as a negative control in order to reinforce the fact that CDKN2A/2B might be a biomarker of response to palbociclib. None of these models harbored another CNV or pathogenic variant of theranostic interest.
Table 1.
Histopathological and molecular features of the three selected PDX models.
| Models | Histology type* | Molecular alteration | Targeted-therapy |
|---|---|---|---|
| CD3 | Conventional | CDKN2A/2B loss | Palbociclib Volasertib |
| CD7 | Conventional | CDKN2A/2B loss | Palbociclib Volasertib |
| CD39 | Conventional | PBRM1 variant (c.599C>G; p.(Ser200Ter)) | Palbociclib Volasertib |
*According to the WHO (World Health Organization) 2020.
CD, cluster of differentiation.
Figure 1.
Genome view profiles of the allele-specific copy number of the CD3 (A), CD7 (B), and CD39 (C) chordoma PDX models. The top graph log-odds-ratio represents the B-Allele Frequency (BAF), and the bottom graph the log depth ratio between tumor and a healthy witness. Both CD3 and CD7 chordoma PDX models harbored a homozygous deletion of CDKN2A/2B (red bar), whereas CD39 did not.
In vivo antitumor efficacy of targeted therapies
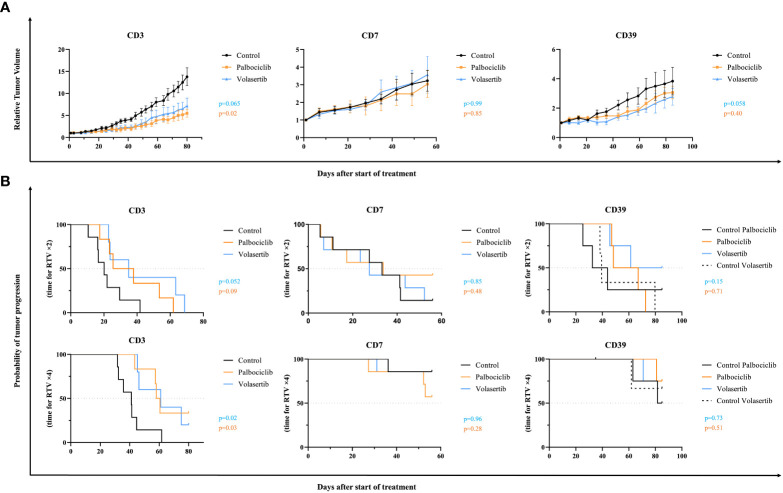
In vivo therapeutic experiments with palbociclib and volasertib were done on these three xenografts. For each model, PDX tumor-bearing mice were randomized into treatment and control groups (n = 4–7 mice per group). For the palbociclib drug, TGI was calculated at 80, 49, and 85 days, for CD3, CD7, and CD39, respectively. For the volasertib drug, TGI was calculated at 77, 28, and 51 days, for CD3, CD7, and CD39, respectively.
In vivo responses to palbociclib and volasertib are shown in the Figure 2 . For the CD3 xenograft, palbociclib induced a significant TGI (p = 0.02) with an optimal TGI of 60%, whereas volasertib showed a slight tumor growth response close to statistical significance (p = 0.065) ( Figure 2A ). The probability of progression-free survival at RTV × 4 was significantly longer in the palbociclib and volasertib treatments groups than in the control (p = 0.03 and p = 0.02, respectively), but not at RTV× 2 (p = 0.09 and p = 0.052, respectively) ( Figure 2B ).
Figure 2.
In vivo efficacy of palbociclib and volasertib, in the CD3, CD7, and CD39 chordoma PDXs. PDX tumor-bearing mice were randomized into each treatment group (n = 4–7 mice per group), and treated with palbociclib 75 mg/kg, 5 days per week (orange), or volasertib 10 mg/kg, 4 days per week (blue). Untreated control is shown in black. (A) Relative tumor volumes. Tumor growth was evaluated by plotting the mean of the relative tumor volume ± SD per group. (B) Probability of tumor progression. The time to reach RTV × 2 and RTV × 4 for each treated mouse has been calculated using Kaplan Meier curves and log-rank test.
For the CD7 xenograft, we did not observe any tumor growth response to both palbociclib and volasertib (p = 0.85 and p > 0.99, respectively), as well as for the CD39 xenograft without CDKN2A/2B deletion (p = 0.40 and 0.058, respectively).
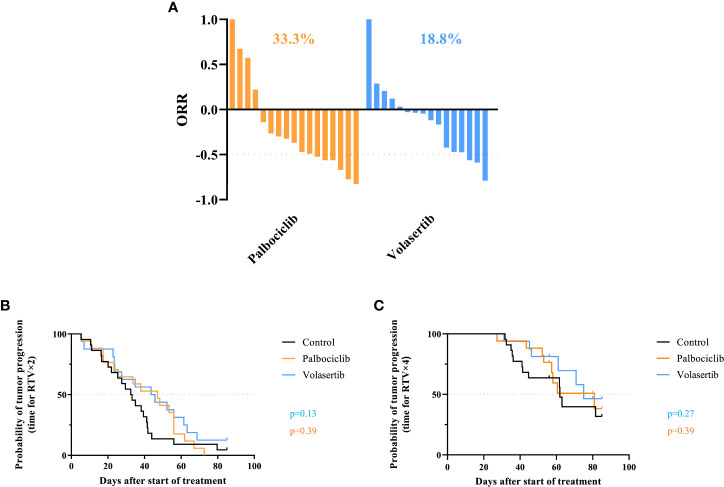
Hence, when we looked at the individual tumor responses in the three treated models ( Figure 3A ), the ORRs lower than -0.5 were 33.3% and 18.8% after palbociclib and volasertib, respectively (p = 0.25). Finally, the study of the progression-free survival across the three PDX models at RTV × 2 and RTV × 4 did not show any significant effect for palbociclib (p = 0.39 and p = 0.39 for RTV × 2 and RTV × 4, respectively) nor with volasertib (p=0.13 and p=0.27 for RTV x 2 and RTV x 4 respectively) ( Figures 3B, C ).
Figure 3.
Individual tumor responses in the three treated models. (A) Overall response rate (ORR) in all treated chordoma PDXs after palbociclib (n = 17) and volasertib (n = 16). A tumor was considered to be responding to treatment if ORR was below -0.5. (B, C) Probability of progression (RTV × 2 and RTV × 4) in all treated chordoma PDXs after palbociclib and volasertib treatments.
Pharmacodynamics study
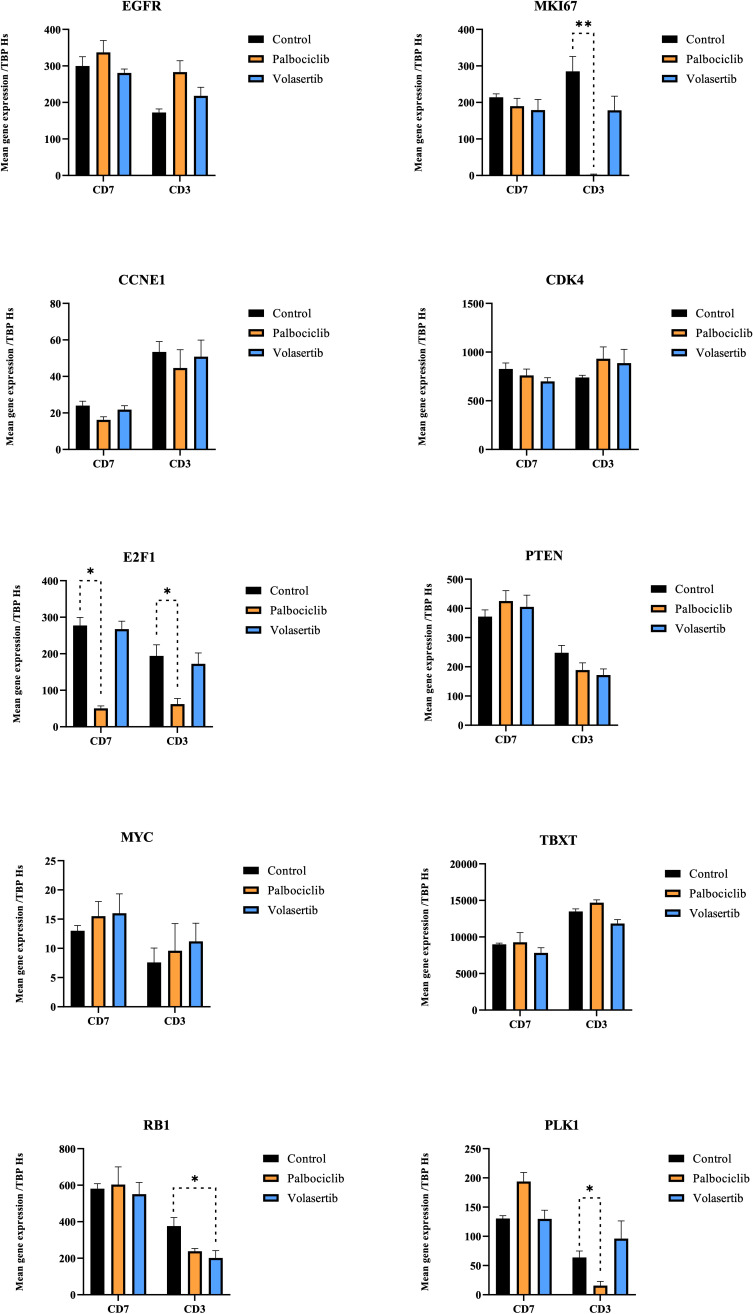
Using RT-PCR analyses, we first looked at modifications of candidate gene expression occurring after palbociclib and volasertib in vivo therapies in CD3 and CD7 models compared to the control groups ( Figure 4 ). For that, we applied RT-qPCR on the genes of interest (described in the materials and methods section) on 28 samples assigned to the control or treatment group. After palbociclib treatment, we observed a significant decrease in human MKI67 (p = 0.008), E2F1 (p = 0.016), and PLK1 (p = 0.016) gene expression in the in vivo responding CD3 xenograft; in contrast, we only observed decreased gene expression in E2F1 (p = 0.029) in the non-responding model CD7. No significant modification of gene expression was noted for CDK4 (palbociclib target), MYC, TBXT, EGFR, CCNE1, and RB1 in both studied models. After volasertib administration, we observed a significant gene expression modification of RB1 (p = 0.03) in the CD3 model. No significant modification of gene expression was noted for the other genes, including PLK1 (volasertib target).
Figure 4.
Gene expression modifications after treatments with palbociclib and volasertib (CD3 and CD7 models). Quantitative RT-PCR was performed to measure the expression of the target genes. Data are presented as level of gene expression compared to the standard level of Homo sapien (Hs) TBP. Genes expressions were compared between mice treated with the indicated agent and the control group. Error bars show the standard error of the mean. * and ** achieve statistical significance compared to control (p < 0.05 and p < 0.01, respectively) by Mann–Whitney U test.
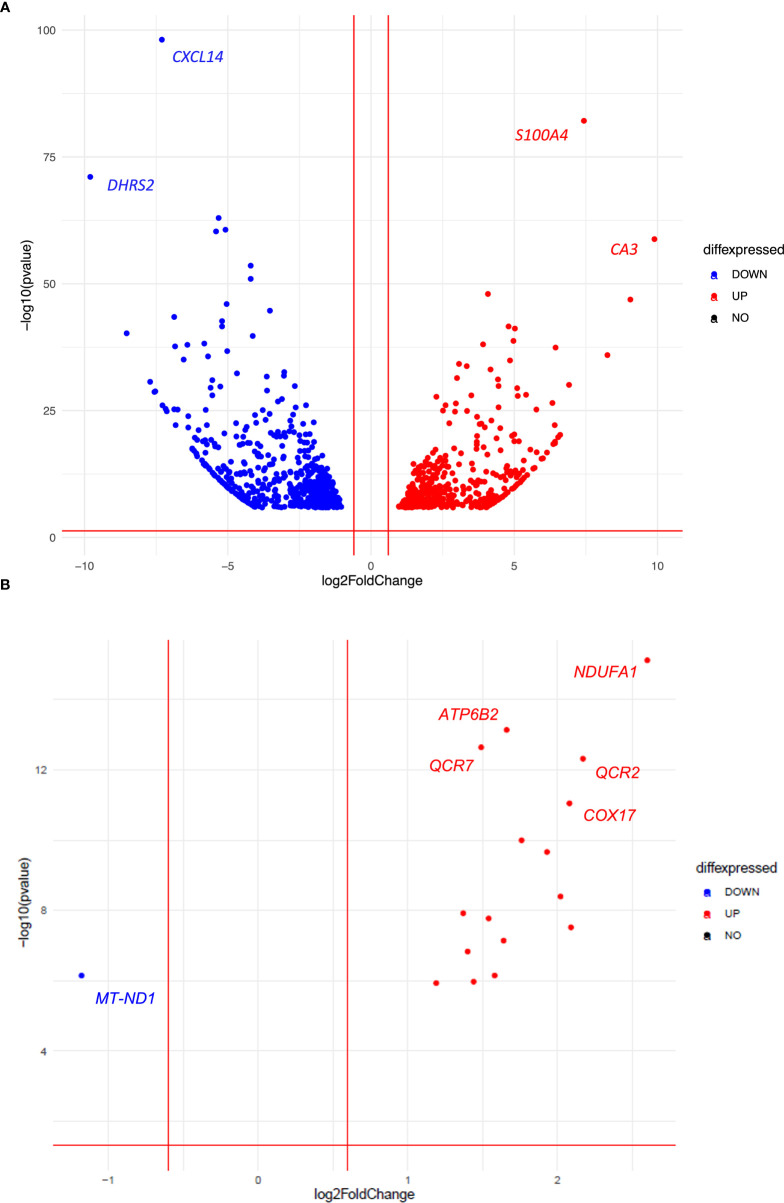
To better understand the opposite efficacy of palbociclib in the two CDKN2A/2B deleted CD3 and CD7 PDXs, we then performed RNAseq analyses in both control and treated groups (three tumors per group). In a differential supervised analysis, comparing control tumors and tumors treated with palbociclib in both PDX models, we observed a more important modification of gene expression in the responding CD3 PDX model (n = 318 genes, Supplementary Table S1 ) compared to the non-responding CD7 PDX model (n = 12 genes; Supplementary Table S2 ), which correlated with in vivo observations. Next, we tested the enrichment of the set of genes resulting from the KEGG database. As expected, we found a significant decrease in the enrichment of cell cycle genes in the responding CD3 tumors (p = 0.04), such as TOP2A (p = 2.2 e-26), MKI67 (p = 4.3 e-20), and AURKB (2.5 e-8). Moreover, we compared in a supervised analysis the gene expression of control untreated CD3 PDX tumors and control untreated CD7 PDX tumors in order to find predictive biomarkers of intrinsic palbociclib resistance ( Supplementary Table S3 ). We observed an upregulation of genes implied in hypoxia such as S100A4 (p = 6.5 e-79) and CA3 (p = 3.8 e-56) in the resistant CD7 PDX model ( Figure 5A ). Among the top 1000 genes in which we observed a significant gene expression variation in both the models (adjusted p-value), we noted an enrichment of the oxidative phosphorylation genes in the resistant CD7 PDX model (p = 0.02) using the KEGG data base, such as NDUFA1 (p = 6.5 e-14), NDUFB2 (p = 1.2 e-07), COX6C (p = 4.4 e-07), COX7A2 (p = 1.9 e-05), COX7C (p = 3.2 e-7), and COX17 (p = 4.2 e-10) ( Figure 5B ). To resume the pharmacodynamic study, we observed a decrease in the expression of genes implied in the cell cycle in the responding CD3 compared to the CD7 PDX model treated by palbociclib. Moreover, we noted a significant enrichment of the expression of genes implied in oxidative phosphorylation (OxPhos) into the non-responding control CD7 PDX model in comparison with the responding control CD3 PDX model.
Figure 5.
Volcanoplots illustrating the top 1000 genes (A) and all genes related to the oxidative phosphorylation (OxPhos) pathway (B), in which we observed a significant gene expression variation in both untreated CD3 and CD7 PDX models (adjusted p-value).
Discussion
In this current study, we explored the antitumor effect of the targeted therapy palbociclib in three PDX chordoma models in order to consider the homozygous deletion of CDKN2A/2B as a theranostic biomarker of response to palbociclib, as suggested in other cancer types (43, 54, 55). We observed various responses to palbociclib concerning both models (CD3, CD7) harboring a loss of CDKN2A/2B, and no tumor efficacy in the non-CDKN2A/2B-deleted PBRM1-mutated PDX (CD39). We also tested the PLK1 inhibitor volasertib in these three PDXs and did not observe any significant anti-tumor response.
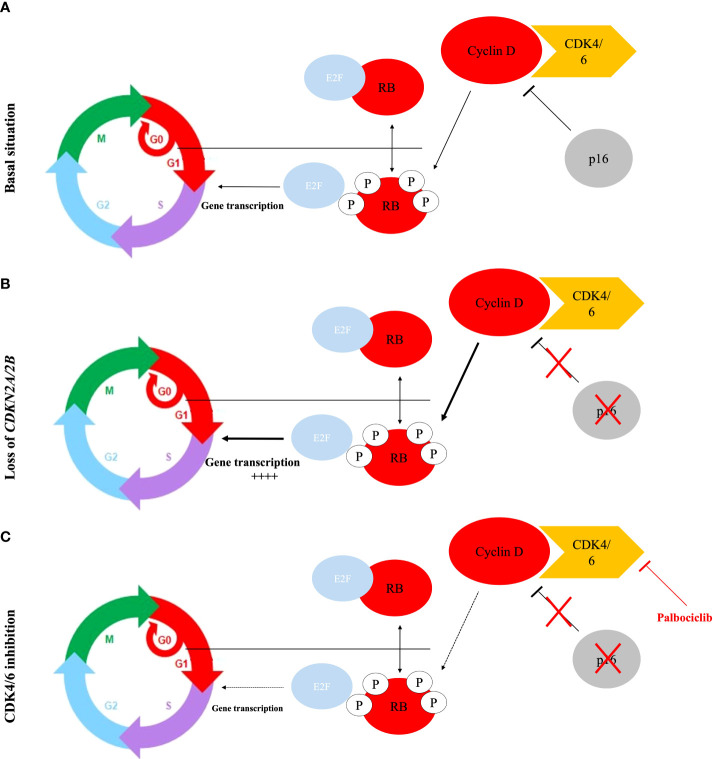
Chordoma tumors are known to overexpress multiple kinases (56) (included PDGFR-α, PDGFR-β, c-Kit, c-Met, pAKT, mTOR, HER2, VEGFR, and EGFR) which are involved in a multitude of cellular functions relevant to cancer pathogenesis. These kinases are well-studied in the field of oncology, and several FDA-approved drugs on the market targeting each kinase can be used as repurposing candidates for advanced chordomas. The first pre- and clinical results of some FDA-approved drugs targeting EGFR, PDGFR-α, PDGFR-β, and c-Kit, have shown partial objective response and clinical benefit in chordomas (57). The lack of data concerning the biology of chordomas (genetic driver events) and theranostic markers hampers the selection of molecularly guided therapies and may explain these sparse results. As kinases, CDK4 and CDK6 play a crucial role in the G1-S phase transition, which is an important restriction point in the normal cell cycle, often reduced in cancer, leading to uncontrolled cell proliferation ( Figure 6 ). Importantly, in chordomas, as well as in our PDX chordoma panel (35), the CDK4/6 regulatory gene CDKN2A (also known as p16) is frequently lost (5, 6), leading to the fact that CDK4/6 may be found in an over-activated state (41). However, it should be noted that other signaling pathways, in addition to CDKN2A loss, are inputted in cyclin D overexpression and promote CDK4/6 activity, leading to uncontrolled cell proliferation (42). Palbociclib, a specific inhibitor of CDK4/6, has recently been tested in in vitro cell line chordoma models harboring the loss of CDKN2A and showed an efficient inhibition of the tumor cell growth (41). Finally, the efficacy of palbociclib has been tested in numerous clinical trials for cancers, and is also currently being tested in a phase II trial, in patients with locally advanced/metastatic chordomas (NCT03110744).
Figure 6.
The classic model for the regulation of the G1/S transition by cyclins and CDK. (A) In basal situation, RB protein phosphorylation performed by cyclin D-CDK4/6 kinase induces E2F releasing and facilitates the expression of E2F target genes, which are critical for initiation of DNA synthesis and entry into S-phase. CDK4/6 activity is repressed by the p16 protein. (B) Loss of p16/CDKN2A/2B activity leads to an overactivation of the CDK4/6 activity and therefore an uncontrolled cell proliferation. (C) The CDK4/6 inhibitor re-establishes the activity of the RB protein activity as an inhibitor of cell division.
In our in vivo pharmacological program, the CD3 model harboring CDKN2A/2B showed a significant tumor growth inhibition (p = 0.02), while the CD7 model showed no tumor response (p = 0.85). Pharmacodynamic study (RT PCR and RNAseq) confirmed the difference between both PDX models harboring both the loss of CDKN2/2B. Indeed, in the responding CD3 PDX model, we observed a decrease in the expression of genes implied in the cell cycle, such as MKI67 (p = 0.008), E2F1 (p = 0.016), PLK1 (p = 0.016), TOP2A (p = 2.2 e-26), and AURKB (p = 2.5 e-8), compared to the non-responding model (CD7). We did not observe tumor response to palbociclib with the xenograft CD39 (p = 0.40) (which did not harbor a loss of CDKN2A/2B); such an observation raises the possibility to consider CDKN2A/2B loss as a theranostic marker of palbociclib antitumor efficacy in chordomas. Considering the various sensitivities to palbociclib, we can formulate the hypothesis on which tumor heterogeneity and preexistence of a resistant clonal subtype to palbociclib in the CD7 PDX model might be present. Indeed, chordoma is known as a heterogeneous tumor (58) with various cell phenotypes. Genetic heterogeneity contributes to selecting a clonal cell population during tumor development and progression, and is well-known as a prominent contributor to therapeutic failure (59). This hypothesis is reinforced by genetic analyses performed on the CD7 chordoma PDX model, which confirmed the loss of CDKN2A/2B in treated and control mice, but not in the primary patient’s tumor (35), suggesting a spatial tumor heterogeneity in which some cells harbor the homozygous deletion of CDKN2A, and others do not. Moreover, as observed in hormone receptor-positive breast cancers, many tumors demonstrate de novo (or intrinsic) resistance to palbociclib, implying genomic alterations (60). When we compared the gene expression in the RNAseq-supervised analysis in non-treated CD3 and CD7 PDX tumors, we observed a significant enrichment of the expression of genes implied in oxidative phosphorylation (OxPhos) into the non-responding CD7 PDX model, as compared to the responding CD3 PDX model. Metabolism reprogramming is well known as a hallmark of cancer (61). While normal cells commonly use mitochondrial oxidative metabolism for energy generation, tumor cells often switch to aerobic glycolysis, described as the Warburg effect (62). Classically, glycolytic metabolism is involved in the induction of acquired drug resistance, such as in estrogen-receptor breast cancer treated with palbociclib (63). However, some authors also suggested that drug resistance could be associated with an upregulation of the oxidative phosphorylation (OxPhos) pathway, which may be explained by the “reverse Warburg effect” (64, 65). Evans et al. (66) recently showed an OxPhos dependence in chemotherapy-resistant triple-negative breast cancer. Moreover, radiation therapy seems to increase in vitro and in vivo OxPhos activity (67). Thus, we can suppose a selection of clonal subtypes to palbociclib in the CD7 PDX model, increased by the history of radiation, leading to a higher OxPhos activity in this model. Interestingly, drug resistance can be reversed by blocking OxPhos (68). In several cancers, new OxPhos inhibitors have already been identified, and are under preclinical and/or clinical evaluation (66, 69). Moreover, favorable results about the synergistic antitumoral effect of the combination of OxPhos inhibitors and palbociclib in a triple-negative breast cancer cell line (66), encourage testing the use of this drug combination or OxPhos inhibitors alone in palbociclib-resistant PDX chordoma models harboring the CDKN2A/2B loss. However, some authors revealed an upregulation of oxidative metabolism (OxPhos) in CDK4/6 inhibitor-tolerant uveal melanomas (70), suggesting further experimental research to conclude a causal association in the Oxphos dependency of palbociclib-resistant chordomas. Although being not fully understood, several other resistance mechanisms of CD4/6 inhibitors exist, as mechanisms bypassing the CDK4/6 inhibition via activating CDK2 signaling, which is another way that phosphorylates RB (52).
The PLK1 inhibitor showed a slight antitumor efficacy without significant activity (p = 0.065) in one model (CD3). These results could be explained by the absence of concomitant amplification or copy number gains of CCND1 and CCNE2 in our chordoma PDXs models (35), both of which are involved in RB inactivation driving tumor cells through the S phase (47). PLK1 inhibition seems to be more promising in a combination of treatments than in monotherapy (46, 71), therefore suggesting the importance of future preclinical trials in order to assess the combination of PLK1 inhibitors with other targeted drugs such as palbociclib. Indeed, PLK1 inhibition results in a G2-M arrest (72). Consequently, the combination of drugs resulting in a dual G1 (palbociclib) and G2-M arrest (volasertib) might therefore be addressed in chordomas.
Study limitations
Considering our disparate results and the low number of tested models (n = 3), there is a lack of support evidence to consider CDKN2A/2B loss as a potential biomarker for CDK4/6 inhibitor sensitivity, suggesting the importance of further studies on more preclinical models. Secondly, we did not perform drug combinations, especially with volasertib and palbociclib, considering the slow-growth of the tumor in PDX models (35). Moreover, we also hypothesized that the enrichment of the expression of genes implied in oxidative phosphorylation (OxPhos) could lead to primary resistance in chordoma. However, our results are insufficient to definitely conclude a causal association considering the few tested models and the absence of additional pharmacogenomic experiments.
Conclusion
Considering the frequency of genomic alterations affecting the CDKN2A/2B gene in chordomas, targeting this biomarker was fundamental. The present study described for the first time the in vivo activity of a CDK4/6 inhibitor and a PLK1 inhibitor in two human chordoma xenografts harboring homozygous deletions of CDKN2A/2B. We also hypothesized that OxPhos activity could lead to as a resistance mechanism to CDK4/6 inhibitors in chordomas. CDK4/6 inhibition may be effective, but further studies in several PDX models are strongly necessary to confirm such an observation, and to identify predictive markers of response or resistance to palbociclib in chordomas and reverse primary or secondary resistances. Investigating volasertib in combination with palbociclib could also provide a novel therapeutic strategy.
Data availability statement
The data presented in the study are deposited in the GEO repository, accession number GSE216419 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE216418).
Ethics statement
The animal study was reviewed and approved by Ethics committee of the Institut Curie CEEA-IC #118 (Authorization APAFiS# 25870-2020060410487032-v1 given by National Authority).
Author contributions
Conceptualization, TP, JM-P, HM, SF, IB and DD. Data curation, TP. Formal analysis, TP. Investigation, TP. Methodology, TP and DD. Project administration, DD. Resources, TP, AD, LC, RB, FN, SV, HA-B. Software, TP. Supervision, JMP, HAB, HM, SF, IB and DD. Validation, SR-R, EM, HM, SF, JM-P and IB. Visualization, TP. Writing – original draft, TP and DD. Writing – review and editing, JM-P and IB. All authors contributed to the article and approved the submitted version.
Acknowledgments
We wish to thank the Animal Platform of the Institut Curie and Raphaëlle Galy for English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.960720/full#supplementary-material
Differential supervised analysis of the gene expression, comparing control tumors and tumors treated with palbociclib for the responding CD3 PDX model.
Differential supervised analysis of the gene expression, comparing control tumors and tumors treated with palbociclib for the non-responding CD7 PDX model.
Differential supervised analysis of the gene expression of control CD3 PDX tumors and control CD7 PDX tumors control PDX tumors.
References
- 1. Stacchiotti S, Sommer J. Chordoma global consensus group. building a global consensus approach to chordoma: A position paper from the medical and patient community. Lancet Oncol (2015) 16:e71–83. doi: 10.1016/S1470-2045(14)71190-8 [DOI] [PubMed] [Google Scholar]
- 2. Zou M-X, Lv G-H, Zhang Q-S, Wang S-F, Li J, Wang X-B. Prognostic factors in skull base chordoma: A systematic literature review and meta-analysis. World Neurosurg (2018) 109:307–27. doi: 10.1016/j.wneu.2017.10.010 [DOI] [PubMed] [Google Scholar]
- 3. Bohman L-E, Koch M, Bailey RL, Alonso-Basanta M, Lee JYK. Skull base chordoma and chondrosarcoma: Influence of clinical and demographic factors on prognosis: a SEER analysis. World Neurosurg (2014) 82:806–14. doi: 10.1016/j.wneu.2014.07.005 [DOI] [PubMed] [Google Scholar]
- 4. Stacchiotti S, Gronchi A, Fossati P, Akiyama T, Alapetite C, Baumann M, et al. Best practices for the management of local-regional recurrent chordoma: A position paper by the chordoma global consensus group. Ann Oncol (2017) 28:1230–42. doi: 10.1093/annonc/mdx054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tarpey PS, Behjati S, Young MD, Martincorena I, Alexandrov LB, Farndon SJ, et al. The driver landscape of sporadic chordoma. Nat Commun (2017) 8. doi: 10.1038/s41467-017-01026-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bai J, Shi J, Li C, Wang S, Zhang T, Hua X, et al. Whole genome sequencing of skull-base chordoma reveals genomic alterations associated with recurrence and chordoma-specific survival. Nat Commun (2021) 12:757. doi: 10.1038/s41467-021-21026-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Le LP, Nielsen GP, Rosenberg AE, Thomas D, Batten JM, Deshpande V, et al. Recurrent chromosomal copy number alterations in sporadic chordomas. PloS One (2011) 6:e18846. doi: 10.1371/journal.pone.0018846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duan Z, Feng Y, Shen J, Hornicek F. Genomic and epigenetic instability in chordoma: Current insights. AGG (2014) 67. doi: 10.2147/AGG.S50523 [DOI] [Google Scholar]
- 9. Wang L, Zehir A, Nafa K, Zhou N, Berger MF, Casanova J, et al. Genomic aberrations frequently alter chromatin regulatory genes in chordoma. Genes Chromosomes Cancer (2016) 55:591–600. doi: 10.1002/gcc.22362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Diaz RJ, Guduk M, Romagnuolo R, Smith CA, Northcott P, Shih D, et al. High-resolution whole-genome analysis of skull base chordomas implicates FHIT loss in chordoma pathogenesis. Neoplasia (2012) 14:788–98. doi: 10.1593/neo.12526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mattox AK, Yang B, Douville C, Lo S-F, Sciubba D, Wolinsky JP, et al. The mutational landscape of spinal chordomas and their sensitive detection using circulating tumor DNA. Neurooncol Adv (2021) 3:vdaa173. doi: 10.1093/noajnl/vdaa173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cottone L, Eden N, Usher I, Lombard P, Ye H, Ligammari L, et al. Frequent alterations in p16/CDKN2A identified by immunohistochemistry and FISH in chordoma. J Pathol Clin Res (2020) 6:113–23. doi: 10.1002/cjp2.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zuccato JA, Patil V, Mansouri S, Liu JC, Nassiri F, Mamatjan Y, et al. DNA Methylation-based prognostic subtypes of chordoma tumors in tissue and plasma. Neuro-Oncology (2022) 24:442–54. doi: 10.1093/neuonc/noab235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang C, Sun J, Yong L, Liang C, Liu T, Xu Y, et al. Deficiency of PTEN and CDKN2A tumor-suppressor genes in conventional and chondroid chordomas: Molecular characteristics and clinical relevance. Onco Targets Ther (2020) 13:4649–63. doi: 10.2147/OTT.S252990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choy E, MacConaill LE, Cote GM, Le LP, Shen JK, Nielsen GP, et al. Genotyping cancer-associated genes in chordoma identifies mutations in oncogenes and areas of chromosomal loss involving CDKN2A, PTEN, and SMARCB1. PloS One (2014) 9:e101283. doi: 10.1371/journal.pone.0101283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Singhal N, Kotasek D, Parnis FX. Response to erlotinib in a patient with treatment refractory chordoma. Anticancer Drugs (2009) 20:953–5. doi: 10.1097/CAD.0b013e328330c7f0 [DOI] [PubMed] [Google Scholar]
- 17. Stacchiotti S, Marrari A, Tamborini E, Palassini E, Virdis E, Messina A, et al. Response to imatinib plus sirolimus in advanced chordoma. Ann Oncol (2009) 20:1886–94. doi: 10.1093/annonc/mdp210 [DOI] [PubMed] [Google Scholar]
- 18. Lindèn O, Stenberg L, Kjellén E. Regression of cervical spinal cord compression in a patient with chordoma following treatment with cetuximab and gefitinib. Acta Oncol (2009) 48:158–9. doi: 10.1080/02841860802266672 [DOI] [PubMed] [Google Scholar]
- 19. Lebellec L, Chauffert B, Blay J-Y, Le Cesne A, Chevreau C, Bompas E, et al. Advanced chordoma treated by first-line molecular targeted therapies: Outcomes and prognostic factors. a retrospective study of the French sarcoma group (GSF/GETO) and the association des neuro-oncologues d’Expression française (ANOCEF). Eur J Cancer (2017) 79:119–28. doi: 10.1016/j.ejca.2017.03.037 [DOI] [PubMed] [Google Scholar]
- 20. Stacchiotti S, Morosi C, Lo Vullo S, Casale A, Palassini E, Frezza AM, et al. Imatinib and everolimus in patients with progressing advanced chordoma: A phase 2 clinical study. Cancer (2018) 124:4056–63. doi: 10.1002/cncr.31685 [DOI] [PubMed] [Google Scholar]
- 21. Hindi N, Casali PG, Morosi C, Messina A, Palassini E, Pilotti S, et al. Imatinib in advanced chordoma: A retrospective case series analysis. Eur J Cancer (2015) 51:2609–14. doi: 10.1016/j.ejca.2015.07.038 [DOI] [PubMed] [Google Scholar]
- 22. George S, Merriam P, Maki RG, Van den Abbeele AD, Yap JT, Akhurst T, et al. Multicenter phase II trial of sunitinib in the treatment of nongastrointestinal stromal tumor sarcomas. J Clin Oncol (2009) 27:3154–60. doi: 10.1200/JCO.2008.20.9890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stacchiotti S, Longhi A, Ferraresi V, Grignani G, Comandone A, Stupp R, et al. Phase II study of imatinib in advanced chordoma. J Clin Oncol (2012) 30:914–20. doi: 10.1200/JCO.2011.35.3656 [DOI] [PubMed] [Google Scholar]
- 24. Stacchiotti S, Tamborini E, Lo Vullo S, Bozzi F, Messina A, Morosi C, et al. Phase II study on lapatinib in advanced EGFR-positive chordoma. Ann Oncol (2013) 24:1931–6. doi: 10.1093/annonc/mdt117 [DOI] [PubMed] [Google Scholar]
- 25. Brüderlein S, Sommer JB, Meltzer PS, Li S, Osada T, Ng D, et al. Molecular characterization of putative chordoma cell lines. Sarcoma (2010) 2010:630129. doi: 10.1155/2010/630129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu X, Nielsen GP, Rosenberg AE, Waterman PR, Yang W, Choy E, et al. Establishment and characterization of a novel chordoma cell line: CH22. J Orthop Res (2012) 30:1666–73. doi: 10.1002/jor.22113 [DOI] [PubMed] [Google Scholar]
- 27. Scheil S, Brüderlein S, Liehr T, Starke H, Herms J, Schulte M, et al. Genome-wide analysis of sixteen chordomas by comparative genomic hybridization and cytogenetics of the first human chordoma cell line, U-CH1. Genes Chromosomes Cancer (2001) 32:203–11. doi: 10.1002/gcc.1184 [DOI] [PubMed] [Google Scholar]
- 28. Bosotti R, Magnaghi P, Di Bella S, Cozzi L, Cusi C, Bozzi F, et al. Establishment and genomic characterization of the new chordoma cell line chor-IN-1. Sci Rep (2017) 7:9226. doi: 10.1038/s41598-017-10044-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rinner B, Froehlich EV, Buerger K, Knausz H, Lohberger B, Scheipl S, et al. Establishment and detailed functional and molecular genetic characterisation of a novel sacral chordoma cell line, MUG-Chor1. Int J Oncol (2012) 40:443–51. doi: 10.3892/ijo.2011.1235 [DOI] [PubMed] [Google Scholar]
- 30. Hsu W, Mohyeldin A, Shah SR, a, Rhys CM, Johnson LF, Sedora-Roman NI, et al. Generation of chordoma cell line JHC7 and the identification of brachyury as a novel molecular target. J Neurosurg (2011) 115:760–9. doi: 10.3171/2011.5.JNS11185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salle H, Pocard M, Lehmann-Che J, Bourthoumieu S, Labrousse F, Pimpie C, et al. Development of a novel orthotopic primary human chordoma xenograft model: A relevant support for future research on chordoma. J Neuropathol Exp Neurol (2020) 79:314–24. doi: 10.1093/jnen/nlz121 [DOI] [PubMed] [Google Scholar]
- 32. Dobrolecki LE, Airhart SD, Alferez DG, Aparicio S, Behbod F, Bentires-Alj M, et al. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev (2016) 35:547–73. doi: 10.1007/s10555-016-9653-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Diaz RJ, Luck A, Bondoc A, Golbourn B, Picard D, Remke M, et al. Characterization of a clival chordoma xenograft model reveals tumor genomic instability. Am J Pathol (2018) 188:2902–11. doi: 10.1016/j.ajpath.2018.08.004 [DOI] [PubMed] [Google Scholar]
- 34. Zhao T, Siu I-M, Williamson T, Zhang H, Ji C, Burger PC, et al. AZD8055 enhances in vivo efficacy of afatinib in chordomas. J Pathol (2021). doi: 10.1002/path.5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Passeri T, Dahmani A, Masliah-Planchon J, Naguez A, Michou M, El Botty R, et al. Dramatic In vivo efficacy of the EZH2-inhibitor tazemetostat in PBRM1-mutated human chordoma xenograft. Cancers (2022) 14:1486. doi: 10.3390/cancers14061486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Masliah-Planchon J, Bièche I, Guinebretière J-M, Bourdeaut F, Delattre O. SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol Mech Dis (2015) 10:145–71. doi: 10.1146/annurev-pathol-012414-040445 [DOI] [PubMed] [Google Scholar]
- 37. Duan R, Du W, Guo W. EZH2: A novel target for cancer treatment. J Hematol Oncol (2020) 13:104. doi: 10.1186/s13045-020-00937-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gounder MM, Zhu G, Roshal L, Lis E, Daigle SR, Blakemore SJ, et al. Immunologic correlates of the abscopal effect in a SMARCB1/INI1-negative poorly differentiated chordoma after EZH2 inhibition and radiotherapy. Clin Cancer Res (2019) 25:2064–71. doi: 10.1158/1078-0432.CCR-18-3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Witkiewicz AK, Knudsen KE, Dicker AP, Knudsen ES. The meaning of p16 ink4a expression in tumors: Functional significance, clinical associations and future developments. Cell Cycle (2011) 10:2497–503. doi: 10.4161/cc.10.15.16776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature (1994) 368:753–6. doi: 10.1038/368753a0 [DOI] [PubMed] [Google Scholar]
- 41. von Witzleben A, Goerttler LT, Marienfeld R, Barth H, Lechel A, Mellert K, et al. Preclinical characterization of novel chordoma cell systems and their targeting by pharmocological inhibitors of the CDK4/6 cell-cycle pathway. Cancer Res (2015) 75:3823–31. doi: 10.1158/0008-5472.CAN-14-3270 [DOI] [PubMed] [Google Scholar]
- 42. Yuan K, Wang X, Dong H, Min W, Hao H, Yang P. Selective inhibition of CDK4/6: A safe and effective strategy for developing anticancer drugs. Acta Pharm Sin B (2021) 11:30–54. doi: 10.1016/j.apsb.2020.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mayer EL, Dueck AC, Martin M, Rubovszky G, Burstein HJ, Bellet-Ezquerra M, et al. Palbociclib with adjuvant endocrine therapy in early breast cancer (PALLAS): Interim analysis of a multicentre, open-label, randomised, phase 3 study. Lancet Oncol (2021) 22:212–22. doi: 10.1016/S1470-2045(20)30642-2 [DOI] [PubMed] [Google Scholar]
- 44. Gjertsen BT, Schöffski P. Discovery and development of the polo-like kinase inhibitor volasertib in cancer therapy. Leukemia (2015) 29:11–9. doi: 10.1038/leu.2014.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Steegmaier M, Hoffmann M, Baum A, Lénárt P, Petronczki M, Krssák M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth. vivo. Curr Biol (2007) 17:316–22. doi: 10.1016/j.cub.2006.12.037 [DOI] [PubMed] [Google Scholar]
- 46. Montaudon E, El Botty R, Vacher S, Déas O, Naguez A, Chateau-Joubert S, et al. High in vitro and in vivo synergistic activity between mTORC1 and PLK1 inhibition in adenocarcinoma NSCLC. Oncotarget (2021) 12:859–72. doi: 10.18632/oncotarget.27930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Montaudon E, Nikitorowicz-Buniak J, Sourd L, Morisset L, El Botty R, Huguet L, et al. PLK1 inhibition exhibits strong anti-tumoral activity in CCND1 -driven breast cancer metastases with acquired palbociclib resistance. Nat Commun (2020) 11:4053. doi: 10.1038/s41467-020-17697-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barber SM, Sadrameli SS, Lee JJ, Fridley JS, Teh BS, Oyelese AA, et al. Chordoma–current understanding and modern treatment paradigms. JCM (2021) 10:1054. doi: 10.3390/jcm10051054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Johnson JI, Decker S, Zaharevitz D, Rubinstein LV, Venditti JM, Schepartz S, et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br J Cancer (2001) 84:1424–31. doi: 10.1054/bjoc.2001.1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bièche I, Parfait B, Tozlu S, Lidereau R, Vidaud M. Quantitation of androgen receptor gene expression in sporadic breast tumors by real-time RT-PCR: evidence that MYC is an AR-regulated gene. Carcinogenesis (2001) 22:1521–6. doi: 10.1093/carcin/22.9.1521 [DOI] [PubMed] [Google Scholar]
- 51. Pandey K, An H, Kim SK, Lee SA, Kim S, Lim SM, et al. Molecular mechanisms of resistance to CDK4/6 inhibitors in breast cancer: A review. Int J Cancer (2019) 145:1179–88. doi: 10.1002/ijc.32020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li Z, Zou W, Zhang J, Zhang Y, Xu Q, Li S, et al. Mechanisms of CDK4/6 inhibitor resistance in luminal breast cancer. Front Pharmacol (2020) 11:580251. doi: 10.3389/fphar.2020.580251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rubino F, Alvarez-Breckenridge C, Akdemir K, Conley AP, Bishop AJ, Wang W-L, et al. Prognostic molecular biomarkers in chordomas: A systematic review and identification of clinically usable biomarker panels. Front Oncol (2022) 12:997506. doi: 10.3389/fonc.2022.997506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Al Baghdadi T, Halabi S, Garrett-Mayer E, Mangat PK, Ahn ER, Sahai V, et al. Palbociclib in patients with pancreatic and biliary cancer with CDKN2A alterations: Results from the targeted agent and profiling utilization registry study. JCO Precis Oncol (2019), 1–8. doi: 10.1200/PO.19.00124 [DOI] [PubMed] [Google Scholar]
- 55. Xue Z, Lui VWY, Li Y, Jia L, You C, Li X, et al. Therapeutic evaluation of palbociclib and its compatibility with other chemotherapies for primary and recurrent nasopharyngeal carcinoma. J Exp Clin Cancer Res (2020) 39:262. doi: 10.1186/s13046-020-01763-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Anderson E, Havener TM, Zorn KM, Foil DH, Lane TR, Capuzzi SJ, et al. Synergistic drug combinations and machine learning for drug repurposing in chordoma. Sci Rep (2020) 10:12982. doi: 10.1038/s41598-020-70026-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gill CM, Fowkes M, Shrivastava RK. Emerging therapeutic targets in chordomas: A review of the literature in the genomic era. Neurosurgery (2019). doi: 10.1093/neuros/nyz342 [DOI] [PubMed] [Google Scholar]
- 58. El-Heliebi A, Kroneis T, Wagner K, Meditz K, Kolb D, Feichtinger J, et al. Resolving tumor heterogeneity: Genes involved in chordoma cell development identified by low-template analysis of morphologically distinct cells. PloS One (2014) 9:e87663. doi: 10.1371/journal.pone.0087663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang J, Späth SS, Marjani SL, Zhang W, Pan X. Characterization of cancer genomic heterogeneity by next-generation sequencing advances precision medicine in cancer treatment. Precis Clin Med (2018) 1:29–48. doi: 10.1093/pcmedi/pby007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wander SA, Cohen O, Gong X, Johnson GN, Buendia-Buendia JE, Lloyd MR, et al. The genomic landscape of intrinsic and acquired resistance to cyclin-dependent kinase 4/6 inhibitors in patients with hormone receptor–positive metastatic breast cancer. Cancer Discovery (2020) 10:1174–93. doi: 10.1158/2159-8290.CD-19-1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 62. Warburg O. On respiratory impairment in cancer cells. Science (1956) 124:269–70. doi: 10.1126/science.124.3215.269 [DOI] [PubMed] [Google Scholar]
- 63. Lorito N, Bacci M, Smiriglia A, Mannelli M, Parri M, Comito G, et al. Glucose metabolic reprogramming of ER breast cancer in acquired resistance to the CDK4/6 inhibitor palbociclib+. Cells (2020) 9:668. doi: 10.3390/cells9030668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zaal EA, Berkers CR. The influence of metabolism on drug response in cancer. Front Oncol (2018) 8:500. doi: 10.3389/fonc.2018.00500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pavlides S, Whitaker-Menezes D, Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, et al. The reverse warburg effect: Aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle (2009) 8:3984–4001. doi: 10.4161/cc.8.23.10238 [DOI] [PubMed] [Google Scholar]
- 66. Evans KW, Yuca E, Scott SS, Zhao M, Paez Arango N, Cruz Pico CX, et al. Oxidative phosphorylation is a metabolic vulnerability in chemotherapy-resistant triple negative breast cancer. Cancer Res (2021). doi: 10.1158/0008-5472.CAN-20-3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen D, Barsoumian HB, Fischer G, Yang L, Verma V, Younes AI, et al. Combination treatment with radiotherapy and a novel oxidative phosphorylation inhibitor overcomes PD-1 resistance and enhances antitumor immunity. J Immunother Cancer (2020) 8:e000289. doi: 10.1136/jitc-2019-000289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee J-S, Lee H, Jang H, Woo SM, Park JB, Lee S-H, et al. Targeting oxidative phosphorylation reverses drug resistance in cancer cells by blocking autophagy recycling. Cells (2020) 9:2013. doi: 10.3390/cells9092013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sica V, Bravo-San Pedro JM, Stoll G, Kroemer G. Oxidative phosphorylation as a potential therapeutic target for cancer therapy. Int J Cancer (2020) 146:10–7. doi: 10.1002/ijc.32616 [DOI] [PubMed] [Google Scholar]
- 70. Teh JLF, Purwin TJ, Han A, Chua V, Patel P, Baqai U, et al. Metabolic adaptations to MEK and CDK4/6 cotargeting in uveal melanoma. Mol Cancer Ther (2020) 19:1719–26. doi: 10.1158/1535-7163.MCT-19-1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Posch C, Cholewa BD, Vujic I, Sanlorenzo M, Ma J, Kim ST, et al. Combined inhibition of MEK and Plk1 has synergistic antitumor activity in NRAS mutant melanoma. J Invest Dermatol (2015) 135:2475–83. doi: 10.1038/jid.2015.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gutteridge REA, Ndiaye MA, Liu X, Ahmad N. Plk1 inhibitors in cancer therapy: From laboratory to clinics. Mol Cancer Ther (2016) 15:1427–35. doi: 10.1158/1535-7163.MCT-15-0897 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differential supervised analysis of the gene expression, comparing control tumors and tumors treated with palbociclib for the responding CD3 PDX model.
Differential supervised analysis of the gene expression, comparing control tumors and tumors treated with palbociclib for the non-responding CD7 PDX model.
Differential supervised analysis of the gene expression of control CD3 PDX tumors and control CD7 PDX tumors control PDX tumors.
Data Availability Statement
The data presented in the study are deposited in the GEO repository, accession number GSE216419 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE216418).