Abstract
Endoreplication is an evolutionarily conserved mechanism for increasing nuclear DNA content (ploidy). Ploidy frequently scales with final cell and organ size, suggesting a key role for endoreplication in these processes. However, exceptions exist, and, consequently, the endoreplication-size nexus remains enigmatic. Here, we show that prolonged tissue folding at the apical hook in Arabidopsis requires endoreplication asymmetry under the control of an auxin gradient. We identify a molecular pathway linking endoreplication levels to cell size through cell wall remodeling and stiffness modulation. We find that endoreplication is not only permissive for growth: Endoreplication reduction enhances wall stiffening, actively reducing cell size. The cell wall integrity kinase THESEUS plays a key role in this feedback loop. Our data thus explain the nonlinearity between ploidy levels and size while also providing a molecular mechanism linking mechanochemical signaling with endoreplication-mediated dynamic control of cell growth.
Endoreplication integrates hormonal and mechanochemical signaling to control tissue bending in Arabidopsis.
INTRODUCTION
In multicellular organisms, spatiotemporal control of growth is critical for shaping and patterning organs and regulating final organ size. Although cell division and elongation both contribute to organ size, several organs in plants and animals undergo substantial increases in size without appreciable cell division (1). Instead, cells in these organs undergo endoreplication to replicate their nuclear DNA several times without mitosis, increasing ploidy and causing their nuclei to enlarge. Endoreplication is thought to allow cells to scale their DNA content to their size (2). However, high endoreplication does not always correlate linearly with faster growth and larger cells (3). For example, during sepal development, polyploid cells grow at the same rate as diploid cells (4). In addition, endoreplication has been associated with the rate of cellular growth but not with growth anisotropy or direction. Hence, the contribution of endoreplication to the control of cell and organ size is a matter of debate, and the mechanism underpinning this contribution thus remains unclear.
Here, we use the nondividing but rapidly elongating hypocotyl cells during apical hook formation as a simple experimental system to investigate the mechanical and biochemical bases for cell size regulation by endoreplication. Under natural conditions, the apical hook forms by bending of the hypocotyl as the seedling emerges from the soil after seed germination. The hypocotyl is an attractive model system for studying the relationship between cell behavior and multicellular morphogenesis because its shape and growth depend primarily on cell elongation rather than cell division (5). This contrasts with the more complex process of gravitropic bending of roots presumably mediated via both cell expansion and cell division. Tissue bending during apical hook formation is governed primarily by rapid cell elongation on the outer side of the hook, with cells on outer side becoming up to threefold or larger than those on the inner side (6). The differential growth that induces hypocotyl bending must be maintained for some time to prevent premature hook opening, necessitating robust spatiotemporal control of growth on opposite sides of the hypocotyl. Previous studies have shown that the differences in growth responsible for apical hook bending depend on asymmetries in the distributions of the phytohormone auxin and cell wall mechanical properties: The inner cells have high auxin concentrations and stiffer walls, suppressing their elongation, while the rapidly growing outer cells have low auxin levels and softer walls (6, 7).
Here, we show that rapid cell growth on the outer side of hook is severely attenuated upon reduction in endoreplication, causing a failure to maintain differential growth between the outer and inner sides and leading to defects in hook development. However, this is not the simple result of differences in ploidy. Rather, we demonstrate the existence of a feedback loop between auxin gradient–dependent spatial control of endoreplication, cell wall mechanics, and wall integrity sensing to regulate cell size via endoreplication. On the basis of this loop, we propose a mechanistic framework where endoreplication acquires an integrated role in dynamic multicellular shape changes.
RESULTS
Endoreplication is required for apical hook maintenance
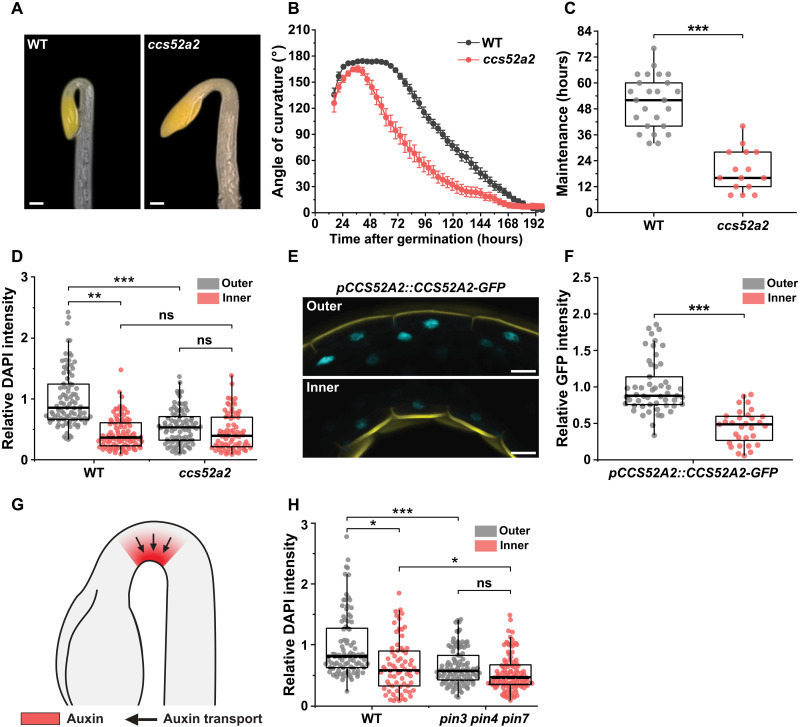
To investigate the role of endoreplication in cell size regulation, we analyzed differential cell elongation during apical hook development in the Arabidopsis cell cycle switch protein 52 a2 (ccs52a2) mutant, which exhibits strongly reduced endoreplication (8) due to inactivation of CCS52A2, a rate-limiting activator of the anaphase-promoting complex/cyclosome E3 ubiquitin ligase (APC/C) (9). Hook development proceeds through three phases: formation, maintenance, and opening (fig. S1) (10). Kinematic analyses of hook development revealed significant hook development defects in ccs52a2: Its maintenance phase is almost 32 hours shorter than that of the wild type (WT) (Fig. 1, A to C).
Fig. 1. Spatial differences in endoreplication regulate hook development in response to auxin.
(A) Representative images of the apical hook of dark-grown WT and ccs52a2 seedlings at 60 hours after germination. Scale bars, 200 μm. (B) Kinematic analysis of hook development in WT (n = 25) and ccs52a2 mutant (n = 15). Graphs represent means ± SEM. (C) The maintenance periods of apical hook in WT and ccs52a2 mutant derived from Fig. 1B. (D) 4′,6-Diamidino-2-phenylindole (DAPI) intensity of epidermal cells on the inner and outer sides overlapping the bent region of the apical hook in WT (n = 6; outer, 90 nuclei; inner, 89 nuclei) and ccs52a2 (n = 6; outer, 86 nuclei; inner, 74 nuclei). (E) Representative confocal images of the outer and inner sides of the pCCS52A2::CCS52A2-GFP seedling. Cyan signal represents CCS52A2-GFP. Propidium iodide (PI) staining was performed to visualize the cell wall (yellow). Scale bars, 20 μm. (F) Quantitative analysis of the CCS52A2 accumulation in the outer and inner sides of pCCS52A2::CCS52A2-GFP in the WT background (outer, n = 53; inner, n = 31). (G) A schematic apical hook in WT showing auxin response maximum (in red) generated by the PIN-dependent polar auxin transport (denoted by black arrows) directed toward the inner side of the hook. This auxin response maximum is disrupted in pin3 pin4 pin7 mutant. (H) DAPI intensity of epidermal cells on the outer and inner sides overlapping the bent region of the apical hook in WT (n = 6; outer, 92 nuclei; inner, 70 nuclei) and pin3 pin4 pin7 (n = 6; outer, 103 nuclei; inner, 98 nuclei). Thick lines in the boxes indicate medians. Statistical significance was calculated using Student’s t test (unpaired, two-tailed) and is indicated as follows: ns (not statistically significant), *P < 0.05, **P < 0.01, and ***P < 0.001.
The proposed correlation between endoreplication and cell size prompted us to investigate whether growth differences between the fast-growing outer side of the apical hook and the slow-growing inner side in the WT are associated with endoreplication asymmetries and whether these asymmetries are disrupted in ccs52a2. Endoreplication quantification based on 4′,6-diamidino-2-phenylindole (DAPI) staining of nuclear DNA (11) in epidermal cells showed that nuclear DNA staining on the hook’s outer side was around 50% more intense than that on the inner side (Fig. 1D and fig. S2), consistent with higher levels of endoreplication in the large and rapidly elongating outer cells. We also validated endoreplication asymmetry using two additional methods. Both DRAQ5 staining of nuclear DNA (12) (fig. S3, A and B) and nuclear volume measurements (13) of epidermal cells on the outer and inner sides of the hook in the WT indicated endoreplication asymmetry (fig. S3, C and D), confirming DAPI-based measurements. Conversely, the DAPI signal on the outer side of the ccs52a2 hook was significantly weaker (by ca. 37%) than that on the outer side of the WT hook, and there was no significant endoreplication asymmetry between the outer and inner sides of the hook in ccs52a2 (Fig. 1D and fig. S2). Our results show that green fluorescent protein (GFP)-tagged CCS52A2 (CCS52A2-GFP) driven by the native promoter (14) is asymmetrically expressed in the hook. CCS52A2 expression is higher on the outer side (relative to the inner side) of the hook, correlating with high endoreplication on the outer side of the hook (Fig. 1, E and F). Thus, our data show that CCS52A2-dependent endoreplication asymmetry leading to elevated endoreplication on the outer side of the hypocotyl is crucial for maintaining the differential growth established during the formation phase of apical hook development.
Auxin asymmetry mediates differential endoreplication during hook development
Apical hook development requires a strong auxin response on the inner side (inhibiting elongation) combined with low auxin levels on the outer side (facilitating elongation) (Fig. 1G) (15). This auxin response asymmetry correlates spatially with the hook’s endoreplication asymmetry (Fig. 1, D and G). We therefore investigated the possibility that the asymmetric auxin response could mediate spatial differences in endoreplication on the hook’s outer and inner sides. The asymmetric auxin response is established by the activity of PIN-FORMED 3 (PIN3)-, 4-, and 7-dependent polar auxin transport (16, 17). We therefore assayed endoreplication in the pin3 pin4 pin7 mutant, which exhibits severe hook defects due to a lack of auxin asymmetry (16, 17). The DAPI intensity on the outer side of the hook in pin3 pin4 pin7 seedlings was significantly lower (by ca. 38%) than that in WT controls (Fig. 1H). Consequently, whereas the WT exhibits significant asymmetry in endoreplication (P = 0.04), the same is not true for pin3 pin4 pin7 (P = 0.11) (Fig. 1H). Last, treatment of WT with polar auxin transport inhibitor N-1-naphthylphthalamic acid (NPA; mimicking pin-triple mutant) disrupted CCS52A2-GFP expression asymmetry (fig. S4). These results show that transport-dependent auxin asymmetry and CCS52A2 expression are crucial mediators of endoreplication asymmetry during hook development. It is worth noting that, in addition to its effect on CCS52A2 expression, polar auxin transport (via PINs) could also affect apical hook via additional pathways.
Growth and mechanical asymmetries in the hook are disrupted in the ccs52a2 mutant
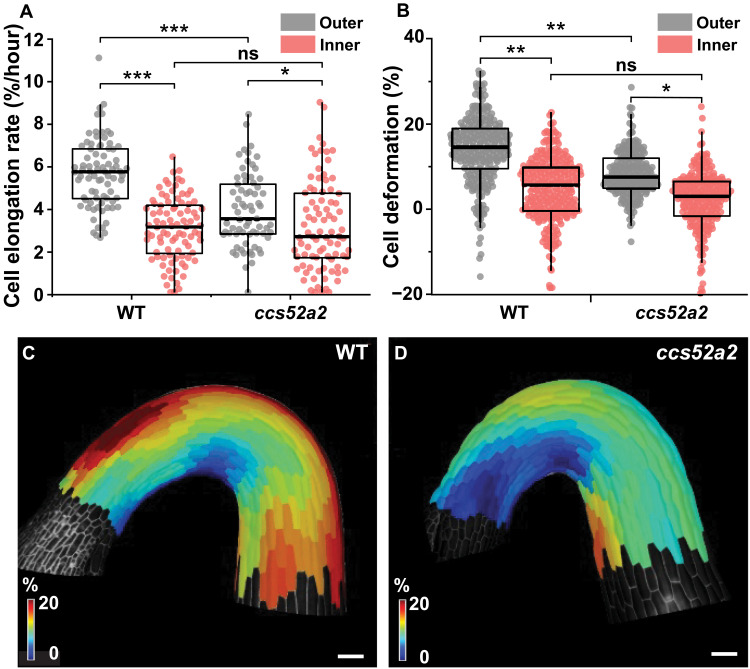
High endoreplication spatially correlates with fast cell elongation on the outer side of the hook. We therefore investigated whether endoreplication attenuation in the ccs52a2 mutant affects cell elongation, leading to hook defects. The cell elongation rate on the outer side of the hook in ccs52a2 (3.84 ± 0.21%/hour) was significantly lower than that in the WT (5.58 ± 0.18%/hour), while there was no significant difference between WT (2.88 ± 0.17%/hour) and ccs52a2 (2.90 ± 0.24%/hour) on the inner side (Fig. 2A and fig. S5). Consequently, cell elongation asymmetry between the outer and inner sides was also notablely attenuated in ccs52a2 (P < 0.01) when compared to the WT (P < 0.001) (Fig. 2A). The attenuation of growth asymmetry in ccs52a2 is thus consistent with its hook maintenance defects.
Fig. 2. Endoreplication mediates in control of differential cell elongation and mechanical properties during hook development.
(A) Cell elongation rate of epidermal cells (%/hour) at 0 to 400 μm from the shoot apex in the WT (n = 5; outer, 80 cells; inner, 88 cells) and ccs52a2 (n = 6; outer, 69 cells; inner, 84 cells). (B) Quantification of the deformation of the epidermal cells in the WT (n = 6; outer, 240 cells; inner, 250 cells) and ccs52a2 (n = 6; outer, 197 cells; inner, 213 cells). Thick lines in the boxes indicate medians. (C and D) Heatmaps of cell deformation following osmotic treatment for the WT (C) and ccs52a2 (D). Statistical significance was calculated using Student’s t test (unpaired, two-tailed) and is indicated as follows: ns, *P < 0.05, **P < 0.01, and ***P < 0.001. Scale bars, 100 μm.
Differential growth on the inner and outer sides of the apical hook relies on mechanical asymmetry: The cell walls are stiffer on the inner side and softer on the outer side (6, 18). The hook developmental and elongation defects of ccs52a2 therefore prompted us to investigate the mechanical properties of its cell walls by measuring cell deformation in response to changes in osmotic pressure, as previously described for the shoot meristem and apical hook (6, 18, 19). In accordance with previous findings, cells on the outer and inner sides of the hook in the WT deformed by 16.7 ± 0.9% and 6.3 ± 1.7%, respectively, indicating that the outer side cells were 2.7 times softer than those on the inner side. In ccs52a2, the outer side cells were significantly (1.8-fold) less deformed (9.5 ± 0.5%) than in the WT. Thus, the outer side cell walls of the ccs52a2 mutant were stiffer than those of the WT, whereas the wall stiffness on the inner side did not differ significantly between WT and ccs52a2 (Fig. 2, B to D). The ccs52a2 mutant thus exhibits weaker mechanical and elongation asymmetry than the WT because the cell walls on the outer side of the apical hook are stiffer, causing slower cell growth (Fig. 2A).
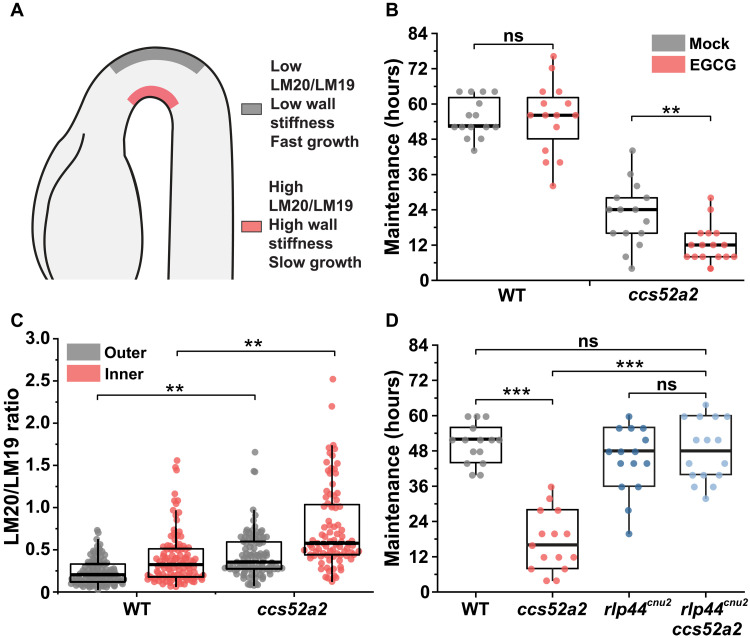
Endoreplication affects pectin methylesterification levels during hook development
Endoreplication has been associated with the regulation of cell wall properties (11, 20). Therefore, we investigated changes in cell wall composition that could explain the increased cell wall stiffness on the outer side of the apical hook in ccs52a2. In the WT, the softness of the outer epidermal cell walls is due to their low content of methylesterified homogalacturonan (HG) pectin, which facilitates rapid cell elongation (Fig. 3A) (6). Moreover, overexpression of PECTIN METHYLESTERASE INHIBITOR 5 (PMEI5) or treatment with exogenous epigallocatechin gallate (EGCG), which inhibits HG demethylesterification, leads to increased wall stiffness and slower elongation of the outer cells, resulting in hook defects (6). To investigate the relationship between endoreplication, HG methylesterification, and hook development, we first analyzed hook development in the WT and ccs52a2 in the presence of EGCG (21). The ccs52a2 mutant was hypersensitive to EGCG: Treatment with 250 μM EGCG had no effect on hook development in the WT but exacerbated the hook maintenance defect of ccs52a2, causing the hook to open more rapidly than in mock-treated controls (Fig. 3B).
Fig. 3. Enhanced HG methylesterification due to reduced endoreplication results in defective apical hook development.
(A) A schematic apical hook in the WT showing high ratio of methylesterified HG (detected by LM20 antibody) to demethylesterified HG (detected by LM19 antibody) on the inner side (with stiffer walls) and the opposite on the fast-growing outer side (with softer walls). (B) Maintenance period of apical hook in WT (mock, n = 15; EGCG, n = 15) and ccs52a2 (mock, n = 15; EGCG, n = 16) after EGCG treatment. (C) LM20/LM19 immunolabeling ratios of longitudinal walls of epidermal cells in the WT (n = 8; outer, 90 cells; inner, 103 cells) and ccs52a2 (n = 6; outer, 95 cells; inner, 87 cells) at 0 to 400 μm from the shoot apical meristem (SAM) (n ≥ 7). (D) Maintenance period of apical hook in WT (n = 15), ccs52a2 (n = 15), rlp44cnu2 (n = 15), and rlp44cnu2 ccs52a2 (n = 16). Thick lines in the boxes indicate medians. Statistical significance was calculated using Student’s t test (unpaired, two-tailed) and is indicated as follows: ns, **P < 0.01, and ***P < 0.001.
We next quantified the levels of HG methylesterification in the outer longitudinal cell wall of the epidermal cells of the apical hook in css52a2 and the WT. In agreement with previously published results (6), the level of HG methylesterification (as indicated by LM20/LM19 ratio) was asymmetric, high on the stiffer inner side relative to the softer outer side of the hook (Fig. 3C and fig. S6). In contrast, the level of HG methylesterification in ccs52a2 was higher, almost twice that of the WT, on both the outer and inner sides of the hypocotyl (the LM20/LM19 ratios for the outer and inner sides in the WT were 0.63 ± 0.10 and 1.12 ± 0.21, respectively, compared to 1.20 ± 0.17 and 2.14 ± 0.34 for ccs52a2) (Fig. 3C and fig. S6). Our results thus show that HG methylesterification is enhanced in ccs52a2, leading to cell wall stiffening, slower cell elongation on the outer side, and hook defects (Fig. 2).
A genetic approach was used to confirm the connection between reduced endoreplication, enhanced HG methylesterification, and hook defects in ccs52a2. Phenotypes resulting from enhanced HG methylesterification can be suppressed by a loss-of-function mutation in RECEPTOR-LIKE PROTEIN 44 (RLP44), a plasma membrane receptor that responds to pectin modification (22). We therefore generated rlp44cnu2 ccs52a2–double mutant and analyzed its hook development. The loss of RLP44 completely suppressed the ccs52a2 mutant’s early hook opening defect because the maintenance phase of rlp44cnu2 ccs52a2 (48.3 ± 2.6 hours) was significantly longer than that of ccs52a2 (17.3 ± 2.6 hours) and very similar to that of the WT (50.7 ± 1.7 hours) (Fig. 3D and fig. S7A). The endoreplication asymmetry was not affected in the rlp44cnu2 mutant (fig. S7B). Together, these results confirm that hook defects in ccs52a2 are linked to enhanced HG methylesterification. This provides a scenario in which differential cell elongation during apical hook formation relies on asymmetric cell wall mechanochemical properties downstream of endoreplication control.
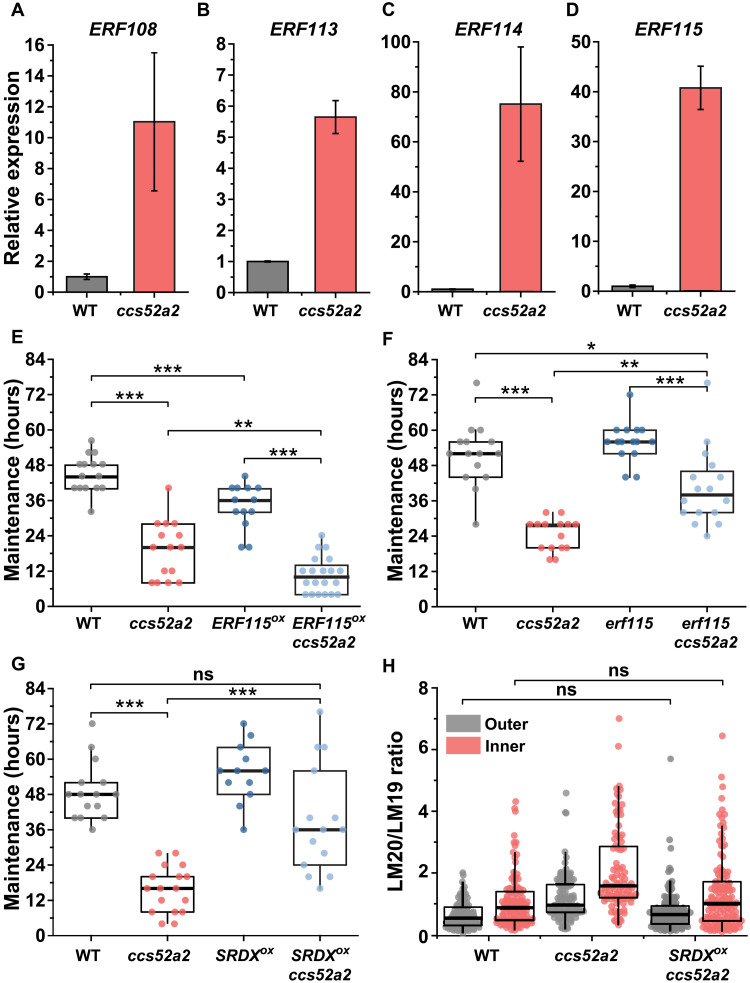
ERF115 is a downstream target of CCS52A2 in hook development
To clarify the molecular mechanism underlying the connection between reduced endoreplication, HG methylesterification, and hook defects in ccs52a2, we investigated the transcription factor ETHYLENE RESPONSE FACTOR 115 (ERF115), a downstream target of APC/CCCS52A2 (23). Expression analysis showed that ERF115 and its closely related homologs ERF108 (RAP2.6), ERF113 (RAP2.6L), and ERF114 (24) were substantially up-regulated in the ccs52a2 mutant relative to the WT (Fig. 4, A to D). Notably, the ERF115-overexpressing mutant ERF115OX (23) displays hook defects similar to those of ccs52a2, with a maintenance phase that is almost 11 hours (ca. 25%) shorter than that of the WT (Fig. 4E and fig. S8A). To determine whether the hook defects of ccs52a2 can be attributed to ERF115 misexpression, we introduced ERF115OX and erf115 loss-of-function mutations into the ccs52a2 mutant background and analyzed their effects on hook development. ERF115 overexpression exacerbated the hook defects of ccs52a2: The maintenance phase in ERF115OX ccs52a2 was almost 9 hours shorter (ca. 45%) than that in the ccs52a2-single mutant (Fig. 4E and fig. S8A). Conversely, the erf115 mutant partially but significantly rescued the ccs52a2 phenotype: The maintenance phase of the erf115 ccs52a2–double mutant was almost 16 hours (ca. 65%) longer than that of the ccs52a2-single mutant (Fig. 4F and fig. S8A). These results indicate that increase in ERF115 activity could contribute to the hook defect of the ccs52a2 mutant.
Fig. 4. ERF115 transcription factor mediates endoreplication defects to modulate hook development.
(A to D) Reverse transcription quantitative polymerase chain reaction (RT-Q-PCR) analysis of the expression of ERF115 subfamily members, ERF108 (A), ERF113 (B), ERF114 (C), and ERF115 (D), in WT and ccs52a2. Expression data were normalized against ubiquitin 10 and the WT value. Graphs represent means ± SEM. (E) Maintenance period of the apical hook in WT (n = 15), ccs52a2 (n = 15), ERF115OX (n = 13), and ERF115OX ccs52a2 (n = 20). (F) Maintenance period of apical hook in WT (n = 15), ccs52a2 (n = 15), erf115 (n = 15), and erf115 ccs52a2 (n = 16). (G) Maintenance period of apical hook in WT (n = 15), ccs52a2 (n = 17), ERF115SRDX (n = 12), and ERF115SRDX ccs52a2 (n = 15). (H) LM20/LM19 immunolabeling ratio of longitudinal walls of epidermal cells in the WT (n = 8; outer, 90 cells; inner, 103 cells), ccs52a2 (n = 6; outer, 95 cells; inner, 87 cells), and ERF115SRDX ccs52a2 (n = 8; outer, 109 cells; inner, 117 cells) lines at 0 to 400 μm from SAM. The data of WT and ccs52a2 are identical to that of Fig. 3C. Lines in the boxes indicate medians. Statistical significance was calculated using Student’s t test (unpaired, two-tailed) and is indicated as follows: ns, *P < 0.05, **P < 0.01, and ***P < 0.001.
However, the suppression of ccs52a2 phenotype by loss of ERF115 is only partial. This may result from redundancy in ERF115 that has three additional members: ERF108, ERF113, and ERF114. Consistently, the expression of ERF108, ERF113, and ERF114 is enhanced in the ccs52a2 mutant (Fig. 4, A to D). To test the hypothesis of a partial compensation by other ERF115 clade members, we introduced a dominant-negative ERF115SRDX fusion (to overcome redundancy and phenocopy erf113, 114, and 115-triple mutants) (25) into the ccs52a2 background. Note that the expression of ERF115SRDX does not affect endoreplication and that ERF115SRDX displays WT-like endoreplication asymmetry in the hook (fig. S8B). Notably, ERF115SRDX almost completely suppressed the hook defect of the ccs52a2 mutant: The maintenance phase of ERF115SRDX ccs52a2 (39.2 ± 4.7 hours) was significantly longer than that of ccs52a2 (15.8 ± 1.8 hours) and did not differ significantly from the WT maintenance period (49.1 ± 2.5 hours; P = 0.07) (Fig. 4G and fig. S8A). Moreover, the growth rate on the outer side in ERF115SRDX ccs52a2 was similar to that in the WT and significantly faster than that of the ccs52a2 mutant (fig. S8C). Thus, ERF115SRDX restored growth rate asymmetry between the outer and inner sides when introduced in ccs52a2 mutant background. Immunolabeling studies showed that ERF115SRDX suppressed the enhancement of HG methylesterification in ccs52a2: The LM20/LM19 ratio for ERF115SRDX ccs52a2 on the outer side of the apical hook (0.65 ± 0.09) was ca. 46% lower than that for ccs52a2 (1.20 ± 0.17), whereas on the inner side, the LM20/LM19 ratios were 1.20 ± 0.31 for ERF115SRDX ccs52a2 and 2.14 ± 0.34 in ccs52a2, a reduction of ca. 44% (Fig. 4H). These results reveal that ERF115 family transcription factors are redundant negative regulators of apical hook development and suggest that asymmetries in cell wall properties (HG methylesterification) during apical hook development are established and maintained via ERF115, downstream of CCS52A2-mediated endoreplication.
Cell wall changes in response to reduced endoreplication are monitored via the cell wall integrity kinase THESEUS1
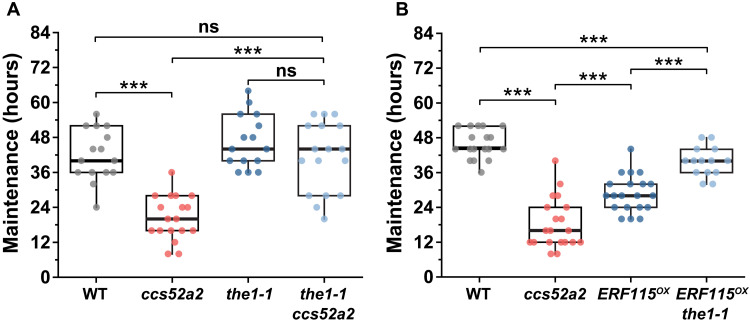
So far, our results provide a molecular pathway relating ploidy to cell elongation through auxin gradient, endoreplication pathway, and cell wall modification. However, mechanical feedbacks can also contribute to hook development (6). Therefore, we next investigated potential mediators of endoreplication responsive mechanochemical feedback in the regulation of cell elongation during hook development. THESEUS1 (THE1), a receptor-like kinase that monitors cell wall integrity, was targeted because it mediates changes in HG methylesterification (26). The loss-of-function mutant allele the1-1 does not cause discernible hook defects (Fig. 5A and fig. S9A), presumably due to redundancy with related receptor-like protein kinases (RLKs) such as HERCULES1 (HERK1), as shown for other responses (26). Consistently, endoreplication asymmetry was not affected by loss of function the1-1 (fig. S9B). However, introducing the the1-1 mutation into the ccs52a2 background largely suppressed the maintenance phase defect of ccs52a2: The maintenance phase in the1-1 ccs52a2 was 41 hours long, ca. 103% longer than that of ccs52a2 and similar to that of the WT (Fig. 5A and fig. S9A). The growth rate of outer side in the1-1 ccs52a2 was similar to that in WT and significantly faster than that in ccs52a2 mutant and resulted in restoration of growth asymmetry between the outer and inner sides in the1-1 ccs52a2 (fig. S9C).
Fig. 5. THE1 monitors cell wall changes in response to defective endoreplication during apical hook development.
(A) Maintenance period of apical hook in WT (n = 15), ccs52a2 (n = 18), the1-1 (n = 15), and the1-1 ccs52a2 (n = 17). (B) Maintenance period of apical hook in WT (n = 18), ccs52a2 (n = 20), ERF115OX (n = 20), and ERF115OX the1-1 (n = 14). Lines in the boxes indicate medians. Statistical significance was calculated using Student’s t test (unpaired, two-tailed) and is indicated as follows: ns and ***P < 0.001.
Because ERF115 misexpression induces mechanical and hook defects similar to those of ccs52a2, we also introduced the the1-1 allele into the ERF115OX to investigate the role of THE1 in the CCS52A2-ERF115 pathway. Introducing the the1-1 allele significantly extended the maintenance phase to 39.7 ± 1.4 hours, compared to 28.8 ± 1.4 hours for ERF115OX (Fig. 5B). This suggests that the perception of increased wall stiffness resulting from elevated HG methylesterification in ccs52a2 occurs via a signaling pathway involving wall receptor kinases such as THE1. Thus, CCS52A2/ERF115-dependent pectin modifications and the monitoring of cell wall status by THE1 appear to be crucial for endoreplication-dependent control of rapid cell elongation and differential growth crucial for hook maintenance.
DISCUSSION
Our results summarized in the model (Fig. 6) provides a mechanistic link between endoreplication and cell and organ size regulation. Although endoreplication correlates positively with cell size, ploidy levels do not scale linearly with cell volume (3). Consequently, the mechanisms through which endoreplication regulates cell size and shape have remained unclear. Two groups (20, 27) recently suggested that the correlation mainly results from a feedback mechanism: Rather than triggering cell enlargement, endoreplication was suggested to occur in response to the demands imposed on cells as their sizes and growth rates increase. For example, an increased DNA content resulting from high endoreplication could enable the high transcriptional, metabolic, and translational output needed to sustain rapid growth and large cells (28). This is consistent with the need to safeguard the mechanical integrity of rapidly growing cells: High endoreplication may be required to deposit enough cell wall material to compensate for the cell wall’s thinning as it stretches during expansion (11). Our data are consistent with that scenario while also providing molecular players involved. Thus, the absence of endoreplication (ccs52a2 mutant) may lead to enhanced mechanical stress or wall damage. In turn, wall integrity sensors such as THE1 would then trigger growth reduction. When endoreplication is reduced but wall integrity sensing is impaired, growth would be restored, explaining the suppression of ccs52a2 phenotype by loss of THE1 kinase. However, the nature of the wall cue that is sensed (e.g., mechanical stress, wall damage, or secondary signals) remains to be investigated.
Fig. 6. A model for endoreplication-mediated mechanochemical control of cell elongation.
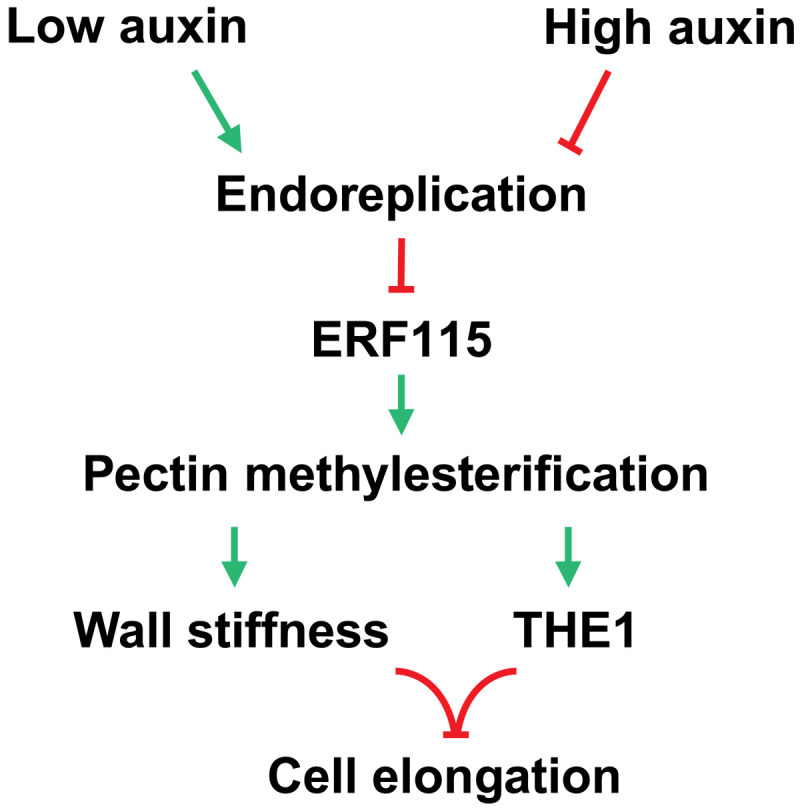
PIN protein–dependent polar auxin transport establishes auxin response asymmetry that controls spatial differences in endoreplication. Endoreplication negatively regulates ERF115 family transcription factors. ERF115 (and related members) regulates cell wall properties via modulation of HG methylesterification levels. Cell elongation is then controlled by mechanical properties of the cell wall (stiffness) and signaling via cell wall integrity kinase THE1, a proposed mediator of HG methylesterification levels, thus linking endoreplication with control of cell size via a mechanochemical feedback.
Another important unresolved question is: If ploidy does not always scale linearly with cell volume, then why are cells always smaller in endoreplication-defective mutants? Endoreplication is classically thought to have a permissive role in cell growth. Thus, endoreplication-associated cell enlargement should not be executed unless the cell wall mechanics is also permissive as suggested by our results while also revealing the components involved. However, our data also unravel a previously unrecognized element: We find that reduced endoreplication causes cell wall remodeling, resulting in wall stiffening. This reveals a feedback mechanism that would limit the cell size following endoreplication attenuation. Our data suggest a scenario where the monitoring of mechanochemical status of the cell wall by the THE1 receptor–like kinase could feed back onto growth, linking reduced endoreplication to slower growth and smaller cells. In other words, when the metabolic rates (or other factors) needed for cell expansion cannot be sustained because endoreplication is attenuated, growth is actively reduced through wall sensing. Although inhibition of endoreplication does not always lead to smaller size as shown by overexpression of KIP-RELATED PROTEIN 1 (KRP1) in tomato (29), endoreplication mutants are frequently smaller in stature. Our findings provide a plausible explanation for the typically small stature of such endoreplication mutants because cell growth would be inhibited in such lines through a mechanism as shown here.
We propose that endoreplication not only facilitates or promotes growth but also acts as a brake on growth: High growth rate is enabled by the provisioning of cell wall material via endoreplication, while, conversely, low endoreplication mediates wall stiffening and slower growth. Previously, a correlation between ploidy and transcription of cell wall–related genes linking these two factors (endoreplication and cell wall) has been noted in yeast (30) and in plants (11), which further supports our findings here. While a role for cell wall modulation was identified here as a component linking endoreplication with growth, additional pathways that link endoreplication with growth regulation may also exist. For example, ERF115 has also been linked with hormonal control of cell elongation, and ERF115 could also mediate turgor effects on growth, being responsive to turgor itself (31, 32).
Our findings thus put forward wall properties and wall sensing at the nexus between endoreplication and size control and reveal a plausible role for endoreplication in the dynamic control of cell size, integrating both biochemical and mechanical cues. Our results suggest a molecular framework in which hormonally patterned spatial differences in endoreplication can modulate growth through and in response to mechanochemical modulation of cell wall properties. Given the consistently reduced stature in endoreplication defective mutants, our findings now open the possibility of investigating the wider applicability of this mechanism in other tissues in the future. This previously unrecognized mechanism for growth regulation by endoreplication implies that cells do not use endoreplication as a simple ploidy-dependent growth amplifier; endoreplication instead appears as an integrator of hormonal and mechanical cues in regulation of cell size and organ shape changes.
MATERIALS AND METHODS
Plant material and growth conditions
The Columbia-0 (Col-0) ecotype of Arabidopsis thaliana and the following transferred DNA insertion and transgenic lines were used in this study: ccs52a2-1 (SALK_001978) (8), ERF115OX (23), ERF115SRDX (23), erf115 (23), pin3 pin4 pin7 (17), rlp44cnu2 (22), the1-1 (33), pCCS52A2::CCS52A2-GFP (14), H2B-YFP (34), and LTI6A-GFP (35). The higher-order mutants used in this study were generated by crossing the lines listed above.
Seeds were stratified at 4°C in the dark for 2 to 4 days on 1/2 Murashige and Skoog (MS) agar medium (Duchefa) (pH 5.8) supplemented with 0.5% (w/v) sucrose and 2.5 mM 2-morpholinoethanesulfonic acid (Sigma-Aldrich). Seeds were then given 6 hours of light and subsequently grown vertically under constant-dark conditions for time-lapse growth kinematics imaging or grown vertically in constant darkness for 48 hours followed by confocal microscopy or reverse transcription quantitative polymerase chain reaction (RT-Q-PCR) analysis. For pharmacological treatments, 250 μM EGCG or 0.1 μM NPA was added to the medium; the same volumes of solvent [dimethyl sulfoxide (DMSO)] were used in the corresponding mock treatments.
Quantification of CCS52A2-GFP
pCCS52A2::CCS52A2-GFP seedlings were grown on 1/2 MS medium vertically under constant-dark conditions for 2 days. The seedlings were then harvested and stained with propidium iodide (PI; 1 mg/ml) in phosphate-buffered saline (PBS) (pH 7.4) for 20 min. The seedlings were then washed twice with PBS buffer for 5 min each and mounted for confocal imaging using a Carl Zeiss LSM880 with a 20× lens (Zeiss Plan-Apochromat 20×/0.8 M27) (excitation: 488 nm for GFP and 561 nm for PI; emission range: 490 to 553 nm for GFP and 600 to 700 nm for PI). Images were stacked, and the GFP fluorescence intensity at bending region was quantified using the Fiji software package [National Center for Biotechnology Information (NCBI)]. The bending region was defined as the region where the longitudinal angle of the neighboring cells was less than 170°. Data were normalized with the value of the outer side of the WT. Data were plotted by PlotsOfData (36).
Immunohistochemistry
Immunolabeling was performed to study cell wall changes as previously described (6). Briefly, seedlings were embedded in LR White Medium grade (TAAB, UK) and sectioned at a thickness of 2.5 μm using a Reichert Ultracut S Wild M3Z microtome (Leica) with a Histo Diamond Knife (8.0 mm, 45° angle) (DiATOME). The sections were immunolabeled with the primary antibodies α-Rat-LM19 (RRID: AB_2734788) and α-Rat-LM20 (diluted 1:50; RRID: AB_2734789, PlantProbes, UK) for 2 hours at room temperature followed by 2 hours of incubation at room temperature with a secondary antibody, α-Rat Cy5 (diluted 1:200; RRID: AB_2340672, Jackson ImmunoResearch, UK). Cy5 fluorescence signals from apical hook cells were imaged using a Carl Zeiss LSM880 microscope with a 32× lens (Zeiss C-Achroplan 32×/0.85-W Corr M27) (excitation, 633 nm; emission range, 638 to 700 nm). Sequential Z-stack images were taken at 1-μm intervals. Images were stacked, and fluorescence intensity was measured using the Fiji software package (NCBI). The LM19 and LM20 immunolabeling signal of the outer epidermal cell walls of cells (0 to 400 μm from Shoot Apical Meristem) was measured individually, and LM20/LM19 ratio of each cell was calculated and plotted for both the outer and inner sides of the hook by PlotsOfData (36).
Gene expression analyses by RT-Q-PCR
Seedlings were grown on 1/2 MS medium vertically under constant-dark conditions for 2 days. The apical hooks were harvested and frozen in liquid nitrogen and stored at −80°C. Total RNA was purified using the RNeasy Plant Mini kit (QIAGEN) or the GenElute Total RNA Purification Kit (Sigma-Aldrich) in accordance with the manufacturer’s recommendations. Genomic DNA was removed using RNase-free Turbo DNase (Invitrogen), and single-stranded cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad) with 1 μg of RNA. Q-PCR was performed with the Light Cycler 480 SYBR Green I Master (Roche) in the 96-well CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Expression data were normalized against ubiquitin 10 using the 2−ΔΔCT method (37). Three biological replicates were performed for each line, and two to three technical replicates were performed for each biological replicate. Primers used in this study are listed in table S1.
Endoploidy analysis
Endoploidy analysis was performed as described earlier (11). Two-day-old seedlings were harvested and fixed in fixation solution (50% methanol and 10% acetic acid) at 4°C for at least 12 hours and stored in the fixative for up to 1 month in darkness (38). The seedlings were incubated in 80% ethanol at 80°C for 5 min and then transferred back to the fixation solution and incubated for 1 hour. The tissue was rehydrated in PBS buffer for 2 hours and stained with DAPI (1 μg/ml) or DRAQ5 (25 μM) for 15 min in PBS buffer (pH 7.4). The tissue was then rinsed extensively and imaged using a Carl Zeiss LSM880 with a 32× lens (Zeiss C-Achroplan 32×/0.85-W Corr M27; excitation, 405 nm; emission range, 410 to 585 nm). Fluorescence signals other than the nuclei of the outer and inner sides at the bending region were removed using MorphoGraphX (39) to facilitate nuclei recognition and measuring. The bending region was defined as the region where the longitudinal angle of the neighboring cells was less than 170°. Images were then stacked, around 10 nuclei within two to four cell files were identified, and fluorescence intensity was measured by Fiji with 3D Objects Counter plugin (40). Data were normalized with the value of the outer side of the WT. Data were plotted for both the outer and inner sides of the hook by PlotsOfData (36). For nuclear size analysis, 2-day-old histone 2B (H2B)–yellow fluorescent protein (YFP) seedlings were harvested and live imaged using a Carl Zeiss LSM880 with a 25× lens (Zeiss LCI Plan-Neofluar 25×/0.8 Imm Korr DIC M27; excitation, 514 and 488 nm; emission range, 526 to 88 nm and 490 to 508 nm, for YFP and GFP respectively). The nuclear size was analyzed by Fiji software (NCBI) with the 3D Objects Counter plugin (40). Data were normalized by the value of the outer side of WT and were plotted by PlotsOfData (36).
Time-lapse imaging of cell elongation
Time-lapse imaging of cell elongation was performed as previously described with minor modifications (6). Col-0 and ccs52a2-1 seedlings were grown for 36 hours on vertical agar plates before treatment. The seedlings were stained with PI (10 mg/ml in liquid 1/2 MS medium) for 1 hour and imaged on vertical agar plates using a Nikon AZ-C2 vertical macro-confocal equipped with a AZ-Plan Apochromat 0.5x (NA 0.05/W.D. 54 mm) macro-objective at 3-hour intervals in a dark chamber. The cell lengths in the two time-lapse images were measured using the segmented line tool in Fiji (NCBI), and the hourly cell elongation rate (%/hour) was calculated.
Growth kinematics analysis of apical hook development
The growth kinematics analysis was performed with a fully automated infrared camera imaging system as previously described (6). Briefly, seeds were illuminated with white light for 4 to 6 hours to promote germination. The plates were then imaged with an infrared camera imaging system in a dark room at 4-hour intervals. The hook curvature angle between the hypocotyl axis vector and cotyledons was measured using Fiji (NCBI). For each line, 12 to 25 seedlings were measured. Data were plotted by PlotsOfData (36). The maintenance period was defined as the period from the hook angle of the seedling reached 95% of its maximum value until the angle decreased by greater than 5% of its maximum value, as previously described (41). For pharmacological treatments, seedlings were germinated and grown on 1/2 MS medium supplemented with 250 μM EGCG. Mock treatments were performed with an equivalent amount of solvent (DMSO).
Analysis of cell deformation
Two-day-old seedlings were immersed in hypo-osmotic medium (H2O) for 30 min to inflate their cells and then incubated in PI solution (1 mg/ml) for another 30 min to stain the cell wall. Samples were imaged using a Carl Zeiss LSM880 with a 32× lens (Zeiss C-Achroplan 32×/0.85-W Corr M27) (excitation, 514 nm; emission range, 566 to 718 nm). Three Z stacks of images were taken for each sample to cover the whole hook region. Subsequently, the samples were incubated in hyperosmotic medium (0.35 M NaCl) for 30 min to deflate cells followed by confocal imaging as above. Z stacks of each seedling were stitched together. The difference in the epidermal cell surface area between hypoosmotic and hyperosmotic conditions (cell deformation) was calculated using MorphoGraphX (39). Cell deformation heatmaps were created by averaging the cell surface changes of each cell with those of its neighbor cells to identify tissue-level patterns (6, 19, 39). Data were plotted for both the outer and inner sides of the hook by PlotsOfData (36).
Statistical analysis
All statistical analyses were based on the two-tailed t test, which was performed using the t.test function in Microsoft Excel. In all figures, statistical significance is indicated as follows: ns (not statistically significant), *P < 0.05, **P < 0.01, and ***P < 0.001.
Accession numbers
Gene information and sequence data can be found in the Arabidopsis Genome Initiative under the following accession numbers: CCS52A2 (AT4G11920), THE1 (AT5G54380), ERF108 (AT1G43160), ERF113 (AT5G13330), ERF114 (AT5G61890), ERF115 (AT5G07310), and RLP44 (AT3G49750).
Acknowledgments
We thank H. Höfte for sharing research materials. We acknowledge the facilities and technical assistance of Umeå Centre for Electron Microscopy (UCEM), part of the National Microscopy Infrastructure NMI (VR-RFI 2019-00217).
Funding: This work was supported by Swedish Research Council (Vetenskapsrådet) grant 2020-03522 (R.P.B.); European Commission Marie Curie Individual Fellowship, 890883 (Y.M.); and Swedish Research Council (Vetenskapsrådet) International Postdoc grant 2020-06442 (K.J.).
Author contributions: Conceptualization: Y.M., O.H., L.D.V., and R.P.B. Methodology: Y.M., K.J., B.A., and R.P.B. Investigation: Y.M., K.J., and B.A. Visualization: Y.M. and K.J. Funding acquisition: Y.M. and R.P.B. Supervision: L.D.V., O.H., and R.P.B. Writing—original draft: Y.M. and R.P.B. Writing—review and editing: Y.M., K.J., B.A., L.D.V., O.H., and R.P.B.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Table S1
REFERENCES AND NOTES
- 1.Orr-Weaver T. L.,When bigger is better: The role of polyploidy in organogenesis. Trends Genet. 31,307–315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Veylder L., Larkin J. C., Schnittger A.,Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 16,624–634 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Tsukaya H.,Has the impact of endoreduplication on cell size been overestimated? New Phytol. 223,11–15 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Roeder A. H. K., Chickarmane V., Cunha A., Obara B., Manjunath B. S., Meyerowitz E. M.,Variability in the control of cell division underlies sepal epidermal patterning in Arabidopsis thaliana. PLoS Biol. 8,e1000367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gendreau E., Traas J., Desnos T., Grandjean O., Caboche M., Hofte H.,Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114,295–305 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jonsson K., Lathe R. S., Kierzkowski D., Routier-Kierzkowska A. L., Hamant O., Bhalerao R. P.,Mechanochemical feedback mediates tissue bending required for seedling emergence. Curr. Biol. 31,1154–1164.e3 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Baral A., Aryal B., Jonsson K., Morris E., Demes E., Takatani S., Verger S., Xu T. D., Bennett M., Hamant O., Bhalerao R. P.,External mechanical cues reveal a katanin-independent mechanism behind auxin-mediated tissue bending in plants. Dev. Cell 56,67–80.e3 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Lammens T., Boudolf V., Kheibarshekan L., Zalmas L. P., Gaamouche T., Maes S., Vanstraelen M., Kondorosi E., La Thangue N. B., Govaerts W., Inze D., De Veylder L.,Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc. Natl. Acad. Sci. U.S.A. 105,14721–14726 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willems A., De Veylder L.,The plant anaphase-promoting complex/cyclosome. Annu. Rev. Cell Dev. Biol. 38,25–48 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Raz V., Ecker J. R.,Regulation of differential growth in the apical hook of Arabidopsis. Development 126,3661–3668 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Bhosale R., Boudolf V., Cuevas F., Lu R., Eekhout T., Hu Z., Van Isterdael G., Lambert G. M., Xu F., Nowack M. K., Smith R. S., Vercauteren I., De Rycke R., Storme V., Beeckman T., Larkin J. C., Kremer A., Hofte H., Galbraith D. W., Kumpf R. P., Maere S., De Veylder L.,A spatiotemporal DNA endoploidy map of the arabidopsis root reveals roles for the endocycle in root development and stress adaptation. Plant Cell 30,2330–2351 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S. Y., Stout J. R., Dharmaiah S., Yde S., Calvi B. R., Walczak C. E.,Transient endoreplication down-regulates the kinesin-14 HSET and contributes to genomic instability. Mol. Biol. Cell 27,2911–2923 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noir S., Marrocco K., Masoud K., Thomann A., Gusti A., Bitrian M., Schnittger A., Genschik P.,The control of arabidopsis thaliana growth by cell proliferation and endoreplication requires the f-box protein FBL17. Plant Cell 27,1461–1476 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanstraelen M., Baloban M., Da Ines O., Cultrone A., Lammens T., Boudolf V., Brown S. C., De Veylder L., Mergaert P., Kondorosi E.,APC/CCCS52A complexes control meristem maintenance in the Arabidopsis root. Proc. Natl. Acad. Sci. U.S.A. 106,11806–11811 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Žádníková P., Wabnik K., Abuzeineh A., Gallemi M., Van Der Straeten D., Smith R. S., Inzé D., Friml J., Prusinkiewicz P., Benková E.,A model of differential growth-guided apical hook formation in plants. Plant Cell 28,2464–2477 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zadnikova P., Petrasek J., Marhavy P., Raz V., Vandenbussche F., Ding Z. J., Schwarzerova K., Morita M. T., Tasaka M., Hejatko J., Van Der Straeten D., Friml J., Benkova E.,Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137,607–617 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Willige B. C., Ahlers S., Zourelidou M., Barbosa I. C., Demarsy E., Trevisan M., Davis P. A., Roelfsema M. R., Hangarter R., Fankhauser C., Schwechheimer C.,D6PK AGCVIII kinases are required for auxin transport and phototropic hypocotyl bending in Arabidopsis. Plant Cell 25,1674–1688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aryal B., Jonsson K., Baral A., Sancho-Andres G., Routier-Kierzkowska A. L., Kierzkowski D., Bhalerao R. P.,Interplay between cell wall and auxin mediates the control of differential cell elongation during apical hook development. Curr. Biol. 30,1733–1739.e3 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Kierzkowski D., Nakayama N., Routier-Kierzkowska A. L., Weber A., Bayer E., Schorderet M., Reinhardt D., Kuhlemeier C., Smith R. S.,Elastic domains regulate growth and organogenesis in the plant shoot apical meristem. Science 335,1096–1099 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Bhosale R., Maere S., De Veylder L.,Endoreplication as a potential driver of cell wall modifications. Curr. Opin. Plant Biol. 51,58–65 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Lewis K. C., Selzer T., Shahar C., Udi Y., Tworowski D., Sagi I.,Inhibition of pectin methyl esterase activity by green tea catechins. Phytochemistry 69,2586–2592 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Wolf S., van der Does D., Ladwig F., Sticht C., Kolbeck A., Schurholz A. K., Augustin S., Keinath N., Rausch T., Greiner S., Schumacher K., Harter K., Zipfel C., Hofte H.,A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc. Natl. Acad. Sci. U.S.A. 111,15261–15266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heyman J., Cools T., Vandenbussche F., Heyndrickx K. S., Van Leene J., Vercauteren I., Vanderauwera S., Vandepoele K., De Jaeger G., Van Der Straeten D., De Veylder L.,ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342,860–863 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Heyman J., Canher B., Bisht A., Christiaens F., De Veylder L.,Emerging role of the plant ERF transcription factors in coordinating wound defense responses and repair. J. Cell Sci. 131,jcs208215 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Lakehal A., Dob A., Rahneshan Z., Novak O., Escamez S., Alallaq S., Strnad M., Tuominen H., Bellini C.,ETHYLENE RESPONSE FACTOR 115 integrates jasmonate and cytokinin signaling machineries to repress adventitious rooting in Arabidopsis. New Phytol. 228,1611–1626 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Gigli-Bisceglia N., van Zelm E., Huo W. Y., Lamers J., Testerink C.,Arabidopsis root responses to salinity depend on pectin modification and cell wall sensing. Development 149,dev200363 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lang L., Schnittger A.,Endoreplication - a means to an end in cell growth and stress response. Curr. Opin. Plant Biol. 54,85–92 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Edgar B. A., Zielke N., Gutierrez C.,Endocycles: A recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell Bio 15,197–210 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Nafati M., Frangne N., Hernould M., Chevalier C., Gévaudant F.,Functional characterization of the tomato cyclin-dependent kinase inhibitor SlKRP1 domains involved in protein-protein interactions. New Phytol. 188,136–149 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Wu C. Y., Rolfe P. A., Gifford D. K., Fink G. R.,Control of transcription by cell size. PLOS Biol. 8,e1000523 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoermayer L., Montesinos J. C., Marhava P., Benková E., Yoshida S., Friml J.,Wounding-induced changes in cellular pressure and localized auxin signalling spatially coordinate restorative divisions in roots. Proc. Natl. Acad. Sci. U.S.A. 117,15322–15331 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canher B., Heyman J., Savina M., Devendran A., Eekhout T., Vercauteren I., Prinsen E., Matosevich R., Xu J., Mironova V., De Veylder L.,Rocks in the auxin stream: Wound-induced auxin accumulation and ERF115 expression synergistically drive stem cell regeneration. Proc. Natl. Acad. Sci. U.S.A. 117,16667–16677 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hematy K., Sado P. E., Van Tuinen A., Rochange S., Desnos T., Balzergue S., Pelletier S., Renou J. P., Hofte H.,A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 17,922–931 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Boisnard-Lorig C., Colon-Carmona A., Bauch W., Hodge S., Doerner P., Bancharel E., Dumas C., Haseloff J., Berger F.,Dynamic analyses of the expression of the HISTONE::YFP fusion protein in arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13,495–509 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cutler S. R., Ehrhardt D. W., Griffitts J. S., Somerville C. R.,Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. U.S.A. 97,3718–3723 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Postma M., Goedhart J.,PlotsOfData–A web app for visualizing data together with their summaries. PLoS Biol. 17,e3000202 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak K. J., Schmittgen T. D.,Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25,402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., Barthélémy J., Palauqui J. C.,High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell 20,1494–1503 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbier de Reuille P., Routier-Kierzkowska A. L., Kierzkowski D., Bassel G. W., Schupbach T., Tauriello G., Bajpai N., Strauss S., Weber A., Kiss A., Burian A., Hofhuis H., Sapala A., Lipowczan M., Heimlicher M. B., Robinson S., Bayer E. M., Basler K., Koumoutsakos P., Roeder A. H., Aegerter-Wilmsen T., Nakayama N., Tsiantis M., Hay A., Kwiatkowska D., Xenarios I., Kuhlemeier C., Smith R. S.,MorphoGraphX: A platform for quantifying morphogenesis in 4D. eLife 4,05864 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolte S., Cordelieres F. P.,A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. Oxford. 224,213–232 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Smet D., Žádníková P., Vandenbussche F., Benkova E., Van Der Straeten D.,Dynamic infrared imaging analysis of apical hook development in Arabidopsis: The case of brassinosteroids. New Phytol. 202,1398–1411 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Table S1