Abstract
Human gut and food microbiomes interact during digestion. The outcome of these interactions influences the taxonomical composition and functional capacity of the resident human gut microbiome, with potential consequential impacts on health and disease. Microbe-microbe interactions between the resident and introduced microbiomes, which likely influence host colonisation, are orchestrated by environmental conditions, elements of the food matrix, host-associated factors as well as social cues from other microorganisms. Quorum sensing is one example of a social cue that allows bacterial communities to regulate genetic expression based on their respective population density and has emerged as an attractive target for therapeutic intervention. By interfering with bacterial quorum sensing, for instance, enzymatic degradation of signalling molecules (quorum quenching) or the application of quorum sensing inhibitory compounds, it may be possible to modulate the microbial composition of communities of interest without incurring negative effects associated with traditional antimicrobial approaches. In this review, we summarise and critically discuss the literature relating to quorum sensing from the perspective of the interactions between the food and human gut microbiome, providing a general overview of the current understanding of the prevalence and influence of quorum sensing in this context, and assessing the potential for therapeutic targeting of quorum sensing mechanisms.
Keywords: quorum sensing, quorum quenching, quorum sensing inhibition, gut microbiome, food microbiome, food matrix
Introduction
The interactions between human, animal and plant microbiomes and their ultimate impact on the assembly and maintenance of community structure and functionality is the focus of intense research efforts (Jagadeesan et al., 2019). In food microbiology, the influence of ingested food microbiomes on the human gut is a particular focus in recent years. The human gut harbours the greatest microbial load of all body sites − estimated to be in the order of 1013 bacterial cells (along with eukaryotic microorganisms and phage; Sender et al., 2016). This resident microbiota plays a significant role in human health and is amenable to outside influences imposed by diet and organisms that are resident in raw and minimally processed foods (Janssens et al., 2018; Walter et al., 2020). Nevertheless, many knowledge gaps persist regarding the dynamics of colonisation, resistance and succession in the food and gut microbiomes and how these may be modulated.
The genetic repertoire of bacteria enables them to perceive and adapt to environmental factors in flux. Environmental sensing and signalling in bacteria is primarily achieved through secondary nucleotide messengers, such as cyclic adenosine monophosphate (cAMP), cyclic diguanylate (c-di-GMP) and the hyperphosphorylated guanosine derivatives collectively referred to as (p)ppGpp (Hengge et al., 2019), as well as Two-Component Systems (TCSs) − pairs of sensory histidine kinases and response regulators that perceive extracellular signals and modulate gene expression accordingly (Liu et al., 2018a). In addition, many bacterial species possess dedicated Quorum Sensing (QS) systems that involve the secretion, extracellular accumulation and subsequent import and processing of specific small chemical signal molecules (Eickhoff and Bassler, 2018). At threshold concentrations, these signal molecules in the extracellular environment induce transcriptional changes that trigger phenotypic adaptation to changing social contexts. These include the coordination of biofilm regulation, expression of virulence or food spoilage traits and social choices such as cooperation, competition and cheating within micro-ecological contexts (Li and Tian, 2012; Allen et al., 2016; Defoirdt, 2018).
QS systems can provide information on the local population of ‘self’ and ‘other’; and it has been also proposed that QS could enable bacteria to detect diffusion-limited situations through an integrated model of efficiency sensing (Hense et al., 2007; Liu et al., 2018a). QS systems are described in both Gram-negative (Papenfort and Bassler, 2016) and Gram-positive bacteria (Verbeke et al., 2017), as well as fungi (Nogueira et al., 2019) and some viruses (Silpe and Bassler, 2019). Furthermore, next-generation sequencing and comparative genomic approaches have identified homologues of known quorum sensing circuits across genus and species boundaries (Whiteley et al., 2017), suggesting its importance in the regulation of survival strategies.
This review summarises the fundamental and translational research to date relating to the targeting of bacterial quorum sensing to produce foods with enhanced safety and quality profiles and the modulation of gut microbiomes for human health. An updated catalogue of quorum sensing activity and its inhibition in foods and the human gut is provided. Finally, we identify and discuss gaps in the literature that can be addressed in order to progress research in this field specifically with a view to the bioprotection of foods and the therapeutic targeting of the gut microbiome. As this review demonstrates, there is great promise for direct applications in food production systems, particularly in the context of functional foods, as well as biotherapeutics in the clinical sphere. On the other hand, some of the gaps identified within have the potential, if addressed, to benefit broader research in the field of microbial ecology in a cross-disciplinary manner.
Quorum sensing in bacteria
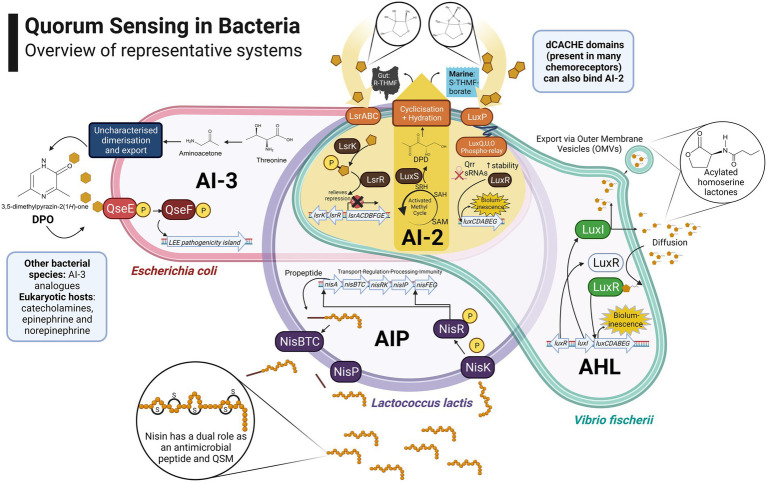
As per the original system first identified in Vibrio fischeri, the simplest QS circuit consists of regulatory gene pairs responsible for the production of signalling molecules (luxI) and transcriptional activation (luxR) of regulated genes responsible for phenotypes such as bioluminescence (Nealson et al., 1970). In addition to global phenotypic regulation based on local population density, QS can also contribute to managing the production of public goods including both the signal molecules themselves (auto-induction) and co-regulated products relevant to the context of foods and the human gut, such as extracellular enzymes, exopolysaccharides, surfactants, antimicrobial compounds, virulence factors and siderophores (Heilmann et al., 2015). An overview of the principal systems found in bacteria is presented in Figure 1. Public goods, in the context of microbiology, refers to molecular components of the secretome produced by individual cells and transferred to the extracellular milieu, potentially benefiting the population as a whole. Public goods are distinct from private goods, which are generally cytosolic or membrane-bound and hence confer an exclusive benefit upon producer cells.
Figure 1.
Overview of representative bacterial quorum sensing systems: Escherichia coli cell illustrates an Autoinducer 3 (AI-3) Quorum Sensing (QS) system which regulates the locus of enterocyte effacement (LEE) pathogenicity island (Walters and Sperandio, 2006; Machado Ribeiro et al., 2021); an Autoinducing Peptide (AIP) system in Lactococcus lactis is presented, in which nisin has a dual role as a Quorum Sensing Molecule (QSM) and antimicrobial peptide (Kleerebezem, 2004); Vibrio fischeri cell illustrates the control of bioluminescence through an Acylated Homoserine Lactone (AHL) system also referred to as Autoinducer 1 (AI-1; Nealson, Platt and Woodland Hastings, 1970); Autoinducer 2 (AI-2) signalling is presented at the intersection due to its role in species- and genera-agnostic communications. The AI-2 biosynthetic pathway is highlighted in yellow alongside the Lsr/Lux pathways which provide for its internalisation/detection in various genera (Pereira, Thompson and Xavier, 2013). Created with BioRender.com.
Understanding the relative contribution of quorum sensing to microbiome structure and functionality is complicated by heterogeneous distribution of QS producers and responders, even within clonal populations (Striednig and Hilbi, 2022). For instance, one study reported that just 68% of V. fischeri cells responded to exogenous QSM (Pérez and Hagen, 2010). Further, receptor promiscuity and divergence in ligand binding of quorum sensing receptors towards non-canonical substrates is suggestive of a system that can be tuned and can adapt over time to changing environmental conditions and competitors (Hawver et al., 2016; Prescott and Decho, 2020). Mapping QS systems in complex ecological environments is complicated by their tendency to follow hierarchical and combinatorial structures (Ng and Bassler, 2009; Cornforth et al., 2014) and to modulate gene expression programs through asymmetry (Hurley and Bassler, 2017), thought to facilitate the management of metabolic trade-offs (Dinh and Prather, 2019). This is perhaps best exemplified by the Qrr sRNAs (Quorum Regulatory sRNAs) of the Vibrio genus and the multifunctional RNAIII which coordinates several stacked regulatory circuits managing the accessory gene regulator (agr) system in Staphylococcus aureus (Bronesky et al., 2016).
Quorum sensing molecules
QS systems involve the production and detection of Quorum Sensing Molecules (QSM) and understanding the synthesis, structure and stability of these compounds is important for developing strategies for therapeutic targeting of these systems. A high-level overview of the principal classes of QSM is provided in Table 1.
Table 1.
Overview of major bacterial Quorum Sensing Molecules (QSM) families, examples, possible modifications, producer and/or responder bacteria utilising the QSM as well as the regulated phenotypes and specificity.
| System | QSM | QSM modification | Producer (and responder) bacteria | Responder bacteria (no production) | Relevant phenotypes regulated | Specificity | Reference |
|---|---|---|---|---|---|---|---|
| Autoinducer 1 (AI-1) | Acylated-homoserine lactones (AHL/HSL) | Homoserine lactone moiety is joined to a variable length (C4-C16) acyl tail, which can bear oxo- or hydroxyl- substitutions. | Gram-negative bacteria, e.g. Aliivibrio fischeri | Species with LuxR solos | las and rhl systems | Generally species-specific due to AHL molecular structure (length of acyl chain; substitutions) | Fuqua, Parsek and Greenberg, (2001); Bez et al., (2021) |
| Autoinducer 2 (AI-2) | S-THMF (2S,4S)-2-methyl-2,3,3,4-tetra-hydroxytetrahydrofuran; R-THMF: (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydro-furan) | S-THMF-borate predominates in marine environments; heptyl modifications are possible | ~Half of sequenced bacterial genomes through luxS | Pseudomonas aeruginosa, Bacillus subtilis and Rhodopseudomonas palustris (through dCACHE domain) | Multiple and varied | Species-agnostic; enteric bacteria detect R-THMF through lsr operon; boronated S-THMF is detected through lux cascade; other bacteria detect through dCACHE domain-containing proteins | Pereira, Thompson and Xavier, (2013) |
| Autoinducer 3 (AI-3) | 3,5-dimethylpyrazin-2-one (DPO) | Various pyrazinone analogues | Enterohemorrhagic Escherichia coli (EHEC) | Many Gram-negative bacteria (through QseC homologues) | Expression of locus of enterocyte effacement (LEE), Shiga-toxin production | Interspecific (differences in relative abundance of AI-3 analogues); Inter-kingdom crosstalk with mammalian hormones epinephrine and norepinephrine. | Clarke et al., (2006); Walters and Sperandio, (2006); Kim et al., (2020); Machado Ribeiro et al., (2021) |
| Autoinducer peptides (AIP) | Post-translationally modified peptides | Length (5–17 amino acids); Linear or cyclical structure | Gram-positive bacteria, e.g. Lactococcus lactis | Sporulation and competence in Bacillus subtilis (phr system); toxin production in C. botulinum and C. perfringens (agr-type system) | Generally species-specific due to the molecular structure | Kleerebezem et al., (1997); Novick and Muir, (1999); Cook and Federle, (2014); Monnet, Juillard and Gardan, (2016) | |
| Small RNAs | miRNAs and siRNAs; Qrr sRNAs (Quorum Regulatory sRNAs) in Vibrio spp.; RNAIII in Staphylococcus aureus | Specificity to targeted mRNA transcripts, which are removed from circulation. | Reported in Escherichia, Salmonella, Streptococcus, Pseudomonas and Vibrio genera | Type III secretion genes in Vibrio harveyi | Specificity to the targeted mRNA transcript | Rutherford et al., (2011); Teng et al., (2011); Fu, Elena and Marquez, (2019) | |
| Diffusible Signal Factor (DSF) | cis-11-methyl-dodecenoic acid Derived from fatty acids with a cis-unsaturated double bond at the 2-position. | Chain length, branched/unbranched structure | Stenotrophomonas maltophilia, Burkholderia cenocepacia and P. aeruginosa | S. aureus, B. cereus, S. enterica, E. coli | Virulence and exopolysaccharide (EPS) production in Xanthomonas oryzae pv. oryzae; Biofilm dispersal in P. aeruginosa | Intraspecific and cross-kingdom crosstalk possible | Sepehr et al., (2014); Zhou et al., (2017); Liu et al., (2018b) |
| Palmitate methyl ester (PAME) | 3-hydroxy palmitic acid methyl ester | Ralstonia solanacearum | Virulence and upregulation of AHL production | Species-specific | Flavier et al., (1997); Mole et al., (2007) | ||
| Diketopiperazines (DKP) | Amino acid-derived cyclic dipeptides | Constituent amino acids; Chirality; | Cronobacter sakazakii, Bacillus cereus, Vibrio spp. | Biofilm formation in C. sakazakii; Increased resistance to oxidative stress in Vibrio spp. | cyclo(Phe-Pro) is detected by both C. sakazakii and B. cereus | Bofinger et al., (2017); Kim et al., (2018); Zink et al., (2021) | |
| Quinolones | 2-heptyl-3-hydroxy-4-quinolone (PQS); 2-alkyl-4(1H)-quinolone (AHQ) |
P. aeruginosa | Virulence factors, including pyocyanin, elastase, lectin, and rhamnolipid production | Species-specific | Rampioni et al., (2016); Lin et al., (2018) | ||
| Dialkylresorcinols | Dialkylresorcinols (DARs) and Cyclohexanediones (CHDs) | Variable R chain | 116 species, including the genera Photorhabdus, Neisseria, Capnocytophaga, Flavobacterium | Pathogenicity against Galleria mellonella | Intraspecific crosstalk possible | Brameyer et al., (2015) | |
| Photopyrones (PPYs) | α-pyrones | Variable R chain | Photorhabdus luminescens | Bacillus atrophaeus | Photorhabdus clumping factor (Pcf); biofilm formation | Intraspecific crosstalk possible | Brachmann et al., (2013); Hickey et al., (2021) |
There are many instances in which QSM fulfil a dual role in addition to QS, such as Autoinducer 2 (AI-2) and Autoinducer Peptides (AIPs). AI-2 systems have been identified in approximately half of sequenced bacterial genomes and are considered to mediate species-agnostic communication, particularly in complex communities, presented at the intersection of the bacterial cells in Figure 1. At the same time, some argue that AI-2 biosynthesis mainly contributes to the detoxification of S-adenosylhomocysteine (SAH) produced during the activated methyl cycle (AMC) and is essential to the metabolism of cysteine and methionine, with an incidental QS role (Kang et al., 2019). Disambiguation of these contrasting theories has been explored through the construction of a LuxS-deficient Streptococcus mutans, followed by restoration of AMC functionality by complementation with S-adenosyl-L-homocysteine hydrolase (sahH from P. aeruginosa; Hu et al., 2018). Whilst biofilm formation and acid tolerance were restored through complementation, homocysteine and lactic acid levels remained aberrant, underlining the broader significance of luxS in the metabolic landscape of the cell.
Similarly, there are numerous examples of AIP QSM which, in addition to facilitating QS, act as antimicrobial peptides such as lantibiotics (bacteriocins are reviewed in greater detail in Section 2.3). In addition to the multifunctionality of QSM themselves, it bears considering that QSM can also be degraded or for instance exported in a non-specific manner through multidrug efflux pumps (Rahmati et al., 2002).
Methods for the analysis of QSM
As raised almost a decade ago (Skandamis and Nychas, 2012), a greater understanding of the relative abundance and significance of QS signalling in microbiomes is required, particularly in providing temporal resolution of QS at significant points in time and space (Donaldson et al., 2016). Accurate and rapid detection, characterisation and quantification of QSM can enhance our understanding of QS in various environments, as well as providing a real-time readout of microbial physiology within a given microbiome.
Conventional methods frequently used to interrogate the physico-chemical properties of QSM include mass spectrometry (MS) and high-performance liquid chromatography (HPLC), whilst biosensor-based systems employing plasmids, chromosomes or enzymes from reporter bacterial strains can rapidly detect a colorimetric, luminescent or fluorescent signal. Chromatographic techniques are more resource-intensive but can detect QSM with a higher degree of accuracy and can also be used to validate novel biosensors (Cutignano, 2019).
Whilst several analytical techniques are available for AI-1 and AI-2 QSM, AIP are generally more difficult to quantify as they are normally found at low concentrations in complex mixtures (Kalkum et al., 2003) whilst AI-3 was only recently characterised and no methods have been developed for its routine analysis yet (Kim et al., 2020). Novel approaches for the detection and characterisation of AIP and AI-3 are needed, in addition to protocols for QSM analysis in the complex matrices associated with food and human gut microbiome samples. In this regard, analytical techniques developed and validated in the context of environmental microbiology, such as the analysis of biofilms in wastewater treatment plants (Wang et al., 2018) could be adapted. These could be combined with emerging fluorescence-based techniques such as Combinatorial Labelling And Spectral Imaging-Fluorescence In-Situ Hybridization (CLASI-FISH) to incorporate the spatial ecology of microbial communities (Wilbert et al., 2020).
Several biosensor strains, often plasmid-based, have been constructed to detect different QSM classes (Wen et al., 2017), however chromatographic methods tend to be preferred to detect mixes of QSM in complex microbial consortia (Gui et al., 2018). Rapid paper-based diagnostic tools containing immobilised whole cell biosensors have also been validated to nanomolar level detection of AHLs and AI-2 QSM (Wynn et al., 2018).
Biochemical analytical techniques are increasingly being paired with in silico studies of quorum sensing-related genes in metagenomic samples to help distinguish QS potential from active QS processes (Rezzonico et al., 2012). In addition to the established Quorumpeps database for AIP peptides (Wynendaele et al., 2013), more recent efforts have generated novel databases including the Omics Database of Fermentative Microbes (ODFM; Whon et al., 2021) and Quorum Sensing of Human Gut Microbes (QSHGM; Wu et al., 2021b). Curated and regularly updated databases are fundamental for accurate bioinformatic predictions, which will assist in modelling the effect of quorum sensing systems in complex communities as part of broader pipelines (Wynendaele et al., 2013). Accurate modelling of quorum sensing in foods and the human gut is necessary in order to establish prevalence, relevance and to evaluate potential interventions. Ex vivo modelling of the human gut, such as through colonic models and gut-on-a-chip approaches, can provide an opportunity to benchmark quorum sensing activity at various points in the digestive process (Garcia-Gutierrez and Cotter, 2021). Fermented foods have also been demonstrated to be tractable and convenient experimental models for studying community assemblage in food microbiomes (Wolfe and Dutton, 2015) and could be evaluated as models for QS studies.
Quorum sensing “eavesdropping”
Whilst QS was originally described in terms of autoinduction, communication through QS can occur across strains, species and even kingdoms (Kendall and Sperandio, 2016). LuxR solos are one such circuit that permit ‘eavesdropping’ by detecting and integrating community-level information to inform the regulation of gene expression without investing in the production of QSM (Subramoni and Venturi, 2009). The metabolic cost of QSM synthesis varies considerably – estimated in the order of 184 ATP per autoinducer peptide (AIP), 8 ATP per acyl-homoserine lactone (AHL) and 0–1 ATP per autoinducer-2 moiety (AI-2; Ruparell et al., 2016), highlighting the likely evolutionary benefit of an eavesdropping strategy.
Notable examples in an enteric context include species of the genera Escherichia, Klebsiella, and Salmonella that possess orthologues of the transcriptional regulatory protein SdiA − detecting AHLs produced by other species – whilst lacking the requisite genes for AHL production (Michael et al., 2001; Lindsay and Ahmer, 2005). This type of QS circuit is implicated in the suppression of traI-mediated conjugation between E. coli and P. aeruginosa (Lu et al., 2017), which could potentially be harnessed in applications to reduce dissemination of genetic determinants of antimicrobial resistance (AMR) by horizontal gene transfer (HGT). Similarly, several instances of food spoilage appear to be augmented by synergy between AHL-QSM producers and non-producers, presumably due to the capacity of the non-producers to detect QSM in the extracellular milieu (Yu et al., 2019; Wang and Xie, 2020).
Quorum sensing inhibition
Due to the wide range of bacterial phenotypes regulated by quorum sensing and their impact in clinical, environmental and agricultural contexts, QS systems have been proposed as a potential therapeutic target for the modulation of microbial communities (Rutherford and Bassler, 2012; Kalia, 2013; Lade et al., 2014). Indeed, the phenomenon of quorum sensing inhibition (QSI; also referred to in the literature as quorum sensing interference to describe broader modulation of QS) has been reported not just amongst bacteria, suggesting an ecological role in governance of cooperation and competition (Murugayah and Gerth, 2019), but also between bacteria and eukaryotic cells including fungi, plants and animals (Hughes and Sperandio, 2008; Joshi et al., 2021). QSI can be accomplished through several mechanisms, such as enzymatic degradation [also referred to as Quorum Quenching (QQ)], production of QSM analogues, signal sequestration or other modulation of signal flux up or downstream.
Whilst various applications in medical and industrial settings have been investigated, the use of QSI to modulate food microbiomes and gut microbiota has received relatively less attention, yet may prove useful in addressing the global threat of antibiotic resistance and enhancing the robustness of probiotic strains (Kareb and Aïder, 2020). It is anticipated that metagenomics and metatranscriptomics will guide studies in this relatively novel area to determine the effect of QSI on microbial communities, whilst genetic engineering can be used to improve and maximise the potential of QQ enzymes derived from natural environments (Billot et al., 2020). Advances in the field of chemistry such as structure–activity studies of synthetic QSM analogues (Rajamani et al., 2008; Lyons et al., 2020) naturally complement such bioprospecting and rational design approaches.
Perhaps some of the most exciting research in the field has emerged at the interdisciplinary juncture between biology and engineering occupied by the field of synthetic biology (Wang et al., 2020). Given the close resemblance between electronic circuits and the intrinsic networks and feedback loops of quorum sensing systems, there is potential for ground-breaking interdisciplinary work. In the field of synthetic ecology, precisely engineered consortia are being used to evaluate the relative importance of quorum sensing pathways and the equilibrium between QS and QSI when targeted for therapeutic modulation (Stephens and Bentley, 2020), as well as advancing the fundamental research in the field (Wu et al., 2021a).
Biofilms as environments which favour QS
Biofilms are aggregations of microbial cells, attached to one another and typically adhered to a surface through a matrix of polymers that are at least partially microbially secreted. They are ubiquitous and considered the predominant form of living adopted by bacteria, fungi, viruses and single-celled eukaryotes (Flemming et al., 2016), with specific relevance to food and food processing environments (Alvarez-Ordóñez et al., 2019) and the human gut (Deng et al., 2020).
As well as physically shielding in-dwelling cells, the structure of the biofilm matrix includes channels and microcolonies that generate nutrient and oxygen gradients. These gradients are thought to contribute to phenotypic and genotypic plasticity and reinforce the heterogeneity generally seen in biofilm-dwelling populations (Penesyan et al., 2019). The distinct microenvironments formed and defined by biofilms are thought to favour interactions with other microorganisms due to their enforced proximity within the matrix and reduced diffusion. The resulting circulation of nutrients, metabolites, QSM and other public goods such as extracellular enzymes and siderophores are considered to influence emergent features in bacteria (Solano et al., 2014; Flemming et al., 2016). Emergent properties refer to characteristics of populations, especially multispecies and biofilm-associated, which differ from those of equivalent planktonic populations. Most commonly studied attributes include social cooperation, nutrient management and antimicrobial tolerance (Flemming et al., 2016).
More specifically, mixed biofilms containing multiple species are commonplace. They may arise due to synergy between different attachment phenotypes, such as specialised fimbriae and exopolysaccharide (EPS) production, with the structure of the latter in particular being influential on the emergent properties of biofilms beyond those of equivalent planktonic populations (Swaggerty et al., 2018).
Quorum sensing in foods and food processing environments
Advances in nucleic acid manipulation and sequencing have helped enhance our understanding of food microbiomes: the microbial communities within and upon food matrices, which become established during primary production and evolve throughout the farm-to-fork continuum (De Filippis et al., 2018; Yap et al., 2022). This field of study receives special attention in light of the emerging, cross-disciplinary approach of One Health, which recognises the interrelationship between human, animal and environmental health [One Health High-Level Expert Panel (OHHLEP) et al., 2022]. These advances have helped the collection of temporal datasets that can better describe the taxonomical and functional evolution of microbiomes across complex food chains, in order to enhance our understanding of their influence on food safety, quality, sustainability and potential impact on human health (Pasolli et al., 2020). Whilst certain species can dominate specific communities at particular times, broadly speaking food microbiomes consist of multiple strains with some degree of interaction, which may affect collective behaviours of microorganisms such as food spoilage (Qian et al., 2018). In regulating phenotypic characteristics and metabolic processes, QS is proposed to enhance microbial adaptation to dynamic environments and has been associated with food quality in both a negative sense with respect to microbial food spoilage (Yuan et al., 2018), as well as enhancing the quality of some fermented foods (Johansen and Jespersen, 2017).
The overall impact of QS in microbial succession dynamics within food microbiomes is difficult to assess due to the frequent heterogeneous responses seen in bacteria to QSM described previously. Further, relative receptor binding affinity studies have demonstrated that many QS receptors can be activated by many other small molecules produced by microorganisms or hosts (Wellington and Greenberg, 2019), as we will outline for various compounds present in foods. Finally, the accumulation, compartmentalisation and stability of QSM are likely to be highly influenced by intrinsic and extrinsic factors of the food matrix and subject to fluctuations at different points of the food production process (Skandamis and Nychas, 2012). For example, studies on AHL-QSM stability in soil established a half-life ranging from hours to days depending on pH and temperature (Delalande, 2005) and the influence of micro-environmental conditions in foods and the human gut remains to be elucidated. Recent technologies have harnessed bacterial QS activity to provide real-time monitoring of microbial activity during food production (Miller and Gilmore, 2020).
Quorum sensing molecules detected in foods
A broad range of QSM have been reported in different foods, as illustrated in Figure 2. Overall, fatty acid-derived QSM such as AHLs and DKPs in foods are predominantly associated with bacteria involved in food spoilage or food-borne illness, whilst associations with AI-2-QSM appear more evenly split between instances of benign bacterial consortia and undesirable ones.
Figure 2.
Overview of Quorum Sensing Processes as involved in food production, adapted from (Blana, Doulgeraki and Nychas, 2011; Plummer, 2012; Nahar et al., 2018; Quintieri et al., 2020; Wang and Xie, 2020). The left hand side of the figure in yellow maps food-associated bacteria reported in the literature to utilise QS for spoilage or pathogenesis, organised into columns based on the class of QSM (AHL, AI-2 and others) and by the type of food in which they are reported. This scheme is repeated on the right hand side in blue for those food-borne bacteria with positive attributes in food with respect to biopreservation and biotransformation. Created with BioRender.com.
Different foods present distinct matrix effects, which will likely impact the stability and mobility of QSM produced by the microbiota within; high AI-2 activity is reported in frozen fish, tomato, cantaloupe, carrots, tofu and cow’s milk, whilst raw poultry, beef and artisanal cheeses appeared to inhibit AI-2 QS activity (Lu et al., 2004). This finding was supported by a more recent study measuring spiked AHL and AI-2 in water, skim milk and meat suspensions via a whole-cell biosensor assay, finding that signal was diminished only in the meat matrix (Wynn et al., 2018). The fluctuating levels of organic acids, pH and other biomolecules during food processing, especially in activities involving fermentation, are likely to influence QSM concentration and stability over time.
AI-2 appears to be particularly significant in the context of fermented food microbiomes. For instance, many bacteria isolated from the surface of smear-ripened cheeses produce AI-2, including Arthrobacter nicotianae, Corynebacterium ammoniagenes, Corynebacterium casei, Microbacterium barkeri, Microbacterium gubbeenense and Staphylococcus equorum subsp. linens (Novak and Fratamico, 2006; Gori et al., 2011). Another study of isolates from West African fermented foods reported AI-2 production in many Aerobic Endospore-producing Bacteria (AEB) such as Bacillus subtilis, B. cereus, B. altitudinis, B. amyloliquefaciens, B. licheniformis, B. aryabhattai, B. safensis, Lysinibacillus macroides and Paenibacillus polymyxa (Qian et al., 2015). This is supported by bioinformatic analysis of the microbiome of cocoa bean fermentations, which revealed a high prevalence of luxS-related genes in the core/soft-core of the lactic acid bacteria (LAB) Limosilactobacillus fermentum, Pediococcus acidilactici and Lactiplantibacillus plantarum. Additional luxS genes were also detected in some accessory genomes (de Almeida et al., 2021) and altogether participation in AI-2 signalling was proposed to help LAB adapt to diverse niches such as food fermentations.
AI-2 QS is also emerging as a key player in supporting desirable biofilms in food processing. For instance, AI-2 is present in high concentrations in Spanish table olive biofilms produced using a sequential yeast-LAB starter culture, which controlled undesirable spontaneous microbiota and maintained organoleptic quality throughout the product shelf life (Benítez-Cabello et al., 2020). In the same study, high AI-2 levels also correlated with high LAB counts on the olive skin surface, supporting the assertion that robust biofilm-forming capacity can enable potential probiotic strains to better survive and become established in fermented food microbiomes. This phenomenon has also been reported in other beneficial members of food microbiomes such as Lactiplantibacillus pentosus in olives (Perpetuini et al., 2016) and Lactiplantibacillus paraplantarum L-ZS9 in kimchi (Park et al., 2016). Thermal treatment is also highly relevant to food processing and whilst its effect on all QSM is not known, AI-2 concentrations are diminished by pasteurisation/ultra-heat-treatment (UHT; Lu et al., 2004).
An important limitation is that many studies report QSM production by strains isolated from foods, rather than quantifying QSM in situ, thereby only estimating the potential prevalence of QS in foods. A further confounding factor is the use of biosensors that may not distinguish between various enantiomers of QSM or between bacterial QSM and host-derived molecular mimics.
Biofilms within food and food industries as environments for QS
Around one quarter of food waste is attributed to microbial processes (Huis in ‘t Veld, 1996), with implications for food safety and quality at a consumer level, economic losses at an industrial level and resource wastage in the context of sustainability. Microbial load and community composition are influenced by the nature and origin of raw materials, processing methods, transport and storage conditions (Jarvis et al., 2018; Van Reckem et al., 2020; Zwirzitz et al., 2020) and may fluctuate in accordance with the conditions of the niche. Food processing typically employs several physico-chemical hurdles to mitigate undesirable microbial growth such as thermal treatment, high pressure processing, use of acids, bacteriocins or other additives as well as regular sanitisation (Khan et al., 2017). Cumulatively, it has been proposed that this process could select for resistant strains that persist in the processing environment, particularly within biofilms, in turn re-contaminating finished foods (Walsh and Fanning, 2008).
Biofilms can be formed at many points in the food production process (Maes et al., 2019; Wagner et al., 2020), beginning with colonisation of raw plant materials and animal tissues during primary production. Biofilms can also form on man-made materials such as plastic or steel food-contact surfaces through preconditioning with biomolecules which facilitate attachment and extracellular matrix production by bacteria. Biofilm dwelling can provide a degree of shielding and protection from physical forces such as desiccation and shear, or biocides, facilitating greater resistance, particularly in mixed biofilms (Giaouris et al., 2015).
Biofilm formation has traditionally been viewed as a negative bacterial trait in the food industry, due to its association with specific spoilage organisms (SSO) and pathogenic bacteria that can re-contaminate foods post-processing, resulting in food spoilage and outbreaks of food-borne illness, as well as acceleration of ablation and corrosion of food-contact surfaces. The impact microbial transmission from biofilms into finished food products is of particular concern given the rising popularity of minimally processed and ready-to-eat foods (Meireles et al., 2018). Further, biofilms are increasingly recognised to promote antimicrobial tolerance (Uruén et al., 2020) as well as act as focal points for HGT between members of the population contained within (Abe, Nomura and Suzuki, 2020), potentially exacerbating the challenge of AMR.
However, biofilms also underpin and drive many key biotransformations in food and QS plays a role in at least some of these. Biofilm formation is thought to be crucial for the growth of milk kefir grains, which is catalysed by autoaggregation between LAB such as L. lactis, Leuconostoc mesenteroides, Lentilactobacillus kefiri, and Lentilactobacillus sunkii followed by synergistic kefiran production (EPS) by LAB and acetic acid bacteria (AAB), with cross-kingdom networking by yeasts such as Kluyveromyces marxianus (Wang et al., 2012; Han et al., 2018). Water kefir has received less attention than its dairy counterpart, but likely recapitulates this paradigm, with dextran-producing LAB thought to play a key role in grain growth (Fels et al., 2018; Guzel-Seydim et al., 2021). QS is likely to receive increased attention in this area, both for its relevance to the production of fermented foods with a biofilm quality such as kefirs, as well as its broader technological relevance in modulating the production of microbially synthesised extracellular polysaccharides.
Freeze-drying survival rates are also a concern in the commercial production of probiotic products. Notably, modulation of osmotic stress exposure in L. plantarum LIP-1 was found to enhance AI-2 production and promote biofilm formation by the strain, enabling it to better withstand the freeze-drying process (Jingjing et al., 2021). As subsequently outlined in the context of the human gut, biofilm formation and linked traits such as EPS production are increasingly being proposed as probiotic traits as they may enhance the survival of probiotic strains throughout gastrointestinal transit (Kelly et al., 2020).
Bacteriocin production
Amongst the bacterial processes regulated by QS in foods, bacteriocin production has received particular attention due to its application in the production of food and feed, as well as antimicrobial therapy. Bacteriocins are a group of ribosomally synthesised peptides with antimicrobial activity, notably mediating interspecies competition due to immunity genes in the producer strain (Cotter et al., 2005). It has been approved as a natural biopreservative in foods by both the FDA in the US and EFSA in the EU and extensively investigated for applications in dairy foods, as well as meat, seafood and vegetable-based foods (Gharsallaoui et al., 2016).
As outlined previously, some AIP fulfil a dual role as both QSM and antimicrobial peptides, including lantibiotics (class I bacteriocins) such as nisin and lacticin 481, produced by some strains of Lactococcus lactis and subtilin, produced by specific strains of B. subtilis (Kleerebezem, 2004; Gobbetti et al., 2007). In the case of class II bacteriocins, such as certain plantaricins in Lactiplantibacillus plantarum, salivaricins in Ligilactobacillus salivarius and some carnobacteriocins produced by members of the genus Carnobacterium, a dedicated peptide pheromone is responsible for regulating transcription of the bacteriocin genes, forming a three-component system (Leisner et al., 2007; Maldonado-Barragán et al., 2009).
Bacteriocin production is by now well-established as an attractive feature in potential probiotic strains. In the context of interspecies interactions and microbial succession, bacteriocin production is proposed to enhance the viability of producer strains, particularly in complex ecosystems (Maldonado-Barragán et al., 2013). In-situ production of certain bacteriocins within foods can control the growth of spoilage and pathogenic microorganisms due to their specificity of action and non-toxic effect on the host (Dobson et al., 2012). Moreover, QS systems offer the potential to fine-tune the dynamics of bacteriocin synthesis. Future studies could examine in greater detail the dynamics of co-culture induction in complex communities, as reported in a strain-specific manner in L. plantarum (Maldonado-Barragán et al., 2013), with a view to optimising in-situ production of bacteriocins.
Quorum sensing in the human gut microbiome
As previously described, the relatively high microbial load in the gut as well as the associations between the gut microbiome and health have made it a point of focus for human microbiome studies. The gut microbiome refers broadly to the gastrointestinal tract, primarily comprising the stomach, small and large intestine, sometimes also including the oral cavity and the microbial communities that become established within. The niches provided by the various compartments are highly dynamic with respect to changes in pH, oxygenation, nutrient availability, host–microbe interactions and microbial load, rendering community assemblage an incredibly complex ecological process (Donaldson et al., 2016). Additional spatial organisation is conferred by the distinct microenvironments provided by the gut lumen, mucin layer, intestinal crypts and mucosal epithelium.
Recent studies have highlighted the biofilm-like structure of mucus-associated bacterial communities in the gut lumen (Duncan et al., 2021), suggesting the likely prevalence of QS considering its previously established link with biofilms. Studies in this field are challenging due to the well-established inter-individual variation present even between ‘healthy’ individuals (Jones et al., 2018). Gaps remain in understanding the relative significance and importance of changes in composition and functionality of microbiomes as well as the ecological forces influencing assembly, maintenance and temporal fluctuation.
Perhaps the clearest instance of an interaction between microbiota present in a consumed food and the gut microbiome is food-borne illness. Many hurdles are mounted against invading bacterial pathogens, both from the resident microbiome (a phenomenon termed ‘colonisation resistance’) as well as host cells and immune mechanisms, all of which must be overcome to establish infection (Buffie and Pamer, 2013). In this delicate balance between colonisation, invasion and regulation, many bacterial pathogens utilise QS, at least in part, to regulate the expression of clinically relevant virulence factors that are metabolically costly and less strategic at low population densities (Rutherford and Bassler, 2012). Indeed, certain components of the human immune system are primed to detect and respond to pathogens and their virulence factors, which will be discussed below.
Quorum sensing molecules detected in the human gut microbiome
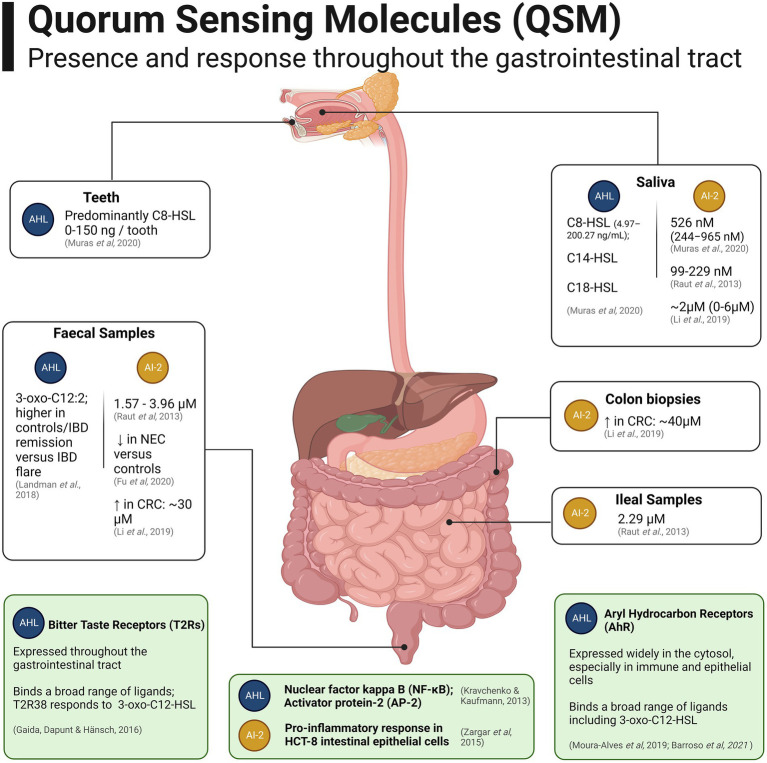
Relatively few studies have evaluated the presence of QSM within the human gut microbiome, due to the technical challenges associated with chromatography on luminal samples and low abundance of the molecules (Xue et al., 2021). Sampling of AHL-QSM at relevant temporal and spatial points is challenging due to their propensity to degrade at biological pH shortly after secretion, either spontaneously to tetramic acid, or by enzymatic hydrolysis of the lactone ring, leading their concentration to reflect temporal rather than historic population information (Peyrottes et al., 2020). Figure 3 provides an overview of the findings reported to date regarding QSM in the human gut.
Figure 3.
Quorum Sensing Molecules (QSM) which have been detected in samples from the human gastrointestinal tract (white boxes). Mammalian receptors present in the human gut capable of binding QSM as ligands are shown in light green boxes towards the bottom of the figure. Created with BioRender.com.
With respect to the oral cavity, high-performance liquid chromatography-coupled mass spectrometry (HPLC-MS) has been used to detect AHL-QSM in extracted teeth and saliva samples, with C8-HSL being most abundant and present in all saliva samples and most of the extracted teeth, whilst C14-HSL and C18-HSL were present at lower levels (Muras et al., 2020).
Mass Spectrometry (MS) has been used to detect the novel QSM 3-oxo-C12:2, a derivative of the AHL 3-oxo-C12 bearing 2 unsaturations, in human faeces (Landman et al., 2018). It was detected at higher levels in both healthy control and Inflammatory Bowel Disease (IBD) patients in remission versus those in active flare and was correlated with higher abundances of Bacillota, especially Faecalibacterium prausnitzii, and lower abundances of E. coli. 3-oxo-C12:2 also displayed anti-inflammatory properties and an ability to limit cytokine-induced tight junction disruption in a Caco-2/TC7 model when compared with the native 3-oxo-C12 from P. aeruginosa (Aguanno et al., 2020). The presence and presumptive bioactivity in the human gut microbiome of AHLs bearing such unsaturations is intriguing, as unsaturated pheromones are generally uncommon and their production had otherwise only been reported in the marine Roseobacter clade (Ziesche et al., 2015; Peyrottes et al., 2020). The producer of the precursor AHL and unsaturations, as well as its precise biological effects, remain unidentified to this date. A novel UPLC-MS/MS method has recently been developed to detect AHL-QSM in caecal, sera and liver samples of germ-free, conventional and Citrobacter rodentium-infected mice (Xue et al., 2021). This study demonstrated the translocation of bacterial AHL-QSM across the intestinal barrier throughout host tissues and the perturbation in AHL profiles provoked by C. rodentium infection.
Relatively few studies have investigated AI-2 levels in the gut microbiome. Liquid chromatography–tandem mass spectrometry was used to detect AI-2 produced by the oral microbiota in saliva samples (Campagna et al., 2009) via the linear precursor molecule 4,5-dihydroxy-2,3-pentanedione (DPD). Concentrations fluctuated considerably between participants (average 526 nM, range 244–965 nM), potentially highlighting inter-individual variation or temporal fluctuations in the oral microbiome (Rickard et al., 2008). AI-2 also appears to mediate inter-kingdom interactions in the human mouth in that, for example, hyphal development in the fungal pathogen Candida albicans is inhibited by AI-2 produced by Aggregatibacter actinomycetemcomitans (Bachtiar et al., 2014). Beyond the oral cavity, AI-2 has been detected in ileal and rectal samples taken during endoscopy from participants with and without IBD (Raut et al., 2013). In the gut, one study found diminished AI-2 levels in the acute phase of Necrotising Enterocolitis (NEC), a serious gastrointestinal disease primarily affecting infants, consequentially proposing the QSM as a potential biomarker for early detection and prophylaxis (Fu et al., 2020). In contrast, AI-2 levels were observed to be significantly higher in faecal samples and biopsies from patients with Colorectal Cancer (CRC) versus those of adenomas/healthy controls (Li et al., 2019a,b).
AI-3 is the most recently described QSM thought to be found in the gut. AI-3 family pyrazinones are produced by a wide range of bacteria relevant to the gut microbiome, as well as in pathogen-free mice colonised with E. coli BW25113, although human microbiome samples have not been interrogated to date using analytical techniques (Kim et al., 2020). Similarly, although direct quantification of other QS systems has not to our knowledge been reported, in silico predictions via the QSHGM database (Wu et al., 2021b) reveal presence of photopyrone, dialkylresorcinol and DSF genetic circuits in a wide range of gut microbes, although this remains to be evaluated using analytical techniques.
Taken together, QS systems appear to be active at various points of the human gut microbiome as are several distinct strategies by the eukaryotic host to monitor and respond to bacterial QSM. Future studies leveraging novel sampling approaches can further clarify the relative importance and clinical significance of the various systems, particularly regarding the interactions between commensals and opportunistic pathogens introduced to the gut microbiome.
Qs influence on collective bacterial behaviours in the gut
Lifestyle switching, through environmental and quorum sensing, has been proposed to favour the survival and colonisation of introduced species in the human gut (Mukherjee and Bassler, 2019). QS can also facilitate the establishment of collective behaviours of microbial populations and communities within the gastrointestinal tract, potentially affecting community assemblage and host physiology. AI-2 systems are most commonly implicated in this context, in both positive and negative senses.
Vibrio cholerae provides one of the clearest examples of lifestyle switching in order to adapt to and survive harsh environmental conditions in the environment and in the gut using QS. It forms aggregates of biofilms when surrounded by other V. cholerae cells and switches to a dispersal strategy to escape the matrix when surrounded by high numbers of ‘other’ non-V. cholerae cells. (March and Bentley, 2004; Bari et al., 2013; Bridges and Bassler, 2021). It has recently been proposed that V. cholerae senses its own quorum via CAI-1 QSM in conjunction with the biogenic amine norspermidine, which is relatively specific to V. cholera, whilst other populations are monitored using AI-2 and spermidine, a ubiquitous biogenic amine found throughout the human gut (Bridges and Bassler, 2021). CAI-1 has also been demonstrated to enhance the virulence of the non-producer Enteropathogenic E. coli (EPEC), possibly contributing to co-infection of the host (Gorelik et al., 2019). AI-2 also modulates sporulation and biofilm formation in the pathogenic Clostridium difficile, facilitating switching between dormancy, vegetative growth, virulence, immune evasion and antibiotic tolerance (Ðapa et al., 2013; Desai and Kenney, 2019; Tijerina-Rodríguez et al., 2019). Additionally, C. difficile uses an agr system mediated by thiolactone QSM to regulate production of the toxins TcdA and TcdB which compromise the host epithelial barrier in C. difficile infection (CDI; Darkoh et al., 2015). Certain C. difficile phage appear to bear and disseminate agr variants amongst their hosts, highlighting the complexity of microbiome dynamics and the difficulty in limiting investigations to single kingdoms (Hargreaves et al., 2014). This also supports the recent implication of bacterial QS as a broad mediator of phage-host interactions in the gut (León-Félix and Villicaña, 2021). Such insights certainly require additional scrutiny, but could prove significant in the development of phage therapy.
There are also many instances where QS activity is associated with positive compositions or outputs from microbial communities. One standout study found that increasing local levels of AI-2 in a mouse model of antibiotic treatment promoted the recovery of Bacillota (previously known as Firmicutes) whilst hindering the expansion of Bacteroidota (previously known as Bacteroidetes; Thompson et al., 2015). As previously described for food-borne probiotic strains, AI-2 QS was also shown to positively influence colonisation of a probiotic Bifidobacterium breve UCC2003 in the gut (Christiaen et al., 2014). Indeed, luxS-deficient Streptococcus mutans mutants incapable of producing AI-2 were found to upregulate carbohydrate metabolism and ABC transporters, highlighting the potential for QS to govern individual behaviours as well as collective ones (Yuan et al., 2021) and underlining the necessity for systems biology approaches when evaluating proposed therapeutic applications of QSI.
Detection of bacterial QS by the human host
Bacterial quorum sensing may also contribute to host–microbe interactions as evidenced by the existence of pathways in eukaryotic multicellular organisms to detect and even interfere with microbial communications (Aframian and Eldar, 2020). Indeed, the broader field of microbial endocrinology abounds with examples of crosstalk between host hormones and bacterial signalling systems, perhaps unsurprisingly considering the hormone-like properties of bacterial QSM (Neuman et al., 2015; Li et al., 2019a). Taken together, this is suggestive of co-evolution and adaptation by the eukaryotic host to monitor for AHL production and coordinate a defensive response.
AHL-QSM are small, lipophilic molecules and although poorly soluble in water, their diffusion into eukaryotic cells has recently been demonstrated, with interaction with intracellular targets also considered possible (Kamaraju et al., 2011; Peyrottes et al., 2020). Molecular self-assembly in aqueous solution into micelles and vesicles has been observed for 3-oxo-C8-AHL, 3-oxo-C12-AHL, C12-AHL and C16-AHL, which may guide our understanding of the kinetics of secreted QSM (Gahan et al., 2020). Bacterial outer membrane vesicles (OMVs) have also been proposed to traffic quorum sensing molecules within and between bacterial microcolonies, circumventing the issue of hydrophobicity (Toyofuku et al., 2017). OMVs have also been detected in host blood, heart and urine samples, being able to cross the mucus layer and epithelium of the gut (Stentz et al., 2015). Whilst OMVs have been studied to the greatest extent in the context of Gram-negative pathogens, attention is turning to their significance in probiotic and commensal strains, including E. coli Nissle 1917 and ECOR63, respectively (Alvarez et al., 2016), however more precise characterisation of their composition and relevance to QS is required.
Bitter taste receptors (T2Rs) belong to the G protein-coupled receptor group and were initially studied in the context of taste, being highly expressed on the tongue. However they have been found to be expressed in a variety of other sites including the upper respiratory tract and gastrointestinal tract, suggesting a broader role (Verbeurgt et al., 2017). Indeed, there is evidence of variable interaction of these receptors with numerous bacterial metabolites including AHLs. T2R38 is one example that is expressed throughout the gastrointestinal tract on intestinal epithelial cells (Behrens and Meyerhof, 2011; Karlsson et al., 2012) and activated by the AHL-QSM 3-oxo-C12-HSL in myeloid cells, enhancing phagocytic activities (Gaida et al., 2016) − although the myeloid cells in the latter study were derived from patient biopsies of osteomyelitis. Yet another study, using a heterologous expression system, found that T2R38 did not respond to 3-oxo-C12 HSL nor the cyclic AIP Agr-D1 thiolactone and CSP-1, but was activated by a range of bacterial metabolites including acetone, 2-butanone, 2-pentanone, 2-methylpropanal, dimethyl disulphide, methylmercaptan, and γ-butyrolactone (Verbeurgt et al., 2017). Methodological differences between the studies may account for this discordance.
Pro-inflammatory cascades have also been reported by AHL interaction with nuclear factor kappa B (NF-κB) and activator protein-2 (AP-2) receptors (Kravchenko and Kaufmann, 2013). The human aryl hydrocarbon receptor (AhR) is present in the human gut and interacts with a wide array of microbial metabolites and was shown to detect and react to relative abundances of AHLs, quinolones and phenazines produced by P. aeruginosa to manage immune response over the course of infection (Moura-Alves et al., 2019; Barroso et al., 2021).
In contrast to AHL-QSM, unmodified AI-2 is hydrophilic and does not appear to penetrate the plasma membrane, although heptyl modifications increase its affinity (Kamaraju et al., 2011). In vitro exposure of HCT-8 intestinal epithelial cells to AI-2 was found to provoke an inflammatory response (Zargar et al., 2015); this interaction could be studied in a more representative model system such as intestinal organoids or in vivo. Recent evidence suggests that mammalian epithelial cells can produce an AI-2 analogue that may mimic and interfere with bacterial QS, although the structure has yet to be determined (Ismail et al., 2016).
Quorum sensing inhibition in food microbiomes
Given the association between QS and bacterial biofilm formation and food spoilage activity, QSI has been proposed as a potential strategy to positively regulate bacterial phenotypes and improve food safety and quality. In fact, many foods themselves exhibit QSI activity associated with defence systems in plant/animal tissue or with the presence of microorganisms that have developed mechanisms to interfere with QS activity of neighbouring species.
Quorum sensing systems are interdependent and complex—the field of QSI can generally benefit from the application of a systems biology approach to understand the network of interactions occurring within a particular microbiome. As applied to food production systems, QSI interventions may need to be timed to suppress bacterial QS at critical control points in the food chain, as opposed to their constitutive deployment, or targeted against specific bacterial species implicated in spoilage or pathogenesis whilst maintaining the structure of the overall food microbiome.
QSI produced in microbe-microbe interactions in food microbiomes
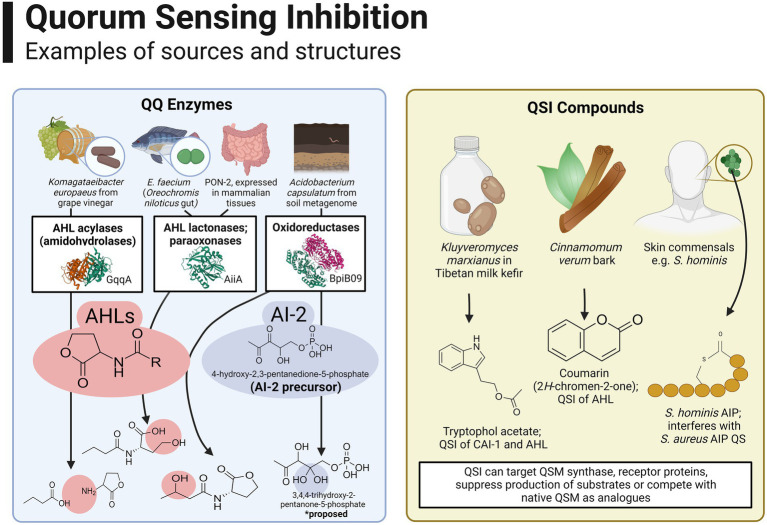
Several instances of QSI activity in foods are attributed to members of food microbiomes. Many quorum quenching (QQ) enzymes belonging to AHL lactonase, acylase and oxidoreductase families have been identified, for the most part being isolated in soil and marine metagenomes, plant symbionts and pathogenic bacteria (Bijtenhoorn et al., 2011; Fetzner, 2015). Figure 4 provides an overview of the source and structures of some prominent examples of QQ enzymes and QSI compounds. The majority of QSI discovered to date are active against AHL-QSM; to our knowledge, only one AI-2 QQ enzyme has been identified to date, originating in a soil metagenomic bank and hypothesised to reduce an AI-2 precursor, 4-hydroxy-2,3-pentanedione-5-phosphate, to the inactive derivative 3,4,4-trihydroxy-2-pentanone-5-phosphate (Weiland-Bräuer et al., 2016). For an extensive list of microbially derived QQ enzymes, the reader is directed to a comprehensive review (Sikdar and Elias, 2020).
Figure 4.
Overview of the source and structures of some prominent examples of QQ enzymes and QSI compounds. Many QQ enzymes have been described for AHLs, such as the AHL acylase GqqA reported in Komagataeibacter europaeus isolated from grape vinegar [Werner et al., 2021; Image from the RCSB PDB (rcsb.org) of PDB ID 7ALZ], AHL lactonases such as AiiA from E. faecium isolated from the gut of Oreochromis niloticus [Vadassery and Pillai, 2020; Image from the RCSB PDB (rcsb.org) of PDB ID 7L5F] and paraoxonase enzymes which are expressed widely in mammalian tissues (Peyrottes et al., 2020). Oxidoreductase enzymes have been isolated from environments such as soil and activity has been reported against AHLs (Bijtenhoorn et al., 2011; Image from the RCSB PDB (rcsb.org) of PDB ID 3RKR) as well as AI-2 (Weiland-Bräuer et al., 2016). Examples of non-enzymatic QSI include tryptophol acetate, active against CAI-1 and AHLs (Malka et al., 2021), coumarin and derivatives, which can interfere with AHL-QS (Gutiérrez-Barranquero et al., 2015) and AIP analogues produced by closely related staphylococcal species (Peng et al., 2019). Created with BioRender.com.
With that being said, food microbiomes have yielded some novel QSI agents in recent years, particularly QQ enzymes as shown in Figure 3. An AHL acylase with was recently characterised in a Komagataeibacter europaeus strain isolated from grape vinegar (Werner et al., 2021), complementing an earlier report of another putative AHL acylase in a S. epidermidis strain isolated from fermented soybean curd (Mukherji and Prabhune, 2015). In the latter, the authors propose QQ as a possible probiotic trait in food fermentations to reduce overall concentrations of AHL-QSM. Another study demonstrated the potential of in silico screening through mining of a Mao-tofu metagenome, discovering a novel AHL lactonase (aii810) active at low temperatures (Fan et al., 2017).
As AHL-QS producers account for a large proportion of the burden of spoilage and pathogenesis in aquaculture, this area has received significant attention and yielded an array of novel AHL-QQ enzymes. Aquaculture and fish gut microbiome isolates including Bacillus sp. QSI-1, Bacillus licheniformis T-1 and Enterococcus faecium QQ12 have been identified as potential probiotic feed ingredients due to their potential modulatory role for the AHL-QSM profile of the fish gut microbiome (Chu et al., 2014) and protecting against A. hydrophila infection through AHL lactonase activity of YtnP (Peng et al., 2021) and autoinducer inactivation (aiiA) homologues (Vadassery and Pillai, 2020). Another study found widespread AHL-QQ activity in bacterial isolates from rainbow trout tissue, including members of the genera Bacillus, Enterobacter, Citrobacter, Acinetobacter, Agrobacterium, Pseudomonas and Stenotrophomonas. Characterisation showed variability in both enzymatic and non-enzymatic modes of action, highlighting the diversity of QQ activity observed in the comparably simple fish gut microbiome, as well as a propensity for cell-bound activity rather than secretion (Torabi Delshad et al., 2018).
Metagenomic studies have revealed the presence of potential quorum quenching determinants in dairy products and production plants (Alexa Oniciuc et al., 2020); food microbiomes may represent in this sense a relatively untapped reservoir for novel QSI determinants. Indeed, there are numerous reports of culture extracts from specific LAB strains exerting anti-biofilm, anti-virulence and QSI effects on other bacterial species; for instance, cell free supernatants of LAB isolated from fermented grape interfere with AI-2 quorum sensing and biofilm formation in Salmonella Typhi and S. Typhimurium (Pelyuntha et al., 2019) and supernatants of LAB strains isolated from beef and cow’s milk inhibited pyocyanin production in a P. aeruginosa isolate of rancid butter and diminished violacein production in C. violaceum (Aman et al., 2021). The molecular mechanism of this widespread yet strain-specific activity remains unelucidated to date, but is thought to involve, at least in part, acid production along with small molecules secreted by the LAB strains. Indeed, another study evaluated both acidic and neutralised supernatants of LAB strains, finding that neutralised supernatants more weakly inhibited biofilm formation in P. aeruginosa compared with acidic fractions; moreover, only acidic supernatants interfered with the las and rhl quorum sensing systems (Rana et al., 2020).
Intriguingly, many examples of QSI arise amongst eukaryotic members of food microbiomes, specifically fungi. AI-2 QSI activity of brown algae has been attributed not to the plant cells but rather to bacterial and fungal endophytes (Tourneroche et al., 2019). Recently, a novel QSI compound (tryptophol acetate) was discovered to be produced by the yeast Kluyveromyces marxianus present in a Tibetan milk kefir (Malka et al., 2021). It was found to interfere with bacterial AHL and CAI-1 signalling, with possible applications for bioprotection of foods and prevention of proliferation of enteric pathogens such as V. cholerae (CAI-1). Again, this highlights the need for systems biology approaches that encompass all relevant kingdoms of life, not solely bacteria, when studying QS in a microbiome context.
Lastly, purified microbially derived enzymes can be incorporated into food formulations or cleaning protocols for food processing plants. To our knowledge, just one study has examined the use of enzymes with quorum quenching activity in foods. In this case, it was found that treatment with a commercial preparation of 7 peptidases and 1 amylase (Flavourzyme) decreased the relative expression of genes involved in quorum sensing (Nahar et al., 2021). However, this study only examined AI-2 related transcription, which could be due to suppression of the AMC (Activated Methyl Cycle) as a result of global metabolic perturbation. Treating raw sturgeon with AiiAAI96 (AHL lactonase) and nisin (bacteriocin) prior to vacuum packaging extended the product shelf life by 5 days (Gui et al., 2021), showing promise as a biopreservative targeting both Gram-positive and negative SSO. Heterologous expression of QQ enzymes in situ offers another potential route for biotechnological applications, as exemplified by the use of a Lacticaseibacillus casei strain, often used in the dairy industry, to express the AHL lactonase AiiK which can attenuate virulence in Aeromonas hydrophila (Dong et al., 2020) and P. aeruginosa (Dong et al., 2018).
QSI By components of food matrices
In addition to microbially produced QSI molecules, it has been established that many components of food matrices can, to some degree, modulate bacterial QS. A brief overview of prominent examples are provided in Table 2. In some cases, such as the organic acids, they may affect the stability of the QSM themselves or downregulate their synthesis by the producer strain due to acid stress. In other instances, such as fatty acids, the QSI phenomenon is thought to arise due to their structural similarity to the fatty-acid-derived QSM including AHLs and DSF. Indeed, in the case of ascorbic acid (vitamin C) the mechanism of action could arise due to its effect on pH or due to the structural similarity between it (5S)-5-[(1R)-1,2-Dihydroxyethyl]-3,4-dihydroxy-2(5H)-furanone) and other furanones – such disambiguation will be critical for harnessing it as a potential QSI agent.
Table 2.
Overview of components of food matrices which can mediate bacterial Quorum Sensing Inhibition (QSI) or related phenotypes, such as biofilm inhibition and sporulation.
| Food component | Examples | QS system or phenotype affected | Bacteria affected | Concentration | References |
|---|---|---|---|---|---|
| Organic acids | Lactic acid | AHL; Anti-biofilm | Chromobacterium violaceum CV026; E. coli; Salmonella sp. | 0.2–1% | Amrutha, Sundar and Shetty, (2017) |
| Acetic acid | AHL; Anti-biofilm | C. violaceum CV026; E. coli; Salmonella sp. | 1–1.5% | ||
| Citric acid | AHL; Anti-biofilm | C. violaceum CV026, E. coli; Salmonella sp. | 1.5–2% | ||
| Malic acid | AI-2 | E. coli O157:H7; S. Typhimurium | 4% | Almasoud et al., (2016) | |
| Ascorbic acid (vitamin C) | AHL | P. aeruginosa | 5–12·5 mg/ml | El-Mowafy, Shaaban and Abd El Galil, (2014); Pandit et al., 2017) | |
| Sporulation; AI-2 | C. perfringens, | 10 to 30 mM (sporulation); 300 mM (AI-2) |
Novak and Fratamico, (2006) | ||
| AI-2 | E. coli EMC17 | 125 mM | Shivaprasad et al., (2021) | ||
| AIP (ComC) | B. subtilis | 10 to 40 mM | Pandit et al., (2017) | ||
| Long Chain Fatty Acids (LCFAs) | Mono-unsaturated palmitoleic myristoleic acids | AHL | Acinetobacter baumannii | 0.02 mg/ml | Nicol et al., (2018) |
| Linoleic acid | DSF | P. aeruginosa | 10 μM | Kim et al., (2021) | |
| Fatty acids produced by fungal endophyte of Coriandrum sativum | Anti-biofilm | S. mutans | 31.3 mg/l | Abdel-Aziz, Emam and Raafat, (2020) | |
| Oleic acid (Cis-9-octadecenoic acid) | Anti-biofilm | S. aureus | 0.1% v/v | Stenz et al., (2008) | |
| AHL | C. violaceum CV026; | 3.696 mg/ml | Singh et al., (2013) | ||
| Palmitic acid (C16:0), Stearic acid (C18:0), Oleic acid (C18:1ω9), Linoleic acid (C18:2ω6) | AI-2 | V. harveyi, E. coli | 1–10 mM | Soni et al., (2008) | |
| Endocannabinoids | Endocannabinoid 2-AG (arachidonic acid derivative) | AI-3 (via QseC) | EHEC, Citrobacter rodentium | Not quantified – 2-AG levels were elevated in intervention group via knockout of monoacylglycerol lipase (Mgll) | Ellermann et al., (2021) |
| Furanones | Brominated, halogenated and natural forms in seaweeds, tomatoes, strawberries and other berries; synthetic derivatives | AHL | P. aeruginosa | 3.125–50 μM | Colin Slaughter, (1999); Manefield et al., (1999); Proctor, McCarron and Ternan, (2020) |
| AI-2 | Vibrio harveyi, Vibrio parahaemolyticus | 20 mg/l | Defoirdt et al., (2006) | ||
| Anti-biofilm | S. Typhimurium; L. monocytogenes; | 10 to 15 μM (S. Typhimurium); 0.05 mmol/l at 24 h, 2 mmol/l at 48 h (L. monocytogenes) | Janssens et al., (2008); Rodríguez-López et al., (2019) | ||
| Coumarin | Dihydrocoumarin | Anti-biofilm | Hafnia alvei | 3.2 mM | Hou et al., (2017) |
| AHL | C. violaceum CV026 | 6.3 mM | |||
| Coumarin from Cinnamomum verum | AHL | P. aeruginosa (pqsA, rhlI, lasI) | 1.36 mM | Gutiérrez-Barranquero et al., (2015) | |
| Vitamin B2 | Riboflavin and derivatives | AHL (LasR system) | P. aeruginosa | 200 μM (effective soluble concentration) | Rajamani et al., (2008) |
| Steviol glycosides | Steviol | AHL (las / rhl systems) | E. coli K802NR-pSB1075 (las); P. aeruginosa PAO-JP2 (pKD-rhlA) (rhl) | 0.26–0.52 mM (las); 0.0325–0.52 mM (rhl) | Markus et al., (2020) |
| Reb A | 0.019375–0.31 mM (rhl) | ||||
| Stevioside | 0.5–1.0 mM (las); 1 mM (rhl) |
Generally, plant-based foods do not appear to support bacterial quorum sensing activity to the same extent as animal-based ones (Medina-Martínez et al., 2006). This phenomenon is hypothesised to arise from co-evolutionary forces between plants and microorganisms and is primarily mediated by inhibitory plant secondary metabolites including furanones, phenolics and other bioactives that can act as QSM analogues (Rodrigues et al., 2016; Zaytseva et al., 2019; Pun et al., 2021). This could suggest the potential for plant-derived ingredients to generally disrupt bacterial QS activity or support their incorporation in food formulations to modulate the food microbiome of the product. However due to the general bioactivity of plant secondary metabolites, possible off-target effects should be assessed and effective QSI activity established in the context of the matrix effects and digestion.
Whilst food matrix-derived peptides may be similar in size and sequence to known QS AIP, no QSI peptides have been described to date in foods. Bioactive peptides are intriguing due to their small size, which allows them to diffuse relatively easily through food matrices and biofilms; additionally, they are amenable to bioengineering against resistance as they are gene-encoded (Bhutia and Maiti, 2008; Field et al., 2019). QSI AIP have been described in coagulase-negative staphylococci in the skin microbiome with activity against S. aureus (Peng et al., 2019) − this could suggest that intra-genus competition within food microbiomes represents an opportunity for future mining and discovery of potential therapeutic agents with direct applications in food production.
This provides a new perspective on food processing and formulation and the impact of ingested foods on the gut microbiome. Whilst many components pertain to raw food ingredients derived from plants and animals, many of these such as organic acids, fatty acids and vitamins can themselves be microbially synthesised and are of interest in their own right in the context of human nutrition. Aromatic and volatile compounds are overrepresented, with many QSI compounds also having a role in flavouring such as Maillard reaction-derived furanones (Wang and Ho, 2008) and lactone derivatives such as coumarins. Understanding the directionality of these effects, particularly with respect to stability and efficacy in food matrices and withstanding transit through the digestive tract, will be of key importance in the development of applications in the food formulation space.
Quorum sensing inhibition in the human gut microbiome
As previously outlined, quorum sensing has been implicated in the onset of pathogenesis in infections and is thought to influence community composition in other human-associated microbiomes such as those of the oral cavity, lung, skin, urinary and reproductive tracts (Bradshaw and Sobel, 2016; Cole et al., 2018; Scoffone et al., 2019; Williams et al., 2019). As some pathogenic bacteria utilise QS to time their deployment of virulence factors, it has been proposed that QS activities of neighbouring bacteria in the gut microbiome could interfere with this timing, either by QSI or by prematurely inducing the quorum level with species-agnostic QSM (Cho et al., 2021). Unsurprisingly, many instances of QSI activity can be observed in the crosstalk between host and microbe and in communications within bacterial communities.
QSI enzymes in host tissue
Paraoxonase (PON) enzymes are expressed widely across human tissues, including the intestines and can inactivate AHL-QSM through hydrolysis of the lactone ring moiety (Camps et al., 2011; Peyrottes et al., 2020). Like many QQ enzymes, PON have a broad substrate specificity and can also inactivate esters, e.g. oestrogen esters, lactones or organophosphates, highlighting the need to consider off-target effects in evaluation of QS-based therapies.
QSI produced in microbe-microbe interactions in the human gut
Microbe-microbe interactions in the human gut can involve cooperation, through sharing of resources and cross-feeding, as well as antagonism, in efforts to outcompete neighbouring cells. Just as in food microbiomes, QS appears to be widespread amongst bacteria in human gut microbiomes and likely, to some degree, mediates interspecies interactions therein. Indeed, AI-2 was shown to orchestrate both cross-feeding and also QSI in co-culture of C. acetobutylicum and D. vulgaris Hildenborough, depending on the extent of nutrient limitation in the system (Ranava et al., 2021), highlighting the conditionality which can be present in bacterial cooperation.
Overall, there are relatively fewer instances of QQ enzymes or QSI compounds being reported from the gut microbiome, possibly due to the focus of bioprospecting efforts on natural environments, as well as the difficulty in successfully culturing gut commensals in vitro. However, QSI potential has been established by some studies, for example, in the oral microbiome 60% of recovered bacteria in dental plaque and saliva exhibited AHL-degradation capacity and treatment of mock communities with the broad-spectrum AHL-lactonase Aii20J reduced biofilm formation and altered the overall community composition (Muras et al., 2020). Penicillin V acylases are microbially produced enzymes within the choloylglycine hydrolase family able to inactivate AHL-QSM due to structural similarity with the functional residues of native AHL acylases (Liu et al., 2012; Mukherji and Prabhune, 2015; Daly et al., 2021). To date, however, there are scarce reports of their presence in human gut microbiomes wherein bile salt hydrolases predominate. This highlights the importance of substrate specificity and receptor promiscuity when potentially targeting QS in complex microbiomes such as those of foods and the gut.
In addition to enzymatic QQ, there are several reports of QSM produced by one species acting as analogues for other species, potentially mediating microbe-microbe interactions. Whilst, to our knowledge, no direct examples exist pertaining to the human gut microbiome, there are examples from the human skin and anaerobic waste digestion microbiomes. AIP produced by skin commensals such as coagulase-negative staphylococci including Staphylococcus hominis can inhibit the agr system of S. aureus, reducing the expression of phenol-soluble modulin α (PSMα) which undermines skin barrier integrity (Williams et al., 2019).
Whilst, to our knowledge, no clinical trials have been conducted to date in humans, engineered probiotics are being evaluated to harness QS to promote human health and wellbeing. An E. coli Nissle 1917 strain designed to respond to the AHL-QSM 3OC12HSL was shown to eliminate and prevent P. aeruginosa gastrointestinal infections in animal models (Hwang et al., 2017). In another study using a rational design approach, expression of small peptides resembling LuxS active sites reduced AI-2 activity of aquaculture pathogens Edwardsiella tarda, A. hydrophila and V. harveyi (Sun and Zhang, 2016). Given the ubiquity of AI-2 signalling, this approach clearly offers broad cross-disciplinary applicability.
Another approach involves direct oral administration of purified QQ enzymes such as AHL-lactonase, which resisted digestion and attenuated virulence of A. hydrophila in a zebrafish model (Cao et al., 2012). Studies examining feasibility of QSI therapies as applied to human gut microbiomes must take digestive processes into account to ensure delivery to the site of action, as well as evaluating the impact on the structure and composition microbiome. Ex vivo modelling of the human gut offers great promise in modelling the effect of QQ enzyme treatment on the functional and taxonomical composition of the gut microbiome.
Conclusion
The field of quorum sensing research has seen a shift from fundamental to translational science in the past two decades. Nevertheless, despite a preponderance of newly discovered and/or bioengineered quorum sensing inhibitors, several important gaps remain to be addressed so that the therapeutic targeting of quorum sensing systems for the modulation of food and human gut microbiomes may expand as a field.
Firstly, analytical techniques must continue to be developed to augment our capacity to monitor quorum sensing activity and efficacy of quorum sensing inhibitory interventions, particularly for AIP-QS systems and QS within complex matrices such as foods and luminal samples from the human gut. Continued innovation with respect to bioinformatic approaches in particular can further potentiate our understanding of the significance of QS within microbiomes generally, not restricted to those discussed in this review. In this sense, an important consideration in future study design will be the disambiguation of QS gene presence/absence and QS functionality in food and gut microbiomes. Whilst biofilms pose challenges with respect to modelling in a meaningful manner, their ubiquity and potential to act as reservoirs for AMR genetic determinants, food-borne pathogens and specific spoilage organisms are likely to drive efforts to better understand, manage and harness biofilms in foods and in the human gut.
Secondly, many interventions such as specific strains, probiotics, prebiotics and phage, are being investigated with a view to the modulation of complex microbiomes such as those of foods and the human gut. As described earlier, quorum sensing-regulated biofilm production has been shown to improve the survival of probiotic strains during food processing, food fermentations and transit through the human gut. We propose that future studies in this field may consider quorum sensing activity in specific strains as a probiotic trait, where supported by evidence and subject to requisite evaluation. Similarly, strains with QS inhibitory activity could be investigated for their potential clinical or biotechnological applications.
Thirdly, our ability to address AMR can be augmented by better understanding the role of QS in disseminating AMR genes, as well as evaluating QSI agents with a view to the development of anti-virulence therapies and combinatorial agents to enhance the efficacy of existing antibiotics. Here, harnessing systems biology and synthetic ecology approaches will be critical to ensure only the most effective and promising hits are brought through to the preclinical stage.
In conclusion, over the span of a little over 50 years, the field of quorum sensing research has been founded and come to be regarded to be of key importance with respect to microbial physiology in the context of social interactions. Whilst food and human gut microbiomes have received relatively less attention, the accumulated literature points to the presence of quorum sensing molecules and regulated behaviours in these contexts. Further, the abundance and diversity of host receptors to survey bacterial QSM, in addition to the numerous examples of QSI systems across kingdoms of life and environmental niches, has likely arisen due to the ubiquity and evolutionary importance of QS within microbiome science. Addressing the knowledge gaps identified above will help in the critical evaluation of proposed QS and QSI interventions in these contexts, with the potential to positively impact human, animal and ecosystem health.
Author contributions
AF, AA-O, CG, and PC conceived the manuscript. KF wrote the manuscript. AA-O, CG, AF, and PC contributed to the final preparation. All authors contributed to the article and approved the submitted version.
Funding
This research was conducted with the financial support of Science Foundation Ireland (SFI) under Grant Number SFI/12/RC/2273 P2.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors acknowledge Gloria Spampinato for her contribution towards the manuscript, as well as the constructive feedback received from reviewers during peer review.
Glossary
| agr | Accessory gene regulator system |
| AHL | Acylated homoserine lactone |
| AhR | Aryl hydrocarbon receptor |
| AI | Autoinducer |
| AIP | Autoinducer peptide |
| AMC | Activated methyl cycle |
| AMR | Antimicrobial resistance |
| ATP | Adenosine triphosphate |
| CAI-1 | Cholera autoinducer |
| cAMP | Cyclic adenosine monophosphate |
| c-di-GMP | Cyclic diguanylate (c-di-GMP) |
| CRC | Colorectal cancer |
| DKP | Diketopiperazines |
| DPD | 4,5-dihydroxy-2,3-pentanedione |
| DPO | 3,5-dimethylpyrazin-2-one |
| DSF | Diffusible signal factor |
| EHEC | Enterohemorrhagic Escherichia coli |
| EPEC | Enteropathogenic E. coli (EPEC) |
| EPS | Exopolysaccharide |
| HGT | Horizontal gene transfer |
| HPLC | High-performance liquid chromatography |
| HSL | Homoserine lactone |
| IBD | Inflammatory bowel disease |
| LAB | Lactic acid bacteria |
| LEE | Locus of enterocyte effacement |
| miRNAs | micro RNAs |
| MS | Mass spectrometry |
| NEC | Necrotising enterocolitis |
| OMV | Outer membrane vesicles |
| Quorum quenching | |
| Qrr sRNAs | Quorum regulatory sRNAs |
| QS | Quorum sensing |
| QSM | Quorum sensing molecules |
| R-THMF | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydro-furan) |
| SAH | S-adenosylhomocysteine |
| SAM | S-adenosyl-L-methionine |
| siRNAs | Small Inhibitory RNAs |
| SRH | S-ribosyl-homocysteine |
| SSO | Specific spoilage organisms |
| S-THMF | (2S,4S)-2-methyl-2,3,3,4-tetra-hydroxytetrahydrofuran |
| TCS | Two-component systems |
References
- Abdel-Aziz M. M., Emam T. M., Raafat M. M. (2020). Hindering of cariogenic biofilm by fatty acid Array derived from an endophytic strain. Biomol. Ther. 10:811. doi: 10.3390/biom10050811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe K., Nomura N., Suzuki S. (2020). Biofilms: hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. FEMS Microbiol. Ecol. 96. doi: 10.1093/femsec/fiaa031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aframian N., Eldar A. (2020). A bacterial tower of babel: quorum-sensing signaling diversity and its evolution. Annu. Rev. Microbiol. 74, 587–606. doi: 10.1146/annurev-micro-012220-063740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguanno D., Coquant G., Postal B. G., Osinski C., Wieckowski M., Stockholm D., et al. (2020). The intestinal quorum sensing 3-oxo-C12:2 acyl homoserine lactone limits cytokine-induced tight junction disruption. Tissure Barr. 8:1832877. doi: 10.1080/21688370.2020.1832877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa Oniciuc E. A., Walsh C. J., Coughlan L. M., Awad A., Simon C. A., Ruiz L., Crispie F.. (2020) ‘Dairy Products and Dairy-Processing Environments as a Reservoir of Antibiotic Resistance and Quorum-Quenching Determinants as Revealed through Functional Metagenomics mSystems, 5, e00723–19. doi: 10.1128/mSystems.00723-19, PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R. C., McNally L., Popat R., Brown S. P. (2016). Quorum sensing protects bacterial co-operation from exploitation by cheats. ISME J. 10, 1706–1716. doi: 10.1038/ismej.2015.232, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasoud A., Hettiarachchy N., Rayaprolu S., Babu D., Kwon Y. M., Mauromoustakos A.. (2016). Inhibitory effects of lactic and malic organic acids on autoinducer type 2 (AI-2) quorum sensing of Escherichia coli O157:H7 and salmonella typhimurium. Science 66, 560–564. doi: 10.1016/j.lwt.2015.11.013 [DOI] [Google Scholar]
- Alvarez C.-S., Badia J., Bosch M., Giménez R., Baldomà L. (2016). Outer membrane vesicles and soluble factors released by probiotic Nissle 1917 and commensal ECOR63 enhance barrier function by regulating expression of tight junction proteins in intestinal epithelial cells. Front. Microbiol. 7:1981. doi: 10.3389/fmicb.2016.01981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Ordóñez A., Coughlan L. M., Briandet R., Cotter P. D. (2019). Biofilms in food processing environments: challenges and opportunities. Annu. Rev. Food Sci. Technol. 10, 173–195. doi: 10.1146/annurev-food-032818-121805, PMID: [DOI] [PubMed] [Google Scholar]
- Aman M., Aneeqha N., Bristi K., Deeksha J., Afza N., Sindhuja V., et al. (2021). Lactic acid bacteria inhibits quorum sensing and biofilm formation of Pseudomonas aeruginosa strain JUPG01 isolated from rancid butter. Biocatal. Agric. Biotechnol. 36. doi: 10.1016/j.bcab.2021.102115 [DOI] [Google Scholar]
- Amrutha B., Sundar K., Shetty P. H. (2017). Study on and biofilms from fresh fruits and vegetables. J. Food Sci. Technol. 54, 1091–1097. doi: 10.1007/s13197-017-2555-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtiar E. W., Bachtiar B. M., Jarosz L. M., Amir L. R., Sunarto H., Ganin H., et al. (2014). AI-2 of Aggregatibacter actinomycetemcomitans inhibits Candida albicans biofilm formation. Front. Cell. Infect. Microbiol. 4:94. doi: 10.3389/fcimb.2014.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari S. M. N., Roky M. K., Mohiuddin M., Kamruzzaman M., Mekalanos J. J., Faruque S. M.. (2013). Quorum-sensing autoinducers resuscitate dormant vibrio cholerae in environmental water samples. Proc. Natl. Acad. Sci. U. S. A. 110, 9926–9931. doi: 10.1073/pnas.1307697110, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso A., Mahler J. V., Fonseca-Castro P. H., Quintana F. J. (2021). The aryl hydrocarbon receptor and the gut-brain axis. Cell. Mol. Immunol. 18, 259–268. doi: 10.1038/s41423-020-00585-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens M., Meyerhof W. (2011). Gustatory and extragustatory functions of mammalian taste receptors. Physiol. Behav. 105, 4–13. doi: 10.1016/j.physbeh.2011.02.010, PMID: [DOI] [PubMed] [Google Scholar]
- Benítez-Cabello A., Calero-Delgado B., Rodríguez-Gómez F., Bautista-Gallego J., Garrido-Fernández A., et al. (2020). The use of multifunctional yeast-lactobacilli starter cultures improves fermentation performance of Spanish-style green table olives. Food Microbiol. 91:103497. doi: 10.1016/j.fm.2020.103497 [DOI] [PubMed] [Google Scholar]
- Bez C., Covaceuszach S., Bertani I., Choudhary K. S., Venturi V. (2021). LuxR Solos from Environmental Fluorescent Pseudomonads. mSphere 6, e01322–e01320. doi: 10.1128/mSphere.01322-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutia S. K, Maiti T. K. (2008). Targeting tumors with peptides from natural sources. Trends Biotechnol 26, 210–217. [DOI] [PubMed] [Google Scholar]
- Bijtenhoorn P., Mayerhofer H., Müller-Dieckmann J., Utpatel C., Schipper C., Hornung C., et al. (2011). A novel metagenomic short-chain dehydrogenase/reductase attenuates Pseudomonas aeruginosa biofilm formation and virulence on Caenorhabditis elegans. PLoS One 6:e26278. doi: 10.1371/journal.pone.0026278, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billot R., Plener L., Jacquet P., Elias M., Chabrière E., Daudé D.. (2020). Engineering acyl-homoserine lactone-interfering enzymes toward bacterial control. J. Biol. Chem. 295, 12993–13007. doi: 10.1074/jbc.REV120.013531, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blana V. A., Doulgeraki A. I., Nychas G.-J. E. (2011). Autoinducer-2-like activity in lactic acid bacteria isolated from minced beef packaged under modified atmospheres. J. Food Prot. 74, 631–635. doi: 10.4315/0362-028X.JFP-10-276, PMID: [DOI] [PubMed] [Google Scholar]
- Bofinger M. R., de Sousa L. S., Fontes J. E. N., Marsaioli A. J. (2017). Diketopiperazines as cross-communication - quorum- sensing signals between Cronobacter sakazakii and Bacillus cereus. ACS Omega 2, 1003–1008. doi: 10.1021/acsomega.6b00513, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann A. O., Brameyer S., Kresovic D., Hitkova I., Kopp Y., Manske C., et al. (2013, 2013). Pyrones as bacterial signaling molecules. Nat. Chem. Biol. 9, 573–578. doi: 10.1038/nchembio.1295 [DOI] [PubMed] [Google Scholar]
- Bradshaw C. S., Sobel J. D. (2016). Current treatment of bacterial vaginosis-limitations and need for innovation. J. Infect. Dis. 214, S14–S20. doi: 10.1093/infdis/jiw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brameyer S., Kresovic D., Bode H. B., Heermann R. (2015). Dialkylresorcinols as bacterial signaling molecules. Proc. Natl. Acad. Sci. U. S. A. 112, 572–577. doi: 10.1073/pnas.1417685112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges A. A., Bassler B. L. (2021). Inverse regulation of biofilm dispersal by polyamine signals. elife 10:e65487. doi: 10.7554/eLife.65487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronesky D., Wu Z., Marzi S., Walter P., Geissmann T., Moreau K., et al. (2016). Staphylococcus aureus RNAIII and its regulon link quorum sensing, stress responses, metabolic adaptation, and regulation of virulence gene expression. Annu. Rev. Microbiol. 70, 299–316. doi: 10.1146/annurev-micro-102215-095708, PMID: [DOI] [PubMed] [Google Scholar]
- Buffie C. G., Pamer E. G. (2013). Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801. doi: 10.1038/nri3535, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagna S. R., Gooding J. R., May A. L. (2009). Direct quantitation of the quorum sensing signal, autoinducer-2, in clinically relevant samples by liquid chromatography-tandem mass spectrometry. Anal. Chem. 81, 6374–6381. doi: 10.1021/ac900824j, PMID: [DOI] [PubMed] [Google Scholar]
- Camps J., Pujol I., Ballester F., Joven J., Simó J. M. (2011). Paraoxonases as potential antibiofilm agents: their relationship with quorum-sensing signals in gram-negative bacteria. Antimicrob. Agents Chemother. 55, 1325–1331. doi: 10.1128/AAC.01502-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., He S., Zhou Z., Zhang M., Mao W., Zhang H., et al. (2012). Orally administered thermostable N-acyl homoserine lactonase from bacillus sp. strain AI96 attenuates Aeromonas hydrophila infection in zebrafish. Appl. Environ. Microbiol. 78, 1899–1908. doi: 10.1128/AEM.06139-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J. Y., Liu R., Macbeth J. C., Hsiao A. (2021). The Interface of and the gut microbiome. Gut Microbes 13:1937015. doi: 10.1080/19490976.2021.1937015, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiaen S. E. A., O’Connell Motherway M., Bottacini F., Lanigan N., Casey P. G., Huys G., et al. (2014). Autoinducer-2 plays a crucial role in gut colonization and probiotic functionality of Bifidobacterium breve UCC2003. PLoS One 9:e98111. doi: 10.1371/journal.pone.0098111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W., Zhou S., Zhu W., Zhuang X. (2014). Quorum quenching bacteria bacillus sp. QSI-1 protect zebrafish (Danio rerio) from Aeromonas hydrophila infection. Sci. Rep. 4:5446. doi: 10.1038/srep05446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M. B., Hughes D. T., Zhu C., Boedeker E. C., Sperandio V. (2006). The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U. S. A. 103, 10420–10425. doi: 10.1073/pnas.0604343103, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. J., Hall C. L., Schniederberend M., Farrow III J. M., Goodson J. R., Pesci E. C., et al. (2018). Host suppression of quorum sensing during catheter-associated urinary tract infections. Nat. Commun. 9:4436. doi: 10.1038/s41467-018-06882-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin Slaughter J. (1999). The naturally occurring furanones: formation and function from pheromone to food. Biol. Rev. Camb. Philos. Soc. 74, 259–276. doi: 10.1017/S0006323199005332, PMID: [DOI] [PubMed] [Google Scholar]
- Cook L. C., Federle M. J. (2014). Peptide pheromone signaling in streptococcus and enterococcus. FEMS Microbiol. Rev. 38, 473–492. doi: 10.1111/1574-6976.12046, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornforth D. M., Popat R., McNally L., Gurney J., Scott-Phillips T. C., Ivens A., et al. (2014). Combinatorial quorum sensing allows bacteria to resolve their social and physical environment. Proc. Natl. Acad. Sci. U. S. A. 111, 4280–4284. doi: 10.1073/pnas.1319175111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P. D., Hill C., Ross R. P. (2005). Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3, 777–788. doi: 10.1038/nrmicro1273, PMID: [DOI] [PubMed] [Google Scholar]
- Cutignano A. (2019). Analytical approaches for the identification of quorum sensing molecules. in Quorum Sensing. ed. G. Tommaro (London: Academic Press), 29–53. [Google Scholar]
- Daly J. W., Keely S. J., Gahan C. G. M. (2021). Functional and phylogenetic diversity of BSH and PVA enzymes. Microorganisms 9:732. doi: 10.3390/microorganisms9040732, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ðapa T., Leuzzi R., Ng Y. K., Baban S. T., Adamo R., Kuehne S. A., et al. (2013). Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J. Bacteriol. 195, 545–555. doi: 10.1128/JB.01980-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darkoh C., DuPont H., Norris S. J., Kaplan H. B. (2015). Toxin synthesis by Clostridium difficile is regulated through quorum signaling. MBio 6:e02569. doi: 10.1128/mBio.02569-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida O. G. G., Vitulo N., de Martinis E. C. P., Felis G. E. (2021). Pangenome analyses of LuxS-coding genes and enzymatic repertoires in cocoa-related lactic acid bacteria. Genomics 113, 1659–1670. doi: 10.1016/j.ygeno.2021.04.010, PMID: [DOI] [PubMed] [Google Scholar]
- De Filippis F., Parente E., Ercolini D. (2018). Recent past, present, and future of the food microbiome. Annu. Rev. Food Sci. Technol. 9, 589–608. doi: 10.1146/annurev-food-030117-012312, PMID: [DOI] [PubMed] [Google Scholar]
- Defoirdt T. (2018). Quorum-sensing systems as targets for Antivirulence therapy. Trends Microbiol. 26, 313–328. doi: 10.1016/j.tim.2017.10.005, PMID: [DOI] [PubMed] [Google Scholar]
- Defoirdt T., Crab R., Wood T. K., Sorgeloos P., Verstraete W., Bossier P.. (2006). Quorum sensing-disrupting brominated furanones protect the gnotobiotic brine shrimp Artemia franciscana from pathogenic Vibrio harveyi, Vibrio campbellii, and Vibrio parahaemolyticus isolates. Appl. Environ. Microbiol. 72, 6419–6423. doi: 10.1128/AEM.00753-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delalande L. (2005). N-hexanoyl-L-homoserine lactone, a mediator of bacterial quorum-sensing regulation, exhibits plant-dependent stability and may be inactivated by germinating Lotus corniculatus seedlings. FEMS Microbiol. Ecol. 52, 13–20. doi: 10.1016/j.femsec.2004.10.005, PMID: [DOI] [PubMed] [Google Scholar]
- Deng Z., Luo X. M., Liu J., Wang H. (2020). Quorum sensing, biofilm, and intestinal mucosal barrier: involvement the role of probiotic. Front. Cell. Infect. Microbiol. 10:538077. doi: 10.3389/fcimb.2020.538077, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai S. K., Kenney L. J. (2019). Switching lifestyles is an adaptive strategy of bacterial pathogens. Front. Cell. Infect. Microbiol. 9:421. doi: 10.3389/fcimb.2019.00421, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh C. V., Prather K. L. J. (2019). Development of an autonomous and bifunctional quorum-sensing circuit for metabolic flux control in engineered Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 116, 25562–25568. doi: 10.1073/pnas.1911144116, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A., Cotter P. D., Ross R. P., Hill C. (2012). Bacteriocin production: a probiotic trait? Appl. Environ. Microbiol. 78, 1–6. doi: 10.1128/AEM.05576-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson G. P., Lee S. M., Mazmanian S. K. (2016). Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32. doi: 10.1038/nrmicro3552, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W., Cai Y., Xu Z., Fu B., Chen Q., Cui Y., et al. (2020). Heterologous expression of AHL lactonase AiiK by lactobacillus casei MCJΔ1 with great quorum quenching ability against Aeromonas hydrophila AH-1 and AH-4. Microb. Cell Factories 19:191. doi: 10.1186/s12934-020-01448-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W., Zhu J., Guo X., Kong D., Zhang Q., Zhou Y., et al. (2018). Characterization of AiiK, an AHL lactonase, from Kurthia huakui LAM0618 and its application in quorum quenching on Pseudomonas aeruginosa PAO1. Sci. Rep. 8:6013. doi: 10.1038/s41598-018-24507-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K., Carey-Ewend K., Vaishnava S. (2021). Spatial analysis of gut microbiome reveals a distinct ecological niche associated with the mucus layer. Gut Microbes 13:1874815. doi: 10.1080/19490976.2021.1874815, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff M. J., Bassler B. L. (2018). SnapShot: bacterial quorum sensing. Cells 174, 1328–1328.e1. doi: 10.1016/j.cell.2018.08.003, PMID: [DOI] [PubMed] [Google Scholar]
- Ellermann M., Jimenez A. G., Pifer R., Ruiz N., Sperandio V. (2021). The canonical long-chain fatty acid sensing machinery processes arachidonic acid to inhibit virulence in Enterohemorrhagic Escherichia coli. MBio 12, e03247–e03220. doi: 10.1128/mBio.03247-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mowafy S. A., Shaaban M. I., Abd El Galil K. H. (2014). Sodium ascorbate as a quorum sensing inhibitor of Pseudomonas aeruginosa. J. Appl. Microbiol. 117, 1388–1399. doi: 10.1111/jam.12631, PMID: [DOI] [PubMed] [Google Scholar]
- Fan X., Liang M., Wang L., Chen R., Li H., Liu X.. (2017). Aii810, a novel cold-adapted -Acylhomoserine lactonase discovered in a metagenome, can strongly attenuate virulence factors and biofilm formation. Front. Microbiol. 8:1950. doi: 10.3389/fmicb.2017.01950, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fels L., Jakob F., Vogel R. F., Wefers D. (2018). Structural characterization of the exopolysaccharides from water kefir. Carbohydr. Polym. 189, 296–303. doi: 10.1016/j.carbpol.2018.02.037, PMID: [DOI] [PubMed] [Google Scholar]
- Field D., et al. (2019). Bioengineering nisin to overcome the nisin resistance protein. Mol. Microbiol. 111, 717–731. [DOI] [PubMed] [Google Scholar]
- Fetzner S. (2015). Quorum quenching enzymes. J. Biotechnol. 201, 2–14. doi: 10.1016/j.jbiotec.2014.09.001 [DOI] [PubMed] [Google Scholar]
- Flavier A. B., Clough S. J., Schell M. A., Denny T. P. (1997). Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 26, 251–259. doi: 10.1046/j.1365-2958.1997.5661945.x, PMID: [DOI] [PubMed] [Google Scholar]
- Flemming H.-C., Wingender J., Szewzyk U., Steinberg P., Rice S. A., Kjelleberg S.. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94, PMID: [DOI] [PubMed] [Google Scholar]
- Fu H., Elena R. C., Marquez P. H. (2019). The roles of small RNAs: insights from bacterial quorum sensing. ExRNA 1:32. doi: 10.1186/s41544-019-0027-8 [DOI] [Google Scholar]
- Fu C.-Y., Li L. Q., Yang T., She X., Ai Q., Wang Z. L.. (2020). Autoinducer-2 May be a new biomarker for monitoring neonatal necrotizing enterocolitis. Front. Cell. Infect. Microbiol. 10:140. doi: 10.3389/fcimb.2020.00140, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C., Parsek M. R., Greenberg E. P. (2001). Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35, 439–468. doi: 10.1146/annurev.genet.35.102401.090913 [DOI] [PubMed] [Google Scholar]
- Gahan C. G., Patel S. J., Boursier M. E., Nyffeler K. E., Jennings J., Abbott N. L., et al. (2020). Bacterial quorum sensing signals self-assemble in aqueous media to form micelles and vesicles: an integrated experimental and molecular dynamics study. J. Phys. Chem. B 124, 3616–3628. doi: 10.1021/acs.jpcb.0c00496, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaida M. M., Dapunt U., Hänsch G. M. (2016). Sensing developing biofilms: the bitter receptor T2R38 on myeloid cells. Patho. Dis. 74:ftw004. doi: 10.1093/femspd/ftw004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gutierrez E., Cotter P. D. (2021). Relevance of organ(s)-on-a-chip systems to the investigation of food-gut microbiota-host interactions. Crit. Rev. Microbiol. 48, 463–488. doi: 10.1080/1040841X.2021.1979933 [DOI] [PubMed] [Google Scholar]
- Gharsallaoui A., Oulahal N., Joly C., Degraeve P. (2016). Nisin as a food preservative: part 1: physicochemical properties, antimicrobial activity, and Main uses. Crit. Rev. Food Sci. Nutr. 56, 1262–1274. doi: 10.1080/10408398.2013.763765, PMID: [DOI] [PubMed] [Google Scholar]
- Giaouris E., Heir E., Desvaux M., Hébraud M., Møretrø T., Langsrud S., et al. (2015). Intra- and inter-species interactions within biofilms of important foodborne bacterial pathogens. Front. Microbiol. 6:841. doi: 10.3389/fmicb.2015.00841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbetti M., de Angelis M., di Cagno R., Minervini F., Limitone A. (2007). Cell-cell communication in food related bacteria. Int. J. Food Microbiol. 120, 34–45. doi: 10.1016/j.ijfoodmicro.2007.06.012, PMID: [DOI] [PubMed] [Google Scholar]
- Gorelik O., Levy N., Shaulov L., Yegodayev K., Meijler M. M., Sal-Man N.. (2019). Vibrio cholerae autoinducer-1 enhances the virulence of enteropathogenic Escherichia coli. Sci. Rep. 9:4122. doi: 10.1038/s41598-019-40859-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori K., Moslehi-Jenabian S., Purrotti M., Jespersen L. (2011). Autoinducer-2 activity produced by bacteria found in smear of surface ripened cheeses. Int. Dairy J. 21, 48–53. doi: 10.1016/j.idairyj.2010.06.009 [DOI] [Google Scholar]
- Gui M., Liu L., Wu R., Hu J., Wang S., Li P.. (2018). Detection of new quorum sensing n-acyl homoserine lactones from aeromonas veronii. Front. Microbiol. 9:1712. doi: 10.3389/fmicb.2018.01712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M., Zhang Y., Gao L., Li P. (2021). Effect of AHL-lactonase and nisin on microbiological, chemical and sensory quality of vacuum packaged sturgeon storage at 4°C. Int. J. Food Prop. 24, 222–232. doi: 10.1080/10942912.2021.1872621 [DOI] [Google Scholar]
- Gutiérrez-Barranquero J. A., Reen F. J., McCarthy R. R., O’Gara F. (2018). Deciphering the role of coumarin as a novel quorum sensing inhibitor suppressing virulence phenotypes in bacterial pathogens. Appl. Microbiol. Biotechnol. 99, 3303–16. doi: 10.1007/s00253-015-6436-1 [DOI] [PubMed] [Google Scholar]
- Guzel-Seydim Z. B., Gökırmaklı Ç., Greene A. K. (2021). A comparison of milk kefir and water kefir: physical, chemical, microbiological and functional properties. Trends Food Sci. Technol. 113, 42–53. doi: 10.1016/j.tifs.2021.04.041 [DOI] [Google Scholar]
- Han X., Zhang L. J., Wu H. Y., Wu Y. F., Zhao S. N. (2018). Investigation of microorganisms involved in kefir biofilm formation. Antonie Van Leeuwenhoek 111, 2361–2370. doi: 10.1007/s10482-018-1125-6, PMID: [DOI] [PubMed] [Google Scholar]
- Hargreaves K. R., Kropinski A. M., Clokie M. R. J. (2014). What does the talking?: quorum sensing signalling genes discovered in a bacteriophage genome. PLoS One 9:e85131. doi: 10.1371/journal.pone.0085131, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawver L. A., Jung S. A., Ng W.-L. (2016). Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol. Rev. 40, 738–752. doi: 10.1093/femsre/fuw014, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann S., Krishna S., Kerr B. (2015). Why do bacteria regulate public goods by quorum sensing?-how the shapes of cost and benefit functions determine the form of optimal regulation. Front. Microbiol. 6:767. doi: 10.3389/fmicb.2015.00767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R., Häussler S., Pruteanu M., Stülke J., Tschowri N., Turgay K.. (2019). Recent advances and current trends in nucleotide second messenger signaling in bacteria. J. Mol. Biol. 431, 908–927. doi: 10.1016/j.jmb.2019.01.014, PMID: [DOI] [PubMed] [Google Scholar]
- Hense B. A., Kuttler C., Müller J., Rothballer M., Hartmann A., Kreft J. U.. (2007). Does efficiency sensing unify diffusion and quorum sensing? Nat. Rev. Microbiol. 5, 230–239. doi: 10.1038/nrmicro1600, PMID: [DOI] [PubMed] [Google Scholar]
- Hickey A., Pardo L. M., Reen F. J., McGlacken G. P. (2021). Pyrones identified as LuxR signal molecules in Photorhabdus and their synthetic analogues can Alter multicellular phenotypic behavior of bacillus atropheaus. ACS Omega 6, 33141–33148. doi: 10.1021/acsomega.1c05508, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H. M., Jiang F., Zhang G. L., Wang J. Y., Zhu Y. H., Liu X. Y.. (2017). Inhibition of hafnia alvei H4 biofilm formation by the food additive Dihydrocoumarin. J. Food Prot. 80, 842–847. doi: 10.4315/0362-028X.JFP-16-460, PMID: [DOI] [PubMed] [Google Scholar]
- Hu X., Wang Y., Gao L., Jiang W., Lin W., Niu C., et al. (2018). The impairment of methyl metabolism from luxS mutation of Streptococcus mutans. Front. Microbiol. 9:404. doi: 10.3389/fmicb.2018.00404, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D. T., Sperandio V. (2008). Inter-kingdom signalling: communication between bacteria and their hosts. Nat. Rev. Microbiol. 6, 111–120. doi: 10.1038/nrmicro1836, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huis in ‘t Veld J. H. (1996). Microbial and biochemical spoilage of foods: an overview. Int. J. Food Microbiol. 33, 1–18. doi: 10.1016/0168-1605(96)01139-7, PMID: [DOI] [PubMed] [Google Scholar]
- Hurley A., Bassler B. L. (2017). Asymmetric regulation of quorum-sensing receptors drives autoinducer-specific gene expression programs in vibrio cholerae. PLoS Genet. 13:e1006826. doi: 10.1371/journal.pgen.1006826, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I. Y., Koh E., Wong A., March J. C., Bentley W. E., Lee Y. S., et al. (2017). Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat. Commun. 8:15028. doi: 10.1038/ncomms15028, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail A. S., Valastyan J. S., Bassler B. L. (2016). A host-produced Autoinducer-2 mimic activates bacterial quorum sensing. Cell Host Microbe 19, 470–480. doi: 10.1016/j.chom.2016.02.020, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeesan B., Gerner-Smidt P., Allard M. W., Leuillet S., Winkler A., Xiao Y., et al. (2019). The use of next generation sequencing for improving food safety: translation into practice. Food Microbiol. 79, 96–115. doi: 10.1016/j.fm.2018.11.005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens Y., Nielandt J., Bronselaer A., Debunne N., Verbeke F., Wynendaele E., et al. (2018). Disbiome database: linking the microbiome to disease. BMC Microbiol. 18:50. doi: 10.1186/s12866-018-1197-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens J. C. A., Steenackers H., Robijns S., Gellens E., Levin J., Zhao H., et al. (2008). Brominated furanones inhibit biofilm formation by salmonella enterica serovar typhimurium. Appl. Environ. Microbiol. 74, 6639–6648. doi: 10.1128/AEM.01262-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis K. G., Daquigan N., White J. R., Morin P. M., Howard L. M., Manetas J. E., et al. (2018). Microbiomes associated with foods from plant and animal sources. Front. Microbiol. 9:2540. doi: 10.3389/fmicb.2018.02540, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jingjing E., Rongze M., Zichao C., Caiqing Y., Ruixue W., Qiaoling Z., et al. (2021). Improving the freeze-drying survival rate of lactobacillus plantarum LIP-1 by increasing biofilm formation based on adjusting the composition of buffer salts in medium. Food Chem. 338:128134. doi: 10.1016/j.foodchem.2020.128134, PMID: [DOI] [PubMed] [Google Scholar]
- Johansen P., Jespersen L. (2017). Impact of quorum sensing on the quality of fermented foods. Curr. Opin. Food Sci. 13, 16–25. doi: 10.1016/j.cofs.2017.01.001 [DOI] [Google Scholar]
- Jones R. B., Zhu X., Moan E., Murff H. J., Ness R. M., Seidner D. L., et al. (2018). Inter-niche and inter-individual variation in gut microbial community assessment using stool, rectal swab, and mucosal samples. Sci. Rep. 8:4139. doi: 10.1038/s41598-018-22408-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi J. R., Khazanov N., Charkowski A., Faigenboim A., Senderowitz H., Yedidia I.. (2021). Interkingdom signaling interference: the effect of plant-derived small molecules on quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 59, 153–190. doi: 10.1146/annurev-phyto-020620-095740, PMID: [DOI] [PubMed] [Google Scholar]
- Kalia V. C. (2013). Quorum sensing inhibitors: an overview. Biotechnol. Adv. 31, 224–245. doi: 10.1016/j.biotechadv.2012.10.004 [DOI] [PubMed] [Google Scholar]
- Kalkum M., Lyon G. J., Chait B. T. (2003). Detection of secreted peptides by using hypothesis-driven multistage mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 100, 2795–2800. doi: 10.1073/pnas.0436605100, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaraju K., Smith J., Wang J., Roy V., Sintim H. O., Bentley W. E., et al. (2011). Effects on membrane lateral pressure suggest permeation mechanisms for bacterial quorum signaling molecules. Biochemistry 50, 6983–6993. doi: 10.1021/bi200684z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Kim H., Goo E., Jeong H., An J. H., Hwang I.. (2019). Unraveling the role of quorum sensing-dependent metabolic homeostasis of the activated methyl cycle in a cooperative population of Burkholderia glumae. Sci. Rep. 9:11038. doi: 10.1038/s41598-019-47460-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareb O., Aïder M. (2020). Quorum sensing circuits in the communicating mechanisms of bacteria and its implication in the biosynthesis of Bacteriocins by lactic acid bacteria: a review. Probiot. Antimicro. Proteins 12, 5–17. doi: 10.1007/s12602-019-09555-4, PMID: [DOI] [PubMed] [Google Scholar]
- Karlsson T., Turkina M. V., Yakymenko O., Magnusson K. E., Vikström E. (2012). The Pseudomonas aeruginosa N-acylhomoserine lactone quorum sensing molecules target IQGAP1 and modulate epithelial cell migration. PLoS Pathog. 8:e1002953. doi: 10.1371/journal.ppat.1002953, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S. M., Lanigan N., O’Neill I. J., Bottacini F., Lugli G. A., Viappiani A., et al. (2020). Bifidobacterial biofilm formation is a multifactorial adaptive phenomenon in response to bile exposure. Sci. Rep. 10:11598. doi: 10.1038/s41598-020-68179-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall M. M., Sperandio V. (2016). What a dinner party! Mechanisms and functions of Interkingdom signaling in host-pathogen associations. MBio 7:e01748. doi: 10.1128/mBio.01748-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I., Tango C. N., Miskeen S., Lee B. H., Oh D. H. (2017). Hurdle technology: a novel approach for enhanced food quality and safety – a review. Food Control 73, 1426–1444. doi: 10.1016/j.foodcont.2016.11.010 [DOI] [Google Scholar]
- Kim H.-S., Cha E., Ham S. Y., Park J. H., Nam S. J., Kwon H., et al. (2021). Linoleic acid inhibits Pseudomonas aeruginosa biofilm formation by activating diffusible signal factor-mediated quorum sensing. Biotechnol. Bioeng. 118, 82–93. doi: 10.1002/bit.27552, PMID: [DOI] [PubMed] [Google Scholar]
- Kim C. S., Gatsios A., Cuesta S., Lam Y. C., Wei Z., Chen H., et al. (2020). Characterization of Autoinducer-3 structure and biosynthesis in E. coli. ACS Central Sci. 6, 197–206. doi: 10.1021/acscentsci.9b01076, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I. H., Kim S. Y., Park N. Y., Wen Y., Lee K. W., Yoon S. Y., et al. (2018). Cyclo-( l -Phe- l -pro), a quorum-sensing signal of Vibrio vulnificus, induces expression of Hydroperoxidase through a ToxR-LeuO-HU-RpoS signaling pathway to confer resistance against oxidative stress. Infect. Immun. 86, e00932–e00917. doi: 10.1128/iai.00932-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem M. (2004). Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides 25, 1405–1414. doi: 10.1016/j.peptides.2003.10.021, PMID: [DOI] [PubMed] [Google Scholar]
- Kleerebezem M., Quadri L. E. N., Kuipers O. P., de Vos W. M. (1997). Quorum sensing by peptide pheromones and two-component signal-transduction systems in gram-positive bacteria. Mol. Microbiol. 24, 895–904. doi: 10.1046/j.1365-2958.1997.4251782.x, PMID: [DOI] [PubMed] [Google Scholar]
- Kravchenko V. V., Kaufmann G. F. (2013). Bacterial inhibition of inflammatory responses via TLR-independent mechanisms. Cell. Microbiol. 15, 527–536. doi: 10.1111/cmi.12109, PMID: [DOI] [PubMed] [Google Scholar]
- Lade H., Paul D., Kweon J. H. (2014). Quorum quenching mediated approaches for control of membrane biofouling. Int. J. Biol. Sci. 10, 550–565. doi: 10.7150/ijbs.9028, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landman C., Grill J. P., Mallet J. M., Marteau P., Humbert L., le Balc’h E., et al. (2018). Inter-kingdom effect on epithelial cells of the N-acyl homoserine lactone 3-oxo-C12:2, a major quorum-sensing molecule from gut microbiota. PLoS One 13:e0202587. doi: 10.1371/journal.pone.0202587, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisner J. J., Laursen B. G., Prévost H., Drider D., Dalgaard P. (2007). Carnobacterium: positive and negative effects in the environment and in foods. FEMS Microbiol. Rev. 31, 592–613. doi: 10.1111/j.1574-6976.2007.00080.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- León-Félix J., Villicaña C. (2021). The impact of quorum sensing on the modulation of phage-host interactions. J. Bacteriol. 203, e00687–e00620. doi: 10.1128/JB.00687-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Peng W., Wu J., Wang X., Ren Y., Li H., et al. (2019a). Autoinducer-2 of gut microbiota, a potential novel marker for human colorectal cancer, is associated with the activation of TNFSF9 signaling in macrophages. Onco. Targets. Ther. 8:e1626192. doi: 10.1080/2162402X.2019.1626192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Ren Y., Fu X. (2019b). Inter-kingdom signaling between gut microbiota and their host. Cell. Mol. Life Sci. 76, 2383–2389. doi: 10.1007/s00018-019-03076-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.-H., Tian X. (2012). Quorum sensing and bacterial social interactions in biofilms. Sensors 12, 2519–2538. doi: 10.3390/s120302519, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Cheng J., Wang Y., Shen X. (2018). The quinolone signal (PQS): not just for quorum sensing anymore. Front. Cell. Infect. Microbiol. 8:230. doi: 10.3389/fcimb.2018.00230, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay A., Ahmer B. M. M. (2005). Effect of sdiA on biosensors of N-acylhomoserine lactones. J. Bacteriol. 187, 5054–5058. doi: 10.1128/JB.187.14.5054-5058.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Chen X., Skogerbø G., Zhang P., Chen R., He S., et al. (2012). The human microbiome: a hot spot of microbial horizontal gene transfer. Genomics 100, 265–270. doi: 10.1016/j.ygeno.2012.07.012, PMID: [DOI] [PubMed] [Google Scholar]
- Liu L., Li T., Cheng X. J., Peng C. T., Li C. C., He L. H., et al. (2018b). Structural and functional studies on Pseudomonas aeruginosa DspI: implications for its role in DSF biosynthesis. Sci. Rep. 8:3928. doi: 10.1038/s41598-018-22300-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Di Sun J. Z., Liu W. (2018a). Two-component signal transduction systems: a major strategy for connecting input stimuli to biofilm formation. Front. Microbiol. 9:3279. doi: 10.3389/fmicb.2018.03279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Hume M. E., Pillai S. D. (2004). Autoinducer-2-like activity associated with foods and its interaction with food additives. J. Food Prot. 67, 1457–1462. doi: 10.4315/0362-028X-67.7.1457, PMID: [DOI] [PubMed] [Google Scholar]
- Lu Y., Zeng J., Wu B., E S., Wang L., Cai R., et al. (2017). Quorum sensing N-acyl Homoserine lactones-SdiA suppresses Escherichia coli- Pseudomonas aeruginosa conjugation through inhibiting traI expression. Front. Cell. Infect. Microbiol. 7:7. doi: 10.3389/fcimb.2017.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons T., Gahan C. G., O’Sullivan T. P. (2020). Structure-activity relationships of furanones, dihydropyrrolones and thiophenones as potential quorum sensing inhibitors. Future Med. Chem. 12, 1925–1943. doi: 10.4155/fmc-2020-0244, PMID: [DOI] [PubMed] [Google Scholar]
- Machado Ribeiro T. R., Salgaço M. K., Adorno M. A. T., da Silva M. A., Piazza R. M. F., Sivieri K., et al. (2021). Human microbiota modulation via QseC sensor kinase mediated in the Escherichia coli O104:H4 outbreak strain infection in microbiome model. BMC Microbiol. 21:163. doi: 10.1186/s12866-021-02220-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes S., Heyndrickx M., Vackier T., Steenackers H., Verplaetse A., Reu K. D. E.. (2019). Identification and spoilage potential of the remaining dominant microbiota on food contact surfaces after cleaning and disinfection in different food industries. J. Food Prot. 82, 262–275. doi: 10.4315/0362-028X.JFP-18-226, PMID: [DOI] [PubMed] [Google Scholar]
- Maldonado-Barragán A., Caballero-Guerrero B., Lucena-Padrós H., Ruiz-Barba J. L. (2013). Induction of bacteriocin production by coculture is widespread among plantaricin-producing lactobacillus plantarum strains with different regulatory operons. Food Microbiol. 33, 40–47. doi: 10.1016/j.fm.2012.08.009, PMID: [DOI] [PubMed] [Google Scholar]
- Maldonado-Barragán A., Ruiz-Barba J. L., Jiménez-Díaz R. (2009). Knockout of three-component regulatory systems reveals that the apparently constitutive plantaricin-production phenotype shown by lactobacillus plantarum on solid medium is regulated via quorum sensing. Int. J. Food Microbiol. 130, 35–42. doi: 10.1016/j.ijfoodmicro.2008.12.033, PMID: [DOI] [PubMed] [Google Scholar]
- Malka O., Kalson D., Yaniv K., Shafir R., Rajendran M., Ben-David O., et al. (2021). Cross-kingdom inhibition of bacterial virulence and communication by probiotic yeast metabolites. Microbiome 9:70. doi: 10.1186/s40168-021-01027-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manefield M., de Nys R., Naresh K., Roger R., Givskov M., Peter S., et al. (1999). Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 145, 283–291. doi: 10.1099/13500872-145-2-283, PMID: [DOI] [PubMed] [Google Scholar]
- March J. C., Bentley W. E. (2004). Quorum sensing and bacterial cross-talk in biotechnology. Curr. Opin. Biotechnol. 15, 495–502. doi: 10.1016/j.copbio.2004.08.013, PMID: [DOI] [PubMed] [Google Scholar]
- Markus V., Share O., Teralı K., Ozer N., Marks R. S., Kushmaro A., et al. (2020). Anti-quorum sensing activity of stevia extract, Stevioside, Rebaudioside a and their Aglycon Steviol. Molecules 25:5480. doi: 10.3390/molecules25225480, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Martínez M. S., Uyttendaele M., Demolder V., Debevere J. (2006). Influence of food system conditions on N-acyl-L-homoserine lactones production by Aeromonas spp. Int. J. Food Microbiol. 112, 244–252. doi: 10.1016/j.ijfoodmicro.2006.04.025, PMID: [DOI] [PubMed] [Google Scholar]
- Meireles A., Faia S., Giaouris E., Simões M. (2018). Antimicrobial susceptibility and sessile behaviour of bacteria isolated from a minimally processed vegetables plant. Biofouling 34, 1150–1160. doi: 10.1080/08927014.2018.1554742, PMID: [DOI] [PubMed] [Google Scholar]
- Michael B., Smith J. N., Swift S., Heffron F., Ahmer B. M. M. (2001). SdiA of salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183, 5733–5742. doi: 10.1128/JB.183.19.5733-5742.2001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C., Gilmore J. (2020). Detection of quorum-sensing molecules for pathogenic molecules using cell-based and cell-free biosensors. Antibiotics 9:259. doi: 10.3390/antibiotics9050259, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mole B. M., Baltrus D. A., Dangl J. L., Grant S. R. (2007). Global virulence regulation networks in phytopathogenic bacteria. Trends Microbiol. 15, 363–371. doi: 10.1016/j.tim.2007.06.005, PMID: [DOI] [PubMed] [Google Scholar]
- Monnet V., Juillard V., Gardan R. (2016). Peptide conversations in gram-positive bacteria. Crit. Rev. Microbiol. 42, 339–351. doi: 10.3109/1040841X.2014.948804, PMID: [DOI] [PubMed] [Google Scholar]
- Moura-Alves P., Puyskens A., Stinn A., Klemm M., Guhlich-Bornhof U., Dorhoi A., et al. (2019). Host monitoring of quorum sensing during infection. Science 366:eaaw1629. doi: 10.1126/science.aaw1629, PMID: [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Bassler B. L. (2019). Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 17, 371–382. doi: 10.1038/s41579-019-0186-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji R., Prabhune A. (2015). Possible correlation between bile salt hydrolysis and AHL Deamidation: Staphylococcus epidermidis RM1, a potent quorum quencher and bile salt hydrolase producer. Appl. Biochem. Biotechnol. 176, 140–150. doi: 10.1007/s12010-015-1563-9, PMID: [DOI] [PubMed] [Google Scholar]
- Muras A., Otero-Casal P., Blanc V., Otero A. (2020). Acyl homoserine lactone-mediated quorum sensing in the oral cavity: a paradigm revisited. Sci. Rep. 10:9800. doi: 10.1038/s41598-020-66704-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugayah S. A., Gerth M. L. (2019). Engineering quorum quenching enzymes: progress and perspectives. Biochem. Soc. Trans. 47, 793–800. doi: 10.1042/BST20180165, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar S., Jeong H. L., Kim Y., Ha A. J. W., Roy P. K., Park S. H., et al. (2021). Inhibitory effects of Flavourzyme on biofilm formation, quorum sensing, and virulence genes of foodborne pathogens salmonella typhimurium and Escherichia coli. Food Res. Int. 147:110461. doi: 10.1016/j.foodres.2021.110461, PMID: [DOI] [PubMed] [Google Scholar]
- Nahar S., Mizan M. F. R., Ha A. J. W., Ha S. D. (2018). Advances and future prospects of enzyme-based biofilm prevention approaches in the food industry. Compr. Rev. Food Sci. Food Saf. 17, 1484–1502. doi: 10.1111/1541-4337.12382, PMID: [DOI] [PubMed] [Google Scholar]
- Nealson K. H., Platt T., Woodland Hastings J. (1970). Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104, 313–322. doi: 10.1128/jb.104.1.313-322.1970, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman H., Debelius J. W., Knight R., Koren O. (2015). Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiol. Rev. 39, 509–521. doi: 10.1093/femsre/fuu010, PMID: [DOI] [PubMed] [Google Scholar]
- Ng W.-L., Bassler B. L. (2009). Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43, 197–222. doi: 10.1146/annurev-genet-102108-134304, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol M., Alexandre S., Luizet J. B., Skogman M., Jouenne T., Salcedo S., et al. (2018). Unsaturated fatty acids affect quorum sensing communication system and inhibit motility and biofilm formation of Acinetobacter baumannii. Int. J. Mol. Sci. 19:214. doi: 10.3390/ijms19010214, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira F., Sharghi S., Kuchler K., Lion T. (2019). Pathogenetic impact of bacterial-fungal interactions. Microorganisms 7:459. doi: 10.3390/microorganisms7100459, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak J. S., Fratamico P. M. (2006). Evaluation of ascorbic acid as a quorum-sensing analogue to control growth, sporulation, and enterotoxin production in Clostridium perfringens. J. Food Sci. 69, FMS72–FMS78. doi: 10.1111/j.1365-2621.2004.tb13374.x [DOI] [Google Scholar]
- Novick R. P., Muir T. W. (1999). Virulence gene regulation by peptides in staphylococci and other gram-positive bacteria. Curr. Opin. Microbiol. 2, 40–45. doi: 10.1016/S1369-5274(99)80007-1, PMID: [DOI] [PubMed] [Google Scholar]
- One Health High-Level Expert Panel (OHHLEP) Adisasmito W. B., Almuhairi S., Behravesh C. B., Bilivogui P., Bukachi S. A., et al. (2022). One health: a new definition for a sustainable and healthy future. PLoS Pathog. 18:e1010537. doi: 10.1371/journal.ppat.1010537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S., Ravikumar V., Abdel-Haleem A. M., Derouiche A., Mokkapati V. R. S. S., Sihlbom C., et al. (2017). Low concentrations of vitamin C reduce the synthesis of extracellular polymers and destabilize bacterial biofilms. Front. Microbiol. 8:2599. doi: 10.3389/fmicb.2017.02599, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K., Bassler B. L. (2016). Quorum sensing signal-response systems in gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Shin H., Lee K., Holzapfel W. (2016). Autoinducer-2 properties of kimchi are associated with lactic acid bacteria involved in its fermentation. Int. J. Food Microbiol. 225, 38–42. doi: 10.1016/j.ijfoodmicro.2016.03.007, PMID: [DOI] [PubMed] [Google Scholar]
- Pasolli E., de Filippis F., Mauriello I. E., Cumbo F., Walsh A. M., Leech J., et al. (2020). Large-scale genome-wide analysis links lactic acid bacteria from food with the gut microbiome. Nat. Commun. 11:2610. doi: 10.1038/s41467-020-16438-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelyuntha W., Chaiyasut C., Kantachote D., Sirilun S. (2019). Cell-free supernatants from cultures of lactic acid bacteria isolated from fermented grape as biocontrol against Typhi and typhimurium virulence via autoinducer-2 and biofilm interference. PeerJ 7:e7555. doi: 10.7717/peerj.7555, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penesyan A., Nagy S. S., Kjelleberg S., Gillings M. R., Paulsen I. T. (2019). Rapid microevolution of biofilm cells in response to antibiotics. NPJ Bio. Microbio. 5:34. doi: 10.1038/s41522-019-0108-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng P., Baldry M., Gless B. H., Bojer M. S., Espinosa-Gongora C., Baig S. J., et al. (2019). Effect of co-inhabiting coagulase negative staphylococci on quorum sensing, host factor binding, and biofilm formation. Front. Microbiol. 10:2212. doi: 10.3389/fmicb.2019.02212, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M., Tong W., Zhao Z., Xiao L., Wang Z., Liu X., et al. (2021). Attenuation of Aeromonas hydrophila infection in Carassius auratus by YtnP, a -acyl Homoserine lactonase from bacillus licheniformis T-1. Antibiotics 10:631. doi: 10.3390/antibiotics10060631, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira C. S., Thompson J. A., Xavier K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37, 156–181. doi: 10.1111/j.1574-6976.2012.00345.x, PMID: [DOI] [PubMed] [Google Scholar]
- Pérez P. D, Hagen S. J. (2010). Heterogeneous response to a quorum-sensing signal in the luminescence of individual Vibrio fischeri. PLoS One, 5: e15473. doi: 10.1371/journal.pone.0015473, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpetuini G., Pham-Hoang B. N., Scornec H., Tofalo R., Schirone M., Suzzi G., et al. (2016). In lactobacillus pentosus, the olive brine adaptation genes are required for biofilm formation. Int. J. Food Microbiol. 216, 104–109. doi: 10.1016/j.ijfoodmicro.2015.10.002, PMID: [DOI] [PubMed] [Google Scholar]
- Peyrottes A., Coquant G., Brot L., Rainteau D., Seksik P., Grill J. P., et al. (2020). Anti-inflammatory effects of analogues of N-acyl Homoserine lactones on eukaryotic cells. Int. J. Mol. Sci. 21:9448. doi: 10.3390/ijms21249448, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer P. J. (2012). LuxS and quorum-sensing in campylobacter. Front. Cell. Infect. Microbiol. 2:22. doi: 10.3389/fcimb.2012.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott R. D., Decho A. W. (2020). Flexibility and adaptability of quorum sensing in nature. Trends Microbiol. 28, 436–444. doi: 10.1016/j.tim.2019.12.004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor C. R., McCarron P. A., Ternan N. G. (2020). Furanone quorum-sensing inhibitors with potential as novel therapeutics against Pseudomonas aeruginosa. J. Med. Microbiol. 69, 195–206. doi: 10.1099/jmm.0.001144, PMID: [DOI] [PubMed] [Google Scholar]
- Pun M., Khazanov N., Galsurker O., Weitman M., Kerem Z., Senderowitz H., et al. (2021). Phloretin, an apple Phytoalexin, affects the virulence and fitness of Pectobacterium brasiliense by interfering with quorum-sensing. Front. Plant Sci. 12:671807. doi: 10.3389/fpls.2021.671807, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y., Kando C. K., Thorsen L., Larsen N., Jespersen L. (2015). Production of autoinducer-2 by aerobic endospore-forming bacteria isolated from the west African fermented foods. FEMS Microbiol. Lett. 362:fnv186. doi: 10.1093/femsle/fnv186, PMID: [DOI] [PubMed] [Google Scholar]
- Qian Y., Ye J.-X., Yang S.-P., Lin Z.-Q., Cao W., Xie J.. (2018). Evaluation of the spoilage potential of Shewanella putrefaciens, Aeromonas hydrophila, and Aeromonas sobria isolated from spoiled Pacific white shrimp (Litopenaeus vannamei) during cold storage. J. Food Saf. 38:e12550. doi: 10.1111/jfs.12550 [DOI] [Google Scholar]
- Quintieri L., Caputo L., de Angelis M., Fanelli F. (2020). Genomic analysis of three cheese-borne pseudomonas lactis with biofilm and spoilage-associated behavior. Microorganisms 8:1208. doi: 10.3390/microorganisms8081208, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmati S., Yang S., Davidson A. L., Zechiedrich E. L. (2002). Control of the AcrAB multidrug efflux pump by quorum-sensing regulator SdiA. Mol. Microbiol 43, 677–85. doi: 10.1046/j.1365-2958.2002.02773.x, PMID: [DOI] [PubMed] [Google Scholar]
- Rajamani S., Bauer W. D., Robinson J. B., Farrow J. M., III, Pesci E. C., Teplitski M., et al. (2008). The vitamin riboflavin and its derivative lumichrome activate the LasR bacterial quorum-sensing receptor. Mol. Plant Microbe Interact. 21, 1184–1192. doi: 10.1094/MPMI-21-9-1184, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampioni G., Falcone M., Heeb S., Frangipani E., Fletcher M. P., Dubern J. F., et al. (2016). Unravelling the genome-wide contributions of specific 2-Alkyl-4-quinolones and PqsE to quorum sensing in Pseudomonas aeruginosa. PLoS Pathog. 12:e1006029. doi: 10.1371/journal.ppat.1006029, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana S., Bhawal S., Kumari A., Kapila S., Kapila R. (2020). pH-dependent inhibition of AHL-mediated quorum sensing by cell-free supernatant of lactic acid bacteria in Pseudomonas aeruginosa PAO1. Microb. Pathog. 142:104105. doi: 10.1016/j.micpath.2020.104105, PMID: [DOI] [PubMed] [Google Scholar]
- Ranava D., Backes C., Karthikeyan G., Ouari O., Soric A., Guiral M., et al. (2021). Metabolic exchange and energetic coupling between nutritionally stressed bacterial species: role of quorum-sensing molecules. MBio 12, e02758–e02720. doi: 10.1128/mBio.02758-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raut N., Pasini P., Daunert S. (2013). Deciphering bacterial universal language by detecting the quorum sensing signal, autoinducer-2, with a whole-cell sensing system. Anal. Chem. 85, 9604–9609. doi: 10.1021/ac401776k, PMID: [DOI] [PubMed] [Google Scholar]
- Rezzonico F., Smits T. H. M., Duffy B. (2012). Detection of AI-2 receptors in genomes of Enterobacteriaceae suggests a role of type-2 quorum sensing in closed ecosystems. Sensors 12, 6645–6665. doi: 10.3390/s120506645, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard A. H., Campagna S. R., Kolenbrander P. E. (2008). Autoinducer-2 is produced in saliva-fed flow conditions relevant to natural oral biofilms. J. Appl. Microbiol. 105, 2096–2103. doi: 10.1111/j.1365-2672.2008.03910.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A. C., D’Ávila de OLIVEIRA B., da SILVA E. R., SACRAMENTO N. T. B., BERTOLDI M. C., PINTO U. M. (2016). Anti-quorum sensing activity of phenolic extract from Eugenia brasiliensis (Brazilian cherry). Food Sci. Tech. 36, 337–343. doi: 10.1590/1678-457X.0089 [DOI] [Google Scholar]
- Rodríguez-López P., Barrenengoa A. E., Pascual-Sáez S., Cabo M. L. (2019). Efficacy of synthetic Furanones on biofilm formation. Foods 8:647. doi: 10.3390/foods8120647, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruparell A., Dubern J. F., Ortori C. A., Harrison F., Halliday N. M., Emtage A., et al. (2016). The fitness burden imposed by synthesising quorum sensing signals. Sci. Rep. 6:33101. doi: 10.1038/srep33101, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S. T., Bassler B. L. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2:a012427. doi: 10.1101/cshperspect.a012427, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S. T., van Kessel J. C., Shao Y., Bassler B. L. (2011). AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 25, 397–408. doi: 10.1101/gad.2015011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoffone V. C., Trespidi G., Chiarelli L. R., Barbieri G., Buroni S. (2019). Quorum sensing as Antivirulence target in cystic fibrosis pathogens. Int. J. Mol. Sci. 20:1838. doi: 10.3390/ijms20081838, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Fuchs S., Milo R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14:e1002533. doi: 10.1371/journal.pbio.1002533, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepehr S., Rahmani-Badi A., Babaie-Naiej H., Soudi M. R. (2014). Unsaturated fatty acid, cis-2-decenoic acid, in combination with disinfectants or antibiotics removes pre-established biofilms formed by food-related bacteria. PLoS One 9:e101677. doi: 10.1371/journal.pone.0101677, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad D. P., Taneja N. K., Lakra A., Sachdev D. (2021). In vitro and in situ abrogation of biofilm formation in E. coli by vitamin C through ROS generation, disruption of quorum sensing and exopolysaccharide production. Food Chem. 341:128171. doi: 10.1016/j.foodchem.2020.128171, PMID: [DOI] [PubMed] [Google Scholar]
- Sikdar R., Elias M. (2020). Quorum quenching enzymes and their effects on virulence, biofilm, and microbiomes: a review of recent advances. Expert Rev. Anti-Infect. Ther. 18, 1221–1233. doi: 10.1080/14787210.2020.1794815, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silpe J. E., Bassler B. L. (2019). A host-produced quorum-sensing autoinducer controls a phage lysis-Lysogeny decision. Cells 176, 268–280.e13. doi: 10.1016/j.cell.2018.10.059, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V. K., Kavita K., Prabhakaran R., Jha B. (2013). Cis-9-octadecenoic acid from the rhizospheric bacterium Stenotrophomonas maltophilia BJ01 shows quorum quenching and anti-biofilm activities. Biofouling 29, 855–867. doi: 10.1080/08927014.2013.807914, PMID: [DOI] [PubMed] [Google Scholar]
- Skandamis P. N., Nychas G.-J. E. (2012). Quorum sensing in the context of food microbiology. Appl. Environ. Microbiol. 78, 5473–5482. doi: 10.1128/AEM.00468-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano C., Echeverz M., Lasa I. (2014). Biofilm dispersion and quorum sensing. Curr. Opin. Microbiol. 18, 96–104. doi: 10.1016/j.mib.2014.02.008 [DOI] [PubMed] [Google Scholar]
- Soni K. A., Jesudhasan P., Cepeda M., Widmer K., Jayaprakasha G. K., Patil B. S., et al. (2008). Identification of ground beef-derived fatty acid inhibitors of autoinducer-2-based cell signaling. J. Food Prot. 71, 134–138. doi: 10.4315/0362-028X-71.1.134, PMID: [DOI] [PubMed] [Google Scholar]
- Stentz R., Horn N., Cross K., Salt L., Brearley C., Livermore D. M., et al. (2015). Cephalosporinases associated with outer membrane vesicles released by Bacteroides spp. protect gut pathogens and commensals against β-lactam antibiotics. J. Antimicrob. Chemother. 70, 701–709. doi: 10.1093/jac/dku466, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenz L., François P., Fischer A., Huyghe A., Tangomo M., Hernandez D., et al. (2008). Impact of oleic acid (cis-9-octadecenoic acid) on bacterial viability and biofilm production in Staphylococcus aureus. FEMS Microbiol. Lett. 287, 149–155. doi: 10.1111/j.1574-6968.2008.01316.x, PMID: [DOI] [PubMed] [Google Scholar]
- Stephens K., Bentley W. E. (2020). Synthetic biology for manipulating quorum sensing in microbial consortia. Trends Microbiol. 28, 633–643. doi: 10.1016/j.tim.2020.03.009, PMID: [DOI] [PubMed] [Google Scholar]
- Striednig B., Hilbi H. (2022). Bacterial quorum sensing and phenotypic heterogeneity: how the collective shapes the individual. Trends Microbiol. 30, 379–389. doi: 10.1016/j.tim.2021.09.001, PMID: [DOI] [PubMed] [Google Scholar]
- Subramoni S., Venturi V. (2009). LuxR-family “solos”: bachelor sensors/regulators of signalling molecules. Microbiology 155, 1377–1385. doi: 10.1099/mic.0.026849-0, PMID: [DOI] [PubMed] [Google Scholar]
- Sun B., Zhang M. (2016). Analysis of the antibacterial effect of an Edwardsiella tarda LuxS inhibitor. Springerplus 5:92. doi: 10.1186/s40064-016-1733-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaggerty C. L., Genovese K. J., He H., Byrd J. A., Jr, Kogut M. H. (2018). Editorial: mechanisms of persistence, survival, and transmission of bacterial foodborne pathogens in production animals. Front. Vet. Sci. 5. doi: 10.3389/fvets.2018.00139, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S.-W., Schaffer J. N., Tu K. C., Mehta P., Lu W., Ong N. P., et al. (2011). Active regulation of receptor ratios controls integration of quorum-sensing signals in Vibrio harveyi. Mol. Syst. Biol. 7:491. doi: 10.1038/msb.2011.30, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. A., Oliveira R. A., Djukovic A., Ubeda C., Xavier K. B. (2015). Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 10, 1861–1871. doi: 10.1016/j.celrep.2015.02.049, PMID: [DOI] [PubMed] [Google Scholar]
- Tijerina-Rodríguez L., Villarreal-Treviño L., Baines S. D., Morfín-Otero R., Camacho-Ortíz A., Flores-Treviño S., et al. (2019). High sporulation and overexpression of virulence factors in biofilms and reduced susceptibility to vancomycin and linezolid in recurrent clostridium [Clostridioides] difficile infection isolates. PLoS One 14:e0220671. doi: 10.1371/journal.pone.0220671, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabi Delshad S., Soltanian S., Sharifiyazdi H., Haghkhah M., Bossier P. (2018). Identification of N-acyl homoserine lactone-degrading bacteria isolated from rainbow trout (Oncorhynchus mykiss). J. Appl. Microbiol. 125, 356–369. doi: 10.1111/jam.13891, PMID: [DOI] [PubMed] [Google Scholar]
- Tourneroche A., Lami R., Hubas C., Blanchet E., Vallet M., Escoubeyrou K., et al. (2019). Bacterial-fungal interactions in the kelp Endomicrobiota drive Autoinducer-2 quorum sensing. Front. Microbiol. 10:1693. doi: 10.3389/fmicb.2019.01693, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku M., Morinaga K., Hashimoto Y., Uhl J., Shimamura H., Inaba H., et al. (2017). Membrane vesicle-mediated bacterial communication. ISME J. 11, 1504–1509. doi: 10.1038/ismej.2017.13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uruén C., Chopo-Escuin G., Tommassen J., Mainar-Jaime R. C., Arenas J. (2020). Biofilms as promoters of bacterial antibiotic resistance and tolerance. Antibiotics 10:3. doi: 10.3390/antibiotics10010003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadassery D. H., Pillai D. (2020). Quorum quenching potential of enterococcus faecium QQ12 isolated from gastrointestinal tract of Oreochromis niloticus and its application as a probiotic for the control of Aeromonas hydrophila infection in goldfish Carassius auratus (Linnaeus 1758). Brazilian J. Microbiol. 51, 1333–1343. doi: 10.1007/s42770-020-00230-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Reckem E., Charmpi C., Van der Veken D., Borremans W., De Vuyst L., Weckx S., et al. (2020). Application of a high-throughput amplicon sequencing method to chart the bacterial communities that are associated with European fermented meats from different origins. Foods 9, 9:1247. doi: 10.3390/foods9091247, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeke F., de Craemer S., Debunne N., Janssens Y., Wynendaele E., van de Wiele C., et al. (2017). Peptides as quorum sensing molecules: measurement techniques and obtained levels in vitro and in vivo. Front. Neurosci. 11:183. doi: 10.3389/fnins.2017.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeurgt C., Veithen A., Carlot S., Tarabichi M., Dumont J. E., Hassid S., et al. (2017). The human bitter taste receptor T2R38 is broadly tuned for bacterial compounds. PLoS One 12:e0181302. doi: 10.1371/journal.pone.0181302, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. M., Pracser N., Thalguter S., Fischel K., Rammer N., Pospíšilová L., et al. (2020). Identification of biofilm hotspots in a meat processing environment: detection of spoilage bacteria in multi-species biofilms. Int. J. Food Microbiol. 328:108668. doi: 10.1016/j.ijfoodmicro.2020.108668, PMID: [DOI] [PubMed] [Google Scholar]
- Walsh C., Fanning S. (2008). Antimicrobial resistance in foodborne pathogens--a cause for concern? Curr. Drug Targets 9, 808–815. doi: 10.2174/138945008785747761, PMID: [DOI] [PubMed] [Google Scholar]
- Walter J., Armet A. M., Finlay B. B., Shanahan F. (2020). Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cells 180, 221–232. doi: 10.1016/j.cell.2019.12.025 [DOI] [PubMed] [Google Scholar]
- Walters M., Sperandio V. (2006). Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect. Immun. 74, 5445–5455. doi: 10.1128/IAI.00099-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-Y., Chen K. N., Lo Y. M., Chiang M. L., Chen H. C., Liu J. R., et al. (2012). Investigation of microorganisms involved in biosynthesis of the kefir grain. Food Microbiol. 32, 274–285. doi: 10.1016/j.fm.2012.07.001, PMID: [DOI] [PubMed] [Google Scholar]
- Wang J., Ding L., Li K., Huang H., Hu H., Geng J., et al. (2018). Estimation of spatial distribution of quorum sensing signaling in sequencing batch biofilm reactor (SBBR) biofilms. Sci. Total Environ. 612, 405–414. doi: 10.1016/j.scitotenv.2017.07.277, PMID: [DOI] [PubMed] [Google Scholar]
- Wang Y., Ho C.-T. (2008). Formation of 2,5-dimethyl-4-hydroxy-3(2H)-furanone through methylglyoxal: a Maillard reaction intermediate. J. Agric. Food Chem. 56, 7405–7409. doi: 10.1021/jf8012025, PMID: [DOI] [PubMed] [Google Scholar]
- Wang S., Payne G. F., Bentley W. E. (2020). Quorum sensing communication: molecularly connecting cells, their neighbors, and even devices. Ann. Rev. Chem. Biomol. Eng. 11, 447–468. doi: 10.1146/annurev-chembioeng-101519-124728, PMID: [DOI] [PubMed] [Google Scholar]
- Wang X.-Y., Xie J. (2020). Quorum sensing system-regulated proteins affect the spoilage potential of co-cultured Acinetobacter johnsonii and Pseudomonas fluorescens from spoiled bigeye tuna (Thunnus obesus) as determined by proteomic analysis. Front. Microbiol. 11:940. doi: 10.3389/fmicb.2020.00940, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiland-Bräuer N., Kisch M. J., Pinnow N., Liese A., Schmitz R. A. (2016, 2016). A highly effective inhibition of biofilm formation by the first metagenome-derived AI-2 quenching enzyme. Front. Microbiol. 7:1098. doi: 10.3389/fmicb.2016.01098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellington S., Greenberg E. P. (2019). Quorum sensing signal selectivity and the potential for interspecies cross talk. MBio 10, e00146–e00119. doi: 10.1128/mBio.00146-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen K. Y., Cameron L., Chappell J., Jensen K., Bell D. J, Kelwick R., et al. (2017). A cell-free biosensor for detecting quorum sensing molecules in P. aeruginosa-infected respiratory samples. ACS Synthetic Biol. 6, 2293–2301. doi: 10.1021/acssynbio.7b00219, PMID: [DOI] [PubMed] [Google Scholar]
- Werner N., Petersen K., Vollstedt C., Garcia P. P., Chow J., Ferrer M., et al. (2021). The Komagataeibacter europaeus GqqA is the prototype of a novel bifunctional N-acyl-homoserine lactone acylase with prephenate dehydratase activity. Sci. Rep. 11:12255. doi: 10.1038/s41598-021-91536-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M., Diggle S. P., Greenberg E. P. (2017). Progress in and promise of bacterial quorum sensing research. Nature 551, 313–320. doi: 10.1038/nature24624, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whon T. W., Ahn S. W., Yang S., Kim J. Y., Kim Y. B., Kim Y., et al. (2021). ODFM, an omics data resource from microorganisms associated with fermented foods. Sci Data 8. doi: 10.1038/s41597-021-00895-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbert S. A., Mark Welch J. L., Borisy G. G. (2020). Spatial ecology of the human tongue dorsum microbiome. Cell Rep. 30, 4003–4015.e3. doi: 10.1016/j.celrep.2020.02.097, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M. R., Costa S. K., Zaramela L. S., Khalil S., Todd D. A., Winter H. L., et al. (2019). Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci. Transl. Med. 11:eaat8329. doi: 10.1126/scitranslmed.aat8329, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe B. E., Dutton R. J. (2015). Fermented foods as experimentally tractable microbial ecosystems. Cells 161, 49–55. doi: 10.1016/j.cell.2015.02.034, PMID: [DOI] [PubMed] [Google Scholar]
- Wu S., Feng J., Liu C., Wu H., Qiu Z., Ge J., et al. (2021b). Machine learning aided construction of the quorum sensing communication network for human gut microbiota. Nat. Commun. 13:3079. doi: 10.1038/s41467-022-30741-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Xu C., Liu J., Liu C., Qiao J. (2021a). Vertical and horizontal quorum-sensing-based multicellular communications. Trends Microbiol. 29, 1130–1142. doi: 10.1016/j.tim.2021.04.006, PMID: [DOI] [PubMed] [Google Scholar]
- Wynendaele E., Bronselaer A., Nielandt J., D’Hondt M., Stalmans S., Bracke N., et al. (2013). Quorumpeps database: chemical space, microbial origin and functionality of quorum sensing peptides. Nucleic Acids Res. 41, D655–D659. doi: 10.1093/nar/gks1137, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn D., Raut N., Joel S., Pasini P., Deo S. K., Daunert S. (2018). Detection of bacterial contamination in food matrices by integration of quorum sensing in a paper-strip test. Analyst 143, 4774–4782. doi: 10.1039/C8AN00878G, PMID: [DOI] [PubMed] [Google Scholar]
- Xue J., Chi L., Tu P., Lai Y., Liu C. W., Ru H., et al. (2021). Detection of gut microbiota and pathogen produced N-acyl homoserine in host circulation and tissues. NPJ Bio. Microbio. 7:53. doi: 10.1038/s41522-021-00224-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap M., Ercolini D., Álvarez-Ordóñez A., O'Toole P. W., O'Sullivan O., Cotter P. D.. (2022). Next-generation food research: use of meta-Omic approaches for characterizing microbial communities along the food chain. Annu. Rev. Food Sci. Technol. 13, 361–384. doi: 10.1146/annurev-food-052720-010751, PMID: [DOI] [PubMed] [Google Scholar]
- Yu H., Li J., Han Y., Shi G., Liu Z., Zeng M. (2019). AHLs-produced bacteria in refrigerated shrimp enhanced the growth and spoilage ability of Shewanella baltica. J. Food Sci. Technol. 56, 114–121. doi: 10.1007/s13197-018-3464-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K., Hou L., Jin Q., Niu C., Mao M., Wang R., et al. (2021). Comparative transcriptomics analysis of Streptococcus mutans with disruption of LuxS/AI-2 quorum sensing and recovery of methyl cycle. Arch. Oral Biol. 127:105137. doi: 10.1016/j.archoralbio.2021.105137, PMID: [DOI] [PubMed] [Google Scholar]
- Yuan L., Sadiq F. A., Burmølle M., Liu T., He G. (2018). Insights into bacterial Milk spoilage with particular emphasis on the roles of heat-stable enzymes, biofilms, and quorum sensing. J. Food Prot. 81, 1651–1660. doi: 10.4315/0362-028X.JFP-18-094, PMID: [DOI] [PubMed] [Google Scholar]
- Zargar A., Quan D. N., Carter K. K., Guo M., Sintim H. O., Payne G. F., et al. (2015). Bacterial secretions of nonpathogenic Escherichia coli elicit inflammatory pathways: a closer investigation of interkingdom signaling. MBio 6:e00025. doi: 10.1128/mBio.00025-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytseva Y. V., Sidorov A. V., Marakaev O. A., Khmel I. A. (2019). Plant-microbial interactions involving quorum sensing regulation. Microbiology 88, 523–533. doi: 10.1134/S0026261719040131 [DOI] [Google Scholar]
- Zhou L., Zhang L. H., Cámara M., He Y. W. (2017). The DSF family of quorum sensing signals: diversity, biosynthesis, and turnover. Trends Microbiol. 25, 293–303. doi: 10.1016/j.tim.2016.11.013, PMID: [DOI] [PubMed] [Google Scholar]
- Ziesche L., Bruns H., Dogs M., Wolter L., Mann F., Wagner-Döbler I., et al. (2015). Homoserine lactones, methyl Oligohydroxybutyrates, and other extracellular metabolites of macroalgae-associated bacteria of the Roseobacter clade: identification and functions. Chembiochem: Eur. J. Chem. Biol. 16, 2094–2107. doi: 10.1002/cbic.201500189, PMID: [DOI] [PubMed] [Google Scholar]
- Zink K. E., Ludvik D. A., Lazzara P. R., Moore T. W., Mandel M. J., Sanchez L. M.. (2021). A small molecule coordinates symbiotic behaviors in a host organ. MBio 12, e03637–e03620. doi: 10.1128/mBio.03637-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwirzitz B., Wetzels S. U., Dixon E. D., Stessl B., Zaiser A., Rabanser I., et al. (2020). The sources and transmission routes of microbial populations throughout a meat processing facility. NPJ Bio. Microbio. 6:26. doi: 10.1038/s41522-020-0136-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]